- 1Department of Traditional Chinese Medical Gynecology, Hangzhou Traditional Chinese Medicine (TCM) Hospital Affiliated to Zhejiang Chinese Medical University, Hangzhou, China
- 2Research Institute of Women’s Reproductive Health, Zhejiang Chinese Medical University, Hangzhou, China
- 3Zhejiang Key Laboratory of Precise Protection and Promotion of Fertility, Hangzhou, China
- 4School of Basic Medical Sciences, Zhejiang Chinese Medical University, Hangzhou, China
- 5Department of Pathology, Zhejiang Cancer Hospital, Hangzhou, China
Background: Among women of childbearing age, polycystic ovary syndrome (PCOS) is the predominant etiology of anovulatory infertility. Recent research has elucidated the role of melatonin as a medicinal agent in PCOS, especially hyperandrogenism. However, the precise mechanisms underlying its therapeutic efficacy remain largely unknown. This study integrated network pharmacology, molecular docking, molecular dynamics simulations, and laboratory confirmation to explore the pharmacological mechanisms of melatonin in PCOS.
Methods: First, we conducted animal studies to evaluate the therapeutic efficacy of melatonin by administering it to circadian disruption-induced PCOS-like rats. Prospective medicinal targets of melatonin were acquired from databases such as DrugBank, Traditional Chinese Medicine Systems Pharmacology, PharmMapper, and SwissTarget Prediction. Targets related to PCOS were extracted from three databases: DisGeNET, GeneCards, and the National Center for Biotechnology Information gene. To visualize the relationships between proteins, a protein-protein interaction network was generated using the STRING database. Further investigation of these targets involved analyzing protein-protein interaction networks and conducting GO/KEGG enrichment analysis. Molecular docking techniques were employed to examine the interactions between melatonin and crucial targets. Molecular dynamics simulations were performed to confirm the stability of the association between the hub targets and the melatonin ligand. Finally, animal studies validated the effect of melatonin on the identified targets.
Results: Animal experiments showed that melatonin ameliorated hyperandrogenism and ovarian dysfunction in constant darkness-induced PCOS-like rats. Network pharmacology analysis demonstrated that melatonin exhibited multiple modulatory effects on circadian rhythm, reproductive processes, metabolic processes, and oocyte maturation. Cytoscape network analysis revealed seven key targets, of which AR and CYP19A1 showed the highest affinity for melatonin by molecular docking. The stability of the AR/CYP19A1-melatonin complex was verified through computational simulations using molecular dynamics techniques. Furthermore, animal experiments have validated that melatonin can regulate key genes associated with hyperandrogenism, including AR and CYP19A1.
Conclusion: Through network pharmacology, molecular docking, and experimental validation, this study reveals how melatonin may ameliorate PCOS and hyperandrogenism. Results suggest melatonin’s effects involve androgen excess mitigation, though further validation is needed. This work provides insight into melatonin’s actions in circadian-associated PCOS.
Introduction
Globally, polycystic ovary syndrome (PCOS) affects 5–18% of women of childbearing age. This complex, chronic condition significantly impacts reproductive, metabolic, and emotional health throughout a woman’s life (1). A hallmark reproductive feature of PCOS is ovarian dysfunction, characterized by hyperandrogenism and irregular or absent ovulation (2). The origins of PCOS are multifactorial and not yet fully elucidated, encompassing genetic predisposition, exposure to environmental toxins, dietary factors, and nutritional status (3). Modern lifestyle changes and erratic sleep habits often lead to disrupted circadian rhythms (4). Critically, substantial evidence now links circadian rhythm disturbances directly to PCOS pathogenesis, highlighting circadian rhythm disruption as a significant environmental risk factor (5).
The molecular basis of circadian rhythms lies in transcriptional-translational feedback loops, primarily involving CLOCK/BMAL1 and PER/CRY proteins. The suprachiasmatic nucleus (SCN) in the hypothalamus acts as the central pacemaker, synchronizing peripheral clocks throughout the body in response to light-dark cycles (6, 7). This circadian system regulates numerous biological processes, including endocrine function, metabolism, and sleep (8, 9). Perturbations in core clock genes, such as Brain and Muscle ARNT-like protein-1 (BMAL1), have been implicated in PCOS pathogenesis, particularly hyperandrogenism (10). Mechanistically, BMAL1 influences the expression of aromatase (CYP19A1), the enzyme converting androgens to estrogens, within estrogen-producing cells (11, 12). Reduced BMAL1 expression correlates strongly with peripheral and ovarian hyperandrogenism (10, 13).
Melatonin, the primary hormone secreted by the pineal gland in response to darkness, is a crucial regulator of circadian rhythms (14). Its effects are mediated primarily through membrane-bound receptors, MTNR1A and MTNR1B (15). Notably, melatonin receptors are expressed within ovarian structures, suggesting a role in regulating sex steroid production during follicular development (16). Polymorphisms in both MTNR1A and MTNR1B genes are associated with PCOS, further strengthening the link between circadian abnormalities and the syndrome (17–19).
Clinical evidence supports that PCOS patients exhibit reduced serum melatonin levels (5) and notably lower follicular melatonin concentrations (5, 20). Importantly, melatonin supplementation significantly reduces testosterone levels in women with PCOS (21, 22). Preclinical studies corroborate this, demonstrating that melatonin improves reproductive and metabolic phenotypes in various PCOS animal models (e.g., Letrozole or DHEA-induced) (23, 24). Collectively, melatonin may prove to be a viable therapeutic option for PCOS management, pending further investigation. However, the precise molecular mechanisms underlying melatonin’s therapeutic efficacy in PCOS remain largely unknown.
Constant darkness exposure provides a robust experimental model of circadian disruption (25). Recent studies show it reliably induces both reproductive (hyperandrogenism, irregular cycles) and metabolic hallmarks of PCOS in rats (10, 26, 27). Crucially, the characteristic downregulation of core clock genes (e.g., Bmal1) observed in these rats mirrors findings in women with PCOS. This model, therefore, offers valuable insights into circadian dysrhythmia-induced PCOS phenotypes and potential therapeutic strategies. Consequently, this study aimed to identify melatonin’s potential targets and elucidate its therapeutic processes in PCOS. We employed an integrated strategy combining network pharmacology, molecular docking, molecular dynamics simulations, and experimental validation in a constant darkness-induced PCOS-like rat model to explore the plausible mechanisms of melatonin action, providing foundational data for future preclinical research.
Methods
Preparing for constant darkness-induced PCOS-like rats
Eighteen female Sprague-Dawley (SD) rats of 5-week-old (Charles River Laboratory Animal Technology, Beijing, China) were housed in Animal Centre of Zhejiang Chinese Medical University (Hangzhou, Zhejiang, China), accredited by Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). The experimental animals were maintained in a specific-pathogen-free (SPF) grade conditions with temperatures ranging from 22°C to 24°C and humidity ranging from 40°C to 65°C. The rats had unrestricted access to regular feed and drinking water in their enclosures at the Laboratory Animal Research Center. The Animal Ethics Committee of Zhejiang Chinese Medical University authorized all experimental procedures (Approval No. 20220815-11).
After adaptive feeding for 1 week, the rats were equally and randomly assigned three groups (by Ying Zhu): control, darkness, and melatonin group (n=6). Figure 1A illustrates the experimental design of the animal study. The control group was kept under a normal 12-hour light/dark (L/D) alternating exposure (illumination beginning at 7:30 AM, Zeitgeber Time 0, ZT0). Both the darkness and melatonin group were housed in constant darkness for 8 weeks. In the melatonin group, rats were intraperitoneally administered melatonin daily at ZT11, with a dosage of 10 mg/kg/day, which was widely preferred in rodent models (10, 28). The control and darkness group were intraperitoneally administered with saline of equal volume. Whole rat body weight was measured weekly throughout the 8-week period. Rats were sacrificed under the effects of 5% isoflurane anesthesia during the diestrus II stage for tissue sample collection.
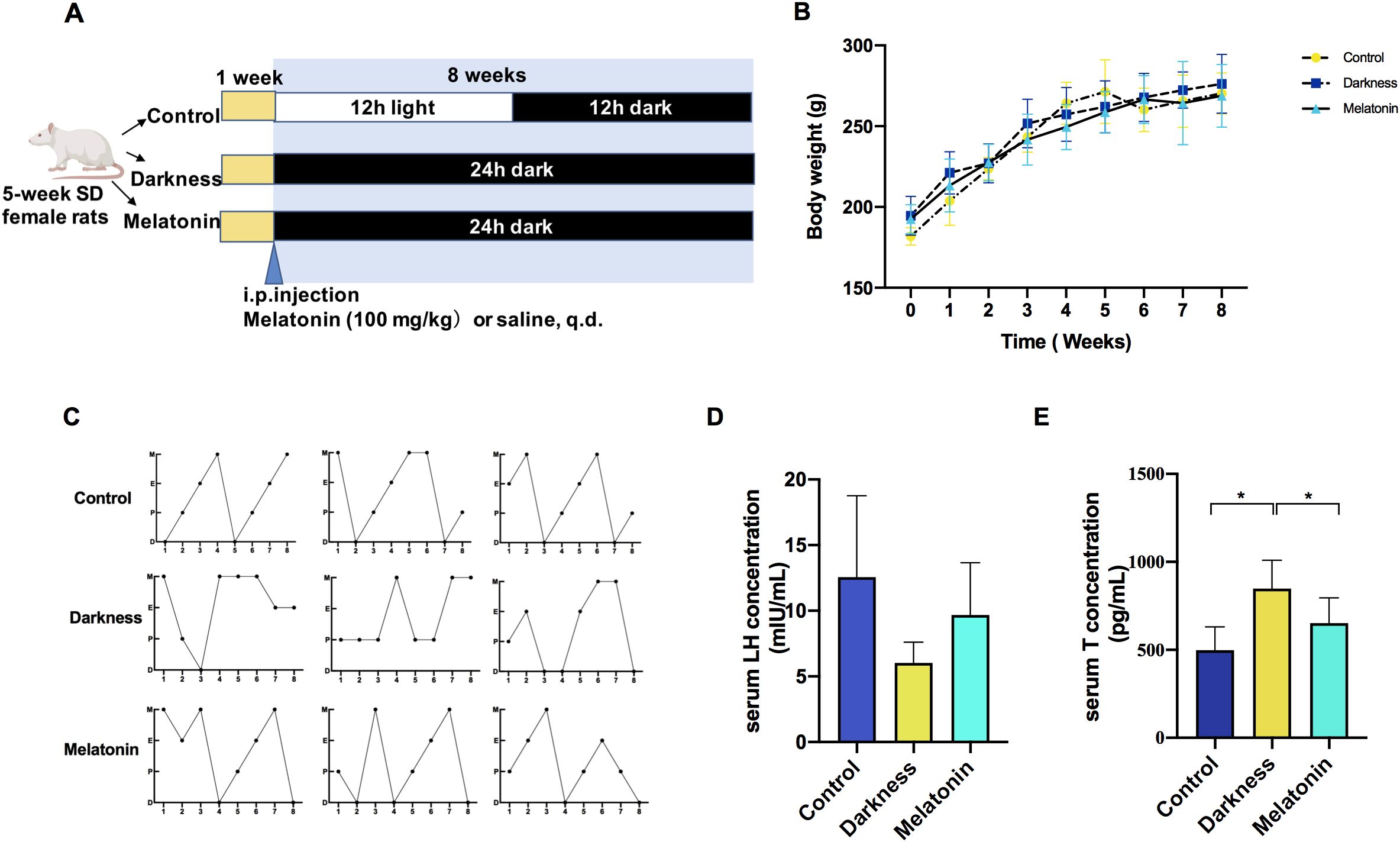
Figure 1. Melatonin treatment ameliorates reproductive hallmarks of PCOS in rats exposed to constant darkness. (A) Timeline of an experiment into darkness-induced PCOS and melatonin treatment. (B) Body weight changes, (C) Representative estrous cycles. M represents for metestrus; D represents for diestrus; P represents for proestrus; E represents for estrus, (D, E) Serum concentrations of LH and testosterone by ELISA. Statistical analysis (D, E) was performed with one-way ANOVA followed by Newman–Keuls multiple comparison test. N=6 per group. Data are expressed as mean± SD. *P < 0.05.
