- 1The First School of Clinical Medicine, Gannan Medical University, Ganzhou, China
- 2Department of Laboratory Medicine, The First Affiliated Hospital of Gannan Medical University, Ganzhou, China
- 3Department of Research and Education, The Second People’s Hospital of Jingdezhen, Jingdezhen, China
- 4School of Medical Technology, Gannan Medical University, Ganzhou, China
Hepatocellular carcinoma (HCC) is a highly malignant epithelial tumor characterized by global high incidence and poor clinical prognosis. Radical surgical resection, as the standard treatment for early-stage HCC patients, has been extensively validated for its therapeutic efficacy. However, epidemiological studies indicate that most patients are already in advanced stages at initial diagnosis, losing eligibility for radical treatment. Notably, HCC pathogenesis exhibits marked etiological heterogeneity, posing significant challenges for clinical management. Although significant breakthroughs have been made in understanding HCC drivers at pathophysiological levels, translational applications of these findings remain hindered by multiple barriers. Currently, elucidating the molecular mechanisms of HCC pathogenesis and identifying effective therapeutic targets constitute major research priorities in this field.Small extracellular vesicles (sEVs) are phospholipid bilayer vesicles (30-150 nm in diameter) carrying functional proteomes and nucleic acids (e.g., miRNAs, lncRNAs) with substantial biological activity. Studies demonstrate that sEVs contribute to malignant phenotype acquisition by modulating key signaling pathways such as PI3K/AKT and Wnt/β-catenin. These molecular cascades ultimately confer hallmark pathological features including aberrant proliferation, apoptosis resistance, and immune evasion to tumor cells. Within multi-network regulatory systems, sEVs serve as crucial intercellular messengers mediating tumor cell interactions with other tumor microenvironment (TME) components (e.g., cancer-associated fibroblasts, immune cells). Such communication facilitates TME reprogramming, pro-angiogenic phenotypic shifts, and therapy resistance development. Nevertheless, the precise molecular mechanisms of sEVs in HCC pathogenesis remain incompletely understood, warranting further exploration of their translational potential in clinical practice.
1 Introduction
Hepatocellular carcinoma (HCC) ranks among the most prevalent and lethal malignancies worldwide, with escalating incidence and mortality rates (1). HCC exhibits multifactorial etiology, with primary risk factors encompassing chronic HBV/HCV infections, alcoholic liver disease, and non-alcoholic fatty liver disease (2). Current therapeutic strategies for HCC merely extend nominal survival curves while inducing broad-spectrum toxicities. This ultimately leads to treatment resistance development in patients (3). Consequently, developing novel therapeutic approaches is imperative. Recent advances in fundamental medical research have progressively unraveled HCC pathogenesis mechanisms. Small extracellular vesicles (sEVs), as critical tumor microenvironment components, have garnered substantial research attention.
sEVs are nanoscale membranous vesicles secreted by diverse cell types, transporting bioactive cargo (proteins, lipids, mRNAs, miRNAs) to mediate intercellular communication and signaling (4). Studies demonstrate HCC-derived sEVs interact not only with tumor cells but also with TME components (fibroblasts, endothelial cells, immune cells), promoting hepatocarcinogenesis and progression via multinetwork fusion mechanisms (5). Although preliminary understanding of sEVs’ mechanistic roles in HCC exists, their complex signaling networks and clinical potential require further exploration.
This review systematically elucidates the multinetwork regulatory mechanisms of sEVs in HCC pathogenesis. Integrating current evidence, we analyze how sEVs drive HCC progression by: (a) modulating pivotal pathways (PI3K/AKT, Wnt/β-catenin); (b) reprogramming TME cellular composition/functionality; (c) enhancing malignant behaviors (proliferation, metastasis). Building upon these mechanisms, we evaluate sEVs’ translational value as precision medicine targets. This review addresses three key questions: (a) sEVs biogenesis/molecular signatures; (b) pathological mechanisms of sEVs-mediated network crosstalk; (c) clinical applications and translational prospects.
2 sEVs
2.1 Definition and classification of sEVs
The International Society for Extracellular Vesicles (ISEVS) defines extracellular vesicles as phospholipid bilayer-enclosed membranous structures ranging from 30–5000 nm in diameter. Their fundamental biological characteristics include cellular origin, lack of replicative capacity, and intercellular communication functions (6). Current classification criteria are based on physical properties and biogenesis pathways: by size as small EVs (sEVs, <200 nm) and large EVs (lEVs, >200 nm); by origin as exosomes (endosomal pathway), microvesicles (plasma membrane budding), and apoptotic bodies (programmed cell death products). Notably, ISEVS recommends using the operational term “small extracellular vesicles” (sEVs) rather than the mechanistically suggestive “exosomes”. This recommendation stems from: technical limitations in distinguishing biogenesis pathways; absence of specific molecular markers; and substantial heterogeneity in clinical samples (6, 7).
2.2 Molecular characteristics and characterization techniques of sEVs
sEVs exhibit characteristic nanoscale size distribution (30–200 nm) and marked morphological heterogeneity. Their bilayer membranes are enriched with tetraspanins (CD63/CD81/CD9) and tissue-specific markers (8, 9). Modern characterization techniques include: (a) Nanoparticle tracking analysis (NTA) for size quantification; (b) Transmission electron microscopy (TEM) for ultrastructure; (c) Super-resolution microscopy overcoming optical limits; (d) Mass spectrometry for molecular profiling (10, 11). Key technical challenges persist: in vitro models are culture-condition dependent (e.g., FBS starvation alters proteomes) (12); xenografts fail to recapitulate full TME interactions (13); clinical samples suffer lipoprotein co-isolation (plasma concentration ~1016/ml) (14). Optimization strategies combine separation techniques (e.g., SEC-density gradients) and surface marker capture, requiring purity-yield tradeoffs (15).
2.3 Biogenesis and uptake of sEVs
Rab GTPases are small GTPases belonging to the Ras superfamily that primarily regulate intracellular membrane trafficking and vesicular transport (16). They cycle between GTP-bound (active) and GDP-bound (inactive) states to modulate functional status, recruiting effector proteins to specific membrane compartments to control vesicle formation, trafficking, and fusion (17). This mechanism is crucial for the biogenesis of small extracellular vesicles (sEVs).
sEVs formation initiates with membrane invagination of early endosomes to generate intraluminal vesicles (ILVs), which subsequently develop into multivesicular bodies (MVBs) (18). Rab GTPases influence sEVs production and release by regulating multiple steps of this process. For instance, Rab27a and Rab27b promote MVB docking with the plasma membrane to enhance sEVs secretion (19), while Rab7 determines whether MVBs undergo degradation or sEVs release (20). Furthermore, Rab11- and Rab35-regulated sEVs secretion appears ESCRT-independent but Rab27-dependent for ILV formation (21), demonstrating the diverse functions of Rab proteins in sEVs biogenesis. Distinct Rab proteins precisely control sEVs generation through specific effector protein networks. Rab5 initiates ILV formation at the early endosome stage (22), while Rab11 affects the recycling endosome pathway (23), collectively ensuring proper sEVs assembly and function.
Selective uptake of sEVs represents a core aspect of intercellular communication, being tightly regulated rather than stochastic. The membrane protein composition of sEVs serves as a key determinant for selective uptake. Integrin family proteins direct sEVs homing to specific tissues (24), explaining why tumor-derived sEVs preferentially target particular organs. Tetraspanins (CD9, CD63, CD81) mediate cell-specific recognition through interactions with receptor cell surface ligands (25). Multiple mechanisms exist for sEVs entry into recipient cells, including clathrin-dependent endocytosis, caveolin-mediated endocytosis, macropinocytosis, and direct membrane fusion (26) (Figure 1).
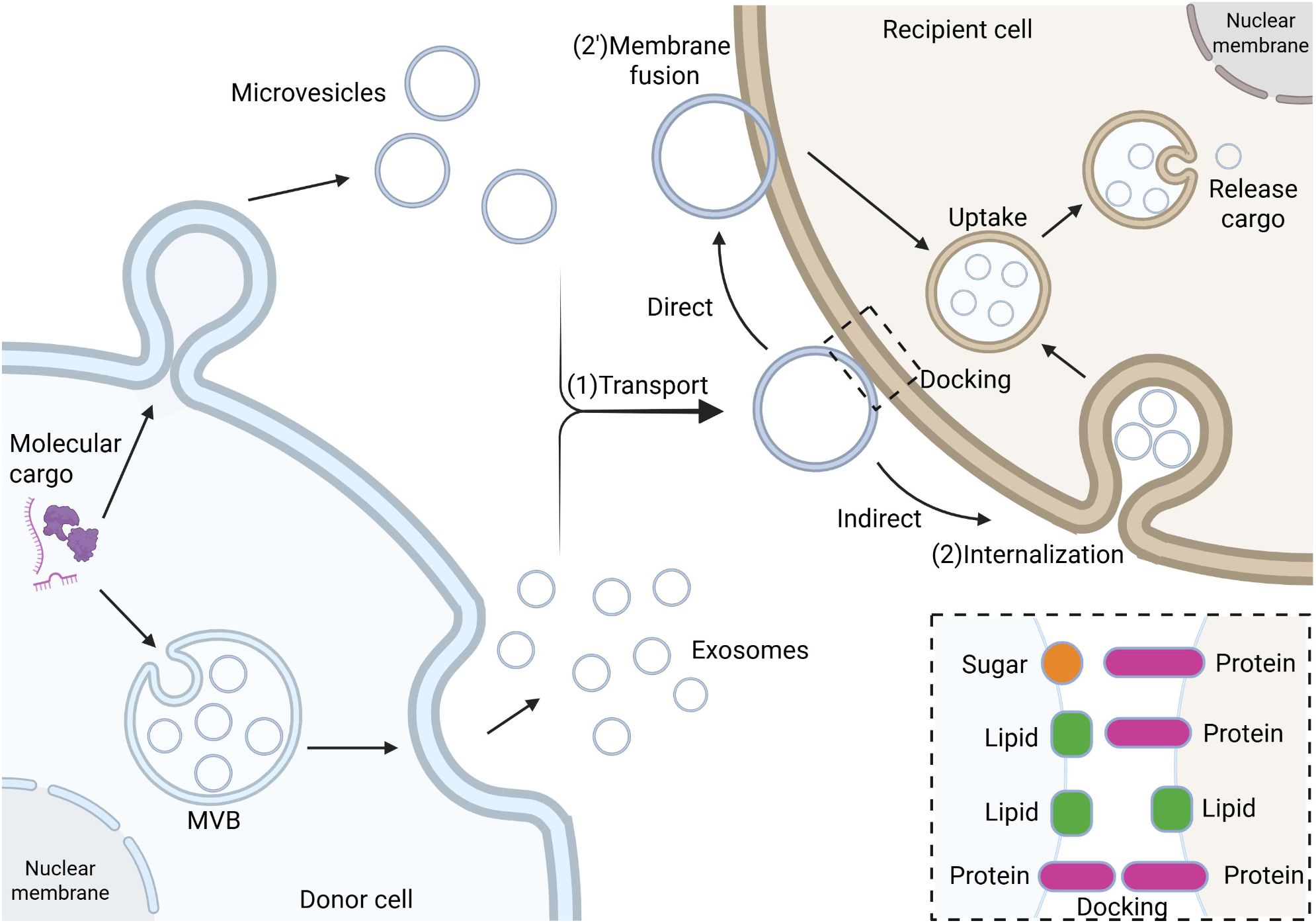
Figure 1. The biogenesis, transport, and internalization mechanisms of sEVs. Biogenesis of sEVs initiates from early endosomes, where the endosomal membrane invaginates to form intraluminal vesicles (ILVs) through membrane budding and sorting mechanisms, which subsequently mature into late endosomes, also known as multivesicular bodies (MVBs). MVBs fuse with the plasma membrane to release their intraluminal sEVs into the extracellular matrix (ECM). During intercellular communication, sEVs bearing surface ligands (e.g., transmembrane proteins or lipids) bind to specific receptors on target cell membranes, followed by internalization via endocytosis (including clathrin-dependent or -independent pathways) or membrane fusion. The newly formed early endosomes undergo maturation in the cytoplasm, ultimately releasing their bioactive cargo (e.g., nucleic acids, proteins) into the target cell cytoplasm, thereby modulating cellular physiological or pathological processes.
2.4 Research limitations of sEVs
Despite established definitions, high-purity sEVs isolation remains challenging due to incomplete biological understanding. In vitro models: Cell line-derived sEVs are experimentally controllable but their biogenesis is altered by artificial conditions (e.g., serum-free media), modifying molecular composition. FBS starvation alters sEVs yield, proteome, protein metabolic regulation, and membrane raft assembly functions (12). Given these effects, sEVs-depleted serum is recommended for in vitro studies. Xenograft models preserve tumor characteristics but fail to replicate dynamic TME interactions during tumor growth. Immunodeficient mice lack complete immune environments and human-mouse cellular interactions, limiting translational studies of sEVs-mediated immunomodulation (13). Clinical samples: Plasma-derived sEVs are clinically relevant but show inter-individual variability and lipoprotein contamination, requiring stringent characterization. Current methods (dUC, ExoQuick) co-isolate plasma proteins/lipoproteins and may induce vesicle aggregation. Notably, plasma lipoproteins (~1016/ml) share size/density characteristics with sEVs (chylomicrons/VLDL/HDL) (14).
