- 1Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Science, Moscow, Russia
- 2Central European Institute of Technology (CEITEC), Masaryk University, Brno, Czech Republic
A commentary on
It has been reported that human TCR repertoires commonly carry so-called public clonotypes – CDR3 variants that are often shared between individuals. Cross-comparison of individual immune repertoires has previously revealed the existence of a population of TCR beta CDR3 variants that are identical at the amino acid level for any two donors (1–3). The lower bound for the total overlap between any two given donors’ TCR beta repertoires within their CD8+ naïve T cell subset has been estimated as ~14,000 identical amino acid CDR3 variants based on comparison of 200,000–600,000 individual TCR beta clonotypes (1). Here, we have used deep profiling data consisting of 1–2 × 106 individual TCR beta clonotypes that we obtained from healthy donors (4) to better estimate the total overlap between TCR beta repertoires for any two individuals.
The apparent paradox is, that the deeper we sequence, the larger is the percentage of observed overlapping clonotypes between the two repertoires, since the number of possible element pairs between the two sets grows geometrically. To demonstrate this, we analyzed TCR beta repertoires for 12 unrelated pairs assembled from a total of nine human donors [adults and children, see Ref. (4) for details]. We plotted the number of identical variants found in samples of increasing size, with up to 106 unique CDR3 sequences randomly drawn from the repertoires of each individual in a given pair (Figure 1). For every pair, the number of shared clonotypes grew geometrically with the arithmetic growth of the sample size (Figures 1A–C, colored lines); at maximum sequencing depth (~1 × 106 unique sequences/donor), we observed an average of ~72,000, 68,000, and 6,000 CDR3 variants that were respectively identical at the amino acid, amino acid only/non-nucleotide and nucleotide level. This exceeds previous estimates (1) by several-fold. The greatest overlap was between two donors from whom we obtained ~1 × 106 and 1.7 × 106 CDR3 variants, where we observed 113,000, 108,000, and 11,000 identical clonotypes at the amino acid, amino acid only/non-nucleotide and nucleotide level, respectively.
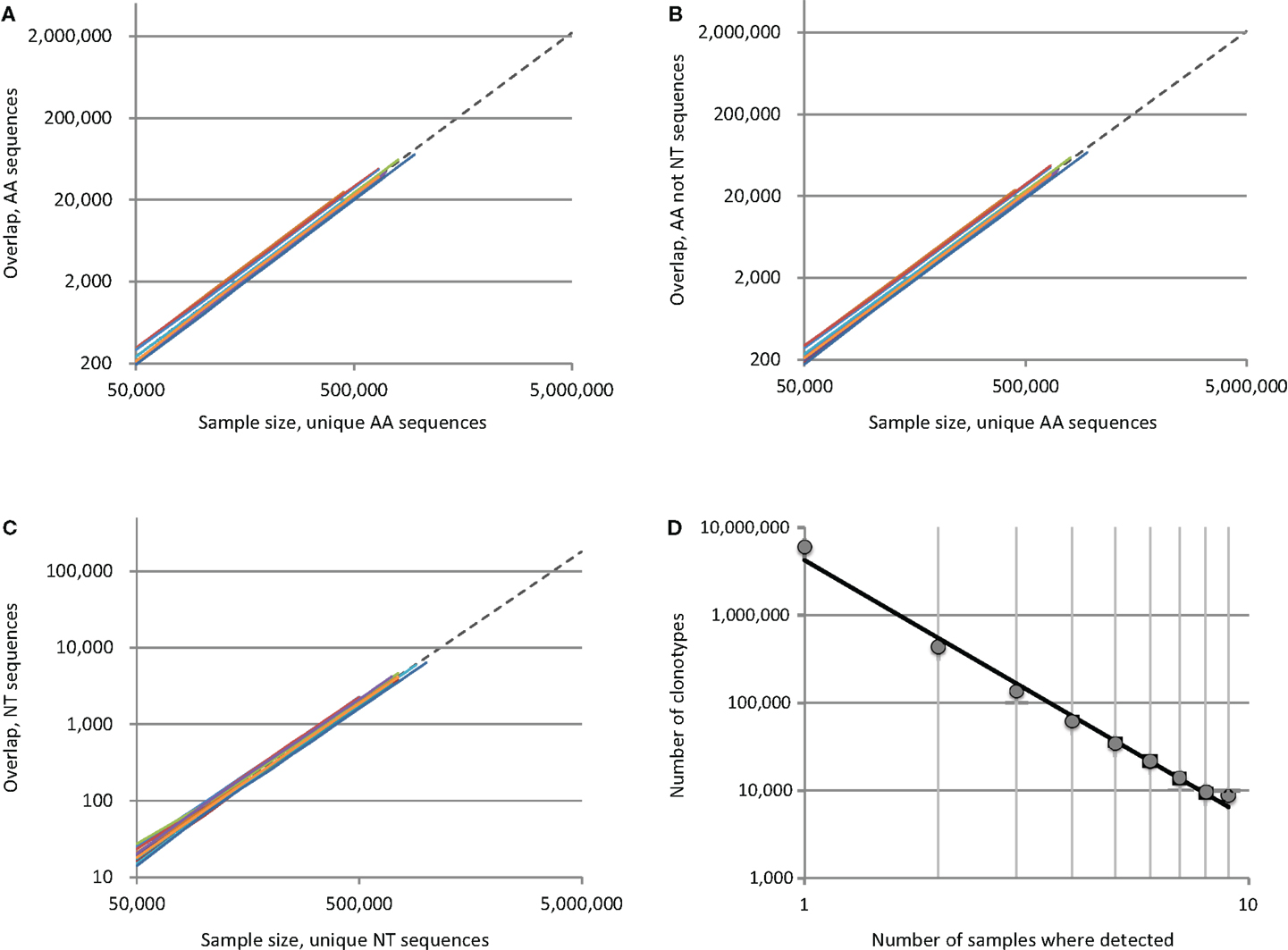
Figure 1. Overlap of individual TCR beta CDR3 repertoires grows geometrically with the number of sequence pairs sampled. Plots indicate the number of shared sequences for 12 unrelated donor pairs in relation to sample size at the level of (A) all amino acid sequences, (B) amino acid sequence only, excluding matches with identical nucleotide sequences, and (C) nucleotide sequences. Each of the 12 colored lines represents the observed overlap between randomly drawn samples of unique CDR3 variants for a different pair of unrelated donors. To extrapolate the predicted level of overlap if the full individual TCR beta repertoires were to be sampled, we plotted fittings of averaged data with a power law (Y = aXb) as dashed lines. (D) We plotted the degree to which unique clonotypes were shared among our nine donors, and found that the frequency with which TCR beta clonotypes occur in human repertoires is distributed according to a power law.
