- 1Liaoning Key Laboratory of Marine Animal Immunology, Dalian Ocean University, Dalian, China
- 2Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 3Liaoning Key Laboratory of Marine Animal Immunology and Disease Control, Dalian Ocean University, Dalian, China
- 4Dalian Key Laboratory of Aquatic Animal Disease Prevention and Control, Dalian Ocean University, Dalian, China
Cluster of differentiation 63 (CD63), a four-transmembrane glycoprotein in the subfamily of tetraspanin, has been widely recognized as a gateway from the infection of foreign invaders to the immune defense of hosts. Its role in Pacific oyster Crassostrea gigas is, however, yet to be discovered. This work makes contributions by identifying CgCD63H, a CD63 homolog with four transmembrane domains and one conservative CCG motif, and establishing its role as a receptor that participates in immune recognition and hemocyte phagocytosis. The presence of CgCD63H messenger RNA (mRNA) in hepatopancreas, labial palps, gill, and hemocytes is confirmed. The expression level of mRNA in hemocytes is found significantly (p < 0.01) upregulated after the injection of Vibrio splendidus. CgCD63H protein, typically distributed over the plasma membrane of oyster hemocytes, is recruited to the Yarrowia lipolytica-containing phagosomes after the stimulation of Y. lipolytica. The recombinant CgCD63H protein expresses binding capacity to glucan (GLU), peptidoglycan (PGN), and lipopolysaccharide (LPS) in the presence of lyophilized hemolymph. The phagocytic rate of hemocytes toward V. splendidus and Y. lipolytica is significantly inhibited (p < 0.01) after incubation with anti-CgCD63H antibody. Our work further suggests that CgCD63H functions as a receptor involved in the immune recognition and hemocyte phagocytosis against invading pathogen, which can be a marker candidate for the hemocyte typing in C. gigas.
Introduction
Tetraspanins establish a conserved superfamily of four-transmembrane glycoproteins involved in a variety of cellular processes, such as cell development, proliferation, activation, adhesion, and motility (1–4). They are ubiquitous in various organisms, sharing a highly conserved architecture with the presence of four transmembrane domains, two extracellular loops containing one Cys–Cys–Gly (CCG) motif, and one intracellular tail containing cysteine palmitoylation sites (5, 6). Tetraspanins are depicted as a gateway for infection because of the roles in uptaking, trafficking, and spread of viruses as well as intracellular bacteria, fungi, and parasites (7). Extensively studied in mammals, 33 members of tetraspanins have been so far characterized and classified into four major subfamilies, namely, cluster of differentiation (CD), CD63, uroplakin (UPK), and retinal degeneration slow (RDS) subfamily (5–9). CD63, the first characterized tetraspanin, constitutes its own subfamily having a more ancient origin than the other tetraspanins (8, 10). CD63 and other tetraspanins interact with the receptors in the plasma membrane as well as themselves to build an interaction network termed as tetraspanin-enriched microdomains (TEMs) (11, 12). TEMs are always achieved by organizing multiple types of receptors and associated components (6, 7), which provide platforms to control pathogen binding, entry, and invasion, and subsequently the immune responses (13, 14). CD63 can interact directly or indirectly with various proteins, for example integrins, other tetraspanins, cell surface receptors, and kinases (15). CD63 regulates the trafficking of its interaction partners and mediates signal transduction events in the regulation of many membrane-associated processes (16). Growing evidences have demonstrated that tetraspanins also participate in the innate immune response, such as pattern recognition and signal transduction (17, 18). For example, CD63 and other tetraspanins such as CD37, CD9, and CD82 are involved in the pathogen recognition, pattern recognition receptor (PRR) complex formation, and antigen-presentation through their cooperation with other receptors (17, 19–21).
The characterization of CD63 has been recently found in invertebrates, for instance, disk abalone Haliotis discus discus (22) and clam Paphia undulate (23). Earlier work has observed an interesting phenomenon: under the stimulation with bacteria, virus, and pathogen-associated molecular patterns (PAMPs), the mRNA expressions of these CD63s change significantly. However, the detailed biological roles of CD63 are still not clear. So far, there are many reports on other tetraspanins in nematode, mollusk, arthropod, and ascidian (24–27), which could offer useful leads to the investigation of CD63 subfamily for invertebrates. For instance, invertebrate tetraspanins could be significantly induced by the stimulations of various pathogens (28–33), and they could function as mediators of innate immune response (34, 35). Tetraspanin D76 from insect (Manduca sexta) was found to play an important role in cell-mediated immune responses by influencing the encapsulation and clearance of bacteria (35). Tetraspanin from Crassostrea ariakensis, Ca-TSP, was also evidenced to associate with phagocytic bodies (25). Although increasing evidences point to the potential roles of invertebrate tetraspanins in innate immunity, most of them are largely limited to the gene diversity and mRNA expression profiles.
As invertebrate, the Pacific oysters (Crassostrea gigas) have been deemed as a model to investigate the innate immunity in mollusks (36, 37). The lack of specific cell markers, however, has greatly impeded the cell typing and innate immune research in invertebrates (38). This work investigates the functions of CD63 in Pacific oysters, aiming to offer new insights to understand the possible role of tetraspanins in the innate immune system of invertebrate, and provides a potential candidate marker for the cell typing. Specifically, a homolog of CD63 (designated CgCD63H) is identified and characterized from C. gigas; the mRNA transcripts of CgCD63H in different tissues as well as the temporal expression pattern after V. splendidus challenge are investigated; the binding ability of CgCD63H to PAMPs is examined; and the roles of CgCD63H during the phagocytosis process of hemocyte are revealed.
Materials and Methods
Oysters and Microbes
Adult oysters C. gigas (average shell length of 13.0 cm) were collected from a local farm in Qingdao, Shandong Province, China, and acclimatized in aerated fresh seawater at 15 ± 2°C for 10 days before processing. The oysters were fed with condensed microalgae, and the water was totally replaced daily.
Bacteria Escherichia coli Transetta (DE3) (Transgen), Staphylococcus aureus (Microbial Culture Collection Center, Beijing, China), V. splendidus (39), and fungi Yarrowia lipolytica (provided by Dr. Chi) were cultured in Luria–Bertani (LB) medium at 37°C, 2216E medium at 28°C, and Yeast Extract–Peptone–Dextrose (YPD) medium at 28°C, respectively. Then, the microorganisms were harvested and resuspended in sterilized seawater (SSW) and adjusted to the final concentration of 2 × 108 CFU ml−1.
Tissue Collection and Immune Challenge
Tissues including the hepatopancreas, mantle, gonad, labial palps, and gills were collected from six oysters as parallel samples. The hemolymphs were aseptically withdrawn from the posterior adductor muscle sinus of these six oysters by using a syringe and immediately centrifuged at 800 × g, 4°C for 10 min to harvest the hemocytes. All these samples were stored at −80°C after addition of 1 ml TRIzol reagent (TaKaRa) for RNA extraction.
For the bacteria challenge experiment, 200 oysters were randomly assigned into control, challenge, and blank groups. Eighty oysters individually received an injection of 100 μl sterilize seawater (SSW) were employed as the control group, while other 80 oysters that received an injection of 100 μl alive V. splendidus suspended in SSW (2 × 108 CFU ml−1) were employed as the challenge group. These treated oysters were maintained in water tanks after injection, and 15 individuals were randomly sampled at 3, 6, 12, 24, and 48 h post-injection. The remaining 40 untreated oysters were employed as the blank group. Hemolymphs collected from three individuals were pooled into one sample, and there were five replicates for each sampling time point. The hemocytes were harvested and stored as described above.
RNA Isolation and cDNA Synthesis
Total RNA was isolated from oyster tissues and hemocytes using Trizol reagent following its protocol (TaKaRa). The first-strand complementary DNA (cDNA) synthesis was carried out based on Promega M-MLV RT Usage information using the DNase I (Promega)-treated total RNA as template and oligo (dT)-adaptor as primer (Table 1). The reaction was performed at 42°C for 1 h, terminated by heating at 95°C for 5 min. The cDNA mix was diluted to 1:100 and stored at −80°C for subsequent gene cloning and SYBR Green fluorescent quantitative real-time PCR (qRT-PCR).
