- 1Key Laboratory of Freshwater Fisheries and Germplasm Resources Utilization, Ministry of Agriculture, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
- 2Wuxi Fisheries College, Nanjing Agricultural University, Wuxi, China
Legumain, also called aspartic endopeptidase (AEP), is a member of the cysteine protease family and is involved in various physiological processes. In this study, we analyzed the characteristics of a novel legumain-like (named Mn-Lel) in the female oriental river prawn, Macrobrachium nipponense, which is involved in ovary maturation. The Mn-Lel is 1,454 bp in length, including a 1,290-bp open reading frame that encodes 430 amino acids. qPCR analysis indicated that Mn-Lel is specifically highly expressed in the hepatopancreas and ovaries of female prawns. It is rarely expressed in embryogenesis, weakly expressed in early larval development stages, and then significantly increased after metamorphosis, which indicated that Mn-Lel is not a maternal gene and mainly plays a role in adults. During the different ovarian stages, Mn-Lel expression in the hepatopancreas had no obvious rules, while its expression in the ovaries had a significant peak in stage III. In situ hybridization studies revealed that Mn-Lel is localized in the oocyte of the ovary. Changes in the gonadosomatic index confirmed the inhibitory effects of Mn-Lel dsRNA on ovary maturation. These results suggest that Mn-Lel has a key role in promoting ovary maturation.
Introduction
Proteolysis in nature is mainly performed by proteases. Protease can be divided into several types according to the corresponding substrates, such as carboxyl peptidase and endopeptidase (1). Endopeptidase, which cleaves from the inside of the peptide chain, includes serine proteases, cysteine proteases, aspartic proteases, threonine proteases, and metalloproteases, and is characterized by its catalytic active central group (2, 3).
Legumain, also called aspartic endopeptidase, is a newly discovered member of the cysteine protease family, first identified in asparagus bean and asparagus bean plants. Like other family members, legumain is mainly located in the lysosome of cells and is involved in various physiological processes such as the degradation of basal egg whites (4). It can specifically hydrolyze the asparagine residue carboxy-terminal peptide bond. In recent years, significant progress has been made in the study of the structure and function of legumain genes and protein in plants, parasites, and mammals (5, 6). Studies in mammals have shown that it is involved in the pathophysiological process of immunity, anti-bone dissolution, and the occurrence and development of tumors (7, 8). The high expression level of legumain is related to tumor differentiation, tumor initiation and development, and tumor invasion (9–12). However, few studies have investigated legumain in normal growth development and reproduction.
Macrobrachium nipponense, also known as the oriental river prawn, is an economically important freshwater species and has been widely farmed not only in China but also in many Asian countries (13). Its production exceeded 240,000 tons in China in 2020 (14). The sexual maturity period of M. nipponense is short. In the breeding season, the ovary’s maturation of prawns is accelerated with the increase of water temperature. It only takes approximately 45 days from hatching to sexual maturity to laying eggs when the water temperature exceeds 28°C (15). Rapid mass self-reproduction in the pond produces a large number of offspring, which leads to a reduction in the resilience and survival rate and an increase in hypoxia risk. High-density farming also affects the market size of prawns and then seriously affects the economic benefits of prawn farming. It has been a problem of prawn farming for many years. To address this, transcriptomes of five ovary stages of M. nipponense were constructed to detect the key pathways and genes that play important roles in ovary maturation (16). The lysosomal pathway was detected to be the most significant enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway in the vitellogenesis stages. A novel legumain-like gene in this pathway showed remarkably expression variation, suggesting that it might have specific roles in M. nipponense ovary maturation.
Therefore, the present study aimed to characterize the structure of this legumain-like gene of M. nipponense and study its expression pattern and subcellular localization. Furthermore, we investigate its function in ovarian maturation. The results of this study were expected to provide a theoretical basis for revealing the regulation mechanism of ovarian development and solving the problem of fast sexual ripening in the M. nipponense aquaculture industry.
Materials and Methods
Sample Collection of Macrobrachium nipponense and Tissue Preparation
No endangered or protected species were involved in this study, and all experimental protocols and methods were approved in July 2021 (Authorization No. 20210715002) by the Animal Care and Use Ethics Committee in the Freshwater Fisheries Research Center (Wuxi, China).
Samples of female M. nipponense (n = 30, BW ± SD: 0.72 ± 0.19 g) used in gene cloning and expression analysis were obtained from Dapu scientific experimental base in Freshwater Fisheries Research Center (Wuxi, China). All prawns were adult and healthy, which were kept in recirculating glass aquarium system under the same environment for at least 1 week. Seven tissues (muscles, M; heart, H; ovary, O; cerebral ganglion, Cg; eyestalks, E; hepatopancreas, He; and gills, G) were collected from adult female prawns (n = 5). Ovarian stage division was according to a previous study (17). The ovaries of living prawn can be distinguished of different stages by color: OI (undeveloped stage, transparent), OII (developing stage, yellow), OIII (nearly-ripe stage, light green), OIV (ripe stage, dark green), and OV (spent stage, gray). Both hepatopancreas and ovaries from 5 stages were collected (n = 5).
The embryos and larvae were sampled according to the criteria of previous studies (17–19). The embryonic stages (n = 20) comprised cleavage stage (CS), blastula stage (BS), gastrulation stage (GS), nauplius stage (NS), and zoea stage (ZS). The larval stages (n = 10) were as follows: L1 (the 1st day after hatching), L5 (the 5th day after hatching), L10 (the 10th day after hatching), L15 (the 15th day after hatching), PL1 (the 1st day after metamorphosis), PL5 (the 5th day after metamorphosis), PL10 (the 10th day after metamorphosis), PL15 (the 15th day after metamorphosis), and PL25 (the 25th day after metamorphosis). All tissues were stored immediately in the liquid nitrogen for further study.
RNA Isolation and cDNA Synthesis
Tissues weighing 0.1 g were used for total RNA isolation. RNAiso Plus Reagent (TaKaRa, Maebashi, Japan) was used for RNA extraction. Agarose electrophoresis (1.5% gel) was used to access RNA quality, and a NanoDrop 1000 was employed to measure the concentration of RNA. An RNA PCR Kit was used to synthesize cDNA (TaKaRa, Japan).
Legumain-Like Sequence Cloning, Phylogenetic, and Structural Analyses
The full-length legumain-like cDNA sequence was obtained from the five ovary stages transcriptomes of M. nipponense (NCBI accession: SAMN11603268–SAMN11603282, Bioproject PRJNA541783). In order to verify the open reading frame sequence (ORF) of legumain-like gene, a pair of primers Mn-Lel-F and Mn-Lel-R was designed (listed in Table 1). PCR products were detected on 1.5% agarose gel and then sent to the company for sequencing (Sangon Biotech Shanghai Co., Shanghai, China).
The legumain-like gene sequence was analyzed using BLAST programs (http://www.ncbi.nlm.nih.gov/BLAST/), and SignalP4.1 was applied in signal peptide predicting (http://www.cbs.dtu.dk/services/SignalP-3.0/). The legumain-like amino acid sequences were used to generate the phylogenetic trees with MEGA5.0 based on the neighbor-joining (NJ) method, and the bootstrapping replications were 1,000 (20, 21). Sequence alignment was built with DNAMAN 6.0 software (18). The known sequences of other animals were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov/). NetNGlyc-1.0 (https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0) was employed to predict potential sites of N-linked glycosylation.
Quantitative Real-Time PCR Analysis
Quantitative Real-Time PCR (qPCR) was used to quantify spatiotemporal mRNA expression levels of the legumain-like gene. EIF gene (eukaryotic translation initiation factor 5A) was used as the internal reference for both tissue distribution and development stage expression analysis (listed in Table 1) (22). The qPCR system was detailed in a previous study (23). The expression levels were calculated based on the 2− ΔΔCT method (24). SPSS 23.0 software was used to do statistical analyses, and one-way ANOVA and two-tailed t-test were used to analyze statistical differences. All data were confirmed to the homogeneity of variance and normal distribution. All qPCR data were described as mean ± SD, and a significant difference was indicated by p < 0.05.
In Situ Hybridization
The ovaries were dissected from mature female prawns and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS; pH 7.4) at 4°C overnight. In situ hybridization (ISH) study was performed on 4-μm-thick formalin-fixed paraffin-embedded sections using Zytofast PLUS CISH implementation kit (ZytoVision GmBH, Bremerhaven, Germany) (25). The blank control groups were routine H&E staining sections without probe poured, and the negative control groups had antisense probes poured. The mRNA locations of Mn-Lel were analyzed with sense probe poured over slides. The anti-sense and sense probes in this study with digoxin signal were designed by Primer5 software based on the cDNA sequence of Mn-Lel and synthesized by Shanghai Sangon Biotech Company (Shanghai, China) (Table 1). All slides were examined under a light microscope for evaluation.
