- 1Key Laboratory of Freshwater Fisheries and Germplasm Resources Utilization, Ministry of Agriculture, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
- 2Wuxi Fisheries College, Nanjing Agricultural University, Wuxi, China
- 3Aquatic Animal Genome Center of Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
- 4Scientific Observing and Experimental Station of Fishery Resources and Environment in the Lower Reaches of the Changjiang River, Ministry of Agriculture and Rural Affairs, Wuxi, China
Commercial fishing of estuarine tapertail anchovy (Coilia nasus), an important anadromous fish species in the Yangtze River of China, has been prohibited due to the serious damage overfishing has caused to the wild population. Research regarding the energy metabolism is important for migratory fish to ensure the continuation of their existence. In this study, we performed, for the first time, a comparative transcriptome analysis of the liver of C. nasus subjected to long-term starvation stress. The results indicated that the damaging effects involved downregulation of the antioxidant capacity and immune response. The positive response to starvation involved upregulation of the anti-allergy and anticancer capacity, which supports the function of starvation in cancer inhibition, as has also been determined for human beings. This study revealed regulatory pathways, differentially expressed genes (DEGs), and mechanisms leading to damage of the liver in C. nasus affected by starvation. This research contributes information for the further study of the energy metabolism mechanism of C. nasus and provides a theoretical reference for starvation metabolism research of other fish species and even human beings.
Introduction
The Yangtze River, winding about 6,387 km, is the largest in China and the third longest river in the world. The Yangtze River is also one of seven rivers that possess abundant aquatic biodiversity. There are 4,300 aquatic organisms, including 400 kinds of fish species; 11 types of national key protection aquatic organisms, such as Chinese sturgeon (Acipenser sinensis), Yangtze sturgeon (Acipenser dabryanus), and Yangtze finless porpoises (Neophocaena asiaeorientalis); and 170 types of fish species particular to the Yangtze River (1).
C. nasus, which belongs to the order Clupeiformes, the family Engraulidae, and the genus Coilia, is an important anadromous fish species found in the Yangtze River of China. It is widely popular due to its delicate and unique flavor and is among the so-called “Three Delicious Fish Species in the Yangtze River” together with the Reeves shad (Tenualosa reevesii) and obscure pufferfish (Takifugu fasciatus) (2, 3). Under the long-term influence of water pollution, overfishing, channel improvement, water project construction, etc., the living environment of aquatic organisms in the Yangtze River is deteriorating day by day, and certain rare species populations have been diminished (4).
As an important anadromous fish species in the Yangtze River, the wild population of C. nasus has been seriously damaged. It has been listed as an endangered fish species in China and the Ministry of Agriculture and Rural Affairs of the People s Republic of China issued a notice prohibiting the commercial fishing of C. nasus in the Yangtze River (5). Therefore, the artificial breeding industry of C. nasus is burdened with the responsibility of providing high-quality fish fry for reproduction and release to restore the wild resources of C. nasus in the Yangtze River while also continuing to enrich people’s dining tables.
Adult individuals of C. nasus, as an anadromous fish in the Yangtze River, undergo reproductive migration along the Yangtze River every February when their gonads have matured. Juvenile C. nasus will settle in the estuarine area until autumn and then undergo seaward migration for growth and fattening (6, 7). During the migration process, starvation is most likely to be caused by climate change, regional change, and uneven food distribution.
Therefore, the regulation of the robust energy metabolism mechanism of C. nasus during its migration is an important topic of research. In addition, in the daily artificial breeding of C. nasus, fish can face starvation due to excessive stocking density, delayed feeding, and uneven feeding. As a result, it is necessary to explore the metabolic mechanisms of C. nasus under starvation stress, which is of positive significance for revealing the regulatory mechanism of the energy metabolism in this species with additional benefits for the restoration of its wild resources in the Yangtze River as well as development of the artificial breeding industry.
The liver is a key organ in fish energy metabolisms (8). At present, there are several reports on the influence of starvation on the livers of fish, including yellow drum (Nibea albiflora) (9), large yellow croaker (Pseudosciaena crocea) (10), red sea bream (Pagrus major) (11), zebrafish (Danio rerio) (12, 13), and common dentex (Dentex dentex) (14). Authors in these studies examined the effects of cold and starvation on the mode of fish metabolism and discussed the metabolic mechanisms of starvation or starvation/refeeding as it relates to liver physiology, nutrient metabolism, and energy-related pathways, such as for the fatty acid metabolism, antioxidant responses, and the immune responses of fish.
The results indicated that crude fat content can be used as a biomarker for the starvation response. In addition, although starvation caused body oxidative stress in the short term, long-term starvation resulted in the failure of cellular antioxidant defenses. Fish have been found to utilize noncarbohydrate substances to produce energy under starvation stress, which causes the enhancement of gluconeogenesis (9–14). On the other hand, comparative transcriptome analyses of fish livers have been carried out to explore the regulatory mechanism of the liver under starvation stress at a broader scope.
Relevant studies have been carried out on large yellow croaker (Larimichthys crocea) (15), Atlantic salmon (Salmo salar) (16), zebrafish (Danio rerio) (17), and rainbow trout (Oncorhynchus mykiss) (18). The effects of starvation or starvation/refeeding on the energy metabolism and the immune response in fish livers were investigated. The most significant involved pathways were mainly those of fat digestion and absorption, the citrate cycle, and glycolysis/gluconeogenesis. There have been few reports of differential pathways relevant to the immune response: in this category, only coagulation cascades were identified (15). Starvation caused the downregulation of many immune-related pathways and regulatory genes in Atlantic salmon (16).
Research on zebrafish indicates that starvation causes significant downregulation of the metabolic activity, including lipid metabolism, protein biosynthesis, proteolysis, and cellular respiration, but results in the upregulation of gluconeogenesis (17). Research on rainbow trout revealed that when fish weight was significantly reduced due to starvation, calpain and 20S proteasome pathways in protein mobilization functioned as an energy source (18). The above studies have shown that, under starvation conditions, the livers of different fish species undergo differential regulation modes.
There have been few reports on the regulatory mechanism of C. nasus under starvation stress. In one of the only studies on this subject, Jin Xin et al. discussed the effect of starvation stress on the chemical composition and blood biochemical parameters of C. nasus (19). Therefore, we conducted the first comparative transcriptome analysis of the liver of near-death C. nasus under long-term starvation stress to reveal key regulatory pathways and genes. We aimed to reveal the mechanisms leading to damage of the liver in C. nasus under starvation stress as well as to lay a theoretical foundation for the investigation of the comprehensive energy metabolism mechanisms of C. nasus providing a theoretical reference for further in-depth research on the restoration of wild C. nasus resources and for the artificial breeding industry.
Materials and Methods
Ethical Statement
This study was approved by the Animal Care and Use Committee of the Freshwater Fisheries Research Center at the Chinese Academy of Fishery Sciences. The experiments were carried out in accordance with the Guidelines for the Care and Use of Laboratory Animals set by the Animal Care and Use Committee of the Freshwater Fisheries Research Center (2003WXEP61, Jan 6th of 2003), and the study was carried out with a field permit (no. 20176CN1619).
Experimental Fish and Starvation Stress
Experimental fish were 5-month-old juvenile C. nasus with an average body weight of 8.6 ± 0.59 g and a body length of 143.6 ± 0.82 mm. The fish were collected from Yixing, an experimental base of the Freshwater Fisheries Research Center of the Chinese Academy of Fishery Sciences. One month before the starvation experiment, juvenile C. nasus were netted and cultured in four aquariums. The aquariums were equipped with circulating water systems. The water quality was measured once each day, the pH was at 7.1, and water temperature was 18 ± 0.62°C. The concentration of dissolved oxygen was 7.8 ± 0.65 mg/L. The experimental fish were fed three times per day.
