- 1Department of Obstetrics and Gynecology, Zhuzhou Central Hospital, Zhuzhou, Hunan, China
- 2Department of Breast Surgery, Zhuzhou Central Hospital, Zhuzhou, Hunan, China
The favorable prognosis of “hot” tumors is widely acknowledged in oncology. Recently, the concept of tertiary lymphoid structures (TLS) has renewed appreciation for local immune cells within tumor tissues. Tissue-resident immune cells, a newly identified subset of tumor-infiltrating lymphocytes, are emerging as potential key players in tumor infiltration and TLS formation, due to their ability to reside indefinitely within tissues and mount effective responses to local antigens. Cervical cancer (CC), the fourth most common cause of cancer-related mortality among women globally, has experienced comparatively limited progress in delineating its tumor immune microenvironment compared to other malignancies. Notably, the role of tissue-resident immune cells within the CC milieu remains inadequately characterized. This comprehensive review aims to synthesize current knowledge and critically evaluate the putative roles of these cells in CC pathogenesis, providing new insights on the intricate dynamics of the local tumor microenvironment.
1 Introduction
Cervical cancer (CC) ranks as the fourth most common gynecological malignancy affecting women globally, representing a significant threat to women’s health (1). According to the GLOBOCAN 2020 report, there were an estimated 604,000 new CC cases worldwide, resulting in approximately 342,000 deaths (2). Despite advancements in multimodal treatment approaches, including surgery, radiotherapy, and chemotherapy, a considerable proportion of patients with advanced-stage CC experience tumor recurrence and poor clinical outcomes. The challenge of effectively reducing disease recurrence and improving long-term survival rates in CC patients remains a critical issue in gynecologic oncology.
The tumor microenvironment (TME) has emerged as a focal point for cancer therapy research, offering potentially safer and more efficacious therapeutic targets (3). Over the past decades, our conceptualization of tumors has evolved from viewing them as simple aggregates of abnormally proliferating cells to recognizing them as complex, highly organized “pseudo-organs”. The TME comprises a heterogeneous population of cells, including immune cells (such as CD8+ and CD4+T lymphocytes, B cells, and natural killer cells), stromal components (including cancer-associated fibroblasts, adipocytes, and extracellular matrix), and vascular elements (both blood and lymphatic endothelial cells). Among these constituents, immune cells have garnered particular attention due to their capacity to recognize and potentially suppress malignant cells, thus emerging as promising targets for immunotherapeutic interventions.
In recent years, the significance of regional immune responses—localized within specific tissues or organs and functioning independently of systemic circulation—has been increasingly recognized in various diseases. It has been observed that immune cells adapt to the microenvironment of tumor tissues, acquiring unique characteristics and functional specializations that may play critical roles in antitumor immunity. This systematic review examines the potential roles of resident immune cells in CC and offers perspectives on their targeting for tumor immunotherapy.
2 Tissue-resident immune cells: definition and subsets
TRICs are a specialized group of immune cells that reside long-term in specific tissues and perform localized immune functions. Unlike circulating immune cells, TRICs remain in tissues under steady-state or non-pathological conditions, establishing local immune memory and rapid response capabilities (4, 5). TRICs primarily include the following subsets:
2.1 Tissue-resident memory T cells
First identified in skin and intestinal tissues, Trm cells were definitively characterized in 2009 through murine models. Trm cells comprise two primary subsets: CD8+ and CD4+Trm. CD8+Trm cells predominantly mediate immune responses in infection and tumor immunity, enabling rapid pathogen and tumor cell elimination (6). CD4+Trm cells have recently gained attention in autoimmune disease research, where they modulate immune responses through cytokine secretion and interactions with other immune cells (7, 8). While CD8+Trm cells focus on direct pathogen and tumor cell clearance, CD4+Trm cells play a regulatory role, supporting local immune responses and tissue repair processes.
2.2 Tissue-resident B cells
Tissue-resident B Cells (also known as resident memory B cells, Brm) represent a recently characterized subset of B lymphocytes, first comprehensively described in 2019 (9). Initially identified in barrier tissues such as the lungs and intestines, these cells have since been discovered in diverse anatomical locations, including the skin, liver, and tumor microenvironments (10). Brm cells exhibit distinct transcriptional and phenotypic profiles that enable long-term persistence within specific tissue compartments and rapid responses to pathogen re-encounter. Unlike circulating memory B cells, they possess unique capabilities for immediate local immune responses, rapidly differentiating into antibody-secreting cells and directly engaging antigens within the tissue microenvironment. Their primary functions encompass local antibody production, immune regulation, and critical contributions to host defense against various pathological conditions, including viral infections, bacterial challenges, autoimmune diseases, and tumor immunity, thereby providing a more targeted and expeditious immune defense.
2.3 Tissue-resident NK cells
Although the discovery of NK cells dates back to the 1970s, it is only in the past two decades that researchers have begun to focus on the tissue-resident characteristics of these cells (11). The tissue-resident NK cells were first identified in the liver of mice and have since been found in various human tissues, including the skin, salivary glands, uterus, adipose tissue, and kidneys (12, 13). These cells possess unique phenotypic and functional properties that enable them to rapidly respond to local infections, tissue damage, or tumors. Their localized presence allows tissue-resident NK cells to exert immediate immune responses through cytokine secretion and direct killing of infected or malignant cells, while also modulating local immune environments and maintaining tissue homeostasis.
3 Potential characteristic molecular markers of TRICs
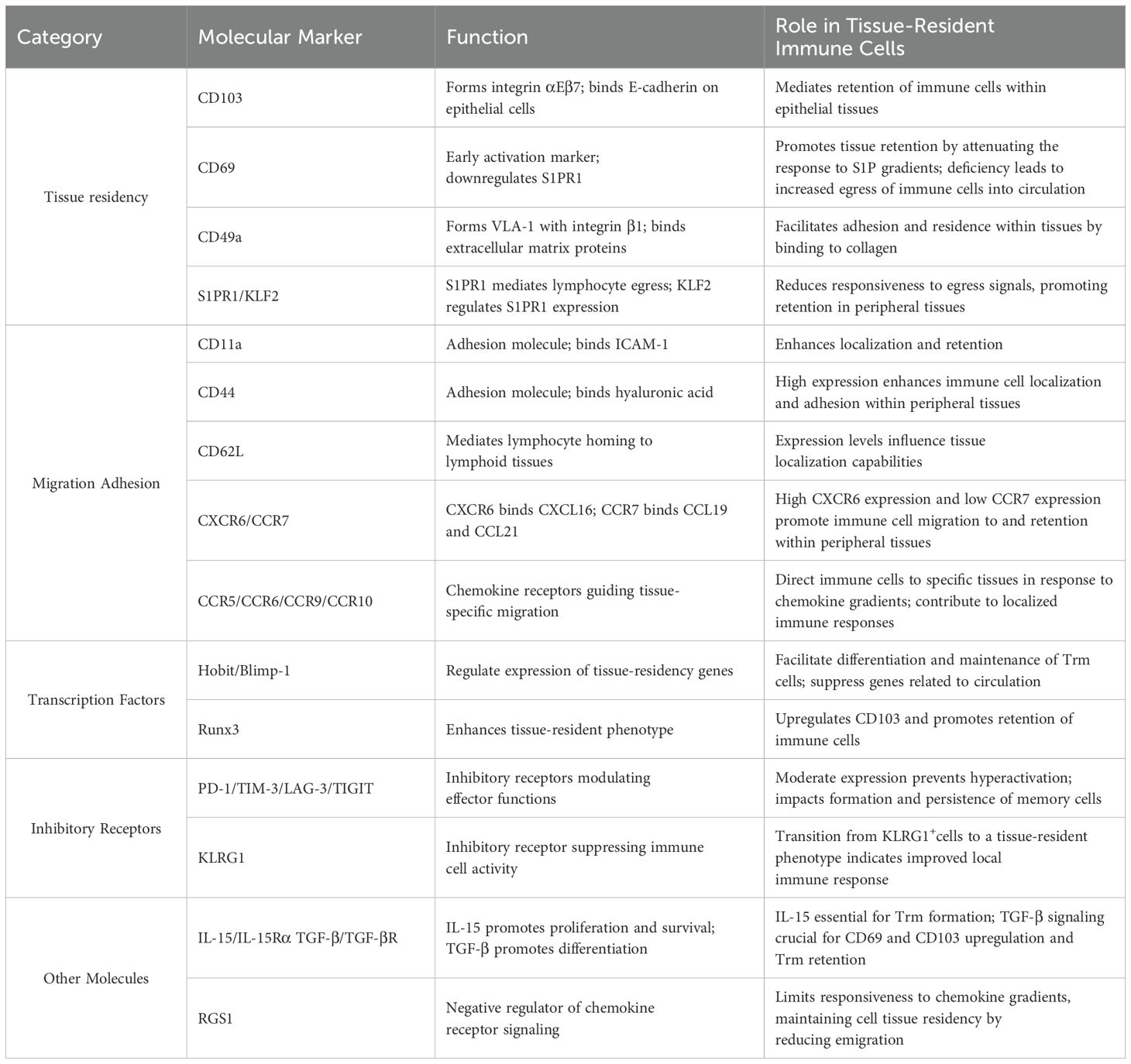
Molecular markers serve as critical tools for defining and investigating tissue-resident immune cells. These markers not only facilitate the identification of distinct cellular subsets but also provide insights into their functional properties and tissue-specific adaptations. While the current understanding of molecular markers is primarily derived from studies on Trm cells, the functional implications of these markers may extend to other tissue-resident immune cell populations. Based on their established roles and potential functions, these molecular markers can be categorized into several groups (Table 1):
3.1 Molecules associated with tissue residency
The establishment and maintenance of tissue residency rely on several key molecular mechanisms. CD103 (Integrin αE), forming the heterodimeric integrin αEβ7, mediates epithelial tissue retention through E-cadherin binding (14). Its expression is indispensable for strategic positioning and long-term maintenance of TRICs within tissues. Similarly, CD49a (integrin α1) combines with integrin β1 to form VLA-1, facilitating adhesion to extracellular matrix components, particularly in organs such as the liver, skin, and female reproductive tract (15).
Tissue retention mechanisms critically involve the CD69-S1PR1 regulatory axis. CD69 promotes tissue residency by downregulating S1P receptor 1 (S1PR1), thereby attenuating cellular response to sphingosine-1-phosphate (S1P) gradients that typically guide lymphocyte egress (16–18). This process is regulated by Krüppel-like factor 2 (KLF2), a transcription factor that modulates S1PR1 expression. Studies have demonstrated that CD69 deficiency or elevated S1PR1/KLF2 expression results in increased susceptibility of TRICs to re-enter circulation, highlighting the crucial role of this regulatory axis in maintaining tissue residency.
3.2 Molecules mediating chemotaxis, migration, and adhesion
Tissue-specific adhesion molecules play distinct roles in different anatomical locations. CD11a serves as a crucial adhesion molecule in organs where CD103 expression is typically absent, such as the liver, spleen, and lymph nodes (19). Enhanced CD11a expression in hepatic tissue-resident CD8+T cells enables their maintenance within hepatic sinusoids (20). CD44, through its interaction with hyaluronic acid, facilitates both cell-matrix adhesion and indirect cell-cell interactions, while CD62L expression levels correlate with tissue localization capabilities, with CD62Llow cells preferentially forming tissue-resident populations (21, 22).
Chemokine receptors play a central role in guiding TRICs to specific tissues and maintaining their localization. CXCR6, a receptor for CXCL16, facilitates the migration and retention of TRIC precursors in peripheral tissues such as the liver, lungs, skin, and intestine (23, 24). In contrast, CCR7, which interacts with CCL19 and CCL21, promotes lymph node homing. Trm cells typically lack CCR7 expression, which prevents their recirculation to lymph nodes and supports their tissue residency (25, 26). The high CXCR6 and low CCR7 expression pattern is a hallmark of Trm cells, ensuring their retention and survival in peripheral tissues.
