- 1Gynecology Department of Traditional Chinese Medicine, The First Affiliated Hospital of Naval Medical University, Shanghai, China
- 2Department of Reproductive Medicine, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine, Shanghai, China
Recent studies have identified that RNA epigenetic modifications, including m6A, m1A, m5C, etc, play pivotal roles in tumor progression. These modifications influence mRNA stability, RNA processing, translational efficiency, and decoding precision. However, comprehensive reviews detailing the connection between m6A RNA modifications and hormone-dependent cancers in both male and female populations remain scarce(breast cancer, ovarian cancer, and endometrial cancer, prostate cancer). In this article, we explore the cellular and molecular roles of various RNA modifications alongside the key elements of the tumor microenvironment. We examine how these RNA modifications influence the development of hormone-dependent cancers through their impact on immune mechanisms. By enhancing our understanding of the function of RNA modifications within the immune systems of four specific tumors, we offer fresh insights for their potential applications in diagnosis and treatment.
1 Introduction
Tumor development is influenced by multiple factors, among which a subset closely associated with hormones, known as hormone-dependent cancers, among the most typical ones are breast cancer, ovarian cancer, endometrial cancer, and prostate cancer (BC, OC, EC, and PC). Their development and treatment are intrinsically linked to hormones, mainly including progesterone receptor (PR), estrogen receptor (ER), androgen receptor (AR) and human epidermal growth factor receptor 2 (HER2). In women, BC is the first malignant tumor with the highest incidence rates and is also the most typical hormone-dependent intermediate utilized (1). In addition, complex molecular bidirectional interactions between hormone receptors (HRs), including ER, PR, and HER2 are present in BC (2, 3). Endocrine therapy (aromatase inhibitors and anti-estrogen therapy or anti-estrogen therapy alone) is the standardized method and is the backbone of adjuvant therapy that significantly reduces the risk of recurrence and mortality (4). Endometrial cancer, on the other hand, is the female malignant tumor with the fastest growing mortality rate, which tends to show elevated ERα levels and promote PR expression (1, 5, 6). Patients are positive for both ERα levels and PR expression tend to have well-differentiated tumors and may be responsive to hormone therapy, resulting in a relatively good prognosis (7). In addition, ovarian cancer is an essential branch of female malignant tumors, and postoperative hormone replacement therapy is necessary to achieve a better quality of living for patients (8). Similar to women, the most prevalent malignant tumor for men, prostate cancer, is significantly affected by androgens. Hence, the basis of prostate cancer treatment is anti-androgen therapy (9, 10). However, their mechanisms of occurrence remain unspecified, and treatment outcomes remain unsatisfactory. This review emphasizes the importance of discovering alternative and targetable molecular pathways that could provide novel therapeutic opportunities.
RNA modifications refer to chemical alterations of RNA nucleobases or ribose molecules. Presently, over 150 distinct modifications have been documented. Pseudouridine Ψ was discovered in the 1950s as the first recognized RNA modification (11). Among the most prevalent mRNA modifications, N6-methyladenosine (m6A) was identified in 2011 (12–14). Other RNA modifications, such as m1A and m5C, have been identified and extensively studied in recent decades (15, 16). RNA methylation has an impact on almost the entire mRNA life cycle - starting from mRNA transcription, mRNA splicing, specific structure, stability and subsequent translation and finally degradation (17–21). Although much of the research has focused on the role of m6A in hormone-dependent cancers, this review also examines the impact of other RNA modifications, such as m1A, m5C, m7G, mcm5s2U, A-to-I, and Ψ. These modifications can be investigated using emerging techniques like RNA immunoprecipitation (RIP), chromatin immunoprecipitation (ChIP), and single-cell omics (22–25).
The role of the immune system in cancer development has attracted increasing attention, particularly concerning the complex immune components within the tumor microenvironment and the adaptable mechanisms of immune evasion. Consequently, immunotherapy has emerged as a novel approach in cancer treatment, aimed at remodeling the immune system and reactivate anti-tumor immune responses to avoid tumor escape (26). Various immunotherapeutic strategies have shown substantial promise in treating a wide range of cancers, predominantly involving immune checkpoint inhibitors like PD1, PD-L1, and CTLA4, antibody-drug couplings, and cancer vaccines (27). However, fewer immunotherapeutic agents have been approved for clinical use in hormone-dependent cancers (28–31). As a distinct regulatory mechanism, RNA modifications, exemplified by m6A, has garnered increasing attention. Tumor-derived intrinsic signals and environmental stimuli can drive aberrant expression and activity-modifying regulators of many RNAs, leading to abnormal RNA modifications, which are essential for shaping the tumor microenvironment and immune escape (32, 33).
Although a substantial body of literature has accumulated on RNA modifications in cancer, the research focus has predominantly centered on m6A modification and its associated enzymatic machinery, with relatively limited exploration of other RNA modification types and a notable paucity of systematic reviews. While tumor immunology remains a prominent research frontier, investigations that integrate RNA modifications with tumor immunity to elucidate their epigenetic regulatory mechanisms remain relatively scarce. Notably, for hormone-dependent tumors, there is a conspicuous lack of comprehensive discussion regarding the potential shared immune regulatory mechanisms and epigenetic modification patterns that may arise from their similar endocrine microenvironment (34). This paper will focus on how RNA modifications play an immunomodulatory role in hormone-dependent cancers, including breast, ovarian, endometrial and prostate cancers (BC, OC, EC, and PC). The mechanisms and implications of prevalent RNA modifications will be explored. Specifically, we aim to elucidate the effects of RNA modifications in diverse immune cell types within hormone-dependent cancers.
2 RNA modification in sex hormone synthesis
2.1 Concepts of different RNA modifications
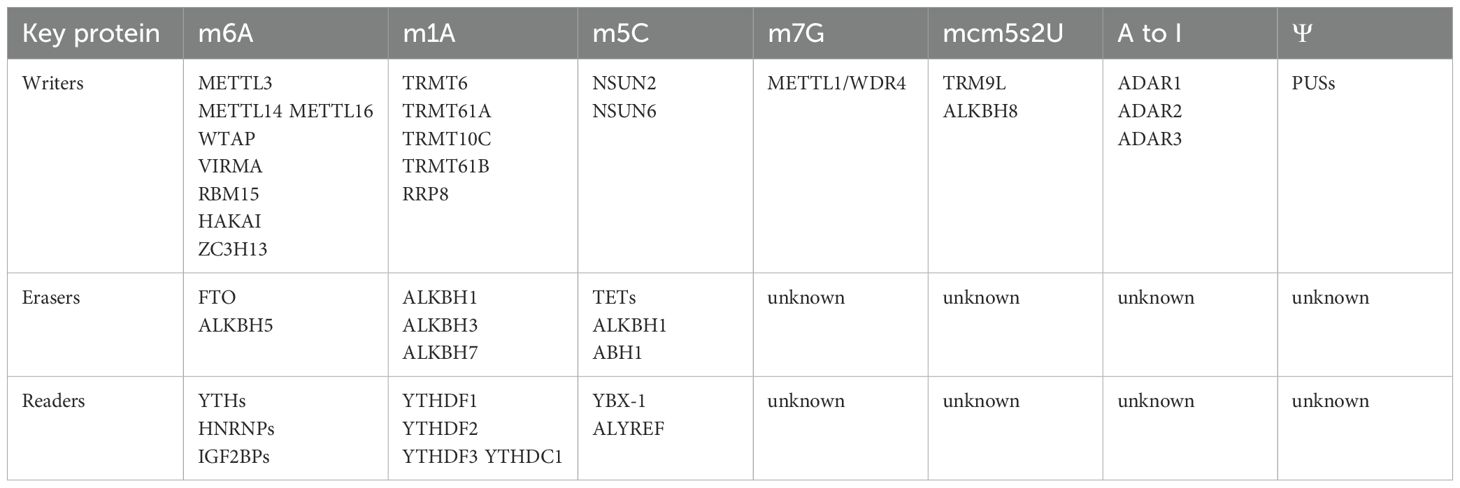
2.1.1 m6A
N6-methyladenosine (m6A) is defined as the methylation of adenine at the N6 position within RNA molecules. It represents the most prevalent modification in eukaryotic lncRNAs and mRNAs, and has also been detected in rRNAs, snRNAs, and tRNAs (Figure 1, 35). The m6A modification is conserved across yeast, mouse, and human mRNA, and is enriched in the RRACH (R = G or A; H = A, C, or U) consensus sequence (36–38). The m6A modification is primarily facilitated by the methyltransferase termed the “writer”, the demethylase known as the “eraser”, and recognition proteins referred to as “reader”. The writer assembles the m6A methyltransferase complex (MTC), which catalyzes site-specific methylation that can be reversed by the eraser. The reader proteins bind to methylated m6A sites and transmit downstream signals, thereby acting as post-transcriptional gene regulators. M6A writers mainly include METTL3, nephroblastoma 1-associated protein (WTAP), KIAA1429 (VIRMA), RBM15, METTL16, METTL14, HAKAI, and ZC3H13 (KIAA0853) (39). To date, only two types of erasers, FTO and ALKBH5, have been recognized (40). The readers involve the YTH family, the HNRNP family, and the IGF2BPs family (39). As an important component of epigenetics, m6A modification and these regulatory proteins are involved in various biological activities in which they play a regulatory role (40–43).
As the predominant “writer,” METTL3 serves as the core catalytic component of MTC, yet it remains inactive without METTL14. Although METTL14 lacks intrinsic methyltransferase activity due to the absence of an S-adenosylmethionine (SAM) binding domain, it aids in substrate RNA recognition and forms methyltransferase structural domains (MTDs) with METTL3 as a heterodimer (44). The MTD structural domains in isolation do not possess methyltransferase activity and necessitate the zinc-finger domain (ZFD) of METTL3 to become enzymatically active (45, 46). Wilms tumor 1-associating protein (WTAP), though not enzymatically active, assists in mRNA methylation by interacting with and recruiting the METTL3-METTL14 complex to target mRNA sites (47, 48). Further studies have identified VIRMA (49), ZC3H13 (50), RBM15/15B (51), and HAKAI (52) as additional cofactors of the METTL3-METTL14 complex. Besides METTL3, three distinct enzymes—METTL16, METTL5, and ZCCHC4—have been recognized as eukaryotic m6A methyltransferases, each responsible for incorporating m6A into U6 small nuclear RNAs (snRNAs) (53), 18S ribosomal RNAs (rRNAs) (54), and 28S rRNAs (55), respectively.
FTO and ALKBH5, known as m6A erasers, act as demethylases that catalyze the conversion of m6A to adenosine. FTO was the first m6A eraser identified, exhibiting specific oxidative demethylation activity against abundant m6A residues on RNA (13), while ALKBH5 was the second eraser discovered (40). FTO demethylates internal m6A residues on mRNAs and U6 RNA, as well as N6,2-O-dimethyladenosine (m6Am) on mRNAs and snRNAs, N1-methyladenosine (m1A) on tRNAs, 3-methylthymine (m3T) on single-stranded DNA (ssDNA), and 3-methyluracil (m3U) on single-stranded RNA (ssRNA) (56, 57). ALKBH5 localizes within nuclear speckles and aids in the assembly of mRNA processing factors, primarily acting on substrates like nuclear nascent RNAs (ssRNAs) (40).
The primary readers of m6A are the YTH family proteins, including YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2, which contain an m6A-binding pocket within their YTH structural domains (58).YTHDF2, the first to be identified, promotes the degradation of cytoplasmic targets by recruiting CCR4-NOT complexes, with its m6A-binding affinity significantly enhanced by SUMOylation (19, 59). Additionally, YTHDF1 and YTHDF3, both cytoplasmic m6A readers, enhance the translation efficiency of target mRNAs and, in some instances, promote their degradation (19, 60, 61). As for nuclear m6A readers, YTHDC1 regulates mRNA fate through multiple mechanisms, such as mRNA splicing (17), nuclear body formation (62), and retrotransposon silencing (63). YTHDC2,localized in both the nucleus and cytoplasm (64), modulates deconjugase activity and influences mRNA decay and translation during spermatogenesis (65–67). Apart from the YTH family, IGF2BPs constitute a distinct group of m6A readers, recognizing m6A through their KH structural domains (20). The IGF2BP proteins (IGF2BP1/2/3) share similar structures, and their binding affinities for various target RNAs may be governed by their KH3 and KH4 structural domains (68).