A control group (C) and a starvation group (S) were established. Each group had two replicates and each aquarium contained ten fish. The stressed group was not fed during the experimental period. On the 26th day of the starvation experiment, we observed that some fish in the stressed group swam laterally and were near death; these fish were caught, placed in 40 mg/L MS-222 solution (Kuer Bioengineering, Beijing, China) for rapid anesthesia and then dissected. We observed that the muscles and internal organs of the sampled fish had atrophied. Then, the remaining excessive starvation fish were also quickly caught and anesthetized. Two fish were sampled from each aquarium. The livers were collected and snap-frozen in liquid nitrogen and then stored at −80°C until further treatment.
Total RNA Extraction, cDNA Library Construction, and Illumina Sequencing
The total RNA for each sample was extracted using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Equal amounts of RNA from fish in the same tank were pooled as one sample to obtain a total of four RNA samples. The qualification and quantification of RNA, cDNA library construction, and Illumina sequencing, were subsequently carried out according to methods in our previously reported study (20). The obtained raw data were submitted to NCBI (NCBI, Bethesda, USA) with accession number PRJNA359698.
Data Filtering and Assembly
Quality control for the raw data was carried out using FASTQC (Babraham Institute, Cambridge, UK). Some primers, adapters, low-quality reads, etc., were processed by Cutadapt (version 1.9.1) (21). The assembly of the transcriptome data was carried out using Trinity (version 2.4.0) with min_kmer_cov set to 2 by default and all other parameters set to default (22).
Function Annotation
Unigenes were aligned on the basis of nonredundant protein (Nr), nonredundant nucleotide (Nt), Swiss-Prot (http://www.uniprot.org/downloads), clusters of orthologous groups for complete eukaryotic genomes (COG, ftp://ftp.ncbi.nih.gov/pub/COG/KOG/kyva), Gene Ontology (GO, http://www.geneontology.org/), and the Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/pathway.html) using BlastX (version 2.2.28+) with an E-value <10–5 (23, 24). GO annotation was performed using Blast2GO (version b2g4pipe_v2.5) (Biobam, Valencia, Spain) (25).
Gene Quantification and Differential Expression Analysis
The assembled unigenes were placed in a constructed library, and the abundance of the expression of each unigene in each sample was measured using Bowtie2 (version 2.1.0) (http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml) (Ben Langmead, Maryland, College Park, USA) (26) and eXpress software (version 1.5.1) (http://www.rna-seqblog.com/express-a-tool-for-quantification-of-rna-seq-data/) (California University, Berkeley, USA) (27). For the software parameters in Bowtie, -k was set to 30, and -t was the default value. In the eXpress software, the strand-specific mode was set to –rf-stranded. The gene expression levels were evaluated as fragments per kilobase of transcript per million mapped reads (FPKM) (28). Differential expression analysis was performed using the DESeq software R package (version no. 1.18.0) (29) (http://bioconductor.org/packages/release/bioc/html/DESeq.html). The resulting p-values were adjusted using Benjamini and Hochberg’s approach to control the false discovery rate. The fold change in gene expression was calculated as the ratio between the expression level of genes in the starvation stress group and in the control group. Genes with an adjusted p-value (padj) < 0.05 and |log2foldchange| > 1 found by DESeq were considered as being differentially expressed. Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) was implemented in the GOseq R packages based on Wallenius’ noncentral hypergeometric distribution (30). The GO terms enriched from DEGs were sequenced with −log10 (p-value), and the top 30 GO terms (including three subcategories, which were biological process (BP), cellular component (CC), and molecular function (MF)) were filtered according to −log10 (p-value) and ordered according to DEG numbers. Finally, we obtained the top 30 GO terms. We used KOBAS software to test the statistical enrichment of differentially expressed genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (31). The KEGG terms were sequenced with the −log10 (p-value), and the top 20 KEGG pathways were filtered according to the −log10 (p-value) and ordered according to the DEG numbers. Finally, we obtained the top 10 KEGG pathways. The above transcriptome data analysis, including function annotation, gene quantification, and differential expression analysis, was carried out following the method reported in our previous study (20).
Quantitative Real-time PCR Validation
To validate the accuracy of high-throughput sequencing results, ten DEGs were randomly selected from the transcriptome data for qRT-PCR analysis. The genes and primers sequences were shown in Supplementary file S1. The qRT-PCR validation experiment was carried out on the ABI 7500 real-time PCR system (ABI, New York, USA). The primers were designed using Primer Premier 5 software (Premier Biosoft, California, USA) (32), and the sequences of the designed primers are shown in Appendix S1. Beta-actin was used as the internal reference. The amplifications were conducted in a 10 μl reactions, which contained 2 ng of total RNA, 5 μl master mixes, and 0.4 μl of each primer. The qRT-PCR reactions were conducted with the following procedure: 95°C for 30 s, then 40 cycles of 95°C for 5 s, 60°C for 34 s, and then 72°C for 50 s. All samples were assayed in triplicate, and the 2−ΔΔCT method was used to calculate the expression level of the ten DEGs. Cycle threshold (CT) refers to the PCR cycle number at which the fluorescence signal crossed a threshold line that was placed in the exponential phase of the amplification curve. ΔΔCT = (CT.S – CT.actin) – (CT.C – CT.actin) where CT.S refers to the CT value of samples in the starvation group, CT.C refers to the CT value of samples in the control group (33).
Statistical Analysis
Statistical analyses were carried out with SPSS 21.1 software (SPSS, Chicago, USA). The values are shown as the means ± SE. Statistical analysis was performed with Student’s t-test, and p < 0.05 was set as the significant difference standard.
Results
Statistics Analysis of Transcriptome Data in the Liver of C. nasus Under Starvation Stress
The statistics of high-throughput sequencing data are shown in Table 1. Q30 refers to the proportion of bases with a base error rate of 0.1%. In this study, the Q30 of each group was higher than 95%, indicating that the transcriptome data had high accuracy.
Top 30 GO Terms Enriched From DEGs
As shown in Figure 1, the top 10 BP terms were mainly involved in the regulation of metabolism, development, and the stress response. Among these were the metabolism involved in the nucleic acid metabolic process (no. 1) and regulation of metabolic process (nos. 2–3); developmental regulation through maintenance of cell number (no. 4), regulation of cell migration (no. 8), cell development (no. 9), tissue morphogenesis (no. 5), tissue structure arrangement regulation (no. 7), regulation of neuron differentiation (no. 6); and stress response regulation, which was mainly related to the immune response (no. 10).