Tissue-specific chemokine receptors orchestrate precise TRIC localization through distinct molecular pathways. CCR5 is critical for maintaining mucosal barrier immunity, with up to 65% of CD4+Trm cells in mucosal tissues expressing high levels of CCR5, essential for their optimal positioning and function within the mucosal barrier (27). The CCR6-CCL20 axis mediates immune cell recruitment to inflammatory sites, with its disruption showing therapeutic potential in conditions like rheumatoid arthritis and inflammatory bowel disease (28). The CCR9-CCL25 pathway specifically promotes Trm cell differentiation and retention in gastrointestinal tissues, where these cells maintain local surveillance while remaining unresponsive to distant antigens (29). Similarly, CCR10-CCL27 signaling supports skin-specific immune responses, with CCR10+CD4+T cells being instrumental in mediating localized skin memory responses, particularly in conditions such as allergic contact dermatitis (30). This sophisticated chemokine receptor network ensures optimal tissue-specific immune surveillance and rapid response to local challenges.
3.3 Transcriptional regulators
Transcriptional regulation of tissue residency involves coordinated actions of key factors. Hobit and Blimp-1 collaboratively establish the tissue-resident program by suppressing circulation-associated genes (CCR7, S1PR1) while upregulating residence-related genes (CD69, CXCR6) (31, 32). Their tissue-specific roles vary, as evidenced in lung influenza-specific CD8+Trm cells, where Blimp-1, but not Hobit, is essential (33, 34). This suggests that Hobit and Blimp-1 employ distinct regulatory mechanisms across different tissues. Runx3 is another critical transcription factor that promotes the tissue-resident phenotype by upregulating CD103 expression (35). Overexpression of Runx3 enhances the persistence and effector functions of CD4+CAR-T cells, improving their tumor-residency capabilities (36). Additionally, Runx3 has been identified as a key regulator in tissue-resident T cells within healthy human breast tissue, highlighting its role in maintaining local barrier immunity (37).
3.4 Immune checkpoint molecules and other regulators
Inhibitory receptors (PD-1, TIM-3, LAG-3, TIGIT) and KLRG1 serve dual functions in TRICs (38, 39). While traditionally associated with T cell exhaustion, these molecules play crucial roles in maintaining tissue residency and preventing hyperactivation-induced tissue damage. For instance, PD-1 signaling promotes long-term memory T cell persistence by modulating metabolic programs (40, 41), while KLRG1 regulation through Bach2 facilitates the transition from effector to tissue-resident phenotypes (42, 43). Understanding this balance between inhibition and maintenance is crucial for therapeutic applications, particularly in cancer immunotherapy.
The development and maintenance of TRICs also depend on cytokine signaling networks. IL-15/IL-15Rα signaling is essential for TRIC formation and survival, promoting their persistence at infection sites (44). TGF-β signaling enhances the expression of tissue-residency markers such as CD103 and CD69, while suppressing circulation-associated genes. Mice lacking TGF-βR exhibit significant reductions in Trm cell numbers, highlighting its essential role in TRIC development (45). Additionally, RGS1, a negative regulator of chemokine receptor signaling, helps maintain tissue residency by limiting responsiveness to peripheral chemokine gradients, thereby reducing tissue egress (46).
4 Mechanisms of TRICs development and retention in tumor
Understanding the development and retention mechanisms of TRICs is crucial for advancing our comprehension of local immune surveillance and tumor immune responses. The molecular mechanisms governing TRIC biology remain largely unexplored. Currently, research on TRICs is predominantly concentrated on Trm cells, which serve as a model system for elucidating the fundamental principles of cellular tissue residency. This section will primarily focus on the detailed molecular mechanisms of Trm cell differentiation and retention (Figure 1), utilizing these insights as a framework to provide broader perspectives on TRIC development in the TME.
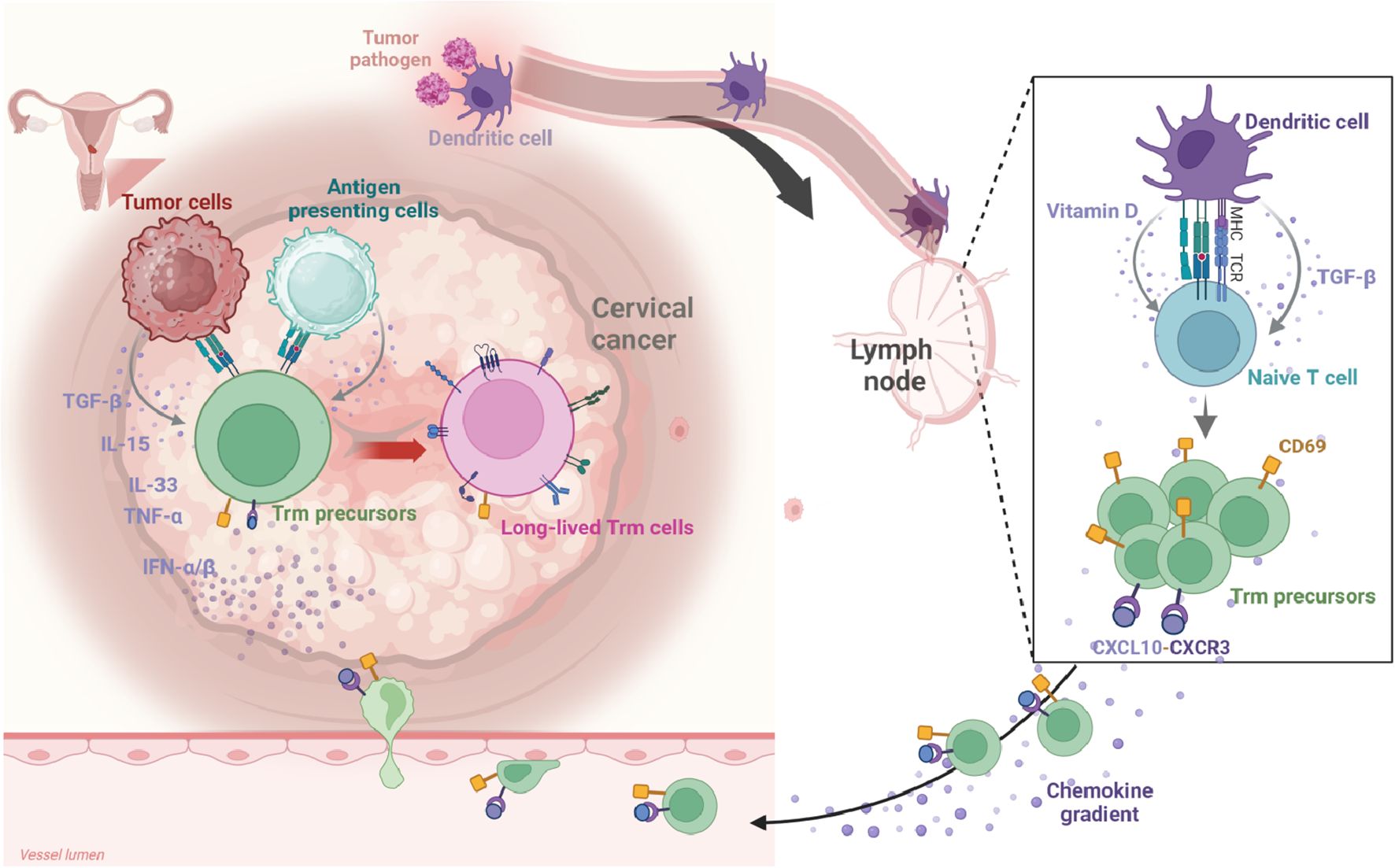
Figure 1. The development of Trm cells in CC. Migratory dendritic cells (DCs) capture tumor antigens within the tumor microenvironment (TME) and migrate to draining lymph nodes. There, they present these antigens to naïve T cells via MHC class molecules, initiating T cell activation and differentiation. Concurrently, DCs secrete cytokines, notably TGF-β, and metabolites like vitamins A and D, which influence T cell fate during the priming phase. Following activation, Trm precursors enter the arterial circulation, guided by chemokines (e.g., CCL5, CXCL9, CXCL10) that direct them toward tumor tissues. The upregulation of CD69 on Trm precursors inhibits S1PR1 signaling, preventing their return to circulation and anchoring them within the TME. Within this environment, Trm precursors undergo final differentiation, driven by a network of inflammatory cytokines and tumor antigen re-stimulation. Created in BioRender. Li, Q. (2025) https://BioRender.com/kfcf5cr.
4.1 Priming and migration of Trm precursors
In tumor immunity, migratory dendritic cells (DCs) and specific subsets such as DC3 capture antigens within tumor tissues before migrating to draining lymph nodes, where they perform crucial antigen presentation functions (47–52). These DCs deliver tumor antigens to naïve T cells via MHC class I or II molecules, triggering T cell activation, proliferation, and functional differentiation. Concurrently, DCs secrete cytokines (particularly TGF-β) and metabolites such as vitamins A and D, which collectively regulate T cell fate determination during this priming phase. During activation, these signals not only stimulate naïve T cell proliferation but also induce expression of specific tissue-homing receptors. This molecular signature equips T cells with the capacity to migrate toward specific peripheral tissues, establishing favorable conditions for their subsequent differentiation into genuine Trm cells within the TME. Consequently, these “pre-programmed” T cells are considered Trm precursors, capable of rapidly converting into functional Trm cells upon receiving local environmental cues.
Following lymph node activation, Trm precursors enter arterial circulation and achieve targeted homing in response to chemotactic signals (such as CCL5, CXCL9 and CXCL10) emanating from tumor or inflammatory sites (53, 54). Chemokines guide activated cells toward inflamed or tumor tissues by binding to corresponding receptors (such as CXCR3 and CCR5) on the cell surface. Ultimately, these Trm precursors traverse the endothelial barrier to enter tumor tissue. Prior to tissue entry, upregulation of surface CD69 plays a decisive role by inhibiting S1PR1 signaling, thereby preventing cells from returning to peripheral circulation and anchoring them within the local microenvironment.
4.2 Cytokine networks in Trm cell differentiation
In the tumor microenvironment, Trm precursor cells undergo a critical final differentiation phase orchestrated by a complex network of inflammatory cytokines and tumor antigen re-stimulation. This process is regulated by a diverse cellular ensemble including tumor cells, endothelial or epithelial cells, macrophages, and DCs that collectively establish a cytokine-rich milieu conducive to Trm development. TGF-β serves as the principal driver of Trm cell differentiation through multiple convergent pathways. Upon receptor binding, TGF-β activates the canonical Smad signaling cascade, whereby phosphorylated Smad2/3 forms complexes with Smad4 that translocate to the nucleus and directly promote CD103 gene transcription (55). Concurrently, TGF-β signaling induces critical transcription factors including Runx3 and Bhlhe40, which further reinforce Trm specific gene expression patterns (56, 57). In synergy with IL-33 and TNF-α, TGF-β also downregulates KLF2 through PI3K/AKT-dependent mechanisms, consequently reducing S1PR1 expression and preventing T cell egress from tissue sites. Additionally, the integration of TGF-β and Notch signaling further shapes the Trm phenotype. TGF-β upregulates Notch ligands on epithelial cells, enhancing Notch signaling in adjacent T cells. Activated Notch intracellular domain (NICD) translocates to the nucleus to regulate Hes1 and Hey1 expression, reinforcing Hobit and Blimp-1 expression while suppressing tissue egress-associated genes (S1PR1, CCR7, KLF2) (32).
IL-15 signaling complements TGF-β functions through interaction with its receptor complex (IL-15Rα/IL-2Rβ/γc), activating the JAK-STAT pathway to promote Trm differentiation and survival (58, 59). Phosphorylated STAT5 induces Hobit and Blimp-1 expression, while simultaneously engaging the PI3K/AKT axis to upregulate anti-apoptotic proteins Bcl-2 and Bcl-xL, substantially enhancing Trm resilience within the hypoxic, nutrient-deprived tumor microenvironment. A critical balance exists between IL-15 and TGF-β in regulating T-bet expression: IL-15 upregulates T-bet, thereby enhancing responsiveness to IL-15 itself through increased IL-15Rβ expression, while TGF-β moderately suppresses T-bet and Eomes to prevent excessive effector differentiation, simultaneously maintaining essential Trm characteristics. Consequently, mature Trm cells maintain T-bet at a “low but sufficient” level—adequate to ensure IL-15 signal sensitivity without compromising CD103 upregulation or allowing effector T cells to outcompete Trm populations.
