- 1College of Bioscience and Biotechnology, Shenyang Agricultural University, Shenyang, Liaoning, China
- 2Key Laboratory of Mariculture & Stock Enhancement in North China’s Sea, Ministry of Agriculture and Rural Affairs, Dalian Ocean University, Dalian, Liaoning, China
To clarify the microRNA (miRNA)-target gene axis is involved in response to pathogen-associated molecular pattern (PAMP)-induced oxidative stress in shellfish, the full-length cDNA of a novel mitogen-activated protein kinase-interacting kinase 1 (MNK1) homolog gene from the scallop Patinopecten yessoensis (PyMNK1) was cloned and characterized. The interaction between miR-1985 and PyMNK1 was verified, and then the responses and possible molecular function of miR-1985, PyMNK1, and miR-1985/PyMNK1 axis to poly(I:C) (a classic virus-related PAMP) stimulation in P. yessoensis were explored and preliminarily dissected. The results indicate: 1) The full-length cDNA of PyMNK1 was 5354 bp, with a high level of sequence conservation across mollusks. 2) MiR-1985 bound to the 3’-UTR of PyMNK1 and negatively regulated the expression of PyMNK1. 3) PyMNK1 may repress the relative expression of superoxide dismutase (SOD) by binding its promoter. 4) Both PyMNK1 silencing and miR-1985 overexpression promoted the expression and enzymatic activity of SOD. 5) The miR-1985/PyMNK1 axis may be involved in the response to poly(I:C) stimulation by elevating the activity of the SOD/catalase axis. To summarize, all observations from this study indicated that P. yessoensis may enhance its redox capability via the miR-1985/PyMNK1/SOD/CAT cascade and thereby alleviate PAMP-induced oxidative stress.
Highlights
● A targeting relationship between miR-1985 and PyMNK1 was verified.
● PyMNK1 regulates superoxide dismutase (SOD) transcription by promoter binding.
● Both PyMNK1 and miR-1985 may affect the expression and enzymatic activity of SOD.
● MiR-1985 enhances the redox capability in response to poly(I:C) stimulation by targeting PyMNK1.
● MiR-1985/PyMNK1 axis may regulate poly(I:C)-induced redox homeostasis alteration by targeting SOD.
1 Introduction
Pathogen-associated molecular patterns (PAMPs) are a class of molecules structurally conserved among many pathogens that elicit a host immune response when bound by germline-encoded host receptors known as pattern-recognition receptors (PRRs) (1). Upon activation by PAMPs, PRR recognition and metabolic reprogramming occurs in activated innate immune cells, which typically increases production of reactive oxygen species (ROS) (2). Although excessive ROS may help the host to eradicate pathogens to some extent, oxidative stress induced by increased ROS production may cause molecular damage, biochemical process disruptions, and cellular dysfunction in the host (3). Therefore, eliminating PAMP-induced oxidative stress as soon as possible while defending against pathogens is an important tradeoff of host innate immune regulation.
MicroRNAs (miRNAs) are a class of small endogenous non-coding RNAs with a length of 18–25 nt that are ubiquitously expressed in metazoan organisms (4). Generally, miRNAs exert their regulatory function by binding to specific sites of their target genes, leading to altered expression of these genes (5). In recent years, the regulatory role of miRNA-target gene pairs has attracted increasing attention, particularly for elucidating the mechanisms of innate immunity regulation and anti-stress responses in many organisms (6, 7). However, the extent to which miRNAs regulate innate immunity and anti-stress responses remains poorly defined, and the regulatory signaling cascades and axes remain unclear.
Shellfish is a common model species for innate immunity research (8), and a high-value seafood with notable popularity worldwide (9). Recently, several studies hypothesized that miR-1985 might be involved in the innate immune response in Biomphalaria tenagophila (10), Argopecten irradians (11), and Pinctada fucata martensii (12). Combined with our preliminary bioinformatics prediction data indicating that miR-1985 may target mitogen-activated protein kinase-interacting kinase 1 (MNK1) in Patinopecten yessoensis, we thus hypothesized that miR-1985 might play a role in the innate immune response in P. yessoensis. To address this hypothesis, we first cloned and characterized the full-length cDNA of an MNK1 homolog from P. yessoensis (PyMNK1) and then verified the interaction between miR-1985 and PyMNK1. Polyinosinic–polycytidylic acid [poly(I:C)] is a synthetic analog of double-stranded RNA (dsRNA) which is a common innate immune stimulator in aquatic animals (such as Danio rerio, Strongylocentrotus intermedius, Scylla paramamosain, Chlamys farreri, Apostichopus japonicus, and Mytilus galloprovincialis) (13–18), we therefore employed poly(I:C) as a virus-mimetics to activate the innate immune system in this study, and the response of both the miR-1985/PyMNK1 axis and its downstream targets were investigated during the stimulation process subsequently. The observations in this study enrich the information pool of miRNA-target gene pairs and their functions, and also provide new clues for elucidating the tradeoff strategy of synchronous pathogen elimination and oxidative stress attenuation during an innate immune response in a host.
2 Materials and methods
2.1 Experimental animals and cell lines
Two hundred 12-month P. yessoensis individuals were collected from the Dalian Lvshun Longwangtang Farm (Dalian, Liaoning Province, China). Prior to experimentation, all specimens were maintained in 1000-L laboratory circulating seawater tanks without feeding for 3 days. P. yessoensis individuals were fed Chlorella vulgaris algae twice a day in vivo experiments following the method of Metian et al. (19). The specimens were temporarily incubated in freshly filtered seawater maintained at 15.0°C, with a salinity of 31.21 ± 0.20, a pH of 8.10 ± 0.03, and a dissolved oxygen concentration of 7.88 mg/L. The water was changed every 2 days.
Human embryonic kidney cells transformed with SV40 large T-antigen (HEK-293T) were used as the vector for verifying the binding sites between the miRNA and its target gene, as these cells exhibit rapid growth, robustness, and strong bioluminescent signals (20). The maintenance of HEK-293T cells (Sangon Biotechnology; Shanghai, China) followed the method of Liu et al. (21).
2.2 Sample collection
Nine healthy adult P. yessoensis were starved for 3 days. Tissues (gills, mantles, hepatopancreas, hemolymph, gonads, muscles) were then carefully collected on ice. The collection of hepatopancreases followed the method of Lyu et al. (22). To collect hemolymph, lymphocytes were centrifuged at 3000 rpm for 10 min at 4°C. All samples were immediately frozen in liquid nitrogen and stored at −80°C for subsequent total RNA extraction, genomic DNA extraction, and expression analysis.
2.3 Full length cDNA cloning of PyMNK1
Total RNA extraction, the determination of RNA integrity and concentration, synthesis of the first-strand cDNA, and rapid amplification of cDNA ends (RACE) were performed according to Ren et al. (23). The obtained cDNA was diluted and stored at −20°C. A SMARTer® RACE 5’/3’ kit (Sangon Biotechnology, China) was used to synthesize a 5’/3’ terminal RACE cDNA template via qualified total RNA synthesis. The reaction and PCR procedure were performed according to the manufacturer’s instructions. The primers used for full length cDNA cloning are listed in Table 1. PCR products were recovered and purified using a SanPrep column DNA gel extraction kit (Sangon Biotechnology, China). The ligation, transformation, and colony PCR experiments were performed according to Ren et al. (23). After colony PCR screening, three positive clones were selected and sent to Sangon Biotechnology (Shanghai, China) for sequencing.
2.4 Superoxide dismutase promoter cloning
Genomic DNA was extracted from gills of P. yessoensis using a DNeasy blood and tissue kit (Qiagen, Germany), following the method of Yoon et al. (24). The DNA sequences for the SOD promoter were cloned based on the National Center for Biotechnology Information database (GenBank No.: 110461622). Primer 5.0 software was used to design primers for sequence amplification of the SOD gene promoter (Table 1). The ligation, transformation, and colony PCR experiments were performed according to Ren et al. (23). All primer synthesis and sequencing were completed by Sangon Biotechnology (Shanghai, China).
2.5 Characterization of PyMNK1 and the SOD promoter sequence
Analyses to characterize MNK1 were performed following the methods of Liu et al. (21), including complete full-length cDNA sequence assembly and annotation, sequence structure analyses, physicochemical property predictions, multiple sequence alignment, and phylogenetic analysis. The information used for the multiple sequence alignment, characteristic domain comparison, and phylogenetic analysis is shown in Supplementary Table S1.
A potential transcription start site of the SOD gene was predicted by Promoter 2.0 (https://services.healthtech.dtu.dk/services/Promoter-2.0/). An analysis of the CpG islands in the SOD gene promoter sequence was performed by Methprimer (http://urogene.org/cgi-bin/methprimer/methprimer.cgi) using the parameters length > 100 bp, Obs/Exp > 0.6, and GC% > 50%.
2.6 Quantitative real-time reverse transcription-polymerase chain reaction and western blotting
The relative expression of miR-1985, PyMNK1, and SOD was analyzed using a LightCycler® 96 real-time quantitative PCR instrument (Roche Life Science, Germany). The primers used for qRT-PCR are listed in Table 1. β-actin and RNAU6B (U6) were used as internal references because they are commonly used in relative transcriptional expression analyses of aquatic animals (25). The relative expression levels of miR-1985, PyMNK1, and SOD were analyzed by the 2−ΔΔCt method (25).
The recombinant protein expression of PyMNK1, polyclonal antibody preparation, and antibody specificity testing were completed at Mai-si Biotechnology (Wuhan, China). Total protein concentration of each sample was determined following the method of Liu et al. (21). Detailed experimental protocol of western blotting was described in the Supplementary File 1.
