- Department of Chemistry, University of Massachusetts, Amherst, MA, United States
Circadian clocks are fundamental, time-tracking systems that allow organisms to adapt to the appropriate time of day and drive many physiological and cellular processes. Altered circadian rhythms can result from night-shift work, chronic jet lag, exposure to bright lights at night, or other conditioning, and have been shown to lead to increased likelihood of cancer, metabolic and cardiovascular diseases, and immune dysregulation. In cases of cancer, worse patient prognoses and drug resistance during treatment have also been observed. Breast, colon, prostate, lung, and ovarian cancers and hepatocellular carcinoma have all been linked in one way or another with altered circadian rhythms. Critical elements at the molecular level of the circadian system have been associated with cancer, but there have been fairly few studies in this regard. In this mini-review, we specifically focus on the role of altered circadian rhythms in breast cancer, providing an overview of studies performed at the epidemiological level through assessments made in animal and cellular models of the disease. We also address the disparities present among studies that take into account the rhythmicity of core clock and other proteins, and those which do not, and offer insights to the use of small molecules for studying the connections between circadian rhythms and cancer. This article will provide the reader with a concise, but thorough account of the research landscape as it pertains to altered circadian rhythms and breast cancer.
Introduction
It was first reported in the 1960s that circadian rhythm disruptions can lead to an increased likelihood of mammary tumor development, and that circadian genes may act as tumor suppressors (1). In previous decades, studies have suggested that alterations to circadian rhythms also accelerate breast epithelial stem-cell proliferation, induce mammary-gland development, and increase the formation of spontaneous breast tumors in mammals (2, 3). Disruptions to circadian rhythms in humans have also been associated with the development of several other cancer types, including prostate (4), endometrial (5), colon (6), lung (7), and ovarian cancers (8) and hepatocellular carcinoma (9). In addition, the rhythmic control of cell fate is believed to affect cancer therapies: the efficacy and/or toxicity of radiotherapy and antitumor therapeutics have been shown to be dependent on the timing of dose administration (10, 11). Thus, understanding the link between biological rhythms and cancers can both assist in the development of new treatments, and in optimization of current therapies.
In mammals, the molecular circadian clock can be divided into three components: input pathways, the central pacemaker, and output pathways. The input pathways transmit information from environmental cues (e.g., light) to the central pacemaker, which is located in the suprachiasmatic nucleus (SCN) of the hypothalamus (12). Within the SCN, multiple single-cell circadian oscillators are synchronized to generate daily circadian outputs (13). Output pathways convert the commands from the central pacemaker into circadian oscillations, which regulate physiological and behavioral functions in peripheral organs and tissues (14).
Circadian oscillations are mainly generated through two transcriptional/translational feedback loops (TTFLs) (15). The core loop involves four core clock genes: Circadian Locomotor Output Cycles Kaput (CLOCK) (16) and brain and muscle Arnt-like protein 1 (BMAL1) (17), which are the activators; and Period (PER1, PER2, and PER3) (18) and Cryptochrome (CRY1 and CRY2) (19), which are the repressors. In the morning, the CLOCK:BMAL1 heterodimer binds to an E-box DNA promoter, activating the transcription of PER, CRY, and other clock controlled genes. Late in the day, PER and CRY proteins dimerize and translocate from the cytoplasm to the nucleus, where they associate with the CLOCK:BMAL1 complex and suppress its transcriptional activity at the E-box site (20). The suppression of CLOCK:BMAL1 is released through the degradation of PER and CRY by ubiquitin-dependent pathways (21, 22) and casein kinases (CKIδ and CKIε) (23), which also control the timing of PER and CRY’s entrance to the nucleus. After PER and CRY are degraded, the cycle begins again with ~24 h periodicity.
The secondary TTFL is mainly driven by transcriptional activation of the retinoid-related orphan receptors (RORs a, b, c) and repression of REV-ERBα/REV-ERBβ (24). To drive the rhythmic oscillation of BMAL1, REV-ERBα binds to the ROR elements in the BMAL1 promoter, suppressing BMAL1 transcription. Conversely, RORa and RORb activate BMAL1 expression (25, 26). The cooperation between the two TTFLs and other kinases and phosphatases, which are critical for regulating period, phase, and amplitude of oscillations, provides robustness against environmental perturbations. This network also helps to maintain accurate circadian timing and adjust phase delays to align with local physiology (27).
Epidemiological Evidence of Altered Clocks’ Effects on Cancer
Lifestyles have dramatically changed since the invention of the light bulb in 1879. Since then, the daily activities of humans have expanded into the night, including “night-shift” occupations (28). According to the U.S. Bureau of Labor Statistics, in 2016, the majority of the employed population worked in the service industry (80.3%), including health care, social assistance, and transportation, followed by manufacturing (7.9%) (29)—areas with high proportions of shift work. Another report published in 2015 found that about 17–24% of the workforce in United States was assigned to irregular or on-call work schedules, including night and rotating shifts (30). These types of schedules can lead to disruption of the sleep–wake cycle and circadian time organization, in addition to exposure to light at night (LAN) for long periods of time (31, 32). Perturbations to sleep and circadian rhythms can cause metabolic changes (33) and immune suppression (34), which can lead to various health problems, including diabetes (35), obesity (36), and cardiovascular disease (37), in addition to cancer (38). As a result, the International Agency for Research on Cancer has classified “shift-work that involves circadian disruption” as a “potential carcinogenic to humans (Group 2A)” (39).
While debated in some instances, epidemiological studies have provided evidence to support the association between shift work and cancer risk (40, 41). Independent cohort studies of night workers and shift workers have observed increased incidence of breast (42), prostate (4), colon (43), and endometrial epithelial malignancies (44) and non-Hodgkin’s lymphoma (45), with risk further increased among individuals who have spent more hours and years working at night (42, 46). A case control study in Western Australia found that there was a 22% increase in breast cancer incidence among those who worked between midnight and 5:00 a.m. (47). Another study in France showed that there was a significant association (OR = 1.95) between breast cancer and women who worked night shifts for more than 4 years before their first full-term pregnancy. At that time their mammary-gland cells were found to be incompletely differentiated, making them more susceptible to circadian disruption effects (48). While it is difficult to eliminate shift work from society, there are some aspects that can be modified, which may decrease the risk of developing adverse health effects. To further understand the contributions of shift work to pathological development, extensive animal and cellular experiments have yielded proposed molecular mechanisms, which will be discussed in Section “Molecular Studies of Circadian Clocks and Breast Cancer.”