2.5 Challenges and optimization in sEVs isolation technology
2.5.1 Inefficient separation of non-vesicular contaminants
Although traditional methods such as ultracentrifugation effectively enrich sEVs, they also co-precipitate contaminants including lipoproteins and protein aggregates (27). The MISEVS2023 guidelines recommend a multi-parametric evaluation strategy, incorporating immunoblotting or mass spectrometry to detect negative markers such as apolipoproteins. Optimization strategies include combining ultracentrifugation with size-exclusion chromatography, employing density gradient centrifugation for enhanced resolution, and exploring emerging technologies such as microfluidics (6). Standardized documentation of isolation methods and contaminant profiles is crucial to ensure reproducibility.
2.5.2 Significant variability in protein marker expression
Commonly used markers such as CD63 and CD9 exhibit heterogeneous distribution across sEVs subpopulations, with expression dynamically influenced by cellular origin and disease state (28). Researchers should employ a combination of universal and cell-specific markers for validation and enhance detection accuracy using advanced techniques such as high-resolution flow cytometry.
2.5.3 Technical bottlenecks in clinical-scale applications
Size-exclusion chromatography suffers from low recovery rates (30-60%) and limited throughput (29), whereas microfluidic technology demonstrates significant advantages, achieving >80% recovery, reducing processing time to minutes, and enabling specific capture of disease-associated sEVs subpopulations (30). Future efforts should focus on standardization through multicenter validation and the development of integrated automated workstations to address scalability challenges.
2.6 Single-vesicle analysis technologies
Conventional bulk analysis methods fail to resolve the high heterogeneity of sEVs, driving the need for single-vesicle detection technologies. Next-generation single-vesicle analysis enables precise characterization of individual vesicles’ physical properties and molecular composition, offering novel insights into sEVs biological functions (31).
Advanced microscopy techniques are revolutionizing sEVs observation. Super-resolution microscopy (STORM/PALM) overcomes the optical diffraction limit, revealing nanoscale structural features of sEVs (32). Cryo-EM preserves native sample states, providing authentic 3D morphological information of sEVs (33).
Single-molecule detection significantly enhances sEVs analysis precision. Single-molecule fluorescence tracks dynamic surface interactions, while nanopore sequencing enables direct RNA detection without amplification (34–36). These approaches offer unique advantages for low-abundance biomarker discovery.
Microfluidic platforms provide high-throughput solutions for sEVs analysis. Integrated with fluorescent labeling or Raman spectroscopy, these chip systems enable rapid sorting and characterization of individual sEVs (37). Digital microfluidics advances further by permitting multiplexed analysis of captured single vesicles.
Machine learning algorithms are transforming sEVs data processing. Deep learning models automatically identify characteristic patterns of sEVs subpopulations, while clustering analysis aids in discovering novel functional classifications (38). These methods are particularly suited for handling massive single-vesicle datasets.
Despite promising prospects, single-vesicle analysis faces sEVseral technical challenges. Key issues requiring resolution include balancing sensitivity with throughput, standardizing detection methods, and ensuring clinical translation feasibility. Overcoming these challenges will determine the technology’s practical utility.
Next-generation technologies will focus on multidimensional integrated analysis. Integrating nanotechnology, biosensing, and advanced computational methods, future single-vesicle analysis may achieve higher-precision multi-omics detection. This will open new possibilities for precision medicine and fundamental research.
3 Network regulation of tumor microenvironment by sEVs in hepatocellular carcinoma
The tumor microenvironment (TME) constitutes a complex ecosystem comprising diverse cell types and their secretory factors (39). This system primarily consists of: (a) Tumor cells - the central component exhibiting uncontrolled proliferative potential and invasiveness (40); (b) Cancer-associated fibroblasts (CAFs) - secreting bioactive factors (growth factors, cytokines, ECM components) to critically regulate tumor progression and metastasis (41); (c) Endothelial and pericytes - forming structural/functional units of tumor vasculature that enhance hematogenous metastasis via angiogenesis (42); (d) Immune cells (T/B cells, TAMs, DCs, MDSCs) - collectively participating in immunesurveillance, immunosuppression and immune evasion (43) (Figure 2).
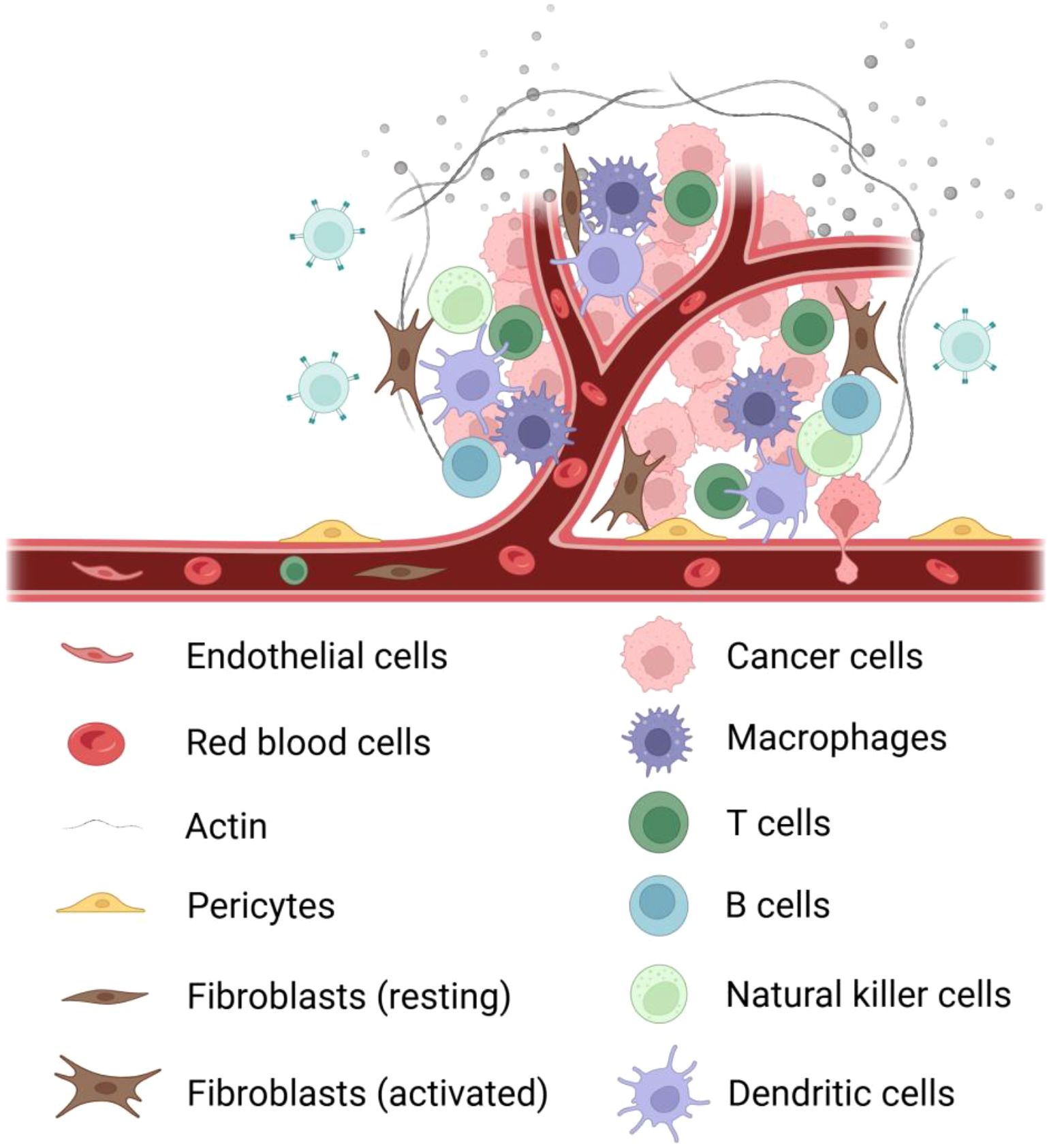
Figure 2. Components of the tumor microenvironment. It mainly contains the following cells: ① tumor cells; ② immune cells: tumor-associated macrophages, T cells, B cells, natural killer cells, dendritic cells; ③ fibroblasts: activated fibroblasts, resting fibroblasts; ④ blood vessels: erythrocytes, pericytes, endothelial cells ; ⑤ extracellular matrix; ⑥ signaling molecules.
sEVs serve as crucial signaling mediators in TME, orchestrating intercellular communication, metabolic reprogramming and immunomodulation (44, 45). By transporting diverse bioactive molecules, sEVs mediate complex crosstalk among tumor, stromal and immune cells to drive malignant progression (46, 47). Metabolically, sEVs remodel TME metabolism by transferring metabolites and regulators to fuel tumor proliferation (48). Notably, sEVs exhibit dual immunoregulatory roles: suppressing effector immune cells while activating immunosuppressive populations to establish an immune-tolerant niche (49). These findings establish sEVs as both essential TME components and promising therapeutic targets, offering novel avenues for treatment optimization and prognostic evaluation.
3.1 sEVs in communication network regulation
3.1.1 sEVs and common oncogenic mechanisms in HCC
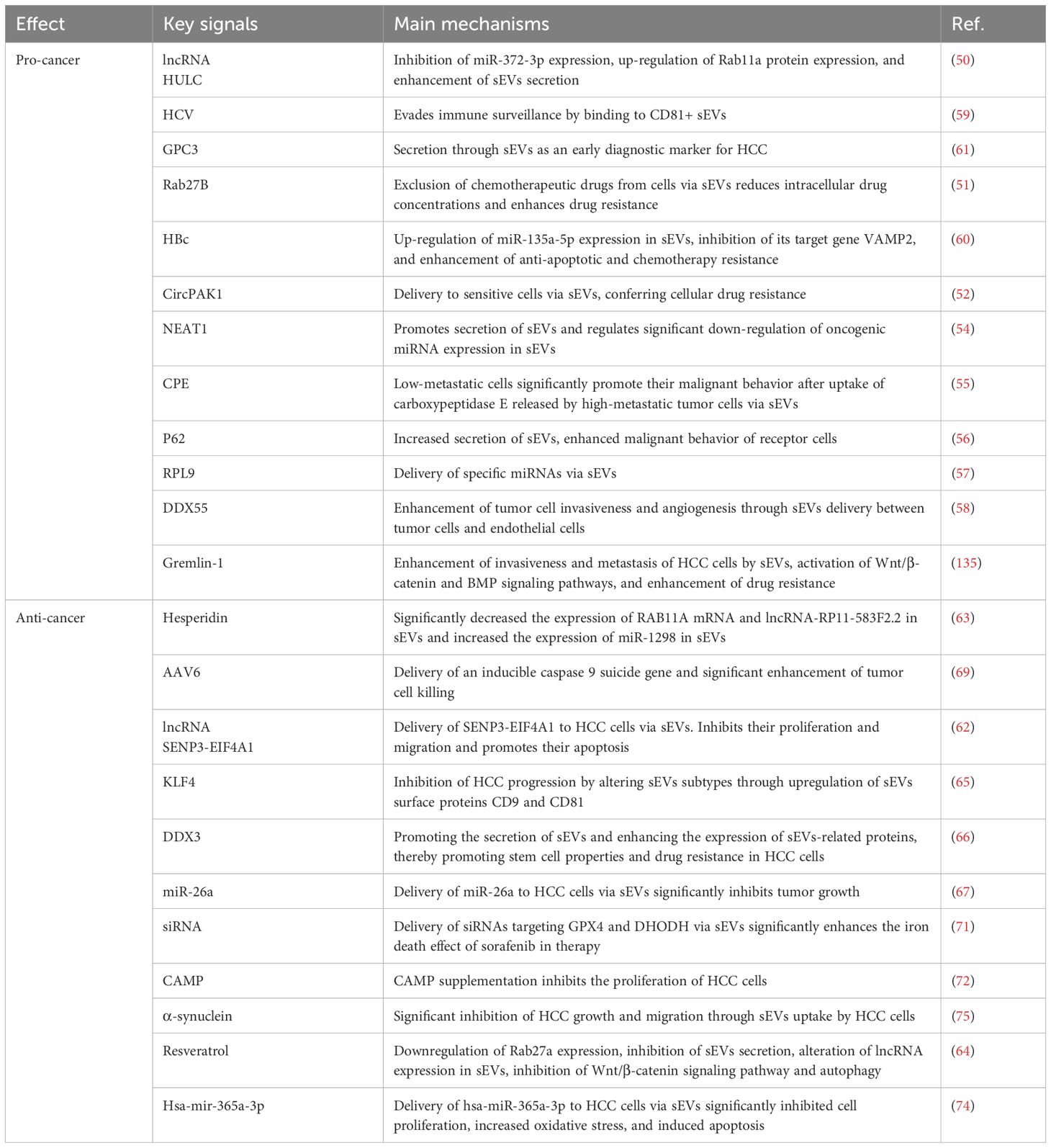
sEVs participate in the common pathological processes of HCC by mediating intercellular communication. At the molecular regulatory level, the long non-coding RNA HULC competitively inhibits miR-372-3p expression, leading to upregulation of Rab11a protein, thereby promoting sEVs secretion and accelerating HCC progression. Notably, the expression level of HULC in serum sEVs of HCC patients is significantly higher than in healthy controls, suggesting its potential as a diagnostic biomarker (50). In drug resistance regulation, upregulated Rab27B expression in drug-resistant HCC cells enhances sEVs secretion, promoting the efflux of chemotherapeutic agents (e.g., 5-fluorouracil) and reducing intracellular drug concentration; genetic knockout of Rab27B reverses this resistant phenotype (51). Furthermore, sEVs-mediated transfer of circPAK1 is a key mechanism of acquired resistance in HCC, as resistant cells transmit circPAK1 to sensitive cells via sEVs, conferring drug resistance (52).
sEVs facilitate malignant behaviors in HCC by transferring specific RNAs and proteins (53). Upregulation of NEAT1 reduces tumor-suppressive miRNAs (e.g., miR-634, miR-638) in sEVs, enhancing the proliferation and invasion of HCC cells (54). Moreover, sEVs secreted by highly metastatic HCC cells carry carboxypeptidase E (CPE), which can be taken up by low-metastatic cells, promoting their malignant transformation, whereas CPE inhibition reverses this effect (55). Overexpression of p62 protein increases sEVs secretion, enhancing the migration and invasion of recipient cells (56). Ribosomal protein L9 (RPL9) transmits miR-24-3p and miR-185-5p via sEVs, further promoting HCC progression (57). DEAD-box helicase 55 (DDX55) is enriched in HCC-derived sEVs and promotes tumor invasion and angiogenesis through intercellular transfer (58). These findings indicate that sEVs drive the malignant phenotype of HCC by regulating key molecular networks, highlighting their importance in targeted therapy.
