- 1Clinical Collaboration Unit Translational Immunology, German Cancer Consortium (DKTK), Department of Internal Medicine, University Hospital Tuebingen, Tuebingen, Germany
- 2DFG Cluster of Excellence 2180 ‘Image-guided and Functional Instructed Tumor Therapy’ (IFIT), University of Tuebingen, Tubingen, Germany
- 3Department of Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States
- 4Departments of Molecular Oncology and Thoracic Oncology, Robert-Bosch-Hospital Stuttgart, Stuttgart, Germany
- 5Department of Hematology, Medical Oncology and Pneumology, University Medical Center of Mainz, Mainz, Germany
Platelets have a crucial function in maintaining hemostasis. However, beyond their role in coagulation and thrombus formation, platelets have been implicated to affect various pathophysiological conditions such as infectious diseases, autoimmune disorders, and cancer. It is well-established that platelets aid local cancer growth by providing growth factors or contributing to cancer angiogenesis. In addition, they promote metastasis, among others by facilitation of tumor cell-extravasation and epithelial-to-mesenchymal-like transition as well as protecting metastasizing cancer cells from immunosurveillance. A variety of membrane-bound and soluble platelet-derived factors are involved in these processes, and many aspects of platelet biology in both health and disease are regulated by platelet-associated metalloproteinases and their inhibitors. Platelets synthesize (i) members of the matrix metalloproteinase (MMP) family and also inhibitors of MMPs such as members of the “tissue inhibitor of metalloproteinases” (TIMP) family as well as (ii) members of the “a disintegrin and metalloproteinase” (ADAM) family including ADAM10. Notably, platelet-associated metalloproteinase activity not only influences functions of platelets themselves: platelets can also induce expression and/or release of metalloproteinases e.g., in leukocytes or cancer cells, and ADAMs are emerging as important components by which platelets directly affect other cell types and function. This review outlines the function of metalloproteinases in platelet biology with a focus on ADAM10 and discusses the role of platelet-derived metalloproteinases in the interaction of platelets with components of the immune system and/or cancer cells.
Introduction
The main function of platelets in the healthy individual is maintenance of hemostasis, i.e., prevention of blood loss and protection of vascular integrity. However, the presence of platelets can, under pathophysiological circumstances, be also unfavorable. Platelets are the main culprits during arterial thrombosis causing tissue ischemia with widespread consequences for the affected individual. Moreover, the presence of platelets is also exploited by cancers to aid both their local progression as well as formation of metastasis. Cancer cells entering the blood stream are rapidly surrounded/coated by platelets, leading to enhanced survival of circulating tumor cells and facilitation of metastasis [reviewed in (1, 2)]. The underlying mechanisms are multifaceted and comprise, among others, supply of growth factors, contribution to endothelial adhesion, and mediation of an epithelial-to-mesenchymal-like transition (EMT-like) (3). This is exemplified by studies in mice where metastasis-formation is inhibited in the absence of platelets (4, 5). Additional depletion of natural killer (NK) cells reverted the anti-metastatic effect of thrombocytopenia, and it was assumed that coating platelets provide mechanical shielding from NK cell attack (6). However, recent evidence revealed the involvement of more complex mechanisms such as conferring of a pseudo-self-phenotype (7, 8). A variety of platelet-derived molecules are implicated in these observations. Among them are members of the metalloproteinase (MP) family such as matrix metalloproteinases (MMPs). While the role of MMPs in platelets in the context of cancer and immunity has been reviewed elsewhere (9), platelets also express other MP family-members such as “a disintegrin and metalloproteinase” (ADAM)s. Given the important role of ADAMs in immunity (10) and cancer (11), the present review highlights the relevance of platelet-derived ADAMs, in particular ADAM10 (herein referred to as pADAMs and pADAM10, respectively) for the role of platelets in cancer and tumor immunology.
ADAMs in Platelets
In humans, 24 ADAM genes including four pseudogenes have been described which give rise to 20 Type-I transmembrane proteins representing the functional members of the ADAM family (excluding the ADAM-like soluble ADAMDEC-1 protease) (12–14). They share a common structure comprising a signal peptide at the N-terminus followed by a prodomain, the (catalytically active) metalloproteinase domain, a disintegrin and a cysteine-rich domain, an EGF-like domain (except in ADAM10 and ADAM17), the transmembrane domain and a cytosplasmic tail at the C-terminus (12). Of note, only 12 ADAM-family members bear a catalytically active site in their metalloproteinase domain, suggesting that ADAM proteins may also exert non-proteolytic functions such as modulating cell-cell interaction through their integrin-binding disintegrin domain (12, 13). Of the proteolytically active ADAMs, ADAM10, and ADAM17 have most extensively been investigated (12) ADAM10 and ADAM17 are vital in a plethora of biological processes, and mice carrying a classical knockout of either protease die during embryonic development (15, 16). Information about the presence of ADAM-family members in platelets is derived from analyses of the platelet-proteome. Studies of the complete platelet-proteome by several investigators point to the presence of pADAM9 and pADAM10 (17, 18), whereas other groups did not detect any ADAM-family members (19, 20). Platelet membrane enrichment techniques ultimately confirmed the presence of pADAM9 and pADAM10 and additionally lead to the identification of pADAM17 (21, 22). Notably, pADAM10 is among the 40 most abundant membrane-associated proteins at ~2,000–4,000 molecules/platelet (22). In addition, RNA-sequencing data revealed that platelets also seem to carry significant amounts of ADAM9 and ADAM10 transcripts, which may serve as an additional pool for translation e.g., upon platelet-activation (23). These data are corroborated by mechanistic studies describing substrates of pADAM10 and pADAM17 in platelets and confirming the expression of proteolytically active proteases. Mice with a platelet-specific knockout of ADAM10 and/or an ADAM17 knockout confined to the hematopoietic system have been generated (24). These mice are viable, have normal platelet counts and represent an ideal model to investigate the role of pADAMs under physiological and pathophysiological circumstances. The function of pADAM9 in platelets, however, is less well-established. Classical ADAM9-knockout mice display no apparent phenotypical changes compared to wildtype mice, arguing against a major role of ADAM9 under physiological circumstances (25). One in-vitro study suggests that the disintegrin-domain of pADAM9 may interact with tumor-cell expressed integrins such as αVβ3, thereby mediating the recruitment of tumor cell-platelet conjugates to collagen (26). Collagen induced platelet activation itself was not dependent on ADAM9 (26). Along this line, studies of the ADAM9-downregulating micro-RNA miR-126 in CD34+ derived platelet-like structures and in the megakaryoblastic cell line MEG-01 confirmed ADAM9-mRNA expression, but also did not identify a major role of pADAM9 in platelet activation (27, 28). This is, as already stated above, in stark contrast to pADAM10 and pADAM17, which play major roles in platelet biology.
