- 1Institute of Plant Protection, Shandong Key Laboratory for Green Prevention and Control of Agricultural Pests, Shandong Academy of Agricultural Sciences, Jinan, Shandong, China
- 2Hebei Provincial Jujube Kernel Utilization Technology Innovation Center, Department of Chemical Engineering and Biotechnology, Xingtai University, Xingtai, China
Introduction: The innate immune system provides defense against invading pathogens in insects and mammals.
Methods: We conducted transcriptomic analyses of the locust Locusta migratoria under Aspergillus oryzae infection to clarify temporal variation in its molecular immune response.
Results: We found that fat body cells and hemocytes play different roles in the immune response of locusts to Aspergillus infection at different time points after inoculation, and melanization was the main process underlying the immune response of female locusts. Most pattern recognition receptors (PRR) genes were up-regulated in fat body cells and down-regulated in hemocytes from 24 h to 72 h after inoculation. This means that fat body cells, but not hemocytes, would be able to precisely recognize invading pathogens. Most serine protease inhibitors (SERPINs) genes and clip domain serine proteinase (CLIP) genes were up-regulated in fat body cells. However, most SERPINs were down-regulated in hemocytes, which indicated that serine proteinases may be inhibited to activate downstream reactions involving the prophenoloxidase (PPO), peroxidase (POD), and Toll pathways. Most lysozymes, PPOs, and peroxiredoxin (PRDX) are effectors that were up-regulated in fat body cells 24 h after inoculation but down-regulated 48 h and 72 h after inoculation. Similar patterns were observed for effectors in hemocytes, which indicates that locust immune genes expression was suppressed by A. oryzae 72 h after inoculation, and might result in the weak melanization of locusts in response to Aspergillus infection.
Discussion: Our findings enhance our understanding of insect–fungi interactions, as well as have implications for the development of more effective microbial control strategies for the management of locust populations.
1 Introduction
The innate immune system of insects and mammals protects hosts against invading pathogens. Non-specific innate immune responses involve many areas of the body and play crucial roles in the prevention of disease and infections (1). There are similarities between the insect immune system and mammalian innate immune system and because the immune system in insects is free of some of the disadvantages of the mammalian immune system, many insects are used in studies of the innate immune system. Some of these insects include Drosophila melanogaster (2, 3), Galleria mellonella (4, 5), and Bombyx mori (6).
Several groups of genes and their pathways have been identified in insect innate immune systems. Pattern recognition receptors (PRRs), effectors, and Toll and IMD pathways are the main immune pathways in insects. PRRs such as peptidoglycan recognition protein (PGRP), β-1,3-glucan binding protein (βGBPs), Calcium-dependent (C-type) lectins, and scavenger receptors (SRs) recognize pathogen infections (7). Toll and Toll-like receptors are initiated through these PRRs and by a cascade of serine protease inhibitors (SERPINs); they play a key role in the development, adhesion, and immune responses of insects (8–10). Antimicrobial effectors are induced via the Toll and IMD (immune deficiency) pathways, as well as by the Janus kinase/signal transducer and activator of transcription (JAK-STAT) pathways to protect hosts against infection (11, 12). The fat body and hemolymph are involved in the humoral immune process, and they play different roles against pathogens (13, 14). The fat body is the main organ responsible for the synthesis of hormones, immune-related proteins, energy compounds, and pheromones (15). Various compounds that are produced in fat body are released in the hemolymph (16); the hemolymph thus plays a key role in humoral defense.
Immune responses to pathogen infections vary among insect species (17). Metarhizium spp. and Beauveria spp. synthesize destruxins and oosporein, respectively, to inhibit prophenoloxidase (PPO) activity and down-regulate the expression of antifungal peptides (18, 19). M. anisopliae and M. robertsii secrete more proteases, chitinases, G protein-coupled receptors, and detoxifying enzymes than M. album and M. acridum (18). Mosquito genomes encode nine to ten POs with different functions (20–22). Previous studies have shown that holometabolous insects, such as Lepidoptera, Coleoptera, and Drosophila, produce antimicrobial peptides (AMPs) through the Toll pathway after infection with fungal pathogens, and AMPs in hemimetabolous pea aphids in the order Hemiptera are produced through the JNK pathway after infection with bacterial pathogens (23). In locusts, many immune-related genes have been identified (24, 25), and Toll and IMD pathways are activated in L. migratoria after infection with Metarhizium acridum (25). However, few studies have examined the molecular immune responses of both the fat body and hemolymph over time in insects (25).
Invasive aspergillosis is a severe fungal infection caused by Aspergillus species in humans, and most studies of the immune response against Aspergillus have focused on mammals (26). The humoral immune response has been poorly studied. The virulence of medically important pathogens has been studied in various insects because of the similarities of insect immune responses with mammalian innate immune responses (27).
Aspergiullus oryzae XJ1 has recently been identified to be highly virulent against L. migratoria and other species of grasshoppers (28, 29). Study of the molecular immune mechanism of locusts against A. oryzae may help promote the development of new biocontrol agents for the management of locust plagues and provide insights into the innate humoral immune response in humans. Here, we conducted transcriptomic analysis and characterized temporal variation in the expression of immune genes in female locust fat body cells and hemocytes under A. oryzae challenge.
2 Materials and methods
2.1 Locust rearing
Locusts, Locusta migratoria manilensis (Orthoptera: Acrididae) were obtained from Institute of Plant Protection, Shandong Academy of Agricultural Sciences. Fifth-instar nymphs were reared in cylindrical steel cages (diameter 15 cm × height 40 cm) with 25-35 locusts per cage. Fresh wheat seedlings were provided as a food source. Observations were made daily to determine the timing of the molting of the fifth-instar nymphs into adults. Newly molted adults were collected and reared in separate cages. All female adults were reared for 2 ± 1 days at 25 ± 1°C with 55 ± 10% relative humidity and a 15 h/9 h light/dark photoperiod.
2.2 A. oryzae strains and L. migratoria inoculation
A. oryzae XJ1 was provided by the Key Laboratory for the Biological Control of Pests of the Ministry of Agriculture, China Agricultural University. The fresh conidia powder was cultured on potato dextrose agar plates at 26.0 ± 1.0°C. The conidia germination rate was 64.25 ± 5.8% (mean ± SEM, n = 4) after 24 h of incubation under standardized conditions, indicating robust conidia viability. A conidial suspension with a concentration of 1 × 107 conidia mL-1 was prepared by suspending conidia in sterile double-distilled water containing 0.3% (vol/vol) Tween-80. The concentration of the conidia was quantified using a Neubauer hemocytometer under a light microscope to ensure accuracy. Healthy adult females were fully immersed in the prepared conidial suspension about 2 seconds to ensure consistent exposure according to previous studies (28, 29). Control adults were inoculated with 0.3% (vol/vol) Tween-80 solution. After treatment, all treated locusts were reared in clear plastic boxes. Adults were housed separately in groups of 3 ± 1 adults in individual clear plastic boxes with a round top (diameter, 14 cm), round bottom (diameter, 9 cm), and height of 14 cm. All treated and control adults were reared for 24 ± 2 h, 48 ± 2 h, and 72 ± 2 h at 25 ± 2°C with 55 ± 10% relative humidity and a 15 h/9 h light/dark photoperiod.
2.3 RNA extraction and sequencing
Female adults were collected from the boxes for RNA extraction. The arthrodial membrane of the hindlegs of treated and control adults was swabbed and air-dried for 15 min for the collection of hemolymph. After cutting the hindleg, female adult hemolymph was collected using a micropipette and transferred to Trizol reagent (Invitrogen, U.S.A). Fat bodies were collected by dissecting the adults on a waxed Petri dish. All collected fat bodies were transferred to Trizol reagent for RNA extraction. RNA was extracted using the Trizol Reagent following the manufacturer instructions. Briefly, the sample was homogenized in 1 mL of Trizol Reagent at room temperature for 5 min. Next, 200 μL of trichloromethane was added and mixed well; the mixture was then left to stand at room temperature for 2 min and centrifuged at 12,000 rpm and 4°C for 15 min. The supernatant was then removed. Subsequently, 500 μL of isopropyl alcohol was added and mixed well; the mixture was then left at room temperature for 10 min and centrifuged at 12,000 rpm and 4°C for 10 min; the precipitate was retained. One mL of 75% ethanol was washed twice (centrifuged at 7,500 rpm and 4°C for 5 min); the precipitate was retained, and an adequate amount of RNase-free water was added after fully drying. The quantity and quality of RNA samples were examined using a Nanodrop spectrophotometer (Thermo Scientific, U.S.A.). A total of four replicates of treated individuals and control individuals were performed for the female fat body samples at each time interval. Fat bodies were collected from 6–10 female adults. A total of four boxes were considered one replicate in the fat body extraction experiment. A total of four replicates of treated individuals and control individuals were performed for the female hemolymph samples 24 h and 72 h after inoculation while three replications of treated and four replications of control were performed 48 h after inoculation. Hemolymph was collected from 10–15 female adults. A total of five boxes were considered one replicate for hemolymph extraction.
mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first-strand cDNA was synthesized using random hexamer primers followed by second-strand cDNA synthesis. The library was ready after end-repair, A-tailing, adapter ligation, size selection, amplification, and purification. The library was checked using a Qubit 2.0 Fluorometer (Thermo Scientific, U.S.A.) and real-time PCR for quantification, and an Agilent 2100 bioanalyzer was used to characterize the size distribution. After library quality control, different libraries were pooled based on the effective concentration (1.5 nM) and targeted amount of data; they were then subjected to Illumina sequencing (a platform provided by Novogene company, China). The basic principle of the sequencing process is “Sequencing by Synthesis,” where fluorescently labeled dNTPs, DNA polymerase, and adapter primers are added to the sequencing flow cell for amplification. The sequencer captures these fluorescence signals and converts them into sequencing peaks through computer software, which yields sequence information of the target fragment.
