- 1College of Life Science and Technology, Wuhan Polytechnic University, Wuhan, Hubei, China
- 2Guangdong Provincial Key Laboratory of Infection Immunity and Inflammation, Department of Pathogen Biology, Shenzhen University Medical School, Shenzhen, China
- 3Guangzhou Medical Research Institute of Infectious Diseases, Infectious Disease Center, Guangzhou Eighth People’s Hospital, Guangzhou Medical University, Guangzhou, Guangdong, China
- 4Center Lab of Longhua Branch and Department of Infectious Disease, Shenzhen People’s Hospital (The Second Clinical Medical College, Jinan University, The First Affiliated Hospital, Southern University of Science and Technology), Shenzhen, Guangdong, China
Ferritin heavy chain 1 (FTH1) is a key iron-storage protein that regulates iron availability, supports immune defense, and prevents iron-induced toxicity. During Mycobacterium tuberculosis (Mtb) infection, macrophages enhance FTH1 expression to sequestrate iron and limit Mtb growth. However, Mtb can exploit the host ferritinophagy pathway to degrade FTH1 and release iron, thereby promoting its survival. Although FTH1 plays an essential role in host–pathogen interaction during Mtb infection, its regulation remains unclear. Previous studies suggest that post-transcriptional mechanism, particularly alternative polyadenylation (APA), are critical in immune responses. We propose that APA, which determines the length of a transcript’s 3′UTR, may regulate FTH1 expression during Mtb infection. Our study demonstrates that Mtb induces APA of FTH1 in macrophages, favoring the production of longer isoforms that enhance protein synthesis. Mechanistically, Mtb disrupts the interaction between NUDT21 and CPSF6, impairing NUDT21’s ability to bind UGUA motifs in the FTH1 3′UTR, a key step in polyadenylation site selection. Silencing NUDT21 reduces macrophage bactericidal activity against Mtb, highlighting its role in immune defense. These findings reveal a novel Mtb-driven mechanism that enhances FTH1 expression via the NUDT21-mediated APA pathway in macrophages, suggesting that Mtb manipulates this process to promote its survival. This study provides new insights into tuberculosis pathogenesis and points to potential avenues for therapeutic exploration.
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), remains one of the world’s most significant infectious diseases (World Health Organization, 2024). Macrophages, key players in the innate immune system, have a paradoxical role in TB pathogenesis (Bo et al., 2023; Weiss and Schaible, 2015). While they can restrict Mtb growth through bactericidal mechanisms, they may also support bacterial persistence by providing a protective niche (Hmama et al., 2015; Pisu et al., 2020; Kurthkoti et al., 2017). Central to macrophage defense against Mtb is iron homeostasis, which supports processes such as oxidative killing, antigen presentation, and phagocytosis (Ganz and Nemeth, 2015). However, excessive intracellular iron can induce oxidative stress, undermining immune function (Ni et al., 2022; Soares and Hamza, 2016).
Ferritin heavy chain 1 (FTH1), a key iron storage protein, helps mitigate iron toxicity by forming a ferritin complex with the light chain (FTL), which sequesters excess iron (Torti and Torti, 2002). FTH1 plays a complex role in the host’s immune response to Mtb. FTH1–deficient bone marrow–derived macrophages (BMDMs) or myeloid cells increase susceptibility to Mtb (Reddy et al., 2018; Khan et al., 2020), suggesting its protective role in host–anti-TB immune response. However, previous research has shown that the upregulation of FTH1 levels in macrophages is associated with TB disease progression (Dai et al., 2023). This paradox arises from Mtb’s ability to exploit ferritinophagy—a selective autophagic process regulated by NCOA4—to degrade FTH1, enhancing its intracellular survival (Dai et al., 2024; Mancias et al., 2014). Taken these together, maintaining a balanced FTH1 expression is essential for supporting immune function and preventing iron overload.
Ferritin expression, particularly FTH1, is tightly regulated at both transcriptional and translational levels (Torti and Torti, 2002). Cytokines such as TNF-α and IL-1 transcriptionally upregulate FTH1 in response to stress and inflammation (Pham et al., 2004; Torti et al., 1988; Wei et al., 1990). Iron regulatory proteins (IRPs) bind to the iron–responsive element (IRE) in the 5′ untranslated region (5′UTR) of FTH1 mRNA, controlling its translation (Hentze et al., 1987; Leibold and Munro, 1988). However, whether there are other molecular mechanisms governing FTH1 expression, particularly in the context of Mtb infection, remain poorly understood.
Given that previous studies have identified two distinct FTH1 transcripts with different 3′UTRs (Joshi et al., 1995), we suggest that alternative polyadenylation (APA) regulation may play a role in Mtb-driven FTH1 expression post-transcriptionally. APA regulation generates mRNA isoforms with distinct 3′UTRs, which affect mRNA stability, translation, and localization (Mitschka and Mayr, 2022). APA is regulated by cleavage and polyadenylation (CPA) machinery and related cis-elements, including the polyadenylation signal (PAS) and downstream (G+U)-rich or U-rich region, which are recognized by factors like CPA specificity factor (CPSF) and cleavage stimulation factor (CSTF). These factors recruit proteins, including cleavage factor I (CFI), CFII and Poly(A) polymerase (PAP), to define functional PASs and facilitate RNA cleavage and polyadenylation. The UGUA motif—a highly prevalent cis-element upstream of polyadenylation site of human and mouse genes—is recognized by NUDT21 (also known as CPSF5, CFIm 25) (Yang et al., 2010). NUDT21, together with CPSF6 (also known as CFIm 68), forms CFI, which regulates the selection between proximal ((pA)p) and distal ((pA)d) polyadenylation sites (Yang et al., 2011). This regulation leads to 3′UTRs of varying lengths, which may contain regulatory elements such as microRNA or RNA-binding protein (RBP) binding sites. Dysregulated poly(A) site usage has been linked to many immune–related diseases—3′UTR shortening is predominantly exhibiting in solid tumors—suggesting that APA may contribute to disease etiology (An et al., 2021; Gruber and Zavolan, 2019; Li et al., 2023).
In this study, we reveal the role of APA in Mtb-driven FTH1 expression in macrophages. Our findings demonstrate that Mtb infection reprograms FTH1 APA, promoting the production of longer isoforms that enhance protein translation. We also highlight the involvement of NUDT21 in this process, underscoring the importance of NUDT21-mediated APA regulation in macrophage responses to Mtb. These insights deepen our understanding of host–pathogen interactions in macrophages during Mtb infection and highlight the need for further exploration of the APA regulation pathway in TB disease, particularly in relation to its regulators and their impact on disease progression.
Materials and methods
Bacteria strains and cell lines
The mycobacterial strains H37Ra (ATCC 25177), its fluorescent engineered version GFP-H37Ra as described previously (Mo et al., 2022), and Bacillus Calmette-Guérin (BCG Danish Strain 1331 sub-strain, NIBSC, SRNC-07/270) were cultured in Middlebrook 7H9 broth (BBL Microbiology Systems) supplemented with 10% oleic acid–albumin–dextrose–catalase (OADC; BD BBL), 0.05% (v/v) Tween-80 (Sigma), and 0.2% (v/v) glycerol (Sigma) or on Middlebrook 7H10 plates (BD Difco) containing 0.5% (v/v) glycerol and 10% OADC. Bone marrow-derived macrophages (BMDMs) were isolated from C57BL/6 mice (The Jackson Laboratory, Stock#000664), as described previously (Dai et al., 2023), and cultured for 1 week in DMEM supplemented with 20% L929-conditioned medium, 1 mM sodium pyruvate, 2 mM L-glutamine, and 10% FBS. Human monocytic THP-1 cells (TIB-202, ATCC) (4x105 cells/mL) were seeded in 6- or 12-well plates (Costar) and differentiated with 20 ng/ml PMA (Sigma-Aldrich) for 24 h. Differentiated THP-1 macrophages and BMDMs were maintained in fresh pre-warmed media until further use. HEK293T and A549 cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), while RAW264.7 cells were a gift from the Center Lab of Longhua Branch, Shenzhen People’s Hospital. These cells were cultured in DMEM (Corning, USA) supplemented with 10% FBS (Gibco, Life Technologies) at 37°C in 5% CO2 until further use.
Mtb infection
Bacteria harvested in the mid-log phase were washed and resuspended in PBS. The multiplicity of infection (MOI) was calculated based on bacterial optical density (OD) and confirmed by plating serial dilutions on Middlebrook 7H10 agar plates to determine the colony-forming units (CFU). PMA-differentiated THP-1 macrophages were infected with Mtb H37Ra or GFP-H37Ra at an MOI of 10 for 6 h, followed by three PBS washes to remove extracellular bacteria. Fresh medium was added, and cells were incubated for the indicated time points. For intracellular survival analysis, CFU assays were performed as described previously (Mo et al., 2022).
Construction of fluorescent reporter plasmids
The 3′UTRs of FTH1 transcripts were cloned into a pEGFP-C1 vector (Addgene, Plasmid #36412) downstream of the GFP ORF to create 3′UTR-Long (L) and 3′UTR-Short (S) plasmids. Scrambled sequences replaced the first or last 50 nucleotides of the upstream sequence element (USE) preceding the proximal (USE1) or distal (USE2) polyadenylation sites, generating pap-USE-F50, pap-USE-R50, pad-USE-F50 and pad-USE-R50 plasmids. A 25-nucleotide region of the downstream sequence element (DSE) was replaced with a scrambled sequence to create pap-DSE-F25 and pap-DSE-R25 plasmids. UGUA motif mutations were introduced by replacing TGTA with altered nucleotides (TAAA for UGUA motif 1; GGCG for UGUA motif 2) based on plasmid 3′UTR-L.