The Cloning and Sequence Analysis of Full-Length cDNA
Sequence information of CgCD63H (XM_011436987.3) was retrieved from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.gov). A pair of gene-specific primers CgCD63H-F and CgCD63H-R (Table 1) was used for cloning of the full-length cDNA sequence. The PCR product was gel purified and cloned into the pMD19-T simple vector (TaKaRa). The resulting sequences were verified and subjected to cluster analysis. The phosphorylation, O-linked glycosylation, N-linked glycosylation, and methylation modifications of CgCD63H were predicted by DISPHOS (http://www.dabi.temple.edu/disphos/), NetOGlyc 4.0 Server (http://www.cbs.dtu.dk/services/NetOGlyc/), NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/), and GPS-MSP Online Service (http://msp.biocuckoo.org/online.php), respectively.
The tetraspanin homologs of CgCD63H from some other species, including Mus musculus, Xenopus tropicalis, Danio rerio, Salmo salar, and Oplegnathus fasciatus, Drosophila melanogaster, Tenebrio molitor, H. discus, Aplysia californica, Biomphalaria glabrata, Pomacea canaliculata, and Mizuhopecten yessoensis were retrieved from NCBI (Supplementary Data 1). The domains of these proteins were predicted using the simple modular architecture research tool (SMART) version 7.0 (http://www.smart.embl-heidelberg.de/). Multiple sequence alignment of CgCD63H and its homologs were performed with the ClustalW multiple alignment program (http://www.ebi.ac.uk/clustalw/). An unrooted phylogenetic tree was constructed based on the sequence alignment by the neighbor-joining (NJ) algorithm using the Mega 6.06 program (http://www.megasoftware.net/).
The qRT-PCR Analysis
The qRT-PCR was carried out to investigate the mRNA expression of CgCD63H. A fragment of 179 bp was amplified using two sequence-specific primers, CgCD63H-rtF and CgCD63H-rtR (Table 1), and the PCR products were sequenced to verify the PCR specificity. Two primers (Table 1) were used to amplify a 200-bp fragment of elongation factors (CgEF1-α) as an internal control to verify the successful reverse transcription and calibrate the cDNA template. The SYBR Green qRT-PCR assay was carried out in an ABI PRISM 7500 Sequence Detection System (Applied Biosystems) as the previous description (40). All data were given in terms of relative mRNA expression using the 2−ΔΔCT method (41).
Preparation of Recombinant Protein and Polyclonal Antibody of CgCD63H
The cDNA fragment encoding the mature peptide of CgCD63H was amplified with specific primers CgCD63H-F and CgCD63H-R (Table 1). A NotI site and a KpnI site were added to the 5′ end of sense primer CgCD63H-exF and antisense primer CgCD63H-exR with the stop codon deletion, respectively. The PCR fragments were cloned into pMD19-T simple vector (TaKaRa), digested completely by restriction enzymes NotI and KpnI (NEB), and then cloned into the NotI/KpnI sites of expression vector pET-30a (Novagen).
The strain E. coli Transetta (DE3) with recombinant plasmid (pET-30a-CgCD63H) was incubated in LB medium (containing 75 μg ml−1 kanamycin), shaken at 220 rpm at 37°C. The control strain with plasmid pET-32a was incubated in the same medium with 100 μg mL−1 ampicillin. When the culture media reached OD600 of 0.5–0.7, the cells were incubated for an additional 4 h with the induction of isopropyl β-D-1-thiogalactopyranoside (IPTG) at the final concentration of 1 mmol L−1. The recombinant protein CgCD63H (designated rCgCD63H) and the recombinant protein Trx (designated rTrx) were purified by a Ni2+ chelating Sepharose column and refolded in gradient urea–Tris-buffered saline (TBS) glycerol buffer as the previous description (40). The resultant proteins were detected by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and their concentration was quantified by bicinchoninic acid (BCA) kit (Beyotime).
For the preparation of polyclonal antibody anti-CgCD63H, rCgCD63H was injected into mice of 6 weeks of age to acquire polyclonal antibody as previously described (42). The serum from the identical mice before immunization was taken as negative control.
Immunocytochemistry of CgCD63H in Hemocytes
Hemolymphs were collected from the oysters cultured in filtered aerated seawater at 18°C for 1 week and immediately centrifuged at 800 × g, 4°C for 10 min to harvest the hemocytes. Modified Leibovitz L-15 media (Gibco) (43) were used to suspend the hemocytes. The hemocyte suspension was added into confocal dishes precoated with gelatin solution [gelatin, 5 g L−1; CrK (SO4)2·12H2O, 0.5 g L−1] and allowed them to adhere to the wall for 3 h. The supernatant was dislodged, and then, 4% paraformaldehyde (PFA; diluted in TBS) was added to fix the hemocytes for 15 min. After rinsing three times with TBST (TBS with 0.1% Tween-20), the dishes were blocked with 200 μl of 3% bovine serum albumin (BSA) [dissolved in phosphate-buffered saline (PBS)] at 37°C for 30 min. The supernatant was removed, and the dishes were incubated with 200 μl anti-CgCD63H (diluted 1:1,000 in blocking buffer) as the primary antibody at 37°C for 1 h. After washing three times with PBST (PBS with 0.1% Tween-20), the dishes were incubated with Alexa Fluer 488-labeled goat-antimouse antibody (diluted 1:1,000 in blocking buffer) as the second antibody at 37°C for 1 h. After another three times of washing with PBST, 4′,6-diamidino-2-phenylindole (DAPI) (diluted 1:10,000 in PBS) was added into the dishes to stain the nucleus. After the last three times of washing, the dishes were mounted in buffered glycerin for observation with a laser scan confocal microscope (ZEISS).
PAMP Binding Assay
The PAMP binding assay was performed according to previous report with modification (44). Briefly, 100 μL (20 mg) of lipopolysaccharides (LPS) from E. coli (Sigma-Aldrich, L2630-10MG), peptidoglycan (PGN) from S. aureus (Sigma-Aldrich, 77140-10MG), β-glucan (GLU) from Saccharomyces cerevisiae (Sigma-Aldrich, 346210-25MG), and mannose (MAN) (Sigma-Aldrich, M2069-25G) were adopted to envelop a 96-well microtiter plate (Costar). The wells were then blocked with 3% BSA (w/v) in PBS at 37°C for 1 h. After washed with PBS-T, 1/2-fold serial dilutions of rCgCD63H in TBS (50 mmol L−1 Tris–HCl, 50 mmol L−1 NaCl, pH 7.6) were added in the presence of 0.1 mg mL−1 BSA or 1 mg lyophilized hemolymph. The same concentration of rTRX was used as negative control. After incubating at 18°C for 2 h followed by three times of washing, 100 μl mouse anti-His tag monoclonal antibody (Genscript, China) diluted to 1:2,000 was added and incubated at 37°C for 1 h. The plate was washed again, and 100 μl of rabbit-anti-mouse Ig-AP conjugate (Sangon Biotech, China) secondary antibody (diluted 1:2,000) was added and incubated at 37°C for 1 h. After the last washing, 100 μl of 0.1% (w/v) p-nitrophenyl phosphate (pNPP, Sigma) in 50 mmol L−1 carbonate bicarbonate buffer (pH 9.8) containing 0.5 mmol L−1 MgCl2 was added and incubated at room temperature in the dark for 30 min. The reaction was stopped by 2 mol L−1 NaOH, and the absorbance was measured at 405 nm. The wells with 100 μl of TBS were used as blank. The assay was repeated at least five times under similar procedures. Samples with Psample – Bblank/Nnegative – Bblank (P/N value) > 2.1 were considered positive (44, 45).
Phagocytosis Assay
Phagocytosis assay was performed according to previous report with modification (46). Microorganisms including S. aureus, V. splendidus, and Y. lipolytica were labeled with fluorescein isothiocyanate (FITC) to investigate the phagocytosis. All the microorganisms were grown to mid-log phase and harvested by centrifugation at 6,000 × g for 15 min. Cells were fixed with 4% PFA for 10 min, washed with 0.1 M NaHCO3 (pH 9.0) for three times, and then mixed with 1 mg mL−1 FITC (Sigma-Aldrich) in 0.1 M NaHCO3 (pH 9.0) buffer at room temperature with continuous gentle stirring overnight.
