- 1Dipartimento di Scienze Biotecnologiche di Base, Cliniche Intensivologiche e Perioperatorie, Università Cattolica del Sacro Cuore, Rome, Italy
- 2Dipartimento di Scienze di Laboratorio ed Ematologiche, Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy
- 3Mater Olbia Hospital, Olbia, Italy
Lactobacillus crispatus is renowned for its antimicrobial properties, and some strains are used to treat vaginal dysbiosis, although the mechanisms underlying the antimicrobial properties remain elusive. We isolated L. crispatus M247 (LcM247) from a commercially available probiotic product Crispact® and tested its antimicrobial activity against selected pathobionts such as Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, Streptococcus agalactiae, Enterococcus faecalis and Candida albicans using both cocultures and testing the antimicrobial activity of cell-free supernatant (CFS) obtained from the culture of the probiotic strain. Furthermore, we demonstrate that CFS antimicrobial activity is pH dependent and that it is not affected by temperature and proteinase K treatment. Proteomic analysis suggests that this activity is mediated by S-layer secreted proteins. In a series of in vitro infection models, we infected Henrietta Lacks’ cervical eukaryotic cancer cells (HeLa) with E. coli, S. agalactiae and C. albicans at specific multiplicities of infection (MOIs) before the administration of LcM247, CFS, gentamicin or fluconazole alone or in combination with LcM247/CFS. We observed a slight decrease in the microbial burden following LcM247 administration, while treatment with CFS significantly reduced microbial growth compared to control and antimicrobial compounds. These results highlight the antimicrobial properties of LcM247 and its CFS and the likely mechanism of action that contributes to the eradication of common pathobionts. We show that actively replicating LcM247 is less efficient than its CFS, so the oral administration of LcM247 may result in treatment failure. Finally, the use of CFS may result in an upswing of the host Lactobacillus strains and promote the engraftment of Lactobacillus probiotic treatments.
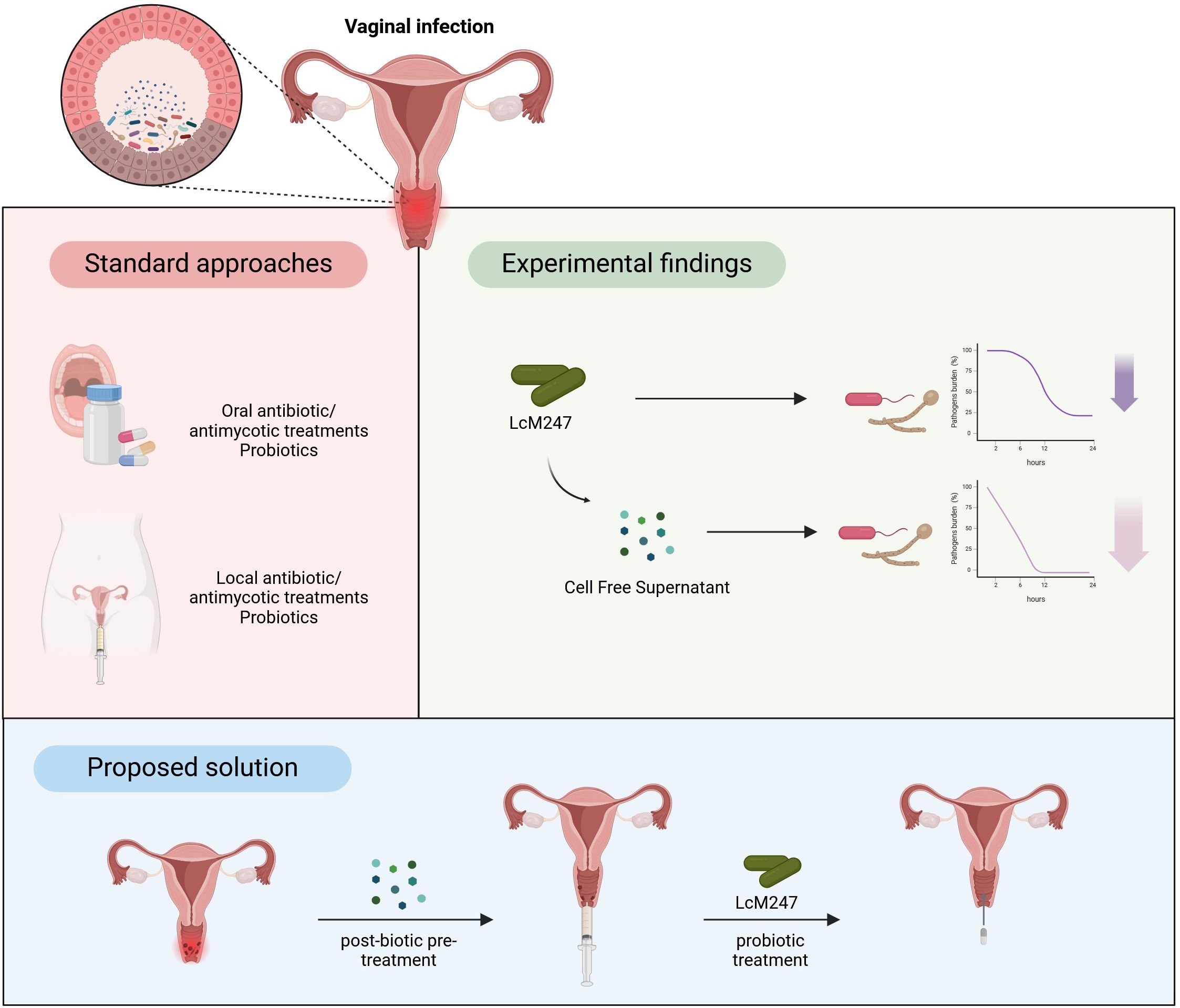
Graphical Abstract. The figure presents a concise three-part model. Firstly, it illustrates the standard approaches commonly used to treat vaginal infections, such as the systemic (oral) or localized administration of antibiotics, antifungal and probiotics. In the subsequent section, the experimental findings panel provides a summary of the in vitro data obtained, which indicated that both live Lactobacillus crispatus M247 (LcM247) and its cell-free supernatant exhibit the capacity to reduce the viability of urogenital pathogens over 24 hours. Notably, the cell-free supernatant (CFS) was found to be more efficacious in causing a more rapid decline in viability. In conclusion, the proposed solution delineates a pioneering two-step therapeutic regime: initially, an intravaginal pre-treatment with post-biotic metabolites (i.e. CFS) should be administered to modify the mucosal environment and reduce pathogen adhesion; subsequent administration of live LcM247 cells would then re-establish and maintain a protective, Lactobacillus-dominated flora.
Introduction
The vagina hosts a complex micro-ecosystem with billions of microorganisms that maintain a symbiotic and mutualistic relationship with the human host that preserve tissue homeostasis (Chen et al., 2021): the host provides a moist, rich in nutrients and warm habitat for several microbes that produce antimicrobial and anti-inflammatory compounds, which provide a first line of defence against harmful pathobionts (Joseph et al., 2021). This microbial balance can be disrupted by internal and/or external factors such as hormonal changes, age, immune system status, infections and antibiotic use, opening the possibility for opportunistic infections (Kumar et al., 2016; Shen et al., 2022). A classic example is bacterial vaginosis (BV), a condition marked by the disruption of the normal vaginal microbiota (Khedkar and Pajai, 2022), characterized by a decrease in lactobacilli and an overgrowth of anaerobic bacteria, including Gardnerella vaginalis and Atopobium vaginae (Abou Chacra et al., 2021; Abbe and Mitchell, 2023). BV potentially increases the risk of sexually transmitted infections (STIs) (van Houdt et al., 2018), pelvic inflammatory disease (PID) (Wiesenfeld et al., 2002; Ravel et al., 2021) and complications during pregnancy (Murphy and Mitchell, 2016; Feehily et al., 2020; Ravel et al., 2021; Masucci et al., 2023). On the contrary, vulvovaginal candidiasis (VVC), a yeast infection, is caused by an overgrowth of Candida species, particularly Candida albicans (Sobel, 2007; Nyirjesy et al., 2022), but unlike BV, it is not typically associated with an imbalance in the bacterial flora.
Under normal conditions, lactobacilli account for 70%–90% of the vaginal bacterial species associated with healthy premenopausal women (Hugenholtz et al., 2022), or they are positively linked to estrogen levels and contraceptive use and negatively linked to childbirth and breastfeeding (Lebeer et al., 2023); they are crucial to vaginal health because of their ability to produce antimicrobial molecules and lactic acid, which maintain the vaginal mucosa at an acidic pH (≈ 4.5) making the environment inhospitable for most microbes (O’Hanlon et al., 2013; France et al., 2020; Lebeer et al., 2023). However, the prevalence and abundance of Lactobacillus species in the vaginal microbiota are subject to a considerable interindividual variability (Greenbaum et al., 2019), although five distinct groups or cluster of vaginal microbial communities have been proposed and named as vaginal community state types (vCSTs) (Smith and Ravel, 2017; France et al., 2020). vCST I, II, III and V are characterized by the high abundance of Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus iners and Lactobacillus jensenii, respectively (Ma and Li, 2017), whereas vCST IV generally reflects a paucity of lactobacilli and an overgrowth of diverse anaerobes, typically associated with symptomatic BV (Norenhag et al., 2020; France et al., 2022). However, vCST IV does not always correspond to symptomatic BV; several studies, particularly in non-Western cohorts, have documented women harboring this diverse community but remain asymptomatic (Fettweis et al., 2014). Moreover, not all Lactobacillus-dominated CSTs confer the same health-promoting properties, with the vCST I showing a stronger association with vaginal healthy status compared to other vCST, suggesting that L. crispatus can be considered a microbial biomarker of a healthy vaginal microbiota (Lepargneur, 2016).
L. crispatus contributes to the vaginal environment by producing both the l-lactic and d-lactic acid isomers (Kaewsrichan et al., 2006; Borges et al., 2014) and synthesizing antimicrobial compounds, such as bacteriocins. Although their role in maintaining vaginal homeostasis remains under investigation, current evidence suggests that these antimicrobial peptides have the potential to regulate microbial communities in the vagina and inhibit the growth of competing microorganisms supporting the maintenance of a balanced microbiome (de Jong et al., 2006; Stoyancheva et al., 2014; Fontana et al., 2020). Furthermore, studies have still demonstrated that L. crispatus modulates the secretion of pro-inflammatory cytokines, which typically increase during bacterial vaginosis (Rose et al., 2012).
To date, the exogenous administration of Lactobacillus-based probiotics in treating BV to restore a healthy vaginal microbiome has attracted more attention from researchers (Cohen et al., 2020; Qi et al., 2023). L. crispatus M247 (LcM247) is a commercial probiotic that, following oral administration, colonizes the gut and vaginal environments, prompting the restoration of the physiological vaginal microbial equilibrium.
The aim of this study was to dissect the activity of LcM247 against various infectious agents, in different experimental in vitro models, using a set of potential vaginal pathobionts. Additionally, we studied the potential application of LcM247 supernatant as a natural antimicrobial agent.
Materials and methods
Bacterial cultures and growth conditions
The probiotic strain used in this study was isolated from the commercial product Crispact® (Pharmextracta S.p.A., Piacenza, Italy), formulated in sachets. Each sachet contained not less than 20 billion colony-forming units (CFUs) of LcM247 (IDA: LMG-P-23257). Diluted powder of the sachet was spread on de Man, Rogosa and Sharpe (MRS) agar plates (Merck, Darmstadt, Germany). Colonies were inoculated in MRS broth and incubated for 48 hours at 37°C. Meanwhile, Lactobacillus were taxonomically identified at the species level using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Bruker, Kontich, Belgium) (Biswas and Rolain, 2013; Anderson et al., 2014).
The indicator strains were selected from our microbial bank or isolated from patients’ samples in the Department of Microbiology and Virology of IRCSS Fondazione Policlinico Agostino Gemelli, Rome (Italy). The pathogens Escherichia coli (ATCC 25922), Klebsiella pneumoniae (clinical isolate from urine), Staphylococcus aureus (ATCC 29213), Enterococcus faecalis (ATCC 29212), Streptococcus agalactiae (clinical isolate from vaginal swab) and C. albicans (ATCC 24433) were used as target microorganisms for the determination of antagonistic activity and cultured in Brain Heart Infusion (BHI) broth (Sigma-Aldrich, St. Louis, MO, USA) for the bacterial strain and in Sabouraud Dextrose broth (Sigma-Aldrich) for C. albicans at 37°C.
Each bacterial culture was mixed with pure sterile glycerol to reach a final concentration of 20% (Sigma-Aldrich), gently mixed and stored at −80°C.
To obtain cell-free supernatant (CFS), LcM247 colonies were inoculated in MRS broth and incubated for 48 hours at 37°C (reaching ~5 × 107 CFU/mL) before centrifuging at 4,000 rpm for 10 min; CFS was harvested, filtered using 0.22-µm cellulose acetate membranes and stored at −80°C until use. An aliquot of CFS was plated onto MRS agar and cultured as previously described to assess that it was free of remaining bacteria.
Assessment of the antimicrobial activity of LcM247 and its CFS
The antagonist effect of LcM247 against the selected pathogens was assessed in coculture assays and compared with the antimicrobial activity of its CFS and with the growth ability of each pathogen in MRS broth. The antagonism experiment was performed in a sterile 96-well plate (Corning® Incorporated Life Sciences, NY, Corning, USA), which was mixed with 100 µL of indicator strains at 5 × 105 CFU/mL [it is widely used as the starting concentration for many antimicrobial assays, including those presented in previously published papers (Rosato et al., 2024)] with 100 µL of sterile MRS broth or 100 µL of LcM247 at 5 × 107 CFU/mL [it is the range of LcM247 CFU/mL reached in a 48-hour incubation to obtain CFS and according to (Rajab et al., 2020)] or 100 µL of CFS. The plate was incubated at 37°C under aerobic conditions and analyzed at 4 and 24 hours. The viable microbial cell counts (CFU/mL) of the indicator strains were reported as log10 reduction of the total count of CFU/mL plating on appropriate media (BHI agar, MacConkey agar, Columbia CNA agar and Sabouraud Dextrose agar), confirming strain identification using MALDI-TOF MS (Bruker). The turbidity of the medium in each well was evaluated using a spectrophotometer (Cytation 5, BioTek, Winooski, VT, USA) at λ = 600 nm.
To assess reciprocal Lactobacillus antagonism, 5 × 105 CFU/mL of L. iners (isolated from a clinical vaginal sample) and LcM247 were incubated with serial dilutions of LcM247 CFS and L. iners CFS, respectively. The evaluation of bacterial viability was measured 24 hours post-incubation by counting CFUs as previously described.
Evaluation of the antimicrobial activity of LcM247 CFS modified
To investigate the CFS antimicrobial effect, the supernatant was alkalinized by adding different volumes of 1 M NaOH until four different conditions were achieved: unaltered CFS (pH 4.5), CFS at pH 5.5, CFS at pH 7 and CFS at pH 9. In addition, the supernatant was diluted in MRS broth to obtain four serial dilutions with the following CFS/MRS concentrations: 0.5, 0.25, 0.12 and 0.06. The pH of each dilution was determined using pH indicator paper (pH range 1.0–10.0) (Sigma-Aldrich, USA). 100ul of the indicator strains with an initial concentration of 5 × 105 CFU/mL were incubated with 100 µL of the above-mentioned conditions for 24 hours at 37°C in a 96-well plate with round-bottom wells. Once incubation was completed, bacterial replication or inhibition was assessed by observing the strain deposited at the bottom of the well as a dark button, and CFUs were determined after serial dilutions of the same wells.
