- 1Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Research Center of Avian Disease, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 3Key Laboratory of Animal Disease and Human Health of Sichuan Province, Sichuan Agricultural University, Chengdu, China
Duck Tembusu virus (TMUV), an emerging avian flavivirus, is highly pathogenic to birds and has the potential to become a zoonotic pathogen. Here, the molecular antiviral mechanism of goose type I, II, and III interferon (goIFNα, goIFNγ, and goIFNλ), the key components of the innate immune pathway, against TMUV was studied. We found that the transcription of goIFNs was obviously driven by TMUV infection in vivo and in vitro, and the titers and copies of TMUV were significantly reduced following treatment with goIFNs. The results of RNA sequencing (RNA-seq) revealed that goIFN stimulation triggered a set of differentially expressed genes at different levels and a positive regulatory feedback loop of IFN release against infection. Two important interferon-stimulated genes, goMx and goOASL, were identified as workhorse IFNs in the inhibition of TMUV replication. The antiviral effects of goMx and goOASL were confirmed by transient overexpression and knockdown assay in vitro. Overall, our findings defined that goose Mx and OASL play key roles in the antiviral effects of type I, II, and III interferon against the TMUV. These results extend our understanding of the transcriptional profile of the goose IFN-mediated signaling pathway and provide insight into the antiviral mechanism of goIFNs against flavivirus infection.
Introduction
In the classical innate immune pathway, incoming viral pathogens are sensed by cytologic host pattern recognition receptors (such as toll-like receptors, RIG-I-like receptors, NOD-like receptors), which activates downstream interferon regulatory factors (IRFs), leading to subsequent IFN production (1, 2). Interferons (IFNs) are cytokines with important antiviral activities and represent a powerful barrier to viral infection (3, 4); IFNs bind their cognate receptors and subsequently initiate a signaling cascade through the Janus kinase signal transducer and activator of transcription (JAK-STAT) pathway, triggering the expression of hundreds of IFN-stimulated genes (ISGs) (5, 6). These ISGs are mainly classified into three groups, positive regulators, negative regulators, and antiviral effectors, based on the numerous regulation mechanisms of innate immune signaling (7). As components of the IFN pathway, positive regulators (e.g., IRF1, cGAS) can promote IFN signaling and the subsequent development of a positive regulatory feedback loop. A small subset of negative regulators, such as SOCS1 and USP18, have the ability to target PRR, IRFs, or JAK/STAT to dampen the response and may even promote or enhance the replication of certain viruses (3, 8–11). Some ISGs were discovered as antiviral effectors that lead to a remarkable antiviral state, effectively intervening in distinct stages of the virus life cycle (12). It is well known that Mx1, OAS1, and PKR are potent antiviral effectors involved in the inhibition of viral infection (13–15).
Duck Tembusu virus (TMUV) is an emerging avian flavivirus, leading to a decrease in heavy egg laying, a sudden decline in feed uptake, and neurological signs in infected egg laying and breeder ducks (16, 17). According to incomplete statistics, multiple types of ducks (Pekin ducks, Cherry Valley ducks, Shaoxing ducks, Jinyun ducks, Longyan ducks, Jinding ducks, Khaki-Campbell ducks, Muscovy ducks, and Domesticated Mallards) have been subjected to TMUV infection (18). In addition to avian cell lines (DEF, GEF, and DF-1), TMUV can infect a range of mammalian cell lines (Vero, BHK21, HeLa, HepG2, and SH-SY5Y) and mosquito cell lines (C6/36 and Aedes albopictus) (19–21). These findings highlight the potential risk of TMUV infection in immunocompromised individuals and warrant studies on cross-species transmission and the pathogenesis of this novel flavivirus (21). Accumulating evidence suggests that IFN-regulated genes or ISGs (RIG-I, MDA5, TLR3, Mx, OAS, PKR, IFN-α, IFN-β, and IFN-γ, etc.) are induced to vastly different levels during TMUV infection in CEF, GEF, and 293 T cells (22–25). Recent studies have verified that TMUV infection triggers the IFN response mainly through MDA5 and TLR3-dependent signaling pathways (26). The expression of duMAVS was significantly upregulated after infection with TMUV, and the overexpression of duMAVS could drive the transcription of IFN-β, NF-κB, and IRF 7, as well as many downstream factors (such as Mx, PKR, OAS, and IL-8), which may partly explain the various viral replications that were significantly reduced by treatment with duMAVS (27). Moreover, RNA-seq analysis of the TMUV-infected goose spleen indicated that goose IFNs were key mediators of the host defense response to TMUV infection (unpublished data). These findings suggested that the IFN signaling pathway was essential in eliciting innate immune responses that restrict TMUV infection, and highly induced genes might be considered good candidates for controlling TMUV replication. However, the molecular antiviral mechanism of IFN against TMUV infection is still unknown.
With those factors in mind, here, we first investigated the expression of goose interferons (IFNα, IFNγ, and IFNλ) after TMUV infection in vivo and in vitro, as well as the antiviral activity of goIFNs against TMUV in vitro. Then, deep RNA sequencing (RNA-seq) (NCBI GEO accession number: GSE101404) was carried out to generate a high-resolution transcriptome map for IFN-treated PBMCs, and subsequently, some ISGs that might be involved in the IFN-dependent antiviral response were identified. Additionally, RNA interference (RNAi) assays were employed to confirm that goMx and goOASL play key roles in the antiviral effects of goIFNs against TMUV. Collectively, these studies provided evidence that the replication of TMUV was controlled through goIFNs, which initiated a positive regulatory feedback loop associated with antiviral and innate immune cellular signaling pathways.
Materials and Methods
Ethics Statement
The animal studies were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University (No. XF2014-18) and followed the National Institutes of Health guidelines for the performance of animal experiments.
Cells and Virus
Blood from geese was collected aseptically with heparin sodium (25 IU/mL). Immediately, PBMCs were isolated with the Goose Lymphocyte Separation Medium Kit (GLSMK) (TBD Sciences, Tianjing, China) according to the manufacturer’s protocol. Subsequently, cells (1 × 108 cells/mL) were maintained in Gibco® RPMI 1640 (Gibco Life Technologies, Shanghai, China) supplemented with 10% fetal bovine serum (FBS). Goose embryo fibroblasts (GEFs) and BHK-21 cells were grown supplemented with 10% FBS and maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco Life Technologies, Shanghai, China). All cells were cultured in 6-well plates at 37°C, 5% CO2. The duck TMUV CQW1 strain (GenBank Accession: KM233707) was isolated by our laboratory (28), and the measured virus titer was 6.3 × 106 TCID50/100 μL, which was reported previously (25).
Animal Experiments and Immunohistochemistry Assay (IHC)
Three-day-old Chinese geese (Anser cygnoides) were purchased from the breeding center of Sichuan Agricultural University (Yaan, Sichuan Province). The goslings were infected with TMUV (i. m. 500 µL) served as the experimental group and those treated with 500 µL of 0.9% NaCl served as the control group. From 1 to 4 dpi, the immune-associated tissues, such as the spleen (SP), liver (LI), brain (B), thymus (T), pancreas (P), and blood (BL), were collected. Subsequently, TMUV antigen, CD4, and CD8α molecule distribution in SP, LI, and B were analyzed by IHC, which was performed according to our previous study (25).
Characterization of GoIFN Expression during TMUV Infections
In an in vitro study, GEFs were infected with 100 µL TMUV (contained 1,000 TCID50), and the control groups were treated with the same dose of PBS. At 12, 24, 36, and 48 h post-infection (hpi), the cells were harvested with 1 mL RNAiso Plus Reagent for RNA extraction, and goIFN (IFNα, IFNγ, and IFNλ) mRNA was detected by quantitative real-time PCR (RT-qPCR). Additionally, in the in vivo study, immune-associated tissues, such as SP, LI, B, T, P, and BL, from TMUV-infected goslings (1–4 dpi), were collected with 1 mL RNAiso Plus reagent for RNA extraction and goIFN mRNA was detected by RT-qPCR.
Reporter Gene Assay
The pGL3-IFNβ-Luc expression plasmid was constructed with the sequence of the duck IFNβ promotor region (GeneBank accession number: KM032183.1). The commercialized pGL4-IRSE-Luc expression plasmid was purchased from Promega (Madison, WI, USA). Originally, GEFs were seeded onto a 48-well plate and transiently transfected with the pGL3-IFNβ-Luc (400 ng/well) or pGL4-IRSE-Luc (400 ng/well). Subsequently, cells were transfected with pRL-TK plasmid (40 ng/well) (Promega, Madison, WI, USA), which acted as an internal control to normalize transfection efficiency. 24 h later, cells were challenged with 100 µL TMUV (contained 1000 TCID50). At 12, 24, 36, and 48 hpi, the cells were harvested for luciferase assays. The luciferase activities were determined with a Dual-GloLuciferase Assay System (Promega) and normalized based on the Renilla luciferase activity.
Antiviral Assay
The recombinant plasmids pcDNA3.1 (+)-goIFN-α, γ, and λ were transfected into BHK-21 cells. Cell lysates from BHK-21 cells were harvested at 24 h post-transfection and clarified by centrifugation at 500 × g for 10 min after freezing and thawing three times. Then, GEFs were incubated with 100 µL goIFN-α, γ, and λ. After 12 h of incubation, cells were infected with 400 µL TMUV (contained 1000 TCID50). At 36, 48, and 60 hpi, cells were collected for the detection of viral copies and viral titers. Samples (200 µL) were extracted with a nucleic acid extraction kit (Tiangen, Shanghai), and then TMUV copies were detected by RT-qPCR using the special primers based on the TMUV-E gene (shown in Table 1). Subsequently, TMUV titers were determined by an endpoint dilution assay in GEFs and the results were analyzed using the Reed–Muench method (TCID50).