Jet lag is another environmental factor associated with altered circadian rhythms and higher incidence of cancers (49). Jet lag (or circadian desynchrony) is a sleep disorder arising from the mismatch between internal body clocks and the environmental light/dark cycle. This condition is typically the result of travel through multiple time zones over a short period of time (50). An early study in Finland showed that flight attendants have significantly higher incidence of breast cancer (81.2/100,000) compared with the general female population (57.4/100,000) (51). A later, follow-up assessment strongly suggested that the increased cancer incidence was related to disruption of sleep rhythms, caused by excess exposure of light during normal sleeping hours, resulting in melatonin dysregulation (52). In addition, a recent study published in 2017, which focused on the effect of exposure to LAN in the United States, showed that there was a 14% increased risk of breast cancer in the highest LAN compared with the lowest LAN (53). Similar results were reported in Israel, where there was a 73% higher incidence of breast cancer in communities with the highest LAN than lowest LAN, across 147 communities (54). All of these epidemiological studies have strongly indicated that the disruption of circadian rhythms contributes to cancer risk.
Molecular Studies of Circadian Clocks and Breast Cancer
The functions of clock genes in each tissue are unique and show specific oscillation patterns (55). Their expression and regulation play important roles in breast biology. It has been shown that the core clock genes exhibit different mRNA expression patterns during mammary-gland development and differentiation in mice (56). Among 14,070 tested genes in human epithelial cells, 1,029 genes showed rhythmic oscillations during lactation. The expression patterns of these genes can be clustered into two groups, one high in the morning and another in the evening, indicating that the expressions change with a period of 24 h (57). Not only are the expression levels of clock genes variable, but they are affected by different developmental stages of breast tissue, and the extracellular microenvironment (58). Thus, it is posited that disruption of clock genes can affect normal breast biology and induce or affect cancerous development.
Breast cancer is heterogeneous and can be classified into subtypes based on histology, tumor grade, lymph node status, and the presence of specific biomarkers (59). The three markers generally used in characterization are estrogen receptor (ER), human epidermal growth factor receptor 2 (HER2), and progesterone receptor (PR) (60, 61). Based on marker status, breast cancer can be grouped into at least four subtypes: luminal A (ER+, PR+/−, HER2−), luminal B (ER+, PR+/−, HER2+), HER2 (ER−, PR−, HER2+), and Basal (ER−, PR−, HER2−) (62, 63). Basal tumors are typically difficult to treat and have poor prognoses. Because they lack ER, PR, and HER2, they are sometimes referred to as “triple-negative.”
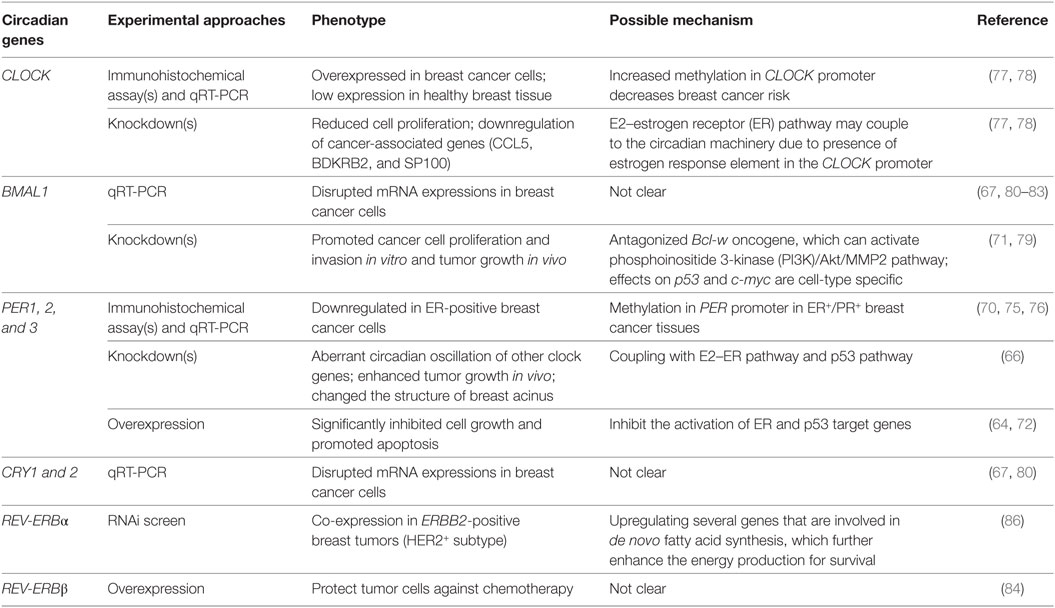
The disruption of nuclear hormone levels and signaling has also been posited to alter circadian rhythms, drawing another connection between rhythms and breast cancer (64). The estrogen receptor-α (ERα) signaling pathway (65) has been linked to the disruption of PER2 in breast cancer (Figure 1) (66, 67). It is known that PER2 is a direct transcriptional target of ERα and its expression is inducible by 17 β-estradiol (E2) simulation (64, 68). In normal human breast epithelial cells, both ERα and PER2 show rhythmic oscillations. The ubiquitous presence or absence of clock proteins has been predominantly used to investigate the relationship between circadian rhythms and breast cancer development (Table 1) (69–71). Knockdown of either PER2 or ERα results in aberrant circadian oscillations of ERα, PER2, BMAL1, and RARA (another direct ERα target gene) and affects breast acinus structures (66). It was first reported in 2007 that suppression of PER2 leads to ERα stabilization, and conversely, overexpression of PER2 in breast cancer cells significantly inhibited cell growth and promoted apoptosis (64, 72). This work was corroborated by showing that complete loss of PER2 mRNA oscillations occurred only in ERα-positive breast cancer cells, while ERα-negative breast cancer cells retained partially rhythmic oscillations (66, 67). In mice, downregulation of PER2 enhanced breast tumor growth, leading to further enhancement of amplitude and phase delay (70). All of these studies have suggested that the expression of clock genes may be disrupted by hormone levels and their signaling circuits (Figure 1) (73, 74). In addition, genome-wide DNA methylation profiling has shown that PER1 is significantly hypomethylated in ER+/PR+ breast cancer tissues (75). A separate study also showed that PER1, 2, and 3 exhibited deviant protein expressions in 55 resected breast cancer tissue sections, when compared with adjacent non-cancerous tissue samples. These fluctuations may be the result of methylation of the PER promoter (76). However, the detailed mechanisms of how hormone signaling affects circadian clocks and vice versa are still unclear.
