- Department of Surgery, the Second Affiliated Hospital of Jiaxing University, Jiaxing, Zhejiang, China
Hepatocellular carcinoma (HCC) represents a major global health challenge, characterized by its complex immune microenvironment that plays a pivotal role in tumor progression and therapeutic response. Long non-coding RNAs (lncRNAs) have emerged as critical regulators of various biological processes, including gene expression and immune cell function. This review explores the multifaceted roles of lncRNAs in modulating the immune microenvironment of HCC. We discuss how lncRNAs influence the infiltration and activation of immune cells, shape cytokine profiles, and regulate immune checkpoint molecules, thereby affecting the tumor’s immunogenicity and response to immunotherapy. Furthermore, we highlight specific lncRNAs implicated in immune evasion mechanisms and their potential as biomarkers and therapeutic targets. By elucidating the intricate interplay between lncRNAs and the immune landscape in HCC, this review aims to provide insights into novel strategies for enhancing immunotherapeutic efficacy and improving patient outcomes.
1 Introduction
HCC is the most common type of primary liver cancer and ranks as one of the leading causes of cancer-related mortality worldwide (1, 2). Its development is often associated with underlying liver diseases, including chronic hepatitis B and C infections, cirrhosis, and metabolic disorders (3–5). Despite advances in surgical and systemic therapies, the prognosis for patients with HCC remains poor, largely due to the tumor’s aggressive nature and its ability to evade the host immune response (6).
The tumor microenvironment (TME) plays a crucial role in HCC progression and therapeutic resistance (7). It is composed of various cell types, including tumor cells, immune cells, fibroblasts, and endothelial cells, all of which interact dynamically to influence tumor behavior (7–9). These immune cells can either promote anti-tumor immunity or facilitate tumor growth through immunosuppressive mechanisms, contributing to the challenges faced in effective treatment (8, 10).
At present, cancer immunotherapies such as immune checkpoint inhibitors (ICIs) (such as anti-PD-1/PD-L1 monoclonal antibody) and CAR-T cell therapy have made breakthroughs in a variety of tumors, but their efficacy in HCC is still limited to a certain extent (11–13). Although checkpoint inhibitors partially improve the survival of patients with advanced HCC, their overall response rate is less than 20% and they are prone to secondary drug resistance (14). CAR-T therapy is effective in hematologic tumors, but progress is slow due to the high heterogeneity of HCC, immunosuppressive microenvironment and physical barrier of solid tumors (15). In addition, HCC is often accompanied by chronic inflammation and cirrhosis, leading to depletion of immune cell function and increased infiltration of immunosuppressive cells (such as Treg and MDSC), further weakening treatment response (8). These limitations highlight the urgent need for in-depth analysis of HCC-specific immune regulatory mechanisms, such as lncrNA-mediated immune escape, to develop combination strategies.
Recent studies have identified lncRNAs as critical regulators of gene expression and cellular processes in cancer (16, 17). These RNA molecules, which are over 200 nucleotides in length and do not code for proteins, have been shown to play diverse roles in modulating cellular behavior, including apoptosis, proliferation, and immune responses (18, 19). Emerging evidence suggests that lncRNAs are involved in shaping the immune microenvironment of various tumors, including HCC, by regulating the functions of immune cells and influencing the expression of immune-related genes (20).
In this review, we will explore the intricate relationships between lncRNAs and the immune microenvironment in HCC. We will discuss how lncRNAs contribute to immune cell infiltration, modulation of cytokine secretion, and regulation of immune checkpoints, thereby impacting the tumor’s immunogenicity and response to therapies. By understanding these mechanisms, we aim to highlight the potential of lncRNAs as biomarkers and therapeutic targets in HCC, paving the way for novel strategies to enhance immunotherapy outcomes.
2 HCC and immune microenvironment
The immune microenvironment in HCC is a complex network that significantly influences tumor progression and therapeutic outcomes (3, 21). Comprised of various immune and stromal cell types, the immune landscape of HCC is characterized by both pro-tumor and anti-tumor activities. A critical feature of this microenvironment is its ability to foster immune evasion, which allows tumors to grow and metastasize (7, 22).
In HCC, diverse immune cell populations, including T cells, natural killer (NK) cells, dendritic cells, and myeloid-derived suppressor cells (MDSCs), contribute to the tumor immune landscape. The balance between these cell types can determine the effectiveness of the immune response against tumors. A high infiltration of MDSCs and regulatory T cells (Tregs) is often associated with poor prognosis, as these cells can suppress the activity of effector T cells and promote an immunosuppressive microenvironment (23, 24). Conversely, a robust presence of activated cytotoxic T lymphocytes (CTLs) and NK cells is linked to better patient outcomes, as these cells can effectively target and destroy tumor cells (25–27).
The immune microenvironment in HCC is heavily influenced by the local cytokine milieu, which can dictate the behavior of immune and tumor cells (28, 29). Pro-inflammatory cytokines such as IL-6 and TNF-α often dominate the microenvironment, promoting tumor proliferation and facilitating immune evasion (30, 31). These cytokines can enhance the recruitment of immunosuppressive cells while inhibiting the function of effector immune cells. Moreover, a dysregulated cytokine profile can lead to chronic inflammation, which is a hallmark of HCC development. This chronic inflammatory state not only supports tumor growth but also contributes to the creation of an immune environment that is unfavorable for effective anti-tumor responses (32, 33).
Immune checkpoint molecules, including PD1, PD-L1, and CTLA-4, play a crucial role in regulating immune responses in HCC (34, 35). The expression of these checkpoints can be upregulated in response to theTME, leading to diminished T cell activation and function (36).
The upregulation of PD-L1 on tumor cells, in particular, has been associated with poor clinical outcomes in HCC, as it enables cancer cells to evade immune detection (37, 38). ICIs targeting PD1 and CTLA-4 have shown promise in clinical settings, offering new avenues for immunotherapy in HCC (39, 40). However, the effectiveness of these therapies can be hindered by the complexity of the immune microenvironment and the various mechanisms of immune evasion employed by HCC.
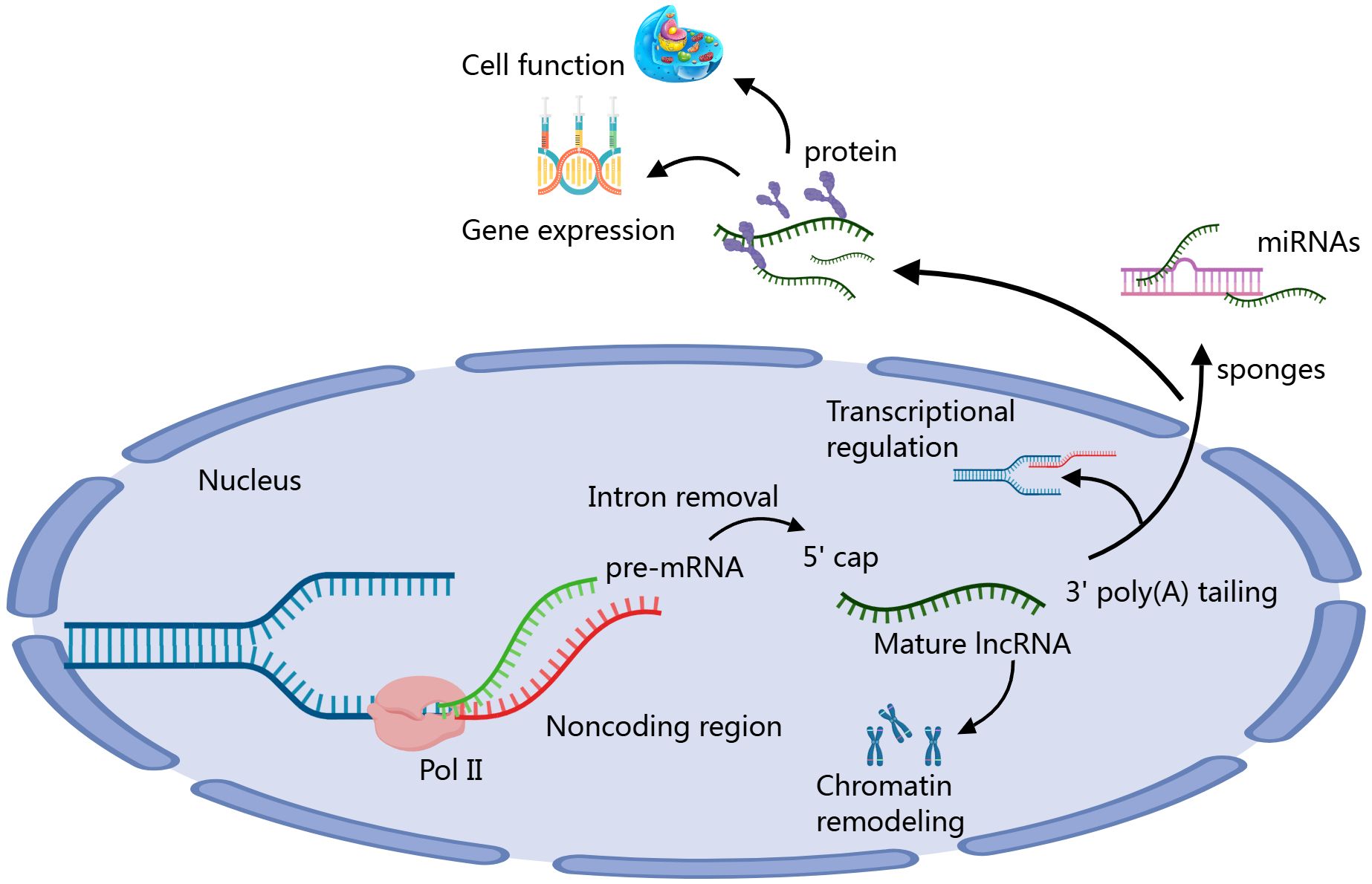
lncRNAs have emerged as important regulators of the immune microenvironment in HCC, influencing various aspects of immune cell behavior and interactions (41). The formation process of lncRNAs involves several steps. First, lncRNAs are transcribed from genomic DNA by RNA polymerase II, typically existing adjacent to or independently from protein-coding genes. After transcription, lncRNAs undergo processing, including 5’ capping, 3’ polyadenylation, and splicing, to become mature. Mature lncRNAs can function in the nucleus or cytoplasm, regulating gene expression and cellular functions through interactions with DNA, RNA, or proteins. They can act as “sponges” to absorb specific miRNAs or participate in transcriptional regulation and chromatin remodeling, thus playing various important biological roles (Figure 1) (42, 43). These lncRNAs can modulate the infiltration and activity of immune cells, alter cytokine production, and regulate immune checkpoint expression (44). For instance, certain lncRNAs have been shown to enhance the recruitment of immunosuppressive cells, such as MDSCs and Tregs, thereby promoting an immunosuppressive environment conducive to tumor growth. Conversely, other lncRNAs may activate effector immune responses by enhancing the function of cytotoxic T cells and NK cells (45–47). By shaping the immune landscape, lncRNAs play a critical role in determining the efficacy of immunotherapies in HCC. Targeting specific lncRNAs could offer a novel approach to reprogram the immune microenvironment, restore anti-tumor immunity, and improve patient responses to existing therapies (48). Thus, further investigation into the functional roles of lncRNAs in HCC’s immune landscape is essential for developing innovative therapeutic strategies.
3 LncRNA regulation of the immune Microenvironment in HCC
3.1 Regulation of immune cells by lncRNAs in HCC
As mentioned above, in the complex immune microenvironment of HCC, lncRNAs play a crucial regulatory role. Recent studies have demonstrated that lncRNAs not only participate in the biological processes of tumor cells but also significantly influence the immune evasion mechanisms of HCC by modulating the functions and interactions of immune cells (Figure 2A). These lncRNAs can shape the TME through various mechanisms, including affecting the differentiation, activation, and functional status of immune cells, as well as regulating intercellular signaling. In this chapter, we will delve into the regulatory mechanisms of lncRNAs on specific immune cells, such as T cells, B cells, macrophages, and dendritic cells, analyze their potential roles in HCC, and discuss the prospects of targeting them for therapeutic interventions.