2.4 Gene identification
Clean reads for subsequent analysis were obtained after filtering raw data and checking the sequencing error rate and GC content distribution. Hisat2 v2.0.5 software (Johns Hopkins University, U.S.A.) was used to quickly and accurately compare clean reads with the reference genome to obtain the reference genome’s location information. Novel transcripts were assembled using String Tie software (Johns Hopkins University, U.S.A.) and annotated using the Pfam database (European Bioinformatics Institute, U.K.).
FeatureCounts v1.5.0-p3 (The Walter and Eliza Hall Institute of Medical Research, Australia) was used to count the number of reads mapped to each gene. Analysis of differentially expressed genes across samples was performed using the DESeq2 R package (1.20.0). DESeq2 provides statistical programs to identify differentially expressed genes by modeling count data using a negative binomial distribution. The significance of differential expression was assessed through hypothesis testing, with P-values calculated based on Wald tests. P-values were subsequently adjusted using the Benjamini and Hochberg’s methods, which controls the false discovery rate (FDR) at a predefined threshold. Genes meeting the criteria of adjusted P-value ≤ 0.05 were considered different-expressed genes, and log2(foldchange) of the gene was calculated according to normalized reads. In this study, genes whose log2(foldchange) > 0 were called up-regulated genes, and whose log2(foldchange) < 0 were called down-regulated genes. Based on gene annotation, Immune-related genes were identified from the differentially expressed genes in control and treated populations. All genes associated with these immune-related genes were further validated using BLAST analysis on the NCBI database, and matched genes were kept. Then we calculated the FPKM of each gene based on the length of the gene and the read count mapped to this gene, and further compared the FPKM values of up/down-regulated genes between treated and control with t-tests, to clarify the significantly changes of their expression levels between treated and control.
2.5 Statistical analysis
According to the principal component analysis, the few samples that were not clustered together were deleted, and the gene expression levels of the remaining samples were compared. Differences in the FPKM values of genes between control and treated populations were analyzed using t-tests. The relative FPKM value of a gene was calculated based on the FPKM value of the gene in the treatment populations subtracted from the average FPKM value of the gene in the control populations. The relative FPKM values of genes varying in expression in the treated populations among time points were analyzed using one-way ANOVA followed by Tukey’s multiple comparison test with the following significance thresholds: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. All Figures were drawn with Microsoft Excel and GraphPad Prism 7 (GraphPad Software, San Diego, CA, United States).
3 Results
3.1 Temporal variation in gene expression in L. migratoria fat body cells and hemocytes after exposure to A. oryzae
The transcriptome of female L. migratoria adult fat body cells and hemocytes from control and infected with A. oryzae were analyzed. Fungal infection induced a marked shift in the genes expression in fat body cells and hemocytes. Differential gene expression analysis revealed 1,048 DEGs, 1,891 DEGs, and 1,037 DEGs in female fat body cells 24 h, 48 h, and 72 h after inoculation, respectively. The fold change of up-regulated and down-regulated genes was higher 48 h after A. oryzae inoculation compared with that 24 h and 72 h after A. oryzae inoculation (Table 1). Differential gene expression analysis revealed 873 DEGs, 881 DEGs, and 840 DEGs in female hemocytes 24 h, 48 h, and 72 h after inoculation, respectively. The number of down-regulated genes in hemocytes was higher than the number of up-regulated genes (Table 1).
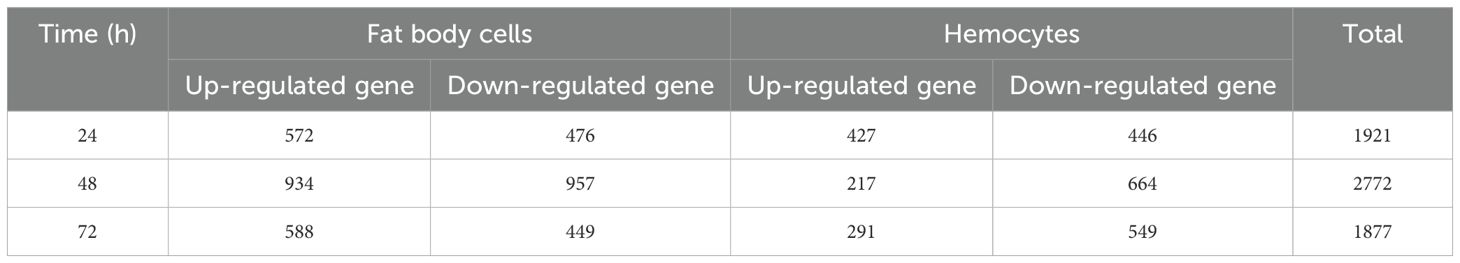
Table 1. Up-regulated and down-regulated genes in female fat body cells and hemocytes in three- time treatment after infection with A. oryzae.
3.2 Differential expression of immune genes
The immune response of female fat body cells and hemocytes varied over time after A. oryzae inoculation. The total number of immune-related genes decreased in female fat body cells over time. In fat body, the total number of up-regulated genes was higher than down-regulated ones; in contrast, in hemocytes the number of up-regulated genes was lower than down-regulated ones at 48 h and 72 h after A. oryzae inoculation. (Table 2). There were 26, 17, and 12 up-regulated immune genes in female fat body cells 24 h, 48 h, and 72 h after A. oryzae inoculation, respectively, and there were 6, 13, and 10 down-regulated immune genes 24 h, 48 h, and 72 h after A. oryzae inoculation, respectively (Table 2). The up-regulated genes were higher than down-regulated genes 24 h after A. oryzae inoculation and up-regulated genes were lower than down-regulated genes 48 h and 72 h after A. oryzae inoculation in female hemocytes (Table 2). To further elucidate the roles of differentially expressed genes in the immune responses of L. migratoria, we examined the expression level of the immune related genes.
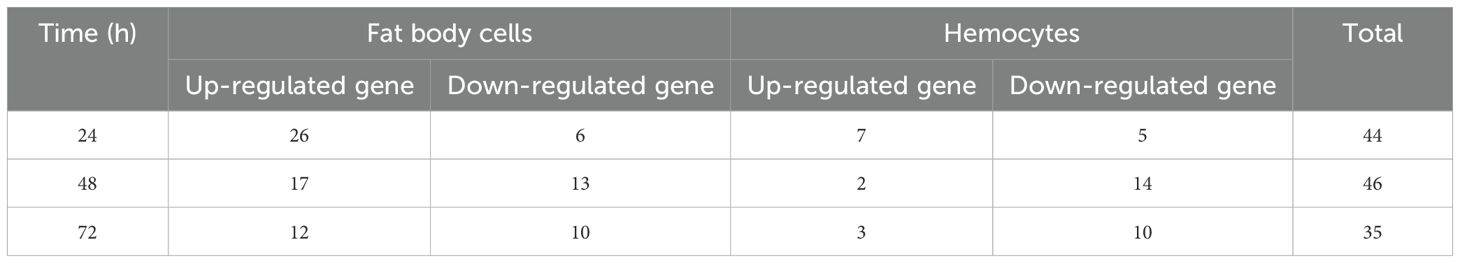
Table 2. Total number of up-regulated and down-regulated immune related genes in female fat body cells and hemocytes after A. oryzae infection.
3.2.1 PRRs
In locust fat body cells, the total number of up/down-regulated PGRP genes decreased from 24 h to 72 h after A. oryzae inoculation; 4, 2, and 3 PGRPs were expressed 24 h, 48 h, and 72 h after A. oryzae inoculation, respectively, and these genes were all up-regulated. The PGRP-LB genes LOCMI01594 and LOCMI16828 were highly up-regulated in female fat body cells compared with other PGRP genes in fat body cells (Figure 1A). The total number of up/down-regulated βGBP genes decreased from 24 to 72 h after A. oryzae inoculation, and 2 and 1 βGBP genes were expressed 24 h and 72 h after A. oryzae inoculation in fat body cells, respectively (Figure 1A). The expression levels of the other PRR genes, including PGRP, βGBP, SRs, and C-type lectins in fat body cells, did not significantly vary over time.
