- 1Center for HIV and STIs, National Institute for Communicable Diseases of the National Health Laboratory Service, Johannesburg, South Africa
- 2Medical Research Council (MRC) Antibody Immunity Research Unit, Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa
- 3Theoretical Division, Los Alamos National Laboratory, Los Alamos, NM, United States
- 4Center for the AIDS Programme of Research in South Africa (CAPRISA), University of KwaZulu-Natal, Durban, South Africa
Broadly neutralizing antibodies (bNAbs) are currently being assessed in clinical trials for their ability to prevent HIV infection. Single chain variable fragments (scFv) of bNAbs have advantages over full antibodies as their smaller size permits improved diffusion into mucosal tissues and facilitates vector-driven gene expression. We have previously shown that scFv of bNAbs individually retain significant breadth and potency. Here we tested combinations of five scFv derived from bNAbs CAP256-VRC26.25 (V2-apex), PGT121 (N332-supersite), 3BNC117 (CD4bs), 8ANC195 (gp120-gp41 interface) and 10E8v4 (MPER). Either two or three scFv were combined in equimolar amounts and tested in the TZM-bl neutralization assay against a multiclade panel of 17 viruses. Experimental IC50 and IC80 data were compared to predicted neutralization titers based on single scFv titers using the Loewe additive and the Bliss-Hill model. Like full-sized antibodies, combinations of scFv showed significantly improved potency and breadth compared to single scFv. Combinations of two or three scFv generally followed an independent action model for breadth and potency with no significant synergy or antagonism observed overall although some exceptions were noted. The Loewe model underestimated potency for some dual and triple combinations while the Bliss-Hill model was better at predicting IC80 titers of triple combinations. Given this, we used the Bliss-Hill model to predict the coverage of scFv against a 45-virus panel at concentrations that correlated with protection in the AMP trials. Using IC80 titers and concentrations of 1μg/mL, there was 93% coverage for one dual scFv combination (3BNC117+10E8v4), and 96% coverage for two of the triple combinations (CAP256.25+3BNC117+10E8v4 and PGT121+3BNC117+10E8v4). Combinations of scFv, therefore, show significantly improved breadth and potency over individual scFv and given their size advantage, have potential for use in passive immunization.
Introduction
Broadly neutralizing antibodies (bNAbs), isolated from a subset of HIV-1 positive individuals, are capable of neutralizing a wide range of HIV viruses. Crucially, bNAbs have been shown to provide protection in non-human primate studies and it is thought that such antibodies are needed for an effective HIV vaccine (1–4). However, to date, no candidate HIV vaccines have been able to elicit bNAbs in humans (5–8). This has led the field to actively explore the possibility of using bNAbs as biological drugs for passive immunization against HIV (9–13).
The results of the first efficacy trials of an antibody for HIV prevention tested in Africa and the Americas have recently been published (14–17). These two Antibody-Mediated Prevention (AMP www.ampstudy.org.za) trials showed that VRC01 had 75% prevention efficacy in high-risk men and women if the infecting virus was sensitive to the antibody at <1μg/ml (IC80). Therefore, to target the extensive envelope diversity, minimize escape, and provide sufficient potency a combination of multiple antibodies will be needed. Several studies have investigated the potential of antibody combinations and observed, as expected, an increase in breadth and potency (18, 19). These studies demonstrate that the complementary neutralization profiles of individual bNAbs can improve the overall breadth and provide higher coverage of multiclade panels of viruses at much lower antibody concentrations (18, 20). By using those antibodies that specifically target the HIV subtypes predominant in a specific area, a geographically relevant set of antibodies may be selected to provide optimal coverage and potency (20). For example, CAP256.25, which shows high potency against clade C viruses, is currently being assessed in combination with PGT121 and VRC07-523LS in dual and triple combination in the South African CAPRISA 012B trial (21, 22). Similarly, there are several ongoing phase 1 trials, testing multispecific antibodies or dual and triple combinations. These trials test the aforementioned antibodies in addition to V3 (10–1074) and V2 (PGDM1400) antibodies and the broadly neutralizing MPER-targeting antibody 10E8v4 (23, 24).
The ability to accurately predict the breadth and potency of antibody combinations without experimental validation enables the rapid identification of optimal combinations. Two models have been used to predict the IC50 and IC80 of antibody combinations based on single antibody titers, the Loewe Additive model, and the Bliss-Hill Independence model. Both models assume that there is no interaction between the different antibodies and that neutralization by combinations of antibodies will be additive, however, owing to different formulations of independence, their predicted results differ (18, 25). These models can also be used to determine whether synergy or antagonism occurs by comparing the predicted data with experimental results. When using the Loewe Additive model most combinations of two antibodies were demonstrated to show additive potency where the experimental potency was close to the predicted IC50 (19). This model in comparison to experimental results has in cases also indicated synergy or antagonism between antibodies targeting specific epitopes such as the CD4 binding site, MPER, and V1/V2 antibodies. However, comprehensive analyses have shown that the Bliss-Hill model tended to be better at predicting IgG combination titers (18, 19) and this model did not predict synergy between these epitopes when predictions were compared to experimental results.
Experimental synergy between anti-HIV antibodies has only been rarely observed, and only in the context of bispecific antibodies. A bispecific antibody that simultaneously engaged the V2 and V3 epitopes showed moderate levels of synergy (26). Another bispecific employing a CAP256.25 scFv and the antibody binding fragment (Fab) of 10-1074, showed moderate levels of improved potency against a few viruses (26). In this case, neutralization was compared to single scFv-Fc or IgG rather than to the predicted combination titers or experimental combinations of the two arms, which may have overestimated the level of synergy (20, 26–30). More convincing evidence of synergy was observed when antibodies targeting a host cell protein and the viral Env protein were combined particularly as part of bispecific or trispecific antibody constructs. This effect is due to the localization of the anti-HIV antibody close to the host cell membrane through CCR5 or CD4 binding, for example, the 10E8-iMab, which targets the MPER region on the HIV virion and the CD4 receptor on the HIV target cell. This bispecific antibody has a geometric mean IC50 potency of 0.002μg/mL compared to 0.4μg/mL for 10E8 and 0.05μg/mL for the iMab indicating a 25-fold improvement over the expected activity (27, 31).
Single chain variable fragments (scFv) are small molecules, which contain the variable heavy and light chain of antibodies connected through a glycine linker. These molecules have some enhanced pharmacokinetic properties such as improved distribution and absorption into mucosal tissues despite a loss of half-life due to lacking an Fc region (32–35). They may also display less steric hindrance when used in combination with other molecules and other bNAbs or scFv (36). This may be especially true for epitopes in close proximity such as the V2 and V3, or V3 and CD4bs. A recent study demonstrated that scFv targeting the V3 and CD4bs could display synergy when combined with fusion inhibitors (37).
We previously demonstrated that scFv of bNAbs retain significant breadth and potency against a multiclade panel of viruses despite potency differences linked to differential affinity for the epitope (38). In particular, 10E8v4 maintained the same breadth and most of its potency as an scFv. Single antibodies may be limited in their ability to prevent HIV infection, however, as evidenced by the recent AMP results. We, therefore, tested combinations of scFv that target major bNAb epitopes on the HIV trimer, namely CAP256.25 (V2 apex), PGT121 (N332-supersite), 3BNC117 (CD4bs), 8ANC195 (gp120-gp41), and 10E8v4 (MPER) and show that they generally follow a model of additive potency and complementary breadth. No significant antagonism or synergy was observed compared to the models, although antibody combinations tested against individual viruses could show variation. Overall, combinations of three scFv antibodies reached considerable breadth and potency indicating that scFv in combinations should be further investigated for passive immunity purposes.
Methods
scFv Construction
scFv were designed and cloned previously as described (38). In short, single constructs containing the variable heavy and light chain interspaced with a 15 or 18 amino acid glycine-serine linker of five HIV-directed bNAbs (CAP256.25, PGT121, 3BNC117, 8ANC195, and 10E8v4) were generated through overlapping PCR or ordered from GenScript (New Jersey, USA) (38). These scFv genes were then cloned into a CMV/R expression plasmid (AIDS Reagent Program, Division of AIDS, NIAID, NIH. For the lambda chain of PGT121, the pBR322 based lambda expression vector was used (AIDS Reagent Program, Division of AIDS, NIAID, NIH).
scFv Protein Expression
The constructs were grown in JM109 bacterial cells and extracted using a plasmid Maxiprep kit (Qiagen, Hilden Germany). Sequences were confirmed using the Applied Biosystems 3500xL Genetic Analyzer. Constructs were expressed as previously described (39). In short, HEK293F suspension cells at 1.5x106 to 2x106 cells/ml were cotransfected with linear Polyethylenimine hydrochloride (molecular weight, 40,000) at a 3:1 ratio with 1μg of plasmid per 1 ml of culture. Supernatants were harvested after 6 days.
scFv proteins were purified using Ni-Sepharose beads (GE Healthcare, Massachusetts USA), washed using a 30mM imidazole–phosphate-buffered saline (PBS) solution and eluted using 400mM imidazole in PBS. Glycerol was added to the elution at a final concentration of 5% to limit aggregation. Eluates were applied to Hiload Superdex 75 or Superdex 200 columns (GE Healthcare) equilibrated with PBS at pH 6.5 (5% glycerol with 0.02% sodium azide). The fractions corresponding to the size of the scFv were collected, pooled, and concentrated using Vivaspin concentrators or Vivapore static concentrators (GE Healthcare). The samples were dialyzed overnight at room temperature to remove sodium azide. Concentrations were measured on a NanoDrop device (Thermofisher, MA, USA), with extinction coefficients at 1% calculated using Expasy ProtParam (40) and characterized by SDS-PAGE. Molar weight was determined by using Expasy ProtParam (CAP256.25: 31.35kDa, PGT121: 28.89kDa, 3BNC117 28.47kDa, 10E8v4: 29.30kDa and 8ANC195 29.00kDa). scFv proteins were stored at -75°C.
IgG Production
IgG constructs were expressed in HEK293F cells as described previously (39). Supernatants were harvested after 6 days and purified using a protein A affinity column. Proteins were eluted using a 0.15M glycine buffer at pH 2.5 buffer into 1M Tris, pH 8, and were concentrated and dialyzed into PBS pH 6.5 containing 5% glycerol. Concentrations were measured on a Nanodrop using an Extinction Coefficient of 13.7 at a 1% solution. The molecular weight of the IgG was calculated using the Expasy ProtParam (40) of CAP256.25 (150.71kDa), 10E8v4 (147.29kDa), 3BNC117: (146.24kDa), PGT121: (146.2kDa), 8ANC195: (147.43kDa). IgG and proteins were stored at -75°C.
Pseudovirus production for TZM-bl Assay
Plasmids containing HIV-1 envelope (gp160) genes cloned in the pcDNA™3.1D/V5-His-TOPO® vector were co-transfected with pSG3Δenv into HEK293T cells and cultured for 48-72hrs at 37°C. Supernatants were harvested and filtered through a 45μm filter and frozen at -80°C. Virus stocks were titrated on TZM-bl cells using a luciferase assay to determine a dilution yielding RLU at least 10-fold above the “cell only” background (40,000-100,000 RLU).
Neutralization Assay
A panel of 43 viruses (41) plus BG505 N332 and CAP256_SU (CAP256.3mo.9C) (42, 43) representing HIV-1 clades A, B, and C was used to compare neutralization titers of IgG and scFv. Neutralization assays were performed in TZM-bl cells as described previously (44–46). Proteins were tested at 200μg/mL for the IgG (~146kDa) and 50μg/mL for the scFv (28-32 kDa). All assays were repeated at least twice. IC50 and IC80 of each antibody tested was calculated and geometric mean potency was calculated for both IgG and scFv using sensitive viruses only.
