- 1Department of Gastroenterology, Qilu Hospital of Shandong University, Jinan, Shandong, China
- 2Shandong Provincial Clinical Research Center for Digestive Disease, Qilu Hospital of Shandong University, Jinan, Shandong, China
- 3Laboratory of Translational Gastroenterology, Qilu Hospital of Shandong University, Jinan, Shandong, China
- 4Robot Engineering Laboratory for Precise Diagnosis and Therapy of Gastrointestinal (GI) Tumor, Qilu Hospital of Shandong University, Jinan, Shandong, China
- 5Shandong Key Laboratory of Oral Tissue Regeneration, Department of Human Microbiome, School and Hospital of Stomatology, Cheeloo College of Medicine, Shandong University, Jinan, Shandong, China
- 6Shandong Engineering Laboratory for Dental Materials and Oral Tissue Regeneration & Shandong Provincial Clinical Research Center for Oral Diseases, Shandong University, Jinan, Shandong, China
Background: To investigate microbial characteristics across multibody sites from chronic pancreatitis (CP), through pancreatic benign tumors, to pancreatic ductal adenocarcinoma (PDAC) at different stages.
Methods: 16S ribosomal RNA (rRNA) amplicon sequencing was conducted on saliva, duodenal fluid, and pancreatic tissue obtained via endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) of patients with CP, pancreatic benign tumors, PDAC in stage I/II, III, and IV. The neutral community model (NCM) assessed the microbial assembly contribution and MaAslin2 identified the differential microbes.
Results: From CP to stage IV PDAC patients, there was a marked surge in influence of salivary and duodenal microbiota on constitution of pancreatic microbial communities. Our observations revealed a successive alteration in microbial species across various bodily sites during PDAC tumorigenesis. Notably, Porphyromonas gingivalis, Treponema denticola, Peptoanaerobacter stomatis, Propionibacterium acidifaciens, Porphyromonas endodontalis, Filifactor alocis, etc., sequentially increased along PDAC progression in pancreatic tissue, whereas bacteria commonly used as probiotics Bifidobacterium breve, Lactiplantibacillus plantarum, etc., declined. Furthermore, the sequentially escalating trends of Peptoanaerobacter stomatis and Propionibacterium acidifaciens during PDAC tumorigenesis were mirrored in duodenal fluid and saliva. Porphyromonas gingivalis, Porphyromonas endodontalis, and Filifactor alocis, which intensified from CP to stage IV PDAC in pancreatic tissue, were also found to be enriched in saliva of patients with short-term survival (STS) compared with those with long-term survival (LTS).
Conclusions: Salivary and duodenal microorganisms were prominent factors in shaping pancreatic microbial landscape in PDAC context. Further exploration of these microbial entities is imperative to unravel their specific roles in PDAC pathogenesis, potentially yielding insights for future therapeutic strategies.
Introduction
Pancreatic cancer, predominantly pancreatic ductal adenocarcinoma (PDAC), is one of the most lethal diseases worldwide. PDAC ranks as the sixth leading cause of cancer-related death in China (1). Due to non-specific early symptoms and a lack of effective screening methods, approximately 80-85% of patients have entered an unresectable or metastatic stage at the time of diagnosis (2), with a 5-year survival rate of only 7.2% (3). This underscores the urgent need to elucidate the pathogenesis of PDAC and identify novel therapeutic targets.
Microbiota residing in diverse anatomical locales, encompassing the oral cavity, duodenum, intestines, and pancreas, among others, have been documented to correlate with various clinical markers indicative of PDAC (4–7). Specifically, the abundance of Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans in salivary fluids is associated with an elevated risk of PDAC (8). Furthermore, Faecalibacterium prausnitzii levels are frequently diminished in the saliva and feces of PDAC patients (6, 9), whereas patients experiencing long-term survival (LTS) display elevated concentrations of Akkermansia muciniphila and Faecalibacterium prausnitzii within the gut microbiome (10). In addition, the intra-tumoral microbiota (IM) within the pancreas plays a pivotal role in PDAC pathogenesis, potentially modulating the progression of the disease (11, 12). Gammaproteobacteria found in PDAC tumors can render the chemotherapeutic agent gemcitabine inactive, thereby contributing to chemotherapy resistance (7).
Furthermore, the human microbiota is inherently dynamic, encompassing fluctuations across short/long timescales (13). Numerous research endeavors have aimed to identify correlations linking temporal shifts in microbiota composition to diseases, notably type I diabetes, necrotizing enterocolitis, and infections (14). While investigations have illuminated disparities in gene expression patterns, metabolic signatures, and therapeutic responses among pancreatic precursor lesions, early-stage, and late-stage PDAC patients (15–18), the intricate microbial dynamics throughout diverse anatomical sites and stages of PDAC remain elusive. This gap in understanding impedes a definitive assessment of whether the distinctive microbial profiles observed between PDAC patients and controls serve as causal agents or merely consequential factors in the pathogenesis of PDAC.
The present study aimed to conduct a comprehensive analysis of the temporal dynamics of the oral, duodenal, and pancreatic microbial communities across pancreatitis, pancreatic benign tumors, and various stages of PDAC. Furthermore, we delved into identifying microbial signatures potentially involved in the initiation and progression of PDAC, thereby providing novel perspectives on its underlying mechanisms and potential therapeutic avenues.
Methods
Subject recruitment
We performed an observational study of 81 PDAC patients who underwent endoscopic ultrasound-guided fine needle aspiration (EUS-FNA) enrolled from January 2022 to December 2022 at the Qilu Hospital of Shandong University. The research adhered strictly to the Declaration of Helsinki principles and secured ethical clearance from the Ethics Committee of the Qilu Hospital, Shandong University, under reference KYLL-202111-018-1. Prior to enrollment, written informed consent was obtained from each participant. Eligible subjects included individuals aged over 18 years with suspected pancreatic lesions necessitating EUS-FNA for diagnostic confirmation.
The exclusion criteria were meticulously defined, encompassing: (I) concurrent presence of other cancers; (II) individuals who had undergone EUS-FNA, endoscopic retrograde cholangiopancreatography (ERCP), X-ray guided punctures, pancreatic surgeries, or received chemotherapy, radiotherapy, or other biological treatments within the preceding period; and (III) recent users (within the past month) of proton-pump inhibitors (PPIs), antibiotics (administered orally or intravenously), probiotics, prebiotics, or synbiotics. For clinicopathological staging, adherence was maintained to the 8th edition of the American Joint Committee on Cancer’s (AJCC) TNM staging system for PDAC. Additionally, PDAC patients were categorized into two distinct groups based on their chemotherapy response: the complete response/partial response (CR/PR) group comprised patients exhibiting complete disappearance of target lesions or a ≥30% reduction in the sum of the longest diameters of baseline lesions following treatment, while the stable disease/progressive disease (SD/PD) group encompassed those whose lesions either decreased slightly yet did not meet PR criteria, increased but fell short of PD status, demonstrated a ≥20% increase in the sum of longest lesion diameters, or manifested new lesions post-chemotherapy. Furthermore, regarding survival outcomes, PDAC patients were differentiated into short-term survival (STS) and LTS groups at the 16-month survival analysis point.
