- 1Department of Pathophysiology, School of Basic Medical Sciences, Anhui Medical University, Hefei, China
- 2Experimental Medicine Center, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 3Experimental Transplantation Surgery, Department of General, Visceral and Vascular Surgery, Friedrich-Schiller-University Jena, Jena, Germany
Inflammation, which is induced by the immune response, is recognized as the driving factor in many diseases, including infections and inflammatory diseases, metabolic disorders and cancers. Genetic variations in pivotal genes associated with the immune response, particularly single nucleotide polymorphisms (SNPs), may account for predisposition and clinical outcome of diseases. Lipopolysaccharide (LPS)-binding protein (LBP) functions as an enhancer of the host response to LPS, the main component of the outer membrane of gram-native bacteria. Given the crucial role of LBP in inflammation, we will review the impact of SNPs in the LBP gene on infections and inflammatory diseases, metabolic disorders and cancers.
Highlights
● Inflammation, which is induced by the immune response, is recognized as the driving factor in many diseases, including infections and inflammatory diseases, metabolic disorders and cancers.
● Genetic variations in pivotal genes associated with the immune response, particularly single nucleotide polymorphisms (SNPs), account for predisposition and clinical outcome of inflammation-related diseases.
● Lipopolysaccharide (LPS)-binding protein (LBP) plays a crucial role in the innate immune response for the development of inflammatory and infectious-related diseases.
● This review aims to summarize the investigation and association of SNPs in the LBP gene with infections and inflammatory diseases, metabolic disorders and cancers.
Introduction
In many diseases, including infections, metabolic disorders and cancers, inflammation is recognized as the driving factor (1–3). Functionally, inflammation is widely defined as a protective response against invading pathogens and endogenous stress signals (4), regulated by various pathways (such as TLR4/NF-kB pathway and IL-6/JAK/STAT3) and a multitude of genes (5–7). Interactions of cells from the innate and the adaptive immune system, together with the corresponding inflammatory mediators, contribute to various aspects of inflammation underlying many organ diseases (3, 8). Although the spectrum of inflammatory diseases is rather wide (ranging from systemic diseases such as sepsis, Type 1 diabetes and organ disease such as hepatitis) the cellular and molecular pathways involved in the inflammatory response remain similar (5).
Lipopolysaccharide, also called endotoxin, is a major component of the outer membrane of gram-negative (GN) bacteria (9). LPS causes endotoxemia when released into blood (10). Release of high levels of LPS into the systemic circulation (or release of lower levels in case of LPS-sensitization), induces a systemic inflammatory response syndrome, also called SIRS. The development of SIRS aggravates any other concomitant inflammatory condition, e.g. contributes to the development of septic shock in cases of sepsis (10).
The inflammatory response to LPS is mediated via LPS binding protein LBP. Lipopolysaccharide (LPS)-binding protein (LBP) is mainly produced in hepatocytes, functions as a secretory class I acute-phase protein (11–13) and plays a pivotal role in the innate immune response. Initially, LPS binds to LBP forming the LPS-LBP complex, which subsequently binds to CD14 and to the MD-4/MD-2 complex (14), ultimately results in the activation of signal transduction pathways and the production of cytokines and other pro-inflammatory mediators (15). Circulating LBP has a concentration-dependent immunologic function. Subnormal levels of LBP protein enhance phagocytosis and clearance of LPS from blood (16). In contrast, at high concentrations, LBP protein attenuates the release of pro-inflammatory cytokines (17). Additionally, LBP transfers LPS to very-low-density lipoproteins, low-density lipoproteins, high-density lipoproteins or chylomicrons, contributing to immune reaction against the infection (18–22). LBP also contributes to the immune response to gram-positive bacteria and fungal infection (23–25).
The physiological concentration of LBP in the serum of healthy human individuals is 5 to 10μg/mL (26). In contrast, during sepsis, up to seven-fold higher levels were observed within 24h (27). Increased LBP levels were also induced by a diet rich in fat and carbohydrates (28). Therefore, individuals suffering from obesity, diabetes, and related metabolic disorders also present with highly elevated LBP-levels (29–31), which was probably attributed to chronic low-grade inflammation induced by alterations of the intestinal flora together with an increased gut permeability to LPS (32, 33).
LBP is encoded by the LBP gene (34). The human LBP gene locates on chromosome 20 and shares an intron-exon pattern with other lipid-binding and transferring proteins on the same chromosome (34, 35). Genetic variations in LBP may contribute to susceptibility and clinical course of infectious diseases (36). On the basis of the crucial function of LPS in the innate immune response, many studies were performed to the genetic variation in the LBP gene in respect to the development of inflammatory diseases.
Single nucleotide polymorphisms (SNPs) are the most common type of genetic variation. The term refers to sequence alternatives (alleles) of a single DNA base with a frequency of more than 1% (37). SNPs influence the risk for the development and the outcome of a disease via various mechanisms but they are not considered to cause diseases (38). In coding regions, nonsynonymous SNPs could generate an amino-acid exchange and in turn affect the structure and biological function of the encoded protein, whereas synonymous SNPs do not lead to amino-acid exchange (39). Gu et al. reported that both synonymous and nonsynonymous SNP led to abnormal gene expression via altering the structure of local messenger RNA (38). In addition, mutations in the promoter region usually result in reduced mRNA levels (37). John C has demonstrated in his previous study that SNPs occurred in less conserved regions more frequently and the lowest occurrence rate was at the splice sites (40).
Evidence accumulated that genetic variations in pivotal genes associated with the immune response, particularly SNPs, may account for predisposition and clinical outcome of many diseases and potentially allow to predict prognosis and therapeutic effects (36, 41, 42).
In this review, we aim to present the current knowledge regarding the occurrence of SNP in the LBP gene and the impact of SNP in the LBP gene on the development and the course of selected infections, inflammatory diseases, metabolic disorders and cancers.
Correlation Between SNP in LBP Gene and Infections, Inflammatory Diseases, Metabolic Disorders and Cancers
LBP-SNPs in Infections
Evidence is accumulating that specific SNPs in the LBP gene are associated with a different course of many infections, including sepsis, infectious complications, and some organs infectious diseases (see in Table 1).
Hubacek et al. reported that a gender-related polymorphism, a nonsynonymous SNP at the 292nd nucleotide of the LBP gene, was associated with an increased predisposition to sepsis in males (p<0.02) but not in female patients (43). Compared with controls, a significantly higher proportion of male individuals in the cohort of patients with sepsis carried at least one Gly98 allele (43). Interestingly, Barber and colleague determined this SNP (T292G) was not exist by pyrosequencing, which occurred due to adjacent nucleotide substitution (T→C) at 291 position. Therefore, the association was thought to be false-positive (52). In addition, Barber et al. reported T291C lacked association with septic complications and survival after trauma (52).
Rs2232582 (T291C) is a synonymous variant located in the coding region, which conferred a nucleotide substitution from T to C. Both variants encoded the amino acid at residue 97 (52). Chien and colleagues used a SNP haplotype tagging approach tagSNP in a retrospectively case-control population with 97 patients and 204 controls (21). They reported the C allele of rs2232582 (tagSNP 6878) was associated with GN bacteremia after hematopoietic cell transplantation (HCT) in multivariate analysis (p=0.001) (21). And in whites, the C allele was identified to be associated with a 2-fold higher risk for GN bacteremia (p=0.002) (21). Additionally, Vollmer et al. studied five LBP-SNPs in 78 infective endocarditis (IE) patients and 156 healthy blood donors, among which T allele of rs2232582 (291C>T) had a weak association with an increased susceptibility to infective endocarditis (p=0.019) (44). However, the significance didn’t remain after Bonferroni correction (291C>T pc=0.095) (44).
Rs2232596 is a synonymous variant and confers A→G nucleotide substitution at the 613rd position. Chien et al. reported rs2232596 (SNP 17002) was associated with GN bacteria in multivariate analysis in HCT (p=0.027) (21). Vollmer et al. determined the Gallele of rs2232596 (613A>G) was associated with an increased susceptibility to infective endocarditis(p=0.018) (44). Similar with rs2232582 in this research,no significance remained after Bonferroni correction (Pc=0.090) (44).
