- Mater Research Institute - University of Queensland, Translational Research Institute, Woolloongabba, QLD, Australia
Sulfonated glycosaminoglycans, such as heparan sulfate and dermatan sulfate, form major components of the cell surface and extracellular matrix, and display vital roles in mammalian physiology, including growth and development. The identification of specific binding to different glycosaminoglycans by a variety of pathogens has led to increased interest in this mechanism for understanding infection. Over the past four decades there have been more than 300 studies on various pathogens that utilize glycosaminoglycans in their infection process. Currently, no articles have collated all known pathogens that use this process. So it is timely that this article provides an overview of all known pathogens that use glycosaminoglycans to enhance their binding and/or infection in human cells. This was done by using the search terms “sulfate/sulphate” “pathogen”, “virus”, “bacteria”, “parasite”, “infection” and “glycosaminoglycans” to curate peer-reviewed and relevant original research articles from PubMed. This search found that glycosaminoglycans are used in the infection process for 59 viruses, 28 bacteria, and 8 other pathogens (i.e. parasitic protozoa, prions). These findings highlight the conserved and widespread use of glycosaminoglycans for enhancing pathogen infection. In addition, the curated list of pathogens in this study provides a resource for future studies to consider potential therapeutic approaches for targeted disruption of the interaction between pathogens and glycosaminoglycans.
1 Introduction
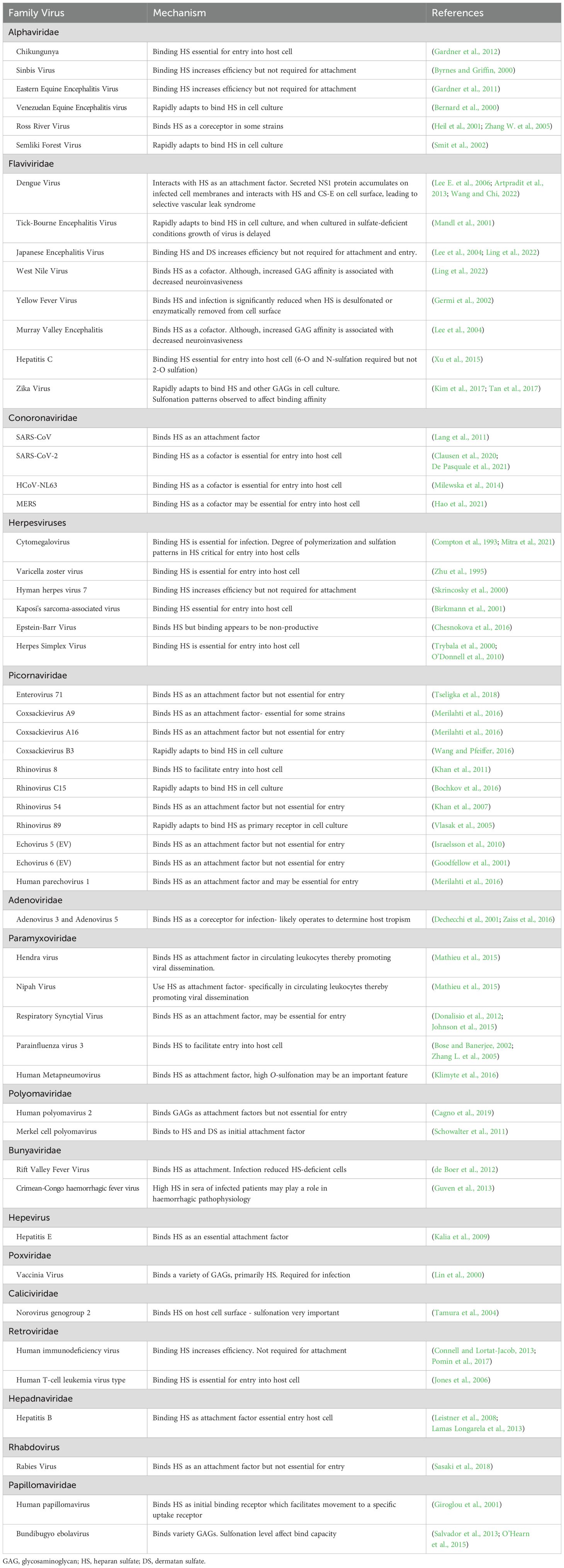
Sulfate (SO42-) plays a critical role in modulating numerous molecular and cellular functions in mammalian physiology (Dawson et al., 2015a). Conjugation of sulfate (sulfonation) to glycosaminoglycans (GAGs) plays an important role in maintaining the structure and function of tissues throughout the body. Several GAGs, including heparan sulfate (HS) and dermatan sulfate (DS), are major components of the cell surface and extracellular matrix (Wang and Chi, 2022). The attachment of numerous pathogens to mammalian host cells is enhanced by the sulfate content of GAGs. Sulfate provides a negative charge, leading to an electrostatic interaction with the basic residues of the pathogen surfaces that increases pathogen concentration at the host cell surface (Figure 1A), thus enhancing more efficient infection (Carvajal-Barriga and Fields, 2023; Lauster et al., 2023). Since the recent COVID pandemic, research into the role of sulfonated GAGs and enhanced pathogen infection has increased with the finding of HS as an attachment receptor for SARS-CoV-2 (De Pasquale et al., 2021).

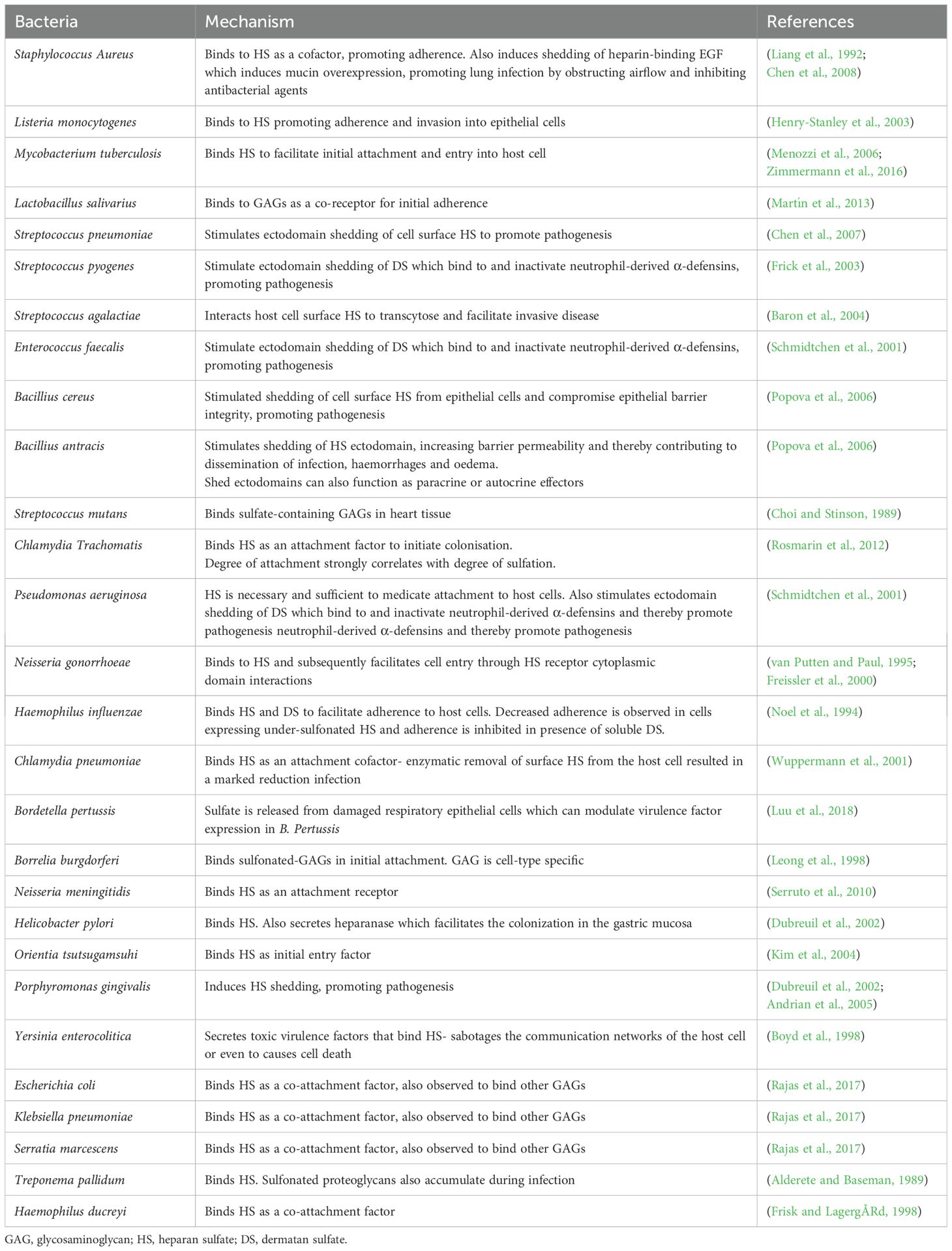
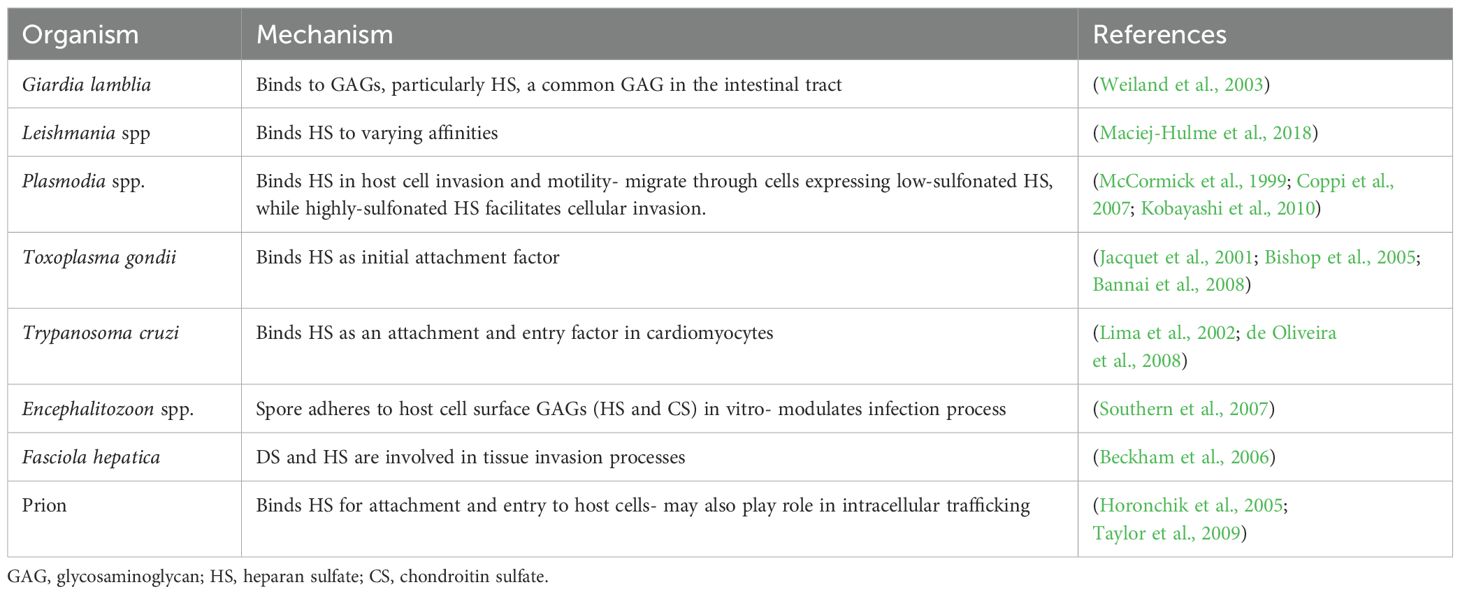
Figure 1. GAG-pathogen interactions. (A) Electrostatic interactions and (B) the functional roles of GAGs in pathogen binding and entry. (C) Summary of pathogens that infect mammalian cells via glycosaminoglycans.
Importantly, a sufficient supply of sulfate is needed to maintain the required sulfate content of GAGs (Cole and Evrovski, 2000; Dawson et al., 2003; Dawson et al., 2009). This is highly relevant when considering the requirement of GAGs for enhancing pathogen binding and entry. Circulating sulfate levels are altered by diet, pharmaceuticals, certain physiological conditions and genetics (Dawson, 2013). By inference, these factors which impact sulfate supply from circulation are proposed to subsequently compromise or enhance infection of GAG-binding pathogens.
Previous studies have focused predominantly on certain pathogens that are known to interact with GAGs. This study aimed to provide an overview of all viral, bacteria and parasitic pathogens that are known to interact with GAGs, leading to enhanced mammalian cell infection. This was done by using the search terms “sulfate/sulphate”, “pathogen”, “virus”, “bacteria”, “parasite”, “infection” and “glycosaminoglycans” to curate peer-reviewed research articles from PubMed, with searches done between February to November 2024. The articles returned from these searches were filtered for English, screened for duplicates and relevance and then reviewed to compile a list of pathogens. It was found that the use of GAGs is a highly conserved feature in the infection process for 95 pathogens (59 viruses, 28 bacteria, 7 parasites and 1 prion). These findings provide information for future studies of pathogen infection and those factors that increase or decrease the sulfate content of GAGs.
2 Sulfate biology
In humans, sulfate is obtained from diet and the intracellular catabolism of sulfur-containing amino acids (Dawson et al., 2015a). Dietary sulfate is absorbed via the intestinal epithelium and supplies approximately a third of daily sulfate requirements (Dawson, 2013). However, intake can vary greatly (1.5–16 mmol/day) depending on types of food consumed and source of drinking water (Dawson, 2013). Circulating sulfate levels are maintained by the kidneys, which filter sulfate in the glomerulus and then reabsorb sulfate in the proximal tubule (Dawson et al., 2015a).
