- 1Department of Neurosciences, Laboratory for Neuroimmunology, Leuven Brain Institute, KU Leuven, Leuven, Belgium
- 2Department of Microbiology, Immunology and Transplantation, Laboratory of Immunobiology, Rega Institute for Medical Research, KU Leuven, Leuven, Belgium
Studies investigating the immunopathology of multiple sclerosis (MS) have largely focused on adaptive T and B lymphocytes. However, in recent years there has been an increased interest in the contribution of innate immune cells, amongst which the natural killer (NK) cells. Apart from their canonical role of controlling viral infections, cell stress and malignancies, NK cells are increasingly being recognized for their modulating effect on the adaptive immune system, both in health and autoimmune disease. From different lines of research there is now evidence that NK cells contribute to MS immunopathology. In this review, we provide an overview of studies that have investigated the role of NK cells in the pathogenesis of MS by use of the experimental autoimmune encephalomyelitis (EAE) animal model, MS genetics or through ex vivo and in vitro work into the immunology of MS patients. With the advent of modern hypothesis-free technologies such as single-cell transcriptomics, we are exposing an unexpected NK cell heterogeneity, increasingly blurring the boundaries between adaptive and innate immunity. We conclude that unravelling this heterogeneity, as well as the mechanistic link between innate and adaptive immune cell functions will lay the foundation for the use of NK cells as prognostic tools and therapeutic targets in MS and a myriad of other currently uncurable autoimmune disorders.
Introduction
Multiple sclerosis (MS) is a chronic autoimmune disorder of the central nervous system (CNS), primarily affecting young adults (1). In this age group, it forms the most common cause of non-traumatic disability (2). The first stages of the disease are characterized by transient inflammation of the brain and spinal cord which damages the myelin and axons, resulting in episodes of neurological dysfunction. As a result of the relatively young age at onset, the lifelong accumulation of disability, the increasing worldwide prevalence and the lack of a curative treatment, MS continues to pose a major socioeconomic burden (3, 4). Current disease-modifying therapies (DMTs) in MS and autoimmune disorders in general are targeting primarily the adaptive immune system, i.e. T and B cells (5–7). However, the contribution of innate immune cells such as natural killer (NK) cells is now gaining increasing attention in a myriad of autoimmune disorders (8–12), including MS (13–16).
NK cells are large granular lymphocytes best known for their capacity to kill virally-infected, malignant and stressed cells (17). They are generally divided into two main subsets, based on the surface expression of CD56 and FcγRIII (CD16): the cytotoxic CD56dimCD16+ (CD56dim) and the regulatory CD56brightCD16- (CD56bright) NK cells (18). In peripheral blood (PB), the CD56dim NK cells account for up to 90% of all NK cells. They are considered the more mature effector subset with variable expression of maturation markers such as CD57 and KLRG1 and high cytotoxic activity mainly through their production of perforin and granzymes A and B (19–21). This subset fulfills the canonical NK cell role of immune surveillance: scanning cells for infection, stress or malignancy. The expression of CD16 allows this subset to engage in antibody dependent cellular cytotoxicity (ADCC) (17). The CD56bright NK cells in contrast constitute the majority of NK cells in secondary lymphoid organs and inflammatory sites (22, 23). These cells are considered less mature and generally more immunoregulatory in nature in part by virtue of their greater cytokine production, most importantly interferon (IFN)-γ (22). The distinct functionalities and distribution of NK cell subsets, both in health and in autoimmune disease, are the result of distinct repertoires of both activating and inhibitory NK cell receptors as well as chemokine receptors (18, 24, 25). For example, whereas CD56dim NK cells express CXCR1, CXCR2, CX3CR1 and S1P5, CD56bright NK cells express CCR7 and CXCR3. In addition, CD56bright NK cells show higher expression of inhibitory major histocompatibility complex (MHC) class 1-binding receptor NKG2A as well as activating receptor NKp46, while CD56dim NK cells are mainly characterized by higher levels of killer immunoglobulin-like receptors (KIR). Activation of NK cells is mediated by a balance of signals received through their inhibitory and activating receptors which are triggered by various ligands present on potential target cells (19). Interestingly, an expanding amount of literature is now describing various NK cell subsets portraying adaptive-like immunological memory and long-lived antigen specificity (26, 27). Three main types of such adaptive NK cells have been described to date: (i) NK cells found in the murine liver displaying long-lived memory to haptens and viral antigens; (ii) human NKG2C+ and murine Ly49H+ NK cells displaying cytomegalovirus (CMV) antigen-specificity and enhanced recall responses; (iii) human and murine cytokine-activated NK cells showing memory-like “trained” features such as enhanced cytotoxicity and production of IFN-γ upon restimulation (28). A more detailed description of all NK cell subtypes and their distinct characteristics falls outside the scope of this review and can be consulted elsewhere (19).
Up until the last decade, NK cells have largely been ignored in the MS field and remain as of today in the shadows of the more studied adaptive T and B cells (Figure 1A). Apart from some exploratory studies in the 1980s (29–34), literature on the contribution of NK cells to MS immunopathogenesis is only recently starting to accumulate (Figure 1B). New methodologies and state-of-the art technologies have allowed for clear evidence being found in different lines of research. In this review, we provide a non-exhaustive overview of such findings from animal models, genetics, ex vivo and in vitro work. We point out the implications of these findings for the use of NK cells as prognostic tools and therapeutic targets in MS and we summarize what work remains and highlight the most pressing open questions that need answering before we can hope to include NK cells in standard clinical practice.
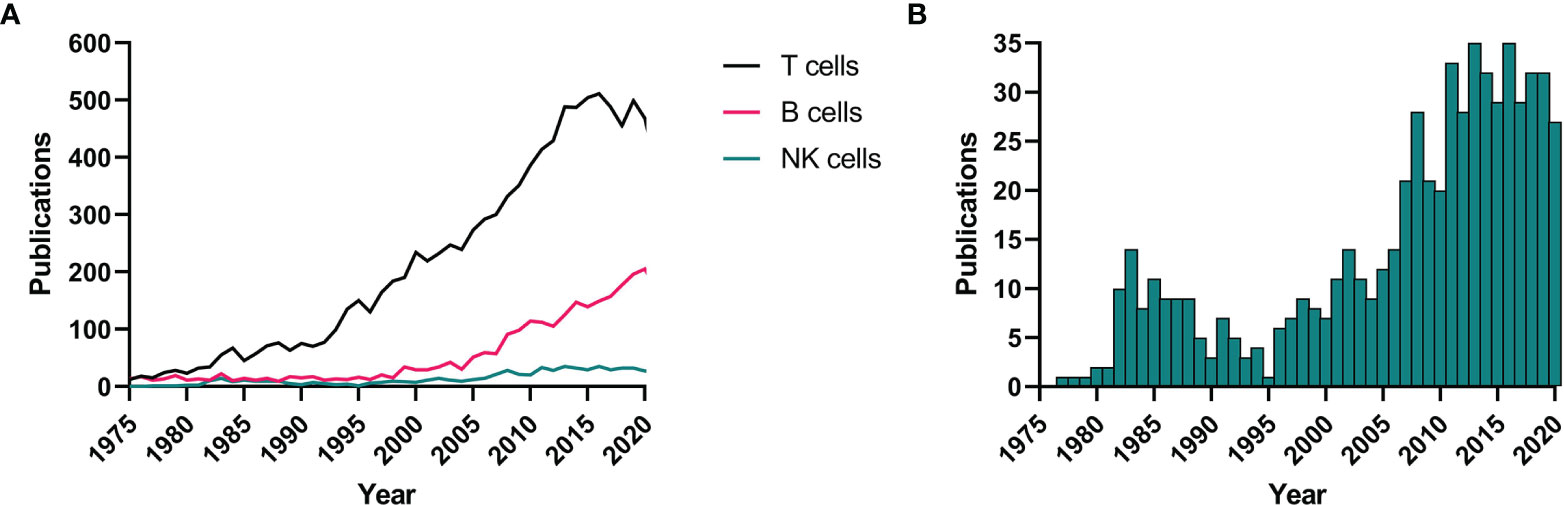
Figure 1 NK cells in MS literature. A systematic literature search in PubMed (NCBI) was performed using the terms “T cells”, “B cells”, or “natural killer cells” + “multiple sclerosis”. Results are shown for all three cell types from 1975 to 2020 (A) as well as a close-up for the NK cell results (B). NK, natural killer; MS, multiple sclerosis.
