- 1Department of Agricultural Microbiology, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
- 2Biochemistry Department, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
- 3Nutrition and Food Sciences Department, National Research Centre, Giza, Egypt
- 4Department of Food Science, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
- 5Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, King Khalid University, Abha, Saudi Arabia
- 6Biology Department, College of Science, King Khalid University, Abha, Saudi Arabia
- 7Blood Products Quality Control and Research Department, National Organization for Research and Control of Biologicals, Cairo, Egypt
- 8Department of Poultry Diseases, Faculty of Veterinary Medicine, Cairo University, Giza, Egypt
- 9Department of Diseases of Birds, Rabbits, Fish & their Care & Wildlife, School of Veterinary Medicine, Badr University in Cairo (BUC), Cairo, Egypt
- 10Molecular Cell Biology Unit, Division of Biochemistry, Department of Chemistry, Faculty of Science, Tanta University, Tanta, Egypt
- 11Division of Biochemistry, Department of Chemistry, Tanta University, Faculty of Science, Tanta, Egypt
- 12Faculty of Control System and Robotics, Information Technologies, Mechanics and Optics University, Saint-Petersburg, Russia
- 13Production Engineering and Mechanical Design Department, Faculty of Engineering, Menofia University, Menofia, Egypt
- 14Soils and Water Science Department, Faculty of Agriculture, Fayoum University, Fayoum, Egypt
- 15Department of Bio-Analytical Science, University of Science and Technology, Daejeon, Republic of Korea
- 16Faculty of Biotechnology, October University for Modern Sciences and Arts, 6th October City, Egypt
- 17Faculty of Medicine, University of Debrecen, Debrecen, Hungary
- 18Department of Biology, College of Science, United Arab Emirates University, Al Ain, United Arab Emirates
- 19Food Microbiology and Biotechnology Laboratory, Food and Nutritional Science Program, North Carolina A&T State University, Greensboro, NC, United States
In recent years, medicinal plants have gained significant attention in modern medicine due to their accessibility, affordability, widespread acceptance, and safety, making herbal remedies highly valued globally. Consequently, ensuring medicinal plants’ quality, efficacy, and safety has become a critical concern for developed and developing nations. The emergence of multidrug-resistant microorganisms poses a serious global health threat, particularly in low-income regions, despite significant advancements in antimicrobial drugs and medical research over the past century. The rapid spread of these multidrug-resistant infections is primarily attributed to improper prescriptions, overuse, and unregulated access to antibiotics. Addressing these challenges, the standardization of plant-derived pharmaceuticals could pave the way for a transformative era in healthcare. Preserving and leveraging the historical knowledge of medicinal plants is essential before such valuable information is lost. Recently, there has been growing interest among natural and pharmaceutical scientists in exploring medicinal plants as potential sources of antimicrobial agents. This current review aims to identify the most common pathogens threatening human health, analyze the factors contributing to the rise of drug-resistant microorganisms, and evaluate the widespread use of medicinal plants across various countries as alternative antibiotics, highlighting their unique mechanisms of antimicrobial resistance.
1 Introduction
In 1928, Alexander Fleming serendipitously discovered penicillin, the first natural antibiotic, marking the onset of antibiotic resistance in the early 20th century (1). This initial discovery catalyzed the advancement of further antibiotics, including streptomycin, chloramphenicol, erythromycin, and chlortetracycline, resulting in a “golden era” of antibiotic development from 1960 to 1980 (2). Nonetheless, the goals of the pharmaceutical sector have changed, resulting in a decrease in the discovery of novel antibiotics and an increase in antimicrobial resistance (AMR) (3). As a result, drug-resistant bacterial infections currently represent a considerable worldwide health risk (3, 4). The comprehensive antibiotic resistance database (CARD) includes more than 5,000 resistance sequences, with a restricted subset linked to notable diseases of concern (4). The problem is exacerbated by the slow pace of new effective antibiotic discovery (5).
AMR is a natural phenomenon in which microorganisms (bacteria, fungi, and protozoa) acquire the capability to endure and proliferate despite the administration of medications such as antibacterial, antifungal, and antiprotozoal antibiotics (6). Antibiotics are essential for addressing bacterial illnesses; however, their extensive application, particularly in resource-limited environments, generates selective pressure that fosters the development of resistance (7, 8). This results in heightened morbidity and death (8). The emergence of “superbugs,” which are resistant to many medications, highlights the gravity of the issue, leading the World Health Organization (WHO) to designate AMR as a significant global health challenge (7). Recent evidence reveals that AMR currently accounts for millions of fatalities annually, with forecasts indicating a substantial rise to 10 million deaths per year by 2050 (9).
The rise of antibiotic resistance poses a significant threat to global health, with methicillin-resistant Staphylococcus aureus (MRSA) serving as a prime example of a “superbug” contributing substantially to mortality from drug-resistant infections (10). Bacteria, among the earliest life forms on Earth, are ubiquitous, inhabiting both domestic and professional environments. While most bacterial species are harmless to humans, specific strains, including S. aureus, Helicobacter pylori, Escherichia coli, and Bacillus anthracis, can overcome host defenses and induce severe illnesses (10, 11). These infections can present as various diseases, including pneumonia, endocarditis, septicemia, and osteomyelitis, underscoring the broad pathogenic capabilities of these organisms (11, 12).
Healthcare-associated infections (HAIs) represent a persistent challenge to patient safety and public health, often leading to serious complications and placing a considerable burden on society (13). Conventional methods for preventing clinical infections have focused on aseptic techniques and systemic antibiotic treatments (13). However, these methods frequently fail to combat established infections effectively (14). A particularly striking example of this challenge is the treatment of infections associated with medical devices. Systemic antibiotic therapy for infections linked to devices such as catheters, artificial prosthetics, subcutaneous sensors, and orthopedic implants demonstrates a disappointingly low success rate, ranging from 22% to 37% (15). The restricted effectiveness highlights the challenge of eliminating established infections when foreign elements are present since they might facilitate bacterial colonization and biofilm development (15).
Furthermore, administering high doses of antibiotics, often necessary to treat localized infections, can harm surrounding tissues. Such high concentrations can lead to cytotoxicity and adverse reactions, further complicating treatment and potentially hindering patient recovery (16). The misuse of antibiotics may be even more alarming due to its contribution to the acceleration of bacterial drug resistance development and dissemination (17). The selective pressure exerted by frequent antibiotic exposure allows resistant strains to thrive, gradually diminishing the effectiveness of these crucial medications (17). This establishes a detrimental loop in which infections become progressively difficult to manage, hence intensifying the demand for elevated antibiotic dosages and aggravating the issue of resistance (17). The convergence of these factors - the rise of superbugs such as MRSA, the difficulties in treating device-related infections, and the adverse effects of high-dose antibiotic therapy - highlights the critical need for novel strategies to prevent and manage bacterial infections in the face of rising AMR (17).
Despite advancements in developing novel antimicrobial agents to address drug-resistant bacteria, such as antibacterial peptides, amphiphiles, and antimicrobial materials, including nanoparticles, hydrogels, engineered surfaces, and coatings, bacterial resistance continues to pose a substantial challenge (18). Current research efforts are, therefore, increasingly focused on developing strategies that can effectively eliminate bacteria without simultaneously driving the evolution and spread of further resistance (18). Consequently, it is imperative to identify novel antimicrobial treatment agents. In recent years, scientific and pharmaceutical communities have exhibited an interest in medicinal plants as potential sources of antimicrobial drugs. Employing selective screening of phytochemicals through medical data offers a dependable approach for identifying innovative medicines (19).
Chinese researchers identified artemisinin from Artemisia annua utilizing insights from traditional Chinese medical literature (20). This discovery has already led to the rescue of millions of individuals suffering from malaria (21). The WHO currently recommends a combination medication based on artemisinin as the treatment for this highly fatal disease. Currently, this medication is extensively utilized worldwide (22). Due to their historical effectiveness as anti-infective agents, plants historically employed for medicinal purposes may be crucial in identifying innovative therapeutics for diverse microbial illnesses (23–25). Ancient herbal treatments have been used for centuries to ease disorders and improve general well-being (26, 27). Typically, medicinal plants’ leaves, bark, roots, and flowers are amalgamated to produce an infusion (28, 29).
For example, compounds from the family Zingiberaceae, such as turmeric (Curcuma longa), and tamarind (Tamarindus indica), have been used to cure several diseases caused by pathogenic microorganisms, including diarrhea and dysentery (30, 31). Few studies have investigated the anti-infective properties of medicinal plants, despite numerous recent ethnobotanical surveys indicating their use by individuals to mitigate infectious ailments (32, 33). Many other studies have conducted targeted studies on the beneficial effects of compounds derived from medicinal plants, such as anti-Candida agents (34), anti-biofilm agents (35), and inhibitors of resistant microbial isolates (36).
The current review comprehensively examines the multidrug-resistant (MDR) microorganisms that present the greatest threat to human health. It analyzes the various causes contributing to antibiotic resistance and emphasizes the potential of medicinal herbs as a safe and efficient treatment.
2 Antimicrobial resistance
In recent decades, microorganisms have increasingly resisted commonly used antibiotics (37, 38). Current medical concerns include not just infectious diseases such as avian influenza, human immunodeficiency virus (HIV), and severe acute respiratory syndrome (SARS) but also the emergence of resistant microorganisms that cause diseases that were previously eradicated, such as tuberculosis and malaria (39). In the past thirty years, the ineffectiveness of antibiotics and the absence of effective vaccinations have led to the demise of more than 25 million individuals, including over 5 million children (7).
Antibiotic resistance occurs when a bacterium becomes unresponsive to previously effective medications. Several frequently utilized antibiotics are now ineffective against 70% of the bacteria that cause nosocomial infections (40). Numerous mechanisms of medication resistance to various illnesses have been proposed, especially for the most commonly utilized pharmaceuticals (40). The modes of action of several antibiotics on Gram-positive and Gram-negative bacteria are illustrated in Figure 1.
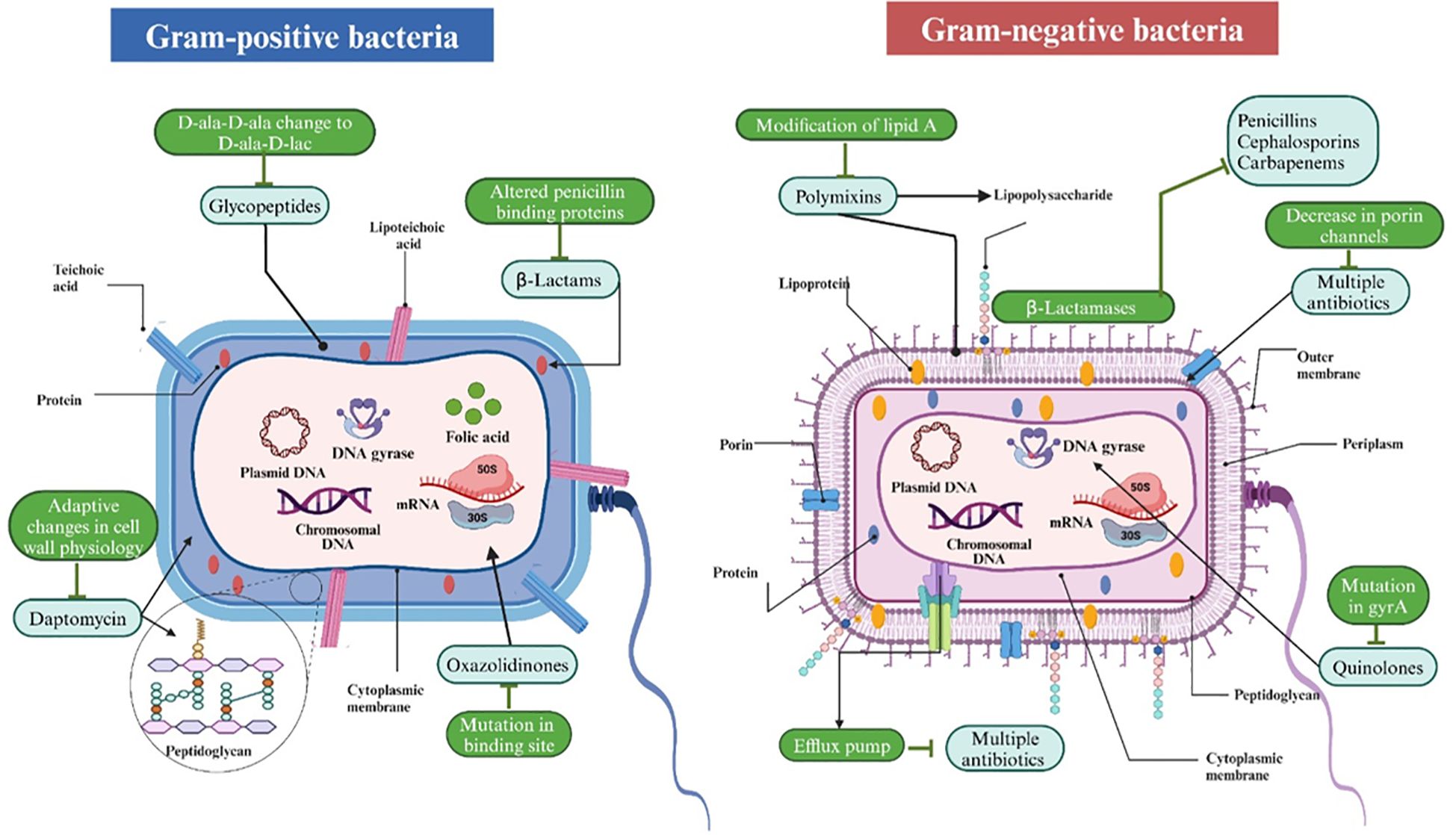
Figure 1. The mechanisms of action of several antibiotics on Gram-positive and Gram-negative bacteria. Gram-positive bacteria, characterized by a thick peptidoglycan layer and teichoic acids, are vulnerable to antibiotics such as glycopeptides (e.g., vancomycin) that impede cell wall formation and daptomycin, which affects membrane potential. Gram-negative bacteria, characterized by an outer membrane and a thinner peptidoglycan layer, exhibit lower permeability yet remain susceptible to polymyxins that compromise the outer membrane and other agents that affect ribosomes (protein synthesis) or DNA gyrase (DNA replication). Both types exhibit analogous vulnerabilities in protein synthesis, folic acid metabolism, and DNA replication, which are targeted by antibiotics such as tetracyclines, sulfonamides, and quinolones, respectively. Resistance mechanisms are also demonstrated, including modified penicillin-binding proteins, beta-lactamase synthesis, efflux pumps, and porin alterations.
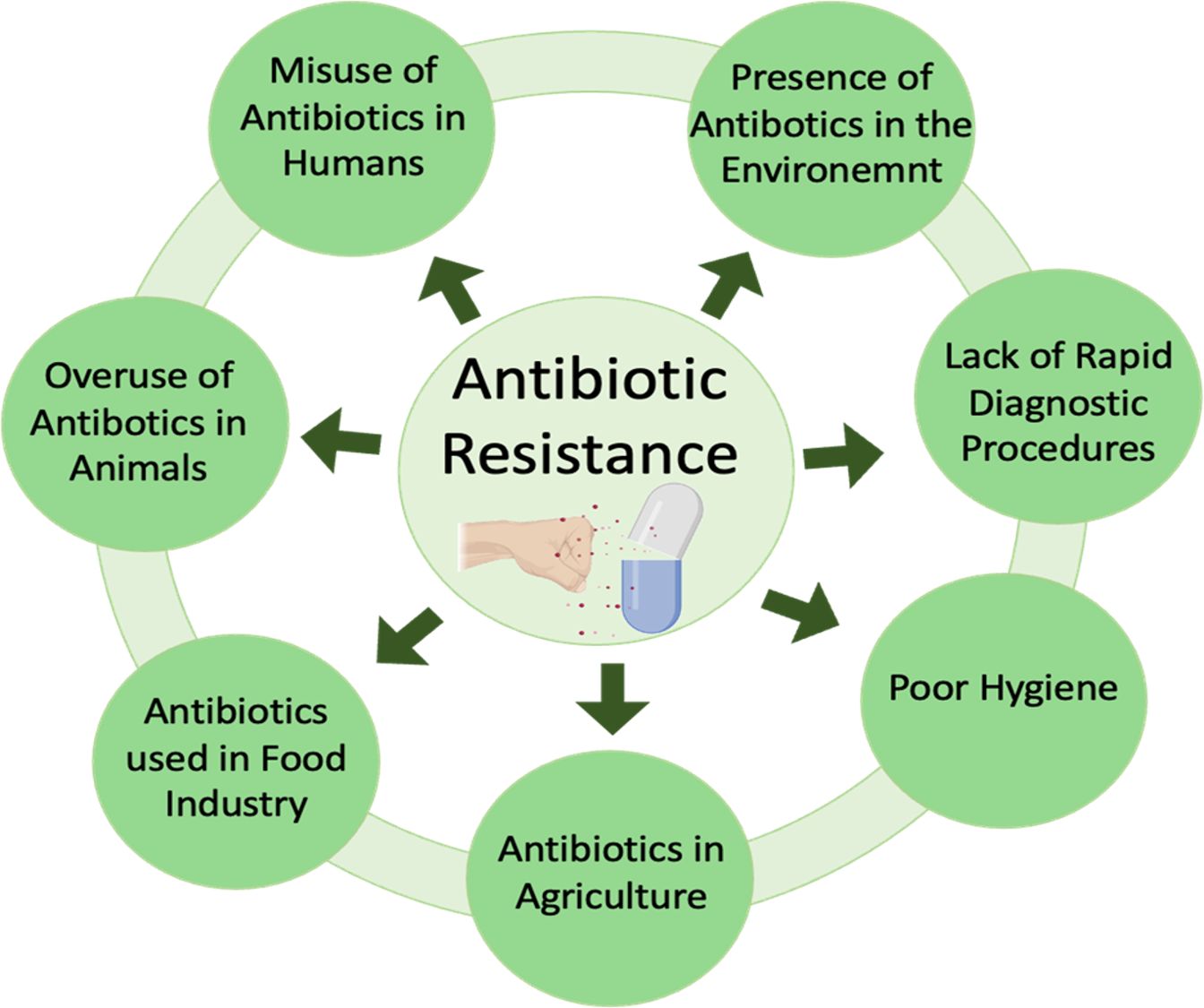
The improper use of antibiotics has been demonstrated to increase the development and spread of drug-resistant microorganisms (41). Inadequate systems for ensuring the quality and continuous supply of medications, coupled with ineffective surveillance and monitoring mechanisms, along with insufficient control and preventive measures, have substantially facilitated the rise of antibiotic resistance, especially in developing nations where policies are deficient (41). Comprehending how these detrimental organisms evade treatment is essential for formulating alternatives or preserving the effectiveness of current antimicrobial therapies. The spread of antibiotic resistance through foods is depicted in Figure 2.
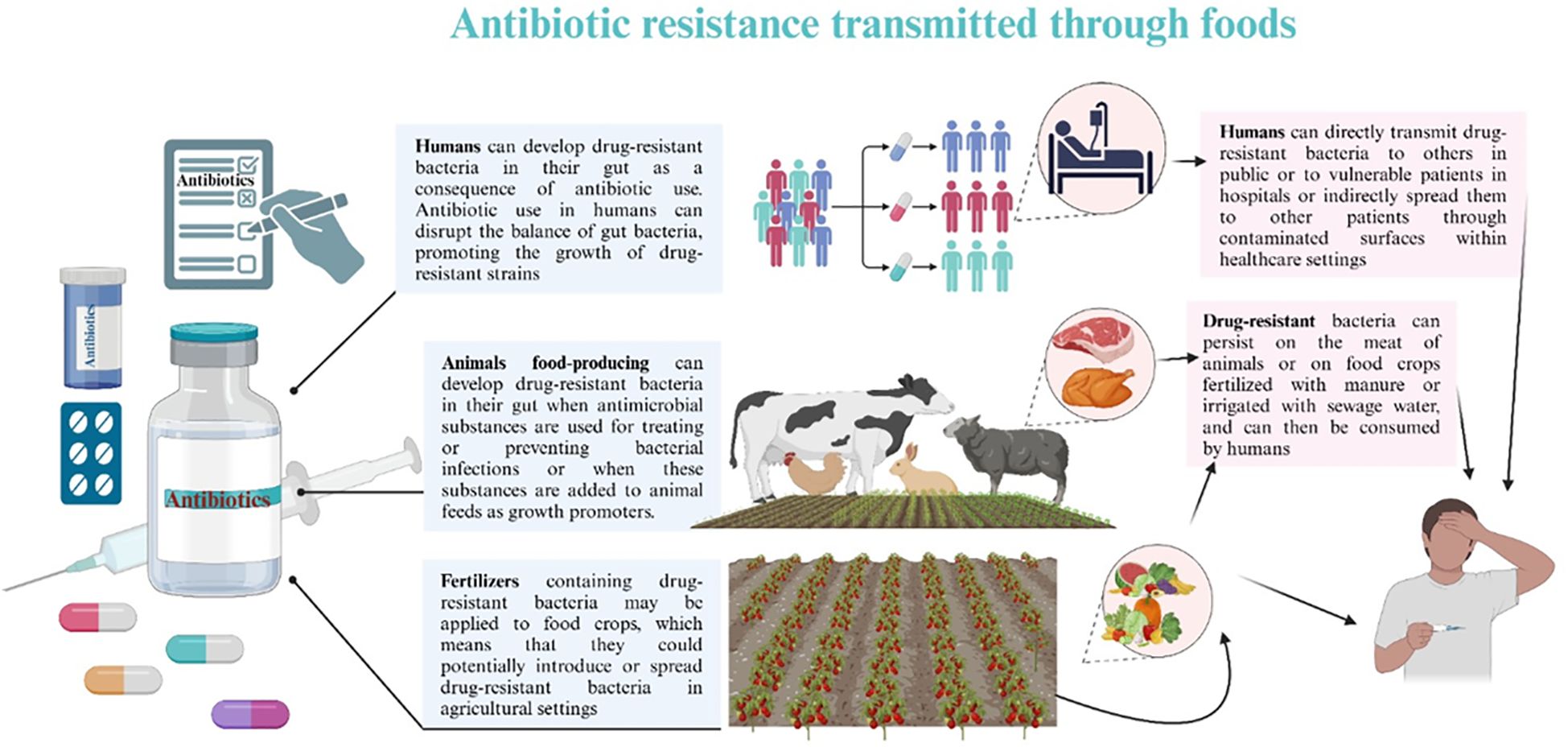
Figure 2. Antibiotic resistance spread through foods. Animal-derived foods, such as dairy, meat, and poultry, provide the principal source of most foodborne microbial diseases. The improper use of antimicrobial agents in animal feed is the principal factor contributing to the increasing public health issue about multi-drug resistant (MDR) microorganisms in these foods. The application of antibiotics in agriculture, livestock production, and human healthcare exerts selection pressure on genes that impart antibiotic resistance. The over-application of antibiotics in animal farming as growth promoters significantly contributes to the development and proliferation of antibiotic resistance. MDR microorganisms, antibiotic residues, and resistance genes can enter agricultural soils via sewage sludge and animal manure, ultimately infiltrating the crop microbiome and the human food chain. Ingesting food can transmit these resistance genes into the human microbiome, posing a significant public health threat.
3 Factors influencing the development of microbial resistance against antibiotics
Genetic alterations can result in the development of antibiotic resistance by mechanisms such as point mutations, deletions, insertions, or the horizontal transfer of resistant genes (42). Moreover, it has been hypothesized that phenotypic drug resistance may arise concurrently with genetic alterations (42). For an antibiotic to be effective, it must possess the capability to penetrate the cell membrane. Consequently, whatever alterations the bacteria induce in these areas will enhance their resistance to treatments (42).
Bacteria can affect antibiotic permeability in many ways, including modifying lipopolysaccharides and obstructing molecular interactions with the drug. Alternative mechanisms encompass the development of membrane vesicles, which increases membrane surface area, hence diminishing the amount of medication entering the cell; alterations in antibiotic transporters (porins); and the utilization of efflux pumps to expel the antibiotic from the cytosol (43). Figure 3 depicts a detailed review of the factors contributing to microorganism antibiotic resistance.
3.1 Genetically based drug resistance
Drug resistance in pathogenic microorganisms may be intrinsic or acquired. Foreign-resistant genes can be introduced into the recipient organism by transformation, transduction, or conjugation to confer resistance (43).
3.2 Mutations
Mutations at many chromosomal locations cause this AMR and can take the following forms: i) spontaneous mutations: this type of mutation often occurs in actively dividing cells, thus the name growth-dependent mutations (43). They arise as random replication mistakes or improper DNA damage repair (43); (ii) hypermutators: these are highly mutable bacteria, and their hypermutable condition results from prolonged exposure to sublethal antibiotic dosages. These cells can escape the hypermutable stage if they acquire a beneficial mutation and begin to divide and multiply (44); and (iii) adaptive mutagenesis: this mutation happens in slow-growing cells exposed to sublethal antibiotic dosages (43).
3.3 Horizontal gene transfer
Horizontal gene transfer allows microorganisms to acquire antibiotic resistance genes from unrelated organisms. This transfer occurs primarily through transformation, transduction, and conjugation. Recombination, a separate process, can also introduce or modify resistance genes within a recipient’s genome (44).
3.4 Phenotypic antibiotic resistance
Phenotypic drug resistance, occurring without genetic changes, arises from a microorganism’s physiological state and its inherent survival strategies (45). This resistance can manifest through various mechanisms, including drug indifference, biofilm formation, persistence, inoculum size effects, and metabolic-state-dependent alterations in antibiotic susceptibility (45).
3.5 Drug indifference
Susceptibility to antimicrobial agents can vary depending on an organism’s physiological state (46). For instance, dormant cells have demonstrated resistance to ampicillin and tetracycline, whereas rapidly dividing cells are more vulnerable. Conversely, while not actively growing, stationary-phase cells are susceptible to ciprofloxacin and streptomycin (46).
3.6 Persistence
The punctate age of dormant cells reveals the occurrence in an antibiotic-resistant microbial culture (47). Persistence comes in two different varieties: Type I and Type II. The first kind is induced by cellular deprivation, whereas the second arises due to the developing cells’ innate biostability (48).
3.7 Biofilms
Biofilm development results from attaching free-living (planktonic) cells to surfaces, as detailed below (49): (i) it involves attaching free-living cells to a pre-conditioned surface by random propulsion, chemotaxis, or motion (49). The initial cell attachment is often reversible and is influenced by various physical and chemical variables (50); (ii) The production of adhesion proteins, such as fimbriae, helps in the permanent attachment of the cells. In addition, internal linkages and extracellular polymeric substances (EPS) emerge after cell adhesion (51). The EPS is formed of polysaccharides, such as colonic acid, chitosan, and alginate, typically generated by the microbial cells and serves as a covering for the sessile cells. Some enzymes, RNA, DNA, nutrients, proteins, and surfactants are additional EPS components (51).
During this stage, the biofilm community experiences microbial proliferation and communication using chemical signaling, leading to micro-colonies growth (52). The biofilm’s resistance to some antimicrobials results from its capacity to impede the antibiotics’ penetration via the EPS layer. For example, Pseudomonas aeruginosa biofilms are resistant to aminoglycosides (often positively charged) due to the organisms’ alginate secretion, which binds to the positively charged antibiotic (53).
3.8 Drug resistance and inoculum size
The efficacy of antibiotic treatment is supposedly affected by inoculum size (54). The elevated importance of the infectious dose of microorganisms in vivo may elucidate the disparity between an antibiotic agent’s minimum inhibitory concentration (MIC) in vitro and in vivo (54). An increased quantity of bacteria that generate enzymes capable of degrading drugs correlates with an escalation in the degradation of medications. The consequence for species devoid of these degrading enzymes remains uncertain (54).
Two factors cause the AMR in these bacteria: (i) a drop in drug concentration resulting from the existence of supplementary drug target sites in both viable and non-viable cells, and (ii) an inadequate ratio of medicinal compounds to a high concentration of microorganisms (55). Based on experimental and mathematical models, both outcomes could apply to drug resistance depending on the size of the inoculums (56).
4 Examples of some common MDR microorganisms and their effects on human health
Infectious diseases contribute to around 15% of the 57 million fatalities globally each year, considered to be one of the 21st century’s critical worldwide human health challenges (57). Infectious diseases continue to be the top causes of death and morbidity, accounting for almost 2.2 million deaths among school-aged children and adolescents in 2019. This is particularly true in countries with low and medium incomes (58). Bacteria, fungi, parasites, and viruses cause human infectious illnesses. In underdeveloped nations, the frequency of microbial infections that cause diarrhea, vomiting, respiratory disorders, urinary tract infections, and hospital-acquired diseases is still high (57).