Type I interferons (IFN-α and IFN-β) play a critical role in shaping Trm differentiation and function through multiple coordinated signaling pathways. These interferons not only upregulate chemokine receptors CXCR3 and CXCR6 via JAK-STAT signaling, thereby enhancing Trm positioning within the tumor microenvironment, but also induce IL-15 and IL-27 expression, creating a positive feedback amplification loop. Type I interferons additionally suppress KLF2 indirectly through PI3K/AKT pathways, reducing S1PR1 levels, while activating NF-κB signaling to upregulate adhesion molecules like CD49a, strengthening interactions between Trm cells and tumor matrix components. Additional cytokines further refine Trm differentiation and functional regulation through diverse pathways. IL-33 independently downregulates transcription of KLF2 and its target gene S1pr1 through PI3K/AKT-dependent mechanisms (17). This pathway converges with and amplifies similar signaling cascades initiated by type I interferons and TGF-β, establishing redundant mechanisms that ensure robust tissue retention. TNF-α predominantly enhances CD69 promoter activity through activation of NF-κB/Rel family members (particularly RelA), significantly increasing CD69 surface expression (60). This likely acts synergistically with adhesion molecules (such as CD49a) upregulated by type I interferons, collectively strengthening Trm cell anchoring within tumor tissues. Concurrently, IL-7 activates the Jak/STAT5 pathway to upregulate Mcl-1, Bcl-2, and Bcl-xL expression (61), forming a survival axis parallel to the IL-15-mediated PI3K/Akt-Bcl2 pathway, jointly maintaining long-term Trm cell persistence.
This multi-pathway cytokine signaling network ultimately converges on the coordinated activation of key transcription factors (including Blimp-1, Hobit, and RUNX3), driving significant upregulation of Trm signature molecules (CD103, CD49a, CXCR6, CD69) while simultaneously repressing circulation-associated gene expression (S1PR1, CCR7, CD62L) and certain effector-related transcription factors (high-level T-bet, Eomes). Through this multi-layered, cooperative signaling network regulation, precursor T cells within the TME complete their transformation into mature Trm cells, acquiring the capacity for long-term residence within tumor tissue.
The long-term maintenance of Trm cells in the TME is supported by a combination of cytokine signaling, metabolic adaptation, and epigenetic regulation. TGF-β and IL-15 remain critical for sustaining Trm survival and function, while hypoxia in the TME synergizes with TGF-β to stabilize the Trm phenotype. Metabolically, Trm cells rely on fatty acid oxidation (FAO) rather than glucose metabolism, enabling them to thrive in the nutrient-deprived, hypoglycemic conditions of the TME (62, 63). Transcription factors such as Bhlhe4 and Runx3 maintain the epigenetic landscape of Trm cells, ensuring the suppression of egress-related genes and the persistence of tissue-retention molecules like CD103 and CD69 (56, 64). Additionally, periodic antigen stimulation within the TME can enhance Trm proliferation and functionality, although excessive stimulation may lead to exhaustion (47).
5 Revisiting inhibitory receptors in TRIC cells: beyond exhaustion paradigms
Tissue-resident immune cells, particularly Trm cells, present a puzzling phenomenon in the tumor microenvironment: despite high expression of inhibitory receptors such as PD-1, TIM-3, LAG-3, and TIGIT, they maintain significant antitumor activity rather than exhibiting the expected functional exhaustion. This observation starkly contrasts with the conventional paradigm that views inhibitory receptors as hallmarks of T cell dysfunction and exhaustion. Understanding this paradox requires in-depth analysis across three dimensions: metabolic reprogramming, signaling pathway equilibrium, and transcriptional regulatory networks.
5.1 Metabolic reprogramming: PD-1-mediated survival advantage
The nutrient-restricted, lactate-rich TME presents significant survival challenges for conventional effector T cells. Under these conditions, the PD-1 signaling pathway unexpectedly confers protective benefits. Upon PD-1/PD-L1 binding, recruitment of SHP-2 and SHP-1 phosphatases inhibits the PI3K/AKT/mTOR pathway downstream of TCR signaling, shifting T cell metabolism from energy-intensive glycolysis toward more economical fatty acid oxidation (FAO) (40, 65). Specifically, PD-1 signaling upregulates carnitine palmitoyltransferase 1A (CPT1A) expression, enhancing FAO while simultaneously attenuating glycolysis and glutaminolysis (66). This metabolic shift particularly benefits Trm cells for several reasons: it delays rapid apoptosis that would otherwise occur in glucose-deprived environments due to excessive glycolytic dependence; it prevents terminal differentiation triggered by high-intensity metabolic activity in effector T cells; and most importantly, it establishes a metabolic profile conducive to long-term survival, thereby maintaining persistent immunosurveillance within the TME. Thus, high PD-1 expression on Trm cells likely represents an adaptive mechanism rather than a marker of functional impairment.
5.2 Signaling pathway balance: antagonistic regulation by IL-15 and TGF-β
Maintenance of Trm functionality depends on a sophisticated cytokine network, particularly the balance between IL-15 and TGF-β. IL-15 activates STAT5, enhancing IFN-γ gene transcription and moderately upregulating T-bet expression. STAT5 directly binds to both the proximal promoter and the CNS1 enhancer of the IFNG gene, while T-bet directly promotes IFNG transcription. Conversely, TGF-β, through the Smad signaling pathway, suppresses high-level expression of T-bet and Eomes, preventing excessive acquisition of effector phenotypes while inducing expression of tissue retention molecules such as CD103. This antagonistic balance is crucial for Trm function: T-bet is maintained at low but sufficient levels (53, 67, 68), which both ensures Trm sensitivity to IL-15 survival signals through upregulation of IL-15Rβ (CD122) and avoids the inhibitory effect of high T-bet levels on Trm formation. Notably, reduced T-bet levels weaken suppression of inhibitory receptors such as PD-1, resulting in increased expression of these molecules without completely blocking effector functions—a key mechanism potentially explaining how Trm cells maintain functional activity despite high inhibitory receptor expression.
5.3 Microenvironmental adaptation: hypoxia and TGF-β synergy
Hypoxic conditions within the TME activate HIF-1α, which enhances TGF-β signaling, establishing another critical regulatory mechanism. HIF-1α not only promotes glucose uptake and metabolism but also potentiates TGF-β/Smad signaling pathway activity. This synergistic interaction reinforces tissue-resident characteristics of Trm cells while maintaining their survival capacity in hypoxic environments (69). Hypoxia-driven metabolic adaptation intersects with PD-1 signaling: PD-1-mediated metabolic shifts favor long-term cellular survival under hypoxic conditions, while interactions between HIF-1α and TGF-β signaling further stabilize the Trm phenotype (70). This multilayered regulation enables Trm cells to maintain sufficient functional activity in response to tumor antigen stimulation despite high inhibitory receptor expression.
Furthermore, after anti-PD-1 treatment, the transcriptional characteristics of Trm cells undergo significant changes, exhibiting functional states similar to effector memory T cells (71). This further suggests that Trm cells expressing inhibitory receptors are not in an irreversible exhausted state, but rather in a highly plastic functional state. However, sustained PD-1 blockade may yield dichotomous consequences: initial function restoration and enhancement, followed by potential terminal differentiation due to intensified glycolysis, ultimately leading to clonal deletion. This phenomenon offers a mechanistic explanation for the observed patterns of therapeutic resistance in clinical settings, where patients initially responding to PD-1 blockade subsequently develop progressive disease (72, 73). The temporal dynamics of Trm functional modulation are characterized by initial reinvigoration followed by potential terminal differentiation and deletion, which closely align with clinical response kinetics. Moreover, this biphasic response pattern suggests that intermittent rather than continuous checkpoint blockade might better preserve the Trm population while maintaining antitumor efficacy.
Despite relatively limited research on tissue-resident immune cells beyond Trm cells, emerging studies have identified high expression of inhibitory receptors in other TRIC subtype like tissue-resident NK cells (74). This observation suggests that similar functional equilibrium and adaptive mechanisms may operate in these cell populations, extending the paradigm of inhibitory receptor-mediated homeostasis across diverse tissue-resident lymphocyte lineages. The phenomenon of TRIC cells maintaining functional activity despite high inhibitory receptor expression reflects an exquisite biological equilibrium. Within this balance, inhibitory receptors serve not merely as functional “brakes” but as “stabilizers” for adaptation to harsh TME conditions. Through these receptors, Trm cells modulate their metabolic state and signal intensity, establishing a balance between terminal differentiation and functional maintenance, thereby achieving long-term residence and preserved functionality within nutrient-depleted, hypoxic, and highly suppressive tumor microenvironments.
6 Tissue-resident memory CD8+T cells in CC: the predominant tumor-reactive population
CD8+Trm cells represent a critical frontier in understanding anti-tumor immunity within CC. Strategically positioned within tumor tissues, these specialized immune sentinels offer a unique mechanism of immune surveillance that transcends traditional circulating T cell responses. Their distinctive molecular profile provides key insights into the complex immunological dynamics of CC progression.
6.1 Molecular markers and phenotypic characteristics
CD8+ tissue-resident memory T cells (CD8+Trms) play a crucial role in anti-tumor immunity within CC microenvironments. Using single-cell RNA sequencing techniques, researchers have conducted comprehensive analyses of Trm cells in CC tissues. Beyond the well-established core markers CD103 and CD69, CD8+Trm cells of CC express significantly higher levels of tissue-residence-associated genes (such as TGFB1 and ITGB7), cytotoxicity genes (including GZMB and PRF1), chemokine genes (notably CXCL13 and CCL5) and inhibitory molecules (LAG3, TIGIT, PDCD1, HAVCR2, and CTLA4), compared to non-Trm cells (75). This distinct transcriptional profile reflects their central role in tumor immune surveillance.
6.2 Clinical prognostic relevance
A large-scale epidemiological study has demonstrated significantly higher diagnosis rates and mortality from advanced CC among women aged over 65 years compared to younger populations (76, 77). This age-related disparity in clinical outcomes correlates with the progressive decline in cervical Trm cells observed in aging populations (78). The depletion of cervical Trm cells in postmenopausal women may represent a crucial mechanism underlying the poor prognosis in elderly CC patients. Notably, CD8+Trm cell density has emerged as a key prognostic indicator across various solid tumors, correlating with tumor size, grade, and overall survival (79–83).
Studies have shown that the expression of the ITGAE gene (encoding CD103) significantly correlates with T cell markers (CD3, CD2), immune checkpoint molecules (PD1, TIGIT), and antigen presentation molecules (HLA-DR, HLA-DQ) (84). High CD103 expression is associated with significantly improved cancer-specific survival, a correlation that remains statistically significant after adjusting for disease stage. Recent investigations have demonstrated that infiltration levels of CD103+CD8+tumor-infiltrating lymphocytes strongly correlate with improved survival outcomes in CC patients (78). This finding has been robustly validated in an independent cohort of 460 CC patients, establishing the expression of these cells as an independent predictive factor for patient prognosis. Furthermore, CD8+Trm cell infiltration is substantially higher in CC tissues compared to normal cervical tissues (75, 85), highlighting their critical role in anti-tumor immune responses.
6.3 Spatial distribution characteristics
Recent spatial transcriptomic analyses of CC have revealed three distinct patterns of CD8+ T cell distribution: the infiltrated pattern, where CD8+T cells effectively penetrate and distribute throughout the tumor epithelium; the excluded pattern, characterized by CD8+T cells predominantly accumulating in the peritumoral stroma without entering the tumor parenchyma; and the desert pattern, marked by an almost complete absence of CD8+T cell infiltration. Importantly, the infiltrated pattern strongly correlates with the MP7 tumor state (epithelial-immune phenotype), which is characterized as a prototypical “immune-hot” tumor where tumor cells highly express antigen-presenting molecules (HLA-DR, HLA-DQ), immune response genes (B2M, CXCL10), and immunomodulatory factors (IDO1, CD274), indicating active antigen presentation and sustained immune activation (86).
