- 1Division of Clinical Immunology, Department of Medicine, Faculty of Medicine of Ribeirão Preto, University of São Paulo, Ribeirão Preto, Brazil
- 2Department of Pathology, School of Medicine of Botucatu, Universidade Estadual Paulista, Botucatu, Brazil
- 3Research Division in Hematology and Immunology, Institute of Emerging Diseases and Innovative Therapies, Saint-Louis Hospital, CEA, Paris, France
Considering that the non-classical HLA-G molecule has well-recognized tolerogenic properties, HLA-G expression is expected to be deleterious when present in tumor cells and in cells chronically infected by viruses, whereas HLA-G expression is expected to be advantageous in autoimmune disorders. The expression of HLA-G on tissue or peripheral blood cells, the levels of soluble HLA-G and polymorphic sites along the gene have been studied in several disorders. In this study, we revised the role of the molecule and polymorphic sites along the HLA-G gene in tumors, viral hepatitis, and parasitic disorders. Overall, several lines of evidence clearly show that the induction of HLA-G expression in tumors has been associated with worse disease outcome and disease spread. In addition, the few studies conducted on hepatitis and parasitic disorders indicate that HLA-G may contribute to disease pathogenesis. Few isolated polymorphic sites, primarily located at the coding or 3′ untranslated HLA-G region, have been evaluated in these disorders, and a complete HLA-G typing together with the study of gene regulatory elements may further help on the understanding of the influence of the genetic background on disease susceptibility.
Introduction
HLA-G is a non-classical class I gene of the human Major Histocompatility Complex (NCBI gene ID: 3135), presenting a restricted tissue expression pattern and encoding molecules with immune modulatory properties. This gene, firstly described by Geraghty and colleagues in 1987 (1), presents a genetic structure that resembles other classical HLA class I genes. However, contrary to that observed for classical class I genes (HLA-A, -B, and -C), the HLA-G gene is quite conserved among different populations and within the same population, presenting only a few non-synonymous mutations and several variation sites characterized as synonymous modifications, intronic variations, or variable sites at the regulatory regions [reviewed at Ref. (2)].
HLA-G does not seem to initiate immune responses as its classical counterparts. Instead, the HLA-G molecule is associated with the induction of inhibitory stimuli for T and B lymphocytes (3, 4), Natural Killer (NK) cells (3), and antigen-presenting cells (APC) (5). The HLA-G molecule may directly interact with multiple inhibitory receptors, including ILT2/CD85j/LILRB1 (ILT2), ILT4/CD85d/LILRB2 (ILT4), and KIR2DL4/CD158d (KIR2DL4).
The HLA-G molecule was firstly detected at the trophoblast in the maternal fetal interface, probably modulating the maternal immune system during pregnancy. Beyond trophoblast expression, HLA-G has been detected in few normal tissues, including cornea (6), thymus (7), and erythroid and endothelial precursors (8), and its upregulation has been detected in several pathological conditions as described in the present review.
Alternative splicing is also an important characteristic of the HLA-G gene. It may produce at least seven protein isoforms generated by alternative splicing of the primary transcript [reviewed at Ref. (2)], in which four isoforms are membrane-bound and three isoforms are soluble due to the lack of a transmembrane domain.
Much effort has been made to evaluate HLA-G worldwide variability. The HLA-G gene seems to present functional polymorphisms mainly in the regulatory regions, probably influencing its expression. Considering data from at least 18 different populations (9–12) the HLA-G locus presents few frequent extended haplotypes. These haplotypes are a combination among a small number of very divergent promoter and 3′ untranslated region (3′UTR) haplotypes (Figures 1 and 2), and a coding allele usually encodes the same HLA-G molecule (Figure 3). The regulatory segments are characterized by the occurrence of several polymorphic sites presenting high heterozygosis. Although there is no consensus regarding where the HLA-G transcription starts (13), the polymorphisms at the 5′ upstream regulatory region (5′URR) have been considered to influence HLA-G expression, mainly because of the fact that polymorphic sites coincides with, or are close to, known transcription factor binding sites (Figure 1) [Reviewed at Ref. (13)]. Likewise, haplotypes at the HLA-G 3′UTR segment have been considered influencing HLA-G expression, mainly because the fact that some polymorphic sites (such as the one at position +3142) may influence the binding of specific microRNAs (14–17) or may influence mRNA stability (such as the one at position +3187) and alternative splicing (such as the 14-bp polymorphism) (Figure 2).
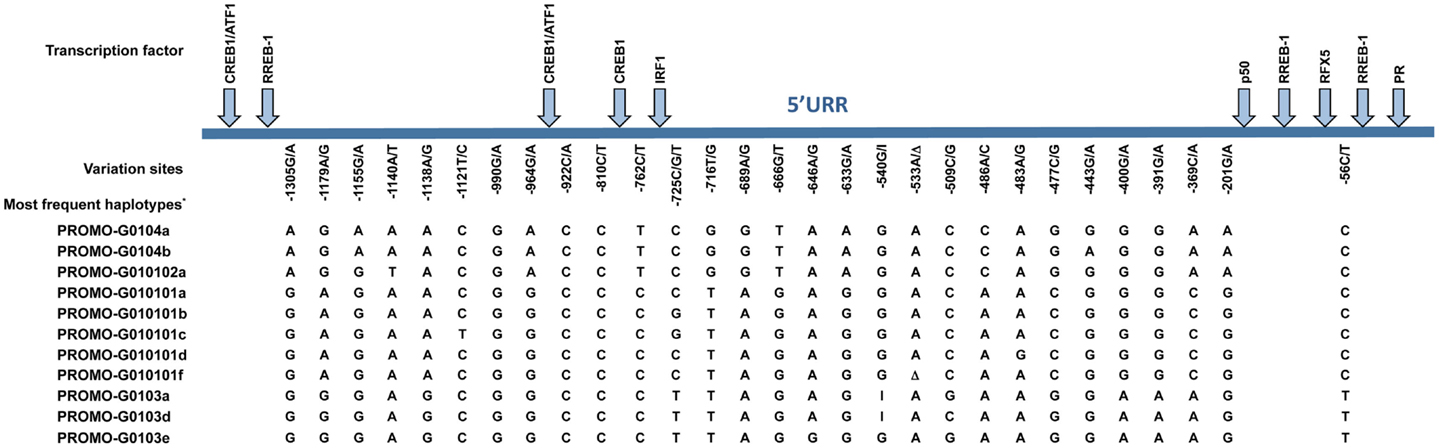
Figure 1. Variation sites at the 5′ upstream regulatory region (5′URR) of the HLA-G gene (1.4 kb upstream of ATG), as well as the target binding sites of the described transcriptional factors. The position of the variation sites is determined in relation to Adenine of the initiation codon ATG. *Since there is no official nomenclature for 5′URR haplotypes, they were designed as previously reported (10). Transcription factors: CREB1, CAMP responsive element binding protein 1; ATF1, cyclic AMP-dependent transcription factor ATF-1; RREB1, Ras responsive element binding protein 1; IRF1, interferon regulatory factor 1; p50, nuclear factor NF-κ-B p105 subunit; RFX5, DNA-binding protein RFX5 (RFX family); PR, progesterone receptor. I, insertion of a guanine at position −540; Δ, deletion of an adenine at position −533.
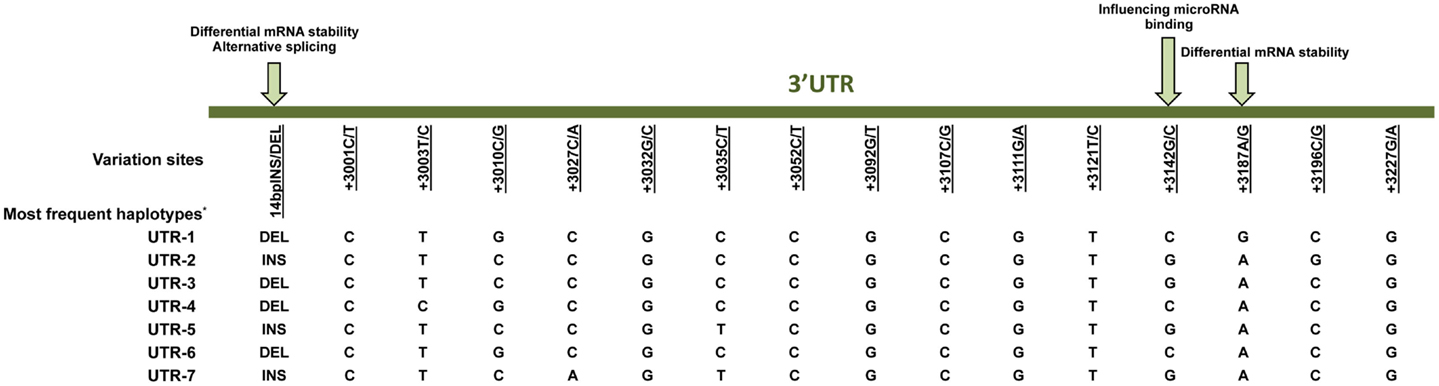
Figure 2. Variation sites at the 3′ untranslated region (3′UTR) of the HLA-G gene that may influence HLA-G expression. Polymorphic sites associated with diseases presented in this review are underlined. Arrows indicate polymorphic sites that have been functionally studied. *Since there is no official nomenclature for 3′UTR haplotypes, they were designed as previously reported (11). DEL, deletion; INS, insertion.