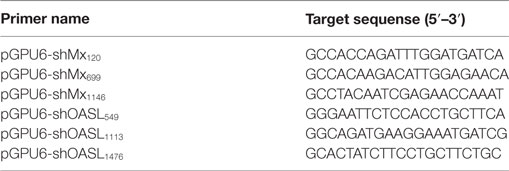
RNA Interference
Small interfering RNAs (siRNAs) targeting goMx and goOASL were chemically synthesized by GenePharma (China). The target sequences for the knockdown of goMx and goOASL are presented in Table 2. The target sequences were inserted into a pGPU6 plasmid to generate pGPU6-shgoMx (shMx120, shMx699, and shM1146) and pGPU6-shgoOASL (shOASL549, shOASL1113, and shOASL1476). A non-targeting shRNA (NC-shRNA) was synthesized as a negative control. The efficiencies of the siRNA were measured by RT-qPCR and Western blot.
Sample Preparation and cDNA Library Construction
PBMCs were stimulated with recombinant goIFN-α, γ, and λ for 3 h, and those treated with cell lysates from pcDNA3.1 (+)-transfected BHK-21 cells were considered as control group. Cells were harvested with 1 mL RNAiso Plus reagent (Takara Bio, Otsu, Japan) for RNA extraction.
Obtained RNA quantity and quality were evaluated using NanoDrop, Qubit, and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The RNA integrity number and purity (28S/18S) of all samples conformed to the requirements of a standard procedure (21). The mRNA of all qualified samples was enriched using oligo (dT) magnetic beads, and fragmented mRNAs were reverse transcribed into single strand cDNA using random hexamer primers. The cDNAs were converted into double strand cDNAs with DNA polymerase I and purified using AMPure XP beads. After qualification and quantification using an Agilent 2100 Bioanalyzer and ABI Step One Plus Real Time PCR System, the libraries were sequenced with Illumina HiSeq™ 2000. To obtain high-quality clean reads for assembly, the raw reads were filtered through the NGS QC TOOLKIT by removing low quality reads. All the clean reads were pooled and assembled using the Trinity de novo assembly program (29). RNA-seq data can be accessed under GSE101404.
Analysis of Differentially Expressed Genes (DEGs)
Comparisons between IFN treatment groups and the control group were performed, and DEGs were analyzed using the DESeq R package (30), a model based on the negative binomial distribution. For the statistical analysis, all read counts were normalized by calculating the FPKM value (31), and further, the FPKM + 1 values were log2 transformed and the means of expression (in log2 FPKMs) were used for further analysis. An P-value <0.05, which was adjusted by the false discovery rate, was defined as the threshold of DEGs, and log2-fold change > 0 (or <0) was defined as upregulated (downregulated genes). A Venn diagram was constructed to show the numbers of DEGs that were either specific or commonly induced among the three IFN treatment groups. Volcano and heat map hierarchical clustering analysis were performed to obtain the distribution and expression pattern of DEFs, respectively. Additionally, DEGs were assigned functional annotations using KEGG pathway enrichment to find immune-related genes (IRGs) for further analysis.
The Validation of RNA-Seq Data by RT-qPCR
According to the results of DEG analysis, we selected 17 IFN-responsive genes from the RNA-seq data for validation by RT-qPCR. In brief, total RNA was extracted using 1 mL RNAiso Plus reagent, and RT-PCR was performed on each sample using a 5X All-In-One RT MasterMix Reagent Kit in accordance with the manufacturer’s instructions (Applied Biological Materials, Richmond, BC, Canada). Finally, the cDNA was stored at −80°C until use. Then, the expression of candidate genes was detected by RT-qPCR performed using the Bio-Rad CFX96 Real-Time Detection System (Bio-Rad, USA), and threshold cycle (Ct) values were normalized to the housekeeping genes duβ-actin or goGAPDH. The relative expression levels of each target gene were calculated with the comparative Ct (2−ΔΔCt) method. Finally, primers were designed using the cDNA in the sequence database (Table 1).
Data Statistics
The statistical analyses were performed with GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA, USA). The differences between values were evaluated by Student’s t-test. P < 0.05 was considered statistically significant, and all values were expressed as the mean ± SEM.
Results
Effects of TMUV in GoIFN Expression In Vivo and In Vitro
To determine the production of goIFNs in response to TMUV challenge, we infected the GEFs and goslings with duck TMUV. In the in vivo study, 3-day-old goslings were infected with TMUV (i. m 500 µL). As shown in Figure 1A, the TMUV antigen was markedly distributed in the SP, LI, and B, which was highly connected with the distribution of the CD8α molecule. We also found that goIFNs were differentially upregulated in all selected tissues during TMUV infection (1–4 dpi), with especially high expression in immune-related tissues, such as LI, SP, and T (Figure 1B, a–d). Notably, significant upregulation of goIFNα was shown in LI and T at all time points, and goIFNγ was always markedly upregulated in T, while the expression level of goIFNλ was almost increased in LI, SP, and T by TMUV. Meanwhile, in the in vitro study, continuous upregulation of goIFNs was detected in GEFs with increasing TMUV infection time (12, 24, 36, and 48 hpi) (Figures 2A–D). Moreover, this upregulation was also detected by reporter assays, and TMUV infection triggered the activation of the IFNβ promoters and IRSE in GEFs at 12, 24, 36, and 48 hpi (Figures 2E,F). Taken together, these results provide evidence that TMUV infection strongly induces the transcription of goIFNs both in vivo and in vitro, which means that goIFNs play an important role in TMUV defense.
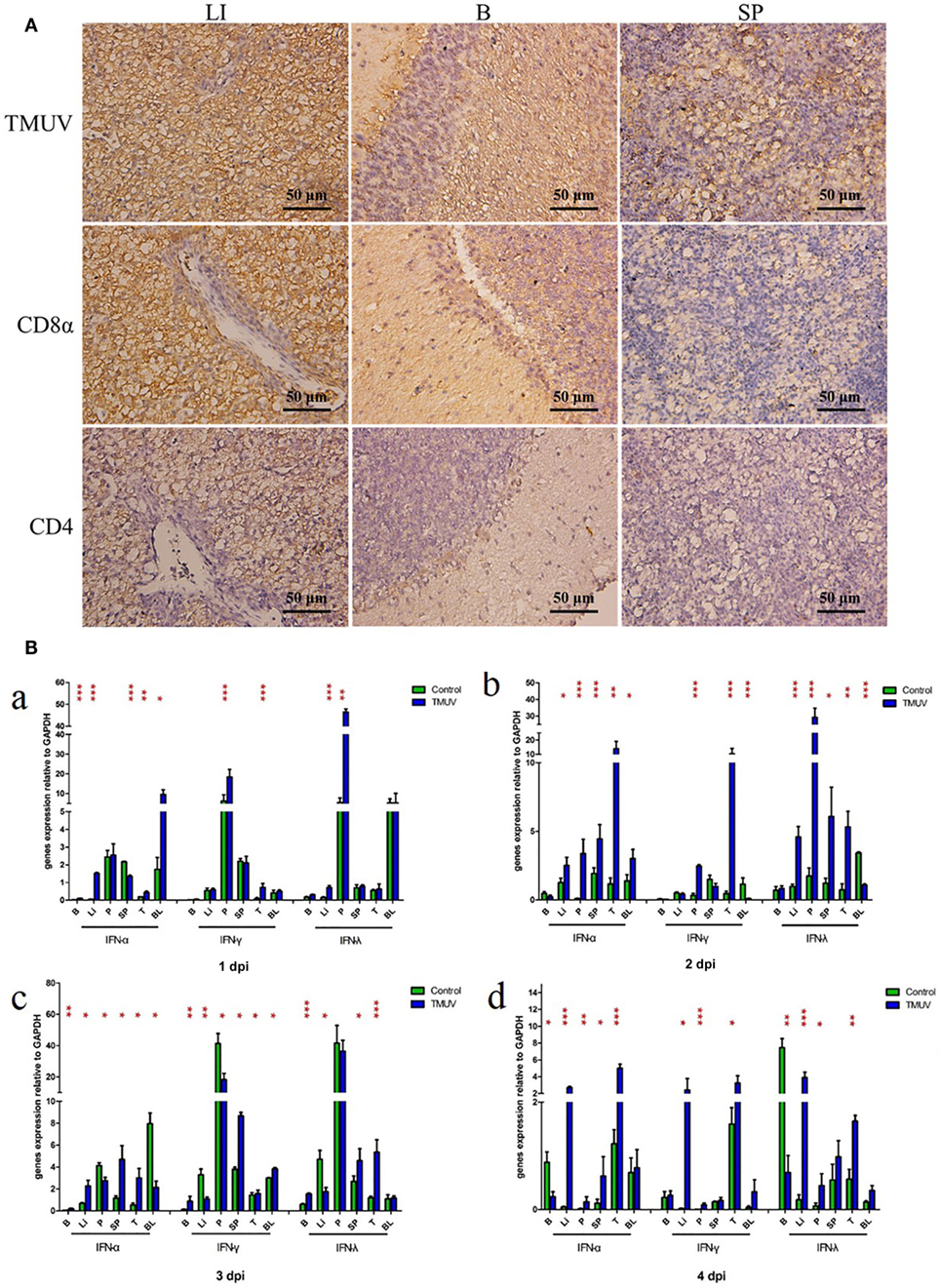
Figure 1. The distribution of viral antigen, CD8α and CD4 as well as the transcription levels of goIFNs in TMUV-infected goslings. (A) The distribution of TMUV antigen and CD8α and CD4 molecules in the liver (LI), brain (B), and spleen (SP) were measured by IHC assay. The dark brown represents positive signaling for TMUV, CD4, and CD8α antigen using immunohistochemical staining, and sections were counterstained with hematoxylin (blue). Rabbit polyclonal antibody against TUMV E protein, mouse anti-duck monoclonal CD4 antibodies (1:100) and mouse anti-goose polyclonal CD8α antibodies (1:100) were used as the primary antibody, while the goat anti-mouse and goat anti-rabbit antibodies were used as the secondary antibody. (B) The mRNA levels of goIFNs in the immune-associated tissues of 1–4 post-infected gosling, such as the spleen (SP), liver (LI), brain (B), thymus (T), pancreas (P), and blood (BL), were detected by quantitative real-time PCR. All results were normalized to GAPDH. Data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.