A Regulatory Role for NK Cells in Animal Models of MS
A commonly studied animal model is experimental autoimmune encephalomyelitis (EAE), in which susceptible rodents develop CNS autoimmunity upon immunization with myelin-derived self-antigens emulsified in complete Freund’s adjuvant (CFA) (35). The majority of EAE studies have reported a regulatory role for NK cells. The mechanisms attributed to such regulatory action of NK cells in EAE include general T cell suppression (36–38), killing of autologous autoreactive T cells (39, 40), inhibition of the differentiation of myelin-reactive T helper type 1 (Th1) or 17 (Th17) cells in the CNS (41, 42) and secretion of neurotrophic factors (43).
In contrast to these studies, some work has been more suggestive of a detrimental role for NK cells. Researchers have postulated that the presence of NK cells, and more specifically NK cell-derived IFN-γ, is a prerequisite for development of EAE pathology (44–46). However, this concept is in strong contrast with the numerous findings that mice in which IFN-γ is depleted (using neutralizing antibodies or through genetic modification) exhibit an exacerbated form of EAE or develop the disease in strains that are normally not susceptible to develop EAE, such as the BALB/c strain (47, 48).
The contradictory reports on the role of NK cells in EAE may be attributed to the use of different animal backgrounds, divergent immunization procedures or NK cell depletion performed at different time points during development of the disease as reported by Winkler-Pickett et al. (46). Moreover, an explanation for these discrepancies might be found in the long-standing underappreciation of NK cell heterogeneity, as has been suggested by Gandhi et al. already in 2010 (49). Yet today, the independent and discrete functions of different NK cell subsets in different physiological compartments remain largely ignored in animal studies (50).
MS Genetics Alludes a Role for NK Cells
In the last decade, the field has seen an expansion in the knowledge on the genetic basis of MS, mainly by virtue of the collaborative efforts of the International Multiple Sclerosis Genetics Consortium (IMSGC) (51–54). The most recent genomic map comprises 200 common genetic risk factors for MS on autosomes, the first one on the X-chromosome and 32 independent effects in the MHC region (55). For the first time, a systematic analysis of these risk factors showed significant enrichment for MS susceptibility loci not only in genes relating to cells of the adaptive immune system, but also in those relating to cells of the innate immune system, including NK cells. In line with this finding, several individual risk genes that appear associated with NK cell activity have been identified in MS. An example is CD226 (55–59) which encodes for DNAX accessory molecule (DNAM)-1, a cell surface glycoprotein expressed on cytotoxic lymphocytes such as NK cells and CD8+ T cells, mediating their activation and killing of target cells (60, 61). The risk haplotype is associated with lower CD226 cell surface expression and MS patients have shown lower CD226 levels than healthy controls (59, 62). Interestingly, CD226 is known to be crucial for the NK cell-mediated elimination of activated T cells (63).
A genome-wide screen for low-frequency risk variants identified three novel associations with MS, including rs35947132 (p.Ala91Val) in the PRF1 gene (54). This variant was previously reported in an Italian candidate-gene study (64), and has been confirmed in an independent cohort in Sardinia (65). PRF1 encodes perforin, a key component of the cytotoxic pathway used by, although not exclusively, NK cells (54, 66). The p.Ala91Val variant is thought to predispose to late-onset familial hemophagocytic lymphohistiocytosis type 2 (FHLH2), a disease resulting from genetic cytotoxic defects in NK cells and CD8+ T cells and involving a massive cytokine storm, including release of IFN-γ (67, 68). Approximately 10% of Europeans carry this variant, which results in protein misfolding, reduced stability and, consequently, partial loss of perforin lytic activity (69). In healthy individuals, it is associated with a 35% reduction in NK cell cytotoxicity, increased overall lymphocyte count and specific augmentation of the cytotoxic memory T-cell compartment (65, 70).
Large-scale genetic studies have shown that genetic risk for MS is dominated by a series of MHC class II risk alleles, while protective effects have been attributed to a number of MHC class I alleles (53). The underlying mechanisms of action of these findings remain elusive. Apart from the classically considered T-cell receptors on CD8+ T cells, NK cells also express receptors that enable them to respond to MHC class I molecules, with the most important ones being the KIRs, C-type lectin-like CD94/NKG2 family and the leukocyte immunoglobulin-like receptors (LILR), such interactions have been put forward as a possible pathway for NK cell involvement in CNS disorders (24, 71). These highly polymorphic genetic regions, including structural variation, are not well captured by genotyping arrays but powerful bioinformatic tools such as large-scale imputation methods (72) and new technologies such as long-read sequencing platforms (73) now enable interrogating their contribution to MS.
Immunoregulatory CD56bright NK Cells in MS Patients
In accordance with findings from animal models and genetics, studies analyzing the cellular composition of PB and cerebrospinal fluid (CSF) in patients and healthy controls mostly point towards an immunoregulatory role for NK cells in MS. Several studies have revealed the presence of NK cells in the CSF and CNS plaques of MS patients (74–79). One of these observed NK cells expressing granzyme K, which is mainly expressed by the CD56bright NK cells (80), in active MS lesions in close proximity to T cells (81). Two studies (81, 82), together investigating 152 MS patients and 42 non-inflammatory controls, described a significant increase of CD56bright NK cells and, to a lower extent, CD56dim NK cells in the CSF of MS patients. This was recently confirmed in periventricular tissue of MS patients versus controls and in periventricular lesions versus normal appearing white matter (83). The first study also reported a significant decrease of peripheral NK cells, predominantly the CD56dim subset (81). As therapeutic modulation of IL-2 receptor signaling in these patients significantly increased the peripheral CD56bright subset but not the CD56dim subset, the authors speculate that NK cell maturation is impaired in MS and NK cells are stuck in the more immature CD56bright stage (81). Even though a deeper mechanical understanding gained by further functional studies is certainly needed, such efforts have put NK cells, specifically the CD56bright subset, on the map of MS immunopathology.
Appreciating NK Cell Heterogeneity in MS
Apart from the classical CD56bright and CD56dim NK cells, more recent work has identified several other distinct NK cells subsets in MS (Figure 2). De Jager et al. described an NK cell subset, defined as CD8dimCD56+CD3-CD4-, which was found to be reduced in PB of untreated MS patients as well as patients with clinically isolated syndrome (CIS) (84). More recently, McKinney et al. also described a population of ‘NK8+ cells’ (CD3−CD56+CD8+) in PB associated with a more favorable clinical outcome in MS (85). Whether or not these two studies are referencing the same NK cell subset is unclear. In another study, a CMV-driven expansion of adaptive-like NKG2C+ NK cells in PB was correlated with a decreased risk of disability progression (86). Such findings are starting to blur the lines between adaptive and innate immune subsets and stress the need for a better appreciation of immune cell plasticity and heterogeneity.