Sulfate reabsorption is mediated by two sulfate transporters; SLC13A1 is located on the apical membrane where it mediates the first step of reabsorption, and SL26A1 which mediates the second step across the basolateral membrane (Karniski et al., 1998). Tissue-specific sulfate transporters mediate the uptake of sulfate from circulation into cells, which is then used to generate 3’-phosphoadenosine 5’-phosphosulfate (PAPS) by PAPS synthetase. The sulfonate group from PAPS is transferred via sulfotransferases to a wide range of endogenous and exogenous molecules (McCarver and Hines, 2002). Sulfate conjugation (sulfonation) alters the physiological properties of molecules including: (i) clearance and detoxification of xenobiotics and certain pharmaceutical drugs (McCarver and Hines, 2002); (ii) inactivation of neurotransmitters, steroids and thyroid hormone (McCarver and Hines, 2002; Dawson, 2012); and (iii) maintenance of tissue structure and function by altering sulfate content of GAGs (Sarrazin et al., 2011). Disturbances within any of these sulfate pathways, and subsequently the balance of sulfonated and unconjugated substrates, has the potential to modify the biophysical properties of cells.
3 Factors impacting circulating sulfate levels
In humans, circulating sulfate level is approximately 300 µmol/L but this can be altered by physiological, environmental and genetic factors (Cole and Evrovski, 2000). Diet is a significant contributing factor to sulfate levels, with food (~0.85 g SO42-/day) and drinking water (~0.78 g SO42-/day) accounting for approximately one third of estimated sulfate requirements (Allen et al., 1989; Florin et al., 1991; Florin et al., 1993). Animal studies have also shown that restricting dietary intake of sulfate intake can lead to hyposulfatemia and reduced sulfonation capacity, which can be reversed by sulfate supplementation (McGarry and Roe, 1973; Price and Jollow, 1989; Hou et al., 2003; Pecora et al., 2006). Additionally, ingestion of some phenolic-based pharmaceuticals that are metabolized by sulfonation are also known to decrease circulating sulfate levels (Kauffman, 2004).
In pregnancy, circulating sulfate concentrations increase significantly with levels peaking in late gestation (Dawson et al., 2015b). This increased sulfatemia is mediated by up-regulation of sulfate reabsorption due to a 2-fold increase in SLC13A1 expression in the maternal kidneys (Dawson et al., 2012; Dawson et al., 2015b). This provides a reservoir to meet the needs of the developing fetus, which has negligible capacity to generate sulfate until late gestation and thereby, is completely reliant on the maternal sulfate supply (Dawson, 2011).
Chronic kidney disease (CKD) is another physiological condition known to affect circulating sulfate levels, increasing by approximately 2-fold due to reduced glomerular filtration rate (Yildirim et al., 2019). Previous studies have shown a reduction in serum sulfate by more than 60% in CKD patients following 6 hours of dialysis (Freeman and Richards, 1979).
More than 90 genes are involved in the maintenance of sulfate homeostasis, including those encoding sulfate transporters (Langford et al., 2017). Previous studies have shown that targeted disruption of Slc13a1 leads to hypersulfaturia, hyposulfatemia and reduced sulfonation capacity in mice (Dawson et al., 2003). Additionally, loss-of-function mutations in human SLC13A1 gene that cause hypersulfaturia and hyposulfatemia have also been identified (Bowling et al., 2012; Tise et al., 2025). To date, 752 validated non-synonymous (ns) single nucleotide polymorphisms (SNPs) in SLC13A1 have been identified, more than 400 of which are predicted to disrupt sulfate transport (Dawson and Markovich, 2007; Langford et al., 2017). SLC13A1 has an uncommonly high ratio (Ka: Ks ≈4:1) of nsSNPs to synonymous SNPs, which is consistent with a strong positive selection for evolutionary change (Kreitman and Comeron, 1999; Dawson and Markovich, 2007). The high Ka: Ks ratio found in SLC13A1, together with the high allelic frequency (range = 22.5 to 40.4%) of N174S which leads to ≈60% loss of sulfate transport function (Lee S. et al., 2006), implies that reduced SLC13A1 function, and subsequent decrease in circulating sulfate level, may have provided a biological benefit to human evolution.
In conclusion, circulating sulfate levels are altered by diet, pharmaceuticals, certain physiological conditions and genetics (Dawson, 2013). Furthermore, low sulfate levels have been linked to a decrease in sulfonation capacity and sulfate content of resulting substrates, including cell-surface GAGs (Dawson et al., 2009). The negative charge conferred by sulfate is an important factor in cellular processes mediated by GAGs, such as the internalization of macromolecules, therefore a decrease in sulfonation capacity has the potential to disrupt these processes (Wadstrom and Ljungh, 1999).
