- 1Fish Health Research Section, Norwegian Veterinary Institute, Ås, Norway
- 2Department of Medical Genetics, Oslo University Hospital, Oslo, Norway
Introduction: In mammals, the ubiquitin–proteasomal pathway plays a key role in the host antiviral response by targeting viral genes for degradation. Here, E1-activating enzymes, E2-conjugating enzymes, and E3 ligases attach the ubiquitin chain to molecules to be functionally modified or destined for degradation. One specialized version of this pathway is performed by ISG15, a ubiquitin-like protein modifier that works through a process called ISGylation. In mammals, ISGylation involves specialized E1–E3 molecules and specialized mammalian ISG15 “deubiquitinases” also exist. Targeting host and viral proteins, ISG15 can inhibit the release of viral particles or hinder viral replication, thereby exerting strong antiviral effects. In Atlantic salmon, endothelial cells from the heart is a main target for infectious salmon anemia virus (ISAV).
Results: Here, we established a new cell line from Atlantic salmon heart tissue denoted ASH2-2, which has endothelial-like characteristics and is permissive for infection with ISAV and other salmonid viruses. We used this cell line as a model to compare the effect of recombinant interferon gamma (rIFNg) and ISAV on genes potentially involved in the ubiquitin–proteasome pathway. ASH2–2 cells have a response profile matching endothelial cells and respond quickly to ISAV infection with upregulation of viral sensors such as DHX58, MDA5, and MX transcripts. Two ISG15 genes are strongly upregulated 48 h post-infection (p.i.) while other ubiquitin genes were unaffected. Related to the mammalian E3 ligases known to be ISGylated, phylogenetic analysis identified two additional teleost-specific HERC8 and HERC9 clusters in addition to the clade previously defined as HERC7. Duplicate genes for Atlantic salmon HERC7 and HERC9 are both more upregulated by virus and rIFNg at 24 h p.i. as opposed to HERC8 genes. Early regulation of the ISGylation process is also indicated by a strong upregulation of one USP18 duplicate already at 4 h p.i.
Conclusion: In conclusion, we expand the list of teleost genes potentially involved in the ubiquitin–proteasome pathway in cells from a main target organ. Our results highlight the need for functional studies to clarify the roles of these candidates.
1 Introduction
Protection against viral infection relies on the host being able to sense infection and induce a protective immune response. In turn, pathogens have evolved multiple strategies to counteract detection by their hosts promoting the evolution of host immune genes. One such example is the ubiquitin(-like) proteasome system, which regulates the innate sensing pathways initiated by pattern recognition receptors (PRRs), ultimately coordinating an effective antiviral immune response.
PRRs detect conserved viral pathogen-associated molecular patterns or so-called PAMPs (1). Zinc finger NFX1-type containing 1 (ZNFX1), stimulator of interferon response cGAMP interactor 1 (STING1), retinoic acid-inducible gene I (RIG-I, alias DDX58), and melanoma differentiation-associated gene 5 (MDA5, alias IFIH1) all express PAMPs that interact with mitochondrial antiviral signaling protein (MAVS), with subsequent induction of interferon-stimulated genes (ISGs) (2–4).
Posttranslational modification of RIG-1 and downstream signaling proteins by different types of ubiquitination regulates their activity. The PAMP LGP2 (Laboratory of Genetics and Physiology 2) does not bind to MAVS but regulates the activities of RIG-I likely through various mechanisms including the prevention of RIG-I binding to MAVS. In peripheral tissues, sensing of viral infection results in expression of interferons and pro-inflammatory cytokines that attract immune cells to the infected region. They also induce expression of a wide array of ISGs that inhibit viral replication and spread (5). One such ISG is the myxovirus-resistance (Mx) gene, an IFN-inducible GTPase. As opposed to humans, Atlantic salmon has nine Mx genes where some respond more to type I IFN while others respond more to type II IFN (6).
Once the virus has entered the cell and starts replicating, the host tries to eliminate viral proteins through the ubiquitin–proteasome pathway [reviewed in (7)]. Ubiquitination is the first step in this protein degradation process. In humans, ubiquitin is encoded by the four genes UBB, UBC, UBA52, and RPS27A where UBB and UBC encode multimeric ubiquitin chains while UBA52 and RPS27A are associated with the ribosomal proteins L40 and S27A, respectively (8). Other ubiquitin-like genes encoding proteins with one or several ubiquitin-like (UbL) domain are NEDD8, SUMO1, and ISG15 (9, 10).
A classical view has been that Ub tagging is required for targeting proteins for 26S proteasomal degradation. Recently, this view has been challenged where multiple authors have shown that alternative Ub-independent proteasome degradation also occurs (11, 12). Additionally, Ub-like domains are shown to bind to 26S proteasomes and activate proteasomal degradation (13, 14). We therefore use Ub/L hereafter to underline that either Ub or Ub-like conjugation can occur.
Initially, a single Ub/L chain is attached to an E1 enzyme, which then transfers this Ub chain to an E2 enzyme and subsequently to an E3 ligation enzyme. The E3 ligation enzyme provides the specificity for further addition of Ub to target proteins prior to proteasomal degradation. In mammals, the ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is attached to various targets in a process called ISGylation (15, 16). ISG15 attachment involves the ubiquitin-activating enzyme E1 (UBE1L), the ubiquitin-binding enzyme E2 (UBCH8), and some E3 ubiquitin ligases such as the ring finger protein (RNF) 213, ARIH1, TRIM25, and HERC5 (16–18). ISGylation not only modifies key molecules in immune signaling pathways but also inhibits viral replication through ISGylation of viral proteins. ISG15 can also exist in a free unconjugated format with various biological functions such as regulating interferon expression or signaling and mediating degradation of RIG-1. In Atlantic salmon, the ISG15 gene responds similarly to both interferon and infection by ISAV (19–21). Atlantic salmon ISG15 has also been shown to bind viral ISAV proteins such as, for instance, segment 8 open reading frame protein 2 (22).
The E3 ubiquitin ligase enzymes provide target specificity and 919 different human E3 enzymes. The various E3 ligases have traditionally been classified into three main families, i.e., RING (really interesting new genes), HECT (homologous to the E6-AP carboxyl terminus), and RBR (ring-between-ring) domain-containing E3 ligases [reviewed in (23)]. RING ligases facilitate the direct transfer of Ub/L chains from the E2 cysteine to the substrate while HECT and RBR ligases transfer the Ub/L chain from the E2 enzyme first to the E3 ligase prior to transferring it to the substrate.
Tripartite motif (TRIM) genes belong to the E3 ligase RING family. Humans harbor 80 TRIM genes further subdivided into nine subgroups depending on their C-terminal domains, subcellular location, and functionality (24). Several human TRIM genes are shown to regulate host antiviral activities against, for instance, influenza virus (25, 26). For instance, TRIM25 has been shown to mediate ubiquitination of RIG-1, leading to type 1 interferon production (27). As a countermeasure, the NS protein of the influenza virus targets TRIM25 to evade RIG-1 recognition (28). In addition to TRIM25 and TRIM39, teleosts have a unique subgroup denoted finTRIMs where many are upregulated upon infection in both zebrafish and rainbow trout (29–32).
Some ring finger protein genes also belong to this RING family of E3 ubiquitin ligases. RNF213 is an interferon upregulated gene that acts as an ISG15 interactor and cellular sensor of ISGylated proteins with broad antimicrobial activity in humans (18, 33). RNF213 is also upregulated by interferon in teleosts (34, 35). Another member of this family (RNF39) activates the ubiquitin–proteasome pathway and thus inhibits RIG-I-like receptor-dependent antiviral immunity (36).
HECT E3 ligase family members are characterized by the presence of a HECT domain and one or more RCC1-like domains. In addition to ubiquitylating proteins for degradation by the 26S proteasome, HECT E3 enzymes regulate the trafficking of many receptors, channels, transporters, and viral proteins [reviewed in (37)]. HERC genes belong to this family with humans having two large genes containing multiple RCC1 domains (HERC1 and HERC2) and four small genes containing single RCC1 domains (HERC3-6) (38). Both HERC5 and HERC6 are upregulated by interferon, where the major ISG15 E3 ligase is human HERC5 and HERC6 in mice (39–41). In humans, HERC5 is shown to attenuate influenza A by catalyzing ISGylation of the viral NS1 protein (42). A teleost-specific HERC gene denoted HERC7 was recently described in carp and zebrafish with four genes in zebrafish (43, 44). Carp HERC7 has an E3 ligase activity for conjugation of both ubiquitin and ISG15 while zebrafish HERC7c only displays the potential to transfer ubiquitin. Zebrafish HERC7c acts as a negative regulator of fish interferon responses by targeting STING, MAVS, and IRF7 for protein degradation.
RBR domain-containing E3 ligases, the third family of E3 ligases, contain three zinc-binding domains termed RING1, in between RING (IBR) and RING2 domains (collectively called the RBR module), and are described as RING/HECT hybrid E3 ligases (45). Humans harbor 14 RBR members including four Ariadne ligases, Parkin, and six RNF ligases [reviewed in (46)]. Several of these RBR ligases affect immunity against viruses. For instance, ARIH1 inhibits influenza A virus replication and facilitates RIG-1-dependent immune signaling (47). RNF144 promotes antiviral responses by modulating STING ubiquitination (33).
Once conjugated to Ub/L chains, some proteins are targeted for degradation by the proteasome. The 26S proteasome consists of a 20S core proteasome and a 19S regulator. In mammals, the 19S regulator contains multiple ubiquitin binding sites, which regulate the proteasome conformation that activates unfolding, deubiquitination, and degradation of various protein substrates [reviewed in (48)]. Three of the 19S subunits function as Ub/L receptors. The subunit Rpn10 functions as the primary ubiquitin chain receptor and the two Rpn13 and Rpn1 subunits can cooperate with Rpn10 to enhance degradation of some proteins (49). Once deubiquitinated and unfolded, the protein is transferred to the 20S core for further degradation.
The 20S core particle has duplicate rings of seven alpha (PSMA) and beta (PSMB) subunits (50). Upon IFN stimulation, the PSMB5–7 subunits are exchanged with the major histocompatibility complex (MHC)-adapted subunits PSMB8–10 in humans. These IFN-induced subunits produce peptides that are optimally suited for binding to the antigen transporter TAP and to classical MHC class I (MHCI) molecules (51). In addition to the PSMB8 and PSMB9 subunits, teleosts have an additional unique PSMB9 subunit denoted PSMB12 as well as an additional PSMB10 subunit denoted PSMB13 (52). In Atlantic salmon, these PSMB8–10-like genes reside in duplicate MHCI regions on chromosome (chr.) 14 and chr.27 (53).
ISGylation is a reversible process where deubiquitinases or DUBs contribute in the deubiquitination process. In mammals, the ubiquitin-specific protease 18 (USP18) is specific for ISG15-conjugated proteins (54–56). In addition to the enzymatic activity, USP18 also interacts with the type I interferon receptor and shuts off downstream signaling.
MHC-compatible protein fragments resulting from proteasomal degradation are then selectively transported into the endoplasmic reticulum through the TAP1/TAP2 channel where they are loaded onto the awaiting MHCI molecule prior to transport to the cell surface for recognition by other immune cells [reviewed in (57)]. MHCI peptide loading and editing are assisted by various chaperones including tapasin-related (TAPBPR) and the aminopeptidase ERAAP molecules (58–61). Additional molecules, such as the tapasin-like (TAPBPL) molecules identified in teleosts and some tetrapods, may have as yet undefined roles in the peptide editing process (53). In Atlantic salmon, TAP2 and TAPBP genes reside within the duplicate MHCI regions on chr.14 containing non-classical MHCI genes and chr.27 harboring the single classical MHCI gene denoted UBA (62). With the recent advances in understanding of the Ub/L-proteasome pathway, it is timely to reinvestigate these responses also in teleosts.
Here, we investigate these responses in a new Atlantic salmon cell model with endothelial-like characteristics originating from the heart, following infection with ISAV, an orthomyxovirus virus with eight negative stranded RNA segments (63). This allows us to assess the interactions between ISAV and host genes in the first model system from a target cell and target organ (64, 65). We compare the effect of viral infection against stimulation with recombinant interferon gamma (rIFNg) on gene expression unaffected by potential viral evasion strategies.
2 Material and methods
2.1 Animal husbandry and ethical considerations
Tissue for the development of primary cells was obtained from a healthy 15-g pre-smolt Atlantic salmon (Salmo salar) at a Norwegian salmon hatchery. A trained fish veterinarian euthanized the fish with an overdose of MS222 (Sigma-Aldrich, St. Louis, MO, USA) of 250 mg/L as recommended in the guidelines, before the whole heart was removed and incubated in medium consisting of Leibovitz-15 (L-15, Lonza, Basel, Switzerland) with the addition of 100 units of potassium penicillin and 100 μg of streptomycin sulfate (1% pen/strep, Lonza) and 20% fetal bovine serum (FBS, Lonza) and processed as described in Section 2.2 on the same day the tissue was harvested.
