- 1Department of Hematology, West China Hospital, Sichuan University, Chengdu, Sichuan, China
- 2Clinic Trial Center, West China Hospital, Sichuan University, Chengdu, Sichuan, China
- 3Department of Thyroid Surgery, National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu, Sichuan, China
- 4State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, and Collaborative Innovation Center for Biotherapy, Chengdu, Sichuan, China
Introduction: Lysosomal-associated protein transmembrane-4 beta (LAPTM4B) protein expression was increased in solid tumors, whereas few studies were performed in hematologic malignancies. We aimed to study the effect of the LAPTM4B gene in pan-cancer and Philadelphia chromosome-positive acute B cell lymphoblastic leukemia (Ph+ B-ALL).
Methods: The differential expression, diagnosis, prognosis, genetic and epigenetic alterations, tumor microenvironment, stemness, immune infiltration cells, function enrichment, single-cell analysis, and drug response across cancers were conducted based on multiple computational tools. Additionally, Ph+ B-ALL transgenic mouse model with Laptm4b knockout was used to analyze the function of LAPTM4B in vivo. BrdU incorporation method, flow cytometry, and Witte-lock Witte culture were used to evaluate the roles of LAPTM4B in vitro.
Results: We identified that LAPTM4B expression was increased in various cancers, with significant associations with clinical outcomes. LAPTM4B expression correlated with DNA and RNA methylation patterns and was associated with drug resistance. It also influenced the tumor immune microenvironment, with implications for immunotherapy response. In leukemia, LAPTM4B was expressed in stem cells and associated with specific subtypes. Knockout of LAPTM4B impeded B-ALL progression in mice and reduced cell proliferation and caused G0/G1 arrest in vitro.
Discussion: Our study elucidated the role LAPTM4B that promoted the development and progression in Ph+ B-ALL. Furthermore, LAPTM4B played a diagnostic, prognostic, and immunological factor.
Introduction
Tumorigenesis is a multifaceted process influenced by a dynamic interplay between internal factors and the tumor microenvironment. Internal factors encompass genetic mutations, epigenetic changes, and the dysregulation of signaling pathways (1, 2). Tumor microenvironment is comprised of various factors including metabolomics, inflammation, angiogenesis, immune system modulation, extracellular matrix (ECM) (3–7). Crucially, membrane proteins play a pivotal role during tumorigenesis by facilitating the transmission of signals between the extracellular environment and the cell’s interior (8). These proteins participate some fundamental cellular processes such as growth, differentiation, and survival (8–10). Therefore, investigating the intricate interplay between internal factors, the tumor microenvironment, and the role of membrane proteins is imperative for comprehending the intricacies of tumorigenesis. Such understanding lays the foundation for the development of targeted strategies aimed at preventing and treating cancer effectively. One of the research interests in our laboratory is to uncover the roles and mechanisms of membrane proteins in the initiation and development of tumors. Building upon the reported biological functions of lysosomal membrane-associated protein transmembrane-4 beta (LAPTM4B) in existing studies, we aim to comprehensively understand its involvement in cancer, especially Philadelphia chromosome-positive acute B cell lymphoblastic leukemia (Ph+ B-ALL).
LAPTM4B is recognized as a late endosomal protein, and it is also distributed in the plasma membrane. It exhibits widespread expression in various tissues throughout the body, with predominant levels observed in the heart, kidney, skeletal muscle, and hematopoietic stem cells (HSCs). In contrast, its expression is relatively lower in peripheral blood leukocytes, spleen, and thymus (11). LAPTM4B involves in multiple biological processes, including cell cycle, cell growth and proliferation, and autophagy. LAPTM4B interacts with and integrin and promotes cell growth and proliferation through a series of enzyme-linked reactions within the membrane (12). LAPTM4B also regulates cell cycle and engages in growth signaling pathways, such as PI3K/AKT and MAPK (13). LAPTM4B also promotes autophagy through the EGFR signaling pathway (13, 14), and loss of LAPTM4B inhibited later stages of autophagy by blocking maturation of the autophagosome (15).
Increased LAPTM4B expression has been observed in various cancers, including breast, liver, lung, ovarian, uterine, and gastric cancers (11, 16–18). Notably, elevated LAPTM4B levels contribute to chemotherapy resistance in breast cancer. The overexpression of LAPTM4B induces resistance to anthracyclines (such as doxorubicin, daunorubicin, and epirubicin) by retaining the drug in the cytoplasm and reducing its nuclear localization, thereby diminishing drug-induced DNA damage (19). In addition to solid tumors, LAPTM4B is also highly expressed in hematologic malignancies. LAPTM4B promoted AML progression by regulating the RPS9/STAT3 axis (20). Elevated LAPTM4B expression is associated with AML patients harboring NPM1 mutations in conjunction with FLT3-ITD mutations (21). In chronic myeloid leukemia (CML) bone marrow (BM) cells, LAPTM4B expression levels were significantly higher than those in normal individuals (22). Similar to observations in solid tumors, CML patients with higher LAPTM4B expression were associated with resistance to tyrosine kinase inhibitor (TKI) treatment (23).
Most studies on LAPTM4B have primarily focused on intracellular signaling in certain types of cancers, and a comprehensive understanding of LAPTM4B in tumorigenesis is still lacking. In this study, we aimed to elucidate the expression, clinical characteristics and immunological characteristics of LAPTM4B across various cancers. In particular, our investigation unveiled a significant correlation between LAPTM4B expression and survival outcomes in Ph+ B-ALL patients. Moreover, it was confirmed that the loss of the Laptm4b impeded BCR-ABL-induced B-ALL progression, both in vitro and in vivo.
Materials and methods
Data acquisition and analysis
The standardized pan-cancer dataset was downloaded from UCSC (https://xenabrowser.net/): TCGA TARGET GTEx (PANCAN, N=19131, G=60499). A log2(x+1) transformation was applied to each expression value, and cancer types with fewer than 3 samples were excluded, resulting in the final expression data for 34 cancer types. Additionally, prognostic data for TCGA were sourced from prior studies (24). Simultaneously, TARGET follow-up data were supplemented from the UCSC database. Samples with a follow-up time of less than 30 days were excluded, and cancer types with fewer than 10 samples were also excluded. The abbreviations section provides the full names and corresponding abbreviations of the tumors.
The Ph+ B-ALL data was downloaded from the GEO database. RMA normalization was performed using the RMA algorithm the NimbleScan 2.5 software. The dataset GSE34861 comprises 191 adult B-ALL samples and 3 normal pre-B samples, and 78 are Ph+ B-ALL samples in B-ALL samples.
Genetic and epigenetic alterations in pan-cancer
Genomic alteration data and methylation data were download from cBioPortal database (https://www.cbioportal.org/). The correlation between LAPTM4B expression and gene promoter methylation was evaluated using Spearman rank correlation. Kaplan−Meier analysis was performed to analyze the relationship between LAPTM4B methylation and the prognosis of patients.
Clinical characteristics LAPTM4B in pan-cancer
We developed the Cox proportional hazards regression model to analyze overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) of LAPTM4B across cancers. Kaplan−Meier analysis was performed to analyze the prognostic significance.
The diagnostic significance of LAPTM4B across cancers was assessed by the Receiver Operator Characteristic (ROC) curve via “pROC” (v1.17.0.1). The diagnosis accuracy was evaluated by the Area under Curve (AUC). The AUC is closer to 1, the diagnosis accuracy is better.
The IC50 values of various compounds in cancer cell lines were obtained from the GDSC dataset (https://www.cancerrxgene.org), to assess the relationship between DLAT and the drug response of tumor cells by the Spearman correlation coefficient. A higher IC50 indicates that cancers are less sensitive to the compounds.
Tumor immune microenvironment analysis
Tumor-infiltrating lymphocytes (TILs) participated in predicting sentinel node status and associated with prognosis (25). ssGSEA scores of the correlation between LAPTM4B and immune cell infiltration for Ph+ B-ALL were calculated using the xCELL algorithm and TILs. Spearman rank correlation was employed to assess the association between LAPTM4B expression and immune cell infiltration in pan-cancer, utilizing the xCELL and CIBERSORT algorithms.
Immune checkpoint-related genes (ICGs) were obtained from a previous study (26). Immune-related genes were downloaded from the TISIDB database (http://cis.hku.hk/TISIDB/index.php). The relationship of immune-related genes and LAPTM4B in Ph+ B-ALL was evaluated using ssGSEA. Immune regulatory genes are distributed in five immune pathways, including chemokine (41 genes), receptor (18 genes), MHC (21 genes), immunoinhibitor (24 genes) and immunostimulator (46 genes). The relationship between immune-related genes and LAPTM4B expression in pan-cancer was evaluated by Spearman rank correlation.
Tumor microenvironment analysis in pan-cancer
We obtained 10,180 tumor samples from a total of 44 tumor types for immune infiltration scores. ESTIMATE was used to reflect the degree of infiltration of stromal or immune cells into tumors. The ESTIMATE algorithm included stromal, immune, and ESTIMATE scores. Spearman rank correlation was used to evaluate the correlation between LAPTM4B expression and these three scores by the R software packages “estimate” and “psych”.
We downloaded all level 4 simple nucleotide variation data of TCGA samples from GDC (https://portal.gdc.cancer.gov/). Tumor mutation burden (TMB) was analyzed using MAftools package (Version 2.8.05) of R software. Tumor stem cell infiltration analysis was performed based on DNA methylation dry score (DNAss) and RNA dry score (RNAss) (27). Spearman rank correlation was used to evaluate the correlation between LAPTM4B expression and TMB, microsatellite instability (MSI), purity, DNAss and RNAss.
Single-cell analysis and enrichment analysis
We conducted the single-cell level expression of LAPTM4B at in leukemia using TISCH2 (28). TISCH2 encompasses 190 tumor scRNA sequence datasets with 6 million cells across 50 cancer types. To assess the functional and signaling aspects with LAPTM4B, we conducted Gene Set Enrichment Analysis (GSEA) on HALLMARK and KEGG pathways. Based on the median expression of LAPTM4B in cancer, the group was divided into high and low expression groups.
The development of LAPTM4B knockout Ph+ B-ALL model and in vitro assay
The B6. LAPTM4Bloxp/loxp mice were generated at Biocytogen Pharmaceuticals (Beijing) Co., Ltd, which were intercrossed with B6.CMV-Cre mice to generate B6. LAPTM4B-/- mice. BCR-ABL induced B-ALL model was developed as previously described (29). Briefly, bone marrow (BM) cells were collected from 8-week-old WT and LAPTM4B-/- mice (n=3) and resuspended with BCR/ABL viral infection medium, centrifugation at 1000g for 90min at 37°C, then cultured at 37°C for 3 h. Then, viral transfected cells were injected into lethally irradicated recipient mice at a dosed of 1x10^5 B cells/mouse via the tail vein.
After the WT or LAPTM4B-/- BM cells were transfected with BCR/ABL, then seeded in DMEM medium containing 10% FBS in 24-well plates at series initial cell numbers of 5x10^5 (500k), 3x10^5 (300k), 1x10^5 (100k), 3x10^4 (30k), 1x10^4 (10k), and 2.5x10^3 (2.5k). Each well cell numbers were adjusted to 1x10^6 cells/well with WT mice BM cells and cultured with DMEM medium containing 10% FBS. The cell number in each well was counted on day 7 post seeding.
