- 1Cancer Center, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2The Medical Department, 3D Medicines Inc., Shanghai, China
- 3Department of Pediatrics, Renmin Hospital of Wuhan University, Wuhan, China
Background: MAP2K1/2 genes are mutated in approximately 8% of melanoma patients; however, the impact of MAP2K1/2 gene alterations on the efficiency of immunotherapy has not been clarified. This study focused on the correlation between MAP2K1/2 gene mutations and the treatment response.
Methods: Six metastatic melanoma clinical cohorts treated with immune checkpoint inhibitors [anti-cytotoxic T lymphocyte antigen-4 (CTLA-4) or anti-programmed cell death-1 (PD-1)] were recruited in this study. RNA expression profiling results from each of these six cohorts and the Cancer Genome Atlas (TCGA) melanoma cohort were analysed to explore the mechanism related to immune activation.
Results: Compared to patients with wild-type MAP2K1/2, those with MAP2K1/2 mutations in an independent anti-CTLA-4-treated cohort had higher objective response rates, longer progression-free survival, and longer overall survival (OS). These findings were further validated in a pooled anti-CTLA-4-treated cohort in terms of the OS. However, there was no correlation between MAP2K1/2 mutations and OS in the anti-PD-1-treated cohort. Subgroup Cox regression analysis suggested that patients with MAP2K1/2 mutations received fewer benefits from anti-PD-1 monotherapy than from anti-CTLA-4 treatment. Furthermore, transcriptome profiling analysis revealed that melanoma tumours with MAP2K mutation was enriched in CD8+ T cells, B cells, and neutrophil cells, also expressed high levels of CD33 and IL10, implying a potential mechanism underlying the benefit of melanoma patients with MAP2K1/2 mutations from anti-CTLA-4 treatment.
Conclusions: MAP2K1/2 mutations were identified as an independent predictive factor for anti-CTLA-4 therapy in melanoma patients. Anti-CTLA-4 treatment might be more effective than anti-PD-1 therapy for patients with MAP2K1/2-mutated melanoma.
Background
Treatment with immune checkpoint inhibitors (ICIs), including antibodies targeting cytotoxic T lymphocyte antigen-4 (CTLA-4) and programmed cell death-1/programmed cell death ligand 1 (PD-1/PD-L1), is becoming a novel therapeutic paradigm for melanomas (1). Ipilimumab, an anti-CTLA-4 monoclonal antibody, has been found to significantly improve overall survival (OS) and progression-free survival (PFS) and increase the long-term survival rate of patients with advanced melanoma (2, 3). Compared to ipilimumab, second-generation ICIs targeting PD-1, namely nivolumab and pembrolizumab, have also been reported to induce an increased response rate, OS, and PFS, with superior toxicity profiles (4, 5). Moreover, ICIs are the standard of care in the systemic treatment of metastatic or unresectable melanomas (6).
Although ICIs significantly increase the survival of melanoma patients, only a subset of patients can benefit from the therapy, and the related mechanisms are still not fully understood. Therefore, the identification of biomarkers to select patients who will be more responsive to ICIs is of utmost importance. Several genomic features, such as high mutational load, high neoantigen load, and tumour clonality have been found to be predictive of a favourable response to anti-CTLA-4 therapy in melanoma patients (7, 8). In addition, it has been reported that aberrations of individual genes, such as SERPINB3/SERPINB4, NRAS, and TP53, are associated with the response to anti-CTLA-4 therapy (9–11). Either high PD-L1 expression or high tumour mutational burden (TMB) has also been recognized as predictors of the effectiveness of anti-PD-1 blockade in melanomas and other solid tumours (12, 13).
The status of driver mutations may also influence the response to ICIs. For example, NRAS-mutated melanoma is a distinct subtype in approximately 5%–20% of patients with melanomas and appears to have a poor prognosis (12, 13). In a retrospective analysis, compared to melanoma patients with wild-type NRAS, those with NRAS mutations were found to have higher objective response rates and prolonged stable disease in response to ICIs (11). NRAS encodes N-Ras, which is a component of the Ras/Raf/MEK/ERK signalling cascade (14). This cascade, also known as the Ras/MAPK signalling cascade, plays an important role in the pathogenesis of melanoma (15). MEK is one of the kinases involved in the Ras/MAPK signalling cascade. Moreover, the encoding genes MAP2K1 and MAP2K2 (MAP2K1/2) are frequently mutated in melanoma, with a frequency of approximately 8% of cases (16, 17). The occurrence of MAP2K1/2 mutations was identified as a mechanism related to BRAF inhibitor resistance in melanoma (18). To date, there are no reports on effective small molecule inhibitors targeting MAP2K1/2 in melanoma. It is also unclear whether mutations in these genes influence the efficacy of immunotherapy.
Preclinical studies have revealed that treatment with MEK inhibitors might improve the sensitivity of immunotherapy in melanoma. MEK inhibition in a melanoma cell line was found to increase the antigen levels, which might potentiate anti-tumour T-cell immunity (19). Moreover, MEK inhibition may reduce the number of Bregs while sparing anti-tumour B-cell function, thereby enhancing anti-tumour immunity (20). In mouse models, MEK inhibitors were found to inhibit tumour growth via increasing the number of intertumoral effector-phenotype CD8+ T cells, and combination therapy with both MEK inhibitors and anti-PD-L1 agents exhibited a synergistic effect on antitumor growth (21).
In this study, through analysis of the sequencing and survival data for several public cohorts with metastatic melanoma, we investigated the association between MAP2K1/2 mutations and the response to anti-CLTA-4 and anti-PD-1 immunotherapies. The influence of MAP2K1/2 mutations on the expression of immunity-related genes was also evaluated by analysing the RNA expression profile data collected from these cohorts and from the Cancer Genome Atlas (TCGA) melanoma cohort. Our aim is to clarify the impact of MAP2K1/2 gene alterations on the efficiency of immunotherapies and to provide guidance for treatment decision-making in MAP2K1/2-mutated melanomas.
Methods
Eligible Literature Search
We performed a systematic computerized search of the MEDLINE (PubMed) database and the Embases database up to November 1, 2020. The search terms were as follows: (Melanomas [MeSH] OR “metastatic melanomas” [Title/Abstract]) AND (“PD-1 blockade”[Title/Abstract] OR “PD-L1 blockade” [Title/Abstract] OR “CTLA‐4 blockade” [Title/Abstract] OR “immune checkpoint inhibitor” [Title/Abstract] OR “immune checkpoint inhibitors” [Title/Abstract] OR “ICI” [Title/Abstract] OR “ICIs” [Title/Abstract] OR “immune checkpoint blockade” [Title/Abstract] OR “immune checkpoint blockades” [Title/Abstract] OR “ICB” [Title/Abstract] OR “ICBs” [Title/Abstract]). Studies with eligible next-generation sequencing data were identified by hand and included if they met the criteria: (a) Clinical trials or study cohorts treated with ICIs; (b) Clinical outcomes of patients were available; (c) The number of evaluable patients was more than 30. We found six cohort studies of metastatic melanoma, specifically the Allen (8), Snyder (7), Hugo (22), and Liu (23) cohorts, and two metastatic pan-cancer cohorts comprising patients with melanoma, namely the Miao (24) and Samstein (25) cohorts.
Study Design and Data Acquisition
In this study, missense, nonsense, and frame-shift mutations in both MAP2K1 and MAP2K2 genes were defined as MAP2K1/2 mutations.
In total, data for 753 melanoma patients were included in our study. Notably, melanoma patients treated with sequential CTLA-4 and PD-1 blockades were excluded from this study. First, we determined the predictive value for CTLA-4 monotherapy of MAP2K1/2 mutations in the Allen cohort and validated this predictive value in a CTLA-4-monotherapy-pooled cohort comprising 239 melanoma samples from the Snyder, Miao, and Samstein cohorts. Thereafter, we explored the impact of MAP2K1/2 mutations on OS in a PD-1-monotherapy-pooled cohort consisting of 285 melanoma samples from the Hugo, Liu, Miao, and Samstein cohorts. The TCGA-skin cutaneous melanoma (SKCM) cohort without ICI treatment (n = 455) and with chemotherapy (n = 73) was analysed to assess the prognostic value of MAP2K1/2 (Figure S1).