Whole-genome sequencing
L. crispatus M247 was grown onto MRS solid medium from frozen stock before being sub-cultured in MRS liquid medium for 48 hours at 37°C (reaching ~5 × 107 CFU/mL) prior to DNA extraction. Highly pure genomic DNA was obtained using DANAGENE Microbial DNA according to the manufacturer’s instructions. A genomic library was prepared using the Illumina DNA Prep Kit (Illumina) and Nextera™ DNA CD Indexes (Illumina). Sequencing was performed using a MiSeq platform (Illumina), generating 250-bp read lengths. FastQ sequences were analyzed using CLC Genomic Workbench v24.0.2 (Qiagen, Valencia, CA, USA) (Nair et al., 2011; Sichtig et al., 2019). Briefly, raw data were quality-checked and then trimmed (using QC for sequencing reads and Trim reads 3.0 plug-in with default parameters, respectively). Trimmed reads were additionally investigated for contamination using Find Best Matches using K-mer Spectra plug-in before de novo assembly using default parameters (minimum contig length of 200 bp). A total of 2.7 million paired-end reads were obtained, while ~2.2 million reads (average of 212 bp) were correctly assembled, obtaining a total of 1,202 contigs (199 > 1 kb, N50 = 11,896, GC 37.5%). Reads mapping to contigs (99.8% to exclude contaminations) showed a mean coverage of 250×, leading to ~2.0-Mb genome length with 100% of the reference genome covered (completeness). A total of 1,064 Coding DNA sequences (CDSs) (69.7%) were annotated with a DIAMOND hit of 1,526 CDS early detections. Genes encoding bacteriocins were detected by submitting the assembly fasta file to the BAGEL4 online software (de Jong et al., 2006; van Heel et al., 2018). Sequencing raw data have been deposited in the National Center for Biotechnology information (NCBI) Sequence Read Archive (BioProject accession number: PRJNA1222580, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1222580).
Enzymatic digestion and mass spectrometry analysis
Protein digestion was performed according to the filter-aided sample preparation (FASP) protocol that combines both purification and digestion (Wisniewski et al., 2009; Distler et al., 2016) of the proteins present in the LcM247 CFS, using MRS broth as a negative control. Briefly, 50 µg of proteins from each sample was reduced (Dithiothreitol (DTT) 8 mM in urea buffer − 8 M urea and 100 mM Tris), alkylated (Indole-3-Acetic Acid (IAA) 50 mM in urea buffer − 8 M urea and 100 mM Tris) and digested by trypsin on filter tubes Microcon® Centrifugal Filter Devices (Merck Millipore Ltd., Cork, Ireland) at a final concentration of 1 μg/μL. Bottom-up proteomic analysis was performed using UltiMate™ 3000RSLCnano–HPLC System (Thermo Fisher Scientific, Waltham, MA, USA) coupled to a high-resolution Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific) with an Electrospray Ionozation (ESI) source. Peptides were separated via an PepMap RSLC C18 column 2 µM, 100 Å, 50 µm × 15 cm (Thermo Fisher Scientific) in gradient elution using an aqueous solution of Formic Acid (FA) (0.1%, v/v) as eluent A and Acetonitrile (ACN)/water (80:20, v/v) with 0.1% (v/v) FA as eluent B. The following step gradient was applied (run time 155 min): 3% eluent B and 97% eluent A (0–110 min), 20% eluent B and 80% eluent A (110–120 min), 40% eluent B and 60% eluent A (120–125 min), 90% eluent B and 10% eluent A (125–145 min) and 3% eluent B and 97% eluent A (145–155 min) (% values, v/v) at a flow rate of 0.300 μL/min. The injection volume was 5 μL (1 μg of peptides), with ion source type Nanospray Ionization (NSI), polarity positive (voltage 1,800 V) and ion transfer tube temperature of 275°C. The following MS parameters were set: the acquisition of high-resolution MS/MS spectra was carried out in data-dependent scan (DDS) mode using Orbitrap as detector, with a resolution of 120,000 in an m/z range of acquisition of 375–1,500– and higher-energy collisional dissociation (HCD) fragmentation. Samples were analyzed in analytical triplicate.
Mass spectrometry data analysis
The bottom-up MS/MS data were elaborated using Proteome Discoverer 2.4.1.15 (Thermo Fisher Scientific) based on the SEQUEST HT algorithm (University of Washington, USA, licensed to Thermo Electron Corp., San Jose, CA, USA) against UniProt databases representing Lactobacillus (https://www.uniprot.org/taxonomy/1578) and L. crispatus (https://www.uniprot.org/uniprotkb/D5H222/entry) proteome. The setting parameters were as follows: minimum precursor mass 350 Da, maximum precursor mass 5,000 Da, total intensity threshold 0.0, minimum peak count 1, signal-to-noise (S/N) threshold 1.5, precursor mass tolerance 10 ppm, fragment mass tolerance 0.02 Da, use average precursor mass False, use average fragment mass False, maximum missed cleavage 2, minimum peptide length 6, maximum peptide length 144, Oxidation/+15.995 Da (M) as dynamic modification, Carbamidomethyl/+57.021 Da (C) as static modification and False Discovery Rate (FDR) at 0.01 (Strict) and 0.05 (Relaxed).
Evaluation of cytotoxicity on eukaryotic cells
To measure cell viability under CFS treatment, Henrietta Lacks’ cervical eukaryotic cancer cells (HeLa) (ATCC® CCL-2.2 TM) were cultured as described above and then plated at a final concentration of 5 × 105 cells/mL in a 96-well plate. The cells were incubated overnight at standard atmosphere conditions (37°C and 5% CO2) until they were incubated with serial dilutions of CFS (CFS, CFS/2, CFS/4, CFS/8 and CFS/16). Untreated cells were used as a negative control, while cells treated with 2% Triton X-100 were used as a positive control. Triton X-100 interacts with lipid bilayers in a non-specific way, solubilizing bio-membranes and causing cell death (Koley and Bard, 2010; Mattei et al., 2017). Treated cells were incubated overnight at standard atmosphere conditions until the MTS Cell Proliferation Assay was performed to evaluate the cellular metabolic activity. Briefly, MTS reagent was added to each well and incubated for 4 hours at 37°C since absorbance was measured at 490 nm. Meanwhile, the cells were also fixed with Paraformaldehyde (PFA) 4% for 30 min and stained with crystal violet for 15 min to assess the monolayer integrity.
Epithelial cell culture and infection
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Euroclone, Pero, Italy) supplemented with 10% inactivated foetal bovine serum (FBS) (Euroclone, Italy), 1% l-glutamine (2 mM) (Euroclone) and 1% streptomycin–penicillin (100 µg/mL and 100 U/mL, respectively) (Euroclone) and were incubated at 37°C and 5% CO2. Adherent cells were washed with sterile warm Dulbecco’s Phosphate-Buffered Saline (DPBS) (Euroclone) and removed for experiments using 1× trypsin (5 μg/mL) in DPBS (Euroclone). Cells were counted and resuspended in DMEM supplemented with 2% Fetal Calf Serum (FCS) and 1% l-glutamine. Finally, cells were seeded in sterile 48-well plates (Euroclone) at a concentration of 5 × 105 cells/mL, filling 0.5 mL/well, and incubated overnight until infection or treatment. HeLa cells were infected with E. coli, S. agalactiae and C. albicans, with a multiplicity of infection (MOI) of 100:1 and a MOI of 10:1 for yeast infection, and resuspended in the cell culture medium. Two hours post-infection, infected cells were washed three times with sterile warm DPBS to remove non-adherent bacteria and treated with L. crispatus (MOI 500:1, e.g. ~2.5 × 108 CFU/mL), CFS (v/v 1:1 in cell medium), gentamicin or fluconazole at MIC and with the combination of antibiotic/antimycotic and LcM247 or CFS. Cells were incubated in standard atmosphere conditions for 24 hours, while CFUs were assessed by harvesting cell monolayers with 0.1 mL of sterile 0.01% Triton X-100 (Sigma-Aldrich, USA) 4 and 24 hours post-infection. Serial dilutions were carried out before plating on selective solid medium. Plates were then incubated at 37°C for 1 day. In the second experimental setting, cells were infected with C. albicans with a MOI of 10:1. Two hours post-infection, infected cells were washed thrice with sterile warm DPBS to remove non-adherent fungi. Subsequently, the cells were treated with fluconazole at the Minimum Inhibitory Concentration (MIC), CFS at v/v of 1:1 or fresh medium. Four hours later, treatments were removed, and cells were washed again to administer all conditions with L. crispatus (MOI 500:1). CFUs were assessed by harvesting cell monolayers with 0.1 mL of sterile 0.01% Triton X-100 at 24 and 48 hours post-infection. Serial dilutions were carried out before plating on selective solid medium. Plates were then incubated at 37°C for 1 day. Concurrently, to investigate LcM247 adherence on cell monolayers, cells were subjected to lysis 48 hours post-infection and plated on MRS agar. The plates were incubated at 37°C for 2 days before CFU counting.
In vivo safety and effectiveness study on Galleria mellonella larva model
To assess the safety of the administered treatments (LcM247, CFS/2, gentamicin, fluconazole, gentamicin–LcM247, gentamicin–CFS/2, fluconazole–LcM247 and fluconazole–CFS/2) also in an in vivo model, healthy G. mellonella larvae measuring 2 to 2.5 cm in length and 0.3 to 0.45 g in body weight and showing no signs of melanization were selected for the experiment. Sterile water and Luria-Bertani (LB) broth (Sigma-Aldrich) were used as negative controls. Ten larvae were used to characterize each group and placed in a separate Petri dish. The larvae’s final left abdomen proleg was injected with 10 µL of each treatment, using 0.5-mL syringes with no dead volume, after sterilizing the region with 70% ethanol. Then, larvae were placed in an incubator at 37°C in the dark until the end of the experiment. The number of dead G. mellonella was scored every 24 hours for 4 days. Larvae were considered dead when they did not respond to touch. Survival curves were plotted using the Kaplan–Meier method, and differences in survival were calculated using the log-rank test (GraphPad Prism 10). Experiments were performed at least twice.
To evaluate the efficacy of the previously mentioned administered compounds, the same experimental setting was used to treat the in vivo infection model of G. mellonella larvae. Each larva’s final left abdomen proleg was injected with 10 µL of E. coli (5 × 105 CFU/mL, that is, 5 × 103 CFU/larvae), S. agalactiae (2 × 108 CFU/mL, that is, 2 × 106– CFU/larvae) and C. albicans (5 × 107 CFU/mL, that is, 5 × 105– CFU/larvae) using 0.5-mL syringes with no dead volume. Prior to treatment, 70% ethanol was used to sterilize the region. After 2 hours of 37°C incubation, 10-µL treatments were administered in the larvae’s final hind limb to investigate the efficacy against the infection. The larvae’s final right abdominal proleg was injected with 10 µL of each treatment. Sterile water was used as negative controls. Ten larvae were used to characterize each group and were separated into different Petri dishes. Infected and treated G. mellonella larvae were kept at 37°C in the dark. The number of dead G. mellonella was scored every 24 hours for 4 days. Larvae were considered dead when they did not respond to touch (Figure 7A). Survival curves were plotted using the Kaplan–Meier method, and differences in survival were calculated using the log-rank test of two nested experiments (GraphPad Prism 10).
Statistical analysis
All data were generated from independent experiments with at least three technical replicates. Statistical significance was indicated using asterisks, with the following thresholds: p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) and p < 0.0001 (****). Microsoft Excel (2024) and Graphpad Prism software v. 10 (GraphPad software) were used to collect and analyze the data. Data were expressed on a representative graph as mean ± SD and analyzed by one-way or two-way ANOVA comparison tests.
Results
L. crispatus M247 and its CFS inhibit the growth of indicator strains
When bacterial vaginosis occurs, including a consequence of antibiotic treatments (Chen et al., 2021; Abbe and Mitchell, 2023; Pendharkar et al., 2023), L. crispatus-based probiotics, including the strain M247 (LcM247) from Crispact® (PharmExtracta S.p.A, Pontenure, Italy), are the most widely used commercially available probiotics to restore healthy microbiota, although their activity is not fully understood (Cesena et al., 2001; Di Pierro et al., 2019).
The LcM247 antagonist effect and its CFS were assessed against six main microbial indicator species: E. coli, K. pneumoniae, S. aureus, E. faecalis, S. agalactiae and C. albicans. Each indicator strain was incubated with LcM247 at a ratio of 1:100 (CFUs/CFUs) or with LcM247 CFS (1:1 v/v). CFUs were assessed at 4 and 24 hours post-incubation. A schematic representation of the experimental setting is reported in Figure 1A.
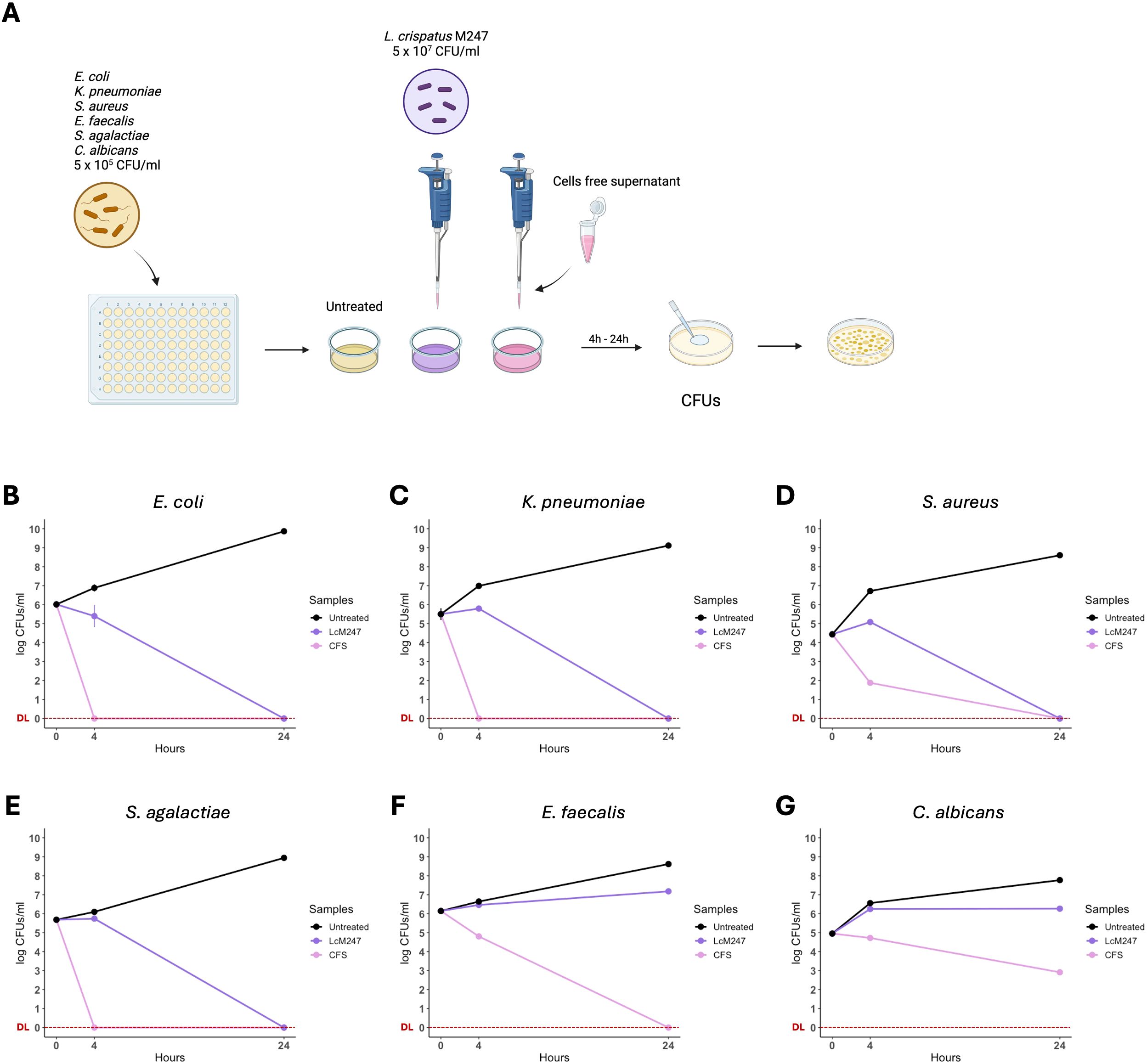
Figure 1. Lactobacillus crispatus M247 and its cell-free supernatant (CFS) differently reduced microbial burden in the short-term period. Antimicrobial properties of L. crispatus M247 (LcM247) and its CFS were assayed by incubating a suspension of 5 × 105 CFU/mL of Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, Streptococcus agalactiae, Enterococcus faecalis and Candida albicans, named as indicator strains. LcM247 was used at the final concentration of 5 × 107 CFU/mL, while CFS was added with a v/v ratio of 1:1 with the microbial cultures (A). Colony-forming units (CFUs) were measured at 4 and 24 hours post-treatment and are presented as line plot reporting the mean ± SD in log10 scale of three repeated experiments (B–G).
