- 1Neuroimmunology Unit, IRCSS Fondazione Santa Lucia, Rome, Italy
- 2Department of Biology and Biotechnology Charles Darwin, Sapienza University, Rome, Italy
T helper (Th) 17 cells are a subtype of CD4 T lymphocytes characterized by the expression of retinoic acid-receptor (RAR)-related orphan receptor (ROR)γt transcription factor, encoded by gene Rorc. These cells are implicated in the pathology of autoimmune inflammatory disorders as well as in the clearance of extracellular infections. The main function of Th17 cells is the production of cytokine called interleukin (IL)-17A. This review highlights recent advances in mechanisms regulating transcription of IL-17A. In particular, we described the lineage defining transcription factor RORγt and other factors that regulate transcription of Il17a or Rorc by interacting with RORγt or by binding their specific DNA regions, which may positively or negatively influence their expression. Moreover, we reported the eventual involvement of those factors in Th17-related diseases, such as multiple sclerosis, rheumatoid arthritis, psoriasis, and Crohn's disease, characterized by an exaggerated Th17 response. Finally, we discussed the potential new therapeutic approaches for Th17-related diseases targeting these transcription factors. The wide knowledge of transcriptional regulators of Th17 cells is crucial for the better understanding of the pathogenic role of these cells and for development of therapeutic strategies aimed at fighting Th17-related diseases.
Introduction
T helper (Th) 17 cells are a subtype of CD4 T lymphocytes, specialized in immune response against fungi and some extracellular bacteria (1–4). The interleukin (IL)-17A, originally named CTLA8, is the most representative cytokine produced by Th17 cells (3, 5, 6), also produced by cytotoxic T lymphocytes, and innate lymphocytes, including γδ T, natural killer T, and group 3 innate lymphoid cells (7).
The binding of IL-17A with its receptor activates the target cells, such as epithelial cells, endothelial cells, and fibroblasts (3, 4, 8) and induces CXCL1, CXCL2, and CXCL8, which attract myeloid cells such as neutrophils to the infected or injured tissue (9); IL-6 and G-CSF, which promote myeloid-driven innate inflammation (10); and β-defensins, S100A8, and lipocalin 2, which protect the host during acute microbial invasion (11).
In addition to IL-17A, Th17 cells produce IL-17F, IL-21, IL-22, and, in human, also IL-26 (3, 5, 6, 12), which collectively ensure an appropriate defense against pathogens. In fact, genetic defects in the Th17–cytokine pathways lead to severe mucocutaneous candidiasis (13–15).
However, a dysregulated activity of Th17 cells has been associated to autoimmune diseases, such as multiple sclerosis (MS), rheumatoid arthritis, psoriasis, and Crohn's disease (8, 16).
Given the relevance of Th17 cells in both physiological and pathological contexts, numerous studies investigated the molecular mechanisms regulating the transcriptional program of Th17 cells.
Majority of the Th17 transcription factors were discovered and validated through analysis of IL-17A expression in mice deficient for specific transcription factors, and mice containing a GFP reporter cDNA knocked-in at the site for initiation of the translation of specific transcription factors (17–21). Similarly, the in vitro expression of IL-17A was assessed in cells cotransfected with constructs overexpressing the specific transcription factors and reporter constructs containing regions upstream of the Il17a transcription start site (17, 19). More recently, modern technologies, such as chromatin immunoprecipation (ChIP) and single-cell RNA-sequencing, were allowed to better explore the functions of transcription factors in Th17 cells (22–24). However, although the expression of Th17 transcription factors was validated in human Th17 cells, most of the studies demonstrating their regulatory mechanism were performed in murine cells.
The first transcription factor discovered, designated as the “lineage defining transcription factor of Th17 cells,” is RORγt, which is essential and sufficient to induce Th17 lineage fate in both human and mouse cells (5, 17, 25).
However, succeeding studies revealed that multiple transcriptional regulators contribute to full Th17 differentiation program through several mechanisms, including binding to specific regions of Il17a and Rorc genes, or interacting and synergizing with RORγt, or facilitating the recruitment of other proteins on Il17a or Rorc promoters.
Collectively, Th17 transcriptional regulators may contribute to Th17 functions in physiological and pathological contexts. Thus, in this review, we reported recent advances on the molecular mechanisms directly regulating transcription of Il17a and Rorc. Moreover, we discussed their involvement in autoimmune disorders associated to an exaggerated Th17 response. Finally, we discussed the recent therapeutic approaches targeting Th17 transcriptional regulators in Th17-related autoimmune diseases.
Retinoic Acid-Receptor-Related Orphan Receptor (ROR) Transcription Factors in Th17 Cells
The retinoic acid-related orphan nuclear receptors (RORs) belong to a superfamily of ligand regulated transcription factors (26, 27). ROR transcription factors bind DNA response elements, called ROR response elements (ROREs) (26, 28), consisting of the consensus core motif AGGTCA preceded by a 5′ A/T-rich sequence located into regulatory regions of target genes (27).
The interaction of ROR factors with their specific ligands allows recruitment of cofactor proteins, which leads to the transcription of their target genes (29).
ROR family is composed of three members, RORα (NR1F1), RORβ (NR1F2), and RORγ (NR1F3) (30–32), encoded by Rora, Rorb, and Rorc genes, respectively. Ror genes may encode different protein isoforms, among which RORα4 and RORγt are the unique isoforms expressed in cells of the immune system (29).
Interestingly, RORγt is expressed in thymocytes at the double-positive stage of T cell development, but is absent in mature thymocytes and in mature naive T cells in spleen and peripheral lymph nodes (33). In 2006, RORγt has been detected in IL-17-producing T cells (17), and it has been shown to play a central role in Th17 differentiation (17, 34).
