- 1Beijing Institute of Hepatology, Beijing Youan Hospital, Capital Medical University, Beijing, China
- 2Integrated Chinese and Western Medicine Center, Beijing Youan Hospital, Capital Medical University, Beijing, China
- 3Beijing Youan Hospital, Capital Medical University, Beijing, China
Exosomes play a crucial role in various biological processes, such as human development, immune responses, and disease occurrence. The membrane proteins on exosomes are pivotal factors for their biological functionality. Currently, numerous membrane proteins have been identified on exosome membranes, participating in intercellular communication, mediating target cell recognition, and regulating immune processes. Furthermore, membrane proteins from exosomes derived from cancer cells can serve as relevant biomarkers for early cancer diagnosis. This article provides a comprehensive review of the composition of exosome membrane proteins and their diverse functions in the organism’s biological processes. Through in-depth exploration of exosome membrane proteins, it is expected to offer essential foundations for the future development of novel biomedical diagnostics and therapies.
1 Introduction
In recent years, as our understanding of exosomes has advanced, the biological roles of exosome membrane proteins in cells and organisms have garnered increasing attention. As a primary constituent of exosomes, exosome membrane proteins not only play a role in the formation and release of exosomes (1–6) but also exhibit diverse functions, including targeting or adhering to receptor cells, anti-apoptotic activities, membrane fusion, signal transduction, metabolism, and structural dynamics (7). Therefore, comprehending the composition and functions of exosome membrane proteins is crucial for understanding the biological characteristics and mechanisms of action of exosomes.
The generation of exosomes involves the inward budding of the plasma membrane and the formation of intraluminal vesicles (ILVs) within multivesicular bodies (MVBs) in the cell. ILVs are eventually secreted as exosomes by the fusion of MVBs with the plasma membrane and released via exocytosis (8–12). The initial inward budding of the plasma membrane forms a cup-shaped structure containing cell surface and soluble proteins related to the extracellular environment. Subsequently, budding of the inner membrane forms ILVs within endosomes, which contain specific proteins, lipids, nucleic acids, and other molecules (13–17). The biogenesis of exosomes is driven by multiple protein-regulated mechanisms, including ESCRT protein complexes, Rab GTPases, Tetraspanins, etc (18). Finally, mature MVBs fuse with the plasma membrane, releasing ILVs as exosomes through exocytosis into the extracellular environment (1, 2). These released exosomes can facilitate intercellular signaling, modulate immune responses, and promote cell-cell communication (18, 19).
In this review, we systematically summarize the composition of exosome membrane proteins and explore their potential applications in mediating target cell recognition, immune regulation, and disease control.
2 Composition and classification of exosome membrane proteins
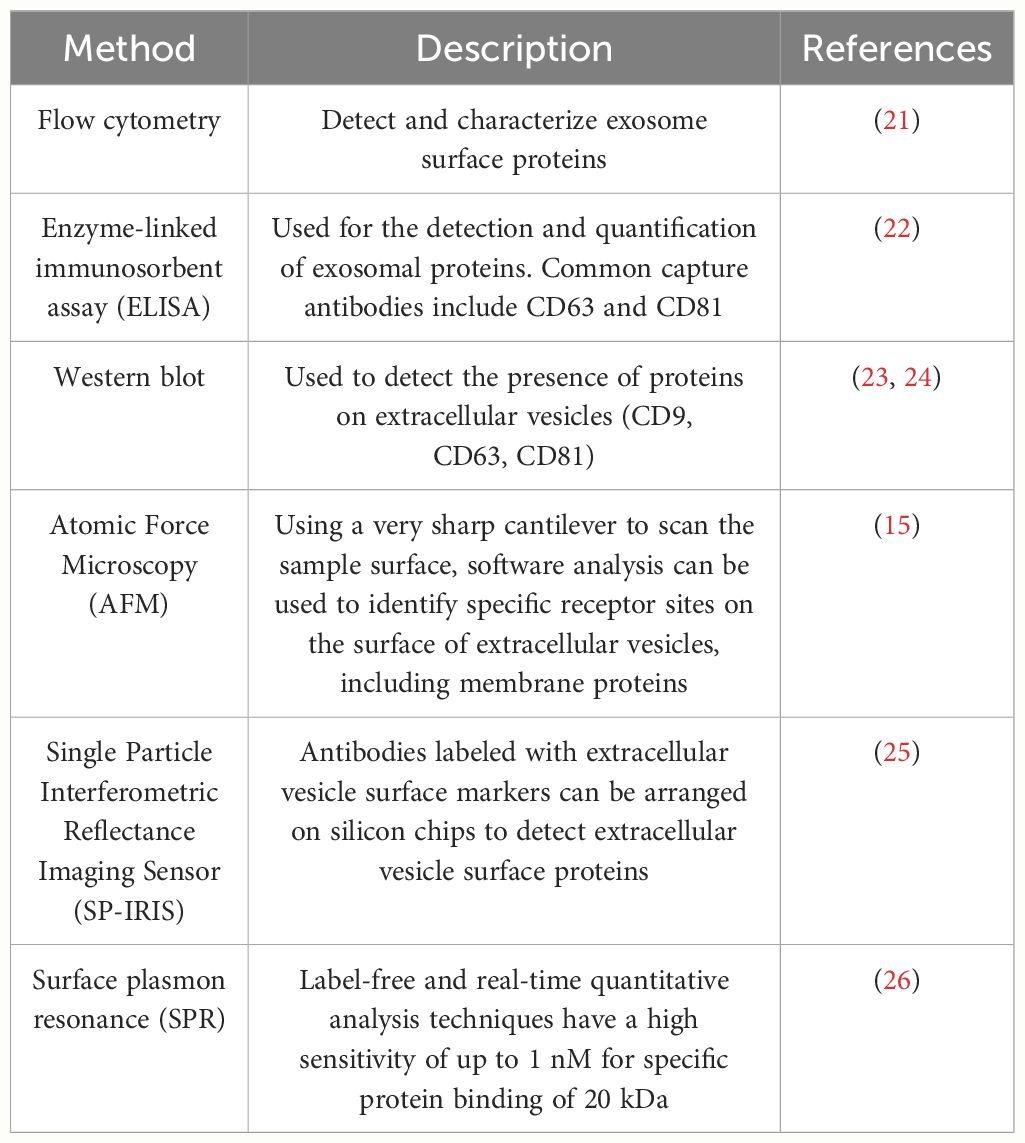
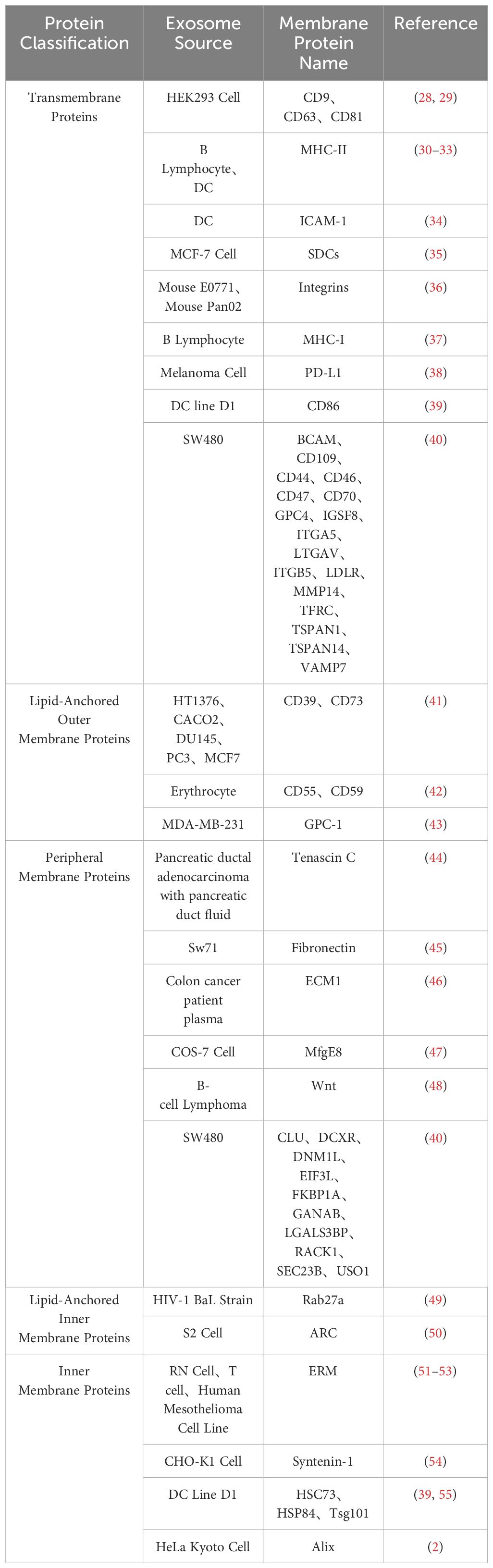
Exosome membrane proteins are classified based on membrane localization into transmembrane proteins, lipid-anchored membrane proteins, peripheral-associated membrane proteins, and inner-associated membrane proteins. According to the current exosome content database, Exocarta (http://www.exocarta.org), 9769 exosome proteins have been identified in exosomes from various cell types and organisms. With the continuous development of modern technology, the detection methods for extracellular vesicle membrane proteins are also constantly being updated. Currently used methods include Western blot, ELISA, Atomic Force Microscopy (AFM), etc. (20). Table 1 summarizes the common methods for detecting extracellular vesicle membrane proteins. Recently, Xiaoni Fang et al. (27), using the integrated GF/PMO platform, identified a total of 334 exosome proteins, including 111 membrane proteins. The GF/PMO platform is an innovative approach that integrates two nanomaterials with different surface properties: hydrophilic macroporous graphene foam (GF) and amphiphilic periodic mesoporous organosilica (PMO). This platform is used for the efficient separation of exosomes from human serum and effective protein analysis, aiding in the identification of more exosome-based disease biomarkers. This method of efficient and specific separation and analysis of exosome proteins holds significant application prospects in biomedical research. Table 2 summarizes some important and noteworthy proteins distributed within the inner membrane, outer membrane, and transmembrane region of exosomes. The arrangement of exosome membrane proteins is illustrated in Figure 1.
A specific class of membrane proteins serves as exosome-specific markers, such as the tetraspanins CD9, CD63, and CD81 (2, 4, 28, 30, 56–63). These proteins have been demonstrated to regulate the transport and function of associated proteins through membrane compartmentalization (64). Lipid-anchored outer membrane proteins, including CD39, CD73, GPC-1, CD55, and CD59, with enzymatic activity, notably CD39 and CD73, have been shown to promote angiogenesis through adenosine A2B receptor signaling (65). Peripheral membrane proteins such as Tenascin C, Fibronectin, ECM1, MfgE8, and Wnt play crucial roles in the functional processes of exosomes. For example, exosomes derived from embryonic stem cells (ESCs) carrying Fibronectin contribute to maintaining their stem cell characteristics (66). Lipid-anchored inner membrane protein Rab27a regulates exosome formation and release (67). Inner membrane proteins Tsg101 and Alix serve as exosome markers and are involved in the biogenesis of multivesicular bodies (MVB) (68). The arrangement of exosome membrane proteins is illustrated in Figure 1.
Exosome membrane proteins vary among different cell sources; for instance, exosomes from antigen-presenting cells (APCs) are rich in transmembrane proteins such as MHC-I, MHC-II, and ICAM-1 (68, 69). The diversity of these membrane proteins determines the versatility of exosome functions (70). Therefore, a focused discussion on the composition and clinical applications of exosome membrane proteins is crucial for guiding future research directions.
3 Roles and functions of exosome membrane proteins
3.1 Diagnostic role of exosome membrane proteins in diseases
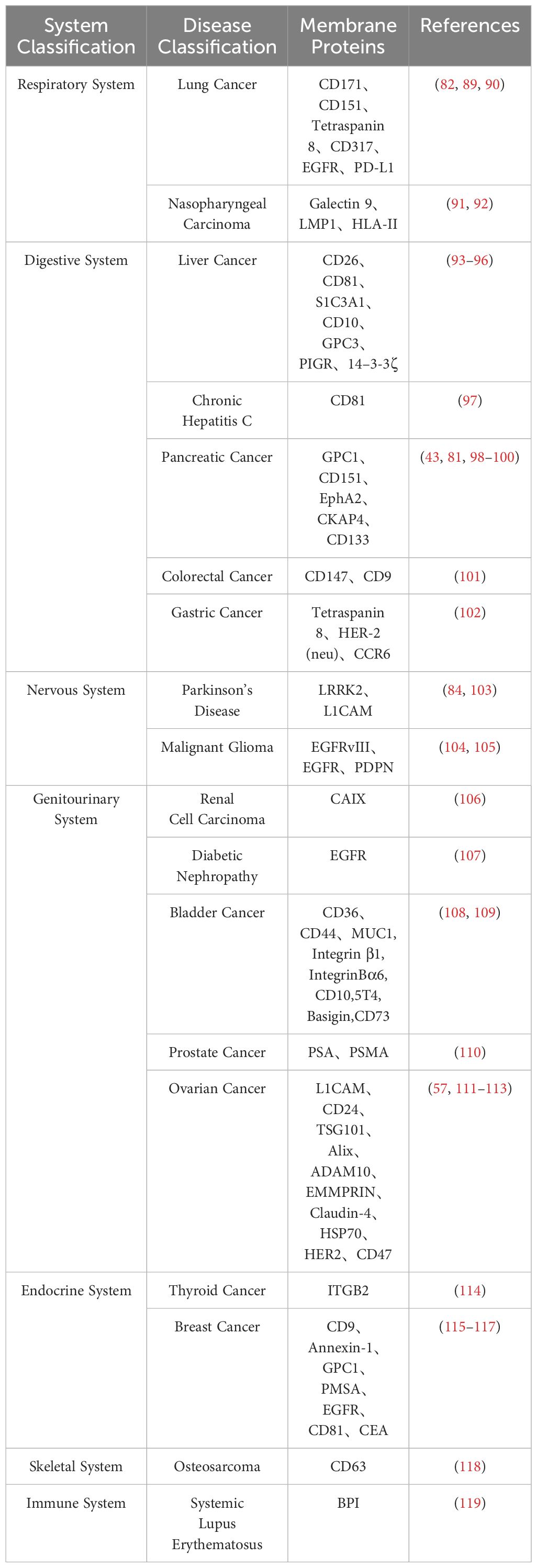
Currently, a substantial body of literature indicates that the molecular components of exosomes, particularly exosome proteins, serve as promising novel markers for the clinical diagnosis of various diseases (71–84). Their application prospects are considerable due to unique advantages: high sensitivity (85), high specificity (43), and high stability (85), making them a preferred option for liquid biopsy. The presence of exosomes can be detected in various bodily fluids (86).
