- 1Center of Excellence for the Oceans, National Taiwan Ocean University, Keelung, Taiwan
- 2Department of Aquaculture, National Pingtung University of Science and Technology, Pingtung, Taiwan
- 3Laboratory Molecular Physiology and Adaptation (PhyMA), National Museum of Natural History (MNHN), National Center of Scientific Research (CNRS), Paris, France
- 4Laboratory Biology of Aquatic Organisms and Ecosystems (BOREA), National Museum of Natural History (MNHN), Sorbonne Université, National Center of Scientific Research (CNRS), Institute of Research for Development (IRD), Paris, France
Eel species are basal teleosts with a unique life cycle including an arrest of sexual maturation before the reproductive oceanic migration. Our early studies showed that this blockade results from a deficient production of pituitary gonadotropins, due in part to a low responsiveness to gonadotropin-releasing hormone (GnRH). Three GnRH receptors have been identified in the eel, among them gnrhr2 is the main pituitary receptor whose expression increases during the sexual maturation induced by gonadotropic treatments. We investigated the role of gonadal hormones in the feedback regulation of gnrhr2 expression in the eel. The effects of steroids and activins were tested in vitro on primary cultures of eel pituitary cells and gnrhr2 transcripts measured by qPCR. In silico analysis of eel gnrhr2 promoter was performed to predict transcription factor binding sites and comparisons were made with gnrhr promoters from other teleosts and mammals. Estradiol and testosterone strongly and dose-dependently increased gnrhr2 transcript levels as measured by qPCR. This stimulatory regulation was not observed with a non-aromatizable androgen, 11 keto-testosterone, and the effect of testosterone was abolished in the presence of an aromatase inhibitor, fadrozole, indicating an estrogen-specific positive control of eel gnrhr2 expression. Other steroids, progesterone and cortisol, had no effect on gnrhr2 expression. Gonadal peptides, activins A and B, were also tested, and showed an inhibitory effect on gnrhr2 expression. Our results show that gonadal steroids exert a positive feedback, mediated by estradiol, on pituitary sensitivity to GnRH in the eel, in line with the regulatory mechanisms of the ovulatory luteinizing hormone (LH) surge in mammals. While investigation on gnrhr promoters is significantly lacking outside mammals, in silico analysis of the eel gnrhr2 promoter allowed us to infer transcription factor binding sites potentially involved in the regulation of gnrhr2 expression. Comparison was made with gnrhr promoters from other teleosts and mammals to discuss their evolutionary conservation. This study in the eel, a basal teleost representative, contributes to our understanding of the regulatory mechanisms of the complex eel life cycle and to raise basic knowledge on the regulation and evolution of pituitary GnRH receptivity in vertebrates.
1 Introduction
In vertebrates, gonadotropin-releasing hormone (GnRH) is the main brain actor controlling the reproductive gonadotropic axis (hypothalamus-pituitary-gonad axis) [for review (1)]. This neurohormone acts on the pituitary via specific G-protein coupled receptors, gonadotropin-releasing hormone receptors (GnRHR), to induce the synthesis and release of gonadotropins, luteinizing hormone (LH) and follicle-stimulating hormone (FSH). In some mammals, two receptor genes (gnrhr1 and gnrhr2) have been characterized. However, in most mammalian species, no functional GnRHR2 is produced due to disruption of gene coding sequence and in the mouse Mus musculus, the gnrhr2 gene is completely absent [for review (2)]. In mammals, GnRHR1 has been largely explored for its involvement in the regulation of pituitary gonadotropins, while GnRHR2 is thought to exert other functions at the brain and peripheral levels [for reviews (2, 3)]. For instance, recent studies in pigs, comparing gnrhr2 knockdown line with control littermates, reported a potent direct action of GnRH2 on gonadal steroidogenesis and gametogenesis in both males and females, without affecting gonadotropins [for review (4)].
In 1997, de Roux and collaborators described the first mutations in the human GnRHR1 in a family with idiopathic hypogonadotropic hypogonadism, characterized by delayed puberty, low gonadotropin and sex steroid levels (5). During childhood, the gonadotropic axis is quiescent, involving a low responsiveness of pituitary gonadotroph cells to GnRH, which partly reflects a reduced expression of the GnRHR1 [for review (6)]. In mice, induced GnRHR1 mutations led to a phenotype similar to the clinical syndrome of hypogonadotropic hypogonadism (7, 8). Ontogeny of pituitary GnRHR1 in male and female rats revealed that its number or expression was maximal in the prepubertal period, when serum, pituitary content and expression levels of both gonadotropins are elevated (9–11).
A number of investigations in mammals reported positive and negative effects of sexual steroids on pituitary responsiveness to GnRH. Orchidectomy and ovariectomy in rats induce drastic increase in pituitary GnRHR number (12, 13) and mRNA levels (14), suggesting a negative feedback of gonadal hormones. Similarly, in castrated adult male Rhesus monkeys Macaca mulatta, an increase of pituitary gnrhr mRNA levels was observed (15). However, species-differences exist in mammals and in contrast to rats and Rhesus monkeys, gonadectomy in mice induces a decrease in GnRHR in both sexes indicating a positive feedback of gonadal steroids (16, 17). No effect of gonadectomy was reported on either the number (18) or the mRNA levels (19) of GnRHR in ewe.
In the female of mammalian species, including mouse, rat, ewe, cow and monkey, estradiol (E2) is considered a major positive regulator of preovulatory LH surge by increasing both pituitary sensitivity to GnRH and GnRH release [for review (20)]. During the estrous cycle, in rat, a positive correlation exists between circulating concentrations of E2 and GnRHR number (21, 22) and mRNA levels (14, 23). Similarly, in the ewe, maximal concentrations or mRNA levels of GnRHR are observed prior to LH surge when plasma E2 concentration are rising (24–26). Early studies demonstrated that E2 stimulates the expression and number of GnRHR by a direct pituitary effect as shown on primary cultures of pituitary cells in rat (27, 28), mouse (29) and sheep (20, 30) [for review (31)].
Activins are peptide hormones produced by the gonads, pituitary and other organs, which were first identified in porcine follicular fluid for their stimulatory role on FSH release by pituitary gonadotrophs (32). Beside their direct stimulatory role on FSH expression and release, activins were also shown to increase the synthesis of GnRHR by cultured rat pituitary cells (33) and the expression of gnrhr by the mouse gonadotroph cell line alphaT3 (34). This supports a role of activins in the pituitary sensitivity to GnRH [for review (35)].
In teleosts, the pioneering studies by Breton and collaborators (36) revealed the presence of a hypothalamic gonadotropin-releasing factor in common carp Cyprinus carpio, and Sherwood and colleagues (37) identified the GnRH sequence in chum salmon Oncorhynchus keta. Since then, teleost GnRH and GnRHR have been and still are the subject of extensive ongoing investigation [for review (38)]. While only one or two GnRHR are present in mammals, up to six gnrhr gene paralogs have been identified in some teleosts such as the Atlantic salmon (39). A recent phylogenetic analysis of vertebrate GnRHRs by Ciani and collaborators divides them into two main types, GnRHR1 and GnRHR2, each divided into further subtypes (39). At sexual maturation, an increase of the pituitary expression of gnrhr was reported in various teleost species, such as in European seabass Dicentrarchus labrax (40), Nile tilapia Oreochromis niloticus (41), pejerrey Odontesthes bonariensis (42), Atlantic cod Gadus morhua (43), Atlantic salmon (39, 44, 45), and chub mackerel Scomber japonicus (46). Our previous studies in the black porgy Acanthopagrus schlegeli showed a specific increase of a pituitary GnRH receptor (gnrhr1, corresponding to gnrhr2bb in Ciani’s nomenclature) during sexual maturation, as well as under in vivo treatments with human chorionic gonadotropin (hCG) or sex steroids (47). We also demonstrated a stimulatory effect of sex steroids on gnrhr1 (gnrhr2bb) expression by black porgy pituitary cells in vitro (47).
Among teleosts, Anguilla species are of special phylogenetic and biological interest. As members of the group of elopomorphs, they are extant representatives of basal teleosts. The eels, Anguilla genus, encompass about nineteen species and subspecies, distributed in the Indian, Pacific and Atlantic Oceans, and all possessing a peculiar life cycle with a reproduction in oceanic area and a juvenile growth in continental watersheds [for review (48)]. Future genitors, named silver eels, migrate downstream towards the ocean, but remain blocked at prepuberty as long as the oceanic migration is prevented [for review (49)]. Experimental sexual maturation of female and male silver eels can be induced by gonadotropic treatments, as first demonstrated by the work of Fontaine and coworkers in the European eel, Anguilla anguilla (50, 51). Such gonadotropic treatments, fish pituitary extract in the female and human chorionic gonadotropin in the male, are currently used to induce experimental maturation in various eel species, and the biological cycle has been successfully closed in the case of the Japanese eel, A. japonica [for review (52)]. As shown by our previous studies in the European eel, the prepubertal blockade is due to a deficiency in pituitary gonadotropin production, itself resulting from a dual brain control: firstly, a teleost species-specific strong inhibition by dopamine (DA); and secondly, similar to the situation in mammals before puberty, a lack of stimulation by GnRH, including both a low production of GnRH and a low pituitary sensitivity to GnRH [ (53); for review (54)]. A triple treatment with sex steroids (E2 or testosterone, T), GnRH agonist and DA antagonist is thus able to stimulate pituitary LH synthesis and release, and subsequent ovarian vitellogenesis (53, 55).
Three GnRH-R genes have been identified by Peñaranda and colleagues in the European eel and named gnrhr1a, gnrhr1b and gnrhr2 (56). During experimental sexual maturation induced by gonadotropic treatments, the expression of pituitary gnrhr2 (corresponding to gnrhr2b in Ciani’s nomenclature) largely increases, suggesting a major role of this receptor in the regulation of gonadotroph cells in male and female eels (56). The increase in eel gnrhr2 expression may likely result from a positive feedback by gonadal hormones, the production of which is stimulated during experimental maturation [for review (49)]. Steroid hormones exert positive feedback on brain GnRH and pituitary LH in the eel as shown by our early studies in A. anguilla [ (57–62); for review (49)] as well as by investigations in A. japonica (63, 64).
We recently investigated the regulation of gnrhr2 expression in primary cultures of eel pituitary cells and showed inhibitory effects of kisspeptins (65), neurokinin B (66) and gonadotropin-inhibitory hormone (67). In the present study, we investigated the effects of sex steroids, corticosteroid, as well as of activins, on the expression of gnrhr2 by European eel pituitary cells in vitro. In order to get more insights on the regulation of eel gnrhr2 expression, we performed in silico analysis of the gnrhr2 proximal promoter in the European eel to infer the presence of potential transcription factor binding sites. Comparative analyses were made with the gnrhr2 promoters of the Japanese eel (Anguilla japonica) and other teleost species (zebrafish Danio rerio and medaka Oryzias latipes), as well as with the gnrhr1 promoters of two mammals (human Homo sapiens and mouse Mus musculus). While investigations on gnrhr promoters are still lacking outside mammals, the present study allowed us to raise the first data on response elements potentially involved in the regulation of gnrhr expression in teleosts. Comparison with gnrhr promoters in mammals led us to infer some evolutionary conserved features across vertebrates.
2 Materials and methods
2.1 Animals
Freshwater female European eels were at the prepubertal “silver” stage, which corresponds to the last continental phase of the eel life cycle, preceding the oceanic reproductive migration. Cloning, tissue distribution and primary cultures were performed using female silver eels purchased from Gebr. Dil import-export BV (Akersloot, The Netherlands) and transferred to MNHN, France. Animals were anesthetized by cooling and then killed by decapitation under the supervision of authorized person (KR, N°R-75UPMC-F1-08) according to the protocol approved by French Cuvier Ethic Committee (N°68-027). Pituitaries were collected in cell serum-free culture medium (CM: Medium 199 with Earle’s salt and sodium bicarbonate buffer, 100 U/ml penicillin, 100 µg/ml streptomycin, 250 ng/ml fungizone; Gibco, Thermo Fisher Scientific, Illkirch, France) just prior to dispersion (15 to 20 eel pituitaries per cell culture).
2.2 Hormones and chemicals
Sex steroids (estradiol, E2; testosterone, T; 11-ketotestosterone, 11-KT; progesterone, P), cortisol (F), and aromatase inhibitor, fadrozole, were all purchased from Sigma-Aldrich (Saint-Quentin Fallavier, France). Recombinant human/mouse/rat activins A and B were purchased from R and D Systems (Lille, France). The recombinant human/mouse/rat activins used in our study have high amino-acid identity with the eel ones, and we previously showed that they were effective in stimulating fshβ expression by primary cultures of European eel pituitary cells (68). Sex steroids and cortisol were dissolved in ultrapure ethanol (Sigma-Aldrich), activins A and B in sterile calcium-free phosphate-buffered saline (PBS) (Gibco), and fadrozole in dimethyl sulfoxide (DMSO, Sigma-Aldrich) to prepare stock solutions that were stored at -20 °C.