Vaginal cytology analysis
For eight consecutive days during rat model development, vaginal smears were examined to determine the estrous cycle stages (by Ran Cheng and Ying Zhu). The procedure involved collecting vaginal secretions from each rat between 9:00 and 10:00 a.m. by rinsing the vagina with a saline solution. Collected secretions were then placed on a glass slide for microscopic analysis. The estrous cycle stages were classified into four categories: metestrus (diestrus I), characterized by a small number of cornified cells and numerous leukocytes; diestrus (diestrus II), primarily composed of leukocytes; proestrus, distinguished by a high concentration of nucleated epithelial cells and estrus, dominated by large, flat, irregularly shaped a nucleated cornified epithelial cells.
PBMC isolation
Peripheral blood mononuclear cells (PBMCs) were obtained from whole blood by standard Ficoll-Hypaque density centrifugation using rat peripheral blood lymphocyte isolation fluid (#KGA3103, KeyGEN Biotechnology) in accordance with the producer’s instructions (by Ran Cheng and Ying Zhu). The mononuclear cells underwent three washing cycles using PBS before being preserved at -80°C for subsequent extraction of total RNA.
Hematoxylin and eosin staining
Rat ovarian tissues were harvested and fixed with 4% paraformaldehyde prior to paraffin embedding (by Sheng-kai Wang). Sections of tissue, 5 μm in thickness, were prepared and subsequently subjected to dewaxing and rehydration using a graded series of ethanol solutions. Tissue samples were colored with hematoxylin for 5 min, and differentiated with hydrochloric acid for 30 s. Finally, the specimens were stained with eosin for 2 minutes and mounted on slides for microscopic examination.
ELISA
The concentrations of LH and testosterone in rat sera were detected using the Rat LH ELISA Kit (#ELK2367, ELK Biotechnology) and Testosterone ELISA Kit (#ELK1332, ELK Biotechnology), respectively (by Chun-xiao Zong). The protocols were performed in accordance with the guidelines.
Determination of quantitative real‐time PCR
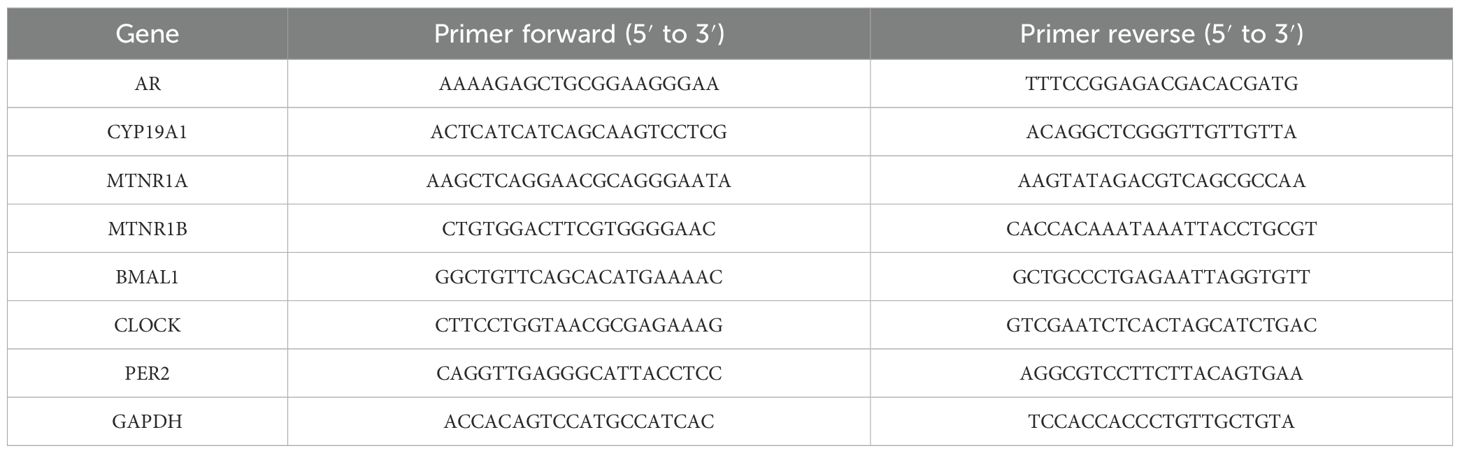
Total RNA from PBMCs and rat ovary tissues was isolated with TRIzol reagent (Invitrogen) and then reverse-transcribed into cDNA (#RR036A, Takara Bio, Dalian, China). RT-qPCR (#RR820A, Takara Bio, Dalian, China) was employed to measure the mRNA expression of target genes (by Ying Zhu). The ΔΔCt method was subsequently utilized for data analysis. The target mRNA level was calculated as the ratio of the target gene to GAPDH. Table 1 tabulates the primer sequences used for the target genes.
Identification of potential medicinal targets of melatonin
To obtain the spatial configuration and Simplified Molecular-Input Line-Entry Specification (SMILES) notation of melatonin, a search was conducted in the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (29). Subsequently, the 3D structure was input into the PharmMapper-server for potential target prediction, whereas SMILES was submitted to SwissTargetPrediction (http://www.swisstargetprediction.ch/) (30, 31). Furthermore, the DrugBank and Traditional Chinese Medicine Systems Pharmacology (TCMSP) (https://tcmspw.com/tcmsp.php) were utilized to recognized melatonin targets (32, 33). The retrieved target names were verified against authorized names in the UniProt database (https://www.uniprot.org/), and duplicate entries were eliminated. Cytoscape 3.8.2 was employed to visualize the results (by Ran Cheng and Chun-xiao Zong).
Identification of PCOS-associated targets
To identify targets associated with PCOS, researchers (by Ran Cheng and Sheng-kai Wang) have utilized three gene databases: GeneCards, DisGeNET, and NCBI (34, 35). The search terms “Polycystic ovary syndrome” and “PCOS” were employed. To ensure result reliability, GeneCards had a minimum relevance score of minimum 0.7, and a gene-disease score of 0.1 or higher, was applied. The STRING database (version 11.5) was used to predict the network of interactions between proteins (PPI) for the given gene sets with a high-confidence interaction score (0.900). Cytoscape software 3.8.2 was then used to visualize the network construction.
Protein-protein interaction network construction and discovery of hub genes
To identify potential medicinal targets, researchers (by Ran Cheng and Sheng-kai Wang) intersected melatonin and PCOS targets using the Venn Diagram tool. The PPI network of the gene lists was predicted using the STRING database with a moderate confidence interaction score (0.400) (36). The Cytoscape plugin “CytoHubba” was employed to identify hub genes using three topological analysis methods: Maximal Clique Centrality (MCC), Maximum Neighborhood Component Centrality (DMNC), and Neighborhood Component Centrality (MNC) (37).
Gene Ontology, Kyoto Encyclopedia of Genes and Genomes enrichment analysis
DAVID (Database for Annotation, Visualization, and Integrated Discovery) equips researchers with an extensive array of functional characterization instruments, enabling them to uncover the biological relevance of substantial gene collections (38). We employed DAVID tools to conduct GO and KEGG pathway enrichment analyses to elucidate the functions of the potential medicinal targets as well as signaling pathways (by Ran Cheng and Sheng-kai Wang). The results of these analyses were visualized using a freely available online bioinformatics platform (https://www.bioinformatics.com.cn/).
Molecular docking
The Protein Data Bank (39) supplied the 3D crystal structures of the target proteins in the PDB format. The PyMOL software (40)was used to eliminate water molecules and excess ligands. Subsequently, AutoDock tools (1.5.6) were utilized to incorporate protons and generate the PDBQT format of the proteins (41).
The 2D SDF structure of melatonin was obtained from PubChem (42). Energy minimization was performed using Chem 3D software (21.0.0) and the mol2 format was created (43). AutoDock tools (version 1.5.6) were then used to create the PDBQT format of melatonin.
Molecular docking with AutoDock Vina (40) was employed to assess melatonin binding and the hub pivot targets. Prior to molecular docking, proteins were represented as secondary structures. AutoDock Vina computed the binding affinity, while PyMol 2.5.4 (open-source version) was displayed to visually render the docking results (by Ran Cheng).
Molecular dynamic simulation
The configurations exhibiting the two leading scores derived through molecular docking were subsequently subjected to molecular dynamics (MD) simulations utilizing Gromacs 2022.3 software (44, 45). The preparation of small molecules involved pretreatment with AmberTools22 and hydrogenation using Gaussian 16 W to determine the response potential. The computed potential data were incorporated into the MD system’s topology file. Throughout the simulation, the conditions were kept constant at 300 K and 1 bar pressure. The simulation employed the amber99sb ildn force field, and Tip3p was chosen as the water model for solvation. To achieve a neutral total charge in the simulated system, an appropriate amount of Na+ ions was introduced. The energy of the simulation system was minimized through a two-step process: first, the steepest descent technique was employed, followed by equilibration procedures. These included 100000 steps each of NVT (constant volume and temperature) and NPT (constant pressure and temperature) equilibration, utilizing a coupling constant of 0.1 ps over a 100ps time period. After equilibration, a free molecular dynamics simulation was performed, comprising five million steps with a 2fs step size, amounting to 100ns. A post-simulation assessment was conducted using the built-in tools of the software. This analysis encompassed the calculations of RMSD, RMSF, and rotational radius of the protein for each amino acid trajectory, along with free energy (MMGBSA) computations and generation of free energy topographic maps, among other relevant data (by Ran Cheng). These structural predictions provide a theoretical basis for the potential mechanism of action, but they need to be comprehensively analyzed in conjunction with the expression changes at the transcriptional level.
