Abstract
Hepatitis B virus (HBV)-associated hepatocellular carcinoma (HBV-HCC) pathogenesis is fueled by persistent HBV infection that stealthily maintains a delicate balance between viral replication and evasion of the host immune system. HBV is remarkably adept at using a combination of both its own, as well as host machinery to ensure its own replication and survival. A key tool in its arsenal, is the HBx protein which can manipulate the epigenetic landscape to decrease its own viral load and enhance persistence, as well as manage host genome epigenetic responses to the presence of viral infection. The HBx protein can initiate epigenetic modifications to dysregulate miRNA expression which, in turn, can regulate downstream epigenetic changes in HBV-HCC pathogenesis. We attempt to link the HBx and miRNA induced epigenetic modulations that influence both the HBV and host genome expression in HBV-HCC pathogenesis. In particular, the review investigates the interplay between CHB infection, the silencing role of miRNA, epigenetic change, immune system expression and HBV-HCC pathogenesis. The review demonstrates exactly how HBx-dysregulated miRNA in HBV-HCC pathogenesis influence and are influenced by epigenetic changes to modulate both viral and host genome expression. In particular, the review identifies a specific subset of HBx induced epigenetic miRNA pathways in HBV-HCC pathogenesis demonstrating the complex interplay between HBV infection, epigenetic change, disease and immune response. The wide-ranging influence of epigenetic change and miRNA modulation offers considerable potential as a therapeutic option in HBV-HCC.
Introduction
In the Globocan 2018 report, 841 080 new liver cancer cases were diagnosed with hepatocellular carcinoma (HCC) (1). Chronic hepatitis B virus (HBV) infection, which has been etiologically implicated in 43% to 80% of total HCC incidence (1–3) remains a primary risk factor and high HBsAg seroprevalence (>5%) levels currently persist in Western Pacific, Africa, East Mediterranean, and East Asia (2). The downstream effect of widespread HBV infection could precipitate 5 million deaths by 2030 from HBV-HCC (3). HBV-HCC pathogenesis is fueled by persistent HBV infection that stealthily ensures a careful balance between replication and the need to remain under the “radar” of the host immune system. HBV is remarkably adept at using a combination of both its own, as well as host machinery to manage viral load and enhance persistence (4, 5). A key tool in this regard, is the manipulation of the epigenetic landscape to modulate the silencing role microRNA (miRNA). The role of HBV dysregulated miRNA at each stage of the HBV-HCC continuum has been well documented from the onset of HBV infection to fibrosis/cirrhosis and the onset of HBV-HCC (6). The regulatory role of HBV dysregulated miRNA has also been extended to specifically examine the interplay between HCC and immune pathways (7).
This review examines the epigenetic role of HBx dysregulated miRNA in HBV-HCC pathogenesis. HBx is a 17 kDa transactivating protein expressed from the X open reading frame of HBV, with little sequence homology to any known genes, hence the name “X”. HBx can modulate several hepatocyte signaling cascades and factors associated with mechanisms that induce cellular transformation. Unlike mammalian hepadnaviruses (HBVs), avian HBVs do not express HBx and do not develop HCC leading to the postulation that the HBx protein has oncogenic potential (8, 9). HBx, can initiate a wide range of epigenetic changes implicated in hepatocarcinogenesis including DNA methylation, histone modifications, chromatin remodeling and microRNA (miRNA) dysregulation (10). One of these epigenetic factors, namely, miRNA are themselves influenced by epigenetic modulation often forming feedback loops that modulate epigenetic change (11, 12).
We attempt to link the epigenetic role of HBx dysregulated miRNA in HBV-HCC pathogenesis in both the HCC and immune pathways. The review commences by describing how the HBx viral protein can modulate both the host, as well as HBV genome expression by influencing the epigenetic landscape. Next a comprehensive list of HBx dysregulated miRNA are presented in Table 1, which are influenced by upstream epigenetic changes, and/or their downstream epigenetic targets. This table also identifies host gene targets in both the HBV-HCC cancer and immune pathways. Four comprehensive examples of epigenetically modified miRNA are then outlined in the next section to demonstrate the complex interplay between viral infection, HBV-HCC pathogenesis and immune system response. The review then examines the potential therapeutic role of miRNA that has yet to be deployed outside of the laboratory setting.
Table 1
| HBx-epi-regulator | miRNA | Epi target | HBV-HCC gene target | Immune gene target | Epi-Ref | HCC Reference |
|---|---|---|---|---|---|---|
| HMT/EZH2 LIN28/tet1/EED/SUZ12 | Let-7c | SUZ12/EED/EZH1/2 | CNKD1/PRICKLE/SFRP5/B-CAT/STAT3/RAS/HMGA2/MYC/ IL-6/IL-10/TLR-4/COL1A2/NGF/BCL-XL/BCL-2/MCL-1z | MYC/STAT3/IFN-b/RAS/TLR4 BCL-XL/SMAD2/SMAD4/NFZ APC2/WNT1/HMGA2/PLZF/IFN/IL-4/IL-17/LIN28B/IGF2BP1 | (13–15) | (16–19) |
| DNMT | miR-1 | HDAC4 | EDN1/PI3K/AKT/METFOXP1 | E2F5/HSP60/HSP70/KCNJ2/GJA1 | (20–22) | (20, 21, 23) |
| HMT/EZH2/EED/C-MYC/SUZ12 | miR-101 | DNMT3A/EZH2/EED/SUZ12/ | GSTP1/FOS/MCL-1/RASSF1A/PRDM2/CNKD1/PRICKLE/SFRP5/B-CAT/AP1/DUSP1/MCL-1/ROCK2/ATG4D/MTOR/SOX9/COX2/RAB5A/STMN1/DNMT3A/FOS/RAP1B/VEGF | ICOS (naïve T-cells)/MCL-1 | (13, 14, 24) | (25) (14, 24, 26–28) |
| DNMT/PPARα | miR-122 | CTNNB1/CCNG1 modulated p53/GLD2/NDRG3/GALNT10/CCNG1/PTTG1/PBF/ADAM10/CCNG1/Igf1R/ADAM 17/BCL-W/NDRG3 | SOCS3/IFN/IP-10/BCL-W | (29, 30) | (31–34) | |
| DNMT1 | miR-124 | EZH2/BMI1 | STAT3/PIK3CA/ROCK2/STAT3/Cyclin D/CDK6, VIM, SMYD3, E2F6, IQGAP1 | STAT3/TRAF6/CYCLIND3/BM11 | (14, 35, 36) | (37) |
| miR-125a | SIRT7 | MMP11/VEGF-A/ERBB2/HBsAg | NF-α/BCL-2/KLF13/BMF | (38, 39) | (40–42) | |
| EZH1/2/HMT/SUZI2/EED/p53 | miR-125b | SIRT7/SUZI2 SUV39H1/EED/EZH1/EZH2/ | SMAD2/4/Sirtuin7/SUV39H1/LIN28 B/PIGF/BCL-2/MCL-1/CNKD1/PRICKLE/SRFRP5/B-CAT/PIGF/MMP2/MMP9 | PRDM1/IRF4/TNF_/BCL-2/MCL-1/LIN28/IRF4/KLF13/BMF/BCL-2/SMAD2/SMAD4 APC2/WNT1//KLF13/TRP5 3INPI/LIN28A/IRF4/BLIMP1 IRF4/BMF | (13, 36, 43, 44) | (45, 46) |
| DNMT3 | miR-132 | p300 | AKT/GSK3/WNT-BCAT | p300/IRAK4/FOXO3/SOX4/ | (47, 48) | (48) |
| HMT/EED/SUZ12/EZH2 | miR-139-5p | EED/SUZ12/EZH1/2 | ZEB1/2/CNKD1/PRICKLE/SFRP5/B-CAT | IL-4/IFN-γ | (13, 14) | (45, 49, 50) |
| miR-140 | DNMT1 | NF-kB/TGFβRI/FIF9/Pin1 | (51) | |||
| miR-145 | HDAC2 | MAP3K/CUL5/ADAM17 | IFN-b/TIRAP/TRAF6 | (52–54) | (55–57) | |
| DNMT1/p53C-MYC | miR-148a | DNMT1 | HPIP/AKT/ERK/FOXO4/ATF5/ERBB3/BCL-2/IRS-1/MTOR/MET/ACVR1/SNAIL/IGF-IR/MIG6/CAND1/CDC25B | CaMKIIα/KIT/MET/SIPI/BACH/PTEN/BIM/GADD45 | (58, 59) | (45, 60–62) |
| miR-152 | DNMT1/DNMT3A | GSTP/CDH1/KIT | CaMKII/KIT | (22, 47, 63) | (42, 63–65) | |
| HDAC-I/EZH2 | miR-155 | PRC2/Phf19/p300/CBP | PTEN/SOX6/ZHX2/SOCS1 | AID/Blimp-1/PRDM1/IFN/SHIP1/SOCSI/BMAL1/PU.1/BACH1/CSFIR/CEBP/ETS1/Th2 induction3/SOCS1/C/EBP/AID/FOXP3 | (43, 66, 67) | (66, 68, 69) |
| HDAC-I SAHA/ C-MYC | miR-17-92 family | DNMT | E2F1, Cyclin G1/PTEN/p21/p27/p57/cccDNA | TNFSF9/CCL-5/IKBKE/c-MAF/AMLI/TP53INPI c-MAF/IFN/CD69/PTEN/TGFBR11/p27/p21/E2F/PHLPP2/BIM/CREB1 | (70, 71) | (72–74) |
| HMT | miR-199a/b | CHC | CD19+ | (75) | (76) | |
| DNMT/p53 | miR-200a | HDAC4 | ZEB1/2/HNF-3β Rho/ROCK/ASB4 | (77, 78) | (45, 79) | |
| EED/SUZ12/EZH1/2 | miR-200b | EED/SUZ12/EZH1/2 | CNKD1/PRICKLE/SFRP5/B-CAT | (13, 78) | (45, 79) | |
| EZH2/BMI1 | miR-203 | RAP1A | SMAD1/BCL11B/RARB/PRKCA/PRKCB1/FMRP | (80) | (81) | |
| DNMT | miR-205 | ACSL4/E2F1/ZEB1/2 | (82) | (45, 82, 83) | ||
| DNMT | miR-221 | HDAC6 | ERα/DDIT4/BMF/p27 p57/PTEN/p21/SOCS3 | PTEN/SOCS3/p57/KIT/p27 | (84, 85) | (86–88) |
| DNMT | Mir-222 | P27kip 1/PTEN/PPP2R2A/p57/p21 | p27 kip 1/PTEN/KIT | (85) | (55, 88–90) | |
| HAT/HDAC1/3/EP300/p50/p65 | miR-224 | PAK4/MMP9 inhibitor-5/SMAD4 | AP15/SMAD4 | (91, 92) | (93–95) | |
| miR-26a | EZH2 | IL-6/IFN_/ER_/Cyclin D2/Cyclin E2/c-JUN/CDK4/6 | IFN-b CDK4/6/MALT1 | (96–98) | (99–101) | |
| miR-29c | DNMT3B/SIRT1 | BCL-2/MCL-1/TNFA1P3 | TCL-1/MCL-1/IFN- | (102–104) | (105, 106) | |
| H3K4ac | miR-29a/b | DNMT1/DNMT3A/B/SETDB1/H3K9/SIRT1 | PTEN/PI3K/AKT/MMP-2 | IFNARI/IFN/T-Bet/EOMES/PTEN/MCL-1/IFN-/SLFN4/CDC42/HBP1/TCL1 | (22, 102, 103, 107, 108) | (55, 109) |
| HBx/P53/DNMT1/3A/3B | miR-34a | SIRTI | CCL22/MAP4K4/SIRT1/CCND1/CDK4/6/MET/C-JUN/CDK2 | IFN-b/FOXP1/CDK2/4/6/CCL22/FOXPN | (110) | (111–115) |
| HDAC | miR-373 | HDAC/SIRT1 | SNAIL-1/CDH1 | MTOR/SIRT1/RELA | (116) | (116) |
| miR-548a | HDAC4 | HBXIP, IFN-λ1 | IFN-λ1 | (117) | (118, 119) |
HBx-dysregulated epi-miRNA and their targets in HBV-HCC.
Column 1 indicates upstream epigenetic proteins that can influence miRNA expression in column 2 (blue miRNA- downregulated and red miRNA-upregulated). Column 3 is downstream epigenetic targets of dysregulated miRNA. Columns 4 and 5 are specific HBV-HCC and immune gene targets. Column 6 refers to epigenetic references only (column 1 and 3) and column 7 to HBV-HCC and immune gene targets.
HBV-HCC Pathogenesis: Role of miRNA
Persistent HBV infection remains a global risk factor that can promote the development of fibrosis/cirrhosis and ultimately the onset of HBV-HCC (120). A significant component of HBV-HCC pathogenesis involves the integration of HBV DNA into the host genome that results in the oncogenic disruption of cellular genes (121). HBV DNA integration can cause host cell deletions, cis/trans-activation, translocations, the increased production of fusion transcripts, aberrant epigenetic changes and generalized genomic instability (122). These changes take place in the form of chronic inflammation and tissue damage that result in the continuous destruction of well differentiated hepatocytes and organized extracellular matrix (ECM). Over time the depletion of hepatocytes and well organized ECM results in their replacement with undifferentiated liver stem cells and poorly organized fibrotic tissue (120) that display changing patterns of apoptosis, regeneration, senescence and survival (123). HBV-HCC pathogenesis involves the deregulation of many cellular signaling pathways including the Wingless-related integration site/Beta-Catenin (WNT/β-CAT), in the Retinoblastoma-Tumor Protein 53 (RB1-TP53) suppressor networks, the Phosphoinositide 3-kinase/mitogen-activated protein kinase (PI3K/MAPK), the Janus kinase/signal transducer (JAK/STAT) pathways and the insulin receptor substrate-1/insulin growth factor (IRS1/IGF) pathways (124–126).
