- 1Livestock and Aquaculture, CSIRO Agriculture and Food, St Lucia, QLD, Australia
- 2Livestock and Aquaculture, CSIRO Agriculture and Food, Woorim, QLD, Australia
- 3Livestock and Aquaculture, CSIRO Agriculture and Food, Hobart, TAS, Australia
Marine farmed Atlantic salmon (Salmo salar) are susceptible to recurrent amoebic gill disease (AGD) caused by the ectoparasite Neoparamoeba perurans over the growout production cycle. The parasite elicits a highly localized response within the gill epithelium resulting in multifocal mucoid patches at the site of parasite attachment. This host-parasite response drives a complex immune reaction, which remains poorly understood. To generate a model for host-parasite interaction during pathogenesis of AGD in Atlantic salmon the local (gill) and systemic transcriptomic response in the host, and the parasite during AGD pathogenesis was explored. A dual RNA-seq approach together with differential gene expression and system-wide statistical analyses of gene and transcription factor networks was employed. A multi-tissue transcriptomic data set was generated from the gill (including both lesioned and non-lesioned tissue), head kidney and spleen tissues naïve and AGD-affected Atlantic salmon sourced from an in vivo AGD challenge trial. Differential gene expression of the salmon host indicates local and systemic upregulation of defense and immune responses. Two transcription factors, znfOZF-like and znf70-like, and their associated gene networks significantly altered with disease state. The majority of genes in these networks are candidates for mediators of the immune response, cellular proliferation and invasion. These include Aurora kinase B-like, rho guanine nucleotide exchange factor 25-like and protein NDNF-like inhibited. Analysis of the N. perurans transcriptome during AGD pathology compared to in vitro cultured N. perurans trophozoites, as a proxy for wild type trophozoites, identified multiple gene candidates for virulence and indicates a potential master regulatory gene system analogous to the two-component PhoP/Q system. Candidate genes identified are associated with invasion of host tissue, evasion of host defense mechanisms and formation of the mucoid lesion. We generated a novel model for host-parasite interaction during AGD pathogenesis through integration of host and parasite functional profiles. Collectively, this dual transcriptomic study provides novel molecular insights into the pathology of AGD and provides alternative theories for future research in a step towards improved management of AGD.
Introduction
Amoebic gill disease (AGD) remains a serious parasitic infection of farmed salmonids globally (1) and has been estimated to increase the cost of production by 20% in Tasmania (2–4). The disease is caused by the marine protozoan parasite Neoparamoeba perurans, which, upon attachment to the mucosal surface of the gill, causes a highly localized host response. The site of amoeba attachment is characterized by epithelial desquamation and edema, epithelial hyperplasia, fusion of secondary lamellae, and interlamellar vesicle formation (5). A reduction in chloride cells is also associated with clinical AGD and is closely linked to epithelial hyperplasia (6). An infiltration of inflammatory immune cells, such as neutrophils and macrophages, can be observed within the central venous sinus adjacent to AGD lesions (5, 7). This cellular response is grossly characterized by raised multifocal lesions on the gill surface (2) leading to inappetence, respiratory stress, and often fatal inflammatory branchialitis.
Previous studies have examined the transcriptomic response of the gill and other immunological organs to AGD using various approaches. While these studies have revealed important insights into the AGD host response, the exact nature of the inflammatory response within AGD-affected tissue has not been fully resolved. Certain studies have identified upregulation of key inflammatory and immune related genes, including TNFα, CD8, CD4, MHCI and MHCIIα (8) within AGD-affected tissue, while other studies have shown either downregulation or no differential expression of these genes (9–11). A number of studies have shown a mRNA upregulation of the pro-inflammatory cytokine interleukin-1 beta (IL-1β) at the site of AGD infection in both rainbow trout (9) and Atlantic salmon (12). Furthermore, in Atlantic salmon the expression of IL-1β mRNA was localized to filament and lamellar epithelium pavement cells within gills (12), and the upregulation of IL-1β appears to largely be restricted to within AGD lesions (11). The highly localized nature of the host response to AGD has also been demonstrated through microarray based-transcriptome profiling where downregulation of the p53 tumor suppressor protein, and associated transcripts were localized to the AGD lesion (13). Downregulation of immune pathways within AGD lesions has also been observed for antigen processing pathways. Indeed, Young et al. (10) identified a coordinated downregulation of the MHCI antigen presentation pathway and interferon-regulatory factor 1 within the lesioned area of the gill. Similarly, a downregulation of transcripts related to apoptosis were also identified within AGD-affected gill (14). More recently, the importance of T helper cells in the immune response to AGD has been described. Indeed, interleukin (IL)-4, a key cytokine involved in the Th2 pathways was significantly upregulated primarily within the interbranchial lymphoid tissue of AGD-affected gill (15) and within AGD lesions (16). In contrast, genes involved in the Th1 pathway, which are primarily responsible for activation of macrophages, were downregulated. The authors go on to postulate that induction of the Th2 response may be associated with an allergic reaction caused by the parasite (15).
While significant effort and progress has been made to understand the host response to AGD, considerably less attention has been paid to understanding the behavior of the parasite. It was not until 2012 when virulent N. perurans was isolated, cultured and shown to generate AGD in a laboratory challenge model (17) that significant effort has been invested into understanding the biology of the parasite (18–23). Perhaps the most striking feature of N. perurans, along with all other Neoparamoeba spp. is that they harbor a eukaryotic endosymbiont phylogenetically related to Perkinsela sp (24). Genome sequencing of the related species Neoparamoeba pemaquidensis and its Perkinsela endosymbiont has demonstrated mosaic biochemical pathways between the two genomes, suggesting an interdependence between host and endosymbiont (25). Furthermore, bacteria and viruses may contribute to the pathogenicity and virulence of N. perurans which has been reported to harbor the pathogenic bacteria, Vibrio within its microenvironment (26). Increased pathogenicity and virulence on passage through amoeba hosts has been reported previously for the amoeba species, Acanthamoeba castellanii and Acanthamoeba astronyxis and the bacteria Legionella pneumophilia (27, 28). Viruses may also contribute to virulence in N. perurans, as amoeba have also been reported to host several giant viruses, including adenoviruses and enteroviruses (28). Despite our increasing knowledge of Neoparamoeba biology, many knowledge gaps exist particularly regarding how the amoeba reacts and behaves when interacting with the host. In many respects, this is due to a lack of specific tools and reagents to investigate amoeba biology during infection. One recent advance was the development of an in vitro model of AGD based on a rainbow trout gill derived cell line which can be stimulated with N. perurans and the host-parasite response profiled (29). While this model successfully generates host responses similar to those observed in vivo, the response of the parasite is yet to be fully integrated.
Transcripts derived from N. perurans as a single-celled eukaryotic organism are polyadenylated like the host mRNA. This means that the gene expression profile of the amoeba during infection can be profiled in parallel to the host using mRNA-based transcriptome sequencing. This approach, commonly referred to as dual RNA-Seq, facilitates gene expression changes to be profiled simultaneously in both the parasite and host (30), and can provide valuable information concerning the expression of virulence factors and immune evasion pathways. With this in mind, the present study applied a dual RNA-Seq approach to characterize the molecular events that occur within an AGD lesion, both from the host and parasite perspective. Compared to in vitro cultured N. perurans trophozoites, wild-type AGD associated N. perurans upregulated multiple gene candidates for virulence factors and a master regulator. Genes associated with invasion of host tissue, evasion of host defense mechanisms and formation of the mucoid lesion were also upregulated. In parallel, localized immune responses were observed in AGD lesions in gill tissue and other immunologically important organs. Finally, drawing both host and parasite transcriptomic responses together we propose a model for host-parasite interaction for AGD in Atlantic salmon.
Materials and Methods
AGD Inoculation and Sample Collection
All animal procedures were approved by the CSIRO Queensland Animal Ethics Committee (project 2017-35, 2018‐09 and 2017-36) under the guidelines of the Australian Code for the Care and Use of Animals for Scientific Purposes (31). Seawater was sourced via offshore spear pumps, filtered (~40 µm), ozonated (100 g O3/h) and ultraviolet treated (80 mJ/cm2) before entering the laboratory. Tasmanian Atlantic salmon were originally imported from the River Philip, Canada in the 1960’s. The all-female Atlantic salmon fry for this project were transported from a commercial Tasmanian hatchery to the Bribie Island Research Centre, Woorim, Australia. Fish were reared in a single cohort in freshwater recirculating systems prior to adaptation to the marine environment. Smolting was achieved by increasing the photoperiod to a 24 h light regime. After 4 weeks, the photoperiod was changed to 12:12 h light/dark and the salinity gradually increased from 3 ppt to 35 ppt overnight. Atlantic salmon smolt (naïve to AGD) with a mean weight of 218 g were habituated in a 5000 L seawater (35 ppt) tank at 16°C, dissolved oxygen (96-100% saturation), pH 7.8, and flow rate of 45 L/min. Fish were fed daily to satiation on a commercial diet (Nutra, Skretting P/L, Cambridge, Australia).
At seven days a subset of 50 animals were transferred to a 1000 L tank to remain naïve to AGD. The other 507 fish in the cohort were subject to an AGD challenge trial (32, 33) by exposure to wild-type gill associated trophozoites of N. perurans. Wild-type (as opposed to laboratory cultured) N. perurans were obtained by the natural exposure of 40 marine adapted Atlantic salmon to wild-type N. perurans through co-habitation with AGD-affected fish maintained in an independent re-circulated marine biosecure experimental tank system dedicated to this purpose. Wild-type N. perurans were originally introduced into the system from AGD-affected gills collected from a commercial farm in Tasmania under animal ethics approval. The day before induction of AGD in experimental fish, 1 L of seawater was collected and subject to centrifugation in 50 mL falcon tubes at 15°C for 20 min at 4500 x g in an Eppendorf5804R (Eppendorf, Germany) to recover N. perurans. The seawater was poured off between subsequent centrifugation rounds until N. perurans were resuspended in a final volume of 10 mL of sterile seawater. The total amoeba in 1 L of seawater was enumerated by averaging repeated trophozoite counts (n = 10) on a hemocytometer (33). To induce AGD in experimental fish for this study, the water flow was stopped, and the water volume halved, before introducing a volume of water from the recirculated system containing a final concentration of 100 N. perurans/L for 2 h before restarting the water flow.
The average gill score for the AGD challenge population was assessed in 36 of the 507 AGD-affected fish using the ordinal ranking methodology on a scale of 0 to 5 of gross AGD pathology across all 16 gill surfaces outlined in Taylor et al. (34). At 21 days post-infection four animals at an average gill score of 3.3 (cohort average 3.1) were sampled from the AGD-affected cohort. AGD was confirmed by presumptive gill scoring and further confirmed by gill histopathology in fish from the same cohort as described by Wynne et al. (32). Gill biopsies were collected using a sterile single-use 2 mm biopsy punch (33), directly from the lesion and a second biopsy approximately 10 mm distal to the lesion from AGD-affected fish. Gill biopsies were also collected from four naïve fish to serve as an AGD-unaffected control. For all fish, primary immune tissues including head kidney and spleen were excised using sterile micro-scissors. A list of all samples collected in this study is provided in Table 1. Approximately 100 mg of tissue was collected and placed in 1.5 mL tubes containing 1 mL of RNAlater (Sigma-Aldrich) before storage at -80°C. For comparison with the lesion biopsy samples, N. perurans wild-type gill associated trophozoites were isolated from the gills of AGD-affected Atlantic salmon and cultured according to the method by English et al. (35) in a 1% malt yeast broth (MYB; 0.01% (w/v) malt extract (Oxoid) and 0.01% (w/v) yeast extract (Oxoid) in filtered, sterile seawater). The floating trophozoite form of N. perurans was isolated from in vitro cultures by centrifugation at 12 000 × g for 8 min at 16°C and the pellet stored at -80°C. The culture acts as a proxy for the wild type N. perurans trophozoites as insufficient N. perurans from the challenge are able to be sourced from the water column. We note that all N. perurans used in this study originated from the same source. However, by contrasting the lesion with the culture condition where there are no host signals, this improves the reliability of identification of N. perurans genes in the host-parasite interaction. The same method was employed by Mohamed et al. (36) in their dual RNA‐seq study of a coral (Acropora tenuis) and its symbiont (Cladocopium goreaui) during the establishment of symbiosis.
Sample Preparation and Sequencing
Total RNA was isolated using RNeasy mini kit (Qiagen). Tissues and the amoeba pellet were lysed in 450 µL of lysis solution on a Precellys 24 homogenizer (Thermo Fisher Scientific) for 2 x 30s at 4.0/ms. RNA was bound to a column and washed twice before elution with 40 µL RNase-free water at room temperature. RNA samples were treated with DNase to remove contaminating DNA using Turbo-DNase (Ambion). RNA quantity and quality were assessed using a NanoDrop ND-1000 spectrometer (Thermo Fisher Scientific) and Agilent 2100 bioanalyzer (Thermo Fisher Scientific). One of the gill lesion samples failed DNA quality analysis and was excluded from RNA-Seq library preparation. This resulted in preparation of a total of 30 RNA-Seq libraries from 11 gill biopsy samples (4 x naïve; 3 x lesion and 4 x distal to the lesion samples from 4 AGD-affected fish), eight head kidney, eight spleen (4 x naïve, 4 x AGD-affected fish) and three in vitro cultured floating N. perurans trophozoites samples. All libraries were prepared using the TruSeq RNA Sample Preparation Kit (Illumina). Host libraries were subject to 150 base pair, paired ended (PE) sequencing on the Illumina Novaseq 4000 sequencing platform at the Australian Genome Research Facility (AGRF) in Melbourne, Australia. In vitro cultured floating N. perurans trophozoites libraries were subject to 150 base pair PE sequencing on the Illumina HiSeq 4000 together with a second independent study (unpublished) undertaken concurrently with this project to conserve resources. Extensive data curation to remove microbial and host contamination from the N. perurans datasets reduces the impact of the different sequencing technologies as described in the section Systemic Host Response. A schematic of the datasets and analyses in this study is reported in Figure 1.
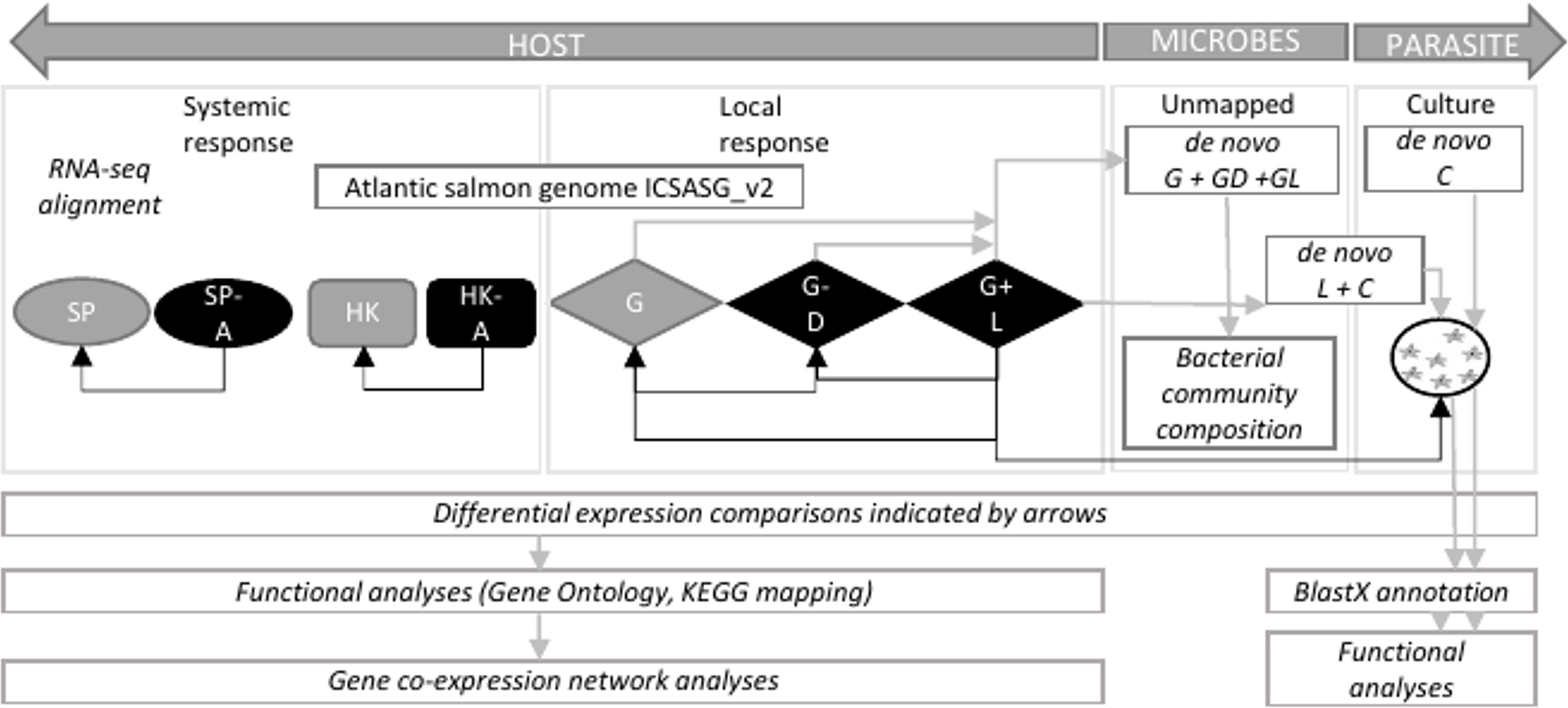
Figure 1 Schematic showing datasets and analytical approach to infer host-parasite interaction in amoebic gill disease (AGD). HOST, Atlantic salmon (Salmo salar); PARASITE, Neoparamoeba perurans; SP, spleen; A, AGD-affected; HK, head kidney; G, gill; D, distal to the lesion; L, lesion; C, cultured floating N. perurans trophozoites.
Dual RNA-Seq Data Analysis
Illumina reads were checked for quality using FastQC version 0.11.8 (37) for all datasets. High quality reads (Q>30) were mapped to the Atlantic salmon genome ICSASG_v2 (38) using HISAT2 version 2.1.0 (39) with default parameters. Alignment files in BAM format were sorted by read name and converted into SAM format using SAMtools version 1.4 (40). Reads uniquely mapped to the salmon genome were extracted from BAM files based on MAPQ > 10 using SAMtools. The Python package HTSeq version 0.7.2 (41) was applied to count unique reads mapped to exons using the reverse parameter for strandedness. The gill lesion samples contained RNA from both the host and the parasite, therefore we used a stepwise in silico mapping approach to separate the reads belonging to each species. Reads were first mapped to the salmon genome to sort unmapped reads into three groups including 1) unmapped read whose mate is mapped; 2) mapped read whose mate is unmapped; and 3) both paired reads are unmapped. Reads were extracted using SAMtools and Bam2fastq options in BEDTools version 2.29.2 (42).