3.1.2 sEVs and unique oncogenic mechanisms in HCC
In virus-associated HCC, sEVs exhibit distinct regulatory features. CD81-positive sEVs mediate viral immune evasion and promote tumor progression in HCV-associated HCC; HCV viral particles exploit CD81-positive sEVs for transmission, and this sEVs subpopulation is significantly enriched in HCV-positive HCC patients, suggesting its potential as a therapeutic target (59). In HBV-associated HCC, hepatitis B virus core antigen (HBcAg) delivers miR-135a-5p via sEVs, suppressing VAMP2 expression, thereby enhancing anti-apoptotic capacity and fostering drug resistance in HCC cells (60). Additionally, dysregulated autophagy in HCC patients leads to aberrant release of Glypican-3 (GPC3) in sEVs, and its high expression profile makes it a candidate molecular marker for early diagnosis (61).
3.1.3 sEVs and universal tumor-suppressive mechanisms in HCC
sEVs suppress HCC malignant progression by delivering tumor-suppressive molecules or regulating key signaling pathways. Studies demonstrate that tumor-suppressive long non-coding RNAs (e.g., SENP3-EIF4A1) delivered by sEVs significantly inhibit HCC cell proliferation and induce apoptosis (62). Hesperidin modulates sEVs molecular composition by reducing oncogenic RNA cargo (e.g., RAB11A mRNA and lncRNA-RP11-583F2.2) while upregulating tumor-suppressive miR-1298 expression, thereby effectively inhibiting hepatic precancerous lesion development (63). Resveratrol downregulates Rab27a to reduce sEVs secretion and alters lncRNA SNHG29 expression in sEVs, consequently inhibiting Wnt/β-catenin signaling and autophagy processes (64). The transcription factor KLF4 suppresses HCC progression by upregulating sEVs surface markers CD9 and CD81, whereas low expression of RNA helicase DDX3 promotes sEVs secretion and enhances stemness features and drug resistance in HCC cells (65, 66).
In therapeutic applications, engineered sEVs demonstrate remarkable targeted delivery potential. For instance, anti-GPC3 antibody-modified sEVs efficiently deliver miR-26a, significantly suppressing HCC growth (67). GalNAc-modified sEVs co-deliver paclitaxel (PTX) and miR122, synergistically enhancing antitumor effects (68). Furthermore, sEVs-based gene editing systems show promising applications; AAV6 vectors effectively deliver suicide genes (e.g., inducible caspase 9), markedly enhancing HCC cell killing (69). The CRISPR-Cas9 ribonucleoprotein system delivered by sEVs also exhibits high-efficiency gene editing capability (70). Advanced studies reveal that multiplex siRNA delivery systems (targeting GPX4 and DHODH) enhance sorafenib-induced ferroptosis to overcome HCC drug resistance (71).
3.1.4 sEVs and HCC-specific tumor-suppressive mechanisms
Certain tumor-suppressive mechanisms exhibit HCC-specific regulatory characteristics. Serum cathelicidin antimicrobial peptide (CAMP) levels are significantly reduced in HCC patients, and CAMP supplementation effectively inhibits HCC cell proliferation, suggesting its potential as a diagnostic biomarker (72). Natural killer (NK) cell-derived sEVs selectively target HCC cells and induce apoptosis (73). Additionally, HEK293 cell-derived sEVs delivering miR-365a-3p significantly suppress HCC proliferation and promote apoptosis (74). Notably, Parkinson’s disease cell-derived sEVs enriched with α-synuclein inhibit HCC growth and migration (75).
In summary, sEVs play pivotal roles in HCC pathogenesis, drug resistance development, and metastasis by participating in complex molecular network regulation. These mechanisms encompass both universal HCC regulatory pathways and virus-specific modes of action, highlighting their translational value as diagnostic biomarkers and targeted therapeutic vehicles (Table 1).
3.2 sEVs in metabolic network regulation
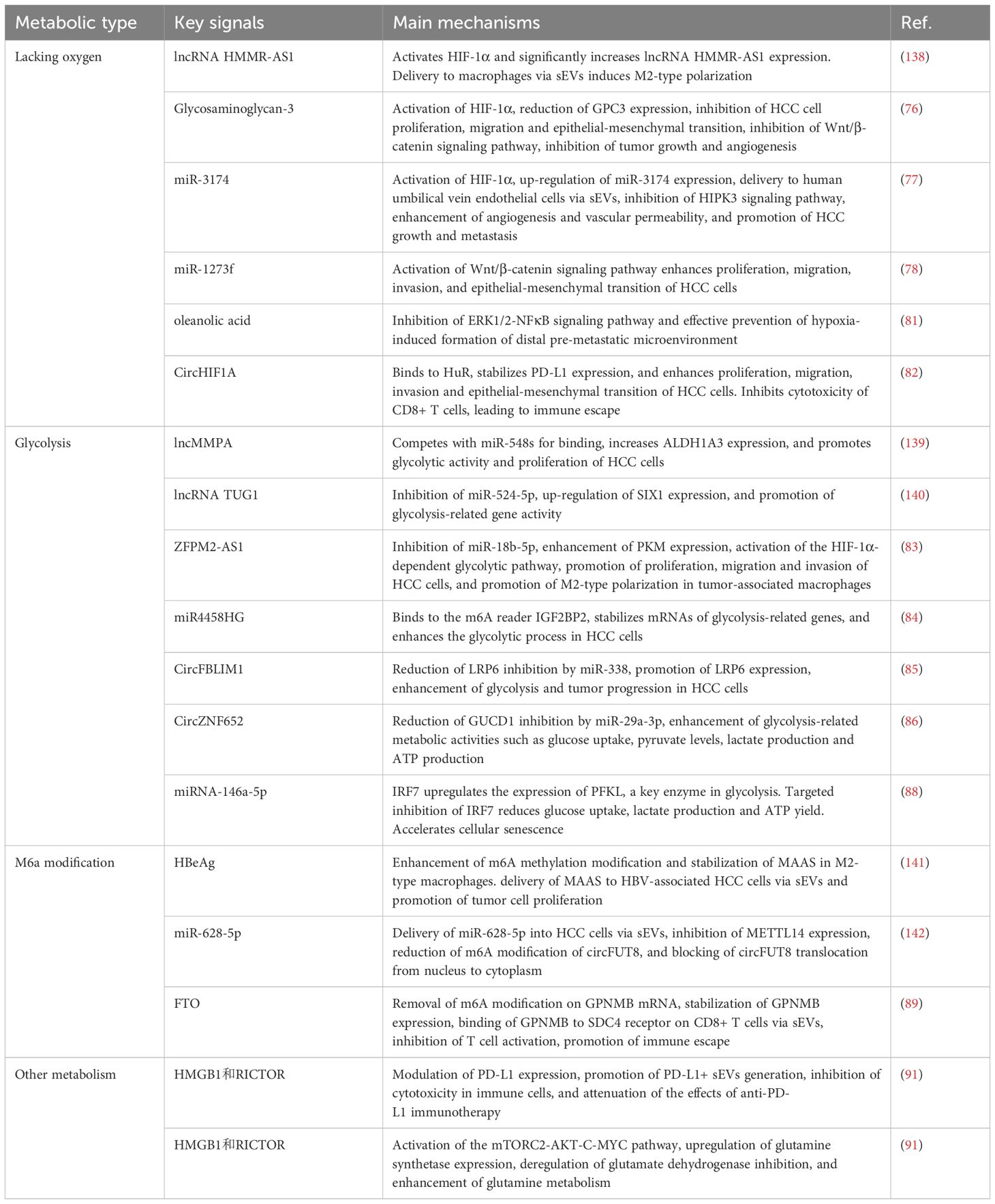
3.2.1 sEVs and hypoxia
The mechanistic role of hypoxic microenvironment in HCC progression has been extensively elucidated. Hypoxia regulates HCC malignancy through sEVs-mediated mechanisms. Hypoxic conditions modulate sEVs secretion via HIF-1α, influencing HCC proliferation, metastasis, and immune evasion. HIF-1α facilitates GPC3 loading into sEVs, reducing intracellular GPC3 to suppress Wnt/β-catenin signaling and tumor growth (76). Hypoxia-regulated sEVs biogenesis promotes angiogenesis through miRNA transfer. HIF-1α upregulates miR-3174 under hypoxia and enhances its packaging into sEVs. These sEVs are delivered to endothelial cells, inhibiting HIPK3 signaling to enhance angiogenesis/vascular permeability and accelerate HCC metastasis (77).
Regarding sEVs-mediated malignant transformation, miR-1273f activates Wnt signaling to promote HCC invasion. Hypoxic HCC-derived sEVs enriched with miR-1273f activate Wnt/β-catenin signaling to enhance proliferation, migration, and EMT (78). Further studies reveal hypoxic sEVs alter hepatocyte mechanical properties. Hypoxic sEVs (H-exos) promote proliferation/migration at lower concentrations than normoxic sEVs, inducing cytoskeletal reorganization and reduced elastic modulus (79). Crucially, hypoxic sEVs induce malignant transformation of normal hepatocytes. Chronic hypoxia enables HCC-sEVs to transform HL-7702 cells, enhancing proliferation/migration, tumor marker expression, and mechanical changes, while promoting tumor growth and liver damage in vivo (80).
Regarding metastasis, sEVs facilitate pre-metastatic niche formation. Hypoxic HCC-sEVs activate fibroblast ERK1/2-NFκB signaling to establish pulmonary PMN. Oleanolic acid (OA) inhibits this pathway to block PMN formation, showing anti-metastatic potential (81). Recent studies show CAF-derived sEVs containing circHIF1A promote immune evasion. Hypoxic CAF-sEVs deliver circHIF1A to stabilize PD-L1, enhance malignancy, and suppress CD8+ T cells, suggesting immunotherapeutic targets (82).
3.2.2 sEVs and glycolysis
HCC-derived sEVs regulate glycolysis via lncRNA transfer to promote progression. sEVs-carried ZFPM2-AS1 suppresses miR-18b-5p to upregulate PKM, activating HIF-1α-dependent glycolysis and enhancing HCC malignancy. ZFPM2-AS1 also promotes M2 macrophage polarization to accelerate progression (83). sEVs-delivered miR4458HG binds IGF2BP2 to stabilize HK2/SLC2A1 mRNAs, enhancing glycolysis and HCC growth (84).
circRNAs modulate HCC glycolysis via miRNA sponging. circFBLIM1 (enriched in HCC-sEVs) sequesters miR-338 to derepress LRP6, promoting glycolysis (85). Similarly, circ-ZNF652 inhibits miR-29a-3p to upregulate GUCD1, enhancing glycolytic flux - its knockout suppresses HCC glycolysis (86).
Highly metastatic HCC cells (e.g., 97H/LM3) secrete sEVs enriched with glycolytic/gluconeogenic/PPP proteins to enhance invasiveness (87). Conversely, senescent HCC cells deliver miR-146a-5p via sEVs to suppress glycolysis. This miRNA targets IRF7 to downregulate PFKL, reducing glucose metabolism and tumor growth (88).
3.2.3 sEVs and other metabolic pathways
FTO demethylates GPNMB mRNA to stabilize its expression and promote sEVs loading. sEVs-delivered GPNMB binds SDC4 on CD8+ T cells to suppress activation, enabling immune evasion. This FTO/m6A/GPNMB axis reveals key HCC mechanisms and therapeutic targets (89).
HCC cells enhance sEVs biogenesis/secretion via ferroptosis to clear misfolded proteins and alleviate ERS. Unsaturated fatty acids (e.g., arachidonic acid) augment this process. Ferroptosis inhibition reduces sEVs release and increases ERS sensitivity, revealing its cytoprotective role (90).
