- 1Department of Virology and Biotechnology, ICMR- National Institute for Research in Tuberculosis, Chennai, India
- 2University of Madras, Chennai, India
- 3Department of Statistics, Epidemiology Unit, ICMR-National Institute for Research in Tuberculosis, Chennai, India
Background: The early diagnosis of tuberculosis using novel non-sputum-based biomarkers is of high priority in the End TB strategy. MicroRNAs (miRNAs) are significant regulators of TB pathogenesis and their differential expression pattern among healthy, latent, and active TB population has revealed their potentiality as biomarkers in recent studies. Thus, we systematically reviewed and performed a meta-analysis on the role of host miRNAs in TB diagnosis. We also reviewed the involvement of miRNAs in the immune response to Mycobacterium tuberculosis (Mtb).
Methods: Pubmed, Ovid and Cochrane databases were searched to retrieve published literature from 2000 to 2020 using predefined keywords. We screened relevant studies based on inclusion and exclusion criteria and the included studies were assessed for their quality using STARD guidelines and QUADAS-2 tool. Funnel plots were constructed to assess the publication bias. The heterogeneity of studies and overall pooled results of sensitivity, specificity and DOR were determined using forest plots.
Results: We retrieved a total of 447 studies collectively from all the databases, out of which 21 studies were included for qualitative analysis. In these studies, miR-29, miR-31, miR-125b, miR146a and miR-155 were consistently reported. The overall sensitivity, specificity and DOR of these miRNAs were found to be 87.9% (81.7-92.2), 81.2% (74.5-86.5) and 43.1(20.3-91.3) respectively. Among these, miR-31 had the maximum diagnostic accuracy, with a sensitivity of 96% (89.7-98.5), specificity of 89% (81.2-93.8) and DOR of 345.9 (90.2-1326.3), meeting the minimal target product profile (TPP) for TB diagnostics.
Conclusion: miRNAs can thus be exploited as potential biomarkers for rapid detection of tuberculosis as evident from their diagnostic performance.
Systematic Review Registration: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42021226559 PROSPERO (CRD42021226559).
Introduction
Tuberculosis (TB) remains as one of the major causes of global morbidity and mortality. Among one-fourth of the world's population latently (asymptomatically) infected with Mycobacterium tuberculosis (Mtb), 5 to 10% bear the risk of developing active TB during their lifetime (1, 2). Identifying and treating this pool of individuals could contribute significantly to TB elimination. An ideal TB biomarker must exhibit explicit identification of subjects with Mtb infection, differentiation between active TB disease and Latent Tuberculosis infection (LTBI), and possibly predict for future progression to active disease (3). Research in the past few years has focused on the identification of a broad range of biomarkers and biosignatures using transcriptomic, metabolomic, and proteomic approaches in various sample types, such as blood, sputum, and urine. Maclean et al. recently published a systematic review encompassing various biomarkers for detecting active TB (4). Independent systematic reviews on the diagnostic accuracy of transcriptomic signatures derived from the whole blood of host for diagnosis of active TB (5, 6) and incipient TB (7), and the utility of biomarkers like IFN-γ (8–10), IP-10 (11, 12), LAM (13, 14), and IL-2 (15) for the detection of TB infection and disease are also available. More recently, systematic reviews on Mtb- specific cytokine biomarkers that can differentiate between active TB and LTBI have been published (16, 17). Reviews on the diagnostic potential of novel Mtb antigens (18) and other informative reviews on current TB biomarker research are also available (19, 20). However, none of these reviews provide accurate data to guide the development of a point-of-care test.
MicroRNAs (miRNAs) are highly conserved, small, non-coding RNAs of ~18-25 nucleotides that are involved in regulating gene expression at the post-transcriptional level (21). The seed region of miRNAs typically spanning six nucleotides (residues 2-8 at the 5’ end) binds to the target mRNA in the 3’ untranslated region (UTR) resulting in translational repression or mRNA degradation (22). In this way, miRNAs modulate several key functions including cell growth, differentiation, and apoptosis. Most importantly, miRNAs regulate the production of a myriad of cytokines and chemokines that modulate the host immune response during various disease states, and aberrant miRNA expression patterns have been associated with the pathogenesis of many diseases (23). Since its binding does not require perfect complementarity, a single mRNA can be acted upon by multiple miRNAs and a single miRNA can target hundreds of mRNAs resulting in enhanced gene regulation (24). Till date, around 2600 functional miRNAs targeting almost 60% of human genes have been identified (25). Beyond their presence in the cellular milieu, miRNAs are also released into circulation and can be identified in different body fluids. Most of the released miRNAs are complexed to proteins like Argonaute2 (AGO2), and are therefore remarkably stable in circulation and resistant to RNases, extreme temperatures, acidic, and alkaline conditions, as well as repeated freeze-thaw cycles (26). With their high stability, ease of recovery, substantial levels of sensitivity and specificity and translational potential as a point of care test, miRNAs are considered to be promising diagnostic biomarkers.
Upon infection with Mtb, the host cells repattern their immune response for the defence arsenal. Mtb on the other hand tries to subvert the host defenses for its intracellular survival by manipulating the host miRNA profile (27). This reprogramming involves the regulation of various biological processes by miRNAs. The first line of host defence to Mtb infection begins when the Pattern Recognition Receptors (PRRs), majorly the Toll-like receptors (TLRs), present on the surface of macrophages and dendritic cells recognise the pathogen-associated molecular patterns (PAMPs). Stimulation of TLRs leads to the induction of miRNA biogenesis. The miRNA coding transcripts are transcribed as primary- miRNA (pri-miRNA >100 nt) by RNA polymerase II. Pri-miRNA is cleaved by a microprocessor complex comprising of an RNase III endonuclease (drosha) and its essential cofactor DGCR8 (Pasha) into precursor miRNA (pre-miRNA) (60-70 nt). The pre-miRNA is loaded onto Exportin 5 and transported to the cytoplasm where it is further processed by Dicer, a ribonuclease III enzyme, with the help of its cofactor TRBP into a duplex of 20-25 nt. A helicase enzyme unwinds the duplex into a mature miRNA and a lesser expressed passenger strand (miRNA*). Under some conditions, mature miRNA can also arise from miRNA*. The mature miRNA is now integrated into a multiprotein complex called RNA-induced silencing complex (RISC) and is equipped for gene regulation (28). Gene regulation by miRNA takes place by three mechanisms, based on the complementarity of the miRNA with its target mRNA. A perfect binding results in mRNA degradation, while a near perfect/partial complementarity leads to translational repression. mRNA deadenylation leading to destability is a third mechanism which accelerates mRNA degradation (29). Through either of these ways, gene regulation by miRNA inside the macrophage leads to the production of multiple cytokines and chemokines that trigger the host innate immune response. TB infection-specific miRNA also arise from other immune cells contained within the granuloma and prime the adaptive immune response. Besides directly controlling gene expression, these miRNA can also be secreted into the extracellular milieu either by passive leakage through cell apoptosis/necrosis or by active secretion via encapsulation within microvesicles/exosomes and by binding to high-density lipoproteins (HDL) or AGO2 proteins. (30) This process can give rise to highly stable Mtb-associated miRNA signatures in various biological fluids (Figure 1).
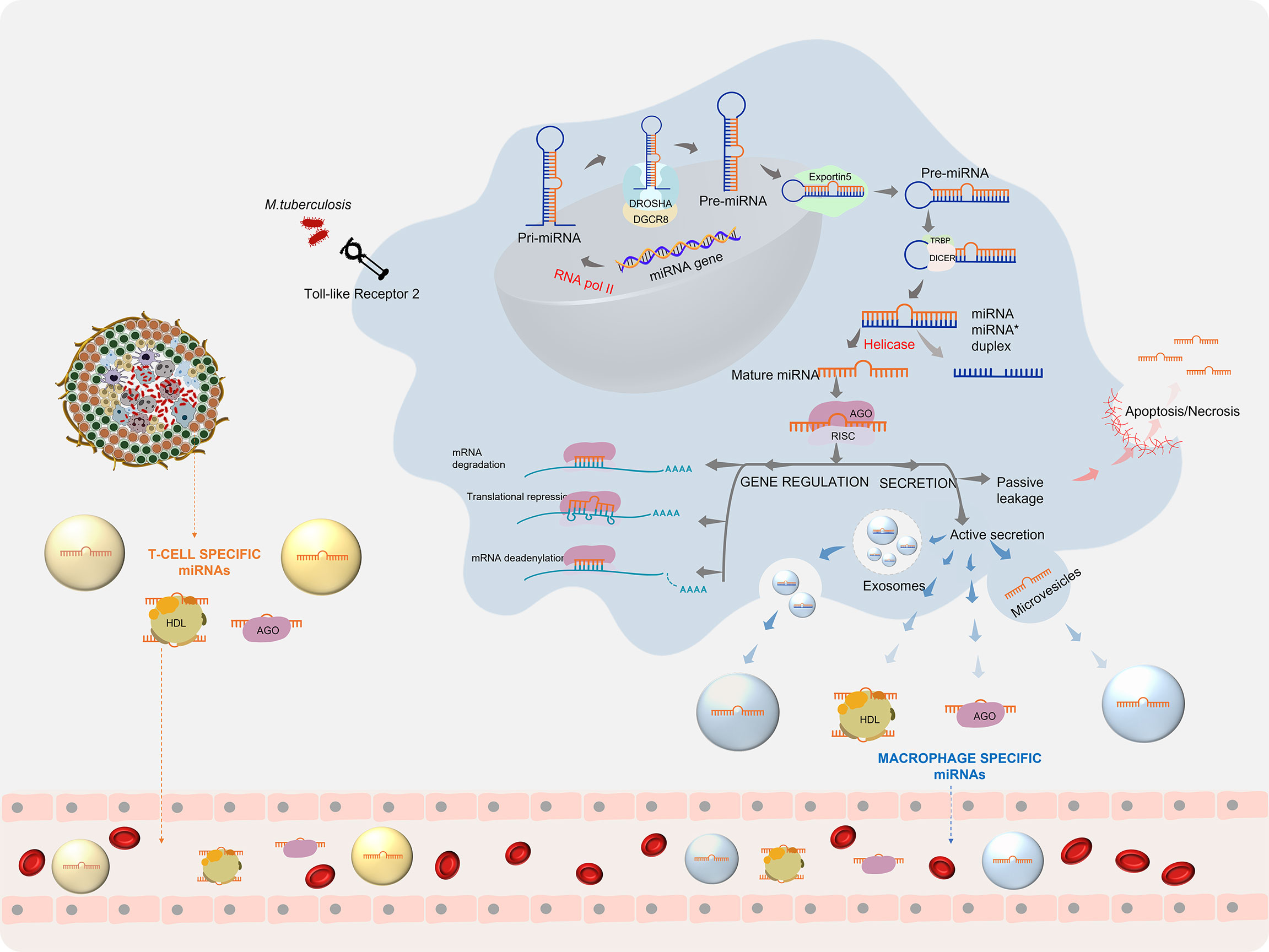
Figure 1 miRNA biogenesis during Mycobacterium tuberculosis infection: Mtb infection leads to the initiation of miRNA production, which after processing gives rise to mature miRNAs. Mature miRNAs can regulate gene expression and can be secreted into the extra cellular environment (30).