In the current stage, many potential targets for cancer treatment are tumor-specific biological markers. Since exosomes derived from cancerous sources carry similar markers on their membrane surfaces, researching exosome membrane protein biomarkers is crucial for the development of targeted cancer therapies (87, 88). The primary component of exosome proteins, membrane proteins (27), offers a reliable choice for developing new disease diagnostic biomarkers. It is gradually becoming a focal point in exosome research. Table 3 summarizes exosome membrane proteins from different disease sources.
Mariantonia Logozzi and colleagues designed an internal sandwich ELISA (Exotest), revealing a significant increase in CD63 and Caveolin-1 in plasma-derived exosomes from melanoma patients. They described a novel non-invasive detection method for assessing the expression of exosome-specific membrane proteins in melanoma patients’ plasma, providing a potential diagnostic tool (120). In 2013, Yusuke Yoshioka and colleagues conducted a comparative analysis of exosome protein markers in different human cancer types. They found elevated levels of CD63 in exosomes derived from malignant cancer cells compared to those from non-cancerous cells, further supporting CD63 as a protein marker for cancer (29, 121). Bingqian Lin et al. developed a specific dual-ligand recognition system based on the exosome membrane, combined with droplet digital PCR (ddPCR) (TRACER), for quantifying tumor-derived exosome PD-L1 (Exo-PD-L1). The tumor-derived Exo-PD-L1 levels detected by TRACER could distinguish cancer patients from healthy blood donors (122). Research indicates that the lipid-anchored outer membrane protein GPC-1 is significantly overexpressed in plasma-derived exosomes from pancreatic ductal adenocarcinoma (PDAC) patients compared to healthy controls, confirming the potential utility of GPC-1 for early PDAC diagnosis (123).
Compared to biomarkers detected directly in conventional specimens (such as serum or urine), exosome biomarkers offer higher specificity and sensitivity due to their superior stability (124). Exosome biomarkers, especially those from easily obtainable biological fluids like saliva, show great potential for clinical applications. In conclusion, exosome biomarkers are still in the early stages of discovery and development, and their potential value in clinical diagnostics requires further exploration. Therefore, if certain membrane proteins are specifically expressed by a particular tumor (125), their expression on circulating exosomes can be utilized as an early diagnostic signal for cancer. The diagnostic potential of exosome membrane proteins in different diseases is depicted in Figure 2.
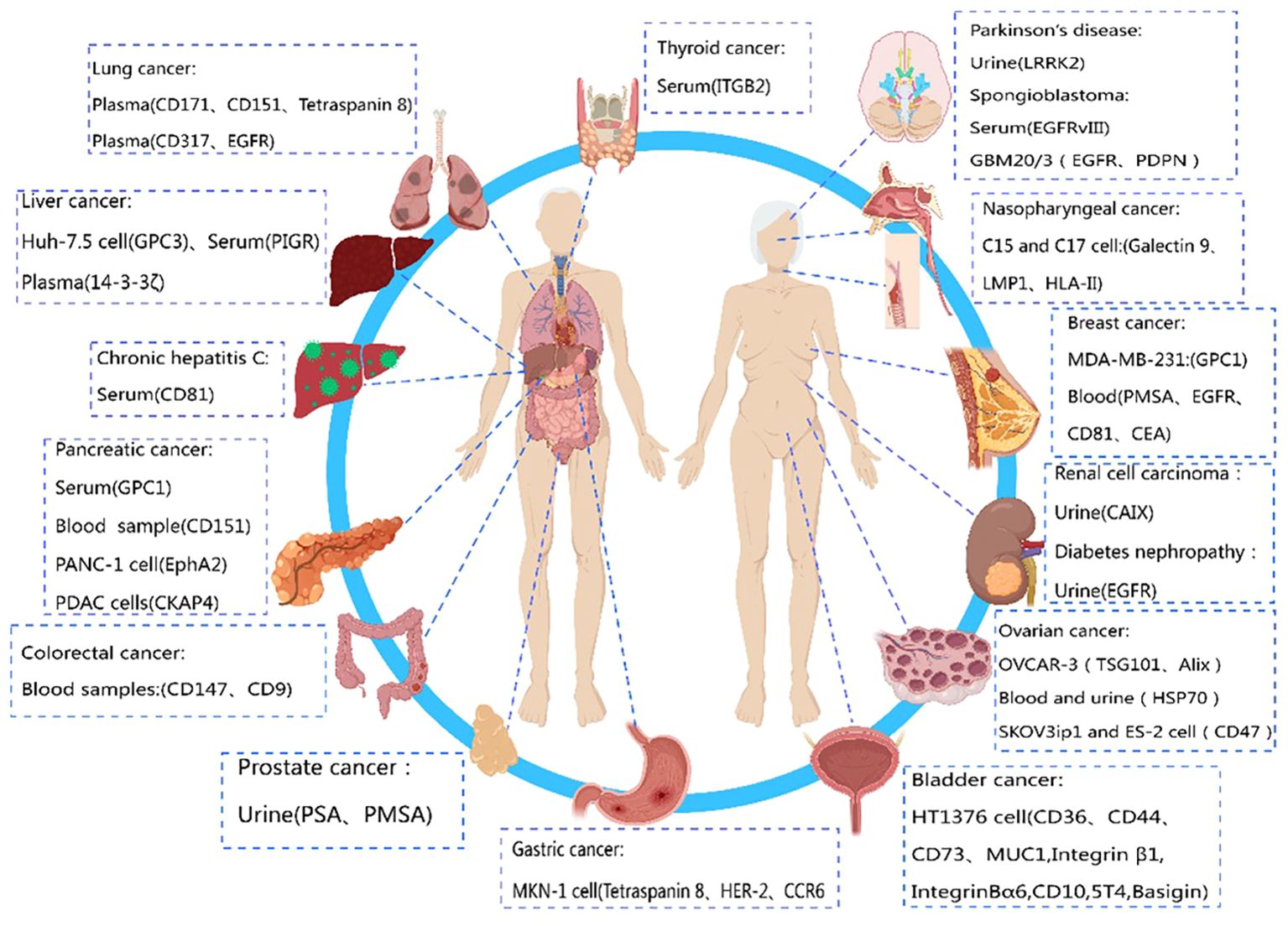
Figure 2 Diagnostic role of exosome membrane proteins in diseases. This figure was created using MedPeer.
3.2 Remote regulatory role of exosome membrane proteins
Current data suggest that exosome membrane proteins can exert regulatory effects on recipient cells (17, 126–132). They identify target cells by binding to surface proteins on recipient cells (133), leading to changes in the recipient cells. Kun Zhao et al. (134) found that exosome tetraspanin protein Tspan8 and CD151 derived from tumor cells can activate the PI3K/Akt signalling pathway by binding to GPCR and RTK proteins on recipient cells, promoting tumor angiogenesis. Similarly, Shi Du et al. demonstrated that tumor cell-derived exosomes carrying tyrosine kinase 2 (TIE2) with an immunoglobulin and epidermal growth factor homology domain deliver TIE2 protein to macrophages. Macrophages carrying TIE2 (TEMs) interact with angiopoietin-2 (ANG2), ultimately promoting cervical cancer angiogenesis (135).
Furthermore, a study detected exosomes in the serum of osteosarcoma patients with lung metastasis and those without lung metastasis. The results revealed a significant expression of PD-L1 and N-cadherin in exosomes from serum of osteosarcoma patients with lung metastasis. This study suggests that exosomes derived from osteosarcoma and carrying PD-L1 and N-cadherin reach the lungs through the circulatory system. The osteosarcoma cells at the lung metastatic site further internalize these exosomes, ultimately promoting the migration and progression of metastatic tumors (136). The regulatory mechanism involves two steps. Firstly, osteosarcoma cells stimulate epithelial cells to transition from an adhesive epithelial state to an active mesenchymal state through the epithelial-mesenchymal transition (EMT) mechanism. This mechanism facilitates the spread of cancer cells at metastatic sites. Secondly, metastatic osteosarcoma cells internalize exosomes derived from primary osteosarcoma, which carry PD-L1 and N-cadherin, promoting lung metastasis. A comprehensive understanding of the complex regulatory mechanisms of exosome membrane proteins in diseases can deepen our understanding of disease development and provide stronger support for the development of innovative treatment methods.
3.3 The role of exosomal membrane proteins in epithelial-mesenchymal transition
EMT is a cellular process that drives the differentiation of epithelial cells into mesenchymal cells. Through specific programs, epithelial cells acquire mesenchymal characteristics, including reduced cell adhesion, loss of cell polarity, and increased cell migration (137–140). Notably, cancer cells that have undergone EMT not only gain distinct molecular characteristics but also develop resistance to chemotherapy and immunotherapy (141–143). Proteins in exosomes significantly influence chemotherapy resistance. Based on their mechanisms of inducing resistance, exosomal proteins are mainly classified into enzymes, transcription factors, membrane proteins, and secreted proteins (144). Laura J. Vellade et al. (145) demonstrated that exosomes carrying PDGFRβ interact with receptors on melanoma cells, leading to dose-dependent activation of the PI3K/AKT signaling pathway and bypassing BRAF inhibition in the MAPK pathway, ultimately resulting in reduced drug sensitivity in melanoma cells.
Reports indicate that tumor-derived exosomes (TEX) carry proteins that promote epithelial-mesenchymal transition, including EMT inducers such as TGF-β, HIF1α, β-catenin, Caveolin-1, and Vimentin. These proteins can enhance the invasion and migration capabilities of recipient cells and contribute to stromal remodeling and the formation of the pre-metastatic niche (146, 147). Research by Mohammad A. Rahman et al. (147) demonstrated that exosomes derived from lung cancer activate the migration process of human bronchial epithelial cells (HBECs) by enhancing their metastatic properties. TEX were isolated from the supernatants of non-metastatic and metastatic lung cancer cell lines via ultracentrifugation, and these exosomes carried epithelial (E-cadherin, ZO-1) and mesenchymal (N-cadherin, Vimentin) markers. Among these, E-cadherin and N-cadherin serve as membrane protein markers.
Furthermore, the exosomal membrane protein CD44 can promote cell migration and invasion by binding to hyaluronic acid and activating EMT-related signaling pathways (148). A recent study by Nakamura and colleagues showed that exosomes derived from ovarian cancer transfer CD44 to human peritoneal mesothelial cells (HPMC), thereby promoting cancer invasion (149). Research by Yao Li et al. (150) found that exosomes carrying the PSGR membrane protein enhanced the migration, invasion, and EMT of low-invasive prostate cancer cells (LNCaP and RWPE-1) and reshaped the mRNA profiles of these cells. Although the morphological, phenotypic, and functional changes associated with EMT have been well described, the molecular and genetic mechanisms by which exosomal membrane proteins drive this process require further investigation (151–154).
3.4 Therapeutic role of exosome membrane proteins
Existing studies indicate that exosome membrane proteins play a crucial role in mediating various disease treatments (125, 133, 155–169). CD47 is usually upregulated on the surface of tumor cells, binding to signal-regulatory protein alpha (SIRPα) on phagocytic cells and inhibiting their phagocytic function, creating a “don’t eat me” signal. Eunee Koh et al. (170) designed engineered exosomes with surfaces carrying SIRPα, disrupting the CD47-SIRPα interaction between cancer cells and macrophages, enhancing the efficiency of phagocytosis of tumor cells. Similarly, Eunji Cho et al. (171) found that exosomes containing SIRPα could more effectively counteract CD47 on cancer cells, enhancing phagocytosis of tumor cells by macrophages and inhibiting the metastatic growth of tumors, offering a new approach to cancer treatment (Figure 3A). Lydia Alvarez-Erviti et al. (172) achieved therapeutic effects for Alzheimer’s disease by modifying exosomes from dendritic cells to deliver therapeutic siRNA drugs, specifically knocking down the expression of beta-amyloid precursor protein 1 (BACE1). LAMP2B fused with a neuron-specific RVG3 peptide mediated the treatment of neurodegenerative diseases, as shown in Figure 3B.