2.3 Primary cultures of eel pituitary cells
Dispersion and primary cultures of pituitary cells were performed using an enzymatic and mechanical method as previously described (69). Briefly, pituitaries from 15 to 20 eels per experiment were incubated at 25 °C in a solution of porcine type II trypsin (Sigma-Aldrich) in PBS. After 1h, the trypsin solution was replaced by a solution of DNase (Sigma-Aldrich) and soya bean trypsin inhibitor (Sigma-Aldrich) for 30 min. Pituitaries were cut in 1mm slices using a McIlwain Tissue Chopper (Thermo Fisher Scientific), and then washed with PBS and mechanically dispersed by repeated passages through a plastic transfer pipette (Falcon, Thermo Fisher scientific, Illkirch, France). After estimating the number of viable cells by Trypan Blue exclusion (Sigma-Aldrich), cells were plated on 96-well plates (62,500 cells/well) pre-coated with poly-L-lysine (Sigma-Aldrich). Cultures were performed in culture medium (CM) at 18 °C under 3% CO2 and saturated humidity.
Treatments were started 24 h after the beginning of culture to allow cell attachment (Day 0). Replicates of 5 wells for control and each treated group were used. Stock solutions were diluted in CM just before addition to the culture wells. The final concentration of ethanol or DMSO in culture wells never exceeded 0.2%; control wells were treated with the same concentration of ethanol and/or DMSO in CM. Culture medium was changed and treatment added to the cells on Day 0, Day 3, and Day 7. Cultures were stopped on Day 8. The effects of treatments were tested in three independent experiments performed on different cell preparations from different batches of eels. Similar responses were observed in the independent experiments.
Total RNA was directly extracted in wells using the Cell-to-cDNA II Kit (Ambion, Thermo Fisher scientific, Illkirch, France) according to the manufacturer’s recommendations. Cells were washed with PBS and lysed with Cell Lysis II Buffer (80 µl/well). The lysates were digested with RNAse-free DNase I (Roche, Thermo Fisher scientific, Illkirch, France). Four µl of RNA solution of each sample was then reverse transcribed with a SuperScript III First Strand cDNA Synthesis Kit (Invitrogen, Thermo Fisher scientific, Illkirch, France). The samples obtained were stored at –20 °C until qPCR.
2.4 Real-time quantitative PCR
Gene specific primers were previously designed based on the nucleotide sequence of the European eel gnrhr2 (56)] and β-actin (70) cDNA, the latter being used as reference gene. Basal gnrhr1a and gnrhr1b expressions were below the threshold of detection in primary cultures of eel pituitary cells (65–67), and none of the treatments tested in the present study could induce their expression above this limit. For this reason, qPCR data were not reported for these two genes.
qPCRs were prepared with 4 µl of diluted cDNA template, 2 µl of PCR grade water, 2 µl of SYBR Green master mix and 1 µl of each forward and reverse primer (500 nM each at final concentration). The protocol was as previously described for β-actin (70) and for gnrhr2 (65, 66). Serial dilutions of cDNA pools of pituitary cells were used as a standard curve. One chosen dilution was also included in each run as a calibrator. Each qPCR run contained a non-template control (cDNA substituted by water) for each primer pairs to confirm that reagents were not contaminated. The specificity of each reaction was assessed by melting curve analysis to ensure the presence of only one product. Each sample was analyzed in duplicate by qPCR. Normalization of data was performed using β-actin mRNA levels.
2.5 Statistics
Results of qPCR are given as mean ± SEM. Means were compared by one-way ANOVA Tukey’s multiple comparison test using Instat (GraphPad Software Inc., San Diego, Calif., USA).
2.6 In silico retrieval of eel gnrhr2 gene sequences for promoter analysis
To investigate the upstream regulatory regions of the gnrhr2 gene, genomic sequences were retrieved from both the European eel and the Japanese eel genomes. The genome assembly for the European eel was obtained from GenBank (GCA_013347855.1), and the gnrhr2 gene was identified on chromosome 16 (accession number: NC_049216). For the Japanese eel, sequence data were collected from genome assembly GCA_025169545.1, with the gnrhr2 gene found to be similarly located on chromosome 16 (accession number: CM_045898). The 5’-flanking regions upstream of the coding sequence (up to about 2 Kb) were extracted for promoter analysis.
2.7 Prediction of transcription factor-binding sites in eel gnrhr2 promoter
Promoter analyses were subsequently performed on the retrieved 5’-flanking sequences to identify putative transcription factor-binding sites that may contribute to the regulation of gnrhr2 gene expression. Predictions were made using the PROMO tool, which is based on the TRANSFAC database, as well as the JASPAR database (https://jaspar.elixir.no) (71). These tools were supplemented by information from previously published studies (72–77).
Several potential regulatory elements were identified based on sequence similarity to motifs previously reported in mouse [for review (77)]: Activating Protein 1 (AP1); cAMP Response Element (CRE); Downstream Activin Regulatory Element (DARE); GnRH Receptor Activating Sequence (GRAS); LIM/homeobox protein LHX3 binding site (LHX3); Steroidogenic Factor 1 (SF1); Sequence Underlying Responsiveness to GnRH (SURG1 and SURG2).
2.8 Comparison with transcription factor-binding sites in the promoters of other teleost gnrhr2 and mammalian gnrhr1
To assess the conservation and divergence of transcriptional regulatory elements across species, we compared the European and Japanese eel gnrhr2 promoter regions with gnrhr promoter sequences from selected vertebrate species. The coding sequence of European eel gnrhr2 was used as a query to perform a translated BLAST (tBLASTn) search against the NCBI nucleotide and genome databases for identification of orthologous genes in other teleosts. The closest sequences retrieved by blasting were in zebrafish, the gnrhr4 gene (GeneID: 100001586) located on chromosome 18 (NC_007129), and in medaka, the gnrhr4 gene (GeneID: 100125529) located on chromosome 6 (NC_019864). The close relationship between eel gnrh2 and these genes in zebrafish and medaka, shown in our study by blasting, is in agreement with Ciani’s phylogeny which clusters them all in the gnrhr2b subtype (39). We also compared to the promoter of the mammalian gnrhr expressed in the pituitary, using human and mice gnrhr1 promoters. The human gnrhr1 (Gene ID: 2798) and mouse gnrhr1 (Gene ID: 14715) are located on chromosome 4 (NC_000004) and chromosome 5 (NC_000071), respectively. For promoter analysis in teleosts and mammals, the gnrhr genomic regions were retrieved to extract up to ~2.0 kb upstream sequences from the ATG start codon, which were analyzed for putative transcription factor-binding sites. Previously published mammalian gnrh1 promoter sequences and response elements [for review (77)] were also used.
3 Results
3.1 Effects of steroid hormones on gnrhr2 transcript levels in eel pituitary cells in vitro
Various concentrations (from 10–11 to 10–7 M) of sex steroids E2, T, P, as well as glucocorticoid, F, were tested over 8 days of culture according to previous experiments (70) (Figure 1). E2 had no effect at 10–11 M but significantly increased gnrhr2 mRNA levels at 10–9 and 10–7 M (x10 and x13, as compared to controls, respectively; P < 0.0001). T had no effect at 10–11 M but significantly increased gnrhr2 mRNA levels at 10–9 and 10–7 M (x6.6, P < 0.01 and x13, P < 0.0001, respectively). No effect of P nor F was observed at the three concentrations tested.
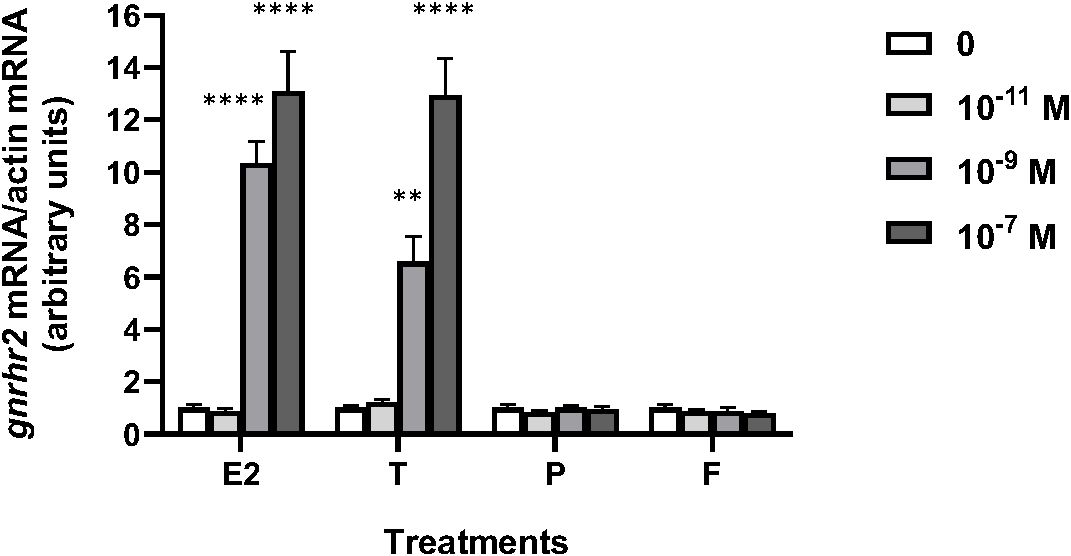
Figure 1. Effects of steroid hormones on gonadotropin-releasing hormone receptor 2 (gnrhr2) transcript levels in primary cultures of eel pituitary cells. Eel pituitary cells were treated with various concentrations of estradiol (E2), testosterone (T), progesterone (P) or cortisol (F) for 8 days. The mRNA levels of gnrhr2 were quantified by qPCR. Data were normalized against β-actin. The Figure displays the results from a representative experiment of three independent cell culture experiments. Mean ± SEM; n=5 well replicates. **, P < 0.01 and ****, P < 0.0001 versus controls, ANOVA.
3.2 Effect of an anti-aromatase, fadrozole, on sex steroid stimulation of gnrhr2 transcript levels in eel pituitary cells in vitro
Fadrozole (10–6 M), an inhibitor of aromatase, was tested alone or in the presence of 10–7 M sex steroids, E2, T or 11-KT (a non-aromatizable androgen) (Figure 2). Fadrozole had no effect alone and did not affect the stimulatory effect of 10–7 M E2 on gnrhr2 mRNA levels (x9.9 in the presence of fadrozole as compared to controls, versus x9.5 in the absence of fadrozole). In contrast, 10–7 M T stimulatory effect (x9.9) on gnrhr2 mRNA levels was completely suppressed by fadrozole (P < 0.0001), reaching control levels. No significant effect of 11-KT on gnrhr2 mRNA levels at 10–7 M was observed in the absence and in the presence of fadrozole.
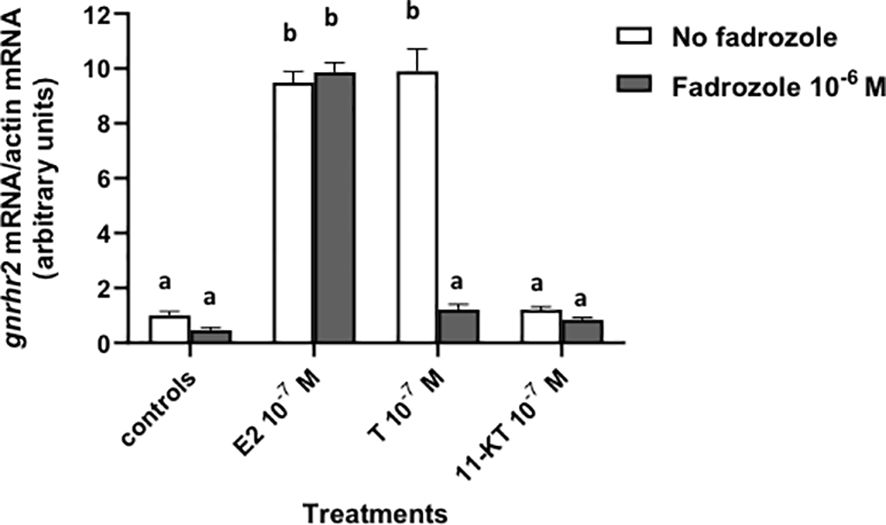
Figure 2. Effect of fadrozole, an aromatase inhibitor, on sex steroid effects on gonadotropin-releasing hormone receptor 2 (gnrhr2) transcript levels in primary cultures of eel pituitary cells. Eel pituitary cells were treated with 10–7 M of estradiol (E2), testosterone (T) or 11-ketotestosterone (11-KT) in the presence or not of 10–6 M fadrozole for 8 days. The mRNA levels of gnrhr2 were quantified by qPCR. Data were normalized against β-actin. The Figure displays the results from a representative experiment of three independent cell culture experiments. Mean ± SEM; n=5 well replicates. Different letters indicate significant differences (P < 0.0001) between groups, ANOVA.
