- 1Division of Immunity and Pathogenesis, Burnett School of Biomedical Sciences, College of Medicine, University of Central Florida, Orlando, FL, United States
- 2Division of Microbiology, College of Science, Oregon State University, Corvallis, OR, United States
Introduction: Chronic bacterial infections are responsible for significant morbidity and mortality in cystic fibrosis (CF) patients. Pseudomonas aeruginosa (Pa), the dominant CF pathogen, and Mycobacterium abscessus (Mab) can individually cause persistent, difficult to treat pulmonary infections. Co-infection by both pathogens leads to severe disease and poor clinical outcomes. Although interactions between Pa and other co-infecting pathogens in CF patients have been the focus of numerous studies, the dynamics of Pa-Mab interactions remain poorly understood.
Methods: To address this knowledge gap, the study examined how Mab and Pa influenced each other through culture-based growth assays and molecular-based dual RNAseq analysis. Growth was measured by CFU determination and luminescence reporter -based readouts.
Results: In initial studies, we noted that the growth of Pa continued unimpeded in a planktonic co-culture model, whereas Pa appeared to exert a bacteriostatic effect on Mab. Strikingly, exposure of Mab to cell-free spent supernatant of Pa resulted in a dramatic, dose-dependent bactericidal effect. Initial characterization indicated that this potent Pa-derived anti-Mab cidal activity was mediated by a heat-sensitive, protease-insensitive soluble factor of >3kDa size. Further analysis demonstrated that expression of this mycobactericidal factor requires LasR, a central regulator of Pa quorum sensing (QS). In contrast, ΔLasR Pa was still able to exert a bacteriostatic effect on Mab during co-culture, pointing to additional LasR-independent factors able to antagonize Mab growth. However, the ability of Mab to adapt during co-culture to counter the cidal effects of a LasR regulated factor suggested complex interspecies dynamics. Dual RNAseq analysis of Mab-Pa co-cultures revealed significant transcriptional remodeling of Mab, with differential expression of 68% of Mab genes compared to minimal transcriptional changes in Pa. Transcriptome analysis reflected slowed Mab growth and metabolic changes akin to a non-replicating persister phase. A tailored Mab response to Pa was evident by enhanced transcript levels of genes predicted to counteract alkylquinolone QS signals, respiratory toxins, and hydrogen cyanide.
Discussion: The study showed Mab is capable of coexisting with Pa despite Pa’s antagonistic effects, eliciting an adaptive molecular response in Mab. This study provides the first transcriptome-level insight into genetic interactions between the two CF pathogens offering potential strategies for disrupting their communities in a CF lung to improve patient clinical performance. Moreover, identification of a novel antimicrobial natural product with potent cidal activity against Mab could lead to new drug targets and therapies for Mab infections.
1 Introduction
Chronic microbial infections are responsible for morbidity and mortality in patients suffering with lung disorders, in particular cystic fibrosis (CF). CF is an autosomal recessive disorder characterized by a defect in cystic fibrosis transmembrane conductance regulator (CFTR) that regulates chloride and bicarbonate secretion across the pulmonary epithelium (Poulsen et al., 1994; Mall and Hartl, 2014; Elborn, 2016; Kunzelmann et al., 2017). This results in improper pH and salt balance leading to accumulation of thick, sticky mucus and defective mucociliary clearance from lung airways. This environment in which innate immune mechanisms are impaired is conducive for opportunistic pathogens to thrive, causing lung deterioration and poor quality of life of CF patients. Among the diverse CF microbial community, notable bacterial pathogens include Pseudomonas aeruginosa (Pa), Staphylococcus aureus, Burkholderia cenocepacia, and nontuberculous mycobacteria (NTM) such as Mycobacterium abscessus (Mab) (Valenza et al., 2008; Thornton and Parkins, 2023).
Pa is a multidrug resistant bacterial pathogen that can colonize the lungs of CF patients as early as the first few years of life (Sader et al., 2018; Recio et al., 2020). Over time, it persists and often becomes the predominant pathogen, outcompeting other colonizers in adulthood (Aebi et al., 1995; Van Daele et al., 2005; Smith et al., 2006; Pittman et al., 2011; Coburn et al., 2015; Bhagirath et al., 2016; Banjar et al., 2022; Thornton and Parkins, 2023). Pa produces several quorum sensing (QS) regulated virulence factors including proteases and secondary metabolites such as phenazines and hydrogen cyanide (HCN) detectable in CF sputum, which facilitate colonization of CF airways (Singh PK et al., 2000; Chambers et al., 2005; Jan, 2017). These QS mediated factors help Pa gain dominance within the CF microbial community by restricting competition. The QS regulatory network of Pa is complex and comprised of four interconnected systems - las, rhl, pqs and iqs (Latifi et al., 1996; Pesci et al., 1997; Wade et al., 2005; Lee et al., 2013). The first two systems are based on acylhomoserine lactone (AHL) signals, while the pqs system requires quinolone signals, and the fourth iqs responds to 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde signal molecules. The las system occupies the top position of the hierarchical QS system and is comprised of AHL signal synthase, LasI, and transcriptional regulator, LasR. This system positively regulates other systems of the QS circuitry; however these downstream pathways can also function independently of LasR under certain conditions (Latifi et al., 1996; Pesci et al., 1997; Diggle et al., 2003; Medina et al., 2003; Dekimpe and Deziel, 2009). LasR controls expression of numerous virulence genes and plays a crucial role in pathogenesis (Latifi et al., 1996; Pesci et al., 1997). Despite extensive study, the role of QS in CF infections remains perplexing. On one hand, las QS-regulated production of redox-active phenazine pigment pyocyanin causes pulmonary exacerbation and lung damage (Lau et al., 2004; Caldwell et al., 2009; Hunter et al., 2012; Rada and Leto, 2013). On the other hand, QS deficient LasR mutants are commonly isolated from CF patients and are responsible for enhanced neutrophil inflammation and disease progression (Smith et al., 2006; Hoffman et al., 2009; Feltner et al., 2016; Hennemann et al., 2021).
Mab, a second bacterial inhabitant of CF airways, is a major threat due to its increased prevalence and a high level of inherent drug resistance (Olivier et al., 2003; Sermet-Gaudelus et al., 2003; Jonsson et al., 2007; Kendall and Winthrop, 2013; Aiello et al., 2018; Thornton and Parkins, 2023; Hu et al., 2024). The prevalence of Mab varies with age, with reports of isolation from pediatric CF patients, but a higher prevalence in older patients over 15 years old (Pierre-Audigier et al., 2005; Roux et al., 2009). Mab exhibits two different colony phenotypes - smooth and rough - and both are commonly isolated from CF patients (Byrd and Lyons, 1999; Howard et al., 2006; Catherinot et al., 2009). Both phenotypes form biofilms, have similar antibiotic sensitivity and macrophage survival though the rough morphotype is considered more virulent (Greendyke and Byrd, 2008; Catherinot et al., 2009; Roux et al., 2016). It is speculated that after initial colonization of airways by smooth strains, a transition to rough phenotype occurs, leading to increased lung lesions and poor clinical outcomes (Howard et al., 2006; Catherinot et al., 2009; Kreutzfeldt et al., 2013; Hedin et al., 2023). Thus, as noted previously for Pa, host-derived selective pressures encountered during prolonged chronic infections may drive Mab evolution, altering Mab interactions with the host as well as with other microbes within the same niche.
There are several lines of evidence that support the relevance of Pa-Mab interspecies interactions. In addition to sharing environmental reservoirs in soil and water (Mangione et al., 2001), within the human host Pa and Mab both occupy the microaerobic respiratory zone in the lung (Bjarnsholt et al., 2009; Qvist et al., 2015) and are isolated from CF sputum samples (De Boeck et al., 2000; Jonsson et al., 2007; Bragonzi et al., 2012). Genomic studies that identified numerous genes shared between these species, attributed to putative horizontal gene transfer (HGT), is further evidence of interactions between Pa and Mab within the lung (Bjarnsholt et al., 2009; Ripoll et al., 2009; Qvist et al., 2015). Highlighting the clinical relevance of interspecies interactions, co-infection by these two opportunistic pathogens is known to increase disease severity and exacerbate the decline in lung function (Adjemian et al., 2018; Hsieh et al., 2018; Takano et al., 2021; Nichols et al., 2023). Yet significant knowledge gaps remain regarding the nature of Mab-Pa interspecies interactions and potential impact on pathogenesis (Birmes et al., 2017; Rodriguez-Sevilla et al., 2018; Rodriguez-Sevilla et al., 2019; Idosa et al., 2022). Birmes et al. demonstrated quenching of quinolone QS signals of Pa by Mab when provided exogenously to Mab cultures (Birmes et al., 2017). Two other research groups examined the coexistence of the two pathogens in an in vitro dual-species biofilm model where Pa exerted a negative effect on Mab (Rodriguez-Sevilla et al., 2019; Idosa et al., 2022). However, Pa-mediated inhibition of Mab was reported to be contact-dependent and restricted to the solid surface biofilm model, with no inhibition in the planktonic co-cultures (Idosa et al., 2022). Extensive analysis of Pa mutants lacking known virulence regulators (e.g. las, rhl, pqs QS systems), secretion systems, motility factors, and iron sequestration genes failed to identify the molecular mechanism responsible for this antagonism (Idosa et al., 2022). It was also noted that targeted killing of Pa with selective antibiotics favored Mab establishment in the dual-species biofilm (Rodriguez-Sevilla et al., 2019). It is crucial to interrogate interactions between these two human CF pathogens for a better understanding of their adaptive responses and impact on disease progression.
This study examined how Mab and Pa influenced each other through culture-based growth assays and molecular-based dual RNAseq analysis. This dual-species interaction study showed that Mab can persist when cultured alongside Pa but experiences substantial loss of viability when cultured in a cell-free Pa extract containing QS-dependent secreted factors. The non-cidal outcome in the co-culture and drastic changes at a molecular level in Mab strongly suggest that Mab employs adaptive mechanisms for survival and coexistence with Pa.
2 Materials and methods
2.1 Bacterial strains and reagents
The bacterial strains Pa WT (PAO1 Ochsner) (Holloway et al., 1979), ΔlasR (in PAO1 Ochsner background) (Wilder et al., 2011), and smooth morphotype Mab 390S (Howard and Byrd, 2000; Greendyke and Byrd, 2008) and Mab-lux (Gupta et al., 2017) were used in this study. For routine culturing and maintenance, Luria Bertani (LB) was used for both Pa and Mab strains. For selection of Mab for species-specific CFU enumeration from co-cultures, LB agar supplemented with 200U/ml of antibiotic polymyxin B sulfate salt (Millipore sigma) was used. No selective media was necessary for Pa as it forms colonies after 24 h unlike Mab, which takes 3–5 days.
2.2 In vitro co-culturing
A planktonic co-culture assay was used to investigate direct interactions between Pa and Mab 390S. LB-Tween 80 (0.05%) media was inoculated from a freshly streaked single colony of Pa strains (WT and the lasR mutant) or Mab culture started from a freezer stock. The bacterial cultures were appropriately diluted to set-up mono and co-cultures in 10 ml LB-Tween in T25 flasks. The starting OD of Pa and Mab was set at 0.05 and 0.15, respectively as used previously to account for growth rate differences (Costa et al., 2015; Birmes et al., 2017). Three monocultures: WT (P), ΔlasR (L), and Mab (MB) and the two co-cultures - Mab+WT Pa (PMB), Mab+ΔlasR (LMB) were incubated at 37°C with shaking at 150 r.p.m. for 24 h. After 24 h, 10-fold serial dilutions were prepared in PBS media and spot-plated (5 µl) onto LB agar and LB agar supplemented with polymyxin B (200U/ml) for growth of Pa and Mab, respectively for colony enumeration.
2.3 Mab growth in Pa supernatant
Mab growth was assessed in spent supernatants of WT (P) and the isogenic ΔlasR (L) strains of Pa to examine the effect of Pa secreted effectors. The Pa strains were grown overnight in LB-Tween media at 37°C, 150 r.p.m. shaking from a freshly streaked isolated colony. The cultures were diluted to OD600 of 0.05 and grown for 24 h. Thereafter, cultures were centrifuged (4000 r.p.m., 30 min., 4°C) and filtered through 0.22 µm pore size cellulose acetate filters (Corning, 43020). The filter-sterilized supernatant was plated (100 µl) onto LB agar plates to ensure that no Pa cells were present in the filtrate. Mab cultures were set at an OD600 of 0.15 in 10 ml LB-Tween (T25 flasks) with 0%, 5%, 25%, and 50% v/v, of Pa spent monoculture supernatant. The cultures were incubated at 37°C with shaking (150 r.p.m.) and after 0, 24, and 48 h 10-fold serial dilutions were spot-plated (5µl) onto LB agar for each time point. The CFU/ml data is an average from three biological experiments each with 4 technical replicates. The same assay was performed with 50% v/v supernatants derived from Pa-Mab co-cultures as well.