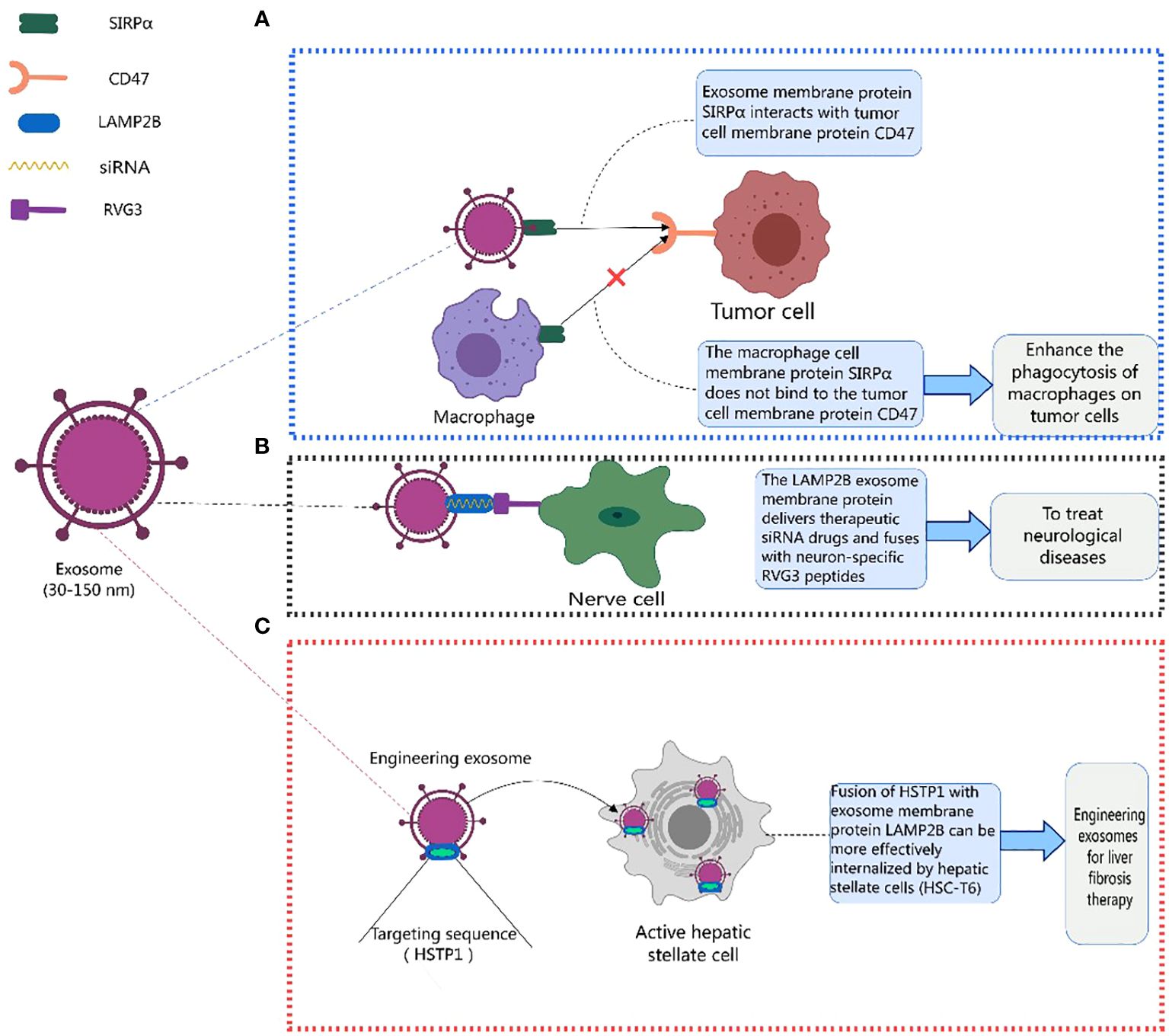
Figure 3 Therapeutic role of exosome membrane proteins in diseases. This figure was created using MedPeer.
Additionally, Yan Lin et al. (173) fused HSTP1 with exosome membrane protein LAMP2B and expressed it on the surface of exosomes through genetic engineering. Engineered exosomes (HSTP1-Exos) were more efficiently internalized by hepatic stellate cells (HSC-T6). HSTP1 is a reliable targeting peptide that specifically binds to activated hepatic stellate cells (aHSC). Exosomes modified with HSTP1 achieved precise treatment of aHSC in complex liver tissues, providing a new approach for the clinical treatment of liver fibrosis (Figure 3C). Currently, preclinical studies on the use of exosomal membrane proteins for disease treatment have achieved many successes (174–179), laying a solid foundation for the further development of clinical trials (178–187). Benjamin Besse et al. conducted a phase II clinical trial using dendritic cell-derived exosomes carrying MHC-I and MHC-II and loaded with IFN-γ (IFN-γ-Dex) to treat non-small cell lung cancer (NSCLC) patients, confirming the ability of Dex to enhance NK cell anti-tumor immunity in advanced NSCLC patients (188). Shengming Dai et al. conducted a phase I clinical trial using exosomes with surface-expressed MHC molecules and heat shock proteins (HSPs) derived from autologous ascites (Aex) combined with granulocyte-macrophage colony-stimulating factor (GM-CSF) to treat colorectal cancer, showing that Aex combined with GM-CSF can induce specific anti-tumor cytotoxic T lymphocyte (CTL) responses (189). Table 4 lists the clinical trials involving exosomal membrane proteins (190, 191).
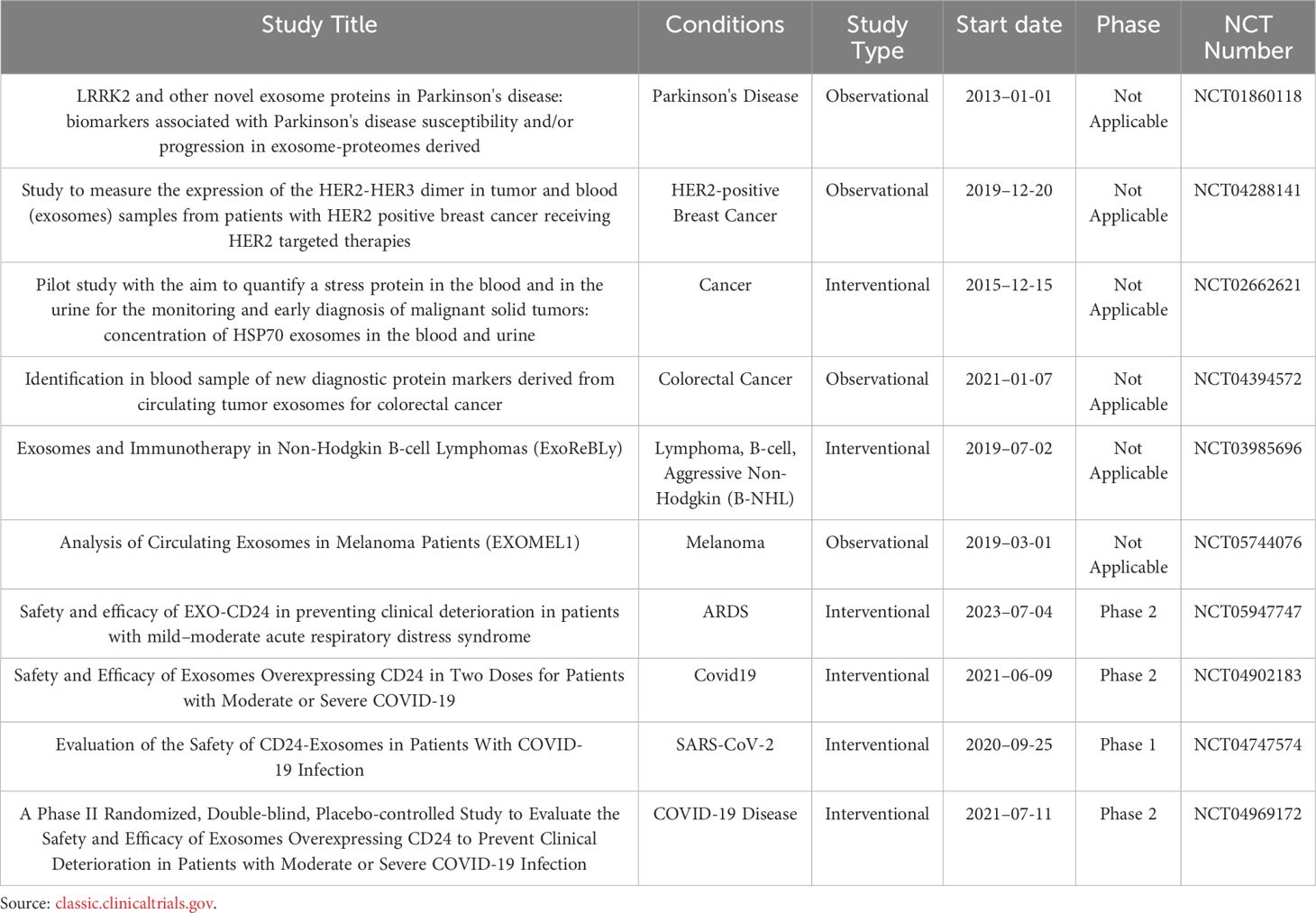
Table 4 Clinical trials using exosomal membrane proteins as primary outcome measures from 2013 to 2024.
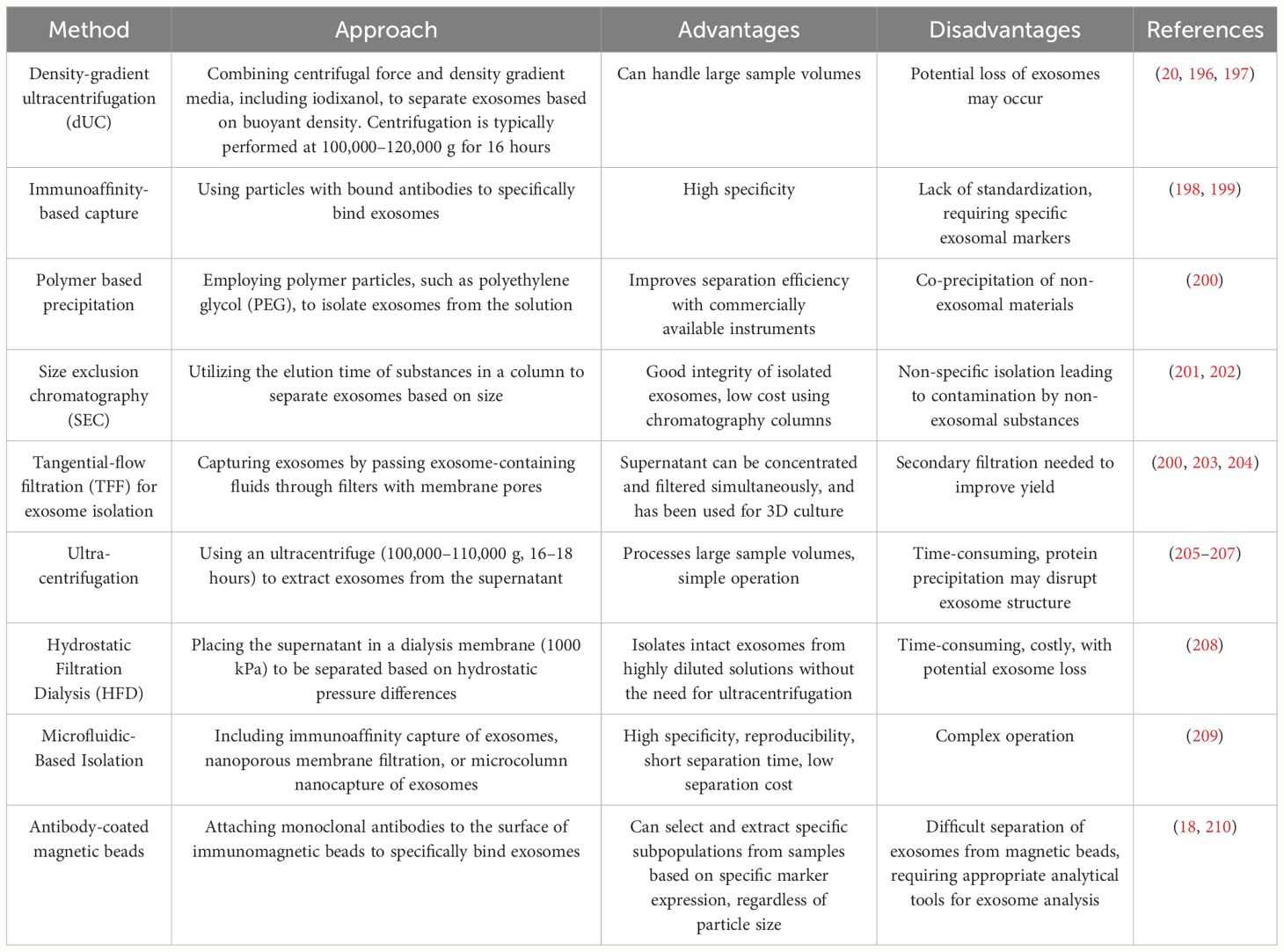
Additionally, before the clinical application of exosomal membrane proteins, issues related to exosome isolation and comprehensive characterization must be addressed (192–194). The lack of standardized procedures for exosome isolation, proper quality control, and consistent characterization methods can hinder the clinical development of exosomes and limit their analysis in standard clinical laboratories (192, 194, 195). Table 5 lists some commonly used methods for exosome isolation and supplements these methods with their advantages and disadvantages.
3.5 Immunomodulatory role of exosome membrane proteins
Previous literature has reported the role of exosomes in immune responses (211–221), primarily mediated by membrane proteins. For instance, the expression of PD-L1 on the surface of exosomes has been confirmed, and its abundance on exosomes is related to the progression of host tumors (38, 222–224). In 2022, Yunxing Lu et al. proposed an integrated microfluidic system for exosome isolation and detection (EXID system) to analyze the abundance of exosome PD-L1 protein markers. The study suggested that the abundance of PD-L1 reflects sensitivity to immune responses, and exosomes containing PD-L1 weaken anti-tumor immunity in the tumor microenvironment (225). Meizhang Li et al. indicated that exosomes derived from Wharton’s Jelly mesenchymal stem cells (WJMSCs) enhance T-cell inhibitory effects through the carried PD-L1, contributing to alleviating immune rejection in organ transplantation, as shown in Figure 4C (226). Furthermore, research results indicate that blocking exosome PD-L1 secretion significantly contributes to anti-tumor immune responses. Inhibiting exosome secretion combined with anti-PD-L1 therapy may enhance clinical anti-tumor effects (227).