Recent reports suggest that most of the cellular immune response elicited by the host and the immune modulation strategies executed by Mtb are regulated by miRNAs (31). Several studies have reported dysregulated expression profiles of miRNAs that are intricately engaged in the host-pathogen interaction in active TB patients as compared to latently infected individuals and healthy controls (Figure 2). In this article, we reviewed the existing literature on the potential of miRNAs to serve as diagnostic biomarkers for TB and evaluated the diagnostic accuracy of reported miRNAs so as to identify a candidate miRNA biomarker that can be used as a reliable TB diagnostic marker. This systematic review has been registered in PROSPERO (CRD42021226559).
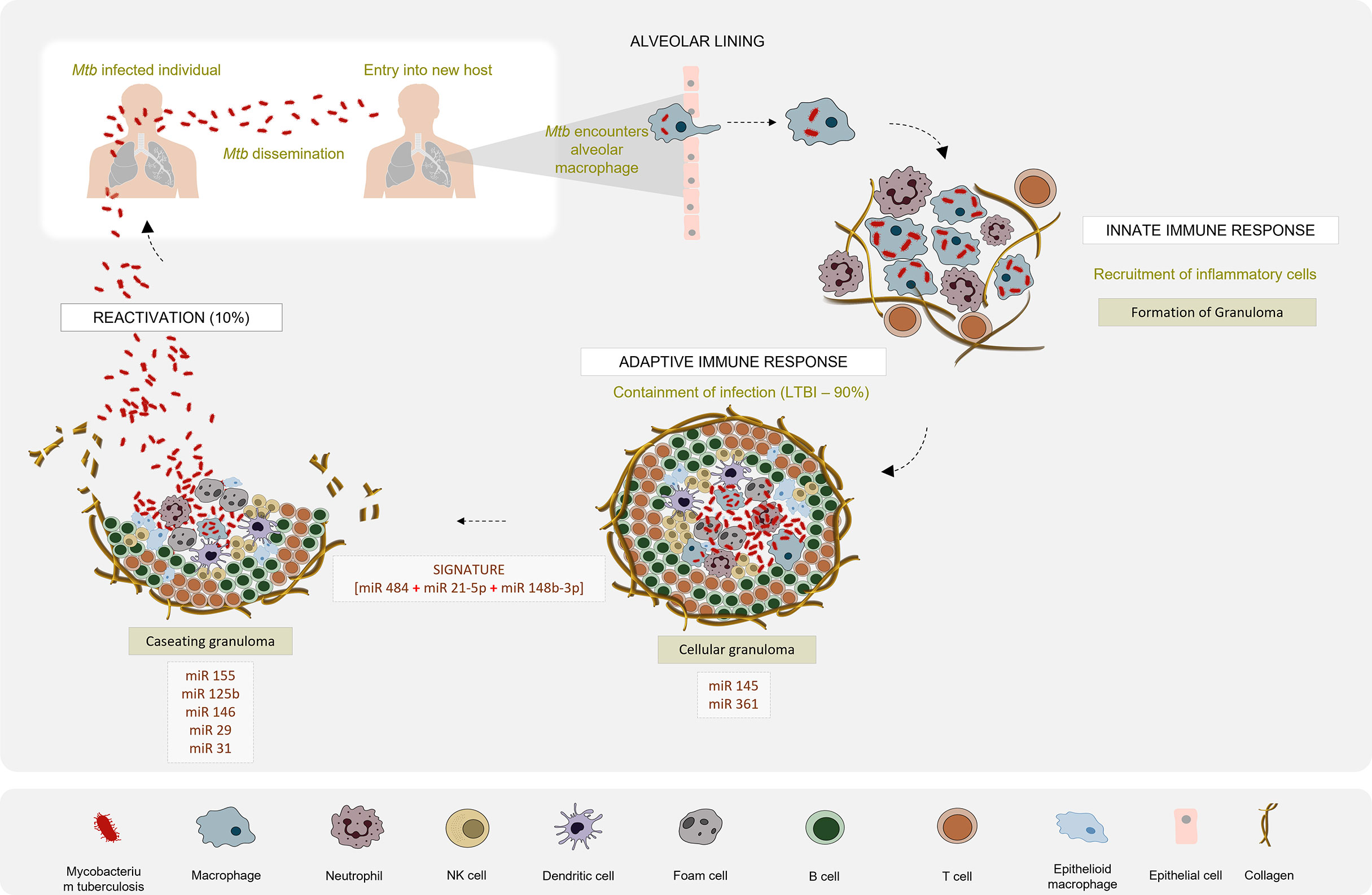
Figure 2 Differential miRNA expression profile in the pathogenesis of Tuberculosis: Mtb infection is initiated when tubercle bacilli from an active TB patient are disseminated through aerosol droplets to a new host. Resident alveolar macrophages in the alveolar space are the first to encounter and phagocytose the ingested Mtb. If Mtb manages to survive this first line of defence, it begins to gain access to the lung interstitial tissue. Dendritic cells also uptake Mtb and transport it to the thoracic lymph nodes for priming a T cell response. These events induce the recruitment of T and B cells and neutrophils, to the site of infection. They aggregate to form a granuloma with the infected macrophage in the centre surrounded by a lymphocytic cuff. Inside the granuloma, the production of a repertoire of pro- and anti-inflammatory cytokines aids the survival of Mtb. Successful evasion of the host immune response leads to a calcified granuloma, and establishment of latency. Active TB disease would ensue if resuscitation occurs and the granuloma starts caseating. Differential expression patterns of miRNAs reported in various studies between the latent and active forms of TB are depicted.
Methods
Search strategy
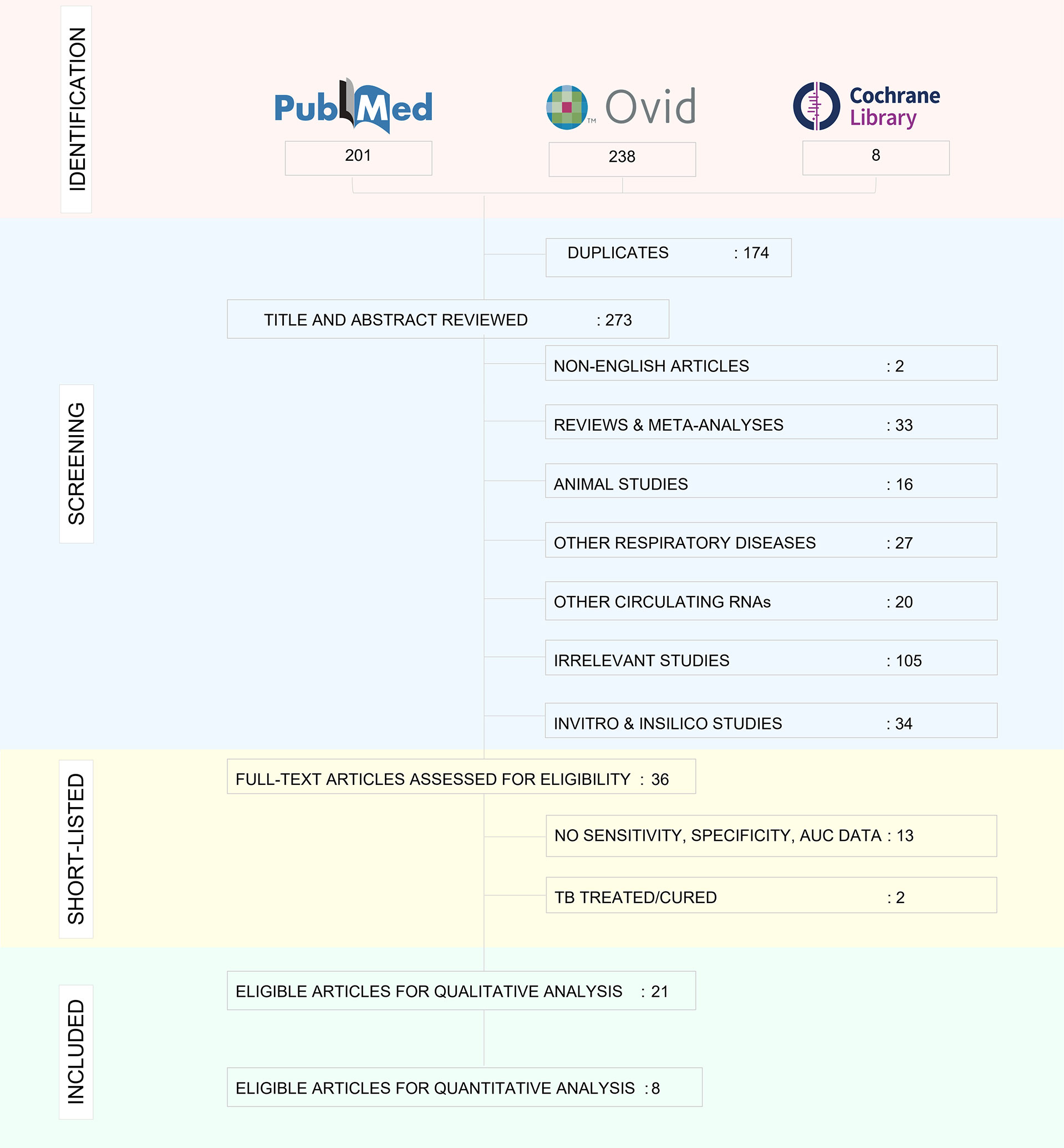
We have performed and reported this systematic review and meta-analysis in compliance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (32). An extensive literature search was executed in PubMed and Ovid interface to retrieve articles from Medline and Embase databases. Cochrane library was also included in the search platform. MeSH terms and key words related to tuberculosis and miRNA were applied for the search and are described in Supplementary Tables 1A–C. We also performed forward and backward reference checking from relevant review articles to identify any additional eligible studies.