These unmapped reads were subject to a de novo transcriptome assembly with Trinity v 2.8.4 (43) to identify amoeba-related genes associated with the lesion. A second de novo transcriptome Trinity assembly was carried out with unmapped gill lesion and in vitro cultured N. perurans trophozoites reads to undertake differential analyses. Both assemblies followed the assembly method recommended by Hass et al. (43) for strand specific RNA-Seq libraries, minimum contig length of 500 base pairs and read normalization set to a depth of 30. Each de novo transcriptome assembly was further curated by annotating all contigs using Diamond version 0.9.31 (44) and blastx to search the NCBI non-redundant database (12/2019) for the top hit with an expected value (e-value) threshold of <1 x 10-5. Contigs were manually curated further based on the taxonomic classification of the annotated sequences using the NCBI taxonomy database (45). The broad categories for curation of each of these transcriptomes were defined as fish, amoeba/kinetoplastid endosymbionts and protozoans, bacteria and other, which were retained for further downstream analyses and interpretation of the host-parasite response. Raw reads were mapped to the de novo unmapped gill lesion transcriptome using RSEM version 1.3.0 (46) to validate transcripts due to the low quantity of amoeba data generated during sequencing.
The mRNA enrichment step prior to library preparation for sequencing should exclude bacterial genes as they are poorly polyadenylated, although this is not always the case (47). In this instance, the ‘contaminants’ are more likely to be endogenous in origin from the microbiome of the host, or N. perurans, or the culture media. Therefore, opportunistic assessment of the microbiome community present in the lesion samples is possible in the context of published data on AGD. Initially the bacterial species associated with gill lesion were identified in the in vitro cultured and gill lesion de novo transcriptome assembly as described above (Figure 1). Following the successful identification of species known to be associated with the gill microbiome of AGD-affected fish, a second de novo transcriptome assembly was generated using Trinity, applying the same parameters as the N. perurans transcriptome to contrast with naïve fish and the gill microbiome distal to the lesion of AGD-affected fish. Unlike the previous N. perurans transcriptome, this assembly included all unmapped raw reads from the gill including those generated from the naïve fish, biopsies distal to AGD lesions, and the AGD lesion data. While the N. perurans transcriptome had the advantage of in vitro cultured N. perurans to increase the depth of coverage for the amoeba, the unmapped (or bacterial microbiome only) transcriptome was limited in its input data to less than 10% of gill sequenced reads. However, the intent of this component of our study was to identify the major bacterial species that presented in conjunction with AGD at significant quantities to bypass the mRNA enrichment step prior to library enrichment, not the complete diversity of the microbiome which is more aptly assessed through microbial 16S sequencing. The de novo transcriptome assembly was curated for bacterial sequences by annotating all contigs using Diamond version 0.9.31 (44) and blastx to search the NCBI non-redundant database (12/2019) for the top hit with an expected value (e-value) threshold of < 1 x 10-5. Contigs were manually curated based on the NCBI taxonomic classification of the bacteria super kingdom. The relative proportions of each gene based on the transcript frequency (TPM) in the overall dataset were mapped for each bacterial candidate in the microbiome of the naïve and AGD-affected fish, including distal to AGD lesions and AGD lesion material.
Differential Gene Expression, Clustering, and Functional Analyses
Differential gene expression among host tissues and curated N. perurans transcriptomes was inferred by analyzing the raw counts using edgeR version 3.20.9 (48) in R version 4.0.2 (49). To understand the local response to AGD pathology three comparisons for the gill biopsy samples were conducted. These were AGD lesion against naïve, distal to the AGD lesion against naïve, and lesion against distal to the AGD lesion. The systemic immune response was evaluated using the head kidney and spleen tissues, where comparisons were between AGD-affected samples and their respective naïve controls. For the parasite, the curated unmapped gill lesion, and the in vitro cultured N. perurans trophozoites transcriptomes were compared. P‐values were corrected using the Benjamini and Hochberg algorithm (50) for multiple testing. Differential gene expression (fold change > 2; P < 0.05) was compared between naïve and disease states. DEGs were hierarchically clustered using the normalized expression (CPM) values that were log2‐transformed and median‐centered by gene (43). Host data was further analyzed to infer function by gene ontology (GO) enrichment analyses. These were performed using the R package clusterProfiler version 3.9 (51) to identify enriched GO terms that belong to the three key GO categories [biological process (BP), cellular component (CC) or molecular functions (MF)] among the DEGs compared to a background set of expressed genes per tissue. GO terms with a corrected P‐value of < 0.05 were considered significant.
Host Master Regulators, Gene Co-expression Networks, and Differential Connectivity Analyses
Key regulatory transcription factors (TF) contributing to differential expression in the AGD host response were assessed using regulatory impact factor (RIF) metrics (52). Data for putative TF genes in Atlantic salmon were obtained as described by Mohamed et al. (53). The normalized counts (CPM) of these TFs were retrieved for all host tissue samples from prior DEG analyses. Genes with a mean expression FPKM < 0.2 were excluded. These TFs were contrasted against the unique list of DEGs previously obtained for each tissue. The RIF approach comprises a set of two metrics designed to assign scores to regulatory genes consistently differentially co-expressed with target genes, and to those able to predict the abundance of target genes. Scores deviating ± 2.57 standard deviation (SD) from the mean were considered significant (corresponding to P < 0.01). Genes from differential expression and RIF analyses were selected based on overlap and mean normalized expression (at least 0.2 FPKM) to construct the networks. For gene network inference, genes were used as nodes and significant connections (edges) between them were identified using the Partial Correlation and Information Theory (PCIT) algorithm (54) to calculate the significance of the correlation between two nodes after accounting for all the other nodes in the network. An initial network was constructed using all samples and gene node connections (2SD; P < 0.05). To explore differential connectivity during AGD, two additional networks were constructed for naïve and AGD-affected samples. The number of connections per gene in relation to the naïve or AGD-affected network was computed enabling identification of differentially connected genes (DCGs), and subsequent review of subnetworks related to host-parasite interaction. Networks were visualized using Cytoscape Version 3.7.2 (55).
Host-Parasite Interaction Pathway Analysis
To explore the host-parasite interaction further, gene pathways for the host and the parasite were independently visualized by submitting genes from differential expression analyses to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps module (56–58). A specific parasitic response pathway from host data was not identified and led to further investigation of the KEGG database. This revealed that when selecting Salmo salar (sasa) as the organism potential human host-pathogen pathways are not automatically searched. Therefore, host and N. perurans genes were converted to KEGG pathway orthologues to enable exploration of the human amoebiasis pathway (hsa05146) associated with the amoeba parasite Entamoeba histolytica to infer a model for host-parasite interaction in AGD.
Results
Dual Transcriptome Sequencing and Assembly
Sequencing produced a total of 3.1 billion PE reads with approximately 81 million PE reads per Atlantic salmon library (Supplementary Table 1). The three lesion samples were sequenced to a higher depth producing approximately 362 M PE reads per library to recover transcriptomic data for N. perurans and the associated bacterial community in the presence of host tissue (Supplementary Table 1). Sequencing of the N. perurans in vitro cultured floating trophozoite samples produced approximately 35 M PE reads per library (Supplementary Table 1). An average mapping rate of 87% (70 M; gill lesion data 313 M) was achieved for all salmon samples to the Atlantic salmon reference genome ICSASG_v2 (38). The remaining 13% (47 M) of PE reads from lesion samples that did not map to the salmon genome were segregated for further analysis.
A de novo transcriptome assembly completed with only the gill lesion data produced 186,310 contigs with a re-mapping rate of 90%. NCBI taxonomy together with a blastx search against the non-redundant NCBI database was then used to classify the longest isoform for each gene (n = 56,657) into categories. A total of 35,306 contigs remained after classification, of which 77%, 2.4%, 0.6% and 20% of contigs contributed to host, N. perurans (and its endosymbiont), bacterial and ‘other’ species respectively. The ‘other’ category consisted of species matches that were considered to be taxonomically too diverse in origin, and of biological significance to provide insight into host-parasite interaction. This is a common issue encountered when working with non-model organisms. The low coverage of N. perurans and endosymbiont genes (n = 833) suggests that the 10% of N. perurans data recovered from the gill lesion was insufficient for a complete transcriptome to be assembled. Therefore, a second de novo transcriptome assembly was completed together with xenic in vitro cultured N. perurans trophozoites (106 M PE reads) and the unmapped gill lesion data (Supplementary Table 1). The initial assembly was 214,381 contigs before taxonomic classification annotated 144,214 contigs. Of these 84%, 6.5%, 1.5% and 8% of contigs contributed to host, N. perurans (and its endosymbiont), bacterial, and other species, respectively. The resultant amoeba and endosymbiont transcriptome contigs were extracted for downstream analyses.
Local Host Response
Differential gene expression analysis was undertaken to compare the local host response in gill tissues of an AGD-affected fish compared to a naïve fish. This included the local response at the lesion and an area distal to the lesion. Hierarchical clustering of the biological replicates using Spearman correlations from a comparison of the normalized gene expression for all gill samples against each other showed that the biological replicates from the AGD lesion were most similar (Supplementary Figure 1A). In contrast, two of the biological replicates for the region distal to the lesion were similar to the lesion samples, while the third sample was similar to the naïve samples (Supplementary Figure 1A), suggesting the stage of disease progression may not have been as advanced for this fish. A total of 1,741 DEGs (fold change > 2; P < 0.01) were identified in the gill samples (Figure 2A). Of these, 644 DEGs were associated with the lesion compared to the control and 25 DEGs had a -log(10)FDR > 20 (Supplementary Figure 1B, Supplementary Table 2). In contrast, the region distal to the lesion exhibited 60 (Supplementary Figure 1C) and 49 DEGs (Supplementary Figure 1D) compared to the lesion and the naïve samples respectively (Figure 2A, Supplementary Table 2).
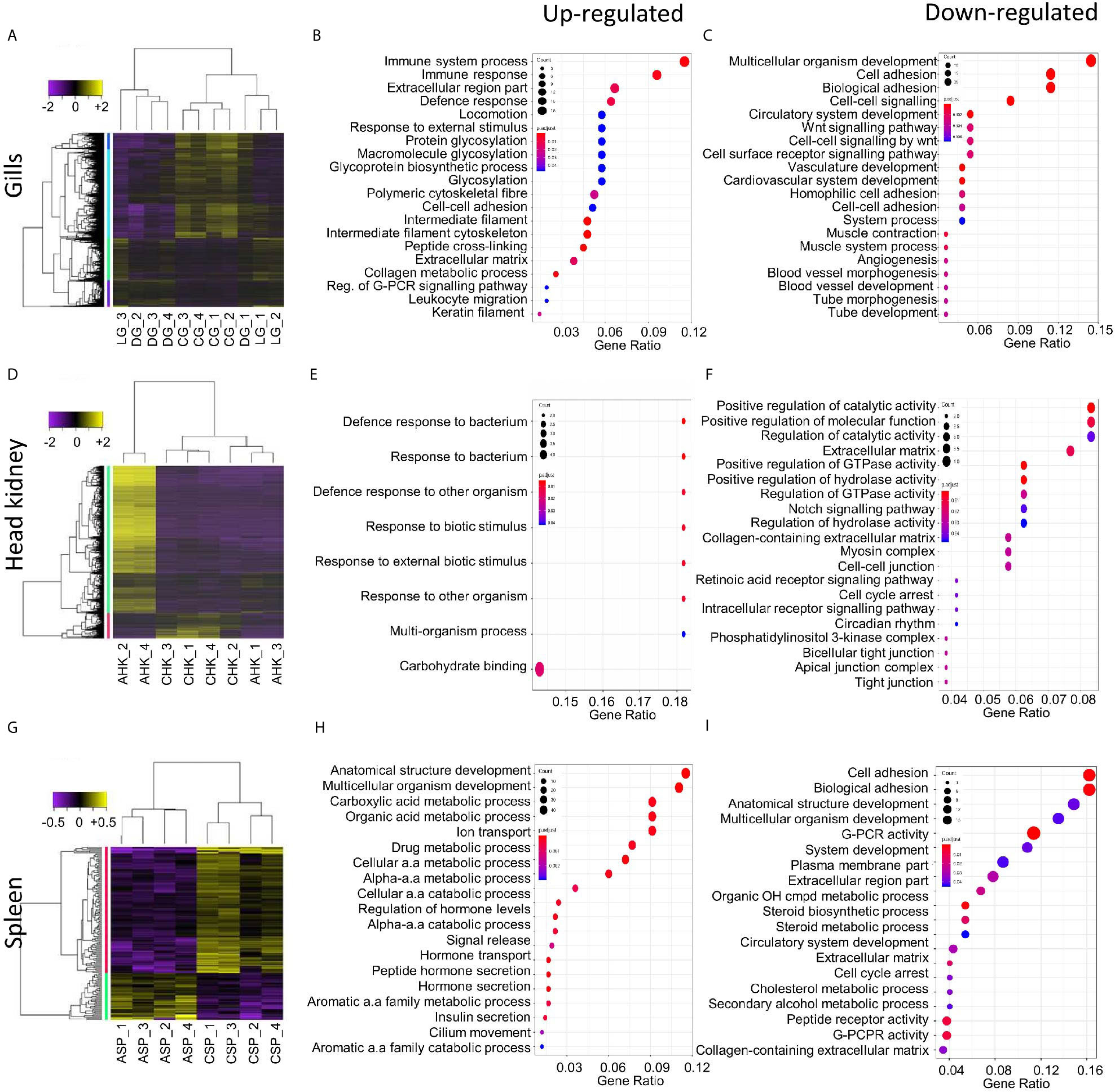
Figure 2 Summary of the host transcriptomic response during amoebic gill disease (AGD) progression. Differential gene expression and significantly enriched gene ontology (GO) terms are shown for the local (A–C) and systemic (D–I) response to AGD in Atlantic salmon. Heat maps show hierarchical clustering of differentially expressed genes (rows) with differential expression among replicates from a naïve (C) and AGD-affected (A) Atlantic salmon. Expression values are log2-transformed and median-centered by gene. (A) The local response is characterized by differential gene expression (fold change > 2; corrected P-value < 0.01) and includes a gill (G) biopsy distal (D) to the lesion (L) of an AGD-affected fish. The systemic response among head kidney (HK) (D) and spleen (SP) (G) replicates is characterized by differential gene expression (fold change > 2; corrected P-value < 0.05). Enriched gene ontology (GO) terms (hypergeometric test, Bonferroni-adjusted P < 0.05) among the differentially expressed genes for the gills (B, C), head kidney (E, F) and spleen (G, H) along with the gene ratio for the genes that map to each term. The majority of the enriched terms are related to host defense and immune response.
GO enrichment analysis was undertaken to identify the function of genes implicated in the local response to N. perurans invasion and subsequent AGD pathology. The top functional enrichment GO categories (BP, MF, CC) are shown in Figures 2B, C and Supplementary Figure 3. The significant GO terms and their corresponding genes are listed in Supplementary Table 3. In brief, enriched GO terms characterized by upregulated DEGs were consistent with activation and mobilization of innate immune system factors, inflammation, host defense and wound healing. While prospective pathogen invasion pathways were connected to the downregulation of genes associated with wnt and integrin-mediated signaling as well as cell adhesion (59–61). GO categories were heavily enriched for downregulated genes in developmental process categories indicative of diverting metabolic resources away from growth.
Systemic Host Response
Differential gene expression analysis was undertaken in key immune organs in four biological replicates to assess the systemic host response of an AGD-affected fish compared to a naïve fish. For the head kidney, hierarchical clustering of the biological replicates based on normalized gene expression indicated that all naïve samples were closely correlated based on Spearman correlation. However, two of the AGD-affected head kidney samples clustered more closely with the naïve samples than their counterparts (Supplementary Figure 2A). In contrast, clustering of the spleen biological replicates clearly resolved AGD-affected from naïve samples (Supplementary Figure 2B). The transcriptomic response in the head kidney involved 1,463 DEGs (fold change >2; P < 0.05) (Figure 2D) of which 21 DEGs had a -log(10)FDR > 5 compared to the naïve samples (Supplementary Figure 2C; Supplementary Table 4). In the spleen, only 155 DEGs (fold change > 2; P < 0.05) were identified (Figure 2G) of which 20 had a -log(10)FDR > 5 compared to the naïve samples (Supplementary Figure 2D; Supplementary Table 5).
GO enrichment analysis was undertaken to identify the function of genes expressed in key immune organs (head kidney and spleen) implicated in the systemic response. The top functional enrichment GO categories (BP, MF, CC) are shown in Figures 2E, F, H, I. The significant GO terms and their corresponding genes are listed in Supplementary Tables 6 and 7. Enriched GO categories in the head kidney were similar to the local gill response with the downregulation of genes that modulate the inflammatory response, as well as signaling receptors and their pathways, that may assist pathogen invasion. A mucin-13-like gene associated with mucosal immunity was also identified in the head kidney (62). In contrast to the gill, GO term categories related to developmental processes were upregulated in the head kidney together with regulatory genes and transcription factors. In the spleen GO enrichment of upregulated genes corresponded to pathogen recognition with subsequent innate immune activation and acute inflammation. While GO enrichment for categories characterized by downregulated genes supported pathogen invasion, prevention of inflammation resolution, moderation of the innate immune response, and lesion suppression.
Gene Co-expression Networks and Key Regulators of the Host Response to AGD
Transcription factors (TFs) are key regulators of gene expression in normal and disease states, however the detection and differential expression of TFs is often obscured by more highly expressed genes (52). Gene co-expression networks were used to characterize the transition between naïve and AGD disease states across all tissues of interest (63–67). Gene co-expression networks essentially seek the points of intersection between a single set of TFs and DEGs from all tissues in the study (e.g. gills, kidney, spleen). In this case, two gene networks are created, one for the naïve and a second for the AGD disease state. The transcription factors are not limited in the number of connections within or across tissues types. RIF metrics assign two scores to TFs (1) a score for TFs that are consistently differentially co-expressed with highly abundant and differentially expressed genes, and (2) TFs that have the ability to predict abundance of differentially expressed genes (52). This study contrasts differential gene expression data against predicted TFs to identify regulators of the local and systemic host response to AGD. A total of 403 TFs consisting of 174, 111 and 118 key TFs were identified in the gill, head kidney and spleen, respectively (Supplementary Table 8). Of these, 30 TFs were also differentially expressed in response to AGD (Supplementary Table 9). The top four included Krueppel-like factor 15 in the head kidney (438 connections), filamin-interacting protein FAM101A-like (435 connections) and peroxisome proliferator-activated receptor alpha (336 connections) in the spleen and early growth response protein 3 (261 connections) from the gill.
