- 1Department of Urology, Second Affiliated Hospital of Dalian Medical University, Dalian, Liaoning, China
- 2Department of Urology, Central Hospital of Dalian, Dalian, Liaoning, China
- 3Institute of Cancer Stem Cell, Dalian Medical University, Dalian, Liaoning, China
- 4Department of Pathology, Central Hospital of Dalian, Dalian, Liaoning, China
- 5Department of Pathology, Dalian Friendship Hospital, Dalian, Liaoning, China
- 6Department of Emergency Medicine, Shengjing Hospital of China Medical University, Shenyang, Liaoning, China
- 7College of Humanities and Social Sciences, Dalian Medical University, Dalian, Liaoning, China
- 8Department of Oncology, Second Affiliated Hospital of Dalian Medical University, Dalian, Liaoning, China
Background: Extranodal natural killer/T-cell lymphoma (ENKTL) involving the adrenal glands is extremely rare, and only a few cases have been reported. However, the genetic alterations, clinicopathological features and prognosis of these patients have not yet been fully elucidated.
Methods: Profiling of tumor mutations in ENKTL patients with adrenal involvement was conducted by whole-genome sequencing, and the predisposing genes and driver mutation gene variants were verified through Sanger sequencing. Immunohistochemical analysis of markers for the diagnosis and tumor microenvironment competent were performed to identify histopathological features. In addition, we searched the Surveillance, Epidemiology, and End Results (SEER), PubMed, Embase, and Scopus databases to perform a population-based study to compare the prognosis between adrenocortical carcinoma (ACC) patients and adrenal ENKTL patients using Kaplan–Meier survival curves and log-rank tests and analyzed the prognostic factors affecting the overall survival (OS) of adrenal ENKTL patients via univariate and multivariate Cox regression analyses.
Results: We screened 15892 somatic single-nucleotide variants (SNVs), 364 somatic insertions and deletions (INDELs), and four driver mutation genes, namely, TET2, STAT3, FAS, and TP53. In addition, immunohistochemical analysis revealed that tumor cells were positive for CD3, CD43, CD56, TIA1, granzyme B, CD2, CD4, and CD7. The immunohistochemistry for detecting components of the tumor microenvironment reveled the infiltration of tumor-associated macrophages (CD68, CD163) and tumor-associated fibroblasts (vimentin, SMA) in the tumor sample. According to our population-based analysis, Kaplan–Meier survival curves revealed that ENKTL patients with adrenal involvement had a significantly poorer prognosis than did patients with ACC (p<0.001), and chemotherapy was a significant prognostic factor for OS in ENKTL patients with adrenal involvement according to Cox multivariate analysis (hazard ratio = 0.318; p=0.027).
Conclusions: The metastasis of ENKTL to the adrenal gland may be due to gene mutations caused by genetic variations, which may provide new therapeutic targets for this disease. The prognosis of adrenal ENKTL patients is markedly worse than that of ACC patients, and chemotherapy may serve as an independent factor of OS in adrenal ENKTL patients. However, our findings still need to be validated in additional studies.
1 Introduction
Extranodal natural killer/T-cell lymphoma (ENKTL) is a particularly aggressive subtype of non-Hodgkin lymphoma that is intimately related to infection with Epstein–Barr virus (EBV) (1). The prevalence of ENKTL is significantly greater in Central and South America and Asia than in the United States and Europe (2, 3). Approximately 80% of ENKTL cases develop in the upper respiratory tract, which includes the nasal cavity, nasopharynx, paranasal sinuses, and palate, with the nasal cavity being the most frequent site; approximately 20% of cases develop in extranasal regions, including the skin, gastrointestinal tract, testes and salivary glands (1, 4). The process of tumorigenesis in ENKTL has multiple stages. There are two widely accepted assumptions about these multistage processes. One hypothesis is that NK/T cells can be infected by EBV and assisted by EBV oncogenes such as EBNA1 and LMP1 to proliferate and avoid apoptosis. In long-term disease processes, cumulative variations in driver genes, such as JAK3 and DDX3X, may contribute to the progression of ENKTL. The other hypothesis is that EBV-infected cells can accumulate mutations due to senescence and chronic exposure to environmental stressors (5, 6). ENKTL always exhibits an invasive course and poor clinical outcome, especially in the presence of extranasal metastases, with a complete remission rate of only approximately 30% and a median survival of 4.3 months (7).
Patients with ENKTL concomitant with adrenal involvement are extremely rare; therefore, the clinicopathological features and genetic characteristics of these patients have not been systematically described. In this study, we reported an eight-year episode of an ENKTL patient with adrenal and multiple site involvement who achieved a complete response (CR) after undergoing surgery, chemotherapy and immunotherapy and remained in good general condition after 101 months of follow-up. Next, we evaluated the genetic alterations of the patient by conducting next-generation sequencing (NGS). Additionally, we reviewed published cases of ENKTL with subsequent adrenal involvement, compared the prognosis of ENKTL patients with adrenal involvement and adrenocortical carcinoma (ACC) patients and explored the risk factors for the prognosis of adrenal ENKTL patients.
2 Materials and methods
2.1 Clinical data
We obtained the patient’s clinical data from the Dalian Central Hospital and the Second Affiliated Hospital of Dalian Medical University. The Ethics Committee of Dalian Central Hospital (ethical approval number: YN2024-029-01) and the Second Affiliated Hospital of Dalian Medical University (ethical approval number: 2023273) approved the research respectively, and the patient provided informed consent. The research complied with the Declaration of Helsinki.
2.2 Immunohistochemical analysis
The formalin-fixed tissues were embedded in paraffin and subsequently cut into 5-µm sections. The sections were dewaxed in xylene and hydrated in ethanol, and antigen repair was performed by placing the slides in sodium citrate buffer and heating them at 95°C for 20 minutes. The slides were incubated in 0.5% hydrogen peroxide for 20 min to block endogenous peroxidase activity, blocked with goat serum for 1 h, and incubated overnight at 4°C with the primary antibodies. Supplementary Table S1 lists the antibodies used in this study. Following the manufacturer’s instructions, the slides were incubated with an avidin-biotin kit. The slides were developed in the chromogen 3,30-diaminobenzidine for 5 minutes at nearly 24°C, and hematoxylin was used for counterstaining for 30 seconds. Microscopy (Leica, Wetzlar, Germany) was used to observe the slides. We detected EBV-encoded small RNAs (EBERs) using an EBV Probe In Situ Hybridization Kit (Zhongshan Golden Bridge Biotechnology Co. Ltd., Beijing, China).
2.3 Population-based study
2.3.1 Data resources and study population
We searched the SEER database (https://seer.cancer.gov/) on June 2023, which consists of 17 registries covering more than 26.5% of the United States population between 2000 and 2020, for information on adrenocortical carcinoma (ACC) patients. Patients with an International Classification of Disease (ICD)-O-3 code of 8370 with malignant behavior were screened. We included a total of 1771 ACC patients in our study after excluding patients with a primary site in the medulla of the adrenal gland, missing histological confirmation or unknown survival time. Patients whose ICD-O-3 code was 9719 and whose primary site was the adrenal gland were screened; 2 ENKTL patients with adrenal involvement were included in our study. Moreover, we searched the PubMed, Embase and Scopus databases using the keywords “natural killer/T cell lymphoma”, “NK/T cell lymphoma” and “adrenal” for ENKTL patients with adrenal involvement up to April 2025. Together with the one patient we reported, a total of 39 patients were included in our study, and the detailed selection flowchart is shown in Supplementary Figure S1. Patient characteristics and survival information, including age, sex, race, primary site, Ann Arbor stage, absence or presence of B symptoms, laterality, tumor size, surgery, radiotherapy and chemotherapy, were summarized. The main endpoint was overall survival (OS).
2.3.2 Statistical analyses
Count data were transformed into categorical variables, and comparisons between ACC patients and ENKTL patients with adrenal involvement were performed using Fisher’s exact probability or Pearson’s chi-square method using SPSS version 26.0 (SPSS Inc., Chicago, IL, USA). We used the ‘survival’ package in R software (version 4.3.1, http://www.R-project.org/) to conduct log-rank tests and plot Kaplan–Meier survival curves. Univariate and multivariate Cox regression analyses were performed to calculate hazard ratios (HRs) and 95% confidence intervals (CIs). A p value <0.05 (two-sided) was considered to indicate statistical significance.
2.4 Whole-genome sequencing
2.4.1 DNA extraction
The FFPE tumor specimens and matched normal specimens were subjected to DNA extraction with a GeneRead DNA FFPE Kit (Qiagen, Germany) according to the manufacturer’s instructions. The extracted specimens were subjected to whole-genome sequencing. We evaluated the DNA quality through agarose gel electrophoresis and a Qubit® DNA Assay Kit in a Qubit® 3.0 Fluorometer (Invitrogen, USA). We performed library construction using DNA samples with concentrations above 20 ng/µL and a total quantity above 0.2 µg.
2.4.2 Library preparation and sequencing
We generated the sequencing libraries using the NEBNext® Ultra™ DNA Library Preparation Kit (NEB, USA) following the manufacturer’s instructions and added an index code to each sample. After the genomic DNA samples were fragmented into 350 bp fragments by sonication, the DNA fragments were polished, a-tailed and ligated to full-length adapters, followed by further PCR amplification. We purified the PCR products with an AMPure XP system (Beckman Coulter, Beverly, USA) and then amplified them using a Qubit® 3.0 Fluorometer (Invitrogen, USA) to detect DNA concentrations and an NGS3K/Caliper to analyze the size distribution of the libraries and quantify them by real-time PCR (3 nM). We clustered the index-coded samples on the cBot Cluster Generation System using the Illumina PE Cluster Kit (Illumina, USA). We then sequenced the DNA library on the Illumina platform to produce 150 bp paired-end reads.
2.4.3 Quality control
Raw data were filtered by removing single-end sequencing reads with adapters, more than 10% of unidentifiable base information, or more than 50% of the read length with low quality (fewer than 5 bases). The sequencing error rate for each base position was than 1%, and the quality of the sequencing data was distributed at a Q30 of no less than 80%.
2.4.4 Bioinformatics analysis
We aligned the validated sequencing data with the reference genome (GRCh37/hg19/GRCh38) by BWA (8) to obtain the initial alignment results in the BAM format. The results were then ranked using SAMtools (8), and Sambamba was used to mark duplicate reads. Finally, the coverage and depth statistics of the marked duplicate reads were calculated.
2.4.5 Variant detection, somatic mutation calling and functional annotation
Based on the initial results (BAM files), SAMtools (9) was used to identify and count the number of single nucleotide polymorphism (SNPs), copy number variations (CNVs) increases and decreases were detected using Control-FREEC (10), which calculates the number of different types of CNV events. The software Lumpy (11) was used to detect structural variations (SVs) and count the number of different types of SVs. MuTect (12), Strelka (13), Control-FREEC (10) and Lumpy (11) were used to detect somatic single-nucleotide variants (SNVs), insertions and deletions (INDELs), CNVs and SVs. Functional annotation of the detected gene variants was performed using ANNOVAR (14).
2.4.6 Identification of potential predisposing genes and driver mutations
We used SAMtools (9) to identify germline mutations (SNPs and INDELs) in the normal tissues of patients from the Cancer Gene Census (CGC) database, Familial Cancer Database, and intOGen database and the reported genes summarized in the Nature literature, and the detected mutations were screened for potential predisposing genes. We examined somatic mutations (SNVs and INDELs) in tumor specimens using MuTect software and then screened for known driver genes by using Bert Vogelstein (15), significant mutated genes (SMGs) (16) and the CGC database (17).
2.4.7 Analysis of tumor purity, tumor ploidy and targeted drug prediction
We used ABSOLUTE to calculate the purity (the proportion of tumor cells to total cells), ploidy (the average copy number of the sample) and cancer DNA fraction (the proportion of tumor DNA to total DNA) of the tumor sample to guarantee analysis quality (18). To identify potentially actionable mutations, tumor-derived genetic alterations were cross-referenced with the NovoDrug knowledgebase for targeted therapy prediction. This comprehensive resource combines validated therapeutic targets from authoritative references including DrugBank, the Pharmacogenomics Knowledge Base (PharmGKB), My Cancer Genome, the Food and Drug Administration (FDA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, supplemented with investigational agents from clinical trial registries like ClinicalTrials.gov. Concurrently, tumor-specific somatic variants were analyzed against the NovoDR database to detect putative resistance markers, providing clinically relevant insights for treatment selection and outcome evaluation (19).
2.4.8 Sanger sequencing
To confirm that the predisposing and driver genes containing mutant bases were detected by NGS, Sanger sequencing was performed. Genomic DNA was extracted according to the manufacturer’s instructions, after which polymerase chain reaction (PCR) was performed. After purification of the PCR products by agarose electrophoresis, the PCR products were sequenced on an Applied Biosystems™ 3730xl sequencer.
2.4.9 Gene mutation analysis across cancers
We used the cBioPortal database (https://www.cbioportal.org/, accessed on 14 August 2023) to analyze the mutant genes confirmed by Sanger sequencing across cancers.
2.4.10 Structure of protein–protein interaction networks
The protein–protein interaction (PPI) network of the genes confirmed by Sanger sequencing was constructed through the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database with a minimum required interaction score of 0.15 and a maximum number of interactors of 50. Then, the PPI network was visualized using Cytoscape software (version 3.9.1).
2.4.11 GO and KEGG pathway enrichment analyses
The biological functions of the node genes in the PPI network were analyzed through Gene Ontology (GO) (20) and KEGG (21) pathway enrichment analyses, setting a statistically significant difference threshold of q value < 0.05 using the “clusterProfiler” R package (version 4.3.1), and the top 20 genes of each analysis were visualized with the “GOplot” R package.
3 Results
3.1 Case presentation
A 57-year-old female patient was admitted to our hospital due to recurrent yellow discharge in the left nasal region and fixed-site headaches secondary to nasal obstruction after upper respiratory tract infections for two years, which were aggravated for two weeks. She underwent surgical resection of a right nasal hemangioma eight years prior. Physical examination revealed right deviation of the nasal septum, right nasal mucosa erosion, crust on the right middle nasal passage, right nasal stenosis, and crust and discharge in the left nasal cavity with unclear nasal structure. Computed tomography (CT) of the paranasal sinus revealed bilateral maxillary and ethmoid sinusitis, thickened right nasal mucosa, partial absence of the middle and inferior turbinate of the right nasal cavity, a growing mass of soft tissue density in the left nasal cavity and left nasal cavity stenosis. Massive amounts of necrotic tissue were removed from the left nasal cavity, and the diagnosis of ENKTL was confirmed by pathological examination of the discharge of the left nasal cavity combined with immunohistochemical results and in situ hybridoma with EBERs. Beginning in October 2016, the patient was treated with 54 Gray local nasal radiotherapy (30 cycles of 1.8 Gray) for 2 months, which resulted in unconfirmed complete remission (uCR), but she did not undergo regular reexamination after radiotherapy.
The patient experienced nasal obstruction for one month and was admitted to the hospital again after four years. Physical examination revealed nasal septum perforation, hypertrophy of the middle and inferior turbinate in the right nasal cavity with sticky discharge on the surface, adhesion of the left nasal cavity and invisible middle nasal meatus. Paranasal sinus CT revealed bilateral maxillary and ethmoidal sinuses filled with soft tissue and a decrease or disappearance of the partial cavity, an enlarged maxillary sinus orifice, partial absence of bilateral turbinate and hyperplasia of soft tissue in the nasal cavity. A nasopharyngoscope revealed a fragile pale mass in the left nasal cavity with a rough surface and unclear nasal structures, and no mass was found in the unobstructed right nasal cavity. Due to the expansive growth of the mass in the left nasal cavity and secondary bone destruction of the lateral nasal wall, the patient underwent endoscopic mass excision of the left nasal cavity. Pathology examination of a 0.4 cm mass resected from the ethmoid sinus and a 0.8 cm mass resected from the maxillary sinus demonstrated a composition of inflammatory granulation tissue and bone tissue with many plasma cells infiltrating the mesenchyme. Based on these findings combined with the immunohistochemical results of IgG (+), IgG4 (–), κ (+), and λ (+) (Figure 1), the patient was diagnosed with left nasal sinusitis rather than ENKTL recurrence.
