- 1Department of Translational Physiology, Infectiology and Public Health, Faculty of Veterinary Medicine, Ghent University, Merelbeke, Belgium
- 2Department of Pharmacology, Weill Medical College, Cornell University, New York, NY, United States
- 3Faculty of Pharmaceutical Sciences, NXTGNT, Ghent University, Ghent, Belgium
- 4Department of Pediatrics, Vagelos College of Physicians and Surgeons, Columbia University Irving Medical Center, New York, NY, United States
Liver-resident NK (lrNK) cells have been studied in humans as well as in mice. Unfortunately, important differences have been observed between murine and human lrNK cells, complicating the extrapolation of data obtained in mice to man. We previously described two NK cell subsets in the porcine liver: A CD8αhigh subset, with a phenotype much like conventional CD8αhigh NK cells found in the peripheral blood, and a specific liver-resident CD8αdim subset which phenotypically strongly resembles human lrNK cells. These data suggest that the pig might be an attractive model for studying lrNK cell biology. In the current study, we used RNA-seq to compare the transcriptome of three porcine NK cell populations: Conventional CD8αhigh NK cells from peripheral blood (cNK cells), CD8αhigh NK cells isolated from the liver, and the liver-specific CD8αdim NK cells. We found that highly expressed transcripts in the CD8αdim lrNK cell population mainly include genes associated with the (adaptive) immune response, whereas transcripts associated with cell migration and extravasation are much less expressed in this subset compared to cNK cells. Overall, our data indicate that CD8αdim lrNK cells show an immature and anti-inflammatory phenotype. Interestingly, we also observed that the CD8αhigh NK cell population that is present in the liver appears to represent a population with an intermediate phenotype. Indeed, while the transcriptome of these cells largely overlaps with that of cNK cells, they also express transcripts associated with liver residency, in particular CXCR6. The current, in-depth characterization of the transcriptome of porcine liver NK cell populations provides a basis to use the pig model for research into liver-resident NK cells.
Introduction
The functional and phenotypical characteristics of peripheral blood natural killer (NK) cells, also referred to as conventional NK (cNK) cells, have been extensively studied since their discovery in the 1970s (1). They were found to predominantly kill virus-infected and malignant cells and produce large amounts of cytokines, particularly IFN-γ, upon stimulation (2). More recently, NK cell populations in non-lymphoid tissues, such as the liver, have been studied and characterized in more detail (3, 4). Liver-resident (lr) NK cells were first described as ‘pit cells’ in the rat liver in 1976 by electron microscopy (5). However, interest in this population was sparked particularly after O’Leary et al. discovered that an intra-hepatic NK cell population in mice could mediate long-lived and antigen-specific adaptive recall responses (6). This led to a better characterization of lrNK cell subsets in mice (7) and humans (8, 9).
Interestingly, lrNK cells are present at a remarkably high frequency as they make up 30-50% of all intrahepatic lymphocytes, compared to cNK cells representing only 5-15% of lymphocytes in the peripheral blood (10, 11). The lrNK population in humans and mice resides primarily in liver sinusoids and interacts with sinusoidal endothelial cells and Kupffer cells, the liver-resident macrophages (4, 12). However, their exact function in the liver is not yet fully understood (13). In mice, lrNK cells are CD49a-positive and express CXCR6 and TNF-related apoptosis-inducing ligand (TRAIL) (14–16). They express high levels of the T-box transcription factor T-bet and low levels of Eomesodermin (Eomes) (7). In contrast, the CD49a+ intrahepatic NK cell population in human represents only a very minor and variable subset of the total lrNK cell population (8). The majority of lrNK cells in human are defined as CD56brightCD49a-CD49e-CXCR6+CD69+ and show an opposite expression of the T-box transcription factors T-bet and Eomes (EomeshighT-betlow) compared to that in murine lrNK cells (9, 17, 18). Also in terms of (potential) plasticity between circulating cNK cells and lrNK cells, there appear to be differences between human and mice. In human, there are indications for such plasticity, e.g. circulating Eomeslow NK cells have been reported to upregulate Eomes expression under the influence of cytokines that are highly expressed in the liver (such as transforming growth factor β (TGF-β) and interleukin (IL)-15) (17). In mice, on the other hand, cNK and lrNK cell populations were shown to arise from separate lineages with only a limited degree of plasticity (19).
The abundant differences between human and mice lrNK cells (20) complicate extrapolation of data generated on murine lrNK cells to man. Interestingly, we recently identified two subpopulations of NK cells in the porcine liver (21). We described a CD8αhigh subpopulation, with a phenotype much like conventional CD8αhigh NK cells found in the peripheral blood, and an abundant liver-resident CD8αdim subpopulation, which strongly resembles human lrNK cells (21). Indeed, like their human counterparts, porcine CD8αdim lrNK cells are EomeshighT-betlowCXCR6+CD49e- and there are indications for partial plasticity between porcine cNK cells and lrNK cells (21).
In the current study, we compared the transcriptome of porcine conventional liver CD8αhigh NK cells, porcine liver-resident CD8αdim NK cells, and porcine peripheral blood cNK cells using RNA-seq, to gain new insights into the potential biological functions of these still largely enigmatic lrNK cells. The in-depth characterization of porcine liver NK cell populations reported here, supports the future use of the pig as an animal model to study lrNK cells, which are difficult to obtain in human. These data form a basis to further characterize the biology and functions of porcine (and human) lrNK cells and possibly target these cells in future vaccination strategies and/or novel therapies against liver-associated diseases.
Materials and methods
Isolation of porcine blood NK cells
Heparinized blood samples (50 units/ml blood, LEO Pharma, Lier, Belgium) were obtained from the external jugular vein of pigs (11–23 weeks old) that were kept as blood donors at the Faculty of Veterinary Medicine, Merelbeke, Belgium. The blood sampling and euthanasia procedures were approved by the Ethical Committee of the Faculty of Veterinary Medicine (EC2017/121). Euthanasia was performed using Euthanimal 20% (natriumpentobarbital 20%, 60mg/2.5kg, IV). Peripheral blood NK cells used for RNA sequencing were isolated from the blood of five six-months-old pigs. Upon blood sampling, the animals were euthanized and the liver was collected to obtain liver NK cells (see section 2.2). NK cells used to perform plasticity experiments were obtained from other animals (11-23 weeks old).
Peripheral blood mononuclear cells (PBMC) were isolated from whole blood by lymphoprep (catalog n. 07851, Axis-Shield, Dundee, UK) density centrifugation. Red blood cells were lysed by a 10 min osmotic shock at room temperature by incubating the collected buffy coat in lysis buffer (composed of 90% NH4Cl 0.83% (w/v) and 10% TRIS 2.06% (w/v) in distilled water at a final pH of 7.2). After a final washing step, PBMC were resuspended in 1 ml phosphate-buffered saline (PBS)- ethylene diamine tetraacetic acid (EDTA) buffer and counted. Afterwards, primary porcine NK cells (CD3−CD172a−CD8α+) were isolated from porcine PBMC as described before (22) by magnetic-activated cell sorting (MACS) depletion of CD3+ and CD172a+ cells (Miltenyi, Bergisch Gladbach, Germany) followed by fluorescence-activated cell sorting (FACS) purification using a BD FACSMelody Cell Sorter (BD Biosciences, Franklin Lakes, NJ, USA) and monoclonal antibodies against porcine CD172a (IgG1, clone 74-22-15a), CD3 (IgG1, clone PPT3), and CD8α (IgG2a, clone 11/295/33) (23, 24). Secondary antibodies used were goat α-mouse IgG2a-AF647 (catalog n. A21241, Invitrogen, Carlsbad, CA, USA) and goat α-mouse IgG1-PE (catalog n. P21129, Invitrogen, Carlsbad, CA, USA). This resulted in a 98.42% ± 0.14% pure population of CD3−CD172a−CD8αhigh blood NK cell population (Supplementary Figures 1, 2). In pigs, NK cells are typically identified based on aCD3−CD172a−CD8+ phenotype, since porcine NK cells do not express CD56 (24). This is in contrast to human NK cells, but similar to what is described in mice and non-human primates such as cynomolgus and rhesus macaques (23). Whether porcine NK cells express NKG2A or NKG2C on their cell surface is unknown and (cross-reactive) antibodies are currently lacking, preventing to use these molecules as NK cell markers in pig.
Isolation of porcine liver NK cells
Blood was collected from five six-months-old pig (see section 2.1), after which the animals were euthanized and the liver was dissected and flushed to obtain liver NK cells from the same pig at the same age for RNA sequencing analysis. Porcine liver NK cells were isolated as described before (21). In short, the portal vein was perfused with 5 l ice-cold PBS, after which a further ice-cold 700 ml PBS was used for a final flush that was collected in different 50 ml centrifuge tubes, followed by concentration of the perfusate by centrifugation. The resulting cell pellet was resuspended in PBS (room temperature), followed by the same protocol described above for primary porcine blood NK cell isolation. FACS purification resulted in a 96.87% ± 0.99% pure population of porcine CD3−CD172a−CD8αhigh liver NK cells and in a 95.14% ± 1.25% pure population of porcine CD3−CD172a−CD8αdim liver NK cells (Supplementary Figure 1). Within the liver lymphocyte population, the CD8αhigh NK cells represented 17.6 ± 8.2% of all cells, whereas the CD8αdim NK cells represented 50.4 ± 16.3% of all cells. After sorting, the post-sort analysis was supplemented with additional control stainings for Tbet and Eomes, as these allow to carefully discriminate the two liver NK cell subpopulations (21) (Supplementary Figure 2).