Function of (p)ADAM10
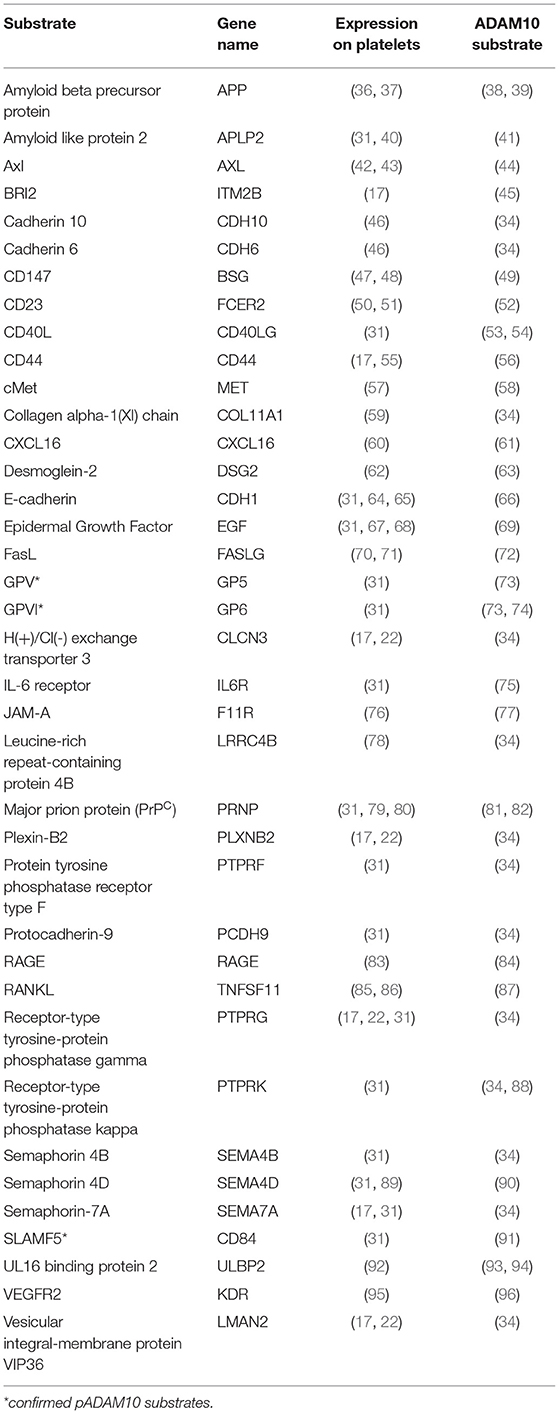
For ADAM10, which is at the center of this review, over 40 different substrates have been described (29, 30). Many of these are expressed on platelets. A comprehensive mass spectrometry-based analysis of proteins in the supernatant of activated platelets identified over 1,000 proteins, of which 69 are membrane anchored and therefore potentially accessible to proteolytic shedding by pADAM10 (31). Of these 69 proteins, shedding of 7 substrates was abrogated by ADAM17 inhibition (31), leaving 62 proteins to be potentially shed by pADAM10. We performed a PubMed literature search for each of these 62 platelet-expressed proteins to select for confirmed substrates of ADAM10. In order to obtain an unbiased list of potential substrates we then started with confirmed ADAM10 substrates (29, 30, 32–35) and individual publications and performed a second PubMed literature search to select for those proteins which are reportedly expressed on platelets. Of note, we here summarize studies deriving from different experimental strategies without applying restrictions e.g., a certain abundance in mass spec studies. Using this approach, we identified a total of 35 putative and 3 confirmed substrates of pADAM10 on platelets (Table 1). The pADAM10 sheddome may still be larger and contain proteins that could not be detected by the mass spectrometry-based techniques employed. Future studies however will be indispensable to validate whether and to which extent these putative substrates are indeed cleaved by pADAM10 as compared to ADAM10 from other sources or different sheddases.
Canonical Shedding Activity of pADAM10
pADAM10's classical mode of action (canonical shedding) is proteolytic cleavage of platelet-expressed substrates by membrane-anchored pADAM10. As of yet, established, i.e., experimentally confirmed, substrates of pADAM10 substrates in platelets primarily comprise molecules relevant for hemostasis (Figure 1A). ADAM10 is the main sheddase of the glycoproteins (GP)V (73) and GPVI (73, 97). GPVI is the collagen receptor of platelets and responsible for collagen-induced platelet activation (98), while GPV is part of the GPIb-IX-V complex, which is responsible for platelet activation upon encounter of vWF or upon binding of P-Selectin or αMβ2 expressed on endothelial cells or on neutrophils, respectively (99). ADAM10 activity is thus regulating key functions of platelet biology, although the role of shedded GP-fragments is not clear, neither under physiological conditions nor in disease (30). Soluble GPVI fragments can be detected in the blood (100) and a synthetic soluble GPVI fragment inhibited in vitro and in vivo platelet activation upon exposure to collagen (101). Of note, GPVI has been shown to promote metastasis in a mouse model (102) and it would thus be interesting to investigate how ADAM10 affects metastasis. Another confirmed substrate of pADAM10 is signaling lymphocyte activation molecule family member 5 (SLAMF5)/CD84, the exact function of which in platelet biology is currently unknown (91). No function of soluble SLAMF5/CD84 has been detected yet. SLAMF5/CD84 displays homophilic interaction and signals via EAT-2 and SAP (103). SLAMF5/CD84 in platelets seems to promote platelet aggregation by homophilic interaction (104), but Slamf5/Cd84−/− mice did not display any phenotype regarding in vitro platelet activation and/or impaired hemostasis in vivo (105). Moreover, platelets express other known ADAM10 substrates which are not yet experimentally confirmed to be shed by pADAM10. These comprise molecules involved in signaling (PTPRF, PTPRK, PTPRG), adhesion (E-cadherin, Cadherin 6/10, CD44) or immunomodulation (CD40L, IL-6 receptor, RANKL, FasL, ULBP2) (see Table 1).
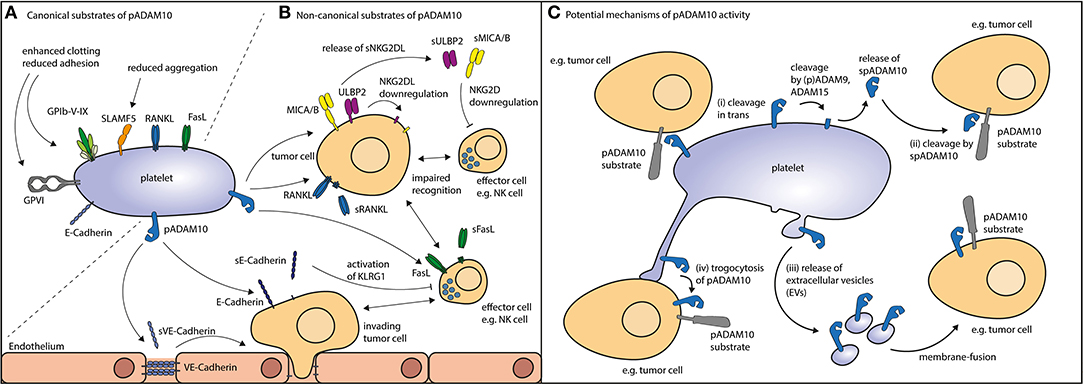
Figure 1. Potential routes of action for pADAM10. (A) Canonical substrates of pADAM10 (see Table 1) not only comprise molecular players involved in platelet function as exemplified by glycoproteins and SLAMF5 but also a plethora of immuoregulatory factors alike FasL and RANKL. (B) Non-canonical substrates of pADAM10 on other cells have been suggested to comprise NKG2DL MICA/B, ULBP2, and ectodomain shedding thereof reportedly dampens NK antitumor reactivity. Proteolytic cleavage of E-Cadherin (resulting in soluble E-Cadherin, sE-Cadherin) may inhibit NK effector functions. Cleavage of VE-Cadherin by pADAM10 may interfere with endothelial junctions thereby facilitating tumor invasion. Moreover, immunomodulation may be mediated by pADAM10 shedding of FasL from the surface of immune effector cells or shedding or RANKL e.g., from the surface of tumor cells. (C) Thus, pADAM10 may besides exerting proteolytic activity against substrates within the same membrane (cis), mediate (i) cleavage in trans (in the membrane of other cellular compounds). Alternatively, pADAM10 may be released as (ii) soluble form (spADAM10), (iii) contained in extracellular vesicles, or (iv) integrate in the membrane of the substrate-expressing cell.
Non-canonical Shedding Activity of pADAM10
pADAM10 may also exert proteolytic activity beyond the canonical shedding, i.e., shedding of platelet-derived substrates by membrane bound pADAM10 which we here summarize as non-canonical shedding of pADAM10. Interestingly, ADAM10 itself is subject to shedding by ADAM9 and ADAM15 (106, 107). Platelets express ADAM9, and analysis of the platelet sheddome has detected soluble pADAM10 (spADAM10) thereby further corroborating these results (31). Whether the soluble form of ADAM10 is proteolytically active is still a matter of debate (108), but clearly opens up a new channel for effects of pADAM10. Analysis of the degradome of murine soluble ADAM10 has been performed and suggested that soluble ADAM10 is in fact biological active (109). Another potential route of action might be cleavage of ADAM substrates in trans, this is when substrate and enzyme are located on different cells. While this is unusual for ADAM-proteases such effects have been described for ADAM10 in the context of Ephrin cleavage (110). In addition, platelets alike leukocytes and tumor cells can release extracellular vesicles (EVs), including exosomes and microvesicles/microparticles, which may be derived from resting as well as activated platelets (111, 112). Interestingly, the main source of EVs found in the plasma are in fact platelets and megakaryocytes (113, 114). Platelet-EVs are increasingly recognized as modulators of immunologic processes (115, 116). In the context of rheumatoid arthritis, platelet-EVs have been found to increase joint inflammation by eliciting an immune response from synovial fibroblasts (115). In addition, recent evidence indicates that platelet-EVs shape the function of regulatory T cells upon P-selectin dependent binding by yet not fully defined mechanisms (116). Of note, ADAM10 may reportedly be active in EVs, thereby being able to exert protease activity (117). Thus, it is tempting to speculate, that platelet-EVs carry active pADAM10 and act on cells other than platelets themselves. Another means to exert shedding on other cells could be facilitated by integration of pADAM10 into the membrane of cellular compounds, e.g., tumor cells (trogocytosis), which we could demonstrate to occur after tumor cell–platelet interaction, albeit not specifically for pADAM10 (7).