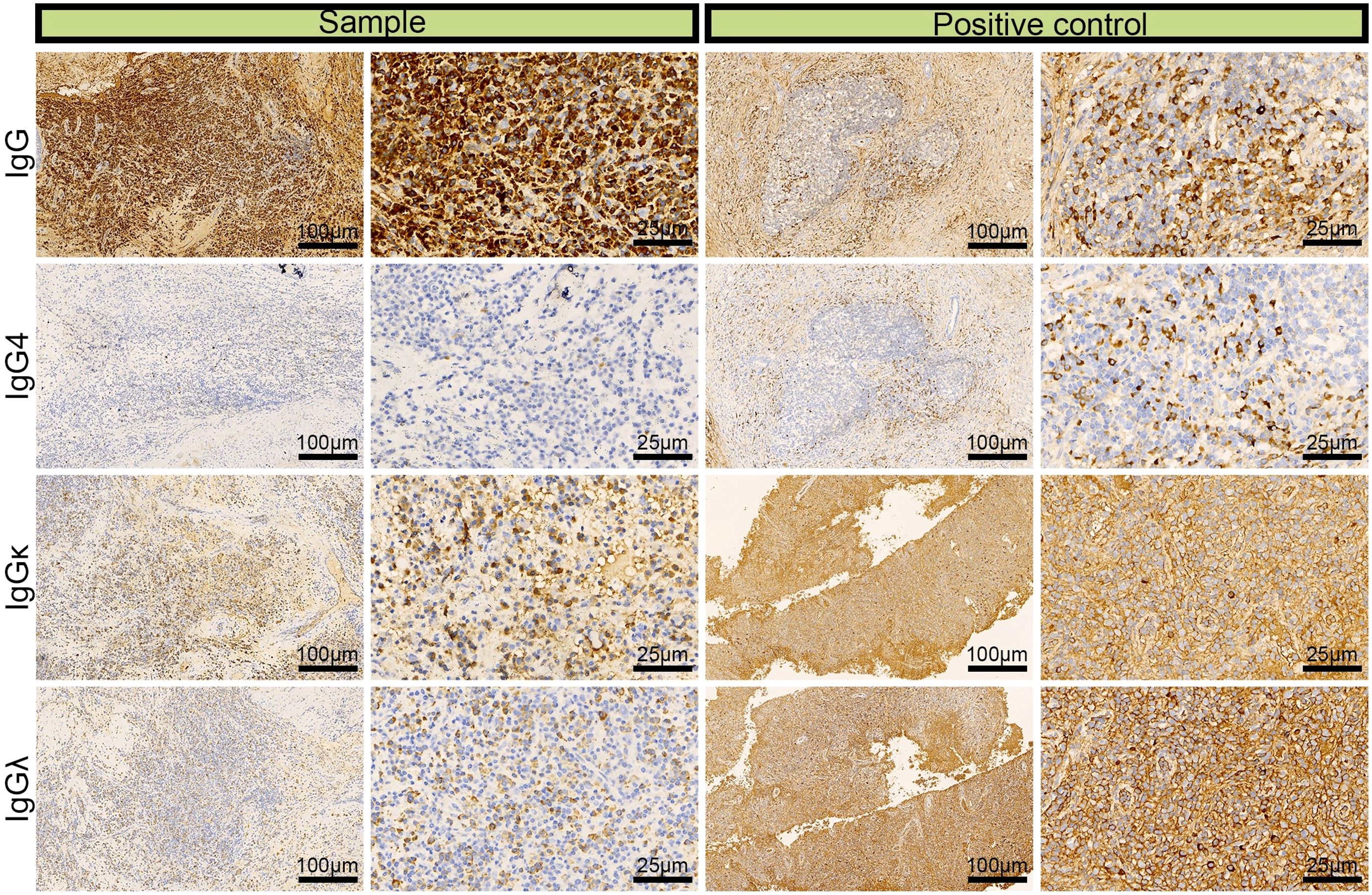
Figure 1. Immunohistochemical images of IgG, lgG4, lgGκ, and lgGλ in patients with left nasal sinusitis (scale bar: 25 μm).
One year later, the patient was referred to our hospital for a third visit and complained of left chest and back pain unrelated to breathing for four days. Chest CT and contrast-enhanced CT of the adrenal glands revealed a left adrenal mass with a diameter of 4.7 cm that showed heterogeneous enhancement, as shown by the three-dimensional structure in Figure 2. We ruled out primary adrenal malignancy owing to normal adrenal hormone levels as follows: epinephrine <0.14 nmol/L (normal 3.24–18 nmol/L), norepinephrine 8.52 nmol/L (normal 0.41-4.43 nmol/L), and dopamine 0.10 nmol/L (normal <0.2 nmol/L). However, the CT findings were indicative of adrenal malignancy; therefore, laparoscopic resection of the tumor was performed. The diagnosis of ENKTL was confirmed by postoperative pathology and immunohistochemistry.
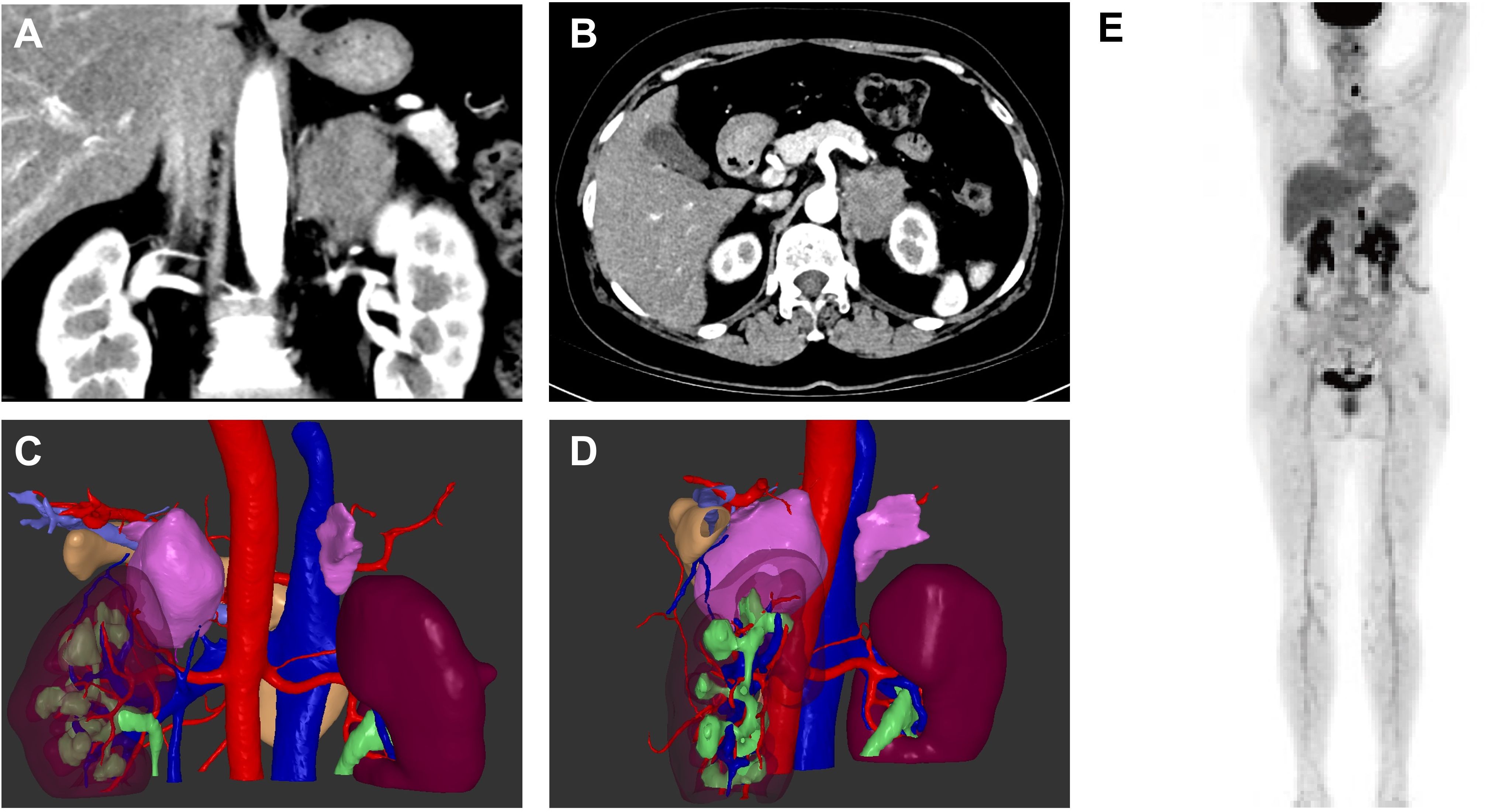
Figure 2. Enhanced computed tomography images of the left adrenal mass in sagittal and coronal views (A, B) and three-dimensional construction images (C, D). 18F-FDG PET/CT of the patient (E).
To help assign the stage and tailor the treatment plan, the patient underwent 18F-fluoro-2-deoxy-D-glucose positron emission tomography-CT examination, as shown in Figure 2E, revealing homogeneous increases in FDG uptake in the mucosa of the nasal tract (maximal standardized uptake value (SUVmax), 3.5-3.7), the left tonsil (SUVmax, 6.5), the left lobe of the thyroid (SUVmax, 23.1), the soft tissue in the left adrenal region (SUVmax, 16.5) and the left supraclavicular lymph node (SUVmax, 3.1). Bone marrow aspirate smears showed no evidence of malignant lymphoma, and the patient had no type B symptoms, such as fever, chills, or night sweats. According to the results, the patient was diagnosed with extranodal natural killer/T-cell lymphoma, nasal type (ENKTL-NT), stage IV. The patient underwent chemotherapy using the P-GEMOX regimen, which included gemcitabine (2 g, Day 1), oxaliplatin (150 mg, Day 1) and pegaspargase (3750 U, Day 1), for a cycle of 21 days. Radiological imaging was used to evaluate the therapeutic effect. After two cycles of chemotherapy, the patient suffered from significant general weakness, nausea, decreased appetite and reduced food intake. Laboratory findings revealed increased alanine aminotransferase (112.86 U/L) and aspartate aminotransferase (59.59 U/L) levels, which was considered to indicate liver damage caused by chemotherapy; therefore, the dose of gemcitabine was reduced to 1.6 g beginning in the third cycle. The patient achieved CR after 6 cycles. Then, PD-1 monoclonal antibody (sintilimab, 200 mg, Day 1) was regularly administered every three weeks. The patient achieved CR after 15 cycles of treatment and continued regular therapy with sintilimab. As of April 2025, the patient had undergone 30 cycles of sintilimab treatment with no signs of recurrence and was in good general condition.
3.2 Pathological features of adrenal ENKTL
3.2.1 Histopathological presentation
The left adrenal surgical specimen measured 4.5*5.0 cm with a complete capsule and a yellow to brown cut surface. HE staining revealed a destroyed adrenal structure with abundant diffusely distributed malignant tumor cells of a medium-large size with an irregular nucleus; light cytoplasmic staining; and aggressively growing tumor cells that invaded thick-walled blood vessels, enveloped the nerves, and infiltrated the adrenal tissue, exhibiting multiple annular foci.
3.2.2 Immunohistochemical profile for diagnosis
We performed IHC analysis, and the following results contributed to the diagnosis of ENKTL: CD3 (+), CD43 (+), CD56 (partial +), TIA (+), granzyme B (+), CD2 (+), CD4 (partial +), CD5 (–), CD7 (+), and CD8 (–) (Figure 3). Moreover, we excluded the diagnosis of B-cell lymphoma based on the CD20 (–) and CD79α (–) results and epithelial-origin tumors based on the CK(AE1/AE3) (–) result. The Ki-67 labeling index was high (90%), as shown in Figure 4. In situ hybridization demonstrated the presence of EBER.
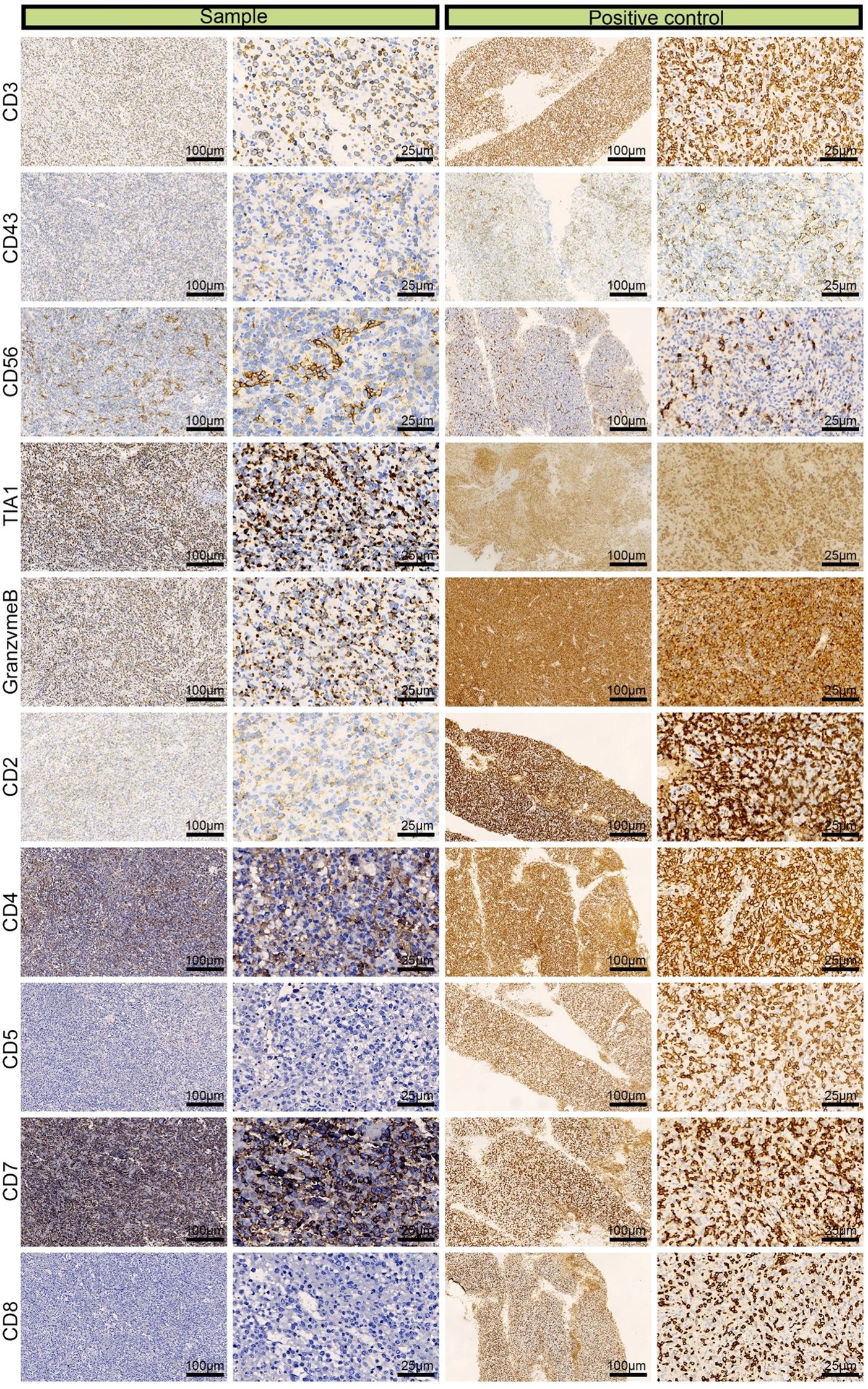
Figure 3. Immunohistochemical images of CD3 (+), CD43 (+), CD56 (partial +), TIA (+), granzyme B (+), CD2 (+), CD4 (partial +), CD5 (–), CD7 (+), and CD8 (–) from a left adrenal sample obtained by laparoscopic resection.