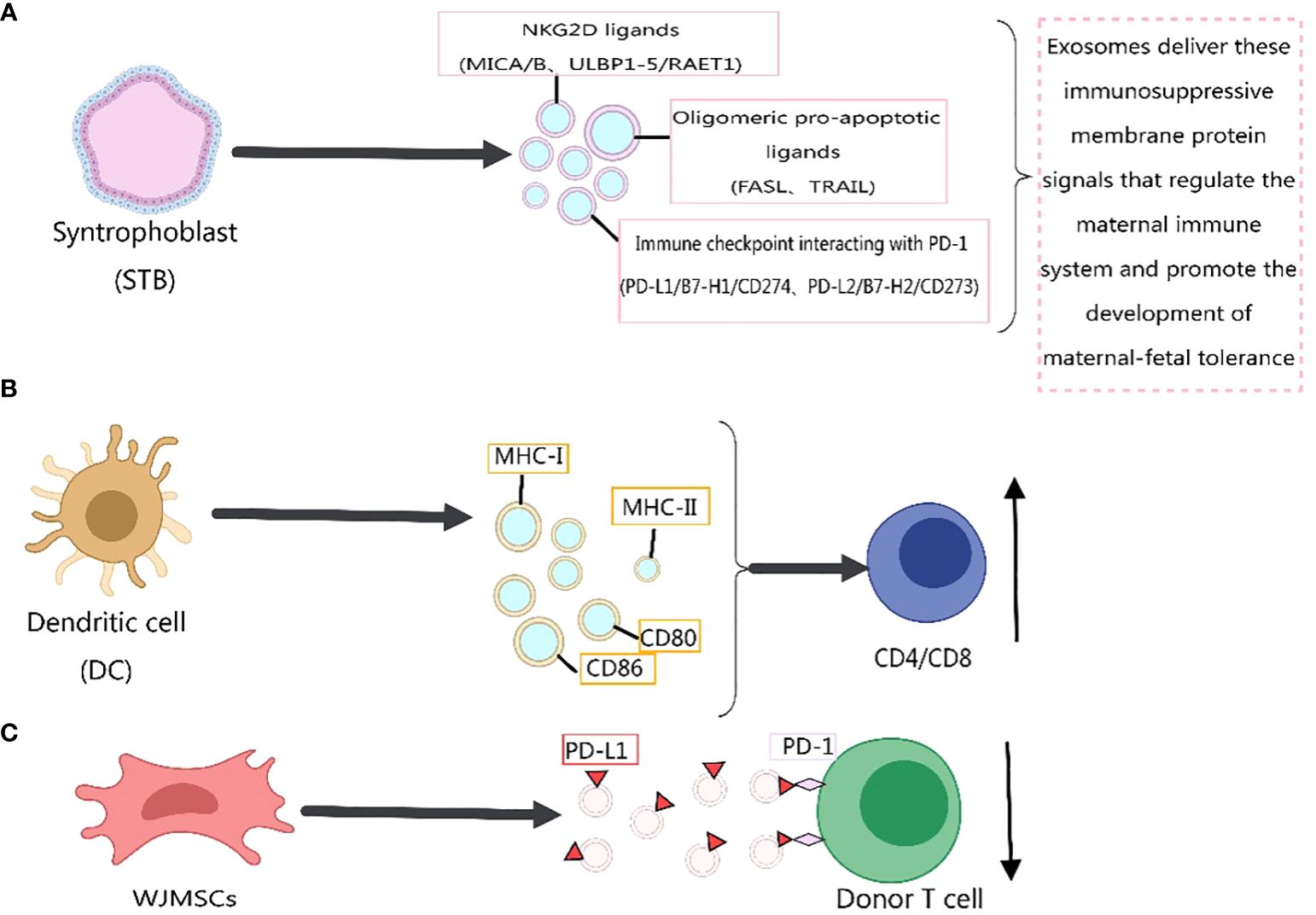
Figure 4 Immunomodulatory effects of exosome membrane proteins on the body. This figure was created using MedPeer.
Recently, Wei Zhang et al. (228) identified three classes of immunosuppressive membrane proteins expressed by syncytiotrophoblast-derived exosomes. These include NKG2D ligands (MICA/B, ULBP1–5/RAET1), oligomerization-induced apoptosis ligands (FASL, TRAIL), and immune checkpoint molecules interacting with PD-1 (PD-L1/B7-H1/CD274, PD-L2/B7-H2/CD273). The delivery of these immunosuppressive membrane protein signals by exosomes regulates the maternal immune system and promotes the development of maternal-fetal tolerance, as depicted in Figure 4A. Exosomes derived from dendritic cells express MHC-I, MHC-II, and immune co-stimulatory molecules CD80 and CD86 on their membrane surfaces, promoting T-cell activation and proliferation and regulating the body’s immune mechanisms (8), as shown in Figure 4B. Previous studies have indicated that MHC-II molecules transferred to recipient dendritic cells through exosomes activate CD4+ T cells. Similarly, MHC-I molecules transferred to dendritic cells through exosomes contribute to the activation of CD8+ T cells (229, 230). In addition, exosome membrane proteins derived from immune cells can influence the development of cancer cells (217), Figure 5. For immunosuppressive molecules expressed on the exosome membrane, blockade can be achieved by incorporating corresponding antibodies, while immune-activating molecules can be applied in clinical therapy.
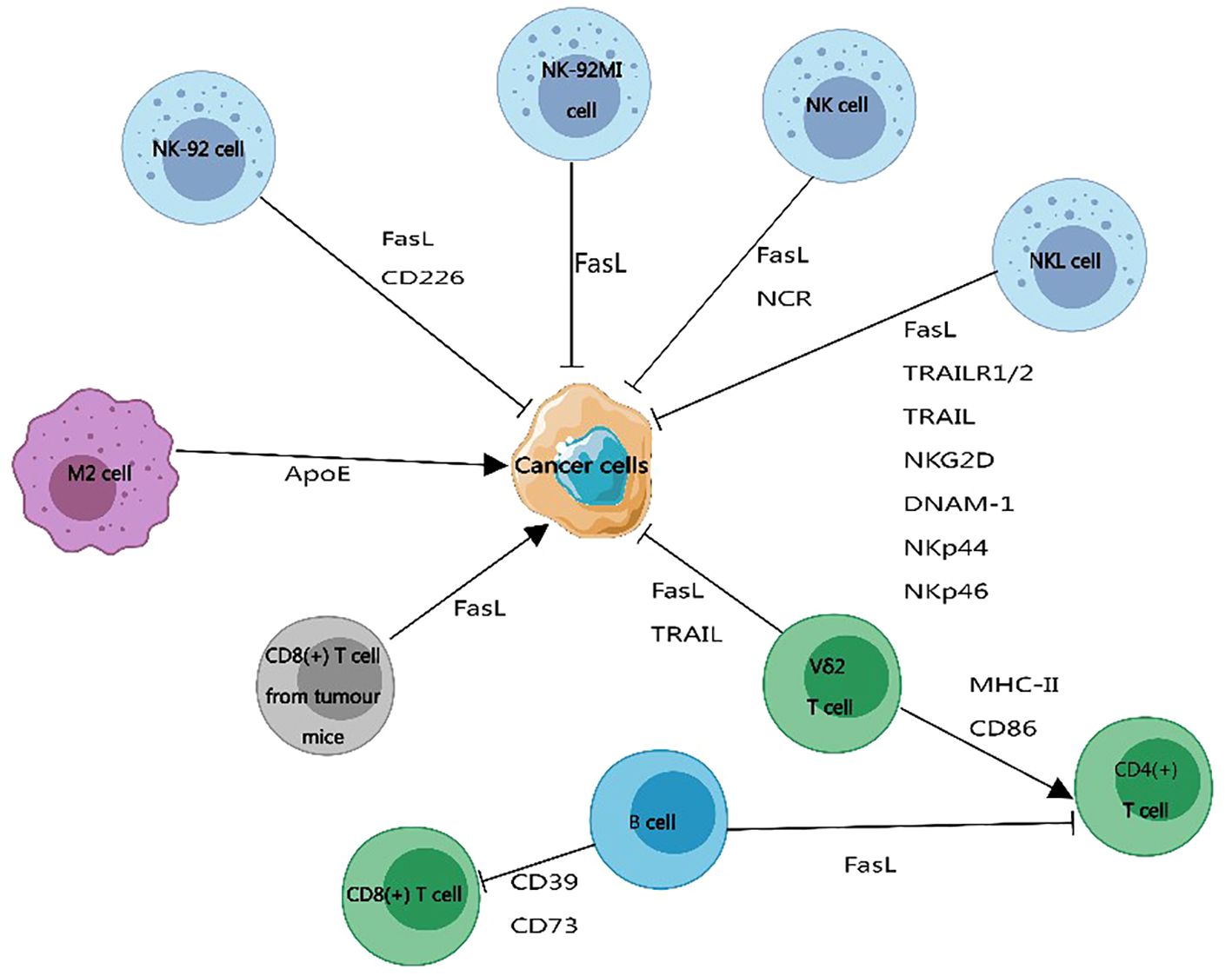
Figure 5 The impact of exosome membrane proteins derived from immune cells on cancer development. Exosome membrane proteins carried by immune cells can promote or inhibit the progression of cancer cells. Exosome membrane proteins produced by B cells, CD8+ T cells from tumor-bearing mice, and M2 macrophages promote cancer cell development. Exosome membrane proteins released by natural killer cells and Vδ2 T cells inhibit the development of cancer. This figure was created using MedPeer.
3.6 Other functions of exosome membrane proteins
In addition to their role in diagnosing diseases, regulating the body’s immune system, and serving as biological carriers targeting receptor cells, exosome membrane proteins also possess other functionalities. Upon generation, exosomes interact with proteins circulating in the surrounding environment, leading to the formation of a “protein corona” (PC). This formation alters the properties of exosomes and influences their functionality within the body (231–233). The protein corona enhances the stability of exosomes, prolonging their circulation lifespan in the body. This protection shields exosomes from degradation and clearance, thereby increasing their survival time in vivo (234, 235).
Furthermore, the presence of the protein corona can impact the interaction between exosomes and target cells. Specific protein coronas may facilitate adhesion and uptake between exosomes and target cells, mediating the entry of biologically active substances released by exosomes into recipient cells (234, 236). In conclusion, research on exosome membrane proteins is ongoing, and the exploration of their functions is expected to deepen.
4 Summary and outlook
With the increasing understanding of exosome membrane proteins, more functionalities of these proteins are gradually coming to light. In addition to the roles mentioned in this article, such as diagnosis and immune regulation, exosome membrane proteins can be redesigned or modified, significantly enriching their functions. This diversity opens up vast potential applications for exosome membrane proteins in the future, making them a focal point of current research. Despite the extensive research on exosome membrane proteins, many proteins on the exosome membrane still have undetermined functions, requiring further in-depth investigation. Moreover, since exosome membrane proteins vary depending on the cell source, it is essential to study them in the context of their origin to obtain more accurate results (125, 133).
Furthermore, membrane proteins of exosomes have garnered significant interest in clinical trials for disease diagnosis and therapy. However, achieving a range of functions in clinical settings remains challenging for researchers (210, 237). To advance the clinical translation of exosomes, several key issues need to be addressed. These include: 1. The need for standardized methods to isolate, characterize, and quantify exosomes to ensure their stability and reproducibility; 2. Developing rigorous preclinical biosafety evaluation protocols to mitigate risks before human trials; 3. Conducting pilot clinical studies to demonstrate feasibility, biological distribution in humans, and preliminary efficacy before large-scale applications (13, 20, 24, 238).
Although researchers from different fields have explored exosome membrane proteins, gaining varying degrees of understanding of protein types and biological functions, the intricate environment within the body poses the need for further exploration and explanation of membrane protein-mediated mechanisms.
Author contributions
FX: Resources, Methodology, Formal analysis, Writing – original draft, Data curation. SL: Writing – original draft, Methodology. PL: Writing – review & editing, Formal analysis. CC: Investigation, Formal analysis, Writing – review & editing. WL: Writing – review & editing, Resources, Funding acquisition. CL: Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Natural Science Foundation of Beijing (7212172), National Natural Science Foundation of China (82274447), High-Level Public Health Technical Talents Project of Beijing (2022–2-024), Beijing Municipal Public Welfare Development and Reform Pilot Project for Medical Research Institutes (JYY2021–10).