In the context of tumor-immunosurveillance, pADAM10 has not only been identified as sheddase of canonical substrates on platelets, but also to be involved in cleavage of the stress-induced NKG2D ligands (NKG2DL) MICA, MICB and ULBP2 from the surface of tumor cells (118, 119) (Figure 1B). Expression of NKG2DL is sensed by cytotoxic lymphocytes such as NK cells that express the cognate activating immunoreceptor NKG2D. Shedding of NKG2DL by ADAM10 (and/or ADAM17) in turn impairs tumor-cell lysis by NK cells. We recently observed reduced surface expression of NKG2DL on tumor cells upon coculture with platelets or exposure to platelet releasate (120). This was mirrored by enhanced detection of soluble NKG2DL, pointing to increased shedding of the surface expressed ligands. These observations were also confirmed by another group evaluating different tumor entities (121). Alike Cluxton and coworkers, we did not find enhanced mRNA-levels of tumor-ADAM10 (or ADAM17) upon platelet coating. We thus concluded that pADAM10 (or pADAM17) might be responsible for the enhanced shedding and may act via one (or more) of the suggested non-canonical shedding pathways: (i) cleavage exerted in trans or (ii) cleavage mediated by soluble (s)pADAM10, (iii) conferred by platelet-derived EVs, or (iv) integration of pADAM10 in the substrate-bearing cell (trogocytosis).
Other interesting non-canonical pADAM10 substrates could be ligands for the epidermal growth factor receptor (EGFR) (122). There are seven well-established EGFR ligands, which are also expressed on tumor cells, and release of their ectodomains via shedding supports tumor progression in an auto/paracrine manner (123). Two (EGF and betacellulin) of the seven EGFR ligands are established substrates of ADAM10 (69) and shedding thereof could thus enhance existing EGFR signaling. Interestingly, EGF itself is expressed on platelets (Table 1) and its release by tumor-coating platelets was shown to support key features of tumor progression (67). In this study, however, release of platelet-derived EGF was shown to depend mainly on platelet derived ADAMDEC-1 rather than pADAM10 (67, 124).
The chemokine CX3CL1 (fractalkine) constitutes another putative target of pADAM10 and exists either as a membrane-anchored form or is released via ectodomain shedding mediated by ADAM10 (125). CX3CL1 is mainly expressed on endothelial cells (126) but can also be found on tumor cell lines of several entities (127, 128). Its cognate receptor CX3CR1 is expressed on NK cells and T cells (129) but also on tumor cells (130). Of note, platelets express CX3CR1 binding of which was shown to mediate platelet activation and enhance adhesion to collagen and fibrinogen (131). In the context of NK cell or T cell mediated immunosurveillance of cancer cells, signaling via the CX3CR1/CX3CL1 axis might either aid or impair cancer progression [reviewed in (129)]. How pADAM10 could modulate the CX3CR1/CX3CL1 axis in this context has not yet been investigated.
Despite putative shedding of EGFR ligands or chemokines, immunomodulation via the above mentioned non-canonical modes of action may as well be executed by pADAM10 on several other substrates alike FasL, RANKL, E-Cadherin, VE-Cadherin (70, 71, 85, 132, 133) (Figure 1C).
Platelet Mediated Activation/Inhibition of (p)ADAM10
The activity of (p)ADAM10 is highly regulated on multiple levels including transcription, translation and posttranslation [reviewed in (108, 134, 135)] Once translated, full-length ADAM10 trafficks through the secretory pathway where it is activated by removal of its prodomain (136). Substrate-access to the catalytic site of mature ADAM10 is still tightly regulated through conformational changes involving its cysteine-rich domain (137). The process of ADAM10 trafficking/maturation is closely connected to the Tetraspanin (Tspan)C8 group (30). This group of six transmembrane proteins includes Tspan5, 10, 14, 15, 17, and 33 which all have been shown to associate with ADAM10, thereby promoting its trafficking to the cell surface as well as its maturation (138). Accumulating evidence indicates that ADAM10 maintains its association with TspanC8 members and that different TspanC8 partners may guide ADAM10-activity toward different substrates suggesting also interesting therapeutic implications (30). The TspanC8 members 14, 15, and 33 have also been identified in human platelets (139, 140) and Tspan33 is among the 40 most frequent platelet-membrane proteins (22). Platelet-expressed TspanC8 members might therefore play an important role in pADAM10 shedding events (30). As yet only indirect evidence, based on transfectants, is available suggesting Tspan14 as a negative regulator of GPVI shedding (140). ADAM10 activity might additionally be modulated by the dynamic composition of the lipid-bilayer by mechanisms such as trapping of substrates within cholesterin-rich lipid-rafts which are usually devoid of ADAM10 [reviewed in (141)]. Moreover, Ca2+ and calmodulin are important regulators of ADAM10 activity (142) also in platelets (135). It was suggested that pro-ADAM10 associates with calmodulin and that Ca2+ influx allows ADAM10-maturation by disrupting this association (56). Likewise, in platelets, Ca2+ influx has been shown to rapidly and potently increase pADAM10 reactivity (74). Of note, calmodulin was also shown to associate with substrates of pADAM10 such as GPVI on platelets thereby putatively negatively regulating their proteolysis (73, 143). Increase of intracellular Ca2+ thus represents a mechanism to rapidly regulate (p)ADAM10-mediated shedding events although the exact underlying processes are not yet fully elucidated.
However, it should be considered that platelet-derived factors could also influence the ADAM10 activity in the platelet-microenvironment. TGFβ has been shown to enhance transcription of Adam10 (and Adam17) in renal cells (144), and platelets can release large amounts of active TGFβ (145). However, neither Cluxton nor our group detected enhanced ADAM10 (or ADAM17) transcription in tumor cells upon exposure to platelets (or platelet releasate). This suggests that specific mechanisms govern transcription of ADAM10 in distinct cell-types. Moreover, it has been shown that thrombin activates PAR-1 receptors on endothelial cells, leading to increased activity of ADAM10 which enhanced cleavage of VE-cadherin leading to facilitation of T cell transmigration (133). Similarly, one might speculate that thrombin released by platelet-covered tumor cells might also facilitate transmigration of tumor cells during metastasis. It should be noted that tumor cells can also express PAR-1, and overexpression of PAR-1 has been implicated in enhanced metastasis (146, 147). PAR-1 signaling in tumor cells has been shown to enhance metastasis in experimental models (148), which is attributed to initiation of an EMT-program (149). The source of thrombin, the main activator of PAR-1, was suggested to be the tumor cells themselves (148), but, in the context of metastasizing tumor cells, coating platelets might additionally contribute to thrombin generation (150). Since ADAM10 is also activated by thrombin as described above (133) and has been implicated in enhanced tumor cell invasiveness (151), it appears possible that in fact activated platelets contribute to tumor cell invasiveness by regulation of ADAM10 via thrombin/PAR-1 signaling (138). ADAM10 is inhibited by TIMP-1 and TIMP-3, with TIMP-1 being inhibitory at lower concentration compared to TIMP-3 (53). Platelets contain, among other TIMPs, TIMP-1, and TIMP-3, which are independently stored in distinct platelet compartments, but are rapidly and completely released upon activation with thrombin (152). Another physiological inhibitor of ADAM10 is the membrane-anchored RECK (153) and has been detected in the platelet membrane proteome (22). The reason for the presence of both ADAM10 and its inhibitors in platelets remains to be thoroughly elucidated and suggests complex context-specific regulation of protease-activity.
Conclusion and Perspective
Platelets are associated with pathophysiology and may influence efficacy of systemic treatment in several diseases. It is thus an appealing idea to take advantage of platelet-derived biomarkers for lipid biopsies (113), especially since platelets may display a malignant phenotype (154). In line, both platelets and platelet-derived EVs have been suggested as putative biomarker (155, 156). The soluble form of ADAM10 may still exert proteolytic activity, however it remains unclear under which instances or toward which targets (108). The potential therapeutic targeting of ADAM10 activity is, while promising in many instances, likely to result also in side effects, given its variety of biological substrates. The closely related ADAM17 which is also expressed on platelets may exert similar activity, and targeting ADAM10 may thus also affect cleavage by ADAM17, especially since (i) hypothetically specific inhibitors still show cross-inhibition of ADAM17 and (ii) ADAM17 may compensate for a lack of ADAM10 shedding events (157). Since distinct tetraspanins are thought to guide ADAM10 substrate shedding, targeting of a specific tetraspanin-ADAM10 complex may thus constitute a promising approach to modulate ADAM10 proteolytic activity toward defined substrates. Further studies unraveling the regulation of ADAM10 activity, also with regard to protease localization (membrane bound, EV, or as soluble truncated form) and shedding of specific substrates are certainly warranted. ADAM10 exerts protease activity on a plethora of factors relevant beyond hemostasis. With this review, we aimed to provide a timely overview about the knowledge of (p)ADAM10, its substrates on platelets and on their microenvironment.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication. Conceptualization: H-GK, HS, KK, and SM. Writing-original draft preparation: KK and SM. Writing-review and editing: H-GK and HS. Visualization: KK.