3′RACE
Total RNA was extracted from Mtb-infected or uninfected macrophages and fluorescent plasmids transfected eukaryotic cells, using RNeasy kits (Omega). First-strand cDNA was synthesized from 1 µg of RNA using the following 3′ap primer:
5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTGtttttttttttttttttt-3′
The 20 µL reaction mix contained 1 mM dNTPs, 1X RT buffer, 0.1 M DTT, and 200 U of MMLV Reverse Transcriptase, and was incubated at 42°C for 60 min before enzyme inactivation at 70°C for 5 min.
For the first PCR amplification, 2 µL of cDNA was amplified using 0.2 µM of each of the following primers:
FTH1 forward: 5′-ATGACCCCCATTTGTGTGACTTCATTGAG-3′;
Fth1 forward: 5′-TACGTCTATCTGTCTATGTCTTGTT-3′;
3′ap-CA reverse: 5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTGttttttttttttttttttCA-3′.
The reaction was completed in 1X PCR buffer, with 0.2 mM dNTPs and 2.5 U Taq polymerase. The PCR conditions were as follows: 95°C for 3–5 min, followed by 35 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min/kb, with a final extension at 72°C for 10 min. A second PCR was performed using 1 µL of the first PCR product and the following primers:
FTH1 forward: 5′-GCCTCGGGCTAATTTCCC-3′;
Fth1 forward: 5′-CTCATGAGGAGAGGGAGCAT-3′;
3′NestedPrimer reverse: 5′-CGCTACGTAACGGCATGACAGTG-3′.
Products were separated by agarose gel electrophoresis and then gel-purified and sequenced.
siRNAs and cell transfection
For siRNA transfection, siFTH1 (targeting all FTH1 transcripts; RiboBio, siG2010191151216466), siFTH1-L (targeting the longer FTH1 isoform; RiboBio, siG2010191151217558), and siNUDT21 (targeting the human NUDT21 gene; RiboBio, stB0001835B) were transfected into THP-1 macrophages, while siNudt21 (targeting the mouse Nudt21 gene; RiboBio, siG170614045522) was transfected into BMDMs, using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer’s protocol. Scrambled siRNAs (siNC) were used as negative controls. For plasmid transfection, cells at 60–80% confluency were transfected using Lipofectamine 3000 (Invitrogen) in serum-free medium, and according to the manufacturer’s instructions. Cells were harvested 24–48 h later for further analyses. Knockdown efficiency was assessed by western blotting 36–48 h post-transfection.
qPCR
Total RNA was extracted using RNeasy kits (Omega) and cDNA was synthesized from 1 µg of total RNA using olig(dT) primer. A total of 1 μl cDNA was then analyzed by quantitative PCR on a 7500 Fast Real-Time PCR System (Thermo Fisher Scientific) using SYBR Green PCR Master Mix (TaKaRa). The following primers were used:
FTH1 forward: 5′-AGAACTACCACCAGGACTCA-3′;
FTH1 reverse: 5′-TCATCGCGGTCAAAGTAGTAAG-3′;
FTH1-L forward: 5′-GCCGTTGTTCAGTTCTAATCACA-3′;
FTH1-L reverse: 5′-CCAAGACAGCCACACCTTAGT-3′;
GFP forward: 5′-AGGACGACGGCAACTACAAG-3′;
GFP reverse: 5′-TTGTACTCCAGCTTGTGCCC-3′;
GAPDH forward: 5′-ACCACAGTCCATGCCATCAC-3′;
GAPDH reverse: 5′-TCCACCACCCTGTTGCTGTA-3′).
Statistical analyses were performed using GraphPad Prism version 8 (GraphPad Software Inc.). Statistical significance between groups was assessed using one-way ANOVA with Tukey’s post hoc test (for two data groups) or two-way ANOVA with Bonferroni’s post hoc test (for three data groups). A p-value < 0.05 was considered statistically significant. Detailed statistical information is provided in the figure legends.
Western blotting
Cells were lysed in ice-cold RIPA buffer (Thermo Fisher Scientific) supplemented with protease inhibitors (Roche). Protein concentrations were determined using a BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of protein (30 µg) were separated by SDS-PAGE, transferred onto PVDF membranes (Merck Millipore), and blocked with 5% non-fat milk in PBST (PBS with 0.1% Tween-20) for 1 hour. Membranes were incubated overnight at 4°C with primary antibodies against FTH1 (Abcam, ab75973; Santa Cruz Biotechnology, sc-376594), NUDT21 (Proteintech, 66335-1-Ig), CPSF6 (Abcam, ab99347), GFP-tag (Proteintech, 50430-2-AP), H3 (Abcam, ab176842), and β-actin (Abcam, ab179467), according to the manufacturer’s instructions. Afterward, membranes were incubated for 1 hour at room temperature with HRP-conjugated anti-rabbit or anti-mouse IgG secondary antibodies (Abcam, ab205718, ab205719). After washing, protein bands were detected using ECL reagent (Thermo Fisher Scientific) following the manufacturer’s protocol and visualized using a Chemiluminescence Image System (Minichem™, China), as previously described (Mo et al., 2022).
For quantitation, protein band intensities were measured using ImageJ software. The integrated density of each target protein band was normalized to its corresponding loading control (e.g., H3 or β-actin) to account for variations in protein loading. The relative band intensities shown below each image were quantified from the specific experiment presented in the figure. Additionally, multiple independent experiments were performed to confirm reproducibility.
In vitro transcription and RNA pull-down
A tRNA scaffolded Streptavidin Aptamer (tRSA) was incorporated into full-length (tRSA-3′UTR-L) and shorter (tRSA-3′UTR-S) 3′UTRs of FTH1 mRNA and prepared using T7 RNA polymerase as described previously (Iioka and Macara, 2015). RNA was purified using TRIzol reagent (Life technologies, cat#15596-026) according to the manufacturer’s protocol. For cell lysate preparation, Mtb-infected or uninfected THP-1–differentiated macrophages were harvested and lysed in lysis buffer (10 mM HEPES pH 7.0, 200 mM NaCl, 10 mM MgCl2, 1% TritonX-100, 1 mM DTT) supplemented with protease inhibitors and an RNase inhibitor (Promega, cat#N2115). The lysate was centrifuged, and the supernatant was collected. Protein concentrations in the lysate were determined using the BCA Protein Assay Kit (Thermo Fisher). The tRSA–tagged RNA (10 µg) was incubated with 200 µg of cell lysate for 2 hours at 4°C with gentle rotation to promote RNA-protein binding. Streptavidin-coated magnetic beads (Invitrogen, 65605D) were then added to capture RNA–protein complexes. After a 1-hour incubation, the beads were washed five times with cold lysis buffer to remove nonspecific binding. The bound RNA–protein complexes were eluted by boiling the beads in 2X SDS-PAGE sample buffer and applied for western blot analysis.
Immunoprecipitation
Cells were lysed in lysis buffer (Beyotime, P0013) containing a protease inhibitor cocktail (Roche). For immunoprecipitation, 1 mg of total protein was incubated overnight at 4°C with 2 µg of anti-NUDT21, anti-CPSF6 or control IgG. The mixture was then incubated for additional 2 hours with 50 µl of protein A/G magnetic beads (Santa Cruz Biotechnology) at 4°C with continuous rotation. The beads were washed, and bound proteins were eluted by boiling the samples in SDS-PAGE sample buffer prior to western blotting analysis.
Immunostaining and confocal microscopy
THP-1 macrophages (2 × 105 cells/ml) infected with GFP-H37Ra (MOI = 10) were fixed with 4% (v/v) formaldehyde for 15 min and permeabilized with 0.2% (v/v) Triton X-100 for 5 min at room temperature. The cells were then blocked with 5% BSA and incubated in the dark with NUDT21 or CPSF6 primary antibodies overnight. After washing, the samples were incubated in the dark for 1 hour with Alexa Fluor-conjugated secondary antibodies (Invitrogen, anti-rabbit Alexa fluor 555, 1:200), and Hoechst (Invitrogen; 10 ng/ml) for 5 min at room temperature before visualization. Images were captured under an Olympus FV1000 confocal microscope (NIKON A1R).
Nuclear isolation
Macrophages were lysed in ice-cold Nuclei Isolation Buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA) with protease inhibitors. After mechanical homogenization, the nuclei were pelleted by centrifugation at 1,000 g for 10 min at 4°C. The supernatant and resuspended nuclei were isolated, boiled in SDS sample buffer, and analyzed by western blotting.
CFU assay
BMDMs were infected with Mtb at an MOI of 10 and incubated for 6 h at 37°C to allow for bacterial uptake. After infection, the macrophages were washed three times with PBS to remove non-internalized bacteria and further incubated at 37 °C in 5% CO2 for 72 h. To quantify intracellular bacteria, macrophages were lysed with 0.1% Triton X-100 (Sigma-Aldrich) in PBS for 15 min at room temperature. The lysates were then serially diluted in PBS and plated in triplicate on Middlebrook 7H10 agar plates. Plates were incubated at 37°C for 3–4 weeks to allow for colony formation. CFU counts were performed at 6-h and 72-h post-infection as described previously (Mo et al., 2022). Statistical analyses were performed using GraphPad Prism version 8 (GraphPad Software Inc.), with statistical significance of differences between groups was determined using a student’s unpaired t test.
Study approval
The present study was approved by the Ethics Committees of Wuhan Polytechnic University (Wuhan, China), Shenzhen University (Shenzhen, China), and Shenzhen People’s Hospital (Shenzhen, China). The study was conducted using BMDMs isolated from animals and did not involve in vivo experiments. However, we acknowledge the ARRIVE guidelines and adhere to ethical principles for animal research. All procedures were performed in compliance with the ethical guidelines set by the Wuhan Polytechnic University Animal Ethics Committee, following relevant institutional and national regulations for the care and use of laboratory animals.
