- 1Department of Medicine, Division of Pulmonary and Critical Care Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL, United States
- 2Canning Thoracic Institute, Northwestern University Feinberg School of Medicine, Chicago, IL, United States
- 3Department of Biochemistry and Molecular Genetics, Northwestern University Feinberg School of Medicine, Chicago, IL, United States
- 4Simpson Querrey Institute for Epigenetics, Northwestern University Feinberg School of Medicine, Chicago, IL, United States
FoxP3+ regulatory T (Treg) cells maintain immune homeostasis, promote self-tolerance, and have an emerging role in resolving acute inflammation, providing tissue protection, and repairing tissue damage. Some data suggest that FoxP3+ T cells are plastic, exhibiting susceptibility to losing their function in inflammatory cytokine-rich microenvironments and paradoxically contributing to inflammatory pathology. As a result, plasticity may represent a barrier to Treg cell immunotherapy. Here, we discuss controversies surrounding Treg cell plasticity and explore determinants of Treg cell stability in inflammatory microenvironments, focusing on epigenetic mechanisms that clinical protocols could leverage to enhance efficacy and limit toxicity of Treg cell-based therapeutics.
Introduction
In health, regulatory T (Treg) cells are essential for maintaining immune homeostasis and promoting self-tolerance. These powerful immuno-modulatory cells, which comprise a subset of CD4+ T cells expressing CD25 (IL-2Rα) and the master transcription factor FoxP3 in humans and mice, suppress immune activation via inhibitory cell surface molecules (e.g., CTLA-4 and PD-1) and secretion of anti-inflammatory cytokines (e.g., IL-10 and TGF-β) to dampen pro-inflammatory effector immune cell functions (1–4). Recent data demonstrate that Treg cells also coordinate resolution of inflammation, provide tissue protection, and orchestrate repair of tissue damage, potentially rendering them useful to treat acute inflammation and tissue injury (5–19). Some animal experiments and observations in humans suggest that FoxP3+ T cells can lose their identity and function following exposure to inflammatory cytokines, resulting in loss of the canonical Treg cell transcriptional signature and acquisition of various helper T (Th) cell pro-inflammatory functions (20–25). Hence, the possibility of Treg cell plasticity represents a barrier to incorporating Treg cells into clinical protocols.
Treg cell development in the thymus involves the establishment of a specific epigenetic landscape, which is independent of, but complimentary to, FoxP3 expression and is required for specification of Treg cell lineage identity and function (26–30). Instability of Treg cell identity and function thus results from the loss of FoxP3 expression or changes in the epigenetic landscape. Natural Treg cells (nTreg cells) originate from the thymus with these transcriptional and epigenetic requirements established, persisting as a self-renewing population in the periphery (31, 32). While nTreg cells possess robust immunosuppressive capabilities, they comprise only 5–10% of human peripheral CD4+ T cells, thus requiring prolonged ex vivo expansion times (~2–5 weeks) to use them in therapeutic transfer protocols targeting acute inflammation. These long culture times thus limit the practicality of nTreg cells to treat acute inflammatory diseases or to promote tissue protection and repair following an acute injury. As naïve T cells are significantly more abundant than nTreg cells in peripheral blood, high numbers of induced Treg (iTreg) cells—naïve CD4+ T cells in which FoxP3 expression and a Treg cell phenotype have been induced by TGF-β in vitro—are rapidly obtainable, presenting a potential alternative to nTreg cells in clinical protocols. Data from murine studies suggest that iTreg cells can be generated within a few days (30, 33–35), possibly facilitating the use of iTreg cells in therapeutic transfer protocols targeting acute inflammation and injury. Induced Treg cells lack nTreg cell-type epigenetic patterns, particularly in DNA methylation, that drive phenotypic stability (27, 29, 30). Thus, defining exploitable epigenetic mechanisms that allow for nTreg cell-level stability in iTreg cells is of particular interest in the pursuit of using iTreg cells as immunotherapy. While minor populations of some immune and non-immune cells can express FoxP3 (36), our review focuses on FoxP3+ T cells.
Treg Cells as Immunotherapy
The therapeutic goals of using Treg cells to induce self-tolerance and mitigate inflammation are to ameliorate immune dysregulation using minimal or no immunosuppressive pharmacotherapy while allowing proper immune responses to take place during the host response to pathogens (37). Pilot trials of Treg cells as cellular immunotherapy in humans have provided proof-of-concept for their use in diseases of auto-reactivity—including type 1 diabetes, graft-versus-host disease, and organ allo-transplantation—with promising results (38–44). In these studies, nTreg cells were isolated from patients for subsequent re-infusion either after storage or ex vivo expansion. Primary strategies of isolation involve obtaining mononuclear cells from leukopheresates, peripheral whole blood, or umbilical cord blood followed by Treg cell sorting using immuno-magnetic systems or flow cytometry cell sorting (45, 46). Ex vivo expansion protocols achieve large, pure, and suppressive cell populations while maintaining good manufacturing practice standards (47–50). Clinical trial protocols have infused dosages as high as 5 x 109 cells, which typically take 2–5 weeks to generate. To enhance Treg cell purity during expansion, several groups have studied the effect of culture in the presence of the mTOR inhibitor rapamycin, as it selectively promotes growth of CD4+CD25+FoxP3+ Treg cells while concomitantly inhibiting CD4+CD25– (non-Treg) effector T cells at low doses (50, 51).
Beyond induction of self-tolerance, emerging evidence demonstrates that Treg cells orchestrate resolution of inflammation, provide tissue protection, and coordinate tissue repair following a growing list of acute insults, including lung injury due to pneumonia, muscle injury, dermal injury, and vascular endothelial injury (5–18). The tissue-protective and -reparative properties of Treg cells appear to be the result of specific inflammatory signals, such as the cytokine IL-18 and the alarmin IL-33. Growth factor receptor ligands such as amphiregulin and keratinocyte growth factor may, in part, mediate these tissue-protective and -reparative functions, which are distinct from canonical T cell receptor (TCR) stimulation-dependent Treg cell suppressive functions. Promising data support broadening the use of Treg cells for the treatment of acute inflammation and tissue injury (19). Nevertheless, some lines of evidence suggest that Treg cells can exhibit plasticity in inflamed and damaged microenvironments, resulting in loss of their identity and the potential to gain pro-inflammatory effector functions (22). Discussed below, manipulating epigenetic determinants of Treg cell stability could aid efforts to maintain their beneficial functions in inflamed and damaged tissue microenvironments while limiting the potential for conversion into pathogenic T cells.
Epigenetic Determinants of Treg Cell Development and Stability
Epigenetic mechanisms include a set of processes that modify transcriptional patterns without altering the underlying DNA sequence, allowing for heritable changes in gene expression. DNA methylation is a dynamic epigenetic modification mediated by a family of DNA methyltransferases (DNMTs) that add methyl groups to the 5’ carbon of cytosine bases to create 5-methylcytosine (5mC), which is associated with chromatin inaccessibility and transcriptional repression (52, 53). The DNMT family member DNMT1 catalyzes maintenance DNA methylation, and ubiquitin-like containing PHD and RING finger domains 1 (UHRF1) recruits DNMT1 to hemi-methylated DNA during DNA replication, serving to maintain DNA methylation patterning in mitotic cells. DNA demethylation occurs either passively during DNA replication or via the catalytic activity of the ten-eleven translocation (TET) family of dioxygenases, which oxidize 5mC to 5-hydroxymethylcytosine (5hmC) and other intermediates that ultimately restore unmethylated cytosine at a given position (54). Histone modifications represent another form of dynamic epigenetic alteration to chromatin, which, in combination with the non-catalytic domains of histone-modifying proteins, modulates transcriptional activity (55). For example, enzymes that promote monomethylation of lysine 4 on histone H3 (H3K4me1) and acetylation of lysine 27 on histone H3 (H3K27ac) mark active enhancer elements and promote transcription. Importantly, cellular metabolism provides substrates for epigenetic writers and erasers (e.g., methyltransferases and demethylases) in Treg cells (56). For example, our group determined that the mitochondrial electron transport chain in Treg cells is required to prevent the accumulation of toxic metabolites such as 2-hydroxyglutarate, which inhibits α-ketoglutarate-dependent enzymes such as the TETs (57). We found that loss of mitochondrial electron transport chain complex III in Treg cells results in increased levels of 2-hydroxyglutarate, altered DNA methylation patterning, and impaired Treg cell suppressive function.
