- 1College of Food and Bioengineering, Zhengzhou University of Light Industry, Zhengzhou, China
- 2Henan Key Laboratory of Cold Chain Food Quality and Safety Control, Zhengzhou University of Light Industry, Zhengzhou, China
The Notch signaling pathway and non-coding RNAs (ncRNAs) play significant roles in regulating key cellular processes such as cancer progression, metastasis, and drug resistance. This article systematically reviews the interactions between the Notch pathway and ncRNAs including miRNAs, lncRNAs, and circRNAs, as well as their overall impact on cancer biology. We focus on the latest research progress on how ncRNAs regulate the Notch pathway through transcriptional regulation, post-transcriptional modifications, and epigenetic mechanisms, and discuss how such interactions affect tumor microenvironment shaping, immune escape mechanisms, and treatment sensitivity. Additionally, this article deeply analyzes potential therapeutic strategies targeting the Notch-ncRNA axis, including its prospects for synergistic application with chemotherapy, radiotherapy, and immunotherapy. By integrating multi-cancer experimental data, we propose individualized diagnosis and treatment strategies based on tumor-specific Notch pathway and ncRNA expression patterns.
1 Introduction
The Notch signaling pathway is an essential cellular communication mechanism involved in various physiological processes, including embryonic development, immune regulation, and cell fate determination. Initiated by ligand-receptor interactions (e.g., Jagged/Delta ligands binding to Notch1–4 receptors), this pathway triggers γ-secretase-mediated cleavage of the receptor, releasing the Notch intracellular domain (NICD). NICD translocates to the nucleus to activate transcription of target genes such as Hes and Hey, thereby regulating cell proliferation, differentiation, migration, and apoptosis (1).
Emerging evidence highlights the critical role of crosstalk between Notch signaling and non-coding RNAs (ncRNAs), including miRNAs, lncRNAs, and circRNAs, in shaping tumor plasticity and therapeutic resistance. For instance, studies have shown that miR-200 family members can inhibit Notch1 expression by targeting its mRNA, thereby reducing the invasiveness of breast cancer cells (2). Conversely, lncRNA PANDA has been identified to enhance Notch signaling activity in gastric cancer by interacting with the chromatin-modifying complex Polycomb repressive complex 2 (PRC2), promoting tumor growth and metastasis (3).
Moreover, circRNAs have demonstrated their ability to regulate Notch signaling through competing endogenous RNA (ceRNA) mechanisms. For example, circZMIZ1 sponges miR-204, which otherwise inhibits the expression of Notch downstream targets, leading to increased cell proliferation and decreased apoptosis in colorectal cancer cells (4). These bidirectional interactions between Notch signaling and ncRNAs create intricate regulatory networks that significantly influence tumor heterogeneity, immune evasion, and therapeutic resistance.
Deciphering these molecular dialogues holds immense potential for developing novel combinatorial therapies. For instance, RNA-based Notch modulators could be coupled with immune checkpoint inhibitors to enhance the efficacy of cancer treatments while addressing the pathway’s paradoxical roles across different cancer subtypes. By integrating insights from ncRNA-Notch crosstalk into precision oncology strategies, researchers can pave the way for more effective and personalized therapeutic approaches.
In summary, the interplay between Notch signaling and ncRNAs represents a promising avenue for understanding tumor biology and developing innovative treatments. With ongoing advancements in RNA sequencing technologies and functional genomics, further exploration of these interactions will undoubtedly yield valuable insights into cancer pathogenesis and therapeutic resistance mechanisms.
1.1 The discovery of the notch signaling pathway
Notch signaling was first discovered in fruit flies (5). In the early 20th century, scientists observed mutants in fruit flies and identified a gene associated with wing defects, which they named “Notch” (6). Later,In 1987, molecular biology studies revealed that the Notch gene encodes a cell surface receptor that participates in cell-to-cell interactions and regulates cell fate determination (7).
In mammals, the Notch signaling pathway consists of four receptors (Notch1-4) and five ligands (Jagged1, Jagged2, Delta-like 1, Delta-like 3, Delta-like 4) (8–12). When a ligand binds to the receptor, the Notch receptor undergoes a conformational change.Itis then cleaved by proteases such as γ-secretase, releases the Notch intracellular domain (NICD) (1, 10–13). NICD enters the cell nucleus and regulates the transcription of target genes, influencing processes like cell proliferation, differentiation, and migration (14). The Notch signaling plays a crucial role in embryonic development, stem cell maintenance, and immune response (Figure 1).
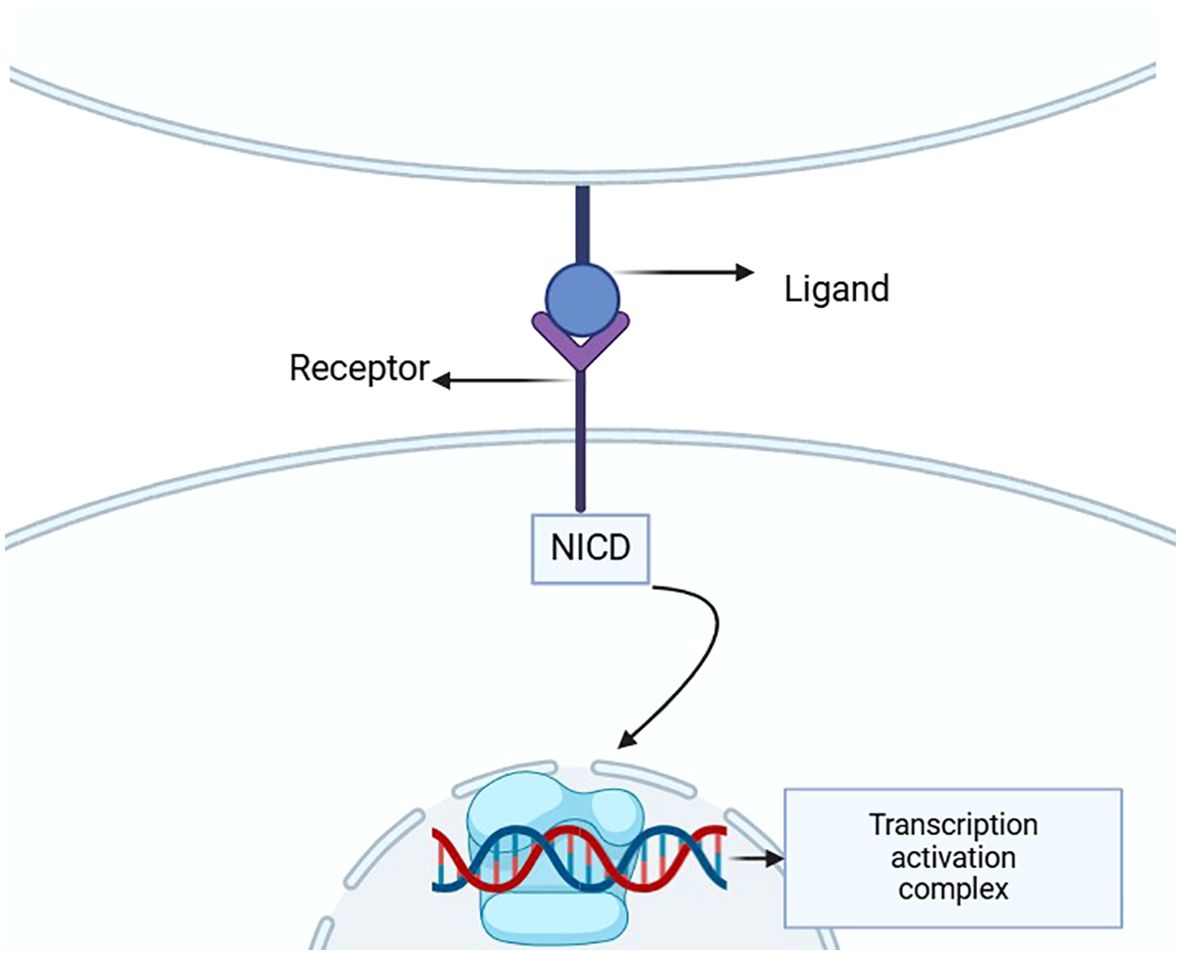
Figure 1. This diagram illustrates the structure of the Notch signaling pathway, highlighting the interaction between the ligand and receptor that triggers the release of NICD. This intracellular domain then translocates to the nucleus, where it activates gene transcription through the transcription activation complex, underscoring its crucial role in developmental processes and disease pathology.
1.2 Introduction to non-coding RNAs
ncRNAs are RNA molecules that do not encode proteins but play essential roles in gene expression regulation, cell function, and disease development (15, 16). They are categorized into small ncRNAs (e.g., miRNAs) and long ncRNAs (e.g., lncRNAs, circRNAs), which regulate Notch signaling through epigenetic modifications (e.g., DNA methylation, histone acetylation), transcriptional interference, or post-transcriptional sponging (17), thereby establishing ncRNAs as critical regulators of the Notch pathway.
1.2.1 miRNA
MicroRNAs are small RNA molecules about 20–24 nucleotides in length. They regulate mRNA stability and translation by binding to the 3’ untranslated region (UTR) of target mRNAs (18). miRNAs play a key role in cell proliferation, differentiation, and tumorigenesis. For example, miR-34a directly targets Notch1 mRNA, suppressing its expression and inhibiting breast cancer stem cell self-renewal.
1.2.2 lncRNA
Long non-coding RNAs are RNA molecules longer than 200 nucleotides that regulate gene transcription and translation, and participate in processes such as tumorigenesis and immune regulation (1). LncRNAs like HOTAIR and MALAT1 interact with the Notch signaling pathway to promote tumor cell proliferation, metastasis, and drug resistance (19).
1.2.3 circRNA
Circular RNAs are stable, covalently closed RNA molecules that act as “sponges” for miRNAs, regulating their inhibitory effect on target genes (20). circHIPK3, for instance, sponges miR-124 to enhance Notch1 signaling in hepatocellular carcinoma.
1.3 Interaction between notch signaling and non-coding RNAs
The reciprocal regulation between Notch signaling and ncRNAs constitutes a self-reinforcing oncogenic circuit, where epigenetic reprogramming and post-transcriptional control converge to drive disease progression. The complex bidirectional crosstalk between Notch signaling and ncRNAs operates through three-tiered regulatory networks: miRNAs (e.g., miR-34a, miR-7) post-transcriptionally degrade Notch receptors/ligands, lncRNAs (e.g., MALAT1, HOTAIR) scaffold epigenetic modifiers to reshape chromatin accessibility, and circRNAs (e.g., CDR1as) sequester miRNAs or directly bind pathway effectors (21). These multilayered controls collectively amplify Notch-driven pathologies, as exemplified by their stage-specific dysregulation in tumor progression and inflammatory cascades. This regulatory hierarchy establishes ncRNAs as molecular rheostats that fine-tune Notch signaling dynamics, creating therapeutic vulnerabilities discussed in later sections.Below we systematically dissect how each RNA subclass mechanistically hijacks Notch signaling.
1.3.1 miRNAs and notch signaling
As the most studied ncRNA subclass, miRNAs provide rapid feedback control of Notch pathway components.Many miRNAs regulate the activity of Notch signaling by targeting Notch receptors, ligands, or downstream genes. For example, miR-34a acts as a tumor-suppressive miRNA by inhibiting Notch1 expression, thus suppressing tumor cell proliferation and metastasis (22). Notably, miRNA-mediated Notch inhibition often synergizes with chemotherapeutic agents, a concept further explored in Section 6.2.
1.3.2 lncRNAs and notch signaling
LncRNAs orchestrate spatial-temporal control of Notch signaling through chromatin remodeling and protein complex assembly. By interacting with key factors in the Notch signaling pathway (such as NICD)lncRNAs like HOTAIR (HOX transcript antisense RNA) and MALAT1 (metastasis-associated long non-coding RNA transcript 1) promote tumor progression through three-dimensional genomic reorganization (23). Their scaffold function makes lncRNAs ideal targets for epigenetic drugs, as detailed in our therapeutic strategy analysis (Section 6.1-6.3).
1.3.3 circRNAs and notch signaling
CircRNAs establish persistent oncogenic loops by sequestering Notch-regulatory miRNAs. CircRNAs influence signal transduction by sequestering miRNAs or directly regulating key molecules in the Notch signaling pathway, contributing to cancer onset and metastasis.Such as circCDR1as sponges miR-7 to upregulate Notch1, promoting angiogenesis in glioblastoma.The closed-loop structure of circRNAs confers exceptional stability in bodily fluids, a property leveraged in liquid biopsy approaches discussed in Section 6.4.