HMGB1/RICTOR upregulate PD-L1 expression and PD-L1+ sEVs release to impair immune function and anti-PD-L1 efficacy. They also enhance glutaminolysis via mTORC2-AKT-c-MYC (upregulating GS) and mTORC1-mediated GDH derepression (91) (Table 2).
sEVs bilayers contain phosphatidylserine, sphingomyelin, and cholesterol, with LPC modulating membrane stability/function. Exogenous LPC/PGD2 activate TGF-β via TLR2/DP1 to promote fibrosis/immunomodulation. Cholesterol-conjugated siRNAs enhance sEVs delivery efficiency, while vitamin E modifications improve cargo loading (92–94). miR-23b-3p is enriched in sEVs from aged mice/FH patients, accelerating senescence/metabolic dysfunction via Tnfaip3 suppression. Targeting miR-23b-3p may treat age-related liver/metabolic disorders (95). While sEVs roles in HCC lipid metabolism require further study, their regulatory functions show significant research value.
3.3 sEVs in immune network regulation
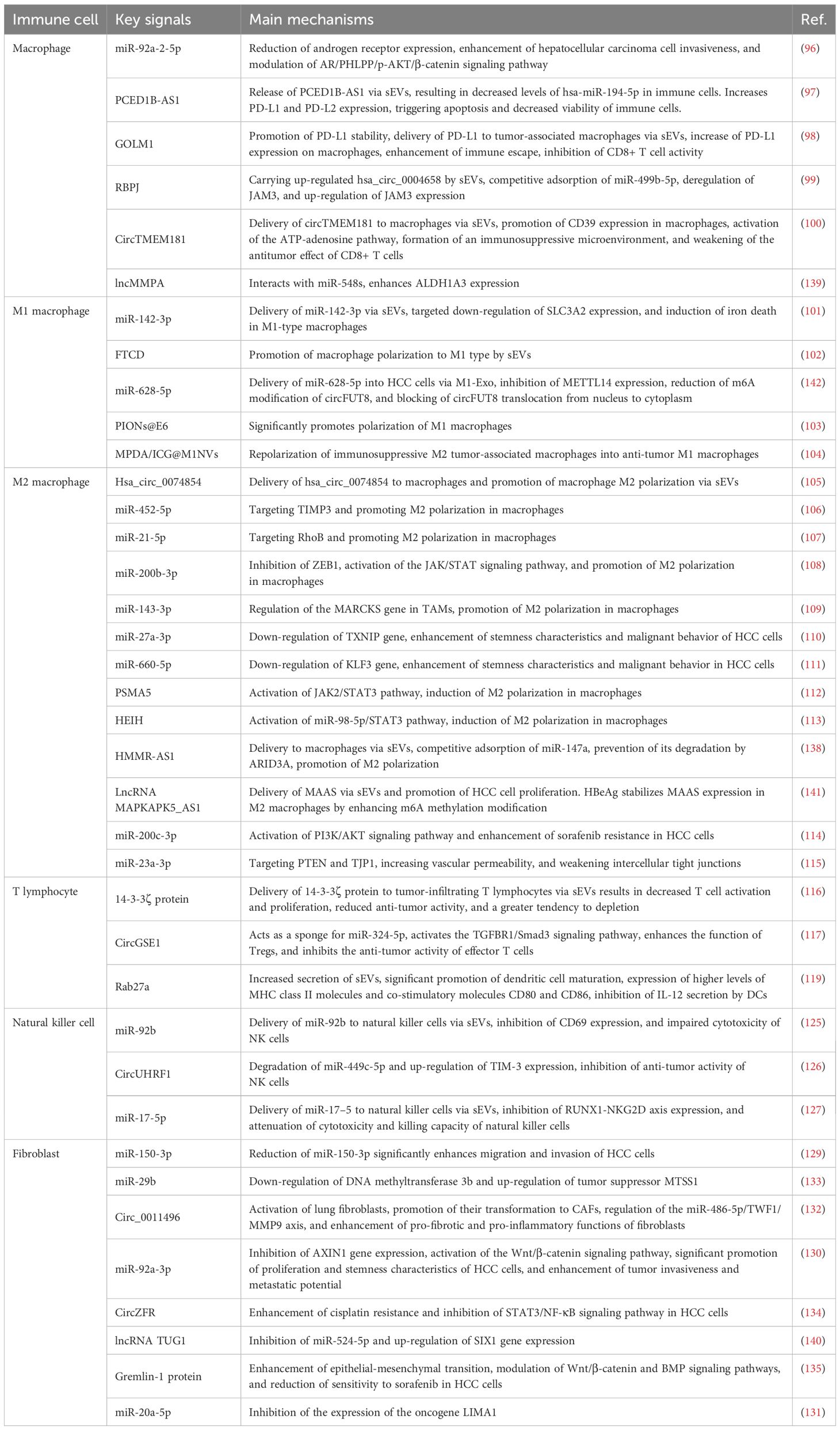
3.3.1 sEVs and macrophages
Macrophage-derived sEVs play crucial regulatory roles in HCC invasive phenotypes. Studies demonstrate macrophages enhance HCC invasiveness by secreting miR-92a-2-5p-enriched sEVs. These sEVs are internalized by HCC cells to downregulate androgen receptor (AR) expression, activating the AR/PHLPP/p-AKT/β-catenin signaling axis and promoting tumor progression. Experimental evidence shows inhibiting sEVs secretion or miR-92a-2-5p knockdown significantly attenuates macrophage-mediated HCC invasion (96).
sEVs-mediated immune evasion and immunosuppression in HCC microenvironment have been extensively investigated. HCC-derived sEVs deliver PCED1B-AS1 to T cells/macrophages, reducing hsa-miR-194-5p to upregulate PD-L1/PD-L2, inducing immune cell apoptosis/dysfunction (97). GOLM1 facilitates PD-L1 transfer via HCC-sEVs to tumor-associated macrophages (TAMs), enhancing immune evasion and CD8+ T cell suppression. Zoledronic acid combined with anti-PD-L1 effectively reverses this immunosuppression (98).
Recent breakthroughs reveal therapeutic potential of macrophage-derived sEVs in HCC. RBPJ-overexpressing macrophage sEVs (RBPJ+/+ Mφ-Exo) deliver hsa_circ_0004658 to suppress HCC proliferation and induce apoptosis. This circRNA sponges miR-499b-5p to derepress JAM3, exerting antitumor effects (99). Conversely, HCC-derived circTMEM181-enriched sEVs upregulate macrophage CD39, activating ATP-adenosine pathway to create immunosuppressive microenvironment and impair anti-PD1 efficacy (100). These findings provide novel directions for sEVs-targeted HCC therapies.
3.3.2 sEVs and M1 macrophages
HBV-associated HCC sEVs exhibit significant miR-142-3p upregulation. These sEVs deliver miR-142-3p to induce M1 macrophage ferroptosis, promoting tumor progression. Mechanistically, miR-142-3p targets SLC3A2 to regulate macrophage ferroptosis. This reveals how sEVs promote HBV+ HCC by modulating macrophage function (101). Additionally, HCC-derived sEVs can drive M1 macrophage polarization. FTCD-mediated sEVs signaling promotes M1 polarization to suppress HCC proliferation (102).
sEVs combined with superparamagnetic iron oxide nanoparticles (PIONs@E6) enhance M1 polarization. This combination increases proinflammatory cytokines (IL-12, TNF-α) and ROS production, effectively suppressing HCC growth in mice. sEVs-nanoparticle conjugates enhance macrophage antitumor immunity (103). A novel sEVs-mimetic nanosystem reprograms immunosuppressive M2 TAMs to antitumor M1 phenotype. Near-infrared irradiation increases M1 macrophages, inhibits tumor growth, and enhances immune activity in TME (104).
3.3.3 sEVs and M2 macrophages
HCC cells regulate macrophage polarization via sEVs secretion to promote tumor development. HCC-sEVs deliver hsa_circ_0074854 and other ncRNAs to induce M2 polarization, enhancing migration/invasion. hsa_circ_0074854 inhibition reverses this effect, confirming its key role in tumor-immune crosstalk (105).
sEVs-delivered miRNAs promote M2 polarization by targeting specific genes. miR-452-5p and miR-21-5p downregulate TIMP3 and RhoB respectively, enhancing HCC malignancy (106, 107). miR-200b-3p reinforces M2 polarization via ZEB1/JAK/STAT pathway (108). IL-6-stimulated HCC cells secrete miR-143-3p-enriched sEVs that promote M2 polarization via MARCKS regulation (109).
M2 macrophage-derived sEVs reciprocally promote tumor progression. Their miR-27a-3p and miR-660-5p suppress TXNIP and KLF3 respectively, enhancing HCC stemness/invasiveness (110, 111). lncRNAs (PSMA5, HEIH) in HCC-sEVs activate JAK2/STAT3 to induce M2 polarization (112, 113).
M2 macrophage sEVs mediate HCC drug resistance and vascular remodeling via specific miRNAs. miR-200c-3p activates PI3K/AKT pathway to induce sorafenib resistance (114). miR-23a-3p targets PTEN/TJP1 to disrupt vascular barriers and promote metastasis (115). These findings reveal multifaceted regulatory roles of sEVs in HCC microenvironment.
3.3.4 sEVs and T lymphocytes
HCC-derived sEVs modulate immune cell functions within the tumor microenvironment through multiple mechanisms, thereby influencing tumor progression. HCC-derived sEVs deliver 14-3-3ζ protein to tumor-infiltrating lymphocytes (TILs), impairing their activation, proliferation, and antitumor functions while accelerating T cell exhaustion. This mechanism demonstrates how sEVs suppress TIL immunocompetence to attenuate antitumor responses and promote HCC progression (116). Furthermore, sEVs play pivotal roles in regulatory T cell (Treg) expansion. HCC-sEVs carrying circGSE1 activate the TGFBR1/Smad3 pathway by sponging miR-324-5p, thereby enhancing Treg-mediated immunosuppression. This process inhibits CD8+ T cell antitumor activity and facilitates HCC immune evasion (117).
Multiple studies have investigated HCC-sEVs regulation of dendritic cells (DCs) and DC-mediated T cell responses. For instance, HCC-sEVs are internalized by DCs to present tumor antigens and activate CD8+ T cells, inducing antitumor immunity. However, sEVs concurrently suppress DC IL-12 secretion, which can be restored by IL-12 supplementation to enhance CTL-mediated tumor killing (118, 119).
The immunomodulatory properties of sEVs confer potential as antitumor vaccines. DC-derived sEVs (Dex) combined with microwave ablation (MWA) enhance CD8+ T cell infiltration while reducing Tregs, remodeling the immunosuppressive microenvironment comparably to DC vaccines (120). Moreover, tumor antigen-loaded sEVs potently enhance T cell function, demonstrating robust antitumor activity both in vitro and in murine models (121, 122).
The synergy between sEVs and immune checkpoint inhibitors has garnered significant attention. Antigen-loaded DC-derived sEVs (DC-TEX) increase intratumoral CD8+ T cells and elevate IFN-γ/IL-2 cytokine levels. Combined with anti-PD-1, they reverse T cell exhaustion and significantly enhance antitumor immunity (123). Beyond antitumor immunity, sEVs exhibit potential in antiviral immunity. HDV antigen-loaded DC-sEVs activate CD8+ T cells and promote Th1 responses via JAK/STAT signaling to suppress HDV replication (124).
3.3.5 sEVs and natural killer cells
HCC-derived sEVs significantly regulate natural killer (NK) cell immune functions. HCC-sEVs deliver miR-92b to NK cells, downregulating CD69 expression and impairing cytotoxicity to facilitate immune evasion (125). Additionally, HCC-sEVs transfer circUHRF1 to downregulate miR-449c-5p and upregulate TIM-3, inducing NK cell exhaustion and impairing anti-PD1 efficacy (126). Another mechanism involves miR-17-5p transfer, which suppresses the RUNX1-NKG2D axis to further compromise NK cell tumoricidal activity (127).
In contrast, NK cell-derived sEVs (NK-exo) enriched with cytotoxic proteins induce HCC apoptosis by inhibiting AKT/ERK1/2 signaling (73). IL-15/IL-21-stimulated NK-exos exhibit enhanced antitumor activity due to elevated cytotoxic protein content (128).
HCC-mediated immunosuppression via sEVs reveals novel immune escape mechanisms, while NK-exos demonstrate therapeutic potential. Future studies should explore blocking protumor sEVs or leveraging NK-exos to enhance antitumor immunity.
3.3.6 sEVs and fibroblasts
Cancer-associated fibroblast (CAF)-derived sEVs regulate HCC migration/invasion via noncoding RNAs. Reduced miR-150-3p in CAF-sEVs enhances HCC migratory/invasive capacities. Low miR-150-3p correlates with poor HCC prognosis, suggesting its regulatory role (129). CAF-sEVs deliver miR-92a-3p to activate Wnt/β-catenin signaling, promoting HCC proliferation/stemness (130). CAFs also modulate tumor suppressors to influence HCC progression. CAF-sEVs transfer miR-20a-5p to suppress LIMA1 and enhance HCC malignancy (131).
During metastasis, B[a]P-treated HCC cells transfer circ_0011496 via sEVs to activate lung fibroblasts into CAFs. This circRNA enhances profibrotic/proinflammatory functions via miR-486-5p/TWF1/MMP9 to drive pulmonary metastasis (132). Conversely, CAF-sEVs-delivered miR-29b suppresses metastasis by downregulating DNMT3b and upregulating MTSS1 (133).
Regarding chemoresistance, CAF-sEVs-circZFR enhances cisplatin resistance by inhibiting STAT3/NF-κB signaling (134). sEVs-transferred Gremlin-1 reduces sorafenib sensitivity via EMT and Wnt/β-catenin/BMP pathway modulation (135). These findings highlight CAF roles in HCC TME and suggest therapeutic strategies (Table 3).