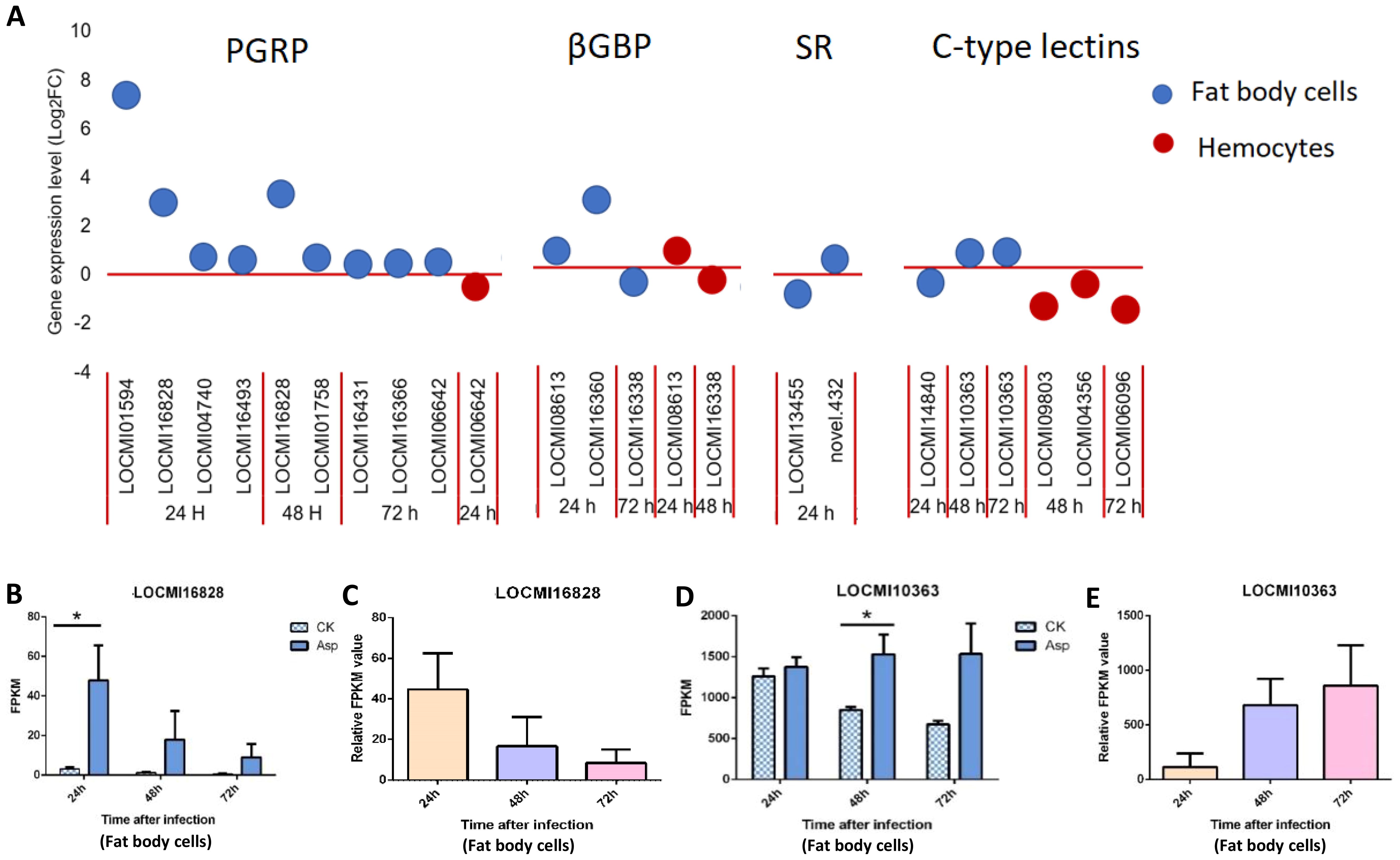
Figure 1. The temporal expression of pattern recognition receptor genes in female locust fat body cells and hemocytes. n=3-4. *p < 0.05. Bar, S.E.M. (A), PRR up-regulated and down-regulated genes after 24 h, 48 h and 72 h inoculation with fungal pathogen, A. oryzae XJ1. (B, D). Differential statistical analysis of expression levels of the gene in fat body between control and infected locusts. CK, control; Asp, A. oryzae. t-test. (C, E). Statistical analysis of temporal expression levels of the same gene in fat body of infected locusts. Ordinary two-way ANOVA. Tukey’s multiple comparisons test.
In hemocytes, the total number of up/down-regulated PRR genes was 6, and this was less than the number of fat body cell genes. In contrast to fat body cell genes, no genes were highly up-regulated in hemocytes, but two C-type lectin genes, LOCMI09803 and LOCMI06096, were highly down-regulated 48 h and 72 h after A. oryzae inoculation (Figure 1A), indicating that these two genes might be inhibited by A. oryzae.
We also analyzed the expression of these same genes expressed at different time intervals. The PGRP-LB gene LOCMI16828 was expressed in fat body cells 24 h and 48 h after A. oryzae inoculation, and its expression 24 h after inoculation was significantly higher in treated individuals than in control individuals (Figure 1B); however, the relative expression of this gene continuously decreased over time in the fat body cells of treated adults (Figure 1C). The expression of the C-type lectin gene LOCMI10363 was significantly higher in treated adults than in control adults 48 h after inoculation (Figure 1D), and its relative expression increased over time in the treated individuals (Figure 1E).
3.2.2 Serine protease, SERPINs and clip domain serine proteinase (CLIP)
In fat body cells, the total number of up/down-regulated SERPIN genes decreased; 7, 6, and 5 genes were expressed in fat body cells 24 h, 48 h, and 72 h after A. oryzae inoculation, respectively. There were 2, 1, and 0 down-regulated SERPIN genes in fat body cells 24 h, 48 h, and 72 h after inoculation, respectively; there were five up-regulated genes 24 h, 48 h, and 72 h after inoculation (Figure 2A). All CLIP genes were up-regulated 24 h after inoculation and down-regulated 48 h after inoculation; they were expressed only in female fat body cells and not in the hemocytes.
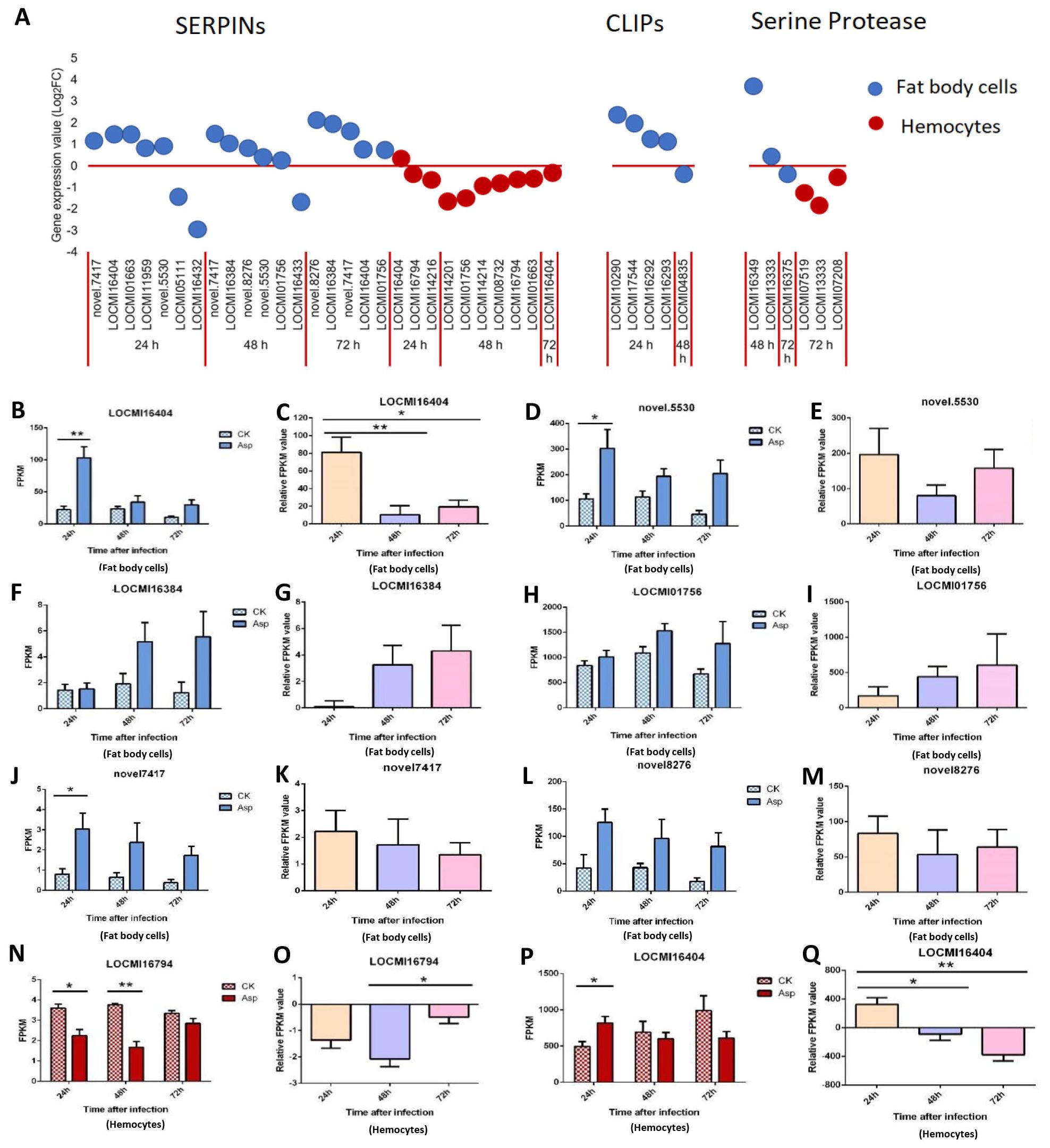
Figure 2. The temporal expression of the genes of SERPINs, CLIP and serine proteases in female locust fat body cells and hemocytes. *p < 0.05. **p < 0.01. n=3-4. Bar, S.E.M. (A), The dynamics of the SERPINs, Serine proteases and CLIPs in female locust fat body and hemocytes after 24 h, 48 h and 72 h inoculation with fungal pathogen, A. oryzae XJ1. (B, D, F, H, J, L), Differential statistical analysis of expression levels of the gene in fat body, and (N, P), in hemocytes between control and infected locusts. t-test. (C, E, G, I, K, M, O), Statistical analysis of temporal expression levels of the same gene in fat body of infected locusts, and (O, Q), in hemocytes. CK, control; Asp, A. oryzae. Ordinary two-way ANOVA. Tukey’s multiple comparisons test.
In hemocytes, the number of up/down-regulated SERPIN genes were 10 at all three time points, and number of SERPIN genes was highest 48 h after inoculation. Among the 10 genes, 9 were down-regulated, and the expression patterns of these genes in hemocytes and fat body cells differed (Figure 2A). Two serine protease genes were up-regulated 48 h after inoculation and one down-regulated 72 h after inoculation in fat body cells while all serine protease was down-regulated in hemocytes 72 h after inoculation (Figure 2A).
The gene LOCMI16404 was expressed in both fat body cells and hemocytes, and it was up-regulated in fat body cells 24 h, 48 h, and 72 h after inoculation in treated adults compared with the findings in control adults (Figure 2B). The expression of this gene was down-regulated in hemocytes 48 h and 72 h after inoculation in treated adults compared with the findings in control adults (Figure 2P). The relative expression level of LOCMI16404 was significantly higher 24 h after inoculation than after 48 h and 72 h of inoculation in fat body cells and hemocytes (Figures 2C, Q).