Immunofluorescence staining
After dissection and flushing of the liver, liver tissue samples were collected. The samples were embedded in 2% Methocel® MC (catalog n. 9004-67-5, Fluka, Buchs, Switserland), snap-frozen in liquid nitrogen, and stored at -80°C until use. Cryosections (5 µm) were cut with a cryotome (Leica CM3050 S), placed on aminopropyltriethoxysilane (APES)-coated glass slides (catalog n. A3648, Sigma-Aldrich, Saint Louis, MO, USA) and fixed in methanol for 30 minutes at -20°C. The slides were dried and stored at -20° until use. Tissue slides were washed 3x 5 minutes with PBS and blocked with PBS + 10% goat serum for 30 minutes in a humid cell at 37°C. Primary antibodies (Table 1) were diluted in PBS and incubated for 1 h in a humid cell at 37°C. Then, slides were washed again 3x 5 minutes with PBS, secondary or primary conjugated antibodies were added (Table 1) and incubated for 1 h in a humid cell at 37°C. Secondary antibodies used were goat-anti-rabbit-Alexa Fluor 488 (catalog n. A-11008, Invitrogen, Carlsbad, CA, USA) and goat-anti-mouse IgG1-Alexa Fluor 647 (catalog n. A-21240, Invitrogen, Carlsbad, CA, USA). For monoclonal antibody stainings, purified mouse IgG1 was used as an isotype control. For the polyclonal antibody stainings, control stainings were performed using only secondary antibody. Finally, slides were counterstained with Hoechst (10 µg/ml) for 10 minutes and mounted on a microscope slide in mounting solution (Dabco). Images were taken with a confocal microscope (TCS SPE DM2500, Leica) using a Leica 63x/1,3 NA oil lens and processed using Fiji (25).
RNA-seq analysis
Three porcine NK cell populations (circulating blood CD8αhigh NK cells, liver CD8αdim NK cells and liver CD8αhigh NK cells) of five six-months-old pigs were FACS-purified. The total number of cells collected for each NK cell subpopulation and each animal ranged between 1.1 million and 2 million cells. Cells were lysed in RLT buffer and total RNA isolation was performed using the RNeasy mini kit (catalog n. 74004, Qiagen, Venlo, The Netherlands). The QuantSeq 3’ mRNA-seq library prep kit (Lexogen, Vienna, Austria) was used to sequence mRNA from the 3’ end on a Nextseq500 (Illumina, San Diego, CA, USA). FastQ files were trimmed using the QuasR package (26), followed by alignment using Rhisat2 (23) to the Sus scrofa reference genome version 11.1. Quality metrics were computed using FastQC. The R-package DESeq2 (24) was used to identify differentially expressed genes between each group (n=5). Gene set enrichment was performed using Enricher from the Clusterprofiler package (27) on differentially expressed genes with an adjusted p-value of <0.05 and a BaseMean >500. Specific pathway information was visualized using the Pathview package (28). In all of the analyses, all three NK cell subpopulations of all five data points (individual pigs) were included.
Porcine blood NK cell culture
For the plasticity experiments, porcine blood NK cells were cultured in 96-well flat-bottomed plates (Nunc, Thermo Fisher Scientific) at a density of 2.5 × 106 cells/ml in 200 µl RPMI (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% (v/v) fetal calf serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 U/ml penicillin (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), 100 μg/ml streptomycin (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) (referred to as porcine NK medium). NK cells were primed with recombinant human interleukin 2 (IL-2) (20 ng/ml; catalog n. PHC0026, Thermo Fisher Scientific, Waltham, MA, USA) or recombinant porcine IL-10 (30 ng/ml; catalog n. 693 PI, R&D systems, Minneapolis, MN, USA) or a cytokine mix containing recombinant human TGF-β1 (10 ng/ml; catalog n. 7754-BH-005, R&D systems, Minneapolis, MN, USA) and IL-10 (30 ng/ml) or TGF-β1 (10 ng/ml) and IL-2 (20 ng/ml) for 60 h with a medium change after 40 h. Viability of fresh porcine NK cells was > 90%, wereas viability after 60 h in cell culture ranged between 55-65% for conditions without IL-2 (IL-10 and TGF-β + IL-10) and between 70-80% for conditions with IL-2 (IL-2 and TGF-β + IL-2).
Flow cytometry
Cell surface marker staining
Freshly isolated NK cells were stored overnight at 4°C in porcine NK medium with 25% fetal calf serum (FCS) and cultured NK cells were collected from the 96-well plates after 60 h of incubation before analysis. NK cells were washed in PBS-EDTA with 1% FCS (further referred to as staining medium) and incubated for 30 minutes with 0.1% Fixable Live/Dead stain (catalog n. L34963, Invitrogen, Carlsbad, CA, USA). Cells were transferred to 96-well conical bottomed plates and incubated for 30 minutes at 4°C with primary antibodies or isotype as listed in Table 2. After incubation, cells were washed, stained with goat anti-mouse IgG1-PE (catalog n. P21129, Invitrogen, Carlsbad, CA, USA) for 30 minutes at 4°C. Cells were washed again and resuspended in 100 µl staining medium for analysis. Flow cytometry was performed using a Beckman Coulter Cytoflex and samples were analyzed using CytExpert software (Beckman Coulter, Brea, CA, USA).
Transcription factor staining
Freshly isolated NK cells were stored overnight at 4°C in porcine NK medium with 25% FCS and cultured NK cells were collected from the 96-well plates after 60 h of incubation before analysis. NK cells were washed in PBS-EDTA with 1% FCS (staining medium) and incubated for 30 minutes with 0.1% Fixable Live/Dead stain (catalog n. L34963, Invitrogen, Carlsbad, CA, USA). Cells were transferred to 96-well conical bottomed plates and fixed and permeabilized using the Foxp3/transcription factor staining buffer set (catalog n. 00-5523-00, Thermo Fisher, Waltham, MA, USA), according to the manufacturer’s instructions. Next, cells were incubated with fluorescently labeled antibodies against the corresponding transcription factors or isotype control antibodies (Table 2) at 4°C for 30 min and washed with permeabilization buffer (catalog n., 00-5523-00, Thermo Fisher, Waltham, MA, USA). Finally, cells were resuspended in 100 µl staining medium for analysis. Flow cytometry was performed using a Beckman Coulter Cytoflex, and samples were analyzed using CytExpert software (Beckman Coulter, Brea, CA, USA).
Statistical analysis
Statistical analysis of plasticity data was performed using Graphpad Prism 9. Data were analyzed for statistical differences with a Kruskal-Wallis test at the 5% significance level. Post hoc analysis was performed using Dunn’s multiple comparisons test.
Results
Porcine liver-resident CD8αdim NK cells are located in the liver sinusoids and display a distinct transcriptomic profile compared to circulating blood NK cells and CD8αhigh liver NK cells
We have previously shown that lrNK cells are abundantly present in the liver of six-months-old pigs (45.5 ± 13.1% of the lymphocytes) compared to the presence of their circulating counterparts in the blood of these pigs at the same age (21 ± 5.2% of the lymphocytes) (21). Since murine and human lrNK cells were described to reside in liver sinusoids (12, 29), we were interested to know whether this was also the case for porcine lrNK cells. To identify a suitable marker to specifically stain lrNK cells on tissue sections for microscopy, lymphocytes obtained from a flushed liver were phenotypically characterized. NKp46 was found to be expressed by all CD8αdim lrNK cells, whereas only ∼3% of the NKp46-expressing cells did not belong to the CD8αdim NK cell population.This small percentage of non-lrNK cells expressing NKp46 were characterized as CD8αhighCD3+ lymphocytes, which might correspond to a rare NKp46-expressing T cell subset and/or a non-conventional lymphocyte subset that was previously described to be enriched in the porcine liver and that displays both T cell and NK cell features, but functionally resembles NK cells (30). In contrast, CD8αhigh NK cells in the liver did not express detectable levels of NKp46. Therefore, NKp46 was chosen as a suitable marker for CD8αdim lrNK cells (Supplementary Figure 3). As shown in Figure 1A, porcine lrNK cells reside in the liver sinusoids, in close contact with the CD31-expressing sinusoidal endothelium. This is in line with their human and mouse counterparts.
