- 1Department of Immunology, Nankai University School of Medicine, Nankai University, Tianjin, China
- 2State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, China
- 3Translational Medicine Institute, Affiliated Tianjin Union Medical Center of Nankai University, Nankai University, Tianjin, China
Tumor is one of the leading causes of death worldwide. The occurrence and development of tumors are related to multiple systems and factors such as the immune system, gut microbiota, and nervous system. The immune system plays a critical role in tumor development. Studies have also found that the gut microbiota can directly or indirectly affect tumorigenesis and tumor development. With increasing attention on the tumor microenvironment in recent years, the nervous system has emerged as a novel regulator of tumor development. Some tumor therapies based on the nervous system have also been tested in clinical trials. However, the nervous system can not only directly interact with tumor cells but also indirectly affect tumor development through the gut microbiota. The nervous system-mediated gut microbiota can regulate tumorigenesis, growth, invasion, and metastasis through the immune system. Here, we mainly explore the potential effects of the nervous system–gut microbiota–immune system axis on tumorigenesis and tumor development. The effects of the nervous system–gut microbiota–immune system axis on tumors involve the nervous system regulating immune cells through the gut microbiota, which can prevent tumor development. Meanwhile, the direct effects of the gut microbiota on tumors and the regulation of the immune system by the nervous system, which can affect tumor development, are also reviewed.
1 Introduction
Tumor is one of the leading causes of death worldwide. The occurrence and development of tumors are related to multiple factors such as depression (1), inhibition of the immune system, and alteration of the gut microbiota (2).
There has been a growing interest in the study of depression in patients with tumors, aiming to assess the association between tumors and depression (3). Although a common underlying pathophysiological mechanism between depression and tumors is yet to be elucidated (4), studies have implicated that there exists an altered gut microbiota in anxiety, stress, and depression (5). Evidence from preclinical and clinical studies has suggested that alterations of the gut microbiota play a key role in the pathophysiology of depression (6). Since tumor and depression are closely interrelated, particularly in patients with advanced cancer, who often present with depression (3), an altered gut microbiota in depression should be also related to tumor development.
The effects of gut microbiota on the occurrence and development of tumors have been widely reviewed (2, 7). Multiple factors associated with the gut microbiota such as gut dysbiosis, metabolites, specific pathogenic microbes, and virulence factors can contribute to the initiation and progression of tumors such as colorectal cancers (CRCs). An altered gut microbiota is found in various CRC stages, including early and advanced CRCs. Metagenomic and metabolomic analyses showed distinct phenotypes in gut microbiota in CRCs (8). In CRC patients, Fusobacterium nucleatum (F. nucleatum) enrichment, short-chain fatty acid (SCFA) depletion, a shift in acetate/acetaldehyde metabolism toward acetyl-CoA production, and reduced microbial GABA (gamma-aminobutyric acid) biosynthesis were noted (8). Furthermore, specific bacteria associated with CRCs were also found. They included Escherichia coli (E. coli), F. nucleatum, Enterococcus faecalis (E. faecalis), Streptococcus gallolyticus (S. gallolyticus), and Bacteroides fragilis (B. fragilis). The metabolites from gut microbiota can not only directly affect tumor development but also play an important role in tumor-associated immune responses such as inflammation and immune tolerance.
The immune system plays a critical role in preventing the occurrence and development of tumors, including immune-effective cells such as CD8 and NK cells; inflammatory cells such as inflammatory macrophages (iMACs), dendritic cells (iDCs), T helper (Th1) cells, and Th17 cells; and immune regulatory cells such as myeloid-derived suppressor cells (MDSCs), immune T regulatory cells (Treg cells), Breg cells, and innate lymphocytes (ILCs). Recent studies involving bio-information analysis also identified some immune cell-associated genes in tumor cells, which could affect tumorigenesis such as DACH1 (9) in benign prostatic hyperplasia; ITGB1 (10), HSP90B1 (11, 12), and pre-mRNA processing factor 19 (PRPF19) (13) in bladder carcinoma; and the monocyte-related gene CCNA2 (14) in prostate adenocarcinoma (PRAD). Importantly, the differentiation and function of these immune cells could be regulated by gut microbiota metabolites (GMMs) such as SCFAs (15–17), tryptophan-derived metabolites (18–20), and bile acid (BA) derivatives (21–24). For example, BAs and their derivatives can affect the differentiation and function of MACs, DCs, MDSCs, Tregs, and T helper (Th) 17 cells (2, 7). Notably, immune cells regulated by the gut microbiota can influence tumor development.
Recent studies have shown that the nervous system, including the central nervous system (CNS) and the peripheral nervous system, plays a role in the progression of various cancers such as gastric, colon, rectal, oral squamous cell, head and neck, prostate, breast, and pancreatic cancers (3), and that the autonomic nerve contributes to prostate cancer progression (25–29). The nervous system can directly affect tumorigenesis, and the nervous system-mediated immune responses also influence the tumor (30, 31). The nervous system–gut microbiota–immune system axis affects tumors where the nervous system regulates immune cells through the gut microbiota to prevent tumor development. This review mainly explores the potential effects of the nervous system–gut microbiota–immune system axis on tumorigenesis and tumor development. Moreover, we also review the direct effects of the gut microbiota on tumors as well as the direct effects of the nervous system on the immune system, which can affect tumor development.
2 The enteric nervous system
The enteric nervous system (ENS), a division of the peripheral nervous system (PNS), is in the gut wall. It includes two main plexuses: the myenteric plexus and the submucosal plexus (32) (Figure 1). The ENS receives inputs from both the sympathetic and parasympathetic/vagal nervous systems (33), which enter the gastrointestinal (GI) tract (5). Motor neurons in the myenteric plexus mainly control muscle movements, whereas neurons in the submucosal plexus regulate secretion and absorption (34). Both neurons in the myenteric plexus and submucosal plexus can affect the composition of gut microbiota. Notably, recent progress in single-cell transcriptomics has enabled the discovery of new neuronal subtypes and improved the previous cell-type classifications (35, 36).
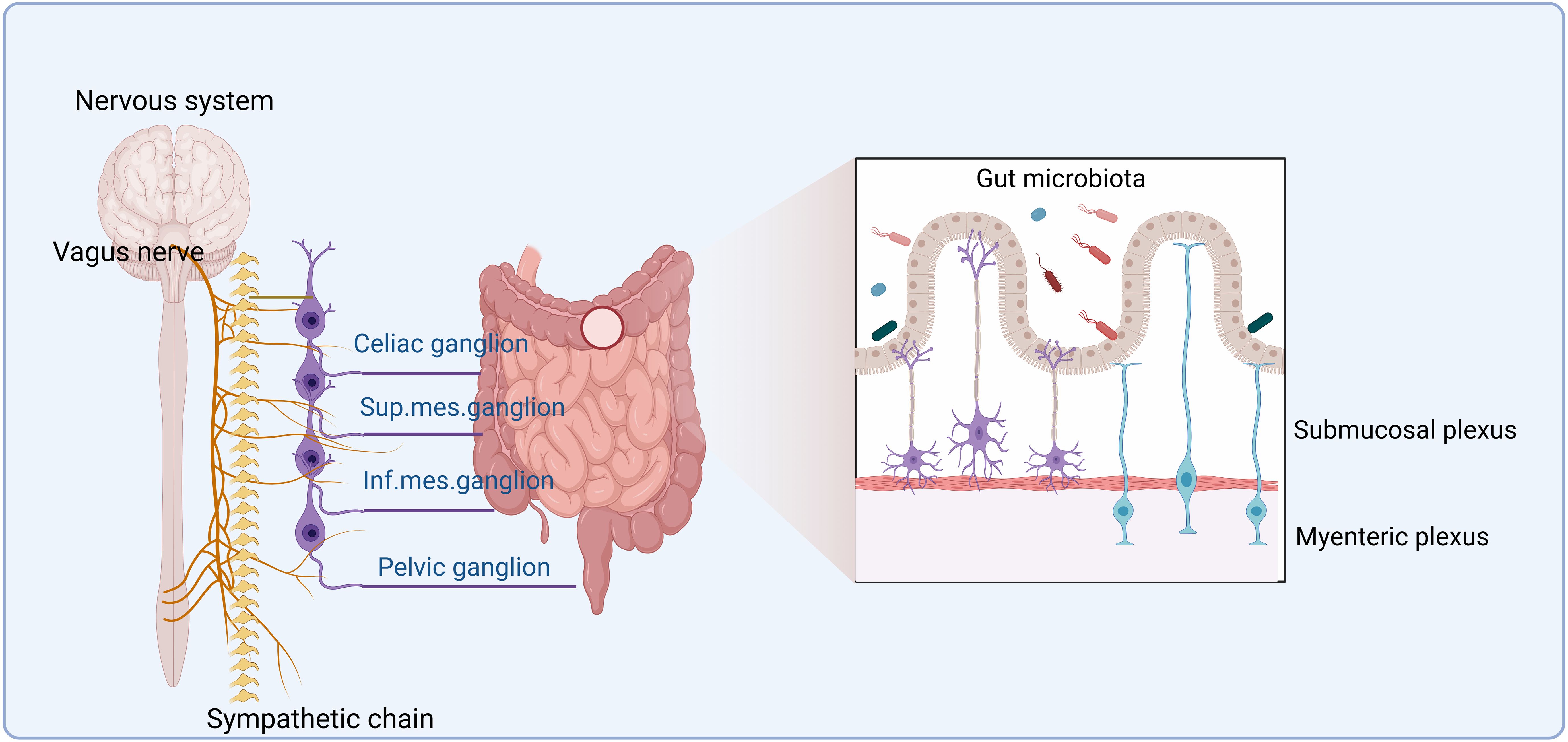
Figure 1. The enteric nervous system. The enteric nervous system (ENS) is formed by the myenteric plexus and submucosal plexus. It receives inputs from both the sympathetic and parasympathetic/vagal nervous systems. The ENS can communicate with the central nervous system (CNS), which encompasses the brain and the spinal cord. After leaving the hindbrain, the parasympathetic vagus nerve travels along the esophagus, through the diaphragm, and ultimately synapses onto the gut tract. On the other hand, the sympathetic nerves originate in the spinal column and synapse onto the sympathetic visceral ganglia, such as the celiac, superior, and inferior mesenteric ganglia. Nervous signals reach the gut microbiota via neurons from the ENS in the submucosa and myenteric plexus of the gut wall.
Although the ENS is capable of functioning autonomously, it also communicates with the CNS, which encompasses the brain and spinal cord (5). The human CNS is the most complex tissue, comprising approximately 86.1 billion neurons in the brain and spinal cord in male individuals, along with a roughly equal number of glial cells (37). CNS canonical activity includes neurotransmitter release at electrochemical synapses and consequent membrane depolarization, which can affect cellular functions (38, 39). Extrinsic connectivity from the CNS to the ENS is dependent on both sympathetic and parasympathetic/vagal nerve fibers (5). After leaving the hindbrain, the parasympathetic vagus nerve can travel along the esophagus onto the GI tract through the diaphragm and ultimately synapses. The sympathetic nerve originates in the spinal column and synapses onto the sympathetic visceral ganglia, such as the celiac, superior, and inferior mesenteric ganglia. Both parasympathetic and sympathetic nerves can directly synapse into the myenteric ganglia (5). Notably, to and from the gut, most neurological signals are through a bidirectional vagus nerve (VN). Signals from the CNS can reach 500 million neurons in the ENS (40) along efferent VN fibers. Bidirectional communication between the ENS and CNS can maintain homeostasis (41). In addition, in the colon, there exist afferent nerves linked to the spinal cord. The pelvic nerves, which are derived from the spinal cord and leave via the sacral spinal nerve, innervate the distal colon and rectum (5).
3 Potential effects of the nervous system–gut microbiota–immune system axis on tumors
Some environmental factors can influence the composition of the gut microbiota, including diet, use of drugs, host genetics, and hygiene (42). However, the gut microbiota is also affected by other factors such as the nervous system. The nervous system–gut microbiota–immune system axis includes the nervous system, gut microbiota, and immune system. The effects of the nervous system–gut microbiota–immune system axis on tumors involve the nervous system regulating immune cells through the gut microbiota, which can prevent tumor development, including the effects of the nervous system on gut microbiota, gut microbiota on the immune system, and immune system on tumors (Figure 2).