Rs2232571, which confers a C→T nucleotide substitution at -778 position in a CAAT box, locates in the promotor region of LBP gene (21) and regulates promoter efficiency (11). Chien et al. described that the SNP is in strong linkage disequilibrium with rs2232582 aforementioned (21). In a prospective cohort of 234 cases, patients of HCT carrying rs2232571 (SNP1683) C allele significantly displayed a 3-fold increase in the risk of death prior to discharge (p=0.001) (21). In the development of GN bacteremia, mortality risk of patients significantly increased five-fold when they carried C allele of rs2232571 (p=0.013) (21). Moreover, rs2232571 C allele carriers exhibited higher median basal serum LBP levels (CC, 17.39μg/mL; TC 10.4μg/mL; TT 8.07μg/mL, p=0.004) (21). In addition, Guinan et al. confirmed previous linkage between rs2232571and plasm LBP levels in HCT (46). Flores and colleagues explored nine polymorphisms of LBP gene in the 5’-flanking region which is the transcription start site from position -1978 to -763. They observed no association between these SNPs along with the incidence of severe sepsis (45). Notably, a common 4-SNP haplotype consisting of allelic combination rs1780616-C/rs5741812-A/rs2232571-T/rs1780617-A was significantly associated with the predisposition to severe sepsis (p=0.016) (45). In comparison with non-carriers, homozygous carriers showed an increased risk of sepsis (p=0.016) and higher mean serum LBP concentrations at inclusion (130.1μg/mL for homozygous carriers and 101.6μg/mL for non-carriers, respectively) and at the seventh day (98.9μg/mL for homozygous carriers and 58.7μg/mL for non-carriers, respectively. p=0.046) (45).
Rs2232613 is a nonsynonymous mutation, which leads to proline changed to leucine at residue 333 (47). Eckert and colleagues described the structure of murine LBP and revealed the presence of rs2232613 affected a “phenylalanine core” in the C-terminal of the LBP and resulted in a dysfunction protein (47). The mutant protein reduced bacterial ligands binding capacity and cytokines-induction function in vitro (47). It was consistent with the results in vivo experiments that the variant T allele carriers displayed lower cytokine levels during experimental LPS stimulation. For example, at 1hr after injection of LPS, the production of TNF-α of heterozygous mutation carriers is significantly lower (p=0.013) (47). Carriers produced lower cytokine concentrations when suffering from ventilator-associated pneumonia (6.2 ± 1.76pg/mL for carriers and 18.4 ± 4.9 pg/mL for WT individuals, respectively. P=0.014) (47). A retrospective clinical analysis of 424 ICU surgical patients indicated that T allele carriers exhibited higher risk for mortality (CT/TT vs. CC, 11.5% vs. 5.8%, p=0.1) (47). Statistically significant difference was also seen in mortality in patients with infectious complications (CT/TT vs. CC, 21.2% vs. 8.9%, p=0.02 for hospital-acquired pneumonia; 33.3% vs. 8.2%,p=0.03 for hospital-acquired pneumonia caused by Gram-negative bacteria) (47). Studies demonstrated that LBP deficiency protected mice from LPS stimulation but exhibited higher lethality in bacterial stimulation, which might account for that carriers of rs2232613 exhibited higher risk of severe clinical infection (47). Guinan et al. also confirmed T allele carriers of rs2232613 displayed lower plasm LBP concentrations but higher efficiency in endotoxin extraction and transfer to MD-2 in HCT patients (46).
Rs2232618, is a functional variant conferring an amino acid substitution from phenylalanine to leucine at residue 436 caused by a T→C nucleotide substitution in the C-terminal domain of the LBP protein (48). According to the research conducted by Hubacek and colleagues, no positive association between rs2232618 with the occurrence of sepsis was described (43). However, Zeng et al. assessed the entire LBP gene in major trauma cohorts in Chinese population via using haplotype tagging SNPs (htSNPs) and detected that among the nine htSNPs studied in LBP, only rs2232618 associated with traumatic sepsis (48). According to 557 trauma patients in Chongqing cohort, C allele carriers showed a higher of sepsis and MODS (TT vs. TC+CC, p<0.0001 for sepsis morbidity rate and p=0.011 for MOD scores, respectively), when compared with T allele carriers (48). TNFα production by peripheral leukocytes of C allele carriers in vitro revealed a significantly increased response to LPS (TT vs. TC, p=0.01). The clinical relevance of genotypic distribution of rs2232618 was further confirmed in Zhejiang cohort with 230 patients (TT vs. TC, p=0.012 for sepsis morbidity rate and p=0.046 for MOD scores, respectively) (48). In vitro experiments, the binding capacity with LPS (p<0.001) and TNFα production (p<0.001) were significantly enhanced with the presence of the variant Leu436 LBP protein. Furthermore, Leu436 LBP protein has stronger binding interactions with CD14 (p=0.043) (48). Lu and their groups confirmed the correlation between genotypic distributions and traumatic sepsis in an enlarged co-located population sample size with 1296 patients in Chongqing (TT vs. TC+CC, p=0.002 for sepsis morbidity rate and p=1.8E-6 for MOD scores, respectively) and 445 patients in Zhejiang (TT vs. TC+CC, p=0.002 for sepsis morbidity rate and p=0.005 for MOD scores, respectively) (50). Combined analysis of two cohorts also showed relevance between rs2232618 and traumatic sepsis (TT vs. TC+CC, p=4.5×10-4 for sepsis morbidity rate and p=1.4E-9 for MOD scores, respectively) (50). In addition, Jabandziev et al. reported rs2232618 in combination with additional four SNPs (BPI rs574350, TLR rs4986790, HSP70 rs2227956, IL-6 rs1800795) could be regarded as a predictor of clinical outcome of sepsis in children (49). The authors defined wild-type homozygotes as A, heterozygotes and mutant homozygotes as B. Among analyses of BPI, TLR and LBP, LBP A + BPI A + TLR A was associated with a high risk of sepsis (p=0.005), LBP B + BPI A + TLR A was a high-risk combination for sepsis with severe condition (p=0.034) (49). On the contrary, LBP A + BPI B + TLR A was a low-risk combination for both sepsis and severe condition (p<0.001 and p=0.003, respectively) (49). In terms of combinations of LBP, IL-6 and TLR, LBP A + IL-6 B +TLR A is a low-risk combination of severe sepsis (p=0.027) (49). Combinations of LBP A + IL-6 A +TLR B and LBP B + IL-6 A +TLR A were regarded to be a high risk for sepsis (p<0.006 and p=0.012, respectively), but the proportion of patients was low (4.2% and 4.8%, respectively) (49). Analysis of five genes showed a combination of wild-type homozygotes was associated with high risk of sepsis. Moreover, BPI A + LBP A + TLR A +HSP70 A +IL-6 B was also a high-risk combination for sepsis (p=0.016) (49). On the contrary, BPI B + LBP A + TLR A +HSP70 A +IL-6 B and BPI B + LBP A + TLR B +HSP70 A +IL-6 B were low-risk combinations for both sepsis (p=0.006 and p=0.041, respectively) and sepsis with severe condition (p=0.001 and p=0.026) (49). In addition, Verstraelen et al. observed rs2232618 was marginally significantly associated with carriage of Atopobium vaginae in bacterial vaginosis during early pregnancy (p=0.056) (51).
LBP-SNPs in Atopy and Asthma
Atopy is viewed as a predisposition to the immune response to diverse antigens/allergens, inducing CD4+ Th2 differentiation and overproduction of immunoglobulin E (IgE) (53). Asthma is a manifestation of atopy, which is a chronic inflammatory airway disease caused by combined influence of genetic and environmental factors (54). Asymptomatic atopic individuals, healthy individuals or atopic individuals without current manifestation of disease exhibited increased serum LBP levels after inhaling LPS. These findings suggest that atopy and asthma related phenotype might be associated with LBP genetic variants (55). Two studies related to asthma reported six SNPs of LBP. In the coding region, rs2232596 is a synonymous mutation and rs2232580 is a missense mutation that confers an amino acid substitution from proline to leucine at residue 9. Moreover, rs6025083 and rs12624843 are located in the intron, rs5741812 is located in the 5’-UTR and rs745144 is located in the 3’-UTR. Reijmerink and colleagues and colleagues demonstrated gene-environment interactions of atopy in children (56). Rs2232596, day-care and one sibling were selected as the best three-factor model (p=0.007) (56). Day-care protected children from developing high total IgE at 1-2 years old who were heterozygotes for rs2232596, but not in children with more than one sibling (56). rs745144, in combination with sibling and CD14 (rs2915863) were selected as the there-factor predictors for total IgE at 6-8 years old (56). Besides, rs6025083, along with TLR1 (rs5743594) and TLR5 (rs851186) were selected as three-factor predictors of gene-gene interaction of specific IgE to indoor allergens 6–8 years old (56). Daley et al. reported rs2232580 and rs5741812 associated with atopy in children with a family history of allergies (p=0.0141 and p=0.423, respectively) (57). In the healthy study cohort, rs12624843 were associated with airway hyperresponsiveness (AHR) (p=0.0443) (57). Additionally, SNP-virus interaction analyses showed rs5741812 was significantly associated with AHR and picornavirus (p=0.048) (57) (see in Table 2).