2.4 lasR complementation
To achieve complementation, low-copy-number plasmids, pJN105 (Newman and Fuqua, 1999) and pJN105.lasR (pJN105.L) (Lee et al., 2006) were transformed into chemically competent lasR mutant cells as described (Wilder et al., 2009). Plasmid pJN105 served as an empty vector control whereas pJN105.lasR expressed lasR from an arabinose -inducible ara-BAD promoter. The spent supernatant from Pa strains (WT/pJN105, ΔlasR/pJN105, and complement ΔlasR/lasR+) was harvested as described earlier except the cultures were induced with 50mM arabinose during growth. Mab growth was evaluated in spent supernatants using a luminescence-based 96-well reporter assay (Gupta et al., 2017). Briefly, Mab-lux was grown in 50% v/v spent supernatant in a total well volume of 100 µl at a final OD600 of 0.01. The plates were incubated at 37°C and the luminescence signal was measured after every 24 h for 2 days. An aliquot was taken for serial dilution and plating to determine CFU for the indicated times.
2.5 Heat, protease sensitivity and size fractionation determination of SPAM
To gain insights into the heat stability, size and chemical nature of Pa-derived anti-mycobacterial factor known as SPAM, Mab growth was examined in heat-denatured, protease treated and size fractionated Pa supernatants. For heat treatment, filter-sterilized supernatants were heated at 95°C for 15 min. in a water bath. For protease treatment, cell-free Pa supernatant was incubated with 200 µg/ml proteinase K for 2 h at 37°C. Size fractionation of the spent supernatant was carried out by centrifuging through 3kDa MWCO filters (Millipore, SIGMA, UFC900308) for 20 min at 4000 r.p.m. Both retentate and flow-through fractionated samples were filter-sterilized using cellulose acetate, low protein binding syringe filters. Mab growth assessment in the heat-denatured supernatant was carried out in flasks as described in the supernatant assay while a high throughput microtiter plate assay described before in lasR complementation was adopted for proteinase K (Prot K) treated, and 3kDa size fractioned Pa supernatants. Controls including no supernatant and unfractionated/untreated supernatant were included in the assay. The data is an average from 3 biological experiments each with a total of 9 technical replicates.
2.6 Total RNA extraction
Total RNA was isolated from mono and co-cultures involving Pa and Mab after 24 h of incubation at 37°C by using previously reported mycobacterial trizol-based RNA isolation technique (Rohde et al., 2012) with modifications. Mab and Pa cultures were mixed with RNA Protect reagent (Qiagen), mixed by inversion, and centrifuged (4300 r.p.m.) for 20 min. The supernatant was discarded, and the pellet was resuspended in 1 ml RNA Protect reagent before storage at -80°C. The pellets were disrupted using a Mini-Bead Beater 16 (Biospec) 2x at max speed for (for 1 min followed by incubation on ice for 1 min. After this, steps were carried out at 4°C centrifuge as described previously using RNeasy kit (Qiagen) (Rohde et al., 2012). Integrity of isolated RNA was determined using Agilent RNA ScreenTape system.
2.7 Dual RNAseq
Transcriptome analysis was carried out with three biological replicates of each sample (mono and co-cultures). Depletion of Ribosomal RNA, mRNA library preparation and sequencing were performed by SeqCenter (earlier MiGS). Library preparation was performed using Illumina’s Stranded Total RNA Prep Ligation with Ribo-Zero Plus kit and 10 bp IDT for Illumina indices. Around Total RNA (275ng) was ribodepleted using oligonucleotide sequences specific for Pa and Mab. These custom probes are listed in Supplementary Table 1. The samples were DNAase treated with Invitrogen DNAase (RNAase-free) and sequencing was performed (NextSeq2000, 2x50bp reads). Sequencing quality control and adapter trimming was performed with bcl-convert (v3.9.3) (https://support-docs.illumina.com/SW/BCL_Convert/Content/SW/FrontPages/BCL_Convert.htm). Read mapping was performed with HISAT2 (Kim et al., 2019) and read quantification was done using Subread’s featureCounts functionality (Liao et al., 2014). Reads for Pseudomonas monoculture samples (P1, P2, P3, L1, L2, L3) were mapped to the P. aeruginosa PAO1 RefSeq genome NC_002516.2, Mab solo culture samples (MB1, MB2, and MB3) to the RefSeq genome GCF_000069185.1 (ASM6918v1), and co-culture samples (PMB1, PMB2, PMB3, LMB1, LMB2, and LMB3) were simultaneously mapped. Differential expression analysis was performed using edgeR’s Quasi-Linear F-Test (qlfTest) functionality against treatment groups. Differentially expressed genes (DEGs) were identified with a cut-off of log2 fold-change (FC) (-1 ≥1 to ≤1) and p-value < 0.05 (Supplementary Table S2). The KEGG pathway analysis was performed using limma’s (Ritchie et al., 2015) “kegga” functionality with genes with FDR<0.05 (adjusted p-value).
3 Results and discussion
3.1 Mab antagonism in a planktonic co-culture with Pa
To gain an initial understanding of the direct interactions between Pa and Mab, we compared growth of individual species when present alone (monoculture) and in the presence of the other species (co-culture). A ΔlasR strain was also included to assess the role of LasR, a master transcriptional regulator of Pa QS in shaping interactions with Mab. As noted above, natural lasR mutants are frequently isolated from chronically infected CF patients, suggesting dysregulation of QS-regulated virulence factors may offer growth advantage to competitors (Hoffman et al., 2009; Feltner et al., 2016). If LasR-dependent factors are key to Pa-Mab interactions, this in vivo evolution of Pa could significantly impact the progression of Mab infections. Using the experimental setup illustrated in Figure 1, we evaluated the growth of non-mucoid Pa and Mab (smooth morphotype) in mono- and co-cultures by CFU enumeration. Co-cultures were started with Pa at a lower density (OD600 = 0.05) compared to Mab (OD600 = 0.15) to compensate for the faster replication rate of Pa. Enumeration of CFU from initial cultures, however, indicated higher CFU/ml of Pa which presents a variable that could affect Pa-Mab interactions. Growth of both WT and ΔlasR Pa strains remained unaffected in the presence of Mab. However, in contrast to the 2–3 log increase in CFU observed when Mab was grown alone, both WT and ΔlasR Pa exerted bacteriostatic antagonism of Mab (Figures 2A, B). There was no change in the smooth colony phenotype of Mab after the co-culturing. The observed antagonism could potentially be attributed to nutrient deprivation of Mab in the presence of fast-growing Pa. To address this, Mab was cultured in 50% water-diluted media to simulate nutrient-limited conditions. After 24 h, Mab showed no significant growth differences between the diluted and the undiluted media (Supplementary Figure 1), indicating that nutrient limitation did not hamper Mab growth. These findings suggest nutrient starvation is unlikely the sole cause of observed Mab antagonism in a co-culture with Pa. Instead, the primary mechanism underlying bacteriostatic inhibition of Mab appears to be a Pa derived, LasR-independent mechanism. This observation is in contrast to a recent report where no Mab antagonism was observed in the planktonic co-culture with Pa, even when different Pa: Mab ratios were tested (Idosa et al., 2022). This discrepancy may be attributable to differences in media, bacterial strains or culture conditions. The observed Mab growth arrest in the presence of Pa in this study indicates that Mab may adopt a non-replicating persister-like state as a strategy to counter Pa antagonism.
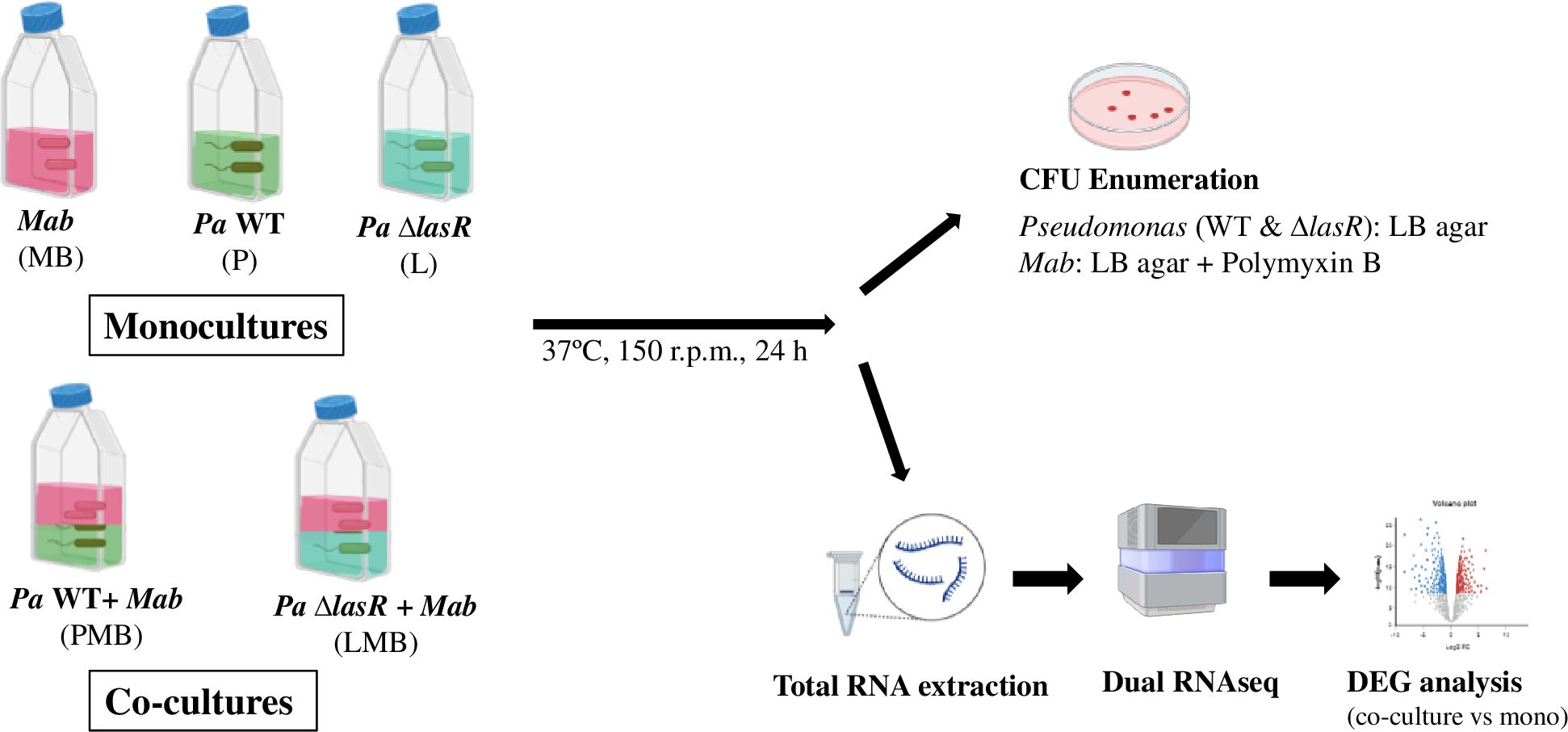
Figure 1. Planktonic co-culture model of Mab-Pa interactions. Monocultures of Mab (MB) and Pa strains (P, L) and co-cultures of Mab + Pa WT (PMB) and Mab + Pa ΔlasR (LMB) were grown for 24 h. Pa and Mab growth was evaluated by plating onto indicated selective media. The mono and co-culture samples were also processed for dual RNAseq analysis.
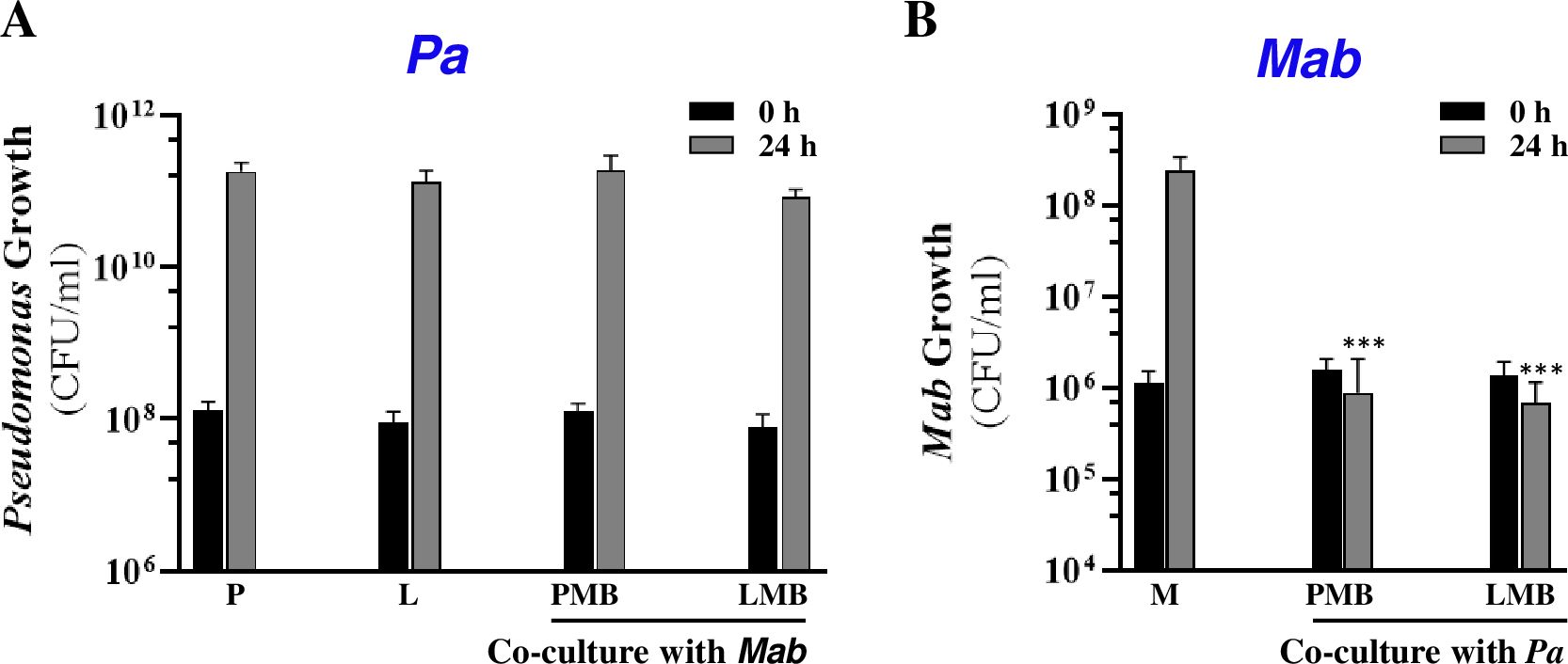
Figure 2. Pa antagonizes Mab. Growth (CFU/ml) of (A) Pa and (B) Mab strains when grown solo and in co-cultures at indicated times. P: Pa WT; L: Pa ΔlasR; MB: Mab; PMB: Pa WT+ Mab; LMB: Pa ΔlasR + Mab. *** denotes statistically significant difference (P<0.0001) (paired t-test) from Mab monoculture after 24 h.