The field has now recognized that stable Treg cell phenotype and function depend on a specific epigenetic landscape to maintain lineage-defining Treg cell gene expression, including at the locus encoding FoxP3 (Table 1) (26–30). Accordingly, nTreg cells can be distinguished from conventional T cells and iTreg cells by characteristic DNA hypomethylation at the Foxp3 promoter and additional elements within Foxp3-associated enhancer regions, such as the Treg cell-specific demethylated region (TSDR), also known as conserved noncoding DNA sequence 2 (CNS2). How the Treg cell lineage establishes and stabilizes its epigenetic signature remains an active area of investigation (Figure 1A). During development in the thymus, TET enzymes and HATs, such as CBP (also known as CREBBP) and p300, are recruited to modify the Foxp3 locus for induction and maintenance of FoxP3 expression, which is followed by establishment of a Treg cell-specific gene expression profile (60, 61). Epigenetic modification at the Foxp3 locus involves TET-mediated 5hmC accumulation at the TSDR and other key regions (59). Importantly, in the absence of these epigenetic modifications, Treg cells can lose FoxP3 expression and gain IL-17 expression.

Table 1 Selected epigenetic modifiers discussed in the text and their role in Treg cell development and maintenance.
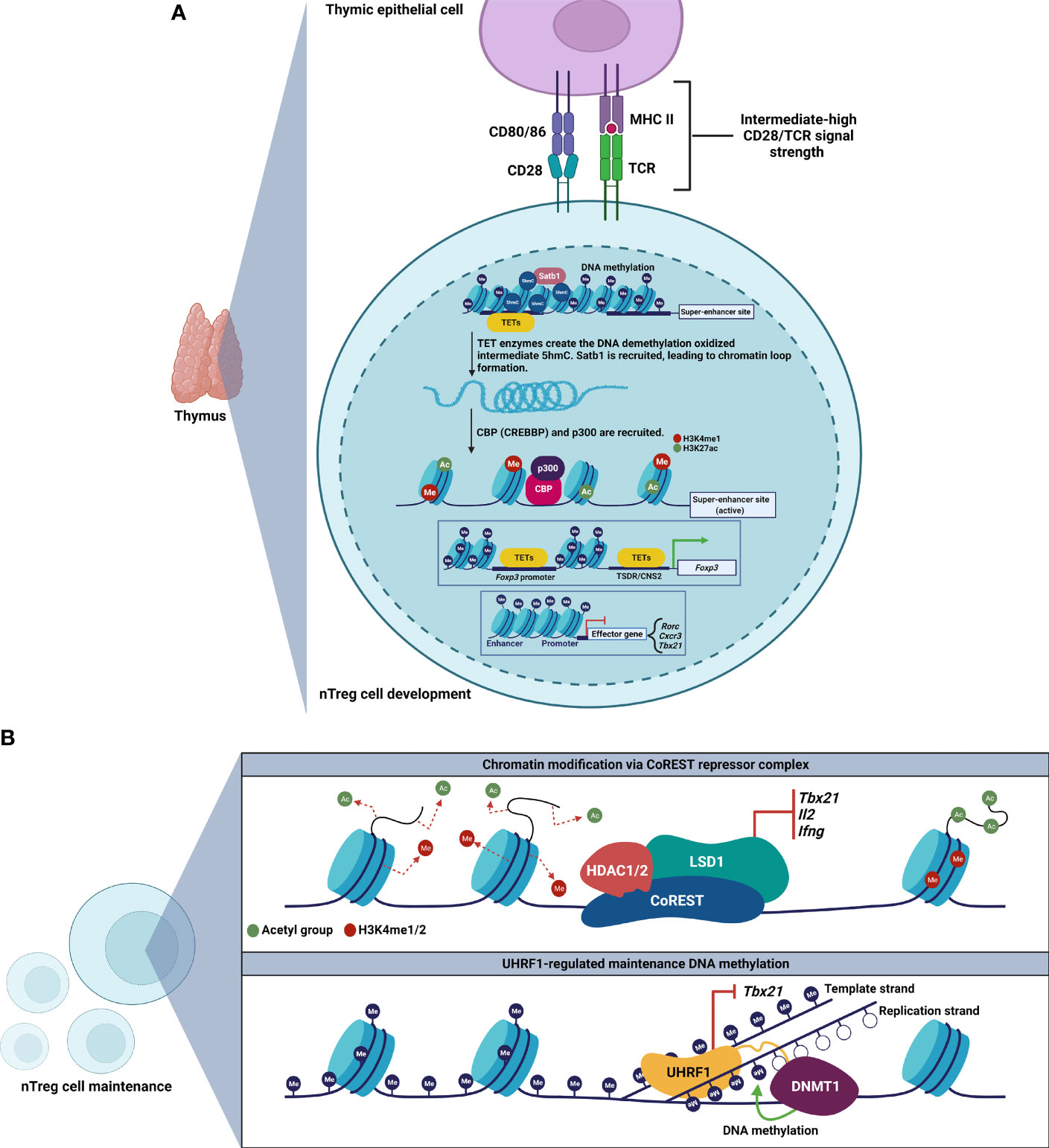
Figure 1 Development and maintenance of Treg cell epigenetic landscapes. (A) Thymic Treg cell development involves establishment of a Treg cell-specific super-enhancer landscape at Foxp3 and other key loci. The chromatin organizer Satb1 establishes a super-enhancer landscape in Treg cells, characterized by active enhancer histone marks, and TET-mediated DNA hypomethylation. Loci encoding effector T cell signature genes are hypermethylated. (B) Maintenance of Treg cell epigenetic patterning requires the CoREST repressor complex (top) and the epigenetic regulator UHRF1 (bottom) to repress loci encoding inflammatory genes.
Beyond the Foxp3 locus, investigators have determined that Treg cell-specific super-enhancers—genomic regions with dense clustering of highly active lineage-defining enhancer elements—regulate Foxp3 and other Treg cell lineage-defining genes (58). In thymic pre-Treg cells, the genome organizer Satb1 binds Treg cell-specific super-enhancer sites, resulting in chromatin loop formation that allows distal regulatory elements to interact with and recruit transcription factors and epigenetic modifiers to activate and stabilize the Treg cell-defining gene regulatory network. Deletion of Satb1 in double-positive thymocytes results in impaired Treg cell-super-enhancer activation and failure to induce Treg cell signature genes, leading to fatal autoimmunity in vivo. These studies also revealed that DNA hypomethylation is a distinguishing feature of the Treg cell-specific super-enhancer landscape in Treg cells. Moreover, our work demonstrated that the Treg cell-specific super-enhancer epigenetic pattern shown to be causally deterministic in mice is also present in Treg cells obtained from the alveolar spaces of patients with severe pneumonia (67). Thus, the Treg cell-specific super-enhancer landscape appears to be a conserved and translationally relevant epigenetic pattern, prompting clinical trials of Treg cell infusions for patients with the acute respiratory distress syndrome due to severe SARS-CoV-2 pneumonia (16, 68, 69).
The role of maintenance of epigenetic marks in stabilizing lineage identity following the initial establishment of epigenetic patterns at Treg cell-specific super-enhancers and at other important non-coding elements remains unclear (Figure 1B). Experimental data suggest that some chromatin organizers necessary for lineage specification are not required for lineage stability. Indeed, deletion of Satb1 in differentiated Treg cells does not lead to any changes in Treg cell numbers or phenotype, indicating that Satb1 is dispensable for Treg cell maintenance (58). In contrast, loss of the chromatin-modifying CoREST repressor complex disrupts FoxP3-driven repression of Th1 cell signature genes encoding T-BET, IL-2, and IFN-γ. Consequently, loss of CoREST results in Treg cell production of IL-2 and IFN-γ, impaired Treg cell function, and enhanced anti-tumor immunity (62).
Maintenance DNA methylation also controls Treg cell stability following FoxP3 induction in nTreg cells. We observed that loss of an epigenetic regulator responsible for maintenance DNA methylation, UHRF1, at the thymic FoxP3+ stage of development in nTreg cells leads to loss of FoxP3 expression and a Scurfy-like phenotype (30). We went on to determine that Treg cell-conditional deletion of UHRF1 results in failure of nTreg cells to persist after FoxP3 induction in the thymus, generating hyperinflammatory ex-FoxP3 cells in which loss of maintenance DNA methylation derepresses Th1 cell signature genes, including Tbx21 (encodes T-BET). Interestingly, UHRF1-deficient ex-FoxP3 cells exhibit downregulation of the TET demethylases and DNA hypermethylation at core Treg cell loci, including Foxp3. These observations support a mechanism in which loss of maintenance DNA methylation unleashes a secondary wave of DNA methylation at core Treg cell loci to generate hyperinflammatory, Th1-skewed, ex-FoxP3 cells. Consistent with these observations, others found that constitutive deletion of the maintenance DNA methyltransferase DNMT1, but not the de novo methyltransferase DNMT3A, results in diminished numbers and suppressive function of Treg cells (65). Interestingly, DNMT1-deficient Treg cells maintain Treg cell-specific DNA hypomethylation patterns at Foxp3, and we determined that UHRF1-deficient Treg cells possess the Treg cell-specific super-enhancer landscape. Additional evidence supports that pan-T cell-specific deficiency of UHRF1 results in defective proliferation and functional maturation of colonic Treg cells (63). Thus, nTreg cells require both a canonical hypomethylation pattern as well as maintenance methylation at loci encoding inflammatory programs to stabilize their lineage identity and function. The role of maintenance DNA methylation in stabilizing iTreg cell identity and function remains less clear. Intriguingly, while we found that UHRF1 is dispensable for induction of FoxP3 expression in iTreg cells (30), others observed augmented suppressive function in iTreg cells generated from UHRF1-deficient naïve CD4+ T cells, even in inflammatory microenvironments (64). As inflammation may drive instability of FoxP3+ T cells, we will explore in the following section how microenvironmental inflammatory signals control T cell plasticity via their influence on epigenetic modifiers.
