- 1Graduate School, Shanghai University of Traditional Chinese Medicine, Shanghai, China
- 2Department of Breast Surgery, Yueyang Hospital of Integrated Traditional Chinese and Western Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, China
Objectives: Previous studies have reported that immunoinflammatory responses have associations with mastitis. Here, we aimed to further figure out whether circulating inflammatory cytokines and immune cells causally impact mastitis liability.
Methods: The two-sample Mendelian randomization made use of genetic variances of 91 inflammatory cytokines from a large publicly available genome-wide association study (GWAS) containing 14,824 participants, 731 immunophenotypes data from 3,757 individuals as exposures separately, and mastitis from a GWAS summary (1880 cases and 211699 controls of European ancestry) as outcome. The primary analysis applied the inverse-variance weighted (IVW) method to estimate causal influences, with MR-Egger, weighted median, weighted mode and simple mode as supplementary approaches. Heterogeneity and pleiotropy were evaluated by the Cochrane Q test, MR-Egger intercept test, and MR-PRESSO global test.
Results: The results indicated that CX3CL1 may be suggestively relevant to the risk of mastitis (odds ratio, OR = 1.434, 95% CI = 1.142~1.800, p = 0.002). Moreover, three immunophenotypes were identified as having a potential causal link to mastitis (p < 0.05). Significantly, CD28- CD8dim %CD8dim (OR = 1.058, 95% CI = 1.024 ~ 1.093, p = 0.0006) and CD45 on CD33br HLA DR+ (OR = 1.097, 95% CI = 1.039 ~ 1.157, p = 0.0008) were found to induce mastitis possibly. Conversely, CD39+ secreting Treg AC (OR = 0.929, 95% CI = 0.884~ 0.978, p = 0.005) pertained to protective factors of mastitis. Cochran’s Q test and MR-Egger intercept test indicated no significant heterogeneity (p > 0.05) or pleiotropy (p > 0.05), supporting the robustness and reliability of our findings.
Conclusion: Our study adds to current knowledge on the causal roles of inflammatory cytokines and immune cells on mastitis by genetic means, thus guiding future clinical research.
1 Introduction
Mastitis is a painful benign breast disease that can be categorized into lactational and non-lactational mastitis. Lactational mastitis (LM) is the most common inflammatory disease of the mammary gland in primiparous women, usually occurring 3-4 weeks postpartum. Global incidence ranges from 3% to 33%, affecting breastfeeding and fetal health (1, 2). Mastitis can also occur in non-breastfeeding women, and rarely in men. Non-lactational mastitis is refractory including periductal mastitis (PDM) and granulomatous lobular mastitis (GLM) (2).
LM happens mostly due to milk stasis or nipple damage, bacteria that colonize the skin invade breasts along the lymphatic and milk duct, and subsequent infections in stagnant lactiferous ducts progress with Staphylococcus aureus being the main pathogen (3). Generally, 5% to 9% of reproductive-aged women will have PDM which often influences the subareolar ducts. The cause of periductal mastitis is still unclear while mammary ductal ectasia, plasma cell infiltration and abscess formation have been implicated as basic pathogenesis (4). In addition, more than one study shows immune responses may play a role (5). Alternatively, GLM is a rare autoimmune inflammatory disorder prone to recur that can be clinically similar to breast cancer with the highest risk in parous women ordinarily within 5 years of parturition (6). The etiology of GLM remains ambiguous while Corynebacterium infection may be closely related to the pathogenesis of GLM (7). The most widely accepted theory indicates that specific triggers such as trauma, bacteria, or exosmotic milk induce autoimmune destruction, giving rise to leakage of ductal secretions into the breast tissue and inflammatory cell infiltration, resulting in a granulomatous response. In such cases, patients may benefit from immunosuppressive therapy (8).
While clinical manifestations and etiology of these disorders are with great heterogeneity, inflammation and immune response appear to be the commonalities in the occurrence and development of such diseases, and their causal relationship remains to be verified. Mendelian randomization (MR) analyses aim to investigate causal correlations between exposures and outcomes via genetic variation such as single nucleotide polymorphisms (SNPs) as instrumental variables (IVs), making them less susceptible to environmental confounders and reverse causation (9). MR uses genetic variants as proxies for exposures to assess causal relationships, minimizing confounding and reverse causation. Although MR has been widely used to study the association between inflammatory factors and immune system disorders (10, 11), evidence on the causality of inflammatory cytokines and immune cells with inflammatory disorders of the breast remains limited. It is unclear whether they play a causal role or are merely a consequence of shared risk factors. However, the causal role of these factors in mastitis has not been fully established. To identify circulating inflammatory cytokines and immune cells as potential risk factors for mastitis, this study conducted a comprehensive two-sample Mendelian randomization (TSMR) analysis which explored their causal connection, providing insights into the role of inflammatory cytokines and immune cells in pathogenesis and potential therapeutic interventions of mastitis.