Cell cycle experiments by the BrdU incorporation method
BrdU was added directly into the prepared cell medium at final concentration of 10 uM, and incubated for 1h. After collection and washing, cells were suspended with PBS containing 0.5% paraformaldehyde on ice for 20 minutes. Then cells were washed with PBS and resuspended with 70% ethanol overnight. Next day, after washing with PBS, cells were resuspended with 2N HCL/0.5% triton X-100 at room temperature for 20 min to denature. After neutralization with 0.1M sodium borate, cells were suspended with PBS containing 0.5% BSA and 0.5%Tween 20 and stained with anti-BrdU antibody-FitC (BD Biosciences) at room temperature for 20min. The cells were resuspended with PBS containing RNase and incubated at 37 ℃. After adding PI, cells were analyzed using flow cytometry.
Statistical analysis
R software (version 4.2.1) was utilized for this analysis. The Wilcoxon’s test and analysis of variance (ANOVA) were applied for comparisons involving two and multiple groups, respectively. Spearman correlation coefficient was employed for correlation analysis. All experiments were conducted in triplicate.
Results
Expressions and alterations of LAPTM4B in human cancers
In order to examine the expression profile of LAPTM4B in pan-cancers, we evaluated its expression across 34 cancer types using data from TCGA, TARGET, and GTEx databases. Our findings revealed high LAPTM4B expression in 28 cancer types compared to normal tissues, including glioblastoma (GBM), lower-grade glioma (LGG), uterine corpus endometrial carcinoma (UCEC), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), lung adenocarcinoma (LUAD), esophageal carcinoma (ESCA), stomach and esophageal carcinoma (STES), colon adenocarcinoma (COAD), colon adenocarcinoma/Rectum adenocarcinoma esophageal carcinoma (COADREAD), stomach adenocarcinoma (STAD), head and neck squamous cell carcinoma (HNSC), lung squamous cell carcinoma (LUSC), liver hepatocellular carcinoma (LIHC), high-risk Wilms tumor (WT), skin cutaneous melanoma (SKCM), bladder urothelial carcinoma (BLCA), thyroid carcinoma (THCA), rectum adenocarcinoma (READ), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), testicular germ cell tumors (TGCT), uterine carcinosarcoma (UCS), acute lymphoblastic leukemia (ALL), acute myeloid leukemia (LAML), Adrenocortical carcinoma (ACC), and cholangiocarcinoma (CHOL). While, low LAPTM4B expression was observed in 4 cancer types, Pan-kidney cohort (KIPAN), prostate adenocarcinoma (PRAD), kidney renal clear cell carcinoma (KIRC), and kidney chromophobe (KICH) (Figure 1A). Specifically, LAPTM4B exhibited high expression in BLCA, BRCA, CHOL, COAD, ESCA, HNSC, LIHC, LUAD, LUSC, READ, STAD, and UCEC, while showing low expression in KICH, KIRC, PRAD, and THCA compared to adjacent paired normal tissues (Figure 1B). These results suggest that elevated LAPTM4B expression is associated with cancer progression in a majority of cases.
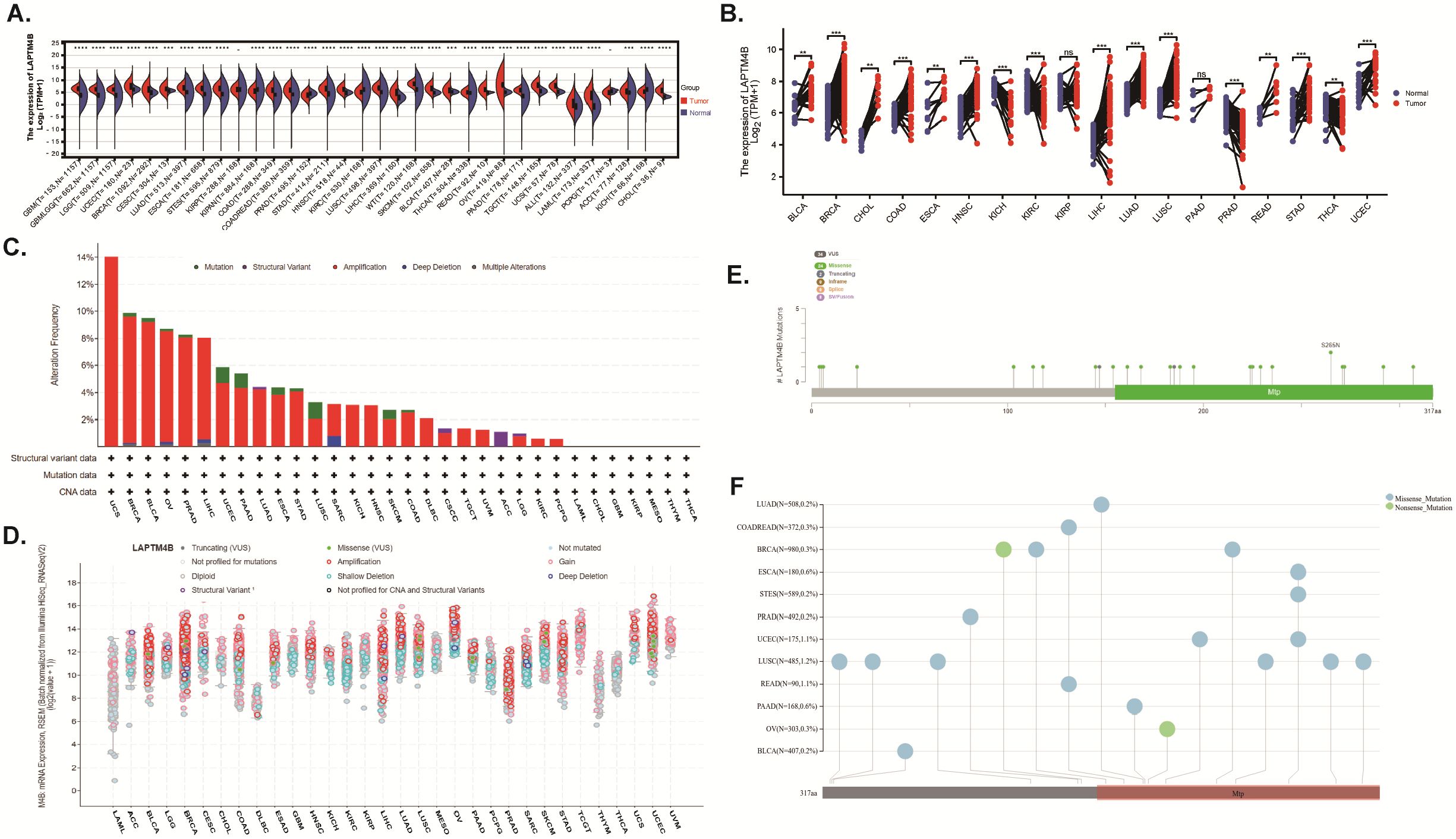
Figure 1. The expression and genetic alteration analysis of LAPTM4B across cancers. (A) LAPTM4B expression levels in tumor and normal samples. (B) Paired differential analysis of LAPTM4B expression in matched tumor and normal samples from TCGA. (C) Bar chart of LAPTM4B mutations across cancers. (D) Mutation counts and types of LAPTM4B across cancers. (E) Mutation diagram of LAPTM4B across protein domains. (F) Landscape of genetic mutation of LAPTM4B across cancers. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns means non significance.
The amplification of LAPTM4B was observed most frequently in UCS, BRCA, BLCA, OV, PRAD, and LIHC, (Figure 1C), and common in most cancers (Figures 1C, D). Moreover, we found 34 mutation sites between amino acids 0 and 317, including 24 missense mutations, 2 truncating, 8 SV/fusion, and S265N as the most frequent mutation sites within LAPTM4B across cancers (Figures 1E, F).
To elucidate potential associations between LAPTM4B and intracellular epigenetic alterations, we examined the status of genomic methylation and the expression of genes involved in mRNA methylation in various types of cancer cells using data from cBioPortal database. We found that there were significant negative correlations between LAPTM4B expression and gene promotor methylation in most tumors (Supplementary Figure 1A). Increased methylation of LAPTM4B mRNA was related to poorer OS in patients with GBM and LGG (Supplementary Figures 1B, C). Furthermore, the relationships between LAPTM4B and genes involved in mRNA m1A, m5C, m6A modifications were evaluated. LAPTM4B expression was significantly positively related to these RNA modification genes in almost all tumors (Supplementary Figure 1D). These results indicated that LAPTM4B could influence tumor development by regulating the repair of RNA and DNA methylation in cancers.
Treatment outcome associated with LAPTM4B alterations in pan-cancers
To evaluate the clinical significance of elevated LAPTM4B expression in various cancers, we conducted a Cox proportional hazards model analysis encompassing OS, DSS, DFI, and PFI. Univariate Cox regression analysis of OS, DSS, PFI, and DFI revealed that LAPTM4B served as a significant risk factor for patients in multiple cancer types, including LIHC, B-ALL, SARC, GBMLGG, SKCM, AML, ACC, UVM, CESC, HNSC, KICH, MESO, UVM, BRCA, and PCPG (Figure 2A). Additionally, Kaplan−Meier survival analyses of OS, DSS, and PFI were further explored across cancers (Figures 2B–D).
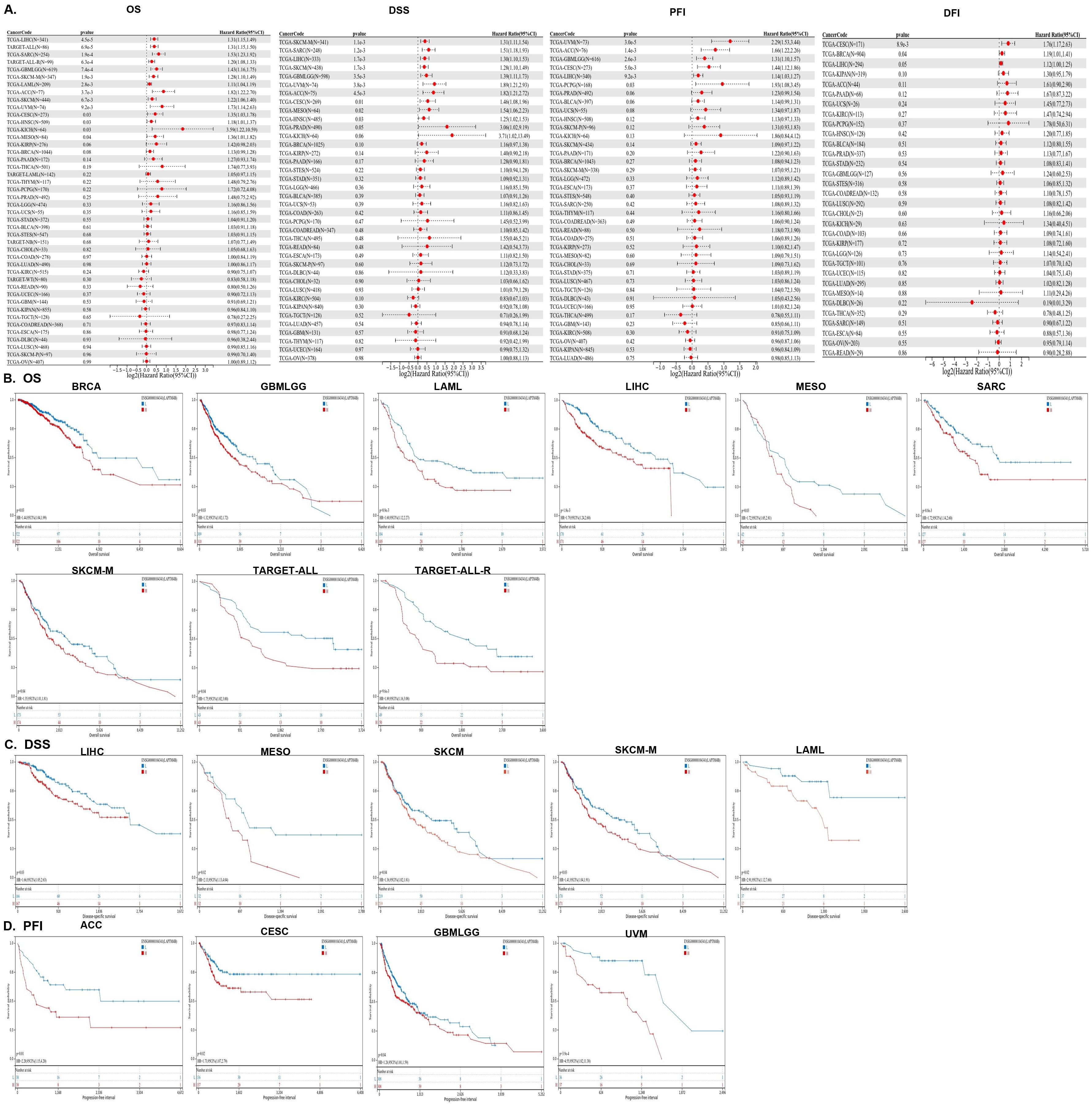
Figure 2. The prognostic analysis of LAMPTM4B across cancers. (A) Forest plots of LAPTM4B by univariate Cox regression analysis across cancers. OS, DSS, PFI, and DFI. Kaplan−Meier curves showing the relationships of LAPTM4B expression with (B) OS. (C) DSS. (D) PFI in pan-cancer.