RNA Interference
The RNA interference (RNAi) primers of legumain-like and green fluorescent protein (GFP) gene, which contained the T7 promoter, were designed for using Snap Dragon (http://www.flyrnai.org/cgibin/RNAifind_primers.pl) (listed in Table 1). The GFP gene was selected as a control (26). Transcript AidTMT7 High Yield Transcription kit (Fermentas, Inc., USA) was used to synthesize in vitro dsRNA of both legumain-like and GFP.
In order to calculate the change degree of gonadosomatic index (GSI) better and more clearly, the ovary development level of experimental prawns should be relatively consistent at the beginning of the experiment. At the beginning of the RNAi experiment, most females were in the ovary stage IV. Therefore, stage IV prawns were selected to ensure a sufficient number of experimental prawns. One hundred healthy female prawns (0.83 ± 0.11 g) in stage IV were randomly divided into an experimental group and a control group (N = 50). A 17-day RNAi experiment was carried out (water temperature 25°C ± 1°C). Both experimental and control groups were injected with 4 μg/g.b.w of ds-legumain-like and ds-GFP each, and the injection site is pericardial cavity membrane (27). Ds-RNA was injected every 5 days, and five prawns in each group were randomly sampled on the 1st, 9th, and 17th days. By silencing the legumain-like gene, interference efficiency in both hepatopancreas and ovary and the GSI (gonadal weight/body weight × 100%) were calculated. Ten prawns were randomly sampled from both groups on the 1st, 9th, and 17th days, and the body and ovary weight were recorded.
Results
Characterization and Structural Analysis of Mn-Lel
The full-length legumain-like cDNA sequence of M. nipponense was 1,454 bp, named as Mn-Lel (GenBank No. OM317599), including 19 and 145 bp of 5′ and 3′ untranslated region (UTR), respectively. It had a polyadenylation signal (AATAAA) and a poly(A) tail in the 3′-UTR, indicating the integrity of the Mn-Lel gene (Figure 1). The ORF of Mn-Lel was 1,290 bp and encoded 430 amino acids, and the estimated molecular mass was 48.05 kDa; the theoretical pI was 4.44. SignalP software analysis showed that Mn-Lel had a signal peptide with a length of 19 amino acid residues, and the predicted tangent point was between Ala19 and Asp20. The results of BLASTP showed that the 21st to 278th amino acid residues constituted a C13 superfamily domain of peptidase. There was a conserved TGD motif (Thr110-Gly111-Asp112) and His140 and Cys181 catalytic dyad in the C13 superfamily domain. Several highly conserved cysteine residues were located in the 212th, 371th, 383th, 403rd, and 420th. We found that there were no potential N-linked glycosylation sites in Mn-Lel based on NetNGlyc-1.0.
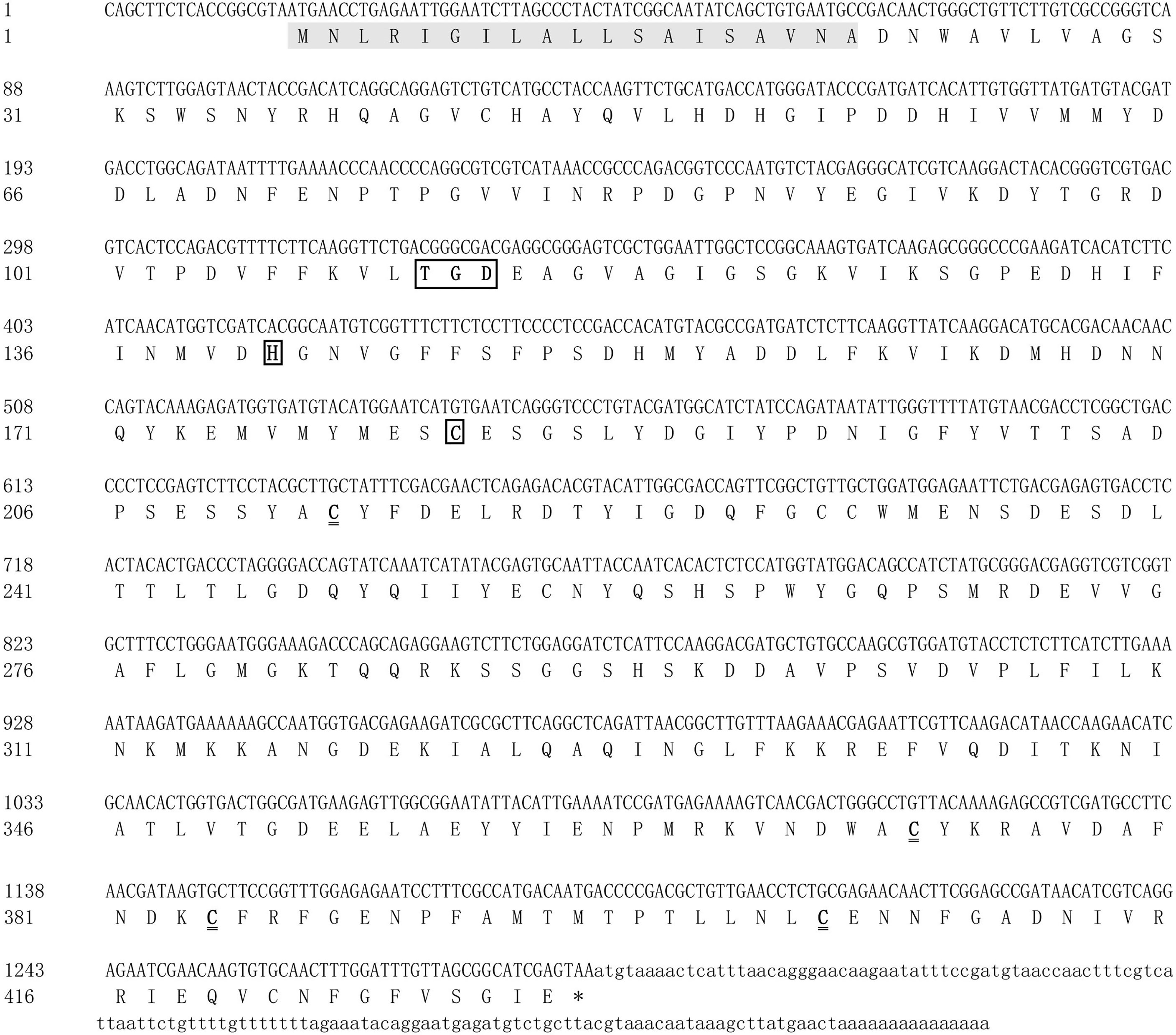
Figure 1 Nucleotide and deduced amino acid sequences of Mn-Lel in Macrobrachium nipponense. The shaded area indicates the signal peptide. The black underlined boxes represent the conserved TGD motif (Thr110-Gly111-Asp112) and His140 and Cys181 catalytic dyad in the C13 superfamily domain. The double underline represents highly conserved cysteine residues were located in the 212th, 371th, 383th, 403rd, and 420th. The asterisk (*) represents the termination codon.
Phylogenetic and Sequence Alignment Analysis of Mn-Lel
Several amino acid sequences of legumain and legumain-like in vertebrates and invertebrates were deduced, and a phylogenetic tree was built to elucidate the phylogenetic relationships based on the NJ method (Figure 2). The results showed that legumain and legumain-like were highly conserved in evolution; all legumain and legumain-like could be divided into two branches. The Mn-Lel and legumain-like protein of Macrobrachium rosenbergii (AJG06865.1) were isolated and clustered into one branch with other crustaceans. The crustacean legumains were then grouped with vertebrates and insects. The legumain from the parasite was isolated to be one branch.
