- 1Department of Veterinary Medicine, College of Agriculture and Veterinary Medicine, United Arab of Emirates University, Al Ain, United Arab Emirates
- 2ASPIRE Research Institute for Food Security in the Drylands (ARIFSID), United Arab Emirates University, Al Ain, United Arab Emirates
Campylobacter spp., primarily C. jejuni and C. coli, are leading causes of bacterial gastroenteritis worldwide. This review provides an overview of literature on the prevalence and distribution of virulence genes in C. jejuni and C. coli isolated from both food samples and humans across GCC countries. The reviewed evidence highlights a gap in our understanding of how differences in the virulence profile affect the pathogenicity of Campylobacter. Research has shown that C. coli is the predominant species found in retail chicken carcasses in the UAE, while C. jejuni is more common in chicken carcasses across other GCC countries. Studies also reveal distinct genotypes of C. jejuni and C. coli, each with varying pathogenicity. These findings underscore the need for further research on the role of virulence genes in shaping the pathogenicity of Campylobacter, which is essential for developing effective intervention and control strategies in the GCC region.
Introduction
Foodborne pathogens remain an emerging threat to food safety and trade at the international level. Globally in Europe and various other places, foodborne infections are a significant burden to the health of humans (EFSA, 2022). Research from the European Centre for Disease Prevention and Control (ECDC) and the Zoonoses Reports of the European Food Safety Authority (EFSA) shows that cases of campylobacteriosis affecting humans, especially through C. jejuni have from time to time in the past decade been higher than those of salmonellosis (Jay-Russell et al., 2013; EFSA, 2019). In the United States of America (USA), the leading microorganisms identified to cause food-borne diseases are C. jejuni, Listeria, and Salmonella (Mohamed et al., 2024, 2025; Mohamed and Habib, 2025). Both Foodborne campylobacteriosis and salmonellosis as monitored by the Foodborne Diseases Active Surveillance Network (FoodNet) of the Centers for Disease Control and Prevention (CDC) have been trending upward in terms of hospitalizations (Tack et al., 2020).
Campylobacter spp., microaerophilic bacteria, thrive in oxygen concentrations of 5% to 10%, along with 10% carbon dioxide (El-Naenaeey et al., 2020; Mohamed et al., 2022). Gram negative with a spiral shaped, curved morphology, they are motile rods with flagella, typically existing as commensals in mammal and bird intestines (Martora et al., 2020). Slow growing, they require specialized growth media with charcoal and antibiotics like cephalothin to suppress competing microorganisms in fecal samples. C. jejuni and C. coli, known for thermotolerance, grow optimally at 42°C (Zhang et al., 2021). However, not all Campylobacter spp. share the same thermotolerance; C. fetus lacks this characteristic (Ibrahim et al., 2018; Baali et al., 2020). Stool cultures for human diarrhea cases follow standard practices at 42°C, using cephalothin containing media to recover C. jejuni and C. coli, although this may not suit other species like C. fetus or C. upsaliensis, which cause illness less frequently (Senok et al., 2007; Tack et al., 2020; Pires and Devleesschauwer, 2021).
The term “virulence genes” generally refers to genes in microorganisms, such as bacteria, that play a role in disease causation and adaptation to stress conditions (Mohamed et al., 2025). These genes are usually related to the pathogenicity of the microorganism or its capacity to exist and create illnesses in the host organism (Mohamed and Habib, 2023). In the case of bacteria such as Campylobacter spp., virulence genes that might be affected by virulence stress may be those related to the process through which the bacteria attaches itself to host cells, enters tissues, and avoids being neutralized by the host’s immune system. Additionally, these genes might also be responsible for the microorganism’s acclimatization to stressful conditions including changes in temperature, pH level, and anything else that may be encountered during the infection process or elsewhere outside the host (Lopes et al., 2021). Examination of virulence stress genes is useful for the knowledge of how bacteria become pathogenic and adapt to existence in various settings. Knowledge of such genes helps in understanding the pathogenicity of the microorganism and may be useful in the search for desirable treatments or protective measures against the disease.