1.4 The role of notch signaling and ncRNAs in cancer and inflammation
Notch signaling and ncRNAs play crucial roles in various diseases, particularly in cancer and inflammatory disorders. Aberrant activation or inhibition of Notch signaling promotes tumor initiation, progression, metastasis, and the development of drug resistance (24). NcRNAs regulate the activity of the Notch pathway, further influencing tumor cell proliferation, metastasis, and immune evasion.
In inflammatory diseases, Notch signaling participates in immune response regulation by modulating immune cell functions. NcRNAs, in turn, regulate immune cell activity, impacting the onset and progression of inflammation. The interplay between Notch signaling and ncRNAs provides new directions and potential targets for treating cancer and inflammatory diseases.
The interactions between Notch signaling and ncRNAs in cancer and inflammation offer fresh perspectives for disease research. A deeper understanding of their interplay is expected to provide new targets and strategies for precision therapy. As the mechanisms of Notch signaling and ncRNAs are further explored, future therapeutic approaches with higher efficacy and specificity may emerge, significantly improving clinical outcomes for cancer and inflammatory disorders.
2 The role of notch signaling pathway in cancer
Previous studies have elucidated the interaction mechanisms between the Notch pathway and ncRNAs, highlighting their bidirectional regulatory roles in disease pathogenesis through synergistic or antagonistic effects on cellular fate and pathological progression. Notably, in cancer, aberrant Notch activation/silencing drives malignant transformation by reprogramming tumor proliferation and remodeling the microenvironment, while ncRNAs act as epigenetic regulators to modulate Notch signaling outputs. The following section will focus on the multidimensional roles of Notch in cancer, delineating the molecular networks underlying tumor initiation, stemness maintenance, metastasis, and therapy resistance. Additionally, therapeutic strategies targeting the Notch-ncRNA axis will be evaluated to advance precision oncology.
The role of the Notch signaling pathway in cancer is complex and diverse, as it can both promote tumorigenesis and progression while also inhibit tumor formationThis dual functionality depends on various factors, including cell type, tumor microenvironment, and the specific activation of the signaling pathway (25). Aberrant regulation of the Notch signaling pathway has been shown to play a critical role in cancer occurrence, metastasis, drug resistance, and immune escape across multiple types of malignancies. This section will review the mechanisms by which Notch signaling operates in different tumor contexts, including its role in maintaining tumor stem cells, facilitating tumor metastasis, and contributing to immune evasion.
2.1 Notch signaling and tumorigenesis
The abnormal activation or inhibition of the Notch signaling pathway plays a critical role in the onset and progression of various cancers. Notch receptors interact with their ligands, such as Jagged or Delta, to initiate downstream signaling cascades. Subsequently,theintracellular domain of Notch (NICD) translocates into the nucleus where it eitheractivates or represses the transcription of specific target genes (26). Under normal physiological conditions, the Notch signaling pathway mainly regulates cell differentiation, proliferation, and apoptosis. However, in tumor cells, abnormal activation or inhibition of Notch signaling can alter cell fate, thus promoting tumorigenesis and progression (27).
For example, in breast cancer, abnormal activation of Notch1 suppresses the expression of differentiation-related genes (such as GATA3) and drives the upregulation of stem cell-related genes (such as SOX2 and OCT4), This sustains the undifferentiated state of tumor cells, enhances their proliferative capacity, and increasing drug resistance (28, 29). Furthermore, activation not only inhibits the expression of key transcription factors involved in differentiation but also induces the transformation of tumor cells into a cancer stem cell phenotype by upregulating stem cell marker genes like SOX2 and OCT4. This increases the tumor’s self-renewal potential (30, 31). These modifications significantly accelerate tumor initiation and progression, particularly fostering metastasis and recurrence. Additionally, Notch1 activation stimulates tumor cell growth and enhances survival by regulating the expression of cyclins (such as Cyclin D1) and anti-apoptotic proteins (such as Bcl-2) (32).
Although activation of the Notch signaling pathway typically promotes tumor cell proliferation and preserves stem cell characteristics, inhibition of Notch signaling may also induce oncogenic effects in some tumor types. For instance, in pancreatic cancer, inhibition of Notch signaling can suppress tumor cell proliferation and promote differentiation (33). Moreover, in some liver cancers, the inhibition of Notch signaling may result in malignant transformation and tumor progression. This suggests that loss or inhibition of Notch signaling may promote tumor cells to transform into a more aggressive and metastatic phenotype by altering their fate and the tumor microenvironment (34). In these cases, inhibition of Notch signaling may trigger the reprogramming of multiple cellular pathways, causing tumor cells to deviate from normal differentiation pathways, resulting in enhanced proliferation, migration, and drug resistance. Meanwhile, the role of different components of the Notch pathway in the tumor microenvironment also shows significant variability. For example, in glioblastoma, inhibition of Notch signaling helps reduce tumor cell survival, promotes differentiation, and suppresses tumor growth (35). Consequently, the regulation of Notch signaling in different tumor types is influenced not only by the intrinsic characteristics of the cells but also by their interactions with other signaling pathways within the tumor microenvironment.
2.2 Notch signaling and cancer stem cells
Cancer stem cells (CSCs) are a special class of cells within tumors that possess self-renewal, differentiation potential, and resistance to therapy. They are the main source of tumor recurrence and metastasis. The Notch signaling pathway plays a key role in maintaining and supporting CSC self-renewal (36). Research has shown that activation of Notch signaling significantly enhances CSC properties, keeping them in an undifferentiated state and providing continuous growth momentum for the tumor (37).
In various types of cancer, including breast cancer, colon cancer, and glioma, abnormal activation or overexpression of Notch signaling is considered a critical factor in maintaining CSC characteristics, promoting tumor growth and conferring therapeutic resistance (38). The Notch pathway, through its classic ligand-receptor interactions (e.g., binding of Notch receptors to Jagged or Delta ligands), activates downstream transcription factors such as Hes1, Hes5, and Hey1, regulating a range of genes related to stem cell fate determination (39). Notch signaling enhances CSC self-renewal and proliferative potential by upregulating stem cell marker genes such as Oct4, Sox2, and Nanog. These genes play essential roles in maintaining the undifferentiated state of stem cells and their proliferation and resistance to therapy (40). Furthermore, Notch signaling may influence cell cycle regulators, anti-apoptotic genes, and DNA repair mechanisms, further enhancing the proliferation capacity of CSCs and their resistance to treatment.
Excessive activation of Notch signaling intersects with other signaling pathways, such as Wnt, Hedgehog, and PI3K/Akt, forming a complex regulatory network that further promotes the survival and functional maintenance of CSCs (41, 42). These signaling pathways act synergistically to enable CSCs to maintain immune escape, proliferation, and drug resistance in the tumor microenvironment, supporting sustained tumor growth.
Therefore, the Notch signaling pathway is a potential therapeutic target for cancer stem cells. Inhibiting Notch signaling activation may help reduce the number of CSCs, thereby slowing tumor recurrence, metastasis, and resistance. Targeting the Notch pathway not only offers a potential strategy for eliminating CSCs but also provides new solutions for addressing drug resistance in cancer treatment.
2.3 Notch signaling and tumor metastasis
Tumor metastasis is one of the leading causes of cancer-related death, and the Notch signaling pathway plays a crucial role in this process. Studies have shown that Notch signaling promotes tumor metastasis by regulating tumor cell migration, invasiveness, and the epithelial-mesenchymal transition (EMT) process (43). EMT is a key step for tumor cells to acquire invasiveness and migratory ability. Notch signaling activates a series of transcription factors such as Snail, Slug, and Twist, triggering EMT, which drives tumor cells to transform into a more invasive and metastatic phenotype (44).
In several types of cancer, including lung, gastric, and pancreatic cancer, activation of Notch signaling has been shown to significantly promote the EMT process, enhancing tumor cell migration and invasiveness (45, 46). Notch signaling, by receptor-ligand binding, activates downstream transcription factors that regulate the expression of genes associated with EMT, such as Snail, Slug, and Twist. The upregulation of these transcription factors leads to the loss of cell polarity, disruption of intercellular junctions, and cytoskeletal remodeling, thereby enabling tumor cells to acquire invasive and migratory properties (47). This process allows tumor cells to break through the boundary of the primary tumor, spread to surrounding tissues, and eventually form metastatic lesions.
Additionally, Notch signaling regulates components of the extracellular matrix (ECM) in the tumor microenvironment, further enhancing tumor cell invasive abilities. Activation of Notch signaling may alter the structure and composition of the ECM, regulating the interaction between tumor cells and the matrix, which in turn enhances tumor cell adhesion, migration, and invasion abilities (48, 49). The close interaction between tumor cells and the matrix is crucial for metastasis; Notch signaling promotes this interaction process, aiding tumor cells in breaking away from the primary tumor and entering the bloodstream or lymphatic system, ultimately leading to distant metastasis (50).
Therefore, inhibiting Notch signaling may weaken tumor cells’ migration and invasion abilities by blocking the EMT process, thus effectively reducing the formation of cancer metastasis. Targeting Notch signaling is expected to provide new therapeutic approaches for the prevention and treatment of cancer metastasis.
2.4 Notch signaling and tumor immune escape
The ability of tumor cells to escape immune system surveillance is one of the key factors contributing to tumor progression, metastasis, and the development of drug resistance (51). Increasing evidence shows that Notch signaling plays an important role in tumor immune escape (52). Notch signaling regulates immune cell function, alters immune responses in the tumor microenvironment, and facilitates immune escape of tumor cells (53, 54).
An important mechanism by which Notch signaling promotes tumor immune escape is by inducing the polarization of tumor-associated macrophages (TAMs) into the M2 phenotype (55). M2 macrophages have strong immunosuppressive effects and can inhibit T cell activation, thereby providing an immune escape advantage for tumor cells. Studies have shown that activation of Notch signaling drives TAM polarization into the M2 phenotype, creating an immunosuppressive microenvironment that enhances tumor cell growth and metastasis (56). This process weakens anti-tumor T cell responses and promotes immune escape of tumor cells.
In addition to regulating immune cells, Notch signaling also promotes immune escape by modulating the expression of immune checkpoint moleculessuch as PD-L1, on the surface of tumor cells (57). Research has shown that activation of Notch signaling significantly upregulates PD-L1 expression in tumors like breast cancer, lung cancer, and melanoma (58). PD-L1 binds to the PD-1 receptor on T cells, inhibiting T-cell immune responses and reducing tumor immune surveillance (59). Through this mechanism, tumor cells can effectively escape recognition and elimination by the host immune system.
Therefore, targeting the Notch signaling pathway may offer novel strategies for enhancing immune system surveillance of tumors and improving immunotherapy outcomes. By inhibiting abnormal Notch signaling activation, it is possible to reduce immunosuppressive elements in the tumor microenvironment, enhance immune cell recognition and cytotoxicity toward tumor cells, and ultimately improve therapeutic efficacy.
2.5 Notch signaling and tumor drug resistance
Tumor cell resistance to therapy is a major challenge in clinical cancer treatment. The abnormal activation of the Notch signaling pathway has been found to be closely related to tumor drug resistance (60). Studies show that Notch signaling enhances tumor cell resistance to chemotherapy and targeted therapy by regulating mechanisms such as cell proliferation, apoptosis, and DNA repair (61). For example, activation of Notch1 can inhibit apoptosis by upregulating anti-apoptotic proteins (such as Bcl-2 and Survivin), thereby increasing tumor cell survival (62). Furthermore, Notch signaling enhance tumor resistance by promoting the proliferation and self-renewal of cancer stem cells.
In cancers such as breast cancer, pancreatic cancer, and non-small-cell lung cancer, abnormal activation of Notch signaling is closely associated with chemotherapy resistance. Therefore, inhibiting Notch signaling or targeting downstream molecules associated with the Notch pathway could become a new strategy to overcome tumor drug resistance.
In summary, the role of Notch signaling in cancer is complex and multifaceted. It directly influences tumor initiation and progression by regulating various aspects of tumor cell behavior, including proliferation, differentiation, migration, immune escape, and drug resistance. As our understanding of the mechanisms underlying Notch signaling deepens, targeting the Notch pathway is expected to offer new therapeutic approaches for cancer treatment. Future research should further explore the interactions between Notch signaling and other cellular pathways to develop treatment strategies with greater clinical application potential.