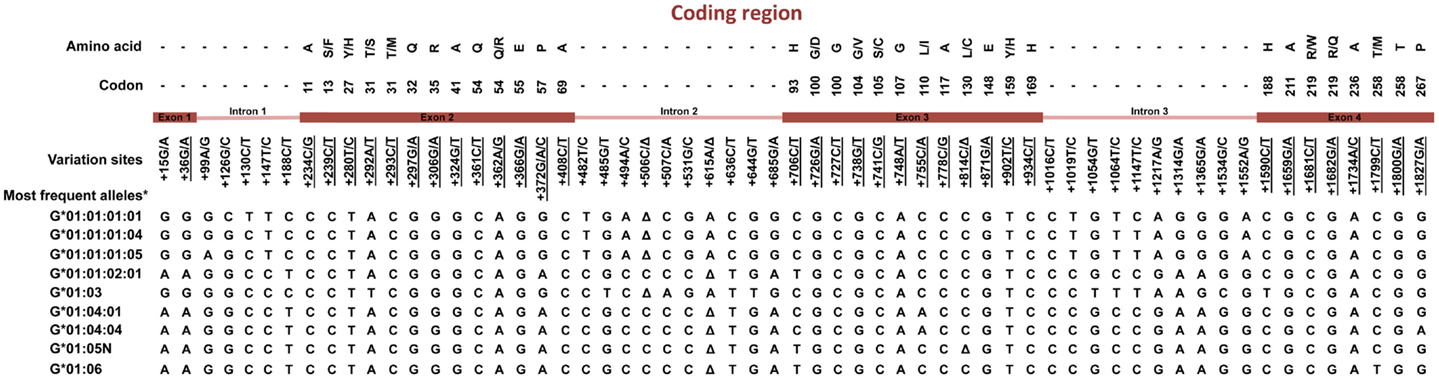
Figure 3. Variation sites at the coding region of the HLA-G gene from exon 1–4. Polymorphic sites associated with diseases presented in this review are underlined. *Haplotypes presenting a global frequency higher than 1% in worldwide populations. Amino acids: A, alanine; S, serine; F, phenylalanine; Y, tyrosine; T, threonine; M, methionine; Q, glutamine; R, arginine; E, glutamic acid; P, proline; H, histidine; G, glycine; D, aspartic acid; V, valine; C, cysteine; L, leucine; I, isoleucine; W, tryptophan. Δ, deletion.
The HLA-G coding region presents mainly synonymous or intronic variation sites. Considering the most frequent HLA-G coding haplotypes found worldwide [reviewed at Ref. (2, 18)], only five different HLA-G full-length molecules are frequently found, in which four are complete molecules encoded by the HLA-G-*01:01, *01:03, *01:04, and *01:06 allele groups, and one is a truncated molecule encoded by the HLA-G*01:05N null allele. Although some different HLA-G molecules were detected worldwide, they are usually quite rare and the same HLA-G coding alleles are usually detected in every population studied so far. Apparently, all these frequently found molecules (exception made to the G*01:05N) present the same modulatory effects described earlier (2). Considering that only a few extended haplotypes are usually found, and considering that most of the HLA-G coding alleles are associated with only one promoter or 3′UTR haplotype, it is possible that most of the associations described so far regarding HLA-G coding polymorphism and pathological conditions are reflecting the presence of specific promoter and 3′UTR sequences and specific HLA-G production capabilities.
In the present review, we report some diseases that have been associated with the modulation of the HLA-G expression, with the presence of specific HLA-G gene variation sites or both, and whenever known, the mechanisms underlying such associations are discussed.
Tumors
The arisen of transformed cells and the spread of cancer cell clones are usually controlled by the immune system cells, particularly by the action of cytototoxic T and NK cells; however, cancer cells have developed several strategies to evade host immune surveillance. Since classical histocompatibility (HLA-A, -B, and -C) molecules present tumor antigens to cytotoxic T cells, tumor cells have developed strategies to escape the cytotoxic effect of T cells by interfering with the expression of these molecules on tumor cell surface. On the other hand, the absence of HLA classical molecules on the surface of tumor cells triggers NK cell activity to eliminate neoplastic cells. If tumor cell expresses HLA-G, the cytotoxic activity of both T and NK cells are inhibited, facilitating tumor cell spread. When the decreased expression of classical HLA molecules is accompanied by an increased expression of immunomodulatory molecules such as HLA-G, the effective cytototoxic immune response against tumor cells is much impoverished [reviewed at Ref. (2)].
Although the study of HLA-G expression in tumor cells has been widely explored [reviewed at Ref. (19–21)], the evaluation of the HLA-G gene polymorphic sites has not been studied at the same extent, and even rarer are the studies evaluating the relationship between HLA-G tumor expression and HLA-G polymorphic sites. Next, we highlight some peculiarities of tumors, for which HLA-G expression (tissue or soluble levels), gene polymorphisms, or both have been evaluated.
HLA-G Expression in Tumors
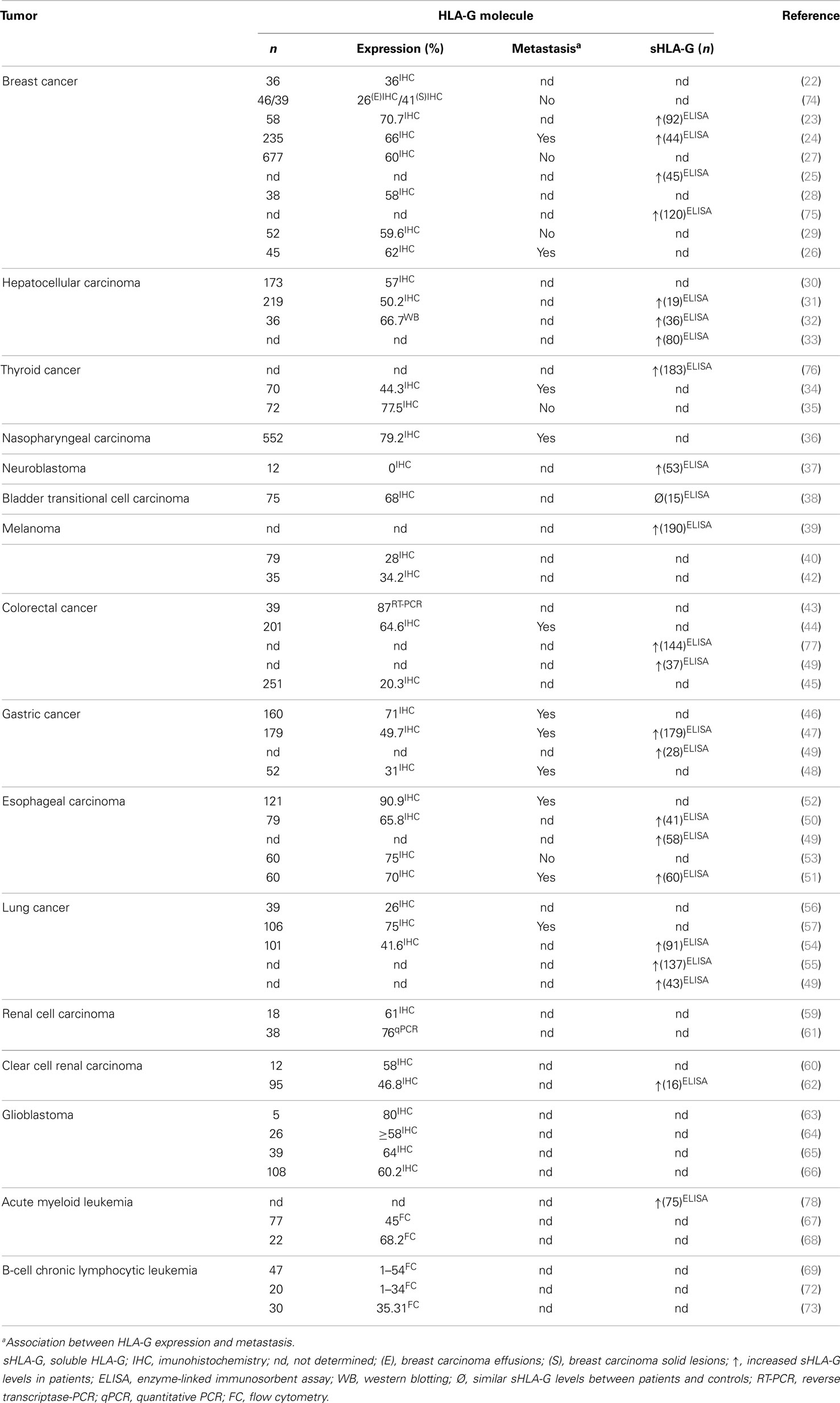
Increased HLA-G expression has been observed in different tumor types, including breast cancer (22–29), hepatocellular carcinoma (30–33), papillary thyroid carcinoma (34, 35), follicular thyroid carcinoma (35), follicular adenoma (35), nasopharyngeal carcinoma (36), neuroblastoma (37), bladder transitional cell carcinoma (TCC) (38), melanoma (39–42), colorectal cancer (43–45), gastric cancer (46–48), esophageal carcinoma (49–53), lung cancer (49, 54–57), renal cell carcinoma (58–62), glioblastoma (63–66), acute myeloid leukemia (67, 68), and B-cell chronic lymphocytic leukemia (69–73). Table 1 summarizes the HLA-G expression in many types of tumors described in this review.