The performance of the gene signature for diagnostic accuracy was evaluated by the ROC curves. Figure 3 showed that 17 types of cancer had high diagnostic accuracy (AUC >0.9), including CHOL, ESCA, GBM, HNSC, LAML, LGG, LUAD, LUSC, OV, PAAD, READ, SKCM, STAD, TGCT, THYM, UCEC and UCS. These results suggested that LAPTM4B had good diagnostic value in a variety of cancers. The detailed results of all cancers were exhibited in the Supplementary Table 1.
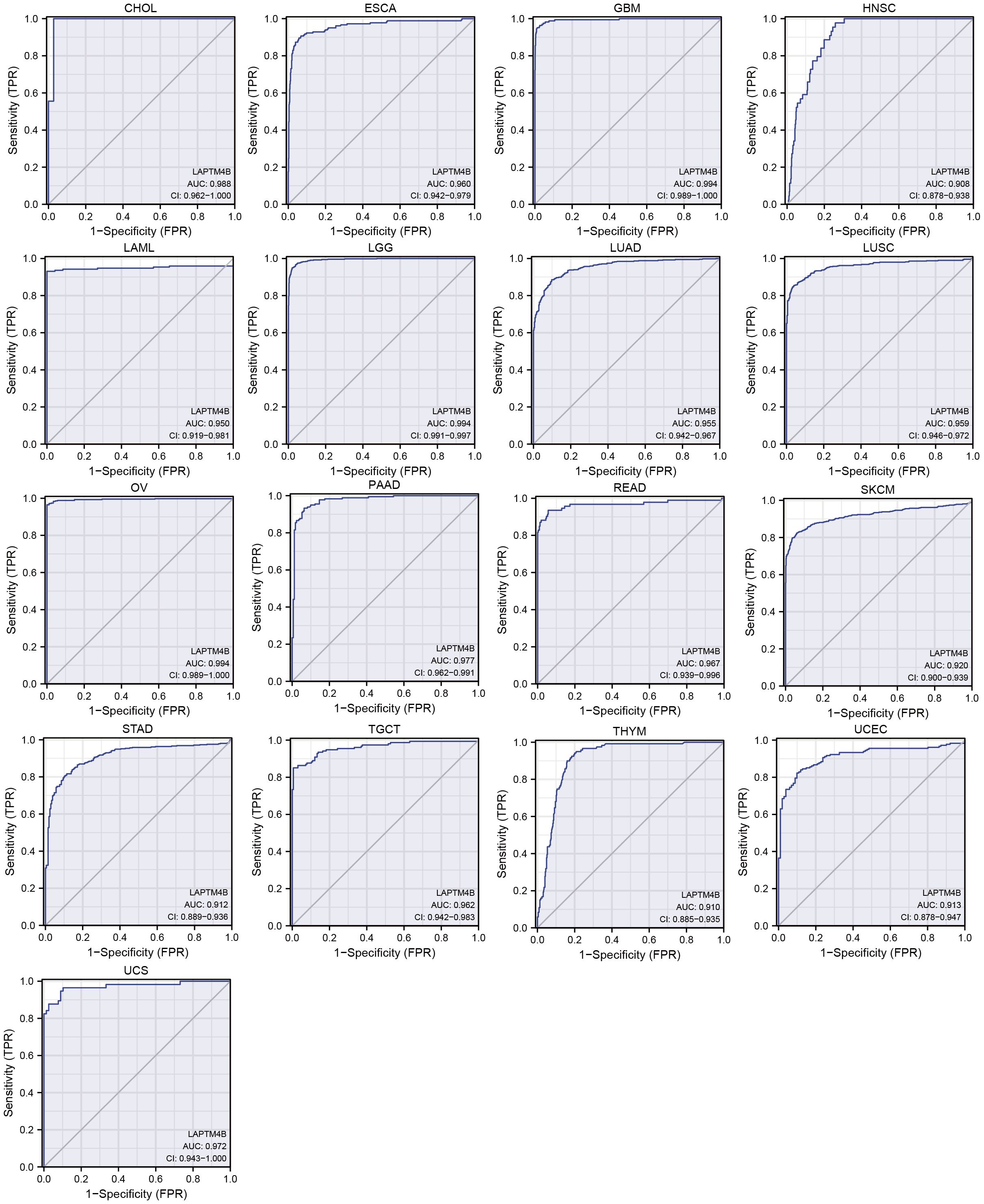
Figure 3. ROC curve of LAPTM4B expression in the TCGA and GTEx database in pan-cancer. Cancers with AUC > 0.9.
To assess the potential correlation between elevated LAPTM4B expression and the drug response of tumor cells, we conducted Spearman correlation coefficient analysis using data from the GDSC dataset. Our findings revealed increased LAPTM4B expression had increased IC50 values of 14 compounds, including rTRAIL, B-Raf inhibitors (PLX-4720, dabrafenib, SB590885), FTI-277 (FTase inhibitor), bexarotene (RXR agonist), dactolisib (PI3K/mTOR inhibitor), luminespib (HSP90 inhibitor), palbociclib (CDK4/6 inhibitor), (5Z)-7-Oxozeaenol (TAK1 inhibitor), QS11 (ARFGAP1 inhibitor), among others, which suggested that increased LAPTM4B lead drug resistance. Conversely, a negative association was observed with elesclomol and afatinib (EGFR/HER2 inhibitor) responses (Table 1). These results suggest that increased LAPTM4B expression may confer resistance to a broad spectrum of therapeutic agents in tumor cells. Moreover, we also found that LAPTM4B was positively correlated with RNAss and DNAss across most of the cancers (Supplementary Figures 2A, B), which indicates that high expression of LAPTM4B might be associated with cancer tumor recurrence and metastasis.
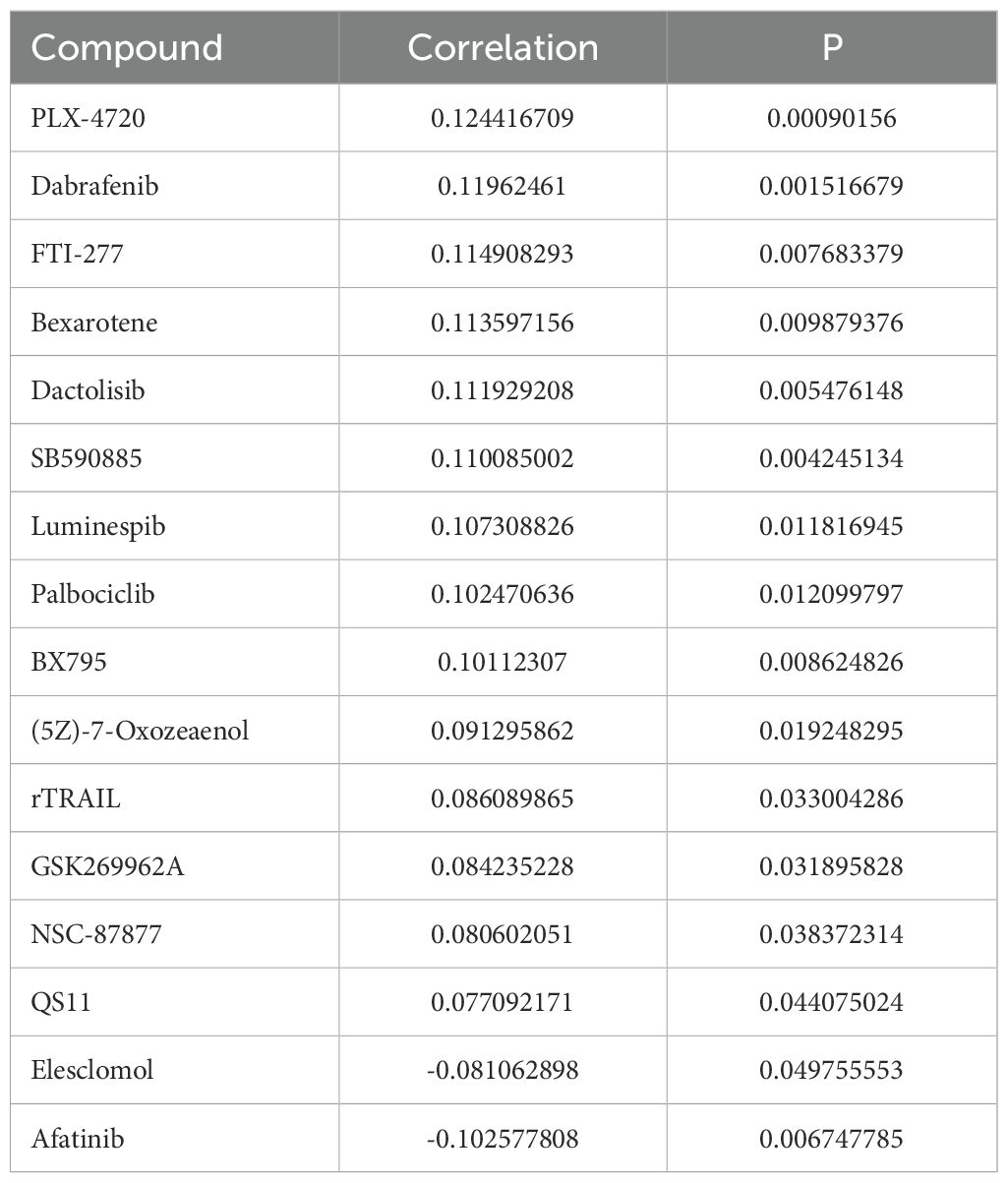
Table 1. Summary of Spearman’s correlation between LAPTM4B expression and drug response (IC50 value) in cancer cell lines based upon the GDSC dataset.