A marked inhibitory effect was only observed against E. coli, in the presence of LcM247, accounting for a reduction by 10% compared to the initial infection solution (p < 0.01) (Figure 1B). Conversely, for the other indicator strains, LcM247 co-treatment was not able to prevent bacterial replication (Figures 1C–F). Finally, no fungistatic effect was still observed on C. albicans replication at 4 hours (Figure 1G).
A significant activity was observed following 24 hours of incubation with LcM247. Indeed, LcM247 exerted a robust bactericidal activity after 24 hours on selected bacteria, leading to the complete elimination of E. coli, K. pneumoniae, S. aureus and S. agalactiae (p < 0.0001) (Figures 1B–E). Conversely, the activity of LcM247 against E. faecalis and C. albicans was negligible in comparison with that of the infection solution at 4 hours post-incubation (Figures 1F, G).
To investigate whether, similar to what is known for lactobacilli, the antimicrobial activity of LcM247 is associated with secreted molecules, the LcM247 CFS was collected, and the microbial indicators chosen with this preparation were incubated. Interestingly, 4-hour incubation with CFS was sufficient to completely inhibit the growth of E. coli, K. pneumoniae and S. agalactiae (p < 0.0001) (Figures 1B, C, E), with a 5-log CFU reduction of S. aureus and a 2-log CFU reduction for both E. faecalis and C. albicans CFUs, compared to untreated controls (Figures 1D, F, G). Notably, no detectable colonies were found after 24 hours of incubation of CFS with all the analyzed bacterial strains (p < 0.0001) (Figures 1B–F), whereas incubation with C. albicans resulted in a 5-log CFU reduction compared to the untreated control (p < 0.0001) (Figure 1G). Our results demonstrate the antimicrobial activity of LcM247, which is conserved in its CFS.
Strengthening these hypotheses, Supplementary Figure S2 illustrates the significant, dose-dependent antimicrobial effect of LcM247 CFS against L. iners bacterial cells. Notably, the reciprocal experiment demonstrated that L. iners CFS failed to exert any detectable inhibitory effect on LcM247, whose growth remained stable across all conditions tested. These results suggest that L. crispatus secretes antimicrobial compounds absent in L. iners supernatant, prompting us to further investigate the bioactive content of LcM247 CFS.
LcM247 CFS antimicrobial activity is pH dependent
Numerous studies have demonstrated that the CFS produced by lactobacilli plays a crucial role in antimicrobial activity, primarily due to its content of lactic acid, which drives the decrease in the environmental pH, and bacteriocins, peptide-based toxins, target bacterial or fungal strains by disrupting their membranes (Abdul-Rahim et al., 2021; Mani-Lopez et al., 2022). Hence, we investigated the antimicrobial effect of CFS to elucidate whether this was concentration- or pH-dependent.
Briefly, four dilutions of CFS, appropriately diluted in MRS medium (v/v CFS/MRS 1:1, 1:4, 1:8 and 1:16 named as CFS/2, CFS/4, CFS/8 and CFS/16, respectively), were obtained and tested against the previously mentioned microbial indicator strains. CFS was also alkalinized by adding different volumes of 1 M NaOH, creating four modified CFS with different pH values (pH 4.5, representing the untreated v/v CFS/MRS 1:1, pH 5.5, pH 7 and pH 9). All modified CFSs were incubated with the indicator strains for 24 hours at 37°C in a 96-well plate with round-bottom wells. At the end of the incubation, the growth was assessed by observing the strain pellet deposited at the bottom of the well and by counting CFUs for each condition (Figure 2A).
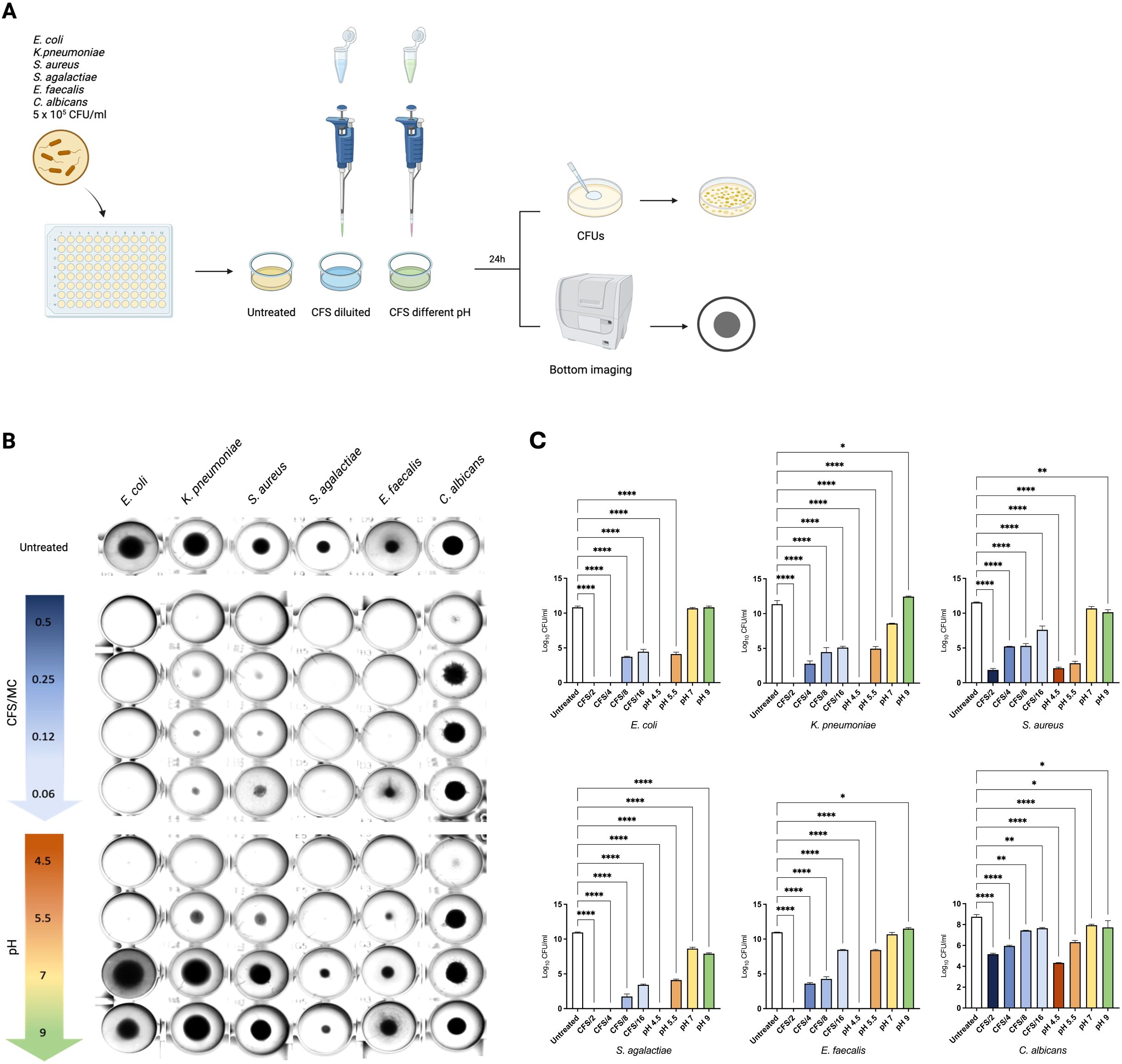
Figure 2. Acidic pH plays a key role in mediating LcM247 CFS antimicrobial activity. CFS antimicrobial activity was assayed using serially diluted CFS and alkalinized CFS by adding different volumes of 1 M NaOH. CFS was diluted in MRS broth to achieve final CFS/MRS v/v ratio of 1:1, 1:2, 1:4 and 1:8 (i.e. CFS/2, CFS/4, CFS/8 and CFS/16, respectively) (as reported in Supplementary Figure S1, pH was maintained at value of 4.5). CFS alkalinization generated unaltered CFS, which corresponded to CFS/2 (pH 4.5), CFS at pH 5.5, CFS at pH 7 and CFS at pH 9. Modified CFS solutions were incubated with a suspension of the indicator strains Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, Streptococcus agalactiae, Enterococcus faecalis and Candida albicans at final concentration of 5 × 105 CFU/mL for 24 hours at 37°C in a 96-well plate with round-bottom wells. Microbial growth in MRS broth was used as a control (A). At the end of the incubation, the microbial growth was assessed by observing the strain pellet deposited at the bottom of the well using the Cytation instrument (B) and measuring colony-forming units (CFUs) determined after serial dilutions of the last well showing microbial growth (C). Data from three repeated experiments are presented as bar plot reporting the mean ± SD on log10 scale. LcM247, Lactobacillus crispatus M247; CFS, cell-free supernatant; MRS, de Man, Rogosa and Sharpe. Statistical signifcance was indicated using asterisks, with the following thresholds: p < 0.05 (*), p < 0.01 (**) and p < 0.0001 (****).
As shown in Figure 2B, the CFS/2, corresponding to CFS showing pH 4.5, significantly inhibited the microbial growth of all indicator strains, compared to the untreated specimens. However, the bacterial pellet at the bottom of the wells appears almost comparable for the other diluted CFSs (CFS/4, CFS/8 and CFS/16), while progressive alkalization was directly associated with microbial growth. Indeed, CFSs at pH 7 and pH 9 lose the ability to block bacterial and fungal replication. These results suggest that acidic pH is necessary to reduce microbial growth.
To corroborate this finding, we measured microbial growth by evaluating CFUs following treatment. As shown in Figure 2C, CFS/2 and CFS at pH 4.5 confirmed a complete growth inhibition of E. coli, K. pneumoniae, S. agalactiae and E. faecalis (p < 0.0001). Conversely, the same treatments did not result in the complete elimination of S. aureus and C. albicans, where the reduction accounted for 9.5 log and 4 log (p < 0.0001), respectively. Similarly, CFS/4 incubated with E. coli and with S. agalactiae led to complete bacterial elimination (p < 0.0001), while a reduced effect was observed for K. pneumoniae, S. aureus and E. faecalis (~8 log, 6 log and 7 log of reduction, respectively, p < 0.0001) and C. albicans (~2 log, p < 0.0001) compared to the untreated controls. CFS/8 and CFS/16 treatments showed a significant but incomplete antimicrobial effect, reducing all gram-negative bacteria and S. agalactiae burden by 9 log and 7.5 log (p < 0.0001), respectively, and S. aureus and E. faecalis by 7 log and 4 log (p < 0.0001), respectively, while reducing C. albicans growth by less than 1 log (p = 0.0024 for CFS/8 and p = 0.0078 for CFS/16). Importantly, the pH values of CFS/2, CFS/4, CFS/8 and CFS/16 were not affected by dilution in MRS liquid medium (Supplementary Figure S1). As previously observed, CFU measurement highlighted that CFS at pH 5.5 maintained a significant antimicrobial activity, reducing bacterial growth from a minimum of 2 log for C. albicans (p < 0.0001) to a maximum of 8 log for S. aureus (p < 0.0001). Importantly, minimal reduction in microbial load was evidenced when the CFS pH was increased to 7 and 9 (p = 0.954 and p > 0.999 for E. coli, p < 0.0001 and p = 0.062 for K. pneumoniae, p = 0.079 and p = 0.005 for S. aureus, p < 0.0001 and p < 0.0001 for S. agalactiae, p = 0.395 and p = 0.045 for E. faecalis and p = 0.395 and p = 0.045 for C. albicans). These results indicate that the LcM247 CFS antimicrobial activity is pH dependent.
Lactobacillus CFS antimicrobial activity is not impaired by heat or proteinase treatment
To elucidate the role of bacteriocins, the antimicrobial activity of LcM247 CFS was assessed through heat inactivation and proteinase K proteolytic enzymatic treatment. After exposure to 98°C for 1 hour and enzymatic treatment at 56°C for 30 min, undiluted CFS was tested against the indicator strains for 24 hours at 37°C before CFU evaluation (Figure 3A). Intriguingly, microbial growth was inhibited even when CFS was heated or enzymatically treated, showing bacterial or fungal pellets comparable to specimens treated with unmodified CFS. Importantly, heated MRS did not affect indicator strains’ growth, unlike acidified MRS, which reduces bacterial pellet size, sometimes making them macroscopically invisible. Conversely, C. albicans appeared full-grown in all conditions (Figure 3B). CFU analysis confirmed that the inhibitory effect of CFS significantly reduced the microbial load from a minimum of 3 log for C. albicans (p < 0.0001) to a maximum of 9 log for E. coli (p < 0.0001), resulting in a complete elimination of E. coli and S. agalactiae (100% reduction) (Figure 3C). These findings were comparable to those observed with heat-inactivated CFS, which showed the same inhibitory effect (p < 0.0001 for each indicator strain and a 100% reduction for E. coli and S. agalactiae). Similarly, proteinase K treatment did not significantly affect the CFS antimicrobial role. Indeed, despite this enzymatic treatment, the CFS still showed a significant ability to decrease microbial growth, which, although slightly less pronounced for E. faecalis and C. albicans, was comparable to the inhibitory effect of unmodified CFS (p < 0.0001). Importantly, the heated MRS had no antimicrobial activity on the indicator strains, which was comparable to that of the normal growth of untreated controls (p = 0.742 for E. coli, p = 0.851 for K. pneumoniae, p = 0.569 for S. aureus, p = 0.013 for S. agalactiae, p = 0.002 for E. faecalis and p = 0.491 for C. albicans). Interestingly, the acidified MRS reduced the microbial burden, especially for those bacteria that were observed to be most sensitive to the acidic microenvironment, in particular E. coli, K. pneumoniae, S. aureus and S. agalactiae (p < 0.0001). Conversely, more resistant strains, such as E. faecalis and C. albicans, did not show a reduction in CFUs after treatment with acidic MRS (p = 0.061 and p = 0.184, respectively). Taken together, these results suggest that CFS pH-dependent antimicrobial properties are mediated by heat- and protease-resistant microbial components.
