- 1Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
- 2Immunology Translational Research Program, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore
- 3Singapore Immunology Network, Agency for Science, Technology and Research, Singapore, Singapore
- 4Department of Rheumatology, Allergy and Immunology, Tan Tock Seng Hospital, Singapore, Singapore
Antinuclear autoantibodies (ANA) are heterogeneous self-reactive antibodies that target the chromatin network, the speckled, the nucleoli, and other nuclear regions. The immunological aberration for ANA production remains partially understood, but ANA are known to be pathogenic, especially, in systemic lupus erythematosus (SLE). Most SLE patients exhibit a highly polygenic disease involving multiple organs, but in rare complement C1q, C1r, or C1s deficiencies, the disease can become largely monogenic. Increasing evidence point to intrinsic autoimmunogenicity of the nuclei. Necrotic cells release fragmented chromatins as nucleosomes and the alarmin HMGB1 is associated with the nucleosomes to activate TLRs and confer anti-chromatin autoimmunogenecity. In speckled regions, the major ANA targets Sm/RNP and SSA/Ro contain snRNAs that confer autoimmunogenecity to Sm/RNP and SSA/Ro antigens. Recently, three GAR/RGG-containing alarmins have been identified in the nucleolus that helps explain its high autoimmunogenicity. Interestingly, C1q binds to the nucleoli exposed by necrotic cells to cause protease C1r and C1s activation. C1s cleaves HMGB1 to inactive its alarmin activity. C1 proteases also degrade many nucleolar autoantigens including nucleolin, a major GAR/RGG-containing autoantigen and alarmin. It appears that the different nuclear regions are intrinsically autoimmunogenic by containing autoantigens and alarmins. However, the extracellular complement C1 complex function to dampen nuclear autoimmunogenecity by degrading these nuclear proteins.
1 Introduction
Our knowledge of autoimmune diseases mostly began with the discovery of the lupus erythematosus (L.E.) cell phenomenon (1). Historically, lupus was considered a skin disease (2). At the juncture of the 19th and 20th centuries, it was found to affect visceral organs with female preponderance (3, 4). L.E. cells are phagocytes in SLE patient bone marrows that contain, besides their endogenous nuclei, additional nuclear fragments (1). Research has found that in the presence of SLE patient sera, L.E. cells could form between normal phagocytes and nuclei (5, 6), and the serum L.E. factors were antinuclear autoantibodies (ANA) of heterogeneous specificities (Table 1) (8, 46). The 1971 and 1982 SLE diagnosis criteria included L.E. cells that are replaced in the 2012 and 2019 criteria by specific ANA (47–50).
ANA are commonly measured by indirect immunofluorescence (IIF) microscopy, giving an overall ANA titer and a fluorescent pattern (8, 51). In the current 2019 criteria, a minimal ANA titer of 1/80 is adopted as the entry criterion (50). Single ANA specificities are also adopted in the diagnosis of SLE and other systemic autoimmunity, e.g., anti-Smith (Sm) antigen (SLE), anti-Ro/SSA and anti-La/SSB (Sjogren’s syndrome or SjS), anti-U1-ribonucleoprotein (U1-snRNP) for mixed connective tissue disease (MCTD), and anti-topoisomerase I (Slc70) for systemic sclerosis (SSc) (8, 46, 52). Early evidence that ANA are pathogenic was the observation that antibodies for double-stranded DNA (anti-dsDNA) appeared in the blood before SLE disease flare and then precipitated out of blood circulation when dsDNA surged and active disease developed (53).
Patient sera can give heterogenous IIF patterns, e.g., homogeneous, speckled, nucleolar, centromere, or others (Table 1) (8, 51). These patterns are systemically named following an international consensus (7). Based on this classification scheme, each pattern is supported by one or more specific ANA-reactive antigens (9–13, 15–20, 25, 26, 28–40, 42–45). The homogeneous pattern is the most common, followed by the speckled and nucleolar patterns (51). Within the nucleolus, ANA can stain homogeneous, clumpy, or punctate patterns. As detailed later, these nucleolar IIF patterns correspond to three distinct nucleolar regions, i.e., the granular component (GC, homogeneous), the dense fibrillar component (DFC, clumpy), and the fibrillar center (FC, punctate) (54–56). Each nucleolar IIF pattern reflects specific antigens targeted by the patient ANA. While certain antigen-specific ANA show disease specificity such as anti-Sm antigen for SLE, IIF patterns are generally shared among different diseases (57). The prevalence of each IIF pattern can vary depending on the study populations, but the homogeneous and speckled patterns consistently dominate these patterns followed by the nucleolar pattern (e.g., 51, 57, 58).
2 The functions of the nucleoli
A nucleolus is formed organically around one or more actively transcribed rRNA genes (56). This includes pre-rRNA transcription by RNA polymerase I (Pol I), its processing by U3-snoRNPs and other snoRNPs, and mature rRNA assembly with 79-80 ribosomal proteins into the 40S and 60S pre-ribosomes (Figure 1) (61, 62). Ribosomes are then transported into the cytoplasm for protein translation (63). An animal cell nucleus usually contains 1-3 nucleoli but faster-growing cells, e.g., cancer cells, have larger and more numerous nucleoli, making rRNA transcription a target in cancer therapy (64).
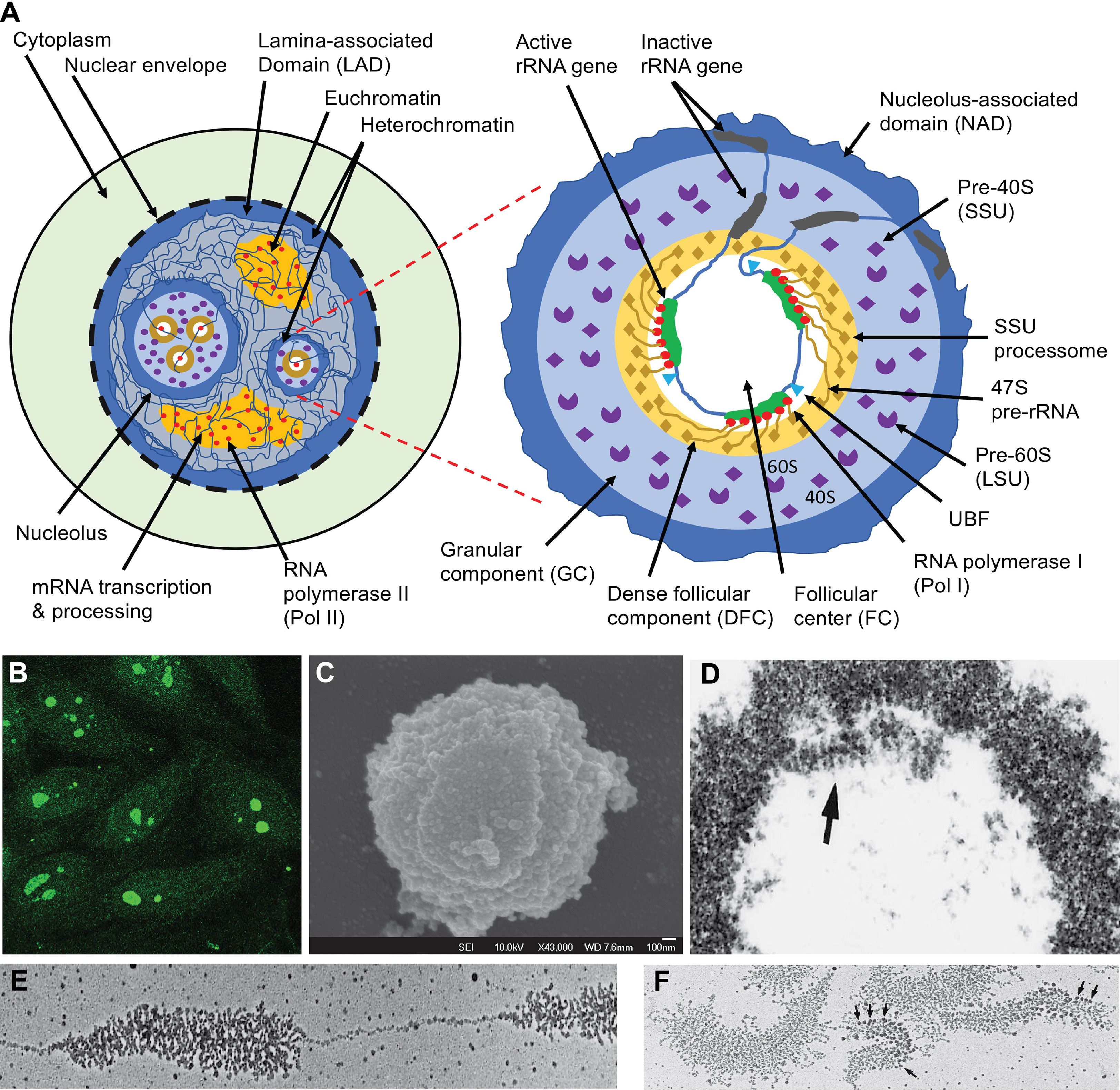
Figure 1 The nucleus, nucleolus, and ribosome biogenesis. (A) Schematic illustration of functional nuclear regions. The left panel highlights the nucleus which is partitioned from the cytoplasm with the nuclear envelope which rests in a dense layer of peripheral nuclear heterochromatin, which is also known as lamina-associated domain (LAD). LAD functions as a major nuclear chromatin scaffold. The nucleoli are distinct nuclear regions that are surrounded by a layer of dense heterochromatin. Between this heterochromatin are loose euchromatin regions where mRNA is transcribed by Pol II and processed by complex machinery. The right panel highlights the structure of a nucleolus. The dense layer of heterochromatin that cover each nucleolus is also known as the nucleolus-associated domain (NAD). Inside the enclosed nucleolar region, there are three distinct regions. The rRNA genes and the transcription machinery (Pol I, UBF, etc.) are localized in the follicular center (FC). The rRNA genes are transcribed; the transcripts (47S pre-rRNA) and their processing machinery form the dense follicular component (DFC), and the finished transcripts complete most of their assembly with 79-80 ribosomal proteins (r-proteins) into the 40S small subunit (SSU) and 60S large subunit (LSU) of ribosomes in the large granular component (GC). (B) Some SLE patients develop ANA which predominantly reacts with the nucleoli (7). (C) Nucleoli can be isolated from the nucleus through sonication, which breaks the chromatin connections between the nucleolar surface heterochromatin layer and the rest of the chromatin network. The image is a nucleolus viewed by scanning electron microscopy (59). (D) Electron micrograph of nucleolus isolated from the locusta oocytes with the arrow pointing to the DFC region. (E) Electron micrograph of a spread locusta oocyte nucleolus to show the tandem rRNA genes and the ~ 100 pre-rRNA transcripts that stem from each rRNA gene. (F) Electron micrographs of multiple rRNA genes and their transcripts. Arrows point to the rRNA processing machinery corresponding to the SSU processomes. Panels (D-F) are reproduced with permission from Scheer et al. (60).
The rRNA genes exist variably in many copies in each eukaryotic cell (56, 65). In human cells, the number of rRNA genes can also vary substantially among individuals (315 ± 104) (66, 67), being tandemly clustered head-to-tail on the short arms of the five acrocentric chromosomes (i.e., chromosomes 13, 14, 15, 21, and 22) (65). During mitosis, rRNA transcription ceases, and the majority of the rRNA processing machinery disperses, leaving only residual transcription machinery on the rRNA genes to form the ‘seed’ nucleolar organizer regions (NOR) (56, 65, 68). Some dispersed nucleolar proteins relocate to the surface cortexes of mitotic chromosomes (69, 70). When cells exit mitosis and the rRNA genes resume transcription, NORs expand de novo into active nucleoli (68). In these interphase cells, in situ NOR-like structures can be induced by inhibiting rRNA transcription (68, 71, 72).
