Abstract
As a prevalent epigenetic modification, the role of m6A has been increasingly highlighted in the alteration of numerous RNAs implicated with multiple biological processes, such as formation, export, translation, and degradation. With further the understanding of m6A, accumulating evidence shows that m6A modification similarly affects metabolic process of non-coding genes. But the specifical interplay of m6A and ncRNAs (non-coding RNAs) in gastrointestinal cancers still lacks complete discussion. Thus, we analyzed and summarized how ncRNAs affect the regulators of m6A and by what means the expression of ncRNAs is altered via m6A in gastrointestinal cancers. We focused on the effect of the interaction of m6A and ncRNAs on the molecular mechanisms of malignant behavior in gastrointestinal cancers, revealing more possibilities of ncRNAs for diagnosis and treatment in term of epigenetic modification.
1 Introduction
Gastrointestinal (GI) cancers are responsible for over one-quarter of cancer-related morbidities and over one-third of mortalities from cancers worldwide (1), seriously affecting human health and wellbeing. GI cancers refer to a group of malignant tumors arising from the digestive tract, which mainly include esophageal cancer, gastric cancer, liver cancer, colorectal cancer, pancreatic cancer, and others (2–6). Smoking, infection, diet, and obesity are common risk factors to initiate the pathogenesis of the majority of GI cancers (7). Although the relevant treatment methods have been improved, the prognosis of patents is still unfavorable. Due to the growing incidence of these cancers in each year, there is a clear necessity of searching for early diagnostic markers and potential therapeutic targets (8, 9).
Recent research on epigenetic modifications has made numerous contributions to the understanding of the pathogenesis and biological characteristics of human diseases (10–13). N6-methyladenosine (m6A), first discovered in 1974 (14, 15), was not only observed as widely distributed in various eukaryotes, such as mammals, plants or yeast (16–19), but also located in bacteria, e.g., mycoplasma (20, 21). Studies on m6A have mainly focused on the consensus sequence RRACH (R = G or A and H = A, C, or U) that is near 3′ untranslated regions (3′-UTRs), 5′-UTR, and internal long exon (22). Methyltransferase and demethylase are responsible for the dynamic alteration of m6A modifications, respectively called “writers” (8, 9, 23) and “erasers” (24, 25). The binding protein (“readers”) (26, 27) can play a role in the occurrence of the methylation process mediated by “writers” and recruit downstream molecules to affect functional signaling. The role of these regulators in GI cancers have gradually attracted increasing attention. Methyltransferase-like 3 drives GLUT1 (Glucose transporter type 1) expression to accelerate glucose utilization and lactate accumulation in colorectal cancer cells (7). Furthermore, AlkBhomolog5 attenuates the stability of LY6/PLAUR domain-containing protein 1 (LYPD1) to constrain the aggressive ability of hepatocellular carcinoma through the elimination of m6A site (28). In addition, Ubiquitin-specific protease 14 (USP14) translation was reported to be upregulated by YTH N6-Methyladenosine RNA Binding Protein 1 (YTHDF1), resulting in carcinogenesis and unfavorable outcome for gastric cancer patients (29). The m6A modification site prevalently appears in mRNA (message RNA) to regulate its metabolism (30), such as biosynthesis, nuclear export and degradation. Increasing evidence has demonstrated that m6A can decorate various ncRNAs (non-coding RNAs) (31, 32), including microRNAs (miRNAs) (33–35), long noncoding RNAs (lncRNAs) (36–38) and circular RNAs (circRNAs) (39–41).
As transcriptomics studies have constantly shown great improvement, the function of non-coding RNAs (ncRNAs) have been gradually noticed and prompted further explorations (42, 43). Emerging evidence proves that several ncRNAs play indispensable roles in the initiation and progression of multiple diseases (44–47), and thus they have a potential value for diagnosis and prognosis. For instance, circACTN4 functions as a sponge to eliminate miR-424-5p to upregulate and recruit Yes-associated protein 1 (YAP1), which exerts a positive effect on inducing Frizzled-7 (FZD7) and in turn facilitates the development of intrahepatic cholangiocarcinoma (48). The phenotypic transdifferentiation of carotid artery smooth muscle cells (SMCs) to macrophage-like cells was reported to be regulated by the activation effect of KLF4 derived from lncRNA MIAT, and then developing into advanced atherosclerotic lesion (49). Hsa-miR-3178 activates the PI3K/Akt cascade to attenuate the sensitivity of PC patients to gemcitabine (50). When m6A modification occurs in the main ncRNAs, their expression and function are altered by specific proteins. The upregulated lncRNA LBX2−AS1 serves as a promoter of tumor by enhancing the proliferation and migration of colorectal cancer cells (51). Methyltransferase-like 3, the known “writers” of m6A, catalyzes and increases the m6A level in lncRNA LBX2−AS1 to enhance its expression by stabilizing LBX2−AS1 in colorectal cancer. Besides, a series of RNAs without translating proteins also affect the m6A modification level of downstream targets by interacting with critical regulators of m6A. Ubiquitin-conjugating enzyme E2T (UBE2T) has been considered as the pivotal target of lncRNA CASC11 to drive the malignant phenotype of hepatocellular carcinoma. On the one hand, lncRNA CASC11 could recruit ALKBH5 to reduce the m6A level in UBE2T mRNA, altering its stability. On the other hand, the interaction between YTHDF2 and UBE2T mRNA was prevented by highly expressed CASC11 (52).
In short, studies on the interplay of m6A and ncRNAs have gradually fallen into the public view, however, understanding the association between them on GI cancers still remains perplexed. Therefore, we summarized several special ncRNAs affecting the m6A level and vital effectors regulating ncRNAs expression in the same way as m6A. We concentrated on the interaction of m6A and ncRNAs in malignant behaviors of GI cancer and hoped that this review could provide promising targets for clinical treatment and promote the further development of epigenetics.
2 Regulators of m6A: “writers”, “erasers” and “readers”
Alternative effects derived from m6A are controlled by methyltransferases (“writers”), demethylases (“erasers”) and m6A-binding proteins (“readers”). The “writers” of m6A mainly consist of methyltransferase-like 3 (METTL3), METTL14, METTL16, METTL5, KIAA1429, Wilms’ tumor-1 associated protein (WTAP), zinc finger CCCH-type containing 13 (ZC3H13), zinc finger CCHC-type-containing 4 (ZCCHC4), and RNA-binding motif protein 15 (RBM15/15B) (23, 53–56). METTL3 was reported to predominantly catalyze the RNA of m6A deposition in 1997 (17), which has a capacity to bind to the methyl donor S-adenosyl-methionine (SAM) and achieve methyl transfer (57, 58). METTL14 is considered as an RNA-binding platform to collectively form a core methyltransferase complex with METTL3, catalyzing and generating m6A modification (54, 59). Meanwhile, WTAP functions as a vital adaptor protein to increase the stability of core methyltransferase complex (23, 60). As for RBM15/15B, it can bind to METTL3 and WTAP to recruit targets of the methyltransferase complex (53, 61). KIAA1429 was found to tend to locate the m6A site near the 3′-UTR and stop codon region (56). Different from the methyltransferase complex, certain reports demonstrated that METTL16, METTL5 and ZCCHC4 independently participate in the process of m6A decoration onto U6 snRNA, 28S rRNA and 18S rRNA (62–66). Abundance of methyltransferases lays the foundation for diversified regulation from m6A modification.
Regarding the research on “erasers”, only two proteins, Fat mass and obesity-associated protein (FTO) and AlkBhomolog5 (ALKBH5), were discovered to dynamically remove the m6A site (67, 68). As the firstly identified demethylase (67), FTO not only mediates m6A demethylation in many cell types but also can remove the m6Am (N6,20-O-dimethyladenosine) demethylase of mRNA (69, 70). ALKBH5 presents higher specificity than FTO and only catalyzes m6A modification to achieve demethylation (71). “Readers” are considered to recognize and bind to m6A sites on RNA, including YT521-B homology (YTH) domain-containing protein family, heterogeneous nuclear ribonucleoprotein (HNRNP) family, and K homology (KH) domain family (IGF2BP1/2/3, also named as IMP1/2/3) (72–74). YTHDC1, which belongs to the YTH domain-containing protein family, was found to be distributed in the nucleus. YTHDC2, YTHDF1, YTHDF2, and YTHDF3 are members of the YTH domain-containing protein family, located in cytoplasm. YTHDF1 was reported to enhance the efficiency of the initiation stage of RNA translation through interplay with initiation factors (75). Meanwhile, YTHDF2 acts in a pivotal role in RNA degradation via binding with mRNA. YTHDF3 also exerts a positive effect on translational efficiency by relying on synergy with YTHDF1 (76). Research on “readers” has been excavating more roles of m6A in hallmark pathways.
3 Interplay between m6A and noncoding RNAs
Noncoding RNAs (ncRNAs) lack the capacity of translating directly into protein but function as crucial transcripts to regulate gene expression (77, 78). On the basis of nucleotide length, ncRNAs are divided into multiple types, such as miRNAs, lncRNAs, circRNAs, snRNAs, rRNAs, and tRNAs, etc (79, 80). The m6A modification in these ncRNAs have been increasingly reported to have a close association with the pathological mechanisms of malignant tumors. One the one hand, several key regulators derived from m6A methylation influence ncRNAs to participate in the initiation and progression of cancers. On the other hand, ncRNAs could also mediate downstream targets to reversely affect the expression of “writers”, “readers” and “erasers” through m6A modification. Thus, further discussions on the interaction between m6A and ncRNAs provide guidance to comprehensively understand the mechanism of malignant behavior in GI cancers.
3.1 lncRNA to m6A
LncRNAs serves as a class of ncRNAs that are over 200 nucleotides in length, and are extensively considered to sponge miRNAs to exert a functional effect (81–83). Recent investigations revealed that lncRNAs could regulate the crucial protein of m6A modification to affect the accumulation of m6A methylation sites (84, 85). In colorectal cancer, lncRNA GAS5 was regarded to have the property of negatively modulating YTHDF3, leading to evidently weakened proliferative and invasive potential of cancer cells (86). Specifically, the negatively mediated effect was mainly achieved through the GAS5/YAP/YTHDF3 axis. YAP belongs to one of most essential components of Hippo signaling, and its phosphorylation could activate the Hippo pathway (87). A dysregulation of the Hippo cascade is required to drive tumorigenesis in colorectal cancer (88, 89). Various kinases are responsible for catalyzing different phosphorylated sites of YAP to regulate its distribution in the cytoplasm and nucleus (90, 91). GAS5 was found to induce ubiquitin-mediated degradation of YAP to inhibit Hippo/YAP signaling. Then, YAP could bind with promoter of YTHDF3 to actively improve transcriptional efficiency of YTHDF3. Overexpression of lncRNA GAS5 could decrease YTHDF3 expression by targeting YAP. But which YTDHF3 could regulate lncRNA GAS5 still remained uncharted.
Furthermore, Wang et al. revealed that lncRNA LINRIS mediated IGF2BP2 to take part in developmental process of colorectal cancer. The increased expression of LINRIS tended to emerge in colorectal cancer tissue compared to adjacent tissue and indicated dismal prognosis for patients. ILINRIS knockdown suppressed the proliferative rate of HCT116 and DLD-1 cells. Further investigation of the promotive effect from LINRIS in CRC showed that LINRIS interacts with IGF2BP2 to block its degradation and maintain stable expression. Numerous studies have reported that IGF2BP2 recognized mRNA with m6A modification to stabilize targeted mRNA, thus participating in the occurrence and development of various cancers (74, 92). When the anti-autophagy gene 5 (ATG5) was rapidly reduced, IGF2BP2 protein level was scarcely altered after knockdown of LINRIS. The result suggested that LINRIS affects IGF2BP2 degradation in the autophagic pathway. Hallmark molecules regulating glycolysis, like MYC, GLUT-1, PKM2 and LDHA, exhibited low levels as LINRIS was depleted. MYC mRNA has been widely acknowledged as the downstream factor of IGF2BP2. Overexpressed IGF2BP2 exerted the reversal effect on the downregulation of glycolysis activity via the LINRIS–IGF2BP2-MYC axis when LINRIS was silenced (93). Thus, it is also necessary to explore the LINRIS–IGF2BP2-MYC axis, which could yield significant improvement in the prognosis of colorectal cancer. The mediation to regulators generally is considered to indirectly effect and relative mechanism need to be deeper discussed.
3.2 m6A to lncRNA
Abundant m6A sites were found in lncRNA, and their expression level was influenced through a specialized protein. Notably, differences in the methylation level of m6A also exerted contradictory effects on the expression of lncRNA owing to the regulation of respective targets. High m6A methylation could induce the downregulation of lncRNAs (Figure 1). For instance, the current study unraveled that lncRNA XIST expression was repressed by the binding of METTL14 and YTDHF2. Specifically, METTL14 elevated the m6A methylation level in XIST to suppress its expression through cooperating with YTDHF2. The corresponding mechanism of lncRNA XIST downregulation was confirmed, that is, YTDHF2 binds with the m6A complex from XIST to participate in the degradation process. In colorectal cancer, METTL14 was expressed lowly in cancer tissue compared with normal tissue, and METTL14 depletion was observed in patients with shorter survival times (94). The knockdown of METTL14 encouraged the biological property of proliferation and invasion by mediating lncRNA XIST.
Figure 1

Interplay between lncRNAs and m6A in gastrointestinal cancers.
In contrast, low methylation in lncRNA NEAT1 gave rise to a low-expression result. ALKBH5, the “eraser” of m6A, could catalyze the appearance of demethylation in lncRNA NEAT1 and its downregulation. In gastric cancer, NEAT1 might interact with EZH2 to reduce its expression, which was significantly related to the invasion and metastasis of tumors. Meanwhile, ALKBH5 silencing could recede the mentioned effect from NEAT1 (95). Similarly, LINC01320 upregulation might be induced by METTL14, and MEETL14 knockdown might trigger the loss of LINC01320; their positive correlation depends on the m6A modification degree. LINC01320 was seen as the activator of gastric cancer to affect its proliferative, invasive and migratory abilities in vitro. Malignant tumor behavior was triggered by LINC01320 overexpression via sponge miR-495-5p. It was acknowledged that RAB19 is downstream factor of miR-495-5p, and LINC01320 takes part in cell growth promotion through the LINC01320/miR-495-5p/RAB19 axis (96). The participation of METTL14 in the LINC01320/miR-495-5p/RAB19 axis deserves further investigation. Taken together, different effects from different m6A methylation levels in lncRNAs chiefly rely on the mediation of diverse targets.