Precursors or derivatives of cholesterol, such as desmosterol (35) and oxysterols (36), respectively, have been identified as activator ligands of RORγt, while bile acid synthesized from cholesterol called 3-oxoLC is an inhibitory ligand of RORγt (37).
RORγt regulates Il17a transcription by binding RORE sequences present in the 2-kb promoter fragment upstream of the transcription start site (38). In addition, the conserved non-coding sequences (CNS)2 (also called CNS5) located in the vicinity of the Il17a gene (approximately 5-kb upstream of promoter) (39) contains two ROREs, which are also conserved in human (39, 40). It has been demonstrated that RORγt binds CNS2 of the Il17a gene (Figure 1) and mediates Il17a transcription by controlling the chromatin remodeling. In fact, CNS2 is also bound by p300 and JmjC domain-containing protein (JMJD)3 that mediate permissive histone acetylation (41, 42) and remove repressive histone marker H3K27me3 (43–45), respectively, resulting in hyperacetylation of histone H3 (46, 47). Moreover, CNS2 interacts with Il17a promoter by forming a loop, and brings CNS2-associated histone remodeling enzymes to the promoter for the activation of Il17a transcription (39).
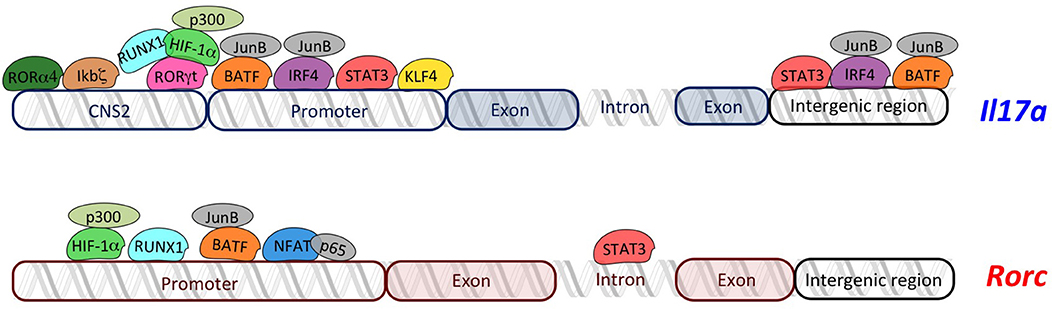
Figure 1. Overview of transcriptional regulators of Il17a and Rorc. The transcriptional regulators of Th17 cells (RORγt, RORα4, ikbζ, RUNX1, HIF-1α, STAT3, IRF4, NFAT, KLF4, and BATF) regulate transcription of Il17a and Rorc by binding specific regions in their loci. Schema does not respect the real organization and structure of each gene locus.
Similarly, it has been demonstrated that RORα4 overexpression promotes, while RORα4 deficiency impairs, Il17a expression (40). Interestingly, coexpression of RORα4 and RORγt causes the synergistic increase in IL-17A, indicating that RORα4 and RORγt work together to regulate Th17 cell differentiation (40, 48).
Given the high similarity of DNA-binding domains between RORα4 and RORγt, they activate Il17a transcription through the same molecular mechanism (40) (Figure 1).
However, RORα4 and RORγt are not sufficient to generate and specify the full Th17 program.
In fact, transcriptional regulators of RORγt, as well as other transcription factors that interact with RORγt, or bind the promoter or the intergenic regions of the Il17a locus, play a crucial role in the generation of Th17 cells.
Other Transcriptional Regulators of Rorc and Il17A
The transcription of RORγt is initiated by activation of the promoter RORC2 into the Rorc locus. RORC2 promoter contains nuclear factor of activated T cells (NFAT)-binding sequences, specific for NFAT and nuclear factor (NF)-kB proteins. Recently, it has been reported that the p65 NF-kB subunit and NFATc2 bind human Rorc promoter and promote a permissive chromatin conformation at RORC2 regulatory regions (49). Consistently, it has been reported that two NF-kB proteins, c-Rel and p65, activate the murine Rorc promoter (50).
Interestingly, the nuclear protein inhibitor of kB (IκB)ζ, which belongs to the IkB kinases and regulates activation of NF-kB pathway, binds CNS2 elements in Il17a locus (Figure 1), thus leading to an efficient recruitment of transcriptional coactivators with histone acetylase activity (18) and promoting Il17a expression without modulating expression of Rorc and Rora (51, 52).
CNS2 region of Il17a is bound by another transcriptional regulator called Runt-related transcription factor (RUNX)1, whose effect is dependent on RORγt. In fact, it has been demonstrated that RUNX1 interacts with RORγt to potentiate Il17a expression and is required for the full effect of RORγt on Il17a expression (38) (Figure 1). Additionally, RUNX1 plays a role in Th17 differentiation, independently of RORγt, by binding the promoter of the gene encoding RORγt through three conserved RUNX1-binding sites (53) (Figure 1).
Hypoxia-inducible factor (HIF)-1α is a key metabolic sensor (19, 54), which binds hypoxia response element (HRE, a conserved HIF)-1α-binding site) located in the proximal region of the Rorc promoter, in both human and mouse (19). Moreover, HIF-1α might physically associate with RORγt, serving as a coactivator for RORγt, thus contributing to Il17a expression without direct DNA binding on Il17a locus (19) (Figure 1). Further studies discovered that HIF-1α activates target genes by recruiting the factor p300, which possesses histone acetyltransferase activity and acetylates histones to “open” the chromatin structure (55). Indeed, the colocalized binding of RORγt, HIF-1α, and p300 occurs at the promoter of the Il17a gene (19).
Signal transducer and activator of transcription (STAT) 3 is another transcription factor regulating RORγt, and IL-17A (56) by interacting with the Stat-binding domains into the Rorc first intron, the Il17a promoter, and the intergenic region of the Il17a locus (Figure 1) (56–58). Moreover, STAT3 regulates positive epigenetic modifications, increasing permissive H3K4me3 marks on its target genes, including Rorc, Rora, and another gene encoding for transcriptional regulator of Th17 cells, called basic leucine zipper ATF-like transcription factor (BATF) (56).