LBP-SNPs in Metabolic Disorders
Increasing evidence suggested gut microbiota and low-grade inflammation are associated with chronic diseases, such as metabolic disorders (66), cardiovascular disease (67) and hypertension (68). Given the involvement of LBP in inflammation (67), some SNPs have been investigated in related diseases (see in Table 2).
Rs1739654 is a synonymous mutation located in the coding region, rs2232578 is located in the 5’-UTR, rs1780627 and rs2232592 are located in intron region. Takeuchi et al. identified this four SNPs of LBP gene on chromosome 20q significantly correlated with susceptibility to type 2 diabetes p=0.001 for rs1739654, p=0.003 for rs1780627, p=0.011 for rs2232592 and p=0.0004 for rs2232578, respectively (58). Further stratified analysis for rs2232592 indicated diabetic patients with the history of obesity and/or earlier age of onset have prominent genetic risk (58).
As previously mentioned, rs2232582 is a synonymous variant and confers T→C nucleotide substitution. Vilade's et al. reported rs2232582 was associated with HIV-1/HAART-associated lipodystrophy syndrome (HALS). Carriage of T allele was significantly associated the higher incidence of HALS whereas carriage of C allele seemed to protect individuals against HALS (p=0.01) (59). Besides, Zhan et al. reported 291T>C (rs2232582) was associated the Carotid intima-media thickness (CIMT) (p<0.05), but not cerebral infarction in Chinese population (61). CIMT is considered to be a risk of cardiovascular and cerebrovascular disease and a surrogate marker to assess AS (69), patients carrying TC genotype exhibited higher mean CIMT compared with TT genotype (61).
As the leading cause of myocardial infarction and stroke (70), atherosclerosis (AS) is a complex disease and involves a series of pathophysiological processes such as inflammation and lipid metabolism (71). Hubacek et al. demonstrated that the frequency of LBP 1306T>C (rs2232618) C allele (Leu436) homozygotes is about threefold in patients with myocardial infarction than in controls (60). But the frequency of this genotype in controls is very low and did not reach statistical significance (2.3% in patients and 0.7% in controls, respectively) (60). In addition, Zhan et al. reported 1306T>C (rs2232618), similar to rs2232582 in the study, was associated with CIMT (p<0.05) (61). Individuals with TC genotype exhibited higher mean CIMT compared with TT genotype (61).
LBP-SNPs in Cancer
Inflammation is closely related to the pathogenesis of cancer (72, 73) and genetic variations of inflammatory regulators were demonstrated to associate with the increased risk or adverse consequence of several tumors (74–79).
Intestinal microbiota dysbiosis leads to more release of LPS and thereby to a higher stimulation of LBP and subsequent production of inflammatory cytokines (75). Chronic low-grade inflammation contributes to carcinogenesis and the development of colorectal carcinoma (CRC) (80). Chen et al. demonstrated that mutant G allele of rs2232596 was significantly associated with the CRC risk in patients compared with healthy controls (GA vs. AA, p=0.003; GG vs. AA, p=0.016; GA+GG vs. AA, p=0.001, respectively). When combined with CD14 rs4914, the risk of CRC increased 3.4 times (p=0.000) (62). Additionally, both smokers carrying GA genotype and drinkers carrying GA or GG genotype had an increased risk for CRC (p=0.005 for smokers and p=0.001 for drinkers, respectively), which revealed the interaction between genes with tobacco and alcohol (62). According to another study conducted by Chang et al, rs1780617 G allele was significantly associated with the risk of CRC and adenoma, a precursor to CRC (p=0.046) (63).
Helicobacter pylori (HP) infection accounts for the progression of gastric cancer (GC) (76). Castaño-Rodríguez et al. demonstrated that rs2232578, which is located in 5’-UTR, induced an increased risk for GC in combination with HP infection in Chinese population (p=0.0292 for AA and p=0.017 for G carriers) (64).
LEE et al. conducted a Genome-wide pathway analysis and identified six SNPs, five genes and nine pathways in glioma. Rs2232580 contributed to the risk of glioma via affecting the interaction of LBP to LPS and immune responses (0.002≤p ≤ 0.013) (65).
The overview of analysis of LBP SNPs which exhibit association in inflammation-related diseases is shown in Table 3 classified with SNP ID. In addition, SNPs that were investigated but did not show correlation are summarized in Table 4.
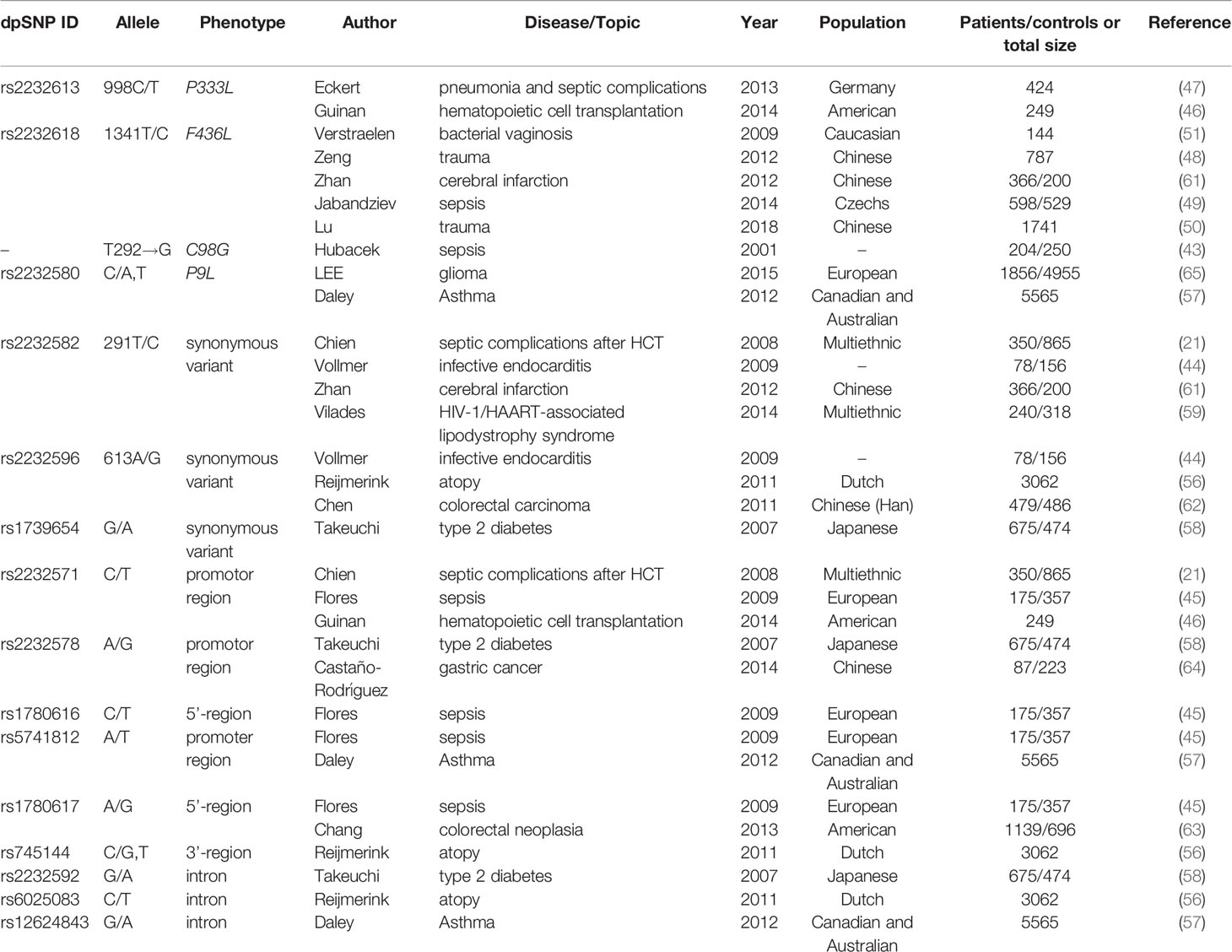
Table 3 Summary of Lipopolysaccharide Binding Protein (LBP) SNPs correlated with corresponding disease.