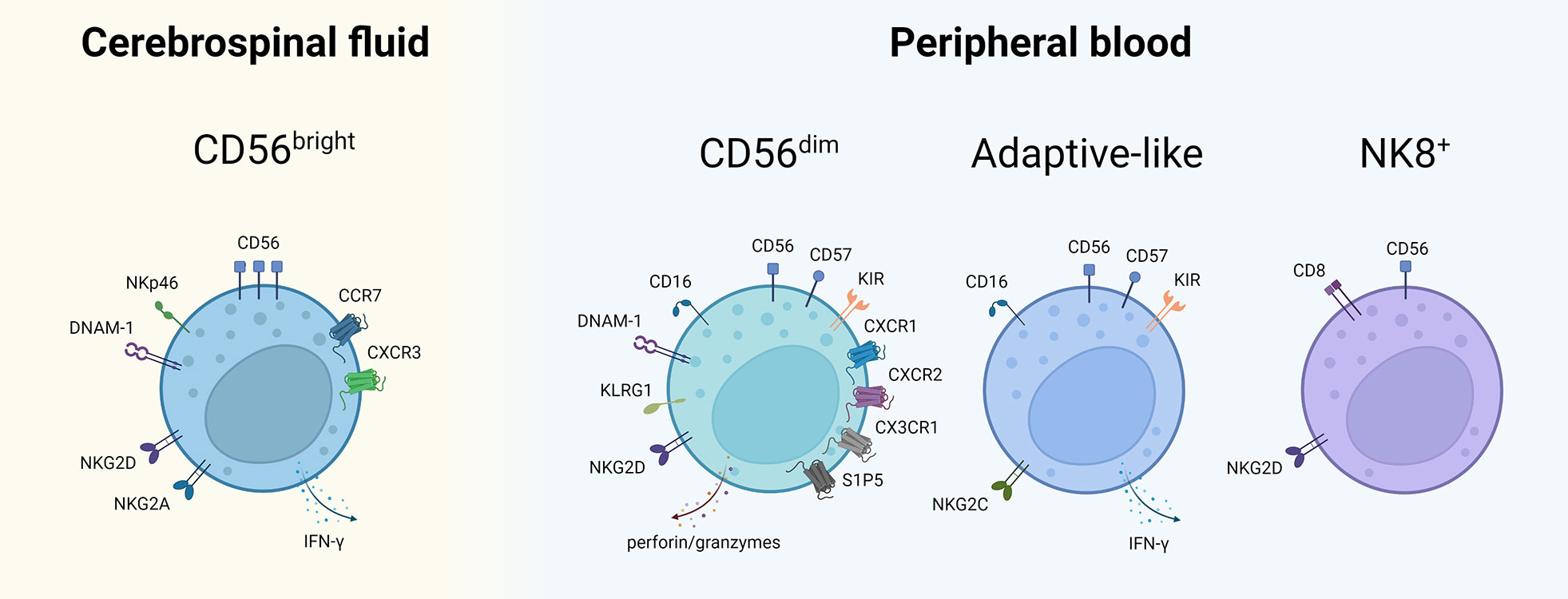
Figure 2 NK cell subsets described in CSF and PB of MS patients. NK cell subsets with their characteristic cell surface markers described thus far in MS. The subsets are placed in the compartment where their putative role in MS has predominantly been described. Of note: the mentioned markers are non-exhaustive, the compartmentalization is not absolute and mutual exchange between CSF and PB is possible. NK, natural killer; CSF, cerebrospinal fluid; PB, peripheral blood; NKT, natural killer T; CD, cluster of differentiation; DNAM-1, DNAX accessory molecule 1; IFN, interferon; CCR, CC-chemokine receptor; CXC, chemokine receptor; KLRG1, killer cell lectin-like receptor G1; KIR, killer immunoglobulin-like receptor; S1P5, sphingosine-1-phosphate receptor 5; NK8+, CD8+ natural killer cell. Created with BioRender.com.
Recent technological advancements in the field of single-cell transcriptomics will be pivotal therein as they exploit hypothesis-free transcriptomic profiles instead of predefined cell marker subsets (87, 88). Such technologies have been able to distinguish up to 7 distinct NK cell subsets, questioning the classical two-subset (CD56bright/dim) view (89–91). However, the available single-cell RNA sequencing (scRNA-seq) studies in MS remain scarce and due to the low number of samples and low resolution, these studies often still limit their analyses of the NK cells to the two main subsets (92–94). In general, scRNA-seq studies confirm the lower abundance of total NK cells in CSF versus PB, with a cluster most likely corresponding to CD56bright NK cells being the predominant NK cell subset in CSF. Schafflick et al. (92) observed an increase in the cluster abundance of two NK cell subsets, most likely corresponding to CD56bright and CD56dim NK cells, in CSF of MS patients (n = 4) as compared to controls (n = 4), in line with previous cytometric analyses described above (81, 82). Ramesh et al. were unable to observe a significant change in total NK cells in the CSF of MS patients (n = 10) as compared to controls (n = 3) and had no further resolution of NK cell subsets (94). More generally, these first studies aptly illustrate the importance of going beyond the canonical immune cell populations and recognizing the potential of formerly unidentified/underappreciated subsets as critical players in MS immunopathology.
More Mechanistic Work Is Needed
The proposed mechanisms by which the aforementioned NK cell subsets exert their immunoregulatory function in MS are manifold. Several in vitro studies have shown that upon activation, T cells upregulate NKG2D ligands, thus becoming more susceptible to NK cell-mediated autologous killing (95–98). In MS, Jiang and colleagues have described a pivotal role for granzyme K in the immunomodulation by CD56bright NK cells (99). These authors showed that eliminating granzyme K profoundly inhibited the ability of CD56bright NK cells to lyse activated syngeneic T cells, thus failing to control autoimmunity. In addition, NK cells have been shown to kill other MS-relevant cell types, such as microglial cells by use of the NKG2D as well as the NKp46 receptor pathways (100), autologous immature myeloid cells via NKp30, NKp46 and DNAM-1 (101–103) and activated macrophages through NKG2D as well (104). Lastly, apart from direct cell-mediated cytotoxicity, NK cells are known to modulate adaptive immunity by cytokine production (105). Evidently, more functional assays will be needed to determine which of these mechanisms is used by exactly which NK cell subset to modulate adaptive immunity and control autoimmunity in MS.
NK Cells in the MS Clinic
Several MS treatments have been described to affect NK cell populations. A systematic overview of the known effects of currently approved DMTs for MS on the NK cell repertoire is beyond the scope of this review and the authors refer to a recent review of Laroni and Uccelli (16). From a historical perspective, the effects of daclizumab, an anti-CD25 mAb that blocks the high-affinity IL-2 receptor on activated T cells, have been particularly interesting. Bielekova et al. showed that the beneficial effects of daclizumab could in part be attributed to an expansion of the CD56bright NK cells, as well as an enhancement of their cytotoxic potential and regulatory capacity (106, 107). The concomitant killing of T cells is mediated by the mechanisms described in previous sections such as increased expression of granzyme K in NK cells and reversal of the diminished expression of DNAM-1 ligand CD155 on T cells (81, 99). Subsequent studies revealed that the expansion of CD56bright NK cells after daclizumab was most likely due to the increased binding of IL-2 to the intermediate IL-2 receptor present on NK cells, which is not targeted by daclizumab (108, 109). Despite the beneficial effects, some MS patients developed autoimmune encephalitis after administration of daclizumab, possibly in part caused by the depletion of regulatory T cells, leading to withdrawal of the drug (110). Fingolimod, a sphingosine-1-phosphate receptor modulator, is a commonly used drug in current MS therapy. A recent study described an expansion of a distinct NK cell subset characterized by an aged phenotype (CD56dimCD16++KIR+/-NKG2A-CD94-CCR7+/-CX3CR1+/-NKG2C-NKG2D+NKp46-DNAM1++CD127+) upon fingolimod treatment (111). Although the authors could not demonstrate an association of any particular NK cell subset with treatment response in their pilot study, they do warn that the observed expansion of a more exhausted and possibly less functional NK cell subset might contribute to the observed adverse events such as infections and malignancies upon long-term fingolimod treatment (112). Cases such as daclizumab and fingolimod stress the importance of isolating the exact immune subsets propagating the established beneficial effects of the treatment and averting the unsolicited effects.
Because of such studies, NK cells are now at last being recognized as interesting targets for biomarker and therapy research in a myriad of autoimmune diseases (113). In MS, studies trying to correlate NK cell titers and functionality with prognosis and therapy outcome are underway (114–118), but more validation work is needed to establish a mechanistic basis for such correlations.