Eligibility criteria
Time Period
Articles published in English from January 2000 till December 2020 were included.
Study types
All study designs (cross-sectional, case-control or cohort studies) were considered regardless of prospective or retrospective sampling. Non-original articles including narratives and systematic reviews, meta-analyses, and conference abstracts were not included for consideration. Animal studies were also excluded.
Biomarker criteria
Studies reporting individual miRNAs or miRNA panels, or both were included. Studies evaluating other small RNAs were not considered. Articles evaluating miRNA as biomarker for other respiratory diseases and diseases other than tuberculosis were excluded as they are beyond the scope of this systematic review. Studies lacking sensitivity, specificity, and area under the curve (AUC) data were not included.
Study population
There were no exclusion criteria for patient characteristics, both adult and paediatric population studies were included.
Screening and data extraction
All study articles retrieved by the search were fetched in Endnote X9 and duplicates were removed. We used Rayyan to manage the selection of studies (33). Publications from the electronic searches were initially subjected to screening of title and abstracts by two reviewers (ED, BS) to identify potentially eligible studies. Following this, the two authors independently performed full text screening to filter relevant studies. For certain articles with insufficient or incomplete information to enable a decision regarding inclusion/exclusion, the authors of the relevant papers were contacted for missing data. We collected details on the publication year, country, sample size (cases and controls separately), male/female proportion, mean or median age, study population (Active TB/LTBI/Healthy), reference test, sample source, screening and validation methods used, miRNA and/or miRNA signatures identified and diagnostic accuracy measures (sensitivity, specificity, AUC). We designed a data table and entered the extracted data into an excel database. We documented the values of True Positives (TP), False Positives (FP), True Negatives (TN), and False Negatives (FN) of the index test results. If not explicitly reported, the values were calculated from the reported sensitivity, specificity, and the sample size. For studies lacking sufficient data to construct a 2 x 2 table, the study authors were contacted requesting the required details. If study authors were unable to provide the information, we did not include the study for meta‐analysis but retained it in the narrative section for qualitative analysis alone. The extracted data were verified by two reviewers (HV and SV), with discrepancies discussed until consensus was reached.
Quality assessment of the included studies
Evaluation using QUADAS-2 tool
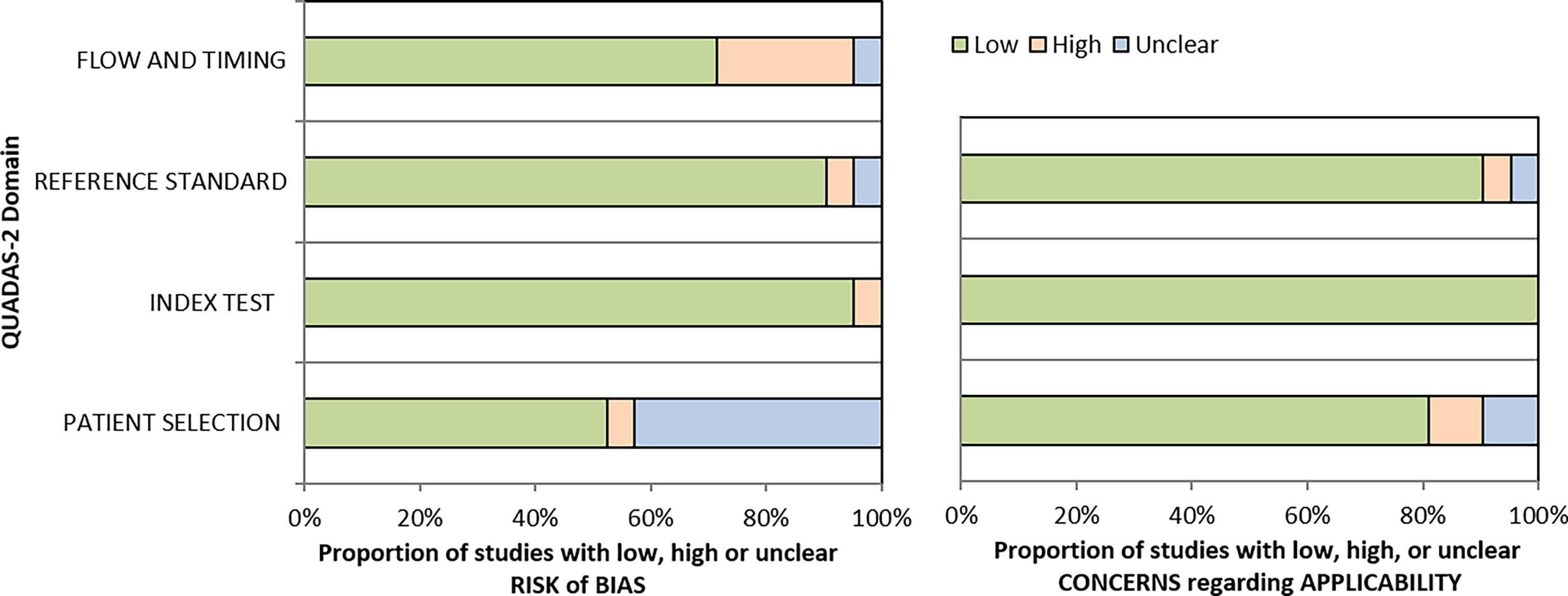
All the included studies were assessed for their methodological quality using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool. The QUADAS-2 tool encompasses four domains - patient selection, index test, reference standard, and flow and timing to assess the risk of bias and clinical applicability of the studies. Each domain consists of signalling questions for which the bias and applicability was judged as “low”, “high”, or “unclear”. If the study provided vital information including the method used for identifying the biomarkers and the proper classification of the study cohort (Active TB or LTBI and control group), then the quality of the study was assessed as good. The seven items in QUADAS-2 were independently answered by three authors (ED, BS, and HV), and discrepancies were resolved by SV.
Adherence to STARD 2015 criteria
The STARD (Standards for Reporting of Diagnostic Accuracy studies) guidelines were framed to improve the transparency, quality, and completeness in reporting diagnostic studies (34, 35). The 30-item checklist consisting of sub-criteria were examined by reviewers BS and SV in tandem and 5 items were removed from the analysis due to lack of relevance in evaluating miRNAs as diagnostic biomarkers. Each criterion in the checklist was answered with “yes”, “no”, or “not applicable” by the two reviewers independently, and then compiled through consensus to provide the final score for each study. The checklist is provided in the Supplementary Data section.
Data synthesis and statistical analysis
All computations were performed using R software (36). KT performed all the statistical analysis. Owing to limited number of studies, a random effects model based on inverse variance approach was applied for the analysis. Funnel plots were generated to visualize biased studies based on Egger’s test. Forest plots were constructed to demonstrate variability of point estimates between studies, their 95% confidence intervals (CI), as well as the weight of the sample sizes. Heterogeneity between studies was evaluated using I square with a cut-off point of ≥ 50% and a p value <0·10 and the degree of heterogeneity was defined by τ square test.
Results
Characteristics of the included studies
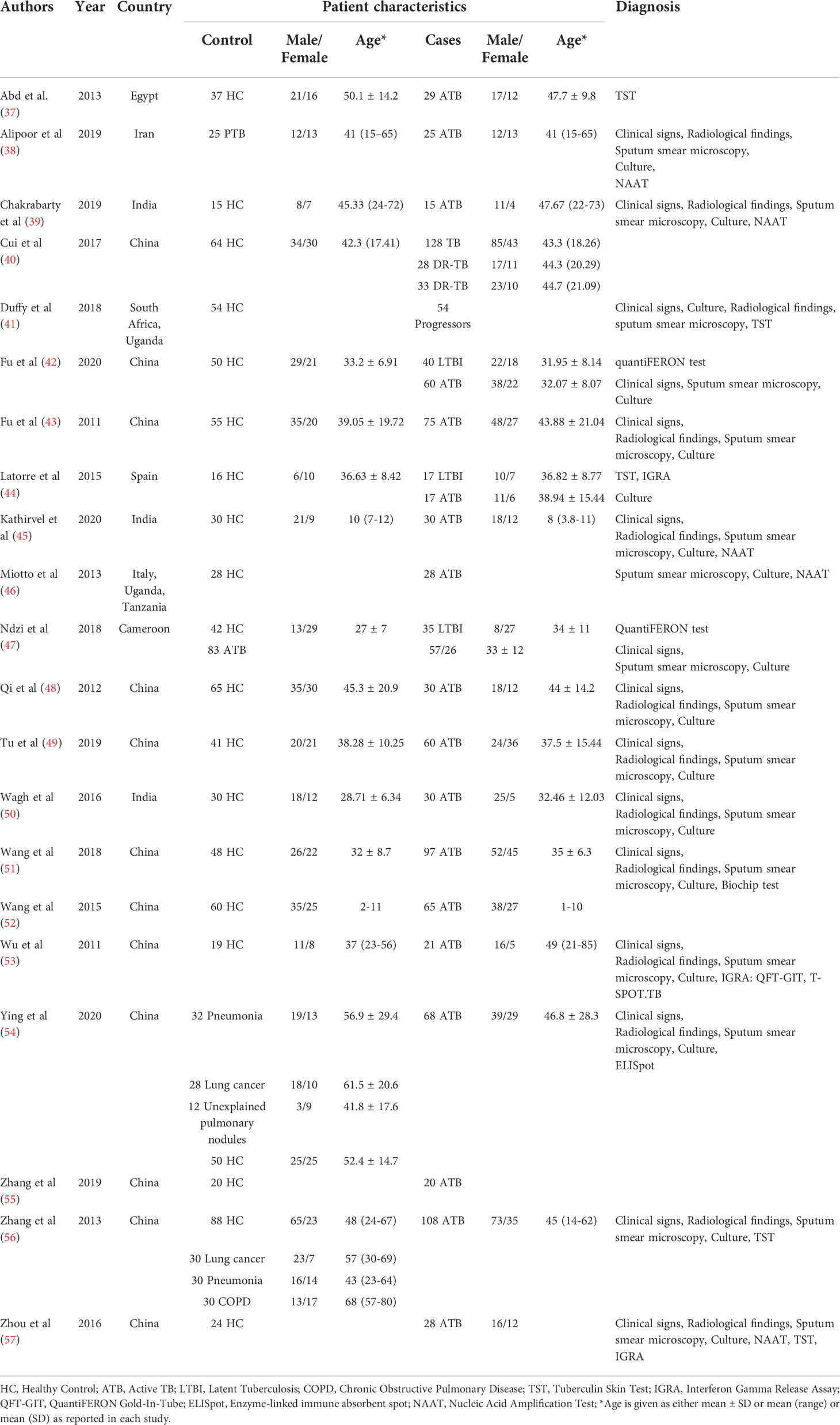
The decision‐making process for including studies in the review is outlined in a PRISMA flowchart (Figure 3) and the study characteristics are tabulated in Table 1. 21 studies were included for qualitative analysis and five studies were excluded due to lack of sensitivity and specificity data. Included articles were published between 2011 and 2020 describing study samples collected between 2011 and 2018. 85·7% of studies were carried out in high TB incident countries (57·1% China; 14·3% Africa; 14·3% India) and 14·3% studies were from countries with low to moderate TB incidence (1 from Spain, 1 from Italy and 1 from Iran). Two studies were multi-centric. Majority of the studies focused on biomarkers for diagnosis of active TB and differentiation from healthy controls and/or latently infected individuals. Two studies specified miRNA biomarkers for LTBI diagnosis. One study provided a miRNA signature for predicting progression from LTBI to active TB.