3.3 Effect of activins A and B on gnrhr2 transcript levels in eel pituitary cells in vitro
Various concentrations of peptide hormones activin A and activin B (from 10–12 to 10–8 M) were tested over 8 days of culture according to previous experiments (68) (Figure 3). Both hormones had no effect at 10–12 M, but gnrhr2 mRNA levels were significantly decreased by activin A at 10–10 and 10–8 M (x0.4, P < 0.05 and x0.34, P < 0.01 as compared to controls, respectively), and by activin B at 10–8 M (x0.36; P < 0.05, as compared to controls).
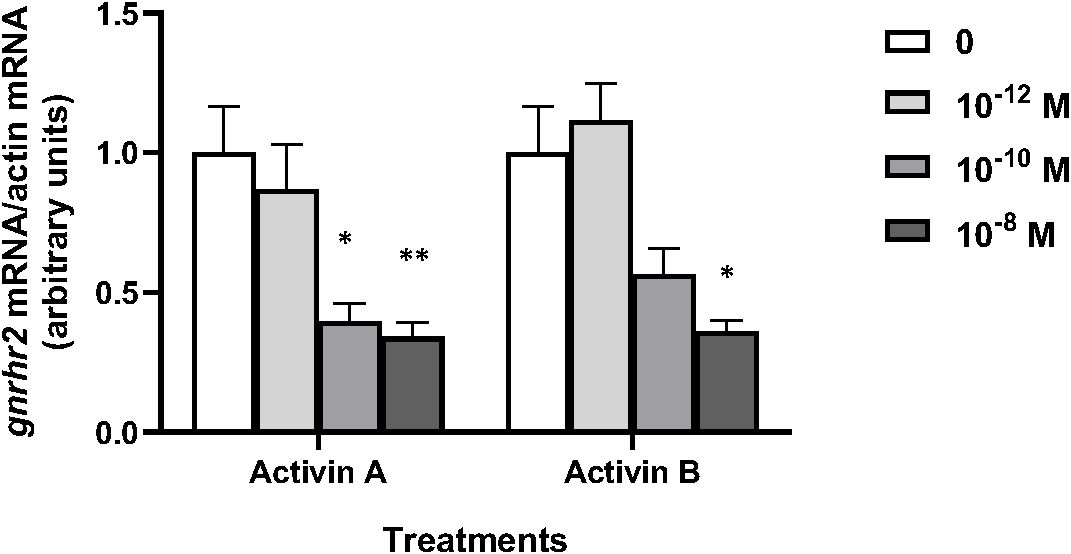
Figure 3. Effects of activins A and B on gonadotropin-releasing hormone receptor 2 (gnrhr2) transcript levels in primary cultures of eel pituitary cells. Eel pituitary cells were treated with various concentrations of activin A or activin B for 8 days. The mRNA levels of gnrhr2 were quantified by qPCR. Data were normalized against β-actin. The Figure displays the results from a representative experiment of three independent cell culture experiments. Mean ± SEM; n=5 well replicates. *, P < 0.05 and **, P < 0.01 versus controls, ANOVA.
3.4 Genomic structure of eel gnrhr2 gene
The gnrhr2 gene sequence of the European eel was retrieved from GenBank genome assembly GCA_013347855.1. The gnrhr2 gene was identified on chromosome 16 (NC_049216), spanning the genomic region from position 12,519,164 to 12,529,023 (Gene ID: 118215598). It is composed of three exons separated by two introns (Figure 4), and the entire gene covers 9,860 bp on the chromosome. The CDS of the gene, registered under accession number XM_035396496, is 1,308 bp in length and encodes a protein of 435 aa, with the corresponding protein sequence recorded under accession number XP_035252387. In the Japanese eel, gnrhr2 was not previously annotated. Gene sequence information was retrieved from GenBank genome assembly GCA_025169545.1, and gnrhr2 was found on chromosome 16 (CM_045898), between positions 23,834,808 and 23,841,537. We predicted exon-intron boundaries following the GT-AG rule. The Japanese eel gnrhr2 gene also comprises three exons and two introns (Figure 4). Its CDS is 1,308 bp long and encodes a protein of 435 aa, showing a high sequence conservation between these two eel species (96.79% identity).
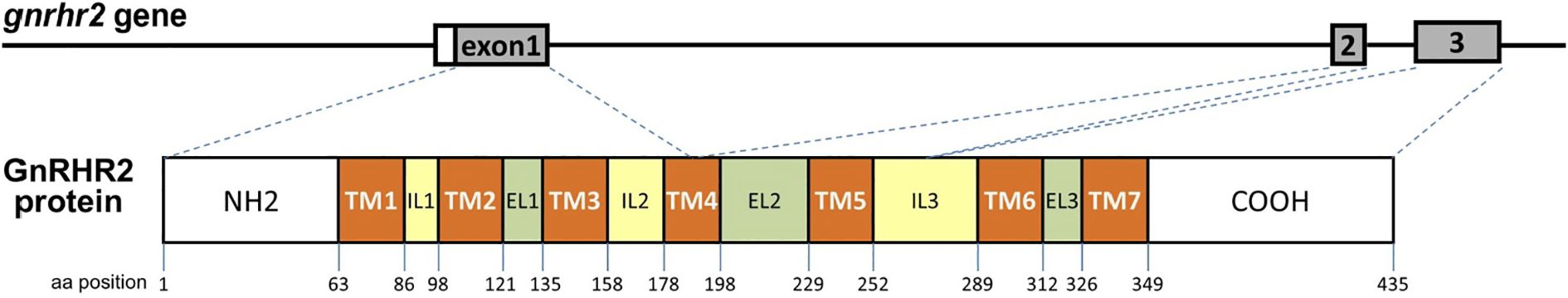
Figure 4. Schematic representation of the European eel gonadotropin-releasing hormone receptor 2 (gnrhr2) gene and its encoded protein structure. The European eel gnrhr2 gene, retrieved from genome assembly GCA_013347855.1, spans 9,860 bp on chromosome 16 (NC_049216) and consists of three exons and two introns. The main transcript (XM_035396496) encodes a 435-amino acid GnRHR2 protein (XP_035252387). The gene structure is shown in the upper panel, with exon numbers indicated. The corresponding GnRHR2 protein structure is illustrated below, showing the seven transmembrane domains (TM1–TM7), the three extracellular loops (EL1–EL3), and the three intracellular loops (IL1–IL3). Exon-protein domain correspondence is indicated by dotted lines. Similar genomic structure was found in the Japanese eel, with retrieved gnrhr2 gene sequence on chromosome 16 (CM_045898) from genome assembly GCA_025169545.1.
The gnrhr2 genes from the European eel and the Japanese eel display a highly conserved exon structure in terms of coding sequence organization and functional domain distribution (Figure 4). In the European eel, exon 1 comprises 582 nucleotides, encoding the first three transmembrane (TM) domains and a portion of the fourth TM domain. Exon 2 comprises 205 nucleotides, encompassing the remaining part of the fourth TM domain and the entire fifth TM domain. Exon 3 consists of 521 nucleotides, encoding the sixth and seventh TM domains along with the remainder of the coding sequence. Similarly, in the Japanese eel, exon 1 contains 582 nucleotides and encodes the same TM domains as in the European eel. Exon 2 includes 204 nucleotides, covering the rest of the fourth TM and the fifth TM domain. Exon 3 spans 522 nucleotides, responsible for encoding the sixth and seventh TM domains and the remaining coding sequence. These findings highlight the strong conservation in the structural organization of the gnrhr2 gene between the two eel species.
3.5 Analysis of eel gnrhr2 promoter region
The predicted response elements are indicated in the promoter sequences of the European eel and the Japanese eel gnrhr2 genes (Figure 5). Sequence analysis up to ~2.0 kb upstream from the ATG start codon in the European eel revealed multiple transcription factor binding sites including a cAMP Response Element (CRE) at position nt -114, that binds CRE-binding protein (CREB), along with a putative binding site for Steroidogenic Factor 1 (SF1) at nt -149. Two TAAT/ATTA motifs were located at nt -187 and nt -1362, representing potential Downstream Activin Regulatory Element (DARE) homeodomain protein binding sites. The binding sites for LHX3, a LIM-homeodomain transcription factor, were found at nt -293 and -572. Two GnRH Receptor Activating Sequence (GRAS) elements known to mediate Suppressor of Mothers Against Decapentaplegic (SMAD) and Forkhead box L2 (FOXL2) binding in mammals, were identified at nt -454 and -1428, and two binding sites for Activating Protein 1 (AP1) were observed at nt -507 and -735. In addition, three Sequence Underlying Responsiveness to GnRH (SURG) – 2 elements (SURG2) were detected at nt -720, -1355, and -1926, which are also AP1 binding sites. A SURG1, an element that was previously reported as interacting with transcription factors such as OCTamer binding transcription factor 1 (OCT1 formally named POU2F1) and Nuclear Factor Y (NFY), was present at nt -1655. These elements were also identified in the promoter sequence of the Japanese eel gnrhr2: a CRE next to the first exon (at nt -114) as in the European eel, but also an additional CRE at -659; two DARE at nt -165 and -1257, two GRAS at nt-350 and -1317, two AP1 at nt -403 and -634, as in the European eel; a single LHX3 at nt -472 corresponding to the second one of the European eel; a SF1 but located further away from the first exon, at nt -604, as compared to the European eel; three SURG2 at nt -619, -1250 and -1815, and a SURG1 at nt -1544, as in the European eel.
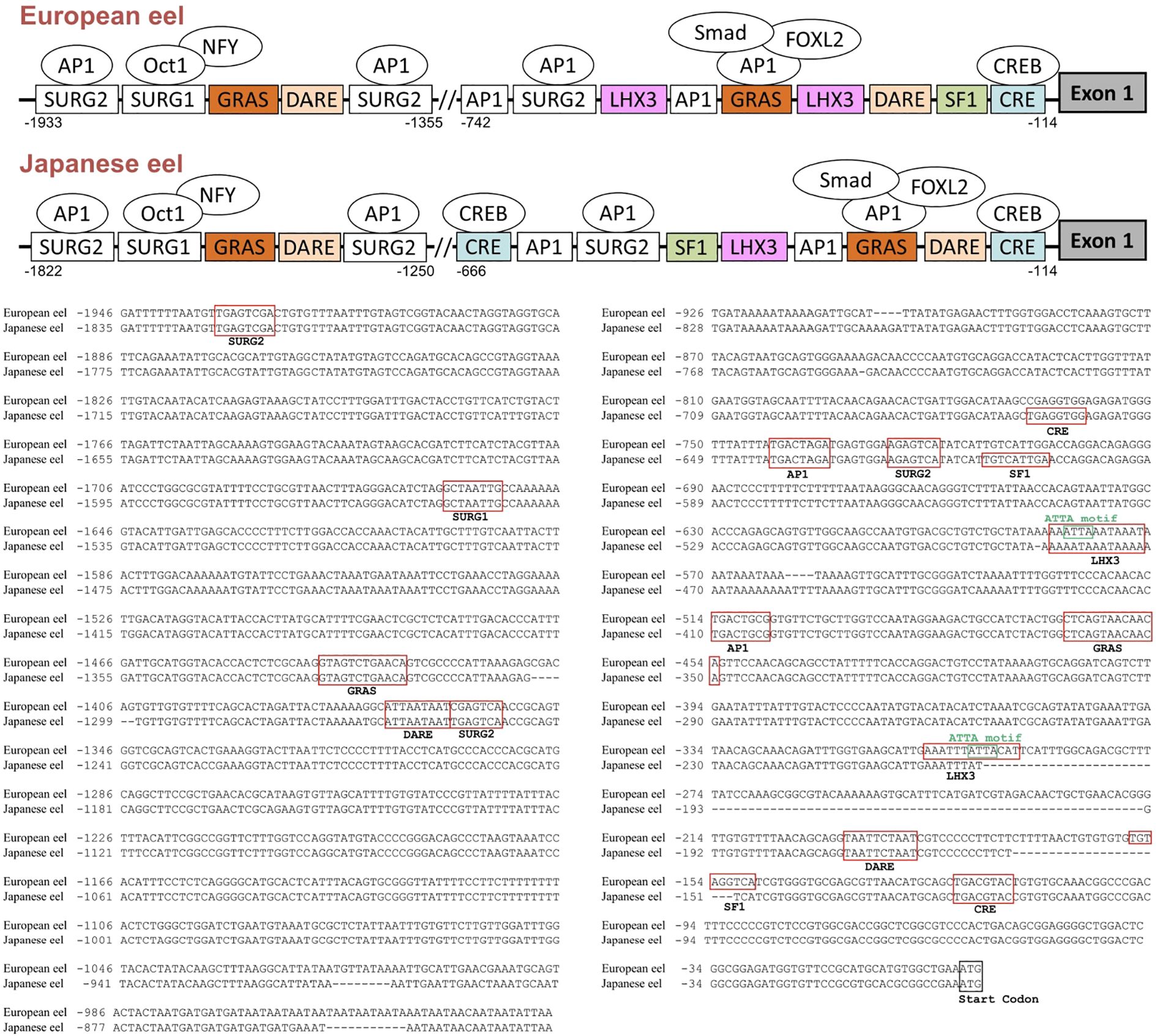
Figure 5. Predicted transcription factor binding sites in the 5′-flanking region of the European and Japanese eel gonadotropin-releasing hormone receptor 2 (gnrhr2) genes. Promoter analysis of the gnrhr2 gene revealed multiple putative transcription factor-binding sites within ~2.0 kb upstream of the ATG start codon. A schematic representation of the gnrhr2 promoter response elements, with some potential binding factors, in both eel species, is provided in the upper panel. The exon 1 coding region is depicted as a dark grey box on the right. The alignment of gnrhr2 promoter sequences between European eel and Japanese eel is shown in the lower panel, with identified response elements boxed in red, and the start codon of exon 1 boxed in black. AP1, activating protein 1; CRE, cAMP response element; CREB, CRE binding protein; DARE, downstream activin regulatory element; FOXL2, Forkhead box L2; GRAS, GnRH receptor activating sequence; LHX3, LIM/homeobox protein LHX3 binding site; NFY, Nuclear Factor Y; OCT1, Octamer binding transcription factor 1; SF1, steroidogenic factor 1; SMAD, Suppressor of mothers against decapentaplegic; SURG, sequence underlying responsiveness to GnRH.