2.7 Binding site predictions and verification of the miR-1985/PyMNK1 axis and PyMNK1/SOD promoter
The target site of miR-1985 in PyMNK1 was identified by using RNA22 v2 microRNA target detection (https://cm.jefferson.edu/rna22/Interactive/) and artificial sequence alignment (Sangon Biotechnology; Shanghai, China). Plasmids expressing the wild-type (WT) PyMNK1 containing a putative target of miR-1985 on the 3’- untranslated region (3’-UTR) and PyMNK1 with a mutated 3’-UTR were synthesized by Sangon Biotechnology (Shanghai, China) (Table 1). The target fragment was inserted between the XhoI and NotI restriction sites of a luciferase plasmid, and the clones were further confirmed by sequencing. In a transfection experiment, HEK-293T cells were inoculated in 96-well white TC plates with a total volume of 100 μL. Then, two solutions were prepared: the first consisted of DMEM (10 µL); either the pmirGLO luciferase plasmid (0.16 µg), or the wild-type PyMNK1 plasmid, or the mutant-type (MT) PyMNK1 3’-UTR plasmid (200 ng); and the miRNA/negative control (NC; 5 pmol). The second solution consisted of 10 μL of DMEM and 0.3 μL of Lipofectamine 2000 (Thermo Fisher Scientific, USA), and was incubated at room temperature for 5 min. Then, 25 μL of the first and 25 μL of the second solutions were combined and incubated at room temperature for 20 min. Then, 100 μL of the medium was added to each well of the 96-well white TC plate containing HEK-293T cells. After transfection (48 h), the cells were collected, and the activity was determined using a dual luciferase reporter assay system (E1980, Promega Biotechnology, Beijing). The luciferase activity was determined by calculating the signal ratio between Renilla luciferase and firefly luciferase using a SpectraMax i3x enzyme marker (Mei Ya Molecular Instruments, China). All experiments were performed with three replicates.
The binding sites of the promoters of SOD and PyMNK1 were identified by using JASPAR (http://jaspar.genereg.net/) and artificial sequence alignment (Sangon Biotechnology; Shanghai, China). Plasmids expressing the wild-type promoter of SOD containing the putative target of PyMNK1 and wild-type PyMNK1 were synthesized by Sangon Biotechnology (Shanghai, China). The target fragment was inserted between the XhoI and NotI sites of the luciferase plasmid, and the clones were further confirmed by sequencing. In a transfection experiment, HEK-293T cells were incubated in 96-well white TC plates with a total volume of 100 μL. Two solutions were prepared: the first consisted of 10 µL of DMEM, 0.16 µg of the pmirGLO luciferase plasmid or 200 ng of the wild-type SOD plasmids or 200 ng of the wild-type PyMNK1 and SOD plasmids, and 5 pmol of the pGL3-control plasmid. Preparation of the second solution and luciferase activity determination are the same as binding site verification of the miR-1985/PyMNK1 axis.
2.8 Functional analysis of miR-1985 and PyMNK1 in vivo
The miR-1985 miRNA agomir, antagomir, and NC were designed and synthesized at Gene-Pharma (Shanghai, China; Table 1) and dissolved in RNase-free water to obtain a working solution of 20 nM. Then, 10 μL of the miR-1985 agomir (or antagomir) or NC (or NC inhibitor) was mixed with 10 μL of Lipofectamine 2000 and 80 μL of phosphate buffered saline (PBS) to serve as the transfection solution. Healthy P. yessoensis were injected with 100 μL of transfection solution or the NC solution mixture. At 24 h post transfection, the gills from each group were collected, immediately frozen in liquid nitrogen, and stored at − 80°C for further analyses, including qRT-PCR, western blotting, SOD and catalase (CAT) enzyme activity assays, and ROS content determination.
Specific small interfering RNAs (siRNAs) targeting PyMNK1 (siMNK1) and RNA interference negative controls (NC-RNAi) were designed and synthesized by Sangon Biotech (Shanghai, China; Table 1). For in vivo PyMNK1 knockdown, 10 μL of siMNK1 (20 nM) or NC-RNAi was mixed with 10 μL of Lipofectamine 2000 and 80 μL of PBS to serve as the transfection solution. Healthy P. yessoensis were injected with 100 μL of the siMNK1 mixture or the NC-RNAi mixture (as a control). The physiological status of all examined P. yessoensis individuals should be the same before and after injection to avoid the stress reaction caused by injection. At 24 h post transfection, the gills from the siMNK1 group and the control group were collected, immediately frozen in liquid nitrogen, and stored at −80°C for further analyses, including qRT-PCR, western blotting, SOD and CAT enzyme activity assay, and ROS content determination.
2.9 Poly(I:C) stimulation and sample collection
A total of 60 healthy P. yessoensis were selected and injected intramuscularly with sterile seawater containing 50 μL of poly(I:C) (InvivoGen, USA; 1.0 mg/mL in sterile seawater) following the method of Yang et al. (26). The injection operation of the P. yessoensis was shown in Supplementary Figure S1. Three individuals from each group were randomly selected at 0 (as control), 6, 12, 24, 48, and 72 h post stimulation (hps) (n = 3). The gills of each individual in each group were collected, immediately frozen in liquid nitrogen, and stored at −80°C for further analyses, including qRT-PCR, western blotting, SOD and CAT enzyme activity assay, and ROS content determination.
2.10 ROS content and enzyme activity determination
Prior to ROS content and enzyme activity analyses, gill samples were homogenized and the protein concentration of the gill homogenates was determined followed the method of Liu et al. (21). ROS content determination was carried out by using a commercial kit (Beyotime Biotechnology, Shanghai, China) following the method of Hao et al. (27). Enzyme activity analyses of SOD and CAT were carried out using commercial kits (BC5160, Solarbio Science & Technology Co., Ltd, Beijing, China; S0038, Beyotime Biotechnology, Shanghai, China) following the method of Yu et al. (28) and Zhang et al. (29).
2.11 Data analysis
All data were expressed as mean ± S.D. Prism 9 (GraphPad, USA) was used for data processing, data collation, and chart drawing. Statistical analysis was performed using independent samples t-tests and one-way analysis of variance (ANOVA) in Prism 9. Significant differences and extremely significant differences were set at P < 0.05 and P < 0.01, respectively.
3 Results
3.1 PyMNK1 cDNA sequence characterization
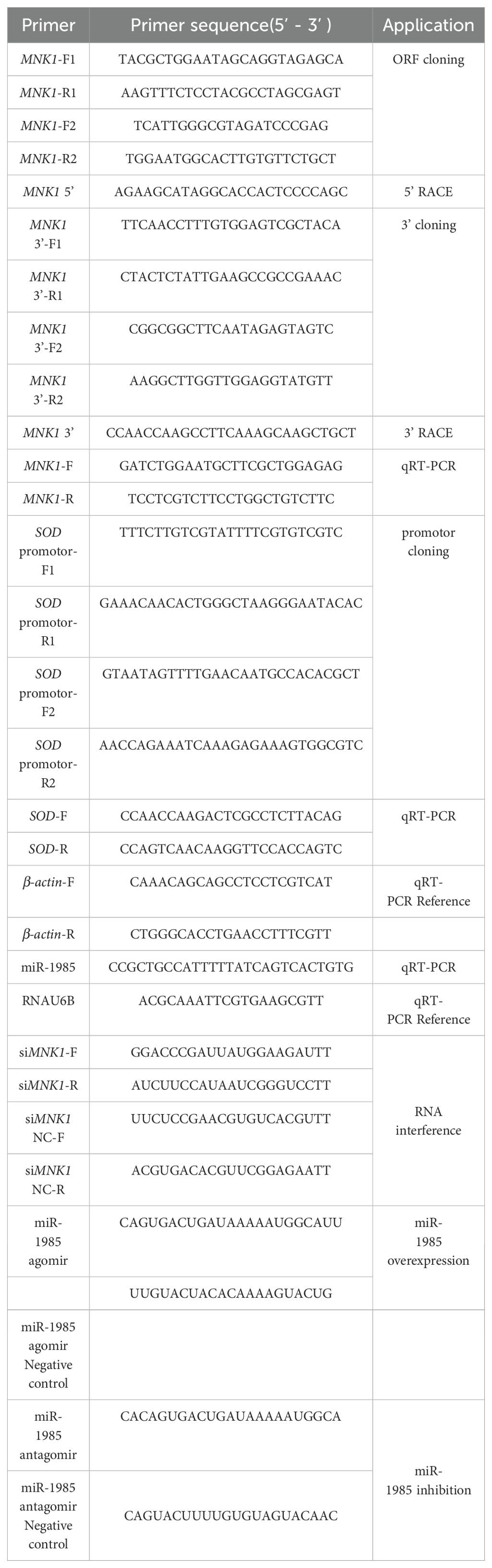
As shown in the results, the cDNA of PyMNK1 (GenBank accession No. PP737163) is 5354 bp, including a 265-bp 5’-UTR, a 1407-bp open reading frame, and a 3682-bp 3’-UTR (Figure 1). The open reading frame of PyMNK1 encoded a 488 aa protein with a predicted molecular weight of 53.40 kDa and theoretical pI of 5.56. Further analyses indicated that the PyMNK1 protein was a non-transmembrane hydrophilic protein (Figures 2A, B). A structure prediction analysis determined that the secondary structure of the PyMNK1 protein contained 22 α-helices, 7 β-strands, and 30 random coils (Figure 2C). A characteristic domain analysis revealed that the PyMNK1 protein contained one S_TKc domain (PAS-A: 285 aa, residues 54–371). The PyMNK1 protein for Homo sapiens (SWISS-MODEL No.: 5wvd.1.A) was used as the template for the 3D structure prediction of the PyMNK1 protein from P. yessoensis and PyMNK1 proteins from five other species. Compared with the template, 3D structure similarities of the MNK1 proteins from the six examined species were 86.75% (Xenopus tropicalis), 82.51% (Danio rerio), 66.10% (Penaeus japonicus), 64.75% (Strongylocentrotus purpuratus), 60.81% (Crassostrea gigas), and 58.76% (P. yessoensis) (Figure 2D). This indicated that the 3D structures of MNK1 homologs were relatively conserved from invertebrates to higher vertebrates, and that they may perform similar functions.

