- 1School of Nursing, Clemson University, Clemson, SC, United States
- 2Illume Fertility Center, Norwalk, CT, United States
Background: The genetic components of the etiologies of ovulatory dysfunction-related infertility (ODRI) are poorly characterized.
Objectives: This paper aimed to comprehensively identify, compile, and categorize published research on relationships between genetics and ovulatory-related infertility in humans.
Methods: A scoping review was performed on research articles relating human genes, ovulatory dysfunction, and infertility retrieved from PubMed and Web of Science databases. A total of 45 articles were included in the study. The data has been organized into three categories based on relevant findings: polycystic ovary syndrome (PCOS), premature ovarian insufficiency (POI), and other diagnoses related to ovulatory dysfunction and infertility.
Results: Sources revealed 235 different genes linked to ovulatory dysfunction and infertility including follicle-stimulating hormone receptor (FSHR), luteinizing hormone/choriogonadotropin receptor (LHCGR), and bone morphogenic protein 15 (BMP15). PCOS-related articles revealed variants in genes with functions focused on androgen production, such as LHCGR and FSHR. POI-related articles revealed variants in genes with functions focused on folliculogenesis and pubertal development, such as BMP15 and STAG3, stromal antigen 3. The “other” category revealed genes resulting in enzyme deficiencies interacting with a wide range of functions.
Conclusions: In this review, we have highlighted the extreme variability in what is known about the genetics of ODRI by compiling and categorizing genes identified in the literature as associated with ODRI and its associated subtypes. We have also provided a comprehensive list of ODRI genes specifically identified in humans. The findings from this review, specifically the list of ODRI genes, can be used for targeted gene panel development in assisted reproductive technology to improve clinical testing and diagnosis, as well as in developing individualized treatment strategies for ODRI patients.
Introduction
Fertility is the ability to achieve a clinical gestation, but the definition of infertility is not as clear (1). The exact constraints used to define infertility were previously convoluted; however, the American Society for Reproductive Medicine (ASRM) recently redefined the condition, as follows: Infertility can be identified in individuals who are unable to conceive, requiring medical intervention. The period of attempted conception before diagnosis of infertility and medical intervention is a standard of one year for female patients under the age of 35 and six months for female patients 35 years of age or older (2).
Ovulation dysfunction is one of the most common conditions associated with infertility (3). It can manifest in different ways. Anovulation is the complete absence of ovulation. Oligoovulation, or infrequent ovulation, is an inconsistent ovulation pattern that is usually paired with irregular menstruation patterns (4). Ovulatory dysfunction is observed in many different disorders and conditions, including the most common one, polycystic ovary syndrome (PCOS), affecting an estimated 10-13% of the female population around the world (5, 6).Certain cardinal components are used to initially diagnose PCOS, specifically, ovulatory dysfunction, clinical or biochemical hyperandrogenism, and polycystic ovarian morphology on a sonographic visualization (5). A positive diagnosis of PCOS is made if the patient has at least two of these components (6). PCOS is a complex disorder, affecting the body at the endocrine and metabolic levels (7, 8). At the endocrine and metabolic level, PCOS patients may suffer from insulin resistance and hyperandrogenism. At the phenotypic level, PCOS patients may suffer from comorbidities such as type two diabetes, hypertension, dyslipidemia, and obesity. It is imperative to highlight that in the classification of the disease, the clinical manifestations vary significantly (8). The variance in manifestations of PCOS has led to subcategorization. Type I classic PCOS is characterized by increased LH and LH/FSH ratio, increased androgens, elevated insulin levels, polycystic ovarian morphology, and increased waist measurements. Type II classic PCOS is characterized by all characteristics of Type I but without polycystic ovarian morphology. Ovulatory PCOS has milder hormonal imbalances and clinical implications. Normoandrogenic PCOS is the rarest form and has no clinical presentation except for a heightened LH and LH/FSH ratio (9).
There are some known gene associations that exist for PCOS. The primary category of genes with a known association with PCOS is involved with the endocrine system. These gene functions are involved with hormone genesis, hormone regulation, hormone action, and hormone secretion (5). Other gene associations to PCOS include the Fat Mass Obesity gene and the PCOS1 gene. While PCOS accounts for an estimated 80% of ovulatory dysfunction-related infertility (ODRI) cases (3), a diagnosis of PCOS does not directly cause infertility (6).
Primary Ovarian Insufficiency (POI) is a condition leading to abnormal or complete lack of ovulation before the age of 40 (10, 11). To confirm a diagnosis of POI, a biologically female patient must be below the age of 40, have repeated elevated levels of Follicle Stimulating Hormone reported on two separate occasions at least four weeks apart, and have menstrual disturbance (11–13). POI is estimated to be present in approximately 1% of the female population (13).
It is known that chromosomal anomalies, such as structural translocations or deletion of an X chromosome, are associated with approximately 15% of POI cases (14), thus our focus is on patients without an obvious cytogenetic association. There are many genes associated with POI. The general functions of these genes include folliculogenesis, follicular development, and ovarian steroidogenesis (14). Identifying and clarifying these gene associations for POI will allow researchers and clinicians to better understand the underlying factors of the condition (11).
Other causes of ovulatory dysfunction and subsequent infertility are obesity, hormone-regulatory disorders, and hypothalamic amenorrhea. A small percentage of this population remains idiopathic, or unexplained (15). Some genes that have documented associations in this subgroup are involved with recombination, synapsis, and DNA mismatch repair (16).
In this scoping review, a systematic search of the literature was performed for genes involved with ovulatory dysfunction and infertility. The aim was to create a record of documented gene associations between ovulatory dysfunction and infertility, and to characterize how these genetic variants may result in female infertility. Secondarily, the aim was to generate a comprehensive list of ODRI-associated genes to serve as a foundation for future research efforts. The research question was: are there associations in the peer-reviewed literature between genetic variants and phenotypic ovulatory dysfunction in infertility patients?
Methods
Search strategy and article selection
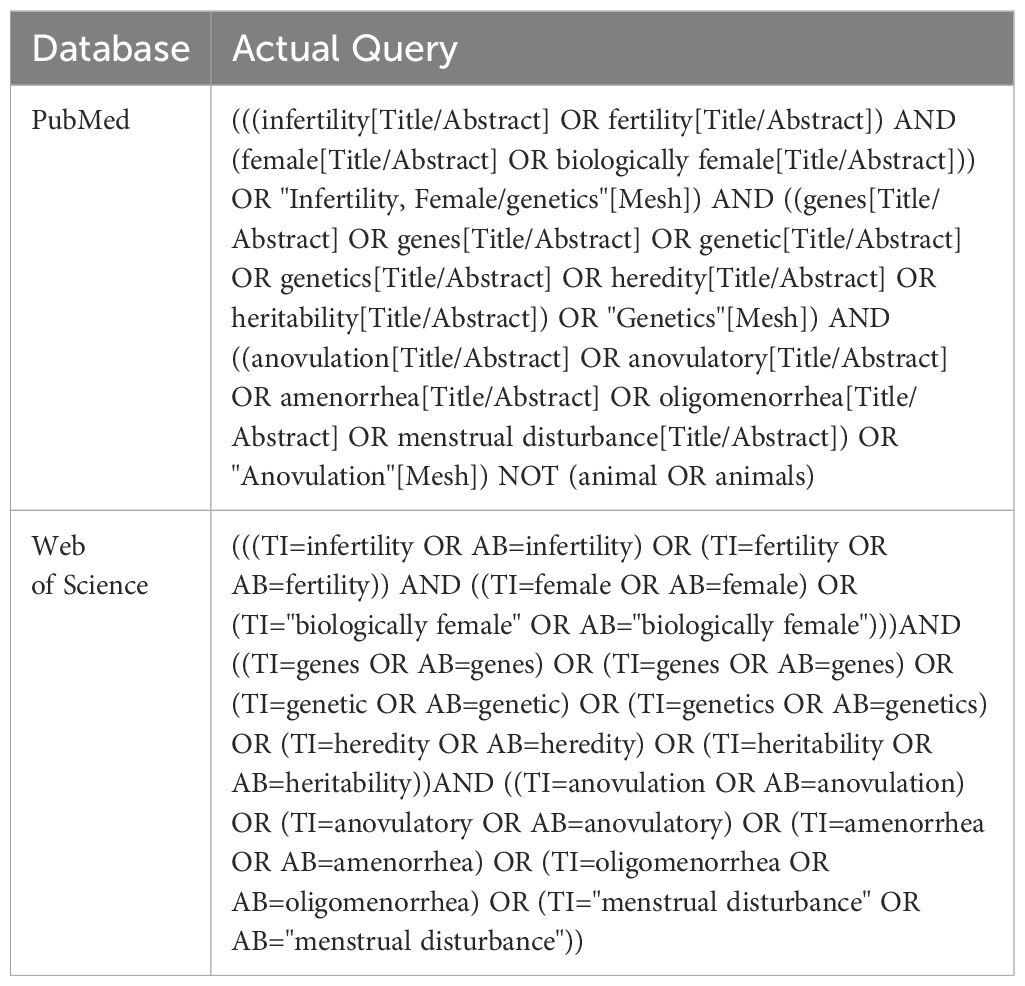
The PRISMA Guideline for Scoping reviews (17) was used to structure the search and analysis of literature included in the study (Figure 1). PubMed and Web of Science databases were queried for papers relevant to the genetics of ovulatory dysfunction and infertility. The search terms used for each database are included in Table 1. Key concepts used to build the queries were female infertility, genes, and ovulatory dysfunction. In addition to the key concepts, all Medical Subject Headings (MeSH) terms were used to increase the breadth of literature retrieved. A single reviewer was responsible for gathering, extracting, and analyzing data in a systematic fashion.