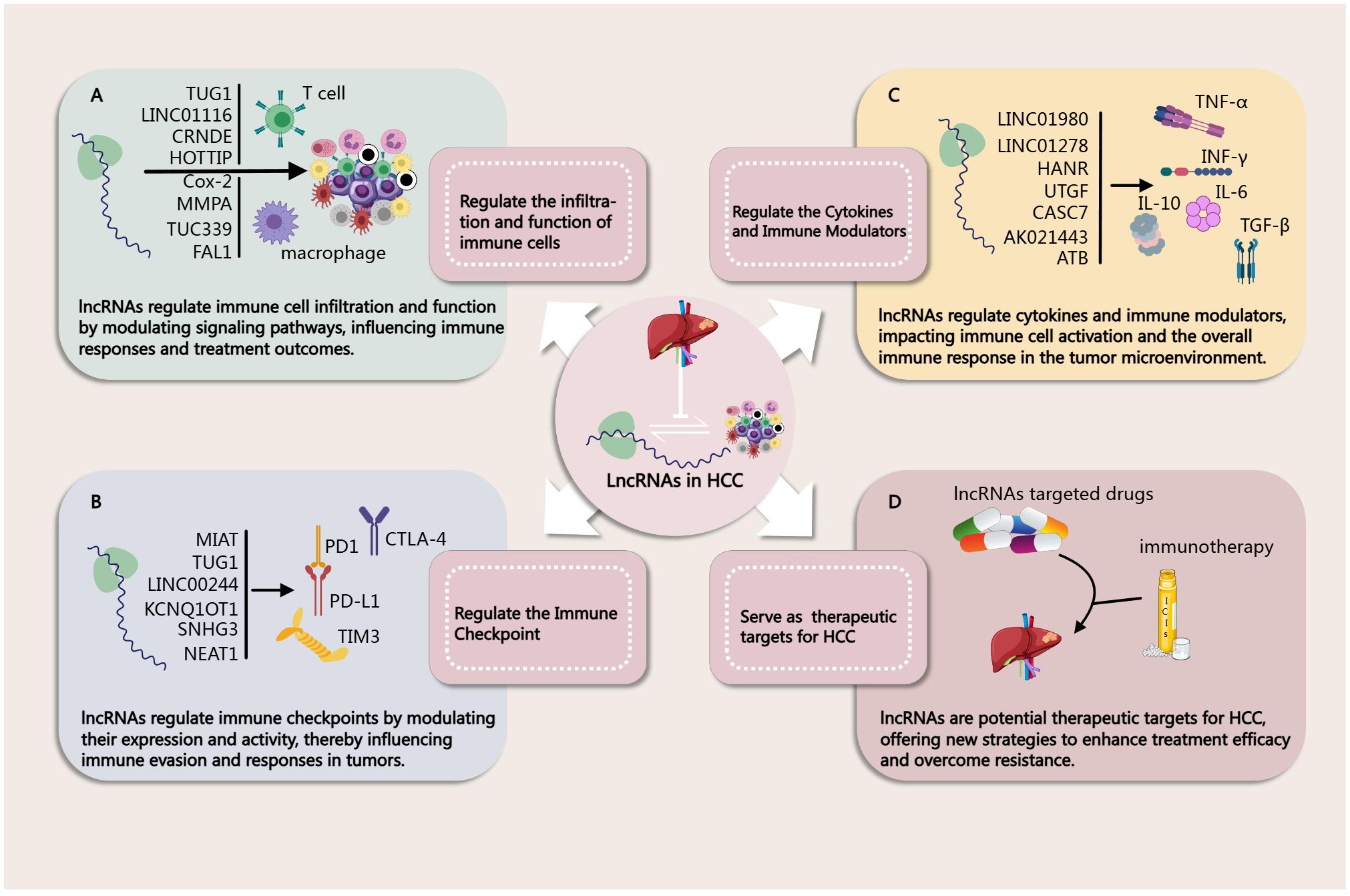
Figure 2. (A) Regulation of immune Cells by lncRNAs in HCC (46, 49, 50, 52, 65–67, 72). (B) Regulation of Immune Checkpoint by lncRNAs in HCC (44, 46, 47, 63, 91, 92). (C) Regulation of Cytokines and Immune Modulators by lncRNAs in HCC (94–96, 98–100, 106). (D) lncRNAs as Biomarkers in HCC Immunotherapy (46, 47, 65, 96).
3.1.1 T cell
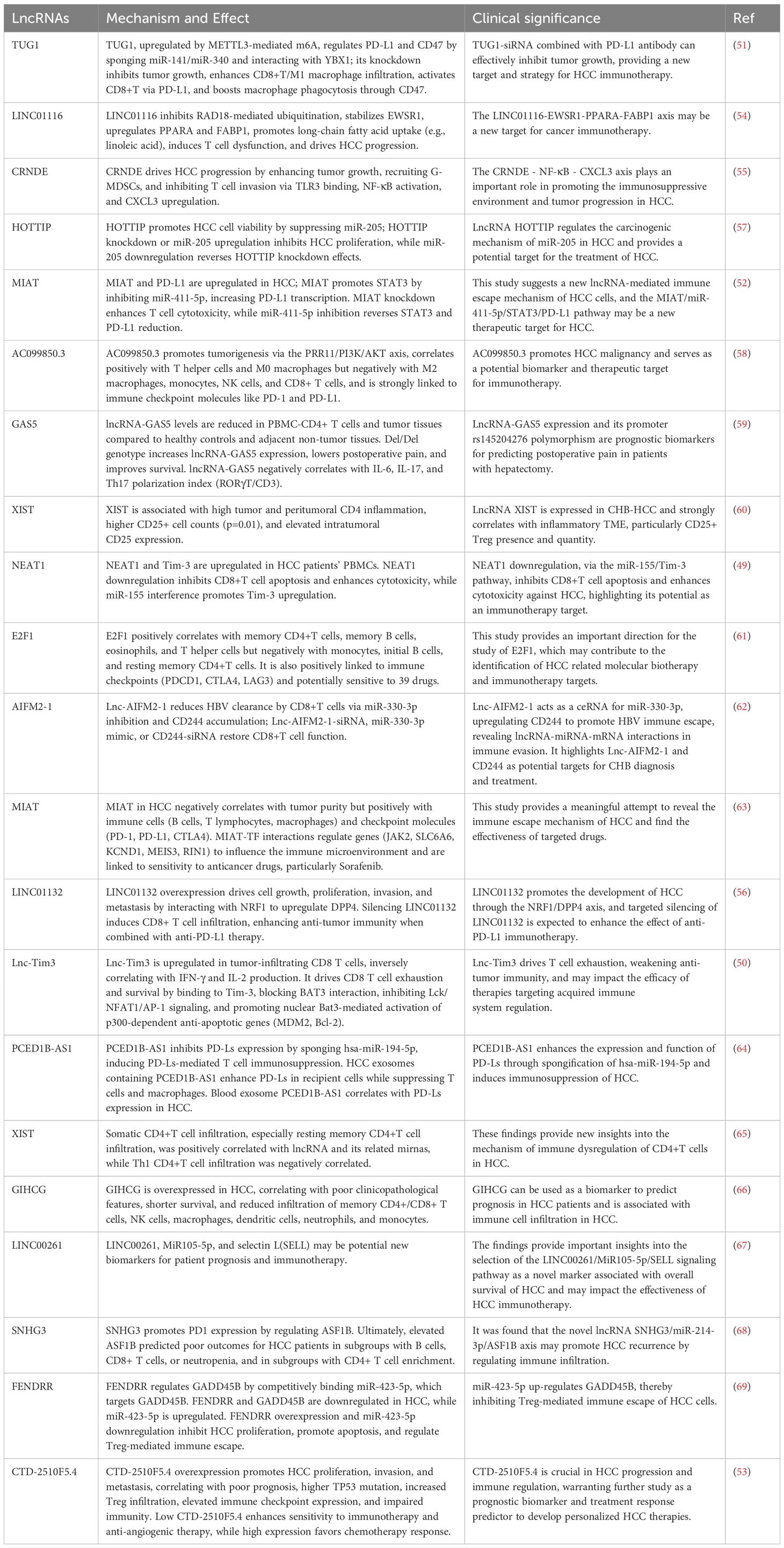
T cells play a crucial role in the immune response of HCC, and lncRNAs regulate T cell function and activity through various mechanisms. They influence T cell development, differentiation, and effector functions, thereby altering the immune balance within the TME. Previous studies have demonstrated the overexpression of several oncogenic lncRNAs in HCC, including TUG1, LINC01116, CRNDE, MIAT, E2F1, LINC01132, and Lnc-Tim3. These lncRNAs can influence T cell activity and function through various pathways or cascading effects, thereby promoting tumor progression. lncRNAs often modulate T cell co-stimulation by regulating downstream miRNAs. Research has shown that NEAT1 and Tim-3 are significantly upregulated in the PBMCs of HCC patients. Downregulation of NEAT1 can inhibit the apoptosis of CD8+ T cells and enhance their cytolytic activity against HCC cells, while interference with miR-155 shows the opposite effect, promoting the upregulation of Tim-3. This study confirms the binding and interaction of NEAT1 with miR-155 in CD8+ T cells. By regulating the miR-155/Tim-3 pathway, downregulation of NEAT1 can inhibit CD8+ T cell apoptosis and enhance their cytolytic activity against HCC, indicating that NEAT1 is an effective target for improving immunotherapy outcomes (49). Lnc-Tim3 can specifically bind to Tim-3, preventing its interaction with Bat3, thereby inhibiting downstream signaling in the Lck/NFAT1/AP-1 pathway, leading to the nuclear localization of Bat3. This enhances the transcriptional activation of pro-apoptotic genes (such as MDM2 and Bcl-2) by p300-dependent p53 and RelA, promoting T cell exhaustion. This suggests that Lnc-Tim3 and its associated signaling pathways may impact the effectiveness of cancer therapies targeting the adaptive immune system (50).
Some lncRNAs can affect T cell function by regulating the expression of immune checkpoints. For instance, TUG1 is upregulated in HCC due to METTL3-mediated m6A modification. Knockdown of TUG1 can inhibit tumor growth and metastasis while increasing the infiltration of CD8+ T cells and M1 macrophages in tumors. TUG1 activates CD8+ T cells through PD-L1 and enhances macrophage phagocytosis via CD47. Mechanistically, TUG1 acts as a “sponge” for miR-141 and miR-340 to regulate the expression of PD-L1 and CD47 while interacting with YBX1 to promote their transcription. Clinical data also indicate a positive correlation between TUG1 and PD-L1 and CD47 in HCC tissues. Co-treatment with Tug1-siRNA and Pdl1 antibodies effectively inhibits tumor growth (51). Additionally, MIAT promotes STAT3 expression by negatively regulating miR-411-5p, thereby increasing the transcription level of PD-L1. Knockdown of MIAT enhances T cell cytotoxicity against HCC cells, and inhibiting miR-411-5p can reverse the decrease in STAT3 and PD-L1 expression caused by MIAT knockdown (52). The lncRNA CTD-2510F5.4 is significantly upregulated under hypoxic conditions and is expressed in Huh7 and HepG2 cells. Overexpression of CTD-2510F5.4 promotes the proliferation, invasion, and metastasis of HCC cells. Patients with high expression of CTD-2510F5.4 have poor prognoses and higher TP53 mutation rates, increased infiltration of immunosuppressive Tregs, and elevated expression of immune checkpoint molecules, leading to impaired immune function. Notably, patients with low expression of CTD-2510F5.4 show higher sensitivity to immunotherapy and anti-angiogenic therapy, while those with high expression respond better to chemotherapy (53).
Some lncRNAs can also influence the immune microenvironment by regulating tumor metabolism. For example, LINC01116 inhibits RAD18-mediated ubiquitination, stabilizing the EWSR1 protein and upregulating the expression of PPARA and long-chain fatty acid transporter FABP1, leading cancer cells to preferentially uptake linoleic acid and other long-chain fatty acids, resulting in T cell dysfunction and malignant progression of HCC. In mouse models, blocking LINC01116 enhances the efficacy of anti-PD1 therapy, increasing the infiltration of cytotoxic T cells and reducing the proportion of exhausted T cells (54).
Additionally, some lncRNAs can influence T cell infiltration and function through various cascading pathways, thereby affecting tumor progression. CRNDE is primarily found in malignant tumor cells and promotes tumor cell growth, recruits a large number of granulocyte-MDSCs (G-MDSCs), and restricts T cell infiltration, thereby facilitating HCC progression. Mechanistically, CRNDE binds to TLR3, activating the NF-κB signaling pathway and increasing the secretion of CXCL3. Knockdown of CRNDE significantly reduces G-MDSC accumulation and enhances T cell infiltration in the TME (55). LINC01132 has been identified as a novel oncogenic lncRNA, with its expression significantly higher in HCC tumor tissues compared to normal tissues, correlating with poor survival rates in patients, primarily driven by copy number amplification. Functional studies indicate that overexpression of LINC01132 promotes cell growth, proliferation, invasion, and metastasis. Mechanistically, LINC01132 enhances DPP4 expression through physical interaction with NRF1, acting as an oncogenic driver. Furthermore, silencing LINC01132 can induce CD8+ T cell infiltration, and when combined with anti-PD-L1 treatment, can improve anti-tumor immunity, suggesting that LINC01132 may serve as a novel immunotherapeutic target in HCC. Therefore, LINC01132 promotes HCC development through the NRF1/DPP4 axis, and targeting LINC01132 may enhance the efficacy of anti-PD-L1 immunotherapy (56).
These findings highlight the complex roles of lncRNAs in the immune microenvironment, suggesting that developing therapeutic strategies targeting specific lncRNAs may provide new avenues to enhance the efficacy of immunotherapy in HCC. Further related studies are summarized in Table 1.
3.1.2 Macrophage
Macrophages play a dual role in the immune microenvironment of HCC, capable of both promoting tumor progression and inhibiting tumor growth. LncRNAs regulate macrophage function through various mechanisms, influencing their polarization states, secretory characteristics, and interactions with tumor cells.
