- 1Diagnostic Department and Public Health Laboratories, Hellenic Pasteur Institute, Athens, Greece
- 2Laboratory of Microbiology, School of Medicine, Democritus University of Thrace, Alexandroupolis, Greece
- 3Laboratory of Clinical Microbiology and Microbial Pathogenesis, School of Medicine, University of Crete, Heraklion, Greece
- 4Bioimaging Unit, Hellenic Pasteur Institute, Athens, Greece
- 5Laboratory of Biochemistry, Department of Chemistry, School of Science and Engineering, University of Crete, Heraklion, Greece
- 6Laboratory of Molecular Genetics, Hellenic Pasteur Institute, Athens, Greece
Introduction: Legionella pneumophila is the causative agent of Legionnaires’ Disease (LD), an atypical pneumonia with potentially fatal outcome. Neutrophils, the first line of defense, infiltrate the lungs during L. pneumophila infection, although the precise immune mechanisms involved remain unclear.
Methods: This study aims to examine in vitro the interaction of neutrophils with L. pneumophila. Neutrophils from healthy individuals were infected with opsonized and non-opsonized bacteria. Phagocytosis was assessed by immunolabeling, and reactive oxygen species (ROS) generation by flow cytometry. The ability of neutrophils to form Neutrophil Extracellular Traps (NETs) in response to L. pneumophila and the impact of these NETs on bacterial proliferation were examined. Immunolabeling and Western blotting were used for specific NET-associated epitope detection.
Results: It was demonstrated that neutrophils phagocytose opsonized L. pneumophila, while non-opsonized bacteria were not phagocytosed. Opsonized bacteria triggered ROS production, unlike non-opsonized bacteria. Neutrophils released NETs upon L. pneumophila interaction in a ROS-independent manner, but these NETs failed to inhibit bacterial proliferation. Notably, IL-1b was detected on NETs.
Discussion: This study provides evidence that neutrophils react to L. pneumophila through phagocytosis, the production of ROS, and NET release. IL-1b on NETs could play a role in complicated LD cases. These findings contribute to the understanding of neutrophil-mediated immune responses in LD.
1 Introduction
Legionella spp. are facultative, intracellular, Gram-negative bacteria which typically inhabit freshwater environments, and their natural hosts are amoebae and protozoa (1, 2). However, they can also multiply in poorly maintained human-made water systems, leading to human infections, with humans considered as accidental hosts (3). Legionella spp. can cause Pontiac fever, which is a relatively mild, self-limiting, flu-like disease, or Legionnaires’ Disease (LD) which is a more severe, life-threatening illness. The primary causative agent of LD is Legionella pneumophila (4). It is an important cause of Community-Acquired Pneumonia (CAP), as well as an occasional cause of Hospital-Acquired Pneumonia (HAP) (5, 6). The severity of LD ranges from a mild cough to a fatal pneumonia with an overall mortality rate of 5-10% (7). Multi-organ failure is a frequent complication correlated with a worse prognosis in LD. Research findings indicate that 61% of individuals experiencing a fatal outcome presented with complications (8).
Human infection occurs through the inhalation of contaminated aerosols, which can be generated by air conditioning systems, cooling towers, spas, fountains, showerheads, etc (9). LD is an atypical pneumonia, manifested with fever, cough, dyspnea, chills, chest pain, headache, and fatigue (6). Moreover, typical clinical manifestation of LD is diarrhea (10). In addition, extrapulmonary manifestations of LD including cardiac, brain, abdominal (gallbladder), joints, and skin involvement have been reported (11). L. pneumophila primarily infects alveolar macrophages and lung epithelial cells; however, it can transmigrate across the lung epithelium barrier (12), leading to organ infiltration, infection of gut epithelial cells, renal dysfunction (13–15), septic shock (15, 16), and ultimately multi-organ failure (17, 18). After invasion into the host cell, the bacterium translocates more than 300 virulence factors using its Type IV Secretion System (T4SS), in order to create a protective niche for intracellular replication (11, 19). For instance, using these effector proteins, the L. pneumophila-containing phagosome is transformed into an endoplasmic reticulum (ER)-like compartment called the Legionella-containing vacuole (LCV) (20), by hijacking protein and membrane material that are involved in vesicle trafficking (21, 22). Additionally, L. pneumophila employs mechanisms to inhibit autophagy (23), as well as to resist the phagosome-lysosome fusion (24, 25), thereby creating a safe replication environment.