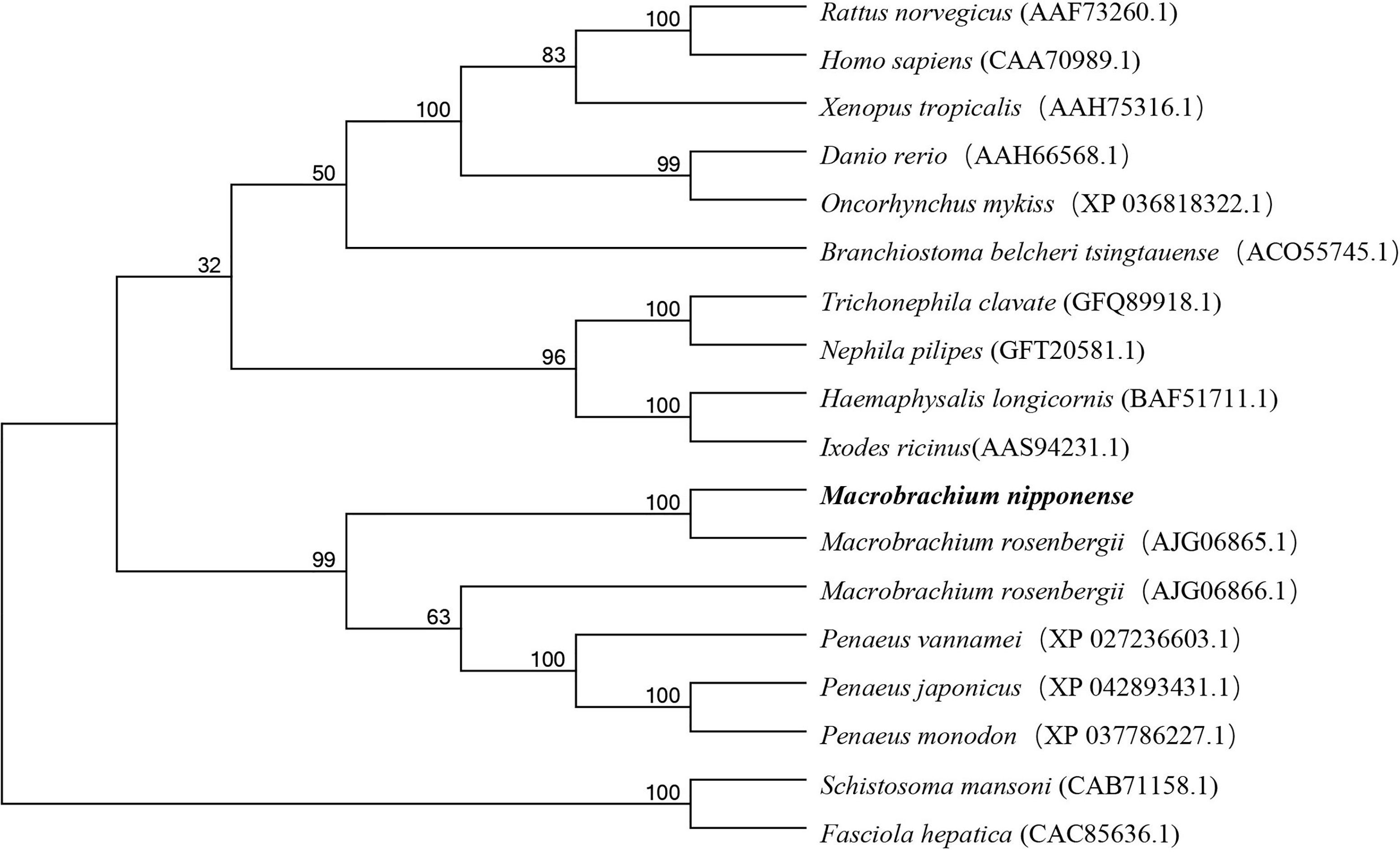
Figure 2 Phylogenetic tree of Mn-Lel amino acid sequence in different groups. The numbers below the node indicate the bootstrap value.
Multiple sequence alignment was performed using DNAMAN 6.0 to compare Mn-Lel gene with legumain and legumain-like of crustaceans and insects in Arthropoda, including M. rosenbergii, Penaeus japonicus, Penaeus monodon, Penaeus vannamei, Haemaphysalis longicornis, and Ixodes ricinus. The homology with these species was 86.06%, 39.60%, 40.71%, 41.15%, 41.81%, 39.96%, and 37.25%, respectively (Figure 3).
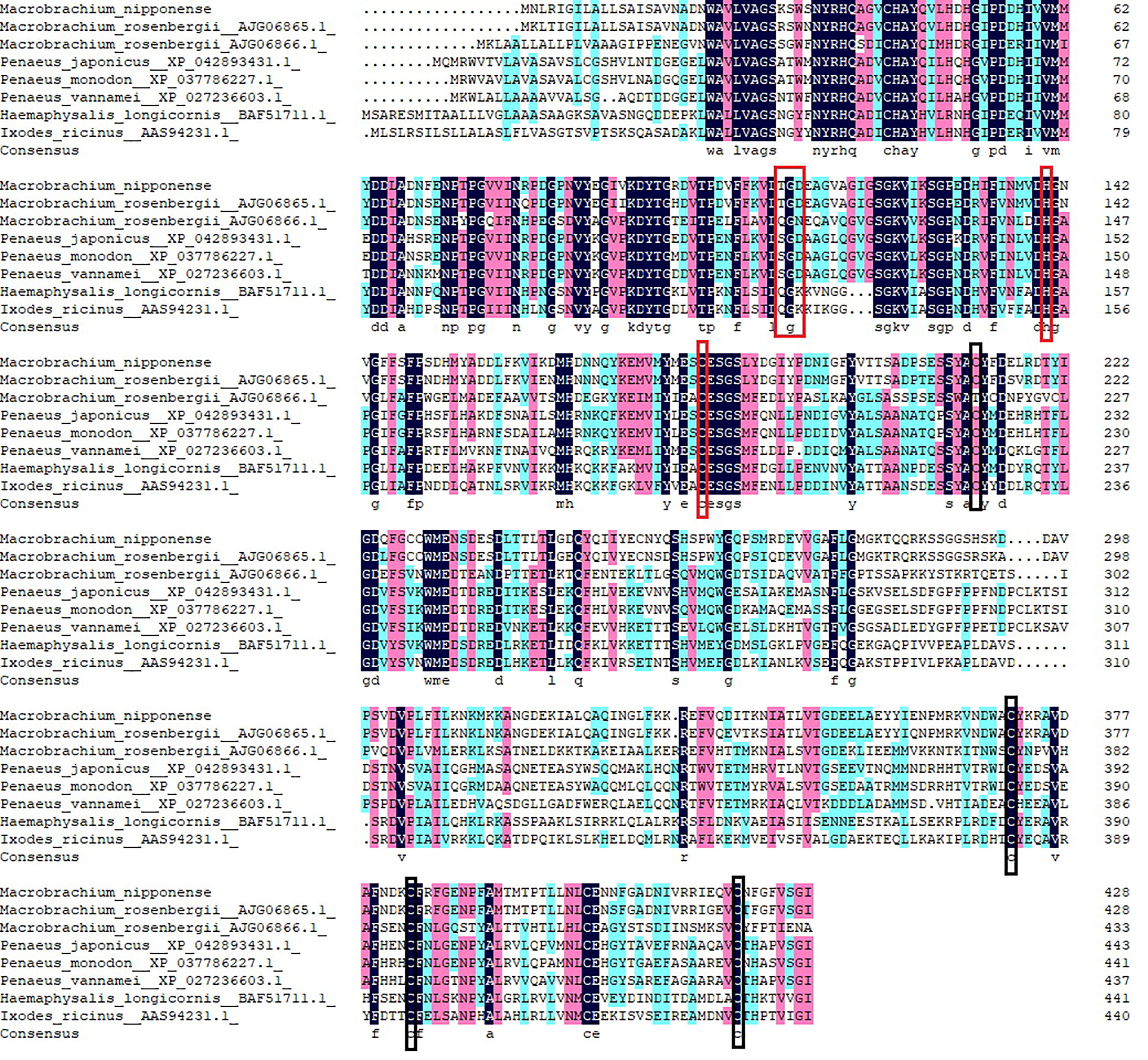
Figure 3 Multiple amino acid alignment and structure prediction of legumains. TGD motif and His-Cys Catalytic dyad are represented by red boxes. Highly conserved cysteine residues are represented by black boxes. Legumain-like protein (Macrobrachium rosenbergii, AJG06865.1), legumain-like protein (M. rosenbergii, AJG06866.1), legumain-like (Penaeus japonicus XP_042893431.1), legumain-like (Penaeus monodon, XP_037786227.1), legumain-like (Penaeus vannamei, XP_027236603.1), legumain (Haemaphysalis longicornis BAF51711.1), and legumain-like (Ixodes ricinus AAS94231.1).
Spatiotemporal Expression Patterns of Mn-Lel
Tissue expression of Mn-Lel was monitored by qPCR using cDNA templates originating from the female muscles (M), heart (H), ovary (O), cerebral ganglion (Cg), eyestalks (E), hepatopancreas (He), and gills (G) tissues as displayed in Figure 4. The Ct value of qPCR revealed that Mn-Lel was hardly expressed in eyestalks, cerebral ganglion, heart, and gills but was weakly expressed in the muscle. High expression levels were detected in the hepatopancreas and ovary. Mn-Lel was highly expressed in both these tissues, but the expression level in the hepatopancreas was nearly 250 times more than that in the ovaries (p < 0.01).
