- 1Fondazione Bruno Kessler, Trento, Italy
- 2Human Inspired Technology Research Centre, University of Padua, Padua, Italy
This paper presents an innovative methodology for addressing the critical issue of data scarcity in clinical research, specifically within emergency departments. Inspired by the recent advancements in the generative abilities of Large Language Models (LLMs), we devised an automated approach based on LLMs to extend an existing publicly available English dataset to new languages. We constructed a pipeline of multiple automated components which first converts an existing annotated dataset from its complex standard format to a simpler inline annotated format, then generates inline annotations in the target language using LLMs, and finally converts the generated target language inline annotations to the dataset's standard format; a manual validation is envisaged for erroneous and missing annotations. By automating the translation and annotation transfer process, the method we propose significantly reduces the resource-intensive task of collecting data and manually annotating them, thus representing a crucial step toward bridging the gap between the need for clinical research and the availability of high-quality data.
1 Introduction
Clinical research in emergency medicine is as difficult as important. It is difficult because the large number of patients to be treated and the chronic shortage of staff make any data collection impractical. It is important so that emergency physicians and nurses can base their practice on evidence produced in the very special context of care in which they work and not, as is predominantly the case today, in contexts far removed from it. The only way to fill the gap between the need for research and the availability of data is to extract them directly from the electronic health records (EHRs) of emergency departments (EDs). This is a difficult task, however, because a large part of the useful information is contained in an unstructured format, in free text notes.
Natural Language Processing (NLP) (1) addresses, among others, the task of automatic extraction of relevant information (e.g., entities and relations among them) from free text. Current technologies for Information Extraction (IE) (2) are based on the use of Large Language Models (LLMs) (3), which, thanks to powerful deep-learning neural networks, are achieving outstanding performance. However, LLMs are pre-trained on huge amounts of general text, mostly taken from the internet, and their performance in more specialized areas, such as the medical domain, may not be optimal. In the paper, we report some of the progress we are obtaining in the context of the eCREAM project (enabling Clinical Research in Emergency and Acute care Medicine), as far as Information Extraction from medical documents is concerned.1
A major objective of eCREAM is to develop state-of-the-art NLP technologies able to interpret EHR content and extract crucial information (metadata) from them, which will then be used to make accurate analysis and prediction of ED effectiveness. The project's significant leaning toward multilinguality is shown by the fact that it addresses six European languages, i.e., English, Italian, Greek, Polish, Slovak, and Slovenian.
Data scarcity poses significant challenges for machine learning projects in the medical domain, particularly within emergency departments. The primary issue is the lack of labeled data, which is crucial for training machine learning models. Labeled data (i.e., data where the correct annotations are provided) is essential in machine learning for several reasons. First, it serves as the foundation for supervised learning, where algorithms learn to make predictions or classify data based on input-output pairs. This process enables models to generalize from seen instances to unseen instances, improving predictions and decision-making. Additionally, labeled data is crucial for evaluating the performance of machine learning models, as it provides benchmarks to qualify the output of the machine learning algorithms. In the medical field, obtaining high-quality, annotated datasets is often difficult due to large patient volume [emergency departments handle a large number of patients daily (4)], time constraints (the urgent nature of emergency medicine means healthcare professionals have limited time to document patient information thoroughly), privacy concerns (patient data is highly sensitive and collecting it implies strict regulations such as GDPR,2 UK GDPR,3 and HIPAA4), data complexity (emergency department records often contain complex and unstructured data, including free-text notes, varied terminologies, and inconsistent formats), resource limitations (emergency departments typically operate with limited resources and staff, who are primarily focused on patient care rather than data management and annotation), and the time-consuming process of manually labeling data.
This problem is exacerbated in emergency departments where the high patient volume and urgent nature of care make systematic data collection and annotation even more impractical. Consequently, the scarcity of labeled data hinders the development and deployment of robust machine learning solutions that could otherwise improve patient outcomes and operational efficiency in emergency settings (5–7).
Annotated data are crucial for training [the process in which a machine learning (ML) algorithm is fed with sufficient training data to learn from] and fine tuning (the supervised learning process where you use a dataset of labeled examples to update the weights of LLM and make the model improve its ability for specific tasks) LLMs. However, currently there are no available training data for most of the languages addressed by eCREAM. In fact, EHRs are being collected from EDs and annotated within the project, but this is an ongoing activity and the data are not yet available for experiments. For the above reasons, we diverted our attention toward publicly available datasets for fine-tuning and testing LLMs. Specifically, we decided to use E3C, i.e., the European Clinical Case Corpus (8), which is available for English and Italian (besides other languages that are outside the scope of the eCREAM project). To the best of our knowledge, however, there are no public datasets available for Slovenian, Slovak, Polish, and Greek.
Producing annotated datasets for a new language from scratch is resource-intensive and time-consuming. The recent advancements in the generative abilities of LLMs inspired us to devise an automated approach based on LLMs to extend E3C to a new language. Despite their strengths in various downstream tasks, LLMs still struggle to comprehend data in complex structured formats like the UIMA CAS XMI format used for the E3C corpus. This limitation prevented us from directly achieving target language annotations of E3C using prompt engineering. Consequently, we constructed a pipeline of multiple automated components to achieve our goal, which can be outlined as follows:
1. Convert an existing annotated dataset from its complex standard format to a simpler inline annotated format.
2. Using the step 1 source language inline annotations, generate inline annotations in the target language using LLM.
3. Convert the generated target language inline annotations to the dataset's standard format.
4. Enforce a manual revision to fix any erroneous annotations or address any missing annotations.
The paper is structured as follows: in Section 2 we discuss some related work, in Section 3 we describe the publicly available annotated corpus that serves as starting point for the generation of our new datasets, in Section 4 we describe the procedure we devised to transfer annotations from one language to another; finally, in Section 5 we discuss our future work.