Data accessibility: data for whole-exome sequencing (WES), copy number alterations (CNAs), gene expression, and clinicopathologic information were collected from the Allen, Snyder, Hugo, and TCGA-SKCM cohorts. Data for targeted-sequencing and OS were obtained from the Samstein cohort using the cBioPortal database. Data for the Liu and Miao cohorts were taken from previous publications.
Evaluation of Clinical Response
In evaluating the clinical response, responders were defined as melanoma patients with different degrees of clinical responses to immunotherapy, characterized as complete response (CR), partial response (PR), or stable disease (SD). Non-responders were defined as those patients with disease progression (PD) following immunotherapy.
Transcriptome Data Normalization and Processing
The transcriptome data were interpreted as fragments per kilobase million mapped reads (FPKM). To compensate for RNA-seq counts within and between samples, the FPKM for every gene was transformed into transcripts per kilobase million (TPM) values by dividing by the sum of FPKM in each sample. The immune filtration cells scores were estimated with the TIMER2.0 website tool using TPM data.
Statistical Analysis
In this study, all data analysis and graphic plotting were performed using the R software (v.3.6.3). Kaplan–Meier curves (log-rank test) of OS were plotted to compare survival outcomes between different subgroups. The proportion of death events of the patients in MAP2K1/2-mutated group and MAP2K1/2-wild-type groups was defined as risk ratio (RR). Pooled analysis was performed using the DerSimonian–Laird random-effects model to compare the RR with 95% of confidence intervals (CI) The degree of heterogeneity between cohorts was assessed via the I2 index. No significant heterogeneity was defined as I2 < 50% and P >0.1. Univariate and multivariate Cox proportional hazard regression analyses were performed to quantify the hazard ratio (HR) of various characteristics. Fisher’s exact tests were used to determine if there were non-random associations between categorical variables. The Wilcoxon–Mann–Whitney test was used to compare the differences in discrete ordinal data between two independent groups.
Results
MAP2K1/2 Mutations as a Biomarker to Predict Favourable Response to Anti-CTLA-4 Therapy and Survival in Metastatic Melanoma
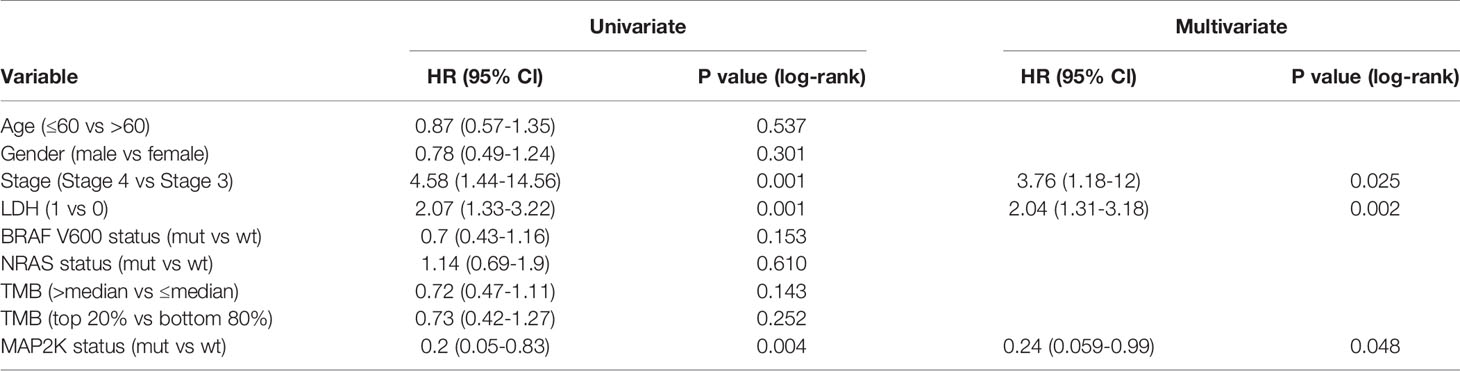
Data analysis was performed using a cohort of 110 patients (Van Allen cohort) with metastatic melanoma treated with ipilimumab. The characteristics of the patients are summarized in Table S1. Seven patients (6.36%) in this cohort harboured MAP2K1 or MAP2K2 mutations and had longer OS (49.2 months vs 8.3 months; HR = 0.2; 95% confidence interval (CI), 0.05–0.83; p = 0.0262; Figure 1A) and PFS (19.4 months vs 2.8 months; HR = 0.37; 95% CI, 0.15–0.91; p = 0.0307; Figure 1B) than those with wild-type MAP2K1/2. Moreover, MAP2K1/2 mutations were more frequent in responders (17.6% vs 1.3%; p = 0.0185; Figure 1C). In univariate analyses, three factors, namely MAP2K1/2 mutations, tumour stage (stage 4 vs stage 3), and the presence of lactate dehydrogenase (LDH; 1 vs 0), were found to be associated with immunotherapeutic OS and PFS (Table 1). Furthermore, in the multivariable Cox proportional hazards regression model adjusted by tumour stage, LDH, and MAP2K1/2 mutations, these three factors were still significantly associated with OS (HR = 0.24; 95% CI, 0.059–0.99; p = 0.048; Table 1) and PFS (HR = 0.39; 95% CI, 0.16–0.99; p = 0.048; Table 1). The results suggested that MAP2K1/2 mutations might be an independent predictor for a favourable clinical response to anti-CTLA4 therapy in patients with melanoma.