Briefly, hemolymph was collected from 15 oysters (5 in each group) and then centrifuged at 800 × g for 10 min to harvest hemocytes. The hemocytes were resuspended with 200 μl modified Leibovitz L-15 media. For hemocyte phagocytic rate assay, the hemocytes suspension was mixed with 20 μl of microbe culture (OD600 = 0.6, suspended in Tris–HCl) together with anti-CgCD63H and incubated in the dark for 1 h. The phagocytosis rate and phagocytosis index (mean FITC fluorescent intensity) were evaluated by FACScan flow cytometry (evaluated by BD, USA) following a previous research (47). PBS was used as blank control, and rTrx and negative serum were employed as negative controls. There were three replicates in each sampling. For the recruitment of CgCD63H to Y. lipolytica-containing phagosomes assay, the hemocyte suspension was mixed with 20 μl of microbe culture (OD600 = 0.6, suspended in Tris–HCl) and incubated in the dark for 1 h. Endogenic CgCD63H was detected by anti-CgCD63H and visualized by DyLight 594-labeled secondary antibody (red). FITC-labeled Y. lipolytica (green) was selected to elicit the phagosome in oyster hemocytes. The nucleus stained by DAPI was observed in blue. The signals were investigated via fluorescence confocal microscopy based on previous description.
Statistical Analysis
All data were given as means ± SE. The data were subjected to one-way analysis of variance (one-way ANOVA) followed by an unpaired, two-tailed t-test. Differences were considered significant at p < 0.05 and extremely significant at p < 0.01.
Results
The Molecular Characters and Phylogeny of CgCD63H
A cDNA fragment of 948 bp nucleotides representing the open reading frame (ORF) of CgCD63H was amplified, which encoded a polypeptide of 315 amino acids with a molecular weight of 35.4 kDa and a theoretical isoelectric point of 5.37. There were four characteristic transmembrane (TM) domains, a short extracellular loop and a large extracellular loop identified in CgCD63H (Supplementary Figures 1, 2). Additionally, conserved CCG motif and Cys residues were revealed in the large extracellular loop of CgCD63H (Figure 1B). In bioinformatics prediction of post-translational modifications, five phosphorylation sites and six O-linked glycosylation sites were detected but no N-linked glycosylation and methylation site (Supplementary Figures 3–6).
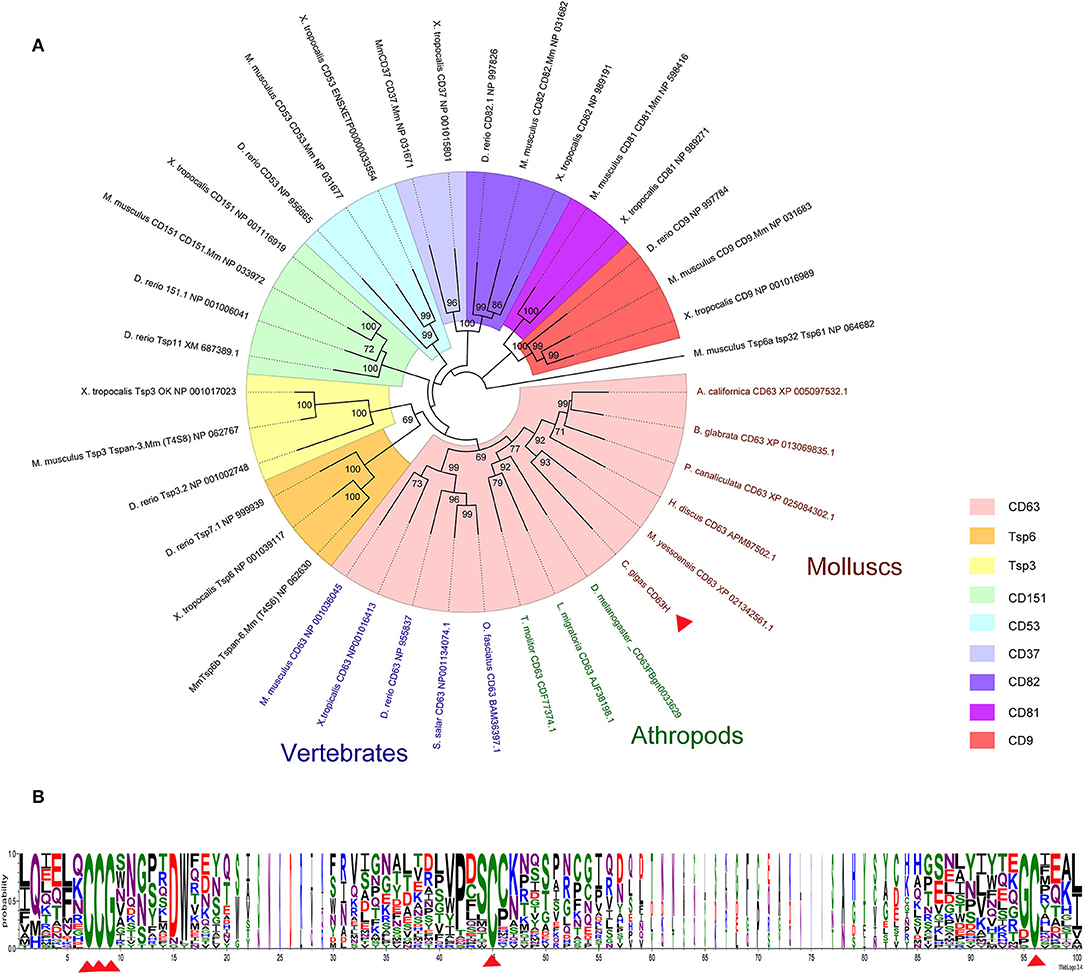
Figure 1. Phylogenetic analysis of CgCD63H. The homologs of CgCD63H from vertebrates (Mus musculus, Xenopus tropicalis, Danio rerio, Salmo salar, and Oplegnathus fasciatus) and invertebrates (Drosophila melanogaster, Tenebrio molitor, Locusta migratoria, Haliotis discus discus, Aplysia californica, Biomphalaria glabrata, Pomacea canaliculata, and Mizuhopecten yessoensis) were retrieved from the National Center for Biotechnology Information (Supplementary Data 1). (A) The unrooted tree was constructed by the neighbor-joining (NJ) algorithm using the Mega 6.06 program based on the multiple sequence alignment by ClustalW. The reliability of the branching was tested by bootstrap (1,000 replicates). Red arrow head indicated CgCD63H. (B) The conserved residues and motifs in large extracellular loop were analyzed based on the multiple sequence alignment.
On the basis of the multiple sequences alignment of tetraspanins, an unrooted phylogenetic tree was constructed by using the neighbor joining (NJ) method (Figure 1A). CgCD63H and 37 homologs from other species were clustered into nine groups, including CD63, CD151, CD53, CD82, CD37, CD81, CD9, TSP3, and TSP6 groups (Figure 1A). CgCD63H was successively bunched together with CD63s from mollusks and arthropods and then assigned into CD63 subgroup in vertebrates.
The Tissue Distribution of CgCD63H mRNA
The qRT-PCR was employed to investigate the distribution of CgCD63H mRNA transcripts in different tissues of oysters. A single peak was revealed in the dissociation curve analysis, indicating the amplification specificity for both CgCD63H and CgEF1-α (data not shown). The CgCD63H mRNA transcripts were detected in all the tested tissues (Figure 2). A significantly higher CgCD63H expression was observed in hepatopancreas, labial palps, gill, and hemocytes, which was 354.9-fold (p < 0.01), 302.7-fold (p < 0.01), 279.7-fold (p < 0.01), and 240.7-fold (p < 0.01) of that in mantle, respectively. The relative expression level of CgCD63H mRNA in the gonad was 1.3-fold (p < 0.01) of that in mantle.
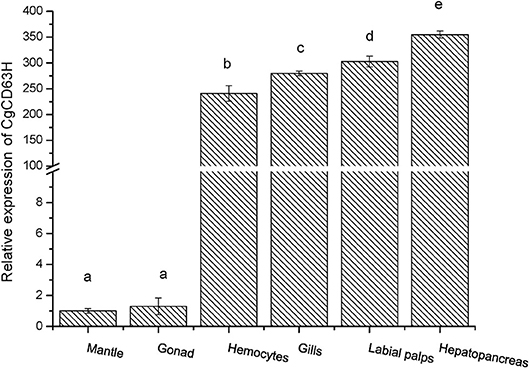
Figure 2. Tissue distribution of the CgCD63H transcripts. CgCD63H relative messenger RNA (mRNA) expression level in mantle, gonad, hemocytes, gills, labial palps, and hepatopancreas was normalized to that of CgEF1-α. Each value was shown as mean ± SE (N = 6), and bars with different letters were significantly different (p < 0.01).