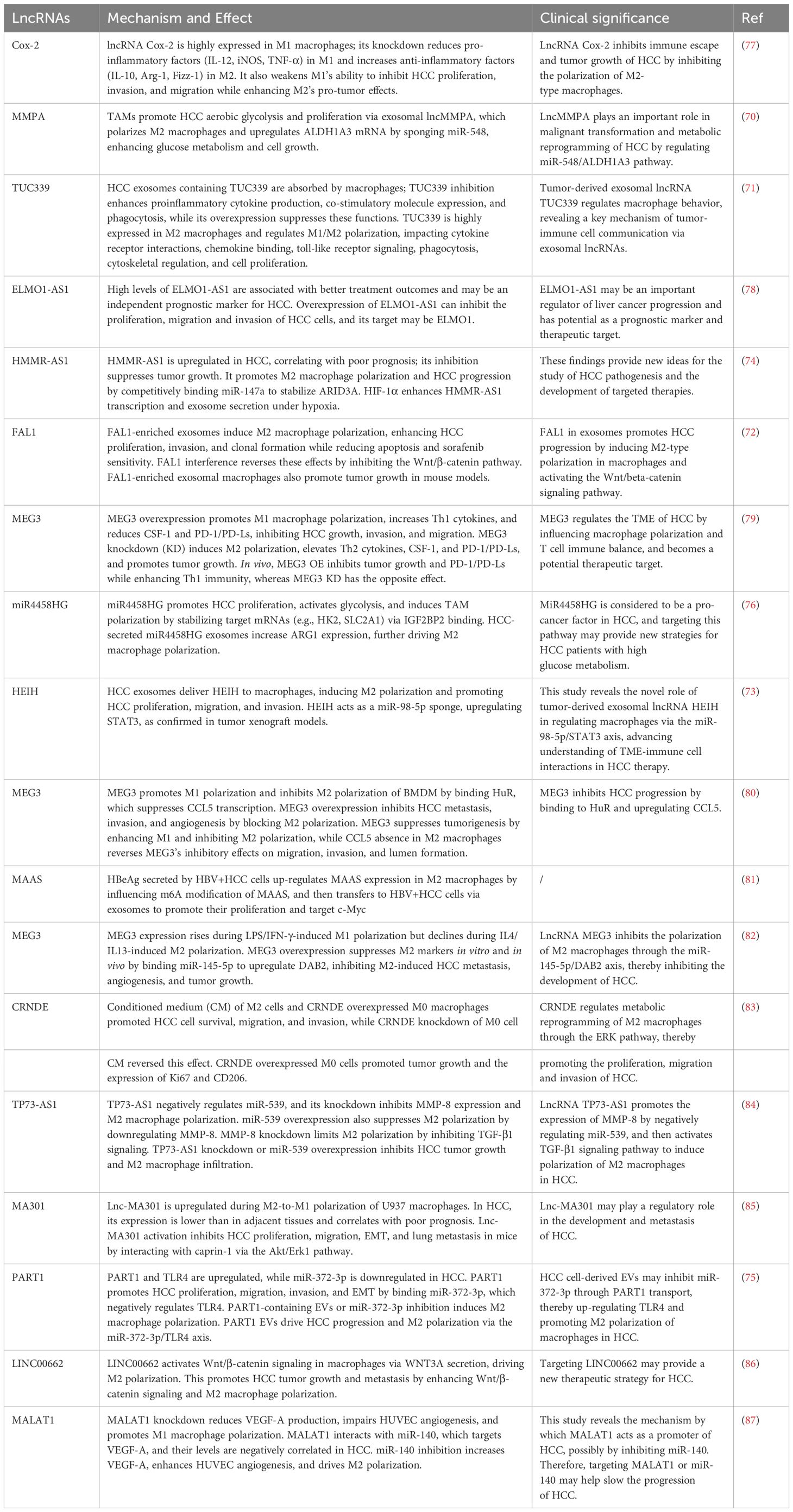
Extracellular vesicles (EVs) in the TME play a crucial role in regulating the tumor immune microenvironment. Studies have shown that tumor-associated macrophages (TAMs) secrete exosomes containing the lncRNA MMPA, enhancing aerobic glycolysis and proliferation in HCC cells. Mechanistically, lncMMPA not only promotes M2 macrophage polarization but also acts as a sponge for miR-548, increasing ALDH1A3 mRNA expression and thereby facilitating glucose metabolism and cell proliferation. Clinical data indicate that lncMMPA expression correlates with TAM glycolysis and decreased survival rates in HCC patients (70). Additionally, HCC cell-derived exosomes are found to contain high levels of TUC339, which can be absorbed by macrophages (THP-1 cells). Inhibition of TUC339 enhances the production of pro-inflammatory factors, expression of co-stimulatory molecules, and phagocytic capacity in macrophages, while overexpression of TUC339 suppresses these functions. TUC339 is highly expressed in M2 macrophages and exhibits distinct regulatory patterns during the polarization switch between M1 and M2 macrophages, indicating its key role in M1/M2 polarization. TUC339 is also associated with cytokine receptor interactions, chemokine receptor binding, Toll-like receptor signaling, FcγR-mediated phagocytosis, cytoskeletal regulation, and cell proliferation (71).
Furthermore, the lncRNA FAL1 is highly enriched in exosomes from the serum of HCC patients and promotes tumor progression. Exosomes enriched with FAL1 can induce M2 polarization in macrophages, enhancing the proliferation, invasion, and clonogenicity of HCC cells while inhibiting apoptosis and sensitivity to sorafenib. Interfering with FAL1 can reverse these effects. FAL1 promotes malignant progression in HCC by activating the Wnt/β-catenin signaling pathway (72). Another study reveals the role of the EZH2-related lncRNA HEIH, which is abnormally upregulated in HCC and correlates with poor prognosis in tumor tissues and cell lines. HEIH is enriched in exosomes found in plasma and cell supernatants and is transported to macrophages by HCC-derived exosomes, inducing M2 polarization and promoting HCC cell proliferation, migration, and invasion. Mechanistically, HEIH acts as a miRNA sponge, binding to miR-98-5p, leading to STAT3 upregulation. This provides evidence for the novel regulatory role of tumor-derived exosomal lncRNA HEIH targeting the miR-98-5p/STAT3 axis in macrophages, aiding the understanding of the complex interactions between the TME and immune cells in HCC treatment (73).
HMMR-AS1, an lncRNA associated with poor prognosis, promotes M2 polarization in macrophages by competitively binding to miR-147a, preventing the degradation of ARID3A, and accelerates HCC progression. Additionally, in hypoxic conditions, HIF-1α enhances HMMR-AS1 transcription by binding to its promoter, increasing exosome secretion (74).
Apart from exosomes, lncRNAs can also influence macrophage polarization and function through EVs. The lncRNA PART1 can be transported to macrophages via tumor-derived EVs, regulating macrophage polarization in HCC. PART1 binds to miR-372-3p, inhibiting its expression, while miR-372-3p negatively regulates TLR4 expression in macrophages. EVs containing PART1, TLR4 overexpression, or miR-372-3p inhibition all induce macrophage polarization toward M2. Thus, EVs from HCC cells may promote M2 polarization by transporting PART1 to inhibit miR-372-3p, leading to upregulation of TLR4 (75).
Metabolic reprogramming is also an important pathway through which lncRNAs influence macrophage status. Research has identified that the lncRNA miR4458HG promotes carcinogenesis in HCC, particularly in glucose metabolism. High expression of miR4458HG enhances HCC cell proliferation, activates glycolytic pathways, and induces polarization of TAMs. Mechanistically, miR4458HG stabilizes its target mRNAs (e.g., HK2 and SLC2A1) by binding to IGF2BP2, a key RNA m6A recognition protein, thereby enhancing glycolysis and altering the physiology of tumor cells. Moreover, miR4458HG secreted by HCC can increase ARG1 expression via exosomes, further promoting M2 polarization in macrophages (76).
The aforementioned studies reveal the complex mechanisms by which lncRNAs regulate macrophage polarization and function within the TME, underscoring their significance in the progression of HCC. Notably, lncRNAs such as lncMMPA, TUC339, and FAL1 influence glucose metabolism, inflammatory responses, and cellular signaling pathways, promoting M2 macrophage polarization and thereby creating a favorable environment for the survival and proliferation of HCC cells. These findings not only deepen our understanding of the roles of lncRNAs in the tumor immune microenvironment but also suggest that future therapeutic strategies should consider targeting these specific lncRNAs to reverse macrophage polarization and enhance the efficacy of immunotherapy for HCC. Further research should focus on the interactions between lncRNAs and other components of the TME to comprehensively elucidate their potential mechanisms in tumor progression. Additional relevant studies are summarized in Table 2.
3.1.3 Others
In addition to T cells and macrophages, other cell types in the tumor immune microenvironment are also regulated by lncrnas. Previous studies have shown that a variety of lncRNAs are associated with tumor invasion of B cells. For example, lncRNA AC099850.3 and E2F1 are positively correlated with the level of memory B cells (58, 61), while RNASEH1-AS1 is negatively correlated with the invasion of B cells (88). NK, as an immune cell responsible for recognizing and killing tumor cells, can also be regulated by LncRNAs through a variety of mechanisms.For example, LINC00707 can interact with YTHDF2, promoting ubiquitination dependent degradation of YTHDF2 protein. It can also influence the cytotoxicity of NK-92MI cells to HCC cells through its interaction with YTHDF2 (89). Treg is an immunosuppressive cell. Previous studies have shown that lncRNA FENDRR up-regulates GADD45B by adsorbing miR-423-5p, and inhibits Treg-mediated immune escape of HCC cells (69).
The previous sections primarily focused on immune-suppressive lncRNAs; however, in addition to those lncRNAs that promote tumor growth and immune suppression, several lncRNAs have been identified as tumor suppressors, playing critical roles in enhancing immune responses and improving survival outcomes in HCC. For example, MEG3 expression increases during LPS/IFNγ-induced M1 polarization, while it decreases during IL4/IL13-induced M2 polarization. Overexpression of MEG3 suppresses M2 polarization markers in mice. Mechanistically, MEG3 regulates DAB2 expression by binding to miR-145-5p. Overexpressing MEG3 inhibits HCC cell metastasis and angiogenesis induced by M2 polarization, and it suppresses tumor growth in vivo (82). Knockdown of MALAT1 inhibited VEGF-A production, impaired the angiogenesis of HUVECs, and promoted the polarization of macrophages to the M1 subgroup. Mechanistic studies confirmed the interaction between MALAT1 and miR-140 and between miR-140 and VEGF-A, revealing a negative correlation between MALAT1 and miR-140 in HCC tissues. In addition, inhibition of miR-140 significantly increased VEGF-A expression, promoted angiogenesis in HUVECs, and polarized macrophages toward the M2 subpopulation (87). LINC00261 has been shown to act as a tumor suppressor in various cancers. Studies indicate that in HCC, LINC00261 expression is downregulated in tumor tissues and is associated with better prognosis. Furthermore, the LINC00261/MiR105-5p/SELL axis is involved in B-cell dysfunction. These findings provide significant insights into selecting the LINC00261/MiR105-5p/SELL signaling pathway as a new biomarker related to overall survival in HCC and may influence the effectiveness of HCC immunotherapy (67). Previous research has shown that JPX-induced XIST inhibits liver cancer progression by sequestering miR-155-5p (90). XIST is more highly expressed in patients with high tumor and peritumoral CD4 inflammation. The number of CD25-positive cells is significantly higher in tumors with XIST expression, suggesting its potential regulatory role in the immune microenvironment (60). It is worth noting that the clinical application of these lncRNAs still faces significant challenges and requires further clinical validation. These relevant lncRNAs are summarized in the corresponding table according to the above classification.
3.2 LncRNAs in immune checkpoint regulation
In the TME, lncRNAs play a crucial role in immune regulation by modulating the expression of PD1 and its ligand PD-L1 (Figure 2B). These lncRNAs are involved in the mechanisms of immune evasion and influence the signaling between tumor cells and immune cells through interactions with miRNAs or transcription factors. Research indicates that the upregulation of certain lncRNAs is closely associated with the expression of PD1/PD-L1, thereby promoting the survival and proliferation of tumor cells.
