- 1Department of Genetics and Developmental Biology, Faculty of Medicine, The Rappaport Family Institute for Research in the Medical Sciences, Technion – Israel Institute of Technology, Haifa, Israel
- 2Department of Human Biology, Faculty of Natural Sciences, University of Haifa, Haifa, Israel
During Drosophila embryogenesis, a large number of apoptotic cells are efficiently engulfed and degraded by professional phagocytes, macrophages. Phagocytic receptors Six-Microns-Under (SIMU), Draper (Drpr) and Croquemort (Crq) are specifically expressed in embryonic macrophages and required for their phagocytic function. However, how this function is established during development remains unclear. Here we demonstrate that the key regulator of Drosophila embryonic hemocyte differentiation, the transcription factor Serpent (Srp), plays a central role in establishing macrophage phagocytic competence. Srp, a homolog of the mammalian GATA factors, is required and sufficient for the specific expression of SIMU, Drpr and Crq receptors in embryonic macrophages. Moreover, we show that each of these receptors can significantly rescue phagocytosis defects of macrophages in srp mutants, including their distribution in the embryo and engulfment of apoptotic cells. This reveals that the proficiency of macrophages to remove apoptotic cells relies on the expression of SIMU, Crq and/or Drpr. However, Glial Cells Missing (GCM) acting downstream of Srp in the differentiation of hemocytes, is dispensable for their phagocytic function during embryogenesis. Taken together, our study discloses the molecular mechanism underlying the development of macrophages as skilled phagocytes of apoptotic cells.
Introduction
During normal development of multicellular organisms superfluous cells are eliminated through apoptosis and subsequent phagocytosis by “professional” phagocytes, macrophages and immature dendritic cells, and “non-professional” tissue-resident neighboring cells (1–3). Phagocytes efficiently remove apoptotic cells with high level of specificity, which is achieved through an ability of transmembrane phagocytic receptors or secreted bridging molecules to recognize “eat me” signals exposed on the surface of apoptotic cells (4–10). Most of the phagocytic receptors are exclusively expressed in phagocytic cells, however, how their specific expression is regulated during development remains poorly understood.
Drosophila “professional” phagocytes macrophages (plasmatocytes) are the most abundant cells in Drosophila hemolymph (~95%), which similarly to mammalian macrophages are responsible for phagocytosis of apoptotic cells, microbes and tissue remodeling (11–15). They originate from the cephalic mesoderm in the embryo and remain in circulation throughout all stages of development (12, 16). The ability of macrophages to phagocytose apoptotic cells is mediated by several receptors such as Croquemort (Crq), a member of the CD36 superfamily (17, 18), Six-Microns-Under (SIMU), Drosophila homolog of Stabilin-2 (19–21) and Draper (Drpr), Drosophila homolog of MEGF10 and Jedi (2, 22–25). During embryogenesis Crq is expressed mostly in macrophages whereas SIMU and Drpr are expressed both in macrophages and in “non-professional” phagocytes glia and ectoderm (19). Our previous study demonstrated that the specific expression of SIMU and Drpr in glia is part of the developmental program responsible for glial cell differentiation (26). However, how the expression of SIMU and Drpr is regulated in macrophages remains unknown.
Serpent (Srp) is a key regulator of macrophage development during embryogenesis (27, 28). Its two isoforms, SrpC and SrpNC, are required for proper differentiation of plasmatocytes (28). srp mutant embryos contain lower number of macrophages, which are abnormally distributed throughout the embryo (27). Transcription factors Glial Cells Missing (GCM) and GCM2 are involved in differentiation of embryonic macrophages downstream of Srp (28). gcm,gcm2 double mutants contain a reduced number of macrophages as well (29). However, we have shown previously that in gcm,gcm2 mutants the expression of the phagocytic receptors SIMU, Drpr and Crq is not altered in the remaining hemocytes (26).
In the work presented here, we demonstrate that Srp is required for apoptotic cell clearance by embryonic macrophages through regulation of SIMU, Drpr and Crq expression in these cells. In addition, we show that Srp is sufficient to drive SIMU and Drpr ectopic expression. We also found that expression of each phagocytic receptor, SIMU, Drpr or Crq, alone in srp mutant macrophages is sufficient to partially rescue their phagocytic skills and distribution, revealing the crucial role each receptor plays in establishment of cell phagocytic ability. However, our data disclose that GCM and GCM2 are dispensable for the phagocytic clearance of apoptotic cells by embryonic macrophages.
Materials and Methods
Fly Strains and Constructs
The following fly strains were used in this work: srpGal4, UAScytGFP (I. R. Evans), UASsrpNC and UASsrpC-FLAG/cyo (J. Casanova, K. Campbell and M. Haenlin), repoGal4 (B. Jones), srp3/TM3 (#2485; Bloomington), UASdrpr (M. Freeman), UASgcm (#5446; Bloomington), UASsimu (30), UAScrq (ORF collection), tubGal80ts (#7019; Bloomington), gcm-lacZ (#5445; Bloomington), simu-cytGFP (19), Df(2L)Exel7042 (#7812; Bloomington). repoGal4::UASsrp; tubGal80ts crosses were placed at 18oC and third instar larvae were transferred to 29°C for 14 hours.
Reporter constructs were generated by cloning different parts of a 2 kb DNA region upstream of the simu ORF, which recapitulates simu embryonic expression in all phagocytic cell populations (glia, macrophages and ectoderm) (19) into the pattB vector containing a cytoplasmic GFP coding sequence. These transgenic constructs were inserted into the attP51C site on chromosome 2R using the QC31 system (31). All strains were raised at 25°C.
Bioinformatic Analysis
The 650 bp sequence was analyzed in Genomatix Mathinspector tool for known Drosophila melanogaster and vertebrate transcription factors binding sites. Only results with matrix similarity greater than 0.7 were selected. Ci value of the results was greater than 60.
Immunohistochemistry and Live Imaging
For immunohistochemistry embryos were fixed and stained according to standard procedures. Guinea pig anti-SIMU (30) and guinea pig anti-Drpr (32) were used at a 1:5000 and 1:100 concentrations, respectively. Rabbit anti-activated caspase 3 (Dcp-1) (Cell Signaling) and mouse anti-GFP (Roche) were used at 1:100 concentration. Rabbit anti-Crq antibody (1:500) is a gift from N. Franc. Rabbit anti-Srp antibody (1:100) is a gift from J. Casanova, K. Campbell and N. Martin. Rabbit anti-Peroxidasin antibody (1:2000) is a gift from Jiwon Shim. Fluorescent secondary antibodies (Cy3/and Cy5/Jackson ImmunoResearch; Alexa Fluor 488/Molecular Probes) were used at 1:200 dilutions. For TUNEL labeling embryos were re-fixed, washed and labeled with the In Situ Cell Death Detection kit (Roche) according to the manufacture instructions. Images were acquired on a confocal microscope Zeiss LSM 700 or on a Zeiss Axio Observer microscope equipped with an Apotome system using the AxioVision software. 75% Glycerol solution was used as the imaging medium.
Live imaging was carried out by dechorionating embryos (stage 15), mounting them under Halocarbon oil, injecting 2–3% egg volume of LysoTracker (Molecular Probes) as described in Ref. (33). Recording started 30 min following injection.
Statistical Analysis
For statistical analysis in each embryo number of apoptotic particles was quantified inside 10 macrophages that contain at least one apoptotic particle. 5–8 embryos of each genotype were tested (n = number of embryos, indicated in each figure legend). The average number of apoptotic particles per macrophage (“phagocytic index”) was calculated per embryo by dividing the total number of apoptotic particles inside labeled macrophages by the number of macrophages taken into account in this embryo. Significance was calculated by an unpaired Student’s t-test or by one-way ANOVA followed by Bonferroni post hoc test.
To count the number of REPO-positive nuclei, apotome stacks (19 µm) were acquired from the whole CNS followed by Image analysis of the designated area using IMARIS (Bitplane) software.
Results
srp Is Required for Expression of SIMU in Embryonic Macrophages
We have previously shown that during embryogenesis simu expression is differentially regulated in macrophages and glia; GCM directly controls simu transcription in glia, but not in macrophages (26). Therefore, how simu expression is regulated in embryonic macrophages remained unclear.
To identify factors responsible for SIMU expression in macrophages, we decided to limit our search to the smallest regulatory unit responsible for SIMU expression in these cells. For that, we reduced a 2 kb DNA region upstream of the simu ORF that directs cytoplasmic GFP expression in all phagocytic cell populations (glia, macrophages and ectoderm) (19) (Figure 1A) to a series of smaller overlying fragments. These fragments of the 2 kb regulatory region, fused to cytoplasmic GFP, were used for transfection in S2 cells and/or for generation of transgenic flies. A 650 bp fragment (Figure 1A) was found as the minimal region that drives GFP expression in S2 cells, as well as in macrophages and glia in the embryo, as shown by a complete overlap of anti-GFP and anti-SIMU labeling (Figures 1C–D’’). The 650 bp fragment contains one GCM binding site (Figure 1B), which explains GFP expression in glia. Smaller fragments were not able to induce any GFP expression in S2 cells. We applied the 650 bp sequence to the Genomatix software to identify binding motifs of known transcription factors.
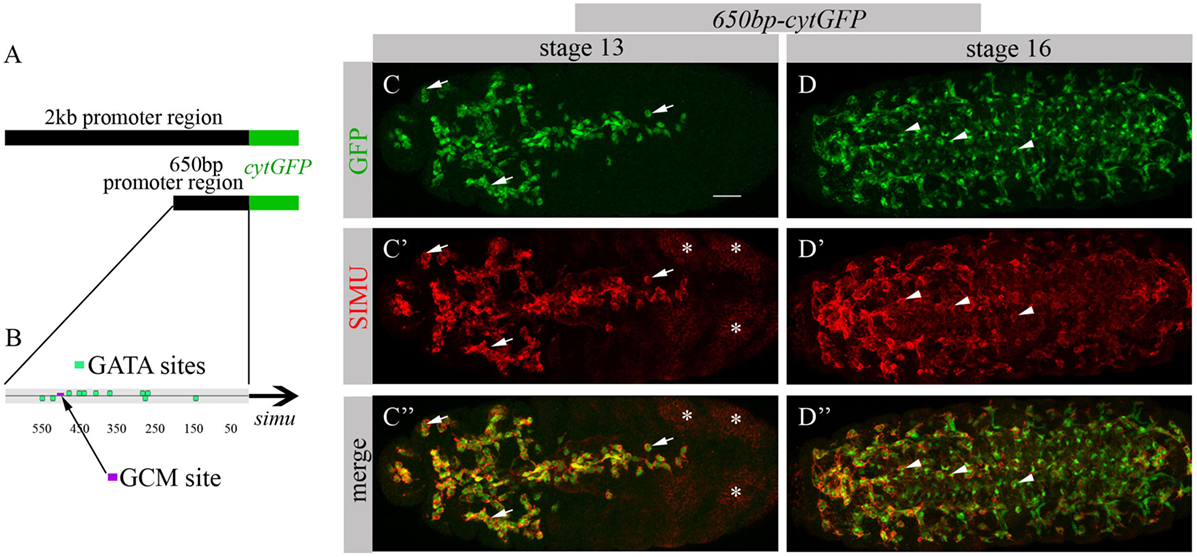
Figure 1. 650 bp region upstream to simu ORF recapitulates simu endogenous embryonic expression and contains multiple GATA binding sites. (A) Schematic of 2 kb region of simu promoter fused to cytoplasmic GFP sequence. (B) Schematic map of 650 bp region of simu promoter fused to cytoplasmic GFP sequence with depicted putative GATA sites and one GCM binding site. (C–D’’) Projections from confocal stacks of the stage 13 (C-C’’) and stage 16 (D–D’’) embryos, ventral view. (C,C’’,D,D’’) Cytoplasmic GFP reporter and (C’,C’’,D’,D’’) SIMU protein as detected on membranes with anti-SIMU antibody. Bar, 20 µm. Note colocalization of GFP and SIMU in macrophages (arrows) and glia (arrowheads) but not in ectoderm (stars).