3.2 Secreted Pa factor(s) are bactericidal toward Mab
Next, we investigated whether Mab inhibition by Pa in co-cultures was mediated by a contact-dependent mechanism, as previously reported, or secreted factors. To address this, we examined the effect of Pa cell-free culture supernatant on Mab growth. When Mab was exposed to a range of concentrations (0%, 5%, 25% and 50% v/v) of Pa WT supernatant (sup), a dose- and time-dependent reduction in CFU was observed (Figure 3A). Exposure to 50% (v/v) Pa WT sup resulted in a 3–4 log decline in CFU compared to the control (no sup) within 24 h and complete sterilization within 48h, a bactericidal effect not even observed with first-line antibiotics used for clinical treatment of Mab infections (Maurer et al., 2014). This indicates the presence of a highly potent mycobactericidal compound. However, this drastic cidal phenotype was lacking with the Pa ΔlasR supernatant, which caused only minor ~1-log reduction in CFU at 48h (Figure 3B). Heterologous expression of the lasR allele from an arabinose inducible promoter in the Pa ΔlasR strain restored the cidal activity as indicated by the decline in Mab CFU and luminescence signal (Figure 3C; Supplementary Figure 2). This suggests that a LasR-dependent secreted factor (directly or indirectly regulated) mediated the potent cidal activity against Mab. We designated this cidal factor as SPAM (Secreted Pseudomonas Antagonist of Mab) throughout the rest of this study.
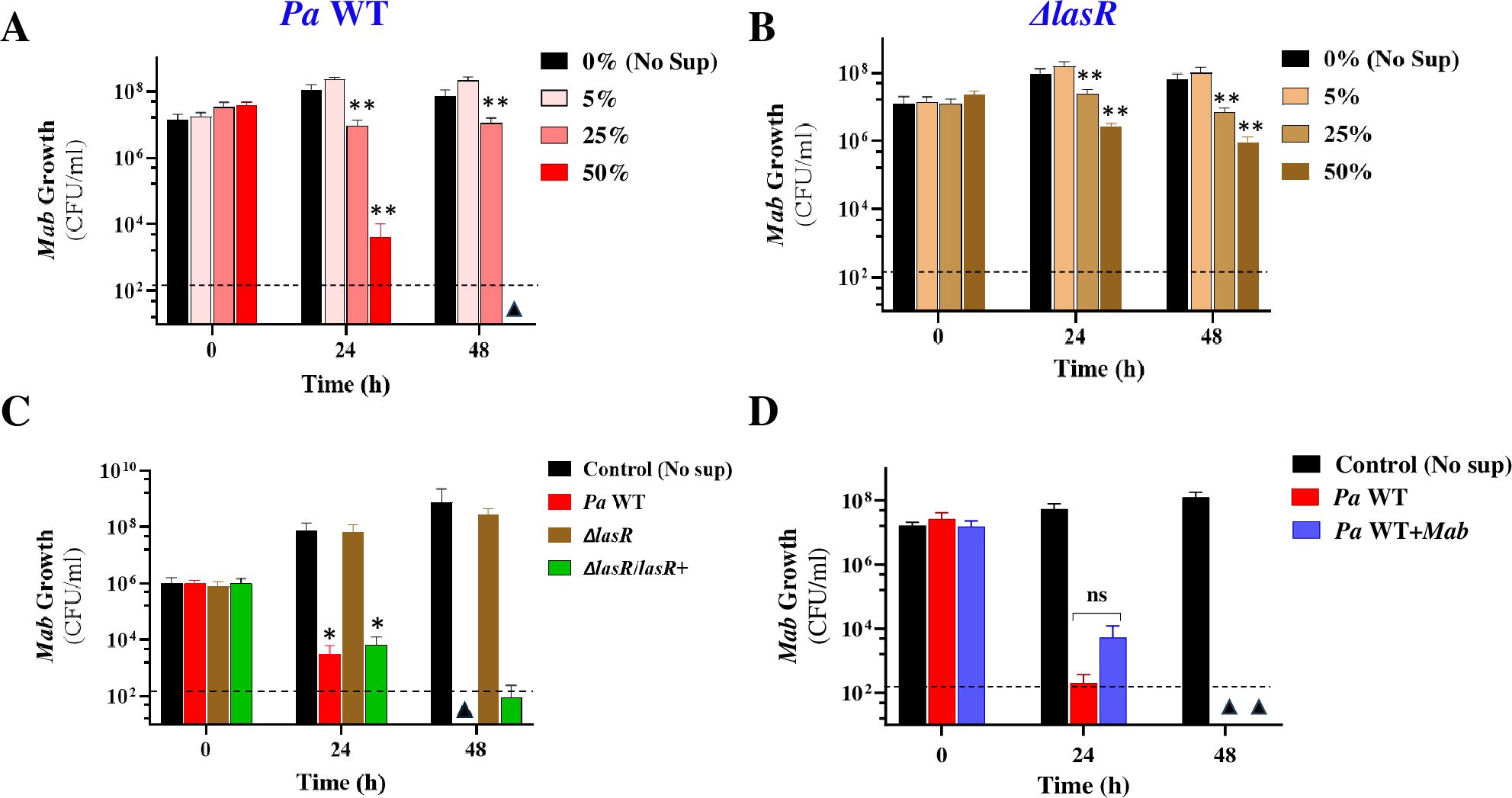
Figure 3. Mab viability in spent Pa supernatants. Mab growth (CFU/ml) in media supplemented with (A) Pa WT and (B) Pa ΔlasR culture supernatants at 0%, 5%, 25% and 50% concentration (v/v) (C) Mab-lux growth in 50%v/v spent Pa supernatants after complementation of lasR mutation. A lasR allele was expressed from an arabinose inducible promoter carrying plasmid, pJN105. Pa WT and Pa Δ lasR strains carry the empty plasmid (D) Mab growth in sup obtained from Pa WT mono and co-cultures with Mab. * P<0.01, ** P<0.001 from control, ns, insignificant (paired t-test). The dotted line denotes the limit of detection (LoD) (2*102 colonies) and triangles denote samples where CFU was below LoD.
The striking susceptibility of “naïve” Mab to killing by secreted Pa factors versus the tolerance of Mab when grown in co-culture with Pa suggested that interspecies interactions have either downregulated the production of cidal factors by Pa or rendered Mab more resistant to their effects, or both. To investigate these hypotheses, we explored the possibility of 1) downregulation of Pa secreted virulence factors and 2) Mab adaptation during co-culturing by evaluating Mab growth in 50% v/v spent supernatant from co-cultures as well. The fact that supernatants from Pa co-cultured with Mab exerted a cidal activity against Mab comparable to that observed with supernatant from Pa monoculture supports the conclusion that Mab-induced downregulation of SPAM did not occur in the co-culture (Figure 3D). The remaining likely explanation is that Mab mounts adaptative responses to counter killing by Pa during their side-by-side growth. The molecular adaptation of Mab in response to Pa presence was explored through dual RNAseq analysis later in the study.
3.3 SPAM characterization
To characterize the potent SPAM factor, we assessed Mab growth in heat-denatured, protease-treated, and size-fractionated spent Pa supernatant. Heat inactivation of Pa (WT) supernatant largely abolished the cidal activity indicating the heat-labile nature of SPAM (Figure 4A). To determine whether SPAM is proteinaceous in nature, Pa sup was pretreated with proteinase K (Prot K) and viability of a Mab-lux reporter strain was evaluated through two readouts - luminescence and CFU. Prot K is a broad-spectrum serine protease that cleaves peptide bonds next to the carboxyl-terminal of aromatic and hydrophobic residues (Bajorath et al., 1988). Two controls, untreated and Prot K treated media were included. The protease treatment did not eliminate the cidal activity of Pa supernatant (Figure 4B; Supplementary Figure 3A). Both the untreated and Prot K digested cultures showed a ~4–5 log decrease in Mab CFU after 48 h compared to controls (Figure 4B). These results strongly suggest that SPAM is not proteinaceous in nature, although we cannot definitively rule out a protein factor involvement because some Pa secreted proteins may resist degradation by Prot K digestion (Bajorath et al., 1988). Size fractionation of supernatant indicated that the potent cidal SPAM factor is >3kDa in size as the retentate fraction killed Mab but not the flow-through fraction (<3kDa) (Figure 4C; Supplementary Figure 3B). This characterization would eliminate small molecules like phenazines or respiratory toxins (e.g. HQNO) as prime candidates, unless they were encapsulated in outer membrane vesicles as reported previously (Vrla et al., 2020). Further investigation will be required to isolate and identify this LasR-controlled interspecies antagonist.
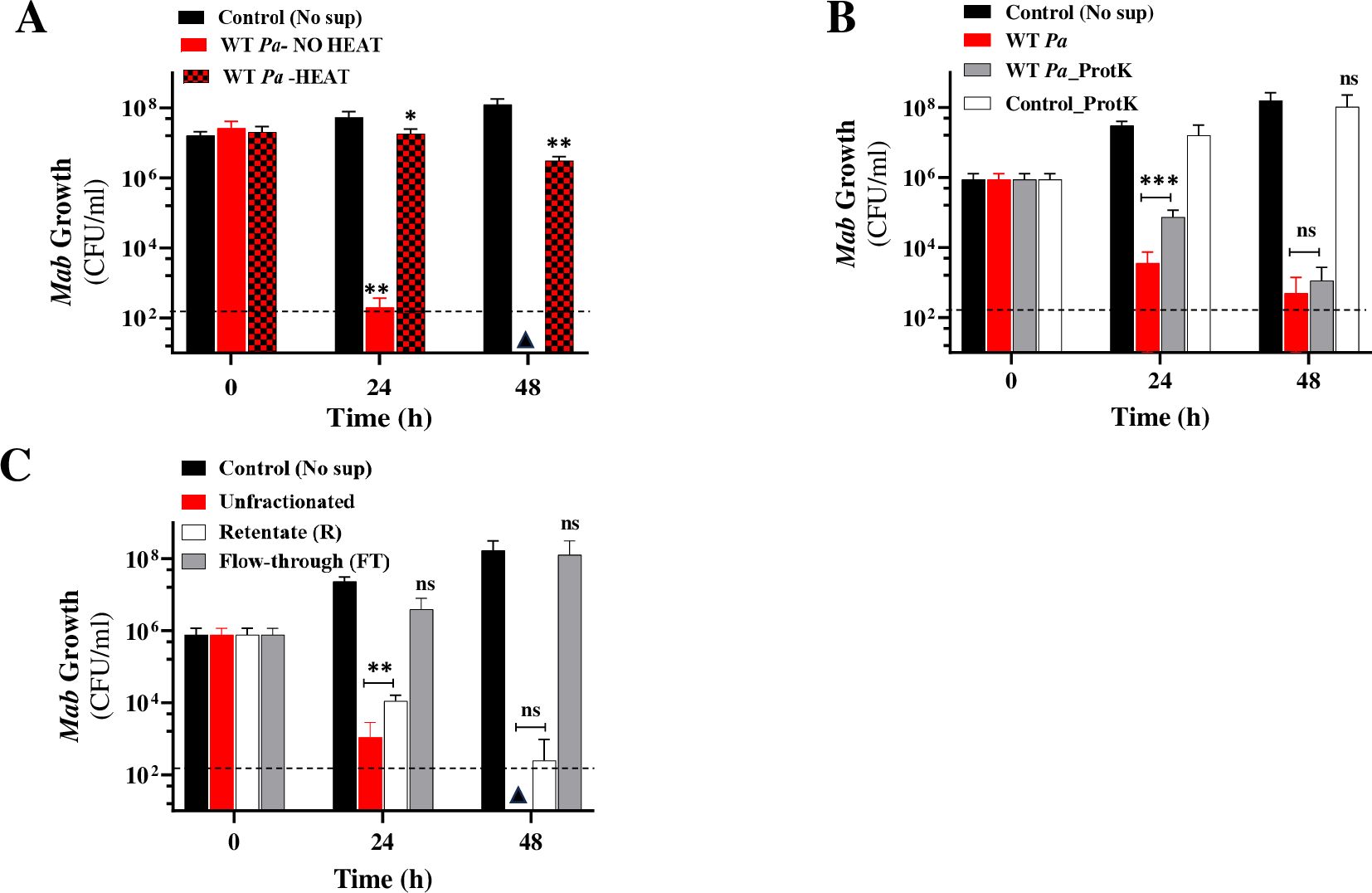
Figure 4. SPAM characterization. Mab growth (CFU/ml) in WT Pa supernatant (50% v/v) (A) with and without heat denaturation (B) proteinase K (ProtK) treated (C) Size fractionated (3kDa MWCO). The data is an average from at least three experiments. Note: For panel B and C, 101 is the lowest dilution plated. * P<0.01, **p <0.001 from control, ***P=0.0001, ns, non-significant (two-tailed paired t-test). The dotted line denotes the limit of detection (LoD) (2*102 colonies) and triangles denote samples where CFU was below LoD.