Experimental Testing of scFv Combinations
Combinations were tested by adding equimolar amounts of two or three scFv proteins in a neutralization assay as described above. A panel of 17 subtype A, B and C viruses were selected based on their sensitivity to at least 2 of the scFv in order to test neutralization of scFv combinations and confirm the Loewe Additivity and Bliss-Hill Independence models. Pre-dilutions containing scFv at 2μM and 10μM each were used to facilitate the assay set up. As a control, the single scFv were diluted to 10μM and 2μM as well and run alongside the combinations as a comparison. The highest concentration tested for the combinations was 30μg/mL or 1000nM. IC50 and IC80 of each experimental antibody combination was calculated, where each antibody is present at the concentration determined at IC50 or IC80.
Loewe Additivity and Bliss-Hill Independence Models
The IC50 and IC80 values from experimental combinations were compared to the predicted IC50 and IC80 based on the Loewe Additive model and the Bliss-Hill model as previously described (18, 19).
The following formula is used to calculate Loewe Additivity.
Post analysis values were recalculated from nM into μg/mL based on the following formula.
Where (n) is the number of antibodies, Mw the molecular weight of the antibodies in the combination, and IC50 the experimental IC50 or theoretical IC50 obtained. The same formula is used for IC80 values replacing the IC50 with IC80 in the formula above. For resistant viruses, the model assumes the titer of the active scFv.
For the Bliss-Hill Independence model, the following formula was used to calculate the Hill function
Where c= bNAb concentration, k = IC50, and
The combination neutralization curve is then calculated using the Bliss Independence model,
with f(A), f(B), etc. being the individual functions of the scFv antibodies. Combination molar IC50 and IC80 titers are calculated by setting f = 0.5 or 0.8 and assuming each scFv is present at the same molarity, and converted to µg/ml using the above formula.
Dual/triple coverage was calculated by considering a virus resistant if less than 2/3 antibodies in the combination were able to neutralize that virus at set concentrations.
We used the following formula for both models to determine which is more accurate in predicting the combination potencies for IC50 and IC80.
Synergy and/or Antagonism Predictions Based on Loewe Additivity and Bliss-Hill Independence Models
Synergy was predicted based on whether the experimental IC50 and IC80 were improved compared to the Loewe Additive model or the Bliss-Hill model. The formula below was used to characterize this effect, with the Bliss-Hill IC50 replacing the Loewe IC50 in the formula below.
Positive values indicate experimental titers lower (i.e. more potent) than predicted and imply synergy, while negative values indicate less potent experimental titers than predicted and imply antagonism. For a given combination of antibodies, the mean and the 95% Confidence interval were calculated using LogFold values for this combination against each virus in the panel.
Statistics
All statistics were done using the GraphPad Prism 8 software. Fold differences of the experimental combinations were calculated with the most potent scFv in the mixture for both IC50 and IC80. A Wilcoxon signed-rank test was performed to test which model was more accurate at predicting the experimental titers for both IC50 and IC80. Pearson’s Correlation between experimental neutralization titers and titers based on the two models was calculated using the GraphPad software. A nonlinear fit model (log-log) was used to predict the slope between the experimental and predicted data, using a robust regression. A log-fold with a 95% confidence interval was used to determine if the fold difference is significantly different from 0, indicating either synergy or antagonism. Breadth-potency curves were drawn using a survival model in GraphPad. Significance was tested using the log-rank Mantel-Cox significance test.
Results
Dual and Triple Combinations of scFv of HIV bNAbs
To assess whether scFv of HIV bNAbs showed increased breadth and potency when used in combination, we tested five different scFv as part of dual and triple combinations. The scFv included those targeting the V2 (CAP256.25), the N332 supersite (PGT121), the CD4bs (3BNC117), and the MPER region (10E8v4), all of which were previously shown to retain significant activity compared to IgG (38). For this study, we added the antibody 8ANC195 that targets the gp120-gp41 interface so that all five major epitopes on the HIV trimer were covered (see single IC50 and IC80 data in nM Supplementary 1A and 1B and in µg/ml 1C and 1D respectively, (38)]. All scFv were expressed and purified by size exclusion columns and, with size and purity confirmed SDS-PAGE gels. scFv were stored in buffers containing 5% glycerol as determined previously to prevent aggregation (38).
Combinations of scFv were tested against a panel of 17 pseudoviruses from subtypes A, B, and C (n=3, 4 and 10, respectively). These were selected based on their sensitivity to at least three of the five scFv under investigation. Equimolar amounts (based on their molecular weights), of each scFv in combinations of two or three were tested, giving a total of 20 different combinations of antibodies (10 dual and 10 triple combinations), which were compared to single scFv neutralization titers. To standardize the output data, the concentration of the single scFv and the scFv combinations in μg/mL were calculated from the nM titers and the molecular weight of the scFvs (Figure 1, Supplementary Figure 2) (38). Combination titers IC50 and IC80 are reported as the concentration of each scFv in the mix in μg/mL or nM.
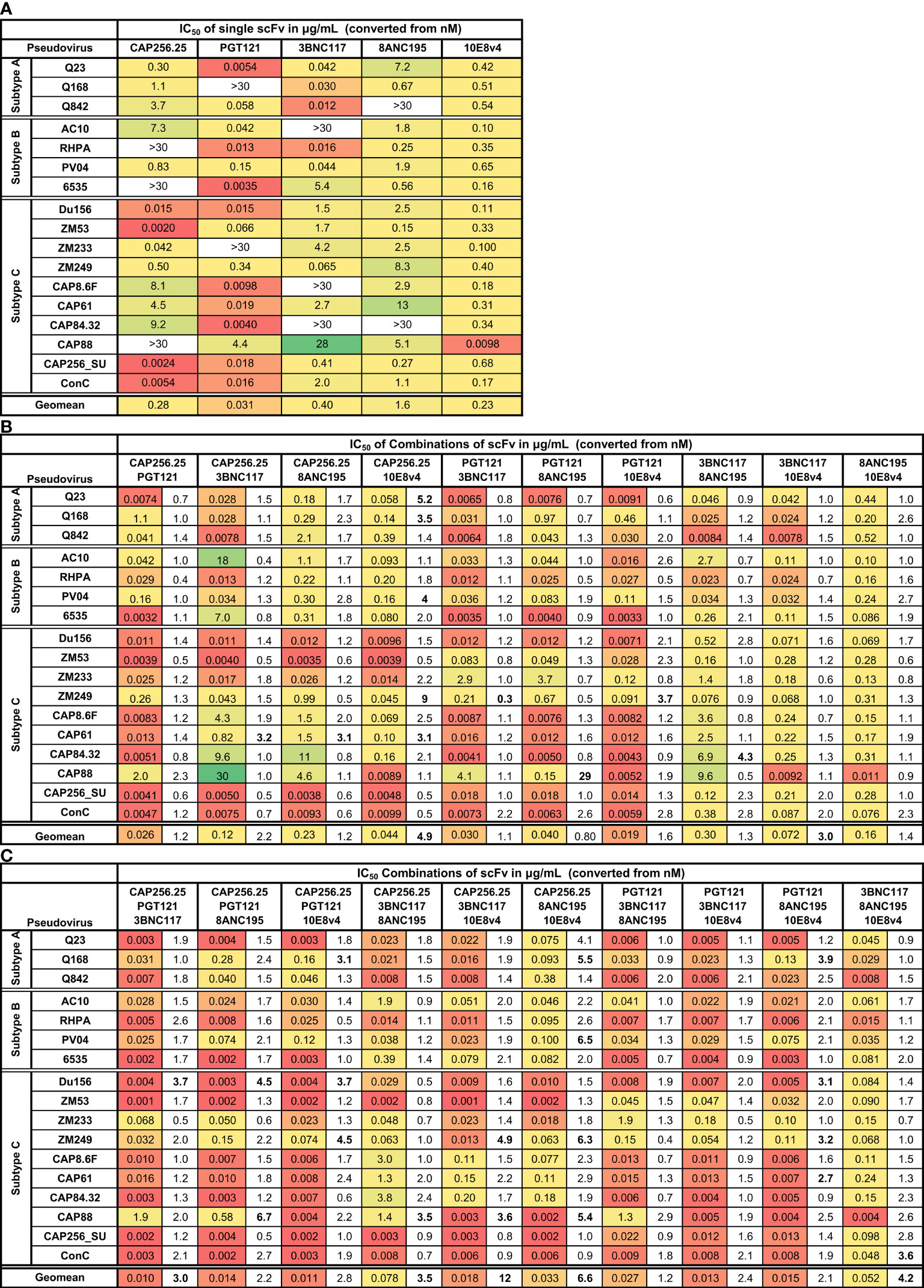
Figure 1 Neutralization titers of single, dual, and triple combinations of scFv. Heat map showing IC50 neutralization titers in μg/mL for single (A), dual (B), and triple (C) combinations of scFv against a panel of 17 subtype A, B, and C viruses. Viruses insensitive to individual bNAbs are shown as >30μg/mL. The fold improvement in IC50 titers of the dual and triple combinations relative to the IC50 of the best scFv in the combination is included in (B, C). Values with >3-fold increase or decrease in neutralization are shown in bold. Geometric mean potency is included at the bottom of each table. Each scFv is present in the combination at the titer indicated.
As expected, combinations of two scFv improved the coverage, with active scFv making up for the inactive scFv. For all dual and triple combinations, breadth reached 100% at IC50 against this 17-virus panel (Figure 1). The neutralization of individual viruses by two or three scFv was usually similar to the IC50 of the most potent scFv in the combination (Figure 1; fold differences are shown in the columns next to the IC50s). Except for a few cases, the virus neutralization titers of the dual and triple combinations fell within 3-fold of the titer of the most potent single scFv in that combination. There was little to no loss of potency noted compared to the most potent scFv for the entire panel for all 20 scFv combinations tested (a total of 340 single test titers). A few instances of potential gain of potency compared to the most potent scFv were noted, with most of these linked to specific scFv combinations (bolded values in Figure 1).(See Supplementary Figure 2 for IC50 titers in nM (A) and μg/mL (B) and for IC80 values in nM (C) and μg/mL (D) respectively).
The combination of CAP256.25+10E8v4 scFv stood out, demonstrating an overall 4.9-fold improvement in geometric mean titer (Figure 1B). This was driven by five viruses of which two were subtype A, one was subtype B and two were subtype C indicating this was not subtype-specific. Five other dual scFv combinations showed improved potency over single scFv for single viruses but this was not linked to any specific combination of antibodies. The 3BNC117+10E8v4 combination showed an overall significant improvement in the geometric mean titer although this was not seen for individual viruses indicating complementarity of neutralization potency of the scFv rather than synergy.
For the triple combinations, five of the ten combinations showed >3-fold improved geometric means compared to the most potent single scFv with a 12-fold improvement for the CAP256.25+3BNC117+10E8v4 combination (Figure 1C). This was higher than for the dual combinations where only two out of ten showed an improvement in geometric mean potency and was driven by improvement in potency against single viruses (16 of the 170 virus-scFv combination pairings). This was due to the scFv (e.g. CAP256.25 and 3BNC117 scFv), in the combinations being potent against different viruses allowing for complementarity in neutralization potency and coverage. Triple combinations overall showed higher improvements in breadth compared to dual combinations or single scFv as a consequence of having more antibodies.
Some viruses appeared to be more sensitive to the effects of combined scFv. For example, a slight potency improvement was observed against the CAP61 and ZM249 viruses for the dual combinations, although for the latter loss of potency was also noted (fold difference <0.33 see Figure 1B). Some enhancement in potency was also seen with these viruses plus Du156 and CAP88 for the triple combinations (Figure 1C).
Overall, we found either improved or similar titers for combinations compared to the most potent single scFv, particularly for triple combinations, indicating that combinations of scFv improve the coverage of a panel of viruses and the potency at which viruses are neutralized. We overlaid neutralization curves of the combinations with the single scFv used in the combination, to determine if there were distinct patterns associated with improved IC50s or where we observed similar IC50 for the combinations compared to the single scFv. For most dual and triple combinations (314/340), an additive effect was seen where the combination curve overlapped with the best scFv in the combination (Figure 2A). Here IC50 did not show a potency improvement compared to the single scFv. A few combinations shifted the curve to the left compared to the curve of the most potent single scFv corresponding with fold potency improvements as seen in Figure 1 and indicating potential synergy (10/170 virus-dual combination pairings and 16/170 in the triple combinations showed improved IC50s) (Figure 2B). In one case we observed decreased potency for the combination compared to the most potent scFv possibly indicating some antagonism (Figure 2C). With the addition of a third antibody this was partially negated (Figure 2C right panel). Overall, most combinations did not show significant potency improvements or loss of potency, suggesting that they largely followed an additive model of potency.