2 Materials and methods
2.1 Study design
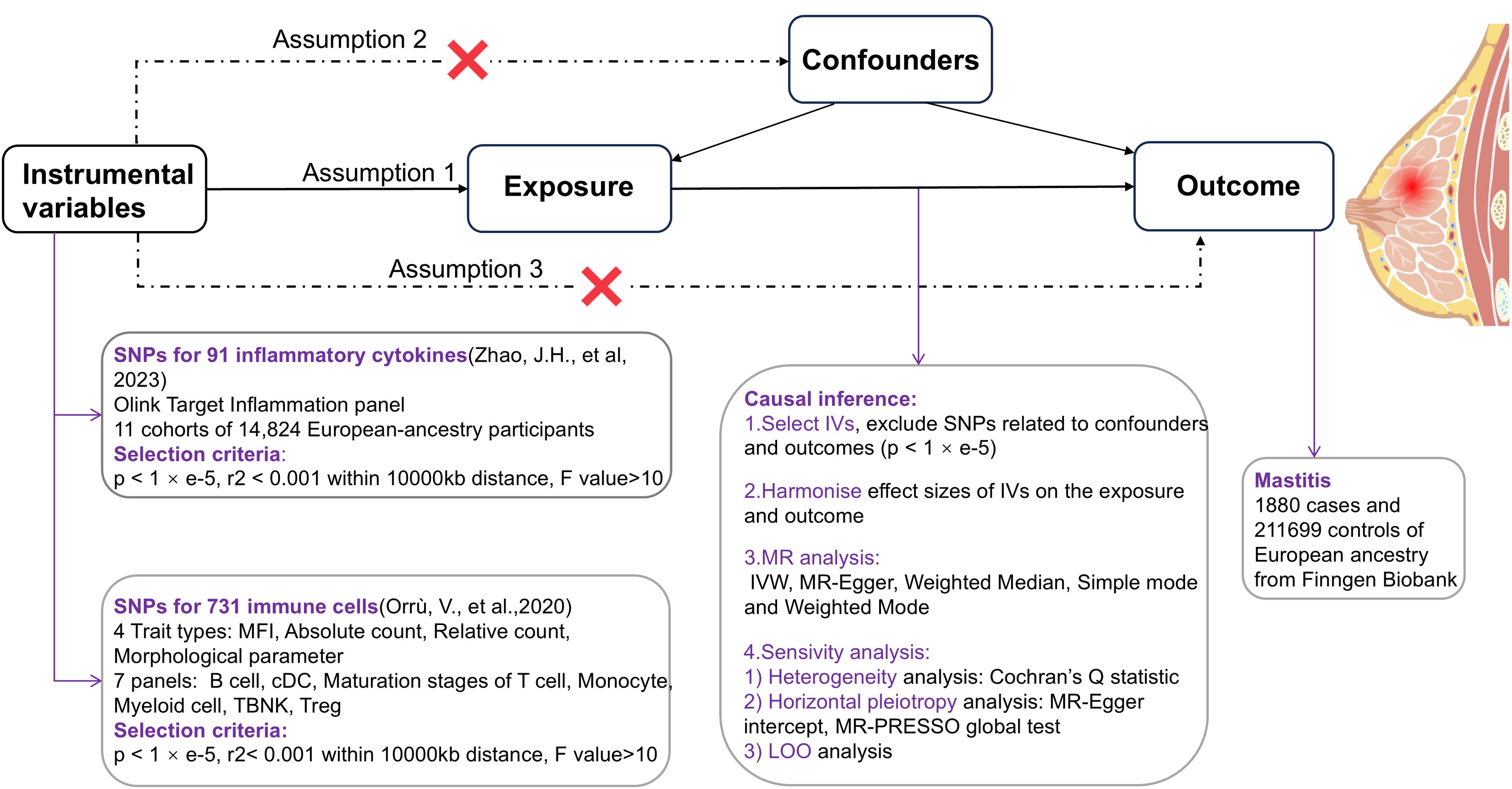
This study employed a TSMR approach, with circulating inflammatory cytokines and immune cells as exposure factors separately, their genetic variant SNPs as IVs respectively, and mastitis as an outcome factor, to examine the causal relationship between exposure and outcome predicted by genes (12). In order to provide a reasonable interpretation of MR analyses, the following 3 core assumptions need to be met (13): ①Relevance - IVs are strongly associated with exposure factors, ②Independence - IVs are uncorrelated with confounders affecting both exposure and outcome, and ③Exclusivity - genetic variant SNPs affect outcomes solely through exposure (Figure 1).
2.2 Data sources
Genetic information on inflammatory factors is derived from the largest known meta-analysis of genome-wide association studies (GWAS) published in 2023 including 11 cohorts of total 14,824 European-ancestry participants (14). Results were meta-analyzed to identify significant protein quantitative trait loci (pQTLs) across 91 plasma proteins using the Olink Target Inflammation panel. 180 significant pQTLs involving 70 proteins were identified, with 33% of these being cis and 67% trans. Additionally, conditional analyses revealed 47 independent pQTL signals, increasing the total number of pQTLs from 180 to 227 (99 cis and 128 trans). Significant pQTL (p ≤5 × 10−10, fixed-effect meta-analysis) findings were validated in an independent cohort (ARISTOTLE) of 1,585 individuals. The analysis involved linear regression of cytokine levels against SNPs, adjusted for age, sex, and BMI, to identify single-variant associations (11). Besides, 72 out of 91 proteins were evaluated for replication in 35,556 participants in the deCODE study, using the aptamer-based SomaScan platform. Overall, 126 (71%) of the 178 testable pQTLs were replicated in either ARISTOTLE or deCODE. The data are publicly available at the website: https://www.phpc.cam.ac.uk/ceu/proteins and the EBI GWAS Catalog (accession numbers GCST90274758 to GCST90274848).
Summary statistics for 731 immune traits are also publicly available in the GWAS catalog database (accession numbers from GCST0001391 to GCST0002121). Among the 731 immune cells, 118 represent absolute cell counts, 389 reflect median fluorescence intensity (MFI), 32 are morphological parameters and 192 pertain to relative cell counts. The initial genome-wide association study on immune traits was conducted using data from 3,757 individuals of European descent, with no overlap between cohorts. A comprehensive set of approximately 22 million markers was genotyped using high-density arrays and subsequently imputed with a Sardinian sequence-based reference panel (15). The study discovered 122 independent signals for 459 cell traits at 70 loci (53 of them novel), revealing molecules and mechanisms involved in cell regulation with significant (p < 1.28 × 10-11) associations. Associations were assessed after adjusting for covariates such as sex, age, and age squared (16).
Genetic information of mastitis as an outcome factor comes from Finngen Biobank (https://r10.finngen.fi), which contains 1880 cases of inflammatory breast disease and 211699 controls of European ancestry (17).
2.3 Selection of instrumental variables
To further screen SNPs with strong correlative associations with inflammatory cytokines as exposed IVs, Firstly, we employed a loose genome-wide significant threshold to enhance the number of available SNPs (p < 1 × e-5) (18). Next, to exclude linkage disequilibrium (LD) and obtain independent IVs, we performed a clumping cutoff (r2 < 0.001 within a 10000-kb distance) (19). Meanwhile, SNPs strongly related to immune cells were selected when the following criteria were satisfied (p < 1 × e-5, r2< 0.001, kb=10000) (20). Then incompatible and palindromic SNPs were eliminated due to the inability to determine whether IV is oriented in the same direction during the harmonization process of the exposure and outcome data (21). After that, for the purpose of erasing the bias of weak IVs, the F value of each SNP was calculated by referring to the formula of F = beta2exposure/se2 exposure and R2 = 2× (1- MAF) ×MAF×beta2 (22) and finally, SNPs with F<10 were removed (23). Additionally, potential pleiotropic confounders were recognized by the Phenoscanner database (http://www.phenoscanner.medschl.cam.ac.uk/) (24).