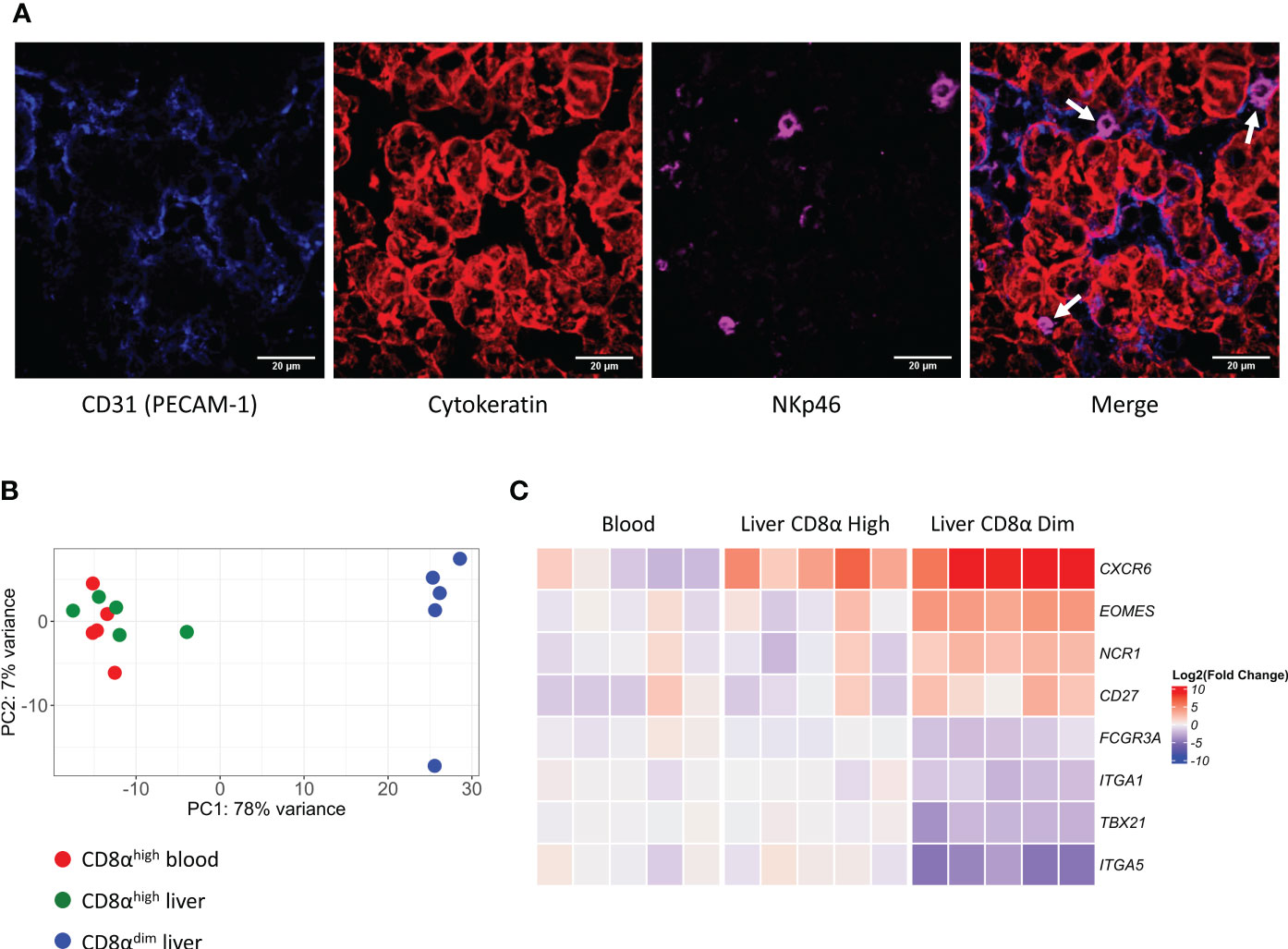
Figure 1 Liver-resident CD8αdim NK cells are located in the liver sinusoids and show a distinct transcriptomic profile compared to liver CD8αhigh NK cells and circulating blood CD8αhigh NK cells. (A) Immunofluorescence staining of liver tissue. NK cells (white arrows) expressing NKp46 (in magenta) are located in the sinusoids, in close contact with the endothelium (PECAM-1, in blue). Hepatocytes stain positive for pan-Cytokeratin (in red). (B) Principal component analysis (PCA) of the transcriptomes of CD8αhigh blood NK cells, CD8αhigh liver NK cells and CD8αdim liver-resident NK cells. (C) Heatmap of gene transcripts of the three described NK cell subsets, displaying the transcription levels of proteins that were previously used to phenotypically characterize liver-resident NK cells.
To further characterize and compare porcine NK cell subsets, we isolated CD8αhigh conventional peripheral blood NK cells, CD8αhigh conventional liver NK cells and CD8αdim liver-resident NK cells from five different animals and performed an RNA-seq analysis on these samples (an overview of the differentially expressed genes (DEGs) can be found in Supplementary Tables 1–3). As visualized by the principal component analysis (PCA) (Figure 1B), CD8αhigh conventional blood NK cells and CD8αhigh conventional liver NK cells cluster together on transcriptional level, whereas the CD8αdim liver-resident NK cells cluster apart from both populations.
We previously described and phenotypically characterized the CD8αdim lrNK cell population by flow cytometry (21). Hence, we compared the transcriptome dataset with this phenotypic characterization on the protein level to determine whether differences in protein levels are reflected by similar differences on transcript level. Interestingly, in line with our previous observations at the protein level, we found that CD8αdim lrNK cells display a significantly increased expression of EOMES mRNA and a significant decrease in TBX21 transcripts, compared to conventional CD8αhigh blood NK cells. In addition, the low expression of ITGA5 (encoding CD49e) transcripts and the high expression levels of CXCR6, NCR1 (encoding NKp46) and CD27 mRNA (Figure 1C) are further in line with our earlier flow cytometric characterization (21), thereby validating both the earlier and the current report. Furthermore, in line with human lrNK cells and in contrast with murine lrNK cells (9, 16), porcine CD8αdim lrNK cells did not show high expression of ITGA1 (encoding CD49a).
CD8αdim lrNK cells display an immature and anti-inflammatory phenotype
In order to obtain a general overview of the most pronounced transcriptional differences between the CD8αdim lrNK cell population and conventional blood NK cells, we first analyzed the genes that were abundantly expressed (BaseMean > 500) and that showed a significant Log2 fold change higher than 2 or lower than -3 (since in general more genes showed a negative fold change) (Supplementary Table 1). Using these criteria, 39 differentially expressed genes were identified between the CD8αdim lrNK cell population and conventional blood NK cells (Figure 2).
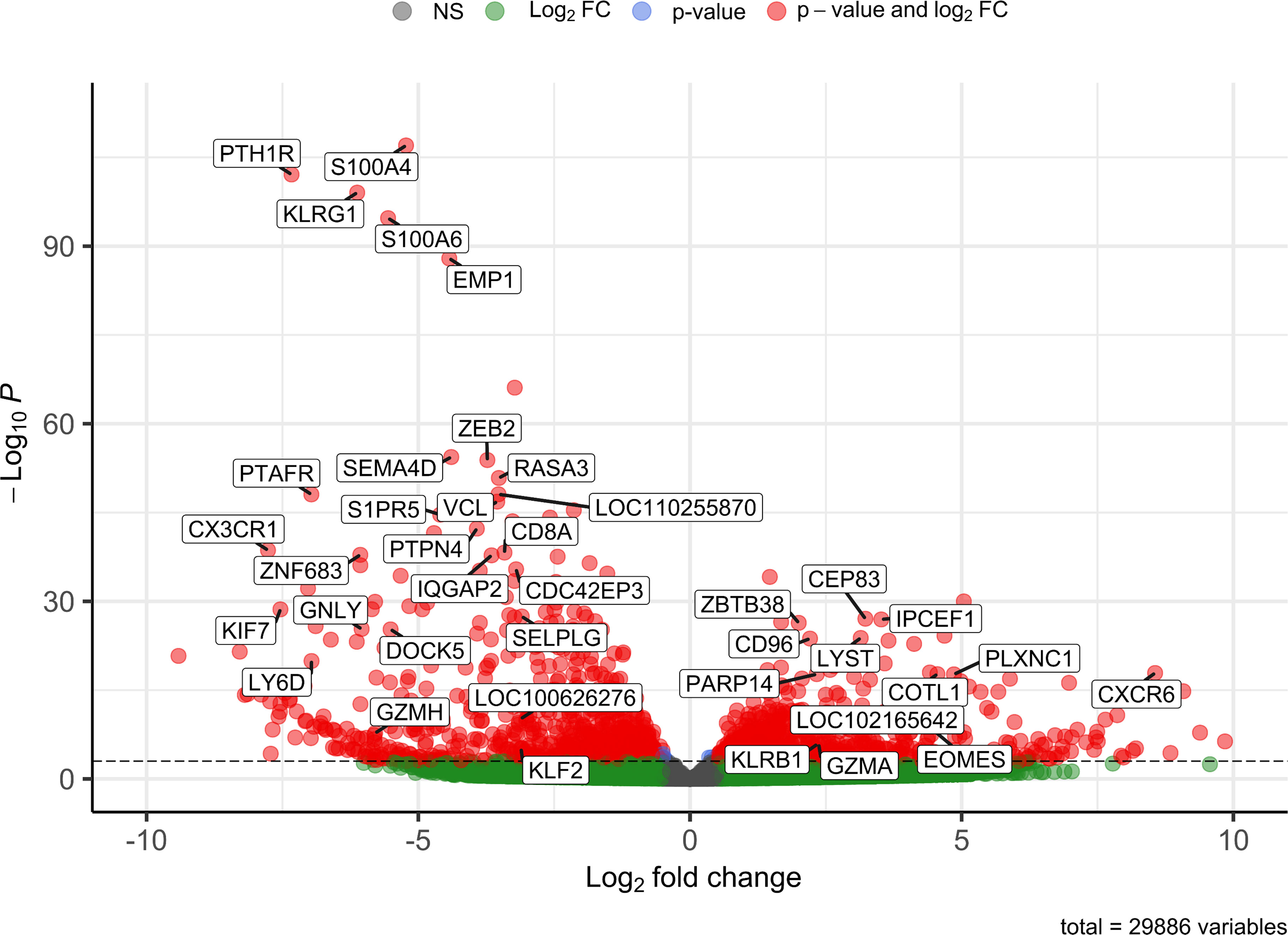
Figure 2 Volcano plot visualizing 39 differentially expressed genes between CD8αdim liver-resident NK cells and conventional blood NK cells. Only transcripts with a BaseMean > 500 and a significant Log2 fold change higher than 2 or lower than -3 are labeled.
A presumably anti-inflammatory function of CD8αdim lrNK cells was suggested by a high expression of CD96 (encoding the activating receptor Tactile) and by a low expression of SEMA4D (encoding CD100, Semaphorin-4D), encoding an immune activation molecule, and of GNLY (Granulysin), encoding a powerful cytotoxic, antimicrobial and pro-inflammatory peptide (31–34). However, abundant expression of genes associated with lytic granules, such as LYST (encoding lysosomal trafficking modulator) and GZMA (encoding granzyme A), and PARP14, encoding a critically important molecule for IFN-γ production, indicate that these cells do have the ability to exert cytotoxic functions and produce cytokines upon activation (35, 36). Nevertheless, granzyme A might preferably act as an immunomodulatory molecule in the pig liver, since human granzyme A has recently been described as a modulator of inflammation rather than a cytotoxic molecule (37).
In addition, CXCR6 transcripts are highly expressed in the CD8αdim lrNK cell population. The ligand for this receptor, CXCL16, is constitutively expressed in the liver and acts as a chemoattractant for CXCR6-expressing cells. Therefore, CXCR6 is regarded as a marker for tissue-resident cells. Besides the high expression of the tissue-resident marker CXCR6, CD8αdim lrNK cells also showed a very low expression of CX3CR1, a marker for circulatory leukocytes, further indicating a tissue-resident phenotype for these cells (38).