2 Background
2.1 Data transfer across languages
The idea of reducing the effort needed to produce new datasets in new languages by transferring semantic annotations from one language to another is already present in the literature (9–11). Annotation projection has often been formulated as the task of transporting, on parallel corpora, the labels pertaining to a given span in the source language into its corresponding span in the target language; it is basically a task consisting of three steps, translation, alignment and annotation transfer. Bentivogli et al. (9) present an approach to the creation of new datasets based on the assumption that annotations can be transferred from a source text to a target text in a different language using word alignment as a bridge; this approach has been used in the creation of the MultiSemCor corpus (10), a multilingual lexical-semantic corpus created for research in natural language processing (NLP), particularly for tasks such as word sense disambiguation (WSD), machine translation, and multilingual NLP. It is an extension of the well-known SemCor corpus, which is a semantically annotated English corpus. In MultiSemCor, texts are aligned at the word level and annotated with a shared inventory of senses.
More recently, García-Ferrero et al. (11) have presented an approach for annotation projection that leverages large pre-trained text-to-text language models and state-of-the-art machine translation technology; their approach divides the label projection task into two subtasks, i.e., a candidate generation step, in which a set of projection candidates using a multilingual T5 model is generated and a candidate selection step, in which the generated candidates are ranked based on translation probabilities.
2.2 Large language models
Large Language Models (LLMs) are AI systems designed to understand, generate, and manipulate human language using deep learning techniques, particularly neural networks inspired by the human brain (12). The development of LLMs has progressed significantly, beginning with relatively small models capable of basic text completion and simple translations. The introduction of the Transformer architecture by Vaswani et al. (13), which uses attention mechanisms to understand context, marked a revolutionary advancement in the field. Scaling up these models with more parameters and training data has led to significant improvements in performance (14), resulting in powerful models like GPT-2 (15) and GPT-3 (16) by OpenAI, and the more recent GPT-4 (17), which excel in multiple linguistic tasks.
In addition to GPT models, other LLMs such as Llama (18, 19), Qwen (20), and Falcon (21) have been developed. These open-source models, while not as performant as GPT models, offer transparency and greater control over fine-tuning and training data, enabling tailored usage for specific needs. LLMs have shown exceptional performance in the biomedical domain, handling tasks such as question answering (22, 23), document classification (24, 25), sequence labeling, relation extraction (26), feature extraction (27), and information extraction (28).
The versatility and robustness of LLMs make them valuable across various applications, including assisting healthcare professionals in writing and summarizing clinical notes (29), analyzing and synthesizing medical literature (30), and enhancing patient communication through chatbots and virtual assistants (31). LLMs have also been widely adopted for translation tasks, with smaller models achieving effective translation with proper adjustments (32), although GPT-4 remains superior in performance (33).
2.3 Language and cultural influences
Language and culture play a pivotal role in shaping the interpretation of medical terms and influencing the diagnostic process. The meaning of symptoms and medical conditions can differ significantly in cultural contexts, affecting how they are documented, communicated, and understood in medical records. This variability presents a unique challenge for artificial intelligence (AI) systems that use natural language processing (NLP) to extract and interpret clinical information. Without a nuanced understanding of language and cultural expressions, these AI models risk misinterpretation, potentially leading to diagnostic inaccuracies or misaligned healthcare interventions (34–36).
2.3.1 AI and NLP in emergency departments
AI systems, particularly those utilizing NLP, are increasingly being deployed to extract critical insights from clinical documentation. By analyzing unstructured text data, such as patient notes and triage notes, these systems aim to enhance diagnostic accuracy and operational efficiency in emergency departments (EDs). However, their success hinges on the ability to process the complex interplay of cultural and linguistic nuances embedded in the data (34, 37, 38).
For example, NLP techniques have been applied to predict patient disposition by analyzing nursing triage notes. These notes, often composed of free text, encapsulate the clinical impressions of nurses and reflect their linguistic and cultural background. To ensure accurate predictions, AI models must be trained to recognize and adapt to these variations, accommodating both the subjective nature of clinical documentation and the diversity of the healthcare workforce (34, 38).
2.3.2 Challenges in information extraction
The process of extracting relevant information from medical records is further complicated by the need for explainable AI models. Clinicians require that AI-derived insights be transparent, traceable, and easily understandable to support evidence-based decision-making. This requires the creation of AI models capable not only of processing various linguistic inputs but also of providing culturally and contextually relevant explanations (36, 39).
In addition, the integration of AI systems into Emergency Departments must account for the wide range of cultural backgrounds among patients and healthcare providers. Cultural diversity affects the way symptoms are reported and interpreted, making it essential for AI to adapt dynamically to these variances. AI solutions that do not consider this diversity risk propagating biases, which could compromise patient safety and care outcomes (40, 41).
3 E3C: European clinical case corpus
Electronic health records (EHRs) are being collected and manually annotated within the eCREAM project, but at the time of writing these data are not yet available so we opted for the exploitation of publicly available datasets, such as E3C, a freely available multilingual corpus encompassing five languages, i.e., English, Italian, French, Spanish, and Basque.5 E3C contains about 25K tokens annotated for each language (plus 1M tokens not annotated). It consists of clinical narratives annotated manually with semantic information, thus allowing for linguistic analysis, benchmarking, and training of information extraction systems.
A clinical case is a statement of a clinical practice focusing on a single patient; E3C focuses on this specific type of clinical narrative because they are rich in clinical entities as well as temporal information, which is almost absent in other clinical documents (e.g., radiology reports).
The E3C's clinical cases typically start presenting the reason for a clinical visit (i.e., the patient's symptoms) and then describe the assessment of the patient's situation; physical exams and laboratory tests play a central role in diagnosing diseases and disorders, therefore they are reported in clinical cases and their results meticulously documented. The final diagnosis might conclude the text, but, more often than not, treatment, outcome, and follow-up are present as well.