The Temporal Expression of CgCD63H mRNA in Hemocytes After V. splendidus Stimulation
The qRT-PCR was also used to detect the expression level of CgCD63H mRNA in oyster hemocytes challenged by alive V. splendidus (Figure 3). The CgCD63H transcripts were significantly upregulated at 9 h after V. splendidus challenge (1.71-fold compared to that in the control group, p < 0.01) and reached the maximum level (1.79-fold, p < 0.01) at 12 h, and then dropped back to the original level at 24 h (1.38-fold, p < 0.01). While no significant change on the expression level of CgCD63H mRNA was observed in the control group during the experiment.
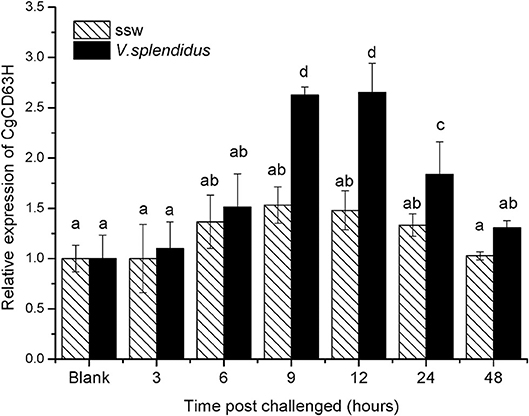
Figure 3. Temporal expression profile of CgCD63H messenger RNA (mRNA) post-V. splendidus challenge. Oyster hemocytes were employed to analyze CgCD63H expression, at 0, 3, 6, 12, 24, and 48 h after V. splendidus challenge. Comparison of the level of CgCD63H mRNA (relative to CgEF1-α) was normalized to 0 h, which was indicated as “relative expression level of CgCD63H.” The values were shown as mean ± SE (n = 6), and bars with different letters were significantly different (p < 0.01).
The Recombinant Protein and Antibody of CgCD63H
The recombinant plasmid (pET-30a-CgCD63H) was transformed into E. coli Transetta (DE3). After IPTG induction for 4 h, the whole cell lysate was analyzed by SDS-PAGE, and a distinct band with a molecular mass of 35 kDa was revealed (lane 3), which was in consistence with the predicted molecular mass of fusion recombinant protein of CgCD63H with His-tag (Figure 4, lane 3). As control, no visible targeted band was detected in the group of cell lysate without IPTG induction. A polyclonal antibody against rCgCD63H was prepared, and Western blotting analysis revealed a distinct single immune-precipitated band with a similar molecular weight predicted by the target sequence. This result suggested a high binding specificity of the polyclonal antibody against CgCD63H (Figure 4, lane 4).
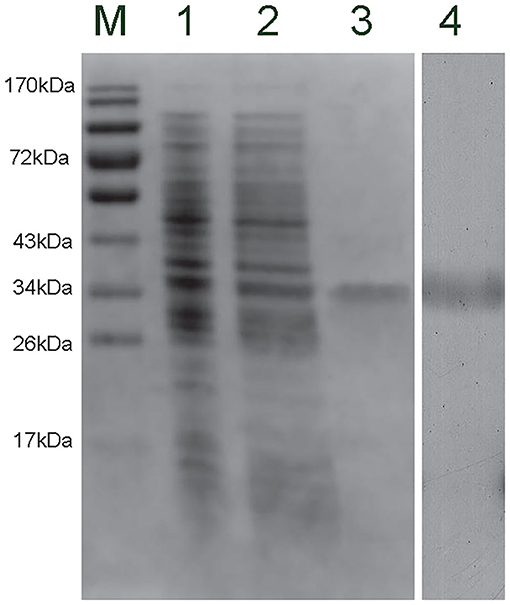
Figure 4. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis of CgCD63H. Lane M, protein molecular standard; lane 1, negative control for CgCD63H (without induction); lane 2, induced rCgCD63H; lane 3, purified rCgCD63H; lane 4, Western blot using purified anti-CgCD63H based on the hemocytes lysate sample.
Subcellular Localization of CgCD63H in Oyster Hemocytes
Fluorescence confocal microscopy was employed to detect the localization of endogenous CgCD63H in oyster hemocytes (Figure 5 and Supplementary Figure 7). The nucleus stained by DAPI was observed in blue, and the plasma membrane stained by DiI was observed in red. The CgCD63H immunoreactive area was stained in green. The positive signal of CgCD63H appeared mainly over the plasma membrane of some oyster hemocytes (Figure 5 and Supplementary Figure 7).
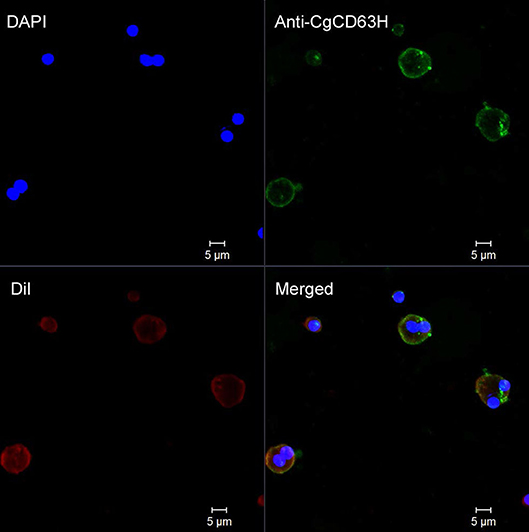
Figure 5. Subcellular localization of CgCD63H protein in oyster hemocytes. Binding of antibody to CgCD63H was visualized by Alexa 488-labeled secondary antibody (green); the nucleus of hemocytes was stained with DAPI (blue), bar = 5 μm.
Binding Capacity of rCgCD63H to Various PAMPs
The binding assay of rCgCD63H to various PAMPs in the presence of lyophilized hemolymph was performed based on the OD405 value. The samples with P/N (Psample – Bblank/Nnegative – Bblank) > 2.1 were considered as positive (Figure 6). No positive value was detected in the absence of lyophilized hemolymph (data not shown). After the addition of lyophilized hemolymph, the P/N values of rCgCD63H toward GLU, PGN, and LPS were all >2.1, under the minimum protein concentration of 1.8375, 1.8375, and 25 μg ml−1, respectively (Figure 6). rCgCD63H possessed affinity to GLU, PGN, and LPS in a dose-dependent manner. The rCgCD63H exhibited relatively higher affinity to GLU and PGN while lower affinity to LPS. The P/N values of rCgCD63H for MAN were all <2.1 under the concentration of 1.8375 to 100 μg ml−1 (Figure 6).
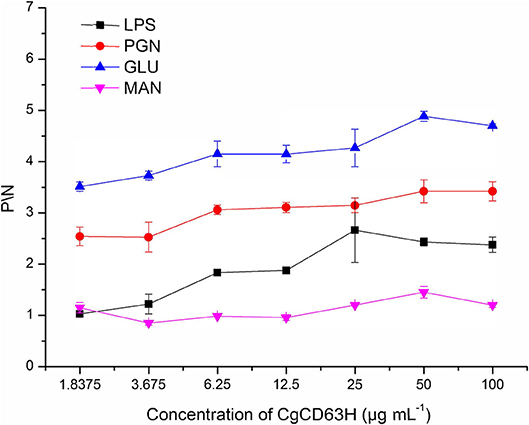
Figure 6. ELISA analysis of the interaction between CgCD63H and the pathogen-associated molecular patterns (PAMPs). Plates were coated with LPS, PGN, GLU, and MAN, and then incubated with a series of concentrations of rCgCD63H and rTrx at the presence of lyophilized hemolymph at 18°C for 2 h. After incubated with anti-CgCD63H, the interaction was detected with goat antimouse Ig-alkaline phosphatase conjugate at 405 nm. Samples with P/N (Psample – Bblank/Nnegative – Bblank) > 2.1 were considered positive. Results are representative of the mean of three replicates ± SE.