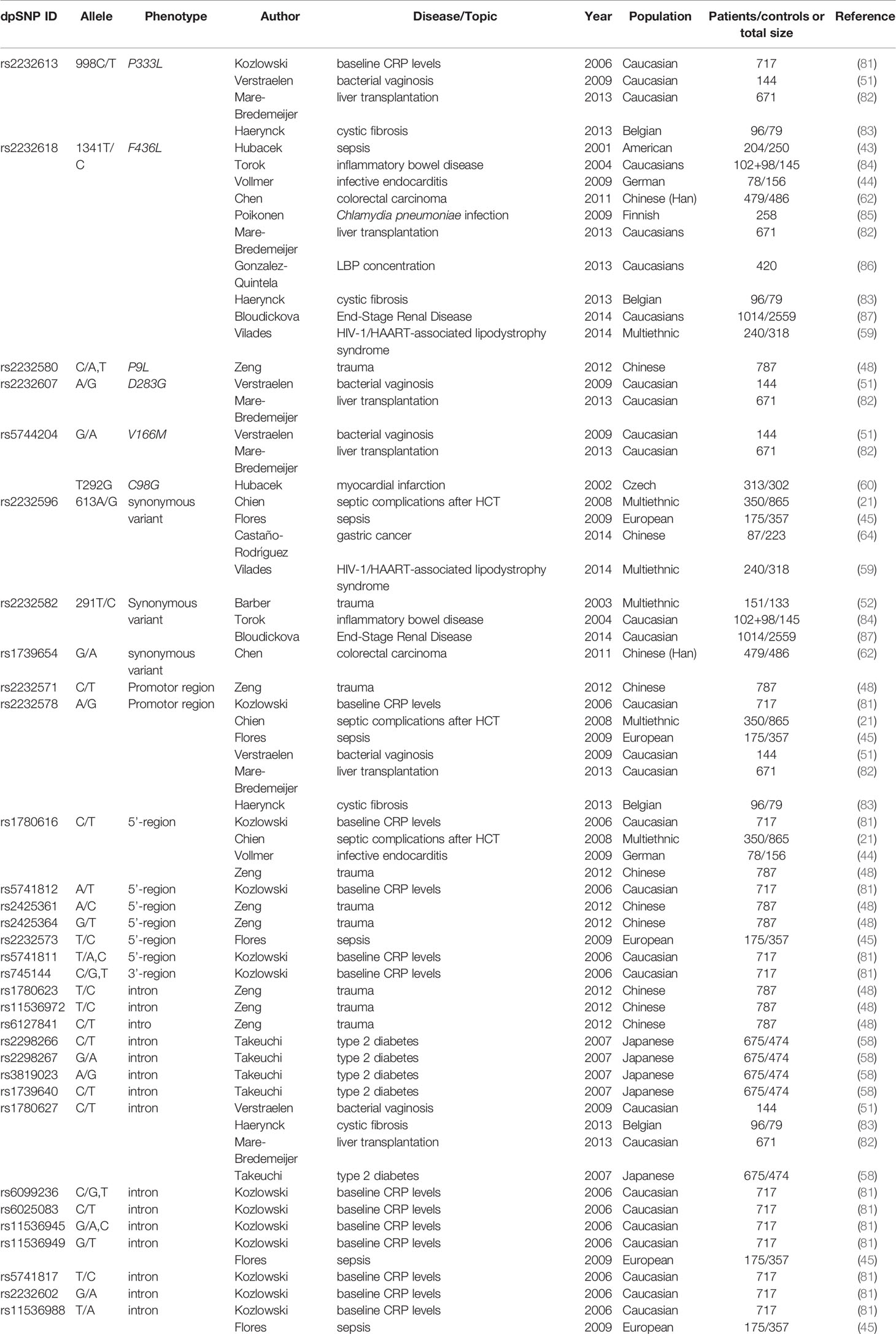
Table 4 Summary of Lipopolysaccharide Binding Protein (LBP) SNPs not associated with corresponding disease.
Results of Meta-analysis
Results of smeta -analysis of susceptibility associated with rs2232618, synonymous 292 and rs2232592 in diseases are performed in Figures 1–3, respectively.
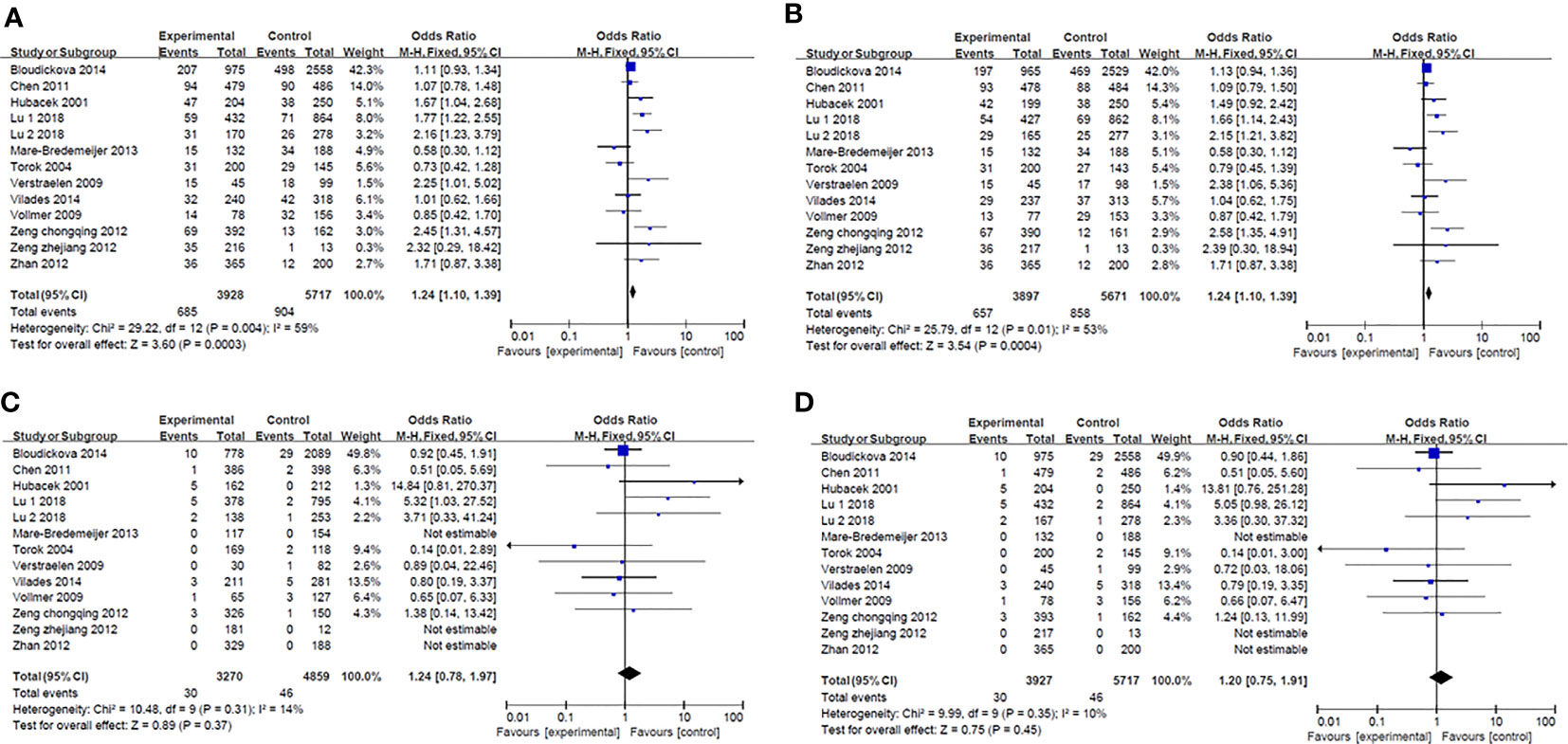
Figure 1 Forest plot in rs2232618. (A) Dominant model (TT+TC vs CC). (B) heterozygote model (TC vs CC). (C) homozygotes models (TT vs CC). (D) recessive models (TT vs TC + CC).