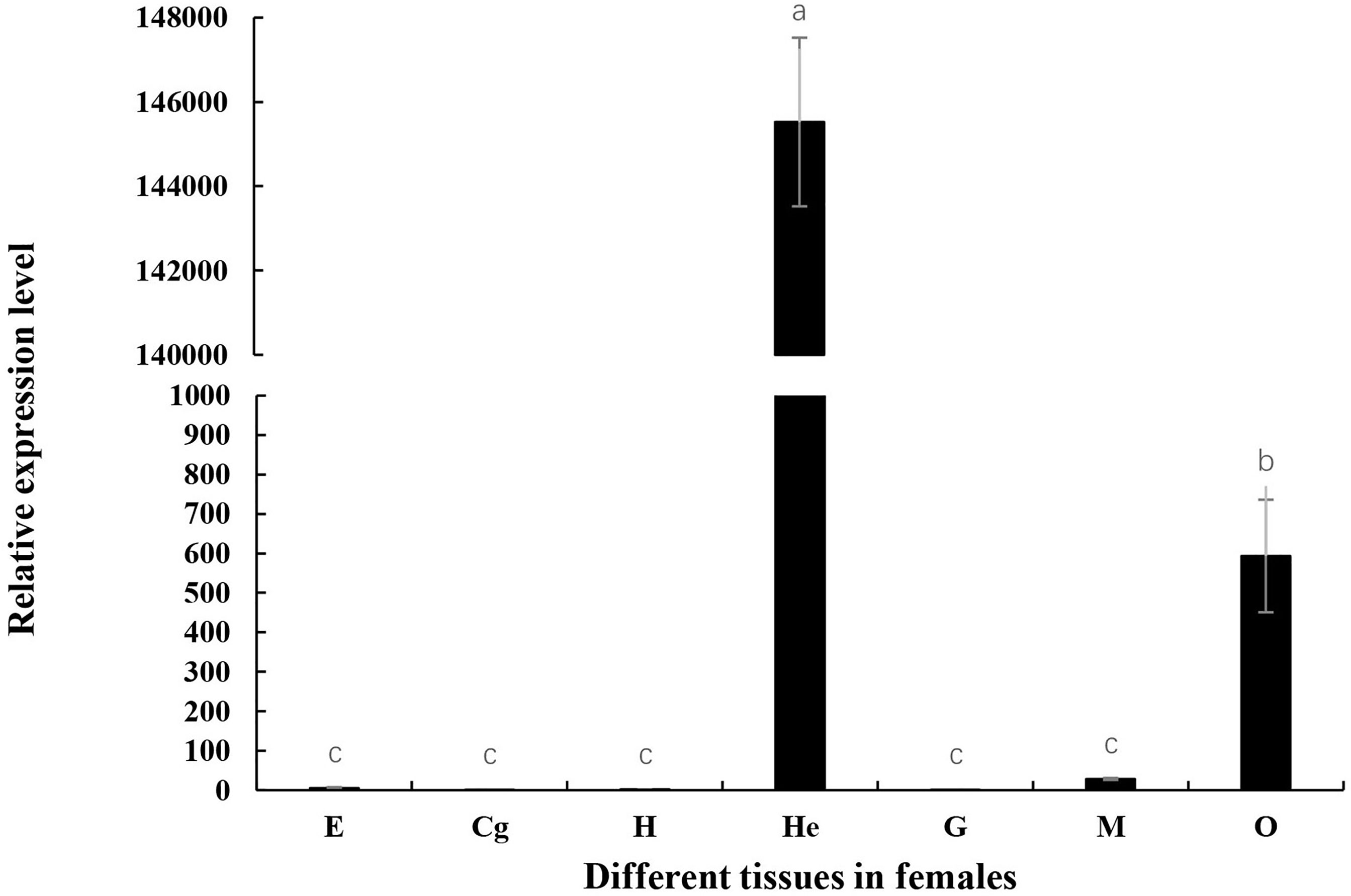
Figure 4 Tissue distribution of Mn-Lel. E, eyestalk; Cg, cerebral ganglion; H, heart; He, hepatopancreas; G, gill; M, muscle; O, ovary. Data are shown as mean ± SD (n = 5). Different letters denote significant differences (p < 0.05). Bars with different letters were considered significant at p < 0.05.
Mn-Lel expression during embryo and larva development was also investigated (Figure 5). According to the Ct value of qPCR, Mn-Lel was hardly expressed in the embryo development stages (from CS stage to ZS stage), and it was also hardly expressed until the 15th day after hatching (L15) (p < 0.05). Mn-Lel expressed significantly from L5 to the 1st day after metamorphosis (PL1) and then dramatically increased PL5 by about 10 times than PL1 (p < 0.05), which reached the top. In later larval development stages (from PL5 stage to PL25 stage), the Mn-Lel showed a significant fluctuation trend (p < 0.05).
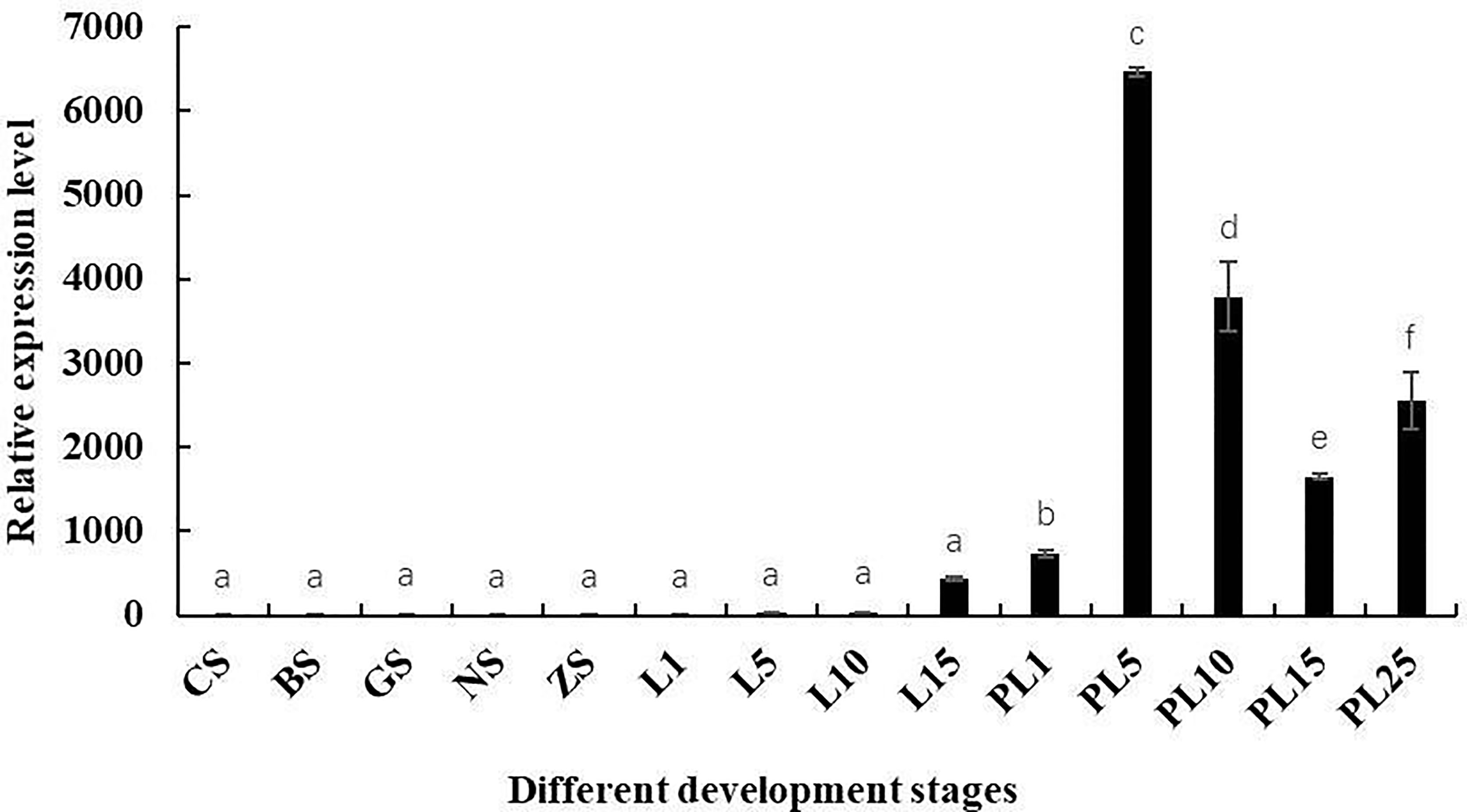
Figure 5 The expression pattern of the Mn-Lel in developmental stages of Macrobrachium nipponense. CS, cleavage-stage; BS, blastocyst stage; GS, gastrulation stage; NS, nauplius stage; ZS, zoea stage; L1, the 1st-day larvae after hatching; L5, the 5th-day larvae after hatching; L10, the 10th-day larvae after hatching; L15, the 15th-day larvae after hatching; PL1, post-larval stage of the 1st day; PL5, post-larval stage of the 5th day; PL10, post-larval stage of the 10th day; PL15, post-larval stage of the 15th day; PL-25M, post-larval stage of the 25th day. Data are presented as the mean ± SD (n = 6). p < 0.05 was considered to be statistically significant. Bars with different letters were considered significant at p < 0.05.