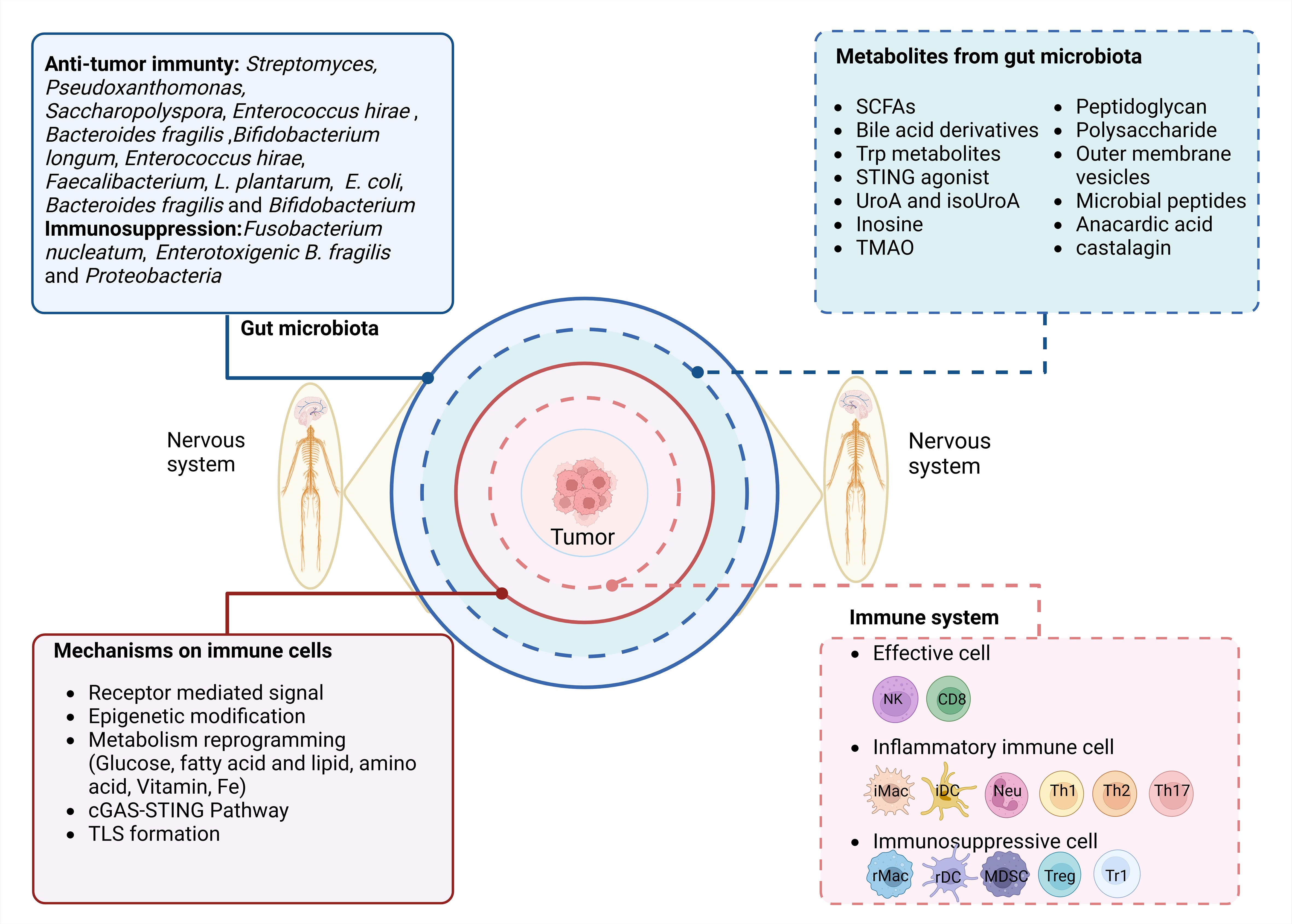
Figure 2. Regulation of the nervous system–gut microbiota–immune axis in tumorigenesis and tumor development. The nervous system–gut microbiota–immune system axis includes the nervous system, the gut microbiota/metabolites, and the immune system. The potential effect of the immune system on the tumor is through nervous system-mediated gut microbiota. They include three parts: 1) effects of the nervous system on gut microbiota/metabolites; 2) regulation of gut microbiota/metabolites in the immune system through multiple pathways such as receptor-mediated signal, epigenetic modification, metabolism reprogramming, cGAS-STING pathway, and tertiary lymphoid structure (TLS) formation; and 3) the effects of the immune system on the tumors.
3.1 Regulation of the enteric nervous system in gut microbiota
The nervous system can affect the occurrence and development of tumors by regulating the gut microbiota.
3.1.1 CNS and gut microbiota
Accumulating evidence from preclinical and clinical studies has shown that there exists altered gut microbiota in anxiety, depression, and stress (5, 43). The disorders were characterized by lower short-chain fatty acid-producing bacteria (e.g., Faecalibacterium) and a higher abundance of pro-inflammatory species (e.g., Enterobacteriaceae and Desulfovibrio) (43). In depressed patients, microbial metabolites such as butyrate decreased. Furthermore, butyrate could also exert antidepressant actions when administered to rodents, reducing stress responsiveness (44). The level of BAs modulated by the microbiota was inversely correlated with the severity of depression symptoms (45). Other microbial metabolites such as lipopolysaccharide (LPS) and trimethylamine-N-oxide from choline were also associated with depression (46). Neurotransmitters [e.g., serotonin or γ-aminobutyric acid (GABA)] from microbes can also regulate depression (44). The abnormal composition of gut microbiota also appeared in different animal models of depression (47). The restoration of gut microbiota with prebiotics or probiotics has gained considerable attention for the management of depression (48). Notably, dissimilar stressors had different effects on the composition of gut microbiota (49, 50).
3.1.2 ENS and gut microbiota
ENS is essential for the regulation of gut microbiota. Multiple molecular constituents from the ENS can potentially affect the composition of gut microbiota such as acetylcholine, catecholamines, neuropeptides, serotonin and histamine, and neurotrophic factors (5). Since ENS neurons do not extend into the intestinal lumen, they play a role in the microbiota through indirect pathways such as microbial molecules, which can penetrate the epithelial barrier (51). In addition, some microbial molecules also affect the composition of gut microbiota by affecting intestinal motility (52, 53). Notably, the effect of the nervous system on the gut microbiota is largely dependent on specific receptors and neuroregulatory substances in bacteria (54).
Notably, the gut microbiota can communicate with the CNS through specific metabolic compounds, e.g., SCFAs, BA derivatives, GABA, glutamate (Glu), norepinephrine (NE), dopamine (DA), serotonin (5-HT), and histamine. Afferent VN fibers transport signals from the gut microbiota to the brain. In response to these stimuli, the brain sends signals back to enteroepithelial cells via efferent VN fibers. There is no direct contact between VN fibers and intestinal microbiota. The nervous signals are via neurons to reach the gut microbiota from the ENS in the submucosa and myenteric plexus of the gut wall (55). So far, several studies have suggested the effects of ETS on gut microbiota (32), e.g., specific subsets of peripherally activated neurons could regulate the gut microbiome in mice without signal involvement from the brain (56). They found that activating gut ChAT+ and TH+ neurons of mice could alter the transcriptional and proteomic landscape of the intestines as well as the gut metagenome and metabolome (56). Deletion of choline acetyltransferase in enteric neurons also caused postnatal intestinal dysmotility and dysbiosis (57).
3.2 Regulation of the gut microbiota in the immune system
Gut microbiota plays a critical role in the differentiation and function of immune cells, which can influence tumorigenesis. In germ-free mice, there indeed exist immunological defects such as smaller Peyer’s patches and lymphoid follicles, abnormal germinal center, and reduced protective IgA (58). Segmented filamentous bacterium can promote Th17 cell differentiation (59), while Clostridium spp. induce Treg cells (60). The commensal bacterial antigens could help shape immunoglobulin repertoires in the gut (61). Thus, microbial signals from both local and distant sources have the ability to regulate innate and adaptive immune responses, causing systemic or tumor local-specific immune regulation (62). They have vast effects on the function and differentiation of immune-effective cells such as effective CD8 and NK cells, immune regulatory (suppressive) cells such as MDSCs, regulatory dendritic cells (rDCs), regulatory macrophages (rMACs), Tregs, Bregs, and also inflammatory cells such as Th1, Th2, and Th17. The regulation of the gut microbiota and its metabolites on immune cells is shown in Figure 3 (2, 7).
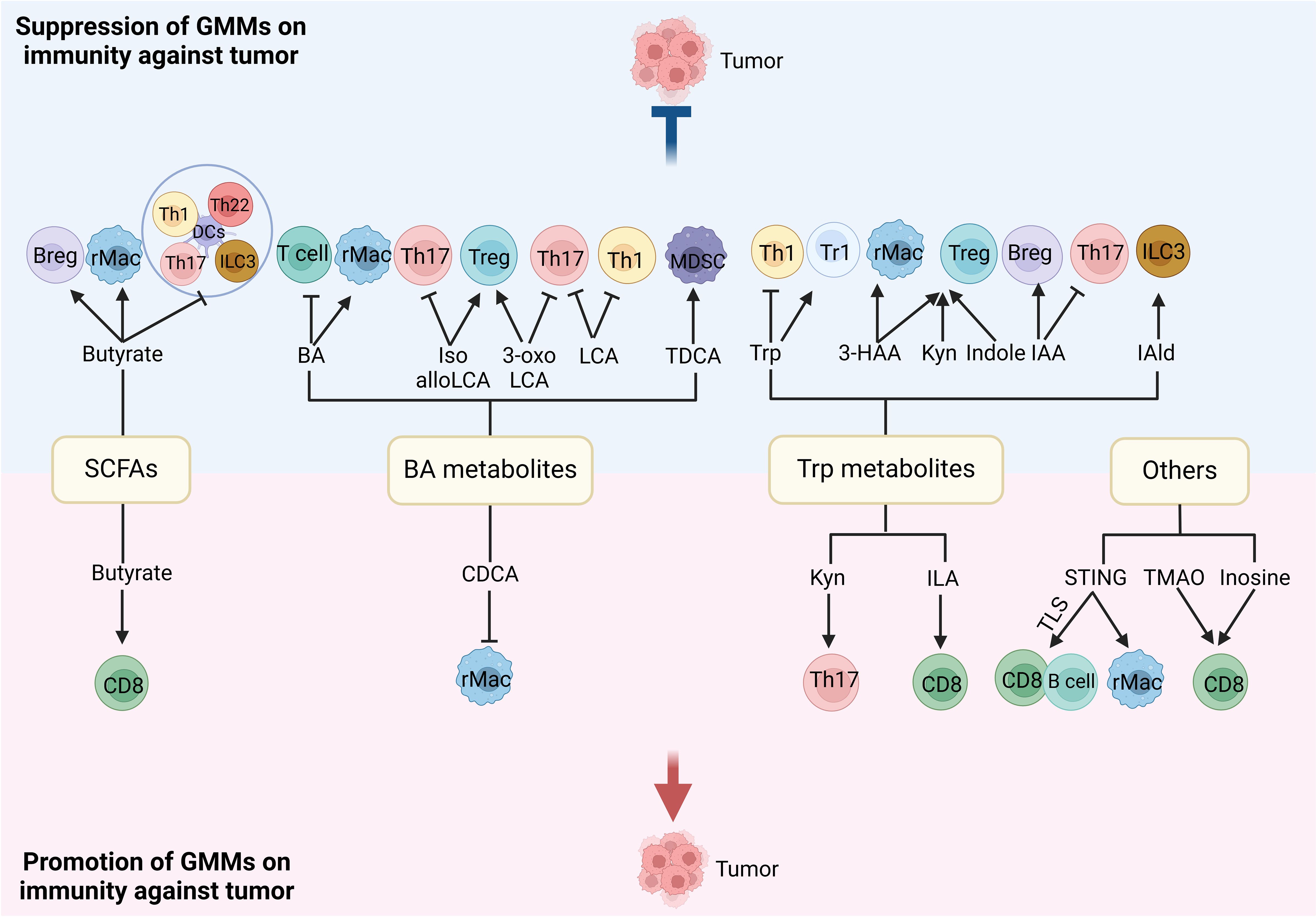
Figure 3. Regulation of gut microbiota metabolites (GMMs) in different immune cells. The gut microbiota/GMMs also have direct negative and positive effects on the function and differentiation of immune cells. Tregs, regulatory T cells; rMACs, regulatory MACs; Bregs, regulatory B cells; MDSCs, myeloid-derived suppressive cells; DCs, dendritic cells; NK cells, natural killer cells; Th cells, T helper cells; ILCs, innate lymphocytes; SCFAs, short-chain fatty acids; CDCA, chenodeoxycholic acid; Trp, tryptophan; Kyn, kynurenine; ILA, indole-3-lactic acid; IPA, indole-3-propionic acid; 3-HAA, 3-hydroxyanthranilic acid; IAA, indole-3-acid-acetic; I3A, indole-3-aldehyde; NorUDCA, norursodeoxycholicacid; STING, stimulator of the interferon gene; TMAO, trimethylamine oxide, TLS, tertiary lymphoid structure.