3.6 Comparison of transcription binding sites in promoter region of eel gnrhr2 and other species gnrhr
The promoter regions of gnrhr2 from the European eel and the Japanese eel were compared to those of gnrhr2 from other teleosts (zebrafish and medaka) and of gnrhr1 from human and mouse. The distribution of consensus transcription factor binding elements identified in these promoter regions are schematically illustrated in Figure 6, including CRE, SF1, DARE, LHX3, GRAS, AP1, as well as SURG1 and SURG2. The sequence alignments of predicted response elements within the gnrhr promoter regions among selected species, with their relative positions, are presented in Figure 7.
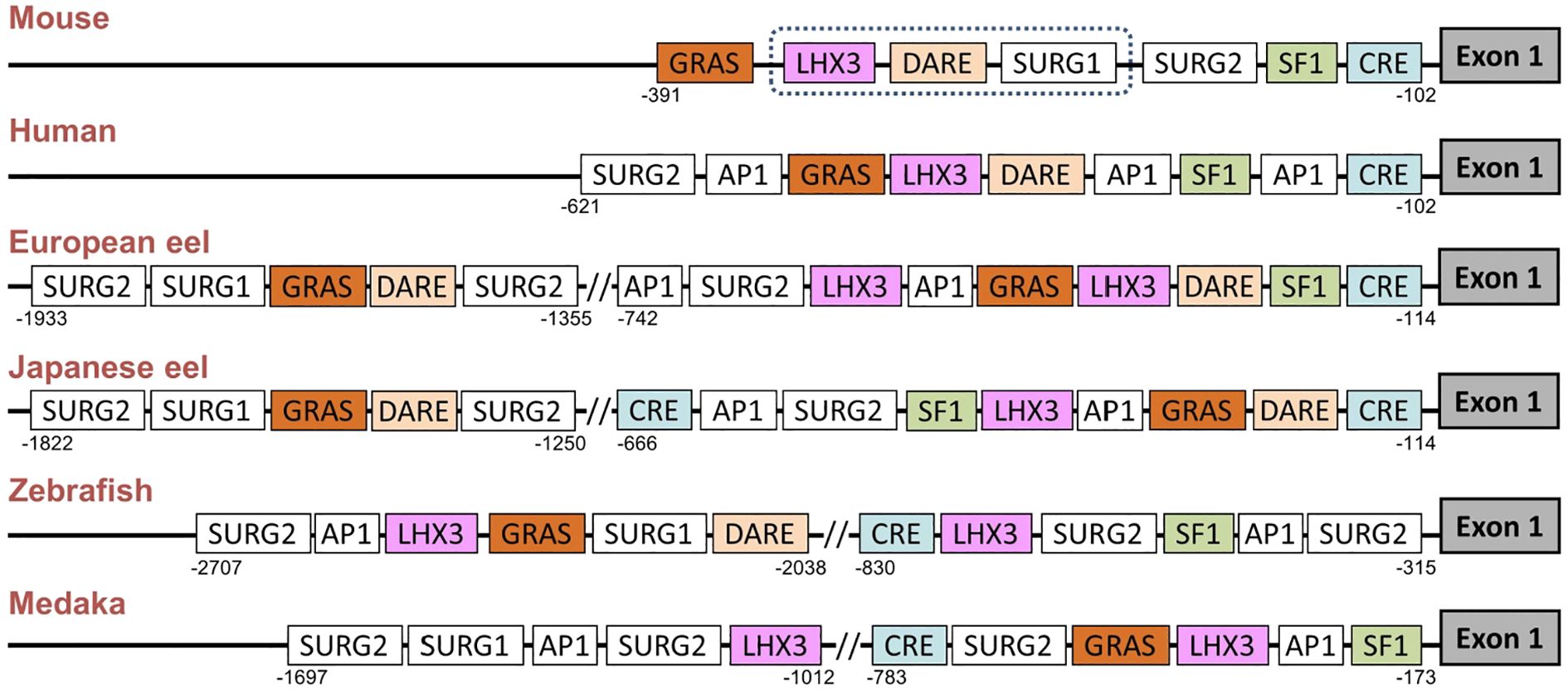
Figure 6. Comparison of predicted transcription factor binding sites in gonadotropin-releasing hormone receptor (gnrhr) promoters between European and Japanese eels and other teleost and mammals. The figure displays a schematic representation of promoter response elements of gnrhr1 from mouse and human and of gnrhr2 from European eel, Japanese eel, zebrafish and medaka. For gene references, see Materials and Methods. The exon 1 coding region is depicted as a dark grey box on the right. The dotted line in the mouse promoter indicates a region with several binding elements overlapping. AP1, activating protein 1; CRE, cAMP response element; DARE, downstream activin regulatory element; GRAS, GnRH receptor activating sequence; LHX3, LIM/homeobox protein LHX3 binding site; SF1, steroidogenic factor 1; SURG, sequence underlying responsiveness to GnRH.

Figure 7. Comparison of predicted transcription factor binding motif sequences in gonadotropin-releasing hormone receptor (gnrhr) promoters between European and Japanese eels and other teleost and mammals. Multiple sequence alignments of predicted response elements were performed using promoter sequences of gnrhr1 from mouse, and human, and gnrhr2 from European eel, Japanese eel, zebrafish, and medaka. Each panel displays a typical response element. When a response element is present in multiple locations in a promoter, the numbers following species names (e.g., European eel.1, Japanese eel.2) correspond to the relative positions, upstream of the ATG start codon. AP1, activating protein 1; CRE, cAMP response element; DARE, downstream activin regulatory element; GRAS, GnRH receptor activating sequence; LHX3, LIM/homeobox protein LHX3 binding site; SF1, steroidogenic factor 1; SURG, sequence underlying responsiveness to GnRH.
Among these predicted response elements, a CRE site was found in each species examined, with an additional one in the Japanese eel. The CRE was located just upstream of exon 1 in the two eel species, as in mammals, while it was positioned more distantly in zebrafish and medaka, at a location corresponding to that of the second CRE of the Japanese eel. A SF1 was retrieved in the promoter of each species investigated.
In the mouse promoter, several binding elements overlap (SURG1, DARE and LHX3), while we found them separated in other species. SURG1 and SURG2, which were originally identified in gnrhr promoter in mammals, were also detected in teleost species. A single SURG1 element was located in all analyzed teleost promoters, whereas multiple SURG2 elements were found. We retrieved DARE in the proximal region of the promoter in human as well as in the European and Japanese eels; an additional DARE was also found in a more distant region of the promoter in both eel species; in zebrafish, a DARE was at a position possibly corresponding to the second DARE of the eels, while no DARE was retrieved in medaka. Concerning LHX3, it was present in all species, with an additional one in teleosts (European eel, zebrafish and medaka).
As previously shown in the mouse gnrhr1 promoter, the GRAS motif was consistently detected in all examined species, with two GRAS found in both eels. The single GRAS motif found in the proximal region of the promoter in medaka may correspond to the first one of the eels, while the one found more distantly in the zebrafish may correspond to the second one of the eels. Repeated AP1 motifs appeared frequently across species, with two sites observed in all analyzed teleosts, and three sites in human.
4 Discussion
4.1 High conservation of gnrhr promoter response elements between teleosts and mammals
The response elements characterized in the eel gnrhr2 promoters are similar to the major ones involved in the tissue-specific activity, and in the response to regulatory factors, for mammalian gnrhr1 promoters in the pituitary gonadotroph cells [ (78–80); for review (77)]. We also identified them in the gnrhr2 promoter of two other teleost model species, zebrafish and medaka. This suggests a high evolutionary conservation of the molecular mechanisms underlying gnrhr gene expression in gonadotroph cells, even between distant species like teleosts and mammals. These shared transcription factor binding motifs include CRE, SF1, DARE, LHX3, GRAS, AP1, SURG1 and SURG2, which have been shown to mediate either basal or regulated expression of gnrh1 in mammal species. Notably, in rodents, mouse and rat, SF1 in the proximal promoter region is essential for gnrhr1 gonadotroph-specific expression and basal transcriptional activity (78, 80). This same factor was also identified in the eel and other teleost gnrhr2 promoters, suggesting conserved pituitary-specific regulation. Various homeobox factor binding sites, which participate in the gonadotroph-specific expression of the mammalian gnrhr1 promoter, are also found in the eel and other teleost gnrhr2, such as SURG1, which binds OCT1 and NFY, both involved in directing basal expression and GnRH-stimulated expression on the gnrhr gene (75). In vivo chromatin immunoprecipitation (ChIP) assays confirmed that OCT1 and NFY bind to the SURG1 element in the mouse, and this binding increases in response to GnRH stimulation (75). In the mouse, SURG2 overlaps with a conserved AP1 consensus binding site and the AP1 binding site is essential for gnrhr gene expression under GnRH stimulation, while GRAS is a composite regulatory element whose functional activity depends on the binding of Smad proteins, AP1, and FOXL2 and mediates both activin and GnRH responsiveness (75). We identified a GRAS motif in the gnrhr promoters of all analyzed species, suggesting a potentially conserved regulatory mechanism across vertebrates. LHX3 binding sites, which directly activate the mouse gnrhr1 promoter through an ATTA core motif (76, 81), were also found in the eel and other teleost gnrhr2 promoters. The DARE motif contains TAAT/ATTA motifs, which were shown to bind homeodomain transcription factors such as LHX3 in the mouse gnrhr1 promoter (82). DARE was also identified in the present study in the promoters of gnrhr1 in human, and gnrhr2 in European eel, Japanese eel, and zebrafish, indicating a conserved regulatory site across vertebrates.
In the present study, the identification of response elements in gnrhr1 promoter of mammalian species (human and mouse) and in gnrhr2 promoter of teleost species (eels, zebrafish and medaka) show a conservation of key regulatory elements in the gnrhr promoter in the osteichthyan lineage, which encompass actinopterygians (such as teleosts) and sarcopterygians (such as mammals). This reveals an ancient origin and evolutionary conservation of transcriptional control mechanisms governing gnrhr expression by pituitary gonadotroph cells. Furthermore, the identification of these conserved elements in eel supports this basal teleost as a valuable comparative model for understanding the evolution of vertebrate reproductive endocrinology. Further comparative analyses in other lineages such as chondrichthyans (cartilaginous fishes) and cyclostomes (jawless vertebrates) would allow the elucidation to whether this regulatory system is already present in early vertebrates, before the emergence of jawed vertebrates.
4.2 In vitro and in silico insights for estrogen-specific stimulation of eel pituitary gnrhr2 expression
In our present study, we showed a stimulatory effect of E2, as well as of T, on gnrhr2 mRNA levels in primary culture of eel pituitary cells. This effect was estrogen-specific, as no such a stimulatory effect was induced by a non-aromatizable androgen 11-KT and the stimulatory effect of T was abolished in the presence of an aromatase inhibitor, fadrozole.
In various teleost species, sex steroids also modulate pituitary responsiveness to GnRH and regulate gnrhr mRNA levels. A study in primary cultured pituitary cells also reported an increase in pituitary gnrhr3 but not gnrhr1 mRNA levels in tilapia after E2 exposure while T was not tested (41). In the black porgy, after both E2 and T treatments, gnrhr1 (gnrhr2bb) mRNA levels were increased in dispersed pituitary cells, while 11-KT did not change them, suggesting that in this species like in the eel, the stimulatory effect of T on gnrhr expression may be mediated by aromatization, thus being estrogen-specific (47). Another in vitro study in Atlantic cod compared the effects of E2, T and dihydrotestosterone DHT [a non-aromatisable androgen but whose metabolite 3β-diol, also named 5α-androstane-3β, 17β-diol, binds to estrogen receptor β (83, 84)] and demonstrated that all increased pituitary gnrhr2a mRNA levels, without affecting gnrhr1b (85). A recent study, using ex vivo whole pituitaries of Atlantic salmon, reported that pituitary gnrhr2bba expression is stimulated by both E2 and 11-KT, indicating both estrogenic and androgenic effects (86).