Characteristics of the study samples
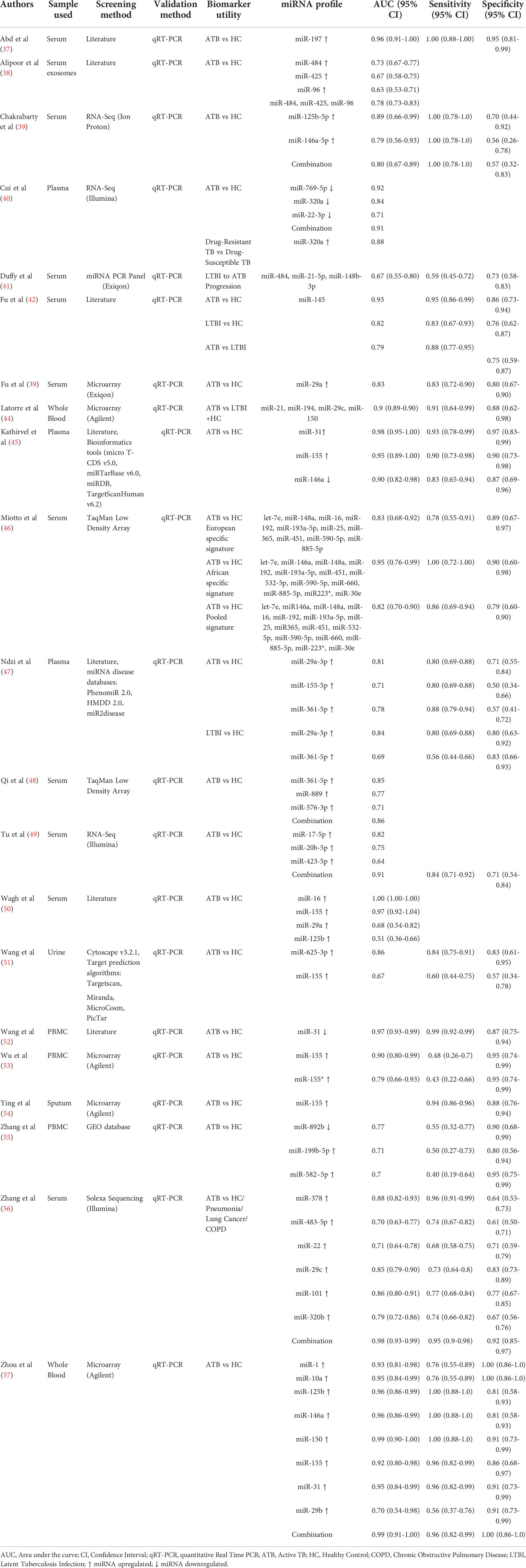
The sample size ranged from 15 to 189 in the ATB cohort, 54 to 118 in the LTBI cohort and 15 to 178 in the healthy control cohort. Three studies comprised paediatric TB patients exclusively, 1 study had a combination of adult and paediatric populations, while the rest were adult patient studies. Ten studies used serum, three used plasma, three used PBMCs, two used whole blood, one used sputum, one used urine, and one used exosomes. miRNA screening was based on either literature or RNA sequencing or microarray, or miRNA databases. Validation of the significant miRNAs was carried out by quantitative real-time polymerase chain reaction (qRT-PCR) in all studies. We identified a total of 31 individual miRNAs and 12 miRNA signatures from 21 eligible studies (Table 2).
Qualitative analysis of the studies
QUADAS-2 and STARD Assessment
The patient flow and selection led to considerable bias, but the overall risk of bias of the included studies assessed by QUADAS was low (<50%) Figure 4. The applicability concerns were very low in all four domains (All 21 included studies exhibited adherence to the STARD guidelines with a moderate score (Supplementary Data).
Quantitative analysis of the studies
The first statistical approach in meta-analysis is to assess the consistency of the reported miRNAs for diagnosis of active TB and LTBI by analysing the heterogeneity. A funnel graph was plotted to assess the likelihood of study bias. An asymmetric distribution of data points was observed with p>0·05 indicating the existence of potential publication bias (Supplementary Figure 1). Forest plots also indicated high heterogeneity (I2>50%). This bias is mainly attributed to inconsistency in the reported miRNAs and small sample size, therefore we selected miRNAs that were reported in at least two studies to ensure reliability of the miRNA as a candidate biomarker. Hence, we included only eight studies for the meta-analysis.
Accuracy of miRNAs as biomarkers of TB diagnosis
Forest plots were constructed for each miRNA group to assess the heterogeneity of the studies and determine the overall pooled values of sensitivity, specificity, and diagnostic odds ratio (DOR). Forest plots for miR-29 family (29a, 29b & 29c) revealed a sensitivity of 73·9% (95% CI 64-81·8), specificity of 80% (95% CI 72·6-85·7), and DOR of 12·3 (95% CI 7·8-19·3). The heterogeneity was less than 50% except for the sensitivity (I2 = 55%). miR-31 exhibited a sensitivity of 96% (95% CI 89·7-98·5), specificity of 89% (95% CI 81·2-93·8), and DOR of 345·9 (95% CI 90·2-1326·3). A sensitivity of 97·6% (95% CI 84·5-99·7), specificity of 77·6% (95% CI 61·1-88·4), and DOR of 126 (95% CI 15·1-1052·7) was shown by miR-125b. Both mir-31 and miR-125b did not show any heterogeneity (I2 = 0%) in the sub-group analysis. miR-146a had a sensitivity of 92·5% (95% CI 72·1-98·3), specificity of 75·8% (95% CI 52·3-89·9), and DOR of 43·7 (95% CI 13·4-142·2) with a heterogeneity (I2) of 66% for specificity. The sensitivity, specificity, and DOR of miR-155 was 89·8% (95% CI 78·5-98·5), 80·9% (95% CI 56·4-93·3), and 43·2 (95% CI 5·2-358·7) respectively with high heterogeneity. The subsequent step was to combine the effect size of each study. The overall pooled sensitivity and specificity of the above miRNAs were 87·9% (95% CI 81·7-92·2) (Figure 5) and 81·2% (95% CI 74·5-86·5) (Figure 6), while the DOR was 43·1 (95%CI 20·3-91·3) (Figure 7).
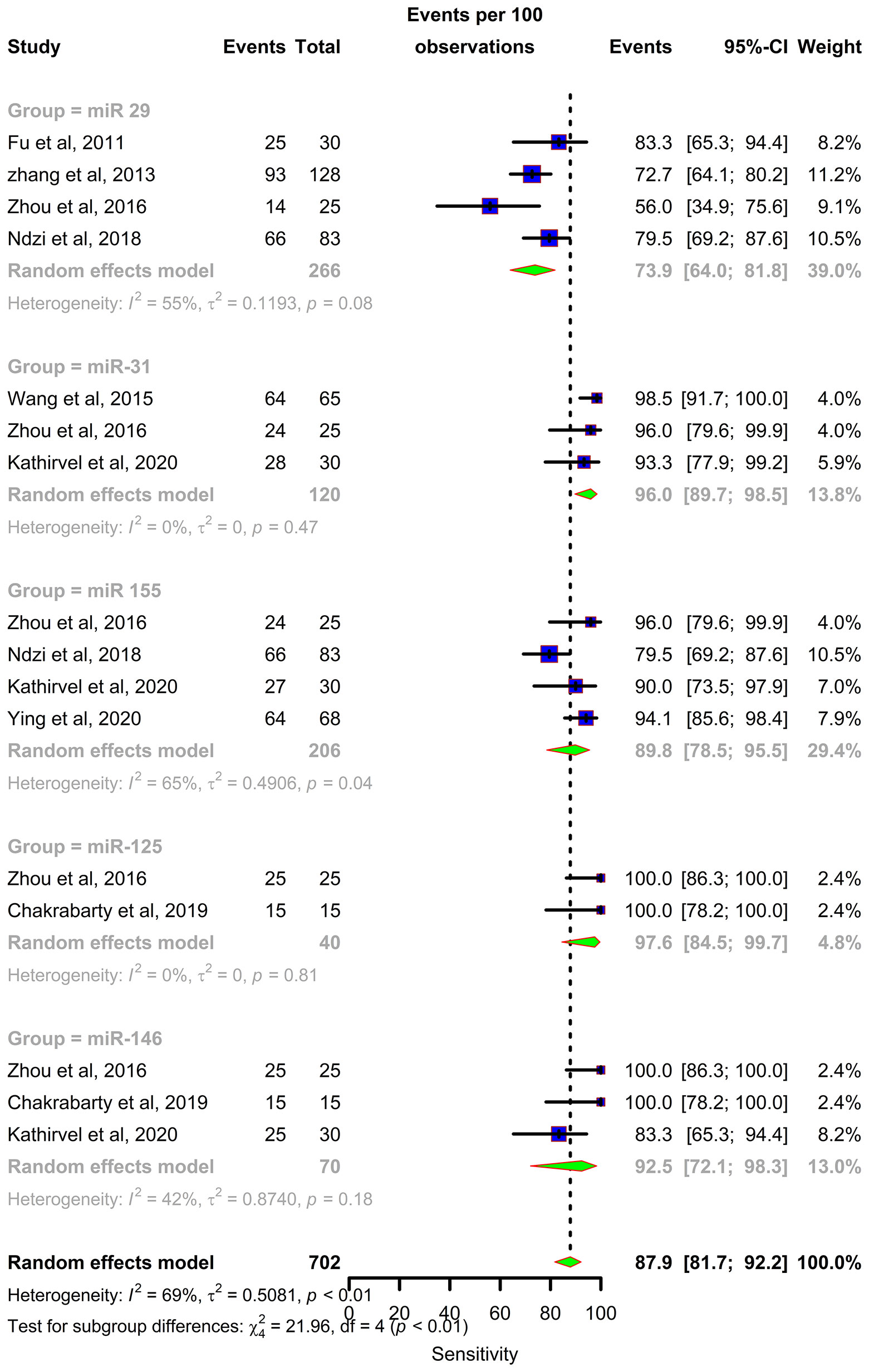
Figure 5 Sensitivity forest plot: Forest plot depicting sensitivity of miR 29 family (miR-29a, 29b, 29c), miR 31, miR 125b, miR 146a and miR 155.