Subsequently, Mn-Lel is expressed in the hepatopancreas and ovaries during the ovary maturation cycle. According to the Ct values, Mn-Lel was highly expressed during the ovary maturation cycle that was examined. The Ct value of Mn-Lel gene in hepatopancreas maintained a remarkably high level in all five ovary stages (Figure 6A). It was found that Mn-Lel was higher in stages I and IV (p < 0.05), and its expression was slightly lower as detected in the other three stages (p < 0.05). In ovaries (Figure 6B), the Ct value of Mn-Lel gene also showed a high expression level. The Mn-Lel expression level doubled obviously from stage I to II (p < 0.05). Then its expression significantly increased from stage II to III approximately five-fold (p < 0.05) and reached the peak in stage III. Subsequently, it decreased dramatically from stage III to IV (p < 0.05).
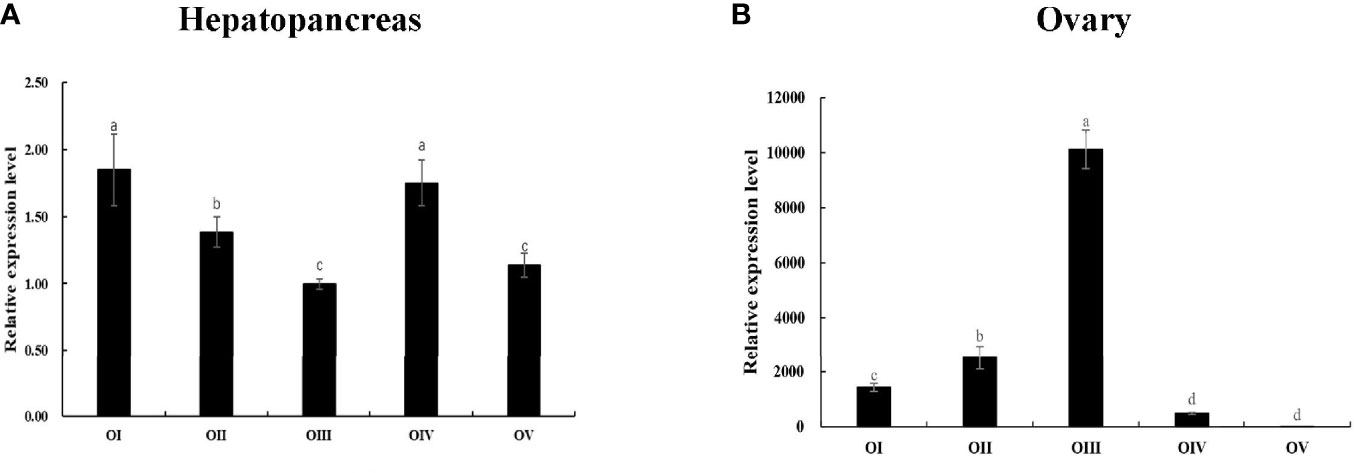
Figure 6 Expression patterns of Mn-Lel in the hepatopancreas (A) and ovary (B) at different ovarian stages. Different ovarian stages expressions: OI, undeveloped stage; OII, developing stage; OIII, nearly-ripe stage; OIV, ripe stage; OV, spent stage. Statistical analyses were performed with one-way ANOVA. Data are shown as mean ± SD (n = 5). Different letters denote significant differences (p < 0.05). Bars with different letters were considered significant at p < 0.05.
Mn-Lel Localization in Five Ovary Stages by In Situ Hybridization
Periodic changes of ovarian development were clearly shown in the paraffin section of Figure 7. In stage I, the space of the ovarian cavity is large, the nucleus of the oocyte is round and large, and the protoplasm is less and transparent. In stage II, the ovarian cavity began to shrink significantly, the oocyte was closely arranged, and the nucleus is relatively small. In stage III, the ovarian cavity becomes narrow, and a layer of vacuoles appears around the oocyte. The oocyte begins to deposit yolk. In stage IV, oocyte development was completed, and the cytoplasm was filled with a large number of oocytes. In stage V, the ovary was filled with empty follicles and a few stage I oocytes. ISH was used to detect the Mn-Lel localization in ovary stages (Figure 7). The results indicated that the Mn-Lel signal was detected in all five stages. The Mn-Lel signal could be found in oocytes including the nucleus, follicle cell, yolk granule, cytoplasmic membrane, and follicle membrane, while follicle membrane, cytoplasmic membrane, and nucleus were the main expressed sites. The color intensity of the signal showed that Mn-Lel is mainly expressed in the membrane of the follicle, cytoplasmic membrane, and nucleus. It showed that the signal of Mn-Lel in the ovary was significantly enhanced during stage II (primary vitellogenesis) and stage III (secondary vitellogenesis) and gradually weakened in other stages.
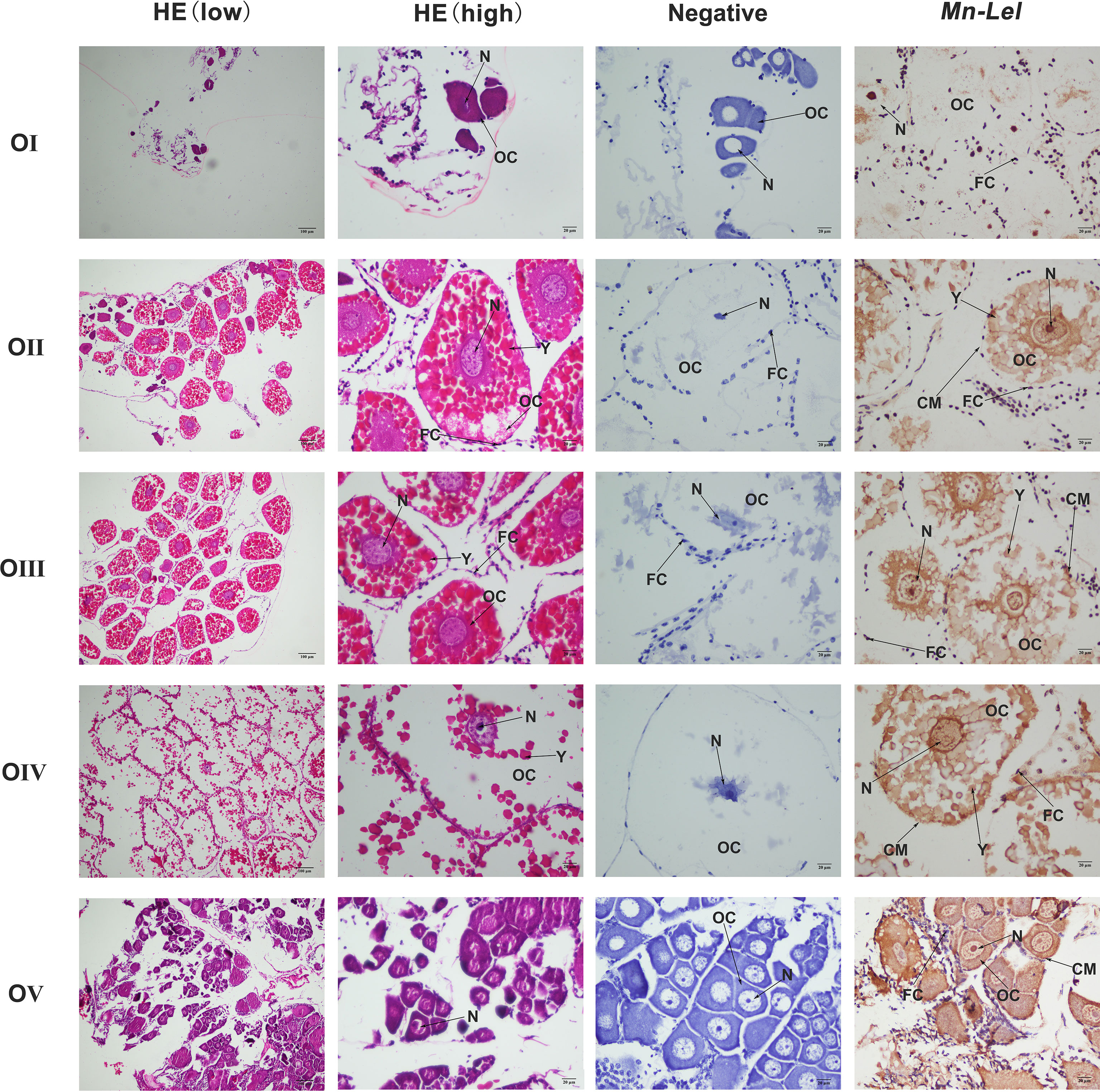
Figure 7 Location of Mn-Lel detected in the ovary by in situ hybridization. A photograph of Macrobrachium nipponense ovary in ovarian cycle. HE, H&E staining paraffin section; OC, oocyte; N, nucleus; CM, cytoplasmic membrane; Y, yolk granule; FC, follicle cell; FM, follicle membrane. Scale bars, ×100 (low), ×400 (high).