4 Sulfonated glycosaminoglycans
All GAGs contain O-sulfonation, while heparan sulfate (HS) also contains N-sulfonation (Rudd et al., 2010). The degree of sulfonation and overall sulfate content of GAGs is dependent on circulating sulfate levels, which are impacted by various factors as described above. Sulfonation of various hydroxyl groups or amino groups present on the glucosamine component determines its ability to interact with various proteins and subsequently its bioactive function (Afratis et al., 2012).
HS consists of repeating disaccharide units of N-acetylglucosamine and hexuronic acid (Casale and Crane, 2025). HS is tethered to a proteoglycan (PG) core protein core via a serine residue connected to a tetrasaccharide (Casale and Crane, 2025). Chondroitin sulfate (CS) and dermatan sulfate are very similar in structural composition to HS, with the primary difference being the presence of N-sulfates present in HS (Rudd et al., 2010). Keratin sulfate (KS) consists of repeating galactose and N-acetylglucosamine disaccharides, with sulfation present on either unit of the disaccharide repeat. Unlike other GAGs, KS is not connected via a tetrasaccharide linker to the PG core. Instead, the three subtypes of KS (KSI, KSII and KSIII) each use a unique mechanism for linkage to the PG core. KSI GAG chains are tethered by a complex glycan structure utilizing an asparagine amino acid link, KSII chains have an N-acetylgalactosamine link via serine or threonine residues, and KSIII has a mannose linker via serine or threonine residues (Prydz, 2015). The molecular structure of individual GAGs determines their resulting properties, including their affinity for binding other molecules (Casale and Crane, 2025).
The negative charge of GAGs is known to enhance the binding and internalization of macromolecules, including various viral, bacterial and parasitic pathogens (Wadstrom and Ljungh, 1999; De Pasquale et al., 2021). Many viruses, including SARS-CoV 2 (Chu et al., 2021), Dengue virus (DENV) (Artpradit et al., 2013) and Herpes Simplex Virus (HSV) (O’Donnell and Shukla, 2008) bind to GAGs as a receptor for their initial attachment to host cells (Figure 1B). Several bacteria, such as Listeria monocytogenes (Henry-Stanley et al., 2003), Mycobacterium tuberculosis (Zimmermann et al., 2016) and Pseudomonas aeruginosa (Bucior et al., 2012), similarly utilize GAGs for attachment to host cells. Additionally, several bacterial pathogens induce the release of DS or HS from cell surface to counteract cationic antimicrobial factors or neutrophil-mediated host defense mechanisms (Park et al., 2001; Schmidtchen et al., 2001; Park et al., 2004; Chen et al., 2007). Furthermore, several pathogens have also been shown to subvert GAGs to prevent detection by immune mechanisms (Chen et al., 2008; Aquino and Park, 2016). Altogether, these studies suggest that GAG–pathogen interactions and subversion of GAG functions are important virulence mechanisms for a wide variety of pathogens.
While GAG-binding occurs in regions of positive charge within the binding proteins of pathogens, it is not simple to predict. Arginine residues are seen to bind more tightly to GAGs than lysine despite having identical net charges (Eilts et al., 2023). It has also been suggested that certain spacing between basic residues may be critical for binding to occur (Eilts et al., 2023). For some GAG-pathogen interactions, the degree and sequence of polymerization and sulfonation have been observed to impact binding affinity (Mitra et al., 2021). For example, CMV has been observed to preferentially bind HS with higher degrees of polymerization and sulfonation (Mitra et al., 2021).
This review brings together all known viruses, bacteria and parasites that utilize GAGs to bind and infect mammalian host cells. It also aims to curate information from those studies exploring the relationship between the sulfate content of GAGs and potential for infection. This knowledge provides a resource for future studies into the role of pathogen invasion into host cells via GAGs and how this may be impacted by those factors which are known to alter circulating sulfate level.
5 Pathogens that utilize sulfonated GAGs for infection
5.1 Viruses
This study identified that binding of GAGs for entry into mammalian cells is conserved across at least 6 virus families; alphaviridae, flaviviridae, coronaviridae, picornaviridae, orthoherpiviridae and paramyxoviridae. In total, 59 viruses were identified as interacting with GAGs for in vivo infection or shown to rapidly adapt to bind GAGs in cultured cell lines (Table 1).
The heavily sulfonated chains of cell-surface GAGs present a global negative charge that can interact electrostatically with basic residues of viral capsid proteins or viral surface glycoproteins of enveloped viruses (Cagno et al., 2019). Viruses utilize these interactions to increase their concentration at the cell surface and increase the chances of binding a more specific entry receptor and initiating the infection process (Rusnati et al., 2009). In some cases, GAGs act directly as the primary attachment receptor (Figure 1B), such as HSV (O’Donnell and Shukla, 2008). HSV-1 envelope glycoproteins gB and/or gC initiates the viral interaction with HS, followed by the binding of gD to a secondary receptor to initiate membrane fusion with the host cell (O’Donnell and Shukla, 2008). Specific positively charged regions of gC interact with 6-O- and 2-O-sulfate groups on HS to confer binding (Feyzi et al., 1997). Additionally, a short lysine-rich region of gB which is required for gB-mediated HSV attachment has been identified as the HS binding domain (Laquerre et al., 1998). GAGs also act as mediators for the initial endocytosis of viral particles (Figure 1B), which controls the virulency and pathogenicity of infection (Bauer et al., 2021). A sufficient sulfate content of GAGs has been shown to be integral in this process, as several studies have shown that treatment with sulfonation inhibitors, enzymatic removal of sulfate or culturing cell lines in sulfate-deficient conditions reduces infection (Trybala et al., 2000; Mandl et al., 2001; Su et al., 2001; Germi et al., 2002; Tamura et al., 2004).
Due to this role in the initial infection process, GAGs have garnered interest in prophylactic and therapeutic antiviral studies. Treating virus particles with GAGs was shown to inhibit binding of surface glycoproteins to host cell receptors, preventing entry and effectively neutralizing the virus (Leonova and Belikov, 2019). Heparinized blood has also been shown to inhibit binding and entry of pathogens known to interact with host cell GAGs (Aquino and Park, 2016). Additionally, some viruses that do not use GAGs in vivo become GAG-dependent after repeated passage in cell culture, resulting in improved viral fitness and out-competing of GAG-independent variants (Cagno et al., 2019). As these viruses can rapidly adapt to utilizing GAGs in cultured cells, similar adaptations have the potential to occur during human infections to promote replication and infection.
5.2 Bacteria
This study identified 28 pathogenic bacteria that bind GAGs or utilize ectodomain shedding of GAGs to promote pathogenesis, of which 11 are gram-positive and 17 are gram-negative (Table 2). GAGs are involved in adhesion and internalization of bacterial pathogens, including both gram-negative and gram-positive bacteria (Garcia et al., 2016a). HS proteoglycans on the cell surface mediate endocytosis of several HS-binding ligands (Figure 1B), although the precise mechanisms leading to ligand internalization are not completely understood (Bartlett and Park, 2011). Certain bacteria have adapted to subvert this mechanism for entry and colonization of host cells. A sufficient degree of sulfonation of these GAGs is required to facilitate this binding, with studies showing that treatment with sulfonation inhibitors or enzymatic removal of sulfate reduces infection (Noel et al., 1994; Rosmarin et al., 2012; Rajas et al., 2017). For example, host cell HS is a receptor for the Group B Streptococcus surface protein ACP. ACP-HS binding was shown to facilitate internalization of Group B Streptococcus via mechanisms requiring rho GTPase-mediated actin polymerization (Kamhi et al., 2013). Higher degree of polymerization and negative charge are also critical to ACP interactions, as infectivity is markedly decreased in host cells deficient in HS polymerases or N-sulfotransferases (Chang et al., 2011).
Additionally, upregulated expression of certain GAGs following tissue injury or epithelial damage is proposed to play a role in increased propensity for bacteria to cause infection in the context of tissue damage and repair (Bartlett and Park, 2011). Studies have shown that the presence of a mixture of GAGs inhibited adhesion to the same extent as when using only HS in gram-positive bacteria. However, the use of a combination of different GAGs significant increased inhibition compared to only HS in gram-negative bacteria, suggesting that HS is the primary GAG used but other GAG species are also involved for these microorganisms (Garcia et al., 2016b).
GAGs are also observed to promote bacterial infection by serving as a soluble inhibitor of innate immunity when released into the extracellular environment via ectodomain shedding (Aquino et al., 2022). Ectodomain shedding via enzymatic cleavage of cell surface GAGs, most commonly the HS proteoglycan sydecan-1, can be induced by certain bacterial pathogens either by hijacking host cell machinery or secreting ectodomain-cleaving enzymes (Bartlett and Park, 2011). Released sydecan-1 ectodomain then binds to and inhibits host immune factors, such as cytokines and antimicrobial peptides, resulting in dysregulation of host immune response and enhancement of pathogenesis (Garcia et al., 2016a).