3 The role of the notch signaling pathway in inflammation
The Notch pathway’s dual regulatory capacity in cancer - governing stemness maintenance and immune evasion - mirrors its pivotal role in inflammatory pathogenesis. Crucially, Notch-mediated control of macrophage polarization and T-cell differentiation, central to creating immunosuppressive tumor microenvironments, is evolutionarily repurposed to drive chronic inflammation through analogous immune cell reprogramming. This functional conservation establishes Notch as a molecular linchpin connecting oncogenic and inflammatory processes through shared mechanisms of cellular fate determination.
The Notch signaling pathway plays a critical role not only in cell development and tumor progression but also in immune regulation and inflammatory responses. As a highly conserved intercellular communication pathway, Notch signaling regulates the differentiation, function, and activity of immune cells through the interactions between its receptors and ligands, thereby influencing the onset and progression of inflammatory responses (Figure 2) (63).
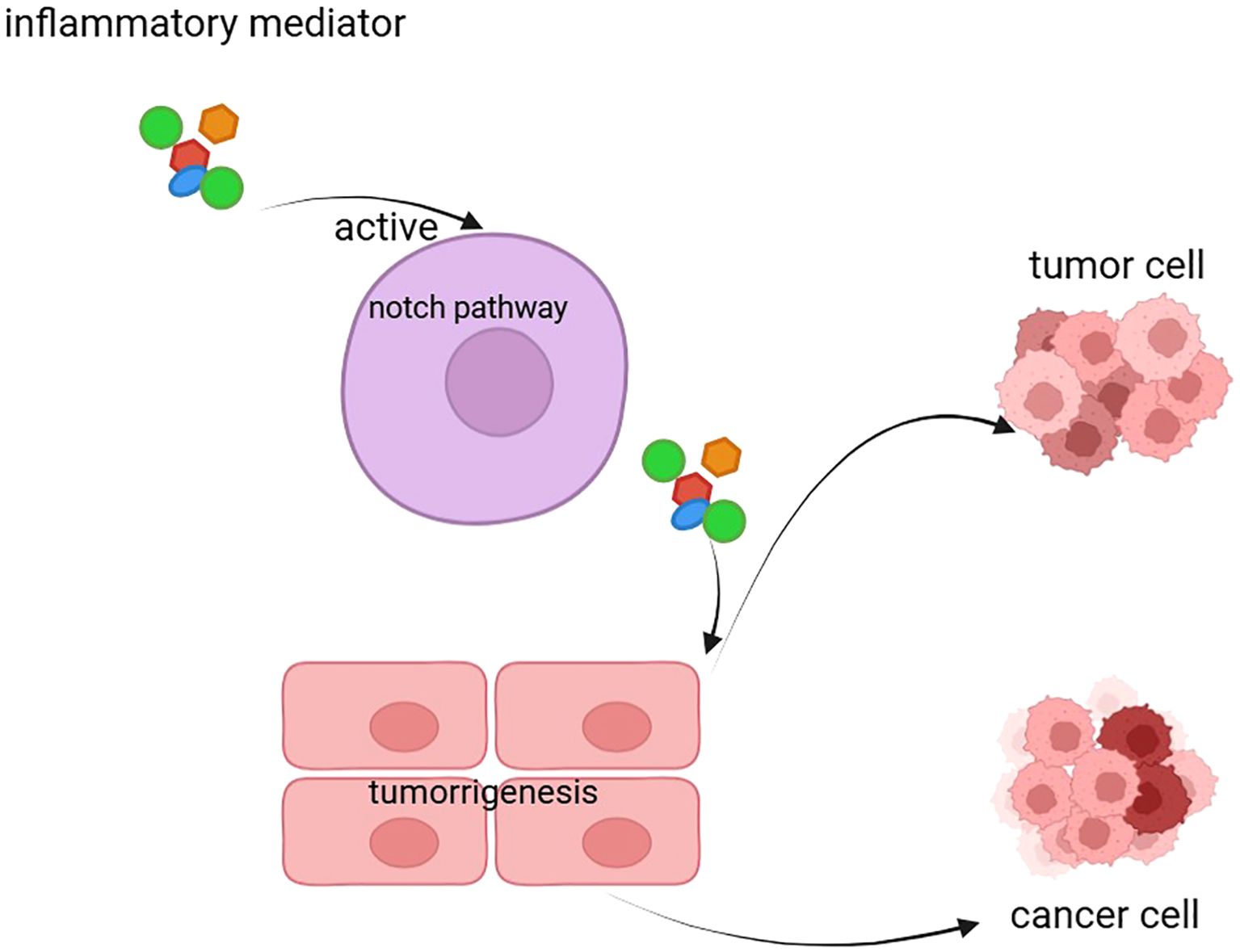
Figure 2. The figure illustrates the inflammation-driven Notch signaling pathway that leads to tumorigenesis and cancer. Inflammatory mediators activate the Notch pathway, which promotes tumor cell proliferation and transformation into cancer cells. This process highlights the role of inflammation in driving the progression from normal cells to malignant states through the Notch signaling mechanism.
Recent studies have demonstrated that the Notch signaling pathway plays pivotal roles in both chronic inflammatory diseases (such as rheumatoid arthritis and inflammatory bowel disease) and acute inflammatory responses (64). This article will explore in detail the role of Notch signaling in inflammation and its potential therapeutic implications.
3.1 The role of the notch signaling pathway in immune cells
The Notch signaling pathway regulates immune responses in various immune cells, with its function depending on the cell type and microenvironment (27). In immune cells such as T cells, B cells, macrophages, and dendritic cells, the activation or inhibition of the Notch pathway directly influences their function and differentiation processes.
3.1.1 Notch signaling in T cells
Notch signaling plays a crucial role in T cell development and function. During T cell development, the Notch pathway regulates the fate of T cell precursors in the thymus, promoting their differentiation into different subsets, such as CD4+ and CD8+ T cells (65). Studies have also shown that Notch signaling is pivotal in T cell activation and differentiation, particularly in regulating the balance between regulatory T cells (Tregs) and effector subsets (Th1/Th17) (66).
In inflammatory processes, Notch signaling contributes to inflammation by promoting Th17 cell differentiation. Th17 cells secrete cytokines such as IL-17, which exacerbate tissue damage and inflammatory responses (67). In chronic inflammatory diseases like rheumatoid arthritis, aberrant activation of the Notch pathway can enhance Th17 cell function, leading to dysregulated immune activation and tissue injury (68). Thus, modulating the activity of the Notch pathway activity can regulate Th17 cell differentiation and alleviate chronic inflammation.
3.1.2 Notch signaling in macrophages
Macrophages are key effector cells in inflammatory responses, capable of regulating local immune reactions through the secretion of cytokines and chemokines. The Notch signaling pathway plays an important role in macrophage polarization, influencing their transition into different subtypes, such as M1 and M2 macrophages (69).
In acute inflammatory responses, M1 macrophages promote inflammation by secreting pro-inflammatory cytokines such as TNF-α and IL-1β. Conversely, in chronic inflammation, M2 macrophages secrete anti-inflammatory cytokines such as IL-10 and TGF-β, exerting immunosuppressive effects (70, 71).
Notch signaling regulates macrophage polarization. For example, Notch1 activation promotes M1 macrophage differentiation, enhancing inflammatory responses. In contrast, Notch2 activation tends to promote M2 macrophage generation, thereby reducing inflammation (72). These findings suggest that targeting the Notch signaling pathway could serve as a crucial target for modulating inflammatory responses.
3.1.3 Notch signaling in dendritic cells
Dendritic cells (DCs) are essential antigen-presenting cells in the immune system that prime T cell-mediated immune responses (73). The Notch signaling pathway plays a critical role in DC development and functional regulation. Notch signaling not only regulates DC maturation but also determines the polarization of T cell responses by influencing their cytokine profiles. For instance, Notch signaling can drive DC polarization towards a Th17 response, thereby exacerbating inflammation (74, 75).
3.2 The role of notch signaling in chronic inflammation
Chronic inflammatory diseases, such as rheumatoid arthritis (RA), inflammatory bowel disease (IBD), and systemic lupus erythematosus (SLE), are closely linked to prolonged immune system activation and tissue damage (76). In these conditions, dysregulated activation or suppression of the Notch signaling pathway may contribute to immune cell imbalance and persistent inflammation.
3.2.1 Rheumatoid arthritis
In RA, abnormal activation of the Notch signaling pathway is strongly associated with sustained inflammation and joint damage. Studies have shown that excessive activation of Notch1 promotes the activation of T cells and macrophages in joint tissues, leading to exacerbated inflammatory responses. Additionally, Notch signaling may participate in joint destruction by regulating stem cell behavior in the bone marrow (77). Targeting the Notch signaling pathway could therefore provide new therapeutic approaches for RA.
3.2.2 Inflammatory bowel disease
IBD encompasses chronic inflammatory disorders involving abnormal activation of the intestinal immune system (78). Research indicates that Notch signaling plays a significant role in the onset and progression of IBD. Activation of Notch1 can enhance Th17 cell function and inflammatory responses, leading to persistent damage to intestinal tissues. Conversely, Notch2 exhibits anti-inflammatory effects by suppressing Th1/Th17 cell differentiation (79). The dual roles of Notch signaling in IBD make it a promising therapeutic target.
3.3 Notch signaling and acute inflammatory response
The acute inflammatory response is a rapid immune reaction to infection or injury aimed at eliminating pathogens and repairing damaged tissues. Notch signaling plays a complex role in acute inflammation, capable of both promoting the inflammatory response and preventing excessive immune activation by regulating immune cell functions (80).
In acute inflammation, Notch signaling modulates immune cell activity, such as that of T cells and macrophages, to rapidly respond to pathological signals. For example, Notch1 activation during acute lung injury enhances macrophage chemotaxis and phagocytosis, aiding in pathogen clearance and tissue repair (81). Simultaneously, Notch signaling regulates immune cell apoptosis and regeneration to prevent tissue damage caused by excessive inflammation.
3.4 Targeted therapies for the notch signaling pathway
Given its critical role in inflammatory responses, targeting the Notch signaling pathway has emerged as a promising therapeutic intervention for treating inflammatory diseases. Studies have explored the potential of alleviating symptoms of chronic inflammatory diseases by inhibiting Notch activity. For instance, Notch inhibitors have demonstrated anti-inflammatory effects in animal models of RA and IBD (82).
However, the pleiotropic regulatory effects of Notch signaling suggest that inhibition might lead to immunosuppression in some cases, increasing the risk of opportunistic infections (83). Thus, precisely modulating Notch signaling, particularly in specific immune cell subsets, remains a crucial challenge in the treatment of inflammatory diseases.
The role of the Notch signaling pathway in inflammation is functionally multifaceted, influencing immune cell differentiation and function as well as the development and progression of chronic inflammatory diseases. Through deeper investigations into the specific mechanisms of Notch signaling in various immune cells and its involvement in inflammatory responses, targeted therapies for inflammatory diseases may become more effective. As understanding of the Notch pathway advances, precise modulation of its role in immune responses will provide novel strategies for clinical treatment of inflammatory conditions.
4 The role of non-coding RNA in cancer
NcRNAs play a crucial role in regulating gene expression, cell proliferation, and apoptosis through their interactions with DNA, RNA, and proteins. These processes are closely related to tumor initiation, progression, metastasis, and drug resistance, making ncRNAs significant in cancer biology and therapy (Table 1) (84).
4.1 miRNAs in cancer
MiRNAs are small ncRNAs that regulate gene expression by binding to the 3’ untranslated regions (UTRs) of target mRNAs, leading to translational repression or degradation (94). Their roles in cancer are diverse, functioning either as tumor suppressors or oncogenes (95).
4.1.1 Tumor-suppressive miRNAs
Some miRNAs are downregulated or lost in cancer cells, playing tumor-suppressive roles. For example, miR-34a directly targets Notch1, inhibiting tumor cell proliferation, migration, and invasion (96). Reduced miR-34a expression is linked to various cancers, including breast, lung, and liver cancers, while restoring miR-34a can significantly inhibit tumor growth and metastasis (97).
miR-143 and miR-145 are also considered tumor-suppressive miRNAs, regulating tumor-related genes like RAS, ERK, and AP-1 to inhibit proliferation and metastasis (98). Their low expression is often associated with increased malignancy, highlighting their critical roles in tumor suppression.