3.2.1 Gut microbiota/its metabolites
The gut microbiome is a fragile ecosystem, composed of bacteria, fungi, protozoans, viruses, and archaea. In many instances, the gut microbiota can regulate tumor progression through antitumor immunity and immunosuppression (63). Studies have also found that B. fragilis (64), B. fragilis (65) and Bifidobacterium (66, 67), B. fragilis and E. coli (68), Enterococcus hirae and Bifidobacterium longum (69), Enterococcus hirae, Faecalibacterium and Lactiplantibacillus plantarum (68), Streptomyces, Pseudoxanthomonas, and Saccharopolyspora (70) are related to antitumor immunity, whereas enterotoxigenic B. fragilis (71), F. nucleatum (72, 73), and Proteobacteria (74) were related to immune tolerance and immunosuppression.
Gut microbiota metabolites such as SCFAs, BA derivatives, and tryptophan metabolites can also regulate the differentiation and function of immune cells. SCFAs, mainly including acetate, propionate, and butyrate, are from dietary fiber fermentation in the cecum and colon by gut bacteria (75, 76). BAs are classified into primary BAs such as cholic acid and chenodeoxycholic acid and secondary BAs such as deoxycholic acid (DCA) and lithocholic acid (LCA). Primary BAs are synthesized in the liver. The secondary BAs are formed by bacteria in the colon. Bacteria can also conjugate chenodeoxycholic acid (CDCA), DCA, or cholic acid (CA) to one or more amino acids such as alanine, arginine, and aspartate (77). Conjugated BAs can be deconjugated in gut bacteria (78) and converted into secondary BAs, i.e., DCA and LCA. Four distinct ways, namely, deconjugation, dehydroxylation, oxidation, and epimerization, are used to transform BAs in humans (79). A range of iso-, oxo-, and epi-derivatives (80) such as oxo-BA metabolites 3-oxoLCA, 7-oxoCDCA, 12-oxoCA, 7-oxoCA, 12-oxoDCA (81), iso-LCA, 3-oxo-LCA, 3-oxoallo-LCA, isoalloLCA, allo-LCA, and 3-ketoLCA are also found in gut bacteria (79, 82, 83). Tryptophan metabolites such as indole (84), indole-3-acid-acetic (IAA) (85–87), indole-3-propionic acid (IPA) (85–87), indole-3-propionic acid (IPA) (88), indoleacrylic acid (IA) (88), indole-3-aldehyde (IAld) (89), skatole (87, 90), and tryptamine can be generated by different gut bacteria. In addition, gut microbiota bacteria also generate Kyn and downstream metabolites such as 3-hydroxyanthranilic acid (3-HAA) (91) through encoding enzymes homologous to those of the eukaryotic kynurenine (Kyn) pathway.
STING agents from Akkermansia muciniphila and Lactobacillus rhamnosus (92, 93), inosine from A. muciniphila and Bifidobacetrium pseudolongum (94), gut microbiota-derived inosine (94), and sulfur-metabolizing bacteria associated with hydrogen sulfide (81) can improve the activation of immune cells. TMAO from the gut microbiota trimethylamine (TMA) promoted CD8+ T-cell-mediated antitumor immunity via pyroptosis in mouse models (95). Other gut microbiota-derived metabolites such as peptidoglycan, polysaccharide, microbial peptides, anacardic acid, outer membrane vesicles, and castalagin could also promote the differentiation and function of inflammatory cells.
Urolithin A may alleviate colitis in mice by triggering AhR activation, improving gut microbiota dysbiosis, and regulating microbial Trp metabolism (96). Bifidobacterium-derived urea (97) was also associated with immunosuppression and tolerance to promote tumor development.
3.2.2 Mechanism(s) of the gut microbiota/its metabolites to regulate immune cells
The regulation of gut microbiota and/or its metabolites in immune cells is through receptor-mediated signals, epigenetic modification, metabolism (glucose, fatty acid and lipid metabolism, amino acid, vitamin, and Fe) reprogramming, cGAS-STING (stimulator of interferon genes) pathway, and formation of tertiary lymphoid structures (TLSs).
3.2.2.1 Receptor-mediated signals
SCFAs exert their role in immune cells through their receptors such as the G-protein-coupled receptor (GPR). BA metabolites depend on receptors such as TGR5 (GPBAR1), FXR (farnesol-X-receptor), VDR (vitamin D receptor), LXR (liver X receptor), PXR (pregnane X receptor), and RORγt (retinoid-related orphan receptor γt) to regulate the differentiation and function of immune cells (20, 98), whereas Trp metabolites are through their receptors such as AhR (99).
3.2.2.2 Epigenetic modification
Epigenetic modifications, including post‐translational modifications on histone and non‐histone proteins, can cause downstream cellular responses through regulating gene expression (100, 101). For example, altered gut microbiota in the body have also been shown to participate in the pathogenesis of bowel diseases by regulating the balance of Th17/Tregs through epigenetic modification (102).
3.2.2.3 Metabolism reprogramming
Glycolysis and mitochondrial oxidative phosphorylation (OXPHOS) are basically metabolic pathways for the generation of ATPs. It can rapidly produce energy and supply biosynthetic precursors to facilitate the function of immune cells such as the survival of proliferating cells (103).
Fatty acid and lipid metabolisms are also involved in the differentiation and function of immune cells. For example, the increased biosynthesis of fatty acids was implicated in macrophage polarization by Bacteroides thetaiotaomicron-derived acetic acid. Fatty acid oxidation contributes to the production of interleukin (IL)-17 and IL-22 in ILC3s. Elevated fatty acid uptake has been observed in activated ILC3s (104).
Amino acid metabolism also affects immune cells, e.g., glutamine metabolism was required for Tfh cells to perform optimally and interact with GC B cells (105). Lactobacillus reuteri with a tryptophan-rich diet could reprogram intraepithelial CD4+ T cells into Treg cells (89).
Vitamin D can regulate innate and adaptive immune cells to exert anticancer effects and can influence cancer cell growth, differentiation, and apoptosis through various mechanisms in the tumor microenvironment (TME) (106).
Recent studies have shown that Fe metabolism is also important in the differentiation and function of immune cells. Fe3+ enters the cell through the transferrin receptor. It is reduced to Fe2+ within the cell, related to ferroptosis. For ferroptosis caused by free iron, lethal lipid peroxides can initiate ferroptosis through catalyzing non-enzymatic Fenton reaction for directly peroxidating polyunsaturated fatty acid-containing phospholipids (107). Ferroptosis, as a new type of cell death, participates in the pathogenesis of diseases. Notably, recent studies have suggested that gut microbiota could regulate ferroptosis in immune cells, e.g., gut microbiota Trp metabolites IDA could inhibit ferroptosis through upregulating ALDH1A3 (aldehyde dehydrogenase 1 family member A3) to generate NADH through AhR. ALDH1A3 is essential for ferroptosis-suppressor protein 1(FSP1)-mediated synthesis of coenzyme Q10 (108).
3.2.2.4 The cGAS-STING pathway
The cGAS-STING pathway can trigger a robust IFN response to enhance therapeutic antitumor immune responses. Numerous studies emphasized the immunostimulatory properties through the cGAS-STING pathway (109).
3.2.2.5 TLS formation
Gut microbiota-mediated regulation in immune cells may also be through TLSs. TLSs are important tissues to generate adaptive T and B cells. Both the gut microbiota and its metabolites can potentially promote mature TLS formation (110). Signatures of mature TLSs have been described in other papers (111, 112). They have typical structures, which are comprised of an internal B-cell zone surrounded by a T-cell-rich area, which includes CD8 cytotoxic T, CD4 Th-1, and Tfh lymphocytes, as well as LAMP3+ DCs (113). However, the composition and distribution of TLSs is different in various tumor types (114) from disorganized cellular aggregates such as early or immature TLSs to mature TLSs (111, 115). Lymphoid aggregates only consisted of a few B cells and T cells without follicular dendritic cells (FDCs) (112). In immature TLS (iTLS), there often are aggregates of immune-suppressive cells such as Treg cells (116, 117), rDCs (116), rMACs (116), PD-1high CD8 T cells (116, 118), and CD4 T cells (118), which can suppress antitumor immunity.
3.3 Regulation of the immune system on tumors
The immune system of individuals influences tumorigenesis. Tumor immune responses, including antitumor immunity and tumor immune tolerance, have been widely reported (119). The role of the immune system in eradicating cancer cells has also been reviewed (120, 121). From preclinical to clinical research, evidence has established that the gut microbiota can modulate immunity against tumors and influence the efficacy of cancer immunotherapies, especially immune checkpoint inhibitors (63, 98, 122).
Taken together, the nervous system–gut microbiota–immune system axis should be a good target for preventing tumors.
4 Direct regulation of the gut microbiota on tumors
Studies have found that gut microbiota or its metabolites can directly affect tumorigenesis and tumor development (Figure 4) (63). Indeed, Heliobacter pylori (H. pylori) made a well-established contribution in carcinogenesis of gastric cancer. Specific bacteria associated with the onset and progression of colorectal carcinoma (CRC) have also been found, such as F. nucleatum, E. faecalis, E. coli, S. gallolyticus, and B. fragilis. They used different mechanisms to cause cancers, e.g., E. coli (123), enterotoxigenic B. fragilis (124), F. nucleatum (125), and Stenotrophomonas elenomonas (126) promoted tumor development through inflammatory responses, whereas colibactin-expressing E. coli (127), cytolethal toxin-expressing Proteobacteria (128), and EspF-expressing E. coli (129, 130) could cause damage of genetic substances in the cells to cause tumors.