Symptoms, tests, observations, treatments, and diseases are all marked in E3C as they are relevant events for the history of a patient, and to understand the evolution of a patient's health it is relevant to place them in chronological order, so temporal relations are also made explicit. Since precision in symptom description and diagnosis is utterly important in the clinical field, clinical findings, body parts, laboratory results, and measurements, etc., are identified as well.
3.1 E3C annotations
E3C foresees two types of semantic annotations that can be used to fine-tune a large language model:
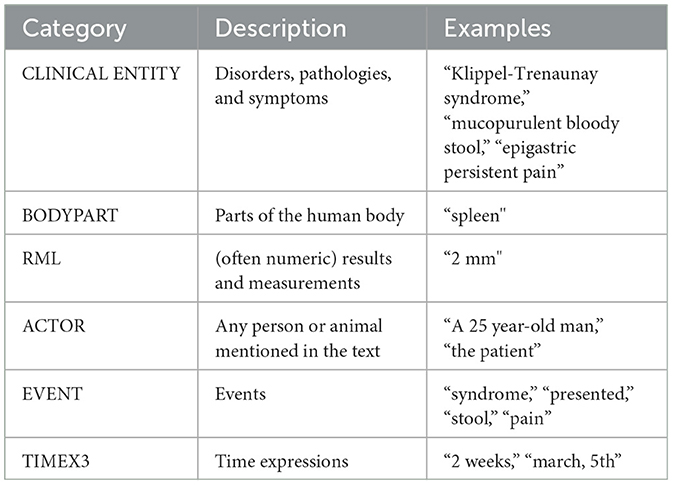
1. clinical entities (such as pathologies and symptoms), body parts, laboratory results (in relation to the test they refer to) and actors.
2. temporal information, i.e., events, time expressions, and relations between them.
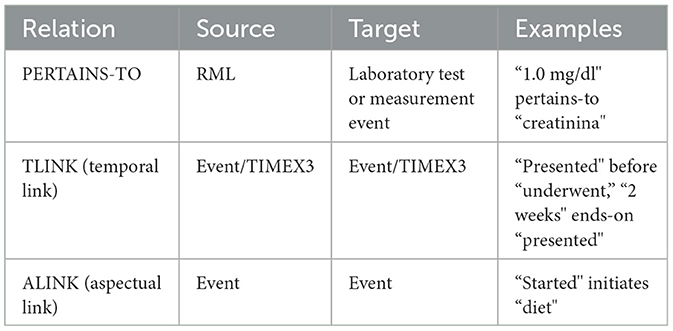
In both cases we have both different categories of span-based annotations and different types of relations, i.e. links between two span-based annotations of the same or different category. E3C's annotation is extremely rich in terms of attributes assigned to the different annotation categories and in terms of relation types (as shown in Figure 1). For the sake of simplicity, we report the annotation categories without specific attributes (see Table 1) and the high level classification of relations (in Table 2).
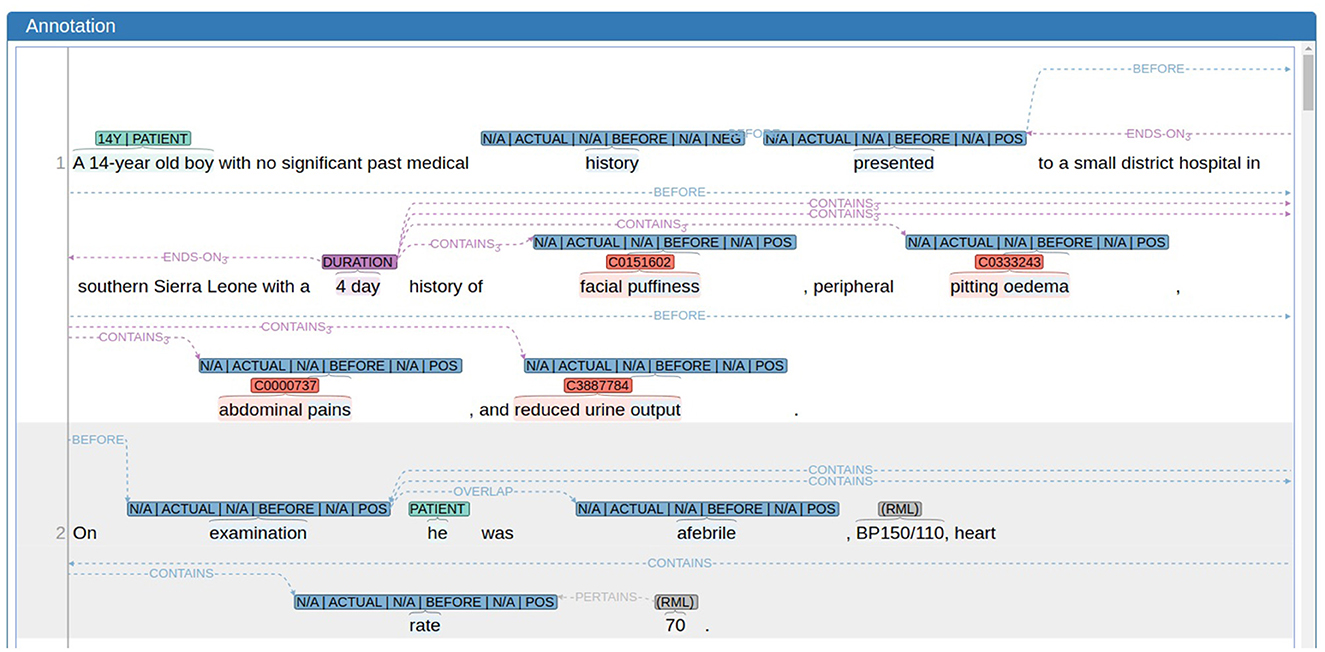
Figure 1. An example of an annotated text in E3C (excerpt from document EN100017.xml) as visualized by the annotation tool WebAnno; annotations are highlighted as follows: clinical entities in red, results and measurements (RMLs) in gray, events in blue, temporal expression in purple, and patients in turquoise.