Several genes involved in F-actin binding and remodeling of the cytoskeleton, such as S100A4, S100A6, PLXNC1 and COTL1 (39–42), were also differentially expressed. For example, expression of S100A4 and S100A6 was strongly reduced in CD8αdim lrNK cells. In contrast, these cells showed a high expression of PLXNC1 and COTL1. Interestingly, COTL1 is known to be induced by TGF-β (43), which was recently reported to cause downregulation of ZEB2, S1PR5 and KLF2 in human and murine T cells and NK cells to enforce tissue retention of these cells (44). In line with this, we also noticed a downregulation of these three TGF-β sensitive genes, implying that also in the pig liver, TGF-β is a driving force for tissue retention of lrNK cells, mediated via downregulation of S1PR5 in these cells. Moreover, ZEB2 and S1PR5 are markers of NK cell maturation, thus low expression of these transcripts indicates a more immature phenotype (45, 46). Also KLRG1, another maturation marker in NK cells (47), was only very weakly expressed by CD8αdim lrNK cells. A low expression of KLRG1 in NK cells is associated with a higher survival and proliferative potential (48, 49). Altogether, these data indicate that lrNK cells show an immature and anti-inflammatory phenotype, but possibly can still react to pathogens when triggered by specific stimuli.
CD8αdim liver NK cells highly express genes associated with the (adaptive) immune response and IFN-γ signaling, whereas lowly expressed genes are associated with cell migration and extravasation
Using gene-set enrichment analysis (GSEA) based on gene ontology (GO), we found that CD8αdim lrNK cells showed a significantly higher expression of gene sets such as “activation of immune response”, “adaptive immune response”, “antigen processing and presentation”, “antigen processing and presentation of peptide antigen” and “interferon gamma mediated signaling pathway” (Figure 3A). In general, these highly expressed gene sets are linked to the regulation of the (adaptive) immune response and IFN-γ signaling. Interestingly, we found that the two porcine MHC II transcripts (SLA-DQ and SLA-DR) showed a significantly higher expression in CD8αdim lrNK cells compared to cNK cells. This might point towards an antigen-presenting function of these cells, since it has been shown that porcine NK cells can indeed upregulate SLA-DR upon vaccination and may process and present antigens via MHC II to T cells (22, 50). Also transcripts encoding proteins that are involved in an efficient IFN-γ response, such as PARP14, JAK1 and STAT1 were more abundantly expressed in CD8αdim lrNK cells compared to cNK cells. On the other hand, CD8αdim lrNK cells showed low expression of transcripts associated with leukocyte migration and extravasation in comparison to cNK cells (Figure 3B). For example, CX3CR1, ITGA1, ITGA4, ITGAL, ITGB2, ITGB7, CD44, SPN and MSN transcripts were significantly less expressed, highlighting again the resident nature of these cells.
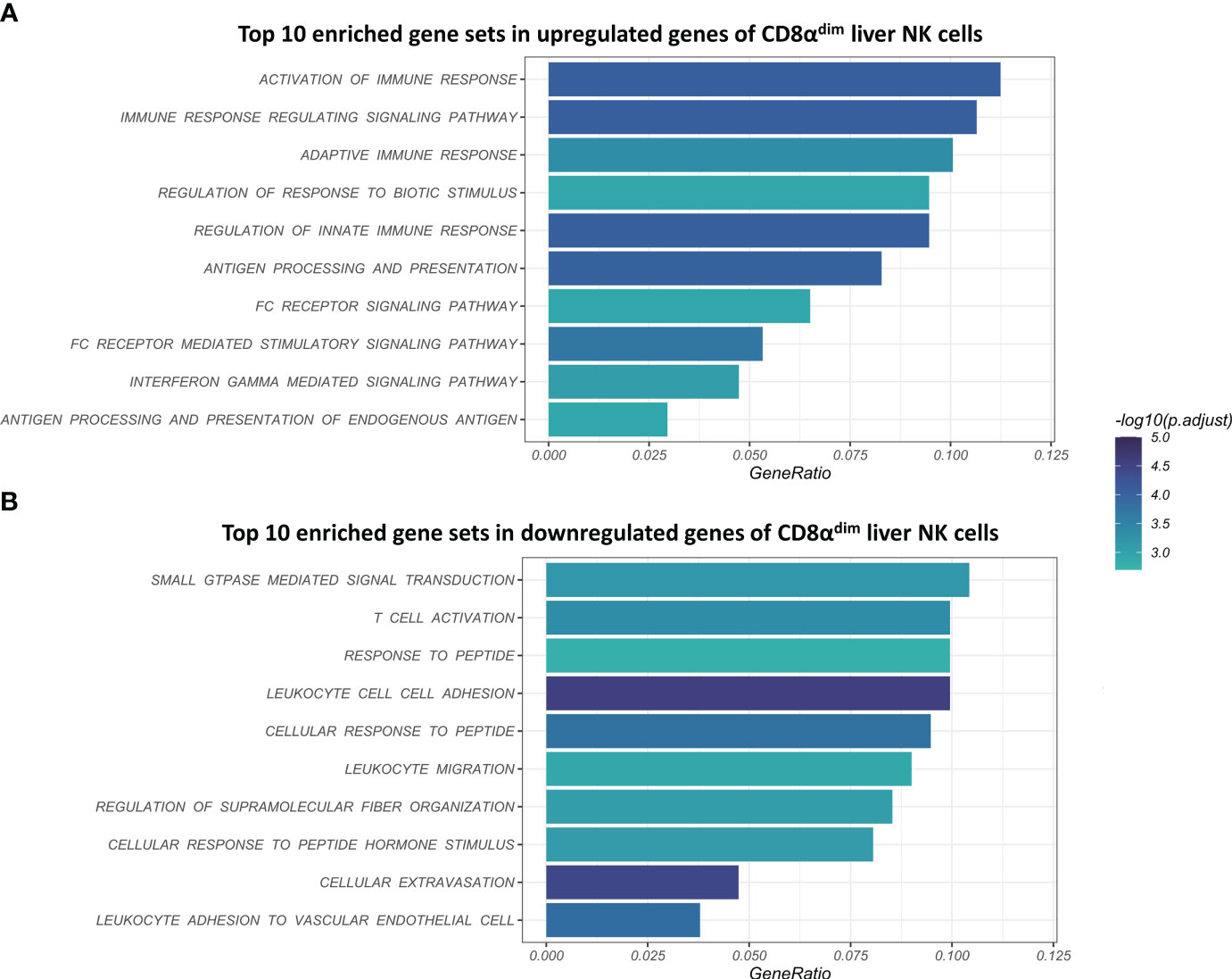
Figure 3 Visualization of the top 10 enriched gene sets of CD8αdim liver-resident NK cells compared to CD8αhigh conventional blood NK cells upon gene set analysis (GSEA) based on gene ontology (GO). (A) Top 10 enriched gene sets in upregulated genes of CD8αdim liver NK cells. (B) Top 10 enriched gene sets in downregulated genes of CD8αdim liver NK cells.
The GSEA pointed towards major changes in the trans-endothelial migration and antigen-presenting capacities of CD8αdim lrNK cells compared to blood NK cells. Therefore, we used Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis to visualize the complete pathways of both processes. KEGG pathways are used to link genomic information with functional cellular processes based on the current knowledge. Figure 4 shows KEGG pathways with transcripts upregulated in CD8αdim lrNK cells compared to cNK cells in green, and downregulated transcripts in red. Regarding antigen processing and presentation, we could confirm that transcripts of several proteins involved in the MHC II pathway were more abundantly expressed in lrNK cells compared to cNK cells (Figure 4A), indicated that this pathway is probably functionally enhanced in lrNK cells. For leukocyte trans-endothelial migration, we observed that all the detected surface proteins involved in trans-endothelial migration, except for ITGB1, were expressed to a much lower extent in CD8αdim lrNK cells compared to cNK cells (Figure 4B), which indicates a less migratory phenotype of these cells, thereby implying a more tissue-resident nature.