Statistical analysis
In the Network Pharmacology section, topological analysis was performed using Cytoscape 3.8.2. The DAVID tools were applied for GO and KEGG enrichment analyses. AutoDock tools (1.5.6) were utilized to construct molecular docking simulations with PyMol 2.5.4, which were constructed using Gromacs 2022.3 software. Data evaluation was performed using GraphPad Prism 8 and SPSS version 25. As for the enrichment analysis, statistical significance was determined by a P -value < 0.05. In the section of experimental validation, results are expressed as mean ± SD. Each experiment was performed three times on an average. Data were first analyzed using the Kolmogorov–Smirnov test to verify the standard distribution. In cases where the data exhibited a normal distribution, statistical analyses were performed using either a paired Student’s t-test or one-way ANOVA, with subsequent Newman-Keuls post hoc testing. As for non-normally distributed data, the Kruskal–Wallis test followed by Dunn’s post-hoc test was applied. The threshold for statistical significance was established at P -value < 0.05(two-sided). This is denoted by *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Melatonin ameliorate hyperandrogenism and ovarian dysfunction in constant darkness-induced PCOS-like rats
To examine how melatonin affects PCOS-like characteristics induced by circadian rhythm disruption, we administered melatonin injections to female Sprague–Dawley rats over an 8-week period while housing them in continuous darkness (Figure 1A). Rat-body-weights in the different groups were identical (Figure 1B). In the PCOS-like rat model subjected to constant darkness, we noted several characteristic features. These included irregular estrous cycles (Figure 1C), an increase in the quantity of abnormally large follicles, downregulation of ovarian corpora lutea (Figure 2), and elevated serum testosterone levels (Figure 1E), which were consistent with previous studies (10, 26, 27). But the serum LH level remained unchanged (Figure 1D). The melatonin administration improved the disrupted estrus cycles and decreased the number of antral follicles in darkness rats. Besides, melatonin treatment also significantly alleviated the high serum testosterone level (Figures 1E, 2).
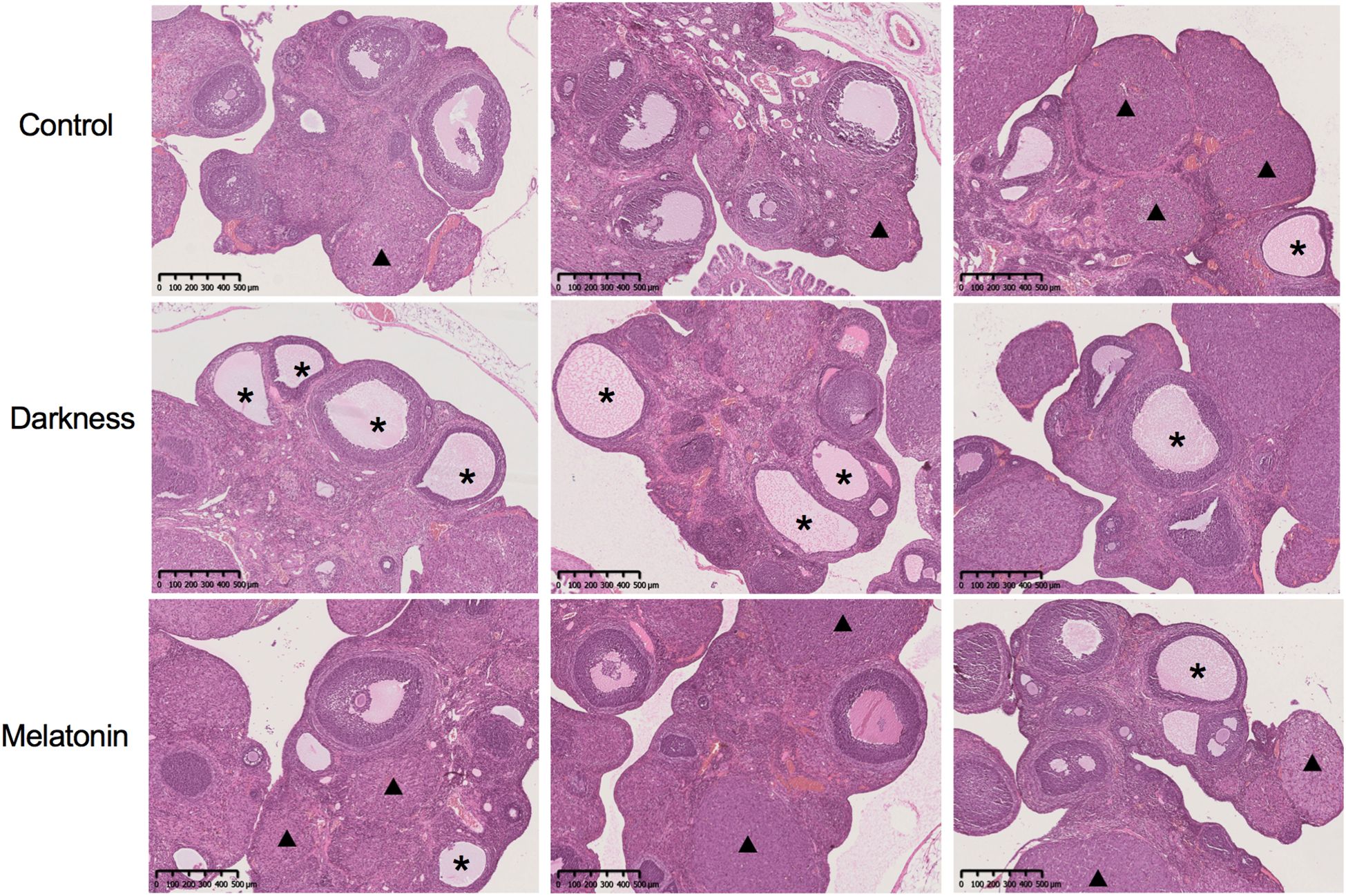
Figure 2. The ovarian and follicular morphologies of rats were assessed by hemoxylin and eosin staining. Asterisk stands for cystic follicles, triangle stands for the corpus luteum. Scale bar = 500 μm. N = 6 per group.
Melatonin targets network
A comprehensive set of 233 melatonin targets was compiled from PharmMapper (119 targets), SwissTargetPrediction (100 targets), DrugBank (17 targets), and TCMSP (7 targets). Among them, 10 duplicated targets were removed and subsequently illustrated using Cytoscape (version 3.8.2) (Supplementary Figure S1).
PPI network of PCOS targets
Based on DisGeNET (262 targets), GeneCards (436 targets), and NCBI Gene (457 targets), 803 PCOS targets were identified. Utilizing Cytoscape’s analysis network feature in the string interaction network, we identified 103 significant targets for PCOS. These targets exhibited above-average values for a degree of centrality of 14.00000, betweenness centrality of 0.00348, and closeness centrality of 0.33856 (Supplementary Figure S2).
PPI network construction
By applying the Venn Diagram tool, researchers (Ran Cheng and Sheng-kai Wang) identified 37 potential targets related to melatonin and PCOS (Figure 3A). Subsequently, we utilized the STRING database to generate a 37-co-targets’ PPI-network (Figure 3B). The MCC, DMNC, and MNC scores were calculated by “CytoHubba” plugin, and the intersection of hub targets [AR, CYP19A1, PGR, IGF1R, MDM2, NR3C1, and CYP1A1] was obtained.
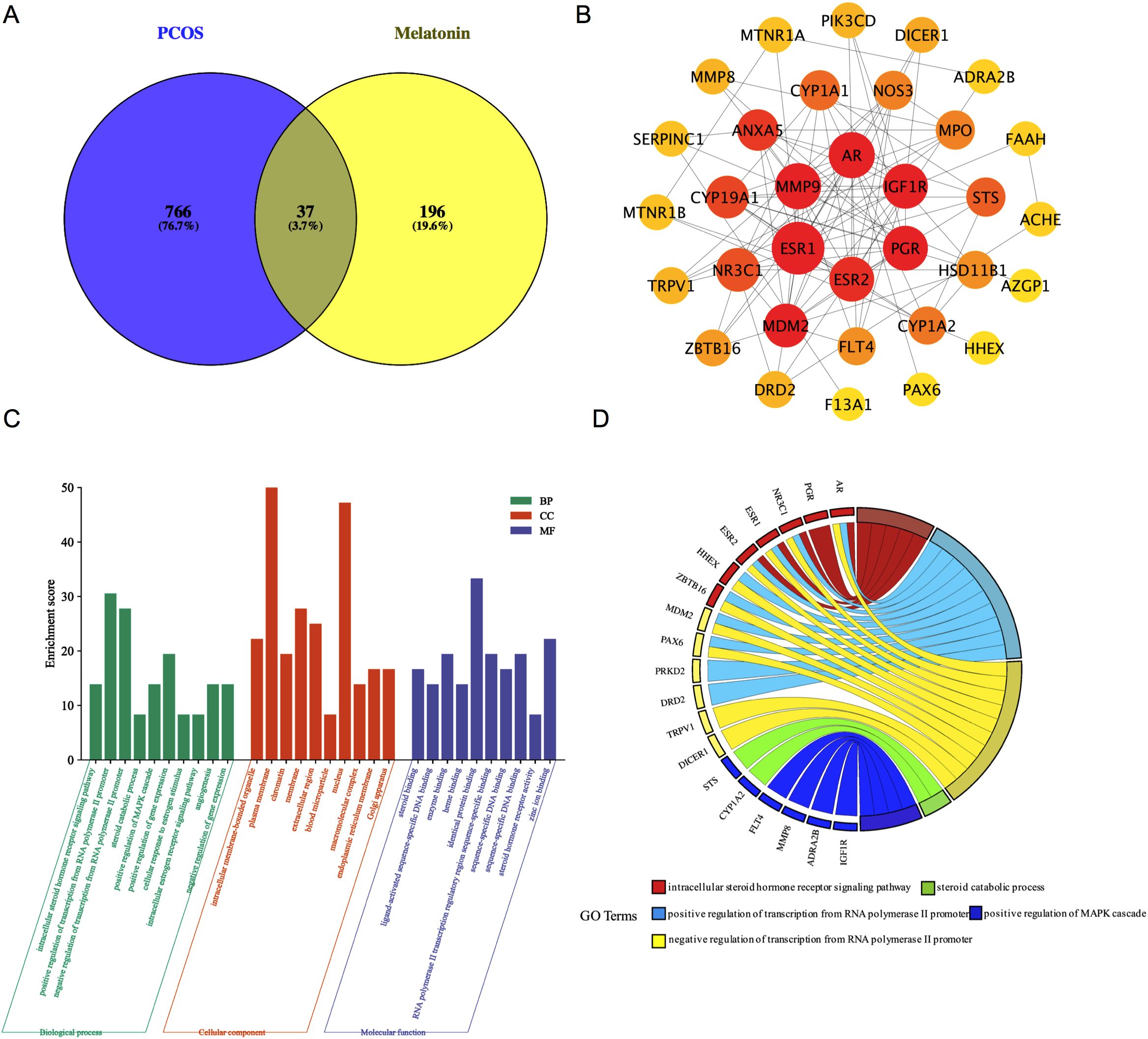
Figure 3. The Venn diagram and PPI network of melatonin predictive targets for PCOS. (A) The common targets of melatonin and PCOS. (B) The PPI network of 37 common target genes with a medium confidence interaction score of 0.400. PPI, protein-protein interaction; PCOS, polycystic ovary syndrome (C) The top 10 GO terms in the enrichment analysis in BP, CC, and MF. (D) The top five enriched biological processes. BP, biological processes; CC, cellular components; MF, molecular function; GO, Gene Ontology.