3.2 Uses of E3C
With respect to the rich set of E3C annotations, the interest of the eCREAM project is currently directed toward two main subtasks:
• the recognition of clinical entities (i.e., disorders, pathologies, and symptoms), and,
• the identification of PERTAINS-TO relations, holding between an RML and the test/measurement it refers to.
It is rather intuitive that clinical entity recognition plays a predominant role in the context of information extraction from medical documents. Additionally, laboratory tests and measurements, along with their results, are also crucial as they provide essential information about a patient's status at specific stages of a disorder's development. By accurately linking laboratory tests to their results, healthcare providers can gain deeper insights into a patient's condition, leading to more precise diagnoses and effective treatment plans. Additionally, implementing these tasks in multiple languages ensures that their benefits reach diverse linguistic regions, thereby enhancing global healthcare standards. Despite its importance, this aspect has been relatively neglected, one significant exception being the twin evaluation tasks, CLinkaRT6 (42) and TESTLINK7 (43), organized in the EVALITA 2023 and IberLEF 2023 events, respectively. CLinkaRT (for Italian) and TESTLINK (for Spanish and Basque) are both based on E3C and focus on evaluating different systems on the task of identifying the RMLs and the laboratory tests or measurement they refer to (PERTAINS-TO relations).
Specifically for the CLinkaRT and TESTLINK evaluations, the organizers of the twin tasks deployed a new version of the Italian, Spanish and Basque sections of E3C where PERTAINS-TO relations had been revised (this revision was performed after releasing the fully annotated dataset). As the English section (the base for our extensions to new languages) has not been used in the evaluation tasks, we took up the task of performing a complete revision of the PERTAINS-TO relations. In addition to this, given the eCREAM's important focus on entity detection, we also looked for clinical entities missing annotations and added them. As a result of this effort we can now rely on a new version8 of the English E3C, with a full revision of PERTAINS-TO relations and a partial revision of clinical entities.
4 Extending the E3C dataset to the eCREAM languages
As explained in detail above, although annotated data for training and fine tuning LLM is crucial, there are currently no available data for a number of languages addressed by eCREAM, i.e., Greek, Polish, Slovak, and Slovenian, so we devised a methodology based on LLMs that allows us to automatically extend E3C to a new language by transferring the annotations. The task we addressed is quite challenging because concepts are expressed in different languages with linguistic constructions that might differ at various linguistic levels, ranging from syntax to morphology.
From the syntactic point of view, for instance, we can have languages (such as English) where the subject of a verb is always expressed and languages (such as Italian, for example) where a pronominal subject can be omitted; in terms of transferring E3C annotations, this is an interesting problem as the pronoun in question (present in the English source text) might refer to the patient therefore be marked as an ACTOR; in the impossibility of finding the pronoun, the automatic system can not possibly transfer the annotation.
Possible problems occurring at the morphological level are represented by the cases in which a single word is translated using a combination of words. This typically happens with lexical gaps (i.e. cases where in the target language there is no single word corresponding to the word in the source language); for instance, the English word “successfully” can not be translated into Italian with one single word, but requires a combination of words (literally corresponding to “with success”). In many cases, however, it is just one of the possible different ways in which a word can be translated (for example, “gently” can be translated into Italian in different ways, one of them being the combination of words corresponding to “in a gentle way”). In both the examples presented above, an adverb is translated using combinations of words belonging to various grammatical categories (prepositions, adjectives, and nouns), which might impact the cross lingual annotation transfer.
The procedure we have implemented for annotation transfer consists of two main phases:
1. an automatic procedure translates the texts of E3C from English to a new language while simultaneously transferring the annotations as well;
2. the erroneous annotations and those that the system has not been able to transfer are corrected/added manually with the help of a native speaker domain expert.
4.1 Automatic porting procedure
To perform the annotation transfer, we begin by using the E3C dataset and generating inline annotations from its original format (as illustrated in Figure 2). E3C data is initially provided in the UIMA CAS XMI format,9 where annotations are maintained separately from the text and linked through begin and end offsets. This structure facilitates machine processing of annotation spans, along with their attributes and relations, using annotation tools (such as WebAnno in our work, see Figure 1). Our objective is to create a parallel XMI representation in the target language based on the source XMI, leveraging LLMs like ChatGPT. However, achieving this is complex due to the intricate nature of XMI (as shown in Figure 3), which encompasses diverse information beyond annotations, complicating direct conversion. The use of XMI is important in our study because it is the original format of the E3C dataset, and understanding its structure is crucial for accurate annotation transfer. It also highlights the challenge of working with complex data serialization formats in NLP tasks and demonstrates our approach to address it by converting XMI to a simpler inline format, enhancing interoperability and simplifying processing. To overcome this, we convert the source XMI to a simpler inline annotation format (depicted in Figure 4), omitting attributes and relations and embedding only span-based annotations within the text using opening and closing tags. A key difficulty lies in the E3C annotation schema's complexity, which includes overlapping and nested entities. We accomplish this conversion by employing an independent Python script that transfers the span-based annotations, preserving all nested entities without any annotation loss.

Figure 3. Example of the stand-off XMI for clinical case EN103007.xml. For the sake of space and simplicity of understanding, the figure presents a reduced content of the actual XMI.

Figure 4. Inline representation of the stand-off XMI from Figure 3.