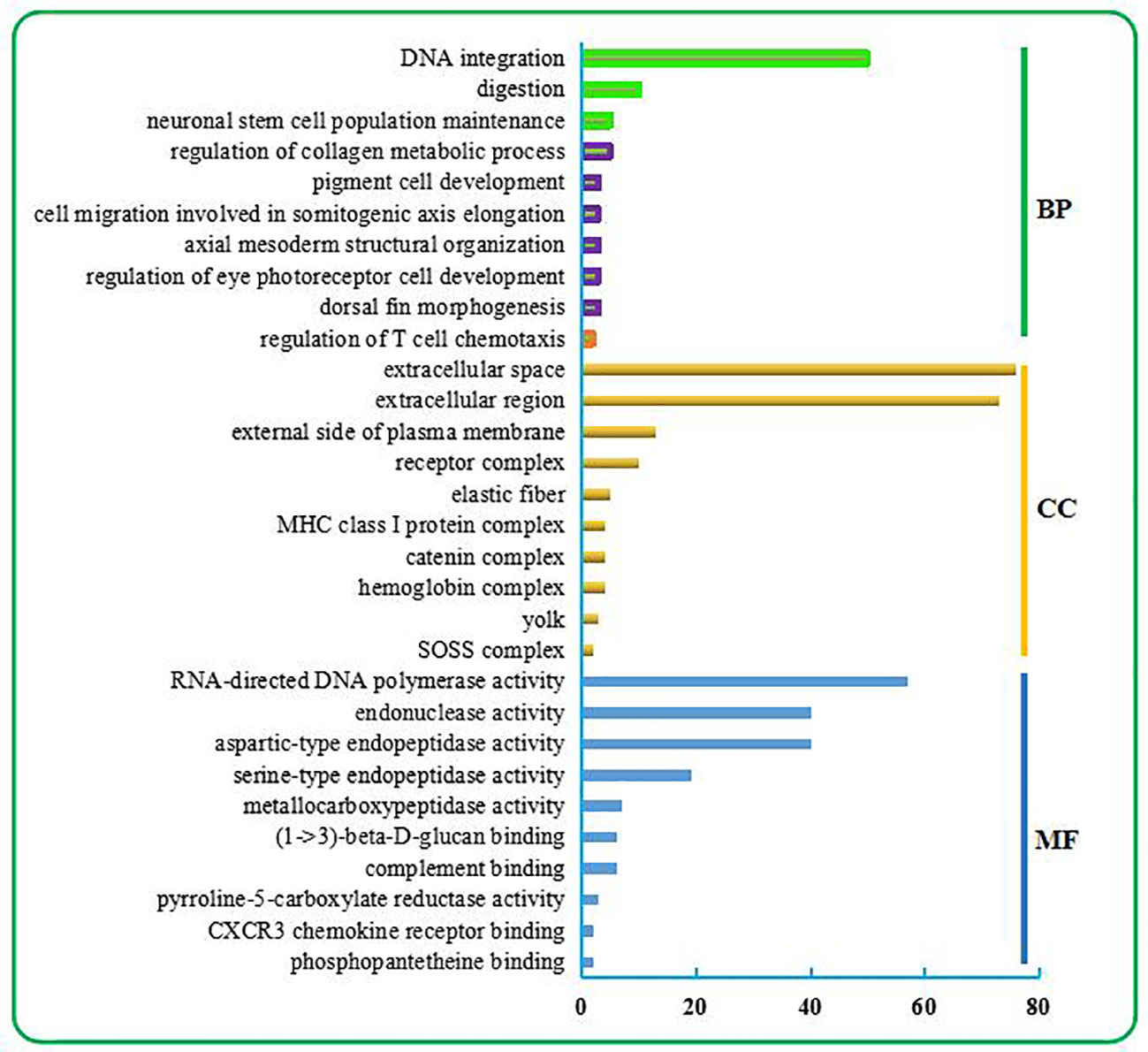
Figure 1 Top 30 GO terms enriched from the liver transcriptome of C. nasus under starvation stress. The top 30 GO terms include the top 10 terms in BP, CC, and MF categories. BP terms were further classified. Three terms relevant to metabolism are marked in the green box. Six terms relevant to development are marked in the purple box. One term relevant to the immune response is marked in the orange box.
Top 10 KEGG Pathways
As shown in Figure 2, the top 10 KEGG pathways were related to the digestion metabolism (nos. 1–3, 5–8) and mainly involved the energy-substance metabolism, including lipids, proteins, and amino acids. In addition, the top 10 KEGG pathways also involved signal transduction regulation, which was mainly related to immune response regulation (nos. 4, 9–10).
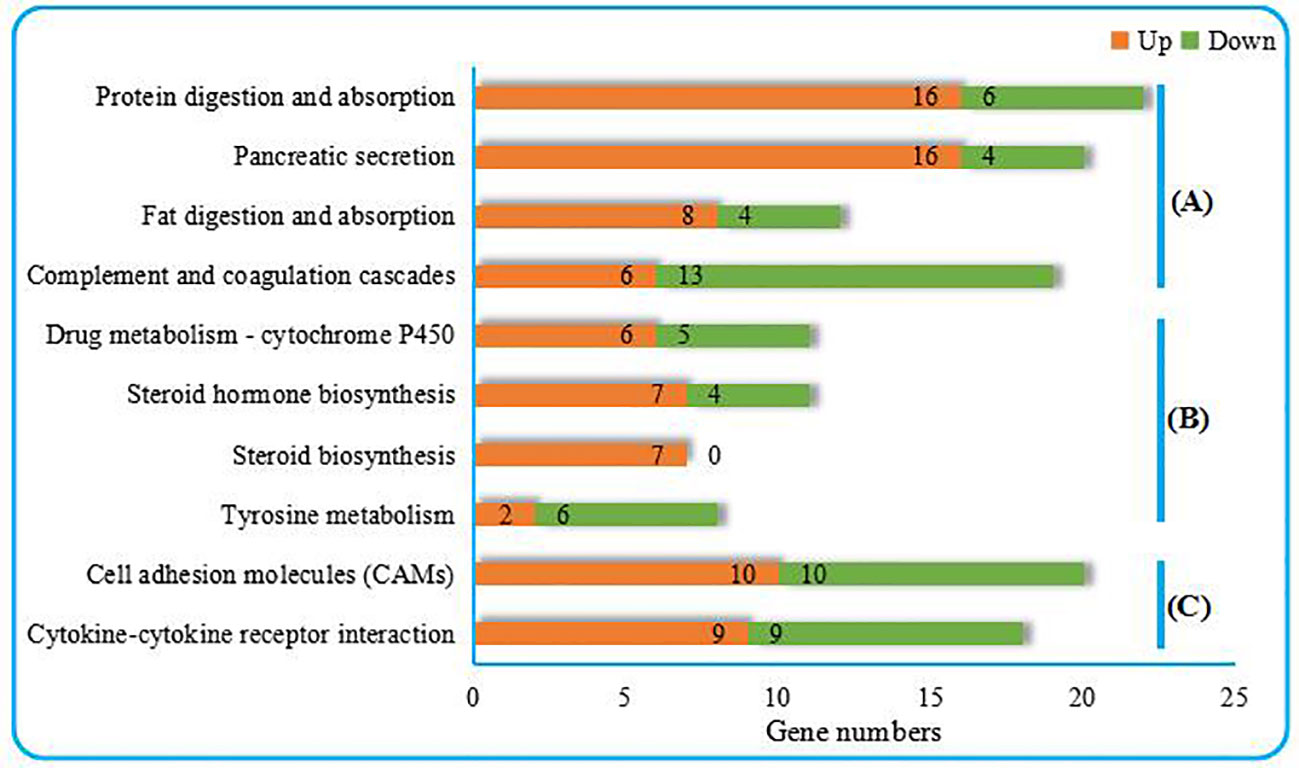
Figure 2 Top 10 KEGG pathways enriched from the liver transcriptome of C. nasus under starvation stress. (A) Organismal systems, (B) Metabolism, (C) Environmental information processing. Numbers marked by orange bars and green bars indicate the numbers of upregulated and downregulated DEGs.
Functional Analysis of Key DEGs
The key DEGs in the top 10 BP and top 10 KEGG pathways were comprehensively analyzed, and they were found to mainly involve the energy-substance metabolism and immune response regulation (Table 2).
Validation of RNA-Seq Data by qPCR
Ten DEGs were randomly selected from the RNA-Seq data. qPCR was performed using the primers shown in Appendix A1. As shown in Figure 3, the directions of changes in the detected DEGs were concordant between the RNA-Seq and qPCR analyses.