GO analysis
DAVID tools were employed to analyze the 37 common targets, with the top 10 Gene Ontology (GO) terms selected based on P-value and count frequency. Figure 3C displays the GO results, which were represented through a platform (free online: https://www.bioinformatics.com.cn/). The five most prominent biological processes (BP) were recognized in line with their count frequency and P-value, as shown in Figure 3D. These key BPs were found to be associated with melatonin treatment for PCOS: the intracellular steroid hormone receptor Signaling pathway, the positive regulation of the MAPK cascade, the steroid catabolic process, and the positive and negative regulation of transcription from the RNA polymerase II promoter. Importantly, four of the seven hub genes (AR, PGR, NR3C1, and MDM2) were enriched among the five leading BPs.
KEGG pathway enrichment analysis
DAVID tools were employed to conduct a KEGG pathway enrichment analysis on the 37 therapeutic targets, identifying 18 pathways with significant associations, as depicted in Figure 4A. The targets enriched within individual pathways are shown in Figure 4B. These 18 pathways were organized into four functional modules, as shown in Figure 4C. Module 1 incorporated two pathways: circadian entrainment (hsa04713) and neuroactive ligand-receptor interaction (hsa04080). Module 2 encompasses six pathways: steroid hormone biosynthesis (hsa00140), ovarian steroidogenesis (hsa04913), prolactin signaling (hsa04917), estrogen signaling (hsa04915), GnRH secretion (hsa04929), and thyroid hormone signaling (hsa04919). Module 3 contained four pathways: diabetic cardiomyopathy (hsa05415), regulation of lipolysis in adipocytes (hsa04923), maturity-onset diabetes of the young (hsa04950), and endocrine resistance (hsa01522). Module 4 was composed of six pathways: the PI3K-Akt Signaling pathway (hsa04151), VEGF Signaling pathway (hsa04370), Rap1 Signaling pathway (hsa04015), FOXO Signaling pathway (hsa04068), oocyte meiosis (hsa04114), and progesterone-mediated oocyte maturation (hsa04914).
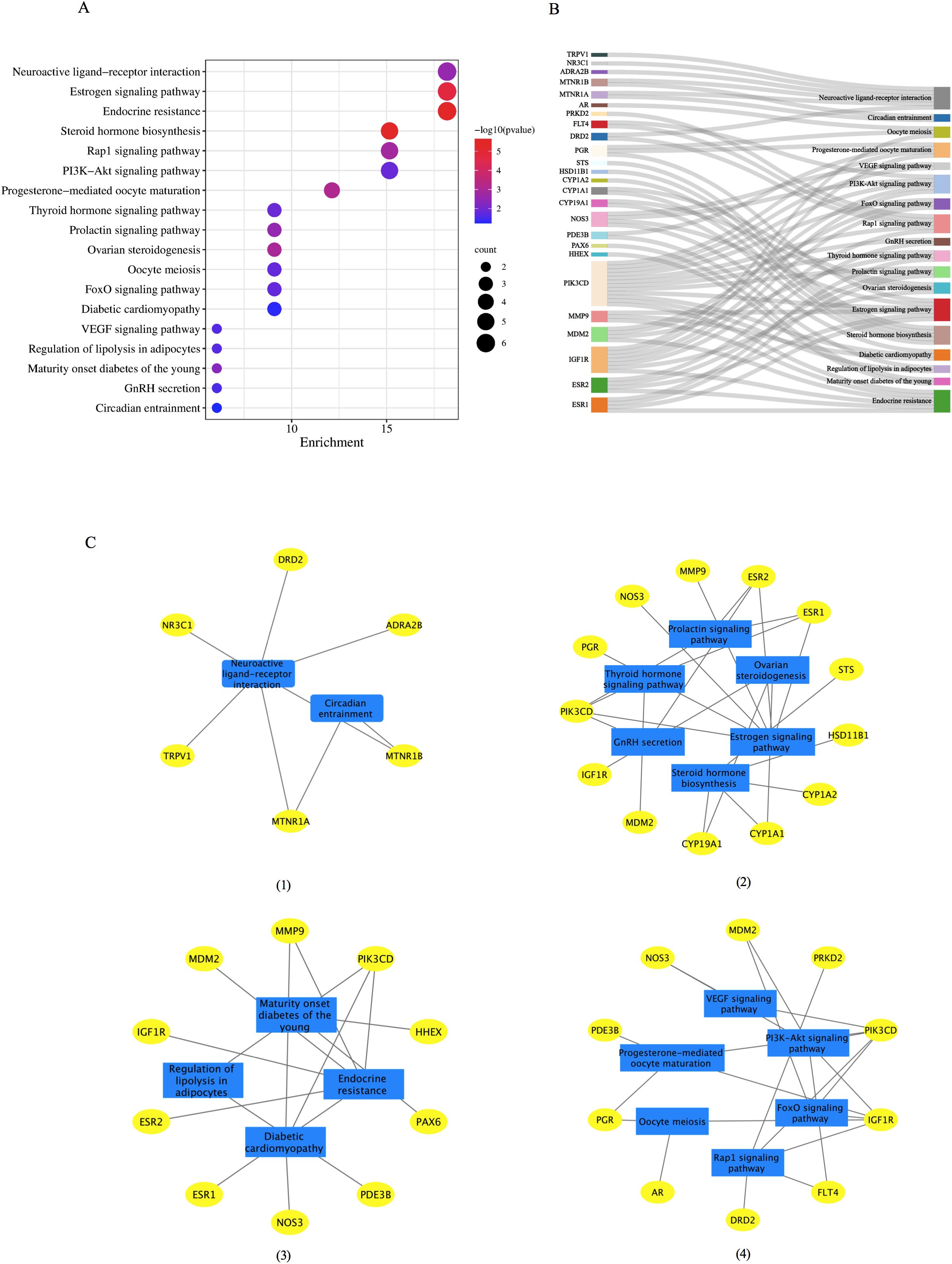
Figure 4. The KEGG functional analysis of the 37 common target genes. (A) The 18 significant pathways. The bubble sizes are indicated from large to small in descending order of the count of the potential target genes enriched in the pathways. The bubble colors are shown from red to blue in descending order of −log10(P value). (B) Target-pathways network. (C) Module analysis of the 18 KEGG pathways. Four modules were obtained according to the function of KEGG: (I) module 1, circadian rhythm; (II) module 2: reproductive process, (III) module 3, metabolic process; (IV) module 4, oocyte maturation. GnRH, gonadotrophin-releasing hormone; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Molecular docking
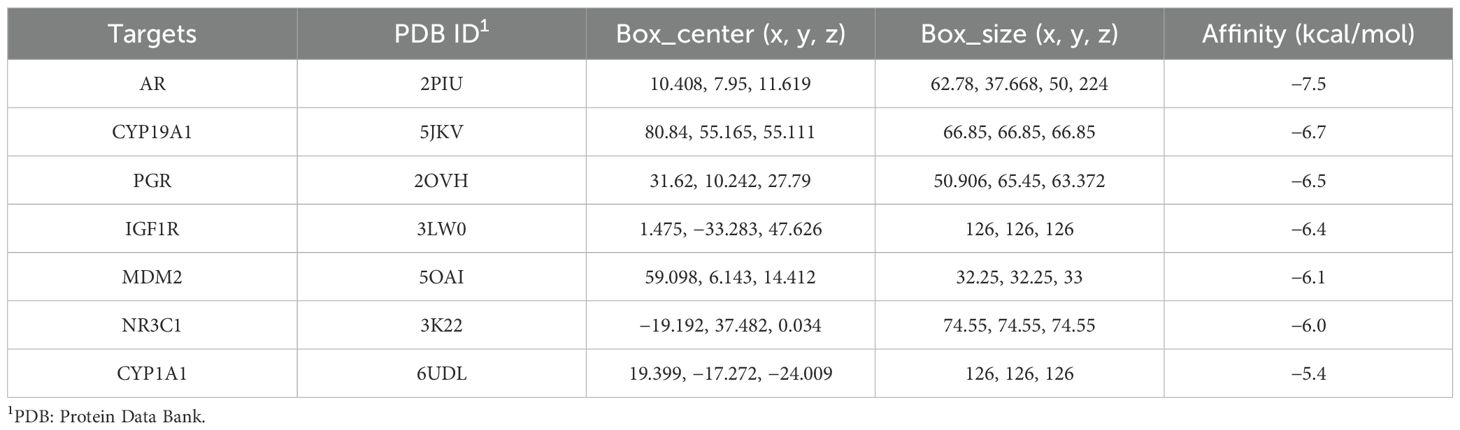
Seven hub genes sorted by MCC, DMNC, and MCN were identified for molecular docking analysis with melatonin (Figure 5). Table 2 outlined the specifications of the molecular docking parameters. A stronger binding ability was indicated by a lower docking affinity. The binding strength between melatonin and its target molecules was substantial, with affinity values ranging from −7.5 to −5.4 kcal/mol. Melatonin may effectively treat PCOS by modulating the activities of these significant targets.
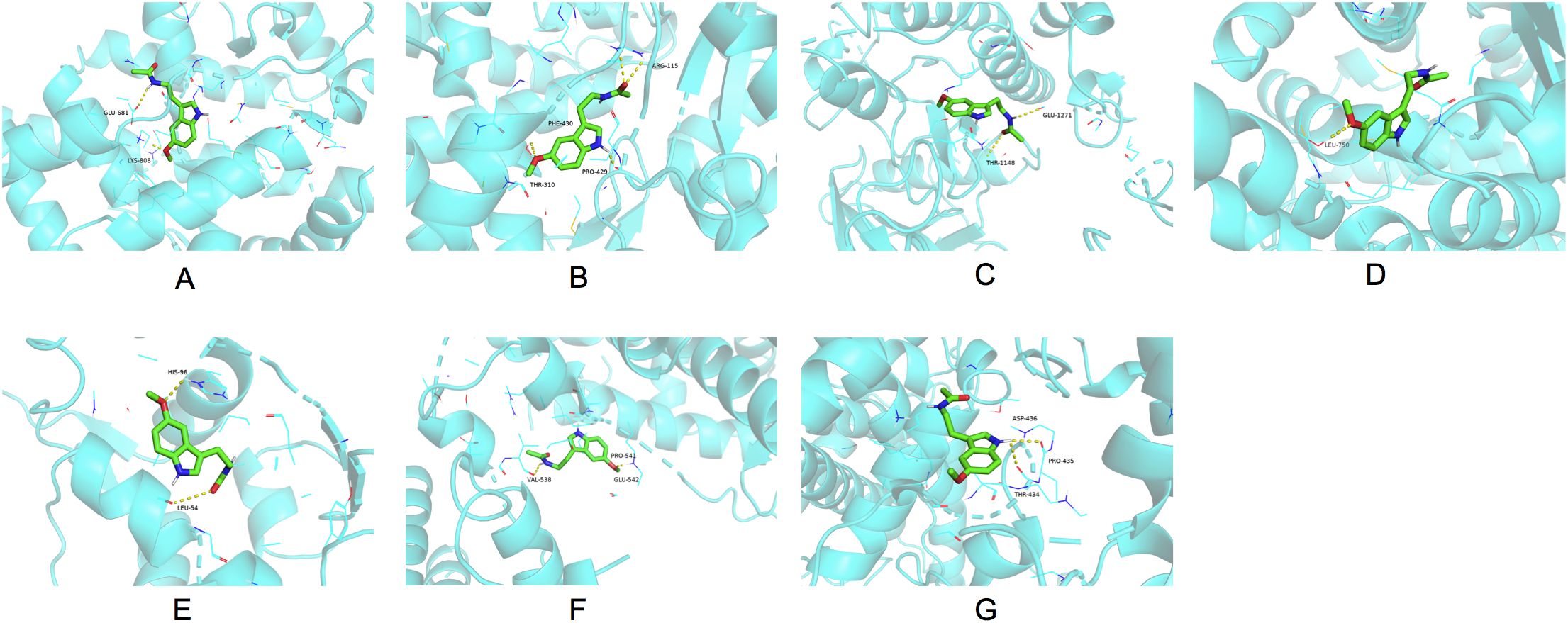
Figure 5. Molecular docking results of melatonin with the seven hub target proteins ranked by the MCC, DMNC and MNC algorithm of the “CytoHubba” plugin using the Cytoscape network analysis. The binding sites of melatonin with AR (A), CYP19A1 (B), PGR (C), IGF1R (D), MDM2 (E), NR3C1 (F), CYP1A1 (G). MCC, Maximal Clique Centrality; DMNC, Maximum Neighborhood Component Centrality; MNC: Neighborhood Component Centrality; AR, androgen receptor; CYP19A1, cytochrome P450 family 19 subfamily A member 1; PGR, progesterone receptor; IGF1R, insulin-like growth factor I receptor; NR3C1, nuclear receptor subfamily 3 group C member 1; CYP1A1, cytochrome P450 family 1 subfamily A member 1.