Figure 2. The transcription levels of goIFNs in TMUV-infected goose embryo fibroblasts (GEFs). (A–C) GEFs were treated with 100 µL TMUV (1,000 TCID50), cells were harvested at 12, 24, 36, and 48 hours post-infection. Then, the expression level of goIFNs mRNA was detected by quantitative real-time PCR. All results were normalized to GAPDH. Data are represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001. (D) Virus copy detection of TMUV-infected GEFs at 12, 24, 36, and 48 h. (E,F) Luciferase activities (IFNβ-luc and IRSE-luc) were determined with a Dual-Glo®Luciferase Assay System (Promega) and normalized based on the Renilla luciferase activities. All luciferase reporter assays were repeated three times.
Antiviral Activity of GoIFNs against TMUV Infection
To explore the effect of goIFNs against TMUV replication, an antiviral activity assay was performed. As Figure S1 in Supplementary Material shows, goIFNs did not show any cytotoxic effect on GEFs. Both viral copies and titers of TMUV were significantly decreased compared with the control group (Figure 3). Therefore, it is believed that pre-treatment with goIFNs prior to infection inhibits the replication of TMUV in GEFs.
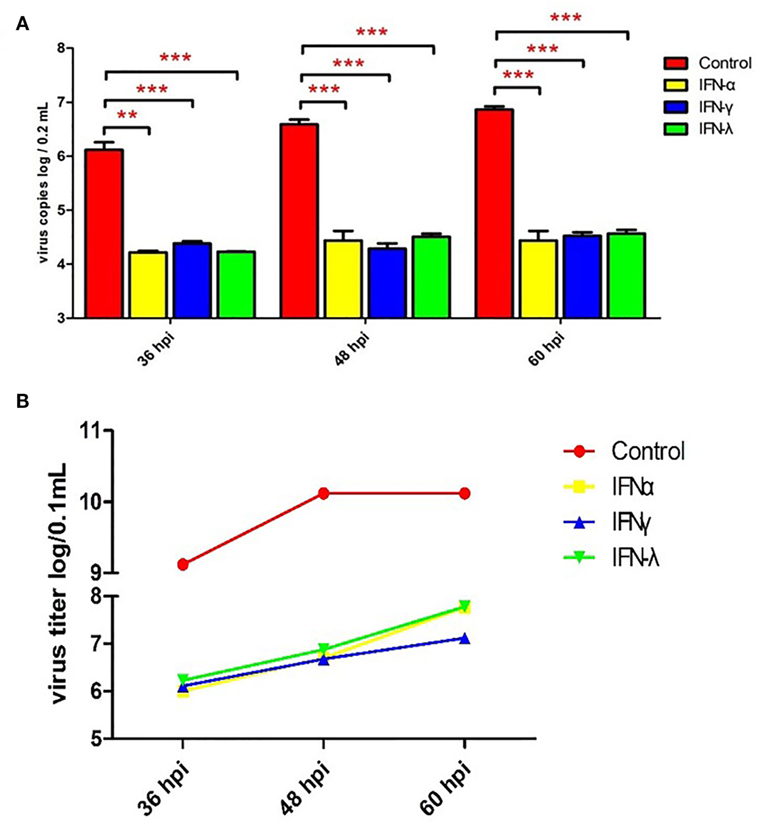
Figure 3. Pre-treating goose embryo fibroblasts (GEFs) with goIFNs reduced the replication of TMUV. GEFs were pre-treated with 100 µL goIFNα, goIFNγ, and goIFNλ (from goIFNs plasmids transfected BHK-21 cells for 12 h). The control group was treated with 100 µL of the cell lysates from pcDNA3.1 (+)-transfected BHK-21 cells. Subsequently, the cells were infected with 400 µL TMUV (contained 1000 TCID50). Cells were harvested at 36, 48, and 60 hpi for the detection of virus copies (A) and titers (B). Four experimental replicates were performed, and all of the results showed the mean ± SEM. Significant differences between the mock groups are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
Effects of GoIFNs on ISG Expression in Goose PBMCs
As already reported by others, IFNs can induce the expression of a large number of ISGs, leading to the induction of the antiviral state to prevent pathogen invasion (32–35). In our previous study, both goIFNα and goIFNγ could trigger the expression of downstream ISGs in GEFs and DEFs, which inhibited DNA virus (duck plague virus) replication in DEFs (36). Here, we assumed that the pre-treatment of GEFs with goIFNs induced the expression of ISGs that protect cells from TMUV infection. Therefore, it is necessary to expand our understanding of the global gene expression profile of goose PBMCs after stimulation with goIFNs. Initially, goIFNα, goIFNγ, and goIFNλ concentrations were assessed based on the results of Western blotting using ImageJ software with GAPDH as a loading control protein. Our finding showed that the protein level of 50 µL of IFNα equal to 70 µL of goIFNγ and 100 µL of goIFNλ (Figure 4A). Therefore, PBMCs were stimulated with equal levels of goIFN-α, γ, and λ. Under the same experimental conditions, Poly (I:C) was chosen as a positive control, and cell lysates from pcDNA3.1 (+)-transfected BHK-21 cells were considered as the experimental control group. At 3, 6, 12, and 24 hpi, cells were harvested for the detection of IFN (IFNα, IFNγ, IFNγ) and ISG (Mx, OASL, SOCS-1, and USP18) mRNAs by RT-qPCR. The highest transcription level of ISGs was shown in the first time point (3 h) following the treatment of IFNs in PBMC (Figure 4B). Therefore, 3-h samples of goIFNs stimulated cells were further subjected to RNA-seq by Illumina HiSeq™ 2000 (Novegene, Beijing).
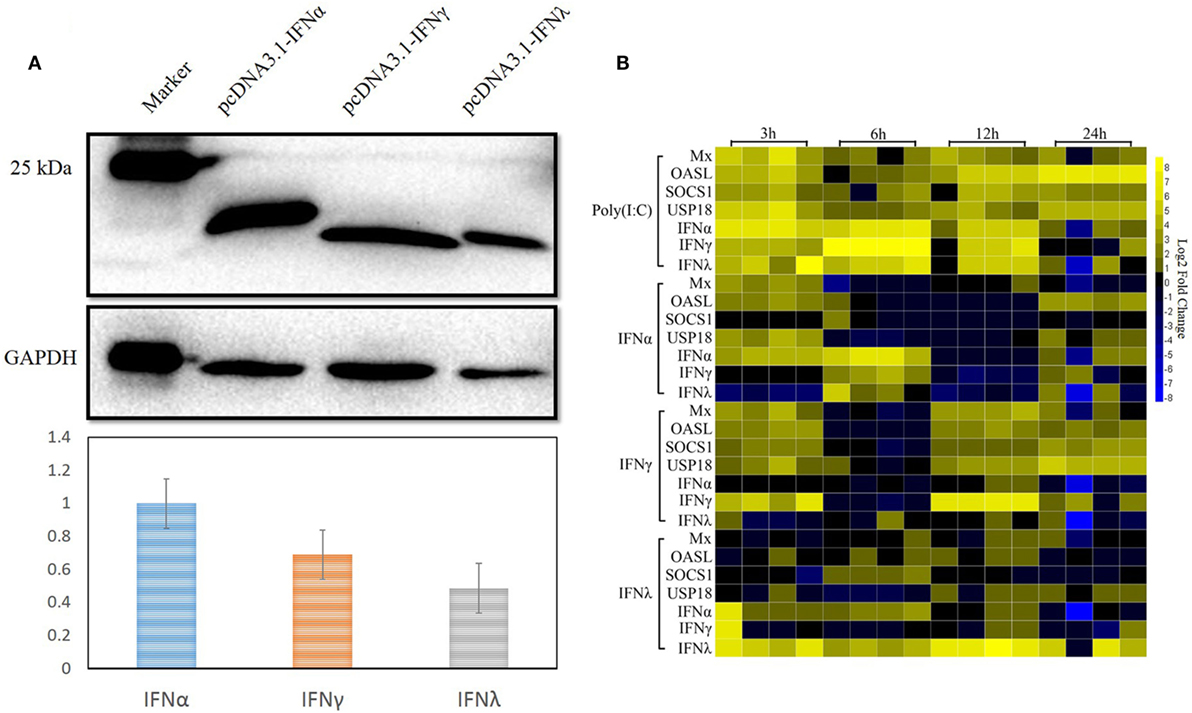
Figure 4. Interferon-stimulated genes were induced by goIFNs in PBMCs. (A) Western blot analysis of goIFNα, goIFNγ, and goIFNλ expressed in the transfected BHK-21 cells. BHK-21 cells were transfected with pcDNA3.1 (+) vector or pcDNA3.1 (+)-goIFNs (α, γ, and λ) recombinant plasmid for 24 h. Cell lysates were measured by Western blot. A monoclonal antibody against his-tag (1:10,000 dilution) and an HRP-labeled goat anti-mouse IgG antibody (1:2,000 dilution) were used as the primary antibody and secondary antibody, respectively. The gray intensity was analyzed by Image J software. (B) Heat map analysis of goIFN-regulated genes in PBMCs. PBMCs were treated with goIFNα, goIFNγ, and goIFNλ, while the poly (I:C)-treated group was considered a positive control. Cells were collected for detection by quantitative real-time PCR. Each column represented a time point (3, 6, 12, and 24 h), and each row represented a unigene (goIFNα, goIFNγ, goIFNλ, goMx, goOASL, goSOCS-1, and goUSP18). The differences in the expression level were shown in distinct colors. Yellow indicated a high expression level, while blue indicated a low expression level.