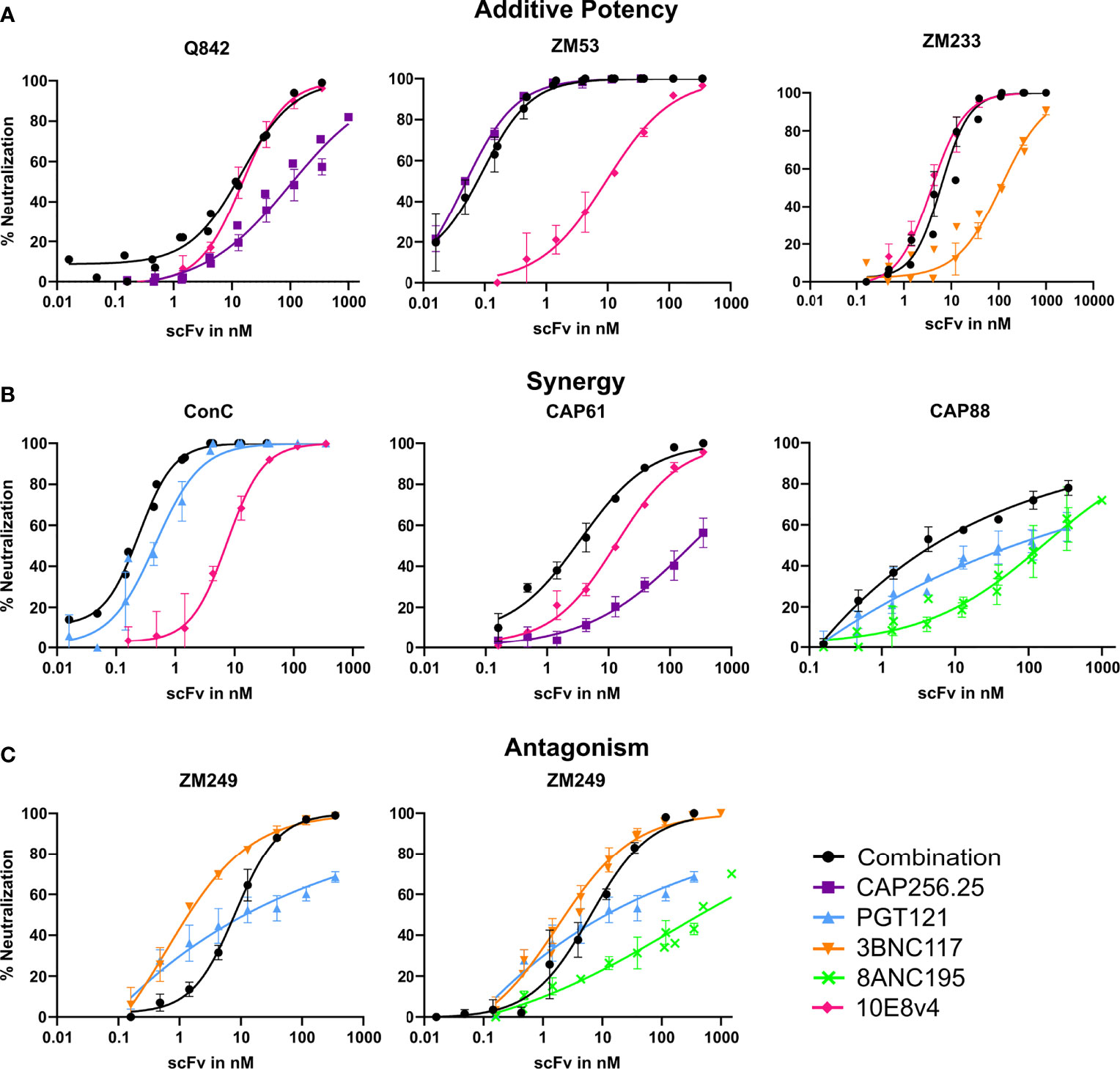
Figure 2 Combinations of scFv that show improved neutralization potency over single scFv. Neutralization curves showing potential additive, synergistic and antagonistic potency effects of combinations of antibodies. Combinations of scFv compared to single scFv neutralization curves with CAP256.25 in purple, PGT121 in light blue, 3BNC117 in orange, 8ANC195 in green and 10E8v4 in pink. The dual or triple combinations are represented in black. Geometric mean values are used for each data point with error bars representing repeat experiments. (A) Examples of combinations where neither synergy nor antagonism is observed for three viruses tested against dual combinations of scFv. Additive potency is represented by the combination curve (black) overlapping with the best scFv in the mixture. (B) Potential synergy as observed in dual combinations of scFv against three different virus strains. Synergy was represented by a left shift of the combination curve (black) relative to the most potent scFv in the mixture, or by steeper neutralization curves resulting in improved IC80. (C) Potential antagonism as represented by a right shift of the combination curve relative to the most potent scFv in the mixture, observed in combinations of two or three scFv tested against ZM249, which was sensitive to all scFv in the combinations.
scFv Follow an Additive Model of Interaction in HIV Neutralization
To further explore whether combinations of scFv follow an additive model of interaction we compared the experimental results with predictions based on the single scFv titers using the Loewe Additive model and the Bliss-Hill models. As mentioned before, both these models assume no synergistic or antagonistic interactions between antibodies (18, 19). We calculated the combination scFv IC50 and IC80 titers based on these 2 models (Methods) and based on the geometric mean IC50 and IC80 of the repeats of single scFv obtained in the experiment. All scFv predicted data titers were compared to the IC50 and IC80 titers of the experimentally tested combinations.
There was a strong significant (p<0.0001) correlation between the experimental IC50 and IC80 and the predicted IC50 and IC80 of the dual combinations for both the Loewe and Bliss Hill model (Loewe, Pearson’s r=0.94 and r=0.79 respectively, Bliss-Hill, r=0.94 and 0.91 respectively) (Figures 3A, B). Similarly, for the triple combinations, there was a strong correlation between the predicted values and the experimental values (Loewe r=0.78 and r=0.79, Bliss-Hill: r=0.70 and r=0.95 respectively) (Figures 3C, D). For each combination, we also determined which model was better by comparing the mean absolute log difference between the experimental values and the predicted values for each individual combination. For the dual combinations, the Bliss Hill model was better at predicting two out of the 10 combinations (10E8v4 combined with CAP256.25 or PGT121), whereas no significant difference between the models was observed for any of the other combinations (Figures 3E, F). Similarly, for the triple combinations, most combinations did not show differences between the two models in the IC50 with the Bliss-Hill model being better at predicting two combinations (8ANC195 + 10E8v4 with either CAP256.25 or PGT121) (Figure 3G). For the IC80 of the triple combinations, 6/10 combinations were significantly better predicted by the Bliss-Hill model compared to the Loewe model (Figure 3H). This indicated that for most combinations both Loewe and Bliss-Hill could predict combination IC50 titers well with a slight advantage for the Bliss-Hill model. However, the Bliss-Hill model is significantly better at predicting triple IC80 titers compared to the Loewe model, with the latter underestimating the potency of the triple combinations at IC80.
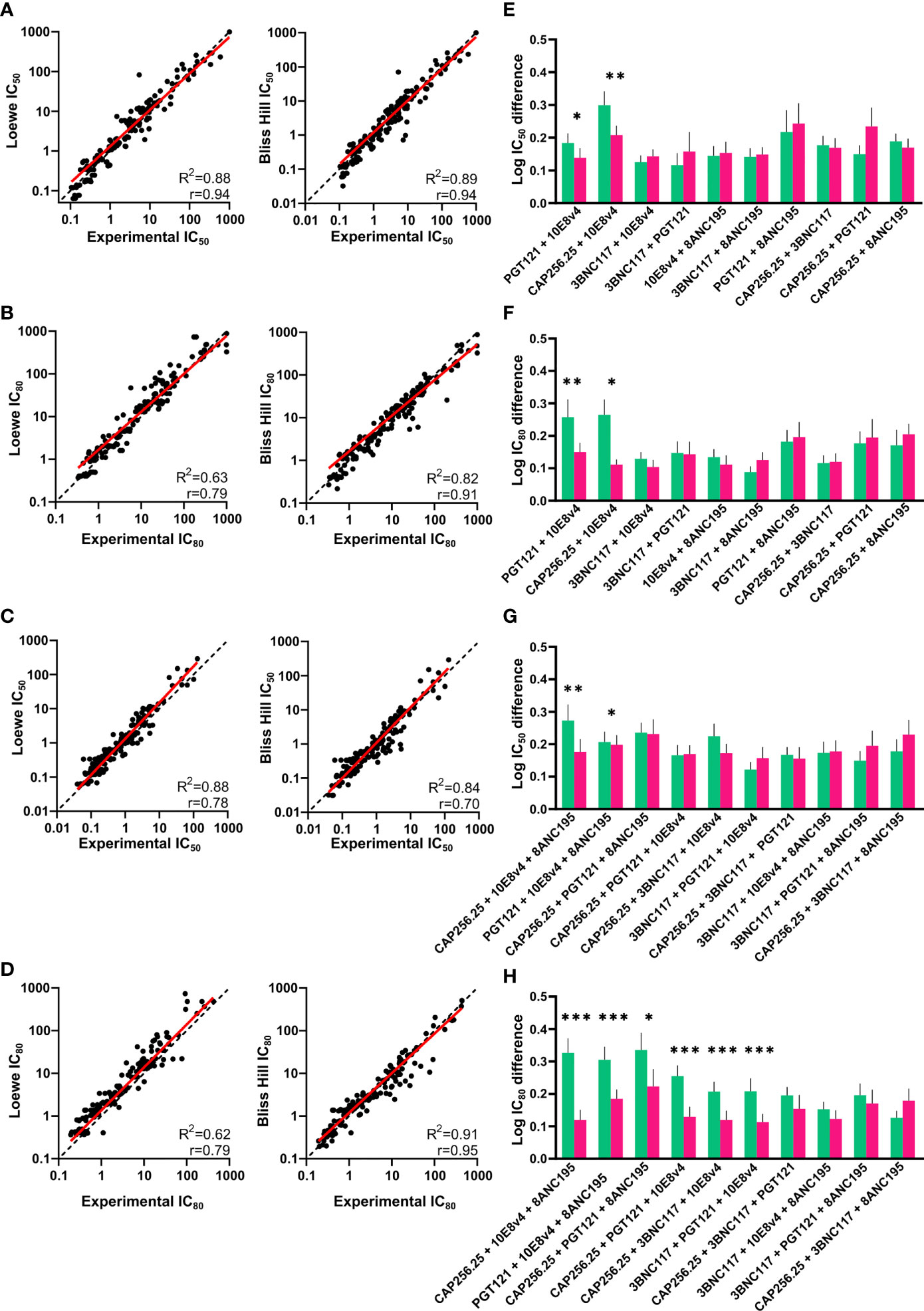
Figure 3 Comparison of experimental and predicted combinations of dual and triple scFv. (A, B) IC50 and IC80 titers of dual antibody combinations plotted against the predicted IC50 and IC80 titers according to the Loewe Additive (left) and Bliss-Hill Independence (right) models. (C, D) Predicted IC50 and IC80 titers of triple combinations versus the experimental IC50 and IC80 titers. Values where both or all titers of single scFv >30μg/mL are excluded. (E, F) Comparison of the absolute Log(IC50) and Log(IC80) difference between the experimental titers and predicted titers according to the Loewe Additive (green) and Bliss-Hill independence (purple) for the dual combinations. (G, H) Comparison of the absolute Log(IC50) and Log(IC80) difference between the experimental titers and predicted titers according to the Loewe Additive (green) and Bliss-Hill independence (purple) models for the triple combinations. For (A, B, E, F), a nonlinear robust regression log-log line (red) and an equity line (black dotted) as well as r, R2, are shown and p<0.0001. For (C, D, G, H), mean values are given with a standard error of the mean. A paired, Wilcoxon t-test was performed on each pair with significant p values above the graphs (p < 0.05 = * p < 0.01 = ** p < 0.001= ***).