Results
Mtb infection alters the FTH1 APA profile in macrophages
We first investigated how Mtb infection affects FTH1 expression in THP-1 macrophages. These cells were infected with the H37Ra Mtb strain or left uninfected, and total RNA were collected at 2, 4, 6 and 24 hours post infection. We observed an increase in FTH1 mRNA levels over time in the infected cells compared with the uninfected cells (Figure 1A), with a corresponding rise in protein abundance (Figure 1B). To explore whether APA contributes to this upregulation, we analyzed the APA profile of FTH1 in Mtb-infected and uninfected THP-1 macrophages using a 3′RACE assay. Interestingly, Mtb infection shifted the APA profile toward the longer FTH1 transcript isoforms (Figure 1C), indicating the potential involvement of APA in this Mtb-driven upregulation of FTH1 expression. To determine if this mechanism is conserved across species, we performed the same 3′RACE assay on RAW264.7 murine macrophages and observed a similar shift toward longer Fth1 isoforms and enhanced protein expression (Figures 1D, E). These findings indicate that Mtb infection triggers FTH1 APA regulation in both human and murine macrophages.
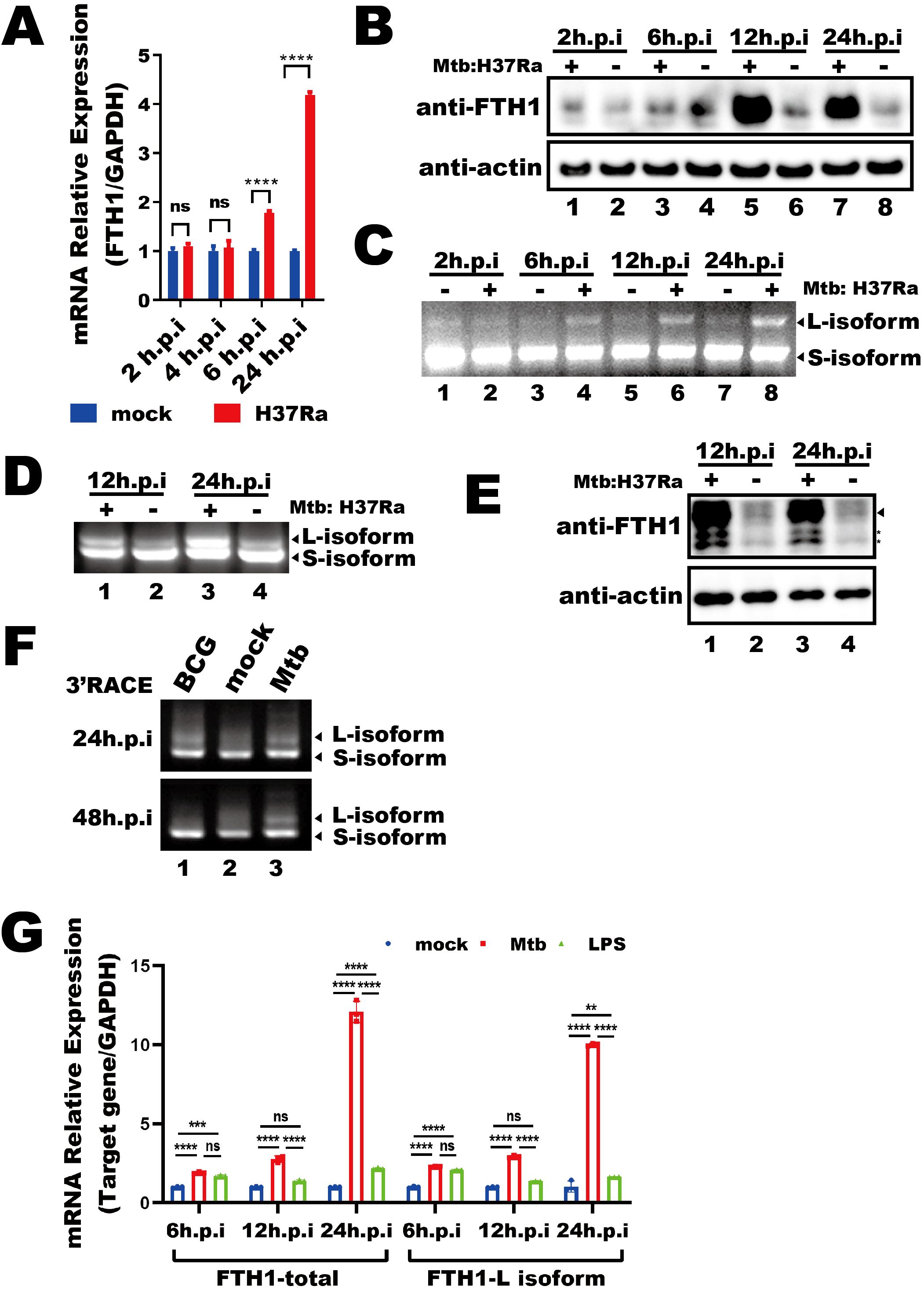
Figure 1. Mtb infection alters the FTH1 APA profile in macrophages. (A) qPCR analysis of FTH1 mRNA in Mtb-infected (H37Ra) or uninfected (mock) macrophages for 2-, 4-, 6-, and 24-hours post-infection. Data are normalized to GAPDH and presented as fold change relative to the control. Error bars represent the mean ± SEM from two independent experiments. Statistical significance was determined by two-way ANOVA with Sidak’s multiple comparisons test; ****P < 0.0001. (B) Western blot analysis of FTH1 protein expression in THP-1 macrophages infected with Mtb at 2, 6, 12, and 24 hours. β-actin was used as a loading control. (C) Agarose gel electrophoresis of 3′RACE products from Mtb-infected or uninfected (control) macrophages. Bands representing different FTH1 transcript isoforms are indicated by arrows. (D) 3′RACE assay of Fth1 transcripts and (E) western blot analysis of mouse FTH1 protein expression in Mtb-infected or uninfected RAW264.7 cells. The specific FTH1 band is marked by an arrow, and nonspecific bands are indicated with stars. β-actin was used as a loading control. (F) Agarose gel electrophoresis of FTH1 3′RACE products from macrophages infected with BCG or Mtb (H37Ra) at 24- and 48-hours post-infection. (G) qPCR analysis of total and longer FTH1 transcripts in THP-1 macrophages infected with Mtb (H37Ra, MOI = 10) or stimulated with LPS (20 ng/mL). GAPDH was used as the reference gene. Error bars represent the mean ± SEM from three independent experiments. Statistical significance was determined by two-way ANOVA with Tukey’s multiple comparisons test; **P < 0.01, ***P < 0.001 ****P < 0.0001. Three independent experiments were performed to ensure reproducibility, and representative images were selected for presentation. ns, not significant.
To evaluate the specificity of the Mtb-driven shift in FTH1 APA, we stimulated THP-1 macrophages with lipopolysaccharide (LPS) or Bacillus Calmette-Guérin (BCG). While BCG stimulation initially increased the expression of longer FTH1 transcript isoforms at 24 hours, this increase diminished by 48 hours post-infection, returning to baseline levels compared with uninfected THP-1 macrophage (Figure 1F, lane 1 vs. lane 2). In contrast, Mtb infection induced a sustained, robust increase in longer FTH1 transcript isoforms at 48 hours in THP-1 macrophages (Figure 1F, lane 3). Additionally, LPS treatment led to only a modest increase in longer FTH1 transcript isoforms (FTH1-L) at 24 hours, much less than the response seen in Mtb-infected THP-1 macrophages (Figure 1G). Together, these results demonstrate that Mtb infection specifically induces a unique FTH1 APA regulation, resulting in enhanced expression of FTH1-L in macrophages.
Mtb-induced longer FTH1 transcripts enhance protein expression
We next investigated the impact of this longer FTH1 transcript on protein expression. The siRNAs targeting the longer FTH1 isoform (FTH1-L) or total FTH1 transcripts were transfected into THP-1 macrophages 24 hours prior to Mtb infection, and FTH1 mRNA levels (Figure 2A) and protein abundance were assessed at 24- and 48-hours post-infection (Figure 2B). Silencing FTH1-L strongly reduced FTH1 protein levels in Mtb-infected THP-1 macrophages at 48 hours (Figure 2B, lane 6 vs. lane 4), highlighting the role of FTH1-L in the upregulation of FTH1 expression in these cells.
