- 1Xiangya Stomatological Hospital and Xiangya School of Stomatology, Central South University, Changsha, China
- 2Hunan Key Laboratory of Oral Health Research, Central South University, Changsha, China
- 3School of Life Sciences, Central South University, Changsha, China
- 4Department of Oral and Maxillofacial Surgery, The Second Xiangya Hospital, Central South University, Changsha, China
Post-translational modifications (PTMs) are integral to the regulation of protein function, stability, and cellular processes. Lysine acetylation, a widespread PTM, has been extensively characterized for its role in eukaryotic cellular functions, particularly in metabolism, gene expression, and disease progression. However, its involvement in oral microbiota remains inadequately explored. This review examines the emerging significance of lysine acetylation in modulating oral microbial communities. The oral cavity, characterized by its unique anatomical and environmental conditions, serves as a dynamic habitat where microbiota interact with host factors such as diet, immune response, pH, and the level of oxygen. Lysine acetylation enables bacterial adaptation to these fluctuating conditions, influencing microbial metabolism, virulence, and stress responses. For example, acetylation of lactate dehydrogenase in Streptococcus mutans reduces its acidogenicity and aciduricity, which decreases its cariogenic potential. In diverse environmental conditions, including hypoxic or anaerobic environments, acetylation regulates energy utilization pathways and enzyme activities, supporting bacterial survival and adaptation. Additionally, acetylation controls the production of extracellular polysaccharides (EPS), which are essential for biofilm formation and bacterial colonization. The acetylation of virulence factors can modulate the pathogenic potential of oral bacteria, either enhancing or inhibiting their activity depending on the specific context and regulatory mechanisms involved. This review also explores the interactions between acetylation and other PTMs, highlighting their synergistic or antagonistic effects on protein function. A deeper understanding of lysine acetylation mechanisms in oral microbiota could provide valuable insights into microbial adaptation and pathogenesis, revealing potential therapeutic targets for oral diseases.
1 Introduction
Post-translational modification (PTM) refers to the chemical modification of proteins following translation, which influences their structure and function. These modifications can impact protein stability, affinity, activity, and subcellular localization, thereby regulating their biological roles. In the context of oral microbiota, PTMs are integral in modulating protein synthesis, metabolism, and virulence. Oral microorganisms utilize PTMs to adapt to external stimuli and control various physiological processes. To date, over 200 distinct PTMs have been cataloged (Minguez et al., 2012), including both minor chemical modifications (e.g., phosphorylation and acetylation) and the addition of complete proteins (e.g., ubiquitination). The most common PTMs observed in oral microbiota include phosphorylation, acetylation, methylation, glycosylation, sumoylation, and lactylation (Wu et al., 2019; Zeng et al., 2020; Ma et al., 2021b).
Lysine is an amphipathic residue, characterized by a hydrophobic side chain and a positively charged ϵN-group at physiological pH. The ϵN-group in the active or binding site of proteins typically engages in salt bridge formation (Moreira et al., 2007). Acylation of lysine neutralizes the amino group’s positive charge, potentially altering the protein’s conformation. Various acylation modifications have been identified, including acetylation (Wang et al., 2010), malonylation (Peng et al., 2011), crotonylation (Montellier et al., 2012), propionylation and butylation (Chen et al., 2007), and succinylation (Lin et al., 2012). These modifications utilize metabolic intermediates as sensors to regulate metabolism and other processes, thereby coordinating metabolic pathways and signal transduction (Wellen and Thompson, 2012).
The most extensively studied lysine modification is acetylation, which occurs through reversible catalysis by protein acetyltransferases and deacetylases. Active acetyl derivatives, such as acetyl phosphate, acetyl CoA, and acetyladenylic acid, are also known to drive protein acetylation (Ramponi et al., 1975; Wagner and Payne, 2013; Weinert et al., 2013; Kuhn et al., 2014). Initially observed in histones (Allfrey et al., 1964), lysine acetylation has since been identified in various eukaryotic non-histone proteins involved in cellular metabolism, the cell cycle, aging, growth, angiogenesis, and oncogenesis (Chuang et al., 2010; Wang et al., 2010; Zhao et al., 2010; Lu et al., 2011; Carafa et al., 2012; Lin et al., 2012). In contrast, research on prokaryotic acetylation remains limited, primarily focusing on a few microbial species. Although lysine acetylation is increasingly recognized as a significant post-translational modification in bacteria, its specific roles and regulatory mechanisms within oral microbiota remain underexplored.
The oral microenvironment is intricate, shaped by the unique anatomy of the oral cavity. Oral microorganisms are highly sensitive to host factors, including diet, immune status, pH, oral hygiene, oxygen levels, and lifestyle choices (Yan et al., 2023). In contrast to slower regulatory mechanisms such as gene expression and protein turnover, lysine acetylation enables bacteria to rapidly adjust their physiological state, offering a mechanism to respond swiftly to environmental changes. For instance, under hypoxic or anaerobic conditions, lysine acetylation modulates bacterial metabolic pathways, optimizing energy utilization and regulating enzymatic activity to mitigate external stressors (Kim et al., 2015; Martín et al., 2021; Toplak et al., 2022; Ma et al., 2024a).
In addition, protein lysine acetylation is a key regulator of microbial virulence. One noteworthy illustration is the production of extracellular polysaccharides (EPS) by oral bacteria via glucosyltransferases. EPS constitutes the primary component of dental plaque biofilms, promoting bacterial colonization and aggregation. Notably, the acetylation status of glucosyltransferase correlates with its enzymatic activity (Ma et al., 2021a). Beyond local effects, oral pathogens can release virulence factors into the bloodstream, leading to systemic infections. Acetylation plays a critical role in the onset and progression of these virulence factors (Ren et al., 2017).
Protein acetylation differs from mRNA or protein synthesis in that it is not templated, instead depending on the recognition and modification by specific enzymes. This process is typically reversible and dynamic, with chemical groups being added or removed from the polypeptide chain by specialized enzymes. Beyond enzymes, certain chemical compounds also contribute to protein acetylation. For instance, acetyl coenzyme A (Ac-CoA) directly enhances the acetylation of RprY in Porphyromonas gingivalis (Wagner and Payne, 2013). Proteins often undergo multiple modifications, with some residues experiencing several modifications simultaneously. For example, in Porphyromonas gingivalis, acetylation and succinylation frequently overlap, and RprY can concurrently undergo both acetylation and phosphorylation (Li et al., 2018). The interplay between acetylation and other PTMs can produce complementary or antagonistic effects, resulting in intricate combinations that influence the structure and function of target proteins, highlighting the complexity and adaptability of regulatory mechanisms.
Recent advances in enrichment strategies for protein lysine acetylation sites have provided substantial evidence supporting the critical role of protein acetylation in regulating both the physiological and pathological processes of oral microorganisms (Figure 1). This review explores the significance and diverse functions of protein lysine acetylation in oral microorganisms, aiming to fill current knowledge gaps and explore the potential of acetylation as a therapeutic target for oral diseases by integrating emerging evidence across major oral microorganisms, including bacteria (e.g., Streptococcus mutans, Porphyromonas gingivalis) and fungi (e.g., Candida albicans), and highlighting its interplay with other post-translational modifications.