3.3.7 sEVs and complement system
sEVs employ complement regulators for self-protection. Surface CD55/CD59 inhibit membrane attack complex (MAC) formation to prevent complement-mediated lysis. This enhances sEVs stability in bodily fluids for prolonged immunomodulation. Antigen-presenting cell-derived sEVs maintain structural integrity via this mechanism (136).
sEVs modulate complement via C3 fragments. B cell/macrophage-derived sEVs containing C3 fragments promote complement activation. This enhances antigen presentation and T cell responses. C3 fragments may also confer additional complement resistance (137).
3.4 sEVs in multi-network regulation
sEVs participate in complex intercellular communication networks by transporting bioactive molecules including proteins, nucleic acids, and lipids. They play pivotal roles in metabolic regulation and immunomodulation: modulating insulin sensitivity, glycolipid metabolic enzyme activity and mitochondrial function to maintain energy homeostasis, while precisely controlling immune responses through antigen presentation, immune receptor interactions and cytokine regulation. This regulation exhibits high specificity depending on sEVs cargo composition and microenvironmental conditions (Figure 3).
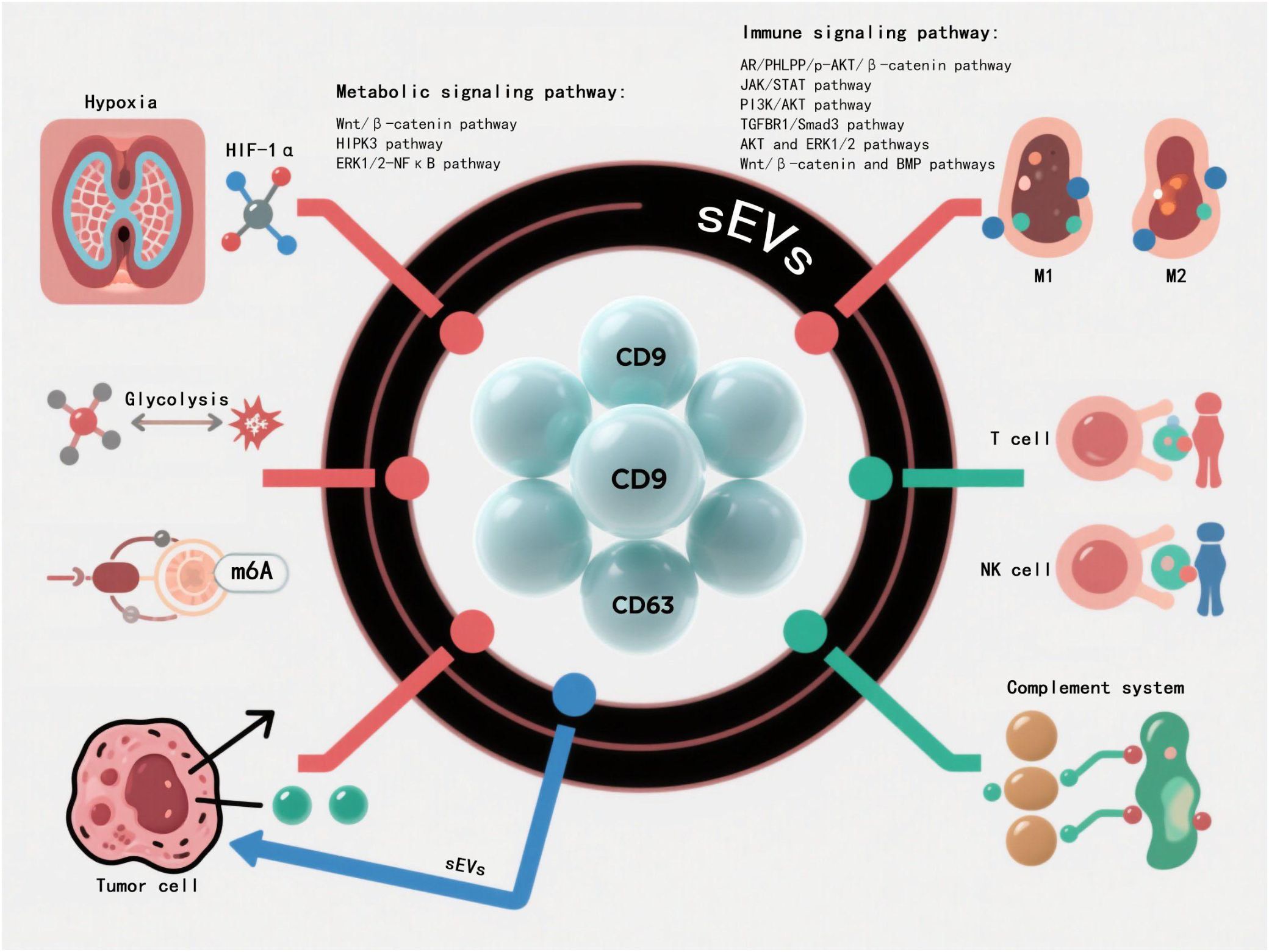
Figure 3. sEVs play a core role in multi-network regulation. sEVs systematically integrate into the highly complex intercellular communication network system through the bioactive molecular libraries such as proteomics,nucleic acid components and lipid groups they carry. The metabolic-immune cross-regulatory network mediated by them shows multi-dimensional regulatory characteristics, among which multiple signal transduction pathways constitute the key molecular hubs of cascade regulation.
In HCC, sEVs-mediated intercellular communication significantly influences tumor progression. Hypoxic conditions activate HIF-1α, promoting HCC cells to secrete HMMR-AS1 lncRNA-enriched sEVs. Upon macrophage uptake, these vesicles competitively bind miR-147a to upregulate ARID3A, inducing M2 polarization that enhances immunosuppression and accelerates tumor progression (138).
Tumor-associated macrophage (TAM)- and cancer-associated fibroblast (CAF)-derived sEVs regulate HCC metabolism through noncoding RNA delivery. TAM-secreted lncMMPA suppresses miR-548s to upregulate ALDH1A3, enhancing glycolysis and tumor proliferation (139). Similarly, CAF-derived TUG1 inhibits miR-524-5p to activate SIX1, promoting glycolysis and invasive capacity (140). These findings demonstrate the central role of sEVs in HCC metabolic reprogramming.
In HBV-associated HCC, HBeAg stabilizes lncRNA MAAS in macrophages via m6A modification, promoting its enrichment in sEVs. MAAS delivery to HCC cells significantly enhances proliferation (141). Conversely, M1 macrophage-derived sEVs deliver miR-628-5p to suppress METTL14-mediated m6A modification of circFUT8, thereby inhibiting tumor growth (142). This contrast highlights the bidirectional regulation of HCC by sEVs through epigenetic mechanisms.
4 Therapeutic applications of sEVs in hepatocellular carcinoma microenvironment
sEVs play a central role in regulating the HCC microenvironment. By mediating the transfer of various oncogenic molecules and signaling pathways, sEVs critically regulate the formation and evolution of the HCC tumor microenvironment. In HCV-associated HCC, CD81+ sEVs significantly impair host immune surveillance through immune evasion mechanisms, establishing them as promising therapeutic targets (59). The Rab27B-dependent sEVs-mediated drug efflux mechanism has been shown to substantially enhance chemoresistance in HCC cells (51). HBV core antigen (HBc) upregulates miR-135a-5p in sEVs to inhibit VAMP2 function, promoting anti-apoptotic properties and drug resistance in HCC cells (60). These studies elucidate the molecular mechanisms of sEVs-mediated therapy resistance in HCC and identify multiple potential targets for therapeutic intervention.
sEVs exhibit multifaceted regulatory functions in HCC immune evasion. sEVs surface-associated immune checkpoint molecules like PD-L1 effectively suppress T cell antitumor activity, reducing clinical response to immunotherapy (82, 91, 97, 98). Under hypoxic conditions, sEVs selectively enrich and deliver specific miRNAs/lncRNAs to enhance immunosuppression and accelerate HCC progression (76–78, 81, 82, 138).
With inherent biocompatibility and targeting capabilities, sEVs offer distinct advantages for drug delivery systems. Nanoengineered sEVs significantly improve targeting precision and bioavailability of therapeutic agents (68, 103, 104, 143). For gene/immunotherapies, sEVs demonstrate remarkable clinical potential by efficiently delivering functional nucleic acids or immunomodulators to enhance T cell activation and antitumor immunity (118, 122).
Artificial intelligence is transforming methodological approaches in sEVs research. Machine learning algorithms significantly enhance TEM and cryo-EM capabilities for sEVs ultrastructural analysis, enabling automated classification and quantification. AI-driven multi-omics integration efficiently identifies sEVs-associated diagnostic biomarkers, with random forest models demonstrating reliability for liquid biopsy applications. Computational biology frameworks integrate sEVs-mediated intercellular networks with tumor ecosystem dynamics, providing novel paradigms for studying oncogenesis.
5 Conclusion
HCC ranks among the most prevalent and lethal malignancies worldwide. Emerging fundamental research demonstrates that sEVs play pivotal regulatory roles in HCC pathogenesis and progression. As crucial intercellular communication vehicles, sEVs orchestrate HCC initiation, progression and malignant transformation through complex molecular networks encompassing cellular communication, metabolic regulation and immunomodulation.
By transporting diverse bioactive molecules (miRNAs, proteins, lipids), sEVs establish sophisticated signaling networks between cancer cells and microenvironmental components. These molecules enhance cancer cell proliferation, invasion and metastatic potential. Specifically, sEVs-enclosed miRNAs can selectively suppress tumor suppressor genes to accelerate HCC malignancy. Furthermore, sEVs reinforce malignant phenotypes by reprogramming cancer cell gene expression profiles.
Regarding metabolic regulation, sEVs modulate HCC metabolic characteristics through multiple mechanisms. They transfer critical metabolic enzymes/regulators and substantially alter glucose, lipid and energy metabolism pathways in HCC cells. Studies show sEVs enhance aerobic glycolysis via specific metabolic enzymes, sustaining proliferative capacity even under hypoxic conditions.
In immunomodulation, sEVs critically contribute to HCC immune evasion. They deliver immune checkpoint molecules (e.g., PD-L1) to impair immune surveillance and tumor clearance. Concurrently, sEVs modulate tumor-associated macrophage polarization while suppressing T/NK cell antitumor activity, establishing an immunosuppressive niche favorable for tumor growth.
In summary, sEVs comprehensively participate in shaping the HCC microenvironment by integrating cellular communication, metabolic reprogramming and immune evasion networks. These findings not only deepen our understanding of HCC pathogenesis but also provide theoretical foundations for novel diagnostic/therapeutic strategies. Future studies should further elucidate sEVs molecular mechanisms in HCC and explore clinical applications of sEVs-based targeted therapies and drug delivery systems. With advancing research, sEVs may emerge as crucial breakthroughs in HCC diagnosis and treatment.