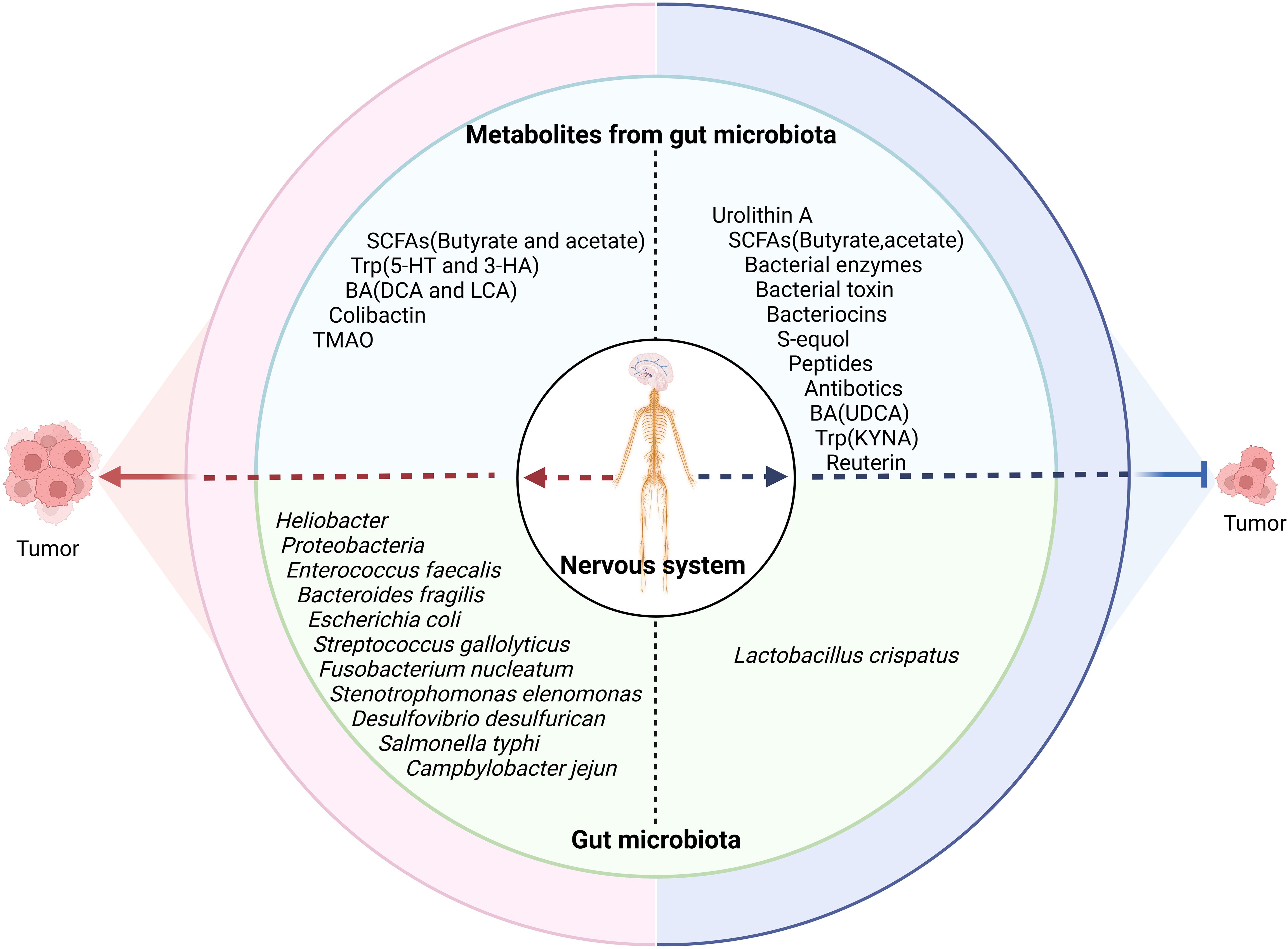
Figure 4. Effects of the nervous system on the tumor through the gut microbiota. The nervous system influences tumor development through gut microbiota/metabolites. SCFAs, short chain fatty acids; Trp, tryptophan; BA, bile acid; DCA, deoxycholic acid; LCA, lithocholic acid; TMAO, trimethylamine N-oxide; UDCA, ursodeoxycholic acid; KYNA, kynurenic acid.
Gut microbiota-derived metabolites can also contribute to tumorigenesis and tumor development such as SCFAs (131), the BA derivatives LCA and DCA (132), and the tryptophan metabolites 5-HT and 3-HAA (133). In addition, the metabolites trimethylamine n-oxide (TMAO) (134) and colibactin from gut commensal pks+ E. coli (135) were also related to tumorigenesis. Colibactin, a mutagenic compound by E. coli, induced DNA double-strand breaks to promote colorectal cancer development (127). Peptidyl aldehydes from E. coli and Bacillus subtilis could enhance carcinogenicity by inhibiting protease activity (136).
However, gut microbiota is also negatively related to tumor development, e.g., the abundance of Lactobacillus crispatus was negatively correlated with the development of ovarian and breast cancers (131). Gut microbiota metabolites such as the secondary BA UDCA, kynurenic acid (137), L. reuteri-derived reuterin (138), purine metabolites from A. muciniphila and B. pseudolongum (139), and urolithin A (UA) (140) could also inhibit tumor development. Gut microbiota-derived bacteriocins, bacterial toxins, peptides, antibiotics, and bacterial enzymes also have direct effects on cancer. The metabolites from thiopeptides by Clostridia and Lactobacillus also displayed anticancer effects through proteasome targeting (141).
5 Regulation of the nervous system-mediated immune system on tumors
Interactions among cancer cells, nerves, and immune cells can regulate overall tumor progression (Figure 5) (142).
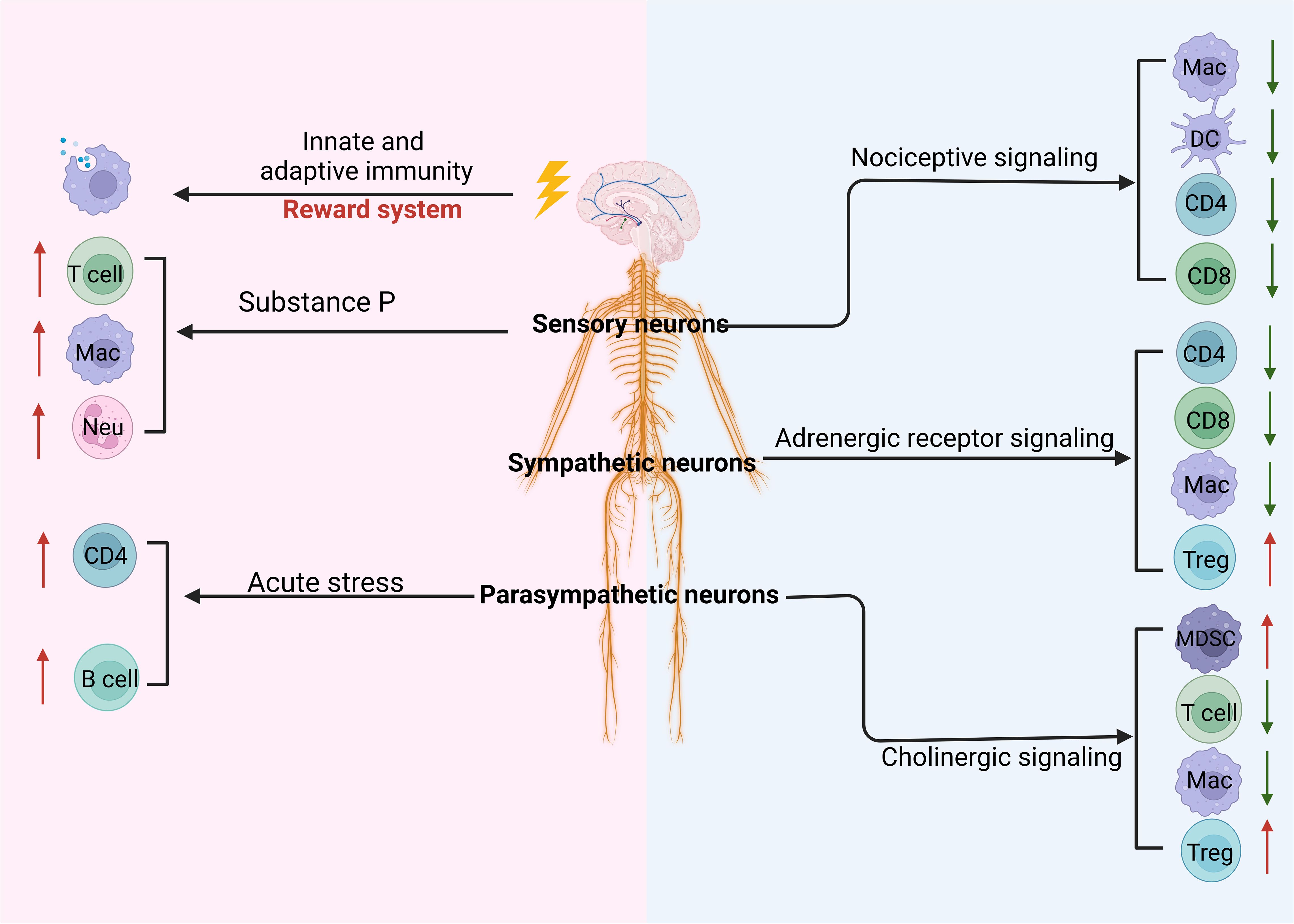
Figure 5. Regulation of the CNS and PNS on the tumor through immune cells. Stimulating the reward system in the central nervous system (CNS) induces innate and adaptive immunity. The peripheral nervous system such as parasympathetic neurons, sympathetic neurons, and sensory neurons can regulate the differentiation and development of immune cells through cholinergic, adrenergic, and nociceptive signaling, respectively. These signaling pathways can inhibit immunity against the tumor through downregulating innate and adaptive immune cells such as macrophages (MACs), dendritic cells (DCs), and CD4 and upregulating immune regulatory cells such as regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs). In addition, sensory neuron-derived substance P can promote the differentiation and function of T cells, MACs, and neutrophils (Neu). Notably, acute stress can also promote CD4 and B-cell immune responses.
5.1 Peripheral nervous system
In tumors, all types of peripheral nerves, namely, sensory, parasympathetic, and sympathetic nerves, have been detected (30, 31). Clinical studies have indicated the potential of targeting the peripheral nervous system for promoting antitumor immune responses (143).
5.1.1 Parasympathetic nervous system
The parasympathetic/vagal nerve is not only involved in the production of inflammatory factors but also in the regulation of the immune system (37). This is mainly achieved through binding of the neurotransmitter acetylcholine to nicotinic-type receptors or muscarinic-type receptors (144). The vagus nerve-derived acetylcholine exerted its effect through the α7 nicotinic acetylcholine receptor (α7nAChR), which was found on macrophages, inhibiting the production and release of pro-inflammatory cytokines like TNF, IL-1β, IL-6, and IL-18 (145, 146). Muscarinic Ach receptors (mAchRs) were also involved in cholinergic regulation of the immune system (147). It could lead to activation of Treg cells and their infiltration into tissues in the periphery (147). The administration of bethanechol, a mAchR agonist, caused suppression of tumor growth via the mAchR-dependent signaling pathway. Cholinergic vagal signaling also suppressed inflammation to promote colon cancer development through the release of trefoil factor 2 from splenic memory T cells (148). Cholinergic nerve deprivation by vagotomy, the surgical removal of the vagus nerve, facilitated the development of gastric and pancreatic tumors, suggesting that cholinergic signals suppressed tumorigenesis (29).
5.1.2 Sympathetic nervous system
Norepinephrine released locally from sympathetic nerves or systemically from the adrenal gland preferentially binds to α-adrenergic receptors (ADRs), such as the α2-ADR (ADRA2) but also the β1-ADR (ADRB1), β2-ADR (ADRB2), and β3-ADR (ADRB3) (149). It could bind to β2-AR on immune cells to exert immune-suppressive effects, including upregulating PD-1 expression (150) and regulating the function of MDSCs and macrophages (26, 151) to limit antitumor immunity (152) and promote T-lymphocyte exhaustion (153). While β2-AR signaling was disrupted, CD8+ T cells exhibited enhanced cytotoxic effector function and migratory capacity (154). During metabolic reprogramming of T cells, β2-AR deficiency also increased mitochondrial membrane potential and biogenesis (155). Through sympathetic innervation, β2-AR signaling also regulated the trafficking of lymphocytes in lymph nodes (156) and egress of hematopoietic stem cells from the bone marrow into the circulation (157), as well as movement of immune cells within tissues (152). The norepinephrine from sympathetic nerves was shown to inhibit NK cell activity via the β-adrenergic receptor (158). Notably, acute stress could trigger adrenergic receptors on CD4+ T lymphocytes and B cells to enhance the production of antibody IgG1 and interferons (IFNs).