Figure 1. Nucleotide and predicted amino acid sequences of PyMNK1. The numbers on the left alternate from top to bottom as the nucleotide sequence and the deduced amino acid sequence numbers; the black font indicates the nucleotide sequence, and the red font indicates the amino acid sequence. The domain is shaded in green. The start codon (ATG) is framed by a blue box; the asterisk (*) represents the termination codon (TGA), and the polyadenylation signal (AATAA) is framed by a box.
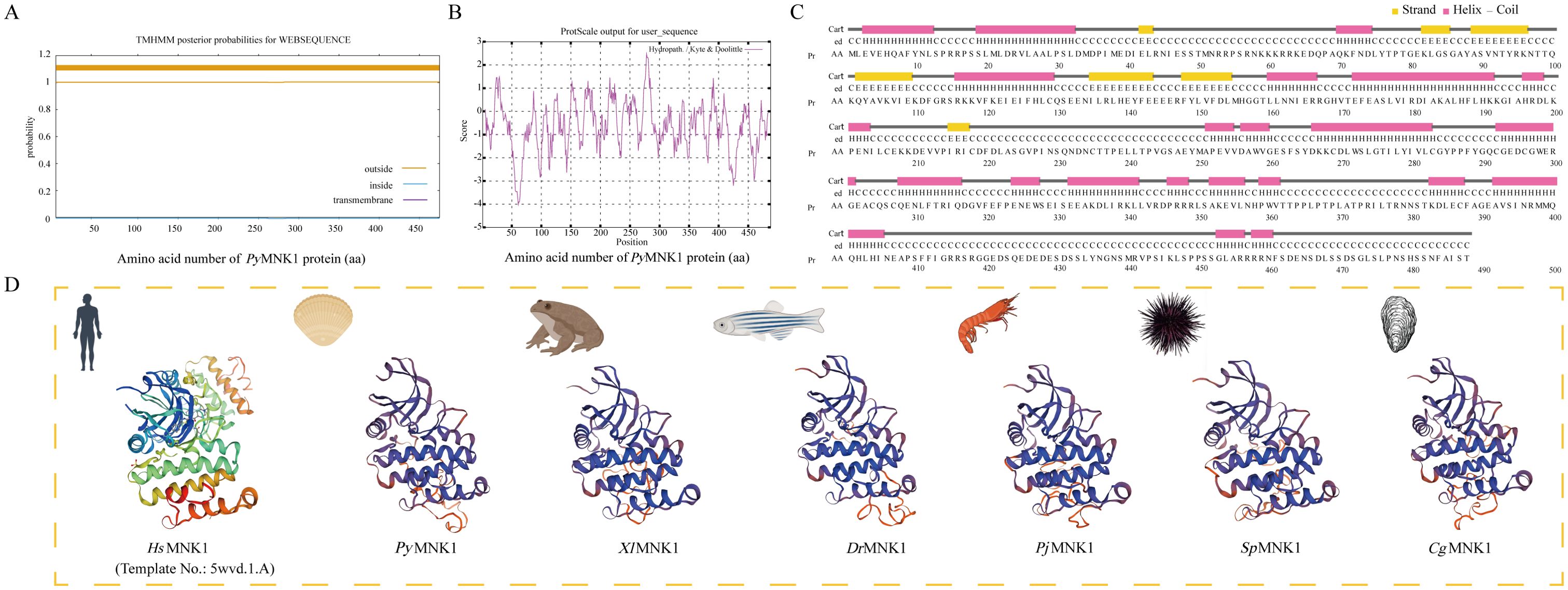
Figure 2. Sequence characteristic analysis of PyMNK1. (A) Transmembrane structure analysis of PyMNK1 protein. (B) Hydrophobic or hydrophilic analysis of PyMNK1 protein. (C) Predicted protein secondary structure of PyMNK1 protein. Yellow, pink and grey highlight strands, helices and coils, respectively. (D) Predicted 3D structure of the PyMNK1 protein and MNK1 proteins from five other species. The 3D structure of Homo sapiens MNK1 protein (HsMNK1; Swiss-model No.: 5wvd.1.A) was used as the template. PyMNK1, XlMNK1, DrMNK1, PjMNK1, SpMNK1, CgMNK1 represent MNK1 proteins from Patinopecten yessoensis, Xenopus laevis, Danio rerio, Penaeus japonicus, Strongylocentrotus purpuratus, Crassostrea gigas, respectively.
A multiple sequence alignment analysis found that the average similarity between the PyMNK1 sequence and MNK1 sequences from 25 other species was 36.98% (Figure 3A). The highest similarity (86.78%) was observed between PyMNK1 and an MNK1 homolog from Pecten maximus. A characteristic domain comparison indicated that the PyMNK1 and MNK1 proteins from 25 other eukaryotic organisms shared relatively high homologies in the S_Tkc domain (75.25%). Moreover, the number and types of characteristic domains of PyMNK1 were the same as those of the MNK1 proteins from the 25 other eukaryotic organisms (Figure 3B). A phylogenetic analysis showed that the PyMNK1 protein and the MNK1 protein from P. maximus were clustered into one branch and belonged to the clade of mollusk, which was consistent with traditional taxonomy (Figure 3C).
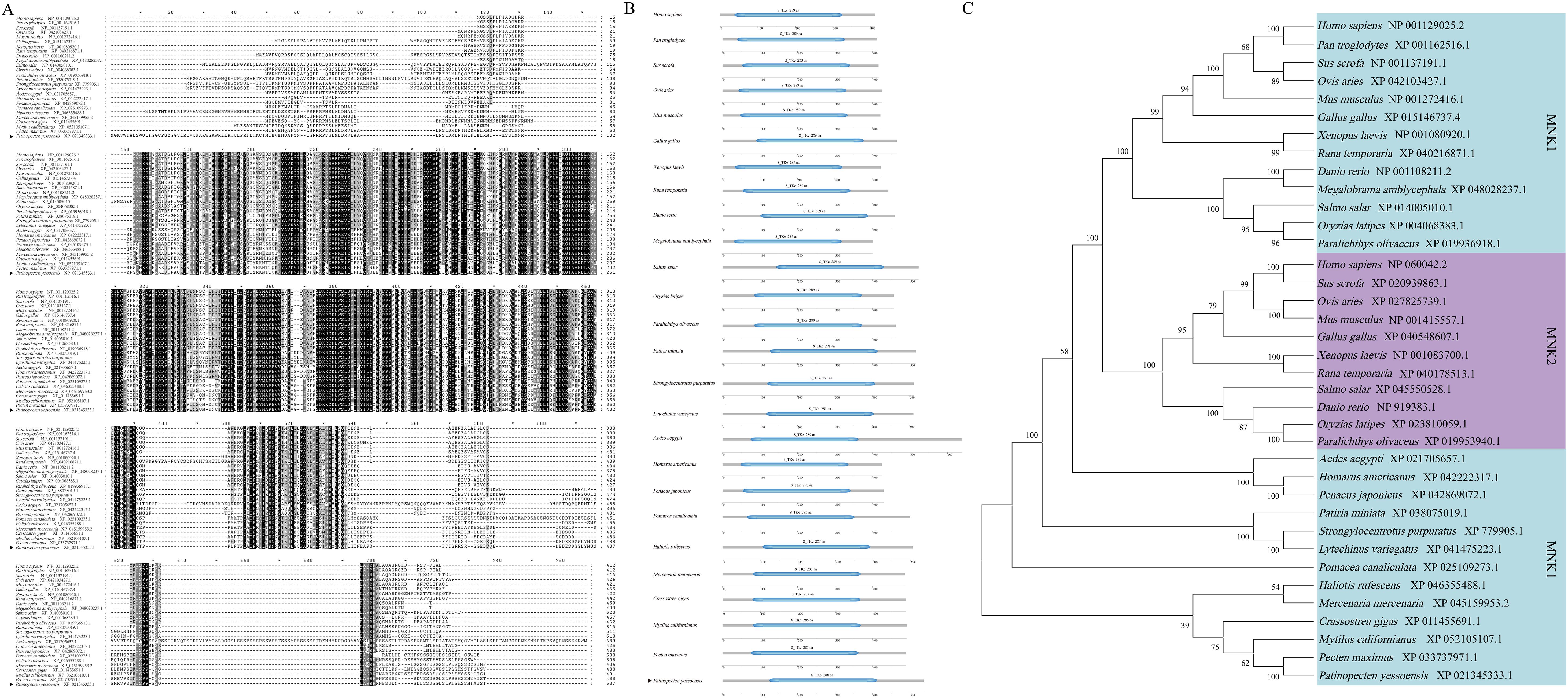
Figure 3. Phylogenetic analysis of PyMNK1. (A) Multiple sequence alignment of MNK1 proteins from Patinopecten yessoensis and 25 other species. (B) Schematic diagram of conserved domains of MNK1 homologs from P. yessoensis and 25 other species. (C) Neighbor-joining phylogenetic tree of MNK1 homologs from P. yessoensis and 36 other species. PyMNK1 is marked with a “◂”; the numbers at the tree nodes indicate bootstrap support values based on 1,000 replicates.