2.1.2 m1A
The m1A modification, similar to m6A, entails the methylation of the first nitrogen on adenosine and is governed by specialized writers, erasers, and readers. This modification predominantly occurs in tRNAs and rRNAs, particularly within GC-rich RNA sequences, impacting ribosomal tertiary structure, RNA stability, and translation efficiency (Figure 1, 69–71). tRNA methyltransferases 6 and 61A (TRMT6/61A) form a complex that exerts MTC-like effects by catalyzing the addition of m1A to t-loop-like RNA structures (72). tRNA methyltransferases 10C and 61B (TRMT10C/61B) respectively catalyze the m1A modification at positions 9 and 58 in mitochondrial tRNAs (73). Moreover, TRMT61B has a similar recognition mechanism for rRNA and tRNA (74). Additionally, NML, also known as RRP8, localizes to the nucleus where it methylates m1A on 28S rRNAs (75). AlkB homologs 1, 3 and 7 act as erasers in charge of m1A demethylation (76–78). YTHDF1, YTHDF2, YTHDF3, and YTHDC1 act as m1A-modified readers to fulfill their biological roles (79).
2.1.3 m5C
m5C is a methylation modification at the 5th carbon atom of cytosine, found in mRNA and lncRNA, and enriched in cytoplasmic and mitochondrial rRNAs and tRNAs (Figure 1, 80, 81). To date, NSUN2 and NSUN6 are the only known m5C methyltransferases within the NSUN family that facilitate mRNA methylation by incorporating m5C, functioning as transcriptional modifiers. NSUN2 regulates the nucleoplasmic transport and RNA-binding affinity of the mRNA export adapter protein ALYREF, which specifically recognizes m5C, thereby influencing mRNA export (81). NSUN6 predominantly targets the 3 untranslated regions (3 UTRs) on the hairpin-like structural loops conserved sequence motif CTCCA, potentially participating in the quality control of translation termination fidelity (82). Two mechanisms for m5C “erasure” have been identified: first, oxidation by the TET family on RNA to produce 5-hydroxymethylcytosine (hm5C); and second, the conversion of 5-formylcytosine (f5C) in mitochondrial tRNA by α-ketoglutarate and iron(II)-dependent dioxygenases ALKBH1 and ABH1 (83–85). BX-1 and the Aly-REF export factor (ALYREF) act as m5C readers, affecting the stability, translation, and transcription of the RNAs they target (81, 86).
2.1.4 m7G
The m7G modification involves the methylation of the 7th nitrogen atom in guanosine and primarily occurs at internal sites of rRNAs, tRNAs, miRNAs, as well as the 5 cap of mRNAs (Figure 1, 87, 88). Although no confirmed erasers or readers have been introduced for m7G yet, the m7G cap can undergo hypermethylation by trimethylguanosine synthase 1 (TGS1) to produce m2,2,7G or may be recognized by the eukaryotic translation initiation factor eIF4E, subsequently affecting RNA maturation, nuclear export, and translation (89, 90).
2.1.5 mcm5s2U
The 5-methoxycarbonylmethyl-2-thiourea modification (mcm⁵s²U), initiated by cm⁵U and mcm⁵U modifications on wobble uridines, is facilitated by human tRNA methyltransferase 9-like protein (TRM9L) and AlkB homologue 8 (ALKBH8) (Figure 1, 91). Wobble uridines, located at the first nucleotide position of the anticodon stem loop in tRNA, are essential for accurate mRNA translation and efficient protein synthesis (92, 93).
2.1.6 A-to-I modification
The A-to-I modification involves the selective hydrolytic deamination of adenosine to inosine (A-to-I editing), a process primarily regulated by the family of double-stranded RNA-specific adenosine deaminases, notably ADAR1, ADAR2, and ADAR3 (Figure 1, 94). ADAR1 and ADAR2 both mediate A-to-I editing in cellular RNA (95).
2.1.7 Pseudouridylation Ψ
The C5-glycosidic isoform of uridine, pseudouridine Ψ, is the most prevalent RNA modification, primarily found in tRNAs and rRNAs (Figure 1, 70, 96). Pseudouridylation occurs via two distinct pathways: RNA-independent pseudouridylation, catalyzed by pseudouridine synthases (PUSs) without a template strand, and RNA-dependent pseudouridylation, which requires the box H/ACA small nuclear ribonucleoprotein RNA-protein complex (82, 97).
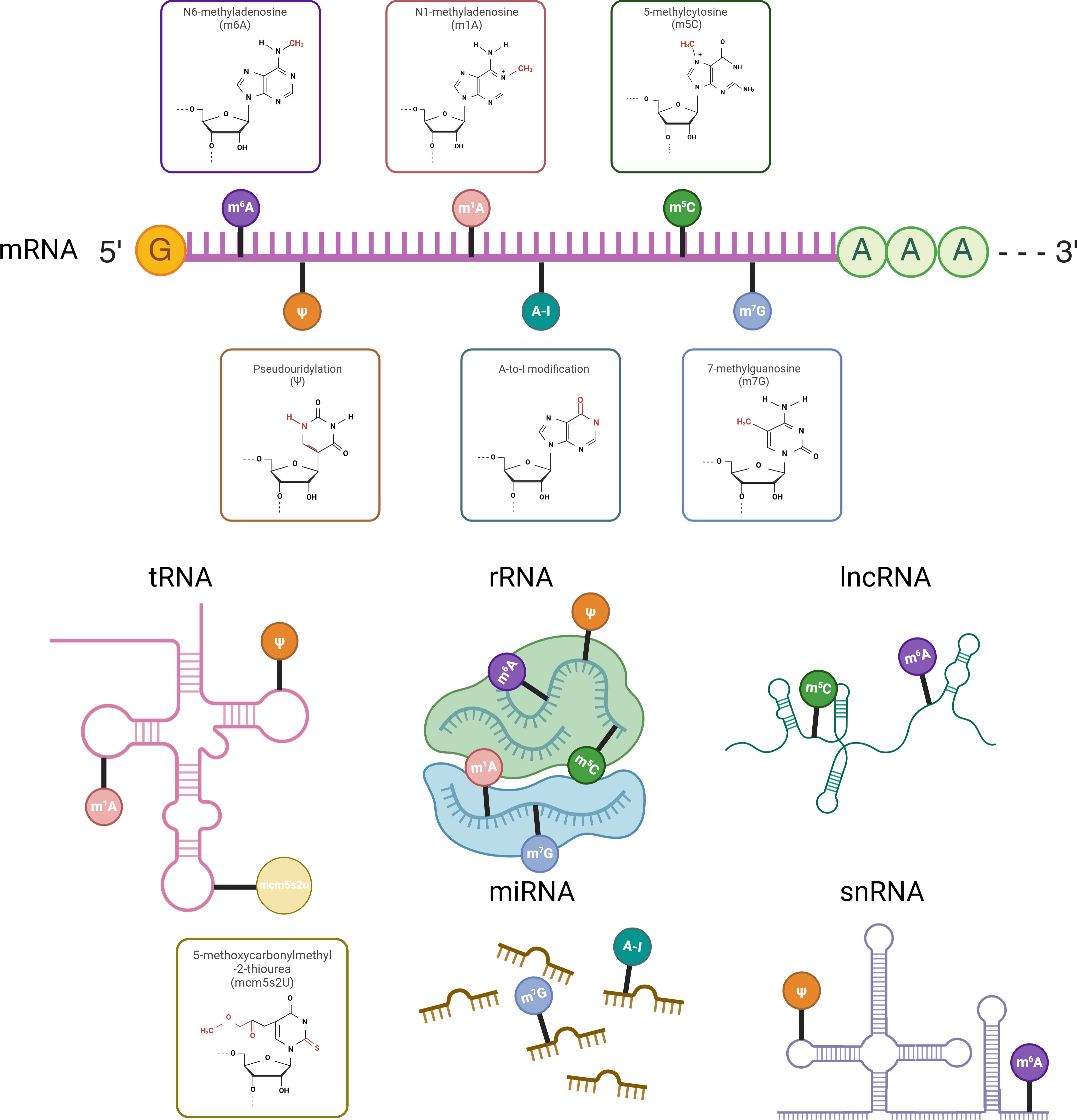
Figure 1. RNA modifications across different RNA types with chemical structures highlighted on the ribose moiety. Different RNA types undergo distinct chemical modifications that influence their stability, processing, and function. mRNA is primarily modified by m⁶A, m¹A, m⁵C, m⁷G, A-to-I, and Ψ, with m⁶A being the most prevalent. tRNA mainly carries m¹A, Ψ, and mcm⁵s²U, while rRNA undergoes m⁶A, m¹A, m⁵C, m⁷G, and Ψ modifications. lncRNA is modified by m⁶A and m⁵C, snRNA by m⁶A and Ψ, and miRNA by m⁷G and A-to-I. The chemical structures of these modifications are marked on the ribose moiety.
2.1.8 Crosstalk between different RNA modification
Multiple RNA modifications are not isolated; rather, they are often interlinked, collaboratively regulating physiological and pathological states in the body. The vast majority of RNA modifications share similar regulatory mechanism, especially writers, allowing different modifications to be controlled by the same class of writers (Table 1), thus forming an interconnected regulatory network. Writers regulating m6A, m1A, and A-to-I modifications are not independent but exhibit significant cross-linking, which is closely associated with colorectal carcinogenesis, the tumor microenvironment (TME), drug sensitivity, and immunotherapy (98). Multiple RNA modifications can act on the same signaling pathway or target RNA to exert either synergistic or antagonistic effects. For example, m6A and m5C both modify FOXC2 mRNA, promoting gastric cancer cell growth (99, 100). Both m6A and A-to-I editing can alter c-MYC mRNA, contributing to the progression of hepatocellular carcinoma (101, 102). Similarly, m6A and Ψ modifications within the RAS pathway have been recognized for their oncogenic impact in colorectal cancer (103, 104). In pancreatic cancer, m6A stimulates the PI3K/Akt/mTOR pathway, driving cancer cell proliferation (105, 106), whereas m1A and m5C are linked with activation of the mTOR pathway and unfavorable prognoses (107, 108). Additionally, interactions between different RNA modifications have been observed. In breast cancer, METTL3-mediated m6A modification is regulated by ADAR1, which subsequently promotes breast cancer progression (109). Interactions among m6A, m5C, m1A, and m7G are also vital for TME regulation, immune infiltration, and immunotherapy in soft tissue sarcoma (STS) (110). The human body is a complex system, where crosstalk among multiple RNA modifications plays an essential role in disease development. Expanding research on RNA modification interactions holds significant clinical promise.
2.2 RNA modification with hormone receptors
Sex hormones in the human body, primarily estrogen, progesterone, and androgens, are regulated by gonadotropins, including gonadotropin-releasing hormone (GnRH), luteinizing hormone (LH), and follicle-stimulating hormone (FSH). Additionally, prolactin (PRL) can reflect the secretion levels of sex hormones in the body. RNA modifications specifically regulate the synthesis, secretion, and ligand-receptor interactions of sex hormones in organisms, thereby influencing physiological and pathological processes.
As the most common m6A methyltransferase, the specific knockdown of METTL3 can alter various biological processes, with diverse and sometimes opposing effects in different cells and molecules. In testicular mesenchymal cells, ambient PM2.5 promotes METTL3-induced m6A modification of SIRT1 mRNA, leading to aberrant cellular autophagy, which inhibits testosterone synthesis and results in impaired spermatogenesis and infertility (111). Knockdown of METTL3 has been observed to significantly promote autophagic flow and increase testosterone production in testicular mesenchymal cells (111). Specific knockdown of the METTL3 gene in the endometrium stabilizes several mRNAs of estrogen-responsive genes, such as Elf3 and Celsr2, while significantly reducing the expression levels of the progesterone receptor (PR) and its target gene Myc (112). Multiple m6A regulatory proteins can act synergistically to regulate hormone levels.