Effect of Knocking Down Mn-Lel on Ovary Maturation of Macrobrachium nipponense
Mn-Lel was knocked down by a 3-week RNAi. The ovaries of both control and experimental groups were in stage IV (ripe stage, dark green) at the beginning, and as the experiment progressed, the ovaries emptied and became full again. The RNAi efficiency of ds-Mn-Lel in hepatopancreas and ovary was tested. In hepatopancreas, the Mn-Lel expression level was significantly downregulated by 92.67% on the 9th day after injection and then by 96.19% on the 17th day after injection (p < 0.01) (Figure 8A). In the ovary, the ds-Mn-Lel showed an obviously decreased efficiency by 81.49% on the 9th day and 99.41% on the 17th day after injection (p < 0.01) (Figure 8B). The GSI in both groups was also tracked and calculated (Figure 8C). In the beginning, the average GSI of control and experiment groups was 12.68% and 12.00%, respectively; however, the experiment progressed, and there were no obvious changes between the two groups (control: 3.91%, ds-Mn-Lel: 4.01%) on the 9th day after injection (p > 0.05). On the 17th day, the average GSI of the control group increased to 10.45%, while it was only 6.94% in the experiment group (p < 0.05).
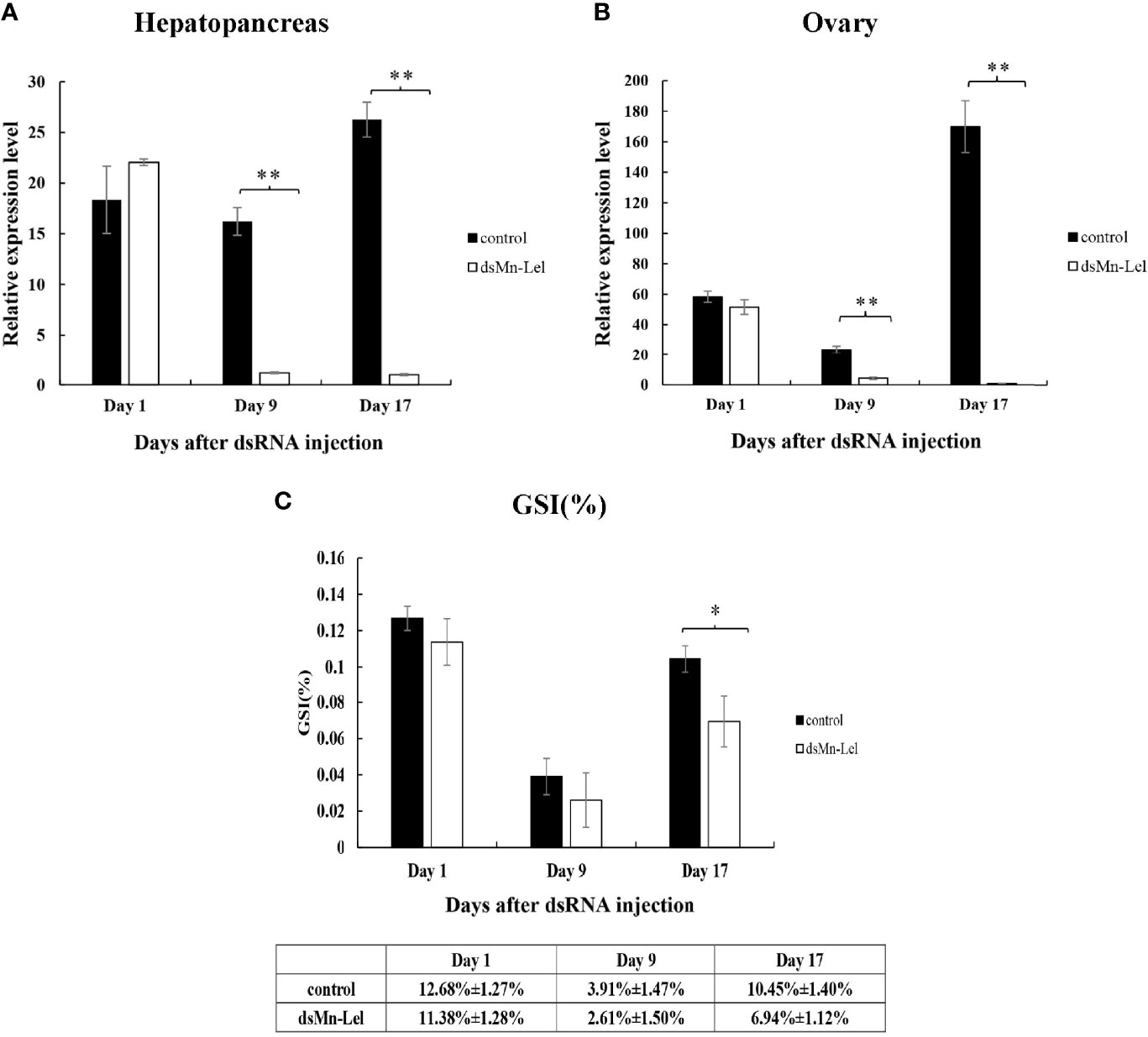
Figure 8 Function analysis of Mn-Lel by RNAi. (A) Efficiency of RNAi-Mn-Lel knockdown in hepatopancreas. (B) Efficiency of RNAi-Mn-Lel knockdown in ovary. (C) Changes in gonadosomatic index (GSI) (%) of female Macrobrachium nipponense after injection with Mn-Lel dsRNA. Data are presented as mean ± SD (n = 6); * denotes statistical significance of p < 0.05. ** denotes statistical significance of p < 0.01.
Discussion
Legumain has been previously reported in parasites and mammals. The precursor enzyme of legumain was relatively conserved and composed of signal-peptide, propeptide, and catalytic domain containing the active center of mature protease. Legumain of Schistosoma mansoni consisted of 429 amino acids (28). H. longicornis has two legumains with a length of 442 amino acids (29, 30). Human and mouse legumain encoded 433 and 435 amino acid polypeptide chains, respectively (5). In this study, Mn-Lel encoded 430 amino acids, with structural characteristics similar to those of humans, arthropods, and schistosoma (5, 28, 31). The sequence included a signal peptide, a C13 superfamily domain of peptidase, containing a His-Cys catalytic dyad, which is necessary for legumain to exert catalytic activity, and was also a required structure for every C13 family member. There was also a conserved TGD motif (Thr110-Gly111-ASP112) in the C13 domain of Mn-Lel, which corresponded to mammalian RGD (Arg-Gly-Asp), Schistosoma japonicum KGD (Lys-Gly-Asp), and tick QGK (Gin-Gly-Lys) (5, 28, 31, 32). It was speculated that the mammalian RGD motif interacts with a component on the cell membrane to help legumain survive in the acidic environment of the lysosome. The largest difference here is that we did not find potential N-linked glycosylation sites in Mn-Lel, which suggested that the role of Mn-Lel might differ from that of conventionally reported legumains. Among vertebrates, the legumain protein sequence of zebrafish has more than 60% homology with legumain in some species; in particular, it has 66.7% homology with legumain in humans, indicating that legumain is highly conserved and plays an important role in evolution (9, 32, 33). Sequence alignment analysis results in the present study showed that the amino acid sequence homology of Mn-Lel, as well as legumain-like protein of M. rosenbergii (AJG06865.1) with other crustacean legumains, was not very high (about 40%). This indicated that these two crustacean legumain-like proteins may play a very important role with others in evolution.
The legumains expression has been studied most extensively in human tumors. In the examination of the normal tissues and a variety of tumor tissue, legumain had a low expression in normal tissue; the high expression was detected in many solid tumors, including breast, colon, lung, prostate, and ovarian tumors and malignant tumors of the central nervous system (34–37). However, there were relatively few studies on legumain expression in normal growth and tissues. legumain in schistosoma, nematodes, ticks, and other parasites is mainly expressed in the intestine and is responsible for the health of host hemoglobin solution (28, 30, 31, 38). Legumain in Amphioxus Branchiostoma belcheri was not widely distributed, and hepatic cecum and hind-gut were the only expression sites (39). In the present, the spatiotemporal expression pattern of Mn-Lel was analyzed by qPCR, and we found that Mn-Lel showed a significant difference in spatiotemporal expression and tissue specificity. In adult prawn tissues, Mn-Lel was specifically highly expressed in hepatopancreas and ovaries but weakly expressed or not expressed in other detected tissues, which were clearly different from immune-related legumains found in other species mentioned above. This suggested that the Mn-Lel might perform a different function. In zebrafish embryo development, legumain was universally expressed and was specifically expressed in hematopoietic tissues 22 h after fertilization, which was consistent with the universal expression profiles in mouse and human hematopoietic tissues (6, 33). Mn-Lel was rarely expressed in embryogenesis and weakly expressed in the early larval development of M. nipponense. It expressed significantly after metamorphosis, which indicated that Mn-Lel was not a maternal gene and mainly plays a role in adults.