BATF forms a heterodimer with JunB, and binds to the Il17a promoter as well as two conserved intergenic elements in the Il17a locus in Th17 cells (Figure 1). Interestingly, BATF synergizes with RORγt by binding to an overlapped conserved region recognized by RORγt into Il17a gene (20). Furthermore, the complex JunB and BATF also promotes the transcription of Rorc and Rora (58, 59) (Figure 1).
Genome-wide JunB-DNA binding analysis, using ChIP sequencing with anti-JunB antibody, revealed that JunB colocalizes in Th17 cells with another transcription factor, called interferon regulatory factor (IRF)4, involved in Th17 differentiation (21). In fact, IRF4 targets sequences enriched for activating protein 1 (AP-1)–IRF composite elements (AICEs) located into regulatory elements of the Il17a promoter (58, 60), which are cobound by BATF, an AP-1 factor (61). Thus, IRF4 and BATF bind cooperatively to structurally divergent AICEs to promote IL-17A activation in Th17 cells (61). Importantly, not only Il17a locus but also Il21, Il22, and Il23r loci contain one or more coincident binding peaks for IRF4 and BATF that were positioned in promoters and/or intronic regions, and ChIP assays verified the binding to these regions of both IRF4 and BATF complexed with JunB (61). The Kruppel-like factor (KLF)4 is another factor involved in the direct regulation of IL-17A, as demonstrated by ChIP analysis. In fact KLF4 binds the Il17a promoter and induces IL-17A expression, independently of RORγt (62).
Altogether, this information reveals a complex interconnected network of transcriptional regulators that finely regulates generation of Th17 cells.
The timing of transcriptional events leading to the full Th17 differentiation remains enigmatic. However, the transcriptional regulators activated upon T cell receptor engagement, such as NFAT, likely initiate the differentiation process by inducing RORγt transcription, and up-regulating receptor for polarizing cytokines, whose ligation leads to activation of other transcription factors. Among them, BATF, IRF4, and STAT3 are considered initiator transcription factors (24, 63). In fact, BATF and IRF4 are responsible of initial chromatin accessibility in Il17a locus and, with STAT3, of initiation of the transcriptional program that is then globally tuned by the lineage-specific transcription factor RORγt, which plays a pivotal deterministic role at key loci (24, 63). Then, RUNX1, HIF1a, and IκBζ can be considered cooperators of ROR nuclear receptors.
Importantly, there is high interconnectivity among transcription factors, including positive feedback loops reinforcing expression of initiator transcription factors BATF, IRF4, and STAT3 (24).
However, a negative feedback loop mediated by c-Maf, which is induced by initiator transcription factors, may limit Th17 response. In particular, c-Maf is a transcriptional regulator that, in Th17 cells, functions as a negative regulator, attenuating the expression of pro-inflammatory loci (e.g., Batf , Rora, Runx1, Il1r1, Ccr6, and Tnf ) and positively regulating few loci linked to attenuating inflammation (e.g., Il9, Il10, Lif , and Ctla4). Another transcription factor known to limit Th17 response is Fosl2 exerting antagonistic effect to BATF, by competing for the same binding sites and by directly repressing BATF (24). STAT1 and STAT5 are known to inhibit Th17 polarization by directly binding Rorc or Il17a loci. In particular, STAT5 represses IL-17A induction by binding the Il17a locus, removing accessible histone marks, and displacing STAT3 occupancy (64, 65); STAT1 has been shown to bind upstream of the Rorc locus in human Hela cells (66).
Th17-Related Transcriptional Regulators in Autoimmune Diseases
Given the crucial role of Th17 cells in autoimmune disorders, the altered expression of Th17 transcriptional regulators may be related to a persistent Th17 cell response typical of diseases, such as psoriasis, rheumatoid arthritis, Crohn's disease, and MS (16). The role of the transcription factors activating a Th17 response has been mainly investigated in the murine model of MS, the experimental autoimmune encephalomyelitis (EAE), where deletion of each specific Th17 transcription factor reduced the disease (17–21, 40, 62, 67, 68). However, the potential involvement of such transcription factors in human autoimmune diseases, as well as their expression in immune cells from patients, has not been largely investigated.
It has been reported that the levels of phosphorylated STAT3 (pSTAT-3) in lymphocytes are up-regulated in MS patients during relapse compared to healthy donors and MS patients in remission phase. Moreover, pSTAT-3 levels positively correlate with magnetic resonance imaging data, indicating that STAT3 activation is associated to disease activity (69). In contrast, the expression of RORγt analyzed at transcriptional (70) and protein level (71) does not differ between MS patients and healthy donors.
However, the activity of RORγt is ligand regulated and the putative natural ligands of RORγt are molecules of the cholesterol pathway. In this context, it has been reported that levels of oxysterols in relapsing-remitting MS patients were associated with conversion to secondary progressive-MS (72).
Moreover, an aberrant activation of STAT3 was found in intestinal T cells of Crohn's disease patients compared to healthy donors (73); the expression of IRF-4 was significantly increased in inflammatory cells of psoriasis patients than that in healthy controls (74); HIF-1α was found strongly expressed by immune cells in the intimal layer of the synovium in rheumatoid arthritis patients (75). However, the lack of correlations with clinical parameters in most part of these studies does not permit the definition of the role of the enhanced expression of those transcriptional regulators in human diseases.
Additionally, genetic abnormalities in Th17 transcriptional regulators may favor Th17 cell response and may influence susceptibility to autoimmune diseases. However, few studies demonstrate association between gene variants of Th17 transcription factors and Th17-related diseases. For instance, single-nucleotide polymorphisms (rs734232) affecting the consensus-binding site for RUNX1, or Runx1 itself, are associated with susceptibility to rheumatoid arthritis and psoriasis (76–78), while Stat3 gene was identified as risk locus for Crohn's disease and MS (79, 80).