METTL3 suppresses the expression of androgen receptors in cardiac fibroblasts by introducing m6A modifications to AR mRNA, which are subsequently recognized by YTHDF2, leading to the degradation of AR-associated mRNA. This m6A modification by METTL3 enhances the binding of YTHDF2 at the modified sites, thereby reducing AR expression. This reduction rescues the inhibitory effects exerted by AR on glycolysis and cardiomyocyte proliferation, ultimately facilitating myocardial fibrogenesis (113). Researchers also found that applying antisense oligonucleotides (ASOs) to target METTL3 can restore Enzalutamide(an effective AR inhibitor)resistance in vitro and in vivo (114). In endometrial cells, METTL3-mediated m6A modification directly influences the mRNA of PR. Specifically, m6A modification in the 5′ untranslated region (5′-UTR) of PR mRNA enhances the translational efficiency of PR proteins in a YTHDF1-dependent manner, a process that is conserved between mice and humans (112). M6A-related proteins such as METTL3 and METTL14 have been reported to increase follicle-stimulating hormone (FSH) levels while decreasing luteinizing hormone (LH) and testosterone (T) levels in PCOS rats, thereby reducing apoptosis and autophagy in ovarian tissue and improving ovarian morphology (115). Additionally, hormones can regulate biological processes by influencing m6A modifications. For instance, FSH can enhance the transcriptional activity of the METTL3 promoter in osteoclasts by inducing the phosphorylation of cyclic AMP response element-binding protein (CREB), which increases the m6A methylation of cathepsin K (CTSK). This methylation enhances the stability of CTSK and promotes osteoclast migration (116).
Other RNA modifications are also associated with hormone receptors, including A-to-I editing and m5C modification. In prostate cancer cells, numerous nucleotide transitions within AR gene transcripts have been identified as mutations that coincide with potential A-to-I, U-to-C, C-to-U, and G-to-A RNA editing sites (117). Furthermore, NSUN2 stabilizes AR mRNA through m5C modification, creating a positive feedback loop that promotes prostate carcinogenesis (118). Research has shown that inhibiting AR leads to the rearrangement of the alternative polyadenylation (APA) subcomplex and disrupts the interaction between the cleavage stimulation factor (CSTF) complex and the cleavage and polyadenylation specificity factor (CPSF) complex (119). In breast cancer, the luminal androgen receptor influences APA subtypes in patients with triple-negative breast cancer (120). In estrogen receptor-positive (ER+) breast cancer, estradiol (E2), a potent proliferative agent, induces APA and 3′-UTR shortening, subsequently activating proto-oncogenes (121).
3 RNA modification in hormone-dependent cancer
3.1 Tumor microenvironment in hormone-dependent cancer
Research indicates that the tumor microenvironment (TME), which consists of infiltrating immune cells such as tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs), along with stromal cells like cancer-associated fibroblasts (CAFs) and endothelial cells (122), plays a pivotal role in tumor development. It fosters tumor progression through complex interactions between cells and the extracellular matrix (ECM) (Figure 2) (123, 124).
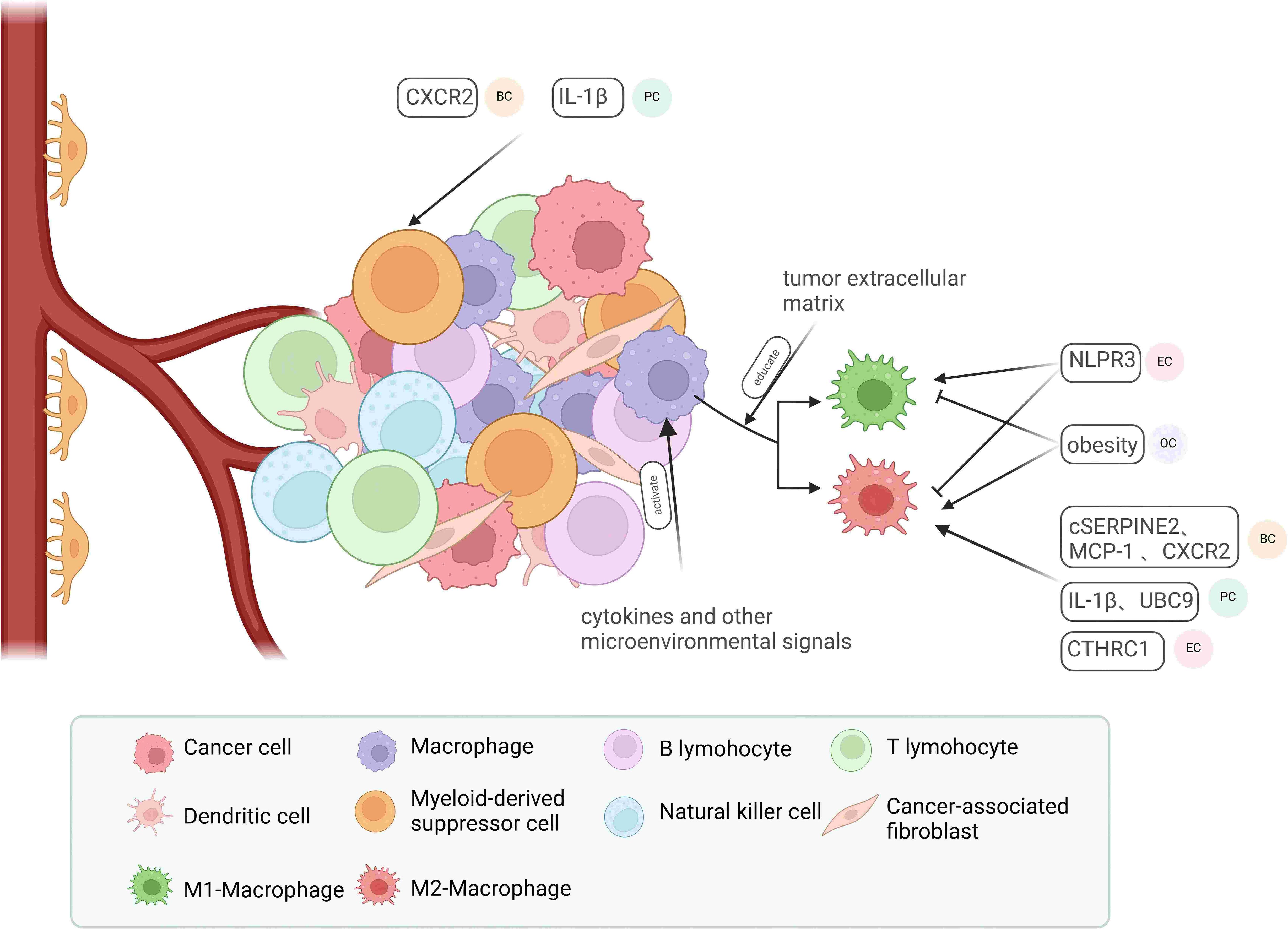
Figure 2. Major components of the tumor microenvironment and their regulatory factors. Macrophages in the tumor microenvironment can be activated by cytokines and various microenvironmental factors, leading to their polarization into M1-type or M2-type macrophages, influenced by multifaceted factors involving the tumor extracellular matrix. In endometrial cancer (EC), NLRP3 promotes M1-type macrophage polarization while inhibiting M2-type polarization. In ovarian cancer (OC), obesity exerts the opposite effect. In breast cancer (BC), pancreatic cancer (PC), and EC, cSERPINE2, MCP-1, CXCR2, UBC9, and CTHRC1 contribute to the recruitment of M2-type macrophages. Additionally, CXCR2 in BC and IL-1β in EC play roles in recruiting myeloid-derived suppressor cells (MDSCs).
Tumor-associated macrophages (TAMs) are the most prevalent immune cells in the tumor microenvironment, crucially supporting tumor progression and immune modulation (125). With their phagocytic and cytotoxic capabilities, macrophages are recognized as immunoreactive cells that can polarize into anti-tumor M1 macrophages or pro-tumor M2 macrophages in response to microenvironmental signals. TAMs closely resemble M2 macrophages and are associated with the Th2 immune response, characterized by high levels of IL-10 and TGF-β production, and they secrete pro-tumorigenic cytokines that promote tumor progression (126–128). Furthermore, TAMs influence angiogenesis and enhance cell proliferation and metastasis by inhibiting CD8+ T cell activity (129, 130). Consequently, various factors can affect tumor development and metastasis by modifying the polarization and recruitment of TAMs (131). In EC, NLRP3 deficiency leads to macrophage polarization into pro-inflammatory M2-type macrophages (132). The tumor exosome cSERPINE2 (133), the chemokines MCP-1 (134) and IL-1β (135), and the secreted protein CTHRC1 (136) facilitate the progression of BC, EC, and pancreatic cancer (PC) by recruiting TAMs. An in vitro study demonstrated that the tumor extracellular matrix (ECM) can directly regulate macrophage populations in ovarian cancer tissues (137). MDSCs are also critical immunosuppressive components in the tumor microenvironment. Two main classes of MDSCs, granulocytic/polymorphonuclear MDSCs (PMN-MDSCs) and monocytic MDSCs (M-MDSCs), can be identified in humans and mice based on their origin, and both significantly suppress immune responses following prolonged exposure to cytokines released during chronic infections, inflammation, autoimmune diseases, and cancer (138). For instance, chronic psychological stress can recruit splenic MDSCs via CXCR2, promoting the formation of a metastatic pre-metastatic niche (PMN) in BC (139). Due to their significant immunosuppressive properties, TAMs and MDSCs are frequently studied as potential targets for tumor therapy. Approaches such as gene knockdown (140), blockade of key molecules (141–143), and remodeling of drug structure (144) aim to inhibit TAMs and MDSCs to achieve clinical benefits.
In most cancers, stromal cells are major components of the TME, playing critical roles in tumor metabolism, growth, and metastasis (145). Cancer-associated fibroblasts (CAFs), key constituents of the stroma, can be activated by various tumor-derived factors (146). CAFs exhibit enhanced expression of several markers, including α-smooth muscle actin (α-SMA), fibroblast activation protein (FAP), fibroblast-specific protein 1 (FSP1), platelet-derived growth factor receptor (PDGFR)-α/β, and poikilodulin (147), and the vast majority display pro-cancer effects (148). CAFs are highly heterogeneous, comprising multiple influential subgroups. In breast cancer, CD26+ and CD26-normal fibroblast populations are transformed into inflammatory CAFs (iCAFs) and myofibroblast CAFs (myCAFs), respectively (149). CD26+ normal fibroblasts (NFs) are converted into pro-tumorigenic iCAFs, which recruit myeloid cells via a CXCL12-dependent mechanism and promote tumor cell invasion through matrix metalloproteinase (MMP) activity (149). MyCAFs, located close to the tumor, are a subtype of CAFs. The molecular and functional diversity of myCAFs arises from diverse sources and activation mechanisms, among which TSPAN8+SIRT6low myCAFs are linked to unfavorable outcomes in breast cancer patients (150). Similarly, in prostate cancer, androgen deprivation therapy (ADT) induces SPP1+ myCAFs, which are critical stromal components driving the progression of castration-resistant prostate cancer (CRPC) (151). Other non-classical subgroups include CD146+ CAFs, which promote endometrial cancer progression by inducing angiogenesis and vasculogenic mimicry (152), and αSMA+VIM+PDGFRβ+CAFs, which are correlated with lower tumor immune infiltration and shorter survival in ovarian cancer patients (153).