Summary of Transcriptome Data
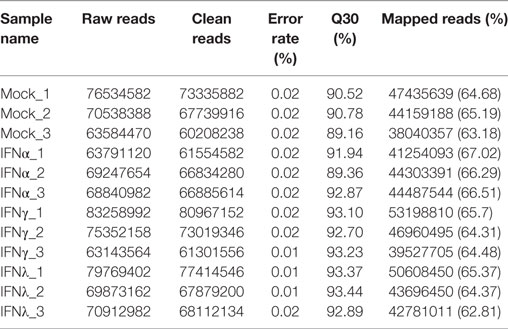
Mock and IFN-stimulated RNA samples were collected for deep sequencing using Illumina HiSeq 2000. The statistics of twelve libraries are shown in Table 3. Briefly, more than 60 million raw reads were generated for each library, with an error rate of 0.01–0.02%. After trimming, more than 95% of the raw reads were clean reads for each library. Additionally, at least 60% of the paired-end reads mapped to the goose (Anser) genome, with a phred quality score (Q30) above 90%. These data suggested that the accuracy and quality of the sequencing data were sufficient for further annotation and expression analysis.
Global Profiling of Gene Expression in Response to GoIFNs in PBMCs
To uncover the innate immune response to goIFNs, we first set out to identify the response of DEGs to goIFNs by pairwise comparisons to mock-treated controls. Overall, based on the criteria that |fold-change| ≥ 2 and P-value ≤ 0.01, we observed 1250 DEGs (4.5% of total genes) and 1296 DEGs (4.6% of total genes) in goIFNα vs. controls and goIFNγ vs. controls, respectively (Figure 5A). In goIFNα vs. control data, 711 genes were upregulated and 539 were downregulated, while in goIFNγ vs. control data, 793 genes were upregulated and 503 were downregulated. Notably, lower DEGs (1% of total genes) were identified in goIFNλ vs. control; among these, 53 were upregulated and 217 were downregulated (Figure 5A). Moreover, a total of 79 co-expressed genes were commonly regulated by all goIFNs (Figure 5B). Of these 79 common genes, 12 were upregulated and 67 were downregulated by goIFNα, 13 were upregulated and 66 were downregulated by goIFNγ, while 2 were upregulated and 77 were downregulated by goIFNλ (Figure 5C). These observations indicated that the goIFNα treatment group displayed similar gene expression to the goIFNγ-treated group, while treatment with equal levels of goIFNλ resulted in mild changes in subsequent gene expression.
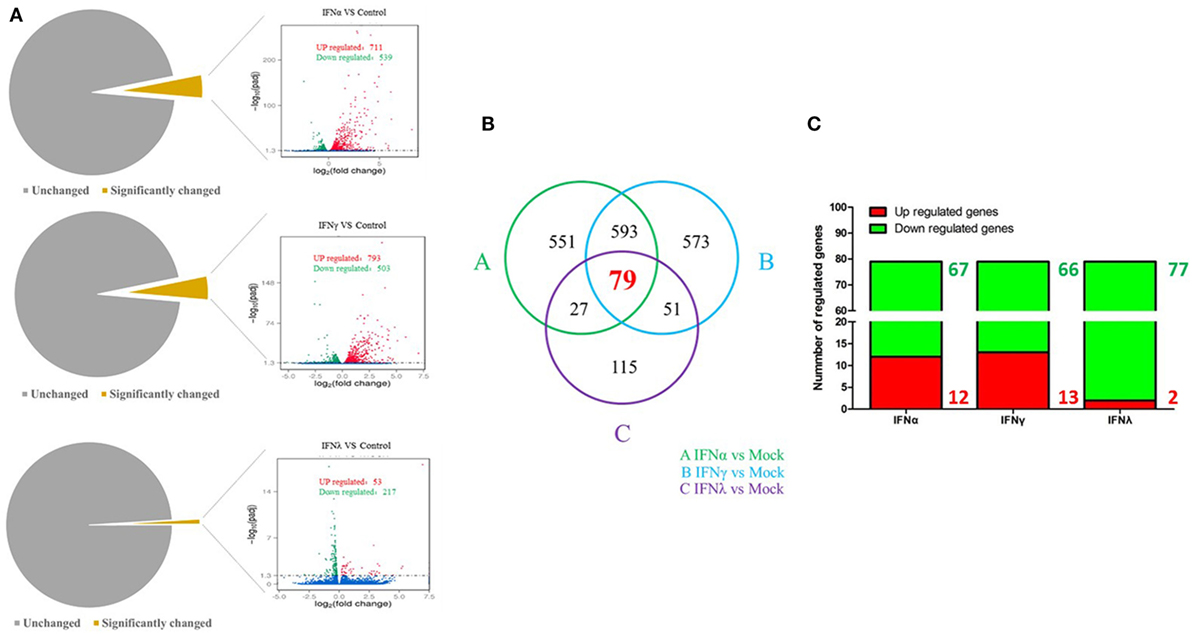
Figure 5. The analysis of differentially expressed genes (DEGs) based on transcriptome analysis. (A) Volcano plot analysis of DEGs. Each plot represents a unigene. Red indicates upregulated genes, while green indicates downregulated genes. (B) Venn diagram analysis of DEGs. Red highlights indicate the numbers of genes commonly regulated by goIFNα, goIFNγ, and goIFNλ. (C) The proportion of up- and downregulated genes commonly regulated by goIFNs. Likewise, red indicates upregulated genes, while green indicates downregulated genes.
Identification and Confirmation of IRGs
To further understand the antiviral mechanism of goIFNs, the IRGs among the identified DEGs were listed. A total of 101 IRGs were defined as ISGs (Figure 6; Table S1 in Supplementary Material), including IFNs, positive regulators (RIG-I, cGAS, PKR), negative regulators (SOCS-1, USP18) and antiviral effectors (Mx, OASL, IFITM5, TRIM25). The top 10 IRGs are presented in Tables S2–4 in Supplementary Material. KEGG analysis showed that the IRGs are mainly involved in the toll-like receptor signaling pathway, JAK-STAT signaling pathway, influenza virus infection pathway and herpes simplex infection pathway. Additionally, we obtained specific regulated genes through the pairwise comparison of goIFNα, goIFNγ, and goIFNλ (Figures S2A–C in Supplementary Material). These findings provided evidence that each type of goIFN induces some unique ISGs, and all goIFNs were required to confer the expression of IRGs for innate immune protection and the restriction of viral infection (Figure 7).
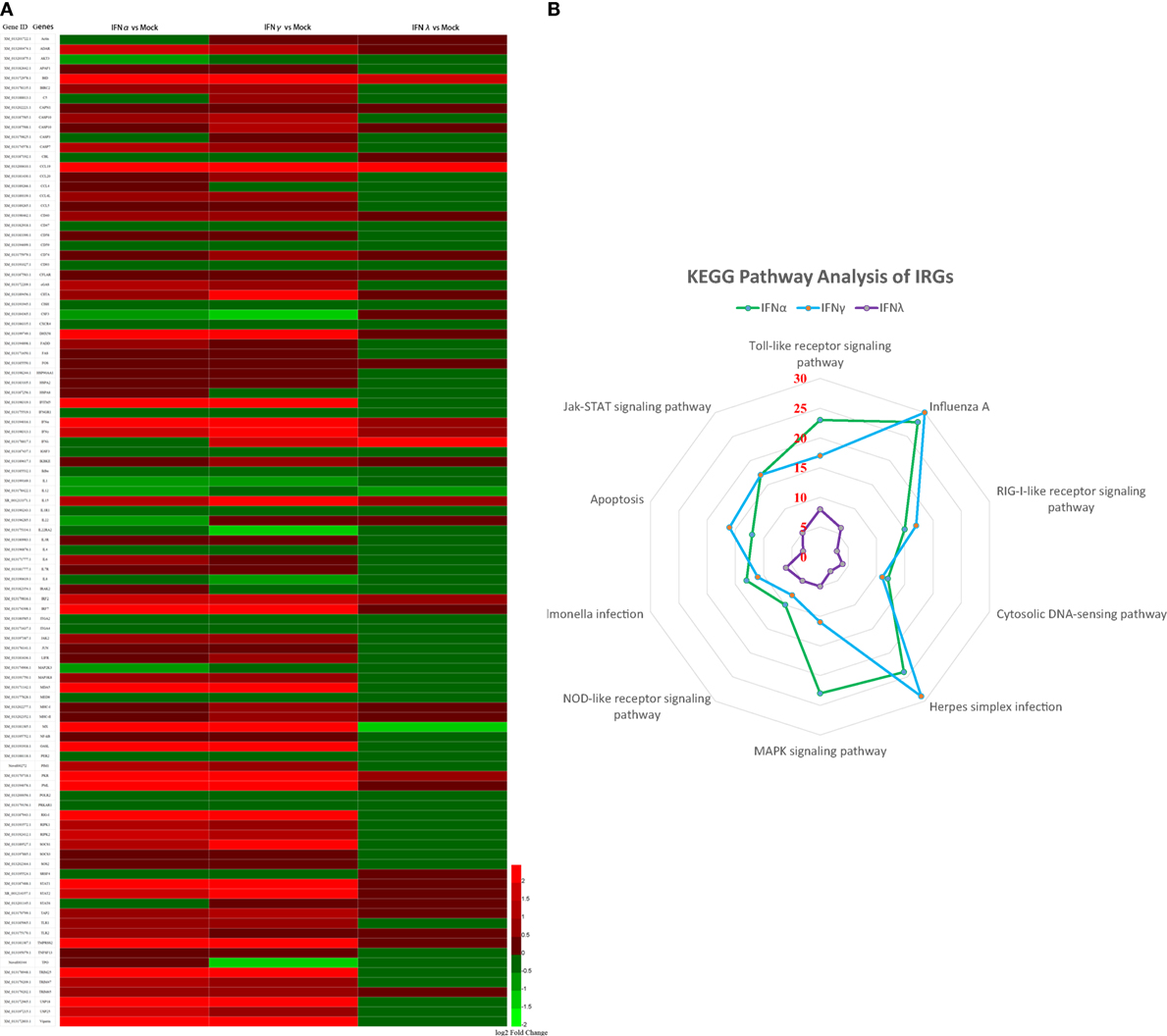
Figure 6. Statistical analysis of immune-related genes (IRGs) based on transcriptome analysis. (A) Expression pattern of IRGs. Each column represents a sample, and each row represents a unigene. Red indicates upregulated genes, while green indicates downregulated genes. (B) KEGG pathway analysis of IRGs. IRGs of goIFNs are mainly involved in the toll-like receptor signaling pathway, Jak-STAT signaling pathway, Influenza A infection pathway and herpes simplex infection pathway.