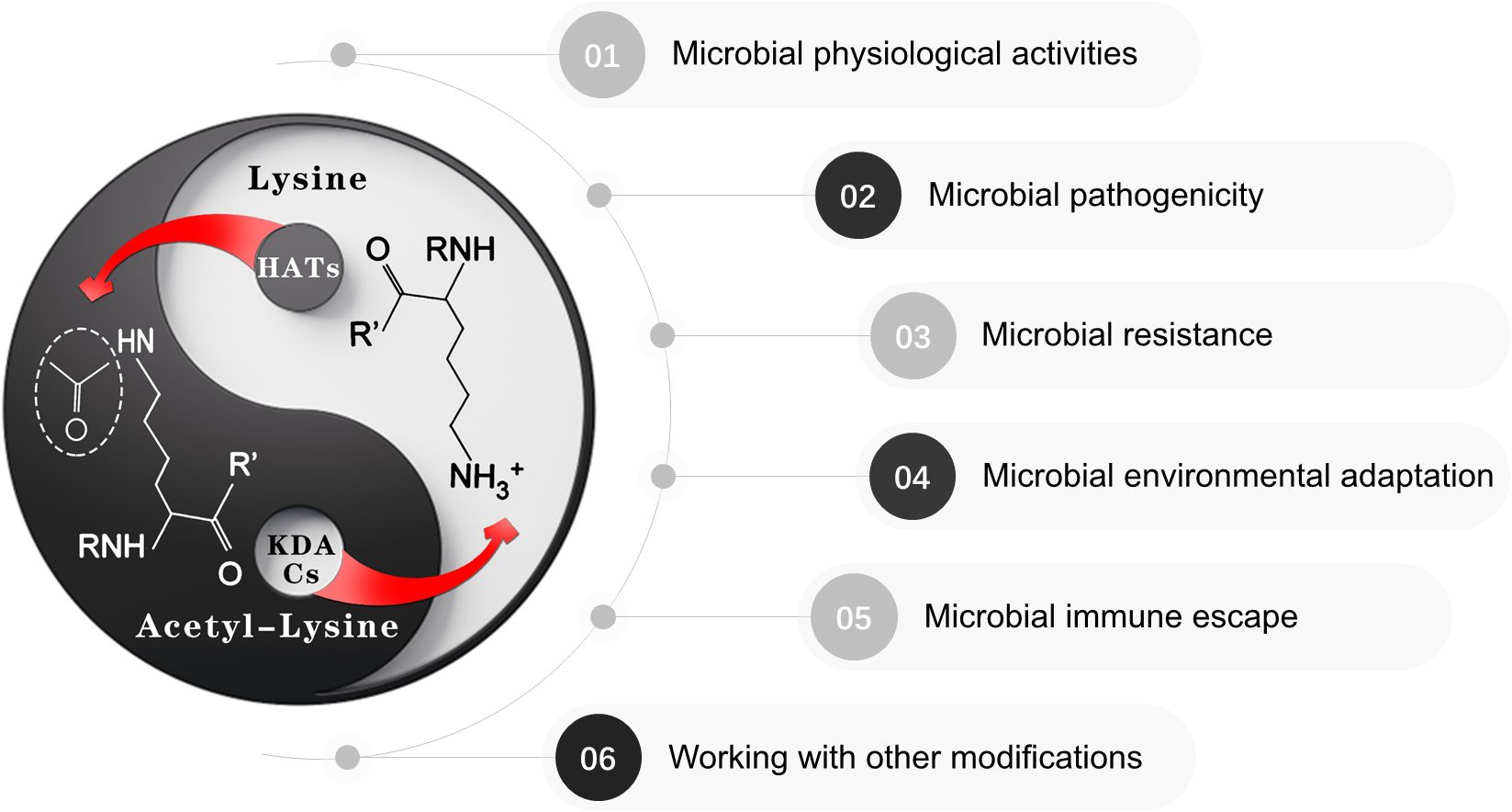
Figure 1. Protein lysine acetylation in oral microbiology. Oral microbiota employs dynamic acetylation/deacetylation modification mechanisms to precisely modulate critical physiological processes including physiological activities, microbial pathogenicity, drug resistance, environmental adaption, immune escape and interaction with other mechanisms. Notably, this dynamically balanced acetylation system endows pathogens with remarkable plasticity, enabling rapid epigenetic-level responses to adaptively transition between host microenvironmental conditions and external stressors. The bidirectional equilibrium inherent in this regulatory network mirrors the Taiji (Yin-Yang) philosophy in Chinese traditional thought – a cyclical interplay of complementary forces that maintains the homeostasis of living systems through dynamic equilibrium.
2 Protein acetylation
2.1 Protein acetylation in oral microorganisms
Lysine acetylation is regulated by two distinct mechanisms: enzymatic and non-enzymatic. The enzymatic process involves the transfer of an acetyl group from acetyl-CoA to the ϵ-amino group of deprotonated lysine, a reaction catalyzed by lysine acetyltransferases (KATs). These enzymes are classified into three primary families: (i) the Gcn5-related N-acetyltransferase (GNAT) family, named after the yeast Gcn5 protein, (ii) the MYST family, which includes human MOZ, yeast Ybf2/Sas3, yeast Sas2, and human Tip60, and (iii) the p300/CBP family, named after human p300 and CBP. While the MYST and p300/CBP families are exclusive to eukaryotic cells, the GNAT family is conserved across all domains of life (Starai and Escalante-Semerena, 2004; Chen et al., 2007; Wang et al., 2010). Several KATs have been identified in various microbial species, all belonging to the GNAT family (Hentchel and Escalante-Semerena, 2015), such as ActA and ActG in Streptococcus mutans (Ma et al., 2021a, 2024a) and VimA and its homolog PG1842 in Porphyromonas gingivalis (Mishra et al., 2018), Among them, the Salmonella enterica protein acetyltransferase Pat (also referred to as YfiQ Escherichia coli) was the first to be identified and remains the most extensively studied (Starai and Escalante-Semerena, 2004).
A non-enzymatic mechanism was identified in Escherichia coli, where acetyl phosphate (AcP) directly transfers its acetyl group to the ϵ-amino group of deprotonated lysine (Weinert et al., 2013; Kuhn et al., 2014), this mechanism may similarly support rapid metabolic adaptation of oral bacteria during nutritional fluctuations or stress conditions which are common in the oral cavity (Di et al., 2023). AcP, a high-energy intermediate in the phosphotransacetylase-acetate kinase (Pta-AckA) pathway, is traditionally recognized as a donor of phosphorylation groups for specific response regulators within two-component signaling systems (Lukat et al., 1992). The yfiQ deletion mutant shows minimal impact on global acetylation levels, while increasing AcP concentrations correspond to elevated global acetylation, indicating that AcP-dependent acetylation is less specific than its enzymatic counterpart and may significantly influence bacterial physiological processes.
2.2 Protein deacetylation in oral microorganisms
Lysine deacetylases (KDACs) enzymatically remove acetyl groups. Currently, two primary KDAC families have been identified and categorized into four groups: the NAD+-dependent sirtuin family (class III) (Blander and Guarente, 2004) and the zinc-dependent Rpd3/Hda1 family (classes I, II, and IV) (Yang and Seto, 2008).
The NAD+-dependent CobB, the most extensively studied bacterial sirtuin, was initially characterized in Streptococcus faecalis (Starai et al., 2002). Its homologs have since been identified in other bacteria, including Escherichia coli (Zhao et al., 2004)and Streptomyces (Mikulik et al., 2012). CobB typically does not exhibit a preference for Pat-dependent or AcP-dependent acetylated lysine, with its substrates participating in diverse cellular processes (AbouElfetouh et al., 2015). For instance, CobB deacetylates both Pat- and AcP-dependent acetylated DnaA (Zhang et al., 2016). Initially regarded as the sole histone deacetylase (HDAC) in Escherichia coli due to the absence of other HDAC homologs, CobB’s role was reassessed upon the identification of a novel deacetylase, YcgC, in Escherichia coli (Tu et al., 2015). YcgC, a member of the serine hydrolase family, lacks significant homology with known KDACs and is neither NAD+- nor Zn2+-dependent. It catalyzes substrate deacetylation through mechanisms distinct from established deacetylases, targeting a different set of acetylated proteins than those regulated by CobB in Escherichia coli. Thus, YcgC and its homologs may constitute a novel bacterial deacetylase family. Additionally, MSMEG_4620, a SIRT4 homolog in Mycobacterium tuberculosis, exhibits both deacetylase and auto-ADP-ribosyltransferase activities (Tan et al., 2016). Consequently, further investigation into novel microbial deacetylases and the interplay between acetylation factors and deacetylases will provide valuable insights.