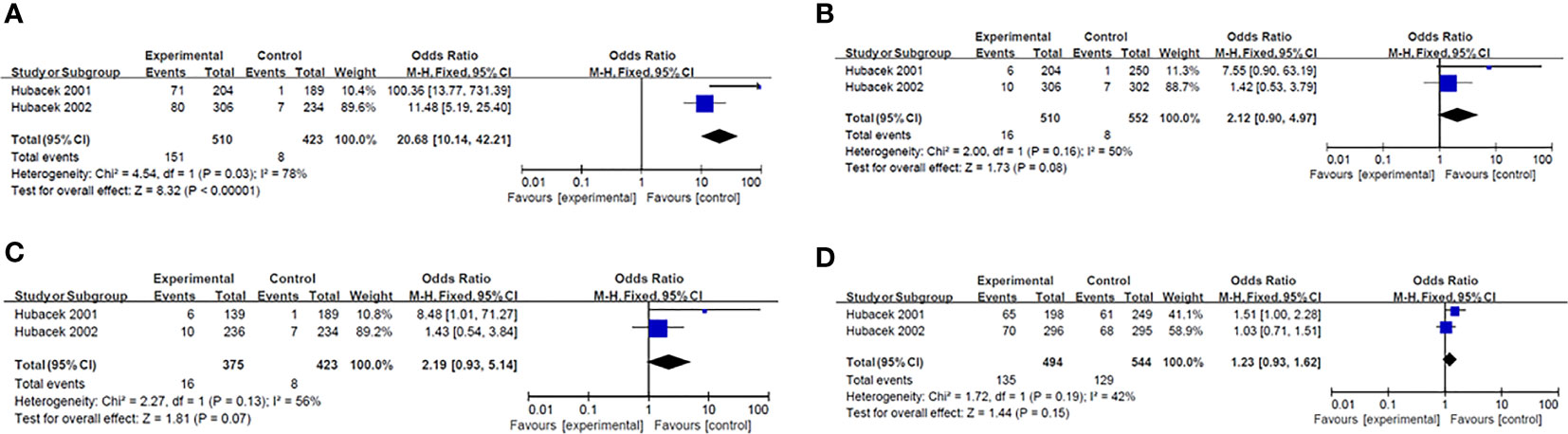
Figure 2 Forest plot in synonymous 292. (A) Dominant model (TT+TG vs GG). (B) heterozygote model (TG vs GG). (C) homozygotes models (TT vs GG). (D) recessive models (TT vs TG +GG).
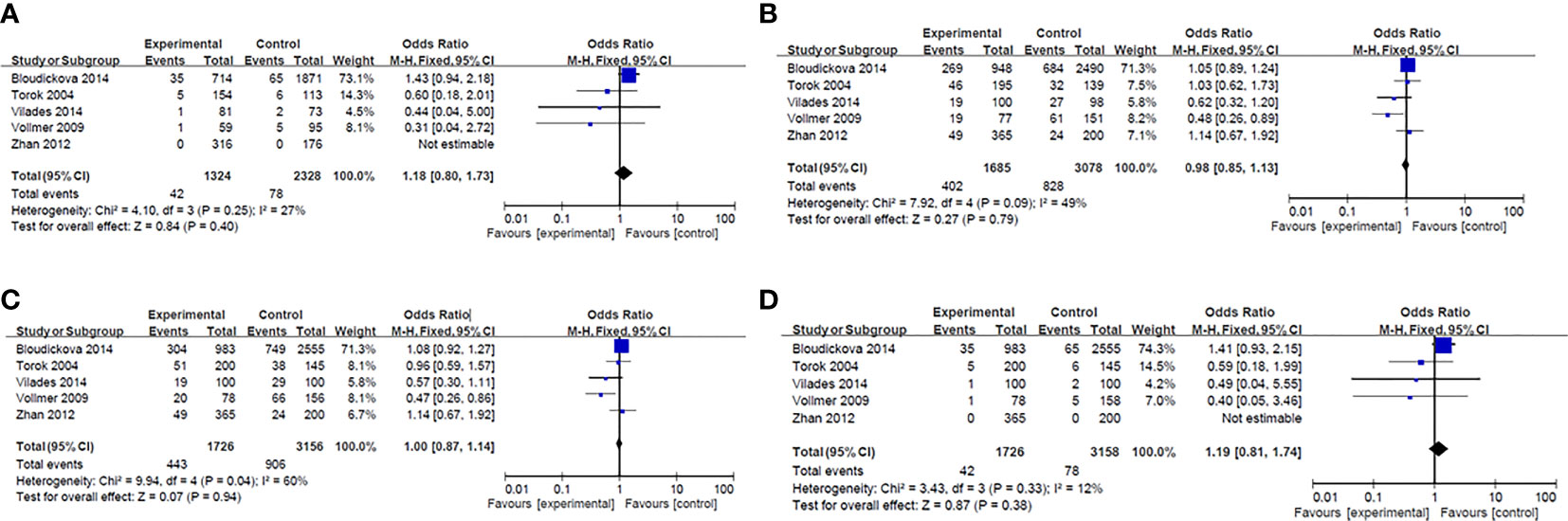
Figure 3 Forest plot in rs2232582. (A) Dominant model (TT+TC vs CC). (B) heterozygote model (TC vs CC). (C) homozygotes models (TT vs CC). (D) recessive models (TT vs TC + CC).
Conclusion
Characterization of genetic predisposition underlying diseases is the prerequisite for identifying patients at risk and for developing specific therapeutic strategies. In this review we give evidence for the relationship between SNPs in the LBP gene and certain inflammatory diseases.
Our finding might help to identify individuals at risk for inflammatory complications by genotyping and thereby contribute to developing more widely applicable diagnostic strategies. Furthermore, our finding might represent one step towards designing therapeutic strategies based on genetic heterogeneity using e.g. KO technologies.
However, more clinical studies including much higher patient numbers are needed to reach statistical significance confirming the reported trends as well as for confirming the postulated relationships in independent patient cohorts.
In addition, identifying high-risk individuals based on genotype and adjusting therapeutic strategies timely in the clinic could improve the prognosis of patients.
However, our findings are limited in three aspects: reproducibility of results and size of patient cohorts. So far, only two of the reported association between LBP-polymorphism and disease association were replicated by an independent study. Thus, additional studies are needed to verify the association between a certain SNP and susceptibility for a given disease.
Some of the reported positive but also negative associations were obtained in small patient cohorts (see in Table 3 and Table 4), which calls for confirmatory studies investigating larger groups of patients. Doing so will lead to sound results which can be used for developing novel therapeutic approaches, e.g., using pharmacological KO strategies.
Author Contributions
Data collection: LM, ZS and HF. Manuscript preparation: LM, AL and HF. Critical revision: LM, UD, XY and HF. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Science Foundation of China (No. 81971875) and the University Natural Science Foundation of Anhui Province (No. KJ2019A0220)
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
LBP, lipopolysaccharide binding protein; LPS, lipopolysaccharide; SNP, Single nucleotide polymorphism; GN, Gram-negative; HCT, hematopoietic cell transplantation; htSNPs, haplotype tagging SNPs; AHR, airway hyperresponsiveness; HALS, HIV-1/HAART-associated lipodystrophy syndrome; AS, atherosclerosis; CIMT, Carotid intima-media thickness; CRC, colorectal carcinoma; HP, Helicobacter pylori; GC, gastric cancer.
References
1. Piya MK, McTernan PG, Kumar S. Adipokine Inflammation and Insulin Resistance: The Role of Glucose, Lipids and Endotoxin. J Endocrinol (2013) 216(1):T1–15. doi: 10.1530/JOE-12-0498