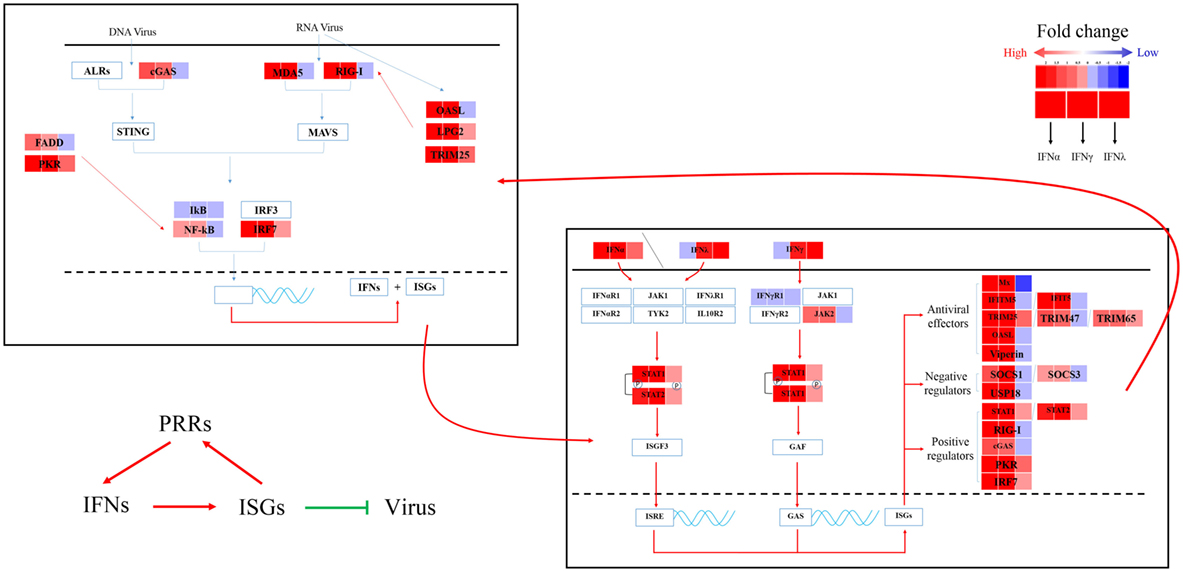
Figure 7. Sketch map of the goose antiviral response against avian TMUV infection in an IFN-dependent way. Red indicates upregulated genes, while blue indicates downregulated genes.
To validate the RNA-Seq results, three types of goIFNs, five antiviral effectors (Mx, OASL, Viperin, TRUM25, and IFITM5), six positive regulators (RIG-I, MDA5, IRF7, NF-kB, STAT1, and JAK2) and three negative regulators (SOCS-1, SOCS-3, and USP18) were chosen for RT-qPCR analysis (Figure 8). Goose PBMCs were treated with goIFNs for 3 h. Meanwhile, PBMCs were treated with pCDNA3.1(+) and Poly(I:C) (30 ng/mL) for 3 h as negative and positive control groups. According to the RT-qPCR analysis results, the expression patterns of most of the genes induced by goIFNα and goIFNγ were highly consistent with those found in the RNA-Seq analysis (Figure 8). However, the RT-qPCR results suggested that a higher expression level of goIFNs induced more genes than RNA-seq (Figure 8), indicating the RNA-seq method is reliable in DEG identification. Moreover, all antiviral effectors (goMx, goOASL, goViperin, goTRIM25, and goIFITM5) were significantly upregulated by goIFNs. As the classical antiviral genes, goMx and goOASL were selected to confirm the roles in the goIFN-induced antiviral immune response.
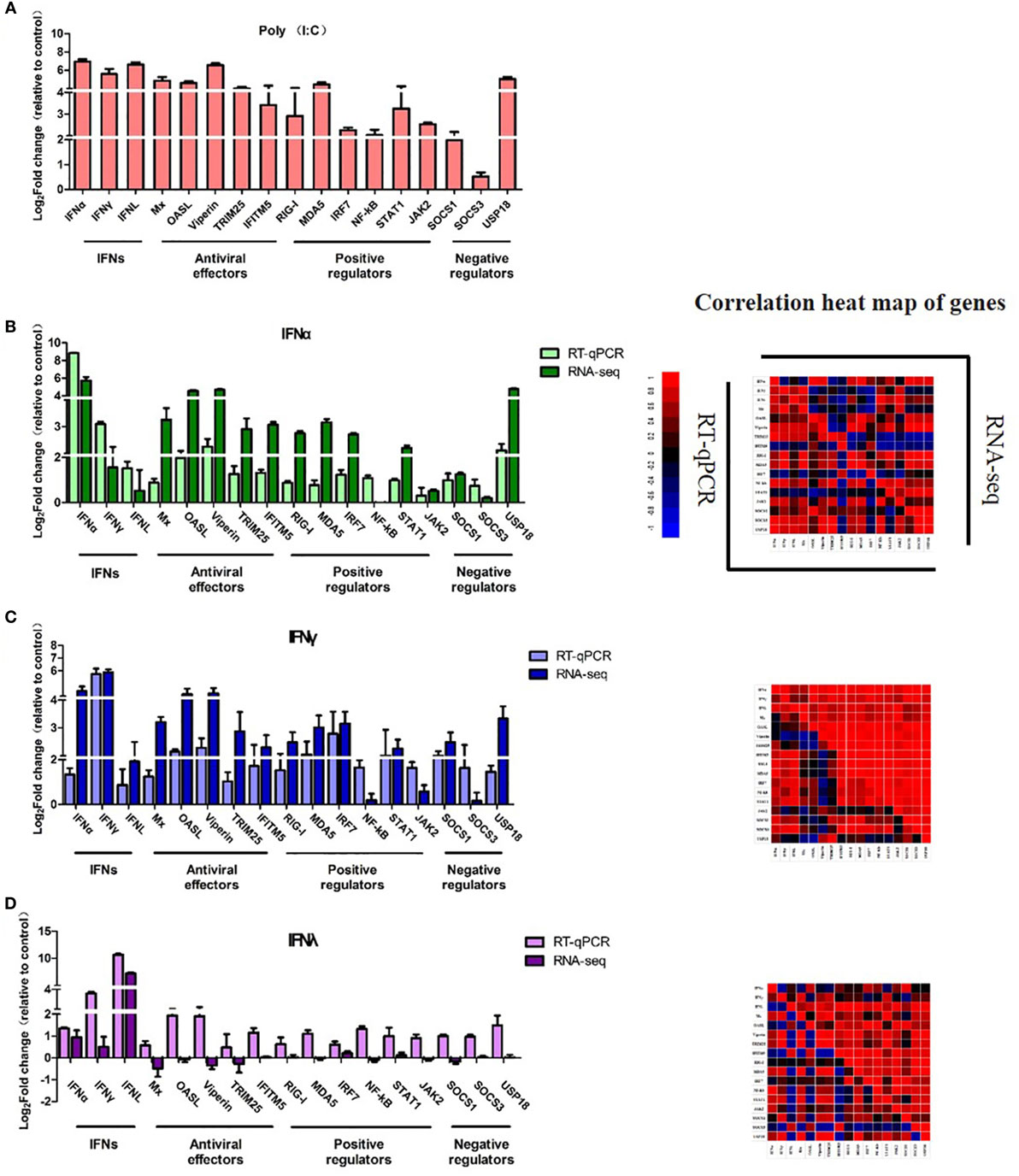
Figure 8. Quantitative Real-time PCR (RT-qPCR) validation of 17 candidate genes. Goose PBMCs were treated with (A) Poly(I:C) (30 ng/mL) and (B) goIFNα, (C) goIFNγ, (D) goIFNλ for 3 h. The mRNA expression level of candidate genes was detected by RT-qPCR. All results were normalized to GAPDH. Correlation analysis of candidate genes from the results of RT-qPCR and RNA-seq was performed by SPSS software. All data were represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
GoMx and GoOASL Play Key Roles in the Antiviral Effects of GoIFNs
To confirm the roles of goMx and goOASL in the goIFN-induced antiviral immune response, overexpression and knockdown by RNAi assay were performed. Initially, we found that cells transfected with shMx1146 and shOASL549 for 24 h were most efficient in knocking down goMx and goOASL expression, respectively (Figure 9). Then, cells were co-transfected with the pcDNA3.1-vector or pcDNA3.1-goMx and pcDNA3.1-goOASL plasmid, together with shMx1146 or shOASL549 and NC-shRNA. After 24 h transfection, cells were infected with TMUV for 36 h, and the viral copies were measured. RT-qPCR analysis showed that the overexpression of goMx or goOASL obviously reduced the TMUV replication, while the antiviral effect was reduced by the silencing of goMx and goOASL expression at 36 hpi, respectively (Figures 10A,B). These results indicated that goMx and goOASL conferred substantial protection against TMUV infection. Therefore, to confirm whether goMx and goOASL were associated with the goIFN-mediated antiviral effect, we pre-treated GEFs with goIFNs and subsequently transfected them with shMx1146 and shOASL549. At 24 h post-transfection, cells were challenged with TMUV, and viral copies were detected by RT-qPCR at 36 hpi. The results demonstrated that knockdown of goMx and goOASL significantly impaired the antiviral response of goIFNs against TMUV infection in vitro (Figures 10C–E). Taken together, our findings suggested that the induction of goMx and goOASL by the goIFN-dependent signaling pathway conferred antiviral and immunomodulatory activities against TMUV infection.