Immune status analysis of LAPTM4B in pan-cancer
To explore the relationship between LAPTM4B expression and immune status in pan-cancer, we conducted a correlation analysis. Overall, we found that LAPTM4B expression was associated with immune subtypes in 19 cancer types and correlated with molecular subtypes in 14 cancer types (Supplementary Figures 3A, B). Additionally, we analyzed stromal and immune cell scores to investigate the relationship between LAPTM4B expression and the tumor immune microenvironment (TIME) across cancers. We observed a positive correlation between LAPTM4B expression and StromalScore, ImmuneScore, and ESTIMATEScore in PAAD, OV, and UVM (Figure 4A). While, LAPTM4B expression showed a negative correlation with these scores in GBM, LGG, LAML, BRCA, CESC, LUAD, STES, SARC, KIRP, KIPAN, STAD, LUSC, WT, SKCM, SKCM-M, THCA, NB, and TCGT (Figure 4A). To explore the correlation between LAPTM4B expression and immune cells, we developed a heat map of LAPTM4B with immune cells by CIBERSORT and xCell. Our result revealed that LAPTM4B was associated with CD8+ T cells, macrophages M2 and Tregs in many cancers, which suggested that high LAPTM4B expression had inhibitory immune microenvironment (Figures 4B, C). Overall, our findings suggested that elevated LAPTM4B expression might be associated with a potential decrease in patients’ immune anti-tumor capabilities.
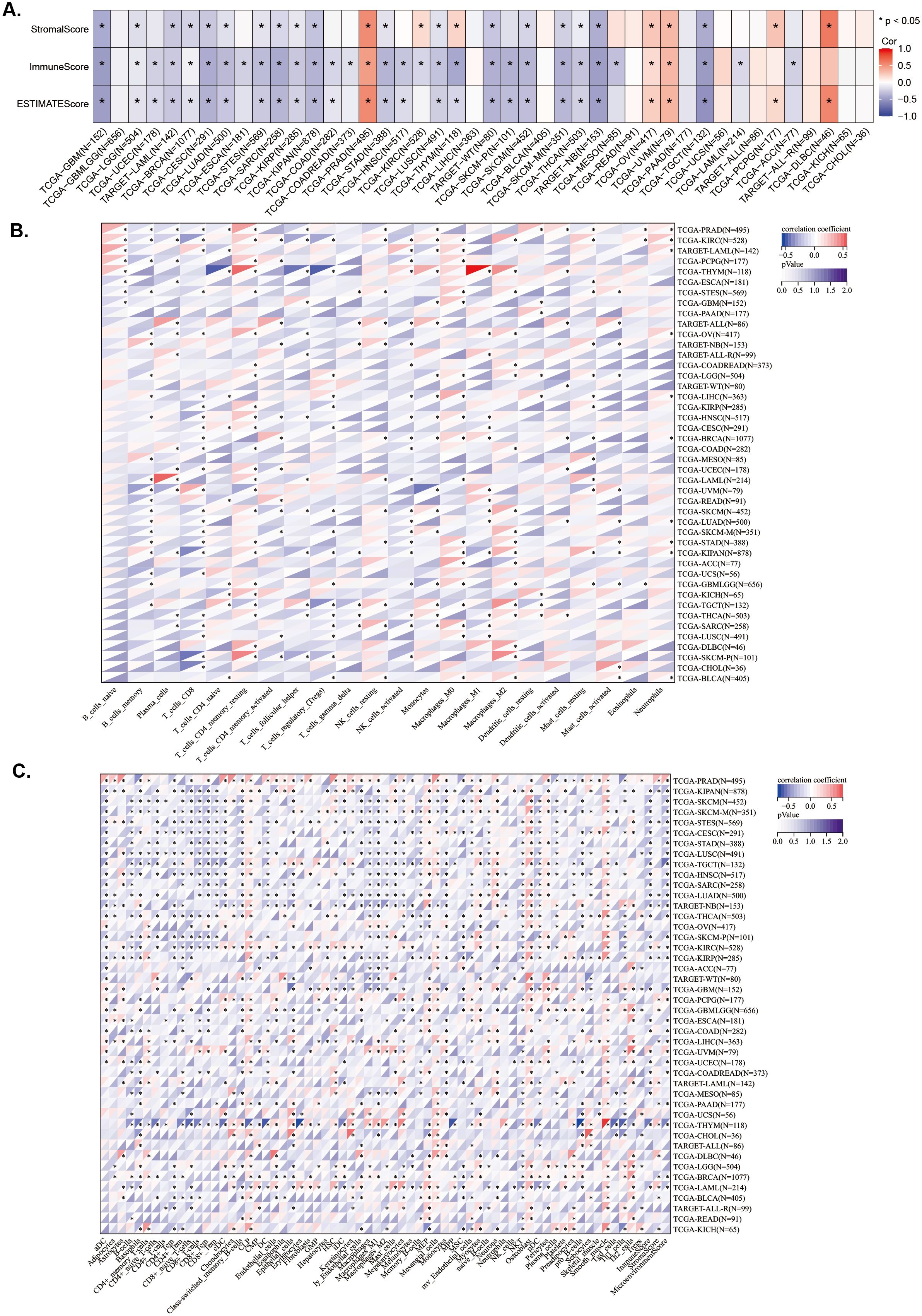
Figure 4. Association between LAPTM4B expression and immune status across cancers. (A) Relationship between LAPTM4B expression and the StromalScore, ImmuneScore, and ESTIMATEScore. Relationships between LAPTM4B expression and the immune cells by CIBERSORT algorithm (B), and the xCell algorithm (C). * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns means non significance.
To investigate whether LAPTM4B expression levels are associated with TMB, MSI, and tumor purity, we conducted analyses using Spearman correlation analysis. The results showed that LAPTM4B expression was positively correlated with TMB in ACC, BRCA, GBMLGG, LAML, LGG, LUAD, PAAD, and THYM, while exhibiting a negative correlation in COAD, COADREAD, ESCA, PRAD, SKCM, and THCA (Figure 5A). The MSI analysis revealed a positive correlation of LAPTM4B expression with MSI in KIPAN, TGCT, and UVM, while a negative correlation in COAD, COADREAD, DLBC, GBMLGG, LGG, PAAD, PRAD, and THCA (Figure 5A). Additionally, LAPTM4B showed a significant correlation with tumor purity, with positive associations in CESC, ESCA, GBM, GBMLGG, HNSC, KIPAN, KIRP, LGG, LUAD, LUSC, SARC, SKCM, STAD, STES, TGCT, and THYM, and negative associations in BLCA, LIHC, OV, PCPG, PRAD, UCS, and UVM (Figures 5A). These findings suggest that LAPTM4B expression might serve as a potential biomarker for immunotherapy.
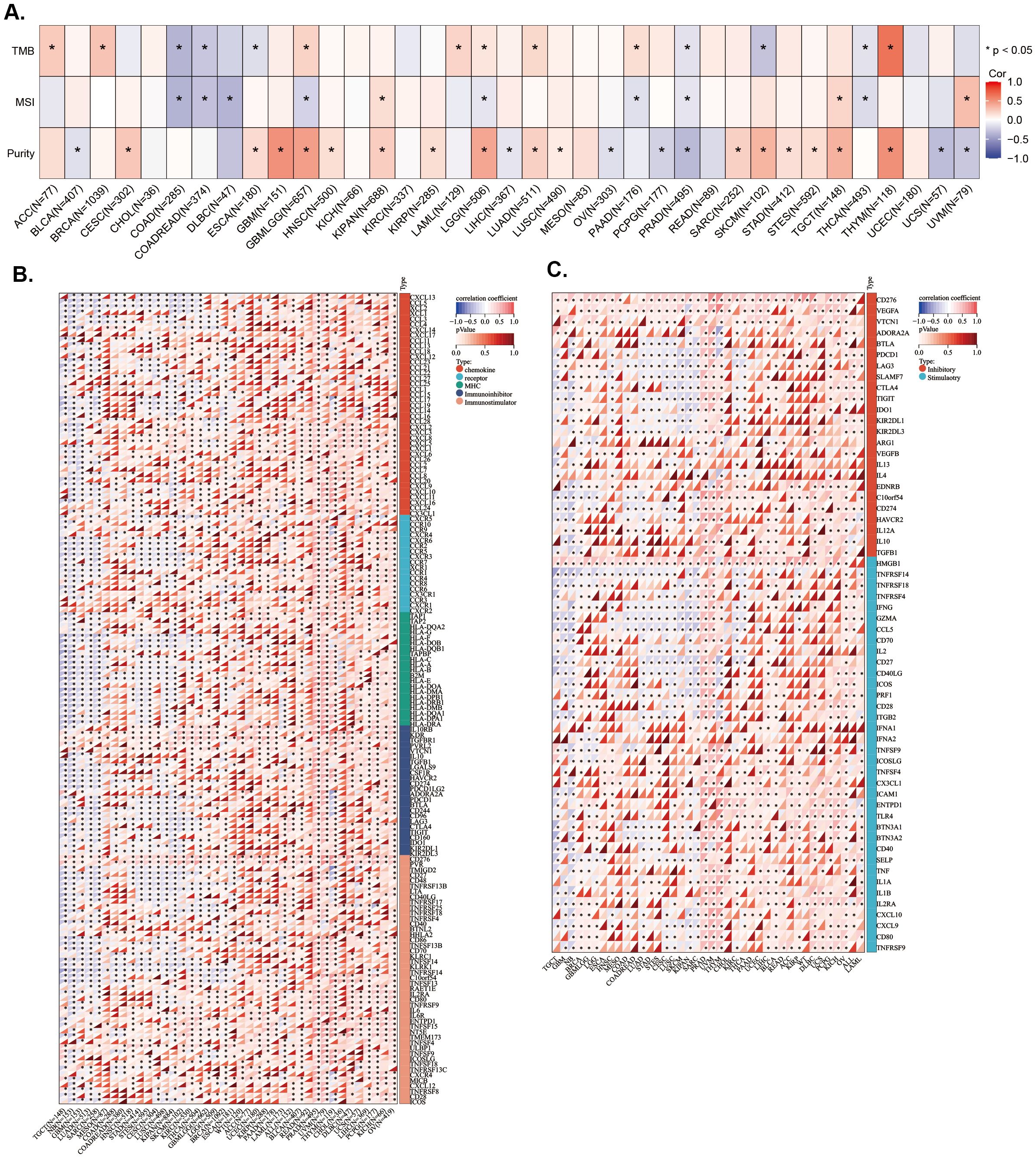
Figure 5. Association between LAPTM4B expression and genes in microenvironment immune cells across cancers. Relationship between LAPTM4B and TBM, MSI, and purity. Correlation between LAPTM4B and immune regulatory genes (A), and immune checkpoint genes (B, C). *p<0.05.
Subsequently, the correlations of expression levels between LAPTM4B and immune checkpoint genes and immune regulatory genes in cancers were also investigated. We found that LAPTM4B expression was positively related to immune regulatory genes in majority tumor types, especially in PRAD, UVM, THYM, LIHC, BLCA, and OV. While, LAPTM4B expression was negatively related to immune regulatory genes in TGCT, GBM, LUAD, SARC, KIPAN, and SKCM (Figure 5B). Additionally, LAPTM4B expression was positively related to immune checkpoint genes in most types of tumors, except for some tumors, which were mainly TGCT, GBM, SKCM, and SARC (Figure 5C). In general, these results suggested that LAPTM4B might regulate immune cell infiltration and immune-related genes functions in most tumor types.