2. Zhu JJ, Wang X. Chronic Inflammation, Autoimmunity and Atherosclerosis. Sheng Li Ke Xue Jin Zhan (2002) 33(4):327–31.
3. Libby P. Inflammatory Mechanisms: The Molecular Basis of Inflammation and Disease. Nutr Rev (2007) 65(12 Pt 2):S140–6. doi: 10.1301/nr.2007.dec.S140-S146
4. Netea MG, Balkwill F, Chonchol M, Cominelli F, Donath MY, Giamarellos–Bourboulis EJ, et al. A Guiding Map for Inflammation. Nat Immunol (2017), 826–31. doi: 10.1038/ni.3790
5. Bayani A, Dunster JL, Crofts JJ, Nelson MR. Mechanisms and Points of Control in the Spread of Inflammation: A Mathematical Investigation. Bull Math Biol (2020) 82(4):45. doi: 10.1007/s11538-020-00709-y
6. Zhu L, Gong X, Gong J, Xuan Y, Fu T, Ni S, et al. Notoginsenoside R1 Upregulates miR-221-3p Expression to Alleviate ox-LDL-Induced Apoptosis, Inflammation, and Oxidative Stress by Inhibiting the TLR4/NF-KappaB Pathway in Huvecs. Braz J Med Biol Res (2020) 53(6):e9346. doi: 10.1590/1414-431x20209346
7. Yeung YT, Aziz F, Guerrero–Castilla A, Arguelles S. Signaling Pathways in Inflammation and Anti-Inflammatory Therapies. Curr Pharm Des (2018) 24(14):1449–84. doi: 10.2174/1381612824666180327165604
8. De Maio A, Torres MB, Reeves RH. Genetic Determinants Influencing the Response to Injury, Inflammation, and Sepsis. Shock (2005) 23(1):11–7. doi: 10.1097/01.shk.0000144134.03598.c5
9. Gnauck A, Lentle RG, Kruger MC. The Characteristics and Function of Bacterial Lipopolysaccharides and Their Endotoxic Potential in Humans. Int Rev Immunol (2016) 35(3):189–218. doi: 10.3109/08830185.2015.1087518
11. Schumann RR, Kirschning CJ, Unbehaun A, Aberle HP, Knope HP, Lamping N, et al. The Lipopolysaccharide-Binding Protein Is a Secretory Class 1 Acute-Phase Protein Whose Gene Is Transcriptionally Activated by APRF/STAT/3 and Other Cytokine-Inducible Nuclear Proteins. Mol Cell Biol (1996) 16(7):3490–503. doi: 10.1128/MCB.16.7.3490
12. Zhou Z, Xu MJ, Gao B. Hepatocytes: A Key Cell Type for Innate Immunity. Cell Mol Immunol (2016) 13(3):301–15. doi: 10.1038/cmi.2015.97
13. Jappe U, Schwager C, Schromm AB, Roldan NG, Stein K, Heine H, et al. Lipophilic Allergens, Different Modes of Allergen-Lipid Interaction and Their Impact on Asthma and Allergy. Front Immunol (2019) 10:122.. doi: 10.3389/fimmu.2019.00122
14. Ryu JK, Kim SJ, Rah SH, Kang JI, Jung HE, Lee D, et al. Reconstruction of LPS Transfer Cascade Reveals Structural Determinants Within LBP, CD14, and TLR4-MD2 for Efficient Lps Recognition and Transfer. Immunity (2017) 46(1):38–50. doi: 10.1016/j.immuni.2016.11.007
15. Guha M, Mackman N. LPS Induction of Gene Expression in Human Monocytes. Cell Signal (2001) 13(2):85–94. doi: 10.1016/S0898-6568(00)00149-2
16. Le Roy D, Di Padova F, Adachi Y, Glauser MP, Calandra T, Heumann D. Critical Role of Lipopolysaccharide-Binding Protein and CD14 in Immune Responses Against Gram-Negative Bacteria. J Immunol (2001) 167(5):2759–65. doi: 10.4049/jimmunol.167.5.2759
17. Zweigner J, Gramm HJ, Singer OC, Wegscheider K, Schumann RR. High Concentrations of Lipopolysaccharide-Binding Protein in Serum of Patients With Severe Sepsis or Septic Shock Inhibit the Lipopolysaccharide Response in Human Monocytes. Blood (2001) 98(13):3800–8. doi: 10.1182/blood.V98.13.3800
18. Vreugdenhil AC, Snoek AM, van't Veer C, Greve JW, Buurman WA. LPS-Binding Protein Circulates in Association With apoB-Containing Lipoproteins and Enhances Endotoxin-LDL/VLDL Interaction. J Clin Invest (2001) 107(2):225–34. doi: 10.1172/JCI10832
19. Wurfel MM, Kunitake ST, Lichenstein H, Kane JP, Wright SD. Lipopolysaccharide (LPS)-Binding Protein Is Carried on Lipoproteins and Acts as a Cofactor in the Neutralization of LPS. J Exp Med (1994) 180(3):1025–35. doi: 10.1084/jem.180.3.1025
20. Hamann L, Alexander C, Stamme C, Zahringer U, Schumann RR. Acute-Phase Concentrations of Lipopolysaccharide (LPS)-Binding Protein Inhibit Innate Immune Cell Activation by Different LPS Chemotypes Via Different Mechanisms. Infect Immun (2005) 73(1):193–200. doi: 10.1128/IAI.73.1.193-200.2005
21. Chien JW, Boeckh MJ, Hansen JA, Clark JG. Lipopolysaccharide Binding Protein Promoter Variants Influence the Risk for Gram-Negative Bacteremia and Mortality After Allogeneic Hematopoietic Cell Transplantation. Blood (2008) 111(4):2462–9. doi: 10.1182/blood-2007-09-101709
22. Levels JHM, Marquart JA, Abraham PR, van den Ende AE, Molhuizen HOF, van Deventer SJH, et al. Lipopolysaccharide Is Transferred From High-Density to Low-Density Lipoproteins by Lipopolysaccharide-Binding Protein and Phospholipid Transfer Protein. Infect Immun (2005) 73(4):2321–6. doi: 10.1128/IAI.73.4.2321-2326.2005
23. Blairon L, Wittebole X, Laterre P-F. Lipopolysaccharide-Binding Protein Serum Levels in Patients With Severe Sepsis Due to Gram-Positive and Fungal Infections. J Infect Dis (2003) 187(2):287–91. doi: 10.1086/346046
24. Schroder NW, Morath S, Alexander C, Hamann L, Hartung T, Zahringer U, et al. Lipoteichoic Acid (LTA) of Streptococcus Pneumoniae and Staphylococcus Aureus Activates Immune Cells Via Toll-Like Receptor (TLR)-2, Lipopolysaccharide-Binding Protein (LBP), and CD14, Whereas TLR-4 and MD-2 Are Not Involved. J Biol Chem (2003) 278(18):15587–94. doi: 10.1074/jbc.M212829200
25. Mueller M, Stamme C, Draing C, Hartung T, Seydel U, Schromm AB, et al. Cell Activation of Human Macrophages by Lipoteichoic Acid Is Strongly Attenuated by Lipopolysaccharide-Binding Protein. J Biol Chem (2006) 281(42):31448–56. doi: 10.1074/jbc.M605966200
26. Sakr Y, Burgett U, Nacul FE, Reinhart K, Brunkhorst F, et al. Lipopolysaccharide Binding Protein in a Surgical Intensive Care Unit: A Marker of Sepsis? Crit Care Med (2008) 36(7):2014–22. doi: 10.1097/CCM.0b013e31817b86e3
27. Chen KF, Chaou CH, Jiang JY, Yu HW, Meng YH, Tang WC, et al. Diagnostic Accuracy of Lipopolysaccharide-Binding Protein as Biomarker for Sepsis in Adult Patients: A Systematic Review and Meta-Analysis. PLoS One (2016) 11(4). doi: 10.1371/journal.pone.0153188
28. Ghanim H, Abuaysheh S, Sia CL, Korzeniewski K, Chaudhuri A, Fernandez–Real JM, et al. Increase in Plasma Endotoxin Concentrations and the Expression of Toll-Like Receptors and Suppressor of Cytokine Signaling-3 in Mononuclear Cells After a High-Fat, High-Carbohydrate Meal: Implications for Insulin Resistance. Diabetes Care (2009) 32(12):2281–7. doi: 10.2337/dc09-0979
29. Liu X, Lu L, Yao P, Ma YW, Wang FJ, Jin QL, et al. Lipopolysaccharide Binding Protein, Obesity Status and Incidence of Metabolic Syndrome: A Prospective Study Among Middle-Aged and Older Chinese. Diabetologia (2014) 57(9):1834–41. doi: 10.1007/s00125-014-3288-7
30. Moreno-Navarrete JM, Ortega F, Serino M, Luche E, Waget A, Pardo G, et al. Circulating Lipopolysaccharide-Binding Protein (LBP) as a Marker of Obesity-Related Insulin Resistance. Int J Obes (2012) 36(11):1442–9. doi: 10.1038/ijo.2011.256