Overall, our co-culturing and supernatant assays investigated direct and indirect Mab-Pa interactions which demonstrated a Pa-derived negative impact on Mab growth. Static growth of Mab when in physical contact with Pa appears to be mediated by either a las independent or a non-QS mechanism whereas cidal activity during indirect interaction with Pa is mediated via Las QS dependent secreted factors.
3.4 Mab adaptation at a molecular level during co-culturing with Pa
To explore molecular cross talk between the two pathogens during co-culturing, we carried out dual RNAseq analysis and compared the changes in gene expression in both Pa and Mab in a co-culture vs monoculture after 24h, corresponding to the planktonic co-culture assay point (Figure 1). We achieved a sequencing depth of 20–80 million total reads per sample meeting the minimum (106) reads requirement for a comprehensive differentially expressed genes (DEGs) analysis (Toung et al., 2011; Westermann et al., 2016). The differentially regulated Mab and Pa genes are listed in Supplementary Table 2 and depicted with heat maps and Venn diagrams (Figure 5; Supplementary Figure 4). Notably, the Mab transcriptome exhibited much more dramatic changes than Pa during co-culture of these two CF pathogens. More than 68% of the coding genes of Mab (3359/4920) were differentially regulated in the presence of WT Pa. Out of a total of 3359 Mab DEGs, 1800 were up-regulated and 1559 were down-regulated whereas a mere 275 and 226 Pa genes showed up and down-regulation, respectively (Figures 5A, C).
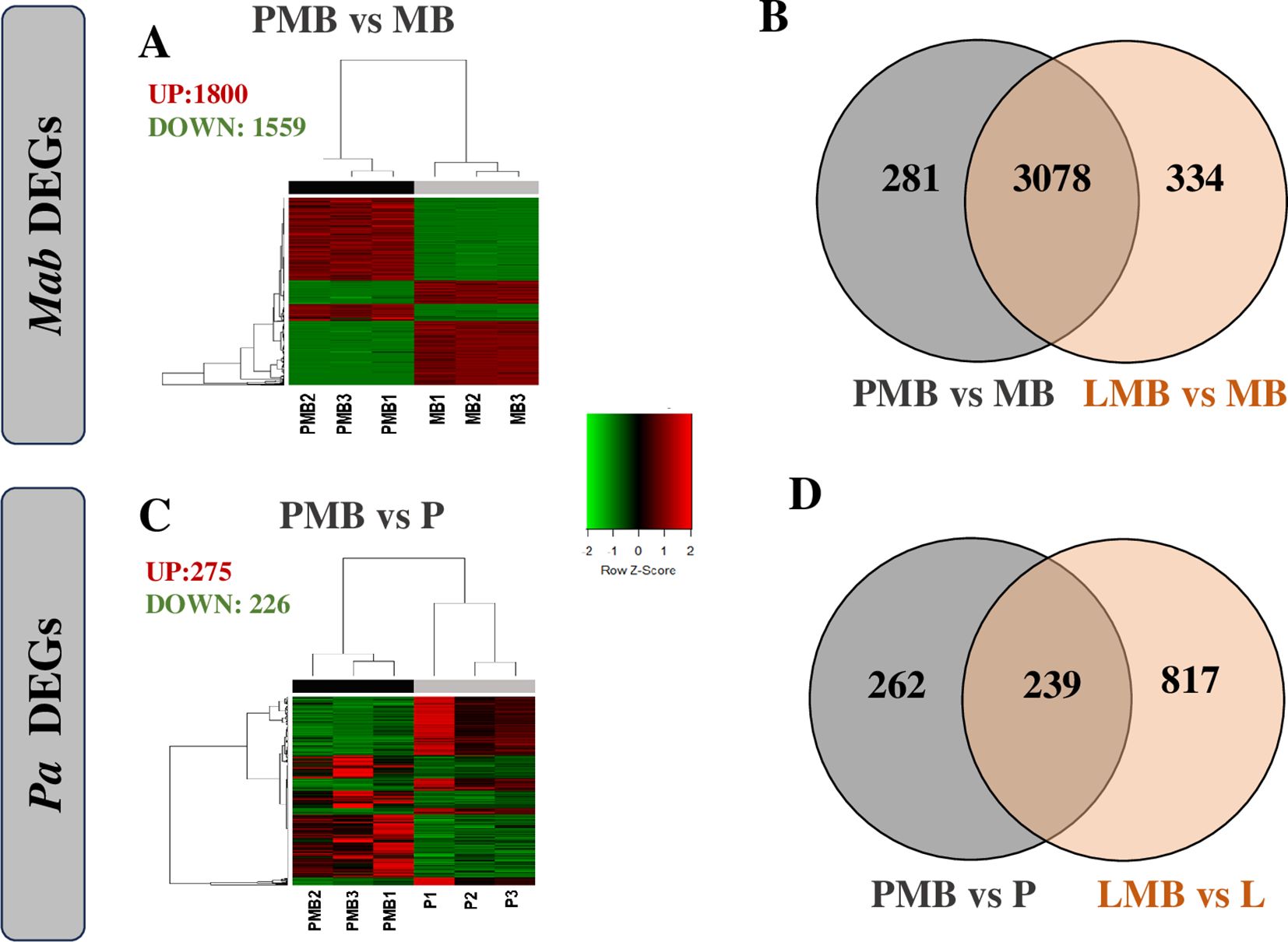
Figure 5. Transcriptomic responses of Mab (A, B) and Pa (C, D). Differentially expressed genes of Mab (A) and Pa (C) normalized counts per million of three replicates are represented by heat map. Venn diagrams represents overlap in Mab expression when cultured with WT and lasR Pa (C) and in Pa expression when cultured with Mab (D). P: Pa WT, L: ΔlasR, MB: Mab, PMB: WT Pa-Mab co-culture, LMB: Δ lasR -Mab co-culture.
The transcriptional profile of Mab and Pa genes did not show any striking differences when the ΔlasR Pa strain was employed. Mab showed altered regulation of 3412 genes with up-regulation of 1866 and down-regulation of 1546 when co-cultured with ΔlasR (Supplementary Figure 4A). Like WT Pa, ΔlasR showed relatively modest changes with 488 and 568 genes up- or down- regulated, respectively (Supplementary Figure 4B). The ΔlasR strain showed downregulation of signature genes including direct LasR-dependent genes (lasI, lasR, lasA, apr, rsaL), phz2 operon, hcnAB, qscR, rsaL, as well as LasR controlled rhl-system (rhlR, rhlAB), and pqs system genes (pqsH) as expected with loss of this QS signal receptor (Supplementary Table 2).
Overall, the extensive transcriptional reprogramming of Mab during co-culture with Pa reflects adaptive responses that likely contributed to Mab’s ability to withstand potentially cidal Pa secreted factors, whereas Pa appears to remain relatively undeterred during this rather one-sided interspecies interaction.
3.5 Co-culture with Pa triggers slow-growing persister-like state in Mab
Persistence, which is characterized by slow or nonreplicating state, leads to antibiotic tolerance requiring prolonged treatment and high chances of treatment failure and recurring infections. Slow growth is marked by down-regulation of DNA replication, transcription and translation machinery, cell-division processes, electron transport, ATP-proton motive force and other cell processes to conserve energy (Beste et al., 2009). Differential gene expression analysis between Mab-Pa co-cultures vs Mab monoculture revealed decreased carbon flux into metabolic pathways, reduced respiration, and energy metabolism (Supplementary Table 3, Supplementary Figure 5). Genes involved in cell division (ftsZ, ftsK, ftsX, ftsE, ftsH, and ftsW), DNA replication (dnaA, dnaN, gyrA, gyrB), and transcription and translation (fusA, tuf, tsf, rpoA, rpoB) were repressed in the presence of Pa. A non-functional LasR did not change this response as Mab co-cultured with the ΔlasR exhibited a similar transcriptomic profile. There was also down-regulation of type-I NADH dehydrogenase encoded by nuoA-N genes, succinate dehydrogenase (sdhABC), cytochrome oxidases, and F0-F1 ATP synthase genes (atpA-E) (Figure 6; Supplementary Table 3). This transcriptional response in Mab is akin to changes observed in M. tuberculosis in its non-replicating persister phase (Shi et al., 2005; Maurya et al., 2018). Entry of Mab into a non-replicating state could be a way to endure hostile conditions generated by Pa under co-culture conditions. This slow-growing state could have clinical implications, by contributing to the disconnect between in vitro and in vivo antibiotic susceptibility and to the ineffectiveness of current treatment regimens.
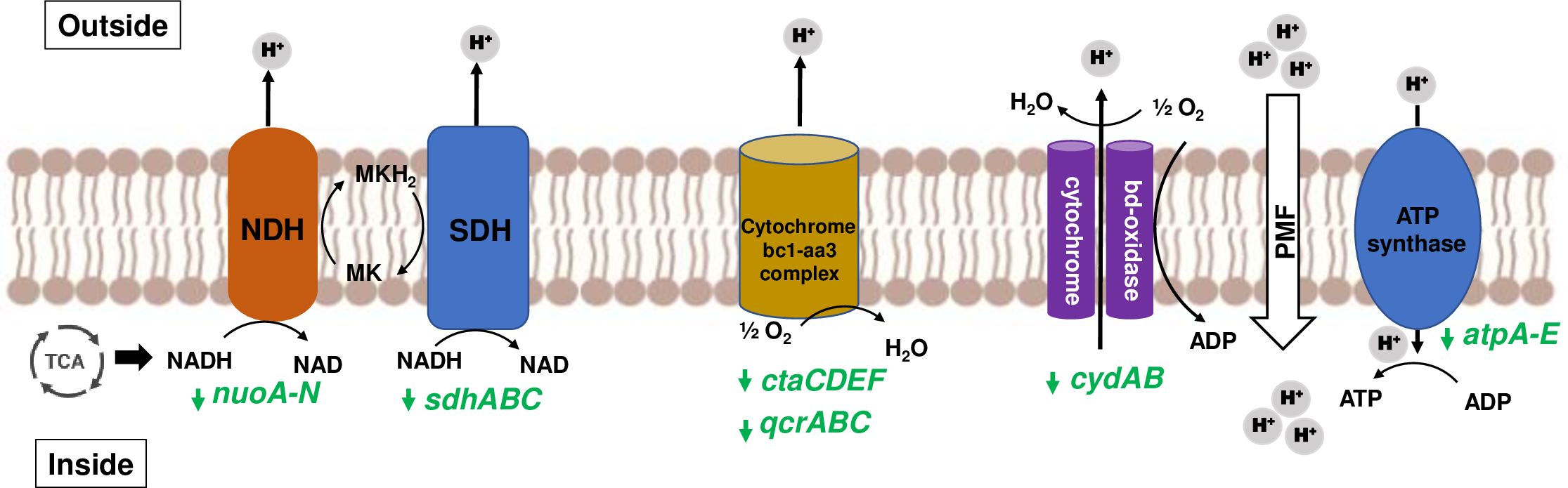
Figure 6. Slowing of Mab respiration during co-culturing. Mycobacterial NADH from TCA cycle enters mycobacterial electron transport chain (ETC) and supplies electrons to NADH dehydrogenase (NDH) and succinate dehydrogenase (SDH) to reduce menaquinone (MK) to menaquinol (MKH2). Cytochrome oxidases (bc1-aa3 complex and bd-type) oxidize back MK by transferring electrons to oxygen. Proton motive force (PMF) generated due to proton translocation is utilized by ATP synthase to produce ATP. Genes encoding ETC components are represented in green, and downward arrow denotes downregulation. FC values of affected genes are listed in Supplementary Table S3 under Electron Transport Chain heading.