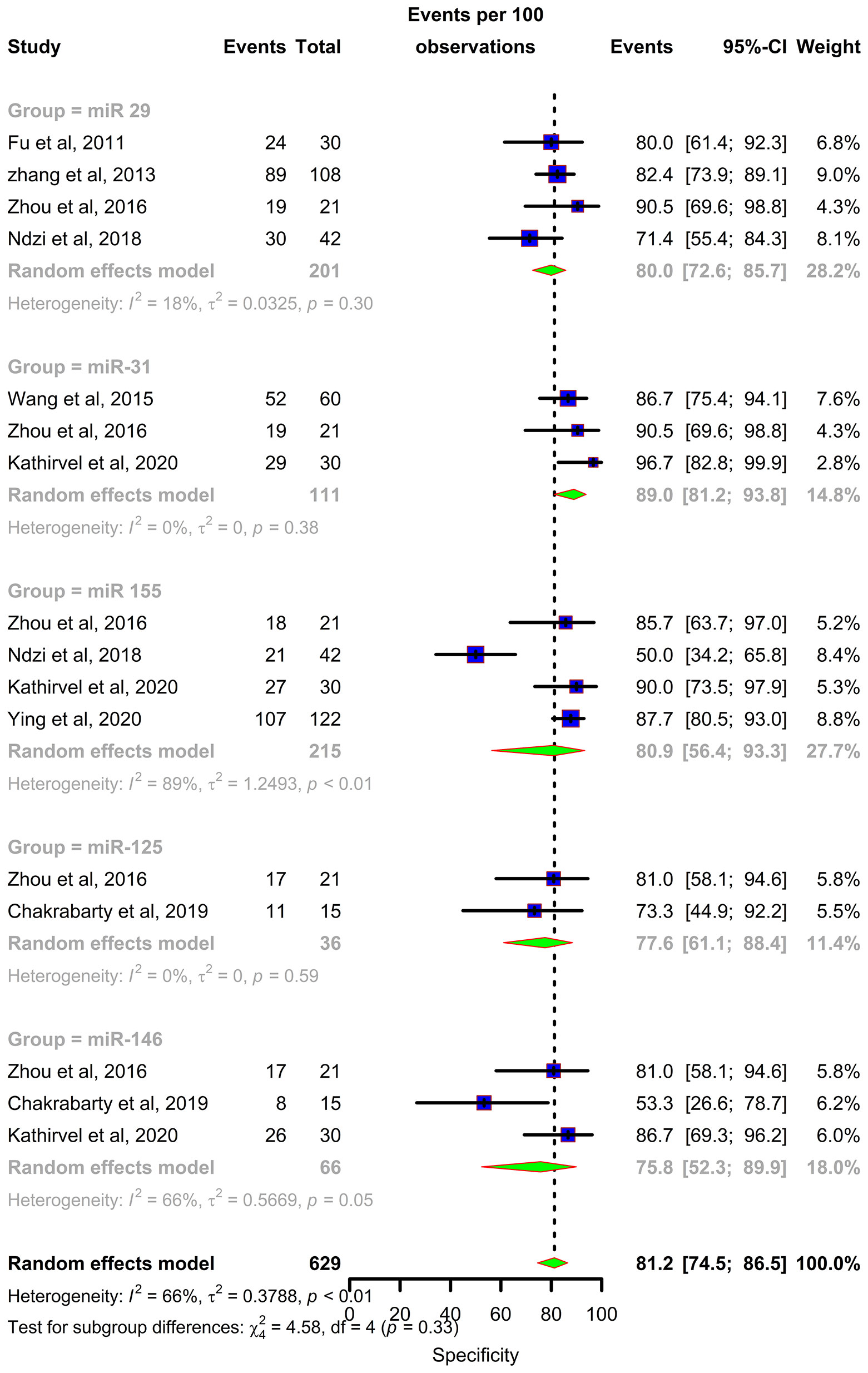
Figure 6 Specificity forest plot: Forest plot depicting specificity of miR 29 family (miR-29a, 29b, 29c), miR 31, miR 125b, miR 146a and miR 155.
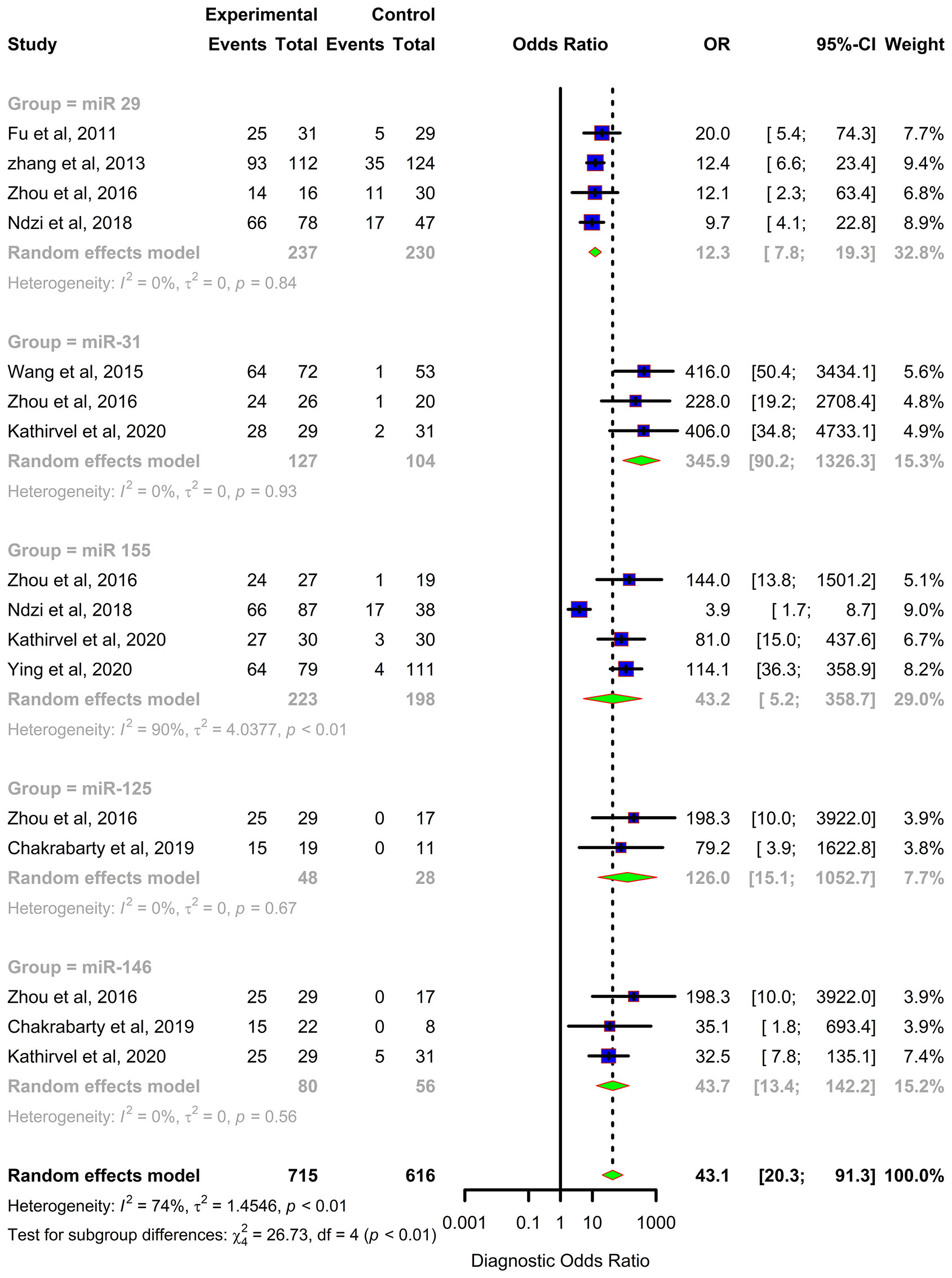
Figure 7 Diagnostic Odds Ratio (DOR) forest plot: Forest plot depicting Diagnostic Odds Ratio (DOR) of miR 29 family (miR-29a, 29b, 29c), miR 31, miR 125b, miR 146a and miR 155.