Additionally, recent studies have found that solid tumors are hypoxic and acidic, with the physiochemical aspects of the TME sustained by chaotic tumor perfusion, resulting in tumor progression and resistance to immunotherapy (154). Apart from recruiting immunosuppressive cells like MDSCs, tumor cells can evade the immune system in various ways. For instance, they modulate T cell responses by altering the levels of immune checkpoint molecules, particularly through the upregulation of PD-L1 (155). Moreover, tumor cells evade recognition and destruction by cytotoxic T cells by reducing MHC-I expression and impairing antigen presentation (156). They may also inhibit the production of CXCL9 and CXCL10, obstructing the infiltration of CXCR3+ effector cells into the tumor, thus facilitating immune evasion and limiting T cell infiltration (157).
3.2 RNA modification in immune system of hormone-dependent cancers
A growing number of studies have demonstrated that human malignancies are correlated with epigenetic alterations in RNA (158, 159). Previous studies have identified RNA modifications, particularly m6A, as playing a pivotal role in hormone-dependent cancers (160–163). These modifications are essential in regulating tumor growth and metastasis (33, 164).In BC, m7G has been linked to immune cell infiltration, including initial B cells, CD4+ memory resting and activated T cells, CD8+ T cells, regulatory T cells, resting and activated natural killer (NK) cells, M1 macrophages, and resting mast cells, with NCBP1 mRNA identified as the most prominent target of m7G (165).The related regulatory enzyme, RBM15B, along with its associated genes TCP1 and ANKRD36, and the RNA demethylase ALKBH family, particularly ALKBH7, are also associated with immune infiltration in breast cancer and are positively correlated with tumor development (166, 167). Furthermore, both m6A and m5C can disrupt DNA replication and affect the tumor immune microenvironment in PC (168, 169). As more relevant studies emerge, the understanding of how RNA modifications govern the immune system in hormone-dependent cancers has been progressively refined at the cellular and molecular levels (Table 2).
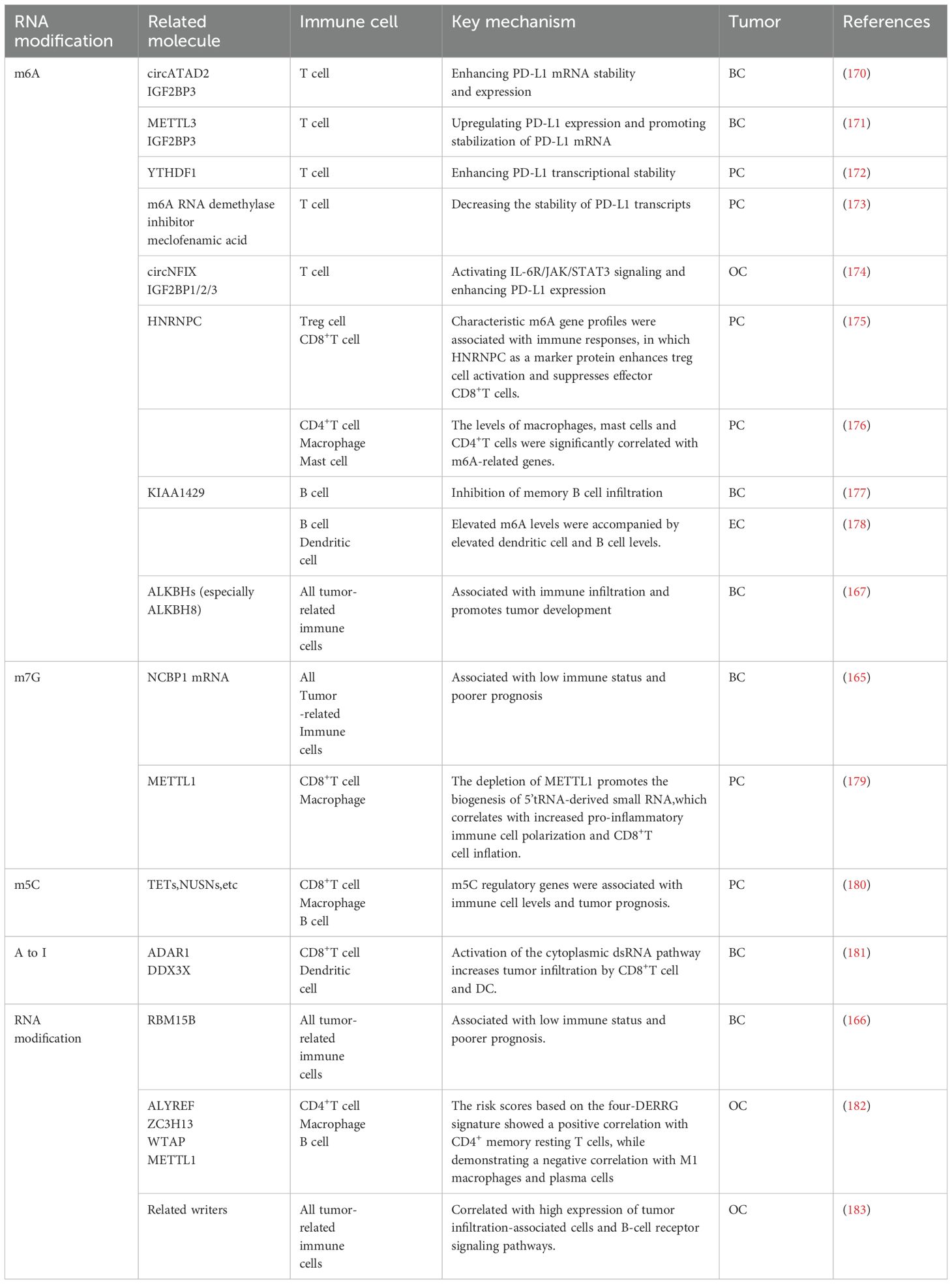
Table 2. The molecules, cells and mechanisms associated with the immunomodulatory role of RNA modifications in hormone-dependent cancers.
3.2.1 T cells
RNA modification promotes tumor immune escape by regulating immune checkpoint molecule expression on T cells. Researchers identified a 4-DERRG signature based on 59 RNA modification-associated regulatory genes (ALYREF, ZC3H13, WTAP, and METTL1) and accordingly categorized the OC patients into two distinct groups, showing significant differences in the immune checkpoint molecule CD276. The regulation of immune checkpoint molecules by m6A is primarily mediated through PD-L1. METTL3-mediated m6A modification occurs in the 3-UTR of PD-L1 mRNA, and circATAD2 can bind to it, enhancing the level of m6A modification (170, 171).The m6A reader IGF2BP3 recognizes this modification, thereby increasing PD-L1 mRNA stability and expression (171). In OC, IGF2BP1/2/3 also recognize m6A modifications, positively regulating circNFIX expression, which activates downstream JAK/STAT3 signaling and enhances PD-L1 expression (174).Thus, m6A-modified PD-L1 may serve as a potential therapeutic target. In BC, ADAR1 synergizes with DEAD-box RNA helicase 3X (DDX3X) to activate the cytoplasmic dsRNA pathway, increasing tumor infiltration of CD8+ T cells and DC cells (181). In PC, the reader YTHDF1 promotes the progression by regulating androgen function-related gene TRIM68 (172). For enhanced photothermal immunotherapy of PC, cyclodextrin-functionalized gold nanorods can deliver the m6A RNA demethylase inhibitor meclofenamic acid, thereby enhancing m6A methylation of mRNAs and decreasing the stability of PD-L1 transcripts (173). Besides, investigators have discovered that the m7G transferase METTL1 is highly expressed in both primary and advanced prostate tumors. Simultaneously, upon METTL1 deletion, the absence of m7G tRNA methylation promotes the generation of a new class of non-coding small RNAs originating from 5 tRNA fragments (179). These small RNAs regulate translation and support the production of key regulators essential for antitumor immune responses (179). These regulators are crucial for promoting CD8+ T cell infiltration and enhancing antitumor effects (179). Similarly, after clustering according to the regulatory genes of m5C (TET1, TET3, DNMT3B, YBX1, NSUN2, NSUN6, NOP2) in patients with PC, significant differences in CD8+ T cell infiltration were observed between the two clusters, with a strong negative correlation to patient prognosis (180).
3.2.2 Macrophages
RNA modification regulates macrophages mainly by altering the number or proportion of M1 and M2 type macrophages. For instance, circITGB6 specifically interacts with the KH1–2 domain of IGF2BP2, leading to increased mRNA stability of FGF9, leading to increased mRNA stability of FGF9 (184). This interaction further encourages the polarization of TAM towards the M2 phenotype, thereby inducing cisplatin resistance in OC (184). In PC, the removal of METTL1 results in the downregulation of anti-inflammatory cytokines, such as macrophage colony-stimulating factor (M-CSF), IL-10, and IL-13, which also promote M2 macrophage polarization (179, 185). In OC and BC, ALKBH3 enhances the half-life of CSF-1 mRNA by removing m1A from the GC-rich region of the 5 UTR of CSF-1 mRNA, facilitating macrophage recruitment and tumor invasion (186).
3.2.3 Other cells
In OC, based on the six lncRNA subgroups of RNA modification-associated writers (m6A, m1A, APA, and A-I), tumor-infiltrating cells such as mast cells, neutrophils, and B-cell receptor signaling pathways were highly expressed in the high-risk group (14). Moreover, m6A writer KIAA1429 was positively correlated with various advanced tumors such as BC, and negatively correlated with memory B-cell infiltration (177). In addition, in EC clusters classified by hypoxia genes, elevated m6A levels were observed alongside increased infiltration of B cells and dendritic cells (DCs) in the high-risk group (178).
3.3 Targeting regulators of RNA modification to treat hormone-dependent cancer
Despite being a relatively new field, drugs targeting RNA modifications are gradually transitioning from the laboratory to the public eye. However, fewer studies have been specifically conducted on the four hormone-dependent cancers, and the following is only a list of drugs of general interest that can potentially be used on hormone-dependent cancers. For m6A, Several inhibitors targeting FTO and ALKBH5 have been developed to impede the progression of various cancers, such as R-2-hydroxyglutarate (R-2HG), FB23-2, IOX1, IOX3, Rhein, Entacapone, and meclofenamic acid (80, 97). Regarding m5C, NSUN2 is upregulated in both BC and PC, and its expression can be reduced by inhibiting sphingosine kinase (SPHK), which maintains sphingolipid balance during cell growth (187–189). Consequently, the SPHK1 inhibitor SK1 emerges as a potential agent for cancer treatment by targeting NSUN2 expression (187–189). Moreover, research on pseudouridine identifies pyrazoline and 5-fluorouracil as common DKC1 inhibitors, employed clinically as anticancer agents (97, 190). In the context of A-to-I editing, 8-azaadenosine and 8-chloroadenosine function as ADAR1 inhibitors, but their limited specificity temporarily precludes clinical application (191, 192).
Furthermore, while no clinical trials have been conducted to date, emerging mechanistic studies suggest that specific RNA modifications may exert dual therapeutic effects in hormone-dependent tumors: either enhancing treatment efficacy or paradoxically promoting drug resistance. In OC, RNA modifications can remodel the tumor microenvironment by upregulating immunogenic RNAs, thereby reversing tumor immune evasion phenotypes and potentially restoring clinical responsiveness to immunotherapy in previously non-responding patients (193). For instance, Y-box binding protein 1 (YBX1) has been shown to enhance homologous recombination proficiency and resistance to platinum-induced stress in OC through m5C modification (194). In breast cancer (BC), METTL3 knockdown significantly increases chemosensitivity to doxorubicin via modulation of the EGF/RAD51 signaling axis (195). Intriguingly, METTL3 depletion has also been found to activate the CDKN1A/EMT pathway and m6A-BAX/caspase-9/-3/-8 cascade, thereby promoting proliferation, migration, and drug resistance in hormone receptor-positive HER2-negative breast cancer (HR+HER2-BC) (196). These findings underscore the complex regulatory networks of RNA modifications in cancer therapeutics, necessitating comprehensive mechanistic elucidation and systematic clinical validation to delineate their therapeutic potential versus risk profiles.