MD simulation
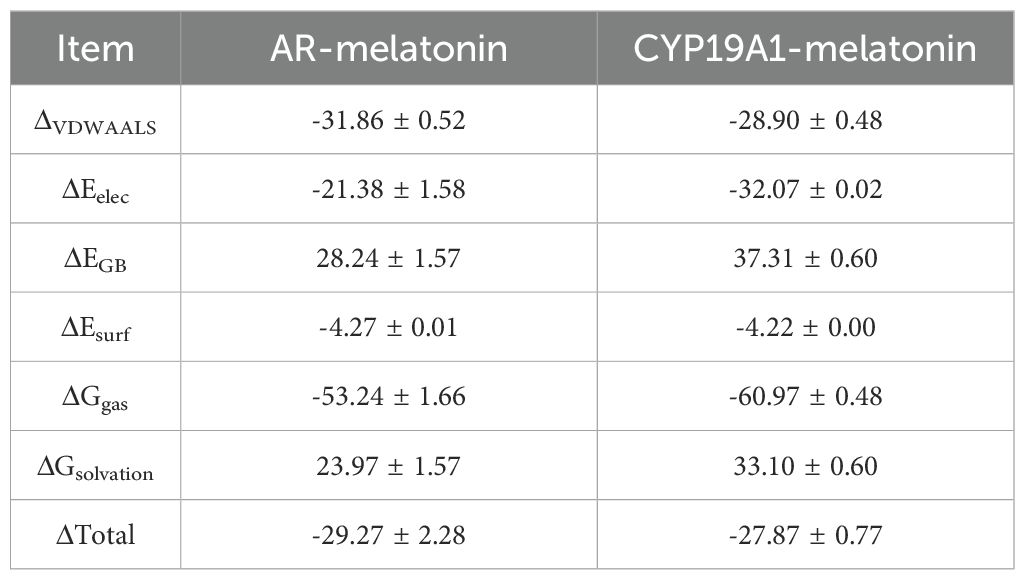
MD simulations were performed to verify the molecular docking simulation results for AR and CYP19A1. The RMSD curves for the AR/CYP19A1-melatonin complexes showed equilibrium after 60 ns, with average RMSD values of 0.2 nm and 0.23 nm (Figure 6A). Evaluation of the molecular stability using RMSD measurements in relation to the starting conformation (Figure 6B) showed that melatonin exhibited the smallest positional shifts when complexed with AR. The Rg values for the two complexes-maintained equilibrium with means of 1.85 nm and 2.25 nm (Figure 6E). The average SASA values remained consistent at 128 nm² and 210 nm² (Figure 6F). Throughout the 100 ns simulation, the count of hydrogen-bonding interactions for the AR/CYP19A1-melatonin complexes ranged from to 1–4 and 1-3, respectively (Figures 6G, H). The RMSF analysis revealed crucial flexible areas within the AR and CYP19A1 proteins, particularly in the vicinity of specific amino acid residues 200–40 for AR-melatonin; 790–800 for CYP19A1-melatonin (Figures 6C, D, I, J). The complex’s Gibbs free energy profiles exhibited clear minimum energy basins (Figures 6A, B, E), accompanied by hydrogen bond counts of 3 and 2, respectively (Figures 6B–D, G–F). Table 3 displayed the binding free energy of the complexes formed between AR/CYP19A1 and melatonin. The MD simulation generated movie-like files depicting the trajectories (Supplementary Movie S1. AR and Supplementary Movie S2. CYP19A1).
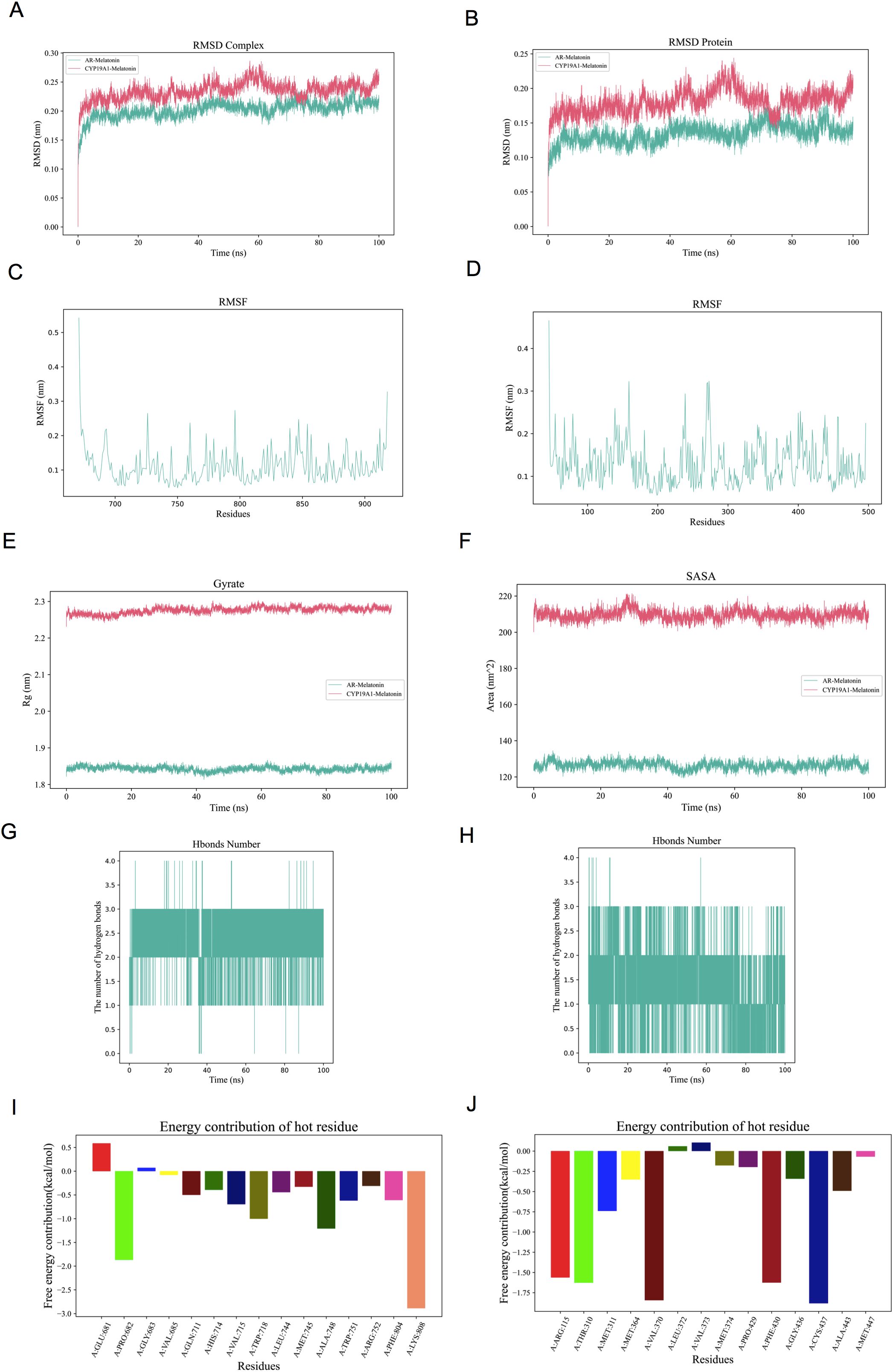
Figure 6. The molecular dynamics’ simulation of AR/CYP19A1–melatonin complex. (A) RMSD curve of the AR/CYP19A1–melatonin complex; (B) RMSD curve of melatonin; (C) RMSF curve of AR; (D) RMSF curve of CYP19A1; (E) Rg curve of the AR/CYP19A1–melatonin complex; (F) SASA curve of the AR/CYP19A1–melatonin complex; (G) Number of hydrogen bonds in the AR–melatonin complex; (H) Number of hydrogen bonds in the CYP19A1–melatonin complex; (I)Amino acid decomposition of the AR–melatonin complex; (J) Amino acid decomposition of the CYP19A1–melatonin complex.
As illustrated in Figure 7, the minimum value of the free energy is indicated by the blue region, while the metastable conformation is represented by the cyan and green regions. A higher prevalence of blue coloration was observed for AR/CYP19A1-melatonin complexes, suggesting favorable binding energetics and conformational stability within the simulation timeframe. Molecular dynamics (MD) simulations thus indicated the potential for stable physical interaction between melatonin and the identified target proteins. While this computational finding supports the plausibility of melatonin binding to these targets, it is important to note that MD simulations primarily assess structural dynamics and binding stability, not functional consequences. The potential biological significance of these interactions, such as modulation of enzymatic activity or transcriptional regulation relevant to hyperandrogenism, remains hypothetical and requires empirical validation through functional assays.