Individuals with immunodeficiency disorders are susceptible to transforming commensal bacteria into pathogens. Candida albicans, Enterococcus faecalis, E. coli, P. aeruginosa, S. aureus, and others have been identified as the most common bacteria and yeast responsible for this infection (57). In addition to causing illnesses, E. coli and S. aureus pose a significant challenge in underdeveloped countries due to their ability to acquire drug resistance (57, 58).
The revival and spread of microbiological infections may be attributed to impoverishment, illiteracy, insufficient sanitation, limited supply of treatments, and deficient health care systems (59). Emerging illnesses and MDR microorganisms have exacerbated the prevalence and severity of infectious diseases worldwide (60). MDR tuberculosis, vancomycin-resistant S. aureus, and enterococci are drug-resistant human infections threatening worldwide public health (61). Given these findings, adequate mitigation strategies must be implemented in the healthcare and public health sectors (61). The Centers for Disease Control and Prevention has proposed numerous solutions, including prevention, increased surveillance, and the development of novel treatments such as antimicrobials derived from natural sources (61).
A microbial infection occurs when a microorganism can enter a host’s tissue, damage cells, and cause illness or death by avoiding detection by the host’s defenses, releasing substances into the bloodstream that facilitate further invasions (such as toxins), and exhibiting other clinical symptoms (62). The skin and mucosal surfaces contain many microorganisms contributing to the human body’s natural flora. They often aid the host by countering possible infections at attachment sites and nutrition and producing harmful antimicrobial chemicals against bacteria (62).
Infectious diseases may be transmitted via several means, such as direct contact between individuals, transfer through the respiratory system, contact with mucous membranes or through sexual activity, injection into the bloodstream, transmission by insects, and contact with contaminated objects (62).
Despite substantial efforts in control and prevention, infectious illnesses persist as the primary cause of disease and mortality worldwide (63). About 9.5% of adult deaths per year are attributed to tuberculosis and lower respiratory diseases (63). Furthermore, children under the age of five are disproportionately affected by contagious diseases, including acute respiratory infections, diarrhea, measles, malaria, and HIV/AIDS, which together contribute to 30% of the total yearly death rate (63). The fast growth of MDR microorganisms has been related to the abuse and overuse of antimicrobial agents with extended-release formulations used to treat illnesses (64). Failure to adhere to prescribed antibiotics elevates the risk of developing antibiotic-resistant pathogens, chronic illnesses, and mortality (65). Second- or third-line drugs, typically more expensive and potentially hazardous, are utilized when resistance to first-line antibiotics develops prevalent (65, 66).
The misuse of antibiotics, along with the swift emergence of microbiological illnesses, is further affected by the following aspects (64): (i) Demographic transitions have led to a growing number of vulnerable populations, particularly the elderly, necessitating hospital-based interventions that expose patients to prevalent drug-resistant pathogens, thereby jeopardizing their health; (ii) Urbanization contributes significantly to the proliferation of diseases such as typhoid, tuberculosis, respiratory infections, and pneumonia, exacerbated by overpopulation and unsanitary conditions; (iii) Environmental degradation, pollution, and variable climatic conditions influence the incidence and transmission of diseases, particularly those vectored by insects, such as malaria; (iv) The AIDS epidemic has markedly increased the population of immunocompromised individuals susceptible to various illnesses; and (v) Annually, millions of cases arise from drug-resistant infectious diseases, including tuberculosis and malaria, due to their recurrent nature (64). Most of this cost is shouldered by low-income countries, mostly due to their insufficient social investments in infrastructure, educational access, training, and other resources necessary for managing and mitigating the development of drug resistance (63, 65, 66).
4.1 E. faecalis
Enterococci are facultative anaerobes, ovoid (0.5–1 µm in diameter), stationary, and non-sporulating, Gram-positive cocci generally regarded as common human commensals (67). They are exceptionally adapted to prosper in nutrient-rich environments that are deficient in oxygen, such as the diverse ecosystems present in the oral cavity, gastrointestinal tract, and vaginal canal (68). The typical quantity of bacterial cells excreted is roughly 1011 per gram of feces. They predominantly constitute the commensal gut microflora (69).
Although there are over 40 physiologically unique species of Enterococcus, only two species account for over 90% of enterococci infections: E. faecalis, and E. faecium (67). Infrequently, enterococci induce infections in healthy and consistent hosts. Endocarditis, sepsis, surgical wound illnesses, urinary system diseases, secondary bacteremia, and inflammatory bowel diseases are among the nosocomial ailments to which E. faecalis is reportedly becoming a significant contributor, according to surveillance data (70).
Enterococci species’ pathogenicity is determined by several characteristics, such as their typical habitat of the gut, how easily they increase there, the ability to detect a range of extracellular matrix proteins, and the availability of collecting material (71). Enterococci rely on biofilm formation to facilitate the development of endodontic and urinary system infections and endocarditis. Sortase C gene, EBPa, EBPb, and EBPc are the components of endocarditis and biofilm-associated pili (EBP) (72). E. faecalis secretes virulence factors that actively suppress host immune responses, including the production of toxins such as cytolysin and extracellular enzymes (73). Antibiotic resistance in enterococci increases the likelihood of colonization and infection and is directly associated with the therapeutic importance of the bacteria. All enterococci have reduced sensitivity to ampicillin, cephalosporins, penicillin, and all semisynthetic penicillins due to the production of penicillin-binding proteins with poor affinity (73).
One of this adaptation’s most obvious effects is the emergence of the genes that cause glycopeptide-vancomycin resistance (74). During the 1960s, MRSA emerged and spread in correlation with the use of vancomycin (74). Conversely, unlike MRSA, enterococci can assemble and sustain a range of gene clusters encoding the metabolic apparatus required for vancomycin resistance (74). Antibiotic-resistant enterococci can transfer their resistance to other pathogens, such as S. aureus and antibiotic-susceptible enterococci, by pheromone-mediated conjugative plasmids or transposons. This significantly threatens public health (74).
Recombinant transmission of dynamic genetic components requires interaction between the donor and recipient cells (74). The interaction occurs inside the microbial biofilm and is encouraged by the release of bacterial sex pheromones, which are small peptides that induce a sexual response and lead to the aggregation or clustering of cells (74). Up to 90% of human enterococcal infections are caused by E. faecalis, which also ranks third among hospital-acquired diseases brought on by resistant bacteria. For these reasons, E. faecalis is a serious hazard to human health (74).
4.2 S. aureus
S. aureus is a Gram-positive nonmotile bacterium which does not produce spores. The diameter of S. aureus ranges from 0.5 to 1.5 µm (75). It constitutes a component of the standard microbial population within the human body, namely in the anterior region of the nostrils, and has the potential to induce infections in individuals with compromised immune systems (75). Approximately 30% of individuals are permanent carriers of this bacterium. S. aureus has a resilient, somewhat amorphous protective coating measuring 20–40 nm in thickness on its cell wall (76). Peptidoglycan, constituting 50% of the staphylococci’s mass, is crucial for forming the thick, multilayered structure of the cell wall and sustaining the elevated intracellular osmotic pressure of the bacterium (77, 78).
When the host’s immune systems are compromised, colonization increases the probability of infection as the pathogen becomes virulent (79, 80). S. aureus is a significant contributor to both hospital-acquired and community-acquired infections, which can result in serious consequences if not addressed. The excessive use of antibiotics intensifies this organism’s resistance to drugs (81, 82).
S. aureus infects by breaching the integrity of the skin or mucous membranes, disseminating locally or systemically to distant organs, leading to severe and life-threatening invasive infections, including bloodstream infections, pneumonia, systemic infections, joint infections, and osteomyelitis (83). It also induces toxin-mediated illnesses when ingested through contaminated food. Food poisoning, and toxic shock are toxin-mediated illnesses caused by S. aureus (84, 85). In certain circumstances, exposure to secretions such as saliva or aerosols emitted during sneezing or coughing may facilitate disease transmission. This bacterium may also be present in unpasteurized milk and other animal products (86).
Pigs, cattle, and poultry may contract S. aureus, resulting in mastitis, arthritis, septicemia, and other conditions (87). The virulence factors of S. aureus are defined by their mechanisms of invasion and proliferation, production of extracellular substances, toxins, and capacity to form biofilms (88, 89). Infection pathogenesis often commences with cellular attachment, succeeded by bacterial invasion and colonization of host tissue. This process releases many adhesion proteins, including elements of the microbial surface that recognize sticky matrix molecules (90, 91).
In contrast to virulence proteins associated with the cell wall, the secreted virulence factors of S. aureus aggressively undermine human defenses by causing harm to host cells and tissues (91). They also impair the host’s immune response by releasing nutrients and promoting the dissemination of pathogens (92). Released virulence factors are classified into four primary categories: superantigens, diverse exoenzymes, random proteins, and pore-forming toxins (88, 93).
Research has shown that staphylococci can acquire resistance to antibiotics such as erythromycin, ampicillin, tetracycline, methicillin, and vancomycin (94). The van gene confers resistance to vancomycin, likely resulting from gene transfer between S. aureus and vancomycin-resistant enterococci (94). The swift global proliferation of methicillin- and vancomycin-resistant S. aureus strains is rendering containment increasingly unfeasible, potentially reverting us to the pre-antibiotic era (94). In regions with minimal methicillin resistance, penicillin, β-lactam antibiotics, cephalosporins, and clindamycin continue to be effective in treating S. aureus infections (95). The utilization of non-lactam antibiotics for the treatment of S. aureus infections has markedly increased owing to the emergence of antibiotic-resistant bacteria, especially MRSA (96).
Methicillin resistance in S. aureus developed swiftly in 1961, within two years after the drug’s clinical launch (97). This bacterium is intrinsically vulnerable to β-lactam agents that impede cell wall synthesis by binding to proteins implicated in peptidoglycan assembly (98). MRSA exhibits resistance to β-lactam antibiotics, including methicillin, isoxazolyl penicillin, and cephalosporins, by modifying its target site. The modification is achieved through the acquisition of penicillin-binding protein 2a, produced by the mecA gene (99). Nosocomial infections, including MRSA, have been acknowledged for numerous years (100). This bacterium can colonize and establish biofilms on biomaterials, leading to a significant prevalence of hospital-acquired infections referred to as HA-MRSA. HA-MRSA is a kind of MRSA infection identified when a positive culture is obtained more than 48 h after a patient’s hospital admission. This infection is managed in a hospital environment (21).
Community-acquired MRSA refers to the transmission of MRSA to the broader community. Various populations, including athletes, inmates, prospective military personnel, nursery attendees, injectable drug users, those often exchanging contaminated commodities, and residents of densely populated areas, have been associated with community-acquired MRSA epidemics (101, 102). Individuals with MRSA who come into contact with animals may transmit the bacterium, leading to livestock-associated infections (LA-MRSA). LA-MRSA has been identified in many animal species (103).
4.3 S. epidermidis
S. epidermidis, a member of the human body’s normal flora, is frequently isolated from various epithelia, including the nose, axillae, and head (104). This organism normally has a symbiotic relationship with its host and prevents pathogenic microorganisms from invading the host (105). Because of its exploitative qualities, particularly in nosocomial infections in immunocompromised patients, the medical community is very interested in controlling this disease (106). Infections produced by coagulase-negative S. epidermidis, possibly related to its proclivity to build biofilms, are prevalent in patients with implanted medical devices (107).
S. epidermidis infections can be caused by normal skin flora at the insertion site, contaminated medical equipment prior to implantation, or healthcare professionals (108). It has also been proposed that these infections were caused by long-term hospitalizations, surgical procedures after implantation, infections in neighboring tissues, and tissue damage following implantation operations (108, 109). Methicillin, one of the principal treatments for Staphylococcus species infections, is resistant to 75-90% of S. epidermidis strains obtained from hospital specimens (110). This bacterium has several antibiotic-resistant forms, including tetracycline, chloramphenicol, erythromycin, sulfonamides, fluoroquinolones, clindamycin, and rifamycin (111).
4.4 L. monocytogenes
L. monocytogenes is a bacterium that causes illness and belongs to the Gram-positive bacteria found in wet environments such as soil, water, and rotting plants and animals (112). It may thrive with refrigeration and other food preservation techniques. People who consume L. monocytogenes-contaminated food may get listeriosis. L. monocytogenes is commonly transmitted through contaminated food processing (112). The symptoms of the condition might persist anywhere from a few days to many weeks, depending on its severity. Fever, muscle cramps, vomiting, diarrhea, and nausea are examples of minor symptoms (112). The most severe form of listeriosis is characterized by headaches, stiff neck, disorientation, loss of balance, and convulsions. Listeriosis can kill infants, the elderly, and immunocompromised people (112, 113).
4.5 E. coli
E. coli, a Gram-negative, rod-shaped, facultatively anaerobic coliform bacterium, is frequently found in the lower intestines of warm-blooded mammals (114). The majority of E. coli strains are typical residents of the gastrointestinal tracts of humans and animals. Furthermore, it was demonstrated that E. coli cells penetrate the neonatal digestive tract immediately after birth (115). The disease-causing groups are classified into two categories: one responsible for intestinal problems and another for infections beyond the intestines, such as sepsis and bladder infections (116).
E. coli is characterized by a diverse array of harmful genotypes, categorized as pathotypes based on their ability to cause disease. Entero-toxigenic E. coli (ETEC) strains have been identified in neonates experiencing diarrhea in developing countries, and they are responsible for secretory diarrheal infections in animals (117). Two serotypes of enteropathogenic E. coli (EPEC) are present: O55:H6 and O127:H6, with atypical strains inducing diarrhea in developed nations. Conversely, the majority of prevalent strains are obtained by fecal contamination (118). Enterohemorrhagic E. coli (EHEC) strains are associated with foodborne infections commonly acquired by the consumption of milk and inadequately cooked animal products, particularly meat (118). Two distinct kinds of Shiga-like toxin producers are found in this group: stx1 and stx2 (119). Entero-invasive E. coli (EIEC) have comparable biochemical, genetic, and pathogenic properties with Shigella species and are known to cause various human diseases (119). Colitis and watery diarrhea are the most common diseases EIEC causes; these infections differ from those brought on by other strains of the same bacteria (119).
Another variant of E. coli is referred to as diffusely adherent E. coli (DAEC) strains. These strains are characterized by the diffusely adherent pattern noted in the HEp-2 adherence assay and are implicated in the etiology of watery diarrhea syndrome in adults (120). Enteroaggregative E. coli (EAEC) is a strain characterized by its remarkable adhesion to HEp-2 cells in culture. They are associated with chronic diarrhea, notably in underprivileged nations and the developed world, particularly among those with HIV/AIDS (120). Human extra-intestinal infections may also be induced by E. coli pathotypes linked to sepsis, meningitis, urinary tract infections, and bloodstream infections. E. coli O18:K1:H7 can cause invasive infections in newborns and urinary tract problems. Certain E. coli strains exhibit resistance to carbapenems, β-lactams, and various other pharmaceuticals (120).
4.6 K. pneumoniae
It is a Gram-negative bacterium that commonly inhabits human lips, skin, and intestines, and has been associated with nosocomial infections. Certain illnesses, such as cancer, diabetes mellitus, liver and biliary tract conditions, and alcoholism, impair the immune system, heightening vulnerability to K. pneumoniae infections (121). This bacterium is also known to cause pneumonia, urinary tract infections, skin infections, and open wounds (121).
Klebsiella infections are frequently managed with cephalosporins, either alone or in conjunction with aminoglycosides (122). Nonetheless, the extensive administration of powerful antibiotics in individuals with Klebsiella infections has resulted in the emergence of drug-resistant bacteria, especially those that produce extended-spectrum beta-lactamase (ESBL) (123). Carbapenems can adversely affect ESBL producers; however, these organisms have evolved novel enzymes known as K. pneumoniae carbapenemases (KPCs) that can degrade carbapenems and have disseminated globally (124, 125). No advancements have been made in synthetic or natural antimicrobials, hence ESBL-producing bacteria remain a significant danger. Bacteria that produce ESBL exhibit increasing resistance to antibiotics, including fluoroquinolones, aminoglycosides, tetracyclines, trimethoprim/sulfamethoxazole, and chloramphenicol (126).
4.7 P. aeruginosa
It is a potentially pathogenic Gram-negative, rod-shaped, spore-forming, and monotrichous bacterium found in aquatic environments and soil surfaces. It possesses an iridescent appearance and emits an aroma reminiscent of grapes or tortillas (127). It exhibits optimal growth within the temperature range of 25°C to 37°C, and its ability to flourish at 42°C sets it apart from numerous other Pseudomonas species. P. aeruginosa is a ubiquitous bacterium capable of persisting in various ecological environments (127).
The bacterium’s intrinsic antibiotic resistance is a significant characteristic that enables its survival under many extreme environmental conditions, both natural and artificial, including biofilms in healthcare settings. This bacterium induces nosocomial infections, especially in immunocompromised persons with neutropenia, severe burns, and cystic fibrosis (128).
Pathogenic P. aeruginosa can cause external and internal infections, including wounds, urinary tract infections, pneumonia, eye infections, sepsis, and endocarditis (129). A surge in MDR Pseudomonas infections has been seen, irrespective of the combination therapy employed (130). The pathogens inflict significant damage to host tissue by cleaving immunoglobulin G and A, modifying the cytoskeletal structure, and disintegrating actin filaments. Specifically, exotoxins, proteases, and exoenzymes, this mechanism facilitates the invasion, spread, and advancement of chronic infections (131).
P. aeruginosa poses a significant challenge in the medical domain for the treatment of infections acquired in hospital environments and the population (132). This bacterium can acquire resistance genes by horizontal gene transfer to aminoglycoside-modifying enzymes and extended-spectrum β-lactamases (132). Resistance may also emerge from modifications in chromosomal genes located at the efflux and target sites, and pathogenic isolates of this or related species have been documented to exhibit antibiotic resistance by innate, acquired, and adaptive mechanisms (132). P. aeruginosa exhibits resistance to several antibiotics, including cephalosporins, carbapenems, aminoglycosides, quinines, ureidopenicillins, quinolones, cefepimes, penicillin, and polymyxins (133, 134).
4.8 Salmonella enterica serovar Typhimurium
Salmonella species, such as Salmonella enterica serovar Typhimurium, Typhi, and Enteritidis, are worldwide dispersed Gram-negative bacteria recognized as significant contributors to several human diseases (135). Salmonella can be disseminated through multiple vectors, including contaminated food and water, human and animal excrement, and deceased animals. Also, S. enterica serovar Typhimurium can be obtained from numerous infected foods, including beef and pig products (136).
Salmonellosis is also linked to contaminated fresh fruits and vegetables, including apples, cantaloupe, alfalfa sprouts, mangoes, lettuce, cilantro, unpasteurized orange juice, tomatoes, melons, celery, and parsley (137). Poultry products, such as chickens, turkeys, geese, and ducks, have been associated with the transmission of Salmonella infections. Salmonella exhibiting resistance to ampicillin, chloramphenicol, and trimethoprim-sulfamethoxazole have been detected in multiple regions, particularly S. enterica serovar Typhimurium (138).
4.9 Shigella flexneri
Shigella is a bacterium classified under the Enterobacteriaceae family. It is distinguished by its Gram-negative characteristics, absence of capsules, and lack of motility (139). S. flexneri is one of the four species of this genus commonly associated with bacillary diarrhea, the principal clinical manifestation of shigellosis, characterized by inflammation of the colon. The symptoms may range from asymptomatic carriage to diarrhea and dysentery (139).
Shigella can reside in humans and captive monkeys, and it is estimated that fewer than 200 bacterial cells are sufficient to induce an infection. Consequently, the proliferation of the bacteria is more probable in highly populated regions with inadequate sanitation, and annual Shigella outbreaks are anticipated to surpass 150 million cases (140). Shigellosis predominantly affects children in developing nations, who are the most severely harmed. Conversely, the epidemic is widespread in congested establishments inside affluent countries, including daycare centers, correctional facilities, and military recruitment camps (141).
Despite the manageable prevalence of shigellosis infections due to the availability of several antibiotic regimens, the emergence of drug-resistant strains is alarming. Antibiotics such sulfonamides, tetracycline, ampicillin, and chloramphenicol are ineffective against these pathogens (141).
4.10 C. albicans
Identifying appropriate treatment for fungal infections without harming the host is the most difficult aspect. This is attributable to the physiological cellular similarities between fungi and humans. Consequently, effective therapies for fungal infections are limited in comparison to those for bacterial disorders (111). Mycoses have become a notable concern in modern medicine owing to the rising population of patients receiving immunosuppressive treatment or experiencing immunodeficiency (142).
Candida species generally exist as unicellular yeasts in a symbiotic connection with their human hosts (143). These yeasts are often confined to the skin, gastrointestinal tract, reproductive organs, and mucosal surfaces of the oral cavity. C. albicans can alternate between two unique forms: a unicellular yeast that reproduces via budding and can also produce elongated filaments, comprising pseudomycelium (143). This capability is essential in infections where the invasive hyphae can disseminate throughout the affected tissue or organ (143).
In contrast, yeast cells disseminate to many regions. This commensal yeast is classified within the genus Candida and is typically located in the oral cavity, dermis, gastrointestinal tract, and vaginal canal of females (144). C. albicans demonstrates a growth pattern marked by yeast cells that are spherical to oval, measuring 3 to 5 µm in width and 5 to 10 µm in length (145).
C. albicans often exists as a nonpathogenic commensal organism. Nonetheless, an excessive presence of this yeast may create a perilous scenario as it can inhabit almost all human tissues and provoke severe systemic infections in individuals with compromised immune systems (146). C. albicans is the sixth most prevalent pathogen in the bloodstream for both acute and chronic yeast infections in humans and is a recognized source of nosocomial infections, particularly in immunocompromised individuals (147).
HIV infection and diabetes are two comorbid conditions frequently linked to this yeast. It is well acknowledged as the etiology of vulvovaginal and oropharyngeal candidiasis. Oral candidiasis, vulvovaginal candidiasis, and invasive candidiasis are illnesses resulting from Candida infections. Oral candidiasis is an opportunistic infection of the oral cavity. It is prevalent among elderly individuals, especially those who utilize dentures (148). This disease is typically transferred to immunocompromised individuals and may signify systemic diseases such as diabetes (149).
In women of reproductive age, particularly those who are HIV-positive, C. albicans is responsible for the heightened prevalence of vulvovaginal candidiasis (150). Nosocomial infections may result from invasive candidiasis, or candidemia, which is the most prevalent opportunistic mycosis worldwide (151). Invasive nosocomial infections can present as urinary tract infections, surgical site infections, bloodstream infections (fungemia), catheter-associated skin abscesses, and myocarditis (152).
C. albicans possesses several acknowledged virulence factors implicated in disease. C. albicans virulence factors bind to endothelial cells in blood vessels and epithelial cells in the respiratory tract, thereby greatly influencing pseudo-hyphae development (152). Surface recognition molecules, hyphal switching, and phenotypic switching facilitate this process. Extracellular hydrolytic enzymes are crucial for the dissemination of microorganisms since they facilitate adhesion, tissue infiltration, invasion, and ultimately, the destruction of host tissue (152).
A common virulence factor that facilitates sickness is hemolysin. Hemolysin-induced erythrocyte lysis enhances iron absorption, an essential component for yeast growth and pathogenic functions (153). Candida lysin contributes to immunological activation, phagocyte recruitment, tissue invasion, and epithelial injury (154, 155).
The azole class is the most often used antifungal for the treatment of Candida infections (156). They are preferred for managing various Candida infections due to their cost-effectiveness, less toxicity, and simple oral administration (156). Currently, there are four primary kinds of antifungal agents utilized for the treatment of severe mycoses, classified according to their mode of action (1): Amphotericin B modifies cell membrane functionality (2); Flucytosine obstructs DNA or RNA synthesis (3); Azoles, including fluconazole, itraconazole, voriconazole, posaconazole, and ravuconazole, impede ergosterol production; and (4) Echinocandins, such as caspofungin, micafungin, and anidulafungin, inhibit glucan synthesis (156, 157).
Over the past decade, there has been a notable rise in the incidence of drug-resistant fungal infections among hospitalized patients. The rise is particularly pronounced with frequently utilized antifungal agents, including fluconazole, miconazole, clotrimazole, tioconazole, amphotericin B, and echinocandins (158). Antifungal resistance mechanisms can be classified into three primary categories: (1) modified drug targets, (2) diminished effective drug concentration, and (3) metabolic bypasses (156).
There are fewer pharmacological options for treating Candida infections compared to bacterial or viral infections, and incidences of treatment resistance are more prevalent (159). The initial report of C. albicans strains exhibiting resistance to azole antifungals occurred in the late 1980s. Since its discovery in the 1990s, fluconazole has been the preferred antifungal agent for unicellular and multicellular fungal infections (159). Nonetheless, despite its initial effectiveness within the medical community, significant instances of medication resistance have arisen, notably among those who are HIV-positive (160).
4.11 Candida tropicalis
C. tropicalis, a species separate from C. albicans. Non-C. albicans Candida (NCAC) is recognized for its ability to induce oral infections (161). The incidence of nosocomial infections, specifically NCAC infections, can be ascribed to several medical interventions and situations, including cancer treatments, surgical implantations, HIV, AIDS, and the overuse of antibiotics (161, 162). C. tropicalis is the most pathogenic among the NCAC species due to its capacity to bind and release proteinases. It is one of the most significant invasive features of pathogenic organisms towards epithelial cells (163). It inhabits the mouth cavity and skin and has been associated with esophageal and nosocomial infections (163).
Candida strains can form biofilms on medical equipment, making them resistant to antifungal agents (164). The incidence of illnesses attributed to C. tropicalis is rising globally, designating this organism as a notable emerging pathogenic yeast. The cause of this organism’s dominance over other NCAC species and its fluconazole resistance remains unidentified (165).
5 Medicinal plants as alternative antibiotics
A variety of drugs exist to address infections and their complex antibiotic-resistance mechanisms (166). The mechanism of action of these antimicrobial agents relies on selective toxicity. Studies have shown that medicinal plants contain a diverse assortment of bioactive chemicals, such as coumarins, flavonoids, phenolics, alkaloids, terpenoids, tannins, essential oils, lectins, polypeptides, and polyacetylenes (167). These chemicals serve as essential precursors for antibiotic synthesis (168). Although synthetic antimicrobial agents have received extensive endorsement, natural compounds sourced from various origins such as plants (169, 170), fungi, lichens, endophytes, and marine organisms including seaweeds, corals, and other microorganisms remain a prominent area of investigation (171, 172).
These natural products exhibit significant potential for addressing antibiotic resistance in bacterial pathogens (173). Plant-derived chemicals are notable for their potential in combating bacterial infections. These naturally occurring phytochemicals have demonstrated diverse advantageous qualities, including antioxidant, antibacterial, and antifungal activity (173). They may also significantly improve the effectiveness of current antibiotics, therefore averting more resistance development (174).
The bioactive compounds in medicinal plant extracts can operate through many mechanisms, including interaction with specific bacterial membrane components, such as anionic phospholipids and lipopolysaccharides, leading to bacterial lysis by membrane disruption (175). Hydrophobicity may affect the reaction’s outcome by interacting with the hydrophobic groups within the membrane. Moreover, the plant extract is administered into the bilayer surface through ionic/electrostatic interactions, resulting in the destabilization and rupture of the cellular membrane (46). Furthermore, the aforementioned processes may function when active components contain hydrophobic and hydrophilic residues (175).
5.1 Extraction of bioactive compounds from medicinal plants
Bioactive components in plant extracts can be isolated, identified, and evaluated by high-performance liquid chromatography (HPLC) (176). HPLC can be classified into two primary categories: analytical HPLC and preparative HPLC (176). Analytical HPLC primarily assesses the qualitative and quantitative characteristics of a given component. Conversely, preparative HPLC is dedicated to the extraction and purification of a valuable chemical (176). The preparative HPLC method is extensively employed to identify physiologically active chemicals (176).
Mass spectrometry (MS) and nuclear magnetic resonance (NMR) are frequently employed to detect pure substances. MS is a scientific method that use ionization and separation to examine ions according to their mass-to-charge ratio. The mass-to-charge ratio serves as the independent variable for plotting the ion signal in a mass spectrum (176). The elemental or isotopic characteristics of a sample, the masses of particles and molecules, and the chemical structures of molecules can all be determined by MS (176). NMR can accurately detect the chemical features of compounds, including the quantities of carbon and proton atoms, bonding patterns, molecular geometries, and relative and absolute stereochemistry’s (176).