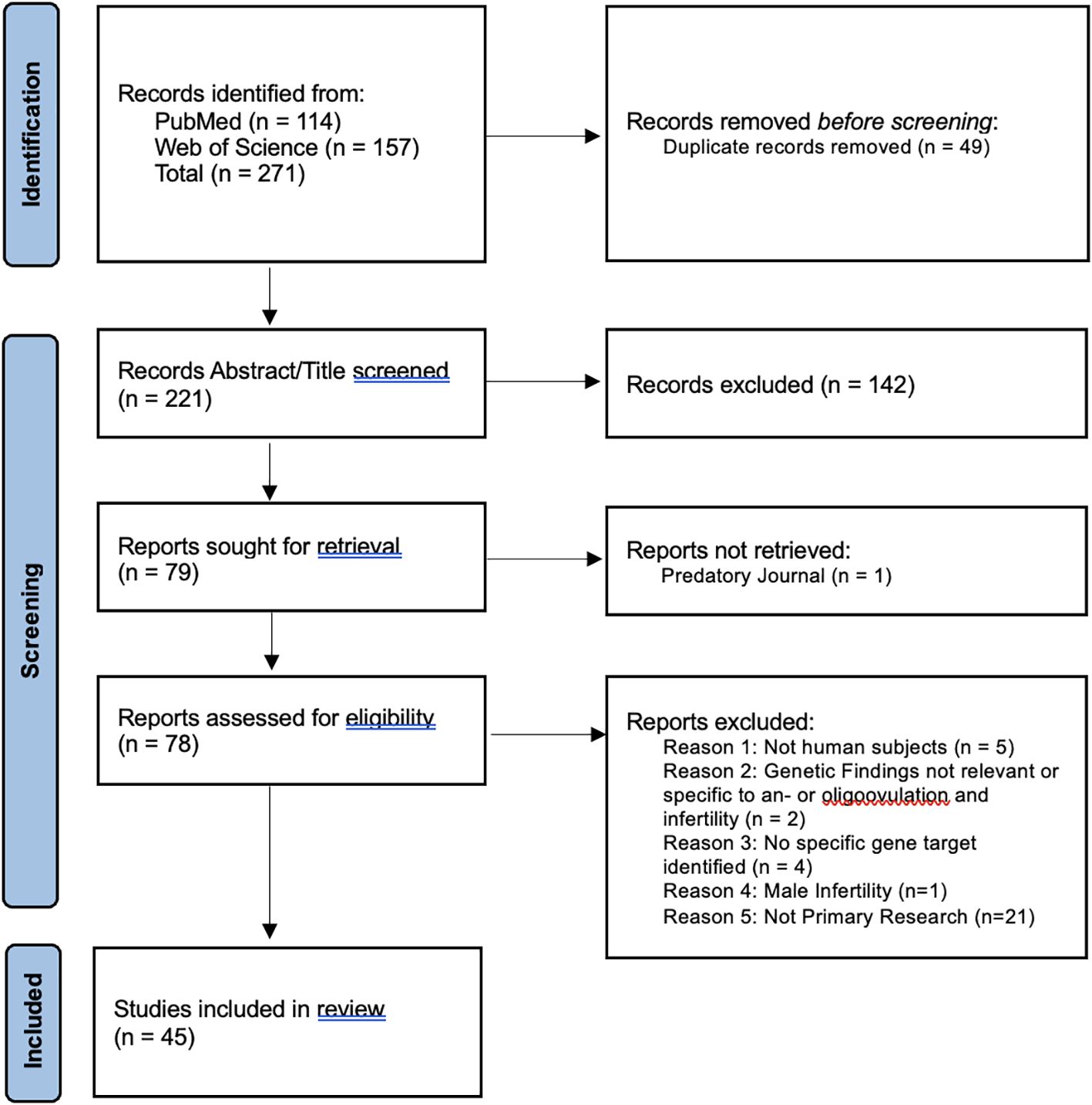
Figure 1. PRISMA 2020 flow diagram chart documenting article retrieval from PubMed and Web of Science and subsequent analysis for eligibility criteria. The primary reviewer participated in all steps, with a second and third reviewer validating the “Records Abstract/Title screened” step.
Duplicate articles were eliminated using a duplicate removal tool in RefWorks (18). A secondary elimination of duplicates was performed manually due to the unique characters of foreign names from the two databases. After duplicate removal, a tab-delimited file containing general article citation information and abstracts was retrieved from RefWorks software.
Article titles and abstracts were reviewed for inclusion and exclusion criteria. Exclusion criteria included male infertility, whole chromosome abnormalities, epigenetic modulation, and chromosomal structural rearrangements. Inclusion criteria, aside from the key concepts, were human subjects, the English language, at least one genetic anomaly identified, and classification as a research article (e. g., not a review or editorial article). There was no start date restriction. The end date restriction was October 4th, 2023.
The primary reviewer was responsible for retrieving articles, duplicate removal, reviewing for inclusion and exclusion criteria, data extraction, and data analysis. A secondary review of the titles and abstracts was split between a second and third reviewer. If the information in the abstract was either insufficient or caused disagreement between the reviewers at the validation of inclusion and exclusion criteria, the paper was evaluated in its entirety by all three reviewers.
Data extraction
A table was created to include the article title, primary author, year, and relevant categories of data. The type of study design and primary study location were extracted from each included article. The category for condition/disease type was broken down into PCOS, POI, or Other, with specific disease affiliation noted. Finally, the type of genetic testing/sequencing was recorded for each article. Any gene with a variant identified as affiliated with ODRI and its projected function was extracted from each article.
Data synthesis and analysis
Gene names used for data inference and analysis were cross-checked using RefSeqGene (19). A complete list of gene names associated with ODRI was extracted from the research articles including the associated condition category, PCOS, POI, or Other. The ClinVar database (20) was queried for genes referenced in a higher number of research articles to identify if existing variants have been previously identified as pathogenic or likely pathogenic. The entire gene list was also used for a Gene Ontology (GO) analysis using the gprofiler2 (21) package in R to identify enriched functions of the gene names extracted from the research articles. The p-value cutoff used for GO-term enrichment analysis was p < 1e-16, adjusted for false positive discovery (21). The GO-term enrichment analysis was used to validate that the construct used in this review for categorization of ODRI was appropriate from a genetic stance.