4.1.2 Oncogenic miRNAs
Conversely, some miRNAs act as oncogenes, being upregulated in cancers. For instance, miR-21 is widely recognized as an oncogene, and its high expression is closely associated with tumor initiation and progression (99).miR-21 promotes proliferation, survival, and invasion by inhibiting target genes like PTEN and PDCD4 (100).
Other oncogenic miRNAs include miR-155 and miR-221/222. miR-155 enhances proliferation and migration by targeting suppressor genes like SOCS1 (101), while miR-221/222 promotes tumor progression by regulating cell cycle inhibitors like p27kip1 (102).
4.2 lncRNAs in cancer
lncRNAs participate in chromatin remodeling, transcriptional control, and signal transduction (103). Their roles in cancer vary, promoting tumor progression in some cases while inhibiting it in others.
4.2.1 Oncogenic lncRNAs
Some lncRNAs are overexpressed in cancer cells, promoting proliferation, metastasis, and drug resistance by regulating tumor suppressor genes, oncogenes, or epigenetic factors. For example, HOTAIR is overexpressed in several cancers, recruiting Polycomb Repressive Complex 2 (PRC2) and other epigenetic regulators to suppress tumor suppressor genes, thereby facilitating proliferation and invasion (104).
MALAT1, another oncogenic lncRNA, is highly expressed in various cancers. It regulates transcription factors and RNA-binding proteins, affecting cell cycle progression, transcription, and migration, thus promoting metastasis (105). High MALAT1 expression correlates with poor prognosis, making it a potential therapeutic target.
4.2.2 Tumor-suppressive lncRNAs
However, the down-regulation of certain lncrnas is associated with the development of cancer. A typical example is TUSC7 (Tumor Suppressor Candidate 7), which has low expression in multiple tumor types, and restoring the expression of TUSC7 can significantly inhibit the proliferation and metastasis of tumor cells (106). Mechanistically, TUSC7 coordinates tumor suppression through direct interaction with p53 protein and sequestration of oncogenic miRNAs (107).
Another tumor-suppressive lncRNA, p53-Activated Noncoding RNA (PANDA), modulates cancer cell fate by stabilizing wild-type p53 protein, thereby inducing cell cycle arrest and apoptotic signaling cascades (108). The low expression of PANDA is associated with the development of various cancers, and restoring its expression is expected to be one of the therapeutic strategies.
4.3 circRNAs in cancer
circRNAs characterized by their unique closed-loop structure, are stable ncRNAs that act as miRNA sponges, regulating gene expression. Abnormal circRNA expression is closely associated with tumor initiation, progression, and metastasis (109).
For instance, circRNA CDR1as binds to miR-7, preventing it from targeting its gene. This interaction promotes tumor proliferation and migration, with high CDR1as levels being linked to poor prognosis (110). Similarly, circHIPK3 regulates cancer cell proliferation and invasion through interactions with miR-124-3p and other molecules, making it a promising biomarker for cancer diagnosis and therapy (111).
4.4 NcRNAs in cancer therapy
The critical roles of ncRNAs in cancer have made them attractive therapeutic targets. Strategies targeting miRNAs, lncRNAs, and circRNAs are being developed. Restoring tumor-suppressive miRNAs or inhibiting oncogenic miRNAs has shown promise in suppressing proliferation and metastasis (112). Similarly, therapies targeting specific lncRNAs and circRNAs are gaining traction in cancer research.
Despite their potential, ncRNA-based clinical applications face challenges. The complexity of ncRNAs and their heterogeneity across tumor types pose difficulties in precise targeting while minimizing side effects. Advancing our understanding of ncRNA mechanisms could lead to more efficient and specific therapies in the future.
In conclusion, ncRNAs play pivotal roles in oncogenesis. miRNAs, lncRNAs, and circRNAs regulate tumor initiation, progression, and metastatic dissemination through diverse mechanisms. Understanding these roles offers new avenues for early diagnosis, prognosis, and individualized therapy. With continued research, ncRNA-based therapies could become vital tools in cancer treatment.
5 Interaction between notch signaling and ncRNAs
NcRNAs serve as molecular hubs in cancer by orchestrating complex regulatory networks, yet their functional impact hinges on dynamic interactions with key signaling pathways. Among these, the Notch signaling pathway—a master regulator of cell fate and tumor progression—has been identified as a critical partner of n cRNAs. Research demonstrates that miRNAs, lncRNAs, and circRNAs directly modulate Notch receptor activity or indirectly influence downstream effectors through epigenetic reprogramming and feedback loops, establishing bidirectional crosstalk. This interplay not only fuels tumor heterogeneity, microenvironment remodeling, and therapy resistance but also unveils opportunities to overcome the limitations of conventional single-target therapies. The following section delves into the Notch-ncRNA axis, dissecting its mechanistic roles in cancer progression and its potential for precision oncology.
The Notch signaling pathway governs fundamental cellular processes, including proliferation, differentiation, and apoptosis. Mounting evidence highlights its intricate crosstalk with ncRNAs, which fine-tune Notch activity at multiple levels. By regulating pathway components or interacting with epigenetic modifiers, ncRNAs shape Notch-driven phenotypes in cancer and inflammatory diseases. These discoveries not only deepen our understanding of disease mechanisms but also pave the way for innovative therapeutic strategies targeting the Notch-ncRNA axis.
5.1 Overview of the notch signaling pathway
Abnormal activation of the Notch signaling pathway is closely associated with various diseases, including cancer, cardiovascular diseases, and immune disorders. In cancer, in particular, Notch activation often leads to tumor proliferation, metastasis, and drug resistance and is also linked to the maintenance of cancer stem cells.
5.2 Interaction between miRNAs and the notch signaling pathway
miRNAs are essential molecules for regulating gene expression. They typically bind to the 3’ untranslated regions (3’ UTRs) of target mRNAs, thereby suppressing their stability and translation. miRNAs play critical roles in regulating the Notch signaling pathway. They can inhibit the expression of Notch receptors to reduce pathway activation or activate the Notch pathway to promote cell proliferation and migration (113).
5.2.1 miRNAs inhibiting notch signaling
For instance, miR-34a targets Notch1, suppressing its expression and negatively regulating the Notch signaling pathway. Low expression of miR-34a is often associated with tumor malignancy, metastasis, and poor prognosis. Thus, miR-34a exhibits potential anti-tumor effects, and restoring miR-34a expression can significantly inhibit cancer cell proliferation and migration (114).
Similarly, miR-7, miR-196a, and miR-129 have also been found to inhibit Notch signaling by directly suppressing the expression of Notch receptors or their transcriptional activators (115). For example, miR-7 targets Notch1 mRNA, suppressing its expression and thereby reducing Notch signaling activity in non-small cell lung cancer (116). Furthermore, miR-196a, which is upregulated in gastric cancer, downregulates Notch2 expression, inhibiting the proliferation and invasive potential of gastric cancer cells (117).
5.2.2 miRNAs activating notch signaling
In contrast to the inhibitory effects of the aforementioned miRNAs, certain miRNAs can promote the activation of Notch signaling, thereby facilitating cancer cell proliferation and metastasis. For instance, miR-221/222 is upregulated in various cancers and promotes tumor cell proliferation and migration by targeting cell cycle regulators such as p27kip1 (118).
Moreover, miR-9 has been shown to regulate Notch signaling, supporting the self-renewal of embryonic stem cells and contributing to tumorigenesis. miR-9 enhances Notch signaling in breast cancer by targeting and upregulating Notch3, which drives breast cancer progression (119).
5.3 Interaction between lncRNAs and notch signaling
LncRNAs play vital roles in gene transcription, epigenetic modification, and cellular biological processes. Many lncRNAs interact with key factors in the Notch signaling pathway to regulate its activity, thereby influencing the onset and progression of cancer.
5.3.1 lncRNAs inhibiting notch signaling
Some lncRNAs suppress the Notch signaling pathway by directly or indirectly regulating the expression of Notch receptors (120). For example, lncRNA HOTAIR binds to the Notch1 receptor and inhibits its signal transduction, thereby promoting malignancy in various cancers. High expression of HOTAIR is closely associated with tumor invasiveness, metastasis, and poor prognosis (121).
Similarly, lncRNA ANRIL (Antisense Noncoding RNA in the INK4 Locus) interacts with critical factors in the Notch signaling pathway to suppress its activity, resulting in tumor cell proliferation and metastasis (122, 123).
5.3.2 lncRNAs activating notch signaling
In contrast to tumor-suppressive lncRNAs, certain long non-coding RNAs function as oncogenic drivers by potentiating Notch signaling transduction (124). A prime example is the metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), which exhibits pathological overexpression across malignancies and directly binds to Notch1 mRNA, thereby stabilizing its transcript and enhancing Notch signaling output. This molecular interplay promotes clonal expansion, metastatic dissemination, and chemoresistance phenotypes in neoplastic cells (125). Mechanistically, MALAT1 orchestrates a multi-layered regulatory network through:1. Transcriptional upregulation of Notch1 via chromatin remodeling complexes. 2. Sponging tumor-suppressive miRNAs (e.g., miR-129-5p). 3.Recruiting epigenetic modifiers (DNMT3A/HDAC1) to reprogram Notch-responsive genes.
Similarly, terminal differentiation-induced non-coding RNA (TINCR) demonstrates oncogenic competence in melanoma, where its ectopic overexpression activates Notch signaling via ligand-independent receptor clustering, consequently augmenting cancer cell motility and matrix-invasion capacity (126).
5.4 Interaction between circRNAs and notch signaling
Circular RNAs (circRNAs) are a class of ncRNAs with a covalently closed loop structure. Due to their stability and regulatory functions, circRNAs have become a focus of cancer research in recent years. CircRNAs regulate the suppression of target genes by acting as miRNA sponges. Additionally, circRNAs can directly interact with key molecules in the Notch signaling pathway, further modulating tumor initiation and progression (127).
5.4.1 circRNAs regulating notch signaling
For example, circRNA CDR1as (Cerebellar Degeneration-Related Protein 1-antisense RNA) acts as a sponge for miR-7, relieving miR-7’s inhibitory effect on Notch1, thereby activating Notch signaling and promoting tumor cell proliferation and migration. The high expression of CDR1as in gliomas exacerbates tumor malignancy through this mechanism (128). Similarly, circHIPK3 enhances Notch signaling activity by regulating molecules like miR-124, driving tumor growth and metastasis (129). The high expression of circRNA circPVT1 in gastric cancer cells activates Notch signaling by binding to miR-125b, promoting cancer cell proliferation (130).
5.5 Bidirectional regulation between notch signaling and ncRNAs
The interaction between Notch signaling and ncRNAs is highly complex. Not only do they mutually regulate each other during signal transduction, but they can also form feedback mechanisms to ensure precise control of cells under various physiological and pathological conditions. Activation of Notch signaling can upregulate the expression of miRNAs and lncRNAs, which in turn influence the regulation of Notch signaling by these ncRNAs. On the other hand, ncRNAs directly bind to Notch receptors or downstream effector molecules, modulating the strength and duration of Notch signaling.
For instance, activation of Notch1 can upregulate the expression of miR-145, which in turn forms a negative feedback loop by inhibiting Notch1’s target genes. In tumor cells, this feedback mechanism helps maintain a balance between cell proliferation and differentiation (131). This bidirectional regulatory mechanism ensures that cells respond accurately to external signals and adapt to changes in their internal environment. In the tumor microenvironment, the interaction between Notch signaling and ncRNAs is especially critical, as they jointly regulate tumor cell proliferation, migration, immune evasion, and drug resistance (Table 2).
6 Therapeutic strategies and prospects
The reciprocal regulatory axis between ncRNAsand the Notch signaling pathway underscore their central role in cancer biology, yet the translational potential of this complex network remains underexplored. Current studies demonstrate that dynamic Notch-ncRNA interactions not only drive malignant phenotypes but also modulate therapeutic responses through epigenetic landscape remodeling and immune microenvironment remodeling. However, translating these mechanisms into clinical applications faces dual challenges: the “double-edged sword” nature of Notch signaling necessitates balancing antitumor efficacy with preservation of normal tissue functions in targeted therapies, while the spatiotemporal specificity of ncRNA expression demands innovations in spatiotemporally controlled delivery platforms and tissue-selective intervention strategies. The sixth section will systematically elaborate on novel therapeutic approaches based on these molecular mechanisms and explore the potential of multimodal combination therapies to overcome therapeutic resistance and enhance efficacy, thereby expanding the horizons of precision oncology.