2.4 Statistical analysis
The main analytical method used to determine the causal relationship between different circulating inflammatory factors and immune cells with mastitis is Inverse variance weighting (IVW) (25). MR-Egger, Weighted Median, Simple mode and Weighted Mode, as a supplement to IVW, can also be utilized to estimate causality (26, 27). It is worth noting that if these methods yield inconsistent results, we will prioritize IVW as the primary outcome. IVW is the major method commonly used in MR studies. It combines all the Wald ratios for each SNP to obtain a pooled estimate. Due to its robustness in providing consistent estimates when all instruments are valid, IVW was chosen as the primary method in this study. Potential heterogeneity was gauged by Cochran’s Q test with IVW and MR-Egger, and pleiotropy was assessed by the MR-PRESSO method and intercept of MR Egger test where all p values exceeding 0.05 signified the absence of both heterogeneity and pleiotropy (28, 29). Simultaneously, a Leave-one-out analysis was applied to evaluate the effect of a single SNP on sensitivity after removing SNPs one by one.
In this study, the odds ratio (OR) and 95% CI were presented. R version 4.3.2, R studio software and R package “TwoSampleMR (version 0.5.8)” were performed for the above analyses with the test criterion α = 0.05. To reduce the interference of correlation-level pleiotropy on the results, we searched the PhenoScanner V2 website to identify and rule out SNPs that were significantly associated (p < 1 × e-8) at the genome-wide level with shared risk factors for mastitis (hyperprolactinemia, nipple injury, smoking, bacterial infection et al.), and to re-run causal inference (1, 5, 30, 31).
3 Results
3.1 Influence of 91 inflammatory cytokines on mastitis
After selecting and coordinating IVs, we used a total of 30 SNPs for MR analysis. All SNPS had F statistics above 10 (ranging from 19.68 to 47.57) and were independent of relevant risk factors, suggesting that they were a powerful instrumental variable (The harmonized data are presented in Supplementary Table 1).
The causal effect estimates of 91 circulating inflammatory cytokines on mastitis susceptibility are summarized in Figure 2. Employing IVW methodology as the primary analytical method, it was revealed that a genetically projected higher abundance of Fractalkine levels (OR: 1.434, 95% CI:1.142~1.800, p = 0.002) was linked to an increased mastitis risk and Weighted Median method also proved the trend (OR: 1.394, 95% CI:1.044~1.862, p = 0.024). Figure 3A illustrates the scatterplot depicting circulating inflammatory cytokines’ causal effects on mastitis. Meanwhile, the sensitivity analyses in this study did not provide any evidence of heterogeneity, based on Cochran’s Q test for CX3CL1 using the IVW (p = 0.156) or MR Egger (p = 0.133) methods (Table 1). Besides, the leave-one-out method did not identify cases where a single SNP significantly influenced the observed outcomes (Figure 3B). What’s more, as shown in Table 1, both the MR-Egger intercept test (p = 0.675) and the MR PRESSO global test (p = 0.176) confirmed the absence of horizontal pleiotropy.
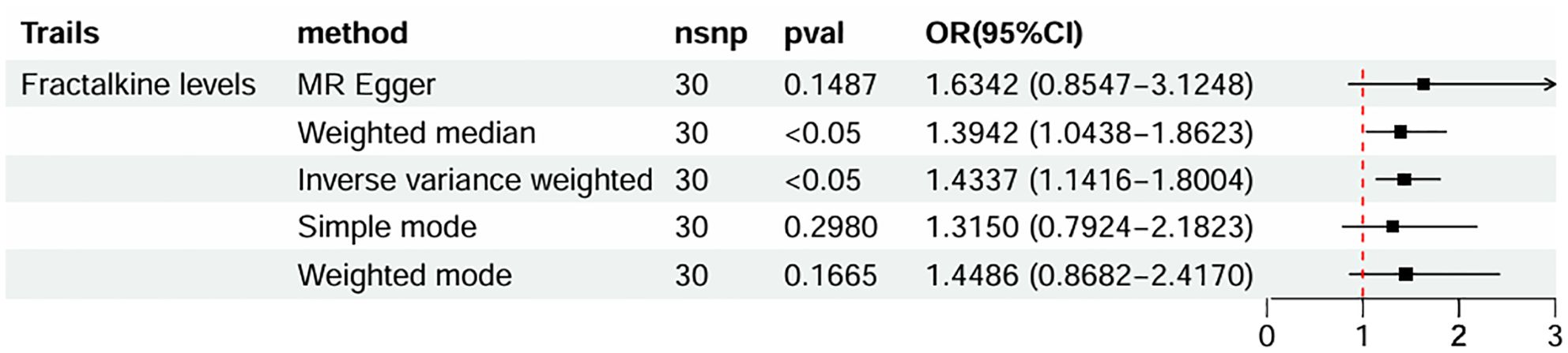
Figure 2. Forest plots of MR results of inflammatory cytokines on mastitis. MR, Mendelian Randomization; nsnp, number of SNPs; OR, odds ratio; CI, confidence interval.