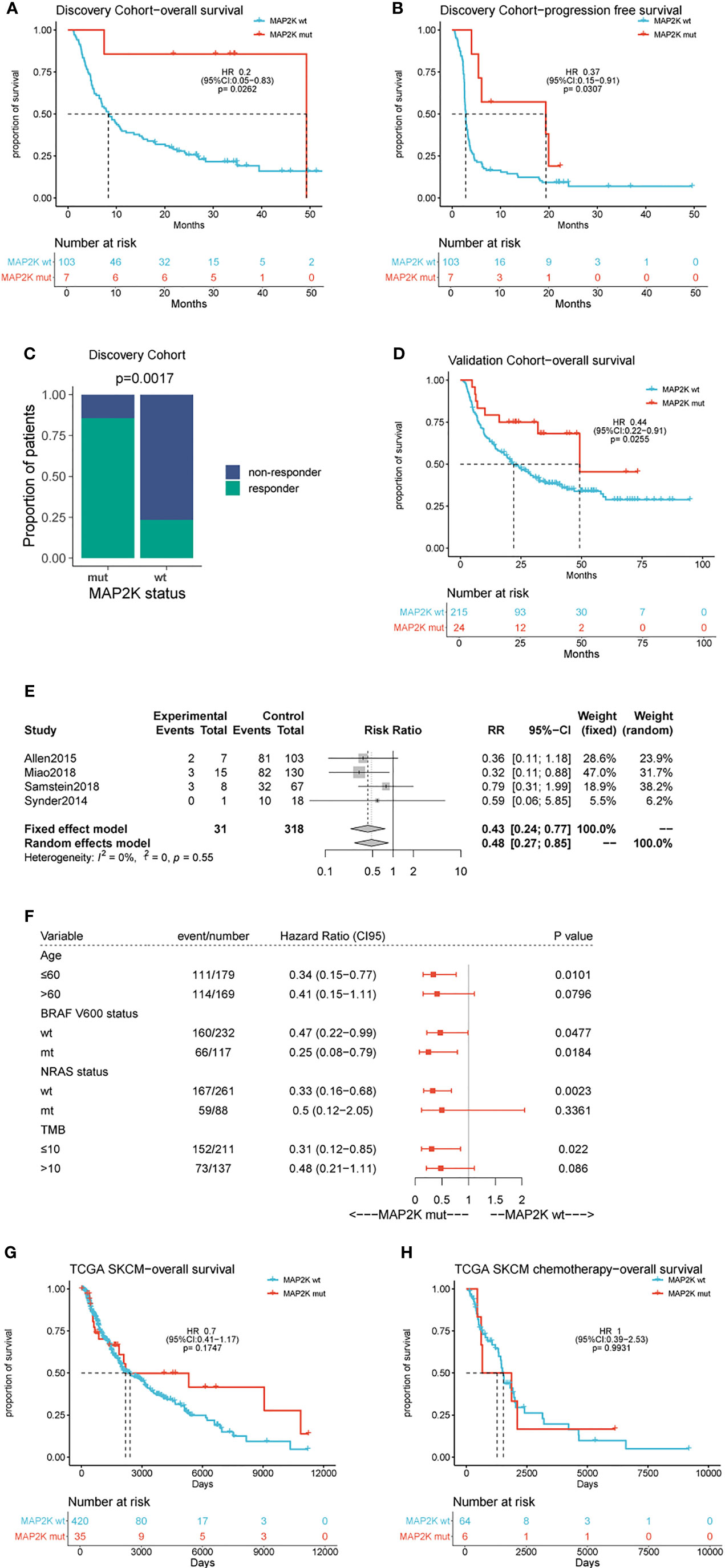
Figure 1 Effect of MAP2K1/2 gene mutations on treatment response in anti-CTLA-4 and non-immunotherapy-treated melanoma. Kaplan-Meier analyses of overall survival (OS) (A), progression-free survival (PFS) (B), and disease-control rate (C) in the anti-CTLA-4-treated discovery cohort. Kaplan-Meier analyses of overall survival (OS) (D) in the anti-CTLA-4-treated validation cohort. Pooled estimates of OS in four anti-CTLA-4-treated cohorts (E). Subgroup Cox analysis of OS in pooled anti-CTLA-4-treated cohorts among patients with and without MAP2K1/2 gene mutations (F). Kaplan-Meier analyses of OS in TCGA melanoma cohort (G) and TCGA chemotherapy-treated cohort (H).
To validate the predictive value of MAP2K1/2 mutations for the efficacy of anti-CTLA4 therapy in melanoma, data analysis was also performed using a pooled cohort, which was a combination of three metastatic melanoma cohorts treated with ipilimumab (Miao, Samstein, and Snyder) (Table S1). This pooled cohort contained 239 patients, among which 24 patients (10%) harboured MAP2K1 or MAP2K2 mutations. According to the data analysis results, MAP2K1/2 mutations were significantly correlated with longer OS in this pooled cohort (49.3 months vs 22.0 months; HR = 0.44; 95% CI, 0.22–0.91; p = 0.0255; Figure 1D) compared to the group with wild-type MAP2K1/2. The meta-analysis also demonstrated that the group with MAP2K1/2 mutations exhibited a significantly reduced risk of death, as compared to the group with wild-type MAP2K1/2 (fixed effects model; relative risk (RR) = 0.43; 95% CI, 0.24–0.77; Figure 1E). No substantial heterogeneity was observed across studies (p = 0.55, Figure 1E), indicating the conclusion was consistent, from data analysis between different cohorts to the association between MAP2K1/2 mutations and the favourable clinical response to anti-CTLA-4 therapy. Further data analysis using a pooled cohort consisting of four cohorts also confirmed that MAP2K1/2 mutations are a predictive factor for superior OS in most subgroups with diverse clinical and molecular characteristics (Figure 1F).
To clarify whether MAP2K1/2 mutations are a predictive or prognostic biomarker, analysis of the data obtained from the TCGA database was performed, based on the MAP2K1/2 mutational status of melanoma patients. No significant difference in OS was observed between the groups with mutated and wild-type MAP2K1/2 in the total population (HR = 0.7; 95% CI, 0.41–1.17; p = 0.1747; Figure 1G) as well as in the chemotherapy population (HR = 1, 95% CI, 0.39–2.53; p = 0.99; Figure 1H). Taken together, these results suggested that MAP2K1/2 gene mutations are a predictor for a favourable clinical response to anti-CTLA-4 therapy in metastatic melanoma rather than a prognostic factor for melanoma.
MAP2K1/2 Mutations Were Not Associated With the Clinical Benefits of Anti-PD-1/L1 Therapy for Metastatic Melanoma
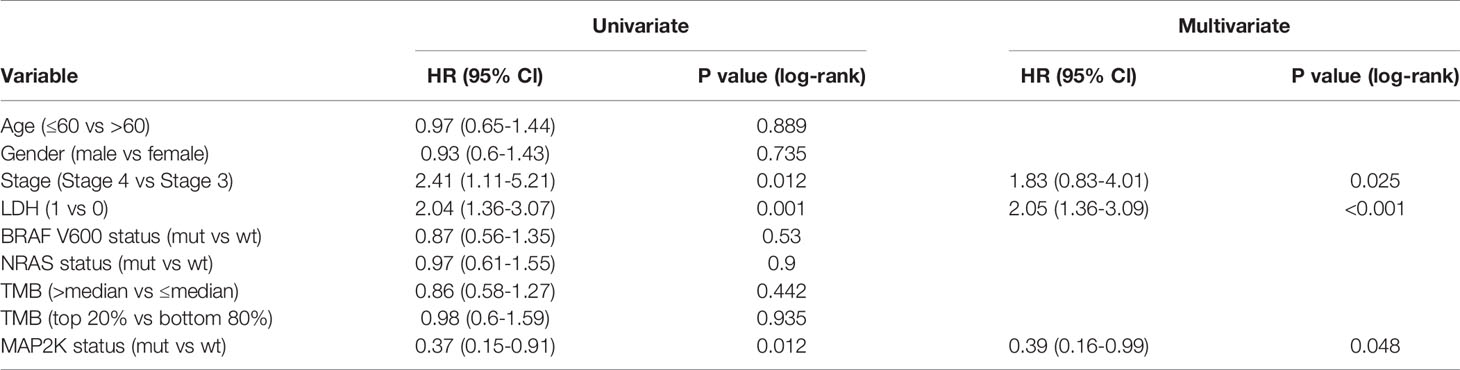
Anti-PD-1 therapy with second-generation ICIs can prolong both PFS and OS in metastatic melanoma patients, with less high-grade toxicity than ipilimumab. To investigate whether MAP2K1/2 mutations are also associated with a favourable clinical response to anti-PD-1/L1 therapy, we performed data analysis using a pooled cohort consisting of three public cohorts of 253 metastatic melanoma patients treated with anti-PD-1 agents (Table S1). Overall, 22 patients (8.7%) harboured MAP2K1/2 mutations in this pooled cohort. The results showed that there was no significant difference in OS between mutated and wild-type MAP2K1/2 groups (27.0 months vs 32.0 months; HR = 1.31; 95% CI, 0.68–2.53; p = 0.4151; Figure 2A). The meta-analysis showed that the risk of death was not reduced in the MAP2K1/2-mutated group as compared to the wild-type MAP2K1/2 group (fixed effects model; RR = 1.13; 95% CI, 0.7–1.84; Figure 2B). Moreover, subgroup analysis indicated that MAP2K1/2 mutations were not a predictor of OS in any subgroup (Figure 2C). These results revealed that MAP2K1/2 gene mutations are not associated with the clinical benefits of anti-PD-1/L1 therapy for metastatic melanoma. Therefore, the predictive value of MAP2K1/2 mutation in metastatic melanoma might be specific to anti-CTLA-4 therapy, rather than anti-PD-1/L1 therapy.