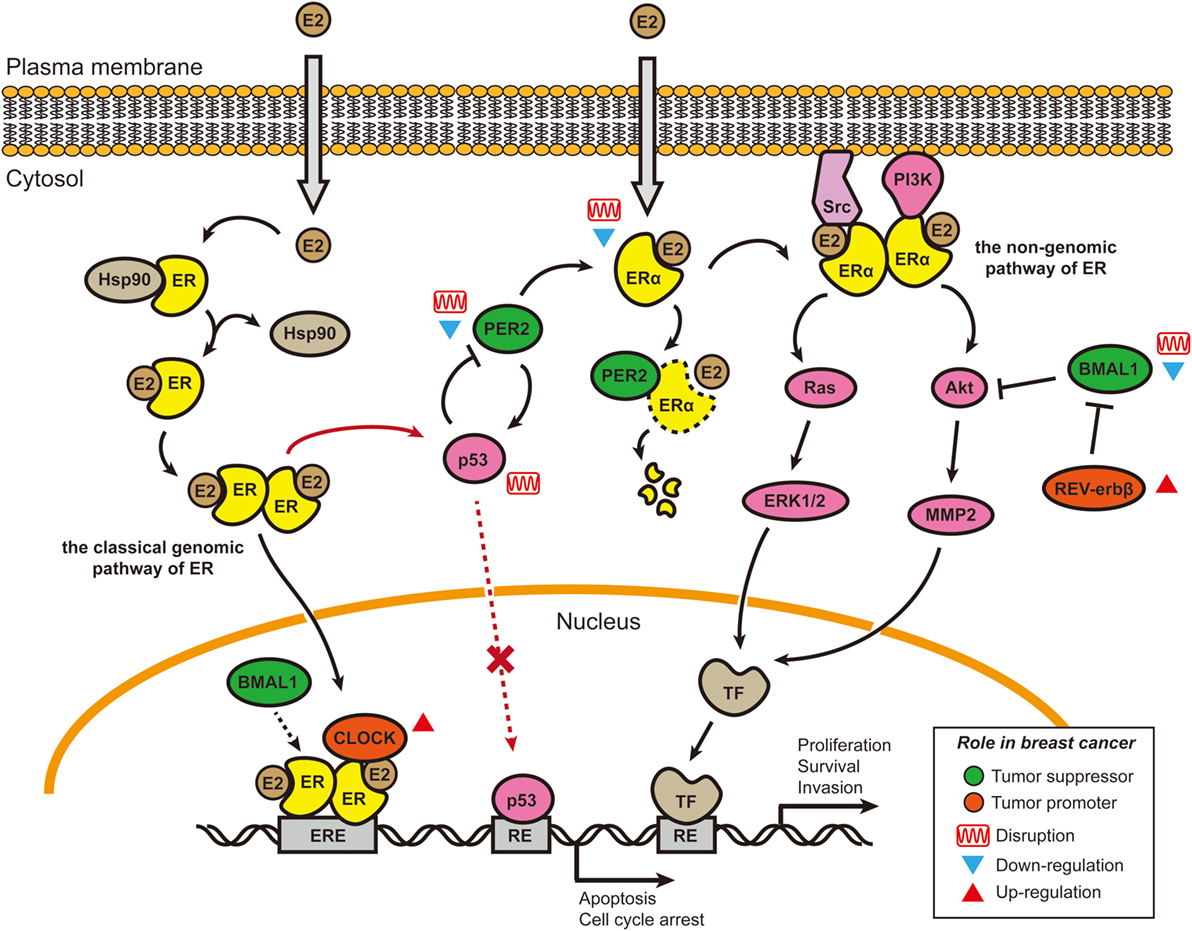
Figure 1. Cross talk between E2–estrogen receptor-α (ERα) signaling pathways and circadian rhythms in breast cancer. Two of the four estrogen signaling pathways involve E2 stimulation and are shown here (65). In the classical genomic pathway, E2-bound estrogen receptor (ER) (either ERα or ERβ) dimerizes, changes conformation, translocates to the nucleus, and binds to the estrogen response elements (EREs). After binding to the EREs, the E2–ER complex recruits other co-activators, including Circadian Locomotor Output Cycles Kaput (CLOCK) and possibly brain and muscle Arnt-like protein 1 (BMAL1) (74), to initiate the transcription of target genes. CLOCK overexpression in breast tumors and promotion of tumor cell proliferation may be caused by co-activation with E2–ER complexes (77, 78). In the non-genomic pathway, E2–ERα complexes accumulate near the membrane and then recruit protein kinases [Src and phosphoinositide 3-kinase (PI3K)] to activate signaling cascades (Akt and Ras/MAPK). BMAL1 has been shown to suppress the Akt/MMP2 pathway and further inhibit cancer cell invasion (79). BMAL1 can suppress cancers, and its expression is downregulated or disrupted in various breast cancer cell lines (67, 80–83). By contrast, REV-ERBβ (a repressor in the secondary transcriptional/translational feedback loop) is generally overexpressed in breast tumor samples; its protective function can allow cancer cells to develop chemotherapy resistance (84). PER2 is a direct ERα target gene and can bind to ERα and cause its degradation. In ERα-positive breast cancer cells, both PER2 and ERα lose their circadian oscillations, the underlying mechanism of which is not well understood. The cancer suppressor p53 can directly bind to the PER2 promoter and inhibit its transcriptional activity (85). E2–ER complexes can block the induction of proapoptotic p53 target genes by binding to p53 protein in ER-positive breast cancer cells, thus helping cancer cells avoid apoptosis (73). Re-introduction of PER2 into the ER-positive breast cancer cells can induce p53 expression (72). Abbreviations: TF, transcriptional factor; RE, response element; E2, 17 β-estradiol.