Conclusion and Future Perspectives
From different lines of research, there is evidence that NK cells are likely to play a role in the pathogenesis of MS. NK cells are present in the CSF of MS patients and although their absolute numbers are relatively low, NK cells have strong cytotoxic activity to target stressed cells which include autologous activated immune cells. Genetic studies reveal polymorphisms in genes important for target cell recognition and for the cytotoxic activity of NK cells. NK cells are also important sources of cytokines that further regulate the immune system. Analysis of NK cells within successful MS treatments shows clear associations with changes in NK cell numbers and phenotype. Overall, the findings reviewed here are indicative of a more regulatory action of NK cells in the pathogenesis of MS, but further research is needed to validate this hypothesis. The importance of NK cells in EAE is also emerging in animal studies, but there is no consensus on their role.
The current state-of-the-art fails to uniformly capture and characterize the complete and intricate heterogeneity of the NK cell repertoire in MS, including both disease-promoting and disease-protective mechanisms, thereby hampering their successful application in the clinic. As current literature has often limited itself to descriptive studies of major NK cell subset frequencies with limited resolution and without many validated functional inferences, our knowledge of the exact mechanistic contribution of specific NK cell subsets to MS pathology remains superficial. The use of more unbiased approaches such as single-cell omics will allow for a more holistic understanding of the immune cell repertoire in MS, uncovering previously unidentified or underappreciated immune players.
As MS is a disease of the CNS, it will be crucial to assess whether specific NK cell subsets infiltrate the CNS by passing the blood-brain barrier from the periphery or whether a tissue-resident innate lymphoid subset, as have been identified in a myriad of other tissues (119), contributes to (the resolution of) inflammatory processes in local MS lesions. Finally, the discovery of clonal expansions of NK cells (120), reminiscent of those described for T and B cells (94, 121), is bridging the gap between the innate and adaptive immune system and merits investigation in the context of MS. In this regard, it is interesting to note that infection with Epstein-Barr virus (EBV) has recently been confirmed as an important contributor to MS risk (122, 123), making it intriguing to speculate that EBV infection also impacts the NK cell repertoire, possibly affecting MS immunopathology, as has been postulated for CMV (124).
In conclusion, there is a clear rationale and need for further investigation of NK cell subsets and their role in MS. As both MS genetics and immunology have shown that this role is clearly intertwined with the adaptive immune system, a fundamentally holistic approach is warranted, integrating both innate and adaptive immune players in MS. Such research will contribute to a better understanding of the complex disease pathogenesis and provide a platform for rational therapy design for this chronic autoimmune disorder.
Author Contributions
JB reviewed the literature, wrote the first draft of the manuscript and produced the figures. AG and PM critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Research of the authors into NK cells in MS is specifically supported by the Belgian Charcot Foundation. Furthermore, research of AG is supported by the Research Fund KU Leuven (C24/16/045), the Research Foundation-Flanders (FWO GOA7219N), MS Liga Vlaanderen, the Queen Elisabeth Medical Foundation, the Progressive MS Alliance, and the Horizon2020 “MultipleMS” consortium (grant EU RIA 733161). Research of PM is supported by grants from the Research Foundation-Flanders (FWO, G0A3218N), by funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 779295 and by C1 grant (C16/17/010) of the KU Leuven.
Conflict of Interest
AG has received consulting/travel fees and/or research funding from Novartis, Roche and Merck.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Compston A, Coles A. Multiple Sclerosis. Lancet (2008) 372(9648):1502–17. doi: 10.1016/S0140-6736(08)61620-7
2. Dobson R, Giovannoni G. Multiple Sclerosis – a Review. Eur J Neurol (2019) 26(1):27–40. doi: 10.1111/ene.13819
3. Kobelt G, Thompson A, Berg J, Gannedahl M, Eriksson J. New Insights Into the Burden and Costs of Multiple Sclerosis in Europe. Mult Scler J (2017) 23(8):1123–36. doi: 10.1177/1352458517694432
4. Oh J, Vidal-Jordana A, Montalban X. Multiple Sclerosis: Clinical Aspects. Curr Opin Neurol (2018) 31(6):752–9. doi: 10.1097/wco.0000000000000622
5. Melamed E, Lee MW. Multiple Sclerosis and Cancer: The Ying-Yang Effect of Disease Modifying Therapies. Front Immunol (2019) 10:2954. doi: 10.3389/fimmu.2019.02954
6. Hauser SL, Cree BAC. Treatment of Multiple Sclerosis: A Review. Am J Med (2020) 133(12):1380–90.e2. doi: 10.1016/j.amjmed.2020.05.049
7. Fugger L, Jensen LT, Rossjohn J. Challenges, Progress, and Prospects of Developing Therapies to Treat Autoimmune Diseases. Cell (2020) 181(1):63–80. doi: 10.1016/j.cell.2020.03.007
8. Flodström-Tullberg M, Bryceson YT, Shi FD, Höglund P, Ljunggren HG. Natural Killer Cells in Human Autoimmunity. Curr Opin Immunol (2009) 21:634–40. doi: 10.1016/j.coi.2009.09.012
9. Zhang C, Tian Z. NK Cell Subsets in Autoimmune Diseases. J Autoimmun (2017) 83:22–30. doi: 10.1016/j.jaut.2017.02.005
10. Gianchecchi E, Delfino DV, Fierabracci A. NK Cells in Autoimmune Diseases: Linking Innate and Adaptive Immune Responses. Autoimmun Rev (2018) 17(2):142–54. doi: 10.1016/j.autrev.2017.11.018
11. Kucuksezer UC, Aktas Cetin E, Esen F, Tahrali I, Akdeniz N, Gelmez MY, et al. The Role of Natural Killer Cells in Autoimmune Diseases. Front Immunol (2021) 12:622306. doi: 10.3389/fimmu.2021.622306
12. Yang Y, Day J, Souza-Fonseca Guimaraes F, Wicks IP, Louis C. Natural Killer Cells in Inflammatory Autoimmune Diseases. Clin Trans Immunol (2021) 10(2):e1250. doi: 10.1002/cti2.1250
13. Chanvillard C, Jacolik R, Infante Duarte C, Nayak R. The Role of Natural Killer Cells in Multiple Sclerosis and Their Therapeutic Implications. Front Immunol (2013) 4:63. doi: 10.3389/fimmu.2013.00063
14. Laroni A. Enhancing Natural Killer Cells Is Beneficial in Multiple Sclerosis – Yes. Mult Scler J (2019) 25(4):510–2. doi: 10.1177/1352458518800776
15. Mimpen M, Smolders J, Hupperts R, Damoiseaux J. Natural Killer Cells in Multiple Sclerosis: A Review. Immunol Lett (2020) 222:1–11. doi: 10.1016/j.imlet.2020.02.012
16. Laroni A, Uccelli A. CD56bright Natural Killer Cells: A Possible Biomarker of Different Treatments in Multiple Sclerosis. J Clin Med (2020) 9(5):1450. doi: 10.3390/jcm9051450
17. Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of Natural Killer Cells. Nat Immunol (2008) 9(5):503–10. doi: 10.1038/ni1582
18. Cooper MA, Fehniger TA, Caligiuri MA. The Biology of Human Natural Killer-Cell Subsets. Trends Immunol (2001) 22(11):633–40. doi: 10.1016/S1471-4906(01)02060-9
19. Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity (2017) 47(5):820–33. doi: 10.1016/j.immuni.2017.10.008
20. Abel AM, Yang C, Thakar MS, Malarkannan S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front Immunol (2018) 9:1869. doi: 10.3389/fimmu.2018.01869
21. Di Vito C, Mikulak J, Mavilio D. On the Way to Become a Natural Killer Cell. Front Immunol (2019) 10:1812. doi: 10.3389/fimmu.2019.01812
22. Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, et al. Human Natural Killer Cells: A Unique Innate Immunoregulatory Role for the CD56(bright) Subset. Blood (2001) 97(10):3146–51. doi: 10.1182/blood.v97.10.3146
23. Poli A, Michel T, Thérésine M, Andrès E, Hentges F, Zimmer J. CD56bright Natural Killer (NK) Cells: An Important NK Cell Subset. Immunology (2009) 126(4):458–65. doi: 10.1111/j.1365-2567.2008.03027.x
24. Kaur G, Trowsdale J, Fugger L. Natural Killer Cells and Their Receptors in Multiple Sclerosis. Brain (2013) 136(9):2657–76. doi: 10.1093/brain/aws159
25. Zitti B, Bryceson YT. Natural Killer Cells in Inflammation and Autoimmunity. Cytokine Growth Factor Rev (2018) 42:37–46. doi: 10.1016/j.cytogfr.2018.08.001
26. Paust S, Von Andrian UH. Natural Killer Cell Memory. Nat Immunol (2011) 12(6):500–8. doi: 10.1038/ni.2032
27. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or Adaptive Immunity? The Example of Natural Killer Cells. Science (2011) 331(6013):44–9. doi: 10.1126/science.1198687
28. Stary V, Stary G. NK Cell-Mediated Recall Responses: Memory-Like, Adaptive, or Antigen-Specific? Front Cell Infect Microbiol (2020) 10:208. doi: 10.3389/fcimb.2020.00208
29. Benczur M, Petrányl GG, Pálffy G, Varga M, Tálas M, Kotsy B, et al. Dysfunction of Natural Killer Cells in Multiple Sclerosis: A Possible Pathogenetic Factor. Clin Exp Immunol (1980) 39(3):657–62.