3.3 miRNA to m6A
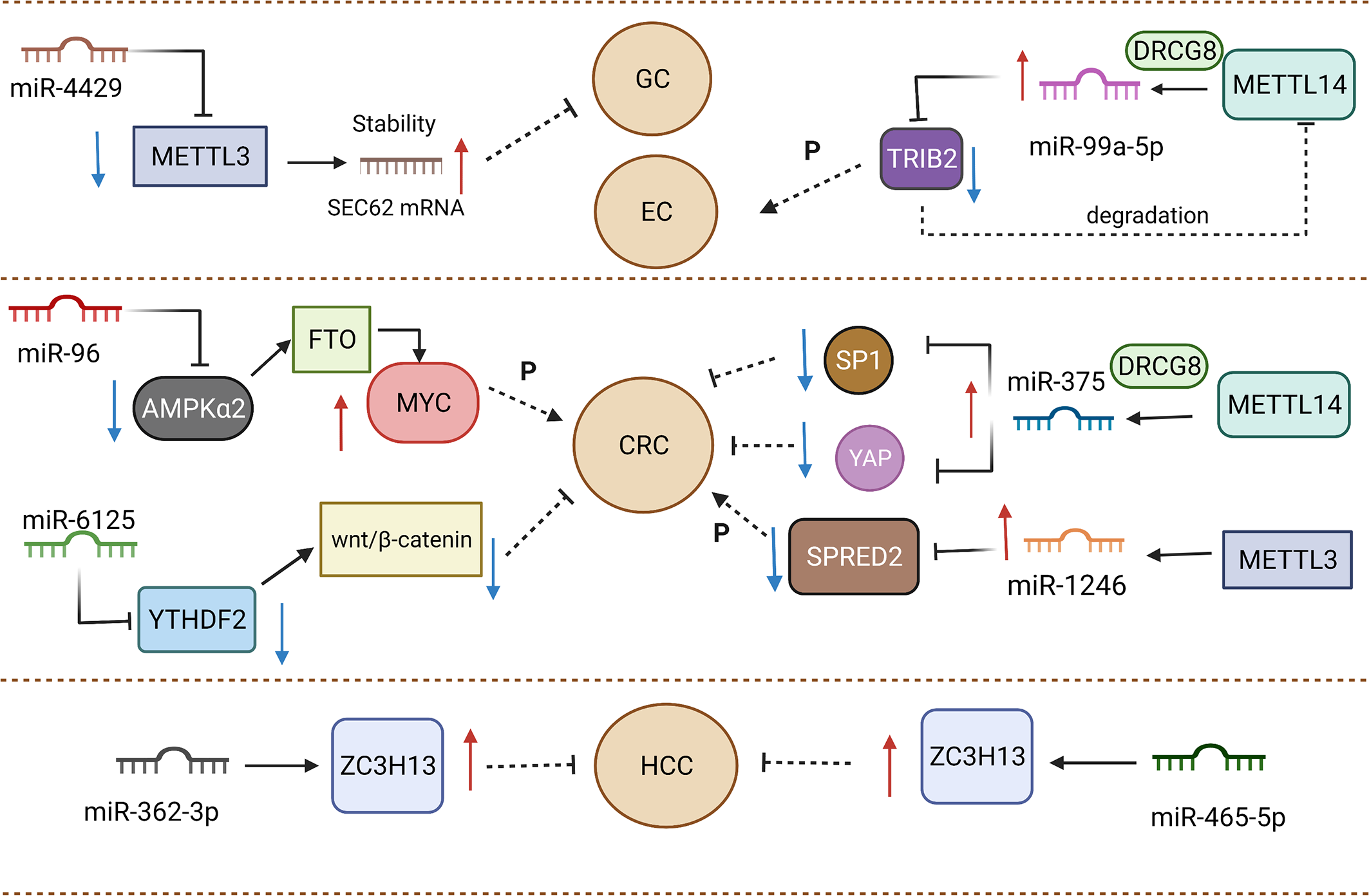
By definition, miRNAs are single-stranded ncRNAs with approximately 21-25 nucleotides, functioning as a RNA-induced silencing complex (RISC) to play a key role in the regulation of gene expression. As post-transcriptional regulators, miRNAs can reduce translation efficiency and impact mRNA degradation through the binding of 3′-UTR region from targeted mRNAs (96). One research reported that miR-4429 could inhibit METTL3 expression to play the role of suppressor in gastric cancer (97). The occurrence of m6A methylation mediated by METTL3 could stabilize SEC62 and exert a positive effect on SEC62 upregulation. Emerging evidence has demonstrated that m6A methylation depends on the promotion of binding between RBP and mRNA to increase mRNA stability (74, 98). METTL3 positively facilitated the binding between IGF2BP1 and SEC62 mRNA, stabilizing SEC62 mRNA in m6A-dependent manner. The lack of METTL3 caused the decline of m6A formation in SEC62. SEC62 was expressed highly in gastric cancer compared to normal tissue, and SEC62 knockdown elevated cell proliferation (99). Meanwhile, METTL3 upregulation was found to exert an analogous effect on participating in gastric cancer progression. MiRNAs not only affect “writers” but also “readers” and “erasers”.
MiR-6125 is considered as a promising biomarker for diagnosis and prognosis, and its decreased expression commonly implies an advanced tumor stage and poorer prognosis for colorectal cancer patients (100). In vitro and in vivo experiments testified that upregulation of miR-6125 prevents the proliferation phenomenon of colorectal cancer through the direct mediation of YTHDF2. MiR-6125 reduces the expression of cyclin D1 to generate the result of arrest at the G0/G1 phase of colorectal cancer cells, which depends on YTHDF2 mediating to take part in Wnt/β-catenin cascades. YTHDF2 serves as an oncogene to impact growth and malignant conversion in cancers (101–103). The ectopic expression of YTHDF2 in SW480 and RKO cells could enhance β-catenin accumulation. Aberrant accumulation from β-catenin gave rise to translocating from the cytoplasm to the nucleus, then modulating the cyclin D1 transcriptional process. The degradation of β-catenin was determined by the complex of Axin, GSK3β and APC. Notably, a deficiency of GSK3β attenuated the suppressive capacity of overexpressed miR-6125 in CRC cells. Therefore, miR-6125 mediated the degradation of β-catenin by affecting GSK3β (100). The role of miR-6125 and YTHDF2 in targeted treatment should be considered and the specific interaction between miR-6125 and YTHDF2 need more investigations and further discussion. MiR-96 could indirectly mediate FTO to affect the aggressive and malignant capacity of colorectal cancer. It was reported that FTO expression was downregulated by AMPKα2. AMPKα2 (104) has been considered as the target of miR-96 to impair proliferation, migration, and invasion in colorectal cancer. FTO could decrease the methylation of MYC mRNA to accelerate MYC accumulation. miR-96 indirectly mediated AMPKα2 to achieve MYC upregulation to enhance the aggressive ability of colorectal cancer, which can be a promising novel target to prolong survival time of patients (105).
3.4 m6A to miRNA
The formation of miRNA genes is commonly considered to initially undergo transcription mediated by RNA polymerase II promoters and maturation by further processing (106). Microprocessors, like the DiGeorge syndrome chromosomal region 8 (DGCR8), take part in processing into precursor miRNAs (pre-miRNAs) and subsequently Dicer carved pre-miRNAs to form mature single-stranded miRNAs (107). METTL14 was reported to regulate miR-375 processing via DGCR8. The number of unprocessed miR-375 increased after the silencing of METTL14. As the writer of m6A, METTL14 served as an antitumor factor of colorectal cancer to affect patient prognosis. The depletion of METTL14 might cause improvement of proliferative, invasive and migrative ability in colorectal cancer cells. The inhibiting effect from METTL14 was found to have close association with miR-375. Overexpression of METTL14 dramatically upregulated miR-375 as well as decreased YAP1 and SP1 expression in HCT116 and HCT8 cells. Therefore, METTL14 could participate in miR-375/YAP1 and miR-375/SP1 to mediate the inhibition of colorectal cancer progression via the manner of m6A modification (108). In addition, Peng et al. demonstrated that silencing METTL3 reduces the expression of mature miR-1246 and increases pri-miR-146 accumulation. When METTL3 was overexpressed, the above result could be reversed, which implied that METTL3 might facilitate the transformation of pri-miR-146 into miR-1246 (109). In colorectal cancer tissue, a clearly elevated m6A modification level was responsible for METTL3. To sum up, effects from regulators of m6A mainly focus on processing program in biogenesis aspect of miRNAs (Figure 2).
Figure 2
Various miRNAs and m6A regulators play promoting or inhibiting roles in gastrointestinal cancers.
3.5 circRNA to m6A
CircRNAs, firstly described in 1976 (110), are regarded to originate from pre-mRNA back-splicing (111) and have a special ring structure (111, 112), the formation of which principally depends on 3′,5′-phosphodiester bond connecting at the 3′-end and 5′-end or an upstream exon (113, 114). As ncRNAs with regulatory potency (115, 116), circRNAs with differential expression have been observed in numerous diseases, and they have become known as significant modulators of pathogenesis (117–120). Wang et al. (120) reported that enhancing circRNA-KIAA1429 activity resulted in accelerating migration, invasion and EMT progression of hepatocellular carcinoma cells. The carcinogenic role of cirRNA-KIAA1429 upregulating the downstream target Zeb1 is yet to be established. The expression of Zeb1 was confirmed to be mediated by YTHDF3 through m6A chemical modification. YTHDF3, acting as a “reader” of m6A, stabilized Zeb1 mRNA and prolonged its lifespan. When YTHDF3 expression was decreased, the effect of overexpressed cirRNA-KIAA1429 also was attenuated via depletion of Zeb1, suggesting that cirRNA-KIAA1429 cooperated with YTHDF3 to affect tumor development.
Emerging evidences have revealed that circRNAs function as a sponge to absorb miRNA, implicating their involvement in the biological process of multiple cancers (121). In fact, several circRNAs establish correlations with miRNA to indirectly mediate the expression of m6A regulators. High abundance of circPTK2 was observed in advanced stages of colorectal cancer patients, and miR-136-5p expression was decreased to induce chemoresistance. YTHDF1 was seen as the effective target of miR-136-5p. At the same time, enforced expression of miR-136-5p could adversely modulate YTHDF1 to suppress the proliferation and improve the sensitivity of colorectal cancer to chemotherapy. CircPTK2 indirectly upregulated YTHDF1 via the elimination of miR-136-5p (122). Additionally, Chi et al. testified that the role of circMAP2K4/hsa-miR-139-5p/YTHDF1 axis in the growth of hepatocellular carcinoma also verified the above-mentioned hypothesis. The inhibition of proliferative ability in hepatocellular carcinoma by hsa-miR-139-5p was partly reversed by highly expressed cirMAP2K4. The promotion of upregulated cirMAP2K4 sponged hsa-miR-139-5p to increase YTHDF1 activity, resulting in stimulating development and progression of hepatocellular carcinoma (123). To sum up, circRNAs are involved in m6A regulation with the aid of miRNA and “readers”, which enriches the existing studies on the molecular mechanism of circRNAs function at the post-transcriptional level.
3.6 m6A to circRNA
Canonical splicing is widely regarded as a mechanism of circRNA formation, depending on the splice signal sites and spliceosomes (124). The participation of reverse complementary sequences in intron flanking sequences was regarded to play a critical role in the process of circRNA biogenesis (125) (Figure 3). A recent study demonstrated that inverted repeated Alu pairs located in flanking introns exerted the effect of fostering extrusion of flanking introns and splicing process (126). Emerging evidence testified that a series of RNA bind proteins (RBPs) bind with flanking introns of circRNA to involve in the alteration of circRNA expression, which contain draw introns (127, 128) and modify the stability of Alu pairs (129, 130) to promote or inhibit circularization. METTL3 was confirmed to form m6A in flanking reverse complementary sequences of circ1662, remarkably elevating the expression of circ1662 in colorectal cancer. Overexpressed circ1662 could induce the invasion, migration and EMT of hepatocellular carcinoma cells in vitro. Increasing the invasive ability of a tumor is considered to easily trigger cancer metastasis. Circ1662 was found to improve the efficiency of nuclear transport of YAP1 to downregulate SMAD3, causing the promotion of tumor aggressiveness. Thus, METTL3 was proved to have an impact on circ1662 formation to drive colorectal cancer metastasis (131). An existing report revealed that knockdown of METTL14 contributed to the reduction of m6A sites in circORC5 and upregulate circORC5 (3). Meanwhile, whether the regulation of METTL14 to circORC5 has a dependency on flanking introns of circRNA remains unclear. In gastric cancer the capabilities of tumorigenesis and invasion from highly expressed METTL14 could be impaired by circORC5 deficiency. CircORC5 functions as a sponge to eliminate miR-30c-2-3p expression to participate in the malignant progression of gastric cancer. Numerous reports have indicated that miR-30c-2-3p downregulation has a negative effect on cell proliferative ability and the normal cycle of pancreatic cancer (132) and breast cancer (133). Collectively, METTL14 alters m6A density of circORC5 to negatively modulate its expression, followed by mediating the miR-30c-2-3p/AKT1S1 axis to induce the occurrence of malignant behavior in gastric cancer. Different from miRNA, regulators of m6A have been reported to affect nuclear export as well as biogenesis.
Figure 3

Association of circRNAs and m6A regulators in gastrointestinal cancers.