Sample collection and processing
All participants underwent rinsing of their mouths with normal saline, adhering strictly to established protocols (19). Following this, the rinse was discarded, and a volume of 3 to 5 milliliters of saliva was promptly collected into a cryopreserved tube. The EUS-FNA procedure, performed by a seasoned endoscopist, utilized linear array echo endoscopes specifically from PENTAX (models EG-3870UTK and EG-3270UK, Tokyo, Japan). As the endoscope navigated to the descending segment of the duodenum, the aspirator delicately drew 5 milliliters of duodenal fluid into a sterile collector, which was then transferred to a cryopreserved tube. Prior to performing the color Doppler-guided puncture, a thorough examination of pancreatic lesions was conducted to ascertain the absence of major blood vessels along the intended needle path. Utilizing a 22-gauge needle sourced from COOK (USA), pancreatic tissue was meticulously collected and deposited into a sterile, cryopreserved tube. Immediately thereafter, all samples underwent rapid freezing in liquid nitrogen, were safely conveyed within liquid nitrogen containers, and stored in lock-in freezer chambers maintained at -80°C until the time of DNA extraction.
DNA extraction and 16S rRNA amplicon sequencing
All samples underwent thawing on ice followed by aliquoting, whereupon microbial DNA extraction was performed utilizing the E.Z.N.A.® soil DNA Kit sourced from Omega Bio-tek, Norcross, Georgia, USA. The DNA extract’s quality was verified both via 1% agarose gel electrophoresis and the NanoDrop 2000 UV-vis spectrophotometer from Thermo Scientific, Wilmington, USA. Subsequently, the hypervariable V3-V4 region of the bacterial 16S rRNA gene was amplified employing primer pairs 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’), facilitated by an ABI GeneAmp® 9700 PCR thermocycler based in California, USA. The purified amplification products were then sequenced on the advanced Illumina MiSeq PE300 or NovaSeq PE250 platform, both originating from Illumina, San Diego, USA.
Paired-end reads were consolidated into sequence tags, adhering to their overlapping characteristics and subject to rigorous quality control measures executed by the UPARSE pipeline. This step excluded subpar reads and chimeric sequences, leveraging vsearch software (with parameters set to maxee = 3 and minlength = 370) for effective filtration. Upon dereplication, the remaining clean reads were clustered into distinct operational taxonomy units (OTUs) using a 97% similarity threshold. Representative reads from 7219 OTUs were aligned against the Ribosomal Database Project, version 18, to ascertain taxonomic identities, requiring an 80% similarity match to species entries within the reference database, thereby providing insights into the microbiome composition.
Statistical analyses
We ran ‘Decontam’ (20) in order to identify and remove contaminant DNA sequences from the dataset. RStudio (R version 4.1.1) was employed for statistical analysis. Adonis2 was used to explore the correlation between the clinical indices and the microbiota. The alpha and beta diversity distance matrices were calculated using the “Vegan” package (version vegan_2.6-4). The Sloan neutral model was used to predict the relationship between occurrence frequency and relative abundance of species (21). The FEAST analysis was used to track the potential sources of the microbial structure (22). Microbial differences between groups were tested using MaAsLin2 analysis (version 2_1.14.1), and we adjusted the effects of age, gender, and body mass index (BMI). Genera or species with a p-value less than 0.05 were retained. At each position, we took the union of the differential microbes between the chronic pancreatitis (CP) group and each of the other four groups and applied the Mfuzz algorithm to cluster the expression trends of the average relative abundance of the differential microbes. According to the similarity of their abundant-variation patterns from CP to stage IV PDAC patients, the differential microbes were divided into four different clusters and named C1-C4. R package ‘ClusterGVis’ was used to generate the fitted trend from CP to stage IV PDAC patients for each cluster. Spearman correlation analysis was employed to establish the correlation network within each module (|Spearman correlation| > 0.6, false discovery rate (FDR)-adjusted p-value < 0.05), as well as to determine the interrelationships between clusters in different groups (|Spearman correlation| > 0.6, p-value < 0.01).
Results
Microbial composition in the multibody sites along the development of PDAC
We enrolled 81 subjects, of whom 6 had CP, 12 had pancreatic benign tumors including serous cystic neoplasm (SCN), mucinous cystic neoplasm (MCN), intraductal papillary mucinous neoplasm (IPMN), and solid pseudopaillary neoplasm (SPN), 20 had PDAC in stage I or II, 30 had PDAC in stage III, and 13 had PDAC in stage IV. Except for carbohydrate antigen 19–9 (CA 19-9), no significant statistical difference was observed in baseline characteristics among the five groups (Supplementary Table S1). We first measured the microbial alpha-diversity among the pancreatic tissue, duodenal fluid, and saliva of the five groups using different methodologies and found there was no significant difference (Supplementary Figure S1). We performed the beta-diversity analysis to compare the microbial composition. Clustering was detected for the Bray-Curtis principal co-ordinates analysis (PCoA) analysis among the five groups, especially in the saliva (Figures 1A–C).
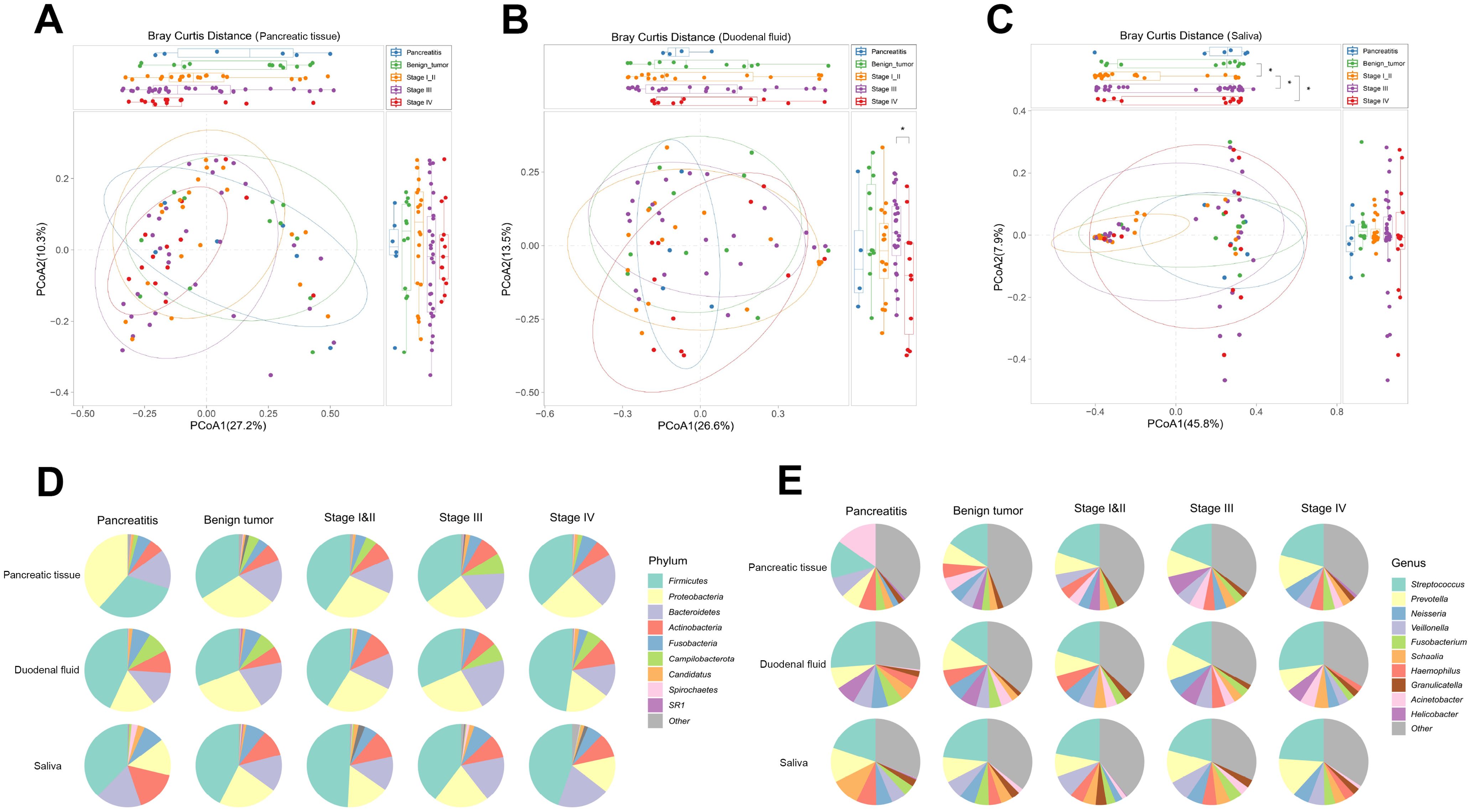
Figure 1. Microbial composition across CP, pancreatic benign tumors, PDAC in stage I/II, III, and IV groups in the multibody sites. (A–C) The principal co-ordinates analysis (PCoA) analysis was used to evaluate the microbial composition in pancreatic tissue (A), duodenal fluid (B), and saliva (C) across the five groups. (D, E) The microbial relative abundance across the five groups of the multibody sites at the phylum (D) and genus (E) levels.