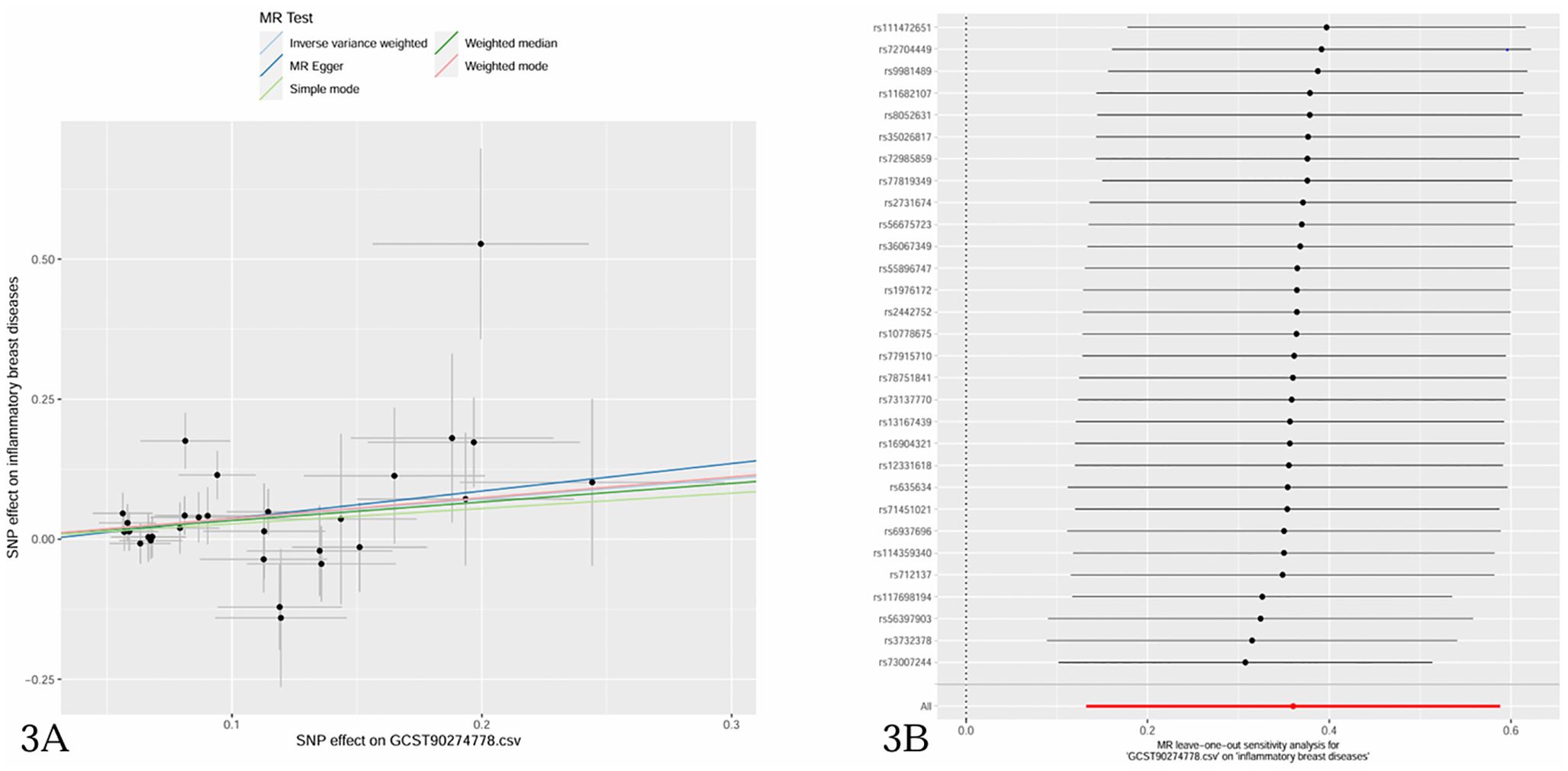
Figure 3. (A) Scatter plots of causal estimates of inflammatory cytokines (Fractalkine levels-CX3CL1) on mastitis. (B) Leave-one-out stability tests causal estimates of inflammatory cytokines (Fractalkine levels-CX3CL1) on mastitis.
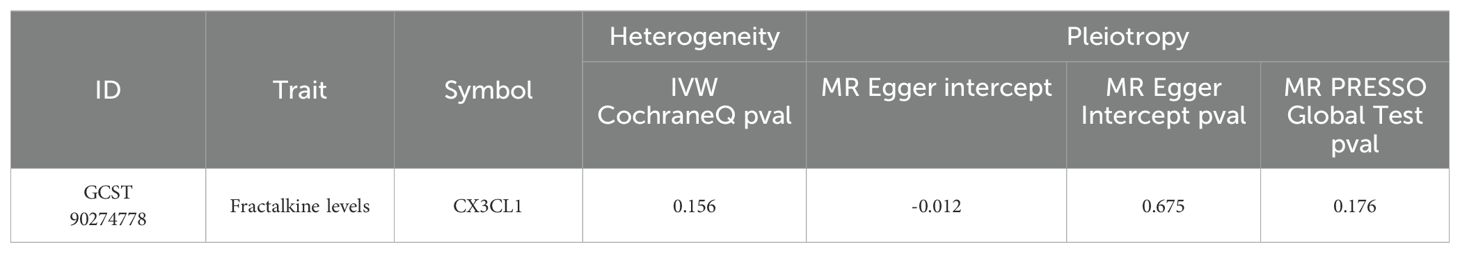
Table 1. Results of the heterogeneity and horizontal pleiotropy for the effect of inflammatory cytokines on mastitis.
3.2 Influence of 731 immunophenotypes on mastitis
The study examined the causal effects of 731 peripheral immune cells on mastitis, as outlined in Supplementary Table 2. Utilizing 57 independent genome-wide significant SNPs as instrumental variables for immunophenotypes, with F-statistics ranging from 19.62 to 2198.81, Mendelian randomization analysis indicated that three immune cells exhibited potential causal relationships with mastitis (p < 0.05), with two showing an increased risk and one showing a negative association. The primary findings of the main Mendelian randomization analyses are depicted in Figure 4, while scatterplots illustrating the causal relationships of immune cells on mastitis are presented in Figures 5A–C. These three immune cells are distributed across three trait types and two panels: relative cell count (Treg panel), absolute cell count (Treg panel), and MFI count (Myeloid cell panel). The respective immunophenotypes were as follows. Strikingly, elevated levels of CD28- CD8dim %CD8dim (OR = 1.058, 95% CI = 1.024 ~ 1.093, p = 0.0006) and CD45 on CD33br HLA DR+ (OR = 1.097, 95% CI = 1.039 ~ 1.157, p = 0.0008) could induce the risk of mastitis. Consistent evidence supported the causal effects of CD28- CD8dim %CD8dim on mastitis across MR Egger (OR = 1.047, 95% CI = 1.013~ 1.082, p = 0.018), Weighted median (OR = 1.051, 95% CI = 1.013~ 1.090, p = 0.009) and Weighted mode methods (OR = 1.054, 95% CI = 1.017~ 1.092, p = 0.011). As to CD45 on CD33br HLA DR+, positive associations with a higher risk of mastitis were also observed by MR Egger (OR = 1.086, 95% CI = 1.014~ 1.163, p = 0.034), Weighted median (OR = 1.099, 95% CI = 1.016~ 1.188, p = 0.018) and Weighted mode methods (OR = 1.100, 95% CI = 1.027~ 1.179, p = 0.016). As shown in Figure 5A, genetically determined higher levels of CD39+ secreting Treg AC (OR = 0.929, 95% CI = 0.884~ 0.978, p = 0.005) were suggestively associated with lower odds of mastitis. Furthermore, neither heterogeneity nor pleiotropy was observed according to the results (Table 2). To detect whether any single instrument disproportionately influenced results, we conducted a leave-one-out analysis by systematically removing each SNP and repeating the MR. No high influence points driving the pooled IVW estimates were recognized in the Leave-one-out analysis (Figures 5D–F).