The expression level of novel.5530 was significantly higher in treated individuals than in control individuals 24 h after inoculation, but no significant difference was observed between treatments and the control at the other two time points (Figures 2D, E). The expression of the SERPIN genes, LOCMI16384, and LOCMI01756, increased over time in fat body cells, but there were no significant differences between treated and control adults or between adults at different time points (Figures 2F–I). Expression levels of novel.7417 and novel.8276 were higher in treated female fat body cells than in control female fat body cells (Figures 2J, L), and the relative expression of treated female adults was higher in female fat body cells 24 h after inoculation than 48 h and 72 h after inoculation (Figures 2K, M). The SERPIN gene LOCMI16794 was significantly down-regulated in female hemocytes 24 h and 48 h after inoculation in treated individuals compared with the findings in control individuals however 72 h after inoculation, no difference was observed in the expression of this gene between infected and control individuals, and this gene was down-regulated in treated adults (Figures 2N, O).
3.2.3 Toll pathway and JAK/STAT pathway
Only one Toll-like receptor was up-regulated in female fat body cells; in hemocytes, 4 Toll-like receptors were up-regulated and 1 was down-regulated compared with the findings in the control (Figure 3A). There was only one Protein toll gene that was up-regulated 48 h after inoculation; two suppressor of cytokine signaling (SOCS) genes in the JAK/STAT pathway were up-regulated 48 h and 72 h after inoculation in female fat body cells (Figure 3A).
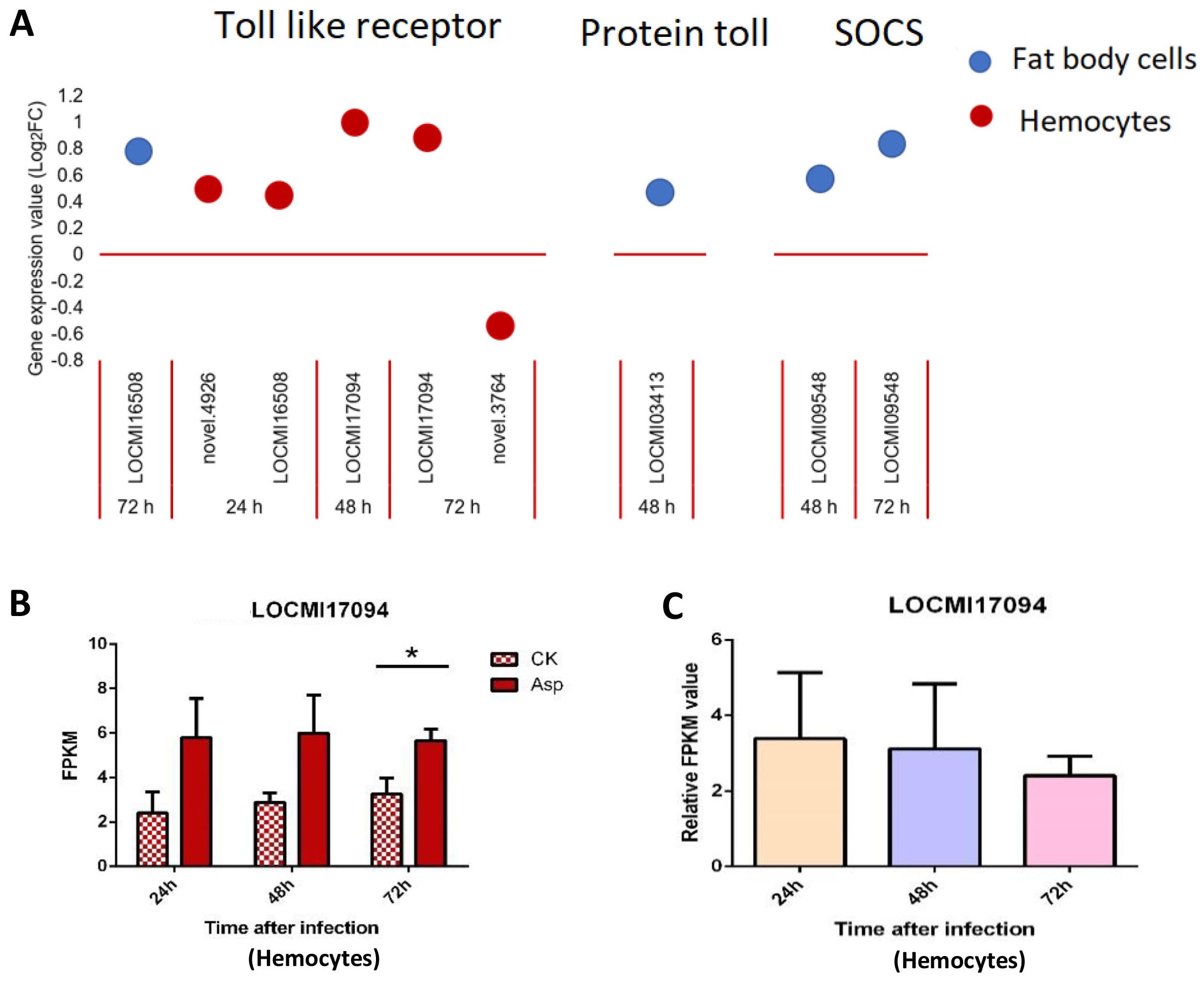
Figure 3. The temporal expression of the genes of Toll-pathway and JAK/STAT in female locust fat body cells and hemocytes. n=3-4. Bar, S.E.M. (A), The dynamics of the Toll-pathway and JAK/STAT genes in female locust fat body cells and hemocytes after 24 h, 48 h and 72 h inoculation with fungal pathogen, A oryzae XJ1. (B), Differential statistical analysis of expression levels of the gene in hemocytes between control and infected locusts. t-test. *p < 0.05. (C), Statistical analysis of temporal expression levels of the same gene in hemocytes of infected locusts. CK, control; Asp, A oryzae. Ordinary two-way ANOVA. Tukey’s multiple comparisons test.
The expression of the Toll-like receptor gene LOCMI17094 significantly differ between treated and control females 72 h after inoculation, and no significant difference was observed between treated and control females 24 h and 48 h after inoculation; no significant differences in the expression of this gene were observed between treated adults at different time points (Figures 3B, C).
3.2.4 AMPs, PPO, prophenoloxidase-activating factor (PPAF) and peroxidase (POD)
In response to A. oryzae infection, four lysozyme genes were up- and down-regulated in fat body cells, and this was higher than the number of genes that were differentially expressed in hemocytes. The lysozyme gene LOCMI2477 was the most highly up-regulated among all lysozyme genes, and its Log2FC value (>8) was highest 24 h after inoculation with A oryzae; the Log2FC values of the other three lysozyme genes in fat body cells were less than 1.5 (Figure 4A).
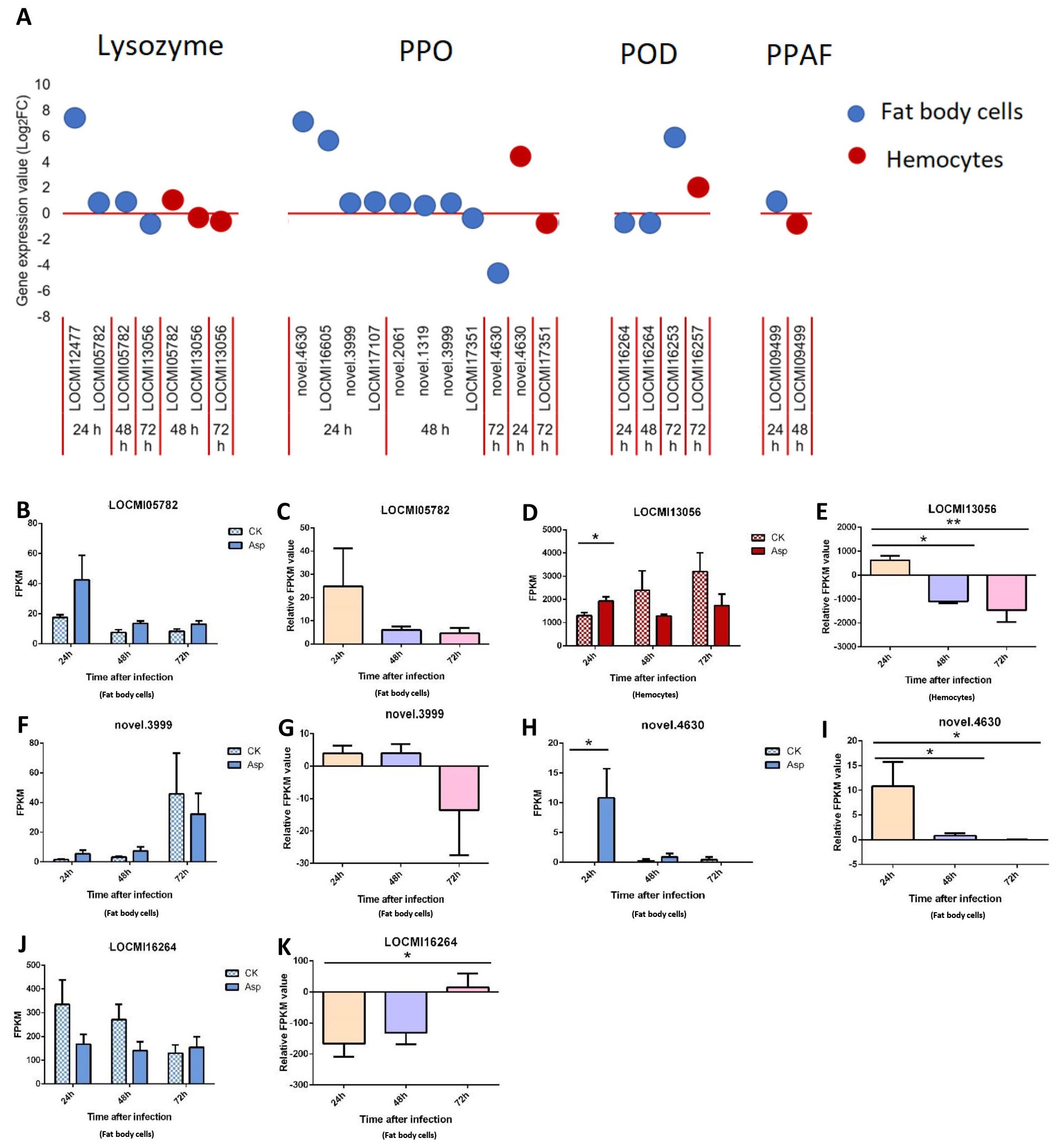
Figure 4. The temporal expression of the genes in effector group in female locust fat body cells and hemocytes. n=3-4. *p < 0.05. **p < 0.01. Bar, S.E.M. (A), The dynamics of the effector genes in female locust fat body and hemocytes after 24 h, 48 h and 72 h inoculation with fungal pathogen, A. oryzae XJ1. (B, F, H, J), Differential statistical analysis of expression levels of the gene in fat body, and D, in hemocytes between control and infected locusts. t-test. (C, G, I, K), Statistical analysis of temporal expression levels of the same gene in fat body of infected locusts, and (E), in hemocytes. CK, control; Asp, A. oryzae. Ordinary two-way ANOVA. Tukey’s multiple comparisons test.