Recruitment of CgCD63H to Y. lipolytica-Containing Phagosomes
The recruitment of CgCD63H after a phagocytic stimulus was investigated via fluorescence confocal microscopy (Figure 7). FITC-labeled Y. lipolytica (green) was selected to elicit the phagosome in oyster hemocytes. The nucleus stained by DAPI was observed in blue. Endogenic CgCD63H was detected by anti-CgCD63H and visualized by DyLight 594-labeled secondary antibody (red). In the absence of phagocytic stimulus, CgCD63H-positive signal was detected on the surface of oyster hemocytes (Figure 5). After 1 h incubation with Y. lipolytica, CgCD63H was highly enriched on the Y. lipolytica-containing phagosomes (Figure 7).
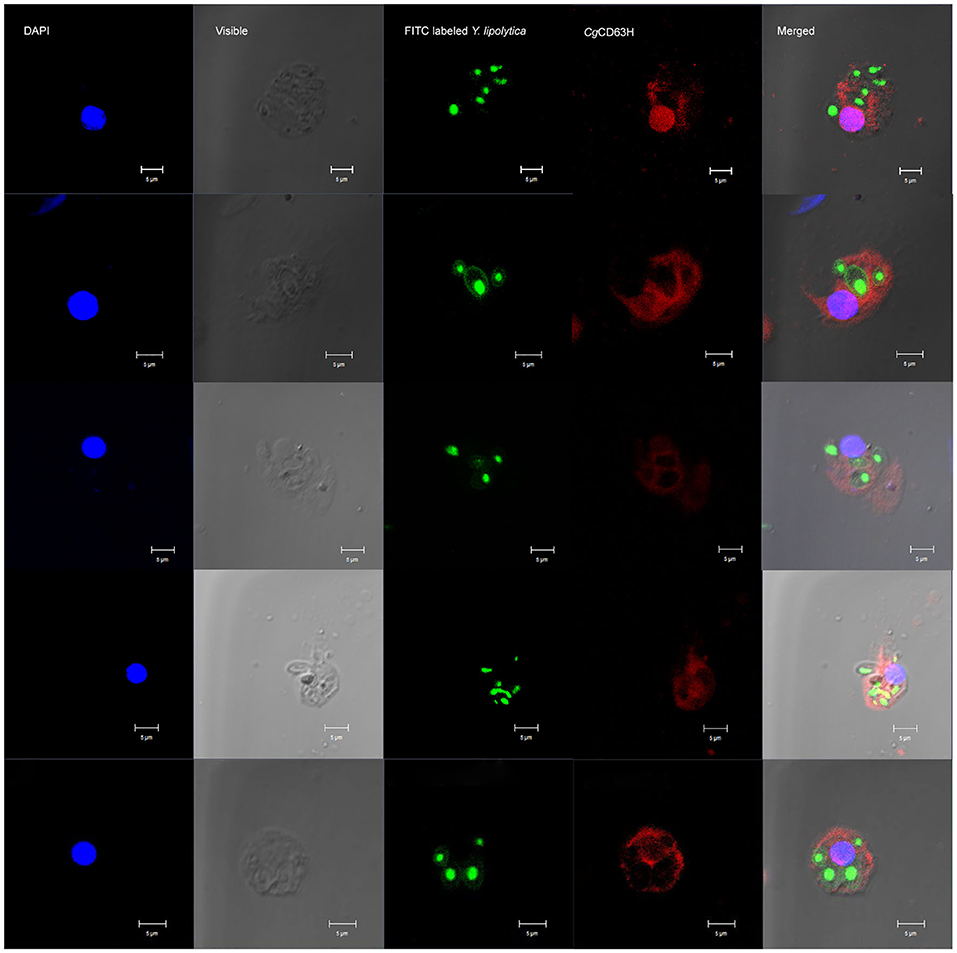
Figure 7. Specific recruitment of CgCD63H to the Y. lipolytica-containing phagosomes. Fluorescein isothiocyanate (FITC)-labeled Y. lipolytica was selected to investigate the phagosome recruitment; the cell membrane and nucleus of Y. lipolytica were indicated with green signal. The hemocyte suspension was mixed with 20 μl of Y. lipolytica (OD600 = 0.6, suspended in Tris–HCl) and incubated in the dark for 1 h. Antibody anti-CgCD63H was employed to detect the endogenic CgCD63H. Binding of antibody to CgCD63H was visualized by DyLight 594-labeled secondary antibody (red); the nucleus of hemocytes was stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue), bar = 5 μm.
The Change of Hemocyte Phagocytic Rate Post-anti-CgCD63H Incubation
Phagocytosis assay was performed on the basis of flow cytometry to test the phagocytic rate and phagocytic index of hemocytes after they were incubated with anti-CgCD63H (Figure 8). The phagocytic rates of hemocyte toward Gram-negative bacteria V. splendidus and fungi Y. lipolytica were significantly down-regulated after they were incubated with anti-CgCD63H, which were 0.60- and 0.86-fold (p < 0.01) compared to that of negative control group, respectively. However, in the Gram-positive bacteria S. aureus group, no significant change was observed after the hemocytes were incubated with anti-CgCD63H compared to the negative control. The change in phagocytic index (mean FITC fluorescent intensity) indicated the same trend (Supplementary Figure 8). The phagocytic indices were 1.67 × 104, 2.15 × 104, and 4.69 × 104 in the negative control group, and 1.78 × 104, 1.30 × 104, and 3.88 × 104 in the anti-CgCD63H blocking group for S. aureus, V. splendidus, and Y. lipolytica, respectively. Significant downregulation of phagocytic index after incubation with anti-CgCD63H was detected in the V. splendidus and Y. lipolytica group, rather than in the S. aureus group.
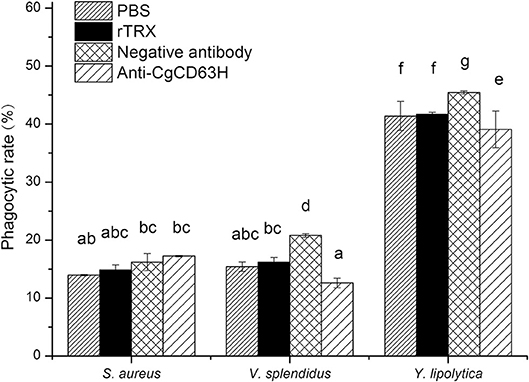
Figure 8. Hemocyte phagocytosis rate post-incubation of anti-CgCD63H. Oyster hemocytes were employed to analyze the change in hemocytes' phagocytosis rate against S. aureus, V. splendidus, and Y. lipolytica post-incubation of anti-CgCD63H. Fluorescein isothiocyanate (FITC)-labeled microbe, anti-CgCD63H, and hemocytes were mixed and incubated for 1 h in dark. Recombinant protein rTRX and negative antibody were employed as negative controls. For each treatment, assay was performed in three different replicates for statistical analysis (p < 0.01).
Discussion
CD63, as a member of tetraspanins, is an evolutionarily conserved protein family involved in multiple aspects of cellular physiological regulation (3, 4, 48). Although tetraspanins has been reported in a wide range of organisms, the functions of CD63 in invertebrate are, however, yet to be discovered (8). In the present study, a homolog of CD63 (designated CgCD63H) was identified from genome database of oyster C. gigas with an open reading frame (ORF) of 948 bp encoding a polypeptide of 315 amino acids. The predicated amino acid sequence of CgCD63H consists of four typical TM domains, a short extracellular loop, and a large extracellular loop. Additionally, a CCG motif and several Cys residues were identified in the large extracellular loop of CgCD63H, which were highly conserved structural features of the previously reported CD63s (8, 22). Coincidentally, CgCD63H was bunched together with CD63 homologs from invertebrates and vertebrates in an unrooted phylogenetic tree. These similar structure characters and reasonable phylogenetic relationships indicated that CgCD63H was a member of the CD63 subfamily in mollusks, and it might function similarly to other CD63s.