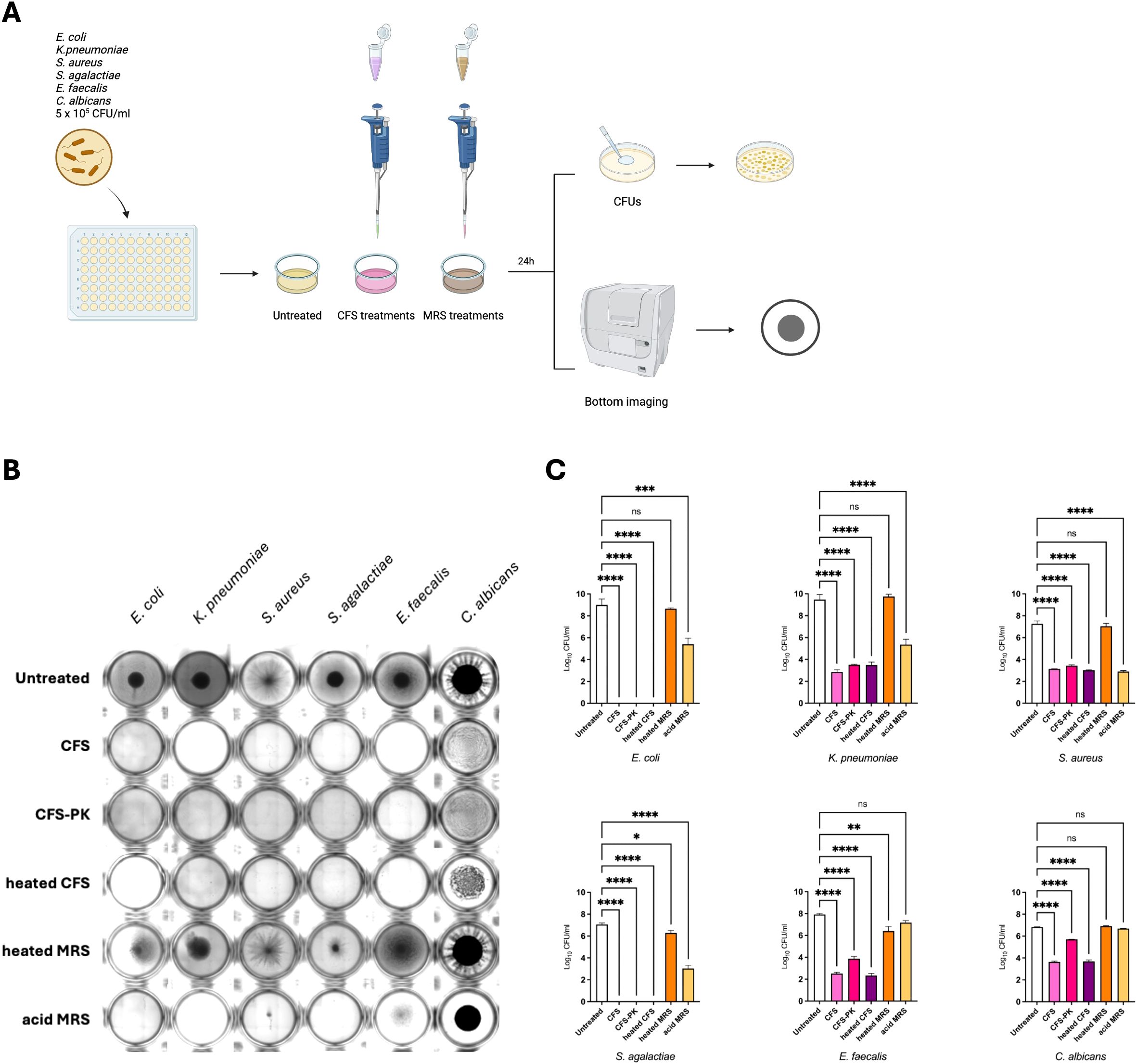
Figure 3. LcM247 CFS maintains antimicrobial activity following heat inactivation and enzymatic treatment. LcM247 CFS was heat inactivated (98°C) or enzymatically treated using proteinase (K) MRS broth previously heated at 98°C for 1 hour and acidified with HCl to reach pH value of 4.5 was used as negative control. Modified CFSs were prepared with 5 × 105 CFU/mL of the indicator strains (Escherichia coli, Klebsiella pneumoniae, Staphylococcus aureus, Streptococcus agalactiae, Enterococcus faecalis and Candida albicans) with a v/v 1:1 ratio and incubated in standard atmosphere conditions for 24 hours (A). At the end of the incubation, the microbial growth was assessed by observing the strain pellet deposited at the bottom of the well using citation instrument (B) and measuring colony-forming units (CFUs) determined after serial dilutions of the last well showing microbial growth (C). Data from three repeated experiments are presented as bar plot reporting the mean ± SD on log10 scale. Statistical signifcance was indicated using asterisks, with the following thresholds: ns: not significant, p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) and p < 0.0001 (****).
Genomic and proteomic analysis of LcM247
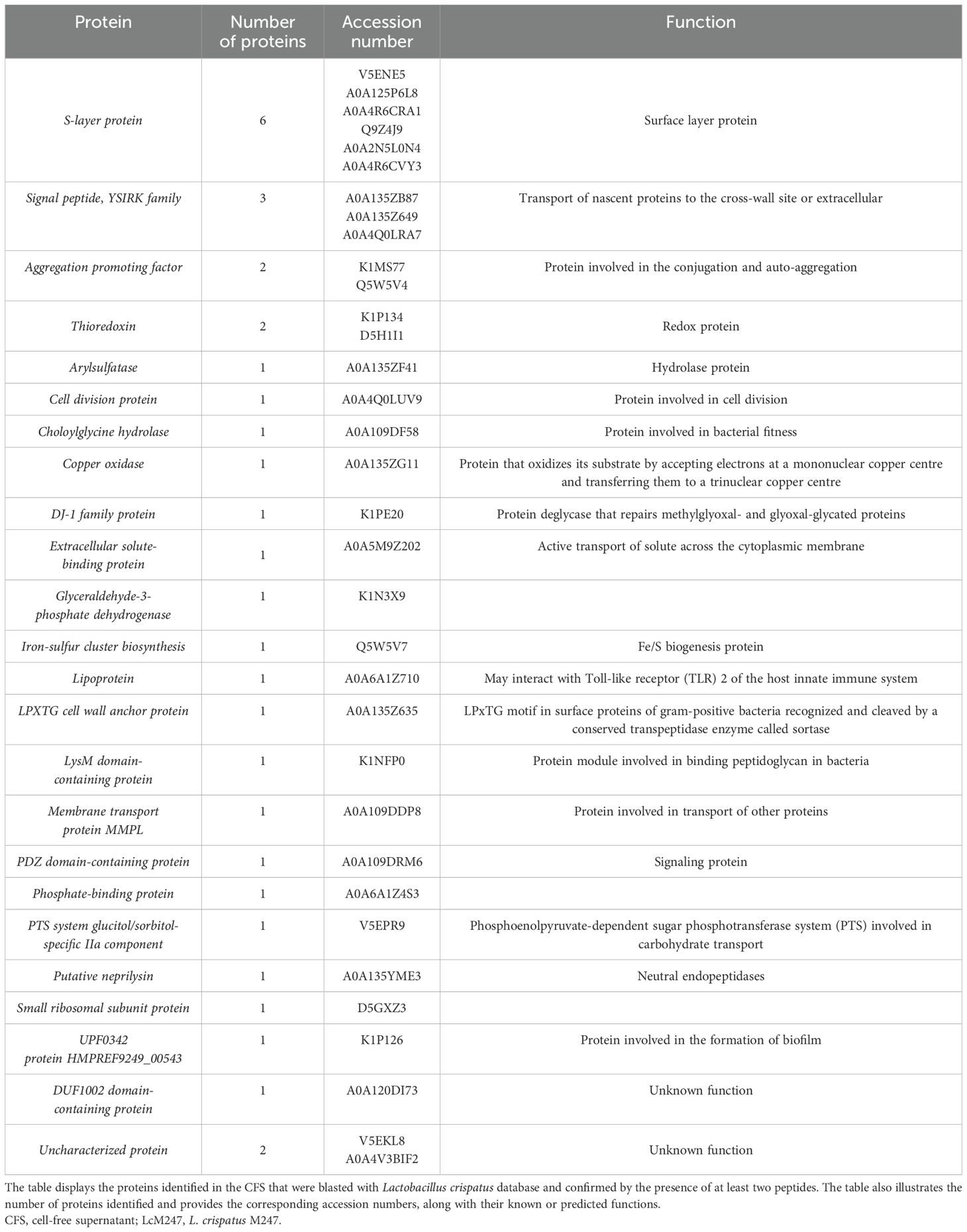
While it is well known that Lactobacillus species produce several metabolites (organic acids and hydrogen peroxide) that are important to maintain a favorable environment, little information is available on the antimicrobial activity of secreted proteins (i.e. bacteriocins). We performed a genomic analysis of the LcM247 genome and a proteomic analysis of its CFS to identify potentially synthesized and secreted peptides/proteins that may mediate the antimicrobial properties. The general genomic characteristics of LcM247 after de novo assembly are reported in the Materials and Methods section. Bacteriocin-encoding genes were predicted by uploading the assembly fasta file into BAGEL4, which is specifically designed for bacteriocin mining in prokaryotic genomes (van Heel et al., 2018). BAGEL4 analyzed the sequence and identified the genome/contig areas of interest (AOIs) that encode bacteriocin-like peptides (Li et al., 2022). The analysis revealed seven AOIs within the genome containing bacteriocin-associated genes (Supplementary Table S1). Putative bacteriocin genes, showing similarity to the bacteriocin genes encoding for Enterolysin A (match 67.299% and 59.615% in comparison with BAGEL4 reference amino acid sequence; for both, no bit score was reported) and Penocin A (match 34.783% with bit score 50.061 and match 36.765% with bit score 39.276 in respect to BAGEL 4 reference amino acid sequence) were detected twice in two separate regions within the LcM247 genome assembly. Of note, the reference strain (CP088015) showed only six AOIs, suggesting that de novo alignment based on the short-read sequencing may be impaired by technical errors and not exactly evidence of gene duplication.
The proteomic composition of the CFS was analyzed to identify secreted peptides that may be potentially responsible for its antimicrobial activity. In Table 1, we summarized the major features revealed in the LcM247 CFS, which were assessed by at least two peptides, which refers to an accurate protein identification (i.e. the detection of at least two unique peptides per protein). Contrary to what we anticipated, the proteomic analysis did not detect any bacteriocin in the Lactobacillus secretome (Table 1). Indeed, we could not detect any bacteriocin when searching for at least two peptides (Supplementary Table S2) in the L. crispatus database or the Lactobacillus genus database (Supplementary Tables S3, S4). Instead, we observed a predominance of surface-associated proteins, many of which appear to be involved in Lactobacillus fitness.
Of note, our analysis highlighted several proteins associated with the S-layer, which is critical for the ability of Lactobacillus to exert antimicrobial effects, or peptides belonging to proteins involved in aggregation-promoting factors and signal peptides of the YSIRK family, suggesting mechanisms related to bacterial adherence and signal transmission (Edelman et al., 2012).
Additionally, structural proteins and enzymes, which are involved in redox reactions and the overall bacterial fitness, were identified. Unfortunately, a label-free analysis measuring the peptide abundance was not possible due to the lack of a reliable Lactobacillus list in databases. Nevertheless, coverage and score measures suggested that S-layer peptides represented the most relevant CFS component. Conversely, the lack of bacteriocins in the CFS points toward a negligible role of these peptides in our experimental settings.
Evaluation of the CFS antimicrobial effect during microbial infection of a cervical epithelial cell line
Our findings on axenic cultures have suggested a rapid and efficient activity of CFS compared to live LcM247, offering a novel potential therapeutic strategy. Hence, we first assessed CFS cytotoxicity on HeLa cells by measuring the metabolic activity and monolayer integrity using MTS and crystal violet assays, respectively (De Maio et al., 2021; Rosato et al., 2024; Santarelli et al., 2024). As reported in Figure 4A, CFS and CFS/2 appeared to impair cell viability, while cells treated with CFS/4, CFS/8 and CFS/16 showed values up to untreated cells. To corroborate these findings, cell monolayer integrity was assessed through staining with a crystal violet solution. This demonstrated that none of the CFS dilutions caused damage to HeLa cells (Figure 4B). The phenomenon under investigation can be attributed to the sensitivity of the MTS reagents to even slight pH variations or certain metabolites present in undiluted or slightly diluted CFS. These factors can lead to an artefact in the MTS signal.
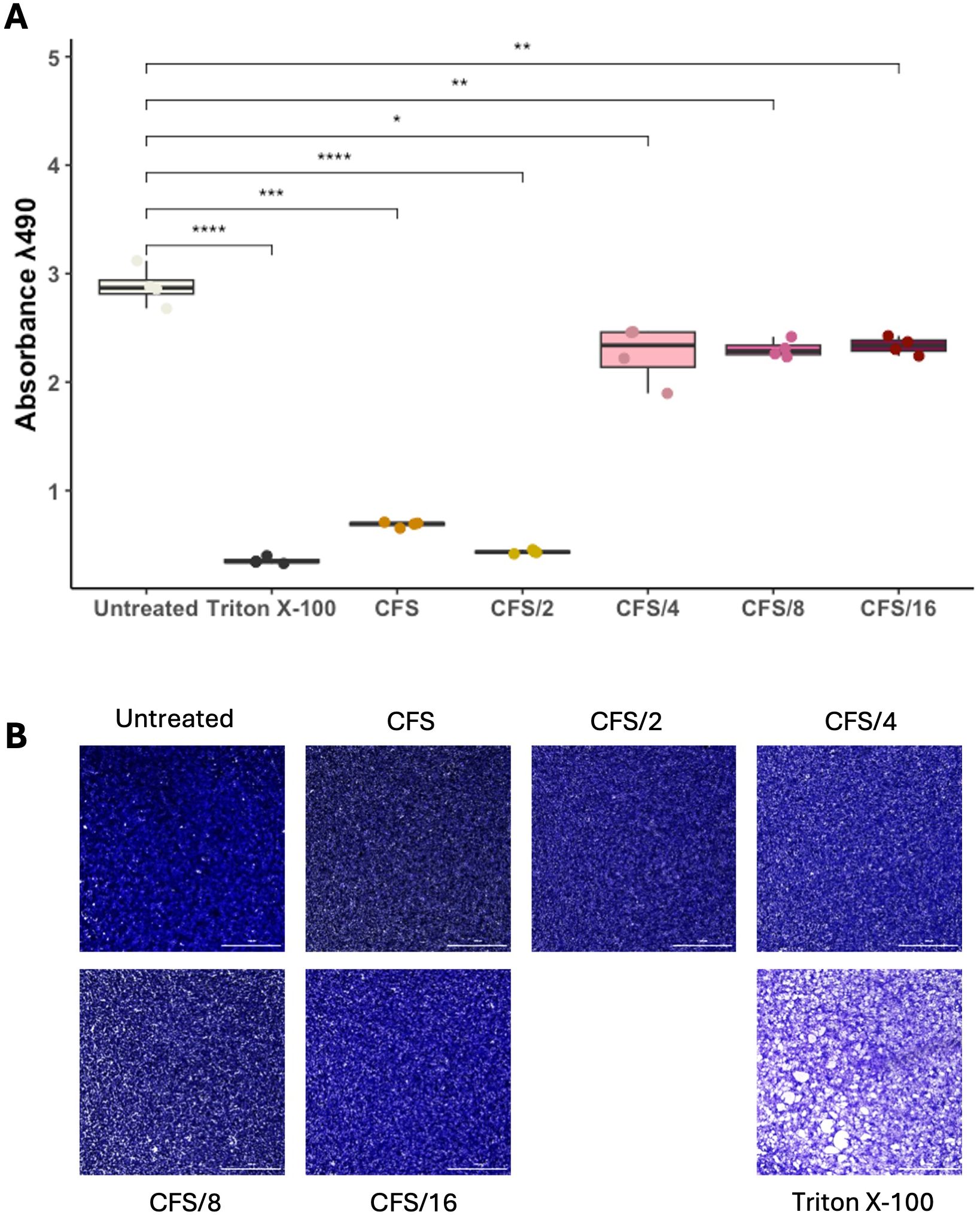
Figure 4. LcM247 CFS shows good biocompatibility on eukaryotic cells. HeLa cells were seeded in a 96-well plate and treated with serial dilutions of CFS (CFS, CFS/2, CFS/4, CFS/8 and CFS/16) when 90% cell monolayer was reached. Untreated cells and cells treated with 2% Triton X-100 were used as negative and positive controls, respectively. One day post-incubation, cells’ metabolic activity was evaluated using MTS assay (A). Results of three repeated experiments were acquired using the Cytation instrument and are reported as box plot chart. Cellular monolayer was fixed and stained using crystal violet staining. Images were acquired using Cytation instrument and processed with ImageJ software to evaluate monolayer’s integrity (B). LcM247, Lactobacillus crispatus M247; CFS, cell-free supernatant. Statistical signifcance was indicated using asterisks, with the following thresholds: p < 0.05 (*), p < 0.01 (**), p < 0.001 (***) and p < 0.0001 (****).