Single-cell and enrichment analysis of LAPTM4B expression in leukemia
LAPTM4B could be a diagnostic, prognostic or therapeutic factor in hepatocellular carcinoma, breast cancer, bladder cancer, renal clear cell carcinoma, nasopharyngeal cancer, lung cancer, osteosarcoma, glioblastoma, gastric cancer, pancreatic ductal adenocarcinoma, ovarian cancer, neck squamous cell carcinomas, prostate cancer, endometrial cancer, colorectal cancer, gallbladder carcinoma, esophageal cancers, cervical carcinoma, melanoma, pancreatic carcinoma and AML and so on (17, 18, 20, 30–51). However, none study was performed in ALL, so we focused on ALL, particularly Ph+ B-ALL, to elucidate and clarify the biological functional characteristics of LAPTM4B. Taking the advantages of single-cell sequencing and open public data, we found that LAPTM4B was expressed mainly in normal HSCs, progenitors, and AML cells (Figures 6A, B). In an ALL sample, we found that LAPTM4B was highly expressed in proerythroblasts, but not malignant cells (Figures 6C, D). Interesting, an analysis based on an expression profiling of 191 B-ALL samples and 3 normal pre-B samples showed that LAPTM4B was more highly expressed in BCR/ABL B-ALL than other subtypes (Figure 6E). Then, we performed the analysis of the function and pathways of LAPTM4B-related genes in Ph+ B-ALL. We found that genes associated with HSCs and leukemia stem cells (LSCs) were up-enriched in high LAPTM4B expression samples (Figures 6F, G), as well as genes associated with cell cycle, DNA replication, MYC target, E2F and G2M checkpoint pathways were also up-enriched in in Ph+ B-ALL (Figures 6H–I).
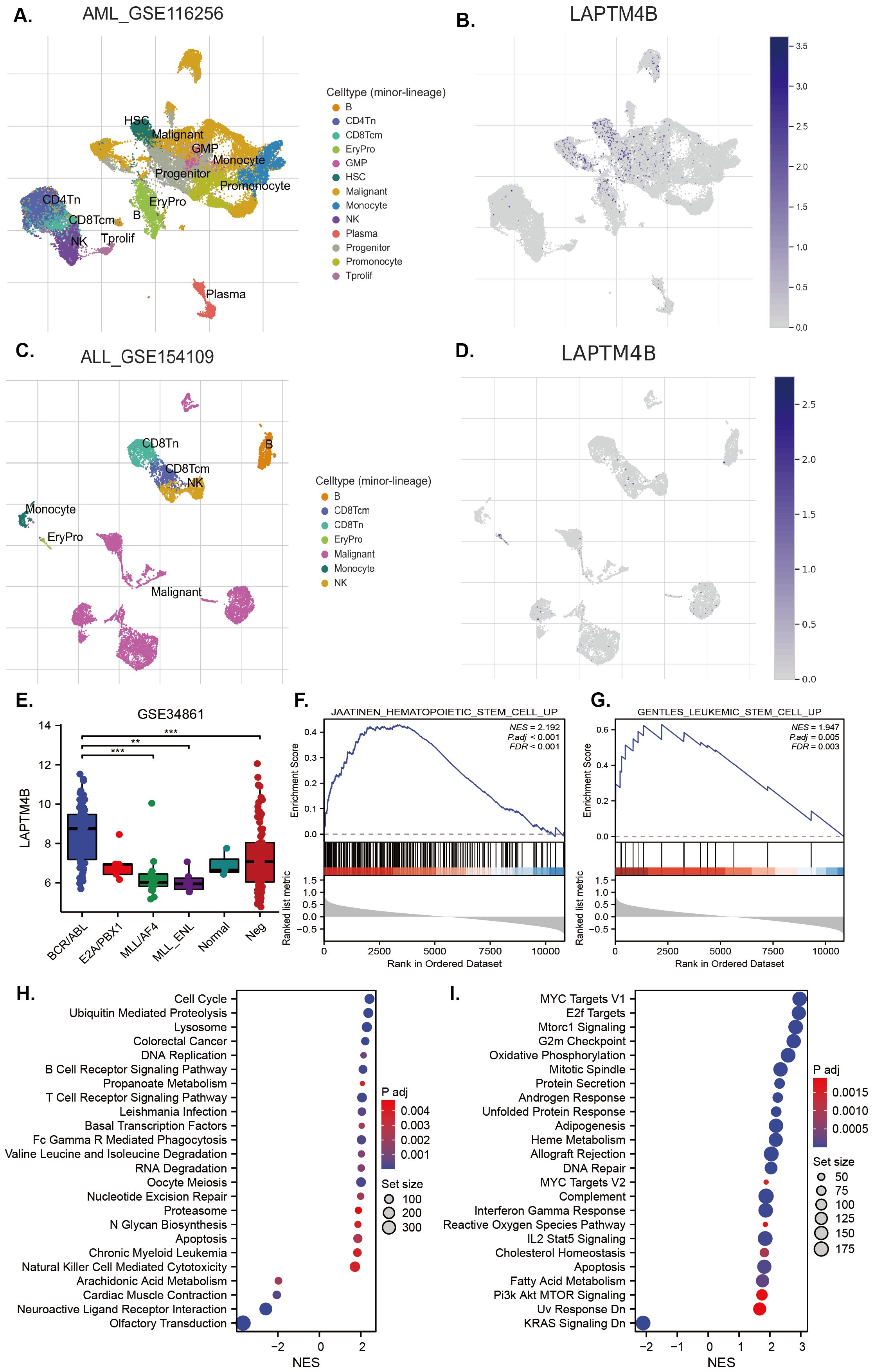
Figure 6. Expression of LAPTM4B at the single-cell level and its related signaling in B-ALL. LAPTM4B expression profiles at single-cell level in AML (A, B), and B-ALL (C, D). The expression of LAPTM4B in different subtypes in B-ALL (E). GSEA analysis of LAPTM4B was related to hematopoietic stem cells (F), and leukemia stem cells (G) in Ph+ B-ALL. KEGG (H) and HALLMARK (I) analysis suggested that LAPTM4B was correlated with the cell cycle, MYC, E2F, and G2M checkpoint pathways in Ph+ B-ALL. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns means non significance.
Relationships between LAPTM4B expression and immune status in Ph+ B-ALL
To evaluate the association of LAPTM4B expression with TME in Ph+ B-ALL, we conducted an ESTIMATE analysis to calculate the stromal score, immune score, ESTIMATE score, and tumor purity within Ph+ B-ALL. We found that LAPTM4B expression was not significantly associated with TME in Ph+ B-ALL (Supplementary Figure 4). Then, the relationship between LAPTM4B and immune cells in Ph+ B-ALL was conducted using xCell algorithm method. The scores of CD4+ memory T cells, CD8+ T cells, HSC, preadipocytes, and Tgd cells were higher; while, the scores of CD4+ Tem, eosinophils, epithelial cells, MSC, and NKT were significantly lower in the high LAPTM4B expression patient samples (Figure 7A). LAPTM4B expression was negatively related to macrophages M2, NKT, mv endothelial cells, and CD4+ Tem; while they were positively correlated with CD4+ memory T cells, Th2 cells, Tgd cells, CD4+ T cells and microenvironment Score (Figure 7B). Moreover, immune infiltration scores of tumor-infiltrating lymphocytes (TILs) type in different LAPTM4B expression groups were also evaluated using ssGSEA. The central memory CD4 T cells, effector memory CD4 T cells, immature B cells, plasmacytoid dendritic cells, and immature dendritic cells were highly expressed in the high LAPTM4B expression patient samples (Figure 7C).
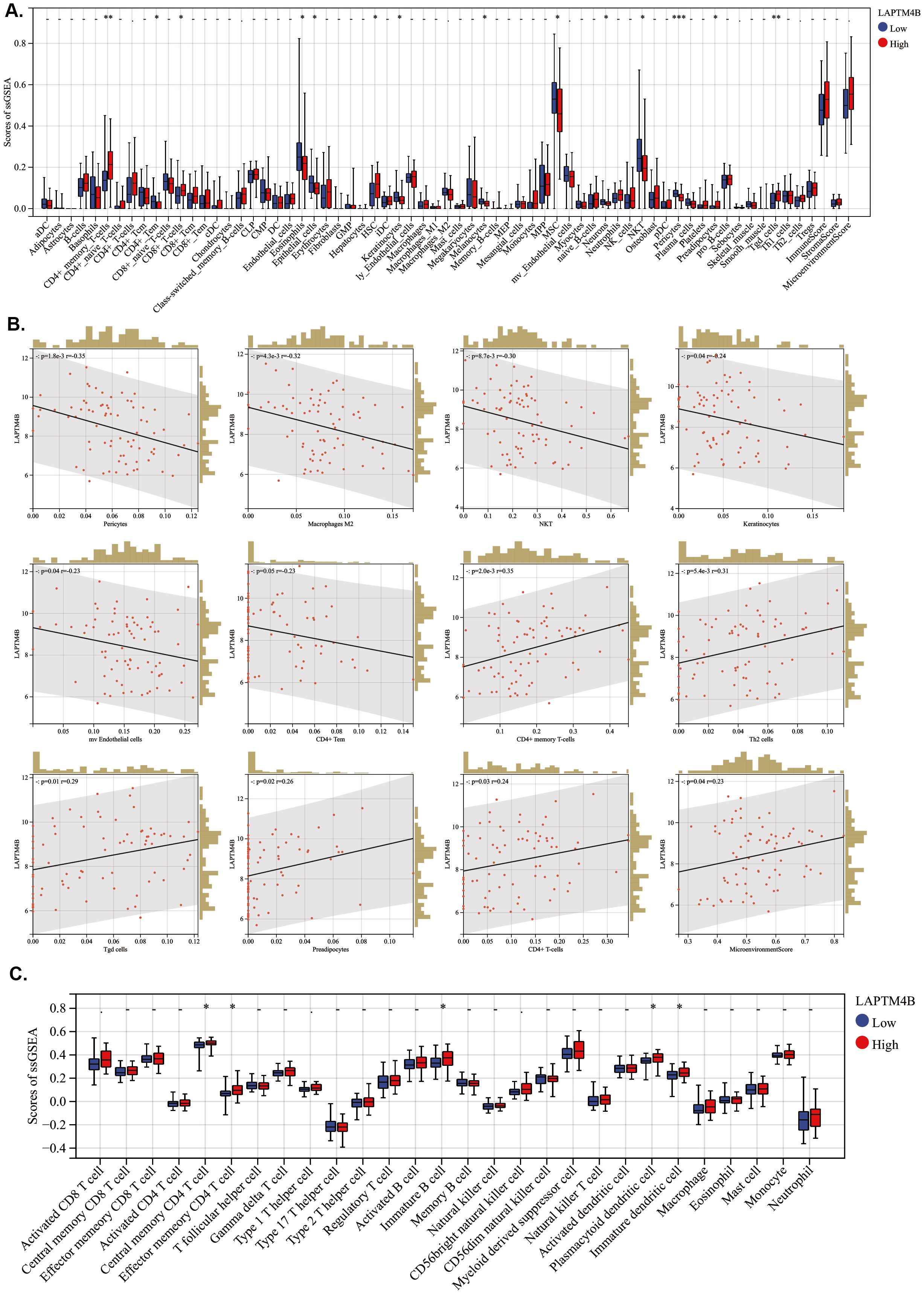
Figure 7. Association between LAPTM4B expression and immune-related cells in ph+ B-ALL. (A) Boxplots of immune cells in different LAPTM4B expression groups by xCell algorithm. (B) Scatterplot of LAPTM4B correlated with immune cells. (C) Boxplots of TILs in different LAPTM4B expression groups. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns means non significance.