The nucleolus also functions as an inner nuclear scaffold for the chromatin network. The peripheral nuclear scaffold is provided by the nuclear lamina (73), which assembles a dense layer of nuclear surface heterochromatin known as the lamina-associated domain (LAD) (74). Each nucleolus is also surrounded by a dense layer of heterochromatin known as nucleolus-associated domain (NAD) (75, 76). These are transcriptionally inactive chromatin regions that are important in chromatin organization or compartmentalization (77, 78).
Thirdly, the nucleolus may also exhibit multiple other functions (79). Many molecules transit through the nucleolus during cellular stress, e.g., viral infection (80), metabolic disruption (81), and UV stimulation (82). Nucleolus-related functions are unknown for most of these proteins (83).
3 The structure of the nucleolus
Nucleoli are dense and visible under light microscopes. By transmission electron microscopy, three distinct regions are found in each nucleolus: one or more FC regions each surrounded by a dense layer of DFC (54, 55), and these are embedded in a greater GC region that borders the outer nucleoplasm through a heterochromatin rim (Figure 1A) (56). The FC region contains the rRNA genes and the RNA polymerase I (Pol I) transcription machinery, including a key transcription factor, the upstream binding factor (UBF) (84–87). An active rRNA gene is simultaneously transcribed by approximately 100 Pol I and, therefore, many pre-rRNA transcripts of varying lengths stem from each active rRNA gene, like tree brunches (Figures 1D–F) (60, 88, 89). At the 5’ end of each pre-rRNA transcript, a complex machinery is attached that processes the transcript into mature 28S, 18S, and 5.8S rRNAs (Figures 1A, F) (54, 90, 91). These pre-rRNAs and their processing machinery form the DFC region. In the GC region, the processed rRNAs assemble with ribosome proteins (r-proteins) to form the 40S and 60S ribosome subunits, and these are transported to the cytoplasm (61, 62).
3.1 The FC region
In this nucleolar region, the rRNA genes are constitutively associated with UBF and the Pol I transcription machinery which, in quiescence, form NORs but they expand into nucleoli during active rRNA transcription (56). The 43-Kb human rRNA gene is first transcribed into a 47S pre-rRNA (92), which is then processed in DFC into mature 18S, 5.8S, and 28S rRNA for assembly with r-proteins (56, 93–95). UBF is a master organizer for these tandem rRNA genes. It binds to an upstream control element (UCE) situated at -156 to -107 bp of each rRNA gene promoter region to initiate the Pol I holoenzyme formation (84). Besides, UBF also binds broadly to other regions in the rRNA gene and organizes rRNA gene configuration with a histone-like function (86, 87).
3.2 The DFC region
The 47S pre-rRNA is simultaneously processed during transcription which includes methylation, pseudouridylation, and cleavage (56, 61, 62, 95). When nucleoli were isolated from the oocytes, they spread like tandem ‘Christmas trees’ along the rRNA gene ‘stem’ under the electron microscope (60, 88, 89). The 5’ processing machinery, including the ribosome small subunit (SSU) processome, were viewed as terminal balls (89). U3-snoRNPs are key SSU elements that methylate and pseudouridylate specific bases in rRNA (Figure 2) (96, 98, 99).
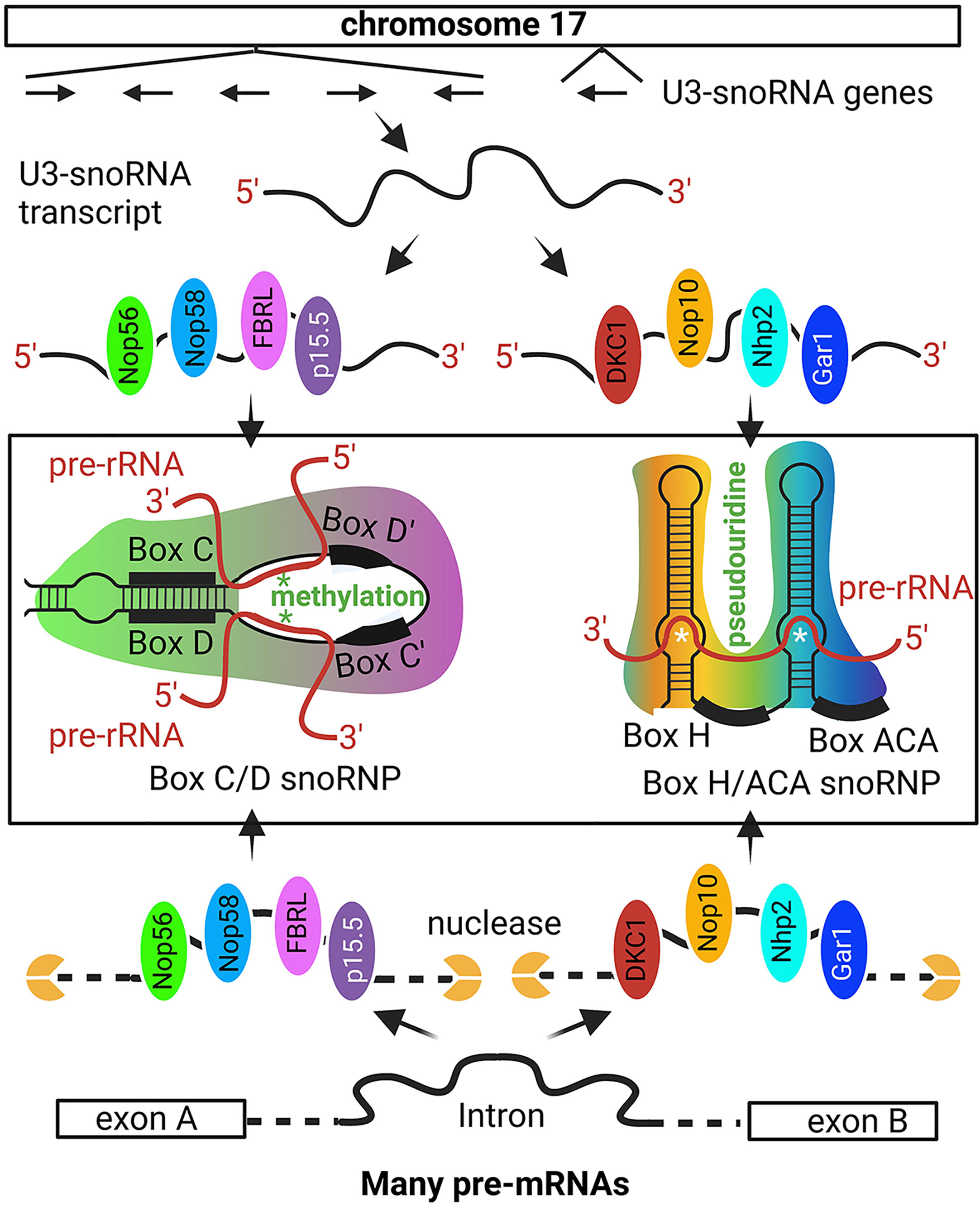
Figure 2 Non-intronic human U3-snoRNA genes on chromosome 17. The majority of snoRNAs are derived from pre-mRNA introns but a small number of U3-snoRNA genes with Pol II promoters are present on chromosome 17. The six snoRNA transcribed from chromosome 17 are bound by distinct protein sets, i.e., the p15.5, Nop56, Nop58, and FBRL set or the Nhp2, Nop10, Gar1, and DKC1 set, to form the C/D and H/ACA box U3 snoRNP, respectively. The C/D box U3 snoRNA binds to specific sequences on pre-rRNA for FBRL to methylate rRNA at specific nucleotides. The H/ACA U3 snoRNA binds to selected pre-rRNA sequences for DKC1 to convert specific uridine into pseudouridine. Besides the six snoRNA genes on chromosome 17, most other snoRNAs are derived from intron sequences spliced out from pr-mRNA. Some intron sequences have the specific features to recruit the p15.5, Nop56, Nop58, and FBRL protein set or the Nhp2, Nop10, Gar1, and DKC1 protein set, which protect these intron regions from nuclease degradation and ultimately form the snoRNPs [Ref (96, 97)]. '*' site of methylation or pseudouridine generation.
U3 snoRNPs include two distinct groups, i.e., box C/D and box H/ACA snoRNPs (96, 100, 101). A box C/D snoRNP contains a guide C/D snoRNA and four core proteins, i.e., SNU13 (NHP2L1), NOP56, NOP58, and fibrillarin (FBRL), and a H/ACA box snoRNP contains a H/ACA box snoRNA and four different core proteins, i.e., NOP10, GAR1, NHP2, and DKC1 (Figure 2) (96, 97). In the C/D box snoRNP, FBRL is a ribose 2’-O-methyltransferase, and in H/ACA snoRNPs, DKC1 is a pseudouridine synthase. The guide RNAs target the SnoRNPs to specific pre-rRNA sites so that specific nucleotides are methylated or specific uridine is converted into pseudouridine (96, 100, 102, 103). FBRL is an autoantigen in SSc patients (104).
Most snoRNAs originate from the introns of pre-mRNAs (97, 105–107), with few being transcribed from their own promoters (Figure 2) (108). snoRNAs are 60-170 bp RNA fragments and more than 1,000 have been predicted in the human genome (100–102). During pre-mRNA splicing, some introns are protected by snoRNA core proteins from exonuclease degradation, and these are further processed into mature snoRNPs (109, 110). Each C/D box or H/ACA box snoRNA is protected by four core proteins (Figure 2) (96, 107, 111).
3.3 The GC region
While the FC region is defined by the tandem rRNA genes (56, 84) and the DFC region is defined by the 47S pre-rRNA (88, 89, 112), a defining scaffold for the GC region is not apparent. NCL and NPM1 are highly abundant in the GC region which could be part of the scaffold (113, 114). The processed rRNAs assemble with r-proteins in the GC region (56, 75, 76). NCL facilitates SSU docking on pre-rRNA (115). NPM1 is a molecular chaperone of the nucleolus (116). Both NCL and NPM1 are autoantigens.
4 ANA, DNA, and RNA
The homogeneous IIF pattern is largely attributed to ANA binding to the chromatin network, e.g., dsDNA or histones (8). The speckled pattern corresponds to sites of mRNA transcription and processing (117, 118). The nucleolus accommodates ribosome biogenesis (56, 63). For these nuclear regions to elicit self-reactive antibodies, they inevitably involve aberrant innate and adaptive immune responses that lead to B cell production of class-switched IgG class ANA (119). In a healthy individual, 5-20% of peripheral naïve B cells are likely to be self-reactive or polyreactive (120, 121). These B cells can become pathogenic ANA-producing B cells in SLE patients (122). This requires nucleus-reactive CD4 T cell help for which adjuvant signals are necessary.
4.1 DNA
In the nucleus, DNA is primarily embedded in the chromatins configurated by histones and additional non-histone DNA-binding proteins. High mobility group box 1 (HMGB1) is a major non-histone DNA-binding protein, and it is also an alarmin that activates innate immunity through Toll-like receptors (TLRs) (123, 124). HMGB1 can be secreted by live cells or passively released by necrotic cells (123). When it is released from secondary necrotic cells in association with fragmented chromatins (nucleosomes), it confers immunogenicity to these known nuclear autoantigens (125).