MicroRNA (miRNA) are a subsidiary subset of epigenetic factors that act as post-transcriptional gene silencers in the HBV-HCC pathways. miRNA collectively attempt to repress target mRNA expression in order to ensure homeostasis and their fluctuating role is explained by the inherently stochastic nature of gene transcription and environmental fluctuations (127). In the HBV-HCC continuum, from asymptomatic HBV infection leading to HCC, an increasing number of miRNA are dysregulated due to the need to respond to viral infection, epigenetic changes (35), inflammation (128), fibrosis (129), cirrhosis (123) and finally, the onset of HCC. One of the most documented HBV tools to modulate both its own, as well as host genome expression is the HBx protein which has been shown to dysregulate multiple miRNA species in key HCC cancer and immune pathways (7).
HBx Induced Epigenetic Changes in HBV-HCC Pathogenesis
HBV DNA and its proteins influence own and host genome expression by employing a range of epigenetic modifications in HBV-HCC pathogenesis, as well as modulating signal transduction, transactivation and transcription to regulate immune response, cell cycle, apoptosis and DNA repair (130). In particular, the HBx protein influences DNA methylation (131), histone modifications (132), chromatin remodeling (133) and miRNA dysregulation (6) which is the central focus of this review (Figure 1).
Figure 1

HBx induced epigenetic expression in HBV-HCC. (A) HBx recruits the CUL4/DDB1 ubiquitination complex to repress SMC5/6 and promote DNA relaxation and cccDNA transcription. (B) HBx recruits CBP/p300 (pCAF) to transactivate HAT (H3K9ac) to promote cccDNA expression. (C) HBx promotes DNMT to repress SOCS1/RASSF1A tumor suppressors that fail to modulate HCC pathogenesis. (D) HBx promotes PLK1 to block HMTs (EZH1/2/EED/SUZ12) that then fail to modulate B-CATENIN induced hepatocarcinogenesis. (E) HBx initiates HMTs (SMYD3) to promote C-MYC induced HCC. (F) HBx promotes HDAC to silence cell cycle controls like p21/p26 and promote HCC.
Regulation of cccDNA Activity
The HBV covalently closed circular DNA (cccDNA), a stable mini-chromosome that is classified as nuclear episomal DNA, serves as the template for viral RNA transcription. In order to enhance the efficiency of HBV replication, this virus co-opts host transcription machinery like HNF1/2/3, C/EBP, CREB and CRTC1 to trigger cccDNA transcription (4). The HBx protein appears to be co-opted to influence its own transcription by triggering histone modifications including histone acetylation and deacetylation (HAT/HDAC), histone methylation and demethylation (HMT/HDMT), DNA methylation (DNMT) and ubiquitination (5). An example in this respect, is when cccDNA transcription is increased the HBx protein co-opts DDB1 and Cul4 to create a ubiquitination complex to repress SMC5/6 which maintains the structure of chromosomes. The repression of SMC5/6, therefore, promotes DNA relaxation to increase the transcription of cccDNA (131, 134) (Figure 1A). The HBx protein can also increase cccDNA transcription by recruiting CBP/p300 (pCAF) to transactivate HATs (H3K9ac) that leads to hyperacetylation (131, 135) (Figure 1B).
DNA Methylation
DNA methylation, the addition of a methyl group by the enzymes DNMT1/2/3A/3B, can silence target gene promoters or enhance target gene expression by silencing transcription regulators in both the HBV and host genome (4, 5). In percentage terms, common host genes targeted for DNA methylation in HBV-HCC include WT1 (54%), SOCS1 (43-65%), SEMA3B (83%), RASSF1A (59-75%), p300 (65%), p27 (48%), p21 (63%), p16 (16-83%), GSTP1 (41-76%), E-cadherin (33-67%), E2F1 (70%), CPS1 (80%) and APC (53-81%) (10). In many cases these epigenetic changes silence tumor suppressor or activate oncogenic proteins and the HBx protein influences DNMT expression to subdue the host immune response, as well as manage its own expression to promote a balance between survival and replication (4). The HBx protein, for example, often promotes DNMT1/3A/3B expression to silence tumor suppressor genes like SOCS1/RASSF1A in HBV-HCC pathogenesis (4, 10) (Figure 1C). It can also use this machinery to suppress cccDNA expression in order to evade the host immune system (136).
Histone Modifications
Histone modifications are post translation modifications to histone proteins that include HAT/HDAC, HMT/HDMT, phosphorylation and ubiquitylation. These post translational modifications primarily occur at the N-terminal of the histone tails and have a fundamental impact on chromatin remodeling which essentially alters the structure of host and HBV DNA in HBV-HCC pathogenesis. For example, the HBx protein can promote HAT by recruiting trans-activator proteins like CBP/p300 complex to induce H3K9ac (135) Conversely, the HBx protein can induce HDAC to promote cell proliferation in HBV-HCC by repressing tumor suppressors like p21/p27 that are important regulators of cell cycle control (132) (see Figure 1F) and promote epithelial-mesenchymal transition (EMT) by repressing CDH1 (137). The HBx protein can also use HDAC to modulate the viral genome expression by recruiting HDAC1/2 to cccDNA to suppress its expression (4, 5). In addition, HBx also induces HMT by upregulation of the SMYD3 gene, which encodes a histone H3-K4-specific methyltransferase to trigger oncogene expression (138). The HBx protein can also induce histone methylation transferases and it has been demonstrated that HBx induced upregulation of SMYD3, that encodes for histone H3-K4-specific methyltransferase (HMT), is linked to the upregulation of the oncogene C-MYC in HCC (138, 139) (Figure 1E). Conversely, the HBx protein can reverse the repressive effect of histone methylation (H3K9me3) on cccDNA by initiating a histone demethylation (HDMT) agent (140).
Polycomb Proteins
The Polycomb repressive complex (PcG) proteins, namely, Polycomb repressive complex 1 (PRC1) and 2 (PRC2) form part of the histone modification machinery that epigenetically regulate chromatin remodeling. PRC2 participates in histone methylation (H3K27me3) and, following histone H2AK119 mono-ubiquitination by PRC1, collaboratively represses target gene transcription (141). In the HBV-HCC continuum, the HBx protein can co-opt the proteins of these two complexes to influence epigenetic changes. For example, HBx upregulates the proto-oncogene PLK1, an enzyme that can block the repressive effect of the PRC2 complex (SUZ12/EED/EZH1/EZH2) to down-regulate WNT antagonists (CNDK1/PRICKLE/SFRP5). The repression of these WNT antagonists leads to increased β-catenin transcription and hepatocarcinogenesis (13, 142) (Figure 1D).
HBx Dysregulated lmiRNA in HBV-HCC and Epigenetic Change
In the HBV-HCC continuum, the HBx protein can influence epigenetic changes like DNA methylation, histone modifications and other non-coding RNA that dysregulate a specific subset of miRNA (called epi-miRNA) that forms the central focus of this review (Table 1). In addition, the HBx protein can dysregulate host genes that modulate miRNA biosynthesis, transcription and translation (47). Simultaneously, miRNA expression modulates downstream epigenetic modulation by targeting epigenetic modifiers suggesting epigenetic feedback loops that directly influence both miRNA and their downstream epigenetic targets (4, 10, 45). In the HBV-HCC continuum, HBx-dysregulated miRNA, therefore, are epigenetic regulators that are themselves epigenetically modulated. In Table 1 we show the HBV-HCC specific gene targets of miRNA identified in the literature, as well as their specific immune targets in HCC. It should be borne in mind that these are dynamic pathways and that the proposed regulatory effect of miRNA is also dynamic.
The HBx protein can upregulate or downregulate specific miRNA expression using specific epigenetic regulation (Table 1). These mechanisms, for instance, include DNA methylation of miR-1/-122/-124/-132/-/148/-200/-205 genes to downregulate miRNA expression (24, 48), histone acetylation or HDAC inhibitors to upregulate miR-224/-29/-155/-17-92 and histone methylation to downregulate Let-7c/miR-101/-125b/-139-5p (45). The HBx protein can also target upstream transcription factors essential for miRNA expression like p53 suppression of miR-23a/-34/-125b/-148a/-192/-200 (143) and C-MYC upregulation of miR-15a/-16/-26a/-101/-148a/-363 (144). This protein also targets p50/65 upregulation of miR-143/-224 (93)and NF-KB upregulation of miR-143/-146a, as well as dysregulating miRNA expression by repressing miRNA biosynthesis machinery like DROSHA (145, 146).
HBx-Dysregulated miRNA in HBV-HCC Pathways
In the HBV-HCC continuum, upregulated miRNA often reduce tumor suppressor expression in the four key HCC cancer pathways, namely, the P13K/MAPK, WNT/β-Catenin, TP53 and JAK/STAT pathways (125). In this section we seek to demonstrate the complexity of the interlocking roles of viral infection, selected epigenetic changes of miRNA in HBV-HCC pathogenesis and the resulting modulation of the host immune system. We illustrate some of the proven epigenetic pathways in HBV-HCC pathogenesis by using four well researched miRNA (miR-29a/b, miR-155, miR-148/152 and miR-101). Two of these miRNA are downregulated (blue) and two are upregulated (red) (see Figures 2–5). These four miRNA are all HBx epigenetically dysregulated in various HBV-HCC pathways, as well as exercise diverse roles in both the innate and adaptive immune pathways. In many cases, the same miRNA plays a significant regulatory role in many other cancers like those of the breast, lung and colon (147). It is important to highlight that the illustrated hypothetical pathways in Figures 2–5 occur in a dynamic context and that the degree of influence of any single path is non constant. It is also important to keep in mind that the HBx protein can modulate miRNA via non epigenetic pathways (e.g. C-MYC/p53) and that in HBV-HCC pathogenesis multiple other factors (e.g. somatic mutations) also influence miRNA expression (45).
Figure 2

Epi-miR-101 in HBV-HCC and immune pathways. HDAC-Histone deacetylation, HAT-histone acetylation, HMT-Histone methylation, DNMT-DNA methylation (pathways in dark red involve identified direct upstream or downstream epigenetic proteins/enzymes). HBx protein can downregulate or initiate HDAC/HAT/DNMT/HMT to repress miR-101 modulation in various HBV-HCC pathways including WNT-B-CATENIN, TP53, and P13K/AKT to influence HCC pathogenesis. HBx epigenetically downregulated miR-101 also regulates macrophage and DC expression in the innate immune system, as well as T-cell and B-cell expression in the adaptive immune system.
Figure 3

Epi-miR-29 in HBV-HCC and immune pathways. HDAC-Histone deacetylation, HAT-histone acetylation, DNMT-DNA methylation, PRC1/2- Polycomb Repressive Complex, SETDB1-SET Domain Bifurcated Histone Lysine Methyltransferase, TET1-Ten-eleven translocation methylcytosine dioxygenase 1 (pathways in dark red involve identified direct upstream or downstream epigenetic proteins/enzymes). HBx protein can downregulate or initiate HDAC/HAT/DNMT/HMT to upregulate miR-29 modulation in various HBV-HCC pathways including AKT/MTOR, TP53, and JAK/STAT to influence HCC pathogenesis. HBx epigenetically upregulated miR-29 also regulates macrophage and DC/NK expression in the innate immune system, as well as T-cell and B-cell expression in the adaptive immune system.
Figure 4

Epi-miR-148/152 in HBV-HCC and immune pathways. HDAC-Histone deacetylation, HAT-histone acetylation, DNMT-DNA methylation, HMT-Histone methylation (pathways in dark red involve identified direct upstream or downstream epigenetic proteins/enzymes). HBx protein can downregulate or initiate HDAC/HAT/DNMT/HMT to repress miR-148/152 modulation in various HBV-HCC pathways including WNT-B-CATENIN, TP53, and AKT/mTOR to influence HCC pathogenesis. HBx epigenetically downregulated miR-148/152 also regulates DC expression in the innate immune system, as well as T-cell and B-cell expression in the adaptive immune system.