BMAL1 has also been proposed to act as a tumor suppressor. In separate studies performed in lung cancer and glioma cells, knockdown of BMAL1 promoted cancer cell proliferation, invasion, and tumor growth, while its overexpression reduced cellular invasiveness (71, 79). Effects occurred in a p53-independent manner (p53 expression was decreased in all BMAL1 knockdowns) and were accompanied by activation of the phosphoinositide 3-kinase (PI3K)–Akt–MMP-2 signaling pathway (79). While these studies used other cancer models to study the role of BMAL1, the findings are likely relevant to breast cancer. p53 mutations in breast cancer are relatively frequent (~20%) (85, 87), and the PI3K/Akt pathway is commonly affected (~70%) (88). However, the same study found that p21 (a p53 target protein) and c-myc exhibited different expression levels in various BMAL1-knockdown colon cancer cells, indicating that the relationships among BMAL1, p21, and c-myc are probably cell-type specific (71).
By contrast, CLOCK has been indicated as a tumor driver. Healthy breast patient tissues showed lower CLOCK expression than breast tumor tissues (77, 78). Knockdown of CLOCK resulted in attenuation of breast cancer proliferation (77) and downregulation of several cancer-associated genes, including ones related to breast tumor progression and metastasis initiation, such as CCL5, BDKRB2, and SP100 (78). Furthermore, increased methylation in the promoter region of CLOCK has been associated with decreased breast cancer risk (78). While these studies provide valuable insight to the involvement of clock proteins in breast cancer development, most of these experiments do not account for the dynamic nature of circadian rhythms, and the fact that they may be altered but not abolished with human behavior and disease.
More recently, a number of in vitro studies have investigated clock gene expression profiles in a time-dependent manner in various breast cancer cell lines, including: MCF7 and T47D (luminal A subtype); HCC-1954 (HER2 positive subtype); MCF10A and MDA-MB-231 (basal-like subtype), and others (67, 80–83). Intrinsic circadian oscillations in cultured cells can be entrained through treatment with high concentrations of serum to serum starved cells (89), or by chemical induction of signaling pathways, such as protein kinase A (via forskolin) (90) or the glucocorticoid receptor (via dexamethasone) (91). After entrainment, the expression patterns of clock genes, including BMAL1, CLOCK, PER1, PER2, CRY1, and CRY2, have largely been analyzed through quantitative real-time PCR, with conflicting results. While some studies revealed rhythmic gene expression in all breast cancer cell lines (67, 82), others did not (80, 83). Major factors contributing to the discrepancies were likely non-uniform cell culture and synchronization methods (i.e., varied serum depletion times before serum shock), which may affect dampening rates over time, due to loss of synchronicity. In addition, the short-term data collection (typically ≤48 h) and insufficient numbers of data points (generally ≥4 h intervals) utilized in these studies are generally insufficient to yield good statistical curve fittings (92, 93), contributing to inaccurate analysis of rhythmic oscillations. However, within each study, it is apparent that oscillations of clock proteins vary across different breast cancer cell models. Application of luciferase reporters and fluorescent proteins (e.g., GFP) can provide better resolution for long-term tracking of circadian oscillations in synchronized cells (14). However, cancer cells can be heterogeneous even in culture conditions (94). Future work should focus on real-time analysis at the single-cell level to reveal how circadian rhythms are involved, disrupted, and deviate from one another in breast cancer. Furthermore, posttranscriptional and -translational modifications to core circadian clock components should also be taken into consideration (95), since many malignant transformations occur posttranscriptionally.
Circadian Chronotherapy and Cancer Treatment
Nearly, all metabolic functions are regulated in a circadian manner: food intake, digestion, detoxification, breakdown, and storage of sugars and fats (96–98). When organs are exposed to xenobiotics (e.g., drugs or environmental toxicants), they undergo classical absorption, distribution, metabolism, and elimination processes, which are all regulated by circadian clocks (11). Hence, accounting for circadian rhythms in the development of treatments and dosing regimens has the potential to improve disease outcomes. Two recent studies reported the effects of chemotherapy on circadian rhythms in patients with metastatic colorectal cancer (99, 100). It was found that chemotherapy-induced disruption was observed in approximately 50% of the patients and was correlated with shortened overall survival rate. Eliminating this perturbation has been suggested to reduce toxicity and enhance efficacy of chemotherapy.
Recently, compounds that specifically target clock components and/or modulate its oscillations have received a great deal of attention (101). There are many advantages to the usage of small molecules in studies of circadian-related diseases: (1) they can help us to better understand the molecular circadian network; (2) they can serve as lead structures for developing drugs; and (3) unlike genetic approaches, which can result in immutable modifications, small molecules can be used in reversible, time- and dose-dependent manners (102, 103). One common example is the amelioration of jet lag via use of the hormone melatonin (104, 105). A double-blind trial showed that melatonin can significantly reduce jet lag and sleep disturbance in an international cabin crew (106). Small molecules can also be used to modify circadian rhythm periods to minimize chronodisruption resulting from shift work. Since the entrained phase is associated with circadian period, the period modification should change the preferred phase of behavior (107). Amplitude enhancement has also been shown to combat metabolic syndromes (108), reverse age-related effects (109), and protect against psychiatric diseases (110).
Small molecules have been used to elucidate the connections between circadian rhythms and breast cancer, for example the role of REV-ERBs in the HER2+ subtype (111). The NR1D1 (REV-ERBα coding gene) is connected to ERBB2 (HER2 coding gene) in the 17q12 amplicon, resulting in their co-expression in ERBB2-positive breast tumors (86). The same study suggested that REV-ERBα serves as a survival factor for HER2+ breast cancer cells. However, more recent work has shown disagreements. By activating REV-ERBs via the synthetic agonist SR9011, decreased cell proliferation was observed in various breast cancer cells, independent of their ER or HER2 status (112). Another study found that dual inhibition of REV-ERBβ and autophagy by ARN5187, a novel REV-ERBβ ligand, can induce cytotoxicity in breast cancer cells (84). It was also shown that REV-ERBβ was dominantly expressed in breast tumor samples, while REV-ERBα was the predominant form in normal tissues. Overexpressed REV-ERBβ appeared to result in protection that made tumor cells resistant to chloroquine, a clinically relevant lysosomotropic agent suppressing autophagy. With ARN5187 treatment, REV-ERB-mediated transcription was inhibited. Grimaldi et al. suggested that this compound has the potential to serve as an anticancer agent (84). Although clock modulators alone may not be sufficient to induce anticancer effects, combined treatment with well-established anticancer drugs should enhance their potency and efficacy, and reduce toxicity of the drugs. Characterization of disrupted circadian patterns in various types of cancer can provide clues for the application of clock modulators in combination with anticancer drugs to achieve the best possible therapeutic results.