CD8+T cells within MP7 regions concurrently express elevated levels of cytotoxic effector molecules (GZMB, PRF1, GNLY) and inhibitory receptors (HAVCR2, TIGIT, LAG3, CTLA4, PDCD1). This molecular signature shows remarkable concordance with CD8+Trm cell markers identified in single-cell sequencing study (75), suggesting that CD8+T cells infiltrating the tumor epithelium are predominantly CD8+Trm subpopulations, further validating the critical role of CD8+Trm cells in immune-hot tumor microenvironments. Spatial interaction analyses further demonstrate that MP7 tumor cells likely actively recruit CD8+T cells to epithelial regions through chemokine-receptor axes, including CXCL9–CXCR3 and CXCL11–CXCR3. Additionally, PROGENy pathway enrichment analysis reveals significant activation of JAK-STAT, MAPK, and NF-κB signaling in the MP7 state, pathways involved in regulating pro-inflammatory cytokine production and immune cell recruitment, establishing the molecular basis for the preferential enrichment of CD8+Trm cells within tumor epithelial regions.
6.4 Molecular regulatory mechanisms of epithelial affinity
The epithelial tropism of CD8+Trm cells is regulated by intricate molecular mechanisms. CD103, a critical adhesion molecule, requires dual signaling for its expression: T cell receptor (TCR) activation and TGF-β signaling. Research by Komdeur et al. revealed that CC exhibits a distinctive TGF-β signaling profile compared to other malignancies (78). In CC, TGF-β signaling is remarkably abundant throughout the tumor microenvironment, evidenced by prominent nuclear pSMAD2/3 expression in tumor nests, surrounding stromal cells, and both CD103+ and CD103-tumor-infiltrating lymphocytes. The characteristic distribution pattern of TGF-β signaling closely relates to HPV biology, as HPV-16 E6 and E7 oncoproteins directly modulate TGF-β1 expression in cervical tumor cells through specific DNA sequence motifs in the TGF-β1 core promoter. Within this TGF-β-enriched microenvironment, direct contact between T cells and tumor cells becomes the decisive factor for CD103 induction, positioning CD103 as an ideal biomarker for identifying tumor-reactive T cells in CC. Beyond TGF-β signaling, as previously described (86), the MP7 tumor state in CC features activated signaling pathways such as JAK-STAT and MAPK that form complex regulatory networks with immune cell recruitment, collectively shaping the tissue localization properties of CD8+Trm cells in CC.
6.5 Cell dynamics in disease progression
Single-cell sequencing analysis has revealed significant gene expression differences in CD8+T cells across various stages of CC progression (87). In early-stage CC, CD8+T cells exhibit robust anti-tumor immune characteristics, marked by highly activated interferon signaling pathways (JAK2, STAT1, IFIH1, ISG15, GBP1, GBP4, GBP5, OAS1, OAS2, OAS3) and enhanced cytotoxic functionality (GZMB, PRF1). Notably, these cells also upregulate RUNX3, CXCR6, CXCL13, and CCL5, a molecular signature strikingly similar to the established profile of Trm identified in previous studies. This suggests that CD8+Trm cells likely constitute the predominant anti-tumor CD8+T cell population in early CC, strategically positioned for immediate immune surveillance and rapid response to tumor antigens.
As the disease progresses to advanced stages, the molecular profile of CD8+T cells undergoes substantial transformation. The previously upregulated immune activation genes show markedly reduced expression levels, replaced by genes associated with tissue remodeling (COL1A1, COL1A2, COL3A2, CYBA, HLA-DPA1) and immunosuppression (S100A8, S100A9). This shift reflects a transition from a highly effective anti-tumor state toward a tissue-adaptive, functionally limited state, likely connected to tumor microenvironment remodeling and immune evasion mechanisms.
Recent spatial transcriptomic analysis of CC has identified distinct tumor molecular phenotypes that correlate with specific immune infiltration patterns. Two predominant states have emerged: MP7, an immune-active tumor state corresponding to the CD8+T cell infiltration pattern within tumor epithelium; and MP6, an immune-suppressive tumor state characterized by high expression of keratinization-associated genes (FABP5, SERPINB3, S100A8/9) and overactivation of the TGF-β signaling pathway. MP6-dominant tumors display “excluded/desert” immune phenotypes, where CD8+T cells are largely prevented from entering tumor epithelial regions. These tumor states correspond to specific spatial distribution patterns of immune cells (infiltrated or excluded/desert pattern), which directly impact anti-tumor immunity and treatment response.
Comparing results from these CC single-cell and spatial transcriptomic studies reveals a striking parallel in molecular characteristics: CD8+T cells in early-stage CC show high similarity in gene expression profiles to CD8+T cells in the MP7 tumor state, while CD8+T cells in late-stage CC exhibit significant commonalities with those in the MP6 tumor state. This correspondence illuminates the tight coupling mechanism between the tumor immune microenvironment and T cell functionality.
This dynamic evolution of CD8+T cell phenotypes, coupled with the corresponding tumor state transitions, provides critical insights into the mechanisms of immune surveillance and escape during CC progression. The early dominance of CD8+Trm cells in MP7-associated “hot” tumors gives way to immune exclusion in MP6-associated advanced tumors, highlighting potential therapeutic targets for different disease stages.
6.6 CD8+Trm dynamics in CC treatment
Notably, patients undergoing chemoradiotherapy typically exhibit lower CD103+Trm infiltration compared to those treated with surgery alone (78). This difference stems from the fact that robust CD103+Trm infiltration signifies an immune-active tumor state, where potent anti-tumor immunity effectively restricts tumor growth, often corresponding to early-stage disease. In such cases, surgery is the preferred treatment option. Conversely, tumors with minimal CD103+Trm infiltration, indicative of an immune-suppressed microenvironment, are more likely to be advanced and thus require chemoradiotherapy for disease control.
Single-cell RNA sequencing provided unprecedented insights into the time-dependent immunological remodeling mechanisms following chemoradiotherapy in CC (88).At three weeks post-treatment, while the absolute number of CD103+ Trm cells temporarily decreased, the tumor microenvironment underwent a series of changes favorable to subsequent immune activation. Most notably, tumor epithelial cells upregulated MHC-II expression, significantly enhancing their antigen presentation capabilities. Concurrently, chemoradiotherapy promoted CD16+ NK cell infiltration and increased their cytotoxicity-related gene expression (GZMB, GZMH, NKG7). Moreover, the proportion of exhausted CD8+ T cells significantly declined, while CD4+ naive T cell proportions increased.
These findings reveal a specific “immune intervention window” at three weeks post-treatment, where innate immune systems are activated, but adaptive immune responses have not yet fully developed. During this period, tumor cells release increased antigens with enhanced presentation capabilities and reduced microenvironmental immunosuppression, creating ideal conditions for targeting HPV-specific T cell activation. Preclinical research provided compelling evidence for leveraging this post-chemoradiotherapy immune window (78). Combining therapeutic vaccination using forest virus expressing E6/E7 with radiotherapy synergistically elevated CD8+CD103+T cell proportions from approximately 10% in untreated tumors to 60% in the combined treatment group. This strategy effectively exploited the “danger signals” generated by chemoradiotherapy alongside increased tumor antigen exposure, thereby enhancing vaccine-mediated anti-HPV adaptive immune responses.
7 Tissue-resident memory CD4+T cells in CC: the key accessory cells for CD8+Trm cells?
While the role of CD8+Trm cells in antitumor immunity is well established, the functional significance of CD4+Trm cells in CC remains less understood and somewhat controversial. Comprehensive single-cell RNA sequencing analyses across multiple cancer types have revealed that tumor-infiltrating Trm cells are predominantly CD8+T cells, characterized by the expression of tissue residency and cytotoxicity-associated genes. In contrast, CD4+T cells within the tumor microenvironment exhibit limited expression of functional activation markers, such as IFIT3, suggesting potential functional impairment (89).
However, emerging evidence challenges this perspective. Studies utilizing TCRα-deficient mouse models have demonstrated that CD4+Trm cells, marked by CXCR6 expression, play a critical role in initiating antitumor immune responses. These cells secrete IFN-γ, which triggers NK cell-mediated tumor elimination through a coordinated immune cascade. These findings highlight the complexity of CD4+Trm cell functions within the tumor microenvironment, where they may act as essential collaborators in enhancing overall tumor immunity by facilitating cytokine secretion and promoting the activation of other immune cells (90).
Recent investigations have demonstrated that CD4+CD103+T cells are the predominant subset producing pro-inflammatory cytokines IFN-γ and TNF-α, with their numbers significantly increased in tumor tissues compared to normal tissues. These cells exhibit a distinctive molecular signature, including high expression of inhibitory receptors like PD-1 and TIM-3, and the tissue-resident marker CD103 (91). Single-cell sequencing analysis of CC has shown significant clonal expansion of CD4+CXCL13+T cells within tumor tissues, indicating antigen-driven local proliferation (92). Although the tissue residency status and functional significance of these expanded clones remain to be fully characterized, insights from melanoma research suggest that CD4+CXCL13+T cells display tumor antigen specificity and express a distinctive molecular signature, including the inhibitory receptor TIM-3, tissue-resident marker CD103, and effector cytokine IFN-γ. Importantly, the frequency of these cells shows a positive correlation with CD8+PD1+CXCL13+T cell infiltration (93), suggesting a coordinated immune response.
These findings collectively suggest that CD4+Trm cells function as critical orchestrators of antitumor immunity through multiple mechanisms. Their local cytokine production appears to support CD8+T cell function, while CXCL13 secretion likely facilitates immune cell recruitment and organization within the tumor microenvironment. This coordinated interaction between CD4+ and CD8+Trm cells may represent a crucial mechanism for effective antitumor immunity, with significant implications for developing more effective immunotherapeutic strategies.
8 Tissue-resident B cells in CC: the potential tumor antigen-specific B cell?
While B cell presence in mucosal tissues has been documented for over two decades (94–97), the formal identification of tissue-resident B cells was not established until 2019, through definitive experiments employing parabiosis and FTY720 blockade (9). Current research has predominantly focused on lung tissue-resident memory B (Brm) cells, which are characterized by high expression of CD69 and specific chemokine receptors, notably CCR6 and CXCR3 (98–100). Mechanistically, CCR6 has been demonstrated to facilitate Brm cell retention within human bronchial-associated lymphoid tissues (100). These cells primarily function to mount rapid, localized humoral responses against respiratory pathogens, including influenza virus.
The discovery of tissue-resident B cells across various organs, including skin, intestines, lungs, and uterus, has sparked interest in their potential roles in tumor immunity. This interest has been further intensified by the established correlation between TLS and favorable clinical outcomes across multiple cancer types. Tumor-infiltrating B cells (TIL-Bs) typically exist within complex immune cell networks, interacting with T cells, NK cells, and other immune components. Notably, their presence is particularly pronounced in highly immunogenic “hot” tumors, where CD4+ and CD8+ Trm cells express elevated levels of CXCL13, the key B cell recruitment factor.
Recent comprehensive analyses across various cancer types, including colon, lung, and kidney cancers, have revealed that memory B cells constitute the predominant TIL-B subset (101). These cells are characterized by high expression of ITGA4, which facilitates their entry into peripheral tissues (102, 103). The persistent presence of tumor antigens appears to drive continuous recruitment and local expansion of memory B cells, maintaining their sustained presence within the tumor microenvironment. One antigen receptor lineage analyze has demonstrated that TIL-Bs exhibit distinct memory B cell characteristics with evidence of localized expansion and differentiation, particularly in breast cancer tissues (104).The presence of memory B cell subsets within tumor tissues positively correlates with improved overall and disease-free survival rates in multiple cancer types, including CC, sarcoma, cutaneous melanoma, and endometrial cancer (93).In lung cancer, CD69+TIL-Bs demonstrate enhanced antigen presentation capabilities to CD4+TILs, modulating their phenotype and function (93).
A recent single-cell transcriptomic analysis of CC patients has revealed distinct B cell populations with tissue-resident characteristics (92). Notably, a CD69+B cell population expressing tissue-residency and memory-associated genes (CCR6, ITGA4, ITGB7, and CD80) was identified, showing enrichment in antigen processing and presentation pathways. Additionally, a distinct CD103+B cell population was observed within well-organized TLS, correlating with enhanced CD8+T cell infiltration and improved clinical outcomes. Trajectory analysis suggests a potential differentiation pathway from CD69+B cells to CD103+B cells, marked by dynamic changes in gene expression patterns, including CCR6 downregulation and Bcl6 upregulation, potentially reflecting their functional evolution from antigen processing to specific anti-tumor responses.