In vivo studies in teleosts also reported that E2 treatment induces an increase in pituitary gnrhr mRNA levels. In tilapia, both gnrhr1 and gnrhr3 mRNA levels increase after E2 treatment (41), while in black porgy, only gnrhr1 (gnrhr2bb) increases (47, 87). The effect of T was tested in Atlantic salmon, showing a stimulatory effect on the pituitary transcripts of gnrhr4 (45). In contrast, in their study on endocrine disrupting chemicals in the hermaphroditic fish Kryptolebias marmoratus, Rhee and colleagues noted that exposure to E2 in water induces a decrease in gnrhr mRNA levels (88).
In rat, E2 treatment of adult female pituitary cells induces an increase in GnRHR number as well as on GnRH-induced LH release (89–91). In ewe, E2 is also able to increase GnRHR number (92) and gnrhr1 mRNA levels by primary cultures of pituitary cells (93, 94). In contrast, E2 has no effect on gnrhr1 mRNA levels expressed in mouse gonadotrope cell line, LβT2 (95).
The positive in vitro effect of E2 or of T after aromatization on gnrhr2 transcripts in the European eel supports our previous in vivo data showing the need to use sex steroids to sensitize the eel pituitary LH response to GnRH (53). This pathway is likely involved in the increase of gnrhr2 expression observed during eel sexual maturation (56) via the positive feedback of sex steroids.
Despite the strong estradiol-specific regulation of eel gnrhr2, we did not evidence a typical estrogen response element (ERE) within the eel gnrhr2 proximal promoter region that we have investigated. This aligns with findings in mammals, in which canonical ERE could not be identified in the gnrhr1 promoter, and thus would not mediate E2 regulatory effect on gnrhr1 expression. Instead, non-classical pathways appear to mediate E2 action on gnrhr1 transcription across mammalian species and cell types [ (20); for reviews (77, 96)]. In human ovarian (OVCAR3) and breast (MCF7) cell lines, it was shown that E2-activated ERα represses gnrhr1 gene transcription via an indirect mechanism involving CBP (CREB binding protein) and AP1 (73). A similar mediation by CREB was also demonstrated to underlie the stimulatory effect of E2 on gnrhr expression by ovine pituitary cells (20). We identified CRE and AP1 in the eel gnrhr2 promoter, as well as in other teleost gnrhr2 promoters, both response elements implicated in E2 signaling in mammalian gnrhr1 promoters. We suggest that the stimulatory effect of E2 on gnrhr2 expression, observed in our study, may be mediated via these two response elements. These findings on estrogen-specific stimulation of gnrhr2 expression levels in eel pituitary cells contribute significantly to the understanding of the conservation of the regulation of gnrhr by estrogens across vertebrates.
The increase in gnrhr expression further highlights the multiple targets of the steroid positive feedback on brain-pituitary gonadotropic axis in the eel, together with the previous demonstration of the stimulatory effects of gonadal steroids on the synthesis of brain GnRH and pituitary LH. We suggest that the positive regulation by E2 of gnrhr2 expression is exerted on LH cells. Differently from the situation in mammals and other tetrapods, where both gonadotropins are produced by the same pituitary cells, LH and FSH are expressed by distinct pituitary cells in teleosts, including in the eel as shown by in situ hybridization (ISH) (97). Furthermore, a recent study proposed a dual neuroendocrine control of gonadotropins in teleosts, with GnRH acting as LH releasing hormone while cholecystokinin as FSH releasing hormone (98). Future experiments, such as double ISH or ISH coupled to immunohistochemistry on dispersed pituitary cells, an approach already set up for DA receptors in the European eel (99), could investigate which pituitary gonadotroph cell(s) express gnrhr2.
4.3 Lack of evidence for progesterone and cortisol regulation of eel pituitary gnrhr2 expression
In the European eel, we observed no direct effect of progesterone on gnrhr2 expression in vitro. To our knowledge, the unique other study regarding the effect of progestogens on gnrhr expression in teleosts was performed in tilapia (41). The authors demonstrated that a progestin, 17α, 20β-dihydroxy-4-pregnen-3-one (DHP), could positively regulate gnrhr1 and gnrhr3 mRNA levels by primary cultures of pituitary cells, while only those of gnrhr1 are elevated in vivo. In contrast, in ewe, an inhibitory effect of progesterone on basal or E2-induced gnrhr1 expression by primary culture of pituitary cells was reported in vitro (92, 94, 100). In vivo, progesterone had no effect on the basal number and/or mRNA levels of pituitary GnRHR of ovariectomized ewes (18, 101) and cows (102), but reduced their E2-induced number [ewes (101); cows (102)]. In contrast, combined treatment of hypogonadic (hpg) female mice with GnRH, E2 and progesterone elevated the pituitary GnRHR number to the same levels as normal mice (103) supporting a positive synergistic effect of progesterone. Further studies may investigate hormone interactions in the regulation of eel gnrhr2 transcript levels.
A functional progesterone response element (PRE) has been characterized in human gnrhr1 promoter, which mediates the inhibitory effect of progesterone in human gonadotroph cells via progesterone receptor isoforms PR-A and PR-B (104). Such a PRE binding site has not been found in the eel gnrhr2 promoter, possibly explaining the absence of progesterone effect on gnrhr2 transcript levels. It should be noted, however, that other signaling mechanisms may mediate the regulatory effects of progesterone, as no PRE were identified in gnrhr1 promoter of some mammalian species such as rodents [for review (77)]. Overall, these findings reveal some species-specific patterns in the hormonal regulation of gnrhr genes, and indicate the importance of promoter structure in hormone responsiveness.
In our study, cortisol did not induce any change in mRNA levels of pituitary gnrhr2 in eel pituitary cells. In contrast, one other recent in vitro study in a teleost, the Atlantic cod, reports an induction of gnrhr2a, but not gnrhr1b, by cortisol (85). In mammals, cortisol has no effect on the basal number of GnRHR and/or gnrhr1 mRNA levels in vitro in ewe (105) and in vivo in castrated sheep (106, 107). However, in vivo it can reduce the stimulatory effect of E2 on the number and mRNA levels of GnRHR in castrated sheep (106, 107), while it increases the stimulatory effect of GnRH on GnRHR number in intact male rats (108). Treatment of mouse gonadotrope cell line, LβT2, with dexamethasone can increase gnrhr1 mRNA levels only in combination with E2 (95). As stress may largely affect fish reproduction, future studies may further address the interaction between corticosteroids, sex steroid and GnRH in the regulation of GnRHR expression in teleosts.
Cortisol response elements, typically referred to as glucocorticoid response elements (GREs), are essential DNA sequences that mediate the transcriptional effects of glucocorticoids via the direct binding of the glucocorticoid receptor (GR). We did not identify a classical GRE in the promoter of eels nor other teleost gnrhr2 genes, in line with the current data in mammals. The lack of GREs in the gnrhr1 promoter across mammalian species suggested more complex mechanisms for glucocorticoid actions. Thus, in the mouse, whose gnrhr1 promoter does not contain a GRE, the transcriptional regulation of the gnrhr1 gene by glucocorticoid is ensured by the recruitment of GR to the AP1 region of this promoter (109, 110).
4.4 In vitro and in silico insights for activin inhibition of eel pituitary gnrhr2 expression
In the European eel, we previously showed that activins oppositely regulate in vitro fshβ and lhβ expression by pituitary cells, with a stimulatory effect on fshβ and an inhibitory effect on lhβ mRNA levels (68). Besides their production by the gonads, localization of activins has been demonstrated within the pituitary, in various cell types [gonadotrophs in mammals (111); somatotrophs in teleosts (112)], suggesting paracrine/autocrine actions at the pituitary level. Activins are known to stimulate FSH production and release from gonadotrope cells in teleosts as in other vertebrates including mammals (113, 114).
In the present study, we show that both activins A and B are able to downregulate gnrhr2 mRNA levels by primary cultures of eel pituitary cells. Few data are available in other teleosts. A recent study using ex vivo whole pituitaries of Atlantic salmon post-smolts, exposed to stimulatory environmental conditions that promote sexual maturation (continuous light and 16 °C), showed that in immature but not maturing males, activin A stimulates the expression of gnrhr2bba (86), the only paralog out of six being stimulated during precocious male parr maturation (39). No effect of activin A on gnrhr2bba expression is observed in immature males exposed to non-stimulatory conditions (86).
Early studies showed that activin A stimulates the synthesis rate (as assayed by density shift technique) of GnRHR by rat pituitary cell cultures (33). This stimulatory effect of activin A on GnRHR is exerted at the transcriptional level as demonstrated in the mouse gonadotrope cell line, αT3-1, using gnrhr1 mRNA assay, run-off experiments, and transfection experiments of gnrhr1 promoter/luciferase reporter gene (34). In contrast, in the ovariectomized ewes, activin A decreases the number of GnRH-R (as assayed by the binding of a GnRH agonist) by primary cultures of pituitary cells but has no effect on their increase induced by E2 (115). This suggests species-specific variations in the positive or negative effects of activin on GnRHR in mammals, as in teleosts.
Extensive promoter studies in mice have identified GRAS as a critical regulatory element mediating activin-induced transcriptional activation of gnrhr1 in gonadotrope-derived cell lines such as αT3–1 and LβT2 cells (79, 81). GRAS functions as a composite enhancer, which recruits SMAD2/3/4 and cooperates with factors like AP1, FOXL2, SF-1 to modulate gene expression via overlapping or adjacent binding motifs [ (74, 79, 116); for review (77)]. We identified GRAS elements in gnrhr promoters of each mammalian and teleost species analyzed in this study. In the gnrhr2 promoters of the European and Japanese eels, we found two putative GRAS motifs, including one in the proximal region near to AP1 binding sites, suggesting a conserved activin-responsive promoter structure.
DARE further enhances responsiveness to activin, coupled with GRAS forming a functionally cooperative “activin-responsive unit (ARU)” within the mouse gnrhr promoter to drive transcriptional activation (81). DARE contains TAAT/ATTA motifs, which serve as binding sites for homeodomain transcription factors. For example, LHX3 has been shown to bind directly to DARE and activate transcription of mouse gnrhr1 when overexpressed (81). Additionally, homeodomain proteins such as Msx1 and Dlx3 also interact with TAAT-rich regions to regulate gnrhr1 transcription during gonadotroph development, with Dlx3 acting as an activator and Msx1 as a repressor, in the mouse (117). We found a DARE in the eel gnrhr2 promoter, but with “TAAT/ATTA” separated by only 2 bp rather than 4 bp spacing of murine DARE. As for the eel, we observed a 2bp spacer for the DARE motif of the human gnrhr1 promoter. In Cherrington et al’s study (81), increasing the spacer length between the tandem TAAT/ATTA motifs from 4 to 5 or 10 bp reduced gnrhr1 promoter activity, but no data are available concerning a shorter space. Whether this shorter spacing affects transcription factor binding or promoter activity remains to be elucidated.
Activin, which we previously showed to stimulate fshβ, while inhibiting lhβ, in the European eel (68), exerts an inhibitory effect on gnrhr2 expression by eel pituitary cells. As discussed above for the effect of steroids, the parallel regulation of lh and gnrhr2 expression by activin supports the hypothesis that the expression and regulation of gnrhr2 concerns mainly LH cells.
5 Conclusion and perspectives
This study gives new information on the regulation of eel pituitary gnrhr2 expression and provides the first insight into the sequence and response elements of gnrhr2 promoter in teleosts. Our results show that while activins inhibit gnrhr2 expression, gonadal steroids exert a positive feedback, mediated by estradiol, on pituitary sensitivity to GnRH in the eel. This may account for the increase in pituitary gnrhr2 mRNA levels reported in female and male eels experimentally matured under gonadotropic treatments (56). This increase in gnrhr expression further highlights the multiple targets of the steroid positive feedback on brain-pituitary gonadotropic axis in the eel, in line with the regulatory mechanisms of the ovulatory LH surge in mammals. Furthermore, the analysis of the eel gnrh2 promoter sequence suggests the absence of a classical ERE and the involvement of non-classical response elements such as CRE and AP1, similarly to the situation in mammals. This regulation by estradiol of GnRH receptivity would be an ancient and conserved mechanism across vertebrates. Currently, final oocyte maturation and ovulation in female eels, matured after chronic gonadotropic treatments, are induced by the administration of a progestogen, acting directly at the ovarian level (118, 119). The present finding of the increase in eel pituitary sensitivity to GnRH as a result of the estradiol positive feedback, further supports the use of alternative treatments to induce an endogenous ovulatory LH peak, by the administration of GnRH- agonist together with dopamine-antagonist. Future studies should also aim at deciphering upstream regulation of endogenous GnRH release in the eel, such as pheromones and environmental factors. This study in the eel, a basal teleost representative, contributes to raise basic and applied knowledge on the regulation and evolution of pituitary GnRH receptivity in vertebrates.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by French Cuvier Ethic Committee, Museum National d’Histoire Naturelle, France. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
C-JL: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. KR: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. C-FC: Funding acquisition, Supervision, Writing – review & editing. SD: Conceptualization, Formal analysis, Funding acquisition, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by grants from the French National Research Agency, France Taiwan ANR NEMO N° ANR-14-CE02-0020, the Yushan Scholar Program MOE, Taiwan, and European Community COST Action EELSUPPORT n°CA22163.