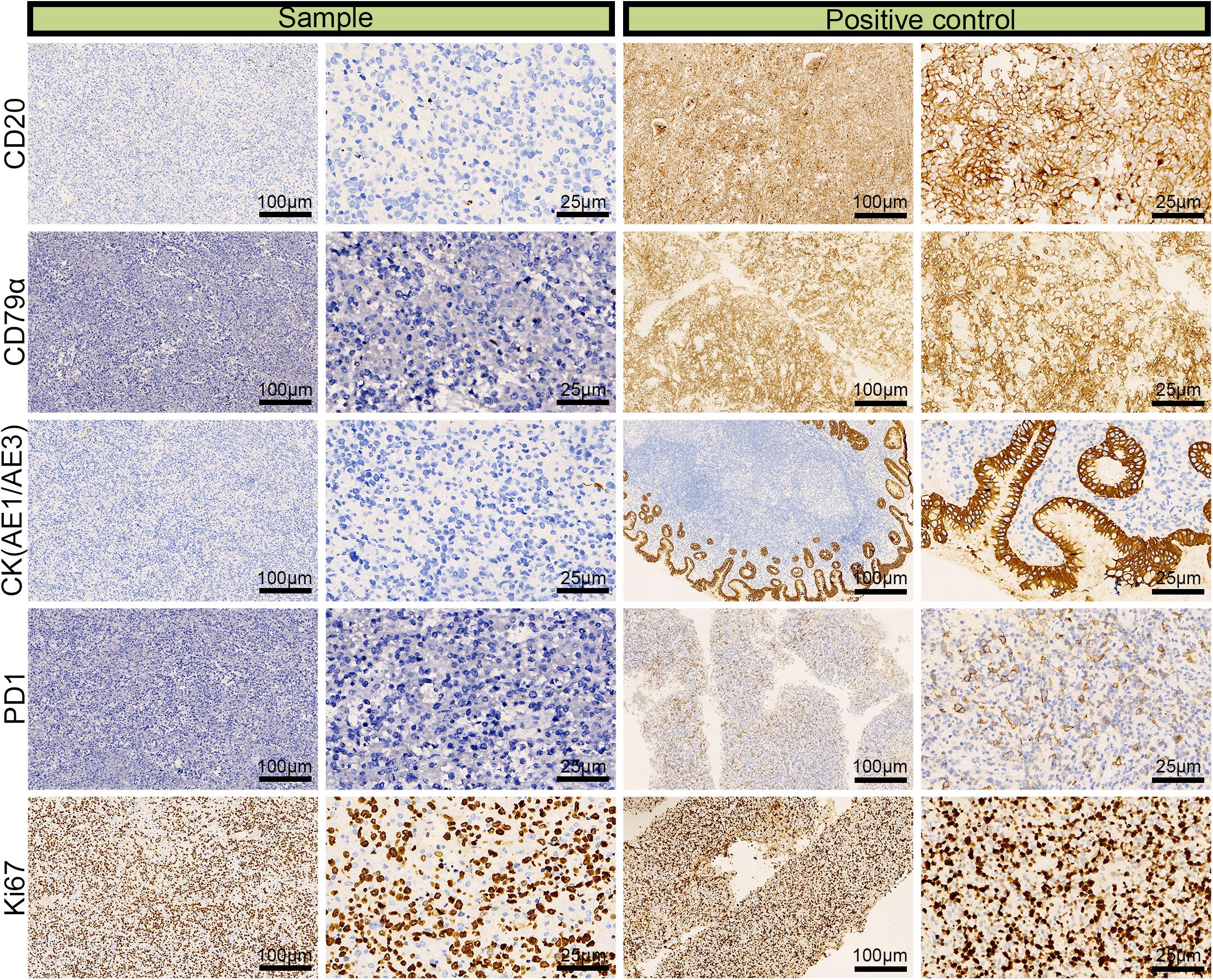
Figure 4. Immunohistochemical images for CD20 (–), CD79α (–), CK (AE1/AE3) (–), and the 90% labeling index of Ki-67 in a left adrenal sample obtained via laparoscopic resection.
3.2.3 Immunohistochemistry for detecting components of the tumor microenvironment
For immunohistochemistry to detect components of the tumor microenvironment, we analyzed the distributions of PD-L1, markers of vascular endothelial cells (CD31 and CD34), markers of TAMs (Tumor-associated macrophages, CD163 and CD68), markers of tumor-associated fibroblasts (vimentin, SMA) in the tumor tissue. As shown in Figure 5, positive staining of PD-L1, CD31 (partial +), CD34, CD68, CD163, vimentin and SMA detected in the tumor tissue, indicating that vascular endothelial cells, TAMs and fibroblasts may participate in the pathogenesis of ENKTL.
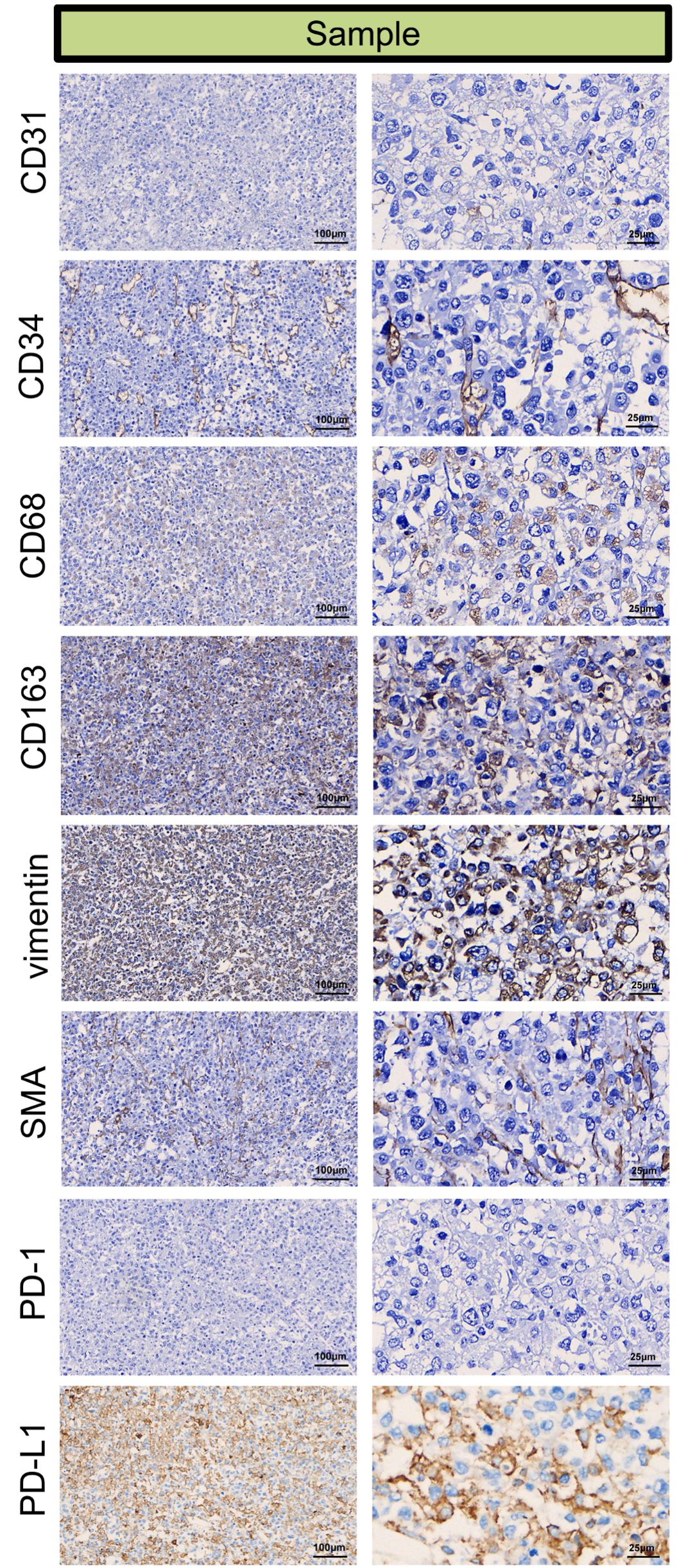
Figure 5. Immunohistochemical microscopic images of CD31 (partial +), CD34 (+), CD68 (+), CD163 (+), vimentin (+), SMA (+), PD-1 (–), PD-L1 (+) for the tumor micro-environment analysis in the tumor sample.
3.3 Population-based study
3.3.1 Patient clinical and demographic characteristics
Until April 2025, 1771 ACC patients in the SEER database and 39 ENKTL patients with adrenal involvement were included in our study. Compared with those of ACC patients, the clinical and demographic characteristics of ENKTL patients with adrenal involvement, except for age and radiotherapy, were highly heterogeneous. The detailed clinical and demographic characteristics are presented in Table 1 and Supplementary Table S2. As shown in the Kaplan–Meier survival plot in Figure 6, ENKTL patients with adrenal involvement exhibited significantly.
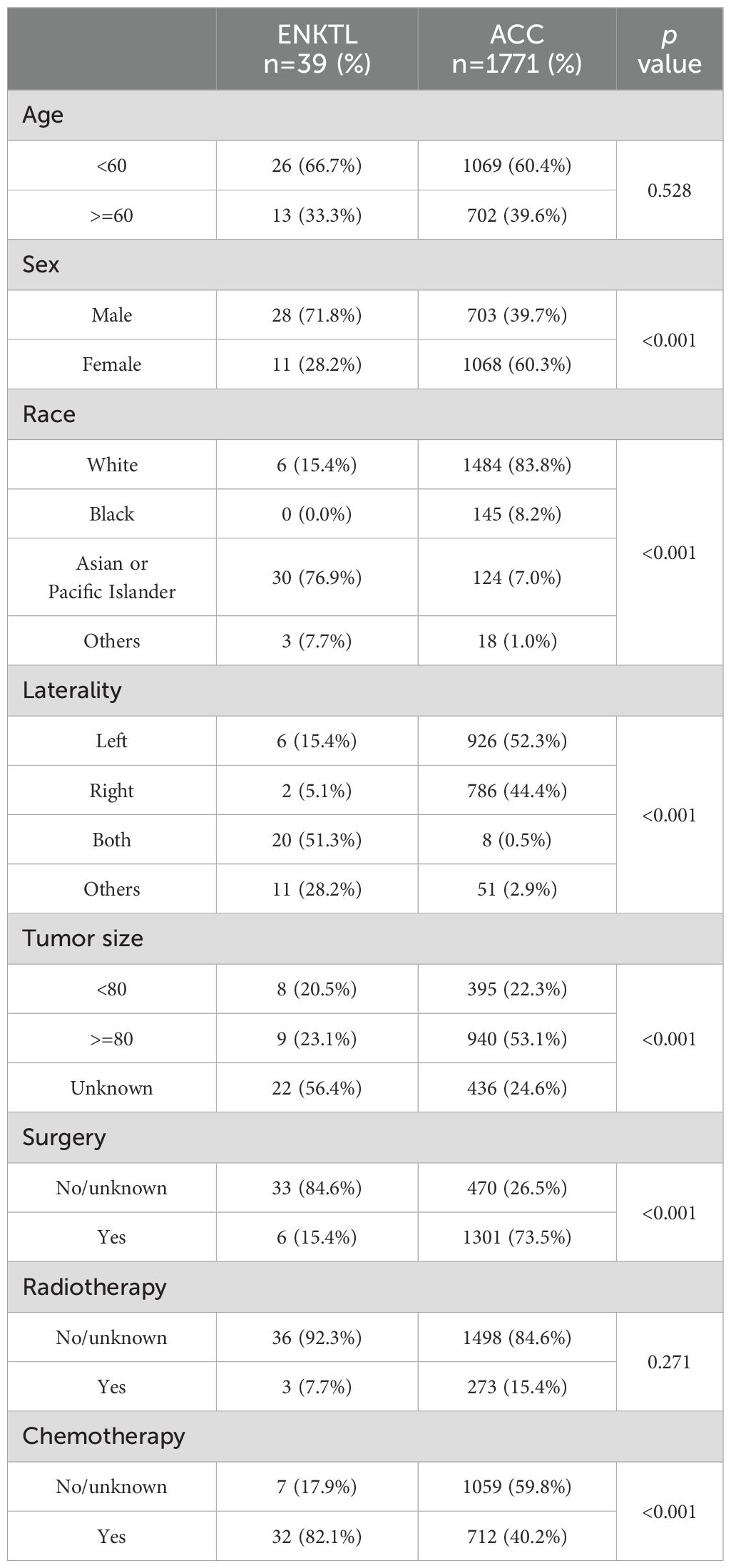
Table 1. Baseline characteristics of patients with adrenal extra-nodal natural killer/T -cell lymphoma (ENKTL) and adrenocortical carcinoma (ACC).
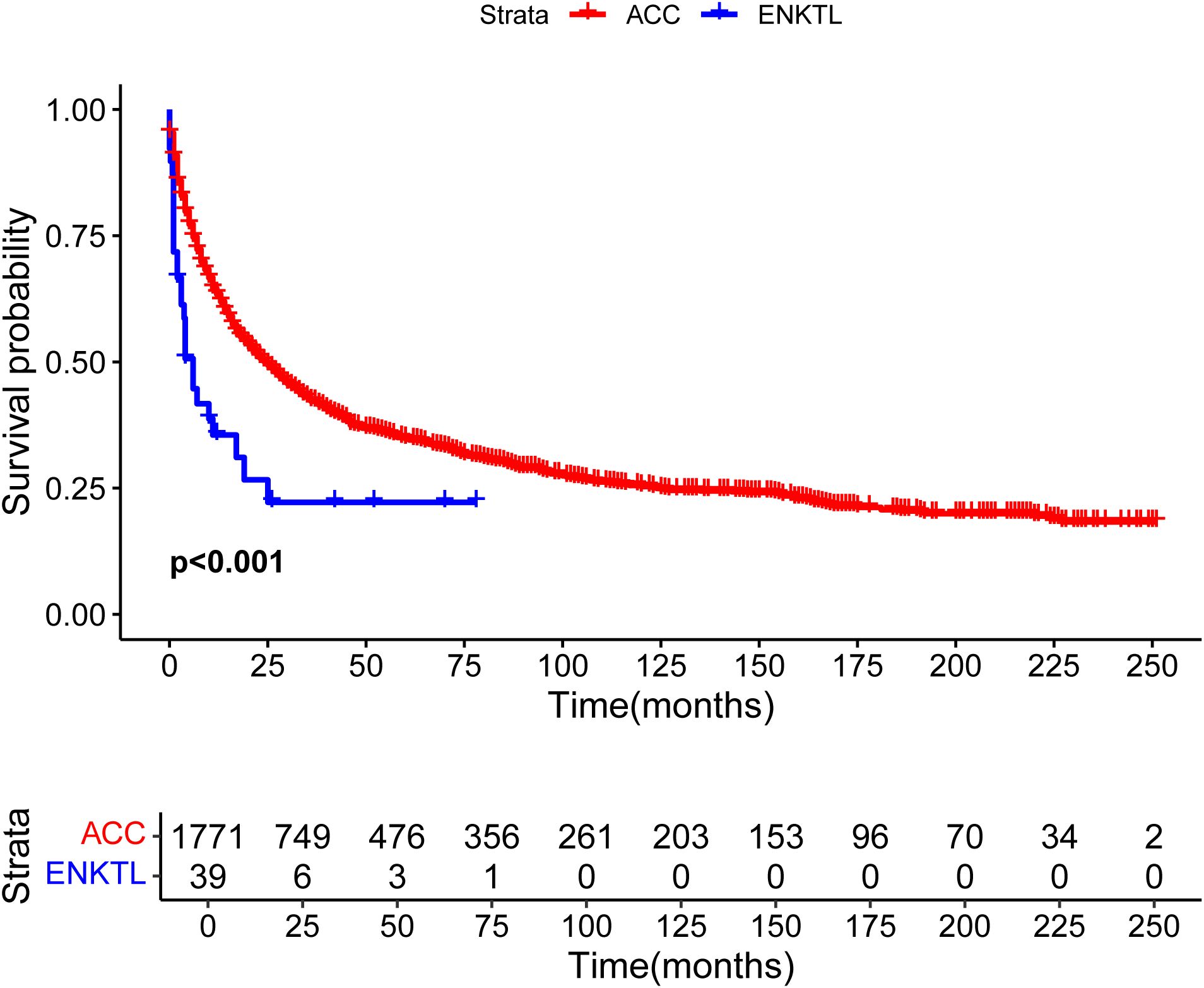
Figure 6. Overall survival curves of patients with extranodal natural killer/T-cell lymphoma (ENKTL) and adrenocortical carcinoma (ACC).