Funding
This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy—EXC 2180—39090067. SM was supported by the Institutional Strategy of the University of Tuebingen (Deutsche Forschungsgemeinschaft, ZUK 63) and the Deutsche Forschungsgemeinschaft, MA 8774/1-1. KK was supported by the Mainz Research School of Translational Biomedicine (TransMed) of the University of Mainz. HS was funded by Deutsche Forschungsgemeinschaft, SA1360/7-3, Wilhelm Sander-Stiftung, 2007.115.3, and Deutsche Krebshilfe 70112914. We acknowledge support by Deutsche Forschungsgemeinschaft and Open Access Publishing Fund of University of Tuebingen.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Gay LJ, Felding-Habermann B. Contribution of platelets to tumour metastasis. Nat Rev Cancer. (2011) 11:123–34. doi: 10.1038/nrc3004
2. Leblanc R, Peyruchaud O. Metastasis: new functional implications of platelets and megakaryocytes. Blood. (2016) 128:24–31. doi: 10.1182/blood-2016-01-636399
3. Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. (2011) 20:576–90. doi: 10.1016/j.ccr.2011.09.009
4. Gasic GJ, Gasic TB, Stewart CC. Antimetastatic effects associated with platelet reduction. Proc Natl Acad Sci USA. (1968) 61:46–52. doi: 10.1073/pnas.61.1.46
5. Camerer E, Qazi AA, Duong DN, Cornelissen I, Advincula R, Coughlin SR. Platelets, protease-activated receptors, and fibrinogen in hematogenous metastasis. Blood. (2004) 104:397–401. doi: 10.1182/blood-2004-02-0434
6. Nieswandt B, Hafner M, Echtenacher B, Mannel DN. Lysis of tumor cells by natural killer cells in mice is impeded by platelets. Cancer Res. (1999) 59:1295–300.
7. Placke T, Orgel M, Schaller M, Jung G, Rammensee HG, Kopp HG, et al. Platelet-derived MHC class I confers a pseudonormal phenotype to cancer cells that subverts the antitumor reactivity of natural killer immune cells. Cancer Res. (2012) 72:440–8. doi: 10.1158/0008-5472.CAN-11-1872
8. Placke T, Kopp HG, Salih HR. Modulation of natural killer cell anti-tumor reactivity by platelets. J Innate Immun. (2011) 3:374–82. doi: 10.1159/000323936
9. Seizer P, May AE. Platelets and matrix metalloproteinases. Thromb Haemost. (2013) 110:903–9. doi: 10.1160/TH13-02-0113
10. Lambrecht BN, Vanderkerken M, Hammad H. The emerging role of ADAM metalloproteinases in immunity. Nat Rev Immunol. (2018) 18:745–58. doi: 10.1038/s41577-018-0068-5
11. Duffy MJ, McKiernan E, O'Donovan N, McGowan PM. Role of ADAMs in cancer formation and progression. Clin Cancer Res. (2009) 15:1140–4. doi: 10.1158/1078-0432.CCR-08-1585
12. Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. (2008) 29:258–89. doi: 10.1016/j.mam.2008.08.001
13. Weber S, Saftig P. Ectodomain shedding and ADAMs in development. Development. (2012) 139:3693–709. doi: 10.1242/dev.076398
14. Brocker CN, Vasiliou V, Nebert DW. Evolutionary divergence and functions of the ADAM and ADAMTS gene families. Hum Genomics. (2009) 4:43–55. doi: 10.1186/1479-7364-4-1-43
15. Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, et al. An essential role for ectodomain shedding in mammalian development. Science. (1998) 282:1281–4. doi: 10.1126/science.282.5392.1281
16. Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, et al. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci. (2010) 30:4833–44. doi: 10.1523/JNEUROSCI.5221-09.2010
17. Burkhart JM, Vaudel M, Gambaryan S, Radau S, Walter U, Martens L, et al. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood. (2012) 120:e73–82. doi: 10.1182/blood-2012-04-416594
18. Qureshi AH, Chaoji V, Maiguel D, Faridi MH, Barth CJ, Salem SM, et al. Proteomic and phospho-proteomic profile of human platelets in basal, resting state: insights into integrin signaling. PLoS ONE. (2009) 4:e7627. doi: 10.1371/journal.pone.0007627
19. O'Neill EE, Brock CJ, von Kriegsheim AF, Pearce AC, Dwek RA, Watson SP, et al. Towards complete analysis of the platelet proteome. Proteomics. (2002) 2:288–305. doi: 10.1002/1615-9861(200203)2:3<288::AID-PROT288>3.0.CO;2-0
20. Garcia A, Prabhakar S, Brock CJ, Pearce AC, Dwek RA, Watson SP, et al. Extensive analysis of the human platelet proteome by two-dimensional gel electrophoresis and mass spectrometry. Proteomics. (2004) 4:656–68. doi: 10.1002/pmic.200300665
21. Senis YA, Tomlinson MG, Garcia A, Dumon S, Heath VL, Herbert J, et al. A comprehensive proteomics and genomics analysis reveals novel transmembrane proteins in human platelets and mouse megakaryocytes including G6b-B, a novel immunoreceptor tyrosine-based inhibitory motif protein. Mol Cell Proteomics. (2007) 6:548–64. doi: 10.1074/mcp.D600007-MCP200
22. Lewandrowski U, Wortelkamp S, Lohrig K, Zahedi RP, Wolters DA, Walter U, et al. Platelet membrane proteomics: a novel repository for functional research. Blood. (2009) 114:e10–9. doi: 10.1182/blood-2009-02-203828
23. Rowley JW, Oler AJ, Tolley ND, Hunter BN, Low EN, Nix DA, et al. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes. Blood. (2011) 118:e101–11. doi: 10.1182/blood-2011-03-339705
24. Bender M, Hofmann S, Stegner D, Chalaris A, Bosl M, Braun A, et al. Differentially regulated GPVI ectodomain shedding by multiple platelet-expressed proteinases. Blood. (2010) 116:3347–55. doi: 10.1182/blood-2010-06-289108
25. Weskamp G, Cai H, Brodie TA, Higashyama S, Manova K, Ludwig T, et al. Mice lacking the metalloprotease-disintegrin MDC9 (ADAM9) have no evident major abnormalities during development or adult life. Mol Cell Biol. (2002) 22:1537–44. doi: 10.1128/MCB.22.5.1537-1544.2002
26. Cominetti MR, Martin AC, Ribeiro JU, Djaafri I, Fauvel-Lafeve F, Crepin M, et al. Inhibition of platelets and tumor cell adhesion by the disintegrin domain of human ADAM9 to collagen I under dynamic flow conditions. Biochimie. (2009) 91:1045–52. doi: 10.1016/j.biochi.2009.05.012
27. Kaudewitz D, Skroblin P, Bender LH, Barwari T, Willeit P, Pechlaner R, et al. Association of microRNAs and YRNAs with platelet function. Circul Res. (2016) 118:420–32. doi: 10.1161/CIRCRESAHA.114.305663
28. Garcia A, Dunoyer-Geindre S, Zapilko V, Nolli S, Reny JL, Fontana P. Functional validation of microRNA-126-3p as a platelet reactivity regulator using human haematopoietic stem cells. Thromb Haemost. (2019) 119:254–63. doi: 10.1055/s-0038-1676802
29. Reiss K, Saftig P. The “a disintegrin and metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol. (2009) 20:126–37. doi: 10.1016/j.semcdb.2008.11.002
30. Matthews AL, Noy PJ, Reyat JS, Tomlinson MG. Regulation of A disintegrin and metalloproteinase (ADAM) family sheddases ADAM10 and ADAM17: the emerging role of tetraspanins and rhomboids. Platelets. (2017) 28:333–41. doi: 10.1080/09537104.2016.1184751
31. Fong KP, Barry C, Tran AN, Traxler EA, Wannemacher KM, Tang HY, et al. Deciphering the human platelet sheddome. Blood. (2011) 117:e15–26. doi: 10.1182/blood-2010-05-283838
32. Crawford HC, Dempsey PJ, Brown G, Adam L, Moss ML. ADAM10 as a therapeutic target for cancer and inflammation. Curr Pharm Des. (2009) 15:2288–99. doi: 10.2174/138161209788682442
33. Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. (2009) 20:164–74. doi: 10.1016/j.semcdb.2008.09.005