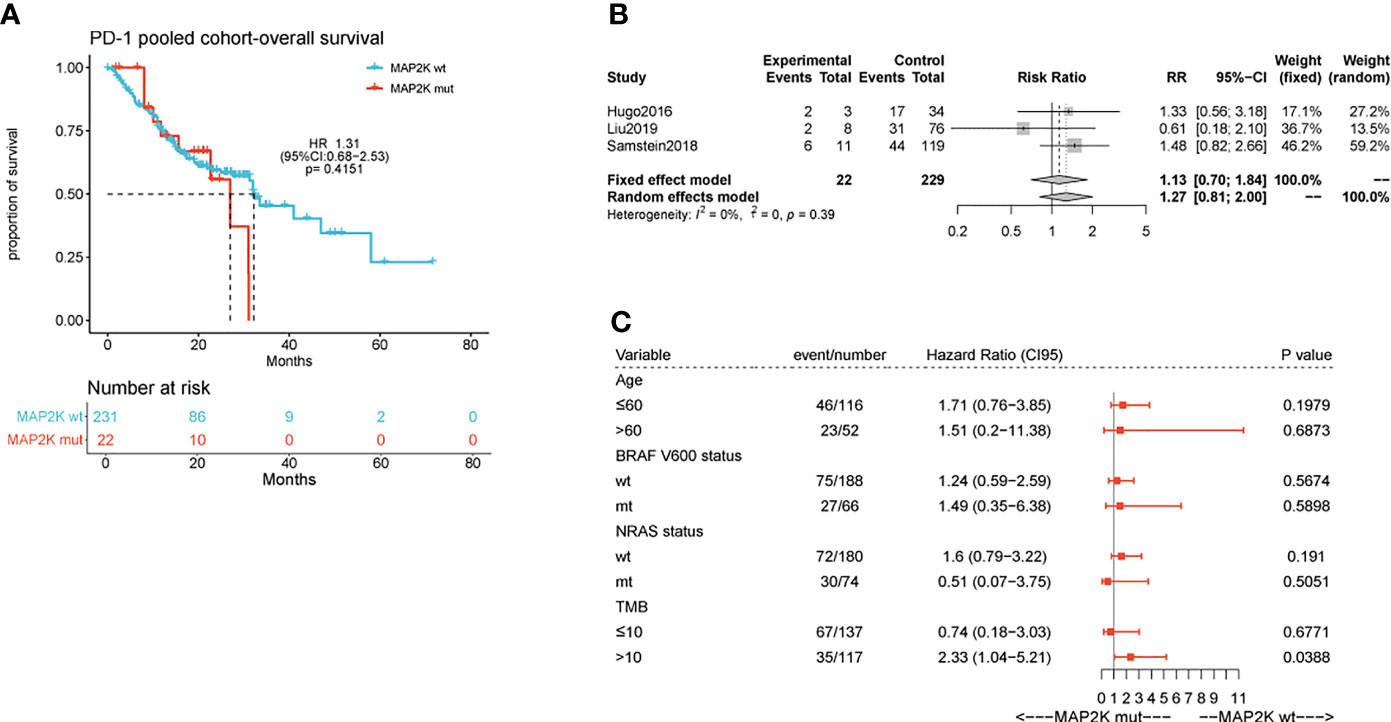
Figure 2 Effect of MAP2K1/2 gene mutations on treatment response in anti-PD-1-treated melanoma. Kaplan-Meier analyses of overall survival (OS) in the anti-PD-1-treated cohort (A). Pooled estimates of OS in three anti-PD-1-treated cohorts (B). Subgroup Cox analysis of OS in pooled anti-PD-1 treated cohorts among patients with and without MAP2K1/2 gene mutations (C).
Mutation Status of MAP2K1/2 Gene May Influence Systemic Treatment Options in Metastatic Melanoma
In this study, the clinical benefits (OS) of anti-CTLA-4 and anti-PD-1 therapies in patients with MAP2K1/2-mutated melanoma were compared in a pooled cohort comprising a combination of six cohorts. Analysis results showed that in the total population, the association of OS with anti-PD-1 therapy was superior to the association with anti-CLTA-4 therapy (median OS, 32.0 months vs 19.2 months; HR = 0.67; 95% CI, 0.53–0.85; p = 0.0009; Figure 3A), consistent with the results of a previous study (5). However, patients harbouring MAP2K1/2 mutations treated with anti-PD-1 monotherapy had significantly poorer OS than those treated with anti-CTLA-4 monotherapy (median OS, 27.0 months vs 49.3 months; HR = 3.26; 95% CI, 1.18–9.02; p = 0.0225; Figure 3B). The difference in OS between the two immunotherapies could be attributed to the remarkably improved survival rate in the MAP2K1/2-mutated group treated with anti-CTLA-4 therapy. Subgroup analysis also revealed that the MAP2K1/2-mutated group was more likely to obtain clinical benefits from anti-CTLA-4 monotherapy, as compared to anti-PD-1 monotherapy (HR = 0.31; 95% CI, 0.11−0.85; Figure 3C), suggesting that for MAP2K1/2-mutated melanomas, the efficacy of anti-CTLA-4 therapy might be superior to that of anti-PD-1 therapy.
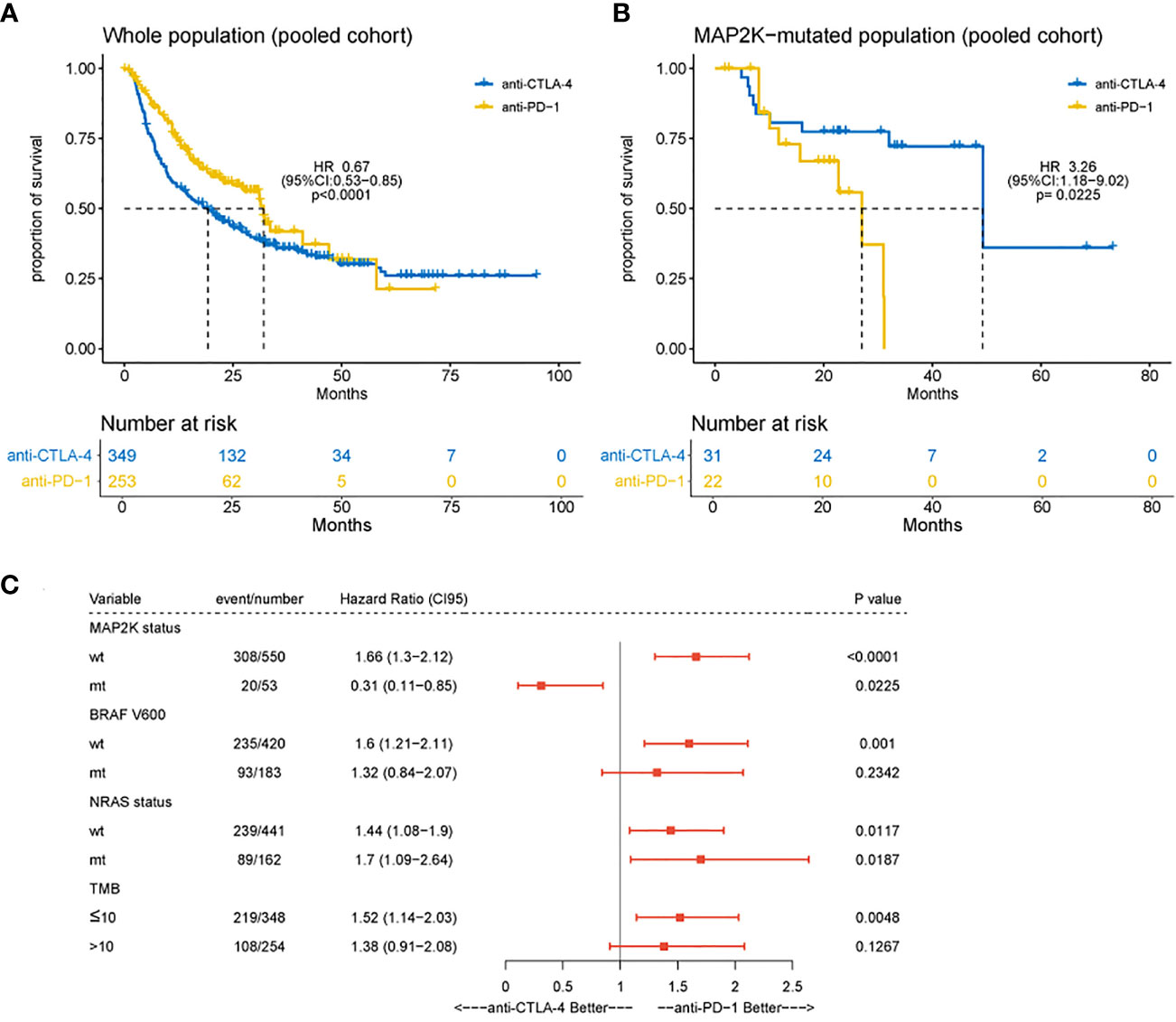
Figure 3 Difference in overall survival between melanoma patients receiving anti-CTLA-4 or anti-PD-1 monotherapy. Kaplan-Meier analyses of overall survival (OS) in overall population (A) and MAP2K1/2-mutated subgroup (B) in the combined cohort of anti-CTLA-4 and anti-PD-1-treated patients. Subgroup Cox analysis of OS among patients receiving anti-CTLA-4 or anti-PD-1 monotherapy (C).