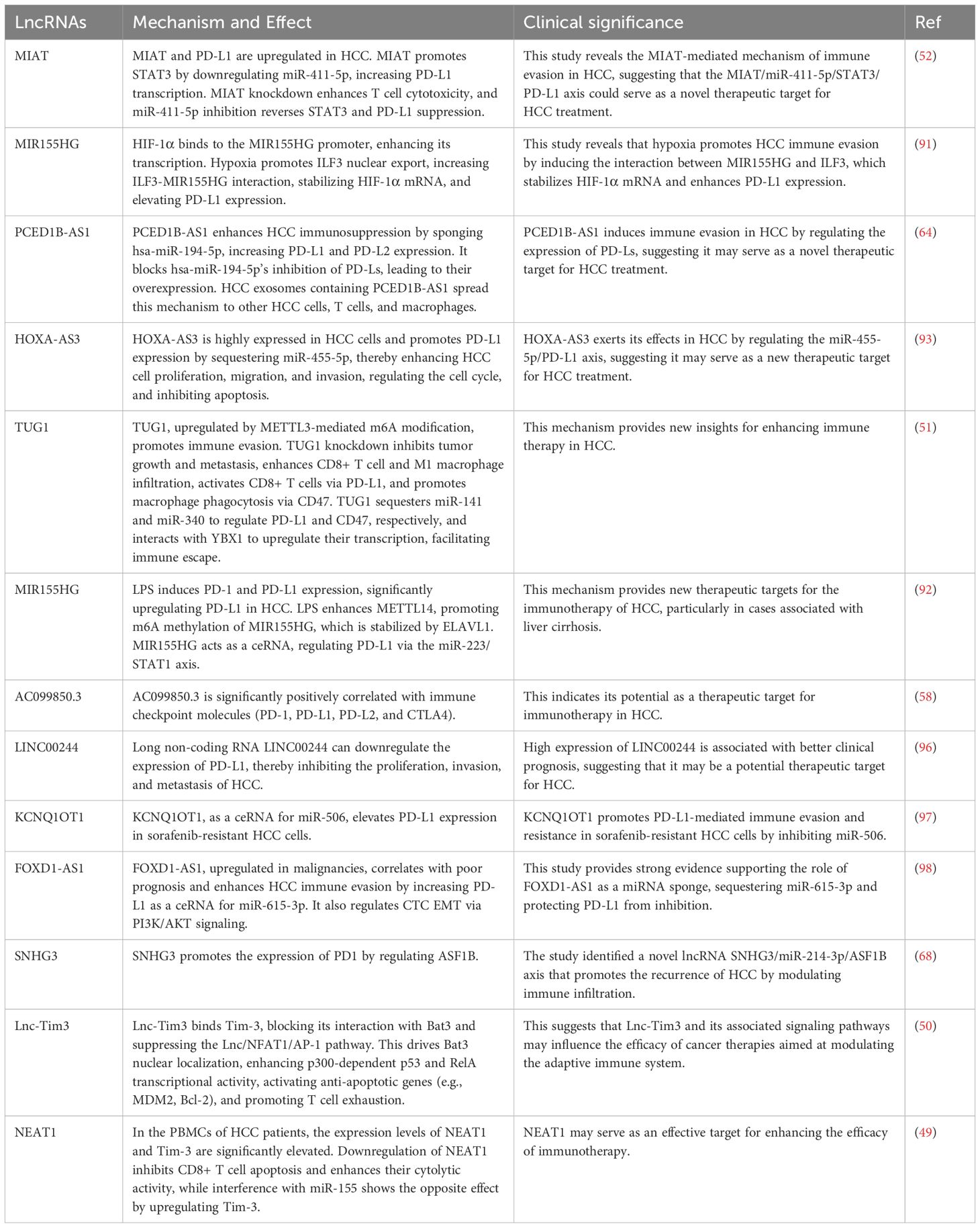
Previous studies have indicated that various lncRNAs are positively correlated with the expression of PD1/PD-L1 or can promote their expression. For example, AC099850.3 is significantly positively correlated with key immune checkpoint molecules (PD1, PD-L1, PD-L2, and CTLA4) (58). MIAT promotes the expression of PD-L1 at the transcriptional level by downregulating miR-411-5p, which in turn enhances STAT3 expression (52). SNHG3 can also promote PD1 expression by regulating ASF1B (68). Hypoxia induces the interaction between MIR155HG and ILF3, stabilizing HIF-1α mRNA and increasing PD-L1 expression, thereby facilitating immune evasion in HCC (91). Additionally, MIR155HG functions as a competitive endogenous RNA (ceRNA) that regulates PD-L1 expression through the miR-223/STAT1 axis (92). HOXA-AS3 is highly expressed in HCC cells and enhances PD-L1 expression by adsorbing miR-455-5p, thereby increasing the proliferation, migration, and invasion abilities of HCC cells, regulating the cell cycle, and inhibiting apoptosis (93).
Multiple mechanisms are involved in the regulation of PD1/PD-L1 by lncRNAs. PCED1B-AS1 enhances the expression of PD-L1 and PD-L2 by adsorbing hsa-miR-194-5p, thereby increasing the immune suppressive ability of HCC cells. Furthermore, HCC cells release exosomes containing PCED1B-AS1, transmitting this immune suppressive mechanism to other HCC cells, T cells, and macrophages. High expression of PCED1B-AS1 is closely related to the overexpression of PD-Ls in HCC and promotes tumor cell proliferation and tumor formation while inhibiting apoptosis (64). METTL3-mediated m6A modification upregulates TUG1, promoting tumor immune evasion. Silencing TUG1 inhibits tumor growth and metastasis, enhances the infiltration of CD8+ T cells and M1 macrophages, and activates CD8+ T cells through PD-L1 while promoting macrophage phagocytosis via CD47. TUG1 regulates the expression of PD-L1 and CD47 by adsorbing miR-141 and miR-340, respectively, and interacts with YBX1 to promote their transcriptional upregulation, ultimately facilitating tumor immune evasion (51).
In addition to PD1/PD-L1, lncRNAs are also associated with other checkpoints such as CTLA-4 and TIM-3, and they can even regulate their levels and functions. For example, Lnc-Tim3 specifically binds to Tim-3, obstructing its interaction with Bat3, which inhibits the downstream Lck/NFAT1/AP-1 signaling pathway, leading to the nuclear localization of Bat3 and enhancing the transcriptional activity of p300-dependent p53 and RelA. This promotes the transcriptional activation of anti-apoptotic genes (such as MDM2 and Bcl-2), contributing to T cell exhaustion, which is linked to diminished anti-tumor immunity (50). Furthermore, a study established a prognostic model for HCC composed of 12 ferroptosis-related lncRNAs, dividing HCC patients into high-risk and low-risk groups. In the high-risk group, the expression of various immune checkpoint molecules, including PD1, CTLA-4, and CD86, was significantly elevated (94). Another study constructed a new feature comprising three ferroptosis-related lncRNAs, which was associated with immune cell infiltration (such as macrophages, myeloid dendritic cells, and neutrophils) and immune checkpoint blockade targets (including PD1, CTLA-4, and TIM-3) (95).
The above studies clearly demonstrate the significant role of lncRNAs in regulating immune checkpoints in HCC. We have summarized the related research in Table 3 for a more intuitive presentation.
3.3 Regulatory roles of LncRNA in cytokines and immune modulators
Gaining a deeper understanding of the role of lncRNAs in cytokines and immune regulatory molecules is also a crucial aspect of providing new strategies and targets for tumor immunotherapy.
TGF-β plays a dual role in HCC. In the early stages, it inhibits cell proliferation and induces apoptosis, serving an anti-tumor role. However, in later stages of HCC, TGF-β promotes tumor cell migration, invasion, and fibrosis in the TME, thereby facilitating tumor progression. Therefore, TGF-β is an important target in HCC treatment. Previous studies have shown that lncRNAs can interact with TGF-β. Research indicates that some lncRNAs can be activated by TGF-β-related pathways in HCC, thereby exerting downstream effects. For instance, lnc-UTGF is transcriptionally induced by TGF-β. Under TGF-β stimulation, SMAD2/3 bind to the lnc-UTGF promoter and activate its expression. Overexpression of lnc-UTGF enhances TGF-β/SMAD signaling, while its knockdown suppresses this signaling. lnc-UTGF interacts with the mRNAs of SMAD2 and SMAD4 through complementary base pairing, thereby increasing the stability of SMAD2/4 mRNA. This indicates the existence of a novel TGF-β/SMAD/lnc-UTGF positive feedback loop (99). Additionally, LINC01980 promotes HCC metastasis via the miR-376b-5p/E2F5 axis under classical TGF-β/SMAD pathway activation (100). TGF-β-activated lncRNA-ATB is upregulated in HCC metastasis and is associated with poor prognosis. lncRNA-ATB induces epithelial-mesenchymal transition (EMT) and cell invasion by competitively binding to the miR-200 family, leading to the upregulation of ZEB1 and ZEB2. Furthermore, lncRNA-ATB promotes the autocrine induction of IL-11 by binding to IL-11 mRNA, activating the STAT3 signaling pathway, and further facilitating the organ colonization of disseminated tumor cells (101). Other studies have shown that some lncRNAs can exert effects by regulating TGF-β secretion. For example, PRR34-AS1 stabilizes the mRNA level of the exosomal protein Rab27a by recruiting DDX3X, thereby promoting the exosomal secretion of VEGF and TGF-β in HCC cells (102). CASC7 can also promote HCC progression by binding to miR-30a-5p, regulating KLF10 and its downstream TGF-β/SMAD3 pathway (103). The downregulation of LINC01278 can reduce HCC cell migration and invasion induced by β-catenin and TGF-β1 (104). HANR and miR-214 jointly regulate EZH2, affecting the level of TGFBR2 (105). Additionally, some lncRNAs participate in TGF-β-induced biological processes. For instance, lncRNA SLC7A11-AS1 and hsa_circ_0006123 are involved in the TGF-β-induced EMT process and may promote HCC metastasis (106).
In addition to TGF-β, lncRNAs also interact with various cytokines and immune regulatory factors such as IL-6, IL-10, INF-γ, and TNF-α (Figure 2C). For instance, lnc-DILC exerts its effects by inhibiting the autocrine IL-6/STAT3 signaling pathway, and there are binding sites between lnc-DILC and the IL-6 promoter. Furthermore, lnc-DILC mediates the interaction between TNF-α/NF-κB signaling and IL-6/STAT3 signaling (107). Both TNF-α and IL-6 can also regulate the expression of lncRNA 00607 (108). The downregulation of lncRNA cox-2 using siRNA significantly decreased the expression of IL-12, iNOS, and TNF-α in M1 macrophages, while increasing the expression of IL-10, Arg-1, and Fizz-1 in M2 macrophages (77). Relevant studies are summarized in Table 4 for a more intuitive presentation.
4 Potential of lncRNAs in HCC immunotherapy
4.1 lncRNAs as biomarkers in HCC immunotherapy
LncRNAs have emerged as promising biomarkers for predicting responses to immunotherapy in HCC. Their unique expression profiles in tumor tissues and circulation provide valuable insights into the TME and immune landscape.
In the previous sections, we discussed specific lncRNAs that are associated with immune cell infiltration, cytokine profiles, and overall immune responses, making them potential prognostic indicators for HCC patients undergoing immunotherapy. For example, lnc-TUG1, lnc-MMPA, lnc-MIAT, and lnc-ATB are lncRNAs whose expression levels are correlated with the state of the tumor immune microenvironment in HCC, suggesting their roles in regulating immune responses (51, 52, 70, 101).
Furthermore, the expression levels of lncRNAs can serve as predictive markers for patient outcomes. By assessing lncRNA profiles, clinicians may better stratify patients, identifying those who are more likely to benefit from immunotherapeutic approaches. This stratification could lead to more personalized treatment strategies, maximizing efficacy while minimizing unnecessary exposure to ineffective therapies. Previous studies have constructed various risk prediction models composed of lncRNAs. For example, one study developed a nine-lncRNA ferroptosis-related signature (CTD-2033A16.3, CTD-2116N20.1, CTD-2510F5.4, DDX11-AS1, LINC00942, LINC01224, LINC01231, LINC01508, and ZFPM2-AS1) to predict the prognosis and immune response in liver cancer. This lncRNA signature may regulate the immune microenvironment of HCC by interfering with TNF-α/NF-κB, IL-2/STAT5 and cytokine/receptor signaling pathways (118). Other studies have also constructed prognostic models using nine copper nephropathy-related lncRNAs. The low-risk group had significantly higher infiltration of immune cells, which primarily have anti-tumor immune functions, and showed a higher sensitivity to ICIs therapy, possibly acting through the AL365361.1/hsa-miR-17-5p/NLRP3 axis. In addition, NLRP3 mutation-sensitive drugs such as VNLG/124, Sunitinib, and rifampicin may achieve better clinical outcomes in high-risk patients (119). More relevant models are summarized in Table 5.
4.2 Combination therapy
In Chapter Three, we discuss how lncRNAs can regulate the TME, influencing the infiltration and function of immune cells, thereby enhancing the efficacy of immunotherapy. Certain lncRNAs can alter the immune status of the TME by inhibiting the expression of immunosuppressive factors or promoting the release of immune-activating factors. This underscores the promising therapeutic prospects of combining lncRNA-targeted therapy with immunotherapy. For example, TUG1 can regulate the expression of PD-L1 and CD47 by acting as a “sponge” for miR-141 and miR-340. Knockdown of TUG1 can inhibit tumor growth and metastasis, increase the infiltration of CD8+ T cells and M1 macrophages in the tumor, and activate CD8+ T cells through PD-L1 while enhancing macrophage phagocytosis through CD47. Therefore, TUG1-siRNA combined with PD-L1 antibodies can effectively suppress tumor growth, providing new targets and strategies for HCCimmunotherapy (51). Additionally, various lncRNAs discussed earlier, such as MIAT (52), LINC01132 (56), MIR155HG (91), PCED1B-AS1 (64), NEAT1 (49), can serve as potential combined treatment targets to enhance the effects of HCC immunotherapy (Figure 2D). However, it is worth noting that, despite the promising therapeutic prospects of combination therapies, large-scale clinical trials of lncRNA-targeted drugs alone or in combination with immunotherapy for treating HCC have not yet been widely conducted.
In addition to the combination with immunotherapy, significant progress has been made in the study of lncRNA targeting drugs in combination with chemotherapy, anti-angiogenic drugs or targeted kinase inhibitors in tumors. The mechanism mainly focuses on the synergistic regulation of tumor drug resistance, angiogenesis and signaling pathways. Studies have shown that lncrnas can affect the expression of apoptosis-related genes by regulating DNA damage repair or adsorbing mirnas as ceRNA, thus reversing chemotherapy resistance (130). In addition, lncrnas can also regulate vascular endothelial growth factor (VEGF) related pathways, such as linc00173.v1, which promotes angiogenesis through the miR-511-5p/VEGFA axis (131). Other studies have shown that lncrnas recruit kinase complexes through scaffold action (e.g., WDR5/KAT2A complex regulates SOD2 gene (132)) or regulate kinase-related signaling pathways (e.g., STAT3/VEGFA axis (133)). However, current research still faces challenges, such as in vivo safety verification of lncRNA targeted delivery systems (such as lipid nanoparticles and viral vectors), as well as dose optimization of combination regimenes and analysis of drug resistance mechanisms. Overall, this combination strategy provides a new direction to overcome the limitations of traditional therapies by regulating the TME and molecular networks in multiple dimensions.