The Genomatix software identified more than 50 different sites, which have been further evaluated by the expression pattern of the corresponding transcription factors. From these potential regulators we focused on three most promising candidates: dSTAT, pangolin and srp, since they are all expressed in embryonic macrophages at stages when simu expression originates (Flybase data base). To examine whether these factors are required for simu expression, we tested SIMU expression in mutant embryos of each candidate, using the anti-SIMU antibody. stat92E and pangolin mutant embryos exhibited normal SIMU staining in embryonic macrophages (results not shown), however, srp mutant embryos containing a strong hypomorph mutation (srp3) (27) did not reveal detectable SIMU staining in embryonic macrophages labeled with a srpGal4,UAScytGFP marker (Figures 2C–D’’). srp mutant embryos exhibited significantly smaller macrophages as evaluated by measuring their diameter (Figure 2E), which were abnormally distributed throughout the embryo compared to control (Figure 2A) and often clustered in the anterior part of the embryo (Figure 2C). Importantly, the CNS of srp mutant embryos was also deformed as visualized with a specific marker for glial cells, an anti-REPO antibody (Figure S1 in Supplementary Material). However, the number of glial cells was not different from control embryos (Figure S1 in Supplementary Material) and SIMU expression was detected in relatively normal levels in glial cells (Figures 2A’–A’’’,C’–C’’’). Together, these data demonstrate that Srp is required for SIMU expression in embryonic macrophages.
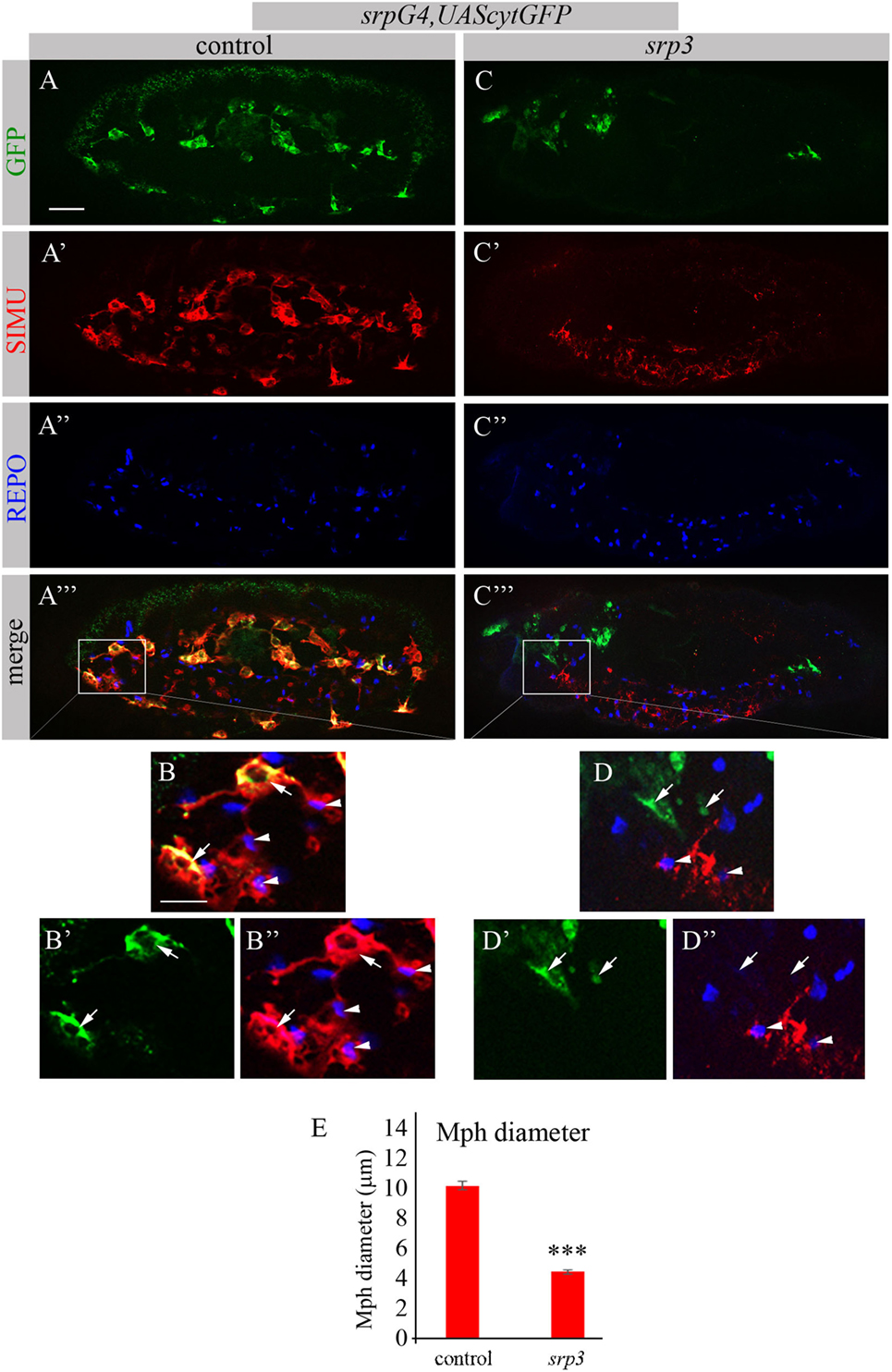
Figure 2. SIMU is not expressed in srp mutant macrophages. (A–D’’) Projections from confocal stacks of the stage 16 embryos, lateral view. Macrophages are labeled with srpGal4,UAScytGFP (green). Anti-SIMU in red. Glial nuclei are labeled with anti-REPO (blue). (A–B’’) Control srpGal4,UAScytGFP embryo. (C–D’’) srpGal4,UAScytGFP; srp3 mutant embryo. (B–B’’,D–D’’) Close up of rectangle areas in (A’’’,C’’’) respectively. All GFP-positive macrophages express SIMU on their membranes in control embryo [(B,B’’), arrows] but no one expresses SIMU in srp mutant embryo [(D,D’’), arrows]. Note SIMU expression in glia (non GFP-positive cells, arrowheads). Bar, 20 µm. (E) Columns represent mean diameter of 10 macrophages in each embryo ± SEM. Control embryos (n = 5). srp3 mutant embryos (n = 7). Asterisks indicate statistical significance versus control, as determined by Student’s t-test, ***p < 0.0001.
srp Is Required for the Phagocytic Function of Embryonic Macrophages
Given that macrophages of srp mutant appear abnormal and do not express SIMU, we tested their ability to phagocytose apoptotic cells. To evaluate their phagocytic capacity, we detected apoptotic particles with an anti-activated Dcp-1 antibody (Drosophila Caspase 3 homolog and a marker of apoptotic cells) (Figures 3A’,A’’,B’,B’’) and labeled macrophages with srpGal4,UAScytGFP (Figures 3A,A’’,B,B’’). We counted the number of apoptotic particles per macrophage, termed “phagocytic index” (explained in Materials and Methods). As expected, apoptotic particles were found inside GFP-positive macrophages in wild type embryos (Figures 3A’’,C). However, we could not detect any apoptotic particles inside macrophages of srp mutant (Figures 3B’’,C), suggesting their abnormal ability to phagocytose apoptotic cells.
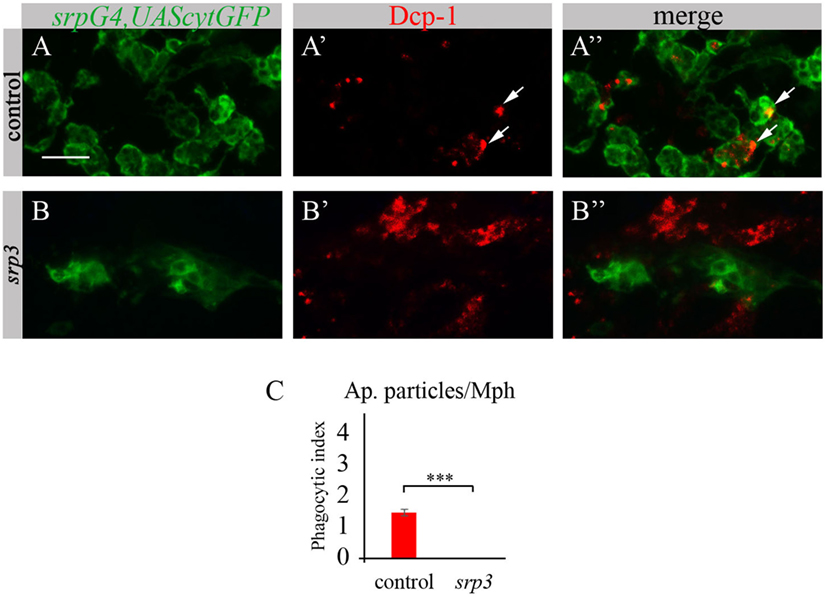
Figure 3. Srp is required for phagocytic ability of embryonic macrophages. (A–B’’) Projections from confocal stacks of the stage 16 embryos, ventral view, anterior region. Macrophages are labeled with srpGal4,UAScytGFP (green) and apoptotic particles are labeled with anti-Dcp-1 (red). In control srpGal4,UAScytGFP embryo (A–A’’) apoptotic particles are mostly inside GFP-positive macrophages (arrows). In srp3 mutant embryo (B–B’’) all apoptotic particles are outside GFP-positive macrophages (B’’). Bar, 20 µm. (C) Quantitation of apoptotic particles in macrophages of described genotypes. Columns represent mean phagocytic index ± SEM. Control embryos (n = 6). srp3 mutant embryos (n = 7). Asterisks indicate statistical significance versus control, as determined by Student’s t-test, ***p < 0.0001.