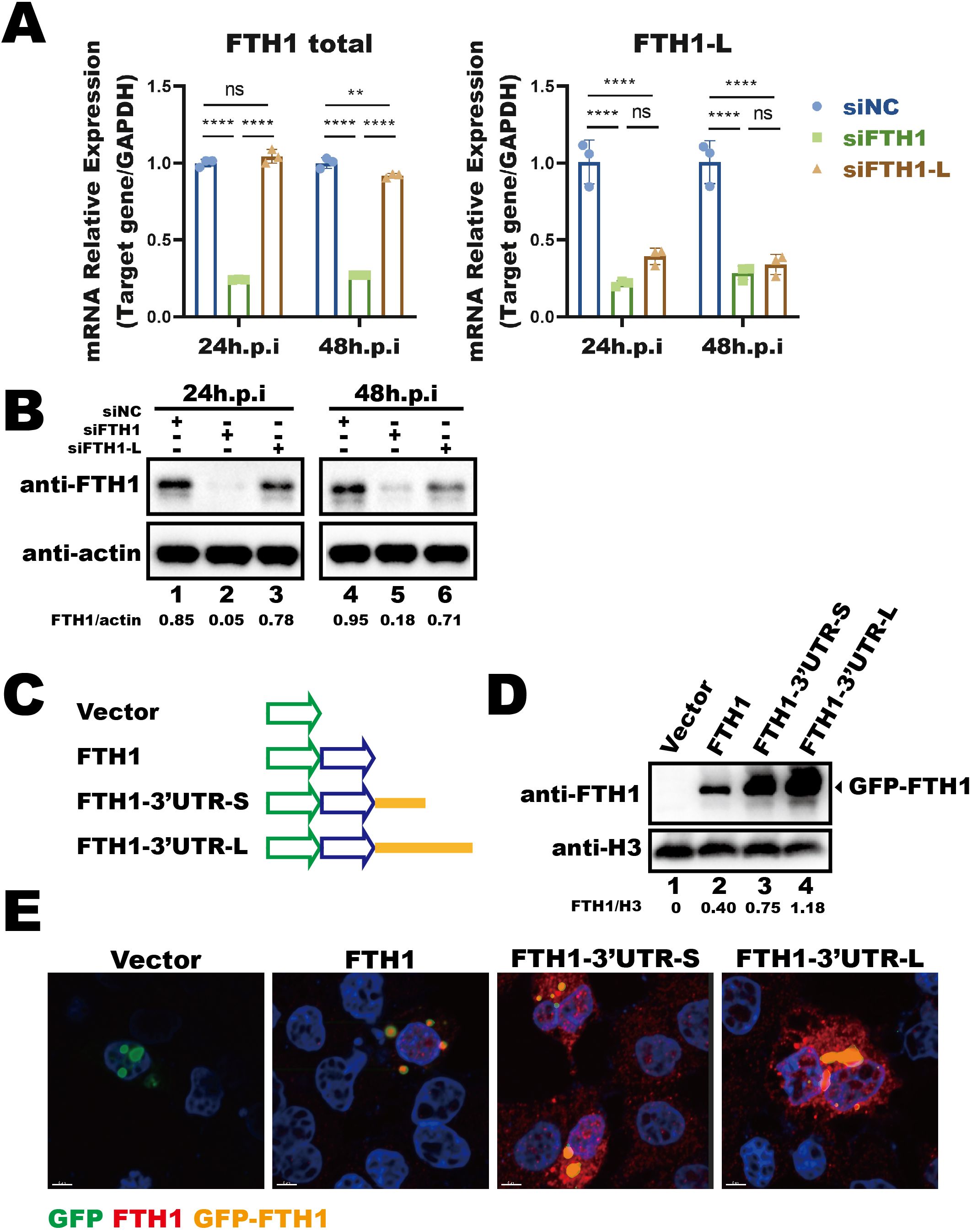
Figure 2. Mtb-induced longer FTH1 transcripts enhance protein expression. (A) qPCR analysis of siRNA knockdown efficiency (siFTH1, green; siFTH1-L, brown) in THP-1 macrophages harvested 24- or 48-hours post-transfection. Total RNA (1 µg) was used for cDNA synthesis, followed by qPCR to measure total FTH1 (left panel) or the longer isoform–specific transcripts (right panel). Data are normalized to GAPDH and presented as fold change relative to the control. Error bars represent the mean ± SEM from three independent experiments. Statistical significance was determined by two-way ANOVA with Tukey’s multiple comparisons test; **P < 0.01, ****P < 0.0001. (B) Western blot analysis of FTH1 expression in THP-1 macrophages transfected with siRNAs targeting total FTH1 mRNA (siFTH1) or the longer isoform (siFTH1-L). β-actin served as a loading control. The relative abundance of FTH1 (normalized to actin) is shown below each lane. The quantification represents the result of a single experiment. (C) Schematic of the GFP-tagged FTH1 expression constructs used to evaluate the 3′UTR’s role in FTH1 expression. (D) Western blot analysis was performed to assess GFP-FTH1 protein expression in A549 cells transfected with 3′UTR-FTH1 overexpression plasmids shown in Figure 3C. Protein levels were quantified using ImageJ, with target protein band intensity normalized to the corresponding loading control. The relative abundance of FTH1 (normalized to H3) is shown below each lane. The quantification represents the result of a single experiment. (E) Confocal microscopy of GFP-FTH1 in A549 cells 24 hours post-transfection to assess its subcellular distribution. Three independent experiments were performed to ensure reproducibility, and representative images were selected for presentation. ns, not significant.
To further investigate the impact of the longer transcript isoform, induced by Mtb-driven APA regulation, on protein translation, we examined the role of 3′UTR in modulating FTH1 translation. We constructed FTH1 overexpression plasmids based on a fluorescent vector, in which the FTH1 coding sequence was fused with either the longer or shorter FTH1 3′UTR isoform (Figure 2C). Upon transfection into A549 cells, both 3′UTRs enhanced translation of the GFP–FTH1 fusion protein compared to the FTH1 plasmid without a 3′UTR (Figure 2D, lane 3 and lane 4 vs. lane 2). Notably, the longer FTH1 3′UTR clearly enhanced protein translation efficiency compared to the shorter isoform, resulting in increased GFP–FTH1 protein abundance (Figure 2D, lane 4 vs. lane 3). These results suggest that the 3′UTR length is linked to the translational efficiency of FTH1 transcripts. However, 3′UTR length did not affect the distribution of FTH1 protein in transfected cells, which was predominantly localized in the cytoplasm (Figure 2E, overexpressed GFP–FTH1 highlighted in orange). Overall, our findings indicate that the Mtb-induced shift in FTH1 APA increases the abundance of FTH1-L isoform, leading to enhanced FTH1 protein production.
The longer FTH1 isoform contains essential APA regulatory cis-elements
To determine the cis-acting elements within the 3′UTR that regulate FTH1-L isoform expression, we examined how specific cis-elements within the 3′UTR influence APA regulation. We cloned either the longer (3′UTR-L) or shorter (3′UTR-S) FTH1 3′UTR into a fluorescent reporter vector (Figure 3A). The upstream element (USE1) of the (pA)p site, which is common to both isoforms, was replaced with an unrelated sequence of the same length. In addition, the upstream element (USE2) of the (pA)d site and the downstream element (DSE), both exclusive to the 3′UTR-L, were substituted with unrelated sequences (Figure 3A).
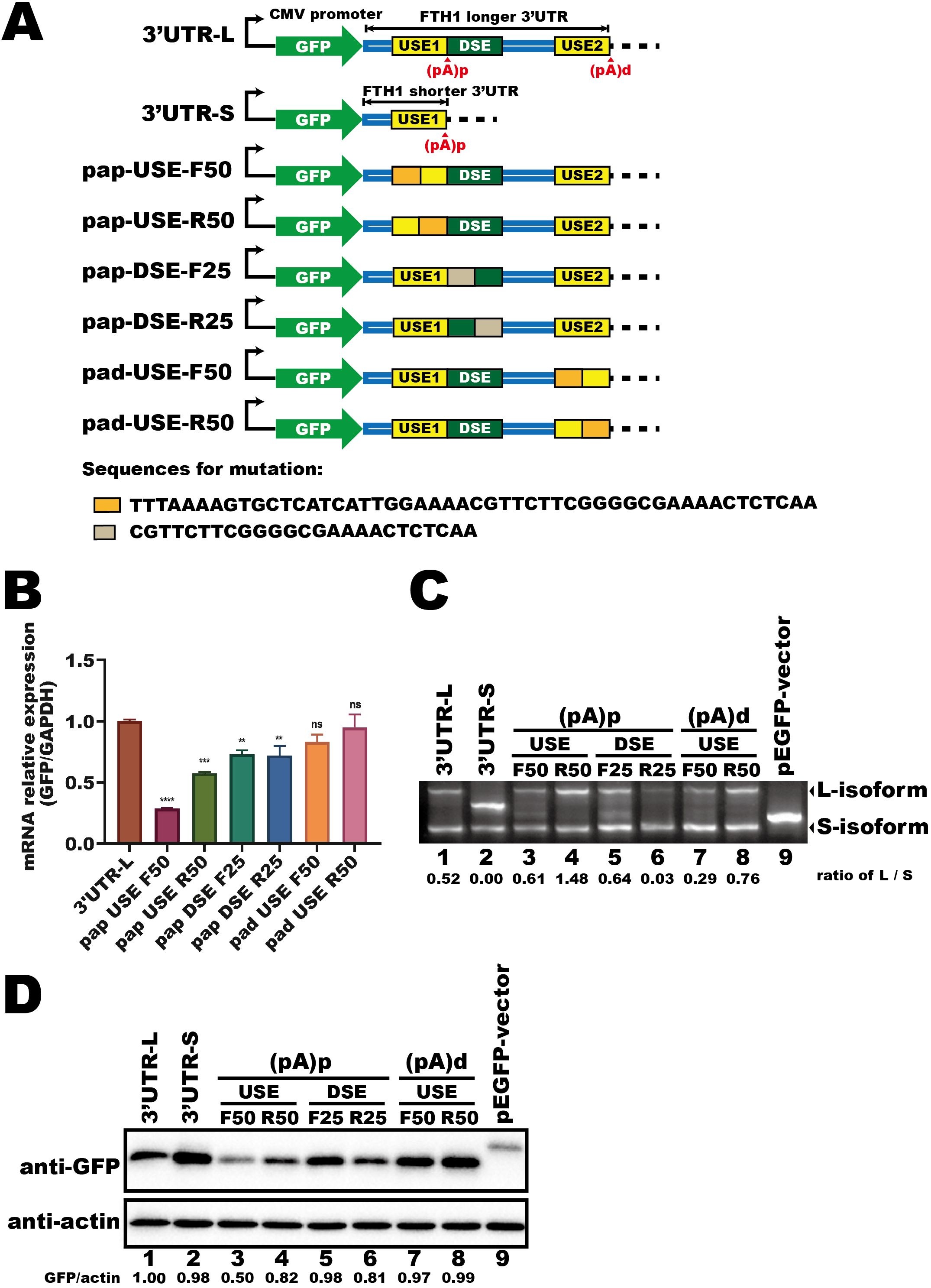
Figure 3. The longer FTH1 isoform contains essential APA regulatory cis-elements. (A) Schematic of the fluorescent reporter plasmids (GFP) containing FTH1 3′UTRs and mutant constructs. Sequences used for replacement are indicated beneath the schematic. (B) qPCR analysis of fluorescent gene expression in HEK293T cells transfected with plasmids as shown in Figure 3A. Data are normalized to GAPDH and presented as fold change relative to the control. Error bars represent the mean ± SEM from three independent experiments. Statistical significance was determined by one-way ANOVA with Dunnett’s multiple comparisons test; **P < 0.01, ***P < 0.001, ****P < 0.0001. (C) 3′RACE assay of fluorescent transcripts in HEK293T cells transfected with plasmids shown in (A). RNA abundances were quantified using ImageJ software, and the numerical values below the blot represent the ratio of the longer isoform (L-isoform) to the shorter isoform (S-isoform). (D) Western blot analysis of GFP protein levels in HEK293T cells transfected with plasmids shown in (A). Protein levels were quantified using ImageJ software. The intensity of the target band was normalized to the corresponding loading control. Numerical values representing the target protein (GFP) abundance relative to the loading control (β-actin) are shown at the bottom of each lane. Three independent experiments were performed to ensure reproducibility, and representative images were selected for presentation. The difference in molecular weight arises from the insertion of the 3′UTR, which introduced a stop codon, producing a shorter ~26 kDa fluorescent protein, whereas the native vector expresses a ~29 kDa protein. ns, not significant.