The lower bound on total individual TCR beta repertoire diversity has previously been estimated to be 5 × 106 unique clonotypes [Ref. (5) and our unpublished data]. With that in mind, we extrapolated our intersection curves by fitting them to a power law model [Y = aXb, as in Ref. (1)], which yielded coefficient “b” close to 2.0 and R2 > 0.999 for all cases (Figures 1A–C, dashed lines). We estimated that the total overlap of the TCR beta CDR3 repertoires for two individuals constitutes ~2,200,000, 2,060,000, and 180,000 variants, i.e. 44.1, 41.3, and 3.6% of a given individual’s sequence diversity at the amino acid, amino acid only/non-nucleotide, and nucleotide level, respectively.
Thus, the real paradox is that nearly half of the TCR beta CDR3 repertoire is functionally identical between any two individuals, in spite of the fact that the theoretical diversity that can be achieved by TCR beta variants has been estimated to be ~5 × 1011 sequences (1, 6). The results from our extrapolation are direct and evident. We took numerous precautions to exclude contamination in our work, including sequencing of pair-analyzed donor repertoires in separate Illumina lanes (4). Even if contaminations were present, these would not affect overlap at the amino acid only/non-nucleotide level (Figure 1B). Furthermore, we performed CDR3 extraction and error correction with MiTCR (http://mitcr.milaboratory.com/) using the stringent ETE algorithm, which eliminates 98% of PCR and sequencing errors with minimal loss of natural TCR beta diversity (7).
Such large overlap between individuals suggests the existence of a rather limited pool of frequently used functional CDR3 sequences. To further investigate this, we calculated the lower and upper bounds of the Chao richness estimate as described in Ref. (8) based on the numbers of singletons and doubletons (sequences observed in one and two individuals, respectively) in 12 paired donors’ samples. From this model, we obtained a confidence interval of 1.2 × 107 to 5.4 × 107 unique amino acid CDR3 sequences, at a significance level of α = 0.001.
These findings represent a shift in our understanding of human adaptive immunity. It now appears likely that recombinatorial biases (3, 9) and thymic selection (4, 10, 11) shape our repertoires so tightly that the majority of TCR beta CDR3 variants expressed by naïve T cells leaving the thymus are chosen from a “short-list” of just under 108 amino acid variants – even shorter than the 2 × 109 “effective sequence space” estimated by Robins and colleagues (1).