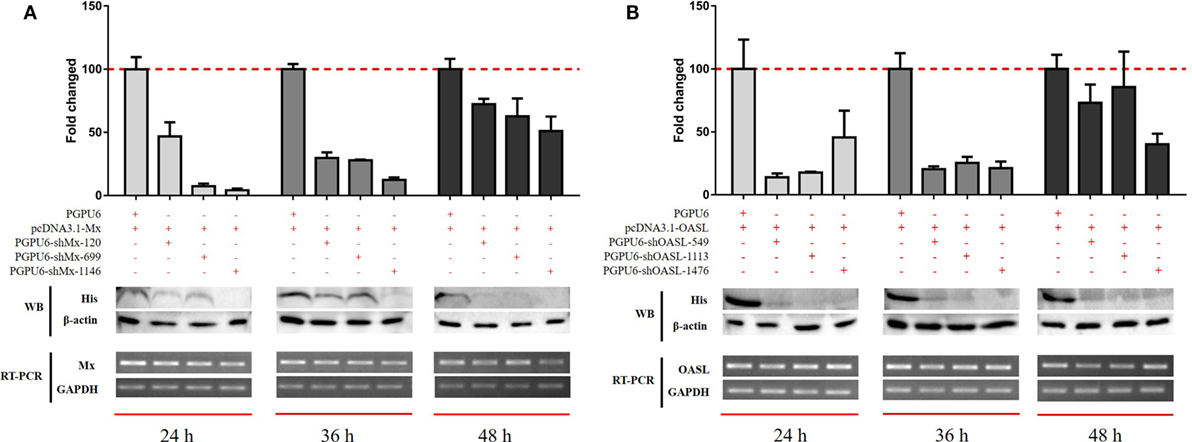
Figure 9. Optimum silencing efficiency of (A) sh-goMx and (B) sh-goOASL. Goose embryo fibroblasts (GEFs) were transfected with pcDNA3.1-goMx and pcDNA3.1-goOASL plasmids, while pcDNA3.1-vector transfected cells were considered as a control group. Meanwhile, cells were cotransfected with sh-goMx (120, 699, 1146) or sh-goOASL (549, 1113, 1476) and cotransfected with non-targeting small RNA as the negative control shRNA. At 24, 36, and 48 h post-transfection, cells were harvested for quantitative real-time PCR and Western blot analysis. All results were normalized to GAPDH and represented as the mean ± SEM (n = 4).
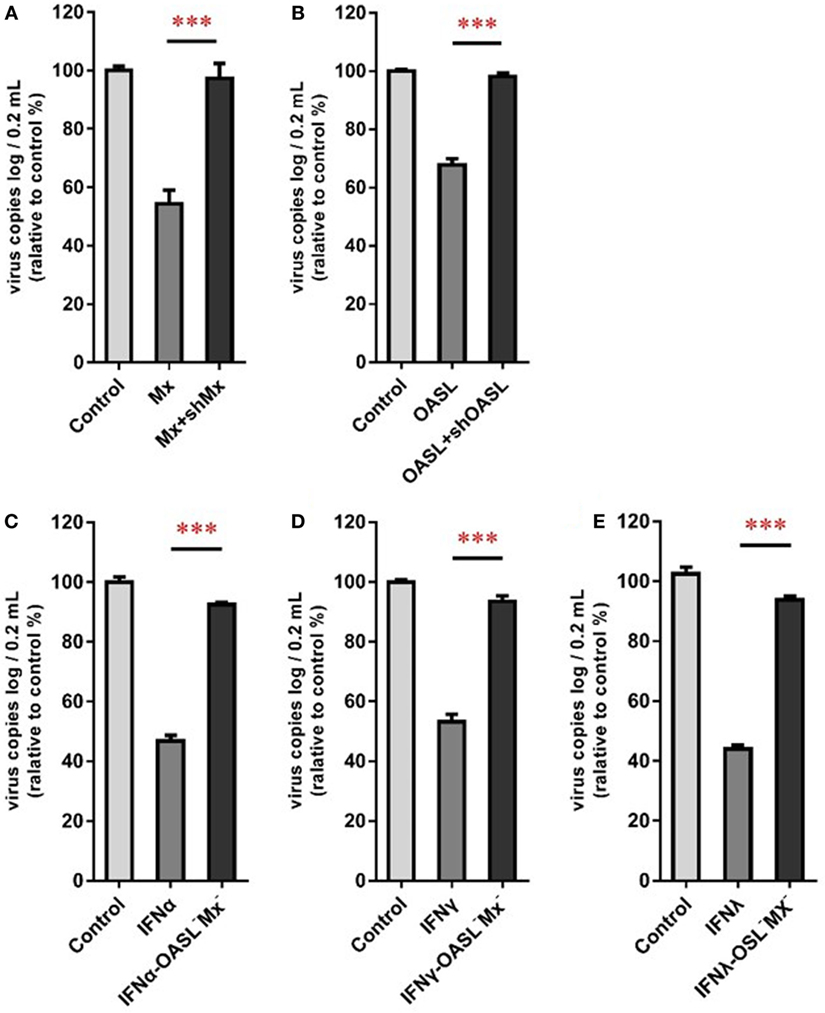
Figure 10. goMx and goOASL play a key role in goIFNs dependent antiviral effects. (A,B) GEFs were co-transfected with the pCDNA3.1-vector or goMx and goOASL plasmid, together with shMx1146, shOASL549, and NC-shRNA. After 24 h transfection, cells were infected with TMUV for 36 h, the virus copies were detected by quantitative real-time PCR (RT-qPCR); (C–E) GEFs were pretreated with goIFNs and subsequently transfected with shMx1146 and shOASL549. At 24 h post-transfection, cells were challenged with TMUV and virus copies were detected by RT-qPCR at 36 h. All data were represented as the mean ± SEM (n = 4). Significant differences from the mock groups are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001.
Discussion
As the first line of defense against invading viruses, interferon is the key component of the innate immune pathway. Generally, IFN-induced signaling cascades are fast and further elicit a positive regulatory feedback loop to enhance the production of IFN or ISGs and boost the antiviral state (35). Moreover, viral infection could effectively trigger the activation of the innate immune response against viral infection. Indeed, a rational hypothesis is that the most highly induced ISGs during infection are the ones that control viral replication (3). In contrast, viruses have the ability to utilize diverse strategies to circumvent the IFN response (37). In this study, the TMUV infection model was first chosen to explore the IFNs involved in the innate immune response in vivo and in vitro, demonstrating that the mRNA expression level of goIFNs was remarkably induced in immune-related tissue (LI, SP, and T) during TMUV infection (1–4 dpi), and the high expression patterns of the CD8α molecule consisted of the viral location. It is noteworthy that CD8+ T cells mainly produce cytokines, such as IFN-γ and TNF-α, which further activate macrophages and inhibit viral replication (38, 39). We speculated that the TMUV-induced IFN response plays a major role in shaping the adaptive immune response (40, 41). A similar IFN response to TMUV infection was observed in the in vitro study, these results suggest that TMUV induced a high expression level of goIFN and activated the IFNβ and IRSE in TMUV-infected GEFs, which was consistent with the previous study (23, 25).
As reported previously, the overexpression of duMAVS increased the expression of IFN and downstream factors and reduced the replication of TMUV (27). However, a study reported that treatment with chicken IFN-α is unable to prevent TMUV infection in DF-1 cells, but mammalian type I IFN can significantly inhibit the replication of TMUV in BHK-21 cells (21). Interestingly, our data indicated that goIFN pre-treatment significantly decreased viral copies and titers, which suggested that goIFNs can inhibit TMUV replication in GEF. This is the first report of avian IFNs that show an inhibition ability in TMUV replication in avian cells. Our previous study revealed that treating GEFs with goIFNs upregulated subsequent ISG expression, such as goIFNs, goMx, and goOASL (36), suggesting that those ISGs may be the workhorses in controlling TMUV infection in vitro. With those facts in mind, here, we further investigated the molecular antiviral mechanism of goose type I, II, and III interferon against TMUV.