Conclusion
Circadian rhythms are essential to the regulation of many physiological and behavioral functions in mammals. Their disruption has been linked to development of many health problems, including breast cancer. This is supported by epidemiological evidence, assessing both shift workers and people exposed to chronic jet lag. The status of core circadian clock components has also been evaluated in cancerous versus healthy tissues, and the significance of these components has been investigated via overexpression or deletions. While more recent studies have addressed changes in oscillations across cancer types, investigations at higher resolutions are required to facilitate more thorough analysis. From the work reviewed here, it is clear that circadian rhythms and proto-oncogenes/signaling pathways (e.g., PI3KCA, p53, or E2–ER) can both affect one another. However, the molecular mechanisms behind these associations are not well understood, and currently very few studies exist that examine the effects of altered rhythms on oncogenic pathways. Future work should also take advantage of existing technologies (including high-resolution confocal microscopy) (113) to track and analyze dynamic circadian oscillations at the single-cell level. While posttranscriptional and -translational modifications are also critical elements of the puzzle, real-time monitoring of these processes remains difficult to achieve. By increasing knowledge of the molecular mechanisms associated with disrupted clocks in cancer, new therapeutics and adjuvants can be developed with enhanced efficacy against the disease.
Author Contributions
H-HL wrote this article and generated the graphic and table, with content and editorial revisions from MF.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer EPS and handling Editor declared their shared affiliation.
Acknowledgments
The authors are grateful to Jessica J. Furtado, L. D. Sujeewa Sampath, and Wei-Ting Wong for helpful discussions.
Funding
H-HL was funded by a Chemistry-Biology Interface Program Fellowship from the University of Massachusetts Amherst.
References
1. Hamilton T. Influence of environmental light and melatonin upon mammary tumour induction. Br J Surg (1969) 56(10):764–6. doi:10.1002/bjs.1800561018
2. Aubert C, Janiaud P, Lecalvez J. Effect of pinealectomy and melatonin on mammary tumor growth in Sprague-Dawley rats under different conditions of lighting. J Neural Transm (1979) 47(2):121–30. doi:10.1007/BF01670163
3. Mhatre MC, Shah PN, Juneja HS. Effect of varying photoperiods on mammary morphology, DNA synthesis, and hormone profile in female rats. J Natl Cancer Inst (1984) 72(6):1411–6. doi:10.1093/jnci/72.6.1411
4. Flynn-Evans EE, Mucci L, Stevens RG, Lockley SW. Shiftwork and prostate-specific antigen in the national health and nutrition examination survey. J Natl Cancer Inst (2013) 105(17):1292–7. doi:10.1093/jnci/djt169
5. Shih MC, Yeh KT, Tang KP, Chen JC, Chang JG. Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Mol Carcinog (2006) 45(10):732–40. doi:10.1002/mc.20198
6. Mostafaie N, Kállay E, Sauerzapf E, Bonner E, Kriwanek S, Cross HS, et al. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog (2009) 48(7):642–7. doi:10.1002/mc.20510
7. Hu S, Shen G, Yin S, Xu W, Hu B. Melatonin and tryptophan circadian profiles in patients with advanced non-small cell lung cancer. Adv Ther (2009) 26(9):886–92. doi:10.1007/s12325-009-0068-8
8. Tokunaga H, Takebayashi Y, Utsunomiya H, Akahira J, Higashimoto M, Mashiko M, et al. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand (2008) 87(10):1060–70. doi:10.1080/00016340802348286
9. Lin YM, Chang JH, Yeh KT, Yang MY, Liu TC, Lin SF, et al. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog (2008) 47(12):925–33. doi:10.1002/mc.20446
10. Roche VP, Mohamad-Djafari A, Innominato PF, Karaboué A, Gorbach A, Lévi FA. Thoracic surface temperature rhythms as circadian biomarkers for cancer chronotherapy. Chronobiol Int (2014) 31(3):409–20. doi:10.3109/07420528.2013.864301
11. Dallmann R, Okyar A, Lévi F. Dosing-time makes the poison: circadian regulation and pharmacotherapy. Trends Mol Med (2016) 22(5):430–45. doi:10.1016/j.molmed.2016.03.004
12. Aschoff J. Circadian rhythms in man. Science (1965) 148(3676):1427–32. doi:10.1126/science.148.3676.1427
13. Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol (2010) 72:551–77. doi:10.1146/annurev-physiol-021909-135919
14. Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol (2004) 14(24):2289–95. doi:10.1016/j.cub.2004.11.057
15. Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol (2014) 24(2):90–9. doi:10.1016/j.tcb.2013.07.002
16. Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science (1998) 280(5369):1564–9. doi:10.1126/science.280.5369.1564
17. Honma S, Ikeda M, Abe H, Tanahashi Y, Namihira M, Honma K, et al. Circadian oscillation of BMAL1, a partner of a mammalian clock gene clock, in rat suprachiasmatic nucleus. Biochem Biophys Res Commun (1998) 250(1):83–7. doi:10.1006/bbrc.1998.9275
18. Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, et al. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell (2009) 36(3):417–30. doi:10.1016/j.molcel.2009.10.012
19. Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science (1998) 282(5393):1490–4. doi:10.1126/science.282.5393.1490
20. Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol (2001) 63:647–76. doi:10.1146/annurev.physiol.63.1.647
21. Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science (2007) 316(5826):900–4. doi:10.1126/science.1141194
22. Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, et al. Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell (2007) 129(5):1011–23. doi:10.1016/j.cell.2007.04.030
23. Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci U S A (2011) 108(39):16451–6. doi:10.1073/pnas.1107178108
24. Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell (2002) 110(2):251–60. doi:10.1016/S0092-8674(02)00825-5
25. Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBα and β and non-essential role for bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet (2008) 4(2):e1000023. doi:10.1371/journal.pgen.1000023
26. Fabienne G, Hugues D, Vincent G, Nicolas C. Differential control of bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms (2005) 20(5):391–403. doi:10.1177/0748730405277232
27. Brown SA, Kowalska E, Dallmann R. (Re)inventing the circadian feedback loop. Dev Cell (2012) 22(3):477–87. doi:10.1016/j.devcel.2012.02.007
28. Haus EL, Smolensky MH. Shift work and cancer risk: potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Med Rev (2013) 17(4):273–84. doi:10.1016/j.smrv.2012.08.003
29. U.S. Bureau of Labor Statistics. Employment by Major Industry Sector. (2017). Available from: https://www.bls.gov/emp/ep_table_201.htm (Accessed: December 29, 2017).