2.2 ASH2–2 cells and study design
The ASH2–2 cells were established by mincing the harvested heart into small pieces with a scalpel. Then, the cells were digested in 0.25% trypsin/EDTA (Lonza) in a sterile container under stirring for 1 h. The solution was then filtered through a cell strainer and transferred to a 15-mL Falcon vial, L-15 Leibovitz medium (#L1518, Merck AS, New Jersey, USA) was added, and the vial was centrifuged at 1,500 rpm for 10 min, to pellet dissolved cells. The cells were washed once with L15 and a new centrifugation was performed. Then, the cells were resuspended in L15 medium with the addition of 100 units of potassium penicillin, 100 μg of streptomycin sulfate (1% pen/strep, Lonza), and 20% FBS (Lonza), and were seeded into Falcon Primaria 6-well plates (Becton Dickson & Company) and incubated at 20°C without CO2 for several weeks until a cell monolayer was established. The medium was changed several times during the incubation period to remove dead and non-adherent cells floating in the medium. The cells were then subcultured as follows: The cell layer was washed with PBS, incubated with a small amount of trypsin/EDTA at room temperature until detachment. L-15 medium with 20% FBS was added and the cells were split 1:1 into new wells. In the next subculture, the cells were passaged 1:2 into a 25-cm2 Falcon Primaria cell flask. After 7–10 passages, the remaining cells represented a relatively homogeneous monolayer of cells. At passage 10, the amount of FBS in the medium was reduced from 20% to 10% for further maintenance, and at this point, several vials for cryopreservation were also secured. Shortly, the cells from a 25-cm2 Falcon Primaria cell flask were treated with trypsin/EDTA, resuspended in L-15 with 2% FBS, and centrifuged at 1,500 rpm for 10 min at 4°C. The cell pellet was dissolved in ice-cold L-15 medium with 50% FBS and placed on ice. Then, a mixture of almost freezing 20% dimethyl sulfoxide (DMSO) in L15 with 50% FBS (stored at −20°C before addition) was carefully added by very gentle mixing. The vials were then stored first for 24 h in a Styrofoam box at −80°C before transferring them to a liquid nitrogen tank.
The cells were demonstrated to be permissive to ISAV through experimental infection. ASH2–2 cells were passaged in 96-well cell plates (Corning) with L15 and 10% FBS as described above and incubated at 20°C without CO2 for 2 days until approximately 80% confluence. A plate of the ISAV-permissive ASK cells (ATCC CRL-2747) was included as a positive reference. Then, standard inoculation of a 10-fold dilution series of ISAV Glesvær was performed as described in the WOAH manual (66). The well plates were then incubated for 7 days. Virus-infected cells were visualized by microscopy following indirect fluorescent antibody test (IFAT) using an anti-ISAV monoclonal antibody (#P10, Aquatic Diagnostics Ltd, UK). In the same setup, the cells were also shown to be permissive to viral hemorrhagic septicemia virus (VHSV) and infectious pancreatic necrosis virus (IPNV) (data not shown).
ISAV Glesvær passage 3 titer 106,3/mL was propagated in ASK cells as described previously for the SHK-1 cells (67) and titered as described in the WOAH manual (66) with some modification. In short, a 10-fold dilution series was performed in 96-well plates using ASK cells. Visualization of positive staining was done as described before, and the titer was calculated with tissue culture infectious dose (TCID) 50 end point titration as describes by Kärber (68).
The rIFNg batch and amount used here have been used previously and shown to provide a proper induction of expected molecules (69). In brief, the mature Atlantic salmon IFNg sequence of NM_001171804.1 inserted into pET15/D-Topo was purchased from GeneArt (Thermo Fisher Scientific, MA, USA) and transfected into Escherichia coli BL21 DE3 (Thermo Fisher Scientific, #C600003). Recombinant protein was isolated and purified following an established protocol (70). The purity of rIFNg was checked on a 4%–12% precast sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel (Thermo Fisher Scientific) stained with SimplyBlue (Thermo Fisher Scientific). Protein concentration was measured with a Qubit Protein Assay kit (#Q33211, Thermo Fisher Scientific).
ASH2–2 cells in passage 27 were seeded into 25 cm2 flasks using trypsin/EDTA totaling 9.4 × 105 cells per 25 flask used in this RNA-Seq study. Cells were grown at 15°C overnight in L-15 Leibovitz medium with L-glutamine (#L1518, Merck AS) supplemented with 5% FBS and gentamicin.
For ISAV infection, cells grown overnight in L15 supplemented media were exchanged with serum-free media containing ISAV isolate Glesvær at 0.1 multiplicity of infection (MOI). Cells were incubated at 15°C for 4 h before they were washed in PBS and returned to serum-supplemented L15 media. Five ASH2–2 flasks infected with ISAV were harvested 4 h post-infection (p.i.) (ISAV-4h samples) and five flasks were harvested 48 h p.i. (ISAV-48h samples). Another five ASH2–2 flasks were stimulated with 1 ng/mL culture media of rIFNg and harvested after 48 h (IFNg-48h samples). Negative controls were five flasks harvested after 4 h (C-4h samples) and five flasks harvested after 48 h (C-48h samples).
2.3 RNA-Seq library preparation and sequencing
Cells from five ASH2–2 flasks from each group were washed twice with ice-cold PBS, trypsinized using 1 mL of 0.25% EDTA-Trypsin and transferred to Eppendorf tubes. Cells were harvested at 2,500 × g for 5 min at 4°C, and resuspended in 350 μl of RLT buffer supplied in the RNeasy Mini kit (#74104, QIAGEN, Hilden, Germany). RNA was isolated including a DNase I treatment step according to the manufacturer’s protocol, (#79254, QIAGEN). RNA quantity and quality were analyzed using a 4150 Tapestation (Thermo Fisher Scientific) according to the manufacturer’s protocols. RNA quantity ranged from 174 to 582 ng/μL with RIN values of 7.4–10.0.
RNA-Seq libraries were prepared using a TruSeq stranded total RNA prep kit (Illumina, San Diego, CA, USA) targeting the poly-A tail to enrich mRNAs. A total of 25 barcoded libraries were pooled together and sequenced on a NovaSeq 6000 SP Flowcell (Illumina) employing 150-bp paired-end sequencing. Library preparation and sequencing were performed at the Norwegian Sequencing Center, Oslo, Norway.
2.4 Illumina data analyses
RNA-Seq analysis: From the raw sequenced data, adapters and low-quality reads were trimmed/removed using BBduk [BBMap v34.56 (71);] using “ktrim = r k = 23 mink = 11 hdist = 1 tbo tpe qtrim = r trimq = 15 maq = 15 minlen = 36 forcetrimright = 149” as parameters. Cleaned reads were aligned against the ENSEMBL Atlantic salmon genome (Salmo_salar.ICSASG_v2) using Hisat2 v2.1.0 (release 104 annotation - Salmo_salar.ICSASG_v2.104.gtf) [GCA_000233375.4 (72);]. Fragments (read pairs) aligning to the 136,077 transcripts were counted using featureCounts v1.4.6-p1 (parameters: “-s 2 -p”) (73). Differential expression analysis was carried out using variance stabilizing transformed data using all cleaned reads in the DESeq2 v1.22.1 (74) package in R v3.5.1. Significance cutoff was set at an adjusted p-value (p-value adjusted for false discovery rate) of 0.05. Sequence data have been submitted to the NCBI Sequence Read Archive (SRA) under the BioProject accession number PRJNA1197162.
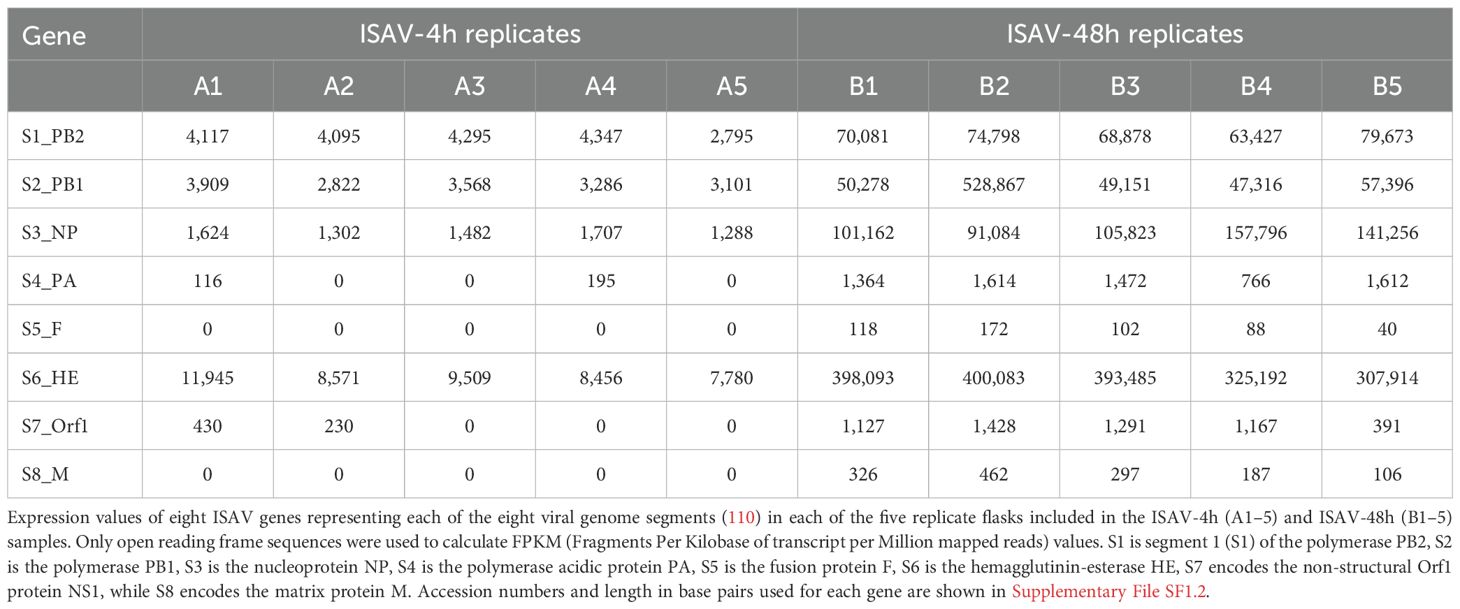
ISAV expression: To calculate the expression of eight ISAV genes, cleaned sequence data were aligned to the above reference using HiSat2 v2.1.0. Total number of reads aligned to each of the 13 genes was counted using featureCounts v1.4.6-p1 using a custom SAF file as defined by the tool guide. In order to compare the samples, FPKM values were calculated from the raw count data using the standard FPKM formula.
2.5 Data mining
Genes of interest were further investigated using the Ensembl accession numbers provided in the RNA-Seq analysis. Gene duplications were identified through tblastn search using translated amino acid sequences against a new Atlantic salmon genome assembly (GCA_905237065.2) as the previous assembly used for RNA-Seq analysis was terminated.
Gene orthology was tested using clustal alignments (75) and phylogenetic analyses using MEGA 7 (76). In phylogenies, the percentage of trees in which the associated taxa clustered together are shown next to the branches. Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with a superior log likelihood value. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position.
Gene homeologs were defined based on regional location (77). We use standardized nomenclature for gene duplicates originating from the fourth salmonid-specific 4R-WGD where one homeolog is given the extension –a and the other homeolog is given the extension –b. Some gene duplicates mostly originating from the 3R-WGD have been given the extension –like or –L, while gene copies of unknown location have consecutive numbers.
2.6 Expression in unstimulated tissues
Reads per kilobase per million mapped reads (RPKM) values were defined using CLC Genomic Workbench 6.0.5 [CLC Genomic workbench] and SRR transcriptome runs from a single individual (77) as follows: HK (head kidney) SRR1422860, gills SRR1422858, spleen SRR1422870, heart SRR1422862, gut SRR1422859, muscle SRR1422866, brain SRR1422856, liver SRR22865, nose SRR1422867, skin SRR1422869, ovary SRR1422871, and testis SRR1422872.1.
3 Results and discussion
Our model system, i.e., the ASH2–2 cells, was made from heart tissue and was shown to be permissive to ISAV (Figure 1). A search for cell-specific markers showed that the ASH2–2 cell line most likely represents cardiac endothelial cells as defined by Andresen et al. (78) [Supplementary File 1 (SF1.1)]. Typically, endothelial expressed genes are cadherin 5, vascular endothelial growth factor receptor, endothelial differentiation-related factor 1, and endothelial lipase.
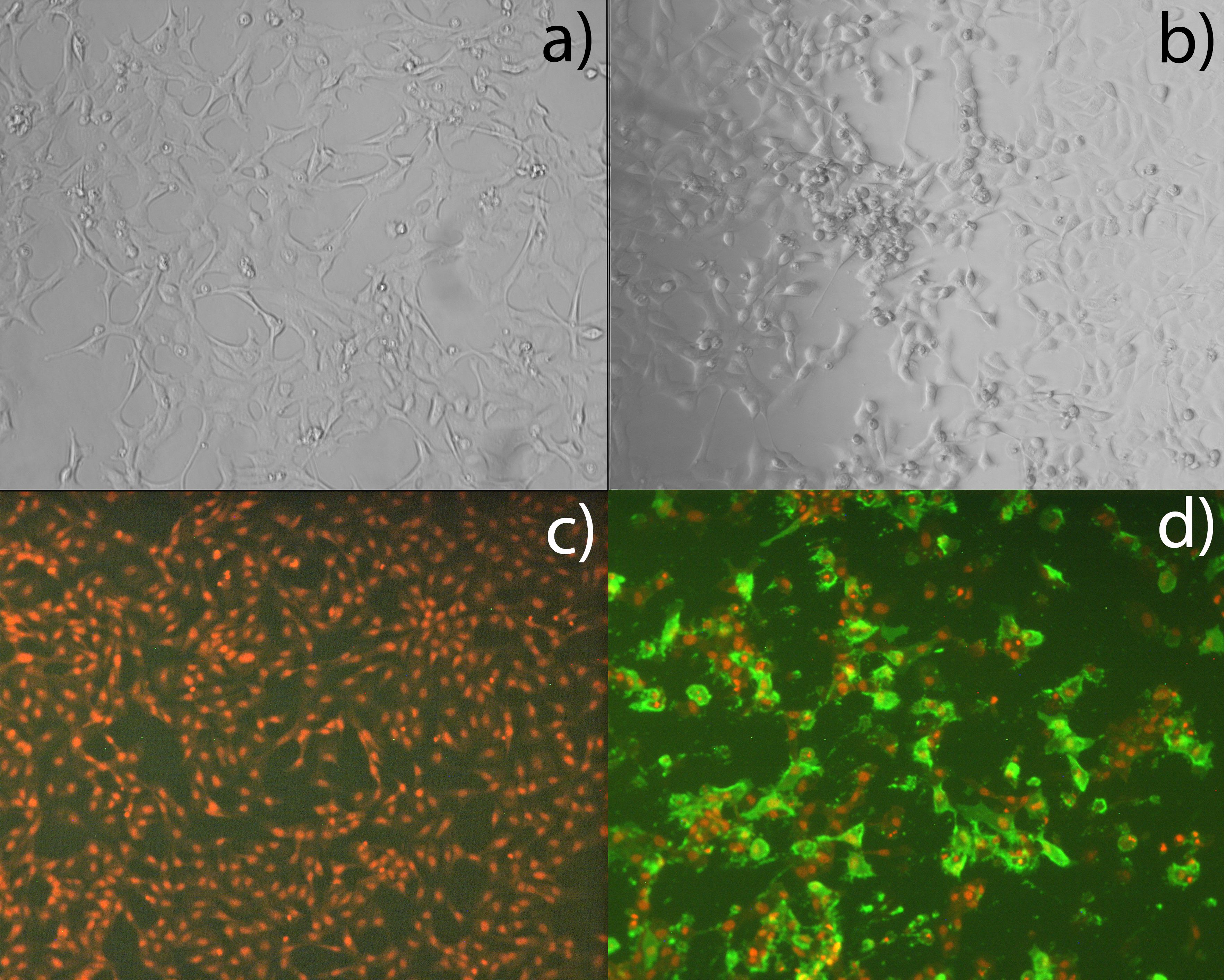
Figure 1. Susceptibility of ASH2–2 cells to ISAV. ASH2–2 passage 12, inoculated with mock control (a, c) and ISAV Glesvær (b, d) and incubated for 7 days. Viral protein visualized by IFAT (green); cell nucleus by propidium iodide (PI, red).