4 Conclusion and perspectives
This review underscores the crucial role of RNA modifications in regulating the progression and immune landscape of hormone-dependent cancers, including breast, ovarian, endometrial, and prostate malignancies. These modifications facilitate tumor growth and metastasis by modulating key immunoregulatory pathways, such as PD-L1 expression, immune cell infiltration, and cytokine signaling, revealing their potential to improve cancer diagnosis and therapy.
Despite increasing recognition of RNA modifications in cancer, the precise molecular mechanisms—especially how these modifications integrate with hormone receptor signaling and shape the immune microenvironment—remain only partially understood. Future studies should elucidate the specific pathways by which RNA modifications influence immune regulation and hormone receptor activity. As RNA modifications affect both hormone receptor function and immunogenic pathways (e.g., PD-L1), there is a compelling rationale for combining hormone therapies (e.g., anti-estrogen, anti-androgen) with immunotherapies or RNA modification inhibitors. Such combination strategies may enhance tumor susceptibility to immune-mediated destruction and mitigate therapeutic resistance.
Although therapeutic applications remain challenging, mounting evidence highlights the significant role of RNA modifications in orchestrating immune regulation and driving hormone-dependent tumor progression. Further investigation into the detailed mechanisms underlying these modifications holds promise for developing more effective and precisely targeted interventions against hormone-dependent cancers.
Author contributions
YJ: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. XL: Supervision, Visualization, Writing – review & editing. HS: Conceptualization, Investigation, Supervision, Validation, Writing – review & editing. PY: Supervision, Writing – review & editing, Software, Funding acquisition. JZ: Funding acquisition, Supervision, Writing – review & editing, Validation, Visualization, Writing – original draft. CY: Supervision, Writing – review & editing, Conceptualization, Funding acquisition, Resources.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82205256, 82104555); Shanghai Famous Elderly Chinese Medicine Doctor Academic Experience Studio (SHGZS-202240); Scientific Mission Statement of the army Logistics Research Project (BHJ23C011).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. (2024) 74:12–49. doi: 10.3322/caac.21820
2. Ouyang D, Su J, Huang P, Li M, Li Q, Zhao P, et al. Identification of lncRNAs via microarray analysis for predicting HER2-negative breast cancer response to neoadjuvant chemotherapy. Int J Clin Exp Pathol. (2018) 11:2621–8.
3. Giuliano M, Trivedi MV, Schiff R. Bidirectional crosstalk between the estrogen receptor and human epidermal growth factor receptor 2 signaling pathways in breast cancer: molecular basis and clinical implications. Breast Care Basel Switz. (2013) 8:256–62. doi: 10.1159/000354253
4. Gradishar WJ, Moran MS, Abraham J, Abramson V, Aft R, Agnese D, et al. NCCN guidelines® Insights: breast cancer, version 4.2023. J Natl Compr Cancer Netw JNCCN. (2023) 21:594–608. doi: 10.6004/jnccn.2023.0031
5. Rodriguez AC, Blanchard Z, Maurer KA, Gertz J. Estrogen signaling in endometrial cancer: a key oncogenic pathway with several open questions. Horm Cancer. (2019) 10:51–63. doi: 10.1007/s12672-019-0358-9
6. Cancer Genome Atlas Research Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, et al. Integrated genomic characterization of endometrial carcinoma. Nature. (2013) 497:67–73. doi: 10.1038/nature12113
7. Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. (1983) 15:10–7. doi: 10.1016/0090-8258(83)90111-7
8. Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet. (2014) 384:1376–88. doi: 10.1016/S0140-6736(13)62146-7
9. Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J Clin. (1972) 22:232–40. doi: 10.3322/canjclin.22.4.232
10. Albertsen P. Androgen deprivation in prostate cancer–step by step. N Engl J Med. (2009) 360:2572–4. doi: 10.1056/NEJMe0901737
11. Cohn WE, Volkin E. Nucleoside-5′-phosphates from ribonucleic acid. Nature. (1951) 167:483–4. doi: 10.1038/167483a0
12. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. (Accessed May 8, 2024).
13. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. (2011) 7:885–7. doi: 10.1038/nchembio.687
15. Wiener D, Schwartz S. The epitranscriptome beyond m6A. Nat Rev Genet. (2021) 22:119–31. doi: 10.1038/s41576-020-00295-8
16. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. (2017) 169:1187–200. doi: 10.1016/j.cell.2017.05.045
17. Xiao W, Adhikari S, Dahal U, Chen Y-S, Hao Y-J, Sun B-F, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. (2016) 61:507–19. doi: 10.1016/j.molcel.2016.01.012
18. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. (2015) 518:560–4. doi: 10.1038/nature14234
19. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. (2014) 505:117–20. doi: 10.1038/nature12730
20. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z
21. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
22. Xu W, Lai Y, Pan Y, Tan M, Ma Y, Sheng H, et al. m6A RNA methylation-mediated NDUFA4 promotes cell proliferation and metabolism in gastric cancer. Cell Death Dis. (2022) 13:715. doi: 10.1038/s41419-022-05132-w
23. Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. (2019) 18:127. doi: 10.1186/s12943-019-1053-8
24. Hamashima K, Wong KW, Sam TW, Teo JHJ, Taneja R, Le MTN, et al. Single-nucleus multiomic mapping of m6A methylomes and transcriptomes in native populations of cells with sn-m6A-CT. Mol Cell. (2023) 25:S1097-2765(23)00649-4. doi: 10.1016/j.molcel.2023.08.010. S1097-2765(23)00649–4
25. Nie W, Wang S, He R, Xu Q, Wang P, Wu Y, et al. A-to-I RNA editing in bacteria increases pathogenicity and tolerance to oxidative stress. PloS Pathog. (2020) 16:e1008740. doi: 10.1371/journal.ppat.1008740
26. Yao Z, Lin Z, Wu W. Global research trends on immunotherapy in cancer: A bibliometric analysis. Hum Vaccines Immunother. (2023) 19:2219191. doi: 10.1080/21645515.2023.2219191
27. Kundu M, Butti R, Panda VK, Malhotra D, Das S, Mitra T, et al. Modulation of the tumor microenvironment and mechanism of immunotherapy-based drug resistance in breast cancer. Mol Cancer. (2024) 23:92. doi: 10.1186/s12943-024-01990-4
28. Xin Q, Chen Y, Sun X, Li R, Wu Y, Huang X. CAR-T therapy for ovarian cancer: Recent advances and future directions. Biochem Pharmacol. (2024) 226:116349. doi: 10.1016/j.bcp.2024.116349
29. He Y, Xu W, Xiao Y-T, Huang H, Gu D, Ren S. Targeting signaling pathways in prostate cancer: mechanisms and clinical trials. Signal Transduct Target Ther. (2022) 7:198. doi: 10.1038/s41392-022-01042-7
30. Bogani G, Monk BJ, Powell MA, Westin SN, Slomovitz B, Moore KN, et al. Adding immunotherapy to first-line treatment of advanced and metastatic endometrial cancer. Ann Oncol Off J Eur Soc Med Oncol. (2024) 35:414–28. doi: 10.1016/j.annonc.2024.02.006
31. Lan H-R, Chen M, Yao S-Y, Chen J-X, Jin K-T. Novel immunotherapies for breast cancer: Focus on 2023 findings. Int Immunopharmacol. (2024) 128:111549. doi: 10.1016/j.intimp.2024.111549
32. Luo Y, Tian W, Kang D, Wu L, Tang H, Wang S, et al. RNA modification gene WDR4 facilitates tumor progression and immunotherapy resistance in breast cancer. J Adv Res. (2024) 2:S2090-1232(24)00266-2. doi: 10.1016/j.jare.2024.06.029. S2090-1232(24)00266–2
33. Han M, Huang Q, Li X, Chen X, Zhu H, Pan Y, et al. M7G-related tumor immunity: novel insights of RNA modification and potential therapeutic targets. Int J Biol Sci. (2024) 20:1238–55. doi: 10.7150/ijbs.90382
34. Chimento A, De Luca A, Avena P, De Amicis F, Casaburi I, Sirianni R, et al. Estrogen receptors-mediated apoptosis in hormone-dependent cancers. Int J Mol Sci. (2022) 23:1242. doi: 10.3390/ijms23031242
35. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. (2014) 15:293–306. doi: 10.1038/nrg3724
36. Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Res. (2010) 38:5327–35. doi: 10.1093/nar/gkq266
37. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. (2012) 485:201–6. doi: 10.1038/nature11112
38. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. (2012) 149:1635–46. doi: 10.1016/j.cell.2012.05.003
39. Zhang C, Liu N. N6-methyladenosine (m6A) modification in gynecological Malignancies. J Cell Physiol. (2022) 237:3465–79. doi: 10.1002/jcp.30828
40. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015
41. Huang J, Jiang B, Li G-W, Zheng D, Li M, Xie X, et al. m6A-modified lincRNA Dubr is required for neuronal development by stabilizing YTHDF1/3 and facilitating mRNA translation. Cell Rep. (2022) 41:111693. doi: 10.1016/j.celrep.2022.111693
42. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. (Accessed May 8, 2024).
43. Vu LP, Cheng Y, Kharas MG. The biology of m6A RNA methylation in normal and Malignant hematopoiesis. Cancer Discov. (2019) 9:25–33. doi: 10.1158/2159-8290.CD-18-0959
44. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. (2016) 534:575–8. doi: 10.1038/nature18298
45. Huang J, Dong X, Gong Z, Qin L-Y, Yang S, Zhu Y-L, et al. Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase. Protein Cell. (2019) 10:272–84. doi: 10.1007/s13238-018-0518-7
46. Śledź P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. eLife. (2016) 5:e18434. doi: 10.7554/eLife.18434
47. Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. (2014) 24:177–89. doi: 10.1038/cr.2014.3
48. Huang Q, Mo J, Liao Z, Chen X, Zhang B. The RNA m6A writer WTAP in diseases: structure, roles, and mechanisms. Cell Death Dis. (2022) 13:852. doi: 10.1038/s41419-022-05268-9