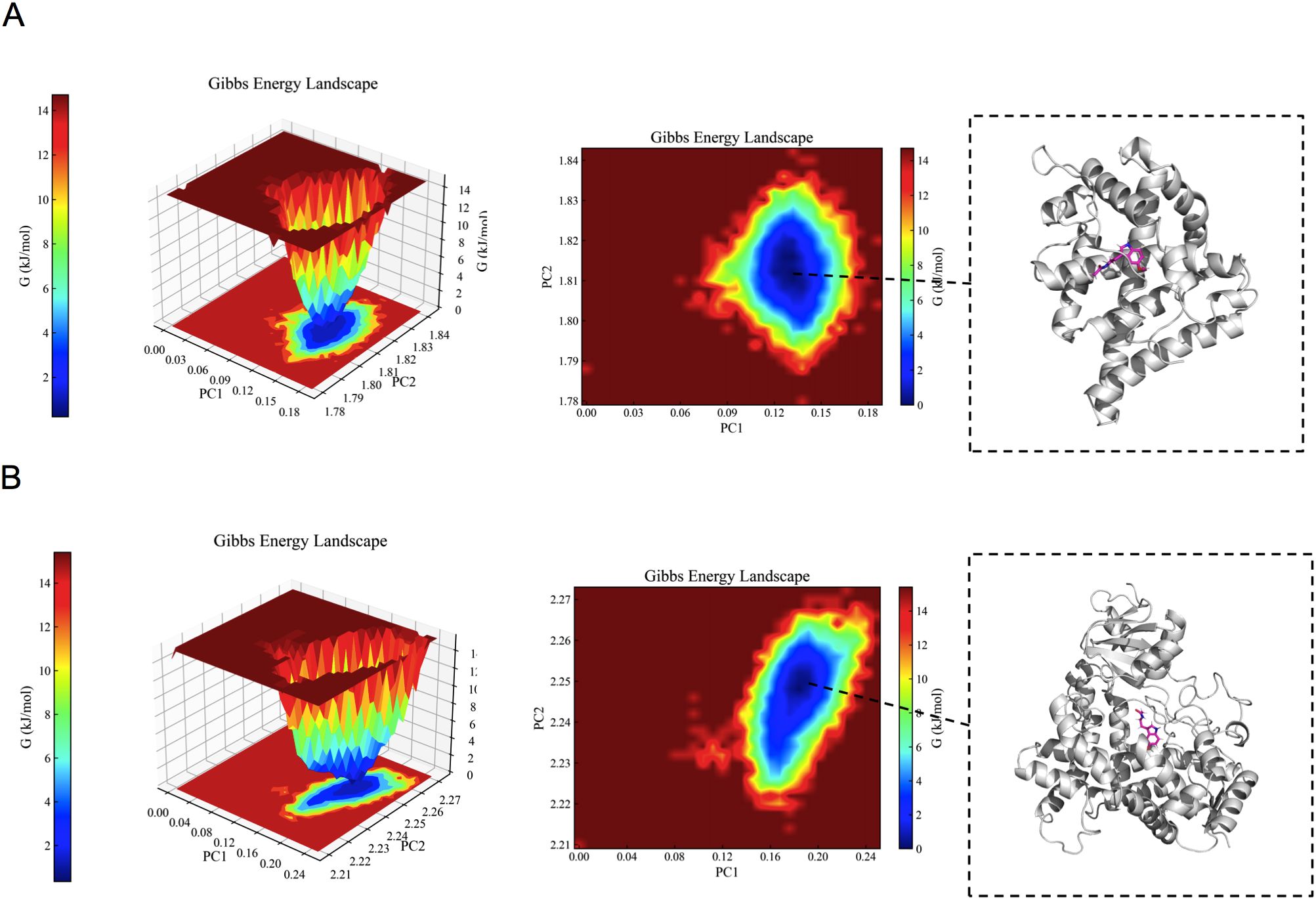
Figure 7. The results of the Gibbs free energy analysis. (A) 3D and 2D contour plots of the AR–melatonin complex. (B) 3D and 2D contour plots of the CYP19A1–melatonin complex.
Effect of melatonin on the mRNA expression of peripheral circadian genes in PBMCs
As illustrated in Figure 8, the mRNA expression of BMAL1 decreased significantly in the darkness contrasting to the control group (P < 0.05). Notably, Melatonin administration substantially reversed the mRNA expression of BMAL1 (P < 0.05). The mRNA expression of CLOCK and PER2 did not differ among the three groups (P < 0.05).
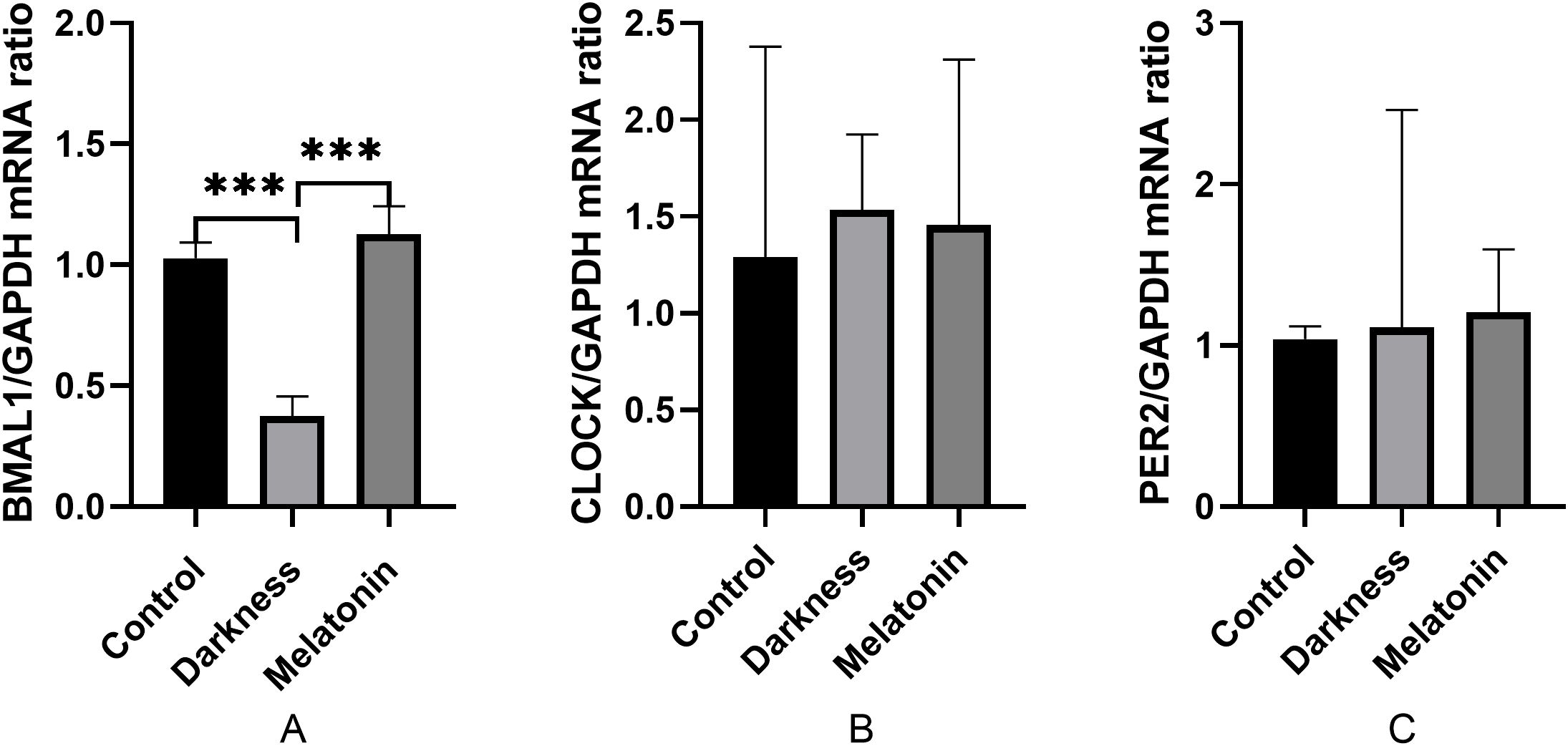
Figure 8. mRNA abundances of BMAL1 (A), CLOCK (B), and PER2 (C) in rat PBMC detected by qPCR. GAPDH was used as a loading control for qPCR analyses. N=6 per group. Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple comparison test. Data are expressed as mean± SD from 3 experiments. ***P < 0.001.
Effect of melatonin on the mRNA expression of MTNR1A, MTNR1B, AR, and CYP19A1 in the ovarian tissues
As illustrated in Figure 9, the mRNA expression of MTNR1A, MTNR1B, and CYP19A1 decreased significantly in the darkness group compared to that in the control group (P < 0.05), whereas the mRNA expression of AR was markedly increased in the darkness group (P < 0.05). Compared to the darkness group, the mRNA expression of MTNR1A, MTNR1B, and CYP19A1 increased significantly in the melatonin group (P < 0.05), whereas the mRNA expression of AR was markedly reduced in the melatonin group (P < 0.05).
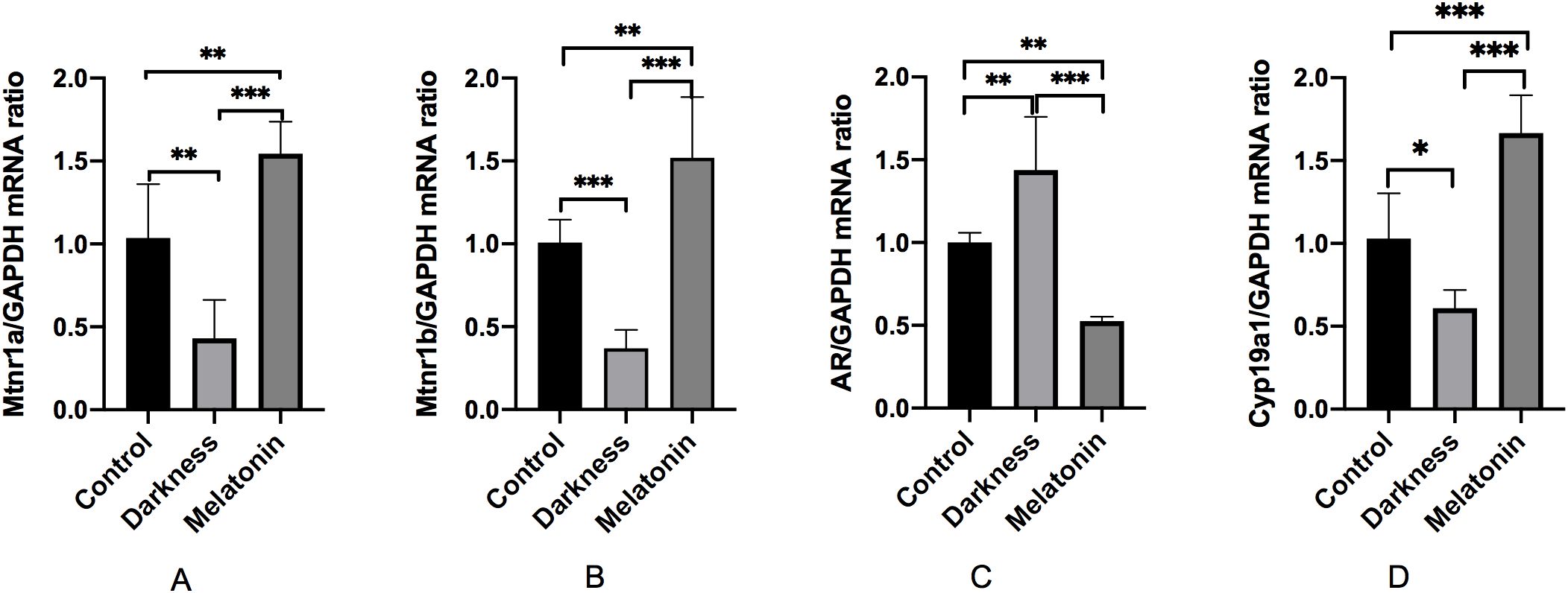
Figure 9. mRNA abundances of MTNR1A (A), MTNR1B (B), AR (C), and CYP19A1 (D) in rat ovary tissues detected by qPCR. GAPDH was used as a loading control for qPCR analyses. N=6 per group. Statistical analysis was performed with one-way ANOVA followed by Newman–Keuls multiple comparison test. Data are expressed as mean± SD from 3 experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
The complex multifactorial etiology of PCOS presents challenges for the formulation of individualized therapeutic approaches. There is a persistent necessity for therapeutic interventions that address the underlying causes and yield efficacious outcomes. Therapeutic strategies targeting hyperandrogenism, a hallmark of PCOS, have been extensively explored. Prolonged darkness leads to hyperandrogenism in PCOS-like rat models (26). One relevant mechanism is related to the downregulation of MTNR1A, which disturbs AR expression and downstream CYP19A1 transcription (26). Studies have also revealed that dysregulation of the expression of some essential clock genes, such as BMAL1, is closely associated with hyperandrogenism (46). CYP19A1 has also been identified as an immediate objective of BMAL1 in PBMCs, and low expression of BMAL1 increases the activity of 5a-reductase, exacerbating hyperandrogenism (13). Recently, promising therapeutic interventions targeting circadian rhythm modifications to mitigate hyperandrogenism have been proposed. Similar to a rhythm synchronizer, melatonin may directly influence both peripheral and central clock characteristics. (12, 47). Clinical investigations have confirmed that melatonin supplementation can enhance sleep quality and significantly alleviate hyperandrogenism in women with PCOS (21, 22). However, the exact mechanism by which melatonin functions remain unclear.