Finally, an in vitro infection model was set up based on HeLa to mimic vaginal dysbiosis and demonstrate the antimicrobial activity of LcM247 and its CFS. HeLa cells were infected with E. coli, S. agalactiae and C. albicans at a MOI of 100:1 for bacterial species and 10:1 for the yeast. Two hours later, infected cells were treated with LcM247 (MOI = 500:1), CFS and antibiotics or antifungals at MICs or in combination (LcM247-CFS/gentamicin or LcM247-CFS/fluconazole). CFUs were performed 4 and 24 hours later (Figure 5A).
The graph in Figure 5B shows that 4 hours after treatment, LcM247 had the worst antibacterial activity against E. coli, different from the other treatments, which showed a significant reduction in the bacterial load (Figure 5B). Interestingly, CFS was confirmed to be the most effective treatment, also in combination with antibiotics, showing a reduction of ~3 log in E. coli CFUs (p < 0.0001). In contrast, gentamicin alone showed moderate bactericidal activity, resulting in a lower efficacy than the co-administration of gentamicin and LcM247. Twenty-four hours after infection, gentamicin showed a strong antibacterial effect, reducing the E. coli load by ~7 log compared to the untreated sample (p < 0.0001) (Figure 5E). LcM247 did not eradicate E. coli, although co-administration with the antibiotic reduced CFUs by ~5 log (p < 0.0001) compared to control. Remarkably, CFS and CFS–gentamicin exhibited complete bacterial elimination (100% reduction compared to the control, p < 0.0001).
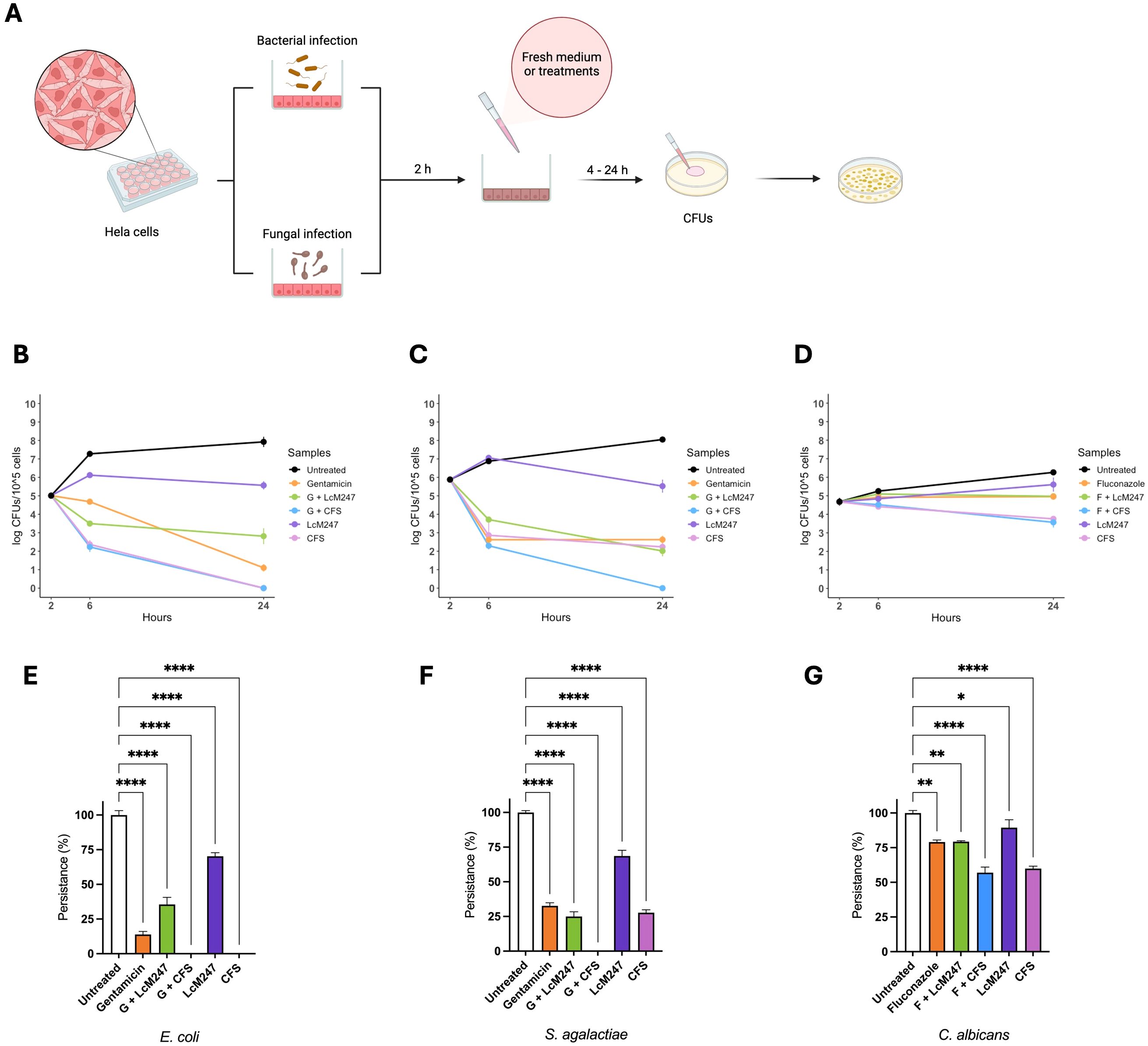
Figure 5. LcM247 CFS reduces microbial burden in cervical epithelial cells. HeLa cells were infected with selected indicator strains Escherichia coli, Streptococcus agalactiae and Candida albicans with multiplicity of infection (MOI) of 100:1, 100:1 and 10:1, respectively. Two hours after infection, cells were washed three times and treated with LcM247 (MOI = 500:1), CFS (CFS v/v 1:1 in culture medium), antibiotic (gentamicin) or antimycotic (fluconazole) at MIC and finally a combination of antibiotic/antimycotic with LcM247 or CFS (A). Four- and twenty-four-hours post-incubation, colony-forming units were assessed. Line plot showing Log CFUs/105 cells generated in three repeated experiments. Persistence at 24 hours was reported as percentage of CFUs obtained for each condition, compared to infected untreated cells. Data from three repeated experiments are presented as bar plot reporting the mean ± SD. Data regarding are reported in panels (B, E) (E. coli), (C, F) (S. agalactiae), and (D, G) (C. albicans). CFS, cell-free supernatant; MOI, multiplicity of infection; LcM247, Lactobacillus crispatus M247; CFU, colony-forming unit. Statistical signifcance was indicated using asterisks, with the following thresholds: p < 0.05 (*), p < 0.01 (**) and p < 0.0001 (****).
As depicted in Figure 5C, 4 hours post-treatment, LcM247 showed no antibacterial activity on S. agalactiae-infected cells, exhibiting a bacterial replication comparable to that of the infected and untreated cells. Interesting, CFS confirmed a significant reduction of ~3 log in the burden of S. agalactiae, equivalent to the antibiotic administration alone and in combination with Lactobacillus or the supernatant. Notably, even after 24 hours of incubation, LcM247 failed to completely eradicate the bacterial load, although it started to exhibit measurable antimicrobial activity relative to the untreated control, reducing CFUs by approximately 3 log (p < 0.0001). The co-administration of gentamicin and CFS emerged as the most effective treatment, exhibiting a complete bactericidal activity (100% of reduction, p < 0.0001) (Figure 5F). The other treatments, including CFS alone, gentamicin alone and the combination of gentamicin with LcM247, showed a limited antibacterial efficacy, resulting in a minimal reduction in microbial load compared to the 4-hour incubation period (~6 log of reduction compared to the control, p < 0.0001).
In the C. albicans infection model, only CFS and CFS in association with fluconazole showed a slight reduction of microbial burden following the 4-hour treatment of infected cells (Figure 5D). Similarly, 24 hours post-incubation, CFS and CFS–fluconazole administration maintained a significant antimicrobial activity, reducing yeast growth by ~1 log compared to the control (p < 0.0001) (Figure 5G). In contrast, fluconazole alone or in combination with LcM247 seemed to only have a fungistatic effect (total reduction of ~1 log compared to the control, p = 0.0019 and p = 0.0020, respectively). Lastly, the LcM247 treatment alone was not able to exert an antimicrobial effect on C. albicans (p = 0.048).
To investigate whether treatment with LcM247 CFS was able to enhance LcM247 engraftment on epithelial cells following an infection, we set up an additional experiment using C. albicans that, based on our previous experimental findings, may require longer treatment to be eradicated. HeLa cells were infected with C. albicans for two hours, followed by administration of fluconazole, LcM247 CFS (at pH 4.5) or fresh medium. Four hours later, LcM247 was added to each condition. CFUs were assessed at 4, 24 and 48 hours post-infection to assess both antifungal activity and the LcM247 burden (Figure 6A).
As depicted in Figure 6B, 4 hours post-infection, CFS treatment significantly reduced C. albicans load in comparison with fluconazole treatment or untreated control. This trend was maintained by 24 hours post-infection, although the co-activity of antimycotic drug or CFS was supported by LcM247.
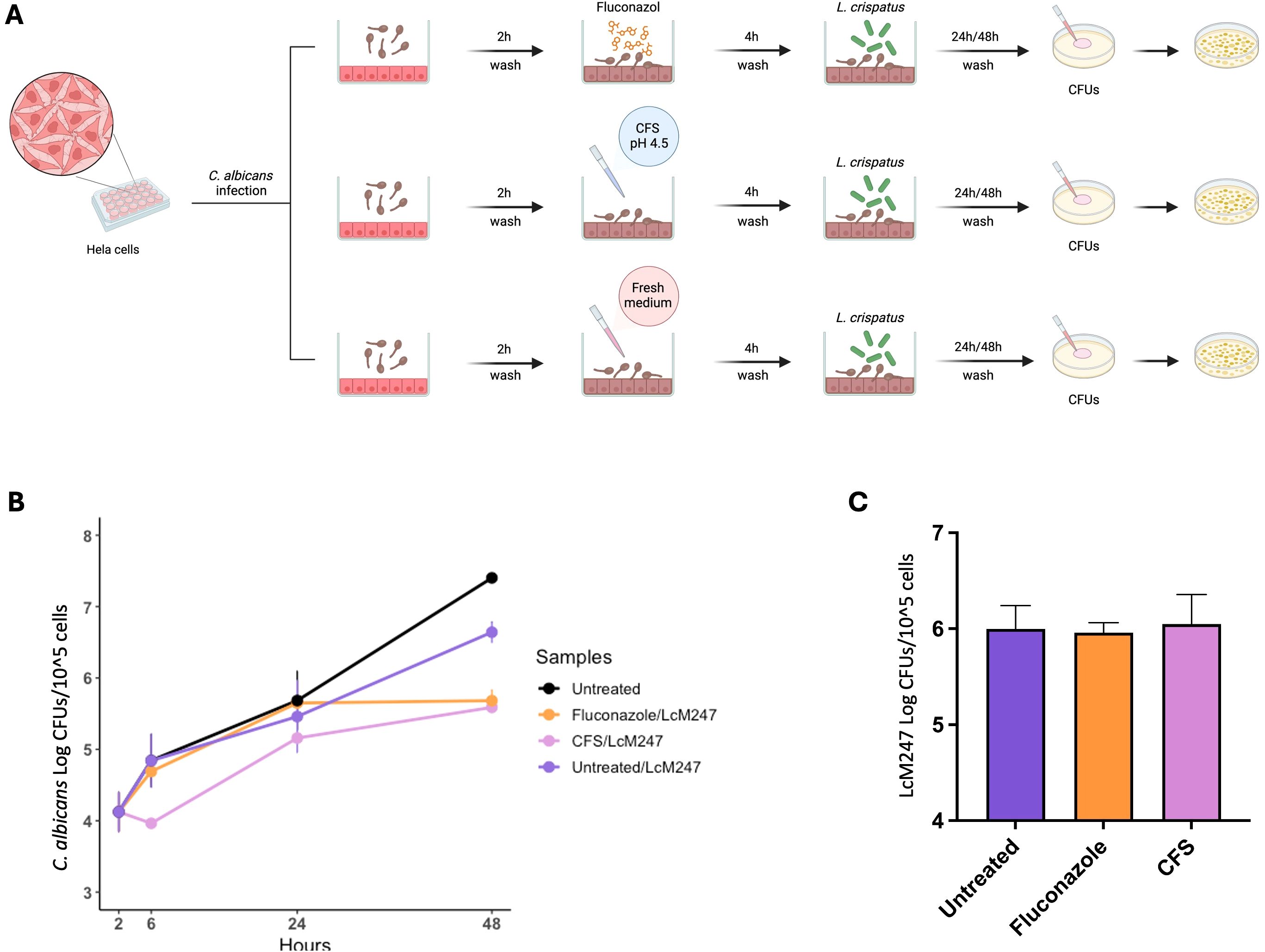
Figure 6. The administration of antimicrobial agents or CFS does not enhance LcM247 engraftment on cells. Schematic representation of the experimental setup. HeLa cells were infected with Candida albicans (MOI 10:1) for 2 hours, followed by washes and treatments with fluconazole (at MIC), CFS (CFS v/v 1:1 in culture medium) or fresh medium for 4 hours. After treatment, LcM247 (MOI 500:1) was added to assess its potential for antifungal activity and its engraftment on epithelial cells. CFU counts were assessed after 4, 24 and 48 hours post-infection (A). Line plot showing Log CFUs/105 cells generated in three repeated experiments (B). LcM247 persistence on cells after 48 hours is presented as bar plot reporting the mean ± SD (C). CFS, cell-free supernatant; LcM247, Lactobacillus crispatus M247; MOI, multiplicity of infection; CFU, colony-forming unit.