4.2 mRNA
While the nucleolar DFC regions are formed from pr-rRNA and its processomes, the speckled regions are formed from pre-mRNA and its processing machinery, e.g., the two ribonucleoprotein complexes Smith antigen (Sm) and SSA/Ro. Purified Sm antigen can induce self-reactive autoantibodies in mice without additional adjuvant (126). This is because its U1-snRNA element is an endogenous adjuvant that activates TLR7 (127). U1-snRNA itself is also an autoantigen (126). U1-snRNP can activate the NOD-like receptor family, pyrin domain-containing 3 (NLRP3) inflammasomes (128). The snRNA components in SSA/Ro60 also activate TLR7 (129). These snRNA alarmins could confer sufficient autoimmunogenecity to these RNPs to cause B and T cell activation.
4.3 rRNA
In the nucleolus, ANA target mostly snoRNP components (21, 59, 130), but snoRNA has not been reported as autoantigens or adjuvants (127, 128). U3-snoRNPs are dominant snoRNPs in the nucleolus (96, 100, 101). Recently, two of the U3-snoRNP protein components have been found to contain alarmin or adjuvant activities, i.e., FBRL and GAR1 (Figure 2) (131). Their alarmin activities are conferred by their GAR/RGG motifs which were first discovered in NCL to activate TLR2 and TLR4 (131). The nucleolus contains the most numerous nuclear autoantigens, and nucleolus-reactive naïve B cells are also prevalent in healthy individuals (120, 121). Whether NCL, FBRL, and GAR1 confer sufficient immunogenicity to nucleolar antigens to induce self-reactive antibodies remains to be determined (131). Some transit extranucleolar molecules could also confer nucleolus autoimmunogenecity (83). For example, the EBV virus appears to confer autoimmunogenicity to the speckled region through cross-reactivity with SSA/Ro, Sm, and DNA (132–135).
5 Antinucleolar autoantibodies (ANoA)
ANoA are frequently found in SSc or scleroderma patients (21, 130, 136, 137). However, they are not sufficiently specific for SSc diagnosis (14). For example, ANoA for Th/To and U3-snoRNP are also developed in other autoimmune diseases (138, 139). ANoA are also prevalent in SLE and SjS patients (48, 49, 140). ANoA frequently target ribonucleoprotein (RNP) or protein complexes (130). NCL, NPM1, and UBF are exceptions (21, 130).
NCL is not a well-studied autoantigen, but its autoantigenicity was shown on a 25-autoantigen array study in which NCL was the 4th most prominent SLE patient autoantigen following dsDNA, ssDNA, and Ro-52/SSA (22). In TLR7hi SLE patients, NCL was the most prominent protein autoantigen after dsDNA and ssDNA (22). NZBxW F1 and MRL/lpr mice spontaneously develop SLE following aging. In these mice, NCL-reactive antibody was detected early before other common autoantibodies (141), implying that NCL could induce its self-reactive antibody. This view is supported by its intramolecular GAR/RGG alarmin motif (131). Likewise, FBRL could also induce its self-reactive antibodies.
NPM1-reactive ANA develop in SSc (23), SLE (142), and various other systemic autoimmune diseases (143, 144). UBF is mostly targeted by autoantibodies in SSc patients (27, 145, 146). Some cancer patients develop ANA that most consistently target NPM1 and UBF (147–149). Coilin is the master organizer of Coiled bodies and is also a well-known autoantigen (41, 150, 151). snRNPs and snoRNPs mature in these small nuclear bodies before being released to the nucleoplasm and nucleoli, respectively (150). Coiled bodies are often conjunct to nucleoli and therefore coilin is also a nucleolar autoantigen (152).
Nucleolar exosomes cleave pre-rRNA during assembly with r-proteins and most members of these protein complexes are autoantigens (24, 153, 154). RNase P and MRP are abundant autoantigenic RNPs in the nucleolus (155, 156). Approximately 60% of nucleolar autoantigens are snoRNPs and the remaining 40% are proteins like NCL, NPM1, and UBF (139). Some nucleolar U8 and U22 snoRNPs are autoantigenic (139). rRNA is also targeted by autoantibodies in MRL/lpr mice and some SLE patients (157, 158). The abundance of nucleolar autoantigens and endogenous alarmins make these nuclear regions potential initiators in ANA production.
6 Genetic predisposition in ANA induction
ANA are a hallmark of SLE (50), making this disease a suitable model for dissecting the molecular causes of these autoantibodies. However, most SLE patients present a polygenic disease for which more than 50 risk genes or non-coding loci have been identified albeit they mostly represent weak SLE risks with low SLE specificity (159–168). These risk genes mostly represent immunological pathways broadly underlying infectious and inflammatory diseases (Figure 3). We selected 13 published SLE risk gene sets for gene ontogeny (GO) analysis (165–177), and found that in the most SLE-specific gene sets, SLE was only ranked the 2nd most significant pathway following the Staphycoccous aureus infection pathway (Figure 3A). The next highest ranking for SLE (5th) was found in the `JA 2015’ gene set (Figure 3B). SLE was ranked outside the top 10 pathways in seven gene sets (Figures 3C, D). With the `COI 2006’ gene set, SLE was not identified as a relevant pathway (Figure 3C).
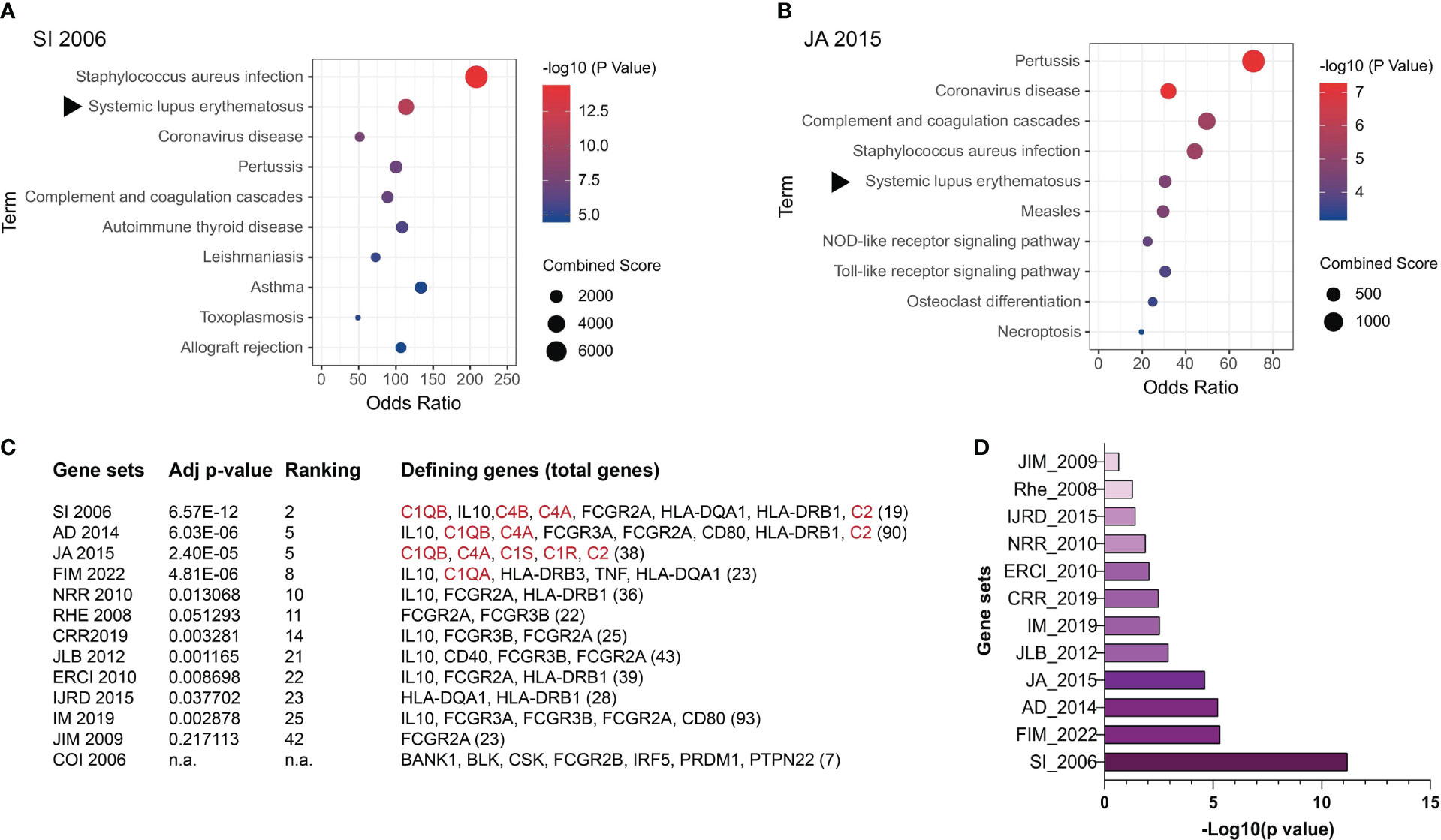
Figure 3 Genetic contributions to SLE pathogenesis. SLE risk genes have been identified based on evidence obtained through genome-wide association studies (GWAS), case reports, and other methods. Here SLE risk gene sets were extracted from 13 articles and their relevance to SLE was assessed through gene ontogeny (GO) analysis. (A) GO analysis of the SI 2006’ SLE risk gene set (165). (B) GO analysis of the `JA 2015’ SLE risk gene set (169). The rest of the gene sets included in this study are AD 2014 (170), FIM 2022 (171), NRR 2010 (172), RHE 2008 (167), CRR 2019 (173), JLB 2012 (174), ERCI 2010 (175), IJRD 2015 (166), IM 2019 (168), JIM 2009 (176), and COI 2006 (177). (C) Relevance of SLE as a disease to the 13 SLE risk gene sets analyzed. Confidence in the level of SLE relevance is indicated by the adjusted p values. The subgroup of SLE risk genes that were identified to derive the p values and SLE ranking positions among other relevant pathways are listed with the total number of SLE risk genes in each set being included at the end of the gene list in the bracket. Complement genes are highlighted in red. n.a. SLE was not identified as one pathway in the COI 2006 gene set. (D) Adjusted p values of the selected SLE risk gene sets except for the COI 2006 gene set.
An important observation was that the four gene sets in which SLE was ranked higher than the 10th position all contained one or more complement proteins, i.e., C1q, C1r, C1s, C4, and C2 (178, 179). The remaining nine gene sets all lacked complement genes.
C1Q, C1R, C1S, and C4 deficiencies are rare, but they often cause monogenic SLE (164, 180–182). Among these strong SLE risk genes, C1q and the two serine proteases C1r and C1s exist as a pentameric C1 complex (C1qC1r2C1s2) (178, 179). When C1q binds to antibodies in immune complexes, it activates C1r and C1s, and the C1s protease then cleaves C4 to trigger the complement classical pathway (183, 184). In SLE, immune complexes are formed between ANA and nuclear antigens which trigger C1-mediated complement activation and inflammatory tissue injuries (Figure 4). The fact that C1 deficiency causes ANA production and SLE pathogenesis was for a long time considered a paradox until research found that C1q not only binds to immune complexes but also binds to apoptotic cells (185).