3 Regulation of oral bacteria by protein acetylation
3.1 Streptococcus mutans
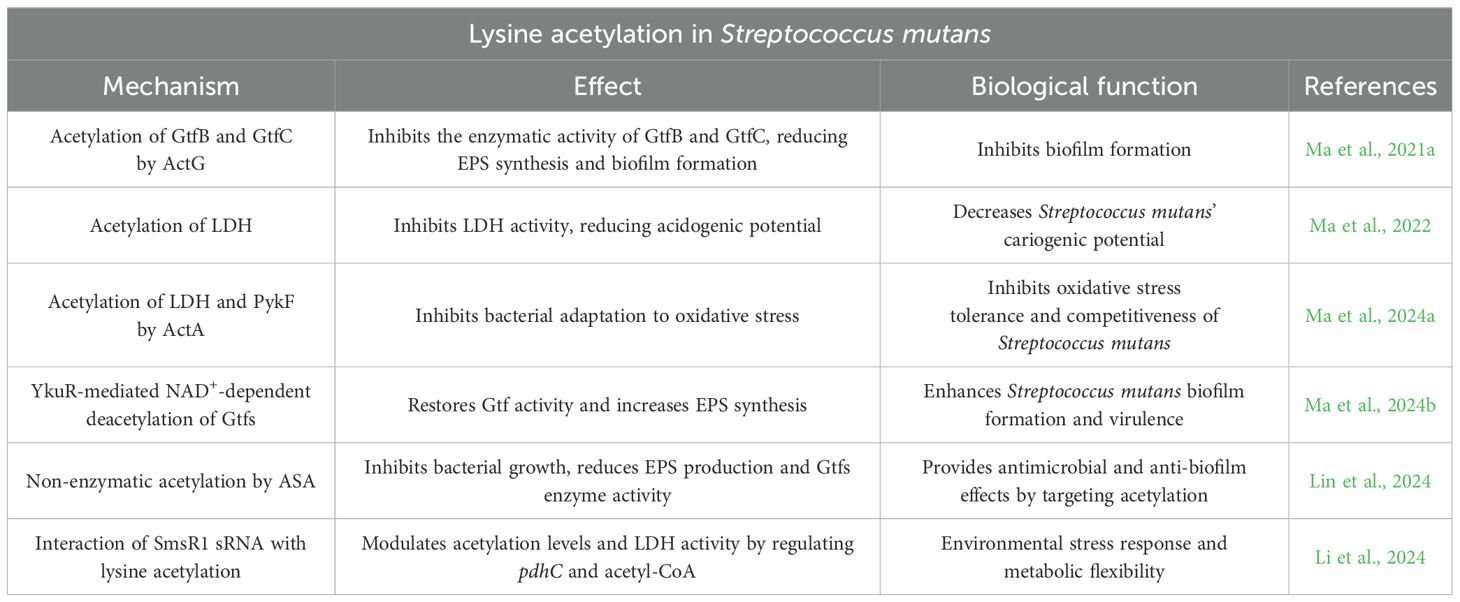
In Streptococcus mutans, lysine acetylation, a widespread and dynamic PTM, is integral to bacterial metabolic regulation and pathogenicity. It has been demonstrated that Streptococcus mutans utilizes sucrose to synthesize exopolysaccharide via glucosyltransferases (Gtfs), a process regulated by lysine acetylation. Analysis of protein acetylation dynamics revealed that 22.7% of proteins were acetylated, with significant enrichment in glycolysis/gluconeogenesis and RNA degradation pathways (Lei et al., 2021).
Acetyltransferase ActG has been shown to acetylate GtfB and GtfC, inhibiting their activity and thus reducing EPS synthesis and biofilm formation. However, site-directed mutagenesis of specific lysine residues was not performed to pinpoint the functional acetylation sites (Ma et al., 2021a). Furthermore, acetylation of lactate dehydrogenase (LDH) also inhibits its enzymatic activity, decreasing the cariogenic potential of Streptococcus mutans (Ma et al., 2022). ActA, a member of the GNAT family in Streptococcus mutans, regulates bacterial adaptation and competitiveness against oxidative stress through acetylation of LDH and pyruvate kinase (PykF) (Ma et al., 2024a). In contrast, the NAD+-dependent deacetylase YkuR reverses Gtfs acetylation and restores their enzymatic activity, thereby promoting EPS production and biofilm formation. Deletion of ykuR elevates global acetylation and attenuates cariogenicity in vivo (Ma et al., 2024b).
Aspirin, a non-enzymatic acetylating agent, inhibits the growth and EPS production of Streptococcus mutans and reduces the enzymatic activity of Gtfs (Lin et al., 2024), further supporting the role of protein acetylation in antimicrobial and anti-biofilm applications.
Additionally, small RNA (sRNA) interacts with lysine acetylation in Streptococcus mutans. SmsR1 sRNA modulates protein acetylation levels and LDH activity by regulating the concentration of the pdhC gene and its metabolite acetyl-CoA (Li et al., 2024), reflecting bacterial adaptability to environmental stress.
These findings highlight the complex regulatory role of lysine acetylation in Streptococcus mutans, influencing bacterial virulence (e.g., biofilm formation) and pathogenicity (e.g., cariogenicity, environmental adaptation) (Table 1), and provide a theoretical foundation for developing new strategies to prevent and treat dental caries.
3.2 Porphyromonas gingivalis
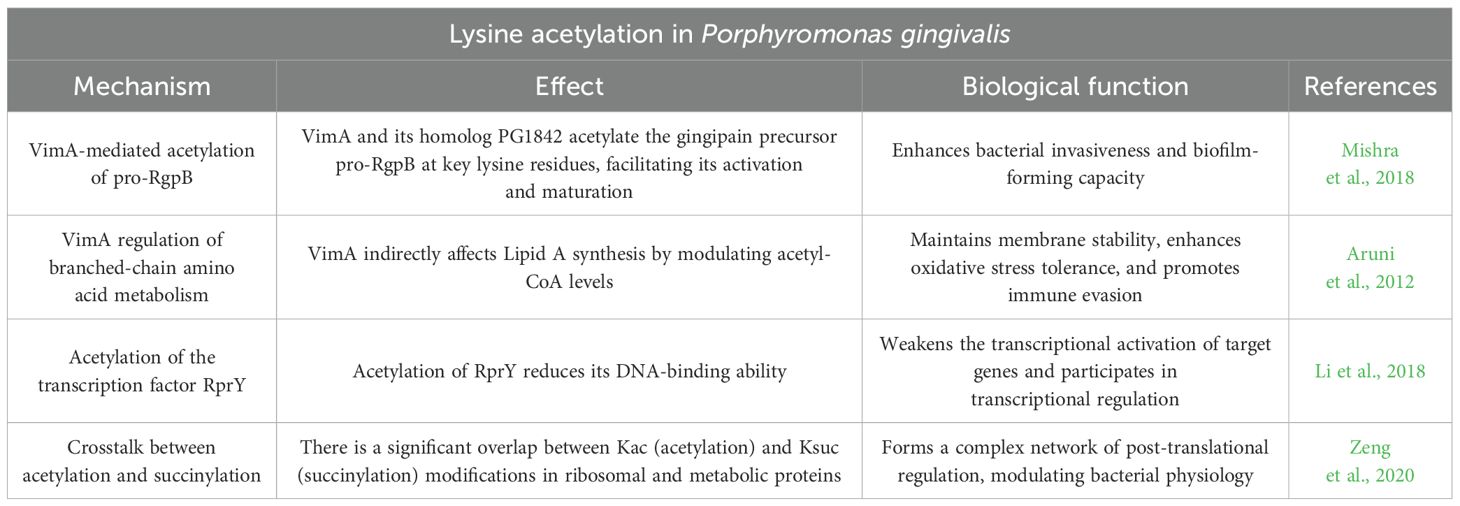
Porphyromonas gingivalis, a Gram-negative anaerobic bacterium, is a key pathogen in chronic periodontitis. Through dynamic analysis of protein acetylation, 130 lysine acetylation sites from 92 Porphyromonas gingivalis proteins were identified, with the majority associated with 45 metabolically active proteins. These proteins are involved in multiple metabolic pathways, where enzymes catalyzing consecutive reactions within the same pathway are frequently acetylated. Notably, 12 enzymes in the anaerobic amino acid fermentation pathway, critical for energy production, also undergo lysine acetylation. This indicates that lysine acetylation plays a central role in the metabolic regulation of Porphyromonas gingivalis, contributing significantly to its survival and metabolic adaptation during infection (Butler et al., 2015).
VimA, a multifunctional protein in Porphyromonas gingivalis, regulates critical biosynthetic pathways through its acetyltransferase activity. It plays a role in the glycosylation and anchoring of bacterial surface proteins, lipid A synthesis, and the maintenance of oxidative stress tolerance. Notably, VimA and its homolog PG1842 acetylate the gingipain precursor pro-RgpB at key lysine residues (Y230, K247, and K248), facilitating its activation and maturation, which enhances both the invasive and biofilm-forming capacities of Porphyromonas gingivalis (Mishra et al., 2018). VimA-deficient mutants, such as FLL92, exhibit reduced invasion efficiency into host cells; however, supplementation with VimA restores invasive potential. This highlights VimA’s essential role in modulating the pathogenicity of Porphyromonas gingivalis. Additionally, VimA influences branched-chain amino acid metabolism by regulating acetyl-CoA levels, thereby affecting lipid A biosynthesis (Aruni et al., 2012). Lipid A, a major component of the outer membrane of Porphyromonas gingivalis, is crucial for immune evasion. Studies have shown that VimA-deficient mutants display impaired survival under oxidative stress, suggesting that VimA contributes to bacterial survival in fluctuating oral environments by enhancing membrane stability and stress tolerance.
Protein acetylation is integral to the transcriptional regulation of Porphyromonas gingivalis. Acetylation of the transcription factor RprY impairs its DNA-binding ability, thereby diminishing its transcriptional activation of target genes (Li et al., 2018).
A significant aspect of lysine acetylation in Porphyromonas gingivalis proteins is its interaction with lysine succinylation (Ksuc). In Porphyromonas gingivalis ATCC 33277, a substantial overlap between Ksuc and Kac occurs, particularly in ribosomal and metabolic proteins, reflecting the complexity of PTMs in bacterial physiological regulation (Zeng et al., 2020).