Therapeutic Approaches Targeting Transcriptional Regulators of Th17 Cells
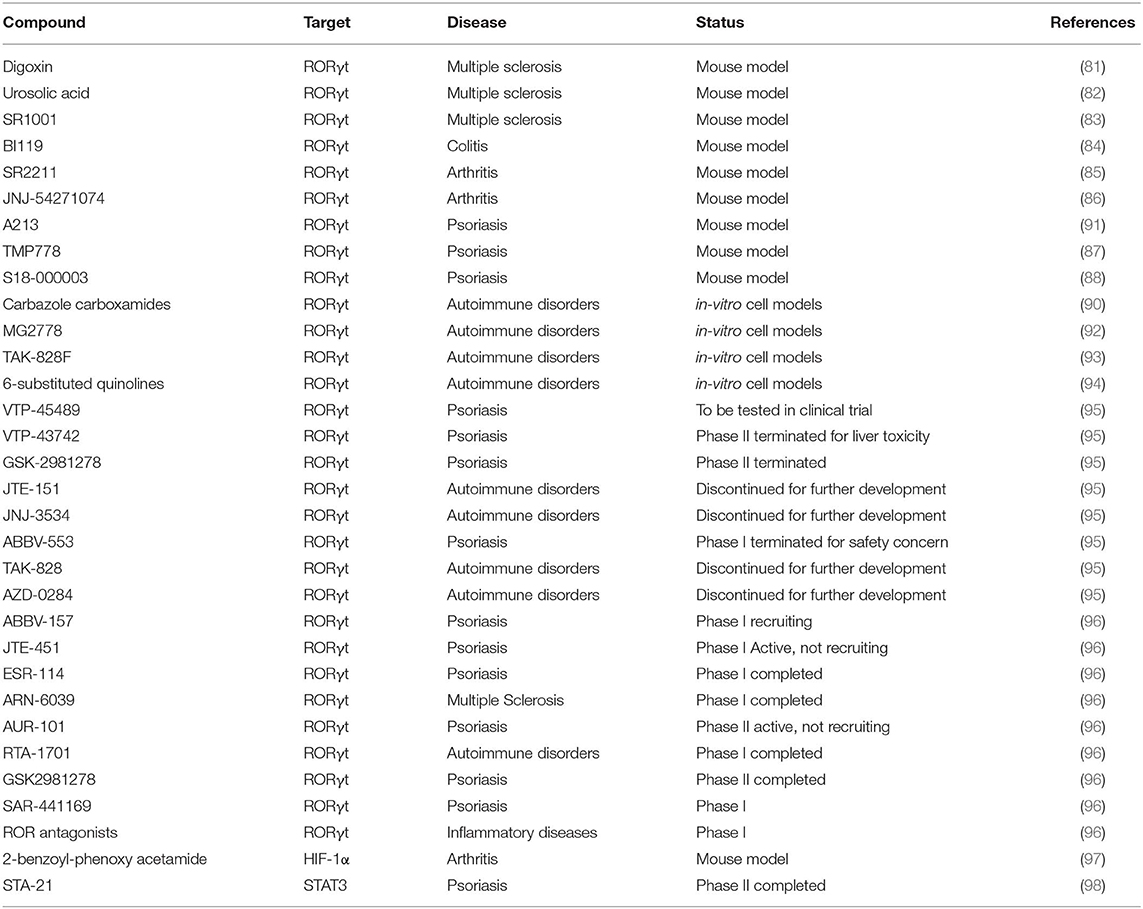
Antibodies targeting IL-17A are approved for the treatment of psoriasis (81), while this approach is ineffective in MS, and deleterious in Crohn's disease (82). Recently, antagonists of Th17 transcriptional regulators have been proposed as potential new treatments of Th17-mediated diseases. Given the high cell specificity, RORγt is the transcription factor representing the ideal target for the manipulation of Th17 cell response. Several molecules targeting RORγt have been discovered and tested in murine models: digoxin, urosolic acid, and SR1001 reduce EAE severity (83–85); BI119 abrogates experimental colitis (86); SR2211 and JNJ-54271074 have therapeutic effect on experimental arthritis (87, 88); TMP778 and S18-000003 show efficacy in a psoriasis-like skin inflammation model (89, 90). In addition, other RORγt inverse agonists have been discovered (carbazole carboxamides, MG2778, TAK-828F, 6-substituted quinolines, A213) and tested as negative regulators of Th17 response (Table 1) (91–96).
Clinical studies testing the actual clinical efficacy and eventual side effects are active or completed. For instance, the oral compound VTP-43742 demonstrated efficacy through the reduction of clinical scores in psoriasis patients (NCT02555709). However, clinical data also showed liver toxicity, and VTP-43742 has been replaced with a new improved molecule VTP-45489 (Table 1). Similarly, other early clinical agents like GSK-2981278, JTE-151, JNJ-3534, ABBV-553, TAK-828, and AZD-0284 were either discontinued or suspended for further development (Table 1) (99). Currently, novel RORγt inhibitors are monitored in the clinical studies: ABBV-157 in psoriasis phase I (NCT03922607); JTE-451 and ESR-114 in psoriasis phase II (NCT03832738 and NCT03630939, respectively); ARN-6039 in MS phase I (NCT03237832); AUR-101 in psoriasis phase II (NCT04207801); RTA-1701 in healthy phase I (NCT03579030); GSK2981278 in psoriasis phase I (NCT03004846 and NCT02548052); SAR-441169 in psoriasis phase I; and ROR antagonists in inflammatory disease phase I (100) (Table 1).