30. Merrill J, Jondal M, Seeley J, Ullberg M, Sidén Å. Decreased NK Killing in Patients With Multiple Sclerosis: An Analysis on the Level of the Single Effector Cell in Peripheral Blood and Cerebrospinal Fluid in Relation to the Activity of the Disease. Clin Exp Immunol (1982) 47(2):419–.
31. Neighbour PA, Grayzel AI, Miller AE. Endogenous and Interferon-Augmented Natural Killer Cell Activity of Human Peripheral Blood Mononuclear Cells In Vitro. Studies of Patients With Multiple Sclerosis, Systemic Lupus Erythematosus or Rheumatoid Arthritis. Clin Exp Immunol (1982) 49(1):11–21.
32. Rice G, Casali P, Merigan T, Oldstone M. Natural Killer Cell Activity in Patients With Multiple Sclerosis Given Alpha Interferon. Ann Neurol (1983) 14(3):333–8. doi: 10.1002/ANA.410140312
33. Hirsch R, Johnson K. Natural Killer Cell Activity in Multiple Sclerosis Patients Treated With Recombinant Interferon-Alpha 2. Clin Immunol Immunopathol (1985) 37(2):236–44. doi: 10.1016/0090-1229(85)90155-2
34. Kastrukoff L, Morgan N, Aziz T, Zecchini D, Berkowitz J, Paty D. Natural Killer (NK) Cells in Chronic Progressive Multiple Sclerosis Patients Treated With Lymphoblastoid Interferon. J Neuroimmunol (1988) 20(1):15–23. doi: 10.1016/0165-5728(88)90109-9
35. Procaccini C, De Rosa V, Pucino V, Formisano L, Matarese G. Animal Models of Multiple Sclerosis. Eur J Pharmacol (2015) 759:182–91. doi: 10.1016/j.ejphar.2015.03.042
36. Matsumoto Y, Kohyama K, Aikawa Y, Shin T, Kawazoe Y, Suzuki Y, et al. Role of Natural Killer Cells and TCR Gamma Delta T Cells in Acute Autoimmune Encephalomyelitis. Eur J Immunol (1998) 28(5):1681–8. doi: 10.1002/(sici)1521-4141(199805)28:05<1681::Aid-immu1681>3.0.Co;2-t
37. Huang D, Shi FD, Jung S, Pien GC, Wang J, Salazar-Mather TP, et al. The Neuronal Chemokine CX3CL1/fractalkine Selectively Recruits NK Cells That Modify Experimental Autoimmune Encephalomyelitis Within the Central Nervous System. FASEB J (2006) 20(7):896–905. doi: 10.1096/fj.05-5465com
38. Galazka G, Jurewicz A, Orlowski W, Stasiolek M, Brosnan CF, Raine CS, et al. EAE Tolerance Induction With Hsp70-Peptide Complexes Depends on H60 and NKG2D Activity. J Immunol (2007) 179(7):4503–12. doi: 10.4049/jimmunol.179.7.4503
39. Xu W, Fazekas G, Hara H, Tabira T. Mechanism of Natural Killer (NK) Cell Regulatory Role in Experimental Autoimmune Encephalomyelitis. J Neuroimmunol (2005) 163(1-2):24–30. doi: 10.1016/j.jneuroim.2005.02.011
40. Hertwig L, Hamann I, Romero-Suarez S, Millward JM, Pietrek R, Chanvillard C, et al. CX3CR1-Dependent Recruitment of Mature NK Cells Into the Central Nervous System Contributes to Control Autoimmune Neuroinflammation. Eur J Immunol (2016) 46(8):1984–96. doi: 10.1002/eji.201546194
41. Hao J, Liu R, Piao W, Zhou Q, Vollmer TL, Campagnolo DI, et al. Central Nervous System (CNS)–resident Natural Killer Cells Suppress Th17 Responses and CNS Autoimmune Pathology. J Exp Med (2010) 207(9):1907–21. doi: 10.1084/jem.20092749
42. Zhang B-N, Yamamura T, Kondo T, Fujiwara M, Tabira T. Regulation of Experimental Autoimmune Encephalomyelitis by Natural Killer (NK) Cells. J Exp Med (1997) 186(10):1677–87. doi: 10.1084/jem.186.10.1677
43. Hammarberg H, Lidman O, Lundberg C, Eltayeb SY, Gielen AW, Muhallab S, et al. Neuroprotection by Encephalomyelitis: Rescue of Mechanically Injured Neurons and Neurotrophin Production by CNS-Infiltrating T and Natural Killer Cells. J Neurosci (2000) 20(14):5283–91. doi: 10.1523/jneurosci.20-14-05283.2000
44. Shi F-D, Takeda K, Akira S, Sarvetnick N, Ljunggren H-G. IL-18 Directs Autoreactive T Cells and Promotes Autodestruction in the Central Nervous System Via Induction of IFN-γ by NK Cells. J Immunol (2000) 165(6):3099–104. doi: 10.4049/jimmunol.165.6.3099
45. Vollmer TL, Liu R, Price M, Rhodes S, La Cava A, Shi F-D. Differential Effects of IL-21 During Initiation and Progression of Autoimmunity Against Neuroantigen. J Immunol (2005) 174(5):2696–701. doi: 10.4049/jimmunol.174.5.2696
46. Winkler-Pickett R, Young HA, Cherry JM, Diehl J, Wine J, Back T, et al. In Vivo Regulation of Experimental Autoimmune Encephalomyelitis by NK Cells: Alteration of Primary Adaptive Responses. J Immunol (2008) 180(7):4495–506. doi: 10.4049/jimmunol.180.7.4495
47. Kelchtermans H, Billiau A, Matthys P. How Interferon-γ Keeps Autoimmune Diseases in Check. Trends Immunol (2008) 29(10):479–86. doi: 10.1016/j.it.2008.07.002
48. Berghmans N, Heremans H, Li S, Martens E, Matthys P, Sorokin L, et al. Rescue From Acute Neuroinflammation by Pharmacological Chemokine-Mediated Deviation of Leukocytes. J Neuroinflamm (2012) 9(1):243. doi: 10.1186/1742-2094-9-243
49. Gandhi R, Laroni A, Weiner HL. Role of the Innate Immune System in the Pathogenesis of Multiple Sclerosis. J Neuroimmunol (2010) 221:7–14. doi: 10.1016/j.jneuroim.2009.10.015
50. Van Kaer L, Postoak JL, Wang C, Yang G, Wu L. Innate, Innate-Like and Adaptive Lymphocytes in the Pathogenesis of MS and EAE. Cell Mol Immunol (2019) 16(6):531–9. doi: 10.1038/s41423-019-0221-5
51. IMSGC, WTCCC2. Genetic Risk and a Primary Role for Cell-Mediated Immune Mechanisms in Multiple Sclerosis. Nature (2011) 476(7359):214–9. doi: 10.1038/nature10251
52. IMSGC. Analysis of Immune-Related Loci Identifies 48 New Susceptibility Variants for Multiple Sclerosis. Nat Genet (2013) 45(11):1353–60. doi: 10.1038/ng.2770
53. IMSGC. Class II HLA Interactions Modulate Genetic Risk for Multiple Sclerosis. Nat Genet (2015) 47(10):1107–13. doi: 10.1038/ng.3395
54. IMSGC. Low-Frequency and Rare-Coding Variation Contributes to Multiple Sclerosis Risk. Cell (2018) 175(6):1679–87.e7. doi: 10.1016/j.cell.2018.09.049