No Synergy or Antagonism Observed for scFv Combinations Against a Panel of HIV Pseudovirus
We next sought to determine if any of the combinations showed significant potency improvement or potency loss in either IC50 or IC80 by comparing the experimental data to the expected titers based on the Bliss Hill model. These analyses allowed us to determine if there is potential synergy (higher potency than predicted), as indicated by a log-fold difference >0.4 (~2.5 fold) and/or antagonism (lower potency than predicted) as indicated by a log-fold difference <-0.4 (Figure 4). The dual combinations had a mean log-fold difference close to 0 (log-fold -0.4<CI<0.4) for both IC50 and IC80 indicating that there was neither antagonism nor synergy, although a few antibody combinations showed some improved potency (red dots, IC50 n=3, 1.7% and IC80: n=0, 0%) or decreased potency (blue dots, IC50 and IC80: n=6, 3.5%) for individual viruses these were relatively few and most showed less than 0.4 log fold difference when compared to model predictions (Figures 4A, B).
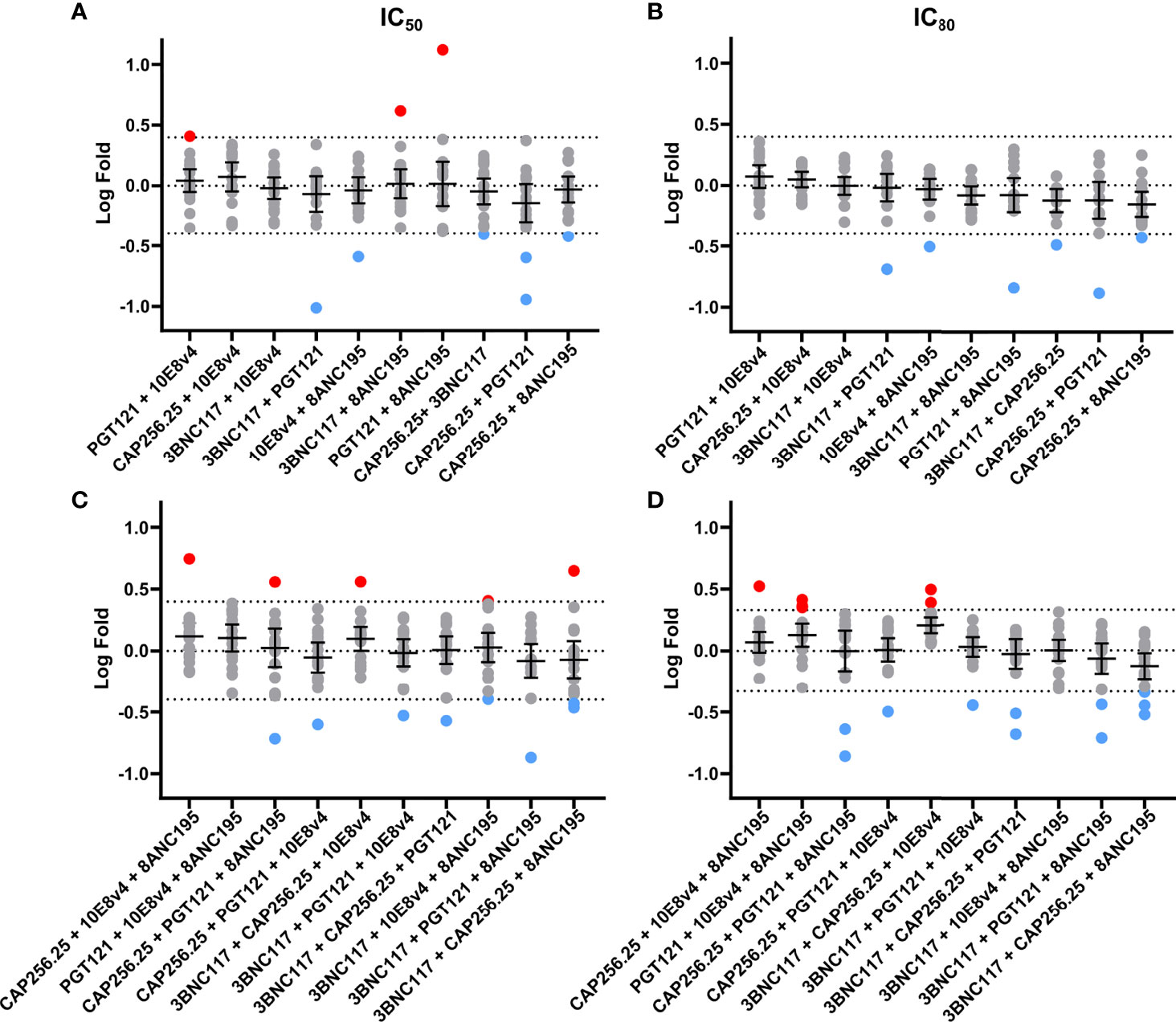
Figure 4 No synergy observed using fold difference between experimental and Bliss-Hill predictions of dual and triple scFv combinations. Log-fold differences of double and triple IC50 (A, C) and IC80 (B, D) experimental combinations to the Bliss-Hill Independence model prediction. Mean and the 95% confidence interval are shown with a dotted line indicating no fold change. Log (2.5) fold difference dotted lines (0.4 or -0.4) are also shown. Red and blue are used to indicate more than a log difference of 0.4 (~2.5x) with red dots indicating potential synergy and blue dots indicating potential antagonism.
For the triple combinations, similar results were observed with 157/170 of the IC50 and 154/170 of the IC80 values showing less than a Log (2.5) difference. The mean on triple combinations was also close to 0 (log-fold -0.4<CI<0.4) for both IC50 and IC80. Similarly, only a few individual viral titers showed a potential loss of potency for the IC50 (n=8, 4.7%) and IC80 (n=11, 6.5%) or a potential gain of potency (IC50 and IC80: n=5, 3%) for triple combinations compared to the model (Figures 4C, D). None of the outliers in dual or triple combinations could be linked to specific viral signatures (data not shown). Notwithstanding these few outliers, we, did not observe any significant synergy nor antagonism for dual or triple combinations of scFv.
Using the Bliss-Hill Model to Predict IC50 and IC80 Titers of scFv Combinations Against a Larger HIV Panel
In order to assess their breadth-potency on a larger virus panel, we used the IC50 and IC80 data of single scFv to predict neutralization titers for combinations of two and three scFv, now including 8ANC195, on a 45-virus panel described in our previous study (38) (see Supplementary Figure 1 for single IC50 in nM (A) and IC80 in nM (B) or μg/mL (C, D) titers) (38). These theoretical scFv combinations were ranked based on their geometric mean potency using both IC50 and IC80 data for dual and triple combinations (Figures 5A, B, E, F). Combinations with the expected highest breadth and potency were plotted using breadth-potency curves (Figures 5C, D, G, H).
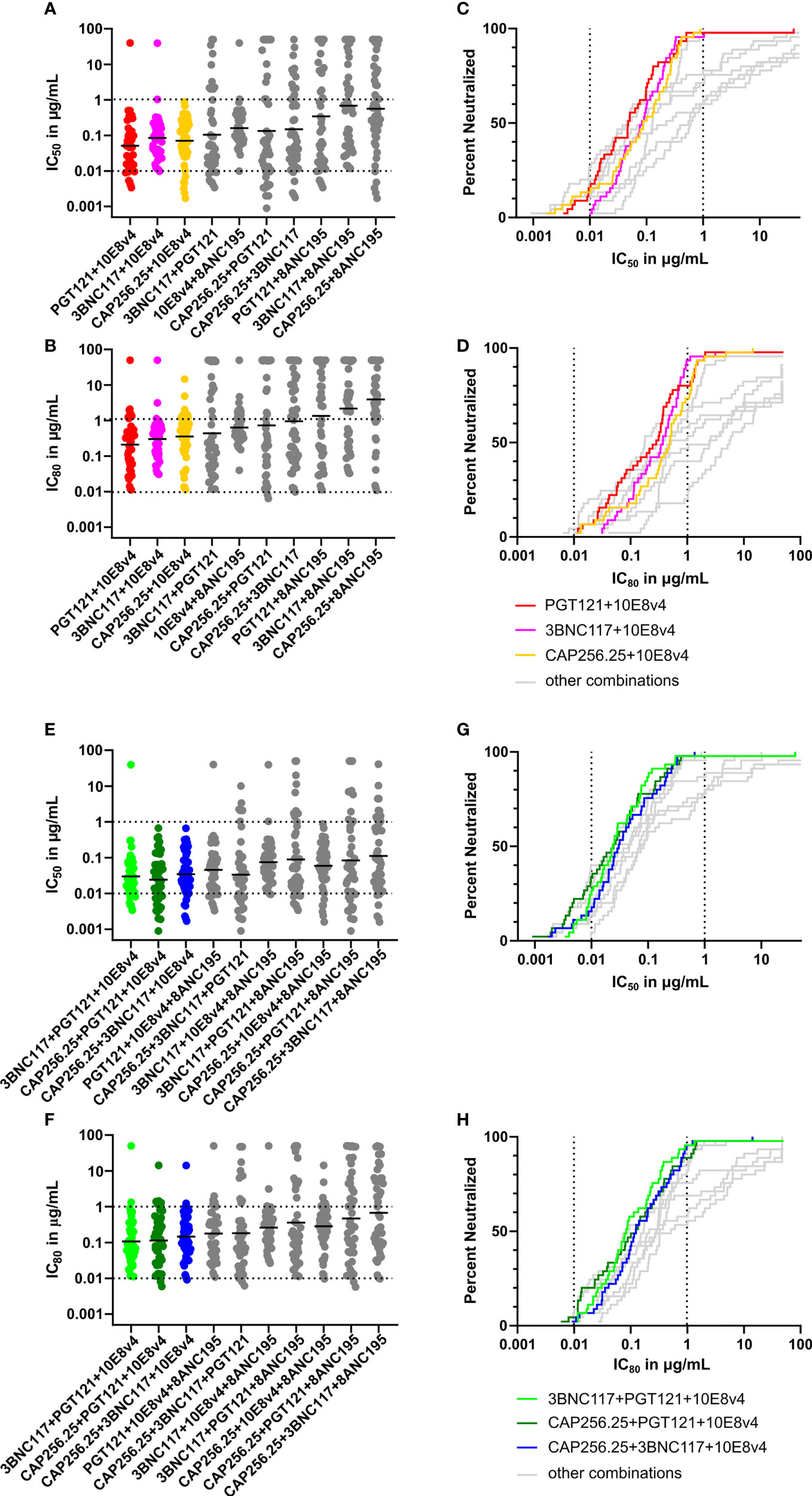
Figure 5 Predicted breadth and potency of scFv combinations against large virus panel. (A) Scatter plot showing predicted IC50 titers of dual combinations based on single scFv data for a panel of 45 viruses using the Bliss-Hill Independence model. The IC50 data is plotted in μg/mL recalculated from nM based on the formula given in the methods section. (B) Predicted IC80 titers of dual combinations based on the Bliss-Hill Independence model ranked by geometric mean. (C) Breadth-potency curves of IC50 titers of most potent and broad dual combinations were plotted for the IC50.with the others in grey. (D) Breadth-potency curves from IC80 data for the most potent and broad dual combinations. (E) IC50 titers for triple combinations based on the Bliss-Hill Independence model. (F) Predicted IC80 titers for the triple combinations ranked based on geometric mean. (G) Breadth potency curves for the IC50 of the most potent and broad triple combinations are plotted. (H) The breadth-potency curves are based on IC80 titers of the most potent and broad combinations. The maximum neutralization for the IC50 and IC80 data is set at 50μg/mL for all plots. Geometric mean is indicated by a black line in A, C, E, and G Dotted lines are shown at 0.01μg/mL and 1μg/mL for breadth-potency plots IC50 and the IC80 (B, D, F, H). Each scFv is present in the combination at the titer indicated.
