A Correction on
Bioinformatic analysis identifies LPL as a critical gene in diabetic kidney disease via lipoprotein metabolism
By Dong Q, Xu H, Xu P, Liu J and Shen Z (2025) Front. Endocrinol. 16:1620032. doi: 10.3389/fendo.2025.1620032
The Figure 5 caption were in the wrong order. The labels for panels (A) and (B) have been swapped. The order has now been corrected.
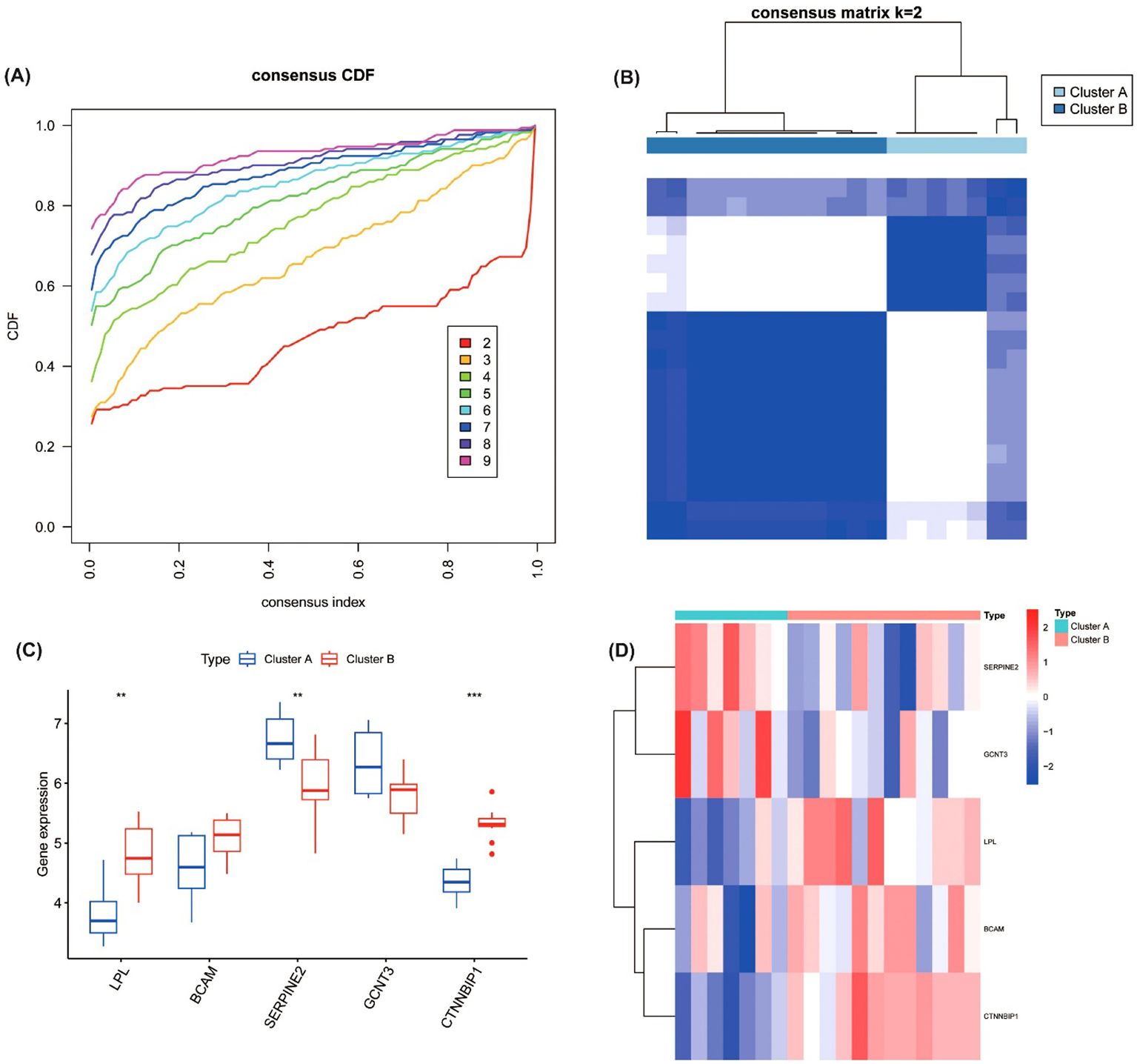
Figure 5. Consensus clustering analysis of five hub genes in DKD patients. (A) Cumulative Distribution Function (CDF) curve for consensus clustering with different values of k. (B) Consensus matrix for clustering, showing optimal stability at k=2, resulting in two molecular subtypes (Cluster A and Cluster B) based on the expression of hub genes. (C) Expression levels of the hub genes (LPL, BCAM, SERPINE2, GCNT3, and CTNNBIP1) in the two identified subtypes. ***P<0.001. (D) Heatmap of gene expression between Cluster A and Cluster B **P<0.01.
The error made was a directional error in the interpretation of the statistical result for the gene CTNNBIP1.
A correction has been made to the section Results, Consensus cluster analysis, Paragraph 2:
“There were significant differences in the expression levels of core genes between the two subtypes (Figures 5C, D). Specifically, compared to Cluster B, Cluster A exhibited significantly reduced expression of LPL (P < 0.01) and CTNNBIP1 (p < 0.001), along with lower expression of BCAM. In contrast, Cluster A showed significantly elevated expression of SERPINE2 (p < 0.01) and higher expression of GCNT3.”
The pathway names and their corresponding P-values have been corrected to match the data presented in Table 1.
A correction has been made to the section Results, Hub gene enrichment analysis, Paragraph 1:
“The results showed that LPL was significantly associated with Cholesterol metabolism (P = 0.012), Glycerolipid metabolism (P = 0.015) and PPAR signaling pathway (P = 0.018, Table 1). These findings suggest that LPL may play a crucial role in lipid metabolism disorders, and through regulating lipidmetabolism and inflammatory responses, it could contribute to the progression of DKD.”
The original version of this article has been updated.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: diabetic kidney disease, lipoprotein lipase, immune cell infiltration, lipid metabolism, bioinformatic
Citation: Dong Q, Xu H, Xu P, Liu J and Shen Z (2025) Correction: Bioinformatic analysis identifies LPL as a critical gene in diabetic kidney disease via lipoprotein metabolism. Front. Endocrinol. 16:1732027. doi: 10.3389/fendo.2025.1732027
Received: 25 October 2025; Accepted: 29 October 2025;
Published: 14 November 2025.
Edited and reviewed by:
Yao-Wu Liu, Xuzhou Medical University, ChinaCopyright © 2025 Dong, Xu, Xu, Liu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhouji Shen, bGhsc2hlbnpob3VqaUBuYnUuZWR1LmNu
†These authors have contributed equally to this work
 Qian Dong
Qian Dong Huan Xu†
Huan Xu†