Another promising target among Th17 transcription factors is HIF-1α. To date, the most advanced HIF pathway-targeted pharmaceuticals in terms of clinical development are cell-permeable prolyl hydroxylase inhibitors, evaluated for treatment of anemia. A number of HIF inhibitors have been developed also for cancer therapy (97) and are considered promising novel treatments for rheumatoid arthritis (101), such as the 2-benzoyl-phenoxy acetamide that acts as anti-arthritic agent in an experimental adjuvant induced arthritis rat model (98) (Table 1). However, none of the compounds targeting HIF-1α has been assessed in clinical trials for rheumatoid arthritis.
STAT3 is another potential drug target currently used for cancer therapy given its aberrant activation in many human tumors (102). Concerning Th17-related diseases, the small STAT3 inhibitor STA-21 has been tested on psoriasis patients in a nonrandomized study, and psoriatic lesions in six of the eight patients showed improvement after topical STA-21 treatment for 2 weeks (NCT01047943) (Table 1) (103). However, this effect is likely related to the inhibition of epidermal keratinocyte proliferation, rather than to immune cell activity (103).
Collectively, these data indicate that Th17 transcriptional regulators are promising targets for Th17-related diseases. However, given their broad expression in different cell types, it is crucial to develop inhibitors highly specific for immune cells to minimize off-target effects.
Conclusions
Since the discovery of Th17 cells, remarkable advances in the understanding of Th17 response have been reported. In particular, the study of the mechanisms regulating the transcription of Rorc and Il17a genes has advanced our understanding of the generation of Th17 cells. Moreover, small molecules interfering with these mechanisms provide promising results in pre-clinical research and clinical trials. Future studies further detailing the transcriptional program of Th17 cells could lead to the identification of pathways or regulators that are specifically activated during diseases. Advances in these points are critical for the development of new compounds that target more accurately the pathogenic effect of Th17 cells, and that could become new therapeutic strategies in Th17-related diseases.
Author Contributions
AC drafted the manuscript. EV critically reviewed the manuscript and finalized the manuscript for submission. AC and EV approved the final version.
Funding
This work was supported by Progetto Giovani Ricercatori Italian Ministry of Health, Italy (cod. GR-2016-02361163) and FISM-Fondazione Italiana Sclerosi Multipla (cod. FISM2016/R/31) to EV.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. (2006) 24:677–88. doi: 10.1016/j.immuni.2006.06.002
2. Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction effector functions of T(H)17 cells. Nature. (2008) 453:1051–7. doi: 10.1038/nature07036
3. Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. (2005) 6:1133–41. doi: 10.1038/ni1261
4. Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. (2009) 27:485–517. doi: 10.1146/annurev.immunol.021908.132710
5. Volpe E, Servant N, Zollinger R, Bogiatzi SI, Hupe P, Barillot E, et al. A critical function for transforming growth factor-beta, interleukin 23 and proinflammatory cytokines in driving and modulating human T(H)-17 responses. Nat Immunol. (2008) 9:650–7. doi: 10.1038/ni.1613
6. Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. (2007) 19:377–82. doi: 10.1016/j.smim.2007.10.009
7. Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol. (2010) 10:479–89. doi: 10.1038/nri2800
8. Maddur MS, Miossec P, Kaveri SV, Bayry J. Th17 cells: biology, pathogenesis of autoimmune and inflammatory diseases, and therapeutic strategies. Am J Pathol. (2012) 181:8–18. doi: 10.1016/j.ajpath.2012.03.044
9. Onishi RM, Gaffen SL. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology. (2010) 129:311–21. doi: 10.1111/j.1365-2567.2009.03240.x
10. Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. (2014) 14:585–600. doi: 10.1038/nri3707
11. McGeachy MJ, Cua DJ, Gaffen SL. The IL-17 family of cytokines in health and disease. Immunity. (2019) 50:892–906. doi: 10.1016/j.immuni.2019.03.021
12. Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. (2007) 8:950–7. doi: 10.1038/ni1497
13. Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, et al. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. (2009) 206:299–311. doi: 10.1084/jem.20081463
14. Drummond RA, Lionakis MS. Organ-specific mechanisms linking innate and adaptive antifungal immunity. Semin Cell Dev Biol. (2019) 89:78–90. doi: 10.1016/j.semcdb.2018.01.008
15. Li J, Vinh DC, Casanova JL, Puel A. Inborn errors of immunity underlying fungal diseases in otherwise healthy individuals. Curr Opin Microbiol. (2017) 40:46–57. doi: 10.1016/j.mib.2017.10.016
16. Patel DD, Kuchroo VK. Th17 Cell Pathway in human immunity: lessons from genetics and therapeutic interventions. Immunity. (2015) 43:1040–51. doi: 10.1016/j.immuni.2015.12.003
17. Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. (2006) 126:1121–33. doi: 10.1016/j.cell.2006.07.035
18. Okamoto K, Iwai Y, Oh-Hora M, Yamamoto M, Morio T, Aoki K, et al. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. (2010) 464:1381–5. doi: 10.1038/nature08922
19. Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. (2011) 146:772–84. doi: 10.1016/j.cell.2011.07.033
20. Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. (2009) 460:405–9. doi: 10.1038/nature08114
21. Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. (2007) 8:958–66. doi: 10.1038/ni1500
22. Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. (2010) 467:967–71. doi: 10.1038/nature09447
23. Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. (2015) 163:1400–12. doi: 10.1016/j.cell.2015.11.009
24. Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, et al. A validated regulatory network for Th17 cell specification. Cell. (2012) 151:289–303. doi: 10.1016/j.cell.2012.09.016
25. Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol. (2008) 9:641–9. doi: 10.1038/ni.1610
26. Giguere V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. (1994) 8:538–53. doi: 10.1101/gad.8.5.538
27. Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. (2009) 7:e003. doi: 10.1621/nrs.07003
28. Medvedev A, Yan ZH, Hirose T, Giguere V, Jetten AM. Cloning of a cDNA encoding the murine orphan receptor RZR/ROR gamma and characterization of its response element. Gene. (1996) 181:199–206. doi: 10.1016/S0378-1119(96)00504-5
29. Zhang Y, Luo XY, Wu DH, Xu Y. ROR nuclear receptors: structures, related diseases, and drug discovery. Acta Pharmacol Sin. (2015) 36:71–87. doi: 10.1038/aps.2014.120
30. Hirose T, Smith RJ, Jetten AM. ROR gamma: the third member of ROR/RZR orphan receptor subfamily that is highly expressed in skeletal muscle. Biochem Biophys Res Commun. (1994) 205:1976–83. doi: 10.1006/bbrc.1994.2902
31. Carlberg C, Hooft van Huijsduijnen R, Staple JK, DeLamarter JF, Becker-Andre RZRs M a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol. (1994). 8:757–70. doi: 10.1210/mend.8.6.7935491
32. Becker-Andre M, Andre E, DeLamarter JF. Identification of nuclear receptor mRNAs by RT-PCR amplification of conserved zinc-finger motif sequences. Biochem Biophys Res Commun. (1993) 194:1371–9. doi: 10.1006/bbrc.1993.1976
33. Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. (2004) 305:248–51. doi: 10.1126/science.1096472
34. Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. (2007) 8:967–74. doi: 10.1038/ni1488
35. Hu X, Wang Y, Hao LY, Liu X, Lesch CA, Sanchez BM, et al. Sterol metabolism controls T(H)17 differentiation by generating endogenous RORgamma agonists. Nat Chem Biol. (2015) 11:141–7. doi: 10.1038/nchembio.1714
36. Soroosh P, Wu J, Xue X, Song J, Sutton SW, Sablad M, et al. Oxysterols are agonist ligands of RORgammat and drive Th17 cell differentiation. Proc Natl Acad Sci USA. (2014) 111:12163–8.
37. Hang S, Paik D, Yao L, Kim E, Jamma T, Lu J, et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
38. Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. (2008) 9:1297–306. doi: 10.1038/ni.1663
39. Wang X, Zhang Y, Yang XO, Nurieva RI, Chang SH, Ojeda SS, et al. Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity. (2012) 36:23–31. doi: 10.1016/j.immuni.2011.10.019
40. Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. (2008) 28:29–39. doi: 10.1016/j.immuni.2007.11.016
41. Wang X, Pan L, Feng Y, Wang Y, Han Q, Han L, et al. P300 plays a role in p16(INK4a) expression and cell cycle arrest. Oncogene. (2008) 27:1894–904. doi: 10.1038/sj.onc.1210821
42. Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature. (2008) 451:846–50. doi: 10.1038/nature06546
43. Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. (2007) 449:731–4. doi: 10.1038/nature06145
44. De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. (2007) 130:1083–94. doi: 10.1016/j.cell.2007.08.019
45. Xiang Y, Zhu Z, Han G, Lin H, Xu L, Chen CD. JMJD3 is a histone H3K27 demethylase. Cell Res. (2007) 17:850–7. doi: 10.1038/cr.2007.83
46. Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. (2009) 9:91–105. doi: 10.1038/nri2487
47. Akimzhanov AM, Yang XO, Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. (2007) 282:5969–72. doi: 10.1074/jbc.C600322200
48. Sundrud MS, Rao A. Regulation of T helper 17 differentiation by orphan nuclear receptors: it's not just ROR gamma t anymore. Immunity. (2008) 28:5–7. doi: 10.1016/j.immuni.2007.12.006
49. Yahia-Cherbal H, Rybczynska M, Lovecchio D, Stephen T, Lescale C, Placek K, et al. NFAT primes the human RORC locus for RORgammat expression in CD4(+) T cells. Nat Commun. (2019) 10:4698. doi: 10.1038/s41467-019-12680-x
50. Ruan Q, Kameswaran V, Zhang Y, Zheng S, Sun J, Wang J, et al. The Th17 immune response is controlled by the Rel-RORgamma-RORgamma T transcriptional axis. J Exp Med. (2011) 208:2321–33. doi: 10.1084/jem.20110462
51. Yamazaki S, Muta T, Takeshige K. A novel IkappaB protein, IkappaB-zeta, induced by proinflammatory stimuli, negatively regulates nuclear factor-kappaB in the nuclei. J Biol Chem. (2001) 276:27657–62. doi: 10.1074/jbc.M103426200
52. Muta T. IkappaB-zeta: an inducible regulator of nuclear factor-kappaB. Vitam Horm. (2006). 74:301–16. doi: 10.1016/S0083-6729(06)74012-2
53. Liu HP, Cao AT, Feng T, Li Q, Zhang W, Yao S, et al. TGF-beta converts Th1 cells into Th17 cells through stimulation of Runx1 expression. Eur J Immunol. (2015) 45:1010–8. doi: 10.1002/eji.201444726
54. Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci STKE. (2007) 2007:cm8. doi: 10.1126/stke.4072007cm8
55. Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. (2004) 11:308–15. doi: 10.1038/nsmb740
56. Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. (2010) 32:605–15. doi: 10.1016/j.immuni.2010.05.003
57. Chen Z, Laurence A, Kanno Y, Pacher-Zavisin M, Zhu BM, Tato C, et al. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci USA. (2006) 103:8137–42. doi: 10.1073/pnas.0600666103
58. Hasan Z, Koizumi SI, Sasaki D, Yamada H, Arakaki N, Fujihara Y, et al. JunB is essential for IL-23-dependent pathogenicity of Th17 cells. Nat Commun. (2017) 8:15628. doi: 10.1038/ncomms15628
59. Yamazaki S, Tanaka Y, Araki H, Kohda A, Sanematsu F, Arasaki T, et al. The AP-1 transcription factor JunB is required for Th17 cell differentiation. Sci Rep. (2017) 7:17402. doi: 10.1038/s41598-017-17597-3
60. Biswas PS, Gupta S, Chang E, Song L, Stirzaker RA, Liao JK, et al. Phosphorylation of IRF4 by ROCK2 regulates IL-17 and IL-21 production and the development of autoimmunity in mice. J Clin Invest. (2010) 120:3280–95. doi: 10.1172/JCI42856
61. Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. (2012) 338:975–80. doi: 10.1126/science.1228309
62. Lebson L, Gocke A, Rosenzweig J, Alder J, Civin C, Calabresi PA, et al. Cutting edge: The transcription factor Kruppel-like factor 4 regulates the differentiation of Th17 cells independently of RORgammat. J Immunol. (2010) 185:7161–4. doi: 10.4049/jimmunol.1002750
63. Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature. (2013) 496:461–8. doi: 10.1038/nature11981
64. Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. (2007) 26:371–81. doi: 10.1016/j.immuni.2007.02.009
65. Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. (2011) 12:247–54. doi: 10.1038/ni.1995
66. Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. (2007) 4:651–7. doi: 10.1038/nmeth1068
67. Liu X, Lee YS, Yu CR, Egwuagu CE. Loss of STAT3 in CD4+ T cells prevents development of experimental autoimmune diseases. J Immunol. (2008) 180:6070–6. doi: 10.4049/jimmunol.180.9.6070
68. Wang Y, Godec J, Ben-Aissa K, Cui K, Zhao K, Pucsek AB, et al. The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-gamma-producing T helper 17 cells. Immunity. (2014) 40:355–66. doi: 10.1016/j.immuni.2014.01.002
69. Frisullo G, Angelucci F, Caggiula M, Nociti V, Iorio R, Patanella AK, et al. pSTAT1, pSTAT3, and T-bet expression in peripheral blood mononuclear cells from relapsing-remitting multiple sclerosis patients correlates with disease activity. J Neurosci Res. (2006) 84:1027–36. doi: 10.1002/jnr.20995
70. Edstrom M, Mellergard J, Mjosberg J, Jenmalm M, Vrethem M, Press R, et al. Transcriptional characteristics of CD4+ T cells in multiple sclerosis: relative lack of suppressive populations in blood. Mult Scler. (2011) 17:57–66. doi: 10.1177/1352458510381256
71. Capone A, Bianco M, Ruocco G, De Bardi M, Battistini L, Ruggieri S, et al. Distinct expression of inflammatory features in T helper 17 cells from multiple sclerosis patients. Cells. (2019) 8:533. doi: 10.3390/cells8060533
72. Fellows Maxwell K, Bhattacharya S, Bodziak ML, Jakimovski D, Hagemeier J, Browne RW, et al. Oxysterols and apolipoproteins in multiple sclerosis: a 5 year follow-up study. J Lipid Res. (2019) 60:1190–8. doi: 10.1194/jlr.M089664
73. Lovato P, Brender C, Agnholt J, Kelsen J, Kaltoft K, Svejgaard A, et al. Constitutive STAT3 activation in intestinal T cells from patients with Crohn's disease. J Biol Chem. (2003) 278:16777–81. doi: 10.1074/jbc.M207999200
74. Ni A, Chen H, Wu Y, Li W, Chen S, Li J. Expression of IRF-4 and IBP in skin lesions of patients with psoriasis vulgaris. J Huazhong Univ Sci Technolog Med Sci. (2012) 32:287–90. doi: 10.1007/s11596-012-0050-6
75. Hollander AP, Corke KP, Freemont AJ, Lewis CE. Expression of hypoxia-inducible factor 1alpha by macrophages in the rheumatoid synovium: implications for targeting of therapeutic genes to the inflamed joint. Arthritis Rheum. (2001) 44:1540–4.
76. Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. (2007) 446:685–9. doi: 10.1038/nature05673
77. Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, Sawada T, et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat Genet. (2003) 35:341–8. doi: 10.1038/ng1267
78. Helms C, Cao L, Krueger JG, Wijsman EM, Chamian F, Gordon D, et al. A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. Nat Genet. (2003) 35:349–56. doi: 10.1038/ng1268
79. International Multiple Sclerosis Genetics C Wellcome Trust Case Control C, Sawcer S, Hellenthal G, Pirinen M, Spencer CC, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. (2011) 476:214–9. doi: 10.1038/nature10251
80. Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. (2008) 40:955–62. doi: 10.1038/ng.175
81. McInnes IB, Mease PJ, Kirkham B, Kavanaugh A, Ritchlin CT, Rahman P, et al. Secukinumab, a human anti-interleukin-17A monoclonal antibody, in patients with psoriatic arthritis (FUTURE 2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. (2015) 386:1137–46. doi: 10.1016/S0140-6736(15)61134-5
82. Hueber W, Sands BE, Lewitzky S, Vandemeulebroecke M, Reinisch W, Higgins PD, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn's disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut. (2012) 61:1693–700. doi: 10.1136/gutjnl-2011-301668
83. Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. (2011) 472:486–90. doi: 10.1038/nature09978