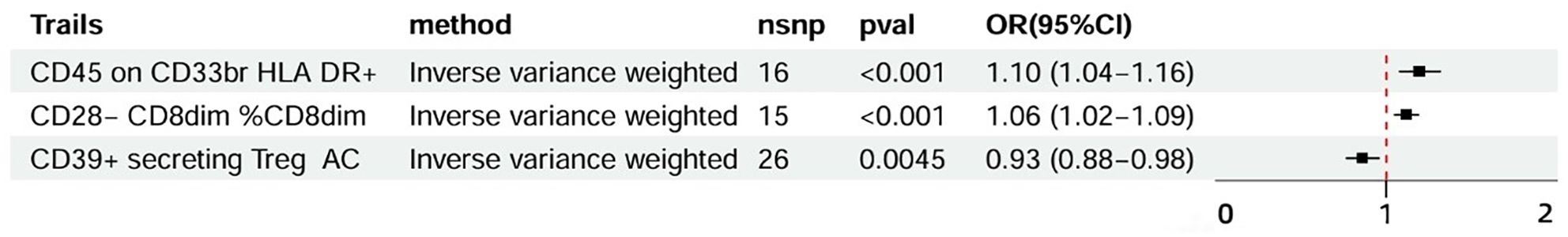
Figure 4. Forest plots of MR results of immune cells on mastitis. MR, Mendelian Randomization; nsnp, number of SNPs; OR, odds ratio; CI, confidence interval.
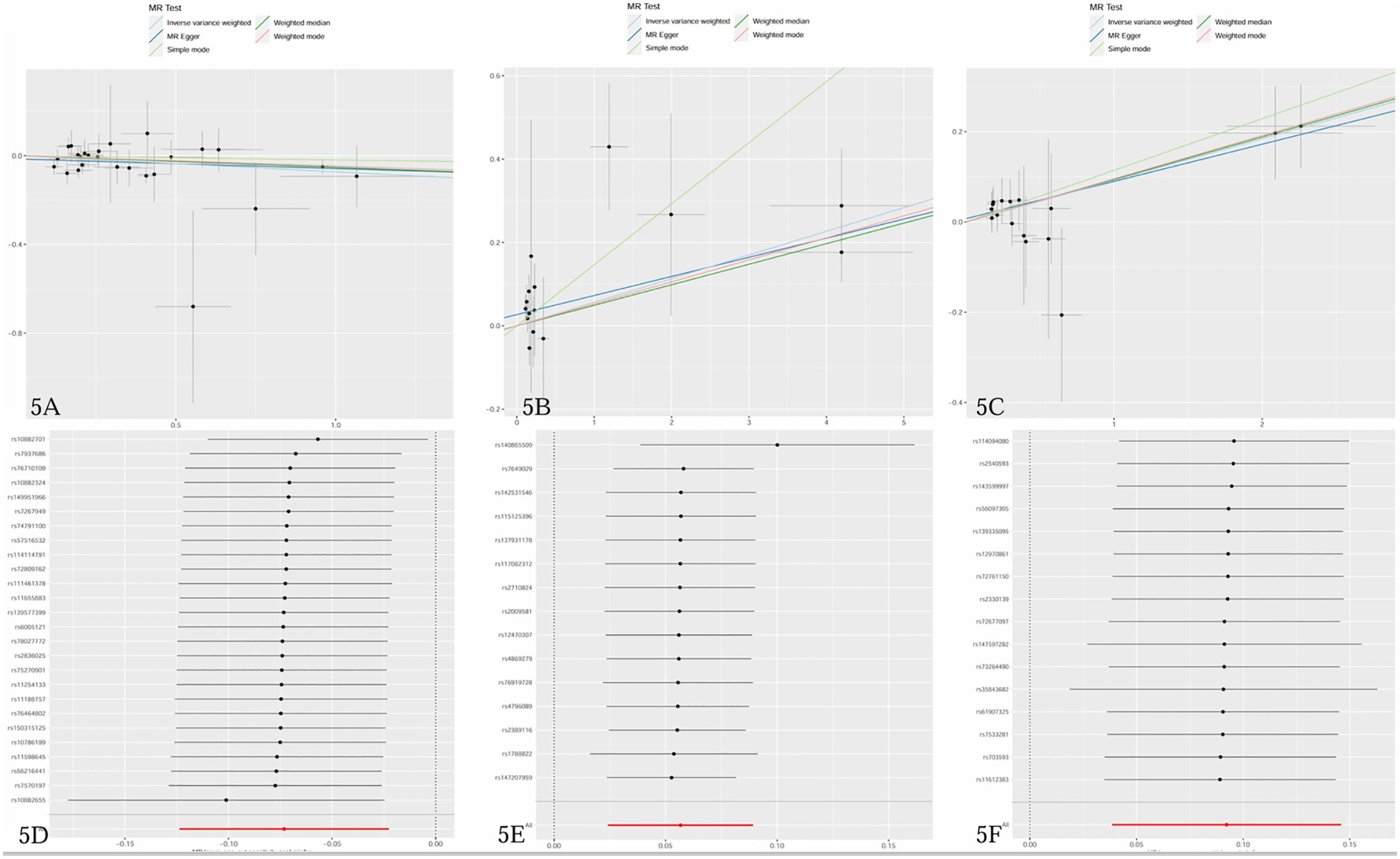
Figure 5. (A–C) Scatter plots of causal estimates of immune cells on mastitis. (D–F) Leave-one-out stability tests causal estimates of immune cells on mastitis. (A, D) CD39+ secreting Treg AC; (B, E) CD28- CD8dim %CD8dim; (C, F) CD45 on CD33br HLA DR+.
