- 1Research Center, The Huizhou Central People’s Hospital, Guangdong Medical University, Huizhou, Guangdong, China
- 2School of Public Health, Benedictine University, Lisle, IL, United States
- 3Department of Hepatobiliary Surgery, Affiliated Hospital of Hebei University, Baoding, China
- 4Department of Ultrasound, Hebei Key Laboratory of Precise Imaging of Inflammation Related Tumors, Affiliated Hospital of Hebei University, Baoding, Hebei, China
- 5Central Laboratory, Hebei Key Laboratory of Cancer Radiotherapy and Chemotherapy, Affiliated Hospital of Hebei University, Baoding, Hebei, China
- 6Department of Pathology, The Huizhou Central People’s Hospital, Guangdong Medical University, Huizhou, Guangdong, China
- 7Center of Stem Cell and Regenerative Medicine, Gaozhou People’s Hospital, Gaozhou, Guangdong, China
Hepatocellular carcinoma (HCC) is a significant contributor to cancer-related deaths in the world. The development and progression of HCC are closely correlated with the abnormal regulation of non-coding RNAs (ncRNAs), such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). Important biological pathways in cancer biology, such as cell proliferation, death, and metastasis, are impacted by these ncRNAs, which modulate gene expression. The abnormal expression of non-coding RNAs in HCC raises the possibility that they could be applied as new biomarkers for diagnosis, prognosis, and treatment targets. Furthermore, by controlling the expression of cancer-related genes, miRNAs can function as either tumor suppressors or oncogenes. On the other hand, lncRNAs play a role in the advancement of cancer by interacting with other molecules within the cell, which, in turn, affects processes such as chromatin remodeling, transcription, and post-transcriptional processes. The importance of ncRNA-driven regulatory systems in HCC is being highlighted by current research, which sheds light on tumor behavior and therapy response. This research highlights the great potential of ncRNAs to improve patient outcomes in this difficult disease landscape by augmenting the present methods of HCC care through the use of precision medicine approaches.
1 Introduction
1.1 Background of hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is now the second deadliest malignancy worldwide, with an estimated 782,500 new cases and 745,500 deaths in 2012 (1, 2). Patients with early-stage HCC may have a better chance of survival after undergoing procedures such as surgical resection, liver transplantation, local ablation, and other curative treatments (3). Even in patients who have undergone potentially curative treatment for HCC, the 5-year recurrence rate may reach 80%–90%. However, there is a rather high recurrence rate (4). The disease had advanced considerably by the time the majority of patients were identified with HCC (5). The drugs sorafenib and regorafenib, which are small-molecule targeted therapies, have been approved by the United States Food and Drug Administration (FDA) as the standard treatments for the advanced stage of the illness. Although sorafenib is currently the sole conventional first-line systemic therapy option for advanced HCC, the observed median survival duration is only 3 months. If patients with HCC were showing improvement during treatment with sorafenib, regorafenib would be used as a second-line medication. Nevertheless, based on a phase 3 clinical research report, the median survival rate remained at a mere 10.6 months. Although sorafenib and regorafenib have the ability to improve the overall survival of patients with HCC, their length is not excessive. The negative consequences of these drugs and the rise of drug resistance are further sources of increasing anxiety. This highlights the critical importance of seeking out new medicines, with a focus on developing more reliable markers for HCC early diagnosis, treatment, and prognosis. Moreover, hepatitis has been considered as a factor in HCC (6, 7), and therefore, the factors in hepatitis should be understood (8, 9).
1.2 Signaling networks in hepatocellular carcinoma
The creation of HCC is caused by the disruption of several signaling pathways, both within and outside the cells (10). Initially, there is a disruption of an intricate network of interconnected pathways that regulate the balance between cell growth and cell death. Cells have the potential to acquire angiogenic, invasive, and metastatic characteristics at the advanced phases of the disease. This is achieved by a process that entails the interactions between neoplastic cells and their surrounding environment. In individuals with HCC, genetic or epigenetic alterations in some elements of the transforming growth factor (TGF)-β pathways, wingless-type (WNT), rat sarcoma virus oncogene (RAS), p53, and retinoblastoma (RB) are frequently observed. These alterations are accountable for the emergence of cancer traits. The activation of the cell membrane receptor tyrosine kinase is caused by several growth factors, the role of which has been demonstrated in chronic liver diseases and the development of HCC (11). Focal growth factor (FGF), hepatocyte growth factor (HGF), platelet-derived growth factor (PDGF), transforming growth factor-beta (TGF-α), epidermal growth factor (EGF), and vascular endothelial growth factor (VEGF) are the molecules that activate these pathways. In the end, these substances boost cell survival and proliferation by attaching to specific receptors on cell surfaces. Angiogenesis, invasion, and metastasis are all processes that they initiate. The met proto-onco-gene (MET), also known as the HGF receptor, was overexpressed in 40%–70% of HCCs (12–15). Moreover, it has been discovered that there is an excessive amount of VEGF present, as evidenced by studies (16, 17), and this has been linked to the development of an advanced cancer phenotype (18–22). There was a correlation between angiogenic and invasive phenotypes and FGF overexpression (23–25). It was discovered that this was true when combined with VEFG. According to some research, overexpression of PDGF and its receptor may contribute to the development of HCC (26, 27). Regarding downstream effectors, it has been established that HCC is marked by RAS overexpression (28), while activating point mutations have only been found extremely infrequently (29, 30).
The Wnt/β-catenin pathway is one of the additional signaling pathways usually involved in hepatocarcinogenesis (31). The stability of β-catenin, which allows its translocation to the nucleus, is the outcome of its activation. It binds to TCF/LEF transcription factors, which are particular to T cells, at that spot. Metalloproteases, cyclin D1, VEFG, c-myc, and c-met are only a few of the numerous target genes that these transcription factors activate. Several mechanisms have been identified as responsible for the aberrant activation of the Wnt-β-catenin pathway in HCC. Approximately 12%–26% of HCC exhibit mutations at the N-terminus of β-catenin that result in increased functionality. Additionally, HCCs may also experience deletions, mutations, or epigenetic changes in the E-cadherin gene. Furthermore, approximately 8%–13% of HCCs are characterized by mutations that lead to a loss of function in the AXIN1 or AXIN2 genes (32, 33). This stability of β-catenin could be explained by the fact that growth hormones phosphorylate and then deactivate GSK-3β, in conjunction with these alterations. Some examples of growth factors are insulin, insulin-like growth factor (IGF)-1, FGF-2, EGF, PDGF, HGF, TGF-β, and tumor necrosis factor (TNF)-α. Another possible reason for the stability of β-catenin could be the use of the Erk-priming mechanism or the activity of the HBV-X protein to promote GSK-3Ω inhibition (34). Recent investigations have revealed that hepatic adenomas are characterized by mutations in the β-catenin gene. These modifications are associated with an increased likelihood of developing cancerous changes (35).
There are a number of downstream effectors that are abnormal in cancer, in addition to the primary consequences of signal transduction pathways, which include cell proliferation and survival. For instance, there has been a lot of study on Rb1, cyclins, CDK inhibitors, cyclin-dependent kinases (CDKs), and their coordination in regulating the cell cycle in HCC. A loss of chromosome 13, found in approximately 30% of HCCs, is one possible mechanism by which the RB1 gene is rendered inactive in these tumors (36, 37), or epigenetic processes (38). Point mutations are very rare. The RB1 protein may be rendered inactive due to an aberrant synthesis of Gankirin, a protein that can bind to RB1, enhance RB1 phosphorylation, and ultimately lead to its proteosome complex fragmentation. It was found that Gankirin was upregulated in all HCC examples (39). In addition, compared to the parenchyma around the tumor, HCC tissue often had reduced levels of cell cycle negative regulators, whereas cyclins were upregulated among the components that control the cell cycle (31). Approximately 60% of HCC exhibit an elevated level of cyclin D1/CDK4 (40). Among the CDK inhibitors, p16/lNIK4A, which targets CDK4 and CDK6 specifically and inactivates CDK4/cyclin D1 complexes (41), is functionally inactivated in a significant portion of HCCs. This is because deletions at the short arm of chromosome 9 occur in approximately 20% of HCCs (32, 36, 37) and methylation of the p16/INK4A promoter occurs in 30% to 70% of cases (42, 43). The CDK inhibitors of the CDK interacting protein (KIP) family, more especially CDKN1B/p27 and CDKN1C/p57, are two more proteins that play a role in HCC tumor suppression and direct cell cycle regulation. Research has demonstrated that compared to the surrounding cirrhosis, HCC tissue exhibits reduced expression levels of CDKN1B/p27 and CDKN1C/p57 (44). Twenty to fifty percent of HCCs show a lack of maternal allele methylation at the KvDMRI imprinted locus at eleven p15.5, where the CDKN1C/p57 gene is located. This has been previously confirmed by us and other researchers. Reductions in CDKN1C/p57 expression have been associated with this mechanism (45–48). The overexpression of miR-221/222, which is found in approximately 70% of HCCs, is one of the main processes that lead to the downregulation of p27 and p57 molecules, as will be shown in the next paragraphs (49–51). Conversely, a high p27 expression level is not always associated with a low cell proliferation rate in head and neck malignancies. Evidence suggests that sequestration into complexes containing cyclinD1 and CDK4 renders p27 inactive in such contexts (52). The current review focuses on understanding the epigenetic modifications in HCC, specifically in relation to non-coding RNAs (ncRNAs), in order to gain insight into the dysregulation of molecular pathways and signaling networks.
2 miRNAs in hepatocellular carcinoma
2.1 miRNA biogenesis and interaction with molecular pathways
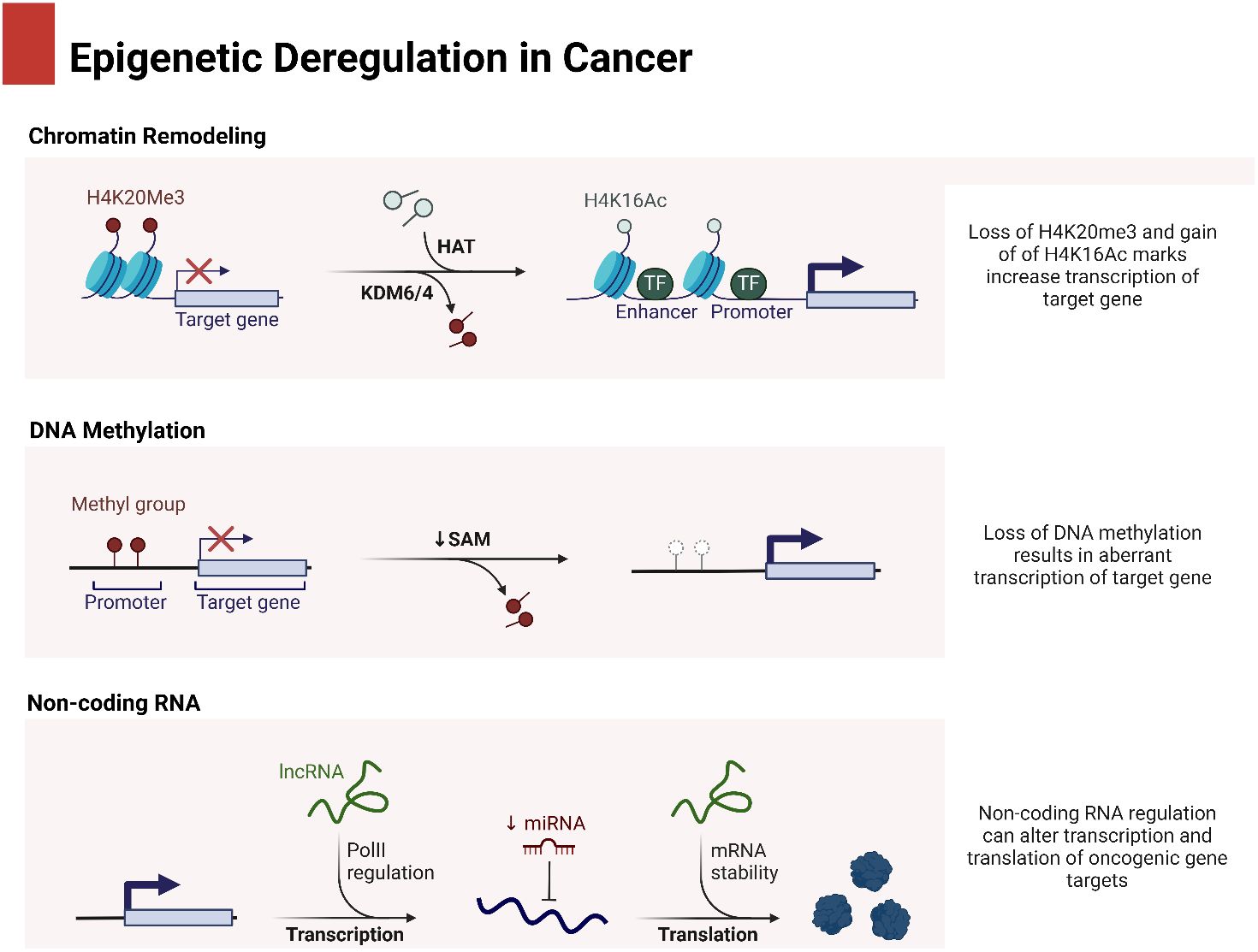
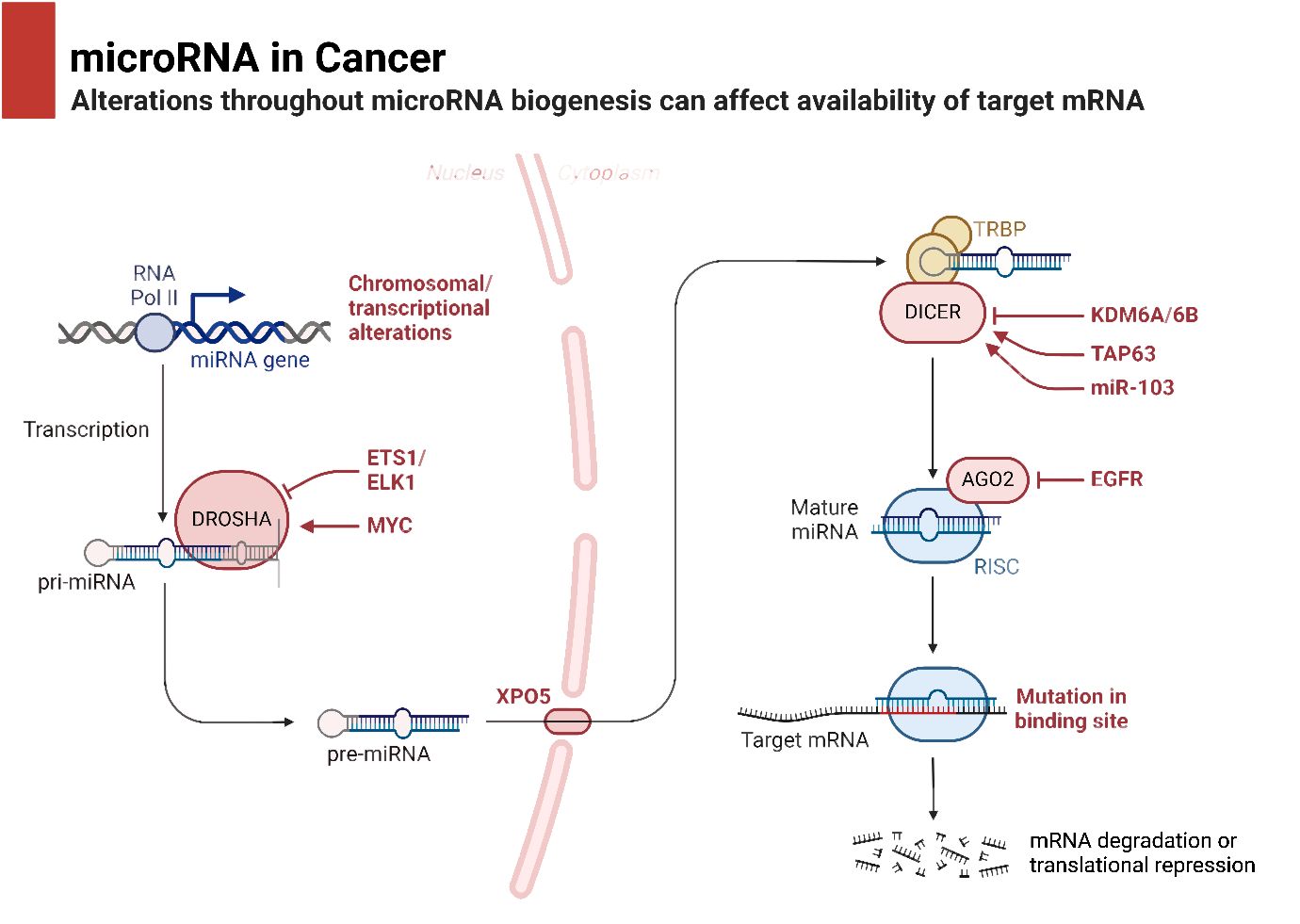
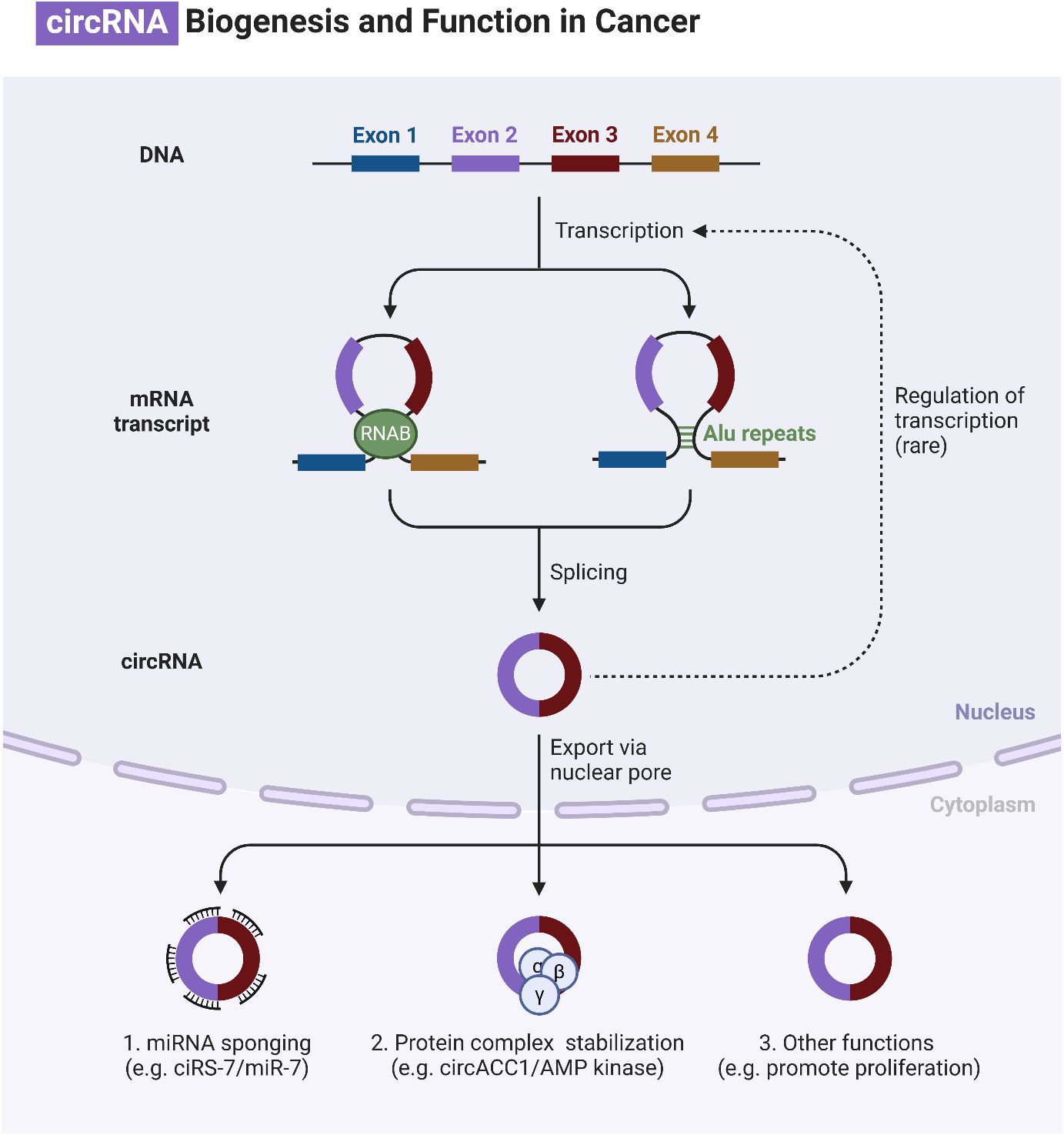
In the process of maturation and decay of RNA molecules, the enzymatic degradation of double-stranded RNA is critical (53). MicroRNAs (miRNAs) are small RNA molecules with one strand that are synthesized naturally by living things. They include approximately 22 nucleotides (54). The miRBase database, accessible at http://www.mirbase.org, now contains 1,492 registered human miRNA sequences. miRNAs, despite lacking coding sequences, have been demonstrated to have a role in the post-transcriptional control of genes that are crucial for fundamental cellular processes and diseases (55). The miRNA gene is typically translated by RNA polymerase II in the nucleus, leading to the formation of a primary transcript called pri-miRNA. This transcript is often between one and four kilobases long (56). Monocistronic transcripts refer to transcripts that contain only a single miRNA gene located downstream of a promoter. On the other hand, the miR-17-92 miRNA polycistron serves as an illustration of a polycistronic transcript, which originates from a single transcript that groups together many miRNA gene products (57). Approximately 50% of miRNA genes, specifically intragenic or mirtrons, are believed to be regulated by the host gene. Intergenic areas are potential sites for them, indicating that they are probably distinct transcriptional units (58). The percentage that changes depends noticeably on where the miRNA genes are located in the genome. The protein DiGeorge syndrome critical region gene 8 (DGCR8) and enzyme nuclease Drosha then come together to form the microprocessor complex. This complex is responsible for converting pri-miRNA, or primary miRNA, into pre-miRNA, or precursor miRNA. It is Exportin-5’s job to deliver the approximately 70-nucleotide pre-miRNA to the cytoplasm. When the miRNA–miRNA duplex reaches its target, another nuclease called Dicer breaks it down into smaller pieces of approximately 18–25 nucleotides (59). Figure 1 is an overview of epigenetic dysregulation in human cancer.
miRNAs demonstrate interaction with various molecular factors in cancer. The miRNAs can dually increase or decrease tumorigenesis, highlighting their function as a double-edged sword. miR-429 impairs the progression of endometrial tumor, and through regulation of DDX53, it enhances drug sensitivity (60). miR-21 and miR-125b are considered as biomarkers in ovarian cancer with overexpression. Moreover, downregulation of miR-125b causes platinum resistance in ovarian cancer (61). miR-1299 is another factor inhibiting cervical tumor. miR-1299 has low expression in cervical cancer, and it is suppressed by KCNQ1OT1 (62). In thyroid cancer, miR-1284 stimulates apoptosis, while it reduces growth and metastasis of tumor cells through E-cadherin upregulation and downregulation of N-cadherin (63). More importantly, miRNAs can develop feedback loop with their targets such as the loop between PAX5 and miR-142 in breast tumor to modulate expression levels of DNMT1 and ZEB1 (64). The plasma levels of miR-1290 and miR-29c-3p in lung cancer are biomarkers (65). Moreover, miR-629 downregulates LATS2 to enhance the growth of prostate tumor (66). The expression of miRNAs can also be regulated by exosomes in human cancers, since the exosomes are able to transfer miRNAs (67). The downregulation of FXYD5 by miR-1180 can suppress the migration and metastasis of pancreatic tumor (68).