3.6 High induction of HGT elements in Mab during co-culture
The Mab genome contains a 23-Kb plasmid, pMAB23, and several other gene clusters predicted to be acquired by horizontal gene transfer (HGT) from closely related mycobacterial species and from non-mycobacterial community (Ripoll et al., 2009). Strikingly, mobile elements, prophage and prophage-like elements and other HGT acquired genes including those putatively derived from Pseudomonas sp. were highly upregulated, with some having a log2 FC as high as 10. All 22 genes (MAB_p01 to MAB_p22c) encoded on the pMAB23 plasmid including the essential gene, repA (MAB_p16c), and putative arsenate resistance conferring genes were induced (Figure 7; Supplementary Table 4). In addition, several Mab genes in a 61 kb prophage region (Dedrick et al., 2021) (MAB_1723 to MAB_1833) exhibited strong transcriptional response in the presence of Pa (both WT and the lasR mutant). Three-prophage like elements (Ripoll et al., 2009) and 17-gene clusters acquired from the non-mycobacterial community were also differentially regulated. Out of the 17-clusters, 13 showed upregulation and only two, involved in degradation of phenylacetic acid (MAB_0899c-0911) and DNA (MAB_1093c-1098), respectively were down-regulated (Figure 7; Supplementary Table 4). The increased expression of mobile elements, prophage and other HGT genes in Mab under co-culture conditions suggest functional importance beyond being just passive elements in the genome. Increased expression of prophage genes is associated with survival under certain environmental conditions such as low pH in M. avium (Miao Zhao et al., 2016). A transcriptomic response including higher expression of transposable elements, plasmid and viral-borne genes has been reported in other organisms such as slow-growing yeast cells for driving diversity for future stressful conditions (van Dijk et al., 2015).
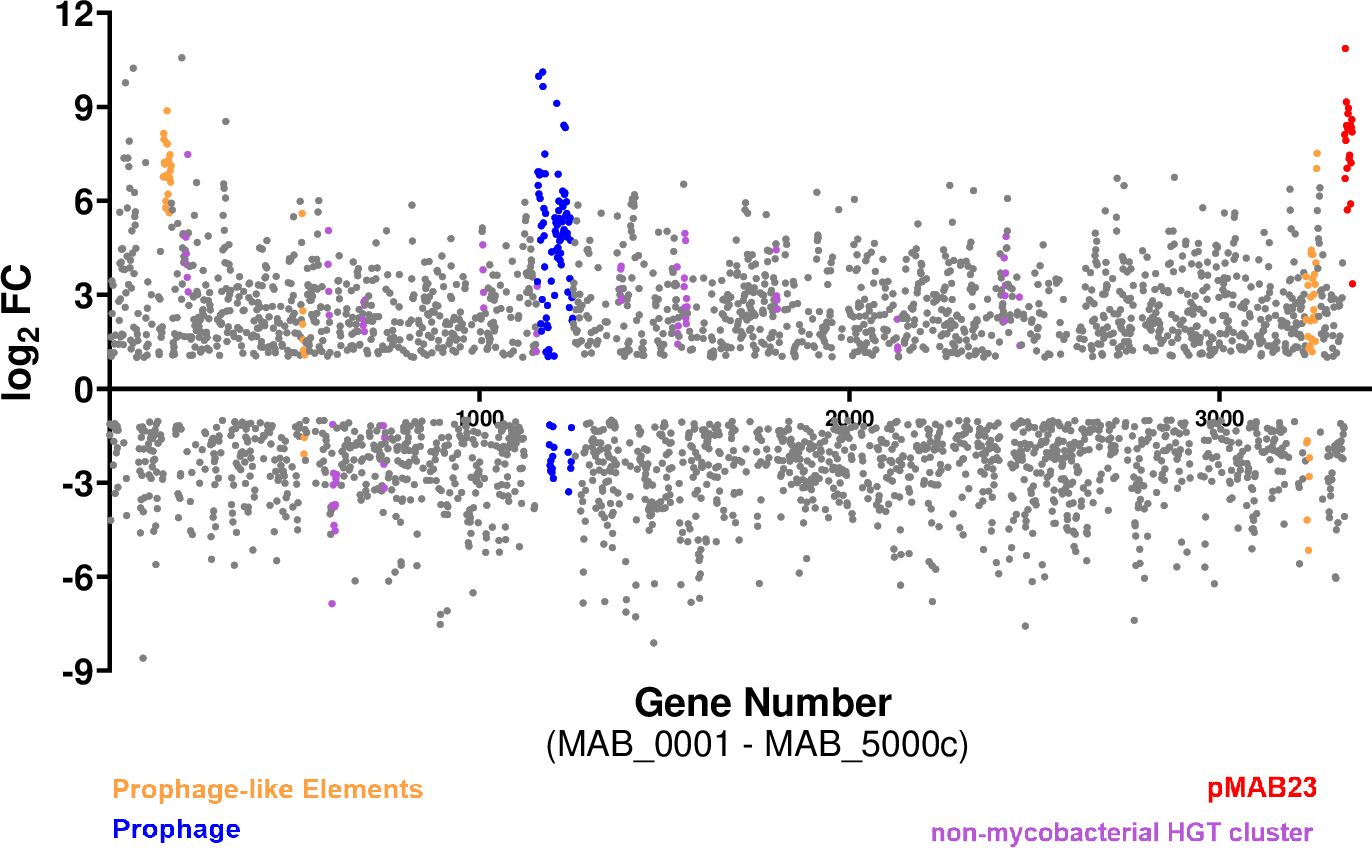
Figure 7. Up regulation of Mab HGT elements in a co-culture. A scatter plot showing differentially regulated genes in Mab (PMB vs MB) where each dot represents a differentially expressed gene. The colored dots are the putative HGT acquired genes.
3.7 Remodeling of central carbon metabolism of Mab in presence of Pa
One of the major transcriptional responses noted in Mab when co-cultured with Pa was in central metabolic pathways. RNAseq data reflected a switch in carbon flow from sugars to fatty acids during co-culturing, with downregulation of glycolysis, gluconeogenesis, and Pentose Phosphate Pathway (PPP) pathways. The key enzymes involved in these pathways such as NADP-dependent dehydrogenase, aldolase, isomerases, and ATP-generating phosphoglycerate kinase, fructose 1,6-bisphoatase (glpX) and PEP carboxykinase (pckA) (Figure 8; Supplementary Table 5) were downregulated. This indicates reduced energy and biosynthetic precursors demand, aligning with the Mab phenotypic shift from replicating to a slow growing phase. Along these same lines, energy-consuming anabolic pathways were downregulated while lipid catabolism increased. There was an upregulation of lipases and fatty acid β-oxidation genes, along with decreased fatty acid synthesis (Supplementary Table 5), which suggest lipid breakdown for membrane biosynthesis or storage as triacylglycerol (TAG) for persistence and future energy sources. The elevated transcripts of methylcitrate cycle (MCC) genes (prpDCB) and methylmalonate-semialdehyde dehydrogenase encoded by mmsa (log2 FC: 4) indicate propionyl CoA accumulation and subsequent detoxification (Munoz-Elias et al., 2006; Savvi et al., 2008) (Figure 8; Supplementary Table 5). Reduced expression of propionyl-CoA carboxylase (accD5), methylmalonyl-CoA mutase (mutB) genes reflect preferential utilization of MCC over methyl malonyl (MM) for propionyl detoxification (Supplementary Table 5). Upregulation of mycolic acid synthesis genes (MAB_2028-MAB_2031 and beta-keto acyl synthases) hints at the incorporation of acetyl and propionyl CoA into mycolic acids. Key enzymes, kasA and kasB with a role in elongation of meromycolate chain showed upregulation (Supplementary Table 5). In addition, upregulation of triacylglycerol synthases (tgs) indicates TAG synthesis. Three out of 7 tgs genes encoded in the Mab genome (MAB_0854, MAB_1278, and MAB_2964), exhibited increased expression. This differential induction of tgs is also noted in M. tuberculosis under different inducing environments. Out of 15 tgs encoded in Mtb, tgs1 was upregulated under hypoxia/nitric oxide stress condition whereas tgs2 was induced by low pH (Viljoen et al., 2016; Maurya et al., 2018). TAG accumulation is a hallmark of persisting Mtb which aids in drug tolerance and survival in the host (Maurya et al., 2018).
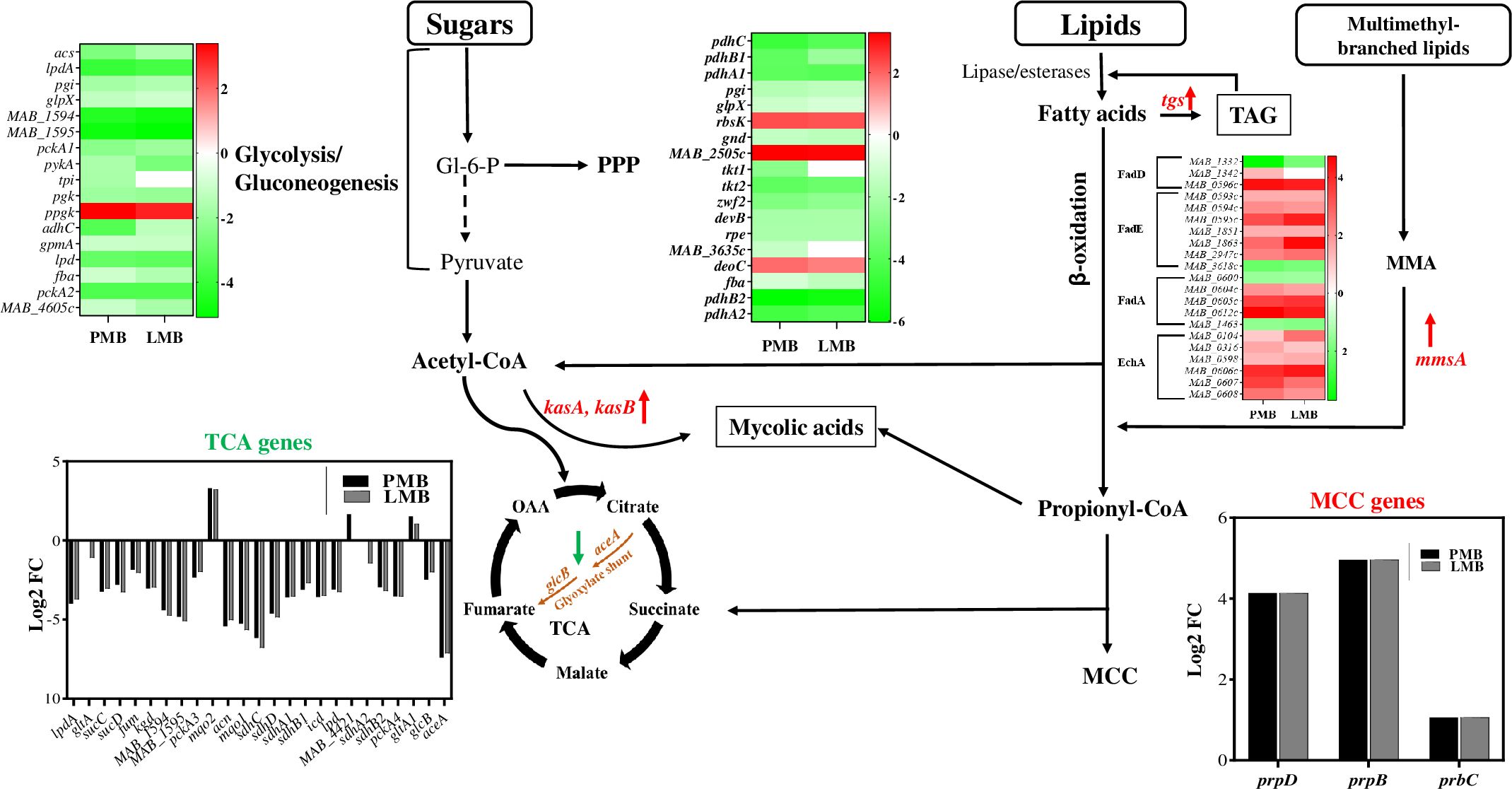
Figure 8. Carbon flow switch from sugars to fatty acids during co-culturing. The figure depicts an integrated view of central carbon metabolic pathways with differential expression of associated genes via heat maps, bar graphs, and by colored up and down arrows. PPP, pentose phosphate pathway; TCA, tricarboxylic acid cycle; OAA, oxaloacetate; MCC, methyl citrate cycle; TAG, triacylglycerol; MMSA, Methylmalonate semialdehyde. Green and red arrow denotes down and upregulation, respectively. Carbon flow shunt to mycolic acids and TAG storage (boxed).
Based on transcriptomic data, we speculate that excess fatty acids are channeled into intracellular lipids accumulation in the form of triacylglycerols (TAG) which can be hydrolyzed back by lipases when needed. TAG synthesis serves as a storage sink for persistence and survival under stress such as co-culture conditions. Resource limitation and competition during co-culture, perhaps enhanced by the lack of ample available sugars and other catabolizable carbon sources in LB medium (Sezonov et al., 2007), may be one driver of fatty acid catabolism and rerouting of carbon flow towards storage and alterations in cell wall composition to maintain cell viability and integrity. Similar carbon flow re-routing has been noted during Mtb infection (Garton et al., 2008; Russell et al., 2009) and has implications in persistence and drug tolerance. The phenotypic growth arrest and persister-like metabolic remodeling may also reflect the impact of respiratory toxins and other antimicrobial metabolites secreted by Pa. Secreted Pa factors like HCN, pyocyanin, and quinolone N-oxides inhibit Staphylococcus aureus growth by blocking respiration, leading to a small-colony persister phenotype (Biswas and Gotz, 2021).