Results
Article selection and inclusion
Figure 1 illustrates how articles were obtained and included in this review. Articles retrieved from the PubMed and Web of Science databases contained 49 duplicates. After duplicate removal, 221 unique titles and abstracts were analyzed for inclusion and exclusion criteria. A total of 142 records were excluded after initial screening for eligibility. One article was not recovered due to predatory publishing.
Out of 78 retrieved articles, 33 did not meet the inclusion criteria upon further analysis. These articles were excluded due to the use of non-human subjects (n=5), genetic findings lacking specific relevance to ovulatory dysfunction and infertility (n=2), no specific gene variant(s) identified (n=4), structural rearrangements (n=4), publication type (n=17), and male infertility (n=1). A total of 45 articles were included in the full article review.
Data extraction
Research article types retrieved and included in this review are case reports (n=20), observational studies (n=22), and experimental studies (n=3). They originate from 21 unique countries. Thirty-eight reports exclusively used DNA sequencing to observe variants in gene loci involved with ovulatory dysfunction. Two articles used transcriptome profiling exclusively and three used both DNA sequencing and RNA profiling to demonstrate how a mutated gene impacts ovulatory outcome.
Out of 45 retrieved research articles (Supplementary Table 1) (22–66), there are 235 unique genes with variants or variable expression levels identified with some association with ODRI. While most gene variants or targets were uniquely identified in only one research article, some were observed at a higher frequency. From the list of 235 genes, 28 gene names were repeated in more than one research article (Figure 2), with 14 having been previously documented in the ClinVar database (20) to have pathogenic or likely pathogenic associations to ovulatory dysfunction, PCOS, or POI. Genes with associated variants mentioned in five articles include follicle-stimulating hormone receptor (FSHR), luteinizing hormone/choriogonadotropin receptor,\ (LHCGR), and bone morphogenic protein 15 (BMP15) (Supplementary Table 2). Genes identified in multiple research articles were considered higher confidence genes and were given increased scrutiny.

Figure 2. A bar graph showing the distribution of genes associated with ovulatory dysfunction and infertility sorted by gene name and stratified by condition type: PCOS, POI, or Other. Gene associations mentioned in one research article or less are excluded from this graph.
Genes associated with polycystic ovary syndrome
PCOS is identified in 10 of the reports included in this review. All articles report genetic variants in nuclear DNA. There were nine observational studies and one experimental study (Figure 3). There were 141 associated genes with PCOS identified in this review. Out of the PCOS-associated gene list, 12 were identified in more than one research article. The 12 higher confidence genes were IGF1, INSR, CYP19A1, FST, PPARG, STAR, HSD17B1, BMP15, FSHR, AR, CYP17A1, and LHCGR. Only four of these higher confidence genes have known pathogenic or likely pathogenic associations to ovulatory dysfunction according to the ClinVar database: PPARG, BMP15, AR, and LHCGR.
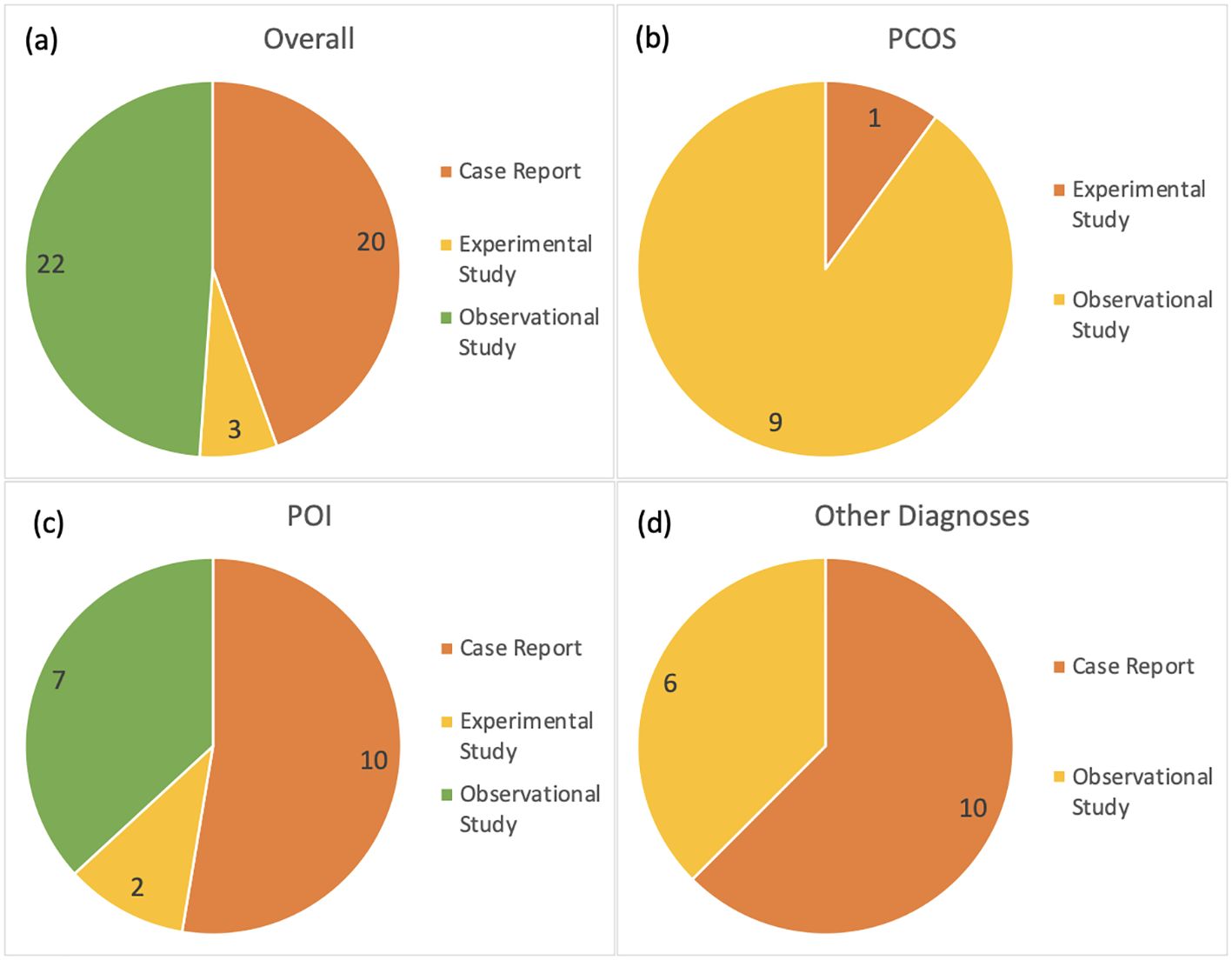
Figure 3. Distribution of research article types included in this scoping review; (a) including all subcategories, (b) articles about PCOS, (c) articles about POI, and (d) remaining articles referencing other categories of disease related to ovulatory dysfunction and infertility. The number of articles is included in the diagrams.
Eight of these articles identified gene variants involved with hormone modulation or synthesis. The earliest report by Urbanek et al. (1999) identified 37 gene loci in a differential gene expression analysis involved with the endocrine system, including androgen synthesis and function. Three papers have identified variants in the LHCGR gene for PCOS associations (Figure 2) (25, 42, 64). The gene is discussed at length by Atoum et al. (2022), discussing how different polymorphisms are associated with specific phenotypic qualities, including hirsutism, obesity, oligomenorrhea, luteinizing hormone, and follicle-stimulating hormone levels.
Ajmal et al. (2021) and Kaur et al. (2018) further identified specific variants in the CYP19A1 and CYP17A1 genes, respectively. The projected function of these genes is in estrogen synthesis, supporting the initial findings of Urbanek et al. (1999). Another gene involved with endocrine function is INSR for insulin sensitivity (59).
The remaining articles identified genes serving basic cellular functions. While one article directly identifies variants in genes involving cell proliferation and growth (35), others identify loss-of-function gene variants associated with cellular death and transcription regulation (22, 42, 60).