Figure 5
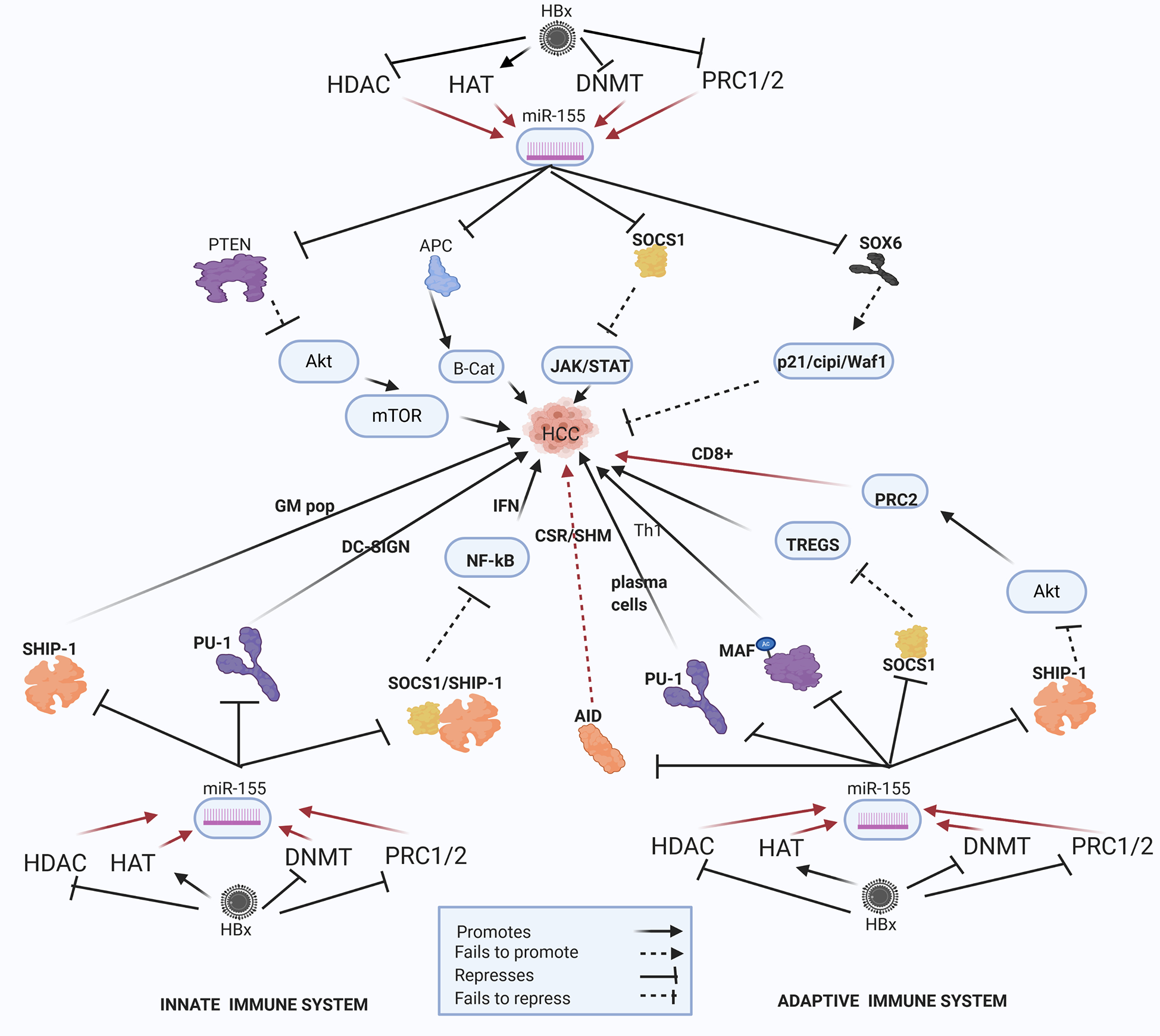
Epi-miR-155 and HBV-HCC and immune pathways. HDAC-Histone deacetylation, HAT-histone acetylation, DNMT-DNA methylation, PRC1/2- Polycomb Repressive Complex 1 and 2 (pathways in dark red involve identified direct upstream or downstream epigenetic proteins/enzymes). HMT-Histone methylation. HBx protein can downregulate or initiate HDAC/HAT/DNMT/HMT to upregulate miR-155 modulation in various HBV-HCC pathways including AKT/MTOR, TP53, and JAK/STAT to influence HCC pathogenesis. HBx epigenetically upregulated miR-155 also regulates germinal matrix (GM) population and NF-kB expression in the innate immune system, as well as T-cell and B-cell expression in the adaptive immune system.
HBx-Dysregulated Epi-miR-101
HBV-HCC Pathogenesis
In HBV-HCC and many other cancers, the tumor suppressor miR-101 is regarded as a key miRNA in epigenetic systems (147) (see Figure 2). This HBx-dysregulated miRNA involves both upstream and downstream epigenetic systems and has been widely reported as downregulated in HBV-HCC (147). The HBx protein can recruit the Polycomb protein EZH2 (148) to downregulate miR-101 but a feedback loop exists because this miRNA also targets EZH2 (149). In HBV-HCC pathogenesis a demonstrated EZH2 downregulated miR-101 pathway promotes HCC progression as a result of failing to modulate COX-2 activated AKT signaling (14, 150). This downregulated miRNA also fails to block the oncogenic MYCN, a member of the MYC family of proteins that are widely cited in many cancers including HCC. The MYC family can directly promote proliferation by promoting CDK/CYC expression (14, 149, 150) as well as promote angiogenesis via promoting VEGFA expression (151). In HBV-HCC pathogenesis, a direct epigenetic target of this miRNA is DNMT3A which targets a range of tumor suppressors. It has been demonstrated that HBx-downregulated miR-101 fails to modulate DNMT3A expression in HBV-HCC and this can contribute to the silencing of tumor suppressor genes like SF1/PRDM2/GSTP1/RUNX3/APC/CDKN2A/STMN1 (24, 149). The silencing of cell cycle inhibitors like CDKN2A in the TP53 pathway, for example promotes cell proliferation in HCC while silencing of the APC tumor suppressor promotes β-catenin expression and the development of EMT (152, 153) HBx (EZH2) downregulated miR-101 can also fail to modulate MCL-1, a key anti-apoptotic member of the BCL-2 family, thus promoting survival in HCC cells as a result of suppressing caspase driven apoptosis (152, 154). This downregulated miRNA also fails to repress the proto-oncogene c-FOS which regulates transcription activity and results in increased invasiveness in HCC pathogenesis (26). EZH2 downregulated miR-101 also fails to modulate CCDC88A that codes for the oncogenic protein GIRDIN which regulates many signal transduction pathways such as AKT/PKB, GAI/S, EGFR and is linked to increased migration and invasiveness in HCC (155). Conversely, downregulated miR-101 also targets the tumor suppressor PTEN in the P13/MAPK pathway that contributes to the activation of this pathway and HBV-HCC pathogenesis (156).
Innate Immune System
Downregulated miR-101 can downregulate the activation of LPS-stimulated macrophages by failing to modulate MKP-1 which de-activates p38 and JUN induction of pro-inflammatory cytokines like TNF-α (157, 158). In another pathway in the innate immune system, LPS-stimulated macrophages are reduced by HBx repressed miR-101 as a result of its failure to modulate DUSP1 which, in turn downregulates ERK1/2/p38/JNK promotion of pro-inflammatory cytokines like TGF-β (159). Reduced DUSP1 is often noted in HCC, however, and it has been demonstrated that it plays a protective role in HCC by lowering the ERK cascade and thus repressing cell proliferation (160). TGF- β plays a major role in the regulation of inflammatory processes that influence immune cell development (159). Alternatively, miR-101 can play a role macrophage expression via activating the NF-KB signally as a result of the upregulation of the KPNB1 transport protein (161–163) which is commonly reported in many cancers including HCC (164). In the innate immune system the activation of dendritic cells (DCs) and neutrophils is also directly influenced by NF-KB signaling suggesting that miR-101 can potentially play a wider role in the activation of leucocytes (165).
Adaptive Immune System
The differential expression of miR-101 has been implicated in modulating T-cell activation. A direct target of miR-101 is the Inducible T-cell co-stimulator (ICOS) mRNA that acts in concert with interaction between T cell receptor(TCR) and MHC class 1 and 2 peptides (166, 167). T-cell function is highly dependent on miR-101 modulation of ICOS mRNA and a reduction in miR-101 mediated regulation can increase ICOS expression on naïve T-cells increases, causing an effector T-cell-like phenotype and that results in autoimmunity (166). HBx-induced stimulation of EZH2 suppresses miR-101 in HBV-HCC (14, 24) and this effect could lead to ICOS upregulation that influences T-cell activation (166). HBx-downregulated miR-101 in HBV-HCC results in a failure to modulate the anti-apoptotic protein MCL-1 thus promoting cell survival (152). Acting alongside this relationship, MCL-1 expression influences an increase in CD8+ T-cell activity (168). A further hypothetical pathway, supported by various studies indicates that miR-101 can initiate T-cell expression, T-cell differentiation and memory T-cells via activating NF-KB signaling by promoting the expression of the KPNB1 transport protein. KPNB1 is often reported as upregulated in many cancers including HCC (161, 162, 169).
B-Cells
HBx-downregulated miR-101 can also influence B-cell development via the regulation of MCL-1 which is regarded as a crucial input to B-cell synthesis (170).
HBx-Dysregulated miRNA-29 Family
HBV-HCC Pathogenesis
HBx upregulated miR-29a/b plays a key epigenetic role in modulating aberrant DNA methylation which is often a key feature of HBV-HCC pathogenesis (171) (Figure 3). HBx-upregulated miR-29 can repress DNMT3A/3B in HBV-HCC (102) acting as a feedback mechanism to modulate DNA methylation. This can influence a range of downstream effects because DNMT3A/3B often targets cell cycle controls in HBV-HCC including CDKN2A, p16(INK4A) and p15(INK4B) (171). By repressing DNMT3A/3B, HBx-upregulated expression of miR-29 acts in support of host cell cycle controls. DNMT3A/3B also silences many tumor suppressors in HBV-HCC pathways like RASSF1/PRDM2/GSTP1/RUNX3/APC therefore, miR-29 induced DNMT3A/3B repression contributes to a secondary support system for tumor suppressor expression to slow HCC progression (172). HBx-upregulated miR-29 family members could increase cell survival by targeting BCL-2 proteins (MCL-1) that retard apoptosis by contributing to the down regulation Caspase 9/3 driven apoptosis in the TP53 cancer pathway (105, 173, 174). The upregulation of the histone methyltransferase (HMT) SETDB1/SIRT1 is a common feature in HCC. HBx-upregulated miR-29, therefore, acts as a tumor suppressor to modulate the histone methylation transferase SETDB1/SIRT1 leading to a reduction in hepatocarcinogenesis possibly because of a reduction in AKT signaling and/or an increase in pro-apoptotic expression (107, 175). The upregulation of miR-29b has been demonstrated to increase HCC carcinogenesis by repressing SOCS1 expression in the JAK/STAT pathway via directly targeting the TET1 DNA demethylation enzyme (176, 177).
Innate Immune System
HBx-upregulated miR-29 upregulates LPS induced macrophage activation by repressing AKT1 suppression of a pro-inflammatory response resulting in increased IL-6, IL-1β and NF-KB signaling (178, 179). In addition, the upregulation of miR-29 can repress Il-12/Il-23 activation of mature DCs (180) and can influence NK production via targeting TBX21/EOMES promotion of IFN-γ (181).
Adaptive Immune System
The miR-29a/b cluster plays a crucial role in the thymic production of T-cells, T-cell differentiation and B-cell oncogenic transformation (182, 183) and miR-29 has been cited as a repressor of the immune system because it directly targets IFN-λ in IFN producing immune cells (184). It has also been demonstrated that miR-29 targets IFNAR1 to promote T-cell production (185). In the presence of infection, type 1 IFN signaling and T-BET/EOMES expression modulate Th1:Th2 differentiation. In turn, miR-29a/b directly targets type 1 IFN/T-BET/EOMES thus playing an important role in Th1:Th2 differentiation. Upregulated miR-29a/b blocks type 1 IFN/T-BET/EOMES to promote Th2 expression and reduce Th1 expression. In the HBV-HCC continuum, miR-29a/b is upregulated by the HBx protein suggesting a viral intervention to promote modulate Th1:Th2 differentiation (182). A similar role is played by miR-29a/b when this miRNA is downregulated by intracellular bacteria and fails to modulate type 1 IFN resulting in an imbalance of the production of CD8+ T-cell (182, 183). The upregulation of miR-29 therefore tilts the Th1:Th2 ratio in favor of Th2 expression that also influences B-cell production (184). In addition, the upregulation of miR-29 can repress Il-12/Il-23 activation of Th17 cells (180).
HBx Epigenetically Dysregulated miR-148/152 Family
HBV-HCC Pathogenesis
HBx downregulation of miR-148/152 has a downstream influence on multiple HBV-HCC pathways (Figure 4). HBx-downregulated miR-152 influences DNMT1 to silence both the CDH1 and GSTP1 tumor suppressors to promote carcinogenesis. The downregulation of CDH1 increases B-CATENIN in the WNT/B-CATENIN pathway and the downregulation of GSTP1 increases cell proliferation (63). Interestingly HBx activation of DNMT1 also represses cell cycle controls like p16INK4A to promote cell proliferation (186) thus further widening the downstream epigenetic change in HBV-HCC pathways. HBx- downregulated miR-148a fails to suppress HPIP induced upregulation of the mTOR pathway contributing to EMT and increased hepatocarcinogenesis (60). The progression of EMT is also less modulated as a result of this downregulated miRNA failing to repress expression in the MET/SNAIL/EMT pathway contributed to progression of HBV-HCC (61). This downregulated family also fails to regulate the production of B-catenin as a result of the reduced repression of WNT1 signaling (187). HBx-downregulated miR-148a increases SMAD pathway expression by failing to directly regulate TGF-B signaling and by failing to repress USP4 induced TGF-B signaling (188, 189).
Innate and Adaptive Immune Pathways
The miR-148/152 family play an especially important role modulating the adaptive immune system. In the adaptive immune system this family targets many genes that influence B and T lymphocyte function. This HBx-downregulated miRNA in HBV-HCC targets BCL2L11/TWIST to influence T-cell differentiation and BACH2/MITF to influence B-cell differentiation (190, 191). This family also plays a role in B-cell tolerance and elevated levels of miR-148a which have been noted in autoimmune disorders. In particular, miR-148a targets GADD45, BIM and PTEN that suppress B-cell tolerance (192). This family also targets CAMK11α to suppress MHC class II levels in antigen stimulated DCs that promote T-cell activation. It can thus be hypothesized that if the HBx protein epigenetically downregulates miR-148/152 then this would result in reduced suppression of MHC class II levels in DC activation of T-cells (193). This family, therefore, plays an important role in the innate system as a result of its ability to modulate antigen presenting (APC) DCs which are regarded as the most important class of APC in the innate immune system. DCs, therefore, link the innate and adaptive immune systems (194). This family also modulates IL-6, TNF-α and IFN-β to repress TLR induced DC activation and it can be hypothesized that downregulated miR-148/152 would fail to repress IL-6/TNF-α/IFN-β induced DC expression (194).