The partial correlation and information theory (PCIT) algorithm is a statistical test used to identify significant correlations between genes and TFs (54). The PCIT algorithm was used to co-analyze key TFs and DEGs to infer gene networks in naïve and AGD disease states. Initially every gene was applied to each disease state to create a network comprising 1,864 genes with 883,408 connections. Visualization of the connections between the gene networks in Cytoscape enabled tissues types to be identified by color, and genes and TFs to be identified by shape and size depending on the number of connections. In the AGD disease state, the genes with the most connections (networks) were identified in the gill (n = 978; 53%) with networks from the head kidney (n = 406; 22%) and spleen (n=450; 25%) contributing equally to the overall host response to AGD. The top four genes based on the number of connections in the gene network in AGD-affected fish were from the gill (n = 3) and head kidney (n = 1) and included aurora kinase B-like, zinc finger protein OZF-like, zinc finger protein 70-like and protein NDNF-like, respectively. Key regulators undergo a substantial change in their number of connections during changes in disease state. Therefore, separate networks were constructed using 12 naïve samples and 15 AGD-affected samples, before identifying genes that underwent the largest change in connectivity (68) (Figure 3). The top 20 differentially connected genes (DCGs) and their putative roles are reported in Table 2. These include eight TFs, of which three belong to the zinc finger (znf) family and were more expressed in the gill (compared to other tissues) indicating a putative role in AGD progression. The most highly connected regulators based on increase (+) or decrease (–) in size of their connected gene networks were znfOZF-like (+537 connections) and znf70-like (+507 connections), while the least connected was znf135-like (-789 connections) in AGD.
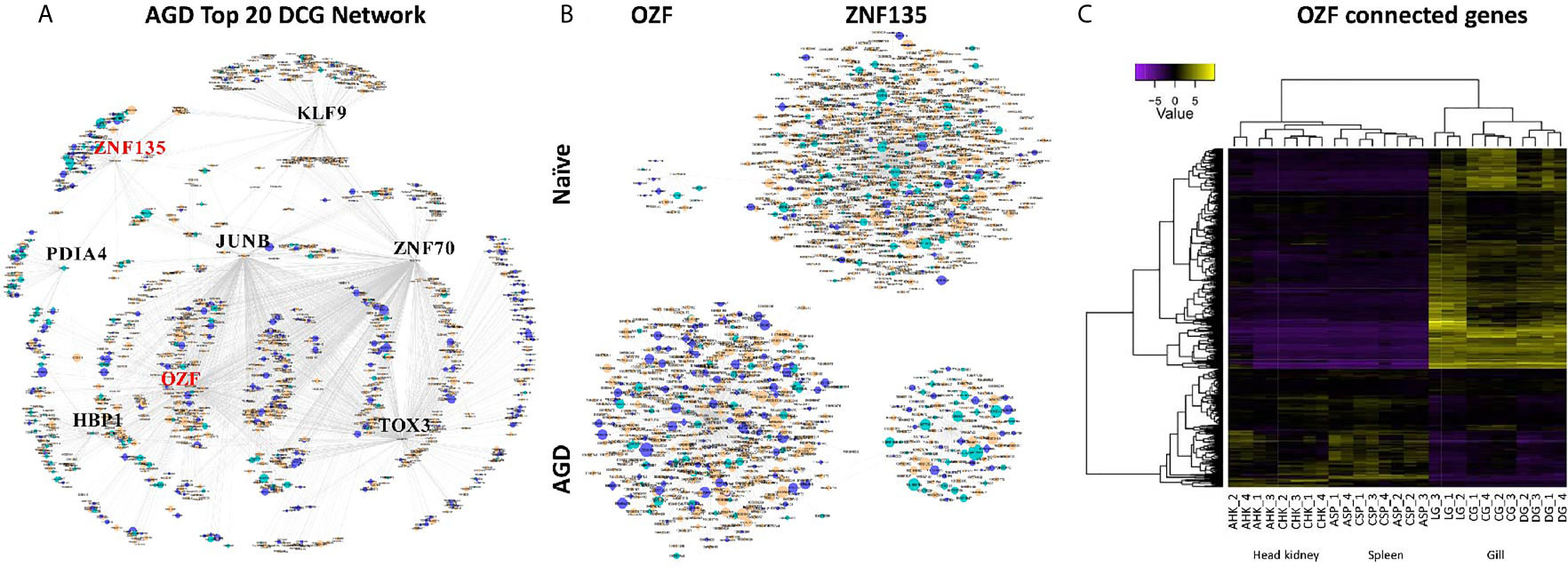
Figure 3 Sub-networks for the top differentially connected genes (DCGs) likely to regulate the transcriptomic response during amoebic gill disease (AGD) in Atlantic salmon. (A) AGD network of 8 transcription factors (TFs) among the top 20 DCGs using the PCIT algorithm. All nodes are represented by ellipses except for genes coding key regulators (TFs) which are diamond shaped. Nodes are orange for gill, green for head kidney and purple for spleen. The size of the nodes is relative to the normalized mean expression values in all samples. (B) Subnetworks of top differentially connected TFs. The networks created with the most differentially connected genes between naïve and AGD-affected networks with zinc finger protein OZF-like (OZF) as the key regulator with the greatest number of connections in the AGD-affected network, while zinc finger 135 (znf135) lost the majority of its connections in the AGD-affected network. (C) Heat map shows hierarchical clustering of differential expression of connected genes (rows) in the OZF network among replicates from a naïve (C) and AGD-affected (A) Atlantic salmon in the head kidney (HK), spleen (SP) and gill (G). The AGD-affected gill data is represented by the lesion (L), a biopsy distal to the lesion (D) and naïve gill (C). Expression values (CPM) are log2-transformed and median-centered by gene.
The sub-network associated with znfOZF-like TF consisted of a total of 558 genes of which 386, 55 and 117 regulatory genes were identified in the gills, head kidney and spleen, respectively. This included a total of 142 TFs with 101 identified in the gill (Supplementary Table 10). Interestingly five (106573105, 106578124, 106586175, 100136550, 106608858) of the top 10 upregulated DCGs were in the znfOZF-like gene network (Tables 2 and 3). Further investigation of the 386 genes expressed in the gill identified 12 genes with potential roles in host-parasite interaction (Table 3). These included hemagglutinin and amebocyte aggregation factor-like isoform X1 (90), macrophage mannose receptor 1-like (91), T-cell immunoreceptor with Ig and ITIM domains, the Ap-1 complex and TFs (jun, fos) (75, 95), cytolysin RTX-A-like (94), mucin-2-like isoform X2 (93), coxsackievirus and adenovirus receptor homolog (92), secreted frizzled-related protein 2-like (85) and T-lymphoma invasion and metastasis-inducing protein 2 (96).
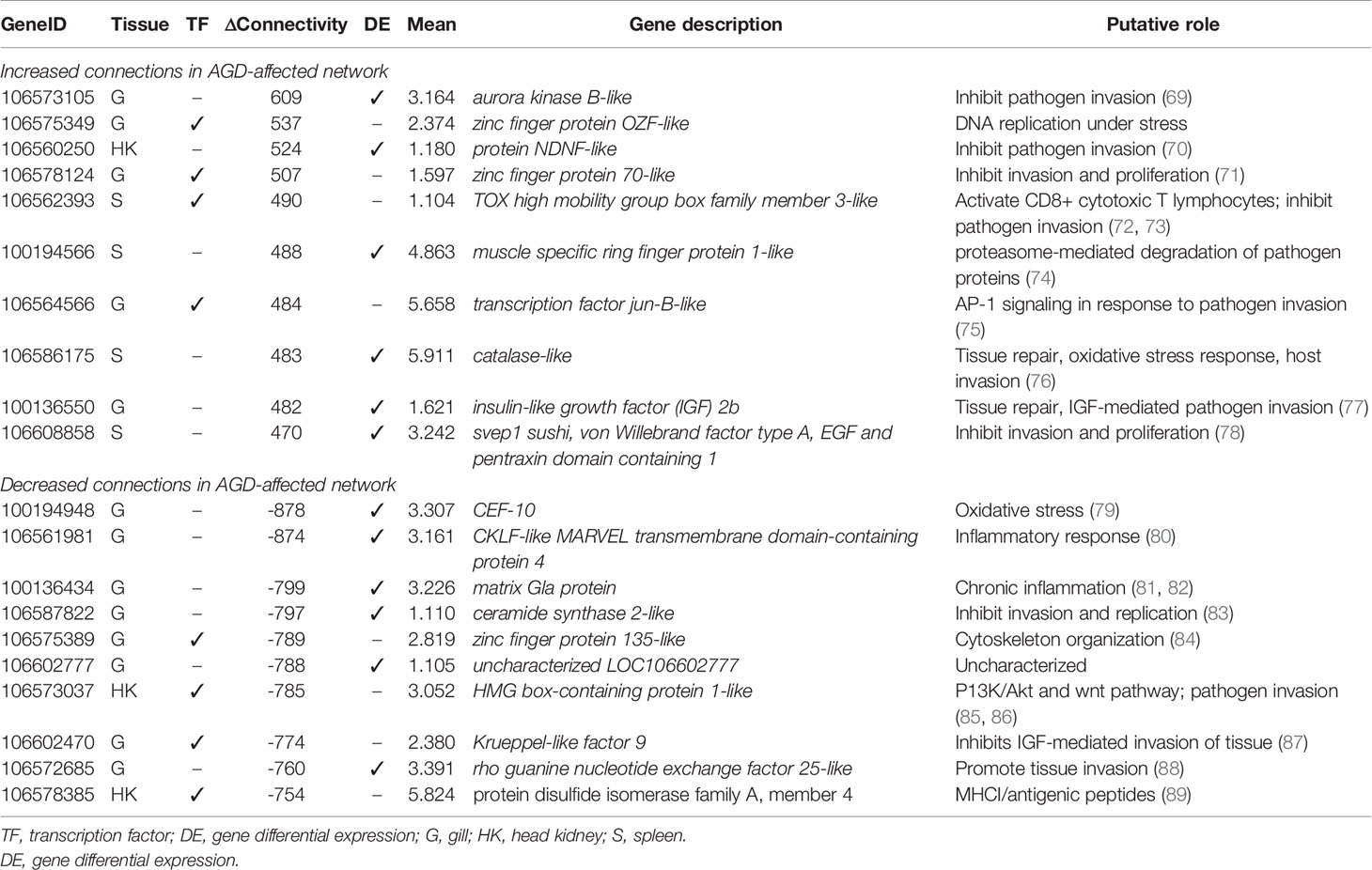
Table 2 Top 20 differentially connected genes associated with amoebic gill disease (AGD) pathology in Atlantic salmon. Data for tissue of maximum expression, differential connectivity between naïve and AGD disease states, and mean expression are shown.

Table 3 Selected genes expressed (fold change > 2; P < 0.05) in the gills from the zinc finger OZF-like gene regulatory network with potential activity in the host-parasite response to amoebic gill disease (AGD).
Neoparamoeba perurans Gene Candidates for Host-Parasite Interaction
To infer candidate genes for host-parasite interaction an initial de novo assembly was generated based on the non-host gill lesion data to produce a total of 833 N. perurans and endosymbiont candidate genes after curation using blastx (e-value <1 x 10-5) to annotate the genes, and species classification to remove host and commensal bacteria genes. The majority of annotated genes from amoeba species matches were represented by Acanthamoeba castellani, Dictyostelium discoideum, Neoparamoeba pemaquidensis, Naegleria gruberi and Tieghemostelium lacteum, while the kinetoplastid endosymbiont was represented by Perkinsela sp. CCAP 1560/4. Pathogenic protozoans were also retained as candidates for virulence and pathogenicity as knowledge of these factors in AGD pathogenesis is highly sought. The low number of transcripts generated by the de novo assembly suggests a low number of raw reads specific to N. perurans were generated during sequencing and may have hindered the assembly of a larger number of transcripts. Assembling gill lesion sequence data together with the in vitro cultured N. perurans floating data increased the number of amoeba species and kinetoplastid transcripts to 9,399 available for differential gene expression. Hierarchical clustering of the biological replicates using Spearman correlation from a comparison of the normalized gene expression for all samples against each other showed that the biological replicates associated with each condition were most similar (Supplementary Figure 4A).
Differential expression of N. perurans in the gill lesion biopsy compared to the in vitro cultured floating trophozoites was undertaken in three biological replicates (Supplementary Figure 4). A total of 375 annotated transcripts were significantly expressed DEG’s (fold change > 2; P < 0.05) and their genes are listed in Supplementary Table 11. Of these DEGs, 28 were expressed by the endosymbiont including two immune suppression genes, Yop effector YopM and cyclophilin. Two stress response genes were also identified chaperone protein DNAj and heat shock protein 70. The remainder were related to cellular processes. A short list of candidate N. perurans genes and their putative roles in host-parasite interaction are provided in Table 4. In brief, candidates for tissue invasion, host immune evasion, pathogenicity, virulence factors and their regulatory systems were identified. The de novo N. perurans transcriptome assembly revealed a further five gene candidates for host-parasite interaction and pathogenicity (Supplementary Table 12). These were extracellular Cu/Zn-superoxide dismutase for defense against the host response and environment (110). AprA, from a master regulatory pathway for virulence that also suppresses host defense (111). Two virulence factors, prokumamolisin activation domain containing protein and prohibitin (112, 113) and lastly a candidate gene export of virulence proteins, vacuolar sorting protein SNF7 (114).
Table 4 Selected genes differentially expressed (fold change > 2; P < 0.05) in Neoparamoeba perurans in the pathogenesis of amoebic gill disease in Atlantic salmon.
Microbial Community Associated With AGD
Although the Illumina libraries used in this study were enriched for polyadenylated transcripts, a considerable number of bacterial reads were obtained and assembled, with functional annotations and taxonomic classifications from the NCBI database as described in the methods (Supplementary Table 13). An opportunistic assessment of bacterial species from these revealed a diverse abundance of many known marine-derived species in both the original N. perurans transcriptome and unmapped gill transcriptome. The relative proportions of each gene based on the transcript frequency (TPM) in the overall dataset were mapped for each bacterial candidate, overall demonstrating a high diversity of species. The most prevalent taxa based on standardized reads in this data are shown in Figure 4. Proportionally the taxa with higher read matrices in the original N. perurans transcriptome included Pseudomonas stutzeri, Halomonas halocynthiae, Rhodopseudomonas palustris and Adhaeribacter aquaticus, which are known marine bacterium (Figure 4A). The remaining top 10 most prevalent identifiable taxa have been identified in a number of aquatic environments, four are known to conform as a commensal species, while Legionella feeleii is a known pathogenic taxon. Interestingly, the Epulopiscium sp. has previously been identified as an intestinal endosymbiont taxon to surgeonfish Naso tonganus (115). The unmapped gill transcriptome data provided a similar story with the addition of naïve fish and AGD-affected data from gill biopsies, from distal to the lesion and the lesion (Supplementary Table 13). The majority of bacteria were detected in all sample types; however, the relative proportions were similar in the bacterial communities of the lesion and distal to the lesion transcripts of the AGD-affected fish compared to the naïve fish (Figure 4B). This is not surprising due to tank/environment effects that may have contributed to different microbial communities after 21 days in the flow through marine system. Known marine pathogenic bacteria were identified in the unmapped data set including Nocardia jejuensis and the Order Flavobacteriales, in addition to those previously mentioned. Interestingly, Nocardia was only identified in AGD-affected fish raising the question of whether it was present before, or after AGD appeared on the gill.
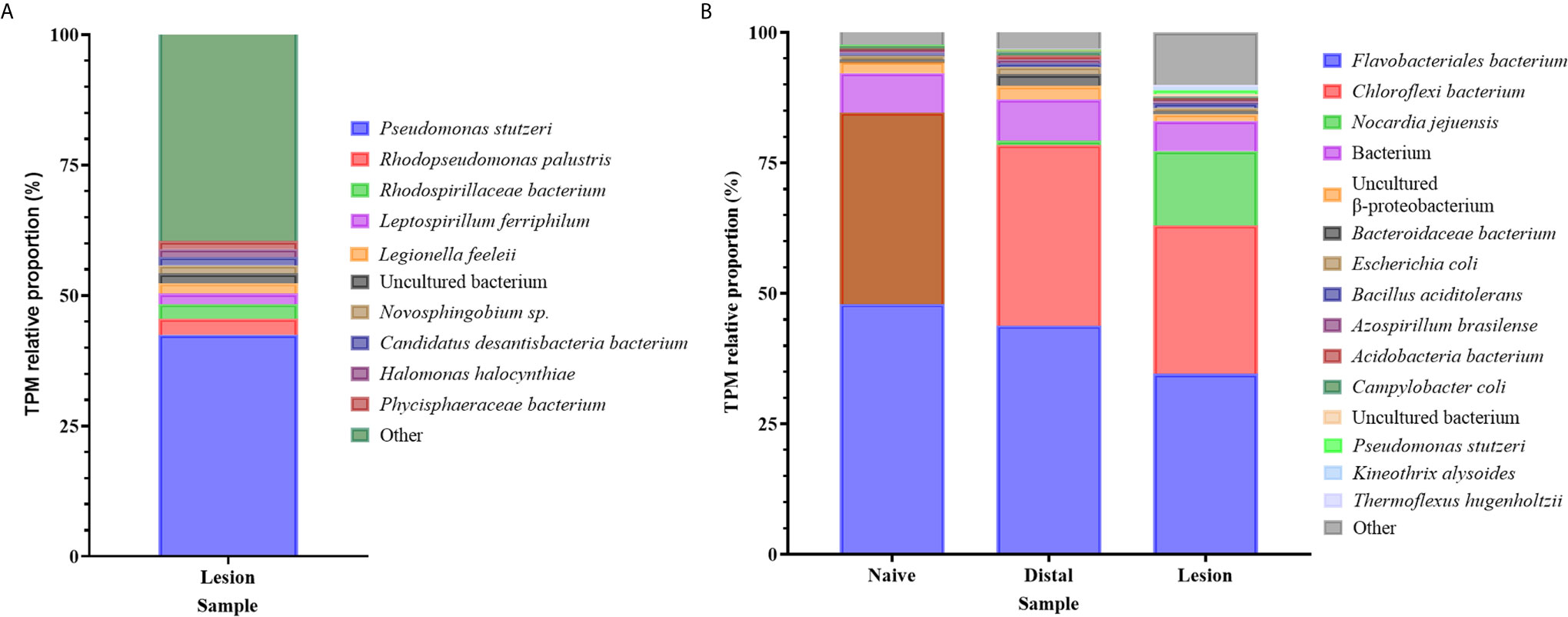
Figure 4 Bar plot of the bacterial taxa identified from sequence reads. (A) Bacterial taxa identified in the unmapped Neoparamoeba perurans transcriptome. While the community diversity of 142 species was plotted to show visual diversity, only the top 12 candidates based on proportional reads (>1% TPM) are denoted in the legend. (B) Bacterial taxa identified in the unmapped Atlantic salmon gill transcriptome from naïve fish, biopsies distal to AGD lesions, and the AGD lesion data. The top 15 candidates based on proportional reads (>1% TPM) are denoted in the legend.