5.3 Parasites and prion
This study identified 7 parasitic organisms and 1 prion particle that interact with GAGs in mammalian infection (Table 3). Various parasitic pathogens have been observed to use GAGs as adhesion receptors to attach to host cells (Kamhi et al., 2013). Mast cells, the primary immune cells involved in protecting against parasitic infections, are particularly rich in highly sulfonated GAGs. These GAGs are released during degranulation in response to parasites (Mulloy et al., 2017). Some parasites, much like bacteria, can synthesize or induce shedding of host GAGs to modulate the host immune response and enhance pathogenicity (Kamhi et al., 2013). HS on the surface of erythrocytes has shown to be important, if not essential, for the binding and entry of Plasmodium falciparum, however the exact mechanisms are not yet known (Kobayashi et al., 2010).
Prion diseases are untreatable and fatal neurodegenerative diseases that result from conversion of a normal cell surface protein into a pathological conformation that is transmissible (Westergard et al., 2007). Enzymatic removal of surface HS, prevention of sulfonation with chlorate or presence of competing sulfonated glycans prevent binding and internalization of infectious prion rods, indicating cell surface HS is required for prion infection (Horonchik et al., 2005). HS is also proposed to play a role in the intracellular trafficking of pathogenic prions (Horonchik et al., 2005).
6 Conclusion
In conclusion, GAGs are involved in the infection process of numerous pathogens and sufficient sulfate content is needed to facilitate these interactions. Circulating sulfate levels are decreased or increased by several factors, leading to altered sulfate content of GAGs which in turn is proposed to subsequently compromise or enhance infection of GAG-binding pathogens. Therapeutic approaches for targeting GAG-pathogen interactions have the potential to reduce pathogen infection. Initial results from in vitro and cell culture studies have increased clinical interest for future prophylactic and therapeutic antipathogen treatments.
Recent studies have focused predominantly on certain pathogens that are known to interact with GAGs. This review brings together all known human pathogens that are known to interact with GAGs in infection. In total 59 viruses, 28 bacteria, 7 parasites and 1 prion were identified, showing that the use of GAGs is a highly conserved feature (Figure 1C). These findings provide a resource for future studies and highlight the need for further studies to investigate the consequences of high or low sulfatemia on pathogen infection.
Author contributions
JM: Writing – original draft, Investigation, Visualization, Formal analysis, Validation, Methodology, Data curation, Conceptualization. PD: Writing – review & editing, Project administration, Resources, Methodology, Validation, Supervision, Investigation, Conceptualization, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by an Ideas grant (2020999) from the Australian National Health and Medical Research Council. We also acknowledge funding support from Mater Research and the Mater Foundation.
Acknowledgments
PD is supported by a Mater Foundation Principal Research Fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Afratis, N., Gialeli, C., Nikitovic, D., Tsegenidis, T., Karousou, E., Theocharis, A. D., et al. (2012). Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J. 279, 1177–1197. doi: 10.1111/j.1742-4658.2012.08529.x
Alderete, J. F. and Baseman, J. B. (1989). Serum lipoprotein binding by Treponema pallidum: possible role for proteoglycans. Sex. Transmit. Infect. 65, 177–182. doi: 10.1136/sti.65.3.177
Allen, H. E., Halley-Henderson, M. A., and Hass, C. N. (1989). Chemical composition of bottled mineral water. Arch. Environ. Health 44, 102–116. doi: 10.1080/00039896.1989.9934383
Andrian, E., Grenier, D., and Rouabhia, M. (2005). Porphyromonas gingivalis lipopolysaccharide induces shedding of syndecan-1 expressed by gingival epithelial cells. J. Cell Physiol. 204, 178–183. doi: 10.1002/jcp.v204:1
Aquino, R. S. and Park, P. W. (2016). Glycosaminoglycans and infection. Front. Biosci. (Landmark Ed) 21, 1260–1277. doi: 10.2741/4455
Aquino, RS, Hayashida, K, Hayashida, A, and Park, PW. (2022). Role of HSPGs in systemic bacterial infections. Methods Mol. Biol. 2303, 605–625. doi: 10.1007/978-1-0716-1398-6_46
Artpradit, C., Robinson, L. N., Gavrilov, B.K., Rurak, T. T., Ruchirawat, M., and and Sasisekharan, R. (2013). Recognition of heparan sulfate by clinical strains of dengue virus serotype 1 using recombinant subviral particles. Virus Res. 176, 69–77. doi: 10.1016/j.virusres.2013.04.017
Bannai, H., Nishikawa, Y., Matsuo, T., Kawase, O., Watanabe, J., Sugimoto, C., et al. (2008). Programmed Cell Death 5 from Toxoplasma gondii: A secreted molecule that exerts a pro-apoptotic effect on host cells. Mol. Biochem. Parasitol. 159, 112–120. doi: 10.1016/j.molbiopara.2008.02.012
Baron, M. J., Bolduc, G. R., Goldberg, M. B., Aupérin, T. C., and Madoff, L. C. (2004). Alpha C protein of group B streptococcus binds host cell surface glycosaminoglycan and enters cells by an actin-dependent mechanism. J. Biol. Chem. 279, 24714–24723. doi: 10.1074/jbc.M402164200
Bartlett, A. H. and Park, P. W. (2011). Heparan sulfate proteoglycans in infection. Glycans Dis. Ther. 19, 31–62. doi: 10.1007/978-3-642-16833-8_2
Bauer, S., Zhang, F., and Linhardt, R. J. (2021). Implications of glycosaminoglycans on viral zoonotic diseases. Diseases 9(4):85. doi: 10.3390/diseases9040085
Beckham, S. A., Law, R. H., Smooker, P. M., Quinsey, N. S., Caffrey, C. R., McKerrow, J. H., et al. (2006). Production and processing of a recombinant Fasciola hepatica cathepsin B-like enzyme (FhcatB1) reveals potential processing mechanisms in the parasite. Biol. Chem. 387, 1053–1061. doi: 10.1515/BC.2006.130
Bernard, K. A., Klimstra, W. B., and Johnston, R. E. (2000). Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276, 93–103. doi: 10.1006/viro.2000.0546
Birkmann, A., Mahr, K., Ensser, A., Yağuboğlu, S., Titgemeyer, F., Fleckenstein, B., et al. (2001). Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75, 11583–11593. doi: 10.1128/JVI.75.23.11583-11593.2001
Bishop, J. R., Crawford, B. E., and Esko, J. D. (2005). Cell surface heparan sulfate promotes replication of toxoplasma gondii. Infect. Immun. 73, 5395–5401. doi: 10.1128/IAI.73.9.5395-5401.2005
Bochkov, Y. A., Watters, K., Basnet, S., Sijapati, S., Hill, M., Palmenberg, A. C., et al. (2016). Mutations in VP1 and 3A proteins improve binding and replication of rhinovirus C15 in HeLa-E8 cells. Virology 499, 350–360. doi: 10.1016/j.virol.2016.09.025
Bose, S. and Banerjee, A. K. (2002). Role of heparan sulfate in human parainfluenza virus type 3 infection. Virology 298, 73–83. doi: 10.1006/viro.2002.1484
Bowling, F. G., Heussler, H. S., McWhinney, A., and Dawson, P. A. (2012). Plasma and urinary sulfate determination in a cohort with autism. Biochem. Genet. 51, 147–153. doi: 10.1007/s10528-012-9550-0
Boyd, A. P., Sory, M. P., Iriarte, M., and Cornelis, G. R. (1998). Heparin interferes with translocation of Yop proteins into HeLa cells and binds to LcrG, a regulatory component of the Yersinia Yop apparatus. Mol. Microbiol. 27, 425–436. doi: 10.1046/j.1365-2958.1998.00691.x
Bucior, I., Pielage, J. F., and Engel, J. N. (2012). Pseudomonas aeruginosa Pili and Flagella Mediate Distinct Binding and Signaling Events at the Apical and Basolateral Surface of Airway Epithelium. PloS Pathog. 8, e1002616. doi: 10.1371/journal.ppat.1002616
Byrnes, A. P. and Griffin, D. E. (2000). Large-plaque mutants of sindbis virus show reduced binding to heparan sulfate, heightened viremia, and slower clearance from the circulation. J. Virol. 74, 644–651. doi: 10.1128/JVI.74.2.644-651.2000
Cagno, V., Tseligka, E. D., Jones, S. T., and Tapparel, C. (2019). Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias? Viruses 11(7):596. doi: 10.3390/v11070596
Carvajal-Barriga, E. J. and Fields, R. D. (2023). Sulfated polysaccharides as multi target molecules to fight COVID 19 and comorbidities. Heliyon 9, e13797. doi: 10.1016/j.heliyon.2023.e13797
Casale, J. and Crane, J. S. (2025). Biochemistry, Glycosaminoglycans (Treasure Island (FL: StatPearls). Available at: https://www.ncbi.nlm.nih.gov/books/NBK544295/.