HBx Epigenetically Dysregulated miR-155
HBV-HCC Pathogenesis
HBx-upregulated miR-155 is a key epi-miRNA that targets both the HBV-HCC immune and cancer pathways (Figure 5). HBx dysregulation of miR-155 can occur due to histone modifications like histone deacetylase inhibitors (HDAC-I) or the repression of polycomb proteins (EZH2) can contribute to upregulated miR-155 expression in HBV-HCC pathways (43, 66, 67). This well researched miRNA is cited as an epigenetic modulator in many cancers including those of the breast, lung and colon (195–197), as well as playing multiple different roles in both the innate and adaptive immune system response (7). In HBV-HCC pathogenesis, upregulated miR-155 typically represses PTEN modulation of AKT/MTOR signaling in the P13K/MAPK pathway that promotes epithelial to mesenchymal transition (EMT) (68, 198). In addition, this miRNA can promote β-catenin expression in the WNT/β-Catenin pathway by repressing the APC/GSK3 destruction complex to thus promoting the transcription of oncogenic proteins like C-MYC (45, 199). This miRNA also represses the SOCS1 tumor suppressor in the JAK/STAT pathway to induce the transcription of CCND1 and c-MYC thus promoting HCC cell proliferation (200, 201). In the TP53 pathway, this key HBx epigenetically upregulated miRNA can repress SOX6 to negate its promotion of p21/Waf1/cip1 modulation of cell cycle controls directly promoting HCC proliferation (6, 69). In a strategy to possibly evade immune system response, this HBx upregulated miR-155 can also subdue HBV replication by blocking the CCAAT/enhancer-binding protein (C/EBP) protein that binds and activates the HBV Enhancer 11/core promoter (199).
Innate Immune System
Epigenetically upregulated miR-155 is a key modulator of pro- and anti-inflammatory responses in the innate immune system (202, 203). It is a particularly important miRNA in the modulation of NF-KB driven induced myelopoiesis as a result of targeting IRAK1/TRAF6 and SHIP1/SOCS1 respectively (204–206) and also targets CSFR to influence myeloid differentiation (207).
Macrophages
SHIP1, an important regulator of the innate system, is a primary target of miR-155 and its repression influences an increase in granulocyte/monocyte cell populations and a reduction in lymphocyte numbers (208, 209) and reduced levels of SHIP1 appears to induce myeloproliferative disorders (208). Interestingly, SHIP-1 is classified as a tumor suppressor in HBV-HCC and reduced levels of SHIP-1 are associated with a poorer prognosis (210). Upregulated mIR-155 in viral infection can induce type 1 IFN induced macrophages via by activating the TLR4/MyD88/JNK/NF-KB dependent pathway. In order to upregulate TLR4 signaling, upregulated miR-155 can suppress both SHIP1 and SOCSI to block their regulation of downstream TLR signaling directly contributing to increased inflammatory signaling and macrophage activation (208). Furthermore, SOCS1 which regulates type I IFN signaling, is targeted by miR-155 in macrophages (211, 212) and the loss of function of SOCS-1 is a common feature in HCC clearly supporting a hypothesis that HBx-upregulated miR-155 promotes the progression of HBV-HCC (200, 201). Finally, it has been demonstrated that AKT signaling can repress miR-155 in macrophages thus indicating a negative feedback loop to fine-tune TLR signaling (213).
Dendritic Cells (DC)
Upregulated-miR-155 modulates the TLR/IL-1 (interleukin-1) inflammation signaling pathway to regulate human monocyte-derived DCs in order to ensure excess damage does not occur (214). TLR/TNF/IFN upregulated miR-155 via AP1/BIC plays a significant homeostatic role in monocytepoiesis by repressing PU.1 which activates DC-SIGN, a C-type lectin receptor to increase pathogen cell surface uptake on DCs (207, 215). Decreased DC-SIGN expression in HCC is related to poor prognosis and PU.I has been identified as a metastasis suppressor possibly relating to the impairment of the antigen presenting capabilities of APCs (216).
Adaptive Immune System: B-Cells
In the adaptive immune system the epigenetically modulated miR-155 can influence B-cell expression by triggering downstream epigenetic changes. Epigenetically upregulated miR-155 (HDAC-I/EZH2) can repress the expression of an important epigenetic regulator like activation induced cytidine de-aminase (AID) which acts as a HDAC inhibitor that binds to specific immunoglobin genes in the nucleus to induce CSR/SHM/antibody diversification (43). The repression of AID by upregulated miR-155 thus leads to a reduction in CSR/SHM/Plasma B cell diversification thus contributing to reduced ability to synthesize pathogen specific antibodies (217, 218). Interestingly, different studies indicate AID is upregulated in both HBV and HCV induced hepatocarcinogenesis (219, 220). This upregulated miRNA also influences B-cell synthesis by targeting Ship-1 which plays an important role in the regulation of immune cell activation in both the innate and adaptive pathways (221). Epigenetically upregulated miR-155 targets PU-1, a critical transcription factor, to block GC B-cell to Plasma cell transition thereby modulating germinal cell B-cell differentiation into memory cells or plasma cells (222).
T-Cells
Epigenetically upregulated miR-155 can target Ship-1 to promote histone building capacity (Phf19) to promote PRC2 expression that promotes histone modifications to repress T-cell senescence and promote CD8+ T-cell expression (67). This miRNA can also modulate IFNγ expression by repressing SHIP1to play a critical role in the reciprocal regulation of CD4+ and CD8+ leukopoiesis (223). MiR-155 also has a role in the generation of exhausted dysfunctional T cells and Fosl2 antagonism of miR-155 can reduce T cell exhaustion during chronic viral infection (224). This upregulated miRNA modulates T helper cell differentiation and the germinal center reaction to synthesize T-cell dependent antibody response. In order to do this, upregulated miR-155 can repress SOCSI to maintain Foxp3+ regulatory T-cell (Treg) generation in order to regulate an autoimmune response (225, 226). It can also enhance Treg and Th17 cells differentiation and IL-17A production by targeting SOCS1 (206). A supporting meta study also confirms that the elevated expression of Tregs can be associated with HCC pathogenesis and Treg upregulation is a feature of the HCC tumor microenvironment (227). Conversely, Tregs can also target miR-155 to provide a negative feedback loop to control Treg expression (228). In the Th1/2 differentiation stage upregulated miR-155 can promote Th1 differentiation as a result of targeting C-MAF (229, 230) and an elevated Th17 to Th1 ratio has been associated with tumor progression in HBV-HCC (231). MiR-155 in Th17 cells can also trigger autoimmune inflammation through a signaling network by targeting the Ets1/IL-23/IL-23R pathway (205).
Clinical Therapeutic Options
The five-year survival rate of advanced HCC remains dismally low and the treatment of advanced stages is limited by a paucity of targeted options despite the fact that HCC cancer pathways and their targeted genes have been well documented. Since the introduction of the multi-kinase inhibitor sorafenib, very little progress has been made in treatment of advanced HCC (232). Novel targeted therapies developed for a range of cancers include the development of immune checkpoint inhibitors like anti-CTLA4 or anti-PD-1/PD-L1 antibodies has introduced new opportunities in clinical oncology (233, 234). Chronic CHB, inflammation and the development of cirrhosis are all hallmarks of HBV-HCC pathogenesis. The question remains as to how miRNA-regulated epigenetic expression can prompt an appropriate immune response.
Our review demonstrates that multiple miRNA can influence epigenetic changes in multiple pathways in HBV-HCC pathogenesis by regulating histone modifications, DNA Methylation and chromatin modelling. For example, we show that HBx- downregulated miR-101 fails to modulate DNMT3A silencing of multiple tumor suppressors in HBV-HCC (24) while simultaneously modulating the expression of macrophages (157), DCs (165), T-cells (166) and B-cells (170). The question remains, however, as to the in vivo clinical potential of deploying miR-101 replacement therapy in HBV-HCC. Current trials using HDAC inhibitors to inhibit cell cycle in HCC have been disappointing (235) and to date there has been no attempt to modulate HDAC expression using miRNA in HCC. In a breast cancer study, for instance, HDAC inhibitors reduced tumorigenesis and apoptosis via microRNA miR-125a-5p in vivo and in vitro (236). Epigenetic targeting of EZH2, a histone-lysine-N-methyltransferase and DNMT1 inhibitor reactivated transcriptionally repressed chemokine genes and augmented T cell response in HCC (237). Our review shows that 29 dysregulated miRNA in HBV-HCC (Table 1) are both regulated and regulate epigenetic changes offering numerous hypotheses to be tested in vitro. In the case of our four HBV-HCC pathways, miR-101 influences PRC2 and DNMT3A silencing, miR-148/152influenced DNMT1/3A silencing, miR-155 repressed PRC2 silencing and miR-29a/b repressed DNMT1/3A silencing in parallel with affecting immune expression in both the innate and adaptive immune systems.
The use of miRNA-led therapeutics is still a work in progress and most likely these therapeutic options would be used as an ancillary form of treatment in support of current options. In theory miRNA-led therapeutics attempt to repress or restore oncogenic and tumor suppressor expression respectively. Currently, miRNA replacement therapy has started investigating whether this could be an adjuvant therapy in support of chemotherapy and radiation (238, 239). This approach relies on the use of synthetic miRNA or miRNA inhibitors to upregulate or downregulate miRNA expression respectively (240). However, this approach has yet to capture the synergistic response generated by multiple miRNA, nor its dynamic homeostatic shifts and this is the puzzle yet to be solved.
Conclusion
This review attempts to link the epigenetic modifications that influence HBV and host genome expression in HBV-HCC pathogenesis in both the hepatocyte and immune pathways. We examine the interplay between CHB infection, the silencing role of miRNA, epigenetic change, immune system expression and HBV-HCC pathogenesis. In particular, we demonstrate how HBx dysregulated miRNA in HBV-HCC pathogenesis influence and are influenced by epigenetic changes to modulate both the HBV and host genome expression. The paper provides useful insights and potential hypotheses of the complex interplay between host gene targets in the principal cancer and immune pathways as a result of HBx dysregulated miRNA, epigenetic change, HBV-HCC pathogenesis and immune response.
This review paper tries to provide a platform for a wide range of evidence-based hypotheses rather than an (exactly) correct snapshot of the role of miRNA in HBV-HCC pathways. Even though HBV-HCC is a specific sub-type of HCC, there are multiple different classes and stages in which the hypothesized figures would operate to greater or lesser degree. This review should contribute to the point of view that our understanding of miRNA-based pathogenesis is far superior to our current ability to translate this knowledge to improve clinical outcomes.
Funding
JM has received funding within the framework of the Basic Research Program at HSE University.
Statements
Author contributions
KS conceptualized review article, performed literature review, wrote up first draft including figures and tables. Rewrote drafts 2-4 after fellow author comment. PA assisted re conceptualization of article, reviewed article. CW reviewed article, added text comments. AC reviewed article, added text comments XL reviewed article, added text comments. AK reviewed article, added text comments. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Bray F Ferlay J Soerjomataram I Siegel RL Torre LA Jemal A . Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2
Ginzberg D Wong RJ Gish R . Global HBV burden: guesstimates and facts. Hepatol Int (2018) 12(4):315–29. doi: 10.1007/s12072-018-9884-8
3
Yang JD Hainaut P Gores GJ Amadou A Plymoth A Roberts LR . A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol (2019) 16(10):589–604. doi: 10.1038/s41575-019-0186-y
4
Dandri M , ed. Epigenetic modulation in chronic hepatitis B virus infection. Seminars in Immunopathology. Springer Berlin Heidelberg: Springer (2020). doi: 10.1007/s00281-020-00780-6
5
Koumbi L Karayiannis P . The epigenetic control of hepatitis B virus modulates the outcome of infection. Front Microbiol (2016) 6:1491. doi: 10.3389/fmicb.2015.01491
6
Sartorius K Makarova J Sartorius B An P Winkler C Chuturgoon A et al . The regulatory role of MicroRNA in Hepatitis-B Virus-associated hepatocellular carcinoma (HBV-HCC) pathogenesis. Cells (2019) 8(12):1504. doi: 10.3390/cells8121504
7
Sartorius K Swadling L An P Makarova J Winkler C Chuturgoon A et al . The multiple roles of hepatitis B virus X protein (HBx) dysregulated MicroRNA in hepatitis B virus-associated hepatocellular carcinoma (HBV-HCC) and immune pathways. Viruses (2020) 12(7):746. doi: 10.3390/v12070746
8
Arbuthnot P Capovilla A Kew M . Putative role of hepatitis B virus X protein in hepatocarcinogenesis: Effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J Gastroenterol Hepatol (2000) 15(4):357–68. doi: 10.1046/j.1440-1746.2000.02069.x
9
Bouchard MJ Schneider RJ . The enigmatic X gene of hepatitis B virus. J Virol (2004) 78(23):12725–34. doi: 10.1128/JVI.78.23.12725-12734.2004
10
Wahid B Ali A Rafique S Idrees M . New Insights Into the Epigenetics of Hepatocellular Carcinoma. BioMed Res Int (2017) 2017:1–26. doi: 10.1155/2017/1609575
11
Zhang X Hou J Lu M . Regulation of hepatitis B virus replication by epigenetic mechanisms and microRNAs. Front Genet (2013) 4:1–6. doi: 10.3389/fgene.2013.00202
12
Xu J An P Winkler CA Yu Y . Dysregulated microRNAs in Hepatitis B Virus-Related Hepatocellular Carcinoma: Potential as Biomarkers and Therapeutic Targets. Front Oncol (2020) 10:1271. doi: 10.3389/fonc.2020.01271
13
Ma L Chua M-S Andrisani O So S . Epigenetics in Hepatocellular Carcinoma: An Update and Future Therapy Perspectives. World J Gastroenterol: WJG (2014) 20(2):333. doi: 10.3748/wjg.v20.i2.333
14
Au SLK Wong CCL Lee JMF Fan DNY Tsang FH Ng IOL et al . Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology (2012) 56(2):622–31. doi: 10.1002/hep.25679
15
Zhu X Wu L Xu J Yang R Wu F . Let-7c microRNA expression and clinical significance in hepatocellular carcinoma. J Int Med Res (2011) 39(6):2323–9. doi: 10.1177/147323001103900631
16
Wu G Huang P Ju X Li Z Wang Y . Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. Int J Clin Exp Med (2015) 8(9):15108.