Discussion
A key component of the interim milestone for 2035 set by WHO for the End TB strategy is early diagnosis of the disease through systematic screening of high-risk groups. This calls for the identification and validation of definite biomarkers that discriminate active and latent TB and also predict progression to disease. Circulating miRNAs are potent biomarkers for TB diagnosis. miRNAs have also been explored as biomarkers for distinguishing between latent and active tuberculosis (47, 58, 59), as well as TB with HIV coinfection (46, 60). Emerging studies also demonstrate their promising role in predicting progression (41). miRNAs elicit an integral role in framing the innate and adaptive immunity during Mtb infection, by influencing gene expression in macrophages, dendritic cell, B and T cells. Multiple in vitro and in vivo studies have demonstrated the role of miRNAs in host immune regulatory mechanisms during TB pathogenesis, including apoptotic pathway trigger, autophagy induction, IFN γ stimulation and TNF α secretion. Mtb employs various strategies to combat these regulatory mechanisms and builds up its survival.
In this systematic review and meta-analysis, we evaluated the diagnostic accuracy of miRNAs as biomarkers for Tuberculosis. The overall analysis demonstrates moderate diagnostic accuracy with a DOR of 43·1. We conducted meta-analysis for miR 29 family (miR-29a, 29b, 29c), miR 31, miR 125b, miR 146a, and miR 155. Among these, miR 31 is specific for diagnosis of paediatric TB with a sensitivity of 96% (95% CI 89·7-98·5) and specificity of 89% (95% CI 81·2-91·2). Elevated levels of miR-31 suppresses MyD88, a TLR-2 adaptor protein and PP2A, thus negatively regulating the host’s innate immunity against Mtb (61). miR-31 also inhibits production of pro-inflammatory cytokines like IL-6, TNF-α, and IFN-γ which facilitate the induction of MTOR-responsive WNT and SHH pathways, resulting in inhibition of IFN γ-mediated autophagy (62). On the contrary, Wang et.al. reported decreased levels of miR-31 expression and significantly higher levels of serum IL-6, TNF-α, NF-κB, and IFN-γ in children with TB (52). However, in vitro studies support the autophagy inhibition role through the upregulation of miR-31. miR-29 was found to be downregulated in IFN-γ producing cells like NK cells and CD4+ and CD8+ T cells after Mycobacterium bovis BCG infection in mice (63). In latent and active TB, miR-29 expression was significantly increased, leading to reduced IFN-γ levels and subsequent signal pathway modifications (64). miR-125b contributes to a suppressed inflammatory response by directly interacting with TRAF6, and subsequently mediates Mtb survival. Additionally, RAF1 inhibition by miR-125b leads to reduced levels of TNF-α, IL-6, etc., indicating miR- mediated immune escape of Mtb (65). miR-146 has a sensitivity of 92·5% (95% CI 72·1-98·3) but a constrained specificity of 75·8% (95% CI 52·3-89·9). Increased levels of miR-146a targets TRAF6 which modulates the suppression of NF-κB and MAPKs pathways thereby hindering iNOS expression and NO synthesis leading to mycobacterial survival (66). On the other hand, it can also lead to TNF-α attenuation and downregulation of potential targets like IRAK1, TRAF6, and PTGS2 (67). Thus, miR-146a has both positive and negative effects on Mtb survival. miR-155 harnesses macrophage and T-cell survival by SHIP1 mediated regulation. This coupled role provides a pro-bacterial environment during the early phase of infection. Increased survival of Mtb-specific T-cells contributes to increased protection against the bacteria (68). Mycobacterium bovis BCG infection is known to upregulate miR-155 expression through TLR2 and other signalling pathways. miR-155 dependent increase in ROS occurs due to the suppression of SHIP1 in BCG-infected macrophages (69). An exhaustive network profile of various miRNAs involved in the pathogenesis and survival of Mtb is depicted in Figure 8.
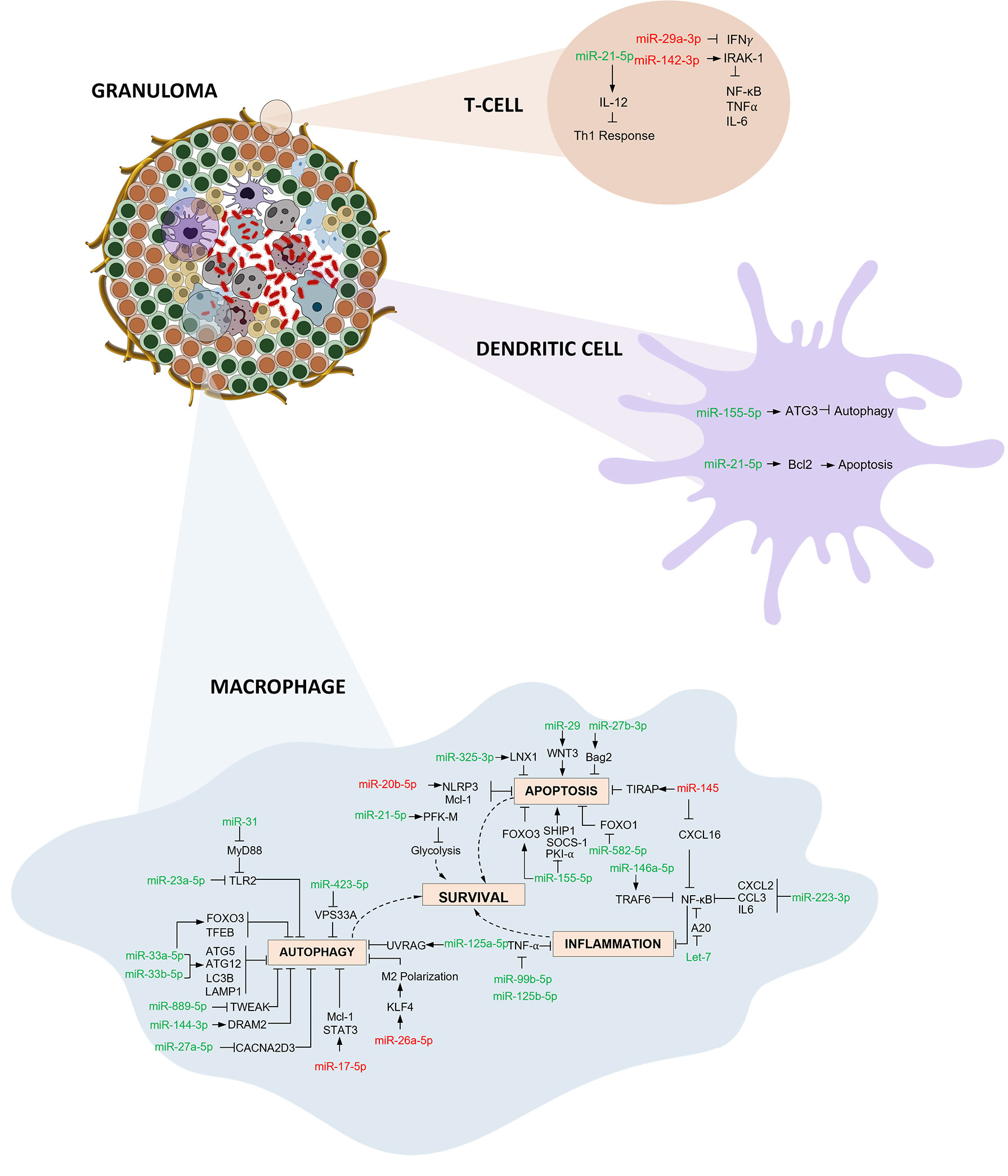
Figure 8 Immunoregulatory mechanisms involving miRNA in macrophages, dendritic cells and T cells: Three primary mechanisms of immune regulation are documented: (i) Autophagy evasion: Autophagy can degrade intracellular Mtb by autolysosomes. Mtb regulates the expression of various miRNAs thereby inhibiting autophagy activation and aiding its survival inside macrophages. (ii) Apoptosis impairment: Host macrophages combat infection by eliminating niche cells for Mtb growth and by packaging tubercle bacilli in apoptotic bodies. Mtb employs multiple strategies to circumvent this programmed cell death. (iii) Inflammation obstruction: Mtb infection initiates the activation of various components of the immune system resulting in recruitment of inflammatory cells. Mtb interferes with various downstream signalling events and inhibits the inflammatory responses thereby thriving within the hostile environment. (miRNAs shown in red are downregulated and green are upregulated).
Despite these promising results, we did observe significant heterogeneity because of discrepancies among the different studies. Various factors starting from the type of sample, its storage and processing, miRNA profiling and validation methods, to data normalization could introduce potential bias and lead to inconsistency in results. Since the number of studies available for analysis was very less, we could not carry out any other subgroup analysis based on sample source or profiling methods. Some studies reported the -3p or -5p variant while some did not, hence we merged the variants. This could have decreased the accuracy of our analysis. Another major limitation is the small sample size in each study. For any biomarker discovery study, the size of the cohort is very crucial and should be representative of the target population. Though reported miRNA signatures had better accuracy than the individual miRNAs, we could not include them for our analysis since no two signatures were the same. Regardless of these limitations, ours is the first meta-analysis that comprehensively aggregated available data on miRNAs and analysed their utility in the field of tuberculosis.
Scientific advancements have paved the way for development of various novel miRNA detection platforms including Bead-Array based profiling, microfluidics, miRNA activity reporter assays and amplification assays [(loop-mediated isothermal amplification (LAMP), exponential isothermal amplification assay (EXPAR), rolling circle amplification (RCA) and strand displacement amplification (SDA)] (70) that can be exploited for the development of sensitive diagnostic tests for TB using the most promising candidate miRNAs. Using miRNAs as diagnostic reagents would help in the development of rapid and cost- effective tests for various infectious diseases, circumventing the disadvantages of conventional diagnostic assays. Yet, detecting weakly expressed miRNAs can be challenging, and this can be overcome by using highly sensitive detection platforms such as small RNA-seq method (71). In this line, higher background signals in fluorescent detection platforms can also pose a challenge and reduce the sensitivity of the test. Nanotechnology-based approaches such as the use of electrochemical biosensors are currently being explored to overcome these challenges (72).