We took an additional approach to evaluate phagocytosis by macrophages using LysoTracker (LT), which specifically labels phagolysosomes/phagosomes (Figures 4A,A’,A’’’,B,B’,B’’’). Macrophages were labeled by srpGal4,UAScytGFP (Figures 4A–A’’,B–B’’) and contained multiple LT-labeled phagolysosomes in wild type embryos (Figures 4A,A’,C). However, in srp mutant embryos we could not detect any LT labeling in GFP-positive cells (Figures 4B,B’,C) once more demonstrating an impaired phagocytic ability of srp mutant macrophages.
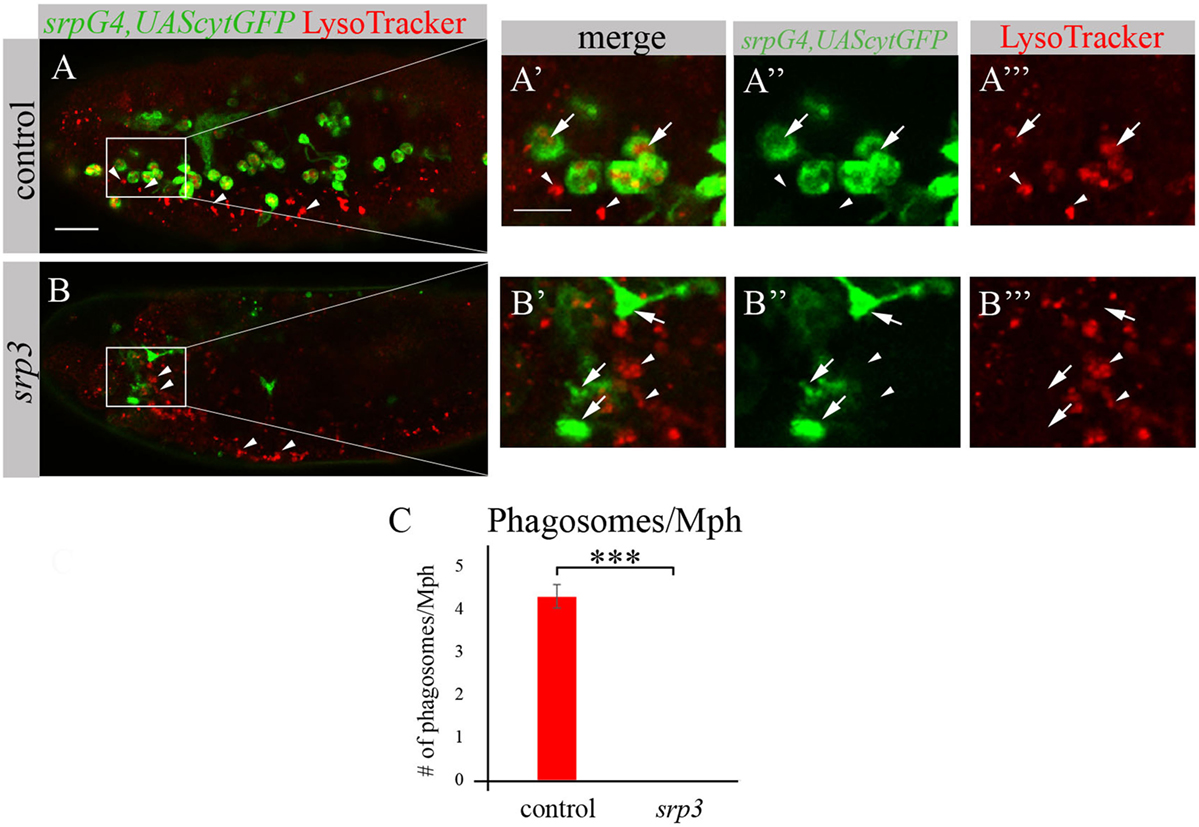
Figure 4. srp mutant macrophages are impaired in phagocytosis. (A–B’’’) Projections from confocal stacks of the stage 16 live embryos, lateral view. srpGal4,UAScytGFP marks macrophages in green. Phagolysosomes are labeled with LysoTracker (LT, red). (A–A’’’) Control srpGal4,UAScytGFP embryo shows numerous LT labeled phagolysosomes inside GFP-positive cells (arrows). (B–B’’’) In srp3 mutant embryo there is no LT labeling in GFP-positive cells (arrows). LT labels glial phagolysosomes in the CNS on the ventral side (arrowheads). Bar, 20 µm. (C) Quantitation of LT-labeled phagolysosomes in macrophages of described genotypes. Columns represent mean number of phagosolysosomes per macrophage ± SEM. Control embryos (n = 5). srp3 mutant embryos (n = 5). Asterisks indicate statistical significance versus control, as determined by Student’s t-test, ***p < 0.0001.
Srp Is Required for Expression of Drpr and Crq in Embryonic Macrophages
The impaired phagocytosis phenotype of srp mutant embryos appears much stronger than simu mutant phenotype (19), suggesting that additional phagocytic receptors may be affected by the absence of srp. To test this, we examined srp mutant embryos for the expression of two additional phagocytic receptors known to participate in apoptotic cell clearance by macrophages, Drpr and Crq (Figure 5). In control embryos, Drpr is specifically expressed in macrophages, glia and ectodermal cells as detected with anti-Drpr antibody (Figures 5A–A’’’). However, we were unable to detect any Drpr protein in macrophages of srp mutant labeled with srpGal4,UAScytGFP, though Drpr expression in the ectoderm remained normal (Figures 5B–B’’’). This reveals that Srp is required for Drpr expression in embryonic macrophages. Similarly, using an anti-Crq antibody (Figures 5C–D’’’) we found that Crq expression was undetectable in srp mutant embryos (Figures 5D–D’’’), indicating that Srp is required for Crq expression in embryonic macrophages as well.
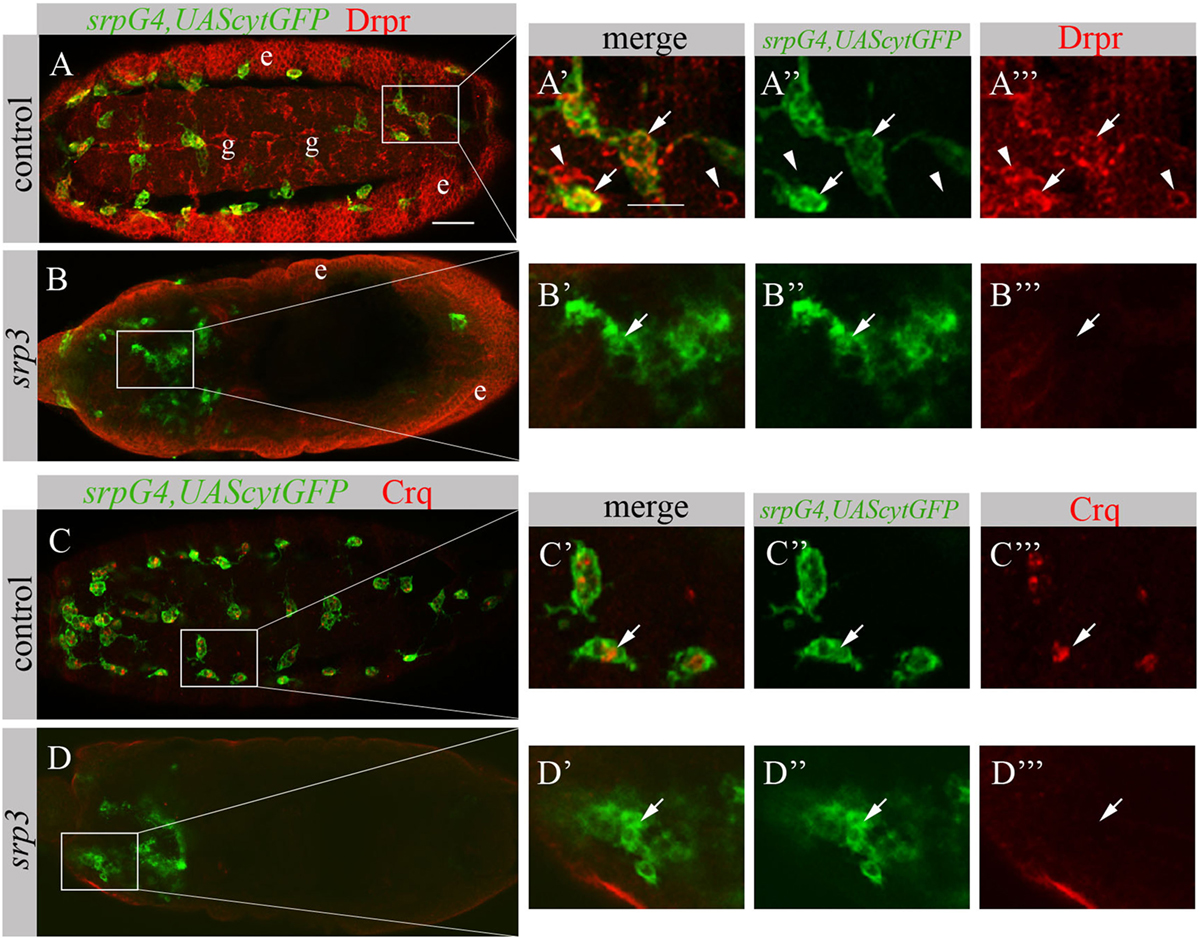
Figure 5. Srp is required for Drpr and Crq expression in embryonic macrophages. (A–D’’’) Projections from confocal stacks of the stage 16 embryos, ventral (A–A’’’) or dorsal view (B–D’’’). Macrophages are labeled with srpGal4,UAScytGFP (green). (A–B’’’) Anti-Drpr in red, ectoderm (e) and glia (g). (C–D’’’) Anti-Crq in red. (A’–A’’’, B’–B’’’, C’–C’’’, D’–D’’’) Close up of rectangle areas in A,B,C and D respectively. (A–A’’’) Control srpGal4,UAScytGFP embryo expressing Drpr in GFP-positive macrophages (arrows) and GFP-negative glia (arrowheads). (C–C’’’) Control srpGal4,UAScytGFP embryo expressing Crq in GFP-positive macrophages (arrows). (B–B’’’, D–D’’’) srp3 mutant embryos show no detectible Drpr (B, B’’’) and Crq (D,D’’’) staining in GFP-positive cells (arrows). Bar, 20 µm.