With the progressive elucidation of Notch-ncRNA interactions, researchers have prioritized therapeutic strategies targeting this axis. Notch signaling and ncRNAs not only show immense promise in cancer therapy but also offer potential therapeutic targets for addressing chronic inflammation, immune evasion, and drug resistance (60). The following sections will delve into these strategies, focusing on advancements in targeted therapy, immunotherapy, and RNA-based interventions.
6.1 Targeting notch signaling pathway in therapy
The abnormal activation or inhibition of Notch signaling plays a critical role in the initiation and progression of various cancers. Therefore, Notch signaling has become an important target in cancer therapy. Current therapeutic strategies mainly include small molecule inhibitors, monoclonal antibodies, and γ-secretase inhibitors.
6.1.1 Small molecule inhibitors
A key regulator of Notch signaling is γ-secretase, which cleaves the Notch receptor, releasing the Notch intracellular domain (NICD) and activating downstream signaling pathways (137). Therefore, γ-secretase inhibitors (such as DAPT, LY3039478, etc.) are considered potential inhibitors of Notch signaling (138). These inhibitors have shown certain therapeutic effects in clinical trials, particularly in hematologic malignancies (e.g., acute myeloid leukemia).
6.1.2 Monoclonal antibodies
Monoclonal antibodies targeting Notch ligands such as Jagged1, Jagged2, and Delta-like can specifically block the binding of Notch receptors to their ligands, thereby inhibiting the activation of Notch signaling (139) Studies have found that monoclonal antibodies targeting Jagged1 and Jagged2 also exhibit tumor growth inhibitory effects in certain cancer types.
6.1.3 Notch receptor targeted
Therapeutic Strategies: Preclinical studies demonstrate that selective anti-Notch1 monoclonal antibodies (e.g., Omp-18) can significantly attenuate tumor cell proliferative capacity and metastatic dissemination through ligand-binding domain blockade. Concurrently, isoform-specific oncogenic roles of Notch3 in hormone receptor-positive breast cancer and Notch4 in non-small cell lung adenocarcinoma have been mechanistically validated, revealing therapeutic vulnerabilities for precision intervention.Notwithstanding these advances, clinical translation faces two fundamental challenges:1.Pathway complexity: The pleiotropic nature of Notch signaling necessitates tumor-specific isoform inhibition to avoid homeostatic disruption in normal tissues.2.Therapeutic window optimization: On-target toxicity arising from conserved receptor-ligand interactions in stem cell niches limits dose escalation
Future investigations must prioritize tumor microenvironment (TME)-responsive delivery systems capable of spatial-temporal modulation and single-cell resolution targeting. Emerging approaches combining γ-secretase modulators with Notch extracellular domain (NECD) decoys may enable context-dependent signaling attenuation while preserving physiological Notch functions.
6.2 Non-coding RNA-based therapeutic strategies
NcRNAs, particularly miRNAs, lncRNAs, and circRNAs, play essential roles in the fine-tuning of gene expression, influencing various biological behaviors of cancer cells, such as proliferation, migration, and immune evasion. Therapeutic strategies based on ncRNAs mainly include miRNA replacement therapy, RNA intervention technologies, and the regulation of lncRNAs and circRNAs to improve the clinical treatment of cancer.
6.2.1 miRNA replacement therapy
Many miRNAs are underexpressed or dysregulated in cancer, leading to abnormal cancer cell proliferation and metastasis. By introducing specific miRNAs (e.g., miR-34a, miR-143) into cancer cells, the normal function of miRNAs can be restored, inhibiting tumor growth and enhancing sensitivity to chemotherapy (140).
6.2.2 RNA intervention technologies
In addition to miRNA replacement therapy, RNA intervention technologies (such as small molecule RNAs, siRNAs, etc.) have become a recent research focus in cancer therapy. By designing specific small molecule RNAs, key gene expressions in cancer cells can be precisely regulated. These technologies can inhibit key molecules in the Notch signaling pathway, such as Jagged and Notch receptors, interfering with cancer cell proliferation and migration. Furthermore, RNA intervention technologies can also target ncRNAs, such as lncRNAs and circRNAs, to regulate intracellular signaling pathways, further controlling cancer development.
6.2.3 lncRNA and circRNA regulatory therapy
Abnormal expression of lncRNAs and circRNAs is widespread in cancer. Therefore, intervening in the expression of these ncRNAs may provide new strategies for cancer therapy. Researchers are developing inhibitors or agonists targeting specific lncRNAs (e.g., HOTAIR, MALAT1) and circRNAs (e.g., circRNA CDR1as) to restore or suppress their functions, inhibiting cancer cell proliferation and metastasis (141).
6.3 Immunotherapy-based strategies
Immunotherapy, as an emerging field in cancer treatment, has made significant progress in recent years. Notch signaling not only regulates cancer cell proliferation and migration but also plays an important role in immune evasion. As a result, immunotherapy targeting the Notch signaling pathway has become a research focus.
6.3.1 Immune checkpoint inhibition
The role of Notch signaling in immune responses suggests that regulating the Notch pathway can improve immune evasion. Studies have found that the Notch pathway plays a significant role in the function of T cells and B cells, and targeting Notch with therapeutic agents may enhance the immune system’s ability to recognize and eliminate tumors (142). Combining immune checkpoint inhibitors with Notch signaling pathway inhibitors may improve the effectiveness of cancer immunotherapy.
6.4 Clinical translation of notch pathway inhibitors: recent advances and challenges
The therapeutic development of Notch pathway inhibitors has achieved substantial clinical progress, with over 30 interventional trials initiated since 2022 demonstrating improved safety and efficacy profiles. Second-generation γ-secretase inhibitors (GSIs) exemplify these advancements, as evidenced by Nirogacestat (NCT05348356) achieving breakthrough designation with a 76% objective response rate in desmoid tumors through optimized pulsatile dosing that mitigates gastrointestinal toxicity (143). This progress extends to receptor-specific biologics, where Brontictuzumab elicited partial responses in 15% of NOTCH1-mutated solid tumors, and Tarextumab combined with chemotherapy extended progression-free survival by 5.1 months in pancreatic cancer (NCT01647828) (144, 145).
Innovative molecular approaches are addressing historical limitations in Notch modulation. The transcriptional inhibitor CB-103 disrupts NICD/RBPJκ complex formation, achieving 40% disease control in adenoid cystic carcinoma while avoiding dose-limiting intestinal toxicity (146). Combination therapies demonstrate synergistic potential, particularly in AL101 plus Pembrolizumab (NCT05608762) for HNSCC, where PD-L1 suppression and T-cell activation drove a 38% overall response rate (147). These therapeutic advances are complemented by liquid biopsy applications, with ctDNA analysis of NOTCH1/FBXW7 mutations achieving 92% concordance with tissue biopsies in T-ALL for real-time treatment monitoring (148).
Persisting challenges in pathway pleiotropy and context-dependency are driving biomarker development. Current strategies integrate 68Ga-Notch4i PET imaging (SUVmax ≥2.5 predictive of response) with single-cell RNA sequencing to delineate tumor-specific Notch activity patterns (149). Pharmaceutical innovation continues with microenvironment-responsive agents, including pH-sensitive GSI formulations and Notch3-targeted CAR T-cell platforms (150). The field is poised for transformative growth, with 18 novel modulators entering clinical trials between 2023-2024, underscoring the evolution toward precision-guided interventions through integrated systems biology and multi-omics profiling (Table 3).
6.5 Future perspectives
The intricate crosstalk between the Notch signaling pathway and non-coding RNAs (ncRNAs) has not only reshaped our understanding of cancer and inflammatory pathogenesis but also opened unprecedented avenues for therapeutic innovation. As we advance, the integration of multidisciplinary tools—from single-cell transcriptomics to spatial omics and artificial intelligence—will be pivotal in unraveling the spatiotemporal dynamics of Notch-ncRNA networks. These approaches could map subtype-specific regulatory landscapes, particularly in tumors where Notch pathway dysregulation converges with ncRNA-driven epigenetic reprogramming, ultimately guiding the design of precision therapies tailored to individual molecular vulnerabilities.
Building on this foundation, overcoming current limitations in therapeutic specificity and delivery remains paramount. Emerging technologies such as nanotechnology and exosome engineering offer promising solutions by enabling targeted modulation of oncogenic ncRNAs (e.g., HOTAIR, MALAT1) or Notch receptors with minimal off-target effects. Simultaneously, combinatorial strategies that pair Notch inhibitors with immune checkpoint blockade or metabolic regulators may disrupt the plasticity of the tumor microenvironment, countering resistance mechanisms rooted in cellular adaptation. The synergy between CRISPR-based functional screening and machine learning could further accelerate the discovery of synthetic lethal nodes within Notch-ncRNA interaction networks, paving the way for novel multidrug regimens that exploit these vulnerabilities.
Translating these insights to the clinic requires bridging cutting-edge science with practical challenges. The development of non-invasive biomarkers—such as liquid biopsy panels tracking Notch-associated ncRNAs—and real-time imaging techniques to monitor pathway activity will revolutionize patient stratification and treatment monitoring. However, balancing therapeutic efficacy with tissue homeostasis demands innovative solutions: tissue-specific delivery systems (e.g., ligand-engineered nanoparticles) or transient epigenetic modulators may mitigate risks to normal stem cell niches while preserving antitumor activity. Beyond oncology, exploring the evolutionary conservation of Notch-ncRNA axes in chronic inflammation and autoimmune disorders could reveal shared pathogenic drivers, expanding therapeutic applications to a broader spectrum of diseases.
Ultimately, the convergence of mechanistic discovery and translational ingenuity promises to transform the Notch-ncRNA interplay from a complex biological paradigm into a cornerstone of clinical practice. By harmonizing molecular insights with advanced engineering and clinical validation, this field holds the potential to deliver durable, personalized therapies that transcend traditional boundaries between cancer and inflammatory diseases, offering new hope for patients worldwide.