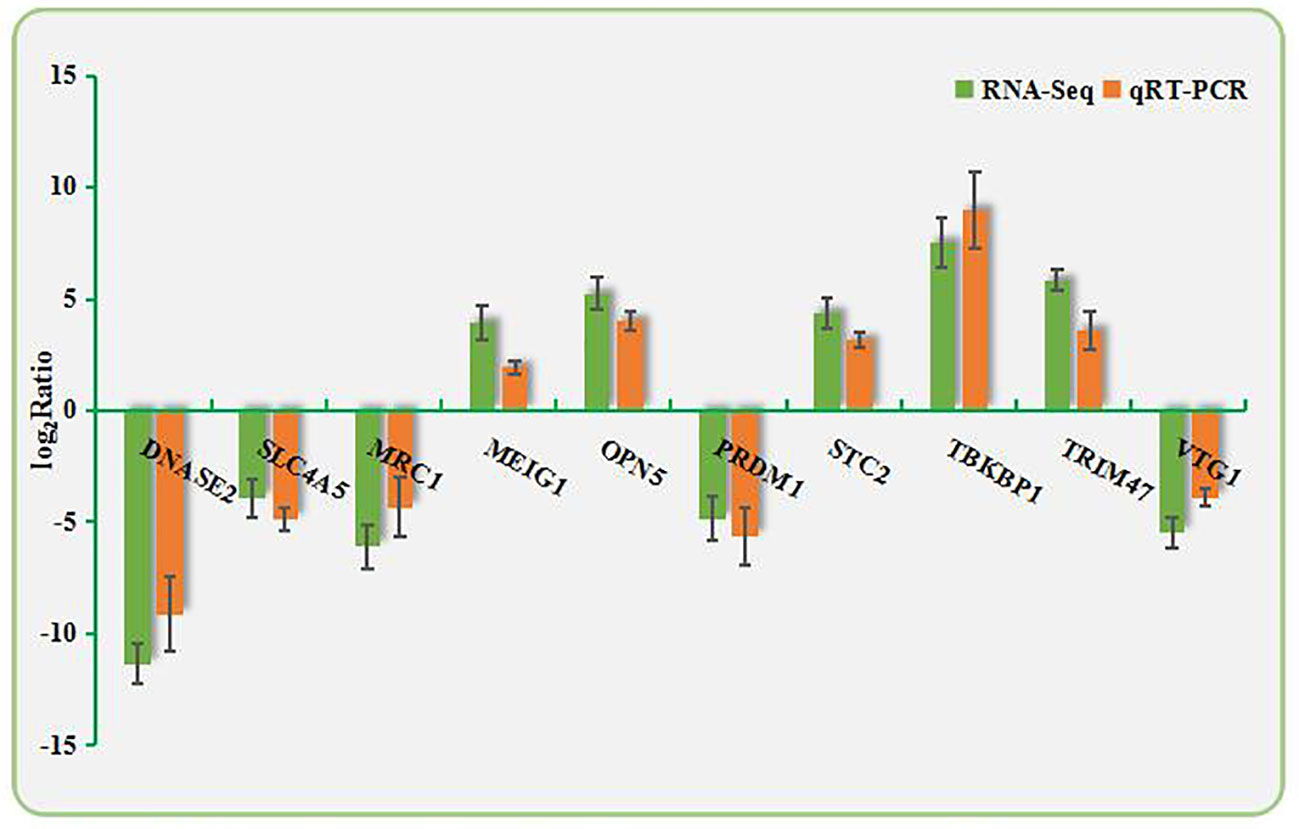
Figure 3 Validation of RNA-Seq data by qPCR. The X-axis shows the names of the assayed gene; the Y-axis is the relative expression level, expressed as log2 (fold change) in gene expression.
Discussion
The top 10 BPs, top 10 KEGG pathways, and the related DEGs were comprehensively analyzed, and the results show that they are mainly involved in the energy-substance metabolism and immune response regulation. The results related to the energy-substance metabolism mainly involved lipid catabolism and transportation, hormone regulation, and antioxidant regulation, and those involving the immune response were mainly related to immune response regulation and antitumor regulation.
Energy-Substance Metabolism
Enhancement of Lipid Catabolism
In this study, many DEGs related to lipid catabolism were upregulated after starvation stress, including bile salt-activated lipase (CEL), ATP-binding cassette subfamily G member 1 (ABCG1), and ATP-binding cassette subfamily G member 5 (ABCG5). CEL, a lipolytic enzyme synthesized and secreted by the pancreas, is conducive to the decomposition and digestion of fat and also participates in the lipoprotein metabolism. CEL can also promote the formation of capillary networks, cell proliferation, cell migration, tissue damage repair, etc. (34, 35).
In this study, CEL was upregulated after starvation stress, thereby, increasing the energy supply and maintaining the energy metabolism balance. ABCG1, which catalyzes the efflux of phospholipids, such as sphingolipids and cholesterol, participates in intracellular lipid transportation (36). ABCG5, which mediates the transport of Mg2+ and ATP-dependent sterols through the cell membrane, plays an important role in the selective transport of cholesterol into and out of intestinal epithelial cells (37). The upregulation of these DEGs promotes the catabolism of fat and lipid transport for energy supply during the starvation period.
The research performed by Lin et al. (38) indicated that feed restriction caused upregulation of the lipid catabolism in the liver of green sturgeon (Acipenser medirostris), and they obtained similar results to this study. The research carried out by Escalante-Rojas et al. (39) showed downregulation of the lipid reserves in the liver of rose spotted snappers (Lutjanus guttatus) under starvation conditions. The research performed by Qian et al. (15) also indicated that many regulatory genes related to lipid catabolism in the liver of large yellow croakers (Larimichthys crocea) were upregulated, which is beneficial for the reallocation of lipid reserves.
Enhancement of Detoxication
In this study, some DEGs relevant to detoxification were significantly upregulated in the liver of C. nasus after starvation stress, including diacylglycerol o-acyltransferase 1 (DGAT1), UDP-glucuronosyltransferase 1A5 (UGT1A5), UDP-glucuronosyltransferase 2A1 (UGT2A1), UDP-glucuronosyltransferase 2B31 (UGT2B31), and UDP-glucuronosyltransferase 2C1 (UGT2C1). DGAT1 plays an important regulatory role in maintaining the homeostasis of retinol (namely, vitamin A) and preventing retinol poisoning (40). UDP-glucuronide transferase (UGT) can catalyze the binding of lipophilic substrates with glucuronic acid to increase the water solubility of metabolites, thus, promoting excretion into the urine or bile. UGT is essential for the elimination and detoxification of drugs, invading organisms, and endogenous compounds (41, 42). In this study, many UGTs, such as UGT1A5, UGT2A1, UGT2B31, and UGT2C1, were upregulated after starvation. They play an important role in binding and subsequently removing potentially toxic exogenous and endogenous compounds. The research conducted by Xu et al. (43) demonstrated that starvation can lead to the upregulation of UDP-glucuronosyltransferase in the liver of mice, which plays an important role in modulating the hormones level and the elimination of harmful substances.
Enhancement of Hormone Regulation
Here, some hormone regulation-relevant DEGs were upregulated, such as catechol o-methyltransferase (COMT), cholesterol 7-alpha-monooxygenase (CYP7A1), 3-beta-hydroxysteroid-delta (8) delta(7)-isomerase (EBP), 7-dehydrocholesterol reductase (DHCR7), lanosterol synthase (LSS), lysosomal acid lipase/cholesteryl ester hydrolase (LIPA), and squalene monooxygenase (SQLE). COMT is beneficial for inactivating the neurotransmitter function of catecholamine, thus, slowing down the metabolism and conserving energy (44).
COMT upregulation is beneficial for the maintenance of energy homeostasis during the starvation period. CYP7A1 is a rate-determining enzyme for bile acid synthesis that is exclusively expressed in the liver, and it can increase the hydrolysis of lipids. Bile acid can save energy and improve its utilization (45). According to this study, its upregulation may be beneficial for energy homeostasis maintenance during starvation stress.
EBP, DHCR7, and LSS are key enzymes in the cholesterol biosynthesis pathway (46–48). LIPA can catalyze the deacylation of the cholesterol core ester to generate free fatty acids and cholesterol (49). SQLE is a rate-limiting enzyme in steride biosynthesis and an important enzyme for catalyzing the formation of cholesterol precursor molecules (50). Free fatty acids and cholesterol are also important energy-supplying substances. According to this study, the upregulation of these DEGs may enhance the energy supply of C. nasus during starvation stress.