Following HEK293T cell transfection, we measured mRNA expression, protein levels, and the APA profile of the fluorescent gene. Mutations near the (pA)d site (pad-USE-F50 and pad-USE-R50) did not markedly alter mRNA levels compared to 3′UTR-L (Figure 3B). However, pad-USE-R50 generated a higher proportion of L-isoform transcripts than pad-USE-F50 (Figure 3C, lane 8 vs. lane 7), leading to a modest increase in GFP protein levels (Figure 3D, lane 8 vs. lane 7). In contrast, DSE mutations caused a ~15% reduction in fluorescent mRNA expression (Figure 3B, pap-DSE-F25 and pap-DSE-R25 vs. 3′UTR-L). Among these, pap-DSE-F25, in which the first 25 nucleotides were replaced with a scrambled sequence, increased L-isoform abundance (Figure 3C, lane 5 vs. lane 6) and enhanced GFP protein expression (Figure 3D, lane 5 vs. lane 6) compared to pap-DSE-R25. Notably, pap-DSE-R25 exhibited the strongest reduction in L-isoform expression (Figure 3C, lane 6 vs. lane 1), while pap-DSE-F25 showed a slight increase (Figure 3C, lane 5 vs. lane 1). These differences further support the idea that the longer isoform has higher translation efficiency.
Mutations in USE1, near the (pA)p site, caused a more pronounced reduction in mRNA expression, particularly in pap-USE-F50 (Figure 3B, pap-USE-F50 and pap-USE-R50 vs. 3′UTR-L). Moreover, mutating the last 50 nucleotides of USE1 (pap-USE-R50) doubled L-isoform abundance (Figure 3C, lane 4 vs. lane 3) and elevated GFP protein levels (Figure 3D, lane 4 vs. lane 3) compared to pap-USE-F50. These findings highlight the importance of cis-elements near the (pA)p site, especially within the final 50 nucleotides of USE1, in regulating FTH1 expression. We propose that trans-acting factors binding to USE1 might mediate Mtb-induced APA regulation of FTH1.
Mtb infection reduces the interaction between NUDT21 and FTH1 transcripts in THP-1 macrophages
Having established that the last 50 nucleotides of the USE1 in FTH1 3′UTR contains key cis-elements involved in regulating gene expression, we next further investigated what is it and how it participates the APA regulation of FTH1 during Mtb infection. We identified two UGUA motifs within this region and assessed their functional significance by creating mutant plasmids based on the 3′UTR-L fluorescent vector (Figure 4A). We found that mutating both motifs altered polyadenylation site selection, resulting in an increased abundance of the longer isoform (Figure 4B, lane 2 vs. lane 1). Mutating the UGUA motif closest to the (pA)p site (UGUA–mut2) also increased longer isoform mRNA (Figure 4B, lane 4 vs. lane 1), while mutation of the other UGUA motif (UGUA–mut1) had no significant effect on the proportion of longer isoform mRNA (Figure 4B, lane 3 vs. lane 1). Moreover, mutations in either UGUA motif or in both motifs resulted in a slight increase in protein expression compared to the wild-type (Figure 4C), suggesting that the UGUA motifs within the FTH1 3′UTR are critical for modulating APA and gene expression.
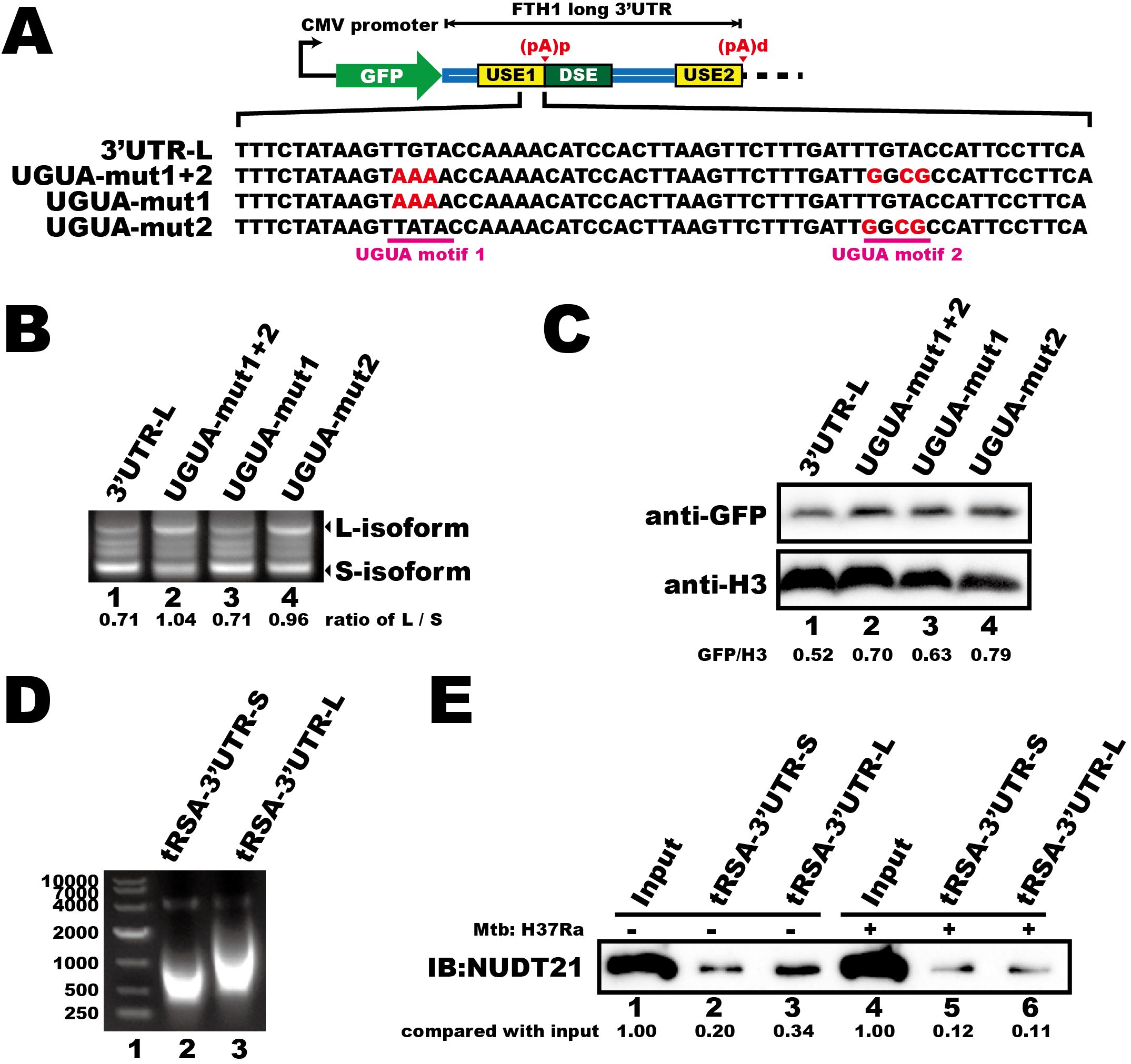
Figure 4. Mtb infection reduces the interaction between NUDT21 and FTH1 transcripts in THP-1 macrophages. (A) Schematic of the fluorescent reporter plasmids (GFP) containing FTH1 3′UTR and UGUA motif–mutated constructs. Mutated nucleotides are highlighted in red. (B) 3′RACE of GFP transcripts and (C) Western blot analysis of GFP protein levels demonstrate how UGUA motif mutations affect the APA profile and protein expression of the fluorescent gene. RNA and protein abundance was quantified using ImageJ software, with normalized numerical values displayed at the bottom of the respective panels. (D) Agarose gel electrophoresis of in vitro–transcribed, tRSA-tagged FTH1 3′UTR constructs representing the short (lane 2) and the long (lane 3) versions. (E) RNA pull-down demonstrating the interaction between NUDT21 and the FTH1 3′UTRs in Mtb-infected or uninfected THP-1 macrophages. Protein abundance was quantified using ImageJ software, and the raw values are shown at the bottom. Three independent experiments were performed to ensure reproducibility, and representative images were selected for presentation.