3.3.2 Identifying prognostic factors for ENKTL patients with adrenal involvement
To determine the prognostic factors influencing the OS of ENKTL patients with adrenal involvement, we conducted univariate and multivariate Cox regression analyses. As shown in the forest plot in Figure 7A, race (p=0.004) and B symptoms (p=0.015) were identified as factors related to prognosis in the univariate Cox regression analysis. The forest plot of the multivariate Cox regression analysis in Figure 7B shows that race (p=0.015) and chemotherapy (p=0.027) was an independent protective factor.
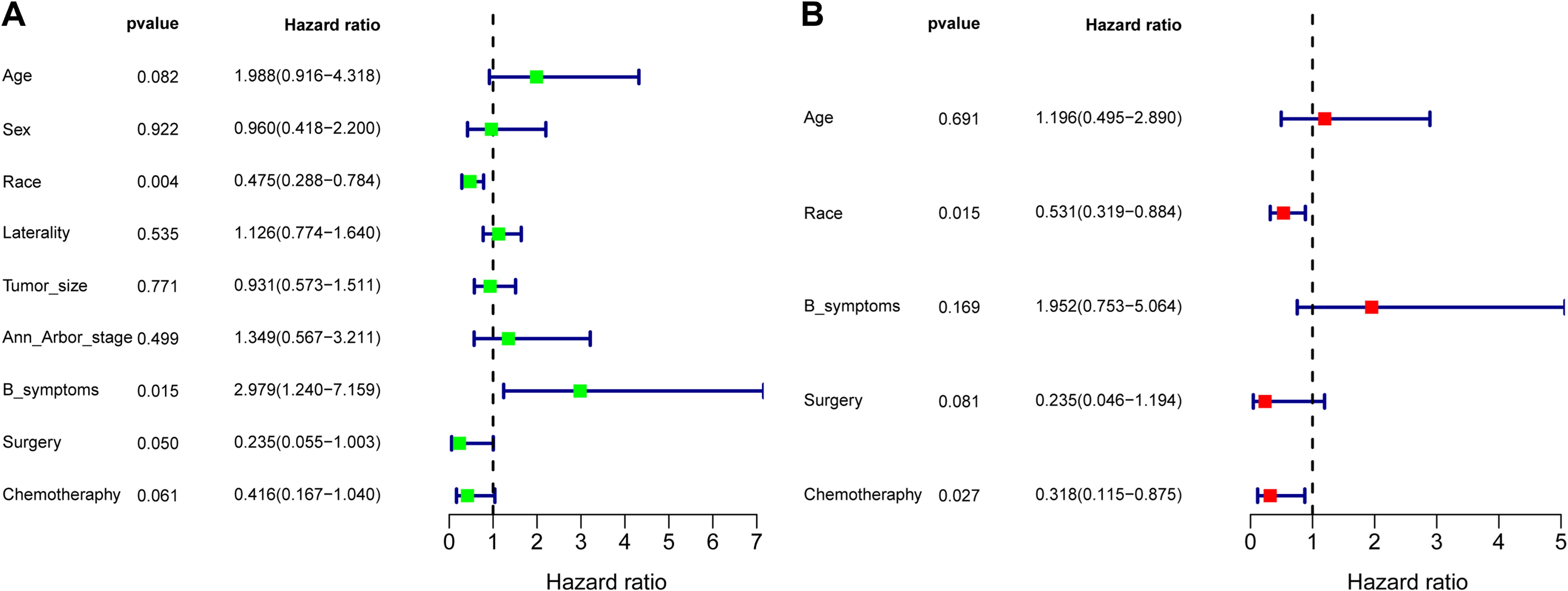
Figure 7. Forest plot of the univariate regression analysis (A) and multivariate regression analysis (B) revealing the relationships between different prognostic factors and overall survival in extranodal natural killer/T-cell lymphoma (ENKTL) patients (n=39). The tetragonal diamonds represent the hazard ratios (HRs), and the horizontal line crossing the diamonds represents the 95% confidence intervals (CIs). The data were analyzed by a Cox proportional hazards model and are shown as HRs and 95% CIs.
3.4 Whole-genome sequencing study
3.4.1 Identification of SNPs and INDELs
The average Q30 of the sequencing samples was 91.76%, and the average error rate was 0.03%, indicating that the sequencing data were of good quality and met the analysis requirements. We sequenced 263,986,447 and 228,437,421 read pairs in the two tumor specimens and 243,399,667 and 212,257,867 read pairs in the two matched normal specimens. A total of 3,400,005 and 3,504,202 SNPs were detected in the tumor specimens and matched normal specimens, respectively. The transformation/inversion ratio of 2.08 reflects the accuracy of the SNP dataset. A total of 497,401 and 535,209 INDELs were verified in the tumor specimens and matched normal specimens, respectively. The details of the INDELs are shown in Figure 8 and Supplementary Tables S3–S6.
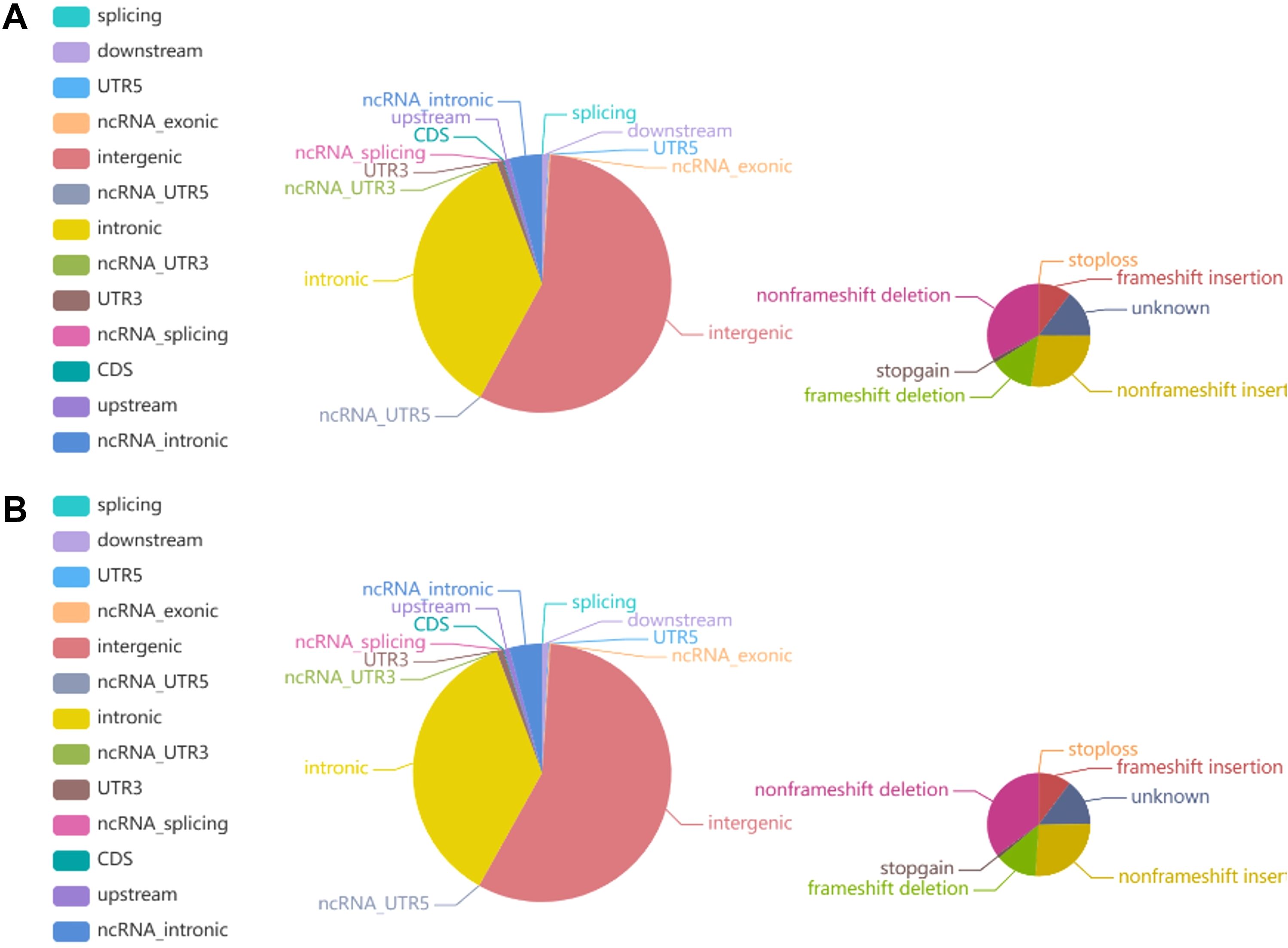
Figure 8. Pie chart representing the distribution of the number of INDELs. (A) The left and right images show the number of INDELs in different regions of the genome and in coding regions in tumor tissue, respectively. (B) The left and right images show the number of INDELs in different regions of the genome and the coding regions in normal tissue, respectively.
3.4.2 Identification of somatic SNVs and INDELs
Somatic mutations have great value in explaining tumorigenesis and progression. We identified a total of 15892 somatic SNVs and 364 somatic INDELs, which were mainly clustered in the intronic and intergenic regions. Detailed information on the somatic SNVs and INDELs is presented in Supplementary Tables S7–S8.
3.4.3 Identification of somatic SVs and CNVs
In the detection of somatic SVs in the tumor specimens, we identified 238 duplicates, 247 deletions, 4 inversions, 24498 interchromosomal translocations and 2930 intrachromosomal translocations. A total of 572 somatic CNVs, including 548 gains and 24 losses, were identified. We show the Circos plot of somatic variations in Figure 9.
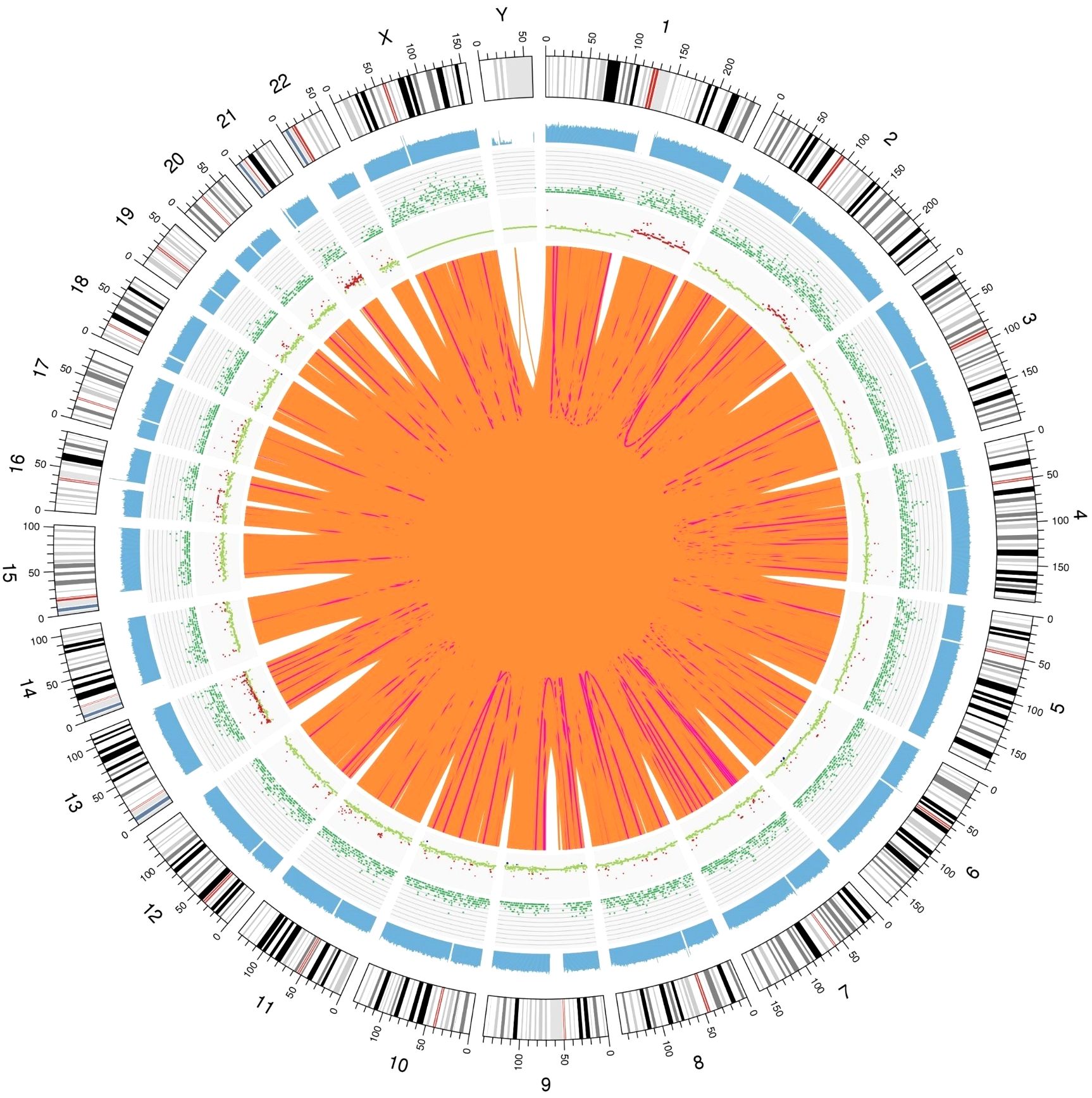
Figure 9. Genomic variation Circos plot. The structure of the three layers that run from the outside to the inside represents the sequence coverage map, SNP and INDEL density, and CNV results, respectively. Red indicates an increase in the copy number, blue indicates a decrease in the copy number, and green indicates a normal copy number.