5.1.3 Sensory neurons
Sensory neurons can activate or suppress host defense and immunity against pathogens, depending on the tissue and disease state. Nociceptive signaling such as the release of calcitonin gene-related peptide (CGRP) and substance P from peripheral nerve fibers could regulate immune responses to affect the development of tumors (159, 160). Substance P (SP), a sensory neurotransmitter, could enhance inflammatory reactions by stimulating the synthesis of pro-inflammatory cytokines (161). SP signaling can promote hematopoietic stem cell activity and hematopoiesis (162) and the survival of active T lymphocytes. In-vivo stimulation with SP has been associated with an increased antitumor immune response via an enhanced function of NK cells (163). CGRP downregulated inflammatory cytokine in macrophages and dendritic cells (DCs) and reduced antigen presentation to effector T cells (164). It also led to the release of anti-inflammatory IL-10 from macrophages and decreased effector CD4+ and CD8+ T lymphocytes via the receptor Ramp1 signaling pathway. Using CGRP-knockout (CGRPKO) mice, reduced tumors were observed in syngeneic oral cancer models (165). The tumor tissue from CGRPKO mice also had a significant increase in tumor-infiltrating CD4+ T cells, cytotoxic CD8+ T cells, and NK1.1+ NK cells as compared to wild-type mice (165). In addition to SP and CGRP, other neurotransmitters such as gamma-aminobutyric acid (GABA), commonly known as serotonin, also play a crucial role in regulating tumor immunity. Studies have shown that supplementing with the neurotransmitter GABA can decrease cAMP levels and inhibit the release of the pro-inflammatory cytokine IL-6.
Notably, the crosstalk between the ENS and immune cells such as macrophages, T cells, and innate lymphoid cells (ILCs) exerts critical roles in maintaining homeostasis.
5.2 Central nervous system
The CNS harbors its own special immune system, which is composed of microglia in the parenchyma, MACs, DCs, monocytes, and the barrier systems within the brain (166). However, the CNS also encodes specific immune responses (167). Activation of the reward system can boost innate and adaptive immunity (168–171). The activation of the ventral tegmental area (VTA), a key component of the reward system, could strengthen the host’s immunological defense (168). The optogenetic stimulation of dopaminergic projections from the VTA to the medial prefrontal cortex could attenuate stress-mediated aversive effects on tumor growth in a breast cancer model (172). The repetitive optogenetic activation of the VTA tyrosine hydroxylase (TH) in the medial prefrontal cortex attenuated stress-induced progression of breast cancers and reduced serum concentration of norepinephrine and corticosterone. In models of melanoma and lung cancer, VTA activation could reduce tumor growth by altering the functional profile of MDSCs (169). The periventricular hypothalamic corticotropin hormone neurons could regulate the trafficking of monocytes and lymphocytes between peripheral tissues and bone marrow by activating the hypothalamic–pituitary–adrenal axis, and they influence the adaptive immune responses by ultimately projecting to the splenic nerve. Central neurotransmitters such as GABA also reduced immunity against tumors (173, 174). In a mouse model of colon cancer, GABA could bind to GABAA receptors on CD8+ T lymphocytes to reduce antitumor immunity and enable tumor growth (173).
Notably, Schwann cells (SCs) in the CNS, specifically nerve repair subtypes, can enhance immune cell chemotaxis by the release of chemokines (175), e.g., SCs can promote the recruitment and immuno-inhibitory function of MDSCs and release C-C motif chemokine ligand 2 (CCL2) to aid in cancer progression. Although cultured with a melanoma-conditioned medium, glial fibrillary acidic protein+ (GFAP+) SCs could also promote mRNA levels of genes, involved in immune surveillance and chemotaxis such as IL-6, transforming growth factor (TGF)-β, and vascular endothelial growth factor (VEGF).
6 Therapeutic potential of the nervous system–gut microbiota–immune system axis on tumors
Tumor-innervating peripheral nerve fibers have recently been found to be associated with tumor development (176). The therapeutic means based on these findings have been developed, e.g., pharmacological blocking of β-adrenergic signaling was shown to enhance CD8+ T-cell infiltration into the TME and to improve the response to immune checkpoint blockade (177). The use of non-selective β1-blockers and β2-blockers has shown effectiveness in improving treatment outcomes across various human cancer types such as melanoma, prostate, breast, pancreatic, ovarian, non-small-cell lung cancer, and colorectal cancer (176).
Since gut microbiome alterations are associated with various cancers and the gut microbiota can communicate with the brain via a bidirectional pathway (178), the nervous system–gut microbiota–immune system axis should also become a target for developing new therapeutic method(s) for preventing tumors. With the advancement in the nervous system–gut microbiota–immune system axis, it is possible to affect nervous signals or nervous transmitters to treat tumors, e.g., the vagus nerve, involved in stress response, can affect the composition of the gut microbiota. Targeting these properties of the vagus nerve could be of interest in preventing tumors. Thus, changing the composition of the gut microbiota through modulating nervous system-associated factors such as neurotransmitters and neuropeptides could potentially improve tumor therapy.
Importantly, specific gut microorganism(s) controlled by the nervous system is identified, and the interventions targeting the gut microbiota, including fecal microbiota transplantation, probiotics, prebiotics, engineered bacteria, and dietary interventions, can be used to treat tumors. The gut microbiome has become an efficient target for making cancer chemotherapy/immunotherapy safer or efficient, which will further improve cancer survival rates in the future.
In addition, specific gut microorganism(s) can regulate the differentiation and function of immune cells through the receptor signal, epigenetic modification, metabolism reprogramming, cGAS-STING pathway, and TLS formation. The inhibitor or promoter, which affects these processes, can also be a potential mean(s) to control tumorigenesis and tumor development.
7 Conclusion and perspective
While existing studies have highlighted the involvement of both sympathetic and parasympathetic nerves in pro- and antitumor immunity, here, we have explored the potential effects of the nervous system–gut microbiota–immune system axis on tumorigenesis and tumor development. The nervous system regulates immune cells through the gut microbiota, which can prevent tumor development, including the effects of the nervous system on gut microbiota, gut microbiota on the immune system, and the immune system on tumors. The ENS is essential for the regulation of gut microbiota. There exists also altered gut microbiota in anxiety, depression, and stress. Gut microbiota has wide effects on the function and differentiation of immune-effective cells such as effective CD8 and NK cells, immune regulatory (suppressive) cells such as Tregs, and also inflammatory cells such as Th1, Th2, and Th17. These gut microbiota-associated immune cells can deeply affect the occurrence and development of tumors.
The regulation in the gut microbiota by the nervous system can potentially become a target for preventing tumor. Thus, the identification of changes in the gut microbiome associated with the nervous system may provide valuable information in the choice of treatment. However, the characteristics, function, and composition of the gut microbiota, which are controlled by the nervous system, remains to be further understood. With the emergence of gut microbiota as an important driver of disease, there is now a unique opportunity to target microbiota in the gut to achieve precision in cancer care (179, 180).
In conclusion, a better understanding of the role of the central and peripheral nervous system and the influence of innervation on the components of the gut microbiota will strengthen the therapeutic armamentarium against tumors.
Author contributions
JW: Investigation, Methodology, Writing – original draft. WC: Investigation, Methodology, Writing – original draft. RY: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by NSFC grants (91842302, 82271779, 81901677, 31470876, 91629102, ISF-NSFC program 31461143010), Tianjin Science and Technology Commission (18JCZDJC35300), CAMS Innovation Fund for Medical Science (CIFMS2017-12M-2-005), a Ministry of Science and Technology grant (2016YFC1303604), the State Key Laboratory of Medicinal Chemical Biology, and The Fundamental Research Funds for the Central University, Nankai University (Grant number 63191724).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Arch JJ, Bright EE, Finkelstein LB, Fink RM, Mitchell JL, Andorsky DJ, et al. Anxiety and depression in metastatic cancer: A critical review of negative impacts on advance care planning and end-of-life decision making with practical recommendations. JCO Oncol Pract. (2023) 19:1097–108. doi: 10.1200/OP.23.00287
2. Xie Y, Liu F. The role of the gut microbiota in tumor, immunity, and immunotherapy. Front Immunol. (2024) 15:1410928. doi: 10.3389/fimmu.2024.1410928
3. Huang JW, Cao CA, Zheng WH, Jia CR, Liu X, Gao SQ, et al. The mechanism of cancer-depression comorbidity. Neuroscience. (2024) 556:25–30. doi: 10.1016/j.neuroscience.2024.07.040
4. Politynska B, Pokorska O, Wojtukiewicz AM, Sawicka M, Mysliwiec M, Honn KV, et al. Is depression the missing link between inflammatory mediators and cancer? Pharmacol Ther. (2022) 240:108293. doi: 10.1016/j.pharmthera.2022.108293
5. Yoo BB, Mazmanian SK. The enteric network: interactions between the immune and nervous systems of the gut. Immunity. (2017) 46:910–26. doi: 10.1016/j.immuni.2017.05.011
6. Chang L, Wei Y, Hashimoto K. Brain-gut-microbiota axis in depression: A historical overview and future directions. Brain Res Bull. (2022) 182:44–56. doi: 10.1016/j.brainresbull.2022.02.004
7. Liu R, Wang J, Liu Y, Gao Y, Yang R. Regulation of gut microbiota on immune cell ferroptosis: A novel insight for immunotherapy against tumor. Cancer Lett. (2024) 598:217115. doi: 10.1016/j.canlet.2024.217115
8. Kong C, Liang L, Liu G, Du L, Yang Y, Liu J, et al. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. (2023) 72:1129–42. doi: 10.1136/gutjnl-2022-327156
9. Wang Y, Wang J, Liu J, Zhu H. Immune-related diagnostic markers for benign prostatic hyperplasia and their potential as drug targets. Front Immunol. (2024) 15:1516362. doi: 10.3389/fimmu.2024.1516362
10. Wang JF, Wang JS, Liu Y, Ji B, Ding BC, Wang YX, et al. Knockdown of integrin beta1 inhibits proliferation and promotes apoptosis in bladder cancer cells. Biofactors. (2025) 51:e2150. doi: 10.1002/biof.2150