Weakening of Water and Salt Metabolism and Antioxidation
Corticosteroid 11-beta-dehydrogenase isozyme 2 (HSD11B2) can catalyze the conversion of cortisol to inactive cortisone and protect nonselective mineralocorticoid receptors from glucocorticoid occupation. HSD11B2 plays a direct regulatory role in the water and salt metabolism in the kidney and skin to maintain blood pressure balance (51). To some extent, its downregulation reflects the downregulation of water and salt metabolism induced by starvation in this study. The results of the research carried out by Lin et al. (38) indicated that restricted feeding caused downregulation of the osmolality in the liver of green sturgeons (Acipenser medirostris).
Acetaldehyde dehydrogenase is the most important enzymatic aldehyde metabolism system in cells. It plays an important role in the substance metabolism, embryo formation, cell proliferation, and differentiation (52). In the presence of NAD(P)+, aldehyde dehydrogenase family 1 member A3 (ALDH1A3) oxidizes acetaldehyde to acetic acid, thereby, participating in the detoxification of aldehydes (53, 54). Aldehyde dehydrogenase family of 3 member B1 (ALDH3B1) can oxidize medium- and long-chain saturated and unsaturated aldehydes and has a strong antioxidant function, allowing it to protect against cytotoxicity caused by lipid peroxidation (55).
Vitamin E, also known as tocopherol, has strong antioxidant functions and plays an important regulatory role in intracellular signal transduction (56). The necessary enzyme for the first step of tocopherol synthesis is 4-hydroxyphenylpyruvate dioxygenase (HPD) (57). Here, the downregulation of the above DEGs hindered the synthesis of vitamin E and other antioxidants and weakened the antioxidant capacity of C. nasus.
A previous study on the liver of zebrafish under starvation stress indicated that starvation caused weakening of the antioxidant capacity of the liver, and ALDH3B1 and HPD were also downregulated (17). Similar conclusions were also obtained in a study on the influence of starvation on the liver of rainbow trout (18), which reflects the significant effect of starvation on the antioxidant system of fish. The research conducted by Varju et al. (58) indicated that excessive starvation caused an impaired antioxidant response in liver of pikeperch (Sander lucioperca L.), and they drew similar conclusions to this research.
Regulation of the Immune Response
Improvement in Anti-Allergy and Anticancer Regulation
Bradykinin can expand blood vessels, increase blood vessel permeability, induce bronchospasm, and promote bronchial contraction and other allergic reactions by binding to bradykinin receptors (BDKRB2) on the surfaces of target cells (59). In this study, bradykinin downregulation was beneficial for decreasing the production of allergic reactions, which is of positive significance to C. nasus. Clusterin (CLU), as an extracellular partner, can block the aggregation of nonnatural proteins, stabilize mitochondrial membrane integrity, and thus inhibit stress-induced apoptosis.
CLU plays an important role in the clearance of immunocomplexes during cell damage (60). In this study, its upregulation was beneficial to the timely clearance of immune complexes after cell injury. Cadherin-2 (CDH2), a calcian-dependent cellular adhesion protein, preferentially mediates intercellular adhesion of homotypic cells, inhibits cell migration, and thus effectively inhibits tumor invasion and metastasis (61). Antitumor studies on human beings (Homo sapiens) demonstrated that starvation may be an effective method in tumor treatment (62, 63). This study also derived the same conclusion.
Leucine-rich repeat-containing protein 4C (LRRC4C) is involved in the inhibition of glioma, induction of the antitumor immune response, and activation of natural killer cells in the destruction of tumor cells (64). Here, LRRC4C upregulation indicated an enhanced antitumor ability (65). On the other hand, many DEGs related to tumor development, such as hepatocyte growth factor (HGF) and hepatocyte growth factor receptor (MET), were downregulated. Upon binding to its receptor MET, HGF can promote the invasion and metastasis of tumor cells (66). In this study, the downregulation of these DEGs implies the weakening of carcinogenicity.
Weakening of the Immune Response
Here, the regulation of the immune response in C. nasus after starvation was weakened. Complement 3 (C3) plays a central role in the activation of the complement system. C3 plays an important role both in classical and bypass activation pathways of complement (67). Complement 6 (C6) plays an important role in cytolysis, the inflammatory response, and immune regulation (68). In a previous study on rainbow trout, C3 was also significantly downregulated after starvation stress, which is consistent with the results of this study (18). Together, these findings demonstrate that starvation can cause the weakening of complement function and, thereby, weaken the immune responses of fish.
In addition, we found some lectins to be downregulated after starvation stress. Integrin alpha-M (ITGAM) participates in various adhesion interactions among monocytes, macrophages, and granulocytes and mediates the uptake of pathogens. It shares a fragment with the C3 receptor and can interact with C3 to jointly mediate the immune response (69). Integrin beta-2 (ITGB2), contributing to natural killer cytotoxicity, participates in leukocyte adhesion and transport, including T cells and neutrophils, and mediates the immune response (70). Integrin alpha-6 (ITGA6), which is a receptor for platelet laminin, plays an important regulatory role in the signal transduction of insulin-like growth factor.
Insulin-like growth factor plays an important regulatory role in the proliferation and function of thymus and immune-competent cells (71). Therefore, ITGA6 also plays an important regulatory role in the immune response. Von Willebrand factor (VWF) plays an important role in hemostasis. VWF is a partner of coagulation factor VIII and can deliver it to the site of injury, stabilize its heterodimeric structure, and protect it from being removed prematurely (72).
Here, its downregulation indicated, in a sense, a decline in coagulation ability. B-cell receptor CD22 (CD22) can mediate the interaction between B cells, participate in the localization of B cells in lymphoid tissues, and play an important regulatory role in signal transduction between antigen and receptor during the immune response (73). Research on the influence of starvation on zebrafish liver also indicated the downregulation of CD22 (17), which is consistent with the results of this study.
Alpha-2-macroglobulin-like protein 1 (A2ML1) affects the activity of proteolytic enzymes, such as chymotrypsin and papain, through direct binding. As a result, the activity of certain proteases can be selectively protected to maintain homeostasis, which is also beneficial for the immune response (74). In this study, its downregulation weakened the protective effect on proteolytic enzymes and was not conducive to the maintenance of homeostasis.
Interleukin-6 receptor subunit beta (IL6ST), which is secreted by macrophages and contributes to cytokine effects, can stimulate and participate in the proliferation and differentiation of immune cells. IL6ST also affects the release of cortisol and plays a regulatory role in homeostasis (75). In this study, its downregulation was associated with a decrease in homeostasis regulation. Stromal cell-derived factor 1 (CXCL12), as a chemokine, performs a regulatory role in the chemotaxis of T cells and monocytes in the immune response (76).
Here, its downregulation reflected a decline in the regulation of the immune response. Tumor necrosis factor receptor superfamily member 14 (TNFRSF14), as a receptor of TNF superfamily members, as well as lymphotoxin and immunoglobulin superfamily members can promote the survival and differentiation of immune cells and the proliferation of T cells. TNFRSF14 plays a leading role in the adaptive immune response and provides a survival signal to effector T cells (77). In this study, its downregulation in the liver of C. nasus after starvation is consistent with the results obtained in previous research on the effect of starvation on the liver of Atlantic salmon (16).
This Study and the Influence of Environmental Pollution and Climate Change on Wild Fish and Aquaculture
Environmental pollution and climate change have had adverse effects on the ecological environment of natural waters including oceans, lakes, and rivers. Climate change effects, such as El Nino, have resulted in abnormal temperature rises, substantial reductions in plankton, mass migrations of fish, and changed distributions of fish. New pathogens are brought to indigenous fish with the invasion of fish. In addition, temperature rises cause a lack of food, which affects the growth of fish, resulting in a decline in fertility and even death (78, 79).
The results of this study indicated that lipid catabolism was upregulated in the liver of C. nasus under starvation, and the antioxidant capability and immunity were downregulated. These adverse physiological alterations in fish were consistent with the unfavorable effects of climate change that lead to declines of the fish population in natural waters and decreases in the aquatic biodiversity. Similarly, climate change also affects the ecological environment of aquaculture ponds and influences the growth, survival, and reproduction of fish species (79). Therefore, to better cope with climate change, marine protection areas should be established to control pollution and overfishing, which will be beneficial for the restoration of the fish population and enhancement of resilience of the waters to climate change.