5 Future perspectives and challenges
Recent advancements in the understanding of lncRNAs have provided valuable insights into their roles within the immune microenvironment of HCC. Research has revealed that lncRNAs are not merely passive players but active regulators of immune cell behavior, influencing tumor progression and response to therapy (134). For instance, studies have highlighted specific lncRNAs that modulate T cell activity, macrophage polarization, and the overall cytokine profile within the TME (135). These findings suggest that lncRNAs can serve as critical mediators of immune evasion and resilience in HCC.
Moreover, the identification of lncRNAs as potential biomarkers for prognosis and treatment response has opened new avenues for personalized immunotherapy approaches in HCC (136). By integrating lncRNA profiling into clinical practice, oncologists may better predict patient responses to immunotherapeutic strategies and tailor treatments accordingly (94, 137). This perspective shift emphasizes the potential for lncRNAs to enhance the efficacy of existing therapies and improve patient outcomes.
Despite the promising insights gained from current research, several challenges remain in translating lncRNA findings into clinical practice. One of the primary obstacles is the complex regulatory mechanisms underlying lncRNA functions. The interactions between lncRNAs and their target genes, as well as the pathways they influence, can be intricate and context-dependent, complicating efforts to target them effectively (138, 139).
Additionally, the identification of specific lncRNA targets poses another challenge. Given the pleiotropic effects of many lncRNAs, achieving specificity in targeting their functions can be difficult (140). This challenge underscores the need for advanced methodologies to elucidate lncRNA interactions and their precise roles within the HCC immune microenvironment.
Drug delivery systems also represent a significant hurdle in lncRNA-based therapies. Ensuring efficient and targeted delivery of lncRNA modulators to tumor tissues while minimizing off-target effects is critical for maximizing therapeutic efficacy (141). Advances in nanotechnology and RNA delivery systems may provide solutions to these challenges, enabling more effective clinical applications of lncRNA therapeutics (142–144).
Although most current studies focus on lncRNAs as biomarkers, some lncRNAs play regulatory roles in the tumor immune microenvironment that render them potential direct therapeutic targets. For example, lncRNA NEAT1 promotes immune evasion by modulating PD-L1 expression, thereby affecting the sensitivity of tumor cells to immunotherapy; inhibition of NEAT1 may enhance immunotherapeutic efficacy (145–147). Similarly, lncRNA MALAT1 influences T cell infiltration and augments tumor immune suppression by modulating the STAT3 signaling pathway (148, 149). In addition, antisense oligonucleotides (ASOs) targeting MALAT1 have demonstrated promising therapeutic potential (150, 151). Moreover, lncRNA UCA1 regulates PD-L1 expression via miR-148a, and its inhibition could improve the efficacy of ICIs (152). Therefore, the design of lncRNA-targeted intervention strategies—such as RNAi, ASOs, or CRISPR-Cas13-mediated lncRNA degradation—could represent a novel immunotherapeutic approach (153–155).
Despite the potential of lncRNAs as therapeutic targets, several challenges remain. Many lncRNAs exhibit tissue- or cell type-specific expression, and strategies to precisely target specific cells without affecting normal tissues need further exploration. Furthermore, the complex structure of lncRNAs, with some harboring highly stable secondary structures, makes them difficult to degrade or inhibit effectively. Although in vitro studies often demonstrate promising intervention outcomes, in vivo delivery of RNA-based therapeutics faces challenges including degradation, immunogenicity, and hepatic clearance.
Various RNA delivery strategies have been developed; however, systems optimized specifically for lncRNAs still require refinement. LNPs, which have been approved by the FDA for mRNA vaccine delivery, can be used for lncRNA modulators but still need improvements in tissue specificity (156, 157). Viral vectors, such as AAVs, can facilitate the stable expression of shRNAs to knock down pathogenic lncRNAs, although they may trigger immune responses (158). Chemical modifications like 2’-O-methylation and locked nucleic acid modifications can enhance the stability of lncRNA interference molecules (159, 160).
Meanwhile, the off-target effects of lncRNA modulators should not be overlooked. Many lncRNAs possess highly homologous sequences, so interference with a specific lncRNA may inadvertently affect other non-target RNAs, leading to unexpected biological effects (161, 162). Additionally, lncRNAs may interact with miRNAs through ceRNA mechanisms, whereby targeting a particular lncRNA could trigger cascade effects in miRNA-associated pathways (160). To evaluate such off-target phenomena, techniques such as single-cell sequencing, RNA pull-down assays, and RBP interaction studies can be employed to assess the specificity of lncRNA modulators (163, 164).
Previous studies, as summarized in this review, have shown that the potential of lncRNAs in HCC immunotherapy has been validated in several preclinical studies, demonstrating their significant potential in regulating the immune microenvironment, promoting immune evasion, and serving as therapeutic targets. However, these findings are still far from mature and lack sufficient clinical validation. Currently, most related studies remain in the basic and preclinical research stages, with a lack of clinical trials targeting lncRNA-based therapies for HCC. For example, ASOs targeting MALAT1 have shown preliminary therapeutic effects in mouse models, but their clinical efficacy still needs further confirmation (151, 165). Similarly, although interventions targeting PVT1 and HOTAIR have shown promising results in preclinical studies, there is still a lack of relevant clinical data to assess their therapeutic potential (166). Furthermore, it is important to acknowledge the many challenges in implementing lncRNA-targeted therapeutic strategies, such as those mentioned earlier regarding the specificity of drug delivery to lncRNAs and the avoidance of systemic toxicity, as well as off-target effects that may cause adverse reactions. Therefore, before advancing lncRNA-targeted therapies into clinical practice, these technical barriers must be addressed, and their safety and efficacy should be validated through more extensive clinical trials. As a result, research on lncRNAs as immune therapy targets remains in its early stages. Despite their potential demonstrated in preclinical experiments, there are still many technical and clinical challenges to overcome before applying them in clinical settings. Future research should place greater emphasis on critically evaluating the feasibility and limitations of these therapeutic strategies and strengthening clinical validation efforts.
Looking ahead, future research directions may focus on the development of lncRNA-based therapies that can be integrated into existing treatment regimens. Exploring combination therapies that leverage the immune-modulating effects of lncRNAs alongside ICIs or targeted therapies could yield synergistic benefits. Furthermore, the continued investigation of lncRNA dynamics in response to various treatment modalities will be crucial for optimizing therapeutic strategies.
In summary, while the field of lncRNAs in HCC presents exciting opportunities for enhancing our understanding of tumor-immune interactions, significant challenges remain. Continued research efforts will be essential to unravel the complexities of lncRNA functions and translate these findings into effective clinical interventions.
Author contributions
SZ: Conceptualization, Writing – original draft, Writing – review & editing. FC: Visualization, Writing – original draft, Writing – review & editing. LH: Visualization, Writing – original draft. XL: Visualization, Writing – original draft. ZG: Visualization, Writing – original draft. MC: Writing – original draft, Writing – review & editing. XW: Conceptualization, Writing – original draft, Writing – review & editing. ZS: Conceptualization, Funding acquisition, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the Medical Health Science and Technology Project of Zhejiang Provincial Health Commission (2022KY1246), and the Science and Technology Bureau of Jiaxing City (2023AZ31002 and 2022AZ10009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviation
HCC, Hepatocellular carcinoma; lncRNAs, Long non-coding RNAs; TME, Tumor microenvironment; NK, Natural killer; MDSCs, Myeloid-derived suppressor cells; Tregs, Regulatory T cells; CTLs, Cytotoxic T lymphocytes; EVs, Extracellular vesicles; TAMs, Tumor-associated macrophages; ceRNA, Competitive endogenous RNA; EMT, Epithelial-mesenchymal transition; ICIs, Immune checkpoint inhibitor; VEGF, Vascular endothelial growth factor; ASOs, Antisense oligonucleotides.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Rajendran L, Ivanics T, Claasen MP, Muaddi H, Sapisochin G. The management of post-transplantation recurrence of hepatocellular carcinoma. Clin Mol Hepatol. (2022) 28:1–16. doi: 10.3350/cmh.2021.0217
3. Llovet JM, Willoughby CE, Singal AG, Greten TF, Heikenwalder M, El-Serag HB, et al. Nonalcoholic steatohepatitis-related hepatocellular carcinoma: pathogenesis and treatment. Nat Rev Gastroenterol Hepatol. (2023) 20:487–503. doi: 10.1038/s41575-023-00754-7
4. Leone V, Ali A, Weber A, Tschaharganeh DF, Heikenwalder M. Liver inflammation and hepatobiliary cancers. Trends Cancer. (2021) 7:606–23. doi: 10.1016/j.trecan.2021.01.012
5. Huang DQ, El-Serag HB, Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. (2021) 18:223–38. doi: 10.1038/s41575-020-00381-6
6. Reig M, Forner A, Rimola J, Ferrer-Fabrega J, Burrel M, Garcia-Criado A, et al. BCLC strategy for prognosis prediction and treatment recommendation: The 2022 update. J Hepatol. (2022) 76:681–93. doi: 10.1016/j.jhep.2021.11.018
7. Shen KY, Zhu Y, Xie SZ, Qin LX. Immunosuppressive tumor microenvironment and immunotherapy of hepatocellular carcinoma: current status and prospectives. J Hematol Oncol. (2024) 17:25. doi: 10.1186/s13045-024-01549-2
8. Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. (2022) 19:151–72. doi: 10.1038/s41571-021-00573-2
9. Donne R, Lujambio A. The liver cancer immune microenvironment: Therapeutic implications for hepatocellular carcinoma. Hepatology. (2023) 77:1773–96. doi: 10.1002/hep.32740
10. Sun G, Liu H, Zhao J, Zhang J, Huang T, Sun G, et al. Macrophage GSK3beta-deficiency inhibits the progression of hepatocellular carcinoma and enhances the sensitivity of anti-PD1 immunotherapy. J Immunother Cancer. (2022) 10. doi: 10.1136/jitc-2022-005655
11. Lin X, Kang K, Chen P, Zeng Z, Li G, Xiong W, et al. Regulatory mechanisms of PD-1/PD-L1 in cancers. Mol Cancer. (2024) 23:108. doi: 10.1186/s12943-024-02023-w
12. Baker DJ, Arany Z, Baur JA, Epstein JA, June CH. CAR T therapy beyond cancer: the evolution of a living drug. Nature. (2023) 619:707–15. doi: 10.1038/s41586-023-06243-w
13. Macek Jilkova Z, Aspord C, Decaens T. Predictive factors for response to PD-1/PD-L1 checkpoint inhibition in the field of hepatocellular carcinoma: current status and challenges. Cancers (Basel). (2019) 11. doi: 10.3390/cancers11101554