Cytokine Signaling and the Epigenetics of Treg Cell Plasticity
Plasticity refers to the capacity of CD4+ T cells to depolarize their specialized functional programs in response to the cytokine milieu of the local microenvironment, resulting in loss of their functional identity and potential for a gained Th-skewed cell phenotype (70). Careful lineage-tracing studies in mice reported the eminent stability of the Treg cell lineage under physiologic and inflammatory conditions (31), and others have argued that the plasticity observed in FoxP3+ T cells in inflammatory or lymphopenic microenvironments results from cellular heterogeneity rather than reprogramming (71). Indeed, minor populations of conventional T cells can transiently express FoxP3 and then differentiate into ex-FoxP3 Th-skewed cells (72, 73). These populations retain the ability to re-express FoxP3 upon activation, a finding correlated with the demethylated status of the TSDR in the conventional T cell population and possibly the Treg cell population.
Nevertheless, several lines of evidence describe plasticity occurring in FoxP3+ T cells to produce ex-FoxP3 cells or FoxP3+ Th-like cells in response to specific signaling events (Figure 2A). For example, Th1-like IFN-γ-secreting FoxP3+ T cells exist in patients with relapsing-remitting multiple sclerosis, a finding recapitulated in vitro when investigators cultured Treg cells from healthy people in the presence of IL-12 (23). IL-4 signaling promotes the development of ex-FoxP3 Th2-like cells in the setting of chronic helminth infection (25). The presence of an IL-6-, IL-21-, and activated B cell-rich environment in the Peyer’s patches of mice results in FoxP3+ T cell transformation into cells with characteristics of follicular Th (Tfh) cells that are capable of promoting germinal center formation (21). Th17 cells express the orphan nuclear receptor RORγt and a characteristic cytokine signature, including IL-17. Regulation of the locus encoding IL-17 via reciprocal actions of STAT3 and STAT5 in part determines Th17-Treg cell plasticity, producing FoxP3+IL-17+ or FoxP3+RORγt+ cells (20, 74, 75). Clinically, the joints of patients with rheumatoid arthritis contain FoxP3+IL-17+ cells, which are also present in mice with experimental joint inflammation (24). These FoxP3+IL-17+ cells may represent a transitional cell population moving toward complete loss of FoxP3, as synovial fibroblast-derived IL-6 can cause CD4+FoxP3+ T cells to lose FoxP3 expression and differentiate into Th17 cells in mice with experimental inflammatory arthritis. Further, treatment of patients with rheumatoid arthritis using the IL-6R inhibitor tocilizumab resulted in significant symptomatic benefit along with decreases in circulating Th17 cells and increases in circulating Treg cells (76). Despite expressing RORγt, FoxP3+RORγt+ cells in the intestines of mice demonstrate transcriptional and epigenetic profiles more similar to FoxP3+RORγt− cells than to FoxP3−RORγt+ cells, including demethylation at Treg cell-characteristic genes encoding FOXP3, CTLA-4, GITR, EOS, and HELIOS. FoxP3+RORγt+ cells retain suppressive function and are more suppressive than FoxP3+RORγt− cells in a T cell transfer colitis model (77, 78). Collectively, these reports identify plasticity within FoxP3+ T cell populations that is induced and modified by specific cytokine microenvironments.
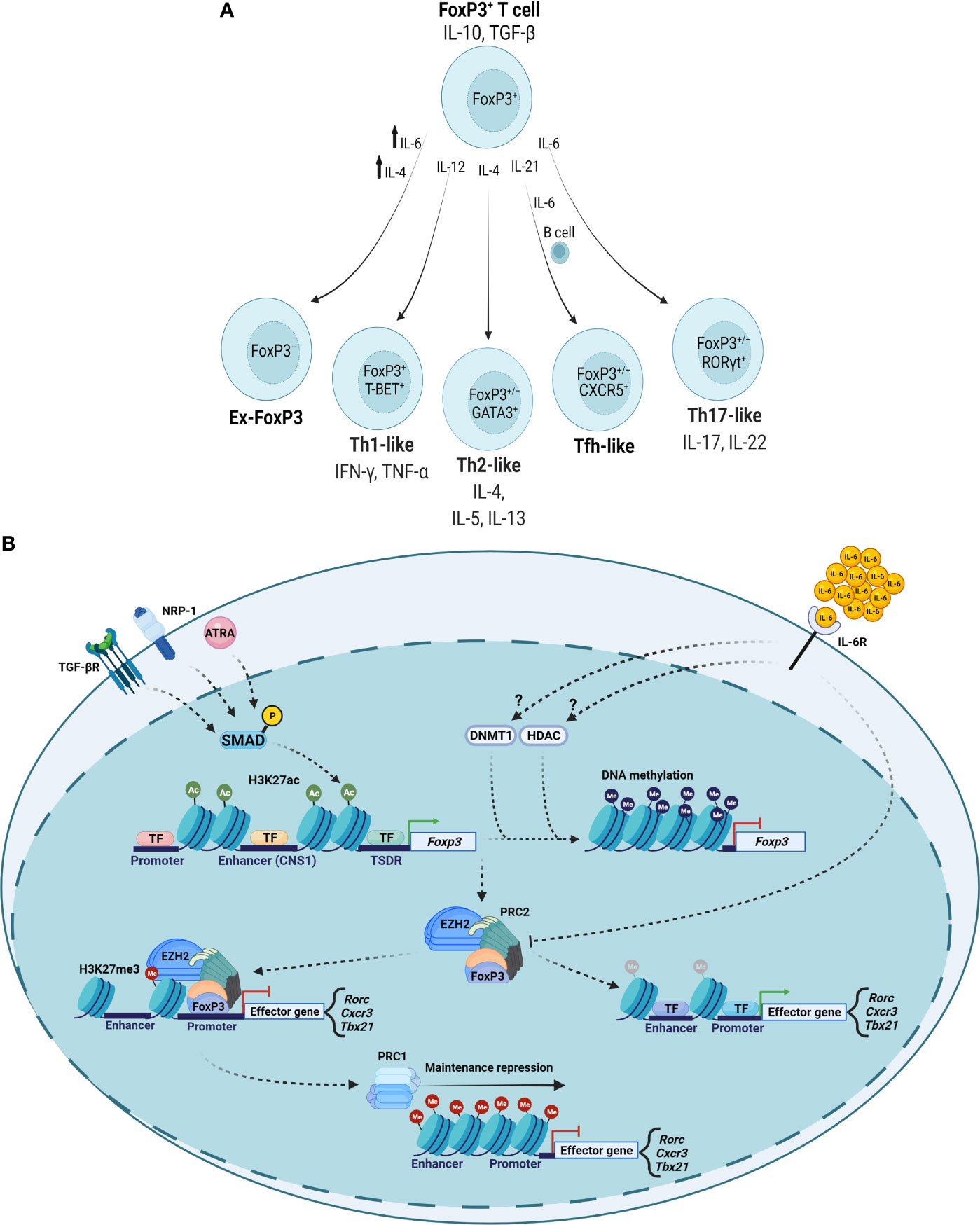
Figure 2 Cytokine-mediated epigenetic reprogramming of FoxP3+ T cell populations. (A) Specific cytokine microenvironments can repolarize FoxP3+ T cells with variable effects on FoxP3 expression and Th cell-like phenotypes. (B) TGF-β, NRP-1, and ATRA signal to maintain Treg cell-type epigenetic patterns. Inflammatory cytokines such as IL-6 can promote DNMT and HDAC activity to result in loss of Foxp3 gene expression and modulate PRC complexes to depress loci encoding inflammatory genes. TF, transcription factor.