To ensure that the observed effect was due to infection with virus, we also investigated expression of ISAV genes 4 h and 48 h p.i. Very few ISAV transcripts were present at 4 h p.i., but increased substantially at 48 h (Table 1; Supplementary File SF1.2).
To compare the difference between viral infection and cytokine stimulation, we infected ASH2–2 cells with ISAV and compared this response to that achieved by stimulating with rIFNg. Viruses such as influenza, another orthomyxovirus, encodes several proteins that actively shut off host gene expression (79). If ISAV uses similar approaches, comparing viral infection to interferon stimulation could detect such effects.
Our sampling time points of 4 and 48 h for ISAV infection with similar negative control time points provided us with an overview of early and late responses. For comparison, we sampled interferon gamma-stimulated samples at 48 h post-stimulation (p.s.). Expression data for all differentially expressed genes for each biological replicate is shown in Supplementary file 2 (Supplementary File SF2). For each of the five samples we had five replicate flasks, i.e., ISAV-4h (A), ISAV-48h (B), negative control 4h (C), negative control 48h (D), and rIFNg-48h (E). All five replicates within each of the five A–E samples showed convincing clustering in principal component analysis and cluster dendrograms; thus, there was limited transcriptional variation between flasks (Figure 2; Supplementary File SF1.2). Expression profiles changed substantially from 4 to 48 h in the ISAV-challenged samples while a similar change was not observed in the negative control 4- to 48-h samples.
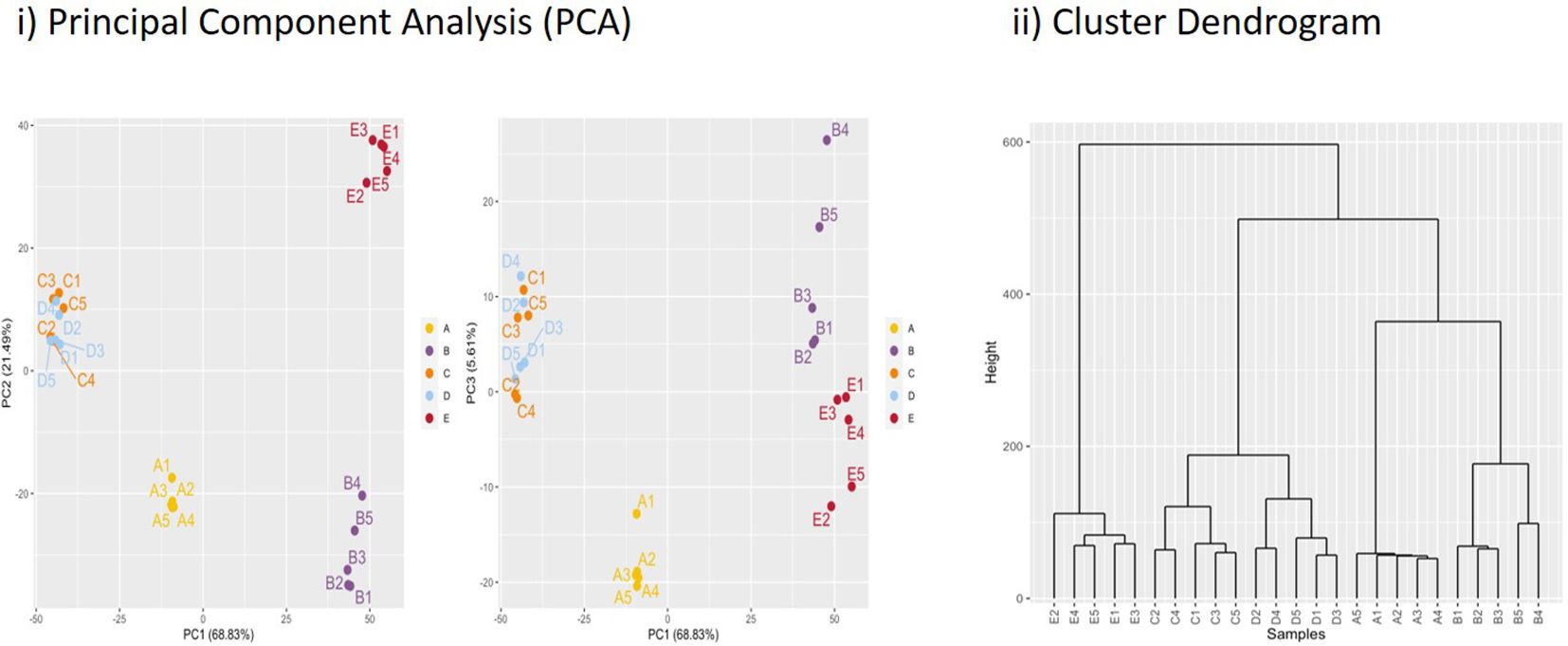
Figure 2. Principal component analysis (PCA) and hierarchical clustering dendrogram. (i) Principal component analysis for data from each replicate flask with percentages of variance associated with each axis. A1–5 (ISAV-4h) and B1–5 (ISAV-48h) are ISAV challenged samples, C (C-4h) and D (C-48h) are negative controls, while E is rIFNg-stimulated flasks (IFNg-48h). (ii) Sample clustering based on normalized data grouping replicates and separating biological conditions. A Euclidian distance is computed between samples and a dendrogram is built upon the Ward criterion.
3.1 Viral sensors and early response genes
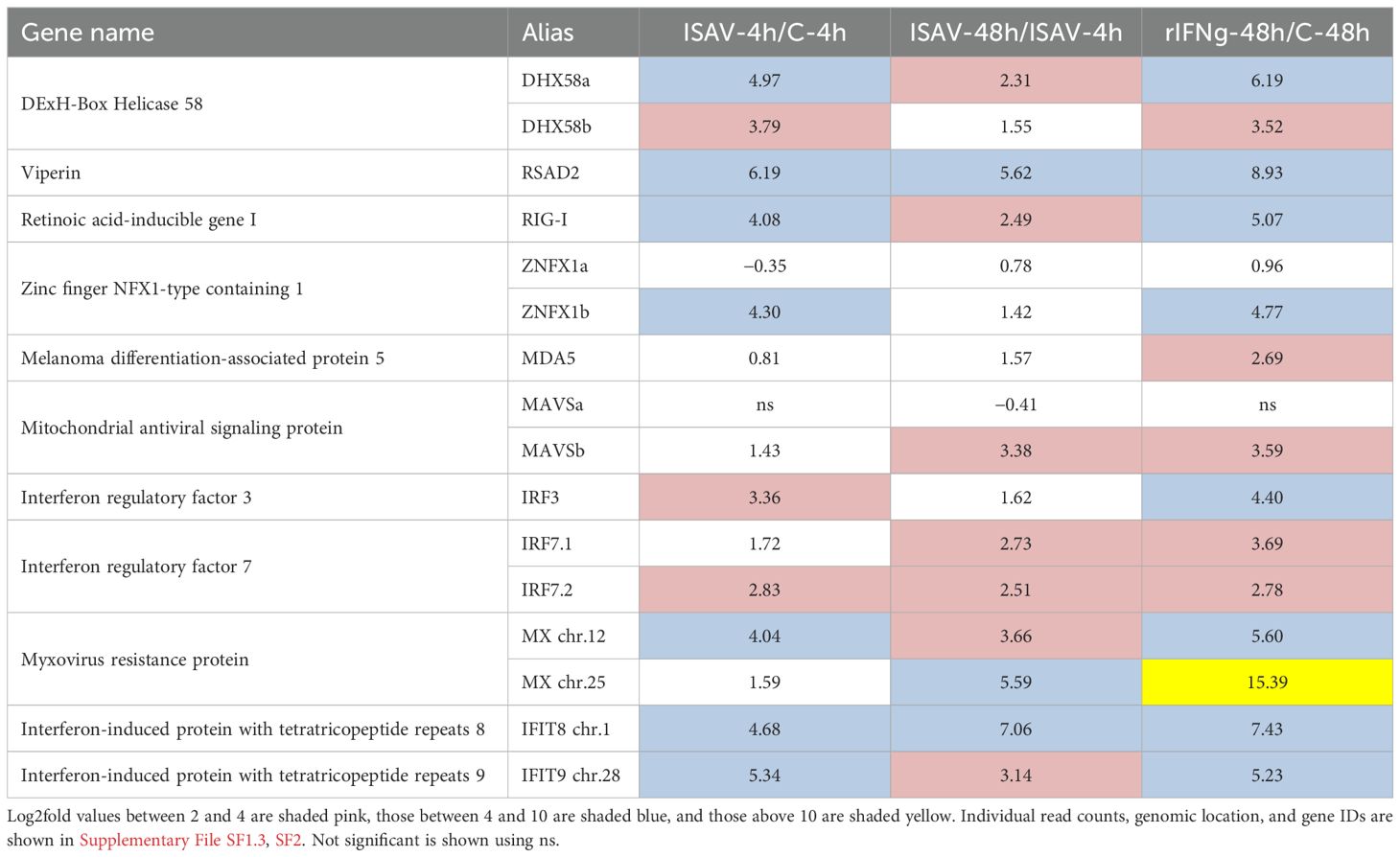
ISAV is a single-stranded RNA virus and should be recognized by various pattern recognition molecules. Based on the observed upregulation of viral sensors, the cell line responded well to infection even at 4 h p.i. despite fairly low expression of viral genes (Table 2; Supplementary File SF1.3). Four viral sensors display above 3.7 log2fold values after ISAV infection at 4 h, i.e., both the homeolog DHX58 (alias LGP2) gene and the RIG-1 (alias DDX58) gene and one of the homeolog ZNFX1 genes. All these genes are more affected by virus than by rIFNg at 48 h. MDA5 displays a similar upregulation in both virus and interferon samples at 48 h.
In mammals, both ZNFX1 and RIG-1 interact with MAVS. This MAVS activation results in the phosphorylation of interferon regulatory factors (IRFs) 3 and 7, which again induces expression of interferon and interferon-stimulated genes [reviewed in (80)]. In ASH2–2 cells, both the single IRF3 and two of the three IRF7 genes are more upregulated by virus than by rIFNg (Table 2). The third IRF7 gene (ENSSSAG00000081805) residing on chr.10 displays no transcripts in our dataset. Of the duplicate MAVS genes, only the one on chr.9 responded with a log2fold increase of more than 3.3 upon ISAV-48h infection as well as rIFNg stimulation.
A comprehensive list of upregulated ISGs in both humans and a teleost is compiled in Levraud et al. (35). Some of these early responders are the myxovirus (Mx) and the IFN-induced protein with tetratrico-peptide repeats (IFIT) genes. Of the nine Mx genes described in Atlantic salmon, genes on chr.12 have been shown to respond more to type I IFN while genes on chr.25 are more affected by type II IFN (6, 81). This is also reflected in our model system where the gene on chr.25 responded more strongly to rIFNg while the gene on chr.12 responded more strongly to viral challenge with a log2fold value of 4 at 4 h p.i. (Table 2; Supplementary File SF1.3). IFIT genes have been implicated in various antiviral responses (82), and in our material, one gene residing on chr.1 and even more so one gene residing on chr.28 responded strongly to ISAV infection with log2fold values above 4.6 at 4 h p.i. (Table 2).
Both ISAV and rIFNg upregulated the transcription of many expected genes in ASH2–2 cells similar to what has been observed in other salmonid cell lines such as SHK-1, ASK, TO, or CHSE cells (69, 83–88). As for differentially expressed early response genes, several viral sensors are strongly affected by ISAV also at 4 h p.i. such as RIG-I, LGP2, and MDA5 (Table 2). These three genes have previously been shown upregulated by various viruses or other stimulants in salmonids (69, 89, 90). Little data exist on ZNFX1 in teleosts, but in humans, this gene is an early sensor of viral RNA (3), similar to what we see in our ISAV-4h samples.