The observed developmental trajectory between CD69+ and CD103+ B cells represents a critical paradigm in understanding B cell-mediated anti-tumor immunity. This transition appears to be orchestrated by a complex interplay of transcriptional regulators and microenvironmental signals. The downregulation of CCR6, traditionally associated with tissue retention and trafficking, coupled with the upregulation of Bcl6, a master regulator of germinal center formation, suggests a programmed transition from tissue surveillance to active participation in local immune responses. This phenotypic evolution may be driven by persistent exposure to tumor antigens and local inflammatory signals within the tumor microenvironment.
9 Tissue-resident NK cells in CC: immune regulators rather than tumor killers?
NK cells represent a distinct arm of innate immunity, characterized by their ability to eliminate tumor cells and virus-infected cells without prior antigen sensitization, positioning them as crucial first-line defenders in anti-tumor immunity (105, 106). Their activation is primarily regulated through a sophisticated balance between inhibitory signals, predominantly mediated by MHC-I recognition, and activating signals. This unique regulatory mechanism enables NK cells to effectively target tumor cells that have downregulated MHC-I expression as an immune evasion strategy, thereby complementing T cell-mediated immunity (107). In CC specifically, NK cell infiltration correlates positively with neoadjuvant chemotherapy efficacy and improved clinical outcomes (108).
CD56brightNK cells emerge as the predominant tissue-resident NK subset across lymphoid and peripheral tissues, demonstrating significant involvement in local tumor immunity (109). In uterine tissue, these cells comprise 30-70% of the total NK cell population, characterized by a CD56brightCD49a+phenotype (109, 110). The tissue-resident signature of these cells is further defined by the expression of specific markers including CD69, CD103, ITGβ7, and CXCR6 (109–111). Functionally, uterine tissue-resident CD56bright NK cells exhibit a unique profile characterized by reduced cytotoxicity but enhanced cytokine production capacity.
Recent comprehensive single-cell transcriptomic analysis across 24 cancer types has revealed distinct functional profiles of CD56brightCD16lo and CD56dimCD16hiNK cells (112). CD56dimCD16hiNK cells demonstrate elevated expression of cytotoxic effector molecules, including perforin and various granzymes (GZMB, GZMA and GZMH). In contrast, CD56brightCD16loNK cells express diverse cytokine genes, particularly IL-8, indicating their prominent role in immune regulation through cytokine production (113, 114). A notable discovery is the exclusive expression of GZMK in CD56brightCD16loNK cells (112), paralleling the identification of GZMK+CD8+T cells as a specialized tissue-resident subset associated with improved survival in various cancers (115, 116).
The pan-cancer analysis has identified RGS1 as a novel marker highly expressed in tumor-associated CD56brightNK cells but absent in circulating NK cells. The combination of RGS1 and CD69 provides superior discrimination between tissue-resident and circulating NK cells compared to conventional markers alone. Mechanistically, RGS1 expression may promote tissue residency through attenuation of G protein-mediated chemotactic signaling (112). While research on tissue-resident NK cells in CC remains limited, recent single-cell sequencing has identified a distinct CD103+GZMK+NK cell population enriched within tumor tissues. Analysis of TCGA data reveals that high expression signatures of this subset correlate with improved overall survival (92), suggesting a potentially crucial role for tissue-resident NK cells in CC immunity.
In the context of CC, the enrichment of CD103+GZMK+NK cells and their correlation with improved survival outcomes raises several important questions. First, what factors within the CC microenvironment promote the accumulation and maintenance of these cells? Second, how do these tissue-resident NK cells interact with other components of the local immune system, particularly tissue-resident T cells and antigen-presenting cells? Understanding these interactions could reveal new therapeutic opportunities and improve current immunotherapy strategies.
10 Therapeutic strategies targeting TRICs in CC
Emerging immunotherapeutic approaches offer new perspectives and possibilities for CC treatment, despite the limitations of traditional surgical, radiotherapeutic, and chemotherapeutic interventions in advanced and recurrent cases. By integrating recent immunotherapy advancements and a comprehensive understanding of tissue-resident immune cells, this manuscript explores innovative targeted therapeutic strategies that may revolutionize CC management.
Immune checkpoint inhibitors have demonstrated remarkable efficacy across multiple cancer types by disrupting immunosuppressive pathways and activating T cell-mediated tumor cell elimination (117). In CC treatment, checkpoint inhibitors such as pembrolizumab and nivolumab have been applied to advanced and recurrent patients, yielding preliminary clinical benefits (118, 119). However, monotherapy remains constrained by limited response rates, with many patients experiencing initial treatment effectiveness followed by progressive disease and immune resistance. Considering the critical role of CD8+Trm cells in CC and the profound impact of high PD-1 expression within the hostile late-stage tumor microenvironment, we propose a novel hypothesis. During initial treatment, Trm cells exhibit effector-like characteristics due to PD-1 signal blockade, yet the maintenance of their effector functions critically depends on sufficient energy supply. The metabolically restricted late-stage tumor environment ultimately compromises these activated Trm cells, leading to functional exhaustion and apoptosis.
Based on our theoretical understanding of Trm cell metabolism and immune checkpoint dynamics, we propose potential strategic approaches that warrant further investigation to enhance immune checkpoint inhibitor responsiveness. Preliminary hypotheses suggest that implementing intermittent anti-PD-1 treatment may alleviate local tumor metabolic burden, potentially enabling T cells to maintain long-term survival through fatty acid oxidation during metabolic stress periods. Additionally, we speculate that combining anti-PD-1 therapy with metabolic environment-modulating agents could potentially offer novel therapeutic strategies. Specifically, agents like metformin, which decrease tumor hypoxia (120, 121), and low-dose bevacizumab, which promotes vascular normalization, might provide innovative approaches to modulating the tumor microenvironment and enhancing immunotherapeutic efficacy (122). However, these approaches remain purely theoretical and require rigorous experimental validation and comprehensive clinical trials to establish their efficacy and safety in cervical cancer treatment.
Immunocellular therapies, including CAR-T cell and tumor-infiltrating lymphocyte (TIL) approaches, offer innovative strategies for directly activating and engineering patient-derived immune cells to enhance antitumor capabilities. TIL therapy, involving extraction, ex vivo expansion, and reinfusion of tumor-infiltrating immune cells, has demonstrated preliminary success in CC treatment. HPV-targeted adoptive T cell therapies have garnered particular attention. One study involving nine patients showed that two cases complete responses at 15 and 22 months post-treatment (123).
CAR-T cell therapy represents a sophisticated genetic engineering approach that introduces chimeric antigen receptors into patient T cells, enabling precise tumor cell recognition and elimination. Given the high expression and stability of HPV E6 and E7 proteins in CC, these antigens have emerged as critical CAR-T therapeutic targets. Preclinical mouse models demonstrated that HPV E7-targeted CAR-T cells successfully induced CC regression, establishing fundamental tumor degradation mechanisms (124). A phase I/II clinical trial targeting HPV-16-positive patients demonstrated the safety and preliminary efficacy of E7-targeted CAR-T cell therapy, with partial tumor remission observed in select patients (125).
Recognizing the unique advantages of CD8+Trm cells—including their tumor antigen-specific killing potential and capacity for sustained tissue residency—targeted immunotherapies focusing on tissue-resident immune cells present extraordinary potential for advanced and metastatic CC treatment. This innovative therapeutic strategy transcends traditional T cell engineering, emphasizing the distinctive biological characteristics of TRICs, such as their persistent presence and specific recognition capabilities within the tumor microenvironment. While targeting TRICs offers a promising new direction in CC immunotherapy, significant challenges remain. The complex acquisition and expansion of tissue-resident immune cells, high CAR-T cell production costs, incomplete understanding of molecular mechanisms, and the intricate process of translating laboratory discoveries into clinical applications necessitate continued rigorous research and interdisciplinary collaboration.
11 Discussion
In this review, we have provided a comprehensive overview of tissue-resident immune cells (TRICs) in cervical cancer (CC), highlighting their potential characteristic markers and functional roles in tumor immunity (Figure 2). While significant progress has been made in understanding these tissue-resident populations, several critical challenges remain to be addressed. First, the prognostic value of tissue-resident immune cells and their correlation with immunotherapy outcomes warrant further investigation through large-scale clinical studies. Understanding how these immune cells influence patient responses to therapies will be essential for developing personalized treatment strategies. Second, the molecular mechanisms governing the differentiation and maintenance of tissue-resident immune cells within the CC microenvironment remain largely undefined. Of particular importance is the identification of key factors that regulate their tissue-residency programs. Future research should focus on developing strategies to enhance the tissue-resident properties of tumor-infiltrating immune cells and increase their density within CC tissues. These advances will be crucial for designing novel therapeutic approaches targeting tissue-resident immune cells.
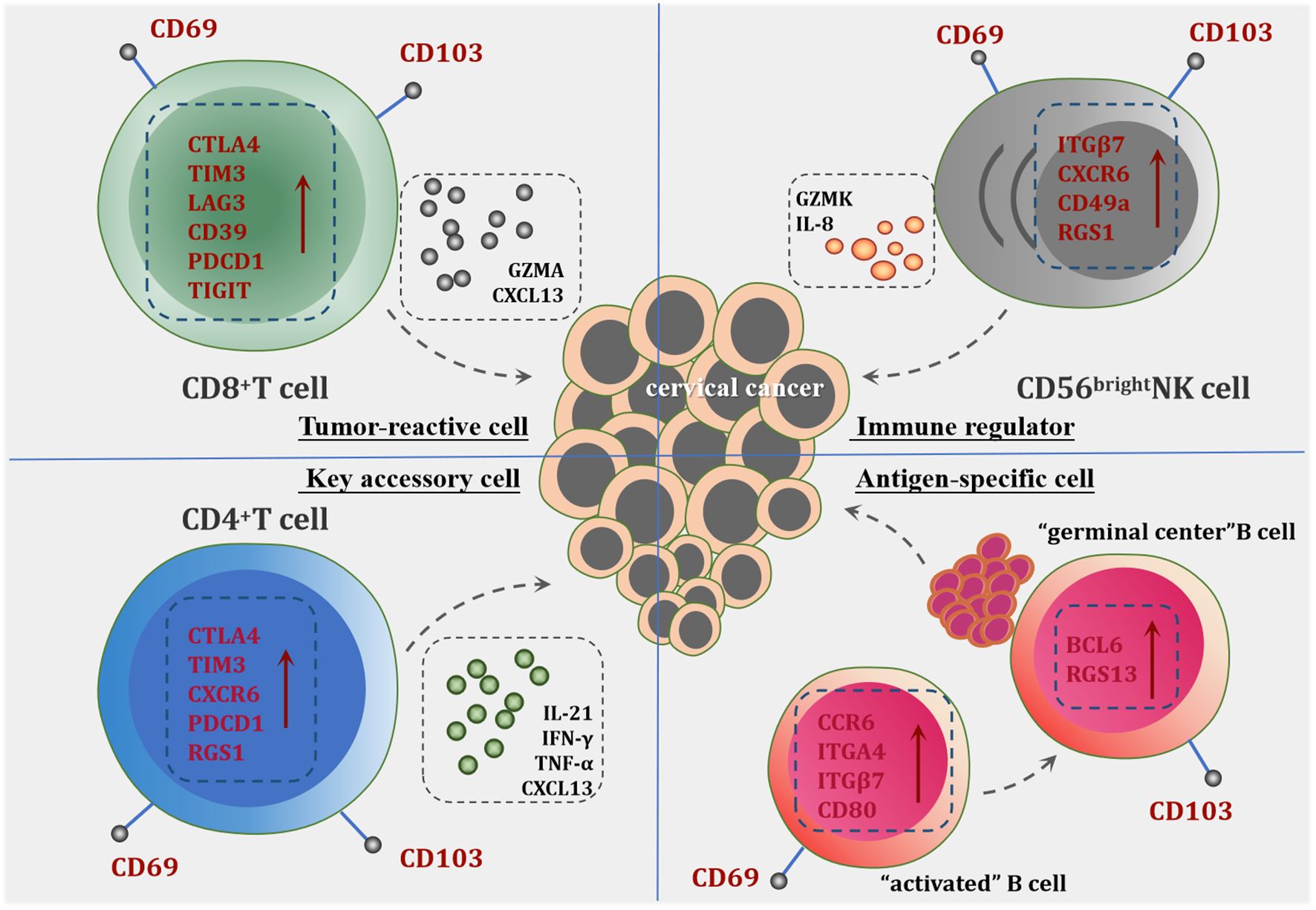
Figure 2. The potential role of tissue-resident immune cells in CC. CD8+Trm cells are characterized by high expression of inhibitory receptors and activation markers, including CD39, LAG3, PD-1 (PDCD1), TIM-3, and TIGIT, indicating their tumor antigen specificity and activation status. They demonstrate significant local expansion within tumor tissues, contributing directly to anti-tumor immunity through cytotoxic activity against tumor cells. CD4+Trm cells play a pivotal role in supporting CD8+Trm cells by the release of cytokines such as IFN-γ, TNF-α and chemokine CXCL13, which facilitate the recruitment and activation of other immune cells. Tissue-resident NK Cells exhibit robust cytokine production capabilities, participating in tumor immune regulation. The significant upregulation of GZMK distinguishes them from conventional NK cells, underscoring a unique functional profile that may contribute to enhanced anti-tumor responses. Tissue-resident B Cells are categorized into CD69+B cells and CD103+B cells. The presence of the CD69+B cell subset indicates the acquisition of local residency and memory capabilities within tumor tissues. The differentiation tendency of CD69+B cells toward CD103+B cells suggests heightened specificity for tumor antigens and a substantial role in TLS formation, thereby amplifying the humoral immune response against the tumor.