CircRNA has been commonly considered to occur in the cytoplasm (134), thus circRNAs located in the nucleus depend on particular proteins to accomplish nuclear export. For instance, it was reported that YTHDC1 could control circNSUN2 translocation (135). YTHDC1 bonds with the GAACU m6A motif of circNSUN2 to stimulate its export from nucleus to cytoplasm. When YTHDC1 expression was decreased, circNSUN2 expression in the nucleus subsequently showed abundant accumulation. Upregulated circNSUN2 was frequently observed in the serum of colorectal cancer patients with liver metastasis and predicted shorter survival times. Notably, the promotion of tumor aggression from circNSUN2 mediated IGF2BP2 and downstream targets to attain, instead of affect, NSUN2 expression. Regulation derived from m6A to circRNA is a better and more comprehensive cognition of circRNA metabolism and function.
4 Association of m6 with non-coding RNAs in various GI cancers
In GI cancers, the m6A modification level is related to clinical characteristics and has a contradictory impact on the prognosis of tumors. Here, according to a variety of GI cancers, we sorted out ncRNAs with m6A modification and further concluded the carcinogenic or anti-cancer effects of different ncRNAs (Table 1; Figure 4).
Table 1
| ncRNAs | Biological function | Molecules | Interaction mechanism | Ref. |
|---|---|---|---|---|
| LINC00022 | Promoting cell proliferation in vitro and tumorigenesis in vivo | FTO | Enhancing LINC00022 expression via reduction of m6A | (2) |
| MiR-194-2 | Promoting cell proliferation, migration in vitro and tumorigenesis in vivo | ALKBH5 | Downregulating miR-194-2 by preventing maturation of pri-miR-194-2 | (136) |
| MiR-99a-5p | Promoting cancer stem cell persistence and radio-resistance | METTL14 | Promoting miR-99a-5p expression by maturing pri-miR-99a-5p | (137) |
| LINC00958 | Promoting cell proliferation, invasion, migration, and lipogenesis | METTL3 | Promoting LINC00958 expression by stabilizing LINC00958 | (138) |
| LINC01273 | Inhibiting cell chemoresistance | METTL3 | Downregulating METTL3 expression via LINC01273 | (138) |
| LncRNA DUXAP8 | Inducing sorafenib resistance | METTL3 | Promoting DUXAP8 expression via METTL3 | (139) |
| MiR-582-3p | Inhibiting tumorigenesis in vivo | / | Promoting the binding of miR-582-3p and YAP by high m6A modification | (140) |
| MiR-362-3p/miR-425-5p | Reducing infiltration of tumor immune cell | ZC3H13 | Downregulating ZC3H13 via MiR-362-3p and miR-425-5p | (141) |
| Has-circ-0058493 | Promoting cell proliferation and invasion in vitro and growth in vivo | METTL3, YTHDF1 | Enhancing has-circ-0058493 expression | (142) |
| CirMAP2K4 | promoting cell proliferation | YTHDF1 | Promoting YTHDF1 expression via cirMAP2K4 | (123) |
| LncRNA ARHGAP5-AS1 | Increasing cell proliferation and chemoresistance and inhibiting cell apoptosis | METTL3 | Recruiting METTL3 to upregulate ARHGAP5 via ARHGAP5-AS1 | (143) |
| LINC00470 | Promoting cell proliferation, invasion, and migration | METTL3 | Preventing the binding of METTL3 and PTEN via LINC00470 | (144) |
| LncRNA NRON | Promoting growth in vitro and vivo | ALKBH5 | Recruiting ALKBH5 to stabilize Nanog mRNA via lncRNA NRON | (144) |
| LncRNA NEAT1 | Inhibiting invasion in vitro | ALKBH5 | Inhibiting NEAT1 expression via downregulation of m6A level | (95) |
| LINC01320 | Promoting cell proliferation migration, and invasion | METTL14 | Enhancing LINC01320 expression | (96) |
| LINC00942 | Promoting chemoresistance in vitro and vivo | MSI2(IGF2BP2) | Promoting MSI2 expression via LINC00942 | (145) |
| MiR-4429 | Preventing cell proliferation | METTL3 | Inhibiting METTL3 expression via MiR-4429 | (97) |
| LINC00857 | Inducing proliferation and impeding apoptosis | / | Stabilizing LINC00857 expression by high m6A level | (146) |
| LncRNA KCNK15-AS1 | Inhibiting proliferation and promoting apoptosis | ALKBH5 | Upregulating KCNK15-AS1 | (147) |
| LncRNA DBH-AS1 | Inhibiting growth and gemcitabine resistance | METTL3 | Promoting DBH-AS1 expression by stabilizing DBH-AS1 | (148) |
| LncRNA ZFAS1 | Promoting cell glycolysis | IMP2 | Enhancing ZFAS1 expression by increasing ZFAS1 stability | (149) |
| LncRNA PTTG3P | Promoting cell glycolysis | METTL3, IGF2BP2 | Promoting PTTG3P expression via METTL3 and IGF2BP2 | (150) |
| LncRNA GAS5 | Inhibiting cell proliferation and invasion invitro and tumorigenesis in vivo | YTHDF3 | Inhibiting YTHDF3 expression via lncRNA GAS5 | (86) |
| LncRNA LINRIS | Inhibiting cell glycolysis | IGF2BP2 | Enhancing stability of IGF2BP2 via lncRNA LINRIS | (93) |
| LncRNA XIST | Promoting proliferation and invasion | METTL14, YTDHF2 | Reducing lncRNA XIST expression by mediating degradation | (151) |
| MiR-6125 | Preventing cell proliferation | YTDHF2 | Inhibited YTDHF2 expression via miR-6125 | (100) |
| MiR-96 | Promoting cell proliferation, migration and invasion and inhibiting cell apoptosis | FTO | Promoted FTO expression via miR-96 | (105) |
| MiR-375 | Inhibiting growth and metastasis in vitro and vivo | METTL14 | Upregulating miR-375 by promoting maturation of pri-miR-375 | (108) |
| MiR-1246 | Promoting invasion and migration in vitro and metastasis | METTL3 | Enhancing miR-1246 expression by promoting maturation of pri-miR-1246 | (109) |
| CircALG1 | Promoting metastasis in vitro and vivo | / | Elevating circALG1 expression and promoting the binding of circALG1 and miR-342-5p via high m6A level | (152) |
| CircPTK2 | Promoting cell proliferation, migration, invasion and chemoresistance | YTDHF1 | Promoting YTDHF1 expression via CircPTK2 | (122) |
| Circ1662 | Promoting cell invasion, migration and EMT in vitro | METTL3 | Upregulating circ1662 | (131) |
| CircORC5 | Inhibiting cell proliferation and invasion in vivo and tumorigenesis in vivo | METTL14 | Reducing circORC5 expression | (3) |
| CircNSUN2 | Promoting cell invasion and migration | YTHDC1 | Upregulating circNSUN2 expression by enhancing circNSUN2 export | (135) |
| Circ3823 | Promoting cell proliferation, invasion, and angiogenesis | YTHDF3, ALKBH5 | Inhibiting circ3823 expression via promoting degradation | (153) |
Mechanism of interaction between main ncRNAs and m6A in gastrointestinal cancers.
Figure 4
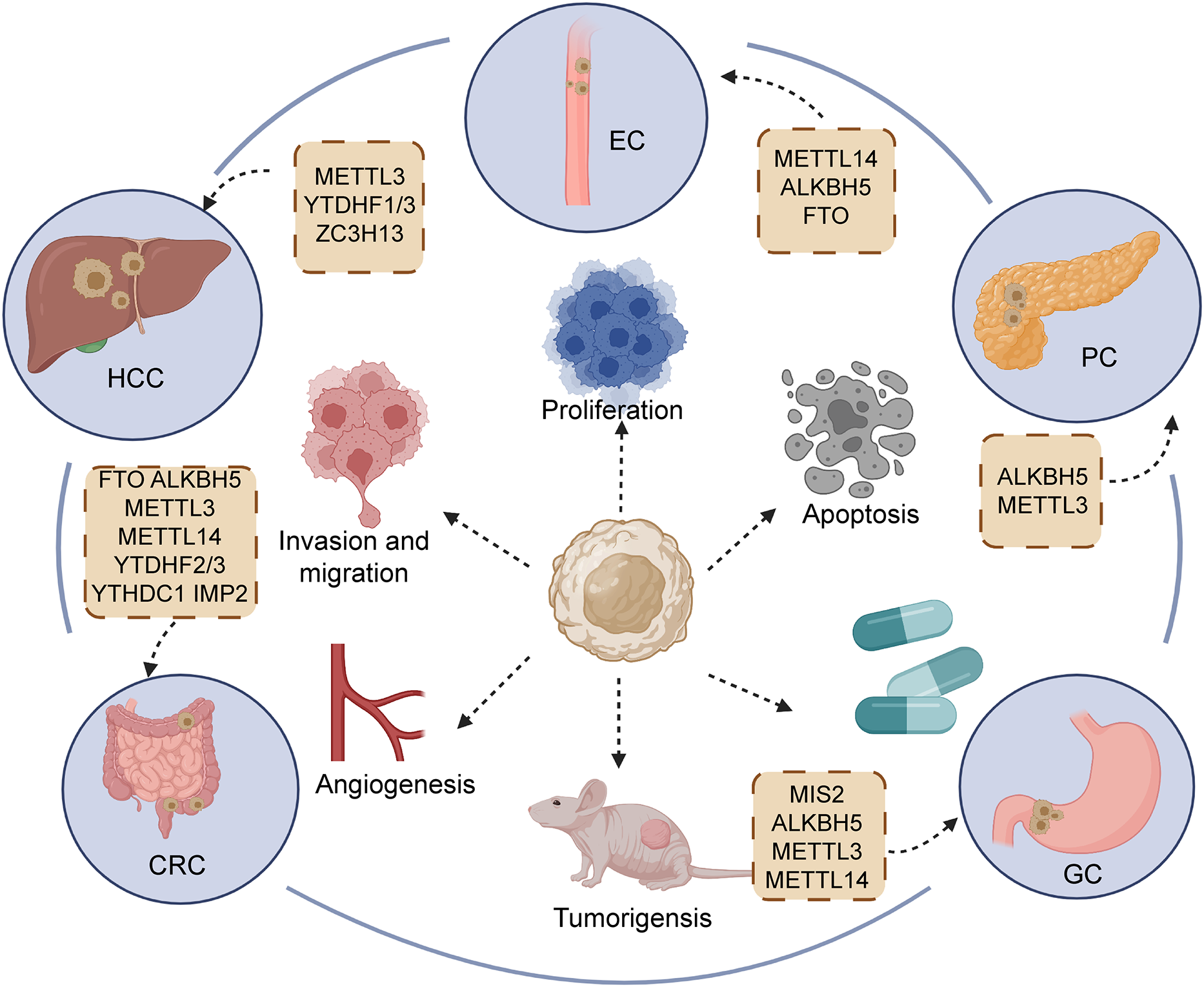
Regulators of m6A participating in the initiation and development of main gastrointestinal cancers, including esophageal cancer (EC), hepatocellular carcinoma (HCC), gastric cancer (GC), pancreatic cancers (PC), and colorectal cancer (CRC).
4.1 Esophageal cancer
In spite of advancements in multidisciplinary antitumor treatment for esophageal cancer (EC), the 5-year survival rate of patients still shows a negative trend, owing to uncharted molecular mechanisms and uncontrolled recurrence (154, 155). Accumulating evidence demonstrates that m6A modification plays a critical role in ncRNA function to affect the biological behavior of EC. Cui et al. (2) uncovered the elevated expression of LINC00022 and shortened survival time in patients with advanced-stage esophageal squamous cell carcinoma (ESCC). FTO overexpression obviously reduced the m6A methylation level in ESCC, and LINC00022 expression exhibited accumulation due to the high expression of FTO. The alteration of cell cycle plays a pivotal role in the proliferative progression of multiple malignancies (2, 156). LINC00022 knockdown was shown to affect the distribution of cell cycle, mainly manifesting in the increase of cells in G0/G1 and reduction of cells in the G2/M phase. In addition, several inhibitors of CDK, including p16, p21 and p53, remained upregulated in KYSE150 and TE1 cell owing to the silencing of LINC00022. Meanwhile, upregulated FTO could positively mediate LINC00022 to reverse the above-mentioned phenomenon. Additionally, the expression of YTHDF2 was also confirmed to influence the FTO/LINC00022 axis. YTHDF2 directly bind with LINC00022 transcript to affect stability and promote degradation. “Eraser” ALKBH5 is fame antitumor factor in hepatocellular and pancreatic cancer (28, 157). In EC, ALKBH5 serves as a positive prognostic factor to predict the survival time of patients (136). DGCR8 is recruited by the demethylase activity of ALKBH5, participating in the processing of pri-miR-194-2. Overexpression of ALKBH5 could prevent DGCR8 from binding with pri-miR-194-2. ALKBH5 repressed the downregulation of RAI1 caused by miR-194-2, contributing to the release of RAI1 and its inhibition of malignancy. RAI1 was confirmed to promote tumor development through participation in the Hippo pathway, which could bind with the DNA region of Hippo signaling to upregulate the expression pathway and block the nuclear translocation of YAP/TAZ (158–160). Thus, the ALKBH5/DGCR8/miR-194-2/Hippo axis to EC targeted treatment opens up a novel orientation. In EC, regulators of m6A modification are able to affect biogenesis and degradation of ncRNAs, which provide a novel orientation to clinical treatment of EC.