In response to A oryzae infection, 9 and 2 PPO genes were up/down-regulated in fat body cells and hemocytes, respectively. Four and three PPO genes were up-regulated 24 h and 48 h after inoculation, respectively, and one PPO gene was down-regulated 72 h after inoculation. The two most highly up-regulated genes in female fat body cells 24 h after inoculation with A oryzae were novel.4630 and LOCMI16605. Two PPO genes were differentially regulated in hemocytes: one was down-regulated, and other was down-regulated (Figure 4A). PPAF was up-regulated in fat body cells 24 h after inoculation and up-regulated in hemocytes 48 h after inoculation (Figure 4A).
A total of four POD genes were up/down-regulated in both fat body cells and hemocytes. Two were up-regulated and one was down-regulated in fat body cells, and only one POD gene was up-regulated in hemocytes.
The expression level of the lysozyme gene LOCMI05782 was higher in treated adults than in control adults, albeit these differences were not significant; the expression of this gene among treated individuals decreased over time (Figures 4B, C). However, the expression of the lysozyme gene LOCMI13056 in hemocytes significantly differed in treated and control adults 24 h after inoculation, and its expression in treated adults at 24 h significantly differed from that at 48 h and 72 h (Figures 4D, E). The expression of the PPO gene novel.3999 did not significantly differ in control and treated adults (Figures 4F, G). The expression of the PPO gene novel.4630 in fat body cells was significantly higher 24 h after inoculation in treated adults than in control adults (Figure 4H), and the relative expression level of this gene was also significantly higher among treated adults 24 h after inoculation than that at 48 h and 72 h after inoculation (Figure 4I). The expression of the POD gene LOCMI16264 did not significantly differ between treated and control adults (Figure 4J) and the relative expression level of this gene was significantly differed between treated adults 24 h and 72 h after inoculation (Figure 4K).
3.2.5 Antioxidant enzymes
The expression of all Dual Oxidase (Duox) genes was altered only in hemocytes. Duox genes were up-regulated 24 h after inoculation and down-regulated 48 h and 72 h after inoculation (Figure 5A). LOCMI16271 was up-regulated 24 h after inoculation and down-regulated 48 h after inoculation.
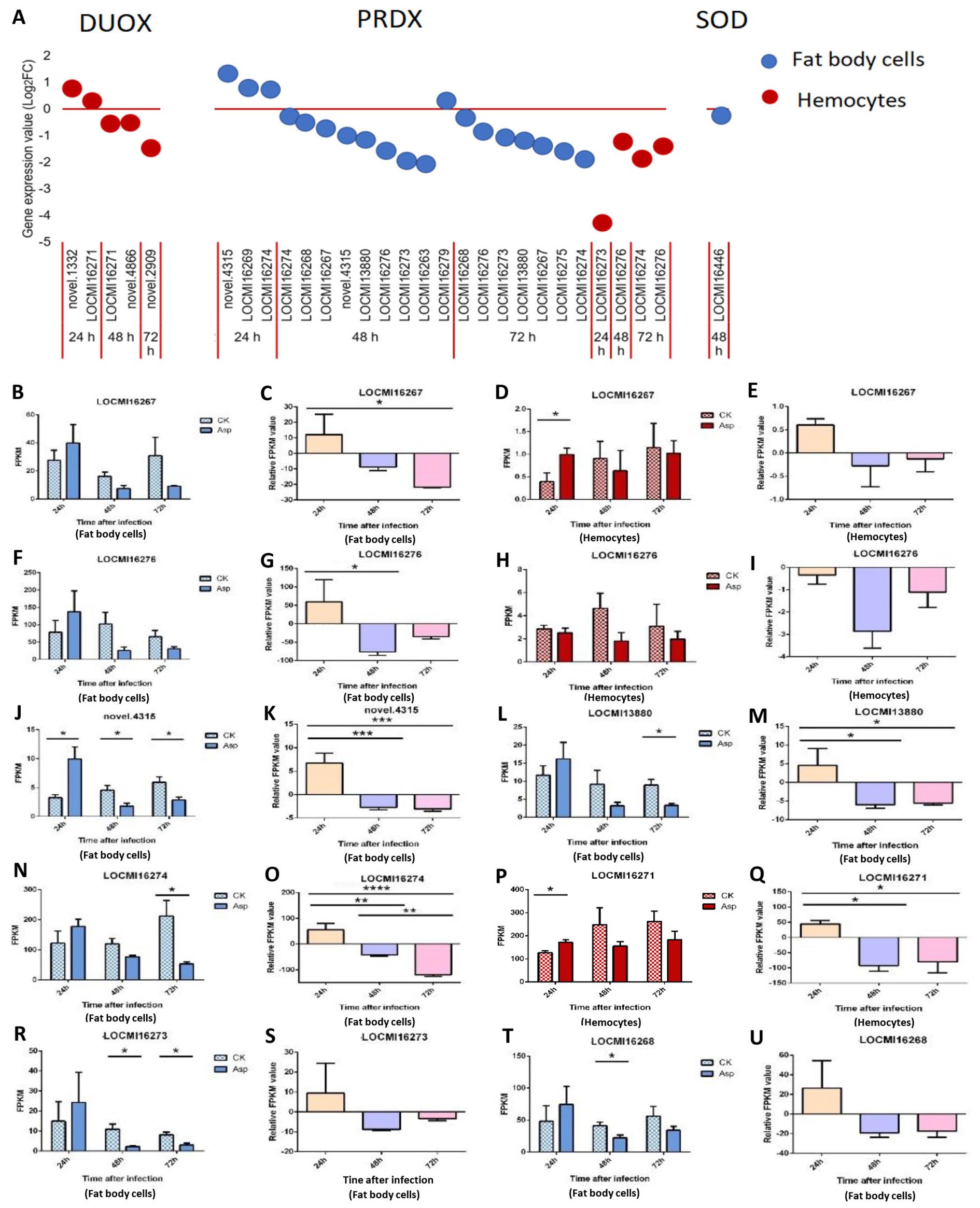
Figure 5. The temporal expression of the genes of effectors in female locust fat body cells and hemocytes. n=3-4. *p < 0.05. **p < 0.01. ***p < 0.001. Bar, S.E.M. (A), The dynamics of the effector genes in female locust fat body cells and hemocytes after 24 h, 48 h, and 72 h inoculation with the fungal pathogen, A. oryzae XJ1. (B, D, F, J, L, N, R, T), Differential statistical analysis of expression levels of the gene in fat body, and (D, H, P), in hemocytes between control and infected locusts. t-test. (C, G, K, M, O, S, U), Statistical analysis of temporal expression levels of the same gene in fat body of infected locusts, and (E, I, Q), in hemocytes. CK, control; Asp, A. oryzae. Ordinary two-way ANOVA. Tukey’s multiple comparisons test.
A total of 23 peroxiredoxin (PRDX) genes were up- and down-regulated in fat body cells and hemocytes (Figure 5A). The novel gene (novel.4315) was up-regulated 24 h after inoculation and down-regulated 48 h after inoculation; LOCMI16274 was up-regulated 24 h after inoculation and down-regulated 48 h and 72 h after inoculation. LOCMI16267 and LOCMI13380 were down-regulated 48 h and 72 h after inoculation in fat body cells. The PRDX genes LOCMI16276 and LOCMI16274 were down-regulated 48 h and 72 h after inoculation in fat body cells and hemocytes. The expression of only one Superoxide dismutase (SOD) gene was altered in female fat body cells following inoculation (Figure 5A).