The best dual combinations, namely PGT121+10E8v4 (red), 3BNC117+10E8v4 (pink), and CAP256.25+10E8v4 (orange) were able to neutralize most viruses barring one at IC50 and IC80 (Figures 5A, B). The breadth-potency curves of the three 10E8v4-containing combinations reached 100% neutralization at the IC50 and 98% neutralization at the IC80 (colored breadth-potency plots versus grey, Figures 5C, D) with most viruses neutralized between 0.1 and 1μg/mL at IC80. The fourth combination of 10E8v4 with 8ANC195 was less effective due to the low potency of both antibodies and the limited breadth of 8ANC195, [see Supplementary Figure 3 (38)]. The other dual combinations showed reduced breadth (up to 30% at 50μg/mL) or reduced potency especially at IC80 (grey dots and lines). Some combinations such as PGT121+3BNC117 had a potent geometric mean below 0.1μg/mL at IC50, but there was a larger spread of neutralization titers and so breadth was lost at IC80 (Figures 5A, B). (See Supplementary Figure 4 for IC50 in nM (A) and μg/mL (B) and IC80 in nM (C) and μg/mL (D) titers of dual, and triple combinations predicted by the Bliss-Hill Independence model).
The triple combinations showed higher levels of potency and complete neutralization with 8/10 combinations showing complete neutralization of the viral panel at IC50, and 6/10 showing near-complete neutralization at IC80 (Figures 5E–H). Moreover, 7/10 triple combinations had a geometric mean potency below 0.1μg/mL at IC50 (Figure 5E, green, dark green, and blue). Of these, three combinations all had a geometric mean IC80 below 1μg/mL and neutralized up to 96% of viruses (Figures 5F, H). This indicated that triple combinations are able to reach nearly 100% neutralization at lower concentrations for both IC50 and IC80 (see Supplementary Figure 4 for IC50 in nM (A) and μg/mL (B) and IC80 in nM (C) and μg/mL (D) titers of dual, and triple combinations predicted by the Bliss-Hill Independence model).
The predicted breadth of each combination was next determined at four concentrations: 0.1, 1, 10, and 50μg/mL in addition to the geometric mean for the combinations tested (with a cutoff at 50μg/mL). Combining these scFv improved the coverage at all concentrations for both IC50 and IC80 (Figure 6). Breadth for dual combinations ranged from 84% to 100% at IC50 < 50µg/ml and 80% to 100% at IC80 < 50µg/ml. For dual combinations, 100% breadth was reached for one combination at IC50 < 1μg/mL, and for four combinations at IC50 < 50μg/mL. For the IC80, 98% breadth was obtained at 10μg/mL for the same combinations. Most combinations of three scFv had 100% breadth at IC50 < 50 µg/ml, except for PGT121+3BNC117+8ANC195 and CAP256.25+PGT121+8ANC195, which had a breadth of 96% and 93% respectively. Breadth was especially improved for lower concentrations at IC80. For example, breadth for single scFv ranged from 0 to 44% for IC50 < 0.1µg/ml and 0 to 24% for IC80 < 0.1µg/ml, whereas dual combinations ranged from 22 to 62% for IC50 and 2 to 29% for IC80. At the same cutoff, the breadth for combinations of 3 antibodies ranged from 47% to 82% for IC50 and 16 to 40% for IC80. Geometric mean potency for the panel also improved from 0.2-4.95 μg/mL for single scFv to 0.27-2.08μg/mL for dual combinations and 0.09-0.61μg/mL for triple combinations at IC80. This was expected as the titers were calculated as containing the same amount of each scFv at that concentration.
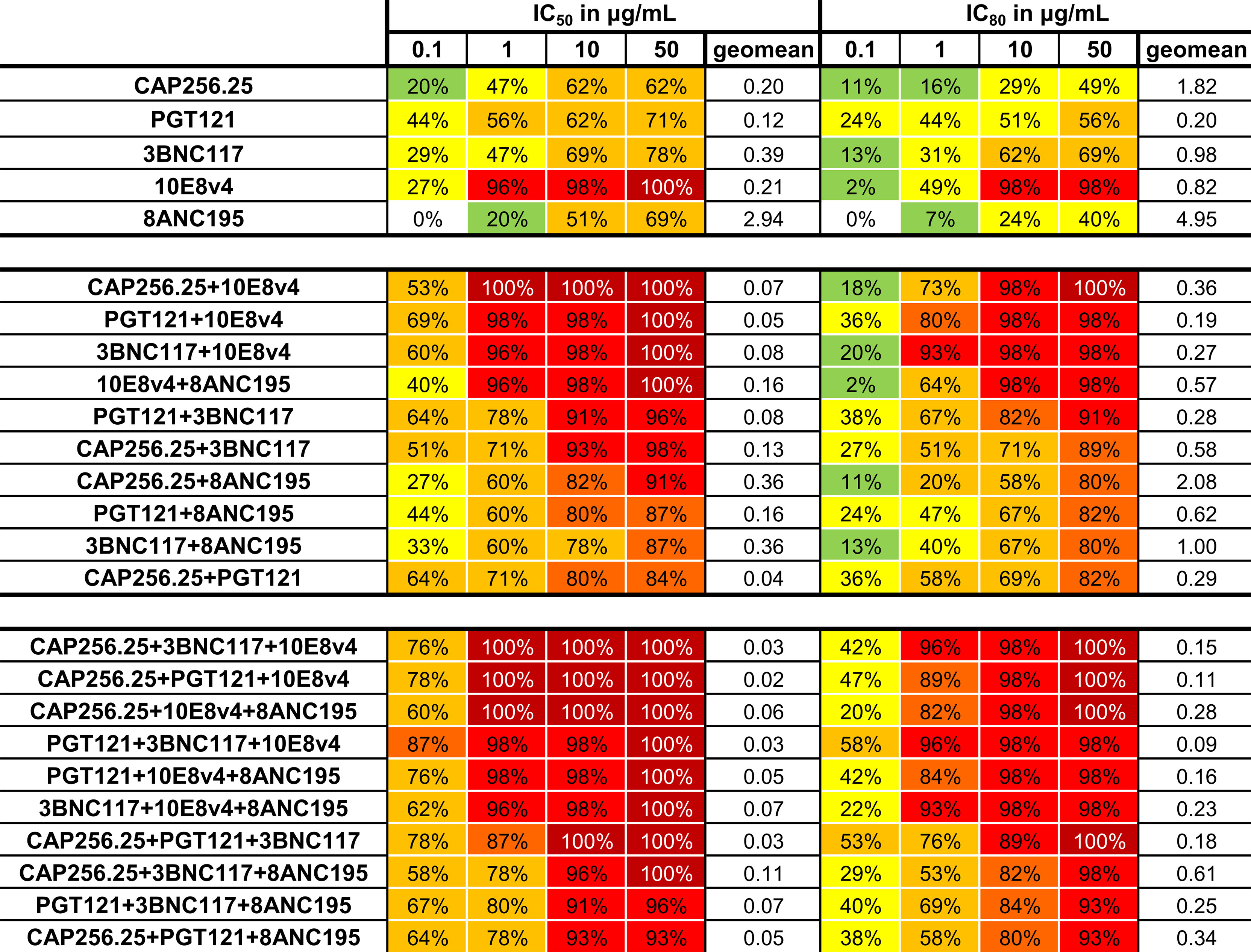
Figure 6 Neutralization breadth for single and combinations of scFv against a 45-virus panel at four concentrations. Percentage neutralization at 0.1, 1, 10, and 50μg/mL was calculated from single scFv titers and from predicted scFv dual and triple combination titers for the IC50 and IC80. Colors indicate percentage neutralization with 1-19% in green, 20-49% yellow, 50-79% yellow-orange, 80-89% orange, 90-99% red and 100% dark red. Geometric mean at 50μg/mL is also given. Each scFv is present in the combinations at the titers indicated.
Since PGT121, 3BNC117, and CAP256.25 all had good potencies as scFv, using these together with 10E8v4 resulted in improved potency of the combination. Moreover, scFv such as CAP256.25 and PGT121 and/or 3BNC117 have complementary profiles, with 3BNC117, for example, being especially potent against subtype B viruses and CAP256.25 being potent against subtype C viruses (see Supplementary Data 1 for the IC50 and IC80 of single scFv) (38). 10E8v4 is the broadest scFv and that resulted in the combinations containing 10E8v4 having the highest breadth. As a result, four of the five triple combinations with the best geometric mean titers included 10E8v4. Conversely, introducing less potent or less broad antibodies such as 8ANC195 resulted in less favorable antibody combinations, where 5 out of 6 combinations with the lowest potency and breadth contained 8ANC195, consistent with the relatively lower potency and breadth of this antibody. Of the top five, three combinations, CAP256.25+PGT121+10E8v4 and CAP256.25+3BNC117+10E8v4 and CAP256.25+8ANC195+10E8v4 neutralized all viruses below 1μg/mL and 98% of viruses at IC80 below 10μg/mL. The best ranked PGT121+3BNC117+10E8v4 combination neutralized 96% of viruses below 1μg/mL and 98% of viruses below 10μg/mL at the IC80 value. This analysis indicated a significant gain of potency and neutralization breadth at lower concentrations than single scFv at both IC50 and IC80. This indicated that combinations of scFv were able to increase the coverage at lower concentrations, which will be vital in clinical settings.
Predicting the Efficacy of scFv Combinations
The AMP trial showed that viral isolates that were sensitive to VRC01 with an IC80<1μg/mL were prevented from establishing infection in 75% of cases. As indicated in Figures 5 and 6, scFv combinations can reach significant coverage by a single antibody at this potency with the best scFv combination potentially reaching a 96% protection level. However, other antibody classes may need higher levels of potency to provide similar protection as VRC01 and viral isolates which are neutralized close to this cut-off may allow for breakthrough. Dual coverage will be able to overcome these issues by neutralizing breakthrough viruses at higher potencies and providing a back-up in case antibody levels drop. Therefore, using this cut-off, we calculated the predicted dual coverage (i.e. by at least two scFv) of the combinations using the thresholds of single scFv IC80 < 1µg/ml and < 10μg/mL. We found for dual combinations, the dual active coverage at 1μg/mL ranged from 20% with improvement at 10μg/mL to 51% for PGT121+10E8v4 (Figures 7A, B), This ranged from 0-20% at 1μg/mL and 9-62% for other combinations of two antibodies (See Supplementary Figure 5).