Acknowledgments
We thank E Ryckelynck and his team from Nodaiwa (Paris, France) for their kind cooperation. We are grateful to Dr MA Virmani (London, UK) for English correction.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
11-KT, 11-ketotestosterone; AP1, activating protein 1; CBP, CREB-binding protein; CRE, cAMP response element; CREB, CRE-binding protein; DA, dopamine; DARE, downstream activin regulatory element; DHP, 17α, 20β-dihydroxy-4-pregnen-3-one; DHT, dihydrotestosterone; E2, estradiol; ERE, estrogen responsive element; F, cortisol; FOXL2, forkhead box L2; FSH, follicle-stimulating hormone; GnRH, gonadotropin-releasing hormone; GnRHR, gonadotropin-releasing hormone receptor; GR, glucocorticoid receptor; GRAS, GnRH receptor activating sequence; GRE, glucocorticoid response element; hCG, human chorionic gonadotropin; LH, luteinizing hormone; LHX3, LIM/homeobox protein LHX3 binding site; NFY, nuclear factor Y; OCT1, octamer binding transcription factor 1; P, progesterone; PR, progesterone receptor; PRE, progesterone responsive element; SF1, steroidogenic factor 1; SMAD, suppressor of mothers against decapentaplegic; SURG, sequence underlying responsiveness to GnRH; T, testosterone.
References
1. Dufour S, Quérat B, Tostivint H, Pasqualini C, Vaudry C, and Rousseau K. Origin and evolution of the neuroendocrine control of reproduction in vertebrates, with special focus on genome and gene duplications. Physiol Rev. (2020) 100:869–943. doi: 10.1037//0033-2909.I26.1.78
2. Desaulniers AT, Cederberg RA, Lents CA, and White BR. Expression and role of gonadotropin-releasing hormone 2 and its receptor in mammals. Front Endocrinol (Lausanne). (2017) 8:269. doi: 10.3389/fendo.2017.00269
3. Hapgood JP, Sadie H, van Biljon W, and Ronacher K. Regulation of expression of mammalian gonadotrophin-releasing hormone receptor genes. J Neuroendocrinol. (2005) 17:619–38. doi: 10.1111/j.1365-2826.2005.01353.x
4. White BR, Cederberg RA, Elsken DH, Ross CE, Lents CA, and Desaulniers AT. Role of gonadotropin-releasing hormone-II and its receptor in swine reproduction. Mol Reprod Dev. (2023) 90:469–79. doi: 10.1002/mrd.23662
5. de Roux N, Young J, Misrahi M, Genet R, Chanson P, Schanson G, et al. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. (1997) 337:1597–602. doi: 10.1111/j.1365-2559.1988.tb01948.x
6. Chevrier L, Guimiot F, and de Roux N. GnRH receptor mutations in isolated gonadotropic deficiency. Mol Cell Endocrinol. (2011) 346:21–8. doi: 10.1016/j.mce.2011.04.018
7. Pask AJ, Kanasaki H, Kaiser UB, Conn PM, Janovick JA, Stockton DW, et al. A novel mouse model of hypogonadotrophic hypogonadism: N-ethyl-N- nitrosourea-induced gonadotropin-releasing hormone receptor gene mutation. Mol Endocrinol. (2005) 19:972–81. doi: 10.1210/me.2004-0192
8. Wu S, Wilson MD, Busby ER, Isaac ER, and Sherwood NM. Disruption of the single copy gonadotropin-releasing hormone receptor in mice by gene trap: Severe reduction of reproductive organs and functions in developing and adult mice. Endocrinology. (2010) 151:1142–52. doi: 10.1210/en.2009-0598
9. Chan V, Clayton R, Knox G, and Catt K. Ontogeny of pituitary GnRH receptors in the rat. Endocrinology. (1981) 108:2086–92. doi: 10.1210/endo-108-6-2086
10. Zapatero-Caballero H, Sanchez-Franco F, Guerra-Perez N, Fernandez-Mendez C, and Fernandez-Vazquez G. Gonadotropin-releasing hormone receptor gene expression during pubertal development of male rats. Biol Reprod. (2003) 68:1764–70. doi: 10.1095/biolreprod.102.008821
11. Zapatero-Caballero H, Sanchez-Franco F, Fernandez-Mendez C, García-San Frutos M, Botella-Cubells LM, and Fernandez-Vazquez G. Gonadotropin-releasing hormone receptor gene expression during pubertal development of female rats. Biol Reprod. (2004) 70:348–55. doi: 10.1095/biolreprod.103.020818
12. Clayton R and Catt K. Regulation of pituitary gonadotropin-releasing hormone receptors by gonadal hormones. Endocrinology. (1981) 108:887–95. doi: 10.1210/endo-108-3-887
13. Frager M, Pieper D, Tonetta S, Duncan J, and Marshall J. Pituitary gonadotropin-releasing hormone receptors. Effects of castration, steroid replacement, and the role of gonadotropin-releasing hormone in modulating receptors in the rat. J Clin Invest. (1981) 67:615–23. doi: 10.1172/JCI110075
14. Kakar SS, Grantham K, Musgrove LC, Devor D, Sellers JC, and Neill JD. Rat gonadotropin-releasing hormone (GnRH) receptor: tissue expression and hormonal regulation of its mRNA. Mol Cell Endocrinol. (1994) 101:151–7. doi: 10.1016/0303-7207(94)90229-1
15. Winters SJ, Kawakami S, Sahu A, and Plant TM. Pituitary follistatin and activin gene expression, and the testicular regulation of FSH in the adult rhesus monkey (Macaca mulatta). Endocrinology. (2001) 142:2874–8. doi: 10.1210/endo.142.7.8234
16. Naik S, Young L, Charlton H, and Clayton R. Pituitary gonadotropin-releasing hormone receptor regulation in mice. I: Males. Endocrinology. (1984) 115:106–13. doi: 10.1210/endo-115-1-106
17. Naik S, Young L, Charlton H, and Clayton R. Pituitary gonadotropin-releasing hormone receptor regulation in mice. II: Females. Endocrinology. (1984) 115:114–20. doi: 10.1210/endo-115-1-114
18. Moss GE, Crowder ME, and Nett TM. GnRH-receptor interaction. VI. Effect of progesterone and estradiol on hypophyseal receptors for GnRH, and serum and hypophyseal concentrations of gonadotropins in ovariectomized ewes. Biol Reprod. (1981) 25:938–44. doi: 10.1095/biolreprod25.5.938
19. Hamernik DL, Clay CM, Turzillo A, Van Kirk EA, and Moss GE. Estradiol increases amounts of messenger ribonucleic acid for gonadotropin-releasing hormone receptors in sheep. Biol Reprod. (1995) 53:179–85. doi: 10.1095/biolreprod53.1.179
20. Davis TL, Whitesell JD, Cantlon JD, Clay CM, and Nett TM. Does a nonclassical signaling mechanism underlie an increase of estradiol-mediated gonadotropin-releasing hormone receptor binding in ovine pituitary cells? Biol Reprod. (2011) 85:770–8. doi: 10.1095/biolreprod.111.091926
21. Savoy-Moore R, Schwartz N, Duncan J, and Marshall J. Pituitary gonadotropin-releasing hormone receptors during the rat estrous cycle. Science. (1980) 209:942–4. doi: 10.1126/science.6250218
22. Clayton R, Solano A, Garcia-Vela A, Dufau M, and Catt K. Regulation of pituitary receptors for gonadotropin-releasing hormone during the rat estrous cycle. Endocrinology. (1980) 107:699–706. doi: 10.1210/endo-107-3-699
23. Bauer-Dantoin A, Hollenberg A, and Jameson J. Dynamic regulation of gonadotropin-releasing hormone receptor mRNA levels in the anterior pituitary gland during the rat estrous cycle. Endocrinology. (1993) 133:1911–4. doi: 10.1210/endo.133.4.8404635
24. Crowder M and Nett T. Pituitary content of gonadotropins and receptors for gonadotropin-releasing hormone (GnRH) and hypothalamic content of GnRH during the periovulatory period of the ewe. Endocrinology. (1984) 114:234–9. doi: 10.1210/endo-114-1-234
25. Brooks J, Taylor PL, Saunders PTK, Eidne KA, Struthers WJ, and Mcneilly AS. Cloning and sequencing of the sheep pituitary gonadotropin-releasing hormone receptor and changes in expression of its mRNA during the estrous cycle. Mol Cell Endocrinol. (1993) 94:23–7. doi: 10.1016/0303-7207(93)90177-L
26. Padmanabhan V, Dalkin A, Yasin M, Haisenleder DJ, Marshall JC, and Landefeld TD. Are immediate early genes involved in gonadotropin-releasing hormone receptor gene regulation? Characterization of changes in GnRH receptor (GnRH- R), c-fos, and c-jun messenger ribonucleic acids during the ovine estrous cycle. Biol Reprod. (1995) 53:263–9. doi: 10.1095/biolreprod53.2.263
27. Drouin J, Lagacé L, and Labrie F. Estradiol-induced increase of the LH responsiveness to LH releasing hormone (LHRH) in rat anterior pituitary cells in culture. Endocrinology. (1976) 99:1477–81. doi: 10.1210/endo-99-6-1477
28. Drouin J and Labrie F. Interactions between 17β-estradiol and progesterone in the control of luteinizing hormone and follicle-stimulating hormone release in rat anterior pituitary cells in culture. Endocrinology. (1981) 108:52–7. doi: 10.1210/endo-108-1-52
29. Naik SI, Young LS, Charlton HM, and Clayton RN. Evidence for a pituitary site of gonadal steroid stimulation of GnRH receptors in female mice. J Reprod Fertil. (1985) 74:615–24. doi: 10.1530/jrf.0.0740615
30. Gregg DW and Nett TM. Direct effects of estradiol-17 beta on the number of gonadotropin-releasing hormone receptors in the ovine pituitary. Biol Reprod. (1989) 40:288–93. doi: 10.1095/biolreprod40.2.288
31. Janjic MM, Stojilkovic SS, and Bjelobaba I. Intrinsic and regulated gonadotropin-releasing hormone receptor gene transcription in mammalian pituitary gonadotrophs. Front Endocrinol (Lausanne). (2017) 8:221. doi: 10.3389/fendo.2017.00221
32. Vale W, Rivier J, Vaughan J, McClintock R, Corrigan A, Woo W, et al. Purification and characterization of an FSH releasing protein from porcine ovarian follicular fluid. Nature. (1986) 321:776–9. doi: 10.22141/2224-0551.0.3.38.2012.100651
33. Braden T and Conn P. Activin-A stimulates the synthesis of gonadotropin-releasing hormone receptors. Endocrinology. (1992) 130:2101–6. doi: 10.1210/endo.130.4.1312442
34. Fernández-Vázquez G, Kaiser U, Albarracin C, and Chin W. Transcriptional activation of the gonadotropin-releasing hormone receptor gene by activin A. Mol Endocrinol. (1996) 10:356–66. doi: 10.1210/me.10.4.356
35. Fortin J, Ongaro L, Li Y, Tran S, Lamba P, Wang Y, et al. Activin signaling in gonadotropes: What does the FOX say … to the SMAD? Mol Endocrinol. (2015) 29:963–77. doi: 10.1210/me.2015-1004
36. Breton B, Jalabert B, Billard R, and Weil C. In vitro stimulation of the release of pituitary gonadotropic hormone by a hypothalamic factor in the carp Cyprinus carpio L. C R Acad Sci Paris D. (1971) 273:2591–4.