Interestingly, 48 hours post-infection, CFS/LcM247 and fluconazole/LcM247 combinations showed a comparable and superior ability to contain C. albicans burden compared to untreated cells or cells treated with LcM247 alone.
The data collected suggest that the administration of CFS and antimycotic has a similar efficacy when administered in combination with the live probiotic strains. Of note, LcM247 engraftment appeared independent from antimicrobial pre-treatment (Figure 6C), suggesting that Lactobacillus’s colonization ability on eukaryotic cells is relatively unaffected by prior exposure to drugs or other treatments.
CFS reduces microbial burden in infected G. mellonella
To corroborate our findings obtained in a monocellular in vitro infection model, we assayed CFS antimicrobial activity in a complex but highly reproducible infection model in G. mellonella. G. mellonella larvae are widely used as a surrogate model of several infectious diseases due to the ease of use and the presence of an innate immune system (Ligeza-Zuber, 2012; Kohler, 2015). LcM247 and CFS cytotoxicity in G. mellonella were assessed, showing a good biocompatibility in this model (Supplementary Figure S3).
G. mellonella was infected with 10 µL of E. coli (5 × 105 CFU/mL), S. agalactiae (2 × 108 CFU/mL) and C. albicans (5 × 107 CFU/mL). Two hours post-infection, LcM247, CFS/2, gentamicin/fluconazole or their combinations were injected, and larvae were incubated at 37°C for 96 hours (Figure 7A). Infected G. mellonella were monitored daily, and live ones were counted.

Figure 7. LcM247 CFS prolongs the survival of infected Galleria mellonella larvae. Schematic representation depicts the experimental in vivo model used to assay antimicrobial activity of LcM247, CFS/2, gentamicin/fluconazole and co-administration of CFS/2 or LcM247 with drugs on infected G. mellonella larvae. Ten microliters of infected solution was injected into the last left proleg. Two hours post-infection, 10-µL treatments were inoculated into the last right proleg. For the control groups, 10 µL of sterile H2O was inoculated. The number of dead G. mellonella was recorded every 24 hours for 4 days (A). Survival curve of G. mellonella infected with Escherichia coli (B), Streptococcus agalactiae (C) and Candida albicans (D) and treated as previously described. LcM247, Lactobacillus crispatus M247; CFS, cell-free supernatant.
As shown in Figure 7B, untreated G. mellonella infected with E. coli had a survival rate of 40% at 96 hours. The same survival rate was observed in infected larvae co-treated with the combination of gentamicin and LcM247. A slightly higher survival rate (50%) was reached when infected larvae were treated with LcM247 alone, gentamicin alone and gentamicin co-administered with CFS/2. Interestingly, CFS/2 treatment demonstrated the strongest antimicrobial effect in the in vivo model, with a minimal mortality of G. mellonella larvae observed until the last day of infection (mortality of 10% at 48 hours, 10% at 72 hours and 20% at 96 hours).
The most striking results were observed for S. agalactiae infection, which was in line with what was previously observed in the in vitro infection model (Figure 7C). Indeed, the untreated group of G. mellonella larvae had a survival rate of 20%, while antibiotic administration increased survival to 40%. Gentamicin in combination with CFS/2 or LcM247 increased the survival rate up to 50% or 40%, respectively. Of note, CFS/2 administration accounted for 90% larval survival, indicating a high activity against S. agalactiae.
Finally, C. albicans infection had a survival rate of approximately 20% in infected larvae (Figure 7D). When infected larvae were treated with LcM247 or a co-administration of fluconazole and LcM247, the survival rate was enhanced to 30% or 40%, respectively. The co-administration of fluconazole and CFS/2 reduced mortality by 50%, while fluconazole and CFS/2 alone were able to decrease mortality up to 40%.
These findings suggested that CFS has a promising antimicrobial activity in in vivo models, even when not co-administered with antimicrobial compounds.
Discussion
The vaginal microbiome is normally dominated by Lactobacillus species, showing low diversity but strong dynamics in changing its composition that can be influenced by various exogenous and endogenous factors (Abou Chacra et al., 2021). Unlike the gut microbiota, the increase in diversity appears associated with a dysbiotic status, such as BV or VVC (Farr et al., 2021; Sun et al., 2023). The clinical aspect of these conditions ranges from an asymptomatic finding in most cases to a clinically symptomatic entity that has a major impact on a woman’s lifestyle (Kroon et al., 2018). At the clinically severe end of the BV spectrum, antibiotic treatment (either systemic or localized) is associated with a 30% recurrence rate within 3 months of initial treatment and up to a 50%–70% recurrence rate within 1 year (Bradshaw et al., 2006). Therapeutic options are very limited in the subpopulation of women who have persistent or recurrent BV despite multiple attempts at antibiotic treatment (Muzny and Kardas, 2020; Muzny et al., 2022). Importantly, the probiotic treatment of symptomatic patients with oral and/or vaginal administration of Lactobacillus strains has produced mixed results (Bohbot et al., 2018), suggesting that a complete change of the vaginal microbiome may be required for an effective cure at the clinically severe end of the BV or VVC spectrum. Indeed, the same with fecal microbiota transplantation (FMT), in which feces from healthy donors are introduced into recipients’ intestines to replace their disease-associated microbiome, vaginal microbiota transplantation (VMT) from healthy female donors has recently been proposed as a therapeutic alternative for patients suffering from symptomatic, intractable and recurrent bacterial vaginosis (Lev-Sagie et al., 2019; Yockey et al., 2022).
We confirmed that the antimicrobial activity of LcM247 showed an early bacteriostatic effect during the first 4 hours of incubation and no significant impact on C. albicans, while it increased their bactericidal activity during the prolonged incubation of 24 hours, eliminating E. coli, K. pneumoniae, S. aureus and S. agalactiae bacteria, pointing out that the growth state of lactobacilli interfere with the adhesion of pathogens. To restore a homeostatic bacterial community, lactobacilli need to properly colonize the mucosa; failure in some treatments may be due to low or insufficient bacterial load due to inaccurate administration or conditions not suitable for proper Lactobacillus colonization (or temporary residence, allowing the recovery of native lactobacilli).
Meanwhile, the antimicrobial properties of CFS showed, after 4 hours, a complete inhibition of E. coli, K. pneumoniae and S. agalactiae and a significant reduction of S. aureus, E. faecalis and C. albicans. Interestingly, after 24 hours of incubation, no colonies were detected for all bacterial strains.
The CFS antimicrobial efficacy is closely tied to the pH of CFS, which exhibited an optimal activity at an acidic pH of 4.5, which significantly decreases when the pH was adjusted to neutral (7) or alkaline (9) levels. While additional molecular factors may contribute to the observed antimicrobial effect, proteomic analyses indicated a limited involvement of bacteriocins, with structural components such as S-layer proteins potentially playing a more prominent role under our experimental conditions.
Recent research suggests that Lactobacillus-based probiotics may help to restore healthy vaginal microbiota by counteracting potential pathogens (Borges et al., 2014). However, there has been growing interest over the past decade in non-living microorganisms, such as microbial extract or cell-free supernatants, which have shown therapeutic potential and beneficial effects (Pique et al., 2019). The lower diversity of vaginal microbiota compared to gut microbiota and the significant dominance of Lactobacillus prompted us to use mono-bacterial probiotics or mono-species probiotics, although recent results obtained via VMT suggest that bacteria together with bacterial products are able to restore a healthy microbial community (Lev-Sagie et al., 2019). This evidence highlights the necessity for further elucidation of the role of probiotics in the treatment of BV/VVC, with a particular focus on determining the most appropriate bacterial strains or products, the optimal timing of administration and the most effective therapeutic strategies. In other words, administrations of probiotics, which need to significantly grow and produce some molecules, may be insufficient to restore vaginal community and to cure the disease. Live bacteria can be administered either topically or orally, often requiring a long time for bacteria to reach the target site, establish themselves and begin to act. This is due to the need for live bacteria to colonize the site, a process that can be slow and subject to host and environmental factors. Indeed, studies have shown that the colonization and activity of probiotics can be inconsistent and slow to manifest (Reid et al., 2011; Borges et al., 2014). In contrast, CFS can be administered directly to the site of the infection, and thanks to its high load of bioactive molecules, including bacteriocins and its acidity, it can exert an antimicrobial effect immediately after application. In addition, non-living bacterial treatments circumvent the safety concerns associated with the use of live microorganisms, particularly in vulnerable populations such as those with compromised immune systems or during pregnancy, such as transmissible antibiotic resistance genes, systemic infections due to the bacterial translocation in human sterile sites, metabolomic disturbances and allergic responses. This makes them a potentially safer and more effective alternative to restoring healthy microbiota and inhibiting pathogenic organisms (Kothari et al., 2019).
In this context, we propose a sequential scheme to assay Lactobacillus activity against a series of indicator strains that represent some of the most etiological agents of BV or VVC. As a model of Lactobacillus, we assessed properties of LcM247, one of the most used Lactobacillus species to treat a wide spectrum of disorders including the dysregulation of the vaginal flora (Cesena et al., 2001; Zhang et al., 2018; Di Pierro et al., 2023b; Di Pierro et al., 2023a).
The use of CFS from lactobacilli for treating vaginal infections offers several advantages over traditional antibiotics or antifungals. Firstly, CFS contains multiple antimicrobial compounds that can target a broad spectrum of pathogens without promoting resistance (O’Hanlon et al., 2013; Fagan and Fairweather, 2014; Kovachev, 2018; Abdul-Rahim et al., 2021; Saha and Saroj, 2022; Assandri et al., 2023; Decout et al., 2024), a significant issue with traditional antibiotics and antifungals. Indeed, the overuse of antibiotics has led to an increase in multidrug-resistant organisms. Secondly, CFS can help to restore and maintain the natural vaginal microbiota, preventing recurring infections, while antibiotics disrupt the delicate balance of the vaginal microbiome, killing also the beneficial bacteria like lactobacilli (Ahrens et al., 2020; Ma et al., 2022).
The proteomic profile of CFS highlights the absence of a significant amount of secreted bacteriocins and, in parallel, the presence of S-layer proteins and metabolic enzymes, which thereby emerge as the most likely candidates for the observed antimicrobial activity of LcM247 CFS. The S-layer proteins form a protective layer that warrants bacterial structural integrity and mediates interactions with the environment (Hynonen and Palva, 2013; Angelescu et al., 2024). The high abundance of S-layer in the CFS results from the non-covalent interaction with bacterial cell surface carbohydrates, and this labile attachment to the bacterial cells may be very important to exert its activity in the surrounding environment (Hynonen and Palva, 2013; Fagan and Fairweather, 2014).
S-layers are made of relatively small protein subunits with a crystalline and regular structure that can quickly and efficiently refold into their active configuration and retain their functional properties, including bactericidal activity (Darbandi et al., 2022; Sugrue et al., 2024). Indeed, S-layer proteins are resistant to heat inactivation at 95°C in the free form (Sara and Sleytr, 2000; Frece et al., 2005); this thermal resistance ensures that the antimicrobial compounds produced by lactobacilli remain effective even after exposure to elevated temperatures. Moreover, S-layer proteins present a high content of acidic and hydrophobic amino acids (isoelectric point in acidic pH range) and lysine as a basic amino acid and a low content of arginine, histidine, methionine and cysteine (Sara and Sleytr, 1996). Of note, S-layer proteins are small proteins that contain few tryptophan residues that are the main target of proteinase K. Hence, the findings that CFS maintains its antimicrobial activity despite heat inactivation and proteinase K treatment are consistent with the possibility that the Lactobacillus S-layer is implicated in this activity.
The biological functions of Lactobacillus S-layer proteins are still not fully elucidated, even though many of these proteins appear to mediate bacterial adherence to host cells or interaction with host immune mediators or show protective or enzymatic functions, at least in non-pathogenic bacteria (Hynonen and Palva, 2013; Klotz et al., 2020; Assandri et al., 2023; Decout et al., 2024). In this context, only one study indicated the S-layer protein of Lactobacillus acidophilus to have murein hydrolase activity against Salmonella enterica (Prado Acosta et al., 2008). Intriguingly, an attempt to exploit these antimicrobial properties was made by Rao and colleagues, who linked the S-layer protein isolated from Lactobacillus buchneri to silver nanoparticles and assayed the antibacterial activity of this preparation against S. enterica and S. aureus. The authors observed a reduction of the MIC of their compound in comparison with silver nanoparticles alone, a higher antibiofilm activity, an increased cell membrane permeability and stronger inhibition of respiratory-chain dehydrogenase activity in treated bacteria (Rao et al., 2022). Unfortunately, the activity of the S-layer alone was not evaluated in the proper environment. It will also be important to assess the antimicrobial activity of S-layers from different Lactobacillus species or strains, considering that S-layer proteins show an overall low amino acid sequence similarity. These heterogeneities in sequence among Lactobacillus S-layer proteins may explain the differential activity of CFS originating from different species, such as the CFS from LcM247 and L. iners CFS or other Lactobacillus CFSs.
While components like S-layer proteins may contribute to antimicrobial effects, the impact of lactic acid should be critically considered (Sablon et al., 2000; Zangl et al., 2020). Indeed, lactic acid exerts antimicrobial activity, primarily in its protonated form at low pH, enabling it to permeabilize microbial membranes and, in its dissociated form, to interfere with the intracellular processes, inhibiting microbial growth or causing cell death (Diez-Gonzalez and Russell, 1997; Alakomi et al., 2000; Sablon et al., 2000). CFS’s pH-dependent activity, along with its heat and protease insensitivity, may be consistent with lactic acid activity; also, the reduced efficacy of the CFS against C. albicans may be explained by its relatively high lactic acid tolerance (Zangl et al., 2020). However, the protonated form predominates when pH < pKa (commonly recognized as approximately 3.8), and previous investigations have explored lactic acid antimicrobial activity only in conditions around this point (pH 3.6 and pH 4.0) and mainly on gram-negative bacteria (Alakomi et al., 2000). Finally, the existence of distinct, individual effects of an initial pH and initial undissociated lactic acid concentration on the bacterial inactivation suggests that lactic acid antimicrobial activity may be characterized by a certain variability (Janssen et al., 2007), an effect that we did not observe in our setting. Although we acknowledge that this may be a limitation of the current study, our findings open to further investigations that may address the complex interaction among pH, lactic acid, bacteriocins and S-layer proteins in their overall synergistic antimicrobial activity in the multifaceted field of lactic acid bacteria (Saha and Saroj, 2022).