Model for Host-Parasite Interaction
A theoretical model for the host-parasite interaction in Atlantic salmon in response to AGD was inferred based on the data generated in this study and visualization in the KEGG pathways module for salmon, Salmon salar (sasa). The model generated from our data is shown in Figure 5 and a list of the genes and their isoforms in the model is provided in Supplementary Table 14. In the local host response to AGD, visualization of KEGG pathways revealed downregulated genes involved in antigen processing and presentation through MHCI/MHCII complexes, the phosphatidylinositol 3’-kinase (PI3K)-Akt signaling pathway and the NF-κβ signaling pathway. In contrast, upregulated genes mapped to the C-type lectin and Toll-like receptor pathways, and the wnt and NOD signaling pathways. KEGG pathway mapping of the systemic response in the head kidney and spleen revealed that the majority of upregulated genes (3,151 and 335, respectively) were involved in pathways for metabolism, focal adhesion, C-type lectins, calcium and toll-like receptor signaling and cellular integrity. However, to a lesser extent wnt signaling, bacterial and viral infection, and immune pathways were also well represented. Downregulated genes in the head kidney and spleen (597 and 221, respectively) mapped predominately to metabolism pathways, followed by a multitude of signaling pathways including calcium, foxO, insulin, MAPK, apelin, Erb8 and p53. Interestingly, the cytokine-cytokine receptor interaction pathway was equally represented by upregulated and downregulated genes in the spleen.
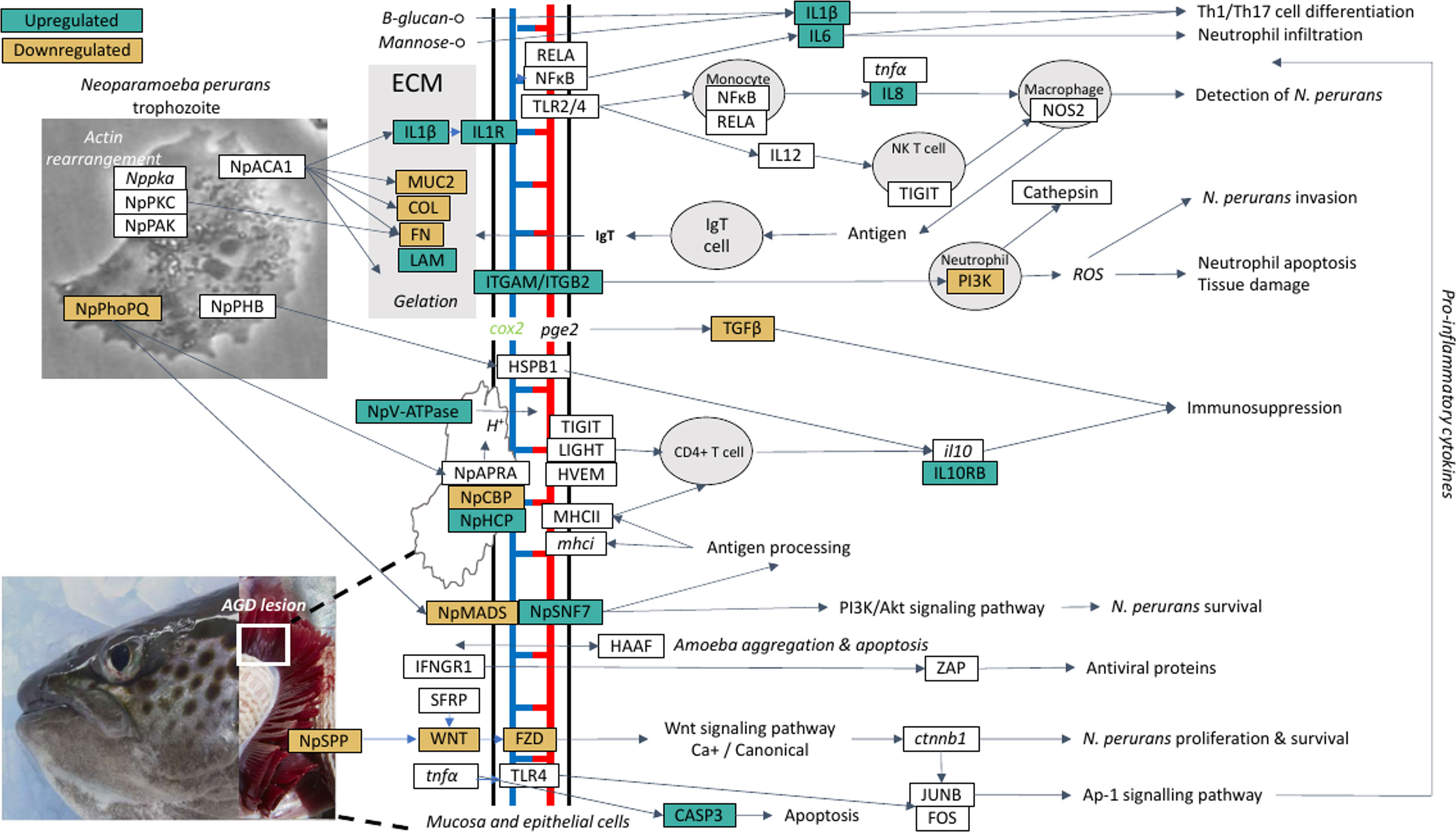
Figure 5 A theoretical model for the host-parasite interaction between Neoparaomoeba perurans candidate genes and Atlantic salmon during the pathogenesis of amoebic gill disease (AGD). The pathway was generated from the differential gene expression and network analyses together with the key KEGG pathways mapped and visualized (sasa04310, sasa05168, sasa05132, hsa04151, hsa05146) for the host and the parasite. Pathogen invasion is facilitated by degradation of host mucus and epithelia. To maintain a pathogen friendly environment on the gill N. perurans releases factors to decrease ammonia and reactive oxygen species released by the host. Actin rearrangement of the pathogen and the host facilitates attachment. Virulence factors are modulated by the pathogen master two component PhoPQ virulence regulatory system. Downregulation of the host wnt, Ap-1 and PI3K/Akt signaling pathways supports pathogen immune evasion, proliferation, and survival. The Th1/17 cell differentiation pathway is upregulated inducing innate and adaptive immune responses in the host. NP signifies N. perurans genes, upregulated genes are green, downregulated brown, not differentially expressed are white, italicized genes were not identified in our dataset. CASP3, caspase-3; COL, collagen (various); cox2, cyclooxygenase 2; ctnnb1, β-catenin; FN, fibronectin; FOS, fos; FZD, frizzled; HAAF, hemagglutinin/amebocyte aggregation factor-like; HSPB1, heat shock protein beta-1-like; HVEM, tumor necrosis factor receptor superfamily; IFNGR1, interferon gamma receptor 1; il10, interleukin 10; IL10RB, interleukin-10 receptor subunit beta-like; IL12, interleukin-12; IL1R1, interleukin-1 receptor type 1-like; IL1β, interleukin-1 beta; IL6, interleukin-6; IL8, interleukin 8; ITGAM, integrin alpha-X-like; ITGB2, integrin beta-2; JUNB, junb; LAM, laminin; LIGHT, tumor necrosis factor ligand superfamily; mhci, major histocompatibility complex class I; MHCII, major histocompatibility complex class II; MUC2, mucin-2-like; NFκB1, nuclear factor NF-κ-β p105 subunit-like; NOS2, nitric oxide synthase 2; NpACA1, prokumamolisin activation domain containing protein; NpAPRA, AprA protease; NpCBP, cathepsin-B; NpHCP, hybrid cluster protein; NpMADS, MADS-box transcription factor; NpPHB, prohibitin; NpPhoPQ, PhoPQ-activated pathogenicity-related protein; NpSNF7, vacuolar sorting protein SNF7; NpSPP, signal peptide peptidase; NpV-ATPase, vacuolar proton ATPase; NpPAK, p-21 activated kinase; Nppka, protein kinase A; NpPKC, protein kinase C; pge2, prostaglandin E2; PI3K, phosphatidylinositol 3’-kinase; RELA, putative transcription factor p65 homolog; SFRP, secreted frizzled-related protein; TGFβ, transforming growth factor beta; TIGIT, T-cell immunoreceptor with Ig and ITIM domains; TLR2/4, toll-like receptor 2 and 4; tnfa, tumor necrosis factor alpha; WNT, protein Wnt; ZAP, zinc finger antiviral protein.
Visualization and mapping of the top 20 DCGs (Table 2) against the KEGG pathways revealed that the top two gill TFs (znfOZF-like, znf70-like) were antiviral proteins in the Herpes simplex virus 1 infection pathway (sasa05168). Further exploration of the znfOZF-like gene network (Figure 3) revealed 26 genes in the Herpes simplex virus 1 infection pathway suggesting a viral-like response may be initiated in response to parasite invasion in AGD. Lastly, a review of genes mapping to components of bacterial and parasite pathways including the Ap-1 and PI3K-Akt signaling pathway (hsa04151) in Salmonella infection (sasa05132), and the human Entamoeba histolytica amoebiasis pathway (hsa05146), provided insight to the proposed host-parasite model for AGD. The model identifies a theoretical pathway for the regulation of virulence in N. perurans, plausible methods for modulating the host environment to support parasite proliferation and evasion of the host immune response.
Discussion
In this study, a dual RNA-Seq approach together with differential gene expression, and system-wide analyses of TF networks was employed to present a theoretical model for the host-parasite interaction during pathogenesis of AGD in Atlantic salmon. A recent review by Marcos-Lopez and Rodger (116) discusses the host response to AGD in detail, therefore we will focus on key findings from our study that provide new insights into the host-parasite interaction, with a particular focus on the factors responsible for virulence, and evasion of the host immune response.
Candidate Pathways for Parasite Propagation and Attachment
Differential gene expression, gene networks and regulatory transcription factors in this study suggest a possible mechanism for N. perurans to evade detection and facilitate propagation on attachment to the gill of AGD-affected Atlantic salmon. The BP GO term ‘Wnt signaling pathway’ was enriched in the gill and the head kidney with downregulated genes including protein wnt-4a-, 5b- and 10a-like, and secreted frizzled related proteins 1-, 2- and 5-like. There are three wnt signaling pathways including the canonical or β-catenin-dependent pathway, and the non-canonical pathways, planar cell polarity and wnt/Ca2+ (calcium) each of which interact with a frizzled transmembrane receptor to promote paracrine or autocrine signaling (117, 118). This pathway is involved in cell cycle control, cytoskeleton reorganization during phagocytosis and cell migration, autophagy, apoptosis, and inflammation (85). The canonical and wnt/Ca2+ signaling pathways were enriched in all host differential expression and network analyses in this study. Both intracellular and extracellular bacteria have evolved to modulate and evade the host innate immune response through subversion of these wnt pathways. This may occur through activation or inhibition of the wnt pathway that results in immune suppression, induction of inflammation, disruption to epithelia and promotion or suppression of cellular proliferation depending on the pathogens intra- or extra-cellular location to the host (85).
In our study, the PhoP/Q system was identified as a candidate for the master regulator of virulence factors in N. perurans through blast similarity with the amoebozoa cellular slime mold, Cavenderia fasciculata (syn. Dictyostelium fasciculatum). This system has been reported to induce pathogenic factors implicated in Salmonella enterica invasion of intestinal epithelium through suppression of the wnt-signaling canonical pathway (119). N. perurans exhibits downregulation of PhoP/Q factors in the lesion, however this may be due to the late stage of the AGD lesion in our study of gill index 3.4. In contrast to the canonical pathway that is activated through multiple wnt proteins, the wnt/Ca2+ pathway is activated only through the binding of wnt5 to frizzled (sasa04310). We propose that the candidate genes for N. perurans virulence factors which are secreted effector proteins, secreted frizzled protein (NpSFRP) and the signal peptide peptidase (NpSPP), may bind to wnt5 subverting the pathway to promote proliferation and survival of N. perurans during invasion. The wnt signaling pathway also interacts with the Ap-1 signaling pathway to induce innate and adaptive immune responses. Likewise, viruses reported to hijack this pathway induce overexpression of the c-Jun and c-Fos proteins. These proteins make up the dimeric Ap-1 TF whose dysregulation promotes carcinogenesis and tumor progression in cancer (75). A multitude of genes identified in this study have been reported to be involved in carcinogenesis or tumorigenesis in cancer, suggesting that dysregulation and overexpression of genes is a common point of intersection with AGD in Atlantic salmon.
Gene Candidates for Host Pathogen Recognition and Sequestration in AGD
Investigation of the genes expressed in the gill identified several candidates for roles in host-parasite interaction (Table 3). Expression of hemagglutinin and amebocyte aggregation factor-like isoform X1 is a candidate gene for sequestration and aggregation of N. perurans into a lesion and agglutination of host blood (90). Pathogens released by the parasite may be recognized by the lectin-C pattern recognition receptor, macrophage mannose receptor 1-like which also stimulates interleukin-1β and the Th1/Th17 pro-inflammatory cytokine response (91). Interestingly, the Th1, Th2 and Th17 cell differentiation pathways reported to be associated with AGD through quantitative trait locus (QTL) and quantitative PCR analyses, were not markedly enriched pathways in our analysis (15, 120). Furthermore, the data were contradictory with previous studies, exhibiting upregulation of Th17 genes and downregulation of several other genes in the Th1, Th2 and Th17 cell differentiation pathways leading to their expression. The genes of the Th1/Th17 pathway identified however suggest that the pathogen may be recognized by a lectin-C pattern recognition receptor, macrophage mannose receptor 1-like or β-glucan receptor which stimulates the Th1/Th17 pro-inflammatory cytokine response (91). The contradictory Th1/Th17 data provides support for using system wide analyses to unravel the mechanisms behind complex disease states. In future studies, the concurrent use of multiple datasets (tissues, timepoints, backgrounds, pathogen strains) and types (transcriptome, methylation) in system wide analyses may prove to be invaluable in progressing our understanding of AGD.
Gene Candidates and Gene Networks in Host Defense Against AGD
The fish in this study had not been exposed to N. perurans, or commensal bacteria or viruses associated with AGD prior to participating in this study. Therefore, the adaptive immune system was not primed to recognize and respond to the specific pathogen/s associated with AGD, in the same manner that the immune system may respond on its second interaction with the disease following successful treatment. Valdenegro-Vega et al. (121) have previously reported that consecutive challenges with N. perurans resulted in elevated IgM gene expression at gill lesions occurring 31 days after infection. While the primary infection does not induce an IgM or IgT transcriptome response. This is suggestive of an inefficient adaptive immune response on first introduction to the N. perurans unrecognized pathogen. With this consideration, the transcriptome differential expression and regulatory transcription factor networks in our study indicate that the host induces the primary defense pathways for bacteria and virus defense at the first interaction with N. perurans (and any viruses or bacteria it may harbor) in AGD in naïve Atlantic salmon.
Gene network analysis is a system wide approach that is able to connect differential gene expression data from all tissues with their regulatory TFs. The gene co-expression network analyses in this study indicate that the TF znfOZF-like regulatory network is the most active in AGD, with five of the top 10 (106573105, 106578124, 106586175, 100136550, 106608858) upregulated DCGs contributing to this gene network (Tables 2 and 3). The znfOZF protein is a Kruppel type of nuclear zinc finger protein whose dysregulation has been implicated in tumor genesis in cancer (71, 122) suggesting its role in AGD is to prevent invasion and proliferation of N. perurans. We identified 26 genes in the Herpes simplex virus 1 pathway from the znfOZF-like regulatory network with a total of 18 genes coding for the anti-viral proteins (ZAP). Furthermore, all of the key regulatory TFs identified in the top 20 DCGs were also ZAP proteins. Each of the ZAP proteins is a zinc finger transcription factor (Supplementary Table 14). The Herpes simplex virus 1 pathway was also consistently identified in our local and systemic KEGG visualizations for the host differential expression data, suggesting a viral-like response may be initiated in response to parasite invasion in AGD. In contrast bacterial defense mechanisms are indicated by genes mapping to the KEGG Ap-1 and PI3K-Akt signaling pathways (hsa04151) in Salmonella infection (sasa05132). These pathways are also closely connected to the wnt signaling pathway, which we propose is a candidate host pathway for manipulation by N. perurans during AGD pathogenesis in our model for host-parasite interaction.
Other host defense mechanisms identified in the znfOZF-like regulatory network are cytolysin RTX-A-like and mucin-2-like isoform X2, which may protect the host cells against bacterial pore-forming toxins and production of an insoluble gel mucus barrier to protect cells against invasion, respectively (93, 94). Of the remaining TFs identified through the network analysis, one was upregulated in the spleen and two were downregulated in the head kidney. TOX high mobility group box family member 3-like (TOX3) in the spleen has been shown to inhibit the proliferation and migration of cancer cells by transcriptional regulation of SNAI1 and SNAI2 to prevent disruption of the epithelial cell layer (72). However, in the spleen TOX3 is a regulator of innate lymphoid cells in particular the pathogen primed CD8+ cytotoxic T lymphocytes suggesting its role in AGD is activation of the immune system (73). In contrast, the HMG box-containing protein 1-like is a regulator of key pathways including the PI3K/Akt and wnt pathways which are downregulated in cancer and hijacked by parasites in the host-parasite interaction during invasion as previously discussed (85, 86). The final key regulatory TF identified in the head kidney is protein disulfide isomerase family A, member 4 (PDIA4). According to KEGG pathway visualization PDIA4 is responsible for the loading of antigenic peptides into MHCI molecules in the endoplasmic reticulum for release at the site of the infection (Sasa04141).
The majority of DCGs associated with AGD exhibited similar activities to those induced by the TFs, acting as mediators of the immune response, or roles in cellular proliferation and invasion. Eight of the 12 gene networks were more expressed in gill tissue with two upregulated and three downregulated in response to AGD. Three have been implicated in invasion of tissue in cancer including Aurora kinase B-like, rho guanine nucleotide exchange factor 25-like, protein NDNF-like inhibited, however, their differential expression suggests a preventative role in this instance (69, 70, 88). Two other genes, svep1 sushi, von Willebrand factor type A EGF and pentraxin domain containing 1, and ceramide synthase 2-like are reported to suppress tumors (78, 83, 123). While muscle specific ring finger protein 1-like, an E3 ubiquitin-protein ligase has been reported to be involved in proteasome-mediated degradation (74) which may be involved in degrading proteins originating from N. perurans in host defense. The inflammatory response to AGD is potentially dampened by the downregulated inflammatory mediated gene networks connected to the gene CKLF-like MARVEL transmembrane domain-containing protein 4 which exhibits similar expression in lung cancer, and matrix Gla protein which has been implicated in chronic inflammatory diseases as well as lung cancer (80–82). Two of the identified regulatory gene networks are associated with tissue repair including insulin-like growth factor 2b and catalase-like (77). The latter is involved in the oxidative stress response and also regulates hydrogen peroxide metabolism (76). Interestingly, Trypanosoma cruzi has been reported to modulate the oxidative stress (FoxO signaling pathway) response to aid invasion of the host (79). Sustained production of reactive oxygen species (ROS) due to the T. cruzi infection coupled with an insufficient antioxidant response leads to long-term oxidative stress in the host. Furthermore, the integrin signaling-associated gene, T. cruzi cyr61, which is a homologue of cef10 in S. salar, is downregulated in the late stages of T. cruzi infection after infiltration of host tissue (79).