Immunological Microenvironment of MAP2K1/2-Mutated Melanoma
To evaluate the impact of MAP2K1/2 mutations on the transcription of immunity-related genes in melanoma, we integrated and analysed the gene expression data for patients from four clinical cohorts and the TCGA-SKCM cohort. To investigate the status of immune cell infiltration in melanoma patients treated with immunotherapies, TIMER2.0, a comprehensive resource for systematic analysis of immune infiltrates across diverse cancer types, was used to analyse the gene expression data of MAP2K1/2-mutated melanoma. Analysis results revealed that MAP2K1/2-mutated melanoma exhibited significantly increased densities of B cells (p = 0.015), CD8+ T cells (p = 0.024), and neutrophils (p = 0.03) and a numerically higher level of myeloid dendritic cells (p = 0.089) compared to those in their wild-type counterparts, implying that melanoma patients with MAP2K1/2 mutations have a favourable microenvironment for tumoral development (Figure 4A). However, there is no difference in macrophages or CD4+ T cells between MAP2K1/2-mutated and wild-type melanomas (Figure 4A). Interestingly, the gene expression levels of CD33, a marker of myeloid-derived suppressor cells (MDSCs), and IL-10, which is mainly secreted by regulatory T cells (Tregs), were higher in MAP2K1/2-mutated melanoma, compared to those in wild-type melanoma (Figure 4B). Moreover, MDSCs and Tregs were reported to be associated with resistance to the PD-1 blockade (26, 27), consistent with the poor prognosis of patients with MAP2K1/2-mutated melanoma who received anti-PD-1 therapy. TMB analysis also revealed that melanoma patients harbouring MAP2K1/2 mutations have a higher level of TMB (Figure 4C). Moreover, except for immunity-related genes, a total of 55 significantly differentially expressed genes were found between MAP2K1/2-mutated and wild-type melanoma in clinical cohort (Table S2), which might be associated with reshape of immunological microenvironment caused by MAPK mutation.
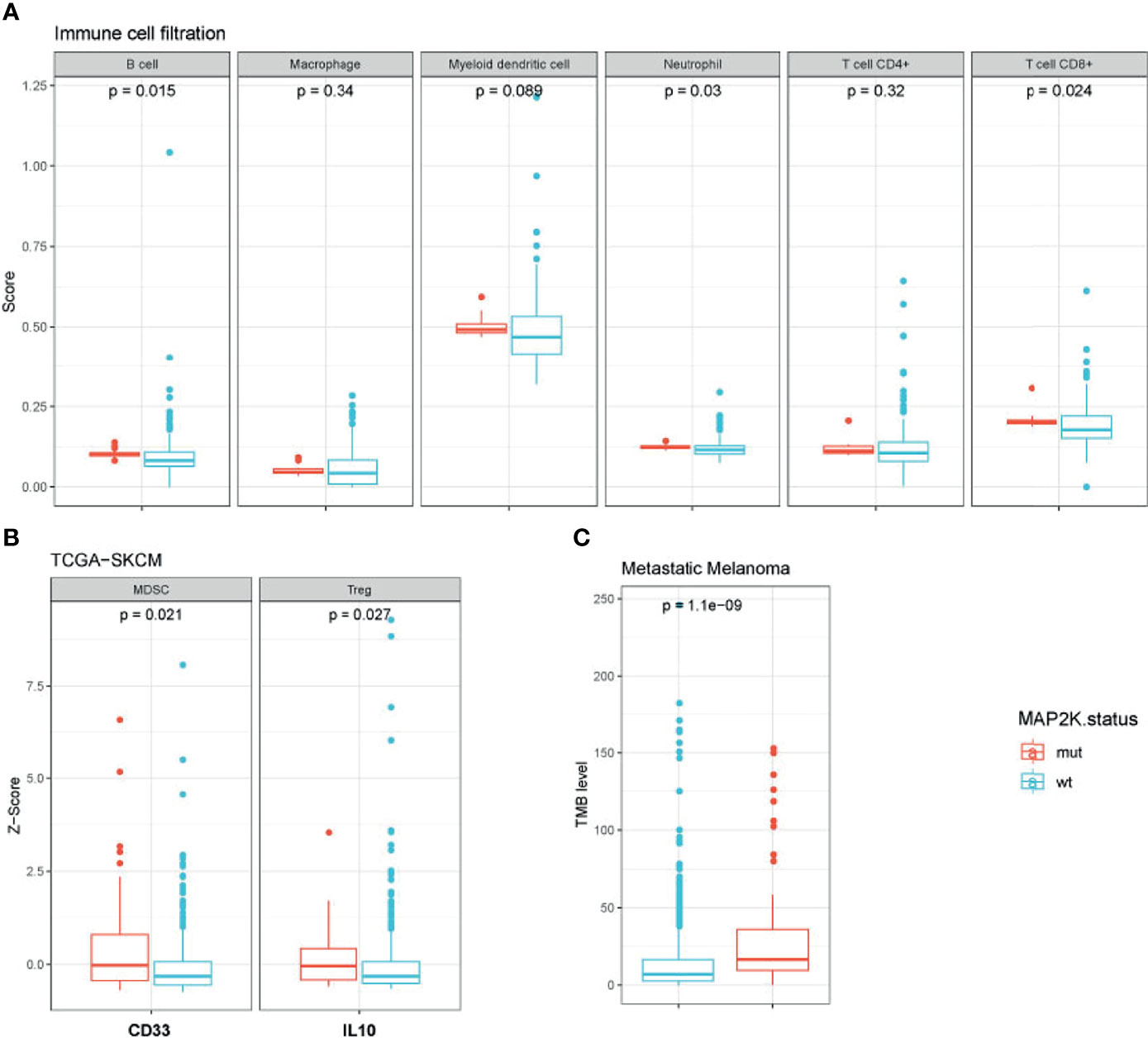
Figure 4 Immunological microenvironment of MAP2K1/2-mutated melanoma. Boxplot comparing immune cell filtration between mutated and wild-type MAP2K1/2 subgroup in metastatic melanoma cohort (A), the expression of immune-related genes between the mutated and wild-type MAP2K1/2 subgroups in TCGA-SKCM cohort (B), and tumour mutational level between mutated and wild-type MAP2K1/2 subgroup in metastatic melanoma cohort (C).