In conclusion, protein acetylation is central to the physiological functions, metabolic regulation, and pathogenicity of Porphyromonas gingivalis. By modulating various metabolic pathways, protein sorting, and surface structure synthesis, acetylation enhances the bacterium’s environmental adaptability, invasiveness, and biofilm formation capabilities (Table 2). These insights present promising molecular targets for the development of novel antimicrobial therapies.
3.3 Actinomycetes
Protein lysine acetylation, a critical PTM, is integral to the regulation of various biological processes in Actinomycetes. In fact, “Amino acid sensing” was specifically observed only in Actinomycetes, based on recent studies identifying ACT domain-containing GNATs (Lu et al., 2017; Lammers, 2021). However, similar to other organisms, such as Streptococcus mutans and Porphyromonas gingivalis, akin regulatory mechanisms such as enzyme activity modulation and metabolic network regulation via acetylation are also present in Actinomycetes (Hesketh et al., 2002).
3.3.1 Amino acid sensing: GNATs and functional properties of the ACT domain
Recent studies have identified a distinct class of GNATs in Actinomycetes, which are integral to acetylation reactions via their unique domains and regulatory mechanisms. Specifically, these enzymes feature two functional domains: the ACT (amino acid binding) domain and the GNAT (N-acetyltransferase) domain. The ACT domain detects specific amino acids, thereby allosterically modulating the acetyltransferase activity of the GNAT domain (Lu et al., 2017). This “amino acid-induced allosteric regulation” implies that amino acids influence acetyltransferase catalytic activity by binding to the ACT domain. GCN5-like acetyltransferases can be classified into two groups based on the type of amino acids induction: Asn-activated PatA and Cys-activated PatA. The former is predominantly found in Streptomyces, while the latter is more widespread across other actinomycete species (Lu et al., 2017). This distinction highlights that acetyltransferase regulation not only fine-tunes metabolic processes through amino acid sensing but also correlates with the physiological traits and ecological adaptability of different actinomycete species.
3.3.2 Enzyme activity regulation: dynamic balance between acetyltransferases and deacetylases
In Actinomycetes, the balance between acetyltransferases (e.g., AcuA) (Gardner et al., 2006), and deacetylases (e.g., Sirtuins) (Tucker and Escalante-Semerena, 2013) plays a pivotal role in fine-tuning enzyme activity, particularly under energy stress or nutrient-limited conditions. This dynamic regulation is critical for adapting metabolic fluxes and optimizing secondary metabolite biosynthesis.Acetylation levels also influence the synthesis of secondary metabolites by modulating the activity of specific transcription factors or metabolic enzymes. By altering transcription factor binding affinity to gene promoters, acetylation can either enhance or suppress DNA binding, thereby regulating the expression of genes involved in secondary metabolite production. This regulatory mechanism is essential for coordinating the biosynthesis of compounds such as antibiotics (e.g., streptomycin) and non-ribosomal peptides in response to environmental or metabolic signals (Martín et al., 2021).
Additionally, acetylation of key signaling enzymes or synthetases, such as acetyl-CoA synthetase (Tucker and Escalante-Semerena, 2013), can alter their catalytic efficiency and affect metabolic flux. This modification not only regulates enzyme activity but also influences the overall metabolic pathway, promoting or inhibiting the synthesis of specific metabolites. For instance, in Streptomyces roseosporus, acetylation of non-ribosomal peptide synthetases suggests a significant role for acetylation in secondary metabolism (Liao et al., 2014). In Streptomyces griseus, acetylation of the StrM enzyme, particularly at lysine site 70, reduces its activity, thereby limiting streptomycin biosynthesis (Ishigaki et al., 2017).
3.3.3 Fine regulation of metabolic networks: from amino acid metabolism to secondary metabolite biosynthesis
The coordinated regulation of amino acid sensing and enzyme activity forms a precise regulatory mechanism within the intricate metabolic network of Actinomycetes. Acetylation is not only involved in central metabolic processes but also significantly influences amino acid metabolic pathways. Studies indicate that AAPatA acetyltransferases, which utilize Asn or Cys as sensing molecules, can modulate cellular metabolic pathways by regulating enzymes involved in aspartate and cysteine metabolism (Xu et al., 2014; Lu et al., 2017).
This acetylation-driven network is particularly responsive to environmental fluctuations. Under changing nutritional conditions or stress, acetylation rapidly adjusts bacterial metabolic pathways by altering the modification states of specific transcription factors and redox enzymes, such as GrhO6 (Toplak et al., 2022). Such regulatory flexibility supports the synthesis of secondary metabolites, enabling Actinomycetes to efficiently adapt to diverse habitats and produce specialized secondary metabolites and antibiotics.
Protein acetylation regulation in Actinomycetes extends beyond traditional cellular functions, including amino acid sensing, enzyme activity modulation, and the precise regulation of metabolic networks (Table 3). Future research should focus on elucidating the specific roles of acetyltransferases in actinomycete antibiotic synthesis and their potential as targets for developing new biological agents or antibiotics. In-depth mechanistic studies could further uncover the complexities of microbial metabolic regulation, offering novel insights for bioengineering and the development of antimicrobial drugs.
4 Regulation of oral Candida albicans by protein acetylation
In recent years, the infection rate of Candida albicans, a common opportunistic fungal pathogen, has risen significantly. The growing prevalence of drug resistance and the limited availability of effective antifungal agents present substantial challenges to clinical management. During investigations into its pathogenic mechanisms and drug resistance, lysine acetylation has emerged as a key epigenetic modification. By regulating chromatin structure, gene expression, and signal transduction, lysine acetylation plays a critical role in the growth, virulence, morphological transformation, and stress response of Candida albicans. As such, it represents a promising target for the development of novel antifungal therapies.
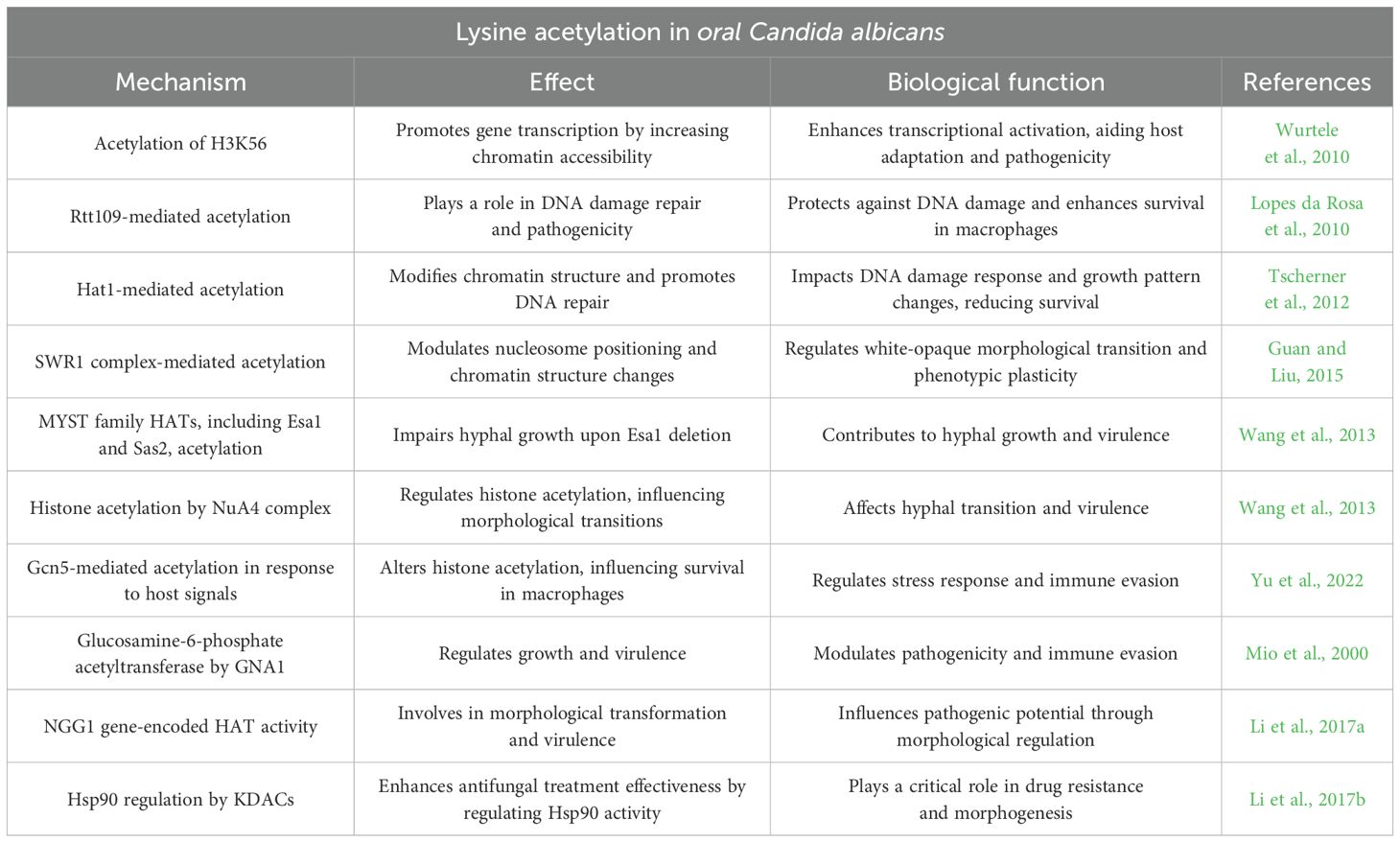
Histone acetylation regulates key physiological processes in Candida albicans, such as DNA replication, transcription, and DNA repair, influencing growth, virulence, drug resistance, and environmental adaptability. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) control the acetylation and deacetylation of histones, respectively. During replication, newly synthesized histones are acetylated by HATs, deposited on DNA, and later deacetylated by HDACs; histone chaperones recognize acetylation patterns during this process (Shahbazian and Grunstein, 2007). Additionally, histone acetylation alters chromatin structure, affecting the recruitment of DNA-binding proteins and transcription factors, which in turn modulates gene transcription (Shahbazian and Grunstein, 2007). For example, in Candida albicans, acetylation of H3K56 promotes gene transcriptional activation by increasing chromatin accessibility, with its level closely linked to transcription factor binding on chromatin (Wurtele et al., 2010). This modification creates a favorable environment for gene transcription, reshaping the transcriptional profile of Candida albicans, aiding host adaptation, and enhancing pathogenicity. Rtt109, another HAT, plays a critical role in DNA damage repair and pathogenicity. Deletion of the rtt109 gene results in heightened endogenous DNA damage and increased susceptibility to host macrophages (Lopes da Rosa et al., 2010). Hat1 also contributes significantly to DNA repair; its loss leads to rapid DNA damage accumulation and shifts the growth pattern from yeast to pseudohyphal form, ultimately reducing survival of Candida albicans (Tscherner et al., 2012). These findings emphasize the essential role of histone acetylation in the physiological and pathogenic processes of Candida albicans.