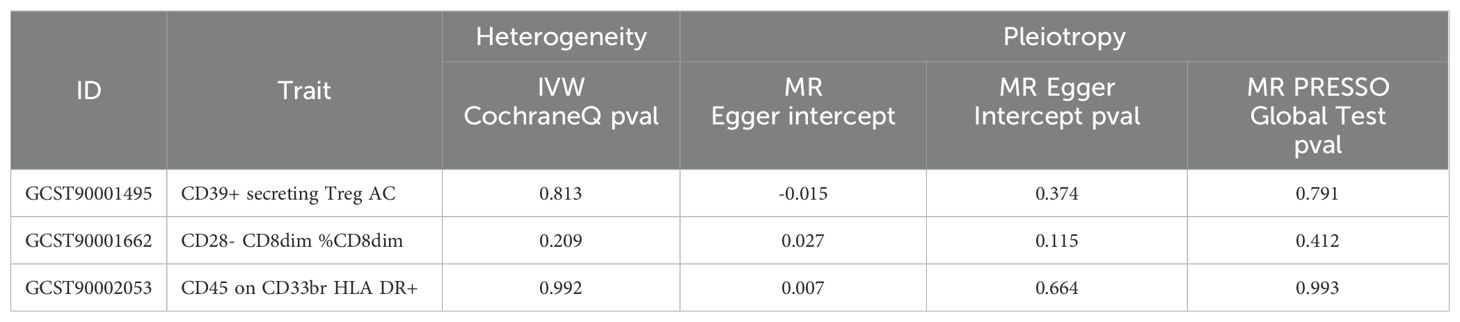
Table 2. Results of the heterogeneity and horizontal pleiotropy for the effect of immune cells on mastitis.
4 Discussion
In the two-sample MR analyses, we first conducted a thorough and comprehensive exploration of the causative links when circulating inflammatory cytokines and immune cells were seen as exposures separately and mastitis as the outcome, utilizing publicly accessible genetic data. And it was shown that CX3CL1, CD28- CD8dim %CD8dim and CD45 on CD33br HLA DR+ may suggestively be the upstream causes of mastitis, while heightened levels of CD39+ secreting Treg AC were connected to a reduced mastitis risk. Among immune cells, two risk factors were found to be related to the Treg panel (CD28- CD8dim %CD8dim) and Myeloid cells (CD45 on CD33br HLA DR+). Additionally, the presence of CD39+ secreting Treg AC from the Treg panel showed a negative association.
The mammary gland is a hormonally responsive exocrine gland. Immunoinflammatory response is a common phenomenon observed in individuals with mastitis (32). Also, it has been the subject of recent research providing compelling evidence of the crucial interplay between immunoinflammatory response with mastitis, including Liu, L., et al., noted that PDM patients exhibit a higher level of IFN- γ, and IL-12A compared to controls (5). These cytokines are secreted by T helper 1 cells aiding in eradicating foreign pathogens. Increased expression pattern of these cytokines suggests that immune responses may play a role in the pathogenesis of periductal mastitis. Similarly, the analysis of immune modulation by L. rhamnosus strains on mastitis identified two strains with significant anti-inflammatory potential, which can strongly induce IL-10 and weakly secrete pro-Th1 cytokine (IL-12 and IFN-γ). Therefore, these strains can be considered as a probiotic candidate for the management of infectious mastitis during lactation (33). After being exposed to lipopolysaccharide (LPS) in vitro experiments, untreated bovine mammary epithelial cells demonstrated a noteworthy reduction in the expression of CXCL8, IL1B, CCL20, and CXCL1 upon inflammatory response (34). Barreto, D.S., et al., also indicated autoimmune response is the most widely accepted etiology of GLM (35), and treatment with immunosuppression (steroids combined with Methotrexate) is an effective breast-conserving option. T lymphocyte subsets play a crucial role in maintaining immune homeostasis, with Treg cells exerting an immunosuppressive function. Likewise, Ucaryilmaz, H., et al., confirmed that there was an intrinsic defect of Tregs in patients with idiopathic granulomatous mastitis through flow cytometry. The expression of Foxp3 in Treg cells, and the frequency of non-suppressive Tregs, were found to be significantly lower in patients with idiopathic granulomatous mastitis when compared to healthy controls (36). However, different immunophenotypes in the Treg panel exhibited distinct roles in our results. Specifically, CD28- CD8dim %CD8dim was involved with increased risk while CD39+ secreting Treg AC demonstrated an opposing effect. Increasing evidence suggests that different subtypes of Tregs appear to have distinct roles in regulating the immune system. CD28 is a key co-stimulatory receptor typically expressed on most CD4+ T cells, binding CD80 and CD86 on antigen-presenting cells to promote T-cell activation. Lack of CD28 expression might impair their functional effectiveness or alter their activation state, leading to increased inflammation or reduced regulatory function. CD4+ CD28- T cells are associated with autoimmune diseases like rheumatic arthritis. Huang Y et al. also discovered that the absence of CD28 expression could identify T cell subsets exhibiting terminally differentiated and senescent characteristics, which were accompanied by the deterioration of the disease (37). Decreased CD28 expression can be an indicator of T-cell exhaustion. While CD39 is an ectoenzyme that converts ATP and ADP to AMP, it helps restrain excessive inflammatory responses (38). The immunoregulatory functions contribute to maintaining immune homeostasis and mitigating tissue damage. Chen C et al. demonstrated the immunosuppressive effect of CD39+ Tregs on acute lung injury via autophagy and the ERK/FOS Pathway (39). Our findings indicate a potential role of Tregs in the pathogenesis of idiopathic granulomatous mastitis and suggest a possible immunological basis for the development of this disease. These insights may inform future therapeutic approaches by targeting Treg cells and addressing the underlying immune dysregulation, which could generate novel therapies in the treatment and management of mastitis. Further investigation into the role of Tregs in this condition is warranted in order to develop more effective diagnostic and treatment strategies for patients with granulomatous mastitis. According to Zhao, J., et al., the expression of Th1 and Th17 cytokines was found to be significantly higher in the breast tissues of patients suffering from nonpuerperal mastitis, whereas the levels of Th2 cytokine and Treg cytokines such as IL-10 and transforming growth factor β were observed to be lower (40). In models of both sterile inflammation and bacterial infection, a diverse recruitment of myeloid cells was observed. The number of CD45+ leukocytes within the mammary gland significantly increased during inflammation and infection, which was in agreement with our studies. Among sterile inflammation models, increased numbers of neutrophils, Ly6C low monocytes and CD11b+F4/80+ macrophages were detected. While numbers of DCs and CD206+ macrophages increased among infection models, which was not seen during the sterile inflammation challenge. Our research also found that CD45 on CD33br HLA DR+ from Myeloid cells was linked to an increased risk of mastitis causally. These findings can provide valuable insights into the different immune responses of the mammary gland during inflammation and infection (41). Through flow cytometric analysis of milk, nonspecific mastitis revealed the highest percentage of CD4+ T lymphocytes, the percentage of CD8+ T lymphocytes was found to be highest in infectious bacterial mastitis (42). Our results showed CD28- CD8dim %CD8dim immunophenotype was significantly linked to the risk of mastitis (p = 0.0006). In the light of these observations, limited research on the relationship between immunoinflammatory response and mastitis may present a new perspective for early detection, prevention and monitoring. It is therefore necessary to conduct further research to explore the specific mechanism of these inflammatory cytokines and immune cells in mastitis.