The correlation between LAPTM4B expression and immune-related genes was also assessed. As previous reports, there were 79 genes related to immune checkpoint (26). We found that TNFRSF14 and TNFSF14 were lowly expressed in the high LAPTM4B expression samples (Supplementary Figure 5A). Additionally, TNFRSF14 was negatively correlated with LAPTM4B expression; whereas, CTLA4, HLA-E, and ICOS were positively associated with LAPTM4B expression (Supplementary Figure 5B). Moreover, the correlation between LAPTM4B and the chemokine genes was also evaluated. We found that CCL1, CCL11, CCL15, CCL19, CCL21, CCL22, CCL24 and CCL25 were lowly expressed in high LAPTM4B expression samples (Supplementary Figure 5C). But, no significant difference was observed on immunoinhibitory genes, immunostimulatory genes, receptor genes, and MHC genes except for IL6, LTA, ULBP1, and XCR1 (Supplementary Figures 6A-D). These results suggested that the high expression of LAPTM4B might affected immune microenvironment in Ph+ B-ALL.
LAPTM4B deletion impairs the development and progression of Ph+ B-ALL
To instigate the involvement of LAPTM4B in the development of Ph+ B-ALL, we employed a Ph+ B-ALL mouse model. Bone marrow (BM) cells from wild type (WT) or LAPTM4B-/- mice were transfected with retrovirus containing BCR/ABL and then injected into lethally irradicated recipients (Figure 8A). Overall, the survival time of recipients receiving LAPTM4B-/- BM cells was significantly longer than that receiving WT BM cells (Figure 8B). We also monitored the number of leukemic cells with BCR/ABL (represented with GFP and B220) in peripheral blood of mice receiving BCR/ABL-transduced WT or LAPTM4B-/- BM cells on the day 10, 20 and 30 post-BM transplantation. We found that the percentages of B-lymphoid leukemic cells were significantly lower in mice receiving BCR/ABL-transduced WT or LAPTM4B-/- BM cells than in those receiving BCR/ABL-transduced WT BM cells at all time points measured (Figure 8C). To investigate the role of LAPTM4B in BCR/ABL-induced leukemogenesis, we conducted an in vitro assay for proliferation of BCR/ABL transformed BM B-lymphoid progenitors, as described in methods. BCR-ABL-transformed B-lymphoid progenitors from LAPTM4B-/- BM cells exhibited much lower number than it transformed those from WT BM cells (Figure 8D). Further, in vitro Brdu assays for cell proliferation rate showed that LAPTM4B deletion impaired Ph+ B-ALL cell proliferation and caused G0/G1 arrest (Figure 8E). These findings demonstrated that LAPTM4B deletion significantly impaired the development and progression of Ph+ B-ALL.
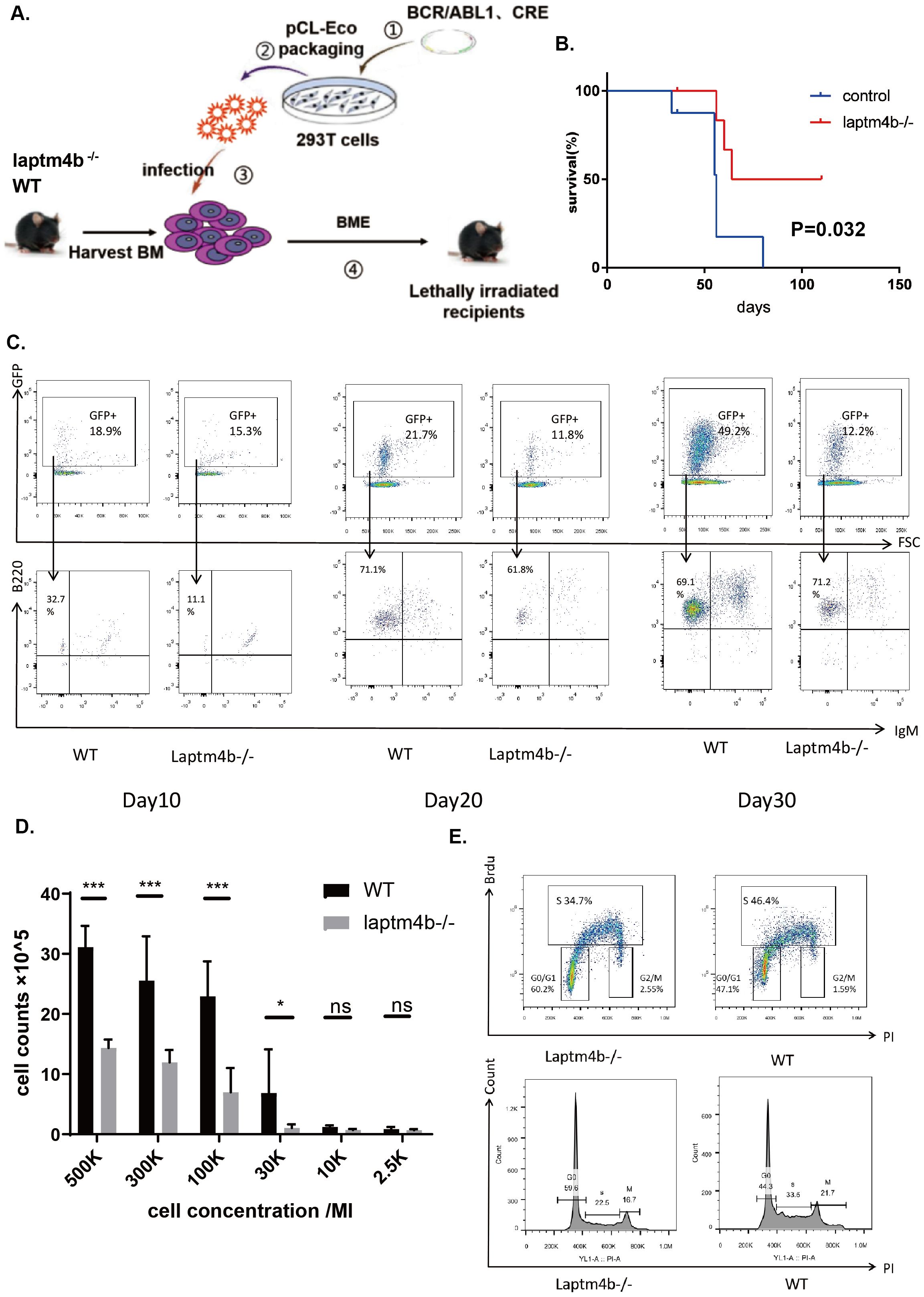
Figure 8. LAPTM4B promoted the development and progression of Ph+ B-ALL. (A) The schematic diagram to establish the mouse model. The Kaplan-Meier curve (B), and the counts of GFP+/B22+/IgM- Ph+ B-ALL cells (C) in LAPTM4B-/- and wild type Ph+ B-ALL mouse model. The cell counting (D), and the cell cycle (E) in LAPTM4B-/- and wild-type Ph+ B-ALL cells in vitro. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, ns means non significance.
Discussion
LAPTM4B is required for lysosomes function, participates in the cell death program, promotes autophagy and tolerance to metabolic stress in cancer cells (52) (53), and is an essential gene for adjuvant drug resistance (15, 54). Our results revealed the role LAPTM4B plays in pan-cancer and Ph+ B-ALL. We determined the expression levels of LATPM4B mRNA in various cancers, and confirmed the most common types of LAPTM4B mutations and their locations. The correlation between epigenetic alterations and LAPTM4B were evaluated. We also assessed the diagnostic, prognostic and therapeutic values of LAPTM4B expression across cancers and differentiated its expression levels across several immune and cellular subtypes of cancers. We associated LAPTM4B expression levels with tumor microenvironment and the infiltration levels of immune cells and genes in various cancers. Besides, the expression of LAPTM4B at the single-cell level and function in B-ALL were explored. We identified that the loss of the LAPTM4B gene impeded BCR-ABL-induced B-ALL progression, both in vitro and in vivo. We also explored the immunological function of LAPTM4B in Ph+ B-ALL.
LAPTM4B was not highly expressed in all tissues, its expression was high in the testis, heart, skeletal muscle and uterus (11), while we found that it was highly expressed in most cancers. And the amplification was the most frequent alteration. However, the expression of LAPTM4B was discovered lowly expressed in KIPAN, KIRC, KICH and PRAD, while the genetic alterations were mainly amplification in these cancers. Therefore, there were other factors affected the expression of LAPTM4B. As we known, abnormal methylation of DNA and RNA promotes various diseases and cancers (55–61). And we found that LAPTM4B expression was significantly related to DNA methylation and RNA modifications in these cancers. Theses might partly explain the inconsistencies between alteration and expression.
A total of 17 types of cancer had high diagnostic accuracy (AUC >0.9), suggesting that LAPTM4B had good diagnostic value. Meanwhile, LAPTM4B was a risk factor in many cancers. Besides, we found that increased LAPTM4B expression led resistance to a broad spectrum of therapeutic agents in tumor cells. These results were consistent with previous studies that LAPTM4B could be a diagnostic, prognostic and therapeutic factor in hepatocellular carcinoma, breast cancer, hepatocellular carcinoma, bladder cancer and renal cell carcinoma and so on (17, 18, 30–38). LAPTM4B expression was different in different molecular or immune subtypes of cancer, which results in different survival in the overall population and particular subtype of cancer. Therefore, immune features should be also considered in the further study.
TMB can reflect the proportion of somatic mutations in tumors (62). MSI refers to the arbitrary length change of microsatellites in tumor tissue due to insertion or deletion of repeat units (63). The purity of the tumor usually related to prognosis (64). TMB, MSI, and tumor purity are emerging biomarkers associated with the immunotherapy response. ESTIMATE reflects the degree of infiltration of stromal or immune cells into tumors. High stemness scores represent the activity of tumor stem cells, and are associated with drug resistance and the continuous proliferation of tumor cells, and are correlated with poorer survival (27). Our results exhibited that LAPTM4B was significant correlated with these indexes. Besides, we showed that LAPTM4B was related to different immune cells in various cancers. In general, the infiltration of activated CD8+ T cells, Tem and Tcm CD8+ cells, and Tem CD4+ cells was associated with good prognosis, whereas MDSCs and Tregs were correlated with bad prognosis (65). A previous study identified that LAPTM4B inhibited human regulatory T cells produced TGF-β1 (66). Besides, LAPTM4B was upregulated in CML TKI-resistant patients (23). Therefore, LAPTM4B might be an immunotherapeutic factor in various cancers.