Histone acetylation influences the morphological plasticity of Candida albicans, determining its environmental adaptability and virulence. Candida albicans can transition between yeast, pseudohyphal, and hyphal forms, with the filamentous forms (pseudohyphae and hyphae) playing a critical role in promoting fungal infection and the formation of drug-resistant biofilms (Odds, 1985; Sudbery, 2011). Various enzymes, including HATs and HADCs, are key regulators of this morphological transition. Altered HAT expression significantly impacts Candida albicans’s ability to adapt to environmental changes. For example, deletion of the SWR1 gene, which is involved in H2A.Z histone variant deposition, induces chromatin structure changes, promoting the transition between white and opaque morphologies. The SWR1 complex also regulates nucleosome positioning at the WOR1 promoter, a master regulator of the white-opaque switch essential for maintaining phenotypic plasticity (Guan and Liu, 2015). Additionally, MYST family HATs, such as Esa1 and Sas2, contribute to hyphal growth. Loss of Esa1 specifically impairs hyphal formation without affecting overall growth, highlighting the significance of HATs in regulating morphology and pathogenicity. Moreover, certain HADCs, like the NuA4 complex, regulate histone acetylation via enzymes such as Yng2, further influencing morphological transitions (Wang et al., 2013). These insights emphasize the critical role of histone acetylation in controlling Candida albicans’s morphological plasticity and its adaptation to various host environments and pathogenic states.
Lysine acetylation is integral to Candida albicans’ ability to respond to host immune defenses. This pathogen evades immune detection by modulating the expression of oxidative stress response genes, a process tightly regulated by dynamic histone acetylation (Kim et al., 2018). In addition to transcriptional regulation, acetylation also influences structural adaptations that facilitate immune evasion, as the histone deacetylase Sir2 has been shown to promote systemic candidiasis by remodeling the fungal cell wall, thereby reducing the exposure of immunogenic components like mannan and β-glucan. This remodeling diminishes recognition by the host’s innate immune system and enhances fungal adhesion to host cells, contributing to increased virulence (Yang et al., 2024). Notably, lysine acetylation at specific sites, such as H3K56, influences Candida albicans’s capacity to tolerate oxidative stress and escape immune surveillance (Conte et al., 2024). By altering chromatin structure, histone acetylation modulates gene expression in response to stressors like reactive oxygen species generated by the host immune system. The role of HATs in this process is critical; for instance, Candida albicans deficient in the lysine acetyltransferase Gcn5 exhibits reduced survival in THP-1 macrophages and heightened susceptibility to various stressors (Yu et al., 2022).
Furthermore, interactions between host-derived signals and epigenetic modifications, such as lysine acetylation, influence pathogen morphology, enhancing its adaptability to the host environment. Although the molecular mechanisms underlying these processes remain under investigation, it is evident that lysine acetylation plays a key mediating role in Candida albicans’ resistance to host immune responses.
Lysine acetyltransferases are essential in mediating drug resistance and pathogenicity. The glucosamine-6-phosphate acetyltransferase encoded by the GNA1 gene influences the growth and virulence of Candida albicans, with its deletion resulting in a marked reduction in pathogenicity (Mio et al., 2000). Similarly, the HAT encoded by the NGG1 gene is involved in morphological transformation and virulence; its knockout substantially diminishes the strain’s pathogenic potential (Li et al., 2017a). Furthermore, Hsp90 plays a critical role in drug resistance and morphogenesis. Research has shown that KDACs exhibit functional redundancy in regulating Hsp90 activity, and their inhibition can enhance the effectiveness of antifungal treatments (Li et al., 2017b).
In conclusion, lysine acetylation contributes significantly to the morphological adaptability, immune evasion, and drug resistance of Candida albicans by modulating chromatin structure, gene expression, and stress responses (Table 4). These epigenetic mechanisms not only enhance understanding of Candida albicans pathogenicity but also open avenues for the development of novel antifungal therapies. Although direct evidence of oral-specific acetylation mechanisms is currently lacking, the unique characteristics of the oral environment and the role of acetylation in gene regulation warrant further investigation into this area.
5 Potential of protein acetylation in the prevention and treatment of oral diseases
Protein lysine acetylation plays a critical role in microbial physiology, influencing both metabolism and pathogenic potential. Recently, it has garnered significant attention in the context of oral diseases. Lysine acetylation influences the onset and progression of conditions such as dental caries, periodontal disease, and oral mucosal disorders by modulating microbial growth, metabolism, and virulence, offering novel insights and therapeutic strategies for their prevention and treatment.
Histone acetyltransferase-based treatments have been utilized in various clinical applications. Many cellular proteins undergo acetylation post-translationally, resulting in alterations to their structure and function. Consequently, HDAC inhibitors have been developed as therapeutic agents, with two currently approved by the US FDA for the treatment of cutaneous T-cell lymphoma (Richon et al., 2009). Moreover, lysine acetyltransferases are emerging as promising drug targets (Folders et al., 2000).
In dental caries research, aspirin, a non-enzymatic acetylating agent, reduces Gtfs activity in Streptococcus mutans. It also inhibits the growth of Streptococcus mutans and the production of EPS, highlighting the potential of protein acetylation in anti-caries therapies (Lin et al., 2024).
In the gingival tissue of periodontitis patients, dysregulated histone acetylation and deacetylation, along with alterations in DNA methylation, are closely linked to immune and inflammatory responses. In experimental periodontitis models, histone acetylation-targeting treatments, such as HDAC inhibitors (HDACi) or acetylated histone mimetics, effectively prevent alveolar bone loss (Cantley et al., 2011; Meng et al., 2014; Li et al., 2020), offering a novel therapeutic approach for periodontal diseases.
In Candida albicans, lysine 56 of histone H3 undergoes acetylation by the acetyltransferase Rtt109p, and pharmacological inhibition or genetic modulation of this enzyme has been proposed as a potential antifungal therapeutic strategy. Acetylation events, often challenging to detect, can significantly impact protein functions, including stability and crystallinity (Mahon et al., 2015). For therapeutic proteins, such modifications may influence immunogenicity and biological activity, thereby affecting safety and efficacy in clinical applications (Walsh and Jefferis, 2006).