14. Rimassa L, Finn RS, Sangro B. Combination immunotherapy for hepatocellular carcinoma. J Hepatol. (2023) 79:506–15. doi: 10.1016/j.jhep.2023.03.003
15. Zhou Y, Wei S, Xu M, Wu X, Dou W, Li H, et al. CAR-T cell therapy for hepatocellular carcinoma: current trends and challenges. Front Immunol. (2024) 15:1489649. doi: 10.3389/fimmu.2024.1489649
16. Chen B, Dragomir MP, Yang C, Li Q, Horst D, Calin GA. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct Target Ther. (2022) 7:121. doi: 10.1038/s41392-022-00975-3
17. Nemeth K, Bayraktar R, Ferracin M, Calin GA. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat Rev Genet. (2024) 25:211–32. doi: 10.1038/s41576-023-00662-1
18. Huang W, Li H, Yu Q, Xiao W, Wang DO. LncRNA-mediated DNA methylation: an emerging mechanism in cancer and beyond. J Exp Clin Cancer Res. (2022) 41:100. doi: 10.1186/s13046-022-02319-z
19. Imamura K, Akimitsu N. Long non-coding RNAs involved in immune responses. Front Immunol. (2014) 5:573. doi: 10.3389/fimmu.2014.00573
20. Mai H, Zhou B, Liu L, Yang F, Conran C, Ji Y, et al. Molecular pattern of lncRNAs in hepatocellular carcinoma. J Exp Clin Cancer Res. (2019) 38:198. doi: 10.1186/s13046-019-1213-0
21. Llovet JM, Pinyol R, Yarchoan M, Singal AG, Marron TU, Schwartz M, et al. Adjuvant and neoadjuvant immunotherapies in hepatocellular carcinoma. Nat Rev Clin Oncol. (2024) 21:294–311. doi: 10.1038/s41571-024-00868-0
22. Giraud J, Chalopin D, Blanc JF, Saleh M. Hepatocellular carcinoma immune landscape and the potential of immunotherapies. Front Immunol. (2021) 12:655697. doi: 10.3389/fimmu.2021.655697
23. Yin S, Jin W, Qiu Y, Fu L, Wang T, Yu H. Solamargine induces hepatocellular carcinoma cell apoptosis and autophagy via inhibiting LIF/miR-192-5p/CYR61/Akt signaling pathways and eliciting immunostimulatory tumor microenvironment. J Hematol Oncol. (2022) 15:32. doi: 10.1186/s13045-022-01248-w
24. Wang H, Zhang H, Wang Y, Brown ZJ, Xia Y, Huang Z, et al. Regulatory T-cell and neutrophil extracellular trap interaction contributes to carcinogenesis in non-alcoholic steatohepatitis. J Hepatol. (2021) 75:1271–83. doi: 10.1016/j.jhep.2021.07.032
25. Liu N, Wang X, Steer CJ, Song G. MicroRNA-206 promotes the recruitment of CD8(+) T cells by driving M1 polarisation of Kupffer cells. Gut. (2022) 71:1642–55. doi: 10.1136/gutjnl-2021-324170
26. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces NK cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. (2020) 19:110. doi: 10.1186/s12943-020-01222-5
27. Wang S, Wu Q, Chen T, Su R, Pan C, Qian J, et al. Blocking CD47 promotes antitumour immunity through CD103(+) dendritic cell-NK cell axis in murine hepatocellular carcinoma model. J Hepatol. (2022) 77:467–78. doi: 10.1016/j.jhep.2022.03.011
28. Finkin S, Yuan D, Stein I, Taniguchi K, Weber A, Unger K, et al. Ectopic lymphoid structures function as microniches for tumor progenitor cells in hepatocellular carcinoma. Nat Immunol. (2015) 16:1235–44. doi: 10.1038/ni.3290
29. Heinrich B, Gertz EM, Schaffer AA, Craig A, Ruf B, Subramanyam V, et al. The tumour microenvironment shapes innate lymphoid cells in patients with hepatocellular carcinoma. Gut. (2022) 71:1161–75. doi: 10.1136/gutjnl-2021-325288
30. Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, et al. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. (2019) 129:3324–38. doi: 10.1172/JCI126022
31. Qi D, Lu M, Xu P, Yao X, Chen Y, Gan L, et al. Transcription factor ETV4 promotes the development of hepatocellular carcinoma by driving hepatic TNF-alpha signaling. Cancer Commun (Lond). (2023) 43:1354–72. doi: 10.1002/cac2.v43.12
32. Kuraji R, Sekino S, Kapila Y, Numabe Y. Periodontal disease-related nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: An emerging concept of oral-liver axis. Periodontol 2000. (2021) 87:204–40. doi: 10.1111/prd.12387
33. Arvanitakis K, Mitroulis I, Germanidis G. Tumor-associated neutrophils in hepatocellular carcinoma pathogenesis, prognosis, and therapy. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13122899
34. Pinter M, Scheiner B, Pinato DJ. Immune checkpoint inhibitors in hepatocellular carcinoma: emerging challenges in clinical practice. Lancet Gastroenterol Hepatol. (2023) 8:760–70. doi: 10.1016/S2468-1253(23)00147-4
35. Sangro B, Chan SL, Meyer T, Reig M, El-Khoueiry A, Galle PR. Diagnosis and management of toxicities of immune checkpoint inhibitors in hepatocellular carcinoma. J Hepatol. (2020) 72:320–41. doi: 10.1016/j.jhep.2019.10.021
36. Wang Z, Wang Y, Gao P, Ding J. Immune checkpoint inhibitor resistance in hepatocellular carcinoma. Cancer Lett. (2023) 555:216038. doi: 10.1016/j.canlet.2022.216038
37. Zhu AX, Abbas AR, de Galarreta MR, Guan Y, Lu S, Koeppen H, et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat Med. (2022) 28:1599–611. doi: 10.1038/s41591-022-01868-2
38. Lu LG, Zhou ZL, Wang XY, Liu BY, Lu JY, Liu S, et al. PD-L1 blockade liberates intrinsic antitumourigenic properties of glycolytic macrophages in hepatocellular carcinoma. Gut. (2022) 71:2551–60. doi: 10.1136/gutjnl-2021-326350
39. Wei CY, Zhu MX, Zhang PF, Huang XY, Wan JK, Yao XZ, et al. PKCalpha/ZFP64/CSF1 axis resets the tumor microenvironment and fuels anti-PD1 resistance in hepatocellular carcinoma. J Hepatol. (2022) 77:163–76. doi: 10.1016/j.jhep.2022.02.019
40. Gao X, Xu N, Li Z, Shen L, Ji K, Zheng Z, et al. Safety and antitumour activity of cadonilimab, an anti-PD-1/CTLA-4 bispecific antibody, for patients with advanced solid tumours (COMPASSION-03): a multicentre, open-label, phase 1b/2 trial. Lancet Oncol. (2023) 24:1134–46. doi: 10.1016/S1470-2045(23)00411-4
41. Ye W, Li H, Zhao J, Lu D, Tao T, Zhu X. Graphene therapy-related lncRNAs as prognostic and immune microenvironmental biomarkers in hepatocellular carcinoma. Transl Oncol. (2024) 43:101915. doi: 10.1016/j.tranon.2024.101915
42. Tzur YB. lncRNAs in fertility: redefining the gene expression paradigm? Trends Genet. (2022) 38:1170–9. doi: 10.1016/j.tig.2022.05.013
43. Herman AB, Tsitsipatis D, Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. (2022) 82:2252–66. doi: 10.1016/j.molcel.2022.05.027
44. Zhang N, Zhang H, Wu W, Zhou R, Li S, Wang Z, et al. Machine learning-based identification of tumor-infiltrating immune cell-associated lncRNAs for improving outcomes and immunotherapy responses in patients with low-grade glioma. Theranostics. (2022) 12:5931–48. doi: 10.7150/thno.74281
45. Safarzadeh E, Asadzadeh Z, Safaei S, Hatefi A, Derakhshani A, Giovannelli F, et al. MicroRNAs and lncRNAs-A new layer of myeloid-derived suppressor cells regulation. Front Immunol. (2020) 11:572323. doi: 10.3389/fimmu.2020.572323
46. Zhang J, Luo Q, Li X, Guo J, Zhu Q, Lu X, et al. Novel role of immune-related non-coding RNAs as potential biomarkers regulating tumour immunoresponse via MICA/NKG2D pathway. Biomark Res. (2023) 11:86. doi: 10.1186/s40364-023-00530-4
47. Gao F, He S, Jin A. MiRNAs and lncRNAs in NK cell biology and NK/T-cell lymphoma. Genes Dis. (2021) 8:590–602. doi: 10.1016/j.gendis.2020.08.006
48. Saadi W, Fatmi A, Pallardo FV, Garcia-Gimenez JL, Mena-Molla S. Long non-coding RNAs as epigenetic regulators of immune checkpoints in cancer immunity. Cancers (Basel). (2022) 15. doi: 10.3390/cancers15010184
49. Yan K, Fu Y, Zhu N, Wang Z, Hong JL, Li Y, et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8(+)T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int J Biochem Cell Biol. (2019) 110:1–8. doi: 10.1016/j.biocel.2019.01.019
50. Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, et al. Long non-coding RNA Lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis. (2018) 9:478. doi: 10.1038/s41419-018-0528-7
51. Xi Q, Yang G, He X, Zhuang H, Li L, Lin B, et al. M(6)A-mediated upregulation of lncRNA TUG1 in liver cancer cells regulates the antitumor response of CD8(+) T cells and phagocytosis of macrophages. Adv Sci (Weinh). (2024) 11:e2400695. doi: 10.1002/advs.202400695
52. Zhang X, Pan B, Qiu J, Ke X, Shen S, Wang X, et al. lncRNA MIAT targets miR-411-5p/STAT3/PD-L1 axis mediating hepatocellular carcinoma immune response. Int J Exp Pathol. (2022) 103:102–11. doi: 10.1111/iep.12440
53. AbuliHaiti Z, Li W, Yang L, Zhang H, Du A, Tang N, et al. Hypoxia-driven lncRNA CTD-2510F5.4: a potential player in hepatocellular carcinoma's prognostic stratification, cellular behavior, tumor microenvironment, and therapeutic response. Mol Biol Rep. (2024) 51:905. doi: 10.1007/s11033-024-09826-6
54. Ma K, Chu J, Liu Y, Sun L, Zhou S, Li X, et al. Hepatocellular carcinoma LINC01116 outcompetes T cells for linoleic acid and accelerates tumor progression. Adv Sci (Weinh). (2024) 11:e2400676. doi: 10.1002/advs.202400676
55. Li X, Liang S, Fei M, Ma K, Sun L, Liu Y, et al. LncRNA CRNDE drives the progression of hepatocellular carcinoma by inducing the immunosuppressive niche. Int J Biol Sci. (2024) 20:718–32. doi: 10.7150/ijbs.85471
56. Zhang J, Pan T, Zhou W, Zhang Y, Xu G, Xu Q, et al. Long noncoding RNA LINC01132 enhances immunosuppression and therapy resistance via NRF1/DPP4 axis in hepatocellular carcinoma. J Exp Clin Cancer Res. (2022) 41:270. doi: 10.1186/s13046-022-02478-z
57. Chen W, Chen F, Gong M, Jin Z, Shu L, Wang ZW, et al. Comprehensive analysis of lncRNA-mediated ceRNA networkfor hepatocellular carcinoma. Front Oncol. (2022) 12:1042928. doi: 10.3389/fonc.2022.1042928
58. Zhong F, Liu S, Hu D, Chen L. LncRNA AC099850.3 promotes hepatocellular carcinoma proliferation and invasion through PRR11/PI3K/AKT axis and is associated with patients prognosis. J Cancer. (2022) 13:1048–60. doi: 10.7150/jca.66092
59. Sha Z, Zhang P. LncRNA-GAS5 and its promoter region polymorphism associate with helper th17 polarization and predict postoperative pain and the prognosis of the patients with hepatocellular carcinoma undergoing hepatectomy. Iran J Immunol. (2022) 19:349–57. doi: 10.22034/IJI.2022.92678.2168
60. Pehlivanoglu B, Aysal A, Agalar C, Egeli T, Ozbilgin M, Unek T, et al. lncRNA XIST interacts with regulatory T cells within the tumor microenvironment in chronic hepatitis B-associated hepatocellular carcinoma. Turk Patoloji Derg. (2024) 40:101–8. doi: 10.5146/tjpath.2023.13161
61. Dong W, Zhan C. Bioinformatic-based mechanism identification of E2F1-related ceRNA and E2F1 immunoassays in hepatocellular carcinoma. J Gastrointest Oncol. (2022) 13:1915–26. doi: 10.21037/jgo-22-674
62. Xie C, Wang S, Zhang H, Zhu Y, Jiang P, Shi S, et al. Lnc-AIFM2-1 promotes HBV immune escape by acting as a ceRNA for miR-330-3p to regulate CD244 expression. Front Immunol. (2023) 14:1121795. doi: 10.3389/fimmu.2023.1121795
63. Peng L, Chen Y, Ou Q, Wang X, Tang N. LncRNA MIAT correlates with immune infiltrates and drug reactions in hepatocellular carcinoma. Int Immunopharmacol. (2020) 89:107071. doi: 10.1016/j.intimp.2020.107071
64. Fan F, Chen K, Lu X, Li A, Liu C, Wu B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol Int. (2021) 15:444–58. doi: 10.1007/s12072-020-10101-6
65. Huang LH, Rau CS, Liu YW, Wu CJ, Chien PC, Lin HP, et al. Exploring the regulatory role of XIST-microRNAs/mRNA network in circulating CD4(+) T cells of hepatocellular carcinoma patients. Biomedicines. (2023) 11. doi: 10.3390/biomedicines11071848
66. Xiao S, Huang S, Yang J. Overexpression of GIHCG is associated with a poor prognosis and immune infiltration in hepatocellular carcinoma. Onco Targets Ther. (2020) 13:11607–19. doi: 10.2147/OTT.S271966
67. Song H, Huang XF, Hu SY, Lu LL, Yang XY. The LINC00261/MiR105-5p/SELL axis is involved in dysfunction of B cell and is associated with overall survival in hepatocellular carcinoma. PeerJ. (2022) 10:e12588. doi: 10.7717/peerj.12588