In line with what has been described by Robertsen et al. (6), an Mx gene on chr.25 was more upregulated by rIFNg than the Mx genes on chr.12 while the gene on chr.12 was more affected by ISAV (Table 2). Owing to sequence divergence between the annotated Ensembl gene sequences and those of Robertsen et al. (6), a further definition of individual Mx genes in either region was not possible (data not shown).
3.2 Ubiquitin-like proteins
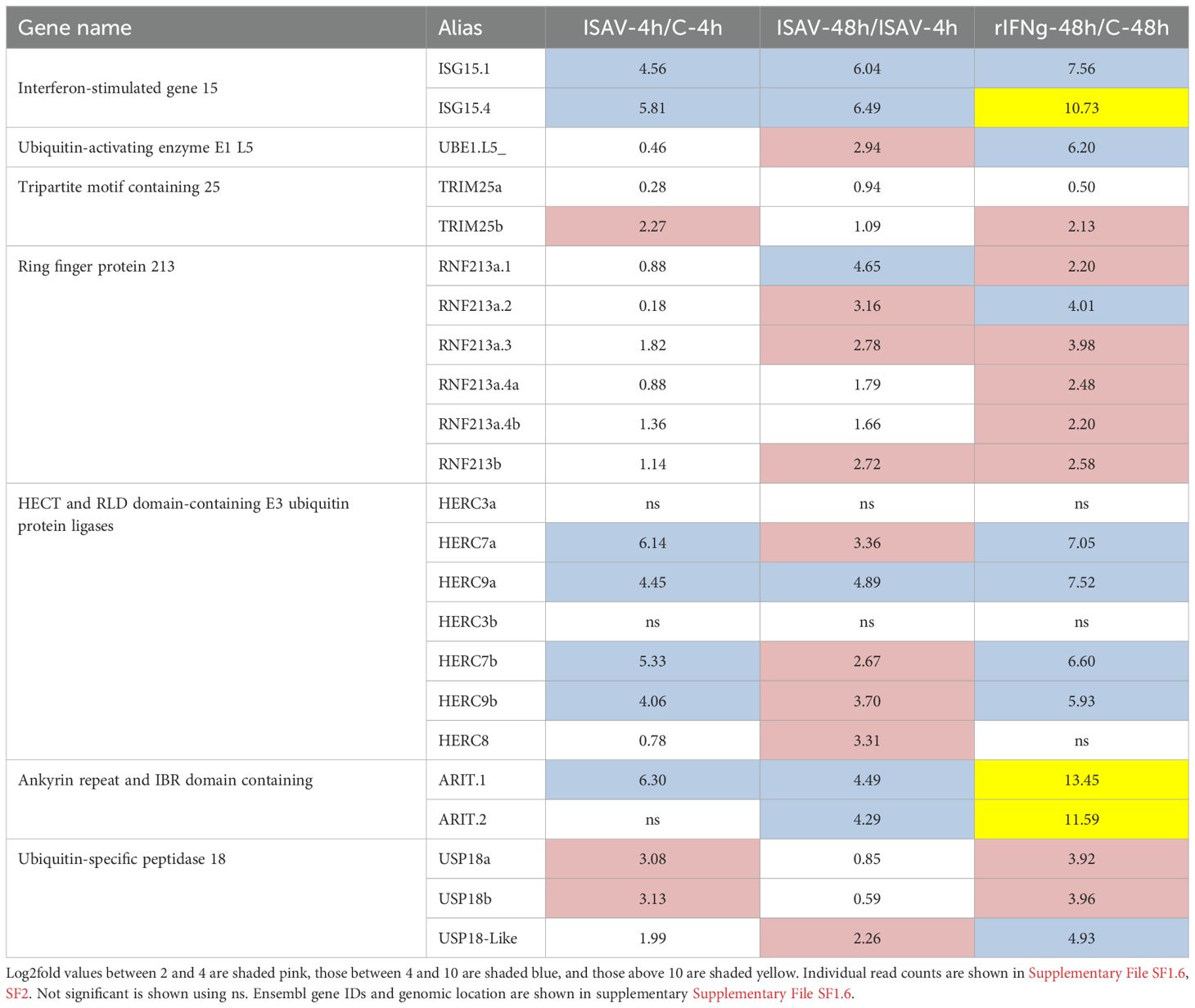
A master regulator induced by interferon or infection is ISG15, a ubiquitin-like protein attached to target proteins through the enzymatic cascade reaction called ISGylation [reviewed in (17)]. There are duplicate closely linked homeolog Atlantic salmon ISG15 genes, two on chr.20 and four on chr.24 (Table 3; Supplementary File SF1.4–1.6) (77). One gene within each region is affected by virus with log2fold values above 4.5 at 4 h p.i. while the remaining ISG15 genes are unaffected (Table 3; Supplementary File SF1.6). Of the other ubiquitin-like genes clustering with human UBB, UBC, UBA52, and RPS27A (Supplementary File SF1), none are significantly upregulated by virus or interferon (Supplementary File SF1.7).
The ISGylation process is mediated by the consecutive action of E1‐activating enzymes, E2‐conjugating enzymes, and E3 ligases that covalently link ISG15 to residues on target proteins (23). Only one E1‐activating enzyme gene displays log2fold values of 2.9 at 48 h p.i. and 6.2 at 48 h p.s. (UBE1.L5, Table 3; Supplementary File SF1.6). This gene is an ortholog to the human known ISG15 E1-activating enzyme UBE1L [ (16) and data not shown]. None of the annotated E2-conjugating enzyme genes display log2fold values above 2 in the infected or stimulated samples (data not shown).
In humans, ISG15 exhibits antiviral activity towards both DNA and RNA viruses, where, for instance, ISGylation of influenza A virus NS1 protein causes protein loss of function and thus inhibition of virus replication (17). Human ISG15 conjugation has been found essential for antiviral IFN response mediated by MDA5, a function that is counteracted by de-ISGylation from the papain-like protease of SARS-CoV-2 (91). Teleost ISG15 has also been shown to be upregulated by a variety of viruses such as ISAV and IPNV in Atlantic salmon (21) and RGNNV in Asian seabass and black seabream (92, 93), supporting a role in antiviral defense also in teleosts. In Atlantic salmon, ISG15 and ubiquitin were shown covalently linked to segment 8 open reading frame 2 of the ISA virus (22). In humans, ISG15 is primarily induced by type 1 interferons (16), while in teleosts, ISG15 is induced by both type I and type II interferons [ (35) and this study]. Whether this difference also reflects downstream differences regarding function or interacting partners remains to be established as functional studies on ubiquitination or ISGylation in teleost are so far limited.
3.3 ISGylation
Attachment of Ub/L chains to proteins involves E1 ubiquitin-activating enzymes, E2 ubiquitin-binding enzymes, and E3 ubiquitin ligases. For human ISG15, the E1-activating enzyme is UBE1L (alias UBA7), and this is the only E1-activating enzyme shown to be affected by infection and interferon stimulation in our material (Table 3). Orthologs of the human ISG15 E2-conjugating enzyme UBCH8 (alias UBE2L6) were not significantly affected by virus or interferon gamma (data not shown).
E3 ubiquitin ligases provide the target specificity for adding ubiquitin or ubiquitin-like residues and are classified into RING, HECT, and RBR domain-containing E3 ligases [reviewed in (23)]. Of the E3 ubiquitin ligases known to be involved in ISGylation, TRIM25, HERC5, and ARIH1 belong to each of these three categories (16).
3.4 RING E3 ligases
TRIM molecules belong to the really interesting new genes or RING family (23). Based on sequence phylogeny, Atlantic salmon harbors two TRIM25 homeologs located on chr.2 and chr.12 in addition to multiple finTRIM genes in which many are partial genes located on unplaced scaffolds (Supplementary File SF1.8). TRIM25b on the chr.12 gene displays a log2fold value above 2 in both the ISAV-4h and the rIFN-48h samples (Table 3; Supplementary File SF1.6). Only a few finTRIMs with known chromosomal location display log2fold values above 2.
In humans, TRIM25 is required for viral RNA sensing performed by the cytoplasmic helicase RIG-I, leading to IFN production (27, 94). TRIM25 has also been implicated in antiviral immunity in teleosts, being a highly expressed ISG in studies of virus-infected cells or tissues in many species (32, 95, 96). In our material, we found one of the duplicate TRIM25 orthologs to have log2fold values above 2 in both the ISAV-4h and the rIFN-48h samples (Table 3). Unfortunately, most of the finTRIMs are too fragmented in the investigated genome to allow a more detailed analysis of these genes.
Ring finger protein 213 (RNF213) also belongs to the RING family of E3 ubiquitin ligases and is upregulated by interferon stimulation and viral challenge in both humans and teleosts (34, 35). Humans only have one RNF213 gene while two genes denoted RNF213a and RNF213b exist in zebrafish (97). We found six Atlantic salmon RNF213 genes where five genes cluster with the zebrafish RNF213a sequence and one clusters with the zebrafish RNF213b sequence (SF1.8). Of the five RNF213a genes, one resides on chr.1, two on chr.3, one on chr.6, and one on chr.22 (SF1.6). All genes are more upregulated by virus at 48 h than by interferon with the gene RNF213a.1 (ENSSSAG00000041408) on chr.1 displaying the strongest upregulation with a log2fold value of 4.6 (Table 3). One gene on chr.3 (RNF213a.4a, ENSSSAG00000043017) and the gene on chr.6 (RNF213a.4b, ENSSSAG00000047562) are homeologs (77) with a sequence identity of 98% (Supplementary SF1.9). The remaining RNF213a gene sequences share between 63% and 83% nucleotide sequence identity. A single Atlantic salmon RNF213b gene (ENSSSAG00000001848) located on chr.1 also displays log2fold values above 2.5 after 48 h p.i. as well as p.s. and shares 49–52 nucleotide sequence identity with the RNF213a sequences. In normal tissues, all salmon RNF213 genes display low expression levels in immune-related tissues and negligible expression in non-immune tissues such as liver and heart, supporting immune gene function (Supplementary SF1.10).
Expression levels identified here may not be representative for all animals, tissues, or cell types. We thus chose to investigate the expression of RNF213 genes in another Atlantic salmon cell line. SHK-1 is an Atlantic salmon cell line developed from head kidney in 1997 (98). This cell line was also defined as endothelial-like and was used to investigate differentially expressed genes following rIFNg stimulation using an old genome assembly from NCBI (GCF_000233375.1_ICSASG_v2) (69). Re-analyzing this dataset for DEGs identified in this study shows that three RNF213a genes in addition to the RNF213b gene were also upregulated in SHK-1, although less so than in the ASH2–2 cells (Supplementary File SF1.11). One gene (RNF213a.1) was not upregulated and RNF213a.3 displayed no simple gene ortholog in the NCBI genome assembly (data not shown). Whether the differences between the two cell lines relate to individual or cell-specific differences remains to be established.
Human RNF213 is upregulated by interferon and has been shown to act as a cellular sensor of ISGylated proteins with broad antimicrobial activity (18, 33). RNF213 genes are also upregulated by stimulation or infection in other teleost species such as grouper or zebrafish (35, 99) suggesting an overall similar role as found for the human RNF213 gene.
3.5 HECT E3 ligases
The second E3 ubiquitin-protein ligase family affected by ISAV and rIFNg is the HERC family. Based on blast search and sequence phylogeny, we found eight small Atlantic salmon HERC genes with two orthologs to human HERC3 and one ortholog to human HERC4 (Figure 3). Two salmon HERC gene sequences cluster with what has been defined as HERC7 from carp and zebrafish (43, 44). The remaining three salmon HERC gene sequences reside in two other strongly supported clades. One clade includes only one sequence from salmon in addition to sequences from Northern pike and neoteleosts, which we here tentatively define as HERC8 sequences. We could not find sequences from spotted gar or cyprinids belonging to this clade, suggesting that it may have arisen in a salmonid predecessor. The other clade includes two sequences from Atlantic salmon in addition to sequences from spotted gar, herring, and neoteleosts, here tentatively defined as HERC9 sequences.