Overall, lysine acetylation plays a key role in regulating the metabolism and pathogenicity of oral pathogens by modulating the activity of critical enzymes. Research in this area not only enhances understanding of the pathogenic mechanisms of oral pathogens but also lays a foundation for the development of novel antibacterial and antifungal therapies. Targeting specific acetylases could substantially improve the efficacy of current treatments and offer potential solutions to manage drug resistance. Future investigations should focus on the role of acetylation in the oral microbiota and its impact on host-pathogen interactions, thus paving the way for new strategies in maintaining oral health and addressing oral diseases.
6 Conclusion and prospect
Acetylation, a key PTM, is central to the functional regulation of oral microorganisms. It modulates various bacterial physiological processes, including metabolism, cell signaling, virulence factor expression, and host-pathogen interactions, by altering protein properties such as structure, activity, localization, and interactions with other biomolecules.
This modification significantly impacts the metabolic state of oral bacteria. While acetylation is typically catalyzed by acetyltransferases, it can also occur non-enzymatically via Ac-CoA (Paik et al., 1970; Yan et al., 2008) or acetyladenylate (Ramponi et al., 1975). As a metabolic intermediate, acetylation enables bacteria to adapt to environmental fluctuations and shifts in metabolic activity, thereby playing a crucial role in the regulation of bacterial physiology and pathogenicity.
Acetylation, along with other PTMs such as succinylation and phosphorylation, collectively regulates bacterial protein functions, forming a complex modification network (Latham and Dent, 2007). This cross-regulation of multiple modifications is essential for controlling bacterial virulence factors, adaptive responses, and metabolic processes. For example, in Porphyromonas gingivalis, substantial overlap has been observed between acetylation and succinylation sites—especially on ribosomal and metabolic proteins—suggesting potential competitive or cooperative regulation of key cellular functions (Zeng et al., 2020). Additionally, the response regulator RprY in Porphyromonas gingivalis is modified by both acetylation and phosphorylation, with each modification exerting distinct effects on its DNA-binding activity and transcriptional regulation (Li et al., 2018). These findings underscore the existence of an intricate PTM crosstalk network that supports bacterial adaptation to fluctuating environmental and host-derived conditions. Nevertheless, systematic studies on the functional interplay among different PTMs in oral microorganisms remain limited. Future research should aim to elucidate the molecular mechanisms and biological consequences of these interactions—particularly the synergistic or antagonistic effects between acetylation, phosphorylation, and succinylation—in governing microbial adaptation, pathogenicity, and immune evasion.
Despite extensive research on acetylation, significant gaps remain in the study of acetylation in oral bacteria. Most investigations have concentrated on a limited number of oral pathogens, such as Porphyromonas gingivalis and Streptococcus mutans. It is important to note that lysine acetylation is a widely conserved modification across diverse oral microbial taxa. Nevertheless, functional studies and comprehensive acetylomic profiling in other clinically relevant oral species—such as Fusobacterium nucleatum, Prevotella intermedia, and Veillonella spp.—remain limited. To address this, future research should broaden the scope of study to include a wider range of oral bacteria and conduct comprehensive acetylomics analyses to better elucidate the acetylation patterns and functions across diverse physiological and pathological states.
From a clinical standpoint, acetylation represents a key mechanism in regulating bacterial virulence and drug resistance, offering potential new targets for the treatment of oral infections. Investigating the role of acetylation in oral microbiota could provide a theoretical foundation for the development of novel antibacterial strategies. Future research should not only examine the fundamental role of acetylation in bacterial function but also explore strategies to mitigate bacterial pathogenicity through acetylation regulation, particularly by targeting the acetylation of key virulence factors to enhance therapeutic outcomes.
In conclusion, acetylation plays a crucial role in the adaptation, virulence, and immune evasion of oral microorganisms. Advances in technologies such as metabolomics, proteomics, and mass spectrometry will further elucidate the role of acetylation in bacterial physiological and pathological processes. This enhanced understanding will not only clarify the mechanisms underlying the virulence of oral bacteria but also open new avenues for the treatment of oral diseases.
Author contributions
YCY: Writing – original draft, Investigation, Funding acquisition, Writing – review & editing. HH: Conceptualization, Supervision, Writing – review & editing. BL: Writing – review & editing. ZL: Supervision, Writing – review & editing. JS: Writing – review & editing. ZZ: Writing – review & editing, Conceptualization, Supervision. YY: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 82301063), Hunan Provincial Natural Science Foundation of China (Grant No. 2023JJ30815), the Science and Technology Innovation Program of Hunan Province (Grant No. 2024RC3069) and the Hunan Provincial Innovation Foundation for Postgraduate (Grant No. 2024ZZTS0278).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
AbouElfetouh, A., Kuhn, M. L., Hu, L. I., Scholle, M. D., Sorensen, D. J., Sahu, A. K., et al. (2015). The E. coli sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites. Microbiologyopen 4, 66–83. doi: 10.1002/mbo3.223
Allfrey, V. G., Faulkner, R., and Mirsky, A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U S A 51, 786–794. doi: 10.1073/pnas.51.5.786
Aruni, A. W., Lee, J., Osbourne, D., Dou, Y., Roy, F., Muthiah, A., et al. (2012). VimA-dependent modulation of acetyl coenzyme A levels and lipid A biosynthesis can alter virulence in Porphyromonas gingivalis. Infect. Immun. 80, 550–564. doi: 10.1128/IAI.06062-11
Blander, G. and Guarente, L. (2004). The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435. doi: 10.1146/annurev.biochem.73.011303.073651
Butler, C. A., Veith, P. D., Nieto, M. F., Dashper, S. G., and Reynolds, E. C. (2015). Lysine acetylation is a common post-translational modification of key metabolic pathway enzymes of the anaerobe Porphyromonas gingivalis. J. Proteomics 128, 352–364. doi: 10.1016/j.jprot.2015.08.015
Cantley, M. D., Bartold, P. M., Marino, V., Fairlie, D. P., Le, G. T., Lucke, A. J., et al. (2011). Histone deacetylase inhibitors and periodontal bone loss. J. Periodontal Res. 46, 697–703. doi: 10.1111/j.1600-0765.2011.01392.x
Carafa, V., Nebbioso, A., and Altucci, L. (2012). Sirtuins and disease: the road ahead. Front. Pharmacol. 3. doi: 10.3389/fphar.2012.00004
Chen, Y., Sprung, R., Tang, Y., Ball, H., Sangras, B., Kim, S. C., et al. (2007). Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell Proteomics 6, 812–819. doi: 10.1074/mcp.M700021-MCP200
Chuang, C., Lin, S. H., Huang, F., Pan, J., Josic, D., and Yu-Lee, L. Y. (2010). Acetylation of RNA processing proteins and cell cycle proteins in mitosis. J. Proteome Res. 9, 4554–4564. doi: 10.1021/pr100281h
Conte, M., Eletto, D., Pannetta, M., Esposito, R., Monti, M. C., Morretta, E., et al. (2024). H3K56 acetylation affects Candida albicans morphology and secreted soluble factors interacting with the host. Biochim. Biophys. Acta Gene Regul. Mech. 1867, 195048. doi: 10.1016/j.bbagrm.2024.195048