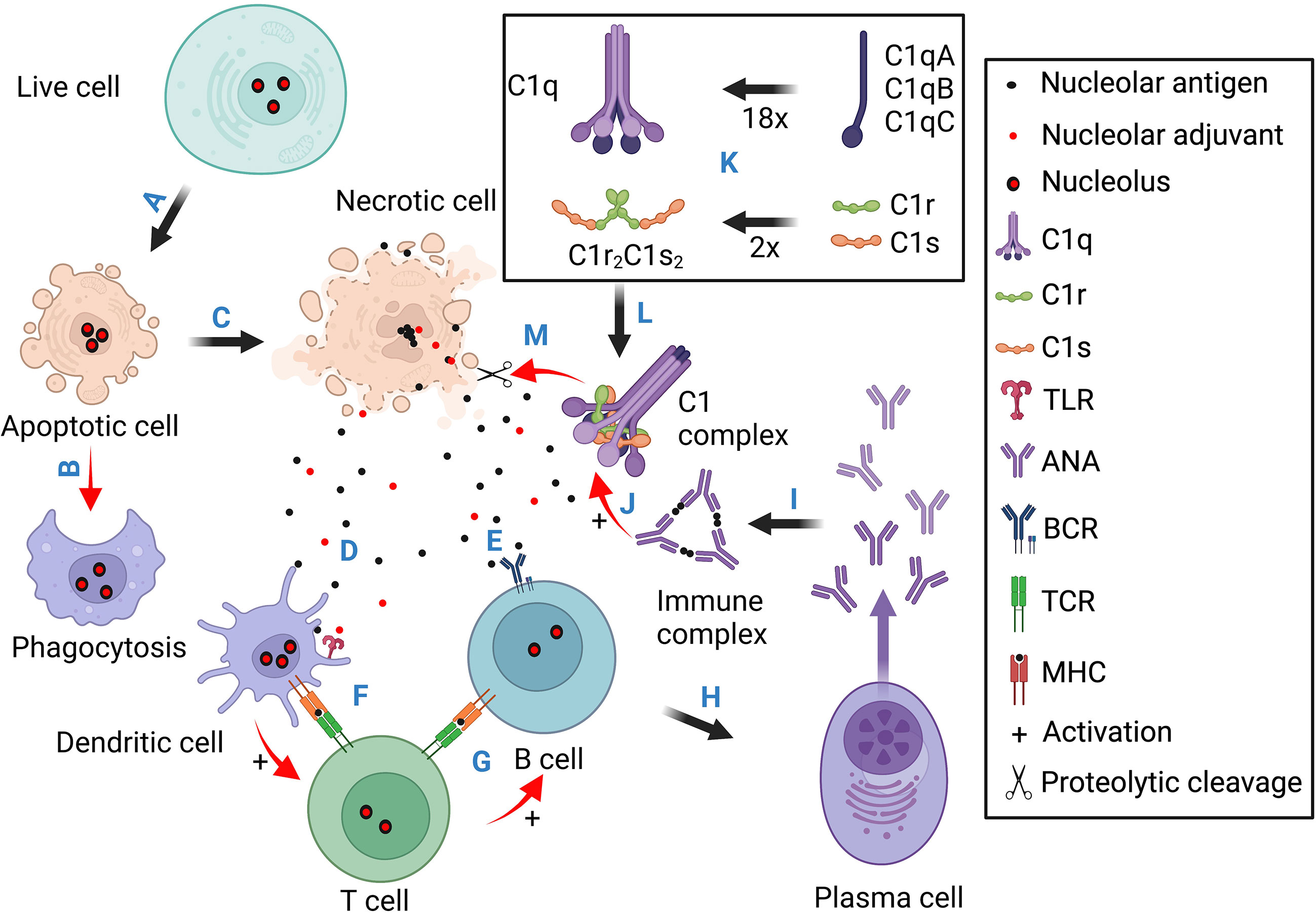
Figure 4 Schematic illustration for the pathogenic contributions of necrotic cell death, the nucleoli, and complement deficiency. This diagram contains four sections. Section 1 (A, B) stresses that normal apoptotic cells are cleared through phagocytosis without eliciting innate and adaptive immune responses. Section 2 (C-I) illustrates the scenario of necrotic cell death. The released nuclear antigens and alarmins activate T cells through dendritic cells which help antigen activation of B cell differentiation into ANA-producing plasma cells. (J) Immune complexes formed between ANA and nuclear. antigens activate Fc receptor- (not shown) and C1/complement-mediated inflammatory tissue injuries. Section 3 (K) illustrates C1q assembly from 18 polypeptide chains and its association with two C1r and two C1s to form the C1 complex. Section 4 (L, M) shows that after C1q binds to necrotic cell debris such as the nucleoli, it activates C1r and C1s into active proteases which then dismantle the exposed nuclear antigens and alarmins to reduce ANA induction. Basically, apoptotic cells are cleared in silence. Necrotic cells can expose both antigens and adjuvants to induce ANA production. When complement C1 is functionally intact, it can degrade nucleolar autoantigens and alarmins (e.g., NCL, FBRL, and GAR1) to reduce ANA induction and C1 deficiency, therefore, causing antinuclear autoimmunity.
7 The dead cell-C1 axis in SLE pathogenesis
The formation of L.E. cells in SLE patients reflects excessive necrotic cell death in the patients, the accumulation of naked nuclei, and nuclear opsonization by ANA for phagocytosis (1, 186). The surge of blood DNA antigen during SLE disease flare also suggests necrotic cell accumulation (53). This status could result from excessive cell death or impaired phagocytic clearance of dying cells (187, 188). For example, necrotic cells release nucleosomes which are rendered autoimmunogenic by the alarmin HMGB1 (125). In mice, injection of UV-induced syngeneic apoptotic cells can cause ANA production (189). This could be partly explained by the ready release of autoantigens and alarmins by UV-induced dead cells, e.g., NCL, NPM1, HMGB1, and FBRL (131).
In 1997, C1q was reported to bind to apoptotic cells via the blebs (185). Subsequent studies focused on the hypothesis that C1q opsonizes apoptotic cells to enhance phagocytosis and regulate phagocyte responses (190, 191). Apoptotic cell disposal is mediated through multiple phagocytic pathways and that mediated by C1q is not dominant (192). On the other hand, C1q exists as a pentameric C1qC1r2C1s2 complex (178, 179), and how C1r/C1s deficiency also leads to monogenic SLE, like C1q deficiency, is not explained by the phagocytosis hypothesis (164, 181, 182). Recent studies suggest that C1r/C1s degrade nuclear autoantigens and alarmin proteins that are exposed by dead cells and bound by C1q.
On necrotic cells, C1q binding is not limited to the surface as it also binds intensely to the nucleoli (Table 1) (21, 130, 193). This activates C1r/C1s into active proteases which cleave numerous nucleolar proteins (59). In the complement system, C1s only cleaves three substrate proteins, but with a peptide library, C1s was found to cleave non-complement peptides that predicted many intracellular protein substrates such as HMGB1 (194, 195). HMGB1 can be released by necrotic cells or secreted by live cells, and it is indeed cleaved by C1s (195). The nucleolar autoantigens NCL and NPM1 and additional other proteins are also cleaved by C1 proteases (59, 193). This makes the C1 complex an extracellular surveillance mechanism over dead cell accumulation, and it functions through phagocytosis and proteolytic dismantling of autoantigens and alarmins to avoid nuclear autoimmunity (59). This helps explain why C1q, C1r, or C1s deficiency often causes monogenic SLE (181, 196).
8 Nucleolar autoimmunogenicity
The strong nucleolar autoantigenicity is characterized by the numerous autoantigens in this nuclear region, and the nucleolus is often the sole ANA-targeted region (51). With isolated nucleolar, nucleoplasmic, and cytoplasmic fractions, nucleolar proteins were found most frequently targeted by SLE patient ANA (59). Besides SLE, hepatocellular carcinoma patients also develop ANA that persistently target nucleolar proteins (148). This is not surprising for the large number of autoantigens in the nucleoli (Table 1) (21, 130). This is further explained by the prevalent (5-20%) nucleus-reactive naïve B cells in healthy individuals that express prominent nucleolus-reactive antigen receptors (120–122). When necrotic cells accumulate, the nucleolar antigens and alarmins could activate these B cells into ANA-producing B cells (122). This has been reported for the major autoantigens in the speckled region, i.e., U1-snRNPs, in which the U1-snRNAs were sufficient adjuvants to confer U1-snRNPs autoimmunogenecity (127).
In the nucleolar DFC region, the C/D box U3-snoRNP component FBRL has dual autoantigen and alarmin activities. In the H/ACA box U3-snoRNPs, the GAR1 component has adjuvant activity albeit autoantigen has not been reported in these complexes. In the nucleolar GC region, NCL also has dual autoantigenic and adjuvant activities (131). It would be interesting to test whether NCL and FBRL induce their self-reactive antibodies and whether these nucleolar alarmins are sufficient to confer autoimmunogenicity to the numerous other nucleolar and nucleoplasmic autoantigens.
In this context, studies on the clone 564 mouse autoantibody suggested an immunological pathway for autoimmunological epitope spreading (197). This antibody is cationic and polyreactive with single-strand DNA/RNA, nucleosomes, La/SSB, etc., and its IIF image showed intense nucleolar and cytoplasmic staining (197–199). Transgenic 564 expressions in C57BL/6 mice (564Igi) produced antibodies that stained the nucleolus (198, 199). In these mice, the transgenic B cells initiate spontaneous germinal centers in which other autoreactive B cells also proliferate to produce ANA of broader specificity (200). Whether NCL- and FBRL-reactive B cells similarly initiate autoreactive germinal centers need to be investigated.
9 Concluding remarks
The collective and individual significance of ANA has been testified by their increasing weightage in SLE diagnosis (50). However, answers remain fragmental with regard to what cause these autoantibodies, e.g., tolerance breakdown, dead cell accumulation, infection, etc. The prevalence of self-reactive naïve B cells in healthy individuals places particular importance on peripheral tolerance (121). The growing number of alarmins in the most autoantigenic nuclear regions, i.e., the chromatin network, the speckled regions, and nucleoli (Table 2), suggests their intrinsic capacity to overwhelm peripheral tolerance after necrotic exposure, and cause ANA production (123–128, 131). Necrotic cells are known to accumulate in SLE patients and release nuclear materials (1, 53). In this context, the four earliest components of the complement classical pathway, i.e., C1q, C1r, C1s, and C4, may be considered as an essential albeit insufficient tolerance mechanism against dead cell-induced autoimmunity (Figure 3) (181). These are rare genetic deficiencies that are not captured in most population studies, and the scarcity of these patients can be explained by the severity and early onset of the disease (166, 172). Nonetheless, these genetic deficiencies have offered a unique pathway of investigation into the causes of ANA and SLE pathogenesis.
The discovery of C1q binding to apoptotic cells formed the cornerstone of an immunological axis in understanding ANA induction and SLE pathogenesis. An initial hypothesis was that C1q opsonizes apoptotic cells for effective clearance to avoid immune exposure (Figure 4) (185, 190, 201). A more recent hypothesis is that C1q targets C1 proteases to dead cells to dismantle autoantigens and alarmins and therefore diminish their immunogenicity and avoid immune responses that lead to ANA production and immune complex-mediated tissue injuries (Figure 4) (178, 179). The observed C1q targeting to the highly autoantigenic nucleoli in necrotic cells (193) and C1s cleavage of nucleolar proteins (59, 131), i.e., autoantigens and alarmins (21, 59, 130), are in line with this hypothesis. Besides nucleolar proteins, the C1 proteases may broadly degrade and inactivate nuclear autoantigens and alarmins like HMGB1 (195).
At present, there is insufficient data to harmonize this hypothesis with how C4 deficiency similarly causes ANA and SLE (181, 182, 202). Based on the complement system, when C1s is activated on dead cells, it is expected to cleave C4 so C4b deposits on dead cells, and C4a is released as a weak anaphylatoxin (183, 184). C4b can target dead cells to phagocytes, B cells, and follicular dendritic cells through the complement receptor CD21/CD35 (183, 184), which is relevant to antibody induction. Carroll and colleagues reported that C4-deficient mice had a defect in transitional autoreactive B cell deletion and tended to form autoreactive germinal centers (198). It is possible that C4b-linked dead cell antigens inhibit autoreactive germinal center reactions and prevent antibody class switch by the prevalent self-reactive naïve B cells (120, 122). It has not been tested whether C4b-linked dead cell antigens are also cleaved more effectively because C2 is only effectively cleaved by C1s when it is associated with C4b. Further study of how C4 is related to this C1-dead cell axis of ANA induction and SLE pathogenesis could reveal more definitive underlying mechanisms for improved diagnosis and therapeutic targeting.