First, we validated the anti-androgen effects of melatonin in a circadian dysrhythmia-induced PCOS-like rat model. PCOS-like rats were induced by long-term environmental exposure to darkness, as previously described (26). We observed high serum levels of total testosterone, estrous cycle irregularity, and polycystic ovarian malformations in the darkness group. These findings indicate that continuous darkness, an unusual condition causing circadian rhythm disruption, leads to changes resembling PCOS morbidity. After 8-week treatment, we found that melatonin reduced serum testosterone levels, restored the estrous cycle, and relieved abnormal ovary morphology.
To explore the underlying mechanism, we conducted network pharmacology analysis. Network pharmacology findings elucidated that melatonin could potentially ameliorate PCOS through multiple pathways and cellular processes. Fundamental biological mechanisms, such as the steroid hormone receptor Signaling pathway within cells, cellular response to estrogen stimulus, steroid catabolic processes, and intracellular ER Signaling pathways, demonstrate the diverse modulatory effects of melatonin on hormone production within cells. Based on the KEGG pathway enrichment analysis, it can be postulated that melatonin may exert beneficial effects by modulating circadian rhythm, reproductive processes, metabolic functions, and oocyte maturation. Perturbations in circadian rhythm can result in reproductive and metabolic disorders. Furthermore, research has demonstrated that melatonin possesses potent antioxidant and anti-inflammatory properties (48). Accordingly, we speculated that melatonin could alleviate reproductive disorders in PCOS by exerting multifaceted effects: melatonin, acting as an essential rhythm synchronizer, could lessen the reproductive and metabolically related symptoms of PCOS. Its strong antioxidant effects also protect oocytes from oxidative damage.
To determine the impact of melatonin on the circadian rhythm of rats, we detected the mRNA expression of some core clock genes in PBMCs. The mRNA expression of BMAL1 was markedly reduced in PBMCs in the dark group, implying that the irregular expression of circadian clock genes in PBMCs resembled that observed in individuals with PCOS (13). After 8-week treatment, we found that melatonin reversed BMAL1 expression. Moreover, we found that PCOS and melatonin share 37 targets. Among the 37 targets, MTNR1A, AR, and CYP19A1 were closely related to hyperandrogenism, as stated above. Molecular docking and MD also showed the highest affinity and stability between AR, CYP19A1 and melatonin, indicating that these targets may serve crucial functions in treating PCOS, but further experimental verification was necessary. Due to the fact that the main and core source of androgens in PCOS patients with hyperandrogenism is the ovaries, so we further employed experimental validation to measure the expression of these genes in the ovarian tissues. We observed downregulated expression of MTNR1A, MTNR1B, and CYP19A1, overexpression of AR in ovary tissues in the darkness group, which were also reversed by melatonin treatment. The results of our study imply that melatonin may relieve hyperandrogenism in darkness-induced PCOS by regulating MTNR1A, MTNR1B, AR, and CYP19A1 expression.
It is worth noting that the 10 mg/kg/day dose of melatonin, commonly used in pre-clinical rodent studies, is substantially higher than typical physiologic doses. In healthy humans, endogenous melatonin production by the pineal gland follows a day-night cycle: secretion begins around 9–10 PM in response to darkness and peaks between 2–4 AM, with serum concentrations (Cmax) typically ranging from 60–100 pg/mL (49). For long-term management of adult insomnia disorder, clinical trials have used oral melatonin doses ranging from 0.3 to 8 mg/day (50). Pharmacokinetic studies indicate that exogenous melatonin administration in humans yields Cmax values ranging from 170 pg/mL to several thousand pg/mL (51). Notably, a 20 mg/day oral dose demonstrated potential neuroprotective benefits and improved sleep quality in breast cancer patients undergoing chemotherapy, whereas a 6 mg/day dose did not show similar cognitive benefits (52, 53). These findings suggest that super-physiological melatonin doses may exert multiple effects, such as circadian rhythm entrainment and anti-inflammatory actions. Considering rats exhibit ~3-fold faster melatonin clearance than humans, the 10 mg/kg/day dose ensures target engagement. Nevertheless, results from our animal experiments still should be interpreted with caution.
Nonetheless, this study had limitations. For instance, while the LH/FSH ratio provides more comprehensive information than LH level alone, we opted not to measure FSH or other laboratory parameters such as AMH. This decision was made primarily due to the study’s focus on verifying melatonin’s efficacy against hyperandrogenism, and also considering funding constraints. We will explore these indicators further in a follow-up study. Furthermore, a limitation of this study was the concurrent administration of melatonin with continuous darkness. Future studies will address this by distinguishing between prophylactic and therapeutic interventions. The pharmacological effects of melatonin cannot be fully anticipated, owing to the scarcity of information in publicly accessible databases. Moreover, it should be noted that the in silico analyses (such as molecular docking and structural prediction) in this study focused on the functional active sites of proteins, while the experimental validation only detected gene expression at the mRNA level. Since there may be differences in post-transcriptional regulation (such as translation efficiency, protein modification, or degradation) between mRNA abundance and protein activity, the direct correlation between the two should be interpreted with caution. This limitation suggests that future studies should combine protein activity assays (such as enzyme activity analysis and Western blot) or cell function experiments to further validate the correspondence between computational predictions and actual biological effects.
Conclusion
Integrating network pharmacology, molecular docking, molecular dynamics simulations, and experimental validation, this study sheds light on potential mechanisms by which melatonin may ameliorate PCOS, especially hyperandrogenism. While these results suggest that melatonin’s therapeutic effects in PCOS involve the mitigation of androgen excess, further studies are warranted to validate these targets in human pathophysiology and explore their clinical translatability. Collectively, this work provides a promising foundation for understanding melatonin’s actions in circadian dysregulation-associated PCOS and highlights novel targets for future mechanistic and therapeutic investigations.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal studies were approved by Animal Ethical and Welfare Committee of ZCMU. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was not obtained from the owners for the participation of their animals in this study because we agree that since the relevant Committee’s authorization was obtained, it may be unnecessary to include the statement about written informed consent from animal owners in the Ethics statements section.
Author contributions
RC: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. YZ: Formal analysis, Methodology, Supervision, Validation, Writing – review & editing. S-KW: Methodology, Supervision, Validation, Writing – review & editing. CX-Z: Methodology, Supervision, Validation, Writing – review & editing. H-LZ: Data curation, Project administration, Supervision, Writing – review & editing. QZ: Data curation, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Ran Cheng of the Project of Zhejiang Provincial Medical and Health Science and Technology Project (grant number: 2024KY215), Ran Cheng of the Project of the Traditional Chinese Medicine Bureau of Zhejiang Province (No. 2024ZR130), Qin Zhang of the Constructional Project of the National Famous and Old TCM Expert Inheritance Studio (No.(2021)270).
Acknowledgments
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. We thank Ziwei Zhou, a research fellow at Griffith University in Australia, for technical guidance in tissue extraction of RNA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2025.1528518/full#supplementary-material
Abbreviations
PCOS, Polycystic ovary syndrome; SD, Sprague-Dawley; SPF, Specific Pathogen Free; PBMCs, Peripheral blood mononuclear cells; HE, Hematoxylin and eosin; qRT‐PCR, quantitative real‐time PCR; TCMSP, Traditional Chinese Medicine Systems Pharmacology; NCBI, National Center for Biotechnology Information; PPI, protein-protein interaction; MCC, Maximal Clique Centrality; DMNC, Maximum Neighborhood Component Centrality; MNC, Neighborhood Component Centrality; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DAVID, Database for Annotation, Visualization and Integrated Discovery; PDB, Protein Data Bank; LH, luteinizing hormone; BP, biological processes.