3.8 Co-culturing of Mab and Pa triggers competition for Fe
Culturing Mab and Pa together forces them to compete for common essential nutrients including Fe to replicate and survive. Pa utilizes several strategies to acquire iron under infection conditions including production and secretion of siderophores (Lamont et al., 2009; Martin et al., 2011). Transcriptional profiles from dual RNAseq showed upregulation of siderophore-mediated iron-uptake, and downregulation of iron-storage in both bacterial pathogens, indicative of a Fe-starvation response (Figure 9; Supplementary Table 6). This pattern of altered expression of iron-responsive genes in both Pa and Mab in our co-culture model is indicative of competition between the interacting pathogens for Fe and likely other essential nutrients.
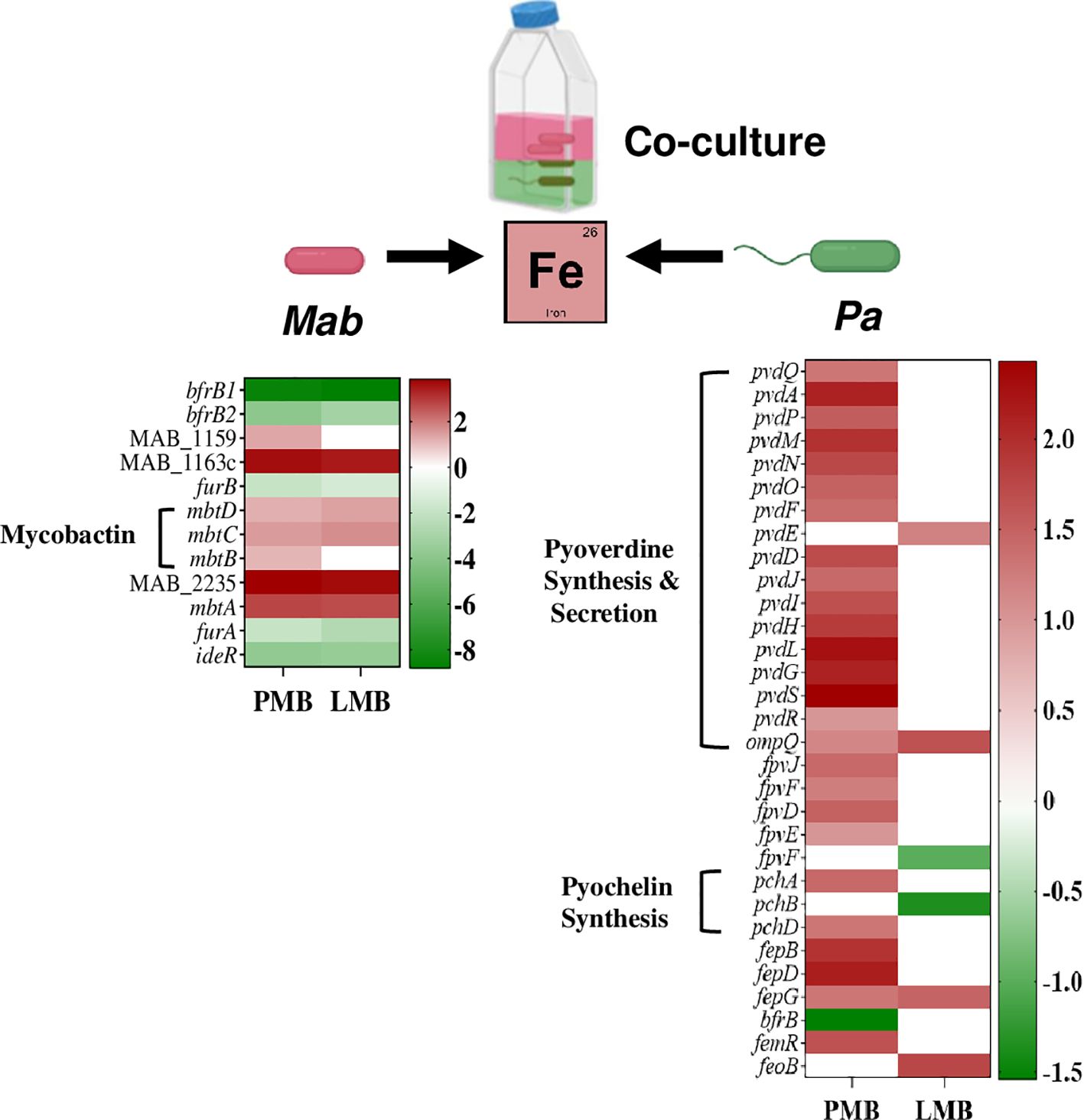
Figure 9. Competition for Fe among Mab and Pa. The heat map shows up and down regulated iron-responsive genes of Mab and Pa (WT and ΔlasR) during co-culture.
Pa produces two extracellular Fe chelating siderophores called pyoverdine and pyochelin with high and low affinity to Fe, respectively. Pyochelin biosynthesis genes, pchA and pchD and the gene cluster involved in synthesis and maturation of the major siderophore, pyoverdine (pvdQAPMNOFEDJIHLG) showed elevated expression in WT Pa when co-cultured with Mab (Figure 9; Supplementary Table 6). Besides synthesis, genes involved in secretion of pyoverdine into the extracellular environment via an ATP-dependent pvdRT-ompQ also had elevated expression. The transcript levels of the regulatory gene pvdS, encoding an iron-starvation sigma factor known for positive regulation of pyoverdine synthesis, was also enhanced. Downregulation of bfrB (bacterioferritin), which is involved in Fe storage, is also consistent with Fe-limiting conditions. Siderophore-mediated Fe uptake and storage genes were not differentially regulated in Pa in the ΔlasR Pa-Mab coculture (Figure 9; Supplementary Table 6) as expected because siderophore production is a QS regulated phenomenon (Stintzi et al., 1998; Venturi, 2006; Lin et al., 2018).
Similar to the Fe-limited transcriptional profile seen for Pa in co-culture, Mab also upregulated genes associated with iron uptake and downregulated iron storage genes. Genes encoding for the synthesis of the mycobacterial siderophore, mycobactin (mbtDCBA) showed higher expression whereas iron-dependent regulators (IdeR, FurA, and FurB) and the two putative Fe storage associated bacterioferritin B genes (MAB_0126, and MAB_0127) were downregulated (Figure 9; Supplementary Table 6).
The iron sequestering response from co-culture partners (Pa WT and Mab) indicates competition and survival efforts. Understanding Fe competition is vital for studying bacterial interactions, pathogenesis, and evolution of microbial communities. The adaption of various Fe –uptake strategies by both Mab and Pa in response to the co-culture environment is reminiscent of the human host infection conditions where the scarce availability of iron forces pathogens to adapt to survive and establish a successful infection. Effective consumption of limited nutrients by pathogens in CF lung will influence interaction dynamics and shape disease progression.
3.9 Interspecies interaction between Mab and Pa alters virulence factor expression
Dual transcriptomic profiling revealed that the well-known virulence factors of Pa including both surface-associated and secreted factors did not show notable up or downregulation except for pyocyanin and siderophores (Supplementary Table 7). Our in vitro assays with Pa spent supernatant from mono and co-cultures also indicated no significant Mab-mediated downregulation of Pa secreted virulence factors.
However, in Mab, the presence of Pa triggered increased expression of several virulence factors such as MmpL/MmpS family lipid transporters, lipoproteins, and drug resistance associated genes involved in efflux and target modification (Supplementary Table 7). Several monooxygenases/dioxygenases implicated in ring cleavage of aromatic compounds were upregulated indicating Mab potential to inactivate Pa aromatic compounds such as pyocyanin and QS signals (Supplementary Table 8). Seventeen out of twenty-three dioxygenases present in Mab genome were up-regulated including the aqd cluster, MAB_0301-MAB_0303, known to degrade Pa AQ signals (Birmes et al., 2017; Birmes et al., 2019) (Supplementary Table 8). Also, elevated expression of cyanate hydratases encoded by MAB_0054c and MAB_2545c may indicate induction of Mab defenses against HCN produced by Pa (Letoffe et al., 2022; Nolan and Allsopp, 2022). This tailored response of Mab to Pa virulence factors indicates Mab adaptation efforts to survive to coexist with Pa in similar niches within CF lung.
4 Conclusion
In this study, we exploited both culture–based and dual RNA transcriptomics strategies to explore interactions between Pa and Mab, two difficult-to-treat pathogens that commonly afflict CF patients. A handful of prior studies utilizing solid surface biofilm models (Rodriguez-Sevilla et al., 2019; Idosa et al., 2022) have reported Pa-mediated antagonism of Mab, although the molecular mechanisms underlying this interaction remain unknown. In contrast to the recent findings of Idosa et al (Idosa et al., 2022), this study reports Pa antagonism of Mab in a planktonic co-culture model, with Pa exerting a strong bacteriostatic effect on Mab. We also discovered the presence of soluble secreted factor(s) in Pa supernatants, which we dubbed SPAM, that exhibited potent bactericidal activity against Mab. Initial efforts to characterize SPAM indicate a protease-resistant, heat-labile factor of >3Kda whose expression is dependent on the LasR QS system. Isolation and identification of this natural product antimycobacterial agent, which is beyond the scope of the current study, may yield a valuable starting point for development of new antimicrobials and a chemical biology tool for identification of novel drug targets in Mab. Several of our key observations were partially corroborated by Nandanwar et al. (2024) in a study of Mab-Pa interactions published during the final preparation of this manuscript, after publication of our corresponding pre-print (Gupta et al., 2024). Specifically, the bacteriostatic effect of Pa on Mab and unperturbed growth of Pa were confirmed by similar co-culture experiments. They noted the inconsistent inhibitory effect of filtered Pa supernatants towards Mab, with either decrease (~1.5log) in CFU over 72h in one figure, or an increase (~1-log) in CFU within 72h in another (vs ~3-log growth of untreated Mab). This contrasts with our data showing potent bactericidal effect on Mab cultures with no recovery of colonies after 48h. The identification of this Pa-derived anti-mycobacterial factor(s) as well as the reason for differences in bactericidal potency between the two studies remain to be elucidated.
The ability of Mab to withstand SPAM-mediated killing and persist alongside Pa in the co-culture model, despite being outnumbered ~100:1 from the outset, serves as evidence that interspecies interactions induce alternations in Mab that facilitate persistence. To gain genome-wide insight into global transcriptional changes triggered by Pa-Mab interactions, we conducted what is to our knowledge the first dual RNAseq analysis of these CF pathogens. Mab was notably more strongly impacted than Pa (based on number of differentially expressed genes), in line with Pa’s dominant role as a “bacterial bully” in its interactions with other species. In addition to Mab genetic signatures consistent with persistence and growth arrest we observed, there was evidence of adaptative responses to thwart known antimicrobial effectors of Pa. This valuable dataset provides novel insights into Pa-Mab interspecies interactions, however, much more work is needed to understand how these interspecies dynamics impact clinical outcomes and treatment. The apparent ability of Mab to withstand potentially bactericidal effectors secreted by Pa is likely critical for Mab to co-exist with Pa during chronic infections of vulnerable patients with CF or other pulmonary conditions like COPD. The well-documented evolution of Pa during chronic infection to yield variants with mucoidy phenotype and altered QS and biofilm response could dramatically affect the dynamic interplay with other species like Mab. Understanding Mab coexistence with Pa phenotypes evolved during CF infection will offer novel insights for strain-specific interventions. Our future work will focus on investigating adaptation of Mab with Pa clinical strains from early and late stages of CF infection. Our discovery of a LasR-dependent bactericidal antagonist of Mab would suggest this loss of LasR signaling in Pa at late stages of infections (Bianconi et al., 2011; Lore et al., 2012; Baldan et al., 2014) may give Mab the upper hand. Finally, the high-level drug resistance and distinct drug susceptibility profiles of Pa and Mab already make treatment of polymicrobial infections challenging. The Pa-induced persister-like phenotype of Mab reflected in transcriptional signatures raises the possibility that interspecies interactions may impact antibiotic tolerance and treatment efficacy.
Data availability statement
Sequencing data generated in this study have been deposited in the NCBI’s SRA database and can be accessed under BioProject ID PRJNA1154599. All other relevant data are within the manuscript and in Supplementary Material.