It is common practice to remove the miRNA passenger strand before loading a mature miRNA guide strand into the RNA-induced silencing complex (RISC). The extent to which this complex regulates a gene depends on how well the miRNA and its target mRNA sequence complement one another in the 3′-untranslated region (3′-UTR). For RISC to cleave and remove mRNA, the mRNA and RISC must be highly complementary. Conversely, if the complementarity is not ideal, it will hinder the translation process (69). Lower levels of messenger RNA (mRNA) were demonstrated to precede a decrease in protein levels in 84% of the cases (70). Functional mRNA target sites typically consist of six or seven nucleotides and exhibit complementarity to miRNA sequences. The sequence is succeeded by an adenosine, which is commonly known as the “seed” sequence of the miRNA. miRNAs that are targeted for destruction are eliminated in cytoplasmic processing bodies, sometimes referred to as P-bodies, which are situated in the cytoplasm (71). The discovery of additional ncRNAs in the last several years deserves note. Possible functions for these RNAs include regulation of gene expression or association with hepatocellular cancer. The conversion of the short nucleolar RNA (snoRNA) ACA45 into RNAs that bind to Argonaute proteins (Ago) and are 20 to 25 nucleotides long by Dicer provides support for the research conducted by Ender et al. Furthermore, luciferase reporter tests provided evidence that ACA45 displayed miRNA-like functionality (72). Yang and colleagues uncovered a long non-coding RNA (lncRNA) known as High Expression in Hepatocellular Carcinoma (lncRNA-HEIH). The expression of this lncRNA is favorably linked to the recurrence of tumors and negatively correlated with survival. Additionally, its expression is changed in HCC. In addition, they demonstrated that the use of shRNA to downregulate lncRNA-HEIH can greatly reduce tumor growth in a mouse model obtained from xenografts (73). Figure 2 is an overview of miRNA biogenesis along with its function in cancer.
The low 5-year survival rate can be linked to two primary factors: a high rate of recurrence and resistance to treatment. The mortality rate is below 10%. Surgical resection is the most effective treatment for HCC; nevertheless, only a small proportion of patients, namely, 10%, are suitable candidates for this therapy at the time of their initial diagnosis. Conversely, the likelihood of survival after removal is approximately 70% if the tumor is solitary and smaller than 2 cm in size. Therefore, obtaining an early diagnosis is imperative in order to improve the prognosis. While AFP (>400 ng/mL) is commonly utilized as a biomarker for breast cancer, its sensitivity and accuracy are very moderate, and it fails to detect HCC in 50% of patients. Also known as prothrombin induced by vitamin K absence-II (PIVKA-II), another marker is referred to as des-γ-carboxy prothrombin (DCP). The aberrant and functionally inactive form of prothrombin is clearly recognizable due to its lack of N-terminal carboxylation prior to release (74, 75). The carboxylase, which typically performs this function, is frequently missing in HCC cells. By combining these biomarkers with AFP and AFP-L3, it is possible to provide a more accurate prediction of the course of HCC in individuals with chronic HBV or HCV. This is particularly useful for assessing portal vein invasion and intrahepatic metastases (75, 76). However, it is important to note that DCP elevation might be caused by other reasons, and a normal DCP level does not always exclude the presence of HCC. Therefore, it is imperative to possess supplementary biomarkers, namely, those associated with first changes, in order to improve the prognosis.
Dysregulation of miRNAs plays a prominent role during hepatocarcinogenesis. miRNA profiles can differentiate between the general population and patients with HCC, as well as other liver illnesses (54, 77). There is a clear difference in miRNA profiles between normal and cancerous tissues, and these differences may even vary by subtype of cancer (78). The presence of miRNAs in liver tumor tissue, serum, plasma, and urine raises the prospect of a less invasive way to track the treatment’s efficacy and predict the prognosis. Several studies have found a connection between miRNAs and HNS cancer. Higher levels of miR-17-92, miR-21, miR-221, miR-222, and miR-224 are frequently observed in HCC tumors (54, 79). In contrast, miR-199a and miR-199b are often downregulated together with let-7, miR-200, miR-29, miR-122, miR-123, and miR-199b (54, 78, 80). Despite miR-122 being downregulated in initial HCC tumors, it is upregulated in the serum of patients with HCC (81). The secretion of miR-122 into the bloodstream from tumors could be the reason behind this. However, miR-199 is highly expressed in healthy liver tissue, despite its downregulation in HCC (82). The downregulation of miR-199a/b is linked to poor survival rates. This is due to the fact that miR-199a/b-3p reduces HCC by blocking the pathways that involve p21-activated kinase 4 (PAK4), Raf, MEK, and ERL. As in the previous instance, miR-99a downregulation is linked to a bad prognosis (83). In contrast, miR-224 is upregulated in HCC (84), and newer research shows that it reflects tumor stage and liver function; greater levels are associated with a worse prognosis (85). Regional variations in the etiology of HCC may impede the creation of effective biomarker panels. Changes in miRNA expression that are linked to the cause can further hinder these efforts. A recent study utilized miRNA expression profiling of liver tissue to identify dysregulated miRNAs that are linked to HBV or HCV-HCC (86). Out of 40 miRNAs, 12 showed dramatically dysregulated expression. All six of these were confirmed in tissue samples; however, plasma levels of miR-126 and miR-142-3p were shown to be elevated in patients with HBV with HCC compared to those without HCC. Both miRNAs improved the area under the curve (AUC) to 0.92 when added to AFP, but neither of them was more effective than AFP alone. Patients with HCV-related HCC and non-viral HCC did not differ in miR-126 levels. This discovery raises the possibility that miRNA biomarkers’ predictive effectiveness can be affected by changes in the underlying causes of HCC. The prevalence of HBV-related HCC is higher in Asia and other regions with a large HBV population, but the reverse is true in Japan. Despite the fact that vaccines are decreasing the frequency of HBV, non-alcoholic steatohepatitis and other non-viral causes of HCC are increasing (87). Thus, it is necessary to assess biomarkers that have been developed and confirmed in one area in other places that have different underlying causes.
2.2 Expression of miRNAs and function in HCC development
A considerable number of investigations have revealed that miRNAs can affect basic cellular functions including differentiation, proliferation, death, invasion, and metastasis (88, 89). Tumors may exhibit distinct miRNA expression patterns compared to normal tissues. These profiles also vary based on the type of tumor that is present. It is noteworthy because miRNA directly targets protein-coding genes related to the cell cycle, death, and metastasis in HCC (90). A recent microarray study has demonstrated that a specific subset of miRNAs undergoes both upregulation and downregulation throughout the progression of HCC. miRNAs are frequently shown to be reduced in their expression in HCC. The downregulated miRNAs may target oncogenes, indicating their possible role in promoting cancer. In contrast, certain miRNAs that are elevated play a role in the progression of cancer in HCC and may be targeted by genes that inhibit tumor growth.
A variety of chronic liver illnesses have been associated with miRNAs, such as alcoholic liver disease, viral hepatitis, non-alcoholic steatohepatitis (NASH) (91–93), and alcoholic liver disease (94–96). On top of that, miRNAs are involved in the onset of HCC and these other chronic diseases. The accumulation of ethanol-induced hepatocyte fat occurs during alcoholic liver disease because miR-217 suppresses the expression of SIRT1 (97). In alcohol-induced HCC, the expression of the miRNAs miR-126, miR-27b, miR-182, miR-183, miR-199, miR-200a, miR-214, and miR-322 was reduced (79, 96). Patients with alcoholic steatohepatitis had a decrease in miR-27 due to epigenetic changes brought about by alcohol usage (98). The risk of HCC is increased in both the onset and progression of non-alcoholic fatty liver disease (NASH), and miRNA is involved in both stages. Recent research has linked miRNAs to the activation of hepatic stellate cells (HSCs), which, in turn, contribute to the advancement of non-alcoholic fatty liver disease (NASH) (99). An increase in the expression of miRNA-33a and sterol regulatory element-binding protein-2 (SREBP2) resulted in the accumulation of unbound cholesterol. This was achieved by activating HSC and interfering with the SREBP2-mediated cholesterol-feedback pathway in HSC, while also inhibiting the signaling of peroxisome proliferator-activated receptor-γ (99). Overexpression of miRNA-21 causes a decrease in expression of tumor suppressor phosphatase and tensin homolog (PTEN) in hepatocytes when unsaturated fatty acids are present (91). The miRNA miR-155 has the ability to decrease the expression of another tumor suppressor gene known as CCAAT-enhancer-binding protein-β. In addition, mice fed a diet low in amino acids including choline had elevated levels (92, 93).
Hepatitis B virus (HBV) and hepatitis C virus (HCV) infections are the leading causes of cirrhosis and HCC (100). Cirrhosis of the liver is linked to a gradual risk of developing liver cancer, known as hepatocarcinogenesis, which ranges from 5% to 30% within a 5-year time frame (100). The experimental model revealed that just two specific miRNAs, miR-210 and miR-199-3p, had a discernible impact on the expression and replication of the HBV gene in individuals with HBV infection. The life cycle of HBV is indirectly controlled by several cellular miRNAs, which impact cellular proteins related to the virus (101). Ura and collaborators studied the expression of 188 miRNA in HCC and the adjacent normal tissues obtained from 12 patients with HBV infection and 14 patients with HCV infection. The expression of six miRNAs was shown to be downregulated in patients with HBV, while the expression of 13 miRNAs was observed to be downregulated in patients with HCV (102). These findings suggest that the miRNA profiles of HBV and HCV infections differ significantly in terms of pattern. The presence of miR-96 or miR-26 is dramatically increased in tissues linked to HBV-related colon cancer, according to multiple studies. Using sequencing techniques, Takizawa and colleagues were able to uncover whole miRNA profiles from 314,000 trustworthy reads collected from HCC tissue and over 268,000 credible reads obtained from the nearby normal liver. These miRNA profiles were constructed using tissues from both HCCs and adjacent normal livers. A study conducted using bioinformatics discovered that HCC was associated with changes in miRNAs such miR-122, miR-21, and miR-34a (103). Given this information, further investigation into the specific miRNA associated with HBV-related HCC could lead to the creation of a viable therapeutic tool for patients with HCC who have HBV infection. Furthermore, miR-196 has a crucial function in HCV-related non-small cell lung cancer by suppressing Bach1, a mammalian transcriptional repressor with a basic leucine zipper structure, and elevating the expression of hemeoxygenase 1, a transcriptional regulator. Furthermore, Diaz and co-workers found that among a total of 2,226 human miRNA, a specific subset of 18 miRNA exhibited expression only in HCV-related HCC (104). One of these 18 miRNAs has been recognized as being the cause of HCC and is correlated with networks that involve retinoic acid, p53, and PTEN (101, 105). The provided findings declare that miRNA pathways have a noticeable influence on the development of HCC during HCV infection.
The miRNAs have shown significant interaction with other molecular pathways in HCC. miR-21-5p has been shown to induce sorafenib resistance in HCC, and through SIRT7 inhibition, it increases USP24 expression to accelerate tumorigenesis (106). The regulation of lipid synthesis and uptake by the miR-3180 can suppress the progression of HCC (107). The enrichment of miRNAs in the exosomes can affect HCC. Exosomal miR-200b-3p has been shown to suppress ZEB1 in increasing M2 polarization of macrophages and stimulation of the JAK/STAT axis (108). On the other hand, miR-424-3p has been shown to enhance the invasion and migration of HCC through reducing the activation of SRF on STAT1/2 (109). miR-17-5p downregulates TGFβR2 to impair the progression of HCC (110). miR-4270 suppresses DNMT3A-induced methylation of the HGFAC promoter to disrupt the progression of HCC (111). The increase in the levels of caspase-3 and caspase-9 is vital for reduction in the viability of tumor cells. miR-767-3p can downregulate caspase-3/-9 to enhance tumorigenesis in HCC (112).
2.3 Circulating miRNAs in HCC development and diagnosis
miRNAs can enter the bloodstream through two distinct mechanisms: passive diffusion, which does not require energy, or active transport, which is selective and occurs in response to certain stimulants (113). The initial stage occurs during cellular disruption in pathological conditions, such as tissue damage, and does not necessitate the presence of adenosine triphosphate (ATP). The active secretion of miRNA is contingent upon the presence of ATP and the temperature, regardless of cellular stimulation. This is the main mechanism by which circulating miRNA (cmiR) is generated for circulation. Microvesicles (MVs) and apoptotic bodies are two potential methods of encapsulating cmiR. In addition, cmiR can be identified using multiproteins or lipoprotein-binding miRNA complexes (114, 115). The conveyance of cmiR is aided by both the aforementioned structures. Based on the findings, Ago 2 is responsible for overseeing the process by which miRNA is loaded into exosomes. Most cell types have the ability to release exosomes, which can be obtained from various physiological fluids such as serum and urine. Given the large quantity of miRNA found in exosomes, it is extremely probable that exosomal miRNA might be used as significant markers for the detection of diseases (116). A certain group of plasma miRNAs does not contain exosomes or MVs. These miRNAs are protected and made stable by binding to Ago 2, Nucleophosmin 1, or high-density lipoprotein (117). The aforementioned research demonstrates that cmiR is both stable and readily obtainable, making it a viable biomarker for the identification of illnesses. Moreover, the expression of miRNA in tumor tissues may not necessarily correspond to its expression in the blood. As a result, tumor-derived miRNA was conceived. The expression of these miRNAs was found to be elevated in tumor tissues and the bloodstream, but decreased dramatically in the bloodstream after tumor excision. Thus, cmiR plays a pivotal role in monitoring the onset and recurrence of tumors. Budhu et al. propose that identifying a specific miRNA signature linked to metastasis could aid in the early identification of HCC (118). This indicates that the dysregulation of miRNA may have diagnostic value. Furthermore, several studies have shown that miRNA signatures may have additional clinical significance in relation to HCC (119, 120). A number of clinical studies have shown that miRNAs are biomarkers for patients during chemotherapy (NCT03779022) and circulating miRNAs can be utilized for the development of diagnostic factors in brain cancer (NCT03630861). The miRNAs are enriched in serum and plasma of ovarian cancer (NCT06329323). For HCC, miRNAs are also potential diagnostic factors based on clinical studies. The hepatic and circulating miR-221 and miR-222 in HCC have clinical significance (NCT02928627). Moreover, in Somali patients, miRNAs are diagnostic factors in HCC (NCT03227510).
2.4 miRNAs and drug resistance in HCC
At present, the pharmacological therapy for HCC includes targeted treatment, chemotherapy, and immunotherapy. Sorafenib, regorafenib, lenvatinib, and tivantinib are drugs that specifically target molecules at the molecular level. Adriamycin, 5-FU, cisplatin, and oxaliplatin are often prescribed chemotherapy drugs in clinical settings. Immunotherapy is a novel approach for the treatment of HCC. The treatment mostly consists of monoclonal antibodies and immune checkpoint inhibitors that target the cytotoxic T lymphocyte antigen-4 (CTLA-4) receptor, programmed cell death protein 1 (PD-1), and PD-1 ligand (PD-L1). This treatment utilizes immunotherapies such as nivolumab, pembrolizumab, MED14736, ipilimumab, and tremelimumab, among other options. miRNAs are short, single-stranded ncRNAs that consist of 19–24 nucleotides. miRNAs have the ability to attach themselves to the 3′-UTRs of specific mRNAs and inhibit their translation or synthesis. miRNAs are involved in various cellular processes such as differentiation, proliferation, death, angiogenesis, and metabolic stress responses (121–123). In the development and advancement of numerous cancers, including HCC, studies have revealed that dysregulated miRNAs can function as tumor suppressors or oncogenes (124–126). It should be noted that compared to drug-sensitive cells, many drug-resistant HCC cells exhibit drastically different miRNA expression. More tailored HCC treatments may be possible as a result of this finding, which opens the door to the potential use of several miRNAs for medication efficacy prediction (127–130). The expression of immune checkpoint molecules in tumor microenvironments can be controlled by miRNAs (131). However, the precise mechanism by which miRNAs contribute to the development of resistance to immune checkpoint blockers remains incompletely known. In addition, there is emerging research suggesting that other miRNAs may play a role in sorafenib resistance. However, only a limited number of miRNAs have been identified to have a role in developing resistance to other advanced targeted treatments.
Sorafenib, an orally administered multikinase inhibitor, was initially demonstrated to decrease cell growth and blood vessel formation by specifically targeting BRAF, Raf-1, Flt3, VEGFR-2/3, and PDGFR-β. Moreover, it was found that sorafenib possesses the capability to selectively affect signaling pathways that are not reliant on Raf, specifically those involved in regulating apoptosis and cell cycle progression (132–134). Sorafenib, an FDA-approved conventional targeted therapy medication for lung cancer, has demonstrated survival benefits in patients with advanced HCC globally. However, the majority of individuals later developed problems that require treatment resistance. Sorafenib resistance is currently caused by various pathways, such as microenvironmental hypoxia, epithelial–mesenchymal transition (EMT), cellular stem cell activation, resistance to apoptosis, autophagy, cell cycle dysregulation, c-jun and/or AKT activation, and abnormal expression of miRNAs and lncRNAs (130, 135–141). Several cancer-causing miRNAs have the ability to induce resistance to sorafenib (139, 142–145). Sorafenib resistance could be caused by miRNAs such miR-93, miR-216a, and miR-217, which target cell cycle protein-dependent kinase 1A (CDKN1A) and impact apoptosis and TGF-β signaling (143, 144). In addition, miR-181 may target and decrease Ras association domain family member 1 (RASSF1), which could lead to sorafenib resistance (145).
Adriamycin, an antibiotic of the anthracycline class, is a powerful inhibitor of DNA and RNA synthesis in rapidly dividing tumor cells. It is a non-specific periodic medicine. Adriamycin has the capability to permeate into the nucleus of HCC cells, where it can potentially engage with DNA and ultimately induce apoptosis. Multiple mechanisms implicated in the development of adriamycin resistance in HCC have been discovered (146). Oncogenic miRNAs can enhance resistance to adriamycin. There are scientific data indicating that Let-7a has the ability to enhance the resistance of HepG2 cells to the antibiotic adriamycin (147). Furthermore, it has been found that the external production of miR-519d specifically affects numerous tumor suppressor genes, including p21 and PTEN. This ultimately leads to an enhanced resistance of HCC cells to adriamycin (148). In contrast, research has shown that numerous tumor suppressor miRNAs can effectively overcome adriamycin resistance in HCC. For instance, a total of 30 separate HCC tissues exhibited a substantial decrease in the expression of MiR-26a/b compared to normal tissues. Moreover, research has discovered that elevated levels of externally introduced miR-26a/b can augment the responsiveness of HCC cells to adriamycin. This is achieved through the specific targeting of ULK1 expression and the autophagy signaling pathway, both in laboratory settings (in vitro) and in living organisms (in vivo) (149).
2.5 miRNA delivery in HCC therapy
Several miRNAs possess the capacity to be utilized in clinical contexts for the therapeutic management of HCC. However, there is still a dilemma about the method of delivering medicinal chemicals in a more concentrated form to certain tissues (150). The intricate network of hepatocytes and non-parenchymal cells, along with the non-specific uptake of therapeutic vesicles, is the primary challenge that needs to be addressed in order to effectively administer miRNA for HCC treatment. Research has been conducted on non-viral distribution mechanisms. An important advantage of a non-viral strategy is the ability to exert precise control over the functionality of therapeutic medications. Conversely, the drawback of non-viral delivery is that its effects are transient and limited in duration after a certain number of administrations. This limitation arises from the fact that therapeutic miRNAs are susceptible to systemic excretion or degradation (sometimes referred to as degradation). miRNAs can be incorporated into stable nucleic acid lipid particles, or SNALPs, due to their capacity to inhibit rapid disintegration. Coating these SNALPs with polyethylene glycol (PEG) increases their circulation period (151, 152). Increased miRNA stability and off-targeting effects are both brought about by 2′-O-methyl modifications (153). As a result of the inherent viral tropism, virus-like particles (VLPs) can also be utilized for the purpose of distribution. Nevertheless, due to the fact that VLP is closely related to a viral external protein, it has the potential to trigger the immune response (154). Moreover, investigations on miR-375 have illustrated that this miRNA is more effectively delivered to the liver when it is conjugated to cholesterol (153). Nevertheless, as previously stated, the therapeutic effect of miRNAs transferred through non-viral means is only transient. Therefore, it is preferable for miRNAs to exhibit consistent and enduring efficacy in the treatment of chronic and inherited illnesses (155). RNAi sponges are responsible for delivering RNA strands that can be cleaved, hence preventing the degradation of siRNA during transportation. Moreover, the application of RNAi sponges presents a distinctive therapeutic approach due to their capacity to effectively inhibit the target miRNAs in a dominant fashion (156). HepG2 cells that were genetically modified with an adenoviral vector containing a miR-21 sponge experienced a significant decrease in the synthesis of miR-21, resulting in a reduction in the expression of MAP2K (157). Preclinical experiments have successfully utilized viruses to deliver miRNAs for the treatment of HCC. For example, when miR-26a was injected into HCC mice via AAV through the bloodstream, it showed a notable ability to inhibit tumor growth (158). It is not surprising that viral vectors have the ability to provide therapeutic effects that continue for a long time. The clinical safety of these vectors affects their use in clinical settings. The liposome technology known as Smarticles®, licensed from Marina Biotech, Inc (159), is utilized for the delivery of MRX34, the initial miRNA mimic currently undergoing clinical trials. At a neutral pH, Smarticles® possess an anionic charge, whereas in an acidic environment, they acquire a cationic charge. Because tumors have a lower pH, their absorption is enhanced while the risk of adverse interactions with normal tissue is reduced. The liposome component of MRX34 consists of different combinations of palmitoyl oleoyl phosphatidyl choline, dioleoyloxytrimethylammonium propane, 1,2-dimyristoylglycerol-3-hemisuccinate, and cholesterol. These mixes are used to inhibit the degradation of miR-34 and improve its distribution efficiency. Figure 3 provides a comprehensive summary of miRNAs in HCC.
Figure 3 The function of miRNAs in HCC, their association with therapy resistance, and their role in cancer diagnosis.
3 Long non-coding RNAs in hepatocellular carcinoma
3.1 Function of lncRNAs and their interaction with molecular pathways
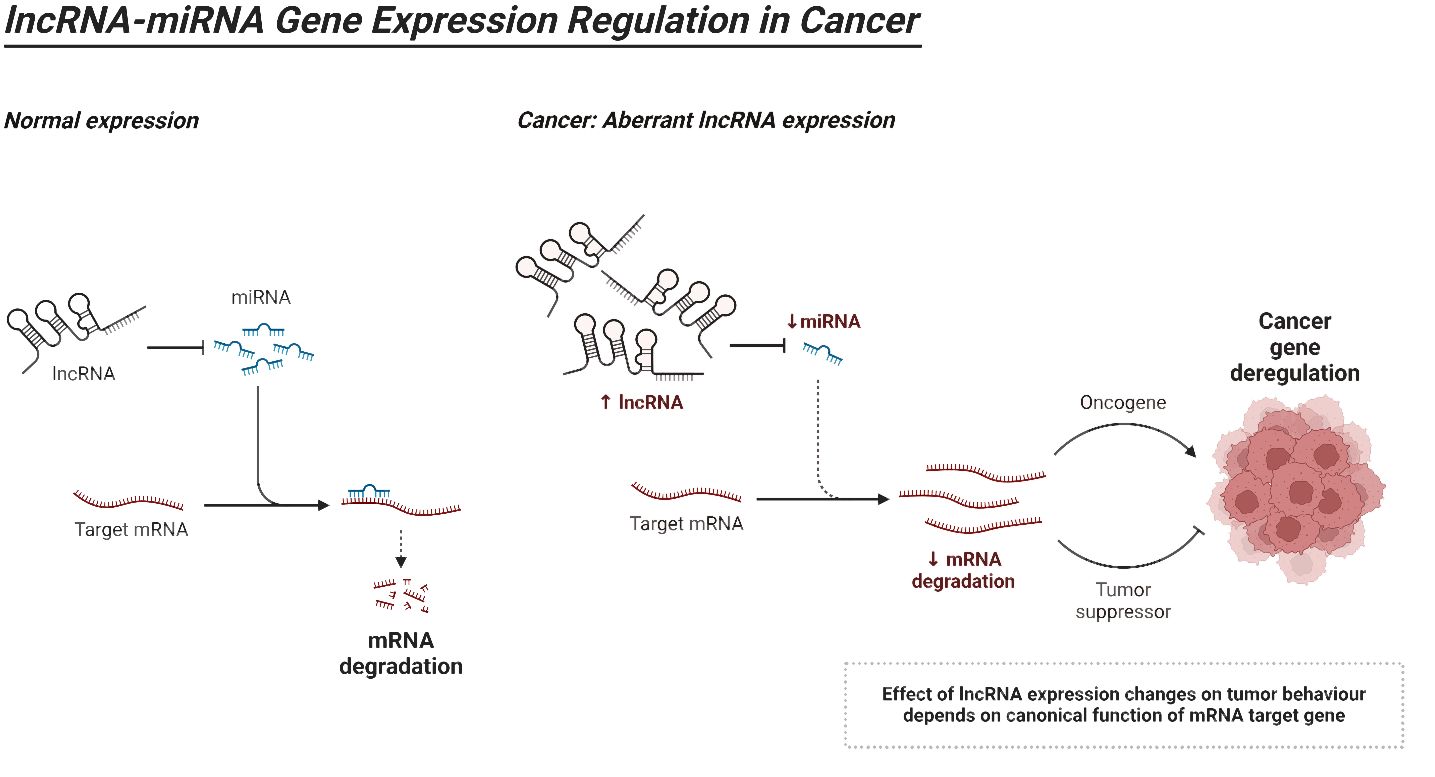
LncRNAs can be classified into five distinct categories according to their proximity to neighboring protein-coding genes: (i) In the sense form, lncRNAs are located in the same region as the sense strand of a protein-coding gene. (ii) In the antisense form, lncRNAs are located in the same region as one or more exons of a protein-coding gene on the opposite strand, and they start at the 3′ end of a protein-coding gene. (iii) In the bidirectional form, both the lncRNA and the protein-coding gene on the opposite strand are expressed within a close distance of less than 1,000 base pairs (bp) from each other in the genome. The intron form of lncRNAs refers to cases when lncRNAs start within an intron of a protein-coding gene, without overlapping exons. On the other hand, the intergenic form, also known as large intervening non-coding RNAs or lincRNAs, are lncRNAs that are located in between genes (160–165). As the investigation of the structure of lncRNAs is still in its early stages, we will provide a summary of the existing structural data on lncRNAs. Regarding the structure and composition of lncRNA, there are three fundamental levels: primary, secondary, and tertiary structures (164). Recent insights into the structural architecture of lncRNAs have enhanced our comprehension of the molecular mechanisms that facilitate the functioning of these RNAs. Furthermore, protein binding sites can be identified inside secondary structures by the presence of duplexes, bulges, hairpins, internal loops, and junctions. These structures serve as the fundamental framework of lncRNAs that control areas without pairing and perform Watson–Crick base pairing (166). In addition, the complex organization of lncRNAs provides sites for interactions and ensures the stability of lncRNAs through the formation of a triple helix at the 3′ end of the lncRNA. The triple helix is used to stabilize lncRNAs that lack a poly(A) tail (167). LncRNAs have a fascinating role in modulating proteins, genes, and ncRNAs. Furthermore, recent research has emphasized the disruption of lncRNAs in different types of human tumors and their connection to the fundamental characteristics and biological processes of cancer (168–170). Figure 4 is an overview of miRNA regulation by lncRNAs in cancer.