References
1. Joham AE, Norman RJ, Stener-Victorin E, Legro RS, Franks S, Moran LJ, et al. Polycystic ovary syndrome. Lancet Diabetes Endocrinol. (2022) 10:668–80. doi: 10.1016/s2213-8587(22)00163-2
2. Rostamtabar M, Esmaeilzadeh S, Tourani M, Rahmani A, Baee M, Shirafkan F, et al. Pathophysiological roles of chronic low-grade inflammation mediators in polycystic ovary syndrome. J Cell Physiol. (2021) 236:824–38. doi: 10.1002/jcp.29912
3. Merkin SS, Phy JL, Sites CK, and Yang D. Environmental determinants of polycystic ovary syndrome. Fertil Steril. (2016) 106:16–24. doi: 10.1016/j.fertnstert.2016.05.011
4. Lee Y, Field JM, and Sehgal A. Circadian rhythms, disease and chronotherapy. J Biol Rhythms. (2021) 36:503–31. doi: 10.1177/07487304211044301
5. Heydari T and Ramdass P. Circadian rhythm disruption and polycystic ovary syndrome: a systematic review and meta-analysis. AJOG Glob Rep. (2025) 5:100479. doi: 10.1016/j.xagr.2025.100479
6. Panda S. Circadian physiology of metabolism. Science. (2016) 354:1008–15. doi: 10.1126/science.aah4967
7. Peng F, Li X, Xiao F, Zhao R, and Sun Z. Circadian clock, diurnal glucose metabolic rhythm, and dawn phenomenon. Trends Neurosci. (2022) 45:471–82. doi: 10.1016/j.tins.2022.03.010
8. la Fleur SE, Blancas-Velazquez AS, Stenvers DJ, and Kalsbeek A. Circadian influences on feeding behavior. Neuropharmacology. (2024) 256:110007. doi: 10.1016/j.neuropharm.2024.110007
9. Luo B, Song J, Zhang J, Han J, Zhou X, and Chen L. The contribution of circadian clock to the biological processes. Front Mol Biosci. (2024) 11:1387576. doi: 10.3389/fmolb.2024.1387576
10. Li S, Zhai J, Chu W, Geng X, Chen ZJ, and Du Y. Altered circadian clock as a novel therapeutic target for constant darkness-induced insulin resistance and hyperandrogenism of polycystic ovary syndrome. Transl Res. (2020) 219:13–29. doi: 10.1016/j.trsl.2020.02.003
11. Holloway KR, Barbieri A, Malyarchuk S, Saxena M, Nedeljkovic-Kurepa A, Cameron Mehl M, et al. SIRT1 positively regulates breast cancer associated human aromatase (CYP19A1) expression. Mol Endocrinol. (2013) 27:480–90. doi: 10.1210/me.2012-1347
12. Sato K, Meng F, Francis H, Wu N, Chen L, Kennedy L, et al. Melatonin and circadian rhythms in liver diseases: Functional roles and potential therapies. J Pineal Res. (2020) 68:e12639. doi: 10.1111/jpi.12639
13. Johnson BS, Krishna MB, Padmanabhan RA, Pillai SM, Jayakrishnan K, and Laloraya M. Derailed peripheral circadian genes in polycystic ovary syndrome patients alters peripheral conversion of androgens synthesis. Hum Reprod. (2022) 37:1835–55. doi: 10.1093/humrep/deac139
14. Moralia MA, Quignon C, Simonneaux M, and Simonneaux V. Environmental disruption of reproductive rhythms. Front Neuroendocrinol. (2022) 66:100990. doi: 10.1016/j.yfrne.2022.100990
15. Stein RM, Kang HJ, McCorvy JD, Glatfelter GC, Jones AJ, Che T, et al. Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature. (2020) 579:609–14. doi: 10.1038/s41586-020-2027-0
16. Brzezinski A, Rai S, Purohit A, and Pandi-Perumal SR. Melatonin, clock genes, and mammalian reproduction: what is the link? Int J Mol Sci. (2021) 22:13240. doi: 10.3390/ijms222413240
17. Li C, Shi Y, You L, Wang L, and Chen ZJ. Melatonin receptor 1A gene polymorphism associated with polycystic ovary syndrome. Gynecol Obstet Invest. (2011) 72:130–4. doi: 10.1159/000323542
18. Song X, Sun X, Ma G, Sun Y, Shi Y, Du Y, et al. Family association study between melatonin receptor gene polymorphisms and polycystic ovary syndrome in Han Chinese. Eur J Obstet Gynecol Reprod Biol. (2015) 195:108–12. doi: 10.1016/j.ejogrb.2015.09.043
19. Yi S, Xu J, Shi H, Li W, Li Q, and Sun YP. Association between melatonin receptor gene polymorphisms and polycystic ovarian syndrome: a systematic review and meta-analysis. Biosci Rep. (2020) 40:BSR20200824–38. doi: 10.1042/bsr20200824
20. Li H, Liu M, and Zhang C. Women with polycystic ovary syndrome (PCOS) have reduced melatonin concentrations in their follicles and have mild sleep disturbances. BMC Womens Health. (2022) 22:79. doi: 10.1186/s12905-022-01661-w
21. Jamilian M, Foroozanfard F, Mirhosseini N, Kavossian E, Aghadavod E, Bahmani F, et al. Effects of melatonin supplementation on hormonal, inflammatory, genetic, and oxidative stress parameters in women with polycystic ovary syndrome. Front Endocrinol (Lausanne). (2019) 10:273. doi: 10.3389/fendo.2019.00273
22. Shabani A, Foroozanfard F, Kavossian E, Aghadavod E, Ostadmohammadi V, Reiter RJ, et al. Effects of melatonin administration on mental health parameters, metabolic and genetic profiles in women with polycystic ovary syndrome: A randomized, double-blind, placebo-controlled trial. J Affect Disord. (2019) 250:51–6. doi: 10.1016/j.jad.2019.02.066
23. Xie F, Zhang J, Zhai M, Liu Y, Hu H, Yu Z, et al. Melatonin ameliorates ovarian dysfunction by regulating autophagy in PCOS via the PI3K-Akt pathway. Reproduction. (2021) 162:73–82. doi: 10.1530/rep-20-0643
24. Basheer M, Bhat AH, Ahmad Hajam Y, Batiha GE, Ataya FS, Fouad D, et al. Melatonin as a promising therapeutic intervention for restoring ovarian function in letrozole-induced polycystic ovary syndrome rats. Heliyon. (2023) 9:e21237. doi: 10.1016/j.heliyon.2023.e21237
25. Zhang J, Kaasik K, Blackburn MR, and Lee CC. Constant darkness is a circadian metabolic signal in mammals. Nature. (2006) 439:340–3. doi: 10.1038/nature04368
26. Chu W, Li S, Geng X, Wang D, Zhai J, Lu G, et al. Long-term environmental exposure of darkness induces hyperandrogenism in PCOS via melatonin receptor 1A and aromatase reduction. Front Cell Dev Biol. (2022) 10:954186. doi: 10.3389/fcell.2022.954186
27. Li S, Zhai J, Chu W, Geng X, Wang D, Jiao L, et al. Alleviation of Limosilactobacillus reuteri in polycystic ovary syndrome protects against circadian dysrhythmia-induced dyslipidemia via capric acid and GALR1 signaling. NPJ Biofilms Microbiomes. (2023) 9:47. doi: 10.1038/s41522-023-00415-2
28. Li Y, Pei T, Zhu H, Wang R, Wu L, Huang X, et al. Melatonin alleviates circadian rhythm disruption-induced enhanced luteinizing hormone pulse frequency and ovarian dysfunction. J Pineal Res. (2025) 77:e70026. doi: 10.1111/jpi.70026
29. Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res. (2016) 44:D1202–1213. doi: 10.1093/nar/gkv951
30. Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. (2017) 45:W356–w360. doi: 10.1093/nar/gkx374
31. Daina A, Michielin O, and Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. (2019) 47:W357–w364. doi: 10.1093/nar/gkz382
32. Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. (2014) 6:13. doi: 10.1186/1758-2946-6-13
33. Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. (2018) 46:D1074–d1082. doi: 10.1093/nar/gkx1037
34. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The geneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinf. (2016) 54:1.30.31–31.30.33. doi: 10.1002/cpbi.5
35. Piñero J, Saüch J, Sanz F, and Furlong LI. The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput Struct Biotechnol J. (2021) 19:2960–7. doi: 10.1016/j.csbj.2021.05.015
36. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. (2021) 49:D605–d612. doi: 10.1093/nar/gkaa1074
37. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, and Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. (2014) 8 Suppl 4:S11. doi: 10.1186/1752-0509-8-s4-s11
38. Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. (2007) 35:W169–175. doi: 10.1093/nar/gkm415
39. Goodsell DS and Burley SK. RCSB Protein Data Bank tools for 3D structure-guided cancer research: human papillomavirus (HPV) case study. Oncogene. (2020) 39:6623–32. doi: 10.1038/s41388-020-01461-2
40. Seeliger D and de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. (2010) 24:417–22. doi: 10.1007/s10822-010-9352-6
41. Tanchuk VY, Tanin VO, Vovk AI, and Poda G. A new, improved hybrid scoring function for molecular docking and scoring based on autoDock and autoDock vina. Chem Biol Drug Des. (2016) 87:618–25. doi: 10.1111/cbdd.12697
42. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. (2021) 49:D1388–d1395. doi: 10.1093/nar/gkaa971
43. McGuire R, Verhoeven S, Vass M, Vriend G, de Esch IJ, Lusher SJ, et al. 3D-e-chem-VM: structural cheminformatics research infrastructure in a freely available virtual machine. J Chem Inf Model. (2017) 57:115–21. doi: 10.1021/acs.jcim.6b00686
44. Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, and Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. (2005) 26:1701–18. doi: 10.1002/jcc.20291
45. Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, and Berk Hess EL. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. (2015) 1-2:19–25. doi: 10.1016/j.softx.2015.06.001
46. Zhai J, Li S, Hu M, Di F, Liu J, and Du Y. Decreased brain and muscle ARNT-like protein 1 expression mediated the contribution of hyperandrogenism to insulin resistance in polycystic ovary syndrome. Reprod Biol Endocrinol. (2020) 18:32. doi: 10.1186/s12958-020-00592-1
47. Vasey C, McBride J, and Penta K. Circadian rhythm dysregulation and restoration: the role of melatonin. Nutrients. (2021) 13:3480. doi: 10.3390/nu13103480
48. Frungieri MB, Calandra RS, Matzkin ME, and Rossi SP. Melatonin as a natural anti-inflammatory and anti-oxidant therapy in the testis: a focus on infertility and aging†. Biol Reprod. (2024) 111:543–56. doi: 10.1093/biolre/ioae087
49. Acuña-Castroviejo D, Escames G, Venegas C, Díaz-Casado ME, Lima-Cabello E, López LC, et al. Extrapineal melatonin: sources, regulation, and potential functions. Cell Mol Life Sci. (2014) 71:2997–3025. doi: 10.1007/s00018-014-1579-2
50. Maruani J, Reynaud E, Chambe J, Palagini L, Bourgin P, and Geoffroy PA. Efficacy of melatonin and ramelteon for the acute and long-term management of insomnia disorder in adults: A systematic review and meta-analysis. J Sleep Res. (2023) 32:e13939. doi: 10.1111/jsr.13939
51. Harpsøe NG, Andersen LP, Gögenur I, and Rosenberg J. Clinical pharmacokinetics of melatonin: a systematic review. Eur J Clin Pharmacol. (2015) 71:901–9. doi: 10.1007/s00228-015-1873-4
52. Hansen MV, Madsen MT, Andersen LT, Hageman I, Rasmussen LS, Bokmand S, et al. Effect of melatonin on cognitive function and sleep in relation to breast cancer surgery: A randomized, double-blind, placebo-controlled trial. Int J Breast Cancer. (2014) 2014:416531. doi: 10.1155/2014/416531
53. Palmer ACS, Zortea M, Souza A, Santos V, Biazús JV, Torres ILS, et al. Clinical impact of melatonin on breast cancer patients undergoing chemotherapy; effects on cognition, sleep and depressive symptoms: A randomized, double-blind, placebo-controlled trial. PloS One. (2020) 15:e0231379. doi: 10.1371/journal.pone.0231379
Keywords: melatonin, polycystic ovary syndrome (PCOS), network pharmacology, circadian rhythm, hyperandrogenism
Citation: Cheng R, Zhu Y, Wang S-k, Zong C-X, Zhao H-L and Zhang Q (2025) Exploration of the molecular mechanism of melatonin against polycystic ovary syndrome based on a network pharmacology approach and experimental validation. Front. Endocrinol. 16:1528518. doi: 10.3389/fendo.2025.1528518
Received: 15 November 2024; Accepted: 14 July 2025;
Published: 05 August 2025.
Edited by:
Andrea Ivonne Loewendorf, ImmunoVation, United StatesReviewed by:
Edgar Ricardo Vázquez-Martínez, National Autonomous University of Mexico, MexicoEdward Cable, Rallidae Therapeutics, United States
Copyright © 2025 Cheng, Zhu, Wang, Zong, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Li Zhao, ejZobEAxNjMuY29t; Qin Zhang, MjAyMWIwMjdAemNtdS5lZHUuY24=
†These authors have contributed equally to this work
‡ORCID: Ran Cheng, orcid.org/0000-0001-8623-4120
Sheng-kai Wang, orcid.org/0000-0002-8342-8648
Chun-Xiao Zong, orcid.org/0000-0002-0838-0306
Qin Zhang, orcid.org/0000-0002-9845-4099
 Ran Cheng1,2,3††
Ran Cheng1,2,3†† Ying Zhu
Ying Zhu Hong-Li Zhao
Hong-Li Zhao