CD63 has been reported to facilitate the immune response of various organisms (49). In the present study, CgCD63H was found to distribute in a wide range of tissues of C. gigas including the hepatopancreas, mantle, gonad, labial palps, gills, as well as hemocytes. The highest expression level of CgCD63H was detected in the hepatopancreas, which was deemed as one of the most important tissues involved in the innate immune defense of mollusks (50). Labial palps, gills, as well as hemocytes were also deemed as important tissues involved in the innate immune defense of mollusks (51–53). The universal distribution of CgCD63H in immune-related tissues led us to further investigate its roles in the immune response. The circulating hemocytes play indispensable roles in the immune response of mollusks against invading pathogens (51). CgCD63H protein was typically distributed over the plasma membrane of oyster hemocytes. Moreover, the expression level of CgCD63H mRNA in hemocytes increased significantly (p < 0.01) and reached the highest levels at 12 h after V. splendidus stimulation. In our previous publication, the transcriptome profiles of C. gigas in response to PAMP treatments were investigated by RNA-seq (54). Based on the published data (NCBI: SRR3110857), the temporal expressions of CgCD63H were found to be significantly upregulated post-LPS stimulation (fold change = 2.07, q = 0), PGN (fold change = 1.74, q = 7.44E-16), and GLU (fold change = 1.57, q = 7.91E-07), indicating the potential role of CgCD63H in the immune response against invading Gram-positive bacteria, Gram-negative bacteria, and fungi. The increased expression of CD63 in response to immune stimulation was depicted as its incipient character in immune response by multiple reports (25, 33). These results suggested that CgCD63H could play important roles in immune defense of oysters against invading pathogens.
Immune recognition is the first step of immune response of mollusk which could discriminate non-self- from self-substances (55). Previous work has shown that mutual recognition between membrane receptors and pathogens leads to receptor clustering and membrane protrusions, which eventually enable microbial engulfment. Several mammalian tetraspanins including CD63 are reported to participate in the recognition of PAMPs through the binding activity with PRRs and tetraspanins traffic at the cell surface (11, 56). In invertebrates, CD63 is also suspected to function in recognition process with evidences of the upregulation of mRNA expression after PAMP stimulation (22). Here, CgCD63H was found to bind GLU, PGN, and LPS only in the presence of lyophilized hemolymph, indicating that CgCD63H could combine PAMPs in an indirect manner, possibly with the assistance of other possible PRRs. Earlier reports have demonstrated that CD63 and CD37 could interact with the C-type lectin Dectin-1, identified as fungi-PRR, through “tetraspanin web” and then initiate the antifungal immune responses in vivo (17, 57). A CD63 homolog from coleopteran beetle was also reported to possess the possible binding ability to PGN and GLU (49). The results supported that CgCD63H could be involved in the regulation of immune recognition through the interaction of other PRRs within the “tetraspanin web,” for instance possible lectins or Ig domain containing cell adhesion molecules (IgCAMs). Since the recognition of the foreign invaders was the first step to initiate the immune response (58), the broad binding spectrum of CgCD63H might endow it with a PRR partner role to organize PRRs on the cell membrane and induce the subsequent downstream immune responses against various intruders in oyster.
The previous reports have confirmed the effect of tetraspanins on phagocytosis in mammals (59–61). As a representative of tetraspanins, CD63 promotes the binding of outer segment particle through the interaction with specific PRRs instead of functioning as a direct receptor (57). In invertebrates, CD63 from calm P. undulata also plays roles in hemocyte-mediated phagocytosis (23). In the present study, CgCD63H was recruited to the Y. lipolytica-containing phagosomes in the hemocytes of oysters after incubation with Y. lipolytica, which was similar to the previous observation in mammalian tetraspanins. The CD63 and CD82 were redistributed after immune stimulation and rapidly recruited to the membrane of nascent C. neoformans-containing phagosomes (19, 21). Additionally, in Figure 5, some positive signal of CgCD63H was found inside of the cell membrane, which might be caused by the phagocytosis of mixed debris in open circulatory system and resident microbiota, and the recruitment of CgCD63H. Meanwhile, the phagocytosis of hemocytes against V. splendidus and Y. lipolytica was significantly inhibited after the incubation with anti-CgCD63H. It was suspected that CgCD63H promoted the phagocytosis through the interaction with multiple possible PRRs with its microdomains instead of acting as a direct invader binding receptor. In mammals, the interaction between tetraspanins and PRRs, such as CD81 and Toll-like receptor 4 (TLR-4) (62), was reported to be necessary in macrophage activation. Likewise, in invertebrates, the integrin–tetraspanin interaction was also observed in M. sexta, and the monoclonal antibody of tetraspanin CD76 could disturb the encapsulation of the invader, Serratia marcescens (35). An increasing body of evidence suggests that phagocytes play important roles in invertebrate innate immune responses. Cells of phagocytes were classified efficiently from the oyster C. gigas, which possess both potent oxidative killing and microbial disintegration capacities (63). Additionally, no significant change of phagocytic rate was found after the hemocytes were incubated with anti-CgCD63H in the Gram-positive bacteria S. aureus group. It has been reported that PFAs have effects on some microbes (64). It was suspected that PFA might have impacts on S. aureus during FITC labeling, which influenced the immune response of oyster. Moreover, there might be other molecules involved in the phagocytosis toward S. aureus, and the phagocytic efficiency did not change significantly even though CgCD63H was blocked by the antibody. As the phagocytosis of oyster hemocytes could be initiated by the recognition of invaders, the recruitment of CgCD63H to the phagosome membrane further suggested the possible interaction between CgCD63H and recognition receptors, as well as the partner roles of CgCD63H in sensing the microbial content of phagosomes and eliciting an appropriate immune response. The difference in the PRRs interacting with CgCD63H may affect the specificity and efficiency in the immune response against different microbes. In the present study, CgCD63H was found to enhance the phagocytosis of V. splendidus and Y. lipolytic other than S. aureus. These results indicated that CgCD63H could interact with the PRRs against V. splendidus and Y. lipolytic with a higher affinity and promote a more efficient cellular immunity.
In conclusion, a member of the molluscan CD63 subfamily, CgCD63H, was identified from C. gigas. CgCD63H mRNA was mainly expressed in immune tissues and induced by the challenge of V. splendidus. The CgCD63H exhibited binding activities to a wide spectrum of PAMPs in the presence of lyophilized hemolymph. Moreover, CgCD63H was recruited to the Y. lipolytica-containing phagosomes of hemocytes after immune stimulation, and the phagocytosis of hemocytes was significantly inhibited after incubation with anti-CgCD63H. All these results collectively indicated that CgCD63H might function as a “gateway” between the pattern recognition of foreign invaders and the subsequent immune responses in oysters. CgCD63H could also be a promising marker in cell typing of phagocytic lines in C. gigas.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
This animal study was reviewed and approved by Ethics Committee of the Institute of Oceanology, Chinese Academy of Sciences.
Author Contributions
CL carried out cloning, expression, and purification of recombinant proteins as well as PAMP binding analyses. CY and SJ performed phagocytosis and flow cell cytometry analysis, while QY, WW, and MW carried out bioinformatics analyses. CL, LW, and LS designed the research and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key R&D Program (2018YFD0900606), grants (Nos. U1706204 and 41961124009) from the National Science Foundation of China, earmarked fund from Modern Agro-Industry Technology Research System (CARS-49) and the Fund for Outstanding Talents and Innovative Team of Agricultural Scientific Research from the Ministry of Agriculture and Rural Affairs in China, the Distinguished Professor of Liaoning (to LS), AoShan Talents Cultivation Program Supported by Qingdao National Laboratory for Marine Science and Technology (No. 2017ASTCP-OS13), Liaoning Climbing Scholar (to LS), the Distinguished Professor of Liaoning (XLYC1902012), Dalian High Level Talent Innovation Support Program (2015R020), and the Research Foundation for Talented Scholars in Dalian Ocean University (to LW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer LZ declared a shared affiliation, with no collaboration, with several of the authors CL, MW, and SJ, to the handling editor at the time of the review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01379/full#supplementary-material
References
1. Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci. (2001) 114(Pt 23):4143–51.