11. Wang Y, Zhu H, Xu H, Qiu Y, Zhu Y, Wang X. Senescence-related gene c-Myc affects bladder cancer cell senescence by interacting with HSP90B1 to regulate cisplatin sensitivity. Aging (Albany NY). (2023) 15:7408–23. doi: 10.18632/aging.204863
12. Wang Y, Zhu H, Wang X. Prognosis and immune infiltration analysis of endoplasmic reticulum stress-related genes in bladder urothelial carcinoma. Front Genet. (2022) 13:965100. doi: 10.3389/fgene.2022.965100
13. Wang Y, Wang J, He J, Ji B, Pang Z, Wang J, et al. Comprehensive analysis of PRPF19 immune infiltrates, DNA methylation, senescence-associated secretory phenotype and ceRNA network in bladder cancer. Front Immunol. (2023) 14:1289198. doi: 10.3389/fimmu.2023.1289198
14. Wang Y, Li C, He J, Zhao Q, Zhou Y, Sun H, et al. Multi-omics analysis and experimental validation of the value of monocyte-associated features in prostate cancer prognosis and immunotherapy. Front Immunol. (2024) 15:1426474. doi: 10.3389/fimmu.2024.1426474
15. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. (2013) 341:569–73. doi: 10.1126/science.1241165
16. Luu M, Visekruna A. Short-chain fatty acids: Bacterial messengers modulating the immunometabolism of T cells. Eur J Immunol. (2019) 49:842–8. doi: 10.1002/eji.201848009
17. Parada Venegas D, de la Fuente MK, Landskron G, Gonzalez MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. (2019) 10:277. doi: 10.3389/fimmu.2019.00277
18. Hezaveh K, Shinde RS, Klotgen A, Halaby MJ, Lamorte S, Ciudad MT, et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity. (2022) 55:324–40 e8. doi: 10.1016/j.immuni.2022.01.006
19. Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab. (2020) 31:837–51 e10. doi: 10.1016/j.cmet.2020.03.003
20. Su X, Gao Y, Yang R. Gut microbiota-derived tryptophan metabolites maintain gut and systemic homeostasis. Cells. (2022) 11. doi: 10.3390/cells11152296
21. Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. (2020) 581:475–9. doi: 10.1038/s41586-020-2193-0
22. Fiorucci S, Biagioli M, Zampella A, Distrutti E. Bile acids activated receptors regulate innate immunity. Front Immunol. (2018) 9:1853. doi: 10.3389/fimmu.2018.01853
23. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. (2019) 576:143–8. doi: 10.1038/s41586-019-1785-z
24. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature. (2020) 577:410–5. doi: 10.1038/s41586-019-1865-0
25. Thaker PH, Han LY, Kamat AA, Arevalo JM, Takahashi R, Lu C, et al. Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nat Med. (2006) 12:939–44. doi: 10.1038/nm1447
26. Sloan EK, Priceman SJ, Cox BF, Yu S, Pimentel MA, Tangkanangnukul V, et al. The sympathetic nervous system induces a metastatic switch in primary breast cancer. Cancer Res. (2010) 70:7042–52. doi: 10.1158/0008-5472.CAN-10-0522
27. Hayakawa Y, Sakitani K, Konishi M, Asfaha S, Niikura R, Tomita H, et al. Nerve growth factor promotes gastric tumorigenesis through aberrant cholinergic signaling. Cancer Cell. (2017) 31:21–34. doi: 10.1016/j.ccell.2016.11.005
28. Renz BW, Takahashi R, Tanaka T, Macchini M, Hayakawa Y, Dantes Z, et al. beta2 adrenergic-neurotrophin feedforward loop promotes pancreatic cancer. Cancer Cell. (2018) 33:75–90 e7. doi: 10.1016/j.ccell.2017.11.007
29. Renz BW, Tanaka T, Sunagawa M, Takahashi R, Jiang Z, Macchini M, et al. Cholinergic signaling via muscarinic receptors directly and indirectly suppresses pancreatic tumorigenesis and cancer stemness. Cancer Discov. (2018) 8:1458–73. doi: 10.1158/2159-8290.CD-18-0046
30. Balood M, Ahmadi M, Eichwald T, Ahmadi A, Majdoubi A, Roversi K, et al. Nociceptor neurons affect cancer immunosurveillance. Nature. (2022) 611:405–12. doi: 10.1038/s41586-022-05374-w
31. Vats K, Kruglov O, Sahoo B, Soman V, Zhang J, Shurin GV, et al. Sensory nerves impede the formation of tertiary lymphoid structures and development of protective antimelanoma immune responses. Cancer Immunol Res. (2022) 10:1141–54. doi: 10.1158/2326-6066.CIR-22-0110
32. Rubio C, Ochoa E, Gatica F, Portilla A, Vazquez D, Rubio-Osornio M. The role of the vagus nerve in the microbiome and digestive system in relation to epilepsy. Curr Med Chem. (2024) 31:6018–31. doi: 10.2174/0109298673260479231010044020
33. Rao M, Gershon MD. Enteric nervous system development: what could possibly go wrong? Nat Rev Neurosci. (2018) 19:552–65. doi: 10.1038/s41583-018-0041-0
34. Dicks LMT. Our mental health is determined by an intrinsic interplay between the central nervous system, enteric nerves, and gut microbiota. Int J Mol Sci. (2023) 25. doi: 10.3390/ijms25010038
35. Kulkarni S, Saha M, Slosberg J, Singh A, Nagaraj S, Becker L, et al. Age-associated changes in lineage composition of the enteric nervous system regulate gut health and disease. Elife. (2023) 12. doi: 10.7554/eLife.88051
36. Virtanen HT, Choopanian P, Porokuokka LL, Forsgard R, Garton DR, Olfat S, et al. Interindividual variation in gut nitrergic neuron density is regulated by GDNF levels and ETV1. Cell Mol Gastroenterol Hepatol. (2024) 18:101405. doi: 10.1016/j.jcmgh.2024.101405
37. Herculano-Houzel S. The human brain in numbers: a linearly scaled-up primate brain. Front Hum Neurosci. (2009) 3:31.2009. doi: 10.3389/neuro.09.031.2009
38. Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. (1996) 16:89–101. doi: 10.1016/s0896-6273(00)80026-4
39. Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. (1998) 392:198–202. doi: 10.1038/32448
40. Bonaz B, Bazin T, Pellissier S. The vagus nerve at the interface of the microbiota-gut-brain axis. Front Neurosci. (2018) 12:49. doi: 10.3389/fnins.2018.00049
41. Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol. (2009) 6:306–14. doi: 10.1038/nrgastro.2009.35
42. Heiss CN, Olofsson LE. The role of the gut microbiota in development, function and disorders of the central nervous system and the enteric nervous system. J Neuroendocrinol. (2019) 31:e12684. doi: 10.1111/jne.12684
43. Simpson CA, Diaz-Arteche C, Eliby D, Schwartz OS, Simmons JG, Cowan CSM. The gut microbiota in anxiety and depression - A systematic review. Clin Psychol Rev. (2021) 83:101943. doi: 10.1016/j.cpr.2020.101943
44. Caspani G, Kennedy S, Foster JA, Swann J. Gut microbial metabolites in depression: understanding the biochemical mechanisms. Microb Cell. (2019) 6:454–81. doi: 10.15698/mic2019.10.693
45. Sun N, Zhang J, Wang J, Liu Z, Wang X, Kang P, et al. Abnormal gut microbiota and bile acids in patients with first-episode major depressive disorder and correlation analysis. Psychiatry Clin Neurosci. (2022) 76:321–8. doi: 10.1111/pcn.13368
46. Ortega MA, Alvarez-Mon MA, Garcia-Montero C, Fraile-Martinez O, Guijarro LG, Lahera G, et al. Gut microbiota metabolites in major depressive disorder-deep insights into their pathophysiological role and potential translational applications. Metabolites. (2022) 12. doi: 10.3390/metabo12010050
47. Zhang Z, Lei A, Xu L, Chen L, Chen Y, Zhang X, et al. Similarity in gene-regulatory networks suggests that cancer cells share characteristics of embryonic neural cells. J Biol Chem. (2017) 292:12842–59. doi: 10.1074/jbc.M117.785865
48. Palepu MSK, Dandekar MP. Remodeling of microbiota gut-brain axis using psychobiotics in depression. Eur J Pharmacol. (2022) 931:175171. doi: 10.1016/j.ejphar.2022.175171
49. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PloS Biol. (2016) 14:e1002533. doi: 10.1371/journal.pbio.1002533
50. Montagnani M, Bottalico L, Potenza MA, Charitos IA, Topi S, Colella M, et al. The crosstalk between gut microbiota and nervous system: A bidirectional interaction between microorganisms and metabolome. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms241210322
51. Macpherson AJ, Pachnis V, Prinz M. Boundaries and integration between microbiota, the nervous system, and immunity. Immunity. (2023) 56:1712–26. doi: 10.1016/j.immuni.2023.07.011
52. Roager HM, Hansen LB, Bahl MI, Frandsen HL, Carvalho V, Gobel RJ, et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol. (2016) 1:16093. doi: 10.1038/nmicrobiol.2016.93
53. Obata Y, Castano A, Boeing S, Bon-Frauches AC, Fung C, Fallesen T, et al. Neuronal programming by microbiota regulates intestinal physiology. Nature. (2020) 578:284–9. doi: 10.1038/s41586-020-1975-8
54. Tian P, Wang G, Zhao J, Zhang H, Chen W. Bifidobacterium with the role of 5-hydroxytryptophan synthesis regulation alleviates the symptom of depression and related microbiota dysbiosis. J Nutr Biochem. (2019) 66:43–51. doi: 10.1016/j.jnutbio.2019.01.007
55. Dicks LMT. Gut bacteria and neurotransmitters. Microorganisms. (2022) 10. doi: 10.3390/microorganisms10091838
56. Griffiths JA, Yoo BB, Thuy-Boun P, Cantu VJ, Weldon KC, Challis C, et al. Peripheral neuronal activation shapes the microbiome and alters gut physiology. Cell Rep. (2024) 43:113953. doi: 10.1016/j.celrep.2024.113953
57. Johnson CD, Barlow-Anacker AJ, Pierre JF, Touw K, Erickson CS, Furness JB, et al. Deletion of choline acetyltransferase in enteric neurons results in postnatal intestinal dysmotility and dysbiosis. FASEB J. (2018) 32:4744–52. doi: 10.1096/fj.201701474RR
58. Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. (2010) 328:1705–9. doi: 10.1126/science.1188454
59. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
60. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
61. Wesemann DR, Portuguese AJ, Meyers RM, Gallagher MP, Cluff-Jones K, Magee JM, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. (2013) 501:112–5. doi: 10.1038/nature12496
62. Cullin N, Azevedo Antunes C, Straussman R, Stein-Thoeringer CK, Elinav E. Microbiome and cancer. Cancer Cell. (2021) 39:1317–41. doi: 10.1016/j.ccell.2021.08.006
63. Park EM, Chelvanambi M, Bhutiani N, Kroemer G, Zitvogel L, Wargo JA. Targeting the gut and tumor microbiota in cancer. Nat Med. (2022) 28:690–703. doi: 10.1038/s41591-022-01779-2
64. Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, et al. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology. (2017) 153:1621–33 e6. doi: 10.1053/j.gastro.2017.08.022
65. Deng H, Li Z, Tan Y, Guo Z, Liu Y, Wang Y, et al. A novel strain of Bacteroides fragilis enhances phagocytosis and polarises M1 macrophages. Sci Rep. (2016) 6:29401. doi: 10.1038/srep29401
66. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. (2015) 350:1084–9. doi: 10.1126/science.aac4255
67. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. (2018) 359:97–103. doi: 10.1126/science.aan4236
68. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. (2018) 359:592–7. doi: 10.1126/science.aah3648
69. Rong Y, Dong Z, Hong Z, Jin Y, Zhang W, Zhang B, et al. Reactivity toward Bifidobacterium longum and Enterococcus hirae demonstrate robust CD8(+) T cell response and better prognosis in HBV-related hepatocellular carcinoma. Exp Cell Res. (2017) 358:352–9. doi: 10.1016/j.yexcr.2017.07.009
70. Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. (2019) 178:795–806 e12. doi: 10.1016/j.cell.2019.07.008
71. Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S, et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. (2017) 10:421–33. doi: 10.1038/mi.2016.53
72. Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. (2016) 22:557–66. doi: 10.3748/wjg.v22.i2.557
73. Galeano Nino JL, Wu H, LaCourse KD, Kempchinsky AG, Baryiames A, Barber B, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. (2022) 611:810–7. doi: 10.1038/s41586-022-05435-0
74. Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. (2018) 8:403–16. doi: 10.1158/2159-8290.CD-17-1134
75. Ratajczak W, Ryl A, Mizerski A, Walczakiewicz K, Sipak O, Laszczynska M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim Pol. (2019) 66:1–12. doi: 10.18388/abp.2018_2648
76. Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
77. Lucas LN, Barrett K, Kerby RL, Zhang Q, Cattaneo LE, Stevenson D, et al. Dominant bacterial phyla from the human gut show widespread ability to transform and conjugate bile acids. mSystems. (2021):e0080521. doi: 10.1128/mSystems.00805-21
78. Bourgin M, Kriaa A, Mkaouar H, Mariaule V, Jablaoui A, Maguin E, et al. Bile salt hydrolases: at the crossroads of microbiota and human health. Microorganisms. (2021) 9. doi: 10.3390/microorganisms9061122
79. Guzior DV, Quinn RA. Review: microbial transformations of human bile acids. Microbiome. (2021) 9:140. doi: 10.1186/s40168-021-01101-1
80. Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol. (2015) 11:685–90. doi: 10.1038/nchembio.1864
81. Fiorucci S, Carino A, Baldoni M, Santucci L, Costanzi E, Graziosi L, et al. Bile acid signaling in inflammatory bowel diseases. Dig Dis Sci. (2021) 66:674–93. doi: 10.1007/s10620-020-06715-3
82. Sato Y, Atarashi K, Plichta DR, Arai Y, Sasajima S, Kearney SM, et al. Novel bile acid biosynthetic pathways are enriched in the microbiome of centenarians. Nature. (2021) 599:458–64. doi: 10.1038/s41586-021-03832-5
83. Paik D, Yao L, Zhang Y, Bae S, D’Agostino GD, Zhang M, et al. Human gut bacteria produce TauEta17-modulating bile acid metabolites. Nature. (2022) 603:907–12. doi: 10.1038/s41586-022-04480-z
84. Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. (2018) 9:3294. doi: 10.1038/s41467-018-05470-4
85. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. (2017) 551:648–52. doi: 10.1038/nature24661
86. Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med. (2016) 22:598–605. doi: 10.1038/nm.4102
87. Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. (2013) 57:523–35. doi: 10.1002/mnfr.201200594
88. Wlodarska M, Luo C, Kolde R, d’Hennezel E, Annand JW, Heim CE, et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. (2017) 22:25–37 e6. doi: 10.1016/j.chom.2017.06.007
89. Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, et al. Lactobacillus reuteri induces gut intraepithelial CD4(+)CD8alphaalpha(+) T cells. Science. (2017) 357:806–10. doi: 10.1126/science.aah5825
90. Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. (1996) 81:288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x
91. Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. (2013) 5:193ra91. doi: 10.1126/scitranslmed.3006438
92. Lam KC, Araya RE, Huang A, Chen Q, Di Modica M, Rodrigues RR, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. (2021) 184:5338–56 e21. doi: 10.1016/j.cell.2021.09.019
93. Si W, Liang H, Bugno J, Xu Q, Ding X, Yang K, et al. Lactobacillus rhamnosus GG induces cGAS/STING- dependent type I interferon and improves response to immune checkpoint blockade. Gut. (2022) 71:521–33. doi: 10.1136/gutjnl-2020-323426
94. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. (2020) 369:1481–9. doi: 10.1126/science.abc3421
95. Wang H, Rong X, Zhao G, Zhou Y, Xiao Y, Ma D, et al. The microbial metabolite trimethylamine N-oxide promotes antitumor immunity in triple-negative breast cancer. Cell Metab. (2022) 34:581–94 e8. doi: 10.1016/j.cmet.2022.02.010
96. Ma M, Wang Y, Fan S, Huang Y, Su X, Lu C. Urolithin A alleviates colitis in mice by improving gut microbiota dysbiosis, modulating microbial tryptophan metabolism, and triggering ahR activation. J Agric Food Chem. (2023) 71:7710–22. doi: 10.1021/acs.jafc.3c00830
97. Chen H, Tong T, Lu SY, Ji L, Xuan B, Zhao G, et al. Urea cycle activation triggered by host-microbiota maladaptation driving colorectal tumorigenesis. Cell Metab. (2023) 35:651–66 e7. doi: 10.1016/j.cmet.2023.03.003
98. Wang J, Zhu N, Su X, Gao Y, Yang R. Gut-microbiota-derived metabolites maintain gut and systemic immune homeostasis. Cells. (2023) 12. doi: 10.3390/cells12050793
99. Goudot C, Coillard A, Villani AC, Gueguen P, Cros A, Sarkizova S, et al. Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity. (2017) 47:582–96 e6. doi: 10.1016/j.immuni.2017.08.016
100. Holscher AS, Schulz WA, Pinkerneil M, Niegisch G, Hoffmann MJ. Combined inhibition of BET proteins and class I HDACs synergistically induces apoptosis in urothelial carcinoma cell lines. Clin Epigenet. (2018) 10:1. doi: 10.1186/s13148-017-0434-3
101. Kaypee S, Sudarshan D, Shanmugam MK, Mukherjee D, Sethi G, Kundu TK. Aberrant lysine acetylation in tumorigenesis: Implications in the development of therapeutics. Pharmacol Ther. (2016) 162:98–119. doi: 10.1016/j.pharmthera.2016.01.011
102. Ma X, Wang X, Zheng G, Tan G, Zhou F, Wei W, et al. Critical role of gut microbiota and epigenetic factors in the pathogenesis of behcet’s disease. Front Cell Dev Biol. (2021) 9:719235. doi: 10.3389/fcell.2021.719235
103. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. (2009) 324:1029–33. doi: 10.1126/science.1160809
104. Di Luccia B, Gilfillan S, Cella M, Colonna M, Huang SC. ILC3s integrate glycolysis and mitochondrial production of reactive oxygen species to fulfill activation demands. J Exp Med. (2019) 216:2231–41. doi: 10.1084/jem.20180549
105. Choi SC, Titov AA, Abboud G, Seay HR, Brusko TM, Roopenian DC, et al. Inhibition of glucose metabolism selectively targets autoreactive follicular helper T cells. Nat Commun. (2018) 9:4369. doi: 10.1038/s41467-018-06686-0
106. Carlberg C, Velleuer E. Vitamin D and the risk for cancer: A molecular analysis. Biochem Pharmacol. (2022) 196:114735. doi: 10.1016/j.bcp.2021.114735
107. Conrad M, Pratt DA. The chemical basis of ferroptosis. Nat Chem Biol. (2019) 15:1137–47. doi: 10.1038/s41589-019-0408-1
108. Cui W, Guo M, Liu D, Xiao P, Yang C, Huang H, et al. Gut microbial metabolite facilitates colorectal cancer development via ferroptosis inhibition. Nat Cell Biol. (2024) 26(1):124–37. doi: 10.1038/s41556-023-01314-6
109. Lin Y, Jin X. Effect of ubiquitin protease system on DNA damage response in prostate cancer (Review). Exp Ther Med. (2024) 27:33. doi: 10.3892/etm.2023.12321
110. Fernandes MR, Aggarwal P, Costa RGF, Cole AM, Trinchieri G. Targeting the gut microbiota for cancer therapy. Nat Rev Cancer. (2022) 22:703–22. doi: 10.1038/s41568-022-00513-x
111. Fridman WH, Meylan M, Pupier G, Calvez A, Hernandez I, Sautes-Fridman C. Tertiary lymphoid structures and B cells: An intratumoral immunity cycle. Immunity. (2023) 56:2254–69. doi: 10.1016/j.immuni.2023.08.009
112. Teillaud JL, Houel A, Panouillot M, Riffard C, Dieu-Nosjean MC. Tertiary lymphoid structures in anticancer immunity. Nat Rev Cancer. (2024) 24(9):629–46. doi: 10.1038/s41568-024-00728-0
113. Schlosser HA, Thelen M, Lechner A, Wennhold K, Garcia-Marquez MA, Rothschild SI, et al. B cells in esophago-gastric adenocarcinoma are highly differentiated, organize in tertiary lymphoid structures and produce tumor-specific antibodies. Oncoimmunology. (2019) 8:e1512458. doi: 10.1080/2162402X.2018.1512458
114. Dominiak A, Chelstowska B, Olejarz W, Nowicka G. Communication in the cancer microenvironment as a target for therapeutic interventions. Cancers (Basel). (2020) 12. doi: 10.3390/cancers12051232
115. Lin JR, Wang S, Coy S, Chen YA, Yapp C, Tyler M, et al. Multiplexed 3D atlas of state transitions and immune interaction in colorectal cancer. Cell. (2023) 186:363–81 e19. doi: 10.1016/j.cell.2022.12.028
116. Xu W, Lu J, Liu WR, Anwaier A, Wu Y, Tian X, et al. Heterogeneity in tertiary lymphoid structures predicts distinct prognosis and immune microenvironment characterizations of clear cell renal cell carcinoma. J Immunother Cancer. (2023) 11. doi: 10.1136/jitc-2023-006667
117. Meylan M, Petitprez F, Lacroix L, Di Tommaso L, Roncalli M, Bougouin A, et al. Early hepatic lesions display immature tertiary lymphoid structures and show elevated expression of immune inhibitory and immunosuppressive molecules. Clin Cancer Res. (2020) 26:4381–9. doi: 10.1158/1078-0432.CCR-19-2929
118. Tietscher S, Wagner J, Anzeneder T, Langwieder C, Rees M, Sobottka B, et al. A comprehensive single-cell map of T cell exhaustion-associated immune environments in human breast cancer. Nat Commun. (2023) 14:98. doi: 10.1038/s41467-022-35238-w
119. Wu B, Zhang B, Li B, Wu H, Jiang M. Cold and hot tumors: from molecular mechanisms to targeted therapy. Signal Transduct Target Ther. (2024) 9:274. doi: 10.1038/s41392-024-01979-x
120. Fujii SI, Shimizu K. Immune networks and therapeutic targeting of iNKT cells in cancer. Trends Immunol. (2019) 40:984–97. doi: 10.1016/j.it.2019.09.008
121. Galluzzi L, Humeau J, Buque A, Zitvogel L, Kroemer G. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat Rev Clin Oncol. (2020) 17:725–41. doi: 10.1038/s41571-020-0413-z
122. Lu Y, Yuan X, Wang M, He Z, Li H, Wang J, et al. Gut microbiota influence immunotherapy responses: mechanisms and therapeutic strategies. J Hematol Oncol. (2022) 15:47. doi: 10.1186/s13045-022-01273-9
123. Lopes A, Billard E, Casse AH, Villeger R, Veziant J, Roche G, et al. Colibactin-positive Escherichia coli induce a procarcinogenic immune environment leading to immunotherapy resistance in colorectal cancer. Int J Cancer. (2020) 146:3147–59. doi: 10.1002/ijc.32920
124. Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. (2018) 23:203–14 e5. doi: 10.1016/j.chom.2018.01.007
125. Lee JA, Yoo SY, Oh HJ, Jeong S, Cho NY, Kang GH, et al. Differential immune microenvironmental features of microsatellite-unstable colorectal cancers according to Fusobacterium nucleatum status. Cancer Immunol Immunother. (2021) 70:47–59. doi: 10.1007/s00262-020-02657-x
126. Ling Z, Shao L, Liu X, Cheng Y, Yan C, Mei Y, et al. Regulatory T cells and plasmacytoid dendritic cells within the tumor microenvironment in gastric cancer are correlated with gastric microbiota dysbiosis: A preliminary study. Front Immunol. (2019) 10:533. doi: 10.3389/fimmu.2019.00533
127. Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci U S A. (2010) 107:11537–42. doi: 10.1073/pnas.1001261107
128. He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. (2019) 68:289–300. doi: 10.1136/gutjnl-2018-317200
129. Maddocks OD, Scanlon KM, Donnenberg MS. An Escherichia coli effector protein promotes host mutation via depletion of DNA mismatch repair proteins. mBio. (2013) 4:e00152–13. doi: 10.1128/mBio.00152-13
130. Kim JJ, Tao H, Carloni E, Leung WK, Graham DY, Sepulveda AR. Helicobacter pylori impairs DNA mismatch repair in gastric epithelial cells. Gastroenterology. (2002) 123:542–53. doi: 10.1053/gast.2002.34751