Srp Is Sufficient to Induce SIMU and Drpr Expression in Larval Glia
To test whether Srp is sufficient to induce SIMU expression, we ectopically expressed different isoforms of Srp, SrpNC (UASsrpNC) or SrpC (UASsrpC), in larval glial cells which normally do not express SIMU (Figure 6A’), using a repoGal4 driver. srp ectopic expression in embryonic glia was prevented by a tubGal80 temperature sensitive (ts) allele until the third instar larval stage. At this stage we moved the progeny (repoGal4,UAScytGFP;tubGal80ts::UASsrpNC or repoGal4,UAScytGFP;tubGal80ts::UASsrpC) from the permissive (18°C) to the restrictive (29°C) temperature of tubGal80ts. Dissected larval brains were stained with anti-Srp (Figures 6A”,A’’’,B’’,B’’’,C’’,C’’’) and anti-SIMU (Figures 6A’,A’’’,B’,B’’’,C’,C’’’) antibodies, which revealed that glial cells ectopically expressing Srp concomitantly expressed SIMU on their membranes (Figures 6B’’’,C’’’). These results demonstrate that srp is sufficient to drive SIMU expression. Both isoforms, SrpNC (Figures 6B–B’’’) and SrpC (Figures 6C–C’’’) were able to induce SIMU expression in larval glia (Figures 6B’,B’’’,C’,C’’’).
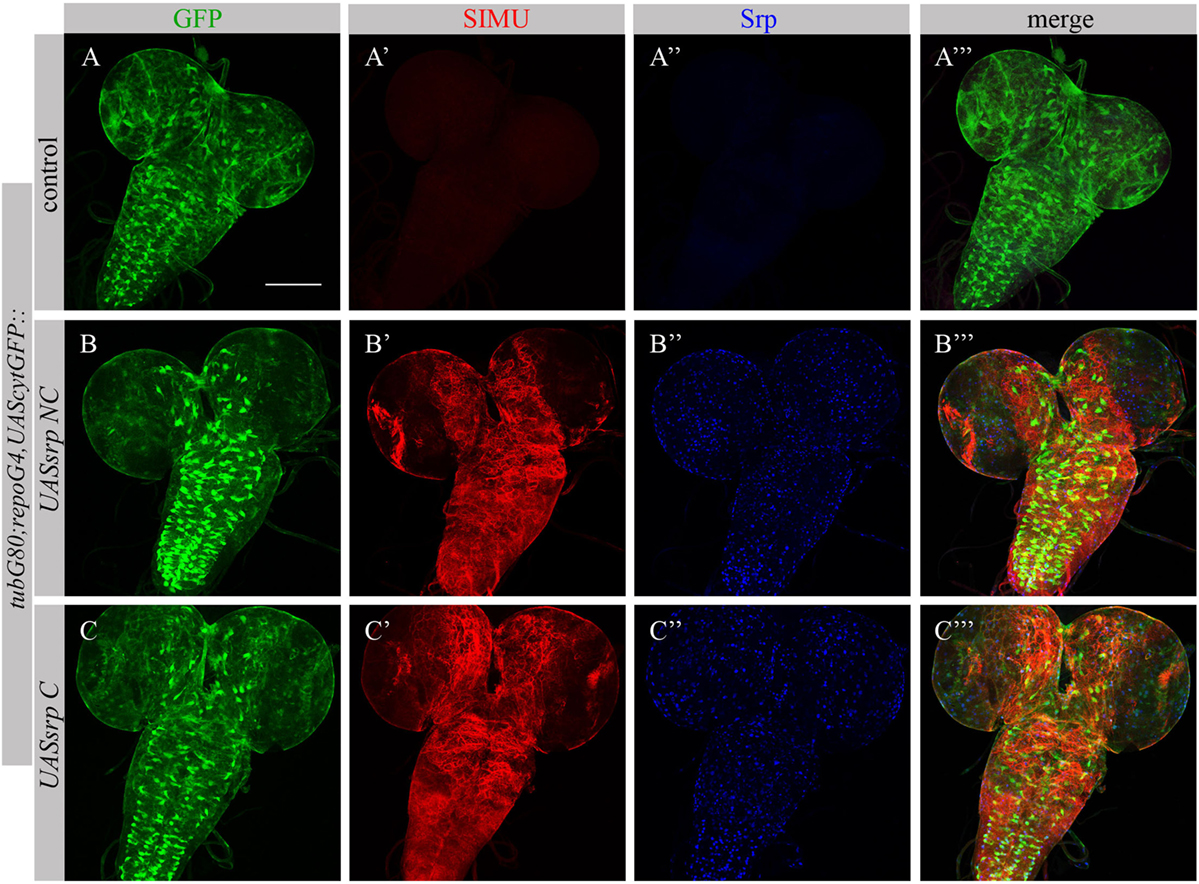
Figure 6. Each Srp isoform (SrpC and SrpNC) is sufficient to drive SIMU expression. (A–C’’’) Projections from confocal stacks of the 3rd instar larval brains stained with anti-SIMU [red, (A’,A’’’,B’,B’’’,C’,C’’’)], glia are labeled with repoGal4,UAScytGFP [green, (A,A’’’,B,B’’’,C,C’’’)], anti-Srp [blue, (A’’,A’’’,B’’,B’’’,C’’,C’’’)]. Bar, 100 µm.
Following ectopic expression of Srp in larval glia, we tested appearance of Drpr in dissected larval brains (repoGal4,UAScytGFP;tubGal80ts::UASsrpNC or repoGal4,UAScytGFP;tubGal80ts::UASsrpC) by staining with anti-Srp and anti-Drpr antibodies. Compared to control glia (Figure 7A’), we detected more Drpr protein on membranes of glial cells ectopically expressing Srp (Figures 7B’,C’). Both isoforms SrpC and SrpNC were able to elevate Drpr expression in larval glia (Figures 7A’,A’’’,B’,B’’’,C’,C’’’), indicating that Srp is sufficient to induce Drpr expression. Importantly, it has been shown previously that SrpC is sufficient to induce Crq ectopic expression whereas SrpNC is not (28). These data suggest that the isoform C of Srp is sufficient to drive Drpr, Crq and SIMU expression, whereas the NC isoform can induce only SIMU and Drpr expression.
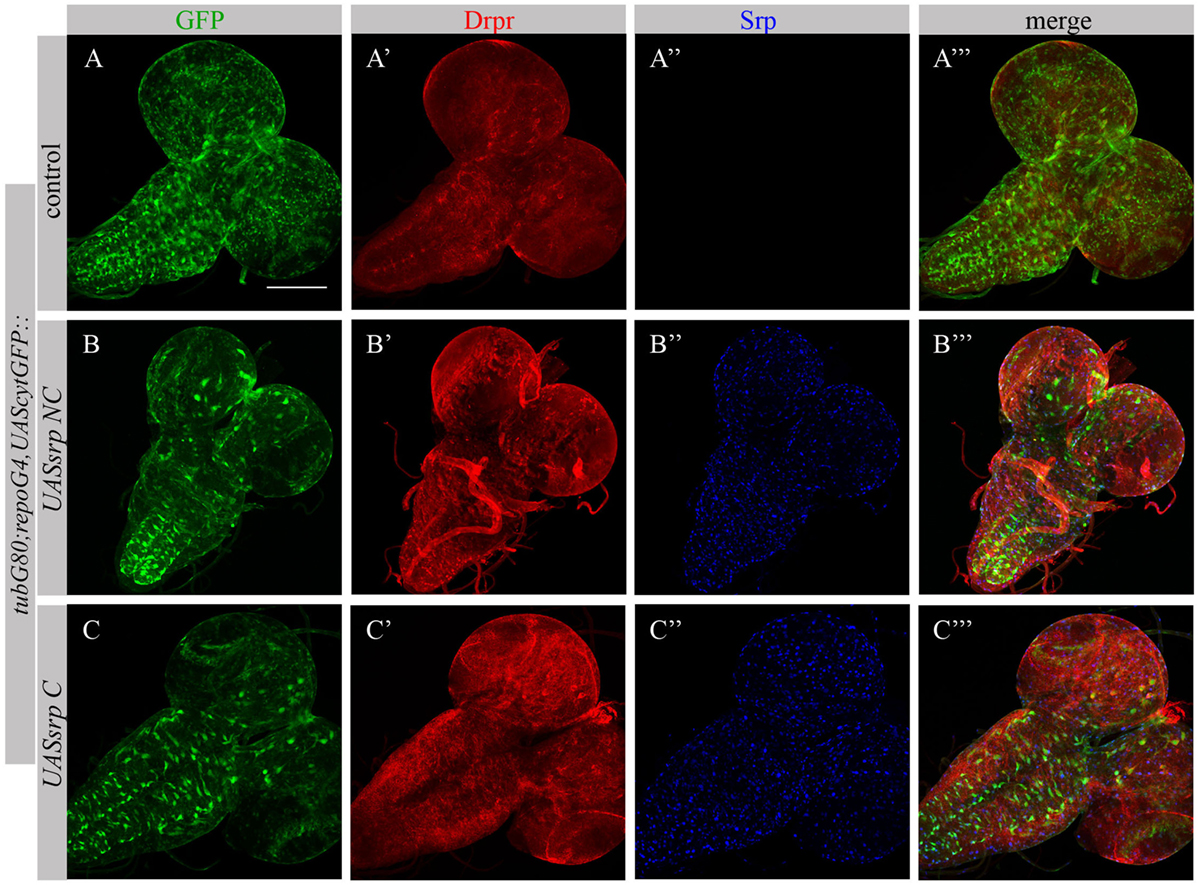
Figure 7. Each Srp isoform (SrpC and SrpNC) is sufficient to drive Drpr expression. (A–C’’’) Projections from confocal stacks of the 3rd instar larval brains stained with anti-Drpr [red, (A’,A’’’,B’,B’’’,C’,C’’’)], glia are labeled with repoGal4,UAScytGFP [green, (A,A’’’,B,B’’’,C,C’’’)], anti-Srp [blue, (A’’,A’’’,B’’,B’’’,C’’,C’’’)]. Bar, 100 µm.
GCM Is Dispensable for the Phagocytic Ability of Embryonic Macrophages
We have previously shown that GCM,GCM2 directly regulate simu expression only in embryonic glia but not in macrophages (26). Moreover, mutant gcm,gcm2 macrophages still express SIMU, Drpr and Crq (26) (Figure 8). However, mutant embryos lacking gcm and gcm2 contain a significantly lower number of embryonic macrophages (29, 34) suggesting that GCM,GCM2 are required for their proliferation, differentiation and/or survival. Nevertheless, whether GCM,GCM2 are essential for phagocytosis of apoptotic cells by macrophages has not been previously established. Using simultaneous labeling of embryonic macrophages with anti-SIMU and apoptotic cells with anti-Dcp-1 antibodies (Figures 8A–B’’’), we observed that gcm,gcm2 mutant macrophages contain apoptotic particles inside them (Figures 8B–B’’’), demonstrating that they are capable of engulfing apoptotic cells. In addition, we performed terminal deoxynucleotidyl transferase dUTP nick and labeling (TUNEL) staining to label DNA fragments, characteristic of apoptotic cells in wild type (Figures 8C–C’’’) and gcm,gcm2 mutant (Figures 8D–D’’’) embryos. Similarly to control embryos, in gcm,gcm2 mutants SIMU-labeled macrophages contain TUNEL-positive particles confirming their ability to phagocytose apoptotic cells (Figures 8C–D’’’). These data demonstrate that GCM,GCM2 are not required for the phagocytic ability of embryonic macrophages.