Data suggest that epigenetic alterations underlie the ability of FoxP3+ T cells to polarize in response to microenvironmental inflammatory signals (Figure 2B). For example, some experiments determined that IL-6 can promote DNMT1-mediated DNA methylation and that histone deacetylase (HDAC) activity can destabilize FoxP3+ T cells (79, 80). EZH2 (enhancer of zeste homolog 2) is the enzymatic subunit of polycomb repressive complex 2 (PRC2), which participates in histone methylation to result in transcriptional repression. PRC1 (polycomb repressive complex 1) maintains the repressed transcriptional state, interacting with chromatin by recognizing PRC2-established H3K27me3 marks in an equilibrium with other histone-modifying complexes and repressive DNA methylation (81, 82). In inflammatory microenvironments, FoxP3-containing complexes incorporate EZH2, which deposits repressive chromatin modifications at FoxP3-bound loci (66). Studies of intestinal inflammation in inflammatory bowel disease suggested a disrupted FoxP3-EZH2 physical interaction that investigators recapitulated by treatment with IL-6 (83). In the context of aging (84), we found that cell-autonomous age-related alterations in DNA methylation drive plasticity in FoxP3+ T cells in the inflamed lungs of aged but not young mice during recovery from influenza pneumonia (18, 85). Our studies in aged hosts revealed co-expression of Th1- and Th17-associated transcription factors (T-BET and RORγt) in lung FoxP3+ T cells 60 days following influenza virus infection along with expression of cognate cytokines (IFN-γ and IL-17). In contrast, other signaling events may stabilize Treg cell-type epigenetic patterns. Transcriptional and epigenetic analyses of human Treg cells from inflamed synovial joints compared with peripheral blood in pediatric patients revealed that Treg cells differentiate into effector Treg (eTreg) cells that are suppressive in vitro and display increased expression of core Treg cell genes (86). Importantly, epigenetic alterations in active enhancer marks, including H3K4me1 and H3K27ac, explained these transcriptional differences. CD103+ intestinal dendritic cells secrete all-trans retinoic acid (ATRA) and TGF-β to induce histone acetylation at the CNS1 region of the FOXP3 locus to promote FoxP3 expression while restricting Th17 polarization (87). Experimental data suggest that neuropilin-1 (NRP-1) also reinforces the stability of Treg cells in inflammatory environments, as silencing of NRP-1 results in diminished FoxP3 expression with a correlative increase in DNA methylation at the TSDR (88). Collectively, several lines of evidence support that epigenetic mechanisms determine FoxP3+ T cell plasticity in inflammatory environments, prompting consideration of leveraging these mechanisms to promote functional stability in clinical Treg cell transfer protocols.
Stabilizing Treg Cell Immunotherapeutic Function via Ex Vivo Epigenetic Modification
Treg cell plasticity may represent an adaptive feature to regulate a given microenvironment. Clinically, however, therapeutic protocols will require a greater understanding of Treg cell plasticity to maximize on-target function and limit unintended toxic inflammation. Recent mouse studies have performed ex vivo modifications to induced and natural Treg cells to enhance their stability in the presence of inflammatory cytokines (Figure 3). As inhibition of DNMTs or HDACs can induce FoxP3 expression and support Treg cell identity, DNMT or HDAC inhibitors could stabilize nTreg cells in culture before therapeutic infusion (11, 27, 79, 89–92). The DNMT adapter protein UHRF1 also remains a drug target of interest. As noted above, adoptive transfer of iTreg cells generated from UHRF1-deficient naïve CD4+ T cells display enhanced suppressive function in colitis models (64). In contrast, UHRF1 overexpression in T cells causes BCL6 downregulation and decreased Tfh cell differentiation, which may serve as a potential therapeutic target in systemic lupus erythematosus (93). Nevertheless, the field requires further data to determine whether modulation of UHRF1 in mature iTreg cells translates into a more stable, suppressive, and reparative state in vivo. Studies of HDAC inhibitors found that they are capable of promoting thymic production of nTreg cells and inducing iTreg cells ex vivo, leading to efficacy in attenuating inflammatory bowel disease and promoting cardiac allograft tolerance (94). Going forward, selective manipulation of HDAC subclass function may stabilize and promote nTreg and iTreg cell function in clinical trial protocols (95).
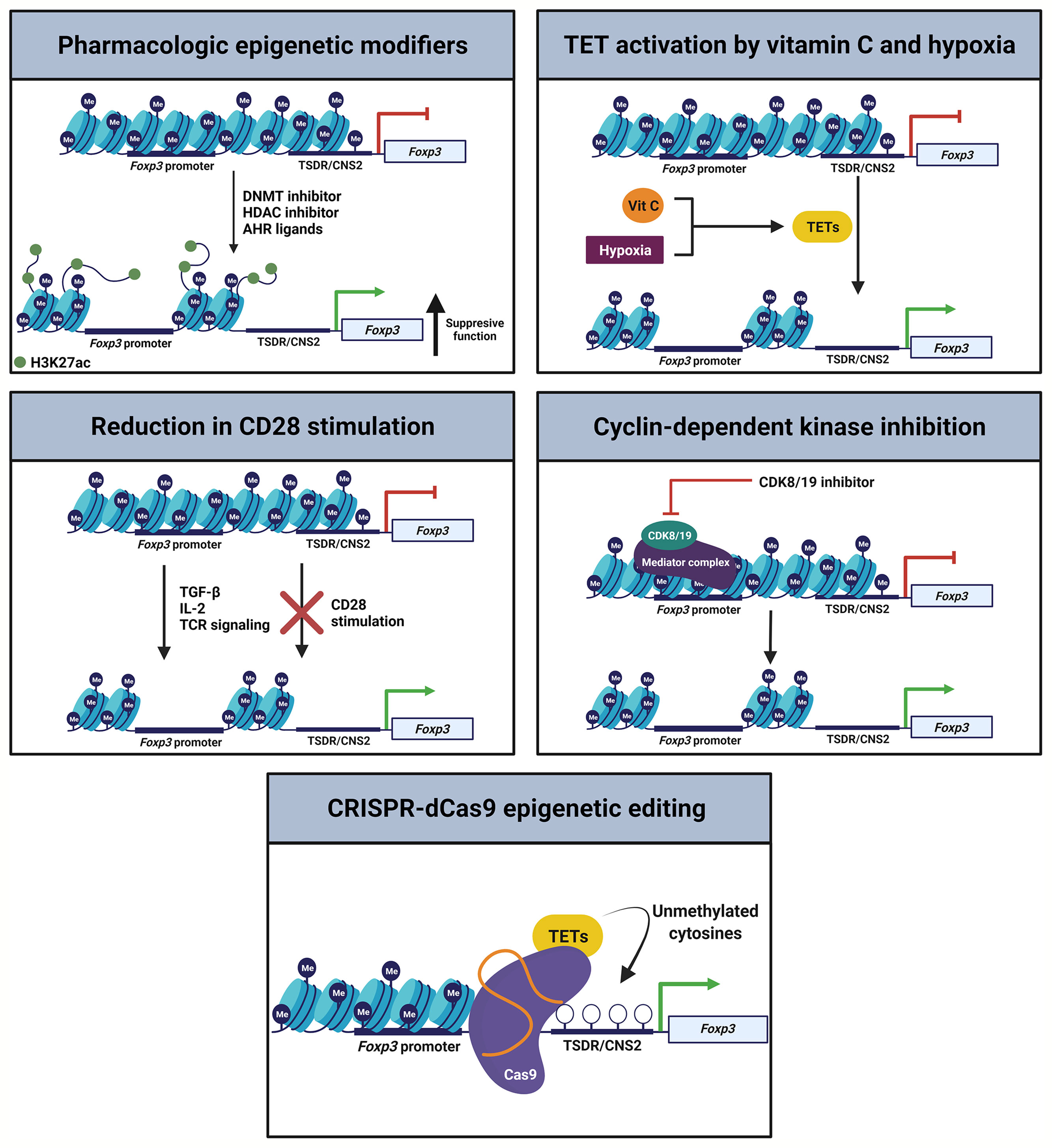
Figure 3 Epigenetic strategies to promote Treg cell stability. Multiple orthogonal pathways could be leveraged during ex vivo generation of iTreg cells or expansion of nTreg cells to promote FoxP3 expression and Treg cell stability, enhancing the efficacy of therapeutic transfer.
The aryl-hydrocarbon receptor (AHR) is a ligand-activated transcription factor that functions in part through interaction with epigenetic regulators, including the mediator complex (96). AHR regulates Treg-Th17 cell plasticity in mice via activation by specific ligands. AHR activation by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) induces functional Treg cells that suppress experimental autoimmune encephalitis. Treatment of mice with TCDD also attenuates delayed hypersensitivity responses, which are associated with induction of Treg cells and suppression of Th17 cells in mesenteric lymph nodes (97). Intriguingly, treatment with AHR ligands such as TCDD or butyrate inhibits pro-inflammatory HDAC classes I and II (97). Hence, generation of iTreg cells or expansion of nTreg cells in the presence of AHR ligands may stabilize the Treg cell epigenetic landscape to maintain their identity following therapeutic transfer.