84. Xu T, Wang X, Zhong B, Nurieva RI, Ding S, Dong C. Ursolic acid suppresses interleukin-17 (IL-17) production by selectively antagonizing the function of RORgamma t protein. J Biol Chem. (2011) 286:22707–10. doi: 10.1074/jbc.C111.250407
85. Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. (2011) 472:491–4. doi: 10.1038/nature10075
86. Withers DR, Hepworth MR, Wang X, Mackley EC, Halford EE, Dutton EE, et al. Transient inhibition of ROR-gammat therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med. (2016) 22:319–23. doi: 10.1038/nm.4046
87. Xue X, Soroosh P, De Leon-Tabaldo A, Luna-Roman R, Sablad M, Rozenkrants N, et al. Pharmacologic modulation of RORgammat translates to efficacy in preclinical and translational models of psoriasis and inflammatory arthritis. Sci Rep. (2016) 6:37977. doi: 10.1038/srep37977
88. Chang MR, Lyda B, Kamenecka TM, Griffin PR. Pharmacologic repression of retinoic acid receptor-related orphan nuclear receptor gamma is therapeutic in the collagen-induced arthritis experimental model. Arthritis Rheumatol. (2014) 66:579–88. doi: 10.1002/art.38272
89. Skepner J, Ramesh R, Trocha M, Schmidt D, Baloglu E, Lobera M, et al. Pharmacologic inhibition of RORgammat regulates Th17 signature gene expression and suppresses cutaneous inflammation in vivo. J Immunol. (2014) 192:2564–75. doi: 10.4049/jimmunol.1302190
90. Imura C, Ueyama A, Sasaki Y, Shimizu M, Furue Y, Tai N, et al. A novel RORgammat inhibitor is a potential therapeutic agent for the topical treatment of psoriasis with low risk of thymic aberrations. J Dermatol Sci. (2019) 93:176–85. doi: 10.1016/j.jdermsci.2019.03.002
91. Shibata A, Uga K, Sato T, Sagara M, Igaki K, Nakamura Y, et al. Pharmacological inhibitory profile of TAK-828F, a potent and selective orally available RORgammat inverse agonist. Biochem Pharmacol. (2018) 150:35–45. doi: 10.1016/j.bcp.2018.01.023
92. Tang L, Yang X, Liang Y, Xie H, Dai Z, Zheng G. Transcription factor retinoid-related orphan receptor gammat: a promising target for the treatment of psoriasis. Front Immunol. (2018) 9:1210. doi: 10.3389/fimmu.2018.01210
93. Huang Y, Yu M, Sun N, Tang T, Yu F, Song X, et al. Discovery of carbazole carboxamides as novel RORgammat inverse agonists. Eur J Med Chem. (2018) 148:465–76. doi: 10.1016/j.ejmech.2018.02.050
94. Dal Pra M, Carta D, Szabadkai G, Suman M, Frion-Herrera Y, Paccagnella N, et al. Targeting RORs nuclear receptors by novel synthetic steroidal inverse agonists for autoimmune disorders. Bioorg Med Chem. (2018) 26:1686–704. doi: 10.1016/j.bmc.2018.02.018
95. Barbay JK, Cummings MD, Abad M, Castro G, Kreutter KD, Kummer DA, et al. 6-Substituted quinolines as RORgammat inverse agonists. Bioorg Med Chem Lett. (2017) 27:5277–83. doi: 10.1016/j.bmcl.2017.10.027
96. Takaishi M, Ishizaki M, Suzuki K, Isobe T, Shimozato T, Sano S. Oral administration of a novel RORgammat antagonist attenuates psoriasis-like skin lesion of two independent mouse models through neutralization of IL-17. J Dermatol Sci. (2017) 85:12–9. doi: 10.1016/j.jdermsci.2016.10.001
97. Ban HS, Uto Y, Won M, Nakamura H. Hypoxia-inducible factor (HIF) inhibitors: a patent survey (2011-2015). Expert Opin Ther Pat. (2016) 26:309–22. doi: 10.1517/13543776.2016.1146252
98. Shankar J, Thippegowda PB, Kanum SA. Inhibition of HIF-1alpha activity by BP-1 ameliorates adjuvant induced arthritis in rats. Biochem Biophys Res Commun. (2009) 387:223–8. doi: 10.1016/j.bbrc.2009.01.086
99. Gege C. RORgammat inhibitors as potential back-ups for the phase II candidate VTP-43742 from vitae pharmaceuticals: patent evaluation of WO2016061160 and US20160122345. Exp Opin Ther Pat. (2017) 27:1–8. doi: 10.1080/13543776.2017.1262350
100. Sun N, Guo H, Wang Y. Retinoic acid receptor-related orphan receptor gamma-t (RORgammat) inhibitors in clinical development for the treatment of autoimmune diseases: a patent review (2016-present). Expert Opin Ther Pat. (2019) 29:663–74. doi: 10.1080/13543776.2019.1655541
101. Hua S, Dias TH. Hypoxia-inducible factor (HIF) as a target for novel therapies in rheumatoid arthritis. Front Pharmacol. (2016) 7:184. doi: 10.3389/fphar.2016.00184
102. Yang L, Lin S, Xu L, Lin J, Zhao C, Huang X. Novel activators and small-molecule inhibitors of STAT3 in cancer. Cytokine Growth Factor Rev. (2019) 49:10–22. doi: 10.1016/j.cytogfr.2019.10.005
Keywords: T helper 17 cells, interleukin-17, retinoic acid receptor related orphan nuclear receptor γt, multiple sclerosis, Crohn's disease, rheumatoid arthritis, psoriasis
Citation: Capone A and Volpe E (2020) Transcriptional Regulators of T Helper 17 Cell Differentiation in Health and Autoimmune Diseases. Front. Immunol. 11:348. doi: 10.3389/fimmu.2020.00348
Received: 13 December 2019; Accepted: 13 February 2020;
Published: 12 March 2020.
Edited by:
Nicola Ivan Lorè, IRCCS San Raffaele Scientific Institute, ItalyReviewed by:
Rami Bechara, University of Pittsburgh, United StatesSamuele Notarbartolo, Istituto Nazionale Genetica Molecolare (INGM), Italy
Laura A. Solt, The Scripps Research Institute, United States
Copyright © 2020 Capone and Volpe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elisabetta Volpe, ZS52b2xwZUBoc2FudGFsdWNpYS5pdA==
 Alessia Capone
Alessia Capone Elisabetta Volpe
Elisabetta Volpe