The dataset is now ready to feed a translation system based on ChatGPT10 that has been developed to translate the clinical cases (English texts), while simultaneously transferring the annotations to the translated texts. To boost translation quality and teach ChatGPT to transfer annotations, a few-shot learning approach (44) has been employed. Consequently, the prompt used consists of three parts, an instruction, an input, and two examples. During the translation, the system not only keeps track of the annotations from the source text that erroneously do not appear in the translated text, but also includes some specific strategies (based on WordNet)11 aimed at identifying possible transfer errors; where the textual span in the source and target languages are not exact translations of each other, in fact, is where an error is most likely to have occurred. The number of annotations not appearing in the translated text and the number of non matching translations together provide an indirect way to automatically quantify the quality of the output of the system. We have tested ChatGPT's ability to translate between English and Italian, using a metric called BERTScore (46) to evaluate the quality. BERTScore compares the original text with its back-translation (translating from English to Italian, then back to English) to see how well the original meaning is preserved. This metric does not just check for word matches but also evaluates the overall meaning and context, making it a reliable way to assess translation quality.
To evaluate overall meaning, BERTScore uses a deep learning model trained on vast amounts of language data. It analyzes the relationships between words and their surrounding context, capturing nuances like synonyms, tone, and sentence structure. For example, it can recognize that “physician” and “doctor” mean the same thing in context, even if the words are different. Additionally, BERTScore assigns a similarity score to each word and phrase by comparing their positions and roles in the sentence, ensuring the generated text aligns with the original in both intent and message. This method ensures that translations are judged not only on surface-level accuracy but also on how well they convey the underlying meaning.
The results showed that ChatGPT consistently produces high-quality translations, as reflected in strong BERTScores (94/100). This means the translations effectively preserve the original meaning, ensuring accurate communication between the two languages. The resultant translations from ChatGPT are the inline annotations in the target language that need to be converted back to the original XMI format. To achieve this, we construct the target XMI by first extracting language-independent information from the source XMI (such as rows that define the file's structure, preamble and closing tags, metadata, etc.). Next, we generate the annotation rows in the target XMI, calculating appropriate offsets for each annotation span based on the target language inline annotations. For each created annotation row in the XMI, we also populate attributes associated with that annotation, retrieving them from the source XMI (as this information was intentionally removed when preparing prompts for translation). Finally, we copy all relations from the source XMI to the newly created target XMI files. Coherency between annotations and relations in the source and target XMIs is maintained using unique XMI identifiers. To facilitate manual validation (discussed in detail in the following section), any missing or mismatched annotation information is intuitively presented in the target XMI through additional metadata rows (for missing cases) and suitable attributes (for mismatches).
4.2 Manual validation
The output produced by the (fully automatic) previous phase intuitively may contain a number of erroneous annotations and cases where the system has not been able to transfer an annotation, which need to be revised/added manually; the person needed for this task has to be a native speaker of the target language, needs to be a domain expert and also be proficient in English.
The manual effort required for the revision is highly reduced by the fact that the procedure does not require revising the whole annotated dataset in the target language, but only the annotations that have been previously selected as potentially transferred to the new language with a wrong textual span. With regard to the selected annotations, in addition, annotators accessing the target dataset are provided with not just the English textual span but all the information about the English source annotations. This means that, while familiarity with the E3C annotation guidelines is still desired, the validation task is strongly simplified as, in the case of errors to be fixed, annotation attributes can be transferred mechanically from the English original version.
In the end, the validation of each annotation consists of three basic steps: checking the textual span provided by the system (this implies comparing a portion of English text with the corresponding portion of text in the translated document), identifying the correct textual span and make a correction if needed, and add the attributes and relations where appropriate.
In a second step, annotators are required to add the missing annotations, i.e., those that, for different reasons, the system has not been able to transfer. In this case, the annotator can rely on the span of the original English annotation and E3C guidelines to correctly identify the span and on the information about the English source annotations, i.e., its attributes and the relations in which it is involved, to complete the annotation.
4.3 The case of Italian
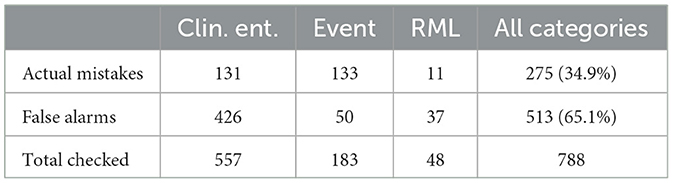
For the first testing of the automatic porting procedure, we chose Italian based on the availability of native speakers able to do the manual revision. Task completion required a total of two working days (in this specific case there was no need for any training sessions). In most cases in fact (almost 35%), the candidate problematic annotations were actually correct, and the annotator was just required to validate them (see Table 3).
Candidate errors can be grouped into two main categories; on the one hand, we have what we call linguistic errors, where ChatGPT's task was made more tough by (sometimes also domain-specific) linguistic issues and on the other hand we have procedural errors, i.e., cases where ChatGPT produced some errors in spite of linear translations between the two languages, maybe because of the excessive nesting of labels. In the following we will focus on linguistic errors.
ChatGPT was able to correctly transfer the textual span even in cases where the translation was not completely straightforward (and therefore selected for manual revision). For instance, the word “headache” (annotated as a clinical entity) was (correctly) translated by means of a three-word expression (“mal di testa,” which has a different syntactic structure) and the expression “60 mm × 50 mm across” (marked as an RML) was translated as “60 mm × 50 mm” (where the omission of the word “across” is more that acceptable); in spite of this, ChatGPT had no problems in transferring both annotations. The same holds for even more complicated cases where we have so-called free translations. A syntactically simple structure like “generalized bodyache” (annotated as a clinical entity) has been translated as “dolori diffusi in tutto il corpo” (which literally corresponds to “pains widespread in the whole body”) and still the annotation transfer worked out well.