Chang, Y. C., Wang, Z., Flax, L. A., Xu, D., Esko, J. D., Nizet, V., et al. (2011). Glycosaminoglycan binding facilitates entry of a bacterial pathogen into central nervous systems. PloS Pathog. 7, e1002082. doi: 10.1371/journal.ppat.1002082
Chen, Y., Hayashida, A., Bennett, A. E., Hollingshead, S. K., and Park, P. W. (2007). Streptococcus pneumoniae sheds syndecan-1 ectodomains through ZmpC, a metalloproteinase virulence factor. J. Biol. Chem. 282, 159–167. doi: 10.1074/jbc.M608542200
Chen, Y., Götte, M., Liu, J., and Park, P. W. (2008). Microbial subversion of heparan sulfate proteoglycans. Mol. Cells 26, 415–426. doi: 10.1016/S1016-8478(23)14017-9
Chesnokova, L. S., Valencia, S. M., and Hutt-Fletcher, L. M. (2016). The BDLF3 gene product of Epstein-Barr virus, gp150, mediates non-productive binding to heparan sulfate on epithelial cells and only the binding domain of CD21 is required for infection. Virology 494, 23–28. doi: 10.1016/j.virol.2016.04.002
Choi, S. H. and Stinson, M. W. (1989). Purification of a Streptococcus mutans protein that binds to heart tissue and glycosaminoglycans. Infect. Immun. 57, 3834–3840. doi: 10.1128/iai.57.12.3834-3840.1989
Chu, H., Hu, B., Huang, X., Chai, Y., Zhou, D., Wang, Y., et al. (2021). Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat. Commun. 12, 134. doi: 10.1038/s41467-020-20457-w
Clausen, T. M., Sandoval, D. R., Spliid, C. B., Pihl, J., Perrett, H. R., Painter, C. D., et al. (2020). SARS-coV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183, 1043–1057.e15. doi: 10.1016/j.cell.2020.09.033
Cole, D. E. and Evrovski, J. (2000). The clinical chemistry of inorganic sulfate. Crit. Rev. Clin. Lab. Sci. 37, 299–344. doi: 10.1080/10408360091174231
Compton, T., Nowlin, D. M., and Cooper, N. R. (1993). Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 193, 834–841. doi: 10.1006/viro.1993.1192
Connell, B. J. and Lortat-Jacob, H. (2013). Human immunodeficiency virus and heparan sulfate: from attachment to entry inhibition. Front. Immunol. 4. doi: 10.3389/fimmu.2013.00385
Coppi, A., Tewari, R., Bishop, J. R., Bennett, B. L., Lawrence, R., Esko, J. D., et al. (2007). Heparan sulfate proteoglycans provide a signal to plasmodium sporozoites to stop migrating and productively invade host cells. Cell Host Microbe 2, 316–327. doi: 10.1016/j.chom.2007.10.002
Dawson, P. A. (2011). Sulfate in fetal development. Semin. Cell Dev. Biol. 22, 653–659. doi: 10.1016/j.semcdb.2011.03.004
Dawson, P. A. (2012). The biological roles of steroid sulfonation. Steroids – From Physiol. to Clin. Med. 2, 45–64. doi: 10.5772/52714
Dawson, P. A. (2013). Role of sulphate in development. Reproduction 146, R81–R89. doi: 10.1530/REP-13-0056
Dawson, P. A., Beck, L., and Markovich, D. (2003). Hyposulfatemia, growth retardation, reduced fertility and seizures in mice lacking a functional NaSi-1 gene. Proc. Natl. Acad. Sci. U.S.A. 100, 13704–13709. doi: 10.1073/pnas.2231298100
Dawson, P. A., Elliott, A., and Bowling, F. G. (2015a). Sulphate in pregnancy. Nutrients 7, 1594–1606. doi: 10.3390/nu7031594
Dawson, P. A., Huxley, S., Gardiner, B., Tran, T., McAuley, J. L., Grimmond, S., et al. (2009). Reduced mucin sulfonation and impaired intestinal barrier function in the hyposulfataemic NaS1 null mouse. Gut 58, 910–919. doi: 10.1136/gut.2007.147595
Dawson, P. A., Petersen, S., Rodwell, R., Johnson, P., Gibbons, K., McWhinney, A., et al. (2015b). Reference intervals for plasma sulfate and urinary sulfate excretion in pregnancy. BMC Preg. Childbirth 15, 96. doi: 10.1186/s12884-015-0526-z
Dawson, P. A. and Markovich, D. (2007). Genetic polymorphisms of human sulfate transporters. CurrPharm 5, 262–274. doi: 10.2174/157016007782793692
Dawson, P. A., Rakoczy, J., and Simmons, D. G. (2012). Placental, renal, and ileal sulfate transporter gene expression in mouse gestation1. Biol. Reprod. 87. doi: 10.1095/biolreprod.111.098749
de Boer, S. M., Kortekaas, J., de Haan, C. A. M., Rottier, P. J. M., Moormann, R. J. M., and Bosch, B. J. (2012). Heparan sulfate facilitates rift valley fever virus entry into the cell. J. Virol. 86, 13767–13771. doi: 10.1128/JVI.01364-12
Dechecchi, M. C., Melotti, P., Bonizzato, A., Santacatterina, M., Chilosi, M., and Cabrini, G. (2001). Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J. Virol. 75, 8772–8780. doi: 10.1128/JVI.75.18.8772-8780.2001
Oliveira Jr., F. O., Alves, C. R., Calvet, C. M., Toma, L., Bouças, R. I., Nader, H. B., et al. (2008). Trypanosoma cruzi heparin-binding proteins and the nature of the host cell heparan sulfate-binding domain. Microb. Pathog. 44, 329–338. doi: 10.1016/j.micpath.2007.10.003
De Pasquale, V., Quiccione, M. S., Tafuri, S., Avallone, L., and Pavone, L. M. (2021). Heparan sulfate proteoglycans in viral infection and treatment: A special focus on SARS-CoV-2. Int. J. Mol. Sci. 22(12):6574. doi: 10.3390/ijms22126574
Donalisio, M., Rusnati, M., Cagno, V., Civra, A., Bugatti, A., Giuliani, A., et al. (2012). Inhibition of human respiratory syncytial virus infectivity by a dendrimeric heparan sulfate-binding peptide. Antimicrob. Agents Chemother. 56, 5278–5288. doi: 10.1128/AAC.00771-12
Dubreuil, J. D., Giudice, G. D., and Rappuoli, R. (2002). Helicobacter pylori interactions with host serum and extracellular matrix proteins: potential role in the infectious process. Microbiol. Mol. Biol. Rev. 66, 617–629. doi: 10.1128/MMBR.66.4.617-629.2002
Eilts, F., Bauer, S., Fraser, K., Dordick, J. S., Wolff, M. W., Linhardt, R. J., et al. (2023). The diverse role of heparan sulfate and other GAGs in SARS-CoV-2 infections and therapeutics. Carbohydr. Polym. 299, 120167. doi: 10.1016/j.carbpol.2022.120167
Feyzi, E., Trybala, E., Bergström, T., Lindahl, U., and Spillmann, D. (1997). Structural requirement of heparan sulfate for interaction with herpes simplex virus type 1 virions and isolated glycoprotein C. J. Biol. Chem. 272, 24850–24857. doi: 10.1074/jbc.272.40.24850
Florin, T., Neale, G., Gibson, G. R., Christl, S. U., and Cummings, J. H. (1991). Metabolism of dietary sulphate: absorption and excretion in humans. Gut 32, 766–773. doi: 10.1136/gut.32.7.766
Florin, T., Neale, G., Goretski, S., and Cummings, J. (1993). The sulfate content of foods and beverages. J. Food Composit. Anal. 6, 140–151. doi: 10.1006/jfca.1993.1016
Freeman, R. M. and Richards, C. J. (1979). Studies on sulfate in end-stage renal disease. Kidney Int. 15, 167–175. doi: 10.1038/ki.1979.22
Freissler, E., Meyer auf der Heyde, A., David, G., Meyer, T. F., and Dehio, C. (2000). Syndecan-1 and syndecan-4 can mediate the invasion of OpaHSPG-expressing Neisseria gonorrhoeae into epithelial cells. Cell. Microbiol. 2, 69–82. doi: 10.1046/j.1462-5822.2000.00036.x
Frick, I. M., Schmidtchen, A., and Sjobring, U. (2003). Interactions between M proteins of Streptococcus pyogenes and glycosaminoglycans promote bacterial adhesion to host cells. Eur. J. Biochem. 270, 2303–2311. doi: 10.1046/j.1432-1033.2003.03600.x
Frisk, A. and LagergÅRd, T. (1998). Characterization of mechanisms involved in adherence of Haemophilus ducreyi to eukaryotic cells. APMIS 106, 539–546. doi: 10.1111/j.1699-0463.1998.tb01382.x
Garcia, B., Merayo-Lloves, J., Martin, C., Alcalde, I., Quiros, L. M., and and Vazquez, F. (2016a). Surface proteoglycans as mediators in bacterial pathogens infections. Front. Microbiol. 7, 220. doi: 10.3389/fmicb.2016.00220
Garcia, B., Merayo-Lloves, J., Rodriguez, D., Alcalde, I., Garcia-Suarez, O., Alfonso, J. F., et al. (2016b). Different use of cell surface glycosaminoglycans as adherence receptors to corneal cells by gram positive and gram negative pathogens. Front. Cell Infect. Microbiol. 6, 173. doi: 10.3389/fcimb.2016.00173