The GO analysis revealed 16 gene functions that were highly enriched for the PCOS and infertility categories. All PCOS enriched terms were categorized in the biological process and included terms related to response to an organic substance (p = 2e-26, Figure 4), hormones, oxygen-containing compounds, as well as cell proliferation.
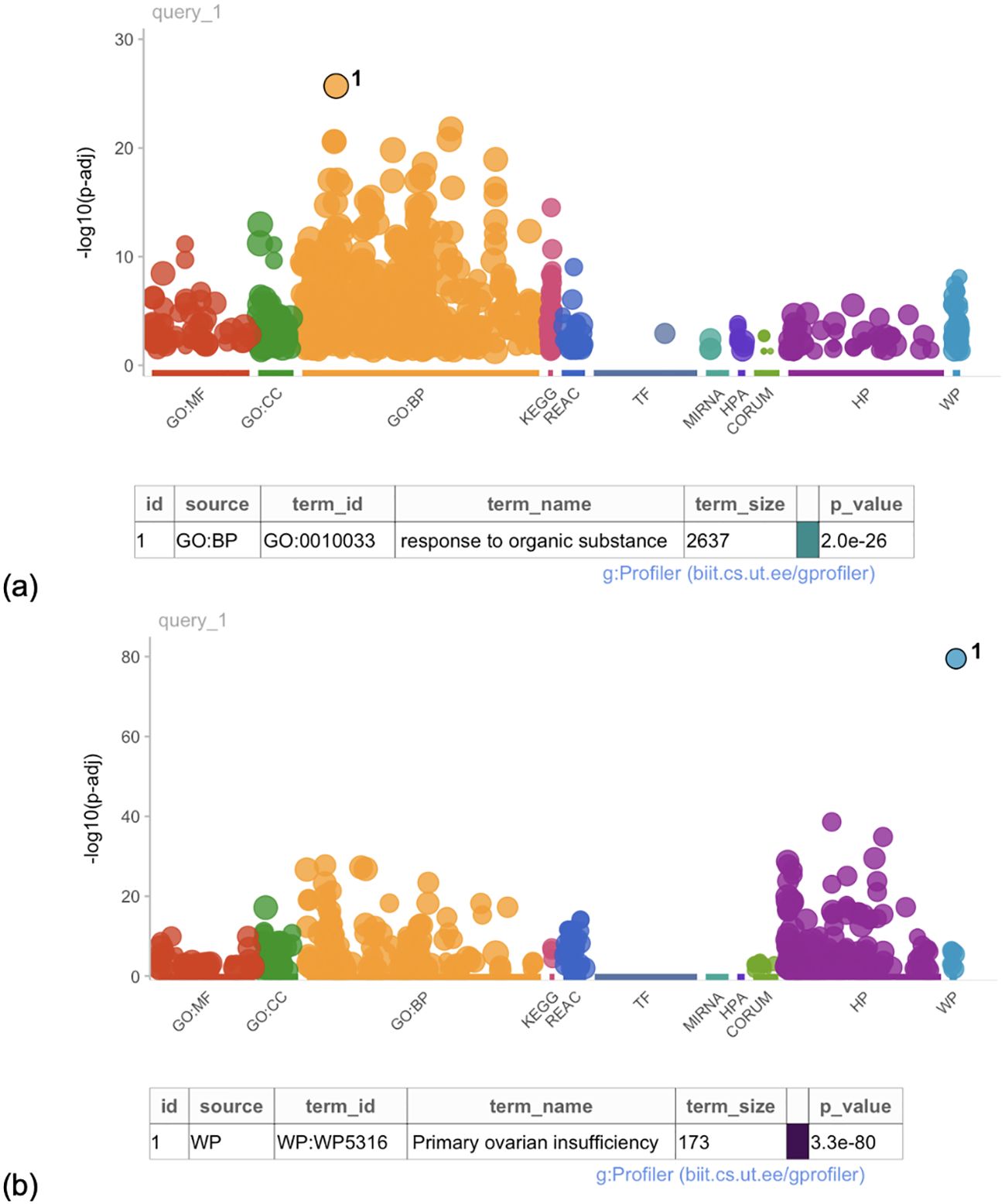
Figure 4. Manhattan plot of the gene ontology analysis for (a) PCOS-associated genes and (b) POI-associated genes. Each dot represents a gene ontology term associated with a gene name extracted from the literature. Gene ontology terms are considered enriched at a p-value < 1e-16.
Genes associated with premature ovarian insufficiency
A total of 19 reports identified genetic targets for POI in this review. Ten of these research articles are case reports. Two articles are experimental studies, and two are observational studies (Figure 3). The number of genes identified in this review associated with POI was 114. Out of these, 16 were identified in more than one research article. The 16 higher confidence genes were BLM, MRPS22, GDF9, NR5A1, MSH4, FIGLA, NOBOX, NUP107, SPIDR, POLG, TP63, STAG3, CLPP, GNAS, CYP21A2, and POR. Majority of the genes identified in this higher confidence gene list are documented in the ClinVar database as having a known association with ovulatory dysfunction of POI. There are nine potentially novel gene targets that are identified as potentially associated with POI in this review: BLM, GDF9, NR5A1, STAR, HSD17B1, FSHR, CLPP, GNAS, and CYP21A2.
One article identified a mitochondrial locus as a contributor to POI: MRPS22 (49). Components of this gene are essential for mitochondrial DNA integrity, mitochondrial ribosomal composition, and ATP production (49, 67).
Gene variants in the POI literature suggest involvement in folliculogenesis and ovarian function. These variants are found in genes such as the GDF9, NOBOX, NUP107, BMP15, NOTCH2, and NR5A1 genes (23, 28, 32, 37, 40, 41, 48, 50, 68). Hormone regulation and synthesis genes were presented by four authors (38, 50, 52, 68).
Variants in FSHR, a gene involved with the cAMP pathway (30) were documented in four POI-specific articles (Figure 2). Two case reports identified novel variants in the FSHR gene in separate populations. In Brazil, a novel homozygous missense mutation caused anovulation for two siblings (30). In Finland, a novel heterozygous mutation impacted the efficacy of the cAMP pathway and downstream caused anovulation (57).
Some gene variants were involved with DNA integrity, transcription, and translation. Yatsenko et al. (2022) reported a variety of variants in the ZSWIM7 gene that are involved with DNA damage response. Franca et al. (2019) described two novel variants in the STAG3 gene involved with genome expression and integrity (Figure 3). Caburet et al. (2014) identified a point mutation in this same gene leading to a premature stop codon, which impacts the development of a protein complex called cohesin. Cohesin plays a critical role in chromosome pairing during meiosis.
The GO term analysis revealed that the most highly enriched GO term across the list of gene names was premature ovarian insufficiency (p = 3.3e-80, Figure 4). In total, however, 43 enriched gene functions were identified as significant for the POI and infertility category. The enriched terms are related to biological process pathways and basic cellular functions. These terms included abnormal morphology of female reproductive physiology and genitalia, DNA damage response and repair, and recombination.
Genes associated with other infertility-related diagnoses
Other diagnoses associated with ovulatory dysfunction and infertility range in type from symptomatic to asymptomatic conditions and account for 16 of the included reports. Ten of these reports are case report studies, and six are observational studies (Figure 3). The disorders include enzyme deficiencies, hormone deficiencies, and unexplained infertility.
The number of genes identified in this review associated with other infertility-related disorders was 21. Out of these, seven were identified in more than one research article. The seven higher confidence genes were CLPP, GNAS, CYP21A2, POR, AR, CYP17A1, and LHCGR. Upon review with the ClinVar database, three genes were identified with pathogenic or likely pathogenic associations to ovulatory dysfunction: POR, AR, and LHCGR. The remaining four genes should be investigated for their potential causal relationship with ODRI.
Lavery et al. (2008) presented a case report on a subject with cortisol reductase deficiency. Other cases of enzyme deficiencies appear to have variants in genes with functions related to hormones and or gonadal development (34, 46, 51).