17
Wang Y Lu Y Toh ST Sung W-K Tan P Chow P et al . Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol (2010) 53(1):57–66. doi: 10.1016/j.jhep.2009.12.043
18
Jiang X Kanda T Wu S Nakamura M Miyamura T Nakamoto S et al . Regulation of microRNA by hepatitis B virus infection and their possible association with control of innate immunity. World J Gastroenterol: WJG (2014) 20(23):7197. doi: 10.3748/wjg.v20.i23.7197
19
Takata A Otsuka M Ohno M Kishikawa T Yoshikawa T Koike K . Mutual antagonism between hepatitis B viral mRNA and host microRNA let-7. Sci Rep (2016) 6:23237. doi: 10.1038/srep23237
20
Datta J Kutay H Nasser MW Nuovo GJ Wang B Majumder S et al . Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res (2008) 68(13):5049–58. doi: 10.1158/0008-5472.CAN-07-6655
21
Zhang X Zhang E Ma Z Pei R Jiang M Schlaak JF et al . Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology (2011) 53(5):1476–85. doi: 10.1002/hep.24195
22
Sarkar N Chakravarty R . Hepatitis B virus infection, microRNAs and liver disease. Int J Mol Sci (2015) 16(8):17746–62. doi: 10.3390/ijms160817746
23
Lu J-W Liao C-Y Yang W-Y Lin Y-M Jin S-LC Wang H-D et al . Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PloS One (2014) 9(1):e85318. doi: 10.1371/journal.pone.0085318
24
Wei X Xiang T Ren G Tan C Liu R Xu X et al . miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal (2013) 25(2):439–46. doi: 10.1016/j.cellsig.2012.10.013
25
Li C-Y Pang Y-Y Yang H Li J Lu H-X Wang H-L et al . Identification of miR-101-3p targets and functional features based on bioinformatics, meta-analysis and experimental verification in hepatocellular carcinoma. Am J Trans Res (2017) 9(5):2088.
26
Li S Fu H Wang Y Tie Y Xing R Zhu J et al . MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology (2009) 49(4):1194–202. doi: 10.1002/hep.22757
27
Xie Y Yao Q Butt AM Guo J Tian Z Bao X et al . Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol Ther (2014) 15(9):1248–55. doi: 10.4161/cbt.29688
28
Fu Y Wei X Tang C Li J Liu R Shen A et al . Circulating microRNA−101 as a potential biomarker for hepatitis B virus−related hepatocellular carcinoma. Oncol Lett (2013) 6(6):1811–5. doi: 10.3892/ol.2013.1638
29
Song K Han C Zhang J Lu D Dash S Feitelson M et al . Epigenetic regulation of MicroRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology (2013) 58(5):1681–92. doi: 10.1002/hep.26514
30
Jung CJ Iyengar S Blahnik KR Ajuha TP Jiang JX Farnham PJ et al . Epigenetic modulation of miR-122 facilitates human embryonic stem cell self-renewal and hepatocellular carcinoma proliferation. PloS One (2011) 6(11):e27740. doi: 10.1371/journal.pone.0027740
31
Fan C-G Wang C-M Tian C Wang Y Li L Sun W-S et al . miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol Rep (2011) 26(5):1281–6. doi: 10.3892/or.2011.1375
32
Fornari F Gramantieri L Giovannini C Veronese A Ferracin M Sabbioni S et al . MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res (2009) 69(14):5761–7. doi: 10.1158/0008-5472.CAN-08-4797
33
Wu Q Liu H-O Liu Y-D Liu W-S Pan D Zhang W-J et al . Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J Biol Chem (2015) 290(2):1170–85. doi: 10.1074/jbc.M114.601203
34
Li C Wang Y Wang S Wu B Hao J Fan H et al . Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol (2013) 87(4):2193–205. doi: 10.1128/JVI.02831-12
35
Furuta M Kozaki K-i Tanaka S Arii S Imoto I Inazawa J . miR-124 and miR-203 are Epigenetically Silenced Tumor-Suppressive microRNAs in Hepatocellular Carcinoma. Carcinogenesis (2009) 31(5):766–76. doi: 10.1093/carcin/bgp250
36
Anwar SL Albat C Krech T Hasemeier B Schipper E Schweitzer N et al . Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int J Cancer (2013) 133(3):660–70. doi: 10.1002/ijc.28068
37
Lu Y Yue X Cui Y Zhang J Wang K . MicroRNA-124 suppresses growth of human hepatocellular carcinoma by targeting STAT3. Biochem Biophys Res Commun (2013) 441(4):873–9. doi: 10.1016/j.bbrc.2013.10.157
38
Coppola N de Stefano G Panella M Onorato L Iodice V Minichini C et al . Lowered expression of microRNA-125a-5p in human hepatocellular carcinoma and up-regulation of its oncogenic targets sirtuin-7, matrix metalloproteinase-11, and c-Raf. Oncotarget (2017) 8(15):25289. doi: 10.18632/oncotarget.15809
39
Kim JK Noh JH Jung KH Eun JW Bae HJ Kim MG et al . Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology (2013) 57(3):1055–67. doi: 10.1002/hep.26101
40
Mosca N Castiello F Coppola N Trotta MC Sagnelli C Pisaturo M et al . Functional interplay between hepatitis B virus X protein and human miR-125a in HBV infection. Biochem Biophys Res Commun (2014) 449(1):141–5. doi: 10.1016/j.bbrc.2014.05.009
41
Potenza N Papa U Mosca N Zerbini F Nobile V Russo A . Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res (2011) 39(12):5157–63. doi: 10.1093/nar/gkr067
42
Chen G Liu M Jiang Z Yu M Wei S . MicroRNAs Play Significant Roles in Pathogenesis of HBV-Related Diseases. J Biomed Sci Eng (2016) 9(10):78. doi: 10.4236/jbise.2016.910B011
43
White CA Pone EJ Lam T Tat C Hayama KL Li G et al . Histone deacetylase inhibitors upregulate B cell microRNAs that silence AID and Blimp-1 expression for epigenetic modulation of antibody and autoantibody responses. J Immunol (2014) 193(12):5933–50. doi: 10.4049/jimmunol.1401702
44
Ngo-Yin Fan D Ho-Ching Tsang F Hoi-Kam Tam A Leung-Kuen Au S Chak-Lui Wong C Wei L et al . Histone lysine methyltransferase, suppressor of variegation 3-9 homolog 1, promotes hepatocellular carcinoma progression and is negatively regulated by microRNA-125b. Hepatology (2013) 57(2):637–47. doi: 10.1002/hep.26083
45
Xie K-L Zhang Y-G Liu J Zeng Y Wu H . MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics (2014) 4(12):1176. doi: 10.7150/thno.8715
46
Liu W Hu J Zhou K Chen F Wang Z Liao B et al . Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. OncoTargets Ther (2017) 10:3843. doi: 10.2147/OTT.S140062
47
Wang Z Wu Z Huang P . The Function of miRNAs in Hepatocarcinogenesis Induced by Hepatitis B Virus X Protein. Oncol Rep (2017) 38(2):652–64. doi: 10.3892/or.2017.5716
48
Wei X Tan C Tang C Ren G Xiang T Qiu Z et al . Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell Signal (2013) 25(5):1037–43. doi: 10.1016/j.cellsig.2013.01.019
49
Wong CCL Wong CM Tung EKK Au SLK Lee JMF Poon RTP et al . The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology (2011) 140(1):322–31. doi: 10.1053/j.gastro.2010.10.006
50
Qiu G Lin Y Zhang H Wu D . miR-139-5p inhibits epithelial–mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochem Biophys Res Commun (2015) 463(3):315–21. doi: 10.1016/j.bbrc.2015.05.062
51
Takata A Otsuka M Yoshikawa T Kishikawa T Hikiba Y Obi S et al . MicroRNA-140 acts as a liver tumor suppressor by controlling NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. Hepatology (2013) 57(1):162–70. doi: 10.1002/hep.26011
52
Noh JH Chang YG Kim MG Jung KH Kim JK Bae HJ et al . MiR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett (2013) 335(2):455–62. doi: 10.1016/j.canlet.2013.03.003
53
Wang W Ding B Lou W Lin S . Promoter Hypomethylation and miR-145-5p Downregulation-Mediated HDAC11 Overexpression Promotes Sorafenib Resistance and Metastasis of Hepatocellular Carcinoma Cells. Front Cell Dev Biol (2020) 8:724. doi: 10.3389/fcell.2020.00724
54
Duan X Hu J Wang Y Gao J Peng D Xia L . MicroRNA-145: A Promising Biomarker for Hepatocellular Carcinoma (HCC). Gene (2014) 541(1):67–8. doi: 10.1016/j.gene.2014.03.018
55
Bandopadhyay M Banerjee A Sarkar N Panigrahi R Datta S Pal A et al . Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by hepatitis B virus X protein in malignant hepatocytes. BMC Cancer (2014) 14(1):721. doi: 10.1186/1471-2407-14-721
56
Gao F Sun X Wang L Tang S Yan C . Downregulation of microRNA-145 Caused by Hepatitis B Virus X Protein Promotes Expression of CUL5 and Contributes to Pathogenesis of Hepatitis B Virus-Associated Hepatocellular Carcinoma. Cell Physiol Biochem (2015) 37(4):1547–59. doi: 10.1159/000438522
57
Yang Xw Zhang Lj Huang Xh Chen Lz Su Q Zeng Wt et al . mi R-145 suppresses cell invasion in hepatocellular carcinoma cells: mi R-145 targets ADAM 17. Hepatol Res (2014) 44(5):551–9. doi: 10.1111/hepr.12152
58
Long X-R He Y Huang C Li J . MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in hepatocellular carcinogenesis. Int J Oncol (2014) 44(6):1915–22. doi: 10.3892/ijo.2014.2373
59
Braconi C Huang N Patel T . MicroRNA-dependent Regulation of DNA Methyltransferase-1 and Tumor Suppressor Gene Expression by Interleukin-6 in Human Malignant Cholangiocytes. Hepatology (2010) 51(3):881–90. doi: 10.1002/hep.23381
60
Xu X Fan Z Kang L Han J Jiang C Zheng X et al . Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest (2013) 123(2):630–45. doi: 10.1172/JCI64265
61
Zhang J Zeng C Xu L Gong J Fang J Zhuang S . MicroRNA-148a Suppresses the Epithelial-Mesenchymal Transition and Metastasis of Hepatoma Cells by Targeting Met/Snail Signaling. Oncogene (2014) 33(31):4069. doi: 10.1038/onc.2013.369
62
Jung KH Zhang J Zhou C Shen H Gagea M Rodriguez-Aguayo C et al . Differentiation therapy for hepatocellular carcinoma: Multifaceted effects of miR-148a on tumor growth and phenotype and liver fibrosis. Hepatology (2016) 63(3):864–79. doi: 10.1002/hep.28367
63
Huang J Wang Y Guo Y Sun S . Down-Regulated microRNA-152 Induces Aberrant DNA Methylation in Hepatitis B Virus–Related Hepatocellular Carcinoma by Targeting DNA Methyltransferase 1. Hepatology (2010) 52(1):60–70. doi: 10.1002/hep.23660
64
Huang H Hu M Li P Lu C Li M . Mir-152 Inhibits Cell Proliferation and Colony Formation of CD133+ Liver Cancer Stem Cells by Targeting KIT. Tumor Biol (2015) 36(2):921–8. doi: 10.1007/s13277-014-2719-x
65
Zhao Z Hu Y Shen X Lao Y Zhang L Qiu X et al . HBx represses RIZ1 expression by DNA methyltransferase 1 involvement in decreased miR-152 in hepatocellular carcinoma. Oncol Rep (2017) 37(5):2811–8. doi: 10.3892/or.2017.5518
66
Song X Tan S Wu Z Xu L Wang Z Xu Y et al . HBV suppresses ZHX2 expression to promote proliferation of HCC through miR-155 activation. Int J Cancer (2018) 143(12):3120–30. doi: 10.1002/ijc.31595
67
Ji Y Fioravanti J Zhu W Wang H Wu T Hu J et al . miR-155 harnesses Phf19 to potentiate cancer immunotherapy through epigenetic reprogramming of CD8+ T cell fate. Nat Commun (2019) 10(1):1–12. doi: 10.1038/s41467-019-09882-8
68
Fu X Wen H Jing L Yang Y Wang W Liang X et al . Micro RNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI 3K/Akt pathway. Cancer Sci (2017) 108(4):620–31. doi: 10.1111/cas.13177
69
Xie Q Chen X Lu F Zhang T Hao M Wang Y et al . Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer (2012) 118(9):2431–42. doi: 10.1002/cncr.26566
70