Author contributions
R-ZN: Writing – original draft, Writing – review & editing. HW: Writing – original draft, Writing – review & editing. S-SW: Investigation, Software, Writing – original draft. CC: Supervision, Validation, Writing – review & editing. H-ML: Formal analysis, Investigation, Writing – original draft. H-KZ: Resources, Visualization, Writing – review & editing. Z-HJ: Project administration, Writing – review & editing. P-FL: Investigation, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Natural Science Foundation of Henan (252300421733), Key Scientific and Technological Project of Henan Province (242102110106), the Key Scientific Research Projects of Colleges and Universities in Henan Province (No. 24A550019), Doctoral Scientific Research Foundation of Zhengzhou University of Light Industry (2022BSJJZK06) and the Youth Cultivation Fund Project of Zhengzhou University of Light Industry (2024XNQNPY15).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. Proofreading of Typographical Errors in the Article
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Kopan R and Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. (2009) 137:216–33. doi: 10.1016/j.cell.2009.03.045
2. Górecki I and Rak B. The role of microRNAs in epithelial to mesenchymal transition and cancers; focusing on mir-200 family. Cancer Treat Res Commun. (2021) 28:100385. doi: 10.1016/j.ctarc.2021.100385
3. De Martino M, Esposito F, and Pallante P. Long non-coding RNAs regulating multiple proliferative pathways in cancer cell. Transl Cancer Res. (2021) 10:3140–57. doi: 10.21037/tcr-21-230
4. Chen J, Wang H, Xu J, Chen E, Meng Q, Wang J, et al. CircZFR promotes colorectal cancer progression via stabilizing BCLAF1 and regulating the miR-3127-5p/RTKN2 axis. Sci China Life Sci. (2024) 67:1881–98. doi: 10.1007/s11427-023-2514-y
5. Metz CW and Bridges CB. Incompatibility of mutant races in drosophila. Proc Natl Acad Sci U.S.A. (1917) 3:673–8. doi: 10.1073/pnas.3.12.673
6. Mohr OL. Character changes caused by mutation of an entire region of a chromosome in drosophila. Genetics. (1919) 4:275–82. doi: 10.1093/genetics/4.3.275
7. Yochem J, Weston K, and Greenwald I. The Caenorhabditis elegans lin- 12 gene encodes a transmembrane protein with overall similarity to Drosophila Notch. Nature. (1988) 335:547–50. doi: 10.1038/335547a0
8. Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, et al. Alagille syndrome is caused by mutations in human Jagged1. which encodes ligand Notch1 Nat Genet. (1997) 16:243–51. doi: 10.1038/ng0797-243
9. Kopan R and Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. (2009) 137:216–33. doi: 10.1016/j.cell.2009.03.045
10. Capaccione KM and Pine SR. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. (2013) 34:1420–30. doi: 10.1093/carcin/bgt127
12. Pitulescu ME, Schmidt I, Giaimo BD, Antoine T, Berkenfeld F, F.Ferrante H, et al. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat Cell Biol. (2017) 19:915–27. doi: 10.1038/ncb3555
13. Siebel C and Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. (2017) 97:1235–94. doi: 10.1152/physrev.00005.2017
14. Majumder S, Crabtree JS, Golde TE, Minter LM, Osborne BA, and Miele L. Targeting Notch in oncology: the path forward. Nat Rev Drug Discov. (2021) 20:125–44. doi: 10.1038/s41573-020-00091-3
15. Mizuno T, Chou MY, and Inouye M. A unique mechanism regulating gene expression:translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci U.S.A. (1984) 81:1966–1970. doi: 10.1073/pnas.81.7.1966
16. Inzulza-Tapia A and Alarcón M. Role of non-coding RNA of human platelet in cardiovascular disease. Curr Med Chem. (2022) 29:3420–3444. doi: 10.2174/0929867329666211230104955
17. Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. (2016) 17:601–614. doi: 10.1038/nrg.2016.85
18. Kim VN, Han J, and Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. (2009) 10:126–39. doi: 10.1038/nrm2632
19. Dakal TC, Philip RR, Bhushan R, Sonar PV, Rajagopal S, and Kumar A. Genetic and epigenetic regulation of non-coding RNAs: Implications in cancer metastasis, stemness and drug resistance. Pathol Res Pract. (2024) 266:155728. doi: 10.1016/j.prp.2024.155728
20. Yi Q, Yue J, Liu Y, Shi H, Sun W, Feng J, et al. Recent advances of exosomal circRNAs in cancer and their potential clinical applications. J Transl Med. (2023) 21:516. doi: 10.1186/s12967-023-04348-4
21. He Z, Zhong Y, Regmi P, Lv T, Ma W, Wang J, et al. Exosomal long non-coding RNA TRPM2-AS promotes angiogenesis in gallbladder cancer through interacting with PABPC1 to activate NOTCH1 signaling pathway. Mol Cancer. (2024) 23:65. doi: 10.1186/s12943-024-01979-z
22. Jiang P, Liu R, Zheng Y, Liu X, Chang L, Xiong S, et al. MiR- 34a inhibits lipopolysaccharide-induced inflammatory response through targeting Notch1 in murine macrophages. Exp Cell Res. (2012) 318:1175–84. doi: 10.1016/j.yexcr.2012.03.018
23. Chen M, Xia Z, Chen C, Hu W, and Yuan Y. LncRNA MALAT1 promotes epithelial-to-mesenchymal transition of esophageal cancer through Ezh2-Notch1 signaling pathway. Anti-Cancer Drugs. (2018) 29:767–73. doi: 10.1097/CAD.0000000000000645
24. Zhang Y, Aodeng G, Liu P, Su W, and Zhao H. Effects of HOX transcript antisense intergenic RNA on the metastasis, epithelial- mesenchymal transition, and Notch signaling pathway in tongue cancer. Transl Cancer Res. (2021) 10:520–8. doi: 10.21037/tcr-20-3452
25. Sun W, Gaykalova DA, Ochs MF, Mambo E, Arnaoutakis D, Liu Y, et al. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. (2014) 74(4):1091–104. doi: 10.1158/0008-5472.CAN-13-1259
26. Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. (2016) 17:722–35. doi: 10.1038/nrm.2016.94
27. Meurette O and Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. (2018) 34:536–48. doi: 10.1016/j.ccell.2018.07.009
28. Fournier M, Javary J, Roh V, Fournier N, Radtke F, and Molecular Medicine EMBO. (2024) 16:3184–3217-3217.
29. Oliphant M, Kong D, Zhou H, Lewis M, and Ford H. Two Sides of the Same Coin: The Role of Developmental pathways and pluripotency factors in normal mammary stem cells and breast cancer metastasis. J mammary gland Biol neoplasia. (2020) 25:85–102. doi: 10.1007/s10911-020-09449-0
30. Chen J-H, Kuo K-T, Bamodu OA, Lin Y-C, Yang R-B, and C.-T.Yeh T-Y. Chao, Upregulated SCUBE2 expression in breast cancer stem cells enhances triple negative breast cancer aggression through modulation of notch signaling and epithelial-to-mesenchymal transition. Exp Cell Res. (2018) 370:444–53. doi: 10.1016/j.yexcr.2018.07.008
31. Majumder M, Xin X, Liu L, Tutunea-Fatan E, Rodriguez-Torres M, Vincent K, et al. COX-2 induces breast cancer stem cells via EP4/PI3K/AKT/NOTCH/WNT axis. Stem Cells. (2016) 34:2290–305. doi: 10.1002/stem.2426
32. Zhou W, Tan W, Huang X, and Yu HG. Doxorubicin combined with Notch1−targeting siRNA for the treatment of gastric cancer. Oncol Lett. (2018) 16:2805–12. doi: 10.3892/ol.2018.9039
33. Wang Z, Zhang Y, Li Y, Banerjee S, Liao J, and Sarkar FH. Down- regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol Cancer Ther. (2006) 5:483–93. doi: 10.1158/1535-7163.MCT-05-0299
34. Zhang Z, Han H, Rong Y, Zhu K, Zhu Z, Tang Z, et al. Hypoxia potentiates gemcitabine-induced stemness in pancreatic cancer cells through AKT/Notch1 signaling. J Exp Clin Cancer Res. (2018) 37:1–13. doi: 10.1186/s13046-018-0972-3
35. Hai L, Zhang C, Li T, Zhou X, Liu B, Li S, et al. Notch1 is a prognostic factor that is distinctly activated in the classical and proneural subtype of glioblastoma and that promotes glioma cell survival via the NF-κB (p65) pathway. Cell Death Dis. (2018) 9:158. doi: 10.1038/s41419-017-0119-z
36. Xiao W, Gao Z, Duan Y, Yuan W, and Ke Y. Notch signaling plays a crucial role in cancer stem-like cells maintaining stemness and mediating chemotaxis in renal cell carcinoma. J Exp Clin Cancer Res. (2017) 36:1–13. doi: 10.1186/s13046-017-0507-3
37. Pannuti A, Foreman K, Rizzo P, Osipo C, Golde T, Osborne B, et al. Targeting Notch to target cancer stem cells. Clin Cancer Res. (2010) 16:3141–52. doi: 10.1158/1078-0432.CCR-09-2823
38. Jin L, Vu T, Yuan G, and Datta PK. STRAP promotes stemness of human colorectal cancer via epigenetic regulation of the NOTCH pathway. Cancer Res. (2017) 77:5464–78. doi: 10.1158/0008-5472.CAN-17-0286
39. Katoh M and Katoh M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int J Oncol. (2007) 31:461–6. doi: 10.3892/ijo.31.2.461
40. Guo Y, Liu S, Wang P, Zhao S, Wang F, Bing L, et al. Expression profile of embryonic stem cell- associated genes Oct4, Sox2 and Nanog in human gliomas. Histopathology. (2011) 59:763–75. doi: 10.1111/j.1365-2559.2011.03993.x
41. El-Habr EA, Levidou G, Trigka E-A, Sakalidou J, Piperi C, Chatziandreou I, et al. Complex interactions between the components of the PI3K/AKT/mTOR pathway, and with components of MAPK, JAK/STAT and Notch-1 pathways, indicate their involvement in meningioma development. Virchows Archiv. (2014) 465:473–85. doi: 10.1007/s00428-014-1641-3
42. Hou H, Xue P, Wang Y, and Li Y. Liraglutide regulates proliferation, differentiation, and apoptosis of preosteoblasts through a signaling network of Notch/Wnt/Hedgehog signaling pathways. Eur Rev Med Pharmacol Sci. (2020) 24:12408–22. doi: 10.26355/eurrev_202012_24037
43. Jeong G-Y, Park MK, Choi H-J, An HW, Park Y-U, Choi H-J, et al. NSD3-induced methylation of H3K36 activates NOTCH signaling to drive breast tumor initiation and metastatic progression. Cancer Res. (2021) 81:77–90. doi: 10.1158/0008-5472.CAN-20-0360
44. Xie M, Zhang L, He CS, Xu F, Liu JL, Hu ZH, et al. Activation of Notch-1 enhances epithelial-mesenchymal transition in gefitinib-acquired resistant lung cancer cells. J Cell Biochem. (2012) 113:1501–13. doi: 10.1002/jcb.v113.5
45. Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. (2015) 527:472–6. doi: 10.1038/nature15748
46. Miao K, Lei JH, Valecha MV, Zhang A, Xu J, Wang L, et al. NOTCH1 activation compensates BRCA1 deficiency and promotes triple-negative breast cancer formation. Nat Commun. (2020) 11:3256. doi: 10.1038/s41467-020-16936-9
47. Ye M, Du J, Wang X, Xiu L, Liu X, Gu Y, et al. Xiaotansanjiefang inhibits the viability of colorectal cancer cells via Jagged 1/Notch 3/Snail signaling pathway. Environ Toxicol. (2022) 37:2957–64. doi: 10.1002/tox.v37.12
48. Lu P, Takai K, Weaver VM, and Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harbor Perspect Biol. (2011) 3:a005058. doi: 10.1101/cshperspect.a005058
49. Jabłońska-Trypuć A, Matejczyk M, and Rosochacki S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J Enzyme Inhib Med Chem. (2016) 31:177–83. doi: 10.3109/14756366.2016.1161620
50. Poltavets V, Kochetkova M, Pitson SM, and Samuel MS. The role of the extracellular matrix and its molecular and cellular regulators in cancer cell plasticity. Front Oncol. (2018) 8:431. doi: 10.3389/fonc.2018.00431
51. Cloughesy TF, Mochizuki AY, Orpilla JR, Hugo W, Lee AH, Davidson TB, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. (2019) 25:477–86. doi: 10.1038/s41591-018-0337-7
52. Zhang R, Boareto M, Engler A, Louvi A, Giachino C, Iber D, et al. Id4 downstream of Notch2 maintains neural stem cell quiescence in the adult hippocampus. Cell Rep. (2019) 28:1485–1498.e1486. doi: 10.1016/j.celrep.2019.07.014