Author contributions
XY: Writing – original draft. DH: Writing – original draft. LP: Writing – original draft. LW: Writing – review & editing. YL: Writing – review & editing. JY: Writing – review & editing. YQ: Writing – review & editing. CS: Writing – review & editing. QW: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Science and Technology Program of Ganzhou Science and Technology Bureau (N2023LNS36838).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Frittoli B, Castaldo A, Santarsiere M, Ascione R, Tanzi G, Ponsiglione A, et al. A unique case of lymphoepithelioma-like HCC with osteoclast-like giant cells: CT imaging features with pathologic correlations. Clin J gastroenterology. (2024) 17:112–7. doi: 10.1007/s12328-023-01871-1
2. Guerriero E, Capone F, Accardo M, Sorice A, Costantini M, Colonna G, et al. GPX4 and GPX7 over-expression in human hepatocellular carcinoma tissues. Eur J histochemistry: EJH. (2015) 59:2540. doi: 10.4081/ejh.2015.2540
3. Chakraborty E and Sarkar D. Emerging therapies for hepatocellular carcinoma (HCC). Cancers. (2022) 14:2798. doi: 10.3390/cancers14112798
4. An Y, Zhu J, Xie Q, Feng J, Gong Y, Fan Q, et al. Tumor Exosomal ENPP1 Hydrolyzes cGAMP to Inhibit cGAS-STING Signaling. Advanced Sci (Weinheim Baden-Wurttemberg Germany). (2024) 11:e2308131. doi: 10.1002/advs.202308131
5. Zeng B, Wang X, Qin Y, Cao L, Zhang C, Meng F, et al. Differences in serum cytokine levels distinguish between clinically noninvasive lung adenocarcinoma and invasive lung adenocarcinoma: A cross-sectional study. Health Sci Rep. (2023) 6:e1522. doi: 10.1002/hsr2.1522
6. Welsh JA, Goberdhan DCI, O’Driscoll L, Buzas EI, Blenkiron C, Bussolati B, et al. Minimal information for studies of extracellular vesicles (MISEVS2023): From basic to advanced approaches. J extracellular vesicles. (2024) 13:e12404. doi: 10.1002/jev2.12451
7. Sana A, Rossi IV, Sabatke B, Bonato LB, Medeiros LCS, and Ramirez MI. An Improved Method to Enrich Large Extracellular Vesicles Derived from Giardia intestinalis through Differential Centrifugation. Life (Basel Switzerland). (2023) 13:1799. doi: 10.3390/life13091799
8. Wu C, Zhou S, Mitchell MI, Hou C, Byers S, Loudig O, et al. Coupling suspension trapping-based sample preparation and data-independent acquisition mass spectrometry for sensitive exosomal proteomic analysis. Analytical bioanalytical Chem. (2022) 414:2585–95. doi: 10.1007/s00216-022-03920-z
9. Tsuchiya A, Terai S, Horiguchi I, Homma Y, Saito A, Nakamura N, et al. Basic points to consider regarding the preparation of extracellular vesicles and their clinical applications in Japan. Regenerative Ther. (2022) 21:19–24. doi: 10.1016/j.reth.2022.05.003
10. Gurunathan S, Kang MH, Jeyaraj M, Qasim M, and Kim JH. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. (2019) 8:307. doi: 10.3390/cells8040307
11. Wang Z, Zhou X, Kong Q, He H, Sun J, Qiu W, et al. Extracellular vesicle preparation and analysis: A state-of-the-art review. Advanced Sci (Weinheim Baden-Wurttemberg Germany). (2024) 11:e2401069. doi: 10.1002/advs.202401069
12. Böröczky T, Dobra G, Bukva M, Gyukity-Sebestyén E, Hunyadi-Gulyás É, Darula Z, et al. Impact of experimental conditions on extracellular vesicles’ Proteome: A comparative study. Life (Basel Switzerland). (2023) 13:206. doi: 10.3390/life13010206
13. HogenEsch H and Nikitin AY. Challenges in pre-clinical testing of anti-cancer drugs in cell culture and in animal models. J Controlled release: Off J Controlled Release Society. (2012) 164:183–6. doi: 10.1016/j.jconrel.2012.02.031
14. Brahmer A, Neuberger EWI, Simon P, and Krämer-Albers EM. Considerations for the analysis of small extracellular vesicles in physical exercise. Front Physiol. (2020) 11:576150. doi: 10.3389/fphys.2020.576150
15. Shami-Shah A, Travis BG, and Walt DR. Advances in extracellular vesicle isolation methods: a path towards cell-type specific EV isolation. Extracellular vesicles circulating Nucleic Acids. (2023) 4:447–60. doi: 10.20517/evcna.2023.14
16. Wang Y, Xiao T, Zhao C, and Li G. The regulation of exosome generation and function in physiological and pathological processes. Int J Mol Sci. (2023) 25:255. doi: 10.3390/ijms25010255
17. Dang XTT, Kavishka JM, Zhang DX, Pirisinu M, and Le MTN. Extracellular vesicles as an efficient and versatile system for drug delivery. Cells. (2020) 9:2191. doi: 10.3390/cells9102191
18. Tan M, Ge Y, Wang X, Wang Y, Liu Y, He F, et al. Extracellular Vesicles (EVs) in tumor diagnosis and therapy. Technol Cancer Res Treat. (2023) 22:15330338231171463. doi: 10.1177/15330338231171463
19. Zou W, Lai M, Zhang Y, Zheng L, Xing Z, Li T, et al. Exosome release is regulated by mTORC1. Adv Sci (Weinheim, Baden-Wurttemberg, Germany). (2019) 6:1801313. doi: 10.1002/advs.201801313
20. He G, Peng X, Wei S, Yang S, Li X, Huang M, et al. Exosomes in the hypoxic TME: from release, uptake and biofunctions to clinical applications. Mol Cancer. (2022) 21:19. doi: 10.1186/s12943-021-01440-5
21. Blanc L and Vidal M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases. (2018) 9:95–106. doi: 10.1080/21541248.2016.1264352
22. Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, and Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. (2008) 121:1649–60. doi: 10.1242/jcs.025726
23. Zobiack N, Rescher U, Ludwig C, Zeuschner D, and Gerke V. The annexin 2/S100A10 complex controls the distribution of transferrin receptor-containing recycling endosomes. Mol Biol Cell. (2003) 14:4896–908. doi: 10.1091/mbc.e03-06-0387
24. Wu J, Wang X, Li Z, Yi X, Hu D, Wang Q, et al. Small extracellular vesicles promote the formation of the pre-metastatic niche through multiple mechanisms in colorectal cancer. Cell Cycle. (2024) 23:131–49. doi: 10.1080/15384101.2024.2311501
25. Liu H, Huang Y, Huang M, Huang Z, Wang Q, Qing L, et al. Current status, opportunities, and challenges of exosomes in oral cancer diagnosis and treatment. Int J Nanomedicine. (2022) 17:2679–705. doi: 10.2147/IJN.S365594
26. Riazifar M, Pone EJ, Lötvall J, and Zhao W. Stem cell extracellular vesicles: extended messages of regeneration. Annu Rev Pharmacol Toxicol. (2017) 57:125–54. doi: 10.1146/annurev-pharmtox-061616-030146
27. Sohal IS and Kasinski AL. Emerging diversity in extracellular vesicles and their roles in cancer. Front Oncol. (2023) 13:1167717. doi: 10.3389/fonc.2023.1167717
28. Palmulli R, Bresteau E, Raposo G, Montagnac G, and van Niel G. In vitro interaction of melanoma-derived extracellular vesicles with collagen. Int J Mol Sci. (2023) 24:3703. doi: 10.3390/ijms24043703
29. Wang Y, Wang Y, Chen Y, Hua Y, Xu L, Zhu M, et al. Circulating microRNAs from plasma small extracellular vesicles as potential diagnostic biomarkers in pediatric epilepsy and drug-resistant epilepsy. Front Mol Neurosci. (2022) 15:823802. doi: 10.3389/fnmol.2022.823802
30. He S, Su L, Hu H, Liu H, Xiong J, Gong X, et al. Immunoregulatory functions and therapeutic potential of natural killer cell-derived extracellular vesicles in chronic diseases. Front Immunol. (2023) 14:1328094. doi: 10.3389/fimmu.2023.1328094
31. Zhang J, Wu J, Wang G, He L, Zheng Z, Wu M, et al. Extracellular vesicles: techniques and biomedical applications related to single vesicle analysis. ACS nano. (2023) 17:17668–98. doi: 10.1021/acsnano.3c03172
32. Urban BE, Dong B, Nguyen TQ, Backman V, Sun C, and Zhang HF. Subsurface super-resolution imaging of unstained polymer nanostructures. Sci Rep. (2016) 6:28156. doi: 10.1038/srep28156
33. Mageswaran SK, Yang WY, Chakrabarty Y, Oikonomou CM, and Jensen GJ. A cryo-electron tomography workflow reveals protrusion-mediated shedding on injured plasma membrane. Sci Adv. (2021) 7:eabc6345. doi: 10.1126/sciadv.abc6345
34. Luchian T, Mereuta L, Park Y, Asandei A, and Schiopu I. Single-molecule, hybridization-based strategies for short nucleic acids detection and recognition with nanopores. Proteomics. (2022) 22:e2100046. doi: 10.1002/pmic.202100046
35. Chen S, Sun Z, Li W, Yu P, Shi Q, Kong F, et al. Digital magnetic detection of biomolecular interactions with single nanoparticles. Nano letters. (2023) 23:2636–43. doi: 10.1021/acs.nanolett.2c04961
36. Isogai T, Hirosawa KM, Kanno M, Sho A, Kasai RS, Komura N, et al. Extracellular vesicles adhere to cells primarily by interactions of integrins and GM1 with laminin. J Cell Biol. (2025) 224:e202404064. doi: 10.1083/jcb.202404064
37. Liu Z, Ng M, Srivastava S, Li T, Liu J, Phu TA, et al. Label-free single-vesicle based surface enhanced Raman spectroscopy: A robust approach for investigating the biomolecular composition of small extracellular vesicles. PloS One. (2024) 19:e0305418. doi: 10.1371/journal.pone.0305418
38. Binotti B, Ninov M, Cepeda AP, Ganzella M, Matti U, Riedel D, et al. ATG9 resides on a unique population of small vesicles in presynaptic nerve terminals. Autophagy. (2024) 20:883–901. doi: 10.1080/15548627.2023.2274204
39. Guo T and Xu J. Cancer-associated fibroblasts: a versatile mediator in tumor progression, metastasis, and targeted therapy. Cancer metastasis Rev. (2024) 43:1095–116. doi: 10.1007/s10555-024-10186-7
40. Stasinopoulos I, Penet MF, Chen Z, Kakkad S, Glunde K, and Bhujwalla ZM. Exploiting the tumor microenvironment for theranostic imaging. NMR biomedicine. (2011) 24:636–47. doi: 10.1002/nbm.1664
41. Kubo N, Araki K, Kuwano H, and Shirabe K. Cancer-associated fibroblasts in hepatocellular carcinoma. World J gastroenterology. (2016) 22:6841–50. doi: 10.3748/wjg.v22.i30.6841
42. Jahroudi N and Greenberger JS. The role of endothelial cells in tumor invasion and metastasis. J neuro-oncology. (1995) 23:99–108. doi: 10.1007/BF01053415
43. Li L, Deng L, Meng X, Gu C, Meng L, Li K, et al. Tumor-targeting anti-EGFR x anti-PD1 bispecific antibody inhibits EGFR-overexpressing tumor growth by combining EGFR blockade and immune activation with direct tumor cell killing. Trans Oncol. (2021) 14:100916. doi: 10.1016/j.tranon.2020.100916
44. Liu C, Zhou X, Zeng H, Yu J, Li W, Zhang W, et al. Endoplasmic reticulum stress potentiates the immunosuppressive microenvironment in hepatocellular carcinoma by promoting the release of SNHG6-enriched small extracellular vesicles. Cancer Immunol Res. (2024) 12:1184–201. doi: 10.1158/2326-6066.CIR-23-0469
45. Shi J, Shen Y, and Zhang J. Emerging roles of small extracellular vesicles in metabolic reprogramming and drug resistance in cancers. Cancer Drug resistance (Alhambra Calif). (2024) 7:38. doi: 10.20517/cdr.2024.81
46. Wei R, Liu S, Zhang S, Min L, and Zhu S. Cellular and extracellular components in tumor microenvironment and their application in early diagnosis of cancers. Analytical Cell Pathol (Amsterdam). (2020) 2020:6283796. doi: 10.1155/2020/6283796
47. Asao T, Tobias GC, Lucotti S, Jones DR, Matei I, and Lyden D. Extracellular vesicles and particles as mediators of long-range communication in cancer: connecting biological function to clinical applications. Extracellular vesicles circulating Nucleic Acids. (2023) 4:461–85. doi: 10.20517/evcna.2023.37
48. Liu Y, Song J, Shi Q, Chen B, Qiu W, Liu Y, et al. Glucose-induced LINC01419 reprograms the glycolytic pathway by recruiting YBX1 to enhance PDK1 mRNA stability in hepatocellular carcinoma. Clin Trans Med. (2024) 14:e70122. doi: 10.1002/ctm2.70122
49. Chang LC, Chiu HM, Wu MS, and Shen TL. The role of small extracellular vesicles in the progression of colorectal cancer and its clinical applications. Int J Mol Sci. (2022) 23:1379. doi: 10.3390/ijms23031379
50. Cao SQ, Zheng H, Sun BC, Wang ZL, Liu T, Guo DH, et al. Long non-coding RNA highly up-regulated in liver cancer promotes exosome secretion. World J gastroenterology. (2019) 25:5283–99. doi: 10.3748/wjg.v25.i35.5283
51. Li R, Dong C, Jiang K, Sun R, Zhou Y, Yin Z, et al. Rab27B enhances drug resistance in hepatocellular carcinoma by promoting exosome-mediated drug efflux. Carcinogenesis. (2020) 41:1583–91. doi: 10.1093/carcin/bgaa029
52. Hao X, Zhang Y, Shi X, Liu H, Zheng Z, Han G, et al. CircPAK1 promotes the progression of hepatocellular carcinoma via modulation of YAP nucleus localization by interacting with 14-3-3ζ. J Exp Clin Cancer research: CR. (2022) 41:281. doi: 10.1186/s13046-022-02494-z
53. Suda T. Targeting therapy for hepatocellular carcinoma by delivering microRNAs as exosomal cargo. World J gastroenterology. (2024) 30:2369–70. doi: 10.3748/wjg.v30.i17.2369
54. Zhang S, Mang Y, Li L, Ran J, Zhao Y, Li L, et al. Long noncoding RNA NEAT1 changes exosome secretion and microRNA expression carried by exosomes in hepatocellular carcinoma cells. J gastrointestinal Oncol. (2021) 12:3033–49. doi: 10.21037/jgo-21-729
55. Hareendran S, Albraidy B, Yang X, Liu A, Breggia A, Chen CC, et al. Exosomal carboxypeptidase E (CPE) and CPE-shRNA-loaded exosomes regulate metastatic phenotype of tumor cells. Int J Mol Sci. (2022) 23:3113. doi: 10.3390/ijms23063113
56. Yang W, Wei J, Lv L, Xie J, Li A, Zheng Z, et al. p62 promotes Malignancy of hepatocellular carcinoma by regulating the secretion of exosomes and the localization of β-catenin. Front bioscience (Landmark edition). (2022) 27:89. doi: 10.31083/j.fbl2703089
57. Li A, Xie J, Lv L, Zheng Z, Yang W, Zhuo W, et al. RPL9 acts as an oncogene by shuttling miRNAs through exosomes in human hepatocellular carcinoma cells. Int J Oncol. (2024) 64:58. doi: 10.3892/ijo.2024.5646
58. Yu B, Zhou S, Long D, Ning Y, Yao H, Zhou E, et al. DDX55 promotes hepatocellular carcinoma progression by interacting with BRD4 and participating in exosome-mediated cell-cell communication. Cancer science. (2022) 113:3002–17. doi: 10.1111/cas.15393
59. Ashraf Malik M, Ishtiyaq Ali Mirza J, Umar M, and Manzoor S. CD81(+) exosomes play a pivotal role in the establishment of hepatitis C persistent infection and contribute toward the progression of hepatocellular carcinoma. Viral Immunol. (2019) 32:453–62. doi: 10.1089/vim.2019.0077
60. Wei XC, Xia YR, Zhou P, Xue X, Ding S, Liu LJ, et al. Hepatitis B core antigen modulates exosomal miR-135a to target vesicle-associated membrane protein 2 promoting chemoresistance in hepatocellular carcinoma. World J gastroenterology. (2021) 27:8302–22. doi: 10.3748/wjg.v27.i48.8302
61. Koksal AR, Thevenot P, Aydin Y, Nunez K, Sandow T, Widmer K, et al. Impaired autophagy response in hepatocellular carcinomas enriches glypican-3 in exosomes, not in the microvesicles. J hepatocellular carcinoma. (2022) 9:959–72. doi: 10.2147/JHC.S376210
62. Wang J, Pu J, Zhang Y, Yao T, Luo Z, Li W, et al. Exosome-transmitted long non-coding RNA SENP3-EIF4A1 suppresses the progression of hepatocellular carcinoma. Aging. (2020) 12:11550–67. doi: 10.18632/aging.103302
63. Hasanin AH, Matboli M, and Seleem HS. Hesperidin suppressed hepatic precancerous lesions via modulation of exophagy in rats. J Cell Biochem. (2020) 121:1295–306. doi: 10.1002/jcb.29363
64. Tong K, Wang P, Li Y, Tong Y, Li X, Yan S, et al. Resveratrol inhibits hepatocellular carcinoma progression through regulating exosome secretion. Curr medicinal Chem. (2024) 31:2107–18. doi: 10.2174/0929867331666230914090053
65. Li Y, Yu S, Li L, Chen J, Quan M, Li Q, et al. KLF4-mediated upregulation of CD9 and CD81 suppresses hepatocellular carcinoma development via JNK signaling. Cell Death disease. (2020) 11:299. doi: 10.1038/s41419-020-2479-z
66. Su YY, Chao CH, Hsu HY, Li HK, Wang YL, Wu Lee YH, et al. DDX3 suppresses hepatocellular carcinoma progression through modulating the secretion and composition of exosome. Am J Cancer Res. (2023) 13:1744–65.