Moreover, CD8+Trm cells represent a core population in the immunobiology of CC, linking HPV-specific adaptive immunity to clinical outcomes. Their unique tissue-residency characteristics, functional adaptability, and therapeutic operability position them as central players in natural tumor control and immunotherapy-mediated responses. A deeper understanding of the heterogeneity of Trm subsets, their tissue localization patterns, and the cellular interaction networks will provide new directions for developing targeted immunotherapeutic strategies.
To further expand the discussion on future research directions, we propose several key areas for investigation. There is a pressing need to identify biomarkers that can predict Trm-mediated immune responses, as these biomarkers could facilitate more precise patient stratification for tailored therapies, enhancing the effectiveness of immunotherapy in cervical cancer. Future studies should also explore the interactions between different immune cell subsets within the tumor microenvironment, as understanding how Trm cells communicate with other immune cells, such as dendritic cells, macrophages, and other T cell populations, will be crucial for elucidating the complex immune landscape in cervical cancer. Investigating combination therapies that integrate checkpoint inhibitors with strategies aimed at enhancing Trm cell function could yield promising results; for instance, exploring how metabolic interventions can restore the effector functions of Trm cells in advanced cervical cancer may provide new therapeutic avenues. Additionally, research should focus on optimizing strategies to exploit the immunological window following chemotherapy and radiotherapy, as understanding the timing and conditions under which Trm cells can be effectively mobilized or expanded will be critical for improving patient outcomes. Lastly, there is a need to develop vaccines or cell therapies that can specifically induce and expand HPV-specific Trm cells, as such approaches could enhance the immune response against cervical cancer and improve long-term patient survival.
By addressing these areas, future research can pave the way for comprehensive immunotherapy strategies that leverage the unique properties of tissue-resident immune cells in cervical cancer. Ultimately, a deeper understanding of these aspects may lead to more effective immunotherapy strategies and improved clinical outcomes for CC patients.
Author contributions
XDL: Data curation, Visualization, Writing – original draft. JD: Supervision, Writing – review & editing. XPL: Supervision, Writing – review & editing. YZ: Supervision, Writing – review & editing. TB: Supervision, Writing – review & editing. JC: Conceptualization, Writing – review & editing. JW: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Natural Science Foundation of Hunan Province of China (no. 2024JJ6729).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lin S, Gao K, Gu S, You L, Qian S, Tang M, et al. Worldwide trends in cervical cancer incidence and mortality, with predictions for the next 15 years. Cancer. (2021) 127:4030–39. doi: 10.1002/cncr.33795
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2020) 70:313. doi: 10.3322/caac.21609
3. Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct Target Ther. (2020) 5:166. doi: 10.1038/s41392-020-00280-x
4. Gray JI, Farber DL. Tissue-resident immune cells in humans. Annu Rev Immunol. (2022) 40:195–220. doi: 10.1146/annurev-immunol-093019-112809
5. Chou C, Li MO. Tissue-resident lymphocytes across innate and adaptive lineages. Front Immunol. (2018) 9:2104. doi: 10.3389/fimmu.2018.02104
6. Milner JJ, Toma C, He Z, Kurd NS, Nguyen QP, McDonald B, et al. Heterogenous populations of tissue-resident CD8(+) T cells are generated in response to infection and Malignancy. Immunity. (2020) 52:808–24. doi: 10.1016/j.immuni.2020.04.007
7. Han JW, Yoon SK. Tissue-resident lymphocytes: implications in immunotherapy for hepatocellular carcinoma. Int J Mol Sci. (2020) 22(1):232. doi: 10.3390/ijms22010232
8. Nguyen QP, Takehara KK, Deng TZ, O’Shea S, Heeg M, Omilusik KD, et al. Transcriptional programming of CD4(+) T(RM) differentiation in viral infection balances effector- and memory-associated gene expression. Sci Immunol. (2023) 8:eabq7486. doi: 10.1126/sciimmunol.abq7486
9. Allie SR, Bradley JE, Mudunuru U, Schultz MD, Graf BA, Lund FE, et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat Immunol. (2019) 20:97–108. doi: 10.1038/s41590-018-0260-6
10. Chen C, Laidlaw BJ. Development and function of tissue-resident memory B cells. Adv Immunol. (2022) 155:1–38. doi: 10.1016/bs.ai.2022.08.001
11. Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, et al. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest. (2013) 123:1444–56. doi: 10.1172/JCI66381
12. Sojka DK, Plougastel-Douglas B, Yang L, Pak-Wittel MA, Artyomov MN, Ivanova Y, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. Elife. (2014) 3:e1659. doi: 10.7554/eLife.01659
13. Peng H, Tian Z. Diversity of tissue-resident NK cells. Semin Immunol. (2017) 31:3–10. doi: 10.1016/j.smim.2017.07.006
14. Xu W, Bergsbaken T, Edelblum KL. The multifunctional nature of CD103 (alphaEbeta7 integrin) signaling in tissue-resident lymphocytes. Am J Physiol Cell Physiol. (2022) 323:C1161–67. doi: 10.1152/ajpcell.00338.2022
15. Cheuk S, Schlums H, Gallais SI, Martini E, Chiang SC, Marquardt N, et al. CD49a expression defines tissue-resident CD8(+) T cells poised for cytotoxic function in human skin. Immunity. (2017) 46:287–300. doi: 10.1016/j.immuni.2017.01.009
16. Zajac AJ, Harrington LE. Tissue-resident T cells lose their S1P1 exit visas. Cell Mol Immunol. (2014) 11:221–23. doi: 10.1038/cmi.2014.7
17. Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol. (2013) 14:1285–93. doi: 10.1038/ni.2745
18. Behr FM, Chuwonpad A, Stark R, van Gisbergen K. Armed and ready: transcriptional regulation of tissue-resident memory CD8 T cells. Front Immunol. (2018) 9:1770. doi: 10.3389/fimmu.2018.01770
19. Topham DJ, Reilly EC. Tissue-resident memory CD8(+) T cells: from phenotype to function. Front Immunol. (2018) 9:515. doi: 10.3389/fimmu.2018.00515
20. McNamara HA, Cai Y, Wagle MV, Sontani Y, Roots CM, Miosge LA, et al. Up-regulation of LFA-1 allows liver-resident memory T cells to patrol and remain in the hepatic sinusoids. Sci Immunol. (2017) 2(9):eaaj1996. doi: 10.1126/sciimmunol.aaj1996
21. Mrass P, Kinjyo I, Ng LG, Reiner SL, Pure E, Weninger W. CD44 mediates successful interstitial navigation by killer T cells and enables efficient antitumor immunity. Immunity. (2008) 29:971–85. doi: 10.1016/j.immuni.2008.10.015
22. Teijaro JR, Turner D, Pham Q, Wherry EJ, Lefrancois L, Farber DL. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J Immunol. (2011) 187:5510–14. doi: 10.4049/jimmunol.1102243
23. Wein AN, McMaster SR, Takamura S, Dunbar PR, Cartwright EK, Hayward SL, et al. CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J Exp Med. (2019) 216:2748–62. doi: 10.1084/jem.20181308
24. Butcher MJ, Wu CI, Waseem T, Galkina EV. CXCR6 regulates the recruitment of pro-inflammatory IL-17A-producing T cells into atherosclerotic aortas. Int Immunol. (2016) 28:255–61. doi: 10.1093/intimm/dxv068
25. Moschovakis GL, Forster R. Multifaceted activities of CCR7 regulate T-cell homeostasis in health and disease. Eur J Immunol. (2012) 42:1949–55. doi: 10.1002/eji.201242614
26. Jung YW, Kim HG, Perry CJ, Kaech SM. CCR7 expression alters memory CD8 T-cell homeostasis by regulating occupancy in IL-7- and IL-15-dependent niches. Proc Natl Acad Sci U.S.A. (2016) 113:8278–83. doi: 10.1073/pnas.1602899113
27. Woodward DA, Roozen HN, Dufort MJ, DeBerg HA, Delaney MA, Mair F, et al. The human tissue-resident CCR5(+) T cell compartment maintains protective and functional properties during inflammation. Sci Transl Med. (2019) 11(521):eaaw8718. doi: 10.1126/scitranslmed.aaw8718
28. Meitei HT, Jadhav N, Lal G. CCR6-CCL20 axis as a therapeutic target for autoimmune diseases. Autoimmun Rev. (2021) 20:102846. doi: 10.1016/j.autrev.2021.102846
29. Fu H, Jangani M, Parmar A, Wang G, Coe D, Spear S, et al. A subset of CCL25-induced gut-homing T cells affects intestinal immunity to infection and cancer. Front Immunol. (2019) 10:271. doi: 10.3389/fimmu.2019.00271
30. Moed H, Boorsma DM, Tensen CP, Flier J, Jonker MJ, Stoof TJ, et al. Increased CCL27-CCR10 expression in allergic contact dermatitis: implications for local skin memory. J Pathol. (2004) 204:39–46. doi: 10.1002/path.1619
31. Parga-Vidal L, Taggenbrock R, Beumer-Chuwonpad A, Aglmous H, Kragten N, Behr FM, et al. Hobit and Blimp-1 regulate T(RM) abundance after LCMV infection by suppressing tissue exit pathways of T(RM) precursors. Eur J Immunol. (2022) 52:1095–111. doi: 10.1002/eji.202149665
32. Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. (2016) 352:459–63. doi: 10.1126/science.aad2035
33. Kragten N, Behr FM, Vieira BF, Remmerswaal E, Wesselink TH, Oja AE, et al. Blimp-1 induces and Hobit maintains the cytotoxic mediator granzyme B in CD8 T cells. Eur J Immunol. (2018) 48:1644–62. doi: 10.1002/eji.201847771
34. Behr FM, Kragten N, Wesselink TH, Nota B, van Lier R, Amsen D, et al. Blimp-1 rather than hobit drives the formation of tissue-resident memory CD8(+) T cells in the lungs. Front Immunol. (2019) 10:400. doi: 10.3389/fimmu.2019.00400
35. Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, et al. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4-/CD8+ T cells. J Immunol. (2005) 175:1694–705. doi: 10.4049/jimmunol.175.3.1694
36. Tang J, Sheng J, Zhang Q, Ji Y, Wang X, Zhang J, et al. Runx3-overexpression cooperates with ex vivo AKT inhibition to generate receptor-engineered T cells with better persistence, tumor-residency, and antitumor ability. J Immunother Cancer. (2023) 11(2):e006119. doi: 10.1136/jitc-2022-006119