Several investigations have examined modulation of TET enzyme activity via treatment with the TET activator vitamin C (ascorbic acid) or culture under low oxygen conditions to enhance Treg cell induction and stability. Vitamin C facilitates demethylation of the Foxp3 CNS2 enhancer region in a TET2/3-dependent manner to increase the stability of FoxP3 expression in TGF-β-induced Treg cells (33, 98). Further, culture of iTreg cells under low oxygen (5%) conditions facilitates CNS2 demethylation and stabilization of FoxP3 both in vitro and in vivo, a finding that correlates with increased TET expression. These post-hypoxia Treg cells exhibit stronger suppressive activity in a colitis model compared with untreated iTreg cells (34), informing potential immunotherapeutic iTreg cell induction protocols. Moreover, activation of TET enzyme activity during ex vivo nTreg cell expansion protocols could likewise support their stability and function.
While CD28 is essential for optimal thymic Treg cell development, CD28 is surprisingly dispensable for Treg cell induction or Treg cell-specific gene hypomethylation in the intestines of mice (35, 99). iTreg cell induction via TGF-β, IL-2, and TCR agonism in the absence of CD28 signaling induces nTreg cell-type DNA hypomethylation in conventional T cells while hindering skewing toward Th cell phenotypes. Data suggest that CD28 acts via the PKC-NF-κB signaling pathway during iTreg cell generation and that inhibition of this pathway enables de novo acquisition of nTreg cell-type DNA hypomethylation. Induced Treg cells generated under conditions of absent CD28 stimulation stably express FoxP3 after adoptive transfer and effectively suppress antigen-specific immune responses in vivo (35). Thus, potential modifications to standard nTreg cell culture practices or iTreg cell induction protocols include relatively straightforward adjustments such as reducing CD28 stimulation during cellular activation.
Cyclin-dependent kinase 8 (CDK8) and CDK19 reversibly associate with the mediator complex as well as regulate epigenetic events such as histone modification and chromatin remodeling (100, 101). Inhibition of CDK8 and CDK19 in conventional T cells induces FoxP3 expression and suppressive function independent of TGF-β signaling in antigen-stimulated effector-memory as well as naïve CD4+ and CD8+ T cells (102). Importantly, inflammatory cytokines do not appear to affect the induction of FoxP3 expression following CDK inhibition. These results suggest that CDK8 and CDK19 physiologically repress FoxP3 expression in activated conventional T cells, prompting consideration of targeting CDK8 and CDK19 in ex vivo iTreg cell generation or nTreg cell expansion protocols.
Finally, CRISPR-dCas9 epigenome editing systems may be of use to enhance FoxP3 stability during and following Treg cell induction or expansion. Kressler and colleagues demonstrated a transient-transfection CRISPR-dCas9-based epigenetic editing method for the selective de-methylation of the TSDR within the endogenous chromatin environment of a living cell (103). The demethylation marks were durable over weeks, including after expression of the editing complex had ceased. Consistent with prior data, however, successful FoxP3 induction was not associated with a switch to a fully functional Treg cell phenotype, highlighting importance of establishing gene expression and methylation patterns at other key loci in the Treg cell genome.
Discussion
FoxP3+ Treg cells represent a powerful cell type capable of inducing self-tolerance, suppressing over-exuberant immune system activation, promoting resolution of inflammation, and effecting protection and repair of damaged tissues. Clinical trial protocols have applied Treg cell immunotherapy to disorders of auto- and allo-reactivity as well as to suppress damaging inflammation and hasten recovery from severe pneumonia. Epigenetic mechanisms, particularly those that regulate DNA methylation, control Treg cell lineage identity, stability, and function. Although the Treg cell lineage displays a strong tendency toward stability, many lines of evidence suggest that FoxP3+ T cells can exhibit plasticity in inflammatory microenvironments, with investigators observing both loss of canonical suppressive function and gain of inflammatory effector functions. Going forward, manipulating the epigenetic state of Treg cells ex vivo prior to infusion could stabilize their identity and function to enhance clinical efficacy while limiting the potential for off-target effects.
Author Contributions
AJ and BS conceptualized the manuscript, wrote the first draft of the manuscript, edited the manuscript, and edited the figures. CR provided essential conceptual input, edited the manuscript, and conceptualized and created the figures. Conceptualization: AJ and BS. Visualization: AJ, CR, and BS. Funding acquisition: BS. Supervision: BS. Writing – original draft: AJ and BS. Writing – review & editing: AJ, CR, and BS. All authors contributed to the article and approved the submitted version.
Funding
National Institutes of Health grant R01HL149883 (BS), National Institutes of Health grant R01HL153122 (BS), National Institutes of Health grant P01HL154998 (BS), National Institutes of Health grant P01AG049665 (BS), National Institutes of Health grant U19AI135964 (BS). The opinions expressed in this article are those of the authors and do not represent any position or policy of the National Institutes of Health.
Conflict of Interest
BS holds United States Patent No. US 10,905,706 B2, “Compositions and Methods to Accelerate Resolution of Acute Lung Inflammation,” and serves on the Scientific Advisory Board of Zoe Biosciences, in which he holds stock options.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Figures created with BioRender.com.
References
1. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 Programs the Development and Function of CD4+CD25+ Regulatory T Cells. Nat Immunol (2003) 4:330–6. doi: 10.1038/ni904
2. Hori S, Nomura T, Sakaguchi S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science (2003) 299:1057–61. doi: 10.1126/science.1079490
3. Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, et al. Foxp3 Transcription-Factor-Dependent and -Independent Regulation of the Regulatory T Cell Transcriptional Signature. Immunity (2007) 27:786–800. doi: 10.1016/j.immuni.2007.09.010
4. Williams LM, Rudensky AY. Maintenance of the Foxp3-Dependent Developmental Program in Mature Regulatory T Cells Requires Continued Expression of Foxp3. Nat Immunol (2007) 8:277–84. doi: 10.1038/ni1437
5. Mckinley L, Logar AJ, Mcallister F, Zheng M, Steele C, Kolls JK. Regulatory T Cells Dampen Pulmonary Inflammation and Lung Injury in an Animal Model of Pneumocystis Pneumonia. J Immunol (2006) 177:6215–26. doi: 10.4049/jimmunol.177.9.6215
6. D'Alessio FR, Tsushima K, Aggarwal NR, West EE, Willett MH, Britos MF, et al. CD4+CD25+Foxp3+ Tregs Resolve Experimental Lung Injury in Mice and are Present in Humans With Acute Lung Injury. J Clin Invest (2009) 119:2898–913. doi: 10.1172/JCI36498
7. Tamosiuniene R, Tian W, Dhillon G, Wang L, Sung YK, Gera L, et al. Regulatory T Cells Limit Vascular Endothelial Injury and Prevent Pulmonary Hypertension. Circ Res (2011) 109:867–79. doi: 10.1161/CIRCRESAHA.110.236927
8. Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, et al. A Special Population of Regulatory T Cells Potentiates Muscle Repair. Cell (2013) 155:1282–95. doi: 10.1016/j.cell.2013.10.054
9. Mock JR, Garibaldi BT, Aggarwal NR, Jenkins J, Limjunyawong N, Singer BD, et al. Foxp3+ Regulatory T Cells Promote Lung Epithelial Proliferation. Mucosal Immunol (2014) 7:1440–51. doi: 10.1038/mi.2014.33
10. Arpaia N, Green JA, Moltedo B, Arvey A, Hemmers S, Yuan S, et al. A Distinct Function of Regulatory T Cells in Tissue Protection. Cell (2015) 162:1078–89. doi: 10.1016/j.cell.2015.08.021
11. Singer BD, Mock JR, Aggarwal NR, Garibaldi BT, Sidhaye VK, Florez MA, et al. Regulatory T Cell DNA Methyltransferase Inhibition Accelerates Resolution of Lung Inflammation. Am J Respir Cell Mol Biol (2015) 52:641–52. doi: 10.1165/rcmb.2014-0327OC
12. Kuswanto W, Burzyn D, Panduro M, Wang KK, Jang YC, Wagers AJ, et al. Poor Repair of Skeletal Muscle in Aging Mice Reflects a Defect in Local, Interleukin-33-Dependent Accumulation of Regulatory T Cells. Immunity (2016) 44:355–67. doi: 10.1016/j.immuni.2016.01.009
13. Nosbaum A, Prevel N, Truong HA, Mehta P, Ettinger M, Scharschmidt TC, et al. Cutting Edge: Regulatory T Cells Facilitate Cutaneous Wound Healing. J Immunol (2016) 196:2010–4. doi: 10.4049/jimmunol.1502139
14. Dial CF, Tune MK, Doerschuk CM, Mock JR. Foxp3(+) Regulatory T Cell Expression of Keratinocyte Growth Factor Enhances Lung Epithelial Proliferation. Am J Respir Cell Mol Biol (2017) 57:162–73. doi: 10.1165/rcmb.2017-0019OC
15. Mock JR, Dial CF, Tune MK, Norton DL, Martin JR, Gomez JC, et al. Transcriptional Analysis of Foxp3+ Tregs and Functions of Two Identified Molecules During Resolution of ALI. JCI Insight (2019) 4:e124958. doi: 10.1172/jci.insight.124958
16. Gladstone DE, Kim BS, Mooney K, Karaba AH, D'Alessio FR. Regulatory T Cells for Treating Patients With COVID-19 and Acute Respiratory Distress Syndrome: Two Case Reports. Ann Intern Med (2020) 173:852–3. doi: 10.7326/L20-0681
17. Mock JR, Dial CF, Tune MK, Gilmore RC, O'neal WK, Dang H, et al. Impact of Regulatory T Cells on Type 2 Alveolar Epithelial Cell Transcriptomes During Resolution of Acute Lung Injury and Contributions of IFN-Gamma. Am J Respir Cell Mol Biol (2020) 63:464–77. doi: 10.1165/rcmb.2019-0399OC
18. Morales-Nebreda L, Helmin KA, Torres Acosta MA, Markov NS, Hu JY, Joudi AM, et al. Aging Imparts Cell-Autonomous Dysfunction to Regulatory T Cells During Recovery From Influenza Pneumonia. JCI Insight (2021) 6:e141690. doi: 10.1172/jci.insight.141690
19. Weinberg SE, Singer BD. Toward a Paradigm to Distinguish Distinct Functions of FOXP3(+) Regulatory T Cells. Immunohorizons (2021) 5:944–52. doi: 10.4049/immunohorizons.2100046
20. Koenen HJ, Smeets RL, Vink PM, Van Rijssen E, Boots AM, Joosten I. Human CD25highFoxp3pos Regulatory T Cells Differentiate Into IL-17-Producing Cells. Blood (2008) 112:2340–52. doi: 10.1182/blood-2008-01-133967
21. Tsuji M, Komatsu N, Kawamoto S, Suzuki K, Kanagawa O, Honjo T, et al. Preferential Generation of Follicular B Helper T Cells From Foxp3+ T Cells in Gut Peyer's Patches. Science (2009) 323:1488–92. doi: 10.1126/science.1169152
22. Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martinez-Llordella M, Ashby M, et al. Instability of the Transcription Factor Foxp3 Leads to the Generation of Pathogenic Memory T Cells In Vivo. Nat Immunol (2009) 10:1000–7. doi: 10.1038/ni.1774
23. Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T Helper Type 1-Like, Foxp3+ Regulatory T Cells in Human Autoimmune Disease. Nat Med (2011) 17:673–5. doi: 10.1038/nm.2389
24. Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-Hora M, Kodama T, et al. Pathogenic Conversion of Foxp3+ T Cells Into TH17 Cells in Autoimmune Arthritis. Nat Med (2014) 20:62–8. doi: 10.1038/nm.3432
25. Pelly VS, Coomes SM, Kannan Y, Gialitakis M, Entwistle LJ, Perez-Lloret J, et al. Interleukin 4 Promotes the Development of Ex-Foxp3 Th2 Cells During Immunity to Intestinal Helminths. J Exp Med (2017) 214:1809–26. doi: 10.1084/jem.20161104
26. Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic Control of the Foxp3 Locus in Regulatory T Cells. PloS Biol (2007) 5:e38. doi: 10.1371/journal.pbio.0050038
27. Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, et al. Epigenetic Regulation of Foxp3 Expression in Regulatory T Cells by DNA Methylation. J Immunol (2009) 182:259–73. doi: 10.4049/jimmunol.182.1.259
28. Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of Conserved Non-Coding DNA Elements in the Foxp3 Gene in Regulatory T-Cell Fate. Nature (2010) 463:808–12. doi: 10.1038/nature08750
29. Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, et al. T Cell Receptor Stimulation-Induced Epigenetic Changes and Foxp3 Expression Are Independent and Complementary Events Required for Treg Cell Development. Immunity (2012) 37:785–99. doi: 10.1016/j.immuni.2012.09.010
30. Helmin KA, Morales-Nebreda L, Torres Acosta MA, Anekalla KR, Chen SY, Abdala-Valencia H, et al. Maintenance DNA Methylation Is Essential for Regulatory T Cell Development and Stability of Suppressive Function. J Clin Invest (2020) 130:6571–87. doi: 10.1172/JCI137712
31. Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, et al. Stability of the Regulatory T Cell Lineage In Vivo. Science (2010) 329:1667–71. doi: 10.1126/science.1191996
32. Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, et al. Foxp3 Exploits a Pre-Existent Enhancer Landscape for Regulatory T Cell Lineage Specification. Cell (2012) 151:153–66. doi: 10.1016/j.cell.2012.06.053
33. Sasidharan Nair V, Song MH, Oh KI. Vitamin C Facilitates Demethylation of the Foxp3 Enhancer in a Tet-Dependent Manner. J Immunol (2016) 196:2119–31. doi: 10.4049/jimmunol.1502352
34. Someya K, Nakatsukasa H, Ito M, Kondo T, Tateda KI, Akanuma T, et al. Improvement of Foxp3 Stability Through CNS2 Demethylation by TET Enzyme Induction and Activation. Int Immunol (2017) 29:365–75. doi: 10.1093/intimm/dxx049
35. Mikami N, Kawakami R, Chen KY, Sugimoto A, Ohkura N, Sakaguchi S. Epigenetic Conversion of Conventional T Cells Into Regulatory T Cells by CD28 Signal Deprivation. Proc Natl Acad Sci USA (2020) 117:12258–68. doi: 10.1073/pnas.1922600117
36. Ziegler SF. FOXP3: Not Just for Regulatory T Cells Anymore. Eur J Immunol (2007) 37:21–3. doi: 10.1002/eji.200636929
37. Singer BD, King LS, D'Alessio FR. Regulatory T Cells as Immunotherapy. Front Immunol (2014) 5:46. doi: 10.3389/fimmu.2014.00046
38. Trzonkowski P, Bieniaszewska M, Juscinska J, Dobyszuk A, Krzystyniak A, Marek N, et al. First-In-Man Clinical Results of the Treatment of Patients With Graft Versus Host Disease With Human Ex Vivo Expanded CD4+CD25+CD127- T Regulatory Cells. Clin Immunol (2009) 133:22–6. doi: 10.1016/j.clim.2009.06.001
39. Di Ianni M, Falzetti F, Carotti A, Terenzi A, Castellino F, Bonifacio E, et al. Tregs Prevent GVHD and Promote Immune Reconstitution in HLA-Haploidentical Transplantation. Blood (2011) 117:3921–8. doi: 10.1182/blood-2010-10-311894
40. Bluestone JA, Buckner JH, Fitch M, Gitelman SE, Gupta S, Hellerstein MK, et al. Type 1 Diabetes Immunotherapy Using Polyclonal Regulatory T Cells. Sci Transl Med (2015) 7:315ra189. doi: 10.1126/scitranslmed.aad4134
41. Brunstein CG, Miller JS, Mckenna DH, Hippen KL, Defor TE, Sumstad D, et al. Umbilical Cord Blood-Derived T Regulatory Cells to Prevent GVHD: Kinetics, Toxicity Profile, and Clinical Effect. Blood (2016) 127:1044–51. doi: 10.1182/blood-2015-06-653667