LncRNAs demonstrate interaction with other molecular targets in human cancers. LncRNA LITATS1 has been shown to suppress EMT and enhances the degradation of TβRI to impair cancer cell plasticity (171). LncRNA MIR17HG is another factor that has been shown to suppress progression of breast cancer, and this factor sponges miR-454-3p to increase levels of FAM135A (172). Similar to miRNAs, the lncRNAs can be enriched in exosomes, and they are main regulators of miRNAs in cancer (173). The SLC31A1 has been considered as a regulator of cuproptosis in breast tumor, and LINC01614 exerts a modulatory impact on this gene (174). The high expression of HOTAIR in breast tumor mediates unfavorable prognosis, and it demonstrates interaction with miR-129-5p (175). Therefore, lncRNAs have an interaction with various molecular pathways in human cancers including ovarian tumor (176, 177).
3.2 LncRNAs in the regulation of proliferation and metastasis
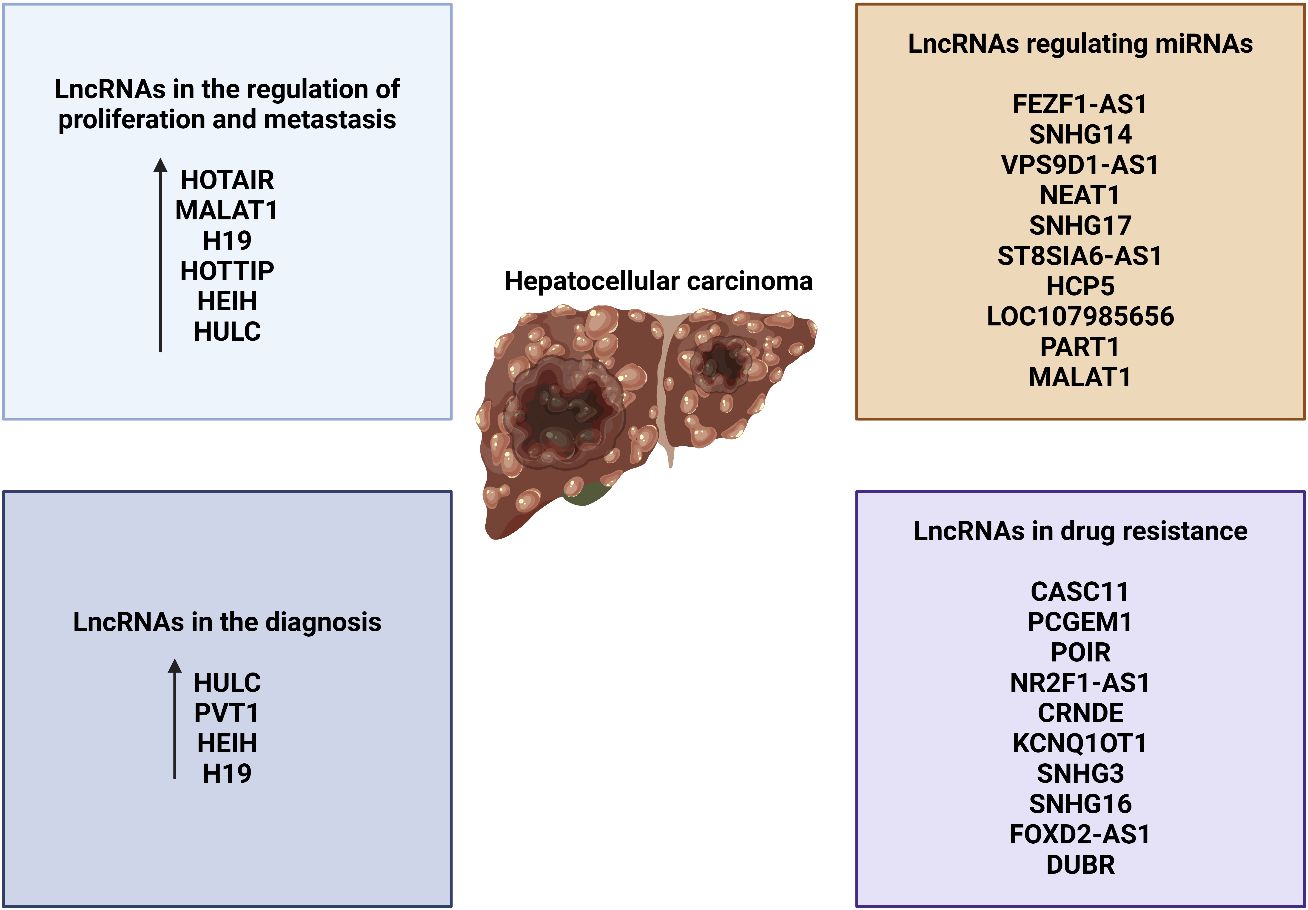
Cancer cells exhibit uncontrolled proliferation in the absence of any external stimulants (161). This is a distinguishing feature exhibited by cancer cells. Cancer cells undergo alterations in the manufacture or function of chemicals that promote or inhibit growth, enabling them to bypass signals that regulate cell proliferation and attain unrestricted growth (89). Investigating the expression of lncRNAs has revealed the existence of lncRNAs that can either stimulate or hinder cell proliferation in many types of cancer, such as prostate cancer. The two lncRNAs that have received the most extensive research attention are HULC and LALR. Both of these lncRNAs have the capacity to impact cell growth by selectively interacting with many essential regulators in separate biological processes. Du and colleagues (178) found that HBV X protein (HBx) can increase the expression of HULC, leading to the promotion of hepatocyte proliferation by reducing the levels of p18 (hyphantria cunea nucleopolyhedrovirus). Their research has demonstrated that HBx can stimulate the HULC promoter via the CREB pathway. By suppressing HULC in a time-dependent manner, there was a significant decrease in cell proliferation in HBx stably transfected cell lines. Conversely, the excessive expression of HULC resulted in an augmentation of cell proliferation in L-O2 cells, which are derived from the parental HepG2 cells. This discovery is of great significance. The study demonstrated that the xenografts derived from same cells in nude mice exhibited equivalent results. Moreover, there is a belief that p18, a tumor suppressor, translocates to the nucleus to initiate the activation of the p53 pathway (179). It is also considered a target gene in human urothelial carcinoma. Subsequent examination uncovered that HULC suppresses the manifestation of p18 in both laboratory and living organism environments, thereby promoting the growth of liver cells. However, it is unclear from this article whether HULC downregulates p18 through direct binding or indirect interaction. Additional validation is necessary now. A further instance of an lncRNA that contributes to cell proliferation is the lncRNA-LALR1 (lncRNA associated with liver regeneration), which consists of approximately 480 bp in mice. According to Xu’s research, LALR1 is specifically increased in hepatocytes after two-thirds partial hepatectomy in mice. The overexpression of this factor can reduce the G0/G1 population in hepatocytes, resulting in an increase in cell proliferation. For a more in-depth examination of the procedure, it is important to mention that LALR1 can inhibit AXIN1 and, thus, stimulate the Wnt/β-catenin pathway in mouse hepatocytes. More specifically, LALR1 can interact with the transcription factor CTCF, causing CTCF to bind to the AXIN1 promoter region and inhibit the production of the gene. The stability of the β-catenin destruction complex decreases when the expression of Axin1 decreases. Meanwhile, proteins like T-cell factor-4 and lymphoid enhancer factor attach to active β-catenin. After being moved to the nucleus, it has an impact on the transcription of particular genes, such as c-myc and cyclin D1. LALR1 regulates the progression of the mouse cell cycle and promotes the growth of hepatocytes. Importantly, research has demonstrated that human liver tissues express a human ortholog of LALR1 (hLALR1), which is a significant discovery. Further investigation is necessary to ascertain the extent to which it exhibits similarities to mouse LALR1 (180).
A growing corpus of studies has demonstrated that the primary tumor is not the primary cause of mortality for many individuals; instead, the metastases are responsible for their demise. The initial stage of both invasion and metastasis involves the process of intravasation, where the cancer cells enter the nearby blood and lymphatic vessels. The following stages involve micrometastases, extravasation, transit through the lymphatic and hematogenous systems, colonization, and intravasation (89, 181). Recent research has demonstrated that a substantial quantity of lncRNAs associated with HCC have important functions in the cellular processes of invasion and metastasis. LncRNA-Dreh, a prototypical example, is an lncRNA that experiences downregulation in expression due to the presence of HBx. It was found that the level of Dreh expression was significantly decreased in HBx-transgenic animals and in mouse liver cells that expressed HBx. Both in laboratory experiments (in vitro) and in living organisms (in vivo), it has been observed that reducing the activity of cellular Dreh is linked to an elevation in the ability of liver cancer cells to migrate and invade surrounding tissues. Studies have demonstrated that Dreh has a higher affinity for vimentin compared to other molecules. Vimentin is the primary structural element of the cytoskeleton in mesenchymal cells and belongs to the type III intermediate filament family. The prevention of HCC metastasis is achieved by altering and reorganizing the expression of vimentin. Transfected cells expressing Dreh exhibit a significant decrease in vimentin levels, accompanied by the development of helical filaments that span from the nuclear membrane to the cellular membrane. However, when subjected to regulation, filament formations withdraw toward the nuclear membrane of the cells. Furthermore, the cytoplasm of cells transfected with Dreh exhibits a substantial presence of filamentous aggregation, together with a considerable quantity of irregular fragmented aggregated structures. Owing to the many alterations made to vimentin, the cells would experience instability, resulting in the facilitation of cell migration. Additionally, a human counterpart of Dreh (hDREH) has frequently been observed to be downregulated in tissues affected by HBV-related HCC. This drop in expression has been strongly associated with lower survival rates among patients with HCC. Given these information, it is crucial to prioritize further investigation of the hDREH in order to further therapeutic approaches (182). Another lncRNA called LET, which stands for low expression in tumor, has been found to have a substantial role in hypoxia-induced metastasis in HCC. LET levels are commonly reduced in various types of cancer, such as squamous cell lung carcinoma, colorectal cancer, and HCC. Through the use of orthotopic tumor models in nude mice, experiments involving both the enhancement and suppression of LET activity have shown that LET effectively reduces the infiltration of the liver and the dissemination of metastases to the abdominal area. Ultimately, Yang and colleagues (183) successfully discovered a pathway that includes hypoxia-inducible factor 1, alpha subunit (HIF-1a), histone deacetylase 3 (HDAC3), and leaflet/NF90. In hypoxic settings, HDAC3, which is controlled by HIF-1a, can inhibit LET via decreasing the histone acetylation-dependent control of the LET promoter region. LET downregulation leads to a decrease in direct interactions between LET and NF90. Consequently, this results in an improvement in the stability of NF90, finally causing a rise in the synthesis of HIF-1a. HIF-1a is a specific mRNA that NF90 targets and is involved in the process of metastasis induced by hypoxia. Thus, the spread of HCC is inhibited by LET through this mechanism of positive feedback. The presence of this feedback loop considerably increases the complexity of the gene regulation network.
The lncRNAs demonstrate interaction with various molecular targets in HCC (184). The lncRNA GBAP1 is stimulated by METTL3, and through the induction of the BMP/SMAD axis, it is able to enhance tumorigenesis (185). The lncRNA SNHG20 has been shown to downregulate miR-5095 and upregulate MBD1 in HCC progression (186). LncRNA SATB2-AS1 is another factor capable of impairing HCC development through suppression of STAT3/HIF-1α and enhancing GRIM-19 levels (187). The lncRNA SNHG1 has the tumor-promoting function in HCC and it can downregulate miR-199a-5p/3p to enhance levels of FANCD2 and G6PD in reducing ferroptosis (188). A number of lncRNAs respond to the hypoxia in the tumor microenvironment. The hypoxia-mediated increase in the levels of lncRNA MRVI10AS1 can recruit the CELF2 as an RNA-binding protein (RBP) to enhance SKA1 stability in increasing HCC progression (189). The cuproptosis-associated lncRNAs can be considered as prognostic factors in HCC (190). The lncRNA HClnc1 is able to induce PKM2 expression and mediates poor survival rate in HCC (191). The lncRNA SLC7A11-AS1 increases the ubiquitination of KLF9 to enhance tumorigenesis in HCC (192). The lncRNA PTOV1-AS1 overexpression modulates miR-505 expression, and it is able to stimulate sorafenib resistance in HCC (193). Taking everything together, the current studies highlight that lncRNAs are versatile factors in HCC and can modulate various hallmarks with different downstream targets (194–196).
3.3 LncRNAs in the regulation of drug resistance in HCC
Considerable progress has been made in the treatment of HCC, thanks to the development of medications. Various anticancer therapies have been employed in this regard. Multiple investigations have been undertaken to investigate the development of treatment resistance in HCC (197–200). Zhang and colleagues (201) discovered that the prevention of miR-15a-5p in HCC leads to resistance to therapy due to the enhanced expression of elF4E. Insufficient radiofrequency ablation in HCC can lead to treatment resistance due to an increase in the expression rate of Hsp70. The suppression of miR-15a-5p in HCC leads to treatment resistance through the overexpression of elF4E. As a result of an increase in the expression level of Hsp70, it has been observed that insufficient radiofrequency ablation can lead to treatment resistance in HCC (202). EZH2 in HCC downregulates the expression of miR-381, resulting in the activation of the Akt signaling pathway, which is accountable for the development of treatment resistance (203). This section discusses the role of lncRNAs in HCC treatment resistance regulation. There are two principal types of lncRNAs in this context: those that operate as chemosensitizers and those that can create drug resistance. That is why it may be possible to elevate HCC cell susceptibility to drugs by hindering carcinogenic lncRNAs. Silencing LINC00173 lowers resistance to cisplatin and is related to a poor outcome in HCC. LINC00173 downregulates miR-641, which, in turn, regulates RAB14 expression, causing cisplatin resistance in HCC (204). Other research looked at how anticancer medications influenced HCC cell survival levels and discovered that these agents instigate cell death (205, 206). Consequently, inhibiting apoptosis in tumor cells might lead to the development of drug resistance, a common trait observed in HCC. ST8SIA6-AS1, a lengthy non-coding RNA, has been demonstrated to hinder apoptosis and expedite the proliferation of HCC cells. The cytoplasmic lncRNA ST8SIA6-AS1 functions as a mediator for the sequestration of miR-4656, resulting in the upregulation of HDAC11 expression and the consequent inhibition of apoptosis in HCC (207). Hence, in order to make HCC cells more responsive to treatment, apoptosis is generally advised.
Growing evidence proposes that the lncRNA NEAT1 has a prominent role in the development of treatment resistance in HCC. During the development of sorafenib resistance in HCC, the lncRNA NEAT1 triggers the activation of protective autophagy. NEAT1 is a lengthy RNA molecule that functions as a mediator for miR-204 sponge, which increases the expression of ATG3 and ultimately triggers autophagy (208). A recent study has demonstrated the involvement of autophagy in the development of treatment resistance in HCC. According to the findings of this study, LINC00160 acts as a decoy by reducing the expression of miR-132, leading to an increase in ATG5 expression. Additionally, it enhances autophagy and promotes treatment resistance in HCC via reducing the expression of p62 (209). The lncRNA NEAT1 is observed to have increased expression in HCC. Considering this, it should have oncogenic activity. The study conducted by Niu and colleagues (210) uncovered that NEAT1 is the causative factor in stimulating the production of miR-149-5p. This, in turn, activates Akt1 signaling and finally leads to the development of resistance to sorafenib in HCC. Increased expression of BLCAF1 has been correlated with enhanced proliferation and invasion of HCC cells, and it plays a considerable role in this process. BLCAF1 is responsible for enhancing NEAT1 expression, which actuates chemoresistance in HCC (211). It can be inferred that suppressing the lncRNA NEAT1 expands the drug sensitivity of HCC cells. The lncRNA/STAT3 axis is involved in modulating the drug susceptibility of HCC cells. An observation has been made that the lncRNA DANCR is excessively produced in HCC. Furthermore, it has been demonstrated that the suppression of this gene is related to the vulnerability of tumor cells to sorafenib. Liu and colleagues’ research (212) reveals that the lncRNA DANCR enhances STAT3 signaling, leading to the development of sorafenib resistance in HCC. Several lncRNAs have represented dual functions as both oncogenes and onco-suppressors in HCC. This specific occurrence is observed in the lncRNA H19 in HCC. Increasing evidence indicates that the lncRNA H19 plays a function in promoting cancer development in HCC. In a study conducted by Wu and colleagues (213), it was discovered that polymorphisms of the lncRNA H19 may increase the risk of developing and beginning HCC.
3.4 Therapeutic targeting of lncRNAs in hepatocellular carcinoma
Presently, the outlook for HCC is often unfavorable, mostly due to the lack of a specific treatment target. Sorafenib, which specifically targets receptor tyrosine kinases (RTKs), is the most commonly prescribed targeted medicine for the treatment of HCC. Resistance to sorafenib is commonly observed in the treatment of HCC (214, 215). In summary, antisense oligonucleotides (ASOs) are specific lncRNAs that bind to short single-stranded DNAs, resulting in the formation of a DNA–RNA complex. RNase H has the ability to identify and break down this particular combination. On the other hand, RISCs are formed when short double-stranded RNAs are attached to the AGO2 protein. Subsequently, it is necessary for them to associate with specific lncRNAs in order to create an RNA–RNA complex, which will subsequently facilitate the suppression of lncRNAs (216). Considering these differences, it is probable that ASOs and RNAi demonstrate varying degrees of effectiveness in silencing, depending on several factors, such as the specific location inside the cell where the targeted lncRNAs are found. Within nuclei, ASOs have demonstrated more efficacy compared to RNA interference (RNAi). However, RNAi has proven to be more effective than ASOs in targeting cytoplasmic lncRNAs (217). This could be due to the fact that RNase H is predominantly located in the nucleus, while RISC largely carries out its duties in the cytoplasm (218, 219). Delivery of tumor-suppressive lncRNAs could potentially serve as an alternative method to knock down oncogenic lncRNAs by delivery. PRAL, for example, is an lncRNA that acts as a tumor suppressor by stabilizing p53. Furthermore, it has been shown that the administration of PRAL by an adenovirus vector may noticeably lessen the growth of HCC in tumor-bearing mice, indicating that it may have potential clinical applications for the treatment of HCC (220). It is important to note that there are a significant number of HCC-targeted ASOs and RNAi therapies available, as they have already been used to treat HBV. In summary, ASOs and siRNAs undergo modifications through chemical conjugates [such as N-acetylgalactosamine (GalNAc)] (221) or are encapsulated in delivery vehicles (such as lipid nanoparticles) (222), leading to an enhanced pharmacokinetic profile. The successful application of ASOs and RNAi in treating HBV establishes a strong foundation for the treatment of HCC and lncRNAi.
3.5 LncRNAs as ceRNA of miRNAs in hepatocellular carcinoma
Furthermore, lncRNAs has the capacity to engage in competition with endogenous RNA and act as “sponges” for miRNAs, thereby regulating the synthesis of target mRNAs. Recent research indicates that several lncRNAs linked to HCC possess the capability to regulate the expression of specific genes by binding to miRNAs (223). This hinders the binding of specific miRNAs to their target mRNAs. The miR-200 family is responsible for inhibiting the process of EMT and the spread of tumors to other parts of the body (224). The human chromosome 11p15.5 serves as the origin of transcription for H19, an lncRNA that is evidently suppressed in HCC. H19 possesses the capability to attach itself to hnRNPU/PCAF/RNAPol II, leading to an augmentation in histone acetylation and the initiation of miRNA-200. The latter hinders the proliferation of malignancies and counteracts the consequences of the EMT (225). In summary, ASOs are specific lncRNAs that bind to short single-stranded DNAs, resulting in the formation of a DNA–RNA complex. RNase H can identify and break down this particular combination. On the other hand, RISCs are formed by attaching short double-stranded RNAs to the AGO2 protein. Subsequently, they must associate with certain lncRNAs to create an RNA–RNA complex, which will subsequently facilitate the suppression of lncRNAs (226). Vertebrate H19 contains both canonical and non-canonical binding sites for let-7. Let-7 is a pivotal protein involved in the pathogenesis of HCC. H19 functions as an in vivo molecular sponge and conducts genome-wide transcriptome profiling. It plays a role in controlling the availability of let-7 (227). It is clear that lncRNAs reduce the regulation of miRNAs by functioning as miRNA sponges. Moreover, miRNAs possess the capacity to directly engage with lncRNAs and inhibit their synthesis (228, 229). The HOXA distal transcript antisense RNA (HOTTIP) is a non-coding antisense transcript that is situated at the farthest end of the HOXA gene cluster. It has the ability to regulate the expression of HOXA genes in close proximity to HCC. An important function of miR-125b in liver cancer is to control the activity of HOTTIP following the completion of transcription. There is compelling evidence indicating a clear link between HOTTIP and miR-125b. It is commonly seen that when miR-125b is downregulated, there is often an increase in HOTTIP expression (230). In the context of liver cancer, miR-192 and miR-204 act as tumor suppressors. Although argonaute 2 (Ago2) is predominantly located in the cytoplasm, additional RNAi factors, including Dicer, TRBP, and TRNC6A/GW182, are also found in the nucleus of the cell. These proteins play a crucial role in the regulation of functional RNAi (231). HOTTIP can partake in physical interaction with Ago2. One notable attribute exhibited by cancer cells is the presence of glutaminolysis (GLS1). As the enzyme that catalyzes the transformation of glutamine to glutamate, mitochondrial GLS1 is significantly critical to the glutaminolysis process (232). Both miR-204 and miR-192 can decrease HOTTIP expression via the Ago2-mediated RNAi pathway. The next step is to silence GLS1 expression in HCC cells, which leads to a drastic decrease in HCC cell viability (233).