Author contributions
JL initiated the article and contributed to the framework and major details of the final version. SW contributed to the details in nucleolar alarmins. JJC contributed to the details on nucleolar structures. BHDT contributed to the details in Figure 4. SYKW contributed to the details on B cells. JZC and JMC helped in bioinformatics that generated Figure 3. KPL contributed to the clinical aspects of this manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work is supported by the Singapore National Medical Research Council Open-funding Individual Research Grants (NMRC/OFIRG/0013/2016; MOH-000958)
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
NCL, nucleolin; NPM1, nucleophosmin 1; ANA, antinuclear autoantibody; upstream binding factor, UBF; FBRL, fibrillarin; HMGB1, high motility group box 1; NOR, nucleolar organizer region.
References
1. Hargraves MM, Richmond H, Morton R. Presentation of two bone marrow elements; the tart cell and the L.E. cell. Proc Staff Meet Mayo Clin (1948) 23(2):25–8.
2. Smith CD, Cyr M. The history of lupus erythematosus. from hippocrates to osler. Rheum Dis Clin North Am (1988) 14(1):1–14. doi: 10.1016/S0889-857X(21)00942-X
4. Baehr G, Klemperer P, Schifrin A. A diffuse disease of the peripheral circulation (usually associated with lupus erythematosus and endocarditis). Am J Med (1952) 13(5):591–6. doi: 10.1016/0002-9343(52)90026-0
5. Hargraves MM. Production in vitro of the L.E. cell phenomenon; use of normal bone marrow elements and blood plasma from patients with acute disseminated lupus erythematosus. Proc Staff Meet Mayo Clin (1949) 24(9):234–7.
6. Holborow EJ, Weir DM, Johnson GD. A serum factor in lupus erythematosus with affinity for tissue nuclei. Br Med J (1957) 2(5047):732–4. doi: 10.1136/bmj.2.5047.732
7. Damoiseaux J, von Muhlen CA, Garcia-De La Torre I, Carballo OG, de Melo Cruvinel W, Francescantonio PL, et al. International consensus on ANA patterns (ICAP): the bumpy road towards a consensus on reporting ANA results. Auto Immun Highlights (2016) 7(1):1. doi: 10.1007/s13317-016-0075-0
8. Nakamura RM, Tan EM. Recent progress in the study of autoantibodies to nuclear antigens. Hum Pathol (1978) 9(1):85–91. doi: 10.1016/S0046-8177(78)80010-0
9. Tan EM. An immunologic precipitin system between soluble nucleoprotein and serum antibody in systemic lupus erythematosus. J Clin Invest (1967) 46(5):735–45. doi: 10.1172/JCI105574
10. Stollar BD. Reactions of systemic lupus erythematosus sera with histone fractions and histone-DNA complexes. Arthritis Rheumatol (1971) 14(4):485–92. doi: 10.1002/art.1780140408
11. Yamane K, Ihn H, Kubo M, Kuwana M, Asano Y, Yazawa N, et al. Anti-U1RNP antibodies in patients with localized scieroderma. Arch Dermatol Res (2001) 293(9):455–9. doi: 10.1007/s004030100254
12. Tan EM, Kunkel HG. Characteristics of a soluble nuclear antigen precipitating with sera of patients with systemic lupus erythematosus. J Immunol (1966) 96(3):464–71. doi: 10.4049/jimmunol.96.3.464
13. Okano Y, Steen VD, Medsger TA Jr. Autoantibody reactive with RNA polymerase III in systemic sclerosis. Ann Intern Med (1993) 119(10):1005–13. doi: 10.7326/0003-4819-119-10-199311150-00007
14. van den Hoogen F, Khanna D, Fransen J, Johnson SR, Baron M, Tyndall A, et al. Classification criteria for systemic sclerosis: an American college of Rheumatology/European league against rheumatism collaborative initiative. Arthritis Rheum (2013) 65(11):2737–47.
15. Chan EK, Tan EM. Human autoantibody-reactive epitopes of SS-B/La are highly conserved in comparison with epitopes recognized by murine monoclonal antibodies. J Exp Med (1987) 166(6):1627–40. doi: 10.1084/jem.166.6.1627
16. Russo K, Hoch S, Dima C, Varga J, Teodorescu M. Circulating anticentromere CENP-a and CENP-b antibodies in patients with diffuse and limited systemic sclerosis, systemic lupus erythematosus, and rheumatoid arthritis. J Rheumatol (2000) 27(1):142–8.
17. Hudson M, Mahler M, Pope J, You D, Tatibouet S, Steele R, et al. Clinical correlates of CENP-a and CENP-b antibodies in a large cohort of patients with systemic sclerosis. J Rheumatol (2012) 39(4):787–94. doi: 10.3899/rheum.111133
18. Fischer A, Pfalzgraf FJ, Feghali-Bostwick CA, Wright TM, Curran-Everett D, West SG, et al. Anti-th/to-positivity in a cohort of patients with idiopathic pulmonary fibrosis. J Rheumatol (2006) 33(8):1600–5.
19. Li XZ, McNeilage LJ, Whittingham S. Autoantibodies to the major nucleolar phosphoprotein B23 define a novel subset of patients with anticardiolipin antibodies. Arthritis Rheumatol (1989) 32(9):1165–9. doi: 10.1002/anr.1780320917
20. Ochs RL, Stein TW Jr., Chan EK, Ruutu M, Tan EM. cDNA cloning and characterization of a novel nucleolar protein. Mol Biol Cell (1996) 7(7):1015–24. doi: 10.1091/mbc.7.7.1015
21. Satoh M, Ceribelli A, Hasegawa T, Tanaka S. Clinical significance of antinucleolar antibodies: biomarkers for autoimmune diseases, malignancies, and others. Clin Rev Allergy Immunol (2022) 63(2):210–39. doi: 10.1007/s12016-022-08931-3
22. Wang T, Marken J, Chen J, Tran VB, Li QZ, Li M, et al. High TLR7 expression drives the expansion of CD19(+)CD24(hi)CD38(hi) transitional b cells and autoantibody production in SLE patients. Front Immunol (2019) 10:1243. doi: 10.3389/fimmu.2019.01243
23. Ulanet DB, Wigley FM, Gelber AC, Rosen A. Autoantibodies against B23, a nucleolar phosphoprotein, occur in scleroderma and are associated with pulmonary hypertension. Arthritis Rheumatol (2003) 49(1):85–92. doi: 10.1002/art.10914
24. Mahler M, Raijmakers R. Novel aspects of autoantibodies to the PM/Scl complex: clinical, genetic and diagnostic insights. Autoimmun Rev (2007) 6(7):432–7. doi: 10.1016/j.autrev.2007.01.013
25. Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell (1985) 54(2):123–33. doi: 10.1111/j.1768-322X.1985.tb00387.x
26. Reimer G, Rose KM, Scheer U, Tan EM. Autoantibody to RNA polymerase I in scleroderma sera. J Clin Invest (1987) 79(1):65–72. doi: 10.1172/JCI112809
27. Chan EK, Imai H, Hamel JC, Tan EM. Human autoantibody to RNA polymerase I transcription factor hUBF. molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J Exp Med (1991) 174(5):1239–44.
28. Konstantinov K, Foisner R, Byrd D, Liu FT, Tsai WM, Wiik A, et al. Integral membrane proteins associated with the nuclear lamina are novel autoimmune antigens of the nuclear envelope. Clin Immunol Immunopathol (1995) 74(1):89–99. doi: 10.1006/clin.1995.1013
29. Konstantinov KN, Galcheva-Gargova Z, Hoier-Madsen M, Wiik A, Ullman S, Halberg P, et al. Autoantibodies to lamins a and c in sera of patients showing peripheral fluorescent antinuclear antibody pattern on HEP-2 cells. J Invest Dermatol (1990) 95(3):304–8. doi: 10.1111/1523-1747.ep12485010
30. Reeves WH, Chaudhary N, Salerno A, Blobel G. Lamin b autoantibodies in sera of certain patients with systemic lupus erythematosus. J Exp Med (1987) 165(3):750–62. doi: 10.1084/jem.165.3.750
31. Coppo P, Clauvel JP, Bengoufa D, Fuentes V, Gouilleux-Gruart V, Courvalin JC, et al. Autoimmune cytopenias associated with autoantibodies to nuclear envelope polypeptides. Am J Hematol (2004) 77(3):241–9. doi: 10.1002/ajh.20188
32. Miyachi K, Shibata M, Onozuka Y, Kikuchi F, Imai N, Horigome T. Primary biliary cirrhosis sera recognize not only gp210 but also proteins of the p62 complex bearing n-acetylglucosamine residues from rat liver nuclear envelope. anti-p62 complex antibody in PBC. Mol Biol Rep (1996) 23(3-4):227–34. doi: 10.1007/BF00351173
33. Wesierska-Gadek J, Klima A, Komina O, Ranftler C, Invernizzi P, Penner E. Characterization of autoantibodies against components of the nuclear pore complexes: high frequency of anti-p62 nucleoporin antibodies. Ann N Y Acad Sci (2007) 1109:519–30. doi: 10.1196/annals.1398.058
34. Watanabe A, Kodera M, Sugiura K, Usuda T, Tan EM, Takasaki Y, et al. Anti-DFS70 antibodies in 597 healthy hospital workers. Arthritis Rheumatol (2004) 50(3):892–900. doi: 10.1002/art.20096
35. Mahler M, Parker T, Peebles CL, Andrade LE, Swart A, Carbone Y, et al. Anti-DFS70/LEDGF antibodies are more prevalent in healthy individuals compared to patients with systemic autoimmune rheumatic diseases. J Rheumatol (2012) 39(11):2104–10. doi: 10.3899/jrheum.120598
36. Vazquez-Del Mercado M, Gomez-Banuelos E, Navarro-Hernandez RE, Pizano-Martinez O, Saldana-Millan A, Chavarria-Avila E, et al. Detection of autoantibodies to DSF70/LEDGFp75 in Mexican hispanics using multiple complementary assay platforms. Auto Immun Highlights (2017) 8(1):1. doi: 10.1007/s13317-016-0089-7
37. Cozzani E, Drosera M, Riva S, Parodi A. Analysis of a multiple nuclear dots pattern in a large cohort of dermatological patients. Clin Lab (2012) 58(3-4):329–32.
38. Granito A, Yang WH, Muratori L, Lim MJ, Nakajima A, Ferri S, et al. PML nuclear body component Sp140 is a novel autoantigen in primary biliary cirrhosis. Am J Gastroenterol (2010) 105(1):125–31. doi: 10.1038/ajg.2009.596
39. Satoh M, Chan JY, Ross SJ, Ceribelli A, Cavazzana I, Franceschini F, et al. Autoantibodies to survival of motor neuron complex in patients with polymyositis: immunoprecipitation of d, e, f, and G proteins without other components of small nuclear ribonucleoproteins. Arthritis Rheumatol (2011) 63(7):1972–8. doi: 10.1002/art.30349
40. Fujimoto M, Kikuchi K, Tamaki T, Yazawa N, Kubo M, Ihn H, et al. Distribution of anti-p80-coilin autoantibody in collagen diseases and various skin diseases. Br J Dermatol (1997) 137(6):916–20. doi: 10.1046/j.1365-2133.1997.19852066.x
41. Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med (1991) 173(6):1407–19. doi: 10.1084/jem.173.6.1407
42. Miyachi K, Fritzler MJ, Tan EM. Autoantibody to a nuclear antigen in proliferating cells. J Immunol (1978) 121(6):2228–34. doi: 10.4049/jimmunol.121.6.2228
43. Mahler M, Miyachi K, Peebles C, Fritzler MJ. The clinical significance of autoantibodies to the proliferating cell nuclear antigen (PCNA). Autoimmun Rev (2012) 11(10):771–5. doi: 10.1016/j.autrev.2012.02.012
44. Casiano CA, Landberg G, Ochs RL, Tan EM. Autoantibodies to a novel cell cycle-regulated protein that accumulates in the nuclear matrix during s phase and is localized in the kinetochores and spindle midzone during mitosis. J Cell Sci (1993) 106(Pt 4):1045–56. doi: 10.1242/jcs.106.4.1045
45. Welner S, Trier NH, Frisch M, Locht H, Hansen PR, Houen G. Correlation between centromere protein-f autoantibodies and cancer analyzed by enzyme-linked immunosorbent assay. Mol Cancer (2013) 12(1):95. doi: 10.1186/1476-4598-12-95
46. Lock RJ, Unsworth DJ. Antibodies to extractable nuclear antigens. has technological drift affected clinical interpretation? J Clin Pathol (2001) 54(3):187–90.