68. Zhan T, Gao X, Wang G, Li F, Shen J, Lu C, et al. Construction of novel lncRNA-miRNA-mRNA network associated with recurrence and identification of immune-related potential regulatory axis in hepatocellular carcinoma. Front Oncol. (2021) 11:626663. doi: 10.3389/fonc.2021.626663
69. Yu Z, Zhao H, Feng X, Li H, Qiu C, Yi X, et al. Long non-coding RNA FENDRR acts as a miR-423-5p sponge to suppress the treg-mediated immune escape of hepatocellular carcinoma cells. Mol Ther Nucleic Acids. (2019) 17:516–29. doi: 10.1016/j.omtn.2019.05.027
70. Xu M, Zhou C, Weng J, Chen Z, Zhou Q, Gao J, et al. Tumor associated macrophages-derived exosomes facilitate hepatocellular carcinoma Malignance by transferring lncMMPA to tumor cells and activating glycolysis pathway. J Exp Clin Cancer Res. (2022) 41:253. doi: 10.1186/s13046-022-02458-3
71. Li X, Lei Y, Wu M, Li N. Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int J Mol Sci. (2018) 19. doi: 10.3390/ijms19102958
72. Lv S, Wang J, Li L. Extracellular vesicular lncRNA FAL1 promotes hepatocellular carcinoma cell proliferation and invasion by inducing macrophage M2 polarization. J Physiol Biochem. (2023) 79:669–82. doi: 10.1007/s13105-022-00922-4
73. Ai JH, Wen YZ, Dai SJ, Zhang LD, Huang ZJ, Shi J. Exosomal lncRNA HEIH, an essential communicator for hepatocellular carcinoma cells and macrophage M2 polarization through the miR-98-5p/STAT3 axis. J Biochem Mol Toxicol. (2024) 38:e23686. doi: 10.1002/jbt.23686
74. Wang X, Zhou Y, Dong K, Zhang H, Gong J, Wang S. Exosomal lncRNA HMMR-AS1 mediates macrophage polarization through miR-147a/ARID3A axis under hypoxia and affects the progression of hepatocellular carcinoma. Environ Toxicol. (2022) 37:1357–72. doi: 10.1002/tox.23489
75. Zhou J, Che J, Xu L, Yang W, Zhou W, Zhou C. Tumor-derived extracellular vesicles containing long noncoding RNA PART1 exert oncogenic effect in hepatocellular carcinoma by polarizing macrophages into M2. Dig Liver Dis. (2022) 54:543–53. doi: 10.1016/j.dld.2021.07.005
76. Ye Y, Wang M, Wang G, Mai Z, Zhou B, Han Y, et al. lncRNA miR4458HG modulates hepatocellular carcinoma progression by activating m6A-dependent glycolysis and promoting the polarization of tumor-associated macrophages. Cell Mol Life Sci. (2023) 80:99. doi: 10.1007/s00018-023-04741-8
77. Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R, et al. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J Cell Biochem. (2018) 119:2951–63. doi: 10.1002/jcb.v119.3
78. Luo T, Chen M, Zhao Y, Wang D, Liu J, Chen J, et al. Macrophage-associated lncRNA ELMO1-AS1: a novel therapeutic target and prognostic biomarker for hepatocellular carcinoma. Onco Targets Ther. (2019) 12:6203–16. doi: 10.2147/OTT.S213833
79. Lin J, Huang J, Tan C, Wu S, Lu X, Pu J. LncRNA MEG3 suppresses hepatocellular carcinoma by stimulating macrophage M1 polarization and modulating immune system via inhibiting CSF-1 in vivo/vitro studies. Int J Biol Macromol. (2024) 281:136459. doi: 10.1016/j.ijbiomac.2024.136459
80. Wei H, Wu X, Huang L, Long C, Lu Q, Huang Z, et al. LncRNA MEG3 reduces the ratio of M2/M1 macrophages through the huR/CCL5 axis in hepatocellular carcinoma. J Hepatocell Carcinoma. (2024) 11:543–62. doi: 10.2147/JHC.S449090
81. Tao L, Li D, Mu S, Tian G, Yan G. LncRNA MAPKAPK5_AS1 facilitates cell proliferation in hepatitis B virus -related hepatocellular carcinoma. Lab Invest. (2022) 102:494–504. doi: 10.1038/s41374-022-00731-9
82. Wei Q, Liu G, Huang Z, Huang Y, Huang L, Huang Z, et al. LncRNA MEG3 Inhibits Tumor Progression by Modulating Macrophage Phenotypic Polarization via miR-145-5p/DAB2 Axis in Hepatocellular Carcinoma. J Hepatocell Carcinoma. (2023) 10:1019–35. doi: 10.2147/JHC.S408800
83. Lin C, Jiang T EC, Wang L, Chen T, Wang X, Xiang Y. LncRNA CRNDE promotes hepatoma cell proliferation by regulating the metabolic reprogramming of M2 macrophages via ERK pathway. Cancer Cell Int. (2024) 24:193. doi: 10.1186/s12935-024-03380-8
84. Chen J, Huang ZB, Liao CJ, Hu XW, Li SL, Qi M, et al. LncRNA TP73-AS1/miR-539/MMP-8 axis modulates M2 macrophage polarization in hepatocellular carcinoma via TGF-beta1 signaling. Cell Signal. (2020) 75:109738. doi: 10.1016/j.cellsig.2020.109738
85. Luo HL, Luo T, Liu JJ, Wu FX, Bai T, Ou C, et al. Macrophage polarization-associated lnc-Ma301 interacts with caprin-1 to inhibit hepatocellular carcinoma metastasis through the Akt/Erk1 pathway. Cancer Cell Int. (2021) 21:422. doi: 10.1186/s12935-021-02133-1
86. Tian X, Wu Y, Yang Y, Wang J, Niu M, Gao S, et al. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/beta-catenin signaling. Mol Oncol. (2020) 14:462–83. doi: 10.1002/1878-0261.12606
87. Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am J Physiol Cell Physiol. (2020) 318:C649–63. doi: 10.1152/ajpcell.00510.2018
88. Sun J, Li Y, Tian H, Chen H, Li J, Li Z. Comprehensive analysis identifies long non-coding RNA RNASEH1-AS1 as a potential prognostic biomarker and oncogenic target in hepatocellular carcinoma. Am J Cancer Res. (2024) 14:996–1014. doi: 10.62347/JPHF4071
89. Wei M, Lu L, Ma J, Luo Z, Tan X, Wang J. LINC00707 impairs the Natural Killer cell antitumour activity in hepatocellular carcinoma through decreasing YTHDF2 stability. J Cell Mol Med. (2024) 28:e18106. doi: 10.1111/jcmm.18106
90. Lin XQ, Huang ZM, Chen X, Wu F, Wu W. XIST induced by JPX suppresses hepatocellular carcinoma by sponging miR-155-5p. Yonsei Med J. (2018) 59:816–26. doi: 10.3349/ymj.2018.59.7.816
91. Qiu J, Zhong F, Zhang Z, Pan B, Ye D, Zhang X, et al. Hypoxia-responsive lncRNA MIR155HG promotes PD-L1 expression in hepatocellular carcinoma cells by enhancing HIF-1alpha mRNA stability. Int Immunopharmacol. (2024) 136:112415. doi: 10.1016/j.intimp.2024.112415
92. Peng L, Pan B, Zhang X, Wang Z, Qiu J, Wang X, et al. Lipopolysaccharide facilitates immune escape of hepatocellular carcinoma cells via m6A modification of lncRNA MIR155HG to upregulate PD-L1 expression. Cell Biol Toxicol. (2022) 38:1159–73. doi: 10.1007/s10565-022-09718-0
93. Zeng C, Ye S, Chen Y, Zhang Q, Luo Y, Gai L, et al. HOXA-AS3 Promotes Proliferation and Migration of Hepatocellular Carcinoma Cells via the miR-455-5p/PD-L1 Axis. J Immunol Res. (2021) 2021:9289719. doi: 10.1155/2021/9289719
94. Dong L, Zhou S, Bai X, He X. Construction of a prognostic model for HCC based on ferroptosis-related lncRNAs expression and its potential to predict the response and irAEs of immunotherapy. Front Pharmacol. (2023) 14:1090895. doi: 10.3389/fphar.2023.1090895
95. Liang J, Zhi Y, Deng W, Zhou W, Li X, Cai Z, et al. Dong Y, et al. Development and validation of ferroptosis-related lncRNAs signature for hepatocellular carcinoma. PeerJ. (2021) 9:e11627. doi: 10.7717/peerj.11627
96. Sun Z, Xue C, Li J, Zhao H, Du Y, Du N. LINC00244 suppresses cell growth and metastasis in hepatocellular carcinoma by downregulating programmed cell death ligand 1. Bioengineered. (2022) 13:7635–47. doi: 10.1080/21655979.2022.2050073
97. Zhang J, Zhao X, Ma X, Yuan Z, Hu M. KCNQ1OT1 contributes to sorafenib resistance and programmed death−ligand−1−mediated immune escape via sponging miR−506 in hepatocellular carcinoma cells. Int J Mol Med. (2020) 46:1794–804. doi: 10.3892/ijmm.2020.4710
98. Guo BL, Zheng QX, Jiang YS, Zhan Y, Huang WJ, Chen ZY. Long non-coding RNAFOXD1-AS1 modulated CTCs epithelial-mesenchymal transition and immune escape in hepatocellular carcinoma in vitro by sponging miR-615-3p. Cancer Rep (Hoboken). (2024) 7:e2050. doi: 10.1002/cnr2.2050
99. Wu MZ, Yuan YC, Huang BY, Chen JX, Li BK, Fang JH, et al. Identification of a TGF-beta/SMAD/lnc-UTGF positive feedback loop and its role in hepatoma metastasis. Signal Transduct Target Ther. (2021) 6:395. doi: 10.1038/s41392-021-00781-3
100. Sheng J, Luo Y, Lv E, Liang H, Tao H, Yu C, et al. LINC01980 induced by TGF-beta promotes hepatocellular carcinoma metastasis via miR-376b-5p/E2F5 axis. Cell Signal. (2023) 112:110923. doi: 10.1016/j.cellsig.2023.110923
101. Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. (2014) 25:666–81. doi: 10.1016/j.ccr.2014.03.010
102. Zhang Z, Zhou Y, Jia Y, Wang C, Zhang M, Xu Z. PRR34-AS1 promotes exosome secretion of VEGF and TGF-beta via recruiting DDX3X to stabilize Rab27a mRNA in hepatocellular carcinoma. J Transl Med. (2022) 20:491. doi: 10.1186/s12967-022-03628-9
103. Li D, Lu L, Liu M, Sun J. Inhibition of long noncoding RNA cancer susceptibility candidate 7 attenuates hepatocellular carcinoma development by targeting microRNA-30a-5p. Bioengineered. (2022) 13:11296–308. doi: 10.1080/21655979.2022.2068289
104. Huang WJ, Tian XP, Bi SX, Zhang SR, He TS, Song LY, et al. The beta-catenin/TCF-4-LINC01278-miR-1258-Smad2/3 axis promotes hepatocellular carcinoma metastasis. Oncogene. (2020) 39:4538–50. doi: 10.1038/s41388-020-1307-3
105. Shi Y, Yang X, Xue X, Sun D, Cai P, Song Q, et al. HANR promotes hepatocellular carcinoma progression via miR-214/EZH2/TGF-beta axis. Biochem Biophys Res Commun. (2018) 506:189–93. doi: 10.1016/j.bbrc.2018.10.038
106. Liang J, Liao J, Liu T, Wang Y, Wen J, Cai N, et al. Comprehensive analysis of TGF-beta-induced mRNAs and ncRNAs in hepatocellular carcinoma. Aging (Albany NY). (2020) 12:19399–420. doi: 10.18632/aging.v12i19
107. Wang X, Sun W, Shen W, Xia M, Chen C, Xiang D, et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J Hepatol. (2016) 64:1283–94. doi: 10.1016/j.jhep.2016.01.019
108. Sun QM, Hu B, Fu PY, Tang WG, Zhang X, Zhan H, et al. Long non-coding RNA 00607 as a tumor suppressor by modulating NF-kappaB p65/p53 signaling axis in hepatocellular carcinoma. Carcinogenesis. (2018) 39:1438–46. doi: 10.1093/carcin/bgy113
109. Li S, Hu X, Yu S, Yi P, Chen R, Huang Z, et al. Hepatic stellate cell-released CXCL1 aggravates HCC Malignant behaviors through the MIR4435-2HG/miR-506-3p/TGFB1 axis. Cancer Sci. (2023) 114:504–20. doi: 10.1111/cas.v114.2
110. Wang Y, Yang L, Dong X, Yang X, Zhang X, Liu Z, et al. Overexpression of NNT-AS1 activates TGF-beta signaling to decrease tumor CD4 lymphocyte infiltration in hepatocellular carcinoma. BioMed Res Int. (2020) 2020:8216541. doi: 10.1155/2020/8216541
111. Yang Y, Min Z. Effect of long non−coding RNA AK021443 on promoting hepatic fibrosis in vitro. Mol Med Rep. (2021) 23. doi: 10.3892/mmr.2021.12518
112. Tang J, Zhuo H, Zhang X, Jiang R, Ji J, Deng L, et al. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. (2014) 5:e1549. doi: 10.1038/cddis.2014.518
113. Liu Y, Chen L, Yuan H, Guo S, Wu G. LncRNA DANCR promotes sorafenib resistance via activation of IL-6/STAT3 signaling in hepatocellular carcinoma cells. Onco Targets Ther. (2020) 13:1145–57. doi: 10.2147/OTT.S229957
114. Lin Y, Jian Z, Jin H, Wei X, Zou X, Guan R, et al. Long non-coding RNA DLGAP1-AS1 facilitates tumorigenesis and epithelial-mesenchymal transition in hepatocellular carcinoma via the feedback loop of miR-26a/b-5p/IL-6/JAK2/STAT3 and Wnt/beta-catenin pathway. Cell Death Dis. (2020) 11:34. doi: 10.1038/s41419-019-2188-7
115. Huang M, Wang H, Hu X, Cao X. lncRNA MALAT1 binds chromatin remodeling subunit BRG1 to epigenetically promote inflammation-related hepatocellular carcinoma progression. Oncoimmunology. (2019) 8:e1518628. doi: 10.1080/2162402X.2018.1518628
116. Ding H, Liu J, Zou R, Cheng P, Su Y. Retraction Note: Long non-coding RNA TPTEP1 inhibits hepatocellular carcinoma progression by suppressing STAT3 phosphorylation. J Exp Clin Cancer Res. (2023) 42:123. doi: 10.1186/s13046-023-02700-6