In most tumors, the increased HLA-G expression has been associated with advanced disease stages, shorter survival time, presence of metastasis, higher tumor grade, weak host immune response, greater tumor size, tumor recurrence, tumor invasion, poor histological grade, lower classical HLA antigen expression, presence of infiltrating T regulatory cells, cancer progression, increased inflammatory cell lesion infiltration, and tumor differentiation (23, 24, 26, 29–32, 34–36, 40–42, 44, 46–48, 50–54, 56, 57, 66, 69, 72, 73, 79). In other tumors, no association between increased HLA-G expression and clinicopathological features has been observed, including bladder TCC (38) and acute myeloid leukemia (67, 68).
Furthermore, increased sHLA-G levels have been reported for breast cancer (23–25, 75), hepatocellular carcinoma (31–33), papillary thyroid carcinoma (76), neuroblastoma (37), melanoma (39), colorectal cancer (49, 77), gastric cancer (47, 49), esophageal carcinoma (49–51), lung cancer (49, 54, 55), renal cell carcinoma (62), and acute myeloid leukemia (78). Higher sHLA-G levels have been associated with: (i) increased number of CD4+ regulatory T (Treg) cells in breast cancer (23), (ii) more aggressive tumor behavior in papillary thyroid carcinoma (76), (iii) local or disseminated relapse in neuroblastoma (37), (iv) advanced stages of disease and tumor load in melanoma (39), (v) higher IL-10 production in esophageal carcinoma (51), (vi) absence of anterior myelodysplasia along with higher leukocytosis in acute myeloid leukemia (78), and (vii) shorter survival time, high-grade tumors, higher IL-10 production, and loss of HLA classical class I molecules in patients with lung cancer (54–56).
Interestingly, sHLA-G levels were significantly decreased in breast cancer patients at 6 and 12 months after surgery (25). In addition, no association between higher sHLA-G levels and clinicopathological features has been observed in hepatocellular carcinoma (33), colorectal cancer (77), gastric cancer (47), esophageal carcinoma (50, 51), and renal cell carcinoma (62). On the other hand, plasma sHLA-G levels were closely similar when bladder TCC patients and healthy controls were compared (38).
Overall, several laboratory (increased HLA-G tumor expression, increased sHLA-G levels, increased levels of IL-10, and a cytokine that induces HLA-G expression) and clinical (advanced disease stages, worse prognosis, and presence of metastasis) findings do corroborate the malefic role of HLA-G in cancer disorders.
Polymorphic Sites at HLA-G Gene and Tumors
Several isolated segments of the HLA-G gene have been studied in tumors, highlighting the 3′ untranslated and coding regions. Certainly, the 14-bpINS/DEL polymorphism is the most studied. In breast cancer patients, the 14-bpDEL allele and 14-bpDEL/DEL genotype were associated with susceptibility to breast cancer in Southeastern Iranian (80) and Korean patients (81); however, no association has been reported for Brazilians (26). In addition, Korean patients exhibiting the 14-bpINS/INS genotype exhibited no HLA-G expression in breast cancer lesions (81). A meta-analysis evaluating the role of the 14-bpINS/DEL polymorphism in breast cancer reports an overall cancer risk in Asian populations (82).
The 14-bpDEL allele was associated with susceptibility to hepatocellular carcinoma in Brazilian (83) and Chinese (84) patients, but not in Korean patients (84). In addition, Chinese patients exhibiting the 14-bpDEL/DEL genotype presented increased HLA-G expression in hepatocellular carcinoma specimens (84). The 14-bpINS/DEL genotype was associated with decreased risk for childhood neuroblastoma development in Australian and New Zealand patients (85). The HLA-G 3′UTR haplotype known as UTR-3 (86) was associated with susceptibility to acute myeloid leukemia development in Italian patients (68).
Considering the HLA-G coding segment, the +755C/A (non-synonymous Leu/Ile substitution at codon 110, which defines the HLA-G*01:04 protein group) was associated with protection against more severe nasopharyngeal carcinoma tumor stages (87).
Regarding the bladder TCC, the HLA-G*01:04:04 allele, and the HLA-G*01:04 allelic group were associated with susceptibility to bladder TCC in smoking patients and the HLA-G*01:03 allele and the HLA-G*01:04 allelic group was associated with protection against bladder TCC development in non-smoking Brazilian patients. In addition, the HLA-G*01:01 allelic group and HLA-G*01:01/G*01:01 genotype were associated with susceptibility to bladder TCC development in non-smokers. Considering the bladder TCC progression, the following associations were observed: (i) the HLA-G*01:03 allele was associated with high-grade tumors among smokers; (ii) the HLA-G*01:01:01/G*01:01:02 genotype was associated with protection against high-grade tumors in the whole group of patients, whereas the same association was observed with the HLA-G*01:01 group, but only among smokers; and (iii) the HLA-G*01:04 allele group was associated with high-grade tumor development in smoker and in the whole group of patients (88).
No association has been observed for: (i) HLA-G coding region alleles in South Korean and Brazilian breast cancer patients (81, 89); (ii) 14-bpINS/DEL polymorphism in Italian patients presenting thyroid cancer (76); (iii) HLA-G*01:03 allele and HLA-G*01:05N null allele in Tunisian patients with nasopharyngeal carcinoma (87); (iv) HLA-G*01:05N null allele with susceptibility to esophagus carcinoma development in Chinese patients (90); (v) 14-bp INS/DEL polymorphic site in Brazilian bladder TCC patients (88); and (vi) +292A/T, +755C/A, and +1799C/T in Australian and New Zealand childhood neuroblastoma patients (85).
To date, HLA-G polymorphisms have not been investigated in the context of melanoma, glioblastoma, colorectal cancer, gastric cancer, lung cancer, and renal cell carcinoma.
Although some polymorphic sites (14-bpDEL allele) and coding region allele groups (HLA-G*01:04) have been previously associated with increased sHLA-G levels, few convincing associations have been reported, exception made to breast cancer for which an extensive meta-analysis has evidenced the role of this polymorphic site in Asiatic patients. Since several polymorphic sites have been described at the HLA-G regulatory regions, exhibiting putative roles on HLA-G expression, the typing of the complete gene and the study of the regulatory elements (transcription factors and microRNAs) produced in the tumor environment may the helpful to understand the mechanisms of tumor evasion mechanisms.