Neutrophils are the most prevalent type of immune cells in human blood and are the first cells to migrate to the site of infection through chemotaxis during the innate immune response (26). They are a type of immune cell that play a key role in combating lung infections and have been reported to continuously patrol the lungs (27). Further, they are equipped with an impressive array of antimicrobial strategies which are tightly regulated to avoid damage in host tissues (28, 29). To counteract an infection, neutrophils use phagocytosis, degranulation, Reactive Oxygen Species (ROS) production and the release of Neutrophil Extracellular Traps (NETs) (30). Indeed, the importance of neutrophils as a first-line defense mechanism in the lungs during L. pneumophila infection has been previously reported (31). During pulmonary infection in mice, alveolar macrophages and neutrophils serve as the primary reservoir cells for L. pneumophila as well as generators of proinflammatory cytokines (32). Nonetheless, exacerbated infiltration of neutrophils has been reported as potential cause of much of the pathology associated with LD but their involvement in Legionella infections has not been thoroughly investigated (33, 34).
NETs are extensive web-like structures consisting of cytoplasmic, granular, and nuclear components, assembled on a scaffold of decondensed chromatin (35). NETs exhibit antimicrobial properties not only through pathogen immobilization via entrapment, but also exert a direct microbicidal activity due to antimicrobial peptides (36, 37), histones (38) and DNA (39). Interestingly, the composition of NETs varies according to the inflammatory environment in which they are formed (40) and it has been reported that NET-bound proteins remain at the site of NET formation even after the DNA backbone has been degraded (41). NETs decorated with Interleukin-1 beta (IL-1β) have been previously identified in patients with inflammatory diseases (42–44). IL-1β is a major proinflammatory cytokine which, in peripheral tissues, is essential for the effective clearance of bacterial infections (45, 46). Nevertheless, IL-1β is also involved in the development of various acute and chronic peripheral diseases (47). For instance, excessive acute activation of the IL-1β system plays a significant role in the orchestration of the cytokine storm, which can lead to severe inflammatory responses and subsequently result in a multi-organ failure associated with sepsis (48–53).
Given this context, the aim of this study was to examine in vitro and understand the neutrophil responses that are orchestrated in a potential L. pneumophila infection e.g phagocytosis, ROS generation, NET release and further identify potential mediators implicated in the pathophysiology of LD infection as specific epitopes decorating these NETs.
2 Materials and methods
2.1 Cultivation of Legionella pneumophila
Legionella pneumophila subsp. pneumophila (ATCC 33152™) was cultivated onto Buffered Charcoal-Yeast Extract (BCYE) agar plates (VWR) for 24 hours at 37°C. Bacteria were collected with a loop and resuspended into 1x Phosphate Buffered Saline (PBS) for OD600 measurement. Opsonization of bacteria was performed by incubation with serum from healthy donors for 30 minutes at 37°C.
2.2 Neutrophil and serum isolation
Peripheral blood neutrophils were isolated from heparinized blood of 10 healthy individuals (five female and five male), from whom written consent was taken, by density double-gradient centrifugation using Histopaque-1119 and Histopaque-1077 (Sigma-Aldrich) (54). After isolation, neutrophils were washed once with 1x PBS and cultured into Roswell Park Memorial Institute (RPMI 1640) medium (Gibco BRL) supplemented with 2% serum from the enrolled healthy donors (54). Neutrophil purity was ≥98% as confirmed by flow cytometry using a PE anti-human CD66b antibody (BioLegend). Healthy serum was collected from healthy blood donors with centrifugation at 1500 × g for 10 min at room temperature and stored in aliquots at -80°C until analysis.