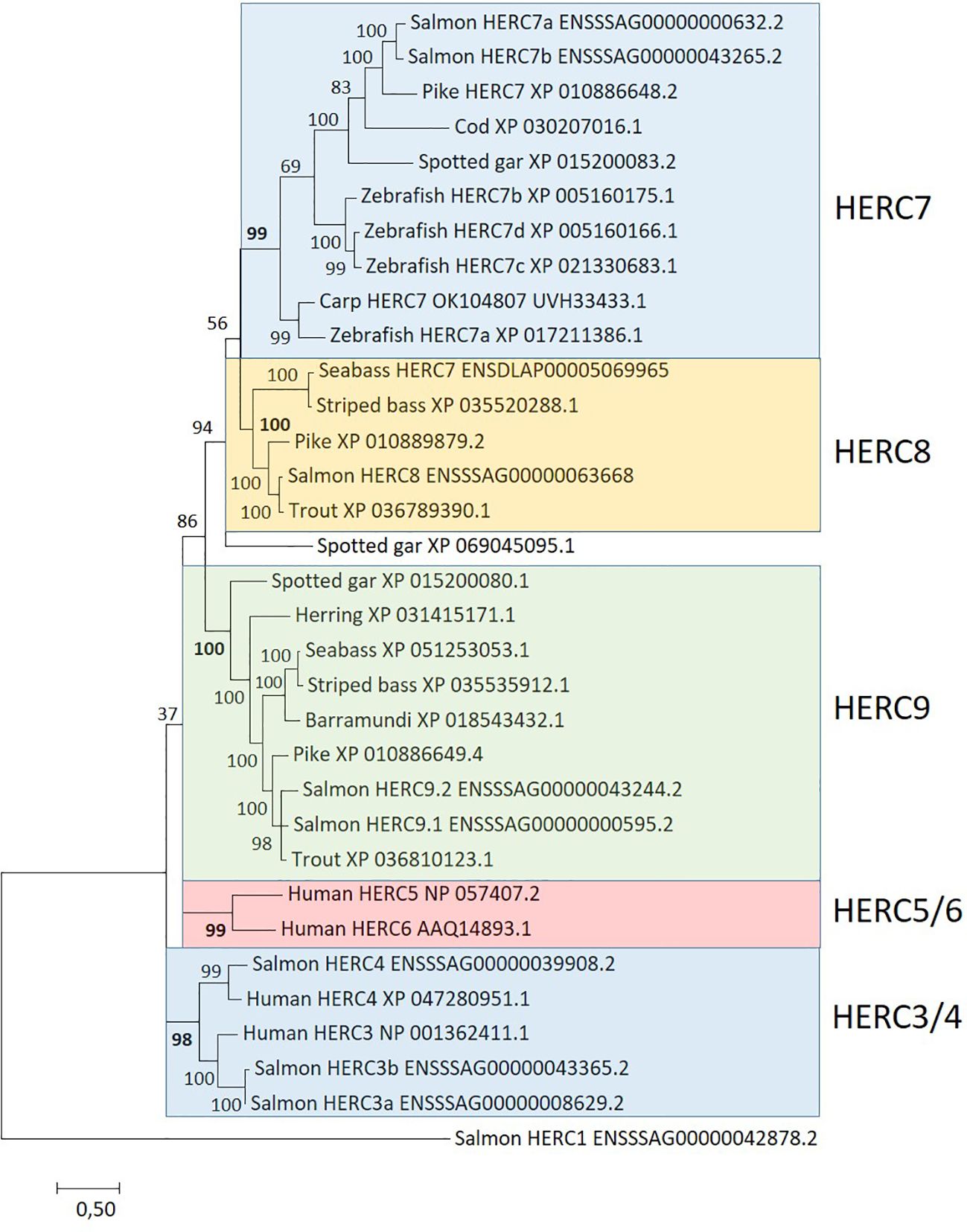
Figure 3. Phylogeny of deduced HERC amino acid sequences from salmon (Salmo salar), trout (Oncorhynchus mykiss), cod (Gadus morhua), striped bass (Morone saxatilis), seabass (Dicentrarchus labrax), barramundi (Lates calcarifer), herring (Clupea harengus), pike (Esox lucius), zebrafish (Danio rerio), spotted gar (Lepisosteus oculatus), and human. Sequence references are shown in the figure where HERC7 sequences from carp, zebrafish, and seabass originate from (43, 44, 107), respectively. Clustering of sequences is highlighted using colored shading. The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix-based model (108). The tree with the highest log likelihood (−29,784.42) is shown. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories [+G, parameter = 1.5507)]. The rate variation model allowed for some sites to be evolutionarily invariable [(+I), 1.29% sites]. The analysis involved 33 amino acid sequences. There were a total of 739 positions in the final dataset.
None of the three Atlantic salmon HERC3 and HERC4 genes were significantly affected by infection or stimulation (Figure 3; Table 3; Supplementary File SF1.7, SF1.8). On the other hand, all four HERC7 and HERC9 genes were strongly upregulated by ISAV with log2fold values above 4 in the ISAV-4h sample with increased transcript levels 48 h p.i. All four genes were also upregulated by interferon gamma, but less affected than by viral infection. The HERC8 gene was only upregulated by ISAV at 48 h, but not affected by rIFNg. Looking at the transcription levels of these HERC7–9 genes in normal tissues, the only gene strongly expressed in immune-related tissues is HERC9a with RPKM values above 50 in both head kidney and spleen and above 10 in gills (Supplementary SF1.11). Expression of the other four HERC7–9 genes were low in immune-relevant tissues and negligible in non-immune organs. Based on sequence alignment and blast against NCBI TSA sequences, the salmon HERC9b gene may be a pseudogene expressing truncated transcripts (data not shown).
To verify expression in another Atlantic salmon sample, we again looked at expression in the SHK-1 cells stimulated with rIFNg (69). In addition, here there were similarities and differences between the two cell lines (Supplementary File SF1.12). Stimulatory effect on HERC3 and HERC4 genes was negligible in both cell lines. For HERC7a/HERC7b and HERC9a, upregulation was much higher in ASH2–2 cells than in SHK-1. These differences between the two cell lines may reflect cell-specific differences with ASH2–2 cells having a stronger response originating from a main target organ.
HERC5 and HERC6 are defined as ISG15-specific E3 ubiquitin ligases with an antiviral effect on humans and mouse, respectively (35, 39–41). Teleosts were not thought to have direct orthologs to the mammalian HERC5 and HERC6, but instead to have a unique teleost-specific sequence defined as HERC7 that cluster with mammalian HERC5 and HERC6 gene sequences (43, 44) (Figure 3). However, this turns out not to be the case. In humans, dogs, and elephants, HERC3, HERC5, and HERC6 genes reside closely linked on chr.4 in between FAM13A (family with sequence similarity 13 member A) and PPM1K (protein phosphatase, Mg2+/Mn2+-dependent 1K) genes whereas HERC4 has translocated elsewhere (100). In Atlantic salmon, closely linked HERC3, HERC7, and HERC9 homeolog genes on chr.4 and 8 are also located in between FAM13A and PPM1K genes (Figure 4). This suggests that the salmon HERC7 and HERC9 genes evolved from their mammalian HERC5 and HERC6 counterparts. As in mammals, HERC4 is located on a different chromosome (Supplementary File SF1.7).
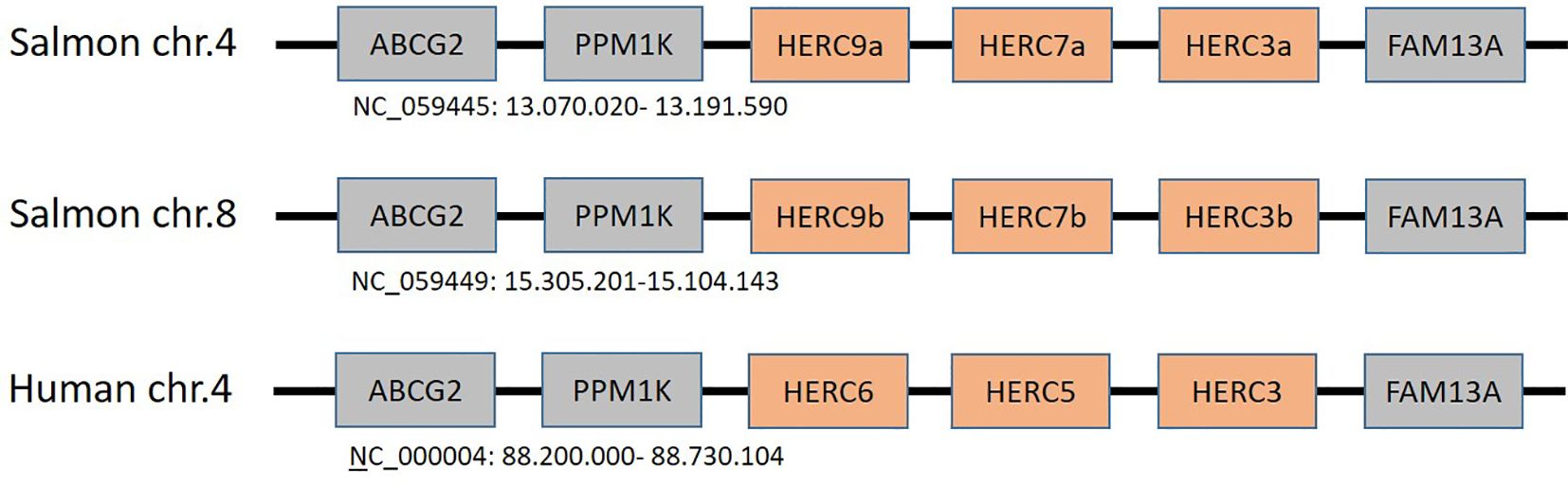
Figure 4. Regional synteny of genes surrounding the human HERC5 and HERC6 genes and the duplicate Atlantic salmon HERC3, HERC7, and HERC9 genes on chromosomes chr.4 and chr.8. NCBI accession numbers with regional chromosomal coordinates are shown below each region.
Crucian carp HERC7 has an E3 ligase activity for conjugation of both ubiquitin and ISG15 while zebrafish HERC7c only displays the potential to transfer ubiquitin (43, 44), while genes in both species are upregulated by viral infection. Based on the difference between the carp and zebrafish HERC7 genes, we cannot predict if one or both salmon genes are involved in ISGylation. Whether salmon HERC8 and HERC9 are also involved in ISGylation will be determined by upcoming functional studies, but with large differences in expression levels of the duplicate HERC9 genes in immune-related tissues, they most likely have different, as yet unknown functions. Being a species with many duplicate functional genes or homeologs following the fourth whole genome duplication, duplicate genes may evolve slightly different or entirely new functions (77).
3.6 RBR E3 ligases
Although also a ring finger protein, RNF144 belongs to the RBR family of E3 ubiquitin ligases. There are nine Atlantic salmon genes annotated as either RNF144A or RNF144B in the Ensembl genome where only two display significant upregulation by virus and even more upon interferon stimulation (ENSSSAG00000044458 and ENSSSAG00000006001). A blast search against other teleost sequences hit multiple teleost sequences annotated as ARI5, a gene only defined as an E3 ubiquitin ligase in arabidopsis (101). A phylogenetic analysis of these gene sequences showed that the two upregulated salmon genes do not cluster with other RNF144 sequences or with ARI5 from arabidopsis or any of the other defined RBR gene sequences (Figure 5). Instead, they form a unique and strongly supported cluster alongside sequences from teleosts, reptiles, and monkeys. A search for similar sequences in primates and humans turned out negative, so the gene has most likely been lost en route to hominoids. Sequences in this clade represent a currently undescribed member of the RBR E3 ligase family, containing the typical RBR domain structure. We tentatively suggest naming sequences belonging to this clade as ARIT, being first defined in a teleost. RBR E3 ubiquitin ligases share the RBR structure, but are classified into distinct subgroups based on 5’ and 3’ domains flanking the RBR domain. ARIT sequences only encode the RBR domain, with no additional 5’ or 3’ sequence, similar to the RNF144 gene. However, ARIT sequences do not include a transmembrane domain as found in salmon and human RNF144A and RNF144B sequences [(102) and Supplementary File SF1.12].
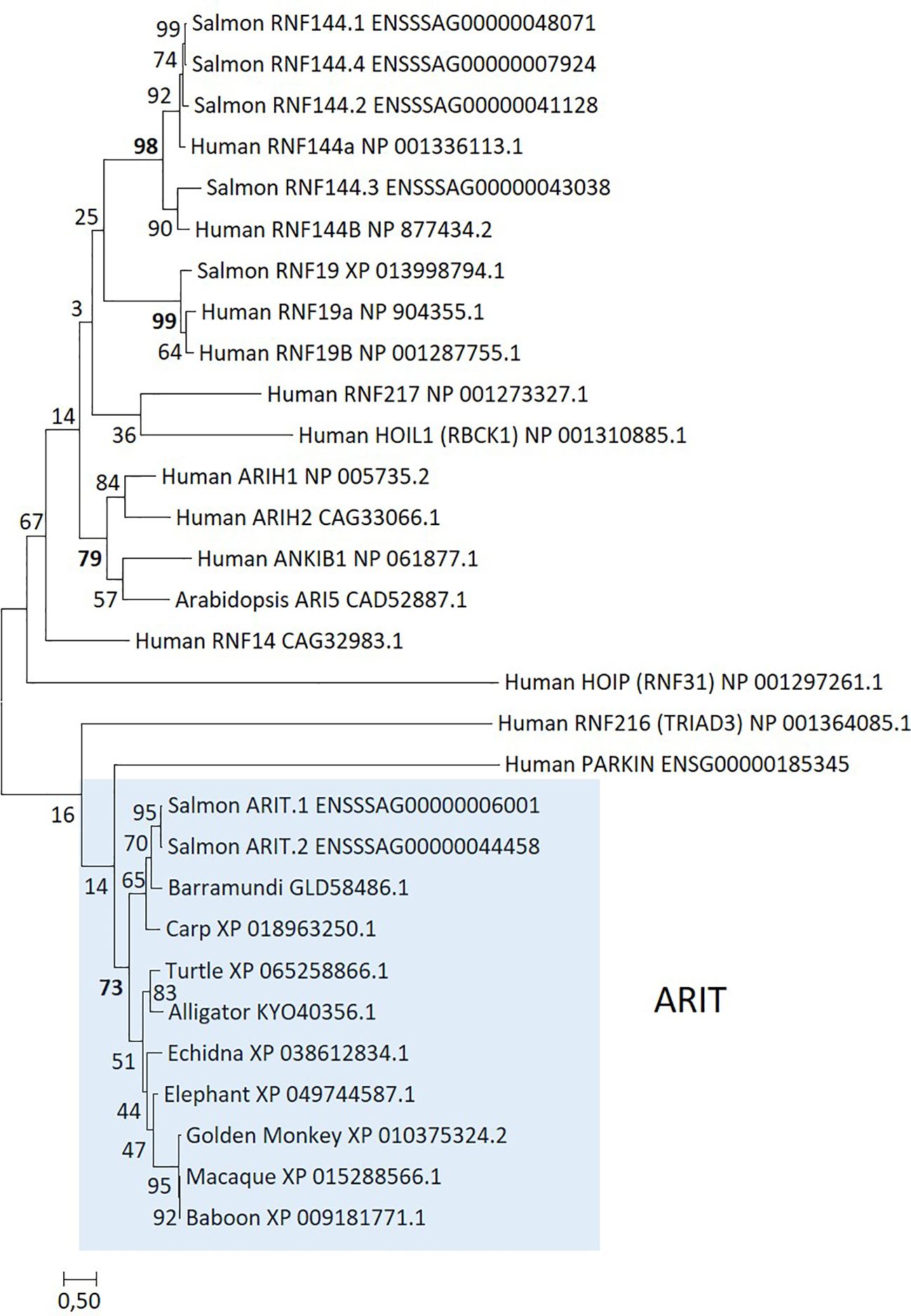
Figure 5. Phylogeny of deduced RBR family amino acid sequences from salmon (Salmo salar), arabidopsis (Arabidopsis thaliana), barramundi (Lates japonicus), carp (Cyprinus carpio), macaque (Macaca fascicularis), baboon (Papio anubis), golden snub-nosed monkey (Rhinopithecus roxellana), elephant (Elephas maximus indicus), echidna (Tachyglossus aculeatus), turtle (Emys orbicularis), alligator (Alligator mississippiensis), and human. Sequence references are shown in the figure and clustering of sequences is highlighted using colored shading. The evolutionary history was inferred by using the maximum likelihood method based on the Whelan and Goldman model (109). The tree with the highest log likelihood (−10,217.29) is shown. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter = 4.0718)]. The analysis involved 30 amino acid sequences. There were a total of 206 positions in the final dataset.