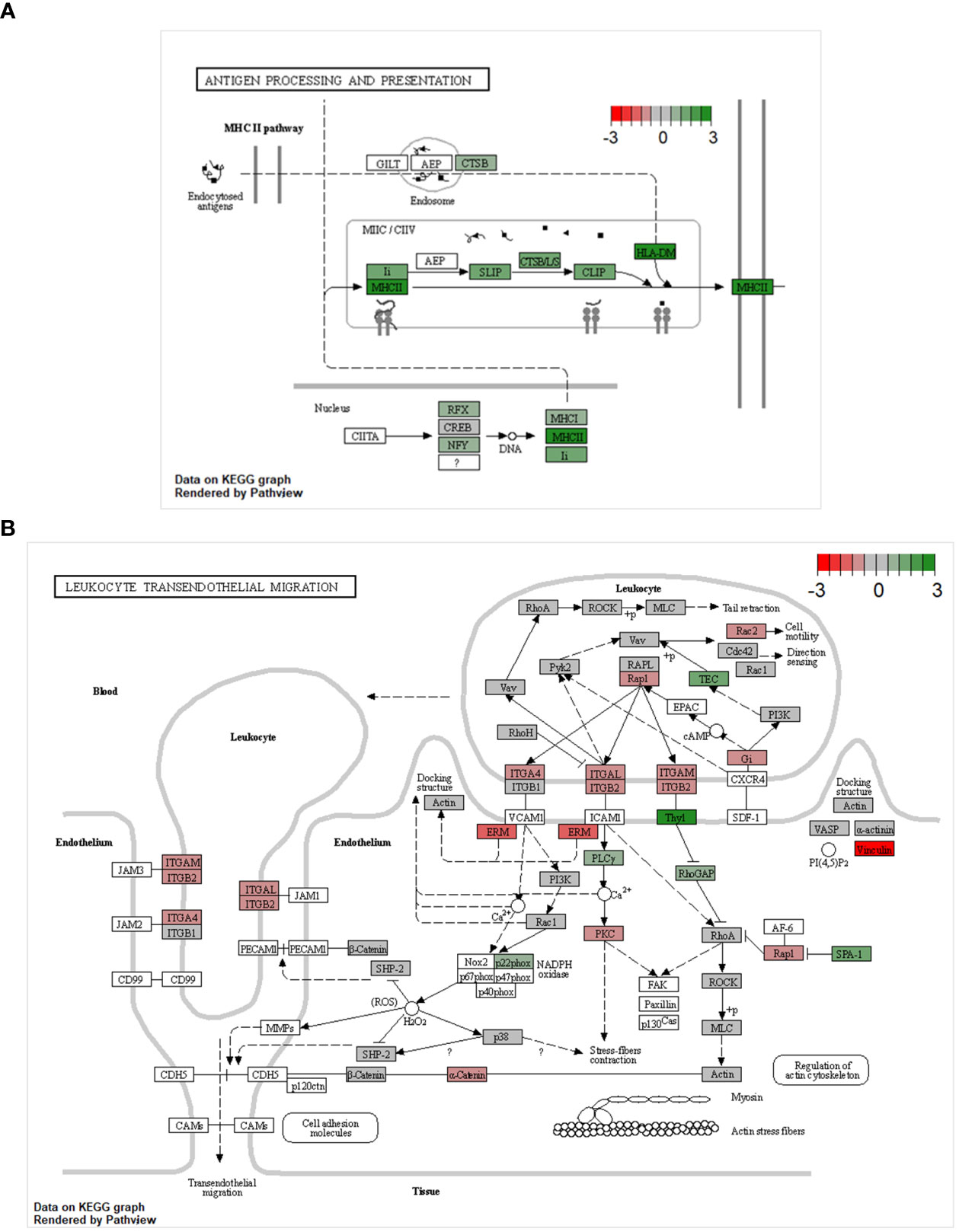
Figure 4 KEGG pathway analysis comparing the transcriptome of CD8αdim liver-resident NK cells to the transcriptome of conventional blood NK cells. KEGG pathways showing transcripts upregulated in CD8αdim liver-resident NK cells compared to conventional blood NK cells in green and downregulated transcripts in red (A) KEGG antigen processing and presentation pathway. Transcripts of several proteins involved in the MHC II pathway are more abundantly expressed in lrNK cells compared to cNK cells. (B) KEGG leukocyte transendothelial migration pathway. All the detected surface proteins involved in trans-endothelial migration, except for ITGB1, are expressed to a much lower extent in lrNK cells compared to cNK cells.
CD8αhigh liver NK cells represent a putative intermediate cell population expressing transcripts associated with conventional blood NK cells as well as with liver resident NK cells
Besides analyzing the CD8αdim lrNK cell population, we were also interested to see whether the CD8αhigh liver NK cell population was either a passer-by population of cNK cells, accidently present in the liver at the moment of liver collection, or rather an intermediate cell population either ‘preparing’ for liver residence or just exiting the liver parenchyma. The former explanation was deemed less likely since the liver was flushed with 5 l ice-cold PBS before collecting the cells, thereby likely removing the vast majority of circulating immune cells. We observed that this CD8αhigh liver NK cell population transcriptionally strongly resembles cNK cells isolated from the peripheral blood, although 22 transcripts were found to be significantly differentially expressed between CD8αhigh liver NK cells and cNK cells (Figure 5) (Supplementary Table 2).
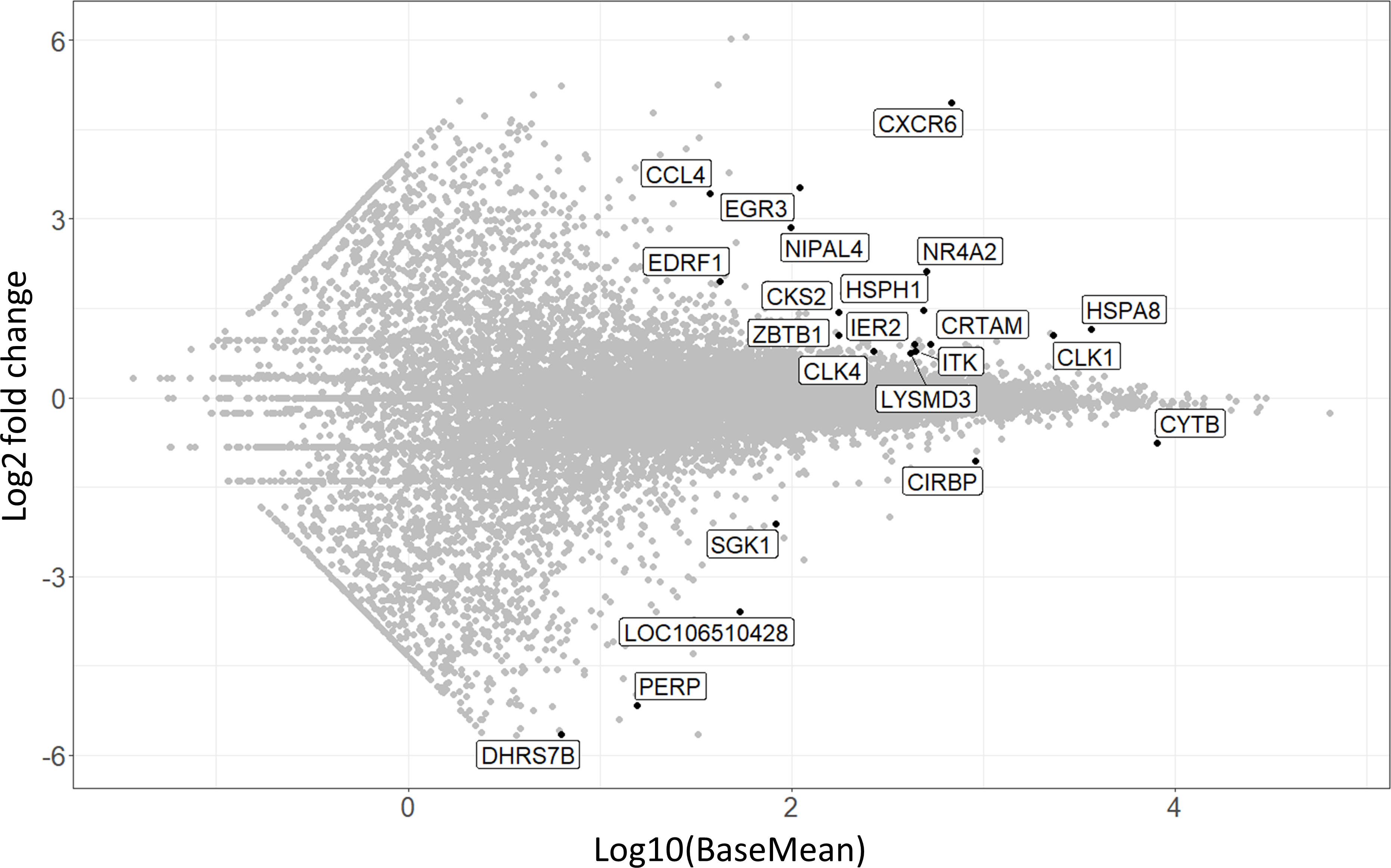
Figure 5 MA-plot visualizing the 22 differentially expressed transcripts in CD8αhigh liver NK cells compared to conventional peripheral blood NK cells. The x-axis shows expression levels, log2 fold change is shown on the y-axis.
Interestingly, CXCR6 was among these 22 transcripts, and showed the highest fold change of all transcripts. This indicates that CD8αhigh liver NK cells isolated from the liver actively upregulate CXCR6 transcription. Intriguingly, these high levels of CXCR6 transcripts do not immediately lead to increased CXCR6 expression at the protein level on the surface of these cells, as we could not detect an increased CXCR6 protein expression on CD8αhigh liver NK cells by flow cytometry (21). Other transcripts that were more abundantly expressed in CD8αhigh liver NK cells compared to cNK cells included EGR3, NR4A2 and HSPA8, which have all been described to be negative immune regulators (51–56). NR4A2, for example, is known as a transcription factor involved in NK cell development and NK cells that express high levels of this transcription factor had a poor cytotoxic activity against tumor cells (53).
Indications for (partial) in vitro plasticity from cNK cells towards CD8αdim lrNK cells
We previously reported limited plasticity between blood cNK cells and CD8αdim lrNK cells, since stimulation of cNK cells with IL-2/12/18 resulted in upregulation of CXCR6 cell surface expression (21). We aimed to further elaborate on these plasticity assays based on the information obtained from this RNA-seq data set. As indicated above, the transcriptomic profile of the CD8αdim lrNK cell population pointed towards a strong influence of TGF-β on these cells. Furthermore, TGF-β and IL-10 were abundantly expressed in healthy human liver (57), although only TGF-β was found to downregulate T-bet expression in human cNK cells (58). TGF-β was thus identified as a critically important regulator of the phenotype of lrNK cells in man (58). Unfortunately, to our knowledge, no studies specifically examining the expression of IL-10 or TGF-β in the pig liver have been performed thus far. The porcine liver cell atlas (59) did show IL-10 transcripts to be expressed by Kupffer cells, the liver-resident macrophages. Since this atlas focuses only on immune cells present in the pig liver, it does not allow to evaluate cytokine expression levels by parenchymal cells.
Therefore, we decided to investigate the effect of TGF-β and IL-10 on porcine cNK cells. In human, plasticity experiments over multiple days are performed in a basal medium containing rhIL-15 to assure NK cell viability (60). In pig, however, rpIL-15 is not sufficient to retain satisfying viability in NK cells and TGF-β treatment was therefore combined with either IL-2 or IL-10, cytokines that are able to maintain porcine cNK cell viability (61, 62). In line with a report in human (58), IL-10 nor TGF-β were able to increase Eomes expression in porcine cNK cells, but TGF-β was able to downregulate T-bet expression in these cells. In contrast to human (58), porcine NK cells also significantly downregulated T-bet expression when treated with IL-10. Furthermore, a trend towards downregulated expression of the integrin CD49e was also observed upon IL-10 treatment of cNK cells, although this difference did not reach statistical significance (Figure 6). These results further support the idea that (partial) plasticity from cNK cells towards lrNK cells may exist in pig, and that TGF-β and IL-10 likely contribute to such process, although on their own they are not sufficient to induce complete transition from cNK cells to lrNK cells.