In the pancreatic tissue, duodenal fluid, and saliva of the five groups, the predominant phyla were Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria, demonstrating a broad phyla-level similarity (Figure 1D). However, differences appeared at the genus level (Figure 1E). In the pancreatic tissue, Streptococcus and Prevotella increased sequentially from CP, to benign tumors, to stage I/II PDAC, to stage III PDAC, finally to stage IV PDAC patients, while Acinetobacter, Veillonella, and Haemophilus decreased sequentially. Helicobacter showed an increasing tendency from CP to stage III PDAC patients. In the duodenal fluid, Fusobacterium reduced sequentially from CP to stage IV PDAC patients. Meanwhile, in the saliva, Prevotella also increased sequentially from benign tumors to stage IV PDAC patients.
Succession of microbes in the multibody sites along the development of PDAC
To figure out the microbes that varied significantly along the development of PDAC, MaAsLin2 analysis was conducted to test the difference of microbes among the five groups. A total of 66, 66, and 72 microbial species in the pancreatic tissue, duodenal fluid, and saliva were identified, respectively. At the genus level, we identified 51, 62, and 46 differently distributed genera in the pancreatic tissue, duodenal fluid, and saliva, as shown in Supplementary Figure S2.
Next, those differently distributed microbial species were clustered into four clusters by the similarity of their abundant-variation pattern and named C1–C4. In the pancreatic tissue, microbes in the C1 including Haematobacter massiliensis, Brevundimonas staleyi, and Acinetobacter junii, maintained a high abundance in CP patients and showed a continuous decrease from CP to stage IV PDAC patients (Figure 2A and Supplementary Table S2). Microbes in C2 preferred to harbor in stage I/II PDAC patients. Microbes in C3 and C4, which involve a group of well-known periodontitis pathogens such as Porphyromonas gingivalis, Treponema denticola, Porphyromonas endodontalis, and Filifactor alocis presented an increasing trend from CP to stage IV PDAC patients. In the duodenal fluid, microbes in the C1 including Schaalia georgiae and Actinomyces massiliensis, maintained a high abundance in CP patients and showed a continuous decrease from CP to stage III PDAC patients (Figure 2B and Supplementary Table S3). However, microbes in the C4 including Treponema amylovorum and Neisseria oralis showed a continuous increase from CP to stage IV PDAC patients. In the saliva, microbes in C1 including Fusobacterium mortiferum, Lactiplantibacillus plantarum, and Loigolactobacillus coryniformis presented a decreasing trend from CP to stage IV PDAC patients (Figure 2C and Supplementary Table S4). Microbes in C2, C3, and C4 including Prevotella fusca, Metamycoplasma salivarium, and Dolosigranulum pigrum maintained a high abundance in stage I/II PDAC patients. Peptoanaerobacter stomatis and Propionibacterium acidifaciens showed sequentially increasing trends along PDAC tumorigenesis all in the pancreatic tissue, duodenal fluid, and saliva.
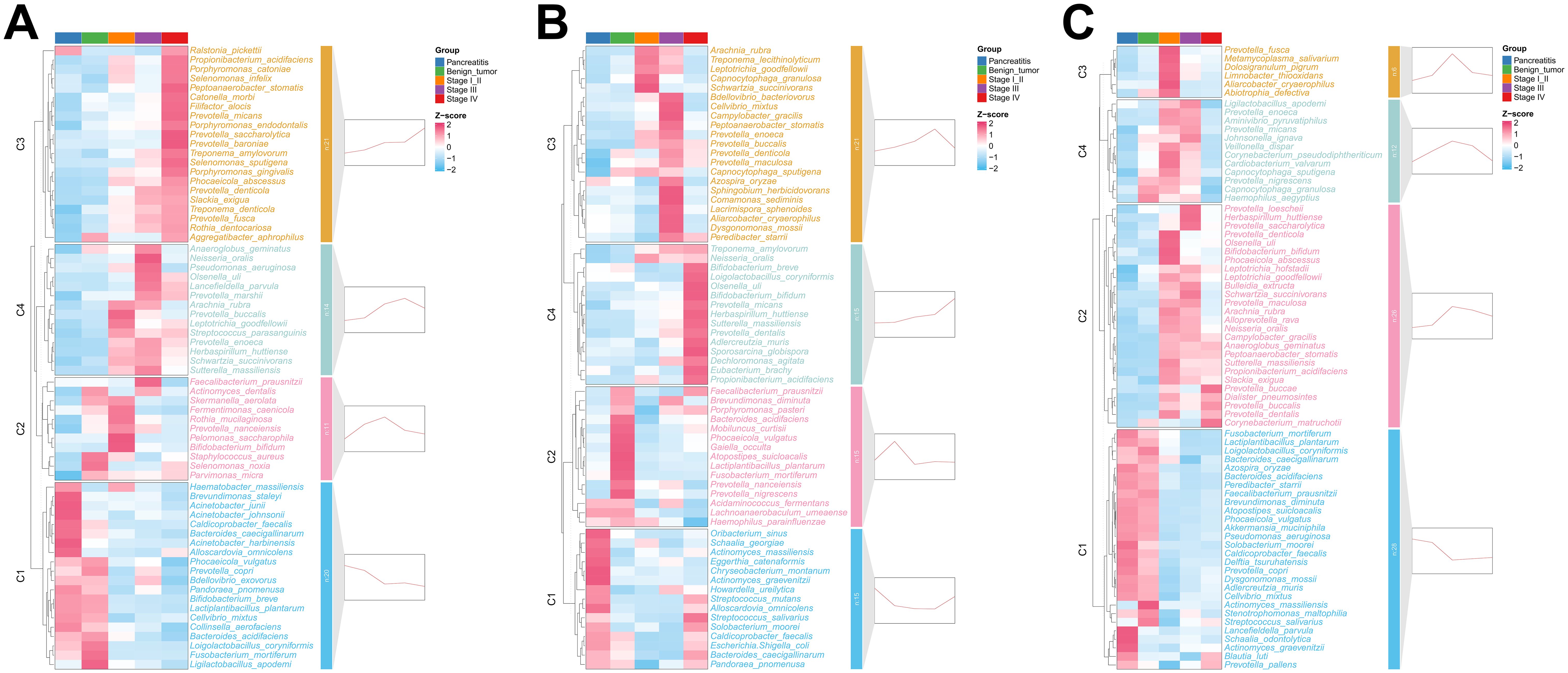
Figure 2. Differential species across the five groups in pancreatic tissue (A), duodenal fluid (B), and saliva (C). The heatmap showed the average relative abundance of differential microbes across the groups, with the z-score representing the row-scaled average relative abundance. The small graphs on the right of each panel displayed the clustering results of the expression trends of the average relative abundance of differential microbes, representing the fitted trend from CP to stage IV PDAC patients for each cluster.