Moon IY Choi JH Chung JW Jang ES Jeong SH Kim JW . MicroRNA−20 induces methylation of hepatitis B virus covalently closed circular DNA in human hepatoma cells. Mol Med Rep (2019) 20(3):2285–93. doi: 10.3892/mmr.2019.10435
71
Yang H Lan P Hou Z Guan Y Zhang J Xu W et al . Histone deacetylase inhibitor SAHA epigenetically regulates miR-17-92 cluster and MCM7 to upregulate MICA expression in hepatoma. Br J Cancer (2015) 112(1):112–21. doi: 10.1038/bjc.2014.547
72
Jung YJ Kim JW Park SJ Min BY Jang ES Kim NY et al . c-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. J Med Virol (2013) 85(6):969–78. doi: 10.1002/jmv.23534
73
Aguda BD Kim Y Piper-Hunter MG Friedman A Marsh CB . MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc Natl Acad Sci (2008) 105(50):19678–83. doi: 10.1073/pnas.0811166106
74
Connolly E Melegari M Landgraf P Tchaikovskaya T Tennant BC Slagle BL et al . Elevated expression of the miR-17–92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol (2008) 173(3):856–64. doi: 10.2353/ajpath.2008.080096
75
Hou J Lin L Zhou W Wang Z Ding G Dong Q et al . Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell (2011) 19(2):232–43. doi: 10.1016/j.ccr.2011.01.001
76
Wang W Zhao LJ Tan Y-X Ren H Qi Z-T . Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol: WJG (2012) 18(38):5442. doi: 10.3748/wjg.v18.i38.5442
77
Yuan Jh Yang F Chen Bf Lu Z Huo Xs Zhou Wp et al . The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology (2011) 54(6):2025–35. doi: 10.1002/hep.24606
78
Zhang L Yang F Yuan J-h Yuan S-x Zhou W-p Huo X-s et al . Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis (2012) 34(3):577–86. doi: 10.1093/carcin/bgs381
79
Hill L Browne G Tulchinsky E . ZEB/miR-200 feedback loop: At the crossroads of signal transduction in cancer. Int J Cancer (2013) 132(4):745–54. doi: 10.1002/ijc.27708
80
Yang F Lv L-Z Cai Q-C Jiang Y . Potential roles of EZH2, Bmi-1 and miR-203 in cell proliferation and invasion in hepatocellular carcinoma cell line Hep3B. World J Gastroenterol (2015) 21(47):13268. doi: 10.3748/wjg.v21.i47.13268
81
Wu A Chen H Xu C Zhou J Chen S Shi Y et al . miR-203a is involved in HBx-induced inflammation by targeting Rap1a. Exp Cell Res (2016) 349(1):191–7. doi: 10.1016/j.yexcr.2016.10.016
82
Zhang T Zhang J Cui M Liu F You X Du Y et al . Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia (2013) 15(11):IN24–IN6. doi: 10.1593/neo.131362
83
Cui M Wang Y Sun B Xiao Z Ye L Zhang X . MiR-205 modulates abnormal lipid metabolism of hepatoma cells via targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) mRNA. Biochem Biophys Res Commun (2014) 444(2):270–5. doi: 10.1016/j.bbrc.2014.01.051
84
Bae HJ Jung KH Eun JW Shen Q Kim HS Park SJ et al . MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J Hepatol (2015) 63(2):408–19. doi: 10.1016/j.jhep.2015.03.019
85
Fornari F Milazzo M Galassi M Callegari E Veronese A Miyaaki H et al . p53/mdm2 feedback loop sustains miR-221 expression and dictates the response to anticancer treatments in hepatocellular carcinoma. Mol Cancer Res (2014) 12(2):203–16. doi: 10.1158/1541-7786.MCR-13-0312-T
86
Chen J-J Tang Y-S Huang S-F Ai J-G Wang H-X Zhang L-P . HBx protein-induced upregulation of microRNA-221 promotes aberrant proliferation in HBV−related hepatocellular carcinoma by targeting estrogen receptor-α. Oncol Rep (2015) 33(2):792–8. doi: 10.3892/or.2014.3647
87
Rong M Chen G Dang Y . Increased miR-221 Expression in Hepatocellular Carcinoma Tissues and its Role in Enhancing Cell Growth and Inhibiting Apoptosis In Vitro. BMC Cancer (2013) 13(1):21. doi: 10.1186/1471-2407-13-21
88
Yang Y-F Wang F Xiao J-J Song Y Zhao Y-Y Cao Y et al . MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27. Int J Clin Exp Med (2014) 7(4):893.
89
Wong QW Ching AK Chan AW Choy K-W To K-F Lai PB et al . MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res (2010) 16(3):867–75. doi: 10.1158/1078-0432.CCR-09-1840
90
Gramantieri L Fornari F Callegari E Sabbioni S Lanza G Croce CM et al . MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med (2008) 12(6a):2189–204. doi: 10.1111/j.1582-4934.2008.00533.x
91
Wang Y Toh HC Chow P Chung AY Meyers DJ Cole PA et al . MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J (2012) 26(7):3032–41. doi: 10.1096/fj.11-201855
92
Wang Y Lee CG . Role of miR-224 in Hepatocellular Carcinoma: A Tool for Possible Therapeutic Intervention? Epigenomics (2011) 3(2):235–43. doi: 10.2217/epi.11.5
93
Wang Y Lee AT Ma JZ Wang J Ren J Yang Y et al . Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem (2008) 283(19):13205–15. doi: 10.1074/jbc.M707629200
94
Lan SH Wu SY Zuchini R Lin XZ Su IJ Tsai TF et al . Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology (2014) 59(2):505–17. doi: 10.1002/hep.26659
95
Scisciani C Vossio S Guerrieri F Schinzari V De Iaco R de Meo PDO et al . Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J Hepatol (2012) 56(4):855–61. doi: 10.1016/j.jhep.2011.11.017
96
Zhuang C Wang P Huang D Xu L Wang X Wang L et al . A double-negative feedback loop between EZH2 and miR-26a regulates tumor cell growth in hepatocellular carcinoma. Int J Oncol (2016) 48(3):1195–204. doi: 10.3892/ijo.2016.3336
97
Zhang X Zhang X Wang T Wang L Tan Z Wei W et al . MicroRNA-26a is a key regulon that inhibits progression and metastasis of c-Myc/EZH2 double high advanced hepatocellular carcinoma. Cancer Lett (2018) 426:98–108. doi: 10.1016/j.canlet.2018.04.005
98
Ma D-N Chai Z-T Zhu X-D Zhang N Zhan D-H Ye B-G et al . MicroRNA-26a suppresses epithelial-mesenchymal transition in human hepatocellular carcinoma by repressing enhancer of zeste homolog 2. J Hematol Oncol (2016) 9(1):1–10. doi: 10.1186/s13045-015-0229-y
99
Chen L Zheng J Zhang Y Yang L Wang J Ni J et al . Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and-independent pathways. Mol Ther (2011) 19(8):1521–8. doi: 10.1038/mt.2011.64
100
Yang X Liang L Zhang XF Jia HL Qin Y Zhu XC et al . MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology (2013) 58(1):158–70. doi: 10.1002/hep.26305
101
Ji J Shi J Budhu A Yu Z Forgues M Roessler S et al . MicroRNA expression, survival, and response to interferon in liver cancer. New Engl J Med (2009) 361(15):1437–47. doi: 10.1056/NEJMoa0901282
102
Parpart S Roessler S Dong F Rao V Takai A Ji J et al . Modulation of miR-29 expression by alpha-fetoprotein is linked to the hepatocellular carcinoma epigenome. Hepatology (2014) 60(3):872–83. doi: 10.1002/hep.27200
103
Kwon JJ Factora TD Dey S Kota J . A Systematic Review of miR-29 in Cancer. Mol Therapy-Oncolytics (2019) 12:173–94. doi: 10.1016/j.omto.2018.12.011
104
Kim HS Shen Q Nam SW . Histone deacetylases and their regulatory microRNAs in hepatocarcinogenesis. J Korean Med Sci (2015) 30(10):1375–80. doi: 10.3346/jkms.2015.30.10.1375
105
Xiong Y Fang JH Yun JP Yang J Zhang Y Jia WH et al . Effects of MicroRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology (2010) 51(3):836–45. doi: 10.1002/hep.23380
106
Wang C-M Wang Y Fan C-G Xu F-F Sun W-S Liu Y-G et al . miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun (2011) 411(3):586–92. doi: 10.1016/j.bbrc.2011.06.191
107
Wong CM Wei L Law CT Ho DWH Tsang FHC Au SLK et al . Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology (2016) 63(2):474–87. doi: 10.1002/hep.28304
108
Morales S Monzo M Navarro A . Epigenetic regulation mechanisms of microRNA expression. Biomol Concepts (2017) 8(5-6):203–12. doi: 10.1515/bmc-2017-0024
109
Kong G Zhang J Zhang S Shan C Ye L Zhang X . Upregulated microRNA-29a by Hepatitis B Virus X Protein Enhances Hepatoma Cell Migration by Targeting PTEN in Cell Culture Model. PloS One (2011) 6(5):e19518. doi: 10.1371/journal.pone.0019518
110
Chamani F Sadeghizadeh M Masoumi M Babashah S . Evaluation of miR-34 Family and DNA Methyltransferases 1, 3A, 3B Gene Expression Levels in Hepatocellular Carcinoma Following Treatment With Dendrosomal Nanocurcumin. Asian Pac J Cancer Prev (2016) 17(S3):219–24. doi: 10.7314/APJCP.2016.17.S3.219
111
Yang P Li Q-J Feng Y Zhang Y Markowitz GJ Ning S et al . TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell (2012) 22(3):291–303. doi: 10.1016/j.ccr.2012.07.023
112
Ou Q Wang G Li B Li W-F . Decreased miR-34a Promotes Growth by Regulating MAP4K4 in Hepatitis B Virus Related Hepatocellular Carcinoma. Int J Clin Exp Med (2017) 10(2):2523–31.
113
Hermeking H . The miR-34 family in cancer and apoptosis. Cell Death Diff (2010) 17(2):193. doi: 10.1038/cdd.2009.56
114
Corney DC Flesken-Nikitin A Godwin AK Wang W Nikitin AY . MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res (2007) 67(18):8433–8. doi: 10.1158/0008-5472.CAN-07-1585
115
Sun F Fu H Liu Q Tie Y Zhu J Xing R et al . Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett (2008) 582(10):1564–8. doi: 10.1016/j.febslet.2008.03.057
116
Arzumanyan A Friedman T Kotei E Ng IO Lian Z Feitelson MA . Epigenetic Repression of E-cadherin Expression by Hepatitis B Virus X Antigen in Liver Cancer. Oncogene (2012) 31(5):563. doi: 10.1038/onc.2011.255
117
Xing T Zhu J Xian J Li A Wang X Wang W et al . miRNA-548ah promotes the replication and expression of hepatitis B virus by targeting histone deacetylase 4. Life Sci (2019) 219:199–208. doi: 10.1016/j.lfs.2018.12.057
118
Hu XM Yan XH Hu YW Huang JL Cao SW Ren TY et al . miRNA-548p suppresses hepatitis B virus X protein associated hepatocellular carcinoma by downregulating oncoprotein hepatitis B x-interacting protein. Hepatol Res (2016) 46(8):804–15. doi: 10.1111/hepr.12618
119
Li Y Xie J Xu X Wang J Ao F Wan Y et al . MicroRNA-548 down-regulates host antiviral response via direct targeting of IFN-λ1. Protein Cell (2013) 4(2):130–41. doi: 10.1007/s13238-012-2081-y
120
Ganem D Prince AM . Hepatitis B Virus Infection—Natural History and Clinical Consequences. New Engl J Med (2004) 350(11):1118–29. doi: 10.1056/NEJMra031087
121
Bréchot C . Pathogenesis of hepatitis B virus—related hepatocellular carcinoma: old and new paradigms. Gastroenterology (2004) 127(5):S56–61. doi: 10.1053/j.gastro.2004.09.016
122
Ringelhan M O’connor T Protzer U Heikenwalder M . The Direct and Indirect Roles of HBV in Liver Cancer: Prospective Markers for HCC Screening and Potential Therapeutic Targets. J Pathol (2015) 235(2):355–67. doi: 10.1002/path.4434
123
Jiang J Gusev Y Aderca I Mettler TA Nagorney DM Brackett DJ et al . Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res (2008) 14(2):419–27. doi: 10.1158/1078-0432.CCR-07-0523
124
Torresi J Tran BM Christiansen D Earnest-Silveira L Schwab RHM Vincan E . HBV-Related Hepatocarcinogenesis: The Role of Signalling Pathways and Innovative Ex Vivo Research Models. BMC Cancer (2019) 19(1):707. doi: 10.1186/s12885-019-5916-6
125
Schulz W . Molecular biology of human cancers: an advanced student’s textbook. Springer Heidelberg: Springer Science & Business Media (2005).