55. IMSGC. Multiple Sclerosis Genomic Map Implicates Peripheral Immune Cells and Microglia in Susceptibility. Science (2019) 365(6460):eaav7188. doi: 10.1126/science.aav7188
56. Hafler JP, Maier LM, Cooper JD, Plagnol V, Hinks A, Simmonds MJ, et al. CD226 Gly307Ser Association With Multiple Autoimmune Diseases. Genes Immun (2009) 10(1):5–10. doi: 10.1038/gene.2008.82
57. Alcina A, Vandenbroeck K, Otaegui D, Saiz A, Gonzalez JR, Fernandez O, et al. The Autoimmune Disease-Associated KIF5A, CD226 and SH2B3 Gene Variants Confer Susceptibility for Multiple Sclerosis. Genes Immun (2010) 11(5):439–45. doi: 10.1038/gene.2010.30
58. Qiu ZX, Zhang K, Qiu XS, Zhou M, Li WM. CD226 Gly307Ser Association With Multiple Autoimmune Diseases: A Meta-Analysis. Hum Immunol (2013) 74(2):249–55. doi: 10.1016/j.humimm.2012.10.009
59. Liu G, Hu Y, Jin S, Jiang Q. Genetic Variant Rs763361 Regulates Multiple Sclerosis CD226gene Expression. Proc Natl Acad Sci USA (2017) 114:E906–7. doi: 10.1073/pnas.1618520114
60. Gilfillan S, Chan CJ, Cella M, Haynes NM, Rapaport AS, Boles KS, et al. DNAM-1 Promotes Activation of Cytotoxic Lymphocytes by Nonprofessional Antigen-Presenting Cells and Tumors. J Exp Med (2008) 205(13):2965–73. doi: 10.1084/jem.20081752
61. Martinet L, Smyth MJ. Balancing Natural Killer Cell Activation Through Paired Receptors. Nat Rev Immunol (2015) 15(4):243–54. doi: 10.1038/nri3799
62. Piédavent-Salomon M, Willing A, Engler JB, Steinbach K, Bauer S, Eggert B, et al. Multiple Sclerosis Associated Genetic Variants Ofcd226impair Regulatory T Cell Function. Brain (2015) 138(11):3263–74. doi: 10.1093/brain/awv256
63. Ardolino M, Zingoni A, Cerboni C, Cecere F, Soriani A, Iannitto ML, et al. DNAM-1 Ligand Expression on Ag-Stimulated T Lymphocytes Is Mediated by ROS-Dependent Activation of DNA-Damage Response: Relevance for NK–T Cell Interaction. Blood (2011) 117(18):4778–86. doi: 10.1182/blood-2010-08-300954
64. Cappellano G, Orilieri E, Comi C, Chiocchetti A, Bocca S, Boggio E, et al. Variations of the Perforin Gene in Patients With Multiple Sclerosis. Genes Immun (2008) 9(5):438–44. doi: 10.1038/gene.2008.35
65. Sidore C, Orrù V, Cocco E, Steri M, Inshaw JR, Pitzalis M, et al. PRF1 Mutation Alters Immune System Activation, Inflammation, and Risk of Autoimmunity. Mult Scler J (2021) 27:1332–40. doi: 10.1177/1352458520963937
66. Voskoboinik I, Smyth MJ, Trapani JA. Perforin-Mediated Target-Cell Death and Immune Homeostasis. Nat Rev Immunol (2006) 6:940–52. doi: 10.1038/nri1983
67. Ueda I, Kurokawa Y, Koike K, Ito S, Sakata A, Matsumora T, et al. Late-Onset Cases of Familial Hemophagocytic Lymphohistiocytosis With Missense Perforin Gene Mutations. Am J Hematol (2007) 82(6):427–32. doi: 10.1002/ajh.20878
68. Brisse E, Wouters CH, Matthys P. Hemophagocytic Lymphohistiocytosis (HLH): A Heterogeneous Spectrum of Cytokine-Driven Immune Disorders. Cytokine Growth Factor Rev (2015) 26(3):263–80. doi: 10.1016/j.cytogfr.2014.10.001
69. Voskoboinik I, Thia M-C, Trapani JA. A Functional Analysis of the Putative Polymorphisms A91V and N252S and 22 Missense Perforin Mutations Associated With Familial Hemophagocytic Lymphohistiocytosis. Blood (2005) 105(12):4700–6. doi: 10.1182/blood-2004-12-4935
70. House IG, Thia K, Brennan AJ, Tothill R, Dobrovic A, Yeh WZ, et al. Heterozygosity for the Common Perforin Mutation, P.A91V, Impairs the Cytotoxicity of Primary Natural Killer Cells From Healthy Individuals. Immunol Cell Biol (2015) 93(6):575–80. doi: 10.1038/icb.2015.1
71. Dendrou CA, Petersen J, Rossjohn J, Fugger L. HLA Variation and Disease. Nat Rev Immunol (2018) 18(5):325–39. doi: 10.1038/nri.2017.143
72. Ahn R, Vukcevic D, Motyer A, Nititham J, Squire DM, Hollenbach JA, et al. Large-Scale Imputation of KIR Copy Number and HLA Alleles in North American and European Psoriasis Case-Control Cohorts Reveals Association of Inhibitory KIR2DL2 With Psoriasis. Front Immunol (2021) 12:684326. doi: 10.3389/fimmu.2021.684326
73. Zhang JY, Roberts H, Flores DSC, Cutler AJ, Brown AC, Whalley JP, et al. Using De Novo Assembly to Identify Structural Variation of Eight Complex Immune System Gene Regions. PloS Comput Biol (2021) 17(8):e1009254. doi: 10.1371/journal.pcbi.1009254
74. Traugott U, Raine CS. Further Lymphocyte Characterization in the Central Nervous System in Multiple Sclerosis. Ann New York Acad Sci (1984) 436:163–80. doi: 10.1111/j.1749-6632.1984.tb14788.x
75. Traugott U. Characterization and Distribution of Lymphocyte Subpopulations in Multiple Sclerosis Plaques Versus Autoimmune Demyelinating Lesions. Springer Semin Immunopathol (1985) 8:1. 1985;8(1):71–95. doi: 10.1007/BF00197248
76. Weber W, Buurman W, Vandermeeren M, Medaer R, Raus J. Fine Analysis of Cytolytic and Natural Killer T Lymphocytes in the CSF in Multiple Sclerosis and Other Neurologic Diseases. Neurology (1987) 37(3):419–25. doi: 10.1212/WNL.37.3.419
77. Hamann I, Dörr J, Glumm R, Chanvillard C, Janssen A, Millward JM, et al. Characterization of Natural Killer Cells in Paired CSF and Blood Samples During Neuroinflammation. J Neuroimmunol (2013) 254(1-2):165–9. doi: 10.1016/j.jneuroim.2012.08.009
78. Liu Q, Sanai N, Jin W-N, La Cava A, Van Kaer L, Shi F-D. Neural Stem Cells Sustain Natural Killer Cells That Dictate Recovery From Brain Inflammation. Nat Neurosci (2016) 19(2):243–52. doi: 10.1038/nn.4211
79. Lagumersindez-Denis N, Wrzos C, Mack M, Winkler A, van der Meer F, Reinert MC, et al. Differential Contribution of Immune Effector Mechanisms to Cortical Demyelination in Multiple Sclerosis. Acta Neuropathol (2017) 134(1):15–34. doi: 10.1007/s00401-017-1706-x
80. Bratke K, Kuepper M, Bade B, Virchow JC, Luttmann W. Differential Expression of Human Granzymes A, B, and K in Natural Killer Cells and During CD8+ T Cell Differentiation in Peripheral Blood. Eur J Immunol (2005) 35(9):2608–16. doi: 10.1002/eji.200526122