Salmon ARIT sequences are strongly upregulated by rIFNg with log2fold values above 11 at 48 h p.s., but less so by ISAV infection with log2fold values above 4 at 48 h p.i. (Table 3; Supplementary File SF1.6). In unstimulated tissues, the ARIT.1 gene displays low expression in immune-relevant tissues, but negligible expression in non-immune tissues (Supplementary File SF1.10). The salmon ARIT.2 gene displays negligible expression in all tissues. Nucleotide sequence identity between the two salmon ARIT sequences is 98% (Supplementary File SF1.9); thus, some reads may have been misplaced for the two genes affecting their observed expression levels. None of the salmon RNF144 sequences are significantly affected by virus or interferon (Supplementary File SF1.7).
We also investigated ARIT expression in the SHK-1 cells stimulated by rIFNg (69). Here, the log2fold values exceeded 7 in SHK-1 while they were above 11 in the ASH2–2 cells (Supplementary File SF1.11). Again, this could reflect differences in tissue origin for the two cell lines, as heart is a main target organ for the virus. Although not an ortholog, ARIH1 has also been listed as an ISG15-specific E3 ubiquitin ligase in mammals (103). Whether the genes here defined as ARIT are ISG15-specific E3 ubiquitin ligases similar to the mammalian ARIH1 gene needs further verification.
3.7 Deubiquitinases
Ubiquitin carboxyl-terminal hydrolases (USPs) assist in removing ubiquitin and ISG15 from proteins (54, 104). Only three of the annotated USPs in our material displayed a significant upregulation by infection or stimulation. In phylogenetic analyses, two of these DEGs cluster convincingly with human USP18 (SF1.13), in mammals known to be specific for removing ISG15 moieties (54, 56). The third salmon DEG sequence clusters with sequences from tetrapods and teleosts mostly annotated as USP47-like and is a sister clade to the USP18 sequences. Both the USP47-like and the USP18 clades are relatives of the human and teleost USP47 sequences. We thus suggest introducing USP18-like as a nomenclature for the sequences belonging to the USP47-like clade. Homeolog salmon USP18 genes display a log2fold upregulation above 3 in ISAV-4h as well as in rIFNg-48h samples (Table 3; Supplementary File SF1.6). The gene here defined as USP18-like displays a log2fold values above 2 in ISAV-48h samples, but above 4 in rIFNg-48h samples.
In humans, USP18 not only specifically de-conjugates ISG15 from target proteins, but also acts as a negative regulator of the type I interferon response (54, 104, 105). Whether salmon USP18 and USP18-like genes have similar dual functions remains to be established.
3.8 Further processing of tagged proteins
When Ub/L molecules are attached by E3 ligases to proteins destined for degradation, they are processed by the 26S proteasome and some suitable fragments will be presented on the cell surface by MHC molecules.
The 26S proteasome complex consists of a 19S lid that in mammals contain multiple binding sites for ubiquitin or ubiquitin-like sequences. None of the salmon 19S subunits are significantly affected by infection or stimulation (data not shown).
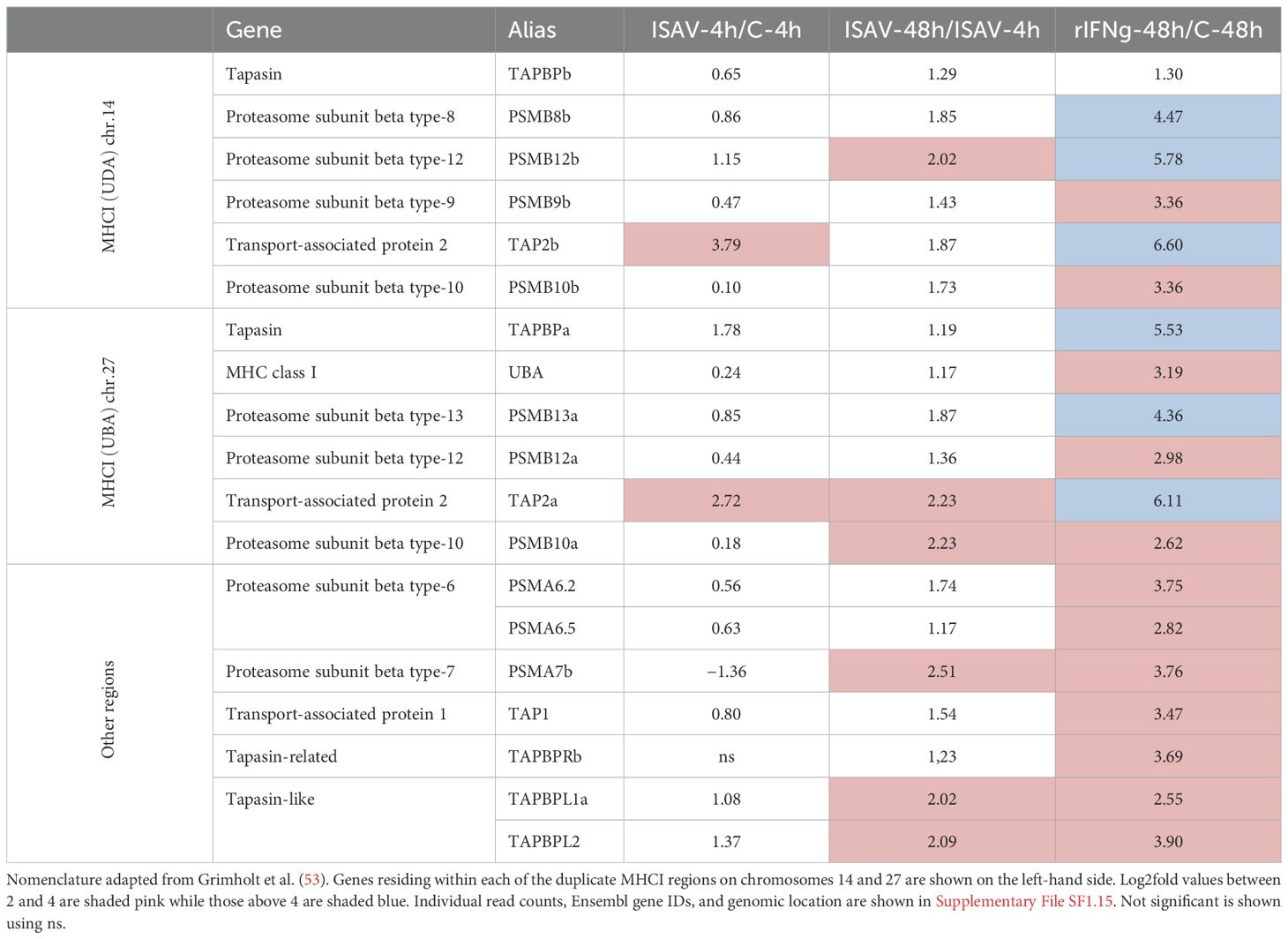
In Atlantic salmon, the 20S core subunits denoted PSMB8, PSMB9, PSMB10, PSMB12, and PSMB13 all reside within the duplicate MHCI regions on chr.14 and chr.27 (53). Of these, PSMB10, PSMB12, and PSMB13 on chr.27 and PSMB9, PSMB10, and PSMB12 on chr.14 display log2fold values above 2 in the rIFN-48h samples (Table 4; Supplementary File SF1.15). In the ISAV-48 samples, only PSMB10 on chr.27 and PSMB12 on chr.14 display log2fold values above 2. Three of the other 20S core subunits, not known to be inducible in mammals, are also affected by stimulation, i.e., PSMA6.2, PSMA6.5, and PSMA7 with log2fold values above 2.8 in rIFNg-48 samples. PSMA7.5 also displays a log2fold value of 2.5 in the ISAV-48h sample.
Degraded MHC-compatible protein fragments are selectively transported into the endoplasmic reticulum (ER) through the TAP1 TAP2 channel. Duplicate TAP2 genes residing on chr.14 and 27 are upregulated by both virus and rIFNg (Table 4, SF1.15). Salmon TAPBP on chr.27 is strongly upregulated by rIFNg with a log2fold value of 5.5 in the rIFNg-48h samples. Two of the TAPBPL genes also display log2fold values above 2 at 48 h in both the virus and the rIFNg samples. In a previous study, we only found one TAPBPR on chr.2 to be expressed and upregulated by stimulation (69). In the current Ensembl genome assembly, a homeolog on chr.2 lacks transcripts while the duplicate on chr.5 is strongly upregulated by rIFNg. Whether this difference relates to genome assembly issues or is a true difference between cell lines remains to be established.
Peptides transported into the ER and loaded into the MHCI groove are transported to the cell surface for recognition by T cells. UBA, a single classical MHCI gene as the main peptide presenter, displays a relatively high expression level also in unstimulated cells, but reaches a log2fold value above 3 in the rIFNg-stimulated sample (Table 4; Supplementary File SF1.15).
PSMB10 and PSMB12 display log2fold values above 2 in both regions while only PSMB8 and PSMB9 on chr.14 display a similar upregulation (Table 4). PSMB10 is located closer to non-classical Z lineage genes and may not affect classical UBA gene peptides. Surprisingly, two PSMA6 genes in addition to one PSMA7 gene were also upregulated by both rIFNg and ISAV. In humans, PSMA6 has been shown to be upregulated during viral hepatitis, with a negative impact on ISG15 gene expression (106), but no such data exist for human PSMB7. The salmon PSMA6.2 gene was also upregulated by rIFNg in the SHK-1 cell line (69), suggesting that it may be a general effect in endothelial cells. Atlantic salmon has five expressed copies of the PSMA6 gene, allowing some to diverge into new functions.
Of the MHCI and proteasome pathway genes, only TAP2 in both duplicated regions displayed log2fold values above 2 at 4 h p.i. with a stronger upregulation by rIFNg at 48 h as compared to ISAV-48h. This may suggest viral interference with MHCI proteasome genes. However, another explanation could be that the transcription machinery is overtaken by viral transcripts. With the single exception of RNF213a.1, expression of all genes shown in Tables 2-4 is higher in the rIFNg-48h sample than in the ISAV-48h sample. This suggests that the host transcriptional machinery shares its capacity between host and viral transcripts.
4 Conclusion
In mammals, ISG15 is a key participant in the host antiviral response through a process called ISGylation where E1-activating enzymes, E2-conjugating enzymes, and E3 ligases attach the UbL chain to molecules to be functionally modified or destined for degradation. Here, we use an endothelial-like cell line originating from a main ISAV target organ to investigate how infection affects expression of genes in this ubiquitin–proteasome pathway. We show that in Atlantic salmon as in mammals, many ortholog genes are upregulated upon viral infection such as ISG15, UBE1L, USP18, and TRIM25. Atlantic salmon, like several other teleosts, have expanded on the human HERC5 and HERC6 genes where we add HERC8 and HERC9 genes to what was previously defined as teleost-specific HERC7 genes. The salmonid-specific whole genome duplication event provided Atlantic salmon with duplicate HERC7 and HERC9 genes that are both strongly upregulated by virus even at 4 h after infection. The HERC8 gene is turned on at 4 h p.i. but quickly downregulated at 48 h p.i. The mammalian ARIH1 gene seems absent from salmon, but we identified a cluster of sequences from both tetrapods and teleosts that we here denote ARIT genes. This gene is present in monkeys, but lost en route to hominids. For Atlantic salmon, one ARIT gene responds strongly at 4 h p.i., while both duplicates are upregulated at 48 h p.i. Functional studies of individual new E3 ubiquitin ligase molecules are needed to understand their biological relevance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1197162.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
UG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing. HS: Formal analysis, Investigation, Methodology, Resources, Visualization, Writing – original draft, Writing – review & editing. AS: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by the Norwegian Research Council (NRC) grant # 274635. Funding for open access publishing is granted by the Norwegian Veterinary Institute.