Variations in the relationship of differential microbes along the development of PDAC
Spearman correlation analysis showed that most of the differential microbes in each cluster had a positive correlation with each other (Supplementary Figure S3). In the pancreatic tissue, the microbes in C1 and C3 had more positive correlations than C4. The microbes in C2 had no correlation. In the duodenal fluid, the microbes in C2 and C3 had more positive correlations than C1 and C4. Microbes in C1 and C2 were more positively correlated than C3 and C4 in the saliva.
The interrelationships between the four clusters (C1–C4) varied along the development of PDAC. In the pancreatic tissue, most microbes in the four clusters were positively correlated with each other (Figure 3A). From CP to stage IV PDAC patients, interrelationships among the four clusters gradually increased and then reduced. Stage I/II PDAC patients had the maximal interrelationships and CP patients had the minimal interrelationships. Microbes in C1 were positively correlated with microbes from C2 but negatively correlated with microbes from C3 and C4 in the benign tumor patients. In the duodenal fluid and saliva, the interrelationships among the four clusters also gradually increased and then reduced along the development of PDAC (Figures 3B, C). These results suggest that the relationship among microbes also changed continuously in addition to the changes in the abundance of individual microorganisms with the development of PDAC.

Figure 3. The relationships of differential species in the four clusters across the five groups in the multibody sites. (A–C) The interrelationships of differential species between the four clusters in pancreatic tissue (A), duodenal fluid (B), and saliva (C) across the five groups. The node sizes represent the mean relative abundance. Lines between nodes represent correlations between the nodes connected by the lines, with red representing positive correlation and green representing negative correlation.
Changes in ecological communities in the multibody sites along the development of PDAC
The neutral community model (NCM) was used to measure the relative contribution of the salivary microbes to the pancreatic microbes assembly. The goodness-of-fit (R2) value of NCM for CP patients was -0.052, that for benign tumor patients was 0.269, that for stage I/II PDAC patients was 0.382, that for stage III PDAC patients was 0.403, and that for stage IV PDAC patients was 0.370 (Figures 4A–E). An increase in R2 along the development of PDAC indicates an increase in the relative contribution of the salivary microbes to the assembly of the pancreatic microbes. We also found an increase in the relative contribution of duodenal microbes to the pancreatic microbial assembly from CP, to benign tumors, to stage I/II PDAC, to stage III PDAC patients (R2 increased from 0.002, to 0.255, to 0.388, to 0.509, Figures 4F–J).
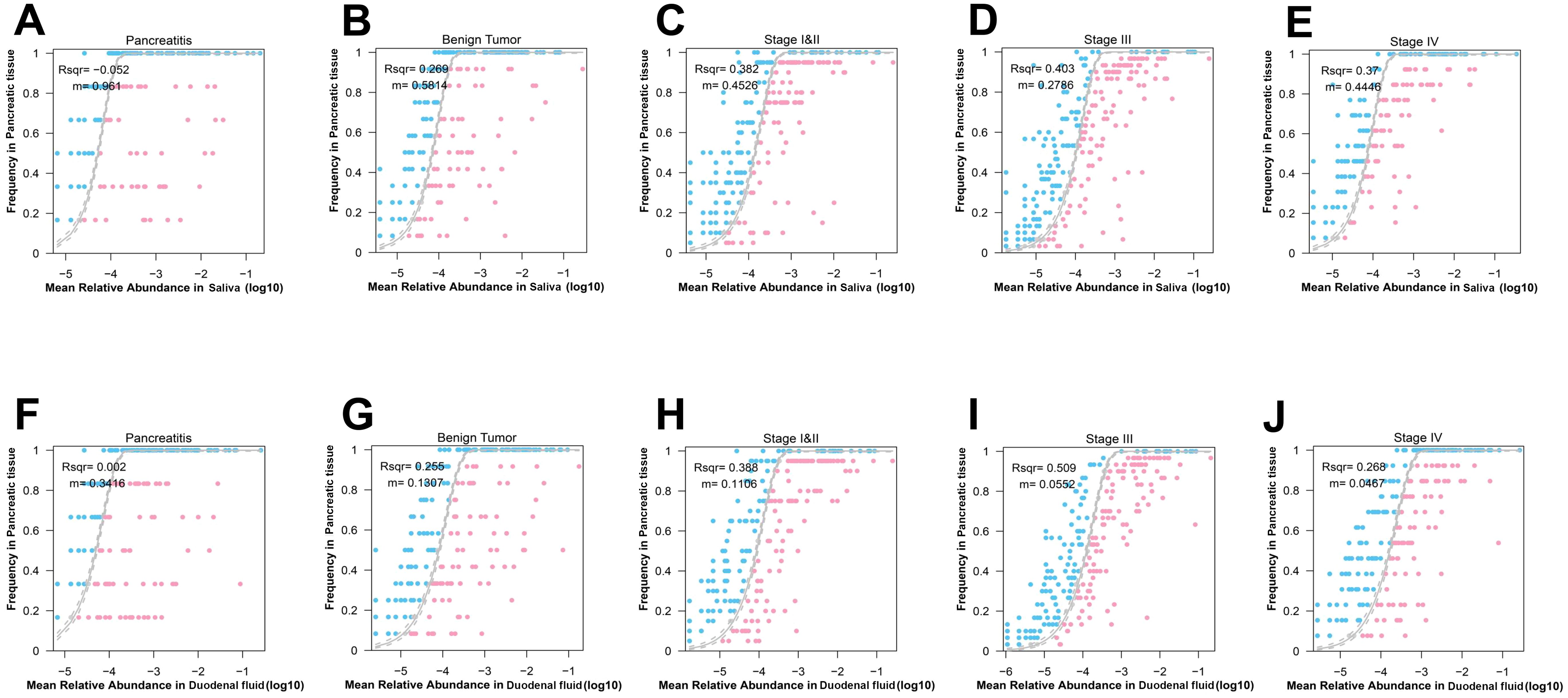
Figure 4. Interactions of the microbes across the five groups in the multibody sites. (A–E) Neutral community model (NCM) of salivary microbes to the pancreatic microbial assembly in patients with CP (A), benign tumor (B), stage I/II PDAC (C), stage III PDAC (D), stage IV PDAC (E). The coefficient of determination (R2) was the goodness of fit of the neutral model. It ranged from ≤ 0 (no fit) to 1 (perfect fit). (F–J) NCM of salivary microbes to the duodenal microbial assembly in patients with CP (F), benign tumor (G), stage I/II PDAC (H), stage III PDAC (I), stage IV PDAC (J).