81. Gross CC, Schulte-Mecklenbeck A, Rünzi A, Kuhlmann T, Posevitz-Fejfár A, Schwab N, et al. Impaired NK-Mediated Regulation of T-Cell Activity in Multiple Sclerosis is Reconstituted by IL-2 Receptor Modulation. Proc Natl Acad Sci USA (2016) 113(21):E2973–82. doi: 10.1073/pnas.1524924113
82. Rodríguez-Martín E, Picón C, Costa-Frossard L, Alenda R, Sainz de la Maza S, Roldán E, et al. Natural Killer Cell Subsets in Cerebrospinal Fluid of Patients With Multiple Sclerosis. Clin Exp Immunol (2015) 180(2):243–9. doi: 10.1111/cei.12580
83. Rodríguez-Lorenzo S, van Olst L, Rodriguez-Mogeda C, Kamermans A, van der Pol SMA, Rodríguez E, et al. Single-Cell Profiling Reveals Periventricular CD56bright NK Cell Accumulation in Multiple Sclerosis. bioRxiv (2021). doi: 10.1101/2021.09.17.460741
84. De Jager PL, Rossin E, Pyne S, Tamayo P, Ottoboni L, Viglietta V, et al. Cytometric Profiling in Multiple Sclerosis Uncovers Patient Population Structure and a Reduction of CD8low Cells. Brain (2008) 131(7):1701–11. doi: 10.1093/brain/awn118
85. McKinney EF, Cuthbertson I, Harris KM, Smilek DE, Connor C, Manferrari G, et al. A CD8+ NK Cell Transcriptomic Signature Associated With Clinical Outcome in Relapsing Remitting Multiple Sclerosis. Nat Commun (2021) 12(1):1–9. doi: 10.1038/s41467-020-20594-2
86. Martínez-Rodríguez JE, Cobo-Calvo A, Villar LM, Munteis E, Blanco Y, Rasal R, et al. Adaptive Natural Killer Cell Response to Cytomegalovirus and Disability Progression in Multiple Sclerosis. Mult Scler J (2016) 22(6):741–52. doi: 10.1177/1352458515601215
87. Papalexi E, Satija R. Single-Cell RNA Sequencing to Explore Immune Cell Heterogeneity. Nat Rev Immunol (2018) 18(1):35–45. doi: 10.1038/nri.2017.76
88. Colonna M, Brioschi S. Neuroinflammation and Neurodegeneration in Human Brain at Single-Cell Resolution. Nat Rev Immunol (2020) 20(2):81–2. doi: 10.1038/s41577-019-0262-0
89. Crinier A, Milpied P, Escalière B, Piperoglou C, Galluso J, Balsamo A, et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity (2018) 49(5):971–86.e5. doi: 10.1016/j.immuni.2018.09.009
90. Yang C, Siebert JR, Burns R, Gerbec ZJ, Bonacci B, Rymaszewski A, et al. Heterogeneity of Human Bone Marrow and Blood Natural Killer Cells Defined by Single-Cell Transcriptome. Nat Commun (2019) 10(1):3931. doi: 10.1038/s41467-019-11947-7
91. Smith SL, Kennedy PR, Stacey KB, Worboys JD, Yarwood A, Seo S, et al. Diversity of Peripheral Blood Human NK Cells Identified by Single-Cell RNA Sequencing. Blood Adv (2020) 4(7):1388–406. doi: 10.1182/bloodadvances.2019000699
92. Schafflick D, Xu CA, Hartlehnert M, Cole M, Schulte-Mecklenbeck A, Lautwein T, et al. Integrated Single Cell Analysis of Blood and Cerebrospinal Fluid Leukocytes in Multiple Sclerosis. Nat Commun (2020) 11(1):247. doi: 10.1038/s41467-019-14118-w
93. Esaulova E, Cantoni C, Shchukina I, Zaitsev K, Bucelli RC, Wu GF, et al. Single-Cell RNA-Seq Analysis of Human CSF Microglia and Myeloid Cells in Neuroinflammation. Neurol - Neuroimmunol Neuroinflamm (2020) 7(4):e732. doi: 10.1212/nxi.0000000000000732
94. Ramesh A, Schubert RD, Greenfield AL, Dandekar R, Loudermilk R, Sabatino JJ Jr, et al. A Pathogenic and Clonally Expanded B Cell Transcriptome in Active Multiple Sclerosis. Proc Natl Acad Sci USA (2020) 117(37):22932–43. doi: 10.1073/pnas.2008523117
95. Rabinovich BA, Li J, Shannon J, Hurren R, Chalupny J, Cosman D, et al. Activated, But Not Resting, T Cells Can Be Recognized and Killed by Syngeneic NK Cells. J Immunol (2003) 170(7):3572–6. doi: 10.4049/jimmunol.170.7.3572
96. Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A. Antigen-Activated Human T Lymphocytes Express Cell-Surface NKG2D Ligands via an ATM/ATR-Dependent Mechanism and Become Susceptible to Autologous NK- Cell Lysis. Blood (2007) 110(2):606–15. doi: 10.1182/blood-2006-10-052720
97. Roy S, Barnes PF, Garg A, Wu S, Cosman D, Vankayalapati R. NK Cells Lyse T Regulatory Cells That Expand in Response to an Intracellular Pathogen. J Immunol (2008) 180(3):1729–36. doi: 10.4049/jimmunol.180.3.1729
98. Zingoni A, Ardolino M, Santoni A, Cerboni C. NKG2D and DNAM-1 Activating Receptors and Their Ligands in NK-T Cell Interactions: Role in the NK Cell-Mediated Negative Regulation of T Cell Responses. Front Immunol (2012) 3:408. doi: 10.3389/fimmu.2012.00408
99. Jiang W, Chai NR, Maric D, Bielekova B. Unexpected Role for Granzyme K in CD56bright NK Cell-Mediated Immunoregulation of Multiple Sclerosis. J Immunol (2011) 187(2):781–90. doi: 10.4049/jimmunol.1100789
100. Lünemann A, Lünemann JD, Roberts S, Messmer B, Da Silva RB, Raine CS, et al. Human NK Cells Kill Resting But Not Activated Microglia via NKG2D- and NKp46-Mediated Recognition. J Immunol (2008) 181(9):6170–7. doi: 10.4049/jimmunol.181.9.6170
101. Spaggiari GM, Carosio R, Pende D, Marcenaro S, Rivera P, Zocchi MR, et al. NK Cell-Mediated Lysis of Autologous Antigen-Presenting Cells Is Triggered by the Engagement of the Phosphatidylinositol 3-Kinase Upon Ligation of the Natural Cytotoxicity Receptors NKp30 and Nkp46. Eur J Immunol (2001) 31(6):1656–65. doi: 10.1002/1521-4141(200106)31:6<1656::aid-immu1656>3.0.co;2-v
102. Ferlazzo G, Tsang ML, Moretta L, Melioli G, Steinman RM, Münz C. Human Dendritic Cells Activate Resting Natural Killer (NK) Cells and Are Recognized via the NKp30 Receptor by Activated NK Cells. J Exp Med (2002) 195(3):343–51. doi: 10.1084/jem.20011149
103. Pende D, Castriconi R, Romagnani P, Spaggiari GM, Marcenaro S, Dondero A, et al. Expression of the DNAM-1 Ligands, Nectin-2 (CD112) and Poliovirus Receptor (CD155), on Dendritic Cells: Relevance for Natural Killer-Dendritic Cell Interaction. Blood (2006) 107(5):2030–6. doi: 10.1182/blood-2005-07-2696