Acknowledgments
Thanks to Inger Böckerman (Norwegian Veterinary Institute, now Norwegian Institute of Public Health, Norway) for assistance in harvesting cells and establishing the ASH2–2 cell line, and the Norwegian Sequencing Centre at Ullevaal for eminent library prep and sequencing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1554680/full#supplementary-material
References
1. Takeuchi O and Akira S. Pattern recognition receptors and inflammation. Cell. (2010) 140:805–20. doi: 10.1016/j.cell.2010.01.022
2. Rehwinkel J and Gack MU. Rig-I-like receptors: their regulation and roles in rna sensing. Nat Rev Immunol. (2020) 20:537–51. doi: 10.1038/s41577-020-0288-3
3. Wang Y, Yuan S, Jia X, Ge Y, Ling T, Nie M, et al. Mitochondria-localised znfx1 functions as a dsrna sensor to initiate antiviral responses through mavs. Nat Cell Biol. (2019) 21:1346–56. doi: 10.1038/s41556-019-0416-0
4. Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, et al. The adaptor protein mita links virus-sensing receptors to irf3 transcription factor activation. Immunity. (2008) 29:538–50. doi: 10.1016/j.immuni.2008.09.003
5. Katze MG, He Y, and Gale M Jr. Viruses and interferon: A fight for supremacy. Nat Rev Immunol. (2002) 2:675–87. doi: 10.1038/nri888
6. Robertsen B, Greiner-Tollersrud L, and Jorgensen LG. Analysis of the atlantic salmon genome reveals a cluster of mx genes that respond more strongly to ifn gamma than to type I ifn. Dev Comp Immunol. (2019) 90:80–9. doi: 10.1016/j.dci.2018.09.004
7. Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. (1998) 17:7151–60. doi: 10.1093/emboj/17.24.7151
8. Redman KL and Rechsteiner M. Identification of the long ubiquitin extension as ribosomal protein S27a. Nature. (1989) 338:438–40. doi: 10.1038/338438a0
9. Cappadocia L and Lima CD. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem Rev. (2018) 118:889–918. doi: 10.1021/acs.chemrev.6b00737
10. Walters KJ, Goh AM, Wang Q, Wagner G, and Howley PM. Ubiquitin family proteins and their relationship to the proteasome: A structural perspective. Biochim Biophys Acta. (2004) 1695:73–87. doi: 10.1016/j.bbamcr.2004.10.005
11. Gu X, Nardone C, Kamitaki N, Mao A, Elledge SJ, and Greenberg ME. The midnolin-proteasome pathway catches proteins for ubiquitination-independent degradation. Science. (2023) 381:eadh5021. doi: 10.1126/science.adh5021
12. Makaros Y, Raiff A, Timms RT, Wagh AR, Gueta MI, Bekturova A, et al. Ubiquitin-independent proteasomal degradation driven by C-degron pathways. Mol Cell. (2023) 83:1921–35 e7. doi: 10.1016/j.molcel.2023.04.023
13. Collins GA and Goldberg AL. Proteins containing ubiquitin-like (Ubl) domains not only bind to 26s proteasomes but also induce their activation. Proc Natl Acad Sci U S A. (2020) 117:4664–74. doi: 10.1073/pnas.1915534117
14. Wang T, Jiang J, Zhang X, Ke X, and Qu Y. Ubiquitin-like modification dependent proteasomal degradation and disease therapy. Trends Mol Med. (2024) 30:1061. doi: 10.1016/j.molmed.2024.05.005
15. Lamotte LA and Tafforeau L. How influenza a virus ns1 deals with the ubiquitin system to evade innate immunity. Viruses. (2021) 13:2309. doi: 10.3390/v13112309
16. Zhang M, Li J, Yan H, Huang J, Wang F, Liu T, et al. Isgylation in innate antiviral immunity and pathogen defense responses: A review. Front Cell Dev Biol. (2021) 9:788410. doi: 10.3389/fcell.2021.788410
17. Perng YC and Lenschow DJ. Isg15 in antiviral immunity and beyond. Nat Rev Microbiol. (2018) 16:423–39. doi: 10.1038/s41579-018-0020-5
18. Thery F, Martina L, Asselman C, Zhang Y, Vessely M, Repo H, et al. Ring finger protein 213 assembles into a sensor for isgylated proteins with antimicrobial activity. Nat Commun. (2021) 12:5772. doi: 10.1038/s41467-021-26061-w
19. Kileng O, Brundtland MI, and Robertsen B. Infectious salmon anemia virus is a powerful inducer of key genes of the type I interferon system of atlantic salmon, but is not inhibited by interferon. Fish Shellfish Immunol. (2007) 23:378–89. doi: 10.1016/j.fsi.2006.11.011
20. Robertsen B. Expression of interferon and interferon-induced genes in salmonids in response to virus infection, interferon-inducing compounds and vaccination. Fish Shellfish Immunol. (2008) 25:351–7. doi: 10.1016/j.fsi.2008.02.004
21. Rokenes TP, Larsen R, and Robertsen B. Atlantic salmon isg15: expression and conjugation to cellular proteins in response to interferon, double-stranded rna and virus infections. Mol Immunol. (2007) 44:950–9. doi: 10.1016/j.molimm.2006.03.016
22. Olsen CM, Markussen T, Thiede B, and Rimstad E. Infectious salmon anaemia virus (Isav) rna binding protein encoded by segment 8 orf2 and its interaction with isav and intracellular proteins. Viruses. (2016) 8:52. doi: 10.3390/v8020052
23. Zheng N and Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. (2017) 86:129–57. doi: 10.1146/annurev-biochem-060815-014922
24. Shen Z, Wei L, Yu ZB, Yao ZY, Cheng J, Wang YT, et al. The roles of trims in antiviral innate immune signaling. Front Cell Infect Microbiol. (2021) 11:628275. doi: 10.3389/fcimb.2021.628275
25. Boudinot P, van der Aa LM, Jouneau L, Du Pasquier L, Pontarotti P, Briolat V, et al. Origin and evolution of trim proteins: new insights from the complete trim repertoire of zebrafish and pufferfish. PloS One. (2011) 6:e22022. doi: 10.1371/journal.pone.0022022
26. Carthagena L, Bergamaschi A, Luna JM, David A, Uchil PD, Margottin-Goguet F, et al. Human trim gene expression in response to interferons. PloS One. (2009) 4:e4894. doi: 10.1371/journal.pone.0004894
27. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. Trim25 ring-finger E3 ubiquitin ligase is essential for rig-I-mediated antiviral activity. Nature. (2007) 446:916–20. doi: 10.1038/nature05732
28. Koliopoulos MG, Lethier M, van der Veen AG, Haubrich K, Hennig J, Kowalinski E, et al. Molecular mechanism of influenza a ns1-mediated trim25 recognition and inhibition. Nat Commun. (2018) 9:1820. doi: 10.1038/s41467-018-04214-8
29. Langevin C, Levraud JP, and Boudinot P. Fish antiviral tripartite motif (Trim) proteins. Fish Shellfish Immunol. (2019) 86:724–33. doi: 10.1016/j.fsi.2018.12.008
30. van der Aa LM, Jouneau L, Laplantine E, Bouchez O, Van Kemenade L, and Boudinot P. Fintrims, fish virus-inducible proteins with E3 ubiquitin ligase activity. Dev Comp Immunol. (2012) 36:433–41. doi: 10.1016/j.dci.2011.08.010
31. van der Aa LM, Levraud JP, Yahmi M, Lauret E, Briolat V, Herbomel P, et al. A large new subset of trim genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. (2009) 7:7. doi: 10.1186/1741-7007-7-7
32. Verrier ER, Genet C, Laloe D, Jaffrezic F, Rau A, Esquerre D, et al. Genetic and transcriptomic analyses provide new insights on the early antiviral response to vhsv in resistant and susceptible rainbow trout. BMC Genomics. (2018) 19:482. doi: 10.1186/s12864-018-4860-1
33. Zhang Y, Yuan Y, Jiang L, Liu Y, and Zhang L. The emerging role of E3 ubiquitin ligase rnf213 as an antimicrobial host determinant. Front Cell Infect Microbiol. (2023) 13:1205355. doi: 10.3389/fcimb.2023.1205355
34. Houzelstein D, Simon-Chazottes D, Batista L, Tokuda S, Langa Vives F, Flamand M, et al. The ring finger protein 213 gene (Rnf213) contributes to rift valley fever resistance in mice. Mamm Genome. (2021) 32:30–7. doi: 10.1007/s00335-020-09856-y
35. Levraud JP, Jouneau L, Briolat V, Laghi V, and Boudinot P. Ifn-stimulated genes in zebrafish and humans define an ancient arsenal of antiviral immunity. J Immunol. (2019) 203:3361–73. doi: 10.4049/jimmunol.1900804
36. Wang W, Jia M, Zhao C, Yu Z, Song H, Qin Y, et al. Rnf39 mediates K48-linked ubiquitination of ddx3x and inhibits rlr-dependent antiviral immunity. Sci Adv. (2021) 7. doi: 10.1126/sciadv.abe5877
37. Rotin D and Kumar S. Physiological functions of the hect family of ubiquitin ligases. Nat Rev Mol Cell Biol. (2009) 10:398–409. doi: 10.1038/nrm2690
38. Hochrainer K, Mayer H, Baranyi U, Binder B, Lipp J, and Kroismayr R. The human herc family of ubiquitin ligases: novel members, genomic organization, expression profiling, and evolutionary aspects. Genomics. (2005) 85:153–64. doi: 10.1016/j.ygeno.2004.10.006
39. Mathieu NA, Paparisto E, Barr SD, and Spratt DE. Herc5 and the isgylation pathway: critical modulators of the antiviral immune response. Viruses. (2021) 13:1102. doi: 10.3390/v13061102
40. Oudshoorn D, van Boheemen S, Sanchez-Aparicio MT, Rajsbaum R, Garcia-Sastre A, and Versteeg GA. Herc6 is the main E3 ligase for global isg15 conjugation in mouse cells. PloS One. (2012) 7:e29870. doi: 10.1371/journal.pone.0029870
41. Zhao X, Perez JM, Faull PA, Chan C, Munting FW, Canadeo LA, et al. Cellular targets and lysine selectivity of the herc5 isg15 ligase. iScience. (2024) 27:108820. doi: 10.1016/j.isci.2024.108820
42. Tang Y, Zhong G, Zhu L, Liu X, Shan Y, Feng H, et al. Herc5 attenuates influenza a virus by catalyzing isgylation of viral ns1 protein. J Immunol. (2010) 184:5777–90. doi: 10.4049/jimmunol.0903588
43. Li YL, Gong XY, Qu ZL, Zhao X, Dan C, Gui JF, et al. A novel non-mammalian-specific herc7 negatively regulates ifn response through degrading rlr signaling factors. J Immunol. (2022) 208:1189–203. doi: 10.4049/jimmunol.2100962
44. Li YL, Gong XY, Qu ZL, Zhao X, Dan C, Sun HY, et al. Zebrafish herc7c acts as an inhibitor of fish ifn response. Int J Mol Sci. (2023) 24:4592. doi: 10.3390/ijms24054592
45. Wenzel DM, Lissounov A, Brzovic PS, and Klevit RE. Ubch7 reactivity profile reveals parkin and hhari to be ring/hect hybrids. Nature. (2011) 474:105–8. doi: 10.1038/nature09966
46. Smit JJ and Sixma TK. Rbr E3-ligases at work. EMBO Rep. (2014) 15:142–54. doi: 10.1002/embr.201338166
47. Wang S, Li Z, Chen Y, Gao S, Qiao J, Liu H, et al. Arih1 inhibits influenza a virus replication and facilitates rig-I dependent immune signaling by interacting with sqstm1/P62. Virol J. (2023) 20:58. doi: 10.1186/s12985-023-02022-1
48. Dikic I and Schulman BA. An expanded lexicon for the ubiquitin code. Nat Rev Mol Cell Biol. (2023) 24:273–87. doi: 10.1038/s41580-022-00543-1
49. Martinez-Fonts K, Davis C, Tomita T, Elsasser S, Nager AR, Shi Y, et al. The proteasome 19s cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat Commun. (2020) 11:477. doi: 10.1038/s41467-019-13906-8
50. Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. (2009) 85:12–36. doi: 10.2183/pjab.85.12
51. Ferrington DA and Gregerson DS. Immunoproteasomes: structure, function, and antigen presentation. Prog Mol Biol Transl Sci. (2012) 109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1
52. McConnell SC, Hernandez KM, Wcisel DJ, Kettleborough RN, Stemple DL, Yoder JA, et al. Alternative haplotypes of antigen processing genes in zebrafish diverged early in vertebrate evolution. Proc Natl Acad Sci U S A. (2016) 113:E5014–23. doi: 10.1073/pnas.1607602113
53. Grimholt U. Whole genome duplications have provided teleosts with many roads to peptide loaded mhc class I molecules. BMC Evol Biol. (2018) 18:25. doi: 10.1186/s12862-018-1138-9
54. Basters A, Knobeloch KP, and Fritz G. How usp18 deals with isg15-modified proteins: structural basis for the specificity of the protease. FEBS J. (2018) 285:1024–9. doi: 10.1111/febs.14260
55. Ketscher L, Hannss R, Morales DJ, Basters A, Guerra S, Goldmann T, et al. Selective inactivation of usp18 isopeptidase activity in vivo enhances isg15 conjugation and viral resistance. Proc Natl Acad Sci U S A. (2015) 112:1577–82. doi: 10.1073/pnas.1412881112
56. Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, and Zhang DE. Ubp43 (Usp18) specifically removes isg15 from conjugated proteins. J Biol Chem. (2002) 277:9976–81. doi: 10.1074/jbc.M109078200
57. Neefjes J, Jongsma ML, Paul P, and Bakke O. Towards a systems understanding of mhc class I and mhc class ii antigen presentation. Nat Rev Immunol. (2011) 11:823–36. doi: 10.1038/nri3084
58. Hammer GE, Gonzalez F, James E, Nolla H, and Shastri N. In the absence of aminopeptidase eraap, mhc class I molecules present many unstable and highly immunogenic peptides. Nat Immunol. (2007) 8:101–8. doi: 10.1038/ni1409
59. Morozov GI, Zhao H, Mage MG, Boyd LF, Jiang J, Dolan MA, et al. Interaction of tapbpr, a tapasin homolog, with mhc-I molecules promotes peptide editing. Proc Natl Acad Sci U S A. (2016) 113:E1006–15. doi: 10.1073/pnas.1519894113
60. Neerincx A, Hermann C, Antrobus R, van Hateren A, Cao H, Trautwein N, et al. Tapbpr bridges udp-glucose: glycoprotein glucosyltransferase 1 onto mhc class I to provide quality control in the antigen presentation pathway. Elife. (2017). doi: 10.7554/eLife.23049
61. Boyle LH, Hermann C, Boname JM, Porter KM, Patel PA, Burr ML, et al. Tapasin-related protein tapbpr is an additional component of the mhc class I presentation pathway. Proc Natl Acad Sci U S A. (2013) 110:3465–70. doi: 10.1073/pnas.1222342110
62. Lukacs MF, Harstad H, Bakke HG, Beetz-Sargent M, McKinnel L, Lubieniecki KP, et al. Comprehensive analysis of mhc class I genes from the U-, S-, and Z-lineages in atlantic salmon. BMC Genomics. (2010) 11:154. doi: 10.1186/1471-2164-11-154
63. Clouthier SC, Rector T, Brown NEC, and Anderson ED. Genomic organization of infectious salmon anaemia virus. J Gen Virol. (2002) 83:421–8. doi: 10.1099/0022-1317-83-2-421
64. Fosse JH, Haraldsen G, Falk K, and Edelmann R. Endothelial cells in emerging viral infections. Front Cardiovasc Med. (2021) 8:619690. doi: 10.3389/fcvm.2021.619690
65. Aamelfot M, Dale OB, Weli SC, Koppang EO, and Falk K. Expression of the infectious salmon anemia virus receptor on atlantic salmon endothelial cells correlates with the cell tropism of the virus. J Virol. (2012) 86:10571–8. doi: 10.1128/JVI.00047-12
66. Aquatic Manual Online Access. World Organisation for Animal Health (2024). Available online at: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/aquatic-manual-online-access/ (Accessed February 26, 2025).