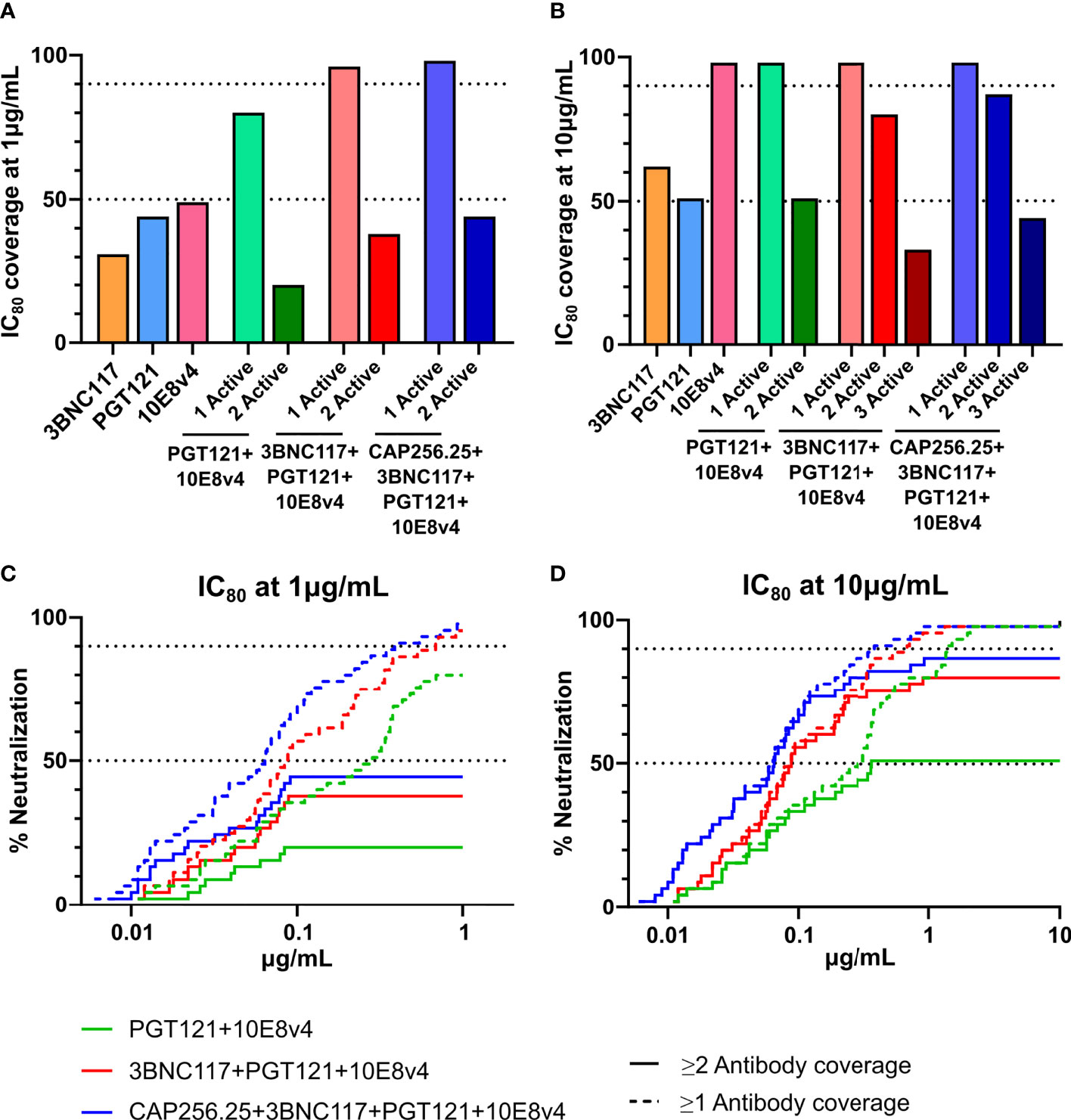
Figure 7 Predicted coverage and estimated efficacy of best scFv combinations. (A, B) Active coverage by one, two, three or four scFv was calculated at 2 concentrations (1μg/mL (A) and 10μg/mL (B) at IC80 based on the predicted titers according to the Bliss-Hill Independence model. The broadest triple and quadruple combinations are shown. (C, D) Breadth Potency curves of active coverage of 1 and 2 scFv are given for the best combinations (3BNC17+10E8v4 and 3BNC17+10E8v4+PGT121, and CAP256.25+3BNC17+10E8v4+PGT121) at 1 μg/mL (C) and 10μg/mL (D). 50% and 90% breadth are indicated by the dotted lines in (C, D).
This was somewhat improved for triple combinations where we observed up to 58-80% dual active coverage at an IC80 of 10μg/mL. The best 3-combinations,3BNC117+PGT121+10E8v4 showing the highest active coverage at 10μg/mL of 80%. Generally, triple combinations showed higher levels of active coverage by 2 antibodies and often included both 10E8v4 and 3BNC117. Similar to the dual combinations, low dual coverage was observed at 1μg/mL up to 38% (See Figure 7A, Supplementary Figure 5. This could not be significantly improved for quadruple combinations which showed 44% and 87% dual coverage respectively at 1 and 10μg/mL. This loss of active coverage by multiple antibodies is also demonstrated in the breadth potency curves (Figures 7C, D). These curves clearly show that although at 10μg/mL most of the dual coverage is maintained there is a significant drop off at 1μg/mL. This was mainly due to antibodies such as 3BNC117, PGT121, and 10E8v4 showing only 31-49% breadth at 1μg/mL and the CAP256.25 scFv losing significant potency compared to the IgG (38). IgG combinations including this antibody could show slightly better active coverage by 2 antibodies, up to 53% at an IC80 of 1μg/mL and up to 91% at 10μg/mL (Supplementary Data 6A for predictions of IgG combinations titers in μg/mL and 6B for neutralization breadth at 4 concentrations and dual coverage by IgG) (38). The other IgG and scFv showed similar levels of potency with specifically the 10E8v4 scFv retaining all of its potency explaining why the IgG combinations did not show higher levels of dual active coverage at 1μg/mL (Figure 7, Supplementary Figure 6B). This indicated that potency of single IgG and scFv needs to be improved to obtain higher levels of active coverage by 2 antibodies at lower antibody levels. The combination data demonstrate that the benefits of combinations of IgG targeting different HIV epitopes apply also to the smaller and more versatile scFv fragments.
Discussion
Combinations of scFv antibodies were able to enhance the breadth and potency of HIV-1 neutralization compared to a single scFv. Specifically, several triple combinations reached 100% coverage at IC50 and IC80 due to complementary neutralization profiles. Dual and triple combinations of scFv demonstrated broad coverage in the potency range that correlated with prevention in the AMP trials (IC80<1μg/mL) and would therefore be expected to be highly effective in combating HIV transmission in high incidence areas.
Generally, dual scFv combinations followed an independent action model rather than a synergistic model of potency. The potency of the combination was usually similar to the potency of the most potent scFv in that mix. Although scFv may show less steric interference between antibodies this did not result in scFv showing significant potency improvements when used in combination. The close positioning of certain epitopes may prevent dual binding such as between V3 and CD4bs targeting scFv indicating that steric hindrance may have still impacted the scFv. Given the distinct MPER epitope located on the gp41 subunit instead of the gp120 subunit, we also expected independent effects for antibodies combined with 10E8v4, which was somewhat indicated in comparing experimental results with the most potent scFv. However, besides a few outliers, this was not apparent using the Bliss-Hill model. It is likely that binding of the MPER antibody confers conformational changes upon binding to its epitope precluding neutralization by other antibodies (47–50). scFv may be especially advantageous in bispecific constructs as their smaller size allows for easier expression. Bispecific products will also increase the local concentration of the second antibody and with the appropriate linker may in such cases allow for synergy. Similarly, minor losses of potency compared to the most potent scFv were overall a rare occurrence and were negated with the addition of a third antibody (in a triple combination with e.g. 10E8v4). The few cases of antagonism noted for scFv combinations may also be due to conformational changes rather than steric hindrance, which would occur after antibody binding. To explore these aspects, scFv-trimer complex crystallization studies would have to be done.
The Loewe model tended to underestimate the potency at IC80 particularly of triple combinations. It also often predicted low-level synergy for combinations containing the MPER antibody 10E8v4 and the V2 antibody CAP256.25. The Bliss-Hill model did not indicate significant antagonism and/or synergy for any combination at either IC50 or IC80. We did observe a few individual outliers where for example the combination showed some improvement in IC50 and IC80 (CAP256.25+3BNC117+10E8v4) or similarly (3BNC117+PGT121) showed loss of potency against individual viruses but no specific viral signature could be identified. Some of these may be limitations of the model itself as these tended to occur more often with viruses where the neutralization curves were not sigmoidal or with a low slope (18). The Bliss-Hill model tended to be better at predicting the IC80 titers and both models were matched for the IC50 titers. This finding is similar to IgG combinations where the Bliss-Hill model was also more accurate at predicting combination titers compared to the Loewe model, therefore the Bliss-Hill model was the preferred model for analyses of larger scFv datasets.
From a clinical perspective, the use of antibody combinations may mitigate the impact of suboptimal antibody levels. Overlapping neutralization can enhance coverage against single viruses, which ensures that complete viral neutralization is achieved at lower concentrations of antibody. The selection of antibody combinations with high potency (IC80) can more effectively counter declining antibody levels associated with passive administration. Although combinations of two scFv neutralized 100% of viruses at the highest concentration tested, the coverage was much reduced at lower concentrations, whereas triple antibody combinations demonstrated much higher coverage at lower concentrations by combining broad antibodies with very potent antibodies. For example, at an IC50 of 1μg/mL, 100% of viruses were neutralized and at an IC80 of 1μg/mL, 96% of viruses were neutralized. This demonstrates that complementary neutralization of the same virus is extremely advantageous, which was previously demonstrated for combinations of IgG (20).
Besides improved coverage, we also observed improved active coverage by 2 antibodies for triple combinations compared to dual combinations (19). In experimental combinations, neutralization slopes were also improved by combinations of antibodies, especially IC80 titers and neutralization plateaus (less than 100% neutralization of a virus) were also less common with antibody combinations (data not shown) (18). It may also be beneficial to use antibodies or scFv that bind trimers in different states; for example, CAP256.25 binds early, whereas 10E8v4 likely binds trimers that are in an intermediate state allowing for complementarity in virus neutralization (51–53). Clinically, active coverage by multiple antibodies will prevent antibody escape when the serum titers drop to close to the neutralization range. The AMP trial demonstrated that if the VRC01 antibody levels dropped, protection against less sensitive viruses (IC80> 1μg/mL) was lost (17). This may differ for other antibodies or for antibodies targeting different epitopes and combinations may also provide dual active coverage of the viral quasispecies further improving this level of protection (54, 55). This highlights the need to use more potent antibodies but also to improve individual antibody potency. In the future, bNAbs and scFv may also need to be assessed in PBMC neutralization assays which may more accurately resemble in vivo neutralization, and could more accurately predict potency (17, 55). We observed that although overall coverage was improved, active coverage by multiple antibodies was low for both IgG and scFv using this set of antibodies. There are multiple efforts ongoing to improve antibody potency and bioavailability. Titers are likely to improve if newer scFv are developed which retain better potency compared to the IgG. Moreover, the selection of antibodies with optimal potency to cover geographically relevant viruses will ensure efficacy in the region.
For long-term antibody-mediated protection, adherence, and maintaining sufficient plasma levels in a wider population is critically important. At lower concentrations, scFv and IgG lose coverage especially in the ability to fully neutralize the virus (concentrations below IC80 titers). This is also observed for IgG, indicating that all the antibodies in the combination must be kept at optimal levels. For IgG, one way to achieve this has been to introduce mutations in the Fc portion of the antibody to increase the half-life by up to 6 months (56, 57). A second option is to introduce them into the body via a stable expression system through a vectored immunoprophylaxis (VIP) approach. Some bNAbs were recently expressed using AAV, showing modest levels in vivo (58, 59). However, due to the space constraints on vectors, constructs with multi-specific modalities would prove difficult. Bispecific products, which would partially overcome this, are also limited by space constraints. scFv in comparison with their much smaller size may be more readily expressed on AAV, allowing for the expression of multiple scFv continuously. scFv have a genetic size approximately 2.6-times smaller than IgG allowing for the expression of minimally three scFv per AAV, and improved expression levels (9, 29, 60–66). AAV based scFv expression can help overcome the short half-life of scFv due to their lack of an Fc region. Some studies have shown success in the use of AAV based scFv for Alzheimer’s disease, and Amyotrophic Lateral Sclerosis amongst others (61, 67, 68). In vivo data will be required to determine the antigenicity and bioavailability of scFv. However, scFv conjugated to short amino acid peptides or linked to targeting molecules were shown to have significant increases in bioavailability and stability (69–72). These strategies mitigate some of the challenges facing scFv.
The scFv studied here demonstrated high breadth and potency when used in combination. Although only a limited number of scFv were tested on a small panel of viruses, we demonstrated their potential against a larger panel of viruses through modeling. By selecting bNAbs that target all five epitopes we also showed the potential for complementary effects. These scFv showed little to no antagonism. As more potent scFv are engineered combinations with even better efficacy may be found. In addition, as scFv are more readily expressed on vectors, this could in future provide, an alternative avenue for the use of bNAbs in passive immunization. The encouraging results of the AMP trials are expected to fast-track the need to consider long-term delivery approaches for those antibodies with clinical benefit.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics Statement
Ethics approval was obtained as per local regulations (M160341). Antibody sequences were obtained from publicly available databases.