3.4.4 Identification of Predisposing and Driver Mutating Genes
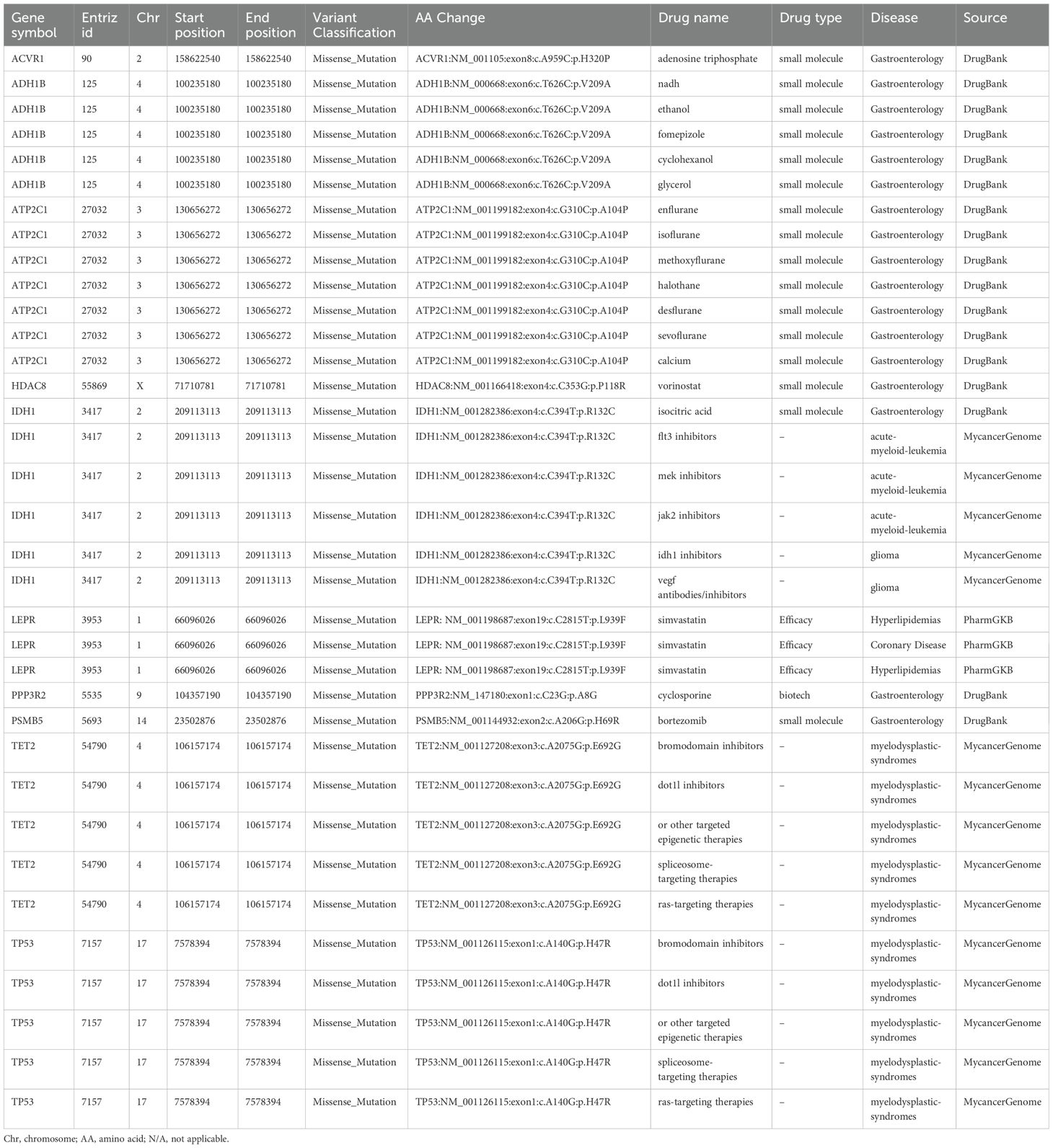
Mutations in cancer predisposition genes (CPGs) significantly increase the risk of cancer in individuals. We identified 24 predisposing genes, including HIP1, ELF1, and ZFHX3, among others, as shown in Supplementary Table S9. Identification of driver mutations and driver mutation genes that give tumors a selective growth advantage is important for understanding cancer pathogenesis. We identified 10 driver mutation genes, namely, LEPR, ACVR1, STAT3, TET2, IDH1, CHD7, STAT3, FAS and TP53, as shown in Supplementary Table S10.
3.4.5 Analysis of tumor purity, ploidy and targeted drug prediction
The tumor purity was 0.4, the tumor ploidy was 3.39, and the cancer DNA fraction was 53%. We identified ten mutation sites associated with targeted drugs, and the detailed information is presented in Table 2. There were one drug (adenosine triphosphate) for ACVR1, five drugs (NADH, ethanol, fomepizole, cyclohexanol, glycerol) for ADH1B, seven drugs (enflurane, isoflurane, methoxyflurane, halothane, desflurane, sevoflurane and calcium) for ATP2C1, one drug (vorinostat) for HDAC8, six drugs (isocitric acid, FLT3 inhibitors, MEK inhibitors, JAK2 inhibitors, IDH1 inhibitors and VEGF antibodies/inhibitors) for IDH1, one drug (simvastatin) for LEPR, one drug (cyclosporine) for PPP3R2, one drug (bortezomib) for PSMB5, five drugs (bromodomain inhibitors, DOT1L inhibitors, other targeted epigenetic therapies, spliceosome-targeting therapies and Ras-targeting therapies) for TET2 and five drugs (bromodomain inhibitors, DOT1L inhibitors, other targeted epigenetic therapies, spliceosome-targeting therapies, and Ras-targeting therapies) for TP53.
3.4.6 Sanger sequencing
To confirm the presence of mutations in the susceptibility and driver genes, we performed Sanger sequencing. We confirmed the mutation of STAT3 at chr17: 40474482 A>T, TET2 at chr4: 106157174 G>A, FAS chr10: 90773993 G>A and TP53 at chr17: 7578394 C>T (Figure 10).
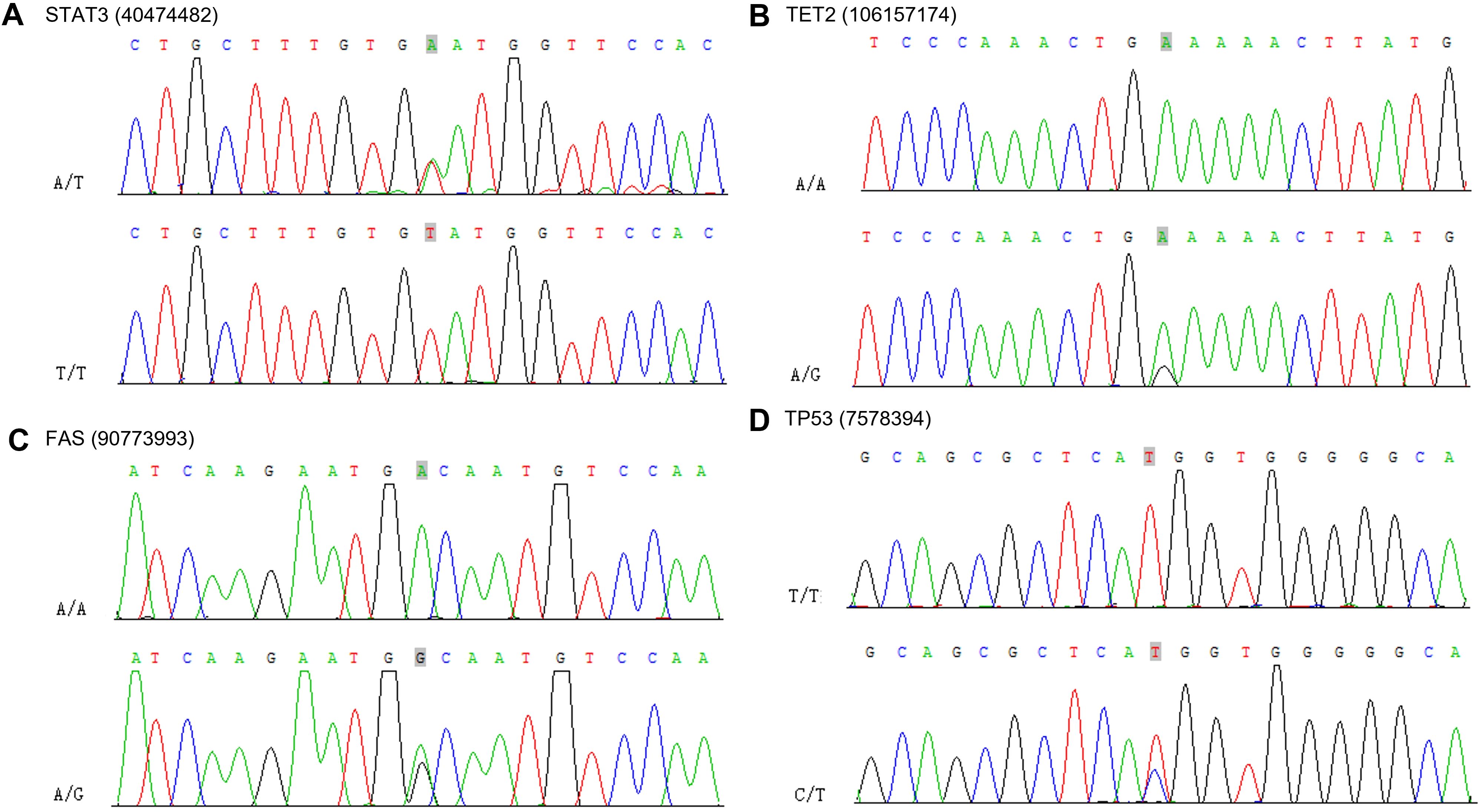
Figure 10. Sanger sequencing electropherograms of the STAT3 mutant at position chr17: 40474482 A>T (A), TET2 mutant at position chr4: 106157174 G>A (B), FAS mutant at position chr10: 90773993 G>A (C) and TP53 mutant at position chr17: 7578394 C>T (D).
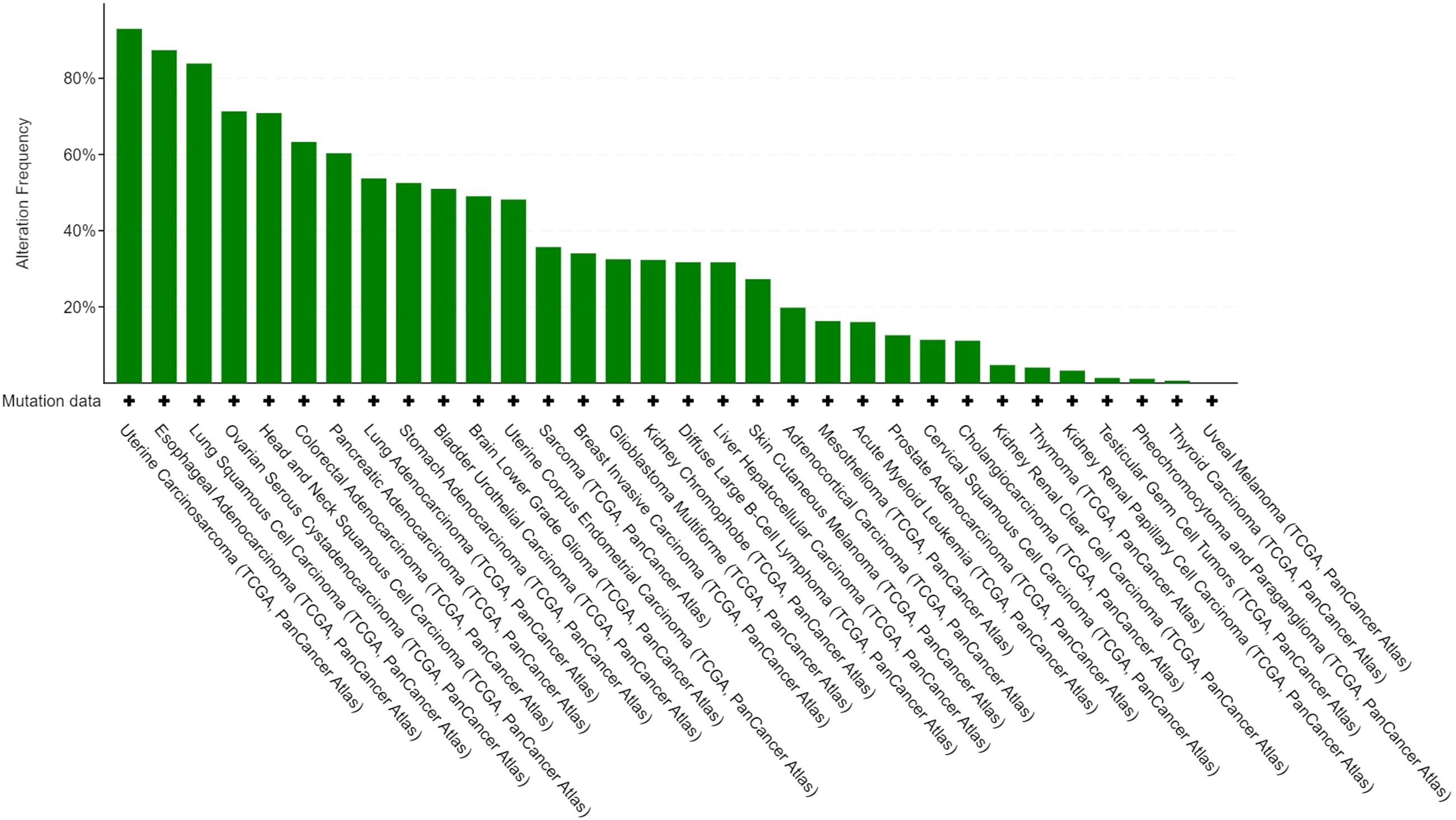
3.4.7 Gene mutation analysis across cancers
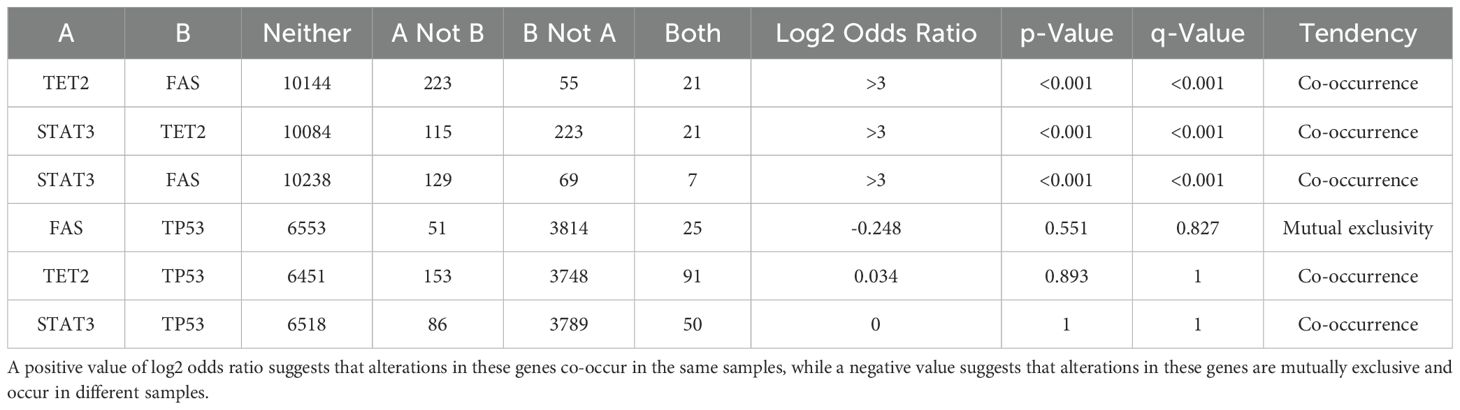
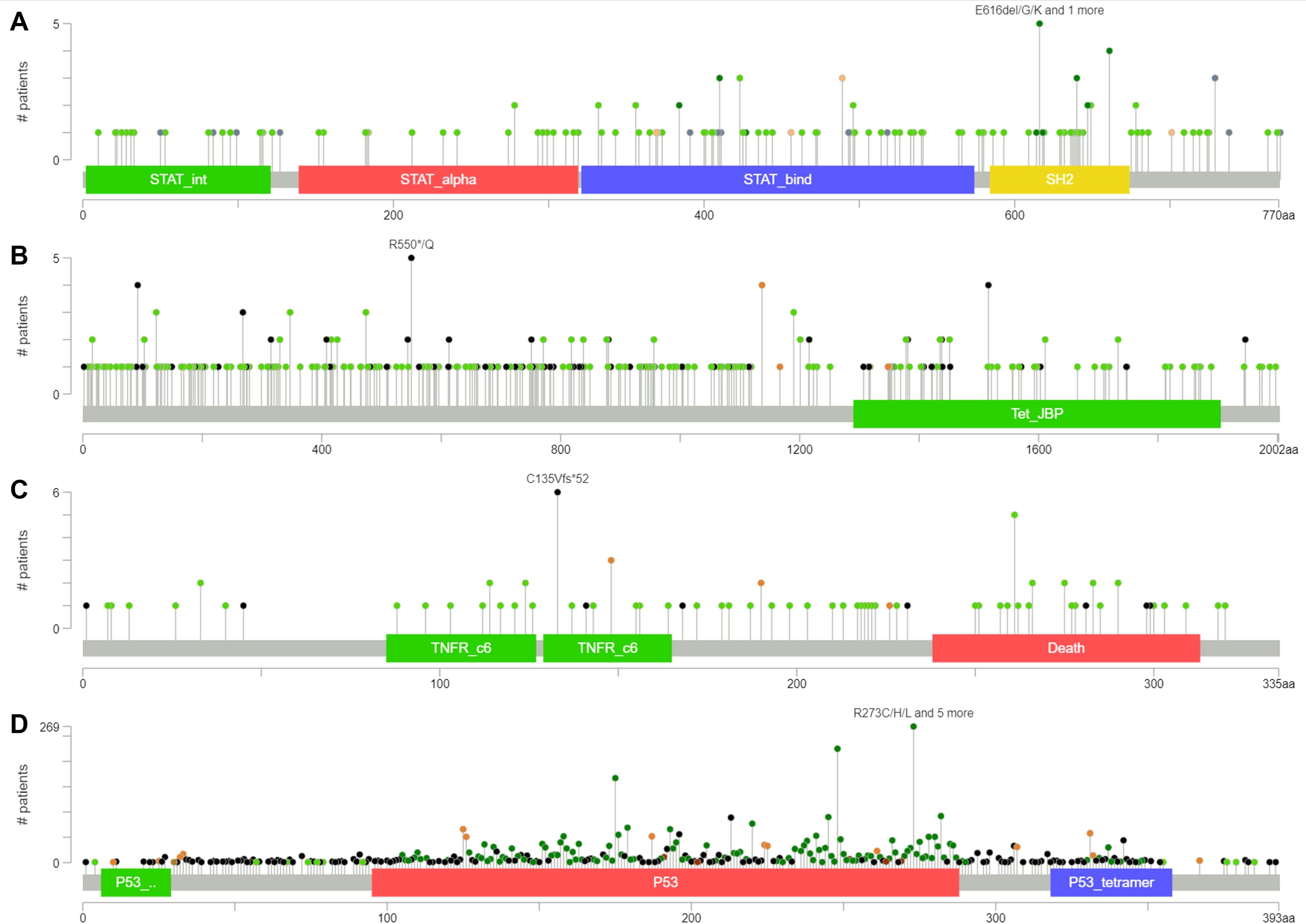
We performed gene mutation analysis of STAT3, TET2, FAS and TP53 in pancancer datasets, including 10967 samples from 32 studies, through the online database cBioPortal. The oncoprints showing the mutation types and cancer type summaries are presented in Figure 11. The mutual exclusivity analysis shown in Table 3 revealed that mutations in STAT3, TET2 and TP53 tended to cooccur between any two genes (p<0.001). The lollipop figures in Figure 12 present the mutation positions of the four genes. Figure 13 shows that mutations in FAS (p=0.0411) and TP53 (p<0.001) were associated with OS.