LcM247 and particularly its CFS have been shown to contribute to the eradication of common pathobionts, suggesting that commercially available probiotics based on this strain may effectively restore a healthy vaginal microbiome. We clearly show that actively replicating LcM247 is less efficient than its CFS, so the oral administration of LcM247 may result in the failure of the treatment. Finally, the use of CFS may result in an upswing of the host L. crispatus strains, which conversely may show a minor tolerance to antibiotic treatment (Costantini et al., 2021). However, our findings together with phylogenetic information regarding L. crispatus strains suggest that the identification of CST-I alone may be not sufficient to describe a healthy vaginal community, prompting for a fine and deep characterization of more strains. As previously demonstrated, significant differences emerged when investigating L. crispatus strains isolated from human hosts or other hosts, as well as isolated from diverse host biological environments, intrinsically displaying different growth patterns and impacting their colonization and persistence (Pan et al., 2019; Hulyalkar et al., 2021). A better understanding of these strains may clarify their role in preserving and regulating vaginal homeostasis during the fertile age.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
GS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. RR: Formal analysis, Investigation, Methodology, Writing – review & editing. MC: Investigation, Methodology, Writing – review & editing. FI: Formal analysis, Visualization, Writing – review & editing. AU: Visualization, Writing – review & editing. MS: Funding acquisition, Visualization, Writing – review & editing. GD: Conceptualization, Supervision, Visualization, Writing – original draft, Writing – review & editing. FD: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. We acknowledge EU funding for the MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project number PE00000007, INF-ACT).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1586442/full#supplementary-material
Supplementary Figure 1 | CFS pH measurement corresponding to serial dilutions and alkalinization. LcM247 CFS was serially diluted at a ratio of 1:1 v/v in MRS broth and pH was measured for each obtained condition. Additionally, CFS was alkalinized by adding different volumes of 1M NaOH until four different conditions were achieved: unaltered CFS (pH 4.5), CFS at pH 5.5, CFS at pH 7, and CFS at pH 9.
Supplementary Figure 2 | LcM247 CFS and L. iners CFS does not have a reciprocal inhibition. Preserving a healthy vaginal microbiota depends on the Lactobacillus species that dominate the vaginal microenvironment. The protective role of LcM247 was assessed against L. iners (typically identifying CST-III) associated with microbiota prone to develop disease. We investigate the interaction between the supernatant of L. crispatus M247 (LcM247) and L. iners cells, and the interaction between the supernatant of L. iners and LcM247. We performed an antimicrobial assay mixing serial dilutions of CFS and Lactobacillus v/v 1:1 and incubating for 24 hours at 37°C. Untreated lactobacilli, incubating in MRS medium v/v 1:1 was used as positive control. Finally, colony forming units (CFUs) were assessed and two bar plots showing Log CFUs/ml of L. iners after incubation with LcM247 CFS (A) and Log CFUs/ml of LcM247 incubating with L. iners CFS (B) were generated by measures of three repeated experiments.
Supplementary Figure 3 | In vivo toxicity. Toxicity survival curve of gentamicin, fluconazole, LcM247 and CFS/2 and their combinations on G. mellonella larvae monitored every 24 hours till 96 hours. G. mellonella larvae were inoculated with 10 µl of 10 µg/ml gentamicin/fluconazole, 5 x 10^7 LcM247, CFS/2 (1:1 v/v) and with the combination of these compounds.
Supplementary Table 1 | Bacteriocins detected in Lactobacillus crispatus M247 from BAGEL 4. The document includes a table listing the Areas of Interest (AOI) for each bacteriocin gene detected from BAGEL 4 in the LcM247 genome, specifying the start and end positions. Additionally, the corresponding nucleotide and protein sequences for every identified bacteriocin are included.
Supplementary Table 2 | Peptides detected by proteomic analysis.
Supplementary Table 3 | Comprehensive Proteomic Analysis of the Cell-Free Supernatant (CFS) from LcM247 founded with Lactobacillus crispatus database.
Supplementary Table 4 | Comprehensive Proteomic Analysis of the Cell-Free Supernatant (CFS) from LcM247 founded with Lactobacillus genus database.
References
Abbe, C. and Mitchell, C. M. (2023). Bacterial vaginosis: a review of approaches to treatment and prevention. Front. Reprod. Health 5, 1100029. doi: 10.3389/frph.2023.1100029
Abdul-Rahim, O., Wu, Q., Price, T. K., Pistone, G., Diebel, K., Bugni, T. S., et al. (2021). Phenyl-lactic acid is an active ingredient in bactericidal supernatants of lactobacillus crispatus. J. Bacteriol. 203, e0036021. doi: 10.1128/JB.00360-21
Abou Chacra, L., Fenollar, F., and Diop, K. (2021). Bacterial vaginosis: what do we currently know? Front. Cell Infect. Microbiol. 11, 672429. doi: 10.3389/fcimb.2021.672429
Ahrens, P., Andersen, L. O., Lilje, B., Johannesen, T. B., Dahl, E. G., Baig, S., et al. (2020). Changes in the vaginal microbiota following antibiotic treatment for Mycoplasma genitalium, Chlamydia trachomatis and bacterial vaginosis. PloS One 15, e0236036. doi: 10.1371/journal.pone.0236036
Alakomi, H. L., Skytta, E., Saarela, M., Mattila-Sandholm, T., Latva-Kala, K., and Helander, I. M. (2000). Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 66, 2001–2005. doi: 10.1128/AEM.66.5.2001-2005.2000
Anderson, A. C., Sanunu, M., Schneider, C., Clad, A., Karygianni, L., Hellwig, E., et al. (2014). Rapid species-level identification of vaginal and oral lactobacilli using MALDI-TOF MS analysis and 16S rDNA sequencing. BMC Microbiol. 14, 312. doi: 10.1186/s12866-014-0312-5
Angelescu, I.-R. Z., Ionetic, E.-C., and Grosu-Tudor, S.-S. (2024). The biological role of the S-Layer produced by lactobacillus helveticus 34.9 in cell protection and its probiotic properties. Fermentation 10, 150. doi: 10.3390/fermentation10030150
Assandri, M. H., Malamud, M., Trejo, F. M., and Serradell, M. L. A. (2023). S-layer proteins as immune players: Tales from pathogenic and non-pathogenic bacteria. Curr. Res. Microb. Sci. 4, 100187. doi: 10.1016/j.crmicr.2023.100187
Biswas, S. and Rolain, J. M. (2013). Use of MALDI-TOF mass spectrometry for identification of bacteria that are difficult to culture. J. Microbiol. Methods 92, 14–24. doi: 10.1016/j.mimet.2012.10.014
Bohbot, J. M., Darai, E., Bretelle, F., Brami, G., Daniel, C., and Cardot, J. M. (2018). Efficacy and safety of vaginally administered lyophilized Lactobacillus crispatus IP 174178 in the prevention of bacterial vaginosis recurrence. J. Gynecol. Obstet. Hum. Reprod. 47, 81–86. doi: 10.1016/j.jogoh.2017.11.005
Borges, S., Silva, J., and Teixeira, P. (2014). The role of lactobacilli and probiotics in maintaining vaginal health. Arch. Gynecol. Obstet. 289, 479–489. doi: 10.1007/s00404-013-3064-9
Bradshaw, C. S., Morton, A. N., Hocking, J., Garland, S. M., Morris, M. B., Moss, L. M., et al. (2006). High recurrence rates of bacterial vaginosis over the course of 12 months after oral metronidazole therapy and factors associated with recurrence. J. Infect. Dis. 193, 1478–1486. doi: 10.1086/jid.2006.193.issue-11
Cesena, C., Morelli, L., Alander, M., Siljander, T., Tuomola, E., Salminen, S., et al. (2001). Lactobacillus crispatus and its nonaggregating mutant in human colonization trials. J. Dairy Sci. 84, 1001–1010. doi: 10.3168/jds.S0022-0302(01)74559-6
Chen, X., Lu, Y., Chen, T., and Li, R. (2021). The female vaginal microbiome in health and bacterial vaginosis. Front. Cell Infect. Microbiol. 11, 631972. doi: 10.3389/fcimb.2021.631972
Cohen, C. R., Wierzbicki, M. R., French, A. L., Morris, S., Newmann, S., Reno, H., et al. (2020). Randomized trial of lactin-V to prevent recurrence of bacterial vaginosis. N. Engl. J. Med. 382, 1906–1915. doi: 10.1056/NEJMoa1915254
Costantini, P. E., Firrincieli, A., Fedi, S., Parolin, C., Viti, C., Cappelletti, M., et al. (2021). Insight into phenotypic and genotypic differences between vaginal Lactobacillus crispatus BC5 and Lactobacillus gasseri BC12 to unravel nutritional and stress factors influencing their metabolic activity. Microb. Genom. 7. doi: 10.1099/mgen.0.000575
Darbandi, A., Asadi, A., Mahdizade Ari, M., Ohadi, E., Talebi, M., Halaj Zadeh, M., et al. (2022). Bacteriocins: Properties and potential use as antimicrobials. J. Clin. Lab. Anal. 36, e24093. doi: 10.1002/jcla.24093
Decout, A., Krasias, I., Roberts, L., Gimeno Molina, B., Charenton, C., Brown Romero, D., et al. (2024). Lactobacillus crispatus S-layer proteins modulate innate immune response and inflammation in the lower female reproductive tract. Nat. Commun. 15, 10879. doi: 10.1038/s41467-024-55233-7
de Jong, A., van Hijum, S. A., Bijlsma, J. J., Kok, J., and Kuipers, O. P. (2006). BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res. 34, W273–W279. doi: 10.1093/nar/gkl237
De Maio, F., Palmieri, V., Babini, G., Augello, A., Palucci, I., Perini, G., et al. (2021). Graphene nanoplatelet and graphene oxide functionalization of face mask materials inhibits infectivity of trapped SARS-CoV-2. iScience 24, 102788. doi: 10.1016/j.isci.2021.102788
Diez-Gonzalez, F. and Russell, J. B. (1997). The ability of Escherichia coli O157:H7 to decrease its intracellular pH and resist the toxicity of acetic acid. Microbiol. (Reading) 143, 1175–1180. doi: 10.1099/00221287-143-4-1175
Di Pierro, F., Bertuccioli, A., Cazzaniga, M., Zerbinati, N., and Guasti, L. (2023a). A clinical report highlighting some factors influencing successful vaginal colonization with probiotic Lactobacillus crispatus. Minerva Med. 114, 883–887. doi: 10.23736/S0026-4806.23.08773-6
Di Pierro, F., Polzonetti, V., Patrone, V., and Morelli, L. (2019). Microbiological assessment of the quality of some commercial products marketed as lactobacillus crispatus-containing probiotic dietary supplements. Microorganisms 7. doi: 10.3390/microorganisms7110524
Di Pierro, F., Sinatra, F., Cester, M., Da Ros, L., Pistolato, M., Da Pare, V., et al. (2023b). Effect of L. crispatus M247 administration on pregnancy outcomes in women undergoing IVF: A controlled, retrospective, observational, and open-Label study. Microorganisms 11. doi: 10.20944/preprints202310.0216.v1
Distler, U., Kuharev, J., Navarro, P., and Tenzer, S. (2016). Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Protoc. 11, 795–812. doi: 10.1038/nprot.2016.042
Edelman, S. M., Lehti, T. A., Kainulainen, V., Antikainen, J., Kylvaja, R., Baumann, M., et al. (2012). Identification of a high-molecular-mass Lactobacillus epithelium adhesin (LEA) of Lactobacillus crispatus ST1 that binds to stratified squamous epithelium. Microbiol. (Reading) 158, 1713–1722. doi: 10.1099/mic.0.057216-0
Fagan, R. P. and Fairweather, N. F. (2014). Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222. doi: 10.1038/nrmicro3213
Farr, A., Effendy, I., Frey Tirri, B., Hof, H., Mayser, P., Petricevic, L., et al. (2021). Guideline: Vulvovaginal candidosis (AWMF 015/072, level S2k). Mycoses 64, 583–602. doi: 10.1111/myc.13248
Feehily, C., Crosby, D., Walsh, C. J., Lawton, E. M., Higgins, S., McAuliffe, F. M., et al. (2020). Shotgun sequencing of the vaginal microbiome reveals both a species and functional potential signature of preterm birth. NPJ Biofilms Microbiomes 6, 50. doi: 10.1038/s41522-020-00162-8
Fettweis, J. M., Brooks, J. P., Serrano, M. G., Sheth, N. U., Girerd, P. H., Edwards, D. J., et al. (2014). Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiol. (Reading) 160, 2272–2282. doi: 10.1099/mic.0.081034-0
Fontana, F., Alessandri, G., Lugli, G. A., Mancabelli, L., Longhi, G., Anzalone, R., et al. (2020). Probiogenomics analysis of 97 lactobacillus crispatus strains as a tool for the identification of promising next-generation probiotics. Microorganisms 9. doi: 10.3390/microorganisms9010073
France, M., Alizadeh, M., Brown, S., Ma, B., and Ravel, J. (2022). Towards a deeper understanding of the vaginal microbiota. Nat. Microbiol. 7, 367–378. doi: 10.1038/s41564-022-01083-2
France, M. T., Ma, B., Gajer, P., Brown, S., Humphrys, M. S., Holm, J. B., et al. (2020). VALENCIA: a nearest centroid classification method for vaginal microbial communities based on composition. Microbiome 8, 166. doi: 10.1186/s40168-020-00934-6
Frece, J., Kos, B., Svetec, I. K., Zgaga, Z., Mrsa, V., and Suskovic, J. (2005). Importance of S-layer proteins in probiotic activity of Lactobacillus acidophilus M92. J. Appl. Microbiol. 98, 285–292. doi: 10.1111/j.1365-2672.2004.02473.x
Greenbaum, S., Greenbaum, G., Moran-Gilad, J., and Weintraub, A. Y. (2019). Ecological dynamics of the vaginal microbiome in relation to health and disease. Am. J. Obstet. Gynecol. 220, 324–335. doi: 10.1016/j.ajog.2018.11.1089
Hugenholtz, F., van der Veer, C., Terpstra, M. L., Borgdorff, H., van Houdt, R., Bruisten, S., et al. (2022). Urine and vaginal microbiota compositions of postmenopausal and premenopausal women differ regardless of recurrent urinary tract infection and renal transplant status. Sci. Rep. 12, 2698. doi: 10.1038/s41598-022-06646-1
Hulyalkar, N. V., Sharon, B. M., Shipman, B. M., Arute, A. P., Zimmern, P. E., and De Nisco, N. J. (2021). Complete genome sequences of three lactobacillus crispatus strains isolated from the urine of postmenopausal women. Microbiol. Resour. Announc. 10, e0101721. doi: 10.1128/MRA.01017-21
Hynonen, U. and Palva, A. (2013). Lactobacillus surface layer proteins: structure, function and applications. Appl. Microbiol. Biotechnol. 97, 5225–5243. doi: 10.1007/s00253-013-4962-2
Janssen, M., Geeraerd, A. H., Cappuyns, A., Garcia-Gonzalez, L., Schockaert, G., Van Houteghem, N., et al. (2007). Individual and combined effects of ph and lactic acid concentration on Listeria innocua inactivation: development of a predictive model and assessment of experimental variability. Appl. Environ. Microbiol. 73, 1601–1611. doi: 10.1128/AEM.02198-06
Joseph, R. J., Ser, H. L., Kuai, Y. H., Tan, L. T., Arasoo, V. J. T., Letchumanan, V., et al. (2021). Finding a balance in the vaginal microbiome: how do we treat and prevent the occurrence of bacterial vaginosis? Antibiot. (Basel) 10. doi: 10.3390/antibiotics10060719
Kaewsrichan, J., Peeyananjarassri, K., and Kongprasertkit, J. (2006). Selection and identification of anaerobic lactobacilli producing inhibitory compounds against vaginal pathogens. FEMS Immunol. Med. Microbiol. 48, 75–83. doi: 10.1111/j.1574-695X.2006.00124.x
Khedkar, R. and Pajai, S. (2022). Bacterial vaginosis: A comprehensive narrative on the etiology, clinical features, and management approach. Cureus 14, e31314. doi: 10.7759/cureus.31314
Klotz, C., Goh, Y. J., O’Flaherty, S., and Barrangou, R. (2020). S-layer associated proteins contribute to the adhesive and immunomodulatory properties of Lactobacillus acidophilus NCFM. BMC Microbiol. 20, 248. doi: 10.1186/s12866-020-01908-2
Kohler, G. (2015). Probiotics research in Galleria mellonella. Virulence 6, 3–5. doi: 10.1080/21505594.2014.998967
Koley, D. and Bard, A. J. (2010). Triton X-100 concentration effects on membrane permeability of a single HeLa cell by scanning electrochemical microscopy (SECM). Proc. Natl. Acad. Sci. U.S.A. 107, 16783–16787. doi: 10.1073/pnas.1011614107
Kothari, D., Patel, S., and Kim, S. K. (2019). Probiotic supplements might not be universally-effective and safe: A review. BioMed. Pharmacother. 111, 537–547. doi: 10.1016/j.biopha.2018.12.104
Kovachev, S. (2018). Defence factors of vaginal lactobacilli. Crit. Rev. Microbiol. 44, 31–39. doi: 10.1080/1040841X.2017.1306688
Kroon, S. J., Ravel, J., and Huston, W. M. (2018). Cervicovaginal microbiota, women’s health, and reproductive outcomes. Fertil. Steril. 110, 327–336. doi: 10.1016/j.fertnstert.2018.06.036
Kumar, M., Yadav, A. K., Verma, V., Singh, B., Mal, G., Nagpal, R., et al. (2016). Bioengineered probiotics as a new hope for health and diseases: an overview of potential and prospects. Future Microbiol. 11, 585–600. doi: 10.2217/fmb.16.4
Lebeer, S., Ahannach, S., Gehrmann, T., Wittouck, S., Eilers, T., Oerlemans, E., et al. (2023). A citizen-science-enabled catalogue of the vaginal microbiome and associated factors. Nat. Microbiol. 8, 2183–2195. doi: 10.1038/s41564-023-01500-0
Lepargneur, J. P. (2016). Lactobacillus crispatus as biomarker of the healthy vaginal tract. Ann. Biol. Clin. (Paris) 74, 421–427. doi: 10.1684/abc.2016.1169
Lev-Sagie, A., Goldman-Wohl, D., Cohen, Y., Dori-Bachash, M., Leshem, A., Mor, U., et al. (2019). Vaginal microbiome transplantation in women with intractable bacterial vaginosis. Nat. Med. 25, 1500–1504. doi: 10.1038/s41591-019-0600-6
Li, H. G., Zhang, X., Mu, H., Sha, S., Lin, Y., Hou, H., et al. (2022). Whole-genome sequencing combined with mass spectrometry to identify bacteriocin and mine silent genes. LWT 169. doi: 10.1016/j.lwt.2022.113975
Ligeza-Zuber, M. (2012). Mechanisms of Galleria mellonella cellular immune response after infection with entomopathogenic fungus Conidiobolus coronatus. Ann. Parasitol. 58, 227–228.