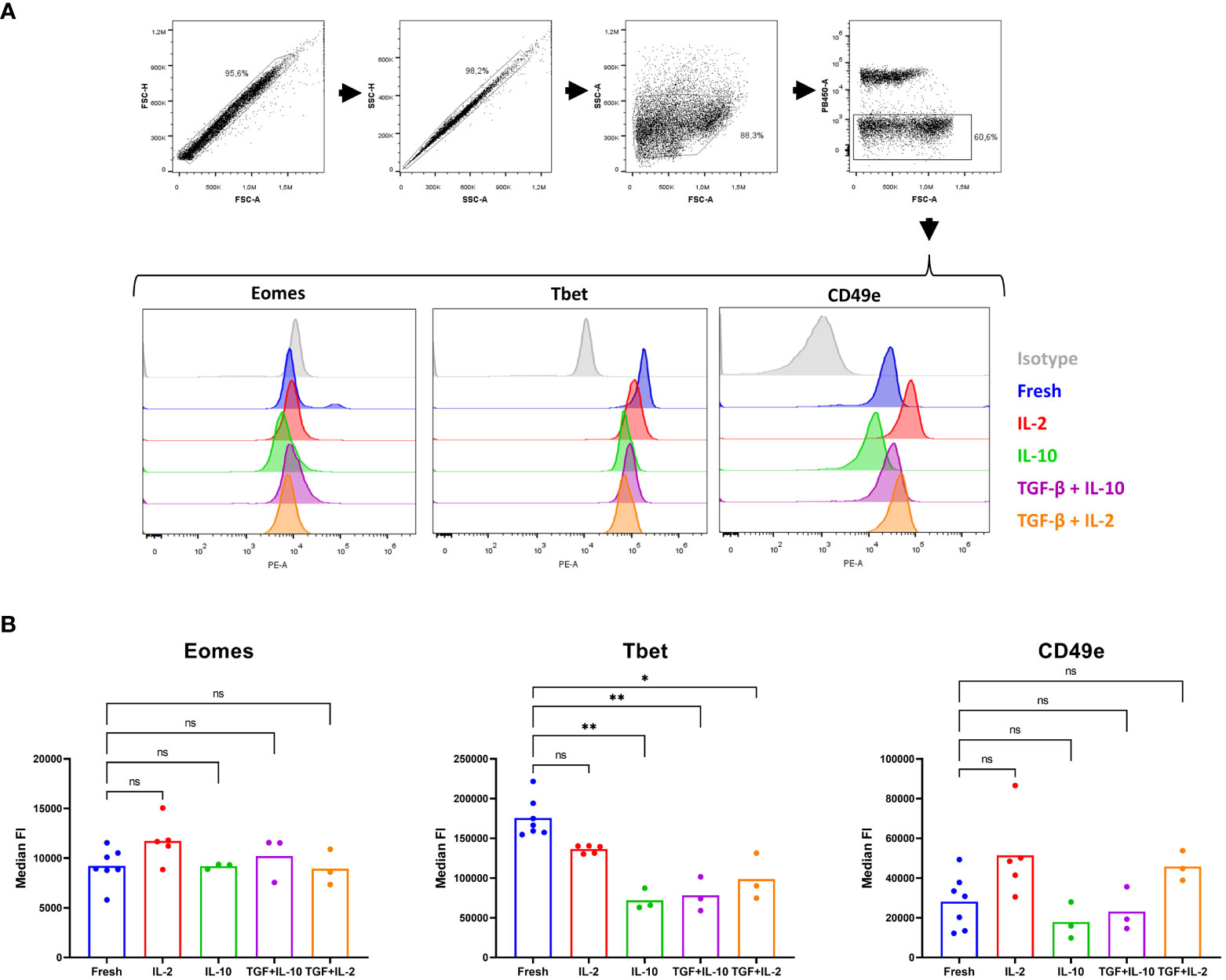
Figure 6 A partial in vitro plasticity is observed upon culturing of conventional blood NK cells with IL-10 and TGF-β. cNK cells were isolated from blood (n = 3 to7) and cultured in the presence of IL-2, IL-10 or combined with TGF-β for 60 h. Subsequently, in freshly isolated cNK cells or cultured cNK cells, expression of three lrNK cell markers were evaluated by flow cytometry. (A) Gating strategy upon staining of fresh or cultured isolated cNK cells. Doublets were excluded based on their forward scatter and side scatter pattern. The light scatter properties of the sorted NK cells were verified. Dead cells were excluded based on a live/dead marker. Histograms show the expression of Eomes, Tbet, CD49e and isotype control for the different conditions. (B) Median fluorescence intensity (FI) of live cells, determined in the PE channel (Eomes, Tbet and CD49e, respectively), is shown. *p < 0.05; **p < 0.01; ns, not significant.
Unfortunately, we could not perform plasticity experiments with CD8αdim lrNK cells to assess their transition towards a cNK cell phenotype, since their viability is severely compromised upon cultivation. Neither IL-2, IL-10, IL-15 nor TGF-β or combinations of these cytokines were able to maintain satisfactory viability of lrNK cells (data not shown).
Discussion
Liver-resident NK cells, as well as other tissue-resident NK cell populations were found to display features that are usually not linked to the innate immune system, such as memory-like responses and longevity (6, 15, 63). Whereas substantial differences exist between lrNK cell populations in mice and human, we have recently identified a porcine CD8αdim lrNK cell population that strongly resembles human lrNK cells (21). In this report, we further characterize this porcine lrNK cell population and compare its transcriptome to that of cNK cells from peripheral blood. Furthermore, we compared both NK cell populations with a second CD8αhigh NK cell population in the pig liver.
We revealed that, in line with data obtained in man, porcine CD8αdim lrNK cells generally display an immature and anti-inflammatory transcriptome. The liver is generally considered as a tolerogenic environment and thus NK cells residing in this milieu seem to adapt to its anti-inflammatory status. In human, TGF-β is thought to play a crucial role in this process (58), and in pig we found that several of the differentially expressed genes between cNK cells and lrNK cells were controlled by TGF-β as well. Moreover, TGF-β was able to downregulate T-bet expression of porcine cNK cells in vitro, indicating a possible plasticity of NK cells between the two compartments.
When studying tissue-resident NK cells, it is important to be aware that another poorly cytotoxic innate immune cell population, highly resembling NK cells, and residing in tissues such as the liver has been described, namely type 1 innate lymphoid cells (ILC1). Due to overlapping phenotypes in the liver, a clear distinction between lrNK cells and ILC1 is controversial, especially in human (64, 65). In contrast, in mice, a liver-specific ILC1 population was identified and is characterized by the expression of CD49b and CD200R1 (66). TGF-β was shown to be involved in the conversion of NK cells towards a less cytotoxic ILC1 population (67, 68). In pig, CD8αdim lrNK cells do not express CD49b or CD200R1, but share the poorly cytotoxic phenotype of murine ILC1. However, like in human, there is no clear evidence for a separate ILC1 population in the pig liver. This is in line with a recently reported liver cell atlas based on single-cell RNA sequencing of livers from different species, where no separate cluster of ILC1 was found in human or pig, whereas in mice a separate ILC1 cluster was identified (59).
In contrast to human (58), in pig, IL-10 also seems to be able to influence T-bet levels in cNK cells, although the biological significance of this cytokine in shaping lrNK cells is currently unclear. Moreover, a cytokine mix containing TGF-β and IL-10 was not sufficient to maintain the viability of porcine lrNK cells in vitro. Additional and currently unknown factor(s) are thus needed for survival and potentially also plasticity of porcine lrNK cells. Whether such factors include chemokines, like for example the CXCR6 ligand CXCL16, and/or survival signal(s) that may depend on cell-cell interactions of lrNK cells with other hepatic cell types is currently unclear and needs further investigation.
Although their origin and development remain elusive, the current transcriptome study allows us to begin to understand the biological functions of lrNK cells in pig. We observed an upregulation of genes associated with antigen presentation, including MHC class II. This indicates that CD8αdim lrNK cells possess immunomodulatory capacities and that it may be warranted to assess whether they may serve as antigen-presenting cells or tissue-resident memory NK cells, as was described previously in mice (6, 63) and human (15). Possibly in line with a potential role in antigen presentation, we previously reported that porcine peripheral blood NK cells can express MHC class II and costimulatory molecules, internalize antigens derived from killed target cells, and may stimulate T cell proliferation (22, 50). Together, these data point towards a very broad and versatile functional repertoire of porcine NK cells. Whether porcine CD8αdim lrNK cells do indeed function in adaptive immune responses – either as antigen-presenting cells, tissue-resident memory NK cells or other – in vivo is still unknown and will likely require vaccination and/or infection trials.
Finally, the CD8αhigh liver NK cell population we characterized in this report, appears to represent an intermediate NK cell population that transcriptionally closely resembles blood cNK cells, but also shows expression of particular transcripts that are associated with the CD8αdim lrNK cell population. This is, to the best of our knowledge, the first description of such a putative intermediate population of liver NK cells in any species and might contribute to further elucidate the origin and development of the lrNK cell population. These CD8αhigh NK cells upregulate transcription of some tissue-resident markers, such as CXCR6, and adopt a more anti-inflammatory transcriptome, seemingly preparing for liver residency. We speculate that under steady-state conditions, these cells may exit the liver again and lose this preparatory phenotype, while, upon encountering specific stimuli, they might adopt a truly tissue-resident phenotype and become tissue-resident cells.