3.6 Clinical importance, diagnosis, and prognosis
The association between several lncRNAs and clinicopathologic characteristics and prognosis in several types of cancers (234–238) highlights the potential of lncRNAs as biomarkers for determining prognosis. Out of the 74 lncRNAs that are not functioning properly in HCC, 63 are linked to the clinicopathologic characteristics of the illness. These characteristics include size, focality, differentiation, encapsulation, invasion, metastasis, disease stage, survival, nontumor traits (such as cirrhosis), serum alpha-fetoprotein (AFP), and the presence or absence of HBV infection. Specifically, it was shown that the majority of lncRNAs were linked to factors such as overall survival, cancer stage, and tumor size. This result enhances the importance of lncRNAs as indicators for prognosis. The rationale for this is that variables such as tumor size, cancer stage, and patient survival rate have a substantial influence on the probability of survival. It is crucial to note that several lncRNAs are strongly linked to many clinicopathologic variables. Cirrhosis, tumor invasiveness, blood AFP levels, HBV status, metastatic disease, HCC stage, and survival are all attributes encompassed within this classification. Six specific lncRNAs—PANDAR, MALAT-1, GPC3-AS1, CARLo-5, UCA1, and LOC90784—have been found to have a substantial correlation with six to eight distinct clinicopathologic characteristics. These findings indicate that these six lncRNAs have the capacity to establish a characteristic of a negative prognosis. Similarly, three lncRNAs (FTX, AOC4P, and linc-p21) that have been reduced in expression have been discovered to be significantly linked to six different clinicopathologic characteristics. Consequently, they have the potential to function as a distinctive indicator of a favorable prognosis. It is important to note that among the lncRNAs that showed either increased or decreased levels, a total of 17 were shown to be clinically associated with distinct tumor characteristics, such as size, location, degree of differentiation, invasion, and spread to other parts of the body. The efficacy of these lncRNAs in modulating cancer-related cellular markers has been confirmed by experiments conducted utilizing in vitro and in vivo mouse models. Cancerous growth is distinguished by specific characteristics, including tumor size, cell cycle, apoptosis, cell transformation, cell migration, invasion, and metastasis. Out of the total of 16 increased lncRNAs, 6 have been proven to play significant roles in the growth of tumors and the spread of metastasis. These six RNAs are GLTC, PCAT-14, SNHG12, MALAT-1, Linc00462, and PDIA3P1. It has been reported that these six lncRNAs modulate key cancer pathways, such as Hedgehog, NF-κB, FGFR1/ERK, mTOR, PI3K/AKT, and p53. On one hand, the downregulation of linc-p21 activates the Notch signaling pathway. On the other hand, the downregulation of uc.134 deactivates the Hippo kinase pathway. The deregulation of these eight lncRNAs, which target crucial cancer pathways and have been validated both clinically and experimentally, showed substantial correlations with tumor characteristics, vascular invasion, metastasis, cancer stage, and survival. This finding reaffirms the potential of these lncRNAs as potential prognostic biomarkers and for the development of drugs (Figure 5) (239). The lncRNAs WRAP53 and UCA-1 are also diagnostic factors in HCC (NCT05088811). However, more clinical studies are required to understand the role of lncRNAs in HCC.
4 CircRNAs in hepatocellular carcinoma
4.1 Biogenesis of circRNAs and their interaction with molecular pathways
Circular RNAs (circRNAs) are generated through lariat-circulating or back-splicing mechanisms, originating from various regions of the genome (240). Exons are the predominant origin of circRNAs, although intergenic, intronic, antisense, and UTR sections can also serve as potential sources, albeit extremely rarely. The designations circRNAs, EIciRNAs, and ciRNAs are used in the order of their origin, specifically from the exon, exon and intron, and intron, respectively. The majority of circRNAs are predominantly localized in the cytoplasm, although they can also be found in other subcellular compartments (241–244). The production of circRNA is affected in many ways by various sequence characteristics. Lengths of introns and exons, repetitive sequences, and RBPs are all examples of such factors. One type of RBP is the quaking protein, another is the heterogeneous nuclear ribonucleoprotein L, and yet another is the muscleblind protein (245–250). Liang and colleagues (251) have recently found, by the use of RNAi screening, that the proportion of linear to circRNA expression is regulated by numerous core spliceosomal and transcription termination factors. How are circRNAs degraded or eliminated? Lately, researchers have been focusing their efforts on exosomes. Exosomes are vesicles with two layers of membranes that contain various functional molecules, such as proteins, miRNAs, lncRNAs, and circRNAs. They have the ability to enhance cell-to-cell communication through either paracrine or endocrine mechanisms (252). CircRNAs can potentially undergo clearance or degradation through exosomal release, a mechanism that involves the removal of circRNAs from the plasma membrane (253). Figure 6 is an overview of circRNAs in cancer.
Understanding the interaction of circRNAs with downstream targets in human cancers can highlight the underlying mechanisms involved in carcinogenesis. In breast tumor, circ_0001667 is able to downregulate miR-6838-5p in increasing CXCL10 expression to accelerate tumorigenesis and mediate angiogenesis (254). Hsa_circ_0063329 is another factor capable of miR-605-5p suppression to increase TGIF2 expression in prostate cancer suppression (255). The circ-hnRNPU is a regulator of tumorigenesis and glycosylation that can suppress NONO-induced c-Myc transactivation (256). circRNA hsa_circ_0067842 has been shown as a regulator of tumorigenesis in breast cancer that enhances invasion and induces immune evasion through PD-L1 upregulation (257). In most cases, the action of circRNAs is related to the modulation of miRNA expression through sponging (258–262). Hsa_circ_0001278 inhibits miR-338-5p to increase AMOTL1 to enhance colorectal cancer progression (263).
4.2 CircRNAs exert dual function in hepatocellular carcinoma
The fact that circRNAs are made in specific tissues makes it likely that they contribute to the development of various diseases (264–267). The consequences revealed that several malignancies, including HCC, exhibit an upregulation of the cancer-causing circRNA known as CDR1as (268). Previous research found that elevated CDR1as levels in HCC tissues were highly associated with invasion of the liver microvasculature and only weakly with HCC development (269). Scientists demonstrated that HCC cell growth and invasion were reduced when CDR1as was inhibited. Recent studies have shown that CDR1as binds to miR-7 and acts as a sponge. HCC cell growth and invasion were inhibited by increasing miRNA-7 expression. Both cyclin E1 (CCNE1) and phosphoinositide 3-kinase catalytic subunit δ (PIK3CD), which are their target genes, saw a decrease in transcription. The PIK3CD/phospho-p70 S6 kinase (p70S6K)/mammalian target of rapamycin (mTOR) signaling pathway is disrupted when CDR1as functions as a miR-7 sponge, enhancing the proliferation and invasion of HCC cells. These results point to a critical function for CDR1as in controlling HCC progression. Furthermore, CDR1as-regulated proteins in HCC cells were identified using a quantitative proteomics approach (270). After conducting the proteomic analysis and functional verification, it was demonstrated that the overexpression of CDR1as has the capacity to enhance the proliferation and progression of the cell cycle in HCC cells. One way to achieve this is by controlling the signaling of epidermal growth factor receptor (EGFR) through the modulation of miR-7 expression. Tumor cells undergo a process called EMT to become metastatic and invasive. During EMT, the tumor cells lose E-cadherin and gain vimentin (271–274). Recent findings have represented that the transcription factor Twist1, known for its ability to trigger EMT, plays a role in upregulating the expression of Cul2 circRNA (circ-10720) (275). Circ-10720 exhibited a significant connection with the occurrence of malignant tumors and a worse prognosis in HCC. Upon conducting additional analysis, it was revealed that circ-10720 promoted the growth, movement, and infiltration of HCC cells. Twist1 may increase the expression of vimentin by increasing the levels of circ-10720, which binds to a group of miRNAs that specifically target vimentin. This implies that the procedure involved in this can be delineated as follows. Therefore, it was discovered that the Twist/circ-10720 pathway has a beneficial impact on the progression of EMT in lung cancer. However, the tumor-promoting function of Twist1 was abolished both in laboratory experiments (in vitro) and in living organisms (in vivo) when circ-10720 was employed to suppress it. The results indicated that circ-10720 plays a role in promoting the development of HCC and suggests that circ-10720 could be a potential target for therapeutic intervention in HCC treatment. This analysis also uncovered some distinctive perspectives on therapy approaches for HCC intervention that are founded on circRNA. The development of cancer and its progression are greatly impacted by aquaporin 3, sometimes referred to as AQP3 (276). Scientists demonstrated an association between high AQP3 expression levels and tumor growth, metastasis, and prognosis of patients with HCC (277, 278). AQP3 levels were markedly increased in the HCC tissues where it was identified. A study revealed that miR-124-3p is noticeably suppressed in HCC and hinders the proliferation and migration of HCC cells by targeting AQP3 (279). Additional study has revealed that circHIPK3 functions as a sponge for miR-124-3p, hence regulating the production of AQP3. CircHIPK3 has the ability to enhance the growth and movement of HCC cells by interacting with the miR-124-3p/AQP3 axis. An in vivo study showed that suppressing circHIPK3 led to a reduction in the progression of HCC. Furthermore, the role of another circRNA known as hsa_circ_0000673 in the progression of HCC has been revealed (280). According to the data, hsa_circ_0000673 was shown to be highly expressed in HCC tissues. Knocking down hsa_circ_0000673 led to a significant reduction in the growth and spread of HCC cells, as well as a prevention of tumor formation in living organisms. Hsa_circ_0000673 functioned as a miR-767-3p sponge, leading to an upregulation of its downstream effector SET. This was achieved through a mechanistic process. Moreover, there is a correlation between the atypical expression of SET and the proliferation of cancer (281, 282). SET has been found to act as an oncogene during the development of cancer. Not only was it found that the levels of SET were increased in HCC, but it was also established that there is a strong correlation between high SET levels and unfavorable clinical outcomes (283). Hsa_circ_0000673 played a role in promoting HCC malignancy via influencing the miR-767-3p/SET pathway.
There was a notable disparity in the expression of circRNA SMAD2 (circSMAD2) between HCC tissues and the adjacent normal tissues (284). A significant association was seen between CircSMAD2 and the extent of differentiation observed in HCC tissues. Overexpression of circSMAD2 has the capacity to inhibit the migratory, invasion, and EMT responses of HCC cells. This study has verified that miR-629 is the specific target of circSMAD2. Moreover, miR-629 has the capacity to mitigate the impact of circSMAD2 on the progression of HCC. Furthermore, it was found that the expression of circC3P1, a different circRNA, was reduced in HCC (285). Overexpressing CircC3P1 significantly suppresses the proliferation, migration, and invasion of colon cancer cells. Moreover, circC3P1 suppressed the progression of HCC and its metastatic potential in live organisms. An important finding is that circC3P1, by acting as a sponge for miR-4641 in HCC cells, resulted in an upregulation of phosphoenolpyruvate carboxykinase 1 (PCK1) expression. The considerable reduction in the proliferation, migration, and invasion of HCC cells was seen when the expression of PCK1 was inhibited by circC3P1. By specifically targeting miR-4641 in HCC, circC3P1 successfully fulfills its role as a tumor suppressor. This was achieved by upregulating the expression of PCK1. Further findings indicated that hsa_circ_0005986 functioned as a tumor suppressor in the progression of HCC (286). The expression of hsa_circ_0005986 was significantly downregulated in HCC tissues compared to healthy tissues. The downregulation of hsa_circ_0005986 led to the release of miR-129-5p, resulting in a decrease in the expression of its target gene, Notch1. The downregulation of hsa_circ_0005986 had a significant impact on the cell cycle transition, leading to an increase in the proliferation of HCC cells. Moreover, there was a direct relationship between the decreased expression level of hsa_circ_0005986 and the clinicopathological characteristics of patients with HCC. These attributes encompassed the dimensions of the tumor, the presence of microvascular invasion (MVI), and the stage of liver cancer as assessed by the Barcelona Clinic Liver Cancer (BCLC) classification. Thus, hsa_circ_0005986 has the potential to not only slow the process of HCC carcinogenesis, but also serve as a possible biomarker for diagnosing HCC. Yu and colleagues (287) utilized RNA-sequencing technology to compare the expression of circRNAs in paired HCC and surrounding non-tumorous tissues from two groups. The cSMARCA5 (hsa_circ_0001445) circRNA has been accurately characterized and originates from exons 15 and 16 of the SMARCA5 gene. The role of cSMARCA5 in the progression of HCC was investigated. The results indicated that the expression of cSMARCA5 was reduced in many HCC tissues. Patients with HCC who underwent surgical removal of the tumor showed a decrease in the expression of cSMARCA5, which was associated with more aggressive clinicopathological characteristics. This indicates that it could serve as a risk factor for both overall survival and recurrence-free survival in those individuals. Research indicates that the overexpression of cSMARCA5 can impede the migration and proliferation of HCC cells. cSMARCA5 increased the expression of the widely recognized tumor suppressor tissue inhibitor of metalloproteinase 3 (TIMP3) by acting as a sponge for miR-17-3p and miR-181b-5p. These findings not only provide new insights into the role of circRNAs in HCC development, but also highlight the importance of cSMARCA5 in promoting HCC growth and metastasis. A separate study (288) discovered that the presence of hsa_circ_0001445 was observed in both HCC and pericancerous tissues that were appropriately paired. The findings revealed a significant decrease in the expression of hsa_circ_0001445 in HCC tissues. This decrease showed an inverse correlation with the number of tumor foci. Based on gain-of-function investigations, overexpression of hsa_circ_0001445 can lead to cell death and limit the ability of HCC cells to move, invade, and multiply in a laboratory setting. The results indicate that hsa_circ_0001445 plays a regulatory role in the progression of HCC.
The circRNAs show interaction with other pathways in HCC. The circ_0002003 overexpression can enhance tumorigenesis through sponging miR-1343-3p to increase DTYMK, DAP3, and STMN1 levels (289). As an anticancer compound, sevoflurance has been shown to upregulate circ_0001649 to upregulate SGTB through miR-19a-3p inhibition in impairing HCC progression (290). Therefore, anticancer compounds can regulate circRNAs in HCC (291, 292). Circ-MBNL3 suppresses miR-873-5p expression to enhance PHF2 levels in impairing HCC progression (293). Hsa_circ_0093335 inhibits miR-338-5p expression to elevate HCC malignancy (294). Hsa_circ_0119412 is another factor that suppresses miR-526b-5p in enhancing STMN1 levels to facilitate tumorigenesis (295). Circ_0124208 downregulates miR-338-3p to upregulate LAMC1 and can be used as a biomarker in HCC (296). Circ_0082319 undergoes upregulation by HuR, and it can sponge miR-505-3p to upregulate PTK2 in tumorigenesis induction (297).
4.3 CircRNAs in the regulation of therapy resistance and therapeutic perspective
Resistance to therapy is a major factor in the reappearance and spread of HCC, and it poses a considerable obstacle to tumor treatment (298, 299). Recently, several studies have found that specific dysregulated circRNAs play a role in the resistance of HCC to chemotherapy and radiation, respectively. CircRBXO11 promotes tumor growth and confers resistance to oxaliplatin in HCC cells by sponging miR-605, which targets FOXO3, and activating ABCB1 (300). Moreover, studies have shown that circ_0003418 functions by suppressing the development of cancer and resistance to cisplatin chemotherapy in HCC through the Wnt/β-catenin pathway (301). Furthermore, circRNA 101505 increases NOR1 expression and absorbs miR-103, making HCC cells less sensitive to cisplatin (302). In addition, cZNF292 has the ability to increase the radiosensitivity of hypoxic HCC cells by decreasing the Wnt/β-catenin pathway and effectively enhancing the nuclear translocation of SOX9 (303). Moreover, Wu and colleagues found that the circRNAs’ profile of HCC could potentially serve as biomarkers for individuals with sorafenib-resistant HCC (304). Sorafenib resistance in HCC is sustained by two primary pathways, according to Xu and colleagues. During the first step, the Wnt/β-catenin pathway is launched and miR-103a-2-5p/miR-660-3p is active. Since the N6-methyladenosine modification increased RNA stability, it was found to improve the expression of circRNA-SORE (305). In addition, circRNA-SORE formed a connection with and stabilized YBX1 by inhibiting PRP19-mediated YBX1 degradation. In addition, HCC cells demonstrated the ability to develop resistance to sorafenib by means of the translocation of circular RNA-SORE through exosomes (306).
The progression of HCC is closely linked to specific circRNAs that show increased expression. Therefore, circRNAs have the potential to be targeted therapeutically for HCC. HCC cells, as well as individuals who have experienced HCC metastasis or recurrence, have demonstrated a heightened level of circASAP1 expression. Studies have shown that circASAP1 promotes the growth of HCC cells, as well as the formation of clusters, movement, infiltration, tumor growth, and spread to the lungs, both in laboratory settings and in living organisms. CircASAP1 has been shown to promote metastasis of HCC through the miR-326/miR-532-5p-MAPK1/CSF-1 signaling pathway. Hence, circASAP1 exhibits the capacity to function as a promising therapeutic target for HCC (307). The heightened expression of circTMEM45A in serum exosomes obtained from patients with HCC has the potential to be utilized as a new diagnostic and therapeutic target for patients with HCC (308). Furthermore, the presence of a regulatory network composed of circ-CDYL-centric ncRNAs, in conjunction with HDGF and HIF1AN, holds promise as valuable biomarkers and targets for the timely identification and management of HCC (309). Therefore, it is believed that increased circRNA expression in HCC aids in the development of cancer, whereas their suppression has the opposite impact in HCC. One way to limit the development of HCC and have an anticancer impact is to use short interfering RNAs that are developed to target the backsplicing junction sites of oncogenic circRNAs (310, 311). Some circRNAs that have their expression downregulated are potentially potential therapeutic targets because of their capacity to restrict tumor growth and prevent the formation of HCC. As an example, cSMARCA5 expression is significantly downregulated in HCC tissues, it is associated with growth and metastasis, and it may serve as a separate prognostic marker for patients with HCC even after hepatectomy. Both in vivo and in vitro investigations have demonstrated that cSMARCA5 inhibits the growth and spread of HCC. This suggests that cSMARCA5 has potential as both a diagnostic and a therapeutic tool for HCC (287). Han and co-workers discovered that circMTO1 expression was associated with a poor outcome for breast cancer patients. Reduced circMTO1 expression intratumorally accelerated HCC development in vivo, suggesting its utility for HCC-targeted therapy. The circMTO1 gene may one day be used as a diagnostic tool and treatment target for people afflicted with HCC (312). Moreover, Zhang and colleagues uncovered that the expression of circDLC1 was reduced in HCC, and this was directly linked to the prognosis of patients with HCC. The overexpression of circDLC1 was observed to restrict the proliferation and metastasis of lung cancer cells in both laboratory and living organism environments. CircDLC1 has the potential to function as both a target for therapy and a biomarker for predicting outcomes in persons with HCC (313). Despite the decrease in the expression of these circRNAs, it is plausible that inducing their overexpression in HCC cells or tissues via transfection could lead to substantial anticancer effects.
4.4 CircRNAs as ceRNA for miRNAs in hepatocellular carcinoma
Studies on the composition of circRNAs have shown that these RNA molecules possess a noticeable abundance of miRNA binding sites, which extend the interaction between circRNA and miRNA (265). Subsequent investigations have illustrated that circRNAs had the capacity to regulate the expression of genes from which they originate through their interaction with miRNA molecules (314, 315). An instance of this can be seen in the correlation between the excessive expression of has_circ_0005075 and the dimensions of the HCC tumor. This discovery revealed that has_circ_0005075 might be responsible for the proliferation of the tumor, and it indicated a remarkable diagnostic capacity (AUROC = 0.94). Moreover, by the use of GO and route analysis, it was anticipated that has_circ_0005075 will be involved in cell adhesion. This pathway exhibited strong associations with cell proliferation, invasion, and metastasis in people diagnosed with HCC. It was postulated that the biological activities of has_circ_0005075 may modulate some components of the tumor cell membrane through biological functions. On top of that, it was found that the gene has_circ_0005075 has the ability to interact with four different miRNAs, namely, hsa-miR-23b-5p, hsa-miR-93-3p, hsa-miR-581, and hsa-miR-23a-5p, resulting in a decrease in their production and functionality. The results demonstrated above indicate that a considerable level of has_circ_0005075 expression in HCC is associated with the development of tumors, suggesting that this gene could serve as a promising biomarker for HCC (316).
Another study discovered that HCC was associated with extremely high rates of the famous circRNA Cdr1as, also known as ciRS7 (317). This circRNA contained a precise match for miR-671 and more than 70 conserved binding sites for miR-7. Blocking Cdr1as can potentially impede the proliferation and spread of hair cancer cells. Furthermore, the expression of miR-7 was decreased throughout this period. Overexpression of miR-7 can potentially suppress the expression of CCNE1 and PIK3CD, two genes that it specifically targets. Suppressing Cdr1as led to a reduction in the expression of several genes. These findings suggest that Cdr1as, apart from its role in targeting miR-7 in HCC, may also function as an oncogene (317). A research conducted by Xu and colleagues showed that the expression of Cdr1as was decreased in HCC. In addition, the study identified a strong association among the expression of Cdr1as and several factors: age below 40 years, serum AFP levels of 400 ng/μL or higher, hepatic MVI, and two specific genes targeted by miR-7, namely, PIK3CD and p70S6K (269). A similar discovery was made, indicating that the presence of Cdr1as was linked to the miR-7 target gene PIK3CD, suggesting that Cdr1as could function as a miR-7 inhibitor. The level of Cdr1as expression is highly correlated with HCC, a relationship that may be substantiated by considering all available information. Further validation is necessary to ascertain whether Cdr1as can function as a potential biomarker for HCC, as its role in this context remains unclear. Through circRNA microarray analysis, we identified the upregulation of circ 100338, which exhibited a robust association with shortened cumulative survival. Furthermore, a positive correlation was found between HBV infection and the progression of metastasis in patients with HCC (318). Circ_100338 acts as a sponge for miR-141-3p, which can be counteracted by miR-141-3p. This, in turn, hinders the progression of cell metastasis in liver cancer. By simultaneously expressing miR-141-3p and circ_100338 in MHCC97H cells, it is feasible to restore the suppression of invasive capacity caused by the excessive production of miR-141-3p. The overexpression of miR-141-3p can reverse the increased migratory and invasive ability of MHCC97H cells caused by circ_100338. These data indicate that circ_100338 could be used as a novel biomarker for diagnosing and assessing patient survival in patients with HBV-related HCC (318). A recent study has shown that the expression of circ_0067934, which is increased in HCC, can promote the Wnt/β-catenin signaling pathway by functioning as a miR-1324 sponge. MiR-1324 has the ability to impede the activation of the Wnt/β-catenin signaling pathway by specifically targeting the 3′-UTR of FZD5. Downregulating the expression of FZD5 and inhibiting the activation of the Wnt/β-catenin signaling pathway can substantially reduce the proliferation, migration, and invasion of Hep3B and HuH7 cells. This can be accomplished by upregulating the expression of miR-1324 or by downregulating the expression of circ_0067934. The study’s findings suggest that targeting the circ_0067934/miR-1324/FZD5/Wnt/β-catenin signaling axis could be a promising approach for treating HCC. Moreover, in this particular instance, cellular demise was seen (319, 320).
4.5 CircRNAs as diagnostic factors in HCC
The overall survival rate for people with HCC is dismal since the disease is typically detected at an advanced stage (321). Early-stage disease is characterized by a small number of symptoms and limited availability of biomarkers (322). Furthermore, the accuracy of the present biomarkers, like α-fetoprotein (AFP) and AFP-L3, in detecting lung cancer is only moderate (322). CircRNAs show high levels of abundance and stability in HCC tissue and bodily fluid, and they are tightly linked to many biological processes in HCC; hence, they have been suggested as possible diagnostic biomarkers for HCC. The ability of hsa_circ_0068669 to serve as a biomarker for the progression of HCC metastasis was demonstrated by Yao and colleagues. There was also a considerable correlation between the amount of microvascular invasion and the expression of hsa_circ_0068669 highlighted by them. The possibility of circRNAs as biomarkers for the diagnosis of HCC has been explored in a recent research (316, 323, 324). The upregulation of Circ-CDYL, a promoter for HCC, may lead to an increase in the expression of several proto-oncogenes. With an AUC of 0.64 [95% confidence interval (CI) = 0.55–0.72], Wei and colleagues (325) revealed the diagnostic performance of circ-CDYL in the early stage of HCC. This remark was made recently. After a thorough analysis of the levels of circ-CDYL expression in relation to HDGF and HIF1A, the results showed an improved diagnostic accuracy, as indicated by an AUC of 0.73 (95% CI = 0.65–0.80), a sensitivity of 75.36%, and a specificity of 66.67%. The AFP only showed an AUC of 0.59 (95% CI = 0.49–0.70), a sensitivity of 50.72%, and a specificity of 83.78% when compared to circ-CDYL with HDGF and HIF1A. According to the results of this experiment, it seems that the combination of circ-CDYL with HDGF and HIF1AN may serve as a more precise diagnostic biomarker compared to AFP. A research on the levels of expression of hsa_circ_0028502 and hsa_circ_0076251 was carried out by Jiang and colleagues (326) in malignant tissues and surrounding para-cancerous tissues. Based on consequences, it was observed that the rates of hsa_circ_0028502 and hsa_circ_0076251 were considerably lower in tissues that were correlated with HCC (p < 0.001). A noticeable correlation was discovered between the level of hsa_circ_0028502 and the stage of metastasis in the tumor nodes (p = 0.015), whereas the expression of hsa_circ_0076251 was associated with the stage of liver cancer at the Barcelona Clinic (p = 0.038). The AUCs of hsa_circ_0028502 and hsa_circ_0076251 were 0.675 and 0.738, respectively, when HCC tissues were differentiated from liver cirrhosis tissues and chronic hepatitis tissues. Additionally, Matboli and collaborators (327) evaluated the diagnostic performance of hsa_circ_001565, hsa_circ_000224, and hsa_circ_000520 for HCC. The results showed that when compared to AFP, these three hsa_circ_samples were more sensitive and specific. A diagnostic performance increase of 80% was achieved with the inclusion of these three biomarkers, leading to a sensitivity of 100% and a specificity of 83.33%. In an ROC curve study including 40 patients with HCC and 40 healthy controls, the AUC for hsa_circ_0016788 was found to be 0.851.59. This discovery expands upon what is already known. Additionally, the hsa_circ_0128298 gene, which was shown to be highly overexpressed in HCC tissues, had a sensitivity of 0.716, a specificity of 0.815, and an AUC of 0.668 (264).