Discussion
In recent years, immunotherapy has greatly improved the survival and quality of life for patients with melanoma, becoming one of the standard treatment regimens for metastatic or advanced melanoma. Although various molecules/antigens have been proposed as possible immunotherapy targets, only anti-CTLA4 antibody and anti-PD-1/L1 antibody are available for immunotherapy against melanoma in clinical practice. A large-scale phase III clinical trial (CheckMate 066) confirmed the efficacy of the anti-PD-1 antibody nivolumab in melanoma treatment. The results showed that nivolumab treatment was superior to standard chemotherapy as the first-line treatment with respect to the OS, PFS, and overall response rate. Later, another trial (Keynote-006) reported the superiority of pembrolizumab to ipilimumab. These studies laid the foundation for the approval of immunotherapy as the first-line treatment for melanoma by the Food and Drug Administration (FDA). Nevertheless, immunotherapy for melanoma treatment is still limited by the fact that only a portion of patients receive the clinical benefits of immunotherapy. For patients who do not respond to immunotherapy, this issue may lead to unnecessary costs (a heavy financial burden for the family) and loss of a valuable period for tumour treatment. Therefore, the identification of biomarkers for predicting the efficacy of immunotherapy would be of great importance.
Among potential biomarkers, CTLA-4 and PD-1 are the leading targets of ICIs in cancer immunotherapy. Previous studies have shown that CTLA-4 and PD-1 have distinct signalling pathways related to the different action mechanisms of immunotherapy (28). Anti-CTLA-4 therapy primarily interferes with the feedback mechanism to improve the proliferation and activation of more T cells, while anti-PD-1 treatment is assumed to attenuate the tumour-induced immunosuppression (29). Retrospective studies have identified different biomarkers that can predict the efficacy of anti-CTLA-4 or anti-PD-1 therapy. For example, loss of major histocompatibility complex (MHC) class I expression is a predictor of resistance to anti-CTLA-4 but not to anti-PD-1 therapy. In contrast, the expression of MHC class II, which is associated with interferon-γ-related signatures, can predict the treatment response to anti-PD-1 therapy, rather than anti-CTLA-4 therapy (23, 28, 30). To further identify more novel predictive biomarkers, we performed data analysis using several public cohorts and found that MAP2K1/2 mutations might be a potential predictor for the clinical benefits of anti-CTLA-4 therapy in advanced melanomas. Our study revealed that patients with MAP2K1/2 mutations had longer PFS and OS than their counterparts without mutations when both groups of patients received ipilimumab treatment. However, the predictive value of MAP2K1/2 mutations is specific to anti-CTLA-4 therapy, rather than anti-PD-1 treatment, because no difference in the survival rate was observed between patients with MAP2K1/2 mutations and their wild-type counterparts after anti-PD-1 treatment. Based on these observations, we investigated the possibility that the difference in clinical benefits between anti-PD-1/L1 and anti-CTLA-4 therapies in patients with MAP2K1/2-mutated melanoma can be used as a biomarker for the selection of the appropriate immunotherapy drug. According to this proposed hypothesis, further data analysis was conducted to compare the efficacy of anti-PD-1/L1 and anti-CTLA-4 therapies in patients with MAP2K1/2-mutated melanoma. Our results indicated that patients with MAP2K1/2-mutated melanoma who received anti-CTLA-4 therapy had better OS. In other words, anti-CTLA-4 therapy was superior to anti-PD-1/L1 therapy for patients with MAP2K1/2-mutated melanoma.
Compared with other proposed efficacy indicators of immunotherapy, such as TMB, microsatellite instability (MSI), and PD-L1 expression, the use of MAP2K1/2 gene mutations as a qualitative biomarker could avoid the dilemma of setting cut-off values. In addition, MAP2K1/2 mutations can be detected in peripheral blood ctDNA, providing a non-invasive approach to identify patients who may receive the benefits of ICI treatment.
In addition, we explored the mechanism underlying the predictive role of MAP2K1/2 in the clinical response to ICIs. TMB, defined as the number of somatic mutations per megabase of interrogated genomic sequences, is considered to be related to the outcome of ICI treatment across multiple tumour types (31–33), though the exact mechanism remaining controversial (34). We found that MAP2K1/2-mutated melanomas exhibited higher TMB levels than MAP2K1/2-wild-type melanomas. This phenomenon could be associated with the predictive effect of TMB on anti-CTLA-4 monotherapy, which was previously reported (7). In addition, we observed an increase in B cells, CD8+ T cells, and neutrophils in MAP2K1/2-mutated melanomas, as compared to MAP2K1/2-wild-type melanomas. Like dendritic cells, B cells can internalize antigens and deliver antigenic peptides to T-cell receptors (35). A previous study showed that high expression of immune cell-derived gene expression signatures in B cells is associated with better response to the anti-CTLA-4 antibody (36). Additionally, the number of MDSCs and Tregs is increased in MAP2K1/2-mutated melanomas. MDSCs are involved in immunosuppression via suppressing the functions of T-cells and natural killer-cells (27). It has been reported that MDSCs can induce the expansion of Tregs and reduce the anti-tumour activity of effector T cells (37), while Tregs can regulate immunosuppression by secreting cytokines, such as IL10, IL35, and TGF-β, thereby suppressing the effector T-cell response. These processes might account for the superiority of anti-CTLA-4 therapy over anti-PD-1 therapy in MAP2K1/2-mutated melanomas. Contrary to our expectations, there was no difference in the expression of MHC class I and II molecules between the MAP2K1/2-mutated and MAP2K1/2-wild-type groups (Figures S2, S3). For 55 significantly differentially expressed genes between MAP2K1/2-mutated and wild-type melanoma in clinical cohort, 14 genes (SMAD9, LRP6, PCDH18, TP53BP2, KDM1A, PKLR, GALNT5, RASGRF2, CTSK, ZNF845, ZNF384, TEK, MTHFD1, TAX1BP1) were associated with the immunological microenvironment as previous reports. Interestingly, TP53BP2, one of the 14 genes, has been proved to activate CD4+ and CD8+ immune and negatively regulate the MAPK signaling pathway in triple-negative breast cancers (38). The impact of MAP2K1/2 gene mutations on the immunological microenvironment should be further assessed through in vitro or in vivo models. MAP2K1/2-mutated cases constitute approximately 8% of all melanoma patients, and the clinical studies of this population has been rarely reported. In fact, the inhibitory drugs of MAP2K1/2, more commonly called MEK inhibitors were identified to be effective in the treatment of melanomas. The combination of MEK inhibitor with BRAF inhibitor has become the standard of care for patients with BRAF-mutated melanoma (39–41). Moreover, in NRAS-mutated melanoma patients, binimetinib has shown its treatment efficacy and represents a treatment option after failure of immunotherapy (42). It has been reported that previous treatment with BRAFi with or without MEKi result in shorter survival in BRAF-mutated melanoma patients when treated with anti-PD-1 antibody (43). Nevertheless, the influence of previous targeted therapies on the therapeutic effect of immunotherapy in MAP2K1/2-mutated melanoma is unknown. Since no effective targeted therapeutic drugs have been reported against MAP2K1/2-mutated melanomas, ICIs are still considered as the preferred systemic treatment for these patients. To the best of our knowledge, the present manuscript is the first report on the investigation of the association between MAP2K1/2 mutations and the clinical response to ICIs. This study can also provide a guideline for making treatment decisions for patients with MAP2K1/2-mutated melanoma. With an innovation-based view, we suggest that for patients with MAP2K1/2-mutated melanoma, anti-CTLA-4 therapy might be more effective than anti-PD-1 monotherapy. Due to the lack of available data, we could not compare the differences in the efficacy of anti-CTLA-4 monotherapy and combination therapy involving both anti-CTLA-4 anti-PD-1 in patients with MAP2K1/2-mutated melanoma in this study. Because of the possibility that combination therapy may increase the incidence of grade 3–4 immune-related adverse events, it would be especially meaningful for patients with poor physical conditions if a comparison study is performed for benefit-risk assessment in ICIs to determine the optimal benefit/risk potential for patients.