Gene Candidates in the Systemic Host Response to Late Stage AGD
The head kidney and spleen, while both lymphoid organs, have different roles in immunity (124). The approximate 9-fold increase in DEGs in the head kidney and spleen is consistent with the head kidneys dual roles in detoxification and the immune response (125). The head kidney responds indirectly to the external parasite through alteration of osmoregulation and excretion at the gill (125) in response to compromised gill physiology from the AGD lesion (5). It also has a direct immune response through the differentiation of leucocytes for general immunity (124). This reduced number of DEGs in the spleen compared to the head kidney reflects the spleens primary role to filter and maintain the red blood cell population in circulation (125), as there is not profuse bleeding at lesions on the gill.
The local host defense at the gill is supported by upregulation of genes coding for cathelicidin antimicrobial peptides and lysozyme C II in the spleen. These gene candidates have previously been identified in Atlantic salmon in response to AGD (14, 126, 127), as well as to the bacterial infection Yersinia ruckerii and the sea louse, Lepeophtheirus salmonis (14, 128, 129). In Atlantic salmon the C-type lectins have been reported to be upregulated during the first five days post infection in AGD (13, 29). While more recently, a glycan and lectin microarray study identified mannobiose and N-acetylgalactosamine as candidates for gill epithelium binding of N. perurans (130). Recombinant mannose-binding proteins have previously been reported to bind N. perurans and produce antibodies in the host (131). While N-acetylgalactosamine has also been reported to be involved in mucosal adherence for the pathogenic amoeba, Entamoeba histolytica (132). Gene candidates for the downregulation of immune BPs with GTPase activity, include the ras-GEF domain-containing family member 1B-B-like and T-cell lymphoma invasion and metastasis 2 which are induced in macrophages in response to Toll-like receptor agonists (133) and participate in proliferation and invasion of tumors (96), respectively. In the spleen the MAPK pathway molecular functions were downregulated suggesting the potential for prevention of resolution of inflammation and lesion suppression in response to N. perurans invasion (134).
The systemic innate immune response in the head kidney was down-regulated through the sphingosine 1-phosphate receptor 4-like receptor (135) and GPCR C3a anaphylatoxin chemotactic receptor-like which causes migration of eosinophils, mast cells and macrophages to the site of injury as part of the complement cascade (136). Several genes coding for GPCRs were downregulated in the systemic response in both the head kidney and spleen including probable CD97 antigen-like GPCR, G-protein coupled receptor 132, 2-oxoglutarate receptor 1-like, G-protein coupled receptor 124-like (wnt pathway), chemokine-like receptor 1, C-C chemokine receptor type 9 and C-X-C chemokine receptor type 4. The P2Y purinoceptor 13-like gene was also downregulated, which has been reported to regulate lung endothelial barrier integrity in humans (137). Multiple MF GO terms associated with DNA binding including binding for transcription and receptor activity in the spleen indicate an active innate inflammatory immune response through modulation of macrophages and hence cytokine activity and histamine release. While increased epithelial proliferation ostensibly at the gill surface is characterized by negative regulation of the delta-like protein 4 and protein jagged-2-like in the Notch signaling pathway in the spleen (138).
Interestingly while the systemic innate immune response shows activation through increased gene expression, the inflammatory response is decreased distal to the AGD lesion at the gill in an inflammatory gradient progressing away from the site of the lesion as indicated by the downregulation of a 5-hydroxytryptamine receptor 7-like, a serotonin receptor on the membranes of immune cells including dendrites, monocytes, macrophages, microglia and lymphocytes (139). Four extracellular and plasma membrane GO terms were enriched among downregulated genes associated with toxin sequestration, humoral immunity through B-cell homeostasis, and tumor invasion. These genes were saxiphilin-like, TNF receptor superfamily member 13B and matrix metalloproteinase-28-like respectively (140–142). Matrix metalloproteinases are also responsible for extracellular matrix degradation involved in tumor invasion and progression (143). A total of 18 metalloproteinases were identified in the enriched GO term ‘metallopeptidase activity’ which included stromelysin-3-like (144). Mucus production and wound healing metabolic processes are also upregulated in the gill, particularly transferases associated with glycoprotein production, a key component of mucus (145). Differential gene expression of mucins in AGD has previously been reported in proximity to gill lesions in AGD in Atlantic salmon (16). Active suppression of parasite invasion is also indicated through upregulation of G-protein coupled receptors including integrin-β-3 (146), and regulatory genes such as G-protein signaling 21-like (147) in the GPCR signaling pathway. Integrins mediate cellular adhesion processes and active immune cells and upregulation of these genes has been reported in AGD-affected Atlantic salmon (148). Collectively, the differential gene expression, KEGG pathway and network analyses show significant gene expression impacts on the host immune response and identified prospective key genes/regulators and pathways that may be modulated by N. perurans to promote AGD pathogenesis.
Parasite Invasion and Contact Dependent Cysteine Proteases
Pathogen tissue invasion may be facilitated through the secretion of proteins including toxins, adhesion molecules, effector proteins and enzymes (149) with pathogenicity inferred by both contact and non-contact mechanisms (150). Figure 5 suggests a model for host parasite interaction based on the differential expression and network analyses in this study where only contact mechanisms appear to be in effect. This includes proteins to facilitate the destruction of the mucus barrier, apoptosis of epithelial cells and re-arrangement of the host actin cytoskeleton to gain access to host tissue. In protozoan and sporozoite parasites cysteine proteases, such as cathepsin B identified in our study are critical for contact dependent host invasion through proteolysis of host extracellular matrix proteins and degradation of host immune proteins, including the amoeba genus Naegleria and E. histolytica (98, 150, 151). Cathepsin B expression in the parasite Giardia duodenalis, is induced by soluble host factors that deter attachment (105). However instead of deterring attachment the interaction results in upregulation of expression of Giardia duodenalis virulence factors, ultimately enabling parasite attachment in human gastroenteritis (105). The interaction between the host and N. perurans cathepsin B needs to be explored further, as it may provide an opportunity to develop alternative treatments for AGD. For example, in Trypanosoma and Toxoplasma sp., cathepsin B has been reported to be essential for survival due to cathepsins role in digesting tissue to provide essential nutrients from the host such as iron (152). Cathepsin B protease inhibition through RNAi, vaccination and chemotherapeutics are widely studied for the prevention of cathepsin B-mediated trematode tissue invasion (fluke) in livestock (153) and may provide a starting point for exploring novel AGD therapeutics. Likewise, tumor invasion in many human cancers is facilitated through over expression of cathepsin (154). A wide variety of cathepsin inhibitors have been developed and investigated in human cancers, however evidence suggests their action may be to potentiate other therapeutics in cancer treatment by facilitating membrane passage to induce apoptosis and necrosis of invading cells (154). Targeted therapeutic strategies that facilitate passage into N. perurans such as cathepsin mediated cell entry may assist in reducing the toxicity of alternative novel treatments to the host by reducing dosage.
Parasite Gene Candidates for Remodeling of Gill Epithelia During Attachment
Histopathology of gill tissue affected by AGD has been reported to show re-modeling of host tissue (5, 7, 155). This is characterized by hyperplasia of the lamellar epithelium, fusion of adjacent lamellae and formation of interlamellar lacunae or vesicles, hyperplasia, and hypertrophy of mucus cells. This results in increased production of gill mucus and proliferation of the lamellar epithelium (5). In the proposed host-parasite interaction model for AGD in Atlantic salmon, we suggest that re-modeling of the gill tissue may be similar to that observed for E. histolytica in amoebiasis in humans (hsa05146). In amoebiasis, E. histolytica facilitates actin rearrangement of the cytoskeleton through binding with host fibronectin (FN) which stimulates protein kinase C (PKC) or protein kinase A (PKA) pathways for rearrangement of actin in the amoebae cytoskeleton in preparation for attachment (156, 157). PKC and several actin and regulatory component genes were isolated in N. perurans (Supplementary Tables 12 and 14), however none were differentially expressed. Similarly, only some of the components necessary for lesion formation (vinculin, α-actinin, tropomyosin, and myosin I) in E. histolytica are present in our dataset (157).
Prokumamolisin was identified in our de novo assembly of unmapped reads through the transcript annotated as prokumamolisin activation domain containing protein which is an orthologue from Acanthamoeba castellanii (112). It is a sedolisin or serine-carboxyl peptidase, which is a proteolytic enzyme that has been reported to be secreted into extracellular space by the pathogenic amoebae Acanthamoeba castellanii, but not by non-pathogenic amoebae (112). Interestingly, prokumamolisin is the inactive form of kumamolisin that acts as a collagenase (158). This suggests prokumamolisin may function in re-modeling of the host epithelia during attachment in N. perurans. Entamoeba histolytica adhesion, migration and phagocytosis is controlled through engagement with a p-21 activated kinase (159) which was also upregulated in our dataset (NpPAK). Interestingly the host gene co-expression network may provide the answer to lesion formation through Atlantic salmon expression of a C-reactive protein, hemagglutinin/amebocyte aggregation factor-like (HAAF). In Limulus polyphemus HAAF induces aggregation of amoebae and binding through interaction with limunectin and an endotoxin binding protein (90). These then induce blood coagulation, complement and adhesion processes by the host to repair the tissue at the affected site.
Gene Candidates for Modulation of the Parasites Local Environment
In our study, we identified candidate genes that suggest the amoeba may defend itself against ROS production thereby circumventing this host defense mechanism. On attachment ubiquitination pathways and ROS detoxification are upregulated in Giardia duodenalis (105). Similar expression patterns are also observed in our study with downregulation of cathepsin B in the lesion and the upregulation of hybrid cluster protein (NpHCP). This host evasion mechanism has also been observed in Entamoeba histolytica during invasion of tissue in amoebiasis in humans (160). In parasites, proteosomes have roles in virulence, toxin production, differentiation, cell cycle, proliferation, and encystation during the invasion of host tissue (104, 161). The ubiquitin proteasome system is represented by the upregulated gene candidate Fbox domain containing protein. Other mechanisms by which amoeba may control their environment include altering the local pH by acidification of the gill tissue creating a safe niche for proliferation. This occurs through the release of protons by expression of a vacuolar H+-ATPase (NpV-ATPase) which has been observed in Leishmania promastigotes and Plasmodium falciparum parasites (102, 103). In AGD, acidification resulting from proton release is likely to benefit N. perurans by reducing ammonium toxicity as it is released from the gills (162) and may also assist in mucus degradation.
Parasite Pathogenicity Gene Candidates
Another form of contact-dependent invasion is observed in Naegleria sp., Acanthamoeba sp. and Entamoeba sp. where cytoplasmic extensions form a phagocytic amoebastome or food cup which enables pathogen actin genes to interact with host heat shock proteins in a pathogenic manner (150, 163). In N. perurans, an alternative theory to the amoebastome for interacting with the host heat shock protein (HPSB1) is the surface protein expressed in this study, prohibitin (NpPHB). This protein has been shown to bind to heat shock protein 70 (Hsp70) in the host-pathogen interaction between Leishmania donovani and macrophages in the human disease leishmaniasis (113). Prohibitin has been reported to increase infectivity by increasing protein surface density and binding to Hsp70 (134, 135). In parasites (and other protozoans) a switch in morphology is also reported to increase pathogenicity (164). To date, N. perurans has been reported to revert to pseudocyst morphology under stress, otherwise it maintains normal trophozoite morphology in in vitro cultured conditions (23). However, conformational changes in morphology may remain a plausible component of virulence for N. perurans in AGD but this theory needs to be explored further.
Gene Candidates for Virulence and Pathogenicity
AprA is a metalloprotease gene known to be a virulence factor in Pseudomonas entomophila however it is also able to suppress induction of host antimicrobial peptides (111, 165). The closest species blast match for NpAprA was Capsaspora owczarzaki, a unicellular amoeba protist rather than a bacterial species suggesting this is a candidate for N. perurans protection against the host immune response, and not a contaminant from the commensal bacteria. AprA expression is regulated by the GacS/GacA two component virulence system of P. entomophila and other species (111). This system is responsible for the production of multiple virulence factors including pore-forming toxins. In N. perurans, NpAprA expression may be regulated by the analogous two component PhoP/Q system (PhoPQ) which was detected in our dataset and is a master regulator of virulence in Salmonella enterica and Pectobacterium versatile (106). In Salmonella enterica, PhoPQ was acquired through lateral gene transfer (106). Both lateral and horizontal gene transfer have been reported between bacteria and eukaryotes including the amoeba, Acanthamoeba castellanii, as well as endosymbionts and their hosts (166–168). In N. perurans, the closest species blast match for NpPhoPQ was the amoebozoan cellular slime mold Cavenderia fasciculata (syn. Dictyostelium fasciculatum) providing support for the hypothesis that the PhoP/Q system is a candidate for master regulator of virulence in N. perurans. Several other gene candidates were identified with potential roles as pathogenic factors including the MADS-box transcription factor (NpMADS) with similarity to the amoebozoan cellular slime mold Tieghemostelium lacteum (syn. Dictyostelium lacteum). These TFs are highly conserved in eukaryotes and have a diversity of functional roles including pathogenicity (107, 108). The downregulation of NpMADS late in AGD in our study is consistent with this proposed gene candidate roles in AGD when virulent protein secretion may not be required. An endosomal sorting complex gene candidate, vacuolar sorting protein SNF7 (NpSNF7) was identified in the de novo lesion with blast match similarity to Acanthamoeba castellanii (114). This gene candidate is reported to export O-polysaccharides without secretion tags from the cytosol (114). The final virulence factor candidate is from the Rab family of GTPases, GTPase activator protein for Ras-like GTPase, which has been reported to regulate amoebapore virulence factors (160). These form pores in host cells to trigger cell death and degradation of the extracellular matrix. This facilitates host invasion whilst evading the subsequent immune response (160).
Do Commensal Bacteria Have a Role in AGD Pathogenicity?
The commensal bacteria associated with the lesion add another layer of complexity to the host-pathogen interaction in AGD (33). Bacterial taxa are ubiquitous throughout the marine aquaculture environment, including on the gill surface of salmon where the assemblage present plays a key role in health and physiological functions (169). As both the gill surface and N. perurans contain a rich community of microbiota, it is pertinent to examine the bacterial species, which may be implicated in AGD progression at the host-pathogen interface. Bacterial reads in the current study were opportunistically assessed for descriptive purposes, as bacterial mRNA is not polyadenylated at the 3’ end of the transcript, a typically necessary feature for successful reverse transcription for sequencing (47). Some consideration should be made to any possible bias of this sequencing chemistry, including potential for increased incidences of species with lower GC content.
A parallel in vivo investigation of the bacterial community present on AGD-affected gill tissue of salmon from the same cohort as the current study determined that a known pathogenic bacterium Tenacibaculum dicentrarchi was heavily abundant within the AGD lesioned tissue (33). Other field and lab-based AGD studies have demonstrated the significant presence of specific taxa, including the genus Psychroserpens (170), Winogradskyella and Staphylococcus (171) implicated in AGD development or linked to the presence of AGD. The Genus Legionella is a known pathogenic clade in mammalian systems. Most prominently the species Legionella feeleii has been shown to harbor intracellularly within amoebae cells (172). It is possible this species may have been present in concert with the AGD causing N. perurans trophozoites, but this requires further substantiation. Pseudomonas stutzeri is a functional denitrifying taxon present in both aquatic and terrestrial soils and sediments (173), and was a dominant taxon taken from the bacterial read data. The functional purpose of denitrifying taxa suits colonization on the surface of teleost fish gills due to the constant secretion of nitrogenous waste products over the gill epithelium (174). The higher proportion of annotated genes based on TPM for P. stutzeri may alternatively reflect microbial dysbiosis at the gill lesion (Supplementary Table 13).
A proportion of identified bacterium were of marine origin, meaning that there is relevance in assessing the species sequenced and their function. The majority of bacterial taxa identified in the gill lesion N. perurans transcriptome were also in the unmapped gill transcriptome. However, several new taxa were identified after addition of naïve and distal to the lesion biopsy raw reads to the lesion transcriptome assembly. Most notably, Nocardia jejuensis only found in AGD-affected fish which is a novel Nocardia species from South Korea with 97.4% similarity to Nocardia salmonicida (175). Nocardia infections are generally thought to be opportunistic, however other species from the Genus including Nocardia seriolae, the causal agent of nocardiosis is known to cause gill nodules together with other clinical disease symptoms in yellowtail and amberjack aquaculture (176). In Atlantic salmon Nocardia asteroides has been isolated from skin lesions in tank reared animals (177). The Flavobacteriales Order contains multiple marine pathogenic bacteria including Fleixbacter and Tenacibaculum (178). The relatively even proportion of this bacterium identified across AGD-affected and naïve fish suggests that these species are part of the normal microbiota and not related specifically to the occurrence of AGD in this study. It is important to note that Genus level identifications may be more relevant than species in the unmapped transcriptome due to the low coverage of contigs greater than 500 bp.
The detection of Halomonas halocynthiae is also pertinent as an exclusive marine inhabiting species, which has been isolated from the gill surface of other marine organisms (179). This genus has also been identified in concert with in vitro cultured N. perurans in amplicon sequencing studies (26). Similarly, Adhaeribacter aquaticus is another species typically associated with aquatic biofilm environments and is likely present in this dataset as a commensal taxon. Another species commonly associated with aquatic bioreactor communities is Leptospirillum ferriphilum, which was identified on the gill (180). This iron-oxidizing species may be a functional commensal taxon utilizing available resources within the cellular respiration and oxygen transfer mechanisms of the gill. The assessment of bacterial reads in the current study has identified several taxa which may play functional roles either on the gill surface, or during AGD pathogenesis. Whether the severity of N. perurans as the primary pathogen responsible for AGD is altered by specific commensal bacteria remains to be determined as future studies examine the interaction between the host, the bacteria and N. perurans in AGD.
In our study we characterized the molecular events 21 days post-AGD induction in the host and parasite to identify gene candidates to propose a model for host-parasite interaction during pathogenesis of AGD in Atlantic salmon. In N. perurans multiple gene candidates are upregulated that are indicators for virulence and regulation of virulence in other species. A comparison of the gene networks between diseases states (AGD v non-AGD) in host tissue indicates that genes expressed in the wnt-pathway are negatively impacted during AGD as observed in disease states in other species where tissue invasion is a factor. Genes candidates associated with evasion of host defense mechanisms and formation of the mucoid lesion were upregulated. This study also presents the first transcriptomic study of the causative agent for AGD, N. perurans providing a new resource to contribute to better understanding of the parasite. These gene candidates, networks and pathways may be explored further to develop new hypotheses for future AGD research, and may lead to new therapeutics for AGD in the future.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166760.
Ethics Statement
The animal study was reviewed and approved by Commonwealth Scientific and Industrial Research Organisation Queensland Animal Ethics Committee under applications 2018-09, 2017-35, and 2017-36.