47. Cohen AS, Canoso JJ. Criteria for the classification of systemic lupus erythematosus–status 1972. Arthritis Rheumatol (1972) 15(5):540–3. doi: 10.1002/art.1780150512
48. Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheumatol (1982) 25(11):1271–7. doi: 10.1002/art.1780251101
49. Petri M, Orbai AM, Alarcon GS, Gordon C, Merrill JT, Fortin PR, et al. Derivation and validation of the systemic lupus international collaborating clinics classification criteria for systemic lupus erythematosus. Arthritis Rheumatol (2012) 64(8):2677–86. doi: 10.1002/art.34473
50. Aringer M, Costenbader K, Daikh D, Brinks R, Mosca M, Ramsey-Goldman R, et al. European League against Rheumatism/American college of rheumatology classification criteria for systemic lupus erythematosus. Arthritis Rheumatol (2019) 71(9):1400–12. doi: 10.1002/art.40930
51. Vermeersch P, Bossuyt X. Prevalence and clinical significance of rare antinuclear antibody patterns. Autoimmun Rev (2013) 12(10):998–1003. doi: 10.1016/j.autrev.2013.03.014
52. Hiepe F, Dorner T, Burmester G. Antinuclear antibody- and extractable nuclear antigen-related diseases. Int Arch Allergy Immunol (2000) 123(1):5–9. doi: 10.1159/000024418
53. Tan EM, Schur PH, Carr RI, Kunkel HG. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J Clin Invest (1966) 45(11):1732–40. doi: 10.1172/JCI105479
54. Reimer G, Raska I, Scheer U, Tan EM. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res (1988) 176(1):117–28. doi: 10.1016/0014-4827(88)90126-7
55. Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA (2010) 1(3):415–31. doi: 10.1002/wrna.39
56. McStay B. Nucleolar organizer regions: genomic 'dark matter' requiring illumination. Genes Dev (2016) 30(14):1598–610. doi: 10.1101/gad.283838.116
57. Andrade LEC, Damoiseaux J, Vergani D, Fritzler MJ. Antinuclear antibodies (ANA) as a criterion for classification and diagnosis of systemic autoimmune diseases. J Transl Autoimmun (2022) 5:100145. doi: 10.1016/j.jtauto.2022.100145
58. Terao C, Ohmura K, Yamada R, Kawaguchi T, Shimizu M, Tabara Y, et al. Association between antinuclear antibodies and the HLA class II locus and heterogeneous characteristics of staining patterns: the nagahama study. Arthritis Rheumatol (2014) 66(12):3395–403. doi: 10.1002/art.38867
59. Cai Y, Wee SYK, Chen J, Teo BHD, Ng YLC, Leong KP, et al. Broad susceptibility of nucleolar proteins and autoantigens to complement C1 protease degradation. J Immunol (2017) 199(12):3981–90. doi: 10.4049/jimmunol.1700728
60. Scheer U, Xia B, Merkert H, Weisenberger D. Looking at Christmas trees in the nucleolus. Chromosoma (1997) 105(7-8):470–80. doi: 10.1007/BF02510484
61. de la Cruz J, Karbstein K, Woolford JL Jr. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu Rev Biochem (2015) 84:93–129. doi: 10.1146/annurev-biochem-060614-033917
62. Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol (2002) 14(3):313–8. doi: 10.1016/S0955-0674(02)00336-8
63. Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol (2003) 13(5):255–63. doi: 10.1016/S0962-8924(03)00054-0
64. Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res (2011) 71(4):1418–30. doi: 10.1158/0008-5472.CAN-10-1728
65. Henderson AS, Warburton D, Atwood KC. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci U S A (1972) 69(11):3394–8. doi: 10.1073/pnas.69.11.3394
66. Parks MM, Kurylo CM, Dass RA, Bojmar L, Lyden D, Vincent CT, et al. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci Adv (2018) 4(2):eaao0665. doi: 10.1126/sciadv.aao0665
67. Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, et al. The complete sequence of a human genome. Science (2022) 376(6588):44–53. doi: 10.1126/science.abj6987
68. Chen J, Teo BHD, Cai Y, Wee SYK, Lu J. The linker histone H1.2 is a novel component of the nucleolar organizer regions. J Biol Chem (2018) 293(7):2358–69.
69. Gautier T, Robert-Nicoud M, Guilly MN, Hernandez-Verdun D. Relocation of nucleolar proteins around chromosomes at mitosis. a study by confocal laser scanning microscopy. J Cell Sci (1992) 102(Pt 4):729–37.
70. Yasuda Y, Maul GG. A nucleolar auto-antigen is part of a major chromosomal surface component. Chromosoma (1990) 99(2):152–60. doi: 10.1007/BF01735332
71. Jordan EG, McGovern JH. The quantitative relationship of the fibrillar centres and other nucleolar components to changes in growth conditions, serum deprivation and low doses of actinomycin d in cultured diploid human fibroblasts (strain MRC-5). J Cell Sci (1981) 52:373–89. doi: 10.1242/jcs.52.1.373
72. Yung BY, Bor AM, Chan PK. Short exposure to actinomycin d induces "reversible" translocation of protein B23 as well as "reversible" inhibition of cell growth and RNA synthesis in HeLa cells. Cancer Res (1990) 50(18):5987–91.
73. Wong X, Melendez-Perez AJ, Reddy KL. The nuclear lamina. Cold Spring Harb Perspect Biol (2022) 14(2):1–25. doi: 10.1101/cshperspect.a040113
74. van Steensel B, Belmont AS. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell (2017) 169(5):780–91. doi: 10.1016/j.cell.2017.04.022
75. Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, et al. Initial genomics of the human nucleolus. PloS Genet (2010) 6(3):e1000889. doi: 10.1371/journal.pgen.1000889
76. van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell (2010) 21(21):3735–48. doi: 10.1091/mbc.e10-06-0508
77. Penagos-Puig A, Furlan-Magaril M. Heterochromatin as an important driver of genome organization. Front Cell Dev Biol (2020) 8:579137. doi: 10.3389/fcell.2020.579137
78. Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature (2019) 570(7761):395–9. doi: 10.1038/s41586-019-1275-3
79. Olson MO, Hingorani K, Szebeni A. Conventional and nonconventional roles of the nucleolus. Int Rev Cytol (2002) 219:199–266. doi: 10.1016/S0074-7696(02)19014-0
80. Lam YW, Evans VC, Heesom KJ, Lamond AI, Matthews DA. Proteomics analysis of the nucleolus in adenovirus-infected cells. Mol Cell Proteomics (2010) 9(1):117–30. doi: 10.1074/mcp.M900338-MCP200
81. Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, et al. Directed proteomic analysis of the human nucleolus. Curr Biol (2002) 12(1):1–11. doi: 10.1016/S0960-9822(01)00650-9
82. Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, et al. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics (2011) 10(10):M111 009241. doi: 10.1074/mcp.M111.009241
83. Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI. NOPdb: nucleolar proteome database–2008 update. Nucleic Acids Res (2009) 37(Database issue):D181–4. doi: 10.1093/nar/gkn804
84. Russell J, Zomerdijk JC. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci (2005) 30(2):87–96. doi: 10.1016/j.tibs.2004.12.008
85. Roussel P, Andre C, Masson C, Geraud G, Hernandez-Verdun D. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J Cell Sci (1993) 104(Pt 2):327–37. doi: 10.1242/jcs.104.2.327
86. O'Sullivan AC, Sullivan GJ, McStay B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol Cell Biol (2002) 22(2):657–68. doi: 10.1128/MCB.22.2.657-668.2002
87. Stefanovsky VY, Pelletier G, Bazett-Jones DP, Crane-Robinson C, Moss T. DNA Looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules. Nucleic Acids Res (2001) 29(15):3241–7. doi: 10.1093/nar/29.15.3241
88. Miller OL Jr., Beatty BR. Visualization of nucleolar genes. Science (1969) 164(3882):955–7. doi: 10.1126/science.164.3882.955
89. Mougey EB, O'Reilly M, Osheim Y, Miller OL Jr., Beyer A, Sollner-Webb B. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev (1993) 7(8):1609–19. doi: 10.1101/gad.7.8.1609
90. Cheutin T, O'Donohue MF, Beorchia A, Vandelaer M, Kaplan H, Defever B, et al. Three-dimensional organization of active rRNA genes within the nucleolus. J Cell Sci (2002) 115(Pt 16):3297–307. doi: 10.1242/jcs.115.16.3297
91. Koberna K, Malinsky J, Pliss A, Masata M, Vecerova J, Fialova M, et al. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of "Christmas trees" in situ. J Cell Biol (2002) 157(5):743–8. doi: 10.1083/jcb.200202007
92. Gonzalez IL, Sylvester JE. Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics (1995) 27(2):320–8. doi: 10.1006/geno.1995.1049
93. Potapova TA, Gerton JL. Ribosomal DNA and the nucleolus in the context of genome organization. Chromosome Res (2019) 27(1-2):109–27. doi: 10.1007/s10577-018-9600-5
94. Lessard F, Igelmann S, Trahan C, Huot G, Saint-Germain E, Mignacca L, et al. Senescence-associated ribosome biogenesis defects contributes to cell cycle arrest through the Rb pathway. Nat Cell Biol (2018) 20(7):789–99. doi: 10.1038/s41556-018-0127-y
95. Lafontaine DL. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol (2015) 22(1):11–9. doi: 10.1038/nsmb.2939
96. Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA (2012) 3(3):397–414. doi: 10.1002/wrna.117
97. Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol (2006) 71:407–17. doi: 10.1101/sqb.2006.71.025
98. Hunziker M, Barandun J, Petfalski E, Tan D, Delan-Forino C, Molloy KR, et al. UtpA and UtpB chaperone nascent pre-ribosomal RNA and U3 snoRNA to initiate eukaryotic ribosome assembly. Nat Commun (2016) 7:12090. doi: 10.1038/ncomms12090
99. Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol (2015) 22(11):920–3. doi: 10.1038/nsmb.3111
100. Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res (2006) 34(Database issue):D158–62. doi: 10.1093/nar/gkj002
101. Bouchard-Bourelle P, Desjardins-Henri C, Mathurin-St-Pierre D, Deschamps-Francoeur G, Fafard-Couture E, Garant JM, et al. snoDB: an interactive database of human snoRNA sequences, abundance and interactions. Nucleic Acids Res (2020) 48(D1):D220–D5. doi: 10.1093/nar/gkz884
102. Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, et al. An updated human snoRNAome. Nucleic Acids Res (2016) 44(11):5068–82. doi: 10.1093/nar/gkw386
103. Krogh N, Jansson MD, Hafner SJ, Tehler D, Birkedal U, Christensen-Dalsgaard M, et al. Profiling of 2'-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res (2016) 44(16):7884–95. doi: 10.1093/nar/gkw482
104. Reimer G, Steen VD, Penning CA, Medsger TA Jr., Tan EM. Correlates between autoantibodies to nucleolar antigens and clinical features in patients with systemic sclerosis (scleroderma). Arthritis Rheumatol (1988) 31(4):525–32. doi: 10.1002/art.1780310409
105. Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics (2009) 94(2):83–8. doi: 10.1016/j.ygeno.2009.05.002
106. Terns M, Terns R. Noncoding RNAs of the H/ACA family. Cold Spring Harb Symp Quant Biol (2006) 71:395–405. doi: 10.1101/sqb.2006.71.034
107. Massenet S, Bertrand E, Verheggen C. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol (2017) 14(6):680–92. doi: 10.1080/15476286.2016.1243646
108. Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res (1997) 25(23):4740–7.