49. VIRMA mediates preferential m6A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. (Accessed May 8, 2024).
50. Zc3h13 Regulates Nuclear RNA m6A Methylation and Mouse Embryonic Stem Cell Self-Renewal. (Accessed May 8, 2024).
51. Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
52. Růžička K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, et al. Identification of factors required for m6 A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. (2017) 215:157–72. doi: 10.1111/nph.14586
53. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. (2017) 169:824–835.e14. doi: 10.1016/j.cell.2017.05.003
54. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. (2019) 47:7719–33. doi: 10.1093/nar/gkz619
55. Pinto R, Vågbø CB, Jakobsson ME, Kim Y, Baltissen MP, O’Donohue M-F, et al. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. (2020) 48:830–46. doi: 10.1093/nar/gkz1147
56. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell. (2018) 71:973–985.e5. doi: 10.1016/j.molcel.2018.08.011
57. Jia G, Yang C-G, Yang S, Jian X, Yi C, Zhou Z, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. (2008) 582:3313–9. doi: 10.1016/j.febslet.2008.08.019
58. Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J. Structural basis for the discriminative recognition of N6-methyladenosine RNA by the human YT521-B homology domain family of proteins. J Biol Chem. (2015) 290:24902–13. doi: 10.1074/jbc.M115.680389
59. Hou G, Zhao X, Li L, Yang Q, Liu X, Huang C, et al. SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic Acids Res. (2021) 49:2859–77. doi: 10.1093/nar/gkab065
60. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. (2017) 27:315–28. doi: 10.1038/cr.2017.15
61. Li J, Chen K, Dong X, Xu Y, Sun Q, Wang H, et al. YTHDF1 promotes mRNA degradation via YTHDF1-AGO2 interaction and phase separation. Cell Prolif. (2022) 55:e13157. doi: 10.1111/cpr.13157
62. Cheng Y, Xie W, Pickering BF, Chu KL, Savino AM, Yang X, et al. N6-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. (2021) 39:958–972.e8. doi: 10.1016/j.ccell.2021.04.017
63. Liu J, Gao M, He J, Wu K, Lin S, Jin L, et al. The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature. (2021) 591:322–6. doi: 10.1038/s41586-021-03313-9
64. Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of m6A transcripts by the 3’→5’ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol Cell. (2017) 68:374–387.e12. doi: 10.1016/j.molcel.2017.09.021
65. Saito Y, Hawley BR, Puno MR, Sarathy SN, Lima CD, Jaffrey SR, et al. YTHDC2 control of gametogenesis requires helicase activity but not m6A binding. Genes Dev. (2022) 36:180–94. doi: 10.1101/gad.349190.121
66. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. (2017) 27:1115–27. doi: 10.1038/cr.2017.99
67. Li L, Krasnykov K, Homolka D, Gos P, Mendel M, Fish RJ, et al. The XRN1-regulated RNA helicase activity of YTHDC2 ensures mouse fertility independently of m6A recognition. Mol Cell. (2022) 82:1678–1690.e12. doi: 10.1016/j.molcel.2022.02.034
68. Ramesh-Kumar D, Guil S. The IGF2BP family of RNA binding proteins links epitranscriptomics to cancer. Semin Cancer Biol. (2022) 86:18–31. doi: 10.1016/j.semcancer.2022.05.009
69. Oerum S, Dégut C, Barraud P, Tisné C. m1A post-transcriptional modification in tRNAs. Biomolecules. (2017) 7:20. doi: 10.3390/biom7010020
70. Shi H, Chai P, Jia R, Fan X. Novel insight into the regulatory roles of diverse RNA modifications: Re-defining the bridge between transcription and translation. Mol Cancer. (2020) 19:78. doi: 10.1186/s12943-020-01194-6
71. Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature. (2016) 530:441–6. doi: 10.1038/nature16998
72. Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. (2017) 551:251–5. doi: 10.1038/nature24456
73. Li J, Zhang H, Wang H. N1-methyladenosine modification in cancer biology: Current status and future perspectives. Comput Struct Biotechnol J. (2022) 20:6578–85. doi: 10.1016/j.csbj.2022.11.045
74. Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, et al. Mitochondrial 16S rRNA Is Methylated by tRNA Methyltransferase TRMT61B in All Vertebrates. PloS Biol. (2016) 14:e1002557. doi: 10.1371/journal.pbio.1002557
75. Sharma S, Hartmann JD, Watzinger P, Klepper A, Peifer C, Kötter P, et al. A single N1-methyladenosine on the large ribosomal subunit rRNA impacts locally its structure and the translation of key metabolic enzymes. Sci Rep. (2018) 8:11904. doi: 10.1038/s41598-018-30383-z
76. Zhang L-S, Xiong Q-P, Peña Perez S, Liu C, Wei J, Le C, et al. ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat Cell Biol. (2021) 23:684–91. doi: 10.1038/s41556-021-00709-7
77. Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. (2016) 167:1897. doi: 10.1016/j.cell.2016.11.045
78. Chen Z, Zhu W, Zhu S, Sun K, Liao J, Liu H, et al. METTL1 promotes hepatocarcinogenesis via m7 G tRNA modification-dependent translation control. Clin Transl Med. (2021) 11:e661. doi: 10.1002/ctm2.661
79. Dai X, Wang T, Gonzalez G, Wang Y. Identification of YTH domain-containing proteins as the readers for N1-methyladenosine in RNA. Anal Chem. (2018) 90:6380–4. doi: 10.1021/acs.analchem.8b01703
80. Han X, Wang M, Zhao Y-L, Yang Y, Yang Y-G. RNA methylations in human cancers. Semin Cancer Biol. (2021) 75:97–115. doi: 10.1016/j.semcancer.2020.11.007
81. Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, Lai W-Y, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. (2017) 27:606–25. doi: 10.1038/cr.2017.55
82. Selmi T, Hussain S, Dietmann S, Heiß M, Borland K, Flad S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. (2021) 49:1006–22. doi: 10.1093/nar/gkaa1193
83. Nakano S, Suzuki T, Kawarada L, Iwata H, Asano K, Suzuki T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat Chem Biol. (2016) 12:546–51. doi: 10.1038/nchembio.2099
84. Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. (2014) 136:11582–5. doi: 10.1021/ja505305z
85. Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. (2016) 35:2104–19. doi: 10.15252/embj.201694885
86. Yang R, Liang X, Wang H, Guo M, Shen H, Shi Y, et al. The RNA methyltransferase NSUN6 suppresses pancreatic cancer development by regulating cell proliferation. EBioMedicine. (2021) 63:103195. doi: 10.1016/j.ebiom.2020.103195
87. Zhang L-S, Liu C, Ma H, Dai Q, Sun H-L, Luo G, et al. Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol Cell. (2019) 74:1304–1316.e8. doi: 10.1016/j.molcel.2019.03.036
88. Enroth C, Poulsen LD, Iversen S, Kirpekar F, Albrechtsen A, Vinther J. Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res. (2019) 47:e126. doi: 10.1093/nar/gkz736
89. Monecke T, Dickmanns A, Ficner R. Structural basis for m7G-cap hypermethylation of small nuclear, small nucleolar and telomerase RNA by the dimethyltransferase TGS1. Nucleic Acids Res. (2009) 37:3865–77. doi: 10.1093/nar/gkp249
90. Culjkovic-Kraljacic B, Skrabanek L, Revuelta MV, Gasiorek J, Cowling VH, Cerchietti L, et al. The eukaryotic translation initiation factor eIF4E elevates steady-state m7G capping of coding and noncoding transcripts. Proc Natl Acad Sci U S A. (2020) 117:26773–83. doi: 10.1073/pnas.2002360117
91. Leihne V, Kirpekar F, Vågbø CB, van den Born E, Krokan HE, Grini PE, et al. Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res. (2011) 39:7688–701. doi: 10.1093/nar/gkr406
92. Lentini JM, Ramos J, Fu D. Monitoring the 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) modification in eukaryotic tRNAs via the γ-toxin endonuclease. RNA N Y N. (2018) 24:749–58. doi: 10.1261/rna.065581.118
93. Monies D, Vågbø CB, Al-Owain M, Alhomaidi S, Alkuraya FS. Recessive truncating mutations in ALKBH8 cause intellectual disability and severe impairment of wobble uridine modification. Am J Hum Genet. (2019) 104:1202–9. doi: 10.1016/j.ajhg.2019.03.026
94. Galeano F, Tomaselli S, Locatelli F, Gallo A. A-to-I RNA editing: the “ADAR” side of human cancer. Semin Cell Dev Biol. (2012) 23:244–50. doi: 10.1016/j.semcdb.2011.09.003
95. Erdmann EA, Mahapatra A, Mukherjee P, Yang B, Hundley HA. To protect and modify double-stranded RNA - the critical roles of ADARs in development, immunity and oncogenesis. Crit Rev Biochem Mol Biol. (2021) 56:54–87. doi: 10.1080/10409238.2020.1856768
96. Guzzi N, Cieśla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. (2018) 173:1204–1216.e26. doi: 10.1016/j.cell.2018.03.008
97. Nombela P, Miguel-López B, Blanco S. The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol Cancer. (2021) 20:18. doi: 10.1186/s12943-020-01263-w
98. Chen H, Yao J, Bao R, Dong Y, Zhang T, Du Y, et al. Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer. (2021) 20:29. doi: 10.1186/s12943-021-01322-w
99. Shen X, Zhao K, Xu L, Cheng G, Zhu J, Gan L, et al. YTHDF2 inhibits gastric cancer cell growth by regulating FOXC2 signaling pathway. Front Genet. (2020) 11:592042. doi: 10.3389/fgene.2020.592042
100. Yan J, Liu J, Huang Z, Huang W, Lv J. FOXC2-AS1 stabilizes FOXC2 mRNA via association with NSUN2 in gastric cancer cells. Hum Cell. (2021) 34:1755–64. doi: 10.1007/s13577-021-00583-3
101. Zeng Z, Wang J, Xu F, Hu P, Hu Y, Zhuo W, et al. The m6A modification-mediated positive feedback between glycolytic lncRNA SLC2A1-DT and c-Myc promotes tumorigenesis of hepatocellular carcinoma. Int J Biol Sci. (2024) 20:1744–62. doi: 10.7150/ijbs.86658
102. Sun Y, Fan J, Wang B, Meng Z, Ren D, Zhao J, et al. The aberrant expression of ADAR1 promotes resistance to BET inhibitors in pancreatic cancer by stabilizing c-Myc. Am J Cancer Res. (2020) 10:148–63.