There were no significant differences in the expression levels of the PRDX gene LOCMI16267 in treated and control adults at all three time points in fat body cells, but significant differences in hemocytes were observed 24 h after inoculation (Figures 5B, D). However, the relative expression levels of this gene 24 h after inoculation were higher than that after 48 h and 72 h inoculation in these two tissues (Figures 5C, E). In fat body, the expression level of PRDX gene LOCMI16276 showed a transient increase 24 h after inoculation, followed by a decline 48 h and 72 h after inoculation (Figure 5G). However, these changes were not statistically significant compared to the control group (Figure 5F). In hemocytes, the expression level of LOCMI16276 decreased at all time points after inoculation, though the reductions were not significant relative to the control (Figures 5H, I). Significant differences in the expression of the PRDX gene novel.4315 was observed between treated and control adults 24 h and 48 h after inoculation (Figure 5J), and significant differences in the expression of LOCMI13880 were observed 72 h after inoculation in fat body cells (Figure 5L). The relative expression levels of the PRDX genes novel.4315 and LOCMI13880 significantly higher 24 h after inoculation than 48 h and 72 h after inoculation (Figures 5K, M). The expression level of the PRDX gene LOCMI16274 was significantly lower in treated adults than in control adults 72 h after inoculation (Figure 5N), and significant differences in the relative expression level of this gene were observed among the all-time points (Figure 5O). The expression of the Duox gene LOCMI16271 24 h after inoculation was significantly higher in treated adults than in control adults, and significant differences in the relative expression level of this gene was significantly higher 24 h than 48 h and 72 h after inoculation (Figures 5P, Q). The expression of PRDX genes LOCMI16273 and LOCMI16268 significantly differ 48 h after inoculation between treatments and the control (Figures 5R, T). The relative expression levels of the PRDX genes LOCMI16273 and LOCMI16268 were higher 24 h after inoculation than 48 h and 72 h after inoculation (Figures 5S, U).
4 Discussion
In insects, SERPINs play a critical role in suppressing the serine protease cascade, thereby negatively regulating the activation of PPOs and Toll signaling pathways. This regulation prevents the overexpression of AMPs and excessive melanization, which could harm host (30, 31). For example, in Drosophila, SERPIN Spn27Ac inhibits PPO activation and suppresses melanization (32). In our study, the expression of most SERPINs and CLIPs was up-regulated in fat body cells (Figure 2A), which was similar to the regulation of SERPIN observed after bacterial infection in silkworm (33), In contrast, the expression of most SERPINs was down-regulated in hemocytes (Figure 2A), suggesting tissue-specific regulation of immune responses. This differential expression pattern highlights the complex and context-dependent roles of SERPINs in modulating immune pathways across different tissues.
In insects, the Toll pathway is essential that mediates resistance to pathogenic fungi (34). This pathway is typically initiated by the recognition of pathogen-associated molecular patterns (PAMPs) through PRRs, followed by the activation of serine protease cascades (35, 36). The canonic components of the Toll pathway include the extracellular cytokine Spätzle, the Toll-like receptor (TLR), the adaptor proteins Tube and MyD88, the kinase Pelle, the inhibitory protein Cactus (the Drosophila homolog of IκB), and the trans activators Dorsal and Dif (37). Furthermore, survival assays have revealed that Toll pathway’s response to entomopathogenic fungi is mediated by an extracellular proteolytic cascade involving the CLIPs, Persephone and the inhibitory serpin Necrotic (38, 39). The Spätzle, a key ligand in the insect Toll pathway, activates downstream immune responses by binding to Toll receptors (40). In our study, although we identified four Spätzle genes in the transcriptome of the treatment group, their expression levels showed no significant differences compared to the control group, indicating that Spätzle was not regulated within 72 h of A. oryzae infection (Supplementary Figure S1).
Studies have demonstrated that the deletion of any of these components (except for Cactus and Dorsal) in this pathway results in a similar immune-deficient phenotype, characterized by the absence of immune genes expression, including antifungal peptide genes, and increased susceptibility to fungal and Gram-positive bacterial infections (40, 41). No significant up-regulation or down-regulation of MyD88, Cactus, or antifungal peptide genes were observed in our study. However, the TLR genes were observed to be up/down-regulated (Figure 3A), sequence alignment revealed that LOCMI16508, novel. 4926, and novel. 3764 all belong to the TLR-2 family, which can be directly activated by chitin present in the fungal cell wall (42); and LOCMI17094 belongs to the TLR-7 family, which is primarily associated with antiviral immunity and also plays a role in the immune response caused by fungal infections (43, 44).
Although these changes were observed, considering the absence of significant changes in the expression of key Toll pathway components and the lack of downstream immune gene activation, we suggested that the Toll immune pathway was not fully activated 72 h after inoculation with A. oryzae. This suggests that other immune mechanisms may be primarily responsible for the observed response at this time point. While our data implicate this hypothesis, we acknowledge that this study did not directly measure key regulatory elements such as Spätzle, a critical upstream protease-activated ligand required for Toll pathway activation in insects. In future work, we will quantify the levels of Spätzle and other key genes in infected hosts to clarify this mechanism.
Melanization, a critical arthropod defense mechanism, plays a pivotal role in processes such as wound healing, encapsulation of pathogens, sequestration of microorganisms, and the production of toxic intermediates that can kill invading microbes (45–47). The melanization cascade is typically triggered by tissue injury or through the recognition of microbial ligands, such as peptidoglycan, β-1,3-glucan binding protein, and lipopolysaccharide (LPS) by PRRs (48–51). Melanization is mediated by the phenoloxidase (PO), which catalyzes the oxidation of mono- and diphenols to orthoquinones. These orthoquinones subsequently polymerize non-enzymatically to form melanin. Enzymatically inactive PPO is cleaved into active PO by prophenoloxidase-activating enzymes, and the activation of PPO is tightly regulated. The CLIPs modulate melanization by regulating PPO activation, a process that is further controlled by SERPINs (52). Survival assays have demonstrated that PPO mutant increased susceptibility to Gram-positive bacterial and fungal infections, underscoring the non-redundant role of melanization in immune defense against fungal pathogens (52).
Notably, we identified up-regulation of PGRPs, CLIPs, SERPINs, and lysozymes, which are known to play critical roles in the melanization reaction through the one Toll-like receptor (Toll-7) and the Protein toll (Figures 1A, 2A). The lysozyme genes (LOCMI13056, LOCMI05782) and PPO genes (novel.4630, LOCMI17351) were appeared together both in fat body cells and hemocytes (Figure 4A). However, the functional roles of these genes in the two tissues remain unknown and warrant further investigation. We further observed a dynamic regulation of immune effectors, including lysozymes, PPOs, and PRDXs, in the fat body cells and hemocytes of locusts following A. oryzae inoculation. Specifically, most of these effectors, particularly lysozyme (LOCMI13056), PPO2 (novel. 4630), and PRDX (novel. 4315), were significantly up-regulated 24 h post-inoculation but showed down-regulation by 72 h (Figures 4A, D, E, H, I, 5A, J, K). This temporal pattern suggests an initial robust immune response, characterized by a strong melanization reaction in L. migratoria 24 h after A. oryzae inoculation. However, by 72 h, PPO, such as PPO2, was down-regulated, indicating a reduction in melanization activity, which likely diminished the defense response against fungal pathogens (53). This temporal pattern is different in other insect species, such as most PPOs or lysozyme were up-regulated after fungus inoculation in C. lectularius and honey bee (17, 54). These findings led us to hypothesize that melanization is a critical defense mechanism in locusts against A. oryzae during the early stages of infection, but may be inhibited by the pathogen over time, and it needs to be demonstrated by further studies.
This also suggest that A. oryzae suppress the locusts’ immune genes expression following infection. Although locusts initially resisted A. oryzae by up-regulating key immune components such as lysozymes, PPOs, and PRDXs 24 h after inoculation, but A. oryzae appeared to suppress the melanization response by inhibiting the expression levels of key immune genes, such as PPOs, 48-72 h after inoculation (Figures 4A, 5A). The suppression of immune-related genes expression may impair locust immunity, facilitating the A. oryzae to establish infection for locusts. The suppression of immune-related genes could be exploited in the development of biological control strategies for locusts using A. oryzae. Such as since PPOs and PRDXs play key roles in defense of locust against Aspergillus, it would be possible to find and develop some inhibitors of these enzymes as enhancing factors for Aspergillus to control locusts. Our results indicated that fat body play a crucial role in defense against Aspergillus infection, this inspires to develop the combination use of Aspergillus and the pathogen, Nosema locustae (Protozoa: Microsporidia) whose main target is fat body, to improve the effects of controlling locusts (55), and it is supported by an recent study (56).
The innate immune systems of insects and mammals share both humoral and cellular responses, with hemocytes in insects and myeloid cells in mammals serving as key cellular mediators (57). Hemocytes, like mammalian neutrophils, target pathogens through mechanisms such as phagocytosis, encapsulation, and superoxide production (58–60). Our results implied that hemocytes in locusts play a critical role in the immune response to fungal pathogens by modulating pathogen recognition and immune signaling while preventing excessive activation. Unlike the fat body, which primarily produces AMPs and effector molecules, hemocytes exhibit fewer up-regulated PRRs and down-regulated specific C-type lectins and SERPIN genes, likely to control protease activity and inflammation. Hemocytes also transiently activate Duox genes to generate reactive oxygen species (ROS), which are later suppressed to avoid oxidative damage, complementing the fat body’s effector role. Locust hemocytes release C-type lectins into the hemolymph, which may mediate immune responses in the fat body just like in B. mori and Tribolium castaneum (61, 62).
In summary, our findings highlight the dynamic interplay between fat body cells and hemocytes during the immune response to fungal pathogens in adult female locusts. The fat body drives humoral responses, including the secretion of AMPs and melanization, and serves as the primary site for immune effector production. In contrast, hemocytes play a complementary role in pathogen recognition, immune regulation, and reactive oxygen species (ROS) management, ensuring a balanced and effective immune response through their regulatory functions (Figure 6). However, our study was limited to analyzing the differential expression of immune genes at the transcriptome level in fat body cells and hemocytes, exploring potential immune responses. In future research, we plan to conduct qRT-PCR, gene editing of selected immune genes, and other related experiments, including chromatographic analyses of hemolymph composition. These efforts will help verify the functions of key genes in the immune response and provide a comprehensive understanding of the systemic immune mechanisms in insects, as well as their temporal and spatial dynamics during pathogen challenges.