Viral Hepatitis
Similar to tumor cells, viruses have also developed several strategies to evade the cytotoxic effect of immune effector cells, including downregulation of HLA classical class I molecules and the upregulation of non-classical molecules, or both. As a corollary, the increased HLA-G expression, induced by the virus itself or by the presence of an inflammatory milieu containing transcription and post-transcription factors that positively modulate HLA-G expression, may exacerbate virus morbidity and/or patient mortality. The influence of HLA-G has been studied in several chronic viral infections; some of them associated with neoplastic transformation, including human immunodeficiency virus (HIV), human papillomavirus (HPV), human cytomegalovirus (hCMV), and hepatitis viruses [reviewed at Ref. (2)].
Increased HLA-G hepatocyte expression in HCV-infected liver specimens has been associated with milder stages of fibrosis and hemosiderin deposit (91). Besides hepatocytes, HLA-G expression was observed on mast cells present in areas of liver fibrosis (92). Increased plasma sHLA-G levels were associated with chronic HCV infection and with increased IL-10 and IFN-γ levels (93). Since the treatment of mast cells with IL-10 and class I interferons induces HLA-G expression (92), infiltrating cells may play an important role on the maintenance of chronic infection and induction of chronic complications.
One study has associated increased HLA-G expression in hepatocytes with the HBV viral load (94). Different studies associated the increased serum/plasma sHLA-G levels with hepatitis B virus infection (33, 95, 96), which were associated with increased percentage of CD4+CD25+FoxP3+ T regulatory and HLA-G+CD14+ monocytes cells in patients exhibiting acute or chronic hepatitis (95), active hepatitis B virus infection (33) and HBeAg negative hepatitis, hepatocellular carcinoma, and increased alanine aminotransferase levels (96).
Regarding the typing of HLA-G 3′UTR polymorphic sites in HCV- and HBV-infected patients, the +3142C allele and 14-bpDEL/+3142C haplotype were underrepresented in Brazilian HCV-infected patients presenting sickle cells disease compared with HCV-negative group (97). On the other hand, the 14-bpINS/INS genotype was overrepresented in African-Brazilian HIV+ patients co-infected with HCV (HIV+/HCV+) compared with HIV+/HCV− patients. Regarding the HLA-G+3142 C/G and 14-bp INS/DEL variants, no significant association has been reported for HIV+/HCV+- (98) and HBV-infected patients (99), respectively, when compared with their respective controls.
Considering that many viruses have developed evasion strategies that are similar to cancer cells and considering that many chronic viral disorders have been associated with cell transformation and malignancy, the expression of HLA-G in these disorders may predict a worse outcome and greater susceptibility to cell transformation.
Protozoan Parasite Infections
Human Malaria Infection
Plasmodium spp. is the etiologic agent of the human malaria and little is known about the role of HLA-G during malaria infection, and all studies have been performed to understand the mother to child transmission. One study reported a decreased HLA-G expression in extravillous trophoblast of Plasmodium falciparum-infected placentas compared to uninfected placentas. If by one hand, HLA-G molecule is almost exclusively expressed in extravillous trophoblast of healthy placenta specimens, on the other hand, HLA-G is detected in intervillous space macrophages of Plasmodium-infected placentas. In addition, NK cells are increased in infected compared to uninfected placentas (100). Furthermore, increased cord plasma levels of sHLA-G have been associated with low birth weight and increased risk of P. falciparum infection in infancy (101).
A family based association study performed on individuals from Niakhar, Senegal, reported that the +3187G allele was associated with higher transmission to children and lower level of parasite density during asymptomatic P. falciparum infection. The HLA-G 3′UTR haplotype known as UTR-1 was associated with a decreased level of parasite density during asymptomatic infection under a dominant model, whereas the HLA-G UTR-3 haplotype was associated with an increased level of parasite density during the follow-up and increased intensity of asymptomatic infection under a recessive model (102).
A second family based association study also conducted on Senegalese population has tested the association of HLA-G 3′UTR variants with acquired anti-malarial humoral immunity. The +3010G and +3142C alleles were overtransmitted to children with increased total IgG and IgG1 antibodies levels against glutamate-rich protein (GLURP) of P. falciparum, and the +3196G allele had a preferential transmission to children with a lower IgG3 response against merozoite surface protein 2 (MSP2). The HLA-G 3′UTR-2 haplotype was associated with a decreased IgG3 response against MSP2, suggesting a role of HLA-G on the regulation of immune humoral response during P. falciparum infection (103).
Human African Trypanosomiasis
Human African trypanosomiasis, also known as sleeping sickness, is caused by protozoan parasites of the Trypanosoma brucei species. Although no studies are available regarding HLA-G expression, genetic studies report associations of HLA-G gene single nucleotide variation sites with the disease. A family based association study reported that the HLA-G 3′UTR-14-bpINS and +3196G alleles had a preferential transmission from heterozygote parents to children and were associated with susceptibility to human African trypanosomiasis (HAT) development. In contrast, the HLA-G 3′UTR +3003C, +3010G, and +3187G alleles showed lower transmission from parents to children and were associated with decreased risk of developing the disease. Regarding HLA-G 3′UTR haplotypes, UTR-2 and UTR-5 haplotypes were associated with higher susceptibility to HAT development, whereas the HLA-G UTR-4 haplotype was associated with decreased risk for HAT development (104).
American Trypanosomiasis
The parasite Trypanosoma cruzi is the etiologic agent of American trypanosomiasis, also known as Chagas disease (105). In the chronic phase, four major clinical forms are observed: (i) cardiac that presents progressive congestive heart failure, various cardiac arrhythmias, thromboembolic events, and sudden death; (ii) digestive that is characterized by clinical signs of megaesophagus, megacolon, or both; (iii) cardiodigestive that comprises clinical and pathological signs of cardiac and digestive involvement; and (iv) indeterminate that develops without evident clinical and pathological signs (106). Recently, our group reported a decreased HLA-G expression on cardiac muscle and colonic cells in patients presenting cardiac or digestive clinical variants, respectively. On the other hand, no significant differences were observed regarding HLA-G expression in the esophagus of patients with digestive form when compared to non-chagasic patients.
Furthermore, we evaluated the polymorphic sites at the HLA-G 3′UTR region in Brazilian chagasic patients. The +3003T allele and +3003TT and +3187GG genotypes were overrepresented, whereas the +3003C allele and +3003CT, +3010GC, and +3042GC genotypes were underrepresented in symptomatic patients. In addition, the +3027CC and +3035CC genotypes, and the +3027C and +3035C alleles were associated with the digestive form of Chagas disease. Regarding HLA-G 3′UTR haplotypes, decreased UTR-4 and UTR-7 frequencies were associated with symptomatic patients and with the digestive form, respectively. On the other hand, UTR-13 was associated with the indeterminate variant and UTR-14 with the cardiac form (107).
Overall, studies on the association between HLA-G and parasitic disorders are still scarce and only the HLA-G 3′UTR has been evaluated.
Conclusion
Considering the tolerogenic properties of HLA-G and considering the aphorism that the induced expression of HLA-G may be detrimental in tumors and chronic viral infection, the overall findings reported is this revision corroborates this idea. Noteworthy, is the induced expression of HLA-G on the surface of tumor cells, which has been associated with greater tumor morbidity, tumor progression, and spreading. In addition, in chronic viral infections associated with pre-neoplastic and neoplastic transformation. On the other hand, the repression of HLA-G expression is less well studied; i.e., the decreased expression of HLA-G in organs or conditions in which a constitutive expression of the molecule is expected. For instance, the decreased expression of HLA-G (placentas of P. falciparum-infected mothers or heart and colonic specimens of Chagas disease) has been associated with morbidity of the chronic parasitic infection.
Studies on the association of the HLA-G gene with diseases of diverse etiology have underestimated the myriad of polymorphic sites present at the various gene segments and have primarily focused on the evaluation of one or few polymorphic sites, particularly at the 3′UTR. Considering that many polymorphic sites along the HLA-G gene can be readily performed and analyzed, and considering the relevant role of isolated polymorphic sites or HLA-G haplotypes on HLA-G expression, HLA-G typing on diseases should add an additional tool on the understanding of the role of HLA-G on disease associations.
Theoretically, polymorphic sites observed along the coding region may modify the encoded protein and consequently the interaction with HLA-G receptors and the formation of HLA-G dimers that may more efficiently bind to HLA-G receptors. Thus, a particular allele and a particular molecule could provide susceptibility or protection against a disease development; however, such associations have not been strong enough to be considered a disease marker, as has been observed for the classical association between HLA-B27 and ankylosing spondylitis. On the other hand, polymorphic sites observed along the HLA-G promoter and 3′UTR gene segments may modify gene expression, accounting for disease morbidity. Unfortunately, few polymorphic sites along regulatory regions have extensively been evaluated regarding their function, and probably a combination of regulatory transcriptional and posttranscriptional elements may account for the final HLA-G production. Therefore, a complete gene evaluation together with the availability of transcription and protein profiles may provide light to the understanding of the mechanisms of HLA-G induction or repression in a specific disorder.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The Review Editor, Joel LeMaoult, declares that despite having co-authored a manuscript and being affiliated with the same institution as author Philippe Moreau, there has been no conflict of interest during the review and handling of this manuscript.