30. Golden L. Irregular Work Scheduling and Its Consequences. Economic Policy Institute (2015). Available from: http://www.epi.org (Accessed: December 29, 2017).
31. Lewy AJ, Wehr TA, Goodwin FK, Newsome DA, Markey SP. Light suppresses melatonin secretion in humans. Science (1980) 210(4475):1267–9. doi:10.1126/science.7434030
32. Dickerson SS, Connors LM, Fayad A, Dean GE. Sleep-wake disturbances in cancer patients: narrative review of literature focusing on improving quality of life outcomes. Nat Sci Sleep (2014) 6:85–100. doi:10.2147/NSS.S34846
33. Arble DM, Ramsey KM, Bass J, Turek FW. Circadian disruption and metabolic disease: findings from animal models. Best Pract Res Clin Endocrinol Metab (2010) 24(5):785–800. doi:10.1016/j.beem.2010.08.003
34. Nathalie L, Nicolas C. Circadian clocks in the immune system. J Biol Rhythms (2015) 30(4):277–90. doi:10.1177/0748730415577723
35. Gale JE, Cox HI, Qian J, Block GD, Colwell CS, Matveyenko AV. Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythms (2011) 26(5):423–33. doi:10.1177/0748730411416341
36. Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH. Circadian disruption leads to insulin resistance and obesity. Curr Biol (2013) 23(5):372–81. doi:10.1016/j.cub.2013.01.048
37. Morris CJ, Purvis TE, Hu K, Scheer FA. Circadian misalignment increases cardiovascular disease risk factors in humans. Proc Natl Acad Sci U S A (2016) 113(10):E1402–11. doi:10.1073/pnas.1516953113
38. Savvidis C, Koutsilieris M. Circadian rhythm disruption in cancer biology. Mol Med (2012) 18:1249–60. doi:10.2119/molmed.2012.00077
39. Baan R, Grosse Y, Straif K, Secretan B, El Ghissassi F, Bouvard V, et al. A review of human carcinogens. Part F: chemical agents and related occupations. Lancet Oncol (2009) 10(12):1143–4. doi:10.1016/S1470-2045(09)70358-4
40. Wegrzyn LR, Tamimi RM, Rosner BA, Brown SB, Stevens RG, Eliassen AH, et al. Rotating night-shift work and the risk of breast cancer in the nurses’ health studies. Am J Epidemiol (2017) 186(5):532–40. doi:10.1093/aje/kwx140
41. Ijaz S, Verbeek J, Seidler A, Lindbohm ML, Ojajärvi A, Orsini N, et al. Night-shift work and breast cancer – a systematic review and meta-analysis. Scand J Work Environ Health (2013) 39(5):431–47. doi:10.5271/sjweh.3371
42. Travis RC, Balkwill A, Fensom GK, Appleby PN, Reeves GK, Wang XS, et al. Night Shift work and breast cancer incidence: three prospective studies and meta-analysis of published studies. J Natl Cancer Inst (2016) 108(12):djw169–169. doi:10.1093/jnci/djw169
43. Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, et al. Night-shift work and risk of colorectal cancer in the nurses’ health study. J Natl Cancer Inst (2003) 95(11):825–8. doi:10.1093/jnci/95.11.825
44. Viswanathan AN, Hankinson SE, Schernhammer ES. Night shift work and the risk of endometrial cancer. Cancer Res (2007) 67(21):10618–22. doi:10.1158/0008-5472.CAN-07-2485
45. Lahti TA, Partonen T, Kyyrönen P, Kauppinen T, Pukkala E. Night-time work predisposes to non-Hodgkin lymphoma. Int J Cancer (2008) 123(9):2148–51. doi:10.1002/ijc.23566
46. Lin X, Chen W, Wei F, Ying M, Wei W, Xie X. Night-shift work increases morbidity of breast cancer and all-cause mortality: a meta-analysis of 16 prospective cohort studies. Sleep Med (2015) 16(11):1381–7. doi:10.1016/j.sleep.2015.02.543
47. Fritschi L, Erren TC, Glass DC, Girschik J, Thomson AK, Saunders C, et al. The association between different night shiftwork factors and breast cancer: a case-control study. Br J Cancer (2013) 109(9):2472–80. doi:10.1038/bjc.2013.544
48. Menegaux F, Truong T, Anger A, Cordina-Duverger E, Lamkarkach F, Arveux P, et al. Night work and breast cancer: a population-based case-control study in France (the CECILE study). Int J Cancer (2013) 132(4):924–31. doi:10.1002/ijc.27669
49. Kettner NM, Voicu H, Finegold MJ, Coarfa C, Sreekumar A, Putluri N, et al. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell (2016) 30(6):909–24. doi:10.1016/j.ccell.2016.10.007
51. Pukkala E, Auvinen A, Wahlberg G. Incidence of cancer among Finnish airline cabin attendants, 1967-92. BMJ (1995) 311(7006):649–52. doi:10.1136/bmj.311.7006.649
52. Kojo K, Pukkala E, Auvinen A. Breast cancer risk among Finnish cabin attendants: a nested case-control study. Occup Environ Med (2005) 62(7):488–93. doi:10.1136/oem.2004.014738
53. James P, Bertrand KA, Hart JE, Schernhammer ES, Tamimi RM, Laden F. Outdoor light at night and breast cancer incidence in the nurses’ health study II. Environ Health Perspect (2017) 125(8):087010. doi:10.1289/EHP935
54. Kloog I, Haim A, Stevens RG, Barchana M, Portnov BA. Light at night co-distributes with incident breast but not lung cancer in the female population of Israel. Chronobiol Int (2008) 25(1):65–81. doi:10.1080/07420520801921572
55. Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A (2004) 101(15):5339–46. doi:10.1073/pnas.0308709101
56. Metz RP, Qu X, Laffin B, Earnest D, Porter WW. Circadian clock and cell cycle gene expression in mouse mammary epithelial cells and in the developing mouse mammary gland. Dev Dyn (2006) 235(1):263–71. doi:10.1002/dvdy.20605
57. Maningat PD, Sen P, Rijnkels M, Sunehag AL, Hadsell DL, Bray M, et al. Gene expression in the human mammary epithelium during lactation: the milk fat globule transcriptome. Physiol Genomics (2009) 37(1):12–22. doi:10.1152/physiolgenomics.90341.2008