4.2 Hepatocellular carcinoma
Hepatocellular carcinoma (HCC), the most common type of primary liver cancer, greatly threatens human health greatly by its striking morbidity and mortality (161). Compared with adjacent normal tissue, increased m6A modification was found in the YAP 3′UTR region of HCC tissue. Notably, m6A methylation plays an indispensable role in inducing the interaction of miR-582-3p with YAP. The overexpression of circ-104075 could elevate the expression of YAP in mRNA and protein level by absorbing miR-582-3p, contributing to the promotion of tumorigenesis in HCC (140). Analogously, METTL3 catalyzes the formation of m6A in has-circ-0058493, followed by causing the binding of YTHDF1 with has-circ-0058493. YTHDF1 dramatically enhanced the exporting efficiency of has-circ-0058493 from the nucleus to the cytoplasm depending on the m6A modification, which promoted the growth and malignant progression of HCC (142). KIAA1429, composed of METTL3, METTL14 and WTAP, is responsible for RNA m6A methylation, mainly including the regions of 3′ UTR and near the stop codon (56, 162). Highly expressed KIAA1429 was found to closely correlate with poorer prognosis for HCC patients. KIAA1429 mediates GATA3 to exert the effect of fostering development and metastasis of HCC both in vivo and in vitro. GATA3 functions as the famous transcript factor to stimulate the activation of multiple tumor suppressors (72, 163). With the help of LncRNA GATA3-AS, KIAA1429 gives rise to the occurrence of m6A methylation in the GATA3 precursor mRNA (pre-mRNA), which causes the interruption of binding between GATA3 pre-mRNA and HuR to enhance the degradation of GATA3 pre-mRNA, subsequently downregulating GATA3 (164). In contrast, KIAA1429 deficiency could recover the interaction of HuR and GATA3 pre-mRNA, leading to maintained stability of GATA3 pre-mRNA to block the malignant behavior of HCC. In short, we can notice that “writers” and “readers” commonly have an impact on phenotype of tumor in most cases.
“Writers” of the m6A modification are also affected by ncRNAs to participate in the complex mechanism of HCC. For instance, the ZC3H13 “writer” is negatively mediated by miR-362-3p/miR-425-5p mimics. Meanwhile, inhibitors of miR-362-3p/miR-425-5p could increase the expression of ZC3H13. In short, ZC3H13 was confirmed as a target of miR-362-3p/miR-425-5p. ZC3H13 might accelerate immune cell infiltration in HCC to affect the prognosis of patients. Downregulated ZC3H13 could predict adverse outcomes in HCC (141). Nevertheless, more research is required to explore which biological behaviors of tumor are affected by ZC3H13 and how miR-362-3p/miR-425-5p mediate ZC3H13. These findings for HCC focused on the fact that “writers” and “readers” are affected by ncRNAs or their mediating effects on ncRNAs. More research is needed to explore the regulators of “erasers” and ncRNAs in hepatocellular carcinoma.
4.3 Gastric cancer
Gastric cancer (GC) is considered as the fifth most frequent category of tumors, and its mortality in cancers ranks fourth globally (1, 165). Various researches have been performed on m6A and ncRNAs to probe the mechanism of malignant phenotype in GC. Wang et al. demonstrated that lncRNA NRON facilitated the proliferative capacity in vitro and in vivo via the modulation of ALKBH5 and Nanog (166). It is widely acknowledged that ALKBH5 strictly regulates the m6A methylation of Nanog (167, 168). LncRNA NRON could recruit ALKBH5 to enhance the stability and prohibit the degradation of Nanog mRNA. In GC, LINC00470 was found to be highly expressed and associated with dismal prognosis. This is because LINC00470 mediates the degradation of PTEN mRNA to decrease the expression of PTEN. Specifically, LINC00470 exerted an impact on the process of binding between METTL3 and PTEN to prevent the occurrence m6A methylation in PTEN (144). PTEN is implicated in the homeostasis of PI3K/AKT cascade to serve as a protective factor of tumors. The reduction of PTEN might induce malignant behavior in human cancers, such as proliferation, angiogenesis and migration (169). Interestingly, YTHDF2 also participates in process of shortening the half-life of PTEN induced by LINC00470. The effect of downregulating PTEN by overexpressing LINC00470 could be restored after silencing YTHDF2 (144). Furthermore, ncRNAs could mediate m6A regulation to influence the formation of chemoresistance. The upregulation of lncRNA ARHGAP5-AS1 was reported to stimulate the expression of ARHGAP5, resulting in attenuating the drug-sensitivity of gastric cells to cisplatin (DDP), ADM and 5-FU. Mechanistically, ARHGAP5-AS1 recruits METTL3 to enhance m6A modification, effectively promoting the transcription of ARHGAP5 and increasing the stability of ARHGAP5 mRNA in the cytoplasm (143). ARHGAP5 depletion reversed the chemotherapy resistance of gastric cells by inducing the activation of autophagy and increasing intracellular drug concentration.
4.4 Pancreatic cancer
As one of top four life-threatening malignant tumors worldwide (170, 171), the 5-year survival rate of pancreatic cancer (PC) of patients is low at approximately 5% (172). The options for PC clinical treatment are mainly surgical resection and chemotherapy approaches (173, 174). Owing to the late diagnosis, a number of patients are unable to attain a satisfactory outcome (172). Thus, new therapeutic targets and early diagnosis are necessary. LINC00857 was discovered to be highly expressed in adverse prognosis of HCC patients. Overexpression LINC00857 could induce proliferation and impede the apoptosis of PC cells by directly absorbing miR-150-5p. It was reported that high m6A methylation rate appeared in LINC00857 and increased its stability in HCC tissue, as compared with cancer adjacent tissue (146). The upregulation of potassium two pore domain channel subfamily K member 15 and WISP2 antisense RNA 1 (KCNK15-AS1) could hamper the growth and stimulate the apoptosis of PC cells. Consistently, the overexpression of KCNK15-AS1 improved epithelial marker (E-cadherin) expression and adversely mediated mesenchymal marker (N-cadherin) expression, remarkably blocking the EMT process. Due to the lack of ALKBH5, the m6A in KCNK15-AS1 presented an elevated level but KCNK15-AS1 expression manifested a decreased level. Collectively, ALKBH5 could mediate m6A methylation to positively upregulate KCNK15-AS1 (147). However, the detailed mechanism of low m6A methylation improving KCNK15-AS1 expression remains unknown. Investigations on m6A and ncRNAs in pancreatic cancer are scarce, while the potential of regulators and ncRNAs should not be ignored.
4.5 Colorectal cancer
Colorectal cancer (CRC), serving as the predominant cause of cancer-related deaths worldwide (175, 176), easily presents parenteral metastasis and thus brings unsatisfactory prognosis (177). Lin et al. (152) observed that circALG1 activity was upregulated in the tumor tissue and peripheral blood of CRC patients. MiR-342-5p could exert a negative effect on regulating placental growth factor (PGF) expression via binding with the 3’UTR region of PGE. CircALG1 facilitated the expression of PGF by sponging miR-342-5p. M6A modification not only elevated circALG1 expression but also reinforced the binding between circALG1 and miR-342-5p. However, the specific mechanism of m6A methylation affecting circALG1 expression remains unknown.
IMP2, a m6A reader expressed in CRC tissues, was discovered to have a close association with lncRNA ZFAS1. The dysfunction of IMP2 contributed to altering ZFAS1 expression. When IMP2 was expressed at a low level, ZFAS1 half-life became shorter in HCT116 and SW620 CRC cells. It was confirmed that IMP2 could directly bind with ZAFAS1 in the KH3-4 domain to activate its expression by intensifying ZAFS1 stability. The m6A modification was regarded to connect IMP2 and ZAFS1. High expression of ZAFS1 could improve the ATPase activity of OLA1 to positively further influence the formation of lactic acid and release of raw material to ATP synthesis, triggering glycolytic signaling to stimulate malignant progression in CRC (149).
Furthermore, METTL3 was validated to enhance m6A modification in lncRNA pituitary tumor-transforming 3 pseudogene (PTTG3P) to promote its expression. The stability of PTTG3P could be strengthened by METTL3 in a m6A-dependent manner. When IGF2BP2 was depleted, its upregulating effect on PTTG3P was also weakened to some extent. METTL3 positively mediated PTTG3P expression with the collaborative participation of IGF2BP2 (150). Overexpression of PTTG3P accelerated glycolysis of CRC. It is well-known that glycolysis occurs faster in malignancy than normal tissue (178–180). The accumulation of tumor lactate has been testified to cause distant metastasis and tumor return (181, 182). Collectively, the activity of METTL3 and IGF2BP2 could have an impact on the malignant phenotype of CRC via PTTG3P. In CRC, more research demonstrated that “writers” and “readers” take part in complex mechanism of initiation and malignancy. These regulators of m6A should be important therapeutic targets regarding the control of ncRNA function.
5 Clinical implications of m6A modification in ncRNAs
5.1 Potential biomarkers in GI cancers
Early diagnostic markers not only identify the type of cancers faster but lay the foundations for radical surgical treatment. Effective prognosis markers can reflect the probability of metastasis and recurrence, better judging the benefits from surgery and monitoring the status of patients post-surgery. Accumulating knowledge has provided new insights into m6A modification in ncRNAs (183, 184), which hopefully offer novel biomarkers for prompt diagnosis and prognosis in gastrointestinal tumors. Liquid biopsy is considered as a non-invasive and low-cost detection method, which could diagnose tumors by analyzing the appropriate biomarkers existing in blood, urine, ascites, cerebrospinal fluid, and so on (184, 185). In the serum of CRC patients, circ3823 (153) was observed to present a high level with remarkable specificity and sensitivity. This finding implies that the accurate and timely diagnosis of colorectal cancer benefits from detecting circ3823 from liquid biopsy. Guo et al. (153) reported that elevated expression of circ3823 had a close association with tumor stage and lymph node metastasis, because circ3823 weakens miR-30c-5p expression to boost tumor proliferation and invasion both in vitro and in vivo. Therefore, circ3823 could also serve as the satisfactory prognosis marker in CRC. Altered circ3823 expression in CRC was affected by YTHDF3 and ALKBH5 mediated by m6A modification, which might be related to the induction of circ3823 degradation, while this presumption needs more experiments to be verified. Analogously, LINC00460 expression also plays an indispensable role in prognosis prediction for patients with CRC (151). Upregulated LINC00460 tended to occur in advanced stages of tumor, because LINC00460 could heighten the proliferative and invasive ability of CRC cells. Tumor growth and lung metastasis would be arrested after silencing of LINC00460 (151). HMGA1 was further proved to be an important downstream regulator of LINC00460. Specifically, LINC00460 interacted with IGF2BP2 and DHX9 to recognize the m6A site mediated by METTL3 in HMGA1, contributing to stabilizing HMGA1 mRNA at the post-transcriptional level (151). With the assistance of m6A modification, the expression of numerous ncRNAs is altered to reflect their differential distribution in malignant and normal diseases. Besides, differences in the expression of special ncRNAs were testified to be significantly correlated to clinical features of cancer, which enlarged the horizons of knowledge on the occurrence and development of tumors.
5.2 Therapeutic targets in GI cancers
The continuous development of nanotherapeutics has brought great hope for the precise and effective treatment of malignant tumors (186–188). For example, taking polymeric nanoparticle (NP) to deliver siRNA might reduce its degradation and maintain its continuous release (189). It was reported that PLGA-based NPs incorporating FAK siRNA obviously improved the sensitivity of ovarian cancer patients to chemotherapy (190). Overexpressed LINC00958 enhances the proliferative capacity of Hep3B and HepG2 cells by mediating miR-3619-5p (138). PLGA-PEG (si-LINC00958) NP was demonstrated to rapidly block tumor growth and development in HCC. Moreover, ETTL3 expression was revealed to have a compact correlation with LINC00958 expression. For instance, LINC00958 expression would present a decline as METTL3 was silenced, which was related to the stabilization of LINC00958 transcripts from m6A induced by METTL3 (138). Thus, the exploration and utilization of interactions between m6A modification and LIC000958 might shed a new light on NP and enrich therapeutic approaches in HCC (138).
Nanotherapeutics has prosper development prospect, but conventional chemotherapy is still the cornerstone of treatment of unresectable HCC. For instance, sorafenib, a tyrosine kinase inhibitor, has been become one of preferred drugs for chemotherapy in patients with an unfavorable stage of tumor (191–193). However, the sorafenib resistance of HCC renders therapeutic effect descending. Current studies confirmed that lncRNA DUXAP8 (139) and LINC01273 (194) are linked to the occurrence of sorafenib resistance. It was discovered that LINC01273 knockdown can impair cell viability and DNA synthesis to increase the sensitivity of tumor to sorafenib (194). At the same time, the above effect could be attenuated due to the downregulation of METTL3. LINC01273 negatively regulated METTL3 expression by stabilizing miR-600. Conversely, METTL3 overexpression also had a suppressing impact on LINC01273 expression. The LINC01273 level was controlled by YTDHF2, and YTDHF2 depletion might enrich LINC01273 to decrease sorafenib sensitivity in a m6A-dependent manner (194). Likewise, DUXAP8 competitively bonded with miR-584-5p to trigger MAPK/ERK signaling, resulting in the chemoresistance of HCC against sorafenib (139). Different from LINC01273, METTL3 exerted an accelerative effect on DUXAP8 expression to affect sorafenib resistance through mediation by m6A (139).
A group of lncRNAs also participate in other complex mechanisms of chemotherapy-drug resistance, including gemcitabine (148) and cisplatin (DDP) (145). It was reported that gemcitabine serves as the first-line drug to treat patients of pancreatic cancer in the malignant stage (195, 196). However, the fact is that the phenomenon of gemcitabine chemoresistance in patients has been prevalent in clinical treatment practice. Decreasing expression of lncRNA DBH-AS1 was observed in gemcitabine-resistant pancreatic cancer tissues and cells (148). Mechanically, it was reported that DBH-AS1 sequestered miR-3163 to induce USP44 upregulation, leading to the enhancement of gemcitabine sensitivity and benefiting prognosis of patients. The study demonstrated that variation of lncRNA DBH-AS1 expression was affected by m6A modification level in the degreed (148). METTL3 catalyzed m6A modification in DBH-AS1 to stabilize DBH-AS1. DBH-AS1 expression would present downregulation when METTL3 was knockdown (148). In gastric cancer, the use of cisplatin alone or combined are predominant chemotherapeutic methods for patients with a lack of surgery opportunity (197, 198). Unlike DBH-AS1, overexpression of LINC00942 presented in gastric cancer cells of DDP resistance. LINC00942 was able to inhibit apoptosis and stimulate stemness, initiating DDP resistance. Specifically, because LINC00942 bind with MSI2 (Musashi2) to hamper degradation from SCFβ‐TRCP E3 ubiquitin ligase, MIS2 stability gained increasing (145). c‐Myc is a well‐known target of chemotherapy resistance in multiple cancers (199). MSI2 stabilized c‐Myc RNA depended on m6A modification. What is noteworthy is that the binding between c‐Myc and MISI was affect by expression of METTL3‐METTL14–WTAP complex (145). LINC00942/MIS2/c‐Myc axis will become the potential targets to elevate DDP treatment efficiency for patients.