Virulence genes such as cadF and flpA fibronectin-binding proteins help bacteria in uptake to the epithelial cells of the small intestine which is essential for colonization and infection. These proteins help to fix bacteria on the host cell so that they cannot be shed off the gastrointestinal tract (Jakee et al., 2015; Talukdar et al., 2020). ciaB belongs to a Campylobacter invasion antigen (Cia) protein subfamily that is involved in the invasion of epithelial cells. To breach the intestinal layer and disease as well as circulate in the body invasion is crucial to bacteria (Gharbi et al., 2022). CDT genes of cdtA, cdtB, and cdtC produce subunits that act on the host cell cycle arrest, and apoptosis resulting in cell death and tissue injury that is connected with gastroenteritis manifestations such as diarrhea and inflammation (Nahar and Rashid, 2018). The flaA and flaB genes are involved in the synthesis of motor flagellin proteins which are required for the bacterial motility to negotiate the mucous layers of GIT for effective colonization and dissemination (Eryildiz et al., 2020). Another stress response gene is htrA, a serine protease which involved in the cleavage of the misfolded proteins and proteins that have been exposed to heat shock and oxidative stress within the host to assist bacteria in surviving within the host (Tegtmeyer et al., 2021). The clpB gene encodes a molecular chaperone that disaggregates and refolds stress-damaged proteins, aiding in survival under heat and osmotic stress and maintaining various cellular functions during infection (Alam et al., 2021). The sodB gene codes for an enzyme that eliminates or reduces oxidation, and enables the bacteria to withstand the attack of reactive oxygen species from the host. Abnormally, the katA gene codes for the enzyme catalase that is responsible for the degradation of hydrogen peroxide to water and oxygen which allows bacteria to combat the host’s immune system (Koolman et al., 2015).
Much as the virulence genes such as cadF and ciaB are responsible for the ability of Campylobacter to adhere and invade host cells and induce a strong immune response, they also instigate the necessity of stress response genes such as htrA and sodB for bacterial survival (Koolman et al., 2015; Nahar and Rashid, 2018). These stress response mechanisms, however, are not only protective of the bacteria and its metabolic processes in stressful conditions, but also toxic to the host since they suppress inflammation in the site of infection that would otherwise hinder bacterial colonization. Campylobacter deploys a multitude of proteins and stress response genes which help it to invade the host, avoid the host’s immune responses, and survive within the hostile milieu of the host organism. The site-specific regulation and interdependence of these genes raise the stakes of Campylobacter’s pathogenic and survival mechanisms (Talukdar et al., 2020).
In this study, we aim to elucidate background knowledge and recent updates, based on published research, on the virulence genes of C. jejuni and C. coli in the GCC countries.
Materials and methods
The study utilized a narrative review of the literature, where each included study serves as the unit of analysis, forming a collective database (Byrne, 2016). Authors extract characteristics from each study, such as the source of Campylobacter, identifying C. jejuni and C. coli as the primary species involved, and virulence factors. To ensure comprehensive coverage, no time frame restrictions were applied to the articles. Searches were conducted on platforms such as PubMed and Google Scholar. Keywords for each country were employed to refine the search “Campylobacter virulence genes in Bahrain”.
Virulence factors in the GCC countries
Campylobacteriosis is considered a significant public health concern, particularly during the summer (Mohamed, 2024). Individuals who work with animals or animal products, such as veterinarians, farmers, poultry processors, slaughterhouse workers, and butchers, face a high risk of infection due to their professions (Habib and Mohamed, 2022). Despite its potential impact on public health, there is a lack of published research on Campylobacter infections in humans within the GCC over the last 20 years, highlighting the need for further studies to address this gap.
Table 1 summarizes the reports of the virulence factors of C. jejuni and C. coli samples isolated from both foods and humans in GCC countries. One key virulence factor in Campylobacter enteritis is the cytolethal distending toxin (CDT), which damages DNA, causing cell cycle arrest and cell death. Inflammation, neutrophil infiltration, and bacteremia suggest invasion as a significant virulence determinant for Campylobacter spp (Wooldridge and Ketley, 1997). Our prior research in the United Arab Emirates found the CDT (governed by the CDT A, B, and C operon) in 88.8% of chicken samples from Abu Dhabi supermarkets, suggesting its potential contribution to the invasion process (Habib et al., 2023). In this study we gathered 45 isolates from chicken meats, employing the Whole-Genome Sequencing technique, each C. coli isolate identified harbored 7 to 11 virulence factors. Of those factors, some are present in all strains studied and connected with adherence (cadF and jlpA), colonization, immune avoidance (capsule synthesis and transport, lipooligosaccharide), and invasion (ciaB). This research report is the first report that features the characterization of C. jejuni and C. coli virulence from retail chicken in the UAE. These results prompted us to conduct additional research (Habib et al., 2024) to study C. jejuni and C. coli prevalence and virulence profiles during the meat processing operations in UAE abattoirs. Campylobacter showed high levels of prevalence throughout the entire poultry processing sequence starting from post-defeathering and continuing through evisceration until final chilling. The whole genome sequencing (WGS) showed major distinctions in virulence factor patterns between the two strains of C. coli and C. jejuni. The virulence factors O-linked protein glycosylation (pglG), pseudaminic acid biosynthesis (pseD and pseI), and flagellin structure (flaA and flaB) together with cytolethal distending toxin (cdtA and cdtC) were found at higher rates in C. jejuni compared to C. coli and this difference reached statistical significance (p < 0.05) through logistic regression analysis. All isolates lacked plasmid-mediated virulence genes and their associated virB-D genes encoding the Type IV secretion system and related virB gene clusters were not detected. Similarly, the motility accessory factor (maf) family genes responsible for flagellar biosynthesis and phase variation were missing from all examined isolates.