Gardner, C. L., Ebel, G. D., Ryman, K. D., and Klimstra, W. B. (2011). Heparan sulfate binding by natural eastern equine encephalitis viruses promotes neurovirulence. Proc. Natl. Acad. Sci. 108, 16026–16031. doi: 10.1073/pnas.1110617108
Gardner, C. L., Burke, C. W., Higgs, S. T., Klimstra, W. B., and Ryman, K. D. (2012). Interferon-alpha/beta deficiency greatly exacerbates arthritogenic disease in mice infected with wild-type chikungunya virus but not with the cell culture-adapted live-attenuated 181/25 vaccine candidate. Virology 425, 103–112. doi: 10.1016/j.virol.2011.12.020
Germi, R., Crance, J. M., Garin, D., Guimet, J., Lortat-Jacob, H., Ruigrok, R. W., et al. (2002). Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology 292, 162–168. doi: 10.1006/viro.2001.1232
Giroglou, T., Florin, L., Schäfer, F., Streeck, R. E., and Sapp, M. (2001). Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 75, 1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001
Goodfellow, I. G., Sioofy, A. B., Powell, R. M., and Evans, D. J. (2001). Echoviruses bind heparan sulfate at the cell surface. J. Virol. 75, 4918–4921. doi: 10.1128/JVI.75.10.4918-4921.2001
Guven, F. M., Aydin, H., Kaya, A., Engin, A., Celik, V. K., Korkmaz, I., et al. (2013). The role of heparan sulphate in pathogenesis of Crimean-Congo hemorrhagic fever disease. J. Vector Borne Dis. 50, 133–136. doi: 10.4103/0972-9062.117485
Hao, W., Ma, B., Li, Z., Wang, X., Gao, X., Li, Y., et al. (2021). Binding of the SARS-CoV-2 spike protein to glycans. Sci. Bull. 66, 1205–1214. doi: 10.1016/j.scib.2021.01.010
Heil, M. L., Albee, A., Strauss, J. H., and Kuhn, R. J. (2001). An amino acid substitution in the coding region of the E2 glycoprotein adapts ross river virus to utilize heparan sulfate as an attachment moiety. J. Virol. 75, 6303–6309. doi: 10.1128/JVI.75.14.6303-6309.2001
Henry-Stanley, M. J., Hess, D. J., Erickson, E. A., Garni, R. M., and Wells, C. L. (2003). Role of heparan sulfate in interactions of Listeria monocytogenes with enterocytes. Med. Microbiol. Immunol. 192, 107–115. doi: 10.1007/s00430-002-0165-7
Horonchik, L., Tzaban, S., Ben-Zaken, O., Yedidia, Y., Rouvinski, A., Papy-Garcia, D., et al. (2005). Heparan sulfate is a cellular receptor for purified infectious prions. J. Biol. Chem. 280, 17062–17067. doi: 10.1074/jbc.M500122200
Hou, C., Wykes, L. J., and Hoffer, L. J. (2003). Urinary sulfur excretion and the nitrogen/sulfur balance ratio reveal nonprotein sulfur amino acid retention in piglets. J. Nutr. 133, 766–772. doi: 10.1093/jn/133.3.766
Israelsson, S., Gullberg, M., Jonsson, N., Roivainen, M., Edman, K., and and Lindberg, A. M. (2010). Studies of Echovirus 5 interactions with the cell surface: Heparan sulfate mediates attachment to the host cell. Virus Res. 151, 170–176. doi: 10.1016/j.virusres.2010.05.001
Jacquet, A., Coulon, L., De Nève, J., Daminet, V., Haumont, M., Garcia, L., et al. (2001). The surface antigen SAG3 mediates the attachment of Toxoplasma gondii to cell-surface proteoglycans. Mol. Biochem. Parasitol. 116, 35–44. doi: 10.1016/S0166-6851(01)00297-3
Johnson, S. M., McNally, B. A., Ioannidis, I., Flano, E., Teng, M. N., Oomens, A. G., et al. (2015). Respiratory syncytial virus uses CX3CR1 as a receptor on primary human airway epithelial cultures. PloS Pathog. 11, e1005318. doi: 10.1371/journal.ppat.1005318
Jones, K. S., Fugo, K., Petrow-Sadowski, C., Huang, Y., Bertolette, D. C., Lisinski, I., et al. (2006). Human T-cell leukemia virus type 1 (HTLV-1) and HTLV-2 use different receptor complexes to enter T cells. J. Virol. 80, 8291–8302. doi: 10.1128/JVI.00389-06
Kalia, M., Chandra, V., Rahman, S. A., Sehgal, D., and Jameel, S. (2009). Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 83, 12714–12724. doi: 10.1128/JVI.00717-09
Kamhi, E., Joo, E. J., Dordick, J. S., and Linhardt, R. J. (2013). Glycosaminoglycans in infectious disease. Biol. Rev. Camb Philos. Soc. 88, 928–943. doi: 10.1111/brv.2013.88.issue-4
Karniski, L. P., Lotscher, M., Fucentese, M., Hilfiker, H., Biber, J., and Murer, H. (1998). Immunolocalization of sat-1 sulfate/oxalate/bicarbonate anion exchanger in the rat kidney. Am. J. Physiol. 275, F79–F87. doi: 10.1152/ajprenal.1998.275.1.F79
Kauffman, F. C. (2004). Sulfonation in pharmacology and toxicology. Drug Metab. Rev. 36, 823–843. doi: 10.1081/DMR-200033496
Khan, A. G., Pichler, J., Rosemann, A., and Blaas, D. (2007). Human Rhinovirus Type 54 Infection via Heparan Sulfate Is Less Efficient and Strictly Dependent on Low Endosomal pH. J. Virol. 81, 4625–4632. doi: 10.1128/JVI.02160-06
Khan, A. G., Pickl-Herk, A., Gajdzik, L., Marlovits, T. C., Fuchs, R., and Blaas, D. (2011). Entry of a heparan sulphate-binding HRV8 variant strictly depends on dynamin but not on clathrin, caveolin, and flotillin. Virology 412, 55–67. doi: 10.1016/j.virol.2010.12.042
Kim, H.-R., Choi, M.-S., and Kim, I.-S. (2004). Role of Syndecan-4 in the cellular invasion of Orientia tsutsugamushi. Microb. Pathog. 36, 219–225. doi: 10.1016/j.micpath.2003.12.005
Kim, S. Y., Zhao, J., Liu, X., Fraser, K., Lin, L., Zhang, X., et al. (2017). Interaction of zika virus envelope protein with glycosaminoglycans. Biochemistry 56, 1151–1162. doi: 10.1021/acs.biochem.6b01056
Klimyte, E. M., Smith, S. E., Oreste, P., Lembo, D., and Dutch, R. E. (2016). Inhibition of human metapneumovirus binding to heparan sulfate blocks infection in human lung cells and airway tissues. J. Virol. 90, 9237–9250. doi: 10.1128/JVI.01362-16
Kobayashi, K., Kato, K., Sugi, T., Takemae, H., Pandey, K., Gong, H., et al. (2010). Plasmodium falciparum BAEBL binds to heparan sulfate proteoglycans on the human erythrocyte surface*. J. Biol. Chem. 285, 1716–1725. doi: 10.1074/jbc.M109.021576
Kreitman, M. and Comeron, J. M. (1999). Coding sequence evolution. Curr. Opin. Genet. Dev. 9, 637–641. doi: 10.1016/S0959-437X(99)00034-9
Lamas Longarela, O., Schmidt, T. T., Schöneweis, K., Romeo, R., Wedemeyer, H., Urban, S., et al. (2013). Proteoglycans act as cellular hepatitis delta virus attachment receptors. PloS One 8, e58340. doi: 10.1371/journal.pone.0058340
Lang, J., Yang, N., Deng, J., Liu, K., Yang, P., Zhang, G., et al. (2011). Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PloS One 6, e23710. doi: 10.1371/journal.pone.0023710
Langford, R., Hurrion, E., and Dawson, P. A. (2017). Genetics and pathophysiology of mammalian sulfate biology. J. Genet. Genomics 44, 7–20. doi: 10.1016/j.jgg.2016.08.001
Laquerre, S., Argnani, R., Anderson, D. B., Zucchini, S., Manservigi, R., and Glorioso, J. C. (1998). Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J. Virol. 72, 6119–6130. doi: 10.1128/JVI.72.7.6119-6130.1998
Lauster, D., Osterrieder, K., Haag, R., Ballauff, M., and Herrmann, A. (2023). Respiratory viruses interacting with cells: the importance of electrostatics. Front. Microbiol. 14, 1169547. doi: 10.3389/fmicb.2023.1169547
Lee, S., Dawson, P. A., Hewavitharana, A. K., Shaw, P. N., and Markovich, D. (2006). Disruption of NaS1 sulfate transport function in mice leads to enhanced acetaminophen-induced hepatotoxicity. Hepatology 43, 1241–1247. doi: 10.1002/hep.21207
Lee, E., Wright, P. J., Davidson, A., and Lobigs, M. (2006). Virulence attenuation of Dengue virus due to augmented glycosaminoglycan-binding affinity and restriction in extraneural dissemination. J. Gen. Virol. 87, 2791–2801. doi: 10.1099/vir.0.82164-0
Lee, E., Hall, R. A., and Lobigs, M. (2004). Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and west Nile viruses. J. Virol. 78, 8271–8280. doi: 10.1128/JVI.78.15.8271-8280.2004