NUDT21 is a key regulator of the APA pathway that interacts with UGUA motifs (Yang et al., 2010). Therefore, we investigated whether Mtb infection interferes with this interaction. We performed RNA pull-down assays using in vitro–transcribed, tRSA–tagged FTH1–3′UTR RNAs (Figure 4D) and lysates from Mtb-infected or uninfected THP-1 macrophages. Mtb infection reduced NUDT21 binding to both FTH1–3′UTRs (Figure 4E, lane 2 vs. lane 5; lane 3 vs. lane 6), with a particularly pronounced decrease in affinity for the longer transcript isoform (Figure 4E, lane 6 vs. lane 3). We propose that this disruption likely contributes to the Mtb-induced FTH1 APA shift and the subsequent upregulation of FTH1-L.
Mtb infection disrupts the interactions between NUDT21 and CPSF6
Given that Mtb infection reduces the interaction between NUDT21 and the FTH1 3′UTR, we sought to investigate the underlying mechanisms. As the CFI complex, composed of NUDT21 and CPSF6, guides precise polyadenylation site selection (Yang et al., 2011), we next investigated whether Mtb infection affects the expression and cellular distribution of these regulatory proteins. During infection, NUDT21 protein levels remained unchanged (Figure 5A), and both western blotting and confocal microscopy showed no noticeable differences in its cellular distribution between infected and uninfected cells (Figures 5B, C). These results suggest that Mtb-driven FTH1 APA regulation is more likely due to functional impairment of NUDT21 rather than alterations in its expression or localization.
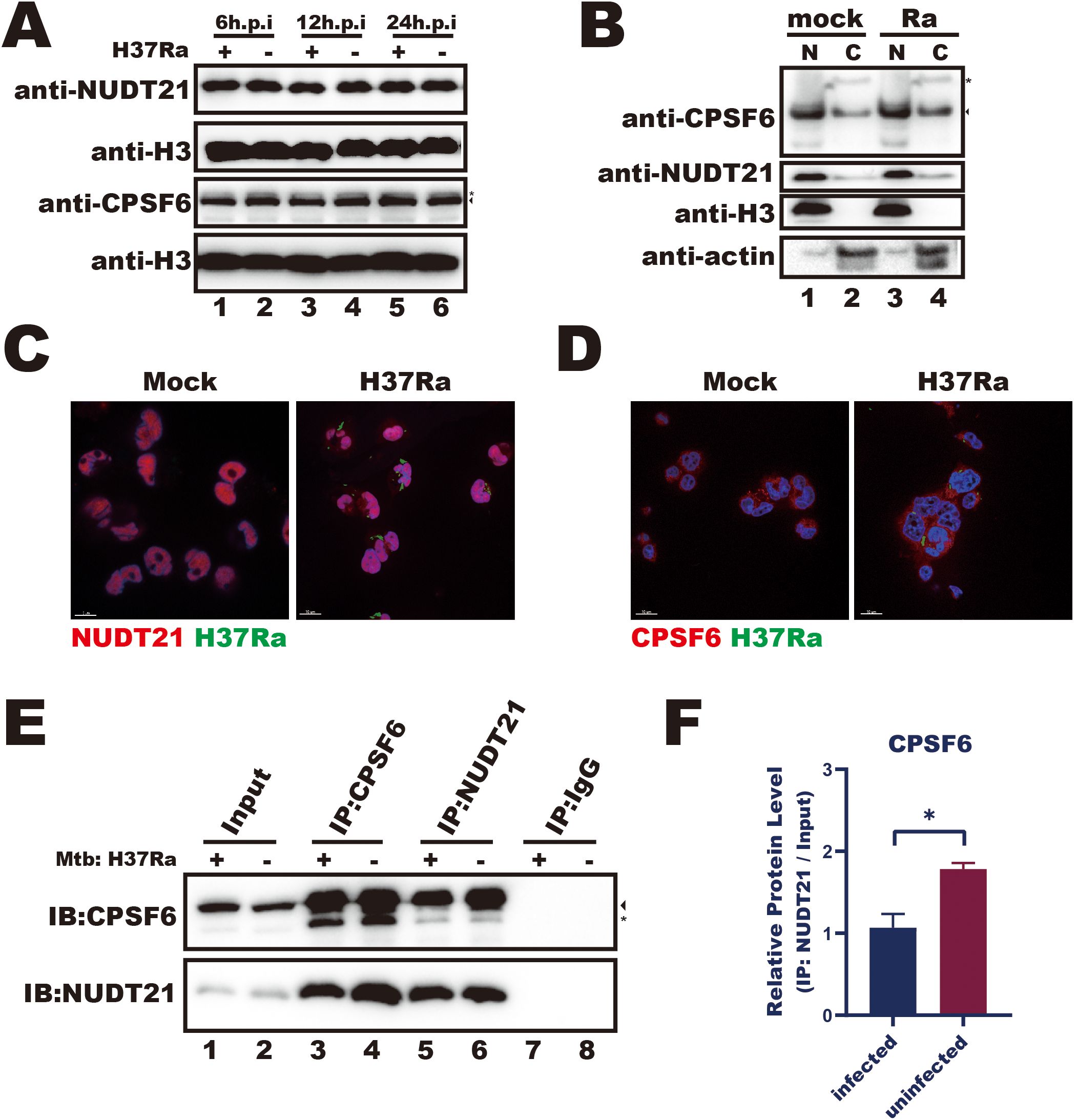
Figure 5. Mtb infection disrupts the interactions between NUDT21 and CPSF6. (A) Western blot analysis of NUDT21 and CPSF6 protein levels in THP-1 macrophages infected with Mtb for 6, 12, and 24 hours or left uninfected (control). H3 served as a loading control. (B) Western blot of nuclear (N) and cytoplasmic (C) fractions showing NUDT21 and CPSF6 distribution in Mtb-infected or uninfected THP-1 macrophages. H3 was used as a nuclear loading control, and β-actin was used as a cytoplasmic loading control. (C) Representative confocal microscopy images of THP-1 macrophages infected with Mtb (green) and immunolabeled for NUDT21 (red) or (D) CPSF6 (red). Hoechst dye was used to stain nuclei (blue). (E) Western blot analysis of immunoprecipitation (IP) products from Mtb-infected and uninfected THP-1 macrophages at 48 hours post-infection. IgG served as a negative control. The target protein band is marked by an arrow, and a nonspecific interaction band is indicated with a star. Representative images from three independent experiments are shown. (F) Quantification of protein band intensities from three independent IP experiments with THP-1 macrophages using ImageJ software. Statistical analysis was performed in GraphPad Prism 8, and differences between infected and uninfected groups were determined using unpaired t test. *P < 0.05.
We then focused on CPSF6, a key RNA-binding partner of NUDT21 within the CFI complex. Similar to NUDT21, Mtb infection did not affect CPSF6 expression or cellular distribution (Figures 5A, B, D). However, reciprocal co-immunoprecipitations assays revealed a significant reduction in the NUDT21–CPSF6 interaction in infected THP-1 macrophages (Figures 5E, F). This dissociation likely compromises the RNA recognition capability of the CFI complex, thereby contributing to APA dysregulation in Mtb-infected macrophages.
siRNA-mediated NUDT21 knockdown impairs macrophage anti-Mtb function
Having shown that Mtb disrupts the interaction between key APA regulators, we next investigated the functional relevance of NUDT21 in macrophage defense against Mtb. To this end, we used siRNA to knock down NUDT21 expression in THP-1 macrophages, which led to a modest increase in FTH1 protein levels (Figure 6A, middle lane). However, the knockdown efficiency in this cell type was suboptimal, and no significant change in intracellular Mtb survival was observed (data not shown).
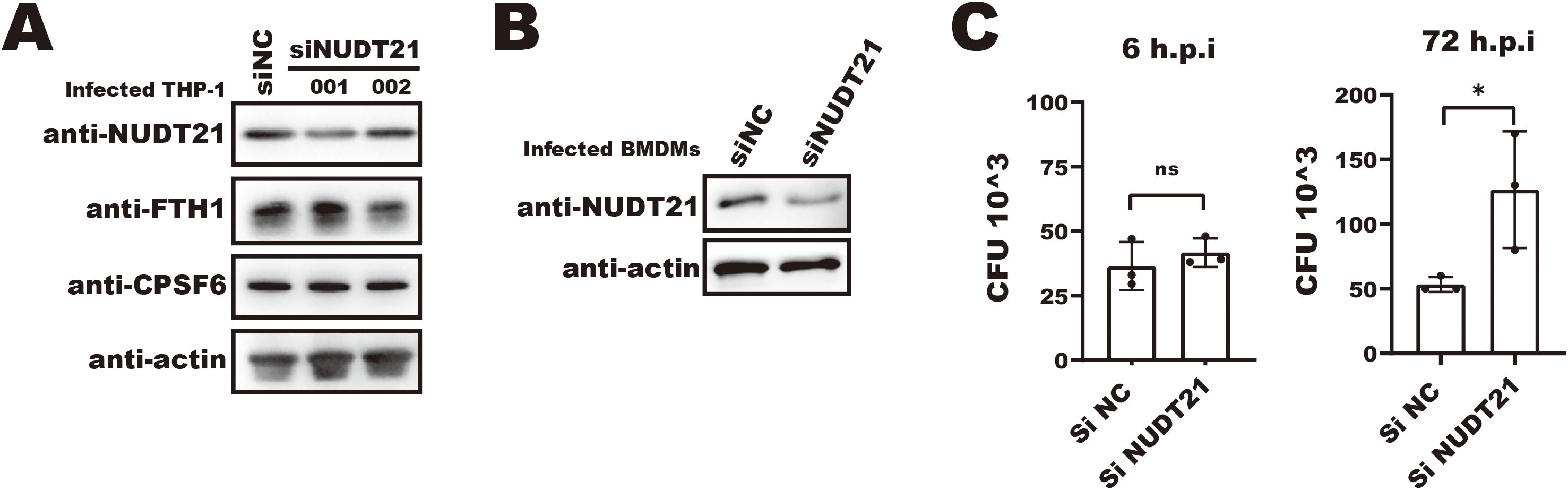
Figure 6. siRNA-mediated NUDT21 knockdown impairs macrophage anti-Mtb function. (A) Western blot analysis of FTH1 and NUDT21 expression in NUDT21 knockdown THP-1 macrophages. β-actin was used as a loading control. (B) Western blot showing siRNA-mediated NUDT21 knockdown in bone marrow–derived macrophages (BMDMs). β-actin was used as a loading control. (C) CFU assays of Mtb infected BMDMs treated with siNUDT21 or control siNC. Cells were infected with Mtb and harvested at 6- and 72-hours post-infection. After serial dilution and plating on 7H10 agar, the plates were incubated at 37°C, and colony-forming units (CFUs) were counted after 3 weeks. *P < 0.05.