3.2 SOD promoter sequence characterization and interaction with PyMNK1
The promoter of the SOD gene is approximately 2027 bp in length, and the predicted transcription start site was the guanine (G; labeled as +1 bp) located 1800 bp upstream of the translation initiation site (ATG) of the SOD gene (Figure 4A). The online software MethPrimer prediction results did not identify any CpG islands in the SOD gene promoter sequence, a result which was similar to five other species (C. gigas, P. japonicus, Mercenaria mercenaria, Mytilus californianus, and Ruditapes philippinarum) (Figure 4B). There were 10 consecutive T nucleotides located at position +1151 to +1160 bp in the SOD promoter of P. yessoensis. In addition, five core promoter regulatory elements were identified in SOD WT1 binding sites: an Abscisic acid responsive element (ABRE) (−47 bp), G-Box (−47 bp), AAGAA motif (−74 bp), TATA box (−111 bp), and Box 4 (−165 bp) (Figure 4C). In addition to these five core promoter transcription elements, three promoter basic regulatory elements were identified in SOD WT2 binding sites: a TGACG motif (+1692 bp), TGA box (+1692 bp), and ABRE (+1786 bp) (Figure 4C).
Figure 4. Sequence and CpG island predictions of the SOD promotor, binding site identification, and verification of the PyMNK1 and SOD promotor. (A) The sequence of the SOD promoter. The transcription initiation site (TSS) is in the red font. The blue box represents the start codon (ATG). (B) CpG island prediction of the SOD promotor from Patinopecten yessoensis and five other species. (C) Schematic representation of putative PyMNK1 targeting sites and regulatory elements in the SOD promotor. (D) An analysis of relative luciferase activity. Different lower-case letters (e.g., a, b) represent the significance levels between different groups (P < 0.05). ** represent P < 0.01 vs. SOD WT1 binding sites group. ## represent P < 0.01 vs. SOD WT2 binding sites group. pGL3-basic, promoter vector; pGL3-control, positive control; WT, wild type.
To verify the binding sites of PyMNK1 on the SOD promoter, luciferase reporter vectors containing WT fragments of PyMNK1 and SOD were constructed (Figure 4C). The results showed that the WT1 SOD group and WT2 SOD group had remarkably higher luciferase activity than the NC group (Figure 4D), indicating that the cloned SOD gene promoter sequence bound to transcription factors to initiate transcription of the SOD gene. The results showed that the WT1 PyMNK1 + WT1 SOD group and WT2 PyMNK1 + WT2 SOD group had remarkably lower luciferase activity than the WT1 SOD group and WT2 SOD group, respectively (Figure 4D), indicating the presence of a binding site between PyMNK1 and miR-1985. These results indicated that PyMNK1 can negatively regulate the promoter of the SOD gene and affect its transcriptional activity.
3.3 Spatial expression and interaction analyses of miR-1985 and PyMNK1
The results of qRT-PCR showed that both miR-1985 and PyMNK1 mRNA were expressed in all the examined tissues. The relative expression of PyMNK1 mRNA in the mantle was the highest of all the examined tissues. In contrast, miR-1985 was highly expressed in gills, followed by the hepatopancreas (Figure 5A). An overall opposite expression trend of miR-1985 and PyMNK1 mRNA was observed across all examined tissues. Specifically, the relative expression of miR-1985 was higher than that of PyMNK1 mRNA in hepatopancreas, gills, mantles, and hemolymph, whereas, lower than that of PyMNK1 mRNA in muscles and gonads. Western blotting results showed that the PyMNK1 protein was expressed in all the examined tissues, with the relative expression level from low to high according to the following: hepatopancreas < hemolymph < gills < gonads < muscles < mantles. This was similar to the pattern of PyMNK1 mRNA expression (Figure 5B).
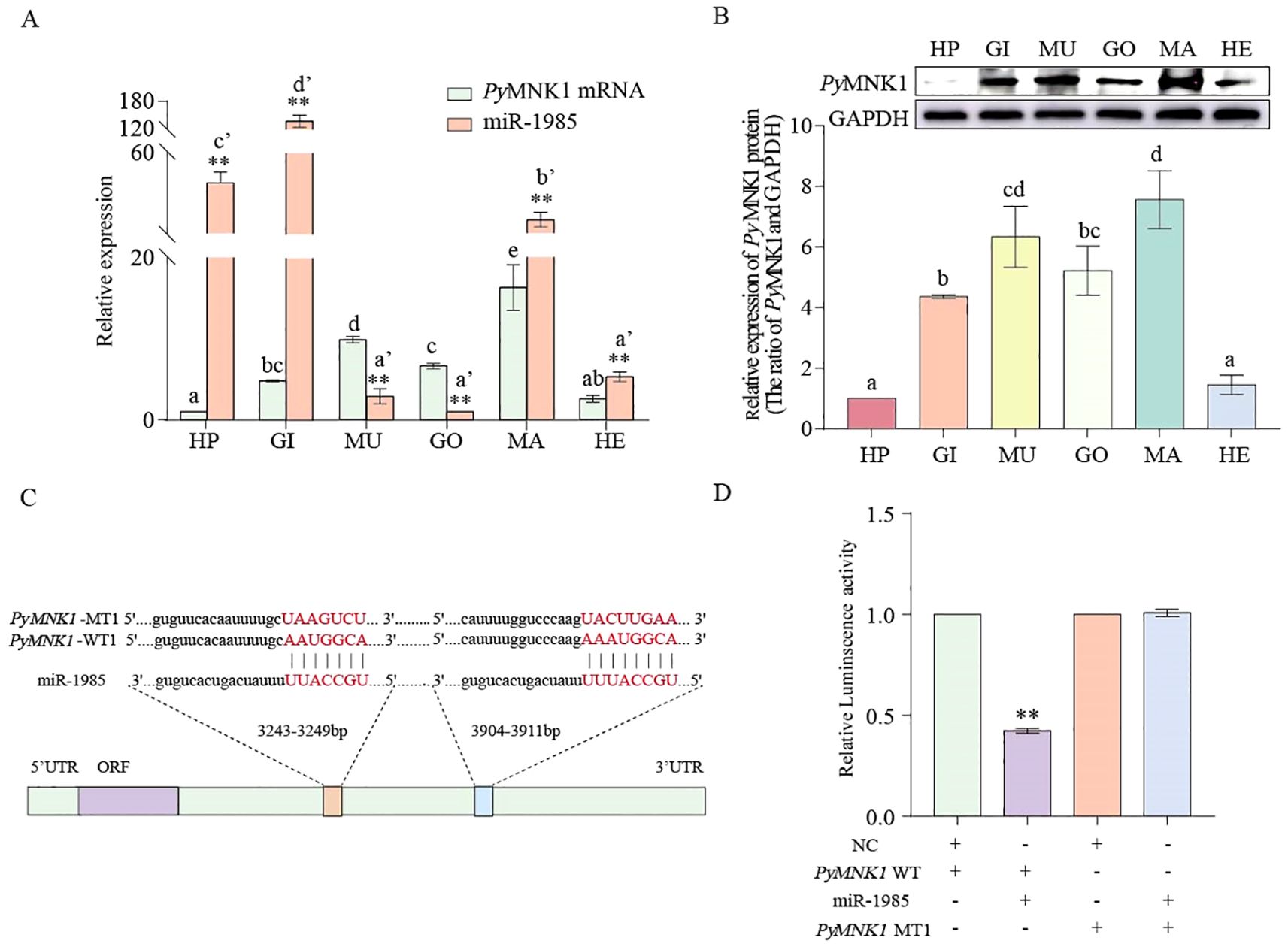
Figure 5. Spatial expression pattern, binding site identification, and verification of miR-1985 and PyMNK1. (A) Spatial expression pattern of miR1985 and PyMNK1. mRNA. HP: hepatopancreas; GO: gonads; MU: muscles; GI: gills; MA: mantles; HE: hemolymph. (B) Spatial expression pattern of PyMNK1 protein. (C, D) Schematic representation of the putative miR-1985 targeting site in PyMNK1 mRNA and an analysis of relative luciferase activity. Different lower-case letters (e.g., a or a’) indicate significant differences between different tissues (P < 0.05). “**” represents an extremely significant difference between miR-1985 and PyMNK1 mRNA in the same tissue or between NC + PyMNK1-WT and miR-1985 + PyMNK1-WT (P < 0.01).
To verify the binding sites of miR-1985 on PyMNK1, luciferase reporter vectors containing WT and MT fragments of the PyMNK1 3’-UTR were constructed (Figure 5C). The results showed that the WT 3’-UTR + miR-1985 mimic group had remarkably lower luciferase activity than the NC group (Figure 5D), indicating the presence of a binding site between PyMNK1 and miR-1985. Further in vivo functional verification experiments revealed that the relative expression levels of PyMNK1 (mRNA and protein) decreased significantly when miR-1985 was overexpressed (P < 0.01), whereas the relative expression of PyMNK1 (mRNA and protein) increased significantly when the expression of miR-1985 was inhibited (P < 0.01), similar to the results of the RNAi experiment for PyMNK1 (Figures 6A, B). These findings indicated that miR-1985 negatively regulated PyMNK1 protein expression post-transcriptionally, and that PyMNK1 siRNA inhibited PyMNK1 expression at the transcriptional level.