The phenomenon of radio-resistance can affect the response to neoadjuvant chemoradiation (200), bringing more unfavorable treatment results in multiple cancers (201, 202), with ESC being no exception. The stemness of cancer cells was reported to participate in the occurrence mechanism of radio-resistance (203). Lowly expressed miR-99a-5p induced stemness maintenance of CSCs to resist radiotherapy in ESC. METTLl4 promoted DGCR8 to process pri-miR-99a-5p, causing mature miR-99a-5p to persist in ESCC. The overexpression of miR-99a-5p could weaken the capacity of DNA repair to increase the radiotherapy sensitivity of ESCC cells by preventing TRIB2 expression. As the downstream factor of miR-99a-5p, TRIB2 in turn negatively affected METTL14 expression. Mechanically, TRIB2 functioned as the bridge to join COP1 and METTL14, forming an ubiquitinated modification of METTL14 to facilitate its degradation. Taken together, METTL14, miR-99a-5p and TRIB2 constitute a special feedback loop to modulate the formation of radio-resistance in ESCC. Activators of METTL14 used in clinical treatment might improve the overall survival times of ESCC patients (137).
These explorations to m6A and ncRNAs deeper uncovered molecular mechanism of resistance of chemotherapy and radiotherapy, providing more possibilities of potential therapeutic targets. And the combination of inhibitors or activators of regulators and chemotherapy drug will ameliorate refractory of gastrointestinal cancers and improve prognosis of patients. But a part of regulators from m6A lack commercial inhibitors or activators. Corresponding side effect from molecular therapy still remains unknown. Due to diverse effects from m6A modification in cancers, only alteration m6A level does not necessarily achieve successfully. More precise target combined with m6A level changes need be considered.
6 Conclusion and prospects
In the past decades, m6A has been testified to occur in DNA, RNA and proteins, and implicated with the biological properties of multiple malignancies. The development of transcriptomics has brought the study of RNA to a climax, and a series of experiments gradually revealed the critical role of m6A on RNA in the regulation of post-transcriptional level. As the most common chemical modification type, m6A not only decorates mRNA but also appears in some non-coding RNAs, such as lncRNAs, miRNAs and circRNAs. The reversible effect of m6A relies on “writers”, “erasers” and “readers”.
These regulators can control the m6A level or directly affect ncRNA metabolism including biogenesis, location and degradation, resulting in positively or negatively altering the expression of ncRNAs. Thus, the abnormal expression of m6A and dysfunction of key regulators in these ncRNAs might contribute to the initiation and development of malignant tumors. Interestingly, non-coding RNAs can also exert an impact on the expression of regulator factors of m6A by indirectly modulating downstream targets or directly regulating particular factors of m6A, which has a close association with the biological function of GI cancers, including proliferation, invasion, migration, apoptosis, angiogenesis, and stemness. Nevertheless, the expression of ncRNAs and regulators would form a feedback loop to reciprocally mediate both sides, inducing chemotherapy resistance and causing dismal prognosis. Further studies on the interaction of m6A and ncRNAs can enrich the knowledge of the function of non-coding RNAs and dig up the comprehensive mechanism of the malignant phenotype of GI cancers.
As technologies and approaches of detecting m6A sites are gradually improved, we can progressively obtain more information on m6A alteration regarding higher-precision transcript segments, which can hopefully render m6A modifications more specific biomarkers in cancers. Regulators of m6A have not only proved as feasible in targeted treatment by numerous theories, but were initially developed by pharmaceutic or biotech companies to step into clinical therapy, with examples of METTL3 and METTL14 inhibitors, and so on. The utilization of potent inhibitors or activators to restore the normal expression level of m6A regulators can interrupt the oncogenic role of upstream or downstream targets, bringing a promising clinical prospect for patients, especially those with resistance to currently available treatments.
Statements
Author contributions
YH and WG designed and guided the study. YX and XY wrote and edited the manuscript. YX helped with reference collection. All authors read and approved the final manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (819028328), Leading Talents of Zhongyuan Science and Technology Innovation (214200510027), Henan Provincial Medical Science and Technology Research Plan (SBGJ2018002 and SBGJ202102117), Science and Technology Innovation Talents in Henan Universities (19HASTIT003), the Gandan Xiangzhao Research Fund (GDXZ2022002), Outstanding Foreign Scientist Studio in Henan Province (GZS2020004) and Henan Provincial Department of Science and Technology Research on Science and Technology (222102310365).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Ferlay J Soerjomataram I Dikshit R Eser S Mathers C Rebelo M et al . Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int J Cancer (2015) 136(5):E359–86. doi: 10.1002/ijc.29210
2
Cui Y Zhang C Ma S Li Z Wang W Li Y et al . Rna M6a demethylase fto-mediated epigenetic up-regulation of Linc00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res (2021) 40(1):294. doi: 10.1186/s13046-021-02096-1
3
Fan HN Chen ZY Chen XY Chen M Yi YC Zhu JS et al . Mettl14-mediated M(6)a modification of Circorc5 suppresses gastric cancer progression by regulating mir-30c-2-3p/Akt1s1 axis. Mol Cancer (2022) 21(1):51. doi: 10.1186/s12943-022-01521-z
4
Shen S Yan J Zhang Y Dong Z Xing J He Y . N6-methyladenosine (M6a)-mediated messenger rna signatures and the tumor immune microenvironment can predict the prognosis of hepatocellular carcinoma. Ann Transl Med (2021) 9(1):59. doi: 10.21037/atm-20-7396
5
Zhou L Jiang J Huang Z Jin P Peng L Luo M et al . Hypoxia-induced lncrna Steap3-As1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing M(6)a-mediated degradation of Steap3 mrna. Mol Cancer (2022) 21(1):168. doi: 10.1186/s12943-022-01638-1
6
Liu Z Zou H Dang Q Xu H Liu L Zhang Y et al . Biological and pharmacological roles of M(6)a modifications in cancer drug resistance. Mol Cancer (2022) 21(1):220. doi: 10.1186/s12943-022-01680-z
7
Chen H Gao S Liu W Wong CC Wu J Wu J et al . Rna N(6)-methyladenosine methyltransferase Mettl3 facilitates colorectal cancer by activating the M(6)a-Glut1-Mtorc1 axis and is a therapeutic target. Gastroenterology (2021) 160(4):1284–300.e16. doi: 10.1053/j.gastro.2020.11.013
8
Oerum S Meynier V Catala M Tisné C . A comprehensive review of M6a/M6am rna methyltransferase structures. Nucleic Acids Res (2021) 49(13):7239–55. doi: 10.1093/nar/gkab378
9
Raj N Wang M Seoane JA Zhao RL Kaiser AM Moonie NA et al . The Mettl3 epitranscriptomic writer amplifies P53 stress responses. Mol Cell (2022) 82(13):2370–84.e10. doi: 10.1016/j.molcel.2022.04.010
10
Wang KC Kang CH Tsai CY Chou NH Tu YT Li GC et al . Ten-eleven translocation 1 dysfunction reduces 5-hydroxymethylcytosine expression levels in gastric cancer cells. Oncol Lett (2018) 15(1):278–84. doi: 10.3892/ol.2017.7264
11
Wu J Chai H Shan H Pan C Xu X Dong W et al . Histone methyltransferase Setd1a induces epithelial-mesenchymal transition to promote invasion and metastasis through epigenetic reprogramming of snail in gastric cancer. Front Cell Dev Biol (2021) 9:657888. doi: 10.3389/fcell.2021.657888
12
Ma Z Wang H Cai Y Wang H Niu K Wu X et al . Epigenetic drift of H3k27me3 in aging links glycolysis to healthy longevity in drosophila. Elife (2018) 7:e35368. doi: 10.7554/eLife.35368
13
Xue C Zhao Y Li L . Advances in rna cytosine-5 methylation: Detection, regulatory mechanisms, biological functions and links to cancer. biomark Res (2020) 8:43. doi: 10.1186/s40364-020-00225-0
14
Schäfer KP . Rna synthesis and processing reactions in a subcellular system from mouse l cells. Hoppe Seylers Z Physiol Chem (1982) 363(1):33–43. doi: 10.1515/bchm2.1982.363.1.33
15
Desrosiers R Friderici K Rottman F . Identification of methylated nucleosides in messenger rna from novikoff hepatoma cells. Proc Natl Acad Sci U.S.A. (1974) 71(10):3971–5. doi: 10.1073/pnas.71.10.3971
16
Luo GZ MacQueen A Zheng G Duan H Dore LC Lu Z et al . Unique features of the M6a methylome in arabidopsis thaliana. Nat Commun (2014) 5:5630. doi: 10.1038/ncomms6630
17
Bokar JA Shambaugh ME Polayes D Matera AG Rottman FM . Purification and cdna cloning of the adomet-binding subunit of the human mrna (N6-Adenosine)-Methyltransferase. Rna (1997) 3(11):1233–47. doi: 10.3390/cancers13051168
18
Bodi Z Bottley A Archer N May ST Fray RG . Yeast M6a methylated mrnas are enriched on translating ribosomes during meiosis, and under rapamycin treatment. PloS One (2015) 10(7):e0132090. doi: 10.1371/journal.pone.0132090
19
Yadav PK Rajvanshi PK Rajasekharan R . The role of yeast M(6)a methyltransferase in peroxisomal fatty acid oxidation. Curr Genet (2018) 64(2):417–22. doi: 10.1007/s00294-017-0769-5
20
Lluch-Senar M Luong K Lloréns-Rico V Delgado J Fang G Spittle K et al . Comprehensive methylome characterization of mycoplasma genitalium and mycoplasma pneumoniae at single-base resolution. PloS Genet (2013) 9(1):e1003191. doi: 10.1371/journal.pgen.1003191
21
Deng X Chen K Luo GZ Weng X Ji Q Zhou T et al . Widespread occurrence of N6-methyladenosine in bacterial mrna. Nucleic Acids Res (2015) 43(13):6557–67. doi: 10.1093/nar/gkv596
22
Wang Y Li Y Toth JI Petroski MD Zhang Z Zhao JC . N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol (2014) 16(2):191–8. doi: 10.1038/ncb2902
23
Ping XL Sun BF Wang L Xiao W Yang X Wang WJ et al . Mammalian wtap is a regulatory subunit of the rna N6-methyladenosine methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
24
Martinez de laCB Markus R Malla S Haig MI Gell C Sang F et al . Modifying the M(6)a brain methylome by Alkbh5-mediated demethylation: A new contender for synaptic tagging. Mol Psychiatry (2021) 26(12):7141–53. doi: 10.1038/s41380-021-01282-z
25
Zhou J Zhang X Hu J Qu R Yu Z Xu H et al . M(6)a demethylase Alkbh5 controls Cd4(+) T cell pathogenicity and promotes autoimmunity. Sci Adv (2021) 7(25):eabg0470. doi: 10.1126/sciadv.abg0470
26
Patil DP Pickering BF Jaffrey SR . Reading M(6)a in the transcriptome: M(6)a-binding proteins. Trends Cell Biol (2018) 28(2):113–27. doi: 10.1016/j.tcb.2017.10.001
27
Bechara R Amatya N Bailey RD Li Y Aggor FEY Li DD et al . The M(6)a reader Imp2 directs autoimmune inflammation through an il-17- and tnfα-dependent C/Ebp transcription factor axis. Sci Immunol (2021) 6(61):eabd1287. doi: 10.1126/sciimmunol.abd1287
28
Chen Y Zhao Y Chen J Peng C Zhang Y Tong R et al . Alkbh5 suppresses malignancy of hepatocellular carcinoma Via M(6)a-guided epigenetic inhibition of Lypd1. Mol Cancer (2020) 19(1):123. doi: 10.1186/s12943-020-01239-w
29
Chen XY Liang R Yi YC Fan HN Chen M Zhang J et al . The M(6)a reader Ythdf1 facilitates the tumorigenesis and metastasis of gastric cancer Via Usp14 translation in an M(6)a-dependent manner. Front Cell Dev Biol (2021) 9:647702. doi: 10.3389/fcell.2021.647702
30
Roignant JY Soller M . M(6)a in mrna: An ancient mechanism for fine-tuning gene expression. Trends Genet (2017) 33(6):380–90. doi: 10.1016/j.tig.2017.04.003
31
Guttman M Rinn JL . Modular regulatory principles of Large non-coding rnas. Nature (2012) 482(7385):339–46. doi: 10.1038/nature10887
32
Xue C Chu Q Zheng Q Jiang S Bao Z Su Y et al . Role of main rna modifications in cancer: N(6)-methyladenosine, 5-methylcytosine, and pseudouridine. Signal Transduct Target Ther (2022) 7(1):142. doi: 10.1038/s41392-022-01003-0
33