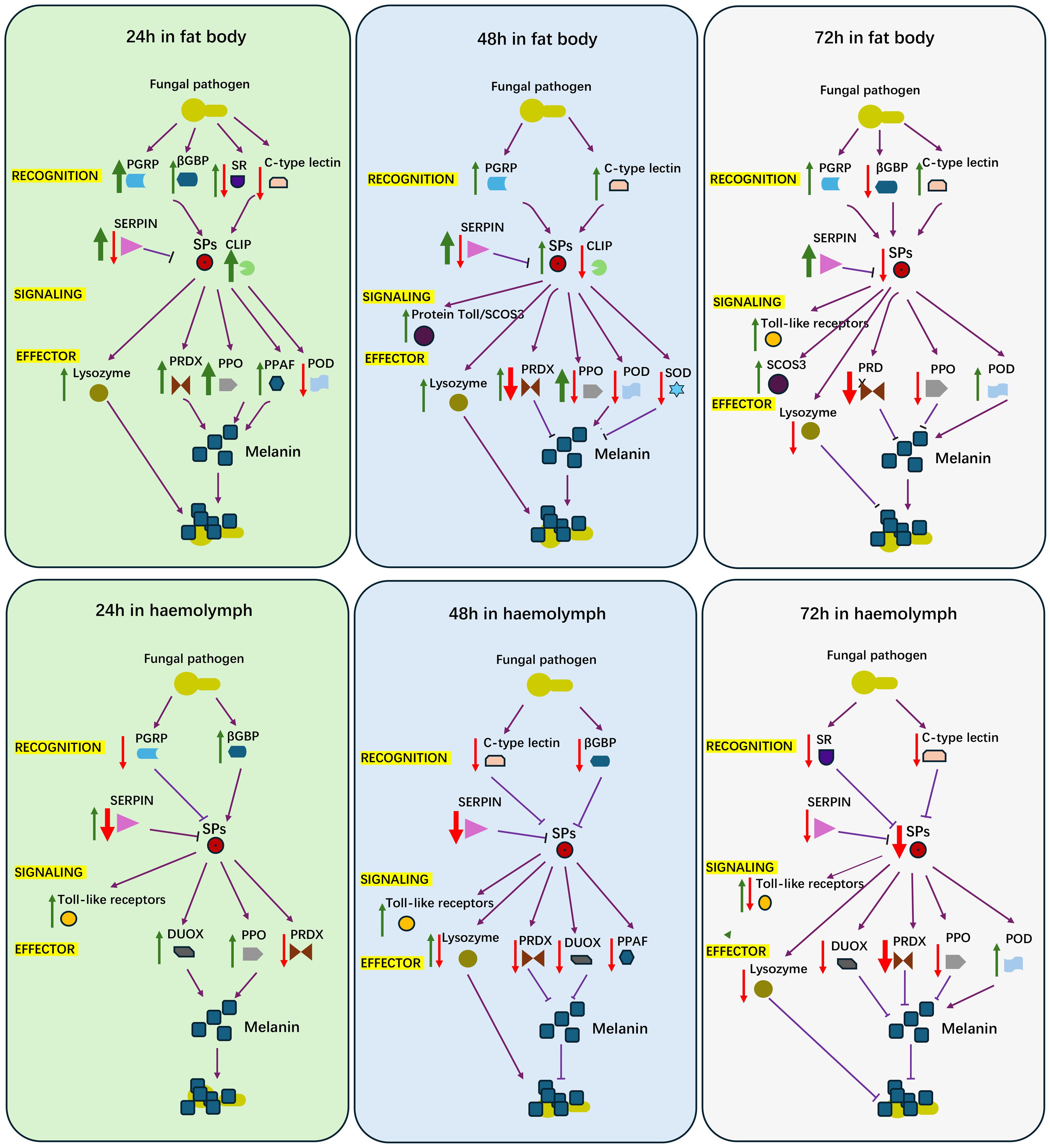
Figure 6. A diagram for proposed features of female locust adult humoral immune response to challenge of Aspergillus oryzae XJ1. Red arrow, down regulated genes; Green arrow, up regulated genes; Purple arrow, the activated relationship; Purple T bar, inhibited relationship; Think arrow, more number of genes; Thin arrow, less number of genes.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1211842, PRJNA1212177, PRJNA1212339, PRJNA1212589, PRJNA1212757, PRJNA1212830, PRJNA1213078, PRJNA1213165, PRJNA1213310, PRJNA1213846, PRJNA1213978, PRJNA1214046, PRJNA1214369, PRJNA1214481, PRJNA1214552.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because my insect is common insect and there is no need to take any ethical approval for this study.
Author contributions
MW: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. XX: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. PZ: Data curation, Formal analysis, Writing – review & editing. JG: Conceptualization, Writing – review & editing, Methodology. SH: Data curation, Software, Writing – review & editing. YY: Supervision, Writing – review & editing. LZ: Conceptualization, Data curation, Funding acquisition, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences (CXGC2024F05, CXGC2024D05), the National Natural Science Foundation of China (No. 32472640), and the postdoctoral research fellowship for Muhammad Saad Waqas from Shandong Academy of Agricultural Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1565964/full#supplementary-material
References
1. Riera Romo M, Pérez-Martínez D, Castillo Ferrer C. Innate immunity in vertebrates: an overview. Immunol. (2016) 148:125–39. doi: 10.1111/imm.12597
2. Cox CR, Gilmore MS. Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect Immun. (2007) 75:1565–76. doi: 10.1128/IAI.01496-06
3. Erickson DL, Lines JL, Pesci EC, Venturi V, Storey DG. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect Immun. (2004) 72:5638–45. doi: 10.1128/IAI.72.10.5638-5645.2004
4. Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. (2005) 73:3842–50. doi: 10.1128/IAI.73.7.3842-3850.2005
5. Mukherjee K, Hain T, Fischer R, Chakraborty T, Vilcinskas A. Brain infection and activation of neuronal repair mechanisms by the human pathogen Listeria monocytogenes in the lepidopteran model host Galleria mellonella. Virulence. (2013) 4:324–32. doi: 10.4161/viru.23629
6. Iiyama K, Chieda Y, Lee JM, Kusakabe T, Yasunaga-Aoki C, Shimizu S. Effect of superoxide dismutase gene inactivation on virulence of Pseudomonas aeruginosa PAO1 toward the silkworm, Bombyx mori. Appl Environ Microbiol. (2007) 73:1569–75. doi: 10.1128/aem.00981-06
7. Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. (2000) 30:95–105. doi: 10.1016/s0965-1748(99)00113-7
8. Aggrawal K, Silverman N. Peptidoglycan recognition in Drosophila. Biochem Soc Trans. (2007) 35:1496–500. doi: 10.1042/BST0351496
9. Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci USA. (2000) 97:10520–5. doi: 10.1073/pnas.180130797
10. Yagi Y, Nishida Y, Ip YT. Functional analysis of Toll-related genes in Drosophila. Dev Growth Differ. (2010) 52:771–83. doi: 10.1111/j.1440-169X.2010.01213.x
11. Yu S, Luo F, Xu Y, Zhang Y, Jin LH. Drosophila innate immunity involves multiple signaling pathways and coordinated communication between different tissues. Front Immunol. (2022) 13:905370. doi: 10.3389/fimmu.2022.905370
12. Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. (2007) 25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615
13. Li S, Yu X, Feng Q. Fat Body Biology in the Last Decade. Annu Rev Entomol. (2019) 64:315–33. doi: 10.1146/annurev-ento-011118-112007
14. Skowronek P, Wojcik L, Strachecka A. Fat body-multifunctional insect tissue. Insects. (2021) 12:547. doi: 10.3390/insects12060547
15. Arrese EL, Soulages LJ. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. (2010) 55:207–25. doi: 10.1146/annurev-ento-112408-085356
16. Paes de Oliveira VT, Cruz-Landim C. Protein content and eletrophoretic profile of fat body and ovary extracts from workers of Melipona quadrifasciata anthidioides (Hymenoptera, Meliponini). Iheringia, Sér. Zool. (2004) 94:417–9. doi: 10.1590/s0073-47212004000400010
17. Meraj S, Salcedo-Porras N, Lowenberger C and Gries G. Activation of immune pathways in common bed bugs, Cimex lectularius, in response to bacterial immune challenges - a transcriptomics analysis. Front Immunol. (2024) 15:1384193. doi: 10.3389/fimmu.2024.1384193
18. Wang B, Kang QJ, Lu YZ, Bai LQ, Wang CS. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc Natl Acad Sci U S A. (2012) 109:1287–92. doi: 10.1073/pnas.1115983109
19. Feng P, Shang Y, Cen K, Wang C. Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc Natl Acad Sci U S A. (2015) 112:11365–70. doi: 10.1073/pnas.1503200112
20. Bartholomay LC, Waterhouse RM, Mayhew GF, Campbell CL, Michel K, Zou Z, et al. Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science. (2010) 330:88–90. doi: 10.1126/science.1193162
21. Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, et al. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. (2015) 347:1258522. doi: 10.1126/science.1258522
22. Waterhouse RM, Kriventseva EV, Meister S, Xi ZY, Alvarez KS, Bartholomay LC, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. (2007) 316:1738–43. doi: 10.1126/science.1139862
23. Ma L, Liu L, Zhao Y, Yang L, Chen C, Li Z, et al. JNK pathway plays a key role in the immune system of the pea aphid and is regulated by microRNA-184. PloS Pathog. (2020) 16:e1008627. doi: 10.1371/journal.ppat.1008627
24. Zhang W, Chen J, Keyhani NO, Zhang Z, Li S, Xia Y. Comparative transcriptomic analysis of immune responses of the migratory locust, Locusta migratoria, to challenge by the fungal insect pathogen, Metarhizium acridum. BMC Genomics. (2015) 16:867. doi: 10.1186/s12864-015-2089-9
25. Zhang W, Zheng X, Chen J, Keyhani NO, Cai K, Xia Y. Spatial and temporal transcriptomic analyses reveal locust initiation of immune responses to Metarhizium acridum at the pre-penetration stage. Dev Comp Immunol. (2020) 104:103524. doi: 10.1016/j.dci.2019.103524
26. Machado M, Fortún J, Muñoz P. Invasive aspergillosis: A comprehensive review. Med Clin (Barc). (2024) 163:189–98. doi: 10.1016/j.medcli.2024.01.045
27. Sheehan G, Farrel G, Kavanagh K. Immmune proming: The secret weapon of the insect world. Virulence. (2020) 11:238–46. doi: 10.1080/21505594.2020.1731137
28. Zhang P, You Y, Song Y, Wang Y, Zhang L. First record of Aspergillus oryzae (Eurotiales: Trichocomaceae) as an entomopathogenic fungus of the locust, Locusta migratoria (Orthoptera: Acrididae). Biocontrol Sci Techn. (2015) 25:1285–98. doi: 10.1080/09583157.2015.1049977
29. You Y, An Z, Zhang X, Liu H, Yang W, Yang M, et al. Virulence of the fungal pathogen, Aspergillus oryzae XJ-1 to adult locusts (Orthoptera: Acrididae) in both laboratory and field trials. Pest Manag Sci. (2023) 79:3767–72. doi: 10.1002/ps.7561
30. Yang F, Wang Y, Sumathipala N, Cao X, Kanost MR, Jiang H. Manduca sexta serpin-12 controls the prophenoloxidase activation system in larval hemolymph. Insect Biochem Mol Biol. (2018) 99:27–36. doi: 10.1016/j.ibmb.2018.05.004