PBMCs are important immune cells and effectively respond to IFN treatment. Therefore, PBMCs were chosen as cell models and were treated with goIFN at 3 h for detecting the transcriptional profile of immune factors using RNA-seq technology. Surprisingly, the numbers of upregulated and downregulated genes by goIFNλ stimulation were much lower than those regulated by goIFNα or goIFNγ stimulation (Figure 5A). Collectively, the stimulation of goIFNs initiated a series of signaling cascades at early stages, leading to the transcriptional regulation of hundreds of DEGs, and each IFN induced a unique and partially overlapping set of ISGs (Figures 5B,C). Most importantly, we identified approximately 101 IRGs, which were mainly involved in the toll-like receptor signaling pathway, JAK-STAT signaling pathway and antiviral pathway (Figure 6). Moreover, these results were verified by RT-qPCR, and the correlation analysis of RNA-Seq data and qRT-PCR results showed that some relevant relationships between candidate genes may be related to the balance of the host immune response, which remains to be further studied in the future. Previously, human bone marrow macrophages were treated with IFNα and IFNγ for 30 min and functionally validated the antiviral activity of 288 type I and type II ISGs against an RNA virus [vesicular stomatitis virus (VSV)] and a DNA virus (murine gammaherpes virus, MHV-68). Ultimately, 34 ISGs, including OAS1, IFITM3, TRIM25, MDA5, and RIG-I, were identified, which elicited an antiviral effect on the replication of either one or both viruses (42). Moreover, 36 ISGs were tested against West Nile virus (WNV) and dengue virus (DENV) infection, and only five ISGs (IFITM2, IFITM3, ISG20, Viprerin, and PKR) efficiently suppressed WNV and/or DENV (43). Based on those and our data, we assume that goIFNs bind to their cognate receptors in goPBMCs, which trigger the Jak/STAT pathway to stimulate the formation of an IFN-stimulated gene factor 3 (ISGF3) trimer or IFN-γ activation factor (GAF). Subsequently, ISGF3 or GAF translocates into the nucleus and binds to IFN-stimulated response elements or gamma-activated sequence (GAS) promoter elements, resulting in the induction of ISGs, including IFNs, positive regulators (RIG-I, cGAS, PKR), negative regulators (SOCS-1, USP18) and antiviral effectors (Mx, OASL, Viperin, IFITM5, TRIM25) (sketch in Figure 7). The positive regulators could further enhance the PRR signaling pathway, which results in the activation of STING/MAVS and leads to the phosphorylation of interferon response factors 3 or 7 (IRF3/7) or the phosphorylation and ubiquitin-mediated degradation of IκB. Phosphorylated dimers of IRF3/7 or NF-κB translocate into the nucleus, where they bind to and activate specific promoters, triggering the subsequent production of IFNs and ISGs as a positive self-regulatory feedback loop. Consequently, the positive self-regulatory feedback loop greatly enhances the innate immune response against viral infection. Here, we hypothesized that goIFN establishes the cellular antiviral state against TMUV through the induction of ISGs. Based on our data, two important ISGs, goMx, and goOASL, were highly upregulated by three types of goIFNs in vitro. With those in mind, goMx and goOASL were considered effective candidates for further antiviral research.
The Mx and OAS families are well-studied proteins in the control of viral infection. Compared with OAS, OASL has additional 2 UBL repeats, which are involved in the initiation of RIG-I-mediated antiviral signaling as a mediator (44). However, only OASL, not OAS, was identified in goose and chicken (45). Our previous research indicated that the overexpression of goOASL significantly reduced the replication of Newcastle disease virus in GEF, and the mRNA expression level of goOASL was significantly increased by TMUV infection in vivo (45). Additionally, the antiviral activity of OAS against viruses in the family flavivirus was reported, including hepatitis C virus (HCV) and WNV (46). Mx family members have various antiviral profiles against a wild range of viruses at unique steps in their life cycle (47). However, the number of antiviral studies of bird Mx is still limited and contradictory. In 1995, chicken Mx (chMx) was first identified with no antiviral activity, but chMx was subsequently reported as an antiviral effector against the influenza virus and VSV (48). Moreover, no antiviral effect of duck Mx has been reported (49). In our study, the over expression of goMx and goOASL obviously reduced the replication of TMUV, and subsequent RNAi assay revealed that the knockdown of endogenous goMx and goOASL decreased the antiviral activity of goIFN in controlling TMUV infection. Therefore, we noted that goMx and goOASL were indispensable in goIFN-mediated innate immune responses that restrict TMUV infection. As one of the first characterized antiviral effector pathways, the antiviral activity of OAS has been widely demonstrated in vitro and in vivo against various viruses (DENV, WNV, HCV, and Japanese Encephalitis virus) through RNase L-dependent and RIG-dependent (RNase L-independent) signaling pathways (44, 50, 51). Reported studies have provided evidence that the Mx GTPase system displayed diverse antiviral activity against many types of viruses at unique steps in their life cycle. Mouse Mx1 inhibited the replication of the influenza virus by blocking viral mRNA synthesis in the nucleus, while human MxA in the cytoplasm prevented the entry of influenza A virus nucleocapsids into the nucleus. However, the molecular mechanism of goMx and goOASL against TMUV infection was uncovered. A further understanding of these ISGs may provide future directions for antiviral therapies against TMUV.
In conclusion, here, goIFN expression was obviously driven by TMUV infection in vivo and in vitro. Additionally, the antiviral effects of goIFNα, goIFNγ, and goIFNλ against the emerging goose flavivirus TMUV in vitro was confirmed for the first time. Based on transcriptome analysis, the gene profile of goIFN-stimulated PBMCs were shown. Subsequently, two important interferon-stimulated genes (ISGs), goMx and goOASL, were identified as the workhorse IFNs in the inhibition of TMUV replication in vitro by transient overexpression and knockdown assay. Collectively, these data have prompted the suggestion that TMUV infection-induced goIFN enhanced the antiviral state through their downstream ISGs, which further form a positive regulatory feedback loop to boost the host antiviral effect.
Ethics Statement
The animal studies were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University (No. XF2014-18) and followed the National Institutes of Health guidelines for the performance of animal experiments.
Author Contributions
SC and WZ designed the experiment. WZ performed the experimental work. WZ and SC wrote the paper. ZW, JZ, MW, RJ, DZ, ML, KS, QY, YW, XC, and AC contributed to analysis the experimental data.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
This work was funded by grants from the National Key Research and Development Program of China (2016YFD0500800, 2017YFD0500800); Integration and Demonstration of Key Technologies for Duck Industrial in Sichuan Province (2014NZ0030); China Agricultural Research System (CARS-43-8).
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.01006/full#supplementary-material.
Figure S1. Cytotoxicity of goIFNα, goIFNγ and goIFNλ in goose embryo fibroblasts (GEFs). GEFs were treated with 10 µL of the cell lysates from pcDNA3.1 (+)-goIFNα, goIFNγ, and goIFNλ-transfected BHK-21 cells for 24 h. Similarly, the control group was treated with 10 µL of the cell lysates from pcDNA3.1 (+)-transfected BHK-21 cells. CCK-8 reagent (10 µL) was added into each well for 2 h at 37°C. After that, the plates were evaluated at the 450 nm wavelength with a multi-detection microplate reader. The results were expressed relative to control cells, which were defined as 100% viable.
Figure S2. Pairwise comparison of goIFNα, goIFNγ, and goIFNλ induced IRGs. (A) goIFNα vs. goIFNγ; (B) goIFNγ vs. goIFNλ; (C) goIFNα vs. goIFNλ. Axes represented fold change in response to goIFNα, goIFNγ, or goIFNλ over untreated cells. Red plots indicated goIFNα specific genes. Green plots indicated goIFNγ specific genes. Purple plots indicated goIFNλ specific genes.