Author Contributions
RD performed all experiments, data interpretation, figures and manuscript generation, and literature review. KW performed data analysis and advised on statistical analysis. PM and LM supervised the project and assisted with data interpretation and manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
We acknowledge research funding from the South African Medical Research Council (SAMRC) Flagship Project, the NIH through a U01 grant (U01AI116086), the Poliomyelitis Research Fund (PRF) through a PRF research grant (17/15), and the Centre for the AIDS Program of Research (CAPRISA). CAPRISA is funded by the South African HIV/AIDS Research and Innovation Platform of the South African Department of Science and Technology and was initially supported by the U.S. NIAID, NIH, U.S. Department of Health and Human Services grant U19 AI51794. R.T.V.D. is supported by a Poliomyelitis Research Foundation Ph.D. bursary (W 16/72). PM is supported by the South African Research Chairs Initiative of the Department of Science and Innovation and National Research Foundation of South Africa (grant no. 98341). KW is supported by the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery grant OPP1032144 (Comprehensive Antibody Vaccine Immune Monitoring Consortium CAVIMC) (https://www.cavd.org/Pages/default.aspx). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank H. Marcotte (Karolinska Institute) for the donation of the CAP256 VH-VL L1 scFv. We thank the AIDS Reagent Program, Division of AIDS, NIAID, NIH, for the use of the pCMVR vector and the pBR322-based IG-lambda expression vector. We also thank J. Mascola, the VRC, USA, for the donation of the PGT121 heavy- and light-chain plasmids.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.734110/full#supplementary-material
References
1. Julg B, Liu P-T, Wagh K, Fischer WM, Abbink P, Mercado NB, et al. Protection Against a Mixed SHIV Challenge by a Broadly Neutralizing Antibody Cocktail. Sci Transl Med (2017) 9:eaao4235. doi: 10.1126/scitranslmed.aao4235
2. Saunders KO, Wang L, Joyce MG, Yang Z-YY, Balazs AB, Cheng C, et al. Broadly Neutralizing Human Immunodeficiency Virus Type 1 Antibody Gene Transfer Protects Nonhuman Primates From Mucosal Simian-Human Immunodeficiency Virus Infection. J Virol (2015) 89:8334–45. doi: 10.1128/jvi.00908-15
3. Martinez-Navio JM, Fuchs SP, Pantry SN, Lauer WA, Duggan NN, Keele BF, et al. Adeno-Associated Virus Delivery of Anti-HIV Monoclonal Antibodies Can Drive Long-Term Virologic Suppression. Immunity (2019) 50:567–575.e5. doi: 10.1016/j.immuni.2019.02.005
4. Parren PW, Marx PA, Hessell AJ, Luckay A, Harouse J, Cheng-Mayer C, et al. Antibody Protects Macaques Against Vaginal Challenge With a Pathogenic R5 Simian/Human Immunodeficiency Virus at Serum Levels Giving Complete Neutralization In Vitro. J Virol (2001) 75:8340–7. doi: 10.1128/JVI.75.17.8340-8347.2001
5. Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 Vaccine Development and Therapy. Science (80-) (2013) 341:1199–204. doi: 10.1126/science.1241144
6. Bekker L-G, Moodie Z, Grunenberg N, Laher F, Tomaras GD, Cohen KW, et al. Subtype C ALVAC-HIV and Bivalent Subtype C Gp120/MF59 HIV-1 Vaccine in Low-Risk, HIV-Uninfected, South African Adults: A Phase 1/2 Trial. Lancet HIV (2018) 5:e366–78. doi: 10.1016/S2352-3018(18)30071-7
7. Karasavvas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, et al. The Thai Phase III HIV Type 1 Vaccine Trial (RV144) Regimen Induces Antibodies That Target Conserved Regions Within the V2 Loop of Gp120. AIDS Res Hum Retroviruses (2012) 28:1444–57. doi: 10.1089/aid.2012.0103
8. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination With ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N Engl J Med (2009) 361:2209–20. doi: 10.1056/NEJMoa0908492
9. Lin A, Balazs AB. Adeno-Associated Virus Gene Delivery of Broadly Neutralizing Antibodies as Prevention and Therapy Against HIV-1 11 Medical and Health Sciences 1103 Clinical Sciences 11 Medical and Health Sciences 1108 Medical Microbiology Marit Van Gils, M.J.Vangils@Amc. Retrovirology (2018) 15:66. doi: 10.1186/s12977-018-0449-7
10. Marcotte H, Hammarström L. Passive Immunization. In: Mucosal Immunology. Elsevier: Cambridge, Massachusetts, United States (2015). 2:1403–34. doi: 10.1016/B978-0-12-415847-4.00071-9
11. Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev Immunol (2016) 34:635–59. doi: 10.1146/annurev-immunol-041015-055515
12. Bhiman JN, Lynch RM. Broadly Neutralizing Antibodies as Treatment: Effects on Virus and Immune System. Curr HIV/AIDS Rep (2017) 14:54–62. doi: 10.1007/s11904-017-0352-1
13. Crowell TA, Colby DJ, Pinyakorn S, Sacdalan C, Pagliuzza A, Intasan J, et al. Safety and Efficacy of VRC01 Broadly Neutralising Antibodies in Adults With Acutely(RV397): A Phase 2, Randomised, Double-Blind, Placebo-Controlled Trial. Lancet HIV (2019) 6:e297–306. doi: 10.1016/S2352-3018(19)30053-0
14. Huang Y, Naidoo L, Zhang L, Carpp LN, Rudnicki E, Randhawa A, et al. Pharmacokinetics and Predicted Neutralisation Coverage of VRC01 in HIV-Uninfected Participants of the Antibody Mediated Prevention (AMP) Trials. EBioMedicine (2021) 64:103203. doi: 10.1016/j.ebiom.2020.103203
15. Edupuganti S, Mgodi N, Karuna ST, Andrew P, Rudnicki E, Kochar N, et al. Feasibility and Successful Enrollment in a Proof-Of-Concept HIV Prevention Trial of VRC01, a Broadly Neutralizing HIV-1 Monoclonal Antibody. J Acquir Immune Defic Syndr (2021) 87:671–9. doi: 10.1097/QAI.0000000000002639
16. Mgodi NM, Takuva S, Edupuganti S, Karuna S, Andrew P, Lazarus E, et al. A Phase 2b Study to Evaluate the Safety and Efficacy of VRC01 Broadly Neutralizing Monoclonal Antibody in Reducing Acquisition of HIV-1 Infection in Women in Sub-Saharan Africa. JAIDS J Acquir Immune Defic Syndr (2021) 87:680–7. doi: 10.1097/QAI.0000000000002649
17. Corey L, Gilbert PB, Juraska M, Montefiori DC, Morris L, Karuna ST, et al. Two Randomized Trials of Neutralizing Antibodies to Prevent HIV-1 Acquisition. N Engl J Med (2021) 384:1003–14. doi: 10.1056/nejmoa2031738
18. Wagh K, Bhattacharya T, Williamson C, Robles A, Bayne MG, Garrity J, et al. Optimal Combinations of Broadly Neutralizing Antibodies for Prevention and Treatment of HIV-1 Clade C Infection. PloS Pathog (2016) 12:e1005520. doi: 10.1371/journal.ppat.1005520
19. Kong R, Louder MK, Wagh K, Bailer RT, DeCamp A, Greene KM, et al. Improving Neutralization Potency and Breadth by Combining Broadly Reactive HIV-1 Antibodies Targeting Major Neutralization Epitopes. J Virol (2015) 89:2659–71. doi: 10.1128/JVI.03136-14
20. Wagh K, Seaman MS, Zingg M, Fitzsimons T, Barouch DH, Burton DR, et al. Potential of Conventional & Bispecific Broadly Neutralizing Antibodies for Prevention of HIV-1 Subtype A, C & D Infections. PloS Pathog (2018) 14:e1006860. doi: 10.1371/journal.ppat.1006860
21. CAPRISA. CAPRISA Studies | CAPRISA (2020). Available at: https://www.caprisa.org/Pages/CAPRISAStudies (Accessed September 3, 2020).
22. Mahomed S, Garrett N, Karim QA, Yende-Zuma N, Capparelli E, Baxter C, et al. Assessing the Safety and Pharmacokinetics of the Anti-HIV Monoclonal Antibody CAP256V2LS Alone and in Combination With VRC07-523LS and PGT121 in South African Women: Study Protocol for the First-in-Human CAPRISA 012B Phase I Clinical Trial. BMJ Open (2020) 10:e042247. doi: 10.1136/bmjopen-2020-042247
23. Karuna ST. Phase 1 Broadly Neutralizing Antibody Efforts(2021). Available at: https://www.hvtn.org/en/community/community-compass/vol20-issue1/phase-1-broadly-neutralizing-antibody-efforts.html (Accessed April 2, 2021).
24. Karuna ST, Corey L. Broadly Neutralizing Antibodies for HIV Prevention. Annu Rev Med (2020) 71:329–46. doi: 10.1146/annurev-med-110118-045506
25. Foucquier J, Guedj M. Analysis of Drug Combinations: Current Methodological Landscape. Pharmacol Res Perspect (2015) 3:e00149. doi: 10.1002/prp2.149
26. Davis-Gardner ME, Alfant B, Weber JA, Gardner MR, Farzan M. A Bispecific Antibody That Simultaneously Recognizes the V2-And V3-Glycan Epitopes of the Hiv-1 Envelope Glycoprotein is Broader and More Potent Than Its Parental Antibodies. MBio (2020) 11(1):e03080–19. doi: 10.1128/mBio.03080-19
27. Huang Y, Yu J, Lanzi A, Yao X, Andrews CD, Tsai L, et al. Engineered Bispecific Antibodies With Exquisite HIV-1-Neutralizing Activity. Cell (2016) 165:1621–31. doi: 10.1016/j.cell.2016.05.024
28. Khan SN, Sok D, Tran K, Movsesyan A, Dubrovskaya V, Burton DR, et al. Targeting the HIV-1 Spike and Coreceptor With Bi- and Trispecific Antibodies for Single-Component Broad Inhibition of Entry. J Virol (2018) 92(18):e00384–18. doi: 10.1128/jvi.00384-18
29. West AP, Galimidi RP, Gnanapragasam PNP, Bjorkman PJ. Single-Chain Fv-Based Anti-HIV Proteins: Potential and Limitations. J Virol (2012) 86:195–202. doi: 10.1128/JVI.05848-11
30. Song R, Pace C, Seaman MS, Fang Q, Sun M, Andrews CD, et al. Distinct HIV-1 Neutralization Potency Profiles of Ibalizumab-Based Bispecific Antibodies. J Acquir Immune Defic Syndr (2016) 73:365–73. doi: 10.1097/QAI.0000000000001119
31. Moshoette T, Ali SA, Papathanasopoulos MA, Killick MA. Engineering and Characterising a Novel, Highly Potent Bispecific Antibody Imab-CAP256 That Targets HIV-1. Retrovirology (2019) 16:1–12. doi: 10.1186/s12977-019-0493-y
32. Colcher D, Pavlinkova G, Beresford G, Booth BJ, Choudhury A, Batra SK. Pharmacokinetics and Biodistribution of Genetically-Engineered Antibodies(1998). Available at: https://search.proquest.com/openview/aad96f811b8e44830fd2f5817ff5b54b/1?pq-origsite=gscholar&cbl=27900 (Accessed August 14, 2018).