104. Nedvetzki S, Sowinski S, Eagle RA, Harris J, VéLy FDR, Pende D, et al. Reciprocal Regulation of Human Natural Killer Cells and Macrophages Associated With Distinct Immune Synapses. Blood (2007) 109(9):3776–85. doi: 10.1182/blood-2006-10-052977
105. Gross CC, Schulte-Mecklenbeck A, Wiendl H, Marcenaro E, de Rosbo NK, Uccelli A, et al. Regulatory Functions of Natural Killer Cells in Multiple Sclerosis. Front Immunol (2016) 7:606. doi: 10.3389/fimmu.2016.00606
106. Bielekova B, Catalfamo M, Reichert-Scrivner S, Packer A, Cerna M, Waldmann T, et al. Regulatory CD56(bright) Natural Killer Cells Mediate Immunomodulatory Effects of IL-2Ralpha-Targeted Therapy (Daclizumab) in Multiple Sclerosis. Proc Natl Acad Sci USA (2006) 103(15):5941–6. doi: 10.1073/PNAS.0601335103
107. Bielekova B, Richert N, Herman M, Ohayon J, Waldmann T, McFarland H, et al. Intrathecal Effects of Daclizumab Treatment of Multiple Sclerosis. Neurology (2011) 77(21):1877–86. doi: 10.1212/WNL.0B013E318239F7EF
108. Martin JF, Perry JSA, Jakhete NR, Wang X, Bielekova B. An IL-2 Paradox: Blocking CD25 on T Cells Induces IL-2–Driven Activation of CD56bright NK Cells. J Immunol (2010) 185(2):1311–20. doi: 10.4049/jimmunol.0902238
109. Sheridan JP, Zhang Y, Riester K, Tang MT, Efros L, Shi J, et al. Intermediate-Affinity Interleukin-2 Receptor Expression Predicts CD56bright Natural Killer Cell Expansion After Daclizumab Treatment in the CHOICE Study of Patients With Multiple Sclerosis. Mult Scler J (2011) 17(12):1441–8. doi: 10.1177/1352458511414755
110. Bianchi A, Ciccarelli O. Daclizumab-Induced Encephalitis in Multiple Sclerosis. Mult Scler J (2019) 25(12):1557–9. doi: 10.1177/1352458519845079
111. Schwichtenberg SC, Wisgalla A, Schroeder-Castagno M, Alvarez-González C, Schlickeiser S, Siebert N, et al. Fingolimod Therapy in Multiple Sclerosis Leads to the Enrichment of a Subpopulation of Aged NK Cells. Neurotherapeutics (2021) 18(3):1783–97. doi: 10.1007/s13311-021-01078-7
112. Cohen JA, Tenenbaum N, Bhatt A, Zhang Y, Kappos L. Extended Treatment With Fingolimod for Relapsing Multiple Sclerosis: The 14-Year LONGTERMS Study Results. Ther Adv Neurol Disord (2019) 12:175628641987832. doi: 10.1177/1756286419878324
113. Gianchecchi E, Delfino DV, Fierabracci A. Natural Killer Cells: Potential Biomarkers and Therapeutic Target in Autoimmune Diseases? Front Immunol (2021) 12:616853. doi: 10.3389/fimmu.2021.616853
114. Gilmore W, Lund BT, Li P, Levy AM, Kelland EE, Akbari O, et al. Repopulation of T, B, and NK Cells Following Alemtuzumab Treatment in Relapsing-Remitting Multiple Sclerosis. J Neuroinflamm (2020) 17(1):189. doi: 10.1186/s12974-020-01847-9
115. Gross CC, Ahmetspahic D, Ruck T, Schulte-Mecklenbeck A, Schwarte K, Jörgens S, et al. Alemtuzumab Treatment Alters Circulating Innate Immune Cells in Multiple Sclerosis. Neurol - Neuroimmunol Neuroinflamm (2016) 3(6):e289. doi: 10.1212/nxi.0000000000000289
116. Kastrukoff LF, Lau A, Wee R, Zecchini D, White R, Paty DW. Clinical Relapses of Multiple Sclerosis Are Associated With ‘Novel’ Valleys in Natural Killer Cell Functional Activity. J Neuroimmunol (2003) 145(1-2):103–14. doi: 10.1016/j.jneuroim.2003.10.001
117. Caruana P, Lemmert K, Ribbons K, Lea R, Lechner-Scott J. Natural Killer Cell Subpopulations Are Associated With MRI Activity in a Relapsing-Remitting Multiple Sclerosis Patient Cohort From Australia. Mult Scler J (2017) 23(11):1479–87. doi: 10.1177/1352458516679267
118. Mimpen M, Muris AH, Rolf L, Gerlach O, Kuhle J, Hupperts R, et al. Prognostic Value of Natural Killer Cell/T Cell Ratios for Disease Activity in Multiple Sclerosis. Eur J Neurol (2021) 28(3):901–9. doi: 10.1111/ene.14680
119. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate Lymphoid Cells: 10 Years on. Cell (2018) 174(5):1054–66. doi: 10.1016/j.cell.2018.07.017
120. Adams NM, Grassmann S, Sun JC. Clonal Expansion of Innate and Adaptive Lymphocytes. Nat Rev Immunol (2020) 20(11):694–707. doi: 10.1038/s41577-020-0307-4
121. Pappalardo JL, Zhang L, Pecsok MK, Perlman K, Zografou C, Raddassi K, et al. Transcriptomic and Clonal Characterization of T Cells in the Human Central Nervous System. Sci Immunol (2020) 5(51):eabb8786. doi: 10.1126/sciimmunol.abb8786
122. Bjornevik K, Cortese M, Healy BC, Kuhle J, Mina MJ, Leng Y, et al. Longitudinal Analysis Reveals High Prevalence of Epstein-Barr Virus Associated With Multiple Sclerosis. Science (2022) 375(6578):296–301. doi: 10.1126/science.abj8222
123. Lanz TV, Brewer RC, Ho PP, Moon J-S, Jude KM, Fernandez D, et al. Clonally Expanded B Cells in Multiple Sclerosis Bind EBV EBNA1 and GlialCAM. Nature (2022) 603:321–7. doi: 10.1038/s41586-022-04432-7
Keywords: multiple sclerosis, natural killer (NK) cells, autoimmune disorders, experimental autoimmune encephalomyelitis (EAE), immune cell heterogeneity, multiple sclerosis genetics
Citation: Beliën J, Goris A and Matthys P (2022) Natural Killer Cells in Multiple Sclerosis: Entering the Stage. Front. Immunol. 13:869447. doi: 10.3389/fimmu.2022.869447
Received: 04 February 2022; Accepted: 14 March 2022;
Published: 06 April 2022.
Edited by:
Carmen Infante-Duarte, Charité Universitätsmedizin Berlin, GermanyReviewed by:
Jan Damoiseaux, Maastricht University Medical Centre, NetherlandsCopyright © 2022 Beliën, Goris and Matthys. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrick Matthys, cGF0cmljay5tYXR0aHlzQGt1bGV1dmVuLmJl
†These authors have contributed equally to this work and share senior authorship
 Jarne Beliën
Jarne Beliën An Goris
An Goris Patrick Matthys
Patrick Matthys