34. Kuhn PH, Colombo AV, Schusser B, Dreymueller D, Wetzel S, Schepers U, et al. Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. Elife. (2016) 5:e12748. doi: 10.7554/eLife.12748
35. Saftig P, Lichtenthaler SF. The alpha secretase ADAM10: A metalloprotease with multiple functions in the brain. Prog Neurobiol. (2015) 135:1–20. doi: 10.1016/j.pneurobio.2015.10.003
36. Canobbio I, Visconte C, Momi S, Guidetti GF, Zara M, Canino J, et al. Platelet amyloid precursor protein is a modulator of venous thromboembolism in mice. Blood. (2017) 130:527–36. doi: 10.1182/blood-2017-01-764910
37. Van Nostrand WE, Schmaier AH, Farrow JS, Cunningham DD. Protease nexin-II (amyloid beta-protein precursor): a platelet alpha-granule protein. Science. (1990) 248:745–8. doi: 10.1126/science.2110384
38. Postina R, Schroeder A, Dewachter I, Bohl J, Schmitt U, Kojro E, et al. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Invest. (2004) 113:1456–64. doi: 10.1172/JCI20864
39. Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, et al. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci USA. (1999) 96:3922–7. doi: 10.1073/pnas.96.7.3922
40. Maynard DM, Heijnen HF, Horne MK, White JG, Gahl WA. Proteomic analysis of platelet alpha-granules using mass spectrometry. J Thromb Haemost. (2007) 5:1945–55. doi: 10.1111/j.1538-7836.2007.02690.x
41. Hogl S, Kuhn PH, Colombo A, Lichtenthaler SF. Determination of the proteolytic cleavage sites of the amyloid precursor-like protein 2 by the proteases ADAM10, BACE1 and gamma-secretase. PLoS ONE. (2011) 6:e21337. doi: 10.1371/journal.pone.0021337
42. Zhou J, Yang A, Wang Y, Chen F, Zhao Z, Davra V, et al. Tyro3, Axl, and Mertk receptors differentially participate in platelet activation and thrombus formation. Cell Commun Signal. (2018) 16:98. doi: 10.1186/s12964-018-0308-0
43. Angelillo-Scherrer A, de Frutos P, Aparicio C, Melis E, Savi P, Lupu F, et al. Deficiency or inhibition of Gas6 causes platelet dysfunction and protects mice against thrombosis. Nat Med. (2001) 7:215–21. doi: 10.1038/84667
44. Miller MA, Oudin MJ, Sullivan RJ, Wang SJ, Meyer AS, Im H, et al. Reduced proteolytic shedding of receptor tyrosine kinases is a post-translational mechanism of kinase inhibitor resistance. Cancer Discov. (2016) 6:382–99. doi: 10.1158/2159-8290.CD-15-0933
45. Martin L, Fluhrer R, Reiss K, Kremmer E, Saftig P, Haass C. Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b. J Biol Chem. (2008) 283:1644–52. doi: 10.1074/jbc.M706661200
46. Maguire PB, Donlon T, Parsons M, Wynne K, Dillon E, Ni Ainle F, et al. Proteomic analysis reveals a strong association of β-catenin with cadherin adherens junctions in resting human platelets. Proteomics. (2018) 18:e1700419. doi: 10.1002/pmic.201700419
47. Pennings GJ, Yong AS, Wong C, Al-Tamimi M, Gardiner EE, Andrews RK, et al. Circulating levels of soluble EMMPRIN (CD147) correlate with levels of soluble glycoprotein VI in human plasma. Platelets. (2014) 25:639–42. doi: 10.3109/09537104.2013.852660
48. Schmidt R, Bultmann A, Fischel S, Gillitzer A, Cullen P, Walch A, et al. Extracellular matrix metalloproteinase inducer (CD147) is a novel receptor on platelets, activates platelets, and augments nuclear factor kappaB-dependent inflammation in monocytes. Circul Res. (2008) 102:302–9. doi: 10.1161/CIRCRESAHA.107.157990
49. Wu B, Cui J, Yang XM, Liu ZY, Song F, Li L, et al. Cytoplasmic fragment of CD147 generated by regulated intramembrane proteolysis contributes to HCC by promoting autophagy. Cell Death Dis. (2017) 8:e2925. doi: 10.1038/cddis.2017.251
50. Capron M, Jouault T, Prin L, Joseph M, Ameisen JC, Butterworth AE, et al. Functional study of a monoclonal antibody to IgE Fc receptor (Fc epsilon R2) of eosinophils, platelets, and macrophages. J Exp Med. (1986) 164:72–89. doi: 10.1084/jem.164.1.72
51. Joseph M, Gounni AS, Kusnierz JP, Vorng H, Sarfati M, Kinet JP, et al. Expression and functions of the high-affinity IgE receptor on human platelets and megakaryocyte precursors. Eur J Immunol. (1997) 27:2212–8. doi: 10.1002/eji.1830270914
52. Weskamp G, Ford JW, Sturgill J, Martin S, Docherty AJ, Swendeman S, et al. ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat Immunol. (2006) 7:1293–8. doi: 10.1038/ni1399
53. Amour A, Knight CG, Webster A, Slocombe PM, Stephens PE, Knauper V, et al. The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. (2000) 473:275–9. doi: 10.1016/S0014-5793(00)01528-3
54. Yacoub D, Benslimane N, Al-Zoobi L, Hassan G, Nadiri A, Mourad W. CD154 is released from T-cells by a disintegrin and metalloproteinase domain-containing protein 10 (ADAM10) and ADAM17 in a CD40 protein-dependent manner. J Biol Chem. (2013) 288:36083–93. doi: 10.1074/jbc.M113.506220
55. Liu G, Liu G, Alzoubi K, Chatterjee M, Walker B, Munzer P, et al. CD44 sensitivity of platelet activation, membrane scrambling and adhesion under high arterial shear rates. Thromb Haemost. (2016) 115:99–108. doi: 10.1160/TH14-10-0847
56. Nagano O, Murakami D, Hartmann D, De Strooper B, Saftig P, Iwatsubo T, et al. Cell-matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca(2+) influx and PKC activation. J Cell Biol. (2004) 165:893–902. doi: 10.1083/jcb.200310024
57. Pietrapiana D, Sala M, Prat M, Sinigaglia F. Met identification on human platelets: role of hepatocyte growth factor in the modulation of platelet activation. FEBS Lett. (2005) 579:4550–4. doi: 10.1016/j.febslet.2005.06.072
58. Kopitz C, Gerg M, Bandapalli OR, Ister D, Pennington CJ, Hauser S, et al. Tissue inhibitor of metalloproteinases-1 promotes liver metastasis by induction of hepatocyte growth factor signaling. Cancer Res. (2007) 67:8615–23. doi: 10.1158/0008-5472.CAN-07-0232
59. Martens L, Van Damme P, Van Damme J, Staes A, Timmerman E, Ghesquiere B, et al. The human platelet proteome mapped by peptide-centric proteomics: a functional protein profile. Proteomics. (2005) 5:3193–204. doi: 10.1002/pmic.200401142
60. Seizer P, Stellos K, Selhorst G, Kramer BF, Lang MR, Gawaz M, et al. CXCL16 is a novel scavenger receptor on platelets and is associated with acute coronary syndrome. Thromb Haemost. (2011) 105:1112–4. doi: 10.1160/TH10-10-0689
61. Abel S, Hundhausen C, Mentlein R, Schulte A, Berkhout TA, Broadway N, et al. The transmembrane CXC-chemokine ligand 16 is induced by IFN-gamma and TNF-alpha and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J Immunol. (2004) 172:6362–72. doi: 10.4049/jimmunol.172.10.6362
62. Wang H, Li ZY, Liu Y, Persson J, Beyer I, Moller T, et al. Desmoglein 2 is a receptor for adenovirus serotypes 3, 7, 11 and 14. Nat Med. (2011) 17:96–104. doi: 10.1038/nm.2270
63. Bech-Serra JJ, Santiago-Josefat B, Esselens C, Saftig P, Baselga J, Arribas J, et al. Proteomic identification of desmoglein 2 and activated leukocyte cell adhesion molecule as substrates of ADAM17 and ADAM10 by difference gel electrophoresis. Mol Cell Biol. (2006) 26:5086–95. doi: 10.1128/MCB.02380-05
64. Elrod JW, Park JH, Oshima T, Sharp CD, Minagar A, Alexander JS. Expression of junctional proteins in human platelets. Platelets. (2003) 14:247–51. doi: 10.1080/0953710031000118894