There are still some limitations in this study. Because the ICI-treated cohorts included in this study came from several research centres, analysis of the data from the pooled-cohort might introduce biases due to differences in the ICI regimen, dose usage, and treatment cycle between institutions. Additionally, there was no specific limitation on the number of previous therapies used in the observed cohorts, which may cause heterogeneity in the survival time and might account for the OS benefit in the whole population not being as obvious as that reported in the phase III clinical trials, although the trend was consistent (44, 45). Because of the limited sample size and lack of molecular information, we could not match the baseline characteristics such as PD-L1 expression and microsatellite stability. Therefore, prospective studies are additionally required to confirm the findings of this study. Further studies are also needed to elucidate the mechanisms underlying the clinical benefits of anti-CTLA-4 therapy in MAP2K1/2-mutated melanoma.
Data Availability Statement
The data of the Allen cohort, Snyder cohort, Hugo cohort, Liu cohort, Miao cohort and Samstein cohort included in this analysis were provided online. The specific accession numbers and repository names are included in Supplementary Table 1.
Author Contributions
Conceptualization: C-ZY and JC. Methodology: M-LH. Validation: X-YL, W-ZX, and Y-XC. Investigation: M-MY. Data curation: Y-HZ and S-YY. Writing—original draft preparation: TY and J-YZ. Writing—review and editing: CG. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81773285 to JC), the Scientific Research Project of Hubei Provincial Health and Family Planning Commission, China (WJ2015MB017 to JC), and the Health Commission of Hubei Province scientific research project, China (WJ2021Z004 to JC).
Conflict of Interest
X-YL, M-MY, W-ZX, CG, Y-XC, and M-LH were employed by 3D Medicines Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2021.785526/full#supplementary-material
Abbreviations
MAP2K, mitogen-activated protein kinase kinase; MAPK, mitogen-activated protein kinase; CTLA-4, cytotoxic T lymphocyte antigen-4; PD-1/PD-L1, programmed cell death-1/ligand-1; ICB, immune checkpoint blockade; ICIs, immune checkpoint inhibitors; CNAs, copy number alterations; WES, whole-exome sequencing; TMB, tumour mutational burden; MSI, microsatellite instability; SKCM, skin cutaneous melanoma; TCGA, The Cancer Genome Atlas; MSKCC, memorial Sloan Kettering cancer center; OS, overall survival; PFS, progression-free survival; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease.
References
1. Robert C, Lanoy E, Besse B. One or Two Immune Checkpoint Inhibitors? Cancer Cell (2019) 36(6):579–81. doi: 10.1016/j.ccell.2019.11.005
2. Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved Survival With Ipilimumab in Patients With Metastatic Melanoma. N Engl J Med (2010) 363(8):711–23. doi: 10.1056/NEJMoa1003466
3. Robert C, Thomas L, Bondarenko I, O'Day S, Weber J, Garbe C, et al. Ipilimumab Plus Dacarbazine for Previously Untreated Metastatic Melanoma. N Engl J Med (2011) 364(26):2517–26. doi: 10.1056/NEJMoa1104621
4. Schachter J, Ribas A, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab Versus Ipilimumab for Advanced Melanoma: Final Overall Survival Results of a Multicentre, Randomised, Open-Label Phase 3 Study (KEYNOTE-006). Lancet (2017) 390(10105):1853–62. doi: 10.1016/S0140-6736(17)31601-X
5. Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, et al. Overall Survival With Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med (2017) 377(14):1345–56. doi: 10.1056/NEJMoa1709684
6. Herrscher H, Robert C. Immune Checkpoint Inhibitors in Melanoma in the Metastatic, Neoadjuvant, and Adjuvant Setting. Curr Opin Oncol (2020) 32(2):106–13. doi: 10.1097/CCO.0000000000000610
7. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med (2014) 371(23):2189–99. doi: 10.1056/NEJMoa1406498
8. Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic Correlates of Response to CTLA-4 Blockade in Metastatic Melanoma. Science (2015) 350(6257):207–11. doi: 10.1126/science.aad0095
9. Riaz N, Havel JJ, Kendall SM, Makarov V, Walsh LA, Desrichard A, et al. Recurrent SERPINB3 and SERPINB4 Mutations in Patients Who Respond to Anti-CTLA4 Immunotherapy. Nat Genet (2016) 48(11):1327–9. doi: 10.1038/ng.3677
10. Xiao W, Du N, Huang T, Guo J, Mo X, Yuan T, et al. TP53 Mutation as Potential Negative Predictor for Response of Anti-CTLA-4 Therapy in Metastatic Melanoma. EBioMedicine (2018) 32:119–24. doi: 10.1016/j.ebiom.2018.05.019
11. Johnson DB, Lovly CM, Flavin M, Panageas KS, Ayers GD, Zhao Z, et al. Impact of NRAS Mutations for Patients With Advanced Melanoma Treated With Immune Therapies. Cancer Immunol Res (2015) 3(3):288–95. doi: 10.1158/2326-6066.CIR-14-0207
12. Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-Tumor Genomic Biomarkers for PD-1 Checkpoint Blockade-Based Immunotherapy. Science (2018) 362(6411):eaar3593. doi: 10.1126/science.aar3593
13. Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab Plus Ipilimumab in Lung Cancer With a High Tumor Mutational Burden. N Engl J Med (2018) 378(22):2093–104. doi: 10.1056/NEJMoa1801946
14. Vu HL, Aplin AE. Targeting Mutant NRAS Signaling Pathways in Melanoma. Pharmacol Res (2016) 107:111–6. doi: 10.1016/j.phrs.2016.03.007
15. Murugan AK, Dong J, Xie J, Xing M. MEK1 Mutations, But Not ERK2 Mutations, Occur in Melanomas and Colon Carcinomas, But None in Thyroid Carcinomas. Cell Cycle (2009) 8(13):2122–4. doi: 10.4161/cc.8.13.8710
16. Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome Sequencing Identifies Recurrent Somatic MAP2K1 and MAP2K2 Mutations in Melanoma. Nat Genet (2011) 44(2):133–9. doi: 10.1038/ng.1026
17. Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-Genome Landscapes of Major Melanoma Subtypes. Nature (2017) 545(7653):175–80. doi: 10.1038/nature22071
18. Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The Genetic Landscape of Clinical Resistance to RAF Inhibition in Metastatic Melanoma. Cancer Discovery (2014) 4(1):94–109. doi: 10.1158/2159-8290.CD-13-0617
19. Kono M, Dunn IS, Durda PJ, Butera D, Rose LB, Haggerty TJ, et al. Role of the Mitogen-Activated Protein Kinase Signaling Pathway in the Regulation of Human Melanocytic Antigen Expression. Mol Cancer Res (2006) 4(10):779–92. doi: 10.1158/1541-7786.MCR-06-0077
20. Yarchoan M, Mohan AA, Dennison L, Vithayathil T, Ruggieri A, Lesinski GB, et al. MEK Inhibition Suppresses B Regulatory Cells and Augments Anti-Tumor Immunity. PloS One (2019) 14(10):e0224600. doi: 10.1371/journal.pone.0224600