Sahakitrungruang et al. (2009) and Papadakis et al. (2020) discuss genes involved with P450 oxidoreductase deficiency (PORD), highlighting the role of the POR and the CYP21A2 genes. CYP21A2 is also affected in Congenital Adrenal Hyperplasia, leading to secondary PORD. The function of these genes is critical for hormonal interaction, gonadal development, and folliculogenesis (34). The remaining articles referencing enzyme deficiencies affecting ovulation and fertility report on variants in genes from the same family of P450 enzymes critical for steroidogenesis: CYP17A1, CYP19A1, and CYP21A2 (34, 46, 51).
For the research articles involving a diagnosis of hormone deficiency, the genes identified are FSHB, FSHR, LHB, LHCGR, and GNRH1. Upon review, the functions of these genes are related to ovulation, pubertal development, and hormonal regulation (43, 44, 53). The LHCGR gene is also identified as pathogenic in some cases of idiopathic infertility. Zhang et al. (2020) and Xu et al. (2023) specifically describe novel variants in the gene being correlated to empty follicle syndrome. All GO terms extracted from the gene list associated with “Other diagnoses” revealed no statistically significant findings.
Discussion
This scoping review of the genetics of ODRI revealed a wide range of genetic variants, which are broadly associated with cell cycle components, inflammation, DNA integrity and endocrine functions (33, 46, 48, 55, 69). Through synthesizing the data from the articles included in this review, we highlighted differences in the gene functions based on the diagnosis categories: PCOS, POI, and “Other.” Performing a GO-term enrichment analysis further validated this observation: functions of genes with associations to each respective category yielded unique functional profiles.
PCOS
Literature documenting gene associations with PCOS and infertility referenced variants in hormone-regulatory regions, consistent with the phenotypic points used in the current diagnostic criteria (22, 25, 46, 70). For example, specific polymorphisms in the LHCGR gene are associated with high levels of luteinizing hormone, sometimes measured as a key feature for PCOS diagnosis. It is theorized that an increase in follicle-stimulating hormone and luteinizing hormone levels explains the excess production of androgens typical of this syndrome (66, 71). Hyperinsulinemia is also theorized to increase the production of androgens in patients with PCOS, though this is through a process independent of the mechanism involving the LHCGR gene (39, 72). Knowledge of genetic underpinnings can help direct treatment options for these patients and help to better classify the subtypes of PCOS in a clinical setting.
POI
Some of the genetic association patterns between PCOS and infertility are akin to those identified between POI and infertility in their function, though the individual genes identified are different. Similar functions observed in the two categories involve general cellular function and DNA repair (22, 31, 60, 61).
Specific variants in the STAG3 gene in POI-related infertility are investigated in depth by Franca et al. (2019). Cohesin is critical for genome expression and integrity and contributes to the proper division of chromosomes during meiosis (54). Patients with complete anovulation carried pathogenic variants in STAG3 in both reports (20).
The FSHR gene, involved with the cAMP pathway, is unique to the POI literature as defined by the constructs of this scoping review. This pathway is vital for pubertal development and ovulation (30). Other genetic associations with POI and infertility reveal a similar theme of pubertal development, folliculogenesis, and follicle maturation. These findings support the unique etiology of ovulatory dysfunction and infertility resulting from POI.
Other diagnoses
The genetics of other diagnoses associated with ovulatory dysfunction and infertility were investigated, and 21 genes were identified. The GO-term enrichment analysis did not reveal specific patterns of functionality or characterization of the genes associated with other diagnoses extracted in this scoping review, which reveals an opportune area for future research and discovery.
Interestingly, the LHCGR gene is identified, as it was with the PCOS category. However, the implications of variants in the “other” category are unique. As a result of the variants described by Zhang et al. (2020) and Xu et al. (2023), patients with LHCGR mutations suffered empty follicle syndrome. Empty follicle syndrome is defined as an inability to retrieve oocytes at the time of surgical removal of follicles during an assisted reproductive treatment cycle (73). The effect that the LHCGR gene has on the LH receptor pathway may prevent downstream signaling, thus preventing changes in the collagen matrix and binding of oocytes, as well as ovulation (27, 33, 73).
On the topic of assisted reproductive technology, Papadakis et al. (2020) note that in patients with PORD, unassisted conception is not reported. This enzyme deficiency is considered an extreme form of CAH due to the severity of complications and presentation of symptoms, including genital ambiguity, amenorrhea, and infertility (29).
Given that the primary outcome of these conditions is ovulatory dysfunction and infertility, the diagnostic classification of disease based on phenotypic observations is suboptimal. Consider the case report presented by Lavery et al. (2008), a Cortisol Reductase Deficiency patient presented with a PCOS-like phenotype. After genetic analysis, the gene variants identified did not match known genetic profiles of PCOS patients. The authors illustrate how gene variants affecting the cortisol pathway, not androgens, can mimic the PCOS phenotype (74). Understanding the genetic underpinnings of ODRI conditions may help clinicians better tailor treatment strategies and opens an opportunity for new genetic technological applications in assisted reproductive technology (75).
Limitations
The utilization of strict inclusion and exclusion criteria was both a limitation and a strength in this scoping review. The inclusion of only English language articles may have resulted in the omission of relevant findings, though necessary due to the article curation methodology employed. A single reviewer was responsible for the review of these articles, which may have contributed to article selection bias. This scoping review, however, employed several strategies to support the effectiveness and uniformity of article selection, such as article validation by a second and third reviewer and the use of strict criteria.
By not limiting the timeframe, genetic testing types varied significantly from Sanger sequencing to whole exome sequencing. All research articles that were identified at initial article collection contained significant clinical genetic findings, suggesting that there is inherent publication bias around the genetics of ODRI. No articles identified revealed evidence that ODRI does not have a genetic cause or association.
Although the choice to omit articles due to chromosomal aberration or epigenetic modulations may have impacted the volume of articles that could have been included, it was done so with intention to focus on single nucleotide polymorphisms and variants. This contributes to ODRI as this is an area that is poorly understood.
Many of the articles included in this review were case reports with few or less subjects involved in the study. Although the small sample sizes in these case studies impact the statistical power and reduce the generalizability of the respective papers, the inclusion of all articles that met criteria was imperative to generating a comprehensive list of ODRI genes. Adding a quality scoring component was not performed to maximize the breadth of this scoping review and to identify a broad spectrum of potential gene targets for ODRI for future use.
A comprehensive review was not performed on all 235 genes, rather this was only performed on genes identified in multiple research articles to focus on genes with a higher confidence. This may have resulted in an incomplete analysis of rare pathogenic genetic variants within the analysis that was performed herein. However, the list of 235 genes was successfully compiled and is available for future research and development efforts.
Though there are limitations, this scoping review recovered germane and interesting data from the two largest literature databases. We looked at primary research articles from around the world, which assures diversity. While a large proportion of articles were case studies, including them allowed for a wider view of an area of research that is poorly studied. Additionally, a systematic approach was employed for this scoping review. Utilization of a systematic approach in combination with a quality assurance analysis increases our confidence in the findings extracted from the literature included in this review.
Future directions
This research presents a large and comprehensive list of genes related to ODRI, which have potential applications ranging from research to clinical utility. In research, a list of vetted genes is particularly useful for targeted bioinformatics analyses on large datasets and can be used to discover potentially pathogenic mechanisms related to ODRI (76). This could contribute to understanding the pathogenic mechanisms underlying other types of infertility and discovering novel roles of the candidate genes. Research that helps to identify genetic underpinnings of ODRI could contribute to future clinical strategies through the development or improvement of tools used in infertility treatment, such as biomarker panels and next generation sequencing panels (77, 78).
Conclusion
In this scoping review, we have highlighted the importance of an ongoing investigation on the genetics of ODRI in the literature. We have identified 235 genes related to ODRI in the literature specific to humans, with 28 genes demonstrating a higher potential relationship to ODRI. Our findings provide a comprehensive list of genes available for use in future research and clinical applications.