Nevertheless, the repertoire has a complex structure and those clonotypes that are characterized as low-complexity [see figure 7 in Ref. (4)] predominantly form the backbone of the shared clonotype pool. Interestingly, when we examined the intersection of all nine donor samples, we found that the number of donors in which a given clonotype can be detected is distributed according to a power law, with a degree of −2.95 and R2 = 0.99 (Figure 1D). These findings confirm the fractal structure of the human TCR beta repertoire that determines the landscape of shared clonotypes (1–3, 12), and may reveal a more complex picture with the deeper profiling experiments.
Acknowledgments
We are grateful to M. Eisenstein for English editing. This work was supported by the Molecular and Cell Biology program RAS, Russian Foundation for Basic Research (12-04-33139, 12-04-00229, 13-04-00998), and European Regional Development Fund (CZ.1.05/1.1.00/02.0068).
References
1. Robins HS, Srivastava SK, Campregher PV, Turtle CJ, Andriesen J, Riddell SR, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med (2010) 2:47ra64. doi:10.1126/scitranslmed.3001442
2. Venturi V, Quigley MF, Greenaway HY, Ng PC, Ende ZS, McIntosh T, et al. A mechanism for TCR sharing between T cell subsets and individuals revealed by pyrosequencing. J Immunol (2011) 186:4285–94. doi:10.4049/jimmunol.1003898
3. Li H, Ye C, Ji G, Wu X, Xiang Z, Li Y, et al. Recombinatorial biases and convergent recombination determine interindividual TCRbeta sharing in murine thymocytes. J Immunol (2012) 189:2404–13. doi:10.4049/jimmunol.1102087
4. Putintseva EV, Britanova OV, Staroverov DB, Merzlyak EM, Turchaninova MA, Shugay M, et al. Mother and child T cell receptor repertoires: deep profiling study. Front Immunol (2013) 4:463. doi:10.3389/fimmu.2013.00463
5. Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood (2009) 114:4099–107. doi:10.1182/blood-2009-04-217604
6. Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature (1988) 334:395–402. doi:10.1038/334395a0
7. Bolotin DA, Shugay M, Mamedov IZ, Putintseva EV, Turchaninova MA, Zvyagin IV, et al. MiTCR: software for T-cell receptor sequencing data analysis. Nat Methods (2013) 10(9):813–4. doi:10.1038/nmeth.2555
8. Eren MI, Chao A, Hwang WH, Colwell RK. Estimating the richness of a population when the maximum number of classes is fixed: a nonparametric solution to an archaeological problem. PLoS One (2012) 7:e34179. doi:10.1371/journal.pone.0034179
9. Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol (2008) 8:231–8. doi:10.1038/nri2260
10. Turner SJ, Doherty PC, McCluskey J, Rossjohn J. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol (2006) 6:883–94. doi:10.1038/nri1977
11. Gras S, Kjer-Nielsen L, Burrows SR, McCluskey J, Rossjohn J. T-cell receptor bias and immunity. Curr Opin Immunol (2008) 20:119–25. doi:10.1016/j.coi.2007.12.001
Keywords: adaptive immunity, TCR repertoire, TCR beta, NGS data analysis, overlap
Citation: Shugay M, Bolotin DA, Putintseva EV, Pogorelyy MV, Mamedov IZ and Chudakov DM (2013) Huge overlap of individual TCR beta repertoires. Front. Immunol. 4:466. doi: 10.3389/fimmu.2013.00466
Received: 30 July 2013; Accepted: 12 September 2013;
Published online: 25 December 2013.
Edited by:
Michal Or-Guil, Humboldt University Berlin, GermanyCopyright: © 2013 Shugay, Bolotin, Putintseva, Pogorelyy, Mamedov and Chudakov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence:Y2h1ZGFrb3ZkbUBtYWlsLnJ1
†Mikhail Shugay and Dmitriy A. Bolotin have contributed equally to this work.
 Mikhail Shugay
Mikhail Shugay Dmitriy A. Bolotin1†
Dmitriy A. Bolotin1† Ekaterina V. Putintseva
Ekaterina V. Putintseva Mikhail V. Pogorelyy
Mikhail V. Pogorelyy Ilgar Z. Mamedov
Ilgar Z. Mamedov Dmitriy M. Chudakov
Dmitriy M. Chudakov