37. Sherwood N, Eiden L, Brownstein M, Spiess J, Rivier J, and Vale W. Characterization of a teleost gonadotropin-releasing hormone. Proc Natl Acad Sci. (1983) 80:2794–8. doi: 10.1073/pnas.80.9.2794
38. Zohar Y, Zmora N, Trudeau V, Muñoz-Cueto J, and Golan M. A half century of fish gonadotropin releasing hormones : breaking paradigms. J Neuroendocr. (2022) 34:e13069. doi: 10.1111/jne.13069
39. Ciani E, Fontaine R, Maugars G, Nourizadeh-Lillabadi R, Andersson E, Bogerd J, et al. Gnrh receptor gnrhr2bbα is expressed exclusively in lhb-expressing cells in Atlantic salmon male parr. Gen Comp Endocrinol. (2020) 285:113293. doi: 10.1016/j.ygcen.2019.113293
40. González-Martínez D, Zmora N, Saligaut D, Zanuy S, Elizur A, Kah O, et al. New insights in developmental origins of different GnRH (gonadotrophin-releasing hormone) systems in perciform fish: An immunohistochemical study in the European sea bass (Dicentrarchus labrax). J Chem Neuroanat. (2004) 28:1–15. doi: 10.1016/j.jchemneu.2004.05.001
41. Levavi-Sivan B, Biran J, and Fireman E. Sex steroids are involved in the regulation of gonadotropin-releasing hormone and dopamine D2 receptors in female tilapia pituitary. Biol Reprod. (2006) 75:642–50. doi: 10.1095/biolreprod.106.051540
42. Guilgur LG, Strüssmann CA, and Somoza GM. mRNA expression of GnRH variants and receptors in the brain, pituitary and ovaries of pejerrey (Odontesthes bonariensis) in relation to the reproductive status. Fish Physiol Biochem. (2009) 35:157–66. doi: 10.1007/s10695-008-9215-4
43. Hildahl J, Sandvik GK, Edvardsen RB, Norberg B, Haug TM, and Weltzien FA. Four gonadotropin releasing hormone receptor genes in Atlantic cod are differentially expressed in the brain and pituitary during puberty. Gen Comp Endocrinol. (2011) 173:333–45. doi: 10.1016/j.ygcen.2011.06.002
44. Melo MC, Andersson E, Fjelldal PG, Bogerd J, França LR, Taranger GL, et al. Salinity and photoperiod modulate pubertal development in Atlantic salmon (Salmo salar). J Endocrinol. (2014) 220:319–32. doi: 10.1530/JOE-13-0240
45. Melo MC, van Dijk P, Andersson E, Nilsen TO, Fjelldal PG, Male R, et al. Androgens directly stimulate spermatogonial differentiation in juvenile Atlantic salmon (Salmo salar). Gen Comp Endocrinol. (2015) 211:52–61. doi: 10.1016/j.ygcen.2014.11.015
46. Lumayno SDP, Ohga H, Selvaraj S, Nyuji M, Yamaguchi A, and Matsuyama M. Molecular characterization and functional analysis of pituitary GnRH receptor in a commercial scombroid fish, chub mackerel (Scomber japonicus). Gen Comp Endocrinol. (2017) 247:143–51. doi: 10.1016/j.ygcen.2017.01.027
47. Lin CJ, Wu GC, Lee MF, Lau EL, Dufour S, and Chang CF. Regulation of two forms of gonadotropin-releasing hormone receptor gene expression in the protandrous black porgy fish, Acanthopagrus schlegeli. Mol Cell Endocrinol. (2010) 323:137–46. doi: 10.1016/j.mce.2010.04.003
48. Tsukamoto K, Kuroki M, and Watanabe S. Common names for all species and subspecies of the genus Anguilla. Env Biol Fish. (2020) 103:985–91. doi: 10.1007/s10641-020-00988-3
49. Rousseau K, Lafont A, Pasquier J, Maugars G, Jolly C, Sébert M, et al. Advances in eel reproductive physiology and endocrinology. In: Trischitta F, Takei Y, Damesceno-Oliveira A, and Sébert P, editors. Eel physiology, vol. eds. Boca Raton, USA: CRC Press (2013). p. 1–43.
50. Fontaine M. Sur la maturation complete des organs génitaux de l’Anguille mâle et l’émission spontanée de ses produits sexuels. C R Acad Sci Paris. (1936) 202:1012–4.
51. Fontaine M, Bertrand E, Lopez E, and Callamand O. Sur la maturation des organes génitaux de l’Anguille femelle (Anguilla Anguilla L.) et l’émission spontanée des oeufs en aquarium. C R Acad Sc Paris. (1964) 259:2907–10.
52. Tanaka H. Progression in artificial seedling production of Japanese eel Anguilla japonica. Fish Sci. (2015) 81:11–9. doi: 10.1007/s12562-014-0821-z
53. Vidal B, Pasqualini C, Le Belle N, Holland M, Sbaihi M, Vernier P, et al. Dopamine inhibits luteinizing hormone synthesis and release in the juvenile European eel: a neuroendocrine lock for the onset of puberty. Biol Reprod. (2004) 71:1491–500. doi: 10.1095/biolreprod.104.030627
54. Dufour S, Weltzien FA, Sebert ME, Le Belle N, Vidal B, Vernier P, et al. Dopaminergic inhibition of reproduction in teleost fishes: Ecophysiological and evolutionary implications. Ann N Y Acad Sci. (2005) 1040:9–21. doi: 10.1196/annals.1327.002
55. Dufour S, Lopez E, Le Menn F, Le Belle N, Baloche S, and Fontaine YA. Stimulation of gonadotropin release and of ovarian development, by the administration of a gonadoliberin agonist and of dopamine antagonists, in female silver eel pretreated with estradiol. Gen Comp Endocrinol. (1988) 70:20–30. doi: 10.1016/0016-6480(88)90090-1
56. Peñaranda DS, Mazzeo I, Hildahl J, Gallego V, Nourizadeh-Lillabadi R, Pérez L, et al. Molecular characterization of three GnRH receptor paralogs in the European eel, Anguilla Anguilla: Tissue-distribution and changes in transcript abundance during artificially induced sexual development. Mol Cell Endocrinol. (2013) 369:1–14. doi: 10.1016/j.mce.2013.01.025
57. Dufour S, Delerue-Le Belle N, and Fontaine YA. Effects of steroid hormones on pituitary immunoreactive gonadotropin in European freshwater eel, Anguilla Anguilla L. Gen Comp Endocrinol. (1983) 52:190–7. doi: 10.1016/0016-6480(83)90112-0
58. Huang YS, Schmitz M, Le Belle N, Chang CF, Quérat B, and Dufour S. Androgens stimulate gonadotropin-II β-subunit in eel pituitary cells in vitro. Mol Cell Endocrinol. (1997) 131:157–66. doi: 10.1016/S0303-7207(97)00100-7
59. Dufour S, Le Belle N, Baloche S, and Fontaine YA. Positive feedback control by the gonads on gonadotropin (GTH) and gonadoliberin (GnRH) levels in experimentally matured female silver eels, Anguilla Anguilla. Fish Physiol Biochem. (1989) 7:157–62. doi: 10.1007/BF00004702
60. Montero M, Le Belle N, King JA, Millar RP, and Dufour S. Differential regulation of the two forms of gonadotropin-releasing hormone (mGnRH and cGnRH-II) by sex steroids in the European female silver eel (Anguilla Anguilla). Neuroendocrinology. (1995) 61:525–35. doi: 10.1159/000126876
61. Dufour S, Fontaine YA, and Kerdelhue B. Increase in brain and pituitary radioimmunoassayable gonadotropin releasing hormone (GnRH) in the European silver eel treated with sexual steroid or human chorionic gonadotropin. Neuropeptides. (1985) 6:495–502. doi: 10.1016/0143-4179(85)90111-8
62. Huang YS, Le Belle N, Schmitz M, Chang CF, Vernier P, Quérat B, et al. In vivo and in vitro effects of sex steroids on gonadotropin (GtH-II) synthesis in the female european eel. Ann New York Acad Sci. (1998) 839:348–50. doi: 10.1111/j.1749-6632.1998.tb10791.x
63. Jeng S, Yueh W, Chen G, Lee Y, Dufour S, and Chang C. Differential expression and regulation of gonadotropins and their receptors in the Japanese eel, Anguilla japonica. Gen Comp Endocrinol. (2007) 154:161–73. doi: 10.1016/j.ygcen.2007.05.026
64. Lai X, Peng S, Bai Z, Cao L, Huang H, Jiang Y, et al. Direct feedback regulation of E2, T, and hCG in the brain-pituitary-gonad axis of Japanese eel (Anguilla japonica) during artificial maturation. Fishes. (2024) 9:265. doi: 10.3390/fishes9070265
65. Pasquier J, Lafont AG, Denis F, Lefranc B, Dubessy C, Moreno-Herrera A, et al. Eel kisspeptins: Identification, functional activity, and inhibition on both pituitary LH and GnRH receptor expression. Front Endocrinol (Lausanne). (2018) 8:353. doi: 10.3389/fendo.2017.00353
66. Campo A, Lafont A-G, Lefranc B, Leprince J, Tostivint H, Kamech N, et al. Tachykinin-3 genes and peptides characterized in a basal teleost, the european eel: Evolutionary perspective and pituitary role. Front Endocrinol (Lausanne). (2018) 9:304. doi: 10.3389/fendo.2018.00304
67. Maugars G, Pasquier J, Atkinson C, Lafont A, Campo A, Kamech N, et al. Gonadotropin-inhibitory hormone in teleosts : New insights from a basal representative, the eel. Gen Comp Endocrinol. (2020) 287:113350. doi: 10.1016/j.ygcen.2019.113350
68. Aroua S, Maugars G, Jeng S-R, Chang C-F, Weltzien F-A, Rousseau K, et al. Pituitary gonadotropins FSH and LH are oppositely regulated by the activin/follistatin system in a basal teleost, the eel. Gen Comp Endocrinol. (2012) 175:82–91. doi: 10.1016/j.ygcen.2011.10.002
69. Montero M, Le Belle N, Vidal B, and Dufour S. Primary cultures of dispersed pituitary cells from estradiol-pretreated female silver eels (Anguilla Anguilla L.): Immunocytochemical characterization of gonadotropic cells and stimulation of gonadotropin release. Gen Comp Endocrinol. (1996) 104:103–15. doi: 10.1006/gcen.1996.0146
70. Aroua S, Weltzien FA, Le BN, and Dufour S. Development of real-time RT-PCR assays for eel gonadotropins and their application to the comparison of in vivo and in vitro effects of sex steroids. Gen Comp Endocrinol. (2007) 153:333–43. doi: 10.1016/j.ygcen.2007.02.027
71. Rauluseviciute I, Riudavets-Puig R, Blanc-Mathieu R, Castro-Mondragon JA, Ferenc K, Kumar V, et al. JASPAR 2024: 20thãnniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. (2024) 52:D174–82. doi: 10.1093/nar/gkad1059
72. Cederberg RA, Smith JE, McDonald EA, Lee C, Perkins AR, and White BR. Activity of the porcine gonadotropin-releasing hormone receptor gene promoter is partially conferred by a distal gonadotrope specific element (GSE) within an upstream enhancing region, two proximal GSEs and a retinoid X receptor binding site. Reprod Biol Endocrinol. (2015) 13:1–16. doi: 10.1186/s12958-015-0033-0
73. Cheng CK, Chow BKC, and Leung PCK. An activator protein 1-like motif mediates 17β-estradiol repression of gonadotropin-releasing hormone receptor promoter via an estrogen receptor α-dependent mechanism in ovarian and breast cancer cells. Mol Endocrinol. (2003) 17:2613–29. doi: 10.1210/me.2003-0217
74. Ellsworth BS, Burns AT, Escudero KW, Duval DL, Nelson SE, and Clay CM. The gonadotropin releasing hormone (GnRH) receptor activating sequence (GRAS) is a composite regulatory element that interacts with multiple classes of transcription factors including Smads, AP-1 and a forkhead DNA binding protein. Mol Cell Endocrinol. (2003) 206:93–111. doi: 10.1016/S0303-7207(03)00235-1
75. Kam KY, Jeong KH, Norwitz ER, Jorgensen EM, and Kaiser UB. Oct-1 and nuclear factor Y bind to the SURG-1 element to direct basal and gonadotropin-releasing hormone (GnRH)-stimulated mouse GnRH receptor gene transcription. Mol Endocrinol. (2005) 19:148–62. doi: 10.1210/me.2004-0025
76. McGillivray SM, Bailey JS, Ramezani R, Kirkwood BJ, and Mellon PL. Mouse GnRH receptor gene expression is mediated by the LHX3 homeodomain protein. Endocrinology. (2005) 146:2180–5. doi: 10.1210/en.2004-1566
77. Schang AL, Quérat B, Simon V, Garrel G, Bleux C, Counis R, et al. Mechanisms underlying the tissue-specific and regulated activity of the Gnrhr promoter in mammals. Front Endocrinol (Lausanne). (2012) 3:162. doi: 10.3389/fendo.2012.00162
78. Duval DL, Nelson SE, and Clay CM. A binding site for steroidogenic factor-1 is part of a complex enhancer that mediates expression of the murine gonadotropin-releasing hormone receptor gene. Biol Reprod. (1997) 56:160–8. doi: 10.1095/biolreprod56.1.160
79. Duval DL, Nelson SE, and Clay CM. The tripartite basal enhancer of the gonadotropin-releasing hormone (GnRH) receptor gene promoter regulates cell-specific expression through a novel GnRH receptor activating sequence. Mol Endocrinol. (1997) 11:1814–21. doi: 10.1210/mend.11.12.0020