37. Longworth AJ, Mallya S, Lev T, Pein M, Adam I, Lincy A, et al. Multiomic and spatial immunophenotyping reveals a prominent Runx3 +resident T cell population in the healthy human breast. J Immunol. (2023) 210:12–63. doi: 10.4049/jimmunol.210.Supp.63.12
38. Park SL, Gebhardt T, Mackay LK. Tissue-resident memory T cells in cancer immunosurveillance. Trends Immunol. (2019) 40:735–47. doi: 10.1016/j.it.2019.06.002
39. Lyu Y, Zhou Y, Shen J. An overview of tissue-resident memory T cells in the intestine: from physiological functions to pathological mechanisms. Front Immunol. (2022) 13:912393. doi: 10.3389/fimmu.2022.912393
40. Kalia V, Yuzefpolskiy Y, Vegaraju A, Xiao H, Baumann F, Jatav S, et al. Metabolic regulation by PD-1 signaling promotes long-lived quiescent CD8 T cell memory in mice. Sci Transl Med. (2021) 13:eaba6006. doi: 10.1126/scitranslmed.aba6006
41. Surojit Sarkar YYAV. PD-1 signals are critical for homeostatic maintenance of memory CD8 T cells. J Immunol. (2020) 204(1_Supplement):81.5. doi: 10.4049/jimmunol.204.Supp.81.5
42. Zhang Y, Chen S, Tang X, Peng Y, Jiang T, Zhang X, et al. The role of KLRG1: a novel biomarker and new therapeutic target. Cell Commun Signal. (2024) 22:337. doi: 10.1186/s12964-024-01714-7
43. Herndler-Brandstetter D, Ishigame H, Shinnakasu R, Plajer V, Stecher C, Zhao J, et al. KLRG1(+) effector CD8(+) T cells lose KLRG1, differentiate into all memory T cell lineages, and convey enhanced protective immunity. Immunity. (2018) 48:716–29. doi: 10.1016/j.immuni.2018.03.015
44. Jabri B, Abadie V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat Rev Immunol. (2015) 15:771–83. doi: 10.1038/nri3919
45. Crowl JT, Heeg M, Ferry A, Milner JJ, Omilusik KD, Toma C, et al. Tissue-resident memory CD8(+) T cells possess unique transcriptional, epigenetic and functional adaptations to different tissue environments. Nat Immunol. (2022) 23:1121–31. doi: 10.1038/s41590-022-01229-8
46. Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. (2017) 20:2921–34. doi: 10.1016/j.celrep.2017.08.078
47. Notarbartolo S, Abrignani S. Human T lymphocytes at tumor sites. Semin Immunopathol. (2022) 44:883–901. doi: 10.1007/s00281-022-00970-4
48. Abdeljaoued S, Arfa S, Kroemer M, Ben KM, Vienot A, Heyd B, et al. Tissue-resident memory T cells in gastrointestinal cancer immunology and immunotherapy: ready for prime time? J Immunother Cancer. (2022) 10(4):e003472. doi: 10.1136/jitc-2021-003472
49. Liu Z, Wang H, Li Z, Dress RJ, Zhu Y, Zhang S, et al. Dendritic cell type 3 arises from Ly6C(+) monocyte-dendritic cell progenitors. Immunity. (2023) 56:1761–77. doi: 10.1016/j.immuni.2023.07.001
50. Leader AM, Grout JA, Maier BB, Nabet BY, Park MD, Tabachnikova A, et al. Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell. (2021) 39:1594–609. doi: 10.1016/j.ccell.2021.10.009
51. Wu TC, Xu K, Banchereau R, Marches F, Yu CI, Martinek J, et al. Reprogramming tumor-infiltrating dendritic cells for CD103+ CD8+ mucosal T-cell differentiation and breast cancer rejection. Cancer Immunol Res. (2014) 2:487–500. doi: 10.1158/2326-6066.CIR-13-0217
52. Bourdely P, Anselmi G, Vaivode K, Ramos RN, Missolo-Koussou Y, Hidalgo S, et al. Transcriptional and functional analysis of CD1c(+) human dendritic cells identifies a CD163(+) subset priming CD8(+)CD103(+) T cells. Immunity. (2020) 53:335–52. doi: 10.1016/j.immuni.2020.06.002
53. Dijkgraaf FE, Kok L, Schumacher T. Formation of tissue-resident CD8(+) T-cell memory. Cold Spring Harb Perspect Biol. (2021) 13(8):a038117. doi: 10.1101/cshperspect.a038117
54. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol. (2013) 14:1294–301. doi: 10.1038/ni.2744
55. Wu B, Zhang G, Guo Z, Wang G, Xu X, Li JL, et al. The SKI proto-oncogene restrains the resident CD103(+)CD8(+) T cell response in viral clearance. Cell Mol Immunol. (2021) 18:2410–21. doi: 10.1038/s41423-020-0495-7
56. Park SL, Mackay LK. Decoding tissue-residency: programming and potential of frontline memory T cells. Cold Spring Harb Perspect Biol. (2021) 13(10):a037960. doi: 10.1101/cshperspect.a037960
57. Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, et al. T-box transcription factors combine with the cytokines TGF-beta and IL-15 to control tissue-resident memory T cell fate. Immunity. (2015) 43:1101–11. doi: 10.1016/j.immuni.2015.11.008
58. Mami-Chouaib F, Blanc C, Corgnac S, Hans S, Malenica I, Granier C, et al. Resident memory T cells, critical components in tumor immunology. J Immunother Cancer. (2018) 6:87. doi: 10.1186/s40425-018-0399-6
59. Tieu R, Zeng Q, Zhao D, Zhang G, Feizi N, Manandhar P, et al. Tissue-resident memory T cell maintenance during antigen persistence requires both cognate antigen and interleukin-15. Sci Immunol. (2023) 8:eadd8454. doi: 10.1126/sciimmunol.add8454
60. Lopez-Cabrera M, Munoz E, Blazquez MV, Ursa MA, Santis AG, Sanchez-Madrid F. Transcriptional regulation of the gene encoding the human C-type lectin leukocyte receptor AIM/CD69 and functional characterization of its tumor necrosis factor-alpha-responsive elements. J Biol Chem. (1995) 270:21545–51. doi: 10.1074/jbc.270.37.21545
61. Carrette F, Surh CD. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin Immunol. (2012) 24:209–17. doi: 10.1016/j.smim.2012.04.010
62. Vander HM, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. (2009) 324:1029–33. doi: 10.1126/science.1160809
63. Whitburn J, Edwards CM. Metabolism in the tumor-bone microenvironment. Curr Osteoporos Rep. (2021) 19:494–99. doi: 10.1007/s11914-021-00695-7
64. Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumors. Nature. (2017) 552:253–57. doi: 10.1038/nature24993
65. Voss K, Larsen SE, Snow AL. Metabolic reprogramming and apoptosis sensitivity: Defining the contours of a T cell response. Cancer Lett. (2017) 408:190–96. doi: 10.1016/j.canlet.2017.08.033
66. Ogando J, Saez ME, Santos J, Nuevo-Tapioles C, Gut M, Esteve-Codina A, et al. PD-1 signaling affects cristae morphology and leads to mitochondrial dysfunction in human CD8(+) T lymphocytes. J Immunother Cancer. (2019) 7:151. doi: 10.1186/s40425-019-0628-7
67. Li G, Yang Q, Zhu Y, Wang HR, Chen X, Zhang X, et al. T-bet and eomes regulate the balance between the effector/central memory T cells versus memory stem like T cells. PloS One. (2013) 8:e67401. doi: 10.1371/journal.pone.0067401
68. Farsakoglu Y, McDonald B, Kaech SM. Motility matters: how CD8(+) T-cell trafficking influences effector and memory cell differentiation. Cold Spring Harb Perspect Biol. (2021) 13(8):a038075. doi: 10.1101/cshperspect.a038075
69. Herbel C, Patsoukis N, Bardhan K, Seth P, Weaver JD, Boussiotis VA. Clinical significance of T cell metabolic reprogramming in cancer. Clin Transl Med. (2016) 5:29. doi: 10.1186/s40169-016-0110-9
70. Hasan F, Chiu Y, Shaw RM, Wang J, Yee C. Hypoxia acts as an environmental cue for the human tissue-resident memory T cell differentiation program. JCI Insight. (2021) 6(10):e138970. doi: 10.1172/jci.insight.138970
71. Liang M, Wang X, Cai D, Guan W, Shen X. Tissue-resident memory T cells in gastrointestinal tumors: turning immune desert into immune oasis. Front Immunol. (2023) 14:1119383. doi: 10.3389/fimmu.2023.1119383
72. Lyu M, Shen Y, Beharee N, Lu J, Deng F, Wang J. The combined use of chemotherapy and radiotherapy with PD-1 inhibitor, pembrolizumab, in advanced cervical cancer: A case report. Onco Targets Ther. (2020) 13:4465–71. doi: 10.2147/OTT.S245190
73. Cheng M, Wang H, Zhao Y, Li G. Efficacy and prognostic factors for response to PD-1 inhibitors in advanced cervical carcinoma: A retrospective study. Drug Des Devel Ther. (2022) 16:887–97. doi: 10.2147/DDDT.S358302
74. Ziegler AE, Fittje P, Muller LM, Ahrenstorf AE, Hagemann K, Hagen SH, et al. The co-inhibitory receptor TIGIT regulates NK cell function and is upregulated in human intrahepatic CD56(bright) NK cells. Front Immunol. (2023) 14:1117320. doi: 10.3389/fimmu.2023.1117320
75. Wang F, Yue S, Huang Q, Lei T, Li X, Wang C, et al. Cellular heterogeneity and key subsets of tissue-resident memory T cells in cervical cancer. NPJ Precis Oncol. (2024) 8:145. doi: 10.1038/s41698-024-00637-3
76. Yost S, Hoekstra A. Cervical cancer in women over 65: An analysis of screening. Gynecol Oncol Rep. (2018) 25:48–51. doi: 10.1016/j.gore.2018.05.010
77. Yang M, Du J, Lu H, Xiang F, Mei H, Xiao H. Global trends and age-specific incidence and mortality of cervical cancer from 1990 to 2019: an international comparative study based on the Global Burden of Disease. BMJ Open. (2022) 12:e55470. doi: 10.1136/bmjopen-2021-055470
78. Komdeur FL, Prins TM, van de Wall S, Plat A, Wisman G, Hollema H, et al. CD103+ tumor-infiltrating lymphocytes are tumor-reactive intraepithelial CD8+ T cells associated with prognostic benefit and therapy response in cervical cancer. Oncoimmunology. (2017) 6:e1338230. doi: 10.1080/2162402X.2017.1338230
79. Park SL, Buzzai A, Rautela J, Hor JL, Hochheiser K, Effern M, et al. Tissue-resident memory CD8(+) T cells promote melanoma-immune equilibrium in skin. Nature. (2019) 565:366–71. doi: 10.1038/s41586-018-0812-9
80. Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med. (2018) 24:986–93. doi: 10.1038/s41591-018-0078-7
81. Ganesan AP, Clarke J, Wood O, Garrido-Martin EM, Chee SJ, Mellows T, et al. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol. (2017) 18:940–50. doi: 10.1038/ni.3775
82. Koh J, Kim S, Kim MY, Go H, Jeon YK, Chung DH. Prognostic implications of intratumoral CD103+ tumor-infiltrating lymphocytes in pulmonary squamous cell carcinoma. Oncotarget. (2017) 8:13762–69. doi: 10.18632/oncotarget.14632
83. Edwards J, Wilmott JS, Madore J, Gide TN, Quek C, Tasker A, et al. CD103(+) tumor-resident CD8(+) T cells are associated with improved survival in immunotherapy-naive melanoma patients and expand significantly during anti-PD-1 treatment. Clin Cancer Res. (2018) 24:3036–45. doi: 10.1158/1078-0432.CCR-17-2257
84. Hu X, Li YQ, Li QG, Ma YL, Peng JJ, Cai SJ. ITGAE defines CD8+ Tumor-infiltrating lymphocytes predicting a better prognostic survival in colorectal cancer. Ebiomedicine. (2018) 35:178–88. doi: 10.1016/j.ebiom.2018.08.003