Leistner, C. M., Gruen-Bernhard, S., and Glebe, D. (2008). Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell. Microbiol. 10, 122–133. doi: 10.1111/j.1462-5822.2007.01023.x
Leong, J. M., Wang, H., Magoun, L., Field, J. A., Morrissey, P. E., Robbins, D., et al. (1998). Different classes of proteoglycans contribute to the attachment of borrelia burgdorferi to cultured endothelial and brain cells. Infect. Immun. 66, 994–999. doi: 10.1128/IAI.66.3.994-999.1998
Leonova, G. N. and Belikov, S. I. (2019). Effect of glycosaminoglycans on pathogenic properties far-eastern tick-borne encephalitis virus. Bull. Exp. Biol. Med. 167, 482–485. doi: 10.1007/s10517-019-04555-4
Liang, O. D., Ascencio, F., Fransson, L. A., and Wadström, T. (1992). Binding of heparan sulfate to Staphylococcus aureus. Infect. Immun. 60, 899–906. doi: 10.1128/iai.60.3.899-906.1992
Lima, A. P.C.A., Almeida, P. C., Tersariol, I. L.S., Schmitz, V., Schmaier, A. H., Juliano, L., et al. (2002). Heparan Sulfate Modulates Kinin Release by Trypanosoma cruzi through the Activity of Cruzipain. J. Biol. Chem. 277, 5875–5881. doi: 10.1074/jbc.M108518200
Lin, C-L., Chung, C-S., Heine, H. G., and Chang, W. (2000). Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74, 3353–3365. doi: 10.1128/JVI.74.7.3353-3365.2000
Ling, J., Li, J., Khan, A., Lundkvist, Å., and Li, J-P. (2022). Is heparan sulfate a target for inhibition of RNA virus infection? Am. J. Physiology-Cell Physiol. 322, C605–C613. doi: 10.1152/ajpcell.00028.2022
Luu, L. D.W., Octavia Zhong, S. L., Raftery, M. J., Sintchenko, V., and Lan, R. (2018). Comparison of the whole cell proteome and secretome of epidemic bordetella pertussis strains from the 2008–2012 Australian epidemic under sulfate-modulating conditions. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02851
Maciej-Hulme, M. L., Skidmore, M. A., and Price, H. P. (2018). The role of heparan sulfate in host macrophage infection by Leishmania species. Biochem. Soc. Trans. 46, 789–796. doi: 10.1042/BST20170398
Mandl, C. W., Kroschewski, H., Allison, S. L., Kofler, R., Holzmann, H., Meixner, T., et al. (2001). Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J. Virol. 75, 5627–5637. doi: 10.1128/JVI.75.12.5627-5637.2001
Martín, R., Martín, C., Escobedo, S., Suárez, J. E., and Quirós, L. M. (2013). Surface glycosaminoglycans mediate adherence between HeLa cells and Lactobacillus salivarius Lv72. BMC Microbiol. 13, 210–210. doi: 10.1186/1471-2180-13-210
Mathieu, C., Dhondt, K. P., Châlons, M., Mély, S., Raoul, H., Negre, D., et al. (2015). Heparan sulfate-dependent enhancement of henipavirus infection. mBio 6(2):e02427. doi: 10.1128/mBio.02427-14
McCarver, D. G. and Hines, R. N. (2002). The ontogeny of human drug-metabolizing enzymes: phase II conjugation enzymes and regulatory mechanisms. J. Pharmacol. Exp. Ther. 300, 361–366. doi: 10.1124/jpet.300.2.361
McCormick, C. J., Tuckwell, D. S., Crisanti, A., Humphries, M. J., and Hollingdale, M. R. (1999). Identification of heparin as a ligand for the A-domain of Plasmodium falciparum thrombospondin-related adhesion protein. Mol. Biochem. Parasitol. 100, 111–124. doi: 10.1016/S0166-6851(99)00052-3
McGarry, P. C. and Roe, D. A. (1973). Development of sulfur depletion in pregnant and fetal rats: interaction of protein restriction and indole or salicylamide administration. J. Nutr. 103, 1279–1290. doi: 10.1093/jn/103.9.1279
Menozzi, F. D., Reddy, V. M., Cayet, D., Raze, D., Debrie, A-S., Dehouck, M-P., et al. (2006). Mycobacterium tuberculosis heparin-binding haemagglutinin adhesin (HBHA) triggers receptor-mediated transcytosis without altering the integrity of tight junctions. Microbes Infect. 8, 1–9. doi: 10.1016/j.micinf.2005.03.023
Merilahti, P., Karelehto, E., and Susi, P. (2016). Role of heparan sulfate in cellular infection of integrin-binding coxsackievirus A9 and human parechovirus 1 isolates. PloS One 11, e0147168. doi: 10.1371/journal.pone.0147168
Milewska, A., Zarebski, M., Nowak, P., Stozek, K., Potempa, J., and Pyrc, K. (2014). Human coronavirus NL63 utilizes heparan sulfate proteoglycans for attachment to target cells. J. Virol. 88, 13221–13230. doi: 10.1128/JVI.02078-14
Mitra, D., Hasan, M. H., Bates, J. T., Bierdeman, M. A., Ederer, D. R., Parmar, R. C., et al. (2021). The degree of polymerization and sulfation patterns in heparan sulfate are critical determinants of cytomegalovirus entry into host cells. PloS Pathog. 17, e1009803. doi: 10.1371/journal.ppat.1009803
Mulloy, B., Lever, R., and Page, C. P. (2017). Mast cell glycosaminoglycans. Glycoconj J. 34, 351–361. doi: 10.1007/s10719-016-9749-0
Noel, G. J., Love, D. C., and Mosser, D. M. (1994). High-molecular-weight proteins of nontypeable Haemophilus influenzae mediate bacterial adhesion to cellular proteoglycans. Infect. Immun. 62, 4028–4033. doi: 10.1128/iai.62.9.4028-4033.1994
O'Donnell, C. D., Kovacs, M., Akhtar, J., Valyi-Nagy, T., and Shukla, D. (2010). Expanding the role of 3-O sulfated heparan sulfate in herpes simplex virus type-1 entry. Virology 397, 389–398. doi: 10.1016/j.virol.2009.11.011
O’Donnell, C. D. and Shukla, D. (2008). The importance of heparan sulfate in herpesvirus infection. Virol. Sin. 23, 383–393. doi: 10.1007/s12250-008-2992-1
O'Hearn, A., Wang, M., Cheng, H., Lear-Rooney, C. M., Koning, K., Rumschlag-Booms, E., et al. (2015). Role of EXT1 and glycosaminoglycans in the early stage of filovirus entry. J. Virol. 89, 5441–5449. doi: 10.1128/JVI.03689-14
Park, P. W., Pier, G. B., Hinkes, M. T., and Bernfield, M. (2001). Exploitation of syndecan-1 shedding by Pseudomonas aeruginosa enhances virulence. Nature 411, 98–102. doi: 10.1038/35075100
Park, P. W., Foster, T. J., Nishi, E., Duncan, S. J., Klagsbrun, M., and Chen, Y. (2004). Activation of syndecan-1 ectodomain shedding by Staphylococcus aureus alpha-toxin and beta-toxin. J. Biol. Chem. 279, 251–258. doi: 10.1074/jbc.M308537200
Pecora, F., Gualeni, B., Forlino, A., Superti-Furga, A., Tenni, R., Cetta, G., et al. (2006). In vivo contribution of amino acid sulfur to cartilage proteoglycan sulfation. Biochem. J. 398, 509–514. doi: 10.1042/BJ20060566
Pomin, V. H., Bezerra, F. F., and Soares, P. A. G. (2017). Sulfated glycans in HIV infection and therapy. Curr. Pharm. Design 23(23), 3405–3414. doi: 10.2174/1381612823666170127113958
Popova, T. G., Millis, B., Bradburne, C., Nazarenko, S., Bailey, C., Chandhoke, V., et al. (2006). Acceleration of epithelial cell syndecan-1 shedding by anthrax hemolytic virulence factors. BMC Microbiol. 6, 8–8. doi: 10.1186/1471-2180-6-8
Price, V. F. and Jollow, D. J. (1989). Effects of sulfur-amino acid-deficient diets on acetaminophen metabolism and hepatotoxicity in rats. Toxicol. Appl. Pharmacol. 101, 356–369. doi: 10.1016/0041-008X(89)90283-4
Prydz, K. (2015). Determinants of glycosaminoglycan (GAG) structure. Biomolecules 5, 2003–2022. doi: 10.3390/biom5032003
Rajas, O., Quirós, L. M., Ortega, M., Vazquez-Espinosa, E., Merayo-Lloves, J., Vazquez, F., et al. (2017). Glycosaminoglycans are involved in bacterial adherence to lung cells. BMC Infect. Dis. 17, 319–319. doi: 10.1186/s12879-017-2418-5
Rosmarin, D. M., Carette, J. E., Olive, A. J., Starnbach, M. N., Brummelkamp, T. R., and Ploegh, H. L. (2012). Attachment of Chlamydia trachomatis L2 to host cells requires sulfation. Proc. Natl. Acad. Sci. 109, 10059–10064. doi: 10.1073/pnas.1120244109
Rudd, T. R., Skidmore, M. A., Guerrini, M., Hricovini, M., Powell, A. K., Siligardi, G., et al. (2010). The conformation and structure of GAGs: recent progress and perspectives. Curr. Opin. Struct. Biol. 20, 567–574. doi: 10.1016/j.sbi.2010.08.004