Acknowledgments
We sincerely appreciate the invaluable guidance and insightful suggestions provided by WL, CL, and CC during the writing process of this article. Their expertise and support greatly enhanced the quality of our work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. (2013) 126:5553–65. doi: 10.1242/jcs.128868
2. Larios J, Mercier V, Roux A, Gruenberg J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J Cell Biol. (2020) 219. doi: 10.1083/jcb.201904113
3. Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. (2010) 12:19–30;sup pp 11-13. doi: 10.1038/ncb2000
4. Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, et al. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. (2021) 31:157–77. doi: 10.1038/s41422-020-00409-1
5. Li X, Li C, Zhang L, Wu M, Cao K, Jiang F, et al. The significance of exosomes in the development and treatment of hepatocellular carcinoma. Mol Cancer. (2020) 19:1. doi: 10.1186/s12943-019-1085-0
6. Noonin C, Thongboonkerd V. Exosome-inflammasome crosstalk and their roles in inflammatory responses. Theranostics. (2021) 11:4436–51. doi: 10.7150/thno.54004
7. Li L, Huang L, Huang C, Xu J, Huang Y, Luo H, et al. The multiomics landscape of serum exosomes during the development of sepsis. J Adv Res. (2022) 39:203–23. doi: 10.1016/j.jare.2021.11.005
8. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. (2020) 367. doi: 10.1126/science.aau6977
9. Sun D, Zhuang X, Zhang S, Deng ZB, Grizzle W, Miller D, et al. Exosomes are endogenous nanoparticles that can deliver biological information between cells. Adv Drug Delivery Rev. (2013) 65:342–7. doi: 10.1016/j.addr.2012.07.002
10. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. (2012) 1820:940–8. doi: 10.1016/j.bbagen.2012.03.017
11. Bang C, Thum T. Exosomes: new players in cell-cell communication. Int J Biochem Cell Biol. (2012) 44:2060–4. doi: 10.1016/j.biocel.2012.08.007
12. EL Andaloussi S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. (2013) 12:347–57. doi: 10.1038/nrd3978
13. Mahl L, Ollig J, Schweihofer V, Wiegand L, Torkler P, Haerteis S, et al. Importance and implications of exosomes in nephrology and urology. Pflugers Arch. (2023) 475:153–66. doi: 10.1007/s00424-022-02771-y
14. Peche VS, Pietka TA, Jacome-Sosa M, Samovski D, Palacios H, Chatterjee-Basu G, et al. Endothelial cell CD36 regulates membrane ceramide formation, exosome fatty acid transfer and circulating fatty acid levels. Nat Commun. (2023) 14:4029. doi: 10.1038/s41467-023-39752-3
15. Gebeyehu A KN, Meckes DG Jr, Sachdeva MS. Role of exosomes for delivery of chemotherapeutic drugs. Crit Rev Ther Drug Carrier Syst. (2021) 38:53–97. doi: 10.1615/CritRevTherDrugCarrierSyst
16. Alenquer M, Amorim MJ. Exosome biogenesis, regulation, and function in viral infection. Viruses. (2015) 7:5066–83. doi: 10.3390/v7092862
17. Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of exosome composition. Cell. (2019) 177:428–445.e418. doi: 10.1016/j.cell.2019.02.029
18. Raghav A, Tripathi P, Mishra BK, Jeong GB, Banday S, Gautam KA, et al. Mesenchymal stromal cell-derived tailored exosomes treat bacteria-associated diabetes foot ulcers: A customized approach from bench to bed. Front Microbiol. (2021) 12:712588. doi: 10.3389/fmicb.2021.712588
19. Gurung S, Perocheau D, Touramanidou L, Baruteau J. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal. (2021) 19:47. doi: 10.1186/s12964-021-00730-1
20. Lopez K, Lai SWT, Lopez Gonzalez EJ, R.G. D, Shuck SC. Extracellular vesicles: A dive into their role in the tumor microenvironment and cancer progression. Front Cell Dev Biol. (2023) 11:1154576. doi: 10.3389/fcell.2023.1154576
21. Chiang CY, Chen C. Toward characterizing extracellular vesicles at a single-particle level. J BioMed Sci. (2019) 26:9. doi: 10.1186/s12929-019-0502-4
22. Hartjes TA, Mytnyk S, Jenster GW, van Steijn V, van Royen ME. Extracellular vesicle quantification and characterization: common methods and emerging approaches. Bioengineering (Basel). (2019) 6:7-32. doi: 10.3390/bioengineering6010007
23. Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. (2018) 7:1535750. doi: 10.1080/20013078.2018.1535750
24. Butreddy A, Kommineni N, Dudhipala N. Exosomes as naturally occurring vehicles for delivery of biopharmaceuticals: insights from drug delivery to clinical perspectives. Nanomaterials (Basel). (2021) 11:1481-523. doi: 10.3390/nano11061481
25. Daaboul GG, Gagni P, Benussi L, Bettotti P, Ciani M, Cretich M, et al. Digital detection of exosomes by interferometric imaging. Sci Rep. (2016) 6:37246. doi: 10.1038/srep37246
26. Miron RJ, Zhang Y. Understanding exosomes: Part 1-Characterization, quantification and isolation techniques. Periodontol 2000. (2024) 94:231–56. doi: 10.1111/prd.12520
27. Fang X, Duan Y, Adkins GB, Pan S, Wang H, Liu Y, et al. Highly efficient exosome isolation and protein analysis by an integrated nanomaterial-based platform. Anal Chem. (2018) 90:2787–95. doi: 10.1021/acs.analchem.7b04861
28. Fordjour FK, Guo C, Ai Y, Daaboul GG, Gould SJ. A shared, stochastic pathway mediates exosome protein budding along plasma and endosome membranes. J Biol Chem. (2022) 298:102394. doi: 10.1016/j.jbc.2022.102394
29. Song Z, Mao J, Barrero RA, Wang P, Zhang F, Wang T. Development of a CD63 aptamer for efficient cancer immunochemistry and immunoaffinity-based exosome isolation. Molecules. (2020) 25:5585-600. doi: 10.3390/molecules25235585
30. Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. (1998) 273:20121–7. doi: 10.1074/jbc.273.32.20121
31. Pitt JM, Andre F, Amigorena S, Soria JC, Eggermont A, Kroemer G, et al. Dendritic cell-derived exosomes for cancer therapy. J Clin Invest. (2016) 126:1224–32. doi: 10.1172/JCI81137
32. Skokos D, Botros HG, Demeure C, Morin J, Peronet R, Birkenmeier G, et al. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J Immunol. (2003) 170:3037–45. doi: 10.4049/jimmunol.170.6.3037
33. Park S, Ahn ES, Kim Y. Neuroblastoma SH-SY5Y cell-derived exosomes stimulate dendrite-like outgrowths and modify the differentiation of A375 melanoma cells. Cell Biol Int. (2015) 39:379–87. doi: 10.1002/cbin.10401
34. Segura E, Nicco C, Lombard B, Veron P, Raposo G, Batteux F, et al. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood. (2005) 106:216–23. doi: 10.1182/blood-2005-01-0220
35. Roucourt B, Meeussen S, Bao J, Zimmermann P, David G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. (2015) 25:412–28. doi: 10.1038/cr.2015.29
36. Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. (2015) 527:329–35. doi: 10.1038/nature15756
37. Kleijmeer MJ, Escola JM, UytdeHaag FG, Jakobson E, Griffith JM, Osterhaus AD, et al. Antigen loading of MHC class I molecules in the endocytic tract. Traffic. (2001) 2:124–37. doi: 10.1034/j.1600-0854.2001.020207.x
38. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. (2018) 560:382–6. doi: 10.1038/s41586-018-0392-8
39. Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, et al. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. (2001) 166:7309–18. doi: 10.4049/jimmunol.166.12.7309
40. Xu R, Greening DW, Chen M, Rai A, Ji H, Takahashi N, et al. Surfaceome of exosomes secreted from the colorectal cancer cell line SW480: peripheral and integral membrane proteins analyzed by proteolysis and TX114. Proteomics. (2019) 19:e1700453. doi: 10.1002/pmic.201700453
41. Clayton A, Al-Taei S, Webber J, Mason MD, Tabi Z. Cancer exosomes express CD39 and CD73, which suppress T cells through adenosine production. J Immunol. (2011) 187:676–83. doi: 10.4049/jimmunol.1003884
42. Rabesandratana H, Toutant J-P, Reggio H, Vidal M. Decay-accelerating factor (CD55) and membrane inhibitor of reactive lysis (CD59) are released within exosomes during in vitro maturation of reticulocytes. Blood. (1998) 91:2573–80. doi: 10.1182/blood.V91.7.2573
43. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. (2015) 523:177–82. doi: 10.1038/nature14581
44. Zheng J, Hernandez JM, Doussot A, Bojmar L, Zambirinis CP, Costa-Silva B, et al. Extracellular matrix proteins and carcinoembryonic antigen-related cell adhesion molecules characterize pancreatic duct fluid exosomes in patients with pancreatic cancer. HPB (Oxford). (2018) 20:597–604. doi: 10.1016/j.hpb.2017.12.010
45. Atay S, Gercel-Taylor C, Taylor DD. Human trophoblast-derived exosomal fibronectin induces pro-inflammatory IL-1beta production by macrophages. Am J Reprod Immunol. (2011) 66:259–69. doi: 10.1111/aji.2011.66.issue-4
46. Santasusagna S, Moreno I, Navarro A, Castellano JJ, Martinez F, Hernandez R, et al. Proteomic analysis of liquid biopsy from tumor-draining vein indicates that high expression of exosomal ECM1 is associated with relapse in stage I-III colon cancer. Transl Oncol. (2018) 11:715–21. doi: 10.1016/j.tranon.2018.03.010
47. Oshima K, Aoki N, Kato T, Kitajima K, Matsuda T. Secretion of a peripheral membrane protein, MFG-E8, as a complex with membrane vesicles. Eur J Biochem. (2002) 269:1209–18. doi: 10.1046/j.1432-1033.2002.02758.x
48. Koch R, Demant M, Aung T, Diering N, Cicholas A, Chapuy B, et al. Populational equilibrium through exosome-mediated Wnt signaling in tumor progression of diffuse large B-cell lymphoma. Blood. (2014) 123:2189–98. doi: 10.1182/blood-2013-08-523886
49. Gerber PP, Cabrini M, Jancic C, Paoletti L, Banchio C, von Bilderling C, et al. Rab27a controls HIV-1 assembly by regulating plasma membrane levels of phosphatidylinositol 4,5-bisphosphate. J Cell Biol. (2015) 209:435–52. doi: 10.1083/jcb.201409082
50. Ashley J, Cordy B, Lucia D, Fradkin LG, Budnik V, Thomson T. Retrovirus-like gag protein arc1 binds RNA and traffics across synaptic boutons. Cell. (2018) 172:262–274.e211. doi: 10.1016/j.cell.2017.12.022
51. Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. (2003) 278:10963–72. doi: 10.1074/jbc.M207550200
52. Shen B, Fang Y, Wu N, Gould SJ. Biogenesis of the posterior pole is mediated by the exosome/microvesicle protein-sorting pathway. J Biol Chem. (2011) 286:44162–76. doi: 10.1074/jbc.M111.274803
53. Hegmans JP, Bard MP, Hemmes A, Luider TM, Kleijmeer MJ, Prins JB, et al. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am J Pathol. (2004) 164:1807–15. doi: 10.1016/S0002-9440(10)63739-X
54. Latysheva N, Muratov G, Rajesh S, Padgett M, Hotchin NA, Overduin M, et al. Syntenin-1 is a new component of tetraspanin-enriched microdomains: mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol Cell Biol. (2006) 26:7707–18. doi: 10.1128/MCB.00849-06
55. Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. (2010) 120:457–71. doi: 10.1172/JCI40483
56. Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. (2010) 4:214–22. doi: 10.1016/j.scr.2009.12.003
57. Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in membranous vesicle and exosome proteomics improving biological understanding and biomarker discovery. Proteomics. (2011) 11:709–20. doi: 10.1002/pmic.201000422
58. He C, Zheng S, Luo Y, Wang B. Exosome theranostics: biology and translational medicine. Theranostics. (2018) 8:237–55. doi: 10.7150/thno.21945
59. Iwai M, Hamatani T, Nakamura A, Kawano N, Kanai S, Kang W, et al. Membrane protein CD9 is repositioned and released to enhance uterine function. Lab Invest. (2019) 99:200–9. doi: 10.1038/s41374-018-0145-1
60. Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA, et al. Proteomic analysis of exosomes isolated from human Malignant pleural effusions. Am J Respir Cell Mol Biol. (2004) 31:114–21. doi: 10.1165/rcmb.2003-0238OC
61. Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. (2009) 6:267–83. doi: 10.1586/epr.09.17
62. Xiao Y, Zhong J, Zhong B, Huang J, Jiang L, Jiang Y, et al. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. (2020) 476:13–22. doi: 10.1016/j.canlet.2020.01.033
63. Ai Y GC, Garcia-Contreras M, Sánchez B LS, Saftics A, Shodubi O, Raghunandan S, et al. Endocytosis blocks the vesicular secretion of exosome marker proteins. Sci Adv. (2024) 10:eadi9156. doi: 10.1126/sciadv.adi9156
64. Charrin S, Jouannet S, Boucheix C, Rubinstein E. Tetraspanins at a glance. J Cell Sci. (2014) 127:3641–8. doi: 10.1242/jcs.154906
65. Ludwig N, Yerneni SS, Azambuja JH, Gillespie DG, Menshikova EV, Jackson EK, et al. Tumor-derived exosomes promote angiogenesis via adenosine A(2B) receptor signaling. Angiogenesis. (2020) 23:599–610. doi: 10.1007/s10456-020-09728-8
66. Hur YH, Feng S, Wilson KF, Cerione RA, Antonyak MA. Embryonic stem cell-derived extracellular vesicles maintain ESC stemness by activating FAK. Dev Cell. (2021) 56:277–291.e276. doi: 10.1016/j.devcel.2020.11.017
67. Song L, Tang S, Han X, Jiang Z, Dong L, Liu C, et al. KIBRA controls exosome secretion via inhibiting the proteasomal degradation of Rab27a. Nat Commun. (2019) 10:1639. doi: 10.1038/s41467-019-09720-x
68. Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. (2012) 83:1484–94. doi: 10.1016/j.bcp.2011.12.037
69. Ghossoub R, Chery M, Audebert S, Leblanc R, Egea-Jimenez AL, Lembo F, et al. Tetraspanin-6 negatively regulates exosome production. Proc Natl Acad Sci U.S.A. (2020) 117:5913–22. doi: 10.1073/pnas.1922447117
70. Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. (2010) 70:1668–78. doi: 10.1158/0008-5472.CAN-09-2470
71. Wu F YZ, Yang L, Fan J, Xu J, Jin Y, Yu J, et al. Smoking induced extracellular vesicles release and their distinct properties in non-small cell lung cancer. J Cancer. (2019) 10:3435–43. doi: 10.7150/jca.30425
72. Lyu Q ZX, Shi LQ, Chen HY, Lu M, Ma XD, Ren L. Exosomes may be the carrier of acupuncture treatment for major depressive disorder. Front Behav Neurosci. (2023) 17:1107265. doi: 10.3389/fnbeh.2023.1107265
73. Hu S MJ, Su C, Chen Y, Shu Y, Qi Z, Zhang B, et al. Engineered exosome-like nanovesicles suppress tumor growth by reprogramming tumor microenvironment and promoting tumor ferroptosis. Acta Biomater. (2021) 135:567–81. doi: 10.1016/j.actbio.2021.09.003
74. Boukouris S MS. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin Appl. (2015) 9:358–67. doi: 10.1002/prca.201400114
75. Penfornis P VK, Whitt J, Pochampally R. Extracellular vesicles as carriers of microRNA, proteins and lipids in tumor microenvironment. Int J Cancer. (2016) 138:14–21. doi: 10.1002/ijc.29417
76. Cheng J WX, Yuan X, Liu G, Chu Q. Emerging roles of exosome-derived biomarkers in cancer theranostics: messages from novel protein targets. Am J Cancer Res. (2022) 12:2226–48.