To the best of our knowledge, this is the first Mendelian randomization research to evaluate the causal relationship between 91 inflammatory cytokines and 731 immune cells with mastitis, which provides genetic evidence emphasizing the strong causal relevance between immunoinflammatory response and mastitis. Nevertheless, it is crucial to acknowledge the restrictions of our study. Firstly, the obtained GWAS summary data predominantly consisted of patients of European descent, potentially generating biased estimates so it should be taken with caution if the conclusions would be applied in diverse ethnic groups. Future research should include diverse populations to determine whether these findings are consistent across different ethnic groups, ultimately enhancing the generalizability of the results and providing a more comprehensive understanding of genetic associations. Secondly, subgroup analyses aimed at attaining more precise correlations were not feasible due to the lack of specific demographic information and clinical records of study patients. Finally, owing to the limitations of the MR analysis, the second and third assumptions could not be properly examined and possible violations against these assumptions may exist. Colocalization analyses could be performed to further explain potential causal variation.
5 Conclusion
To sum up, our TSMR findings suggest that circulating inflammatory cytokines and immune cells might causally influence the risk of mastitis. It can be concluded an upregulation of CX3CL1, CD28- CD8dim %CD8dim and CD45 on CD33br HLA DR+ may contribute to an increased risk of mastitis, while raised levels of CD39+ secreting Treg AC are associated with a lower risk. Furthermore, these biomarkers may act as the initiatives in the onset and progression of mastitis, which could potentially serve as a viable approach for earlier diagnosis and more effective treatment options. They can also serve as candidate molecules for future mechanism exploration.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
JC: Conceptualization, Data curation, Investigation, Methodology, Project administration, Software, Writing – original draft, Writing – review & editing. BS: Data curation, Investigation, Formal analysis, Methodology, Software, Writing – review & editing. XZ: Formal analysis, Methodology, Project administration, Supervision, Writing – review & editing. CG: Data curation, Investigation, Methodology, Project administration, Writing – review & editing. YJ: Funding acquisition, Resources, Supervision, Validation, Writing – review & editing. XX: Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. Shanghai Science and Technology Committee (No. 22Y11923000 and No. 23Y11920900); National Natural Science Foundation of China (No. 82004446).
Acknowledgments
The authors would like to thank the investigators of the GWAS catalog database. And they wish to express their gratitude to the participants and researchers of the FinnGen study as well.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1409545/full#supplementary-material
References
1. Louis-Jacques AF, Berwick M, Mitchell KB. Risk factors, symptoms, and treatment of lactational mastitis. JAMA: J Am Med Assoc. (2023) 329:588–9. doi: 10.1001/jama.2023.0004
2. Scott DM. Inflammatory diseases of the breast. Best Pract Res Clin Obstet Gynaecol. (2022) 83:72–87. doi: 10.1016/j.bpobgyn.2021.11.013
3. Angelopoulou A, Field D, Ryan CA, Stanton C, Hill C, Ross RP. The microbiology and treatment of human mastitis. Med Microbiol Immun. (2018) 207:83–94. doi: 10.1007/s00430-017-0532-z
4. Xu H, Liu R, Lv Y, Fan Z, Mu W, Yang Q, et al. Treatments for periductal mastitis: systematic review and meta-analysis. Breast Care. (2022) 17:55–62. doi: 10.1159/000514419
5. Liu L, Zhou F, Wang P, Yu L, Ma Z, Li Y, et al. Periductal mastitis: an inflammatory disease related to bacterial infection and consequent immune responses? Mediat Inflammation. (2017) 2017:1–9. doi: 10.1155/2017/5309081
6. Shi L, Wu J, Hu Y, Zhang X, Li Z, Xi P, et al. Biomedical indicators of patients with non-puerperal mastitis: A retrospective study. Nutrients. (2022) 14:4816. doi: 10.3390/nu14224816
7. Jiao Y, Chang K, Jiang Y, Zhang J. Identification of periductal mastitis and granulomatous lobular mastitis: a literature review. Ann Transl Med. (2023) 11:158. doi: 10.21037/atm-22-6473
8. Parperis K, Achilleos S, Costi E, Vardas M. Autoimmune rheumatic diseases associated with granulomatous mastitis. Rheumatol Int. (2023) 43:399–407. doi: 10.1007/s00296-022-05251-9
9. Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. (2017) 318:1925–6. doi: 10.1001/jama.2017.17219
10. Gu J, Yan GM, Kong XL, Zhang YY, Huang LH, Lu HM. Assessing the causal relationship between immune traits and systemic lupus erythematosus by bi-directional Mendelian randomization analysis. Mol Genet Genomics. (2023) 298:1493–503. doi: 10.1007/s00438-023-02071-9
11. Liu C, Liu X, Xin H, Li X. Associations of inflammatory cytokines with palmoplantar pustulosis: a bidirectional Mendelian randomization study. Front Med (Lausanne). (2024) 11:1387210. doi: 10.3389/fmed.2024.1387210
12. Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. (2016) 27:3253–65. doi: 10.1681/ASN.2016010098
13. Bowden J, Holmes MV. Meta-analysis and Mendelian randomization: A review. Res Synth Methods. (2019) 10:486–96. doi: 10.1002/jrsm.1346
14. Zhao JH, Stacey D, Eriksson N, Macdonald-Dunlop E, Hedman ÅK, Kalnapenkis A, et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Immunol. (2023) 24:1540–51. doi: 10.1038/s41590-023-01588-w