Taken together, we showed that lrNK cells in the pig express high levels of genes related to antigen presentation and generally display an anti-inflammatory and immature phenotype. This confirms that lrNK cells are valuable novel targets for future vaccination strategies or novel therapies against liver-associated diseases. Furthermore, we suggest that, as in human, TGF-β is important for plasticity between blood and liver NK cell subsets, although IL-10 might provide additional support in pig. We also characterized the CD8αhigh liver NK cell population as a putative intermediate subset that might be exploited to provide additional information regarding plasticity in the future. Porcine lrNK cells display poor survival in vitro, even when supplementing with different cytokines, indicating that one or more cardinal factor(s) in porcine lrNK viability and cell biology remain elusive and need to be determined in the future. Although the CXCL16-CXCR6 signaling axis has been described in many studies as a potentially important survival factor for CXCR6-expressing cells (15, 69), we could not enhance the viability of porcine lrNK cells by adding porcine recombinant CXCL16 to the culture medium (data not shown). Potentially additional chemokines might be involved, since our current transcriptome study shows that lrNK cells display increased expression of e.g. transcripts encoding CCR6 and CCR9. In addition, cell-cell interactions might be crucial for lrNK cells in order to survive. Therefore, we believe that establishing live organoids in which CXCL16 and potential other relevant factors can be expressed by sinusoidal cells as is the case in vivo, might be a promising future step to further unravel the functional and biological role of lrNK cells. Such studies will allow to further address the value of the pig as an animal model to translate features of lrNK cells into innovative clinical applications, both in animal and man.
Data availability statement
The datasets generated and analyzed for this study can be found in NCBI’s Gene Expression Omnibus (GEO) repository and are accessible through GEO Series accession number GSE229497 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE229497).
Ethics statement
The animal study was reviewed and approved by Ethical Committee of the Faculty of Veterinary Medicine, Ghent University (EC2017/121).
Author contributions
LH, SDP, SD, and EH-S performed the experiments. LH, EM, EC, HF, and BD designed the research and wrote the manuscript. LH and RJ made the figures. LH, RJ, DD, and FV analyzed the results. All authors contributed to the article and approved the submitted version.
Funding
LH is supported by a Ghent University assistantship, a travel grant from FWO-Vlaanderen (grant number K204921N) and the Ghent University mobility fund. SD is supported by a Ph.D. grant from FWO-Vlaanderen (grant number FWO.SPB.2019.002.01). RJ was supported by a fellowship from the Belgian American Education Foundation (BAEF). This research is supported by research projects from the FWO-Vlaanderen (grant number G.0176.15), the Special Research Fund of Ghent University (BOF24Y2021000101 and BOFBAS2018) and the Hercules Foundation (grant AUGE-035).
Acknowledgments
The authors would like to thank Charlotte Helsmoortel, Cliff Van Waesberghe, and Simon Brabant for excellent technical assistance and assistance, and Rudy Cooman and Jan Clement for animal caretaking.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1219078/full#supplementary-material
References
1. Yokoyama WM, Sojka DK, Peng H, Tian Z. Tissue-resident natural killer cells. Cold Spring Harb Symp Quant Biol (2013) 78:149–56. doi: 10.1101/sqb.2013.78.020354
2. Caligiuri MA. Human natural killer cells. Blood (2008) 112:461–9. doi: 10.1182/blood-2007-09-077438
3. Sojka DK, Tian Z, Yokoyama WM. Tissue-resident natural killer cells and their potential diversity. Semin Immunol (2014) 26:127–31. doi: 10.1016/j.smim.2014.01.010
4. Hashemi E, Malarkannan S. Tissue-resident NK cells: development, maturation, and clinical relevance. Cancers (Basel) (2020) 12. doi: 10.3390/cancers12061553
5. Wisse E, van’t Noordende JM, van der Meulen J, Daems WT. The pit cell: Description of a new type of cell occurring in rat liver sinusoids and peripheral blood. Cell Tissue Res (1976) 173:423–35. doi: 10.1007/BF00224305
6. O’Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol (2006) 7:507–16. doi: 10.1038/ni1332
7. Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity (2012) 36:55–67. doi: 10.1016/j.immuni.2011.11.016
8. Marquardt N, Béziat V, Nyström S, Hengst J, Ivarsson MA, Kekäläinen E, et al. Cutting edge: identification and characterization of human intrahepatic CD49a + NK cells. J Immunol (2015) 194:2467–71. doi: 10.4049/jimmunol.1402756
9. Stegmann KA, Robertson F, Hansi N, Gill U, Pallant C, Christophides T, et al. CXCR6 marks a novel subset of T-betloEomeshi natural killer cells residing in human liver. Sci Rep (2016) 6:26157. doi: 10.1038/srep26157
10. Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology (2006) 43:S54–62. doi: 10.1002/hep.21060
11. Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol (2009) 27:147–63. doi: 10.1146/annurev.immunol.021908.132629
12. Hudspeth K, Donadon M, Cimino M, Pontarini E, Tentorio P, Preti M, et al. Human liver-resident CD56bright/CD16neg NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimmun (2016) 66:40–50. doi: 10.1016/j.jaut.2015.08.011
13. Björkström NK, Ljunggren H-G, Michaëlsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol (2016) 16:310–20. doi: 10.1038/nri.2016.34
14. Takeda K, Cretney E, Hayakawa Y, Ota T, Akiba H, Ogasawara K, et al. TRAIL identifies immature natural killer cells in newborn mice and adult mouse liver. Blood (2005) 105:2082–9. doi: 10.1182/blood-2004-08-3262
15. Paust S, Gill HS, Wang B-Z, Flynn MP, Moseman EA, Senman B, et al. Critical role for the chemokine receptor CXCR6 in NK cell–mediated antigen-specific memory of haptens and viruses. Nat Immunol (2010) 11:1127–35. doi: 10.1038/ni.1953
16. Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest (2013) 123:1444–56. doi: 10.1172/JCI66381
17. Cuff AO, Robertson FP, Stegmann KA, Pallett LJ, Maini MK, Davidson BR, et al. Eomeshi NK cells in human liver are long-lived and do not recirculate but can be replenished from the circulation. J Immunol (2016) 197:4283–91. doi: 10.4049/jimmunol.1601424
18. Harmon C, Robinson MW, Fahey R, Whelan S, Houlihan DD, Geoghegan J, et al. Tissue-resident Eomes(hi) T-bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol (2016) 46:2111–20. doi: 10.1002/eji.201646559
19. Daussy C, Faure F, Mayol K, Viel S, Gasteiger G, Charrier E, et al. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J Exp Med (2014) 211:563–77. doi: 10.1084/jem.20131560
20. Jameson G, Robinson MW. Insights into human intrahepatic NK cell function from single cell RNA sequencing datasets. Front Immunol (2021) 12:649311. doi: 10.3389/fimmu.2021.649311
21. De Pelsmaeker S, Denaeghel S, Hermans L, Favoreel HW. Identification of a porcine liver eomeshigh t-betlow nk cell subset that resembles human liver resident nk cells. Front Immunol (2019) 10:2561. doi: 10.3389/fimmu.2019.02561
22. De Pelsmaeker S, Devriendt B, Leclercq G, Favoreel HW. Porcine NK cells display features associated with antigen-presenting cells. J Leukoc Biol (2018) 103:129–40. doi: 10.1002/JLB.4A0417-163RR
23. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol (2019) 37:907–15. doi: 10.1038/s41587-019-0201-4
24. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol (2014) 15:550. doi: 10.1186/s13059-014-0550-8
25. Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods (2012) 9:676–82. doi: 10.1038/nmeth.2019
26. Gaidatzis D, Lerch A, Hahne F, Stadler MB. QuasR: quantification and annotation of short reads in R. Bioinformatics (2015) 31:1130–2. doi: 10.1093/bioinformatics/btu781
27. Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS (2012) 16:284–7. doi: 10.1089/omi.2011.0118
28. Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics (2013) 29:1830–1. doi: 10.1093/bioinformatics/btt285
29. Peng H, Wisse E, Tian Z. Liver natural killer cells: Subsets and roles in liver immunity. Cell Mol Immunol (2016) 13:328–36. doi: 10.1038/cmi.2015.96
30. Mair KH, Stadler M, Talker SC, Forberg H, Storset AK, Müllebner A, et al. Porcine CD3(+)NKp46(+) lymphocytes have NK-cell characteristics and are present in increased frequencies in the lungs of influenza-infected animals. Front Immunol (2016) 7:263. doi: 10.3389/fimmu.2016.00263
31. Chan CJ, Martinet L, Gilfillan S, Souza-Fonseca-Guimaraes F, Chow MT, Town L, et al. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat Immunol (2014) 15:431–8. doi: 10.1038/ni.2850
32. Kurioka A, Cosgrove C, Simoni Y, van Wilgenburg B, Geremia A, Björkander S, et al. CD161 defines a functionally distinct subset of pro-inflammatory natural killer cells. . Front Immunol (2018) 9:486. doi: 10.3389/fimmu.2018.00486
33. He Y, Guo Y, Fan C, Lei Y, Zhou Y, Zhang M, et al. Interferon-α-enhanced CD100/plexin-B1/B2 interactions promote natural killer cell functions in patients with chronic hepatitis C virus infection. Front Immunol (2017) 8:1435. doi: 10.3389/fimmu.2017.01435
34. Krensky AM, Clayberger C. Biology and clinical relevance of granulysin. Tissue Antigens (2009) 73:193–8. doi: 10.1111/j.1399-0039.2008.01218.x
35. Gil-Krzewska A, Wood SM, Murakami Y, Nguyen V, Chiang SCC, Cullinane AR, et al. Chediak-Higashi syndrome: Lysosomal trafficking regulator domains regulate exocytosis of lytic granules but not cytokine secretion by natural killer cells. J Allergy Clin Immunol (2016) 137:1165–77. doi: 10.1016/j.jaci.2015.08.039
36. Grunewald ME, Chen Y, Kuny C, Maejima T, Lease R, Ferraris D, et al. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PloS Pathog (2019) 15:e1007756. doi: 10.1371/journal.ppat.1007756
37. van Daalen KR, Reijneveld JF, Bovenschen N. Modulation of inflammation by extracellular granzyme A. Front Immunol (2020) 11:931. doi: 10.3389/fimmu.2020.00931
38. Yamauchi T, Hoki T, Oba T, Jain V, Chen H, Attwood K, et al. T-cell CX3CR1 expression as a dynamic blood-based biomarker of response to immune checkpoint inhibitors. Nat Commun (2021) 12:1402. doi: 10.1038/s41467-021-21619-0
39. Lazova R, Gould Rothberg BE, Rimm D, Scott G. The semaphorin 7A receptor Plexin C1 is lost during melanoma metastasis. Am J Dermatopathol (2009) 31:177–81. doi: 10.1097/DAD.0b013e318196672d
40. Scott GA, McClelland LA, Fricke AF, Fender A. Plexin C1, a receptor for semaphorin 7a, inactivates cofilin and is a potential tumor suppressor for melanoma progression. J Invest Dermatol (2009) 129:954–63. doi: 10.1038/jid.2008.329
41. Provost P, Doucet J, Stock A, Gerisch G, Samuelsson B, Rådmark O. Coactosin-like protein, a human F-actin-binding protein: critical role of lysine-75. Biochem J (2001) 359:255–63. doi: 10.1042/0264-6021:3590255
42. Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, et al. Functions of S100 proteins. Curr Mol Med (2013) 13:24–57.