80. Pincas H, Amoyel K, Counis R, and Laverrière JN. Proximal cis-acting elements, including steroidogenic factor 1, mediate the efficiency of a distal enhancer in the promoter of the rat gonadotropin-releasing hormone receptor gene. Mol Endocrinol. (2001) 15:319–37. doi: 10.1210/mend.15.2.0593
81. Cherrington BD, Farmerie TA, and Clay CM. A specific helical orientation underlies the functional contribution of the activin responsive unit to transcriptional activity of the murine gonadotropin-releasing hormone receptor gene promoter. Endocrine. (2006) 29:425–33. doi: 10.1385/ENDO:29:3:425
82. Cherrington BD, Farmerie TA, Lents CA, Cantlon JD, Roberson MS, and Clay CM. Activin responsiveness of the murine gonadotropin-releasing hormone receptor gene is mediated by a composite enhancer containing spatially distinct regulatory elements. Mol Endocrinol. (2005) 19:898–912. doi: 10.1210/me.2004-0214
83. Kuiper GGJM, Shughrue PJ, Merchenthaler I, and Gustafsson JÅ. The estrogen receptor β subtype: A novel mediator of estrogen action in neuroendocrine systems. Front Neuroendocrinol. (1998) 19:253–86. doi: 10.1006/frne.1998.0170
84. Oliveira AG, Coelho PH, Guedes FD, Mahecha GAB, Hess RA, and Oliveira CA. 5α-Androstane-3β,17β-diol (3β-diol), an estrogenic metabolite of 5α-dihydrotestosterone, is a potent modulator of estrogen receptor ERβ expression in the ventral prostrate of adult rats. Steroids. (2007) 72:914–22. doi: 10.1016/j.steroids.2007.08.001
85. Von Krogh K, Bjørndal GT, Nourizadeh-Lillabadi R, Hodne K, Ropstad E, Haug TM, et al. Sex steroids differentially regulate fshb, lhb and gnrhr expression in Atlantic cod (Gadus morhua) pituitary. Reproduction. (2017) 154:581–94. doi: 10.1530/REP-17-0208
86. Crespo D, Skaftnesmo KO, Kjærner-Semb E, Yilmaz O, Norberg B, Olausson S, et al. Pituitary gonadotropin gene expression during induced onset of postsmolt maturation in male atlantic salmon: in vivo and tissue culture studies. Front Endocrinol (Lausanne). (2022) 13:826920. doi: 10.3389/fendo.2022.826920
87. Lin CJ, Wu GC, Dufour S, and Chang CF. Activation of the brain-pituitary-gonadotropic axis in the black porgy Acanthopagrus schlegelii during gonadal differentiation and testis development and effect of estradiol treatment. Gen Comp Endocrinol. (2019) 281:17–29. doi: 10.1016/j.ygcen.2019.05.008
88. Rhee JS, Seo JS, Raisuddin S, Ki JS, Lee KW, Kim IC, et al. Gonadotropin-releasing hormone receptor (GnRHR) gene expression is differently modulated in gender types of the hermaphroditic fish Kryptolebias marmoratus by endocrine disrupting chemicals. Comp Biochem Physiol - C Toxicol Pharmacol. (2008) 147:357–65. doi: 10.1016/j.cbpc.2008.01.007
89. Emons G, Nill J, Sturm R, and Ortmann O. Effects of progesterone on gonadotropin-releasing hormone receptor concentration in cultured estrogen-primed female rat pituitary cells. J Steroid Biochem Mol Biol. (1992) 42:831–9. doi: 10.1016/0960-0760(92)90091-V
90. Emons G, Hoffmann HG, Brack C, Ortmann O, Sturm R, Ball P, et al. Modulation of gonadotropin-releasing hormone receptor concentration in cultured female rat pituitary cells by estradiol treatment. J Steroid Biochem. (1988) 31:751–6. doi: 10.1016/0022-4731(88)90282-8
91. Menon M, Peegel H, and Katta V. Estradiol potentiation of gonadotropin-releasing hormone responsiveness in the anterior pituitary is mediated by an increase in gonadotropin-releasing hormone receptors. Am J Obstet Gynecol. (1985) 151:534–40. doi: 10.1016/0002-9378(85)90284-4
92. Sealfon SC, Laws SC, Wu JC, Gillo B, and Miller WL. Hormonal regulation of gonadotropin-releasing hormone receptors and messenger RNA activity in ovine pituitary culture. Mol Endocrinol. (1990) 4:1980–7. doi: 10.1210/mend-4-12-1980
93. Laws SC, Webster JC, and Miller WL. Estradiol alters the effectiveness of gonadotropin-releasing hormone (GnRH) in ovine pituitary cultures: GnRH receptors versus responsiveness to GnRH. Endocrinology. (1990) 127:381–6. doi: 10.1210/endo-127-1-381
94. Wu JC, Sealfon SC, and Miller WL. Gonadal hormones and gonadotropin-releasing hormone (GnRH) alter messenger ribonucleic acid levels for GnRH receptors in sheep. Endocrinology. (1994) 134:1846–50. doi: 10.1210/endo.134.4.8137751
95. Turgeon JL, Kimura Y, Waring DW, and Mellon PL. Steroid and pulsatile gonadotropin-releasing hormone (GnRH) regulation of luteinizing hormone and GnRH receptor in a novel gonadotrope cell line. Mol Endocrinol. (1996) 10:439–50. doi: 10.1210/me.10.4.439
96. Cheng CK and Leung PC. Molecular biology of gonadotropin-releasing hormone (GnRH)-I, GnRH-II, and their receptors in humans. Endocr Rev. (2005) 26:283–30. doi: 10.1210/er.2003-0039
97. Schmitz M, Aroua S, Vidal B, Le Belle N, Elie P, and Dufour S. Differential regulation of luteinizing hormone and follicle-stimulating hormone expression during ovarian development and under sexual steroid feedback in the European eel. Neuroendocrinology. (2005) 81:107–19. doi: 10.1159/000086404
98. Uehara S, Nishike Y, Maeda K, Karigo T, Kuraku S, Okubo K, et al. Identification of the FSH-RH as the other gonadotropin-releasing hormone. Nat Commun. (2024) 15:5342. doi: 10.1038/s41467-024-49564-8
99. Jolly C, Rousseau K, Prézeau L, Vol C, Tomkiewicz J, Dufour S, et al. Functional characterisation of eel dopamine D2 receptors and involvement in the direct inhibition of pituitary gonadotrophins. J Neuroendocrinol. (2016) 28. doi: 10.1111/jne.12411
100. Laws SC, Beggs MJ, Webster JC, Miller WL, and Miller WL. Inhibin increases and progesterone decreases receptors for gonadotropin-releasing hormone in ovine pituitary culture. Endocrinology. (1990) 127:373–80. doi: 10.1210/endo-127-1-373
101. Turzillo AM, Clapper JA, Moss GE, and Nett TM. Regulation of ovine GnRH receptor gene expression by progesterone and oestradiol. J Reprod Fertil. (1998) 113:251–6. doi: 10.1530/jrf.0.1130251
102. Looper ML, Vizcarra JA, Wettemann RP, Malayer JR, Braden TD, Geisert RD, et al. Influence of estradiol, progesterone, and nutrition on concentrations of gonadotropins and GnRH receptors, and abundance of mRNA for GnRH receptors and gonadotropin subunits in pituitary glands of beef cows. J Anim Sci. (2003) 81:269–78. doi: 10.2527/2003.811269x
103. Naik SI, Saade G, Detta A, and Clayton RN. Homologous ligand regulation of gonadotrophin-releasing hormone receptors in vivo: relationship to gonadotrophin secretion and gonadal steroids. J Endocrinol. (1985) 107:41–7. doi: 10.1677/joe.0.1070041
104. Cheng KW, Cheng CK, and Leung PCK. Differential role of PR-A and -B isoforms in transcription regulation of human GnRH receptor gene. Mol Endocrinol. (2001) 15:2078–92. doi: 10.1210/mend.15.12.0739
105. Gregg DW, Allen MC, and Nett TM. Estradiol-induced increase in number of gonadotropin-releasing hormone receptors in cultured ovine pituitary cells. Biol Reprod. (1990) 43:1032–6. doi: 10.1095/biolreprod43.6.1032
106. Daley CA, Sakurai H, Adams BM, and Adams TE. Effect of stress-like concentrations of cortisol on gonadotroph function in orchidectomized sheep. Biol Reprod. (1999) 60:158–63. doi: 10.1016/S0378-4320(00)00141-X
107. Daley CA, Sakurai H, Adams BM, and Adams TE. Effect of stress-like concentrations of cortisol on the feedback potency of oestradiol in orchidectomized sheep. Anim Reprod Sci. (2000) 59:167–78. doi: 10.1016/S0378-4320(00)00141-X
108. Rosen H, Jameel ML, and Barkan AL. Dexamethasone suppresses gonadotropin-releasing hormone (GnRH) secretion and has direct pituitary effects in male rats: Differential regulation of gnrh receptor and gonadotropin responses to GnRH. Endocrinology. (1988) 122:2873–80. doi: 10.1210/endo-122-6-2873
109. Maya-Núñez G and Conn PM. Transcriptional regulation of the GnRH receptor gene by glucocorticoids. Mol Cell Endocrinol. (2003) 200:89–98. doi: 10.1016/S0303-7207(02)00419-7
110. Kotitschke A, Sadie-Van Gijsen H, Avenant C, Fernandes S, and Hapgood JP. Genomic and nongenomic cross talk between the gonadotropin-releasing hormone receptor and glucocorticoid receptor signaling pathways. Mol Endocrinol. (2009) 23:1726–45. doi: 10.1210/me.2008-0462
111. Corrigan A, Bilezikjian L, Carroll R, Bald L, Schmelzer C, Fendly B, et al. Evidence for an autocrine role of activin B within rat anterior pituitary cultures. Endocrinology. (1991) 128:1682–4. doi: 10.1210/endo-128-3-1682
112. Ge W and Peter R. Activin-like peptides in somatotrophs and activin stimulation of growth hormone release in goldfish. Gen Comp Endocrinol. (1994) 95:213–21. doi: 10.1006/gcen.1994.1118
113. Bilezikjian LM, Blount AL, Leal AMO, Donaldson CJ, Fischer WH, and Vale WW. Autocrine/paracrine regulation of pituitary function by activin, inhibin and follistatin. Mol Cell Endocrinol. (2004) 225:29–36. doi: 10.1016/j.mce.2004.02.010
114. Xia Y and Schneyer AL. The biology of activin: Recent advances in structure, regulation and function. J Endocrinol. (2009) 202:1–12. doi: 10.1677/JOE-08-0549
115. Gregg DW, Schwall RH, and Nett TM. Regulation of gonadotropin secretion and number of gonadotropin-releasing hormone receptors by inhibin, activin-A, and estradiol. Biol Reprod. (1991) 44:725–32. doi: 10.1095/biolreprod44.4.725
116. Duval DL, Ellsworth BS, and Clay CM. Is gonadotrope expression of the gonadotropin releasing hormone receptor gene mediated by autocrine/paracrine stimulation of an activin response element? Endocrinology. (1999) 140:1949–52. doi: 10.1210/endo.140.4.6780
117. Xie H, Cherrington BD, Meadows JD, Witham EA, and Mellon PL. Msx1 homeodomain protein represses the αGSU and GnRH receptor genes during gonadotrope development. Mol Endocrinol. (2013) 27:422–36. doi: 10.1210/me.2012-1289
118. Ohta H, Kagawa H, Tanaka H, Okuzawa K, and Hirose K. Changes in fertilization and hatching rates with time after ovulation induced by 17, 20β-dihydroxy-4-pregnen-3-one in the Japanese eel. Anguilla japonica. Aquaculture. (1996) 139:291–301. doi: 10.1016/0044-8486(95)01167-6
119. Jéhannet P, Palstra AP, Meijerhof M, Schipper H, Giménez IN, Dirks RP, et al. The induction of oocyte maturation and ovulation in the European eel (Anguilla Anguilla): in vitro and in vivo comparison of progesterone with 17α,20β-dihydroxy-4-pregnen-3-one. Front Physiol. (2023) 14:1207542. doi: 10.3389/fphys.2023.1207542
Keywords: GnRH receptor, promoter, sex steroids, activins, pituitary cells, Anguilla, teleosts, mammals
Citation: Lin C-J, Rousseau K, Chang C-F and Dufour S (2025) In vitro and in silico insights on the regulation by gonadal hormones of pituitary GnRH receptor expression in a basal teleost, the European eel. Front. Endocrinol. 16:1673260. doi: 10.3389/fendo.2025.1673260
Received: 25 July 2025; Accepted: 08 September 2025;
Published: 29 September 2025.
Edited by:
Hamid R. Habibi, University of Calgary, CanadaReviewed by:
Juan F. Asturiano, Universitat Politècnica de València, SpainQi Yin, Carnegie Institution for Science, United States
Pamela Navarrete-Ramírez, Michoacana University of San Nicolás de Hidalgo, Mexico
Copyright © 2025 Lin, Rousseau, Chang and Dufour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sylvie Dufour, c3lsdmllLmR1Zm91ckBtbmhuLmZy
 Chien-Ju Lin
Chien-Ju Lin Karine Rousseau
Karine Rousseau Ching-Fong Chang
Ching-Fong Chang Sylvie Dufour
Sylvie Dufour