Author contributions
RG: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. MS: Resources, Writing – review & editing. KR: Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1569331/full#supplementary-material
References
Adjemian, J., Olivier, K. N., Prevots, D. R. (2018). Epidemiology of pulmonary nontuberculous mycobacterial sputum positivity in patients with cystic fibrosis in the United States, 2010-2014. Ann. Am. Thorac. Soc 15, 817–826. doi: 10.1513/AnnalsATS.201709-727OC
Aebi, C., Bracher, R., Liechtigallati, S., Tschappeler, H., Rudeberg, A., Kraemer, R. (1995). The age at onset of chronic pseudomonas-aeruginosa colonization in cystic-fibrosis - prognostic-significance. Eur. J. Pediatr. 154, S69–S73. doi: 10.1007/BF02191510
Aiello, T. B., Levy, C. E., Zaccariotto, T. R., Paschoal, I. A., Pereira, M. C., da Silva, M. T. N., et al. (2018). Prevalence and clinical outcomes of nontuberculous mycobacteria in a Brazilian cystic fibrosis reference center. Pathog Dis. 76. doi: 10.1093/femspd/fty051
Bajorath, J., Hinrichs, W., Saenger, W. (1988). The enzymatic activity of proteinase K is controlled by calcium. Eur. J. Biochem. 176, 441–447. doi: 10.1111/j.1432-1033.1988.tb14301.x
Baldan, R., Cigana, C., Testa, F., Bianconi, I., De Simone, M., Pellin, D., et al. (2014). Adaptation of Pseudomonas aeruginosa in Cystic Fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PloS One 9, e89614. doi: 10.1371/journal.pone.0089614
Banjar, H., Ghawi, A., AlMogarri, I., Alhaider, S., Alomran, H., Hejazi, A., et al. (2022). First report on the prevalence of bacteria in cystic fibrosis patients (CF) in a tertiary care center in Saudi Arabia. Int. J. Pediatr. Adolesc. Med. 9, 108–112. doi: 10.1016/j.ijpam.2021.07.001
Beste, D. J. V., Espasa, M., Bonde, B., Kierzek, A. M., Stewart, G. R., McFadden, J. (2009). The genetic requirements for fast and slow growth in mycobacteria. PloS One 4. doi: 10.1371/journal.pone.0005349
Bhagirath, A. Y., Li, Y. Q., Somayajula, D., Dadashi, M., Badr, S., Duan, K. M. (2016). Cystic fibrosis lung environment and Pseudomonas aeruginosa infection. BMC Pulm Med. 16. doi: 10.1186/s12890-016-0339-5
Bianconi, I., Milani, A., Cigana, C., Paroni, M., Levesque, R. C., Bertoni, G., et al. (2011). Positive signature-tagged mutagenesis in Pseudomonas aeruginosa: tracking patho-adaptive mutations promoting airways chronic infection. PloS Pathog. 7, e1001270. doi: 10.1371/journal.ppat.1001270
Birmes, F. S., Saring, R., Hauke, M. C., Ritzmann, N. H., Drees, S. L., Daniel, J., et al. (2019). Interference with pseudomonas aeruginosa quorum sensing and virulence by the mycobacterial pseudomonas quinolone signal dioxygenase aqdC in combination with the N-acylhomoserine lactone lactonase qsdA. Infect. Immun. 87. doi: 10.1128/IAI.00278-19
Birmes, F. S., Wolf, T., Kohl, T. A., Ruger, K., Bange, F., Kalinowski, J., et al. (2017). Mycobacterium abscessus subsp abscessus Is Capable of Degrading Pseudomonas aeruginosa Quinolone Signals. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00339
Biswas, L., Gotz, F. (2021). Molecular mechanisms of staphylococcus and pseudomonas interactions in cystic fibrosis. Front. Cell Infect. Microbiol. 11, 824042. doi: 10.3389/fcimb.2021.824042
Bjarnsholt, T., Jensen, P. O., Fiandaca, M. J., Pedersen, J., Hansen, C. R., Andersen, C. B., et al. (2009). Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44, 547–558. doi: 10.1002/ppul.21011
Bragonzi, A., Farulla, I., Paroni, M., Twomey, K. B., Pirone, L., Lore, N. I., et al. (2012). Modelling co-infection of the cystic fibrosis lung by Pseudomonas aeruginosa and Burkholderia cenocepacia reveals influences on biofilm formation and host response. PloS One 7, e52330. doi: 10.1371/journal.pone.0052330
Byrd, T. F., Lyons, C. R. (1999). Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infect. Immun. 67, 4700–4707. doi: 10.1128/IAI.67.9.4700-4707.1999
Caldwell, C. C., Chen, Y., Goetzmann, H. S., Hao, Y., Borchers, M. T., Hassett, D. J., et al. (2009). Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am. J. Pathol. 175, 2473–2488. doi: 10.2353/ajpath.2009.090166
Catherinot, E., Roux, A. L., Macheras, E., Hubert, D., Matmar, M., Dannhoffer, L., et al. (2009). Acute respiratory failure involving an R variant of Mycobacterium abscessus. J. Clin. Microbiol. 47, 271–274. doi: 10.1128/JCM.01478-08
Chambers, D., Scott, F., Bangur, R., Davies, R., Lim, A., Walters, S., et al. (2005). Factors associated with infection by Pseudomonas aeruginosa in adult cystic fibrosis. Eur. Respir. J. 26, 651–656. doi: 10.1183/09031936.05.00126704
Coburn, B., Wang, P. W., Diaz Caballero, J., Clark, S. T., Brahma, V., Donaldson, S., et al. (2015). Lung microbiota across age and disease stage in cystic fibrosis. Sci. Rep. 5, 10241. doi: 10.1038/srep10241
Costa, K. C., Bergkessel, M., Saunders, S., Korlach, J., Newman, D. K. (2015). Enzymatic degradation of phenazines can generate energy and protect sensitive organisms from toxicity. Mbio. 6. doi: 10.1128/mBio.01520-15
De Boeck, K., Alifier, M., Vandeputte, S. (2000). Sputum induction in young cystic fibrosis patients. Eur. Respir. J. 16, 91–94. doi: 10.1034/j.1399-3003.2000.16a16.x
Dedrick, R. M., Aull, H. G., Jacobs-Sera, D., Garlena, R. A., Russell, D. A., Smith, B. E., et al. (2021). The prophage and plasmid mobilome as a likely driver of mycobacterium abscessus diversity. Mbio 12. doi: 10.1128/mBio.03441-20
Dekimpe, V., Deziel, E. (2009). Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiol. (Reading). 155, 712–723. doi: 10.1099/mic.0.022764-0
Diggle, S. P., Winzer, K., Chhabra, S. R., Worrall, K. E., Camara, M., Williams, P. (2003). The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50, 29–43. doi: 10.1046/j.1365-2958.2003.03672.x
Feltner, J. B., Wolter, D. J., Pope, C. E., Groleau, M. C., Smalley, N. E., Greenberg, E. P., et al. (2016). LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in pseudomonas aeruginosa. mBio 7. doi: 10.1128/mBio.01513-16
Garton, N. J., Waddell, S. J., Sherratt, A. L., Lee, S. M., Smith, R. J., Senner, C., et al. (2008). Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PloS Med. 5, e75. doi: 10.1371/journal.pmed.0050075
Greendyke, R., Byrd, T. F. (2008). Differential antibiotic susceptibility of Mycobacterium abscessus variants in biofilms and macrophages compared to that of planktonic bacteria. Antimicrob Agents Chemother. 52, 2019–2026. doi: 10.1128/AAC.00986-07
Gupta, R., Netherton, M., Byrd, T. F., Rohde, K. H. (2017). Reporter-Based Assays for High-Throughput Drug Screening against Mycobacterium abscessus. Front. Microbiol. 8, 2204. doi: 10.3389/fmicb.2017.02204
Gupta, R., Schuster, M., Rohde, K. H. (2024). Mycobacterium abscessus persistence in the face of Pseudomonas aeruginosa antagonism. bioRxiv. doi: 10.1101/2024.09.23.614414
Hedin, W., Froberg, G., Fredman, K., Chryssanthou, E., Selmeryd, I., Gillman, A., et al. (2023). A rough colony morphology of mycobacterium abscessus is associated with cavitary pulmonary disease and poor clinical outcome. J. Infect. Dis. 227, 820–827. doi: 10.1093/infdis/jiad007
Hennemann, L. C., LaFayette, S. L., Malet, J. K., Bortolotti, P., Yang, T., McKay, G. A., et al. (2021). LasR-deficient Pseudomonas aeruginosa variants increase airway epithelial mICAM-1 expression and enhance neutrophilic lung inflammation. PloS Pathog. 17, e1009375. doi: 10.1371/journal.ppat.1009375
Hoffman, L. R., Kulasekara, H. D., Emerson, J., Houston, L. S., Burns, J. L., Ramsey, B. W., et al. (2009). Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst Fibros. 8, 66–70. doi: 10.1016/j.jcf.2008.09.006
Holloway, B. W., Krishnapillai, V., Morgan, A. F. (1979). Chromosomal genetics of pseudomonas. Microbiol Rev. 43, 73–102. doi: 10.1128/mr.43.1.73-102.1979
Howard, S. T., Byrd, T. F. (2000). The rapidly growing mycobacteria: saprophytes and parasites. Microbes Infect. 2, 1845–1853. doi: 10.1016/S1286-4579(00)01338-1
Howard, S. T., Rhoades, E., Recht, J., Pang, X. H., Alsup, A., Kolter, R., et al. (2006). Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiol-Sgm. 152, 1581–1590. doi: 10.1099/mic.0.28625-0
Hsieh, M. H., Lin, C. Y., Wang, C. Y., Fang, Y. F., Lo, Y. L., Lin, S. M., et al. (2018). Impact of concomitant nontuberculous mycobacteria and Pseudomonas aeruginosa isolates in non-cystic fibrosis bronchiectasis. Infect. Drug Resist. 11, 1137–1143. doi: 10.2147/IDR.S169789
Hu, Y., Li, T., Liu, W., Zhu, D., Feng, X., Chen, Y., et al. (2024). Prevalence and antimicrobial susceptibility pattern of Mycobacterium abscessus complex isolates in Chongqing, Southwest China. Heliyon. 10, e34546. doi: 10.1016/j.heliyon.2024.e34546
Hunter, R. C., Klepac-Ceraj, V., Lorenzi, M. M., Grotzinger, H., Martin, T. R., Newman, D. K. (2012). Phenazine content in the cystic fibrosis respiratory tract negatively correlates with lung function and microbial complexity. Am. J. Respir. Cell Mol. Biol. 47, 738–745. doi: 10.1165/rcmb.2012-0088OC
Idosa, A. W., Wozniak, D. J., Hall-Stoodley, L. (2022). Surface Dependent Inhibition of Mycobacterium abscessus by Diverse Pseudomonas aeruginosa Strains. Microbiol Spectr. 10 (6), e0247122. doi: 10.1128/spectrum.02471-22
Jan, A. T. (2017). Outer membrane vesicles (OMVs) of gram-negative bacteria: A perspective update. Front. Microbiol. 8, 1053. doi: 10.3389/fmicb.2017.01053
Jonsson, B. E., Gilljam, M., Lindblad, A., Ridell, M., Wold, A. E., Welinder-Olsson, C. (2007). Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J. Clin. Microbiol. 45, 1497–1504. doi: 10.1128/JCM.02592-06
Kendall, B. A., Winthrop, K. L. (2013). Update on the epidemiology of pulmonary nontuberculous mycobacterial infections. Semin. Respir. Crit. Care Med. 34, 87–94. doi: 10.1055/s-0033-1333567
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907. doi: 10.1038/s41587-019-0201-4
Kreutzfeldt, K. M., McAdam, P. R., Claxton, P., Holmes, A., Seagar, A. L., Laurenson, I. F., et al. (2013). Molecular Longitudinal Tracking of Mycobacterium abscessus spp. during Chronic Infection of the Human Lung. PloS One 8, e63237. doi: 10.1371/journal.pone.0063237
Kunzelmann, K., Schreiber, R., Hadorn, H. B. (2017). Bicarbonate in cystic fibrosis. J. Cyst Fibros. 16, 653–662. doi: 10.1016/j.jcf.2017.06.005
Lamont, I. L., Konings, A. F., Reid, D. W. (2009). Iron acquisition by Pseudomonas aeruginosa in the lungs of patients with cystic fibrosis. Biometals. 22, 53–60. doi: 10.1007/s10534-008-9197-9
Latifi, A., Foglino, M., Tanaka, K., Williams, P., Lazdunski, A. (1996). A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21, 1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x
Lau, G. W., Ran, H. M., Kong, F. S., Hassett, D. J., Mavrodi, D. (2004). Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 72, 4275–4278. doi: 10.1128/IAI.72.7.4275-4278.2004
Lee, J. H., Lequette, Y., Greenberg, E. P. (2006). Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59, 602–609. doi: 10.1111/j.1365-2958.2005.04960.x
Lee, J., Wu, J., Deng, Y., Wang, J., Wang, C., Wang, J., et al. (2013). A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 9, 339–343. doi: 10.1038/nchembio.1225
Letoffe, S., Wu, Y., Darch, S. E., Beloin, C., Whiteley, M., Touqui, L., et al. (2022). Pseudomonas aeruginosa Production of Hydrogen Cyanide Leads to Airborne Control of Staphylococcus aureus Growth in Biofilm and In Vivo Lung Environments. mBio. 13, e0215422. doi: 10.1128/mbio.02154-22
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lin, J., Cheng, J., Wang, Y., Shen, X. (2018). The pseudomonas quinolone signal (PQS): not just for quorum sensing anymore. Front. Cell Infect. Microbiol. 8, 230. doi: 10.3389/fcimb.2018.00230