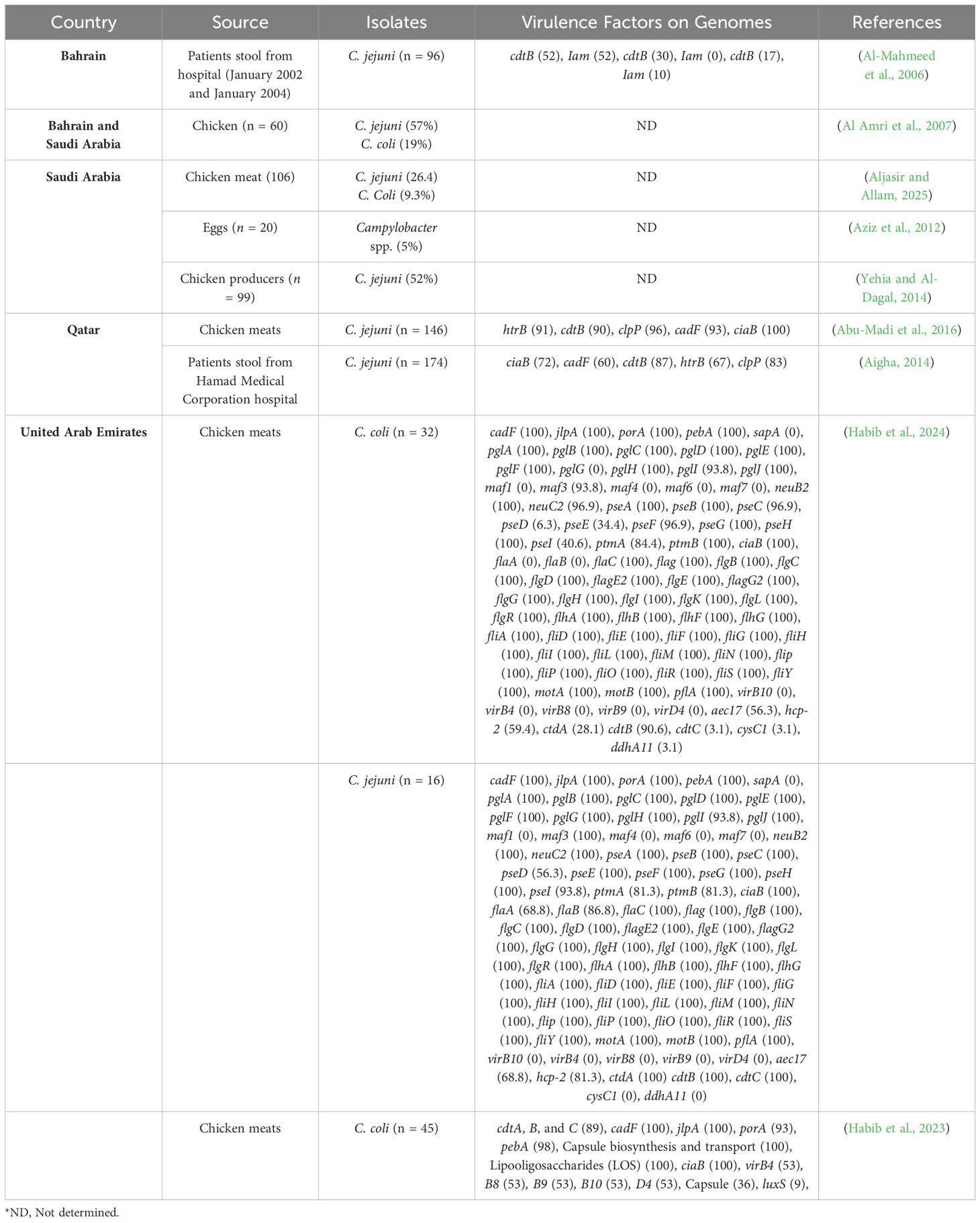
Table 1. Prevalence of virulence factors in C. jejuni and C. coli found in food samples and humans across GCC countries.