131. Rossi T, Vergara D, Fanini F, Maffia M, Bravaccini S, Pirini F. Microbiota-derived metabolites in tumor progression and metastasis. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21165786
132. Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol. (2009) 15:3329–40. doi: 10.3748/wjg.15.3329
133. Liu D, Liang CH, Huang B, Zhuang X, Cui W, Yang L, et al. Tryptophan metabolism acts as a new anti-ferroptotic pathway to mediate tumor growth. Adv Sci (Weinh). (2023) 10:e2204006. doi: 10.1002/advs.202204006
134. Jalandra R, Dalal N, Yadav AK, Verma D, Sharma M, Singh R, et al. Emerging role of trimethylamine-N-oxide (TMAO) in colorectal cancer. Appl Microbiol Biotechnol. (2021) 105:7651–60. doi: 10.1007/s00253-021-11582-7
135. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J, et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature. (2020) 580:269–73. doi: 10.1038/s41586-020-2080-8
136. Guo CJ, Chang FY, Wyche TP, Backus KM, Acker TM, Funabashi M, et al. Discovery of reactive microbiota-derived metabolites that inhibit host proteases. Cell. (2017) 168:517–26 e18. doi: 10.1016/j.cell.2016.12.021
137. Walczak K, Turski WA, Rajtar G. Kynurenic acid inhibits colon cancer proliferation in vitro: effects on signaling pathways. Amino Acids. (2014) 46:2393–401. doi: 10.1007/s00726-014-1790-3
138. Bell HN, Rebernick RJ, Goyert J, Singhal R, Kuljanin M, Kerk SA, et al. Reuterin in the healthy gut microbiome suppresses colorectal cancer growth through altering redox balance. Cancer Cell. (2022) 40:185–200 e6. doi: 10.1016/j.ccell.2021.12.001
139. Liu J, Hong S, Yang J, Zhang X, Wang Y, Wang H, et al. Targeting purine metabolism in ovarian cancer. J Ovarian Res. (2022) 15:93. doi: 10.1186/s13048-022-01022-z
140. Totiger TM, Srinivasan S, Jala VR, Lamichhane P, Dosch AR, Gaidarski AA 3rd, et al. Urolithin A, a novel natural compound to target PI3K/AKT/mTOR pathway in pancreatic cancer. Mol Cancer Ther. (2019) 18:301–11. doi: 10.1158/1535-7163.MCT-18-0464
141. Vinogradov AA, Suga H. Introduction to thiopeptides: biological activity, biosynthesis, and strategies for functional reprogramming. Cell Chem Biol. (2020) 27:1032–51. doi: 10.1016/j.chembiol.2020.07.003
142. Mancusi R, Monje M. The neuroscience of cancer. Nature. (2023) 618:467–79. doi: 10.1038/s41586-023-05968-y
143. Kizil B, De Virgiliis F, Scheiermann C. Neural control of tumor immunity. FEBS J. (2024) 291:4670–9. doi: 10.1111/febs.17280
144. Borovikova LV, Ivanova S, Zhang M, Yang H, Botchkina GI, Watkins LR, et al. Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature. (2000) 405:458–62. doi: 10.1038/35013070
145. Wang H, Yu M, Ochani M, Amella CA, Tanovic M, Susarla S, et al. Nicotinic acetylcholine receptor alpha7 subunit is an essential regulator of inflammation. Nature. (2003) 421:384–8. doi: 10.1038/nature01339
146. Rosas-Ballina M, Olofsson PS, Ochani M, Valdes-Ferrer SI, Levine YA, Reardon C, et al. Acetylcholine-synthesizing T cells relay neural signals in a vagus nerve circuit. Science. (2011) 334:98–101. doi: 10.1126/science.1209985
147. Teratani T, Mikami Y, Nakamoto N, Suzuki T, Harada Y, Okabayashi K, et al. The liver-brain-gut neural arc maintains the T(reg) cell niche in the gut. Nature. (2020) 585:591–6. doi: 10.1038/s41586-020-2425-3
148. Dubeykovskaya Z, Si Y, Chen X, Worthley DL, Renz BW, Urbanska AM, et al. Neural innervation stimulates splenic TFF2 to arrest myeloid cell expansion and cancer. Nat Commun. (2016) 7:10517. doi: 10.1038/ncomms10517
149. Zhu J, Naulaerts S, Boudhan L, Martin M, Gatto L, Van den Eynde BJ. Tumour immune rejection triggered by activation of alpha2-adrenergic receptors. Nature. (2023) 618:607–15. doi: 10.1038/s41586-023-06110-8
150. Yang H, Xia L, Chen J, Zhang S, Martin V, Li Q, et al. Stress-glucocorticoid-TSC22D3 axis compromises therapy-induced antitumor immunity. Nat Med. (2019) 25:1428–41. doi: 10.1038/s41591-019-0566-4
151. Mohammadpour H, MacDonald CR, Qiao G, Chen M, Dong B, Hylander BL, et al. beta2 adrenergic receptor-mediated signaling regulates the immunosuppressive potential of myeloid-derived suppressor cells. J Clin Invest. (2019) 129:5537–52. doi: 10.1172/JCI129502
152. Devi S, Alexandre YO, Loi JK, Gillis R, Ghazanfari N, Creed SJ, et al. Adrenergic regulation of the vasculature impairs leukocyte interstitial migration and suppresses immune responses. Immunity. (2021) 54:1219–30 e7. doi: 10.1016/j.immuni.2021.03.025
153. Qiao G, Chen M, Mohammadpour H, MacDonald CR, Bucsek MJ, Hylander BL, et al. Chronic adrenergic stress contributes to metabolic dysfunction and an exhausted phenotype in T cells in the tumor microenvironment. Cancer Immunol Res. (2021) 9:651–64. doi: 10.1158/2326-6066.CIR-20-0445
154. Qiao G, Chen M, Bucsek MJ, Repasky EA, Hylander BL. Adrenergic signaling: A targetable checkpoint limiting development of the antitumor immune response. Front Immunol. (2018) 9:164. doi: 10.3389/fimmu.2018.00164
155. Farooq MA, Ajmal I, Hui X, Chen Y, Ren Y, Jiang W. beta2-adrenergic receptor mediated inhibition of T cell function and its implications for CAR-T cell therapy. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms241612837
156. Suzuki K, Hayano Y, Nakai A, Furuta F, Noda M. Adrenergic control of the adaptive immune response by diurnal lymphocyte recirculation through lymph nodes. J Exp Med. (2016) 213:2567–74. doi: 10.1084/jem.20160723
157. Mendez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. (2008) 452:442–7. doi: 10.1038/nature06685
158. Ben-Eliyahu S, Shakhar G, Page GG, Stefanski V, Shakhar K. Suppression of NK cell activity and of resistance to metastasis by stress: a role for adrenal catecholamines and beta-adrenoceptors. Neuroimmunomodulation. (2000) 8:154–64. doi: 10.1159/000054276
159. Foster SL, Seehus CR, Woolf CJ, Talbot S. Sense and immunity: context-dependent neuro-immune interplay. Front Immunol. (2017) 8:1463. doi: 10.3389/fimmu.2017.01463
160. Baral P, Udit S, Chiu IM. Pain and immunity: implications for host defence. Nat Rev Immunol. (2019) 19:433–47. doi: 10.1038/s41577-019-0147-2
161. Douglas SD, Leeman SE. Neurokinin-1 receptor: functional significance in the immune system in reference to selected infections and inflammation. Ann N Y Acad Sci. (2011) 1217:83–95. doi: 10.1111/j.1749-6632.2010.05826.x
162. Nowicki M, Ostalska-Nowicka D, Kondraciuk B, Miskowiak B. The significance of substance P in physiological and Malignant haematopoiesis. J Clin Pathol. (2007) 60:749–55. doi: 10.1136/jcp.2006.041475
163. Manske JM, Hanson SE. Substance-P-mediated immunomodulation of tumor growth in a murine model. Neuroimmunomodulation. (2005) 12:201–10. doi: 10.1159/000085652
164. Duan JX, Zhou Y, Zhou AY, Guan XX, Liu T, Yang HH, et al. Calcitonin gene-related peptide exerts anti-inflammatory property through regulating murine macrophages polarization in vitro. Mol Immunol. (2017) 91:105–13. doi: 10.1016/j.molimm.2017.08.020
165. McIlvried LA, Atherton MA, Horan NL, Goch TN, Scheff NN. Sensory neurotransmitter calcitonin gene-related peptide modulates tumor growth and lymphocyte infiltration in oral squamous cell carcinoma. Adv Biol (Weinh). (2022) 6:e2200019. doi: 10.1002/adbi.202200019
166. Ma Q, Chen J, Kong X, Zeng Y, Chen Z, Liu H, et al. Interactions between CNS and immune cells in tuberculous meningitis. Front Immunol. (2024) 15:1326859. doi: 10.3389/fimmu.2024.1326859
167. Koren T, Yifa R, Amer M, Krot M, Boshnak N, Ben-Shaanan TL, et al. Insular cortex neurons encode and retrieve specific immune responses. Cell. (2021) 184:5902–15 e17. doi: 10.1016/j.cell.2021.10.013
168. Ben-Shaanan TL, Azulay-Debby H, Dubovik T, Starosvetsky E, Korin B, Schiller M, et al. Activation of the reward system boosts innate and adaptive immunity. Nat Med. (2016) 22:940–4. doi: 10.1038/nm.4133
169. Ben-Shaanan TL, Schiller M, Azulay-Debby H, Korin B, Boshnak N, Koren T, et al. Modulation of anti-tumor immunity by the brain’s reward system. Nat Commun. (2018) 9:2723. doi: 10.1038/s41467-018-05283-5
170. Lei Y, Liao F, Tian Y, Wang Y, Xia F, Wang J. Investigating the crosstalk between chronic stress and immune cells: implications for enhanced cancer therapy. Front Neurosci. (2023) 17:1321176. doi: 10.3389/fnins.2023.1321176
171. Liu Y, Tian S, Ning B, Huang T, Li Y, Wei Y. Stress and cancer: The mechanisms of immune dysregulation and management. Front Immunol. (2022) 13:1032294. doi: 10.3389/fimmu.2022.1032294
172. Xu XR, Xiao Q, Hong YC, Liu YH, Liu Y, Tu J. Activation of dopaminergic VTA inputs to the mPFC ameliorates chronic stress-induced breast tumor progression. CNS Neurosci Ther. (2021) 27:206–19. doi: 10.1111/cns.13465
173. Zhang B, Vogelzang A, Miyajima M, Sugiura Y, Wu Y, Chamoto K, et al. B cell-derived GABA elicits IL-10(+) macrophages to limit anti-tumour immunity. Nature. (2021) 599:471–6. doi: 10.1038/s41586-021-04082-1
174. Schneider MA, Heeb L, Beffinger MM, Pantelyushin S, Linecker M, Roth L, et al. Attenuation of peripheral serotonin inhibits tumor growth and enhances immune checkpoint blockade therapy in murine tumor models. Sci Transl Med. (2021) 13:eabc8188. doi: 10.1126/scitranslmed.abc8188
175. Alpizar SA, Baker AL, Gulledge AT, Hoppa MB. Loss of neurofascin-186 disrupts alignment of ankyrinG relative to its binding partners in the axon initial segment. Front Cell Neurosci. (2019) 13:1. doi: 10.3389/fncel.2019.00001
176. Magnon C, Hondermarck H. The neural addiction of cancer. Nat Rev Cancer. (2023) 23:317–34. doi: 10.1038/s41568-023-00556-8
177. Bucsek MJ, Qiao G, MacDonald CR, Giridharan T, Evans L, Niedzwecki B, et al. beta-adrenergic signaling in mice housed at standard temperatures suppresses an effector phenotype in CD8(+) T cells and undermines checkpoint inhibitor therapy. Cancer Res. (2017) 77:5639–51. doi: 10.1158/0008-5472.CAN-17-0546
178. Aljarrah D, Chalour N, Zorgani A, Nissan T, Pranjol MZI. Exploring the gut microbiota and its potential as a biomarker in gliomas. BioMed Pharmacother. (2024) 173:116420. doi: 10.1016/j.biopha.2024.116420
179. Beam A, Clinger E, Hao L. Effect of diet and dietary components on the composition of the gut microbiota. Nutrients. (2021) 13. doi: 10.3390/nu13082795
Keywords: nervous system, gut microbiota, immune cells, tumor environment, signal transduction
Citation: Wang J, Cheng W and Yang R (2025) Nervous system–gut microbiota–immune system axis: future directions for preventing tumor. Front. Immunol. 16:1535955. doi: 10.3389/fimmu.2025.1535955
Received: 28 November 2024; Accepted: 01 April 2025;
Published: 01 May 2025.
Edited by:
Jeni Prosperi, Indiana University School of Medicine, South Bend, United StatesReviewed by:
Xiaopan Xu, Air Force Medical University, ChinaYaXuan Wang, First Affiliated Hospital of Harbin Medical University, China
Copyright © 2025 Wang, Cheng and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rongcun Yang, cnlhbmdAbmFua2FpLmVkdS5jbg==
 Juanjuan Wang
Juanjuan Wang Wenyue Cheng1
Wenyue Cheng1 Rongcun Yang
Rongcun Yang