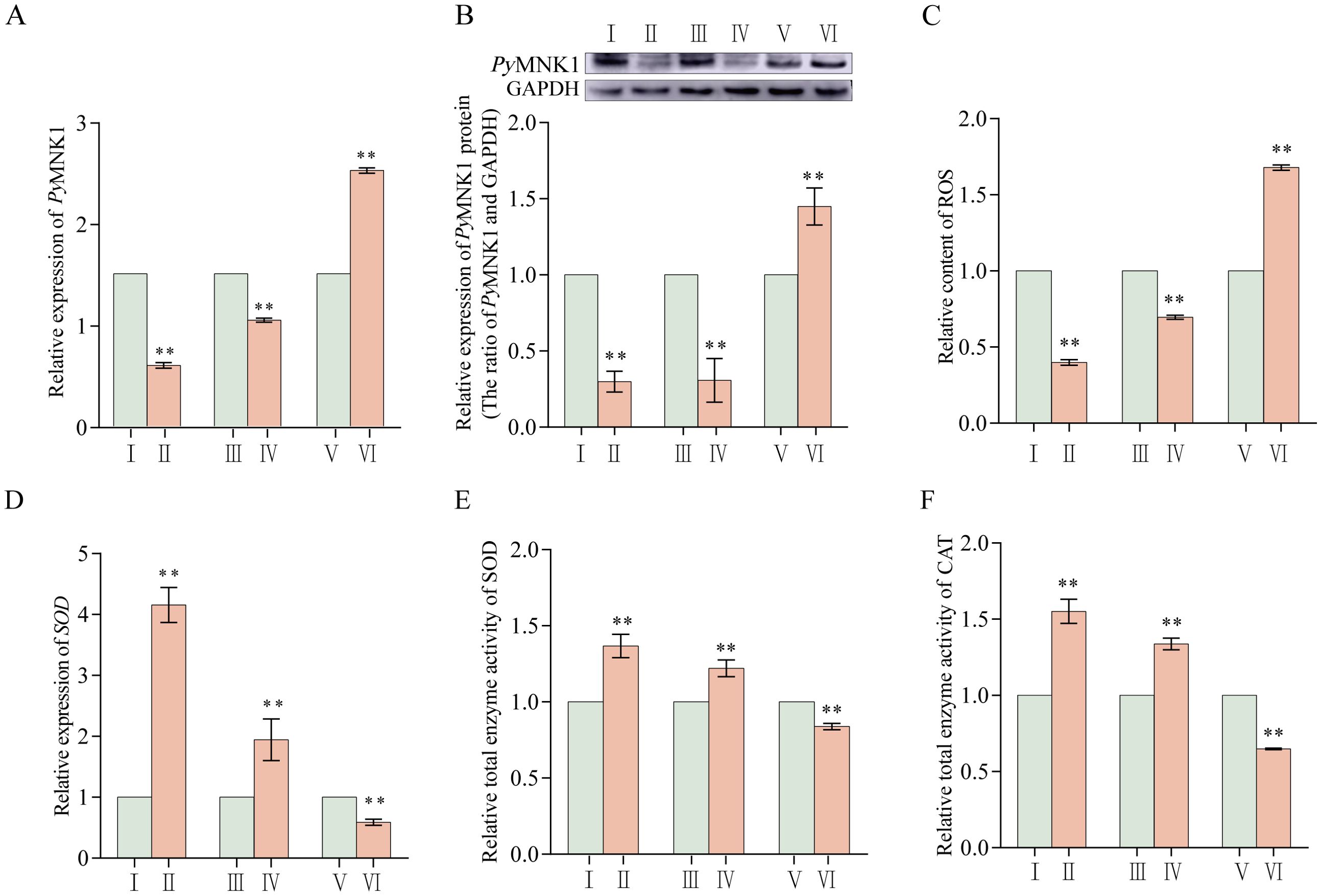
Figure 6. Effects of PyMNK1 and miR-1985 on mRNA expression, ROS content, and total SOD and CAT enzymatic activity changes in gills of Patinopecten yessoensis. (A, B) Effects of PyMNK1 and miR-1985 on PyMNK1 mRNA and protein expression. (C) Effects of PyMNK1 and miR-1985 on ROS content. (D-F) Effects of PyMNK1 and miR-1985 on SOD mRNA expression and enzymatic activity of SOD and CAT. I: negative control of PyMNK1 silencing group (NC-RNAi); II: PyMNK1 silencing group (RNAi); III: negative control of miR-1985 overexpression group (NC-Agomir); IV: miR-1985 overexpression group (Agomir); V: negative control of miR-1985 inhibition group (NC-Antagomir); VI: miR-1985 inhibition group (Antagomir). * represents P < 0.05; ** represents P < 0.01.
3.4 Effects of miR-1985 and PyMNK1 on mRNA expression and enzymatic activity of SOD in P. yessoensis
The relative expression (mRNA and protein) of PyMNK1 significantly decreased after PyMNK1 silencing and miR-1985 overexpression, indicating that both RNAi and miR-1985 overexpression significantly inhibited the relative expression of PyMNK1 (Figures 6A, B). At the same time, the relative content of ROS after PyMNK1 silencing or miR-1985 overexpression was significantly reduced (Figure 6C). Moreover, after PyMNK1 silencing or miR-1985 overexpression, the relative expression of SOD mRNA significantly increased and the enzymatic activity of SOD and CAT also significantly increased (Figures 6D–F).
3.5 Changes in ROS, miR-1985, PyMNK1 expression (mRNA and protein), and SOD and CAT activity after poly(I:C) stimulation
After P. yessoensis individuals were stimulated with poly(I:C), the relative expression level of miR-1985 in gills was upregulated at 6 hps, downregulated at 12 hps, and then upregulated from 36 to 72 hps (Figure 7A). The relative expression of PyMNK1 mRNA in gills of P. yessoensis initially increased, then decreased, then increased again, then decreased, and finally increased after poly(I:C) stimulation. The three peaks of mRNA expression appeared at 6, 24, and 72 hps (Figure 7B). The trend of the relative expression of PyMNK1 protein in gills of P. yessoensis after poly(I:C) stimulation was similar to the trend of mRNA expression (Figure 7C).
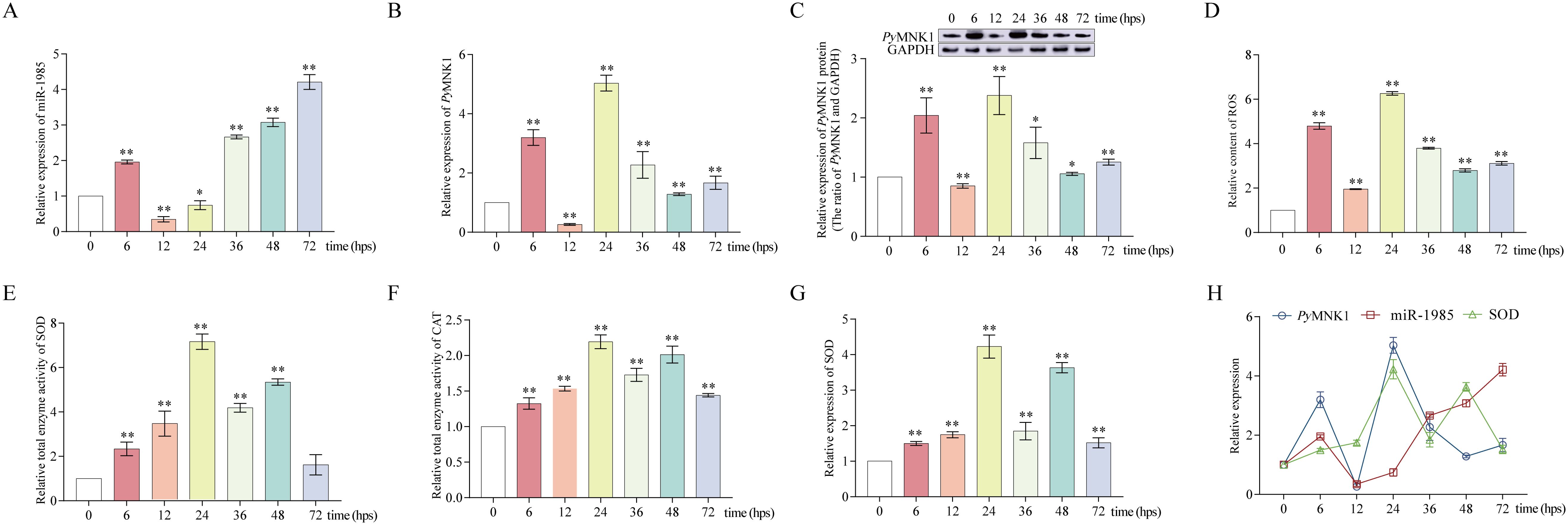
Figure 7. Poly(I:C) stimulation altered miR-1985, PyMNK1 (mRNA and protein), and SOD mRNA expression, ROS content, and total SOD and CAT enzymatic activity in gills of Patinopecten yessoensis. (A-C) Relative expression of miR-1985 and PyMNK1 (mRNA and protein) in gills of P. yessoensis after poly(I:C) stimulation. (D–F) Changes in the ROS content and total SOD and CAT enzymatic activity in gills of P. yessoensis after poly(I:C) stimulation. (G) Relative expression of SOD mRNA in gills of P. yessoensis after poly(I:C) stimulation. (H) Relative expression of miR-1985, PyMNK1 mRNA, and SOD mRNA after poly(I:C) stimulation. * represents P < 0.05 vs. 0 hps; ** represent P < 0.01 vs. 0 hps.