43. Ducimetière L, Lucchiari G, Litscher G, Nater M, Heeb L, Nuñez NG, et al. Conventional NK cells and tissue-resident ILC1s join forces to control liver metastasis. Proc Natl Acad Sci (2021) 118:e2026271118. doi: 10.1073/pnas.2026271118
44. Evrard M, Wynne-Jones E, Peng C, Kato Y, Christo SN, Fonseca R, et al. Sphingosine 1-phosphate receptor 5 (S1PR5) regulates the peripheral retention of tissue-resident lymphocytes. J Exp Med (2021) 219:e20210116. doi: 10.1084/jem.20210116
45. van Helden MJ, Goossens S, Daussy C, Mathieu A-L, Faure F, Marçais A, et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J Exp Med (2015) 212:2015–25. doi: 10.1084/jem.20150809
46. Walzer T, Chiossone L, Chaix J, Calver A, Carozzo C, Garrigue-Antar L, et al. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat Immunol (2007) 8:1337–44. doi: 10.1038/ni1523
47. Huntington ND, Tabarias H, Fairfax K, Brady J, Hayakawa Y, Degli-Esposti MA, et al. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J Immunol (2007) 178:4764–70. doi: 10.4049/jimmunol.178.8.4764
48. Scheiter M, Lau U, van Ham M, Bulitta B, Gröbe L, Garritsen H, et al. Proteome analysis of distinct developmental stages of human natural killer (NK) cells. Mol Cell Proteomics (2013) 12:1099–114. doi: 10.1074/mcp.M112.024596
49. Nie Y, Liu D, Yang W, Li Y, Zhang L, Cheng X, et al. Increased expression of TIGIT and KLRG1 correlates with impaired CD56bright NK cell immunity in HPV16-related cervical intraepithelial neoplasia. Virol J (2022) 19:68. doi: 10.1186/s12985-022-01776-4
50. De Pelsmaeker S, Devriendt B, De Regge N, Favoreel HW. Porcine NK cells stimulate proliferation of pseudorabies virus-experienced CD8+ and CD4+CD8+ T cells. Front Immunol (2019) 9:3188. doi: 10.3389/fimmu.2018.03188
51. Taefehshokr S, Key YA, Khakpour M, Dadebighlu P, Oveisi A. Early growth response 2 and Egr3 are unique regulators in immune system. Cent Eur J of Immunology (2017) 42:205–9. doi: 10.5114/ceji.2017.69363
52. Safford M, Collins S, Lutz MA, Allen A, Huang C-T, Kowalski J, et al. Egr-2 and Egr-3 are negative regulators of T cell activation. Nat Immunol (2005) 6:472–80. doi: 10.1038/ni1193
53. Moreno-Nieves UY, Tay JK, Saumyaa S, Horowitz NB, Shin JH, Mohammad IA, et al. Landscape of innate lymphoid cells in human head and neck cancer reveals divergent NK cell states in the tumor microenvironment. Proc Natl Acad Sci (2021) 118:e2101169118. doi: 10.1073/pnas.2101169118
54. Chen J, López-Moyado IF, Seo H, Lio C-WJ, Hempleman LJ, Sekiya T, et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature (2019) 567:530–4. doi: 10.1038/s41586-019-0985-x
55. Krause M, Heck TG, Bittencourt A, Scomazzon SP, Newsholme P, Curi R, et al. The chaperone balance hypothesis: the importance of the extracellular to intracellular HSP70 ratio to inflammation-driven type 2 diabetes, the effect of exercise, and the implications for clinical management. Mediators Inflammation (2015) 2015:249205. doi: 10.1155/2015/249205
56. Garrido C, Gurbuxani S, Ravagnan L, Kroemer G. Heat shock proteins: endogenous modulators of apoptotic cell death. Biochem Biophys Res Commun (2001) 286:433–42. doi: 10.1006/bbrc.2001.5427
57. Kelly AM, Golden-Mason L, Traynor O, Geoghegan J, McEntee G, Hegarty JE, et al. Changes in hepatic immunoregulatory cytokines in patients with metastatic colorectal carcinoma: Implications for hepatic anti-tumour immunity. Cytokine (2006) 35:171–9. doi: 10.1016/j.cyto.2006.07.019
58. Harmon C, Jameson G, Almuaili D, Houlihan DD, Hoti E, Geoghegan J, et al. Liver-derived TGF-β Maintains the eomeshiTbetlo phenotype of liver resident natural killer cells. Front Immunol (2019) 10:1502. doi: 10.3389/fimmu.2019.01502
59. Guilliams M, Bonnardel J, Haest B, Vanderborght B, Wagner C, Remmerie A, et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell (2022) 185:379–396.e38. doi: 10.1016/j.cell.2021.12.018
60. Dunne J, Lynch S, O’Farrelly C, Todryk S, Hegarty JE, Feighery C, et al. Selective expansion and partial activation of human NK cells and NK receptor-positive T cells by IL-2 and IL-151. J Immunol (2001) 167:3129–38. doi: 10.4049/jimmunol.167.6.3129
61. Charley B, Fradelizi D. Differential effects of human and porcine interleukin 2 on natural killing (NK) activity of newborn piglets and adult pigs lymphocytes. Ann Rech Vet (1987) 18:227–32.
62. Hermans L, De Pelsmaeker S, Denaeghel S, Cox E, Favoreel HW, Devriendt B. β-glucan-induced IL-10 secretion by monocytes triggers porcine NK cell cytotoxicity. Front Immunol (2021) 12:634402. doi: 10.3389/fimmu.2021.634402
63. Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature (2009) 457:557–61. doi: 10.1038/nature07665
64. Meininger I, Carrasco A, Rao A, Soini T, Kokkinou E, Mjösberg J. Tissue-specific features of innate lymphoid cells. Trends Immunol (2020) 41:902–17. doi: 10.1016/j.it.2020.08.009
65. Bernink JH, Mjösberg J, Spits H. Human ILC1: to be or not to be. Immunity (2017) 46:756–7. doi: 10.1016/j.immuni.2017.05.001
66. Nabekura T, Riggan L, Hildreth AD, O’Sullivan TE, Shibuya A. Type 1 innate lymphoid cells protect mice from acute liver injury via interferon-γ Secretion for upregulating bcl-xL expression in hepatocytes. Immunity (2020) 52:96–108.e9. doi: 10.1016/j.immuni.2019.11.004
67. Cortez VS, Ulland TK, Cervantes-Barragan L, Bando JK, Robinette ML, Wang Q, et al. SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat Immunol (2017) 18:995–1003. doi: 10.1038/ni.3809
68. Gao Y, Souza-Fonseca-Guimaraes F, Bald T, Ng SS, Young A, Ngiow SF, et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol (2017) 18:1004–15. doi: 10.1038/ni.3800
Keywords: liver-resident, NK cells, transcriptome, pig, plasticity
Citation: Hermans L, Denaeghel S, Jansens RJJ, De Pelsmaeker S, Van Nieuwerburgh F, Deforce D, Hegewisch-Solloa E, Mace EM, Cox E, Devriendt B and Favoreel HW (2023) Comparative transcriptomics of porcine liver-resident CD8αdim, liver CD8αhigh and circulating blood CD8αhigh NK cells reveals an intermediate phenotype of liver CD8αhigh NK cells. Front. Immunol. 14:1219078. doi: 10.3389/fimmu.2023.1219078
Received: 08 May 2023; Accepted: 02 August 2023;
Published: 18 August 2023.
Edited by:
Falko Steinbach, University of Surrey, United KingdomReviewed by:
Friederike Ebner, Free University of Berlin, GermanyHarikrishnan Balachandran, Duke University, United States
Copyright © 2023 Hermans, Denaeghel, Jansens, De Pelsmaeker, Van Nieuwerburgh, Deforce, Hegewisch-Solloa, Mace, Cox, Devriendt and Favoreel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bert Devriendt, Yi5kZXZyaWVuZHRAdWdlbnQuYmU=; Herman W. Favoreel, SGVybWFuLkZhdm9yZWVsQHVnZW50LmJl
†These authors share first authorship
‡These authors share senior authorship
 Leen Hermans
Leen Hermans Sofie Denaeghel
Sofie Denaeghel Robert J. J. Jansens2†
Robert J. J. Jansens2† Steffi De Pelsmaeker
Steffi De Pelsmaeker Filip Van Nieuwerburgh
Filip Van Nieuwerburgh Dieter Deforce
Dieter Deforce Emily M. Mace
Emily M. Mace Eric Cox
Eric Cox Bert Devriendt
Bert Devriendt Herman W. Favoreel
Herman W. Favoreel