Acknowledgments
This work was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior [grant number CAPES/COFECUB 653/09], Conselho Nacional de Desenvolvimento Científico e Tecnológico [CNPq Science Without Borders Program, grant number 236754/2012-2; Special Visiting Researcher, grant number 406594/2013-9; Young Talents, grant number 401641/2013-9; CNPq edital 71/2013, grant number 406594/2013-9; CNPq Universal, grant number 466036/2013-5; CNPq/MS/SCTIE/DECIT No. 31/2014 – Pesquisas sobre Doença de Chagas, grant number 467157/2014-6], and Núcleo de Apoio a Pesquisa em Doenças Inflamatórias (NAP-DIN).
References
1. Geraghty DE, Koller BH, Orr HT. A human major histocompatibility complex class I gene that encodes a protein with a shortened cytoplasmic segment. Proc Natl Acad Sci U S A (1987) 84(24):9145–9. doi:10.1073/pnas.84.24.9145
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
2. Donadi EA, Castelli EC, Arnaiz-Villena A, Roger M, Rey D, Moreau P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci (2011) 68(3):369–95. doi:10.1007/s00018-010-0580-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
3. Rouas-Freiss N, Goncalves RM, Menier C, Dausset J, Carosella ED. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc Natl Acad Sci U S A (1997) 94(21):11520–5. doi:10.1073/pnas.94.21.11520
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
4. Naji A, Menier C, Morandi F, Agaugue S, Maki G, Ferretti E, et al. Binding of HLA-G to ITIM-bearing Ig-like transcript 2 receptor suppresses B cell responses. J Immunol (2014) 192(4):1536–46. doi:10.4049/jimmunol.1300438
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
5. Horuzsko A, Lenfant F, Munn DH, Mellor AL. Maturation of antigen-presenting cells is compromised in HLA-G transgenic mice. Int Immunol (2001) 13(3):385–94. doi:10.1093/intimm/13.3.385
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
6. Le Discorde M, Moreau P, Sabatier P, Legeais JM, Carosella ED. Expression of HLA-G in human cornea, an immune-privileged tissue. Hum Immunol (2003) 64(11):1039–44. doi:10.1016/j.humimm.2003.08.346
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
7. Lefebvre S, Adrian F, Moreau P, Gourand L, Dausset J, Berrih-Aknin S, et al. Modulation of HLA-G expression in human thymic and amniotic epithelial cells. Hum Immunol (2000) 61(11):1095–101. doi:10.1016/S0198-8859(00)00192-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
8. Menier C, Rabreau M, Challier JC, Le Discorde M, Carosella ED, Rouas-Freiss N. Erythroblasts secrete the nonclassical HLA-G molecule from primitive to definitive hematopoiesis. Blood (2004) 104(10):3153–60. doi:10.1182/blood-2004-03-0809
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
9. Tan Z, Shon AM, Ober C. Evidence of balancing selection at the HLA-G promoter region. Hum Mol Genet (2005) 14(23):3619–28. doi:10.1093/hmg/ddi389
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
10. Castelli EC, Mendes-Junior CT, Veiga-Castelli LC, Roger M, Moreau P, Donadi EA. A comprehensive study of polymorphic sites along the HLA-G gene: implication for gene regulation and evolution. Mol Biol Evol (2011) 28(11):3069–86. doi:10.1093/molbev/msr138
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
11. Sabbagh A, Luisi P, Castelli EC, Gineau L, Courtin D, Milet J, et al. Worldwide genetic variation at the 3’ untranslated region of the HLA-G gene: balancing selection influencing genetic diversity. Genes Immun (2014) 15(2):95–106. doi:10.1038/gene.2013.67
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
12. Santos KE, Lima TH, Felicio LP, Massaro JD, Palomino GM, Silva AC, et al. Insights on the HLA-G evolutionary history provided by a nearby Alu insertion. Mol Biol Evol (2013) 30(11):2423–34. doi:10.1093/molbev/mst142
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
13. Castelli EC, Veiga-Castelli LC, Yaghi L, Moreau P, Donadi EA. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res (2014) 2014:734068. doi:10.1155/2014/734068
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
14. Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet (2007) 81(4):829–34. doi:10.1086/521200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
15. Manaster I, Goldman-Wohl D, Greenfield C, Nachmani D, Tsukerman P, Hamani Y, et al. MiRNA-mediated control of HLA-G expression and function. PLoS One (2012) 7(3):e33395. doi:10.1371/journal.pone.0033395
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
16. Castelli EC, Moreau P, Oya e Chiromatzo A, Mendes-Junior CT, Veiga-Castelli LC, Yaghi L, et al. In silico analysis of microRNAS targeting the HLA-G 3’ untranslated region alleles and haplotypes. Hum Immunol (2009) 70(12):1020–5. doi:10.1016/j.humimm.2009.07.028
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
17. Martelli-Palomino G, Pancotto JA, Muniz YC, Mendes-Junior CT, Castelli EC, Massaro JD, et al. Polymorphic sites at the 3’ untranslated region of the HLA-G gene are associated with differential HLA-G soluble levels in the Brazilian and French population. PLoS One (2013) 8(10):e71742. doi:10.1371/journal.pone.0071742
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
18. Castelli EC, Ramalho J, Porto IO, Lima TH, Felicio LP, Sabbagh A, et al. Insights into HLA-G genetics provided by worldwide haplotype diversity. Front Immunol (2014) 5:476. doi:10.3389/fimmu.2014.00476
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
19. Rouas-Freiss N, Moreau P, Ferrone S, Carosella ED. HLA-G proteins in cancer: do they provide tumor cells with an escape mechanism? Cancer Res (2005) 65(22):10139–44. doi:10.1158/0008-5472.CAN-05-0097
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
20. Carosella ED, Moreau P, Lemaoult J, Rouas-Freiss N. HLA-G: from biology to clinical benefits. Trends Immunol (2008) 29(3):125–32. doi:10.1016/j.it.2007.11.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
21. Bukur J, Jasinski S, Seliger B. The role of classical and non-classical HLA class I antigens in human tumors. Semin Cancer Biol (2012) 22(4):350–8. doi:10.1016/j.semcancer.2012.03.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
22. Lefebvre S, Antoine M, Uzan S, McMaster M, Dausset J, Carosella ED, et al. Specific activation of the non-classical class I histocompatibility HLA-G antigen and expression of the ILT2 inhibitory receptor in human breast cancer. J Pathol (2002) 196(3):266–74. doi:10.1002/path.1039
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
23. Chen HX, Lin A, Shen CJ, Zhen R, Chen BG, Zhang X, et al. Upregulation of human leukocyte antigen-G expression and its clinical significance in ductal breast cancer. Hum Immunol (2010) 71(9):892–8. doi:10.1016/j.humimm.2010.06.009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
24. He X, Dong DD, Yie SM, Yang H, Cao M, Ye SR, et al. HLA-G expression in human breast cancer: implications for diagnosis and prognosis, and effect on allocytotoxic lymphocyte response after hormone treatment in vitro. Ann Surg Oncol (2010) 17(5):1459–69. doi:10.1245/s10434-009-0891-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
25. Sayed D, Badr G, Maximous D, Mikhail NN, Abu-Tarboush F, Alhazza IM. HLA-G and its relation to proliferation index in detection and monitoring breast cancer patients. Tissue Antigens (2010) 75(1):40–7. doi:10.1111/j.1399-0039.2009.01393.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
26. Ramos CS, Goncalves AS, Marinho LC, Gomes Avelino MA, Saddi VA, Lopes AC, et al. Analysis of HLA-G gene polymorphism and protein expression in invasive breast ductal carcinoma. Hum Immunol (2014) 75(7):667–72. doi:10.1016/j.humimm.2014.04.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
27. de Kruijf EM, Sajet A, van Nes JG, Natanov R, Putter H, Smit VT, et al. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol (2010) 185(12):7452–9. doi:10.4049/jimmunol.1002629
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
28. Elliott RL, Jiang XP, Phillips JT, Barnett BG, Head JF. Human leukocyte antigen G expression in breast cancer: role in immunosuppression. Cancer Biother Radiopharm (2011) 26(2):153–7. doi:10.1089/cbr.2010.0924
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
29. da Silva GB, Silva TG, Duarte RA, Neto NL, Carrara HH, Donadi EA, et al. Expression of the classical and nonclassical HLA molecules in breast cancer. Int J Breast Cancer (2013) 2013:250435. doi:10.1155/2013/250435