Microbial differences in different chemotherapy responses of PDAC patients
Among the 63 PDAC patients, 60 received standard chemotherapy, of whom 14 were in the CR/PR group and 46 were in the SD/PD group. No significant statistical difference was observed in baseline characteristics between the two groups (Supplementary Table S5). We first measured the microbial alpha-diversity among the three sites between the two groups and found the Chao1 index in the saliva of SD/PD patients was significantly higher than that of CR/PR patients (Figures 5A, B).
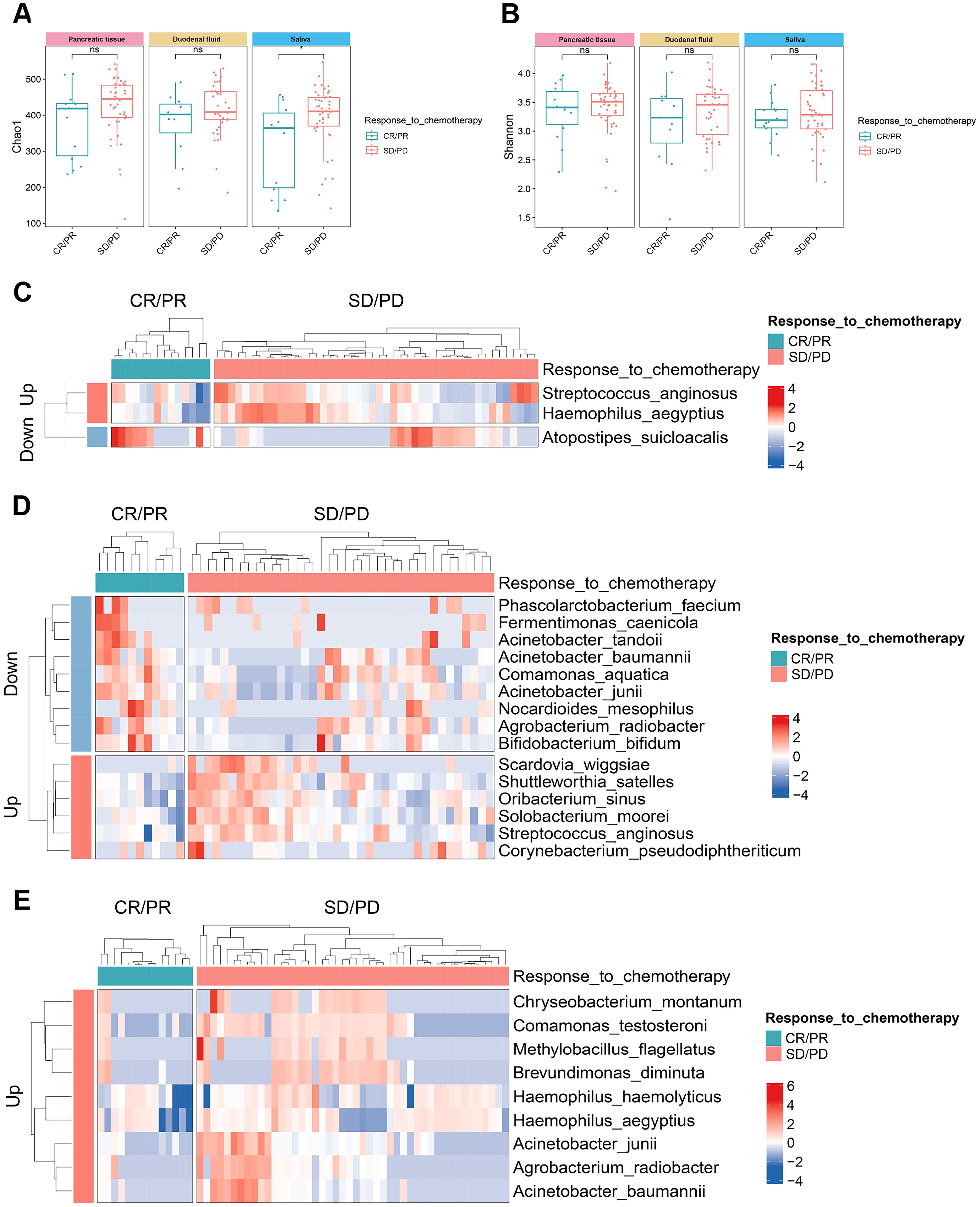
Figure 5. The microbial alpha-diversity and differential species between the groups with different chemotherapy responses in the multibody sites. (A, B) The Chao1 (A) and Shannon (B) indices of alpha-diversity between CR/PR group and SD/PD group in the multibody sites. (C–E) Differential species between CR/PR group and SD/PD group in pancreatic tissue (C), duodenal fluid (D), and saliva (E). CR/PR, complete response/partial response; SD/PD, stable disease/progressive disease.
MaAsLin2 analysis detected the differential microbes between the two groups. At the species level, Streptococcus anginosus and Haemophilus aegyptius were significantly enriched in the pancreatic tissue of the SD/PD patients compared with the CR/PR patients, while Atopostipes suicloacalis was significantly reduced (Figure 5C). In the duodenal fluid, Scardovia wiggsiae, Shuttleworthia satelles, and Oribacterium sinus increased in the SD/PD patients, whereas Acinetobacter junii, Fermentimonas caenicola, and Phascolarctobacterium faecium decreased (Figure 5D). In the saliva, Chryseobacterium montanum, Comamonas testosteroni, and Methylobacillus flagellates were enriched in the SD/PD patients compared with the CR/PR patients (Figure 5E). Similar differences were also found across the five stages of PDAC tumorigenesis. Acinetobacter junii also presented a decreasing trend from CP to stage IV PDAC patients in the pancreatic tissue. Fermentimonas caenicola was also reduced in the pancreatic tissue of stage IV PDAC patients compared with CP patients.
Microbial differences in PDAC patients with different survival
Among the 63 PDAC patients, 35 were in the STS group and 28 were in the LTS group. No significant statistical difference was observed in baseline characteristics between the two groups (Supplementary Table S6). We also measured the microbial alpha-diversity in the pancreatic tissue, duodenal fluid, and saliva between the two groups using different methodologies and found there was no significant difference (Supplementary Figure S4). At the species level, MaAsLin2 analysis detected that Eggerthia catenaformis and Cardiobacterium hominis were significantly enriched, while Fermentimonas caenicola, Skermanella aerolata, Limosilactobacillus reuter, etc. were significantly reduced in the pancreatic tissue of the STS patients compared with the LTS patients (Figure 6A). In the duodenal fluid, Caldicoprobacter faecalis, Herbaspirillum huttiense, etc. increased in the STS patients, whereas Filifactor alocis and Prevotella nanceiensis decreased (Figure 6B). In the saliva, Porphyromonas gingivalis, Porphyromonas endodontalis, and Filifactor alocis were enriched in the STS patients compared with the LTS patients, while Fermentimonas caenicola was reduced (Figure 6C). Similar differences were also found across the five stages of PDAC tumorigenesis. Porphyromonas gingivalis, Porphyromonas endodontalis, and Filifactor alocis also presented an increasing trend from CP to stage IV PDAC patients in the pancreatic tissue. Herbaspirillum huttiense also increased in the duodenual fluid across the five stages of PDAC tumorigenesis. Fermentimonas caenicola was also reduced in the pancreatic tissue of stage IV PDAC patients compared with CP patients. Prevotella nanceiensis also decreased in the duodenual fluid of stage IV PDAC patients compared with CP patients.