In other cases, inevitably, the automatically produced output actually required some manual intervention.12 For instance, in “calcified masses” (translated as “masse calcificate”), we had two distinct clinical entities (“calcified” and “masses”) but the Italian output erroneously resulted in one single entity whose textual span covered the whole expression; this is probably due to the different word order in the two languages as in Italian, unlike in English, an adjective generally comes after the noun it refers to (and in fact “masse calcificate” literally corresponds to “masses calcified”). The impact of this linguistic phenomenon is even more evident with more complex (yet quite frequent) syntactic structures encompassing entities with overlapping spans. Let's take, as an example, the expression “anterior and posterior capsular rupture,” where we have two clinical entities: E1 “posterior capsular rupture” (whose textual span is straightforward) and E2 “anterior [...] capsular rupture” (whose extent “anterior and posterior capsular rupture” also includes the two extra words “and posterior”).13 In the Italian output E1 was missing, so we had to add the corresponding annotation (“rottura capsulare anteriore e posteriore”),14 while for E2, which was transferred as “rottura capsulare anteriore e posteriore,” we had to reduce the textual span to obtain “rottura capsulare anteriore.”
5 Conclusion
This research introduces an innovative method to address the critical issue of data scarcity in clinical research, specifically within emergency departments. The method involves converting complex annotated datasets into simpler formats, using LLMs for translation and annotation transfer, and reconverting the annotations into standard formats. By automating the translation and annotation transfer process, we have significantly reduced the resource-intensive task of manually creating annotated datasets in new languages. This methodology not only improves the ability to perform robust information extraction in multiple languages but also ensures that emergency medical research can be based on accurate, context-specific data. The benefits of this approach include improved data availability for clinical research, improved predictive analytics in emergency medicine, and a robust framework to extend NLP capabilities to underserved languages. This work represents a crucial step toward bridging the gap between the need for clinical research and the availability of high-quality data in emergency departments.
The paper highlights the potential impact on clinical research. Further validation through real-world applications and clinical pilot studies would strengthen its findings. Providing evidence of how this approach enhances decision-making in emergency departments and integrating it with Electronic Health Records (EHRs) would demonstrate its practical utility. Addressing potential biases in LLM-generated annotations and ensuring high translation accuracy across various medical terminologies will be critical to its adoption in clinical practice.
Ongoing efforts are directed toward extending the E3C corpus to additional European languages such as Greek, Slovak, and Polish using the developed pipeline. This expansion aims to further enhance the multilingual capacity and accessibility of clinical data, facilitating broader and more inclusive research opportunities across diverse linguistic communities.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://huggingface.co/collections/NLP-FBK/e3c-projected-676a7d6221608d60e4e9fd89.
Author contributions
BM: Writing – original draft, Writing – review & editing. SF: Writing – original draft, Writing – review & editing. PF: Writing – original draft, Writing – review & editing. SG: Writing – original draft, Writing – review & editing. AL: Writing – review & editing, Writing – original draft. GM: Writing – review & editing, Writing – original draft. MS: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work has been partially funded by the European Union under the Horizon Europe eCREAM Project (Grant Agreement No. 101057726) and IDEA4RC Project (Grant Agreement No. 101057048).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
Views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the European Health and Digital Executive Agency (HADEA). Neither the European Union nor the granting authority can be held responsible for them.
Footnotes
1. ^eCREAM (https://ecreamproject.eu) is a European project coordinated by the Laboratory of Clinical Epidemiology at the Istituto di Ricerche Farmacologiche Mario Negri IRCCS (IRFMN). It involves 11 partners in 8 countries (France, Greece, Italy, Poland, Slovakia, Slovenia, Switzerland, and the United Kingdom).
2. ^General Data Protection Regulation in EU, see here.
3. ^UK General Data Protection Regulation here.
4. ^Health Insurance Portability and Accountability Act in the US, See here.
5. ^The European Clinical Case Corpus (E3C) is available for download from the project's website: https://e3c.fbk.eu/data.
6. ^https://e3c.fbk.eu/clinkart
7. ^https://e3c.fbk.eu/testlinkiberlef
8. ^Link to the dataset is available in the ‘Data Availability Statement' section of this paper.
9. ^The Apache UIMA framework employs the XMI (eXtensible Markup Language) within its CAS (Common Analysis Structure) format for standard data serialization. Specifically, the CAS XMI format is a serialization of the Common Analysis Structure (CAS), which is a data model used by UIMA to represent unstructured information and associated metadata. It can represent various data types, including annotations (e.g., named entities, part-of-speech tags), type systems (definitions of annotation types and their features), and Sofa data (the subject of analysis, such as the text itself) https://uima.apache.org/.
10. ^Here, gpt-4-0125 from Open AI platform has been employed to translate texts.
11. ^WordNet is a comprehensive lexical database of the English language developed at Princeton University, which organizes words into sets of synonyms called synsets. Each synset represents a single concept and includes definitions and usage examples (45).
12. ^The link to both the unrevised and revised projections of the Italian E3C is available in the “Data Availability Statement” section of this paper.
13. ^According to the E3C guidelines, it is not admitted to mark discontinuous text spans like “anterior and posterior capsular rupture;” if it is the case, all the words between those interested by the annotation must also be included, and the annotation is marked as “discontinuous.”
14. ^Notice that it contains the extra words “anteriore e” and it is marked as “discontinuous.”
References
1. Chowdhary KR. Natural Language Processing. New Delhi: Springer India (2020). p. 603–649. doi: 10.1007/978-81-322-3972-7_19
2. Grishman R. Information extraction: techniques and challenges. In:Pazienza MT, , editor. Information Extraction A Multidisciplinary Approach to an Emerging Information Technology. Berlin, Heidelberg: Springer Berlin Heidelberg (1997). p. 10–27. doi: 10.1007/3-540-63438-X_2
3. Naveed H, Khan AU, Qiu S, Saqib M, Anwar S, Usman M, et al. A comprehensive overview of large language models. CoRR.abs/2307.06435. (2023).