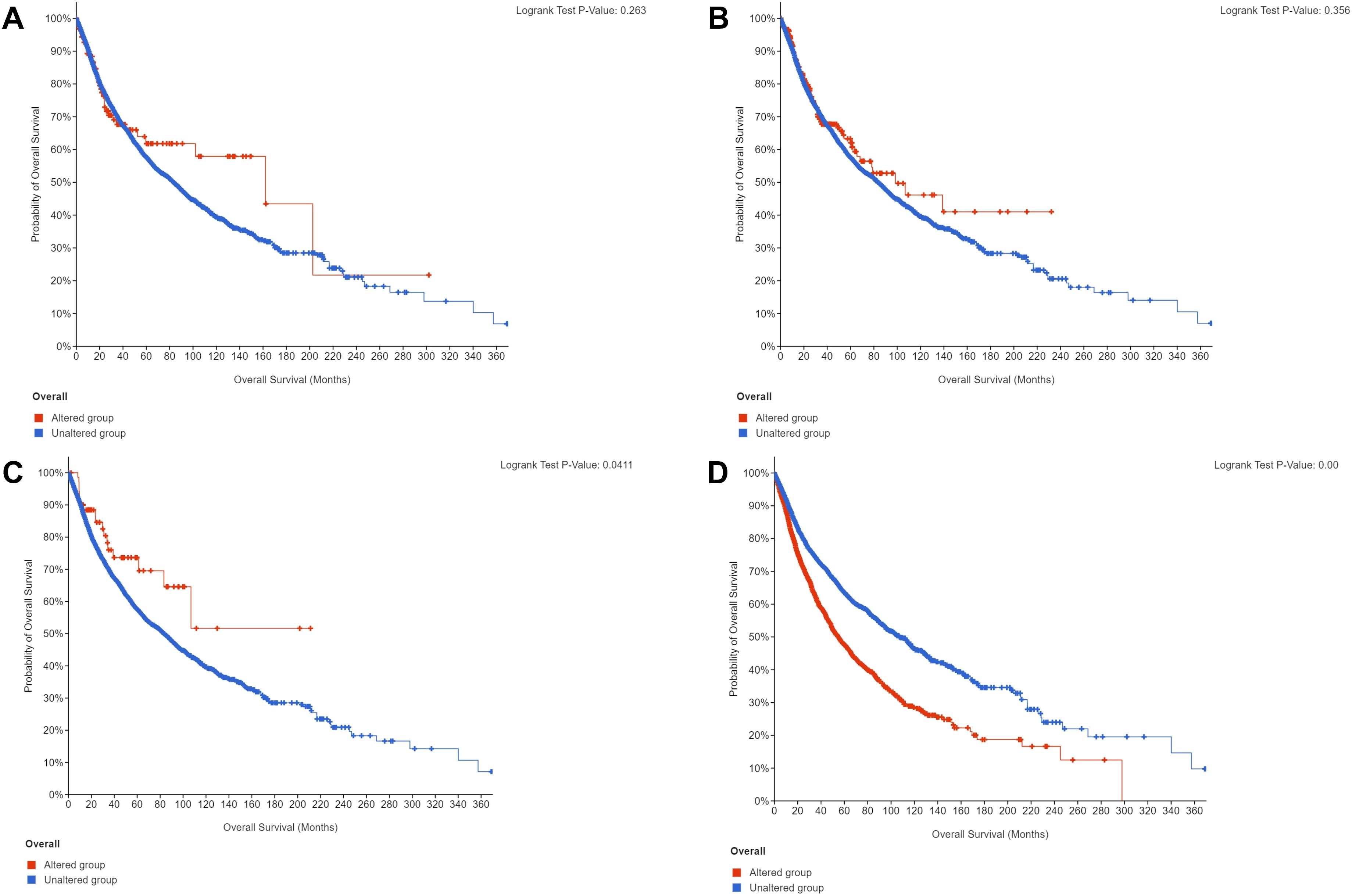
Figure 13. Overall survival curves according to gene mutation status. (A) STAT3. (B) TET2. (C) FAS. (D) TP53.
3.4.8 Structure of the protein–protein interaction network and functional enrichment
The PPI network (Figure 14) contained 54 nodes and 119 edges. The GO analysis results revealed that the node genes were markedly enriched in the intrinsic apoptotic signaling pathway and extracellular matrix organization in the biological process category, the PML body and membrane raft in the cellular component category, and p53 binding in the molecular function category (Figure 15). According to the KEGG analysis, the genes were significantly enriched in the p53 signaling pathway (Figure 15).
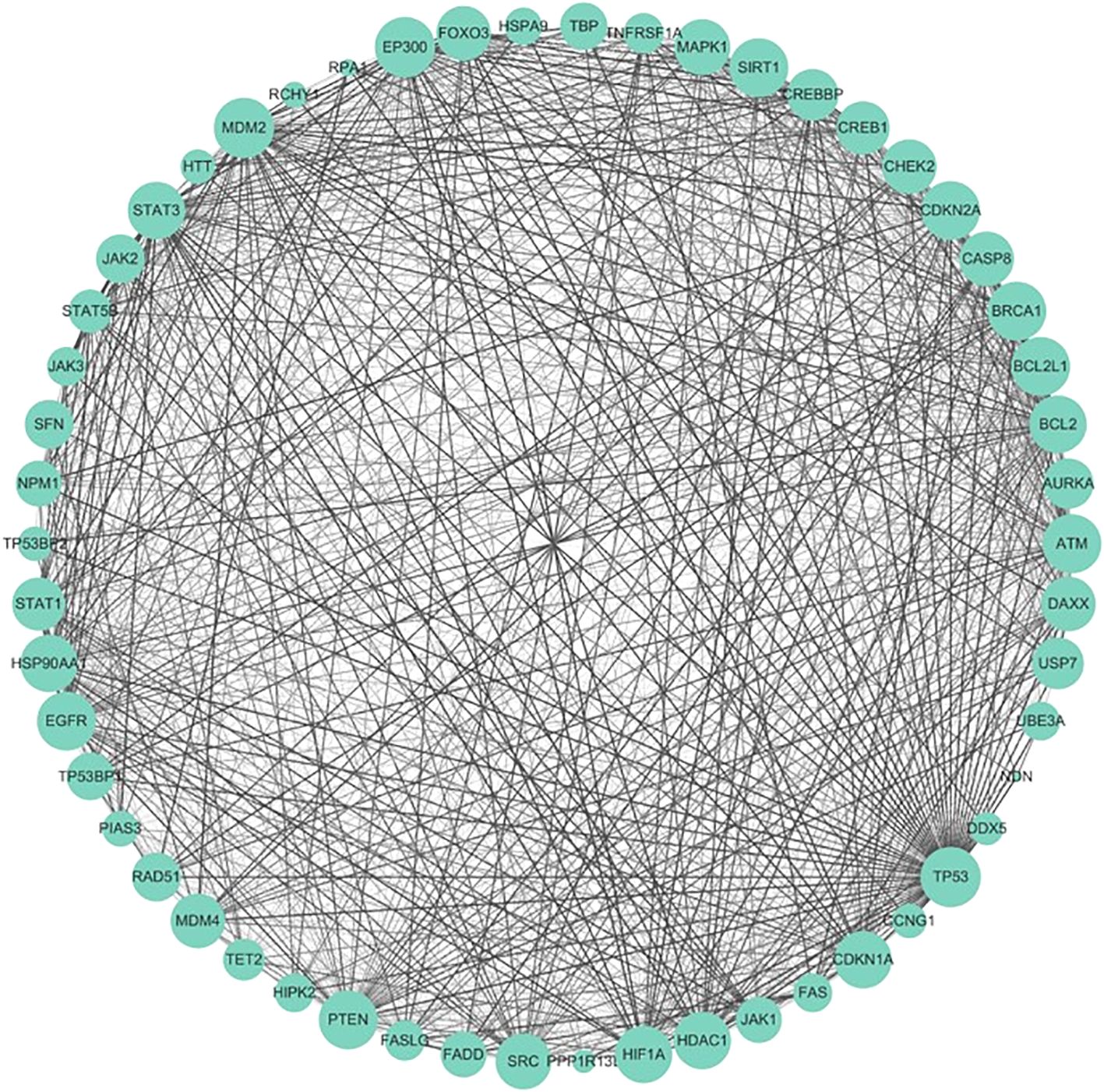
Figure 14. Protein–protein interaction network (PPI). The size of the nodes represents the degree of the nodes, and the thickness and color of the edge represents the score.
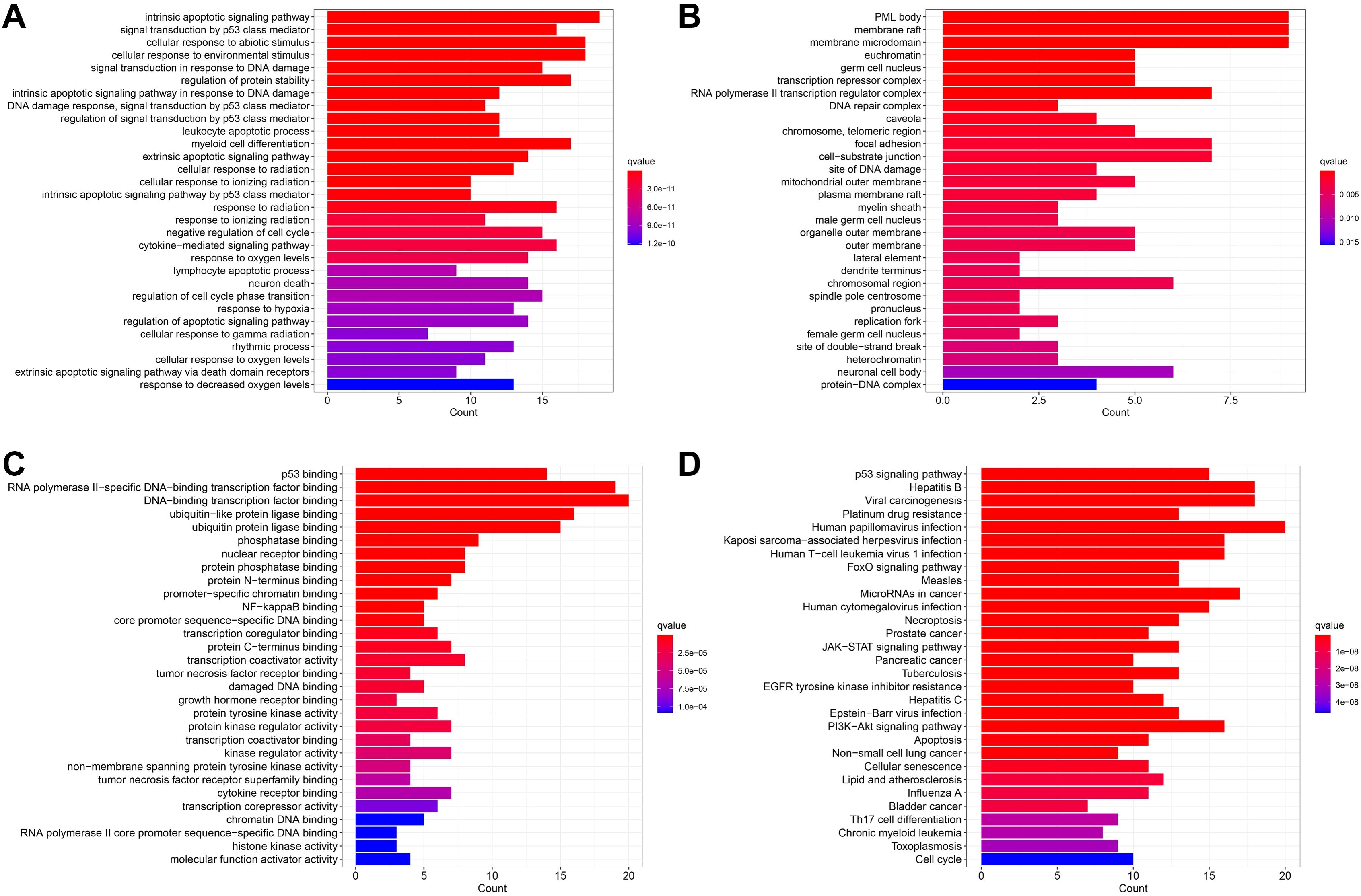
Figure 15. Enrichment analysis results. (A) The BP category of GO analysis. (B) The CC category of GO analysis. (C) The MF category of GO analysis. (D) KEGG pathway enrichment analysis.
4 Discussion
Adrenal tumors diagnosed as ENKTL are very rare, and patients tend to have a poor prognosis. In our study, we present a patient whose primary tumor site was nasal with secondary adrenal and systemic metastases. Despite being Ann Arbor stage IV and EBV positive as well as having a very high-risk NRI (age >60 years, stage IV, primary tumor invasion), after surgery, systemic therapy and immunotherapy, the patient was in good general condition with no signs of recurrence during 78 months of regular follow-up. To the authors’ knowledge, this is the longest surviving ENKTL patient with adrenal gland involvement according to a review of the literature on adrenal ENKTL patients. To determine the clinicopathological characteristics and prognosis of adrenal ENKTL patients, we enrolled 1771 ACC patients and 31 adrenal ENKTL patients after searching the PubMed, Medline, Cochrane Library, Web of Science, Embase, Scopus and SEER databases. The sex (p<0.001), race (p<0.001), laterality (p<0.001), tumor size (p<0.001), surgery (p<0.001) and chemotherapy (p<0.001) characteristics of the ENKTL patients were different from those of the ACC patients. According to the prognosis analyses, compared with ACC patients, ENKTL patients had a significantly poorer prognosis (p<0.001). The prognostic analyses of OS in patients with ENKTL showed that chemotherapy was an independent factor influencing the prognosis of ENKTL patients, which suggested that routine chemotherapy for ENKTL patients can improve prognosis and prolong patient survival. In the immunohistochemistry for detecting components of the tumor microenvironment, the infiltration of vascular endothelial cells and TAMs was detected, indicating the participation of vascular endothelial cells and TAMs in the pathogenesis of ENKTL. In the complex tumor microenvironment, inflammatory cells exert multifaceted and context-dependent effects on cancer progression, with TAMs representing a critical cellular component that exhibits distinct phenotypic plasticity and pro-tumorigenic functional properties across diverse malignancies (22–25). In the immunohistochemical analysis of CD68+ TAMs in 70 ENKTL patients Wang et al. performed demonstrated that high TAMs infiltration (>60/high-power field) was significantly associated with adverse clinical features, including B symptoms, elevated LDH levels, high Ki-67 expression, and lower complete response rates (46.7% vs 77.5%, p=0.008). Subsequent survival analysis revealed that patients in the high-TAM group had significantly worse 5-year overall survival (OS: 43.3% vs 79.5%) and progression-free survival (PFS: 30.0% vs 60.9%) compared to the low-TAM group (all p<0.01). Multivariate Cox regression analysis further identified CD68+ TAMs as an independent poor prognostic factor (OS: HR=3.195; PFS: HR=2.890) (26). TAMs may be involved in ENKTL progression by promoting tumor proliferation, inhibiting apoptosis, and mediating chemoresistance, suggesting that targeting TAMs may be a novel strategy for ENKTL patients.