In addition, Yao and colleagues (328) conducted an analysis on the expression of circZKSCAN1 (hsa_circ_0001727) in a cohort of 102 patients who were diagnosed with HCC. The gene expression in tumor tissues was markedly lower compared to the non-tumorous samples that were matched to the tumor tissues (p < 0.05), as indicated by the results. The expression rate of circZKSCAN1 was represented to be remarkably associated with many clinical indicators, including tumor grade (p < 0.001), vascular invasion (p = 0.002), microvascular invasion (p = 0.002), the existence of cirrhosis (p = 0.031), and the number of tumors (p < 0.01). When used as a diagnostic biomarker, circZKSCAN1 consistently illustrated strong performance with an AUC of 0.834, a sensitivity of 82.2%, and a specificity of 72.4%. Plasma circRNAs can be utilized as diagnostic biomarkers for HCC, together with alterations observed in HCC tissue. In a comprehensive study conducted by Yu and co-workers (329), a plasma circRNA panel (circPanel) was developed and evaluated. This panel included three specific circRNAs: hsa_circ_0000976, hsa_circ_0007750, and hsa_circ_0139897. The main objective of this large-scale multicenter inquiry was to determine the effectiveness of this panel in identifying HCC that is connected to the HBV. The circPanel, a newly created diagnostic tool, outperformed AFP in terms of diagnostic performance in a validation sample of 306 individuals. The circPanel had an AUC of 0.843 (95% CI = 0.796–0.890), while AFP had an AUC of 0.747 (95% CI = 0.691–0.804). The circPanel demonstrated a reliable performance in diagnosing small HCC (solitary < 3 cm) and AFP-negative HCC compared to other approaches, with AUC values of 0.838 (95% CI = 0.776–0.900) and 0.857 (95% CI = 0.793–0.921), respectively. Thus, growing data emphasize that circRNAs play a significant role in the control of tumor formation and can be regarded as potential therapeutic and diagnostic factors in HCC (330–337). A clinical study has also shown that circ_0004001 can be utilized for the diagnosis of HCC (NCT06042842). Figure 7 is an overview of circRNAs in HCC. Table 1 is an overview of the ncRNAs in the regulation of tumorigenesis in HCC.
5 Conclusion, future perspectives, and challenges
The diverse biomolecular properties of HCC are evident in its wide range of clinical outcomes and progression, as well as its morphological features, which are evaluated by radiologists and pathologists (368). The foundation of both cirrhotic nodules and hepatocarcinogenesis is controlled by different molecular mechanisms. This development is linked to the up- or downregulation of many miRNAs. The current research illustrates that mTOR is the most frequently observed miRNA-regulated pathway in HCC. Moreover, the rates of miRNA expression also modulate other pathways, such as apoptosis, Wnt, and MAPK. In addition, there are other miRNAs that are expected to have therapeutic significance, particularly in relation to post-surgical recurrence and responsiveness to systemic therapy. These miRNAs are involved in at least one of the main molecular pathways that contribute to the development of hepatocarcinogenesis. The identification of specific tissue and serum miRNAs that can accurately predict the development of HCC in patients with cirrhosis, anticipate the recurrence of HCC after surgery, or forecast the response to systemic treatment has the potential to greatly enhance the management of these patients. The clinical application of miRNAs has consistently posed challenges, mostly due to the variation among laboratories and the identification of an appropriate control for normalization. A recent study conducted by our team revealed the alteration in the miRNA profile of HCC compared to a group of healthy livers and a group of cirrhotic tissues, which were used as non-tumor controls. Some authors have proposed utilizing stable miRNAs as controls when investigating the expression of other miRNAs. In order to gain a deeper understanding of the potential role of these molecules as theragnostic indicators in HCC, it would be beneficial to provide further details regarding the most commonly observed miRNA in live animal models, such as rats and mice. Based on the existing knowledge, this would be carried out in a manner that is appropriate and in line with the current circumstances. A diagnostic method for liver nodules based on miRNA and a treatment that regulates miRNA are still being developed. However, it is necessary to address the issue by establishing standardized protocols for miRNA analysis in different laboratories. These two approaches of diagnosis and treatment are essential.
The primary goals of this review are to provide a foundational understanding of liver cancer genesis, to discuss the role of lncRNA in this process, to examine the impact of tumor angiogenesis on HCC progression, and to explain how lncRNA controls tumor angiogenesis (369). Investigating how lncRNAs regulate tumor cell phenotypic alterations that promote HCC cell invasion and metastasis was the next step. Once in the circulation, lncRNA helps HCC cells avoid detection by the immune system by changing how susceptible tumor cells are to anoikis and by giving the immune system a way to avoid cancer cells in general. Finally, we will go over two questions that still have not been answered regarding cancer metastasis: targeted metastasis and tumor dormancy. These two matters hold a lot of potential. Additionally, the pro-metastasis niche, vascular dormancy, and immunological dormancy are the conventional explanations for these two inquiries. A lot of questions remain unanswered, despite the fact that new research has shed light on the role that lncRNA plays in HCC metastasis. Even though the best way to treat HCC is to remove the tumor surgically when it is still small, most individuals are found to have the disease far advanced when they have surgery. The good news is that there are now very few medications that can be used as a first line of defense against HCC. The multi-targeted tyrosine kinase inhibitors doxorubicin and sorafenib are two such examples of such drugs. It was found that many essential signaling pathways are involved at the start of HCC. The MEK pathway, the PI3K/AKT/mTOR system, the Wnt/β-catenin pathway, the JAK/STAT route, and the IGF pathway are all pathways that fall under this category (370). There is evidence suggesting that lncRNAs play a role in controlling the signal pathway mentioned earlier and offer new strategies for the clinical diagnosis and treatment of HCC (223). There has been substantial progress in tumor immunotherapy in recent years, leading to the widespread use of tumor immunovaccine in clinical settings. HCC is a common tumor that is linked to inflammation, and immunotherapy for this specific malignancy has reached an advanced stage. Tremelimumab, a CTLA-4 inhibitor, pembrolizumab, a PD-1/PD-L1 inhibitor, and nivolumab have all been used in clinical therapy (371). A recent study has shown that lncRNAs play a role in regulating the immune system in cancer. Special attention should be given to the investigation of immunotherapy that is linked to lncRNA in future investigations. Furthermore, the investigation of focused spread of cancerous growths is a captivating and auspicious research pursuit. The preferred sites for the spread of different types of tumors are unique from each other. Enhancing our comprehension of the underlying mechanisms that trigger the dissemination of tumor metastasis to particular sites may facilitate the amelioration of their treatment. The phenomenon of tumor dormancy has attracted growing attention, leading to several experimental investigations. Conversely, the study of tumor dormancy is currently in its initial phases, and numerous crucial inquiries remain unresolved. Is it accurate to say that all cancers in the human body contain dormant tumor cells? What is the duration of tumor dormancy? Which types of cancer cells are in a state of dormancy? Can latent tumor cells always become active? What is the method for reviving them? Ultimately, future research on lncRNAs should prioritize investigating their role in modulating the immune system, targeting metastasis, and regulating dormancy in liver cancer, to comprehend its role in metastasis and as a promising target for pharmaceutical intervention.
Emerging evidence indicates that some circRNAs show varying levels of expression in the tissues of patients with HCC. Furthermore, the disruption of these circRNAs is linked to clinicopathological features in patients with HCC (372). CircRNAs play a regulatory role in transcription by acting as sponges for miRNAs and RBPs. This enables them to control the production of certain target proteins and miRNAs. miRNAs and proteins contribute to the advancement of HCC and are linked to cell invasion, metastasis, and proliferation. The unique benefits of circulatory RNAs, including their high quantity, durability, and occurrence in many physiological fluids, render them appealing as diagnostic and prognostic markers and as targets for therapy in cases of HCC. circRNAs are very stable and plentiful in exosomes generated from serum. Exosomes carry a cargo of RNAs and proteins that tumor cells release and transfer to other cells in order to affect their behavior. Hence, circRNAs can play a dual role in promoting cancer advancement and serving as non-invasive biomarkers for cancer detection. Moreover, the presence of fusion circRNA contributes to the cancer cells’ capacity to withstand treatment. This resistance is a major obstacle in the use of chemotherapy for cancer treatment, and it could be a contributing factor to the ineffectiveness of current treatments in eradicating malignant tumors. However, it remains uncertain whether any circRNA, including fusion circRNA, is involved in the resistance of HCC to treatment. The investigation of circRNAs in HCC is still in its early stages, unlike the research on coding RNAs, mRNAs, and lncRNAs. Currently, there is a lack of knowledge about the prevalence of functional circRNAs in HCC. The majority of these studies have primarily examined the functions of these circRNAs as miRNA sponges in promoting the proliferation of HCC cells. However, only a limited number of these circRNAs have been thoroughly researched in terms of their mechanisms of action. A comprehensive understanding of the synthesis, degradation, cellular localization, functions, and mechanisms of action of the vast majority of circRNAs that are currently awaiting investigation is still lacking. A more comprehensive study of the relationship between circRNAs and the causes, behaviors, and molecular processes of HCC would enhance our understanding of human disorders. On top of that, this would provide new opportunities for improving the diagnosis, prognosis, and treatment of HCC. During hypoxia, the exosomes can enhance the progression of lung pre-metastatic niche in HCC through the delivery of miR-4508. The upregulation of exosomal miR-4508 is observed in HCC, and it can modulate the p38 MAPK–NF-κB axis (373).
The current studies highlighted the fact that dysregulation of ncRNAs commonly occurs in HCC. The major ncRNAs including miRNAs, lncRNAs, and circRNAs are therapeutic, diagnostic, and prognostic factors. From a therapeutic aspect, the ncRNAs can be targeted by the drugs in HCC therapy. However, the drugs have a poor pharmacokinetic profile and their clinical importance is limited. Therefore, the researchers are suggested to focus on the application of genetic tools for the regulation of ncRNAs. Moreover, the epigenetic drugs have a low ability in binding to the ncRNAs as epigenetic factors and regulate cancer progression. Although a high number of studies have evaluated the function of ncRNAs in the modulation of tumorigenesis, one of the largest limitations is the lack of focus on the application of nanoparticles for the regulation of ncRNAs.
Author contributions
YT: Validation, Software, Project administration, Methodology, Investigation, Formal Analysis, Data curation, Conceptualization, Writing – review & editing, Writing – original draft. MZ: Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Methodology, Formal Analysis, Data curation, Conceptualization. L-XL: Writing – original draft, Supervision, Project administration, Methodology, Investigation, Formal Analysis, Data curation, Conceptualization. Z-CW: Writing – review & editing, Validation, Software, Methodology, Investigation, Data curation. BL: Writing – review & editing, Visualization, Validation, Resources, Project administration, Methodology, Investigation, Formal Analysis, Data curation. YH: Writing – original draft, Project administration, Methodology, Data curation, Conceptualization. XW: Writing – review & editing, Validation, Supervision, Project administration, Methodology, Investigation, Formal Analysis, Data curation. Y-ZL: Writing – review & editing, Validation, Project administration, Methodology, Investigation, Formal Analysis, Data curation. FW: Writing – review & editing, Writing – original draft, Software, Methodology, Investigation, Formal Analysis, Data curation, Conceptualization. XF: Writing – review & editing, Writing – original draft, Supervision, Project administration, Methodology, Data curation. YYT: Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal Analysis, Data curation.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Guangdong Basic and Applied Basic Research Fund (No. 2023B1515120085).
Acknowledgments
The figures were drawn by Biorender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Xu X, Tao Y, Shan L, Chen R, Jiang H, Qian Z, et al. The role of microRNAs in hepatocellular carcinoma. J Cancer. (2018) 9:3557–69. doi: 10.7150/jca.26350
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. (2015) 136:E359–86. doi: 10.1002/ijc.29210
3. Tunissiolli NM, Castanhole-Nunes MMU, Biselli-Chicote PM, Pavarino EC, da Silva RF, da Silva RC, et al. Hepatocellular carcinoma: a comprehensive review of biomarkers, clinical aspects, and therapy. Asian Pac J Cancer Prev. (2017) 18:863–72. doi: 10.22034/APJCP.2017.18.4.863
4. Xia F, Wu LL, Lau WY, Huan HB, Wen XD, Ma KS, et al. Adjuvant sorafenib after heptectomy for Barcelona Clinic Liver Cancer-stage C hepatocellular carcinoma patients. World J Gastroenterol. (2016) 22:5384–92. doi: 10.3748/wjg.v22.i23.5384
5. Gosalia AJ, Martin P, Jones PD. Advances and future directions in the treatment of hepatocellular carcinoma. Gastroenterol Hepatol (N Y). (2017) 13:398–410.
6. Goble S, et al. Hepatitis C and hepatocellular carcinoma in Latin America: Elimination as a path to cancer prevention. Ann Hepatol. (2023) 28:101149. doi: 10.1016/j.aohep.2023.101149
7. Boora S, Sharma V, Kaushik S, Bhupatiraju AV, Singh S, Kaushik S. Hepatitis B virus-induced hepatocellular carcinoma: a persistent global problem. Braz J Microbiol. (2023) 54:679–89. doi: 10.1007/s42770-023-00970-y
8. Li XM, Ma L, Yang YB, Shi ZJ, Zhou SS. Prognostic factors of fulminant hepatitis in pregnancy. Chin Med J (Engl). (2005) 118:1754–7.
9. Yang Y, Deng L, Li X, Shi Z, Chen D, Chen X, et al. Evaluation of the prognosis of fulminant viral hepatitis in late pregnancy by the MELD scoring system. Eur J Clin Microbiol Infect Dis. (2012) 31:2673–8. doi: 10.1007/s10096-012-1613-y
10. Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. (2008) 12:2189–204. doi: 10.1111/j.1582-4934.2008.00533.x
11. Ito Y, Sasaki Y, Horimoto M, Wada S, Tanaka Y, Kasahara A, et al. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology. (1998) 27:951–8. doi: 10.1002/(ISSN)1527-3350
12. Boix L, Rosa JL, Ventura F, Castells A, Bruix J, Rodés J, et al. c-met mRNA overexpression in human hepatocellular carcinoma. Hepatology. (1994) 19:88–91. doi: 10.1016/0270-9139(94)90057-4
13. Suzuki K, Hayashi N, Yamada Y, Yoshihara H, Miyamoto Y, Ito Y, et al. Expression of the c-met protooncogene in human hepatocellular carcinoma. Hepatology. (1994) 20:1231–6. doi: 10.1016/0270-9139(94)90762-5
14. Grigioni WF, Fiorentino M, D'Errico A, Ponzetto A, Crepaldi T, Prat M, et al. Overexpression of c-met protooncogene product and raised Ki67 index in hepatocellular carcinomas with respect to benign liver conditions. Hepatology. (1995) 21:1543–6. doi: 10.1002/(ISSN)1527-3350
15. Ueki T, Fujimoto J, Suzuki T, Yamamoto H, Okamoto E. Expression of hepatocyte growth factor and its receptor, the c-met proto-oncogene, in hepatocellular carcinoma. Hepatology. (1997) 25:619–23. doi: 10.1002/(ISSN)1527-3350
16. Sugano Y, Matsuzaki K, Tahashi Y, Furukawa F, Mori S, Yamagata H, et al. Distortion of autocrine transforming growth factor beta signal accelerates Malignant potential by enhancing cell growth as well as PAI-1 and VEGF production in human hepatocellular carcinoma cells. Oncogene. (2003) 22:2309–21. doi: 10.1038/sj.onc.1206305
17. Moon WS, Rhyu KH, Kang MJ, Lee DG, Yu HC, Yeum JH, et al. Overexpression of VEGF and angiopoietin 2: a key to high vascularity of hepatocellular carcinoma? Mod Pathol. (2003) 16:552–7. doi: 10.1097/01.MP.0000071841.17900.69
18. Hirohashi K, Yamamoto T, Uenishi T, Ogawa M, Sakabe K, Takemura S, et al. CD44 and VEGF expression in extrahepatic metastasis of human hepatocellular carcinoma. Hepatogastroenterology. (2004) 51:1121–3.
19. Ferrero S, Ragni N. Prognostic value of plasma and serum VEGF levels in patients with resectable hepatocellular carcinoma. Ann Surg Oncol. (2003) 10:1123. doi: 10.1245/ASO.2003.08.924
20. Iguchi H, Yokota M, Fukutomi M, Uchimura K, Yonemasu H, Hachitanda Y, et al. A possible role of VEGF in osteolytic bone metastasis of hepatocellular carcinoma. J Exp Clin Cancer Res. (2002) 21:309–13.
21. Schmitt M, Horbach A, Kubitz R, Frilling A, Häussinger D. Disruption of hepatocellular tight junctions by vascular endothelial growth factor (VEGF): a novel mechanism for tumor invasion. J Hepatol. (2004) 41:274–83. doi: 10.1016/j.jhep.2004.04.035
22. Dhar DK, Naora H, Yamanoi A, Ono T, Kohno H, Otani H, et al. Requisite role of VEGF receptors in angiogenesis of hepatocellular carcinoma: a comparison with angiopoietin/Tie pathway. Anticancer Res. (2002) 22:379–86.
23. Motoo Y, Sawabu N, Yamaguchi Y, Terada T, Nakanuma Y. Sinusoidal capillarization of human hepatocellular carcinoma: possible promotion by fibroblast growth factor. Oncology. (1993) 50:270–4. doi: 10.1159/000227194
24. Rosmorduc O, Wendum D, Corpechot C, Galy B, Sebbagh N, Raleigh J, et al. Hepatocellular hypoxia-induced vascular endothelial growth factor expression and angiogenesis in experimental biliary cirrhosis. Am J Pathol. (1999) 155:1065–73. doi: 10.1016/S0002-9440(10)65209-1
25. Uematsu S, Higashi T, Nouso K, Kariyama K, Nakamura S, Suzuki M, et al. Altered expression of vascular endothelial growth factor, fibroblast growth factor-2 and endostatin in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. (2005) 20:583–8. doi: 10.1111/j.1440-1746.2005.03726.x
26. Fischer AN, Fuchs E, Mikula M, Huber H, Beug H, Mikulits W. PDGF essentially links TGF-beta signaling to nuclear beta-catenin accumulation in hepatocellular carcinoma progression. Oncogene. (2007) 26:3395–405. doi: 10.1038/sj.onc.1210121
27. Campbell JS, Hughes SD, Gilbertson DG, Palmer TE, Holdren MS, Haran AC, et al. Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc Natl Acad Sci U.S.A. (2005) 102:3389–94. doi: 10.1073/pnas.0409722102
28. Jagirdar J, Nonomura A, Patil J, Thor A, Paronetto F. ras oncogene p21 expression in hepatocellular carcinoma. J Exp Pathol. (1989) 4:37–46.
29. Takada S, Koike K. Activated N-ras gene was found in human hepatoma tissue but only in a small fraction of the tumor cells. Oncogene. (1989) 4:189–93.
30. Challen C, Guo K, Collier JD, Cavanagh D, Bassendine MF. Infrequent point mutations in codons 12 and 61 of ras oncogenes in human hepatocellular carcinomas. J Hepatol. (1992) 14:342–6. doi: 10.1016/0168-8278(92)90181-N
31. Xu XR, Huang J, Xu ZG, Qian BZ, Zhu ZD, Yan Q, et al. Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci U.S.A. (2001) 98:15089–94. doi: 10.1073/pnas.241522398
32. Laurent-Puig P, Legoix P, Bluteau O, Belghiti J, Franco D, Binot F, et al. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology. (2001) 120:1763–73. doi: 10.1053/gast.2001.24798
33. Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. (2000) 24:245–50. doi: 10.1038/73448
34. Ding Q, Xia W, Liu JC, Yang JY, Lee DF, Xia J, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. (2005) 19:159–70. doi: 10.1016/j.molcel.2005.06.009
35. Zucman-Rossi J, Jeannot E, Nhieu JT, Scoazec JY, Guettier C, Rebouissou S, et al. Genotype-phenotype correlation in hepatocellular adenoma: new classification and relationship with HCC. Hepatology. (2006) 43:515–24. doi: 10.1002/(ISSN)1527-3350
36. Boige V, Laurent-Puig P, Fouchet P, Fléjou JF, Monges G, Bedossa P, et al. Concerted nonsyntenic allelic losses in hyperploid hepatocellular carcinoma as determined by a high-resolution allelotype. Cancer Res. (1997) 57:1986–90.
37. Nagai H, Pineau P, Tiollais P, Buendia MA, Dejean A. Comprehensive allelotyping of human hepatocellular carcinoma. Oncogene. (1997) 14:2927–33. doi: 10.1038/sj.onc.1201136
38. Lin Y, Shi CY, Li B, Soo BH, Mohammed-Ali S, Wee A, et al. Tumour suppressor p53 and Rb genes in human hepatocellular carcinoma. Ann Acad Med Singap. (1996) 25:22–30.