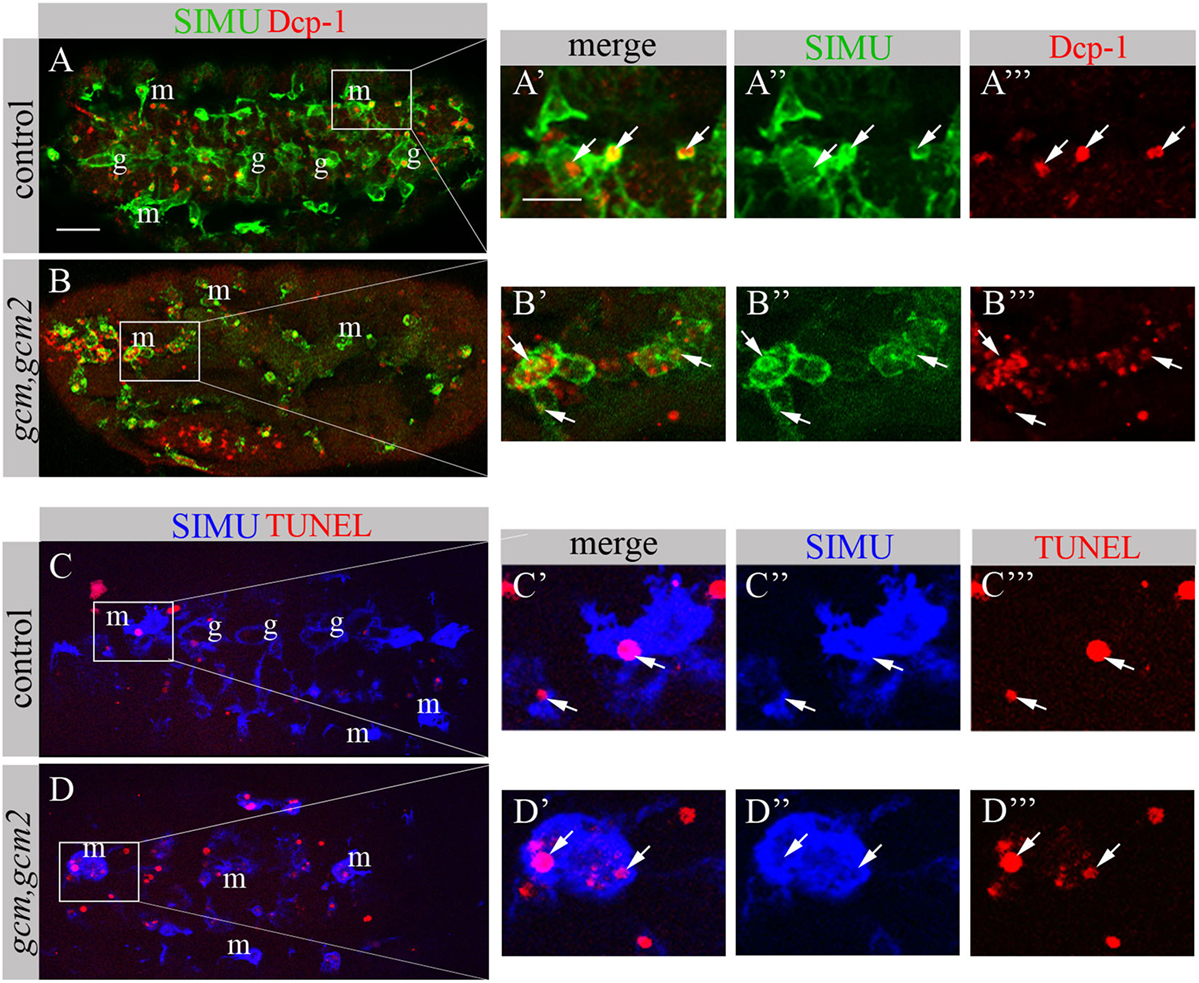
Figure 8. gcm,gcm2 mutant macrophages phagocytose apoptotic cells. (A–D’’’) Projections from confocal stacks of the stage 16 embryo; (A–B’’’) apoptotic cells are in red (Dcp-1), SIMU protein with anti-SIMU in green. (C–D’’’) TUNEL in red, SIMU protein with anti-SIMU in blue. (A–A’’’, C–C’’’) Ventral view of control embryos displays SIMU expression in glia (g) and macrophages (m). Dcp-1 and TUNEL staining are found inside SIMU-labeled cells [(A-A’’’, C–C’’’) arrows]. (B–B’’’, D–D’’’) gcm,gcm2 deficient embryos; no glia are labeled with SIMU but macrophages are (m). Dcp-1 and TUNEL staining are found inside SIMU-positive macrophages (arrows). Bar, 20 µm.
Based on our previous study showing that GCM,GCM2 directly regulate simu expression in embryonic glia (26) we assumed that GCM,GCM2 may also induce simu expression in macrophages. However, since Srp binding sites in simu promoter are located in close proximity to the GCM binding site (L. Waltzer—personal communication) we hypothesized that it may sterically prevent GCM,GCM2 binding. To test this we aimed to examine whether GCM,GCM2 are able to induce SIMU expression in the absence of Srp (srp mutant). Normally GCM expression is not detected in srp mutants (Figures 9B’’’,b). Therefore, we expressed GCM (UASgcm) in srp mutant macrophages using the srpGal4 driver (srpGal4,UAScytGFP;srp3::UASgcm;srp3) and tested whether it induces SIMU expression (Figure 9). No evident appearance of SIMU has been detected in srp mutant macrophages expressing GCM (Figures 9C’’’,c), indicating that GCM is not sufficient to induce simu in the absence of Srp. Moreover, in these embryos no Drpr expression was noticed in macrophages as well (Figures 9C’’’,c) demonstrating that GCM is also not sufficient to induce Drpr expression in embryonic macrophages.
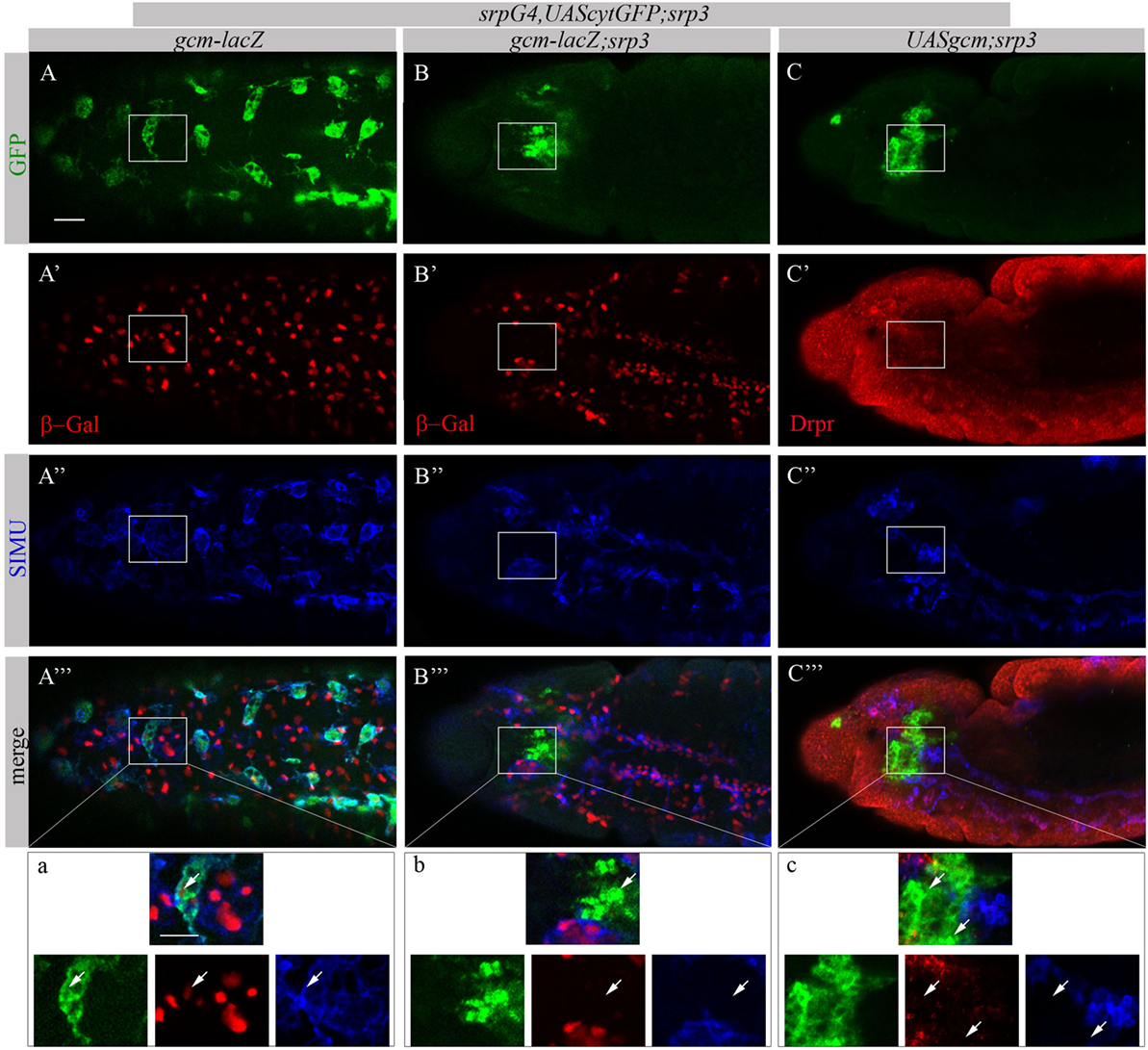
Figure 9. GCM is not sufficient to drive SIMU and Drpr expression in embryonic macrophages. (A–c) Projections from confocal stacks of the stage 16 embryos, (A–a) ventral view and (B–c) lateral view. Macrophages are labeled with srpGal4,UAScytGFP [green, (A,A’’’,a,B,B’’’,b,C,C’’’,c)] and SIMU protein with anti-SIMU [blue, (A’’,A’’’,a,B’’,B’’’,b,C’’,C’’’,c)]. (A–b) gcm-lacZ reporter in red and (C–c) Drpr with anti-Drpr in red. (A–a) Control srpGal4,UAScytGFP embryo. β-Gal and SIMU are expressed in GFP-positive macrophages (arrows). (B–b) srp3 mutant embryo. No β-Gal and SIMU are detected in GFP-positive cells (arrows). (C–c) srp3 mutant carrying GCM (srpGal4::UASgcm) in macrophages. No SIMU and Drpr are detected in GFP-positive cells (arrows). Bar, 20 µm.