21. Ebert PJR, Cheung J, Yang Y, McNamara E, Hong R, Moskalenko M, et al. MAP Kinase Inhibition Promotes T Cell and Anti-Tumor Activity in Combination With PD-L1 Checkpoint Blockade. Immunity (2016) 44(3):609–21. doi: 10.1016/j.immuni.2016.01.024
22. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell (2016) 165(1):35–44. doi: 10.1016/j.cell.2016.02.065
23. Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Arnon L, et al. Integrative Molecular and Clinical Modeling of Clinical Outcomes to PD1 Blockade In Patients With Metastatic Melanoma. Nat Med (2019) 25(12):1916–27. doi: 10.1038/s41591-019-0654-5
24. Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, et al. Genomic Correlates of Response to Immune Checkpoint Therapies in Clear Cell Renal Cell Carcinoma. Science (2018) 359(6377):801–6. doi: 10.1126/science.aan5951
25. Samstein RM, Lee CH, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, et al. Tumor Mutational Load Predicts Survival After Immunotherapy Across Multiple Cancer Types. Nat Genet (2019) 51(2):202–6. doi: 10.1038/s41588-018-0312-8
26. Dyck L, Wilk MM, Raverdeau M, Misiak A, Boon L, Mills KH. Anti-PD-1 Inhibits Foxp3(+) Treg Cell Conversion and Unleashes Intratumoural Effector T Cells Thereby Enhancing the Efficacy of a Cancer Vaccine in a Mouse Model. Cancer Immunol Immunother (2016) 65(12):1491–8. doi: 10.1007/s00262-016-1906-6
27. Najjar YG, Finke JH. Clinical Perspectives on Targeting of Myeloid Derived Suppressor Cells in the Treatment of Cancer. Front Oncol (2013) 3:49. doi: 10.3389/fonc.2013.00049
28. Rodig SJ, Gusenleitner D, Jackson DG, Gjini E, Giobbie-Hurder A, Jin C, et al. MHC Proteins Confer Differential Sensitivity to CTLA-4 and PD-1 Blockade in Untreated Metastatic Melanoma. Sci Transl Med (2018) 10(450):eaar3342. doi: 10.1126/scitranslmed.aar3342
29. Topalian SL, Drake CG, Pardoll DM. Immune Checkpoint Blockade: A Common Denominator Approach to Cancer Therapy. Cancer Cell (2015) 27(4):450–61. doi: 10.1016/j.ccell.2015.03.001
30. Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, et al. Resistance to Checkpoint Blockade Therapy Through Inactivation of Antigen Presentation. Nat Commun (2017) 8(1):1136. doi: 10.1038/s41467-017-01062-w
31. Ott PA, Bang YJ, Piha-Paul SA, Razak ARA, Bennouna J, Soria JC, et al. T-Cell-Inflamed Gene-Expression Profile, Programmed Death Ligand 1 Expression, and Tumor Mutational Burden Predict Efficacy in Patients Treated With Pembrolizumab Across 20 Cancers: KEYNOTE-028. J Clin Oncol (2019) 37(4):318–27. doi: 10.1200/JCO.2018.78.2276
32. Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and Microenvironment Evolution During Immunotherapy With Nivolumab. Cell (2017) 171(4):934–949.e16. doi: 10.1016/j.cell.2017.09.028
33. Carbone DP, Reck M, Paz-Ares L, Creelan B, Horn L, Steins M, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med (2017) 376(25):2415–26. doi: 10.1056/NEJMoa1613493
34. McGrail DJ, Pilie PG, Rashid NU, Voorwerk L, Slagter M, Kok M, et al. High Tumor Mutation Burden Fails to Predict Immune Checkpoint Blockade Response Across All Cancer Types. Ann Oncol (2021) 32(5):661–72. doi: 10.1016/j.annonc.2021.02.006
35. Schroeder HW. B-Cell Development and Differentiation. In: Clinical Immunology, Book, 5th Edition Elsevier, (2019) pp. 107–18.e1. doi: 10.1016/B978-0-7020-6896-6.00007-7
36. Varn FS, Wang Y, Cheng C. A B Cell-Derived Gene Expression Signature Associates With an Immunologically Active Tumor Microenvironment and Response to Immune Checkpoint Blockade Therapy. Oncoimmunology (2019) 8(1):e1513440. doi: 10.1080/2162402X.2018.1513440
37. Pistoia V, Morandi F, Bianchi G, Pezzolo A, Prigione I, Raffaghello L. Immunosuppressive Microenvironment in Neuroblastoma. Front Oncol (2013) 3:167. doi: 10.3389/fonc.2013.00167
38. Quist J, Mirza H, Cheang MCU, Telli ML, O'Shaughnessy JA, Lord CJ, et al. A Four-Gene Decision Tree Signature Classification of Triple-Negative Breast Cancer: Implications for Targeted Therapeutics. Mol Cancer Ther (2019) 18(1):204–12. doi: 10.1158/1535-7163.MCT-18-0243
39. Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Dabrafenib and Trametinib Versus Dabrafenib and Placebo for Val600 BRAF-Mutant Melanoma: A Multicentre, Double-Blind, Phase 3 Randomised Controlled Trial. Lancet (2015) 386(9992):444–51. doi: 10.1016/S0140-6736(15)60898-4
40. Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined Vemurafenib and Cobimetinib in BRAF-Mutated Melanoma. N Engl J Med (2014) 371(20):1867–76. doi: 10.1056/NEJMoa1408868
41. Dummer R, Ascierto PA, Gogas HJ, Arance A, Mandala M, Liszkay G, et al. Encorafenib Plus Binimetinib Versus Vemurafenib or Encorafenib in Patients With BRAF-Mutant Melanoma (COLUMBUS): A Multicentre, Open-Label, Randomised Phase 3 Trial. Lancet Oncol (2018) 19(5):603–15. doi: 10.1016/S1470-2045(18)30142-6
42. Dummer R, Schadendorf D, Ascierto PA, Arance A, Dutriaux C, Di Giacomo AM, et al. Binimetinib Versus Dacarbazine in Patients With Advanced NRAS-Mutant Melanoma (NEMO): A Multicentre, Open-Label, Randomised, Phase 3 Trial. Lancet Oncol (2017) 18(4):435–45. doi: 10.1016/S1470-2045(17)30180-8
43. Puzanov I, Ribas A, Robert C, Schachter J, Nyakas M, Daud A, et al. Association of BRAF V600E/K Mutation Status and Prior BRAF/MEK Inhibition With Pembrolizumab Outcomes in Advanced Melanoma: Pooled Analysis of 3 Clinical Trials. JAMA Oncol (2020) 6(8):1256–64. doi: 10.1001/jamaoncol.2020.2288
44. Hodi FS, Chesney J, Pavlick AC, Robert C, Grossmann KF, McDermott DF, et al. Combined Nivolumab and Ipilimumab Versus Ipilimumab Alone in Patients With Advanced Melanoma: 2-Year Overall Survival Outcomes in a Multicentre, Randomised, Controlled, Phase 2 Trial. Lancet Oncol (2016) 17(11):1558–68. doi: 10.1016/S1470-2045(16)30366-7
Keywords: melanoma, CTLA-4 blockade, PD-1 blockade, MAPK pathway, immunotherapy
Citation: Ye T, Zhang J-Y, Liu X-Y, Zhou Y-H, Yuan S-Y, Yang M-M, Xie W-Z, Gao C, Chen Y-X, Huang M-L, Ye C-Z and Chen J (2022) The Predictive Value of MAP2K1/2 Mutations on Efficiency of Immunotherapy in Melanoma. Front. Immunol. 12:785526. doi: 10.3389/fimmu.2021.785526
Received: 29 September 2021; Accepted: 07 December 2021;
Published: 06 January 2022.
Edited by:
Zong Sheng Guo, Roswell Park Comprehensive Cancer Center, United StatesReviewed by:
Qingyue Zheng, Peking Union Medical College Hospital (CAMS), ChinaYuanzhuo Wang, Chinese Academy of Medical Sciences and Peking Union Medical College, China
Copyright © 2022 Ye, Zhang, Liu, Zhou, Yuan, Yang, Xie, Gao, Chen, Huang, Ye and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Chen, Y2hlbmppbmd3aEBodXN0LmVkdS5jbg==; Cheng-Zhi Ye, eWN6MTUzOTE1Njk5NzBAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Ting Ye1†
Ting Ye1† Meng-Mei Yang
Meng-Mei Yang Jing Chen
Jing Chen