We conducted WGS to detect genetic alterations in the patient, and validation by Sanger sequencing confirmed that STAT3, TET2, FAS and TP53 were the 4 main mutant driver genes. TP53 encodes the protein P53, which regulates the expression of the corresponding target genes in response to various cellular stresses, inducing the regulation of DNA repair, apoptosis, senescence, and metabolism (27). More than 50% of cancer patients carry TP53 mutations (28), and the most common type of mutation in TP53 is a missense mutation, which encodes a mutant P53 protein that not only inhibits the oncogenic effects of wild-type P53 but also exhibits new oncogenic functions, such as the promotion of cell proliferation, evasion of apoptosis, metabolic changes, and migration (27). TET2 is an epigenetic regulator that mediates DNA demethylation, promotes gene transcription and is crucial for host hematopoiesis and immunity (29). TET2 is frequently mutated in the hematopoietic system, eventually leading to the development of a series of blood malignancies (30). STAT3 is usually activated by IL-6 family cytokines via JAK and is involved in multiple cellular pathways, which in turn affects the proliferation, progression and immune dysfunction of cancer cells. STAT3 is usually considered an oncogene but may act as a tumor suppressor under specific circumstances. The aberrant activation of STAT3 occurs in approximately 70% of tumors. Inhibition of STAT3 in cancer not only decreases cell survival but can also reactivate the anticancer immune response, providing dual benefits (31). Accumulating evidence shows that STAT3 mutations participate in cancer pathogenesis, including in ENKTL. STAT3 mutations were first identified in hyper-IgE syndrome (32), and STAT3 mutations in solid tumors were detected in the research of Pilati et al. on inflammatory hepatocellular adenomas (33). The JAK-STAT signaling pathway is an evolutionarily conserved mechanism that mediates cellular responses to cytokines, interferons, and growth factors, regulating key processes such as immune function, inflammation, cell proliferation, and differentiation. Dysregulation of this pathway is strongly linked to autoimmune disorders fox example rheumatoid arthritis, atopic dermatitis and cancers including myeloproliferative neoplasms, prostate and breast cancer (34, 35). In next-generation sequencing of 34 ENKL samples, the JAK/STAT pathway was altered in 55.9% of cases, predominantly through STAT3 mutations in the SH2 domain that likely induce constitutive pathway activation. The genetic alterations may be further amplified by EBV infection, which enhances STAT3 signaling and reinforces the pathway oncogenic role in ENKTL (36). The key role of JAK/STAT signaling pathway in the pathogenesis of ENKTL was also been suggested by the previous researches (37, 38). Targeting JAK-STAT with inhibitors for example ruxolitinib shows therapeutic potential, particularly in myeloproliferative neoplasms and leukemias, though challenges remain in understanding its full regulatory mechanisms and optimizing clinical applications (39). The binding of FAS, a transmembrane receptor, to its natural ligand (FasL) plays a pivotal role in immune homeostasis and surveillance. Mutations in FAS (40), mainly at the site of the fine death receptor structural domain involved in the recruitment of the connexin FAS-associated protein to the death structural domain, have been found to be associated with the disruption of self-tolerance in patients with autoimmune lymphoproliferative syndrome. FAS mutations have also been found in lymphomas, suggesting that FAS may be a tumor suppressor gene. Death receptor structural domain site mutations promote tumor progression by rendering tumor cells resistant to apoptotic responses but may also promote tumor progression by promoting tumorigenesis and inflammatory or autoimmune mechanisms (41). Takakuwa et al. found that approximately 50% of ENKTL patients harbored FAS gene deletions or mutations, which impaired the formation of the death-inducing signaling complex, leading to inactivation of the FAS signaling pathway and enabling tumor cells to evade apoptosis (42, 43). Moreover, the EBV-encoded LMP1 protein activates both NF-κB and PI3K/Akt pathways, leading to upregulation of anti-apoptotic proteins (such as c-FLIP and Bcl-2), which indirectly suppresses FAS-mediated apoptosis. The combined effects of FAS functional defects and EBV oncoproteins in promoting immune evasion may contribute to disease progression in ENKTL (44, 45). FAS mutations result in defective apoptotic signaling in ENKTL cells, serving as a critical mechanism for immune evasion and tumor cell survival. Targeting the pathway through combinatorial approaches, such as synergizing immune checkpoint inhibitors with apoptosis-inducing therapeutics, represents a promising treatment strategy for ENKTL patients. In ENKTL patients, mutations in TP53, STAT3 and TET2 are frequent, although the proportion of these mutations is heterogeneous across studies. TP53 mutations are associated with increased mortality in ENKTL patients (27) and is the most frequent mutation in relapsed patients (46). Recurrent mutations in genes in the JAK-STAT pathway, including STAT3, play an important role in ENKTL survival and proliferation and the development of precise therapeutic approaches for JAK/STAT inhibition (47). Moreover, STAT3 mutation has been investigated in EBV-positive inflammatory follicular dendritic cell sarcoma and EBV-positive HIV-associated diffuse large B-cell lymphoma (48, 49), and we believe that chronic active EBV infection may play an important role in the role of STAT3 mutation in activating tumor growth. As an epigenetic biomarker of ENKTL, TET2 mutation is significantly associated with the loss of protein expression and is correlated with shorter overall survival, suggesting that TET2 may be involved in the cancer-driving process of ENKTL (50).
For patients with stage III/IV or relapsed/refractory disease, asparaginase‐based chemotherapy is the primary treatment (51). Regimens containing L-asparaginase, including SMILE and AspaMetDex (L-asparaginase, methotrexate, and dexamethasone), exhibited similar efficiency, while patients showed better tolerance to the AspaMetDex regimen (52). The P-GEMOX and DDGP (dexamethasone, cisplatin, gemcitabine, and pegaspargase) regimens involving the substitution of L-asparaginase, such as pegylated asparaginase, have shown promising results. Compared with SMILE, P-GEMOX and DDGP resulted in a better treatment response and survival benefit and exhibited reduced toxicity (53). Moreover, the GDP regimen (gemcitabine, dexamethasone, and cisplatin) had equally superior efficacy and reduced toxicity, including a lower incidence of myelosuppression, prolonged activated partial thromboplastin time, pancreatitis, and anaphylactic reaction, in comparison to L-asparaginase treatment (54). The therapeutic efficacy and good tolerance of monotherapy with PD-1/PD-L1 antibodies in ENKTL patients who did not respond to asparaginase-based treatment have been demonstrated in early-phase trials and retrospective studies (55, 56). The efficacy of PD-1 antibody (sintilimab) combined with P-GEMOX was highly promising, achieving an 85% complete response rate and 100% overall response rate in the research of Tian et al. (57), consistent with previous reports demonstrating an 88.9% objective response rate (58), further confirming the robust antitumor activity of this regimen in advanced ENKTL with durable survival benefits and a manageable safety profile. A retrospective study by Wang X et al. also revealed that immunochemotherapy with an anti-PD-1 antibody was an independent prognostic factor for longer PFS in ENKTL patients (p<0.001) (59). What’s more, PD-L1 monoclonal antibodies sugemalimab demonstrate promising efficacy in relapsed/refractory ENKTL, with a 35.9% complete response rate and durable survival benefits, while maintaining a manageable safety profile compared to PD-1 inhibitors including sintilimab, pembrolizumab, nivolumab and tislelizumab (60). LMP, which is expressed in the tumor cells of EBV-associated malignancies, is a natural candidate target for cytotoxic T lymphocytes (61), and patients who received LMP-specific cytotoxic T cells achieved better PFS and OS and less toxicity (62, 63). Some new therapeutic targets explored by NGS have also shown excellent potential for the treatment of ENKTL. Daratumumab, a monoclonal antibody against CD38, and brentuximab vedotin, an antibody targeting CD30 conjugated to the cytotoxic agent monomethyl auristatin E, both showed promising activity in ENKTL patients (62, 64, 65). The efficacy of ruxolitinib, a JAK3-STAT pathway inhibitor, in peripheral T-cell lymphoma patients has been confirmed (66), while a clinical trial evaluating ruxolitinib in ENKTL patients is ongoing (NCT02974647) (6). Our WGS analysis confirmed that the STAT3 mutation may be a potential therapeutic target for this patient. The benefit of hematopoietic stem cell transplantation in patients with advanced-stage or relapsed disease has been demonstrated in several small, retrospective studies, but prospective, randomized clinical trials are needed (67–69). Moreover, several potential targets, such as EZH2, BCOR, MLL2, NF-kappaB, NOTCH, Bcl-xL, TET1/TET2, EP200, ASXL3, CREBBP and ARID1A, have also been investigated (69–71).
There were some limitations in our study. Given the rarity of ENKTL with adrenal involvement, the statistical power of our WGS findings may be constrained, sequencing studies with large samples may be helpful for systematically elucidating the genetic alterations of adrenal ENKTL to enhance the generalizability and reproducibility of our conclusions. What’s more, the cells and animal experiments are also required to perform to validate the role of genetic alterations in the ENKTL involved adrenal mechanisms. In the analyses of prognostic and clinicopathological factors, the results may have been affected by confounding factors due to the limited sample size, which restricts the ability to match ENKTL patients and ACC patients by propensity matching scores. Moreover, the limited information available for patients in the SEER database hindered comprehensive analysis of significant factors and the efficacy of immunotherapy and therapeutic targets. Studies with larger samples may be more helpful in comparing survival between ENKTL patients and ACC patients and in exploring risk factors affecting survival in ENKTL patients.
5 Conclusion
In conclusion, adrenal ENKTL is very rare, and this study described a patient with primary nasal tumors and secondary adrenal and multiple systemic metastases. Four mutation driver genes, STAT3, TET2, FAS and TP53, were identified using WES and Sanger sequencing, which may contribute to targeted therapy for this disease. According to our prognostic analysis, patients with adrenal ENKTL presented a significantly worse prognosis than patients with ACC, and administering chemotherapy improved the prognosis of adrenal ENKTL patients.
Data availability statement
To protect the privacy of the sampled individual, the SNP data presented in this article are not readily available. Requests to access the dataset should be directed to the corresponding author Bo Fan.
Ethics statement
The studies involving humans were approved by the Ethics Committee of Dalian Central Hospital (ethical approval number: YN2024-029-01) and the Second Affiliated Hospital of Dalian Medical University (ethical approval number: 2023273). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
LZ: Data curation, Formal analysis, Software, Writing – original draft. HaG: Validation, Writing – original draft. SM: Investigation, Writing – original draft. XF: Methodology, Software, Writing – review & editing. HuG: Methodology, Resources, Writing – review & editing. MS: Conceptualization, Data curation, Writing – review & editing. SW: Software, Supervision, Writing – review & editing. TL: Investigation, Supervision, Writing – review & editing. GY: Project administration, Writing – review & editing. XY: Project administration, Writing – review & editing. XS: Project administration, Writing – review & editing. BF: Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The present study was supported by the National Natural Science Foundation of China (NSFC32100928), the Joint Fund Project of Liaoning Provincial Science and Technology Programme (2023-MSLH-021), the Interdisciplinary Research Cooperation Project Team Funding of Dalian Medical University Youth-specific category of free exploration (JCHZ2023020), the United Foundation for Medico-engineering Cooperation from Dalian Neusoft University of Information and the Second Hospital of Dalian Medical University (LH-JSRZ-202201), “1+X” Program for Clinical Competency Enhancement–interdisciplinary Innovation Project, the Second Hospital of Dalian Medical University (2022JCXKYB15), the Young Reserve Talent Project of the Second Hospital of Dalian Medical University (Grant no. dy2yhbrc202010) and the Scientific Research Project of Ministry of Education of Liaoning Province (LJKQZ20222382), the Scientific Research Project of Ministry of Education of Liaoning Province (LJ212410161046), Open Fund Project of State Key Laboratory of Structural Analysis of Industrial Equipment, Dalian University of Technology (GZ23102), Chinese Medicine Scientific Research Program Project of Dalian Municipal Health Commission (23Z12002), Industry-University Cooperation Collaborative Education Program of Ministry of Education(231005073090218) and “1+X” Program for Upgrading Clinical Skills of Young Physicians and Nurses, the Second Affiliated Hospital of Dalian Medical University’s (2024LCJSYL06).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1566794/full#supplementary-material
Abbreviations
ENKTL, Extranodal natural killer/T-cell lymphoma; SEER, Surveillance, Epidemiology, and End Results; ACC, adrenocortical carcinoma; OS, overall survival; SNVs, single-nucleotide variants; INDELs, insertions and deletions; EBV, Epstein–Barr virus; CR, complete response; NGS, next-generation sequencing; EBER, EBV-encoded small RNA; ICD, International Classification of Disease; HR, hazard ratio; CI, confidence interval; SNP, single nucleotide polymorphism; CNV, copy number variation; SV, structural variation; CGC, Cancer Gene Census; SMG, significant mutated gene; PharmGKB, Pharmacogenomics Knowledge Base; FDA, Food and Drug Administration; KEGG, Kyoto Encyclopedia of Genes and Genomes; PCR, polymerase chain reaction; PPI, protein–protein interaction; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins; GO, Gene Ontology; CT, computed tomography; SUVmax, maximal standardized uptake value; TAM, tumor-associated macrophage; CPG, cancer predisposition gene; PFS, progression-free survival.