65. Scanlon VM, Teixeira AM, Tyagi T, Zou S, Zhang PX, Booth CJ, et al. Epithelial (E)-cadherin is a novel mediator of platelet aggregation and clot stability. Thromb Haemost. (2019) 119:744–57. doi: 10.1055/s-0039-1679908
66. Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, et al. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci USA. (2005) 102:9182–7. doi: 10.1073/pnas.0500918102
67. Chen R, Jin G, Li W, McIntyre TM. Epidermal growth factor (EGF) autocrine activation of human platelets promotes EGF receptor-dependent oral squamous cell carcinoma invasion, migration, and epithelial mesenchymal transition. J Immunol. (2018) 201:2154–64. doi: 10.4049/jimmunol.1800124
68. Oka Y, Orth DN. Human plasma epidermal growth factor/beta-urogastrone is associated with blood platelets. J Clin Invest. (1983) 72:249–59. doi: 10.1172/JCI110964
69. Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, et al. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. (2004) 164:769–79. doi: 10.1083/jcb.200307137
70. Schleicher RI, Reichenbach F, Kraft P, Kumar A, Lescan M, Todt F, et al. Platelets induce apoptosis via membrane-bound FasL. Blood. (2015) 126:1483–93. doi: 10.1182/blood-2013-12-544445
71. Klatt C, Kruger I, Zey S, Krott KJ, Spelleken M, Gowert NS, et al. Platelet-RBC interaction mediated by FasL/FasR induces procoagulant activity important for thrombosis. J Clin Invest. (2018) 128:3906–25. doi: 10.1172/JCI92077
72. Schulte M, Reiss K, Lettau M, Maretzky T, Ludwig A, Hartmann D, et al. ADAM10 regulates FasL cell surface expression and modulates FasL-induced cytotoxicity and activation-induced cell death. Cell Death Differ. (2007) 14:1040–9. doi: 10.1038/sj.cdd.4402101
73. Gardiner EE, Karunakaran D, Shen Y, Arthur JF, Andrews RK, Berndt MC. Controlled shedding of platelet glycoprotein (GP)VI and GPIb-IX-V by ADAM family metalloproteinases. J Thromb Haemost. (2007) 5:1530–7. doi: 10.1111/j.1538-7836.2007.02590.x
74. Baaten C, Swieringa F, Misztal T, Mastenbroek TG, Feijge MAH, Bock PE, et al. Platelet heterogeneity in activation-induced glycoprotein shedding: functional effects. Blood Adv. (2018) 2:2320–31. doi: 10.1182/bloodadvances.2017011544
75. Matthews V, Schuster B, Schutze S, Bussmeyer I, Ludwig A, Hundhausen C, et al. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE). J Biol Chem. (2003) 278:38829–39. doi: 10.1074/jbc.M210584200
76. Kornecki E, Walkowiak B, Naik UP, Ehrlich YH. Activation of human platelets by a stimulatory monoclonal antibody. J Biol Chem. (1990) 265:10042–8.
77. Koenen RR, Pruessmeyer J, Soehnlein O, Fraemohs L, Zernecke A, Schwarz N, et al. Regulated release and functional modulation of junctional adhesion molecule A by disintegrin metalloproteinases. Blood. (2009) 113:4799–809. doi: 10.1182/blood-2008-04-152330
78. Wong JW, McRedmond JP, Cagney G. Activity profiling of platelets by chemical proteomics. Proteomics. (2009) 9:40–50. doi: 10.1002/pmic.200800185
79. Barclay GR, Hope J, Birkett CR, Turner ML. Distribution of cell-associated prion protein in normal adult blood determined by flow cytometry. Br J Haematol. (1999) 107:804–14. doi: 10.1046/j.1365-2141.1999.01789.x
80. Robertson C, Booth SA, Beniac DR, Coulthart MB, Booth TF, McNicol A. Cellular prion protein is released on exosomes from activated platelets. Blood. (2006) 107:3907–11. doi: 10.1182/blood-2005-02-0802
81. Linsenmeier L, Mohammadi B, Wetzel S, Puig B, Jackson WS, Hartmann A, et al. Structural and mechanistic aspects influencing the ADAM10-mediated shedding of the prion protein. Mol Neurodegener. (2018) 13:18. doi: 10.1186/s13024-018-0248-6
82. Jarosz-Griffiths HH, Corbett NJ, Rowland HA, Fisher K, Jones AC, Baron J, et al. Proteolytic shedding of the prion protein via activation of metallopeptidase ADAM10 reduces cellular binding and toxicity of amyloid-beta oligomers. J Biol Chem. (2019) 294:7085–97. doi: 10.1074/jbc.RA118.005364
83. Gawlowski T, Stratmann B, Ruetter R, Buenting CE, Menart B, Weiss J, et al. Advanced glycation end products strongly activate platelets. Eur J Nutr. (2009) 48:475–81. doi: 10.1007/s00394-009-0038-6
84. Raucci A, Cugusi S, Antonelli A, Barabino SM, Monti L, Bierhaus A, et al. A soluble form of the receptor for advanced glycation endproducts (RAGE) is produced by proteolytic cleavage of the membrane-bound form by the sheddase a disintegrin and metalloprotease 10 (ADAM10). FASEB J. (2008) 22:3716–27. doi: 10.1096/fj.08-109033
85. Clar KL, Hinterleitner C, Schneider P, Salih HR, Maurer S. Inhibition of NK reactivity against solid tumors by platelet-derived RANKL. Cancers. (2019) 11:277. doi: 10.3390/cancers11030277
86. Nakanishi T, Inaba M, Inagaki-Katashiba N, Tanaka A, Vien PT, Kibata K, et al. Platelet-derived RANK ligand enhances CCL17 secretion from dendritic cells mediated by thymic stromal lymphopoietin. Platelets. (2015) 26:425–31. doi: 10.3109/09537104.2014.920081
87. Hikita A, Yana I, Wakeyama H, Nakamura M, Kadono Y, Oshima Y, et al. Negative regulation of osteoclastogenesis by ectodomain shedding of receptor activator of NF-kappaB ligand. J Biol Chem. (2006) 281:36846–55. doi: 10.1074/jbc.M606656200
88. Anders L, Mertins P, Lammich S, Murgia M, Hartmann D, Saftig P, et al. Furin-, ADAM 10-, and gamma-secretase-mediated cleavage of a receptor tyrosine phosphatase and regulation of beta-catenin's transcriptional activity. Mol Cell Biol. (2006) 26:3917–34. doi: 10.1128/MCB.26.10.3917-3934.2006
89. Mou P, Zeng Z, Li Q, Liu X, Xin X, Wannemacher KM, et al. Identification of a calmodulin-binding domain in Sema4D that regulates its exodomain shedding in platelets. Blood. (2013) 121:4221–30. doi: 10.1182/blood-2012-11-470609
90. Shen S, Ke Y, Dang E, Fang H, Chang Y, Zhang J, et al. Semaphorin 4D from CD15(+) granulocytes via ADAM10-induced cleavage contributes to antibody production in bullous pemphigoid. J Invest Dermatol. (2018) 138:588–97. doi: 10.1016/j.jid.2017.09.037
91. Hofmann S, Vogtle T, Bender M, Rose-John S, Nieswandt B. The SLAM family member CD84 is regulated by ADAM10 and calpain in platelets. J Thromb Haemost. (2012) 10:2581–92. doi: 10.1111/jth.12013
92. Nowbakht P, Ionescu MC, Rohner A, Kalberer CP, Rossy E, Mori L, et al. Ligands for natural killer cell-activating receptors are expressed upon the maturation of normal myelomonocytic cells but at low levels in acute myeloid leukemias. Blood. (2005) 105:3615–22. doi: 10.1182/blood-2004-07-2585
93. Waldhauer I, Steinle A. Proteolytic release of soluble UL16-binding protein 2 from tumor cells. Cancer Res. (2006) 66:2520–6. doi: 10.1158/0008-5472.CAN-05-2520
94. Wolpert F, Tritschler I, Steinle A, Weller M, Eisele G. A disintegrin and metalloproteinases 10 and 17 modulate the immunogenicity of glioblastoma-initiating cells. Neuro Oncol. (2014) 16:382–91. doi: 10.1093/neuonc/not232
95. Selheim F, Holmsen H, Vassbotn FS. Identification of functional VEGF receptors on human platelets. FEBS Lett. (2002) 512:107–10. doi: 10.1016/S0014-5793(02)02232-9