85. Maskey N, Thapa N, Maharjan M, Shrestha G, Maharjan N, Cai H, et al. Infiltrating CD4 and CD8 lymphocytes in HPV infected uterine cervical milieu. Cancer Manag Res. (2019) 11:7647–55. doi: 10.2147/CMAR.S217264
86. Fan J, Lu F, Qin T, Peng W, Zhuang X, Li Y, et al. Multiomic analysis of cervical squamous cell carcinoma identifies cellular ecosystems with biological and clinical relevance. Nat Genet. (2023) 55:2175–88. doi: 10.1038/s41588-023-01570-0
87. Liu X, Ni G, Zhang P, Li H, Li J, Cavallazzi SB, et al. Single-nucleus RNA sequencing and deep tissue proteomics reveal distinct tumor microenvironment in stage-I and II cervical cancer. J Exp Clin Cancer Res. (2023) 42:28. doi: 10.1186/s13046-023-02598-0
88. Liu C, Li X, Huang Q, Zhang M, Lei T, Wang F, et al. Single-cell RNA-sequencing reveals radiochemotherapy-induced innate immune activation and MHC-II upregulation in cervical cancer. Signal Transduct Target Ther. (2023) 8:44. doi: 10.1038/s41392-022-01264-9
89. Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat Commun. (2019) 10:4706. doi: 10.1038/s41467-019-12464-3
90. Zhang H, Zhu Z, Modrak S, Little A. Tissue-resident memory CD4(+) T cells play a dominant role in the initiation of antitumor immunity. J Immunol. (2022) 208:2837–46. doi: 10.4049/jimmunol.2100852
91. Oja AE, Piet B, van der Zwan D, Blaauwgeers H, Mensink M, de Kivit S, et al. Functional heterogeneity of CD4(+) tumor-infiltrating lymphocytes with a resident memory phenotype in NSCLC. Front Immunol. (2018) 9:2654. doi: 10.3389/fimmu.2018.02654
92. Cao G, Yue J, Ruan Y, Han Y, Zhi Y, Lu J, et al. Single-cell dissection of cervical cancer reveals key subsets of the tumor immune microenvironment. EMBO J. (2023) 42:e110757. doi: 10.15252/embj.2022110757
93. Veatch JR, Lee SM, Shasha C, Singhi N, Szeto JL, Moshiri AS, et al. Neoantigen-specific CD4(+) T cells in human melanoma have diverse differentiation states and correlate with CD8(+) T cell, macrophage, and B cell function. Cancer Cell. (2022) 40:393–409. doi: 10.1016/j.ccell.2022.03.006
94. Liu AH, Jena PK, Wysocki LJ. Tracing the development of single memory-lineage B cells in a highly defined immune response. J Exp Med. (1996) 183:2053–63. doi: 10.1084/jem.183.5.2053
95. Onodera T, Takahashi Y, Yokoi Y, Ato M, Kodama Y, Hachimura S, et al. Memory B cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc Natl Acad Sci U.S.A. (2012) 109:2485–90. doi: 10.1073/pnas.1115369109
96. Adachi Y, Onodera T, Yamada Y, Daio R, Tsuiji M, Inoue T, et al. Distinct germinal center selection at local sites shapes memory B cell response to viral escape. J Exp Med. (2015) 212:1709–23. doi: 10.1084/jem.20142284
97. Weisel NM, Weisel FJ, Farber DL, Borghesi LA, Shen Y, Ma W, et al. Comprehensive analyses of B-cell compartments across the human body reveal novel subsets and a gut-resident memory phenotype. Blood. (2020) 136:2774–85. doi: 10.1182/blood.2019002782
98. Mathew NR, Jayanthan JK, Smirnov IV, Robinson JL, Axelsson H, Nakka SS, et al. Single-cell BCR and transcriptome analysis after influenza infection reveals spatiotemporal dynamics of antigen-specific B cells. Cell Rep. (2021) 35:109286. doi: 10.1016/j.celrep.2021.109286
99. Gregoire C, Spinelli L, Villazala-Merino S, Gil L, Holgado MP, Moussa M, et al. Viral infection engenders bona fide and bystander subsets of lung-resident memory B cells through a permissive mechanism. Immunity. (2022) 55:1216–33. doi: 10.1016/j.immuni.2022.06.002
100. Tan HX, Juno JA, Esterbauer R, Kelly HG, Wragg KM, Konstandopoulos P, et al. Lung-resident memory B cells established after pulmonary influenza infection display distinct transcriptional and phenotypic profiles. Sci Immunol. (2022) 7:eabf5314. doi: 10.1126/sciimmunol.abf5314
101. Hu Q, Hong Y, Qi P, Lu G, Mai X, Xu S, et al. Atlas of breast cancer infiltrated B-lymphocytes revealed by paired single-cell RNA-sequencing and antigen receptor profiling. Nat Commun. (2021) 12:2186. doi: 10.1038/s41467-021-22300-2
102. Camponeschi A, Gerasimcik N, Wang Y, Fredriksson T, Chen D, Farroni C, et al. Dissecting integrin expression and function on memory B cells in mice and humans in autoimmunity. Front Immunol. (2019) 10:534. doi: 10.3389/fimmu.2019.00534
103. Lehmann-Horn K, Sagan SA, Winger RC, Spencer CM, Bernard CC, Sobel RA, et al. CNS accumulation of regulatory B cells is VLA-4-dependent. Neurol Neuroimmunol Neuroinflamm. (2016) 3:e212. doi: 10.1212/NXI.0000000000000212
104. Ho DW, Tsui YM, Chan LK, Sze KM, Zhang X, Cheu JW, et al. Single-cell RNA sequencing shows the immunosuppressive landscape and tumor heterogeneity of HBV-associated hepatocellular carcinoma. Nat Commun. (2021) 12:3684. doi: 10.1038/s41467-021-24010-1
105. Davis ZB, Felices M, Verneris MR, Miller JS. Natural killer cell adoptive transfer therapy: exploiting the first line of defense against cancer. Cancer J. (2015) 21:486–91. doi: 10.1097/PPO.0000000000000156
106. Wolf NK, Kissiov DU, Raulet DH. Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat Rev Immunol. (2023) 23:90–105. doi: 10.1038/s41577-022-00732-1
107. Kyrysyuk O, Wucherpfennig KW. Designing cancer immunotherapies that engage T cells and NK cells. Annu Rev Immunol. (2023) 41:17–38. doi: 10.1146/annurev-immunol-101921-044122
108. Zhang Y, Yu M, Jing Y, Cheng J, Zhang C, Cheng L, et al. Baseline immunity and impact of chemotherapy on immune microenvironment in cervical cancer. Br J Cancer. (2021) 124:414–24. doi: 10.1038/s41416-020-01123-w
109. Melsen JE, Lugthart G, Lankester AC, Schilham MW. Human circulating and tissue-resident CD56bright natural killer cell populations. Front Immunol. (2016) 7:262. doi: 10.3389/fimmu.2016.00262
110. Montaldo E, Vacca P, Chiossone L, Croxatto D, Loiacono F, Martini S, et al. Unique eomes(+) NK cell subsets are present in uterus and decidua during early pregnancy. Front Immunol. (2015) 6:646. doi: 10.3389/fimmu.2015.00646
111. Wang F, Zhou Y, Fu B, Wu Y, Zhang R, Sun R, et al. Molecular signatures and transcriptional regulatory networks of human immature decidual NK and mature peripheral NK cells. Eur J Immunol. (2014) 44:2771–84. doi: 10.1002/eji.201344183
112. Tang F, Li J, Qi L, Liu D, Bo Y, Qin S, et al. A pan-cancer single-cell panorama of human natural killer cells. Cell. (2023) 186:4235–51. doi: 10.1016/j.cell.2023.07.034
113. Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. (2001) 97:3146–51. doi: 10.1182/blood.v97.10.3146
114. Jacobs R, Hintzen G, Kemper A, Beul K, Kempf S, Behrens G, et al. CD56bright cells differ in their KIR repertoire and cytotoxic features from CD56dim NK cells. Eur J Immunol. (2001) 31:3121–27. doi: 10.1002/1521-4141(2001010)31:10<3121::aid-immu3121>3.0.co;2-4
115. Guo X, Zhang Y, Zheng L, Zheng C, Song J, Zhang Q, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med. (2018) 24:978–85. doi: 10.1038/s41591-018-0045-3
116. Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. (2018) 175:998–1013. doi: 10.1016/j.cell.2018.10.038
117. Sherer MV, Kotha NV, Williamson C, Mayadev J. Advances in immunotherapy for cervical cancer: recent developments and future directions. Int J Gynecol Cancer. (2022) 32:281–87. doi: 10.1136/ijgc-2021-002492
118. Lorusso D, Xiang Y, Hasegawa K, Scambia G, Leiva M, Ramos-Elias P, et al. Pembrolizumab or placebo with chemoradiotherapy followed by pembrolizumab or placebo for newly diagnosed, high-risk, locally advanced cervical cancer (ENGOT-cx11/GOG-3047/KEYNOTE-A18): a randomized, double-blind, phase 3 clinical trial. Lancet. (2024) 403:1341–50. doi: 10.1016/S0140-6736(24)00317-9
119. Petre I, Vernic C, Petre I, Vlad CS, Sipos SI, Bordianu A, et al. Systematic review on the effectiveness and outcomes of nivolumab treatment schemes in advanced and metastatic cervical cancer. Diseases. (2024) 12(4):77. doi: 10.3390/diseases12040077
120. Tseng CH. Metformin use and cervical cancer risk in female patients with type 2 diabetes. Oncotarget. (2016) 7:59548–55. doi: 10.18632/oncotarget.10934
121. Han K, Fyles A, Shek T, Croke J, Dhani N, D’Souza D, et al. A phase II randomized trial of chemoradiation with or without metformin in locally advanced cervical cancer. Clin Cancer Res. (2022) 28:5263–71. doi: 10.1158/1078-0432.CCR-22-1665
122. Zak K, Satora M, Skrabalak I, Tarkowski R, Ostrowska-Lesko M, Bobinski M. The potential influence of residual or recurrent disease on bevacizumab treatment efficacy in ovarian cancer: current evidence and future perspectives. Cancers (Basel). (2024) 16(5):1063. doi: 10.3390/cancers16051063
123. Stevanovic S, Draper LM, Langhan MM, Campbell TE, Kwong ML, Wunderlich JR, et al. Complete regression of metastatic cervical cancer after treatment with human papillomavirus-targeted tumor-infiltrating T cells. J Clin Oncol. (2015) 33:1543–50. doi: 10.1200/JCO.2014.58.9093
124. Jin BY, Campbell TE, Draper LM, Stevanovic S, Weissbrich B, Yu Z, et al. Engineered T cells targeting E7 mediate regression of human papillomavirus cancers in a murine model. JCI Insight. (2018) 3(8):e99488. doi: 10.1172/jci.insight.99488
Keywords: cervical cancer, tissue-resident immune cells, local tumor microenvironment, Trm cell differentiation, tissue-resident memory T cells
Citation: Li X, Deng J, Liu X, Zhou Y, Bi T, Chen J and Wang J (2025) Tissue-resident immune cells in cervical cancer: emerging roles and therapeutic implications. Front. Immunol. 16:1541950. doi: 10.3389/fimmu.2025.1541950
Received: 09 December 2024; Accepted: 02 April 2025;
Published: 22 April 2025.
Edited by:
Wen-Hao Xu, Fudan University, ChinaReviewed by:
Jayasri Das Sarma, Indian Institute of Science Education and Research Kolkata, IndiaXiaosong Liu, First People’s Hospital of Foshan, China
Copyright © 2025 Li, Deng, Liu, Zhou, Bi, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jingjing Chen, amVuc2VuYTEwMzBAMTYzLmNvbQ==; Jinjin Wang, amluamlud2FuZzQxMjAwMEBzaW5hLmNvbQ==
 Xidie Li1
Xidie Li1 Jinjin Wang
Jinjin Wang