The list of ODRI genes can be used for targeted gene panel development in assisted reproductive technology to improve clinical testing and diagnosis. More specifically, the use of genetic profiling for patients with ovulatory dysfunction and infertility has the potential to improve patient experience and develop individualized treatments in the future. Our findings support the idea that utilizing genetic testing in infertility treatment may also allow for better classification and treatment of infertility disorders (5, 12, 14, 16), particularly in the realm of ODRI.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ED: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. SS: Supervision, Validation, Writing – review & editing. CH: Supervision, Validation, Writing – review & editing. SR: Conceptualization, Writing – review & editing. LB: Writing – review & editing. JH: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Funding support was provided by Illume Fertility Center exclusively for publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2025.1458711/full#supplementary-material
Supplementary Table 1 | Data extraction table containing journal article demographics and categorical data. See attached File “OvDys_DataExtraction_DiPietro.xlsx”.
Supplementary Table 2 | A list of the 28 gene names with associated variants included in more than one research article with associations to ovulatory dysfunction and infertility where “N” is equal to the number of research articles that mention variants in that specific gene. Genes with a (*) indicate at least one likely pathogenic or pathogenic variant related to ovarian dysfunction, or the respective conditions is present in the ClinVar database.
References
1. Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, de Mouzon J, Sokol R, et al. The international glossary on infertility and fertility care. Fertil Steril. (2017) 32:1786–801. doi: 10.1093/humrep/dex234
2. Gracia C, Anderson J, Amato P, Flyckt R, Hansen K, Hill M, et al. Medicine PC of the AS for R. Definition of infertility: a committee opinion. Fertil Steril. (2023) 120:1170. doi: 10.1016/S0015-0282(23)01971-4
3. Lawrenz B, Melado L, and Fatemi HM. Ovulation induction in anovulatory infertility is obsolete. Reprod Biomed Online. (2023) 46:221–4. doi: 10.1016/j.rbmo.2022.08.102
4. Munro MG, Balen AH, Cho S, Critchley HOD, Díaz I, Ferriani R, et al. FIGO ovulatory disorders classification system. Hum Reprod. (2022) 37:2446–64. doi: 10.1093/humrep/deac180
5. Khan MJ, Ullah A, and Basit S. Genetic basis of polycystic ovary syndrome (PCOS): current perspectives. Appl Clin Genet. (2019) 12:249–60. doi: 10.2147/TACG.S200341
6. Teede HJ, Tay CT, Laven J, Dokras A, Moran LJ, Piltonen TT, et al. Recommendations from the 2023 international evidence-based guideline for the assessment and management of polycystic ovary syndrome. J Clin Endocrinol Metab. (2023) 120:767–93. doi: 10.1016/j.fertnstert.2023.07.025
7. Mumusoglu S and Yildiz BO. Polycystic ovary syndrome phenotypes and prevalence: Differential impact of diagnostic criteria and clinical versus unselected population. Endocr Metab Res. (2020) 12:66–71. doi: 10.1016/j.coemr.2020.03.004
8. Lizneva D, Suturina L, Walker W, Brakta S, Gavrilova-Jordan L, Azziz R, et al. Criteria, prevalence, and phenotypes of polycystic ovary syndrome. Fertil Steril. (2016) 106:6–15. doi: 10.1016/j.fertnstert.2016.05.003
9. Guastella E, Longo RA, and Carmina E. Clinical and endocrine characteristics of the main polycystic ovary syndrome phenotypes. Fertil Steril. (2010) 94:2197–201. doi: 10.1016/j.fertnstert.2010.02.014
10. Rebar RW and Keator CS. Expanding our knowledge of premature ovarian insufficiency. Fertil Steril. (2021) 115:328–9. doi: 10.1016/j.fertnstert.2020.09.145
11. Jo HC, Park JK, Baek JC, Park JE, Kang MY, and Cho IA. Clinicopathological features of premature ovarian insufficiency associated with chromosome abnormalities. J Genet Med. (2019) 16:10–4. doi: 10.5734/JGM.2019.16.1.10
12. Ishizuka B. Current understanding of the etiology, symptomatology, and treatment options in premature ovarian insufficiency (POI). Front Endocrinol. (2021) 12:626924. doi: 10.3389/fendo.2021.626924
13. Webber L, Davies M, Anderson R, Bartlett J, Braat D, Cartwright B, et al. ESHRE Guideline: management of women with premature ovarian insufficiency. Hum Reprod (Oxford, England). (2016) 31:926–37. doi: 10.1093/humrep/dew027
14. Yang Q, Mumusoglu S, Qin Y, Sun Y, and Hsueh AJ. A kaleidoscopic view of ovarian genes associated with premature ovarian insufficiency and senescence. The FASEB journal. (2021) 35:e21753. doi: 10.1096/fj.202100756R
15. Carson SA and Kallen AN. Diagnosis and management of infertility: A review. JAMA. (2021) 326:65–76. doi: 10.1001/jama.2021.4788
16. Zeng Y, Li L, Li Q, Hu J, Zhang N, Wu L, et al. Genetic screening in patients with ovarian dysfunction. Clin Genet. (2023) 103:352–7. doi: 10.1111/cge.14267
17. Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, et al. PRISMA extension for scoping reviews (PRISMA-scR): checklist and explanation. Ann Intern Med. (2018) 169:467–73. doi: 10.7326/M18-0850
18. RefWorks: finding duplicate references in your account . Available online at: www.refworks.comhttps://www.refworks.com/Refworks/help/508help/Finding_Duplicate_References_In_Your_Account.htm.
19. O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. (2015) 44:D733–745. doi: 10.1093/nar/gkv1189
20. Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. (2018) 46:D1062–1067. doi: 10.1093/nar/gkx1153
21. Kolberg L, Raudvere U, Kuzmin I, Vilo J, and Peterson H. gprofiler2 – an R package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler [version 2; peer review: 2 approved. F1000Research. (2020) 9:709. doi: 10.12688/f1000research.24956.2
22. Jansen E, Laven JSE, Dommerholt HBR, Polman J, van Rijt C, van den Hurk C, et al. Abnormal gene expression profiles in human ovaries from polycystic ovary syndrome patients. Mol Endocrinol. (2004) 18:3050–63. doi: 10.1210/me.2004-0074
23. Di Pasquale E, Rossetti R, Marozzi A, Bodega B, Borgato S, Cavallo L, et al. Identification of new variants of human BMP15 gene in a large cohort of women with premature ovarian failure. J Clin Endocrinol Metab. (2006) 91:1976–9. doi: 10.1210/jc.2005-2650
24. Lledó B, Llácer J, Turienzo A, Ortiz JA, Guerrero J, Morales R, et al. Androgen receptor CAG repeat length is associated with ovarian reserve but not with ovarian response. Reprod Biomed Online. (2014) 29:509–15. doi: 10.1016/j.rbmo.2014.06.012
25. Urbanek M, Legro RS, Driscoll DA, Azziz R, Ehrmann DA, Norman RJ, et al. Thirty-seven candidate genes for polycystic ovary syndrome: strongest evidence for linkage is with follistatin. Proc Natl Acad Sci USA. (1999) 96:8573–8. doi: 10.1073/pnas.96.15.8573