67. Mahati S, Fu X, Ma X, Zhang H, and Xiao L. Delivery of miR-26a using an exosomes-based nanosystem inhibited proliferation of hepatocellular carcinoma. Front Mol biosciences. (2021) 8:738219. doi: 10.3389/fmolb.2021.738219
68. Ellipilli S, Wang H, Binzel DW, Shu D, and Guo P. Ligand-displaying-exosomes using RNA nanotechnology for targeted delivery of multi-specific drugs for liver cancer regression. Nanomedicine: nanotechnology biology Med. (2023) 50:102667. doi: 10.1016/j.nano.2023.102667
69. Khan N, Maurya S, Bammidi S, and Jayandharan GR. AAV6 vexosomes mediate robust suicide gene delivery in a murine model of hepatocellular carcinoma. Mol Ther Methods Clin Dev. (2020) 17:497–504. doi: 10.1016/j.omtm.2020.03.006
70. Wan T, Zhong J, Pan Q, Zhou T, Ping Y, and Liu X. Exosome-mediated delivery of Cas9 ribonucleoprotein complexes for tissue-specific gene therapy of liver diseases. Sci Adv. (2022) 8:eabp9435. doi: 10.1126/sciadv.abp9435
71. Li X, Yu Q, Zhao R, Guo X, Liu C, Zhang K, et al. Designer exosomes for targeted delivery of a novel therapeutic cargo to enhance sorafenib-mediated ferroptosis in hepatocellular carcinoma. Front Oncol. (2022) 12:898156. doi: 10.3389/fonc.2022.898156
72. Huang LH, Rau CS, Liu YW, Lin HP, Wu YC, Tsai CW, et al. Cathelicidin antimicrobial peptide acts as a tumor suppressor in hepatocellular carcinoma. Int J Mol Sci. (2023) 24:15652. doi: 10.3390/ijms242115652
73. Kim HY, Min HK, Song HW, Yoo A, Lee S, Kim KP, et al. Delivery of human natural killer cell-derived exosomes for liver cancer therapy: an in vivo study in subcutaneous and orthotopic animal models. Drug delivery. (2022) 29:2897–911. doi: 10.1080/10717544.2022.2118898
74. Ghotaslou A, Azizsoltani A, Baghaei K, and Alizadeh E. Harnessing HEK293 cell-derived exosomes for hsa-miR-365a-3p delivery: Potential application in hepatocellular carcinoma therapy. Heliyon. (2024) 10:e29333. doi: 10.1016/j.heliyon.2024.e29333
75. Hou TZ, Yang HM, Cheng YZ, Gu L, Zhang JN, and Zhang H. The Parkinson’s disease-associated protein α-synuclein inhibits hepatoma by exosome delivery. Mol carcinogenesis. (2023) 62:1163–75. doi: 10.1002/mc.23553
76. Wang P, Tong K, Li Y, Li X, Zhang Y, Gu J, et al. The role and mechanism of HIF-1α-mediated glypican-3 secretion in hypoxia-induced tumor progression in hepatocellular carcinoma. Cell signalling. (2024) 114:111007. doi: 10.1016/j.cellsig.2023.111007
77. Yang X, Wu M, Kong X, Wang Y, Hu C, Zhu D, et al. Exosomal miR-3174 induced by hypoxia promotes angiogenesis and metastasis of hepatocellular carcinoma by inhibiting HIPK3. iScience. (2024) 27:108955. doi: 10.1016/j.isci.2024.108955
78. Yu Y, Min Z, Zhou Z, Linhong M, Tao R, Yan L, et al. Hypoxia-induced exosomes promote hepatocellular carcinoma proliferation and metastasis via miR-1273f transfer. Exp Cell Res. (2019) 385:111649. doi: 10.1016/j.yexcr.2019.111649
79. Ju T, Yang F, Wang S, Wang J, Song Z, Xu H, et al. Nanoscopic characterization of hepatocytes treated with normoxic and hypoxic tumor-derived exosomes. Micron (Oxford England: 1993). (2022) 158:103283. doi: 10.1016/j.micron.2022.103283
80. Ju T, Dong J, Wang B, Qu K, Cheng C, He X, et al. Cancer development in hepatocytes by long-term induction of hypoxic hepatocellular carcinoma cell (HCC)-derived exosomes in vivo and in vitro. Mol pharmaceutics. (2023) 20:5579–92. doi: 10.1021/acs.molpharmaceut.3c00488
81. Jia W, Liang S, Jin M, Li S, Yuan J, Zhang J, et al. Oleanolic acid inhibits hypoxic tumor-derived exosomes-induced premetastatic niche formation in hepatocellular carcinoma by targeting ERK1/2-NFκB signaling. Phytomedicine: Int J phytotherapy phytopharmacology. (2024) 126:155208. doi: 10.1016/j.phymed.2023.155208
82. Shang H, Lu L, Fan M, Lu Y, Shi X, and Lu H. Exosomal circHIF1A derived from hypoxic-induced carcinoma-associated fibroblasts promotes hepatocellular carcinoma cell Malignant phenotypes and immune escape. Int immunopharmacology. (2024) 138:112282. doi: 10.1016/j.intimp.2024.112282
83. Ji W, Bai J, and Ke Y. Exosomal ZFPM2-AS1 contributes to tumorigenesis, metastasis, stemness, macrophage polarization, and infiltration in hepatocellular carcinoma through PKM mediated glycolysis. Environ toxicology. (2023) 38:1332–46. doi: 10.1002/tox.23767
84. Ye Y, Wang M, Wang G, Mai Z, Zhou B, Han Y, et al. lncRNA miR4458HG modulates hepatocellular carcinoma progression by activating m6A-dependent glycolysis and promoting the polarization of tumor-associated macrophages. Cell Mol Life sciences: CMLS. (2023) 80:99. doi: 10.1007/s00018-023-04741-8
85. Lai Z, Wei T, Li Q, Wang X, Zhang Y, and Zhang S. Exosomal circFBLIM1 Promotes Hepatocellular Carcinoma Progression and Glycolysis by Regulating the miR-338/LRP6 Axis. Cancer biotherapy radiopharmaceuticals. (2023) 38:674–83. doi: 10.1089/cbr.2020.3564
86. Li Y, Zang H, Zhang X, and Huang G. Exosomal Circ-ZNF652 Promotes Cell Proliferation, Migration, Invasion and Glycolysis in Hepatocellular Carcinoma via miR-29a-3p/GUCD1 Axis. Cancer Manage Res. (2020) 12:7739–51. doi: 10.2147/CMAR.S259424
87. Zhang J, Lu S, Zhou Y, Meng K, Chen Z, Cui Y, et al. Motile hepatocellular carcinoma cells preferentially secret sugar metabolism regulatory proteins via exosomes. Proteomics. (2017) 17. doi: 10.1002/pmic.201700103
88. Yang S, Li A, Lv L, Zheng Z, Liu P, Min J, et al. Exosomal miRNA-146a-5p Derived from Senescent Hepatocellular Carcinoma Cells Promotes Aging and Inhibits Aerobic Glycolysis in Liver Cells via Targeting IRF7. J Cancer. (2024) 15:4448–66. doi: 10.7150/jca.96500
89. Chen A, Zhang VX, Zhang Q, Sze KM, Tian L, Huang H, et al. Targeting the oncogenic m6A demethylase FTO suppresses tumourigenesis and potentiates immune response in hepatocellular carcinoma. Gut. (2024) 74:90–102. doi: 10.1136/gutjnl-2024-331903
90. Yang J, Xu H, Wu W, Huang H, Zhang C, Tang W, et al. Ferroptosis signaling promotes the release of misfolded proteins via exosomes to rescue ER stress in hepatocellular carcinoma. Free Radical Biol Med. (2023) 202:110–20. doi: 10.1016/j.freeradbiomed.2023.03.027
91. Wei Y, Tang X, Ren Y, Yang Y, Song F, Fu J, et al. An RNA-RNA crosstalk network involving HMGB1 and RICTOR facilitates hepatocellular carcinoma tumorigenesis by promoting glutamine metabolism and impedes immunotherapy by PD-L1+ exosomes activity. Signal transduction targeted Ther. (2021) 6:421. doi: 10.1038/s41392-021-00801-2
92. Liu P, Zhu W, Chen C, Yan B, Zhu L, Chen X, et al. The mechanisms of lysophosphatidylcholine in the development of diseases. Life Sci. (2020) 247:117443. doi: 10.1016/j.lfs.2020.117443
93. Ramos CC, Pires J, Gonzalez E, Garcia-Vallicrosa C, Reis CA, Falcon-Perez JM, et al. Extracellular vesicles in tumor-adipose tissue crosstalk: key drivers and therapeutic targets in cancer cachexia. Extracellular vesicles circulating Nucleic Acids. (2024) 5:371–96. doi: 10.20517/evcna.2024.36
94. Biscans A, Haraszti RA, Echeverria D, Miller R, Didiot MC, Nikan M, et al. Hydrophobicity of lipid-conjugated siRNAs predicts productive loading to small extracellular vesicles. Mol therapy: J Am Soc Gene Ther. (2018) 26:1520–8. doi: 10.1016/j.ymthe.2018.03.019
95. Jin Y, Sun G, Chen B, Feng S, Tang M, Wang H, et al. Delivering miR-23b-3p by small extracellular vesicles to promote cell senescence and aberrant lipid metabolism. BMC Biol. (2025) 23:41. doi: 10.1186/s12915-025-02143-9
96. Liu G, Ouyang X, Sun Y, Xiao Y, You B, Gao Y, et al. The miR-92a-2-5p in exosomes from macrophages increases liver cancer cells invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling. Cell Death differentiation. (2020) 27:3258–72. doi: 10.1038/s41418-020-0575-3
97. Fan F, Chen K, Lu X, Li A, Liu C, and Wu B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol Int. (2021) 15:444–58. doi: 10.1007/s12072-020-10101-6
98. Chen J, Lin Z, Liu L, Zhang R, Geng Y, Fan M, et al. GOLM1 exacerbates CD8(+) T cell suppression in hepatocellular carcinoma by promoting exosomal PD-L1 transport into tumor-associated macrophages. Signal transduction targeted Ther. (2021) 6:397. doi: 10.1038/s41392-021-00784-0
99. Zhang L, Zhang J, Li P, Li T, Zhou Z, and Wu H. Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death disease. (2022) 13:32. doi: 10.1038/s41419-021-04345-9
100. Lu JC, Zhang PF, Huang XY, Guo XJ, Gao C, Zeng HY, et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J Hematol Oncol. (2021) 14:200. doi: 10.1186/s13045-021-01207-x
101. Hu Z, Zhang H, Liu W, Yin Y, Jiang J, Yan C, et al. Mechanism of HBV-positive liver cancer cell exosomal miR-142-3p by inducing ferroptosis of M1 macrophages to promote liver cancer progression. Trans Cancer Res. (2022) 11:1173–87. doi: 10.21037/tcr-22-96
102. Liu Y, Tang Y, Jiang H, Zhang X, Chen X, Guo J, et al. Exosome-related FTCD facilitates M1 macrophage polarization and impacts the prognosis of hepatocellular carcinoma. Biomolecules. (2023) 14:41. doi: 10.3390/biom14010041
103. Chen H, Jiang S, Zhang P, Ren Z, and Wen J. Exosomes synergized with PIONs@E6 enhance their immunity against hepatocellular carcinoma via promoting M1 macrophages polarization. Int immunopharmacology. (2021) 99:107960. doi: 10.1016/j.intimp.2021.107960
104. Chen Y, Li X, Shang H, Sun Y, Wang C, Wang X, et al. Mechanism exploration of synergistic photo-immunotherapy strategy based on a novel exosome-like nanosystem for remodeling the immune microenvironment of HCC. Nano convergence. (2024) 11:31. doi: 10.1186/s40580-024-00441-6
105. Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization. Int J nanomedicine. (2021) 16:2803–18. doi: 10.2147/IJN.S284560
106. Zongqiang H, Jiapeng C, Yingpeng Z, Chuntao Y, Yiting W, Jiashun Z, et al. Exosomal miR-452-5p induce M2 macrophage polarization to accelerate hepatocellular carcinoma progression by targeting TIMP3. J Immunol Res. (2022) 2022:1032106. doi: 10.1155/2022/1032106