58. Casey T, Patel O, Dykema K, Dover H, Furge K, Plaut K. Molecular signatures reveal circadian clocks may orchestrate the homeorhetic response to lactation. PLoS One (2009) 4(10):e7395. doi:10.1371/journal.pone.0007395
59. Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res (2011) 13:215. doi:10.1186/bcr2889
60. Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A (1999) 96(16):9212–7. doi:10.1073/pnas.96.16.9212
61. Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature (2000) 406(6797):747–52. doi:10.1038/35021093
62. Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell (2006) 10(6):515–27. doi:10.1016/j.ccr.2006.10.008
63. Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res (2010) 12(5):R68. doi:10.1186/bcr2635
64. Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene (2007) 26(57):7916–20. doi:10.1038/sj.onc.1210585
65. Le Romancer M, Poulard C, Cohen P, Sentis S, Renoir JM, Corbo L. Cracking the Estrogen receptor’s posttranslational code in breast tumors. Endocr Rev (2011) 32(5):597–622. doi:10.1210/er.2010-0016
66. Rossetti S, Corlazzoli F, Gregorski A, Azmi NH, Sacchi N. Identification of an estrogen-regulated circadian mechanism necessary for breast acinar morphogenesis. Cell Cycle (2012) 11(19):3691–700. doi:10.4161/cc.21946
67. Rossetti S, Esposito J, Corlazzoli F, Gregorski A, Sacchi N. Entrainment of breast (cancer) epithelial cells detects distinct circadian oscillation patterns for clock and hormone receptor genes. Cell Cycle (2012) 11(2):350–60. doi:10.4161/cc.11.2.18792
68. Nakamura TJ, Sellix MT, Menaker M, Block GD. Estrogen directly modulates circadian rhythms of PER2 expression in the uterus. Am J Physiol Endocrinol Metab (2008) 295(5):E1025–31. doi:10.1152/ajpendo.90392.2008
69. Hua H, Wang Y, Wan C, Liu Y, Zhu B, Yang C, et al. Circadian gene mPer2 overexpression induces cancer cell apoptosis. Cancer Sci (2006) 97(7):589–96. doi:10.1111/j.1349-7006.2006.00225.x
70. Yang X, Wood PA, Oh EY, Du-Quiton J, Ansell CM, Hrushesky WJ. Down regulation of circadian clock gene Period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat (2009) 117(2):423–31. doi:10.1007/s10549-008-0133-z
71. Zeng ZL, Wu MW, Sun J, Sun YL, Cai YC, Huang YJ, et al. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J Biochem (2010) 148(3):319–26. doi:10.1093/jb/mvq069
72. Xiang S, Coffelt SB, Mao L, Yuan L, Cheng Q, Hill SM. Period-2: a tumor suppressor gene in breast cancer. J Circadian Rhythms (2008) 6:4. doi:10.1186/1740-3391-6-4
73. Bailey ST, Shin H, Westerling T, Liu XS, Brown M. Estrogen receptor prevents p53-dependent apoptosis in breast cancer. Proc Natl Acad Sci U S A (2012) 109(44):18060–5. doi:10.1073/pnas.1018858109
74. Li S, Wang M, Ao X, Chang AK, Yang C, Zhao F, et al. CLOCK is a substrate of SUMO and sumoylation of CLOCK upregulates the transcriptional activity of estrogen receptor-α. Oncogene (2012) 32(41):4883–91. doi:10.1038/onc.2012.518
75. Li L, Lee KM, Han W, Choi JY, Lee JY, Kang GH, et al. Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum Mol Genet (2010) 19(21):4273–7. doi:10.1093/hmg/ddq351
76. Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ, Chang JG. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis (2005) 26(7):1241–6. doi:10.1093/carcin/bgi075
77. Xiao L, Chang AK, Zang MX, Bi H, Li S, Wang M, et al. Induction of the CLOCK gene by E2-ERα signaling promotes the proliferation of breast cancer cells. PLoS One (2014) 9(5):e95878. doi:10.1371/journal.pone.0095878
78. Hoffman AE, Yi CH, Zheng T, Stevens RG, Leaderer D, Zhang Y, et al. CLOCK in breast tumorigenesis: evidence from genetic, epigenetic, and transcriptional profiling analyses. Cancer Res (2010) 70(4):1459–68. doi:10.1158/0008-5472.CAN-09-3798
79. Jung CH, Kim EM, Park JK, Hwang SG, Moon SK, Kim WJ, et al. Bmal1 suppresses cancer cell invasion by blocking the phosphoinositide 3-kinase-Akt-MMP-2 signaling pathway. Oncol Rep (2013) 29(6):2109–13. doi:10.3892/or.2013.2381
80. Xiang S, Mao L, Duplessis T, Yuan L, Dauchy R, Dauchy E, et al. Oscillation of clock and clock controlled genes induced by serum shock in human breast epithelial and breast cancer cells: regulation by melatonin. Breast Cancer (Auckl) (2012) 6:137–50. doi:10.4137/bcbcr.s9673
81. Cadenas C, van de Sandt L, Edlund K, Lohr M, Hellwig B, Marchan R, et al. Loss of circadian clock gene expression is associated with tumor progression in breast cancer. Cell Cycle (2014) 13(20):3282–91. doi:10.4161/15384101.2014.954454
82. Gutiérrez-Monreal MA, Treviño V, Moreno-Cuevas JE, Scott SP. Identification of circadian-related gene expression profiles in entrained breast cancer cell lines. Chronobiol Int (2016) 33(4):392–405. doi:10.3109/07420528.2016.1152976
83. Chacolla-Huaringa R, Moreno-Cuevas J, Trevino V, Scott SP. Entrainment of breast cell lines results in rhythmic fluctuations of microRNAs. Int J Mol Sci (2017) 18(7):1499. doi:10.3390/ijms18071499
84. De Mei C, Ercolani L, Parodi C, Veronesi M, Lo Vecchio C, Bottegoni G, et al. Dual inhibition of REV-ERBβ and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene (2014) 34(20):2597–608. doi:10.1038/onc.2014.203
85. Miki T, Matsumoto T, Zhao Z, Lee CC. p53 regulates period2 expression and the circadian clock. Nat Commun (2013) 4:2444. doi:10.1038/ncomms3444
86. Kourtidis A, Jain R, Carkner RD, Eifert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Res (2010) 70(5):1783–92. doi:10.1158/0008-5472.CAN-09-1550