4. Jones SS, Thomas A, Evans RS, Welch SJ, Haug PJ, Snow GL. Forecasting daily patient volumes in the emergency department. Acad Emerg Med. (2008) 15:159–70. doi: 10.1111/j.1553-2712.2007.00032.x
5. Lin MP, Sharma D, Venkatesh A, Epstein SK, Janke A, Genes N, et al. The clinical emergency data registry: structure, use, and limitations for research. Ann Emerg Med. (2024) 83:467–76. doi: 10.1016/j.annemergmed.2023.12.014
6. Farley HL, Baumlin KM, Hamedani AG, et al. Quality and safety implications of emergency department information systems. Ann Emerg Med. (2013) 62:399–407. doi: 10.1016/j.annemergmed.2013.05.019
7. Yamamoto LG, Khan ANGA. Challenges of electronic medical record implementation in the emergency department. Pediatr Emerg Care. (2006) 22:184–91.
8. Magnini B, Altuna B, Lavelli A, Speranza M, Zanoli R. The E3C project: collection and annotation of a multilingual corpus of clinical cases. In: Proceedings of the Seventh Italian Conference on Computational Linguistics, CLiC-It 2020. Bologna, Italy (2021). doi: 10.4000/books.aaccademia.8663
9. Bentivogli L, Forner P, Pianta E. Evaluating cross-language annotation transfer in the MultiSemCor corpus. In: Proceedings of the 20th International Conference on Computational Linguistics. Geneva, Switzerland: COLING (2004). doi: 10.3115/1220355.1220408
10. Bentivogli L, Pianta E. Exploiting parallel texts in the creation of multilingual semantically annotated resources: the MultiSemCor Corpus. Nat Lang Eng. (2005) 11:247–61. doi: 10.1017/S1351324905003839
11. García-Ferrero I, Agerri R, Rigau G. T-Projection: high quality annotation projection for sequence labeling tasks. In:Bouamor H, Pino J, Bali K, , editors. Findings of the Association for Computational Linguistics: EMNLP 2023. Singapore: Association for Computational Linguistics (2023). p. 15203–15217. doi: 10.18653/v1/2023.findings-emnlp.1015
12. Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev. (1958) 65:386. doi: 10.1037/h0042519
13. Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. In: Proceedings of the 31st International Conference on Neural Information Processing Systems. NIPS'17. Red Hook, NY, USA: Curran Associates Inc. (2017). p. 6000–6010.
14. Hoffmann J, Borgeaud S, Mensch A, Buchatskaya E, Cai T, Rutherford E, et al. Training compute-optimal large language models. arXiv preprint arXiv:220315556. (2022).
15. Radford A, Wu J, Child R, Luan D, Amodei D, Sutskever I, et al. Language models are unsupervised multitask learners. OpenAI blog. (2019) 1:9.
16. Brown T, Mann B, Ryder N, Subbiah M, Kaplan JD, Dhariwal P, et al. Language models are few-shot learners. In:Larochelle H, Ranzato M, Hadsell R, Balcan MF, Lin H, , editors. Advances in Neural Information Processing Systems. Curran Associates, Inc. (2020). p. 1877–1901.
17. OpenAI, Achiam J, Adler S, Agarwal S, Ahmad L, Akkaya I, et al. GPT-4 Technical Report. arXiv:2303.08774. (2024).
18. Touvron H, Lavril T, Izacard G, Martinet X, Lachaux MA, Lacroix T, et al. Llama: open and efficient foundation language models. arXiv preprint arXiv:230213971. (2023).
19. Touvron H, Martin L, Stone K, Albert P, Almahairi A, Babaei Y, et al. Llama 2: open foundation and fine-tuned chat models. arXiv preprint arXiv:230709288. (2023).
20. Bai J, Bai S, Chu Y, Cui Z, Dang K, Deng X, et al. Qwen technical report. arXiv preprint arXiv:230916609. (2023).
21. Almazrouei E, Alobeidli H, Alshamsi A, Cappelli A, Cojocaru R, Debbah M, et al. The falcon series of open language models. arXiv preprint arXiv:231116867. (2023).
22. Singhal K, Azizi S, Tu T, Mahdavi SS, Wei J, Chung HW, et al. Large language models encode clinical knowledge. Nature. (2023) 620:172–80. doi: 10.1038/s41586-023-06291-2
23. Chen Z, Cano AH, Romanou A, Bonnet A, Matoba K, Salvi F, et al. Meditron-70b: scaling medical pretraining for large language models. arXiv preprint arXiv:231116079. (2023).
24. Luo R, Sun L, Xia Y, Qin T, Zhang S, Poon H, et al. BioGPT: generative pre-trained transformer for biomedical text generation and mining. Brief Bioinform. (2022) 23:bbac409. doi: 10.1093/bib/bbac409
25. Lewis P, Ott M, Du J, Stoyanov V. Pretrained language models for biomedical and clinical tasks: understanding and extending the state-of-the-art. In: Proceedings of the 3rd Clinical Natural Language Processing Workshop. Online: Association for Computational Linguistics (2020). p. 146–57. doi: 10.18653/v1/2020.clinicalnlp-1.17
26. Gururangan S, Marasović A, Swayamdipta S, Lo K, Beltagy I, Downey D, et al. Don't stop pretraining: adapt language models to domains and tasks. In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. Online: Association for Computational Linguistics (2020). p. 8342–8360. doi: 10.18653/v1/2020.acl-main.740
27. Huang K, Altosaar J, Ranganath R. ClinicalBERT: modeling clinical notes and predicting hospital readmission. arXiv preprint arXiv:190405342. (2020).