117. Ma A, Sun Y, Ogbodu RO, Xiao L, Deng H, Zhou H. Identification of biomarkers of hepatocellular carcinoma gene prognosis based on the immune-related lncRNA signature of transcriptome data. Funct Integr Genomics. (2023) 23:104. doi: 10.1007/s10142-023-01019-x
118. Xu Z, Peng B, Liang Q, Chen X, Cai Y, Zeng S, et al. Construction of a ferroptosis-Related nine-lncRNA signature for predicting prognosis and immune response in hepatocellular carcinoma. Front Immunol. (2021) 12:719175. doi: 10.3389/fimmu.2021.719175
119. Mao Z, Nie Y, Jia W, Wang Y, Li J, Zhang T, et al. Revealing prognostic and immunotherapy-sensitive characteristics of a novel cuproptosis-related lncRNA model in hepatocellular carcinoma patients by genomic analysis. Cancers (Basel). (2023) 15. doi: 10.3390/cancers15020544
120. Wang X, Chen J, Lin L, Li Y, Tao Q, Lang Z, et al. Machine learning integrations develop an antigen-presenting-cells and T-Cells-Infiltration derived LncRNA signature for improving clinical outcomes in hepatocellular carcinoma. BMC Cancer. (2023) 23:284. doi: 10.1186/s12885-023-10766-w
121. Xie S, Zhong J, Zhang Z, Huang W, Lin X, Pan Y, et al. Novel risk model based on angiogenesis-related lncRNAs for prognosis prediction of hepatocellular carcinoma. Cancer Cell Int. (2023) 23:159. doi: 10.1186/s12935-023-02975-x
122. Zhang L, Hu S, Chen J, Ma S, Liu F, Liu C, et al. Comprehensive analysis of the MIR4435-2HG/miR-1-3p/MMP9/miR-29-3p/DUXAP8 ceRNA network axis in hepatocellular carcinoma. Discov Oncol. (2021) 12:38. doi: 10.1007/s12672-021-00436-3
123. Hu S, Zhang J, Guo G, Zhang L, Dai J, Gao Y. Comprehensive analysis of GSEC/miR-101-3p/SNX16/PAPOLG axis in hepatocellular carcinoma. PloS One. (2022) 17:e0267117. doi: 10.1371/journal.pone.0267117
124. Li X, Zhang Z, Liu M, Fu X, A J, Chen G, et al. Establishment of a lncRNA-based prognostic gene signature associated with altered immune responses in HCC. Front Immunol. (2022) 13:880288. doi: 10.3389/fimmu.2022.880288
125. Huang F, Zhang C, Yang W, Zhou Y, Yang Y, Yang X, et al. Identification of a DNA damage repair-related LncRNA signature for predicting the prognosis and immunotherapy response of hepatocellular carcinoma. BMC Genomics. (2024) 25:155. doi: 10.1186/s12864-024-10055-1
126. Xia W, Zeng C, Zheng Z, Huang C, Zhou Y, Bai L. Development and validation of a novel mitochondrion and ferroptosis-related long non-coding RNA prognostic signature in hepatocellular carcinoma. Front Cell Dev Biol. (2022) 10:844759. doi: 10.3389/fcell.2022.844759
127. Yu ZL, Zhu ZM. Comprehensive analysis of N6-methyladenosine -related long non-coding RNAs and immune cell infiltration in hepatocellular carcinoma. Bioengineered. (2021) 12:1708–24. doi: 10.1080/21655979.2021.1923381
128. Lu D, Liao J, Cheng H, Ma Q, Wu F, Xie F, et al. Construction and systematic evaluation of a machine learning-based cuproptosis-related lncRNA score signature to predict the response to immunotherapy in hepatocellular carcinoma. Front Immunol. (2023) 14:1097075. doi: 10.3389/fimmu.2023.1097075
129. Huang EM, Ma N, Ma T, Zhou JY, Yang WS, Liu CX, et al. Cuproptosis-related long non-coding RNAs model that effectively predicts prognosis in hepatocellular carcinoma. World J Gastrointest Oncol. (2022) 14:1981–2003. doi: 10.4251/wjgo.v14.i10.1981
130. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. (2009) 23:1494–504. doi: 10.1101/gad.1800909
131. Chen J, Liu A, Wang Z, Wang B, Chai X, Lu W, et al. LINC00173.v1 promotes angiogenesis and progression of lung squamous cell carcinoma by sponging miR-511-5p to regulate VEGFA expression. Mol Cancer. (2020) 19:98. doi: 10.1186/s12943-020-01217-2
132. Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. (2016) 6:784–801. doi: 10.1158/2159-8290.CD-15-0921
133. Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. (2018) 37:4094–109. doi: 10.1038/s41388-018-0250-z
134. Li Y, Jiang T, Zhou W, Li J, Li X, Wang Q, et al. Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun. (2020) 11:1000. doi: 10.1038/s41467-020-14802-2
135. Xu Z, Chen Y, Ma L, Chen Y, Liu J, Guo Y, et al. Role of exosomal non-coding RNAs from tumor cells and tumor-associated macrophages in the tumor microenvironment. Mol Ther. (2022) 30:3133–54. doi: 10.1016/j.ymthe.2022.01.046
136. Nakagawa C, Kadlera Nagaraj M, Hernandez JC, Uthay Kumar DB, Shukla V, Machida R, et al. beta-CATENIN stabilizes HIF2 through lncRNA and inhibits intravenous immunoglobulin immunotherapy. Front Immunol. (2023) 14:1204907. doi: 10.3389/fimmu.2023.1204907
137. Jia X, Wang Y, Yang Y, Fu Y, Liu Y. Constructed risk prognosis model associated with disulfidptosis lncRNAs in HCC. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms242417626
138. Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. (2018) 75:467–84. doi: 10.1007/s00018-017-2626-6
139. Zhang L, Zou J, Wang Z, Li L. A subpathway and target gene cluster-based approach uncovers lncRNAs associated with human primordial follicle activation. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms241310525
140. Lim LJ, Wong SYS, Huang F, Lim S, Chong SS, Ooi LL, et al. Roles and regulation of long noncoding RNAs in hepatocellular carcinoma. Cancer Res. (2019) 79:5131–9. doi: 10.1158/0008-5472.CAN-19-0255
141. Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. (2020) 13:5. doi: 10.1186/s13045-019-0839-x
142. Vilaca A, de Windt LJ, Fernandes H, Ferreira L. Strategies and challenges for non-viral delivery of non-coding RNAs to the heart. Trends Mol Med. (2023) 29:70–91. doi: 10.1016/j.molmed.2022.10.002
143. Machyna M, Kiefer L, Simon MD. Enhanced nucleotide chemistry and toehold nanotechnology reveals lncRNA spreading on chromatin. Nat Struct Mol Biol. (2020) 27:297–304. doi: 10.1038/s41594-020-0390-z
144. Horns F, Martinez JA, Fan C, Haque M, Linton JM, Tobin V, et al. Engineering RNA export for measurement and manipulation of living cells. Cell. (2023) 186:3642–3658 e3632. doi: 10.1016/j.cell.2023.06.013
145. Yuan J, Yang J, Wang R, Hao H, Li J. LncRNA NEAT1 regulate diffuse large B-cell lymphoma by targeting miR-495-3p/PD-L1 axis. Immunopharmacol Immunotoxicol. (2022) 44:429–36. doi: 10.1080/08923973.2022.2052896
146. Yin L, Wang Y. Extracellular vesicles derived from M2-polarized tumor-associated macrophages promote immune escape in ovarian cancer through NEAT1/miR-101-3p/ZEB1/PD-L1 axis. Cancer Immunol Immunother. (2023) 72:743–58. doi: 10.1007/s00262-022-03305-2
147. Toker J, Iorgulescu JB, Ling AL, Villa GR, Gadet J, Parida L, et al. Clinical importance of the lncRNA NEAT1 in cancer patients treated with immune checkpoint inhibitors. Clin Cancer Res. (2023) 29:2226–38. doi: 10.1158/1078-0432.CCR-22-3714
148. Yang Q, Chen W, Xu Y, Lv X, Zhang M, Jiang H. Polyphyllin I modulates MALAT1/STAT3 signaling to induce apoptosis in gefitinib-resistant non-small cell lung cancer. Toxicol Appl Pharmacol. (2018) 356:1–7. doi: 10.1016/j.taap.2018.07.031
149. Jing Z, Liu Q, Xie W, Wei Y, Liu J, Zhang Y, et al. NCAPD3 promotes prostate cancer progression by up-regulating EZH2 and MALAT1 through STAT3 and E2F1. Cell Signal. (2022) 92:110265. doi: 10.1016/j.cellsig.2022.110265
150. Goyal B, Yadav SRM, Awasthee N, Gupta S, Kunnumakkara AB, Gupta SC. Diagnostic, prognostic, and therapeutic significance of long non-coding RNA MALAT1 in cancer. Biochim Biophys Acta Rev Cancer. (2021) 1875:188502. doi: 10.1016/j.bbcan.2021.188502
151. Barker SJ, Thayer MB, Kim C, Tatarakis D, Simon MJ, Dial R, et al. Targeting the transferrin receptor to transport antisense oligonucleotides across the mammalian blood-brain barrier. Sci Transl Med. (2024) 16:eadi2245. doi: 10.1126/scitranslmed.adi2245
152. Wang X, Zhang Y, Zheng J, Yao C, Lu X. LncRNA UCA1 attenuated the killing effect of cytotoxic CD8 + T cells on anaplastic thyroid carcinoma via miR-148a/PD-L1 pathway. Cancer Immunol Immunother. (2021) 70:2235–45. doi: 10.1007/s00262-020-02753-y
153. Yang LZ, Wang Y, Li SQ, Yao RW, Luan PF, Wu H, et al. Dynamic imaging of RNA in living cells by CRISPR-cas13 systems. Mol Cell. (2019) 76:981–997 e987. doi: 10.1016/j.molcel.2019.10.024
154. Chen Y, Li Z, Chen X, Zhang S. Long non-coding RNAs: From disease code to drug role. Acta Pharm Sin B. (2021) 11:340–54. doi: 10.1016/j.apsb.2020.10.001
155. Zhang L, Xu W, Gao X, Li W, Qi S, Guo D, et al. lncRNA sensing of a viral suppressor of RNAi activates non-canonical innate immune signaling in drosophila. Cell Host Microbe. (2020) 27:115–128 e118. doi: 10.1016/j.chom.2019.12.006
156. Han S, Chen X, Huang L. The tumor therapeutic potential of long non-coding RNA delivery and targeting. Acta Pharm Sin B. (2023) 13:1371–82. doi: 10.1016/j.apsb.2022.12.005
157. Niu X, Zhang J, Zhang J, Bai L, Hu S, Zhang Z, et al. Lipid nanoparticle-mediated oip5-as1 delivery preserves mitochondrial function in myocardial ischemia/reperfusion injury by inhibiting the p53 pathway. ACS Appl Mater Interfaces. (2024) 16:61565–82. doi: 10.1021/acsami.4c10032
158. Sarangi P, Senthilkumar MB, Amit S, Kumar N, Jayandharan GR. AAV mediated repression of Neat1 lncRNA combined with F8 gene augmentation mitigates pathological mediators of joint disease in haemophilia. J Cell Mol Med. (2024) 28:e18460. doi: 10.1111/jcmm.v28.11
159. Wu H, Qin W, Lu S, Wang X, Zhang J, Sun T, et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2'-O-methylation via NOP58 recruitment in colorectal cancer. Mol Cancer. (2020) 19:95. doi: 10.1186/s12943-020-01201-w
160. Amodio N, Stamato MA, Juli G, Morelli E, Fulciniti M, Manzoni M, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. (2018) 32:1948–57. doi: 10.1038/s41375-018-0067-3
161. Gao P, Lyu Q, Ghanam AR, Lazzarotto CR, Newby GA, Zhang W, et al. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome Biol. (2021) 22:83. doi: 10.1186/s13059-021-02304-3
162. Sehgal A, Vaishnaw A, Fitzgerald K. Liver as a target for oligonucleotide therapeutics. J Hepatol. (2013) 59:1354–9. doi: 10.1016/j.jhep.2013.05.045
163. Li G, Zhu X, Wang Y, Ma H, Wang Y, Wu H, et al. Transcription-wide impact by RESCUE-induced off-target single-nucleotide variants in mammalian cells. J Mol Cell Biol. (2023) 15. doi: 10.1093/jmcb/mjad011
164. Zhang L, Liang D, Chen C, Wang Y, Amu G, Yang J, et al. Circular siRNAs for reducing off-target effects and enhancing long-term gene silencing in cells and mice. Mol Ther Nucleic Acids. (2018) 10:237–44. doi: 10.1016/j.omtn.2017.12.007
165. Mortberg MA, Gentile JE, Nadaf NM, Vanderburg C, Simmons S, Dubinsky D, et al. A single-cell map of antisense oligonucleotide activity in the brain. Nucleic Acids Res. (2023) 51:7109–24. doi: 10.1093/nar/gkad371
Keywords: lncRNAs, HCC, immune microenvironment, immunotherapy, immune evasion
Citation: Zhao S, Chen F, Hu L, Li X, Gao Z, Chen M, Wang X and Song Z (2025) Long non-coding rnas as key modulators of the immune microenvironment in hepatocellular carcinoma: implications for Immunotherapy. Front. Immunol. 16:1523190. doi: 10.3389/fimmu.2025.1523190
Received: 05 November 2024; Accepted: 02 April 2025;
Published: 25 April 2025.
Edited by:
Alice Conigliaro, University of Palermo, ItalyReviewed by:
Jin-Fen Xiao, University of Texas MD Anderson Cancer Center, United StatesMengsha Shi, Nanjing Medical University, China
Baojie Mao, Zhejiang University, China
Copyright © 2025 Zhao, Chen, Hu, Li, Gao, Chen, Wang and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minjie Chen, Y2hlbm1pbmppZWRvY3RvckAxNjMuY29t; Xiaoguang Wang, eGlhb2d1YW5nd2FuZ3NAMTYzLmNvbQ==; Zhengwei Song, ZG9jdG9yc29uZ3p3QHpqeHUuZWR1LmNu
†These authors share first authorship
 Siqi Zhao
Siqi Zhao Fei Chen†
Fei Chen† Lingyu Hu
Lingyu Hu Zhengwei Song
Zhengwei Song