26. França MM, Nishi MY, Funari MFA, Lerario AM, Baracat EC, Hayashida SAY, et al. Two rare loss-of-function variants in the STAG3 gene leading to primary ovarian insufficiency. Eur J Med Genet. (2019) 62:186–9. doi: 10.1016/j.ejmg.2018.07.008
27. Zhang Z, Wu L, Diao F, Chen B, Fu J, Mao X, et al. Novel mutations in LHCGR (luteinizing hormone/choriogonadotropin receptor): expanding the spectrum of mutations responsible for human empty follicle syndrome. J Assist Reprod Genet. (2020) 37:2861–8. doi: 10.1007/s10815-020-01931-2
28. Sassi A, Désir J, Duerinckx S, Soblet J, Van Dooren S, Bonduelle M, et al. Compound heterozygous null mutations of NOBOX in sisters with delayed puberty and primary amenorrhea. Mol Genet Genomic Med. (2021) 9:e1776. doi: 10.1002/mgg3.v9.10
29. Sahakitrungruang T, Huang N, Tee MK, Agrawal V, Russell WE, Crock P, et al. Clinical, genetic, and enzymatic characterization of P450 oxidoreductase deficiency in four patients. J Clin Endocrinol Metab. (2009) 94:4992–5000. doi: 10.1210/jc.2009-1460
30. França MM, Lerario AM, Funari MFA, Nishi MY, Narcizo AM, de Mello MP, et al. A novel homozygous missense FSHR variant associated with hypergonadotropic hypogonadism in two siblings from a Brazilian family. Sex Dev. (2017) 11:137–42. doi: 10.1159/000477193
31. Li L, Feng F, Zhao M, Li T, Yue W, Ma X, et al. NOTCH2 variant D1853H is mutated in two non-syndromic premature ovarian insufficiency patients from a Chinese pedigree. J Ovarian Res. (2020) 13 doi: 10.1186/s13048-020-00645-4
32. Sakka R, Abdelhedi F, Sellami H, Pichon B, Lajmi Y, Mnif M, et al. An unusual familial Xp22.12 microduplication including EIF1AX: A novel candidate dosage-sensitive gene for premature ovarian insufficiency. Eur J Med Genet. (2022) 65:104613. doi: 10.1016/j.ejmg.2022.104613
33. Xu Y, Wang E, Liu T, Wang S, Wu F, Zhao X, et al. Whole exome sequencing identifies a novel homozygous missense mutation of LHCGR gene in primary infertile women with empty follicle syndrome. J Obstet Gynaecol Res. (2023) 49:2436–45. doi: 10.1111/jog.v49.10
34. Holmes-Walker DJ, Conway GS, Honour JW, Rumsby G, and Jacobs HS. Menstrual disturbance and hypersecretion of progesterone in women with congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Clin Endocrinol. (1995) 43:291–6. doi: 10.1111/j.1365-2265.1995.tb02034.x
35. Gónzalez A, Ramírez-Lorca R, Calatayud C, Mendoza N, Ruiz A, Sáez ME, et al. Association of genetic markers within the BMP15 gene with anovulation and infertility in women with polycystic ovary syndrome. Fertil Steril. (2008) 90:447–9. doi: 10.1016/j.fertnstert.2007.06.083
36. Kaur R, Kaur T, and Kaur A. Genetic association study from North India to analyze association of CYP19A1 and CYP17A1 with polycystic ovary syndrome. J Assist Reprod Genet. (2018) 35:1123–9. doi: 10.1007/s10815-018-1162-0
37. Zhang T, Ma Q, Shen Q, Jiang C, Zou F, Shen Y, et al. Identification of novel biallelic variants in BMP15 in two siblings with premature ovarian insufficiency. J Assist Reprod Genet. (2022) 39:2125–34. doi: 10.1007/s10815-022-02574-1
38. Bachelot A, Rouxel A, Massin N, Dulon J, Courtillot C, Matuchansky C, et al. Phenotyping and genetic studies of 357 consecutive patients presenting with premature ovarian failure. Eur J Endocrinol. (2009) 161:179–87. doi: 10.1530/EJE-09-0231
39. Zhang Y, Ho K, Keaton JM, Hartzel DN, Day F, Justice AE, et al. A genome-wide association study of polycystic ovary syndrome identified from electronic health records. Am J Obstet Gynecol. (2020) 223:559.e1–559.e21. doi: 10.1016/j.ajog.2020.04.004
40. Tucker EJ, Gutfreund N, Belaud-Rotureau MA, Gilot D, Brun T, Kline BL, et al. Dominant TP63 missense variants lead to constitutive activation and premature ovarian insufficiency. (2022) 43:1443–53. doi: 10.1002/humu.v43.10
41. Marinakis NM, Tsoutsou E, Sofocleous C, Veltra D, Papaefthimiou P, Lytras A, et al. Ovarian insufficiency and secondary amenorrhea in a patient with a novel variant within GDF9 gene. Menopause (New York, N.Y.). (2022) 29:491–5. doi: 10.1097/GME.0000000000001928
42. Vishnubotla DS, Shek AP, and Madireddi S. Pooled genetic analysis identifies variants that confer enhanced susceptibility to PCOS in Indian ethnicity. Gene. (2020) 752:144760. doi: 10.1016/j.gene.2020.144760
43. Sagi SV, Joshi H, Whiles E, Hikmat M, Puthi VR, MacDougall J, et al. Normosmic idiopathic hypogonadotropic hypogonadism due to a novel GNRH1 variant in two siblings. Endocrinol Diabetes Metab Case Rep. (2020) 2020:19-0145. doi: 10.1530/EDM-19–0145
44. Zhu L, Xiao N, Zhang T, Kong P, Xu B, Fang Z, et al. Clinical and genetic analysis of an isolated follicle-stimulating hormone deficiency female patient. J Assist Reprod Genet. (2020) 37:1441–8. doi: 10.1007/s10815-020-01786-7
45. Papadakis GE, Dumont A, Bouligand J, Chasseloup F, Raggi A, Catteau-Jonard S, et al. Non-classic cytochrome P450 oxidoreductase deficiency strongly linked with menstrual cycle disorders and female infertility as primary manifestations. Hum Reprod (Oxford, England). (2020) 35:939–49. doi: 10.1093/humrep/deaa020
46. Zhang D, Yao F, Luo M, Wang Y, Tian T, Deng S, et al. Clinical characteristics and molecular etiology of partial 17α-hydroxylase deficiency diagnosed in 46,XX patients. Front Endocrinol. (2022) 13:978026. doi: 10.3389/fendo.2022.978026
47. Yatsenko SA, Gurbuz F, Topaloglu AK, Berman AJ, Martin PM, Rodrigue-Escribà M, et al. Pathogenic variants in ZSWIM7 cause primary ovarian insufficiency. J Clin Endocrinol Metab. (2022) 107:e2359–64. doi: 10.1210/clinem/dgac090
48. Ke H, Tang S, Guo T, Hou D, Jiao X, Li S, et al. Landscape of pathogenic mutations in premature ovarian insufficiency. Nat Med. (2023) 29:483–92. doi: 10.1038/s41591-022-02194-3
49. Chen AL, Tiosano D, Guran T, Baris HN, Bayram Y, Mory A, et al. Mutations in the mitochondrial ribosomal protein MRPS22 lead to primary ovarian insufficiency. Human Mol Genet. (2018) 27:1913–26. doi: 10.1093/hmg/ddy098
50. Domenice S, MaChado AZ, Ferreira FM, Ferraz-de-Souza B, Lerario AM, Lin L, et al. Wide spectrum of NR5A1-related phenotypes in 46,XY and 46,XX individuals. Birth Defects Res C: Embryo Today: Reviews. (2016) 108:309–20. doi: 10.1002/bdrc.v108.4
51. Xu Y, Jiang ST, Yan Z, Niu Y, Du WH, Liu BL, et al. Phenotypic heterogeneity and fertility potential of patients with 17-hydroxylase/17,20-lyase deficiency. J Clin Endocrinol Metab. (2022) 107:E2610–8. doi: 10.1210/clinem/dgac029
52. Brayman MJ, Pepa PA, Berdy SE, and Mellon PL. Androgen receptor repression of gnRH gene transcription. Mol Endocrinol. (2012) 26:2–13. doi: 10.1210/me.2011-1015
53. Lofrano-Porto A, Barra GB, Giacomini LA, Nascimento PP, Latronico AC, Casulari LA, et al. Luteinizing hormone beta mutation and hypogonadism in men and women. N Engl J Med. (2007) 357:897–904. doi: 10.1056/NEJMoa071999