Gas chromatography and mass spectrometry (GC–MS) can be employed to analyze essential oils. This device employs an inert gas stream to introduce the sample into the mobile phase (177). The vaporized sample is upheld by inert material while it traverses the stationary phase of the capillary column. The specific analytes/compounds are transmitted through a MS detector, and the resultant data is presented on a computer or recording apparatus (177).
Utilizing an ethnopharmacological strategy for the screening of bioactive components provides multiple advantages (176). It facilitates the pre-screening of gathered species according to their ethnomedicinal applications and preliminary safety standards, so substantially reducing the time and cost of the process (176, 177). Numerous natural compounds derived from medicinal plants have been effectively identified by the ethnopharmacological method, and these compounds have provided a foundation for innovative methods in the pharmaceutical sector. An example of this is the method employed to extract artemisinin, an antimalarial compound, from the plant A. annua (177).
5.2 Bioactive compounds from medicinal plants
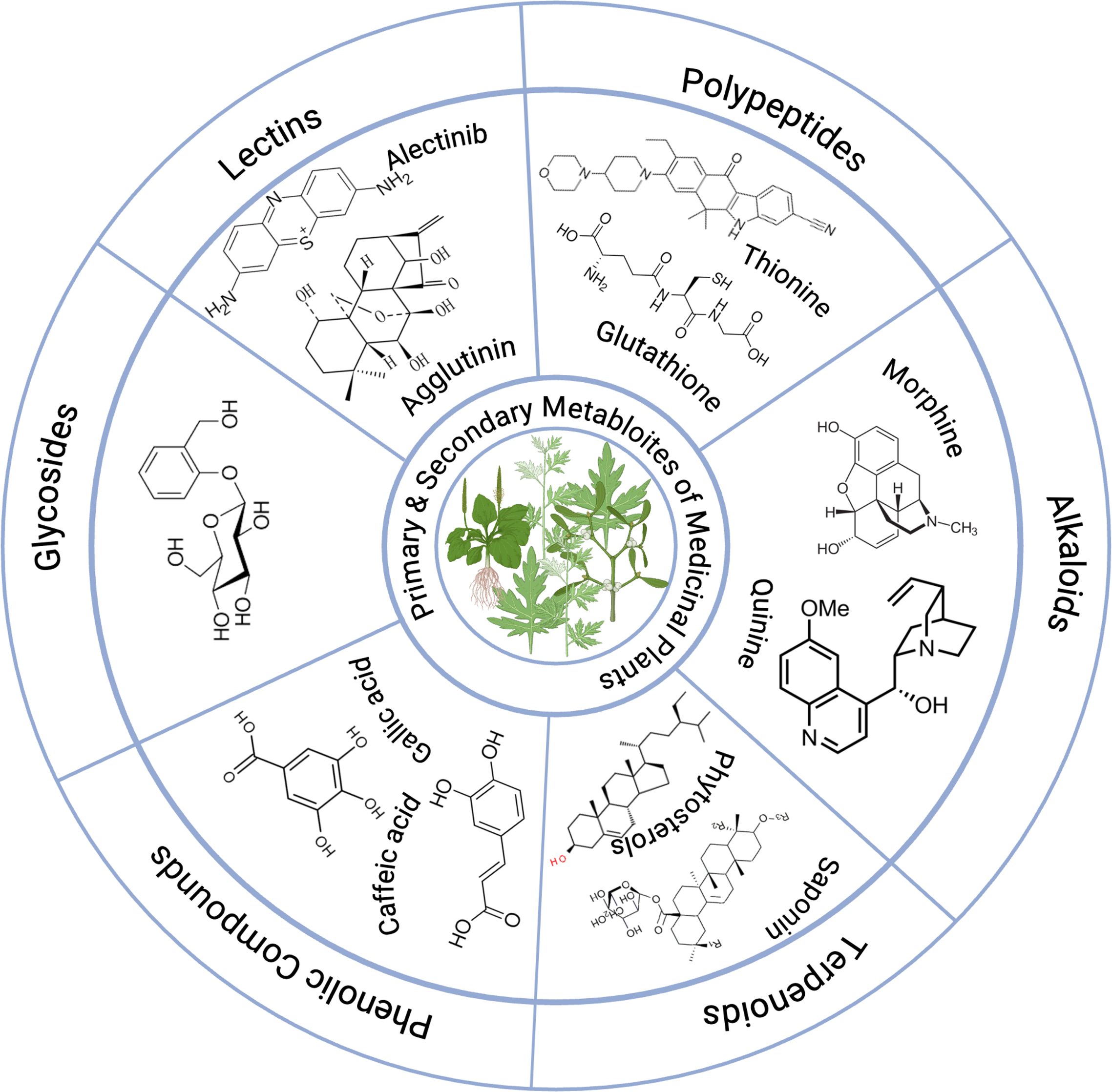
The bioactive components of various medicinal plants against certain MDR microorganisms are illustrated in Table 1, which demonstrates their effectiveness against specific MDR microorganisms. Furthermore, examples of primary and secondary metabolites found in medicinal plants are presented in Figure 4.
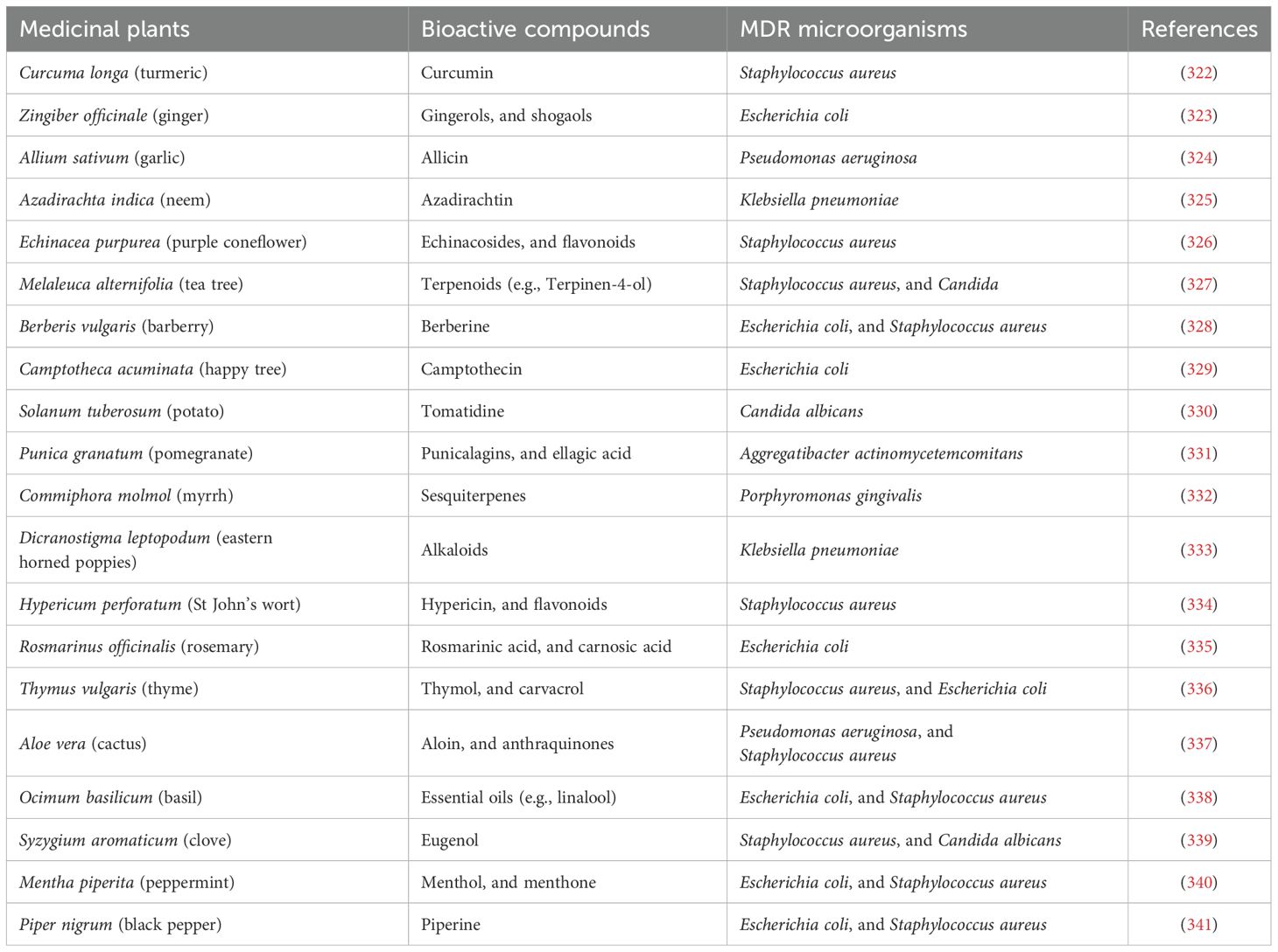
Table 1. Bioactive compounds of some medicinal plants against certain multi-drug resistant (MDR) microorganisms.
5.2.1 Phenolic compounds
These secondary metabolites comprise phenolic and hydroxyl functional groups linked to an aromatic ring known as phenol. Phenol itself is made up of chemically varied groupings. Gautam et al. (178) assert that phenolic chemicals serve multiple protective functions in nature, including facilitating healthy plant development and reproduction, enhancing seed germination prior to harvest, and providing protection against diseases and predators (178). Four distinct categories of polyphenols comprise phenolic acids, flavonoids, tannins, and stilbenes (179). Alkyl esters of hydroxybenzoic acid parabens are widely utilized as antimicrobials and preservatives in pharmaceuticals, cosmetics, food, and drinks (179, 180).
Flavonoids are a significant subset of polyphenolic compounds, encompassing over 9,000 recognized natural molecules. Various kinds of flavonoids have been ascribed with antiallergic, antidiabetic, anti-inflammatory, antiviral, antibacterial, antifungal, antiproliferative, anticarcinogenic, hepatoprotective, and antioxidant properties (181). Moreover, numerous investigations have demonstrated the effectiveness of these compounds in addressing MDR bacteria of both Gram-negative and Gram-positive types (182). In reaction to microbial infection, plants synthesize secondary metabolites; thus, the extensive in vitro data on their broad-spectrum antibacterial efficacy aligns effectively (182). Tannins are extensively distributed in various plant parts, and it has been suggested that they serve as a natural defense against microbial invasion. Tannins exhibit antibacterial effects against fungi, yeasts, bacteria, and viruses. Tannic acid and propyl gallate exhibit inhibitory effect against food-borne bacteria, aquatic bacteria, and microorganisms that affect flavor (182, 183).
Aristri et al. (183) reported that tannins had antibacterial properties, which appear to be linked to the cleavage of the ester linkage between gallic acid and polyols. The disruption of DNA synthesis, the inactivation of cellular enzymes, the alteration of membrane fluidity, and the inhibition of protein synthesis have all been associated with the antimicrobial effects of various phenolic compounds (183).
5.2.2 Terpenoids
Terpenoids are the predominant secondary metabolites, originating metabolically from acetyl-CoA or glycolytic intermediates. Plants produce several terpenoids as secondary metabolites, which are thought to provide protection against insect and mammalian herbivores (184). The molecular structure common to these metabolites is (C10H16). The antibacterial properties of several terpenes against bacteria, fungi, viruses, and protozoa have been recorded (184). Oxygenated terpenoids with varied structures exhibit antibacterial activity against numerous bacteria and fungi, especially those with alcohol functional groups. These molecules demonstrate superior action relative to aldehydes and ketones. While the precise mechanism of action of these terpenoids remains unclear, it is hypothesized that they may compromise cell membranes (185).
Farha et al. (186) investigated the efficacy of (+)-nootkatone against biofilms of MDR S. aureus and analyzed its hypothesized molecular mechanism. (+)-Nootkatone shown bactericidal efficacy against S. aureus strains SJTUF 20758 and ATCC 25343 (186). Light microscopy and confocal laser scanning microscopy demonstrated that (+)-nootkatone significantly diminished biofilm thickness. The bacterial growth curve indicated that the antibacterial efficacy of (+)-nootkatone was dosage-dependent, with dosages below the MIC failing to impede the proliferation of free-floating bacterial cells (186).
Additionally, (+)-nootkatone decreased the motility of S. aureus. At a concentration of 200 grams/ml, (+)-nootkatone decreased biofilm by 50% and eradicated 80% of bacterial cells. The transcriptional analysis demonstrated that (+)-nootkatone inhibited the expression of genes associated with biofilm development, including Sara, icaA, Agra, RNAIII, and spa (186). Moreover, the MTT assay indicated that (+)-nootkatone displayed no toxicity to human foreskin fibroblast (HFF) cells. Consequently, (+)-nootkatone is a potentially safe phytochemical for application in the food industry (186).
5.2.3 Alkaloids
Nitrogen constitutes a component of alkaloids, which are secondary metabolites prevalent in numerous plants. These metabolites comprising organic acids, exhibit hemolytic activity, and are toxic to bacteria (187). For 4000 years, people have cohabited with and employed alkaloid-rich plants. In the early 1800s, the therapeutically active constituents of this medication class were found (187). The analgesic and narcotic effects of the morphine component obtained from Papaver somniferum (opium poppy) have been utilized. After several years, researchers identified substances like strychnine, emetine, brucine, piperine, caffeine, quinine, cinchonine, and colchicine. To far, over 10,000 alkaloids have been recorded (188).
Alkaloids exhibit significant biological activity owing to their capacity to establish hydrogen bonds with enzymes, receptors, and proteins. This capability is enabled by a nitrogen atom that absorbs protons and one or more amine hydrogen atoms that contribute protons (188). Alkaloids have gained significance in the treatment of infectious diseases exhibiting MDR due to their antibacterial capabilities. Consequently, scientists focus on these intriguing secondary plant metabolites (188, 189). To obtain pure alkaloids, it is essential to devise alternate extraction procedures, as their natural sources typically yield very minimal quantities (189).
5.2.4 Glycosides
These secondary metabolites comprise glucose or an alternative sugar combined with non-sugar molecules, such as terpenes or phenolic compounds. Cyanogenic glycosides, saponins, solanines, and mustard oil glycosides are the toxic glycosides implicated in plant poisonings (190). Nonetheless, several glycosides, including cardiac glycosides, have been empirically documented to offer therapeutic benefits, especially in the management of heart disease and cancer (191).
MRSA and MDR Acinetobacter baumannii (MDRAB) pose substantial challenges owing to their ability to acquire resistance to commonly utilized antibiotics. This resistance enables them to induce chronic infections and impede the healing process (192). El-Shiekh et al. (192) investigated the efficacy of Caralluma quadrangular extracts (MeOH, CH2Cl2, and n-butanol) against MDR MRSA USA300 and A. baumannii AB5057. The MeOH extract and both fractions of C. quadrangular demonstrated a significant decrease in biofilm formation in vitro. All doses (0.625, 0.313, and 0.156 mg/ml) eliminated pre-existing MRSA and MDRAB biofilms (192).
C. quadrangular extracts significantly decreased bacterial loads in MRSA-infected dermal lesions in murine models (192). Four pregnane glycosides and one flavone glycoside were isolated from the bioactive n-butanol fraction. The biofilm inhibition and detachment properties of the isolated compounds (Rus A–E) were assessed. Rus C demonstrated the greatest activity level among the compounds, with an IC50 value of 0.139 mmole, whilst Rus E exhibited the lowest activity level, with an IC50 value of 0.818 mmole (192). The results demonstrated that extracts from C. quadrangular or its constituents could diminish biofilm adherence and the pathogenicity of MRSA and MDRAB. Furthermore, they may serve as a topical antibacterial treatment for MRSA skin infections (192).
Strophanthin, a steroidal glycoside derived from Moringa oleifera seeds, surpasses alum in its efficacy to aggregate and precipitate inorganic and organic materials in wastewater (193). Furthermore, it has been demonstrated to decrease microbial load by 55% and coliform burden by 65% (193, 194). The exact mechanism of cardiac glycosides is not fully understood; however, it is posited that they inhibit Na+/K+-ATPase by binding to its receptor, resulting in elevated intracellular sodium ion concentrations. Consequently, the regulation of Na+/Ca2+ transport across the cell membrane is modified, leading to advantageous inotropic effects (195).
5.2.5 Lectins and polypeptides
Lectins are proteins that have a great attraction for carbohydrates, namely the glycosidic bonds found in the seeds and tubers of various plants, such as wheat, potatoes, and beans (196). More significant lectin molecules, such as MAP30 from bitter melon and GAP31 from Gelonium multiflorum, have demonstrated the ability to inhibit viral replication (HIV, CMV) by interfering with viral interactions with crucial host cell components (197). Peptides exhibit a cationic charge and disulfide bonds, allowing them to inhibit microbial development. Thionins, often found in barley and wheat, inhibit the proliferation of yeasts and both Gram-negative and Gram-positive bacteria (198).
Pandey and Srivastava (199) demonstrated that the sugar beet thionins AX1 and AX2 had antifungal properties but lacked antibacterial activity. In a study by Bilal et al. (200), fabatin was found to have effects comparable to g-thionins, diminishing antibacterial efficacy against E. coli, P. aeruginosa, and Enterococcus hirae. The sulfur-containing secondary metabolites may enhance their activity by promoting the development of ion channels in the cell membrane (196).
6 Biological activities of medicinal plants
The benefits of medicinal plants, including their antibacterial, antioxidant, and anticancer characteristics, are illustrated in Table 2.
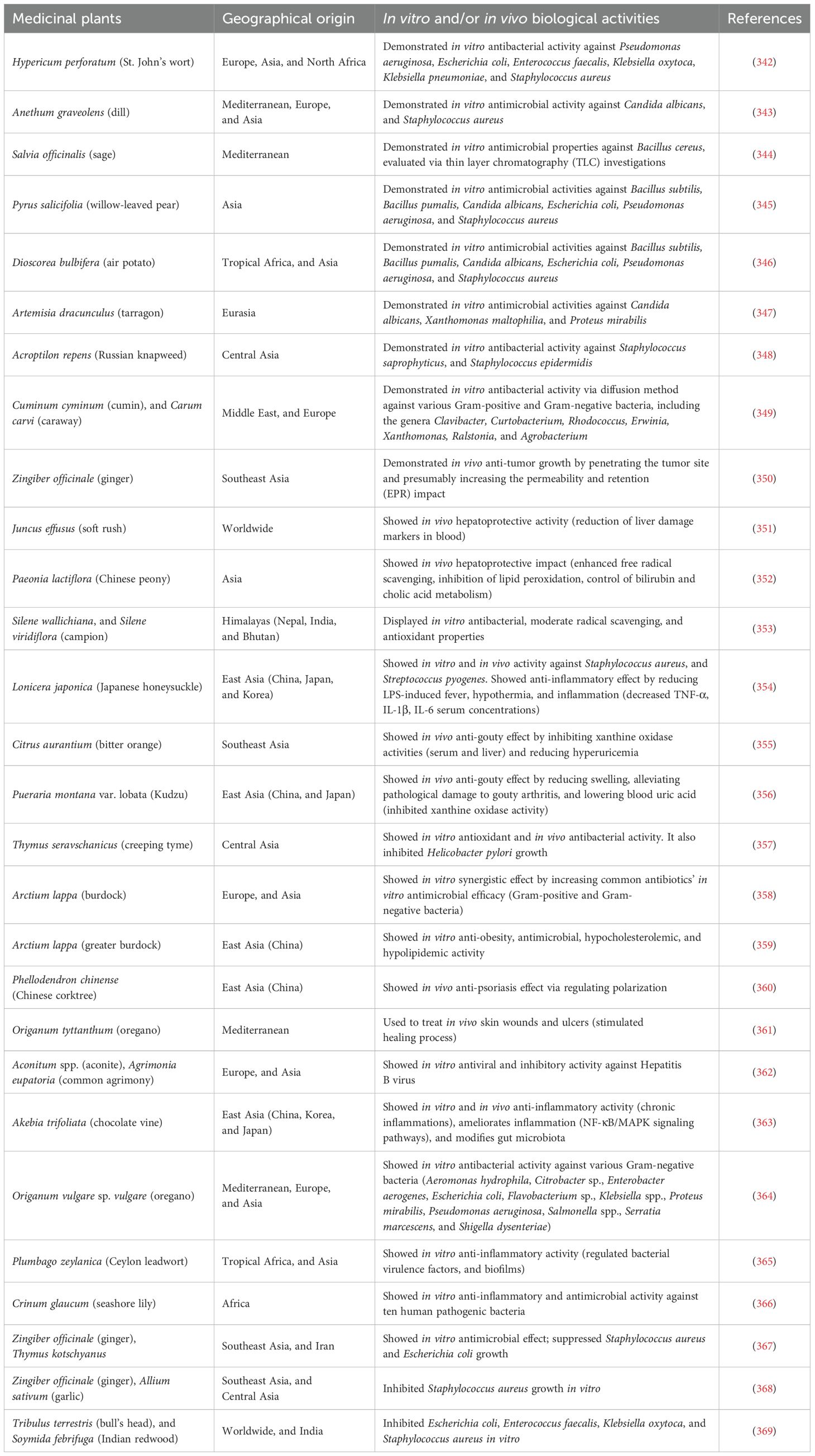
Table 2. Benefits of medicinal plants and their antibacterial, antioxidant, and anticancer properties.
6.1 Antioxidant and anticancer activities
Medicinal herbs include many active compounds, whether fresh, dried, or crushed. These constitute the essential principles of phytotherapy. Conversely, the majority of conventional pharmaceutical drugs, including those with components derived from medicinal plants, function as a chemical marker with a singular active ingredient (201). Cinnamon exemplifies these two processes effectively. Historically, infectious diseases were treated with an infusion of cinnamon bark. The primary secondary metabolite of the bark, cinnamaldehyde, demonstrated efficacy as an antibacterial agent and may serve as a traditional remedy comparable to synthetic antimicrobials (201).
About 67% of anti-cancer pharmaceuticals comprise chemicals derived from or primarily produced from flora or fauna (201). Many are employed in the treatment of cancer, particularly as chemotherapeutics. The FDA has included taxol (Paclitaxel), a phenolic chemical sourced from the bark of Taxus brevifolia, in its roster of authorized cancer medications (201). Moreover, several extracts are available as commercially sold goods. Xuancheng Baicao Plants Industry and Trade Ltd., situated in China, offers powdered Lonicera (honeysuckles) extract containing 25% chlorogenic acid and Ganoderma extract with a 30% polysaccharide content (202).
The efficacy of solvents employed to extract active compounds from medicinal plants varies. Moreover, preparations of medicinal plants and essential oils exhibit significant antioxidant and antiviral activities. Clove extract exhibits superior antioxidant action compared to vitamin C against 2,2-diphenyl-1-picrylhydrazyl (DPPH), 2,2′-casino-bis (3-ethylbenzothiazole-6-sulfonic acid) (ABTS), and superoxide radicals. Moreover, clove extract and essential oil have antiviral properties (202). The chemical composition of medicinal plants, especially the occurrence of phenolic compounds, is linked to their notable antioxidant and antibacterial properties in extracts and essential oils (113).
6.2 Antimicrobial activity of medicinal plants against MDR microorganisms
Before the discovery of microorganisms and their functions, humanity has traditionally recognized plants as a source of medical efficacy (202, 203). Medicinal plants are effective against various viruses, including herpes, adenovirus, poliovirus, and coxsackievirus, and possess anticancer and anti-inflammatory properties (203). In light of the increasing use of herbal therapy as an alternative to traditional antibiotics and the proliferation of antibiotic-resistant bacteria, numerous studies have been conducted on the antibacterial properties of medicinal herbs (204, 205). These medicinal plants are acknowledged to possess significant untapped bioactive chemical potential (205–208).
Numerous therapeutic plant extracts possess secondary metabolites exhibiting significant efficacy against MDR microorganisms. Panda et al. (209) demonstrated a range of ethnobotanical plants possessing MDR characteristics. The acetone extract of the leaves of Smilax zeylanica and Syzygium praecox, known for their efficacy in treating ulcers and skin infections, showed antibacterial activity against MRSA by compromising cellular membranes via ionic and electrostatic interactions (209). Moreover, the ethanol extract of long-leaf varnish tree fruit exhibits MDR activity against E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species via ionic interactions between terpenoids and steroid compounds in the extract and the bacterial cell membrane (209).
The ethanol extract of climbing acacia leaves is utilized ethnobotanically to alleviate dysentery and diarrhea, as well as to reduce cholesterol levels, whilst the acetone extract of java cedar leaves is employed to address skin ailments. The antimicrobial activity of both plants against K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species is attributed to electrostatic interactions between positively charged secondary metabolites and negatively charged bacterial cell membranes. The amphipathic properties of Ceylon olive blossom aqua extract, Buddha coconut bark ethanol extract, and charcoal tree leaves acetone extract facilitate concurrent interaction with MDR bacterial cells, including K. pneumoniae, A. baumannii, P. aeruginosa, and Enterobacter species (209). Furthermore, the extracts employed in traditional medicine for the treatment of cardiovascular diseases, dermatological infections, and wound infections (209).
Richwagen et al. (210) indicated that the ethanol extract of lavender woody stems may alleviate pain through ionic interactions between the cell membranes of MDR S. aureus, A. baumannii, and P. aeruginosa and the flavonoid extract. The methanol extract of coastal golden leaf and bark is traditionally recognized for its efficacy in addressing digestive disorders, ocular issues, and infertility, exhibiting hepatoprotective, antioxidant, anticancer, antiviral, and antibacterial attributes (210). The amphipathic characteristics of the plant contribute to its antibacterial efficacy against MDR S. aureus, K. pneumoniae, P. aeruginosa, Enterobacter species, and MRSA (210).
Pallah et al. (211) demonstrated a range of plants exhibiting MDR activities. Cherry plum, plum, red currant, and methanol extracts exhibited antioxidant, anti-inflammatory, and antibacterial activities against MDR species, including S. aureus, K. pneumoniae, and P. aeruginosa, through electrostatic interactions (211). The Jostaberry methanol extract demonstrated inhibitory zones against S. aureus and P. aeruginosa using the same method (211). Additionally, methanol extract of sweet cherry fruit is conventionally employed to alleviate osteoarthritis and address cardiovascular illnesses. Sweet cherry exhibited antibacterial action against MDR organisms S. aureus, K. pneumoniae, and P. aeruginosa owing to its amphipathic characteristics (211).
The complete plant ethanol extract of Maidenhair fern and Chirayita is effective against S. aureus, and E. faecium. The extract of maidenhair fern possesses astringent, demulcent, expectorant, and diuretic characteristics, successfully addressing urinary tract infections (212). Chirayita extract is utilized for inflammation, hepatitis, digestive disorders, chronic fever, malaria, dermatological conditions, and bronchial infections. Both extracts combat infections via amphipathicity (212). The extract from aerial portions addresses pruritus, viral inflammatory dermatoses, malaria, and typhoid, specifically targeting E. faecium and P. aeruginosa through electrostatic interactions. Timber fruit extract addresses cancer, gastrointestinal disorders such as cholera and dysentery, and oral infections including toothaches and mouth ulcers. It additionally targets E. faecium and S. aureus through electrostatic interactions (212).
Shobha et al. (213) investigated the antibacterial properties of an ethanol extract derived from cashew leaves. The herb is conventionally utilized for venereal disorders, gastrointestinal issues, dermatological conditions, stomatitis, bronchitis, psoriasis, dental pain, and periodontal infections. The extract comprises hydrophobic and hydrophilic residues, hence demonstrating MDR antibacterial action against S. aureus, K. pneumoniae, and P. aeruginosa (213).
Chandrasekharan et al. (214) investigated the therapeutic applications and mechanisms of several medicinal plant extracts against MDR microorganisms. The pipli root aqueous extract and the ethanol extract of Bermuda grass leaves function via electrostatic interactions. Pipli root extract is recognized for its antioxidant and anti-inflammatory activities, especially against E. faecium and A. baumannii (214). The ethanol extract of Bermuda grass leaves serves as a laxative, tonic for the brain and heart, and a cooling agent to eliminate S. aureus (214). The hexane extract of lemongrass modulates hypertension, epilepsy, and gastrointestinal and central nervous system problems, specifically targeting P. aeruginosa and E. faecium via ionic interactions (214). Aloe extract facilitates wound healing, mitigates inflammation, and addresses burn-related skin injuries by rebuilding compromised skin. It targets P. aeruginosa and Enterococcus casseliflavus through amphipathicity, encompassing hydrophobic and hydrophilic residues (214).
Nayim et al. (215) indicate that certain plant extracts exhibit significant antibacterial efficacy against MDR bacteria due to ionic interactions. Methanol extracts from cocoa seeds, bush butter tree, and okra leaves suppress MDR K. pneumoniae, but sweet potato leaf extract targets the same bacteria through amphipathicity (215). These plants possess traditional therapeutic applications: cocoa seeds for disorders such as dermal injuries and diarrhea, bush butter tree for tonsillitis, okra leaves for infections, and sweet potato leaves for conditions including diabetes and inflammation (215).