The study by Abu-Madi, (Abu-Madi et al., 2016) conducted in Qatar focused on Campylobacter spp. isolates obtained from supermarkets, including those from Saudi and Qatari producers. The research aimed to assess the presence of virulence stress genes, specifically htrB, cdtB, clpP, cadF, and ciaB. These findings shed light on the multiple virulence factors associated with adherence, invasion capabilities, and toxin production, all crucial for Campylobacter spp.’s ability to colonize and cause disease. The examination of these selected virulence genes in this study holds significant implications for understanding disease pathogenesis. The high prevalence of these genes among the isolates may be attributed to their importance in aiding C. jejuni survival in the high temperatures experienced during the summer months in the GCC countries. In another study in Qatar by Aigha (Aigha, 2014), a relatively significant number of C. jejuni isolates, showing the presence of the main virulence and stress response genes in isolates collected from Hamad Medical Corporation hospital.
A molecular investigation targeting the CDT (cdtB) and invasion-associated marker (iam) genes was conducted on C. jejuni strains isolated from stool samples obtained between January 2002 and January 2004 in Bahrain. This molecular characterization aimed to establish correlations with socio-demographic and clinical parameters of the patients. Out of the 96 tested C. jejuni strains, 50 (52%) exhibited the presence of both cdtB and iam genes, 30 (31%) showed the cdtB gene presence but not the iam gene, and 16 (17%) lacked both cdtB and iam genes. Notably, a significant majority of patients (66 out of 96, constituting 69%) were under the age of 3 years, with markedly higher detection rates of cdtB+/iam+ and cdtB+/iam− strains in this particular age group. Strains lacking both virulence genes (cdtB and iam) led to symptomatic infections in this age group, in stark contrast to older patients who experienced asymptomatic infections when infected with these strains (Al-Mahmeed et al., 2006).
(Senok and Botta, 2009) conducted a study delving into the molecular characterization of C. jejuni isolates from Bahrain. Their research presented data that illuminated the link between the presence of combinations of potential virulence genes, the invasive behavior of the bacteria, and the severity of clinical infections.
These studies reflect valuable research work in molecular identification of virulence genes in C. jejuni and C. coli strains isolated from the GCC countries. They reveal different genotypes in C. jejuni and C. coli that have different pathogenicity (Senok and Botta, 2009; Aigha, 2014; Habib et al., 2023). Strains positive for cia, iam, and cdtB genes are found to be more invasive, especially in severe infections. The ciaB gene is associated with increased invasiveness and the severity of the infection. In strains devoid of all three genes, the in vitro invasiveness remains high, which points toward the presence of other unidentified virulence factors (Johansson et al., 2021). Campylobacter enteritis affects children below the age of three years (Asuming-Bediako et al., 2019). emphasizing how multiple virulence factors are involved in Campylobacter infections in the GCC countries.
Conclusions
The review demonstrates the intricate virulence mechanisms of C. jejuni and C. coli in GCC nations with a focus on CDT as the major toxin that causes cellular destruction while triggering inflammation and promoting cell invasiveness. A high occurrence of CDT-associated genes was identified in retail chicken samples from the UAE research which might serve to advance C. jejuni and C. coli pathogenicity. The virulence determinants found in C. jejuni through WGS analysis surpass those of C. coli by showing higher numbers of attachment, colonization, and immune-escape factors while displaying distinct flagellar differences glycosylation patterns, and toxin production levels. Research in Qatar and Bahrain establishes that disease severity heavily relies on virulence genes including cdtB, iam, ciaB, htrB, and cadF. Analyzing C. jejuni and C. coli isolates from chicken meat reveals significant prevalence rates of these genes which indicates their important position as infection reservoirs for human beings. The pathogenic bacteria are transmitted across different points of the meat supply chain during processing and retail distribution. However, the persistence of invasiveness in strains lacking well-characterized virulence factors indicates the involvement of additional, yet unidentified, pathogenic mechanisms. Collectively, these findings highlight the need for continued molecular surveillance and in-depth investigations into the genetic diversity of Campylobacter across the GCC region. Knowledge about virulence mechanisms will help develop specific preventive measures to diminish foodborne Campylobacter infections across the GCC.