The relative ROS content exhibited an overall increasing trend after poly(I:C) stimulation with three peaks appearing at 6, 24, and 72 hps (Figure 7D), consistent with the relative expression trend of MNK1 mRNA and protein. Although the relative enzyme activity of both SOD and CAT in P. yessoensis gills exhibited an generally increasing trend after poly(I:C) stimulation, the alteration trend was opposite to that of the relative ROS content at 6–12 and 36–72 hps (Figures 7E, F). In addition, the relative expression trend of SOD mRNA was consistent with the alteration change of SOD relative enzyme activity (Figures 7G, H).
3.6 Effects of miR-1985 on the SOD/CAT axis by regulating PyMNK1 in gills after poly(I:C) stimulation
Because the trend of the ROS content was consistent with the trend of MNK1 expression, we further investigated whether miR-1985 downregulated ROS production through the SOD/CAT axis by regulating PyMNK1 in the gills of P. yessoensis after poly(I:C) stimulation (Figures 8A–G).
In PyMNK1 expression inhibition (RNAi) group or miR-1985 overexpression (agomir) group, the relative ROS content decreased in the gills of P. yessoensis after poly(I:C) stimulation compared with that of the control group (Figure 8A). Moreover, the relative total SOD and CAT enzyme activity increased higher than that of the control group in the gills of P. yessoensis after poly(I:C) stimulation when knocking down the expression of PyMNK1 or overexpressing miR-1985 (Figures 8B, C).
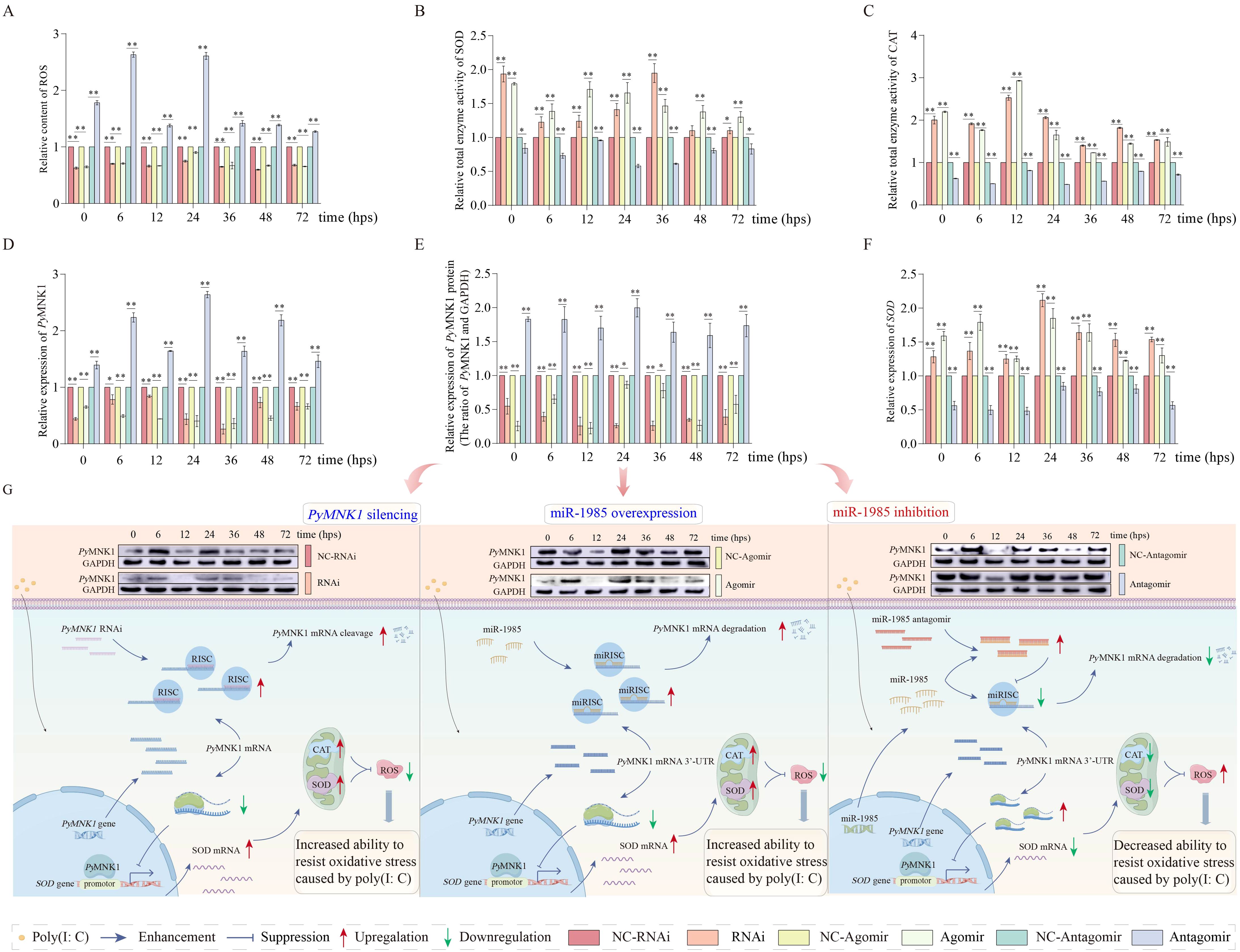
Figure 8. Effects of miR-1985 expression and PyMNK1 silencing on PyMNK1 (mRNA and protein) expression, ROS content, and total enzymatic activity of SOD and CAT in gills of Patinopecten yessoensis after poly(I:C) stimulation. (A–C) Effects of miR-1985 expression and PyMNK1 silencing on ROS content, total SOD and CAT enzyme activity in gills of P. yessoensis after poly(I:C) stimulation. (D–E) Effects of miR-1985 expression and PyMNK1 silencing on PyMNK1 (mRNA and protein) expression in gills of P. yessoensis after poly(I:C) stimulation. (F) Effects of miR-1985 expression and PyMNK1 silencing on SOD mRNA expression in gills of P. yessoensis after poly(I:C) stimulation. (G) Schematic diagrams of miR-1985 and PyMNK1 involved in responding to poly(I:C) stimulation in P. yessoensis. Schematic images were drawn by Figdraw (http://www.figdraw.com). NC-RNAi, negative control of PyMNK1 silencing group; RNAi, PyMNK1 silencing group; NC-Agomir, negative control of miR-1985 overexpression group; Agomir, miR-1985 overexpression group; NC-Antagomir, negative control of miR-1985 inhibition group; Antagomir, miR-1985 inhibition group. * represents P < 0.05; ** represents P < 0.01.
In the miR-1985 inhibition (antagomir) group, the relative ROS content extremely significantly increased in the gills of P. yessoensis after poly(I:C) stimulation (Figure 8A). The relative expression of PyMNK1 (mRNA and protein) exhibited generally increasing trends in the gills of P. yessoensis after poly(I:C) stimulation (Figures 8D, E). The relative total SOD and CAT enzyme activity exhibited generally decreasing trends in the gills of P. yessoensis after poly(I:C) stimulation (Figures 8B, C).
4 Discussion
Regulation of gene expression at the post-transcriptional or translational level to balance the production of ROS is increasingly being studied in the field of redox biology and innate immunology (30). In this study, we focused on clarifying the regulatory mechanism of miR-1985 on PyMNK1 gene expression and the function of the miR1985/PyMNK1 axis in response to PAMP-induced oxidative stress (ROS production) in shellfish.
As we expected, sequence identification data indicated that there were two binding sites for miR-1985 on the 3’-UTR of PyMNK1 mRNA. A luciferase reporter assay confirmed that miR-1985 may attenuate PyMNK1 expression by binding its 3’-UTR. Moreover, the trends of the relative expression trends of miR-1985 and the PyMNK1 gene were opposite of each other, based on a spatial expression analysis. The binding sites, interaction relationship, and spatial expression pattern between miR-1985 and the PyMNK1 gene were consistent with the conserved binding and interaction mechanism generally present between an miRNA and its target genes (31).