Ma, Z. S. and Li, L. (2017). Quantifying the human vaginal community state types (CSTs) with the species specificity index. PeerJ 5, e3366. doi: 10.7717/peerj.3366
Ma, X., Wu, M., Wang, C., Li, H., Fan, A., Wang, Y., et al. (2022). The pathogenesis of prevalent aerobic bacteria in aerobic vaginitis and adverse pregnancy outcomes: a narrative review. Reprod. Health 19, 21. doi: 10.1186/s12978-021-01292-8
Mani-Lopez, E., Arrioja-Breton, D., and Lopez-Malo, A. (2022). The impacts of antimicrobial and antifungal activity of cell-free supernatants from lactic acid bacteria in vitro and foods. Compr. Rev. Food Sci. Food Saf. 21, 604–641. doi: 10.1111/1541-4337.12872
Masucci, L., D’Ippolito, S., De Maio, F., Quaranta, G., Mazzarella, R., Bianco, D. M., et al. (2023). Celiac disease predisposition and genital tract microbiota in women affected by recurrent pregnancy loss. Nutrients 15. doi: 10.3390/nu15010221
Mattei, B., Lira, R. B., Perez, K. R., and Riske, K. A. (2017). Membrane permeabilization induced by Triton X-100: The role of membrane phase state and edge tension. Chem. Phys. Lipids 202, 28–37. doi: 10.1016/j.chemphyslip.2016.11.009
Murphy, K. and Mitchell, C. M. (2016). The interplay of host immunity, environment and the risk of bacterial vaginosis and associated reproductive health outcomes. J. Infect. Dis. 214 Suppl 1, S29–S35. doi: 10.1093/infdis/jiw140
Muzny, C. A., Balkus, J., Mitchell, C., Sobel, J. D., Workowski, K., Marrazzo, J., et al. (2022). Diagnosis and management of bacterial vaginosis: summary of evidence reviewed for the 2021 centers for disease control and prevention sexually transmitted infections treatment guidelines. Clin. Infect. Dis. 74, S144–S151. doi: 10.1093/cid/ciac021
Muzny, C. A. and Kardas, P. (2020). A narrative review of current challenges in the diagnosis and management of bacterial vaginosis. Sex Transm. Dis. 47, 441–446. doi: 10.1097/OLQ.0000000000001178
Nair, D., Memmi, G., Hernandez, D., Bard, J., Beaume, M., Gill, S., et al. (2011). Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J. Bacteriol. 193, 2332–2335. doi: 10.1128/JB.00027-11
Norenhag, J., Du, J., Olovsson, M., Verstraelen, H., Engstrand, L., and Brusselaers, N. (2020). The vaginal microbiota, human papillomavirus and cervical dysplasia: a systematic review and network meta-analysis. BJOG 127, 171–180. doi: 10.1111/1471-0528.15854
Nyirjesy, P., Brookhart, C., Lazenby, G., Schwebke, J., and Sobel, J. D. (2022). Vulvovaginal candidiasis: A review of the evidence for the 2021 centers for disease control and prevention of sexually transmitted infections treatment guidelines. Clin. Infect. Dis. 74, S162–S168. doi: 10.1093/cid/ciab1057
O’Hanlon, D. E., Moench, T. R., and Cone, R. A. (2013). Vaginal pH and microbicidal lactic acid when lactobacilli dominate the microbiota. PloS One 8, e80074. doi: 10.1371/journal.pone.0080074
Pan, M., Hidalgo-Cantabrana, C., Goh, Y. J., Sanozky-Dawes, R., and Barrangou, R. (2019). Comparative Analysis of Lactobacillus gasseri and Lactobacillus crispatus Isolated From Human Urogenital and Gastrointestinal Tracts. Front. Microbiol. 10, 3146. doi: 10.3389/fmicb.2019.03146
Pendharkar, S., Skafte-Holm, A., Simsek, G., and Haahr, T. (2023). Lactobacilli and their probiotic effects in the vagina of reproductive age women. Microorganisms 11. doi: 10.3390/microorganisms11030636
Pique, N., Berlanga, M., and Minana-Galbis, D. (2019). Health benefits of heat-killed (Tyndallized) probiotics: an overview. Int. J. Mol. Sci. 20. doi: 10.3390/ijms20102534
Prado Acosta, M., Mercedes Palomino, M., Allievi, M. C., Sanchez Rivas, C., and Ruzal, S. M. (2008). Murein hydrolase activity in the surface layer of Lactobacillus acidophilus ATCC 4356. Appl. Environ. Microbiol. 74, 7824–7827. doi: 10.1128/AEM.01712-08
Qi, F., Fan, S., Fang, C., Ge, L., Lyu, J., Huang, Z., et al. (2023). Orally administrated Lactobacillus gasseri TM13 and Lactobacillus crispatus LG55 can restore the vaginal health of patients recovering from bacterial vaginosis. Front. Immunol. 14, 1125239. doi: 10.3389/fimmu.2023.1125239
Rajab, S., Tabandeh, F., Shahraky, M. K., and Alahyaribeik, S. (2020). The effect of lactobacillus cell size on its probiotic characteristics. Anaerobe 62, 102103. doi: 10.1016/j.anaerobe.2019.102103
Rao, S. Q., Zhang, R. Y., Chen, R., Gao, Y. J., Gao, L., and Yang, Z. Q. (2022). Nanoarchitectonics for enhanced antibacterial activity with Lactobacillus buchneri S-layer proteins-coated silver nanoparticles. J. Hazard. Mater. 426, 128029. doi: 10.1016/j.jhazmat.2021.128029
Ravel, J., Moreno, I., and Simon, C. (2021). Bacterial vaginosis and its association with infertility, endometritis, and pelvic inflammatory disease. Am. J. Obstet. Gynecol. 224, 251–257. doi: 10.1016/j.ajog.2020.10.019
Reid, G., Younes, J. A., van der Mei, H. C., Gloor, G. B., Knight, R., and Busscher, H. J. (2011). Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat. Rev. Microbiol. 9, 27–38. doi: 10.1038/nrmicro2473
Rosato, R., Santarelli, G., Augello, A., Perini, G., De Spirito, M., Sanguinetti, M., et al. (2024). Exploration of the graphene quantum dots-blue light combination: A promising treatment against bacterial infection. Int. J. Mol. Sci. 25. doi: 10.3390/ijms25158033
Rose, W. A., 2nd, McGowin, C. L., Spagnuolo, R. A., Eaves-Pyles, T. D., Popov, V. L., and Pyles, R. B. (2012). : Commensal bacteria modulate innate immune responses of vaginal epithelial cell multilayer cultures. PloS One 7, e32728. doi: 10.1371/journal.pone.0032728
Sablon, E., Contreras, B., and Vandamme, E. (2000). Antimicrobial peptides of lactic acid bacteria: mode of action, genetics and biosynthesis. Adv. Biochem. Eng. Biotechnol. 68, 21–60. doi: 10.1007/3-540-45564-7_2
Saha, U. B. and Saroj, S. D. (2022). Lactic acid bacteria: prominent player in the fight against human pathogens. Expert Rev. Anti Infect. Ther. 20, 1435–1453. doi: 10.1080/14787210.2022.2128765
Santarelli, G., Perini, G., Salustri, A., Palucci, I., Rosato, R., Palmieri, V., et al. (2024). Unraveling the potential of graphene quantum dots against Mycobacterium tuberculosis infection. Front. Microbiol. 15, 1395815. doi: 10.3389/fmicb.2024.1395815
Sara, M. and Sleytr, U. B. (1996). Crystalline bacterial cell surface layers (S-layers): from cell structure to biomimetics. Prog. Biophys. Mol. Biol. 65, 83–111. doi: 10.1016/S0079-6107(96)00007-7
Sara, M. and Sleytr, U. B. (2000). S-layer proteins. J. Bacteriol. 182, 859–868. doi: 10.1128/JB.182.4.859-868.2000
Shen, L., Zhang, W., Yuan, Y., Zhu, W., and Shang, A. (2022). Vaginal microecological characteristics of women in different physiological and pathological period. Front. Cell Infect. Microbiol. 12, 959793. doi: 10.3389/fcimb.2022.959793
Sichtig, H., Minogue, T., Yan, Y., Stefan, C., Hall, A., Tallon, L., et al. (2019). FDA-ARGOS is a database with public quality-controlled reference genomes for diagnostic use and regulatory science. Nat. Commun. 10, 3313. doi: 10.1038/s41467-019-11306-6
Smith, S. B. and Ravel, J. (2017). The vaginal microbiota, host defence and reproductive physiology. J. Physiol. 595, 451–463. doi: 10.1113/tjp.2017.595.issue-2
Sobel, J. D. (2007). Vulvovaginal candidosis. Lancet 369, 1961–1971. doi: 10.1016/S0140-6736(07)60917-9
Stoyancheva, G., Marzotto, M., Dellaglio, F., and Torriani, S. (2014). Bacteriocin production and gene sequencing analysis from vaginal Lactobacillus strains. Arch. Microbiol. 196, 645–653. doi: 10.1007/s00203-014-1003-1
Sugrue, I., Ross, R. P., and Hill, C. (2024). Bacteriocin diversity, function, discovery and application as antimicrobials. Nat. Rev. Microbiol. 22, 556–571. doi: 10.1038/s41579-024-01045-x
Sun, Z., Ge, X., Qiu, B., Xiang, Z., Jiang, C., Wu, J., et al. (2023). Vulvovaginal candidiasis and vaginal microflora interaction: Microflora changes and probiotic therapy. Front. Cell Infect. Microbiol. 13, 1123026. doi: 10.3389/fcimb.2023.1123026
van Heel, A. J., de Jong, A., Song, C., Viel, J. H., Kok, J., and Kuipers, O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281. doi: 10.1093/nar/gky383
van Houdt, R., Ma, B., Bruisten, S. M., Speksnijder, A., Ravel, J., and de Vries, H. J. C. (2018). Lactobacillus iners-dominated vaginal microbiota is associated with increased susceptibility to Chlamydia trachomatis infection in Dutch women: a case-control study. Sex Transm. Infect. 94, 117–123. doi: 10.1136/sextrans-2017-053133
Wiesenfeld, H. C., Hillier, S. L., Krohn, M. A., Amortegui, A. J., Heine, R. P., Landers, D. V., et al. (2002). Lower genital tract infection and endometritis: insight into subclinical pelvic inflammatory disease. Obstet. Gynecol. 100, 456–463. doi: 10.1097/00006250-200209000-00011
Wisniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Yockey, L. J., Hussain, F. A., Bergerat, A., Reissis, A., Worrall, D., Xu, J., et al. (2022). Screening and characterization of vaginal fluid donations for vaginal microbiota transplantation. Sci. Rep. 12, 17948. doi: 10.1038/s41598-022-22873-y
Zangl, I., Beyer, R., Pap, I. J., Strauss, J., Aspock, C., Willinger, B., et al. (2020). Human pathogenic candida species respond distinctively to lactic acid stress. J. Fungi (Basel) 6. doi: 10.3390/jof6040348
Keywords: Lactobacillus crispatus, vaginal infections, probiotics, cell-free supernatant, LcM247
Citation: Santarelli G, Rosato R, Cicchinelli M, Iavarone F, Urbani A, Sanguinetti M, Delogu G and De Maio F (2025) The activity of cell-free supernatant of Lactobacillus crispatus M247: a promising treatment against vaginal infections. Front. Cell. Infect. Microbiol. 15:1586442. doi: 10.3389/fcimb.2025.1586442
Received: 02 March 2025; Accepted: 12 May 2025;
Published: 11 June 2025.
Edited by:
Carola Parolin, University of Bologna, ItalyReviewed by:
Lorenzo Colombini, University of Siena, ItalyJelle Dillen, University of Antwerp, Belgium
Copyright © 2025 Santarelli, Rosato, Cicchinelli, Iavarone, Urbani, Sanguinetti, Delogu and De Maio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Flavio De Maio, ZmxhdmlvLmRlbWFpb0Bwb2xpY2xpbmljb2dlbWVsbGkuaXQ=
†These authors have contributed equally to this work
 Giulia Santarelli
Giulia Santarelli Roberto Rosato
Roberto Rosato Michela Cicchinelli2
Michela Cicchinelli2 Federica Iavarone
Federica Iavarone Maurizio Sanguinetti
Maurizio Sanguinetti Giovanni Delogu
Giovanni Delogu Flavio De Maio
Flavio De Maio