54. Caburet S, Arboleda VA, Llano E, Overbeek PA, Barbero JL, Oka K, et al. Mutant cohesin in premature ovarian failure. N Engl J Med. (2014) 370:943–9. doi: 10.1056/NEJMoa1309635
55. Pirtea P, Haggerty E, Hagege E, Tran C, de Ziegler D, Farabet C, et al. Successful ART outcome in a woman with McCune-Albright syndrome: a case report and literature review. J Assist Reprod Genet. (2023) 40:1669–75. doi: 10.1007/s10815-023-02844-6
56. Ren Y, Diao FY, Katari S, Yatsenko S, Jiang HY, Wood-Trageser MA, et al. Functional study of a novel missense single-nucleotide variant of NUP107 in two daughters of Mexican origin with premature ovarian insufficiency. Mol Genet Genomic Med. (2018) 6:276–81. doi: 10.1002/mgg3.2018.6.issue-2
57. Doherty E, Pakarinen P, Tiitinen A, Kiilavuori A, Huhtaniemi I, Forrest S, et al. A novel mutation in the FSH receptor inhibiting signal transduction and causing primary ovarian failure. J Clin Endocrinol Metab. (2002) 87:1151–5. doi: 10.1210/jcem.87.3.8319
58. Lavery GG, Walker EA, Tiganescu A, Ride JP, Shackleton C, Tomlinson JW, et al. Steroid biomarkers and genetic studies reveal inactivating mutations in hexose-6-phosphate dehydrogenase in patients with cortisone reductase deficiency. J Clin Endocrinol Metab. (2008) 93:3827–32. doi: 10.1210/jc.2008-0743
59. Sinha S, Kar K, Chowdhury PP, and Dasgupta A. Single nucleotide polymorphism of insulin receptor gene rs2059806 in polycystic ovary syndrome- A case-control study. J Clin Diagn Res. (2021) 15:GC07–10. doi: 10.7860/JCDR/2021/49653.15395
60. Amjadi F, Zandieh Z, Mehdizadeh M, Ajdary M, Aghamajidi A, Raoufi E, et al. Molecular signature of immunological mechanism behind impaired endometrial receptivity in polycystic ovarian syndrome. Arch Endocrinol Metab. (2022) 66:303–11. doi: 10.20945/2359-3997000000476
61. Rossetti R, Moleri S, Guizzardi F, Gentilini D, Libera L, Marozzi A, et al. Targeted next-generation sequencing indicates a frequent oligogenic involvement in primary ovarian insufficiency onset. Front Endocrinol. (2021) 12. doi: 10.3389/fendo.2021.664645
62. Cyranska-Chyrek E, Filipowicz D, Szczepanek-Parulska E, Nowaczyk M, Ambroziak U, Toutounchi S, et al. Primary pigmented nodular adrenocortical disease (PPNAD) as an underlying cause of symptoms in a patient presenting with hirsutism and secondary amenorrhea: case report and literature review. Gynaecol Endocrinol. (2018) 34:1022–6. doi: 10.1080/09513590.2018.1493101
63. Dursun F, Mohamoud H, Karim N, Naeem M, Jelani M, and Kirmizibekmez H. A novel missense mutation in the CLPP gene causing perrault syndrome type 3 in a turkish family. J Clin Res Pediatr Endocrinol. (2016) 8:472–7. doi: 10.4274/jcrpe.2717
64. Ajmal N, Khan SZ, Asmat R, Khan P, Sadia H, Sajjad N, et al. Association of SNP rs700519 in CYP19A1 Gene with Polycystic Ovary Syndrome (PCOS) among Females of Quetta, Pakistan. Int J Hum Genet. (2021) 21:96–101. doi: 10.31901/24566330.2021/21.02.782
65. Caronia LM, Martin C, Welt CK, Sykiotis GP, Quinton R, Thambundit A, et al. A genetic basis for functional hypothalamic amenorrhea. N Engl J Med. (2011) 66:618–9. doi: 10.1097/OGX.0b013e31822f94c4
66. Atoum MF, Alajlouni MM, and Alzoughool F. A case-control study of the luteinizing hormone level in luteinizing hormone receptor gene (rs2293275) polymorphism in polycystic ovarian syndrome females. Public Health Genomics. (2022) 25:89–97. doi: 10.1159/000521971
67. Khodamoradi K, Khosravizadeh Z, Rashidi Z, Talebi A, and Hassanzadeh G. The role of mitochondria in premature ovarian failure: A review. J Contemp Med Sci. (2020) 6:1–7. doi: 10.22317/jcms.v6i1.712
68. Rossetti R, Ferrari I, Bonomi M, and Persani L. Genetics of primary ovarian insufficiency. Clin Genet. (2017) 91:183–98. doi: 10.1111/cge.2017.91.issue-2
69. Saeed S, Hassan J, Javed SM, Shan S, and Naz M. A familial case of robertsonian translocation 13;14: case report. Cureus. (2022) 14:e29430. doi: 10.7759/cureus.29430
70. Lizneva D, Suturina L, Walker W, Brakta S, Gavrilova-Jordan L, Azziz R., et al. Criteria, prevalence, and phenotypes of polycystic ovary syndrome. Fertil Steril. (2016) 106:6–15. doi: 10.1016/j.fertnstert.2016.05.003
71. Hawkins SM and Matzuk MM. The menstrual cycle: basic biology. Ann NY Acad Sci. (2008) 1135:10–8. doi: 10.1196/nyas.2008.1135.issue-1
72. Unluhizarci K, Karaca Z, and Kelestimur F. Role of insulin and insulin resistance in androgen excess disorders. World J Diabetes. (2021) 12:616–29. doi: 10.4239/wjd.v12.i5.616
73. Revelli A, Carosso A, Grassi G, Gennarelli G, Canosa S, and Benedetto C. Empty follicle syndrome revisited: definition, incidence, aetiology, early diagnosis and treatment. Reprod Biomed Online. (2017) 35:132–8. doi: 10.1016/j.rbmo.2017.04.012
74. Lavery GG, Walker EA, Tiganescu A, Ride JP, Shackleton C, Tomlinson JW, et al. Steroid biomarkers and genetic studies reveal inactivating mutations in hexose-6-phosphate dehydrogenase in patients with cortisone reductase deficiency. J Clin Endocrinol Metab. (2008) 93:3827–32. doi: 10.1210/jc.2008-0743
75. Yang Q, Mumusoglu S, Qin Y, Sun Y, and Hsueh AJ. A kaleidoscopic view of ovarian genes associated with premature ovarian insufficiency and senescence. FASEB Journal: Official Publication of The Federation of American Societies For Experimental Biology. (2021) 35:. doi: 10.1096/fj.202100756R
76. Uffelmann E, Huang QQ, Munung NS, de Vries J, Okada Y, Martin AR, et al. Genome-wide association studies. (2021). Available online at: https://explore.openaire.eu/search/result?id=doi_dedup:_::b520aa26b836ff5258fc2ff56d1254a1 (Accessed November 10, 2023).
77. Agharezaee N, Hashemi M, Shahani M, and Gilany K. Male infertility, precision medicine and systems proteomics. J Reprod Infertil. (2018) 19:185–92.
Keywords: ovulatory and anovulatory cycles, ovulatory dysfunction, genetics, infertility, ovulatory dysfunctional infertility, genes
Citation: DiPietro EE, Sarasua SM, Hopkins CS, Ganakammal SR, Boccuto L and Hurwitz J (2025) Genetics of ovulatory dysfunction and infertility: a scoping review and gene ontology analysis. Front. Endocrinol. 16:1458711. doi: 10.3389/fendo.2025.1458711
Received: 02 July 2024; Accepted: 01 May 2025;
Published: 04 June 2025.
Edited by:
Vikas Kumar Roy, Mizoram University, IndiaReviewed by:
Ashraf Akintayo Akintola, Kyungpook National University, Republic of KoreaXiangping Peng, Waseda University, Japan
Copyright © 2025 DiPietro, Sarasua, Hopkins, Ganakammal, Boccuto and Hurwitz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erin E. DiPietro, ZWRpcGlldEBnLmNsZW1zb24uZWR1
 Erin E. DiPietro
Erin E. DiPietro Sara M. Sarasua
Sara M. Sarasua Casey S. Hopkins1
Casey S. Hopkins1 Satishkumar Ranganathan Ganakammal
Satishkumar Ranganathan Ganakammal Luigi Boccuto
Luigi Boccuto