39. Higashitsuji H, Itoh K, Nagao T, Dawson S, Nonoguchi K, Kido T, et al. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Med. (2000) 6:96–9. doi: 10.1038/71600
40. Joo M, Kang YK, Kim MR, Lee HK, Jang JJ. Cyclin D1 overexpression in hepatocellular carcinoma. Liver. (2001) 21:89–95. doi: 10.1034/j.1600-0676.2001.021002089.x
41. Hickman ES, Moroni MC, Helin K. The role of p53 and pRB in apoptosis and cancer. Curr Opin Genet Dev. (2002) 12:60–6. doi: 10.1016/S0959-437X(01)00265-9
42. Weihrauch M, Benicke M, Lehnert G, Wittekind C, Wrbitzky R, Tannapfel A. Frequent k- ras -2 mutations and p16(INK4A)methylation in hepatocellular carcinomas in workers exposed to vinyl chloride. Br J Cancer. (2001) 84:982–9. doi: 10.1054/bjoc.2000.1675
43. Matsuda Y, Ichida T, Matsuzawa J, Sugimura K, Asakura H. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology. (1999) 116:394–400. doi: 10.1016/S0016-5085(99)70137-X
44. Ito Y, Takeda T, Sakon M, Tsujimoto M, Monden M, Matsuura N. Expression of p57/Kip2 protein in hepatocellular carcinoma. Oncology. (2001) 61:221–5. doi: 10.1159/000055378
45. Scelfo RA, Schwienbacher C, Veronese A, Gramantieri L, Bolondi L, Querzoli P, et al. Loss of methylation at chromosome 11p15.5 is common in human adult tumors. Oncogene. (2002) 21:2564–72. doi: 10.1038/sj.onc.1205336
46. Schwienbacher C, Gramantieri L, Scelfo R, Veronese A, Calin GA, Bolondi L, et al. Gain of imprinting at chromosome 11p15: A pathogenetic mechanism identified in human hepatocarcinomas. Proc Natl Acad Sci U.S.A. (2000) 97:5445–9. doi: 10.1073/pnas.090087497
47. Diaz-Meyer N, Day CD, Khatod K, Maher ER, Cooper W, Reik W, et al. Silencing of CDKN1C (p57KIP2) is associated with hypomethylation at KvDMR1 in Beckwith-Wiedemann syndrome. J Med Genet. (2003) 40:797–801. doi: 10.1136/jmg.40.11.797
48. Soejima H, Nakagawachi T, Zhao W, Higashimoto K, Urano T, Matsukura S, et al. Silencing of imprinted CDKN1C gene expression is associated with loss of CpG and histone H3 lysine 9 methylation at DMR-LIT1 in esophageal cancer. Oncogene. (2004) 23:4380–8. doi: 10.1038/sj.onc.1207576
49. Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. (2008) 27:5651–61. doi: 10.1038/onc.2008.178
50. Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, et al. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. (2008) 68:2773–80. doi: 10.1158/0008-5472.CAN-07-6754
51. le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. (2007) 26:3699–708. doi: 10.1038/sj.emboj.7601790
52. Matsuda Y, Ichida T. p16 and p27 are functionally correlated during the progress of hepatocarcinogenesis. Med Mol Morphol. (2006) 39:169–75. doi: 10.1007/s00795-006-0339-2
53. Shi Z, Nicholson RH, Jaggi R, Nicholson AW. Characterization of Aquifex aeolicus ribonuclease III and the reactivity epitopes of its pre-ribosomal RNA substrates. Nucleic Acids Res. (2011) 39:2756–68. doi: 10.1093/nar/gkq1030
54. Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. (2012) 56:1371–83. doi: 10.1016/j.jhep.2011.11.026
55. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. (2007) 23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406
56. Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci. (2007) 104:17719–24. doi: 10.1073/pnas.0703890104
57. Lee Y, Jeon K, Lee J-T, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. (2002) 21:4663–70. doi: 10.1093/emboj/cdf476
58. Griffiths-Jones S. Annotating noncoding RNA genes. Annu Rev Genomics Hum Genet. (2007) 8:279–98. doi: 10.1146/annurev.genom.8.080706.092419
59. Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. (2000) 404:293–6. doi: 10.1038/35005107
60. Lee KJ, Singh N, Bizuneh M, Kim NH, Kim HS, Kim Y, et al. miR-429 suppresses endometrial cancer progression and drug resistance via DDX53. J Pers Med. (2023) 13. doi: 10.3390/jpm13091302
61. Habel A, Nassar F, Itani M, Bouaziz H, Hadj-Ahmed M, Msheik Z, et al. Mir-21 and Mir-125b as theranostic biomarkers for epithelial ovarian cancer in Tunisian women. Afr Health Sci. (2023) 23:256–64. doi: 10.4314/ahs.v23i2.29
62. Yang Q, Fu J, Wang M, Fang Y, Fu J. MiR-1299 is regulated by KCNQ1OT1 and inhibits cervical cancer progression. Cell Mol Biol (Noisy-le-grand). (2023) 69:166–73. doi: 10.14715/cmb/2023.69.10.24
63. Zhang L, Ge R, Cheng A, Hu T. MiR-1284 suppresses the proliferation and migration of thyroid cancer. Cell Mol Biol (Noisy-le-grand). (2023) 69:246–9. doi: 10.14715/cmb/2023.69.8.38
64. Chen ZH, Chen YB, Yue HR, Zhou XJ, Ma HY, Wang X, et al. PAX5-miR-142 feedback loop promotes breast cancer proliferation by regulating DNMT1 and ZEB1. Mol Med. (2023) 29:89. doi: 10.1186/s10020-023-00681-y
65. Zhang Q, Zheng K, Gao Y, Zhao S, Zhao Y, Li W, et al. Plasma exosomal miR-1290 and miR-29c-3p as diagnostic biomarkers for lung cancer. Heliyon. (2023) 9:e21059. doi: 10.1016/j.heliyon.2023.e21059
66. Li Y, Zeng S, Cao L. Mir-629 repressed LATS2 expression and promoted the proliferation of prostate cancer cells. Horm Metab Res. (2023) 55:573–9.
67. Liu MX, Chu KM. MiR-410-3p suppresses primary gastric cancer and exosomes regulate endogenous expression of miR-410-3p. Am J Cancer Res. (2023) 13:2670–80.
68. Xie H, Li J, Lu M, Zhang R, Mao H. miR-1180 targets FXYD5 to regulate pancreatic cancer cells migration and invasion. Mol Biotechnol. (2023). doi: 10.1007/s12033-023-00923-8
69. Bartel DP. MicroRNAs: target recognition and regulatory functions. cell. (2009) 136:215–33. doi: 10.1016/j.cell.2009.01.002
70. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. (2010) 466:835–40. doi: 10.1038/nature09267
71. Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. (2005) 7:719–23. doi: 10.1038/ncb1274
72. Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell. (2008) 32:519–28. doi: 10.1016/j.molcel.2008.10.017
73. Yang F, Zhang L, Huo X, Yuan J, Xu D, Yuan S, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. (2011) 54:1679–89. doi: 10.1002/hep.24563
74. Hayes CN, Chayama K. MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. Int J Mol Sci. (2016) 17:280. doi: 10.3390/ijms17030280
75. Bertino G, Ardiri AM, Calvagno GS, Bertino N, Boemi PM. Prognostic and diagnostic value of des-γ-carboxy prothrombin in liver cancer. Drug News Perspect. (2010) 23:498–508.
76. Durazo FA, Blatt LM, Corey WG, Lin JH, Han S, Saab S, et al. Des-gamma-carboxyprothrombin, alpha-fetoprotein and AFP-L3 in patients with chronic hepatitis, cirrhosis and hepatocellular carcinoma. J Gastroenterol Hepatol. (2008) 23:1541–8. doi: 10.1111/j.1440-1746.2008.05395.x
77. Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. (2013) 14:1–14. doi: 10.1186/1471-2164-14-319
78. Anwar SL, Lehmann U. MicroRNAs: emerging novel clinical biomarkers for hepatocellular carcinomas. J Clin Med. (2015) 4:1631–50. doi: 10.3390/jcm4081631
79. Ladeiro Y, Couchy G, Balabaud C, Bioulac‐Sage P, Pelletier L, Rebouissou S, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. (2008) 47:1955–63. doi: 10.1002/(ISSN)1527-3350
80. Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. (2011) 104:235–40. doi: 10.1038/sj.bjc.6606010
81. Qi P, Cheng S, Wang H, Li N, Chen Y, Gao C. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PloS One. (2011) 6:e28486. doi: 10.1371/journal.pone.0028486
82. Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. (2011) 19:232–43. doi: 10.1016/j.ccr.2011.01.001
83. Li D, Liu X, Lin L, Hou J, Li N, Wang C, et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J Biol Chem. (2011) 286:36677–85. doi: 10.1074/jbc.M111.270561
84. Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang Y, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. (2008) 283:13205–15. doi: 10.1074/jbc.M707629200
85. Zhuang L-p, Meng Z-q. Serum miR-224 reflects stage of hepatocellular carcinoma and predicts survival. BioMed Res Int. (2015) 2015. doi: 10.1155/2015/731781
86. Ghosh A, Ghosh A, Datta S, Dasgupta D, Das S, Ray S, et al. Hepatic mi R-126 is a potential plasma biomarker for detection of hepatitis B virus infected hepatocellular carcinoma. Int J Cancer. (2016) 138:2732–44. doi: 10.1002/ijc.29999
87. Chen Y, Shen A, Rider PJ, Yu Y, Wu K, Mu Y, et al. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. (2011) 25:4511. doi: 10.1096/fj.11-187781
88. Morishita A, Masaki T. miRNA in hepatocellular carcinoma. Hepatol Res. (2015) 45:128–41. doi: 10.1111/hepr.12386
89. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
90. Liu WR, Shi YH, Peng YF, Fan J. Epigenetics of hepatocellular carcinoma: a new horizon. Chin Med J (Engl). (2012) 125:2349–60.
91. Vinciguerra M, Sgroi A, Veyrat-Durebex C, Rubbia-Brandt L, Buhler LH, Foti M. Unsaturated fatty acids inhibit the expression of tumor suppressor phosphatase and tensin homolog (PTEN) via microRNA-21 up-regulation in hepatocytes. Hepatology. (2009) 49:1176–84. doi: 10.1002/hep.22737
92. O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. (2008) 205:585–94. doi: 10.1084/jem.20072108
93. Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. (2009) 50:1152–61. doi: 10.1002/hep.23100
94. Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY, et al. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology. (2009) 136:683–93. doi: 10.1053/j.gastro.2008.10.029
95. Wang Z, Lin S, Li JJ, Xu Z, Yao H, Zhu X, et al. MYC protein inhibits transcription of the microRNA cluster MC-let-7a-1~let-7d via noncanonical E-box. J Biol Chem. (2011) 286:39703–14. doi: 10.1074/jbc.M111.293126
96. Au SL, Wong CC, Lee JM, Fan DN, Tsang FH, Ng IO, et al. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. (2012) 56:622–31. doi: 10.1002/hep.25679
97. Yin H, Hu M, Zhang R, Shen Z, Flatow L, You M. MicroRNA-217 promotes ethanol-induced fat accumulation in hepatocytes by down-regulating SIRT1. J Biol Chem. (2012) 287:9817–26. doi: 10.1074/jbc.M111.333534
98. Choudhury M, Shukla SD. Surrogate alcohols and their metabolites modify histone H3 acetylation: involvement of histone acetyl transferase and histone deacetylase. Alcohol Clin Exp Res. (2008) 32:829–39. doi: 10.1111/j.1530-0277.2008.00630.x
99. Tomita K, Teratani T, Suzuki T, Shimizu M, Sato H, Narimatsu K, et al. Free cholesterol accumulation in hepatic stellate cells: mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology. (2014) 59:154–69. doi: 10.1002/hep.26604
100. El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. (2012) 142:1264–1273.e1. doi: 10.1053/j.gastro.2011.12.061
101. Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. (2009) 361:1437–47. doi: 10.1056/NEJMoa0901282
102. Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. (2009) 49:1098–112. doi: 10.1002/hep.22749
103. Mizuguchi Y, Mishima T, Yokomuro S, Arima Y, Kawahigashi Y, Shigehara K, et al. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PloS One. (2011) 6:e15304. doi: 10.1371/journal.pone.0015304
104. Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, et al. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. (2013) 133:816–24. doi: 10.1002/ijc.28075
105. Hsu HC, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F. Expression of p53 gene in 184 unifocal hepatocellular carcinomas: association with tumor growth and invasiveness. Cancer Res. (1993) 53:4691–4.
106. Hu Z, Zhao Y, Mang Y, Zhu J, Yu L, Li L, et al. MiR-21-5p promotes sorafenib resistance and hepatocellular carcinoma progression by regulating SIRT7 ubiquitination through USP24. Life Sci. (2023) 325:121773. doi: 10.1016/j.lfs.2023.121773
107. Hong J, Liu J, Zhang Y, Ding L, Ye Q. MiR-3180 inhibits hepatocellular carcinoma growth and metastasis by targeting lipid synthesis and uptake. Cancer Cell Int. (2023) 23:66. doi: 10.1186/s12935-023-02915-9
108. Xu Y, Luan G, Liu F, Zhang Y, Li Z, Liu Z, et al. Exosomal miR-200b-3p induce macrophage polarization by regulating transcriptional repressor ZEB1 in hepatocellular carcinoma. Hepatol Int. (2023) 17:889–903. doi: 10.1007/s12072-023-10507-y
109. Feng L, Chen X, Li P, Li Y, Zhai Y, Liu X, et al. miR-424-3p promotes metastasis of hepatocellular carcinoma via targeting the SRF-STAT1/2 axis. Carcinogenesis. (2023) 44:610–25. doi: 10.1093/carcin/bgad037
110. Liu HT, Luo CP, Jiang MJ, Deng ZJ, Teng YX, Su JY, et al. miR-17-5p slows progression of hepatocellular carcinoma by downregulating TGFβR2. Clin Transl Oncol. (2023) 25:2960–71. doi: 10.1007/s12094-023-03164-y
111. Zou Q, Cao S. miR-4270 suppresses hepatocellular carcinoma progression by inhibiting DNMT3A-mediated methylation of HGFAC promoter. PeerJ. (2023) 11:e16566. doi: 10.7717/peerj.16566
112. Wu M, Deng J, Yao D, Li S. MiR-767-3p promotes the progression of hepatocellular carcinoma via targeting CASP-3/-9. Am J Transl Res. (2023) 15:2926–38.
113. Wang Y, Tian Y. miRNA for diagnosis and clinical implications of human hepatocellular carcinoma. Hepatol Res. (2016) 46:89–99. doi: 10.1111/hepr.12571
114. Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. (2012) 32:326–48. doi: 10.1002/med.20215
115. Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. (2002) 2:569–79. doi: 10.1038/nri855
116. Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. BioMed Res Int. (2014) 2014. doi: 10.1155/2014/864894
117. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U.S.A. (2011) 108:5003–8. doi: 10.1073/pnas.1019055108
118. Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. (2008) 47:897–907. doi: 10.1002/hep.22160
119. Wei R, Huang GL, Zhang MY, Li BK, Zhang HZ, Shi M, et al. Clinical significance and prognostic value of microRNA expression signatures in hepatocellular carcinoma. Clin Cancer Res. (2013) 19:4780–91. doi: 10.1158/1078-0432.CCR-12-2728
120. Liang Z, Gao Y, Shi W, Zhai D, Li S, Jing L, et al. Expression and significance of microRNA-183 in hepatocellular carcinoma. Sci World J. (2013) 2013. doi: 10.1155/2013/381874
121. Yuan G, Wu H, Du Y, He F. Tumor suppressor role of microRNA-545 in oral squamous cell carcinoma. Oncol Lett. (2019) 17:2063–8.
122. Chen L, Gao H, Liang J, Qiao J, Duan J, Shi H, et al. miR-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Am J Cancer Res. (2018) 8:2387–401.
123. Xu B, Lu X, Zhao Y, Liu C, Huang X, Chen S, et al. MicroRNA-135a induces prostate cancer cell apoptosis via inhibition of STAT6. Oncol Lett. (2019) 17:1889–95.
124. Ni JS, Zheng H, Huang ZP, Hong YG, Ou YL, Tao YP, et al. MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol Lett. (2019) 17:2317–27.
125. Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, et al. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. (2018) 37:324. doi: 10.1186/s13046-018-0965-2
126. Wang Y, Chang W, Chang W, Chang X, Zhai S, Pan G, et al. MicroRNA-376c-3p facilitates human hepatocellular carcinoma progression via repressing AT-rich interaction domain 2. J Cancer. (2018) 9:4187–96. doi: 10.7150/jca.27939
127. Jiang XM, Yu XN, Liu TT, Zhu HR, Shi X, Bilegsaikhan E, et al. microRNA-19a-3p promotes tumor metastasis and chemoresistance through the PTEN/Akt pathway in hepatocellular carcinoma. BioMed Pharmacother. (2018) 105:1147–54. doi: 10.1016/j.biopha.2018.06.097
128. Tian T, Fu X, Lu J, Ruan Z, Nan K, Yao Y, et al. MicroRNA-760 inhibits doxorubicin resistance in hepatocellular carcinoma through regulating notch1/hes1-PTEN/akt signaling pathway. J Biochem Mol Toxicol. (2018) 32:e22167. doi: 10.1002/jbt.22167
129. Huang Y, Chen G, Wang Y, He R, Du J, Jiao X, et al. Inhibition of microRNA-16 facilitates the paclitaxel resistance by targeting IKBKB via NF-κB signaling pathway in hepatocellular carcinoma. Biochem Biophys Res Commun. (2018) 503:1035–41. doi: 10.1016/j.bbrc.2018.06.113
130. Zhang K, Chen J, Zhou H, Chen Y, Zhi Y, Zhang B, et al. PU.1/microRNA-142-3p targets ATG5/ATG16L1 to inactivate autophagy and sensitize hepatocellular carcinoma cells to sorafenib. Cell Death Dis. (2018) 9:312. doi: 10.1038/s41419-018-0344-0
131. Zhang G, Li N, Li Z, Zhu Q, Li F, Yang C, et al. microRNA-4717 differentially interacts with its polymorphic target in the PD1 3’ untranslated region: A mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget. (2015) 6:18933–44. doi: 10.18632/oncotarget.v6i22
132. Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. (2004) 64:7099–109. doi: 10.1158/0008-5472.CAN-04-1443
133. Yu C, Bruzek LM, Meng XW, Gores GJ, Carter CA, Kaufmann SH, et al. The role of Mcl-1 downregulation in the proapoptotic activity of the multikinase inhibitor BAY 43-9006. Oncogene. (2005) 24:6861–9. doi: 10.1038/sj.onc.1208841
134. Sonntag R, Gassler N, Bangen JM, Trautwein C, Liedtke C. Pro-apoptotic Sorafenib signaling in murine hepatocytes depends on Malignancy and is associated with PUMA expression in vitro and in vivo. Cell Death Dis. (2014) 5:e1030. doi: 10.1038/cddis.2013.557
135. Wang Z, Hu P, Tang F, Xie C. HDAC6-mediated EGFR stabilization and activation restrict cell response to sorafenib in non-small cell lung cancer cells. Med Oncol. (2016) 33:50. doi: 10.1007/s12032-016-0765-5
136. Xiang QF, Zhan MX, Li Y, Liang H, Hu C, Huang YM, et al. Activation of MET promotes resistance to sorafenib in hepatocellular carcinoma cells via the AKT/ERK1/2-EGR1 pathway. Artif Cells Nanomed Biotechnol. (2019) 47:83–9. doi: 10.1080/21691401.2018.1543195
137. Xu H, Zhao L, Fang Q, Sun J, Zhang S, Zhan C, et al. MiR-338-3p inhibits hepatocarcinoma cells and sensitizes these cells to sorafenib by targeting hypoxia-induced factor 1α. PloS One. (2014) 9:e115565. doi: 10.1371/journal.pone.0115565
138. Liu H, Wang M, Liang N, Guan L. PDCD2 sensitizes HepG2 cells to sorafenib by suppressing epithelial−mesenchymal transition. Mol Med Rep. (2019) 19:2173–9. doi: 10.3892/mmr
139. Pollutri D, Patrizi C, Marinelli S, Giovannini C, Trombetta E, Giannone FA, et al. The epigenetically regulated miR-494 associates with stem-cell phenotype and induces sorafenib resistance in hepatocellular carcinoma. Cell Death Dis. (2018) 9:4. doi: 10.1038/s41419-017-0076-6
140. Fornari F, Pollutri D, Patrizi C, La Bella T, Marinelli S, Casadei Gardini A, et al. In Hepatocellular Carcinoma miR-221 Modulates Sorafenib Resistance through Inhibition of Caspase-3-Mediated Apoptosis. Clin Cancer Res. (2017) 23:3953–65. doi: 10.1158/1078-0432.CCR-16-1464
141. Zhang PF, Wang F, Wu J, Wu Y, Huang W, Liu D, et al. LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J Cell Physiol. (2019) 234:2788–94. doi: 10.1002/jcp.27095
142. He C, Dong X, Zhai B, Jiang X, Dong D, Li B, et al. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. (2015) 6:28867–81. doi: 10.18632/oncotarget.v6i30
143. Ohta K, Hoshino H, Wang J, Ono S, Iida Y, Hata K, et al. MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget. (2015) 6:3211–24. doi: 10.18632/oncotarget.v6i5
144. Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. (2013) 58:629–41. doi: 10.1002/hep.26369
145. Azumi J, Tsubota T, Sakabe T, Shiota G. miR-181a induces sorafenib resistance of hepatocellular carcinoma cells through downregulation of RASSF1 expression. Cancer Sci. (2016) 107:1256–62. doi: 10.1111/cas.13006
146. Wei L, Wang X, Lv L, Liu J, Xing H, Song Y, et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. (2019) 18:147. doi: 10.1186/s12943-019-1086-z
147. Tsang WP, Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. (2008) 13:1215–22. doi: 10.1007/s10495-008-0256-z
148. Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G, et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. (2012) 227:275–85. doi: 10.1002/path.3995
149. Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, et al. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. (2017) 8:e2540. doi: 10.1038/cddis.2016.461
150. Yang N, Ekanem NR, Sakyi CA, Ray SD. Hepatocellular carcinoma and microRNA: New perspectives on therapeutics and diagnostics. Advanced Drug Delivery Rev. (2015) 81:62–74. doi: 10.1016/j.addr.2014.10.029
151. Lee Y-H, Andersen JB, Song H-T, Judge AD, Seo D, Ishikawa T, et al. Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer Res. (2010) 70:8264–9. doi: 10.1158/0008-5472.CAN-10-0749
152. Hsu S-h, Yu B, Wang X, Lu Y, Schmidt CR, Lee RJ, et al. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomed: Nanotechnol Biol Med. (2013) 9:1169–80. doi: 10.1016/j.nano.2013.05.007
153. He X, Chang Y, Meng F, Wang M, Xie Q, Tang F, et al. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. (2012) 31:3357–69. doi: 10.1038/onc.2011.500
154. Takamura S, Niikura M, Li T, Takeda N, Kusagawa S, Takebe Y, et al. DNA vaccine-encapsulated virus-like particles derived from an orally transmissible virus stimulate mucosal and systemic immune responses by oral administration. Gene Ther. (2004) 11:628–35. doi: 10.1038/sj.gt.3302193
155. Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat materials. (2012) 11:316–22. doi: 10.1038/nmat3253
156. Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. (2010) 20:R858–61. doi: 10.1016/j.cub.2010.08.052
157. Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z, et al. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer. (2013) 13:1–9. doi: 10.1186/1471-2407-13-469
158. Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang H-W, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. (2009) 137:1005–17. doi: 10.1016/j.cell.2009.04.021
159. Austin T. First microRNA mimic enters clinic. Nat Biotechnol. (2013) 31:577. doi: 10.1038/nbt0713-577
160. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. (2009) 136:629–41. doi: 10.1016/j.cell.2009.02.006
161. Huang J-L, Zheng L, Hu Y-W, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. (2014) 35:507–14. doi: 10.1093/carcin/bgt405
162. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. (2013) 339:159–66. doi: 10.1016/j.canlet.2013.06.013
164. Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. (2013) 45:1895–910. doi: 10.1016/j.biocel.2013.05.030
165. Li C, Chen J, Zhang K, Feng B, Wang R, Chen L, et al. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. (2015) 36:423–34. doi: 10.1159/000430109
166. Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat Rev Genet. (2011) 12:641–55. doi: 10.1038/nrg3049
167. Jayaraj GG, Pandey S, Scaria V, Maiti S. Potential G-quadruplexes in the human long non-coding transcriptome. RNA Biol. (2012) 9:81–9. doi: 10.4161/rna.9.1.18047
168. Xue W, Yang L, Chen C, Ashrafizadeh M, Tian Y, Sun R. Wnt/β-catenin-driven EMT regulation in human cancers. Cell Mol Life Sci. (2024) 81:79. doi: 10.1007/s00018-023-05099-7
169. Ashrafizadeh M, Zhang W, Tian Y, Sethi G, Zhang X, Qiu A. Molecular panorama of therapy resistance in prostate cancer: a pre-clinical and bioinformatics analysis for clinical translation. Cancer Metastasis Rev. (2024). doi: 10.1007/s10555-024-10168-9
170. Guo Z, Ashrafizadeh M, Zhang W, Zou R, Sethi G, Zhang X. Molecular profile of metastasis, cell plasticity and EMT in pancreatic cancer: a pre-clinical connection to aggressiveness and drug resistance. Cancer Metastasis Rev. (2023). doi: 10.1007/s10555-023-10125-y
171. Fan C, Wang Q, Kuipers TB, Cats D, Iyengar PV, Hagenaars SC, et al. LncRNA LITATS1 suppresses TGF-β-induced EMT and cancer cell plasticity by potentiating TβRI degradation. EMBO J. (2023) 42:e112806. doi: 10.15252/embj.2022112806
172. Xu J, Hu M, Gao Y, Wang Y, Yuan X, Yang Y, et al. LncRNA MIR17HG Suppresses Breast Cancer Proliferation and Migration as ceRNA to Target FAM135A by Sponging miR-454-3p. Mol Biotechnol. (2023) 65:2071–85. doi: 10.1007/s12033-023-00706-1
173. Shi L, Li B, Zhang Y, Chen Y, Tan J, Chen Y, et al. Exosomal lncRNA Mir100hg derived from cancer stem cells enhance glycolysis and promote metastasis of lung adenocarcinoma through mircroRNA-15a-5p/31-5p. Cell Commun Signal. (2023) 21:248. doi: 10.1186/s12964-023-01281-3
174. Wu JH, Cheng TC, Zhu B, Gao HY, Zheng L, Chen WX. Identification of cuproptosis-related gene SLC31A1 and upstream LncRNA-miRNA regulatory axis in breast cancer. Sci Rep. (2023) 13:18390. doi: 10.1038/s41598-023-45761-5
175. Qian L, Li L, Li Y, Li S, Zhang B, Zhu Y, et al. LncRNA HOTAIR as a ceRNA is related to breast cancer risk and prognosis. Breast Cancer Res Treat. (2023) 200:375–90. doi: 10.1007/s10549-023-06982-4
176. Huang K, Chen X, Geng Z, Xiong X, Cong Y, Pan X, et al. LncRNA SLC25A21-AS1 increases the chemosensitivity and inhibits the progression of ovarian cancer by upregulating the expression of KCNK4. Funct Integr Genomics. (2023) 23:110. doi: 10.1007/s10142-023-01035-x
177. Jin Y, Qiu J, Lu X, Ma Y, Li G. LncRNA CACNA1G-AS1 up-regulates FTH1 to inhibit ferroptosis and promote Malignant phenotypes in ovarian cancer cells. Oncol Res. (2023) 31:169–79. doi: 10.32604/or.2023.027815
178. Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, et al. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. (2012) 287:26302–11. doi: 10.1074/jbc.M112.342113
179. Park BJ, Kang JW, Lee SW, Choi SJ, Shin YK, Ahn YH, et al. The haploinsufficient tumor suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell. (2005) 120:209–21. doi: 10.1016/j.cell.2004.11.054
180. Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC, et al. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/β-catenin signaling. Hepatology. (2013) 58:739–51. doi: 10.1002/hep.26361
181. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. (2012) 9:703–19. doi: 10.4161/rna.20481
182. Huang JF, Guo YJ, Zhao CX, Yuan SX, Wang Y, Tang GN, et al. Hepatitis B virus X protein (HBx)-related long noncoding RNA (lncRNA) down-regulated expression by HBx (Dreh) inhibits hepatocellular carcinoma metastasis by targeting the intermediate filament protein vimentin. Hepatology. (2013) 57:1882–92. doi: 10.1002/hep.26195
183. Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. (2013) 49:1083–96. doi: 10.1016/j.molcel.2013.01.010
184. Jiang J, Dong W, Zhang W, Wang Q, Wang R, Wang J, et al. LncRNA SLC1A5-AS/MZF1/ASCT2 axis contributes to Malignant progression of hepatocellular carcinoma. Discovery Med. (2023) 35:995–1014. doi: 10.24976/Discov.Med.202335179.96
185. Liu R, Yin G, Tuo H, Guo Y, Zhu Y, Zhang L, et al. METTL3-induced lncRNA GBAP1 promotes hepatocellular carcinoma progression by activating BMP/SMAD pathway. Biol Direct. (2023) 18:53. doi: 10.1186/s13062-023-00409-2
186. Xu B, Li C, Yang B, Zhou F, Tang K, Wang L, et al. LncRNA SNHG20 silencing inhibits hepatocellular carcinoma progression by sponging miR-5095 from MBD1. Am J Transl Res. (2023) 15:4314–31.