Water corridors can be established in the Yangtze River, and conservation areas can be networked to help fish find suitable habitats more safely under variable water temperatures and chemical environments. On the other hand, scientific research on selective breeding should be further intensified. Individuals with good characteristics, including fast growth and strong resistance (strong resistance to diseases and extreme weather), should be selected out. Transgenic techniques can be further improved and utilized to obtain genetically improved new varieties and, finally, to improve the germplasm of aquatic organisms in natural waters and aquaculture ponds to better cope with environmental pollution and climate change.
Conclusions
We performed a comparative transcriptome analysis of the livers of near-death C. nasus after long-term starvation stress. The results showed that the main damaging effects involved the decreased regulation of water and salt metabolism, decreased antioxidant capacity, and the weakening of the immune response. The positive responses in C. nasus were the upregulation of certain detoxification regulatory genes and, more importantly, the upregulation of anti-allergy and anti-cancer regulatory genes as well as the downregulation of DEGs related to tumor development.
This research on C. nasus supports the important function of starvation in cancer inhibition. The results of this study revealed key regulatory pathways, DEGs, and the mechanisms leading to damage of the liver in C. nasus caused by starvation, which contributes essential information for further study of the energy metabolism mechanism of C. nasus as well as provides a theoretical reference for starvation metabolism research in other fish species and even human beings.
Data Availability Statement
The data sets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of the Freshwater Fisheries Research Center at the Chinese Academy of Fishery Sciences.
Author Contributions
GX supervised the research. MW designed, performed experiments, analyzed the transcriptomic data, and wrote the manuscript. ZZ and YT assisted the sampling. SS and YW made contritions to the revised manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Key R & D Program of China (2019YFD0901203), the National Natural Science Foundations of China (31672643), the Central Public-Interest Scientific Institution Basal Research Fund, Freshwater Fisheries Research Center, CAFS (2018JBFR03), and the National Infrastructure of Fishery Germplasm Resources (2017DKA3047-003).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Mr. Cui and Mr. Yang for their assistance in sampling in the Yixing experimental base of the Freshwater Fisheries Research Center of the Chinese Academy of Fishery Sciences.
Supplementary Material
The Supplementary Material for this article online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.622315/full#supplementary-material
Abbreviations
C. nasus, Coilia nasus; KOG, clusters of orthologous groups for eukaryotic complete genomes; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes; BP, Biological process; CC, Cellular component; MF, Molecular function; qPCR, quantitative real-time polymerase chain reaction; CT, cycle threshold.
References
1. Li JH, Chen XC, Zhang XL, Huang ZJ, Xiao L, Huang LL, et al. Fish biodiversity conservation and restoration, Yangtze river basin, China, urgently needs ‘scientific’ and ‘ecological’ action. Water (2020) 12:3043. doi: 10.3390/w12113043
2. Gu HL, Feng YM, You HB, Fan CJ. Research development on resource survey and artificial culture of Coilia nasus (in Chinese). Jiangsu Agr Sci (2016) 44:265–7. doi: 10.15889/j.issn.1002-1302.2016.03.075
3. Zhang MY, Xu DP, Liu K, Shi WG. Studies on biological characteristics and change of resource of Coilia nasus Schlegel in the lower reaches of the Yangtze River (in Chinese). Resour Environ Yangtze Basin (2005) 14:694–8. doi: 10.1007/s11769-005-0033-7
4. Chen MM, Yu DP, Lian YX, Liu ZG. Population abundance and habitat preference of the Yangtze finless porpoise in the highest density section of the Yangtze River. Aquat Conserv (2020) 30:1088–97. doi: 10.1002/aqc.3299
5. Mao CZ, Jiao XM, Zhong JS, Hua WH, Zhang XY, Wu JX. Research progress on resource status and protection of Coilia nasus in Yangtze river estuary (in Chinese). J Huaihai Inst Tech (2015) 24:78–83. doi: 10.3969/j.issn.1672-6685.2015.03.018
6. Ma CY, Cheng QQ, Zhang QY. Development of 12 polymorphic microsatellite markers in Coilia ectenes Jordan and Seale, 1905 (Clupeiformes: Engraulidae) and cross-species amplification in Coilia mystus Linnaeus, 1758. Environ Biol Fish (2011) 91:243–9. doi: 10.1007/s10641-011-9766-6
7. Cheng Q, Han J. Morphological variations and discriminant analysis of two populations of Coilia ectenes (in Chinese). J Lake Sci (2004) 16:356–64.
8. Conde-Sieira M, Capelli V, Alvarez-Otero R, Diaz-Rua A, Velasco C, Comesana S, et al. Hypothalamic AMPK alpha 2 regulates liver energy metabolism in rainbow trout through vagal innervation. Am J Physiol-Reg I (2020) 318:R122–34. doi: 10.1152/ajpregu.00264.2019
9. Jiao S, Nie MM, Song HB, Xu DD, You F. Physiological responses to cold and starvation stresses in the liver of yellow drum (Nibea albiflora) revealed by LC-MS metabolomics. Sci Total Environ (2020) 715:136940. doi: 10.1016/j.scitotenv.2020.136940
10. Zheng JL, Zhu QL, Shen B, Zeng L, Zhu AY, Wu CW. Effects of starvation on lipid accumulation and antioxidant response in the right and left lobes of liver in large yellow croaker. Pseudosciaena Crocea Ecol Indic (2016) 66:269–74. doi: 10.1016/j.ecolind.2016.01.037
11. Mohapatra S, Chakraborty T, Shimizu S, Urasaki S, Matsubara T, Nagahama Y, et al. Starvation beneficially influences the liver physiology and nutrient metabolism in Edwardsiella tarda infected red sea bream (Pagrus major). Comp Biochem Phys A (2015) 189:1–10. doi: 10.1016/j.cbpa.2015.07.003
12. Olmez A, Bayir M, Wang CF, Bayir A. Effects of long-term starvation and refeeding on fatty acid metabolism-related gene expressions in the liver of zebrafish, Danio rerio. Turk J Vet Anim Sci (2015) 39:654–60. doi: 10.3906/vet-1507-54
13. Enyu YL, Shu-Chien AC. Proteomics analysis of mitochondrial extract from liver of female zebrafish undergoing starvation and refeeding. Aquacult Nutr (2011) 17:E413–23. doi: 10.1111/j.1365-2095.2010.00776.x
14. Morales AE, Perez-Jimenez A, Hidalgo MC, Abellan E, Cardenete G. Oxidative stress and antioxidant defenses after prolonged starvation in Dentex dentex liver. Comp Biochem Phys C (2004) 139:153–61. doi: 10.1016/j.cca.2004.10.008
15. Qian B, Xue L, Huang H. Liver transcriptome analysis of the large yellow croaker (Larimichthys crocea) during fasting by using RNASeq. PLoS One (2016) 1:e0150240. doi: 10.1371/journal.pone.0150240
16. Martin SAMS, Douglas A, Houlihan DF, Secombes CJ. Starvation alters the liver transcriptome of the innate immune response in Atlantic salmon (Salmo salar). BMC Genomics (2010) 11:418. doi: 10.1186/1471-2164-11-418
17. Drew RE, Rodnick KJ, Settles M, Wacyk J, Churchill E, Powell MS, et al. Effect of starvation on transcriptomes of brain and liver in adult female zebrafish (Danio rerio). Physiol Genomics (2008) 35:283–95. doi: 10.1152/physiolgenomics.90213.2008
18. Salem M, Silverstein J, Rexroad III C.E., Yao JB. Effect of starvation on global gene expression and proteolysis in rainbow trout (Oncorhynchus mykiss). BMC Genomics (2007) 8:328. doi: 10.1186/1471-2164-8-328
19. Jin X, Xu GC, Du FK, Xu P, Gu R,B. Impacts of starvation stress on morphological change, chemical composition and blood biochemical parameters in Coilia nasus. Chin J Zool (2014) 49:897–903. doi: 10.13859/j.cjz.201406013
20. Wang MY, Tang YK, Yu JH, Su SY, Li JL, Yu F, et al. Molecular insights into the sex-differential regulation of signal transduction in the cerebral ganglion and metabolism in the hepatopancreas of Eriocheir sinensis during reproduction. Genomics (2020) 112:71–81. doi: 10.1016/j.ygeno.2019.10.014
21. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J (2011) 17:1. doi: 10.14806/ej.17.1.200
22. Grabherr M, Haas B, Yassour M, Levin J, Thompson D, Amit I, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol (2013) 29:644–52. doi: 10.1038/nbt.1883
23. Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
24. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res (2008) 36:D480–484. doi: 10.1093/nar/gkm882
25. Conesa A, Gotz S, Garcia J. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics (2005) 21:3674–6. doi: 10.1093/bioinformatics/bti610
26. Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods (2012) 9:357–9. doi: 10.1038/NMETH.1923
27. Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods (2013) 10:71–3. doi: 10.1038/NMETH.2251
28. Trapnell C, Williams B, Pertea G, Mortazavi A, Kwan G, Van B, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol (2010) 28:511–5. doi: 10.1038/nbt.1621