Author contributions
M-YM: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. HK: Project administration, Supervision, Writing – review & editing. IH: Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The author(s) declare that the publication fee was covered by the Research and Sponsored Projects Office, United Arab Emirates University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abu-Madi, M., Behnke, J. M., Sharma, A., Bearden, R., Al-Banna, N. (2016). Prevalence of virulence/stress genes in Campylobacter jejuni from chicken meat sold in Qatari retail outlets. PLoS One 11. doi: 10.1371/journal.pone.0156938
Aigha, I. I. (2014). Molecular analysis of Campylobacter jejuni virulence genes from clinical isolates in Qatar. (Thesis). Qatar University.
Alam, A., Bröms, J. E., Kumar, R., Sjöstedt, A. (2021). The role of ClpB in bacterial stress responses and virulence. Front. Mol. Biosci 8. doi: 10.3389/fmolb.2021.668910
Al Amri, A., Senok, A. C., Ismaeel, A. Y., Al-Mahmeed, A. E., Botta, G. A. (2007). Multiplex PCR for direct identification of Campylobacter spp. in human and chicken stools. J. Med. Microbiol 56, 1350–1355. doi: 10.1099/jmm.0.47220-0
Aljasir, S. F., Allam, S. A. (2025). Prevalence and antibiotic resistance of Salmonella spp. and Campylobacter spp. isolated from retail chickens in Saudi Arabia. Microbiol Res. (Pavia) 16, 27. doi: 10.3390/microbiolres16010027
Al-Mahmeed, A., Senok, A. C., Ismaeel, A. Y., Bindayna, K. M., Tabbara, K. S., Botta, G. A. (2006). Clinical relevance of virulence genes in Campylobacter jejuni isolates in Bahrain. J. Med. Microbiol 55, 839–843. doi: 10.1099/jmm.0.46500-0
Asuming-Bediako, N., Kunadu, A. P. H., Abraham, S., Habib, I. (2019). Campylobacter at the human–food interface: the african perspective. Pathogens 8. doi: 10.3390/pathogens8020087
Aziz, A., Bahobail, S., Hassan, S. A., El-Deeb, B. A. (2012). Microbial quality and content aflatoxins of commercially available eggs in Taif, Saudi Arabia. Afr J. Microbiol Res. 6, 3337–3342. doi: 10.5897/AJMR12.229
Baali, M., Lounis, M., Al Amir, H. L., Ayachi, A., Hakem, A., Kassah-Laouar, A. (2020). Prevalence, seasonality, and antimicrobial resistance of thermotolerant Campylobacter isolated from broiler farms and slaughterhouses in East Algeria. Vet World 13, 1221–1228. doi: 10.14202/vetworld.2020.1221-1228
Byrne, J. A. (2016). Improving the peer review of narrative literature reviews. Res. Integr. Peer Rev. 1, 12. doi: 10.1186/s41073-016-0019-2
EFSA (2019). The european union one health 2018 zoonoses report. EFSA J. 17, 14–19. doi: 10.2903/j.efsa.2019.5926
EFSA (2022). The european union one health 2021 zoonoses report. EFSA J. 20, 22–35. doi: 10.2903/j.efsa.2022.7666
El-Naenaeey, E., Abd El-Hamid, M., Khalifa, E. (2020). Foodborne campylobacter species: taxonomy, isolation, virulence attributes and antimicrobial resistance. Zagazig Vet J. 48, 414–432. doi: 10.21608/zvjz.2020.40250.1118
Eryildiz, C., Tabakcioglu, K., Kuyucuklu, G., Sakru, N. (2020). Investigation of antimicrobial resistance and virulence genes of Campylobacter isolates from patients in a tertiary hospital in Edirne, Turkey. Indian J. Med. Microbiol 38, 157–161. doi: 10.4103/ijmm.IJMM_20_78