References
1. Tse E, Kwong YL. The diagnosis and management of NK/T-cell lymphomas. J Hematol Oncol. (2017) 10:85. doi: 10.1186/s13045-017-0452-9
2. Tian Z, Tian J, Liao J. NK/T-cell lymphoma with rash and peripheral neuropathy as the first manifestation: a case report and literature review. Diagn Pathol. (2023) 18:2. doi: 10.1186/s13000-023-01286-z
3. Tse E, Zhao WL, Xiong J, Kwong YL. How we treat NK/T-cell lymphomas. J Hematol Oncol. (2022) 15:74. doi: 10.1186/s13045-022-01293-5
4. Yang Y, Zhang YJ, Zhu Y, Cao JZ, Yuan ZY, Xu LM, et al. Prognostic nomogram for overall survival in previously untreated patients with extranodal NK/T-cell lymphoma, nasal-type: a multicenter study. Leukemia. (2015) 29:1571–7. doi: 10.1038/leu.2015.44
5. Kimura H. EBV in T-/NK-cell tumorigenesis. Adv Exp Med Biol. (2018) 1045:459–75. doi: 10.1007/978-981-10-7230-7_21
6. He X, Gao Y, Li Z, Huang H. Review on natural killer/T-cell lymphoma. Hematol Oncol. (2023) 41:221–9. doi: 10.1002/hon.v41.2
7. Au WY, Weisenburger DD, Intragumtornchai T, Nakamura S, Kim WS, Sng I, et al. Clinical differences between nasal and extranasal natural killer/T-cell lymphoma: a study of 136 cases from the International Peripheral T-Cell Lymphoma Project. Blood. (2009) 113:3931–7. doi: 10.1182/blood-2008-10-185256
8. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. (2010) 26:589–95. doi: 10.1093/bioinformatics/btp698
9. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. (2009) 25:2078–9. doi: 10.1093/bioinformatics/btp352
10. Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. (2012) 28:423–5. doi: 10.1093/bioinformatics/btr670
11. Layer RM, Chiang C, Quinlan AR, Hall IM. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. (2014) 15:R84. doi: 10.1186/gb-2014-15-6-r84
12. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. (2013) 31:213–9. doi: 10.1038/nbt.2514
13. Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. (2012) 28:1811–7. doi: 10.1093/bioinformatics/bts271
14. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. (2010) 38:e164. doi: 10.1093/nar/gkq603
15. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. (2013) 339:1546–58. doi: 10.1126/science.1235122
16. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, et al. Mutational landscape and significance across 12 major cancer types. Nature. (2013) 502:333–9. doi: 10.1038/nature12634
17. Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. (2013) 3:2650. doi: 10.1038/srep02650
18. Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. (2012) 30:413–21. doi: 10.1038/nbt.2203
19. Madaj R, Geoffrey B, Sanker A, Valluri PP. Target2DeNovoDrug: a novel programmatic tool for in silico-deep learning based de novo drug design for any target of interest. J Biomol Struct Dyn. (2022) 40:7511–6. doi: 10.1080/07391102.2021.1898474
20. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. (2000) 25:25–9. doi: 10.1038/75556
21. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. (2000) 28:27–30. doi: 10.1093/nar/28.1.27
22. Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer. (2007) 121:2381–6. doi: 10.1002/ijc.23192
23. Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Cell. (2005) 7:513–20. doi: 10.1016/j.ccr.2005.05.024
24. Komohara Y, Niino D, Ohnishi K, Ohshima K, Takeya M. Role of tumor-associated macrophages in hematological Malignancies. Pathol Int. (2015) 65:170–6. doi: 10.1111/pin.12259
25. Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel). (2014) 6:1670–90. doi: 10.3390/cancers6031670
26. Wang H, Li P, Wang L, Xia Z, Huang H, Lu Y, et al. High numbers of CD68+ tumor-associated macrophages correlate with poor prognosis in extranodal NK/T-cell lymphoma, nasal type. Ann Hematol. (2015) 94:1535–44. doi: 10.1007/s00277-015-2401-4
27. Gao Y, Li Y, Ma G, Zhao G, Liu H. KMT2D and TP53 mutation status improve the prognostic value of the International Prognostic Index (IPI) stratification in ENKTL patients. Neoplasma. (2020) 67:636–43. doi: 10.4149/neo_2020_190307N194
28. Wang J, Liu W, Zhang L, Zhang J. Targeting mutant p53 stabilization for cancer therapy. Front Pharmacol. (2023) 14:1215995. doi: 10.3389/fphar.2023.1215995
29. Hu Y, Chen X, Ling Y, Zhou K, Han M, Wang X, et al. Influenza A virus inhibits TET2 expression by endoribonuclease PA-X to attenuate type I interferon signaling and promote viral replication. PloS Pathog. (2023) 19:e1011550. doi: 10.1371/journal.ppat.1011550
30. Lazarenkov A, Sardina JL. Dissecting TET2 regulatory networks in blood differentiation and cancer. Cancers (Basel). (2022) 14:830. doi: 10.3390/cancers14030830
31. Arena A, Romeo MA, Benedetti R, Gilardini Montani MS, Santarelli R, Gonnella R, et al. NRF2 and STAT3: friends or foes in carcinogenesis. Discov Oncol. (2023) 14:37. doi: 10.1007/s12672-023-00644-z
32. Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. (2007) 357:1608–19. doi: 10.1056/NEJMoa073687
33. Pilati C, Amessou M, Bihl MP, Balabaud C, Nhieu JT, Paradis V, et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J Exp Med. (2011) 208:1359–66. doi: 10.1084/jem.20110283
34. Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. (2018) 15:234–48. doi: 10.1038/nrclinonc.2018.8
35. Xue C, Yao Q, Gu X, Shi Q, Yuan X, Chu Q, et al. Evolving cognition of the JAK-STAT signaling pathway: autoimmune disorders and cancer. Signal Transduct Target Ther. (2023) 8:204. doi: 10.1038/s41392-023-01468-7
36. Lee S, Park HY, Kang SY, Kim SJ, Hwang J, Lee S, et al. Genetic alterations of JAK/STAT cascade and histone modification in extranodal NK/T-cell lymphoma nasal type. Oncotarget. (2015) 6:17764–76. doi: 10.18632/oncotarget.3776
37. Koo GC, Tan SY, Tang T, Poon SL, Allen GE, Tan L, et al. Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov. (2012) 2:591–7. doi: 10.1158/2159-8290.CD-12-0028
38. Bouchekioua A, Scourzic L, de Wever O, Zhang Y, Cervera P, Aline-Fardin A, et al. JAK3 deregulation by activating mutations confers invasive growth advantage in extranodal nasal-type natural killer cell lymphoma. Leukemia. (2014) 28:338–48. doi: 10.1038/leu.2013.157
39. Liang D, Wang Q, Zhang W, Tang H, Song C, Yan Z, et al. JAK/STAT in leukemia: a clinical update. Mol Cancer. (2024) 23:25. doi: 10.1186/s12943-023-01929-1
40. Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, Sameshima M, et al. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell. (1991) 66:233–43. doi: 10.1016/0092-8674(91)90614-5
41. Devel L, Guedeney N, Bregant S, Chowdhury A, Jean M, Legembre P. Role of metalloproteases in the CD95 signaling pathways. Front Immunol. (2022) 13:1074099. doi: 10.3389/fimmu.2022.1074099
42. Takakuwa T, Dong Z, Nakatsuka S, Kojya S, Harabuchi Y, Yang WI, et al. Frequent mutations of Fas gene in nasal NK/T cell lymphoma. Oncogene. (2002) 21:4702–5. doi: 10.1038/sj.onc.1205571
43. Yim J, Koh J, Kim S, Song SG, Bae JM, Yun H, et al. Clinicopathologic and genetic features of primary T-cell lymphomas of the central nervous system: an analysis of 11 cases using targeted gene sequencing. Am J Surg Pathol. (2022) 46:486–97. doi: 10.1097/PAS.0000000000001859
44. Jeon YK, Kim H, Park SO, Choi HY, Kim YA, Park SS, et al. Resistance to Fas-mediated apoptosis is restored by cycloheximide through the downregulation of cellular FLIPL in NK/T-cell lymphoma. Lab Invest. (2005) 85:874–84. doi: 10.1038/labinvest.3700291
45. Kataoka K, Miyoshi H, Sakata S, Hwang SY, Jeong H, Choi JR, et al. Frequent structural variations involving programmed death ligands in Epstein-Barr virus-associated lymphomas. Leukemia. (2019) 33:1687–99. doi: 10.1038/s41375-019-0380-5
46. Kim JJ, Kim HY, Choi Z, Hwang SY, Jeong H, Choi JR, et al. In-depth circulating tumor DNA sequencing for prognostication and monitoring in natural killer/T-cell lymphomas. Front Oncol. (2023) 13:1109715. doi: 10.3389/fonc.2023.1109715
47. Dobashi A, Tsuyama N, Asaka R, Togashi Y, Ueda K, Sakata S, et al. Frequent BCOR aberrations in extranodal NK/T-Cell lymphoma, nasal type. Genes Chromosomes Cancer. (2016) 55:460–71. doi: 10.1002/gcc.22348
48. Ramsey MC, Sabatini P, Watson G, Chawla T, Ko M, Sakhdari A. Case Report: Identification of a novel STAT3 mutation in EBV-positive inflammatory follicular dendritic cell sarcoma. Front Oncol. (2023) 13:1266897. doi: 10.3389/fonc.2023.1266897
49. Chapman JR, Bouska AC, Zhang W, Alderuccio JP, Lossos IS, Rimsza LM, et al. EBV-positive HIV-associated diffuse large B cell lymphomas are characterized by JAK/STAT (STAT3) pathway mutations and unique clinicopathologic features. Br J Haematol. (2021) 194:870–8. doi: 10.1111/bjh.v194.5
50. Gao LM, Zhao S, Zhang WY, Wang M, Li HF, Lizaso A, et al. Somatic mutations in KMT2D and TET2 associated with worse prognosis in Epstein-Barr virus-associated T or natural killer-cell lymphoproliferative disorders. Cancer Biol Ther. (2019) 20:1319–27. doi: 10.1080/15384047.2019.1638670
51. Bi XW, Jiang WQ, Zhang WW, Huang JJ, Xia Y, Wang Y, et al. Treatment outcome of patients with advanced stage natural killer/T-cell lymphoma: elucidating the effects of asparaginase and postchemotherapeutic radiotherapy. Ann Hematol. (2015) 94:1175–84. doi: 10.1007/s00277-015-2336-9
52. Jaccard A, Gachard N, Marin B, Rogez S, Audrain M, Suarez F, et al. Efficacy of L-asparaginase with methotrexate and dexamethasone (AspaMetDex regimen) in patients with refractory or relapsing extranodal NK/T-cell lymphoma, a phase 2 study. Blood. (2011) 117:1834–9. doi: 10.1182/blood-2010-09-307454
53. Li X, Cui Y, Sun Z, Zhang L, Li L, Wang X, et al. DDGP versus SMILE in newly diagnosed advanced natural killer/T-cell lymphoma: A randomized controlled, multicenter, open-label study in China. Clin Cancer Res. (2016) 22:5223–8. doi: 10.1158/1078-0432.CCR-16-0153
54. Wang JJ, Dong M, He XH, Li YX, Wang WH, Liu P, et al. GDP (Gemcitabine, dexamethasone, and cisplatin) is highly effective and well-tolerated for newly diagnosed stage IV and relapsed/refractory extranodal natural killer/T-cell lymphoma, nasal type. Med (Baltimore). (2016) 95:e2787. doi: 10.1097/MD.0000000000002787
55. Li X, Cheng Y, Zhang M, Yan J, Li L, Fu X, et al. Activity of pembrolizumab in relapsed/refractory NK/T-cell lymphoma. J Hematol Oncol. (2018) 11:15. doi: 10.1186/s13045-018-0559-7
56. Kwong YL, Chan T, Tan D, Kim SJ, Poon LM, Mow B, et al. PD1 blockade with pembrolizumab is highly effective in relapsed or refractory NK/T-cell lymphoma failing l-asparaginase. Blood. (2017) 129:2437–42. doi: 10.1182/blood-2016-12-756841
57. Tian XP, Cai J, Xia Y, Zhang YC, Wang L, Liu PP, et al. First-line sintilimab with pegaspargase, gemcitabine, and oxaliplatin in advanced extranodal natural killer/T cell lymphoma (SPIRIT): a multicentre, single-arm, phase 2 trial. Lancet Haematol. (2024) 11:e336–336e344. doi: 10.1016/S2352-3026(24)00066-8
58. Cai J, Liu P, Huang H, Li Y, Ma S, Zhou H, et al. Combination of anti-PD-1 antibody with P-GEMOX as a potentially effective immunochemotherapy for advanced natural killer/T cell lymphoma. Signal Transduct Target Ther. (2020) 5:289. doi: 10.1038/s41392-020-00331-3
59. Wang X, Wen L, Liao J, Feng Y, Li Y, Zhou Z, et al. First-line immunotherapy with anti-PD-1 antibody for extranodal NK/T-cell lymphoma: A retrospective study. Br J Haematol. (2023) 202:812–24. doi: 10.1111/bjh.18957
60. Feng Y, Liu X, Yu J, Song Z, Li L, Qiu L, et al. An evaluation of sugemalimab for the treatment of relapsed or refractory extranodal natural killer T-cell lymphoma. Expert Opin Biol Ther. (2025) 25:9–14. doi: 10.1080/14712598.2024.2444400
61. Yamaguchi M, Suzuki R, Oguchi M. Advances in the treatment of extranodal NK/T-cell lymphoma, nasal type. Blood. (2018) 131:2528–40. doi: 10.1182/blood-2017-12-791418
62. Bollard CM, Gottschalk S, Torrano V, Diouf O, Ku S, Hazrat Y, et al. Sustained complete responses in patients with lymphoma receiving autologous cytotoxic T lymphocytes targeting Epstein-Barr virus latent membrane proteins. J Clin Oncol. (2014) 32:798–808. doi: 10.1200/JCO.2013.51.5304
63. Cho SG, Kim N, Sohn HJ, Lee SK, Oh ST, Lee HJ, et al. Long-term outcome of extranodal NK/T cell lymphoma patients treated with postremission therapy using EBV LMP1 and LMP2a-specific CTLs. Mol Ther. (2015) 23:1401–9. doi: 10.1038/mt.2015.91
64. Hari P, Raj RV, Olteanu H. Targeting CD38 in refractory extranodal natural killer cell-T-cell lymphoma. N Engl J Med. (2016) 375:1501–2. doi: 10.1056/NEJMc1605684
65. Piedra-Quintero ZL, Wilson Z, Nava P, Guerau-de-Arellano M. CD38: an immunomodulatory molecule in inflammation and autoimmunity. Front Immunol. (2020) 11:597959. doi: 10.3389/fimmu.2020.597959
66. Moskowitz AJ, Ghione P, Jacobsen E, Ruan J, Schatz JH, Noor S, et al. A phase 2 biomarker-driven study of ruxolitinib demonstrates effectiveness of JAK/STAT targeting in T-cell lymphomas. Blood. (2021) 138:2828–37. doi: 10.1182/blood.2021013379
67. Kim SJ, Park S, Kang ES, Choi JY, Lim DH, Ko YH, et al. Induction treatment with SMILE and consolidation with autologous stem cell transplantation for newly diagnosed stage IV extranodal natural killer/T-cell lymphoma patients. Ann Hematol. (2015) 94:71–8. doi: 10.1007/s00277-014-2171-4
68. Yhim HY, Kim JS, Mun YC, Moon JH, Chae YS, Park Y, et al. Clinical outcomes and prognostic factors of up-front autologous stem cell transplantation in patients with extranodal natural killer/T cell lymphoma. Biol Blood Marrow Transplant. (2015) 21:1597–604. doi: 10.1016/j.bbmt.2015.05.003
69. Fox CP, Boumendil A, Schmitz N, Finel H, Luan JJ, Sucak G, et al. High-dose therapy and autologous stem cell transplantation for extra-nodal NK/T lymphoma in patients from the Western hemisphere: a study from the European Society for Blood and Marrow Transplantation. Leuk Lymphoma. (2015) 56:3295–300. doi: 10.3109/10428194.2015.1037764
70. Liu C, Ding X, Li G, Zhang Y, Shao Y, Liu L, et al. Targeting Bcl-xL is a potential therapeutic strategy for extranodal NK/T cell lymphoma. iScience. (2023) 26:107369. doi: 10.1016/j.isci.2023.107369
Keywords: extranodal natural killer/T-cell lymphoma, adrenal, whole-genome sequencing, gene mutations, population-based study
Citation: Zhang L, Gao H, Ma S, Fan X, Guo H, Sun M, Wen S, Liu T, Yu G, Yuan X, Sun X and Fan B (2025) Functional screening of somatic mutant events in extranodal natural killer/T-cell lymphoma with adrenal involvement. Front. Immunol. 16:1566794. doi: 10.3389/fimmu.2025.1566794
Received: 25 January 2025; Accepted: 22 April 2025;
Published: 13 May 2025.
Edited by:
Lan Wu, Vanderbilt University Medical Center, United StatesReviewed by:
Hua Wang, Sun Yat-sen University Cancer Center (SYSUCC), ChinaWeiping Liu, Peking University, China
Copyright © 2025 Zhang, Gao, Ma, Fan, Guo, Sun, Wen, Liu, Yu, Yuan, Sun and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Fan, ZmFuYm9AZG11LmVkdS5jbg==; Xiuhua Sun, MzAzODY2OEB2aXAuc2luYS5jb20=; Xiaoying Yuan, eWluZzE5ODdtZWR1QDE2My5jb20=; Guanghai Yu, ZGx6eHlnaEAxMjYuY29t
†These authors have contributed equally to this work
 Luxin Zhang
Luxin Zhang Haifeng Gao2†
Haifeng Gao2† Man Sun
Man Sun Xiaoying Yuan
Xiaoying Yuan Bo Fan
Bo Fan