109. Kufel J, Allmang C, Chanfreau G, Petfalski E, Lafontaine DL, Tollervey D. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by la binding. Mol Cell Biol (2000) 20(15):5415–24. doi: 10.1128/MCB.20.15.5415-5424.2000
110. Samarsky DA, Fournier MJ, Singer RH, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J (1998) 17(13):3747–57. doi: 10.1093/emboj/17.13.3747
111. Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol (2007) 8(3):209–20. doi: 10.1038/nrm2124
112. Barandun J, Chaker-Margot M, Hunziker M, Molloy KR, Chait BT, Klinge S. The complete structure of the small-subunit processome. Nat Struct Mol Biol (2017) 24(11):944–53. doi: 10.1038/nsmb.3472
113. Lischwe MA, Smetana K, Olson MO, Busch H. Proteins C23 and B23 are the major nucleolar silver staining proteins. Life Sci (1979) 25(8):701–8. doi: 10.1016/0024-3205(79)90512-5
114. Li YP, Busch RK, Valdez BC, Busch H. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur J Biochem (1996) 237(1):153–8. doi: 10.1111/j.1432-1033.1996.0153n.x
115. Turner AJ, Knox AA, Prieto JL, McStay B, Watkins NJ. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol Cell Biol (2009) 29(11):3007–17. doi: 10.1128/MCB.00029-09
116. Lindstrom MS. NPM1/B23: a multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem Res Int (2011) 2011:195209. doi: 10.1155/2011/195209
117. Girard C, Will CL, Peng J, Makarov EM, Kastner B, Lemm I, et al. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun (2012) 3:994. doi: 10.1038/ncomms1998
118. Dias AP, Dufu K, Lei H, Reed R. A role for TREX components in the release of spliced mRNA from nuclear speckle domains. Nat Commun (2010) 1:97. doi: 10.1038/ncomms1103
119. Silverman GJ, Vas J, Gronwall C. Protective autoantibodies in the rheumatic diseases: lessons for therapy. Nat Rev Rheumatol (2013) 9(5):291–300. doi: 10.1038/nrrheum.2013.30
120. Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human b cell precursors. Science (2003) 301(5638):1374–7. doi: 10.1126/science.1086907
121. Yurasov S, Wardemann H, Hammersen J, Tsuiji M, Meffre E, Pascual V, et al. Defective b cell tolerance checkpoints in systemic lupus erythematosus. J Exp Med (2005) 201(5):703–11. doi: 10.1084/jem.20042251
122. Mietzner B, Tsuiji M, Scheid J, Velinzon K, Tiller T, Abraham K, et al. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc Natl Acad Sci U S A (2008) 105(28):9727–32. doi: 10.1073/pnas.0803644105
123. Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol (2010) 28:367–88. doi: 10.1146/annurev.immunol.021908.132603
124. Das N, Dewan V, Grace PM, Gunn RJ, Tamura R, Tzarum N, et al. HMGB1 activates proinflammatory signaling via TLR5 leading to allodynia. Cell Rep (2016) 17(4):1128–40. doi: 10.1016/j.celrep.2016.09.076
125. Urbonaviciute V, Furnrohr BG, Meister S, Munoz L, Heyder P, De Marchis F, et al. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J Exp Med (2008) 205(13):3007–18. doi: 10.1084/jem.20081165
126. Reuter R, Luhrmann R. Immunization of mice with purified U1 small nuclear ribonucleoprotein (RNP) induces a pattern of antibody specificities characteristic of the anti-Sm and anti-RNP autoimmune response of patients with lupus erythematosus, as measured by monoclonal antibodies. Proc Natl Acad Sci U S A (1986) 83(22):8689–93. doi: 10.1073/pnas.83.22.8689
127. Kelly-Scumpia KM, Nacionales DC, Scumpia PO, Weinstein JS, Narain S, Moldawer LL, et al. In vivo adjuvant activity of the RNA component of the Sm/RNP lupus autoantigen. Arthritis Rheumatol (2007) 56(10):3379–86. doi: 10.1002/art.22946
128. Shin MS, Kang Y, Lee N, Kim SH, Kang KS, Lazova R, et al. U1-small nuclear ribonucleoprotein activates the NLRP3 inflammasome in human monocytes. J Immunol (2012) 188(10):4769–75. doi: 10.4049/jimmunol.1103355
129. Kelly KM, Zhuang H, Nacionales DC, Scumpia PO, Lyons R, Akaogi J, et al. "Endogenous adjuvant" activity of the RNA components of lupus autoantigens Sm/RNP and ro 60. Arthritis Rheumatol (2006) 54(5):1557–67.
130. Welting TJ, Raijmakers R, Pruijn GJ. Autoantigenicity of nucleolar complexes. Autoimmun Rev (2003) 2(6):313–21. doi: 10.1016/S1568-9972(03)00029-6
131. Wu S, Teo BHD, Wee SYK, Chen J, Lu J. The GAR/RGG motif defines a family of nuclear alarmins. Cell Death Dis (2021) 12(5):477. doi: 10.1038/s41419-021-03766-w
132. McClain MT, Heinlen LD, Dennis GJ, Roebuck J, Harley JB, James JA. Early events in lupus humoral autoimmunity suggest initiation through molecular mimicry. Nat Med (2005) 11(1):85–9. doi: 10.1038/nm1167
133. Poole BD, Gross T, Maier S, Harley JB, James JA. Lupus-like autoantibody development in rabbits and mice after immunization with EBNA-1 fragments. J Autoimmun (2008) 31(4):362–71. doi: 10.1016/j.jaut.2008.08.007
134. Singh D, Oudit O, Hajtovic S, Sarbaugh D, Salis R, Adebowale T, et al. Antibodies to an Epstein Barr virus protein that cross-react with dsDNA have pathogenic potential. Mol Immunol (2021) 132:41–52. doi: 10.1016/j.molimm.2021.01.013
135. Ayoubian H, Frohlich T, Pogodski D, Flatley A, Kremmer E, Schepers A, et al. Antibodies against the mono-methylated arginine-glycine repeat (MMA-RG) of the Epstein-Barr virus nuclear antigen 2 (EBNA2) identify potential cellular proteins targeted in viral transformation. J Gen Virol (2017) 98(8):2128–42. doi: 10.1099/jgv.0.000870
136. Ho KT, Reveille JD. The clinical relevance of autoantibodies in scleroderma. Arthritis Res Ther (2003) 5(2):80–93.
137. Yang JM, Hildebrandt B, Luderschmidt C, Pollard KM. Human scleroderma sera contain autoantibodies to protein components specific to the U3 small nucleolar RNP complex. Arthritis Rheumatol (2003) 48(1):210–7. doi: 10.1002/art.10729
138. Kuwana M, Kimura K, Hirakata M, Kawakami Y, Ikeda Y. Differences in autoantibody response to Th/To between systemic sclerosis and other autoimmune diseases. Ann Rheum Dis (2002) 61(9):842–6. doi: 10.1136/ard.61.9.842
139. Van Eenennaam H, Vogelzangs JH, Bisschops L, Te Boome LC, Seelig HP, Renz M, et al. Autoantibodies against small nucleolar ribonucleoprotein complexes and their clinical associations. Clin Exp Immunol (2002) 130(3):532–40. doi: 10.1046/j.1365-2249.2002.01991.x
140. Shiboski CH, Shiboski SC, Seror R, Criswell LA, Labetoulle M, Lietman TM, et al. 2016 American College of Rheumatology/European league against rheumatism classification criteria for primary sjogren's syndrome: a consensus and data-driven methodology involving three international patient cohorts. Arthritis Rheumatol (2017) 69(1):35–45. doi: 10.1002/art.39859
141. Hirata D, Iwamoto M, Yoshio T, Okazaki H, Masuyama J, Mimori A, et al. Nucleolin as the earliest target molecule of autoantibodies produced in MRL/lpr lupus-prone mice. Clin Immunol (2000) 97(1):50–8. doi: 10.1006/clim.2000.4916
142. Kindas-Mugge I. Human autoantibodies against a nucleolar protein. Biochem Biophys Res Commun (1989) 163(2):1119–27. doi: 10.1016/0006-291X(89)92337-1
143. Pfeifle J, Anderer FA, Franke M. Characterisation of nucleolar proteins as autoantigens using human autoimmune sera. Ann Rheum Dis (1986) 45(12):978–86. doi: 10.1136/ard.45.12.978
144. Ritchie RF. Antinucleolar antibodies. their frequency and diagnostic association. N Engl J Med (1970) 282(21):1174–8. doi: 10.1056/NEJM197005212822104
145. Rodriguez-Sanchez JL, Gelpi C, Juarez C, Hardin JA. Anti-NOR 90. a new autoantibody in scleroderma that recognizes a 90-kDa component of the nucleolus-organizing region of chromatin. J Immunol (1987) 139(8):2579–84.
146. Dagher JH, Scheer U, Voit R, Grummt I, Lonzetti L, Raymond Y, et al. Autoantibodies to NOR 90/hUBF: longterm clinical and serological followup in a patient with limited systemic sclerosis suggests an antigen driven immune response. J Rheumatol (2002) 29(7):1543–7.
147. Brankin B, Skaar TC, Brotzman M, Trock B, Clarke R. Autoantibodies to the nuclear phosphoprotein nucleophosmin in breast cancer patients. Cancer Epidemiol Biomarkers Prev (1998) 7(12):1109–15.
148. Imai H, Ochs RL, Kiyosawa K, Nakamura RM, Tan EM. Nucleolar antigens and autoantibodies in hepatocellular carcinoma and other malignancies. Am J Pathol (1992) 140(4):859–70.
149. Zhang JY, Wang X, Peng XX, Chan EK. Autoantibody responses in Chinese hepatocellular carcinoma. J Clin Immunol (2002) 22(2):98–105. doi: 10.1023/A:1014483803483
150. Ogg SC, Lamond AI. Cajal bodies and coilin–moving towards function. J Cell Biol (2002) 159(1):17–21. doi: 10.1083/jcb.200206111
151. Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, et al. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through cajal bodies. Mol Cell (2014) 56(3):389–99. doi: 10.1016/j.molcel.2014.10.004
152. Trinkle-Mulcahy L, Sleeman JE. The cajal body and the nucleolus: "In a relationship" or "It's complicated"? RNA Biol (2017) 14(6):739–51. doi: 10.1080/15476286.2016.1236169
153. Schilders G, van Dijk E, Raijmakers R, Pruijn GJ. Cell and molecular biology of the exosome: how to make or break an RNA. Int Rev Cytol (2006) 251:159–208. doi: 10.1016/S0074-7696(06)51005-8
154. Brouwer R, Pruijn GJ, van Venrooij WJ. The human exosome: an autoantigenic complex of exoribonucleases in myositis and scleroderma. Arthritis Res (2001) 3(2):102–6. doi: 10.1186/ar147
155. Jacobson MR, Cao LG, Wang YL, Pederson T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol (1995) 131(Pt 2):1649–58. doi: 10.1083/jcb.131.6.1649
156. Jarrous N, Wolenski JS, Wesolowski D, Lee C, Altman S. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease p. J Cell Biol (1999) 146(3):559–72. doi: 10.1083/jcb.146.3.559
157. Lerner EA, Lerner MR, Janeway CA Jr., Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci U S A (1981) 78(5):2737–41. doi: 10.1073/pnas.78.5.2737
158. Sato T, Uchiumi T, Arakawa M, Kominami R. Serological association of lupus autoantibodies to a limited functional domain of 28S ribosomal RNA and to the ribosomal proteins bound to the domain. Clin Exp Immunol (1994) 98(1):35–9.