29. Anders S, Huber W. Differential expression of RNA-Seq data at the gene level-the DESeq package. EMBL (2013) 22:1–24.
30. Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol (2005) 11:R14. doi: 10.1186/gb-2010-11-2-r14
31. Mao X, Cai T, Olyarchuk JG, Wei LP. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics (2005) 21:3787–93. doi: 10.1093/bioinformatics/bti430
32. DeCaria JE, Montero-Odasso M, Wolfe D, Chesworth BM, Petrella RJ. The effect of intra-articular hyaluronic acid treatment on gait velocity in older knee osteoarthritis patients: A randomized, controlled study. Arch Gerontol Geriat (2012) 55:310–5. doi: 10.1016/j.archger.2011.11.007
33. Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
34. Kurtovic I, Marshall SN, Zhao X. Hydrophobic immobilization of a bile salt activated lipase from Chinook salmon (Oncorhynchus tshawytscha). J Mol Catal B-Enzym (2011) 72:168–74. doi: 10.1016/j.molcatb.2011.06.001
35. Gjellesvik DR, Lombardo D, Walther BT. Pancreatic bile salt dependent lipase from cod (Gadus morhua): purification and properties. Biochim Et Biophys Acta (1992) 1124:123–34. doi: 10.1016/0005-2760(92)90088-D
36. Gu HM, Wang FQ, Zhang DW. Caveolin-1 interacts with ATP binding cassette transporter G1 (ABCG1) and regulates ABCG1-mediated cholesterol efflux. Biochim Biophys Acta (2014) 1841:847–58. doi: 10.1016/j.bbalip.2014.02.002
37. Schmitz G, Langmann T, Heimerl S. Role of ABCG1 and other ABCG family members in lipid metabolism. J Lipid Res (2001) 42:1513–20.
38. Lin CY, Huang LH, Deng DF, Lee SH, Liang HJ, Hung SSO. Metabolic adaptation to feed restriction on the green sturgeon (Acipenser medirostris) fingerlings. Sci Total Environ (2019) 684:78–88. doi: 10.1016/j.scitotenv.2019.05.044
39. Escalante-Rojas M, Martínez-Brown JM, Ibarra-Castro L, Llera-Herrera R, García-Gasca A. Effects of feed restriction on growth performance, lipid mobilization, and gene expression in rose spotted snapper (Lutjanus guttatus). J Comp Physiol B (2020) 190:275–86. doi: 10.1007/s00360-020-01268-3
40. Ma Z, Onorato JM, Chen L, Nelson DW, Yen CE, Cheng D. Synthesis of neutral ether lipid monoalkyl-diacylglycerol by lipid acyltransferases. J Lipid Res (2017) 58:1091–9. doi: 10.1194/jlr.M073445
41. Alonen A, Finel M, Kostiainen R. The human UDP-glucuronosyltransferase UGT1A3 is highly selective towards N2 in the tetrazole ring of losartan, candesartan, and zolarsartan. Biochem Pharmacol (2008) 76:763–72. doi: 10.1016/j.bcp.2008.07.006
42. Sneitz N, Vahermo M, Mosorin J, Laakkonen L, Poirier D, Finel M. Regiospecificity and stereospecificity of human UDP-glucuronosyltransferases in the glucuronidation of estriol, 16-epiestriol, 17-epiestriol, and 13-epiestradiol. Drug Metab Dispos (2013) 41:582–91. doi: 10.1124/dmd.112.049072
43. Xu JL, Kulkarni SR, Li LY, Slitt AL. UDP-glucuronosyltransferase expression in mouse liver is increased in obesity- and fasting-induced steatosis. Drug Metab Dispos (2012) 40:259–66. doi: 10.1124/dmd.111.039925
44. Palma PN, Rodrigues ML, Archer M, Bonifacio MJ, Loureiro AI, Learmonth DA, et al. Comparative study of ortho- and meta-nitrated inhibitors of catechol-O-methyltransferase: interactions with the active site and regioselectivity of O-methylation. Mol Pharmacol (2006) 70:143–53. doi: 10.1124/mol.106.023119
45. Cheema SK, Agellon LB. The muirne and human cholesteorl 7 alpha hydorxylase gene promoters are differentially responsive to regulation by fatty acids mediated via peroxisome proliferator activated receptor alpha. J Biol Chem (2000) 275:12530–6. doi: 10.1074/jbc.275.17.12530
46. Moebius FF, Fitzky BU, Wietzorrek G, Haidekker A, Eder A, Glossmann H. Cloning of an emopamil-binding protein (EBP)-like protein that lacks sterol delta 8-delta7 isomerase activity. Biochem J (2003) 374:229–37. doi: 10.1042/BJ20030465
47. Luu W, Hart-Smith G, Sharpe LJ, Brown AJ. The terminal enzymes of cholesterol synthesis, DHCR24 and DHCR7, interact physically and functionally. J Lipid Res (2015) 56:888–97. doi: 10.1194/jlr.M056986
48. Zhao L, Chen XJ, Zhu J, Xi YB, Yang X, Hu LD, et al. Lanosterol reverses protein aggregation in cataracts. Nature (2015) 523:607–11. doi: 10.1038/nature14650
49. Zschenker O, Oezden D, Ameis D. Lysosomal acid lipase as a preproprotein. J Biochem (2004) 136:65–72. doi: 10.1093/jb/mvh093
50. Padyana AK, Gross S, Jin L, Cianchetta G, Narayanaswamy R, Wang F, et al. Structure and inhibition mechanism of the catalytic domain of human squalene epoxidase. Nat Commun (2019) 10:97. doi: 10.1038/s41467-018-07928-x
51. van Uum SHM, Hermu ARMM, Smits P, Thien T, Lenders JWM. The role of 11 beta-hydroxysteroid dehydrogenase in the pathogenesis of hypertension. Cardiovasc Res (1998) 38:16–24. doi: 10.1016/S0008-6363(97)00299-X
52. Marcato P, Dean CA, Giacomantonio CA, Lee PWK. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle (2011) 10:1378–84. doi: 10.4161/cc.10.9.15486
53. Marcato P, Dean CA, Giacomantonio CA, Lee PWK. Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD(+) and retinoic acid. Sci Rep (2016) 6:35710. doi: 10.1038/srep35710
54. Outhwaite JE, Natale BV, Natale DRC, Simmons DG. Expression of aldehyde dehydrogenase family 1, member A3 in glycogen trophoblast cells of the murine placenta. Placenta (2015) 36:304–11. doi: 10.1016/j.placenta.2014.12.002
55. Kitamura T, Naganuma T, Abe K, Nakahara K, Ohno Y, Kihara A. Substrate specificity, plasma membrane localization, and lipid modification of the aldehyde dehydrogenase ALDH3B1. Biochim Biophys Acta (2013) 1831:1395–401. doi: 10.1016/j.bbalip.2013.05.007
56. Liang WK, Zhang WW, Lv ZM, Li CH. 4-Hydroxyphenylpyruvate dioxygenase from sea cucumber Apostichopus japonicus negatively regulates reactive oxygen species production. Fish Shellfish Immun (2020) 101:261–8. doi: 10.1016/j.fsi.2020.04.013