Author Contributions
AM, NB, and JW contributed to conception and design of the study. JS organized and collected the samples. NB, PL, and AM performed the data acquisition. AM performed the host bioinformatic/systems biology analyses. NB performed all pathway visualization/interpretation and the parasite bioinformatic analyses. NB interpreted the data and wrote the first draft of the manuscript. NB, JW, JS, and AM wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was funded by CSIRO Agriculture and Food Science Investment Project 252 (OD-206222).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the support of Richard Taylor as the principle investigator of the animal ethics application under which these samples were collected as part of a larger project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.672700/full#supplementary-material
Supplementary Table 1 | Illumina sequencing statistics. Data yield, number of obtained raw paired-end reads, number of mapped fragments for each RNA-Seq library [Excel table].
Supplementary Table 2 | Top differentially expressed genes (-log10FDR > 5) in the gill. (A) Gill lesion (L) of an AGD-affected fish compared to a gill biopsy in a naïve (C) Atlantic salmon. (B) Gill lesion biopsy compared to a biopsy distal (D) to the lesion of an AGDaffected fish. (C) Gill biopsy distal to the lesion of an AGD-affected fish compared to a gill biopsy in a naïve Atlantic salmon. [Excel table]
Supplementary Table 3 | Enriched gene ontology (GO) terms for the local response to AGD at the gills of Atlantic salmon at the lesion, distal to the lesion and in naïve fish. [Excel table]
Supplementary Table 4 | Top differentially expressed genes (-log10FDR > 5) in the head kidney of an AGD-affected fish (A) compared to a naïve (C) Atlantic salmon. [Excel table].
Supplementary Table 5 | Top differentially expressed genes (-log10FDR > 5) in the spleen of an AGD-affected fish (A) compared to a naïve (C) Atlantic salmon. [Excel table].
Supplementary Table 6 | Enriched gene ontology (fold change > 2; P < 0.05) for the systemic response to amoebic gill disease in the head kidney of Atlantic salmon affected by AGD in contrast to naïve fish. [Excel table].
Supplementary Table 7 | Enriched gene ontology (fold change > 2; P < 0.05) for the systemic response to amoebic gill disease in the spleen of Atlantic salmon affected by AGD in contrast to naïve fish. [Excel table].
Supplementary Table 8 | Key transcription factors (TFs) and their regulatory impact factor (RIF) score in amoebic gill disease in Atlantic salmon. [Excel table].
Supplementary Table 9 | Differential expression of host transcription factors (TFs) (fold change > 2; P < 0.05) in response to amoebic gill disease pathology in Atlantic salmon. [Excel table].
Supplementary Table 10 | Differential expression of genes and transcription factors (TFs) (fold change > 2; P < 0.05) in the zinc finger OZF-like gene network. [Excel table].
Supplementary Table 11 | Differential expression of Neoparamoeba perurans genes (fold change < 2; P < 0.05) in the lesion of an amoebic gill disease affected fish in comparison to cultured trophozoites. [Excel table].
Supplementary Table 12 | Neoparamoeba perurans genes identified in the de novo assembly of the unmapped Atlantic salmon amoebic gill disease lesion data. [Excel table].
Supplementary Table 13 | Bacterial species and their genes identified in the unmapped Atlantic salmon amoebic gill disease data. [Excel table].
Supplementary Table 14 | Genes identified in the host-parasite response to amoebic gill disease in Atlantic salmon (host) and Neoparamoeba perurans (pathogen). The model is shown in Figure 4. [Excel table].
Supplementary Figure S1 | Differential gene expression in the gills in response to amoebic gill disease. (A) Level of agreement among the biological replicates of a naïve (n=4; red, CG), AGD-affected tissue distal to the AGD lesion (n=4; green, DG), and a lesion on a gill affected by AGD (n=3; blue, LG). The heat map shows the hierarchically clustered Spearmancorrelations resulting from comparing normalized expression for all gill samples against each other. Sample clustering indicates the consistency between the biological replicates. (B–D) Volcano plots of differentially expressed genes in the gill, highlighting genes with a minimum 2-fold change in expression and false discovery rate greater than five comparing AGD lesion to a naïve fish (B); the lesion compared to the region distal to the lesion of an AGD-affected fish (C); and the region distal to the lesion of an AGD-affected fish compared to a naïve fish (D). [word doc]
Supplementary Figure S2 | Differential gene expression in the head kidney and spleen in response to amoebic gill disease. Level of agreement among the head kidney (HK) (A) and spleen (SP) (B) biological replicates of a naïve (n=4; C; red) and an AGD-affected Atlantic salmon (n=4; A; blue). The heat map shows the hierarchically clustered Spearman correlations resulting from comparing normalized expression for HK or SP samples against each other. Sample clustering indicates consistency between biological replicates. Volcano plots of differentially expressed genes in the HK (C) and SP (D), highlighting genes with a minimum 2-fold change in expression and false discovery rate greater than five comparing AGD lesion to a naïve fish. [word doc]
Supplementary Figure S3 | Enriched gene ontology (GO) terms (hypergeometric test, Bonferroni-adjusted P < 0.05) among the upregulated (A) and downregulated (B) differentially expressed genes in the biopsy distal to the lesion. The majority of the upregulated enriched terms are related to host and immune response and downregulated to organism developmental processes. [word doc]
Supplementary Figure S4 | Differential gene expression in Neoparamoeba perurans in the lesion of amoebic gill disease in Atlantic salmon compared to cultured N. perurans trophozoites. (A) Heat map shows hierarchical clustering of differentially expressed genes (rows) with differential expression among replicates from a gill biopsy of the lesion (LES) and cultured trophozoites (CUL) from N. perurans. Expression values are log2-transformed and median-centered by gene. (B) Volcano plot of differentially expressed genes in the gill biopsy lesion, highlighting genes with a minimum 2-fold change in expression (P < 0.05) compared to in vitro cultured N. perurans trophozoites. [word doc]
References
1. Shinn AP, Pratoomyot J, Bron JE, Paladini G, Brooker EE, Brooker AJ. Economic Costs of Protistan and Metazoan Parasites to Global Mariculture. Parasitology (2015) 142:196–270. doi: 10.1017/S0031182014001437
2. Munday BL, Zilberg D, Findlay V. Gill Disease of Marine Fish Caused by Infection With Neoparamoeba Pemaquidensis. J Fish Dis (2001) 24:497–507. doi: 10.1046/j.1365-2761.2001.00329.x
3. Powell MD, Reynolds P, Kristensen T. Freshwater Treatment of Amoebic Gill Disease and Sea-Lice in Seawater Salmon Production: Considerations of Water Chemistry and Fish Welfare in Norway. Aquaculture (2015) 448:18–28. doi: 10.1016/j.aquaculture.2015.05.027
4. Kube PD, Taylor RS, Elliott NG. Genetic Variation in Parasite Resistance of Atlantic Salmon to Amoebic Gill Disease Over Multiple Infections. Aquaculture (2012) 364–365:165–72. doi: 10.1016/j.aquaculture.2012.08.026
5. Adams MB, Nowak BF. Amoebic Gill Disease: Sequential Pathology in Cultured Atlantic Salmon, Salmo Salar L. J Fish Dis (2003) 26:601–14. doi: 10.1046/j.1365-2761.2003.00496.x
6. Chang YC, Hamlin-Wright H, Monaghan S, Herath T, Baily J, del Pozo J, et al. Changes in Distribution, Morphology and Ultrastructure of Chloride Cell in Atlantic Salmon During an AGD Infection. J Fish Dis (2019) 42:1433–46. doi: 10.1111/jfd.13073
7. Adams MB, Nowak BF. Distribution and Structure of Lesions in the Gills of Atlantic Salmon, Salmo Salar L., Affected With Amoebic Gill Disease. J Fish Dis (2001) 24:535–42. doi: 10.1046/j.1365-2761.2001.00330.x
8. Pennacchi Y, Leef MJ, Crosbie PBB, Nowak BF, Bridle AR. Evidence of Immune and Inflammatory Processes in the Gills of AGD-affected Atlantic Salmon, Salmo Salar L. Fish Shellfish Immunol (2014) 36:563–70. doi: 10.1016/j.fsi.2013.12.013
9. Bridle AR, Morrison RN, Nowak BF. The Expression of Immune-Regulatory Genes in Rainbow Trout, Oncorhynchus Mykiss, During Amoebic Gill Disease (AGD). Fish Shellfish Immunol (2006) 20:346–64. doi: 10.1016/j.fsi.2005.05.014
10. Young ND, Cooper GA, Nowak BF, Koop BF, Morrison RN. Coordinated Down-Regulation of the Antigen Processing Machinery in the Gills of Amoebic Gill Disease-Affected Atlantic Salmon (Salmo Salar L.). Mol Immunol (2008) 45:2581–97. doi: 10.1016/j.molimm.2007.12.023
11. Morrison RN, Zou J, Secombes CJ, Scapigliati G, Adams MB, Nowak BF. Molecular Cloning and Expression Analysis of Tumour Necrosis Factor-α in Amoebic Gill Disease (AGD)-Affected Atlantic Salmon (Salmo Salar L.). Fish Shellfish Immunol (2007) 23:1015–31. doi: 10.1016/j.fsi.2007.04.003
12. Bridle AR, Morrison RN, Cupit Cunningham PM, Nowak BF. Quantitation of Immune Response Gene Expression and Cellular Localisation of Interleukin-1β mRNA in Atlantic Salmon, Salmo Salar L., Affected by Amoebic Gill Disease (AGD). Vet Immunol Immunopathol (2006) 114:121–34. doi: 10.1016/j.vetimm.2006.08.002
13. Morrison RN, Cooper GA, Koop BF, Rise ML, Bridle AR, Adams MB, et al. Transcriptome Profiling the Gills of Amoebic Gill Disease (AGD)-Affected Atlantic Salmon (Salmo Salar L.): A Role for Tumor Suppressor p53 in AGD Pathogenesis? Physiol Genomics (2006) 26:15–34. doi: 10.1152/physiolgenomics.00320.2005
14. Wynne JW, O’Sullivan MG, Cook MT, Stone G, Nowak BF, Lovell DR, et al. Transcriptome Analyses of Amoebic Gill Disease-Affected Atlantic Salmon (Salmo Salar) Tissues Reveal Localized Host Gene Suppression. Mar Biotechnol (2008) 10:388–403. doi: 10.1007/s10126-007-9075-4
15. Benedicenti O, Collins C, Wang T, McCarthy U, Secombes CJ. Which Th Pathway is Involved During Late Stage Amoebic Gill Disease? Fish Shellfish Immunol (2015) 46:417–25. doi: 10.1016/j.fsi.2015.07.002
16. Marcos-López M, Calduch-Giner JA, Mirimin L, MacCarthy E, Rodger HD, O’Connor I, et al. Gene Expression Analysis of Atlantic Salmon Gills Reveals Mucin 5 and Interleukin 4/13 as Key Molecules During Amoebic Gill Disease. Sci Rep (2018) 8:1–15. doi: 10.1038/s41598-018-32019-8
17. Bridle AR, Davenport DL, Crosbie PBB, Polinski M, Nowak BF. Neoparamoeba Perurans Loses Virulence During Clonal Culture. Int J Parasitol (2015) 45:575–8. doi: 10.1016/j.ijpara.2015.04.005
18. Dhufaigh KN, Botwright N, Talbot A, O’connor I, Maccarthy E, Slattery O. Comparative Proteomic Proling of Newly Acquired, Virulent and Attenuated Neoparamoeba Perurans Proteins Associated With Amoebic Gill Disease. Sci Rep (2021) 11:683. doi: 10.21203/rs.3.rs-108031/v1
19. Dhufaigh K, MacCarthy E, Slattery O. Biochemical and Proteomic Characterisation of Secretory Proteins From the Protozoan Parasite Paramoeba Perurans Revealed by an In Vitro Model. In: 19th International Conference on Diseases of Fish and Shellfish. Porto, Portugal: European Association of Fish Pathologists (2019). p. 324.
20. Hooper P, Bateman K, Santos E, Paley R, Cano I. Characterising Association of Neoparamoeba Perurans to RTGILL-W1 Cells in an In Vitro Model for Amoebic Gill Disease (AGD). In: 19th International Conference on Diseases of Fish and Shellfish. Porto, Portugal: European Association of Fish Pathologists (2019). p. 389.
21. Botwright NA, Greenfield P, McWilliam S, Wynne JW. The Hidden Genome of a Marine Amoeba Neoparamoeba Perurans. In: . 19th International Conference on Diseases of Fish and Shellfish. Porto, Portugal: European Association of Fish Pathologists (2019). p. 23.
22. Blindheim S, Anderson L, Trosse C, Karlsbakk E, Nylund A. Phenotypic Characterization of Paramoeba Perurans Clones Obtained From Different Populations of Atlantic Salmon and Ballan Wrasse. In: 19th International Conference on Diseases of Fish and Shellfish. Porto, Portugal: European Association of Fish Pathologists (2019). p. 24.
23. Lima PC, Taylor RS, Cook M. Pseudocyst Formation in the Marine Parasitic Amoeba Neoparamoeba Perurans: A Short-Term Survival Strategy to Abrupt Salinity Variation. J Fish Dis (2017) 40:1109–13. doi: 10.1111/jfd.12588
24. Sibbald SJ, Cenci U, Colp M, Eglit Y, Kelly CJO, John M. Diversity and Evolution of Paramoeba Spp. and Their Kinetoplastid Endosymbionts. J Eukaryot Microbiol (2017) 64:598–607. doi: 10.1111/jeu.12394
25. Tanifuji G, Cenci U, Moog D, Dean S, Nakayama T, David V, et al. Genome Sequencing Reveals Metabolic and Cellular Interdependence in an Amoeba-Kinetoplastid Symbiosis. Sci Rep (2017) 7:1–13. doi: 10.1038/s41598-017-11866-x
26. MacPhail DPC, Koppenstein R, Maciver SK, Paley R, Longshaw M, Henriquez FL. Vibrio Species are Predominantly Intracellular Within Cultures of Neoparamoeba Perurans, Causative Agent of Amoebic Gill Disease (Agd). Aquaculture (2021) 532:736083. doi: 10.1016/j.aquaculture.2020.736083
27. Boamah DK, Zhou G, Ensminger AW, O’Connor TJ. From Many Hosts, One Accidental Pathogen: The Diverse Protozoan Hosts of Legionella. Front Cell Infect Microbiol (2017) 7:477. doi: 10.3389/fcimb.2017.00477
28. Guimaraes AJ, Gomes KX, Cortines JR, Peralta JM, Peralta RHS. Acanthamoeba Spp. As a Universal Host for Pathogenic Microorganisms: One Bridge From Environment to Host Virulence. Microbiol Res (2016) 193:30–8. doi: 10.1016/j.micres.2016.08.001
29. Cano I, Taylor NG, Bayley A, Gunning S, McCullough R, Bateman K, et al. In Vitro Gill Cell Monolayer Successfully Reproduces In Vivo Atlantic Salmon Host Responses to Neoparamoeba Perurans Infection. Fish Shellfish Immunol (2019) 86:287–300. doi: 10.1016/j.fsi.2018.11.029
30. Westermann AJ, Gorski SA, Vogel J. Dual RNA-seq of Pathogen and Host. Nat Rev Microbiol (2012) 10:618–30. doi: 10.1038/nrmicro2852
31. National Health and Medical Research Council. Australian Code for the Care and Use of Animals for Scientific Purposes. 8th ed. Canberra: National Health and Medical Research Council (2013). doi: 10.1111/j.1751-0813.1998.tb10161.x
32. Wynne JW, Stratford C, Slinger J, Samsing F, Rigby M, McCulloch R, et al. The Interaction Between Temperature and Dose on the Efficacy and Biochemical Response of Atlantic Salmon to Hydrogen Peroxide Treatment for Amoebic Gill Disease. J Fish Dis (2020) 43:39–48. doi: 10.1111/jfd.13110
33. Slinger J, Adams MB, Wynne JW. Bacteriomic Profiling of Branchial Lesions Induced by Neoparamoeba Perurans Challenge Reveals Commensal Dysbiosis and an Association With Tenacibaculum Dicentrarchi in AGD-affected Atlantic Salmon (Salmo Salar L.). Microorganisms (2020) 8:1189. doi: 10.3390/microorganisms8081189
34. Taylor RS, Muller WJ, Cook MT, Kube PD, Elliott NG. Gill Observations in Atlantic Salmon (Salmo Salar, L.) During Repeated Amoebic Gill Disease (AGD) Field Exposure and Survival Challenge. Aquaculture (2009) 290:1–8. doi: 10.1016/j.aquaculture.2009.01.030
35. English CJ, Tyml T, Botwright NA, Barnes AC, Wynne JW, Lima PC, et al. A Diversity of Amoebae Colonise the Gills of Farmed Atlantic Salmon (Salmo Salar) With Amoebic Gill Disease (AGD). Eur J Protistol (2019) 67:27–45. doi: 10.1016/j.ejop.2018.10.003
36. Mohamed AR, Andrade N, Moya A, Chan CX, Negri AP, Bourne DG, et al. Dual RNA-sequencing Analyses of a Coral and its Native Symbiont During the Establishment of Symbiosis. Mol Ecol (2020) 29:3921–37. doi: 10.1111/mec.15612
37. Andrews S. FastQC A Quality Control tool for High Throughput Sequence Data. (2010). Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
38. Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, et al. The Atlantic Salmon Genome Provides Insights Into Rediploidization. Nature (2016) 533:200–5. doi: 10.1038/nature17164
39. Kim D, Langmead B, Salzberg SL. Hisat: A Fast Spliced Aligner With Low Memory Requirements. Nat Methods (2015) 12:357–60. doi: 10.1038/nmeth.3317
40. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map Format and Samtools. Bioinformatics (2009) 25:2078–9. doi: 10.1093/bioinformatics/btp352
41. Anders S, Pyl PT, Huber W. Htseq-a Python Framework to Work With High-Throughput Sequencing Data. Bioinformatics (2015) 31:166–9. doi: 10.1093/bioinformatics/btu638
42. Quinlan AR, Hall IM. Bedtools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics (2010) 26:841–2. doi: 10.1093/bioinformatics/btq033
43. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De Novo Transcript Sequence Reconstruction From RNA-seq Using the Trinity Platform for Reference Generation and Analysis. Nat Protoc (2013) 8:1494–512. doi: 10.1038/nprot.2013.084
44. Buchfink B, Xie C, Huson DH. Fast and Sensitive Protein Alignment Using DIAMOND. Nat Methods (2015) 12:59–60. doi: 10.1038/nmeth.3176
45. Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, et al. Ncbi Taxonomy: A Comprehensive Update on Curation, Resources and Tools. Database (Oxford) (2020) 2020:baaa062. doi: 10.1093/database/baaa062
46. Li B, Dewey CN. Rsem: Accurate Transcript Quantification From RNA-Seq Data With or Without a Reference Genome. BMC Bioinf (2011) 12:323. doi: 10.1186/1471-2105-12-323
47. Sangiovanni M, Granata I, Thind AS, Guarracino MR. From Trash to Treasure: Detecting Unexpected Contamination in Unmapped NGS Data. BMC Bioinf (2019) 20:168. doi: 10.1186/s12859-019-2684-x
48. Robinson MD, McCarthy DJ, Smyth GK. Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
49. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing (2020). Available at: https://www.r-project.org/.
50. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Hypthesis Testing. J R Stat Soc Ser B (1995) 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
51. Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omi A J Integr Biol (2012) 16:284–7. doi: 10.1089/omi.2011.0118
52. Reverter A, Hudson NJ, Nagaraj SH, Pérez-Enciso M, Dalrymple BP. Regulatory Impact Factors: Unraveling the Transcriptional Regulation of Complex Traits From Expression Data. Bioinformatics (2010) 26:896–904. doi: 10.1093/bioinformatics/btq051
53. Mohamed AR, King H, Evans B, Reverter A, Kijas JW. Multi-Tissue Transcriptome Profiling of North American Derived Atlantic Salmon. Front Genet (2018) 9:369. doi: 10.3389/fgene.2018.00369
54. Reverter A, Chan EKF. Combining Partial Correlation and an Information Theory Approach to the Reversed Engineering of Gene Co-Expression Networks. Bioinformatics (2008) 24:2491–7. doi: 10.1093/bioinformatics/btn482
55. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res (2003) 13:2498–504. doi: 10.1101/gr.1239303
56. Kanehisa M, Goto S. Kegg: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res (2000) 28:27–30. doi: 10.1093/nar/28.1.27
57. Kanehisa M. Toward Understanding the Origin and Evolution of Cellular Organisms. Protein Sci (2019) 28:1947–51. doi: 10.1002/pro.3715
58. Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. Kegg: Integrating Viruses and Cellular Organisms. Nucleic Acids Res (2021) 49:D545–51. doi: 10.1093/nar/gkaa970
59. Cosin-Roger J, Ortiz-Masià MD, Barrachina MD. Macrophages as an Emerging Source of Wnt Ligands: Relevance in Mucosal Integrity. Front Immunol (2019) 10:2297. doi: 10.3389/fimmu.2019.02297
60. Jati S, Sarraf TR, Naskar D, Sen M. Wnt Signaling: Pathogen Incursion and Immune Defense. Front Immunol (2019) 10:2551. doi: 10.3389/fimmu.2019.02551
61. Voorneveld PW, Kodach LL, Jacobs RJ, Van Noesel CJMM, Peppelenbosch MP, Korkmaz KS, et al. The BMP Pathway Either Enhances or Inhibits the Wnt Pathway Depending on the SMAD4 and p53 Status in CRC. Br J Cancer (2015) 112:122–30. doi: 10.1038/bjc.2014.560
62. LaMonte GM, Orjuela-Sanchez P, Calla J, Wang LT, Li S, Swann J, et al. Dual RNA-seq Identifies Human Mucosal Immunity Protein Mucin-13 as a Hallmark of Plasmodium Exoerythrocytic Infection. Nat Commun (2019) 10:488. doi: 10.1038/s41467-019-08349-0
63. De Paiva Daibert RM, De Biagi Junior CAO, De Oliveira Vieira F, Da Silva MVGB, Hottz ED, Pinheiro MBM, et al. Lipopolysaccharide Triggers Different Transcriptional Signatures in Taurine and Indicine Cattle Macrophages: Reactive Oxygen Species and Potential Outcomes to the Development of Immune Response to Infections. PloS One (2020) 15:e0241861. doi: 10.1371/journal.pone.0241861
64. Hendrickx DM, Glaab E. Comparative Transcriptome Analysis of Parkinson’s Disease and Hutchinson-Gilford Progeria Syndrome Reveals Shared Susceptible Cellular Network Processes. BMC Med Genomics (2020) 13:114. doi: 10.1186/s12920-020-00761-6
65. Deng SP, Zhu L, Huang DS. Mining the Bladder Cancer-Associated Genes by an Integrated Strategy for the Construction and Analysis of Differential Co-Expression Networks. BMC Genomics (2015) 16:S4. doi: 10.1186/1471-2164-16-S3-S4
66. Ingham AB, Osborne SA, Menzies M, Briscoe S, Chen W, Kongsuwan K, et al. RNF14 is a Regulator of Mitochondrial and Immune Function in Muscle. BMC Syst Biol (2014) 8:10. doi: 10.1186/1752-0509-8-10
67. Fortes MRS, Reverter A, Nagaraj SH, Zhang Y, Jonsson NN, Barris W, et al. A Single Nucleotide Polymorphism-Derived Regulatory Gene Network Underlying Puberty in 2 Tropical Breeds of Beef Cattle. J Anim Sci (2011) 89:1669–83. doi: 10.2527/jas.2010-3681
68. Mohamed AR, Naval-Sanchez M, Menzies M, Evans B, King H, Reverter A, et al. Integrated Transcriptome, DNA Methylome and Chromatin State Accessibility Landscapes Reveal Regulators of Atlantic Salmon Maturation. bioRxiv (2020), 2020.08.28.272286. doi: 10.1101/2020.08.28.272286
69. Sharma A, Madhunapantula SV, Gowda R, Berg A, Neves RI, Robertson GP. Identification of Aurora Kinase B and wee1-like Protein Kinase as Downstream Targets of V600EB-RAF in Melanoma. Am J Pathol (2013) 182:1151–62. doi: 10.1016/j.ajpath.2012.12.019
70. Xia L, Li S, Liu Y, Huang Y, Ni B, Wan L, et al. NDNF Inhibits the Migration and Invasion of Human Renal Cancer Cells Through Epithelial-Mesenchymal Transition. Oncol Lett (2019) 17:2969–75. doi: 10.3892/ol.2019.9937
71. Li W, Zhang H, Min P, Zhu J, Xu D, Jiang W, et al. Downregulated miRNA-1269a Variant (rs73239138) Decreases the Susceptibility to Gastric Cancer Via Targeting ZNF70. Oncol Lett (2017) 14:6345–54. doi: 10.3892/ol.2017.7091
72. Jiang B, Chen W, Qin H, Diao W, Li B, Cao W, et al. TOX3 Inhibits Cancer Cell Migration and Invasion Via Transcriptional Regulation of SNAI1 and SNAI2 in Clear Cell Renal Cell Carcinoma. Cancer Lett (2019) 449:76–86. doi: 10.1016/j.canlet.2019.02.020
73. Page N, Klimek B, De Roo M, Steinbach K, Soldati H, Lemeille S, et al. Expression of the DNA-binding Factor TOX Promotes the Encephalitogenic Potential of Microbe-Induced Autoreactive CD8+ T Cells. Immunity (2018) 48:937–50. doi: 10.1016/j.immuni.2018.04.005
74. Napolitano LM, Jaffray EG, Hay RT, Meroni G. Functional Interactions Between Ubiquitin E2 Enzymes and TRIM Proteins. Biochem J (2011) 434:309–19. doi: 10.1042/BJ20101487
75. Gazon H, Barbeau B, Mesnard JM, Peloponese JM. Hijacking of the AP-1 Signaling Pathway During Development of ATL. Front Microbiol (2018) 8:2686. doi: 10.3389/fmicb.2017.02686
76. Góth L, Rass P, Páy A. Catalase Enzyme Mutations and Their Association With Diseases. Mol Diagn (2004) 8:141–9. doi: 10.2165/00066982-200408030-00001
77. Chablais F. Jaźwińska A. IGF Signaling Between Blastema and Wound Epidermis is Required for Fin Regeneration. Development (2010) 137:871–9. doi: 10.1242/dev.043885
78. Chen L, Liu D, Yi X, Qi L, Tian X, Sun B, et al. The Novel miR-1269b-regulated Protein SVEP1 Induces Hepatocellular Carcinoma Proliferation and Metastasis Likely Through the PI3K/Akt Pathway. Cell Death Dis (2020) 11:320. doi: 10.1038/s41419-020-2535-8
79. Manque PA, Probst C, Pereira MCS, Rampazzo RCP, Ozaki LS, Pavoni DP, et al. Trypanosoma Cruzi Infection Induces a Global Host Cell Response in Cardiomyocytes. Infect Immun (2011) 79:1855–62. doi: 10.1128/IAI.00643-10
80. Li M, Luo F, Tian X, Yin S, Zhou L, Zheng S. Chemokine-Like Factor-Like MARVEL Transmembrane Domain-Containing Family in Hepatocellular Carcinoma: Latest Advances. Front Oncol (2020) 10:595973. doi: 10.3389/fonc.2020.595973
81. Viegas CSB, Costa RM, Santos L, Videira PA, Silva Z, Araújo N, et al. Gla-Rich Protein Function as an Anti-Inflammatory Agent in Monocytes/Macrophages: Implications for Calcification-Related Chronic Inflammatory Diseases. PloS One (2017) 12:e0177829. doi: 10.1371/journal.pone.0177829
82. Bianchi F, Hu J, Pelosi G, Cirincione R, Ferguson M, Ratcliffe C, et al. Lung Cancers Detected by Screening With Spiral Computed Tomography Have a Malignant Phenotype When Analyzed by cDNA Microarray. Clin Cancer Res (2004) 10:6023–8. doi: 10.1158/1078-0432.CCR-04-0619
83. Zhang Q, Wang J, Yan W, dan Wang D, Yang S, Zhou S, et al. Clinical and Pathological Significance of Homo Sapiens Ceramide Synthase 2 (CerS-2) in Diverse Human Cancers. Biosci Rep (2019) 39:BSR20181743. doi: 10.1042/BSR20181743
84. Ochoa D, Hercules A, Carmona M, Suveges D, Gonzalez-Uriarte A, Malangone C, et al. Open Targets Platform: Supporting Systematic Drug-Target Identification and Prioritisation. Nucleic Acids Res (2021) 49:D1302–10. doi: 10.1093/nar/gkaa1027
85. Rogan MR, Patterson LL, Wang JY, McBride JW. Bacterial Manipulation of Wnt Signaling: A Host-Pathogen Tug-of-Wnt. Front Immunol (2019) 10:2390. doi: 10.3389/fimmu.2019.02390
86. Bollaert E, de Rocca Serra A, Demoulin JB. The HMG Box Transcription Factor HBP1: A Cell Cycle Inhibitor At the Crossroads of Cancer Signaling Pathways. Cell Mol Life Sci (2019) 76:1529–39. doi: 10.1007/s00018-019-03012-9
87. Simmen FA, Xiao R, Velarde MC, Nicholson RD, Bowman MT, Fujii-Kuriyama Y, et al. Dysregulation of Intestinal Crypt Cell Proliferation and Villus Cell Migration in Mice Lacking Krüppel-like Factor 9. Am J Physiol Gastrointest Liver Physiol (2007) 292:G1757–69. doi: 10.1152/ajpgi.00013.2007
88. Cook DR, Rossman KL, Der CJ. Rho Guanine Nucleotide Exchange Factors: Regulators of Rho Gtpase Activity in Development and Disease. Oncogene (2014) 33:4021–35. doi: 10.1038/onc.2013.362
89. Wang Z, Zhang H, Cheng Q. Pdia4: The Basic Characteristics, Functions and its Potential Connection With Cancer. BioMed Pharmacother (2020) 122:109688. doi: 10.1016/j.biopha.2019.109688
90. Fujii N, Minetti CASA, Nakhasi HL, Chen SW, Barbehenn E, Nunes PH, et al. Isolation, cDNA Cloning, and Characterization of an 18-kDa Hemagglutinin and Amebocyte Aggregation Factor From Limulus Polyphemus. J Biol Chem (1992) 267:22452–9. doi: 10.1016/s0021-9258(18)41693-6
91. Gazi U, Martinez-Pomares L. Influence of the Mannose Receptor in Host Immune Responses. Immunobiology (2009) 214:554–61. doi: 10.1016/j.imbio.2008.11.004
92. Pinkert S, Röger C, Kurreck J, Bergelson JM, Fechner H. The Coxsackievirus and Aenovirus Receptor: Glycosylation and the Extracellular D2 Domain are Not Required for Coxsackievirus B3 Infection. J Virol (2016) 90:5601–10. doi: 10.1128/jvi.00315-16
93. Sveen LR, Grammes FT, Ytteborg E, Takle H, Jørgensen SM. Genome-Wide Analysis of Atlantic Salmon (Salmo Salar) Mucin Genes and Their Role as Biomarkers. PloS One (2017) 12:e0189103. doi: 10.1371/journal.pone.0189103
94. Brito C, Cabanes D, Sarmento Mesquita F, Sousa S. Mechanisms Protecting Host Cells Against Bacterial Pore-Forming Toxins. Cell Mol Life Sci (2019) 76:1319–39. doi: 10.1007/s00018-018-2992-8
95. Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, et al. The Immunoreceptor TIGIT Regulates Antitumor and Antiviral CD8+ T Cell Effector Function. Cancer Cell (2014) 26:923–37. doi: 10.1016/j.ccell.2014.10.018
96. Chen JS, Su IJ, Leu YW, Young KC, Sun HS. Expression of T-cell Lymphoma Invasion and Metastasis 2 (TIAM2) Promotes Proliferation and Invasion of Liver Cancer. Int J Cancer (2012) 130:1302–13. doi: 10.1002/ijc.26117
97. Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. Ankyrin-Repeat Containing Proteins of Microbes: A Conserved Structure With Functional Diversity. Trends Microbiol (2010) 18:132–9. doi: 10.1016/j.tim.2009.11.004
98. Thaí TL, Kang JM, Lê HG, Lee J, Yoo WG, Shin HJ, et al. Fowlerstefin, a Cysteine Protease Inhibitor of Naegleria Fowleri, Induces Inflammatory Responses in BV-2 Microglial Cells In Vitro. Parasites Vectors (2020) 13:41. doi: 10.1186/s13071-020-3909-6
99. Mohanam S, Jasti SL, Kondraganti SR, Chandrasekar N, Lakka SS, Kin Y, et al. Down-Regulation of Cathepsin B Expression Impairs the Invasive and Tumorigenic Potential of Human Glioblastoma Cells. Oncogene (2001) 20:3665–73. doi: 10.1038/sj.onc.1204480
100. Canova MJ, Molle V. Bacterial Serine/Threonine Protein Kinases in Host-Pathogen Interactions. J Biol Chem (2014) 289:9473–9. doi: 10.1074/jbc.R113.529917
101. Pinter N, Hach CA, Hampel M, Rekhter D, Zienkiewicz K, Feussner I, et al. Signal Peptide Peptidase Activity Connects the Unfolded Protein Response to Plant Defense Suppression by Ustilago Maydis. PloS Pathog (2019) 15:e1007734. doi: 10.1371/journal.ppat.1007734
102. Moradin N, Descoteaux A. Leishmania Promastigotes: Building a Safe Niche Within Macrophages. Front Cell Infect Microbiol (2012) 2:121. doi: 10.3389/fcimb.2012.00121
103. Hayashi M, Yamada H, Mitamura T, Horii T, Yamamoto A, Moriyama Y. Vacuolar H+-ATPase Localized in Plasma Membranes of Malaria Parasite Cells, Plasmodium Falciparum, is Involved in Regional Acidification of Parasitized Erythrocytes. J Biol Chem (2000) 275:P34353–34358. doi: 10.1074/jbc.M003323200
104. Jöhnk B, Bayram Ö, Abelmann A, Heinekamp T, Mattern DJ, Brakhage AA, et al. SCF Ubiquitin Ligase F-box Protein Fbx15 Controls Nuclear Co-Repressor Localization, Stress Response and Virulence of the Human Pathogen Aspergillus Fumigatus. PloS Pathog (2016) 12:e1005899. doi: 10.1371/journal.ppat.1005899
105. Emery SJ, Mirzaei M, Vuong D, Pascovici D, Chick JM, Lacey E, et al. Induction of Virulence Factors in Giardia Duodenalis Independent of Host Attachment. Sci Rep (2016) 6:20765. doi: 10.1038/srep20765
106. García-Calderón CB, Casadesús J, Ramos-Morales F. Rcs and PhoPQ Regulatory Overlap in the Control of Salmonella Enterica Virulence. J Bacteriol (2007) 189:6635–44. doi: 10.1128/JB.00640-07
107. Zhang Z, Li H, Qin G, He C, Li B, Tian S. The MADS-Box Transcription Factor Bcmads1 is Required for Growth, Sclerotia Production and Pathogenicity of. Botrytis Cinerea Sci Rep (2016) 6:33901. doi: 10.1038/srep33901
108. Zhao X, Yang X, Lu Z, Wang H, He Z, Zhou G, et al. MADS-Box Transcription Factor Mcm1 Controls Cell Cycle, Fungal Development, Cell Integrity and Virulence in the Filamentous Insect Pathogenic Fungus Beauveria Bassiana. Environ Microbiol (2019) 21:3392–416. doi: 10.1111/1462-2920.14629
109. Pentland DR, Piper-Brown E, Mühlschlegel FA, Gourlay CW. Ras Signalling in Pathogenic Yeasts. Microb Cell (2018) 5:63–73. doi: 10.15698/mic2018.02.612
110. Mondola P, Damiano S, Sasso A, Santillo M. The Cu, Zn Superoxide Dismutase: Not Only a Dismutase Enzyme. Front Physiol (2016) 7:594. doi: 10.3389/fphys.2016.00594
111. Keebaugh ES, Schlenke TA. Insights From Natural Host-Parasite Interactions: The Drosophila Model. Dev Comp Immunol (2014) 42:111–23. doi: 10.1016/j.dci.2013.06.001
112. Moon EK, Choi HS, Park SM, Kong HH, Quan FS. Comparison of Proteins Secreted Into Extracellular Space of Pathogenic and non-Pathogenic Acanthamoeba Castellanii. Korean J Parasitol (2018) 56:553–8. doi: 10.3347/kjp.2018.56.6.553
113. Jain R, Ghoshal A, Mandal C, Shaha C. Leishmania Cell Surface Prohibitin: Role in Host-Parasite Interaction. Cell Microbiol (2010) 12:432–52. doi: 10.1111/j.1462-5822.2009.01406.x
114. Rodrigo RM, Crestani J, Kmetzsch L, De S, Araujo G, Frases S, et al. The Vacuolar-Sorting Protein Snf7 is Required for Export of Virulence Determinants in Members of the Cryptococcus Neoformans Complex. Sci Rep (2014) 4:6198. doi: 10.1038/srep06198
115. Miller DA, Suen G, Clements KD, Angert ER. The Genomic Basis for the Evolution of a Novel Form of Cellular Reproduction in the Bacterium Epulopiscium. BMC Genomics (2012) 13:265. doi: 10.1186/1471-2164-13-265
116. Marcos-López M, Rodger HD, Marcos-lópez M, Rodger HD, Marcos-López M, Rodger HD. Amoebic Gill Disease and Host Response in Atlantic Salmon ( Salmo Salar L .): A Review. Parasite Immunol (2020) 42:e12766. doi: 10.1111/pim.12766
117. Nusse R, Clevers H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell (2017) 169:985–99. doi: 10.1016/j.cell.2017.05.016