Likewise, extracts from neem tree bark and pomelo leaves demonstrate efficacy against MDR Enterobacter species via electrostatic interactions (215). Neem bark is typically utilized for dermatological conditions and febrile illnesses, whereas pomelo leaves are employed for neurological problems and hemorrhagic conditions (215). Winter squash bean extract, utilized in traditional therapy for renal and gastrointestinal ailments, also addresses MDR Enterobacter and K. pneumoniae via amphipathicity (215). Ultimately, common bean leaf extract suppresses MDR E. casseliflavus and is commonly utilized for diabetes and weight management (215).
Subramani et al. (216) identified multiple plants demonstrating MDR activities. The methanol extract of Lantana leaves yields secondary compounds that mitigate inflammation, combat malaria, alleviate spasms, suppress tumor proliferation, avert ulcer development, and specifically target Enterobacter species and P. aeruginosa through electrostatic interactions (216). The alcoholic extract of myrtle leaves showed efficacy against S. aureus and P. aeruginosa via electrostatic interactions (216). It addresses cancer, inflammation, diabetes, ulcers, hypertension, diarrhea, and rheumatism. Extracts of Palash (Butea monosperma) leaves (ethanol, hexane, and water) enhanced diuresis and menstrual flow while suppressing Enterobacter species through ionic interactions (216). The ethanol extract of Burflower (Neolamarckia cadamba) tree leaves and bark alleviated fever, uterine ailments, skin problems, and inflammation, while also functioning as a febrifuge, antidiarrheal, antihyperglycemic, and antibacterial agent against A. baumannii and P. aeruginosa, attributed to their amphipathic characteristics (216).
Masoumian and Zandi (217) demonstrated the therapeutic efficacy of various plant extracts against MDR species. The extract of cinnamon bark water is efficacious in managing diabetes, reducing oxidative stress, and mitigating inflammation. This extract shows efficacy against S. aureus and P. aeruginosa due to ionic interactions (217). Similarly, medicinal Aloe leaf water extracts precisely target S. aureus and P. aeruginosa, facilitating the treatment of digestive issues and burns via ionic interactions. The aqueous extract of mint leaves has shown efficacy against acid reflux, irritable bowel syndrome, and the common cold (217). It demonstrated efficacy against S. aureus and P. aeruginosa owing to its amphipathic metabolites, enabling the extract to engage concurrently with hydrophobic and hydrophilic residues (217). The aqueous extract of henna leaves exhibited antibacterial activity against S. aureus and P. aeruginosa via electrostatic interactions. It proficiently addressed burns, wounds, and cutaneous infections while enhancing hair vitality (217).
Likewise, the chloroform extract of snake flower leaves effectively addressed skin and wound infections due to the hydrophobic properties of the plant extract (218). Ghuman et al. (219) showed that chloroform extract from bitter Aloe leaves can address skin problems by targeting S. aureus, P. aeruginosa, and A. baumannii. The extract’s efficacy stemmed from its capacity to adhere to the hydrophobic groups located in the bacterial membrane (219). Furthermore, the ethanol extract of hrse mint (Mentha longifolia) aerial parts significantly mitigated throat and oral discomfort, addressing sore throats and targeting Mycobacterium tuberculosis through its amphipathic characteristics (219).
Agarwal et al. (220) indicated that the ethanol extract of Phyllanthus emblica (Indian gooseberry) aerial parts could effectively target M. tuberculosis and MRSA via electrostatic interactions (220). Moreover, the dichloromethane extract of tree Aloe leaves demonstrated effectiveness in addressing skin, gastrointestinal, and respiratory ailments, countering S. aureus and P. aeruginosa owing to their hydrophobic properties (220). Bacha et al. (221) examined the antibacterial efficacy of methanol extract from cardamom (Elettaria cardamomum) fruit against S. aureus. The mechanism of action entails ionic and electrostatic interactions that result in the deposition of the plant extract on the bacterial cell membrane, inducing instability and disintegration of the cellular membrane (221).
Additionally, Arullappan et al. (222) demonstrated that the leaf extract of Clinacanthus nutans had a notable antifungal activity against C. albicans in a laboratory environment, with MIC above 1 mg/ml. Numerous research, including those by Ortega‐Ramirez et al. (223), Ma et al. (224), and Shanmugam et al. (225), have elucidated the antibacterial characteristics of Allium cepa (onion). Zhou et al. (226) demonstrated that a gel including Durio zibethinus polysaccharide at a dose of 100 mg/mL efficiently suppresses the proliferation of oral infections. Diris et al. (227), and Apridamayanti et al. (228) reported the antibacterial properties of Malabar melastome (Melastoma malabathricum). The inhibitory activity of this plant extract may be ascribed to its phenolic constituents, such as flavonols and ellagic acid (229).
Under laboratory conditions, the fruit extract of Cucurbita maxima (Buttercup squash) has demonstrated antibacterial efficacy against S. aureus, Bacillus subtilis, and Micrococcus luteus (230). The constituent of this fruit, which comprises caffeic acid and p-coumaric acid, possesses antibacterial properties (230). Numerous research studies have demonstrated the antibacterial efficacy of Zingiber cassumunar (cassumunar ginger) essential oil against dermatophytes (231, 232). Research conducted by (33) demonstrated that the traditional herbal preparation known as “Loloh,” commonly prepared as a decoction on Bali Island, is effective in treating several ailments caused by pathogens, including diarrhea and dysentery. Numerous investigations have demonstrated the antibacterial efficacy of Andrographis paniculata (green chiretta) under in vitro conditions against Gram-positive and Gram-negative bacteria (233–235).
Andrographolide, the principal active constituent from the stem and leaves of Andrographis paniculata, has demonstrated significant efficacy as an anti-diarrheal agent in clinical trials and is accountable for the plant’s antibacterial properties (236, 237). The antibacterial properties of Blumea balsamifera (ngai camphor), Jatropha curcas (barbados nut), and Lablab purpureus (hyacinth bean) have been recorded in the literature reviewed by Priya and Karthika (238) and Rampadarath et al. (239).
Women in Minahasa, North Sulawesi, Indonesia, frequently utilize Baker, a traditional herbal steam bath, for thermotherapy, aromatherapy, and postpartum recuperation. Alexa et al. (240) indicated that 31 species utilized in “Baker” produce essential oils, including Citrus hystrix, Curcuma xanthorrhiza, Cymbopogon citratus, Cymbopogon nardus, Myristica fragrans, and Syzygium aromaticum. The volatile oil constituents (eugenol, β-pinene, sabinene, and zingiberene) linked to the medicinal benefits of these plants have been identified (240).
Eugenol derived from Syzygium aromaticum (clove) exhibited antibacterial properties against S. aureus ATCC 29213 and MRSA clinical isolates. Furthermore, it impeded the production of biofilms, hence diminishing the colonization of S. aureus in vivo (241). It efficiently eradicates existing biofilms, diminishes the expression of genes linked to biofilms and enterotoxins, disrupts the cell membrane, and results in the release of cellular contents (241). Moreover, eugenol has demonstrated antifungal capabilities and the capacity to inhibit oral microorganisms associated with periodontal disease and dental caries (242–244). Eugenol is readily accessible and utilized as a counterirritant, topical antiseptic, and in dentistry, frequently coupled with zinc oxide for analgesia and root canal obturation (244, 245).
7 Antimicrobial mechanism of medicinal plants against MDR
Medicinal plants have gained significant attention for their potential to combat MDR microorganisms due to their diverse biochemical and molecular mechanisms (173, 182, 208). The action mechanism of certain medicinal herbs against MDR microorganisms is illustrated in Table 3. Furthermore, Figure 5 illustrates the antibacterial mechanism they employ to combat MDR. These mechanisms often differ from conventional antibiotics, making them promising alternatives in the fight against AMR (Figure 5).
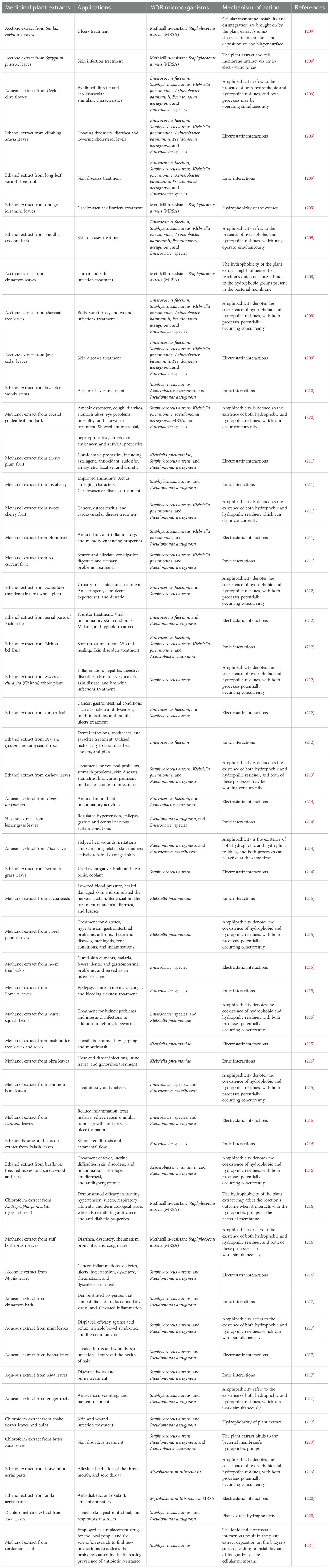
Table 3. Mechanism of action of some medicinal plants against multi-drug resistant (MDR) microorganisms.
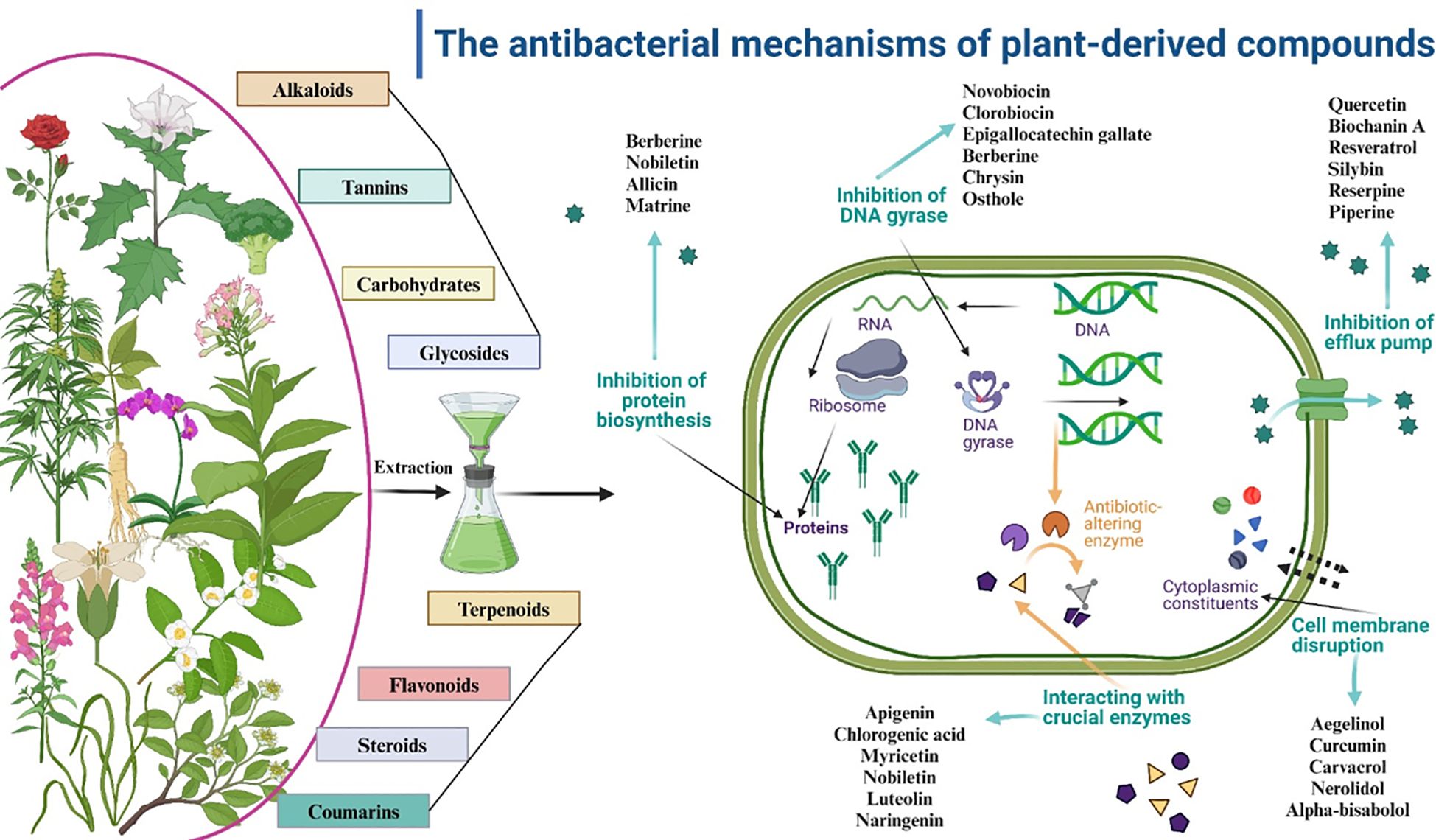
Figure 5. Antibacterial mechanism of medicinal plants against multi-drug resistance (MDR) as natural alternatives to antibiotics.
7.1 Disruption and inhibition of microbial cell membranes
Numerous bioactive chemicals in medicinal plants, including terpenoids, flavonoids, and alkaloids, engage with microbial cell membranes, resulting in structural impairment. These chemicals can enhance membrane permeability, resulting in the efflux of cellular constituents and subsequent cell death (173, 182, 208). Compounds such as carvacrol (derived from oregano) and thymol (extracted from thyme) compromise the lipid bilayer of bacterial membranes, undermining their structural integrity (173, 208).
Moghrovyan and Sahakyan (246) revealed that essential oils derived from Origanum vulgare compromised the membrane potential of E. coli and S. aureus, resulting in cell lysis (246). Alkaloids derived from Dicranostigma leptopodum enhance the permeability of K. pneumoniae cells, impeding their development (246). Due to their lipophilic characteristics, terpenes easily infiltrate bacterial cell membranes, resulting in damage and demonstrating antibacterial or bactericidal properties, as evidenced by carvacrol, thymol, and eugenol against several bacterial strains (247). Flavonoids similarly compromise the cell membranes of Candida species, increasing permeability and disrupting membrane protein functionality (247). Furthermore, the alkaloid tomatidine, present in potatoes, inhibits the enzymes involved in ergosterol production in many species of Candida and Saccharomyces (248).
Flavonoids exhibit potential as antimicrobials by blocking bacterial fatty acid biosynthesis enzymes, such as quercetin, apigenin, and sakuranetin from Plukenetia volubilis leaves, which target specific enzymes in H. pylori (249). These flavonoids, such as quercetin and apigenin, inhibit peptidoglycan production, an essential component of the bacterial cell wall (249). In fungi, acidic terpenoids such as enfumafungin block β (1, 3)-D-glucan synthase, an enzyme essential for glucan production in the cell wall, by a non-competitive mechanism (250). Furthermore, the flavonoid glabridin compromises the structural integrity of fungal cell walls, illustrating an alternative mechanism for antifungal action (251).
7.2 Inhibition of efflux pumps
Plant-derived chemicals provide many techniques to address microbial resistance, especially via inhibiting efflux pumps (252). Efflux pumps constitute a primary mechanism of antibiotic resistance in MDR bacteria. Medicinal herbs possess chemicals that can obstruct these pumps, so limiting the evacuation of antibiotics and augmenting their effectiveness (252). The many methods, such as membrane rupture, interference with biosynthesis and metabolism, biofilm inhibition, and reactive oxygen species formation, underscore the potential of plant-derived chemicals, particularly those from the Caucasus region, to address antimicrobial resistance (252). Flavonoids and alkaloids, including berberine (derived from Berberis species), interact with efflux pump proteins, inhibiting their activity (182, 208, 252).
Yu et al. (252) emphasized that berberine blocked the NorA efflux pump in MRSA, hence reversing resistance to ciprofloxacin. Alkaloids, including ellagic and tannic acids, together with different flavonoids, impede efflux pump activity in bacteria such as A. baumannii (253). Terpenes, notably carvacrol, serve as effective inhibitors of efflux pumps, particularly in S. aureus, possibly via influencing gene expression or binding to efflux proteins (254). Curcumin, a polyphenol, regulates efflux pump activity in yeast, whereas many flavonoids exhibit comparable inhibitory effects in fungi, resulting in cellular mortality (254).
7.3 Inhibition of biofilm formation
Biofilms are protective matrices created by MDR microorganisms, rendering them resistant to antibiotics. Compounds derived from medicinal plants can impede biofilm growth or dismantle pre-existing biofilms (255). Polyphenols and tannins disrupt quorum sensing, a critical communication mechanism for biofilm development (255). Flavonoids and alkaloids exhibited notable antibiofilm efficacy against several microbial species. Flavonoids, including phloretin, can impede biofilm development in E. coli by downregulating curli gene expression, hence limiting fimbriae formation (255). They can also elicit aberrant multicellular aggregation in S. aureus, inhibiting the formation of structured biofilm formations (155). Alkaloids such as anguinarine and berberine derived from apple significantly inhibit biofilm formation in Proteus rettgeri and MDR E. coli, respectively, by disrupting the synthesis of biofilm-related compounds (256).
Moreover, some flavonoids (quercetin, apigenin) and phenolic acids (ellagic, caffeic) impede biofilm formation in Candida species, including the drug-resistant Candida auris (257). Prenyflavanone 8PP inhibits C. albicans biofilms by enhancing the generation of reactive oxygen and nitrogen species (257). Curcumin from Curcuma longa suppresses biofilm formation in P. aeruginosa via downregulating quorum sensing-related genes (258).
7.4 Interference with microbial enzymes
Compounds from medicinal plants can inhibit essential enzymes critical for microbial survival, including DNA gyrase, topoisomerase, and β-lactamase. Alkaloids and phenolic acids attach to the active sites of these enzymes, hindering DNA replication and cellular division (259). Bioactive phytochemicals interfere with bacterial metabolism via multiple methods. Bioflavonoids can impede the F1 or membrane-associated F1Fo ATPases of E. coli (259). Catechin and epigallocatechin gallate inhibit F1Fo-ATPase and lactate dehydrogenase in Streptococcus mutans (259). Flavonoids derived from Chimonanthus salicifolius SY also influence ATPase activity in P. aeruginosa and S. aureus, impeding their development (259).
Berberine derived from Corydalis turtschaninovii modifies glucose metabolism in Streptococcus pyogenes via regulating carbohydrate absorption and transformation (260). Moreover, terpenes have antifungal properties against several fungi by obstructing respiratory enzymes such as succinate dehydrogenase and NADH oxidase. Moreover, epigallocatechin gallate (EGCG) derived from green tea decreased β-lactamase activity in K. pneumoniae, hence reinstating susceptibility to β-lactam drugs (261).
7.5 Induction of oxidative stress
Numerous plant-derived chemicals, including quinones and polyphenols, produce reactive oxygen species in microbial cells, surpassing their antioxidant defenses and inducing oxidative damage (262). Reactive oxygen species inflict damage on DNA, proteins, and lipids, resulting in cellular apoptosis (262). The antibacterial efficacy of catechin against Gram-positive and Gram-negative bacteria is ascribed to the induction of reactive oxygen species subsequent to membrane permeabilization. Likewise, berberine induces elevated reactive oxygen species generation in S. pyogenes (262).
In Candida species, the flavonoid baicalein induces reactive oxygen species buildup and consequent death. Moreover, honey flavonoids and quercetin impede the growth of C. albicans by increasing intracellular reactive oxygen species levels (263). Santra et al. (264) demonstrated that plumbagin, derived from Plumbago zeylanica, produced oxidative stress in C. albicans, leading to fungal cell death (264).
7.6 Modulation of host immune responses
Certain medicinal plants augment the host immune system, enhancing its capacity to combat MDR microorganisms. Polysaccharides and saponins activate immune cells, including macrophages and neutrophils, to phagocytize infections (265). Natural immunostimulants, such as probiotics and traditional Chinese medicines, are progressively utilized as animal feed additives to manage Salmonella pullorum infections, owing to their environmentally favorable characteristics and absence of chemical residues (265). Zanthoxylum bungeanum meal, a byproduct of oil production, possesses bioactive chemicals and exhibits extensive antibacterial activity (265).
Likewise, Schisandra chinensis and Scutellaria baicalensis exhibit anti-inflammatory, antioxidant, and antibacterial characteristics and are recognized for their ability to improve animal development and productivity (265, 266). Conversely, Abdelmotaleb et al. (267) reported that Echinacea purpurea extracts augmented macrophage function against MDR M. tuberculosis.
7.7 Synergistic effects with conventional antibiotics
Herbal medicines frequently demonstrate synergistic effects when used in conjunction with antibiotics, hence augmenting their effectiveness against MDR pathogens (208, 268). Phytochemicals can diminish microbial defenses, rendering them more vulnerable to drugs. Hasna et al. (268) examined the synergistic effects of methanolic extracts from four Algerian medicinal plants (garlic, red onion, cumin, and fenugreek) in conjunction with several strains of lactic acid bacteria against H. pylori. The in vivo antibacterial efficacy of fenugreek extract in conjunction with Bifidobacterium breve was assessed to validate the synergistic impact of this particular combination on H. pylori colonization (268). All evaluated combinations of plant extracts and probiotics exhibited inhibitory activity against H. pylori, but with differing degrees of efficacy (268).
Saquib et al. (269) examined the synergistic antibacterial properties of ethanolic extracts from Punica granatum (pericarp), Commiphora molmol, and Azadirachta indica (bark) in conjunction with standard antibiotics (amoxicillin, metronidazole, tetracycline, and azithromycin) against principal periodontopathic bacteria (Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, and Aggregatibacter actinomycetemcomitans). Synergistic antibacterial action was noted when plant extracts were coupled with antibiotics, with the most effective synergy observed between P. granatum extract and amoxicillin against A. actinomycetemcomitans (269). Magryś et al. (270) observed that the combination of Allium sativum (garlic) extract with amoxicillin considerably decreased the MIC against MDR H. pylori.
Ayaz et al. (271) conducted a study to investigate the antibacterial efficacy and minimum bactericidal concentration (MBC) of three particular agents (thymol, EDTA, and vancomycin) against both susceptible and resistant bacterial strains. The research also examined the synergistic effects of thymol and EDTA, as well as the interactions of vancomycin with these two compounds (271). Thymol exhibited significant bactericidal activity, with MBC between 60 and 4000 g ml/1. Conversely, EDTA exhibited bacteriostatic activity solely beyond the MBC range of 60-4000 g ml/1 (271).
Gram-positive bacteria demonstrated MICs between 0.125 and 16 g ml/, whereas Gram-negative bacteria displayed MICs from 32 to 128 g ml/1. The interaction mechanism between various chemical combinations and MRSA and E. coli was examined by checkerboard dilution and time-kill curve assays (271).
7.8 Inhibition of virulence factors
Medicinal plants can inhibit the synthesis of virulence components, including toxins and adhesion molecules, hence diminishing the pathogenicity of MDR microorganisms (208, 272). Compounds such as resveratrol and quercetin inhibit the expression of genes that encode virulence factors. Resveratrol, derived from grapes, suppressed the synthesis of staphylococcal enterotoxins in MRSA (272). El-Mahdy et al. (273) suggested that resveratrol and curcumin may serve as alternative treatment strategies for treating MRSA infections by diminishing toxin production.
8 Medicinal plants as a complementary therapy
Humans and animals have used medicinal plants to treat a variety of diseases (274). Nonetheless, understanding of the causes of diseases and the curative qualities of specific plants remained severely limited; all information was based on empirical experimentation. Following the acquisition of suitable knowledge, transactions were based on insights gained via vast experience (274, 275). Prior to the advent of iatrochemistry in the 16th century, these herbs served as a means of treatment and prophylaxis (275, 276). The practice of Ayurvedic medicine, which demonstrates the healing properties of plants, indicates that old approaches have not been forgotten despite the introduction of antibiotics (275).
Although not all plants display these traits, the primary bioresource of active chemicals is found in esteemed medicinal herbs (277, 278). Various established bioactive chemicals have been employed in Ayurvedic practices, modern medicine, nutraceuticals, and pharmaceutical intermediates, serving as lead molecules for synthetic drugs and are currently accessible for purchase (277–279). The function of these molecules in development, photosynthesis, reproduction, or other fundamental activities remains undetermined. Secondary metabolites are synthesized to defend against biotic and abiotic stressors, including fungal and bacterial infections (280). Since antiquity, botanical products have served as the principal resource for human welfare and the cornerstone of all life on Earth (278, 280). Karunamoorthi et al. (281) and Jamshidi-Kia et al. (282) indicated that humans have utilized plants with medicinal properties as the foundation of intricate traditional medicine to treat diseases worldwide.
The utilization of medicinal plants can be dated to antiquity through various historical texts (283, 284), for example, records from Ancient Egypt date to 1500 BCE, the Chinese medical text “Shen Nong Ben Cao” originates from 200 CE, and Dioscorides’ De Materia Medica chronicles the Mediterranean pharmacopeia between 50 and 70 CE (283). WHO estimates indicate that 80% of individuals in poor nations primarily rely on traditional herbal remedies for their medicinal needs (284).
Research undertaken from 1981 to 2007 on the development of new pharmaceuticals, including antimicrobials, indicated that over 50% of the drugs licensed since 1994 are derived from natural sources (23, 285, 286). It is estimated that almost 80% of the chemicals utilized in pharmaceuticals globally are derived from natural sources, and now, there are more than a 100 novel candidates based on raw materials in the clinical development phase (285, 286). Moreover, despite substantial advancements in chemical synthesis, at least 25% of the medications currently utilized in contemporary medicine may be traced back to plant roots (286). This encompasses 60% of anticancer therapy and 75% of antiinfective medicines (287).
The choice of plant species is essential for the success of novel anti-infective discoveries in pharmaceutical development. Random selection, chemotaxonomic, and ethnopharmacological methodologies are employed to identify plants for biological and phytochemical investigation (288). The indiscriminate method sometimes entails collecting plants from several regions to generate a substantial quantity of plant samples, ensuring considerable chemical diversity and enhancing the likelihood of discovering a biologically active component (289). The sample of plant components is frequently selected and examined without the benefit of ethnobotanical and chemotaxonomic expertise (288). The presence of certain chemotaxonomic markers within each family or genus is a determinant in the selection of plants utilizing the chemotaxonomic method (289). This technique entails phytochemical screening to detect particular active biological markers in the plant, including flavonoids and alkaloids. The foundation for selection and evaluation in the ethnopharmacological method is the oral or written knowledge of a plant’s historic therapeutic application (290).
Ethnopharmacology is a versatile approach for discovering novel antibiotics. This subject is an interdisciplinary domain that integrates diverse fields like botany, chemistry, biochemistry, pharmacology, anthropology, archeology, history, and languages (290, 291). It specifically emphasizes the systematic observation, description, and experimental investigation of traditional medicine and the traits that hinder organism development (290, 291). Numerous resources exist for studying organized conventional medicinal systems, such as traditional Chinese medicine, Unani, Kampo, and Ayurveda. These resources encompass computer databases, fieldwork, literature, herbals, and review articles that frequently provide evaluations of medicinal plants according to geographic region or ethnic culture (291).
This approach has demonstrated remarkable efficacy in screening and identifying plants containing bioactive compounds potentially useful for the development of anti-infective pharmaceuticals (290). The technique involves choosing plant species according to the preferences of specific demographic groups for the treatment of infectious diseases (e.g., diarrhea, malaria) (291). The selected plants will be subjected to extraction processes, such as the acquisition of a crude extract or essential oil, which will then be evaluated for their ability to inhibit the growth of harmful microorganisms (290, 291).
Following the confirmation of the extract’s inhibitory activity, characterized by dose dependence (MIC), a targeted assay may be performed to identify the active constituents (292). This procedure entails the selection and refinement of the most active fraction(s) (293). Chemical testing methodologies and contemporary pharmacognosy are employed to ascertain the composition of the final fraction. The identified chemicals may be utilized to test and validate biological targets specific to disease (292, 293).