The following observations were made during this comprehensive systematic review and meta-analysis: (i) A substantial between-platform discordance is inevitable since the miRNA spectrum between various sample sources are different, (ii) miRNA signatures have better diagnostic performance than single miRNAs, (iii) miRNA expression profile is age-specific; miR 31 specifically finds application in diagnosis of Paediatric TB (iv) Distinct miRNA profiles for different stages of TB is emerging. This can lead to the identification of a promising biomarker panel to predict progression from Latent Tuberculosis infection to Active TB disease.
Conclusion
Early diagnosis of tuberculosis is crucial for achieving the WHO global End TB milestone. miRNAs are known to have potential diagnostic abilities for various diseases with demonstrated accuracy. Here we present a comprehensive overview of the immunological role of miRNAs in tuberculosis by systematically reviewing the available literature. We also performed a meta-analysis of five miRNAs and demonstrate their promising role in TB diagnosis. The overall diagnostic odds ratio (DOR) of the five miRNAs was 43·1. miR-31 individually exhibited the maximum DOR of 345·9 portraying very high diagnostic accuracy.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ED, BS, SV and HV collected and interpreted data. KT performed statistical analysis. LH designed the study. All authors contributed to the article and approved the submitted version.
Funding
ED thanks Department of Science and Technology (DST), India for providing INSPIRE fellowship.
Acknowledgments
The authors thank Mr. Bennett Henzeler, National Institute for Research in Tuberculosis for his help in designing the figures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.954396/full#supplementary-material
References
1. Cohen A, Mathiasen VD, Schön T, Wejse C. The global prevalence of latent tuberculosis: A systematic review and meta-analysis. Eur Respir J (2019) 54:1–14. doi: 10.1183/13993003.00655-2019
2. Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, et al. The growing burden of tuberculosis: Global trends and interactions with the HIV epidemic. Arch Intern Med (2003) 163:1009–21. doi: 10.1001/archinte.163.9.1009
3. McNerney R, Maeurer M, Abubakar I, Marais B, Mchugh TD, Ford N, et al. Tuberculosis diagnostics and biomarkers: Needs, challenges, recent advances, and opportunities. J Infect Dis (2012) 205:S147–58. doi: 10.1093/infdis/jir860
4. MacLean E, Broger T, Yerlikaya S, Fernandez-Carballo BL, Pai M, Denkinger CM. A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol (2019) 4:748–58. doi: 10.1038/s41564-019-0380-2
5. Mulenga H, Zauchenberger C-Z, Bunyasi EW, Mbandi SK, Mendelsohn SC, Kagina B, et al. Performance of diagnostic and predictive host blood transcriptomic signatures for tuberculosis disease: A systematic review and meta-analysis. PloS One (2020) 15:1–17. doi: 10.1371/journal.pone.0237574
6. Warsinske H, Vashisht R, Khatri P. Host-response-based gene signatures for tuberculosis diagnosis: A systematic comparison of 16 signatures. PloS Med (2019) 16:1–19. doi: 10.1371/journal.pmed.1002786
7. Gupta RK, Turner CT, Venturini C, Esmail H, Rangaka MX, Copas A, et al. Concise whole blood transcriptional signatures for incipient tuberculosis: a systematic review and patient-level pooled meta-analysis. Lancet Respir Med (2020) 8:395–406. doi: 10.1016/S2213-2600(19)30282-6
8. Diel R, Loddenkemper R, Nienhaus A. Predictive value of interferon-γ release assays and tuberculin skin testing for progression from latent TB infection to disease state: a meta-analysis. Chest (2012) 142:63–75. doi: 10.1378/chest.11-3157
9. Pai M, Denkinger CM, Kik SV, Rangaka MX, Zwerling A, Oxlade O, et al. Gamma interferon release assays for detection of mycobacterium tuberculosis infection. Clin Microbiol Rev (2014) 27:3–20. doi: 10.1128/CMR.00034-13
10. Metcalfe JZ, Everett CK, Steingart KR, Cattamanchi A, Huang L, Hopewell PC, et al. Interferon-γ release assays for active pulmonary tuberculosis diagnosis in adults in low- and middle-income countries: systematic review and meta-analysis. J Infect Dis (2011) 204 Suppl 4:S1120–1129. doi: 10.1093/infdis/jir410
11. Qiu X, Tang Y, Yue Y, Zeng Y, Li W, Qu Y, et al. Accuracy of interferon-γ-induced protein 10 for diagnosing latent tuberculosis infection: a systematic review and meta-analysis. Clin Microbiol Infect (2019) 25:667–72. doi: 10.1016/j.cmi.2018.12.006
12. Santos VS, Goletti D, Kontogianni K, Adams ER, Molina-Moya B, Dominguez J, et al. Acute phase proteins and IP-10 as triage tests for the diagnosis of tuberculosis: systematic review and meta-analysis. Clin Microbiol Infect (2019) 25:169–77. doi: 10.1016/j.cmi.2018.07.017
13. Gupta-Wright A, Peters JA, Flach C, Lawn SD. Detection of lipoarabinomannan (LAM) in urine is an independent predictor of mortality risk in patients receiving treatment for HIV-associated tuberculosis in sub-Saharan Africa: a systematic review and meta-analysis. BMC Med (2016) 14:53. doi: 10.1186/s12916-016-0603-9
14. Minion J, Leung E, Talbot E, Dheda K, Pai M, Menzies D. Diagnosing tuberculosis with urine lipoarabinomannan: systematic review and meta-analysis. Eur Respir J (2011) 38:1398–405. doi: 10.1183/09031936.00025711
15. Mamishi S, Pourakbari B, Teymuri M, Rubbo P-A, Tuaillon E, Keshtkar AA, et al. Diagnostic accuracy of IL-2 for the diagnosis of latent tuberculosis: a systematic review and meta-analysis. Eur J Clin Microbiol Infect Dis (2014) 33:2111–9. doi: 10.1007/s10096-014-2190-z
16. Sudbury EL, Clifford V, Messina NL, Song R, Curtis N. Mycobacterium tuberculosis-specific cytokine biomarkers to differentiate active TB and LTBI: A systematic review. J Infect (2020) 81:873–81. doi: 10.1016/j.jinf.2020.09.032
17. Qiu B, Liu Q, Li Z, Song H, Xu D, Ji Y, et al. Evaluation of cytokines as a biomarker to distinguish active tuberculosis from latent tuberculosis infection: a diagnostic meta-analysis. BMJ Open (2020) 10:e039501. doi: 10.1136/bmjopen-2020-039501
18. Meier NR, Jacobsen M, Ottenhoff THM, Ritz N. A systematic review on novel mycobacterium tuberculosis antigens and their discriminatory potential for the diagnosis of latent and active tuberculosis. Front Immunol (2018) 9:2476. doi: 10.3389/fimmu.2018.02476
19. Yong YK, Tan HY, Saeidi A, Wong WF, Vignesh R, Velu V, et al. Immune biomarkers for diagnosis and treatment monitoring of tuberculosis: Current developments and future prospects. Front Microbiol (2019) 10:2789. doi: 10.3389/fmicb.2019.02789
20. Goletti D, Petruccioli E, Joosten SA, Ottenhoff THM. Tuberculosis biomarkers: From diagnosis to protection. Infect Dis Rep (2016) 8:24–32. doi: 10.4081/idr.2016.6568
21. Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta (BBA) - Mol Cell Res (2010) 1803:1231–43. doi: 10.1016/j.bbamcr.2010.06.013
22. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA–target recognition. PloS Biol (2005) 3:0404–18. doi: 10.1371/journal.pbio.0030085
23. Ardekani AM, Naeini MM. The role of MicroRNAs in human diseases. Avicenna J Med Biotechnol (2010) 2:161–79.
24. Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol (2007) 17:118–26. doi: 10.1016/j.tcb.2006.12.007
25. Plotnikova O, Baranova A, Skoblov M. Comprehensive analysis of human microRNA–mRNA interactome. Front Genet (2019) 10:933. doi: 10.3389/fgene.2019.00933
26. Sohel MH. Extracellular/Circulating MicroRNAs: Release mechanisms, functions and challenges. Achiev Life Sci (2016) 10:175–86. doi: 10.1016/j.als.2016.11.007
27. Alipoor SD, Adcock IM, Tabarsi P, Folkerts G, Mortaz E. MiRNAs in tuberculosis: Their decisive role in the fate of TB. Eur J Pharmacol (2020) 886:173529. doi: 10.1016/j.ejphar.2020.173529
28. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (2018) 9:402. doi: 10.3389/fendo.2018.00402
29. Iwakawa H, Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol (2015) 25:651–65. doi: 10.1016/j.tcb.2015.07.011
30. Correia CN, Nalpas NC, McLoughlin KE, Browne JA, Gordon SV, MacHugh DE, et al. Circulating microRNAs as potential biomarkers of infectious disease. Front Immunol (2017) 8:118. doi: 10.3389/fimmu.2017.00118
31. Behrouzi A, Alimohammadi M, Nafari AH, Yousefi MH, Riazi Rad F, Vaziri F, et al. The role of host miRNAs on mycobacterium tuberculosis. ExRNA (2019) 1:40. doi: 10.1186/s41544-019-0040-y
32. Moher D, Liberati A, Tetzlaff J, Altman DG, Group TP. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PloS Med (2009) 6:e1000097. doi: 10.1371/journal.pmed.1000097
33. Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan–a web and mobile app for systematic reviews. Syst Rev (2016) 5:1–10. doi: 10.1186/s13643-016-0384-4
34. Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ (2015) 351:h5527. doi: 10.1136/bmj.h5527
35. Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open (2016) 6:e012799. doi: 10.1136/bmjopen-2016-012799
36. Core Team. R. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing (2021). Available at: https://www.R-project.org/.