Then, we found that LAPTM4B is expressed primarily in stem cells at single-cell level in leukemia, and highly expressed in BCR/ABL subtype of B-ALL, and upregulated stem cell pathway in Ph+ B-ALL. A previous study also identified LAPTM4B as a candidate gene related to stemness, which was the downstream target of HOXB4 in hematopoietic progenitor cells (67). Similarly, the study verified that LAPTM4B was closely related to the stemness of HCC (16). These studies illustrated that LAPTM4B might regulate stem cell-related genes. Additionally, LAPTM4B might participated in the signaling pathways of MYC, E2F, cell cycle, and T/B cell receptor. And in the Ph+ B-ALL mouse model, LAPTM4B knockout prolonged survival, inhibited cell proliferation and arrest of G0/G1. The results were similar to the previous study, which exhibited that LAPTM4B promoted the entry of cells from the G1 into the S phase in breast cancer (68).
In B-cell malignancies, leukemic cells can alter the normal microenvironment, in favor of their growth, survival and resistance to cytotoxic therapies (69). Immune cells are important constituents of the tumor stroma and play a crucial role in tumor development and progression60 (70, 71). Our study showed that LAPTM4B expression was associated with HSC, preadipocytes, immature B cells, and immature dendritic cells in Ph+ B-ALL. These results validated that LAPTM4B was related to stem cell pathways as before analyzed. Moreover, the results demonstrated that high LAPTM4B expression had low TNFRSF14 and TNFSF14 expression, and was positively correlated with the CTLA4 and ICOS checkpoint gene. TNFSF14 was an immune-activating gene, mainly expressed in activated T cells, activated natural killer cells and immature dendritic cells (72). TNFRSF14 is a receptor for BTLA, TNFSF14/LIGHT, and homotrimeric TNFSF1, is involved in lymphocyte activation. CTLA4 also known as CD152, is a protein receptor that acts as an immune checkpoint and negatively regulates the immune response (73). ICOS, also called CD278, ICOS were immune checkpoint proteins expressed on activated T cells. Tumor-infiltrating Tregs expressed high levels of cell surface molecules associated with T-cell activation, such as CTLA4, PD-1, LAG3, TIGIT, ICOS, and TNF receptor superfamily members (74). These results suggested that LAPTM4B might influence the efficacy of immunotherapy.
In summary, our study systematically performed a comprehensive pan-cancer analysis of LAPTM4B, and explored the expression, immunological features, and functions of the LAPTM4B gene in the Ph+ B-ALL mouse model. We demonstrated the abnormal expression profiles of LAPTM4B and it was related to clinical diagnosis, prognosis, genetic and epigenetic alterations, immunological features, and drug response. Additionally, we identified that increased LAPTM4B expression was associated with an unfavorable prognosis and promoted the development and progression in Ph+ B-ALL, and was related to immune status. These results could help illustrate the underlying oncogenic role and immunological function of LAPTM4B in cancers.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the Institutional Animal Care and Use Committee (IACUC), West China Hospital, Sichuan University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
HZ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Software, Writing – original draft. YY: Conceptualization, Methodology, Validation, Writing – original draft. WH: Data curation, Methodology, Writing – original draft. LZ: Methodology, Validation, Writing – original draft. YH: Writing – review & editing. TN: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Natural Science Foundation of Sichuan Province (NO. 24NSFSC3632), 1.3.5 Project for Disciplines of Excellence (No. ZYJC21007), 1.3.5 Project of High Altitude Medicine (No. GYYX24003), 1.3.5 Project for Artificial Intelligence (No. ZYAI24039), West China Hospital, Sichuan University, Key Research and Development Program of Sichuan Province (No. 2023YFS0031), National Key Research and Development Program of China (No. 2022YFC2502600, 2022YFC2502603), and National Natural Science Foundation of China (No. 82370192, 82404734, U24A20680).
Acknowledgments
The authors acknowledge the databases in this article for providing their platforms and those contributors for uploading their valuable datasets.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1522293/full#supplementary-material
References
1. Fujii M, Sekine S, Sato T. Decoding the basis of histological variation in human cancer. Nat Rev Cancer. (2023) 24(2):141–58. doi: 10.1038/s41568-023-00648-5
2. Fulton-Ward T, Middleton G. The impact of genomic context on outcomes of solid cancer patients treated with genotype-matched targeted therapies: a comprehensive review. Ann Oncol. (2023) 34:1113–30. doi: 10.1016/j.annonc.2023.10.124
3. Sleeboom JJF, van Tienderen GS, Schenke-Layland K, van der Laan LJW, Khalil AA, Verstegen MMA. The extracellular matrix as hallmark of cancer and metastasis: From biomechanics to therapeutic targets. Sci Trans Med. (2024) 16:eadg3840. doi: 10.1126/scitranslmed.adg3840
4. Mempel TR, Lill JK, Altenburger LM. How chemokines organize the tumour microenvironment. Nat Rev Cancer. (2024) 24:28–50. doi: 10.1038/s41568-023-00635-w
5. Berrell N, Sadeghirad H, Blick T, Bidgood C, Leggatt GR, O'Byrne K, et al. Metabolomics at the tumor microenvironment interface: Decoding cellular conversations. Medicinal Res Rev. (2023) 44(3):1121–46. doi: 10.1002/med.22010
6. Zhang Z, Li X, Wang Y, Wei Y, Wei X. Involvement of inflammasomes in tumor microenvironment and tumor therapies. J Hematol Oncol. (2023) 16:24. doi: 10.1186/s13045-023-01407-7
7. Choi Y, Jung K. Normalization of the tumor microenvironment by harnessing vascular and immune modulation to achieve enhanced cancer therapy. Exp Mol Med. (2023) 55:2308–19. doi: 10.1038/s12276-023-01114-w
8. Lu S, Gan L, Lu T, Zhang K, Zhang J, Wu X, et al. Endosialin in cancer: expression patterns, mechanistic insights, and therapeutic approaches. Theranostics. (2024) 14:379–91. doi: 10.7150/thno.89495
9. Wang C, Zhang Y, Chen W, Wang Y, Xing D. Epidermal growth factor receptor PROTACs as an effective strategy for cancer therapy: A review. Biochim Biophys Acta Rev cancer. (2023) 1878:188927. doi: 10.1016/j.bbcan.2023.188927
10. Malla R, Marni R, Chakraborty A. Exploring the role of CD151 in the tumor immune microenvironment: Therapeutic and clinical perspectives. Biochim Biophys Acta Rev cancer. (2023) 1878:188898. doi: 10.1016/j.bbcan.2023.188898
11. Meng Y, Wang L, Chen D, Chang Y, Zhang M, Xu JJ, et al. LAPTM4B: an oncogene in various solid tumors and its functions. Oncogene. (2016) 35:6359–65. doi: 10.1038/onc.2016.189
12. Peng C, Zhou RL, Shao GZ, Rui JA, Wang SB, Lin M, et al. Expression of lysosome-associated protein transmembrane 4B-35 in cancer and its correlation with the differentiation status of hepatocellular carcinoma. World J gastroenterology. (2005) 11:2704–8. doi: 10.3748/wjg.v11.i18.2704
13. Ji X, Ma H, Du Y. Role and mechanism of action of LAPTM4B in EGFR-mediated autophagy. Oncol letters. (2022) 23:109. doi: 10.3892/ol.2022.13229
14. Wu M, Zhang P. EGFR-mediated autophagy in tumourigenesis and therapeutic resistance. Cancer letters. (2020) 469:207–16. doi: 10.1016/j.canlet.2019.10.030
15. Li Y, Zhang Q, Tian R, Wang Q, Zhao JJ, Iglehart JD, et al. Lysosomal transmembrane protein LAPTM4B promotes autophagy and tolerance to metabolic stress in cancer cells. Cancer Res. (2011) 71:7481–9. doi: 10.1158/0008-5472.CAN-11-0940
16. Liao J, Wang J, Xu Y, Wu Y, Wang M, Zhao Q, et al. LAPTM4B-YAP loop feedback amplification enhances the stemness of hepatocellular carcinoma. iScience. (2023) 26:106754. doi: 10.1016/j.isci.2023.106754
17. Wang L, Wang Y, Zhang Q. Serum LAPTM4B as a potential diagnostic and prognostic biomarker for breast cancer. BioMed Res Int. (2022) 2022:6786351. doi: 10.1155/2022/6786351
18. Su Q, Luo H, Zhang M, Gao L, Zhao F. LAPTM4B promotes the progression of nasopharyngeal cancer. Bosnian J basic Med Sci. (2021) 21:305–12. doi: 10.17305/bjbms.2020.4738
19. Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Li Y, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. (2010) 16:214–8. doi: 10.1038/nm.2090
20. Huang Y, Peng M, Qin H, Li Y, Pei L, Liu X, et al. LAPTM4B promotes AML progression through regulating RPS9/STAT3 axis. Cell signalling. (2023) 106:110623. doi: 10.1016/j.cellsig.2023.110623
21. Huang L, Zhou K, Yang Y, Shang Z, Wang J, Wang D, et al. FLT3-ITD-associated gene-expression signatures in NPM1-mutated cytogenetically normal acute myeloid leukemia. Int J hematology. (2012) 96:234–40. doi: 10.1007/s12185-012-1115-9
22. Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Béné MC, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. (2010) 28:2529–37. doi: 10.1200/JCO.2009.23.4732
23. Singh N, Tripathi AK, Sahu DK, Mishra A, Linan M, Argente B, et al. Differential genomics and transcriptomics between tyrosine kinase inhibitor-sensitive and -resistant BCR-ABL-dependent chronic myeloid leukemia. Oncotarget. (2018) 9:30385–418. doi: 10.18632/oncotarget.25752
24. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. (2018) 173:400–16.e11. doi: 10.1016/j.cell.2018.02.052
25. Hall M, Liu H, Malafa M, Centeno B, Hodul PJ, Pimiento J, et al. Expansion of tumor-infiltrating lymphocytes (TIL) from human pancreatic tumors. J immunotherapy cancer. (2016) 4:61. doi: 10.1186/s40425-016-0164-7
26. Hu FF, Liu CJ, Liu LL, Zhang Q, Guo AY. Expression profile of immune checkpoint genes and their roles in predicting immunotherapy response. Briefings Bioinf. (2021) 22(3). doi: 10.1093/bib/bbaa176
27. Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. (2018) 173:338–54.e15. doi: 10.1016/j.cell.2018.03.034
28. Han Y, Wang Y, Dong X, Sun D, Liu Z, Yue J, et al. TISCH2: expanded datasets and new tools for single-cell transcriptome analyses of the tumor microenvironment. Nucleic Acids Res. (2023) 51:D1425–d31. doi: 10.1093/nar/gkac959