As epigenetic factors, miRNAs affect phenotypes by altering the function of their target genes (32). Therefore, a functional exploration of the PyMNK1 gene then become one of the concerns of this study after confirming the existence of the targeted regulatory relationships between miR-1985 and the PyMNK1 gene. A sequence characterization data analysis revealed that MNK1 shares average similarities of 42.96% and 60.48% with MNK1 homologs from invertebrates and vertebrates, respectively, suggesting that MNK1 homologs share similar functions from invertebrates to vertebrates. Expression analysis data also showed that PyMNK1 was ubiquitously expressed in all tissues examined in this study, and showed a similar expression pattern to that of H. sapiens (33), suggesting a relatively conserved expression pattern of MNK1 homologs from invertebrates to vertebrates. It has been well documented that MNK1 homologs correlate closely with survival signals in response to a wide variety of stimuli (34–37). In particular, MNK1 is mainly known as a protein that promotes viral infection (37). In this study, overall upregulation of PyMNK1 (both mRNA and protein) was observed after virus-related PAMP [poly(I:C)] stimulation (except 12 hps) accompanied with an obvious and continuous increase of the ROS content in gills of P. yessoensis. This observation was consistent with the ROS production rule of viral infections (38), and also suggests that PyMNK1 may be involved in the immune response to viral infections in P. yessoensis, similar to the function of MNK1 homologs documented in mammals (37). Furthermore, similar trends of PyMNK1 relative expression and relative ROS content suggest a possible tight linkage between these two. Further RNAi data provided evidence that the relative ROS level decreased when the expression of PyMNK1 was knocked down in gills of P. yessoensis. What is the possible mechanism of PyMNK1-induced ROS content changes? Commonly, the SOD/CAT axis is the most prominent redox cascade that catalyzes ROS to H2O and O2, in which SOD first catalyzes the dismutation of O2− to hydrogen peroxide (H2O2), and CAT in turn detoxifies H2O2 to H2O and O2 (39). It is acknowledged that the stronger the enzymatic activity of the SOD/CAT axis, the better the scavenging effect of ROS (40). Typically, the activity of a given enzyme can be enhanced by increasing its concentration (41). In this study, we noticed that after knocking down the expression of PyMNK1, the total enzymatic activity of the SOD/CAT axis was enhanced, and SOD mRNA expression increased. We thus hypothesized that PyMNK1 may suppress the enzymatic activity of the SOD/CAT axis by reducing the SOD concentration, through targeting the transcription of the SOD gene. As we expected, a dual-luciferase reporter assay revealed that PyMNK1 directly bound to the promoter of the SOD gene and attenuated its transcription. It has been well documented that MNK1 homologs often bind to eukaryotic initiation factor 4E (eIF-4E) to perform their biological functions (42). Although we did not explore the role of the MNK1/eIF-4E axis in this study, our results at least confirmed that MNK1 directly bound to the promoter of the SOD gene, thus acting as a transcriptional suppressor. This observation also suggests that MNK1 homologs may be involved in cellular process regulation in both a direct and in-direct manner. Taken together, we conclude that inhibition of PyMNK1 may increase the total enzymatic activity of the SOD/CAT axis by enhancing SOD transcription, consequently reducing ROS content and enhancing the immune response. Similarly, it was proposed by Xu et al. (43) that MNK1 suppression may be used as an efficient immunotherapy route to block cancer invasion and metastasis by enhancing the immune response (44). We thus speculated that the immune-regulatory function of MNK1 homologs is highly conserved from invertebrates to vertebrates.
Finally, in the context that PyMNK1 is one target gene of miR-1985 and PyMNK1 may affect the ROS content by regulating the SOD/CAT axis, we thus speculated that miR-1985 may affect the ROS content by targeting PyMNK1. As we thought, overexpression of miR-1985 inhibited the expression of PyMNK1, enhancing the total enzymatic activity of the SOD/CAT axis and reducing ROS content in gills of P. yessoensis after poly(I:C) stimulation, which was consistent with the results of PyMNK1 RNAi (PyMNK1 expression knock down) in this study. Moreover, we also noticed that the miR-1985/PyMNK1 axis did function during the entire poly(I:C) stimulation period. Specifically, the expression of both miR-1985 and PyMNK1 in gills of P. yessoensis fluctuated and were generally similar from 0 to 24 hps, which was inconsistent with the interaction relationship obtained in this study, suggesting that miR-1985 and PyMNK1 may not form a functional axis during this period, and the regulatory mechanism of total enzymatic activity of the SOD/CAT axis and the ROS content require further elucidation. However, from 36 to 48 hps, opposite fluctuations of miR-1985 and PyMNK1 expression in gills of P. yessoensis were observed, which was consistent with the interaction identified in this study. Moreover, the changes in the relative expression of SOD, total enzymatic activity of the SOD/CAT axis, and the ROS content were also consistent with the altered expression of PyMNK1. These observations suggest that miR-1985 may bind to PyMNK1 to form miR-1985/PyMNK1, and PyMNK1 may in turn bind to the SOD promoter, and thus a cascade involving miR-1985/PyMNK1/SOD/CAT perform the function of poly(I:C)-induced ROS scavenging in gills of P. yessoensis from 36 to 48 hps. At 72 hps, changes of the relative mRNA expression of SOD and total enzymatic activity of the SOD/CAT axis conform to the regulatory rule of PyMNK1 as determined in this study, although consistent trends of miR-1985 and PyMNK1 expression in gills of P. yessoensis were still observed. This observation indicated that the miR-1985/PyMNK1 axis may disintegrate at 72 hps, although the miR-1985/PyMNK1 axis may not regulate ROS, PyMNK1 may affect the total enzymatic activity of the SOD/CAT axis to achieve ROS reduction by regulating transcription of the SOD gene. As for the mechanism of ROS regulation at 72 hps, further investigation is required. Taken together, we conclude that miR-1985 may reduce poly(I:C)-induced ROS production by regulating the total enzymatic activity of the SOD/CAT axis by negatively targeting PyMNK1.
Conclusions
In this study, we preliminarily clarified the function and mechanism of the miR-1985/PyMNK1 axis in response to PAMP-induced oxidative stress in shellfish for the first time. The results revealed that miR-1985 may negatively regulate the expression of PyMNK1; furthermore, PyMNK1 may bind to the promoter of SOD, thus attenuating the expression of SOD and reducing the activity of the SOD/CAT axis. We thus conclude that shellfish may enhance their redox capability via the miR-1985/PyMNK1/SOD/CAT axis and thereby alleviate PAMP-induced oxidative stress. Moreover, we can also take advantage of the observations in this study to not only enhance disease prevention and control of cultured shellfish, but also to improve the molecular selective breeding of shellfish in the future. Specifically, we could design or mine miR-1985 activators or MNK1 homolog inhibitors in shellfish aquaculture, and we could also mine valuable SNPs from MNK1 homolog genes or SOD promoter sequences to develop new molecular assistive selective breeding markers for shellfish cultivation. In general, the observations from this study enrich our knowledge of miRNA/mRNA axis functions in redox homeostasis of aquatic invertebrates, and provide new clues for developing disease control strategies for economical shellfish aquaculture at the molecular level.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by the ethics committee of Dalian Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LY: Data curation, Formal Analysis, Writing – original draft, Writing – review & editing. HD: Formal Analysis, Writing – original draft. SL: Data curation, Writing – original draft. JX: Data curation, Writing – original draft. ZH: Data curation, Writing – original draft. DY: Data curation, Writing – original draft. YZ: Formal Analysis, Methodology, Writing – review & editing, Writing – original draft. YC: Formal Analysis, Methodology, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key R & D Program of China (No. 2022YFD2400302).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1556591/full#supplementary-material
References
1. Remick BC, Gaidt MM, Vance RE. Effector-triggered immunity. Annu Rev Immunol. (2023) 41:453–81. doi: 10.1146/annurev-immunol-101721-031732
2. Sun L, Wang X, Saredy J, Yuan Z, Yang X, Wang H. Innate-adaptive immunity interplay and redox regulation in immune response. Redox Biol. (2020) 37:101759. doi: 10.1016/j.redox.2020.101759
3. Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. (2014) 24:R453–62. doi: 10.1016/j.cub.2014.03.034
4. Zhou W, Xie Y, Li Y, Xie M, Zhang Z, Yang Y, et al. Research progress on the regulation of nutrition and immunity by microRNAs in fish. Fish Shellfish Immunol. (2021) 113:1–8. doi: 10.1016/j.fsi.2021.03.011
5. Farhana A, Alsrhani A, Alghsham RS, Derafa W, Khan YS, Rasheed Z. Gold nanoparticles downregulate IL-6 expression/Production by upregulating microRNA-26a-5p and deactivating the RelA and NF-κBp50 transcription pathways in activated breast cancer cells. Int J Mol Sci. (2024) 25:1404. doi: 10.3390/ijms25031404