31. Sakura T, Morioka T, Shioi A, Kakutani Y, Miki Y, Yamazaki Y, et al. Lipopolysaccharide-Binding Protein Is Associated With Arterial Stiffness in Patients With Type 2 Diabetes: A Cross-Sectional Study. Cardiovasc Diabetol (2017) 16. doi: 10.1186/s12933-017-0545-3
32. Cani PD, Osto M, Geurts L, Everard A. Involvement of Gut Microbiota in the Development of Low-Grade Inflammation and Type 2 Diabetes Associated With Obesity. Gut Microbes (2012) 3(4):279–88. doi: 10.4161/gmic.19625
33. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in Gut Microbiota Control Metabolic Endotoxemia-Induced Inflammation in High-Fat Diet-Induced Obesity and Diabetes in Mice. Diabetes (2008) 57(6):1470–81. doi: 10.2337/db07-1403
34. Bingle CD, Craven CJ. Meet the Relatives: A Family of BPI- and LBP-Related Proteins. Trends Immunol (2004) 25(2):53–5. doi: 10.1016/j.it.2003.11.007
35. Kirschning CJ, Au–Young J, Lamping N, Reuter D, Pfeil D, Seilhamer JJ, et al. Similar Organization of the Lipopolysaccharide-Binding Protein (LBP) and Phospholipid Transfer Protein (PLTP) Genes Suggests a Common Gene Family of Lipid-Binding Proteins. Genomics (1997) 46(3):416–25. doi: 10.1006/geno.1997.5030
36. Sorensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and Environmental Influences on Premature Death in Adult Adoptees. N Engl J Med (1988) 318(12):727–32. doi: 10.1056/NEJM198803243181202
37. Barnes MR. SNP and Mutation Data on the Web - Hidden Treasures for Uncovering. Comp Funct Genomics (2002) 3(1):67–74. doi: 10.1002/cfg.131
38. Hildebrand F, Mommsen P, Frink M, van Griensven M, Krettek C. Genetic Predisposition for Development of Complications in Multiple Trauma Patients. Shock (2011) 35(5):440–8. doi: 10.1097/SHK.0b013e31820e2152
39. Gu W, Gurguis CI, Zhou JJ, Zhu Y, Ko EA, Ko JH, et al. Functional and Structural Consequence of Rare Exonic Single Nucleotide Polymorphisms: One Story, Two Tales. Genome Biol Evol (2015) 7(10):2929–40. doi: 10.1093/gbe/evv191
40. Castle JC. Snps Occur in Regions With Less Genomic Sequence Conservation. PLoS One (2011) 6(6):e20660. doi: 10.1371/journal.pone.0020660
42. Schumann RR. Old and New Findings on Lipopolysaccharide-Binding Protein: A Soluble Pattern-Recognition Molecule. Biochem Soc Trans (2011) 39(4):989–93. doi: 10.1042/BST0390989
43. Hubacek JA, Stuber F, Frohlich D, Book M, Wetegrove S, Ritter M, et al. Gene Variants of the Bactericidal/Permeability Increasing Protein and Lipopolysaccharide Binding Protein in Sepsis Patients: Gender-Specific Genetic Predisposition to Sepsis. Crit Care Med (2001) 29(3):557–61. doi: 10.1097/00003246-200103000-00015
44. Vollmer T, Kleesiek K, Dreier J. Lipopolysaccharide-Binding Protein (LBP) Gene Polymorphisms: Rapid Genotyping by Real-Time PCR and Association With Infective Endocarditis. Clin Biochem (2009) 42(13-14):1413–9. doi: 10.1016/j.clinbiochem.2009.06.010
45. Flores C, Perez–Mendez L, Maca–Meyer N, Muriel A, Espinosa E, Blanco J, et al. A Common Haplotype of the LBP Gene Predisposes to Severe Sepsis. Crit Care Med (2009) 37(10):2759–66. doi: 10.1097/00003246-200910000-00013
46. Guinan EC, Palmer CD, Mancuso CJ, Brennan L, Stoler–Barak L, Kalish LA, et al. Identification of Single Nucleotide Polymorphisms in Hematopoietic Cell Transplant Patients Affecting Early Recognition of, and Response to, Endotoxin. Innate Immun (2014) 20(7):697–711. doi: 10.1177/1753425913505122
47. Eckert JK, Kim YJ, Kim JI, Gurtler K, Oh DY, Sur S, et al. The Crystal Structure of Lipopolysaccharide Binding Protein Reveals the Location of a Frequent Mutation That Impairs Innate Immunity. Immunity (2013) 39(4):647–60. doi: 10.1016/j.immuni.2013.09.005
48. Zeng L, Gu W, Zhang AQ, Zhang M, Zhang LY, Du DY, et al. A Functional Variant of Lipopolysaccharide Binding Protein Predisposes to Sepsis and Organ Dysfunction in Patients With Major Trauma. Ann Surg (2012) 255(1):147–57. doi: 10.1097/SLA.0b013e3182389515
49. Jabandziev P, Smerek M, Michalek J, Fedora M, Kosinova L, Hubacek JA, et al. Multiple Gene-to-Gene Interactions in Children With Sepsis: A Combination of Five Gene Variants Predicts Outcome of Life-Threatening Sepsis. Crit Care (2014) 18(1):R1. doi: 10.1186/cc13174
50. Lu HX, Sun JH, Wen DL, Du J, Zeng L, Zhang AQ, et al. LBP RS2232618 Polymorphism Contributes to Risk of Sepsis After Trauma. World J Emerg Surg (2018) 13:52. doi: 10.1186/s13017-018-0214-1
51. Verstraelen H, Verhelst R, Nuytinck L, Roelens K, De Meester E, De Vos D, et al. Gene Polymorphisms of Toll-Like and Related Recognition Receptors in Relation to the Vaginal Carriage of Gardnerella Vaginalis and Atopobium Vaginae. J Reprod Immunol (2009) 79(2):163–73. doi: 10.1016/j.jri.2008.10.006
52. Barber RC, O’Keefe GE. Characterization of a Single Nucleotide Polymorphism in the Lipopolysaccharide Binding Protein and Its Association With Sepsis. Am J Respir Crit Care Med (2003) 167(10):1316–20. doi: 10.1164/rccm.200209-1064OC
54. Villeneuve T, Guilleminault L. Asthma and Obesity in Adults. Rev Mal Respir (2020) 37(1):60–74. doi: 10.1016/j.rmr.2019.03.016
55. Michel O, Dentener M, Corazza F, Buurman W, Rylander R. Healthy Subjects Express Differences in Clinical Responses to Inhaled Lipopolysaccharide That Are Related With Inflammation and With Atopy. J Allergy Clin Immunol (2001) 107(5):797–804. doi: 10.1067/mai.2001.114249
56. Reijmerink NE, Kerkhof M, Bottema RW, Gerritsen J, Stelma FF, Thijs C, et al. Toll-Like Receptors and Microbial Exposure: Gene-Gene and Gene-Environment Interaction in the Development of Atopy. Eur Respir J (2011) 38(4):833–40. doi: 10.1183/09031936.00099210
57. Daley D, Park JE, He JQ, Yan J, Akhabir L, Stefanowicz D, et al. Associations and Interactions of Genetic Polymorphisms in Innate Immunity Genes With Early Viral Infections and Susceptibility to Asthma and Asthma-Related Phenotypes. J Allergy Clin Immunol (2012) 130(6):1284–93. doi: 10.1016/j.jaci.2012.07.051
58. Takeuchi F, Yanai K, Inomata H, Kuzuya N, Kajio H, Honjo S, et al. Search of Type 2 Diabetes Susceptibility Gene on Chromosome 20q. Biochem Biophys Res Commun (2007) 357(4):1100–6. doi: 10.1016/j.bbrc.2007.04.063
59. Vilades C, Escote X, Lopez–Dupla M, Martinez E, Domingo P, Asensi V, et al. Involvement of the LPS-LPB-CD14-MD2-TLR4 Inflammation Pathway in HIV-1/HAART-Associated Lipodystrophy Syndrome (HALS). J Antimicrob Chemother (2014) 69(6):1653–9. doi: 10.1093/jac/dku032
60. Hubacek JA, Pitha J, Skodova Z, Adamkova V, Podrapska I, Schmitz G, et al. Polymorphisms in the Lipopolysaccharide-Binding Protein and Bactericidal/Permeability-Increasing Protein in Patients With Myocardial Infarction. Clin Chem Lab Med (2002) 40(11):1097–100. doi: 10.1515/cclm.2002.191
61. Zhan Q, Yuan M, Wang XH, Duan XM, Yang QD, Xia J. Association of Lipopolysaccharide-Binding Protein Gene Polymorphisms With Cerebral Infarction in a Chinese Population. J Thromb Thrombolysis (2012) 34(2):260–8. doi: 10.1007/s11239-012-0701-2