29. Hu Y, Swerdlow S, Duffy TM, Weinmann R, Lee FY, Li S. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc Natl Acad Sci United States America. (2006) 103:16870–5. doi: 10.1073/pnas.0606509103
30. Ren Y, Hu K, Bi L, Wu H, Li Y, Han Y, et al. Noninvasively visualize the expression of LAPTM4B protein using a novel (18)F-labeled peptide PET probe in hepatocellular carcinoma. Nucl Med Biol. (2021) 100-101:52–60. doi: 10.1016/j.nucmedbio.2021.06.003
31. Zhong H, Yuan C, He J, Yu Y, Jin Y, Huang Y, et al. Engineering peptide-functionalized biomimetic nanointerfaces for synergetic capture of circulating tumor cells in an epCAM-independent manner. Analytical Chem. (2021) 93:9778–87. doi: 10.1021/acs.analchem.1c01254
32. Wang L, Meng Y, Zhang QY. LAPTM4B is a novel diagnostic and prognostic marker for lung adenocarcinoma and associated with mutant EGFR. BMC cancer. (2019) 19:293. doi: 10.1186/s12885-019-5506-7
33. Yang Y, Xu J, Zhang Q. Detection of urinary survivin using a magnetic particles-based chemiluminescence immunoassay for the preliminary diagnosis of bladder cancer and renal cell carcinoma combined with LAPTM4B. Oncol letters. (2018) 15:7923–33. doi: 10.3892/ol.2018.8317
34. Gan X, Li S, Wang Y, Du H, Hu Y, Xing X, et al. Aspartate β-hydroxylase serves as a prognostic biomarker for neoadjuvant chemotherapy in gastric cancer. Int J Mol Sci. (2023) 24(6). doi: 10.3390/ijms24065482
35. Meng Y, Wang L, Xu J, Zhang Q. AP4 positively regulates LAPTM4B to promote hepatocellular carcinoma growth and metastasis, while reducing chemotherapy sensitivity. Mol Oncol. (2018) 12:373–90. doi: 10.1002/mol2.2018.12.issue-3
36. Dong X, Tamura K, Kobayashi D, Ando N, Sumita K, Maehara T. LAPTM4B-35 is a novel prognostic factor for glioblastoma. J neuro-oncology. (2017) 132:295–303. doi: 10.1007/s11060-017-2369-0
37. Yang Z, Senninger N, Flammang I, Ye Q, Dhayat SA. Clinical impact of circulating LAPTM4B-35 in pancreatic ductal adenocarcinoma. J Cancer Res Clin Oncol. (2019) 145:1165–78. doi: 10.1007/s00432-019-02863-w
38. Maki Y, Fujimoto J, Lang W, Xu L, Behrens C, Wistuba II, et al. LAPTM4B is associated with poor prognosis in NSCLC and promotes the NRF2-mediated stress response pathway in lung cancer cells. Sci Rep. (2015) 5:13846. doi: 10.1038/srep13846
39. Yan R, Liu D, Guo H, Liu M, Lv D, Björkblom B, et al. LAPTM4B counteracts ferroptosis via suppressing the ubiquitin-proteasome degradation of SLC7A11 in non-small cell lung cancer. Cell Death disease. (2024) 15:436. doi: 10.1038/s41419-024-06836-x
40. Yan R, Liu D, Wang J, Liu M, Guo H, Bai J, et al. miR-137-LAPTM4B regulates cytoskeleton organization and cancer metastasis via the RhoA-LIMK-Cofilin pathway in osteosarcoma. Oncogenesis. (2023) 12:25. doi: 10.1038/s41389-023-00471-5
41. Wang H, Wang Q, Wu Y, Lou J, Zhu S, Xu Y. Autophagy-related gene LAPTM4B promotes the progression of renal clear cell carcinoma and is associated with immunity. Front Pharmacol. (2023) 14:1118217. doi: 10.3389/fphar.2023.1118217
42. Xu Y, Liu Y, Zhou R, Meng F, Gao Y, Yang S, et al. LAPTM4B polymorphisms is associated with ovarian cancer susceptibility and its prognosis. Japanese J Clin Oncol. (2012) 42:413–9. doi: 10.1093/jjco/hys026
43. Kotowski U, Kadletz L, Schneider S, Oberndorfer F, Schnoell J, Gurnhofer E, et al. Overexpression of LAPTM4B-35 is a negative prognostic factor in head and neck squamous cell carcinoma. Sci Rep. (2019) 9:18866. doi: 10.1038/s41598-019-55319-z
44. Zhang H, Wei Q, Liu R, Qi S, Liang P, Qi C, et al. Overexpression of LAPTM4B-35: a novel marker of poor prognosis of prostate cancer. PloS One. (2014) 9:e91069. doi: 10.1371/journal.pone.0091069
45. Meng FL, Yin MZ, Song HT, Yang H, Lou G, Zhou RL. LAPTM4B-35 overexpression is an independent prognostic marker in endometrial carcinoma. Int J gynecological Cancer. (2010) 20:745–50. doi: 10.1111/IGC.0b013e3181e02f90
46. Kang Y, Yin M, Jiang W, Zhang H, Xia B, Xue Y, et al. Overexpression of LAPTM4B-35 is associated with poor prognosis in colorectal carcinoma. Am J surgery. (2012) 204:677–83. doi: 10.1016/j.amjsurg.2012.02.003
47. Zhai G, Yan K, Ji X, Xu W, Yang J, Xiong F, et al. LAPTM4B allele *2 is a marker of poor prognosis for gallbladder carcinoma. PloS One. (2012) 7:e45290. doi: 10.1371/journal.pone.0045290
48. Cheng XJ, Xu W, Zhang QY, Zhou RL. Relationship between LAPTM4B gene polymorphism and susceptibility of colorectal and esophageal cancers. Ann Oncol. (2008) 19:527–32. doi: 10.1093/annonc/mdm469
49. Meng F, Song H, Luo C, Yin M, Xu Y, Liu H, et al. Correlation of LAPTM4B polymorphisms with cervical carcinoma. Cancer. (2011) 117:2652–8. doi: 10.1002/cncr.v117.12
50. Zhang M, Zhou R, Xu J, Zhang Q. Relationship between LAPTM4B gene polymorphism and susceptibility of Malignant melanoma in chinese patients. Trans Oncol. (2014) 7:638–43. doi: 10.1016/j.tranon.2014.07.001
51. Zhang G, Liang Y, Huang Y, Chen Y, Zhou R. Elevated lysosome-associated protein transmembrane-4β-35 is an independent prognostic marker in pancreatic carcinoma. J Int Med Res. (2012) 40:1275–83. doi: 10.1177/147323001204000406
52. Vergarajauregui S, Martina JA, Puertollano R. LAPTMs regulate lysosomal function and interact with mucolipin 1: new clues for understanding mucolipidosis type IV. J Cell Sci. (2011) 124:459–68. doi: 10.1242/jcs.076240
53. Blom T, Li S, Dichlberger A, Bäck N, Kim YA, Loizides-Mangold U, et al. LAPTM4B facilitates late endosomal ceramide export to control cell death pathways. Nat Chem Biol. (2015) 11:799–806. doi: 10.1038/nchembio.1889
54. Li Y, Iglehart JD, Richardson AL, Wang ZC. The amplified cancer gene LAPTM4B promotes tumor growth and tolerance to stress through the induction of autophagy. Autophagy. (2012) 8:273–4. doi: 10.4161/auto.8.2.18941
55. Zhao A, Zhou H, Yang J, Li M, Niu T. Epigenetic regulation in hematopoiesis and its implications in the targeted therapy of hematologic Malignancies. Signal transduction targeted Ther. (2023) 8:71. doi: 10.1038/s41392-023-01342-6
56. Xu X, Peng Q, Jiang X, Tan S, Yang Y, Yang W, et al. Metabolic reprogramming and epigenetic modifications in cancer: from the impacts and mechanisms to the treatment potential. Exp Mol Med. (2023) 55:1357–70. doi: 10.1038/s12276-023-01020-1
57. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. (2017) 18:31–42. doi: 10.1038/nrm.2016.132
58. Qu X, Zhang Y, Sang X, Ren D, Zhao H, Wong STC. Methyladenosine modification in RNAs: from regulatory roles to therapeutic implications in cancer. Cancers. (2022) 14(13). doi: 10.3390/cancers14133195
59. Li M, Tao Z, Zhao Y, Li L, Zheng J, Li Z, et al. 5-methylcytosine RNA methyltransferases and their potential roles in cancer. J Trans Med. (2022) 20:214. doi: 10.1186/s12967-022-03427-2
60. Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. (2016) 76:3446–50. doi: 10.1158/0008-5472.CAN-15-3278
61. Jones PA, Baylin SB. The epigenomics of cancer. Cell. (2007) 128:683–92. doi: 10.1016/j.cell.2007.01.029
62. Choucair K, Morand S, Stanbery L, Edelman G, Dworkin L, Nemunaitis J. TMB: a promising immune-response biomarker, and potential spearhead in advancing targeted therapy trials. Cancer Gene Ther. (2020) 27:841–53. doi: 10.1038/s41417-020-0174-y
63. Bonneville R, Krook MA, Kautto EA, Miya J, Wing MR, Chen HZ, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. (2017) 2017. doi: 10.1200/PO.17.00073
64. Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The immune landscape of cancer. Immunity. (2018) 48:812–30.e14. doi: 10.1016/j.immuni.2018.03.023
65. Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. (2017) 18:248–62. doi: 10.1016/j.celrep.2016.12.019
66. Huygens C, Liénart S, Dedobbeleer O, Stockis J, Gauthy E, Coulie PG, et al. Lysosomal-associated transmembrane protein 4B (LAPTM4B) decreases transforming growth factor β1 (TGF-β1) production in human regulatory T cells. J Biol Chem. (2015) 290:20105–16. doi: 10.1074/jbc.M115.655340
67. Lee HM, Zhang H, Schulz V, Tuck DP, Forget BG. Downstream targets of HOXB4 in a cell line model of primitive hematopoietic progenitor cells. Blood. (2010) 116:720–30. doi: 10.1182/blood-2009-11-253872
68. Tao D, Liang J, Pan Y, Zhou Y, Feng Y, Zhang L, et al. In vitro and in vivo study on the effect of lysosome-associated protein transmembrane 4 beta on the progression of breast cancer. J Breast cancer. (2019) 22:375–86. doi: 10.4048/jbc.2019.22.e43
69. Hughes AM, Kuek V, Kotecha RS, Cheung LC. The bone marrow microenvironment in B-cell development and Malignancy. Cancers. (2022) 14(9). doi: 10.3390/cancers14092089
70. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. (2019) 79:4557–66. doi: 10.1158/0008-5472.CAN-18-3962
71. Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer letters. (2020) 470:126–33. doi: 10.1016/j.canlet.2019.11.009
72. Ware CF, Sedý JR. TNF Superfamily Networks: bidirectional and interference pathways of the herpesvirus entry mediator (TNFSF14). Curr Opin Immunol. (2011) 23(5):627–31. doi: 10.1016/j.coi.2011.08.008
73. Patwekar M, Sehar N, Patwekar F, Medikeri A, Ali S, Aldossri RM, et al. Novel immune checkpoint targets: A promising therapy for cancer treatments. Int Immunopharmacol. (2024). doi: 10.1016/j.intimp.2023.111186
Keywords: LAPTM4B, Ph+ B-ALL, pan-cancer, diagnosis, prognosis
Citation: Zhou H, Yi Y, He W, Zheng L, Hu Y and Niu T (2025) A comprehensive prognostic and immune analysis of LAPTM4B in pan-cancer and Philadelphia chromosome-positive acute lymphoblastic leukemia. Front. Immunol. 16:1522293. doi: 10.3389/fimmu.2025.1522293
Received: 04 November 2024; Accepted: 11 February 2025;
Published: 28 February 2025.
Edited by:
Hai Fang, Shanghai Jiao Tong University, ChinaReviewed by:
Jing-dong Zhou, Jiangsu University Affiliated People’s Hospital, ChinaLin Fu, The Second Affiliated Hospital of Guangzhou Medical University, China
Copyright © 2025 Zhou, Yi, He, Zheng, Hu and Niu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Niu, bml1dGluZ0B3Y2hzY3UuY24=; Yiguo Hu, aHV5aWd1b0BzY3UuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Hui Zhou
Hui Zhou Yuyao Yi
Yuyao Yi Wei He3,4
Wei He3,4 Li Zheng
Li Zheng Yiguo Hu
Yiguo Hu Ting Niu
Ting Niu