87. Pharoah PD, Day NE, Caldas C. Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer (1999) 80(12):1968–73. doi:10.1038/sj.bjc.6690628
88. Bhat-Nakshatri P, Goswami CP, Badve S, Magnani L, Lupien M, Nakshatri H. Molecular insights of pathways resulting from two common PIK3CA mutations in breast cancer. Cancer Res (2016) 76(13):3989–4001. doi:10.1158/0008-5472.CAN-15-3174
89. Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell (1998) 93(6):929–37. doi:10.1016/S0092-8674(00)81199-X
90. Yagita K, Okamura H. Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett (2000) 465(1):79–82. doi:10.1016/S0014-5793(99)01724-X
91. Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science (2000) 289(5488):2344–7. doi:10.1126/science.289.5488.2344
92. Izumo M, Sato TR, Straume M, Johnson CH. Quantitative analyses of circadian gene expression in mammalian cell cultures. PLoS Comput Biol (2006) 2(10):e136. doi:10.1371/journal.pcbi.0020136
93. Refinetti R, Lissen GC, Halberg F. Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res (2007) 38(4):275–325. doi:10.1080/09291010600903692
94. Kiessling S, Beaulieu-Laroche L, Blum ID, Landgraf D, Welsh DK, Storch KF, et al. Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol (2017) 15(1):13. doi:10.1186/s12915-017-0349-7
95. Jang C, Lahens NF, Hogenesch JB, Sehgal A. Ribosome profiling reveals an important role for translational control in circadian gene expression. Genome Res (2015) 25(12):1836–47. doi:10.1101/gr.191296.115
96. Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci U S A (2012) 109(7):2625–9. doi:10.1073/pnas.1114410109
97. Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A (2012) 109(14):5541–6. doi:10.1073/pnas.1118726109
98. Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A (2009) 106(24):9890–5. doi:10.1073/pnas.0900617106
99. Innominato PF, Giacchetti S, Bjarnason GA, Focan C, Garufi C, Coudert B, et al. Prediction of overall survival through circadian rest-activity monitoring during chemotherapy for metastatic colorectal cancer. Int J Cancer (2012) 131(11):2684–92. doi:10.1002/ijc.27574
100. Lévi F, Dugué PA, Innominato P, Karaboué A, Dispersyn G, Parganiha A, et al. Wrist actimetry circadian rhythm as a robust predictor of colorectal cancer patients survival. Chronobiol Int (2014) 31(8):891–900. doi:10.3109/07420528.2014.924523
101. Wallach T, Kramer A. Chemical chronobiology: toward drugs manipulating time. FEBS Lett (2015) 589(14):1530–8. doi:10.1016/j.febslet.2015.04.059
102. Antoch MP, Kondratov RV. Pharmacological modulators of the circadian clock as potential therapeutic drugs: focus on genotoxic/anticancer therapy. Handb Exp Pharmacol (2013) 217:289–309. doi:10.1007/978-3-642-25950-0_12
103. Chen Z, Yoo SH, Takahashi JS. Small molecule modifiers of circadian clocks. Cell Mol Life Sci (2013) 70(16):2985–98. doi:10.1007/s00018-012-1207-y
104. Herxheimer A, Petrie KJ. Melatonin for the prevention and treatment of jet lag. Cochrane Database Syst Rev (2002) (2):CD001520. doi:10.1002/14651858.cd001520
105. Arendt J. Melatonin: characteristics, concerns, and prospects. J Biol Rhythms (2005) 20(4):291–303. doi:10.1177/0748730405277492
106. Petrie K, Dawson AG, Thompson L, Brook R. A double-blind trial of melatonin as a treatment for jet lag in international cabin crew. Biol Psychiatry (1993) 33(7):526–30. doi:10.1016/0006-3223(93)90007-Z
107. Duffy JF, Rimmer DW, Czeisler CA. Association of intrinsic circadian period with morningness-eveningness, usual wake time, and circadian phase. Behav Neurosci (2001) 115(4):895–9. doi:10.1037/0735-7044.115.4.895
108. He B, Nohara K, Park N, Park YS, Guillory B, Zhao Z, et al. The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab (2016) 23(4):610–21. doi:10.1016/j.cmet.2016.03.007
109. Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell (2013) 153(7):1448–60. doi:10.1016/j.cell.2013.05.027
110. Li J, Lu WQ, Beesley S, Loudon AS, Meng QJ. Lithium impacts on the amplitude and period of the molecular circadian clockwork. PLoS One (2012) 7(3):e33292. doi:10.1371/journal.pone.0033292
111. Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov (2014) 13(3):197–216. doi:10.1038/nrd4100
112. Wang Y, Kojetin D, Burris TP. Anti-proliferative actions of a synthetic REV-ERBα/β agonist in breast cancer cells. Biochem Pharmacol (2015) 96(4):315–22. doi:10.1016/j.bcp.2015.06.010
Keywords: altered circadian rhythms, shift work, breast cancer, molecular mechanism, hormone pathways, small molecule modulators
Citation: Lin H-H and Farkas ME (2018) Altered Circadian Rhythms and Breast Cancer: From the Human to the Molecular Level. Front. Endocrinol. 9:219. doi: 10.3389/fendo.2018.00219
Received: 31 December 2017; Accepted: 18 April 2018;
Published: 04 May 2018
Edited by:
Arturo Ortega, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional (CINVESTAV-IPN), MexicoReviewed by:
Wei-jiang Zhao, Shantou University Medical College, ChinaEduardo Perez-Salazar, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional (CINVESTAV-IPN), Mexico
Edyta Reszka, Nofer Institute of Occupational Medicine, Poland
Copyright: © 2018 Lin and Farkas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michelle E. Farkas, ZmFya2FzQGNoZW0udW1hc3MuZWR1
 Hui-Hsien Lin
Hui-Hsien Lin Michelle E. Farkas
Michelle E. Farkas