96. Donners MM, Wolfs IM, Olieslagers S, Mohammadi-Motahhari Z, Tchaikovski V, Heeneman S, et al. A disintegrin and metalloprotease 10 is a novel mediator of vascular endothelial growth factor-induced endothelial cell function in angiogenesis and is associated with atherosclerosis. Arterioscler Thromb Vasc Biol. (2010) 30:2188–95. doi: 10.1161/ATVBAHA.110.213124
97. Mammadova-Bach E, Ollivier V, Loyau S, Schaff M, Dumont B, Favier R, et al. Platelet glycoprotein VI binds to polymerized fibrin and promotes thrombin generation. Blood. (2015) 126:683–91. doi: 10.1182/blood-2015-02-629717
98. Moroi M, Jung SM. Platelet glycoprotein VI: its structure and function. Thromb Res. (2004) 114:221–33. doi: 10.1016/j.thromres.2004.06.046
99. Andrews RK, Gardiner EE, Shen Y, Whisstock JC, Berndt MC. Glycoprotein Ib-IX-V. Int J Biochem Cell Biol. (2003) 35:1170–4. doi: 10.1016/S1357-2725(02)00280-7
100. Gardiner EE, Andrews RK. Platelet receptor expression and shedding: glycoprotein Ib-IX-V and glycoprotein VI. Transfus Med Rev. (2014) 28:56–60. doi: 10.1016/j.tmrv.2014.03.001
101. Massberg S, Konrad I, Bultmann A, Schulz C, Munch G, Peluso M, et al. Soluble glycoprotein VI dimer inhibits platelet adhesion and aggregation to the injured vessel wall in vivo. FASEB J. (2004) 18:397–9. doi: 10.1096/fj.03-0464fje
102. Jain S, Russell S, Ware J. Platelet glycoprotein VI facilitates experimental lung metastasis in syngenic mouse models. J Thromb Haemost. (2009) 7:1713–7. doi: 10.1111/j.1538-7836.2009.03559.x
103. Yan Q, Malashkevich VN, Fedorov A, Fedorov E, Cao E, Lary JW, et al. Structure of CD84 provides insight into SLAM family function. Proc Natl Acad Sci USA. (2007) 104:10583–8. doi: 10.1073/pnas.0703893104
104. Nanda N, Andre P, Bao M, Clauser K, Deguzman F, Howie D, et al. Platelet aggregation induces platelet aggregate stability via SLAM family receptor signaling. Blood. (2005) 106:3028–34. doi: 10.1182/blood-2005-01-0333
105. Hofmann S, Braun A, Pozgaj R, Morowski M, Vogtle T, Nieswandt B. Mice lacking the SLAM family member CD84 display unaltered platelet function in hemostasis and thrombosis. PLoS ONE. (2014) 9:e115306. doi: 10.1371/journal.pone.0115306
106. Parkin E, Harris B. A disintegrin and metalloproteinase (ADAM)-mediated ectodomain shedding of ADAM10. J Neurochem. (2009) 108:1464–79. doi: 10.1111/j.1471-4159.2009.05907.x
107. Tousseyn T, Thathiah A, Jorissen E, Raemaekers T, Konietzko U, Reiss K, et al. ADAM10, the rate-limiting protease of regulated intramembrane proteolysis of Notch and other proteins, is processed by ADAMS-9, ADAMS-15, and the gamma-secretase. J Biol Chem. (2009) 284:11738–47. doi: 10.1074/jbc.M805894200
108. Endres K, Deller T. Regulation of alpha-secretase ADAM10 in vitro and in vivo: genetic, epigenetic, and protein-based mechanisms. Front Mol Neurosci. (2017) 10:56. doi: 10.3389/fnmol.2017.00056
109. Scharfenberg F, Helbig A, Sammel M, Benzel J, Schlomann U, Peters F, et al. Degradome of soluble ADAM10 and ADAM17 metalloproteases. Cell Mol Life Sci. (2019). doi: 10.1007/s00018-019-03184-4. [Epub ahead of print].
110. Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, et al. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. (2005) 123:291–304. doi: 10.1016/j.cell.2005.08.014
111. Goubran H, Sabry W, Kotb R, Seghatchian J, Burnouf T. Platelet microparticles and cancer: an intimate cross-talk. Transfus Apher Sci. (2015) 53:168–72. doi: 10.1016/j.transci.2015.10.014
112. Cauwenberghs S, Feijge MA, Harper AG, Sage SO, Curvers J, Heemskerk JW. Shedding of procoagulant microparticles from unstimulated platelets by integrin-mediated destabilization of actin cytoskeleton. FEBS Lett. (2006) 580:5313–20. doi: 10.1016/j.febslet.2006.08.082
113. Haemmerle M, Stone RL, Menter DG, Afshar-Kharghan V, Sood AK. The platelet lifeline to cancer: challenges and opportunities. Cancer Cell. (2018) 33:965–83. doi: 10.1016/j.ccell.2018.03.002
114. Flaumenhaft R, Dilks JR, Richardson J, Alden E, Patel-Hett SR, Battinelli E, et al. Megakaryocyte-derived microparticles: direct visualization and distinction from platelet-derived microparticles. Blood. (2009) 113:1112–21. doi: 10.1182/blood-2008-06-163832
115. Boilard E, Nigrovic PA, Larabee K, Watts GF, Coblyn JS, Weinblatt ME, et al. Platelets amplify inflammation in arthritis via collagen-dependent microparticle production. Science. (2010) 327:580–3. doi: 10.1126/science.1181928
116. Dinkla S, van Cranenbroek B, van der Heijden WA, He X, Wallbrecher R, Dumitriu IE, et al. Platelet microparticles inhibit IL-17 production by regulatory T cells through P-selectin. Blood. (2016) 127:1976–86. doi: 10.1182/blood-2015-04-640300
117. Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, et al. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. (2003) 17:292–4. doi: 10.1096/fj.02-0430fje
118. Waldhauer I, Goehlsdorf D, Gieseke F, Weinschenk T, Wittenbrink M, Ludwig A, et al. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. (2008) 68:6368–76. doi: 10.1158/0008-5472.CAN-07-6768
119. Chitadze G, Lettau M, Bhat J, Wesch D, Steinle A, Furst D, et al. Shedding of endogenous MHC class I-related chain molecules A and B from different human tumor entities: heterogeneous involvement of the “a disintegrin and metalloproteases” 10 and 17. Int J Cancer. (2013) 133:1557–66. doi: 10.1002/ijc.28174
120. Maurer S, Kropp KN, Klein G, Steinle A, Haen SP, Walz JS, et al. Platelet-mediated shedding of NKG2D ligands impairs NK cell immune-surveillance of tumor cells. Oncoimmunology. (2018) 7:e1364827. doi: 10.1080/2162402X.2017.1364827
121. Cluxton CD, Spillane C, O'Toole SA, Sheils O, Gardiner CM, O'Leary JJ. Suppression of natural killer cell NKG2D and CD226 anti-tumour cascades by platelet cloaked cancer cells: Implications for the metastatic cascade. PLoS ONE. (2019) 14:e0211538. doi: 10.1371/journal.pone.0211538
122. Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. (2005) 6:32–43. doi: 10.1038/nrm1548
123. Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. (2005) 5:341–54. doi: 10.1038/nrc1609
124. Chen R, Jin G, McIntyre TM. The soluble protease ADAMDEC1 released from activated platelets hydrolyzes platelet membrane pro-epidermal growth factor (EGF) to active high-molecular-weight EGF. J Biol Chem. (2017) 292:10112–22. doi: 10.1074/jbc.M116.771642
125. Hundhausen C, Misztela D, Berkhout TA, Broadway N, Saftig P, Reiss K, et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood. (2003) 102:1186–95. doi: 10.1182/blood-2002-12-3775
126. Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, et al. A new class of membrane-bound chemokine with a CX3C motif. Nature. (1997) 385:640–4. doi: 10.1038/385640a0
127. Siddiqui I, Erreni M, van Brakel M, Debets R, Allavena P. Enhanced recruitment of genetically modified CX3CR1-positive human T cells into Fractalkine/CX3CL1 expressing tumors: importance of the chemokine gradient. J Immunother Cancer. (2016) 4:21. doi: 10.1186/s40425-016-0125-1
128. Gaudin F, Nasreddine S, Donnadieu AC, Emilie D, Combadiere C, Prevot S, et al. Identification of the chemokine CX3CL1 as a new regulator of malignant cell proliferation in epithelial ovarian cancer. PLoS ONE. (2011) 6:e21546. doi: 10.1371/journal.pone.0021546