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
30. Cai MY, Xu YF, Qiu SJ, Ju MJ, Gao Q, Li YW, et al. Human leukocyte antigen-G protein expression is an unfavorable prognostic predictor of hepatocellular carcinoma following curative resection. Clin Cancer Res (2009) 15(14):4686–93. doi:10.1158/1078-0432.CCR-09-0463
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
31. Lin A, Chen HX, Zhu CC, Zhang X, Xu HH, Zhang JG, et al. Aberrant human leukocyte antigen-G expression and its clinical relevance in hepatocellular carcinoma. J Cell Mol Med (2010) 14(8):2162–71. doi:10.1111/j.1582-4934.2009.00917.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
32. Wang Y, Ye Z, Meng XQ, Zheng SS. Expression of HLA-G in patients with hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int (2011) 10(2):158–63. doi:10.1016/S1499-3872(11)60025-8
33. Park Y, Park Y, Lim HS, Kim YS, Hong DJ, Kim HS. Soluble human leukocyte antigen-G expression in hepatitis B virus infection and hepatocellular carcinoma. Tissue Antigens (2012) 79(2):97–103. doi:10.1111/j.1399-0039.2011.01814.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
34. Nunes LM, Ayres FM, Francescantonio IC, Saddi VA, Avelino MA, Alencar Rde C, et al. Association between the HLA-G molecule and lymph node metastasis in papillary thyroid cancer. Hum Immunol (2013) 74(4):447–51. doi:10.1016/j.humimm.2012.12.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
35. de Figueiredo Feitosa NL, Crispim JC, Zanetti BR, Magalhaes PK, Soares CP, Soares EG, et al. HLA-G is differentially expressed in thyroid tissues. Thyroid (2014) 24(3):585–92. doi:10.1089/thy.2013.0246
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
36. Cai MB, Han HQ, Bei JX, Liu CC, Lei JJ, Cui Q, et al. Expression of human leukocyte antigen G is associated with prognosis in nasopharyngeal carcinoma. Int J Biol Sci (2012) 8(6):891–900. doi:10.7150/ijbs.4383
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
37. Morandi F, Levreri I, Bocca P, Galleni B, Raffaghello L, Ferrone S, et al. Human neuroblastoma cells trigger an immunosuppressive program in monocytes by stimulating soluble HLA-G release. Cancer Res (2007) 67(13):6433–41. doi:10.1158/0008-5472.CAN-06-4588
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
38. Gan LH, Huang LF, Zhang X, Lin A, Xu DP, Wang Q, et al. Tumor-specific upregulation of human leukocyte antigen-G expression in bladder transitional cell carcinoma. Hum Immunol (2010) 71(9):899–904. doi:10.1016/j.humimm.2010.06.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
39. Ugurel S, Rebmann V, Ferrone S, Tilgen W, Grosse-Wilde H, Reinhold U. Soluble human leukocyte antigen – G serum level is elevated in melanoma patients and is further increased by interferon-alpha immunotherapy. Cancer (2001) 92(2):369–76. doi:10.1002/1097-0142(20010715)92:2<369::AID-CNCR1332>3.0.CO;2-U
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
40. Ibrahim EC, Aractingi S, Allory Y, Borrini F, Dupuy A, Duvillard P, et al. Analysis of HLA antigen expression in benign and malignant melanocytic lesions reveals that upregulation of HLA-G expression correlates with malignant transformation, high inflammatory infiltration and HLA-A1 genotype. Int J Cancer (2004) 108(2):243–50. doi:10.1002/ijc.11456
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
41. Bezuhly M, Howlett A, Colp P, Conrad DM, Walsh N, Rowden G, et al. Quantitative HLA-G expression in metastasising and non-metastasising primary thin cutaneous melanomas. Dermatology (2008) 217(3):281–3. doi:10.1159/000150602
42. Fang X, Zhang X, Li J. Up-regulation of human leukocyte antigen G expression in primary cutaneous malignant melanoma associated with host-vs-tumor immune response. J Huazhong Univ Sci Technolog Med Sci (2008) 28(2):219–21. doi:10.1007/s11596-008-0227-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
43. Fukushima Y, Oshika Y, Nakamura M, Tokunaga T, Hatanaka H, Abe Y, et al. Increased expression of human histocompatibility leukocyte antigen-G in colorectal cancer cells. Int J Mol Med (1998) 2(3):349–51.
44. Ye SR, Yang H, Li K, Dong DD, Lin XM, Yie SM. Human leukocyte antigen G expression: as a significant prognostic indicator for patients with colorectal cancer. Mod Pathol (2007) 20(3):375–83. doi:10.1038/modpathol.3800751
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
45. Zeestraten EC, Reimers MS, Saadatmand S, Dekker JW, Liefers GJ, van den Elsen PJ, et al. Combined analysis of HLA class I, HLA-E and HLA-G predicts prognosis in colon cancer patients. Br J Cancer (2014) 110(2):459–68. doi:10.1038/bjc.2013.696
46. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of human leukocyte antigen G (HLA-G) correlates with poor prognosis in gastric carcinoma. Ann Surg Oncol (2007) 14(10):2721–9. doi:10.1245/s10434-007-9464-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
47. Du L, Xiao X, Wang C, Zhang X, Zheng N, Wang L, et al. Human leukocyte antigen-G is closely associated with tumor immune escape in gastric cancer by increasing local regulatory T cells. Cancer Sci (2011) 102(7):1272–80. doi:10.1111/j.1349-7006.2011.01951.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
48. Tuncel T, Karagoz B, Haholu A, Ozgun A, Emirzeoglu L, Bilgi O, et al. Immunoregulatory function of HLA-G in gastric cancer. Asian Pac J Cancer Prev (2013) 14(12):7681–4. doi:10.7314/APJCP.2013.14.12.7681
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
49. Cao M, Yie SM, Liu J, Ye SR, Xia D, Gao E. Plasma soluble HLA-G is a potential biomarker for diagnosis of colorectal, gastric, esophageal and lung cancer. Tissue Antigens (2011) 78(2):120–8. doi:10.1111/j.1399-0039.2011.01716.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
50. Lin A, Zhang X, Zhou WJ, Ruan YY, Xu DP, Wang Q, et al. Human leukocyte antigen-G expression is associated with a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Cancer (2011) 129(6):1382–90. doi:10.1002/ijc.25807
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
51. Zheng J, Xu C, Chu D, Zhang X, Li J, Ji G, et al. Human leukocyte antigen G is associated with esophageal squamous cell carcinoma progression and poor prognosis. Immunol Lett (2014) 161(1):13–9. doi:10.1016/j.imlet.2014.04.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
52. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of HLA-G is associated with prognosis in esophageal squamous cell carcinoma. Am J Clin Pathol (2007) 128(6):1002–9. doi:10.1309/JNCW1QLDFB6AM9WE
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
53. Hu J, Li L, Liu Y, Chen Y, Liu C, Liang W, et al. Overexpression of HLA-G Is positively associated with Kazakh esophageal squamous cell carcinoma in Xinjiang, China. Viral Immunol (2013) 26(3):180–4. doi:10.1089/vim.2012.0085
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
54. Lin A, Zhu CC, Chen HX, Chen BF, Zhang X, Zhang JG, et al. Clinical relevance and functional implications for human leukocyte antigen-g expression in non-small-cell lung cancer. J Cell Mol Med (2010) 14(9):2318–29. doi:10.1111/j.1582-4934.2009.00858.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
55. Schutt P, Schutt B, Switala M, Bauer S, Stamatis G, Opalka B, et al. Prognostic relevance of soluble human leukocyte antigen-G and total human leukocyte antigen class I molecules in lung cancer patients. Hum Immunol (2010) 71(5):489–95. doi:10.1016/j.humimm.2010.02.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
56. Urosevic M, Kurrer MO, Kamarashev J, Mueller B, Weder W, Burg G, et al. Human leukocyte antigen G up-regulation in lung cancer associates with high-grade histology, human leukocyte antigen class I loss and interleukin-10 production. Am J Pathol (2001) 159(3):817–24. doi:10.1016/S0002-9440(10)61756-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
57. Yie SM, Yang H, Ye SR, Li K, Dong DD, Lin XM. Expression of human leukocyte antigen G (HLA-G) is associated with prognosis in non-small cell lung cancer. Lung Cancer (2007) 58(2):267–74. doi:10.1016/j.lungcan.2007.06.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
58. Hanak L, Slaby O, Lauerova L, Kren L, Nenutil R, Michalek J. Expression pattern of HLA class I antigens in renal cell carcinoma and primary cell line cultures: methodological implications for immunotherapy. Med Sci Monit (2009) 15(12):CR638–43.