15. Orrù V, Steri M, Sidore C, Marongiu M, Serra V, Olla S, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. (2020) 52:1036–45. doi: 10.1038/s41588-020-0684-4
16. Sidore C, Busonero F, Maschio A, Porcu E, Naitza S, Zoledziewska M, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. (2015) 47:1272–81. doi: 10.1038/ng.3368
17. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. (2023) 613:508–18. doi: 10.1038/s41586-022-05473-8
18. Zhang S, Zhang W, Ren H, Xue R, Wang Z, Wang Z, et al. Mendelian randomization analysis revealed a gut microbiota–mammary axis in breast cancer. Front Microbiol. (2023) 14:1193725. doi: 10.3389/fmicb.2023.1193725
19. Bouras E, Karhunen V, Gill D, Huang J, Haycock PC, Gunter MJ, et al. Circulating inflammatory cytokines and risk of five cancers: a Mendelian randomization analysis. BMC Med. (2022) 20:3. doi: 10.1186/s12916-021-02193-0
20. Li J, Niu Q, Wu A, Zhang Y, Hong L, Wang H. Causal relationship between circulating immune cells and the risk of type 2 diabetes: a Mendelian randomization study. Front Endocrinol (Lausanne). (2023) 14:1210415. doi: 10.3389/fendo.2023.1210415
21. Xiang M, Wang Y, Gao Z, Wang J, Chen Q, Sun Z, et al. Exploring causal correlations between inflammatory cytokines and systemic lupus erythematosus: A Mendelian randomization. Front Immunol. (2023) 13:985729. doi: 10.3389/fimmu.2022.985729
22. Chen H, Peng L, Wang Z, He Y, Zhang X. Exploring the causal relationship between periodontitis and gut microbiome: Unveiling the oral-gut and gut-oral axes through bidirectional Mendelian randomization. J Clin Periodontol. (2024) 51:417–30. doi: 10.1111/jcpe.13906
23. Pierce BL, Ahsan H, VanderWeele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. (2011) 40:740–52. doi: 10.1093/ije/dyq151
24. Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. (2019) 35:4851–3. doi: 10.1093/bioinformatics/btz469
25. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. (2017) 46:1734–9. doi: 10.1093/ije/dyx034
26. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
27. Bowden J, Davey SG, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
28. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
29. Verbanck M, Chen C, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
30. Fattahi AS, Amini G, Sajedi F, Mehrad-Majd H. Factors affecting recurrence of idiopathic granulomatous mastitis: A systematic review. Breast J. (2023) 2023:9947797. doi: 10.1155/2023/9947797
31. Ong SS, Xu J, Sim CK, Khng AJ, Ho PJ, Kwan P, et al. Profiling microbial communities in idiopathic granulomatous mastitis. Int J Mol Sci. (2023) 24(2):1042. doi: 10.3390/ijms24021042
32. Michie C, Lockie F, Lynn W. The challenge of mastitis. Arch Dis Child. (2003) 88:818–21. doi: 10.1136/adc.88.9.818
33. Bousmaha-Marroki L, Boutillier D, Marroki A, Grangette C. In vitro anti-staphylococcal and anti-inflammatory abilities of lacticaseibacillus rhamnosus from infant gut microbiota as potential probiotic against infectious women mastitis. Probiotics Antimicro. (2021) 13:970–81. doi: 10.1007/s12602-021-09755-x
34. Kurz JP, Richards MP, Garcia M, Wang Z. Exogenous phospholipase A2 affects inflammatory gene expression in primary bovine mammary epithelial cells. J Dairy Res. (2019) 86:177–80. doi: 10.1017/S0022029919000232
35. Barreto DS, Sedgwick EL, Nagi CS, Benveniste AP. Granulomatous mastitis: etiology, imaging, pathology, treatment, and clinical findings. Breast Cancer Res Tr. (2018) 171:527–34. doi: 10.1007/s10549-018-4870-3
36. Ucaryilmaz H, Koksal H, Emsen A, Kadoglou N, Dixon JM, Artac H. The role of regulatory T and B cells in the etiopathogenesis of idiopathic granulomatous mastitis. Immunol Invest. (2022) 51:357–67. doi: 10.1080/08820139.2020.1832114
37. Huang Y, Zheng H, Zhu Y, Hong Y, Zha J, Lin Z, et al. Loss of CD28 expression associates with severe T-cell exhaustion in acute myeloid leukemia. Front Immunol. (2023) 14:1139517. doi: 10.3389/fimmu.2023.1139517
38. Borsellino G, Kleinewietfeld M, Di Mitri D, Sternjak A, Diamantini A, Giometto R, et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood. (2007) 110:1225–32. doi: 10.1182/blood-2006-12-064527
39. Chen C, Li X, Li C, Jin J, Wang D, Zhao Y, et al. CD39+ Regulatory T cells attenuate lipopolysaccharide-induced acute lung injury via autophagy and the ERK/FOS pathway. Front Immunol. (2021) 11:602605. doi: 10.3389/fimmu.2020.602605
40. Zhao J, Ji H, Wang X, Wang Y, Xia Z. Association of nonpuerperal mastitis with cytokines related to helper T cells TH1/TH2 and TH17/treg. Altern Ther Health M. (2023) 29(8):150–5.
41. Wilson GJ, Fukuoka A, Vidler F, Graham GJ. Diverse myeloid cells are recruited to the developing and inflamed mammary gland. Immunology. (2022) 165:206–18. doi: 10.1111/imm.13430
Keywords: mastitis, cytokines, Mendelian randomization, genome-wide association study (GWAS), inflammation, immunity
Citation: Chen J, Su B, Zhang X, Gao C, Ji Y and Xue X (2024) Mendelian randomization suggests causal correlations between inflammatory cytokines and immune cells with mastitis. Front. Immunol. 15:1409545. doi: 10.3389/fimmu.2024.1409545
Received: 05 April 2024; Accepted: 09 September 2024;
Published: 27 September 2024.
Edited by:
Efe Sezgin, Izmir Institute of Technology, TürkiyeReviewed by:
Tarik Safak, Kastamonu University, TürkiyeXiaonan Zhang, Chongqing Medical University, China
Copyright © 2024 Chen, Su, Zhang, Gao, Ji and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaohong Xue, eGlhb2hvbmdfeHVlQDEyNi5jb20=
†These authors have contributed equally to this work and shared the first authorship
 Jiaying Chen
Jiaying Chen Ben Su
Ben Su Xinyue Zhang2
Xinyue Zhang2