126
Chung W Kim M de la Monte S Longato L Carlson R Slagle BL et al . Activation of signal transduction pathways during hepatic oncogenesis. Cancer Lett (2016) 370(1):1–9. doi: 10.1016/j.canlet.2015.09.016
127
Vidigal JA Ventura A . The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol (2015) 25(3):137–47. doi: 10.1016/j.tcb.2014.11.004
128
Hatziapostolou M Polytarchou C Aggelidou E Drakaki A Poultsides GA Jaeger SA et al . An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell (2011) 147(6):1233–47. doi: 10.1016/j.cell.2011.10.043
129
Murakami Y Yasuda T Saigo K Urashima T Toyoda H Okanoue T et al . Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene (2006) 25(17):2537. doi: 10.1038/sj.onc.1209283
130
Ali A Abdel-Hafiz H Suhail M Al-Mars A Zakaria MK Fatima K et al . Hepatitis B virus, HBx mutants and their role in hepatocellular carcinoma. World J Gastroenterol: WJG (2014) 20(30):10238. doi: 10.3748/wjg.v20.i30.10238
131
Belloni L Pollicino T De Nicola F Guerrieri F Raffa G Fanciulli M et al . Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci (2009) 106(47):19975–9. doi: 10.1073/pnas.0908365106
132
Tian Y Ou J-h . Genetic and epigenetic alterations in hepatitis B virus-associated hepatocellular carcinoma. Virol Sin (2015) 30(2):85–91. doi: 10.1007/s12250-015-3582-7
133
Luo L Chen S Gong Q Luo N Lei Y Guo J et al . Hepatitis B virus X protein modulates remodelling of minichromosomes related to hepatitis B virus replication in HepG2 cells. Int J Mol Med (2013) 31(1):197–204. doi: 10.3892/ijmm.2012.1165
134
Niu C Livingston CM Li L Beran RK Daffis S Ramakrishnan D et al . The Smc5/6 complex restricts HBV when localized to ND10 without inducing an innate immune response and is counteracted by the HBV X protein shortly after infection. PloS One (2017) 12(1):e0169648. doi: 10.1371/journal.pone.0169648
135
Jin Q Yu LR Wang L Zhang Z Kasper LH Lee JE et al . Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J (2011) 30(2):249–62. doi: 10.1038/emboj.2010.318
136
Guo Y Li Y Mu S Zhang J Yan Z . Evidence that methylation of hepatitis B virus covalently closed circular DNA in liver tissues of patients with chronic hepatitis B modulates HBV replication. J Med Virol (2009) 81(7):1177–83. doi: 10.1002/jmv.21525
137
Tian Y Yang W Song J Wu Y Ni B . Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol Cell Biol (2013) 33(15):2810–6. doi: 10.1128/MCB.00205-13
138
Hamamoto R Furukawa Y Morita M Iimura Y Silva FP Li M et al . SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol (2004) 6(8):731–40. doi: 10.1038/ncb1151
139
Yang L He J Chen L Wang G . Hepatitis B Virus X Protein Upregulates Expression of SMYD3 and C-MYC in HepG2 Cells. Med Oncol (2009) 26(4):445. doi: 10.1007/s12032-008-9144-1
140
Rivière L Gerossier L Ducroux A Dion S Deng Q Michel M-L et al . HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol (2015) 63(5):1093–102. doi: 10.1016/j.jhep.2015.06.023
141
Kitagawa M Kitagawa K Kotake Y Niida H Ohhata T . Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci (2013) 70(24):4785–94. doi: 10.1007/s00018-013-1423-0
142
Sanna L Marchesi I Melone MA Bagella L . The role of enhancer of zeste homolog 2: From viral epigenetics to the carcinogenesis of hepatocellular carcinoma. J Cell Physiol (2018) 233(9):6508–17. doi: 10.1002/jcp.26545
143
Lee SG Rho HM . Transcriptional Repression of the Human p53 Gene by Hepatitis B Viral X Protein. Oncogene (2000) 19(3):468–71. doi: 10.1038/sj.onc.1203312
144
Han H Sun D Li W Shen H Zhu Y Li C et al . A c-Myc-MicroRNA functional feedback loop affects hepatocarcinogenesis. Hepatology (2013) 57(6):2378–89. doi: 10.1002/hep.26302
145
Ren M Qin D Li K Qu J Wang L Wang Z et al . Correlation between hepatitis B virus protein and microRNA processor Drosha in cells expressing HBV. Antiviral Res (2012) 94(3):225–31. doi: 10.1016/j.antiviral.2012.04.004
146
Liu AM Zhang C Burchard J Fan S Wong K-F Dai H et al . Global Regulation on microRNA in Hepatitis B Virus-Associated Hepatocellular Carcinoma. Omics: J Integr Biol (2011) 15(3):187–91. doi: 10.1089/omi.2010.0098
147
Arif K Elliott EK Haupt LM Griffiths LR . Regulatory Mechanisms of Epigenetic miRNA Relationships in Human Cancer and Potential as Therapeutic Targets. Cancers (2020) 12(10):2922. doi: 10.3390/cancers12102922
148
Hu J-J Song W Zhang S-D Shen X-H Qiu X-M Wu H-Z et al . HBx-upregulated lncRNA UCA1 promotes cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling. Sci Rep (2016) 6(1):1–13. doi: 10.1038/srep23521
149
Zheng F Liao Y-J Cai M-Y Liu T-H Chen S-P Wu P-H et al . Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PloS Genet (2015) 11(2):e1004873. doi: 10.1371/journal.pgen.1004873
150
Leng J Han C Demetris AJ Michalopoulos GK Wu T . Cyclooxygenase-2 Promotes Hepatocellular Carcinoma Cell Growth Through Akt Activation: Evidence for Akt Inhibition in Celecoxib-Induced Apoptosis. Hepatology (2003) 38(3):756–68. doi: 10.1053/jhep.2003.50380
151
Lin C-P Liu C-R Lee C-N Chan T-S Liu HE . Targeting c-Myc as a novel approach for hepatocellular carcinoma. World J Hepatol (2010) 2(1):16. doi: 10.4254/wjh.v2.i1.16
152
He H Tian W Chen H Deng Y . MicroRNA-101 sensitizes hepatocellular carcinoma cells to doxorubicin-induced apoptosis via targeting Mcl-1. Mol Med Rep (2016) 13(2):1923–9. doi: 10.3892/mmr.2015.4727
153
Schreiber S Rignall B Braeuning A Marx-Stoelting P Ott T Buchmann A et al . Phenotype of single hepatocytes expressing an activated version of β-catenin in liver of transgenic mice. J Mol Histol (2011) 42(5):393. doi: 10.1007/s10735-011-9342-6
154
Su H Yang J-R Xu T Huang J Xu L Yuan Y et al . MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res (2009) 69(3):1135–42. doi: 10.1158/0008-5472.CAN-08-2886
155
Cao K Li J Zhao Y Wang Q Zeng Q He S et al . miR-101 inhibiting cell proliferation, migration and invasion in hepatocellular carcinoma through downregulating Girdin. Mol Cells (2016) 39(2):96. doi: 10.14348/molcells.2016.2161
156
Lv X Li J Yang B . Clinical effects of miR-101 on prognosis of hepatocellular carcinoma and carcinogenic mechanism of anti-miR-101. Oncol Rep (2016) 36(4):2184–92. doi: 10.3892/or.2016.4980
157
Zhu Q-Y Liu Q Chen J-X Lan K Ge B-X . MicroRNA-101 Targets MAPK Phosphatase-1 to Regulate the Activation of MAPKs in Macrophages. J Immunol (2010) 185(12):7435–42. doi: 10.4049/jimmunol.1000798
158
Wang X Liu Y . Regulation of innate immune response by MAP kinase phosphatase-1. Cell Signal (2007) 19(7):1372–82. doi: 10.1016/j.cellsig.2007.03.013
159
Wei X Tang C Lu X Liu R Zhou M He D et al . MiR-101 targets DUSP1 to regulate the TGF-β secretion in sorafenib inhibits macrophage-induced growth of hepatocarcinoma. Oncotarget (2015) 6(21):18389. doi: 10.18632/oncotarget.4089
160
Shen J Zhang Y Yu H Shen B Liang Y Jin R et al . Role of DUSP1/MKP1 in tumorigenesis, tumor progression and therapy. Cancer Med (2016) 5(8):2061–8. doi: 10.1002/cam4.772
161
Liu T Zhang L Joo D Sun S-C . NF-κB signaling in inflammation. Signal Transduct Targeted Ther (2017) 2(1):1–9. doi: 10.1038/sigtrans.2017.23
162
Liu JC Xue DF Wang XQ Ai DB Qin PJ . MiR-101 Relates to Chronic Peripheral Neuropathic Pain Through Targeting KPNB1 and Regulating NF-κb Signaling. Kaohsiung J Med Sci (2019) 35(3):139–45. doi: 10.1002/kjm2.12025
163
Hume DA . The many alternative faces of macrophage activation. Front Immunol (2015) 6:370. doi: 10.3389/fimmu.2015.00370
164
Yang L Hu B Zhang Y Qiang S Cai J Huang W et al . Suppression of the nuclear transporter-KPNβ1 expression inhibits tumor proliferation in hepatocellular carcinoma. Med Oncol (2015) 32(4):128. doi: 10.1007/s12032-015-0559-1
165
Newton K Dixit VM . Signaling in innate immunity and inflammation. Cold Spring Harbor Perspect Biol (2012) 4(3):a006049. doi: 10.1101/cshperspect.a006049
166
Yu D Tan AH-M Hu X Athanasopoulos V Simpson N Silva DG et al . Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature (2007) 450(7167):299–303. doi: 10.1038/nature06253
167
Wikenheiser DJ Stumhofer JS . ICOS co-stimulation: friend or foe? Front Immunol (2016) 7:304. doi: 10.3389/fimmu.2016.00304
168
Pfannenstiel LW Gastman BR . Mcl-1 expression influcences CD8+ anti-tumor immunity. J ImmunoTher Cancer (2015) 3(S2):P323. doi: 10.1186/2051-1426-3-S2-P323
169
Oh H Grinberg-Bleyer Y Liao W Maloney D Wang P Wu Z et al . An NF-κB transcription-factor-dependent lineage-specific transcriptional program promotes regulatory T cell identity and function. Immunity (2017) 47(3):450–65. doi: 10.1016/j.immuni.2017.08.010
170
Vikström IB Slomp A Carrington EM Moesbergen LM Chang C Kelly GL et al . MCL-1 is required throughout B-cell development and its loss sensitizes specific B-cell subsets to inhibition of BCL-2 or BCL-XL. Cell Death Dis (2016) 7(8):e2345–e. doi: 10.1038/cddis.2016.237
171
Dong Y Wang A . Aberrant DNA methylation in hepatocellular carcinoma tumor suppression. Oncol Lett (2014) 8(3):963–8. doi: 10.3892/ol.2014.2301
172
Wu H Zhang W Wu Z Liu Y Shi Y Gong J et al . miR-29c-3p regulates DNMT3B and LATS1 methylation to inhibit tumor progression in hepatocellular carcinoma. Cell Death Dis (2019) 10(2):1–18. doi: 10.1038/s41419-018-1281-7
173
Xu J Zhu X Wu L Yang R Yang Z Wang Q et al . MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/β-catenin pathway. Liver Int (2012) 32(5):752–60. doi: 10.1111/j.1478-3231.2011.02750.x
174
Mott JL Kobayashi S Bronk SF Gores GJ . mir-29 Regulates Mcl-1 Protein Expression and Apoptosis. Oncogene (2007) 26(42):6133–40. doi: 10.1038/sj.onc.1210436
175
Torrano J Al Emran A Hammerlindl H Schaider H . Emerging roles of H3K9me3, SETDB1 and SETDB2 in therapy-induced cellular reprogramming. Clin Epigenet (2019) 11(1):43. doi: 10.1186/s13148-019-0644-y
176
Lin LL Wang W Hu Z Wang LW Chang J Qian H . Negative Feedback of miR-29 Family TET1 Involves in Hepatocellular Cancer. Med Oncol (2014) 31(12):291. doi: 10.1007/s12032-014-0291-2
177
Chen Q Yin D Zhang Y Yu L Li X-D Zhou Z-J et al . MicroRNA-29a induces loss of 5-hydroxymethylcytosine and promotes metastasis of hepatocellular carcinoma through a TET–SOCS1–MMP9 signaling axis. Cell Death Dis (2017) 8(6):e2906–e. doi: 10.1038/cddis.2017.142
178
Tang B Li X Ren Y Wang J Xu D Hang Y et al . MicroRNA-29a regulates lipopolysaccharide (LPS)-induced inflammatory responses in murine macrophages through the Akt1/NF-κB pathway. Exp Cell Res (2017) 360(2):74–80. doi: 10.1016/j.yexcr.2017.08.013
179
Troutman TD Bazan JF Pasare C . Toll-like receptors, signaling adapters and regulation of the pro-inflammatory response by PI3K. Cell Cycle (2012) 11(19):3559–67. doi: 10.4161/cc.21572
180
Brain O Owens BM Pichulik T Allan P Khatamzas E Leslie A et al . The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity (2013) 39(3):521–36. doi: 10.1016/j.immuni.2013.08.035
181
Leong JW Sullivan RP Fehniger TA . Natural Killer Cell Regulation by microRNAs in Health and Disease. BioMed Res Int (2012) 2012:1–12. doi: 10.1155/2012/632329
182
Liston A Papadopoulou AS Danso-Abeam D Dooley J . MicroRNA-29 in the adaptive immune system: setting the threshold. Cell Mol Life Sci (2012) 69(21):3533–41. doi: 10.1007/s00018-012-1124-0
183
Lu LF Liston A . MicroRNA in the Immune System, microRNA as an Immune System. Immunology (2009) 127(3):291–8. doi: 10.1111/j.1365-2567.2009.03092.x
184
Ma F Xu S Liu X Zhang Q Xu X Liu M et al . The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol (2011) 12(9):861–9. doi: 10.1038/ni.2073
185
Papadopoulou AS Dooley J Linterman MA Pierson W Ucar O Kyewski B et al . The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat Immunol (2012) 13(2):181–7. doi: 10.1038/ni.2193
186
Jung JK Arora P Pagano JS Jang KL . Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res (2007) 67(12):5771–8. doi: 10.1158/0008-5472.CAN-07-0529
187