103. Kan G, Wang Z, Sheng C, Chen G, Yao C, Mao Y, et al. Dual inhibition of DKC1 and MEK1/2 synergistically restrains the growth of colorectal cancer cells. Adv Sci Weinh Baden-Wurtt Ger. (2021) 8:2004344. doi: 10.1002/advs.202004344
104. Liu T, Li H, Du G, Ma J, Shen J, Zhao H, et al. Comprehensive analysis of m6A regulator-based methylation modification patterns characterized by distinct immune profiles in colon adenocarcinomas. Gene. (2022) 821:146250. doi: 10.1016/j.gene.2022.146250
105. Chen J, Ye M, Bai J, Gong Z, Yan L, Gu D, et al. ALKBH5 enhances lipid metabolism reprogramming by increasing stability of FABP5 to promote pancreatic neuroendocrine neoplasms progression in an m6A-IGF2BP2-dependent manner. J Transl Med. (2023) 21:741. doi: 10.1186/s12967-023-04578-6
106. Zhang Y, Liu X, Wang Y, Lai S, Wang Z, Yang Y, et al. The m6A demethylase ALKBH5-mediated upregulation of DDIT4-AS1 maintains pancreatic cancer stemness and suppresses chemosensitivity by activating the mTOR pathway. Mol Cancer. (2022) 21:174. doi: 10.1186/s12943-022-01647-0
107. Yun D, Yang Z, Zhang S, Yang H, Liu D, Grützmann R, et al. An m5C methylation regulator-associated signature predicts prognosis and therapy response in pancreatic cancer. Front Cell Dev Biol. (2022) 10:975684. doi: 10.3389/fcell.2022.975684
108. Zheng Q, Yu X, Zhang Q, He Y, Guo W. Genetic characteristics and prognostic implications of m1A regulators in pancreatic cancer. Biosci Rep. (2021) 41:BSR20210337. doi: 10.1042/BSR20210337
109. Li Y, Wang N-X, Yin C, Jiang S-S, Li J-C, Yang S-Y. RNA editing enzyme ADAR1 regulates METTL3 in an editing dependent manner to promote breast cancer progression via METTL3/ARHGAP5/YTHDF1 axis. Int J Mol Sci. (2022) 23:9656. doi: 10.3390/ijms23179656
110. Qi L, Zhang W, Ren X, Xu R, Yang Z, Chen R, et al. Cross-talk of multiple types of RNA modification regulators uncovers the tumor microenvironment and immune infiltrates in soft tissue sarcoma. Front Immunol. (2022) 13:921223. doi: 10.3389/fimmu.2022.921223
111. Jiang L, Lin X, Jiang J, Qiu C, Zheng S, Zhao N, et al. METTL3-m6A-SIRT1 axis affects autophagic flux contributing to PM2.5-induced inhibition of testosterone production in Leydig cells. Sci Total Environ. (2024) 918:170701. doi: 10.1016/j.scitotenv.2024.170701
112. Wan S, Sun Y, Zong J, Meng W, Yan J, Chen K, et al. METTL3-dependent m6A methylation facilitates uterine receptivity and female fertility via balancing estrogen and progesterone signaling. Cell Death Dis. (2023) 14:349. doi: 10.1038/s41419-023-05866-1
113. Zhou Y, Song K, Tu B, Sun H, Ding J-F, Luo Y, et al. METTL3 boosts glycolysis and cardiac fibroblast proliferation by increasing AR methylation. Int J Biol Macromol. (2022) 223:899–915. doi: 10.1016/j.ijbiomac.2022.11.042
114. Li Y, Zhu S, Chen Y, Ma Q, Kan D, Yu W, et al. Post-transcriptional modification of m6A methylase METTL3 regulates ERK-induced androgen-deprived treatment resistance prostate cancer. Cell Death Dis. (2023) 14:289. doi: 10.1038/s41419-023-05773-5
115. Zhang Y, Zhou H, Ding C. The ameliorative effect of CangFu Daotan Decoction on polycystic ovary syndrome of rodent model is associated with m6A methylation and Wnt/β-catenin pathway. Gynecol Endocrinol. (2023) 39:2181637. doi: 10.1080/09513590.2023.2181637
116. Li X, Fan C, Wang J, Li P, Xu X, Guo R, et al. Follicle-stimulating hormone accelerates osteoclast migration by enhancing methyltransferase-like 3-mediated m6A methylation of cathepsin K. J Mol Endocrinol. (2024) 72:e230130. doi: 10.1530/JME-23-0130
117. Martinez HD, Jasavala RJ, Hinkson I, Fitzgerald LD, Trimmer JS, Kung H-J, et al. RNA editing of androgen receptor gene transcripts in prostate cancer cells. J Biol Chem. (2008) 283:29938–49. doi: 10.1074/jbc.M800534200
118. Zhu W, Wan F, Xu W, Liu Z, Wang J, Zhang H, et al. Positive epigenetic regulation loop between AR and NSUN2 promotes prostate cancer progression. Clin Transl Med. (2022) 12:e1028. doi: 10.1002/ctm2.1028
119. Caggiano C, Pieraccioli M, Pitolli C, Babini G, Zheng D, Tian B, et al. The androgen receptor couples promoter recruitment of RNA processing factors to regulation of alternative polyadenylation at the 3’ end of transcripts. Nucleic Acids Res. (2022) 50:9780–96. doi: 10.1093/nar/gkac737
120. Wang L, Lang G-T, Xue M-Z, Yang L, Chen L, Yao L, et al. Dissecting the heterogeneity of the alternative polyadenylation profiles in triple-negative breast cancers. Theranostics. (2020) 10:10531–47. doi: 10.7150/thno.40944
121. Akman BH, Can T, Erson-Bensan AE. Estrogen-induced upregulation and 3’-UTR shortening of CDC6. Nucleic Acids Res. (2012) 40:10679–88. doi: 10.1093/nar/gks855
122. Bader JE, Voss K, Rathmell JC. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell. (2020) 78:1019–33. doi: 10.1016/j.molcel.2020.05.034
123. Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, et al. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun Signal CCS. (2020) 18:59. doi: 10.1186/s12964-020-0530-4
124. Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer. (2021) 20:131. doi: 10.1186/s12943-021-01428-1
125. Qiu S-Q, Waaijer SJH, Zwager MC, de Vries EGE, van der Vegt B, Schröder CP. Tumor-associated macrophages in breast cancer: Innocent bystander or important player? Cancer Treat Rev. (2018) 70:178–89. doi: 10.1016/j.ctrv.2018.08.010
126. Su S, Liu Q, Chen J, Chen J, Chen F, He C, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. (2014) 25:605–20. doi: 10.1016/j.ccr.2014.03.021
127. Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. (2002) 23:549–55. doi: 10.1016/s1471-4906(02)02302-5
128. Sousa S, Brion R, Lintunen M, Kronqvist P, Sandholm J, Mönkkönen J, et al. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res BCR. (2015) 17:101. doi: 10.1186/s13058-015-0621-0
129. Xu M, Liu M, Du X, Li S, Li H, Li X, et al. Intratumoral delivery of IL-21 overcomes anti-her2/neu resistance through shifting tumor-associated macrophages from M2 to M1 phenotype. J Immunol Baltim Md. (2015) 194:4997–5006. doi: 10.4049/jimmunol.1402603
130. Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. (2015) 27:462–72. doi: 10.1016/j.ccell.2015.02.015
131. Liu Y, Yang J, Hilliard TS, Wang Z, Johnson J, Wang W, et al. Host obesity alters the ovarian tumor immune microenvironment and impacts response to standard of care chemotherapy. J Exp Clin Cancer Res CR. (2023) 42:165. doi: 10.1186/s13046-023-02740-y
132. Zhu X, Xu Y, Wang J, Xue Z, Qiu T, Chen J. Loss of NLRP3 reduces oxidative stress and polarizes intratumor macrophages to attenuate immune attack on endometrial cancer. Front Immunol. (2023) 14:1165602. doi: 10.3389/fimmu.2023.1165602
133. Zhou B, Mo Z, Lai G, Chen X, Li R, Wu R, et al. Targeting tumor exosomal circular RNA cSERPINE2 suppresses breast cancer progression by modulating MALT1-NF-κB-IL-6 axis of tumor-associated macrophages. J Exp Clin Cancer Res CR. (2023) 42:48. doi: 10.1186/s13046-023-02620-5
134. Yoshimura T, Li C, Wang Y, Matsukawa A. The chemokine monocyte chemoattractant protein-1/CCL2 is a promoter of breast cancer metastasis. Cell Mol Immunol. (2023) 20:714–38. doi: 10.1038/s41423-023-01013-0
135. Wang D, Cheng C, Chen X, Wang J, Liu K, Jing N, et al. IL-1β Is an androgen-responsive target in macrophages for immunotherapy of prostate cancer. Adv Sci Weinh Baden-Wurtt Ger. (2023) 10:e2206889. doi: 10.1002/advs.202206889
136. Li L-Y, Yin K-M, Bai Y-H, Zhang Z-G, Di W, Zhang S. CTHRC1 promotes M2-like macrophage recruitment and myometrial invasion in endometrial carcinoma by integrin-Akt signaling pathway. Clin Exp Metastasis. (2019) 36:351–63. doi: 10.1007/s10585-019-09971-4
137. Puttock EH, Tyler EJ, Manni M, Maniati E, Butterworth C, Burger Ramos M, et al. Extracellular matrix educates an immunoregulatory tumor macrophage phenotype found in ovarian cancer metastasis. Nat Commun. (2023) 14:2514. doi: 10.1038/s41467-023-38093-5
138. Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. (2021) 21:485–98. doi: 10.1038/s41577-020-00490-y
139. Zheng Y, Wang N, Wang S, Zhang J, Yang B, Wang Z. Chronic psychological stress promotes breast cancer pre-metastatic niche formation by mobilizing splenic MDSCs via TAM/CXCL1 signaling. J Exp Clin Cancer Res CR. (2023) 42:129. doi: 10.1186/s13046-023-02696-z
140. Xiao J, Sun F, Wang Y-N, Liu B, Zhou P, Wang F-X, et al. UBC9 deficiency enhances immunostimulatory macrophage activation and subsequent antitumor T cell response in prostate cancer. J Clin Invest. (2023) 133:e158352. doi: 10.1172/JCI158352
141. Chung H, Gyu-Mi P, Na YR, Lee Y-S, Choi H, Seok SH. Comprehensive characterization of early-programmed tumor microenvironment by tumor-associated macrophages reveals galectin-1 as an immune modulatory target in breast cancer. Theranostics. (2024) 14:843–60. doi: 10.7150/thno.88917
142. Cen Y, Chen Y, Li X, Chen X, Yu B, Yan M, et al. Optical controlled and nuclear targeted CECR2 competitor to downregulate CSF-1 for metastatic breast cancer immunotherapy. Biomaterials. (2024) 308:122568. doi: 10.1016/j.biomaterials.2024.122568
143. Ducarouge B, Redavid A-R, Victoor C, Chira R, Fonseca A, Hervieu M, et al. Netrin-1 blockade inhibits tumor associated Myeloid-derived suppressor cells, cancer stemness and alleviates resistance to chemotherapy and immune checkpoint inhibitor. Cell Death Differ. (2023) 30:2201–12. doi: 10.1038/s41418-023-01209-x
144. Huang T, Zhang Q, Yi J, Wang R, Zhang Z, Luo P, et al. PEG-sheddable nanodrug remodels tumor microenvironment to promote effector T cell infiltration and revise their exhaustion for breast cancer immunotherapy. Small Weinh Bergstr Ger. (2023) 19:e2301749. doi: 10.1002/smll.202301749
145. Andersson P, Yang Y, Hosaka K, Zhang Y, Fischer C, Braun H, et al. Molecular mechanisms of IL-33-mediated stromal interactions in cancer metastasis. JCI Insight. (2018) 3:e122375. doi: 10.1172/jci.insight.122375
146. Butti R, Nimma R, Kundu G, Bulbule A, Kumar TVS, Gunasekaran VP, et al. Tumor-derived osteopontin drives the resident fibroblast to myofibroblast differentiation through Twist1 to promote breast cancer progression. Oncogene. (2021) 40:2002–17. doi: 10.1038/s41388-021-01663-2
147. Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov. (2019) 18:99–115. doi: 10.1038/s41573-018-0004-1
148. Mizutani Y, Kobayashi H, Iida T, Asai N, Masamune A, Hara A, et al. Meflin-positive cancer-associated fibroblasts inhibit pancreatic carcinogenesis. Cancer Res. (2019) 79:5367–81. doi: 10.1158/0008-5472.CAN-19-0454
149. Houthuijzen JM, de Bruijn R, van der Burg E, Drenth AP, Wientjens E, Filipovic T, et al. CD26-negative and CD26-positive tissue-resident fibroblasts contribute to functionally distinct CAF subpopulations in breast cancer. Nat Commun. (2023) 14:183. doi: 10.1038/s41467-023-35793-w
150. Fan G, Yu B, Tang L, Zhu R, Chen J, Zhu Y, et al. TSPAN8+ myofibroblastic cancer-associated fibroblasts promote chemoresistance in patients with breast cancer. Sci Transl Med. (2024) 16:eadj5705. doi: 10.1126/scitranslmed.adj5705
151. Wang H, Li N, Liu Q, Guo J, Pan Q, Cheng B, et al. Antiandrogen treatment induces stromal cell reprogramming to promote castration resistance in prostate cancer. Cancer Cell. (2023) 41:1345–1362.e9. doi: 10.1016/j.ccell.2023.05.016