Researchers evaluated almost 2000 traditional Chinese herbal formulas derived from ancient texts of traditional Chinese medicine (294). Clinical investigations demonstrated that the ether extract of A. annua effectively inhibits the growth of Plasmodium falciparum, the causative cause of malaria (294). Subsequently, the researchers synthesized the derivative dihydroartemisinin and isolated the pure component artemisinin (294). The WHO advocates for artemisinin-based combination therapy as the primary treatment for malaria, which is extensively utilized worldwide (22).
Traditional Chinese medicine uses the roots and rhizomes of Coptidis spp. (Ranunculaceae) to treat ailments such as dysentery and diarrhea attributed to damp heat. The Chinese Pharmacopeia identifies the three Coptis species constituting “Huanglian” as Coptis chinensis, Coptis deltoidea, and Coptis teeta. Qi et al. (295) assert that Huanglian comprises berberine, an isoquinoline alkaloid, which is the principal constituent accountable for its antidiarrheal effects (295).
Numerous researchers have delineated the MIC range of berberine against diarrhoea-associated bacteria, such as S. aureus, MRSA, Vibrio cholera, Bacillus cereus, and E. coli, as 12.5–469 µg/ml (296–298). Numerous clinical investigations including pediatric patients have demonstrated that berberine and its derivatives (berberine tannate, hydrochloride, and berberine sulfate) are useful in treating diarrhea. Acute infectious diarrhea was effectively managed with berberine tannate and its combination with sulfadimidine and neomycin, as reported by Khameneh et al. (243). Clinical studies have demonstrated that berberine tannate is efficacious, including in pediatric patients with gastroenteritis (299). Research indicates that berberine hydrochloride and berberine sulfate effectively treat diarrhea in comparison to conventional antibiotics like chloramphenicol and streptomycin. They can significantly diminish the volume of liquid stool and the frequency of urgent bowel motions (300, 301).
Several pharmaceutical companies have promoted berberine as a therapeutic agent for diarrhea and intestinal infections (302). It is obtainable as an extract, exemplified by Huang Lian Su Tablets derived from Coptis chinensis extract, or in its pure form, such as berberine hydrochloride tablets produced by Northeast Pharmaceutical Group in Shenyang, China (302). A further example of effective ethnopharmacological research is the extraction of essential oil from Melaleuca alternifolia (Myrtaceae), a procedure equally grounded in ethnopharmacological principles. The tree is a shrub with delicate, parchment-like bark native to the humid coastal regions of northern New South Wales, Australia (302). The Bundjalung Aboriginal people utilized leaf poultices on wounds and ailments, inhaling vapors from burned leaves to alleviate coughs and colds (302). Numerous antimicrobial studies have demonstrated that tea tree oil exhibits antibacterial activity against S. aureus and MRSA in both in vitro and human trials (277). Numerous monoterpenes elucidate its inhibitory impact, including 1,8-cineole, β-terpinene, and terpinene-4-ol (302). Tea tree oil has been extensively marketed and utilized in inhalation therapy for the treatment of bacterial and fungal pneumonia (303).
Notwithstanding these facts, research on the screening, identification, and characterization of plant constituents is still regarded as crucial for discovering novel phytochemicals, particularly those with antimicrobial activities (304, 305). The work is ongoing, and further plants with antibacterial actiivites have been uncovered, including garlic (306), cinnamon (307), Glycyrrhiza glabra (liquorice), Viola odoratta, Amomum subulatum, Elettaria cardamomum (308), among numerous others. Thus, it indicates considerable potential for the development of new antibacterial agents from underexploited plants (309). This assertion is corroborated by other articles indicating that medicinal plants are the principal sources of antibacterial secondary metabolites (309). It is important to note that hardly a minuscule percentage of the estimated 750,000 plant species on Earth have been studied and utilized (310).
9 The long-term use of medicinal plants and the potential microbial resistance
For generations, medicinal plants have been utilized to address numerous ailments. Nonetheless, the prolonged utilization of these plants may foster the emergence of microbial resistance (311). Microbial resistance refers to the capacity of microorganisms, including bacteria, fungi, viruses, and parasites, to withstand the effects of medications that were previously effective in treating infections caused by these organisms (312). This resistance can complicate infection treatment, resulting in prolonged illnesses, elevated healthcare expenses, and sometimes fatal outcomes (311, 312).
The excessive usage and improper use of therapeutic herbs might generate selective pressure that promotes the survival of resistant bacteria. This parallels the phenomenon where excessive antibiotic use results in antibiotic resistance. Nakamoto et al. (313) asserted that garlic possesses several chemicals exhibiting antibacterial properties. Nonetheless, excessive use of garlic may result in the emergence of resistance to these chemicals. Echinacea (coneflower) is a widely utilized herbal treatment for colds and influenza. Nevertheless, research indicates that prolonged usage of Echinacea may diminish the efficacy of the immune system (313, 314).
Additionally, Hydrastis canadensis (goldenseal) comprises a component known as berberine, which exhibits antimicrobial properties. Excessive use of goldenseal may result in the development of resistance to berberine (314). Conversely, sub-lethal doses of medicinal herbs may fail to eradicate all microorganisms responsible for infection. The remaining organisms may subsequently acquire resistance to the medicinal plant (315). Moreover, resistance genes can be disseminated among microorganisms via other means, including plasmids and transposons. This indicates that a microorganism can obtain resistance genes from other bacteria, even without direct exposure to a medicinal plant (316, 317). Not all therapeutic herbs contribute to microbial resistance. Certain medicinal herbs may aid in the prevention of resistance development (318). Research indicates that cranberry juice may aid in the prevention of urinary tract infections by inhibiting bacterial adhesion to the urinary tract walls (319).
9.1 Factors influencing the development of resistance
In contrast to single-molecule antibiotics, medicinal plants comprise a complex array of bioactive chemicals, such as alkaloids, terpenoids, flavonoids, phenolics, and others. The “cocktail” effect poses a distinct challenge to bacteria striving to acquire resistance (320). Unlike conventional medicines that typically target a singular bacterial route, plant extracts frequently demonstrate antimicrobial activity via many pathways (320). This complex assault substantially complicates the ability of microorganisms to evolve resistance, as they are required to create several resistance mechanisms simultaneously (320). Several factors influence the development of resistance (320), as outlined below:-
i. Complexity of the extract: A higher diversity of bioactive chemicals in the plant extract correlates with reduced selective pressure for resistance. A diverse array of chemicals that target various biological systems complicates the adaptation of microorganisms (320).
ii. Concentration of active compounds: The conventional application of medicinal plants typically entails lesser amounts of active constituents compared to pure antibiotics. Reduced concentrations can diminish selective pressure, when microorganisms encounter sublethal dosages that may impede growth without causing mortality, so facilitating adaptation (320, 321). It is crucial to acknowledge that certain plant extracts may include elevated levels of certain chemicals, thereby intensifying selective pressure (321).
iii. Duration and frequency of exposure: Prolonged and repeated exposure to plant extracts may elevate the chance of resistance development, even at low dosages. Extended exposure offers increased opportunity for microorganisms to adapt and evolve (320, 321).
iv. Specific compounds versus whole extracts: Resistance is more probable to emerge against isolated plant chemicals, particularly when utilized as single-molecule antibiotics. The utilization of entire plant extracts, characterized by their intricate compositions, is typically seen as presenting a diminished risk of resistance (320).
v. Microbial species: Diverse microbial species possess differing abilities to acquire resistance. Certain species possess more adaptability and a higher propensity for developing resistance than others.
vi. Presence of resistance genes: Microorganisms can obtain resistance genes via horizontal gene transfer. The existence of pre-existing resistance genes within a microbial community can elevate the probability of resistance emergence against plant-derived antimicrobials, primarily if these compounds target the exact mechanisms as traditional antibiotics (316, 317).
10 Future concerns
The potential for employing medicinal plants against MDR bacteria is significant; nonetheless, it requires thorough research and development. Despite the extended history of medicinal plant utilization, there is a significant gap in our understanding of the sustained evolution of resistance to these complicated combinations. Research should focus on investigating the effects of extended exposure to medicinal plant extracts on microbial communities; determining the specific mechanisms by which bacteria may develop resistance to plant-derived antimicrobials; assessing the rate of resistance emergence to botanical extracts relative to conventional antibiotics; establishing standardized methodologies for evaluating antimicrobial efficacy and resistance emergence to plant extracts; and monitoring for signs of reduced efficacy of herbal remedies in clinical settings.
10.1 Unveiling the potential of medicinal plants
Medicinal plants contain a substantial array of bioactive chemicals with various modes of action. These chemicals may potentially surmount the resistance mechanisms established by MDR bacteria. The intricate amalgamation of phytochemicals in medicinal plants can demonstrate synergistic effects, wherein the collective activity surpasses the sum of its individual constituents. This combination can augment antibacterial efficacy and diminish the probability of resistance emergence.
10.2 Overcoming challenges
Maintaining uniform quality and effectiveness of plant-derived pharmaceuticals is essential. This necessitates uniform growing procedures, extraction techniques, and quality control protocols to ensure the consistency of active chemicals. Comprehensive scientific research is essential to confirm the effectiveness and safety of therapeutic plants against MDR bacteria. This includes in vitro and in vivo investigations to ascertain the mechanisms of action, ideal doses, and possible toxicity. Furthermore, the overexploitation of therapeutic plants may result in their depletion. Sustainable harvesting methods and conservation initiatives are crucial for guaranteeing the enduring availability of these precious resources.
10.3 Future directions
Medicinal plants can provide innovative lead chemicals for the development of new antibiotics. Advanced methodologies like genomics, proteomics, and metabolomics help expedite the identification and characterization of bioactive compounds. Integrating plant-derived antimicrobials with current antibiotics can augment their efficacy and address resistance. This method can help decrease the dosage of traditional antibiotics, hence reducing adverse effects and the emergence of further resistance. Comprehending the interplay between phytochemicals and specific patients can facilitate tailored therapeutic approaches. This method considers personal genetic composition, gut microbiota, and additional variables to enhance the efficacy of phytotherapeutics.
10.4 Cooperation and knowledge exchange
Combining traditional knowledge of medicinal plant usage with contemporary scientific studies can yield significant insights into their effectiveness and safety. Collaboration among scientists, healthcare practitioners, and traditional healers is crucial for converting research findings into applicable clinical practices. Moreover, exchanging information and resources among nations helps expedite the discovery and development of novel plant-based pharmaceuticals to address MDR bacteria globally. By confronting the problems and seizing the opportunities, medicinal plants can significantly contribute to combating MDR bacteria, providing a sustainable and potentially transformative healthcare solution.
11 Conclusion
Medicinal plants serve multiple purposes due to their efficacy, few side effects, and phytochemical constituents that can effectively treat numerous ailments. Utilizing potent medicinal herbs to address specific ailments mitigates or diminishes infections. Approximately 85% of the populace in developing nations utilizes traditional medicine, predominantly herbal medicines. Addressing antibiotic resistance is a crucial global health concern. This review examines research from 2014 to 2025 regarding the potential of medicinal plants as sources of novel antimicrobial agents. Due to the escalating issue of antibiotic resistance, these plant extracts present viable alternatives to traditional antibiotics, which are progressively losing efficacy.
Author contributions
ME-S: Resources, Writing – original draft, Writing – review & editing. AhS: Methodology, Writing – original draft, Writing – review & editing. DM: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. SK: Formal analysis, Writing – original draft, Writing – review & editing. MA: Writing – original draft, Writing – review & editing, Formal analysis. AA: Writing – original draft, Resources, Writing – review & editing. EI: Software, Writing – original draft, Methodology. HS: Writing – original draft, Writing – review & editing, Formal analysis, Software. SA: Data curation, Writing – original draft, Writing – review & editing. AbS: Writing – original draft, Writing – review & editing, Data curation. SE: Formal analysis, Writing – original draft, Writing – review & editing, Software. MF: Data curation, Writing – original draft, Writing – review & editing, Formal analysis. TA-E: Data curation, Software, Writing – original draft, Writing – review & editing. MA: Writing – original draft, Data curation, Formal analysis, Software, Writing – review & editing. HA: Methodology, Writing – original draft. ME-T: Writing – review & editing, Data curation. BM: Writing – original draft, Writing – review & editing, Formal analysis, Methodology. SA: Writing – original draft, Writing – review & editing, Project administration, Resources. KE-T: Writing – original draft, Writing – review & editing, Software. SI: Formal analysis, Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This publication was made possible by grant number NC.X- 267-5-12-170-1 from the National Institute of Food and Agriculture (NIFA) and the Department of Family and Consumer Sciences and the Agriculture Research Station at North Carolina Agriculture and Technical State University (Greensboro, NC, USA 27411). This work was also supported, in part, by 1890 Capacity Building Program grant no. (2020-38821-31113/project accession no.021765). The authors extend their appreciation to the Deanship of Research and Graduate Studies at King Khalid University for funding this work through Large Research Project under grant number (R. G. P. 1/104/45).
Conflict of interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Uddin TM, Chakraborty AJ, Khusro A, Zidan BRM, Mitra S, Emran TB, et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J Infect Public Health. (2021) 14:1750–66. doi: 10.1016/j.jiph.2021.10.020
2. Abdallah EM, Alhatlani BY, de Paula Menezes R, Martins CHG. Back to Nature: Medicinal plants as promising sources for antibacterial drugs in the post-antibiotic era. Plants. (2023) 12:3077. doi: 10.3390/plants12173077
3. Podolsky SH. The evolving response to antibiotic resistance (1945–2018). Palgrave Commun. (2018) 4:124. doi: 10.1057/s41599-018-0181-x
4. Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. (2023) 51:D690–9. doi: 10.1093/nar/gkac920
5. Muteeb G, Rehman MT, Shahwan M, Aatif M. Origin of antibiotics and antibiotic resistance, and their impacts on drug development: A narrative review. Pharmaceuticals. (2023) 16:1615. doi: 10.3390/ph16111615
6. Aggarwal R, Mahajan P, Pandiya S, Bajaj A, Verma SK, Yadav P, et al. Antibiotic resistance: a global crisis, problems and solutions. Crit Rev Microbiol. (2024) 50:896–921. doi: 10.1080/1040841X.2024.2313024
7. Ahmed SK, Hussein S, Qurbani K, Ibrahim RH, Fareeq A, Mahmood KA, et al. Antimicrobial resistance: impacts, challenges, and future prospects. J Med Surg Public Health. (2024) 2:100081. doi: 10.1016/j.glmedi.2024.100081
8. Salam MA, Al-Amin MY, Salam MT, Pawar JS, Akhter N, Rabaan AA, et al. Antimicrobial resistance: A growing serious threat for global public health. Healthcare. (2023) 11:1946. doi: 10.3390/healthcare11131946
9. Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A, et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. (2022) 399:629–55. doi: 10.1016/S0140-6736(21)02724-0
10. Wang M. Surveying the immuno-surfacome of Staphylococcus aureus with focus on peptidoglycan hydrolases. (2022). PhD thesis. University of Groningen, Groningen, The Netherlands. (2022). doi: 10.33612/diss.202979659
11. Wu R, Payne M, Zhang L, Lan R. Uncovering the boundaries of Campylobacter species through large-scale phylogenetic and nucleotide identity analyses. mSystems. (2024) 9:e01218–e01223. doi: 10.1128/msystems.01218-23
12. Soni J, Sinha S, Pandey R. Understanding bacterial pathogenicity: A closer look at the journey of harmful microbe. Front Microbiol. (2024) 15:1370818. doi: 10.3389/fmicb.2024.1370818
13. Lv Y, Huang X, Wu J, Xiao X, Ma C, Jiang X, et al. Economic burden attributable to healthcare-associated infections at western China hospitals: 6 Year, prospective cohort study. J Infect. (2024) 88:112–22. doi: 10.1016/j.jinf.2023.12.008
14. Payne V, Johnson MJ, Hunt K, Prieto J. Investigating the implementation of a complex intervention to reduce central line-associated bloodstream infections in the neonatal intensive care unit, using normalisation process theory. J Neonatal Nurs. (2025) 31:186–95. doi: 10.1016/j.jnn.2024.07.026
15. Tettey F, Parupelli SK, Desai S. A review of biomedical devices: classification, regulatory guidelines, human factors, software as a medical device, and cybersecurity. BioMed Mater. (2024) 2:316–41. doi: 10.1007/s44174-023-00113-9
16. Hall S, Grayson J, Grant G, Vertullo C, Anoopkumar-Dukie S. In vitro evaluation of vancomycin-induced toxicity in human primary knee chondrocytes. Int J Toxicol. (2024) 43:177–83. doi: 10.1177/10915818231216413
17. World Health Organization (WHO). Global research agenda for antimicrobial resistance in human health. Geneva, Switzerland. (2024). Licence: CC BY-NC-SA 3.0 IGO. p. 95. Available online at: https://www.who.int/publications/i/item/9789240102309.
18. da Silva WP, Lopes GV, Ramires T, Kleinubing NR. May phenolics mitigate the antimicrobial resistance in foodborne pathogens? Curr Opin Food Sci. (2024) 55:101107. doi: 10.1016/j.cofs.2023.101107
19. Thomford NE, Senthebane DA, Rowe A, Munro D, Seele P, Maroyi A, et al. Natural products for drug discovery in the 21st century: innovations for novel drug discovery. Int J Mol Sci. (2018) 19:1578. doi: 10.3390/ijms19061578
20. Feng X, Cao S, Qiu F, Zhang B. Traditional application and modern pharmacological research of Artemisia annua L. Pharmacol Ther. (2020) 216:107650. doi: 10.1016/j.pharmthera.2020.107650
21. Bhattacharya S, Bir R, Majumdar T. Evaluation of multidrug resistant Staphylococcus aureus and their association with biofilm production in a Tertiary Care Hospital, Tripura, Northeast India. J Clin Diagn Res. (2015) 9:DC01–OC4. doi: 10.7860/JCDR/2015/13965.6417
22. World Health Organization (WHO). Guidelines for the treatment of malaria: Third Editon. World Health Organization. Geneva, Switzerland: WHO Library Cataloguing-in-Publication Data (2015). p. 313.
23. Anand U, Jacobo-Herrera N, Altemimi A, Lakhssassi N. A comprehensive review on medicinal plants as antimicrobial therapeutics: potential avenues of biocompatible drug discovery. Metabolites. (2019) 9:258. doi: 10.3390/metabo9110258
24. Pei Z, Li F, Gong J, Zou L, Ding L, Qiu F. Research progress on nano-drug delivery systems for antitumor active components of traditional Chinese medicine. Chin Tradit Herb Drugs. (2021) 52:7658–67. doi: 10.7501/j.issn.0253-2670.2021.24.029
25. Sabry BA, Badr AN, Mohammed DM, Desoukey MA, Farouk A. Validating the protective role of orange and tangerine peel extracts foramending food safety against microorganisms’ contamination using molecular docking. Heliyon. (2024) 10:e27737. doi: 10.1016/j.heliyon.2024.e27737
26. Jamal A. Embracing nature’s therapeutic potential: Herbal medicine. Int J Multidiscip Sci Arts. (2023) 2:117–26. doi: 10.47709/ijmdsa.v2i1.2620
27. Alshafei MM, Mabrouk AM, Hanafi EM, Ramadan MM, Korany RM, Kassem SS, et al. Prophylactic supplementation of microencapsulated Boswellia serrata and probiotic bacteria in metabolic syndrome rats. Food Biosci. (2023) 51:102325. doi: 10.1016/j.fbio.2022.102325
28. Chaachouay N, Douira A, Zidane L. Herbal medicine used in the treatment of human diseases in the Rif, Northern Morocco. Arab J Sci Eng. (2022) 47:131–53. doi: 10.1007/s13369-021-05501-1
29. Abdoon ASS, Hegazy AM, Abdel-Azeem AS, Al-Atrash AM, Mohammed DM. The protective effects of some herbs on mitigating HFD-induced obesity via enhancing biochemical indicators and fertility in female rats. Heliyon. (2024) 10:e30249. doi: 10.1016/j.heliyon.2024.e30249
30. Muktar Y, Mamo G, Tesfaye B, Belina D. A Rev major bacterial causes calf diarrhea its Diagn method. J Vet Med Anim Health. (2015) 7:173–85. doi: 10.5897/JVMAH2014.0351
31. Li D, Xu K-Y, Zhao W-P, Liu M-F, Feng R, Li D-Q, et al. Chinese medicinal herb-derived carbon dots for common diseases: efficacies and potential mechanisms. Front Pharmacol. (2022) 13:815479. doi: 10.3389/fphar.2022.815479
32. Silalahi M, Walujo EB, Supriatna J, Mangunwardoyo W. The local knowledge of medicinal plants trader and diversity of medicinal plants in the Kabanjahe traditional market, North Sumatra, Indonesia. J Ethnopharmacol. (2015) 175:432–43. doi: 10.1016/j.jep.2015.09.009
33. Sujarwo W, Keim AP, Savo V, Guarrera PM, Caneva G. Ethnobotanical study of Loloh: Traditional herbal drinks from Bali (Indonesia). J Ethnopharmacol. (2015) 169:34–48. doi: 10.1016/j.jep.2015.03.079
34. Teodoro GR, Ellepola K, Seneviratne CJ, Koga-Ito CY. Potential use of phenolic acids as anti-Candida agents: A review. Front Microbiol. (2015) 6:1420. doi: 10.3389/fmicb.2015.01420
35. Pratiwi SU, Lagendijk EL, Hertiani T, De Weert S, Cornellius A, Van Den Hondel J. Antimicrobial effects of Indonesian medicinal plants extracts on planktonic and biofilm growth of Pseudomonas aeruginosa and Staphylococcus aureus. Int J Pharm Pharm Sci. (2015) 7:183–91. doi: 10.4172/2376-0354.1000119
36. Wikaningtyas P, Sukandar EY. The antibacterial activity of selected plants towards resistant bacteria isolated from clinical specimens. Asian Pac J Trop Biomed. (2016) 6:16–9. doi: 10.1016/j.apjtb.2015.08.003
37. Selvarajan R, Obize C, Sibanda T, Abia ALK, Long H. Evolution and emergence of antibiotic resistance in given ecosystems: possible strategies for addressing the challenge of antibiotic resistance. Antibiotics. (2022) 12:28. doi: 10.3390/antibiotics12010028
38. Sarmah P, Dan MM, Adapa D, Sarangi T. A review on common pathogenic microorganisms and their impact on human health. Electronic J Biol. (2018) 14:50–8.
39. Makam P, Matsa R. Big Three” infectious diseases: Tuberculosis, Malaria and HIV/AIDS. Curr Top Med Chem. (2021) 21:2779–99. doi: 10.2174/1568026621666210916170417
40. Vasan N, Baselga J, Hyman DM. A view on drug resistance in cancer. Nature. (2019) 575:299–309. doi: 10.1038/s41586-019-1730-1
41. Iskandar K, Molinier L, Hallit S, Sartelli M, Catena F, Coccolini F, et al. Drivers of antibiotic resistance transmission in low-and middle-income countries from a “one health” perspective—a review. Antibiotics. (2020) 9:372. doi: 10.3390/antibiotics9070372
42. Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat Rev Microbiol. (2017) 15:453–64. doi: 10.1038/nrmicro.2017.42
43. Chandra H, Bishnoi P, Yadav A, Patni B, Mishra AP, Nautiyal AR. Antimicrobial resistance and the alternative resources with special emphasis on plant-based antimicrobials—a review. Plants. (2017) 6:16. doi: 10.3390/plants6020016
44. Garte S. Targeted hypermutation as a survival strategy: A theoretical approach. Acta Biotheor. (2023) 71:20. doi: 10.1007/s10441-023-09471-9
45. Rojewska M, Smułek W, Kaczorek E, Prochaska K. Langmuir monolayer techniques for the investigation of model bacterial membranes and antibiotic biodegradation mechanisms. Membranes. (2021) 11:707. doi: 10.3390/membranes11090707
46. Li T, Feng J, Xiao S, Shi W, Sullivan D, Zhang Y. Identification of FDA-approved drugs with activity against stationary phase Bartonella henselae. Antibiotics. (2019) 8:50. doi: 10.3390/antibiotics8020050
47. Wang F, Fu Y, Lin Z, Zhang B, Se J, Guo X, et al. Neglected drivers of antibiotic resistance: Survival of extended-spectrum β-lactamase-producing pathogenic Escherichia coli from livestock waste through dormancy and release of transformable extracellular antibiotic resistance genes under heat treatment. Environ Sci Technol. (2023) 57:9955–64. doi: 10.1021/acs.est.3c02377
48. Soundy J, Day D. Delivery of antibacterial silver nanoclusters to Pseudomonas aeruginosa using species-specific DNA aptamers. J Med Microbiol. (2020) 69:640–52. doi: 10.1099/jmm.0.001174
49. Alotaibi GF, Bukhari MA. Factors influencing bacterial biofilm formation and development. Am J BioMed Sci Res. (2021) 12:617–26. doi: 10.34297/AJBSR.2021.12.001820
50. Berne C, Ellison CK, Ducret A, Brun YV. Bacterial adhesion at the single-cell level. Nat Rev Microbiol. (2018) 16:616–27. doi: 10.1038/s41579-018-0057-5
51. Yesankar P, Pal M, Patil A, Qureshi A. Microbial exopolymeric substances and biosurfactants as ‘bioavailability enhancers’ for polycyclic aromatic hydrocarbons biodegradation. Int J Environ Sci Technol. (2023) 20:5823–44. doi: 10.1007/s13762-022-04068-0
52. Thaarup IC, Iversen AKS, Lichtenberg M, Bjarnsholt T, Jakobsen TH. Biofilm survival strategies in chronic wounds. Microorganisms. (2022) 10:775. doi: 10.3390/microorganisms10040775
53. Yin R, Cheng J, Wang J, Li P, Lin J. Treatment of Pseudomonas aeruginosa infectious biofilms: Challenges and strategies. Front Microbiol. (2022) 13:955286. doi: 10.3389/fmicb.2022.955286
54. Kowalska-Krochmal B, Dudek-Wicher R. The minimum inhibitory concentration of antibiotics: Methods, interpretation, clinical relevance. Pathogens. (2021) 10:165. doi: 10.3390/pathogens10020165
55. Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. (2017) 179:204–22. doi: 10.1016/j.trsl.2016.08.002
56. Dilbato T, Begna F, Joshi RK. Reviews on challenges, opportunities and future prospects of antimicrobial activities of medicinal plants: Alternative solutions to combat antimicrobial resistance. Int J Herb Med. (2019) 7:10–8.