59. Ibrahim EC, Guerra N, Lacombe MJ, Angevin E, Chouaib S, Carosella ED, et al. Tumor-specific up-regulation of the nonclassical class I HLA-G antigen expression in renal carcinoma. Cancer Res (2001) 61(18):6838–45.
60. Ibrahim EC, Allory Y, Commo F, Gattegno B, Callard P, Paul P. Altered pattern of major histocompatibility complex expression in renal carcinoma: tumor-specific expression of the nonclassical human leukocyte antigen-G molecule is restricted to clear cell carcinoma while up-regulation of other major histocompatibility complex antigens is primarily distributed in all subtypes of renal carcinoma. Am J Pathol (2003) 162(2):501–8. doi:10.1016/S0002-9440(10)63844-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
61. Kren L, Valkovsky I, Dolezel J, Capak I, Pacik D, Poprach A, et al. HLA-G and HLA-E specific mRNAs connote opposite prognostic significance in renal cell carcinoma. Diagn Pathol (2012) 7:58. doi:10.1186/1746-1596-7-58
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
62. Li BL, Lin A, Zhang XJ, Zhang X, Zhang JG, Wang Q, et al. Characterization of HLA-G expression in renal cell carcinoma. Tissue Antigens (2009) 74(3):213–21. doi:10.1111/j.1399-0039.2009.01302.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
63. Wiendl H, Mitsdoerffer M, Hofmeister V, Wischhusen J, Bornemann A, Meyermann R, et al. A functional role of HLA-G expression in human gliomas: an alternative strategy of immune escape. J Immunol (2002) 168(9):4772–80. doi:10.4049/jimmunol.168.9.4772
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
64. Kren L, Muckova K, Lzicarova E, Sova M, Vybihal V, Svoboda T, et al. Production of immune-modulatory nonclassical molecules HLA-G and HLA-E by tumor infiltrating ameboid microglia/macrophages in glioblastomas: a role in innate immunity? J Neuroimmunol (2010) 220(1–2):131–5. doi:10.1016/j.jneuroim.2010.01.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
65. Kren L, Slaby O, Muckova K, Lzicarova E, Sova M, Vybihal V, et al. Expression of immune-modulatory molecules HLA-G and HLA-E by tumor cells in glioblastomas: an unexpected prognostic significance? Neuropathology (2011) 31(2):129–34. doi:10.1111/j.1440-1789.2010.01149.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
66. Wastowski IJ, Simoes RT, Yaghi L, Donadi EA, Pancoto JT, Poras I, et al. Human leukocyte antigen-G is frequently expressed in glioblastoma and may be induced in vitro by combined 5-aza-2’-deoxycytidine and interferon-gamma treatments: results from a multicentric study. Am J Pathol (2013) 182(2):540–52. doi:10.1016/j.ajpath.2012.10.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
67. Guo QY, Chen BG, Ruan YY, Lin A, Yan WH. HLA-G expression is irrelevant to prognosis in patients with acute myeloid leukemia. Leuk Res (2011) 35(10):1350–4. doi:10.1016/j.leukres.2011.05.036
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
68. Locafaro G, Amodio G, Tomasoni D, Tresoldi C, Ciceri F, Gregori S. HLA-G expression on blasts and tolerogenic cells in patients affected by acute myeloid leukemia. J Immunol Res (2014) 2014:636292. doi:10.1155/2014/636292
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
69. Nuckel H, Rebmann V, Durig J, Duhrsen U, Grosse-Wilde H. HLA-G expression is associated with an unfavorable outcome and immunodeficiency in chronic lymphocytic leukemia. Blood (2005) 105(4):1694–8. doi:10.1182/blood-2004-08-3335
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
70. Rebmann V, Nuckel H, Duhrsen U, Grosse-Wilde H. HLA-G in B-chronic lymphocytic leukaemia: clinical relevance and functional implications. Semin Cancer Biol (2007) 17(6):430–5. doi:10.1016/j.semcancer.2007.06.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
71. Giannopoulos K, Dmoszynska A, Bojarska-Junak A, Schmitt M, Rolinski J. Expression of HLA-G in patients with B-cell chronic lymphocytic leukemia (B-CLL). Folia Histochem Cytobiol (2008) 46(4):457–60. doi:10.2478/v10042-008-0072-x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
72. Erikci AA, Karagoz B, Ozyurt M, Ozturk A, Kilic S, Bilgi O. HLA-G expression in B chronic lymphocytic leukemia: a new prognostic marker? Hematology (2009) 14(2):101–5. doi:10.1179/102453309X385197
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
73. Attia MA, Nosair NA, Gawally A, Elnagar G, Elshafey EM. HLA-G expression as a prognostic indicator in B-cell chronic lymphocytic leukemia. Acta Haematol (2014) 132(1):53–8. doi:10.1159/000353757
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
74. Kleinberg L, Florenes VA, Skrede M, Dong HP, Nielsen S, McMaster MT, et al. Expression of HLA-G in malignant mesothelioma and clinically aggressive breast carcinoma. Virchows Arch (2006) 449(1):31–9. doi:10.1007/s00428-005-0144-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
75. Provatopoulou X, Kalogera E, Sagkriotis A, Zagouri F, Nonni A, Zografos GC, et al. Soluble human leukocyte antigen-G expression in patients with ductal and lobular breast malignancy. Anticancer Res (2012) 32(3):1021–6.