152. Yu Z, Zhang Q, Wei S, Zhang Y, Zhou T, Zhang Q, et al. CD146+CAFs promote progression of endometrial cancer by inducing angiogenesis and vasculogenic mimicry via IL-10/JAK1/STAT3 pathway. Cell Commun Signal CCS. (2024) 22:170. doi: 10.1186/s12964-024-01550-9
153. Xu AM, Haro M, Walts AE, Hu Y, John J, Karlan BY, et al. Spatiotemporal architecture of immune cells and cancer-associated fibroblasts in high-grade serous ovarian carcinoma. Sci Adv. (2024) 10:eadk8805. doi: 10.1126/sciadv.adk8805
154. Kopecka J, Salaroglio IC, Perez-Ruiz E, Sarmento-Ribeiro AB, Saponara S, De Las Rivas J, et al. Hypoxia as a driver of resistance to immunotherapy. Drug Resist Update Rev Comment Antimicrob Anticancer Chemother. (2021) 59:100787. doi: 10.1016/j.drup.2021.100787
155. de Boniface J, Mao Y, Schmidt-Mende J, Kiessling R, Poschke I. Expression patterns of the immunomodulatory enzyme arginase 1 in blood, lymph nodes and tumor tissue of early-stage breast cancer patients. Oncoimmunology. (2012) 1:1305–12. doi: 10.4161/onci.21678
156. van de Weijer ML, Samanta K, Sergejevs N, Jiang L, Dueñas ME, Heunis T, et al. Tapasin assembly surveillance by the RNF185/Membralin ubiquitin ligase complex regulates MHC-I surface expression. Nat Commun. (2024) 15:8508. doi: 10.1038/s41467-024-52772-x
157. Santoni M, Romagnoli E, Saladino T, Foghini L, Guarino S, Capponi M, et al. Triple negative breast cancer: Key role of Tumor-Associated Macrophages in regulating the activity of anti-PD-1/PD-L1 agents. Biochim Biophys Acta Rev Cancer. (2018) 1869:78–84. doi: 10.1016/j.bbcan.2017.10.007
158. Deng X, Qing Y, Horne D, Huang H, Chen J. The roles and implications of RNA m6A modification in cancer. Nat Rev Clin Oncol. (2023) 20:507–26. doi: 10.1038/s41571-023-00774-x
159. Zhuang H, Yu B, Tao D, Xu X, Xu Y, Wang J, et al. The role of m6A methylation in therapy resistance in cancer. Mol Cancer. (2023) 22:91. doi: 10.1186/s12943-023-01782-2
160. Ying Y, Wu Y, Zhang F, Tang Y, Yi J, Ma X, et al. Co-transcriptional R-loops-mediated epigenetic regulation drives growth retardation and docetaxel chemosensitivity enhancement in advanced prostate cancer. Mol Cancer. (2024) 23:79. doi: 10.1186/s12943-024-01994-0
161. Xu X, Zhuang X, Yu H, Li P, Li X, Lin H, et al. FSH induces EMT in ovarian cancer via ALKBH5-regulated Snail m6A demethylation. Theranostics. (2024) 14:2151–66. doi: 10.7150/thno.94161
162. Luo F, Zhang M, Sun B, Xu C, Yang Y, Zhang Y, et al. LINC00115 promotes chemoresistant breast cancer stem-like cell stemness and metastasis through SETDB1/PLK3/HIF1α signaling. Mol Cancer. (2024) 23:60. doi: 10.1186/s12943-024-01975-3
163. Chen S-J, Zhang J, Zhou T, Rao S-S, Li Q, Xiao L-Y, et al. Epigenetically upregulated NSUN2 confers ferroptosis resistance in endometrial cancer via m5C modification of SLC7A11 mRNA. Redox Biol. (2024) 69:102975. doi: 10.1016/j.redox.2023.102975
164. Gu J, Cao H, Chen X, Zhang XD, Thorne RF, Liu X. RNA m6A modifications regulate crosstalk between tumor metabolism and immunity. Wiley Interdiscip Rev RNA. (2024) 15:e1829. doi: 10.1002/wrna.1829
165. Li J, Zheng L, Song L, Dong Z, Bai W, Qi L. Identification and validation of N7 -methylguanosine-associated gene NCBP1 as prognostic and immune-associated biomarkers in breast cancer patients. J Cell Mol Med. (2024) 28:e18067. doi: 10.1111/jcmm.18067
166. Zhang X, Shen L, Zhu Y, Zhai C, Zeng H, Liu X, et al. Crosstalk of RNA methylation writers defines tumor microenvironment and alisertib resistance in breast cancer. Front Endocrinol. (2023) 14:1166939. doi: 10.3389/fendo.2023.1166939
167. Chen H, Zhou L, Li J, Hu K. ALKBH family members as novel biomarkers and prognostic factors in human breast cancer. Aging. (2022) 14:6579–93. doi: 10.18632/aging.204231
168. Xu Z, Chen S, Zhang Y, Liu R, Chen M. Roles of m5C RNA modification patterns in biochemical recurrence and tumor microenvironment characterization of prostate adenocarcinoma. Front Immunol. (2022) 13:869759. doi: 10.3389/fimmu.2022.869759
169. Liu Z, Zhong J, Zeng J, Duan X, Lu J, Sun X, et al. Characterization of the m6A-associated tumor immune microenvironment in prostate cancer to aid immunotherapy. Front Immunol. (2021) 12:735170. doi: 10.3389/fimmu.2021.735170
170. Zhang Z, Huo W, Li J. circATAD2 mitigates CD8+ T cells antitumor immune surveillance in breast cancer via IGF2BP3/m6A/PD-L1 manner. Cancer Immunol Immunother. (2024) 73:130. doi: 10.1007/s00262-024-03705-6
171. Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W, et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N6-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. (2022) 21:60. doi: 10.1186/s12943-021-01447-y
172. Wang Y, Jin P, Wang X. N6-methyladenosine regulator YTHDF1 represses the CD8 + T cell-mediated antitumor immunity and ferroptosis in prostate cancer via m6A/PD-L1 manner. Apoptosis. (2024) 29:142–53. doi: 10.1007/s10495-023-01885-7
173. Liu J, Song Y, Wang Y, Han M, Wang C, Yan F. Cyclodextrin-functionalized gold nanorods loaded with meclofenamic acid for improving N6-methyladenosine-mediated second near-infrared photothermal immunotherapy. ACS Appl Mater Interfaces. (2022) 14:40612–23. doi: 10.1021/acsami.2c09978
174. Wang R, Ye H, Yang B, Ao M, Yu X, Wu Y, et al. m6A-modified circNFIX promotes ovarian cancer progression and immune escape via activating IL-6R/JAK1/STAT3 signaling by sponging miR-647. Int Immunopharmacol. (2023) 124:110879. doi: 10.1016/j.intimp.2023.110879
175. Cheng Y, Li L, Wei X, Xu F, Huang X, Qi F, et al. HNRNPC suppresses tumor immune microenvironment by activating Treg cells promoting the progression of prostate cancer. Cancer Sci. (2023) 114:1830–45. doi: 10.1111/cas.15745
176. Li C, Peng D, Gan Y, Zhou L, Hou W, Wang B, et al. The m6A methylation landscape, molecular characterization and clinical relevance in prostate adenocarcinoma. Front Immunol. (2023) 14:1086907. doi: 10.3389/fimmu.2023.1086907
177. Ma C, Zheng Q, Wang Y, Li G, Zhao M, Sun Z. Pan-cancer analysis and experimental validation revealed the m6A methyltransferase KIAA1429 as a potential biomarker for diagnosis, prognosis, and immunotherapy. Aging. (2023) 15:8664–91. doi: 10.18632/aging.204968
178. Jiao Y, Geng R, Zhong Z, Ni S, Liu W, He Z, et al. A hypoxia molecular signature-based prognostic model for endometrial cancer patients. Int J Mol Sci. (2023) 24:1675. doi: 10.3390/ijms24021675
179. García-Vílchez R, Añazco-Guenkova AM, Dietmann S, López J, Morón-Calvente V, D’Ambrosi S, et al. METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer. Mol Cancer. (2023) 22:119. doi: 10.1186/s12943-023-01809-8
180. Yu G, Bao J, Zhan M, Wang J, Li X, Gu X, et al. Comprehensive analysis of m5C methylation regulatory genes and tumor microenvironment in prostate cancer. Front Immunol. (2022) 13:914577. doi: 10.3389/fimmu.2022.914577
181. Choi H, Kwon J, Cho MS, Sun Y, Zheng X, Wang J, et al. Targeting DDX3X Triggers Antitumor Immunity via a dsRNA-Mediated Tumor-Intrinsic Type I Interferon Response. Cancer Res. (2021) 81:3607–20. doi: 10.1158/0008-5472.CAN-20-3790
182. Zheng P, Li N, Zhan X. Ovarian cancer subtypes based on the regulatory genes of RNA modifications: Novel prediction model of prognosis. Front Endocrinol. (2022) 13:972341. doi: 10.3389/fendo.2022.972341
183. Ye L, Pan K, Fang S, Wu S-N, Chen S, Tang S, et al. Four types of RNA modification writer-related lncRNAs are effective predictors of prognosis and immunotherapy response in serous ovarian carcinoma. Front Immunol. (2022) 13:863484. doi: 10.3389/fimmu.2022.863484
184. Li H, Luo F, Jiang X, Zhang W, Xiang T, Pan Q, et al. CircITGB6 promotes ovarian cancer cisplatin resistance by resetting tumor-associated macrophage polarization toward the M2 phenotype. J Immunother Cancer. (2022) 10:e004029. doi: 10.1136/jitc-2021-004029
185. DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. (2019) 19:369–82. doi: 10.1038/s41577-019-0127-6
186. Woo H-H, Chambers SK. Human ALKBH3-induced m1A demethylation increases the CSF-1 mRNA stability in breast and ovarian cancer cells. Biochim Biophys Acta Gene Regul Mech. (2019) 1862:35–46. doi: 10.1016/j.bbagrm.2018.10.008
187. Alshaker H, Thrower H, Pchejetski D. Sphingosine kinase 1 in breast cancer-A new molecular marker and a therapy target. Front Oncol. (2020) 10:289. doi: 10.3389/fonc.2020.00289
188. Zheng X, Li W, Ren L, Liu J, Pang X, Chen X, et al. The sphingosine kinase-1/sphingosine-1-phosphate axis in cancer: Potential target for anticancer therapy. Pharmacol Ther. (2019) 195:85–99. doi: 10.1016/j.pharmthera.2018.10.011
189. Alshaker H, Wang Q, Brewer D, Pchejetski D. Transcriptome-wide effects of sphingosine kinases knockdown in metastatic prostate and breast cancer cells: implications for therapeutic targeting. Front Pharmacol. (2019) 10:303. doi: 10.3389/fphar.2019.00303
190. Xue C, Chu Q, Zheng Q, Jiang S, Bao Z, Su Y, et al. Role of main RNA modifications in cancer: N6-methyladenosine, 5-methylcytosine, and pseudouridine. Signal Transduct Target Ther. (2022) 7:142. doi: 10.1038/s41392-022-01003-0
191. Cottrell KA, Torres LS, Dizon MG, Weber JD. 8-azaadenosine and 8-chloroadenosine are not selective inhibitors of ADAR. Cancer Res Commun. (2021) 1:56–64. doi: 10.1158/2767-9764.crc-21-0027
192. Goncharov AO, Shender VO, Kuznetsova KG, Kliuchnikova AA, Moshkovskii SA. Interplay between A-to-I editing and splicing of RNA: A potential point of application for cancer therapy. Int J Mol Sci. (2022) 23:5240. doi: 10.3390/ijms23095240
193. Gomez S, Cox OL, Walker RR, Rentia U, Hadley M, Arthofer E, et al. Inhibiting DNA methylation and RNA editing upregulates immunogenic RNA to transform the tumor microenvironment and prolong survival in ovarian cancer. J Immunother Cancer. (2022) 10:e004974. doi: 10.1136/jitc-2022-004974
194. Meng H, Miao H, Zhang Y, Chen T, Yuan L, Wan Y, et al. YBX1 promotes homologous recombination and resistance to platinum-induced stress in ovarian cancer by recognizing m5C modification. Cancer Lett. (2024) 597:217064. doi: 10.1016/j.canlet.2024.217064
195. Li E, Xia M, Du Y, Long K, Ji F, Pan F, et al. METTL3 promotes homologous recombination repair and modulates chemotherapeutic response in breast cancer by regulating the EGF/RAD51 axis. eLife. (2022) 11:e75231. doi: 10.7554/eLife.75231
Keywords: RNA modification, sex hormone-dependent cancer, tumor microenvironment, tumor-immunology, epigenetics
Citation: Jiang Y, Liang X, Sun H, Yin P, Zhou J and Yu C (2025) Immunomodulatory role of RNA modifications in sex hormone-dependent cancers. Front. Immunol. 16:1513037. doi: 10.3389/fimmu.2025.1513037
Received: 17 October 2024; Accepted: 14 April 2025;
Published: 08 May 2025.
Edited by:
Zhihao Wang, Wuhan University, ChinaCopyright © 2025 Jiang, Liang, Sun, Yin, Zhou and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Zhou, c2h1dGNtempAMTI2LmNvbQ==; Chaoqin Yu, Y3F5dUBzbW11LmVkdS5jbg==
†These authors have contributed equally to this work
‡ORCID: Jing Zhou, orcid.org/0000-0002-9115-0623
Chaoqin Yu, orcid.org/0000-0003-1614-2655
 Yujia Jiang
Yujia Jiang Xiaolan Liang
Xiaolan Liang Hongyi Sun
Hongyi Sun Ping Yin
Ping Yin Jing Zhou1*‡
Jing Zhou1*‡