37. Abd-El-Fattah AA, Sadik NAH, Shaker OG, Aboulftouh ML. Differential MicroRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem Biophys (2013) 67:875–84. doi: 10.1007/s12013-013-9575-y
38. Alipoor SD, Tabarsi P, Varahram M, Movassaghi M, Dizaji MK, Folkerts G, et al. Serum exosomal miRNAs are associated with active pulmonary tuberculosis. Dis Markers (2019) 2019:1907426. doi: 10.1155/2019/1907426
39. Chakrabarty S, Kumar A, Raviprasad K, Mallya S, Satyamoorthy K, Chawla K. Host and MTB genome encoded miRNA markers for diagnosis of tuberculosis. Tuberculosis (2019) 116:37–43. doi: 10.1016/j.tube.2019.04.002
40. Cui J-Y, Liang H-W, Pan X-L, Li D, Jiao N, Liu Y-H, et al. Characterization of a novel panel of plasma microRNAs that discriminates between mycobacterium tuberculosis infection and healthy individuals. PloS One (2017) 12:e0184113. doi: 10.1371/journal.pone.0184113
41. Duffy FJ, Thompson E, Downing K, Suliman S, Mayanja-Kizza H, Henry Boom W, et al. A serum circulating miRNA signature for short-term risk of progression to active tuberculosis among household contacts. Front Immunol (2018) 9:661. doi: 10.3389/fimmu.2018.00661
42. Fu Y, Yang X, Chen H, Lu Y, Fu Y, Yang X, et al. Diagnostic value of miR-145 and its regulatory role in macrophage immune response in tuberculosis. Genet Mol Biol (2020) 43:1–9. doi: 10.1590/1678-4685-gmb-2019-0238
43. Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol (2011) 49:4246–51. doi: 10.1128/JCM.05459-11
44. Latorre I, Leidinger P, Backes C, Domínguez J, de Souza-Galvão ML, Maldonado J, et al. A novel whole-blood miRNA signature for a rapid diagnosis of pulmonary tuberculosis. Eur Respir J (2015) 45:1173–6. doi: 10.1183/09031936.00221514
45. Kathirvel M, Saranya S, Mahadevan S. Expression levels of candidate circulating microRNAs in pediatric tuberculosis. Pathog Glob Health (2020) 114:262–70. doi: 10.1080/20477724.2020.1761140
46. Miotto P, Mwangoka G, Valente IC, Norbis L, Sotgiu G, Bosu R, et al. miRNA signatures in sera of patients with active pulmonary tuberculosis. PloS One (2013) 8:e80149. doi: 10.1371/journal.pone.0080149
47. Ndzi EN, Nkenfou CN, Mekue LM, Zentilin L, Tamgue O, Pefura EWY, et al. MicroRNA hsa-miR-29a-3p is a plasma biomarker for the differential diagnosis and monitoring of tuberculosis. Tuberc (Edinb) (2019) 114:69–76. doi: 10.1016/j.tube.2018.12.001
48. Qi Y, Cui L, Ge Y, Shi Z, Zhao K, Guo X, et al. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect Dis (2012) 12:384. doi: 10.1186/1471-2334-12-384
49. Tu H, Yang S, Jiang T, Wei L, Shi L, Liu C, et al. Elevated pulmonary tuberculosis biomarker miR-423-5p plays critical role in the occurrence of active TB by inhibiting autophagosome-lysosome fusion. Emerg Microbes Infect (2019) 8:448–60. doi: 10.1080/22221751.2019.1590129
50. Wagh V, Urhekar A, Modi D. Levels of microRNA miR-16 and miR-155 are altered in serum of patients with tuberculosis and associate with responses to therapy. Tuberc (Edinb) (2017) 102:24–30. doi: 10.1016/j.tube.2016.10.007
51. Wang J, Zhu X, Xiong X, Ge P, Liu H, Ren N, et al. Identification of potential urine proteins and microRNA biomarkers for the diagnosis of pulmonary tuberculosis patients. Emerg Microbes Infect (2018) 7:63. doi: 10.1038/s41426-018-0066-5
52. Wang JX, Xu J, Han YF, Zhu YB, Zhang WJ. Diagnostic values of microRNA-31 in peripheral blood mononuclear cells for pediatric pulmonary tuberculosis in Chinese patients. Genet Mol Res (2015) 14:17235–43. doi: 10.4238/2015.December.16.23
53. Wu J, Lu C, Diao N, Zhang S, Wang S, Wang F, et al. Analysis of microRNA expression profiling identifies miR-155 and miR-155* as potential diagnostic markers for active tuberculosis: a preliminary study. Hum Immunol (2012) 73:31–7. doi: 10.1016/j.humimm.2011.10.003
54. Ying H, FengYing S, YanHong W, YouMing H, FaYou Z, HongXiang Z, et al. MicroRNA-155 from sputum as noninvasive biomarker for diagnosis of active pulmonary tuberculosis. Iran J Basic Med Sci (2020) 23:1419–25. doi: 10.22038/ijbms.2020.44029.10324
55. Zhang Y, Zhang X, Zhao Z, Zheng Y, Xiao Z, Li F. Integrated bioinformatics analysis and validation revealed potential immune-regulatory miR-892b, miR-199b-5p and miR-582-5p as diagnostic biomarkers in active tuberculosis. Microb Pathog (2019) 134:103563. doi: 10.1016/j.micpath.2019.103563
56. Zhang X, Guo J, Fan S, Li Y, Wei L, Yang X, et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PloS One (2013) 8:1–11. doi: 10.1371/journal.pone.0081076
57. Zhou M, Yu G, Yang X, Zhu C, Zhang Z, Zhan X. Circulating microRNAs as biomarkers for the early diagnosis of childhood tuberculosis infection. Mol Med Rep (2016) 13:4620–6. doi: 10.3892/mmr.2016.5097
58. Lyu L, Zhang X, Li C, Yang T, Wang J, Pan L, et al. Small RNA profiles of serum exosomes derived from individuals with latent and active tuberculosis. Front Microbiol (2019) 10:1174. doi: 10.3389/fmicb.2019.01174
59. Zhang H, Sun Z, Wei W, Liu Z, Fleming J, Zhang S, et al. Identification of serum microRNA biomarkers for tuberculosis using RNA-seq. PloS One (2014) 9:1–7. doi: 10.1371/journal.pone.0088909
60. Olsson O, Tesfaye F, Søkilde R, Mazurek J, Abebe M, Yeba H, et al. Expression of MicroRNAs is dysregulated by HIV while mycobacterium tuberculosis drives alterations of small nucleolar RNAs in HIV positive adults with active tuberculosis. Front Microbiol (2022) 12:808250. doi: 10.3389/fmicb.2021.808250
61. Ghorpade DS, Holla S, Kaveri SV, Bayry J, Patil SA, Balaji KN. Sonic hedgehog-dependent induction of microRNA 31 and microRNA 150 regulates mycobacterium bovis BCG-driven toll-like receptor 2 signaling. Mol Cell Biol (2013) 33:543–56. doi: 10.1128/MCB.01108-12
62. Holla S, Kurowska-Stolarska M, Bayry J, Balaji KN. Selective inhibition of IFNG-induced autophagy by Mir155- and Mir31-responsive WNT5A and SHH signaling. Autophagy (2014) 10:311–30. doi: 10.4161/auto.27225
63. Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat Immunol (2011) 12:861–9. doi: 10.1038/ni.2073
64. Yi Z, Fu Y, Li J, Zhang B. [Expression and bioinformatic analysis of miR-29 family, target gene IFN-γ in CD4(+) T cells from subjects with latent tuberculosis infection]. Zhonghua Yu Fang Yi Xue Za Zhi (2013) 47:632–6.
65. Niu W, Sun B, Li M, Cui J, Huang J, Zhang L. TLR-4/microRNA-125a/NF-κB signaling modulates the immune response to mycobacterium tuberculosis infection. Cell Cycle (2018) 17:1931–45. doi: 10.1080/15384101.2018.1509636
66. Li M, Wang J, Fang Y, Gong S, Li M, Wu M, et al. microRNA-146a promotes mycobacterial survival in macrophages through suppressing nitric oxide production. Sci Rep (2016) 6:23351. doi: 10.1038/srep23351
67. Liu Z, Zhou G, Deng X, Yu Q, Hu Y, Sun H, et al. Analysis of miRNA expression profiling in human macrophages responding to mycobacterium infection: Induction of the immune regulator miR-146a. J Infect (2014) 68:553–61. doi: 10.1016/j.jinf.2013.12.017
68. Rothchild AC, Sissons JR, Shafiani S, Plaisier C, Min D, Mai D, et al. MiR-155–regulated molecular network orchestrates cell fate in the innate and adaptive immune response to mycobacterium tuberculosis. PNAS (2016) 113:E6172–81. doi: 10.1073/pnas.1608255113
69. Wang J, Wu M, Wen J, Yang K, Li M, Zhan X, et al. MicroRNA-155 induction by mycobacterium bovis BCG enhances ROS production through targeting SHIP1. Mol Immunol (2014) 62:29–36. doi: 10.1016/j.molimm.2014.05.012
70. Siddika T, Heinemann IU. Bringing MicroRNAs to light: Methods for MicroRNA quantification and visualization in live cells(2021) (Accessed August 12, 2022).
71. Godoy PM, Barczak AJ, DeHoff P, Srinivasan S, Etheridge A, Galas D, et al. Comparison of reproducibility, accuracy, sensitivity, and specificity of miRNA quantification platforms. Cell Rep (2019) 29:4212–4222.e5. doi: 10.1016/j.celrep.2019.11.078
Keywords: tuberculosis, miRNA, diagnosis, biomarker, immune modulation, mycobacterium tuberculosis
Citation: Daniel EA, Sathiyamani B, Thiruvengadam K, Vivekanandan S, Vembuli H and Hanna LE (2022) MicroRNAs as diagnostic biomarkers for Tuberculosis: A systematic review and meta- analysis. Front. Immunol. 13:954396. doi: 10.3389/fimmu.2022.954396
Received: 27 May 2022; Accepted: 31 August 2022;
Published: 27 September 2022.
Edited by:
Pere-Joan Cardona, Hospital Universitari Germans Trias i Pujol, SpainReviewed by:
Yogesh Singh, University of Tübingen, GermanyShakila Harshavardhan, Madurai Kamaraj University, India
Copyright © 2022 Daniel, Sathiyamani, Thiruvengadam, Vivekanandan, Vembuli and Hanna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luke Elizabeth Hanna, aGFubmF0cmNAeWFob28uY29t
 Evangeline Ann Daniel
Evangeline Ann Daniel Balakumaran Sathiyamani
Balakumaran Sathiyamani Kannan Thiruvengadam
Kannan Thiruvengadam Sandhya Vivekanandan
Sandhya Vivekanandan Hemanathan Vembuli
Hemanathan Vembuli Luke Elizabeth Hanna
Luke Elizabeth Hanna