28. Farzi S, Ghosh S, Lavelli A, Magnini B. Get the Best out of 1B LLMs: insights from information extraction on clinical documents. In: Proceedings of the 23rd Workshop on Biomedical Natural Language Processing, BioNLP@ACL 2024, Bangkok, Thailand, August 16, 2024. Association for Computational Linguistics (2024). p. 266–276. doi: 10.18653/v1/2024.bionlp-1.21
29. Van Veen D, Van Uden C, Blankemeier L, Delbrouck JB, Aali A, Bluethgen C, et al. Clinical text summarization: adapting large language models can outperform human experts. Res Squ. (2023) 30:rs.3.rs-3483777. doi: 10.21203/rs.3.rs-3483777/v1
30. Doneva SE, Qin S, Sick B, Ellendorff T, Goldman JP, Schneider G, et al. Large Language Models to process, analyze, and synthesize biomedical texts-a scoping review. bioRxiv. (2024). p. 2024-04. doi: 10.1101/2024.04.19.588095
31. Shaikh A, Haritha C, Mullick A, Gadia V. Enhancing patient care with AI chatbots and virtual assistants. Int J Pharm Sci. (2024) 2:1–1.
32. Xu H, Kim YJ, Sharaf A, Awadalla HH. A paradigm shift in machine translation: boosting translation performance of large language models. arXiv preprint arXiv:230911674. (2024).
33. Wang L, Lyu C, Ji T, Zhang Z, Yu D, Shi S, et al. Document-level machine translation with large language models. arXiv preprint arXiv:230402210. (2023). doi: 10.18653/v1/2023.emnlp-main.1036
34. Sterling NW, Patzer RE Di M, Schrager JD. Prediction of emergency department patient disposition based on natural language processing of triage notes. Int J Med Inform. (2019) 129:184–8. doi: 10.1016/j.ijmedinf.2019.06.008
35. Strafford H, Fonferko-Shadrach B, Pickrell WO, Lyons J, Lacey A, Evans J, et al. Improving surveillance of emergency department activity through natural language processing. Int J Popul Data Sci. (2024) 9:2667. doi: 10.23889/ijpds.v9i5.2667
36. Ho TK, Luo YF, Guido RC. Explainability of methods for critical information extraction from clinical documents: a survey of representative works. IEEE Signal Process Mag. (2022) 39:96–106. doi: 10.1109/MSP.2022.3155906
37. Liang H, Tsui BY, Ni H, Valentim CCS, Baxter SL, Liu G, et al. Evaluation and accurate diagnoses of pediatric diseases using artificial intelligence. Nat Med. (2019) 25:433–438. doi: 10.1038/s41591-018-0335-9
38. Ben-Haim G, Yosef M, Rowand E, Ben-Yosef J, Berman A, Sina S, et al. Combination of machine learning algorithms with natural language processing may increase the probability of bacteremia detection in the emergency department: a retrospective, big-data analysis of 94,482 patients. Digital Health. (2024) 10:20552076241277673. doi: 10.1177/20552076241277673
39. Daniel M, Park S, Seifert CM, Chandanabhumma PP, Fetters MD, Wilson E, et al. Understanding diagnostic processes in emergency departments: a mixed methods case study protocol. BMJ Open. (2021) 11:e044194. doi: 10.1136/bmjopen-2020-044194
40. Goenaga I, Atutxa A, Gojenola K, Oronoz M, Agerri R. Explanatory argument extraction of correct answers in resident medical exams. Artif Intell Med. (2024) 157:102985. doi: 10.1016/j.artmed.2024.102985
41. Zou X, He W, Huang Y, Ouyang Y, Zhang Z, Wu Y, et al. AI-driven diagnostic assistance in medical inquiry: reinforcement learning algorithm development and validation. J Med Internet Res. (2024) 26:e54616. doi: 10.2196/54616
42. Altuna B, Karunakaran G, Lavelli A, Magnini B, Speranza M, Zanoli R. CLinkaRT at EVALITA 2023: overview of the task on linking a lab result to its test event in the clinical domain. In: Proceedings of the Eighth Evaluation Campaign of Natural Language Processing and Speech Tools for Italian. Final Workshop (EVALITA 2023), CEUR. org, Parma, Italy (2023).
43. Altuna B, Agerri R, Salas-Espejo L, Saiz JJ, Lavelli A, Magnini B, et al. Overview of TESTLINK at IberLEF 2023: linking results to clinical laboratory tests and measurements. Procesam Lenguaje Nat. (2023) 71:313–20.
44. Mann B, Ryder N, Subbiah M, Kaplan J, Dhariwal P, Neelakantan A, et al. Language models are few-shot learners. arXiv preprint arXiv:200514165. (2020).
45. Miller GA. WordNet: a lexical database for English. Commun ACM. (1995) 38:39–41. doi: 10.1145/219717.219748
Keywords: clinical data scarcity, multilingual dataset expansion, large language models, automated annotation transfer, emergency department data, low-resource settings, clinical multilingual NLP
Citation: Magnini B, Farzi S, Ferrazzi P, Ghosh S, Lavelli A, Mezzanotte G and Speranza M (2025) A cost-effective approach to counterbalance the scarcity of medical datasets. Front. Disaster Emerg. Med. 3:1558200. doi: 10.3389/femer.2025.1558200
Received: 09 January 2025; Accepted: 10 April 2025;
Published: 09 May 2025.
Edited by:
Göksu Bozdereli Berikol, Ufuk University, TürkiyeReviewed by:
Tamorish Kole, University of South Wales, United KingdomJan-Cedric Hansen, Centre d'Hébergement et d'Accompagnement Gérontologique (CHAG), France
Copyright © 2025 Magnini, Farzi, Ferrazzi, Ghosh, Lavelli, Mezzanotte and Speranza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bernardo Magnini, bWFnbmluaUBmYmsuZXU=
 Bernardo Magnini
Bernardo Magnini Saeed Farzi1
Saeed Farzi1 Soumitra Ghosh
Soumitra Ghosh Alberto Lavelli
Alberto Lavelli