2. Wright M, Moseley G, Van Spriel A. Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue Antigens. (2004) 64:533–42. doi: 10.1111/j.1399-0039.2004.00321.x
3. Wright MD, Ni J, Rudy GB. The L6 membrane proteins a new four-transmembrane superfamily. Protein Sci. (2000) 9:1594–600. doi: 10.1110/ps.9.8.1594
4. Wright MD, Tomlinson MG. The ins and outs of the transmembrane 4 superfamily. Immunol Today. (1994) 15:588–94. doi: 10.1016/0167-5699(94)90222-4
5. Charrin S, Le Naour F, Silvie O, Milhiet P, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. (2009) 420:133–54. doi: 10.1042/BJ20082422
6. Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Bio. (2005) 6:801–11. doi: 10.1038/nrm1736
7. Monk PN, Partridge LJ. Tetraspanins-gateways for infection. Infect Disord Drug Targets. (2012) 12:4–17. doi: 10.2174/187152612798994957
8. Garcia-España A, Chung PJ, Sarkar IN, Stiner E, Sun TT, DeSalle R. Appearance of new tetraspanin genes during vertebrate evolution. Genomics. (2008) 91:326–34. doi: 10.1016/j.ygeno.2007.12.005
9. Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, et al. The phylogenetic analysis of tetraspanins projects the evolution of cell-cell interactions from unicellular to multicellular organisms. Genomics. (2005) 86:674–84. doi: 10.1016/j.ygeno.2005.08.004
10. Hemler ME. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu Rev Cell Dev Biol. (2003) 19:397–422. doi: 10.1146/annurev.cellbio.19.111301.153609
11. Rubinstein E, Le Naour F, Lagaudrière-Gesbert C, Billard M, Conjeaud H, Boucheix C. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur J Immunol. (1996) 26:2657–65. doi: 10.1002/eji.1830261117
12. Sanyal M, Fernandez R, Levy S. Enhanced B cell activation in the absence of CD81. Int Immunol. (2009) 21:1225–37. doi: 10.1093/intimm/dxp090
13. Figdor CG, van Spriel AB. Fungal pattern-recognition receptors and tetraspanins: partners on antigen-presenting cells. Trends Immunol. (2010) 31:91–6. doi: 10.1016/j.it.2009.11.005
14. Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol. (2005) 5:136–48. doi: 10.1038/nri1548
15. Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp cell Res. (2009) 315:1584–92. doi: 10.1016/j.yexcr.2008.09.020
16. Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Ann Rev Biochem. (2003) 72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800
17. Meyer-Wentrup F, Figdor CG, Ansems M, Brossart P, Wright MD, Adema GJ, et al. Dectin-1 interaction with tetraspanin CD37 inhibits IL-6 production. J Immunol. (2007) 178:154–62. doi: 10.4049/jimmunol.178.1.154
18. Jones E, Demaria M, Wright M. Tetraspanins in cellular immunity. Biochem Soc Trans. (2011) 39:506. doi: 10.1042/BST0390506
19. Artavanis-Tsakonas K, Love JC, Ploegh HL, Vyas JM. Recruitment of CD63 to Cryptococcus neoformans phagosomes requires acidification. Proc Natl Acad Sci USA. (2006) 103:15945–50. doi: 10.1073/pnas.0607528103
20. Suzuki M, Tachibana I, Takeda Y, He P, Minami S, Iwasaki T, et al. Tetraspanin CD9 negatively regulates lipopolysaccharide-induced macrophage activation and lung inflammation. J Immunol. (2009) 182:6485–93. doi: 10.4049/jimmunol.0802797
21. Artavanis-Tsakonas K, Kasperkovitz PV, Papa E, Cardenas ML, Khan NS, Van der Veen AG, et al. The tetraspanin CD82 is specifically recruited to fungal and bacterial phagosomes prior to acidification. Infect Immun. (2011) 79:1098–106. doi: 10.1128/IAI.01135-10
22. Priyathilaka TT, Bathige SDNK, Herath HMLPB, Lee S, Lee J. Molecular identification of disk abalone (Haliotis discus discus) tetraspanin 33 and CD63: Insights into potent players in the disk abalone host defense system. Fish Shellfish Immunol. (2017) 69:173–84. doi: 10.1016/j.fsi.2017.08.020
23. Yu M, Yang S, Sun H, Xia Q. CD63 promotes Hemocyte-mediated phagocytosis in the clam, Paphia undulata. J. Immunol. Res. (2016) 2016:7893490. doi: 10.1155/2016/7893490
24. Moribe H, Yochem J, Yamada H, Tabuse Y, Fujimoto T, Mekada E. Tetraspanin protein (TSP-15) is required for epidermal integrity in Caenorhabditis elegans. J Cell Sci. (2004) 117:5209–20. doi: 10.1242/jcs.01403
25. Luo M, Ye S, Xu T, Wu X, Yang P. Molecular characterization of a novel tetraspanin from the oyster, Crassostrea ariakensis: variation, localization and relationship to oyster host defense. Fish Shellfish Immunol. (2012) 33:294–304. doi: 10.1016/j.fsi.2012.05.009
26. Todres E, Nardi J, Robertson H. The tetraspanin superfamily in insects. Insect Mol Biol. (2000) 9:581–90. doi: 10.1046/j.1365-2583.2000.00222.x
27. Konno A, Padma P, Ushimaru Y, Inaba K. Multidimensional analysis of uncharacterized sperm proteins in Ciona intestinalis: EST-based analysis and functional immunoscreening of testis-expressed genes. Zool Sci. (2010) 27:204–15. doi: 10.2108/zsj.27.204
28. Wang Y, Wang M, Wang B, Liu M, Jiang K, Hou X, et al. A preliminary attempt to explore the potential functions of a tetraspanin gene (MmTSPAN) in the innate immunity of hard clam Meretrix meretrix: sequence features and expression profiles. Fish Shellfish Immunol. (2019) 88:135–41. doi: 10.1016/j.fsi.2019.01.048
29. Zhu XJ, Yang X, He W, Xiong Y, Liu J, Dai ZM. Involvement of tetraspanin 8 in the innate immune response of the giant prawn, Macrobrachium rosenbergii. Fish Shellfish Immunol. (2019) 86:459–64. doi: 10.1016/j.fsi.2018.11.055
30. Lau YT, Gambino L, Santos B, Espinosa EP, Allam B. Regulation of oyster (Crassostrea virginica) hemocyte motility by the intracellular parasite Perkinsus marinus: a possible mechanism for host infection. Fish Shellfish Immunol. (2018) 78:18–25. doi: 10.1016/j.fsi.2018.04.019
31. Morga B, Arzul I, Faury N, Segarra A, Chollet B, Renault T. Molecular responses of Ostrea edulis haemocytes to an in vitro infection with Bonamia ostreae. Dev Comp Immunol. (2011) 35:323–33. doi: 10.1016/j.dci.2010.10.005
32. Tanguy A, Guo X, Ford SE. Discovery of genes expressed in response to Perkinsus marinus challenge in Eastern (Crassostrea virginica) and Pacific (C. gigas) oysters. Gene. (2004) 338:121–31. doi: 10.1016/j.gene.2004.05.019
33. Wang B, Li F, Xiang J, Gui L, Luo Z, Yan H. Three tetraspanins from Chinese shrimp, Fenneropenaeus chinensis, may play important roles in WSSV infection. J Fish Dis. (2010) 33:15–29. doi: 10.1111/j.1365-2761.2009.01079.x
34. Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. (2001) 58:1189–205. doi: 10.1007/PL00000933
35. Zhuang S, Kelo L, Nardi JB, Kanost MR. An integrin-tetraspanin interaction required for cellular innate immune responses of an insect, Manduca sexta. J Biol Chem. (2007) 282:22563–72. doi: 10.1074/jbc.M700341200
36. Gueguen Y, Herpin A, Aumelas A, Garnier J, Fievet J, Escoubas JM, et al. Characterization of a defensin from the oyster Crassostrea gigas recombinant production, folding, solution structure, antimicrobial activities, and gene expression. J Biol Chem. (2006) 281:313–23. doi: 10.1074/jbc.M510850200
37. Yang S, Wu X. Identification and functional characterization of a human sTRAIL homolog, CasTRAIL, in an invertebrate oyster Crassostrea ariakensis. Dev Comp Immunol. (2010) 34:538–45. doi: 10.1016/j.dci.2009.12.014