31. Liu H, Xu J, Wang L, Guo P, Tang Z, Sun X, et al. Serpin-1a and serpin-6 regulate the Toll pathway immune homeostasis by synergistically inhibiting the Spätzle-processing enzyme CLIP2 in silkworm, Bombyx mori. PloS Pathog. (2023) 19:e1011740. doi: 10.1371/journal.ppat.1011740
32. Ligoxygakis P, Pelte N, Ji C, Leclerc V, Duvic B, Belvin M, et al. A serpin mutant links Toll activation to melanization in the host defense of Drosophila. EMBO J. (2002) 21:6330–7. doi: 10.1093/emboj/cdf661
33. Qie X, Yan X, Wang W, Liu Y, Zhang L, Hao C, et al. Serpin-4 negatively regulates prophenoloxidase activation and antimicrobial peptide synthesis in the silkworm, Bombyx mori. Int J Mol Sci. (2023) 25:313. doi: 10.3390/ijms25010313
34. Kojour MAM, Han YS, Jo YH. An overview of insect innate immunity. Entomol Res. (2020) 50:282–91. doi: 10.1111/1748-5967.12437
35. Valanne S, Wang JH, Rämet M. The Drosophila Toll signaling pathway. J Immunol. (2011) 186:649–56. doi: 10.4049/jimmunol.1002302
36. Valanne S, Vesala L, Maasdorp MK, Salminen Ts Rämet M. The Drosophila Toll pathway in innate immunity: From the core pathway toward effector functions. J Immunol. (2022) 209:1817–25. doi: 10.4049/jimmunol.2200476
37. Ferrandon D. The complementary facets of epithelial host defenses in the genetic model organism Drosophila melanogaster: From resistance to resilience. Curr Opin Immunol. (2013) 25:59–70. doi: 10.1016/j.coi.2012.11.008
38. Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. (2002) 297:114–16. doi: 10.1126/science.1072391
39. Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffman JA, et al. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. (1999) 285:1917–19. doi: 10.1126/science.285.5435.1917
40. Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. (1996) 86:973–83. doi: 10.1016/s0092-8674(00)80172-5
41. Rutschmann S, Kilinc A, Ferrandon D. Cutting edge: the toll pathway is required for resistance to gram-positive bacterial infections in Drosophila. Immunol. (2002) 168:1542–46. doi: 10.4049/jimmunol.168.4.1542
42. Shen P, Li W, Wang Y, He X, He L. Binding mode of chitin and TLR2 via molecular docking and dynamics simulation. Mol Simul. (2016) 42:936–41. doi: 10.1080/08927022.2015.1124102
43. Gupta CL, Akhtar S, Sayyed U, Pathak N, Bajpai P. In silico analysis of human Toll-like receptor 7 ligand binding domain. Biotechnol Appl Biochem. (2016) 63:441–50. doi: 10.1002/bab.1377
44. Geng T, Huang Y, Hou C, Qin G, Lv D, Guo X. Inductive expression patterns of genes related to Toll signaling pathway in silkworm (Bombyx mori) upon Beauveria bassiana infection. J Asia-pacific Entomol. (2016) 19:861–8. doi: 10.1016/j.aspen.2016.08.001
45. Nappi AJ, Vass E. Melanogenesis and the generation of cytotoxic molecules during insect cellular immune reactions. Pigment Cell Res. (1993) 6:117–26. doi: 10.1111/j.1600-0749.1993.tb00590.x
46. Ashida M. The prophenoloxidase cascade in insect immunity. Res Immunol. (1990) 141:908–10. doi: 10.1016/0923-2494(90)90191-z
47. Söderhäll K, Cerenius L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr Opin Immunol. (1998) 10:23–8. doi: 10.1016/s0952-7915(98)80026-5
48. Ochiai M, Ashida M. A pattern recognition protein for peptidoglycan. Cloning the cDNA and the gene of the silkworm, Bombyx mori. J Biol Chem. (1999) 274:11854–58. doi: 10.1074/jbc.274.17.11854
49. Ochiai M, Ashida M. A pattern-recognition protein for beta-1,3-glucan. The binding domain and the cDNA cloning of beta-1,3-glucan recognition protein from the silkworm, Bombyx mori. J Biol Chem. (2000) 275:4995–5002. doi: 10.1074/jbc.275.7.4995
50. Ma C, Kanost MR. A beta1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J Biol Chem. (2000) 275:7505–14. doi: 10.1074/jbc.275.11.7505
51. Nakhleh J, Moussawi LE, Osta MA. The melanization response in insect immunity. Adv Insect Physiol. (2017) 52:83–109. doi: 10.1016/bs.aiip.2016.11.002
52. Lee MH, Osaki T, Lee JY, Baek MJ, Zhang R, Park JW, et al. Peptidoglycan recognition proteins involved in 1,3-beta-D-glucan-dependent prophenoloxidase activation system of insect. J Biol Chem. (2004) 279:3218–27. doi: 10.1074/jbc.M309821200
53. Yu SJ, Ding LL, Luo R, Yang J, Liu HQ, Cong L, et al. Identification of immunity-related genes in Dialeurodes citri against entomopathogenic fungus Lecanicillium attenuatum by RNA-Seq analysis. PloS One. (2016) 11:e0162659. doi: 10.1371/journal.pone.0162659
54. Aronstein KA, Murray KD, Saldivar E. Transcriptional responses in Honey Bee larvae infected with chalkbrood fungus. BMC Genomics. (2010) 11:391. doi: 10.1186/1471-2164-11-391
55. Zhang L, Lecoq M. Nosema locustae (Protozoa, Microsporidia), a biological agent for locust and grasshopper control. Agronomy. (2021) 11:711. doi: 10.3390/agronomy11040711
56. Zhang P, Yang M, You Y, Zhang L. Synergistic effect of mixture of Microsporidium Nosema locustae (Protista: Microsporidia) and novel fungus Aspergillus oryzae XJ-1 (Eurotiales: Trichocomaceae) against adult Locusta migratoria (Orthoptera: Acrididae) in laboratory. Agronomy. (2025) 15:364. doi: 10.3390/agronomy15020364
57. Sheehan G, Garvey A, Croke M, Kavanagh K. Innate humoral immune defenses in mammals and insects: The same, with differences? Virulence. (2018) 9:1625–39. doi: 10.1080/21505594.2018.1526531
58. Bergin D, Reeves EP, Renwick J, Wientjes FB, Kavanagh K. Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect Immun. (2005) 73:4161–70. doi: 10.1128/IAI.73.7.4161-4170.2005
59. Renwick J, Reeves EP, Wientjes FB, Kavanagh K. Translocation of proteins homologous to human neutrophil p47phox and p67phox to the cell membrane in activated hemocytes of Galleria mellonella. Dev Comp Immunol. (2007) 31:347–59. doi: 10.1016/j.dci.2006.06.007
60. Browne N, Heelan M, Kavanagh K. An analysis of the structural and functional similarities of insect hemocytes and mammalian phagocytes. Virulence. (2013) 4:597–603. doi: 10.4161/viru.25906
61. Zhan M, Shahzad T, Yang P, Liu S, Yu X, Rao X. A single-CRD C-type lectin is important for bacterial clearance in the silkworm. Dev Comp Immunol. (2016) 65:330–9. doi: 10.1016/j.dci.2016.08.004
Keywords: innate immune, immune genes, temporal expression patterns, female locust (Locusta migratoria), infection, fungal pathogen (Aspergillus oryzae)
Citation: Waqas MS, Xu X, Zhang P, Guo J, Hu S, You Y and Zhang L (2025) Characterization of temporal expression of immune genes in female locust challenged by fungal pathogen, Aspergillus sp.. Front. Immunol. 16:1565964. doi: 10.3389/fimmu.2025.1565964
Received: 24 January 2025; Accepted: 31 March 2025;
Published: 28 April 2025.
Edited by:
Liang Jiang, Southwest University, ChinaReviewed by:
Ilias Kounatidis, Technology, Engineering and Mathematics, The Open University, United KingdomAmr A Mohamed, Cairo University, Egypt
Copyright © 2025 Waqas, Xu, Zhang, Guo, Hu, You and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Long Zhang, bG9jdXN0QGNhdS5lZHUuY24=; Yinwei You, eXl3MzAwMDBAMTYzLmNvbQ==
†These authors share first authorship
 Muhammad Saad Waqas
Muhammad Saad Waqas Xiao Xu
Xiao Xu Pengfei Zhang2
Pengfei Zhang2 Yinwei You
Yinwei You Long Zhang
Long Zhang