57. Bloom DE, Cadarette D. Infectious disease threats in the twenty-first century: strengthening the global response. Front Immunol. (2019) 10:549. doi: 10.3389/fimmu.2019.00549
59. El-Sayed A, Kamel M. Climatic changes and their role in emergence and re-emergence of diseases. Environ Sci pollut Res. (2020) 27:22336–52. doi: 10.1007/s11356-020-08896-w
60. Reynolds D, Burnham JP, Guillamet CV, McCabe M, Yuenger V, Betthauser K, et al. The threat of multidrug-resistant/extensively drug-resistant Gram-negative respiratory infections: Another pandemic. Eur Respir J. (2022) 31:220068. doi: 10.1183/16000617.0068-2022
61. Vivas R, Barbosa AAT, Dolabela SS, Jain S. Multidrug-resistant bacteria and alternative methods to control them: An overview. Microb Drug Resist. (2019) 25:890–908. doi: 10.1089/mdr.2018.0319
62. Thakur A, Mikkelsen H, Jungersen G. Intracellular pathogens: host immunity and microbial persistence strategies. J Immunol Res. (2019) 2019:1356540. doi: 10.1155/2019/1356540
63. Amere GA, Nayak P, Salindri AD, Narayan KV, Magee MJ. Contribution of smoking to tuberculosis incidence and mortality in high-tuberculosis-burden countries. Am J Epidemiol. (2018) 187:1846–55. doi: 10.1093/aje/kwy081
64. Abo-zeid Y, Amer A, El-Houssieny B, Mahmoud M, Sakran W. Overview on bacterial resistance and nanoparticles to overcome bacterial resistance. J Adv Pharm Res. (2021) 5:312–26. doi: 10.21608/aprh.2021.76488.1131
65. Pulingam T, Parumasivam T, Gazzali AM, Sulaiman AM, Chee JY, Lakshmanan M, et al. Antimicrobial resistance: Prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur J Pharm Sci. (2022) 170:106103. doi: 10.1016/j.ejps.2021.106103
66. Saha M, Sarkar A. Review on multiple facets of drug resistance: A rising challenge in the 21st century. J Xenobiot. (2021) 11:197–214. doi: 10.3390/jox11040013
67. Krawczyk B, Wityk P, Gałęcka M, Michalik M. The many faces of Enterococcus spp.-commensal, probiotic and opportunistic pathogen. Microorganisms. (2021) 9:1900. doi: 10.3390/microorganisms9091900
68. Breitburg D, Levin LA, Oschlies A, Grégoire M, Chavez FP, Conley DJ, et al. Declining oxygen in the global ocean and coastal waters. Science. (2018) 359:eaam7240. doi: 10.1126/science.aam7240
69. Bojanova DP, Bordenstein SR. Fecal transplants: What is being transferred? PloS Biol. (2016) 14:e1002503. doi: 10.1371/journal.pbio.1002503
70. Williamson AJ, Jacobson R, van Praagh JB, Gaines S, Koo HY, Lee B, et al. Enterococcus faecalis promotes a migratory and invasive phenotype in colon cancer cells. Neoplasia. (2022) 27:100787. doi: 10.1016/j.neo.2022.100787
71. Fiore E, Van Tyne D, Gilmore MS. Pathogenicity of enterococci. Microbiol Spectr. (2019) 7:4. doi: 10.1128/microbiolspec
72. Ho FK, Lam LN, Matysik A, Watts TD, Wong JJ, Chong KKL, et al. Role of sortase-assembled Ebp pili in Enterococcus faecalis adhesion to iron oxides and its impact on extracellular electron transfer. Microbiol Spectr. (2025) 0:e02337–24. doi: 10.1128/spectrum.02337-24
73. Vale de Macedo GH, Costa GD, Oliveira ER, Damasceno GV, Mendonça JS, Silva LD, et al. Interplay between ESKAPE pathogens and immunity in skin infections: An overview of the major determinants of virulence and antibiotic resistance. Pathogens. (2021) 10:148. doi: 10.3390/pathogens10020148
74. Wiśniewski P, Zakrzewski A, Chajęcka-Wierzchowska W, Zadernowska A. Possibility of transfer and activation of ‘silent’ tetracycline resistance genes among Enterococcus faecalis under high-pressure processing. Food Microbiol. (2024) 120:104481. doi: 10.1016/j.fm.2024.104481
75. Tigabu A, Getaneh A. Staphylococcus aureus, ESKAPE bacteria challenging current health care and community settings: A literature review. Clin Lab. (2021) 67:7. doi: 10.7754/Clin.Lab.2020.200930
76. Ganesan N, Mishra B, Felix L, Mylonakis E. Antimicrobial peptides and small molecules targeting the cell membrane of Staphylococcus aureus. Microbiol Mol Biol Rev. (2023) 87:e0003722. doi: 10.1128/mmbr.00037-22
77. Chen X, Schneewind O, Missiakas D. Engineered human antibodies for the opsonization and killing of Staphylococcus aureus. Proc Natl Acad Sci. (2022) 119:e2114478119. doi: 10.1073/pnas.2114478119
78. Walter A, Unsleber S, Rismondo J, Jorge AM, Peschel A, Gründling A, et al. Phosphoglycerol-type wall and lipoteichoic acids are enantiomeric polymers differentiated by the stereospecific glycerophosphodiesterase GlpQ. J Biol Chem. (2020) 295:4024–34. doi: 10.1074/jbc.RA120.012566
79. Sauer K, Stoodley P, Goeres DM, Hall-Stoodley L, Burmølle M, Stewart PS, et al. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat Rev Microbiol. (2022) 20:608–20. doi: 10.1038/s41579-022-00767-0
80. Stevens EJ, Bates KA, King KC. Host microbiota can facilitate pathogen infection. PloS Pathog. (2021) 17:e1009514. doi: 10.1371/journal.ppat.1009514
81. Tsouklidis N, Kumar R, Heindl SE, Soni R, Khan S. Understanding the fight against resistance: hospital-acquired methicillin-resistant Staphylococcus aureus vs community-acquired methicillin-resistant Staphylococcus aureus. Cureus. (2020) 12:e8867. doi: 10.7759/cureus.8867
83. Corrado A, Donato P, Maccari S, Cecchi R, Spadafina T, Arcidiacono L, et al. Staphylococcus aureus-dependent septic arthritis in murine knee joints: Local immune response and beneficial effects of vaccination. Sci Rep. (2016) 6:38043. doi: 10.1038/srep38043
84. Mishra AK, Yadav P, Mishra A. A systemic review on staphylococcal scalded skin syndrome (SSSS): A rare and critical disease of neonates. Open Microbiol J. (2016) 10:150–9. doi: 10.2174/1874285801610010150
85. Boswihi SS, Udo EE. Methicillin-resistant Staphylococcus aureus: An update on the epidemiology, treatment options and infection control. Curr Med Res Pract. (2018) 8:18–24. doi: 10.1016/j.cmrp.2018.01.001
86. Sebastianski M, Bridger NA, Featherstone RM, Robinson JL. Disease outbreaks linked to pasteurized and unpasteurized dairy products in Canada and the United States: Asystematic review. Can J Public Health. (2022) 113:569–78. doi: 10.17269/s41997-022-00614-y
87. Algammal AM, Hetta HF, Elkelish A, Alkhalifah DH, Hozzein WN, Batiha GE, et al. Methicillin-resistant Staphylococcus aureus (MRSA): One health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist. (2020) 13:3255–65. doi: 10.2147/IDR.S272733
88. Kong C, Neoh HM, Nathan S. Targeting Staphylococcus aureus toxins: A potential form of anti-virulence therapy. Toxins. (2016) 8:72. doi: 10.3390/toxins8030072
89. Lacey KA, Geoghegan JA, McLoughlin RM. The role of Staphylococcus aureus virulence factors in skin infection and their potential as vaccine antigens. Pathogens. (2016) 5:22. doi: 10.3390/pathogens5010022
90. Ramírez-Larrota JS, Eckhard U. An introduction to bacterial biofilms and their proteases, and their roles in host infection and immune evasion. Biomolecules. (2022) 12:306. doi: 10.3390/biom12020306
91. Al-Mebairik NF, El-Kersh TA, Al-Sheikh YA, Marie MAM. A review of virulence factors, pathogenesis, and antibiotic resistance in Staphylococcus aureus. Rev Res Med Microbiol. (2016) 27:50–6. doi: 10.1097/MRM.0000000000000067
92. Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. (2021) 12:547–69. doi: 10.1080/21505594.2021.1878688
93. Łoś J, Zielińska S, Krajewska A, Zalewska M, Małachowska A, Kwaśnicka K, et al. Temperate phages, prophages, and lysogeny. In: Harper DR, Abedon ST, Burrowes BH, McConville ML, editors. Bacteriophages: biology, technology, therapy. Cham, Switzerland: Springer (2021) p. 119–50. doi: 10.1007/978-3-319-41986-2_3
94. Gajdács M. The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics. (2019) 8:52. doi: 10.3390/antibiotics8020052
95. Moellering RC Jr. MRSA: the first half century. J Antimicrob Chemother. (2012) 67:4–11. doi: 10.1093/jac/dkr437
96. Lu T, DeLeo FR, Singh SK. Pathogenesis of Staphylococcus aureus in humans. In: Human emerging and re-emerging infections: viral and parasitic infections, John Wiley & Sons, Inc, Hoboken (2015). p. 711–48. doi: 10.1002/9781118644843.ch39
97. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. (2019) 17:203–18. doi: 10.1038/s41579-018-0147-4
98. Dabhi M, Patel R, Shah V, Soni R, Saraf M, Rawal R, et al. Penicillin-binding proteins: the master builders and breakers of bacterial cell walls and its interaction with β-lactam antibiotics. J Proteins Proteom. (2024) 15:215–32. doi: 10.1007/s42485-024-00135-x
99. Zhan X-Y, Zhu Q-Y. Evolution of methicillin-resistant Staphylococcus aureus: evidence of positive selection in a penicillin-binding protein (PBP) 2a coding gene mecA. Infect Genet Evol. (2018) 59:16–22. doi: 10.1016/j.meegid.2018.01.018
100. Samia NI, Robicsek A, Heesterbeek H, Peterson LR. Methicillin-resistant Staphylococcus aureus nosocomial infection has a distinct epidemiological position and acts as a marker for overall hospital-acquired infection trends. Sci Rep. (2022) 12:17007. doi: 10.1038/s41598-022-21300-6
101. Braun T, Kahanov L, Dannelly K, Lauber C. CA-MRSA infection incidence and care in high school and intercollegiate athletics. Med Sci Sports Exerc. (2016) 48:1530–8. doi: 10.1249/mss.0000000000000940
102. Junnila J, Hirvioja T, Rintala E, Auranen K, Rantakokko-Jalava K, Silvola J, et al. Changing epidemiology of methicillin-resistant Staphylococcus aureus in a low endemicity area—new challenges for MRSA control. Eur J Clin Microbiol Infect Dis. (2020) 39:2299–307. doi: 10.1007/s10096-020-03824-9
103. Sørensen AIV, Toft N, Boklund A, Espinosa-Gongora C, Græsbøll K, Larsen J, et al. A mechanistic model for spread of livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) within a pig herd. PloS One. (2017) 12:e0188429. doi: 10.1371/journal.pone.0188429
104. Siciliano V, Passerotto RA, Chiuchiarelli M, Leanza GM, Ojetti V. Difficult-to-treat pathogens: a review on the management of multidrug-resistant Staphylococcus epidermidis. Life. (2023) 13:1126. doi: 10.3390/life13051126
105. Ortega-Peña S, Rodríguez-Martínez S, Cancino-Diaz ME, Cancino-Diaz JC. Staphylococcus epidermidis controls opportunistic pathogens in the nose, could it help to regulate SARS-CoV-2 (COVID-19) infection? Life. (2022) 12:341. doi: 10.3390/life12030341
106. Severn MM, Horswill AR. Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nat Rev Microbiol. (2023) 21:97–111. doi: 10.1038/s41579-022-00780-3
107. Becker K, Both A, Weißelberg S, Heilmann C, Rohde H. Emergence of coagulase-negative staphylococci. Expert Rev Anti Infect Ther. (2020) 18:349–66. doi: 10.1080/14787210.2020.1730813
108. Oliveira WF, Silva PMS, Silva RCS, Silva GMM, MaChado G, Coelho LCBB, et al. Staphylococcus aureus and Staphylococcus epidermidis infections on implants. J Hosp Infect. (2018) 98:111–7. doi: 10.1016/j.jhin.2017.11.008
109. Kao MS, Yang JH, Balasubramaniam A, Traisaeng S, Yang AG, Yang JJ, et al. Colonization of nasal cavities by Staphylococcus epidermidis mitigates SARS-CoV-2 nucleocapsid phosphoprotein-induced interleukin (IL)-6 in the lung. Microb Biotechnol. (2022) 15:1984–94. doi: 10.1111/1751-7915.13994
110. Peixoto PB, Massinhani FH, Dos Santos KR, Chamon RC, Silva RB, Correa FE, et al. Methicillin-resistant Staphylococcus epidermidis isolates with reduced vancomycin susceptibility from bloodstream infections in a neonatal intensive care unit. J Med Microbiol. (2020) 69:41–5. doi: 10.1099/jmm.0.001117
111. Urban-Chmiel R, Marek A, Stępień-Pyśniak D, Wieczorek K, Dec M, Nowaczek A, et al. Antibiotic resistance in bacteria—a review. Antibiotics. (2022) 11:1079. doi: 10.3390/antibiotics11081079
112. McMurray R, Ball M, Tunney M, Corcionivoschi N, Situ C. Antibacterial activity of four plant extracts extracted from traditional Chinese medicinal plants against Listeria monocytogenes, Escherichia coli, and Salmonella enterica subsp enterica serovar Enteritidis. Microorganisms. (2020) 8:962. doi: 10.3390/microorganisms8060962
113. Parham S, Kharazi AZ, Bakhsheshi-Rad HR, Nur H, Ismail AF, Sharif S, et al. Antioxidant, antimicrobial and antiviral properties of herbal materials. Antioxidants. (2020) 9:1309. doi: 10.3390/antiox9121309
114. Beshiru A, Okoh AI, Igbinosa EO. Processed ready-to-eat (RTE) foods sold in Yenagoa Nigeria were colonized by diarrheagenic Escherichia coli which constitute a probable hazard to human health. PloS One. (2022) 17:e0266059. doi: 10.1371/journal.pone.0266059
115. He L, Wang C, Simujide H, Aricha H, Zhang J, Liu B, et al. Effect of early pathogenic Escherichia coli infection on the intestinal barrier and immune function in newborn calves. Front Cell Infect Microbiol. (2022) 12:818276. doi: 10.3389/fcimb.2022.818276
116. Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, Petersen AM. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin Microbiol Rev. (2019) 32:e00060–18. doi: 10.1128/cmr.00060-18
117. Zil-e-Huma, Tareen AM, Kaleem Ullah KU, Asmat TM, Abdul Samad AS, Iqbal AI, et al. Incidence of diarrheagenic Escherichia coli pathotypes in children suffering from diarrhea in tertiary care hospitals, Quetta, Pakistan. Pak J Zool. (2019) 51:2015–21. doi: 10.17582/journal.pjz/2019.51.6.2015.2021
118. Mulu BM, Belete MA, Demlie TB, Tassew H, Sisay Tessema T. Characteristics of pathogenic Escherichia coli associated with diarrhea in children under five years in northwestern Ethiopia. Trop Med Infect Dis. (2024) 9:65. doi: 10.3390/tropicalmed9030065
119. Wang X, Yu D, Chui L, Zhou T, Feng Y, Cao Y, et al. A comprehensive review on shiga toxin subtypes and their niche-related distribution characteristics in shiga-toxin-producing E. coli and other bacterial hosts. Microorganisms. (2024) 12:687. doi: 10.3390/microorganisms12040687
120. Modgil V, Kaur H, Mohan B, Taneja N. Association of putative virulence genes with HEp-2 cell adherence and biofilm production in enteroaggregative E. coli strains isolated from acute diarrheal and healthy children, India. Indian J Microbiol. (2024) 64:186–97. doi: 10.1007/s12088-023-01157-7
121. Chang D, Sharma L, Dela Cruz CS, Zhang D. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front Microbiol. (2021) 12:750662. doi: 10.3389/fmicb.2021.750662
122. Wentao N, Guobao L, Jin Z, Junchang C, Rui W, Zhancheng G, et al. In vitro activity of minocycline combined with aminoglycosides against Klebsiella pneumoniae carbapenemase-producing K. pneumoniae. J Antibiot. (2018) 71:506–13. doi: 10.1038/s41429-017-0024-9
123. Priyanka A, Akshatha K, Deekshit VK, Prarthana J, Akhila DS. Klebsiella pneumoniae infections and antimicrobial drug resistance. In: Siddhardha B, Dyavaiah M, Syed A, editors. Model organisms for microbial pathogenesis, biofilm formation and antimicrobial drug discovery, Springer, Singapore (2020). p. 195–225. doi: 10.1007/978-981-15-1695-5_12
124. Vaneci-Silva D, Assane IM, de Oliveira AL, Gomes FC, Moro EB, Kotzent S, et al. Klebsiella pneumoniae causing mass mortality in juvenile Nile tilapia in Brazil: isolation, characterization, pathogenicity and phylogenetic relationship with other environmental and pathogenic strains from livestock and human sources. Aquaculture. (2022) 546:737376. doi: 10.1016/j.aquaculture.2021.737376
125. Ding L, Shen S, Chen J, Tian Z, Shi Q, Han R, et al. Klebsiella pneumoniae carbapenemase variants: the new threat to global public health. Clin Microbiol Rev. (2023) 36:e0000823. doi: 10.1128/cmr.00008-23
126. Kimera ZI, Mgaya FX, Mshana SE, Karimuribo ED, Matee MI. Occurrence of extended spectrum beta lactamase (ESBL) producers, quinolone and carbapenem resistant Enterobacteriaceae isolated from environmental samples along Msimbazi River basin ecosystem in Tanzania. Int J Environ Res Public Health. (2021) 18:8264. doi: 10.3390/ijerph18168264
127. Wu W, Jin Y, Bai F, Jin S. Pseudomonas aeruginosa. In: Tang YW, Liu D, Sussman M, Schwartzman J, Poxton I, editors. Molecular medical microbiology. Cambridge, Massachusetts, USA: Academic Press (2015) p. 753–67. doi: 10.1016/B978-0-12-397169-2.00041-X
128. Frieri M, Kumar K, Boutin A. Antibiotic resistance. J Infect Public Health. (2017) 10:369–78. doi: 10.1016/j.jiph.2016.08.007
130. Kunz Coyne AJ, El Ghali A, Holger D, Rebold N, Rybak MJ. Therapeutic strategies for emerging multidrug-resistant. Pseudomonas aeruginosa. Infect Dis Ther. (2022) 11:661–82. doi: 10.1007/s40121-022-00591-2
131. Chatterjee M, Anju C, Biswas L, Kumar VA, Mohan CG, Biswas R. Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int J Med Microbiol. (2016) 306:48–58. doi: 10.1016/j.ijmm.2015.11.004
132. Lorusso AB, Carrara JA, Barroso CDN, Tuon FF, Faoro H. Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. Int J Mol Sci. (2022) 23:15779. doi: 10.3390/ijms232415779
133. Silva A, Silva V, Igrejas G, Poeta P. Carbapenems and Pseudomonas aeruginosa: mechanisms and epidemiology. In: Hashmi MZ, editor. Antibiotics and antimicrobial resistance genes in the environment. Amsterdam, Netherlands: Elsevier (2020) p. 253–68. doi: 10.1016/B978-0-12-818882-8.00017-6
134. Gharieb R, Saad M, Khedr M, El Gohary A, Ibrahim H. Occurrence, virulence, carbapenem resistance, susceptibility to disinfectants, and public health hazard of Pseudomonas aeruginosa isolated from animals, humans, and the environment in intensive farms. J Appl Microbiol. (2022) 132:256–67. doi: 10.1111/jam.15191
135. Yin X, Fu Y, Tate H, Pinto C, Dudley EG, M’ikanatha NM. Genomic analysis of Salmonella Typhimurium from humans and food sources accurately predicts phenotypic multi-drug resistance. Food Microbiol. (2022) 103:103957. doi: 10.1016/j.fm.2021.103957
136. Mkangara M. Prevention and control of human Salmonella enterica infections: An implication in food safety. Int J Food Sci. (2023) 2023:8899596. doi: 10.1155/2023/8899596
137. Balali GI, Yar DD, Afua Dela VG, Adjei-Kusi P. Microbial contamination: an increasing threat to the consumption of fresh fruits and vegetables in today’s world. Int J Microbiol. (2020) 2020:3029295. doi: 10.1155/2020/3029295
138. Peruzy MF, Capuano F, Proroga YTR, Cristiano D, Carullo MR, Murru N. Antimicrobial susceptibility testing for Salmonella serovars isolated from food samples: Five-year monitoring (2015–2019). Antibiotics. (2020) 9:365. doi: 10.3390/antibiotics9070365
139. Van Hauwermeiren F, Lamkanfi M. How to dodge pyroptosis: lessons from. Shigella flexneri. Cell Res. (2022) 32:227–8. doi: 10.1038/s41422-021-00607-5
140. Odo SE, Uchechukwu CF, Ezemadu UR. Foodborne diseases and intoxication in Nigeria: prevalence of Escherichia coli O157:H7 Salmonella, Shigella and Staphylococcus aureus. J Adv Microbiol. (2021) 20:84–94. doi: 10.9734/jamb/2020/v20i1230312
141. Edward S, Mureithi E, Shaban N. Shigellosis dynamics: Modelling the effects of treatment, sanitation, and education in the presence of carriers. Int J Math Math Sci. (2020) 2020:3476458. doi: 10.1155/2020/3476458
142. Abd Elaziz D, Abd-El-Ghany M, Meshaal S, El Hawary R, Lotfy S, Galal N, et al. Fungal infections in primary immunodeficiency diseases. Clin Immunol. (2020) 219:108553. doi: 10.1016/j.clim.2020.108553
143. Halder LD, Babych S, Palme DI, Mansouri-Ghahnavieh E, Ivanov L, Ashonibare V, et al. Candida albicans induces cross-kingdom miRNA trafficking in human monocytes to promote fungal growth. mBio. (2022) 13:e03563–21. doi: 10.1128/mbio.03563-21
144. Gow NA, Yadav B. Microbe profile: Candida albicans: a shape-changing, opportunistic pathogenic fungus of humans. Microbiology. (2017) 163:1145–7. doi: 10.1099/mic.0.000499
145. Le PH, Nguyen DHK, Aburto Medina A, Linklater DP, Loebbe C, Crawford RJ, et al. Surface architecture influences the rigidity of Candida albicans cells. Nanomaterials. (2022) 12:567. doi: 10.3390/nano12030567
146. Tsui C, Kong EF, Jabra-Rizk MA. Pathogenesis of Candida albicans biofilm. Pathog Dis. (2016) 74:ftw018. doi: 10.1093/femspd/ftw018
147. Zhong L, Dong Z, Liu F, Li H, Tang K, Zheng C, et al. Incidence, clinical characteristics, risk factors, and outcomes of patients with mixed Candida/bacterial bloodstream infections: a retrospective study. Ann Clin Microbiol Antimicrob. (2022) 21:45. doi: 10.1186/s12941-022-00538-y
148. Patel M. Oral cavity and Candida albicans: Colonization to the development of infection. Pathogens. (2022) 11:335. doi: 10.3390/pathogens11030335
149. Vila T, Sultan AS, Montelongo-Jauregui D, Jabra-Rizk MA. Oral candidiasis: a disease of opportunity. J Fungi. (2020) 6:15. doi: 10.3390/jof6010015
150. Rodríguez-Cerdeira C, Martínez-Herrera E, Carnero-Gregorio M, López-Barcenas A, Fabbrocini G, Fida M, et al. Pathogenesis and clinical relevance of Candida biofilms in vulvovaginal candidiasis. Front Microbiol. (2020) 11:544480. doi: 10.3389/fmicb.2020.544480
151. Riera FO, Caeiro JP, Angiolini SC, Vigezzi C, Rodriguez E, Icely PA, et al. Invasive candidiasis: update and current challenges in the management of this mycosis in South America. Antibiotics. (2022) 11:877. doi: 10.3390/antibiotics11070877
152. Motbainor H, Bereded F, Mulu W. Multi-drug resistance of blood stream, urinary tract, and surgical site nosocomial infections of Acinetobacter baumannii and Pseudomonas aeruginosa among patients hospitalized at Felegehiwot Referral Hospital, Northwest Ethiopia: a cross-sectional study. BMC Infect Dis. (2020) 20:29. doi: 10.1186/s12879-020-4811-8
153. Todd OA. Identification of effectors of synergistic lethality in Candida albicans-Staphylococcus aureus polymicrobial intra-abdominal infection. PhD Thesis. Memphis, Tennessee, USA: The University of Tennessee Health Science Center (2021). doi: 10.21007/etd.cghs.2021.0537
154. Richardson JP, Willems HME, Moyes DL, Shoaie S, Barker KS, Tan SL, et al. Candidalysin drives epithelial signaling, neutrophil recruitment, and immunopathology at the vaginal mucosa. Infect Immun. (2018) 86:e00645–17. doi: 10.1128/IAI.00645-17
155. Li X, Leonardi I, Iliev ID. Candidalysin sets off the innate alarm. Sci Immunol. (2017) 2:17. doi: 10.1126/sciimmunol.aao5703
156. Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol. (2017) 7:2173. doi: 10.3389/fmicb.2016.02173
157. Szymański M, Chmielewska S, Czyżewska U, Malinowska M, Tylicki A. Echinocandins–structure, mechanism of action and use in antifungal therapy. J Enzyme Inhib Med Chem. (2022) 37:876–94. doi: 10.1080/14756366.2022.2050224
158. Perlin DS. Echinocandin resistance in Candida. Clin Infect Dis. (2015) 61:S612–7. doi: 10.1093/cid/civ791
159. Bhattacharya S, Sae-Tia S, Fries BC. Candidiasis and mechanisms of antifungal resistance. Antibiotics. (2020) 9:312. doi: 10.3390/antibiotics9060312
160. Zuo L, Liu K, Liu H, Hu Y, Zhang Z, Qin J, et al. Trend of HIV-1 drug resistance in China: a systematic review and meta-analysis of data accumulated over 17 years (2001–2017). EClinical Med. (2020) 18:100238. doi: 10.1016/j.eclinm.2019.100238
161. Chen Y, Chen Q, Wang X, Sun F, Fan Y, Liu X, et al. Hemostatic action of lotus leaf charcoal is probably due to transformation of flavonol aglycones from flavonol glycosides in traditional Chinese medicine. J Ethnopharmacol. (2020) 249:112364. doi: 10.1016/j.jep.2019.112364
162. Chen F, Li J, Wang H, Ba Q. Anti-tumor effects of Chinese medicine compounds by regulating immune cells in the microenvironment. Front Oncol. (2021) 11:746917. doi: 10.3389/fonc.2021.746917
163. Zuza-Alves DL, Silva-Rocha WP, Chaves GM. An update on Candida tropicalis based on basic and clinical approaches. Front Microbiol. (2017) 8:1927. doi: 10.3389/fmicb.2017.01927
164. Khalifa HO, Watanabe A, Kamei K. Azole and echinocandin resistance mechanisms and genotyping of Candida tropicalis in Japan: Cross-boundary dissemination and animal–human transmission of C. tropicalis infection. Clin Microbiol Infect. (2022) 28:302. doi: 10.1016/j.cmi.2021.10.004
165. Posteraro B, De Carolis E, Criscuolo M, Ballanti S, De Angeli G, Del Principe MI, et al. Candidaemia in haematological malignancy patients from a SEIFEM study: epidemiological patterns according to antifungal prophylaxis. Mycoses. (2020) 63:900–10. doi: 10.1111/myc.13130
166. Dowling A, O’Dwyer J, Adley C. Antibiotics: mode of action and mechanisms of resistance. In: Méndez-Vilas A, editor. Antimicrobial research: novel bioknowledge and educational programs, Formatex Research Center, PA, USA (2017). p. 536–45.