42. Mathew JM, J HV, Lefever A, Konieczna I, Stratton C, He J, et al. A Phase I Clinical Trial With Ex Vivo Expanded Recipient Regulatory T Cells in Living Donor Kidney Transplants. Sci Rep (2018) 8:7428. doi: 10.1038/s41598-018-25574-7
43. Sawitzki B, Harden PN, Reinke P, Moreau A, Hutchinson JA, Game DS, et al. Regulatory Cell Therapy in Kidney Transplantation (The ONE Study): A Harmonised Design and Analysis of Seven Non-Randomised, Single-Arm, Phase 1/2A Trials. Lancet (2020) 395:1627–39. doi: 10.1016/S0140-6736(20)30167-7
44. Harden PN, Game DS, Sawitzki B, van der Net JB, Hester J, Bushell A, et al. Feasibility, Long-Term Safety, and Immune Monitoring of Regulatory T Cell Therapy in Living Donor Kidney Transplant Recipients. Am J Transplant (2021) 21:1603–11. doi: 10.1111/ajt.16395
45. Di Ianni M, Del Papa B, Cecchini D, Bonifacio E, Moretti L, Zei T, et al. Immunomagnetic Isolation of CD4+CD25+FoxP3+ Natural T Regulatory Lymphocytes for Clinical Applications. Clin Exp Immunol (2009) 156:246–53. doi: 10.1111/j.1365-2249.2009.03901.x
46. Fritsche E, Volk HD, Reinke P, Abou-El-Enein M. Toward an Optimized Process for Clinical Manufacturing of CAR-Treg Cell Therapy. Trends Biotechnol (2020) 38:1099–112. doi: 10.1016/j.tibtech.2019.12.009
47. Putnam AL, Brusko TM, Lee MR, Liu W, Szot GL, Ghosh T, et al. Expansion of Human Regulatory T-Cells From Patients With Type 1 Diabetes. Diabetes (2009) 58:652–62. doi: 10.2337/db08-1168
48. Trzonkowski P, Szarynska M, Mysliwska J, Mysliwski A. Ex Vivo Expansion of CD4(+)CD25(+) T Regulatory Cells for Immunosuppressive Therapy. Cytometry A (2009) 75:175–88. doi: 10.1002/cyto.a.20659
49. Putnam AL, Safinia N, Medvec A, Laszkowska M, Wray M, Mintz MA, et al. Clinical Grade Manufacturing of Human Alloantigen-Reactive Regulatory T Cells for Use in Transplantation. Am J Transplant (2013) 13:3010–20. doi: 10.1111/ajt.12433
50. Fraser H, Safinia N, Grageda N, Thirkell S, Lowe K, Fry LJ, et al. A Rapamycin-Based GMP-Compatible Process for the Isolation and Expansion of Regulatory T Cells for Clinical Trials. Mol Ther Methods Clin Dev (2018) 8:198–209. doi: 10.1016/j.omtm.2018.01.006
51. Strauss L, Whiteside TL, Knights A, Bergmann C, Knuth A, Zippelius A. Selective Survival of Naturally Occurring Human CD4+CD25+Foxp3+ Regulatory T Cells Cultured With Rapamycin. J Immunol (2007) 178:320–9. doi: 10.4049/jimmunol.178.1.320
52. Morales-Nebreda L, Mclafferty FS, Singer BD. DNA Methylation as a Transcriptional Regulator of the Immune System. Transl Res (2019) 204:1–18. doi: 10.1016/j.trsl.2018.08.001
53. Singer BD. A Practical Guide to the Measurement and Analysis of DNA Methylation. Am J Respir Cell Mol Biol (2019) 61:417–28. doi: 10.1165/rcmb.2019-0150TR
54. Wang L, Ozark PA, Smith ER, Zhao Z, Marshall SA, Rendleman EJ, et al. TET2 Coactivates Gene Expression Through Demethylation of Enhancers. Sci Adv (2018) 4:eaau6986. doi: 10.1126/sciadv.aau6986
55. Morgan MAJ, Shilatifard A. Reevaluating the Roles of Histone-Modifying Enzymes and Their Associated Chromatin Modifications in Transcriptional Regulation. Nat Genet (2020) 52:1271–81. doi: 10.1038/s41588-020-00736-4
56. Singer BD, Chandel NS. Immunometabolism of Pro-Repair Cells. J Clin Invest (2019) 129:2597–607. doi: 10.1172/JCI124613
57. Weinberg SE, Singer BD, Steinert EM, Martinez CA, Mehta MM, Martinez-Reyes I, et al. Mitochondrial Complex III is Essential for Suppressive Function of Regulatory T Cells. Nature (2019) 565:495–9. doi: 10.1038/s41586-018-0846-z
58. Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, et al. Guidance of Regulatory T Cell Development by Satb1-Dependent Super-Enhancer Establishment. Nat Immunol (2017) 18:173–83. doi: 10.1038/ni.3646
59. Toker A, Engelbert D, Garg G, Polansky JK, Floess S, Miyao T, et al. Active Demethylation of the Foxp3 Locus Leads to the Generation of Stable Regulatory T Cells Within the Thymus. J Immunol (2013) 190:3180–8. doi: 10.4049/jimmunol.1203473
60. Yang R, Qu C, Zhou Y, Konkel JE, Shi S, Liu Y, et al. Hydrogen Sulfide Promotes Tet1- and Tet2-Mediated Foxp3 Demethylation to Drive Regulatory T Cell Differentiation and Maintain Immune Homeostasis. Immunity (2015) 43:251–63. doi: 10.1016/j.immuni.2015.07.017
61. Liu Y, Wang L, Han R, Beier UH, Akimova T, Bhatti T, et al. Two Histone/Protein Acetyltransferases, CBP and P300, are Indispensable for Foxp3+ T-Regulatory Cell Development and Function. Mol Cell Biol (2014) 34:3993–4007. doi: 10.1128/MCB.00919-14
62. Xiong Y, Wang L, Di Giorgio E, Akimova T, Beier UH, Han R, et al. Inhibiting the Coregulator CoREST Impairs Foxp3+ Treg Function and Promotes Antitumor Immunity. J Clin Invest (2020) 130:1830–42. doi: 10.1172/JCI131375
63. Obata Y, Furusawa Y, Endo TA, Sharif J, Takahashi D, Atarashi K, et al. The Epigenetic Regulator Uhrf1 Facilitates the Proliferation and Maturation of Colonic Regulatory T Cells. Nat Immunol (2014) 15:571–9. doi: 10.1038/ni.2886
64. Sun X, Cui Y, Feng H, Liu H, Liu X. TGF-Beta Signaling Controls Foxp3 Methylation and T Reg Cell Differentiation by Modulating Uhrf1 Activity. J Exp Med (2019) 216:2819–37. doi: 10.1084/jem.20190550
65. Wang L, Liu Y, Beier UH, Han R, Bhatti TR, Akimova T, et al. Foxp3+ T-Regulatory Cells Require DNA Methyltransferase 1 Expression to Prevent Development of Lethal Autoimmunity. Blood (2013) 121:3631–9. doi: 10.1182/blood-2012-08-451765
66. Arvey A, van der Veeken J, Samstein RM, Feng Y, Stamatoyannopoulos JA, Rudensky AY. Inflammation-Induced Repression of Chromatin Bound by the Transcription Factor Foxp3 in Regulatory T Cells. Nat Immunol (2014) 15:580–7. doi: 10.1038/ni.2868
67. Walter JM, Helmin KA, Abdala-Valencia H, Wunderink RG, Singer BD. Multidimensional Assessment of Alveolar T Cells in Critically Ill Patients. JCI Insight (2018) 3:e123287. doi: 10.1172/jci.insight.123287
68. Singer BD. COVID-19 and the Next Influenza Season. Sci Adv (2020) 6:eabd0086. doi: 10.1126/sciadv.abd0086
69. Budinger GRS, Misharin AV, Ridge KM, Singer BD, Wunderink RG. Distinctive Features of Severe SARS-CoV-2 Pneumonia. J Clin Invest (2021) 131:e149412. doi: 10.1172/JCI149412
70. Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T Cell Lineage Differentiation. Immunity (2009) 30:646–55. doi: 10.1016/j.immuni.2009.05.001
71. Hori S. Regulatory T Cell Plasticity: Beyond the Controversies. Trends Immunol (2011) 32:295–300. doi: 10.1016/j.it.2011.04.004
72. Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of Natural Foxp3+ T Cells: A Committed Regulatory T-Cell Lineage and an Uncommitted Minor Population Retaining Plasticity. Proc Natl Acad Sci USA (2009) 106:1903–8. doi: 10.1073/pnas.0811556106
73. Miyao T, Floess S, Setoguchi R, Luche H, Fehling HJ, Waldmann H, et al. Plasticity of Foxp3(+) T Cells Reflects Promiscuous Foxp3 Expression in Conventional T Cells But Not Reprogramming of Regulatory T Cells. Immunity (2012) 36:262–75. doi: 10.1016/j.immuni.2011.12.012
74. Lochner M, Peduto L, Cherrier M, Sawa S, Langa F, Varona R, et al. In Vivo Equilibrium of Proinflammatory IL-17+ and Regulatory IL-10+ Foxp3+ RORgamma T+ T Cells. J Exp Med (2008) 205:1381–93. doi: 10.1084/jem.20080034
75. Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, et al. Opposing Regulation of the Locus Encoding IL-17 Through Direct, Reciprocal Actions of STAT3 and STAT5. Nat Immunol (2011) 12:247–54. doi: 10.1038/ni.1995