Di, Y., Xu, S., Chi, M., Hu, Y., Zhang, X., Wang, H., et al. (2023). Acetylation of cyclic AMP receptor protein by acetyl phosphate modulates mycobacterial virulence. Microbiol Spectr. 11, e0400222. doi: 10.1128/spectrum.04002-22
Folders, J., Tommassen, J., van Loon, L. C., and Bitter, W. (2000). Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J. Bacteriol 182, 1257–1263. doi: 10.1128/JB.182.5.1257-1263.2000
Gardner, J. G., Grundy, F. J., Henkin, T. M., and Escalante-Semerena, J. C. (2006). Control of acetyl-coenzyme A synthetase (AcsA) activity by acetylation/deacetylation without NAD(+) involvement in Bacillus subtilis. J. Bacteriol 188, 5460–5468. doi: 10.1128/JB.00215-06
Guan, Z. and Liu, H. (2015). Overlapping functions between SWR1 deletion and H3K56 acetylation in candida albicans. Eukaryot Cell 14, 578–587. doi: 10.1128/EC.00002-15
Hentchel, K. L. and Escalante-Semerena, J. C. (2015). Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol Mol. Biol. Rev. 79, 321–346. doi: 10.1128/MMBR.00020-15
Hesketh, A. R., Chandra, G., Shaw, A. D., Rowland, J. J., Kell, D. B., Bibb, M. J., et al. (2002). Primary and secondary metabolism, and post-translational protein modifications, as portrayed by proteomic analysis of Streptomyces coelicolor. Mol. Microbiol 46, 917–932. doi: 10.1046/j.1365-2958.2002.03219.x
Ishigaki, Y., Akanuma, G., Yoshida, M., Horinouchi, S., Kosono, S., and Ohnishi, Y. (2017). Protein acetylation involved in streptomycin biosynthesis in Streptomyces griseus. J. Proteomics 155, 63–72. doi: 10.1016/j.jprot.2016.12.006
Kim, J., Lee, J. E., and Lee, J. S. (2015). Histone deacetylase-mediated morphological transition in Candida albicans. J. Microbiol 53, 805–811. doi: 10.1007/s12275-015-5488-3
Kim, J., Park, S., and Lee, J. S. (2018). Epigenetic control of oxidative stresses by histone acetyltransferases in candida albicans. J. Microbiol Biotechnol. 28, 181–189. doi: 10.4014/jmb.1707.07029
Kuhn, M. L., Zemaitaitis, B., Hu, L. I., Sahu, A., Sorensen, D., Minasov, G., et al. (2014). Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PloS One 9, e94816. doi: 10.1371/journal.pone.0094816
Lammers, M. (2021). Post-translational lysine ac(et)ylation in bacteria: A biochemical, structural, and synthetic biological perspective. Front. Microbiol 12. doi: 10.3389/fmicb.2021.757179
Latham, J. A. and Dent, S. Y. (2007). Cross-regulation of histone modifications. Nat. Struct. Mol. Biol. 14, 1017–1024. doi: 10.1038/nsmb1307
Lei, L., Zeng, J., Wang, L., Gong, T., Zheng, X., Qiu, W., et al. (2021). Quantitative acetylome analysis reveals involvement of glucosyltransferase acetylation in Streptococcus mutans biofilm formation. Environ. Microbiol Rep. 13, 86–97. doi: 10.1111/1758-2229.12907
Li, D. D., Fuchs, B. B., Wang, Y., Huang, X. W., Hu, D. D., Sun, Y., et al. (2017a). Histone acetyltransferase encoded by NGG1 is required for morphological conversion and virulence of Candida albicans. Future Microbiol 12, 1497–1510. doi: 10.2217/fmb-2017-0084
Li, Y., Krishnan, K., and Duncan, M. J. (2018). Post-translational regulation of a Porphyromonas gingivalis regulator. J. Microbiol 10, 1487743. doi: 10.1080/20002297.2018.1487743
Li, Q., Liu, F., Dang, R., Feng, C., Xiao, R., Hua, Y., et al. (2020). Epigenetic modifier trichostatin A enhanced osteogenic differentiation of mesenchymal stem cells by inhibiting NF-kappaB (p65) DNA binding and promoted periodontal repair in rats. J. Cell Physiol. 235, 9691–9701. doi: 10.1002/jcp.29780
Li, J., Ma, Q., Huang, J., Liu, Y., Zhou, J., Yu, S., et al. (2024). Small RNA SmsR1 modulates acidogenicity and cariogenic virulence by affecting protein acetylation in Streptococcus mutans. PloS Pathog 20, e1012147. doi: 10.1371/journal.ppat.1012147
Li, X., Robbins, N., O’Meara, T. R., and Cowen, L. E. (2017b). Extensive functional redundancy in the regulation of Candida albicans drug resistance and morphogenesis by lysine deacetylases Hos2, Hda1, Rpd3 and Rpd31. Mol. Microbiol 103, 635–656. doi: 10.1111/mmi.13578
Liao, G., Xie, L., Li, X., Cheng, Z., and Xie, J. (2014). Unexpected extensive lysine acetylation in the trump-card antibiotic producer Streptomyces roseosporus revealed by proteome-wide profiling. J. Proteomics 106, 260–269. doi: 10.1016/j.jprot.2014.04.017
Lin, Y., Ma, Q., Yan, J., Gong, T., Huang, J., Chen, J., et al. (2024). Inhibition of Streptococcus mutans growth and biofilm formation through protein acetylation. Mol. Microbiol 39, 334–343. doi: 10.1111/omi.12452
Lin, H., Su, X., and He, B. (2012). Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol. 7, 947–960. doi: 10.1021/cb3001793
Lopes da Rosa, J., Boyartchuk, V. L., Zhu, L. J., and Kaufman, P. D. (2010). Histone acetyltransferase Rtt109 is required for Candida albicans pathogenesis. Proc. Natl. Acad. Sci. U S A 107, 1594–1599. doi: 10.1073/pnas.0912427107
Lu, J. Y., Lin, Y. Y., Sheu, J. C., Wu, J. T., Lee, F. J., Chen, Y., et al. (2011). Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell 146, 969–979. doi: 10.1016/j.cell.2011.07.044
Lu, Y. X., Liu, X. X., Liu, W. B., and Ye, B. C. (2017). Identification and characterization of two types of amino acid-regulated acetyltransferases in actinobacteria. Biosci Rep. 37 (4), BSR20170157. doi: 10.1042/BSR20170157
Lukat, G. S., McCleary, W. R., Stock, A. M., and Stock, J. B. (1992). Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc. Natl. Acad. Sci. U S A 89, 718–722. doi: 10.1073/pnas.89.2.718
Ma, Q., Li, J., Yu, S., Liu, Y., Zhou, J., Wang, X., et al. (2024a). ActA-mediated PykF acetylation negatively regulates oxidative stress adaptability of Streptococcus mutans. mBio 15, e0183924. doi: 10.1128/mbio.01839-24
Ma, Q., Li, J., Yu, S., Zhou, J., Liu, Y., Wang, X., et al. (2024b). YkuR functions as a protein deacetylase in Streptococcus mutans. Proc. Natl. Acad. Sci. U S A 121, e2407820121. doi: 10.1073/pnas.2407820121
Ma, Q., Pan, Y., Chen, Y., Yu, S., Huang, J., Liu, Y., et al. (2021a). Acetylation of glucosyltransferases regulates Streptococcus mutans biofilm formation and virulence. PloS Pathog 17, e1010134. doi: 10.1371/journal.ppat.1010134
Ma, Q., Pan, Y., Chen, Y., Yu, S., Huang, J., Liu, Y., et al. (2022). Acetylation of lactate dehydrogenase negatively regulates the acidogenicity of streptococcus mutans. mBio 13, e0201322. doi: 10.1128/mbio.02013-22
Ma, Q., Zhang, Q., Chen, Y., Yu, S., Huang, J., Liu, Y., et al. (2021b). Post-translational modifications in oral bacteria and their functional impact. Front. Microbiol 12. doi: 10.3389/fmicb.2021.784923
Mahon, B. P., Lomelino, C. L., Salguero, A. L., Driscoll, J. M., Pinard, M. A., and McKenna, R. (2015). Observed surface lysine acetylation of human carbonic anhydrase II expressed in Escherichia coli. Protein Sci. 24, 1800–1807. doi: 10.1002/pro.2771
Martín, J. F., Liras, P., and Sanchez, S. (2021). Modulation of gene expression in actinobacteria by translational modification of transcriptional factors and secondary metabolite biosynthetic enzymes. Front. Microbiol 12. doi: 10.3389/fmicb.2021.630694
Meng, S., Zhang, L., Tang, Y., Tu, Q., Zheng, L., Yu, L., et al. (2014). BET inhibitor JQ1 blocks inflammation and bone destruction. J. Dent. Res. 93, 657–662. doi: 10.1177/0022034514534261
Mikulik, K., Felsberg, J., Kudrnacova, E., Bezouskova, S., Setinova, D., Stodulkova, E., et al. (2012). CobB1 deacetylase activity in Streptomyces coelicolor. Biochem. Cell Biol. 90, 179–187. doi: 10.1139/o11-086
Minguez, P., Parca, L., Diella, F., Mende, D. R., Kumar, R., Helmer-Citterich, M., et al. (2012). Deciphering a global network of functionally associated post-translational modifications. Mol. Syst. Biol. 8, 599. doi: 10.1038/msb.2012.31
Mio, T., Kokado, M., Arisawa, M., and Yamada-Okabe, H. (2000). Reduced virulence of Candida albicans mutants lacking the GNA1 gene encoding glucosamine-6-phosphate acetyltransferase. Microbiol. (Reading) 146, 1753–1758. doi: 10.1099/00221287-146-7-1753