77. Araujo-Abad S, Saceda M, de Juan Romero C. Biomedical application of small extracellular vesicles in cancer treatment. Adv Drug Delivery Rev. (2022) 182:114117. doi: 10.1016/j.addr.2022.114117
78. Choi E-S, Faruque HA, Kim J-H, Kim KJ, Choi JE, Kim BA, et al. CD5L as an extracellular vesicle-derived biomarker for liquid biopsy of lung cancer. Diagnostics. (2021) 11:620-37. doi: 10.3390/diagnostics11040620
79. Khan NA AM, El-Menyar A, Biswas KH, Rizoli S, Al-Thani H. The evolving role of extracellular vesicles (exosomes) as biomarkers in traumatic brain injury: Clinical perspectives and therapeutic implications. Front Aging Neurosci. (2022) 14:933434. doi: 10.3389/fnagi.2022.933434
80. Li Y, Zhang Y, Qiu F, Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. (2011) 32:1976–83. doi: 10.1002/elps.201000598
81. Sakaue T, Koga H, Iwamoto H, Nakamura T, Ikezono Y, Abe M, et al. Glycosylation of ascites-derived exosomal CD133: a potential prognostic biomarker in patients with advanced pancreatic cancer. Med Mol Morphol. (2019) 52:198–208. doi: 10.1007/s00795-019-00218-5
82. Sandfeld-Paulsen B, Jakobsen KR, Baek R, Folkersen BH, Rasmussen TR, Meldgaard P, et al. Exosomal proteins as diagnostic biomarkers in lung cancer. J Thorac Oncol. (2016) 11:1701–10. doi: 10.1016/j.jtho.2016.05.034
83. Sandúa A AE, González Á. Exosomes in lung cancer: actors and heralds of tumor development. Cancers (Basel). (2021) 13:4330. doi: 10.3390/cancers13174330
84. Xylaki M, Chopra A, Weber S, Bartl M, Outeiro TF, Mollenhauer B. Extracellular vesicles for the diagnosis of parkinson’s disease: systematic review and meta-analysis. Mov Disord. (2023) 38:1585–97. doi: 10.1002/mds.29497
85. Chen IH, Xue L, Hsu CC, Paez JS, Pan L, Andaluz H, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci USA. (2017) 114:3175–80. doi: 10.1073/pnas.1618088114
86. Jiang L, Dong H, Cao H, Ji X, Luan S, Liu J. Exosomes in pathogenesis, diagnosis, and treatment of alzheimer’s disease. Med Sci Monit. (2019) 25:3329–35. doi: 10.12659/MSM.914027
87. Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery - a novel application for the mesenchymal stem cell. Biotechnol Adv. (2013) 31:543–51. doi: 10.1016/j.biotechadv.2012.08.008
88. Min L, Zhu S, Chen L, Liu X, Wei R, Zhao L, et al. Evaluation of circulating small extracellular vesicles derived miRNAs as biomarkers of early colon cancer: a comparison with plasma total miRNAs. J Extracell Vesicles. (2019) 8:1643670. doi: 10.1080/20013078.2019.1643670
89. Jakobsen KR, Paulsen BS, Baek R, Varming K, Sorensen BS, Jorgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles. (2015) 4:26659. doi: 10.3402/jev.v4.26659
90. Chang L, Wu H, Chen R, Sun X, Yang Y, Huang C, et al. Microporous PdCuB nanotag-based electrochemical aptasensor with Au@CuCl(2) nanowires interface for ultrasensitive detection of PD-L1-positive exosomes in the serum of lung cancer patients. J Nanobiotechnology. (2023) 21:86. doi: 10.1186/s12951-023-01845-y
91. Keryer-Bibens C, Pioche-Durieu C, Villemant C, Souquere S, Nishi N, Hirashima M, et al. Exosomes released by EBV-infected nasopharyngeal carcinoma cells convey the viral latent membrane protein 1 and the immunomodulatory protein galectin 9. BMC Cancer. (2006) 6:283. doi: 10.1186/1471-2407-6-283
92. Houali K, Wang X, Shimizu Y, Djennaoui D, Nicholls J, Fiorini S, et al. A new diagnostic marker for secreted Epstein-Barr virus encoded LMP1 and BARF1 oncoproteins in the serum and saliva of patients with nasopharyngeal carcinoma. Clin Cancer Res. (2007) 13:4993–5000. doi: 10.1158/1078-0432.CCR-06-2945
93. Deng F, Miller J. A review on protein markers of exosome from different bio-resources and the antibodies used for characterization. J Histotechnol. (2019) 42:226–39. doi: 10.1080/01478885.2019.1646984
94. Koksal AR, Thevenot P, Aydin Y, Nunez K, Sandow T, Widmer K, et al. Impaired autophagy response in hepatocellular carcinomas enriches glypican-3 in exosomes, not in the microvesicles. J Hepatocell Carcinoma. (2022) 9:959–72. doi: 10.2147/JHC.S376210
95. Arbelaiz A, Azkargorta M, Santos-Laso Á, Perugorria MJ, Erice O, Gonzalez E, et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. J Hepatol. (2017) 66:S207–8. doi: 10.1016/S0168-8278(17)30712-2
96. Wang X, Shen H, Zhangyuan G, Huang R, Zhang W, He Q, et al. 14-3-3zeta delivered by hepatocellular carcinoma-derived exosomes impaired anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death Dis. (2018) 9:159. doi: 10.1038/s41419-017-0180-7
97. Welker MW, Reichert D, Susser S, Sarrazin C, Martinez Y, Herrmann E, et al. Soluble serum CD81 is elevated in patients with chronic hepatitis C and correlates with alanine aminotransferase serum activity. PloS One. (2012) 7:e30796. doi: 10.1371/journal.pone.0030796
98. Castillo J, Bernard V, San Lucas FA, Allenson K, Capello M, Kim DU, et al. Surfaceome profiling enables isolation of cancer-specific exosomal cargo in liquid biopsies from pancreatic cancer patients. Ann Oncol. (2018) 29:223–9. doi: 10.1093/annonc/mdx542
99. Fan J, Wei Q, Koay EJ, Liu Y, Ning B, Bernard PW, et al. Chemoresistance transmission via exosome-mediated ephA2 transfer in pancreatic cancer. Theranostics. (2018) 8:5986–94. doi: 10.7150/thno.26650
100. Kimura H, Yamamoto H, Harada T, Fumoto K, Osugi Y, Sada R, et al. CKAP4, a DKK1 receptor, is a biomarker in exosomes derived from pancreatic cancer and a molecular target for therapy. Clin Cancer Res. (2019) 25:1936–47. doi: 10.1158/1078-0432.CCR-18-2124
101. Yoshioka Y, Kosaka N, Konishi Y, Ohta H, Okamoto H, Sonoda H, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. (2014) 5:3591. doi: 10.1038/ncomms4591
102. Anami K, Oue N, Noguchi T, Sakamoto N, Sentani K, Hayashi T, et al. TSPAN8, identified by Escherichia coli ampicillin secretion trap, is associated with cell growth and invasion in gastric cancer. Gastric Cancer. (2016) 19:370–80. doi: 10.1007/s10120-015-0478-z
103. Fraser KB MM, Alcalay RN, West AB. Urinary LRRK2 phosphorylation predicts parkinsonian phenotypes in G2019S LRRK2 carriers. Neurology. (2016) 86:994–9. doi: 10.1212/WNL.0000000000002436
104. Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. (2008) 10:1470–6. doi: 10.1038/ncb1800
105. Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. (2012) 18:1835–40. doi: 10.1038/nm.2994
106. Raimondo F, Morosi L, Corbetta S, Chinello C, Brambilla P, Della Mina P, et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst. (2013) 9:1220–33. doi: 10.1039/c3mb25582d
107. Li T, Liu TC, Liu N, Li MJ, Zhang M. Urinary exosome proteins PAK6 and EGFR as noninvasive diagnostic biomarkers of diabetic nephropathy. BMC Nephrol. (2023) 24:291. doi: 10.1186/s12882-023-03343-7
108. Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. (2010) 9:1324–38. doi: 10.1074/mcp.M000063-MCP201
109. Wang YT, Shi T, Srivastava S, Kagan J, Liu T, Rodland KD. Proteomic analysis of exosomes for discovery of protein biomarkers for prostate and bladder cancer. Cancers (Basel). (2020) 12:2335-53. doi: 10.3390/cancers12092335
110. Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. (2009) 100:1603–7. doi: 10.1038/sj.bjc.6605058
111. Liang B, Peng P, Chen S, Li L, Zhang M, Cao D, et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. (2013) 80:171–82. doi: 10.1016/j.jprot.2012.12.029
112. Chanteloup G, Cordonnier M, Isambert N, Bertaut A, Marcion G, Garrido C, et al. Membrane-bound exosomal HSP70 as a biomarker for detection and monitoring of Malignant solid tumours: a pilot study. Pilot Feasibility Stud. (2020) 6:35. doi: 10.1186/s40814-020-00577-2
113. Shimizu A, Sawada K, Kobayashi M, Yamamoto M, Yagi T, Kinose Y, et al. Exosomal CD47 plays an essential role in immune evasion in ovarian cancer. Mol Cancer Res. (2021) 19:1583–95. doi: 10.1158/1541-7786.MCR-20-0956
114. Luo D ZS, Xia W, Huang L, Ge W, Wang T. Proteomics study of serum exosomes from papillary thyroid cancer patients. Endocr Relat Cancer. (2018) 25:879–91. doi: 10.1530/ERC-17-0547
115. Risha Y, Minic Z, Ghobadloo SM, Berezovski MV. The proteomic analysis of breast cell line exosomes reveals disease patterns and potential biomarkers. Sci Rep. (2020) 10:13572. doi: 10.1038/s41598-020-70393-4
116. Wang X, Huang J, Chen W, Li G, Li Z, Lei J. The updated role of exosomal proteins in the diagnosis, prognosis, and treatment of cancer. Exp Mol Med. (2022) 54:1390–400. doi: 10.1038/s12276-022-00855-4
117. Wang Y, Gao W, Sun M, Feng B, Shen H, Zhu J, et al. A filter-electrochemical microfluidic chip for multiple surface protein analysis of exosomes to detect and classify breast cancer. Biosens Bioelectron. (2023) 239:115590. doi: 10.1016/j.bios.2023.115590
118. Han Z, Peng X, Yang Y, Yi J, Zhao D, Bao Q, et al. Integrated microfluidic-SERS for exosome biomarker profiling and osteosarcoma diagnosis. Biosens Bioelectron. (2022) 217:114709. doi: 10.1016/j.bios.2022.114709
119. Chuang HC, Chen MH, Chen YM, Yang HY, Ciou YR, Hsueh CH, et al. BPI overexpression suppresses Treg differentiation and induces exosome-mediated inflammation in systemic lupus erythematosus. Theranostics. (2021) 11:9953–66. doi: 10.7150/thno.63743
120. Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PloS One. (2009) 4:e5219. doi: 10.1371/journal.pone.0005219
121. Yoshioka Y, Konishi Y, Kosaka N, Katsuda T, Kato T, Ochiya. T. Comparative marker analysis of extracellular vesicles in different human cancer types. J Extracell Vesicles. (2013) 2. doi: 10.3402/jev.v2i0.20424
122. Lin B, Tian T, Lu Y, Liu D, Huang M, Zhu L, et al. Tracing tumor-derived exosomal PD-L1 by dual-aptamer activated proximity-induced droplet digital PCR. Angew Chem Int Ed Engl. (2021) 60:7582–6. doi: 10.1002/anie.202015628
123. Yin T, Xu L, Gil B, Merali N, Sokolikova MS, Gaboriau DCA, et al. Graphene sensor arrays for rapid and accurate detection of pancreatic cancer exosomes in patients’ Blood plasma samples. ACS Nano. (2023) 17:14619–31. doi: 10.1021/acsnano.3c01812
124. Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, et al. Exosomes: novel biomarkers for clinical diagnosis. ScientificWorldJournal. (2015) 2015:657086. doi: 10.1155/2015/657086
125. Salunkhe S, Dheeraj, Basak M, Chitkara D, Mittal A. Surface functionalization of exosomes for target-specific delivery and in vivo imaging & tracking: Strategies and significance. J Control Release. (2020) 326:599–614. doi: 10.1016/j.jconrel.2020.07.042
126. Liang Y, Duan L, Lu J, Xia J. Engineering exosomes for targeted drug delivery. Theranostics. (2021) 11:3183–95. doi: 10.7150/thno.52570
127. Tashima T. Smart Strategies for Therapeutic Agent Delivery into Brain across the Blood-Brain Barrier Using Receptor-Mediated Transcytosis. Chem Pharm Bull (Tokyo). (2020) 68:316–25. doi: 10.1248/cpb.c19-00854
128. Creeden JF, Sevier J, Zhang JT, Lapitsky Y, Brunicardi FC, Jin G, et al. Smart exosomes enhance PDAC targeted therapy. J Control Release. (2024) 368:413–29. doi: 10.1016/j.jconrel.2024.02.037
129. Ghaffari K, Moradi-Hasanabad A, Sobhani-Nasab A, Javaheri J, Ghasemi A. Application of cell-derived exosomes in the hematological Malignancies therapy. Front Pharmacol. (2023) 14:1263834. doi: 10.3389/fphar.2023.1263834
130. Hermann DM, Peruzzotti-Jametti L, Giebel B, Pluchino S. Extracellular vesicles set the stage for brain plasticity and recovery by multimodal signalling. Brain. (2024) 147:372–89. doi: 10.1093/brain/awad332
131. Jang Y, Park J, Kim P, Park EJ, Sun H, Baek Y, et al. Development of exosome membrane materials-fused microbubbles for enhanced stability and efficient drug delivery of ultrasound contrast agent. Acta Pharm Sin B. (2023) 13:4983–98. doi: 10.1016/j.apsb.2023.08.022
132. Vandergriff A, Huang K, Shen D, Hu S, Hensley MT, Caranasos TG, et al. Targeting regenerative exosomes to myocardial infarction using cardiac homing peptide. Theranostics. (2018) 8:1869–78. doi: 10.7150/thno.20524