129. D'Haese JG, Demir IE, Friess H, Ceyhan GO. Fractalkine/CX3CR1: why a single chemokine-receptor duo bears a major and unique therapeutic potential. Expert Opin Ther Targets. (2010) 14:207–19. doi: 10.1517/14728220903540265
130. Tardaguila M, Mira E, Garcia-Cabezas MA, Feijoo AM, Quintela-Fandino M, Azcoitia I, et al. CX3CL1 promotes breast cancer via transactivation of the EGF pathway. Cancer Res. (2013) 73:4461–73. doi: 10.1158/0008-5472.CAN-12-3828
131. Schafer A, Schulz C, Eigenthaler M, Fraccarollo D, Kobsar A, Gawaz M, et al. Novel role of the membrane-bound chemokine fractalkine in platelet activation and adhesion. Blood. (2004) 103:407–12. doi: 10.1182/blood-2002-10-3260
132. Hu QP, Kuang JY, Yang QK, Bian XW, Yu SC. Beyond a tumor suppressor: Soluble E-cadherin promotes the progression of cancer. Int J Cancer. (2016) 138:2804–12. doi: 10.1002/ijc.29982
133. Schulz B, Pruessmeyer J, Maretzky T, Ludwig A, Blobel CP, Saftig P, et al. ADAM10 regulates endothelial permeability and T-Cell transmigration by proteolysis of vascular endothelial cadherin. Circul Res. (2008) 102:1192–201. doi: 10.1161/CIRCRESAHA.107.169805
134. Peron R, Vatanabe IP, Manzine PR, Camins A, Cominetti MR. Alpha-secretase ADAM10 regulation: insights into Alzheimer's disease treatment. Pharmaceuticals. (2018) 11:E12. doi: 10.3390/ph11010012
135. Montague SJ, Andrews RK, Gardiner EE. Mechanisms of receptor shedding in platelets. Blood. (2018) 132:2535–45. doi: 10.1182/blood-2018-03-742668
136. Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. (2001) 15:1837–9. doi: 10.1096/fj.01-0007fje
137. Seegar TCM, Killingsworth LB, Saha N, Meyer PA, Patra D, Zimmerman B, et al. Structural basis for regulated proteolysis by the alpha-secretase ADAM10. Cell. (2017) 171:1638–48.e7. doi: 10.1016/j.cell.2017.11.014
138. Haining EJ, Yang J, Bailey RL, Khan K, Collier R, Tsai S, et al. The TspanC8 subgroup of tetraspanins interacts with A disintegrin and metalloprotease 10 (ADAM10) and regulates its maturation and cell surface expression. J Biol Chem. (2012) 287:39753–65. doi: 10.1074/jbc.M112.416503
139. Tomlinson MG. Platelet tetraspanins: small but interesting. J Thromb Haemost. (2009) 7:2070–3. doi: 10.1111/j.1538-7836.2009.03613.x
140. Noy PJ, Yang J, Reyat JS, Matthews AL, Charlton AE, Furmston J, et al. TspanC8 tetraspanins and a disintegrin and metalloprotease 10 (ADAM10) interact via their extracellular regions: Evidence for distinct binding mechanisms for different TspanC8 proteins. J Biol Chem. (2016) 291:3145–57. doi: 10.1074/jbc.M115.703058
141. Reiss K, Bhakdi S. The plasma membrane: penultimate regulator of ADAM sheddase function. Biochim Biophys Acta Mol Cell Res. (2017) 1864(11 Pt B):2082–7. doi: 10.1016/j.bbamcr.2017.06.006
142. Horiuchi K, Le Gall S, Schulte M, Yamaguchi T, Reiss K, Murphy G, et al. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell. (2007) 18:176–88. doi: 10.1091/mbc.e06-01-0014
143. Andrews RK, Suzuki-Inoue K, Shen Y, Tulasne D, Watson SP, Berndt MC. Interaction of calmodulin with the cytoplasmic domain of platelet glycoprotein VI. Blood. (2002) 99:4219–21. doi: 10.1182/blood-2001-11-0008
144. Ramdas V, McBride M, Denby L, Baker AH. Canonical transforming growth factor-beta signaling regulates disintegrin metalloprotease expression in experimental renal fibrosis via miR-29. Am J Pathol. (2013) 183:1885–96. doi: 10.1016/j.ajpath.2013.08.027
145. Kopp HG, Placke T, Salih HR. Platelet-derived transforming growth factor-beta down-regulates NKG2D thereby inhibiting natural killer cell antitumor reactivity. Cancer Res. (2009) 69:7775–83. doi: 10.1158/0008-5472.CAN-09-2123
146. Massi D, Naldini A, Ardinghi C, Carraro F, Franchi A, Paglierani M, et al. Expression of protease-activated receptors 1 and 2 in melanocytic nevi and malignant melanoma. Hum Pathol. (2005) 36:676–85. doi: 10.1016/j.humpath.2005.04.008
147. Darmoul D, Gratio V, Devaud H, Lehy T, Laburthe M. Aberrant expression and activation of the thrombin receptor protease-activated receptor-1 induces cell proliferation and motility in human colon cancer cells. Am J Pathol. (2003) 162:1503–13. doi: 10.1016/S0002-9440(10)64283-6
148. Nierodzik ML, Chen K, Takeshita K, Li JJ, Huang YQ, Feng XS, et al. Protease-activated receptor 1 (PAR-1) is required and rate-limiting for thrombin-enhanced experimental pulmonary metastasis. Blood. (1998) 92:3694–700. doi: 10.1182/blood.V92.10.3694
149. Yang E, Cisowski J, Nguyen N, O'Callaghan K, Xu J, Agarwal A, et al. Dysregulated protease activated receptor 1 (PAR1) promotes metastatic phenotype in breast cancer through HMGA2. Oncogene. (2016) 35:1529–40. doi: 10.1038/onc.2015.217
150. Zhao L, Thorsheim CL, Suzuki A, Stalker TJ, Min SH, Lian L, et al. Phosphatidylinositol transfer protein-alpha in platelets is inconsequential for thrombosis yet is utilized for tumor metastasis. Nat Commun. (2017) 8:1216. doi: 10.1038/s41467-017-01181-4
151. Sepult C, Bellefroid M, Rocks N, Donati K, Gerard C, Gilles C, et al. ADAM10 mediates malignant pleural mesothelioma invasiveness. Oncogene. (2019) 38:3521–34. doi: 10.1038/s41388-018-0669-2
152. Villeneuve J, Block A, Le Bousse-Kerdiles MC, Lepreux S, Nurden P, Ripoche J, et al. Tissue inhibitors of matrix metalloproteinases in platelets and megakaryocytes: a novel organization for these secreted proteins. Exp Hematol. (2009) 37:849–56. doi: 10.1016/j.exphem.2009.03.009
153. Muraguchi T, Takegami Y, Ohtsuka T, Kitajima S, Chandana EP, Omura A, et al. RECK modulates Notch signaling during cortical neurogenesis by regulating ADAM10 activity. Nat Neurosci. (2007) 10:838–45. doi: 10.1038/nn1922
154. Wiesner T, Bugl S, Mayer F, Hartmann JT, Kopp HG. Differential changes in platelet VEGF, Tsp, CXCL12, and CXCL4 in patients with metastatic cancer. Clin Exp Metastasis. (2010) 27:141–9. doi: 10.1007/s10585-010-9311-6
155. Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. (2015) 28:666–76. doi: 10.1016/j.ccell.2015.09.018
156. Mege D, Panicot-Dubois L, Ouaissi M, Robert S, Sielezneff I, Sastre B, et al. The origin and concentration of circulating microparticles differ according to cancer type and evolution: a prospective single-center study. Int J Cancer. (2016) 138:939–48. doi: 10.1002/ijc.29837
Keywords: platelets, tumor, immune evasion, ADAM10, ectodomain shedding
Citation: Maurer S, Kopp H-G, Salih HR and Kropp KN (2020) Modulation of Immune Responses by Platelet-Derived ADAM10. Front. Immunol. 11:44. doi: 10.3389/fimmu.2020.00044
Received: 09 December 2019; Accepted: 09 January 2020;
Published: 05 February 2020.
Edited by:
Armando Rossello, University of Pisa, ItalyReviewed by:
Andreas Ludwig, RWTH Aachen University, GermanyWilliam Parks, Cedars-Sinai Medical Center, United States
Copyright © 2020 Maurer, Kopp, Salih and Kropp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefanie Maurer, c3RlZmFuaWUubWF1cmVyQG1lZC51bmktdHVlYmluZ2VuLmRl
 Stefanie Maurer
Stefanie Maurer Hans-Georg Kopp4
Hans-Georg Kopp4 Helmut R. Salih
Helmut R. Salih Korbinian N. Kropp
Korbinian N. Kropp