Rusnati, M., Vicenzi, E., Donalisio, M., Oreste, P., Landolfo, S., and Lembo, D. (2009). Sulfated K5 Escherichia coli polysaccharide derivatives: A novel class of candidate antiviral microbicides. Pharmacol. Ther. 123, 310–322. doi: 10.1016/j.pharmthera.2009.05.001
Salvador, B., Sexton, N. R., Carrion, R., Nunneley, J., Patterson, J. L., Steffen, I., et al. (2013). Filoviruses utilize glycosaminoglycans for their attachment to target cells. J. Virol. 87, 3295–3304. doi: 10.1128/JVI.01621-12
Sarrazin, S., Lamanna, W. C., and Esko, J. D. (2011). Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3(7), a004952. doi: 10.1101/cshperspect.a004952
Sasaki, M., Anindita, P. D., Ito, N., Sugiyama, M., Carr, M., Fukuhara, H., et al. (2018). The role of heparan sulfate proteoglycans as an attachment factor for rabies virus entry and infection. J. Infect. Dis. 217, 1740–1749. doi: 10.1093/infdis/jiy081
Schmidtchen, A., Frick, I. M., and Björck, L. (2001). Dermatan sulphate is released by proteinases of common pathogenic bacteria and inactivates antibacterial alpha-defensin. Mol. Microbiol. 39, 708–713. doi: 10.1046/j.1365-2958.2001.02251.x
Schowalter, R. M., Pastrana, D. V., and Buck, C. B. (2011). Glycosaminoglycans and sialylated glycans sequentially facilitate merkel cell polyomavirus infectious entry. PloS Pathog. 7, e1002161. doi: 10.1371/journal.ppat.1002161
Serruto, D., Spadafina, T., Ciucchi, L., Lewis, L. A., Ram, S., Tontini, M., et al. (2010). Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc. Natl. Acad. Sci. 107, 3770–3775. doi: 10.1073/pnas.0915162107
Skrincosky, D., Hocknell, P., Whetter, L., Secchiero, P., Chandran, B., and Dewhurst, S. (2000). Identification and analysis of a novel heparin-binding glycoprotein encoded by human herpesvirus 7. J. Virol. 74, 4530–4540. doi: 10.1128/JVI.74.10.4530-4540.2000
Smit, J. M., Waarts, B-L., Kimata, K., Klimstra, W. B., Bittman, R., and Wilschut, J. (2002). Adaptation of alphaviruses to heparan sulfate: interaction of sindbis and semliki forest viruses with liposomes containing lipid-conjugated heparin. J. Virol. 76, 10128–10137. doi: 10.1128/JVI.76.20.10128-10137.2002
Southern, T. R., Jolly, C. E., Lester, M. E., and Hayman, J. R. (2007). EnP1, a microsporidian spore wall protein that enables spores to adhere to and infect host cells in vitro. Eukary. Cell 6, 1354–1362. doi: 10.1128/EC.00113-07
Su, C. M., Liao, C. L., Lee, Y. L., and Lin, Y. L. (2001). Highly sulfated forms of heparin sulfate are involved in Japanese encephalitis virus infection. Virology 286, 206–215. doi: 10.1006/viro.2001.0986
Tamura, M., Natori, K., Kobayashi, M., Miyamura, T., and Takeda, N. (2004). Genogroup II noroviruses efficiently bind to heparan sulfate proteoglycan associated with the cellular membrane. J. Virol. 78, 3817–3826. doi: 10.1128/JVI.78.8.3817-3826.2004
Tan, C. W., Sam, I. C., Chong, W. L., Lee, V. S., and Chan, Y. F. (2017). Polysulfonate suramin inhibits Zika virus infection. Antiviral Res. 143, 186–194. doi: 10.1016/j.antiviral.2017.04.017
Taylor, D. R., Whitehouse, I. J., and Hooper, N. M. (2009). Glypican-1 mediates both prion protein lipid raft association and disease isoform formation. PloS Pathog. 5, e1000666. doi: 10.1371/journal.ppat.1000666
Tise, C. G., Ashton, K., de Hayr, L., Lee, K. D., Patkar, O. L., Krzesinski, E., et al. (2025). Biallelic SLC13A1 loss-of-function variants result in impaired sulfate transport and skeletal phenotypes, including short stature, scoliosis, and skeletal dysplasia. Genet. Med. Open 3, 101958. doi: 10.1016/j.gimo.2024.101958
Trybala, E., Liljeqvist, J. A., Svennerholm, B., and Bergstrom, T. (2000). Herpes simplex virus types 1 and 2 differ in their interaction with heparan sulfate. J. Virol. 74, 9106–9114. doi: 10.1128/JVI.74.19.9106-9114.2000
Tseligka, E. D., Sobo, K., Stoppini, L., Cagno, V., Abdul, F., Piuz, I., et al. (2018). A VP1 mutation acquired during an enterovirus 71 disseminated infection confers heparan sulfate binding ability and modulates ex vivo tropism. PloS Pathog. 14, e1007190. doi: 10.1371/journal.ppat.1007190
van Putten, J. P. and Paul, S. M. (1995). Binding of syndecan-like cell surface proteoglycan receptors is required for Neisseria gonorrhoeae entry into human mucosal cells. EMBO J. 14, 2144–2154. doi: 10.1002/j.1460-2075.1995.tb07208.x
Vlasak, M., Goesler, I., and Blaas, D. (2005). Human rhinovirus type 89 variants use heparan sulfate proteoglycan for cell attachment. J. Virol. 79, 5963–5970. doi: 10.1128/JVI.79.10.5963-5970.2005
Wadstrom, T. and Ljungh, Å. (1999). Glycosaminoglycan-binding microbial proteins in tissue adhesion and invasion: key events in microbial pathogenicity. J. Med. Microbiol. 48, 223–233. doi: 10.1099/00222615-48-3-223
Wang, Q. and Chi, L. (2022). The alterations and roles of glycosaminoglycans in human diseases. Polym. (Basel) 14(22), 5014. doi: 10.3390/polym14225014
Wang, Y. and Pfeiffer, J. K. (2016). Emergence of a large-plaque variant in mice infected with coxsackievirus B3. mBio 7(2), e00119. doi: 10.1128/mBio.00119-16
Weiland, M. E.L., Palm, J. E.D., Griffiths, W. J., McCaffery, J. M., and Svärd, S. G. (2003). Characterisation of alpha-1 giardin: an immunodominant Giardia lamblia annexin with glycosaminoglycan-binding activity. Int. J. Parasitol. 33, 1341–1351. doi: 10.1016/S0020-7519(03)00201-7
Westergard, L., Christensen, H. M., and Harris, D. A. (2007). The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim. Biophys. Acta 1772, 629–644. doi: 10.1016/j.bbadis.2007.02.011
Wuppermann, F. N., Hegemann, J. H., and Jantos, C. A. (2001). Heparan sulfate–like glycosaminoglycan is a cellular receptor for chlamydia pneumoniae. J. Infect. Dis. 184, 181–187. doi: 10.1086/jid.2001.184.issue-2
Xu, Y., Martinez, P., Séron, K., Luo, G., Allain, F., Dubuisson, J., et al. (2015). Characterization of hepatitis C virus interaction with heparan sulfate proteoglycans. J. Virol. 89, 3846–3858. doi: 10.1128/JVI.03647-14
Yildirim, I., Hur, E., Magden, K., Ilikhan, S., Engin, H., Can, M., et al. (2019). Serum sulphate levels in hemodialysis patients. Int. J. Nephrol. 2019, 1063514. doi: 10.1155/2019/1063514
Zaiss, A. K., Foley, E. M., Lawrence, R., Schneider, L. S., Hoveida, H., Secrest, P., et al. (2016). Hepatocyte heparan sulfate is required for adeno-associated virus 2 but dispensable for adenovirus 5 liver transduction in vivo. J. Virol. 90, 412–420. doi: 10.1128/JVI.01939-15
Zhang, W., Heil, M., Kuhn, R. J., and Baker, T. S. (2005). Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology 332, 511–518. doi: 10.1016/j.virol.2004.11.043
Zhang, L., Bukreyev, A., Thompson, C. I., Watson, B., Peeples, M. E., Collins, P. L., et al. (2005). Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79, 1113–1124. doi: 10.1128/JVI.79.2.1113-1124.2005
Zhu, Z., Gershon, M. D., Ambron, R., Gabel, C., and Gershon, A. A. (1995). Infection of cells by varicella zoster virus: inhibition of viral entry by mannose 6-phosphate and heparin. Proc. Natl. Acad. Sci. 92, 3546–3550. doi: 10.1073/pnas.92.8.3546
Keywords: sulfate, virus, bacteria, parasite, infection, glycosaminoglycan, proteoglycan
Citation: Morris JS and Dawson PA (2025) Pathogens that infect mammalian cells via sulfonated glycosaminoglycans. Front. Cell. Infect. Microbiol. 15:1613923. doi: 10.3389/fcimb.2025.1613923
Received: 18 April 2025; Accepted: 22 May 2025;
Published: 10 June 2025.
Edited by:
Antoinette van der Kuyl, University of Amsterdam, NetherlandsCopyright © 2025 Morris and Dawson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul A. Dawson, cGF1bC5kYXdzb25AbWF0ZXIudXEuZWR1LmF1
 Jessica S. Morris
Jessica S. Morris Paul A. Dawson
Paul A. Dawson