Gharbi, M., Béjaoui, A., Ben Hamda, C., Ghedira, K., Ghram, A., Maaroufi, A. (2022). Distribution of virulence and antibiotic resistance genes in Campylobacter jejuni and Campylobacter coli isolated from broiler chickens in Tunisia. J. Microbiology Immunol. Infection 55, 1273–1282. doi: 10.1016/j.jmii.2021.07.001
Habib, I., Ibrahim Mohamed, M. Y., Ghazawi, A., Lakshmi, G. B., Khan, M., Li, D., et al. (2023). Genomic characterization of molecular markers associated with antimicrobial resistance and virulence of the prevalent Campylobacter coli isolated from retail chicken meat in the United Arab Emirates. Curr. Res. Food Sci. 6. doi: 10.1016/j.crfs.2023.100434
Habib, I., Mohamed, M. Y. I. (2022). “Foodborne infections in the middle east,” in Food Safety in the Middle East (Elsevier), 71–107. doi: 10.1016/B978-0-12-822417-5.00005-2
Habib, I., Mohamed, M.-Y. I., Lakshmi, G. B., Al Marzooqi, H. M., Afifi, H. S., Shehata, M. G., et al. (2024). Quantitative assessment and genomic profiling of Campylobacter dynamics in poultry processing: a case study in the United Arab Emirates integrated abattoir system. Front. Microbiol 15. doi: 10.3389/fmicb.2024.1439424
Ibrahim, M. J., Abdul-Aziz, S., Bitrus, A. A., Mohammed, D. G., Abu, J., Bejo, S. K., et al. (2018). Occurrence of multidrug resistant (MDR) Campylobacter species isolated from retail chicken meats in Selangor, Malaysia and their associated risk factors. Malays J. Microbiol 14, 261–272.
Jakee, J.--., Ata, N. S., Hakim, A. S., Syame, S. M., Omara, S. T. (2015). Prevalence of virulence genes and antimicrobial resistance patterns of Campylobacter species isolated from chicken in Egypt. Asian J. Poultry Sci. 9, 250–261. doi: 10.3923/ajpsaj.2015.250.261
Jay-Russell, M. T., Mandrell, R. E., Yuan, J., Bates, A., Manalac, R., Mohle-Boetani, J., et al. (2013). Using major outer membrane protein typing as an epidemiological tool to investigate outbreaks caused by milk-borne Campylobacter jejuni isolates in California. J. Clin. Microbiol 51, 195–201. doi: 10.1128/JCM.01845-12
Johansson, C., Kampmann, C., Nilsson, A., Dicksved, J., Engstrand, L., Rautelin, H. (2021). Genomic and phenotypic characteristics in geographically separated clinical Campylobacter jejuni ST 353 CC isolates. Microorganisms 9. doi: 10.3390/microorganisms9122540
Koolman, L., Whyte, P., Burgess, C., Bolton, D. (2015). Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog Dis. 12, 424–432. doi: 10.1089/fpd.2014.1883
Lopes, G. V., Ramires, T., Kleinubing, N. R., Scheik, L. K., Fiorentini, Â.M., Padilha da Silva, W. (2021). Virulence factors of foodborne pathogen Campylobacter jejuni. Microb Pathog 161, 105265. doi: 10.1016/j.micpath.2021.105265
Martora, F., Pagliuca, C., Della Pepa, M. E., Teresa, M., Rocca, D., Curto, S., et al. (2020). Campylobacter jejuni bacteremia in Italian pediatric patients with acute lymphoblastic leukemia: Report of two cases. New Microbiol 43, 1121–7138.