Mishra, A., Roy, F., Dou, Y., Zhang, K., Tang, H., Fletcher, H. M., et al. (2018). Role of acetyltransferase PG1842 in gingipain biogenesis in porphyromonas gingivalis. J. Bacteriology 200 (21), e00385-18. doi: 10.1128/jb.00385-18
Montellier, E., Rousseaux, S., Zhao, Y., and Khochbin, S. (2012). Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression. Bioessays 34, 187–193. doi: 10.1002/bies.201100141
Moreira, I. S., Fernandes, P. A., and Ramos, M. J. (2007). Hot spots–a review of the protein-protein interface determinant amino-acid residues. Proteins 68, 803–812. doi: 10.1002/prot.21396
Odds, F. C. (1985). Morphogenesis in candida albicans. Crit. Rev. Microbiol 12, 45–93. doi: 10.3109/10408418509104425
Paik, W. K., Pearson, D., Lee, H. W., and Kim, S. (1970). Nonenzymatic acetylation of histones with acetyl-CoA. Biochim. Biophys. Acta 213, 513–522. doi: 10.1016/0005-2787(70)90058-4
Peng, C., Lu, Z., Xie, Z., Cheng, Z., Chen, Y., Tan, M., et al. (2011). The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell Proteomics 10, M111 012658. doi: 10.1074/mcp.M111.012658
Ramponi, G., Manao, G., and Camici, G. (1975). Nonenzymatic acetylation of histones with acetyl phosphate and acetyl adenylate. Biochemistry 14, 2681–2685. doi: 10.1021/bi00683a018
Ren, J., Sang, Y., Lu, J., and Yao, Y. F. (2017). Protein acetylation and its role in bacterial virulence. Trends Microbiol 25, 768–779. doi: 10.1016/j.tim.2017.04.001
Richon, V. M., Garcia-Vargas, J., and Hardwick, J. S. (2009). Development of vorinostat: current applications and future perspectives for cancer therapy. Cancer Lett. 280, 201–210. doi: 10.1016/j.canlet.2009.01.002
Shahbazian, M. D. and Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100. doi: 10.1146/annurev.biochem.76.052705.162114
Starai, V. J., Celic, I., Cole, R. N., Boeke, J. D., and Escalante-Semerena, J. C. (2002). Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390–2392. doi: 10.1126/science.1077650
Starai, V. J. and Escalante-Semerena, J. C. (2004). Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J. Mol. Biol. 340, 1005–1012. doi: 10.1016/j.jmb.2004.05.010
Sudbery, P. E. (2011). Growth of Candida albicans hyphae. Nat. Rev. Microbiol 9, 737–748. doi: 10.1038/nrmicro2636
Tan, Y., Xu, Z., Tao, J., Ni, J., Zhao, W., Lu, J., et al. (2016). A SIRT4-like auto ADP-ribosyltransferase is essential for the environmental growth of Mycobacterium smegmatis. Acta Biochim. Biophys. Sin. (Shanghai) 48, 145–152. doi: 10.1093/abbs/gmv121
Toplak, M., Nagel, A., Frensch, B., Lechtenberg, T., and Teufel, R. (2022). An acetyltransferase controls the metabolic flux in rubromycin polyketide biosynthesis by direct modulation of redox tailoring enzymes. Chem. Sci. 13, 7157–7164. doi: 10.1039/d2sc01952c
Tscherner, M., Stappler, E., Hnisz, D., and Kuchler, K. (2012). The histone acetyltransferase Hat1 facilitates DNA damage repair and morphogenesis in Candida albicans. Mol. Microbiol 86, 1197–1214. doi: 10.1111/mmi.12051
Tu, S., Guo, S. J., Chen, C. S., Liu, C. X., Jiang, H. W., Ge, F., et al. (2015). YcgC represents a new protein deacetylase family in prokaryotes. Elife 4, e05322. doi: 10.7554/eLife.05322
Tucker, A. C. and Escalante-Semerena, J. C. (2013). Acetoacetyl-CoA synthetase activity is controlled by a protein acetyltransferase with unique domain organization in Streptomyces lividans. Mol. Microbiol 87, 152–167. doi: 10.1111/mmi.12088
Wagner, G. R. and Payne, R. M. (2013). Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 288, 29036–29045. doi: 10.1074/jbc.M113.486753
Walsh, G. and Jefferis, R. (2006). Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 24, 1241–1252. doi: 10.1038/nbt1252
Wang, X., Chang, P., Ding, J., and Chen, J. (2013). Distinct and redundant roles of the two MYST histone acetyltransferases Esa1 and Sas2 in cell growth and morphogenesis of Candida albicans. Eukaryot Cell 12, 438–449. doi: 10.1128/EC.00275-12
Wang, Q., Zhang, Y., Yang, C., Xiong, H., Lin, Y., Yao, J., et al. (2010). Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007. doi: 10.1126/science.1179687
Weinert, B. T., Iesmantavicius, V., Wagner, S. A., Scholz, C., Gummesson, B., Beli, P., et al. (2013). Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51, 265–272. doi: 10.1016/j.molcel.2013.06.003
Wellen, K. E. and Thompson, C. B. (2012). A two-way street: reciprocal regulation of metabolism and signalling. Nat. Rev. Mol. Cell Biol. 13, 270–276. doi: 10.1038/nrm3305
Wu, L., Gong, T., Zhou, X., Zeng, J., Huang, R., Wu, Y., et al. (2019). Global analysis of lysine succinylome in the periodontal pathogen Porphyromonas gingivalis. Mol. Microbiol 34, 74–83. doi: 10.1111/omi.12255
Wurtele, H., Tsao, S., Lepine, G., Mullick, A., Tremblay, J., Drogaris, P., et al. (2010). Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat. Med. 16, 774–780. doi: 10.1038/nm.2175
Xu, J. Y., You, D., Leng, P. Q., and Ye, B. C. (2014). Allosteric regulation of a protein acetyltransferase in Micromonospora aurantiaca by the amino acids cysteine and arginine. J. Biol. Chem. 289, 27034–27045. doi: 10.1074/jbc.M114.579078
Yan, J., Barak, R., Liarzi, O., Shainskaya, A., and Eisenbach, M. (2008). In vivo acetylation of CheY, a response regulator in chemotaxis of Escherichia coli. J. Mol. Biol. 376, 1260–1271. doi: 10.1016/j.jmb.2007.12.070
Yan, Y., Hailun, H., Fenghui, Y., Pingting, L., Lei, L., Zhili, Z., et al. (2023). Streptococcus mutans dexA affects exopolysaccharides production and biofilm homeostasis. Mol. Microbiol 38, 134–144. doi: 10.1111/omi.12395
Yang, C., Li, G., Zhang, Q., Bai, W., Li, Q., Zhang, P., et al. (2024). Histone deacetylase Sir2 promotes the systemic Candida albicans infection by facilitating its immune escape via remodeling the cell wall and maintaining the metabolic activity. mBio 15, e0044524. doi: 10.1128/mbio.00445-24
Yang, X. J. and Seto, E. (2008). The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206–218. doi: 10.1038/nrm2346
Yu, S., Paderu, P., Lee, A., Eirekat, S., Healey, K., Chen, L., et al. (2022). Histone acetylation regulator gcn5 mediates drug resistance and virulence of candida glabrata. Microbiol Spectr. 10, e0096322. doi: 10.1128/spectrum.00963-22
Zeng, J., Wu, L., Chen, Q., Wang, L., Qiu, W., Zheng, X., et al. (2020). Comprehensive profiling of protein lysine acetylation and its overlap with lysine succinylation in the Porphyromonas gingivalis fimbriated strain ATCC 33277. Mol. Microbiol 35, 240–250. doi: 10.1111/omi.12312
Zhang, Q., Zhou, A., Li, S., Ni, J., Tao, J., Lu, J., et al. (2016). Reversible lysine acetylation is involved in DNA replication initiation by regulating activities of initiator DnaA in Escherichia coli. Sci. Rep. 6, 30837. doi: 10.1038/srep30837
Zhao, K., Chai, X., and Marmorstein, R. (2004). Structure and substrate binding properties of cobB, a Sir2 homolog protein deacetylase from Escherichia coli. J. Mol. Biol. 337, 731–741. doi: 10.1016/j.jmb.2004.01.060
Keywords: lysine acetylation, oral microorganisms, bacterial virulence, environmental adaptation, immune escape
Citation: Yang Y, He H, Liu B, Li Z, Sun J, Zhao Z and Yang Y (2025) Protein lysine acetylation regulates oral microorganisms. Front. Cell. Infect. Microbiol. 15:1594947. doi: 10.3389/fcimb.2025.1594947
Received: 17 March 2025; Accepted: 28 April 2025;
Published: 15 May 2025.
Edited by:
Keke Zhang, Wenzhou Medical University, ChinaReviewed by:
Yuqing Li, Sichuan University, ChinaHong Chen, Chongqing Medical University, China
Bin Zhang, Xi’an Jiaotong University, China
Copyright © 2025 Yang, He, Liu, Li, Sun, Zhao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Yang, eWFuZ3lhbmNzdUBzY3UuZWR1LmNu; Zhili Zhao, emhpbGktemhhb0Bjc3UuZWR1LmNu
 Yuanchao Yang
Yuanchao Yang Hailun He
Hailun He Bingshi Liu
Bingshi Liu Zhuoyue Li
Zhuoyue Li Jiaman Sun
Jiaman Sun Zhili Zhao
Zhili Zhao Yan Yang
Yan Yang