Ferragut Cardoso AP Banerjee M Nail AN Lykoudi A States JC . Mirna dysregulation is an emerging modulator of genomic instability. Semin Cancer Biol (2021) 76:120–31. doi: 10.1016/j.semcancer.2021.05.004
34
DeVeale B Swindlehurst-Chan J Blelloch R . The roles of micrornas in mouse development. Nat Rev Genet (2021) 22(5):307–23. doi: 10.1038/s41576-020-00309-5
35
Rupaimoole R Calin GA Lopez-Berestein G Sood AK . Mirna deregulation in cancer cells and the tumor microenvironment. Cancer Discovery (2016) 6(3):235–46. doi: 10.1158/2159-8290.Cd-15-0893
36
Quinn JJ Chang HY . Unique features of long non-coding rna biogenesis and function. Nat Rev Genet (2016) 17(1):47–62. doi: 10.1038/nrg.2015.10
37
Liu SJ Dang HX Lim DA Feng FY Maher CA . Long noncoding rnas in cancer metastasis. Nat Rev Cancer (2021) 21(7):446–60. doi: 10.1038/s41568-021-00353-1
38
Winkle M El-Daly SM Fabbri M Calin GA . Noncoding rna therapeutics - challenges and potential solutions. Nat Rev Drug Discovery (2021) 20(8):629–51. doi: 10.1038/s41573-021-00219-z
39
Zhang M Xin Y . Circular rnas: A new frontier for cancer diagnosis and therapy. J Hematol Oncol (2018) 11(1):21. doi: 10.1186/s13045-018-0569-5
40
Zhou WY Cai ZR Liu J Wang DS Ju HQ Xu RH . Circular rna: Metabolism, functions and interactions with proteins. Mol Cancer (2020) 19(1):172. doi: 10.1186/s12943-020-01286-3
41
Kristensen LS Jakobsen T Hager H Kjems J . The emerging roles of circrnas in cancer and oncology. Nat Rev Clin Oncol (2022) 19(3):188–206. doi: 10.1038/s41571-021-00585-y
42
Xue C Li G Zheng Q Gu X Bao Z Lu J et al . The functional roles of the Circrna/Wnt axis in cancer. Mol Cancer (2022) 21(1):108. doi: 10.1186/s12943-022-01582-0
43
Xue C Li G Lu J Li L . Crosstalk between circrnas and the Pi3k/Akt signaling pathway in cancer progression. Signal Transduct Target Ther (2021) 6(1):400. doi: 10.1038/s41392-021-00788-w
44
Dube U Del-Aguila JL Li Z Budde JP Jiang S Hsu S et al . An atlas of cortical circular rna expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci (2019) 22(11):1903–12. doi: 10.1038/s41593-019-0501-5
45
Yu J Xu QG Wang ZG Yang Y Zhang L Ma JZ et al . Circular rna Csmarca5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol (2018) 68(6):1214–27. doi: 10.1016/j.jhep.2018.01.012
46
Li H Trager LE Liu X Hastings MH Xiao C Guerra J et al . Lncexact1 and Dchs2 regulate physiological and pathological cardiac growth. Circulation (2022) 145(16):1218–33. doi: 10.1161/circulationaha.121.056850
47
Wang Y Wang P Zhang Y Xu J Li Z Li Z et al . Decreased expression of the host long-noncoding rna-gm facilitates viral escape by inhibiting the kinase activity Tbk1 Via s-glutathionylation. Immunity (2020) 53(6):1168–81.e7. doi: 10.1016/j.immuni.2020.11.010
48
Chen Q Wang H Li Z Li F Liang L Zou Y et al . Circular rna Actn4 promotes intrahepatic cholangiocarcinoma progression by recruiting Ybx1 to initiate Fzd7 transcription. J Hepatol (2022) 76(1):135–47. doi: 10.1016/j.jhep.2021.08.027
49
Fasolo F Jin H Winski G Chernogubova E Pauli J Winter H et al . Long noncoding rna miat controls advanced atherosclerotic lesion formation and plaque destabilization. Circulation (2021) 144(19):1567–83. doi: 10.1161/circulationaha.120.052023
50
Gu J Huang W Wang X Zhang J Tao T Zheng Y et al . Hsa-Mir-3178/Rhob/Pi3k/Akt, a novel signaling pathway regulates abc transporters to reverse gemcitabine resistance in pancreatic cancer. Mol Cancer (2022) 21(1):112. doi: 10.1186/s12943-022-01587-9
51
Ma YN Hong YG Yu GY Jiang SY Zhao BL Guo A et al . Lncrna Lbx2-As1 promotes colorectal cancer progression and 5-fluorouracil resistance. Cancer Cell Int (2021) 21(1):501. doi: 10.1186/s12935-021-02209-y
52
Chen F Li M Wang L . Lncrna Casc11 promotes hepatocellular carcinoma progression Via upregulation of Ube2t in a M(6)a-dependent manner. Front Oncol (2021) 11:772671. doi: 10.3389/fonc.2021.772671
53
Knuckles P Lence T Haussmann IU Jacob D Kreim N Carl SH et al . Zc3h13/Flacc is required for adenosine methylation by bridging the mrna-binding factor Rbm15/Spenito to the M(6)a machinery component Wtap/Fl(2)D. Genes Dev (2018) 32(5-6):415–29. doi: 10.1101/gad.309146.117
54
Liu J Yue Y Han D Wang X Fu Y Zhang L et al . A Mettl3-Mettl14 complex mediates mammalian nuclear rna N6-adenosine methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
55
Wen J Lv R Ma H Shen H He C Wang J et al . Zc3h13 regulates nuclear rna M(6)a methylation and mouse embryonic stem cell self-renewal. Mol Cell (2018) 69(6):1028–38.e6. doi: 10.1016/j.molcel.2018.02.015
56
Yue Y Liu J Cui X Cao J Luo G Zhang Z et al . Virma mediates preferential M(6)a mrna methylation in 3'utr and near stop codon and associates with alternative polyadenylation. Cell Discovery (2018) 4:10. doi: 10.1038/s41421-018-0019-0
57
Wang X Feng J Xue Y Guan Z Zhang D Liu Z et al . Structural basis of N(6)-adenosine methylation by the Mettl3-Mettl14 complex. Nature (2016) 534(7608):575–8. doi: 10.1038/nature18298
58
Śledź P Jinek M . Structural insights into the molecular mechanism of the M(6)a writer complex. Elife (2016) 5:e18434. doi: 10.7554/eLife.18434
59
Wang P Doxtader KA Nam Y . Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell (2016) 63(2):306–17. doi: 10.1016/j.molcel.2016.05.041
60
Schöller E Weichmann F Treiber T Ringle S Treiber N Flatley A et al . Interactions, localization, and phosphorylation of the M(6)a generating Mettl3-Mettl14-Wtap complex. Rna (2018) 24(4):499–512. doi: 10.1261/rna.064063.117
61
Patil DP Chen CK Pickering BF Chow A Jackson C Guttman M et al . M(6)a rna methylation promotes xist-mediated transcriptional repression. Nature (2016) 537(7620):369–73. doi: 10.1038/nature19342
62
Brown JA Kinzig CG DeGregorio SJ Steitz JA . Methyltransferase-like protein 16 binds the 3'-terminal triple helix of Malat1 long noncoding rna. Proc Natl Acad Sci U.S.A. (2016) 113(49):14013–8. doi: 10.1073/pnas.1614759113
63
Ma JZ Yang F Zhou CC Liu F Yuan JH Wang F et al . Mettl14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -Methyladenosine-Dependent primary microrna processing. Hepatology (2017) 65(2):529–43. doi: 10.1002/hep.28885
64
van Tran N Ernst FGM Hawley BR Zorbas C Ulryck N Hackert P et al . The human 18s rrna M6a methyltransferase Mettl5 is stabilized by Trmt112. Nucleic Acids Res (2019) 47(15):7719–33. doi: 10.1093/nar/gkz619
65
Pendleton KE Chen B Liu K Hunter OV Xie Y Tu BP et al . The U6 snrna M(6)a methyltransferase Mettl16 regulates Sam synthetase intron retention. Cell (2017) 169(5):824–35.e14. doi: 10.1016/j.cell.2017.05.003
66
Clark SJ Molloy PL . Smoke-induced changes to the epigenome provide fertile ground for oncogenic mutation. Cancer Cell (2017) 32(3):278–80. doi: 10.1016/j.ccell.2017.08.016
67
Jia G Fu Y Zhao X Dai Q Zheng G Yang Y et al . N6-methyladenosine in nuclear rna is a major substrate of the obesity-associated fto. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687
68
Zheng G Dahl JA Niu Y Fedorcsak P Huang CM Li CJ et al . Alkbh5 is a mammalian rna demethylase that impacts rna metabolism and mouse fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
69
Mauer J Sindelar M Despic V Guez T Hawley BR Vasseur JJ et al . Fto controls reversible M(6)Am rna methylation during snrna biogenesis. Nat Chem Biol (2019) 15(4):340–7. doi: 10.1038/s41589-019-0231-8
70
Mauer J Luo X Blanjoie A Jiao X Grozhik AV Patil DP et al . Reversible methylation of M(6)a(M) in the 5' cap controls mrna stability. Nature (2017) 541(7637):371–5. doi: 10.1038/nature21022
71
Huang H Weng H Chen J . M(6)a modification in coding and non-coding rnas: Roles and therapeutic implications in cancer. Cancer Cell (2020) 37(3):270–88. doi: 10.1016/j.ccell.2020.02.004
72
Haussmann IU Bodi Z Sanchez-Moran E Mongan NP Archer N Fray RG et al . M(6)a potentiates sxl alternative pre-mrna splicing for robust drosophila sex determination. Nature (2016) 540(7632):301–4. doi: 10.1038/nature20577
73
Li Y Sun Z Liu B Shan Y Zhao L Jia L . Tumor-suppressive mir-26a and mir-26b inhibit cell aggressiveness by regulating Fut4 in colorectal cancer. Cell Death Dis (2017) 8(6):e2892. doi: 10.1038/cddis.2017.281
74
Huang H Weng H Sun W Qin X Shi H Wu H et al . Recognition of rna N(6)-methyladenosine by Igf2bp proteins enhances mrna stability and translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
75
Wang X Zhao BS Roundtree IA Lu Z Han D Ma H et al . N(6)-methyladenosine modulates messenger rna translation efficiency. Cell (2015) 161(6):1388–99. doi: 10.1016/j.cell.2015.05.014
76
Wang X Lu Z Gomez A Hon GC Yue Y Han D et al . N6-Methyladenosine-Dependent regulation of messenger rna stability. Nature (2014) 505(7481):117–20. doi: 10.1038/nature12730
77
Kopp F Mendell JT . Functional classification and experimental dissection of long noncoding rnas. Cell (2018) 172(3):393–407. doi: 10.1016/j.cell.2018.01.011
78
Yan H Bu P . Non-coding rna in cancer. Essays Biochem (2021) 65(4):625–39. doi: 10.1042/ebc20200032
79
Qu S Yang X Li X Wang J Gao Y Shang R et al . Circular rna: A new star of noncoding rnas. Cancer Lett (2015) 365(2):141–8. doi: 10.1016/j.canlet.2015.06.003
80
Zhang Y Mao Q Xia Q Cheng J Huang Z Li Y et al . Noncoding rnas link metabolic reprogramming to immune microenvironment in cancers. J Hematol Oncol (2021) 14(1):169. doi: 10.1186/s13045-021-01179-y
81
Bridges MC Daulagala AC Kourtidis A . Lnccation: Lncrna localization and function. J Cell Biol (2021) 220(2):e202009045. doi: 10.1083/jcb.202009045
82
Nojima T Proudfoot NJ . Mechanisms of lncrna biogenesis as revealed by nascent transcriptomics. Nat Rev Mol Cell Biol (2022) 23(6):389–406. doi: 10.1038/s41580-021-00447-6
83
Ferrè F Colantoni A Helmer-Citterich M . Revealing protein-lncrna interaction. Brief Bioinform (2016) 17(1):106–16. doi: 10.1093/bib/bbv031
84
Yang H Hu Y Weng M Liu X Wan P Hu Y et al . Hypoxia inducible lncrna-cbslr modulates ferroptosis through M6a-Ythdf2-Dependent modulation of cbs in gastric cancer. J Adv Res (2022) 37:91–106. doi: 10.1016/j.jare.2021.10.001
85
Liu H Xu Y Yao B Sui T Lai L Li Z . A novel N6-methyladenosine (M6a)-dependent fate decision for the lncrna Thor. Cell Death Dis (2020) 11(8):613. doi: 10.1038/s41419-020-02833-y
86
Ni W Yao S Zhou Y Liu Y Huang P Zhou A et al . Long noncoding rna Gas5 inhibits progression of colorectal cancer by interacting with and triggering yap phosphorylation and degradation and is negatively regulated by the M(6)a reader Ythdf3. Mol Cancer (2019) 18(1):143. doi: 10.1186/s12943-019-1079-y
87
Huang J Wu S Barrera J Matthews K Pan D . The hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating yorkie, the drosophila homolog of yap. Cell (2005) 122(3):421–34. doi: 10.1016/j.cell.2005.06.007
88
Zhao B Li L Lei Q Guan KL . The hippo-yap pathway in organ size control and tumorigenesis: An updated version. Genes Dev (2010) 24(9):862–74. doi: 10.1101/gad.1909210
89
Guo PD Lu XX Gan WJ Li XM He XS Zhang S et al . Rarγ downregulation contributes to colorectal tumorigenesis and metastasis by derepressing the hippo-yap pathway. Cancer Res (2016) 76(13):3813–25. doi: 10.1158/0008-5472.Can-15-2882
90
Hong AW Meng Z Yuan HX Plouffe SW Moon S Kim W et al . Osmotic stress-induced phosphorylation by nlk at Ser128 activates yap. EMBO Rep (2017) 18(1):72–86. doi: 10.15252/embr.201642681
91
Moon S Kim W Kim S Kim Y Song Y Bilousov O et al . Phosphorylation by nlk inhibits yap-14-3-3-Interactions and induces its nuclear localization. EMBO Rep (2017) 18(1):61–71. doi: 10.15252/embr.201642683
92
Li Z Zhang J Liu X Li S Wang Q Di C et al . The Linc01138 drives malignancies Via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun (2018) 9(1):1572. doi: 10.1038/s41467-018-04006-0
93
Wang Y Lu JH Wu QN Jin Y Wang DS Chen YX et al . Lncrna linris stabilizes Igf2bp2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer (2019) 18(1):174. doi: 10.1186/s12943-019-1105-0
94