6. Zhan Y, Liu L, Zhao T, Sun J, Cui D, Li Y, et al. MicroRNAs involved in innate immunity regulation in the sea cucumber: A review. Fish Shellfish Immunol. (2019) 95:297–304. doi: 10.1016/j.fsi.2019.10.049
7. Gilyazova I, Asadullina D, Kagirova E, Sikka R, Mustafin A, Ivanova E, et al. MiRNA-146a-A key player in immunity and diseases. Int J Mol Sci. (2023) 24:12767. doi: 10.3390/ijms241612767
8. Ortega-Villaizan M, Mercado L, Chico V. Editorial: Antiviral immune response in fish and shellfish. Front Immunol. (2023) 14:1155538. doi: 10.3389/fimmu.2023.1155538
9. Sharifinia M, Bahmanbeigloo ZA, Keshavarzifard M, Khanjani MH, Lyons BP. Microplastic pollution as a grand challenge in marine research: A closer look at their adverse impacts on the immune and reproductive systems. Ecotoxicol Environ Saf. (2020) 204:111109. doi: 10.1016/j.ecoenv.2020.111109
10. Alves TC, Queiroz FR, de Melo Neto AB, da Rocha Fernandes G, Pais FS, de Jesus Jeremias W, et al. Identification and characterization of microRNAs in Biomphalaria tenagophila and comparative analysis of their expression in Schistosoma mansoni-resistant and -susceptible snail populations. Gene. (2023) 884:147742. doi: 10.1016/j.gene.2023.147742
11. Li X, Lin S, Fan S, Huang X, Zhang Z, Qin Z. Identification and characterization of microRNAs involving in initial sex differentiation of Chlamys farreri gonads. Biol (Basel). (2022) 11:456. doi: 10.3390/biology11030456
12. Zhang YT. Characterization and function analysis of miRNA in the transplantation immunity in Pinctada fucata martensii. Guangdong Ocean University (2022). doi: 10.27788/d.cnki.ggdhy.2021.000222
13. Muire PJ, Hanson LA, Petrie-Hanson L. Rapid natural killer cell gene responses, generated by TLR ligand-induced trained immunity, provide protection to bacterial infection in rag1-/- mutant zebrafish (Danio rerio). Int J Mol Sci. (2025) 26:962. doi: 10.3390/ijms26030962
14. Huang B, Qu Y, Zhang H, Ma J, Chen J, Jie C, et al. The characterization of a novel IRF8-like homolog and its role in the immune modulation of the sea urchin Strongylocentrotus intermedius. Fish Shellfish Immunol. (2025) 159:110179. doi: 10.1016/j.fsi.2025.110179
15. Peng B, Lin J, Wan H, Zou P, Zhang Z, Wang Y. Identification of toll-like receptor family and the immune function of new Sptlr-6 gene of Scylla paramamosain. Fish Shellfish Immunol. (2024) 149:109609. doi: 10.1016/j.fsi.2024.109609
16. Liu W, Ma J, Chen J, Huang B, Liu F, Li L, et al. A novel TBK1/IKKϵ is involved in immune response and interacts with MyD88 and MAVS in the scallop Chlamys farreri. Front Immunol. (2023) 13:1091419. doi: 10.3389/fimmu.2022.1091419
17. Zhao Y, Guo M, Lv Z, Zhang W, Shao Y, Zhao X, et al. Fas-associated death domain (FADD) in sea cucumber (Apostichopus japonicus): Molecular cloning, characterization and pro-apoptotic function analysis. Dev Comp Immunol. (2020) 108:103673. doi: 10.1016/j.dci.2020.103673
18. Romero A, Dios S, Poisa-Beiro L, Costa MM, Posada D, Figueras A, et al. Individual sequence variability and functional activities of fibrinogen-related proteins (FREPs) in the mediterranean mussel (Mytilus galloprovincialis) suggest ancient and complex immune recognition models in invertebrates. Dev Comp Immunol. (2011) 35:334–44. doi: 10.1016/j.dci.2010.10.007
19. Metian M, Warnau M, Cosson RP, Oberhänsli F, Bustamante P. Bioaccumulation and detoxification processes of Hg in the king scallop Pecten maximus: field and laboratory investigations. Aquat Toxicol. (2008) 90:204–13. doi: 10.1016/j.aquatox.2008.08.014
20. Rio DC, Clark SG, Tjian R. A mammalian host-vector system that regulates expression and amplification of transfected genes by temperature induction. Science. (1985) 227:23–8. doi: 10.1126/science.2981116
21. Liu L, Zhao T, Lin K, Zou Y, Yan H, Zhan Y, et al. Identification of a novel RhoA gene in the sea cucumber Apostichopus japonicus and its immune regulatory function via interacting with miR-2012-5p. Int J Biol Macromol. (2022) 203:572–82. doi: 10.1016/j.ijbiomac.2022.01.176
22. Lyu Q, Jiao W, Zhang K, Bao Z, Wang S, Liu W. Proteomic analysis of scallop hepatopancreatic extract provides insights into marine polysaccharide digestion. Sci Rep. (2016) 6:34866. doi: 10.1038/srep34866
23. Ren L, Li K, Zhan Y, Wang Y, Yao Y, Chen Y, et al. Isolation of a New PAK1 Gene from Sea Cucumber (Apostichopus japonicus) and its expression analysis and function characterization. J Ocean Univ China. (2019) 18:1147–57. doi: 10.1007/s11802-019-4034-z
24. Yoon J, Gu WB, Konuma M, Kobayashi M, Yokoi H, Osada M, et al. Gene delivery available in molluscan cells by strong promoter discovered from bivalve-infectious virus. Proc Natl Acad Sci USA. (2022) 119:e2209910119. doi: 10.1073/pnas.2209910119
25. Zhao T, Ding S, Hao Z, Wang X, Zhan Y, Chang Y. Integrated miRNA-mRNA analysis provides potential biomarkers for selective breeding in bay scallop (Argopecten irradians). Genomics. (2021) 113:2744–55. doi: 10.1016/j.ygeno.2021.05.040
26. Yang J, Huang M, Zhang H, Wang L, Wang H, Wang L, et al. CfLec-3 from scallop: an entrance to non-self recognition mechanism of invertebrate C-type lectin. Sci Rep. (2015) 5:10068. doi: 10.1038/srep10068
27. Hao H, Cao L, Jiang C, Che Y, Zhang S, Takahashi S, et al. Farnesoid X receptor regulation of the NLRP3 inflammasome underlies cholestasis-associated sepsis. Cell Metab. (2017) 25:856–867.e5. doi: 10.1016/j.cmet.2017.03.007
28. Yu SY, Dong B, Fang ZF, Hu XQ, Tang L, Zhou SH. Knockdown of lncRNA AK139328 alleviates myocardial ischemia/reperfusion injury in diabetic mice via modulating miR-204-3p and inhibiting autophagy. J Cell Mol Med. (2018) 22:4886–98. doi: 10.1111/jcmm.13754
29. Zhang S, Li Y, Sun S, Liu L, Mu X, Liu S, et al. Single-atom nanozymes catalytically surpassing naturally occurring enzymes as sustained stitching for brain trauma. Nat Commun. (2022) 13:4744. doi: 10.1038/s41467-022-32411-z
30. Lennicke C, Cochemé HM. Redox metabolism: ROS as specific molecular regulators of cell signaling and function. Mol Cell. (2021) 81:3691–707. doi: 10.1016/j.molcel.2021.08.018
31. Li L, Veksler-Lublinsky I, Zinovyeva A. HRPK-1, a conserved KH-domain protein, modulates microRNA activity during Caenorhabditis elegans development. PloS Genet. (2019) 15:e1008067. doi: 10.1371/journal.pgen.1008067
32. Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat Rev Genet. (2014) 15:599–612. doi: 10.1038/nrg3765
33. O’Loghlen A, González VM, Piñeiro D, Pérez-Morgado MI, Salinas M, Martín ME. Identification and molecular characterization of Mnk1b, a splice variant of human MAP kinase-interacting kinase Mnk1. Exp Cell Res. (2004) 299:343–55. doi: 10.1016/j.yexcr.2004.06.006
34. Koval L, Proshkina E, Shaposhnikov M, Moskalev A. The role of DNA repair genes in radiation-induced adaptive response in Drosophila melanogaster is differential and conditional. Biogerontology. (2020) 21:45–56. doi: 10.1007/s10522-019-09842-1
35. Mihail SM, Wangzhou A, Kunjilwar KK, Moy JK, Dussor G, Walters ET, et al. MNK-eIF4E signaling is a highly conserved mechanism for sensory neuron axonal plasticity: evidence from Aplysia californica. Philos Trans R Soc Lond B Biol Sci. (2019) 374:20190289. doi: 10.1098/rstb.2019.0289
36. Franco-Enzástiga Ú, Natarajan K, David ET, Patel K, Ravirala A, Price TJ. Vinorelbine causes a neuropathic pain-like state in mice via STING and MNK1 signaling associated with type I interferon induction. iScience. (2024) 27:108808. doi: 10.1016/j.isci.2024.108808
37. Kumar R, Khandelwal N, Thachamvally R, Tripathi BN, Barua S, Kashyap SK, et al. Role of MAPK/MNK1 signaling in virus replication. Virus Res. (2018) 253:48–61. doi: 10.1016/j.virusres.2018.05.028
38. Herb M, Schramm M. Functions of ROS in macrophages and antimicrobial immunity. Antioxid (Basel). (2021) 10:313. doi: 10.3390/antiox10020313
39. Weydert CJ, Cullen JJ. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat Protoc. (2010) 5:51–66. doi: 10.1038/nprot.2009.197
40. Wang Y, Branicky R, Noë A, Hekimi S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J Cell Biol. (2018) 217:1915–28. doi: 10.1083/jcb.201708007
41. Ding S, Cargill AA, Medintz IL, Claussen JC. Increasing the activity of immobilized enzymes with nanoparticle conjugation. Curr Opin Biotechnol. (2015) 34:242–50. doi: 10.1016/j.copbio.2015.04.005
42. Proud CG. Mnks, eIF4E phosphorylation and cancer. Biochim Biophys Acta. (2015) 1849:766–73. doi: 10.1016/j.bbagrm.2014.10.003
43. Xu Y, Poggio M, Jin HY, Shi Z, Forester CM, Wang Y, et al. Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat Med. (2019) 25:301–11. doi: 10.1038/s41591-018-0321-2
Keywords: miR-1985, mitogen-activated protein kinase-interacting kinase 1, Patinopecten yessoensis, oxidative stress, innate immunity
Citation: Yu L, Deng H, Liu S, Xia J, Hao Z, Yin D, Zhan Y and Chang Y (2025) MicroRNA-1985 enhances the redox capability of scallop (Patinopecten yessoensis) in response to poly(I:C) stimulation by targeting MNK1. Front. Immunol. 16:1556591. doi: 10.3389/fimmu.2025.1556591
Received: 07 January 2025; Accepted: 15 April 2025;
Published: 08 May 2025.
Edited by:
Fulong Nan, Qingdao Agricultural University, ChinaCopyright © 2025 Yu, Deng, Liu, Xia, Hao, Yin, Zhan and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoyao Zhan, enl5ZGxvdUBob3RtYWlsLmNvbQ==; Yaqing Chang, eXFrZXlsYWJAaG90bWFpbC5jb20=
 Linghui Yu1,2
Linghui Yu1,2 Zhenlin Hao
Zhenlin Hao Yaoyao Zhan
Yaoyao Zhan Yaqing Chang
Yaqing Chang