Each Phagocytic Receptor (SIMU, Drpr or Crq) Partially Rescues Distribution of srp Mutant Macrophages and Their Defects in Phagocytosis
To investigate whether the impaired phagocytic ability of srp mutant macrophages results merely from the absence of the receptor expression, we performed rescue experiments. We expressed either SIMU (Figures 10C–C’’’), Drpr (Figures 10D–D’’’) or Crq (Figures 10E–E’’’) specifically in srp mutant macrophages using the srpGal4 driver and tested their ability to phagocytose apoptotic cells by immunostaining with the anti-Dcp-1 antibody (Figure 10). Surprisingly, we found that srp mutant macrophages expressing SIMU, Drpr or Crq (srpGal4,UAScytGFP;srp3::UASsimu;srp3 or srpGal4,UAScytGFP;srp3::UASdrpr;srp3 or srpGal4,UAScytGFP;srp3::UAScrq;srp3) did not appear in clusters in the anterior part of the embryo like in srp mutants (Figures 9B–10B–B”) but were distributed throughout the embryo (Figures 10C–E). Moreover, their diameter was significantly bigger as compared to srp mutant macrophages (Figure 10G) and we found engulfed apoptotic cells inside these macrophages (Figures 10C’,D’,E’), indicating that they are capable of apoptotic cell clearance. We counted the number of apoptotic cells per macrophage (phagocytic index) in control, srp mutant and embryos carrying different rescue constructs (Figures 10A–E,H). These data revealed a significantly higher phagocytic index in srp mutant macrophages that express each receptor alone (Figure 10H), demonstrating that each phagocytic receptor, SIMU, Drpr or Crq is able by itself to partially rescue srp mutant phagocytosis phenotype. However, interestingly, in these rescued embryos significantly more apoptotic cells were detected inside macrophages compared to control embryos, demonstrating apoptotic cell accumulation. Importantly, we tested the effect of overexpression of each receptor in wild type macrophages using srpGal4 driver (srpGal4::UASsimu or srpGal4::UASdrpr or srpGal4::UAScrq). Compared to control no significant difference was detected in phagocytic index of macrophages overexpressing each receptor (Figure S2 in Supplementary Material), suggesting that in wild type embryo phagocytic ability of macrophages is not affected by overexpression of phagocytic receptors and might be limited by the overall amount of apoptotic cells in the embryo.
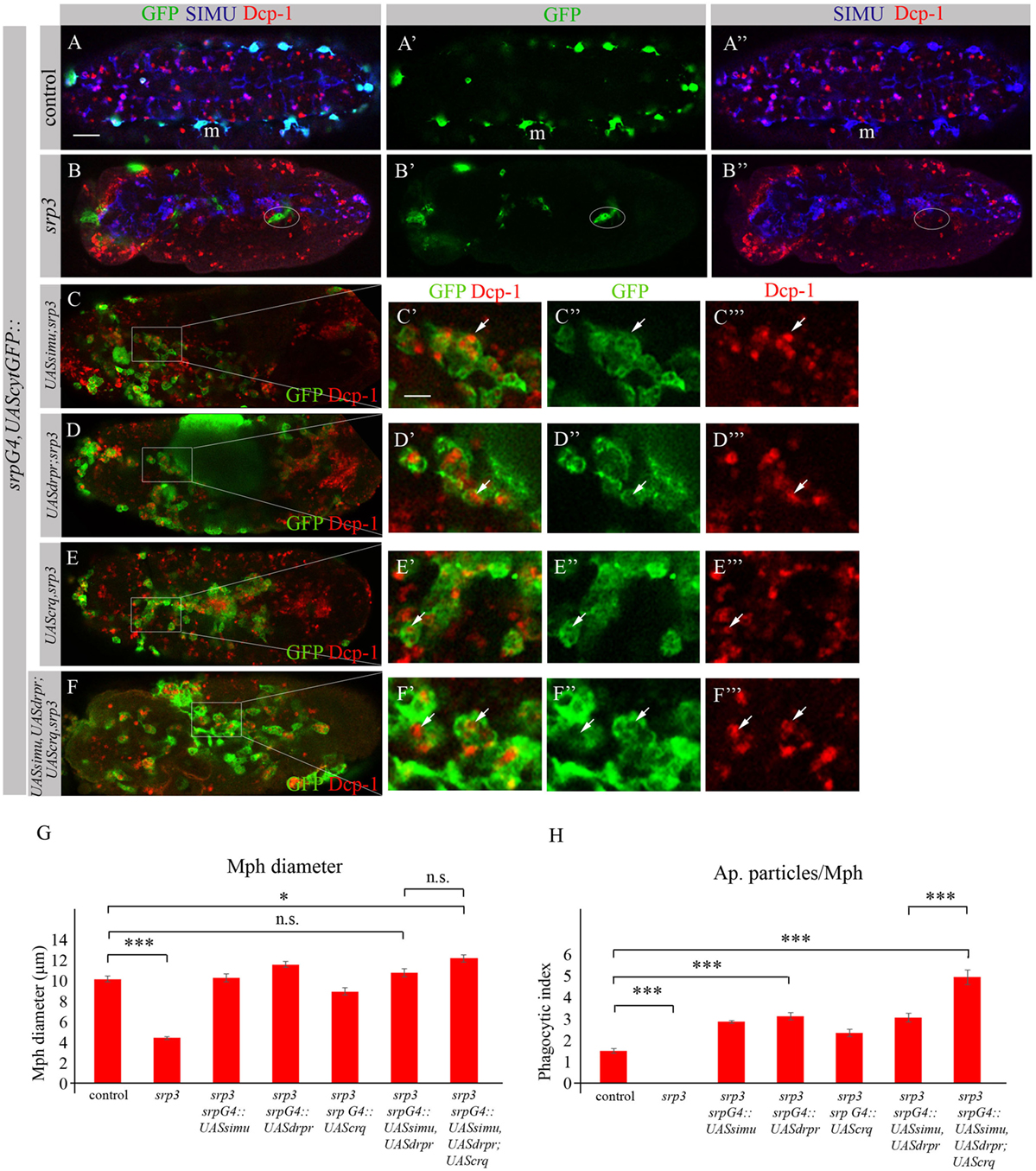
Figure 10. Each phagocytic receptor SIMU, Drpr or Crq rescues phagocytosis defects and distribution of srp mutant macrophages. (A–F’’’) Projections from confocal stacks of the stage 16 embryos. Macrophages are labeled with srpGal4,UAScytGFP [green, (A,A’,B,B’,C–C’’,D–D’’,E–E’’,F–F’’)], SIMU protein with anti-SIMU [blue, (A,A’’,B,B’’)] and apoptotic cells with anti-Dcp-1 [red, (A,A’’,B,B’’,C,C’,C’’’,D,D’,D’’’,E,E’,E’’’,F,F’,F’’’)]. (A–A’’) Control srpGal4,UAScytGFP embryo. (B–B’’) srp3 mutant embryo. (C–C’’’) srpGal4,UAScytGFP; srp3::UASsimu;srp3. (D–D’’’) srpGal4,UAScytGFP; srp3::UASdrpr;srp3. (E–E’’’) srpGal4,UAScytGFP; srp3::UAScrq,srp3. (F–F’’’) srpGal4,UAScytGFP; srp3::UASsimu,UASdrpr;UAScrq,srp3. Bar, 20 µm. (G) Columns represent mean diameter of 10 macrophages in each embryo ± SEM of following genotypes: control embryos (n = 5), srp3 mutant embryos (n = 7), srpGal4,UAScytGFP; srp3::UASsimu;srp3 (n = 6), srpGal4,UAScytGFP; srp3::UASdrpr;srp3 (n = 5), srpGal4,UAScytGFP; srp3::UAScrq,srp3 (n = 8), srpGal4,UAScytGFP; srp3::UASsimu,UASdrpr;srp3 (n = 8), srpGal4,UAScytGFP; srp3::UASsimu,UASdrpr;UAScrq,srp3 (n = 6). Asterisks indicate statistical significance versus control, as determined by one-way ANOVA followed by Bonferroni post hoc test, ***p < 0.0001, *p < 0.05, n.s. >0.05. (H) Columns represent mean phagocytic index ± SEM of following genotypes: control embryos (n = 6), srp3 mutant embryos (n = 7), srpGal4,UAScytGFP; srp3::UASsimu;srp3 (n = 6), srpGal4,UAScytGFP; srp3::UASdrpr;srp3 (n = 6), srpGal4,UAScytGFP; srp3::UAScrq,srp3 (n = 8), srpGal4,UAScytGFP; srp3::UASsimu,UASdrpr;srp3 (n = 8), srpGal4,UAScytGFP; srp3::UASsimu,UASdrpr;UAScrq,srp3 (n = 6). Asterisks indicate statistical significance versus control, as determined by one-way ANOVA followed by Bonferroni post hoc test, ***p < 0.0001.
The situation is different in srp mutant where compared to wild type much more apoptotic particles are present in the embryo (Figures 10A,B). When we tested co-expression of simu and drpr simultaneously in srp mutant macrophages using the srpGal4 driver (srpGal4,UAScytGFP;srp3::UASsimu,UASdrpr;srp3) we obtained a similar amount of cells inside the macrophages as with each receptor alone (Figure 10H), suggesting the same engulfment/degradation ratio in clearance of apoptotic cells. However, when all three receptors SIMU, Drpr and Crq were expressed in srp mutant macrophages (srpGal4,UAScytGFP;srp3::UASsimu,UASdrpr;UAScrq,srp3), we observed a significantly higher phagocytic index as compared to each receptor alone (Figures 10F–F’’’,H), which indicates additional accumulation of apoptotic cells inside macrophages. This result may designate a higher engulfment/degradation ratio in macrophages expressing all three phagocytic receptors.
To test this assumption we evaluated degradation ability of srp mutant macrophages expressing SIMU and Drpr only or all three receptors SIMU, Drpr and Crq by quantifying LT-positive phagolysosomes in macrophages labeled with srpGal4,cytGFP (Figures 11A–E). No significant difference in the number of LT-positive phagosolysosomes was found between control macrophages (Figures 11A–A’’’,E) and srp mutant macrophages expressing two receptors (srpGal4,UAScytGFP;srp3::UASsimu,UASdrpr;srp3) (Figures 11C–C’’’,E) or three receptors together (srpGal4,UAScytGFP;srp3::UASsimu,UASdrpr;UAScrq,srp3) (Figures 11D–D’’’,E) indicating the similar degradation rate. These data strongly support our suggestion that the higher phagocytic index and bigger diameter of srp mutant macrophages expressing all three receptors than in srp mutant macrophages expressing only SIMU and Drpr is a result of the higher engulfment/degradation ratio and accumulation of apoptotic particles inside them.