57. Munné-Bosch S, Alegre L. The function of tocopherols and tocotrienols in plant. Crit Rev Plant Sci (2002) 21:31–57. doi: 10.1080/0735-260291044179
58. Varju M, Müller T, Bokor Z, Zarski D, Mézes M, Balogh K. The effects of excessive starvation on antioxidant defence and lipid peroxidation in intensively reared, commercial-size pikeperch (Sander lucioperca L.). Egyptian J Aquat Res (2018) 44:349–52. doi: 10.1016/j.ejar.2018.11.003
59. Maurer M, Bader M, Bas M, Bossi F, Cicardi M, Cugno M, et al. New topics in bradykinin research. Allergy (2011) 66:1397–406. doi: 10.1111/j.1398-9995.2011.02686.x
60. Jeong YM, Jin TE, Choi JH, Lee MS, Kim HT, Hwang KS, et al. Induction of clusterin Expression by neuronal cell death in zebrafish J. Genet. Genomics (2014) 41:583–9. doi: 10.1016/j.jgg.2014.08.007
61. Rebman JK, Kirchoff KE, Walsh GS. Cadherin-2 Is Required Cell Autonomously for Collective Migration of Facial Branchiomotor Neurons. PLoS One (2016) 11:e0164433. doi: 10.1371/journal.pone.0164433
62. Xie W, Deng WW, Zan MH, Rao L, Yu GT, Zhu DM, et al. Cancer Cell Membrane Camouflaged Nanoparticles to Realize Starvation Therapy Together with Checkpoint Blockade for Enhancing Cancer Therapy. ACS Nano (2019) 13:2849–57. doi: 10.1021/acsnano.8b03788
63. Yang BW, Ding L, Chen Y, Shi JL. Augmenting Tumor-Starvation Therapy by Cancer Cell Autophagy Inhibition. Adv Sci (2020) 7:1902847. doi: 10.1002/advs.201902847
64. Lin J, Ho WH, Gurney AL, Rosenthal A. The netrin-G1 ligand NGL-1 promotes the outgrowth of thalamocortical axons. Nat Neurosci (2013) 6:1270–6. doi: 10.1890/03-5087
65. Wang L, Fraser CC, Kikly K, Wells AD, Han R, Coyle AJ, et al. B7-H3 promotes acute and chronic allograft rejection. Eur J Immunol (2005) 35:428–38. doi: 10.1002/eji.200425518
66. Matsumoto K, Umitsu M, Dinuka M, Silva D, Roy A, Bottaro DP. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci (2017) 108:296–307. doi: 10.1111/cas.13156
67. Xiong ZQ, Qian WH, Suzuki K, McNamara JO. Formation of complement membrane attack complex in mammalian cerebral cortex evokes seizures and neurodegeneration. J Neurosci (2003) 23:955–60. doi: 10.1152/jn.00779.2002
68. Pilzer D, Gasser O, Moskovich O, Schifferli JA, Fishelson Z. Emission of membrane vesicles: roles in complement resistance, immunity and cancer. Springer Semin Immunopathol (2005) 27375–87. doi: 10.1007/s00281-005-0004-1
69. Bai M, Grieshaber-Bouyer R, Wang J, Schmider AB, Wilson ZS, Zeng L, et al. CD177 modulates human neutrophil migration through activation-mediated integrin and chemoreceptor regulation. Blood (2017) 130:2092–100. doi: 10.1182/blood-2017-03-768507
70. Swaim CD, Scott AF, Canadeo LA, Huibregtse JM. Extracellular ISG15 signals cytokine secretion through the LFA-1 integrin receptor. Mol Cell (2017) 68:581–90. doi: 10.1016/j.molcel.2017.10.003
71. Cedano Prieto DM, Cheng Y, Chang CC, Yu J, Takada YK, Takada Y. Direct integrin binding to insulin-like growth factor-2 through the C-domain is required for insulin-like growth factor receptor type 1 (IGF1R) signaling. PLoS One (2017) 12:E0184285–E0184285. doi: 10.1371/journal.pone.0184285
72. Ni H, Denis CV, Subbarao S, Degen JL, Sato TN, Hynes RO, et al. Persistence of platelet thrombus formation in arterioles of mice lacking both von Willebrand factor and fibrinogen. J Clin Invest (2000) 106:385–92. doi: 10.1172/JCI9896
73. Fernandes VE, Ercoli G, Benard A, Brandl C, Fahnenstiel H, Muller-Winkler J, et al. The B-cell inhibitory receptor CD22 is a major factor in host resistance to Streptococcus pneumoniae infection. PLoS Pathog (2020) 16:e1008464. doi: 10.1371/journal.ppat.1008464
74. Galliano MF, Toulza E, Gallinaro H, Jonca N, Ishida-Yamamoto A, Serre G, et al. A novel protease inhibitor of the alpha2-macroglobulin family expressed in the human epidermis. J Biol Chem (2006) 281:5780–9. doi: 10.1074/jbc.M508017200
75. Shahin T, Aschenbrenner D, Cagdas D, Bal SK, Conde CD, Garncarz W, et al. Selective loss of function variants in IL6ST cause Hyper-IgE syndrome with distinct impairments of T-cell phenotype and function. Haematologica (2019) 104:609–21. doi: 10.3324/haematol.2018.194233
76. Fujita M, Davari P, Takada YK, Takada Y. Stromal cell-derived factor-1 (CXCL12) activates integrins by direct binding to an allosteric ligand-binding site (site 2) of integrins without CXCR4. Biochem J (2018) 475:723–32. doi: 10.1042/BCJ20170867
77. Sedy JR, Bjordahl RL, Bekiaris V, Macauley MG, Ware BC, Norris PS. CD160 activation by herpesvirus entry mediator augments inflammatory cytokine production and cytolytic function by NK cells. J Immunol (2013) 191:828–36. doi: 10.4049/jimmunol.1300894
78. Van Putten I, Metcalf S, Frusher S, Marshall N, Tull M. Fishing for the impacts of climate change in the marine sector: a case study. Int J Clim Change Str (2014) 6:421–41. doi: 10.1108/IJCCSM-01-2013-0002
Keywords: Coilia nasus, liver, comparative transcriptome, starvation, immune response
Citation: Wang M, Xu G, Tang Y, Su S, Wang Y and Zhu Z (2021) Investigation of the Molecular Mechanisms of Antioxidant Damage and Immune Response Downregulation in Liver of Coilia nasus Under Starvation Stress. Front. Endocrinol. 12:622315. doi: 10.3389/fendo.2021.622315
Received: 28 October 2020; Accepted: 08 January 2021;
Published: 26 February 2021.
Edited by:
Oliana Carnevali, Marche Polytechnic University, ItalyReviewed by:
Takashi Yada, Japan Fisheries Research and Education Agency (FRA), JapanElisabeth Eppler, University of Bern, Switzerland
Hui Zhang, South China Agricultural University, China
Copyright © 2021 Wang, Xu, Tang, Su, Wang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gangchun Xu, eHVnY0BmZnJjLmNu; Yongkai Tang, dGFuZ3lrQGZmcmMuY24=
 Meiyao Wang
Meiyao Wang Gangchun Xu1,2*
Gangchun Xu1,2*