133. Choi H, Choi Y, Yim HY, Mirzaaghasi A, Yoo JK, Choi C. Biodistribution of exosomes and engineering strategies for targeted delivery of therapeutic exosomes. Tissue Eng Regener Med. (2021) 18:499–511. doi: 10.1007/s13770-021-00361-0
134. Zhao K, Wang Z, Hackert T, Pitzer C, Zoller M. Tspan8 and Tspan8/CD151 knockout mice unravel the contribution of tumor and host exosomes to tumor progression. J Exp Clin Cancer Res. (2018) 37:312. doi: 10.1186/s13046-018-0961-6
135. Du S, Qian J, Tan S, Li W, Liu P, Zhao J, et al. Tumor cell-derived exosomes deliver TIE2 protein to macrophages to promote angiogenesis in cervical cancer. Cancer Lett. (2022) 529:168–79. doi: 10.1016/j.canlet.2022.01.005
136. Wang J, Zhang H, Sun X, Wang X, Ren T, Huang Y, et al. Exosomal PD-L1 and N-cadherin predict pulmonary metastasis progression for osteosarcoma patients. J Nanobiotechnology. (2020) 18:149-67. doi: 10.1186/s12951-020-00710-6
137. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res. (2020) 39:149. doi: 10.1186/s13046-020-01648-1
138. Luan W, Ding Y, Xi H, Ruan H, Lu F, Ma S, et al. Exosomal miR-106b-5p derived from melanoma cell promotes primary melanocytes epithelial-mesenchymal transition through targeting EphA4. J Exp Clin Cancer Res. (2021) 40:107. doi: 10.1186/s13046-021-01906-w
139. Paul I, Bolzan D, Youssef A, Gagnon KA, Hook H, Karemore G, et al. Parallelized multidimensional analytic framework applied to mammary epithelial cells uncovers regulatory principles in EMT. Nat Commun. (2023) 14:688. doi: 10.1038/s41467-023-36122-x
140. Zhou WJ, Zhang J, Xie F, Wu JN, Ye JF, Wang J, et al. CD45RO(-)CD8(+) T cell-derived exosomes restrict estrogen-driven endometrial cancer development via the ERbeta/miR-765/PLP2/Notch axis. Theranostics. (2021) 11:5330–45. doi: 10.7150/thno.58337
141. Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. (2010) 29:4741–51. doi: 10.1038/onc.2010.215
142. Milman N, Ginini L, Gil Z. Exosomes and their role in tumorigenesis and anticancer drug resistance. Drug Resist Update. (2019) 45:1–12. doi: 10.1016/j.drup.2019.07.003
143. Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. (2017) 141:220–30. doi: 10.1002/ijc.30669
144. Li S, Yi M, Dong B, Jiao Y, Luo S, Wu K. The roles of exosomes in cancer drug resistance and its therapeutic application. Clin Transl Med. (2020) 10:e257. doi: 10.1002/ctm2.257
145. Vella LJ, Behren A, Coleman B, Greening DW, Hill AF, Cebon J. Intercellular resistance to BRAF inhibition can be mediated by extracellular vesicle-associated PDGFRbeta. Neoplasia. (2017) 19:932–40. doi: 10.1016/j.neo.2017.07.002
146. Syn N, Wang L, Sethi G, Thiery JP, Goh BC. Exosome-mediated metastasis: from epithelial-mesenchymal transition to escape from immunosurveillance. Trends Pharmacol Sci. (2016) 37:606–17. doi: 10.1016/j.tips.2016.04.006
147. Rahman MA BJ, Lovat F, Gao M, Otterson GA, Nana-Sinkam P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget. (2016) 7:54852–66. doi: 10.18632/oncotarget.v7i34
148. Skandalis SS, Karalis TT, Chatzopoulos A, Karamanos NK. Hyaluronan-CD44 axis orchestrates cancer stem cell functions. Cell Signal. (2019) 63:109377. doi: 10.1016/j.cellsig.2019.109377
149. Nakamura K, Sawada K, Kinose Y, Yoshimura A, Toda A, Nakatsuka E, et al. Exosomes promote ovarian cancer cell invasion through transfer of CD44 to peritoneal mesothelial cells. Mol Cancer Res. (2017) 15:78–92. doi: 10.1158/1541-7786.MCR-16-0191
150. Li Y, Li Q, Li D, Gu J, Qian D, Qin X, et al. Exosome carrying PSGR promotes stemness and epithelial-mesenchymal transition of low aggressive prostate cancer cells. Life Sci. (2021) 264:118638. doi: 10.1016/j.lfs.2020.118638
151. Whiteside TL. The role of tumor-derived exosomes in epithelial mesenchymal transition (EMT). Transl Cancer Res. (2017) 6:S90–2. doi: 10.21037/tcr.2017.02.13
152. Jiao T HY, Sun H, Yang L. Exosomal lnc-CDHR derived from human umbilical cord mesenchymal stem cells attenuates peritoneal epithelial-mesenchymal transition through AKT/FOXO pathway. Aging (Albany NY). (2023) 15:6921–32. doi: 10.18632/aging.v15i14
153. Lin Q, Chen X, Meng F, Ogawa K, Li M, Song R, et al. ASPH-notch Axis guided Exosomal delivery of Prometastatic Secretome renders breast Cancer multi-organ metastasis. Mol Cancer. (2019) 18:156. doi: 10.1186/s12943-019-1077-0
154. Yao Y, Chen R, Wang G, Zhang Y, Liu F. Exosomes derived from mesenchymal stem cells reverse EMT via TGF-beta1/Smad pathway and promote repair of damaged endometrium. Stem Cell Res Ther. (2019) 10:225. doi: 10.1186/s13287-019-1332-8
155. Gao X RN, Dong X, Zuo B, Yang R, Zhou Q, Moulton HM, et al. Anchor peptide captures, targets, and loads exosomes of diverse origins for diagnostics and therapy. Sci Transl Med. (2018) 10:1–14. doi: 10.1126/scitranslmed.aat0195
156. Li Z, Zhou X, Wei M, Gao X, Zhao L, Shi R, et al. In vitro and in vivo RNA inhibition by CD9-huR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Lett. (2019) 19:19–28. doi: 10.1021/acs.nanolett.8b02689
157. Wang X, Chen Y, Zhao Z, Meng Q, Yu Y, Sun J, et al. Engineered exosomes with ischemic myocardium-targeting peptide for targeted therapy in myocardial infarction. J Am Heart Assoc. (2018) 7:e008737. doi: 10.1161/JAHA.118.008737
158. Wang C, Li N, Li Y, Hou S, Zhang W, Meng Z, et al. Engineering a HEK-293T exosome-based delivery platform for efficient tumor-targeting chemotherapy/internal irradiation combination therapy. J Nanobiotechnology. (2022) 20:247. doi: 10.1186/s12951-022-01462-1
159. Liang Y, Xu X, Xu L, Iqbal Z, Ouyang K, Zhang H, et al. Chondrocyte-specific genomic editing enabled by hybrid exosomes for osteoarthritis treatment. Theranostics. (2022) 12:4866–78. doi: 10.7150/thno.69368
160. Chen SY, Lin MC, Tsai JS, He PL, Luo WT, Herschman H, et al. EP(4) antagonist-elicited extracellular vesicles from mesenchymal stem cells rescue cognition/learning deficiencies by restoring brain cellular functions. Stem Cells Transl Med. (2019) 8:707–23. doi: 10.1002/sctm.18-0284
161. Elashiry M, Elsayed R, Elashiry MM, Rashid MH, Ara R, Arbab AS, et al. Proteomic characterization, biodistribution, and functional studies of immune-therapeutic exosomes: implications for inflammatory lung diseases. Front Immunol. (2021) 12:636222. doi: 10.3389/fimmu.2021.636222
162. Fang Y, Wu F, Shang G, Yin C. SCARA5 in bone marrow stromal cell-derived exosomes inhibits colorectal cancer progression by inactivating the PI3K/Akt pathway. Genomics. (2023) 115:110636. doi: 10.1016/j.ygeno.2023.110636
163. Hu W, Huang F, Ning L, Hao J, Wan J, Hao S. Enhanced immunogenicity of leukemia-derived exosomes via transfection with lentiviral vectors encoding costimulatory molecules. Cell Oncol (Dordr). (2020) 43:889–900. doi: 10.1007/s13402-020-00535-3
164. Iyaswamy A, Thakur A, Guan XJ, Krishnamoorthi S, Fung TY, Lu K, et al. Fe65-engineered neuronal exosomes encapsulating corynoxine-B ameliorate cognition and pathology of Alzheimer’s disease. Signal Transduct Target Ther. (2023) 8:404. doi: 10.1038/s41392-023-01657-4
165. Li K, Xue W, Lu Z, Wang S, Zheng J, Lu K, et al. Tumor-derived exosomal ADAM17 promotes pre-metastatic niche formation by enhancing vascular permeability in colorectal cancer. J Exp Clin Cancer Res. (2024) 43:59. doi: 10.1186/s13046-024-02991-3
166. Liu C, Liu D, Wang S, Gan L, Yang X, Ma C. Identification of the SNARE complex that mediates the fusion of multivesicular bodies with the plasma membrane in exosome secretion. J Extracell Vesicles. (2023) 12:e12356. doi: 10.1002/jev2.12356
167. Liu C, Xia C, Xia C. Biology and function of exosomes in tumor immunotherapy. BioMed Pharmacother. (2023) 169:115853. doi: 10.1016/j.biopha.2023.115853
168. Rossi F, Noren H, Jove R, Beljanski V, Grinnemo KH. Differences and similarities between cancer and somatic stem cells: therapeutic implications. Stem Cell Res Ther. (2020) 11:489. doi: 10.1186/s13287-020-02018-6
169. Yin P, Jiang Y, Fang X, Wang D, Li Y, Chen M, et al. Cell-based therapies for degenerative musculoskeletal diseases. Adv Sci (Weinh). (2023) 10:e2207050. doi: 10.1002/advs.202207050
170. Koh E, Lee EJ, Nam GH, Hong Y, Cho E, Yang Y, et al. Exosome-SIRPalpha, a CD47 blockade increases cancer cell phagocytosis. Biomaterials. (2017) 121:121–9. doi: 10.1016/j.biomaterials.2017.01.004
171. Cho E, Nam GH, Hong Y, Kim YK, Kim DH, Yang Y, et al. Comparison of exosomes and ferritin protein nanocages for the delivery of membrane protein therapeutics. J Control Release. (2018) 279:326–35. doi: 10.1016/j.jconrel.2018.04.037
172. Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. (2011) 29:341–5. doi: 10.1038/nbt.1807
173. Lin Y, Yan M, Bai Z, Xie Y, Ren L, Wei J, et al. Huc-MSC-derived exosomes modified with the targeting peptide of aHSCs for liver fibrosis therapy. J Nanobiotechnology. (2022) 20:432. doi: 10.1186/s12951-022-01636-x
174. Liu Q, Wu J, Wang H, Jia Z, Li G. Human infrapatellar fat pad mesenchymal stem cell-derived extracellular vesicles purified by anion exchange chromatography suppress osteoarthritis progression in a mouse model. Clin Orthop Relat Res. (2024). 482:1246-62. doi: 10.1097/CORR.0000000000003067
175. Gao D, Shang J, Sun R, Shi Y, Jiang H, Ma M, et al. Changes in the morphology, number, and protein levels of plasma exosomes in CADASIL patients. J Alzheimers Dis. (2021) 81:221–9. doi: 10.3233/JAD-210101
176. Ala M. The beneficial effects of mesenchymal stem cells and their exosomes on myocardial infarction and critical considerations for enhancing their efficacy. Ageing Res Rev. (2023) 89:101980. doi: 10.1016/j.arr.2023.101980
177. Cook N, Hansen AR, Siu LL, Abdul Razak AR. Early phase clinical trials to identify optimal dosing and safety. Mol Oncol. (2015) 9:997–1007. doi: 10.1016/j.molonc.2014.07.025
178. Fusco C, De Rosa G, Spatocco I, Vitiello E, Procaccini C, Frige C, et al. Extracellular vesicles as human therapeutics: A scoping review of the literature. J Extracell Vesicles. (2024) 13:e12433. doi: 10.1002/jev2.12433
179. Barile L, Marban E. Injury minimization after myocardial infarction: focus on extracellular vesicles. Eur Heart J. (2024) 45:1602–9. doi: 10.1093/eurheartj/ehae089
180. Nakamori Y, Park EJ, Shimaoka M. Immune deregulation in sepsis and septic shock: reversing immune paralysis by targeting PD-1/PD-L1 pathway. Front Immunol. (2020) 11:624279. doi: 10.3389/fimmu.2020.624279
181. Awal HB, Nandula SR, Domingues CC, Dore FJ, Kundu N, Brichacek B, et al. Linagliptin, when compared to placebo, improves CD34+ve endothelial progenitor cells in type 2 diabetes subjects with chronic kidney disease taking metformin and/or insulin: a randomized controlled trial. Cardiovasc Diabetol. (2020) 19:72. doi: 10.1186/s12933-020-01046-z
182. Propper DJ, Gao F, Saunders MP, Sarker D, Hartley JA, Spanswick VJ, et al. PANTHER: AZD8931, inhibitor of EGFR, ERBB2 and ERBB3 signalling, combined with FOLFIRI: a Phase I/II study to determine the importance of schedule and activity in colorectal cancer. Br J Cancer. (2023) 128:245–54. doi: 10.1038/s41416-022-02015-x
183. Achour B, Al-Majdoub ZM, Grybos-Gajniak A, Lea K, Kilford P, Zhang M, et al. Liquid biopsy enables quantification of the abundance and interindividual variability of hepatic enzymes and transporters. Clin Pharmacol Ther. (2021) 109:222–32. doi: 10.1002/cpt.2102
184. Allegra A, Petrarca C, Di Gioacchino M, Casciaro M, Musolino C, Gangemi S. Exosome-mediated therapeutic strategies for management of solid and hematological Malignancies. Cells. (2022) 11:1128-60. doi: 10.3390/cells11071128
185. Johnson J, Law SQK, Shojaee M, Hall AS, Bhuiyan S, Lim MBL, et al. First-in-human clinical trial of allogeneic, platelet-derived extracellular vesicles as a potential therapeutic for delayed wound healing. J Extracell Vesicles. (2023) 12:e12332. doi: 10.1002/jev2.12332