Yang X Zhang S He C Xue P Zhang L He Z et al . METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer (2020) 19(1):46. doi: 10.1186/s12943-020-1146-4
95
Zhang J Guo S Piao HY Wang Y Wu Y Meng XY et al . Alkbh5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncrna Neat1. J Physiol Biochem (2019) 75(3):379–89. doi: 10.1007/s13105-019-00690-8
96
Hu N Ji H . N6-methyladenosine (M6a)-mediated up-regulation of long noncoding rna Linc01320 promotes the proliferation, migration, and invasion of gastric cancer Via Mir495-5p/Rab19 axis. Bioengineered (2021) 12(1):4081–91. doi: 10.1080/21655979.2021.1953210
97
He H Wu W Sun Z Chai L . Mir-4429 prevented gastric cancer progression through targeting Mettl3 to inhibit M(6)a-caused stabilization of Sec62. Biochem Biophys Res Commun (2019) 517(4):581–7. doi: 10.1016/j.bbrc.2019.07.058
98
Müller S Glaß M Singh AK Haase J Bley N Fuchs T et al . Igf2bp1 promotes srf-dependent transcription in cancer in a M6a- and mirna-dependent manner. Nucleic Acids Res (2019) 47(1):375–90. doi: 10.1093/nar/gky1012
99
Lin S Liu J Jiang W Wang P Sun C Wang X et al . Mettl3 promotes the proliferation and mobility of gastric cancer cells. Open Med (Wars) (2019) 14:25–31. doi: 10.1515/med-2019-0005
100
Li H Zhang N Jiao X Wang C Sun W He Y et al . Downregulation of microrna-6125 promotes colorectal cancer growth through Ythdf2-dependent recognition of N6-Methyladenosine-Modified Gsk3β. Clin Transl Med (2021) 11(10):e602. doi: 10.1002/ctm2.602
101
Zhong L Liao D Zhang M Zeng C Li X Zhang R et al . Ythdf2 suppresses cell proliferation and growth Via destabilizing the egfr mrna in hepatocellular carcinoma. Cancer Lett (2019) 442:252–61. doi: 10.1016/j.canlet.2018.11.006
102
Zhang C Huang S Zhuang H Ruan S Zhou Z Huang K et al . Ythdf2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating Oct4 expression Via M6a rna methylation. Oncogene (2020) 39(23):4507–18. doi: 10.1038/s41388-020-1303-7
103
Chen J Sun Y Xu X Wang D He J Zhou H et al . Yth domain family 2 orchestrates epithelial-mesenchymal Transition/Proliferation dichotomy in pancreatic cancer cells. Cell Cycle (2017) 16(23):2259–71. doi: 10.1080/15384101.2017.1380125
104
Wang Y Buyse J Song Z Decuypere E Everaert N . Ampk is involved in the differential neonatal performance of chicks hatching at different time. Gen Comp Endocrinol (2016) 228:53–9. doi: 10.1016/j.ygcen.2016.02.008
105
Yue C Chen J Li Z Li L Chen J Guo Y . Microrna-96 promotes occurrence and progression of colorectal cancer Via regulation of the Ampkα2-Fto-M6a/Myc axis. J Exp Clin Cancer Res (2020) 39(1):240. doi: 10.1186/s13046-020-01731-7
106
Li Z Rana TM . Therapeutic targeting of micrornas: Current status and future challenges. Nat Rev Drug Discovery (2014) 13(8):622–38. doi: 10.1038/nrd4359
107
Nguyen TA Jo MH Choi YG Park J Kwon SC Hohng S et al . Functional anatomy of the human microprocessor. Cell (2015) 161(6):1374–87. doi: 10.1016/j.cell.2015.05.010
108
Chen X Xu M Xu X Zeng K Liu X Sun L et al . Retracted: Mettl14 suppresses crc progression Via regulating N6-Methyladenosine-Dependent primary mir-375 processing. Mol Ther (2020) 28(2):599–612. doi: 10.1016/j.ymthe.2019.11.016
109
Peng W Li J Chen R Gu Q Yang P Qian W et al . Upregulated Mettl3 promotes metastasis of colorectal cancer Via mir-1246/Spred2/Mapk signaling pathway. J Exp Clin Cancer Res (2019) 38(1):393. doi: 10.1186/s13046-019-1408-4
110
Sanger HL Klotz G Riesner D Gross HJ Kleinschmidt AK . Viroids are single-stranded covalently closed circular rna molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U.S.A. (1976) 73(11):3852–6. doi: 10.1073/pnas.73.11.3852
111
Xiao MS Ai Y Wilusz JE . Biogenesis and functions of circular rnas come into focus. Trends Cell Biol (2020) 30(3):226–40. doi: 10.1016/j.tcb.2019.12.004
112
Jeck WR Sorrentino JA Wang K Slevin MK Burd CE Liu J et al . Circular rnas are abundant, conserved, and associated with alu repeats. Rna (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
113
Chen LL Yang L . Regulation of circrna biogenesis. RNA Biol (2015) 12(4):381–8. doi: 10.1080/15476286.2015.1020271
114
Chen LL . The expanding regulatory mechanisms and cellular functions of circular rnas. Nat Rev Mol Cell Biol (2020) 21(8):475–90. doi: 10.1038/s41580-020-0243-y
115
Memczak S Jens M Elefsinioti A Torti F Krueger J Rybak A et al . Circular rnas are a Large class of animal rnas with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
116
Guo JU Agarwal V Guo H Bartel DP . Expanded identification and characterization of mammalian circular rnas. Genome Biol (2014) 15(7):409. doi: 10.1186/s13059-014-0409-z
117
Aufiero S Reckman YJ Pinto YM Creemers EE . Circular rnas open a new chapter in cardiovascular biology. Nat Rev Cardiol (2019) 16(8):503–14. doi: 10.1038/s41569-019-0185-2
118
Li J Sun D Pu W Wang J Peng Y . Circular rnas in cancer: Biogenesis, function, and clinical significance. Trends Cancer (2020) 6(4):319–36. doi: 10.1016/j.trecan.2020.01.012
119
Mehta SL Dempsey RJ Vemuganti R . Role of circular rnas in brain development and cns diseases. Prog Neurobiol (2020) 186:101746. doi: 10.1016/j.pneurobio.2020.101746
120
Zhou Z Sun B Huang S Zhao L . Roles of circular rnas in immune regulation and autoimmune diseases. Cell Death Dis (2019) 10(7):503. doi: 10.1038/s41419-019-1744-5
121
Fu L Jiang Z Li T Hu Y Guo J . Circular rnas in hepatocellular carcinoma: Functions and implications. Cancer Med (2018) 7(7):3101–9. doi: 10.1002/cam4.1574
122
Jiang Z Hou Z Liu W Yu Z Liang Z Chen S . Circular rna protein tyrosine kinase 2 (Circptk2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance. Bioengineered (2022) 13(1):810–23. doi: 10.1080/21655979.2021.2012952
123
Chi F Cao Y Chen Y . Analysis and validation of circrna-mirna network in regulating M(6)a rna methylation modulators reveals Circmap2k4/Mir-139-5p/Ythdf1 axis involving the proliferation of hepatocellular carcinoma. Front Oncol (2021) 11:560506. doi: 10.3389/fonc.2021.560506
124
Starke S Jost I Rossbach O Schneider T Schreiner S Hung LH et al . Exon circularization requires canonical splice signals. Cell Rep (2015) 10(1):103–11. doi: 10.1016/j.celrep.2014.12.002
125
Chen LL . The biogenesis and emerging roles of circular rnas. Nat Rev Mol Cell Biol (2016) 17(4):205–11. doi: 10.1038/nrm.2015.32
126
Zhang XO Wang HB Zhang Y Lu X Chen LL Yang L . Complementary sequence-mediated exon circularization. Cell (2014) 159(1):134–47. doi: 10.1016/j.cell.2014.09.001
127
Conn SJ Pillman KA Toubia J Conn VM Salmanidis M Phillips CA et al . The rna binding protein quaking regulates formation of circrnas. Cell (2015) 160(6):1125–34. doi: 10.1016/j.cell.2015.02.014
128
Ashwal-Fluss R Meyer M Pamudurti NR Ivanov A Bartok O Hanan M et al . Circrna biogenesis competes with pre-mrna splicing. Mol Cell (2014) 56(1):55–66. doi: 10.1016/j.molcel.2014.08.019
129
Aktaş T Avşar Ilık İ Maticzka D Bhardwaj V Pessoa Rodrigues C Mittler G et al . Dhx9 suppresses rna processing defects originating from the alu invasion of the human genome. Nature (2017) 544(7648):115–9. doi: 10.1038/nature21715
130
Li X Liu CX Xue W Zhang Y Jiang S Yin QF et al . Coordinated circrna biogenesis and function with Nf90/Nf110 in viral infection. Mol Cell (2017) 67(2):214–27.e7. doi: 10.1016/j.molcel.2017.05.023
131
Chen C Yuan W Zhou Q Shao B Guo Y Wang W et al . N6-Methyladenosine-Induced Circ1662 promotes metastasis of colorectal cancer by accelerating Yap1 nuclear localization. Theranostics (2021) 11(9):4298–315. doi: 10.7150/thno.51342
132
Tanaka T Okada R Hozaka Y Wada M Moriya S Satake S et al . Molecular pathogenesis of pancreatic ductal adenocarcinoma: Impact of mir-30c-5p and mir-30c-2-3p regulation on oncogenic genes. Cancers (Basel) (2020) 12(10):1106–19. doi: 10.3390/cancers12102731
133
Shukla K Sharma AK Ward A Will R Hielscher T Balwierz A et al . Microrna-30c-2-3p negatively regulates nf-κb signaling and cell cycle progression through downregulation of tradd and Ccne1 in breast cancer. Mol Oncol (2015) 9(6):1106–19. doi: 10.1016/j.molonc.2015.01.008
134
Salzman J Gawad C Wang PL Lacayo N Brown PO . Circular rnas are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One (2012) 7(2):e30733. doi: 10.1371/journal.pone.0030733
135
Chen RX Chen X Xia LP Zhang JX Pan ZZ Ma XD et al . N(6)-methyladenosine modification of Circnsun2 facilitates cytoplasmic export and stabilizes Hmga2 to promote colorectal liver metastasis. Nat Commun (2019) 10(1):4695. doi: 10.1038/s41467-019-12651-2
136
Chen P Li S Zhang K Zhao R Cui J Zhou W et al . N(6)-methyladenosine demethylase Alkbh5 suppresses malignancy of esophageal cancer by regulating microrna biogenesis and Rai1 expression. Oncogene (2021) 40(37):5600–12. doi: 10.1038/s41388-021-01966-4
137
Liu Z Wu K Gu S Wang W Xie S Lu T et al . A methyltransferase-like 14/Mir-99a-5p/Tribble 2 positive feedback circuit promotes cancer stem cell persistence and radioresistance Via histone deacetylase 2-mediated epigenetic modulation in esophageal squamous cell carcinoma. Clin Transl Med (2021) 11(9):e545. doi: 10.1002/ctm2.545
138
Zuo X Chen Z Gao W Zhang Y Wang J Wang J et al . M6a-mediated upregulation of Linc00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol (2020) 13(1):5. doi: 10.1186/s13045-019-0839-x
139
Liu Z Lu J Fang H Sheng J Cui M Yang Y et al . M6a modification-mediated Duxap8 regulation of malignant phenotype and chemotherapy resistance of hepatocellular carcinoma through mir-584-5p/Mapk1/Erk pathway axis. Front Cell Dev Biol (2021) 9:783385. doi: 10.3389/fcell.2021.783385
140
Zhang X Xu Y Qian Z Zheng W Wu Q Chen Y et al . Circrna_104075 stimulates yap-dependent tumorigenesis through the regulation of Hnf4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis (2018) 9(11):1091. doi: 10.1038/s41419-018-1132-6
141
Wu S Liu S Cao Y Chao G Wang P Pan H . Downregulation of Zc3h13 by mir-362-3p/Mir-425-5p is associated with a poor prognosis and adverse outcomes in hepatocellular carcinoma. Aging (Albany NY) (2022) 14(5):2304–19. doi: 10.18632/aging.203939
142
Wu A Hu Y Xu Y Xu J Wang X Cai A et al . Methyltransferase-like 3-mediated M6a methylation of Hsa_Circ_0058493 accelerates hepatocellular carcinoma progression by binding to yth domain-containing protein 1. Front Cell Dev Biol (2021) 9:762588. doi: 10.3389/fcell.2021.762588
143
Zhu L Zhu Y Han S Chen M Song P Dai D et al . Impaired autophagic degradation of lncrna Arhgap5-As1 promotes chemoresistance in gastric cancer. Cell Death Dis (2019) 10(6):383. doi: 10.1038/s41419-019-1585-2
144
Yan J Huang X Zhang X Chen Z Ye C Xiang W et al . Lncrna Linc00470 promotes the degradation of pten mrna to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun (2020) 521(4):887–93. doi: 10.1016/j.bbrc.2019.11.016
145
Zhu Y Zhou B Hu X Ying S Zhou Q Xu W et al . Lncrna Linc00942 promotes chemoresistance in gastric cancer by suppressing Msi2 degradation to enhance c-myc mrna stability. Clin Transl Med (2022) 12(1):e703. doi: 10.1002/ctm2.703
146
Meng X Deng Y He S Niu L Zhu H . M(6)a-mediated upregulation of Linc00857 promotes pancreatic cancer tumorigenesis by regulating the mir-150-5p/E2f3 axis. Front Oncol (2021) 11:629947. doi: 10.3389/fonc.2021.629947
147
He Y Yue H Cheng Y Ding Z Xu Z Lv C et al . Alkbh5-mediated M(6)a demethylation of Kcnk15-As1 inhibits pancreatic cancer progression Via regulating Kcnk15 and Pten/Akt signaling. Cell Death Dis (2021) 12(12):1121. doi: 10.1038/s41419-021-04401-4
148
Ye X Wang LP Han C Hu H Ni CM Qiao GL et al . Increased M(6)a modification of lncrna dbh-As1 suppresses pancreatic cancer growth and gemcitabine resistance Via the mir-3163/Usp44 axis. Ann Transl Med (2022) 10(6):304. doi: 10.21037/atm-22-556
149
Lu S Han L Hu X Sun T Xu D Li Y et al . N6-methyladenosine reader Imp2 stabilizes the Zfas1/Ola1 axis and activates the warburg effect: Implication in colorectal cancer. J Hematol Oncol (2021) 14(1):188. doi: 10.1186/s13045-021-01204-0