76. Samson M, Audia S, Janikashvili N, Ciudad M, Trad M, Fraszczak J, et al. Brief Report: Inhibition of Interleukin-6 Function Corrects Th17/Treg Cell Imbalance in Patients With Rheumatoid Arthritis. Arthritis Rheum (2012) 64:2499–503. doi: 10.1002/art.34477
77. Beriou G, Costantino CM, Ashley CW, Yang L, Kuchroo VK, Baecher-Allan C, et al. IL-17-Producing Human Peripheral Regulatory T Cells Retain Suppressive Function. Blood (2009) 113:4240–9. doi: 10.1182/blood-2008-10-183251
78. Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, et al. Foxp3(+) T Cells Expressing RORgammat Represent a Stable Regulatory T-Cell Effector Lineage With Enhanced Suppressive Capacity During Intestinal Inflammation. Mucosal Immunol (2016) 9:444–57. doi: 10.1038/mi.2015.74
79. Samanta A, Li B, Song X, Bembas K, Zhang G, Katsumata M, et al. TGF-Beta and IL-6 Signals Modulate Chromatin Binding and Promoter Occupancy by Acetylated FOXP3. Proc Natl Acad Sci USA (2008) 105:14023–7. doi: 10.1073/pnas.0806726105
80. Li Y, Deuring J, Peppelenbosch MP, Kuipers EJ, De Haar C, van der Woude CJ. IL-6-Induced DNMT1 Activity Mediates SOCS3 Promoter Hypermethylation in Ulcerative Colitis-Related Colorectal Cancer. Carcinogenesis (2012) 33:1889–96. doi: 10.1093/carcin/bgs214
81. Piunti A, Smith ER, Morgan MAJ, Ugarenko M, Khaltyan N, Helmin KA, et al. CATACOMB: An Endogenous Inducible Gene That Antagonizes H3K27 Methylation Activity of Polycomb Repressive Complex 2 via an H3K27M-Like Mechanism. Sci Adv (2019) 5:eaax2887. doi: 10.1126/sciadv.aax2887
82. Douillet D, Sze CC, Ryan C, Piunti A, Shah AP, Ugarenko M, et al. Uncoupling Histone H3K4 Trimethylation From Developmental Gene Expression via an Equilibrium of COMPASS, Polycomb and DNA Methylation. Nat Genet (2020) 52:615–25. doi: 10.1038/s41588-020-0618-1
83. Bamidele AO, Svingen PA, Sagstetter MR, Sarmento OF, Gonzalez M, Braga Neto MB, et al. Disruption of FOXP3-EZH2 Interaction Represents a Pathobiological Mechanism in Intestinal Inflammation. Cell Mol Gastroenterol Hepatol (2019) 7:55–71. doi: 10.1016/j.jcmgh.2018.08.009
84. Torres Acosta MA, Singer BD. Pathogenesis of COVID-19-Induced ARDS: Implications for an Ageing Population. Eur Respir J (2020) 56:2002049. doi: 10.1183/13993003.02049-2020
85. Mcquattie-Pimentel AC, Ren Z, Joshi N, Watanabe S, Stoeger T, Chi M, et al. The Lung Microenvironment Shapes a Dysfunctional Response of Alveolar Macrophages in Aging. J Clin Invest (2021) 131:e140299. doi: 10.1172/JCI140299
86. Mijnheer G, Lutter L, Mokry M, van der Wal M, Scholman R, Fleskens V, et al. Conserved Human Effector Treg Cell Transcriptomic and Epigenetic Signature in Arthritic Joint Inflammation. Nat Commun (2021) 12:2710. doi: 10.1038/s41467-021-22975-7
87. Oliveira LM, Teixeira FME, Sato MN. Impact of Retinoic Acid on Immune Cells and Inflammatory Diseases. Mediators Inflamm (2018) 2018:3067126. doi: 10.1155/2018/3067126
88. Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, et al. Neuropilin-1 Distinguishes Natural and Inducible Regulatory T Cells Among Regulatory T Cell Subsets In Vivo. J Exp Med (2012) 209:1713–1722, S1711-1719. doi: 10.1084/jem.20120822
89. Chan MW, Chang CB, Tung CH, Sun J, Suen JL, Wu SF. Low-Dose 5-Aza-2'-Deoxycytidine Pretreatment Inhibits Experimental Autoimmune Encephalomyelitis by Induction of Regulatory T Cells. Mol Med (2014) 20:248–56. doi: 10.2119/molmed.2013.00159
90. Lu CH, Wu CJ, Chan CC, Nguyen DT, Lin KR, Lin SJ, et al. DNA Methyltransferase Inhibitor Promotes Human CD4(+)CD25(h)FOXP3(+) Regulatory T Lymphocyte Induction Under Suboptimal TCR Stimulation. Front Immunol (2016) 7:488. doi: 10.3389/fimmu.2016.00488
91. Mcgrath-Morrow SA, Ndeh R, Helmin KA, Chen SY, Anekalla KR, Abdala-Valencia H, et al. DNA Methylation Regulates the Neonatal CD4(+) T-Cell Response to Pneumonia in Mice. J Biol Chem (2018) 293:11772–83. doi: 10.1074/jbc.RA118.003589
92. Han P, Hou Y, Zhao Y, Liu Y, Yu T, Sun Y, et al. Low-Dose Decitabine Modulates T-Cell Homeostasis and Restores Immune Tolerance in Immune Thrombocytopenia. Blood (2021) 138:674–88. doi: 10.1182/blood.2020008477
93. Liu L, Hu L, Yang L, Jia S, Du P, Min X, et al. UHRF1 Downregulation Promotes T Follicular Helper Cell Differentiation by Increasing BCL6 Expression in SLE. Clin Epigenet (2021) 13:31. doi: 10.1186/s13148-021-01007-7
94. Tao R, De Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase Inhibition Promotes the Generation and Function of Regulatory T Cells. Nat Med (2007) 13:1299–307. doi: 10.1038/nm1652
95. Wang L, Beier UH, Akimova T, Dahiya S, Han R, Samanta A, et al. Histone/protein Deacetylase Inhibitor Therapy for Enhancement of Foxp3+ T-Regulatory Cell Function Posttransplantation. Am J Transplant (2018) 18:1596–603. doi: 10.1111/ajt.14749
96. Rothhammer V, Quintana FJ. The Aryl Hydrocarbon Receptor: An Environmental Sensor Integrating Immune Responses in Health and Disease. Nat Rev Immunol (2019) 19:184–97. doi: 10.1038/s41577-019-0125-8
97. Abdulla OA, Neamah W, Sultan M, Alghetaa HK, Singh N, Busbee PB, et al. The Ability of AhR Ligands to Attenuate Delayed Type Hypersensitivity Reaction Is Associated With Alterations in the Gut Microbiota. Front Immunol (2021) 12:684727. doi: 10.3389/fimmu.2021.684727
98. Yue X, Trifari S, Aijo T, Tsagaratou A, Pastor WA, Zepeda-Martinez JA, et al. Control of Foxp3 Stability Through Modulation of TET Activity. J Exp Med (2016) 213:377–97. doi: 10.1084/jem.20151438
99. Wakamatsu E, Omori H, Tabata Y, Akieda Y, Watanabe S, Ogawa S, et al. CD28 Co-Stimulation is Dispensable for the Steady State Homeostasis of Intestinal Regulatory T Cells. Int Immunol (2018) 30:171–80. doi: 10.1093/intimm/dxy013
100. Tsutsui T, Fukasawa R, Shinmyouzu K, Nakagawa R, Tobe K, Tanaka A, et al. Mediator Complex Recruits Epigenetic Regulators via its Two Cyclin-Dependent Kinase Subunits to Repress Transcription of Immune Response Genes. J Biol Chem (2013) 288:20955–65. doi: 10.1074/jbc.M113.486746
101. Allen BL, Taatjes DJ. The Mediator Complex: A Central Integrator of Transcription. Nat Rev Mol Cell Biol (2015) 16:155–66. doi: 10.1038/nrm3951
102. Akamatsu M, Mikami N, Ohkura N, Kawakami R, Kitagawa Y, Sugimoto A, et al. Conversion of Antigen-Specific Effector/Memory T Cells Into Foxp3-Expressing Treg Cells by Inhibition of CDK8/19. Sci Immunol (2019) 4:eaaw2707. doi: 10.1126/sciimmunol.aaw2707
Keywords: regulatory T cells, plasticity, inflammation, epigenetics, DNA methylation, therapeutics
Citation: Joudi AM, Reyes Flores CP and Singer BD (2022) Epigenetic Control of Regulatory T Cell Stability and Function: Implications for Translation. Front. Immunol. 13:861607. doi: 10.3389/fimmu.2022.861607
Received: 24 January 2022; Accepted: 14 February 2022;
Published: 02 March 2022.
Edited by:
Thomas Wekerle, Medical University of Vienna, AustriaReviewed by:
Wayne William Hancock, University of Pennsylvania, United StatesQizhi Tang, University of California, San Francisco, United States
Wenji Piao, University of Maryland, Baltimore, United States
Copyright © 2022 Joudi, Reyes Flores and Singer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin D. Singer, YmVuamFtaW4tc2luZ2VyQG5vcnRod2VzdGVybi5lZHU=
 Anthony M. Joudi1,2
Anthony M. Joudi1,2 Carla P. Reyes Flores
Carla P. Reyes Flores Benjamin D. Singer
Benjamin D. Singer