187. Huang J, Yang Y, Zhao F, Zhang Z, Deng J, Lu W, et al. LncRNA SATB2-AS1 overexpression represses the development of hepatocellular carcinoma through regulating the miR-3678-3p/GRIM-19 axis. Cancer Cell Int. (2023) 23:82. doi: 10.1186/s12935-023-02901-1
188. Zhou L, Zhang Q, Cheng J, Shen X, Li J, Chen M, et al. LncRNA SNHG1 upregulates FANCD2 and G6PD to suppress ferroptosis by sponging miR-199a-5p/3p in hepatocellular carcinoma. Drug Discovery Ther. (2023) 17:248–56. doi: 10.5582/ddt.2023.01035
189. Tuo H, Liu R, Wang Y, Yang W, Liu Q. Hypoxia-induced lncRNA MRVI1-AS1 accelerates hepatocellular carcinoma progression by recruiting RNA-binding protein CELF2 to stabilize SKA1 mRNA. World J Surg Oncol. (2023) 21:111. doi: 10.1186/s12957-023-02993-z
190. Liangyu Z, Bochao Z, Guoquan Y, Yuan Z, Heng L, Hanyu Z. Bioinformatics prediction and experimental verification identify cuproptosis-related lncRNA as prognosis biomarkers of hepatocellular carcinoma. Biochem Biophys Rep. (2023) 35:101502. doi: 10.1016/j.bbrep.2023.101502
191. Zhu Q, Lei Z, Xu C, Zhang Z, Yu Z, Cheng Z, et al. LncRNA HClnc1 facilitates hepatocellular carcinoma progression by regulating PKM2 signaling and indicates poor survival outcome after hepatectomy. Cancer Med. (2023) 12:14526–44. doi: 10.1002/cam4.6117
192. Zeng FL, Lin J, Xie X, Xie YK, Zhang JH, Xu D, et al. LncRNA SLC7A11-AS1 promotes the progression of hepatocellular carcinoma by mediating KLF9 ubiquitination. Neoplasma. (2023) 70:361–74. doi: 10.4149/neo_2023_230323N162
193. Chi X, Chen Z, Chen Y, Hong H, Yu J, Lv L. Upregulation of lncRNA PTOV1-AS1 in hepatocellular carcinoma contributes to disease progression and sorafenib resistance through regulating miR-505. J Biochem Mol Toxicol. (2023) 37:e23437. doi: 10.1002/jbt.23437
194. Gu D, Tong M, Wang J, Zhang B, Liu J, Song G, et al. Overexpression of the lncRNA HOTAIRM1 promotes lenvatinib resistance by downregulating miR-34a and activating autophagy in hepatocellular carcinoma. Discovery Oncol. (2023) 14:66. doi: 10.1007/s12672-023-00673-8
195. Sun J, Li Y, Shi M, Tian H, Li J, Zhu K, et al. A Positive Feedback Loop of lncRNA HOXD-AS2 and SMYD3 Facilitates Hepatocellular Carcinoma Progression via the MEK/ERK Pathway. J Hepatocell Carcinoma. (2023) 10:1237–56. doi: 10.2147/JHC.S416946
196. Quan B, Liu W, Yao F, Li M, Tang B, Li J, et al. LINC02362/hsa-miR-18a-5p/FDX1 axis suppresses proliferation and drives cuproptosis and oxaliplatin sensitivity of hepatocellular carcinoma. Am J Cancer Res. (2023) 13:5590–609.
197. Hashemi M, Mirzaei S, Zandieh MA, Rezaei S, Amirabbas K, Dehghanpour A, et al. Long non-coding RNAs (lncRNAs) in hepatocellular carcinoma progression: Biological functions and new therapeutic targets. Prog Biophysics Mol Biol. (2023) 177:207–28. doi: 10.1016/j.pbiomolbio.2022.12.004
198. Jia X, Li L, Wang F, Xue Y, Wu T, Jia Q, et al. DUB3/KLF4 combats tumor growth and chemoresistance in hepatocellular carcinoma. Cell Death Discovery. (2022) 8:166. doi: 10.1038/s41420-022-00988-5
199. Wang Y, Li N, Zheng Y, Wang A, Yu C, Song Z, et al. KIAA1217 promotes epithelial-mesenchymal transition and hepatocellular carcinoma metastasis by interacting with and activating STAT3. Int J Mol Sci. (2021) 23:104. doi: 10.3390/ijms23010104
200. Sun J, Zheng X, Wang B, Cai Y, Zheng L, Hu L, et al. LncRNA LIMT (LINC01089) contributes to sorafenib chemoresistance via regulation of miR-665 and epithelial to mesenchymal transition in hepatocellular carcinoma cells: LIMT contributes to sorafenib chemoresistance in HCC. Acta Biochim Biophys Sin. (2022) 54:261. doi: 10.3724/abbs.2021019
201. Zhang Y, Tie Q, Bao Z, Shao Z, Zhang L. Inhibition of miR-15a-5p promotes the chemoresistance to pirarubicin in hepatocellular carcinoma via targeting eIF4E. Comput Math Methods Med. (2021) 2021. doi: 10.1155/2021/6468405
202. Wang F, Xu C, Li G, Lv P, Gu J. Incomplete radiofrequency ablation induced chemoresistance by up-regulating heat shock protein 70 in hepatocellular carcinoma. Exp Cell Res. (2021) 409:112910. doi: 10.1016/j.yexcr.2021.112910
203. Zhou J, Che J, Xu L, Yang W, Li Y, Zhou W, et al. Enhancer of zeste homolog 2 promotes hepatocellular cancer progression and chemoresistance by enhancing protein kinase B activation through microRNA-381-mediated SET domain bifurcated 1. Bioengineered. (2022) 13:5737–55. doi: 10.1080/21655979.2021.2023792
204. Zhao G, Zhang A, Sun S, Ding Y. Long non−coding RNA LINC00173 enhances cisplatin resistance in hepatocellular carcinoma via the microRNA−641/RAB14 axis. Oncol Lett. (2021) 21:1–9. doi: 10.3892/ol
205. Rajendran P, Li F, Manu KA, Shanmugam MK, Loo SY, Kumar AP, et al. γ-.,endran,ra is a novel inhibitor of constitutive and inducible STAT3 signalling pathway in human hepatocellular carcinoma: potential role as an antiproliferative, proiprolifera and chemosensitizing agent. Br J Pharmacol. (2011) 163:283–98. doi: 10.1111/j.1476-5381.2010.01187.x
206. Arora L, Mohan CD, Yang MH, Rangappa S, Deivasigamani A, Kumar AP, et al. Tris (dibenzylideneacetone) dipalladium (0)(Tris DBA) abrogates tumor progression in hepatocellular carcinoma and multiple myeloma preclinical models by regulating the STAT3 signaling pathway. Cancers. (2021) 13:5479. doi: 10.3390/cancers13215479
207. Fei Q, Song F, Jiang X, Hong H, Xu X, Jin Z, et al. LncRNA ST8SIA6-AS1 promotes hepatocellular carcinoma cell proliferation and resistance to apoptosis by targeting miR-4656/HDAC11 axis. Cancer Cell Int. (2020) 20:1–10. doi: 10.1186/s12935-020-01325-5
208. Li X, Zhou Y, Yang L, Ma Y, Peng X, Yang S, et al. LncRNA NEAT1 promotes autophagy via regulating miRulatingna and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J Cell Physiol. (2020) 235:3402–13. doi: 10.1002/jcp.29230
209. Zhang W, Liu Y, Fu Y, Han W, Xu H, Wen L, et al. Long non-coding RNA LINC00160 functions as a decoy of microRNA-132 to mediate autophagy and drug resistance in hepatocellular carcinoma via inhibition of PIK3R3. Cancer Lett. (2020) 478:22–33. doi: 10.1016/j.canlet.2020.02.014
210. Niu Y, Tang G, Wu X, Wu C. LncRNA NEAT1 modulates sorafenib resistance in hepatocellular carcinoma through regulating the miR-149-5p/AKT1 axis. Saudi J Gastroenterol. (2020) 26:194–203. doi: 10.4103/sjg.SJG_4_20
211. Mou S-J, Yang P-F, Liu Y-P, Xu N, Jiang W-W, Yue W-J. BCLAF1 promotes cell proliferation, invasion and drug-resistance though targeting lncRNA NEAT1 in hepatocellular carcinoma. Life Sci. (2020) 242:117177. doi: 10.1016/j.lfs.2019.117177
212. Liu Y, Chen L, Yuan H, Guo S, Wu G. LncRNA DANCR promotes sorafenib resistance via activation of IL-6/STAT3 signaling in hepatocellular carcinoma cells. OncoTargets Ther. (2020) p:1145–57. doi: 10.2147/OTT
213. Wu E-R, Chou Y-E, Liu Y-F, Hsueh K-C, Lee H-L, Yang S-F, et al. Association of lncRNA H19 gene polymorphisms with the occurrence of hepatocellular carcinoma. Genes. (2019) 10:506. doi: 10.3390/genes10070506
214. Huang Z, Zhou J-K, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. (2020) 19:77. doi: 10.1186/s12943-020-01188-4
215. Llovet JM, Montal R, Sia D, Finn RS. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. (2018) 15:599–616. doi: 10.1038/s41571-018-0073-4
216. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discovery. (2017) 16:167–79. doi: 10.1038/nrd.2016.117
217. Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. (2016) 44:863–77. doi: 10.1093/nar/gkv1206
218. Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. (2012) 488:111–5. doi: 10.1038/nature11362
219. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
220. Zhou CC, Yang F, Yuan SX, Ma JZ, Liu F, Yuan JH, et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology. (2016) 63:850–63. doi: 10.1002/hep.28393
221. Javanbakht H, Mueller H, Walther J, Zhou X, Lopez A, Pattupara T, et al. Liver-targeted anti-HBV single-stranded oligonucleotides with locked nucleic acid potently reduce HBV gene expression in vivo. Mol Ther Nucleic Acids. (2018) 11:441–54. doi: 10.1016/j.omtn.2018.02.005
222. Thi EP, Dhillon AP, Ardzinski A, Bidirici-Ertekin L, Cobarrubias KD, Cuconati A, et al. ARB-1740, a RNA interference therapeutic for chronic hepatitis B infection. ACS Infect Dis. (2019) 5:725–37. doi: 10.1021/acsinfecdis.8b00191
223. Li C, Yang J, Liu C, Wang X, Zhang L. Long non-coding RNAs in hepatocellular carcinoma: Ordering of the complicated lncRNA regulatory network and novel strategies for HCC clinical diagnosis and treatment. Pharmacol Res. (2020) 158:104848. doi: 10.1016/j.phrs.2020.104848
224. Hung C-S, Liu H-H, Liu J-J, Yeh C-T, Chang T-C, Wu C-H, et al. MicroRNA-200a and-200b mediated hepatocellular carcinoma cell migration through the epithelial to mesenchymal transition markers. Ann Surg Oncol. (2013) 20:360–8. doi: 10.1245/s10434-012-2482-4
225. Zhang L, Yang F, Yuan J-h, Yuan S-x, Zhou W-p, Huo X-s, et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. (2013) 34:577–86. doi: 10.1093/carcin/bgs381
226. Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. (2012) 19:586–93. doi: 10.1038/nsmb.2296
227. Kallen AN, Zhou X-B, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. (2013) 52:101–12. doi: 10.1016/j.molcel.2013.08.027
228. Fernandes JC, Acuña SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long non-coding RNAs in the regulation of gene expression: physiology and disease. Non-coding RNA. (2019) 5:17. doi: 10.3390/ncrna5010017
229. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta (BBA)-Gene Regul Mech. (2016) 1859:169–76. doi: 10.1016/j.bbagrm.2015.06.015
230. Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, et al. Long nong,g,ang RNA HOTTIP is frequently upequently,. in hepatocellular carcinoma and is targeted by tumour suppressive miRpressi. Liver Int. (2015) 35:1597–606. doi: 10.1111/liv.12746
231. Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep. (2014) 6:211–21. doi: 10.1016/j.celrep.2013.12.013
232. Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang X-Y, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci. (2008) 105:18782–7. doi: 10.1073/pnas.0810199105
233. Ge Y, Yan X, Jin Y, Yang X, Yu X, Zhou L, et al. fMiRNA-192 and miRNA-204 directly suppress lncRNA HOTTIP and interrupt GLS1-mediated glutaminolysis in hepatocellular carcinoma. PloS Genet. (2015) 11:e1005726. doi: 10.1371/journal.pgen.1005726
234. Lu R, Chen J, Kong L, Zhu H. Prognostic value of lncRNA ROR expression in various cancers: a meta-analysis. Bioscience Rep. (2018) 38:BSR20181095. doi: 10.1042/BSR20181095
235. Tian T, Wang M, Lin S, Guo Y, Dai Z, Liu K, et al. The impact of lncRNA dysregulation on clinicopathology and survival of breast cancer: a systematic review and meta-analysis. Mol Therapy-Nucleic Acids. (2018) 12:359–69. doi: 10.1016/j.omtn.2018.05.018
236. Yang H, Huang C, Cao M, Wang Y, Liu Y. Long non-coding RNA CRNDE may be associated with poor prognosis by promoting proliferation and inhibiting apoptosis of cervical cancer cells through targeting PI3K/AKT. Neoplasma. (2018) 65:872–80. doi: 10.4149/neo_2018_171225N841
237. Xie H, Ma B, Gao Q, Zhan H, Liu Y, Chen Z, et al. Long non-coding RNA CRNDE in cancer prognosis: Review and meta-analysis. Clinica chimica Acta. (2018) 485:262–71. doi: 10.1016/j.cca.2018.07.003
238. Shen X, Piao L, Zhang S, Cui Y, Cui Y, Quan X, et al. Long non-coding RNA activated by TGF-β expression in cancer prognosis: a meta-analysis. Int J Surg. (2018) 58:37–45. doi: 10.1016/j.ijsu.2018.08.004
239. Lim LJ, Wong SY, Huang F, Lim S, Chong SS, Ooi LL, et al. Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. (2019) 79:5131–9. doi: 10.1158/0008-5472.CAN-19-0255
240. Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: Functions and implications. Cancer Med. (2018) 7:3101–9. doi: 10.1002/cam4.1574
241. Tian M, Chen R, Li T, Xiao B. Reduced expression of circRNA hsa_circ_0003159 in gastric cancer and its clinical significance. J Clin Lab Anal. (2018) 32. doi: 10.1002/jcla.22281
242. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. (2016) 17:679–92. doi: 10.1038/nrg.2016.114
243. Shao Y, Li J, Lu R, Li T, Yang Y, Xiao B, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. (2017) 6:1173–80. doi: 10.1002/cam4.1055
244. Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. (2017) 466:167–71. doi: 10.1016/j.cca.2017.01.025
245. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. (2015) 160:1125–34. doi: 10.1016/j.cell.2015.02.014
246. Shi L, Yan P, Liang Y, Sun Y, Shen J, Zhou S, et al. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. (2017) 8:e3171. doi: 10.1038/cddis.2017.556
247. Meyer M, Pamudurti N, Ivanov A, Bartok O, Hanan M, Evantal N, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. (2014) 56:55–66. doi: 10.1016/j.molcel.2014.08.019
248. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. (2017) 67:214–227.e7. doi: 10.1016/j.molcel.2017.05.023
249. Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci U.S.A. (2017) 114:E5207–e5215. doi: 10.1073/pnas.1617467114
250. Gupta SK, Garg A, Bär C, Chatterjee S, Foinquinos A, Milting H, et al. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res. (2018) 122:246–54. doi: 10.1161/CIRCRESAHA.117.311335
251. Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen LL, et al. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell. (2017) 68:940–954.e3. doi: 10.1016/j.molcel.2017.10.034
252. Yu X, Odenthal M, Fries JW. Exosomes as miRNA carriers: formation–function–future. Int J Mol Sci. (2016) 17:2028. doi: 10.3390/ijms17122028
253. Lasda E, Parker R. Circular RNAs co-precipitate with extracellular vesicles: A possible mechanism for circRNA clearance. PloS One. (2016) 11:e0148407. doi: 10.1371/journal.pone.0148407
254. Zhang X, Huang J, Wang J, Li Y, Hu G, Li H. Circ_0001667 accelerates breast cancer proliferation and angiogenesis through regulating CXCL10 expression by sponging miR-6838-5p. Thorac Cancer. (2023) 14:881–92. doi: 10.1111/1759-7714.14820
255. Lv D, Cen S, Yang S, Zou Z, Zhong J, Pan Z, et al. Hsa_circ_0063329 inhibits prostate cancer growth and metastasis by modulating the miR-605-5p/tgif2 axis. Cell Cycle. (2023) 22:1101–15. doi: 10.1080/15384101.2023.2174658
256. Li H, Jiao W, Song J, Wang J, Chen G, Li D, et al. circ-hnRNPU inhibits NONO-mediated c-Myc transactivation and mRNA stabilization essential for glycosylation and cancer progression. J Exp Clin Cancer Res. (2023) 42:313. doi: 10.1186/s13046-023-02898-5
257. Li J, Esteller M. Circular RNA hsa_circ_0067842 facilitates tumor metastasis and immune escape in breast cancer through HuR/CMTM6/PD-L1 axis. Biol Direct. (2023) 18:48. doi: 10.1186/s13062-023-00397-3
258. Xiao W, Li P. Circ_0087862 promotes tumorigenesis and glycolysis in colorectal cancer by sponging miR-296-3p to regulate PGK1 expression. Pathol Res Pract. (2023) 248:154695. doi: 10.1016/j.prp.2023.154695
259. Luo X, Liu J, Wang X, Wang Y, Yuan J, Zhang Y. Circ_0005615 promotes cervical cancer cell growth and metastasis by modulating the miR-138-5p/KDM2A axis. J Biochem Mol Toxicol. (2023) 37:e23410. doi: 10.1002/jbt.23410
260. Liu C, Mei T. Circ_0001821 potentiates cell growth, metastasis, and stemness in colorectal cancer by regulating miR-339-3p/CST1. Biochem Genet. (2023) 61:1451–69. doi: 10.1007/s10528-022-10329-x
261. Wu CC, Hou BC, Yang YH, Li XF, Ma HC, Li BX. Circ_0084188 promotes colorectal cancer progression by sponging miR-654-3p and regulating kruppel-like factor 12. Kaohsiung J Med Sci. (2023) 39:1062–76. doi: 10.1002/kjm2.12749
262. Yu F, Luo F, Zhang X, Huang Q. Circ_0006646 accelerates the growth and metastasis of cervical cancer by elevating RRM2 through miR-758-3p. Biochem Genet. (2023) 61:1300–18. doi: 10.1007/s10528-022-10320-6
263. Ge H, Yan Y, Wang H, Bian J, Deng Z, Su X, et al. Hsa_circ_0001278 Facilitates Colorectal Cancer Progression via Sponging miR-338-5p and Regulating AMOTL1 Expression. Comb Chem High Throughput Screen. (2023). doi: 10.2174/0113862073265207231108052536
264. Chen D, Zhang C, Lin J, Song X, Wang H. Screening differential circular RNA expression profiles reveal that hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res. (2018) 10:1275–83. doi: 10.2147/CMAR
265. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. (2016) 238:42–51. doi: 10.1016/j.jbiotec.2016.09.011
266. Abu N, Jamal R. Circular RNAs as promising biomarkers: a mini-review. Front Physiol. (2016) 7:213360. doi: 10.3389/fphys.2016.00355
267. Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. (2016) 6:1167.
268. Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (Review). Oncol Rep. (2015) 33:2669–74. doi: 10.3892/or.2015.3904
269. Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. (2017) 143:17–27. doi: 10.1007/s00432-016-2256-7
270. Yang X, Xiong Q, Wu Y, Li S, Ge F. Quantitative proteomics reveals the regulatory networks of circular RNA CDR1as in hepatocellular carcinoma cells. J Proteome Res. (2017) 16:3891–902. doi: 10.1021/acs.jproteome.7b00519
271. Eastham AM, Spencer H, Soncin F, Ritson S, Merry CL, Stern PL, et al. Epithelial-mesenchymal transition events during human embryonic stem cell differentiation. Cancer Res. (2007) 67:11254–62. doi: 10.1158/0008-5472.CAN-07-2253
272. Sidhu K, Kapoor NR, Pandey V, Kumar V. The “Macro” World of microRNAs in hepatocellular carcinoma. Front Oncol. (2015) 5:68. doi: 10.3389/fonc.2015.00068
273. Ashrafizadeh M, Mirzaei S, Hashemi F, Zarrabi A, Zabolian A, Saleki H, et al. New insight towards development of paclitaxel and docetaxel resistance in cancer cells: EMT as a novel molecular mechanism and therapeutic possibilities. Biomed Pharmacother. (2021) 141:111824. doi: 10.1016/j.biopha.2021.111824
274. Ashrafizadeh M, Zarrabi A, Hushmandi K, Kalantari M, Mohammadinejad R, Javaheri T, et al. Association of the epithelial-mesenchymal transition (EMT) with cisplatin resistance. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21114002
275. Meng J, Chen S, Han JX, Qian B, Wang XR, Zhong WL, et al. Twist1 regulates vimentin through cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. (2018) 78:4150–62. doi: 10.1158/0008-5472.CAN-17-3009
276. Marlar S, Jensen HH, Login FH, Nejsum LN. Aquaporin-3 in cancer. Int J Mol Sci. (2017) 18. doi: 10.3390/ijms18102106
277. Guo X, Sun T, Yang M, Li Z, Li Z, Gao Y. Prognostic value of combined aquaporin 3 and aquaporin 5 overexpression in hepatocellular carcinoma. BioMed Res Int. (2013) 2013:206525. doi: 10.1155/2013/206525
278. Chen XF, Li CF, Lü L, Mei ZC. Expression and clinical significance of aquaglyceroporins in human hepatocellular carcinoma. Mol Med Rep. (2016) 13:5283–9. doi: 10.3892/mmr.2016.5184
279. Chen G, Shi Y, Liu M, Sun . circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. (2018) 9:175. doi: 10.1038/s41419-017-0204-3
280. Jiang W, Wen D, Gong L, Wang Y, Liu Z, Yin F. Circular RNA hsa_circ_0000673 promotes hepatocellular carcinoma Malignance by decreasing miR-767-3p targeting SET. Biochem Biophys Res Commun. (2018) 500:211–6. doi: 10.1016/j.bbrc.2018.04.041
281. Hung MH, Wang CY, Chen YL, Chu PY, Hsiao YJ, Tai WT, et al. SET antagonist enhances the chemosensitivity of non-small cell lung cancer cells by reactivating protein phosphatase 2A. Oncotarget. (2016) 7:638–55. doi: 10.18632/oncotarget.v7i1
282. Rincón R, Cristóbal I, Zazo S, Arpí O, Menéndez S, Manso R, et al. PP2A inhibition determines poor outcome and doxorubicin resistance in early breast cancer and its activation shows promising therapeutic effects. Oncotarget. (2015) 6:4299–314. doi: 10.18632/oncotarget.v6i6
283. Hung MH, Chen YL, Chu PY, Shih CT, Yu HC, Tai WT, et al. Upregulation of the oncoprotein SET determines poor clinical outcomes in hepatocellular carcinoma and shows therapeutic potential. Oncogene. (2016) 35:4891–902. doi: 10.1038/onc.2016.21
284. Zhang X, Luo P, Jing W, Zhou H, Liang C, Tu J. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther. (2018) 11:2853–63. doi: 10.2147/OTT
285. Zhong L, Wang Y, Cheng Y, Wang W, Lu B, Zhu L, et al. Circular RNA circC3P1 suppresses hepatocellular carcinoma growth and metastasis through miR-4641/PCK1 pathway. Biochem Biophys Res Commun. (2018) 499:1044–9. doi: 10.1016/j.bbrc.2018.03.221
286. Fu L, Chen Q, Yao T, Li T, Ying S, Hu Y, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. (2017) 8:43878–88. doi: 10.18632/oncotarget.v8i27
287. Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma JZ, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. (2018) 68:1214–27. doi: 10.1016/j.jhep.2018.01.012
288. Zhang X, Zhou H, Jing W, Luo P, Qiu S, Liu X, et al. The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Dis Markers 2018. (2018), 3073467. doi: 10.1155/2018/3073467
289. Zhou L, Wang Q, Hou J, Wu X, Wang L, Chen X. Upregulation of hsa_circ_0002003 promotes hepatocellular carcinoma progression. BMC Cancer. (2023) 23:611. doi: 10.1186/s12885-023-11086-9
290. Sun N, Gong J, Zhang W, Yang X, Liu J. Sevoflurane suppresses hepatocellular carcinoma cell progression via circ_0001649/miR-19a-3p/SGTB axis. Histol Histopathol. (2023) 38:537–47.
291. Chang X, Huang Z, Zhang Z, Pan W, Song C. Matrine inhibits hepatocellular carcinoma cell Malignancy through the circ_0013290/miR-139-5p/MMP16 pathway. Histol Histopathol. (2023) 38:1179–92.
292. Chang X, Li H, Huang Z, Song C, Zhang Z, Pan W, et al. Matrine suppresses hepatocellular carcinoma tumorigenesis by modulating circ_0055976/miR-1179/lactate dehydrogenase A axis. Environ Toxicol. (2024) 39:1481–93. doi: 10.1002/tox.24041
293. Peng L, Chen J, Li M, Wang R. Circ_MBNL3 restrains hepatocellular carcinoma progression by sponging miR-873-5p to release PHF2. Biochem Genet. (2023) 61:1015–34. doi: 10.1007/s10528-022-10295-4
294. Qin X, Zhou L, Shen Y, Gu Y, Tang J, Qian J, et al. CircularRNA Hsa_circ_0093335 promotes hepatocellular carcinoma progression via sponging miR-338-5p. J Cell Mol Med. (2023) 27:4080–92. doi: 10.1111/jcmm.17991
295. Wu S, Tang T, Zhou H, Huang J, Kang X, Zhang J, et al. Hsa_circ_0119412 is a tumor promoter in hepatocellular carcinoma by inhibiting miR-526b-5p to upregulate STMN1. Cancer Biol Ther. (2023) 24:2256951. doi: 10.1080/15384047.2023.2256951
296. Chen J, Liu Z, Zhong Y, Chen H, Xie L. Circ_0124208 promotes the progression of hepatocellular carcinoma by regulating the miR-338-3p/LAMC1 axis. Mol Biotechnol. (2023) 65:1750–63. doi: 10.1007/s12033-023-00686-2
297. Qin C, Liu S, Chen W, Xue D, Guo T, Wu B. HuR-induced circ_0082319 contributes to hepatocellular carcinoma by elevating PTK2 through miR-505-3p. Naunyn Schmiedebergs Arch Pharmacol. (2023). doi: 10.1007/s00210-023-02793-y
298. Zhang Y, Wang Y. Circular RNAs in hepatocellular carcinoma: Emerging functions to clinical significances. Front Oncol. (2021) 11:667428. doi: 10.3389/fonc.2021.667428
299. Cui C, Yang J, Li X, Liu D, Fu L, Wang X. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer. (2020) 19:1–16. doi: 10.1186/s12943-020-01180-y
300. Li J, Qin X, Wu R, Wan L, Zhang L, Liu R. Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miRoughncenlarC1-11 axis. J Cell Mol Med. (2020) 24:5152–61. doi: 10.1111/jcmm.15162
301. Chen H, Liu S, Li M, Huang P, Li X. circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance through Wnt/β-catenin pathway in hepatocellular carcinoma. OncoTargets Ther. (2019), 9539–49. doi: 10.2147/OTT
302. Luo Y, Fu Y, Huang R, Gao M, Liu F, Gui R, et al. CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR-103 and promotes oxidored-nitro domain-containing protein 1 expression. Cell Death Discovery. (2019) 5:121. doi: 10.1038/s41420-019-0202-6
303. Yang W, Liu Y, Gao R, Xiu Z, Sun T. Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721 cell proliferation, vasculogenic mimicry, and radioresistance. Cell signalling. (2019) 60:122–35. doi: 10.1016/j.cellsig.2019.04.011
304. Wu M-y, Tang Y-p, Liu J-j, Liang R, Luo X-l. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J Cancer. (2020) 11:2993. doi: 10.7150/jca.39854
305. Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N 6-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. (2020) 19:1–16. doi: 10.1186/s12943-020-01281-8
306. Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal transduction targeted Ther. (2020) 5:298. doi: 10.1038/s41392-020-00375-5
307. Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. (2020) 72:906–22. doi: 10.1002/hep.31068
308. Zhang T, Jing B, Bai Y, Zhang Y, Yu H. Circular RNA circTMEM45A acts as the sponge of MicroRNA-665 to promote hepatocellular carcinoma progression. Mol Therapy-Nucleic Acids. (2020) 22:285–97. doi: 10.1016/j.omtn.2020.08.011
309. Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A noncoding regulatory RNAs network driven by Circenkor acts specifically in the early stages hepatocellular carcinoma. Hepatology. (2020) 71:130–47. doi: 10.1002/hep.30795
310. Wan B, Hu H, Wang R, Liu W, Chen D. Therapeutic potential of circular RNAs in osteosarcoma. Front Oncol. (2020) 10:370. doi: 10.3389/fonc.2020.00370
311. Long F, Lin Z, Li L, Ma M, Lu Z, Jing L, et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer. (2021) 20:26. doi: 10.1186/s12943-021-01318-6
312. Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA,. to suppress hepatocellular carcinoma progression. Hepatology. (2017) 66:1151–64. doi: 10.1002/hep.29270
313. Liu H, Lan T, Li H, Xu L, Chen X, Liao H, et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics. (2021) 11:1396. doi: 10.7150/thno.53227
314. Cortés-López M, Miura P. Focus: epigenetics: emerging functions of circular RNAs. Yale J Biol Med. (2016) 89:527.
315. Fan X, Weng X, Zhao Y, Chen W, Gan T, Xu D. Circular RNAs in cardiovascular disease: an overview. BioMed Res Int. (2017) 2017. doi: 10.1155/2017/5135781
316. Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, et al. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine. (2016) 95:e3811. doi: 10.1097/MD.0000000000003811
317. Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PloS One. (2016) 11:e0158347. doi: 10.1371/journal.pone.0158347
318. Huang X-Y, Huang Z-L, Xu Y-H, Zheng Q, Chen Z, Song W, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. (2017) 7:5428. doi: 10.1038/s41598-017-05432-8
319. Huang X-Y, Huang Z-L, Xu Y-H, Zheng Q, Chen Z, Song W, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/β-catenin axis. Biochem Biophys Res Commun. (2018) 497:626–32. doi: 10.1016/j.bbrc.2018.02.119
320. Yao R, Zou H, Liao W. Prospect of circular RNA in hepatocellular carcinoma: a novel potential biomarker and therapeutic target. Front Oncol. (2018) 8:332. doi: 10.3389/fonc.2018.00332
321. Sun J-Y, Zhang X-Y, Cao Y-Z, Zhou X, Gu J, Mu X-X. Diagnostic and prognostic value of circular RNAs in hepatocellular carcinoma. J Cell Mol Med. (2020) 24:5438–45. doi: 10.1111/jcmm.15258
322. Juárez-Hernández E, Motola-Kuba D, Chávez-Tapia NC, Uribe M, Barbero Becerra V. Biomarkers in hepatocellular carcinoma: an overview. Expert Rev Gastroenterol Hepatol. (2017) 11:549–58. doi: 10.1080/17474124.2017.1311785
323. Zhu K, Zhan H, Peng Y, Yang L, Gao Q, Jia H, et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. (2020) 41:296–302. doi: 10.1093/carcin/bgz154
324. Qiao GL, Chen L, Jiang WH, Yang C, Yang CM, Song LN, et al. Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. Onco Targets Ther. (2019) 12:5849–60. doi: 10.2147/OTT
325. Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. (2020) 71:130–47. doi: 10.1002/hep.30795
326. Jiang Z, Shen L, Wang S, Wu S, Hu Y, Guo J, et al. Hsa_circ_0028502 and hsa_circ_0076251 are potential novel biomarkers for hepatocellular carcinoma. Cancer Med. (2019) 8:7278–87.
327. Matboli M, Shafei AE, Ali MA, Ashry AM, Kamal KM, Agag MA, et al. circRNAs (hsa_circ_00156, hsa_circ _000224, and hsa_circ _000520) are novel potential biomarkers in hepatocellular carcinoma. J Cell Biochem. (2019) 120:7711–24.
328. Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. (2017) 11:422–37.
329. Yu J, Ding WB, Wang MC, Guo XG, Xu J, Xu QG, et al. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int J Cancer. (2020) 146:1754–63. doi: 10.1002/ijc.32647
330. Aishanjiang K, Wei X-d, Fu Y, Lin X, Ma Y, Le J, et al. Circular RNAs and hepatocellular carcinoma: new epigenetic players with diagnostic and prognostic roles. Front Oncol. (2021) 11:653717. doi: 10.3389/fonc.2021.653717
331. Zhou Y, Mao X, Peng R, Bai . CircRNAs in hepatocellular carcinoma: characteristic, functions and clinical significance. Int J Med Sci. (2022) 19:2033. doi: 10.7150/ijms.74713
332. Xiong D, He R, Dang Y, Wu H, Feng Z, Chen G. The latest overview of circRNA in the progression, diagnosis, prognosis, treatment, and drug resistance of hepatocellular carcinoma. Front Oncol. (2021) 10:608257. doi: 10.3389/fonc.2020.608257
333. Rao G, Peng X, Fu X. Circular RNAs in hepatocellular carcinoma: biogenesis, function, and pathology. Front Genet. (2023) 14:1106665. doi: 10.3389/fgene.2023.1106665
334. Huang Z, Xia H, Liu S, Zhao X, He R, Wang Z, et al. The mechanism and clinical significance of circular RNAs in hepatocellular carcinoma. Front Oncol. (2021) 11:714665. doi: 10.3389/fonc.2021.714665
335. Niu Z-S, Wang W-H. Circular RNAs in hepatocellular carcinoma: Recent advances. World J Gastrointestinal Oncol. (2022) 14:1067. doi: 10.4251/wjgo.v14.i6.1067
336. Hu M, Li X, Jiang Z, Xia Q, Hu Y, Guo J, et al. Exosomes and circular RNAs: promising partners in hepatocellular carcinoma from bench to bedside. Discover Oncol. (2023) 14:60. doi: 10.1007/s12672-023-00672-9
337. Li Z-d, Li Y-l, Lu J, Liang S, Zhang C, Zeng L-h. Recent research progress of circular RNAs in hepatocellular carcinoma. Front Oncol. (2024) 13:1192386. doi: 10.3389/fonc.2023.1192386
338. Wu M, Sun T, Xing L. Circ_0004913 inhibits cell growth, metastasis, and glycolysis by absorbing miR-184 to regulate HAMP in hepatocellular carcinoma. Cancer Biother Radiopharm. (2023) 38:708–19. doi: 10.1089/cbr.2020.3779
339. Zhang CP, Huang XY. Circular RNA circ_KIAA1429 accelerates hepatocellular carcinoma progression via the miR-133a-3p/high mobility group AT-hook 2 (HMGA2) axis in an m6A-dependent manner. Hum Cell. (2023) 36:1741–54. doi: 10.1007/s13577-023-00933-3
340. Wang XB, Luo T, Lu SL, Lu HZ, Zhao TY, Jiang ZJ, et al. Circular RNA hsa_circ_0005218 promotes the early recurrence of hepatocellular carcinoma by targeting the miR-31-5p/CDK1 pathway. Heliyon. (2023) 9:e14816. doi: 10.1016/j.heliyon.2023.e14816
341. Yang M, Yu T, Han L. Hsa_circ_0010882 facilitates hepatocellular carcinoma progression by modulating M1/M2 macrophage polarization. J Viral Hepat. (2024) 31:189–96. doi: 10.1111/jvh.13917
342. Gao P, Yang Y, Li X, Zhao Q, Liu Y, Dong C, et al. Circular RNA hsa_circ_0098181 inhibits metastasis in hepatocellular carcinoma by activating the Hippo signaling pathway via interaction with eEF2. Ann Hepatol. (2023) 28:101124. doi: 10.1016/j.aohep.2023.101124
343. Zhang T, Sun Q, Shen C, Qian Y. Circular RNA circ_0003028 regulates cell development through modulating miR-498/ornithine decarboxylase 1 axis in hepatocellular carcinoma. Anticancer Drugs. (2023) 34:507–18. doi: 10.1097/CAD.0000000000001457
344. Bai L, Yu Z, Li Q, Hu X, Liu X, Li F, et al. Circ_0073228 serves as a competitive endogenous ribonucleic acid to facilitate proliferation and inhibit apoptosis of hepatocellular carcinoma cells by the miR-139-5p/deoxyribonuclease II axis. J Gene Med. (2023) 25:e3507. doi: 10.1002/jgm.3507
345. Ai J, Zhang W, Deng W, Yan L, Zhang L, Huang Z, et al. A hsa_circ_001726 axis regulated by E2F6 contributes to metastasis of hepatocellular carcinoma. BMC Cancer. (2024) 24:14. doi: 10.1186/s12885-023-11703-7
346. Pu J, Wang J, Li W, Lu Y, Wu X, Long X, et al. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J Cell Mol Med. (2024) 28:e15010. doi: 10.1111/jcmm.15010
347. Wang K, Lu Q, Luo Y, Yu G, Wang Z, Lin J, et al. Circ_MAPK9 promotes STAT3 and LDHA expression by silencing miR-642b-3p and affects the progression of hepatocellular carcinoma. Biol Direct. (2024) 19:4. doi: 10.1186/s13062-023-00442-1
348. Zhang S, Ma J, Yu T, Song Z, Lau WY, Zha Y. circ_HMGCS1 modulates hepatocellular carcinoma chemoresistance via miR-338-5p/IL-7 pathway. J Cell Mol Med. (2024) 28:e18137. doi: 10.1111/jcmm.18137
349. Ni L, Gao Q, Zhao Q, Dai K, Jin M, Fu C, et al. Circ-EIF3I promotes hepatocellular carcinoma progression through modulating miR-361-3p/DUSP2 axis. DNA Cell Biol. (2024). doi: 10.1089/dna.2023.0400
350. Yin J, Wang M, Chen J, Li H, Zhuo J, Lu B, et al. CircZCCHC2 (hsa_circ_0000854) promotes hepatocellular carcinoma progression through modulating miR-936/BTBD7 axis and activating Rho/ROCK2 pathway. Noncoding RNA Res. (2024) 9:437–46. doi: 10.1016/j.ncrna.2023.12.004
351. Zhu Y, Xiao B, Liu M, Chen M, Xia N, Guo H, et al. N6-methyladenosine-modified oncofetal lncRNA MIR4435-2HG contributed to stemness features of hepatocellular carcinoma cells by regulating rRNA 2’-O methylation. Cell Mol Biol Lett. (2023) 28:89. doi: 10.1186/s11658-023-00493-2
352. Xu G, Ban K, Mu H, Wang B. Human umbilical cord mesenchymal stem cells-derived exosomal lncRNA FAM99B represses hepatocellular carcinoma cell Malignancy. Mol Biotechnol. (2023). doi: 10.1007/s12033-023-00795-y
353. Ye D, Ma J, Yu T, Ran F, Zha Y. LncRNA FAM13A-AS1, transcriptionally regulated by PHOX2B, modulates hepatocellular carcinoma chemoresistance via stabilizing PPARγ. Gene. (2023) 877:147570. doi: 10.1016/j.gene.2023.147570
354. Xu Y, Luan G, Li Z, Liu Z, Qin G, Chu Y. Tumour-derived exosomal lncRNA SNHG16 induces telocytes to promote metastasis of hepatocellular carcinoma via the miR-942-3p/MMP9 axis. Cell Oncol (Dordr). (2023) 46:251–64. doi: 10.1007/s13402-023-00782-0
355. Zeng Z, Wang J, Xu F, Hu P, Hu Y, Zhuo W, et al. The m6A modification-mediated positive feedback between glycolytic lncRNA SLC2A1-DT and c-Myc promotes tumorigenesis of hepatocellular carcinoma. Int J Biol Sci. (2024) 20:1744–62. doi: 10.7150/ijbs.86658
356. Huang Y, Li D, Lu L, Song D, Li P. LncRNA HEIH modulates the proliferation, migration, and invasion of hepatocellular carcinoma cells by regulating the miR-193a-5p/CDK8 axis. Transl Cancer Res. (2024) 13:423–36. doi: 10.21037/tcr
357. Liu C, Xu K, Liu J, He C, Liu P, Fu Q, et al. LncRNA RP11-620J15.3 promotes HCC cell proliferation and metastasis by targeting miR-326/GPI to enhance glycolysis. Biol Direct. (2023) 18:15. doi: 10.1186/s13062-023-00370-0
358. Zhu X, Yu H, Li H, Zhang W, Sun L, Dou T, et al. lncRNA SNHG1 promotes the progression of hepatocellular carcinoma by regulating the miR-7-5p/IGF2BP2 axis. Heliyon. (2024) 10:e27631. doi: 10.1016/j.heliyon.2024.e27631
359. Xing K, Bian X, Shi D, Dong S, Zhou H, Xiao S, et al. miR-612 enhances RSL3-induced ferroptosis of hepatocellular carcinoma cells via mevalonate pathway. J Hepatocell Carcinoma. (2023) 10:2173–85. doi: 10.2147/JHC.S433332
360. Wang D, Yang J. MiR-375 attenuates sorafenib resistance of hepatocellular carcinoma cells by inhibiting cell autophagy. Acta Biochim Pol. (2023) 70:239–46. doi: 10.18388/abp.2020_5657
361. Dong S, Wang W, Liao Z, Fan Y, Wang Q, Zhang L. MYC-activated LINC00607 promotes hepatocellular carcinoma progression by regulating the miR-584-3p/ROCK1 axis. J Gene Med. (2023) 25:e3477. doi: 10.1002/jgm.3477
362. Lu F, Gao J, Luo Y, Jin WL, Wang H, Li CX, et al. CircCPSF6 promotes hepatocellular carcinoma cancer progression by regulating MAP4K4 through sponging miR-145-5p. Mol Cell Probes. (2023) 71:101920. doi: 10.1016/j.mcp.2023.101920
363. Hu J, Liu WF, Zhang XY, Shi GM, Yang XR, Zhou KQ, et al. Synthetic miR-26a mimics delivered by tumor exosomes repress hepatocellular carcinoma through downregulating lymphoid enhancer factor 1. Hepatol Int. (2023) 17:1265–78. doi: 10.1007/s12072-023-10527-8
364. Chen B, Li Q, Li Y, Li Q, Lai H, Huang S, et al. circTMEM181 upregulates ARHGAP29 to inhibit hepatocellular carcinoma migration and invasion by sponging miR-519a-5p. Hepatol Res. (2023) 53:334–43. doi: 10.1111/hepr.13870
365. Feng Z, Wu J. hsa_circ_0129047 upregulates LYVE1 to inhibit hepatocellular carcinoma progression by sponging miR-492. Dis Markers 2023. (2023), 6978234. doi: 10.1155/2023/6978234
366. Zheng C, Liang J, Yu S, Xu H, Dai L, Xu D. Circ-SNX27 sponging miR-375/RPN1 axis contributes to hepatocellular carcinoma progression. Korean J Physiol Pharmacol. (2023) 27:333–44. doi: 10.4196/kjpp.2023.27.4.333
367. Cheng Y. Hsa-circ-0000098 promotes the progression of hepatocellular carcinoma by regulation of miR-136-5p/MMP2 axis. Adv Clin Exp Med. (2023) 32:689–700. doi: 10.17219/acem/157063
368. Vasuri F, Visani M, Acquaviva G, Brand T, Fiorentino M, Pession A, et al. Role of microRNAs in the main molecular pathways of hepatocellular carcinoma. World J Gastroenterol. (2018) 24:2647–60. doi: 10.3748/wjg.v24.i25.2647
369. Lv E, Sheng J, Yu C, Rao D, Huang W. LncRNA influence sequential steps of hepatocellular carcinoma metastasis. Biomed Pharmacother. (2021) 136:111224. doi: 10.1016/j.biopha.2021.111224
370. Swamy SG, Kameshwar VH, Shubha PB, Looi CY, Shanmugam MK, Arfuso F, et al. Targeting multiple oncogenic pathways for the treatment of hepatocellular carcinoma. Targeted Oncol. (2017) 12:1–10. doi: 10.1007/s11523-016-0452-7
371. Fu Y, Liu S, Zeng S, Shen H. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J Exp Clin Cancer Res. (2019) 38:1–21. doi: 10.1186/s13046-019-1396-4
372. Qiu LP, Wu YH, Yu XF, Tang Q, Chen L, Chen KP, et al. The emerging role of circular RNAs in hepatocellular carcinoma. J Cancer. (2018) 9:1548–59. doi: 10.7150/jca.24566
Keywords: hepatocellular carcinoma, miRNA sponge, diagnosis and prognosis, therapy resistance, epigenetic factors
Citation: Tian Y, Zhang M, Liu L-x, Wang Z-c, Liu B, Huang Y, Wang X, Ling Y-z, Wang F, Feng X and Tu Y (2024) Exploring non-coding RNA mechanisms in hepatocellular carcinoma: implications for therapy and prognosis. Front. Immunol. 15:1400744. doi: 10.3389/fimmu.2024.1400744
Received: 14 March 2024; Accepted: 03 April 2024;
Published: 10 May 2024.
Edited by:
Fan Feng, The 302th Hospital of PLA, ChinaReviewed by:
Jin Zhang, University of Mississippi Medical Center, United StatesZhongjie Shi, Wayne State University, United States
Copyright © 2024 Tian, Zhang, Liu, Wang, Liu, Huang, Wang, Ling, Wang, Feng and Tu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanyang Tu, dHVmbW11QDE4OC5jb20=; Xiaoqiang Feng, ZmVuZ3hxOTNAMTYzLmNvbQ==; Furong Wang, bGR5eV93YW5nZnJAbHp1LmVkdS5jb20=
†These authors have contributed equally to this work
 Yu Tian
Yu Tian Meng Zhang3†
Meng Zhang3† Li-xia Liu
Li-xia Liu Bin Liu
Bin Liu Yun-zhi Ling
Yun-zhi Ling Xiaoqiang Feng
Xiaoqiang Feng Yanyang Tu
Yanyang Tu