76. Dardano A, Rizzo R, Polini A, Stignani M, Tognini S, Pasqualetti G, et al. Soluble human leukocyte antigen-g and its insertion/deletion polymorphism in papillary thyroid carcinoma: novel potential biomarkers of disease? J Clin Endocrinol Metab (2012) 97(11):4080–6. doi:10.1210/jc.2012-2231
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
77. Zhu CB, Wang CX, Zhang X, Zhang J, Li W. Serum sHLA-G levels: a useful indicator in distinguishing colorectal cancer from benign colorectal diseases. Int J Cancer (2011) 128(3):617–22. doi:10.1002/ijc.25372
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
78. Gros F, Sebti Y, de Guibert S, Branger B, Bernard M, Fauchet R, et al. Soluble HLA-G molecules increase during acute leukemia, especially in subtypes affecting monocytic and lymphoid lineages. Neoplasia (2006) 8(3):223–30. doi:10.1593/neo.05703
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
79. Morandi F, Scaruffi P, Gallo F, Stigliani S, Moretti S, Bonassi S, et al. Bone marrow-infiltrating human neuroblastoma cells express high levels of calprotectin and HLA-G proteins. PLoS One (2012) 7(1):e29922. doi:10.1371/journal.pone.0029922
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
80. Eskandari-Nasab E, Hashemi M, Hasani SS, Omrani M, Taheri M, Mashhadi MA. Association between HLA-G 3’UTR 14-bp ins/del polymorphism and susceptibility to breast cancer. Cancer Biomark (2013) 13(4):253–9. doi:10.3233/CBM-130364
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
81. Jeong S, Park S, Park BW, Park Y, Kwon OJ, Kim HS. Human leukocyte antigen-G (HLA-G) polymorphism and expression in breast cancer patients. PLoS One (2014) 9(5):e98284. doi:10.1371/journal.pone.0098284
82. Ge YZ, Ge Q, Li MH, Shi GM, Xu X, Xu LW, et al. Association between human leukocyte antigen-G 14-bp insertion/deletion polymorphism and cancer risk: a meta-analysis and systematic review. Hum Immunol (2014) 75(8):827–32. doi:10.1016/j.humimm.2014.06.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
83. Teixeira AC, Mendes-Junior CT, Souza FF, Marano LA, Deghaide NH, Ferreira SC, et al. The 14bp-deletion allele in the HLA-G gene confers susceptibility to the development of hepatocellular carcinoma in the Brazilian population. Tissue Antigens (2013) 81(6):408–13. doi:10.1111/tan.12097
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
84. Jiang Y, Chen S, Jia S, Zhu Z, Gao X, Dong D, et al. Association of HLA-G 3’ UTR 14-bp insertion/deletion polymorphism with hepatocellular carcinoma susceptibility in a Chinese population. DNA Cell Biol (2011) 30(12):1027–32. doi:10.1089/dna.2011.1238
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
85. Lau DT, Norris MD, Marshall GM, Haber M, Ashton LJ. HLA-G polymorphisms, genetic susceptibility, and clinical outcome in childhood neuroblastoma. Tissue Antigens (2011) 78(6):421–7. doi:10.1111/j.1399-0039.2011.01781.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
86. Castelli EC, Mendes-Junior CT, Deghaide NH, de Albuquerque RS, Muniz YC, Simoes RT, et al. The genetic structure of 3’untranslated region of the HLA-G gene: polymorphisms and haplotypes. Genes Immun (2010) 11(2):134–41. doi:10.1038/gene.2009.74
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
87. Ghandri N, Gabbouj S, Farhat K, Bouaouina N, Abdelaziz H, Nouri A, et al. Association of HLA-G polymorphisms with nasopharyngeal carcinoma risk and clinical outcome. Hum Immunol (2011) 72(2):150–8. doi:10.1016/j.humimm.2010.10.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
88. Castelli EC, Mendes-Junior CT, Viana de Camargo JL, Donadi EA. HLA-G polymorphism and transitional cell carcinoma of the bladder in a Brazilian population. Tissue Antigens (2008) 72(2):149–57. doi:10.1111/j.1399-0039.2008.01091.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
89. Rolfsen GB, Castelli EC, Donadi EA, Duarte RA, Soares CP. HLA-G polymorphism and breast cancer. Int J Immunogenet (2014) 41(2):143–8. doi:10.1111/iji.12092
90. Chen Y, Gao XJ, Deng YC, Zhang HX. Relationship between HLA-G gene polymorphism and the susceptibility of esophageal cancer in Kazakh and Han nationality in Xinjiang. Biomarkers (2012) 17(1):9–15. doi:10.3109/1354750X.2011.633242
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
91. de Oliveira Crispim JC, Silva TG, Souto FJ, Souza FF, Bassi CL, Soares CP, et al. Upregulation of soluble and membrane-bound human leukocyte antigen G expression is primarily observed in the milder histopathological stages of chronic hepatitis C virus infection. Hum Immunol (2012) 73(3):258–62. doi:10.1016/j.humimm.2011.12.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
92. Amiot L, Vu N, Rauch M, L’Helgoualc’h A, Chalmel F, Gascan H, et al. Expression of HLA-G by mast cells is associated with hepatitis C virus-induced liver fibrosis. J Hepatol (2014) 60(2):245–52. doi:10.1016/j.jhep.2013.09.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
93. Weng PJ, Fu YM, Ding SX, Xu DP, Lin A, Yan WH. Elevation of plasma soluble human leukocyte antigen-G in patients with chronic hepatitis C virus infection. Hum Immunol (2011) 72(5):406–11. doi:10.1016/j.humimm.2011.02.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
94. Souto FJ, Crispim JC, Ferreira SC, da Silva AS, Bassi CL, Soares CP, et al. Liver HLA-G expression is associated with multiple clinical and histopathological forms of chronic hepatitis B virus infection. J Viral Hepat (2011) 18(2):102–5. doi:10.1111/j.1365-2893.2010.01286.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
95. Shi WW, Lin A, Xu DP, Bao WG, Zhang JG, Chen SY, et al. Plasma soluble human leukocyte antigen-G expression is a potential clinical biomarker in patients with hepatitis B virus infection. Hum Immunol (2011) 72(11):1068–73. doi:10.1016/j.humimm.2011.06.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
96. Han Q, Li N, Zhu Q, Li Z, Zhang G, Chen J, et al. Association of serum soluble human leukocyte antigen-G levels with chronic hepatitis B virus infection. Clin Exp Med (2014) 14(1):35–43. doi:10.1007/s10238-012-0214-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
97. Cordero EA, Veit TD, da Silva MA, Jacques SM, Silla LM, Chies JA. HLA-G polymorphism influences the susceptibility to HCV infection in sickle cell disease patients. Tissue Antigens (2009) 74(4):308–13. doi:10.1111/j.1399-0039.2009.01331.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
98. da Silva GK, Vianna P, Veit TD, Crovella S, Catamo E, Cordero EA, et al. Influence of HLA-G polymorphisms in human immunodeficiency virus infection and hepatitis C virus co-infection in Brazilian and Italian individuals. Infect Genet Evol (2014) 21:418–23. doi:10.1016/j.meegid.2013.12.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
99. Kim SK, Chung JH, Jeon JW, Park JJ, Cha JM, Joo KR, et al. Association between HLA-G 14-bp insertion/deletion polymorphism and hepatocellular carcinoma in Korean patients with chronic hepatitis B viral infection. Hepatogastroenterology (2013) 60(124):796–8. doi:10.5754/hge11180
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
100. Sartelet H, Schleiermacher D, Le-Hesran JY, Graesslin O, Gaillard D, Fe M, et al. Less HLA-G expression in Plasmodium falciparum-infected third trimester placentas is associated with more natural killer cells. Placenta (2005) 26(6):505–11. doi:10.1016/j.placenta.2004.08.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
101. Sadissou I, d’Almeida T, Cottrell G, Luty A, Krawice-Radanne I, Massougbodji A, et al. High plasma levels of HLA-G are associated with low birth weight and with an increased risk of malaria in infancy. Malar J (2014) 13(1):312. doi:10.1186/1475-2875-13-312
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
102. Garcia A, Milet J, Courtin D, Sabbagh A, Massaro JD, Castelli EC, et al. Association of HLA-G 3’UTR polymorphisms with response to malaria infection: a first insight. Infect Genet Evol (2013) 16:263–9. doi:10.1016/j.meegid.2013.02.021
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
103. Sabbagh A, Courtin D, Milet J, Massaro JD, Castelli EC, Migot-Nabias F, et al. Association of HLA-G 3’ untranslated region polymorphisms with antibody response against Plasmodium falciparum antigens: preliminary results. Tissue Antigens (2013) 82(1):53–8. doi:10.1111/tan.12140
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
104. Courtin D, Milet J, Sabbagh A, Massaro JD, Castelli EC, Jamonneau V, et al. HLA-G 3’ UTR-2 haplotype is associated with human African trypanosomiasis susceptibility. Infect Genet Evol (2013) 17:1–7. doi:10.1016/j.meegid.2013.03.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
105. Ayo CM, Dalalio MM, Visentainer JE, Reis PG, Sippert EA, Jarduli LR, et al. Genetic susceptibility to Chagas disease: an overview about the infection and about the association between disease and the immune response genes. Biomed Res Int (2013) 2013:284729. doi:10.1155/2013/284729
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
106. Marin-Neto JA, Rassi A Jr. Update on Chagas heart disease on the first centennial of its discovery. Rev Esp Cardiol (2009) 62(11):1211–6. doi:10.1016/S1885-5857(09)73346-8
Keywords: HLA-G, tumors, viral hepatitis, parasitic disorders, polymorphism
Citation: Dias FC, Castelli EC, Collares CVA, Moreau P and Donadi EA (2015) The role of HLA-G molecule and HLA-G gene polymorphisms in tumors, viral hepatitis, and parasitic diseases. Front. Immunol. 6:9. doi: 10.3389/fimmu.2015.00009
Received: 03 October 2014; Accepted: 07 January 2015;
Published online: 02 February 2015.
Edited by:
Silvia Gregori, San Raffaele Telethon Institute for Gene Therapy, ItalyReviewed by:
Giada Amodio, San Raffaele Telethon Institute for Gene Therapy, ItalyJoel LeMaoult, Commissariat à l’Energie Atomique et aux Energies Alternatives, France
Copyright: © 2015 Dias, Castelli, Collares, Moreau and Donadi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eduardo A. Donadi, Universidade de São Paulo, Avenida Bandeirantes 3900, Bairro Monte Alegre, Ribeirão Preto, São Paulo 14049-900, Brazil e-mail:ZWFkb25hZGlAZm1ycC51c3AuYnI=
 Fabrício C. Dias
Fabrício C. Dias Erick C. Castelli
Erick C. Castelli Cristhianna V. A. Collares
Cristhianna V. A. Collares Philippe Moreau
Philippe Moreau Eduardo A. Donadi
Eduardo A. Donadi