Lore, N. I., Cigana, C., De Fino, I., Riva, C., Juhas, M., Schwager, S., et al. (2012). Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PloS One 7, e35648. doi: 10.1371/journal.pone.0035648
Mall, M. A., Hartl, D. (2014). CFTR: cystic fibrosis and beyond. Eur. Respir. J. 44, 1042–1054. doi: 10.1183/09031936.00228013
Mangione, E. J., Huitt, G., Lenaway, D., Beebe, J., Bailey, A., Figoski, M., et al. (2001). Nontuberculous mycobacterial disease following hot tub exposure. Emerg Infect. Dis. 7, 1039–1042. doi: 10.3201/eid0706.010623
Martin, L. W., Reid, D. W., Sharples, K. J., Lamont, I. L. (2011). Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals. 24, 1059–1067. doi: 10.1007/s10534-011-9464-z
Maurer, F. P., Bruderer, V. L., Ritter, C., Castelberg, C., Bloemberg, G. V., Bottger, E. C. (2014). Lack of antimicrobial bactericidal activity in mycobacterium abscessus. Antimicrob Agents Chemother. 58, 3828–3836. doi: 10.1128/AAC.02448-14
Maurya, R. K., Bharti, S., Krishnan, M. Y. (2018). Triacylglycerols: fuelling the hibernating mycobacterium tuberculosis. Front. Cell Infect. Microbiol. 8, 450. doi: 10.3389/fcimb.2018.00450
Medina, G., Juarez, K., Diaz, R., Soberon-Chavez, G. (2003). Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiol. (Reading). 149, 3073–3081. doi: 10.1099/mic.0.26282-0
Miao Zhao, K. G., Danelishvili, L., Jeffrey, B., Bermudez, L. E. (2016). Identification of Prophages within the Mycobacterium avium 104 Genome and the Link of Their Function Regarding to Environment Survival. Adv. Microbiol. 6, 927–941. doi: 10.4236/aim.2016.613087
Munoz-Elias, E. J., Upton, A. M., Cherian, J., McKinney, J. D. (2006). Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 60, 1109–1122. doi: 10.1111/j.1365-2958.2006.05155.x
Nandanwar, N., Gu, G., Gibson, J. E., Neely, M. N. (2024). Polymicrobial interactions influence Mycobacterium abscessus co-existence and biofilm forming capabilities. Front. Microbiol. 15, 1484510. doi: 10.3389/fmicb.2024.1484510
Newman, J. R., Fuqua, C. (1999). Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene. 227, 197–203. doi: 10.1016/S0378-1119(98)00601-5
Nichols, D. P., Morgan, S. J., Skalland, M., Vo, A. T., Van Dalfsen, J. M., Singh, S. B., et al. (2023). Pharmacologic improvement of CFTR function rapidly decreases sputum pathogen density, but lung infections generally persist. J. Clin. Invest. 133. doi: 10.1172/JCI167957
Nolan, L. M., Allsopp, L. P. (2022). Antimicrobial weapons of pseudomonas aeruginosa. Adv. Exp. Med. Biol. 1386, 223–256. doi: 10.1007/978-3-031-08491-1_8
Olivier, K. N., Weber, D. J., Wallace, R. J., Faiz, A. R., Lee, J. H., Zhang, Y. S., et al. (2003). Nontuberculous mycobacteria I: Multicenter prevalence study in cystic fibrosis. Am. J. Resp Crit. Care 167, 828–834. doi: 10.1164/rccm.200207-678OC
Pesci, E. C., Pearson, J. P., Seed, P. C., Iglewski, B. H. (1997). Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179, 3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997
Pierre-Audigier, C., Ferroni, A., Sermet-Gaudelus, I., Le Bourgeois, M., Offredo, C., Vu-Thien, H., et al. (2005). Age-related prevalence and distribution of nontuberculous mycobacterial species among patients with cystic fibrosis. J. Clin. Microbiol. 43, 3467–3470. doi: 10.1128/JCM.43.7.3467-3470.2005
Pittman, J. E., Calloway, E. H., Kiser, M., Yeatts, J., Davis, S. D., Drumm, M. L., et al. (2011). Age of pseudomonas aeruginosa acquisition and subsequent severity of cystic fibrosis lung disease. Pediatr. Pulm. 46, 497–504. doi: 10.1002/ppul.21397
Poulsen, J. H., Fischer, H., Illek, B., Machen, T. E. (1994). Bicarbonate conductance and pH regulatory capability of cystic fibrosis transmembrane conductance regulator. Proc. Natl. Acad. Sci. U S A. 91, 5340–5344. doi: 10.1073/pnas.91.12.5340
Qvist, T., Eickhardt, S., Kragh, K. N., Andersen, C. B., Iversen, M., Hoiby, N., et al. (2015). Chronic pulmonary disease with Mycobacterium abscessus complex is a biofilm infection. Eur. Respir. J. 46, 1823–1826. doi: 10.1183/13993003.01102-2015
Rada, B., Leto, T. L. (2013). Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol. 21, 73–81. doi: 10.1016/j.tim.2012.10.004
Recio, R., Mancheno, M., Viedma, E., Villa, J., Orellana, M. A., Lora-Tamayo, J., et al. (2020). Predictors of mortality in bloodstream infections caused by pseudomonas aeruginosa and impact of antimicrobial resistance and bacterial virulence. Antimicrob Agents Chemother. 64. doi: 10.1128/AAC.01759-19
Ripoll, F., Pasek, S., Schenowitz, C., Dossat, C., Barbe, V., Rottman, M., et al. (2009). Non mycobacterial virulence genes in the genome of the emerging pathogen mycobacterium abscessus. PloS One 4. doi: 10.1371/journal.pone.0005660
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. doi: 10.1093/nar/gkv007
Rodriguez-Sevilla, G., Crabbe, A., Garcia-Coca, M., Aguilera-Correa, J. J., Esteban, J., Perez-Jorge, C. (2019). Antimicrobial Treatment Provides a Competitive Advantage to Mycobacterium abscessus in a Dual-Species Biofilm with Pseudomonas aeruginosa. Antimicrob Agents Chemother. 63. doi: 10.1128/AAC.01547-19
Rodriguez-Sevilla, G., Garcia-Coca, M., Romera-Garcia, D., Aguilera-Correa, J. J., Mahillo-Fernandez, I., Esteban, J., et al. (2018). Non-Tuberculous Mycobacteria multispecies biofilms in cystic fibrosis: development of an in vitro Mycobacterium abscessus and Pseudomonas aeruginosa dual species biofilm model. Int. J. Med. Microbiol. 308, 413–423. doi: 10.1016/j.ijmm.2018.03.003
Rohde, K. H., Veiga, D. F. T., Caldwell, S., Balazsi, G., Russell, D. G. (2012). Linking the Transcriptional Profiles and the Physiological States of Mycobacterium tuberculosis during an Extended Intracellular Infection. PloS Pathogens. 8. doi: 10.1371/journal.ppat.1002769
Roux, A. L., Catherinot, E., Ripoll, F., Soismier, N., Macheras, E., Ravilly, S., et al. (2009). Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J. Clin. Microbiol. 47, 4124–4128. doi: 10.1128/JCM.01257-09
Roux, A. L., Viljoen, A., Bah, A., Simeone, R., Bernut, A., Laencina, L., et al. (2016). The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol. 6. doi: 10.1098/rsob.160185
Russell, D. G., Cardona, P. J., Kim, M. J., Allain, S., Altare, F. (2009). Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 10, 943–948. doi: 10.1038/ni.1781
Sader, H. S., Castanheira, M., Duncan, L. R., Flamm, R. K. (2018). Antimicrobial susceptibility of enterobacteriaceae and pseudomonas aeruginosa isolates from United States medical centers stratified by infection type: results from the international network for optimal resistance monitoring (INFORM) surveillance program, 2015-2016. Diagn. Microbiol Infect. Dis. 92, 69–74. doi: 10.1016/j.diagmicrobio.2018.04.012
Savvi, S., Warner, D. F., Kana, B. D., McKinney, J. D., Mizrahi, V., Dawes, S. S. (2008). Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids. J. Bacteriol. 190, 3886–3895. doi: 10.1128/JB.01767-07
Sermet-Gaudelus, I., Le Bourgeois, M., Pierre-Audigier, C., Offredo, C., Guillemot, D., Halley, S., et al. (2003). Mycobacterium abscessus and children with cystic fibrosis. Emerg Infect. Dis. 9, 1587–1591. doi: 10.3201/eid0912.020774
Sezonov, G., Joseleau-Petit, D., D’Ari, R. (2007). Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189, 8746–8749. doi: 10.1128/JB.01368-07
Shi, L., Sohaskey, C. D., Kana, B. D., Dawes, S., North, R. J., Mizrahi, V., et al. (2005). Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. U S A. 102, 15629–15634. doi: 10.1073/pnas.0507850102
Singh PK, S. A., Parsek, M. R., Moninger, T. O., Welsh, M. J., Greenberg, E. P. (2000). Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 407, 762–764. doi: 10.1038/35037627
Smith, E. E., Buckley, D. G., Wu, Z. N., Saenphimmachak, C., Hoffman, L. R., D’Argenio, D. A., et al. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. P Natl. Acad. Sci. USA. 103, 8487–8492. doi: 10.1073/pnas.0602138103
Stintzi, A., Evans, K., Meyer, J. M., Poole, K. (1998). Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol Lett. 166, 341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x
Takano, K., Shimada, D., Kashiwagura, S., Kamioka, Y., Hariu, M., Watanabe, Y., et al. (2021). Severe pulmonary mycobacterium abscessus cases due to co-infection with other microorganisms well treated by clarithromycin and sitafloxacin in Japan. Int. Med. Case Rep. J. 14, 465–470. doi: 10.2147/IMCRJ.S321969
Thornton, C. S., Parkins, M. D. (2023). Microbial epidemiology of the cystic fibrosis airways: past, present, and future. Semin. Respir. Crit. Care Med. 44, 269–286. doi: 10.1055/s-0042-1758732
Toung, J. M., Morley, M., Li, M. Y., Cheung, V. G. (2011). RNA-sequence analysis of human B-cells. Genome Res. 21, 991–998. doi: 10.1101/gr.116335.110
Valenza, G., Tappe, D., Turnwald, D., Frosch, M., Konig, C., Hebestreit, H., et al. (2008). Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J. Cyst Fibros. 7, 123–127. doi: 10.1016/j.jcf.2007.06.006
Van Daele, S. G., Franckx, H., Verhelst, R., Schelstraete, P., Haerynck, F., Van Simaey, L., et al. (2005). Epidemiology of Pseudomonas aeruginosa in a cystic fibrosis rehabilitation centre. Eur. Respir. J. 25, 474–481. doi: 10.1183/09031936.05.00050304
van Dijk, D., Dhar, R., Missarova, A. M., Espinar, L., Blevins, W. R., Lehner, B., et al. (2015). Slow-growing cells within isogenic populations have increased RNA polymerase error rates and DNA damage. Nat. Commun. 6. doi: 10.1038/ncomms8972
Venturi, V. (2006). Regulation of quorum sensing in Pseudomonas. FEMS Microbiol Rev. 30, 274–291. doi: 10.1111/j.1574-6976.2005.00012.x
Viljoen, A., Blaise, M., de Chastellier, C., Kremer, L. (2016). MAB_3551c encodes the primary triacylglycerol synthase involved in lipid accumulation in Mycobacterium abscessus. Mol. Microbiol. 102, 611–627. doi: 10.1111/mmi.2016.102.issue-4
Vrla, G. D., Esposito, M., Zhang, C., Kang, Y., Seyedsayamdost, M. R., Gitai, Z. (2020). Cytotoxic alkyl-quinolones mediate surface-induced virulence in Pseudomonas aeruginosa. PloS Pathog. 16, e1008867. doi: 10.1371/journal.ppat.1008867
Wade, D. S., Calfee, M. W., Rocha, E. R., Ling, E. A., Engstrom, E., Coleman, J. P., et al. (2005). Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J. Bacteriol. 187, 4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005
Westermann, A. J., Forstner, K. U., Amman, F., Barquist, L., Chao, Y. J., Schulte, L. N., et al. (2016). Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 529, 496. doi: 10.1038/nature16547
Wilder, C. N., Allada, G., Schuster, M. (2009). Instantaneous within-patient diversity of Pseudomonas aeruginosa quorum-sensing populations from cystic fibrosis lung infections. Infect. Immun. 77, 5631–5639. doi: 10.1128/IAI.00755-09
Keywords: Mycobacterium abscessus, Pseudomonas aeruginosa, mixed microbial interactions, dual RNAseq, secreted factors, quorum sensing
Citation: Gupta R, Schuster M and Rohde KH (2025) Mycobacterium abscessus persistence in the face of Pseudomonas aeruginosa antagonism. Front. Cell. Infect. Microbiol. 15:1569331. doi: 10.3389/fcimb.2025.1569331
Received: 31 January 2025; Accepted: 17 April 2025;
Published: 09 May 2025.
Edited by:
Hongwei David Yu, Marshall University, United StatesReviewed by:
Megan R. Kiedrowski, University of Alabama at Birmingham, United StatesBrahmchetna Bedi, Emory University, United States
Copyright © 2025 Gupta, Schuster and Rohde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyle H. Rohde, a3lsZS5yb2hkZUB1Y2YuZWR1; Rashmi Gupta, cmc3MDAwQGdtYWlsLmNvbQ==
 Rashmi Gupta
Rashmi Gupta Martin Schuster
Martin Schuster Kyle H. Rohde
Kyle H. Rohde