186. Hu D, Yang R, Wang G, Li H, Fan X, Liang G. Emerging strategies to overcome current CAR-T therapy dilemmas - exosomes derived from CAR-T cells. Int J Nanomedicine. (2024) 19:2773–91. doi: 10.2147/IJN.S445101
187. Cheng B, Li C, Li J, Gong L, Liang P, Chen Y, et al. The activity and immune dynamics of PD-1 inhibition on high-risk pulmonary ground glass opacity lesions: insights from a single-arm, phase II trial. Signal Transduct Target Ther. (2024) 9:93. doi: 10.1038/s41392-024-01799-z
188. Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology. (2016) 5:e1071008. doi: 10.1080/2162402X.2015.1071008
189. Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, et al. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. (2008) 16:782–90. doi: 10.1038/mt.2008.1
190. Poinsot V, Pizzinat N, Ong-Meang V. Engineered and mimicked extracellular nanovesicles for therapeutic delivery. Nanomaterials (Basel). (2024) 14:639-71. doi: 10.3390/nano14070639
191. Aljuhani A, Albalawi O, Albalawi R, Alsalama R, Alatawi S, Altemani R, et al. Exosomes in COVID-19 infection: Focus on role in diagnosis, pathogenesis, immunity, and clinical trials. Cell Biol Int. (2023) 47:1049–67. doi: 10.1002/cbin.12014
192. Khan NA, Asim M, Biswas KH, Alansari AN, Saman H, Sarwar MZ, et al. Exosome nanovesicles as potential biomarkers and immune checkpoint signaling modulators in lung cancer microenvironment: recent advances and emerging concepts. J Exp Clin Cancer Res. (2023) 42:221. doi: 10.1186/s13046-023-02753-7
193. Vrablova V, Kosutova N, Blsakova A, Bertokova A, Kasak P, Bertok T, et al. Glycosylation in extracellular vesicles: Isolation, characterization, composition, analysis and clinical applications. Biotechnol Adv. (2023) 67:108196. doi: 10.1016/j.biotechadv.2023.108196
194. Treton G, Sayer C, Schurz M, Jaritsch M, Muller A, Matea CT, et al. Quantitative and functional characterisation of extracellular vesicles after passive loading with hydrophobic or cholesterol-tagged small molecules. J Control Release. (2023) 361:694–716. doi: 10.1016/j.jconrel.2023.08.010
195. Vahabi M, Comandatore A, Centra C, Blandino G, Morelli L, Giovannetti E. Thinking small to win big? A critical review on the potential application of extracellular vesicles for biomarker discovery and new therapeutic approaches in pancreatic cancer. Semin Cancer Biol. (2023) 97:50–67. doi: 10.1016/j.semcancer.2023.11.003
196. Zhang Z, Wang C, Li T, Liu Z, Li L. Comparison of ultracentrifugation and density gradient separation methods for isolating Tca8113 human tongue cancer cell line-derived exosomes. Oncol Lett. (2014) 8:1701–6. doi: 10.3892/ol.2014.2373
197. Brennan K, Martin K, FitzGerald SP, O’Sullivan J, Wu Y, Blanco A, et al. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci Rep. (2020) 10:1039. doi: 10.1038/s41598-020-57497-7
198. Brett SI, Lucien F, Guo C, Williams KC, Kim Y, Durfee PN, et al. Immunoaffinity based methods are superior to kits for purification of prostate derived extracellular vesicles from plasma samples. Prostate. (2017) 77:1335–43. doi: 10.1002/pros.23393
199. Morales-Kastresana A, Musich TA, Welsh JA, Telford W, Demberg T, Wood JCS, et al. High-fidelity detection and sorting of nanoscale vesicles in viral disease and cancer. J Extracell Vesicles. (2019) 8:1597603. doi: 10.1080/20013078.2019.1597603
200. Kuo WP. Extracellular vesicles methods and protocols. Methods Mol Biol. (2017) 1660:1–441. doi: 10.1007/978-1-4939-7253-1
201. Palmieri V, Lucchetti D, Gatto I, Maiorana A, Marcantoni M, Maulucci G, et al. Dynamic light scattering for the characterization and counting of extracellular vesicles: a powerful noninvasive tool. J Nanoparticle Res. (2014) 16:2583-90. doi: 10.1007/s11051-014-2583-z
202. Gamez-Valero A, Monguio-Tortajada M, Carreras-Planella L, Franquesa M, Beyer K, Borras FE. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci Rep. (2016) 6:33641. doi: 10.1038/srep33641
203. Busatto S, Vilanilam G, Ticer T, Lin WL, Dickson DW, Shapiro S, et al. Tangential flow filtration for highly efficient concentration of extracellular vesicles from large volumes of fluid. Cells. (2018) 7:273-83. doi: 10.3390/cells7120273
204. Paterna A, Rao E, Adamo G, Raccosta S, Picciotto S, Romancino D, et al. Isolation of extracellular vesicles from microalgae: A renewable and scalable bioprocess. Front Bioeng Biotechnol. (2022) 10:836747. doi: 10.3389/fbioe.2022.836747
205. Momen-Heravi F. Isolation of extracellular vesicles by ultracentrifugation. Methods Mol Biol. (2017) 1660:25–32. doi: 10.1007/978-1-4939-7253-1_3
206. Gardiner C, Di Vizio D, Sahoo S, Thery C, Witwer KW, Wauben M, et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. (2016) 5:32945. doi: 10.3402/jev.v5.32945
207. Takov K, Yellon DM, Davidson SM. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: yield, purity and functional potential. J Extracell Vesicles. (2019) 8:1560809. doi: 10.1080/20013078.2018.1560809
208. Musante L, Tataruch DE, Holthofer H. Use and isolation of urinary exosomes as biomarkers for diabetic nephropathy. Front Endocrinol (Lausanne). (2014) 5:149. doi: 10.3389/fendo.2014.00149
209. Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip. (2012) 12:5202–10. doi: 10.1039/c2lc41006k
210. Rashidi M, Bijari S, Khazaei AH, Shojaei-Ghahrizjani F, Rezakhani L. The role of milk-derived exosomes in the treatment of diseases. Front Genet. (2022) 13:1009338. doi: 10.3389/fgene.2022.1009338
211. Gehrmann U, Naslund TI, Hiltbrunner S, Larssen P, Gabrielsson S. Harnessing the exosome-induced immune response for cancer immunotherapy. Semin Cancer Biol. (2014) 28:58–67. doi: 10.1016/j.semcancer.2014.05.003
212. Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. (2014) 14:195–208. doi: 10.1038/nri3622
213. Kurywchak P, Tavormina J, Kalluri R. The emerging roles of exosomes in the modulation of immune responses in cancer. Genome Med. (2018) 10:23. doi: 10.1186/s13073-018-0535-4
214. Ayala-Mar S, Donoso-Quezada J, Gonzalez-Valdez J. Clinical implications of exosomal PD-L1 in cancer immunotherapy. J Immunol Res. (2021) 2021:8839978. doi: 10.1155/2021/8839978
215. Fan Y, Che X, Qu J, Hou K, Wen T, Li Z, et al. Exosomal PD-L1 retains immunosuppressive activity and is associated with gastric cancer prognosis. Ann Surg Oncol. (2019) 26:3745–55. doi: 10.1245/s10434-019-07431-7
216. Huang SH, Li Y, Zhang J, Rong J, Ye S. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest. (2013) 31:330–5. doi: 10.3109/07357907.2013.789905
217. Lin WC, Lin YT, Chao HC, Lin YY, Hwang WL. Haematopoietic cell-derived exosomes in cancer development and therapeutics: From basic science to clinical practice. Clin Transl Med. (2023) 13:e1448. doi: 10.1002/ctm2.1448
218. Liu J, Liu J, Qin G, Li J, Fu Z, Li J, et al. MDSCs-derived GPR84 induces CD8(+) T-cell senescence via p53 activation to suppress the antitumor response. J Immunother Cancer. (2023) 11. doi: 10.1136/jitc-2023-007802
219. Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, et al. Suppression of exosomal PD-L1 induces systemic anti-tumor immunity and memory. Cell. (2019) 177:414–427.e413. doi: 10.1016/j.cell.2019.02.016
220. Qin S, Xu L, Yi M, Yu S, Wu K, Luo S. Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol Cancer. (2019) 18:155. doi: 10.1186/s12943-019-1091-2
221. Xing C, Li H, Li RJ, Yin L, Zhang HF, Huang ZN, et al. The roles of exosomal immune checkpoint proteins in tumors. Mil Med Res. (2021) 8:56. doi: 10.1186/s40779-021-00350-3
222. Timaner M, Kotsofruk R, Raviv Z, Magidey K, Shechter D, Kan T, et al. Microparticles from tumors exposed to radiation promote immune evasion in part by PD-L1. Oncogene. (2020) 39:187–203. doi: 10.1038/s41388-019-0971-7
223. Del Re M, Marconcini R, Pasquini G, Rofi E, Vivaldi C, Bloise F, et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br J Cancer. (2018) 118:820–4. doi: 10.1038/bjc.2018.9
224. Ning Y, Shen K, Wu Q, Sun X, Bai Y, Xie Y, et al. Tumor exosomes block dendritic cells maturation to decrease the T cell immune response. Immunol Lett. (2018) 199:36–43. doi: 10.1016/j.imlet.2018.05.002
225. Lu Y, Ye L, Jian X, Yang D, Zhang H, Tong Z, et al. Integrated microfluidic system for isolating exosome and analyzing protein marker PD-L1. Biosens Bioelectron. (2022) 204:113879. doi: 10.1016/j.bios.2021.113879
226. Li M, Soder R, Abhyankar S, Abdelhakim H, Braun MW, Trinidad CV, et al. WJMSC-derived small extracellular vesicle enhance T cell suppression through PD-L1. J Extracellular Vesicles. (2021) 10. doi: 10.1002/jev2.12067
227. Yang Y, Li CW, Chan LC, Wei Y, Hsu JM, Xia W, et al. Exosomal PD-L1 harbors active defense function to suppress T cell killing of breast cancer cells and promote tumor growth. Cell Res. (2018) 28:862–4. doi: 10.1038/s41422-018-0060-4
228. Zhang W, Zhong W, Wang B, Yang J, Yang J, Yu Z, et al. ICAM-1-mediated adhesion is a prerequisite for exosome-induced T cell suppression. Dev Cell. (2022) 57:329–343.e327. doi: 10.1016/j.devcel.2022.01.002
229. Thery C, Duban L, Segura E, Veron P, Lantz O, Amigorena S. Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat Immunol. (2002) 3:1156–62. doi: 10.1038/ni854
230. Andre F, Chaput N, Schartz NE, Flament C, Aubert N, Bernard J, et al. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J Immunol. (2004) 172:2126–36. doi: 10.4049/jimmunol.172.4.2126
231. Yang J, Zhang X, Liu C, Wang Z, Deng L, Feng C, et al. Biologically modified nanoparticles as theranostic bionanomaterials. Prog Materials Sci. (2021) 118. doi: 10.1016/j.pmatsci.2020.100768
232. Wu J-Y, Li Y-J, Wang J, Hu X-B, Huang S, Luo S, et al. Multifunctional exosome-mimetics for targeted anti-glioblastoma therapy by manipulating protein corona. J Nanobiotechnology. (2021) 19:405-19. doi: 10.1186/s12951-021-01153-3
233. Toth EA, Turiak L, Visnovitz T, Cserep C, Mazlo A, Sodar BW, et al. Formation of a protein corona on the surface of extracellular vesicles in blood plasma. J Extracell Vesicles. (2021) 10:e12140. doi: 10.1002/jev2.12140
234. Heidarzadeh M, Zarebkohan A, Rahbarghazi R, Sokullu E. Protein corona and exosomes: new challenges and prospects. Cell Commun Signal. (2023) 21:64. doi: 10.1186/s12964-023-01089-1
235. Chiesa M, Garg J, Kang YT, Chen G. Thermal conductivity and viscosity of water-in-oil nanoemulsions. Colloids Surfaces A: Physicochemical Eng Aspects. (2008) 326:67–72. doi: 10.1016/j.colsurfa.2008.05.028
236. Park DJ, Yun WS, Kim WC, Park JE, Lee SH, Ha S, et al. Improvement of stem cell-derived exosome release efficiency by surface-modified nanoparticles. J Nanobiotechnology. (2020) 18:178. doi: 10.1186/s12951-020-00739-7
237. Giovannelli L, Bari E, Jommi C, Tartara F, Armocida D, Garbossa D, et al. Mesenchymal stem cell secretome and extracellular vesicles for neurodegenerative diseases: Risk-benefit profile and next steps for the market access. Bioact Mater. (2023) 29:16–35. doi: 10.1016/j.bioactmat.2023.06.013
Keywords: exosomes, membrane proteins, functions, applications, immunoregulation
Citation: Xu F, Luo S, Lu P, Cai C, Li W and Li C (2024) Composition, functions, and applications of exosomal membrane proteins. Front. Immunol. 15:1408415. doi: 10.3389/fimmu.2024.1408415
Received: 28 March 2024; Accepted: 15 July 2024;
Published: 01 August 2024.
Edited by:
Muhammad Babar Khawar, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Paul Engeroff, Bern University Hospital, SwitzerlandAnish Chakkumkal, Pharmaceutical Companies of Johnson and Johnson, Netherlands
Copyright © 2024 Xu, Luo, Lu, Cai, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Cai, ZmFuZ3plY2FpQDEyNi5jb20=; Weihua Li, bGl3ZWlodWFAY2NtdS5lZHUuY24=; Chuanyun Li, bGljaHVhbnkwMzg4QDE2My5jb20=
 Fang Xu
Fang Xu Shumin Luo
Shumin Luo Pengpeng Lu
Pengpeng Lu Chao Cai2*
Chao Cai2* Weihua Li
Weihua Li