62. Chen R, Luo FK, Wang YL, Tang JL, Liu YS. LBP and CD14 Polymorphisms Correlate With Increased Colorectal Carcinoma Risk in Han Chinese. World J Gastroenterol (2011) 17(18):2326–31. doi: 10.3748/wjg.v17.i18.2326
63. Chang CM, Chia VM, Gunter MJ, Zanetti KA, Ryan BM, Goodman JE, et al. Innate Immunity Gene Polymorphisms and the Risk of Colorectal Neoplasia. Carcinogenesis (2013) 34(11):2512–20. doi: 10.1093/carcin/bgt228
64. Castano-Rodriguez N, Kaakoush NO, Pardo AL, Goh KL, Fock KM, Mitchell HM. Genetic Polymorphisms in the Toll-Like Receptor Signalling Pathway in Helicobacter Pylori Infection and Related Gastric Cancer. Hum Immunol (2014) 75(8):808–15. doi: 10.1016/j.humimm.2014.06.001
65. Lee YH, Song GG. Genome-Wide Pathway Analysis in Glioma. Neoplasma (2015) 62(2):230–8. doi: 10.4149/neo_2015_028
66. Laugerette F, Furet JP, Debard C, Daira P, Loizon E, Geloen A, et al. Oil Composition of High-Fat Diet Affects Metabolic Inflammation Differently in Connection With Endotoxin Receptors in Mice. Am J Physiol Endocrinol Metab (2012) 302(3):E374–86. doi: 10.1152/ajpendo.00314.2011
67. Kazemian N, Mahmoudi M, Halperin F, Wu JC, Pakpour S. Gut Microbiota and Cardiovascular Disease: Opportunities and Challenges. Microbiome (2020) 8(1):36. doi: 10.1186/s40168-020-00821-0
68. Donertas Ayaz B, Zubcevic J. Gut Microbiota and Neuroinflammation in Pathogenesis of Hypertension: A Potential Role for Hydrogen Sulfide. Pharmacol Res (2020) 153:104677. doi: 10.1016/j.phrs.2020.104677
69. Jang AY, Ryu J, Oh PC, Moon J, Chung WJ. Feasibility and Applicability of Wireless Handheld Ultrasound Measurement of Carotid Intima-Media Thickness in Patients with Cardiac Symptoms. Yonsei Med J (2020) 61:129–36. doi: 10.3349/ymj.2020.61.2.129
70. Poller WC, Nahrendorf M, Swirski FK. Hematopoiesis and Cardiovascular Disease. Circ Res (2020) 126(8):1061–85. doi: 10.1161/CIRCRESAHA.120.315895
71. Zakirov FH, Zhang D, Grechko AV, Wu WK, Poznyak AV, Orekhov AN, et al. Lipid-Based Gene Delivery to Macrophage Mitochondria for Atherosclerosis Therapy. Pharmacol Res Perspect (2020) 8(2):e00584. doi: 10.1002/prp2.584
72. Karimzadeh MR, Zarin M, Ehtesham N, Khosravi S, Soosanabadi M, Mosallaei M, et al. MicroRNA Binding Site Polymorphism in Inflammatory Genes Associated With Colorectal Cancer: Literature Review and Bioinformatics Analysis. Cancer Gene Ther (2020) 27(10-11):739–53. doi: 10.1038/s41417-020-0172-0
73. Sowers JL, Johnson KM, Conrad C, Patterson JT, Sowers LC, et al. The Role of Inflammation in Brain Cancer. Adv Exp Med Biol (2014) 816:75–105. doi: 10.1007/978-3-0348-0837-8_4
74. Kaanane H, Senhaji N, Berradi H, Benchakroun N, Benider A, Karkouri M, et al. Association of Variants in IL6-Related Genes With Lung Cancer Risk in Moroccan Population. Lung (2019) 197(5):601–8. doi: 10.1007/s00408-019-00261-0
75. Kuzma J, Chmelar D, Hajek M, Lochmanova A, Ciznar I, Rozloznik M, et al. The Role of Intestinal Microbiota in the Pathogenesis of Colorectal Carcinoma. Folia Microbiol (Praha) (2020) 65(1):17–24. doi: 10.1007/s12223-019-00706-2
76. Rudnicka K, Backert S, Chmiela M. Genetic Polymorphisms in Inflammatory and Other Regulators in Gastric Cancer: Risks and Clinical Consequences. Curr Top Microbiol Immunol (2019) 421:53–76. doi: 10.1007/978-3-030-15138-6_3
77. Martinez-Campos C, Torres–Poveda K, Camorlinga–Ponce M, Flores–Luna L, Maldonado–Bernal C, Madrid–Marina V, et al. Polymorphisms in IL-10 and TGF-Beta Gene Promoter Are Associated With Lower Risk to Gastric Cancer in a Mexican Population. BMC Cancer (2019) 19(1):453. doi: 10.1186/s12885-019-5627-z
78. Razzaghi MR, Mazloomfard MM, Malekian S, Razzaghi Z. Association of Macrophage Inhibitory Factor -173 Gene Polymorphism With Biological Behavior of Prostate Cancer. Urol J (2019) 16(1):32–6. doi: 10.22037/uj.v0i0.3968
79. Zmorzynski S, Popek–Marciniec S, Szudy–Szczyrek A, Wojcierowska–Litwin M, Korszen–Pilecka I, Chocholska S, et al. The Association of GSTT1, GSTM1, and TNF-Alpha Polymorphisms With the Risk and Outcome in Multiple Myeloma. Front Oncol (2019) 9:1056. doi: 10.3389/fonc.2019.01056
80. Hnatyszyn A, Hryhorowicz S, Kaczmarek–Rys M, Lis E, Slomski R, Scott RJ, et al. Colorectal Carcinoma in the Course of Inflammatory Bowel Diseases. Hered Cancer Clin Pract (2019) 17:18. doi: 10.1186/s13053-019-0118-4
81. Kozlowski P, Miller DT, Zee RYL, Danik JS, Chasman DI, Lazarus R, et al. Lack of Association Between Genetic Variation in 9 Innate Immunity Genes and Baseline Crp Levels. Ann Hum Genet (2006) 70(5):574–86. doi: 10.1111/j.1469-1809.2005.00256.x
82. de Mare-Bredemeijer EL, Mancham S, Utomo WK, de Canck I, van Thielen M, de Meester E, et al. Genetic Polymorphisms in Innate Immunity Receptors Do Not Predict the Risk of Bacterial and Fungal Infections and Acute Rejection After Liver Transplantation. Transpl Infect Dis (2013) 15(2):120–33. doi: 10.1111/tid.12034
83. Haerynck F, Mahachie John JM, Van Steen K, Schelstraete P, Van daele S, Loeys B, et al. Genetic Variations in Toll-Like Receptor Pathway and Lung Function Decline in Cystic Fibrosis Patients. Hum Immunol (2013) 74(12):1649–55. doi: 10.1016/j.humimm.2013.08.282
84. Torok HP, Glas J, Tonenchi L, Mussack T, Folwaczny C. Polymorphisms of the Lipopolysaccharide-Signaling Complex in Inflammatory Bowel Disease: Association of a Mutation in the Toll-Like Receptor 4 Gene With Ulcerative Colitis. Clin Immunol (2004) 112(1):85–91. doi: 10.1016/j.clim.2004.03.002
85. Poikonen K, Lajunen T, Silvennoinen–Kassinen S, Leinonen M, Saikku P. Effects of CD14, Tlr2, TLR4, LPB, and IL-6 Gene Polymorphisms on Chlamydia Pneumoniae Growth in Human Macrophages In Vitro. Scand J Immunol (2009) 70(1):34–9. doi: 10.1111/j.1365-3083.2009.02267.x
86. Gonzalez-Quintela A, Alonso M, Campos J, Vizcaino L, Loidi L, Gude F. Determinants of Serum Concentrations of Lipopolysaccharide-Binding Protein (LBP) in the Adult Population: The Role of Obesity. PLoS One (2013) 8(1):e54600. doi: 10.1371/journal.pone.0054600
Keywords: inflammation, single nucleotide polymorphisms, lipopolysaccharide-binding protein, infections, inflammatory diseases, metabolic disorders, cancers
Citation: Meng L, Song Z, Liu A, Dahmen U, Yang X and Fang H (2021) Effects of Lipopolysaccharide-Binding Protein (LBP) Single Nucleotide Polymorphism (SNP) in Infections, Inflammatory Diseases, Metabolic Disorders and Cancers. Front. Immunol. 12:681810. doi: 10.3389/fimmu.2021.681810
Received: 18 March 2021; Accepted: 08 June 2021;
Published: 06 July 2021.
Edited by:
Anna Di Nardo, University of California, San Diego, United StatesReviewed by:
Maurizio Zanetti, University of California, San Diego, United StatesChangjiang Dong, University of East Anglia, United Kingdom
Copyright © 2021 Meng, Song, Liu, Dahmen, Yang and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao Yang, eXgxOTkyMDcxNkB5ZWFoLm5ldA==; Haoshu Fang, ZmFuZ2hhb3NodUBhaG11LmVkdS5jbg==
 Leilei Meng1
Leilei Meng1 Zichen Song
Zichen Song Anding Liu
Anding Liu Uta Dahmen
Uta Dahmen Xiao Yang
Xiao Yang Haoshu Fang
Haoshu Fang