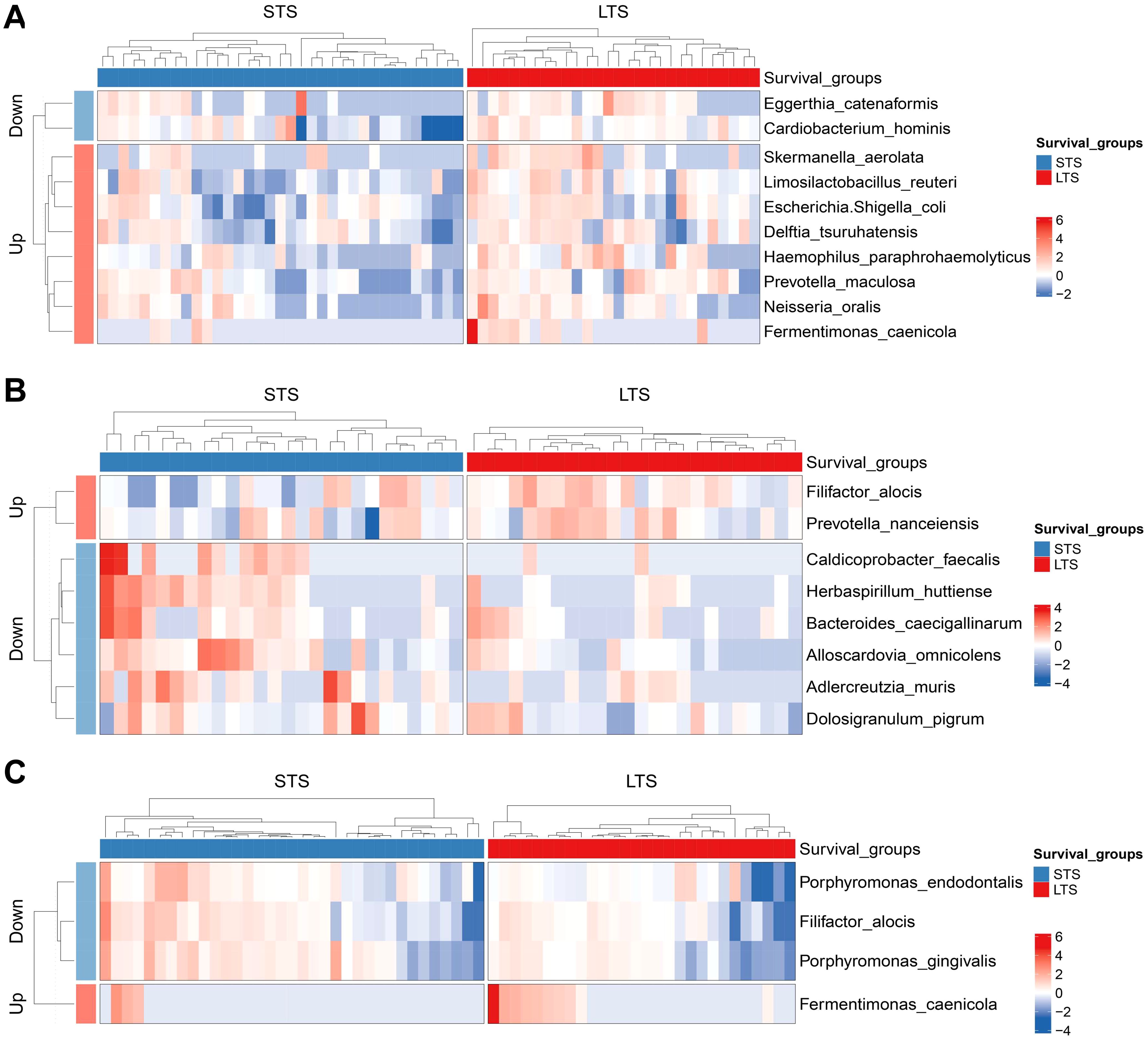
Figure 6. Differential species between the groups with different survival in pancreatic tissue (A), duodenal fluid (B), and saliva (C). LTS, long-term survival; STS, short-term survival.
Discussion
The pathogenesis and early detection of PDAC remain formidable challenges central to ongoing efforts to alleviate the burden of this disease. To our knowledge, this is the first time series analysis of microbiota in pancreatic tissue, duodenal fluid, and saliva across the five stages of PDAC tumorigenesis using pancreatic tissue obtained via EUS-FNA. Our key findings highlighted the significant role of salivary and duodenal microbes in shaping the pancreatic microbial composition associated with PDAC. Notably, we observed continuous increase or decrease of specific species along PDAC tumorigenesis. Furthermore, we found Porphyromonas gingivalis, Porphyromonas endodontalis, and Filifactor alocis presented an increasing trend from CP to stage IV PDAC patients in the pancreatic tissue and were also enriched in the saliva of STS patients. In the duodenal fluid, Herbaspirillum huttiense increased sequentially along PDAC tumorigenesis and also increased in the STS patients compared with the LTS patients.
The increased R2 of NCM along PDAC tumorigenesis indicated that salivary and duodenal microbes were significant contributors to the pancreatic microbial composition associated with PDAC. Studies have found microbial translocation active in some diseases, particularly involving the gut microbiota, which can affect various organs through a damaged intestinal barrier (23, 24). In addition, from CP to stage IV PDAC patients, interrelationships among the four clusters gradually increased and then reduced. Previous studies pointed out that communities in which a large proportion of members are connected through positive links tend to be unstable due to positive feedback and co-oscillation in response to environmental fluctuations (25). Therefore, community instability and microbial translocation may happen along PDAC tumorigenesis. However, the precise mechanisms require further elucidation.
Our study confirmed a continuous increase or decrease of specific species along PDAC tumorigenesis. Interestingly, the red complex (Porphyromonas gingivalis and Treponema denticola), major periodontitis-causing pathogens, presented an increasing trend along PDAC tumorigenesis in the pancreatic tissue (26). And Porphyromonas gingivalis was also enriched in the saliva of STS patients compared with the LTS patients. The red complex can secrete peptidyl-arginine deiminase (PAD) to cause the release of neutrophil extracellular traps (NETs). NETs are elevated in both human and murine models of PDAC and associated with poor patient outcomes, metastasis, fibrosis, proliferation, and immune evasion (27, 28). Tan et al. also demonstrated accelerated tumor growth in PDAC murine models when exposed to Porphyromonas gingivalis via oral gavage. They confirmed the localization of bacteria to the pancreas and a resultant pro-inflammatory tumor microenvironment (TME) characterized by a neutrophil-dominated milieu (29). Other periodontal pathogens, including Peptoanaerobacter stomatis, Slackia exigua, Propionibacterium acidifaciens, Catonella morbi, Filifactor alocis, and Rothia dentocariosa, related to oral diseases like periodontitis or oral cavity squamous cell carcinoma (OSCC), were also increased sequentially from CP to stage IV PDAC patients in the pancreatic tissue (30–35). The sequentially increasing trends along PDAC tumorigenesis of Peptoanaerobacter stomatis and Propionibacterium acidifaciens were also observed in the duodenal fluid and saliva. In addition, Prevotella dentalis can promote the development of colorectal cancer (CRC) (36), while Slackia exigua is associated with gastric cancer (GC) (37). Porphyromonas endodontalis was identified as the new CRC driver species and the top microbial biomarker to differentiate esophageal squamous carcinoma (ESC) from controls (38, 39). In contrast, bacteria commonly used as probiotics Bifidobacterium breve and Lactiplantibacillus plantarum presented a decreasing trend along PDAC tumorigenesis in the pancreatic tissue. Indole-3-lactic acid derived from Bifidobacterium breve and Lactobacillus plantarum can ameliorate colorectal tumorigenesis via epigenetic regulation of CD8+ T cell immunity (40, 41). Whether these microbes are involved in the occurrence of PDAC needs to be further studied.