167. Olaleye OO, Kim D-H, Spriggs KA. Antiproliferative activities of some selected Nigerian medicinal plants against breast, liver, and cervical cancer cells. BMC Complement Altern Med. (2024) 24:110. doi: 10.1186/s12906-024-04365-w
168. Humayun Akhter SM, Naeem S, Ansari US, Siddiqui VU, Bharti SS, Ahmad S, et al. Green Synthesis of metal oxide nanoparticles using Plumbago zeylanica root extract, spectrochemical characterization, and antibacterial activity against common pathogen. Appl Res. (2025) 4:e202400200. doi: 10.1002/appl.202400200
169. Halim N, Saidin J, Zin NAM, Vigneswari S. Antibacterial potential of marine organisms and coastal plants in combating antibacterial resistance: A review. Malays J Microbiol. (2024) 20:244–62. doi: 10.21161/mjm.230485
170. Mayer AM, Mayer VA, Swanson-Mungerson M, Pierce ML, Rodríguez AD, Nakamura F, et al. Marine pharmacology in 2019–2021: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar Drugs. (2024) 22:309. doi: 10.3390/md22070309
171. Rousta N, Aslan M, Yesilcimen AM, Ozcan F, Sar T, Taherzadeh MJ. Effects of fungal based bioactive compounds on human health. Crit Rev Food Sci Nutr. (2024) 64:7004–27. doi: 10.1080/10408398.2023.2178379
172. Djilianov D, Moyankova D, Mladenov P, Topouzova-Hristova T, Kostadinova A, Staneva G, et al. Resurrection Plants-A valuable source of natural bioactive compounds: from word-of-mouth to scientifically proven sustainable use. Metabolites. (2024) 14:113. doi: 10.3390/metabo14020113
173. Abass S, Parveen R, Irfan M, Malik Z, Husain SA, Ahmad S. Mechanism of antibacterial phytoconstituents: An updated review. Arch Microbiol. (2024) 206:325. doi: 10.1007/s00203-024-04035-y
174. Xiang ZD, Guan HD, Zhao X, Xie Q, Cai FJ, Xie ZJ, et al. Protoberberine alkaloids: a review of the gastroprotective effects, pharmacokinetics, and toxicity. Phytomedicine. (2024) 126:155444. doi: 10.1016/j.phymed.2024.155444
175. Seyfi R, Kahaki FA, Ebrahimi T, Montazersaheb S, Eyvazi S, Babaeipour V, et al. Antimicrobial peptides (AMPs): roles, functions and mechanism of action. Int J Pept Res Ther. (2020) 26:1451–63. doi: 10.1007/s10989-019-09946-9
176. Žuvela P, Skoczylas M, Jay Liu J, Ba̧czek T, Kaliszan R, Wong MW, et al. Column characterization and selection systems in reversed-phase high-performance liquid chromatography. Chem Rev. (2019) 119:3674–729. doi: 10.1021/acs.chemrev.8b00246
177. Soni R, Shankar G, Mukhopadhyay P, Gupta V. A concise review on Artemisia annua L.: A major source of diverse medicinal compounds. Ind Crops Prod. (2022) 184:115072. doi: 10.1016/j.indcrop.2022.115072
178. Gautam AK, Singh PK, Aravind M. Defensive role of plant phenolics against pathogenic microbes for sustainable agriculture. In: Lone R, Shuab R, Kamili AN, editors. Plant phenolics in sustainable agriculture, Springer, Singapore (2020). p. 579–94. doi: 10.1007/978-981-15-4890-1_25
179. Saad AM, Mohamed AS, El-Saadony MT, Sitohy MZ. Palatable functional cucumber juices supplemented with polyphenols-rich herbal extracts. LWT. (2021) 148:111668. doi: 10.1016/j.lwt.2021.111668
180. El-Saadony MT, Saad AM, Elakkad HA, El-Tahan AM, Alshahrani OA, Alshilawi MS, et al. Flavoring and extending the shelf life of cucumber juice with aroma compounds-rich herbal extracts at 4°C through controlling chemical and microbial fluctuations. Saudi J Biol Sci. (2022) 29:346–54. doi: 10.1016/j.sjbs.2021.08.092
181. Ullah A, Munir S, Badshah SL, Khan N, Ghani L, Poulson BG, et al. Important flavonoids and their role as a therapeutic agent. Molecules. (2020) 25:5243. doi: 10.3390/molecules25225243
182. Jubair N, Rajagopal M, Chinnappan S, Abdullah NB, Fatima A. Review on the antibacterial mechanism of plant-derived compounds against multidrug-resistant bacteria (MDR). Evid-based Complement Altern Med. (2021) 2021:3663315. doi: 10.1155/2021/3663315
183. Aristri MA, Lubis MAR, Iswanto AH, Fatriasari W, Sari RK, Antov P, et al. Bio-based polyurethane resins derived from tannin: Source, synthesis, characterisation, and application. Forests. (2021) 12:1516. doi: 10.3390/f12111516
184. Zulhendri F, Chandrasekaran K, Kowacz M, Ravalia M, Kripal K, Fearnley J, et al. Antiviral, antibacterial, antifungal, and antiparasitic properties of propolis: A review. Foods. (2021) 10:1360. doi: 10.3390/foods10061360
185. King JP, Wang A. Putative roles of terpenoids in primitive membranes. Front Ecol Evol. (2023) 11:1272163. doi: 10.3389/fevo.2023.1272163
186. Farha AK, Yang QQ, Kim G, Zhang D, Mavumengwana V, Habimana O, et al. Inhibition of multidrug-resistant foodborne Staphylococcus aureus biofilms by a natural terpenoid (+)-nootkatone and related molecular mechanism. Food Control. (2020) 112:107154. doi: 10.1016/j.foodcont.2020.107154
187. Kandar CC. Secondary metabolites from plant sources. In: Pal D, Nayak AK, editors. Bioactive natural products for pharmaceutical applications. Advanced structured materials. Cham, Switzerland: Springer (2021) p. 329–77. doi: 10.1007/978-3-030-54027-2_10
188. Adibah KZM, Azzreena MA. Plant toxins: alkaloids and their toxicities. GSC Biol Pharm Sci. (2019) 6:021–9. doi: 10.30574/gscbps.2019.6.2.0003
189. Khameneh B, Iranshahy M, Ghandadi M, Ghoochi Atashbeyk D, Fazly Bazzaz BS, Iranshahi M. Investigation of the antibacterial activity and efflux pump inhibitory effect of co-loaded piperine and gentamicin nanoliposomes in methicillin-resistant Staphylococcus aureus. Drug Dev Ind Pharm. (2015) 41:989–94. doi: 10.3109/03639045.2014.920025
190. Sultan A, Afroza B, Mufti S, Malik AA, Sakib N, Akhter A, et al. Vegetable food toxicants and their harmful effects on health. Int J Curr Microbiol Appl Sci. (2020) 11:1923–36.
191. Ayogu JI, Odoh AS. Prospects and therapeutic applications of cardiac glycosides in cancer remediation. ACS Comb Sci. (2020) 22:543–53. doi: 10.1021/acscombsci.0c00082
192. El-Shiekh RA, Hassan M, Hashem RA, Abdel-Sattar E. Bioguided isolation of antibiofilm and antibacterial pregnane glycosides from Caralluma quadrangula: Disarming multidrug-resistant pathogens. Antibiotics. (2021) 10:811. doi: 10.3390/antibiotics10070811
193. Alamgir ANM. Therapeutic use of medicinal plants and their extracts. Cham, Switzerland: Springer (2017). p. 546. doi: 10.1007/978-3-319-63862-1
194. Mohammed DM, Salem MB, Elzallat M, Hammam OA, Suliman AA. Moringa oleifera L. mediated zinc oxide nano-biofertilizer alleviates non-alcoholic steatohepatitis via modulating de novo lipogenesis pathway and miRNA-122 expression. Food Biosci. (2024) 60:104286. doi: 10.1016/j.fbio.2024.104286
195. Xie M, Huang HL, Zhang WH, Gao L, Wang YW, Zhu XJ, et al. Increased sarcoplasmic/endoplasmic reticulum calcium ATPase 2a activity underlies the mechanism of the positive inotropic effect of ivabradine. Exp Physiol. (2020) 105:477–88. doi: 10.1113/EP087964
196. Jiang Y, Chen Y, Song Z, Tan Z, Cheng J. Recent advances in design of antimicrobial peptides and polypeptides toward clinical translation. Adv Drug Delivery Rev. (2021) 170:261–80. doi: 10.1016/j.addr.2020.12.016
197. Mekloy P, Sakchaisri K, Jiratchariyakul W, Bunsupa S. Investigation of genes coding for anti-HIV proteins MAP30 and MRK29 from Thai bitter melon (Momordica charantia L.). Pharm Sci Asia. (2022) 49:59–68. doi: 10.29090/psa.2022.01.21.104
198. Mandal A, Biswas D, Hazra B. Natural products from plants with prospective anti-HIV activity and relevant mechanisms of action. Stud Nat Prod Chem. (2020) 66:225–71. doi: 10.1016/B978-0-12-817907-9.00009-X
199. Pandey R, Srivastava S. Plant thionins: the green antimicrobial agents. In: Pandey R, Srivastava S, editors. Integrative approaches to biotechnology. CRC Press, Boca Raton, USA (2024). p. 93–110.
200. Bilal T, Mushtaq T, Ahmad PI, Gangoo SA, Behar B, Ayoob B, et al. Botanicals their use as antimicrobial, antifungal and anti-insecticides. Pharma Innov. (2022) 11:1521–8.
201. Cardoso JC, Oliveira MEBD, Cardoso FDC. Advances and challenges on the in vitro production of secondary metabolites from medicinal plants. Hortic Bras. (2019) 37:124–32. doi: 10.1590/S0102-053620190201
202. Yin H, Jiang N, Shi W, Chi X, Liu S, Chen J-L, et al. Development and effects of influenza antiviral drugs. Molecules. (2021) 26:810. doi: 10.3390/molecules26040810
203. Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. (2020) 83:770–803. doi: 10.1021/acs.jnatprod.9b01285
204. Gadisa E, Weldearegay G, Desta K, Tsegaye G, Hailu S, Jote K, et al. Combined antibacterial effect of essential oils from three most commonly used Ethiopian traditional medicinal plants on multidrug resistant bacteria. BMC Complement Altern Med. (2019) 19:24. doi: 10.1186/s12906-019-2429-4
205. El-Saadony MT, Zabermawi NM, Zabermawi NM, Burollus MA, Shafi ME, Alagawany M, et al. Nutritional aspects and health benefits of bioactive plant compounds against infectious diseases: a review. Food Rev Int. (2023) 39:2138–60. doi: 10.1080/87559129.2021.1944183
206. Kebede T, Gadisa E, Tufa A. Antimicrobial activities evaluation and phytochemical screening of some selected medicinal plants: A possible alternative in the treatment of multidrug-resistant microbes. PloS One. (2021) 16:e0249253. doi: 10.1371/journal.pone.0249253
207. Yeshiwas Y, Tadele ET, Tiruneh W. The dynamics of medicinal plants utilization practice nexus its health and economic role in Ethiopia: a review paper. Int J Biodivers Conserv. (2019) 11:31–47. doi: 10.5897/IJBC2018.1201
208. Gonelimali FD, Lin J, Miao W, Xuan J, Charles F, Chen M, et al. Antimicrobial properties and mechanism of action of some plant extracts against food pathogens and spoilage microorganisms. Front Microbiol. (2018) 9:1639. doi: 10.3389/fmicb.2018.01639
209. Panda SK, Das R, Lavigne R, Luyten W. Indian medicinal plant extracts to control multidrug-resistant S. aureus, including in biofilms. S Afr J Bot. (2020) 128:283–91. doi: 10.1016/j.sajb.2019.11.019
210. Richwagen N, Lyles JT, Dale BL, Quave CL. Antibacterial activity of Kalanchoe mortagei and K. fedtschenkoi against ESKAPE pathogens. Front Pharmacol. (2019) 10:67. doi: 10.3389/fphar.2019.00067
211. Pallah O, Meleshko T, Tymoshchuk S, Yusko L, Bugyna L. How to escape’the ESKAPE pathogens’ using plant extracts. ScienceRise Biol Sci. (2019) 20-21:30–7. doi: 10.15587/2519-8025.2019.193155
212. Khan MF, Tang H, Lyles JT, Pineau R, Mashwani Z-U-R, Quave CL. Antibacterial properties of medicinal plants from Pakistan against multidrug-resistant ESKAPE pathogens. Front Pharmacol. (2018) 9:815. doi: 10.3389/fphar.2018.00815
213. Shobha K, Rao AS, Pai KSR, Bhat S. Antimicrobial activity of aqueous and ethanolic leaf extracts of Anacardium occidentale. Asian J Pharm Clin Res. (2018) 11:474–6. doi: 10.22159/ajpcr.2018.v11i12.29073
214. Chandrasekharan D, Ballal R, Ballal BB, Khetmalas MB. evaluation of selected medicinal plants for their potential antimicrobial activities against ESKAPE pathogens and the study of p-glycoprotein related antibiosis; an indirect approach to assess efflux mechanism. Int J Recent Sci Res. (2020) 9:9461–29466. doi: 10.24327/IJRSR
215. Nayim P, Mbaveng AT, Wamba BE, Fankam AG, Dzotam JK, Kuete V. Antibacterial and antibiotic-potentiating activities of thirteen Cameroonian edible plants against gram-negative resistant phenotypes. Sci World J. (2018) 2018:4020294. doi: 10.1155/2018/4020294
216. Subramani R, Narayanasamy M, Feussner K-D. Plant-derived antimicrobials to fight against multi-drug-resistant human pathogens. 3 Biotech. (2017) 7:172. doi: 10.1007/s13205-017-0848-9
217. Masoumian M, Zandi M. Antimicrobial activity of some medicinal plant extracts against multidrug resistant bacteria. Zahedan J Res Med Sci. (2017) 19:e10080. doi: 10.5812/zjrms.10080
218. Mustapha A, AlSharksi AN, Eze UA, Samaila RK, Ukwah BN, Anyiam AF, et al. Phytochemical composition, in silico molecular docking analysis and antibacterial activity of Lawsonia inermis linn leaves extracts against extended spectrum beta-lactamases-producing strains of Klebsiella pneumoniae. BioMed. (2024) 4:277–92. doi: 10.3390/biomed4030022
219. Ghuman S, Ncube B, Finnie JF, McGaw LJ, Coopoosamy RM, Van Staden J. Antimicrobial activity, phenolic content, and cytotoxicity of medicinal plant extracts used for treating dermatological diseases and wound healing in KwaZulu-Natal, South Africa. Front Pharmacol. (2016) 7:320. doi: 10.3389/fphar.2016.00320
220. Agarwal P, Agarwal N, Gupta R, Gupta M, Sharma B. Antibacterial activity of plants extracts against methicillin-resistant Staphylococcus aureus vancomycin-resistant Enterococcus faecalis. J Microb Biochem Technol. (2016) 8:404–7. doi: 10.4172/1948-5948.1000316
221. Bacha K, Tariku Y, Gebreyesus F, Zerihun S, Mohammed A, Weiland-Bräuer N, et al. Antimicrobial and anti-quorum sensing activities of selected medicinal plants of Ethiopia: Implication for development of potent antimicrobial agents. BMC Microbiol. (2016) 16:139. doi: 10.1186/s12866-016-0765-9
222. Arullappan S, Rajamanickam P, Thevar N, Kodimani CC. In vitro screening of cytotoxic, antimicrobial and antioxidant activities of Clinacanthus nutans (Acanthaceae) leaf extracts. Trop J Pharm Res. (2014) 13:1455–61. doi: 10.4314/tjpr.v13i9.11
223. Ortega-Ramirez LA, Silva-Espinoza BA, Vargas-Arispuro I, Gonzalez-Aguilar GA, Cruz-Valenzuela MR, Nazzaro F, et al. Combination of Cymbopogon citratus and Allium cepa essential oils increased antibacterial activity in leafy vegetables. J Sci Food Agric. (2017) 97:2166–73. doi: 10.1002/jsfa.8025
224. Ma Y-L, Zhu D-Y, Thakur K, Wang C-H, Wang H, Ren Y-F, et al. Antioxidant and antibacterial evaluation of polysaccharides sequentially extracted from onion (Allium cepa L.). Int J Biol Macromol. (2018) 111:92–101. doi: 10.1016/j.ijbiomac.2017.12.154
225. Shanmugam J, Dhayalan M, Savaas Umar MR, Gopal M, Ali Khan M, Simal-Gandara J, et al. Green synthesis of silver nanoparticles using Allium cepa var. aggregatum natural extract: antibacterial and cytotoxic properties. Nanomaterials. (2022) 12:1725. doi: 10.3390/nano12101725
226. Zhou Y, Chen X, Chen T, Chen X. A review of the antibacterial activity and mechanisms of plant polysaccharides. Trends Food Sci Technol. (2022) 123:264–80. doi: 10.1016/j.tifs.2022.03.020
227. Diris M, Basri A, Metali F, Ahmad N, Taha H. Phytochemicals and antimicrobial activities of Melastoma malabathricum and Melastoma beccarianum leaf crude extracts. Res J Phytochem. (2017) 11:35–41.
228. Apridamayanti P, Sari R, Rachmaningtyas A, Aranthi V. Antioxidant, antibacterial activity and FICI (Fractional Inhibitory Concentration Index) of ethanolic extract of Melastoma malabathricum leaves with amoxicillin against pathogenic bacteria. Nus Biosci. (2021) 13:2. doi: 10.13057/nusbiosci/n130202
229. Wong K-C, Hag Ali DM, Boey P-L. Chemical constituents and antibacterial activity of Melastoma malabathricum L. Nat Prod Res. (2012) 26:609–18. doi: 10.1080/14786419.2010.538395
230. Świderski G, Gołębiewska E, Kowalczyk N, Kalinowska M, Świsłocka R, Wołejko E, et al. Structure, antioxidant activity and antimicrobial study of light lanthanide complexes with p-coumaric acid. Materials. (2024) 17:1324. doi: 10.3390/ma17061324
231. Boonyanugomol W, Kraisriwattana K, Rukseree K, Boonsam K, Narachai P. In vitro synergistic antibacterial activity of the essential oil from Zingiber cassumunar Roxb against extensively drug-resistant Acinetobacter baumannii strains. J Infect Public Health. (2017) 10:586–92. doi: 10.1016/j.jiph.2017.01.008
232. Han A-R, Kim H, Piao D, Jung C-H, Seo EK. Phytochemicals and bioactivities of Zingiber cassumunar Roxb. Molecules. (2021) 26:2377. doi: 10.3390/molecules26082377
233. Malahubban M, Alimon A, Sazili A, Fakurazi S, Zakry F. Phytochemical analysis of Andrographis paniculata and Orthosiphon stamineus leaf extracts for their antibacterial and antioxidant potential. Trop Biomed. (2013) 30:467–80.
234. Mishra PK, Singh RK, Gupta A, Chaturvedi A, Pandey R, Tiwari SP, et al. Antibacterial activity of Andrographis paniculata (Burm. f.) Wall ex Nees leaves against clinical pathogens. J Pharm Res. (2013) 7:459–62. doi: 10.1016/j.jopr.2013.05.009
235. Al-Snafi AE. The pharmacology and medical importance of Dolichos lablab (Lablab purpureus)-A review. IOSR J Pharm. (2017) 7:22–30. doi: 10.9790/3013-0702012230
236. Jayakumar T, Hsieh C-Y, Lee J-J, Sheu J-R. Experimental and clinical pharmacology of Andrographis paniculata and its major bioactive phytoconstituent andrographolide. Evid Based Complementary Altern Med. (2013) 2013:846740. doi: 10.1155/2013/846740
237. Banerjee M, Parai D, Chattopadhyay S, Mukherjee SK. Andrographolide: antibacterial activity against common bacteria of human health concern and possible mechanism of action. Folia Microbiol. (2017) 62:237–44. doi: 10.1007/s12223-017-0496-9
238. Priya S, Karthika M. Antibacterial activity of the volatile oil of lablab purpureus against some burn isolates. Int J Pharm Sci Res. (2013) 4:54–6.
239. Rampadarath S, Puchooa D, Jeewon R. Jatropha curcas L: Phytochemical, antimicrobial and larvicidal properties. Asian Pac J Trop Biomed. (2016) 6:858–65. doi: 10.1016/j.apjtb.2016.01.019
240. Alexa E, Bota V, Sumălan RM, Obistioiu D, Negrea M, Cocan I, et al. Natural emulsions based on essential oils as antifungal and antimycotoxicogenic agents on wheat for bakery industry. Foods. (2022) 11:2926. doi: 10.3390/foods11182926
241. Yadav MK, Chae S-W, Im GJ, Chung J-W, Song J-J. Eugenol: a phyto-compound effective against methicillin-resistant and methicillin-sensitive Staphylococcus aureus clinical strain biofilms. PloS One. (2015) 10:e0119564. doi: 10.1371/journal.pone.0119564
242. Marchese A, Barbieri R, Coppo E, Orhan IE, Daglia M, Nabavi SF, et al. Antimicrobial activity of eugenol and essential oils containing eugenol: A mechanistic viewpoint. Crit Rev Microbiol. (2017) 43:668–89. doi: 10.1080/1040841X.2017.1295225
243. Khameneh B, Eskin NM, Iranshahy M, Fazly Bazzaz BS. Phytochemicals: a promising weapon in the arsenal against antibiotic-resistant bacteria. Antibiotics. (2021) 10:1044. doi: 10.3390/antibiotics10091044
244. Kumar M, Prakash S, Radha, Kumari N, Pundir A, Punia S, et al. Beneficial role of antioxidant secondary metabolites from medicinal plants in maintaining oral health. Antioxidants. (2021) 10:1061. doi: 10.3390/antiox10071061
245. Vachhani K, Rao D, Panwar S, Gupta S. An in vivo comparative clinical and radiographic evaluation of a mixture of zinc oxide–Ocimum sanctum extract, zinc oxide–ozonated oil, and zinc oxide–eugenol as obturating materials in primary molars. J Indian Soc Pedod Prev Dent. (2022) 40:398–403. doi: 10.4103/jisppd.jisppd_204_22
246. Moghrovyan A, Sahakyan N. Antimicrobial activity and mechanisms of action of Origanum vulgare L. essential oil: effects on membrane-associated properties. AIMS Biophys. (2024) 11:508–26. doi: 10.3934/biophy.2024027
247. Yan Y, Li X, Zhang C, Lv L, Gao B, Li M. Research progress on antibacterial activities and mechanisms of natural alkaloids: A review. Antibiotics. (2021) 10:318. doi: 10.3390/antibiotics10030318
248. Bailly C. The steroidal alkaloids α-tomatine and tomatidine: Panorama of their mode of action and pharmacological properties. Steroids. (2021) 176:108933. doi: 10.1016/j.steroids.2021.108933
249. Tan A, Scortecci KC, Cabral De Medeiros NM, Kukula-Koch W, Butler TJ, Smith SM, et al. Plukenetia volubilis leaves as source of anti-Helicobacter pylori agents. Front Pharmacol. (2024) 15:1461447. doi: 10.3389/fphar.2024.1461447
250. Emrick D. Characterization of the fungicidal activity and biochemical impact of occidiofungin, a novel antifungal compound derived from Burkholderia contaminans Doctoral thesis. Mississippi State University, Mississippi, MS, USA (2019). p. 1031.
251. Kalli S, Vallieres C, Violet J, Sanders J-W, Chapman J, Vincken JP, et al. Cellular Responses and targets in food spoilage yeasts exposed to antifungal prenylated isoflavonoids. Microbiol Spectr. (2023) 11:e01327–23. doi: 10.1128/spectrum.01327-23
252. Yu X, Hao Z, Liu P, Liu M, Zhao L, Zhao X. Increased expression of efflux pump NorA drives the rapid evolutionary trajectory from tolerance to resistance against ciprofloxacin in Staphylococcus aureus. Antimicrob Agents Chemother. (2022) 66:e0059422. doi: 10.1128/aac.00594-22
253. Rana T, Farooq U, Kaur N, Khan A, Khan A. Plants derived efflux pump inhibitors: An approach against multidrug-resistant gram-negative bacteria Klebsiella pneumoniae. Pharmacophore. (2023) 14:71–9. doi: 10.51847/7FQXFNYnT5
254. Aksoy CS. Evaluation of thymol and carvacrol as efflux pumps inhibitors in Staphylococcus aureus. Master thesis, (2019) Marmara University, Istanbul, Turkey. p. 90.
255. Buchmann D, Schwabe M, Weiss R, Kuss AW, Schaufler K, Schlüter R, et al. Natural phenolic compounds as biofilm inhibitors of multidrug-resistant Escherichia coli–the role of similar biological processes despite structural diversity. Front Microbiol. (2023) 14:1232039. doi: 10.3389/fmicb.2023.1232039
256. Birru RL, Bein K, Bondarchuk N, Wells H, Lin Q, Di YP, et al. Antimicrobial and anti-inflammatory activity of apple polyphenol phloretin on respiratory pathogens associated with chronic obstructive pulmonary disease. Front Cell Infect Microbiol. (2021) 11:652944. doi: 10.3389/fcimb.2021.652944
257. Ogidi OI, Emaikwu NG. Plant phenolic compound isolation and its bioinformatics approaches to molecular mechanisms in antimicrobial activities and resistance. In: Pathak PD, Raut R, Jaramillo-Isaza S, Borkar P, Jhaveri RH, editors. Computational approaches in biotechnology and bioinformatics. CRC Press, Boca Raton, FL, USA (2024). p. 188–215. doi: 10.1201/9781003354437
258. Vadakkan K, Sathishkumar K, Mapranathukaran VO, Ngangbam AK, Nongmaithem BD, Hemapriya J, et al. Critical review on plant-derived quorum sensing signaling inhibitors in Pseudomonas aeruginosa. Bioorg Chem. (2024) 151:107649. doi: 10.1016/j.bioorg.2024.107649
259. Zhou H, Chen L, Ouyang K, Zhang Q, Wang W. Antibacterial activity and mechanism of flavonoids from Chimonanthus salicifolius SY Hu. and its transcriptome analysis against Staphylococcus aureus. Front Microbiol. (2023) 13:1103476. doi: 10.3389/fmicb.2022.1103476
260. Kim G, Xu Y, Zhang J, Sui Z, Corke H. Antibacterial activity and multi-targeting mechanism of dehydrocorydaline from Corydalis turtschaninovii Bess. against Listeria monocytogenes. Front Microbiol. (2022) 12:799094. doi: 10.3389/fmicb.2021.799094
261. Nisa FU, Javid A, Ahmed M. Green tea (Camellia sinensis) leaf extract inhibits the activity of β-lactamases. Int J Appl Exp Biol. (2025) 4:41–8. doi: 10.56612/ijaaeb.v1i1.111
262. Kosalec I, Jembrek MJ, Vlainić J. The spectrum of berberine antibacterial and antifungal activities. In: Rai M, Kosalec I, editors. Promising antimicrobials from natural products. Springer, Cham, Switzerland. (2022) p. 119–32. doi: 10.1007/978-3-030-83504-0_7
263. Sarkar C, Chaudhary P, Jamaddar S, Janmeda P, Mondal M, Mubarak MS, et al. Redox activity of flavonoids: Impact on human health, therapeutics, and chemical safety. Chem Res Toxicol. (2022) 35:140–62. doi: 10.1021/acs.chemrestox.1c00348
264. Santra I, Chakraborty A, Ghosh B. Evaluation of plumbagin synthesis: mimicking in vivo plant systems through the application of elicitors inducing stress on in vitro regenerated Plumbago zeylanica L. Plant Cell Tiss Organ Cult (PCTOC). (2024) 157:11. doi: 10.1007/s11240-024-02732-6
265. Shen J, Liu T, Qian Y, Yan S, Liu Z, Jia F. Therapeutic effect of probiotic-fermented herbal blend as antibiotic alternative on salmonellosis by multi-drug resistant Salmonella pullorum. Food Biosci. (2024) 57:103585. doi: 10.1016/j.fbio.2024.103585
266. Hooda P, Malik R, Bhatia S, Al-Harrasi A, Najmi A, Zoghebi K, et al. Phytoimmunomodulators: a review of natural modulators for complex immune system. Heliyon. (2024) 10:e23790. doi: 10.1016/j.heliyon.2023.e23790
267. Abdelmotaleb MM, Elshikh HH, Abdel-Aziz MM, Elaasser MM, Yosri M. Evaluation of antibacterial efficacy and phytochemical analysis of EChinacea purpurea towards MDR strains with clinical origins. Al-Azhar Bull Sci. (2023) 34:3. doi: 10.58675/2636-3305.1643
268. Hasna B, Houari H, Koula D, Marina S, Emilia U, Assia B. In vitro and in vivo study of combined effect of some Algerian medicinal plants and probiotics against Helicobacter pylori. Microorganisms. (2023) 11:1242. doi: 10.3390/microorganisms11051242
269. Saquib SA, AlQahtani NA, Ahmad I, Arora S, Asif SM, Javali MA, et al. Synergistic antibacterial activity of herbal extracts with antibiotics on bacteria responsible for periodontitis. J Infect Dev Ctries. (2021) 15:1685–93. doi: 10.3855/jidc.14904
270. Magryś A, Olender A, Tchórzewska D. Antibacterial properties of Allium sativum L. against the most emerging multidrug-resistant bacteria and its synergy with antibiotics. Arch Microbiol. (2021) 203:2257–68. doi: 10.1007/s00203-021-02248-z
271. Ayaz M, Ullah F, Sadiq A, Ullah F, Ovais M, Ahmed J, et al. Synergistic interactions of phytochemicals with antimicrobial agents: Potential strategy to counteract drug resistance. Chem Biol Interact. (2019) 308:294–303. doi: 10.1016/j.cbi.2019.05.050
272. Cui W, Wang Y, Zhang L, Liu F, Duan G, Chen S, et al. Recent advances in the use of resveratrol against Staphylococcus aureus infections. Med Int. (2024) 4:67. doi: 10.3892/mi.2024.191
273. El-Mahdy AM, Alqahtani M, Almukainzi M, Alghoribi MF, Abdel-Rhman SH. Effect of resveratrol and curcumin on gene expression of methicillin-resistant Staphylococcus aureus (MRSA) toxins. J Microbiol Biotechnol. (2023) 34:141. doi: 10.4014/jmb.2309.09001
274. Reda F, El-Saadony M, El-Rayes T, Farahat M, Attia G, Alagawany M. Dietary effect of licorice (Glycyrrhiza glabra) on quail performance, carcass, blood metabolites and intestinal microbiota. Poult Sci. (2021) 100:101266. doi: 10.1016/j.psj.2021.101266
275. Kamkin V, Kamarova A, Shalabayev B, Kussainov A, Anuarbekov M, Abeuov S. Comparative analysis of the efficiency of medicinal plants for the treatment and prevention of COVID-19. Int J Biomaterials. (2022) 2022:5943649. doi: 10.1155/2022/5943649
276. Hanafi EM, Korany R, Tawfeek M, Hozyen HF, El-Nattat WS, Ramadan MM, et al. Effect of frankincense (Boswellia carterii) on animal reproductive performance. Egypt J Chem. (2023) 66:213–23. doi: 10.21608/ejchem.2022.138276.6093
277. Hammer KA, Carson CF, Riley TV. Effects of Melaleuca alternifolia (tea tree) essential oil and the major monoterpene component terpinen-4-ol on the development of single-and multistep antibiotic resistance and antimicrobial susceptibility. Antimicrob Agents Chemother. (2012) 56:909–15. doi: 10.1128/aac.05741-11
278. Elmoslemany AM, Elzallat M, Abd-Elfatah MH, Mohammed DM, Abd Elhady EE. Possible therapeutic effect of frankincense (Gum olibanum) and myrrh (Commiphora myrrha) resins extracts on DEN/CCL4 induced hepatocellular carcinoma in rats. Phytomedicine Plus. (2024) 4:100517. doi: 10.1016/j.phyplu.2023.100517
279. El-Saadony MT, Elsadek MF, Mohamed AS, Taha AE, Ahmed BM, Saad AM. Effects of chemical and natural additives on cucumber juice’s quality, shelf life, and safety. Foods. (2020) 9:639. doi: 10.3390/foods9050639
280. Rosado-Aguilar JA, Arjona-Cambranes K, Torres-Acosta JFJ, Rodríguez-Vivas RI, Bolio-González ME, Ortega-Pacheco A, et al. Plant products and secondary metabolites with acaricide activity against ticks. Vet Parasitol. (2017) 238:66–76. doi: 10.1016/j.vetpar.2017.03.023
281. Karunamoorthi K, Jegajeevanram K, Vijayalakshmi J, Mengistie E. Traditional medicinal plants: a source of phytotherapeutic modality in resource-constrained health care settings. J Evid Based Complementary Altern Med. (2013) 18:67–74. doi: 10.1177/2156587212460241
282. Jamshidi-Kia F, Lorigooini Z, Amini-Khoei H. Medicinal plants: Past history and future perspective. J Herbmed Pharmacol. (2017) 7:1–7. doi: 10.15171/jhp.2018.01
283. Quave CL. Antibiotics from nature: traditional medicine as a source of new solutions for combating antimicrobial resistance. AMR Control. (2016) 98:102.