38. Jiang S, Jia Z, Zhang T, Wang L, Qiu L, Sun J, et al. Functional characterisation of phagocytes in the Pacific oyster Crassostrea gigas. PeerJ. (2016) 4:e2590. doi: 10.7717/peerj.2590
39. Liu R, Qiu L, Yu Z, Zi J, Yue F, Wang L, et al. Identification and characterisation of pathogenic Vibrio splendidus from Yesso scallop (Patinopecten yessoensis) cultured in a low temperature environment. J Invertebr Pathol. (2013) 114:144–50. doi: 10.1016/j.jip.2013.07.005
40. Zhang H, Kong P, Wang L, Zhou Z, Yang J, Zhang Y, et al. Cflec-5, a pattern recognition receptor in scallop Chlamys farreri agglutinating yeast Pichia pastoris. Fish Shellfish Immunol. (2010) 29:149–56. doi: 10.1016/j.fsi.2010.02.024
41. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
42. Cheng S, Zhan W, Xing J, Sheng X. Development and characterization of monoclonal antibody to the lymphocystis disease virus of Japanese flounder Paralichthys olivaceus isolated from China. J Virol Methods. (2006) 135:173–80. doi: 10.1016/j.jviromet.2006.03.016
43. Cao A, Mercado L, Ramos-Martinez JI, Barcia R. Primary cultures of hemocytes from Mytilus galloprovincialis Lmk.: expression of IL-2Rα subunit. Aquaculture. (2003) 216:1–8. doi: 10.1016/S0044-8486(02)00140-0
44. Yu Y, Yu Y, Huang H, Feng K, Pan M, Yuan S, et al. A short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J Immunol. (2007) 179:8425–34. doi: 10.4049/jimmunol.179.12.8425
45. Xu J, Jiang S, Li Y, Li M, Cheng Q, Zhao D, et al. Caspase-3 serves as an intracellular immune receptor specific for lipopolysaccharide in oyster Crassostrea gigas. Dev Comp Immunol. (2016) 61:1–12. doi: 10.1016/j.dci.2016.03.015
46. Wootton E, Dyrynda E, Ratcliffe N. Bivalve immunity: comparisons between the marine mussel (Mytilus edulis), the edible cockle (Cerastoderma edule) and the razor-shell (Ensis siliqua). Fish Shellfish Immunol. (2003) 15:195–210. doi: 10.1016/S1050-4648(02)00161-4
47. Lv Z, Wang L, Jia Z, Sun J, Wang W, Liu Z, et al. Hemolymph C1qDC promotes the phagocytosis of oyster Crassostrea gigas hemocytes by interacting with the membrane receptor β-integrin. Dev Comp Immunol. (2019) 98:42–53. doi: 10.1016/j.dci.2019.04.004
48. Green LR, Monk PN, Partridge LJ, Morris P, Gorringe AR, Read RC. Cooperative role for tetraspanins in adhesin-mediated attachment of bacterial species to human epithelial cells. Infect Immun. (2011) 79:2241–9. doi: 10.1128/IAI.01354-10
49. Patnaik BB, Kang SM, Seo GW, Lee HJ, Patnaik HH, Jo YH, et al. Molecular cloning, sequence characterization and expression analysis of a CD63 homologue from the coleopteran beetle, Tenebrio molitor. Int J Mol Sci. (2013) 14:20744–67. doi: 10.3390/ijms141020744
50. Zhang H, Song X, Wang L, Kong P, Yang J, Liu L, et al. AiCTL-6, a novel C-type lectin from bay scallop Argopecten irradians with a long C-type lectin-like domain. Fish Shellfish Immunol. (2011) 30:17–26. doi: 10.1016/j.fsi.2009.12.019
51. Costa M, Prado-Álvarez M, Gestal C, Li H, Roch P, Novoa B, et al. Functional and molecular immune response of Mediterranean mussel (Mytilus galloprovincialis) haemocytes against pathogen-associated molecular patterns and bacteria. Fish Shellfish Immunol. (2009) 26:515–23. doi: 10.1016/j.fsi.2009.02.001
52. Yang C, Wang L, Zhang H, Wang L, Huang M, Sun Z, et al. A new fibrinogen-related protein from Argopecten irradians (Aifrep-2) with broad recognition spectrum and bacteria agglutination activity. Fish Shellfish Immunol. (2014) 38:221–9. doi: 10.1016/j.fsi.2014.03.025
53. Jing X, Espinosa EP, Perrigault M, Allam B. Identification, molecular characterization and expression analysis of a mucosal c-type lectin in the eastern oyster, crassostrea virginica. Fish Shellfish Immunol. (2011) 30:0–858. doi: 10.1016/j.fsi.2011.01.007
54. Wang L, Zhang H, Wang M, Zhou Z, Wang W, Liu R, et al. The transcriptomic expression of pattern recognition receptors: insight into molecular recognition of various invading pathogens in Oyster Crassostrea gigas. Dev Comp Immunol. (2019) 91:1–7. doi: 10.1016/j.dci.2018.09.021
55. Song L, Wang L, Zhang H, Wang M. The immune system and its modulation mechanism in scallop. Fish Shellfish Immunol. (2015) 46:65–78. doi: 10.1016/j.fsi.2015.03.013
56. Little KD, Hemler ME, Stipp CS. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-Gαq/11 association. Mol Biol Cell. (2004) 15:2375–87. doi: 10.1091/mbc.e03-12-0886
57. Mantegazza AR, Barrio MM, Moutel S, Bover L, Weck M, Brossart P, et al. CD63 tetraspanin slows down cell migration and translocates to the endosomal-lysosomal-MIICs route after extracellular stimuli in human immature dendritic cells. Blood. (2004) 104:1183–90. doi: 10.1182/blood-2004-01-0104
58. Wang XW, Wang JX. Diversity and multiple functions of lectins in shrimp immunity. Dev Comp Immunol. (2013) 39:27–38. doi: 10.1016/j.dci.2012.04.009
59. Dijkstra S, Geisert E, Dijkstra C, Bär P, Joosten E. CD81 and microglial activation in vitro: proliferation, phagocytosis and nitric oxide production. J Neuroimmunol. (2001) 114:151–9. doi: 10.1016/S0165-5728(01)00240-5
60. Chang Y, Finnemann SC. Tetraspanin CD81 is required for the αvβ5-integrin-dependent particle-binding step of RPE phagocytosis. J Cell Sci. (2007) 120:3053–63. doi: 10.1242/jcs.006361
61. Takeda Y, Tachibana I, Miyado K, Kobayashi M, Miyazaki T, Funakoshi T, et al. Tetraspanins CD9 and CD81 function to prevent the fusion of mononuclear phagocytes. J Cell Biol. (2003) 161:945–56. doi: 10.1083/jcb.200212031
62. Pfeiffer A, Böttcher A, Ors ó E, Kapinsky M, Nagy P, Bodnár A, et al. Lipopolysaccharide and ceramide docking to CD14 provokes ligand-specific receptor clustering in rafts. Eur J Immunol. (2001) 31:3153–64. doi: 10.1002/1521-4141(200111)31:11<3153::AID-IMMU3153>3.0.CO;2-0
63. Jiang S, Qiu L, Wang L, Jia Z, Lv Z, Wang M, et al. Transcriptomic and quantitative proteomic analyses provide insights into the phagocytic killing of hemocytes in the oyster Crassostrea gigas. Front Immunol. (2018) 9:1280. doi: 10.3389/fimmu.2018.01280
Keywords: tetraspanin, phagosomes recruitment, receptor, innate immune response, Crassostrea gigas
Citation: Liu C, Yang C, Wang M, Jiang S, Yi Q, Wang W, Wang L and Song L (2020) A CD63 Homolog Specially Recruited to the Fungi-Contained Phagosomes Is Involved in the Cellular Immune Response of Oyster Crassostrea gigas. Front. Immunol. 11:1379. doi: 10.3389/fimmu.2020.01379
Received: 29 February 2020; Accepted: 29 May 2020;
Published: 22 July 2020.
Edited by:
Xinjiang Lu, Ningbo University, ChinaReviewed by:
Zhen Xu, Huazhong Agricultural University, ChinaLinlin Zhang, Institute of Oceanology (CAS), China
Xian-Wei Wang, Shandong University, China
Copyright © 2020 Liu, Yang, Wang, Jiang, Yi, Wang, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingling Wang, d2FuZ2xpbmdsaW5nQGRsb3UuZWR1LmNu; Linsheng Song, bHNoc29uZ0BkbG91LmVkdS5jbg==
 Conghui Liu
Conghui Liu Chuanyan Yang1,3,4
Chuanyan Yang1,3,4 Shuai Jiang
Shuai Jiang Weilin Wang
Weilin Wang Lingling Wang
Lingling Wang Linsheng Song
Linsheng Song