33. Unverdorben F, Richter F, Hutt M, Seifert O, Malinge P, Fischer N, et al. Pharmacokinetic Properties of IgG and Various Fc Fusion Proteins in Mice. MAbs (2016) 8:120–8. doi: 10.1080/19420862.2015.1113360
34. Yokota T, Milenic DE, Whitlow M, Schlom J. Rapid Tumor Penetration of a Single-Chain Fv and Comparison With Other Immunoglobulin Forms. Cancer Res (1992) 52:3402–8. doi: 10.1007/s00429-013-0597-4
35. Unverdorben F, Färber-Schwarz A, Richter F, Hutt M, Kontermann RE. Half-Life Extension of a Single-Chain Diabody by Fusion to Domain B of Staphylococcal Protein A. Protein Eng Des Sel (2012) 25:81–8. doi: 10.1093/protein/gzr061
36. Kumar S, Kumar R, Khan L, Makhdoomi MA, Thiruvengadam R, Mohata M, et al. CD4-Binding Site Directed Cross-Neutralizing scFv Monoclonals From HIV-1 Subtype C Infected Indian Children. Front Immunol (2017) 8:1568. doi: 10.3389/fimmu.2017.01568
37. Alam MMM, Kuwata T, Tanaka K, Alam MMM, Takahama S, Shimura K, et al. Synergistic Inhibition of Cell-to-Cell HIV-1 Infection by Combinations of Single Chain Variable Fragments and Fusion Inhibitors. Biochem Biophys Rep (2019) 20:100687. doi: 10.1016/j.bbrep.2019.100687
38. van Dorsten RT, Lambson BE, Wibmer CK, Weinberg MS, Moore PL, Morris L. Neutralization Breadth and Potency of Single-Chain Variable Fragments Derived From Broadly Neutralizing Antibodies Targeting Multiple Epitopes on the HIV-1 Envelope. J Virol (2020) 94(2):e01533–19. doi: 10.1128/JVI.01533-19
39. Portolano N, Watson PJ, Fairall L, Millard CJ, Milano CP, Song Y, et al. Recombinant Protein Expression for Structural Biology in HEK 293F Suspension Cells: A Novel and Accessible Approach. J Vis Exp (2014) 92:e51897. doi: 10.3791/51897
40. Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein Identification and Analysis Tools on the ExPASy Server. In: The Proteomics Protocols Handbook. Totowa, NJ: Humana Press (2005). 571–607. doi: 10.1385/1-59259-890-0:571
41. Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, et al. The Neutralization Breadth of HIV-1 Develops Incrementally Over Four Years and Is Associated With CD4+ T Cell Decline and High Viral Load During Acute Infection. J Virol (2011) 85:4828–40. doi: 10.1128/JVI.00198-11
42. Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, et al. Developmental Pathway for Potent V1V2-Directed HIV-Neutralizing Antibodies. Nature (2014) 509:55–62. doi: 10.1038/nature13036
43. Moore PL, Sheward DJ, Nonyane M, Ranchobe N, Hermanus T, Gray ES, et al. Multiple Pathways of Escape From HIV Broadly Cross-Neutralizing V2-Dependent Antibodies. J Virol (2013) 87:4882–94. doi: 10.1128/JVI.03424-12
44. Todd CA, Greene KM, Yu X, Ozaki DA, Gao H, Huang Y, et al. Development and Implementation of an International Proficiency Testing Program for a Neutralizing Antibody Assay for HIV-1 in TZM-Bl Cells. J Immunol Methods (2012) 375:57–67. doi: 10.1016/j.jim.2011.09.007
45. Sarzotti-Kelsoe M, Bailer RT, Turk E, li LC, Bilska M, Greene KM, et al. Optimization and Validation of the TZM-Bl Assay for Standardized Assessments of Neutralizing Antibodies Against HIV-1. J Immunol Methods (2014) 409:131–46. doi: 10.1016/j.jim.2013.11.022
46. Mascola JR, D’Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, et al. Recommendations for the Design and Use of Standard Virus Panels To Assess Neutralizing Antibody Responses Elicited by Candidate Human Immunodeficiency Virus Type 1 Vaccines. J Virol (2005) 79:10103–7. doi: 10.1128/jvi.79.16.10103-10107.2005
47. Rujas E, Caaveiro JMM, Partida-Hanon A, Gulzar N, Morante K, Apellániz B, et al. Structural Basis for Broad Neutralization of HIV-1 Through the Molecular Recognition of 10E8 Helical Epitope at the Membrane Interface. Sci Rep (2016) 6:1–13. doi: 10.1038/srep38177
48. Wang Y, Kaur P, Sun Z-YJ, Elbahnasawy MA, Hayati Z, Qiao Z-S, et al. Topological Analysis of the Gp41 MPER on Lipid Bilayers Relevant to the Metastable HIV-1 Envelope Prefusion State. Proc Natl Acad Sci (2019) 116:22556–66. doi: 10.1073/PNAS.1912427116
49. Fu Q, Shaik MM, Cai Y, Ghantous F, Piai A, Peng H, et al. Structure of the Membrane Proximal External Region of HIV-1 Envelope Glycoprotein. Proc Natl Acad Sci (2018) 115:E8892–9. doi: 10.1073/pnas.1807259115
50. Kim AS, Leaman DP, Zwick MB. Antibody to Gp41 MPER Alters Functional Properties of HIV-1 Env Without Complete Neutralization. PloS Pathog (2014) 10:e1004271. doi: 10.1371/JOURNAL.PPAT.1004271
51. Pinto D, Fenwick C, Caillat C, Silacci C, Guseva S, Dehez F, et al. Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody Ln01. Cell Host Microbe (2019) 26:623–37.e8. doi: 10.1016/j.chom.2019.09.016
52. Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang G-Y, Chambers M, et al. Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation. Cell Rep (2017) 19:719–32. doi: 10.1016/j.celrep.2017.04.013
53. Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, et al. New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency. J Virol (2015) 90:76–91. doi: 10.1128/JVI.01791-15
54. Lorenzi JCC, Mendoza P, Cohen YZ, Nogueira L, Lavine C, Sapiente J, et al. Neutralizing Activity of Broadly Neutralizing Anti-HIV-1 Antibodies Against Primary African Isolates. J Virol (2021) 95(5):e01909–20. doi: 10.1128/jvi.01909-20
55. Cohen YZ, Lorenzi JCC, Seaman MS, Nogueira L, Schoofs T, Krassnig L, et al. Neutralizing Activity of Broadly Neutralizing Anti-HIV-1 Antibodies Against Clade B Clinical Isolates Produced in Peripheral Blood Mononuclear Cells. J Virol (2017) 92:e01883–17. doi: 10.1128/JVI.01883-17
56. Liu Q, Lai YT, Zhang P, Louder MK, Pegu A, Rawi R, et al. Improvement of Antibody Functionality by Structure-Guided Paratope Engraftment. Nat Commun (2019) 10:741. doi: 10.1038/s41467-019-08658-4
57. Rosenberg YJ, Lewis GK, Montefiori DC, LaBranche CC, Lewis MG, Urban LA, et al. Introduction of the YTE Mutation Into the Non-Immunogenic HIV bnAb PGT121 Induces Anti-Drug Antibodies in Macaques. PloS One (2019) 14:e0212649. doi: 10.1371/journal.pone.0212649
58. van den Berg FT, Makoah NA, Ali SA, Scott TA, Mapengo RE, Mutsvunguma LZ, et al. AAV-Mediated Expression of Broadly Neutralizing and Vaccine-Like Antibodies Targeting the HIV-1 Envelope V2 Region. Mol Ther - Methods Clin Dev (2019) 14:100–12. doi: 10.1016/j.omtm.2019.06.002
59. Brady JM, Baltimore D, Balazs AB. Antibody Gene Transfer With Adeno-Associated Viral Vectors as a Method for HIV Prevention. Immunol Rev (2017) 275:324–33. doi: 10.1111/imr.12478
60. Monnier P, Vigouroux R, Tassew N. In Vivo Applications of Single Chain Fv (Variable Domain) (scFv) Fragments. Antibodies (2013) 2:193–208. doi: 10.3390/antib2020193
61. Hay CE, Gonzalez GA, Ewing LE, Reichard EE, Hambuchen MD, Nanaware-Kharade N, et al. Development and Testing of AAV-Delivered Single-Chain Variable Fragments for the Treatment of Methamphetamine Abuse. PloS One (2018) 13:e0200060. doi: 10.1371/journal.pone.0200060
62. Gruenert AK, Czugala M, Mueller C, Schmeer M, Schleef M, Kruse FE, et al. Self-Complementary Adeno-Associated Virus Vectors Improve Transduction Efficiency of Corneal Endothelial Cells. PloS One (2016) 11:e0152589. doi: 10.1371/journal.pone.0152589
63. Dong J-Y, Fan P-D, Frizzell RA. Quantitative Analysis of the Packaging Capacity of Recombinant Adeno-Associated Virus. Hum Gene Ther (2008) 7:2101–12. doi: 10.1089/hum.1996.7.17-2101
64. Wu Z, Yang H, Colosi P. Effect of Genome Size on AAV Vector Packaging. Mol Ther (2010) 18:80–6. doi: 10.1038/mt.2009.255
65. Liu Z, Chen O, Wall JBJ, Zheng M, Zhou Y, Wang L, et al. Systematic Comparison of 2A Peptides for Cloning Multi-Genes in a Polycistronic Vector. Sci Rep (2017) 7:2193. doi: 10.1038/s41598-017-02460-2
66. Raj D, Davidoff AM, Nathwani AC. Self-Complementary Adeno-Associated Viral Vectors for Gene Therapy of Hemophilia B: Progress and Challenges. Expert Rev Hematol (2011) 4:539–49. doi: 10.1586/ehm.11.48
67. Patel P, Kriz J, Gravel M, Soucy G, Bareil C, Gravel C, et al. Adeno-Associated Virus-Mediated Delivery of a Recombinant Single-Chain Antibody Against Misfolded Superoxide Dismutase for Treatment of Amyotrophic Lateral Sclerosis. Mol Ther (2014) 22:498–510. doi: 10.1038/mt.2013.239
68. Krishnaswamy S, Huang HW, Marchal IS, Ryoo HD, Sigurdsson EM. Neuronally Expressed Anti-Tau scFv Prevents Tauopathy-Induced Phenotypes in Drosophila Models. Neurobiol Dis (2020) 137:104770. doi: 10.1016/j.nbd.2020.104770
69. Morita S, Noguchi H, Horii T, Nakabayashi K, Kimura M, Okamura K, et al. Targeted DNA Demethylation In Vivo Using Dcas9-Peptide Repeat and scFv-TET1 Catalytic Domain Fusions. Nat Biotechnol (2016) 34:1060–5. doi: 10.1038/nbt.3658
70. Phoolcharoen W, Banyard AC, Prehaud C, Selden D, Wu G, Birch CPD, et al. In Vitro and In Vivo Evaluation of a Single Chain Antibody Fragment Generated in Planta With Potent Rabies Neutralisation Activity. Vaccine (2019) 37:4673–80. doi: 10.1016/j.vaccine.2018.02.057
71. Greineder CF, Villa CH, Walsh LR, Kiseleva RY, Hood ED, Khoshnejad M, et al. Site-Specific Modification of Single-Chain Antibody Fragments for Bioconjugation and Vascular Immunotargeting. Bioconjug Chem (2018) 29:56–66. doi: 10.1021/acs.bioconjchem.7b00592
Keywords: HIV, broadly neutralizing antibodies, single chain variable fragments, combinations of scFv, HIV prevention
Citation: van Dorsten RT, Wagh K, Moore PL and Morris L (2021) Combinations of Single Chain Variable Fragments From HIV Broadly Neutralizing Antibodies Demonstrate High Potency and Breadth. Front. Immunol. 12:734110. doi: 10.3389/fimmu.2021.734110
Received: 30 June 2021; Accepted: 31 August 2021;
Published: 16 September 2021.
Edited by:
Susu M. Zughaier, Qatar University, QatarReviewed by:
Christopher Andrew Cottrell, The Scripps Research Institute, United StatesMichael Zwick, The Scripps Research Institute, United States
Copyright © 2021 van Dorsten, Wagh, Moore and Morris. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lynn Morris, bHlubm1AbmljZC5hYy56YQ==
 Rebecca T. van Dorsten
Rebecca T. van Dorsten Kshitij Wagh
Kshitij Wagh Penny L. Moore
Penny L. Moore Lynn Morris
Lynn Morris