In contrast, siRNA-mediated NUDT21 knockdown in BMDMs resulted in a significantly higher intracellular Mtb burden at 72 hours post-infection (Figures 6B, C), underscoring the importance of NUDT21 in maintaining macrophage anti-Mtb function. Together, these findings suggest that Mtb-induced disruption of NUDT21 activity contributes to dysregulated APA of immune-relevant transcripts, which may in turn impair macrophage-mediated host defense.
Discussion
FTH1 is crucial for maintaining iron homeostasis in macrophages and plays a vital role in the immune response to Mtb infection (Reddy et al., 2018). By sequestering excess intracellular iron, FTH1 limits its availability to Mtb, thereby hindering bacterial growth (Cassat and Skaar, 2013). However, Mtb can degrade FTH1 through the host ferritinophagy pathway, releasing iron to support its survival (Dai et al., 2024). These findings position FTH1 as a central mediator in balancing iron homeostasis within host–pathogen interactions during Mtb infection. In this study, we reveal a novel Mtb-driven NUDT21-mediated APA mechanism that enhances FTH1 expression in macrophages, adding a new layer of complexity to the regulation of iron homeostasis in these cells during Mtb infection.
The expression of FTH1 is regulated through multiple pathways in response to infection. Notably, the transcription factor Nrf2 promotes iron storage by increasing ferritin levels (Kerins and Ooi, 2018), while pro-inflammatory cytokines such as IL-6, IL-1β, and TNF-α upregulate FTH1 transcription (Rogers, 1996; Torti and Torti, 2002). However, the levels of these factors typically return to baseline due to host immune regulation, which aims to reduced excessive inflammation, or as a result of Mtb’s immune evasion mechanisms. Consequently, these pathways alone do not fully explain the sustained expression of FTH1 in Mtb-infected macrophages. At the translational level, the well-established IRP–IRE system regulates FTH1 expression in response to intracellular iron levels (Wilkinson and Pantopoulos, 2014; Rouault, 2006). Yet, Mtb-infected macrophages exhibit significantly lower intracellular iron concentrations (Dai et al., 2019), creating a paradox of reduced iron and sustained FTH1 expression. This discrepancy raised the question of whether IRP–IRE-mediated control alone is sufficient to account for the observed increase in FTH1 expression, prompting us to explore alternative regulatory mechanisms.
Most human genes contain multiple polyadenylation sites (PASs), allowing cells to regulate gene expression by selecting different PASs in response to various signals (Tian et al., 2005). NUDT21, a critical component of the APA machinery, guides PAS selection during mRNA 3′-end processing, influencing cellular functions and fate decisions (Chen et al., 2024; Brumbaugh et al., 2018). In this study, we found that Mtb infection impairs NUDT21’s recognition of the FTH1 3′UTR (Figure 4) due to its dissociation from CPSF6 (Figure 5). We hypothesize that this NUDT21–CPSF6 dissociation may extend beyond FTH1, affecting the expression of other macrophage genes involved in the response to Mtb. Supporting this, NUDT21 knockdown significantly increased Mtb intracellular survival in BMDMs (Figure 6), highlighting its role in sustaining macrophage immunity. However, we did not assess whether this effect is FTH1-dependent. Given that Mtb exploits host iron metabolism for survival (Dai et al., 2023), further studies are needed to clarify the underlying mechanism.
Interestingly, our investigation of other genes involved in host–pathogen interactions, such as IDO1, revealed that APA reprogramming does not always favor the longer isoform. These gene-specific shifts in APA underscore the complexity of host gene regulation during Mtb infection and highlight the importance of APA in shaping the host’s immune response to the pathogen. Future studies should further explore changes in host transcript profiles during Mtb infection to identify additional APA-regulated genes, which could serve as molecular biomarkers or therapeutic targets.
Our study also demonstrated that Mtb infection impedes the recognition of UGUA motifs within the FTH1 transcript in macrophages (Figure 4). Since UGUA motif are crucial for polyadenylation site selection (Yang et al., 2010), disruption of this interaction leads to the production of longer FTH1 transcripts, which exhibit enhanced translational efficiency (Figure 2). This finding emphasizes the importance of cis-elements within the FTH1 3′UTR, and their corresponding trans-factors, in regulating gene expression during Mtb infection. Furthermore, we identified that the first 25 nucleotides in the DSE region influence polyadenylation site selection and protein expression in a fluorescent reporter system (Figure 3C, lane 5; 3D, lane 5). These results suggest that other cis-elements within the DSE, along with their corresponding trans-factors, contribute to the regulation of FTH1 APA in Mtb-infected macrophages. While our study implicates these cis-elements, the potential role of trans-factors, such as CstF64, in the Mtb-induced FTH1 APA shift remains speculative and warrants further investigation (Yao et al., 2012; Mukherjee et al., 2023).
Despite providing significant insights into the regulation of FTH1 expression in Mtb-infected macrophages, our study has several limitations that should be considered. First, we used the avirulent H37Ra strain instead of the virulent H37Rv strain. H37Ra is commonly used in host-pathogen interaction studies due to its genetic similarity to H37Rv and its suitability for biosafety level 2 (BSL-2) laboratories. However, its attenuated virulence may lead to differences in macrophage responses and APA regulation. Future studies using virulent strains will be necessary to confirm whether the APA-mediated regulation of FTH1 expression is conserved.
Additionally, while our study focuses on Mtb infection, FTH1 upregulation has been observed in response to various intracellular pathogens and inflammatory conditions, suggesting a broader role in iron homeostasis and immune regulation. However, the extent and regulatory mechanisms of FTH1 induction likely differ by pathogen and immune context. Further investigation is needed to delineate these mechanisms and their implications in different infectious contexts.
Furthermore, given the established role of CPSF6 in HIV-1 infection (Bejarano et al., 2019; Achuthan et al., 2018), the Mtb-induced release of CPSF6 from NUDT21 in co-infected individuals could potentially exacerbate HIV-1 replication, posing a public health concern, particularly in regions with high co-prevalence of both TB and HIV. While the clinical relevance of NUDT21–CPSF6 dysregulation in co-infection remains speculative, this pathway warrants investigation in cohorts with dual TB/HIV infections. We propose that targeting the NUDT21–CPSF6 axis could serve as a therapeutic strategy to enhance macrophage function and strengthen host defenses in these high-risk populations.
In summary, our study uncovers a novel APA mechanism that regulates FTH1 expression in macrophages during Mtb infection. By disrupting the interaction between NUDT21 and CPSF6, Mtb shifts polyadenylation toward longer FTH1 transcripts, leading to increased protein production, which ultimately compromises macrophage immunity. These findings emphasize the critical role of post-transcriptional regulation in host–pathogen interactions and open up new avenues for therapeutic strategies aimed at enhancing macrophage defenses against Mtb. Future research should further explore the roles of additional trans-factors in APA regulation and investigate potential approaches to modulate this pathway for better infection control.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://figshare.com/, https://doi.org/10.6084/m9.figshare.28428101.
Ethics statement
The animal study was approved by Ethics Committee of Wuhan Polytechnic University, Shenzhen University, and Shenzhen People’s Hospital. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. NC: Formal Analysis, Writing – review & editing, Investigation. YD: Formal Analysis, Investigation, Writing – review & editing, Methodology. XC: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. XZ: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Shenzhen Municipal Fundamental Research Project under Grant.JCYJ20220818102609021; China Postdoctoral Science Foundation under Grant.2021M691237; and Research Funding of Wuhan Polytechnic University under Grant.2024RZ013.