To date, no legumain reports have been related to reproduction. In view of the high specific expression of Mn-Lel in the hepatopancreas and ovaries of M. nipponense, we subsequently studied its expression patterns in hepatopancreas and ovaries at five different ovary stages. Mn-Lel, as well as the other two cathepsin L genes discovered in transcriptomes of 5 ovary stages of M. nipponense, were significantly enriched in the lysosomal pathway in vitellogenesis stages (stage II to stage III) (16). Studies in crustaceans showed that vitellogenin was transported from the hepatopancreas to the oocyte through the hemolymph (40, 41). Ovary stage III was a rapid period of growth and proliferation of oocytes, which provided nutrients and energy. Previous studies (42, 43) suggested that two cathepsin L genes are mainly expressed in the hepatopancreas, and their expression levels in the ovary reached the maximum in stage III, which was similar to Mn-Lel. ISH results also showed that a clear Mn-Lel signal was detected in all five stages. The Mn-Lel signal could be found in oocytes including the nucleus, follicle cell, yolk granule, cytoplasmic membrane, and follicle membrane. It was well distributed in the nucleus and around the yolk. These results gave evidence that Mn-Lel plays a special role in ovary maturation like other cathepsin L genes. Legumain, as a protease, not only specifically hydrolyzes peptide bonds of asparagine residues. In addition to protein degradation, it could also hydrolyze and activate some precursor proteins (proenzyme and prohormone), or activate other proteolytic enzyme systems to participate in various physiological activities of the body (6). In parasites, studies on legumain are mainly confined to schistosoma, nematodes, and ticks. They were mainly involved in host hemoglobin degradation (28, 30, 38). Mammalian legumains are involved in many pathophysiological processes such as immunity, bone dissolution, and tumorigenesis (44–46). In addition, few functional studies on legumain have been conducted. In the present study, RNAi was used to illustrate the special roles of Mn-Lel in the ovary maturation of M. nipponense. The ds-Mn-Lel showed extreme inhibitory effects on Mn-Lel expression. GSI result intuitively showed the inhibitory effect on ovary maturation. The results demonstrate that Mn-Lel played an important role in yolk accumulation. Inhibiting the expression of Mn-Lel can effectively inhibit ovarian maturation.
Conclusion
In the present study, a novel function legumain-like protease from ovary transcriptomes of M. nipponense was characterized and named as Mn-Lel. It was the first report about a reproduction that involved legumain gene in crustaceans. The expression, distribution, and function of Mn-Lel gene in M. nipponense were systematically analyzed by qRT-PCR, ISH, and RNAi. Our research results strongly prove that Mn-Lel plays a pivotal role in the ovarian maturation process of M. nipponense. This study enriched the molecular mechanisms of ovary maturation during the reproduction period of female M. nipponense and provided new insights for reproductive regulation mechanisms in crustaceans.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by Animal Care and Use Ethics Committee in the Freshwater Fisheries Research Center (Wuxi, China).
Author Contributions
Designed the study: HQ and HF. Carried out the experiments and wrote the original draft: SJ and YX. Provided technical assistance: WZ and JZ. Methodology and data curation: SJ and YX. Resources: DC and YG. Software: YW. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This research was supported by grants from Jiangsu Agricultural Industry Technology System (JATS [2021] 515); the National Key R&D Program of China (2018YFD0901303); Central Public-interest Scientific Institution Basal Research Fund CAFS (2020TD36); the China Agriculture Research System-48 (CARS-48); the New cultivar breeding Major Project of Jiangsu province (PZCZ201745); and the seed industry revitalization project of Jiangsu province (JBGS [2021] 118). Thanks to the Jiangsu Province Platform for the conservation and utilization of agricultural germplasm.
References
1. Wiederanders B. Structure-Function Relationships in Class CA1 Cysteine Peptidase Propeptides. Acta Biochim Pol (2003) 50:691–713. doi: 10.18388/abp.2003_3661
2. Barrett AJ, Rawlings ND. Evolutionary Lines of Cysteine Peptidases. Biol Chem (2001) 382:727–34. doi: 10.1515/BC.2001.088
3. Dickinson DP. Cysteine Peptidases of Mammals: Their Biological Roles and Potential Effects in the Oral Cavity and Other Tissues in Health and Disease. Crit Rev Oral Biol Med (2002) 13:238–75. doi: 10.1177/154411130201300304
4. Hara-Nishimura I, Takeuchi Y, Nishimura M. Molecular Characterization of a Vacuolar Processing Enzyme Related to a Putative Cysteine Proteinase of Schistosoma mansoni. Plant Cell (1993) 5:1651–9. doi: 10.2307/3869746
5. Chen JM, Dando PM, Rawlings ND, Brown MA, Young NE, Stevens RA, et al. Cloning, Isolation, and Characterization of Mammalian Legumain, an Asparaginyl Endopeptidase. J Biol Chem (1997) 272:8090–8. doi: 10.1074/jbc.272.12.8090
6. Dall E, Brandstetter H. Structure and Function of Legumain in Health and Disease. Biochimie (2016) 122:126–50. doi: 10.1016/j.biochi.2015.09.022
7. Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, et al. Targeting Tumor-Associated Macrophages as a Novel Strategy Against Breast Cancer. J Clin Invest (2006) 116:2132–41. doi: 10.1172/JCI27648
8. Haugen MH, Boye K, Nesland JM, Pettersen SJ, Egeland EV, Tamhane T, et al. High Expression of the Cysteine Proteinase Legumain in Colorectal Cancer–Implications for Therapeutic Targeting. Eur J Cancer (2015) 51:9–17. doi: 10.1158/1078-0432.CCR-05-0141
9. Wu M, Shao GR, Zhang FX, Wu WX, Xu P, Ruan ZM. Legumain Protein as a Potential Predictive Biomarker for Asian Patients With Breast Carcinoma. Asian Pac J Cancer Prev (2015) 15:10773–7. doi: 10.7314/APJCP.2014.15.24.10773
10. Wu T, Sun L, Wu Y, Xiang R, Li Y, Rong W, et al. Prognostic Value of Legumain in Uveal Melanoma. Mol Med Rep (2016) 13:2377–84. doi: 10.3892/mmr.2016.4838
11. Meng F, Liu W. Knockdown of Legumain Suppresses Cervical Cancer Cell Migration and Invasion. Oncol Res (2016) 23:7. doi: 10.3727/096504015X14410238486649
12. Zhen Y, Chunlei G, Wenzhi S, Shuangtao Z, Na L, Rongrong W, et al. Clinicopathologic Significance of Legumain Overexpression in Cancer: A Systematic Review and Meta-Analysis. Sci Rep (2015) 5:1–9. doi: 10.1038/srep16599
13. Kong Y, Ding Z, Zhang Y, Zhou P, Wu C, Zhu M, et al. Types of Carbohydrate in Feed Affect the Growth Performance, Antioxidant Capacity, Immunity, and Activity of Digestive and Carbohydrate Metabolism Enzymes in Juvenile Macrobrachium nipponense. Aquaculture (2019) 512:734282. doi: 10.1016/j.aquaculture.2019.734282
14. Ministry of Agriculture Fisheries Bureau. China Fishery Statistical Yearbook. Beijing, China: Chinese Agricultural Press (2020).