118. De A. Wnt/Ca 2 Signaling Pathway: A Brief Overview. Acta Biochim Biophys Sin (Shanghai) (2011) 43:745–56. doi: 10.1093/abbs/gmr079
119. Sun J, Hobert ME, Rao AS, Neish AS, Madara JL. Bacterial Activation of β-Catenin Signaling in Human Epithelia. Am J Physiol Liver Physiol (2004) 287:G220–7. doi: 10.1152/ajpgi.00498.2003
120. Robledo D, Hamilton A, Gutiérrez AP, Bron JE, Houston RD. Characterising the Mechanisms Underlying Genetic Resistance to Amoebic Gill Disease in Atlantic Salmon Using RNA Sequencing. BMC Genomics (2020) 21:271. doi: 10.1186/s12864-020-6694-x
121. Valdenegro-Vega VA, Polinski M, Bridle A, Crosbie P, Leef M, Nowak BF. Effects of Single and Repeated Infections With Neoparamoeba Perurans on Antibody Levels and Immune Gene Expression in Atlantic Salmon (Salmo Salar). Fish Shellfish Immunol (2015) 42:522–9. doi: 10.1016/j.fsi.2014.11.031
122. Ferbus D, Bovin C, Validire P, Goubin G. The Zinc Finger Protein OZF (ZNF146) in Overexpressed in Colorectal Cancer. J Pathol (2003) 200:177–82. doi: 10.1002/path.1337
123. Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in Cancer: Mechanisms and Advances in Clinical Trials. Mol Cancer (2019) 18:26. doi: 10.1186/s12943-019-0954-x
124. Secombes CJ, Ellis AE. The Immunology of Teleosts. In: Roberts RJ, editor. Fish Pathology. Oxford, UK: Blackwell Publishing Ltd. (2012). p. 144–66. doi: 10.1002/9781118222942.ch4
125. Roberts RJ, Ellis AE. “The Anatomy and Physiology of Teleosts”. In: Roberts RJ, editor. Fish Pathology. Oxford, UK: Blackwell Publishing Ltd. (2012). p. 17–61. doi: 10.1002/9781118222942.ch2
126. Marcos-López M, Espinosa Ruiz C, Rodger HD, O’Connor I, MacCarthy E, Esteban MÁ. Local and Systemic Humoral Immune Response in Farmed Atlantic Salmon (Salmo Salar L.) Under a Natural Amoebic Gill Disease Outbreak. Fish Shellfish Immunol (2017) 66:207–16. doi: 10.1016/j.fsi.2017.05.029
127. Gross KA, Powell MD, Butler R, Morrison RN, Nowak BF. Changes in the Innate Immune Response of Atlantic Salmon, Salmo Salar L., Exposed to Experimental Infection With Neoparamoeba Sp. J Fish Dis (2005) 28:293–9. doi: 10.1111/j.1365-2761.2005.00633.x
128. Núñez-Acuña G, Gallardo-Escárate C, Fields DM, Shema S, Skiftesvik AB, Ormazábal I, et al. The Atlantic Salmon (Salmo Salar) Antimicrobial Peptide Cathelicidin-2 is a Molecular Host-Associated Cue for the Salmon Louse (Lepeophtheirus Salmonis). Sci Rep (2018) 8:13738. doi: 10.1038/s41598-018-31885-6
129. Bridle A, Nosworthy E, Polinski M, Nowak B. Evidence of an Antimicrobial-Immunomodulatory Role of Atlantic Salmon Cathelicidins During Infection With Yersinia Ruckeri. PloS One (2011) 6:e23417. doi: 10.1371/journal.pone.0023417
130. Lima PC, Hartley-Tassell L, Cooper O, Wynne JW. Searching for the Sweet Spot of Amoebic Gill Disease of Farmed Atlantic Salmon: The Potential Role of Glycan-Lectin Interactions in the Adhesion of Neoparamoeba Perurans. Int J Parasitol (2021) 51:545–57. doi: 10.1016/j.ijpara.2020.11.009
131. Valdenegro-Vega VA, Crosbie PBB, Cook MT, Vincent BN, Nowak BF. Administration of Recombinant Attachment Protein (r22C03) of Neoparamoeba Perurans Induces Humoral Immune Response Against the Parasite in Atlantic Salmon (Salmo Salar). Fish Shellfish Immunol (2014) 38:294–302. doi: 10.1016/j.fsi.2014.03.034
132. Petri WA, Haque R, Mann BJ. The Bittersweet Interface of Parasite and Host: Lectin-carbohydrate Interactions During Human Invasion by the Parasite Entamoeba Histolytica. Annu Rev Microbiol (2002) 56:39–64. doi: 10.1146/annurev.micro.56.012302.160959
133. Andrade WA, Silva AM, Alves VS, Salgado APC, Melo MB, Andrade HM, et al. Early Endosome Localization and Activity of RasGEF1b, a Toll-Like Receptor-Inducible Ras Guanine-Nucleotide Exchange Factor. Genes Immun (2010) 11:447–57. doi: 10.1038/gene.2009.107
134. Hoppstädter J, Ammit AJ. Role of Dual-Specificity Phosphatase 1 in Glucocorticoid-Driven Antiinflammatory Responses. Front Immunol (2019) 10:1446. doi: 10.3389/fimmu.2019.01446
135. Olesch C, Ringel C, Brüne B, Weigert A. Beyond Immune Cell Migration: The Emerging Role of the sphingosine-1-phosphate Receptor S1PR4 as a Modulator of Innate Immune Cell Activation. Mediators Inflammation (2017) 2017:6059203. doi: 10.1155/2017/6059203
136. Muenstermann M, Strobel L, Klos A, Wetsel RA, Woodruff TM, Köhl J, et al. Distinct Roles of the Anaphylatoxin Receptors C3aR, C5aR1 and C5aR2 in Experimental Meningococcal Infections. Virulence (2019) 10:677–94. doi: 10.1080/21505594.2019.1640035
137. Zemskov E, Lucas R, Verin AD, Umapathy NS. P2Y Receptors as Regulators of Lung Endothelial Barrier Integrity. J Cardiovasc Dis Res (2011) 2:14–22. doi: 10.4103/0975-3583.78582
138. Wang Z, Li Y, Kong D, H. Sarkar F. The Role of Notch Signaling Pathway in Epithelial-Mesenchymal Transition (EMT) During Development and Tumor Aggressiveness. Curr Drug Targets (2010) 11:745–51. doi: 10.2174/138945010791170860
139. Quintero-Villegas A, Valdés-Ferrer SI. Role of 5-HT7 Receptors in the Immune System in Health and Disease. Mol Med (2020) 26:2. doi: 10.1186/s10020-019-0126-x
140. Zhang J, Pan Q, Yan W, Wang Y, He X, Zhao Z. Overexpression of MMP21 and MMP28 is Associated With Gastric Cancer Progression and Poor Prognosis. Oncol Lett (2018) 15:7776–82. doi: 10.3892/ol.2018.8328
141. Yen TJ, Lolicato M, Thomas-Tran R, Du Bois J, Minor DL. Structure of the Saxiphilin:Saxitoxin (STX) Complex Reveals a Convergent Molecular Recognition Strategy for Paralytic Toxins. Sci Adv (2019) 5:eaax2650. doi: 10.1126/sciadv.aax2650
142. Sazzini M, Zuntini R, Farjadian S, Quinti I, Ricci G, Romeo G, et al. An Evolutionary Approach to the Medical Implications of the Tumor Necrosis Factor Receptor Superfamily Member 13B (TNFRSF13B) Gene. Genes Immun (2009) 10:566–78. doi: 10.1038/gene.2009.43
143. Lu P, Takai K, Weaver VM, Werb Z. Extracellular Matrix Degradation and Remodeling in Development and Disease. Cold Spring Harb Perspect Biol (2011) 3:a005058. doi: 10.1101/cshperspect.a005058
144. Nakopoulou L, Panayotopoulou EG, Giannopoulou I, Alexandrou P, Katsarou S, Athanassiadou P, et al. Stromelysin-3 Protein Expression in Invasive Breast Cancer: Relation to Proliferation, Cell Survival and Patients’ Outcome. Mod Pathol (2002) 15:1154–61. doi: 10.1097/01.MP.0000037317.84782.CD
145. Benktander J, Padra JT, Maynard B, Birchenough G, Botwright NA, McCulloch R, et al. Gill Mucus and Gill Mucin O-glycosylation in Healthy and Amoebic Gill Disease-Affected Atlantic Salmon. Microorganisms (2020) 8:1871. doi: 10.3390/microorganisms8121871
146. Jakus Z, Fodor S, Abram CL, Lowell CA, Mócsai A. Immunoreceptor-Like Signaling by β2 and β3 Integrins. Trends Cell Biol (2007) 17:493–501. doi: 10.1016/j.tcb.2007.09.001
147. Kim Y, Ghil S. Regulators of G-protein Signaling, RGS2 and RGS4, Inhibit Protease-Activated Receptor 4-Mediated Signaling by Forming a Complex With the Receptor and Gα in Live Cells. Cell Commun Signal (2020) 18:86. doi: 10.1186/s12964-020-00552-7
148. Boison SA, Gjerde B, Hillestad B, Makvandi-Nejad S, Moghadam HK. Genomic and Transcriptomic Analysis of Amoebic Gill Disease Resistance in Atlantic Salmon (Salmo Salar L.). Front Genet (2019) 10:68. doi: 10.3389/fgene.2019.00068
149. Sen R, Nayak L, De RK. A Review on Host–Pathogen Interactions: Classification and Prediction. Eur J Clin Microbiol Infect Dis (2016) 35:1581–99. doi: 10.1007/s10096-016-2716-7
150. Lee J, Kim JH, Sohn HJ, Yang HJ, Na BK, Chwae YJ, et al. Novel Cathepsin B and Cathepsin B-like Cysteine Protease of Naegleria Fowleri Excretory-Secretory Proteins and Their Biochemical Properties. Parasitol Res (2014) 113:2765–76. doi: 10.1007/s00436-014-3936-3
151. Keene WE, Petitt MG, Allen S, McKerrow JH. The Major Neutral Proteinase of Entamoeba Histolytica. J Exp Med (1986) 163:536–49. doi: 10.1084/jem.163.3.536
152. Dou Z, Carruthers VB. Cathepsin Proteases in Toxoplasma Gondii. Adv Exp Med Biol (2011) 712:49–61. doi: 10.1007/978-1-4419-8414-2_4
153. Smooker PM, Jayaraj R, Pike RN, Spithill TW. Cathepsin B Proteases of Flukes: The Key to Facilitating Parasite Control? Trends Parasitol (2010) 26:506–14. doi: 10.1016/j.pt.2010.06.001
154. Ruan H, Hao S, Young P, Zhang H. Targeting Cathepsin B for Cancer Therapies. Horizons Cancer Res (2015) 56:23–40.
155. Adams MB, Ellard K, Nowak BF. Gross Pathology and its Relationship With Histopathology of Amoebic Gill Disease (AGD) in Farmed Atlantic Salmon, Salmo Salar L. J Fish Dis (2004) 27:151–61. doi: 10.1111/j.1365-2761.2004.00526.x
156. Santiago A, Carbajal ME, Benítez-King G, Meza I. Entamoeba Histolytica: PKC Transduction Pathway Activation in the Trophozoite-Fibronectin Interaction. Exp Parasitol (1994) 79:436–44. doi: 10.1006/expr.1994.1105
157. Espinosa-Cantellano M, Martínez-Palomo A. Pathogenesis of Intestinal Amebiasis: From Molecules to Disease. Clin Microbiol Rev (2000) 13:318–31. doi: 10.1128/cmr.13.2.318
158. Wlodawer A, Li M, Gustchina A, Oyama H, Dunn BM, Oda K. Structural and Enzymatic Properties of the Sedolisin Family of Serine-Carboxyl Peptidases. Acta Biochim Pol (2003) 50:81–102. doi: 10.18388/abp.2003_3716
159. Labruyère E, Zimmer C, Galy V, Olivo-Marin JC, Guillén N. EhPAK, a Member of the p21-activated Kinase Family, is Involved in the Control of Entamoeba Histolytica Migration and Phagocytosis. J Cell Sci (2003) 116:61–71. doi: 10.1242/jcs.00190
160. Vicente JB, Ehrenkaufer GM, Saraiva LM, Teixeira M, Singh U. Entamoeba Histolytica Modulates a Complex Repertoire of Novel Genes in Response to Oxidative and Nitrosative Stresses: Implications for Amebic Pathogenesis. Cell Microbiol (2009) 11:51–69. doi: 10.1111/j.1462-5822.2008.01236.x
161. Muñoz C, San Francisco J, Gutiérrez B, González J. Role of the Ubiquitin-Proteasome Systems in the Biology and Virulence of Protozoan Parasites. BioMed Res Int (2015) 2015:141526. doi: 10.1155/2015/141526
162. Randall D, Tsui T. Tribute to R. G. Boutilier: Acid–Base Transfer Across Fish Gills. J Exp Biol (2006) 209:1179–84. doi: 10.1242/jeb.02100
163. Sohn HJ, Kim JH, Shin MH, Song KJ, Shin HJ. The Nf-actin Gene is an Important Factor for Food-Cup Formation and Cytotoxicity of Pathogenic Naegleria Fowleri. Parasitol Res (2010) 106:917–24. doi: 10.1007/s00436-010-1760-y
164. Sinha A, Sarkar S. Ubiquitin-Proteasome System - A Target to Control Pathogenic Protozoa. In: Mendez-Vilas A, editor. Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education Vol. 1. Badajoz: Formate Research Centerx (2013). p. 764–73.
165. Liehl P, Blight M, Vodovar N, Boccard F, Lemaitre B. Prevalence of Local Immune Response Against Oral Infection in a Drosophila/Pseudomonas Infection Model. PloS Pathog (2006) 2:e56. doi: 10.1371/journal.ppat.0020056
166. Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, et al. Genome of Acanthamoeba Castellanii Highlights Extensive Lateral Gene Transfer and Early Evolution of Tyrosine Kinase Signaling. Genome Biol (2013) 14:R11. doi: 10.1186/gb-2013-14-2-r11
167. Moliner C, Raoult D, Fournier PE. Evidence of Horizontal Gene Transfer Between Amoeba and Bacteria. Clin Microbiol Infect (2009) 15:178–80. doi: 10.1111/j.1469-0691.2008.02216.x
168. Thomas V, Greub G. Amoeba/Amoebal Symbiont Genetic Transfers: Lessons From Giant Virus Neighbours. Intervirology (2010) 53:254–67. doi: 10.1159/000312910
169. Llewellyn MS, Boutin S, Hoseinifar SH, Derome N. Teleost Microbiomes: The State of the Art in Their Characterization, Manipulation and Importance in Aquaculture and Fisheries. Front Microbiol (2014) 5:207. doi: 10.3389/fmicb.2014.00207
170. Bowman JP, Nowak B. Salmonid Gill Bacteria and Their Relationship to Amoebic Gill Disease. J Fish Dis (2004) 27:483–92. doi: 10.1111/j.1365-2761.2004.00569.x
171. Embar-Gopinath S, Butler R, Nowak B. Influence of Salmonid Gill Bacteria on Development and Severity of Amoebic Gill Disease. Dis Aquat Organ (2005) 67:55–60. doi: 10.3354/dao067055
172. Wang C, Chuai X, Liang M. Legionella Feeleii: Pneumonia or Pontiac Fever? Bacterial Virulence Traits and Host Immune Response. Med Microbiol Immunol (2019) 208:25–32. doi: 10.1007/s00430-018-0571-0
173. Lalucat J, Bennasar A, Bosch R, García-Valdés E, Palleroni NJ. Biology of Pseudomonas Stutzeri. Microbiol Mol Biol Rev (2006) 70:510–47. doi: 10.1128/mmbr.00047-05
174. Evans DH, Piermarini PM, Choe KP. The Multifunctional Fish Gill: Dominant Site of Gas Exchange, Osmoregulation, Acid-Base Regulation, and Excretion of Nitrogenous Waste. Physiol Rev (2005) 85:97–177. doi: 10.1152/physrev.00050.2003
175. Lee SD. Nocardia Jejuensis Sp. Nov., a Novel Actinomycete Isolated From a Natural Cave on Jeju Island, Republic of Korea. Int J Syst Evol Microbiol (2006) 56:559–62. doi: 10.1099/ijs.0.63866-0
176. Labrie L, Ng J, Tan Z, Komar C, Ho E, Grisez L. Nocardial Infections in Fish: An Emerging Problem in Both Freshwater and Marine Aquaculture Systems in Asia. In: Diseases in Asian Aquaculture Vi. Manila: Diseases in Asian Aquaculture VI. Fish Health Section, Asian Fisheries Society (2008).
177. Bransden MP, Carson J, Munday BL, Handlinger JH, Carter CG, Nowak BF. Nocardiosis in Tank-Reared Atlantic Salmon, Salmo Salar L. J Fish Dis (2000) 23:83–5. doi: 10.1046/j.1365-2761.2000.00201.x
178. McBride MJ. The Family Flavobacteriaceae. In: Rosenberg E, editor. The Prokaryotes – Other Major Lineages of Bacteria and the Archaea. Springer-Verlag Berlin Heidelberg (2014). doi: 10.1007/978-3-642-38954-2_130
179. Romanenko LA, Schumann P, Rohde M, Mikhailov VV, Stackebrandt E. Halomonas Halocynthiae Sp. Nov., Isolated From the Marine Ascidian Halocynthia Aurantium. Int J Syst Evol Microbiol (2002) 52:1767–72. doi: 10.1099/00207713-52-5-1767
Keywords: Atlantic salmon (Salmo salar), Neoparamoeba perurans, amoebic gill disease, host-parasite interaction, immunity, aquaculture, dual RNA-Seq
Citation: Botwright NA, Mohamed AR, Slinger J, Lima PC and Wynne JW (2021) Host-Parasite Interaction of Atlantic salmon (Salmo salar) and the Ectoparasite Neoparamoeba perurans in Amoebic Gill Disease. Front. Immunol. 12:672700. doi: 10.3389/fimmu.2021.672700
Received: 26 February 2021; Accepted: 05 May 2021;
Published: 31 May 2021.
Edited by:
Samuel A. M. Martin, University of Aberdeen, United KingdomReviewed by:
Diego Robledo, University of Edinburgh, United KingdomMaria K. Dahle, Norwegian Veterinary Institute (NVI), Norway
Copyright © 2021 Botwright, Mohamed, Slinger, Lima and Wynne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Natasha A. Botwright, TmF0YXNoYS5Cb3R3cmlnaHRAY3Npcm8uYXU=
†Present address: Amin R. Mohamed, Zoology Department, Faculty of Science, Benha University, Benha, Egypt
 Natasha A. Botwright
Natasha A. Botwright Amin R. Mohamed
Amin R. Mohamed Joel Slinger
Joel Slinger Paula C. Lima
Paula C. Lima James W. Wynne
James W. Wynne