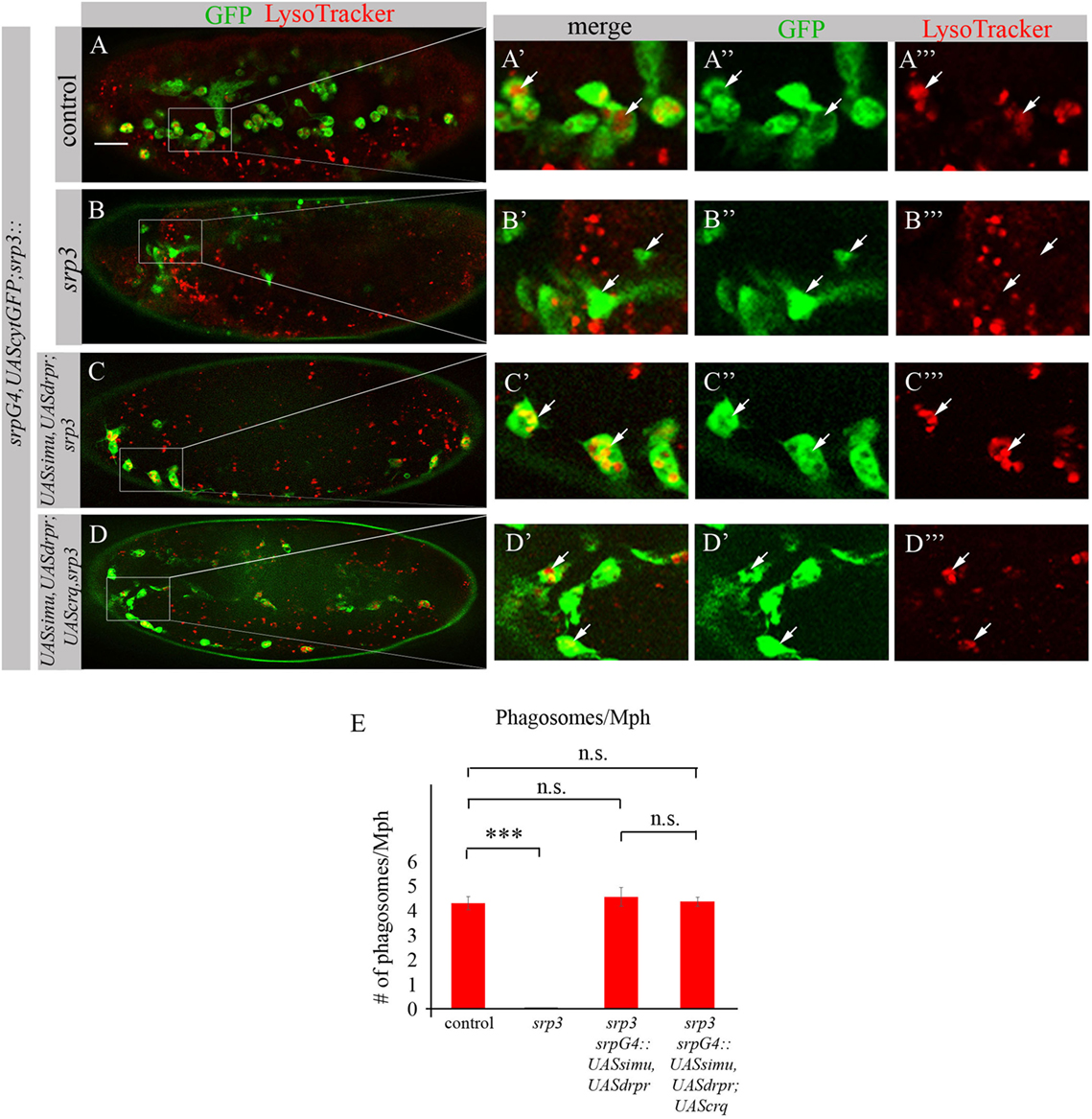
Figure 11. Phagocytic receptors SIMU, Drpr and/or Crq rescue phagocytosis defects of srp mutant macrophages. (A–D’’’) Stage 16 embryos. Macrophages are labeled with srpGal4,UAScytGFP [green, (A–A’’,B–B’’,C–C’’,D–D’’)], Phagolysosomes are labeled with LysoTracker (LT, red). (A–A’’’) Control srpGal4,UAScytGFP embryo. (B–B’’’) srp3 mutant embryo. (C–C’’’) srpGal4,UAScytGFP; srp3::UASsimu,UASdrpr;srp3. (D–D’’’) srpGal4,UAScytGFP; srp3::UASsimu,UASdrpr;UAScrq,srp3. Bar, 20 µm. (E) Quantitation of LT-labeled phagolysosomes per macrophage of described genotypes. Columns represent mean number of phagosolysosomes ± SEM of following genotypes: control embryos (n = 5), srp3 mutant embryos (n = 5), srpGal4,UAScytGFP; srp3::UASsimu,UASdrpr;srp3 (n = 5), srpGal4,UAScytGFP; srp3::UASsimu,UASdrpr;UAScrq,srp3 (n = 5). Asterisks indicate statistical significance versus control, as determined by one-way ANOVA followed by Bonferroni post hoc test, ***p < 0.0001, n.s. >0.05.
Discussion
Apoptotic cell clearance by “professional” and “non-professional” phagocytes plays a critical role during development of multicellular organisms. How the phagocytes acquire their ability to phagocytose apoptotic cells remains poorly understood. Key regulators of this process are phagocytic receptors for apoptotic cells that are specifically expressed on plasma membranes of phagocytes. However, the molecular mechanisms controlling expression of phagocytic receptors and therefore creating phagocytic ability of embryonic macrophages were unknown.
Using Drosophila embryonic macrophages as a model for development of “professional” phagocytes, we discovered that the GATA factor Srp is necessary for the specific expression of the phagocytic receptors SIMU, Drpr and Crq in these cells and sufficient to induce their expression in ectopic places. Therefore, the absence of Srp results in formation of abnormal macrophages lacking phagocytic receptors and thus incapable of apoptotic cell clearance. The defects in clearance can be substantially rescued by specific expression of each of the phagocytic receptors alone in embryonic macrophages. Surprisingly, we found that the presence of phagocytic receptors in srp mutant macrophages could also partially rescue their abnormal distribution. Interestingly, expression of each receptor, SIMU, Drpr or Crq resulted in comparable rescue of phagocytosis defects evaluated by phagocytic index. Similar phagocytic capacity of srp mutant macrophages expressing only one receptor suggests that each receptor is capable of persuading engulfment of apoptotic cells by macrophages. However, strikingly less apoptotic cells per macrophage are detected in the wild type embryos even if they overexpress the phagocytic receptors SIMU or Drpr or Crq. This could be explained by, in general, higher number of apoptotic cells present in srp mutant embryos and/or by their slower or impaired degradation inside phagolysosomes. Our results from the experiments with LT labeling of phagosomes suggest that higher number of engulfed apoptotic cells in the rescued macrophages is not accompanied by higher number of LT-positive phagolysosomes and therefore indicates slower degradation of engulfed apoptotic particles. This suggests that Srp may regulate expression of factors involved in the phagosome maturation process and therefore the degradation step in apoptotic cell clearance might be affected by its absence.
Furthermore, since SIMU and Crq are tethering receptors that are required for recognition and engulfment of apoptotic cells, expression of each receptor in srp mutant macrophages leads to the similar phenotype of engulfment and accumulation of apoptotic cells inside macrophages. However, we have previously shown that Drpr is mostly involved in degradation of apoptotic cells when SIMU and Crq are present (19). Our current results suggest that Drpr is capable of both engulfment and degradation of apoptotic particles when other receptors are missing, which is revealed by comparable phagocytic index in srp mutant macrophages that express Drpr alone with those that express SIMU or Crq. However, surprisingly, SIMU and Drpr joint expression demonstrates no additive effect on the phagocytic index. The possible explanation for this finding is that while SIMU allows more efficient engulfment compared to Drpr alone, Drpr itself permits faster degradation of the engulfed material. This is finally resulting in the similar phagocytic index of SIMU and Drpr joint expression to the expression of each one of them by itself. Interestingly though, when all three receptors are expressed (SIMU, Drpr and Crq), the amount of apoptotic cells per macrophage is significantly increased compared to SIMU and Drpr joint expression. These data suggest increased engulfment (by two tethering receptors SIMU and Crq) but limited degradation, which is mediated only by Drpr. Further confirmation of this conclusion comes from the same number of LT-positive phagolysosomes in the rescued macrophages expressing two receptors (SIMU and Drpr) and expressing all three receptors (SIMU, Drpr and Crq) demonstrating the same degradation rate and accumulation of more apoptotic cells in the macrophages expressing all three receptors. Taken together we demonstrate here that Srp creates phagocytic ability of embryonic macrophages by inducing balanced expression of the tethering receptors SIMU and Crq and the signaling receptor Drpr.
Our previous results revealed that GCM was not required for SIMU, Drpr and Crq expression in embryonic macrophages (26). Here we expanded our analysis on GCM role in apoptotic cell clearance by macrophages and demonstrate that GCM,GCM2 are not required for their function in phagocytosis of apoptotic cells. Significantly lower number of macrophages has been previously reported in gcm or gcm,gcm2 double mutants compared to wild type (29, 34). Our data exhibit that the remaining macrophages express SIMU, Drpr and Crq. This finding suggests two possible scenarios: (1) the lack of gcm,gcm2 may lead to apoptosis of macrophages resulting in the reduction of their number; Increased volume of apoptotic particles detected in gcm,gcm2 mutants may also outcome from increased apoptosis of macrophages in addition to the abnormal apoptotic cell clearance by glial cells (26).
Another possibility (2) could be as shown in Figure 12. It has been demonstrated previously that GCM,GCM2 repress Lozenge (Lz)—a fate determinant factor of crystal cell development (35, 36). Two waves of plasmatocyte development were proposed: first starts from general Srp-positive hemocyte precursors and second develops from Lz-positive crystal cell precursors (CCPs) (35, 36). We suggest that during the first wave Srp regulates SIMU, Drpr and Crq expression in plasmatocytes independently of GCM,GCM2 and Lz. However, later on Lz-positive CCPs differentiate to crystal cells that do not express SIMU, Drpr and Crq, which may result from Lz function in these cells (Figure 12). The second wave of plasmatocyte formation evolving from CCPs requires GCM/GCM2, which repress Lz expression in part of CCPs that become macrophages (36) and express all three phagocytic receptors (Figure 12). If gcm,gcm2 are absent, the second wave does not occur resulting in the reduced number of macrophages that express SIMU, Drpr and Crq compared to wild type embryos. We suggest that both possibilities can lead to the reduced number of macrophages in the gcm,gcm2 mutant embryos.