107. Yu H, Pan J, Zheng S, Cai D, Luo A, Xia Z, et al. Hepatocellular carcinoma cell-derived exosomal miR-21-5p induces macrophage M2 polarization by targeting rhoB. Int J Mol Sci. (2023) 24:4593. doi: 10.3390/ijms24054593
108. Xu Y, Luan G, Liu F, Zhang Y, Li Z, Liu Z, et al. Exosomal miR-200b-3p induce macrophage polarization by regulating transcriptional repressor ZEB1 in hepatocellular carcinoma. Hepatol Int. (2023) 17:889–903. doi: 10.1007/s12072-023-10507-y
109. Ren X, Ju Y, Wang C, Wei R, Sun H, and Zhang Q. MARCKS on tumor-associated macrophages is correlated with immune infiltrates and poor prognosis in hepatocellular carcinoma. Cancer Invest. (2021) 39:756–68. doi: 10.1080/07357907.2021.1950757
110. Li W, Xin X, Li X, Geng J, and Sun Y. Exosomes secreted by M2 macrophages promote cancer stemness of hepatocellular carcinoma via the miR-27a-3p/TXNIP pathways. Int Immunopharmacol. (2021) 101:107585. doi: 10.1016/j.intimp.2021.107585
111. Tian B, Zhou L, Wang J, and Yang P. miR-660-5p-loaded M2 macrophages-derived exosomes augment hepatocellular carcinoma development through regulating KLF3. Int Immunopharmacol. (2021) 101:108157. doi: 10.1016/j.intimp.2021.108157
112. Xie S, Li X, Yan J, Yu H, Chen S, and Chen K. Knockdown of liver cancer cell-secreted exosomal PSMA5 controls macrophage polarization to restrain cancer progression by blocking JAK2/STAT3 signaling. Immunity Inflammation disease. (2024) 12:e1146. doi: 10.1002/iid3.1146
113. Ai JH, Wen YZ, Dai SJ, Zhang LD, Huang ZJ, and Shi J. Exosomal lncRNA HEIH, an essential communicator for hepatocellular carcinoma cells and macrophage M2 polarization through the miR-98-5p/STAT3 axis. J Biochem Mol toxicology. (2024) 38:e23686. doi: 10.1002/jbt.23686
114. Li W, Zhao B, Wang Q, Lu J, Wu X, and Chen X. M2 macrophage exosomes promote resistance to sorafenib in hepatocellular carcinoma cells via miR-200c-3p. Int immunopharmacology. (2024) 139:112807. doi: 10.1016/j.intimp.2024.112807
115. Lu Y, Han G, Zhang Y, Zhang L, Li Z, Wang Q, et al. M2 macrophage-secreted exosomes promote metastasis and increase vascular permeability in hepatocellular carcinoma. Cell communication signaling: CCS. (2023) 21:299. doi: 10.1186/s12964-022-00872-w
116. Wang X, Shen H, Zhangyuan G, Huang R, Zhang W, He Q, et al. 14-3-3ζ delivered by hepatocellular carcinoma-derived exosomes impaired anti-tumor function of tumor-infiltrating T lymphocytes. Cell Death disease. (2018) 9:159. doi: 10.1038/s41419-017-0180-7
117. Huang M, Huang X, and Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer science. (2022) 113:1968–83. doi: 10.1111/cas.15365
118. Rao Q, Zuo B, Lu Z, Gao X, You A, Wu C, et al. Tumor-derived exosomes elicit tumor suppression in murine hepatocellular carcinoma models and humans in vitro. Hepatol (Baltimore Md). (2016) 64:456–72. doi: 10.1002/hep.28549
119. Li J, Lin W, Huang T, Chen M, and Lin Q. IL-12 improves the anti-HCC efficacy of dendritic cells loaded with exosomes from overexpressing Rab27a tumor cells. Exp Cell Res. (2024) 439:114073. doi: 10.1016/j.yexcr.2024.114073
120. Zhong X, Zhou Y, Cao Y, Ding J, Wang P, Luo Y, et al. Enhanced antitumor efficacy through microwave ablation combined with a dendritic cell-derived exosome vaccine in hepatocellular carcinoma. Int J hyperthermia: Off J Eur Soc Hyperthermic Oncology North Am Hyperthermia Group. (2020) 37:1210–8. doi: 10.1080/02656736.2020.1836406
121. Shi S, Wang L, Wang C, Xu J, and Niu Z. Serum-derived exosomes function as tumor antigens in patients with advanced hepatocellular carcinoma. Mol Immunol. (2021) 134:210–7. doi: 10.1016/j.molimm.2021.03.017
122. Lu Z, Zuo B, Jing R, Gao X, Rao Q, Liu Z, et al. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J hepatology. (2017) 67:739–48. doi: 10.1016/j.jhep.2017.05.019
123. Chang C, Pei Y, Zhang C, Zhang W, Qin Y, and Shi S. Combination therapy with dendritic cell loaded-exosomes supplemented with PD-1 inhibition at different time points have superior antitumor effect in hepatocellular carcinoma. Cancer immunology immunotherapy: CII. (2023) 72:3727–38. doi: 10.1007/s00262-023-03525-0
124. Yao T, Lv M, Ma S, Chen J, Zhang Y, Yu Y, et al. Ubiquitinated hepatitis D antigen-loaded microvesicles induce a potent specific cellular immune response to inhibit HDV replication in vivo. Microbiol spectrum. (2021) 9:e0102421. doi: 10.1128/Spectrum.01024-21
125. Nakano T, Chen IH, Wang CC, Chen PJ, Tseng HP, Huang KT, et al. Circulating exosomal miR-92b: Its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am J transplantation: Off J Am Soc Transplant Am Soc Transplant Surgeons. (2019) 19:3250–62. doi: 10.1111/ajt.15490
126. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol cancer. (2020) 19:110. doi: 10.1186/s12943-020-01222-5
127. Zhou Z, Li T, Li J, Lin W, and Zheng Q. Exosomal transfer of HCC-derived miR-17-5p downregulates NK cell function by targeting RUNX1-NKG2D axis. Int immunopharmacology. (2024) 136:112361. doi: 10.1016/j.intimp.2024.112361
128. Kim IY, Kim HY, Song HW, Park JO, Choi YH, and Choi E. Functional enhancement of exosomes derived from NK cells by IL-15 and IL-21 synergy against hepatocellular carcinoma cells: The cytotoxicity and apoptosis in vitro study. Heliyon. (2023) 9:e16962. doi: 10.1016/j.heliyon.2023.e16962
129. Yugawa K, Yoshizumi T, Mano Y, Itoh S, Harada N, Ikegami T, et al. Cancer-associated fibroblasts promote hepatocellular carcinoma progression through downregulation of exosomal miR-150-3p. Eur J Surg oncology: J Eur Soc Surg Oncol Br Assoc Surg Oncol. (2021) 47:384–93. doi: 10.1016/j.ejso.2020.08.002
130. Su Z, Lu C, Zhang F, Liu H, Li M, Qiao M, et al. Cancer-associated fibroblasts-secreted exosomal miR-92a-3p promotes tumor growth and stemness in hepatocellular carcinoma through activation of Wnt/β-catenin signaling pathway by suppressing AXIN1. J Cell Physiol. (2024) 239:e31344. doi: 10.1002/jcp.31344
131. Qi Y, Wang H, Zhang Q, Liu Z, Wang T, Wu Z, et al. CAF-Released Exosomal miR-20a-5p Facilitates HCC Progression via the LIMA1-Mediated β-Catenin Pathway. Cells. (2022) 11:3857. doi: 10.3390/cells11233857
132. Mu W, Gu P, Li H, Zhou J, Jian Y, Jia W, et al. Exposure of benzo[a]pyrene induces HCC exosome-circular RNA to activate lung fibroblasts and trigger organotropic metastasis. Cancer Commun (London England). (2024) 44:718–38. doi: 10.1002/cac2.12574
133. Liu X, Wang H, Yang M, Hou Y, Chen Y, and Bie P. Exosomal miR-29b from cancer-associated fibroblasts inhibits the migration and invasion of hepatocellular carcinoma cells. Trans Cancer Res. (2020) 9:2576–87. doi: 10.21037/tcr.2020.02.68
134. Zhou Y, Tang W, Zhuo H, Zhu D, Rong D, Sun J, et al. Cancer-associated fibroblast exosomes promote chemoresistance to cisplatin in hepatocellular carcinoma through circZFR targeting signal transducers and activators of transcription (STAT3)/nuclear factor -kappa B (NF-κB) pathway. Bioengineered. (2022) 13:4786–97. doi: 10.1080/21655979.2022.2032972
135. Qin W, Wang L, Tian H, Wu X, Xiao C, Pan Y, et al. CAF-derived exosomes transmitted Gremlin-1 promotes cancer progression and decreases the sensitivity of hepatoma cells to sorafenib. Mol carcinogenesis. (2022) 61:764–75. doi: 10.1002/mc.23416
136. Clayton A, Harris CL, Court J, Mason MD, and Morgan BP. Antigen-presenting cell exosomes are protected from complement-mediated lysis by expression of CD55 and CD59. Eur J Immunol. (2003) 33:522–31. doi: 10.1002/immu.200310028
137. Papp K, Végh P, Prechl J, Kerekes K, Kovács J, Csikós G, et al. B lymphocytes and macrophages release cell membrane deposited C3-fragments on exosomes with T cell response-enhancing capacity. Mol Immunol. (2008) 45:2343–51. doi: 10.1016/j.molimm.2007.11.021
138. Wang X, Zhou Y, Dong K, Zhang H, Gong J, and Wang S. Exosomal lncRNA HMMR-AS1 mediates macrophage polarization through miR-147a/ARID3A axis under hypoxia and affects the progression of hepatocellular carcinoma. Environ toxicology. (2022) 37:1357–72. doi: 10.1002/tox.23489
139. Xu M, Zhou C, Weng J, Chen Z, Zhou Q, Gao J, et al. Tumor associated macrophages-derived exosomes facilitate hepatocellular carcinoma Malignance by transferring lncMMPA to tumor cells and activating glycolysis pathway. J Exp Clin Cancer research: CR. (2022) 41:253. doi: 10.1186/s13046-022-02458-3
140. Lu L, Huang J, Mo J, Da X, Li Q, Fan M, et al. Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol Biol letters. (2022) 27:17. doi: 10.1186/s11658-022-00309-9
141. Tao L, Li D, Mu S, Tian G, and Yan G. LncRNA MAPKAPK5_AS1 facilitates cell proliferation in hepatitis B virus -related hepatocellular carcinoma. Lab investigation; J Tech Methods pathology. (2022) 102:494–504. doi: 10.1038/s41374-022-00731-9
142. Wang L, Yi X, Xiao X, Zheng Q, Ma L, and Li B. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell Mol Biol letters. (2022) 27:106. doi: 10.1186/s11658-022-00406-9
Keywords: hepatocellular carcinoma, small extracellular vesicles, multi-network fusion, mechanism, therapy
Citation: Yuan X, Huang D, Peng L, Lin Y, Wang L, Yan J, Qiu Y, Song C and Wang Q (2025) Advances in the mechanism of small extracellular vesicles promoting the development of hepatocellular carcinoma through multi-network fusion. Front. Immunol. 16:1558468. doi: 10.3389/fimmu.2025.1558468
Received: 10 January 2025; Accepted: 17 June 2025;
Published: 09 July 2025.
Edited by:
Anirban Ganguly, All India Institute of Medical Sciences Deoghar, IndiaReviewed by:
Dwijendra K. Gupta, Allahabad University, IndiaZhijin Fan, Sun Yat-sen University, China
Copyright © 2025 Yuan, Huang, Peng, Lin, Wang, Yan, Qiu, Song and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Wang, cXdhbmdrQG5hdmVyLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Xiaoying Yuan
Xiaoying Yuan Defa Huang
Defa Huang Liang Peng
Liang Peng Yilong Lin
Yilong Lin Lijuan Wang
Lijuan Wang Jiawei Yan4
Jiawei Yan4