Zhang J-g Shi Y Hong D-f Song M Huang D Wang C-y et al . MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting WNT1/β-catenin pathway. Sci Rep (2015) 5:8087. doi: 10.1038/srep08087
188
Heo MJ Kim YM Koo JH Yang YM An J Lee S-K et al . microRNA-148a dysregulation discriminates poor prognosis of hepatocellular carcinoma in association with USP4 overexpression. Oncotarget (2014) 5(9):2792. doi: 10.18632/oncotarget.1920
189
Jiang F Mu J Wang X Ye X Si L Ning S et al . The repressive effect of miR-148a on TGF beta-SMADs signal pathway is involved in the glabridin-induced inhibition of the cancer stem cells-like properties in hepatocellular carcinoma cells. PloS One (2014) 9(5):e96698. doi: 10.1371/journal.pone.0096698
190
Friedrich M Pracht K Mashreghi MF Jäck HM Radbruch A Seliger B . The role of the miR-148/-152 family in physiology and disease. Eur J Immunol (2017) 47(12):2026–38. doi: 10.1002/eji.201747132
191
Porstner M Winkelmann R Daum P Schmid J Pracht K Côrte-Real J et al . miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur J Immunol (2015) 45(4):1206–15. doi: 10.1002/eji.201444637
192
Gonzalez-Martin A Adams BD Lai M Shepherd J Salvador-Bernaldez M Salvador JM et al . The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat Immunol (2016) 17(4):433–40. doi: 10.1038/ni.3385
193
Herrmann TL Agrawal RS Connolly SF McCaffrey RL Schlomann J Kusner DJ . MHC Class II levels and intracellular localization in human dendritic cells are regulated by calmodulin kinase II. J leukocyte Biol (2007) 82(3):686–99. doi: 10.1189/jlb.0107045
194
Liu X Zhan Z Xu L Ma F Li D Guo Z et al . MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIα. J Immunol (2010) 185(12):7244–51. doi: 10.4049/jimmunol.1001573
195
Yang M Shen H Qiu C Ni Y Wang L Dong W et al . High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer (2013) 49(3):604–15. doi: 10.1016/j.ejca.2012.09.031
196
Mattiske S Suetani RJ Neilsen PM Callen DF . The Oncogenic Role of miR-155 in Breast Cancer. Cancer Epidemiol Prev Biomarkers (2012) 21(8):1236–43. doi: 10.1158/1055-9965.EPI-12-0173
197
Pu J Bai D Yang X Lu X Xu L Lu J . Adrenaline promotes cell proliferation and increases chemoresistance in colon cancer HT29 cells through induction of miR-155. Biochem Biophys Res Commun (2012) 428(2):210–5. doi: 10.1016/j.bbrc.2012.09.126
198
Kong X Liu F Gao J . MiR-155 Promotes Epithelial-Mesenchymal Transition in Hepatocellular Carcinoma Cells Through the Activation of PI3K/SGK3/β-Catenin Signaling Pathways. Oncotarget (2016) 7(40):66051. doi: 10.18632/oncotarget.11800
199
Wang B Majumder S Nuovo G Kutay H Volinia S Patel T et al . Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid–defined diet in C57BL/6 mice. Hepatology (2009) 50(4):1152–61. doi: 10.1002/hep.23100
200
Chen Z Ma T Huang C Hu T Li J . The Pivotal Role of microRNA-155 in the Control of Cancer. J Cell Physiol (2014) 229(5):545–50. doi: 10.1002/jcp.24492
201
Gui Y Yeganeh M Donates Y Tobelaim W Chababi W Mayhue M et al . Regulation of MET receptor tyrosine kinase signaling by suppressor of cytokine signaling 1 in hepatocellular carcinoma. Oncogene (2015) 34(46):5718. doi: 10.1038/onc.2015.20
202
Schulte LN Westermann AJ Vogel J . Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res (2013) 41(1):542–53. doi: 10.1093/nar/gks1030
203
Curtis AM Fagundes CT Yang G Palsson-McDermott EM Wochal P McGettrick AF et al . Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc Natl Acad Sci (2015) 112(23):7231–6. doi: 10.1073/pnas.1501327112
204
Yang L Boldin MP Yu Y Liu CS Ea C-K Ramakrishnan P et al . miR-146a controls the resolution of T cell responses in mice. J Exp Med (2012) 209(9):1655–70. doi: 10.1084/jem.20112218
205
Hu R Huffaker TB Kagele DA Runtsch MC Bake E Chaudhuri AA et al . MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J Immunol (2013) 190(12):5972–80. doi: 10.4049/jimmunol.1300351
206
Yao R Ma Y-L Liang W Li H-H Ma Z-J Yu X et al . MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PloS One (2012) 7(10):e46082. doi: 10.1371/journal.pone.0046082
207
O’connell RM Rao DS Chaudhuri AA Baltimore D . Physiological and Pathological Roles for microRNAs in the Immune System. Nat Rev Immunol (2010) 10(2):111–22. doi: 10.1038/nri2708
208
O’Connell RM Chaudhuri AA Rao DS Baltimore D . Inositol phosphatase SHIP1 is a primary target of miR-155. Proc Natl Acad Sci (2009) 106(17):7113–8. doi: 10.1073/pnas.0902636106
209
Gulyaeva LF Kushlinskiy NE . Regulatory mechanisms of microRNA expression. J Trans Med (2016) 14(1):143. doi: 10.1186/s12967-016-0893-x
210
Wen L-Z Ding K Wang Z-R Ding C-H Lei S-J Liu J-P et al . SHP-1 acts as a tumor suppressor in hepatocarcinogenesis and HCC progression. Cancer Res (2018) 78(16):4680–91. doi: 10.1158/0008-5472.CAN-17-3896
211
Wang P Hou J Lin L Wang C Liu X Li D et al . Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J Immunol (2010) 185(10):6226–33. doi: 10.4049/jimmunol.1000491
212
Sasi W Sharma AK Mokbel K . The role of suppressors of cytokine signalling in human neoplasms. Mol Biol Int (2014) 2014:1–24. doi: 10.1155/2014/630797
213
Androulidaki A Iliopoulos D Arranz A Doxaki C Schworer S Zacharioudaki V et al . The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity (2009) 31(2):220–31. doi: 10.1016/j.immuni.2009.06.024
214
Ceppi M Pereira PM Dunand-Sauthier I Barras E Reith W Santos MA et al . MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci (2009) 106(8):2735–40. doi: 10.1073/pnas.0811073106
215
Gerloff D Grundler R Wurm A Bräuer-Hartmann D Katzerke C Hartmann J et al . NF-κB/STAT5/miR-155 network targets PU. 1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia (2015) 29(3):535–47. doi: 10.1038/leu.2014.231
216
Xia HB Wang HJ Song SS Zhang JG He XL Hu ZM et al . Decreased DC−SIGNR expression in hepatocellular carcinoma predicts poor patient prognosis. Oncol Lett (2020) 19(1):69–76. doi: 10.3892/ol.2019.11074
217
Choudhary M Tamrakar A Singh AK Jain M Jaiswal A Kodgire P . AID Biology: A pathological and clinical perspective. Int Rev Immunol (2018) 37(1):37–56. doi: 10.1080/08830185.2017.1369980
218
Borchert GM Holton NW Larson ED . Repression of Human Activation Induced Cytidine Deaminase by miR-93 and Mir-155. BMC Cancer (2011) 11(1):1–9. doi: 10.1186/1471-2407-11-347
219
Kou T Marusawa H Kinoshita K Endo Y Im O Ueda Y et al . Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. Int J Cancer (2007) 120(3):469–76. doi: 10.1002/ijc.22292
220
Endo Y Marusawa H Kinoshita K Morisawa T Sakurai T Okazaki I et al . Expression of activation-induced cytidine deaminase in human hepatocytes via nf-κ b signaling. Oncogene (2007) 26(38):5587–95. doi: 10.1038/sj.onc.1210344
221
Pauls SD Marshall AJ . Regulation of immune cell signaling by SHIP1: A phosphatase, scaffold protein, and potential therapeutic target. Eur J Immunol (2017) 47(6):932–45. doi: 10.1002/eji.201646795
222
Vigorito E Kohlhaas S Lu D Leyland R . miR-155: An ancient regulator of the immune system. Immunol Rev (2013) 253(1):146–57. doi: 10.1111/imr.12057
223
Huffaker TB Hu R Runtsch MC Bake E Chen X Zhao J et al . Epistasis between microRNAs 155 and 146a during T cell-mediated antitumor immunity. Cell Rep (2012) 2(6):1697–709. doi: 10.1016/j.celrep.2012.10.025
224
Stelekati E Chen Z Manne S Kurachi M Ali M-A Lewy K et al . Long-term persistence of exhausted CD8 T cells in chronic infection is regulated by MicroRNA-155. Cell Rep (2018) 23(7):2142–56. doi: 10.1016/j.celrep.2018.04.038
225
Zhou X Jeker LT Fife BT Zhu S Anderson MS McManus MT et al . Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med (2008) 205(9):1983–91. doi: 10.1084/jem.20080707
226
Lu L-F Thai T-H Calado DP Chaudhry A Kubo M Tanaka K et al . Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity (2009) 30(1):80–91. doi: 10.1016/j.immuni.2008.11.010
227
Zhao H-Q Li W-M Lu Z-Q Yao Y-M . Roles of Tregs in Development of Hepatocellular Carcinoma: A Meta-Analysis. World J Gastroenterol: WJG (2014) 20(24):7971. doi: 10.3748/wjg.v20.i24.7971
228
Kohlhaas S Garden OA Scudamore C Turner M Okkenhaug K Vigorito E . Cutting Edge: The Foxp3 Target miR-155 Contributes to the Development of Regulatory T Cells. J Immunol (2009) 182(5):2578–82. doi: 10.4049/jimmunol.0803162
229
Thai T-H Calado DP Casola S Ansel KM Xiao C Xue Y et al . Regulation of the germinal center response by microRNA-155. Science (2007) 316(5824):604–8. doi: 10.1126/science.1141229
230
Rodriguez A Vigorito E Clare S Warren MV Couttet P Soond DR et al . Requirement of bic/microRNA-155 for normal immune function. Science (2007) 316(5824):608–11. doi: 10.1126/science.1139253
231
Yan J Liu X-L Xiao G Li N-L Deng Y-N Han L-Z et al . Prevalence and Clinical Relevance of T-helper Cells, Th17 and Th1, in Hepatitis B Virus-Related Hepatocellular Carcinoma. PloS One (2014) 9(5):e96080. doi: 10.1371/journal.pone.0096080
232
Neureiter D Stintzing S Kiesslich T Ocker M . Hepatocellular carcinoma: therapeutic advances in signaling, epigenetic and immune targets. World J Gastroenterol (2019) 25(25):3136. doi: 10.3748/wjg.v25.i25.3136
233
Wu K Kryczek I Chen L Zou W Welling TH . Kupffer cell suppression of CD8+ T cells in human hepatocellular carcinoma is mediated by B7-H1/programmed death-1 interactions. Cancer Res (2009) 69(20):8067–75. doi: 10.1158/0008-5472.CAN-09-0901
234
Calderaro J Rousseau B Amaddeo G Mercey M Charpy C Costentin C et al . Programmed death ligand 1 expression in hepatocellular carcinoma: relationship with clinical and pathological features. Hepatology (2016) 64(6):2038–46. doi: 10.1002/hep.28710
235
Yeo W Chung HC Chan SL Wang LZ Lim R Picus J et al . Epigenetic therapy using belinostat for patients with unresectable hepatocellular carcinoma: a multicenter phase I/II study with biomarker and pharmacokinetic analysis of tumors from patients in the Mayo Phase II Consortium and the Cancer Therapeutics Research Group. J Clin Oncol (2012) 30(27):3361. doi: 10.1200/JCO.2011.41.2395
236
Hsieh T-H Hsu C-Y Tsai C-F Long C-Y Wu C-H Wu D-C et al . HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol Ther (2015) 23(4):656–66. doi: 10.1038/mt.2014.247
237
Hong YK Li Y Pandit H Li S Pulliam Z Zheng Q et al . Epigenetic modulation enhances immunotherapy for hepatocellular carcinoma. Cell Immunol (2019) 336:66–74. doi: 10.1016/j.cellimm.2018.12.010
238
Petrovic N Ergun S . miRNAs as potential treatment targets and treatment options in cancer. Mol Diagnosis Ther (2018) 22(2):157–68. doi: 10.1007/s40291-017-0314-8
239
Mollaei H Safaralizadeh R Rostami Z . MicroRNA replacement therapy in cancer. J Cell Physiol (2019) 234(8):12369–84. doi: 10.1002/jcp.28058
240
Hosseinahli N Aghapour M Duijf PH Baradaran B . Treating cancer with microRNA replacement therapy: A literature review. J Cell Physiol (2018) 233(8):5574–88. doi: 10.1002/jcp.26514
Summary
Keywords
hepatitis B virus-associated hepatocellular carcinoma, epigenetic modulation, HBx, microRNA, immunology
Citation
Sartorius K, An P, Winkler C, Chuturgoon A, Li X, Makarova J and Kramvis A (2021) The Epigenetic Modulation of Cancer and Immune Pathways in Hepatitis B Virus-Associated Hepatocellular Carcinoma: The Influence of HBx and miRNA Dysregulation. Front. Immunol. 12:661204. doi: 10.3389/fimmu.2021.661204
Received
30 January 2021
Accepted
15 April 2021
Published
29 April 2021
Volume
12 - 2021
Edited by
Paul Laszlo Bollyky, Stanford University, United States
Reviewed by
Rebecca Leigh Schmidt, Upper Iowa University, United States; Yean Kong Yong, Xiamen University, Malaysia, Malaysia
Updates
Copyright
© 2021 Sartorius, An, Winkler, Chuturgoon, Li, Makarova and Kramvis.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kurt Sartorius, Kurt.Sartorius@wits.ac.za
This article was submitted to Microbial Immunology, a section of the journal Frontiers in Immunology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.