Our study had several limitations. First, the sample size was limited. Second, it remains unclear whether the observed microbial differences are the causes or consequences of PDAC. Further in vivo and in vitro research is needed to substantiate our findings.
To our knowledge, this is the first time series analysis to describe microbiota in pancreatic tissue, duodenal fluid, and saliva across the five stages of PDAC tumorigenesis. Further studies regarding the functions of the above-mentioned microbes were warranted as the results may reveal the mechanisms underlying the occurrence and development of PDAC and provide a basis for future treatments.
Conclusions
Salivary and duodenal microorganisms play a significant role in shaping the pancreatic microbial environment in the context of PDAC. Notably, Porphyromonas gingivalis, Porphyromonas endodontalis, and Filifactor alocis, which sequentially increased in pancreatic tissue from CP to stage IV PDAC patients, were also enriched in saliva of patients with STS compared with those with LTS. Further investigation into these microbial entities is essential for elucidating their precise roles in PDAC development, which may provide valuable insights for future therapeutic interventions.
Data availability statement
The data presented in the study are deposited in the Figshare repository, DOI number 10.6084/m9.figshare.26798644.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the Qilu Hospital of Shandong University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
YZ: Writing – original draft. XL: Writing – original draft. MZ: Writing – review & editing. LL: Writing – review & editing. GZ: Writing – review & editing. CC: Writing – review & editing. LW: Writing – review & editing. PW: Writing – review & editing. NZ: Writing – review & editing. QF: Writing – review & editing. ZL: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study is supported by the National Natural Science Foundation of China (No. 82261160396), the Taishan Scholars Program of Shandong Province (No. tsqn202306344), and the ECCM Program of Clinical Research Center of Shandong University (No. 2021SDUCRCB004).
Acknowledgments
We would like to thank all our participants and investigators as well as the staff of all participating hospitals, laboratories and research institutes for their contributions to the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2024.1487242/full#supplementary-material
Abbreviations
AJCC, American Joint Committee on Cancer; BMI, body mass index; CA 19-9, carbohydrate antigen 19–9; CP, chronic pancreatitis; CRC, colorectal cancer; CR/PR, complete response/partial response; ERCP, endoscopic retrograde cholangiopancreatography; ESC, esophageal squamous carcinoma; EUS-FNA, endoscopic ultrasound-guided fine needle aspiration; FDR, false discovery rate; GC, gastric cancer; IM, intra-tumoral microbiota; IPMN, intraductal papillary mucinous neoplasm; LTS, long-term survival; MCN, mucinous cystic neoplasm; NCM, neutral community model; NET, neutrophil extracellular trap; OSCC, oral cavity squamous cell carcinoma; OUT, Operational taxonomic unit; PAD, peptidyl-arginine deiminase; PCoA, principal coordinate analysis; PDAC, pancreatic ductal adenocarcinoma; PPI, proton-pump inhibitor; SCN, serous cystic neoplasm; SD/PD, stable disease/progressive disease; SPN, solid pseudopaillary neoplasm; STS, short-term survival; TME, tumor microenvironment.
References
1. Xia C, Dong X, Li H, Cao M, Sun D, He S, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). (2022) 135:584–90. doi: 10.1097/cm9.0000000000002108
2. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. (2020) 395:2008–20. doi: 10.1016/s0140-6736(20)30974-0
3. Zeng H, Chen W, Zheng R, Zhang S, Ji JS, X-n Z, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Global Health. (2018) 6 5:e555–e67. doi: 10.1016/S2214-109X(18)30127-X
4. Bracci PM. Oral health and the oral microbiome in pancreatic cancer: an overview of epidemiological studies. Cancer J. (2017) 23:310–4. doi: 10.1097/PPO.0000000000000287
5. Mei QX, Huang CL, Luo SZ, Zhang XM, Zeng Y, Lu YY. Characterization of the duodenal bacterial microbiota in patients with pancreatic head cancer vs. healthy controls. Pancreatology. (2018) 18:438–45. doi: 10.1016/j.pan.2018.03.005
6. Zhou W, Zhang D, Li Z, Jiang H, Li J, Ren R, et al. The fecal microbiota of patients with pancreatic ductal adenocarcinoma and autoimmune pancreatitis characterized by metagenomic sequencing. J Transl Med. (2021) 19:215. doi: 10.1186/s12967-021-02882-7
7. Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. (2017) 357:1156–60. doi: 10.1126/science.aah5043
8. Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. (2016) 67:120–7. doi: 10.1136/gutjnl-2016-312580
9. Nagata N, Nishijima S, Kojima Y, Hisada Y, Imbe K, Miyoshi-Akiyama T, et al. Metagenomic identification of microbial signatures predicting pancreatic cancer from a multinational study. Gastroenterology. (2022) 163:222–38. doi: 10.1053/j.gastro.2022.03.054
10. Kharofa J, Haslam D, Wilkinson R, Weiss A, Patel S, Wang K, et al. Analysis of the fecal metagenome in long-term survivors of pancreas cancer. Cancer. (2023) 129:1986–94. doi: 10.1002/cncr.34748
11. Riquelme EM, Zhang Y, Zhang L, Montiel MF, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. (2019) 178:795–806.e12. doi: 10.1016/j.cell.2019.07.008
12. Li X, Saxena D. The mycobiome-immune axis: The next frontier in pancreatic cancer. Cancer Cell. (2022) 40 2:120–2. doi: 10.1016/j.ccell.2022.01.009
13. Bucci V, Tzen B, Li N, Simmons M, Tanoue T, Bogart E, et al. MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses. Genome Biol. (2016) 17:121. doi: 10.1186/s13059-016-0980-6
14. Bogart E, Creswell R, Gerber GK. MITRE: inferring features from microbiota time-series data linked to host status. Genome Biol. (2019) 20:186. doi: 10.1186/s13059-019-1788-y
15. Takano S, Fukasawa M, Shindo H, Takahashi E, Fukasawa Y, Kawakami S, et al. Next-generation sequencing of endoscopically obtained tissues from patients with all stages of pancreatic cancer. Cancer Sci. (2022) 113:1069–77. doi: 10.1111/cas.15249
16. Roy N, Malik S, Villanueva KE, Urano A, Lu X, Von Figura G, et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. (2015) 29:658–71. doi: 10.1101/gad.256628.114