150
Zheng Y Wang Y Liu Y Xie L Ge J Yu G et al . N6-methyladenosine modification of Pttg3p contributes to colorectal cancer proliferation Via Yap1. Front Oncol (2021) 11:669731. doi: 10.3389/fonc.2021.669731
151
Hou P Meng S Li M Lin T Chu S Li Z et al . Linc00460/Dhx9/Igf2bp2 complex promotes colorectal cancer proliferation and metastasis by mediating Hmga1 mrna stability depending on M6a modification. J Exp Clin Cancer Res (2021) 40(1):52. doi: 10.1186/s13046-021-01857-2
152
Lin C Ma M Zhang Y Li L Long F Xie C et al . The N(6)-methyladenosine modification of Circalg1 promotes the metastasis of colorectal cancer mediated by the mir-342-5p/Pgf signalling pathway. Mol Cancer (2022) 21(1):80. doi: 10.1186/s12943-022-01560-6
153
Guo Y Guo Y Chen C Fan D Wu X Zhao L et al . Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: Involvement of mir-30c-5p/Tcf7 axis. Mol Cancer (2021) 20(1):93. doi: 10.1186/s12943-021-01372-0
154
Kelly RJ Ajani JA Kuzdzal J Zander T Van Cutsem E Piessen G et al . Adjuvant nivolumab in resected esophageal or gastroesophageal junction cancer. N Engl J Med (2021) 384(13):1191–203. doi: 10.1056/NEJMoa2032125
155
Thrumurthy SG Chaudry MA Thrumurthy SSD Mughal M . Oesophageal cancer: Risks, prevention, and diagnosis. Bmj (2019) 366:l4373. doi: 10.1136/bmj.l4373
156
Majumdar A Burban DJ Muretta JM Thompson AR Engel TA Rasmussen DM et al . Allostery governs Cdk2 activation and differential recognition of cdk inhibitors. Nat Chem Biol (2021) 17(4):456–64. doi: 10.1038/s41589-020-00725-y
157
Tang B Yang Y Kang M Wang Y Wang Y Bi Y et al . M(6)a demethylase Alkbh5 inhibits pancreatic cancer tumorigenesis by decreasing wif-1 rna methylation and mediating wnt signaling. Mol Cancer (2020) 19(1):3. doi: 10.1186/s12943-019-1128-6
158
Cui M Li Z . Downregulation of yap inhibits proliferation and induces apoptosis in eca-109 cells. Exp Ther Med (2018) 15(1):1048–52. doi: 10.3892/etm.2017.5492
159
Qu Y Zhang L Wang J Chen P Jia Y Wang C et al . Yes-associated protein (Yap) predicts poor prognosis and regulates progression of esophageal squamous cell cancer through epithelial-mesenchymal transition. Exp Ther Med (2019) 18(4):2993–3001. doi: 10.3892/etm.2019.7896
160
Zhang W Nandakumar N Shi Y Manzano M Smith A Graham G et al . Downstream of mutant kras, the transcription regulator yap is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal (2014) 7(324):ra42. doi: 10.1126/scisignal.2005049
161
Xue C Jia J Gu X Zhou L Lu J Zheng Q et al . Intratumoral bacteria interact with metabolites and genetic alterations in hepatocellular carcinoma. Signal Transduct Target Ther (2022) 7(1):335. doi: 10.1038/s41392-022-01159-9
162
Qian JY Gao J Sun X Cao MD Shi L Xia TS et al . Kiaa1429 acts as an oncogenic factor in breast cancer by regulating Cdk1 in an N6-Methyladenosine-Independent manner. Oncogene (2019) 38(33):6123–41. doi: 10.1038/s41388-019-0861-z
163
Chou J Lin JH Brenot A Kim JW Provot S Werb Z . Gata3 suppresses metastasis and modulates the tumour microenvironment by regulating microrna-29b expression. Nat Cell Biol (2013) 15(2):201–13. doi: 10.1038/ncb2672
164
Lan T Li H Zhang D Xu L Liu H Hao X et al . Kiaa1429 contributes to liver cancer progression through N6-Methyladenosine-Dependent post-transcriptional modification of Gata3. Mol Cancer (2019) 18(1):186. doi: 10.1186/s12943-019-1106-z
165
Sung H Ferlay J Siegel RL Laversanne M Soerjomataram I Jemal A et al . Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
166
Wang S Wang Y Zhang Z Zhu C Wang C Yu F et al . Long non-coding rna nron promotes tumor proliferation by regulating Alkbh5 and nanog in gastric cancer. J Cancer (2021) 12(22):6861–72. doi: 10.7150/jca.60737
167
Shriwas O Priyadarshini M Samal SK Rath R Panda S Das Majumdar SK et al . Ddx3 modulates cisplatin resistance in oscc through Alkbh5-mediated M(6)a-demethylation of Foxm1 and nanog. Apoptosis (2020) 25(3-4):233–46. doi: 10.1007/s10495-020-01591-8
168
Zhang C Samanta D Lu H Bullen JW Zhang H Chen I et al . Hypoxia induces the breast cancer stem cell phenotype by hif-dependent and Alkbh5-mediated M6a-demethylation of nanog mrna. Proc Natl Acad Sci U.S.A. (2016) 113(14):E2047–56. doi: 10.1073/pnas.1602883113
169
Gkountakos A Sartori G Falcone I Piro G Ciuffreda L Carbone C et al . Pten in lung cancer: Dealing with the problem, building on new knowledge and turning the game around. Cancers (Basel) (2019) 11(8):1141. doi: 10.3390/cancers11081141
170
Ren B Cui M Yang G Wang H Feng M You L et al . Tumor microenvironment participates in metastasis of pancreatic cancer. Mol Cancer (2018) 17(1):108. doi: 10.1186/s12943-018-0858-1
171
Goral V . Pancreatic cancer: Pathogenesis and diagnosis. Asian Pac J Cancer Prev (2015) 16(14):5619–24. doi: 10.7314/apjcp.2015.16.14.5619
172
Gillen S Schuster T Meyer Zum Büschenfelde C Friess H Kleeff J . Preoperative/Neoadjuvant therapy in pancreatic cancer: A systematic review and meta-analysis of response and resection percentages. PloS Med (2010) 7(4):e1000267. doi: 10.1371/journal.pmed.1000267
173
Springfeld C Jäger D Büchler MW Strobel O Hackert T Palmer DH et al . Chemotherapy for pancreatic cancer. Presse Med (2019) 48(3 Pt 2):e159–e74. doi: 10.1016/j.lpm.2019.02.025
174
Loveday BPT Lipton L Thomson BN . Pancreatic cancer: An update on diagnosis and management. Aust J Gen Pract (2019) 48(12):826–31. doi: 10.31128/ajgp-06-19-4957
175
Dekker E Tanis PJ Vleugels JLA Kasi PM Wallace MB . Colorectal cancer. Lancet (2019) 394(10207):1467–80. doi: 10.1016/s0140-6736(19)32319-0
176
Siegel RL Miller KD Jemal A . Cancer statistics, 2019. CA Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
177
Luo XJ Zhao Q Liu J Zheng JB Qiu MZ Ju HQ et al . Novel genetic and epigenetic biomarkers of prognostic and predictive significance in stage Ii/Iii colorectal cancer. Mol Ther (2021) 29(2):587–96. doi: 10.1016/j.ymthe.2020.12.017
178
Ganapathy-Kanniappan S Geschwind JF . Tumor glycolysis as a target for cancer therapy: Progress and prospects. Mol Cancer (2013) 12:152. doi: 10.1186/1476-4598-12-152
179
Abbaszadeh Z Çeşmeli S Biray Avcı Ç. Crucial players in glycolysis: Cancer progress. Gene (2020) 726:144158. doi: 10.1016/j.gene.2019.144158
180
Liu R Wang X Shen Y He A . Long non-coding rna-based glycolysis-targeted cancer therapy: Feasibility, progression and limitations. Mol Biol Rep (2021) 48(3):2713–27. doi: 10.1007/s11033-021-06247-7
181
Yang J Ren B Yang G Wang H Chen G You L et al . The enhancement of glycolysis regulates pancreatic cancer metastasis. Cell Mol Life Sci (2020) 77(2):305–21. doi: 10.1007/s00018-019-03278-z
182
Li Z Zhang H . Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci (2016) 73(2):377–92. doi: 10.1007/s00018-015-2070-4
183
Wen S Wei Y Zen C Xiong W Niu Y Zhao Y . Long non-coding rna Neat1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol Cancer (2020) 19(1):171. doi: 10.1186/s12943-020-01293-4
184
Yang D Qiao J Wang G Lan Y Li G Guo X et al . N6-methyladenosine modification of lincrna 1281 is critically required for mesc differentiation potential. Nucleic Acids Res (2018) 46(8):3906–20. doi: 10.1093/nar/gky130
185
Ye Q Ling S Zheng S Xu X . Liquid biopsy in hepatocellular carcinoma: Circulating tumor cells and circulating tumor DNA. Mol Cancer (2019) 18(1):114. doi: 10.1186/s12943-019-1043-x
186
Song W Anselmo AC Huang L . Nanotechnology intervention of the microbiome for cancer therapy. Nat Nanotechnol (2019) 14(12):1093–103. doi: 10.1038/s41565-019-0589-5
187
Zhang Y Li M Gao X Chen Y Liu T . Nanotechnology in cancer diagnosis: Progress, challenges and opportunities. J Hematol Oncol (2019) 12(1):137. doi: 10.1186/s13045-019-0833-3
188
Asadujjaman M Cho KH Jang DJ Kim JE Jee JP . Nanotechnology in the arena of cancer immunotherapy. Arch Pharm Res (2020) 43(1):58–79. doi: 10.1007/s12272-020-01207-4
189
Sousa AR Oliveira AV Oliveira MJ Sarmento B . Nanotechnology-based sirna delivery strategies for metastatic colorectal cancer therapy. Int J Pharm (2019) 568:118530. doi: 10.1016/j.ijpharm.2019.118530
190
Byeon Y Lee JW Choi WS Won JE Kim GH Kim MG et al . Cd44-targeting plga nanoparticles incorporating paclitaxel and fak sirna overcome chemoresistance in epithelial ovarian cancer. Cancer Res (2018) 78(21):6247–56. doi: 10.1158/0008-5472.Can-17-3871
191
Bruix J da Fonseca LG Reig M . Insights into the success and failure of systemic therapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol (2019) 16(10):617–30. doi: 10.1038/s41575-019-0179-x
192
Kudo M Finn RS Qin S Han KH Ikeda K Piscaglia F et al . Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet (2018) 391(10126):1163–73. doi: 10.1016/s0140-6736(18)30207-1
193
Llovet JM Montal R Sia D Finn RS . Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol (2018) 15(10):599–616. doi: 10.1038/s41571-018-0073-4
194
Kong H Sun J Zhang W Zhang H Li H . Long intergenic non-protein coding rna 1273 confers sorafenib resistance in hepatocellular carcinoma Via regulation of methyltransferase 3. Bioengineered (2022) 13(2):3108–21. doi: 10.1080/21655979.2022.2025701
195
Ansari D Tingstedt B Andersson B Holmquist F Sturesson C Williamsson C et al . Pancreatic cancer: Yesterday, today and tomorrow. Future Oncol (2016) 12(16):1929–46. doi: 10.2217/fon-2016-0010
196
Vincent A Herman J Schulick R Hruban RH Goggins M . Pancreatic cancer. Lancet (2011) 378(9791):607–20. doi: 10.1016/s0140-6736(10)62307-0
197
Kang YK Chin K Chung HC Kadowaki S Oh SC Nakayama N et al . S-1 plus leucovorin and oxaliplatin versus s-1 plus cisplatin as first-line therapy in patients with advanced gastric cancer (Solar): A randomised, open-label, phase 3 trial. Lancet Oncol (2020) 21(8):1045–56. doi: 10.1016/s1470-2045(20)30315-6
198
Das M . Neoadjuvant chemotherapy: Survival benefit in gastric cancer. Lancet Oncol (2017) 18(6):e307. doi: 10.1016/s1470-2045(17)30321-2
199
Fatma H Maurya SK Siddique HR . Epigenetic modifications of c-myc: Role in cancer cell reprogramming, progression and chemoresistance. Semin Cancer Biol (2022) 83:166–76. doi: 10.1016/j.semcancer.2020.11.008
200
van Hagen P Hulshof MC van Lanschot JJ Steyerberg EW van Berge Henegouwen MI Wijnhoven BP et al . Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med (2012) 366(22):2074–84. doi: 10.1056/NEJMoa1112088
201
Cardama GA Alonso DF Gonzalez N Maggio J Gomez DE Rolfo C et al . Relevance of small gtpase Rac1 pathway in drug and radio-resistance mechanisms: Opportunities in cancer therapeutics. Crit Rev Oncol Hematol (2018) 124:29–36. doi: 10.1016/j.critrevonc.2018.01.012
202
Masadah R Rauf S Pratama MY Tiribelli C Pascut D . The role of micrornas in the cisplatin- and radio-resistance of cervical cancer. Cancers (Basel) (2021) 13(5):1168. doi: 10.3390/cancers13051168
203
Liu L Zhang Z Zhou L Hu L Yin C Qing D et al . Cancer associated fibroblasts-derived exosomes contribute to radioresistance through promoting colorectal cancer stem cells phenotype. Exp Cell Res (2020) 391(2):111956. doi: 10.1016/j.yexcr.2020.111956
Summary
Keywords
gastrointestinal cancers, m6A, lncRNA, miRNA, circRNA
Citation
Xu Y, Yu X, Guo W and He Y (2023) Emerging role of interaction between m6A and main ncRNAs in gastrointestinal (GI) cancers. Front. Immunol. 14:1129298. doi: 10.3389/fimmu.2023.1129298
Received
21 December 2022
Accepted
06 February 2023
Published
17 February 2023
Volume
14 - 2023
Edited by
Qihui Shi, Fudan University, China
Reviewed by
Zhuo Wang, Fudan University, China; Chunying Wang, Fudan University, China
Updates
Copyright
© 2023 Xu, Yu, Guo and He.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenzhi Guo, fccguowz@zzu.edu.cn; Yuting He, fccheyt1@zzu.edu.cn
†These authors have contributed equally to this work
This article was submitted to Cancer Immunity and Immunotherapy, a section of the journal Frontiers in Immunology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.