159. Deng Y, Tsao BP. Advances in lupus genetics and epigenetics. Curr Opin Rheumatol (2014) 26(5):482–92. doi: 10.1097/BOR.0000000000000086
160. Cui Y, Sheng Y, Zhang X. Genetic susceptibility to SLE: recent progress from GWAS. J Autoimmun (2013) 41:25–33. doi: 10.1016/j.jaut.2013.01.008
161. Chen L, Morris DL, Vyse TJ. Genetic advances in systemic lupus erythematosus: an update. Curr Opin Rheumatol (2017) 29(5):423–33. doi: 10.1097/BOR.0000000000000411
162. Langefeld CD, Ainsworth HC, Cunninghame Graham DS, Kelly JA, Comeau ME, Marion MC, et al. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat Commun (2017) 8:16021. doi: 10.1038/ncomms16021
163. Yin X, Kim K, Suetsugu H, Bang SY, Wen L, Koido M, et al. Meta-analysis of 208370 East asians identifies 113 susceptibility loci for systemic lupus erythematosus. Ann Rheum Dis (2021) 80(5):632–40. doi: 10.1136/annrheumdis-2020-219209
164. Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun (2009) 10(5):373–9. doi: 10.1038/gene.2009.39
165. Harley JB, Kelly JA, Kaufman KM. Unraveling the genetics of systemic lupus erythematosus. Springer Semin Immunopathol (2006) 28(2):119–30. doi: 10.1007/s00281-006-0040-5
166. Lee HS, Bae SC. Recent advances in systemic lupus erythematosus genetics in an Asian population. Int J Rheum Dis (2015) 18(2):192–9. doi: 10.1111/1756-185X.12498
167. Rhodes B, Vyse TJ. The genetics of SLE: an update in the light of genome-wide association studies. Rheumatol (Oxford) (2008) 47(11):1603–11. doi: 10.1093/rheumatology/ken247
168. Sawada T, Fujimori D, Yamamoto Y. Systemic lupus erythematosus and immunodeficiency. Immunol Med (2019) 42(1):1–9. doi: 10.1080/25785826.2019.1628466
169. Ghodke-Puranik Y, Niewold TB. Immunogenetics of systemic lupus erythematosus: a comprehensive review. J Autoimmun (2015) 64:125–36. doi: 10.1016/j.jaut.2015.08.004
170. Ramos PS, Shaftman SR, Ward RC, Langefeld CD. Genes associated with SLE are targets of recent positive selection. Autoimmune Dis (2014) 2014:203435. doi: 10.1155/2014/203435
171. Chen HW, Barber G, Chong BF. The genetic landscape of cutaneous lupus erythematosus. Front Med (Lausanne) (2022) 9:916011. doi: 10.3389/fmed.2022.916011
172. Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol (2010) 6(12):683–92. doi: 10.1038/nrrheum.2010.176
173. Fike AJ, Elcheva I, Rahman ZSM. The post-GWAS era: how to validate the contribution of gene variants in lupus. Curr Rheumatol Rep (2019) 21(1):3. doi: 10.1007/s11926-019-0801-5
174. Vaughn SE, Kottyan LC, Munroe ME, Harley JB. Genetic susceptibility to lupus: the biological basis of genetic risk found in b cell signaling pathways. J Leukoc Biol (2012) 92(3):577–91. doi: 10.1189/jlb.0212095
175. Flesher DL, Sun X, Behrens TW, Graham RR, Criswell LA. Recent advances in the genetics of systemic lupus erythematosus. Expert Rev Clin Immunol (2010) 6(3):461–79. doi: 10.1586/eci.10.8
176. Graham RR, Hom G, Ortmann W, Behrens TW. Review of recent genome-wide association scans in lupus. J Intern Med (2009) 265(6):680–8. doi: 10.1111/j.1365-2796.2009.02096.x
177. Suurmond J, Calise J, Malkiel S, Diamond B. DNA-Reactive b cells in lupus. Curr Opin Immunol (2016) 43:1–7. doi: 10.1016/j.coi.2016.07.002
178. Reid KBM. Complement component C1q: historical perspective of a functionally versatile, and structurally unusual, serum protein. Front Immunol (2018) 9:764. doi: 10.3389/fimmu.2018.00764
179. Lu J, Kishore U. C1 complex: an adaptable proteolytic module for complement and non-complement functions. Front Immunol (2017) 8:592. doi: 10.3389/fimmu.2017.00592
180. Alperin JM, Ortiz-Fernandez L, Sawalha AH. Monogenic lupus: a developing paradigm of disease. Front Immunol (2018) 9:2496. doi: 10.3389/fimmu.2018.02496
181. Pickering MC, Botto M, Taylor PR, Lachmann PJ, Walport MJ. Systemic lupus erythematosus, complement deficiency, and apoptosis. Adv Immunol (2000) 76:227–324. doi: 10.1016/S0065-2776(01)76021-X
182. Lewis MJ, Botto M. Complement deficiencies in humans and animals: links to autoimmunity. Autoimmunity (2006) 39(5):367–78. doi: 10.1080/08916930600739233
184. Walport MJ. Complement. First Two Parts N Engl J Med (2001) 344(14):1058–66. doi: 10.1056/NEJM200104053441406
185. Korb LC, Ahearn JM. C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes: complement deficiency and systemic lupus erythematosus revisited. J Immunol (1997) 158(10):4525–8. doi: 10.4049/jimmunol.158.10.4525
186. Nagata S. Apoptosis and clearance of apoptotic cells. Annu Rev Immunol (2018) 36:489–517. doi: 10.1146/annurev-immunol-042617-053010
187. Schulze C, Munoz LE, Franz S, Sarter K, Chaurio RA, Gaipl US, et al. Clearance deficiency–a potential link between infections and autoimmunity. Autoimmun Rev (2008) 8(1):5–8. doi: 10.1016/j.autrev.2008.07.049
188. Mackay IR, Leskovsek NV, Rose NR. Cell damage and autoimmunity: a critical appraisal. J Autoimmun (2008) 30(1-2):5–11. doi: 10.1016/j.jaut.2007.11.009
189. Mevorach D, Zhou JL, Song X, Elkon KB. Systemic exposure to irradiated apoptotic cells induces autoantibody production. J Exp Med (1998) 188(2):387–92. doi: 10.1084/jem.188.2.387
190. Ogden CA, deCathelineau A, Hoffmann PR, Bratton D, Ghebrehiwet B, Fadok VA, et al. C1q and mannose binding lectin engagement of cell surface calreticulin and CD91 initiates macropinocytosis and uptake of apoptotic cells. J Exp Med (2001) 194(6):781–95. doi: 10.1084/jem.194.6.781
191. Benoit ME, Clarke EV, Morgado P, Fraser DA, Tenner AJ. Complement protein C1q directs macrophage polarization and limits inflammasome activity during the uptake of apoptotic cells. J Immunol (2012) 188(11):5682–93. doi: 10.4049/jimmunol.1103760
192. Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol (2014) 14(3):166–80. doi: 10.1038/nri3607
193. Cai Y, Teo BH, Yeo JG, Lu J. C1q protein binds to the apoptotic nucleolus and causes C1 protease degradation of nucleolar proteins. J Biol Chem (2015) 290(37):22570–80. doi: 10.1074/jbc.M115.670661
194. Kerr FK, O'Brien G, Quinsey NS, Whisstock JC, Boyd S, de la Banda MG, et al. Elucidation of the substrate specificity of the C1s protease of the classical complement pathway. J Biol Chem (2005) 280(47):39510–4. doi: 10.1074/jbc.M506131200
195. Yeo JG, Leong J, Arkachaisri T, Cai Y, Teo BH, Tan JH, et al. Proteolytic inactivation of nuclear alarmin high-mobility group box 1 by complement protease C1s during apoptosis. Cell Death Discovery (2016) 2:16069. doi: 10.1038/cddiscovery.2016.69
196. Macedo AC, Isaac L. Systemic lupus erythematosus and deficiencies of early components of the complement classical pathway. Front Immunol (2016) 7:55. doi: 10.3389/fimmu.2016.00055
197. Gavalchin J, Seder RA, Datta SK. The NZB X SWR model of lupus nephritis. i. cross-reactive idiotypes of monoclonal anti-DNA antibodies in relation to antigenic specificity, charge, and allotype. identification of interconnected idiotype families inherited from the normal SWR and the autoimmune NZB parents. J Immunol (1987) 138(1):128–37.
198. Chatterjee P, Agyemang AF, Alimzhanov MB, Degn S, Tsiftsoglou SA, Alicot E, et al. Complement C4 maintains peripheral b-cell tolerance in a myeloid cell dependent manner. Eur J Immunol (2013) 43(9):2441–50. doi: 10.1002/eji.201343412
199. Berland R, Fernandez L, Kari E, Han JH, Lomakin I, Akira S, et al. Toll-like receptor 7-dependent loss of b cell tolerance in pathogenic autoantibody knockin mice. Immunity (2006) 25(3):429–40. doi: 10.1016/j.immuni.2006.07.014
200. Degn SE, van der Poel CE, Firl DJ, Ayoglu B, Al Qureshah FA, Bajic G, et al. Clonal evolution of autoreactive germinal centers. Cell (2017) 170(5):913–26 e19. doi: 10.1016/j.cell.2017.07.026
201. Fraser DA, Pisalyaput K, Tenner AJ. C1q enhances microglial clearance of apoptotic neurons and neuronal blebs, and modulates subsequent inflammatory cytokine production. J Neurochem (2010) 112(3):733–43. doi: 10.1111/j.1471-4159.2009.06494.x
Keywords: ANA, SLE, nucleolin, GAR/RGG, alarmin, nucleolus autoimmunity, complement C1
Citation: Wu S, Chen J, Teo BHD, Wee SYK, Wong MHM, Cui J, Chen J, Leong KP and Lu J (2023) The axis of complement C1 and nucleolus in antinuclear autoimmunity. Front. Immunol. 14:1196544. doi: 10.3389/fimmu.2023.1196544
Received: 30 March 2023; Accepted: 19 May 2023;
Published: 09 June 2023.
Edited by:
Nicole Thielens, UMR5075 Institut de Biologie Structurale (IBS), FranceReviewed by:
Myoungsun Son, Feinstein Institute for Medical Research, United StatesMarie-Agnes Dragon-Durey, Université Paris Cité, France
Copyright © 2023 Wu, Chen, Teo, Wee, Wong, Cui, Chen, Leong and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhua Lu, bWljbHVqaEBudXMuZWR1LnNn
 Shan Wu
Shan Wu Junjie Chen
Junjie Chen Boon Heng Dennis Teo
Boon Heng Dennis Teo Seng Yin Kelly Wee
Seng Yin Kelly Wee Ming Hui Millie Wong
Ming Hui Millie Wong Jianzhou Cui
Jianzhou Cui Jinmiao Chen
Jinmiao Chen Khai Pang Leong
Khai Pang Leong Jinhua Lu
Jinhua Lu