17. Chang CK, Shih TT, Tien YW, Chang MC, Chang YT, Yang SH, et al. Metabolic alterations in pancreatic cancer detected by in vivo (1)H-MR spectroscopy: correlation with normal pancreas, PET metabolic activity, clinical stages, and survival outcome. Diagnostics (Basel). (2021) 11:1541. doi: 10.3390/diagnostics11091541
18. Li XH, Zhou EL, Dong XY, Zhao CY, Han YX, Cui BK, et al. Adjuvant chemotherapy in pancreatic cancer at different AJCC stages: a propensity score matching analysis. Eur J Med Res. (2023) 28:606. doi: 10.1186/s40001-023-01572-y
19. Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi AS, et al. Characterization of the oral fungal microbiome (Mycobiome) in healthy individuals. PLoS Pathog. (2010) 6:e1000713. doi: 10.1371/journal.ppat.1000713
20. Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. (2018) 6:226. doi: 10.1186/s40168-018-0605-2
21. Zhang J, Wu Y, Liu J, Yang Y, Li H, Wu X, et al. Differential oral microbial input determines two microbiota pneumo-types associated with health status. Advanced Science. (2022) 9:2203115. doi: 10.1002/advs.202203115
22. Shenhav L, Thompson M, Joseph TA, Briscoe L, Furman O, Bogumil D, et al. FEAST: fast expectation-maximization for microbial source tracking. Nat Methods. (2019) 16:627–32. doi: 10.1038/s41592-019-0431-x
23. Bernard-Raichon L, Venzon M, Klein J, Axelrad JE, Zhang CZ, Sullivan AP, et al. Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nat Commun. (2022) 13:5926. doi: 10.1038/s41467-022-33395-6
24. Hsu CL, Schnabl B. The gut-liver axis and gut microbiota in health and liver disease. Nat Rev Microbiol. (2023) 21:719–33. doi: 10.1038/s41579-023-00904-3
25. Coyte KZ, Schluter J, Foster KR. The ecology of the microbiome: Networks, competition, and stability. Science. (2015) 350:663–6. doi: 10.1126/science.aad2602
26. Suzuki N, Yoneda M, Hirofuji T. Mixed red-complex bacterial infection in periodontitis. Int J Dent. (2013) 2013:587279. doi: 10.1155/2013/587279
27. Cools-Lartigue J, Spicer J, McDonald B, Gowing S, Chow S, Giannias B, et al. Neutrophil extracellular traps sequester circulating tumor cells and promote metastasis. J Clin Invest. (2013) 123:3446–58. doi: 10.1172/jci67484
28. Pieterse E, Rother N, Garsen M, Hofstra JM, Satchell SC, Hoffmann M, et al. Neutrophil extracellular traps drive endothelial-to-mesenchymal transition. Arterioscler Thromb Vasc Biol. (2017) 37:1371–9. doi: 10.1161/atvbaha.117.309002
29. Tan Q, Ma X, Yang B, Liu Y, Xie Y, Wang X, et al. Periodontitis pathogen Porphyromonas gingivalis promotes pancreatic tumorigenesis via neutrophil elastase from tumor-associated neutrophils. Gut Microbes. (2022) 14:2073785. doi: 10.1080/19490976.2022.2073785
30. Padial-Molina M, Montalvo-Acosta S, Martín-Morales N, Pérez-Carrasco V, Magan-Fernandez A, Mesa F, et al. Correlation between inflammasomes and microbiota in peri-implantitis. Int J Mol Sci. (2024) 25:961. doi: 10.3390/ijms25020961
31. Shen C, Simpson J, Clawson JB, Lam S, Kingsley K. Prevalence of Oral Pathogen Slackia exigua among Clinical Orthodontic and Non-Orthodontic Saliva Samples. Microorganisms. (2023) 11:867. doi: 10.3390/microorganisms11040867
32. Obata J, Fujishima K, Nagata E, Oho T. Pathogenic mechanisms of cariogenic Propionibacterium acidifaciens. Arch Oral Biol. (2019) 105:46–51. doi: 10.1016/j.archoralbio.2019.06.005
33. Cai L, Zhu H, Mou Q, Wong PY, Lan L, Ng CWK, et al. Integrative analysis reveals associations between oral microbiota dysbiosis and host genetic and epigenetic aberrations in oral cavity squamous cell carcinoma. NPJ Biofilms Microbiomes. (2024) 10:39. doi: 10.1038/s41522-024-00511-x
34. Razooqi Z, Tjellström I, Höglund Åberg C, Kwamin F, Claesson R, Haubek D, et al. Association of Filifactor alocis and its RTX toxin gene ftxA with periodontal attachment loss, and in synergy with Aggregatibacter actinomycetemcomitans. Front Cell Infect Microbiol. (2024) 14:1376358. doi: 10.3389/fcimb.2024.1376358
35. Perera M, Al-Hebshi NN, Perera I, Ipe D, Ulett GC, Speicher DJ, et al. Inflammatory bacteriome and oral squamous cell carcinoma. J Dent Res. (2018) 97:725–32. doi: 10.1177/0022034518767118
36. Shi F, Liu G, Lin Y, Guo CL, Han J, Chu ESH, et al. Altered gut microbiome composition by appendectomy contributes to colorectal cancer. Oncogene. (2023) 42:530–40. doi: 10.1038/s41388-022-02569-3
37. Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. (2018) 67:1024–32. doi: 10.1136/gutjnl-2017-314281
38. Cai P, Xiong J, Sha H, Dai X, Lu J. Tumor bacterial markers diagnose the initiation and four stages of colorectal cancer. Front Cell Infect Microbiol. (2023) 13:1123544. doi: 10.3389/fcimb.2023.1123544
39. Chen H, Jiang X, Zhu F, Yang R, Yu X, Zhou X, et al. Characteristics of the oral and gastric microbiome in patients with early-stage intramucosal esophageal squamous cell carcinoma. BMC Microbiol. (2024) 24:88. doi: 10.1186/s12866-024-03233-4
40. Li Y, Li Q, Yuan R, Wang Y, Guo C, Wang L. Bifidobacterium breve-derived indole-3-lactic acid ameliorates colitis-associated tumorigenesis by directing the differentiation of immature colonic macrophages. Theranostics. (2024) 14:2719–35. doi: 10.7150/thno.92350
Keywords: pancreatic neoplasms, pancreatitis, endoscopic ultrasound-guided fine needle aspiration, microbiota, time series analysis
Citation: Zhu Y, Liang X, Zhi M, Li L, Zhang G, Chen C, Wang L, Wang P, Zhong N, Feng Q and Li Z (2024) Succession of the multi-site microbiome along pancreatic ductal adenocarcinoma tumorigenesis. Front. Immunol. 15:1487242. doi: 10.3389/fimmu.2024.1487242
Received: 27 August 2024; Accepted: 14 October 2024;
Published: 07 November 2024.
Edited by:
Theresa Marie Rossouw, University of Pretoria, South AfricaReviewed by:
Sara Federici, The University of Manchester, United KingdomAncuta Lupu, Grigore T. Popa University of Medicine and Pharmacy, Romania
Copyright © 2024 Zhu, Liang, Zhi, Li, Zhang, Chen, Wang, Wang, Zhong, Feng and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Li, cWlsdWxpemhlbkBzZHUuZWR1LmNu; Qiang Feng, ZmVuZ3FpYW5nQHNkdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Yiqing Zhu1,2,3,4†
Yiqing Zhu1,2,3,4† Lixiang Li
Lixiang Li Guoming Zhang
Guoming Zhang Ning Zhong
Ning Zhong Qiang Feng
Qiang Feng Zhen Li
Zhen Li