15. Qiao H, Fu H, Xiong Y, Jiang S, Sun S, Jin S, et al. Molecular Insights Into Reproduction Regulation of Female Oriental River Prawns Macrobrachium Nipponense Through Comparative Transcriptomic Analysis. Sci Rep (2017) 7:1–11. doi: 10.1038/s41598-017-10439-2
16. Zhang Y, Jiang S, Qiao H, Xiong Y, Fu H, Zhang W, et al. Transcriptome Analysis of Five Ovarian Stages Reveals Gonad Maturation in Female Macrobrachium nipponense. BMC Genomics (2021) 22:1–11. doi: 10.1186/s12864-021-07737-5
17. Qiao H, Xiong Y, Zhang W, Fu H, Jiang S, Sun S, et al. Characterization, Expression, and Function Analysis of Gonad-Inhibiting Hormone in Oriental River Prawn, Macrobrachium nipponense and its Induced Expression by Temperature. Comp Biochem Physiol Part A: Mol Integr Physiol (2015) 185:1–8. doi: 10.1371/journal.pone.0198861
18. Wang Y, Jin S, Fu H, Qiao H, Sun S, Zhang W, et al. Identification and Characterization of the DMRT11E Gene in the Oriental River Prawn Macrobrachium nipponense. Int J Mol Sci (2019) 20:1734. doi: 10.3390/ijms20071734
19. Müller Y, Ammar D, Nazari E. Embryonic Development of Four Species of Palaemonid Prawns (Crustacea, Decapoda): Pre-Naupliar, Naupliar and Post-Naupliar Periods. Rev Bras Zool (2004) 21:27–32. doi: 10.1007/BF02366257
20. Bairoch A, Bucher P, Hofmann K. The PROSITE Database, its Status in 1997. Nucleic Acids Res (1997) 25:217–21. doi: 10.1093/nar/25.1.217
21. Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: Network Protein Sequence Analysis. Trends Biochem Sci (2000) 25:47–150. doi: 10.1016/s0968-0004(99)01540-6
22. Hu Y, Fu H, Qiao H, Sun S, Zhang W, Jin S, et al. Validation and Evaluation of Reference Genes for Quantitative Real-Time PCR in Macrobrachium nipponense. Int J Mol Sci (2018) 19:2258. doi: 10.3390/ijms19082258
23. Yuan H, Qiao H, Fu Y, Fu H, Zhang W, Jin S, et al. RNA Interference Shows That Spook, the Precursor Gene of 20-Hydroxyecdysone (20E), Regulates the Molting of Macrobrachium nipponense. J Steroid Biochem Mol Biol (2021) 213:105976. doi: 10.1016/j.jsbmb.2021.105976
24. Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2– ΔΔCT Method. Nethods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
25. Li F, Qiao H, Fu HT, Sun SM, Zhang WY, Jin SB, et al. Identification and Characterization of Opsin Gene and Its Role in Ovarian Maturation in the Oriental River Prawn Macrobrachium nipponense. Comp Biochem Physiol B-Biochem Mol Biol (2018) 218:1–12. doi: 10.1016/j.cbpb.2017.12.016
26. Zhang SB, Jiang P, Wang ZQ, Long SR, Liu RD, Zhang X, et al. Dsrna-Mediated Silencing of Nudix Hydrolase in Trichinella Spiralis Inhibits the Larval Invasion and Survival in Mice. Exp Parasitol (2016) 162:35–42. doi: 10.1016/j.exppara.2016.01.005
27. Qiao H, Jiang F, Xiong Y, Jiang S, Fu H, Li F, et al. Characterization, Expression Patterns of Molt-Inhibiting Hormone Gene of Macrobrachium nipponense and Its Roles in Molting and Growth. PloS One (2018) 13(6):e0198861. doi: 10.1371/journal.pone.0198861
28. Caffrey CR, Mathieu MA, Gaffney AM, Salter JP, Sajid M, Lucas KD. Et al. Identification of a cDNA Encoding an Active Asparaginyl Endopeptidase of Schistosoma mansoni and its Expression in Pichia Pastoris. FEBS Lett (2000) 466:244–8. doi: 10.1016/S0014-5793(99)01798-6
29. Alim MA, Tsuji N, Miyoshi T, Islam MK, Huang X, Motobu M, et al. HlLgm2, a Member of Asparaginyl Endopeptidases/Legumains in the Midgut of the Ixodid Tick Haemaphysalis longicornis, is Involved in Blood-Meal Digestion. J Insect Physiol (2008) 54:573–85. doi: 10.1016/j.jinsphys.2007.12.006
30. Alim MA, Tsuji N, Miyoshi T, Islam MK, Huang X, Motobu M, et al. Characterization of Asparaginyl Endopeptidase, Legumain Induced by Blood Feeding in the Ixodid Tick Haemaphysalis longicornis. Insect Biochem Mol Biol (2007) 37:911–22. doi: 10.1016/j.ibmb.2007.04.010
31. Sojka D, Hajdušek O, Dvořák J, Sajid M, Franta Z, Schneider EL, et al. IrAE–An Asparaginyl Endopeptidase (Legumain) in the Gut of the Hard Tick Ixodes Ricinus. Int J Parasitol (2007) 37(7):713–24. doi: 10.1016/j.ijpara.2006.12.020
32. Yuan Y, Ge S, Sun H, Dong X, Zhao H, An L, et al. Intracellular Self-Assembly and Disassembly of 19F Nanoparticles Confer Respective “Off” and “On” 19f NMR/MRI Signals for Legumain Activity Detection in Zebrafish. ACS Nano (2015) 9:5117–24. doi: 10.1021/acsnano.5b00287
33. Ma L, Shen YQ, Khatri HP, Schachner M. The Asparaginyl Endopeptidase Legumain is Essential for Functional Recovery After Spinal Cord Injury in Adult Zebrafish. PloS One (2014) 9:e95098. doi: 10.1371/journal.pone.0095098
34. Liu C, Sun C, Huang H, Janda K, Edgington T. Overexpression of Legumain in Tumors is Significant for Invasion/Metastasis and a Candidate Enzymatic Target for Prodrug Therapy. Cancer Res (2003) 63:2957–64. doi: 10.1097/00002820-200306000-00012
35. Gawenda J, Traub F, Lück HJ, Kreipe H, Wasielewski R. Legumain Expression as a Prognostic Factor in Breast Cancer Patients. Breast Cancer Res Treat (2007) 102:1–6. doi: 10.1007/s10549-006-9311-z
36. Wang L, Chen S, Zhang M, Li N, Chen Y, Su W, et al. Legumain: A Biomarker for Diagnosis and Prognosis of Human Ovarian Cancer. J Cell Biochem (2012) 113:2679–86. doi: 10.1002/jcb.24143
37. Mai CW, Chung FF, Leong CO. Targeting Legumain as a Novel Therapeutic Strategy in Cancers. Curr Drug Targets (2017) 18(11):1259–68. doi: 10.2174/1389450117666161216125344
38. Oliver EM, Skuce PJ, McNair CM, Knox DP. Identification and Characterization of an Asparaginyl Proteinase (Legumain) From the Parasitic Nematode, Haemonchus contortus. Parasitology (2006) 133(2):237–44. doi: 10.1017/S0031182006000229
39. Teng L, Wada H, Zhang S. Identification and Functional Characterization of Legumain in Amphioxus Branchiostoma Belcheri. Biosci Rep (2010) 30:177–86. doi: 10.1042/bsr20090049
40. Ara F, Damrongphol P. Vitellogenin Gene Expression at Different Ovarian Stages in the Giant Freshwater Prawn, Macrobrachium rosenbergii, and Stimulation by 4-Nonylphenol. Aquac Res (2014) 45:320–6. doi: 10.1111/j.1365-2109.2012.03229.x
41. Urtgam S, Treerattrakool S, Roytrakul S, Wongtripop S, Prommoon J, Panyim S, et al. Correlation Between Gonad-Inhibiting Hormone and Vitellogenin During Ovarian Maturation in the Domesticated Penaeus monodon. Aquaculture (2015) 437:1–9. doi: 10.1016/j.aquaculture.2014.11.014
42. Zhu J, Fu H, Qiao H, Jin S, Zhang W, Jiang S, et al. Expression and Functional Analysis of Cathepsin L1 in Ovarian Development of the Oriental River Prawn, Macrobrachium nipponense. Aquac Rep (2021) 20:100724. doi: 10.1016/j.aqrep.2021.100724
43. Jiang S, Xiong Y, Zhang W, Zhu J, Cheng D, Gong Y. Et al. Molecular Characterization of a Novel Cathepsin L in Macrobrachium nipponense and its Function in Ovary Maturation. Front Endocrinol (2022) . 12:816813:816813. doi: 10.3389/fendo.2021.816813
44. Liang DS, Wen ZJ, Wang JH, Zhu FF, Guo F, Zhou JL, et al. Legumain Protease-Sheddable PEGylated, Tuftsin-Modified Nanoparticles for Selective Targeting to Tumour-Associated Macrophages. J Drug Target (2022) 30:82–93. doi: 10.1080/1061186X.2021.1906886
45. Reddy BD, Beeraka NM, Chitturi CH, Madhunapantula SV. An Overview of Targeting Legumain for Inhibiting Cancers. Curr Pharm Design (2021) 27(31):3337–48. doi: 10.2174/1381612826666201125111625
Keywords: Macrobrachium nipponense, legumain-like gene, ovary maturation, mRNA expression, RNA interference
Citation: Jiang S, Xiong Y, Zhang W, Zhu J, Cheng D, Gong Y, Wu Y, Qiao H and Fu H (2022) A Novel Legumain-Like Protease in Macrobrachium nipponense: Identification, Characterization, and Function Analysis in Ovary Maturation. Front. Endocrinol. 13:858726. doi: 10.3389/fendo.2022.858726
Received: 20 January 2022; Accepted: 28 February 2022;
Published: 24 March 2022.
Edited by:
Chao Bian, BGI Academy of Marine Sciences, ChinaReviewed by:
Tsuyoshi Kawada, Suntory Foundation for Life Sciences, JapanShi Li Liu, Zhejiang Institute of Freshwater Fisheries, China
Copyright © 2022 Jiang, Xiong, Zhang, Zhu, Cheng, Gong, Wu, Qiao and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongtuo Fu, ZnVodEBmZnJjLmNu; Hui Qiao, cWlhb2hAZmZyYy5jbg==
†These authors have contributed equally to this work and share first authorship
 Sufei Jiang
Sufei Jiang Yiwei Xiong1†
Yiwei Xiong1† Hui Qiao
Hui Qiao Hongtuo Fu
Hongtuo Fu