References
1. Beachboard DC, Horner SM. Innate immune evasion strategies of DNA and RNA viruses. Curr Opin Microbiol (2016) 32:113–9. doi:10.1016/j.mib.2016.05.015
2. Radoshevich L, Dussurget O. Cytosolic innate immune sensing and signaling upon infection. Front Microbiol (2016) 7:313. doi:10.3389/fmicb.2016.00313
3. Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol (2011) 1:519–25. doi:10.1016/j.coviro.2011.10.008
4. Lin F, Young HA. Interferons: success in anti-viral immunotherapy. Cytokine Growth Factor Rev (2014) 25:369. doi:10.1016/j.cytogfr.2014.07.015
5. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell (2010) 140:805–20. doi:10.1016/j.cell.2010.01.022
6. Nallar SC, Kalvakolanu DV. Interferons, signal transduction pathways, and the central nervous system. J Interferon Cytokine Res (2014) 34:559–76. doi:10.1089/jir.2014.0021
7. Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol (2014) 32:513–45. doi:10.1146/annurev-immunol-032713-120231
8. Randall G, Chen L, Panis M, Fischer AK, Lindenbach BD, Sun J, et al. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology (2006) 131:1584–91. doi:10.1053/j.gastro.2006.08.043
9. Francois-Newton V, Magno de Freitas Almeida G, Payelle-Brogard B, Monneron D, Pichard-Garcia L, Piehler J, et al. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon alpha response. PLoS One (2011) 6:e22200. doi:10.1371/journal.pone.0022200
10. Delgado-Ortega M, Marc D, Dupont J, Trapp S, Berri M, Meurens F. SOCS proteins in infectious diseases of mammals. Vet Immunol Immunopathol (2013) 151:1–19. doi:10.1016/j.vetimm.2012.11.008
11. Kershaw NJ, Murphy JM, Lucet IS, Nicola NA, Babon JJ. Regulation of Janus kinases by SOCS proteins. Biochem Soc Trans (2013) 41:1042–7. doi:10.1042/BST20130077
12. Sen GC. Viruses and interferons. Annu Rev Microbiol (2001) 55:255–81. doi:10.1146/annurev.micro.55.1.255
13. Chakrabarti A, Jha BK, Silverman RH. New insights into the role of RNase L in innate immunity. J Interferon Cytokine Res (2011) 31:49–57. doi:10.1089/jir.2010.0120
14. Haller O, Kochs G. Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res (2011) 31:79–87. doi:10.1089/jir.2010.0076
15. Munir M, Berg M. The multiple faces of proteinkinase R in antiviral defense. Virulence (2013) 4:85–9. doi:10.4161/viru.23134
16. Su J, Li S, Hu X, Yu X, Wang Y, Liu P, et al. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS One (2011) 6:e18106. doi:10.1371/journal.pone.0018106
17. Liu P, Lu H, Li S, Wu Y, Gao GF, Su J. Duck egg drop syndrome virus: an emerging Tembusu-related flavivirus in China. Sci China Life Sci (2013) 56:701–10. doi:10.1007/s11427-013-4515-z
18. Tang Y, Diao Y, Yu C, Gao X, Ju X, Xue C, et al. Characterization of a Tembusu virus isolated from naturally infected house sparrows (Passer domesticus) in Northern China. Transbound Emerg Dis (2013) 60:152–8. doi:10.1111/j.1865-1682.2012.01328.x
19. Petz LN, Turell MJ, Padilla S, Long LS, Reinbold-Wasson DD, Smith DR, et al. Development of conventional and real-time reverse transcription polymerase chain reaction assays to detect Tembusu virus in Culex tarsalis mosquitoes. Am J Trop Med Hyg (2014) 91:666–71. doi:10.4269/ajtmh.13-0218
20. Tang Y, Diao Y, Chen H, Ou Q, Liu X, Gao X, et al. Isolation and genetic characterization of a Tembusu virus strain isolated from mosquitoes in Shandong, China. Transbound Emerg Dis (2015) 62:209–16. doi:10.1111/tbed.12111
21. Wang HJ, Li XF, Liu L, Xu YP, Ye Q, Deng YQ, et al. The emerging duck flavivirus is not pathogenic for primates and is highly sensitive to mammalian interferon antiviral signaling. J Virol (2016) 90:6538–48. doi:10.1128/JVI.00197-16
22. Wu L, Liu J, Chen P, Jiang Y, Ding L, Lin Y, et al. The sequential tissue distribution of duck Tembusu virus in adult ducks. Biomed Res Int (2014) 2014:703930. doi:10.1155/2014/703930
23. Li N, Wang Y, Li R, Liu J, Zhang J, Cai Y, et al. Immune responses of ducks infected with duck Tembusu virus. Front Microbiol (2015) 6:425. doi:10.3389/fmicb.2015.00425
24. Fu G, Chen C, Huang Y, Cheng L, Fu Q, Wan C, et al. Comparative analysis of transcriptional profiles of retinoic-acid-induced gene I-like receptors and interferons in seven tissues from ducks infected with avian Tembusu virus. Arch Virol (2016) 161:11–8. doi:10.1007/s00705-015-2621-x
25. Zhou H, Chen S, Wang M, Jia R, Zhu D, Liu M, et al. Antigen distribution of TMUV and GPV are coincident with the expression profiles of CD8alpha-positive cells and goose IFNgamma. Sci Rep (2016) 6:25545. doi:10.1038/srep25545
26. Chen S, Luo G, Yang Z, Lin S, Chen S, Wang S, et al. Avian Tembusu virus infection effectively triggers host innate immune response through MDA5 and TLR3-dependent signaling pathways. Vet Res (2016) 47:74. doi:10.1186/s13567-016-0358-5
27. Li N, Hong T, Li R, Wang Y, Guo M, Cao Z, et al. Cherry valley ducks mitochondrial antiviral-signaling protein-mediated signaling pathway and antiviral activity research. Front Immunol (2016) 7:377. doi:10.3389/fimmu.2016.00377
28. Zhu K, Huang J, Jia R, Zhang B, Wang M, Zhu D, et al. Identification and molecular characterization of a novel duck Tembusu virus isolate from Southwest China. Arch Virol (2015) 160:1–10. doi:10.1007/s00705-015-2513-0
29. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol (2011) 29:644–52. doi:10.1038/nbt.1883
30. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol (2010) 11:R106. doi:10.1186/gb-2010-11-10-r106
31. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol (2010) 28:511–5. doi:10.1038/nbt.1621
32. Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev (2004) 202:8. doi:10.1111/j.0105-2896.2004.00204.x
33. Al-Yahya S, Mahmoud L, Al-Zoghaibi F, Al-Tuhami A, Amer H, Almajhdi FN, et al. Human cytokinome analysis for interferon response. J Virol (2015) 89:7108–19. doi:10.1128/JVI.03729-14
34. Grunvogel O, Esser-Nobis K, Windisch MP, Frese M, Trippler M, Bartenschlager R, et al. Type I and type II interferon responses in two human liver cell lines (Huh-7 and HuH6). Genom Data (2016) 7:166–70. doi:10.1016/j.gdata.2015.12.017
35. Mostafavi S, Yoshida H, Moodley D, LeBoite H, Rothamel K, Raj T, et al. Parsing the interferon transcriptional network and its disease associations. Cell (2016) 164:564–78. doi:10.1016/j.cell.2015.12.032
36. Zhou H, Chen S, Zhou Q, Wei YN, Wang MS, Jia RY, et al. Cross-species antiviral activity of goose interferons against duck plague virus is related to its positive self-feedback regulation and subsequent interferon stimulated genes induction. Viruses (2016) 8:E195. doi:10.3390/v8070195
37. Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol (2008) 89:1–47. doi:10.1099/vir.0.83391-0
38. Whitmire JK. Induction and function of virus-specific CD4+ T cell responses. Virology (2011) 411:216–28. doi:10.1016/j.virol.2010.12.015
39. Zhao Q, Liu F, Chen S, Yan X, Qi Y, Wang M, et al. Chinese goose (Anser cygnoides) CD8a: cloning, tissue distribution and immunobiological in splenic mononuclear cells. Gene (2013) 529:332–9. doi:10.1016/j.gene.2013.07.104
40. Kolumam GA, Thomas S, Thompson LJ, Sprent J, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med (2005) 202:637–50. doi:10.1084/jem.20050821
41. Havenar-Daughton C, Kolumam GA, Murali-Krishna K. Cutting edge: the direct action of type I IFN on CD4 T cells is critical for sustaining clonal expansion in response to a viral but not a bacterial infection. J Immunol (2006) 176:3315–9. doi:10.4049/jimmunol.176.6.3315
42. Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A (2012) 109:4239–44. doi:10.1073/pnas.1114981109
43. Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, et al. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol (2010) 84:8332–41. doi:10.1128/JVI.02199-09
44. Zhu J, Ghosh A, Sarkar SN. OASL – a new player in controlling antiviral innate immunity. Curr Opin Virol (2015) 12:15–9. doi:10.1016/j.coviro.2015.01.010
45. Yang C, Liu F, Chen S, Wang M, Jia R, Zhu D, et al. Identification of 2′-5′-oligoadenylate synthetase-like gene in goose: gene structure, expression patterns, and antiviral activity against Newcastle disease virus. J Interferon Cytokine Res (2016) 36:563–72. doi:10.1089/jir.2015.0167
46. Drappier M, Michiels T. Inhibition of the OAS/RNase L pathway by viruses. Curr Opin Virol (2015) 15:19–26. doi:10.1016/j.coviro.2015.07.002
47. Haller O, Staeheli P, Schwemmle M, Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol (2015) 23:154–63. doi:10.1016/j.tim.2014.12.003
48. Ko JH, Jin HK, Asano A, Takada A, Ninomiya A, Kida H, et al. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res (2002) 12:595–601. doi:10.1101/gr.210702.%20Article%20published%20online%20before%20print%20in%20March%202002
49. Bazzigher L, Schwarz A, Staeheli P. No enhanced influenza virus resistance of murine and avian cells expressing cloned duck mx protein. Virology (1993) 195:100–12. doi:10.1006/viro.1993.1350
50. Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, et al. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylatedependent RNase L. EMBO J (1997) 16:6355–63. doi:10.1093/emboj/16.21.6355
Keywords: goose interferons, Tembusu virus, antiviral response, RNA sequencing, Mx, OASL
Citation: Chen S, Zhang W, Wu Z, Zhang J, Wang M, Jia R, Zhu D, Liu M, Sun K, Yang Q, Wu Y, Chen X and Cheng A (2017) Goose Mx and OASL Play Vital Roles in the Antiviral Effects of Type I, II, and III Interferon against Newly Emerging Avian Flavivirus. Front. Immunol. 8:1006. doi: 10.3389/fimmu.2017.01006
Received: 27 March 2017; Accepted: 07 August 2017;
Published: 23 August 2017
Edited by:
Charlotte Odendall, King’s College London, United KingdomReviewed by:
Xiaoyu Hu, Tsinghua University, ChinaSungjin Kim, Michigan State University, United States
Copyright: © 2017 Chen, Zhang, Wu, Zhang, Wang, Jia, Zhu, Liu, Sun, Yang, Wu, Chen and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shun Chen, c2h1bmNoZW5Ac2ljYXUuZWR1LmNu;
Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally as co-first authors of this work.
 Shun Chen
Shun Chen Wei Zhang
Wei Zhang Zhen Wu1
Zhen Wu1 Dekang Zhu
Dekang Zhu Mafeng Liu
Mafeng Liu Anchun Cheng
Anchun Cheng