Acknowledgments
The authors thank Siwei Mo (Shenzhen University) for support during the confocal microscopy assays, Haoqiang Wan (Shenzhen people’s hospital) for kindly providing the RAW264.7 cell line, and Jessica Tamanini (Shenzhen University and ET Editing) for editing the manuscript before submission.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Achuthan, V., Perreira, J. M., Sowd, G. A., Puray-Chavez, M., Mcdougall, W. M., Paulucci-Holthauzen, A., et al. (2018). Capsid-CPSF6 interaction licenses nuclear HIV-1 trafficking to sites of viral DNA integration. Cell Host Microbe 24, 392–404 e8. doi: 10.1016/j.chom.2018.08.002
An, S., Li, Y., Lin, Y., Chu, J., Su, J., Chen, Q., et al. (2021). Genome-wide profiling reveals alternative polyadenylation of innate immune-related mRNA in patients with COVID-19. Front. Immunol. 12, 756288. doi: 10.3389/fimmu.2021.756288
Bejarano, D. A., Peng, K., Laketa, V., Börner, K., Jost, K. L., Lucic, B., et al. (2019). HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. eLife 8, e41800. doi: 10.7554/eLife.41800
Bo, H., Moure, U. A. E., Yang, Y., Pan, J., Li, L., Wang, M., et al. (2023). Mycobacterium tuberculosis-macrophage interaction: Molecular updates. Front. Cell Infect. Microbiol. 13, 1062963. doi: 10.3389/fcimb.2023.1062963
Brumbaugh, J., Di Stefano, B., Wang, X., Borkent, M., Forouzmand, E., Clowers, K. J., et al. (2018). Nudt21 controls cell fate by connecting alternative polyadenylation to chromatin signaling. Cell 172, 106–120 e21. doi: 10.1016/j.cell.2017.11.023
Cassat, J. E., Skaar, E. P. (2013). Iron in infection and immunity. Cell Host Microbe 13, 509–519. doi: 10.1016/j.chom.2013.04.010
Chen, Y., Chen, B., Li, J., Li, H., Wang, G., Cai, X., et al. (2024). Alternative mRNA polyadenylation regulates macrophage hyperactivation via the autophagy pathway. Cell Mol. Immunol. 21, 1522–1534. doi: 10.1038/s41423-024-01237-8
Dai, Y., Shan, W., Yang, Q., Guo, J., Zhai, R., Tang, X., et al. (2019). Biomarkers of iron metabolism facilitate clinical diagnosis in Mycobacterium tuberculosis infection. Thorax 74, 1161–1167. doi: 10.1136/thoraxjnl-2018-212557
Dai, Y., Zhu, C., Xiao, W., Chen, X., Cai, Y. (2024). Mycobacterium tuberculosis induces host autophagic ferritin degradation for enhanced iron bioavailability and bacterial growth. Autophagy 20, 943–945. doi: 10.1080/15548627.2023.2213983
Dai, Y., Zhu, C., Xiao, W., Huang, K., Wang, X., Shi, C., et al. (2023). Mycobacterium tuberculosis hijacks host TRIM21- and NCOA4-dependent ferritinophagy to enhance intracellular growth. J. Clin. Invest. 133 (8), e159941. doi: 10.1172/JCI159941
Ganz, T., Nemeth, E. (2015). Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 15, 500–510. doi: 10.1038/nri3863
Gruber, A. J., Zavolan, M. (2019). Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 20, 599–614. doi: 10.1038/s41576-019-0145-z
Hentze, M. W., Caughman, S. W., Rouault, T. A., Barriocanal, J. G., Dancis, A., Harford, J. B., et al. (1987). Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science 238, 1570–1573. doi: 10.1126/science.3685996
Hmama, Z., Pena-Diaz, S., Joseph, S., Av-Gay, Y. (2015). Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunol. Rev. 264, 220–232. doi: 10.1111/imr.2015.264.issue-1
Iioka, H., Macara, I. G. (2015). Detection of RNA-protein interactions using tethered RNA affinity capture. Methods Mol. Biol. 1316, 67–73. doi: 10.1007/978-1-4939-2730-2_6
Joshi, J. G., Fleming, J. T., Dhar, M., Chauthaiwale, V. (1995). A novel ferritin heavy chain messenger ribonucleic acid in the human brain. J. Neurol. Sci. 134 Suppl, 52–56. doi: 10.1016/0022-510X(95)00208-J
Kerins, M. J., Ooi, A. (2018). The roles of NRF2 in modulating cellular iron homeostasis. Antioxid Redox Signal 29, 1756–1773. doi: 10.1089/ars.2017.7176
Khan, N., Downey, J., Sanz, J., Kaufmann, E., Blankenhaus, B., Pacis, A., et al. (2020). M. tuberculosis reprograms hematopoietic stem cells to limit myelopoiesis and impair trained immunity. Cell 183, 752–770 e22. doi: 10.1016/j.cell.2020.09.062
Kurthkoti, K., Amin, H., Marakalala, M. J., Ghanny, S., Subbian, S., Sakatos, A., et al. (2017). The capacity of mycobacterium tuberculosis to survive iron starvation might enable it to persist in iron-deprived microenvironments of human granulomas. mBio 8 (4), e01092–17. doi: 10.1128/mBio.01092-17
Leibold, E. A., Munro, H. N. (1988). Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5’ untranslated region of ferritin heavy- and light-subunit mRNAs. Proc. Natl. Acad. Sci. U.S.A. 85, 2171–2175. doi: 10.1073/pnas.85.7.2171
Li, L., Ma, X., Cui, Y., Rotival, M., Chen, W., Zou, X., et al. (2023). Immune-response 3’UTR alternative polyadenylation quantitative trait loci contribute to variation in human complex traits and diseases. Nat. Commun. 14, 8347. doi: 10.1038/s41467-023-44191-1
Mancias, J. D., Wang, X., Gygi, S. P., Harper, J. W., Kimmelman, A. C. (2014). Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109. doi: 10.1038/nature13148
Mitschka, S., Mayr, C. (2022). Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell Biol. 23, 779–796. doi: 10.1038/s41580-022-00507-5
Mo, S., Liu, X., Zhang, K., Wang, W., Cai, Y., Ouyang, Q., et al. (2022). Flunarizine suppresses Mycobacterium tuberculosis growth via calmodulin-dependent phagosome maturation. J. Leukoc. Biol. 111, 1021–1029. doi: 10.1002/JLB.4A0221-119RR
Mukherjee, S., Graber, J. H., Moore, C. L. (2023). Macrophage differentiation is marked by increased abundance of the mRNA 3’ end processing machinery, altered poly(A) site usage, and sensitivity to the level of CstF64. Front. Immunol. 14, 1091403. doi: 10.3389/fimmu.2023.1091403
Ni, S., Yuan, Y., Kuang, Y., Li, X. (2022). Iron metabolism and immune regulation. Front. Immunol. 13, 816282. doi: 10.3389/fimmu.2022.816282
Pham, C. G., Bubici, C., Zazzeroni, F., Papa, S., Jones, J., Alvarez, K., et al. (2004). Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell 119, 529–542. doi: 10.1016/j.cell.2004.10.017
Pisu, D., Huang, L., Grenier, J. K., Russell, D. G. (2020). Dual RNA-seq of mtb-infected macrophages in vivo reveals ontologically distinct host-pathogen interactions. Cell Rep. 30, 335–350 e4. doi: 10.1016/j.celrep.2019.12.033
Reddy, V. P., Chinta, K. C., Saini, V., Glasgow, J. N., Hull, T. D., Traylor, A., et al. (2018). Ferritin H deficiency in myeloid compartments dysregulates host energy metabolism and increases susceptibility to mycobacterium tuberculosis infection. Front. Immunol. 9, 860. doi: 10.3389/fimmu.2018.00860
Rogers, J. T. (1996). Ferritin translation by interleukin-1and interleukin-6: the role of sequences upstream of the start codons of the heavy and light subunit genes. Blood 87, 2525–2537. doi: 10.1182/blood.V87.6.2525.bloodjournal8762525
Rouault, T. A. (2006). The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2, 406–414. doi: 10.1038/nchembio807
Soares, M. P., Hamza, I. (2016). Macrophages and iron metabolism. Immunity 44, 492–504. doi: 10.1016/j.immuni.2016.02.016
Tian, B., Hu, J., Zhang, H., Lutz, C. S. (2005). A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212. doi: 10.1093/nar/gki158
Torti, S. V., Kwak, E. L., Miller, S. C., Miller, L. L., Ringold, G. M., Myambo, K. B., et al. (1988). The molecular cloning and characterization of murine ferritin heavy chain, a tumor necrosis factor-inducible gene. J. Biol. Chem. 263, 12638–12644. doi: 10.1016/S0021-9258(18)37801-3
Torti, F. M., Torti, S. V. (2002). Regulation of ferritin genes and protein. Blood 99, 3505–3516. doi: 10.1182/blood.V99.10.3505
Wei, Y., Miller, S. C., Tsuji, Y., Torti, S. V., Torti, F. M. (1990). Interleukin 1 induces ferritin heavy chain in human muscle cells. Biochem. Biophys. Res. Commun. 169, 289–296. doi: 10.1016/0006-291X(90)91466-6
Weiss, G., Schaible, U. E. (2015). Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 264, 182–203. doi: 10.1111/imr.2015.264.issue-1
World Health Organization. (2024). Global tuberculosis report 2024. Geneva: World Health Organization.
Wilkinson, N., Pantopoulos, K. (2014). The IRP/IRE system in vivo: insights from mouse models. Front. Pharmacol. 5, 176. doi: 10.3389/fphar.2014.00176
Yang, Q., Coseno, M., Gilmartin, G. M., Doublié, S. (2011). Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure. 19 (3), 368–77. doi: 10.1016/j.str.2010.12.021
Yang, Q., Gilmartin, G. M., Doublie, S. (2010). Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3’ processing. Proc. Natl. Acad. Sci. U.S.A. 107, 10062–10067. doi: 10.1073/pnas.1000848107
Keywords: Mycobacterium tuberculosis, alternative polyadenylation, FTH1, macrophage, NUDT21
Citation: Liu X, Cai N, Dai Y, Chen X and Zeng X (2025) Mycobacterium tuberculosis modulates NUDT21-mediated alternative polyadenylation to enhance FTH1 expression in macrophages and promotes intracellular growth. Front. Cell. Infect. Microbiol. 15:1578163. doi: 10.3389/fcimb.2025.1578163
Received: 17 February 2025; Accepted: 07 April 2025;
Published: 02 May 2025.
Edited by:
Paige Allen, Virginia Commonwealth University, United StatesReviewed by:
Avishek Mitra, Oklahoma State University, United StatesAndrea J. Wolf, Cedars Sinai Medical Center, United States
Copyright © 2025 Liu, Cai, Dai, Chen and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaobin Zeng, emVuZy54aWFvYmluQHN6aG9zcGl0YWwuY29t; Xinchun Chen, Y2hlbnhpbmNodW5Ac3p1LmVkdS5jbg==
 Xiaoqian Liu
Xiaoqian Liu Ningjian Cai
Ningjian Cai Youchao Dai3
Youchao Dai3 Xinchun Chen
Xinchun Chen Xiaobin Zeng
Xiaobin Zeng