67. Dannevig BH, Falk K, and Press CM. Propagation of infectious salmon anaemia (Isa) virus in cell culture. Vet Res. (1995) 26:438–42.
68. Kärber G. Beitrag zur kollektiven behandlung pharmakologiseher reihenversuche. Arch Exp Path Pharmacol. (1931) 162:4. doi: 10.1007/BF01863914
69. Grimholt U, Fosse JH, and Sundaram AYM. Selective stimulation of duplicated atlantic salmon mhc pathway genes by interferon-gamma. Front Immunol. (2020) 11:571650. doi: 10.3389/fimmu.2020.571650
70. Sun B, Skjaeveland I, Svingerud T, Zou J, Jorgensen J, and Robertsen B. Antiviral activity of salmonid gamma interferon against infectious pancreatic necrosis virus and salmonid alphavirus and its dependency on type I interferon. J Virol. (2011) 85:9188–98. doi: 10.1128/JVI.00319-11
71. Bushnell B. Bbmap: A fast, accurate, splice-aware aligner. (2014) Berkeley, CA., USA: Lawrence Berkeley National Lab. Available at: https://www.osti.gov/biblio/1241166 (Accessed January 2024).
72. Kim D, Langmead B, and Salzberg SL. Hisat: A fast spliced aligner with low memory requirements. Nat Methods. (2015) 12:357–60. doi: 10.1038/nmeth.3317
73. Liao Y, Smyth GK, and Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. (2014) 30:923–30. doi: 10.1093/bioinformatics/btt656
74. Love MI, Huber W, and Anders S. Moderated estimation of fold change and dispersion for rna-seq data with deseq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
75. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and clustal X version 2.0. Bioinformatics. (2007) 23:2947–8. doi: 10.1093/bioinformatics/btm404
76. Kumar S, Stecher G, and Tamura K. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. (2016) 33:1870–4. doi: 10.1093/molbev/msw054
77. Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, et al. The atlantic salmon genome provides insights into rediploidization. Nature. (2016) 533:200–5. doi: 10.1038/nature17164
78. Andresen AMS, Taylor RS, Grimholt U, Daniels RR, Sun J, Dobie R, et al. Mapping the cellular landscape of atlantic salmon head kidney by single cell and single nucleus transcriptomics. Fish Shellfish Immunol. (2024) 146:109357. doi: 10.1016/j.fsi.2024.109357
79. Gaucherand L and Gaglia MM. The role of viral rna degrading factors in shutoff of host gene expression. Annu Rev Virol. (2022) 9:213–38. doi: 10.1146/annurev-virology-100120-012345
80. Schneider WM, Chevillotte MD, and Rice CM. Interferon-stimulated genes: A complex web of host defenses. Annu Rev Immunol. (2014) 32:513–45. doi: 10.1146/annurev-immunol-032713-120231
81. Robertsen B, Trobridge G, and Leong JA. Molecular cloning of double-stranded rna inducible mx genes from atlantic salmon (Salmo salar L.). Dev Comp Immunol. (1997) 21:397–412. doi: 10.1016/s0145-305x(97)00019-0
82. Diamond MS and Farzan M. The broad-spectrum antiviral functions of ifit and ifitm proteins. Nat Rev Immunol. (2013) 13:46–57. doi: 10.1038/nri3344
83. Andresen AMS, Boudinot P, and Gjoen T. Kinetics of transcriptional response against poly (I:C) and infectious salmon anemia virus (Isav) in atlantic salmon kidney (Ask) cell line. Dev Comp Immunol. (2020) 110:103716. doi: 10.1016/j.dci.2020.103716
84. Gervais O, Penaloza C, Gratacap R, Papadopoulou A, Beltran M, Henderson NC, et al. Understanding host response to infectious salmon anaemia virus in an atlantic salmon cell line using single-cell rna sequencing. BMC Genomics. (2023) 24:161. doi: 10.1186/s12864-023-09254-z
85. Kibenge MJ, Munir K, and Kibenge FS. Constitutive expression of atlantic salmon mx1 protein in chse-214 cells confers resistance to infectious salmon anaemia virus. Virol J. (2005) 2:75. doi: 10.1186/1743-422X-2-75
86. Schiotz BL, Jorgensen SM, Rexroad C, Gjoen T, and Krasnov A. Transcriptomic analysis of responses to infectious salmon anemia virus infection in macrophage-like cells. Virus Res. (2008) 136:65–74. doi: 10.1016/j.virusres.2008.04.019
87. Workenhe ST, Hori TS, Rise ML, Kibenge MJ, and Kibenge FS. Infectious salmon anaemia virus (Isav) isolates induce distinct gene expression responses in the atlantic salmon (Salmo salar) macrophage/dendritic-like cell line to, assessed using genomic techniques. Mol Immunol. (2009) 46:2955–74. doi: 10.1016/j.molimm.2009.06.015
88. Xu C, Evensen O, and Munang’andu HM. Transcriptome analysis shows that ifn-I treatment and concurrent sav3 infection enriches mhc-I antigen processing and presentation pathways in atlantic salmon-derived macrophage/dendritic cells. Viruses. (2019) 11:464. doi: 10.3390/v11050464
89. Chang M, Collet B, Nie P, Lester K, Campbell S, Secombes CJ, et al. Expression and functional characterization of the rig-I-like receptors mda5 and lgp2 in rainbow trout (Oncorhynchus mykiss). J Virol. (2011) 85:8403–12. doi: 10.1128/JVI.00445-10
90. Nerbovik IG, Solheim MA, Eggestol HO, Ronneseth A, Jakobsen RA, Wergeland HI, et al. Molecular cloning of mda5, phylogenetic analysis of rig-I-like receptors (Rlrs) and differential gene expression of rlrs, interferons and proinflammatory cytokines after in vitro challenge with ipnv, isav and sav in the salmonid cell line to. J Fish Dis. (2017) 40:1529–44. doi: 10.1111/jfd.12622
91. Liu G, Lee JH, Parker ZM, Acharya D, Chiang JJ, van Gent M, et al. Isg15-dependent activation of the sensor mda5 is antagonized by the sars-cov-2 papain-like protease to evade host innate immunity. Nat Microbiol. (2021) 6:467–78. doi: 10.1038/s41564-021-00884-1
92. Krishna Priya RS, Premraj A, Sivakumar KC, and Sajeevan TP. Identification of two isg15 homologues involved in host immune response against rgnnv in asian seabass (Lates calcarifer). Fish Shellfish Immunol Rep. (2022) 3:100054. doi: 10.1016/j.fsirep.2022.100054
93. Liu W, Xiang Y, Zhang W, Jia P, Yi M, and Jia K. Expression pattern, antiviral role and regulation analysis of interferon-stimulated gene 15 in black seabream, acanthopagrus schlegelii. Fish Shellfish Immunol. (2018) 82:60–7. doi: 10.1016/j.fsi.2018.07.041
94. Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, et al. Influenza a virus ns1 targets the ubiquitin ligase trim25 to evade recognition by the host viral rna sensor rig-I. Cell Host Microbe. (2009) 5:439–49. doi: 10.1016/j.chom.2009.04.006
95. Briolat V, Jouneau L, Carvalho R, Palha N, Langevin C, Herbomel P, et al. Contrasted innate responses to two viruses in zebrafish: insights into the ancestral repertoire of vertebrate ifn-stimulated genes. J Immunol. (2014) 192:4328–41. doi: 10.4049/jimmunol.1302611
96. Xu C, Evensen O, and Munang’andu HM. De novo transcriptome analysis shows that sav-3 infection upregulates pattern recognition receptors of the endosomal toll-like and rig-I-like receptor signaling pathways in macrophage/dendritic like to-cells. Viruses. (2016) 8:114. doi: 10.3390/v8040114
97. Kotani Y, Morito D, Yamazaki S, Ogino K, Kawakami K, Takashima S, et al. Neuromuscular regulation in zebrafish by a large aaa+ Atpase/ubiquitin ligase, mysterin/rnf213. Sci Rep. (2015) 5:16161. doi: 10.1038/srep16161
98. Dannevig BH, Brudeseth BE, Gjoen T, Rode M, Wergeland HI, Evensen O, et al. Characterisation of a long-term cell line (Shk-1) developed from the head kidney of atlantic salmon (Salmo salar L). Fish Shellfish Immun. (1997) 7:213–26. doi: 10.1006/fsim.1996.0076
99. Kim JO, Kim JO, Kim WS, and Oh MJ. Characterization of the transcriptome and gene expression of brain tissue in sevenband grouper (Hyporthodus septemfasciatus) in response to nnv infection. Genes (Basel). (2017) 8:31. doi: 10.3390/genes8010031
100. Paparisto E, Woods MW, Coleman MD, Moghadasi SA, Kochar DS, Tom SK, et al. Evolution-guided structural and functional analyses of the herc family reveal an ancient marine origin and determinants of antiviral activity. J Virol. (2018) 92. doi: 10.1128/JVI.00528-18
101. Mladek C, Guger K, and Hauser MT. Identification and characterization of the ariadne gene family in arabidopsis. A Group of Putative E3 Ligases. Plant Physiol. (2003) 131:27–40. doi: 10.1104/pp.012781
102. Marin I, Lucas JI, Gradilla AC, and Ferrus A. Parkin and relatives: the rbr family of ubiquitin ligases. Physiol Genomics. (2004) 17:253–63. doi: 10.1152/physiolgenomics.00226.2003
103. Xiong TC, Wei MC, Li FX, Shi M, Gan H, Tang Z, et al. The E3 ubiquitin ligase arih1 promotes antiviral immunity and autoimmunity by inducing mono-isgylation and oligomerization of cgas. Nat Commun. (2022) 13:5973. doi: 10.1038/s41467-022-33671-5
104. Basters A, Knobeloch KP, and Fritz G. Usp18 - a multifunctional component in the interferon response. Biosci Rep. (2018) 38. doi: 10.1042/BSR20180250
105. Honke N, Shaabani N, Zhang DE, Hardt C, and Lang KS. Multiple functions of usp18. Cell Death Dis. (2016) 7:e2444. doi: 10.1038/cddis.2016.326
106. Broering R, Trippler M, Werner M, Real CI, Megger DA, Bracht T, et al. Hepatic expression of proteasome subunit alpha type-6 is upregulated during viral hepatitis and putatively regulates the expression of isg15 ubiquitin-like modifier, a proviral host gene in hepatitis C virus infection. J Viral Hepat. (2016) 23:375–86. doi: 10.1111/jvh.12508
107. Valero Y, Chaves-Pozo E, and Cuesta A. Fish herc7: phylogeny, characterization, and potential implications for antiviral immunity in european sea bass. Int J Mol Sci. (2024) 25:7751. doi: 10.3390/ijms25147751
108. Jones DT, Taylor WR, and Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. (1992) 8:275–82. doi: 10.1093/bioinformatics/8.3.275
109. Whelan S and Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. (2001) 18:691–9. doi: 10.1093/oxfordjournals.molbev.a003851
Keywords: Atlantic salmon, ISAV, IFN gamma, ISG15, ISGylation, E3 ubiquitin ligases, HERC, RBR family
Citation: Grimholt U, Sindre H and Sundaram AYM (2025) ISGylation and E3 ubiquitin ligases: an Atlantic salmon genetic perspective. Front. Immunol. 16:1554680. doi: 10.3389/fimmu.2025.1554680
Received: 02 January 2025; Accepted: 15 May 2025;
Published: 24 June 2025.
Edited by:
Jia Cai, Guangdong Ocean University, ChinaReviewed by:
Johannes M. Dijkstra, Fujita Health University, JapanVenkatesh Kumaresan, University of Texas at San Antonio, United States
Copyright © 2025 Grimholt, Sindre and Sundaram. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Unni Grimholt, dW5uaS5ncmltaG9sdEB2ZXRpbnN0Lm5v
 Unni Grimholt
Unni Grimholt Hilde Sindre
Hilde Sindre Arvind Y. M. Sundaram
Arvind Y. M. Sundaram