Mohamed, M.-Y. I. (2024). Campylobacteriosis in North Africa. AIMS Agric. Food 9, 801–821. doi: 10.3934/agrfood.2024043
Mohamed, M.-Y. I., Abu, J., Aziz, S. A., Zakaria, Z., Khan, A. R., Habib, I. (2022). Occurrence of antibiotic resistant C. jejuni and E. coli in wild birds, chickens, humans, and the environment in Malay villages, Kedah, Malaysia. Vet Med. (Praha) 67, 298–308. doi: 10.17221/102/2021-VETMED
Mohamed, M.-Y. I., Habib, I. (2023). Pathogenic E. coli in the Food Chain across the Arab Countries: A Descriptive Review. Foods 12, 3726. doi: 10.3390/foods12203726
Mohamed, M.-Y. I., Habib, I. (2025). Listeria monocytogenes in food products, and its virulence in North Africa. AIMS Agric. Food 10, 97–128. doi: 10.3934/agrfood.2025006
Mohamed, M.-Y. I., Habib, I., Khalifa, H. O. (2024). Salmonella in the food chain within the Gulf Cooperation Council countries. AIMS Microbiol 10, 468–488. doi: 10.3934/microbiol.2024023
Mohamed, M.-Y. I., Khalifa, H. O., Habib, I. (2025). Food pathways of Salmonella and Its ability to cause gastroenteritis in North Africa. Foods 14, 253. doi: 10.3390/foods14020253
Nahar, N., Rashid, R. B. (2018). Genotypic analysis of the virulence and antibiotic resistance genes in Campylobacter species in silico. J. Bioanal BioMed. 10, 13–23. doi: 10.4172/1948-593X.1000199
Pires, S. M., Devleesschauwer, B. (2021). “Estimates of global disease burden associated with foodborne pathogens,” in Foodborne Infections and Intoxications (Elsevier), 3–17. doi: 10.1016/B978-0-12-819519-2.00020-7
Senok, A. C., Botta, G. A. (2009). Campylobacter enteritis in the arabian gulf. J. Infect. Developing Countries 3, 74–82. doi: 10.3855/jidc.52
Senok, A., Yousif, A., Mazi, W., Sharaf, E., Bindayna, K., Elnima, E.-A., et al. (2007). Pattern of antibiotic susceptibility in Campylobacter jejuni isolates of human and poultry origin. Jpn. J. Infect. Dis. 60, 1–4. doi: 10.7883/yoken.JJID.2007.1
Tack, D. M., Ray, L., Griffin, P. M., Cieslak, P. R., Dunn, J., Rissman, T., et al. (2020). Preliminary incidence and trends of infections with pathogens transmitted commonly through food — foodborne diseases active surveillance network, 10 U.S. Sites 2016–2019. MMWR Morb Mortal Wkly Rep. 69, 509–514. Available at: https://www.cdc.gov/mmwr/mmwr_continuingEducation.html (Accessed December 22, 2024).
Talukdar, P. K., Negretti, N. M., Turner, K. L., Konkel, M. E. (2020). Molecular dissection of the Campylobacter jejuni CadF and FlpA virulence proteins in binding to host cell fibronectin. Microorganisms 8, 389. doi: 10.3390/microorganisms8030389
Tegtmeyer, N., Sharafutdinov, I., Harrer, A., Soltan Esmaeili, D., Linz, B., Backert, S. (2021). Chapter Campylobacter virulence factors and molecular host-pathogen interactions, in Fighting Campylobacter Infections. Curr Top. Microbiol. Immunol. 169–202. doi: 10.1007/978-3-030-65481-8_7
Wooldridge, K. G., Ketley, J. M. (1997). Campylobacter-host cell interactions. Trends Microbiol 5, 96–102. doi: 10.1016/S0966-842X(97)01004-4
Yehia, H. M., Al-Dagal, M. (2014). Prevalence of Campylobacter jejuni in chicken produced by major poultry companies in Saudi Arabia. Int. J. Food Contam 1. doi: 10.1186/s40550-014-0002-y
Keywords: Campylobacter, virulence genes, foodborne infection, one health, Gulf Cooperation Council countries
Citation: Mohamed M-YI, Khalifa HO and Habib I (2025) The role of virulence genes in Campylobacter pathogenicity: a perspective from the Gulf Cooperation Council countries. Front. Cell. Infect. Microbiol. 15:1584835. doi: 10.3389/fcimb.2025.1584835
Received: 27 February 2025; Accepted: 02 April 2025;
Published: 25 April 2025.
Edited by:
Valerie J. Carabetta, Cooper Medical School of Rowan University, United StatesReviewed by:
Muhammad Akbar Shahid, Bahauddin Zakariya University, PakistanCopyright © 2025 Mohamed, Khalifa and Habib. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohamed-Yousif Ibrahim Mohamed, bW9oYW1lZC15b3VzaWYtaUB1YWV1LmFjLmFl
†ORCID: Mohamed-Yousif Ibrahim Mohamed, orcid.org/0000-0002-9563-4393
Hazim O. Khalifa, orcid.org/0000-0001-9861-9693
Ihab Habib, orcid.org/0000-0002-0938-0607
 Mohamed-Yousif Ibrahim Mohamed
Mohamed-Yousif Ibrahim Mohamed Hazim O. Khalifa1†
Hazim O. Khalifa1†