284. Ekor M. The growing use of herbal medicines: issues relating to adverse reactions and challenges in monitoring safety. Front Pharmacol. (2014) 4:177. doi: 10.3389/fphar.2013.00177
285. Moloney MG. Natural products as a source for novel antibiotics. Trends Pharmacol Sci. (2016) 37:689–701. doi: 10.1016/j.tips.2016.05.001
286. Bernardini S, Tiezzi A, Laghezza Masci V, Ovidi E. Natural products for human health: an historical overview of the drug discovery approaches. Nat Prod Res. (2018) 32:1926–50. doi: 10.1080/14786419.2017.1356838
287. Nahar L, Sarker SD. Medicinal natural products—An introduction. In: Sarker SD, Nahar L, editors. Annual reports in medicinal chemistry. Amsterdam, Netherlands: Elsevier (2020). p. 1–44. doi: 10.1016/bs.armc.2020.02.008
288. Bora PS, Suresh PS, Kumari S, Anmol L, Puri S, Sharma U. Integrated approach for the quality assurance of commercially important himalayan medicinal plants. In: Ekiert HM, Ramawat KG, Arora J, editors. Medicinal plants. Sustainable development and biodiversity. Springer, Cham, Switzerland. (2021) p. 721–68. doi: 10.1007/978-3-030-74779-4_22
289. Lautié E, Russo O, Ducrot P, Boutin JA. Unraveling plant natural chemical diversity for drug discovery purposes. Front Pharmacol. (2020) 11:397. doi: 10.3389/fphar.2020.00397
290. Mussin J, Giusiano G. Ethno–phytopharmacology: product validation process based on traditional knowledge of medicinal plants. In: Chong P, Newman D, Steinmacher D, editors. Agricultural, forestry and bioindustry biotechnology and biodiscovery. Springer, Cham, Switzerland. (2020) p. 331–53. doi: 10.1007/978-3-030-51358-0_17
291. Süntar I. Importance of ethnopharmacological studies in drug discovery: role of medicinal plants. Phytochem Rev. (2020) 19:1199–209. doi: 10.1007/s11101-019-09629-9
292. Moore N, Hamza N, Berke B, Umar A. News from Tartary: an ethnopharmacological approach to drug and therapeutic discovery. Br J Clin Pharmacol. (2017) 83:33–7. doi: 10.1111/bcp.13042
293. El-Shall NA, Abd-El-Hack ME, Albaqami NM, Khafaga AF, Taha AE, Swelum AA, et al. Phytochemical control of poultry coccidiosis: a review. Poult Sci. (2022) 101:101542. doi: 10.1016/j.psj.2021.101542
294. Liu Q, Meng X, Li Y, Zhao C-N, Tang G-Y, Li H-B. Antibacterial and antifungal activities of spices. Int J Mol Sci. (2017) 18:1283. doi: 10.3390/ijms18061283
295. Qi Y, Zhang Q, Zhu H. Huang-Lian Jie-Du decoction: a review on phytochemical, pharmacological and pharmacokinetic investigations. Chin Med. (2019) 14:57. doi: 10.1186/s13020-019-0277-2
296. Kokoska L, Kloucek P, Leuner O, Novy P. Plant-derived products as antibacterial and antifungal agents in human health care. Curr Med Chem. (2019) 26:5501–41. doi: 10.2174/0929867325666180831144344
297. Dehau T, Cherlet M, Croubels S, Van Immerseel F, Goossens E. A high dose of dietary berberine improves gut wall morphology, despite an expansion of Enterobacteriaceae and a reduction in beneficial microbiota in broiler chickens. Msystems. (2023) 8:e0123922. doi: 10.1128/msystems.01239-22
298. Purwaningsih I, Maksum IP, Sumiarsa D, Sriwidodo S. A review of Fibraurea tinctoria and its component, berberine, as an antidiabetic and antioxidant. Molecules. (2023) 28:1294. doi: 10.3390/molecules28031294
299. Belwal T, Bisht A, Devkota HP, Ullah H, Khan H, Pandey A, et al. Phytopharmacology and clinical updates of Berberis species against diabetes and other metabolic diseases. Front Pharmacol. (2020) 11:41. doi: 10.3389/fphar.2020.00041
300. Mittal RP, Jaitak V. Plant-derived natural alkaloids as new antimicrobial and adjuvant agents in existing antimicrobial therapy. Curr Drug Targets. (2019) 20:1409–33. doi: 10.2174/1389450120666190618124224
301. Qassadi FI, Zhu Z, Monaghan TM. Plant-Derived products with therapeutic potential against gastrointestinal bacteria. Pathogens. (2023) 12:333. doi: 10.3390/pathogens12020333
302. Ray AS, Mandal SK, Rahaman CH. Phytochemistry, pharmacolog, and safety issues of essential oils: Applications in aromatherapy. In: Goyal MR, Suleria HAR, Ayeleso AO, Joel TJ, Panda SK, editors. The therapeutic properties of medicinal plants. New Jersey, USA: Apple Academic Press (2019). p. 35–72.
303. Li M, Zhu L, Liu B, Du L, Jia X, Han L, et al. Tea tree oil nanoemulsions for inhalation therapies of bacterial and fungal pneumonia. Colloids Surf B Biointerfaces. (2016) 141:408–16. doi: 10.1016/j.colsurfb.(2016).02.017
304. Wang C-H, Hsieh Y-H, Powers ZM, Kao C-Y. Defeating antibiotic-resistant bacteria: exploring alternative therapies for a post-antibiotic era. Int J Mol Sci. (2020) 1:1061. doi: 10.3390/ijms21031061
305. Nwobodo DC, Ugwu MC, Anie CO, Al-Ouqaili MT, Ikem JC, Chigozie UV, et al. Antibiotic resistance: The challenges and some emerging strategies for tackling a global menace. J Clin Lab Anal. (2022) 36:e24655. doi: 10.1002/jcla.24655
306. Bhatwalkar SB, Mondal R, Krishna SBN, Adam JK, Govender P, Anupam R. Antibacterial properties of organosulfur compounds of garlic (Allium sativum). Front Microbiol. (2021) 12:613077. doi: 10.3389/fmicb.2021.613077
307. El Atki Y, Aouam I, El Kamari F, Taroq A, Nayme K, Timinouni M, et al. Antibacterial activity of cinnamon essential oils and their synergistic potential with antibiotics. J Adv Pharm Technol Res. (2019) 10:63–7. doi: 10.4103/japtr.JAPTR_366_18
308. Bhatia M, Chaudhary J, Jain A, Sonam, Dhingra A, Chopra B. A recent update on ayurvedic anti-asthmatic formulations: Highlighting the role of major anti-asthmatic plants. Curr Tradit Med. (2024) 10:22–33. doi: 10.2174/2215083810666230622111859
309. Hussein RA, El-Anssary AA. Plants secondary metabolites: the key drivers of the pharmacological actions of medicinal plants. In: Builders PF, editor. Herbal medicine. IntechOpen Limited, London, United Kingdom (2019). p. 11–30. doi: 10.5772/intechopen.76139
310. Chassagne F, Samarakoon T, Porras G, Lyles JT, Dettweiler M, Marquez L, et al. A systematic review of plants with antibacterial activities: A taxonomic and phylogenetic perspective. Front Pharmacol. (2021) 11:586548. doi: 10.3389/fphar.2020.586548
311. WHO (World Health Organization). Traditional medicine has a long history of contributing to conventional medicine and continues to hold promise (2023). Available online at: https://www.who.int/news-room/feature-stories/detail/traditional-medicine-has-a-long-history-of-contributing-to-conventional-medicine-and-continues-to-hold-promise (Accessed August 10, 2023).
312. WHO (World Health Organization). Antimicrobial resistance (2023). Available online at: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (Accessed November 21, 2023).
313. Nakamoto M, Kunimura K, Suzuki JI, Kodera yl. Antimicrobial properties of hydrophobic compounds in garlic: Allicin, vinyldithiin, ajoene and diallyl polysulfides. Exp Ther Med. (2020) 19:1550–3. doi: 10.3892/etm.2019.8388
314. Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc Natl Acad Sci. (2000) 97:1433–7. doi: 10.1073/pnas.030540597
315. Song J, Zhao J, Cai X, Qin S, Chen Z, Huang X, et al. Lianhuaqingwen capsule inhibits non-lethal doses of influenza virus-induced secondary Staphylococcus aureus infection in mice. J Ethnopharmacol. (2022) 298:115653. doi: 10.1016/j.jep.2022.115653
316. Michaelis C, Grohmann E. Horizontal gene transfer of antibiotic resistance genes in biofilms. Antibiotics. (2023) 12:328. doi: 10.3390/antibiotics12020328
317. Wang Y, Batra A, Schulenburg H, Dagan T. Gene sharing among plasmids and chromosomes reveals barriers for antibiotic resistance gene transfer. Philos Trans R Soc B. (2022) 377:20200467. doi: 10.1098/rstb.2020.0467
318. Vaou N, Stavropoulou E, Voidarou C, Tsigalou C, Bezirtzoglou E. Towards advances in medicinal plant antimicrobial activity: A review study on challenges and future perspectives. Microorganisms. (2021) 9:2041. doi: 10.3390/microorganisms9102041
319. Williams G, Stothart CI, Hahn D, Stephens JH, Craig JC, Hodson EM. Cranberries for preventing urinary tract infections. Cochrane Database Syst Rev. (2023) 4:CD001321. doi: 10.1002/14651858.CD001321.pub7
320. Suganya T, Packiavathy IASV, Aseervatham GSB, Carmona A, Rashmi V, Mariappan S, et al. Tackling multiple-drug-resistant bacteria with conventional and complex phytochemicals. Front Cell Infect Microbiol. (2022) 12:883839. doi: 10.3389/fcimb.2022.883839
321. Vaou N, Stavropoulou E, Voidarou C, Tsakris Z, Rozos G, Tsigalou C, et al. Interactions between medical plant-derived bioactive compounds: focus on antimicrobial combination effects. Antibiotics. (2022) 11:1014. doi: 10.3390/antibiotics11081014
322. Khaleghian M, Sahrayi H, Hafezi Y, Mirshafeeyan M, Moghaddam ZS, Farasati Far B, et al. In silico design and mechanistic study of niosome-encapsulated curcumin against multidrug-resistant Staphylococcus aureus biofilms. Front Microbiol. (2023) 14:1277533. doi: 10.3389/fmicb.2023.1277533
323. Sari NKY, Permatasari AAAP, Wahyuningsih SPA, Geraldi A, Wiradana PA, Widhiantara IG, et al. Mechanism of antimicrobial resistance and red ginger as the solution for source of natural antioxidant: A brief review. Indones J Pharm. (2023) 34:1–23. doi: 10.22146/ijp.5307
324. Ashrit P, Sadanandan B, Shetty K, Vaniyamparambath V. Polymicrobial biofilm dynamics of multidrug-resistant Candida albicans and ampicillin-resistant Escherichia coli and antimicrobial inhibition by aqueous garlic extract. Antibiotics. (2022) 11:573. doi: 10.3390/antibiotics11050573
325. Omar MM, Hosseny EN, Handak EM, al-Sarraj F, El-Hejin AM. Detection of (CTX-M) resistance genes in extended spectrum beta-lactamases bacterial isolates of Escherichia coli and Klebsiella pneumonia and the antibacterial effect of ethanolic extract of neem plant (Azadirachta indica) against these bacteria. Egypt J Chem. (2024) 67:337–45. doi: 10.21608/ejchem.2023.223851.8284
326. Taghiloo S, Ghajari G, Zand Z, Kabiri-Samani S, Kabiri H, Rajaei N, et al. Designing alginate/chitosan nanoparticles containing Echinacea angustifolia: a novel candidate for combating multidrug-resistant Staphylococcus aureus. Chem Biodiversity. (2023) 20:e202201008. doi: 10.1002/cbdv.202201008
327. Cai D, Ye Y, Song J, Liu S, Zeng X, Zhu B, et al. A chitosan-modified tea tree oil self-nanoemulsion improves antibacterial effects in vivo and in vitro and promotes wound healing by influencing metabolic processes against multidrug-resistant bacterial infections. Int J Biol Macromol. (2024) 281:136404. doi: 10.1016/j.ijbiomac.2024.136404
328. Li Y, Ge X. Role of berberine as a potential efflux pump inhibitor against MdfA from Escherichia coli: in vitro and in silico studies. Microbiol Spectr. (2023) 11:e03324–e03322. doi: 10.1128/spectrum.03324-22
329. Wang H, Nie C, Luo M, Bai Q, Yao Z, Lv H, et al. Novel GSH-responsive prodrugs derived from indole-chalcone and camptothecin trigger apoptosis and autophagy in colon cancer. Bioorg Chem. (2024) 143:107056. doi: 10.1016/j.bioorg.2023.107056
330. Liu Z, Li Y, Wei D, Wang J, Qiao C, G-y Li, et al. Biosensor-enabled discovery of CaERG6 inhibitors and their antifungal mode of action against Candida albicans. ACS Infect Dis. (2023) 9:785–800. doi: 10.1021/acsinfecdis.2c00490
331. Kiran S, Tariq A, Iqbal S, Naseem Z, Siddique W, Jabeen S, et al. Punicalagin, a pomegranate polyphenol sensitizes the activity of antibiotics against three MDR pathogens of the Enterobacteriaceae. BMC Complement Med Ther. (2024) 24:93. doi: 10.1186/s12906-024-04376-7
332. Elhaj YHA. A laboratory evaluation of antibacterial activity of Commiphora myrrha gum resins from Kingdom of Saudi Arabia 2023. Egypt Acad J Biol Sci C Physiol Mol Biol. (2023) 15:675–81. doi: 10.21608/eajbsc.2023.326666
333. Tian Y, Wang Z, Xu X, Guo Y, Ma Y, Lu Y, et al. Natural alkaloids from Dicranostigma leptopodum (Maxim.) fedde and their G5. NHAc-PBA dendrimer-alkaloid complexes for targeting chemotherapy in breast cancer MCF-7 cells. Nat Prod Res. (2024) 8:1–18. doi: 10.1080/14786419.2024.2335669
334. Sherif MM, Elshikh HH, Abdel-Aziz MM, Elaasser MM, Yosri M. In vitro antibacterial and phytochemical screening of Hypericum perforatum extract as potential antimicrobial agents against multi-drug-resistant (MDR) strains of clinical origin. BioMed Res Int. (2023) 2023:6934398. doi: 10.1155/2023/6934398
335. Magtalas TJ, Anapi GR. Evaluation of bacteriostatic effect of rosemary and oregano essential oils against a non-pathogenic surrogate of Salmonella spp. (E. coli ATCC 9637). Biol Life Sci Forum. (2025) 40:7. doi: 10.3390/blsf2024040007
336. Farhadi K, Rajabi E, Varpaei HA, Iranzadasl M, Khodaparast S, Salehi M. Thymol and carvacrol against Klebsiella: anti-bacterial, anti-biofilm, and synergistic activities—a systematic review. Front Pharmacol. (2024) 15:1487083. doi: 10.1155/2023/6934398
337. Abdelgader LMA, Yousif EA, Eltaib HA, Kwami WS, Mahjaf GM, Altaher TA, et al. In vitro antibacterial activity of Aloe Barbadensis Millar (Aloe vera) against Pseudomonas aeruginosa isolates in Khartoum State, Sudan. Middle East Res J Microbiol Biotechnol. (2024) 04:1–5. doi: 10.36348/merjmb.2024.v04i03.001
338. Oliveira GDS, McManus C, Sousa HAF, Santos PHGS, Dos Santos VM. A mini-review of the main effects of essential oils from Citrus aurantifolia, Ocimum basilicum, and Allium sativum as safe antimicrobial activity in poultry. Animals. (2024) 14:382. doi: 10.3390/ani14030382
339. Al-Tawalbeh DM, Alawneh JM, Momani W, Mayyas A. Comparative antibacterial activity of clove extract against Pseudomonas aeruginosa. BMC Complement Med Ther. (2025) 25:7. doi: 10.1186/s12906-024-04740-7
340. Kafa AHT, Aslan R, Celik C, Hasbek M. Antimicrobial synergism and antibiofilm activities of Pelargonium graveolens, Rosemary officinalis, and Mentha piperita essential oils against extreme drug-resistant Acinetobacter baumannii clinical isolates. Z für Naturforschung C. (2022) 77:95–104. doi: 10.1515/znc-2021-0079
341. Abd-El-Hack ME, El-Shall NA, El-Kasrawy NI, El-Saadony MT, Shafi ME, Zabermawi NM, et al. The use of black pepper (Piper guineense) as an ecofriendly antimicrobial agent to fight foodborne microorganisms. Environ Sci pollut Res. (2022) 29:10894–907. doi: 10.1007/s11356-021-17806-7
342. Egamberdieva D, Mamadalieva N, Khodjimatov O, Tiezzi A. Medicinal plants from Chatkal Biosphere Reserve used for folk medicine in Uzbekistan. Med Aromat Plant Sci Biotechnol. (2013) 7:56–64.
343. Yili A, Aisa H, Maksimov V, Veshkurova O, Salikhov S. Chemical composition and antimicrobial activity of essential oil from seeds of Anethum graveolens growing in Uzbekistan. J Chem Nat Compd. (2009) 45:280–1. doi: 10.1007/s10600-009-9275-4
344. Egamberdieva D, Wirth S, Behrendt U, Ahmad P, Berg G. Antimicrobial activity of medicinal plants correlates with the proportion of antagonistic endophytes. Front Microbiol. (2017) 8:199. doi: 10.3389/fmicb.2017.00199
345. Climati E, Mastrogiovanni F, Valeri M, Salvini L, Bonechi C, Mamadalieva NZ, et al. Methyl carnosate, an antibacterial diterpene isolated from Salvia officinalis leaves. Nat Prod Commun. (2013) 8:1934578X1300800402. doi: 10.1177/1934578X1300800402
346. Mamadalieva NZ, Youssef FS, Ashour ML, Sasmakov SA, Tiezzi A, Azimova SS. Chemical composition, antimicrobial and antioxidant activities of the essential oils of three Uzbek Lamiaceae species. Nat Prod Res. (2019) 33:2394–7. doi: 10.1080/14786419.2018.1443088
347. Salunke CA, Satpute RA. Phytochemical screening and in vitro antimicrobial activity of extracts from tubers of wild dioscorea species. J Root Crops. (2018) 44:61–5.
348. Curini M, Epifano F, Genovese S, Tammaro F, Menghini L. Composition and antimicrobial activity of the essential oil of Artemisia dracunculus “piemontese” from Italy. Chem Nat Compd. (2006) 42:738–9. doi: 10.1007/s10600-006-0268-2
349. Norouzi-Arasi H, Yavari I, Chalabian F, Kiarostami V, Ghaffarzadeh F, Nasirian A. Chemical constituents and antimicrobial activities of the essential oil of Acroptilon repens (L.) DC. Flavour Fragr J. (2006) 21:247–9. doi: 10.1002/ffj.1568
350. Iacobellis NS, Lo Cantore P, Capasso F, Senatore F. Antibacterial activity of Cuminum cyminum L. Carum carvi L. essential oils. J Agric Food Chem. (2005) 53:57–61. doi: 10.1021/jf0487351
351. Li CL, Ou CM, Huang CC, Wu WC, Chen YP, Lin TE, et al. Carbon dots prepared from ginger exhibiting efficient inhibition of human hepatocellular carcinoma cells. J Mater Chem B. (2014) 2:45644571. doi: 10.1039/C4TB00216D
352. Cheng J, Zhang M, Sun Z, Lu F, Xiong W, Luo J, et al. Hemostatic and hepatoprotective bioactivity of junci medulla carbonisata-derived carbon dots. Nanomedicine. (2019) 14:431446. doi: 10.2217/nnm-2018-0285
353. Zhao Y, Zhang Y, Kong H, Zhang M, Cheng J, Wu J, et al. Carbon dots from Paeoniae Radix Alba carbonisata: hepatoprotective effect. Int J Nanomedicine. (2020) 15:90499059. doi: 10.2147/IJN.S281976
354. Mamadalieva NZ. Phytoecdysteroids from Silene plants: distribution, diversity and biological (antitumour, antibacterial and antioxidant) activities. Bol Latinoam Caribe Plant Med Aromat. (2012) 11:47497.
355. Wu J, Zhang M, Cheng J, Zhang Y, Luo J, Liu Y, et al. Effect of Lonicerae japonicae flos carbonisata-derived carbon dots on rat models of fever and hypothermia induced by lipopolysaccharide. Int J Nanomedicine. (2020) 15:4139. doi: 10.2147/IJN.S248467
356. Wang X, Zhang Y, Zhang M, Kong H, Wang S, Cheng J, et al. Novel carbon dots derived from Puerariae Lobatae Radix and their anti-gout effects. Molecules. (2019) 24:4152. doi: 10.3390/molecules24224152
357. Wang S, Zhang Y, Kong H, Zhang M, Cheng J, Wang X, et al. Antihyperuricemic and anti-gouty arthritis activities of Aurantii fructus immaturus carbonisata-derived carbon dots. Nanomedicine. (2019) 14:2925–39. doi: 10.2217/nnm-2019-0255
358. Zhumakanova BS, Korona-Głowniak I, Skalicka-Woźniak K, Ludwiczuk A, Baj T, Wojtanowski KK, et al. Phytochemical fingerprinting and in vitro antimicrobial and antioxidant activity of the aerial parts of Thymus marschallianus Willd. and Thymus seravschanicus Klokov growing widely in southern Kazakhstan. Molecules. (2021) 26:3193. doi: 10.3390/molecules26113193
359. Pirvu L, Nicorescu I, Hlevca C, Albu B, Nicorescu V. Burdock (Arctium lappa) leaf extracts increase the in vitro antimicrobial efficacy of common antibiotics on gram-positive and gram-negative bacteria. Open Chem. (2017) 15:92102. doi: 10.1515/chem-2017-0012
360. Nuraliyev YN. Hypocholesterinemic and hypolipidemic effects of ether oil of Origanum tyttanthum Gontsck. Khimiko-Farm Zh. (1994) 28:63–64.
361. Zhang M, Cheng J, Hu J, Luo J, Zhang Y, Lu F, et al. Green Phellodendri Chinensis cortex-based carbon dots for ameliorating imiquimod-induced psoriasis-like inflammation in mice. J Nanobiotechnol. (2021) 19:114. doi: 10.1186/s12951-021-00847-y
362. Qasem AM, Zeng Z, Rowan MG, Blagbrough IS. Norditerpenoid alkaloids from Aconitum and Delphinium: structural relevance in medicine, toxicology, and metabolism. Nat Prod Rep. (2022) 39:460473. doi: 10.1039/D1NP00029B
363. Kwon DH, Kwon HY, Kim HJ, Chang EJ, Kim MB, Yoon SK, et al. Inhibition of hepatitis B virus by an aqueous extract of Agrimonia eupatoria L. Phytother Res. (2005) 19:3558. doi: 10.1002/ptr.1689
364. Wang X, Yu N, Wang Z, Qiu T, Jiang L, Zhu X, et al. Akebia trifoliata pericarp extract ameliorates inflammation through NF-κB/MAPK signaling pathways and modifies gut microbiota. Food Funct. (2019) 11:468296. doi: 10.1039/C9FO02917F
365. Şahin F, Güllüce M, Daferera D, Sökmen A, Sökmen M, Polissiou M, et al. Biological activities of the essential oils and methanol extract of Origanum vulgare ssp. vulgare in the Eastern Anatolia region Turkey. Food Control. (2004) 15:549557. doi: 10.1016/j.foodcont.2003.08.009
366. Qais FA, Ahmad I, Husain FM, Alomar SY, Ahmad N, Albalawi F, et al. Interference of quorum sensing regulated bacterial virulence factors and biofilms by Plumbago zeylanica extract. Microsc Res Tech. (2021) 84:3150–60. doi: 10.1002/jemt.23872
367. Kianfé B, Kühlborn J, Tchuenguem R, Tchegnitegni B, Ponou B, Groß J, et al. Antimicrobial secondary metabolites from the medicinal plant Crinum glaucum A. Chev. (Amaryllidaceae). S Afr J Bot. (2020) 133:161–6. doi: 10.1016/j.sajb.2020.07.026
368. Qader MK, Khalid NS, Abdullah AM. Antibacterial activity of some plant extracts against clinical pathogens. Int J Microbiol Immunol Res. (2013) 1:53–6.
369. Sapkota R, Dasgupta R, Rawat D, Nancy N, Rawat DS. Antibacterial effects of plants extracts on human microbial pathogens and microbial limit tests. Int J Res Pharm Chem. (2012) 2:926–36.
Keywords: antibiotics, antimicrobial agents, herbal medicine, human health, mechanism of action, multidrug resistance, pathogens
Citation: El-Saadony MT, Saad AM, Mohammed DM, Korma SA, Alshahrani MY, Ahmed AE, Ibrahim EH, Salem HM, Alkafaas SS, Saif AM, Elkafas SS, Fahmy MA, Abd El-Mageed TA, Abady MM, Assal HY, El-Tarabily MK, Mathew BT, AbuQamar SF, El-Tarabily KA and Ibrahim SA (2025) Medicinal plants: bioactive compounds, biological activities, combating multidrug-resistant microorganisms, and human health benefits - a comprehensive review. Front. Immunol. 16:1491777. doi: 10.3389/fimmu.2025.1491777
Received: 05 September 2024; Accepted: 13 March 2025;
Published: 28 April 2025.
Edited by:
Nemat Ali, King Saud University, Saudi ArabiaReviewed by:
Hirak Jyoti Chakraborty, ICAR-CIFRI, Kolkata, IndiaPir Tariq Shah, Dalian University of Technology, China
Copyright © 2025 El-Saadony, Saad, Mohammed, Korma, Alshahrani, Ahmed, Ibrahim, Salem, Alkafaas, Saif, Elkafas, Fahmy, Abd El-Mageed, Abady, Assal, El-Tarabily, Mathew, AbuQamar, El-Tarabily and Ibrahim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Synan F. AbuQamar, c2FidXFhbWFyQHVhZXUuYWMuYWU=; Khaled A. El-Tarabily, a3RhcmFiaWx5QHVhZXUuYWMuYWU=
 Mohamed T. El-Saadony
Mohamed T. El-Saadony Ahmed M. Saad2
Ahmed M. Saad2 Dina Mostafa Mohammed
Dina Mostafa Mohammed Sameh A. Korma
Sameh A. Korma Heba M. Salem
Heba M. Salem Mohamed A. Fahmy
Mohamed A. Fahmy Taia A. Abd El-Mageed
Taia A. Abd El-Mageed Synan F. AbuQamar
Synan F. AbuQamar Khaled A. El-Tarabily
Khaled A. El-Tarabily