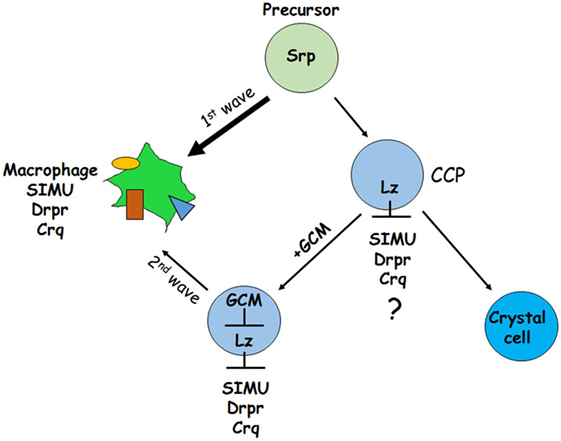
Figure 12. Schematic representation of two waves regulating development of embryonic macrophages. First wave starts from general Srp-positive hemocyte precursors and second develops from Lz-positive crystal cell precursors (CCPs). During the first wave Srp regulates SIMU, Drpr and Crq expression in plasmatocytes with no involvement of GCM,GCM2 and Lz. In the CCPs Lz expression likely (?) inhibits SIMU, Drpr and Crq expression resulting in formation of crystal cells that do not express these receptors. The second wave evolves from CCPs where GCM,GCM2 repress Lz, which allows expression of SIMU, Drpr and Crq and formation of macrophages. Srp may regulate additional factors involved in macrophage differentiation.
The question why GCM,GCM2 do not regulate SIMU expression in embryonic macrophages through their binding sites remains open. We suggest that a repressor of GCM activity may act at early stages of embryogenesis in hemocyte precursors. During later stages of embryogenesis GCM,GCM2 directly induces simu expression in glial cells (26). Intriguingly, the same transcription factors GCM,GCM2 behave differently in two phagocytic cell populations glia and macrophages. This finding demonstrates that the phagocytic competence of different cell populations is determined by specific expression of phagocytic receptors that is regulated by diverse developmental programs. Using the Drosophila embryo as a model, we were able to expose basic molecular mechanisms essential for establishment of embryonic macrophages as potent phagocytes during development.
Author Contributions
Conceived and designed the experiments: ES, BS, KH-M, FL-A, and EK. Performed the experiments: ES, BS, KH-M, and FL-A. Analyzed the data: ES, BS, KH-M, FL-A, and EK. Wrote the paper: EK.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank I. R. Evans, M. Freeman, J. Casanova, K. Campbell, N. Martin, M. Haenlin, N. Franc, J. Shim, the Developmental Studies Hybridoma Bank and the Bloomington Stock Center for generously providing fly strains and antibodies. We thank L. Waltzer for sharing unpublished data. We thank H. Toledano for comments on the manuscript and the Kurant laboratory members for constructive criticism and support. We also thank E. Suss-Toby and L. Liba at the Technion Interdepartmental Bioimaging facility for excellent technical support. We gratefully acknowledge financial support from Israel Science Foundation (grant 1872/15).
Supplementary Material
The Supplementary Material for this article can be found online at http://www.frontiersin.org/articles/10.3389/fimmu.2018.00266/full#supplementary-material.
References
1. Henson PM, Hume DA. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol (2006) 27(5):244–50. doi:10.1016/j.it.2006.03.005
2. Scheib JL, Sullivan CS, Carter BD. Jedi-1 and MEGF10 signal engulfment of apoptotic neurons through the tyrosine kinase Syk. J Neurosci (2012) 32(38):13022–31. doi:10.1523/JNEUROSCI.6350-11.2012
3. Elliott MR, Ravichandran KS. Death in the CNS: six-microns-under. Cell (2008) 133(3):393–5. doi:10.1016/j.cell.2008.04.014
4. Lauber K, Blumenthal SG, Waibel M, Wesselborg S. Clearance of apoptotic cells: getting rid of the corpses. Mol Cell (2004) 14(3):277–87. doi:10.1016/S1097-2765(04)00237-0
5. Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol (2007) 7(12):964–74. doi:10.1038/nri2214
6. Stuart LM, Ezekowitz RA. Phagocytosis: elegant complexity. Immunity (2005) 22(5):539–50. doi:10.1016/j.immuni.2005.05.002
7. Kinchen JM, Ravichandran KS. Journey to the grave: signaling events regulating removal of apoptotic cells. J Cell Sci (2007) 120(Pt 13):2143–9. doi:10.1242/jcs.03463
8. Kinchen JM, Ravichandran KS. Phagocytic signaling: you can touch, but you can’t eat. Curr Biol (2008) 18(12):R521–4. doi:10.1016/j.cub.2008.04.058
9. Ravichandran KS. Beginnings of a good apoptotic meal: the find-me and eat-me signaling pathways. Immunity (2011) 35(4):445–55. doi:10.1016/j.immuni.2011.09.004
10. Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol (2008) 9(10):781–95. doi:10.1038/nrm2515
11. Ulvila J, Vanha-Aho LM, Ramet M. Drosophila phagocytosis – still many unknowns under the surface. APMIS (2011) 119(10):651–62. doi:10.1111/j.1600-0463.2011.02792.x
12. Gold KS, Bruckner K. Macrophages and cellular immunity in Drosophila melanogaster. Semin Immunol (2015) 27(6):357–68. doi:10.1016/j.smim.2016.03.010
13. Ratheesh A, Belyaeva V, Siekhaus DE. Drosophila immune cell migration and adhesion during embryonic development and larval immune responses. Curr Opin Cell Biol (2015) 36:71–9. doi:10.1016/j.ceb.2015.07.003
14. Evans IR, Wood W. Drosophila blood cell chemotaxis. Curr Opin Cell Biol (2014) 30:1–8. doi:10.1016/j.ceb.2014.04.002
15. Tepass U, Fessler LI, Aziz A, Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development (1994) 120(7):1829–37.
16. Holz A, Bossinger B, Strasser T, Janning W, Klapper R. The two origins of hemocytes in Drosophila. Development (2003) 130(20):4955–62. doi:10.1242/dev.00702
17. Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity (1996) 4(5):431–43. doi:10.1016/S1074-7613(00)80410-0
18. Fadok VA, Warner ML, Bratton DL, Henson PM. CD36 is required for phagocytosis of apoptotic cells by human macrophages that use either a phosphatidylserine receptor or the vitronectin receptor (alpha v beta 3). J Immunol (1998) 161(11):6250–7.
19. Kurant E, Axelrod S, Leaman D, Gaul U. Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell (2008) 133(3):498–509. doi:10.1016/j.cell.2008.02.052
20. Park SY, Jung MY, Kim HJ, Lee SJ, Kim SY, Lee BH, et al. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ (2008) 15(1):192–201. doi:10.1038/sj.cdd.4402242
21. Kim S, Park SY, Kim SY, Bae DJ, Pyo JH, Hong M, et al. Cross talk between engulfment receptors stabilin-2 and integrin alphavbeta5 orchestrates engulfment of phosphatidylserine-exposed erythrocytes. Mol Cell Biol (2012) 32(14):2698–708. doi:10.1128/MCB.06743-11
22. Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron (2003) 38(4):567–80. doi:10.1016/S0896-6273(03)00289-7
23. Wu HH, Bellmunt E, Scheib JL, Venegas V, Burkert C, Reichardt LF, et al. Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nat Neurosci (2009) 12(12):1534–41. doi:10.1038/nn.2446
24. Iram T, Ramirez-Ortiz Z, Byrne MH, Coleman UA, Kingery ND, Means TK, et al. Megf10 is a receptor for C1Q that mediates clearance of apoptotic cells by astrocytes. J Neurosci (2016) 36(19):5185–92. doi:10.1523/JNEUROSCI.3850-15.2016
25. Purice MD, Speese SD, Logan MA. Delayed glial clearance of degenerating axons in aged Drosophila is due to reduced PI3K/Draper activity. Nat Commun (2016) 7:12871. doi:10.1038/ncomms12871
26. Shklyar B, Sellman Y, Shklover J, Mishnaevski K, Levy-Adam F, Kurant E. Developmental regulation of glial cell phagocytic function during Drosophila embryogenesis. Dev Biol (2014) 393(2):255–69. doi:10.1016/j.ydbio.2014.07.005
27. Rehorn KP, Thelen H, Michelson AM, Reuter R. A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development (1996) 122(12):4023–31.
28. Waltzer L, Bataille L, Peyrefitte S, Haenlin M. Two isoforms of Serpent containing either one or two GATA zinc fingers have different roles in Drosophila haematopoiesis. EMBO J (2002) 21(20):5477–86. doi:10.1093/emboj/cdf545
29. Bernardoni R, Vivancos V, Giangrande A. glide/gcm is expressed and required in the scavenger cell lineage. Dev Biol (1997) 191(1):118–30. doi:10.1006/dbio.1997.8702
30. Shklyar B, Levy-Adam F, Mishnaevski K, Kurant E. Caspase activity is required for engulfment of apoptotic cells. Mol Cell Biol (2013) 33(16):3191–201. doi:10.1128/MCB.00233-13
31. Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science (2006) 314(5806):1747–51. doi:10.1126/science.1134426
32. Shklover J, Mishnaevski K, Levy-Adam F, Kurant E. JNK pathway activation is able to synchronize neuronal death and glial phagocytosis in Drosophila. Cell Death Dis (2015) 6:e1649. doi:10.1038/cddis.2015.27
33. Shklyar B, Shklover J, Kurant E. Live imaging of apoptotic cell clearance during Drosophila embryogenesis. J Vis Exp (2013) 78:e50151. doi:10.3791/50151
34. Alfonso TB, Jones BW. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol (2002) 248(2):369–83. doi:10.1006/dbio.2002.0740
35. Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science (2000) 288(5463):146–9. doi:10.1126/science.288.5463.146
Keywords: Drosophila, macrophages, phagocytosis, apoptosis, SIMU, Serpent, GATA, development
Citation: Shlyakhover E, Shklyar B, Hakim-Mishnaevski K, Levy-Adam F and Kurant E (2018) Drosophila GATA Factor Serpent Establishes Phagocytic Ability of Embryonic Macrophages. Front. Immunol. 9:266. doi: 10.3389/fimmu.2018.00266
Received: 11 September 2017; Accepted: 30 January 2018;
Published: 08 March 2018
Edited by:
Francesca Granucci, Università degli studi di Milano Bicocca, ItalyReviewed by:
Alessandra Mancino, San Raffaele Hospital (IRCCS), ItalyCarole Poon, Peter MacCallum Cancer Centre, Australia
Copyright: © 2018 Shlyakhover, Shklyar, Hakim-Mishnaevski, Levy-Adam and Kurant. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Estee Kurant, ZWt1cmFudEB1bml2LmhhaWZhLmFjLmls
†These authors have contributed equally to this work.
 Evgeny Shlyakhover
Evgeny Shlyakhover Boris Shklyar2†
Boris Shklyar2† Estee Kurant
Estee Kurant