53. Engler A, Rolando C, Giachino C, Saotome I, Erni A, Brien C, et al. Notch2 signaling maintains NSC quiescence in the murine ventricular- subventricular zone. Cell Rep. (2018) 22:992–1002. doi: 10.1016/j.celrep.2017.12.094
54. Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, and Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. (2010) 30:3489–98. doi: 10.1523/JNEUROSCI.4987-09.2010
55. Bowman RL, Klemm F, Akkari L, Pyonteck SM, Sevenich L, Quail DF, et al. Macrophage ontogeny underlies differences in tumor-specific education in brain Malignancies. Cell Rep. (2016) 17:2445–59. doi: 10.1016/j.celrep.2016.10.052
56. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. (2013) 4:2612. doi: 10.1038/ncomms3612
57. Wang H, Yi X, Wang X, Yang Y, Zhang H, Wang H, et al. Nucleo-cytosolic acetyl-CoA drives tumor immune evasion by regulating PD-L1 in melanoma. Cell Reports. (2024) 43. doi: 10.1016/j.celrep.2024.115015
58. Wang J, Deng R, Chen S, Deng S, Hu Q, Xu B, et al. Helicobacter pylori CagA promotes immune evasion of gastric cancer by upregulating PD-L1 level in exosomes. iScience. (2023) 26(12):108414. doi: 10.1016/j.isci.2023.108414
59. Liu Y, Peng Y, Du W, Yu C, Peng Z, Qin L, et al. PD-L1-mediated immune evasion in triple- negative breast cancer is linked to the loss of ZNF652. Cell Rep. (2023) 42(11):113343. doi: 10.1016/j.celrep.2023.113343
60. Wang Z, Li Y, Ahmad A, Azmi AS, Banerjee S, Kong D, et al. Targeting Notch signaling pathway to overcome drug resistance for cancer therapy. Biochim Biophys Acta (BBA) - Rev Cancer. (2010) 1806:258–67. doi: 10.1016/j.bbcan.2010.06.001
61. Feng M, Santhanam RK, Xing H, Zhou M, and Jia H. Inhibition of γ- secretase/Notch pathway as a potential therapy for reversing cancer drug resistance. Biochem Pharmacol. (2024) 220:115991. doi: 10.1016/j.bcp.2023.115991
62. Li M, Chen F, Clifton N, Sullivan DM, Dalton WS, Gabrilovich DI, et al. Combined inhibition of Notch signaling and Bcl-2/Bcl-xL results in synergistic antimyeloma effect. Mol Cancer Ther. (2010) 9:3200–9. doi: 10.1158/1535-7163.MCT-10-0372
63. Mantovani A, Allavena P, Sica A, and Balkwill F. Cancer-related inflammation. nature. (2008) 454:436–44. doi: 10.1038/nature07205
64. Pu Z, Yang F, Wang L, Diao Y, and Chen D. Advancements of compounds targeting Wnt and Notch signalling pathways in the treatment of inflammatory bowel disease and colon cancer. J Drug Targeting. (2021) 29:507–19. doi: 10.1080/1061186X.2020.1864741
65. Fridman WH, Zitvogel L, Sautès-Fridman C, and Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. (2017) 14:717–34. doi: 10.1038/nrclinonc.2017.101
66. Jiao Z, Wang W, Hua S, Liu M, Wang H, Wang X, et al. Blockade of notch signaling ameliorates murine collagen- induced arthritis via suppressing th1 and th17 cell responses. Am J Pathol. (2014) 184:1085–93. doi: 10.1016/j.ajpath.2013.12.010
67. van Hamburg JP, Asmawidjaja P, Davelaar N, Mus A, Colin E, Hazes J, et al. Th17 cells, but not Th1 cells, from patients with early rheumatoid arthritis are potent inducers of matrix metalloproteinases and proinflammatory cytokines upon synovial fibroblast interaction, including autocrine interleukin - 17Aproduction. Arthritis Rheumatism. (2011) 63:73–83. doi: 10.1002/art.30093
68. Wei K, Korsunsky I, Marshall JL, Gao A, Watts GFM, Major T, et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature. (2020) 582:259–64. doi: 10.1038/s41586-020-2222-z
69. Khoshnavay Foumani M, Amirshahrokhi K, Namjoo Z, and Niapour A. Carvedilol attenuates inflammatory reactions of lipopolysaccharide- stimulated BV2 cells and modulates M1/M2 polarization of microglia via regulating NLRP3, Notch, and PPAR-γ signaling pathways. Naunyn Schmiedebergs Arch Pharmacol. (2024) 397:4727–36. doi: 10.1007/s00210-023-02914-7
70. Li T, Zhang YS, Wan M, Wu W, Yao YF, and Li WJ. Ganoderma atrum polysaccharide modulates the M1/M2 polarization of macrophages linked to the Notch signaling pathway. Food Funct. (2022) 13:4216–28. doi: 10.1039/D1FO04309A
71. Li QQ, Ding DH, Wang XY, Sun YY, and Wu J. Lipoxin A4 regulates microglial M1/M2 polarization after cerebral ischemia- reperfusion injury via the Notch signaling pathway. Exp Neurol. (2021) 339:113645. doi: 10.1016/j.expneurol.2021.113645
72. Huntzicker EG, Hötzel K, Choy L, Che L, Ross J, Pau G, et al. Differential effects of targeting Notch receptors in a mouse model of liver cancer. Hepatology. (2015) 61:942–52. doi: 10.1002/hep.27566
73. Wang M, Shang Z, Qiao F, Hei J, Ma X, and Wang Y. Notch signaling pathway involved in Echinococcus granulosus infection regulates dendritic cell development and differentiation. Front Cell Infect Microbiol. (2023) 13:1147025. doi: 10.3389/fcimb.2023.1147025
74. Liu Q, Chen C, He Y, Mai W, Ruan S, Ning Y, et al. Notch signaling regulates the function and phenotype of dendritic cells in helicobacter pylori infection. Microorganisms. (2023) 11(11):2818. doi: 10.3390/microorganisms11112818
75. Meng L, Hu S, Wang J, He S, and Zhang Y. DLL4(+) dendritic cells: Key regulators of Notch Signaling in effector T cell responses. Pharmacol Res. (2016) 113:449–57. doi: 10.1016/j.phrs.2016.09.001
76. Smolen JS, Aletaha D, Barton A, Burmester GR, Emery P, Firestein GS, et al. Rheumatoid arthritis. Nat Rev Dis Primers. (2018) 4:18001. doi: 10.1038/nrdp.2018.1
77. Scott DL, Wolfe F, and Huizinga TWJ. Rheumatoid arthritis. Lancet. (2010) 376:1094–108. doi: 10.1016/S0140-6736(10)60826-4
78. Lin J-C, Wu J-Q, Wang F, Tang F-Y, Sun J, Xu B, et al. QingBai decoction regulates intestinal permeability of dextran sulphate sodium-induced colitis through the modulation of notch and NF-κB signalling. Cell Proliferation. (2019) 52:e12547. doi: 10.1111/cpr.2019.52.issue-2
79. Ghorbaninejad M, Heydari R, Mohammadi P, Shahrokh S, Haghazali M, Khanabadi B, et al. Contribution of NOTCH signaling pathway along with TNF-α in the intestinal inflammation of ulcerative colitis. Gastroenterol Hepatol Bed Bench. (2019) 12:S80–s86.
80. Ayaz F and Osborne BA. Non-canonical notch signaling in cancer and immunity. Front Oncol. (2014) 4:345. doi: 10.3389/fonc.2014.00345
81. Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, et al. Leukemia-Associated Mutations within the NOTCH1 Heterodimerization Domain Fall into at Least Two Distinct Mechanistic Classes. Mol Cell Biol. (2006) 26:4642–51. doi: 10.1128/MCB.01655-05
82. Yabuuchi S, Pai SG, Campbell NR, de Wilde RF, Oliveira E, Korangath P, et al. Notch signaling pathway targeted therapy suppresses tumor progression and metastatic spread in pancreatic cancer. Cancer Lett. (2013) 335:41–51. doi: 10.1016/j.canlet.2013.01.054
83. Dufraine J, Funahashi Y, and Kitajewski J. Notch signaling regulates tumor angiogenesis by diverse mechanisms. Oncogene. (2008) 27:5132–7. doi: 10.1038/onc.2008.227
84. Wang J, Zhu S, Meng N, He Y, Lu R, and Yan G-R. ncRNA-encoded peptides or proteins and cancer. Mol Ther. (2019) 27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001
85. Su G, Sun G, Liu H, Shu L, and Liang Z. Downregulation of miR-34a promotes endothelial cell growth and suppresses apoptosis in atherosclerosis by regulating Bcl-2. Heart Vessels. (2018) 33:1185–94. doi: 10.1007/s00380-018-1169-6
86. Dong G, Liang X, Wang D, Gao H, Wang L, Wang L, et al. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol. (2014) 31:1–10. doi: 10.1007/s12032-014-0057-x
87. Sun X-J, Liu H, Zhang P, Zhang X-D, Jiang Z-W, and Jiang C-C. miR-10b promotes migration and invasion in nasopharyngeal carcinoma cells. Asian Pacific J Cancer Prev. (2013) 14:5533–7. doi: 10.7314/APJCP.2013.14.9.5533
88. Hu J, Xu Y, Hao J, Wang S, Li C, and Meng S. MiR-122 in hepatic function and liver diseases. Protein Cell. (2012) 3:364–71. doi: 10.1007/s13238-012-2036-3
89. Ding Y, Zhang C, Zhang J, Zhang N, Li T, Fang J, et al. miR-145 inhibits proliferation and migration of breast cancer cells by directly or indirectly regulating TGF-β1 expression. Int J Oncol. (2017) 50:1701–10. doi: 10.3892/ijo.2017.3945
90. Tao S-f, He H-f, and Chen Q. Quercetin inhibits proliferation and invasion acts by up-regulating miR-146a in human breast cancer cells. Mol Cell Biochem. (2015) 402:93–100. doi: 10.1007/s11010-014-2317-7
91. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. nature. (2010) 464:1071–6. doi: 10.1038/nature08975
92. Sun R, Qin C, Jiang B, Fang S, Pan X, Peng L, et al. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial–mesenchymal transition. Mol Biosyst. (2016) 12:952–62. doi: 10.1039/C5MB00685F
93. Chen ZG, Zhao HJ, Lin L, Liu JB, Bai JZ, and Wang GS. Circular RNA CirCHIPK3 promotes cell proliferation and invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis. Thorac Cancer. (2020) 11:2660–71. doi: 10.1111/1759-7714.13603
94. O’Hara SP, Mott JL, Splinter PL, Gores GJ, and LaRusso NF. MicroRNAs: key modulators of posttranscriptional gene expression. Gastroenterology. (2009) 136:17–25. doi: 10.1053/j.gastro.2008.11.028
95. Zhang B, Zhang Y, Wang J, Zhang Y, Chen J, Pan Y, et al. Screening and identification of a targeting peptide to hepatocarcinoma from a phage display peptide library. Mol Med. (2007) 13:246–54. doi: 10.2119/2006-00115.Zhang
96. Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differentiation. (2010) 17:193–9. doi: 10.1038/cdd.2009.56
97. Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, et al. Inactivation of miR- 34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. (2008) 7:2591–600. doi: 10.4161/cc.7.16.6533
98. Galoian K, Guettouche T, Issac B, Navarro L, and Temple H. Lost miRNA surveillance of Notch. IGFR pathway—road to sarcomagenesis Tumor Biol. (2014) 35:483–92. doi: 10.1007/s13277-013-1068-5
99. Xiong Y, Zhang Y-Y, Wu Y-Y, Wang X-D, Wan L-H, and Zhou L-M. Correlation of over-expressions of miR-21 and Notch-1 in human colorectal cancer with clinical stages. Life Sci. (2014) 106:19–24. doi: 10.1016/j.lfs.2014.04.017
100. Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. (2006) 24:4677–84. doi: 10.1200/JCO.2005.05.5194
101. Zheng X, Huang H, Liu J, Li M, Liu M, and Luo T. Propofol attenuates inflammatory response in LPS-activated microglia by regulating the miR-155/SOCS1 pathway. Inflammation. (2018) 41:11–9. doi: 10.1007/s10753-017-0658-6
102. Wang L, Zhang H, Rodriguez S, Cao L, Parish J, Mumaw C, et al. Notch-dependent repression of miR-155 in the bone marrow niche regulates hematopoiesis in an NF-κB-dependent manner. Cell Stem Cell. (2014) 15:51–65. doi: 10.1016/j.stem.2014.04.021
103. Cheetham SW, Gruhl F, Mattick JS, and Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. (2013) 108:2419–25. doi: 10.1038/bjc.2013.233
104. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
105. Yoshimoto R, Mayeda A, Yoshida M, and Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta (BBA) - Gene Regul Mech. (2016) 1859:192–9. doi: 10.1016/j.bbagrm.2015.09.012
106. Yue L and Guo J. LncRNA TUSC7 suppresses pancreatic carcinoma progression by modulating miR-371a-5p expression. J Cell Physiol. (2019) 234:15911–21. doi: 10.1002/jcp.v234.9
107. Shang C, Guo Y, Hong Y, and Xue YX. Long non-coding RNA TUSC7, a target of miR-23b, plays tumor-suppressing roles in human gliomas. Front Cell Neurosci. (2016) 10:235. doi: 10.3389/fncel.2016.00235
108. Kotake Y, Kitagawa K, Ohhata T, Sakai S, Uchida C, Niida H, et al. Long non-coding RNA, PANDA, contributes to the stabilization of p53 tumor suppressor protein. Anticancer Res. (2016) 36:1605–11.
109. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. (2013) 495:384–8. doi: 10.1038/nature11993
110. Hanniford D, Ulloa-Morales A, Karz A, Berzoti-Coelho MG, Moubarak RS, Sánchez-Sendra B, et al. Epigenetic silencing of CDR1as drives IGF2BP3- mediated melanoma invasion and metastasis. Cancer Cell. (2020) 37:55–70.e15. doi: 10.1016/j.ccell.2019.12.007
111. Zhang Y, Liu Q, and Liao Q. CircHIPK3: a promising cancer-related circular RNA. Am J Transl Res. (2020) 12:6694–704.
112. Xue C, Li G, Zheng Q, Gu X, Bao Z, Lu J, et al. The functional roles of the circRNA/Wnt axis in cancer. Mol Cancer. (2022) 21:108. doi: 10.1186/s12943-022-01582-0
113. Wang Z, Li Y, Kong D, Ahmad A, Banerjee S, and Sarkar FH. Cross-talk between miRNA and Notch signaling pathways in tumor development and progression. Cancer Lett. (2010) 292:141–8. doi: 10.1016/j.canlet.2009.11.012
114. Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. (2007) 17:1298–307. doi: 10.1016/j.cub.2007.06.068
115. Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. (2007) 297:1901–8. doi: 10.1001/jama.297.17.1901
116. Korać P, Antica M, and Matulić M. MiR-7 in cancer development. Biomedicines. (2021) 9:325. doi: 10.3390/biomedicines9030325
117. Rastgar Rezaei Y, Zarezadeh R, Nikanfar S, Oghbaei H, Nazdikbin N, Bahrami-Asl Z, et al. microRNAs in the pathogenesis of non-obstructive azoospermia: the underlying mechanisms and therapeutic potentials. Syst Biol Reprod Med. (2021) 67:337–53. doi: 10.1080/19396368.2021.1951890
118. Liang Y-K, Lin H-Y, Dou X-W, Chen M, Wei X-L, Zhang Y-Q, et al. MiR-221/222 promote epithelial-mesenchymal transition by targeting Notch3 in breast cancer cell lines. NPJ Breast Cancer. (2018) 4:20. doi: 10.1038/s41523-018-0073-7
119. Mohammadi-Yeganeh S, Mansouri A, and Paryan M. Targeting of miR9/NOTCH1 interaction reduces metastatic behavior in triple- negative breast cancer. Chem Biol Drug design. (2015) 86:1185–91. doi: 10.1111/cbdd.12584
120. Guo Q, Qian Z, Yan D, Li L, and Huang L. LncRNA-MEG3inhibits cell proliferation of endometrial carcinoma by repressing Notch signaling. Biomedicine Pharmacotherapy. (2016) 82:589–94. doi: 10.1016/j.biopha.2016.02.049
121. Lee M, Kim HJ, Kim SW, Park S-A, Chun K-H, Cho NH, et al. The long non-coding RNA HOTAIR increases tumour growth and invasion in cervical cancer by targeting the Notch pathway. Oncotarget. (2016) 7:44558. doi: 10.18632/oncotarget.10065
122. Sanchez A, Lhuillier J, Grosjean G, Ayadi L, and Maenner S. The long non-coding RNA ANRIL in cancers. Cancers. (2023) 15:4160. doi: 10.3390/cancers15164160
123. Ghafouri-Fard S, Glassy MC, Abak A, Hussen BM, Niazi V, and Taheri M. The interaction between miRNAs/lncRNAs and Notch pathway in human disorders. Biomedicine Pharmacotherapy. (2021) 138:111496. doi: 10.1016/j.biopha.2021.111496
124. Wu Q, Lu S, Zhang L, and Zhao L. LncRNA HOXA-AS2 activates the notch pathway to promote cervical cancer cell proliferation and migration. Reprod Sci. (2021) 28:3000–9. doi: 10.1007/s43032-021-00626-y
125. Mazarei M, Shahabi Rabori V, Ghasemi N, Salehi M, Rayatpisheh N, Jahangiri N, et al. LncRNA MALAT1 signaling pathway and clinical applications in overcome on cancers metastasis. Clin Exp Med. (2023) 23:4457–72. doi: 10.1007/s10238-023-01179-x
126. Shi J, Guo C, Li Y, and Ma J. The long noncoding RNA TINCR promotes self-renewal of human liver cancer stem cells through autophagy activation. Cell Death Dis. (2022) 13:961. doi: 10.1038/s41419-022-05424-1
127. Farooqi AA, Naureen H, and Attar R. Regulation of cell signaling pathways by circular RNAs and microRNAs in different cancers: Spotlight on Wnt/β-catenin, JAK/STAT, TGF/SMAD, SHH/GLI, NOTCH and Hippo pathways. Semin Cell Dev Biol. (2022) 124:72–81. doi: 10.1016/j.semcdb.2021.04.002
128. Pan Y, Mao Y, Jin R, and Jiang L. Crosstalk between the Notch signaling pathway and non−coding RNAs in gastrointestinal cancers. Oncol Lett. (2018) 15:31–40. doi: 10.3892/ol.2017.7294
129. Liu WG and Xu Q. Upregulation of circHIPK3 promotes the progression of gastric cancer via Wnt/β-catenin pathway and indicates a poor prognosis. Eur Rev Med Pharmacol Sci. (2019) 23(18):7905–12. doi: 10.26355/eurrev_201909_19004
130. Jia Y and Gu W. Up-regulation of circPVT1 in T cell acute lymphoblastic leukemia promoted cell proliferation via miR- 30e/DLL4 induced activating NOTCH signaling. Pathology-Research Pract. (2021) 224:153536. doi: 10.1016/j.prp.2021.153536
131. Girdauskas E, Petersen J, Neumann N, Ungelenk M, Kurth I, Reichenspurner H, et al. MiR-145 expression and rare NOTCH1 variants in bicuspid aortic valve-associated aortopathy. PloS One. (2018) 13:e0200205. doi: 10.1371/journal.pone.0200205
132. Hashemi M, Hasani S, Hajimazdarany S, Mirmazloomi SR, Makvandy S, Zabihi A, et al. Non-coding RNAs targeting notch signaling pathway in cancer: From proliferation to cancer therapy resistance. Int J Biol macromolecules. (2022) 222:1151–67. doi: 10.1016/j.ijbiomac.2022.09.203
133. Xue Y-Z, Li Z-J, Liu W-T, Shan J-J, Wang L, and Su Q. Down-regulation of lncRNA MALAT1 alleviates vascular lesion and vascular remodeling of rats with hypertension. Aging (Albany NY). (2019) 11:5192. doi: 10.18632/aging.102113
134. Feng Y, Yang F, Zheng J, Shi L, Xie T, Lin Y, et al. Circular RNA HIPK3 mediates epithelial–mesenchymal transition of retinal pigment epithelial cells by sponging multiple microRNAs. Sci Rep. (2024) 14:28872. doi: 10.1038/s41598-024-71119-6
135. Zheng L, Cao J, Liu L, Xu H, Chen L, Kang L, et al. Long noncoding RNA LINC00982 upregulates CTSF expression to inhibit gastric cancer progression via the transcription factor HEY1, American Journal of Physiology-Gastrointestinal and Liver. Physiology. (2021) 320:G816–28. doi: 10.1152/ajpgi.00209.2020
136. Proença C, Freitas M, Ribeiro D, Rufino AT, Fernandes E, and Ferreira de Oliveira JMP. The role of flavonoids in the regulation of epithelial-mesenchymal transition in cancer: A review on targeting signaling pathways and metastasis. Medicinal Res Rev. (2023) 43:1878–945. doi: 10.1002/med.21966
137. Meng RD, Shelton CC, Li Y-M, Qin L-X, Notterman D, Paty PB, et al. γ-Secretase inhibitors abrogate oxaliplatin- induced activation of the Notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer Res. (2009) 69:573–82. doi: 10.1158/0008-5472.CAN-08-2088
138. Feng J, Wang J, Liu Q, Li J, Zhang Q, Zhuang Z, et al. DAPT, a γ-secretase inhibitor, suppresses tumorigenesis, and progression of growth hormone-producing adenomas by targeting notch signaling. Front Oncol. (2019) 9:809. doi: 10.3389/fonc.2019.00809
139. Wang M, Xue L, Cao Q, Lin Y, Ding Y, Yang P, et al. Expression of Notch1, Jagged1 and beta-catenin and their clinicopathological significance in hepatocellular carcinoma. Neoplasma. (2009) 56:533–41. doi: 10.4149/neo_2009_06_533
140. Hu Y, Ou Y, Wu K, Chen Y, and Sun W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumor Biol. (2012) 33:1863–70. doi: 10.1007/s13277-012-0446-8
141. Abdi E, Latifi-Navid S, Kholghi-Oskooei V, Pourfarzi F, and Yazdanbod A. Interaction between lncRNAs HOTAIR and MALAT1 tagSNPs in gastric cancer. Br J Biomed Sci. (2021) 78:147–50. doi: 10.1080/09674845.2020.1866260
142. Radtke F, Wilson A, and MacDonald HR. Notch signaling in T-and B-cell development. Curr Opin Immunol. (2004) 16:174–9. doi: 10.1016/j.coi.2004.01.002
143. Hanna GJ, Stathis A, Lopez-Miranda E, Racca F, Quon D, Leyvraz S, et al. A phase I study of the pan-notch inhibitor CB-103 for patients with advanced adenoid cystic carcinoma and other tumors. Cancer Res Commun. (2023) 3:1853–61. doi: 10.1158/2767-9764.CRC-23-0333
144. Li R, Hu Z, Qiao Q, Zhou D, and Sun M. Anti-NOTCH1 therapy with OMP-52 M51 inhibits salivary adenoid cystic carcinoma by depressing epithelial-mesenchymal transition (EMT) process and inducing ferroptosis. Toxicol Appl Pharmacol. (2024) 484:116825. doi: 10.1016/j.taap.2024.116825
145. Hu ZI, Bendell JC, Bullock A, LoConte NK, Hatoum H, Ritch P, et al. A randomized phase II trial of nab-paclitaxel and gemcitabine with tarextumab or placebo in patients with untreated metastatic pancreatic cancer. Cancer Med. (2019) 8:5148–57. doi: 10.1002/cam4.v8.11
146. Lehal R, Zaric J, Vigolo M, Urech C, Frismantas V, Zangger N, et al. Pharmacological disruption of the Notch transcription factor complex. Proc Natl Acad Sci U.S.A. (2020) 117:16292–301. doi: 10.1073/pnas.1922606117
147. Ferrarotto R, Mishra V, Herz E, Yaacov A, Solomon O, Rauch R, et al. AL101, a gamma-secretase inhibitor, has potent antitumor activity against adenoid cystic carcinoma with activated NOTCH signaling. Cell Death Dis. (2022) 13:678. doi: 10.1038/s41419-022-05133-9
148. Yuan Y, Li J, Xue TL, Hu HR, Lin W, Liu SG, et al. Prognostic significance of NOTCH1/FBXW7 mutations in pediatric T cell acute lymphoblastic leukemia: a study of minimal residual disease risk-directed CCLG-ALL 2008 treatment protocol. Leuk Lymphoma. (2022) 63:1624–33. doi: 10.1080/10428194.2022.2032033
149. Shen X, Zhou H, Zhou X, Liu Z, Meng X, Zhang L, et al. 68Ga-grazytracer PET for noninvasive assessment of response to immunotherapy in solid tumors and lymphomas: a phase 1/2 clinical trial. Nat Commun. (2024) 15:8791. doi: 10.1038/s41467-024-53197-2
Keywords: notch signaling pathway, precision medicine, cancer treatment, immunotherapy, targeted therapy
Citation: Nie R-Z, Wang H, Wang S-S, Chen C, Luo H-M, Zhang H-K, Jing Z-H and Li P-F (2025) The role of notch signaling pathway and non-coding RNAs in cancer and inflammation: progress, therapeutic insights, and future directions. Front. Immunol. 16:1567040. doi: 10.3389/fimmu.2025.1567040
Received: 26 January 2025; Accepted: 27 May 2025;
Published: 20 June 2025.
Edited by:
Jere W. McBride, University of Texas Medical Branch at Galveston, United StatesCopyright © 2025 Nie, Wang, Wang, Chen, Luo, Zhang, Jing and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei-Feng Li, cGVpZmVuZ2xpQHp6dWxpLmVkdS5jbg==
 Rong-Zu Nie1,2
Rong-Zu Nie1,2 Hang Wang
Hang Wang Chen Chen
Chen Chen