2.3 Stimulation studies with opsonized and non-opsonized L. pneumophila
Neutrophils were seeded and cultured in 24-well plates at a density of 1–2 × 105 cells per well in RPMI 1640 medium supplemented with 2% healthy serum, in a 5% CO2 atmosphere, at 37°C as previously described (42). For opsonized conditions, the cultures were supplemented with 2% healthy serum, while for non-opsonized conditions, cultures were maintained in serum-free medium. Neutrophils were incubated for 20 minutes in order to settle and were then stimulated with ionomycin as a positive control and opsonized L. pneumophila at a concentration of ~10 bacteria per neutrophil. For the ROS inhibition studies, neutrophils were pretreated for 15 min at 37°C with 10 μM diphenyleneiodonium chloride (DPI) (Sigma-Aldrich) before the infection. The inhibitory effect of DPI was confirmed by FACS analysis using the FagoFlowEx Kit (exbio) according to manufacturer’s recommendations with neutrophils infected by Escherichia coli as a positive control (Supplementary Figure 1). Different duration of stimulation was used depending on the assay according to optimization experiments: 60 minutes for phagocytosis assay, 30 minutes for ROS release and 3 hours for NET generation.
2.4 Phagocytosis determination
Phagocytosis quantification by immunofluorescence microscopy was performed as previously described (55). Peripheral blood neutrophils were seeded on poly-L-lysine coated coverslips and after stimulation, cells were fixed with 4% paraformaldehyde (PFA). Immunolabeling was conducted with the following antibodies: a rabbit anti-L. pneumophila polyclonal Ab (1/150; Life Technologies), a donkey anti-rabbit IgG (H+L) antibody conjugated to a fluorophore with an emission peak at 568 nm (1/500; Biotium), and DNA was counterstained by 4′,6-diamidino-2-phenylindole (DAPI) (1/10.000; Sigma-Aldrich). Images were acquired using a Leica TCS-SP8 confocal microscope with a 63x objective. Phagocytosis quantification was performed with Imaris v.9.3.1 (Oxford Instruments). Initially, the number of nuclei per image was calculated using the surfaces module. Following, a surface was created for the neutrophils (green channel - MPO), which was used as a mask for the red channel (L. pneumophila). In this way, Imaris generates a new channel, which contains only the L. pneumophila bacteria inside the neutrophils. Finally, a surface for the masked red channel was created and the number of L. pneumophila bacteria inside the neutrophils was calculated. Phagocytosis was quantified as the percentage of the number of neutrophils with L. pneumophila bacteria inside them against the total number of neutrophils (number of nuclei) per image.
2.5 NET visualization and quantification
NET visualization was performed by immunofluorescence microscopy as previously described (55). Peripheral blood neutrophils were seeded, stimulated and fixed as previously described. Immunolabeling was conducted with the following antibodies: a rabbit anti-citrullinated H3 (R2+R8+R17) (1/500 dilution; Abcam) polyclonal Ab, a goat anti-myeloperoxidase (MPO)-specific mAb (1/150 dilution; R&D systems) and a mouse anti-IL-1β antibody (1/200 dilution; OriGene). A donkey anti-goat IgG (H+L) antibody conjugated to a fluorophore with an emission peak at 647 nm (1/500; Biotium), a donkey anti-rabbit IgG (H+L) antibody conjugated to a fluorophore with an emission peak at 568 nm (1/500; Biotium), and a donkey anti-mouse IgG (H+L) antibody conjugated to a fluorophore with an emission peak at 488 nm (1/500; Biotium) were utilized as secondary antibodies. DNA was counterstained by DAPI (1/10.000; Sigma-Aldrich). Images were acquired using a Leica TCS-SP8 confocal microscope with a 20x objective. The percentage of NET-releasing cells was determined by eye examination of several fields from every sample. Quantification of NET release was conducted with a MPO/DNA complex ELISA as previously demonstrated (56, 57). Briefly, 50 μl of a 5 μg/ml anti-MPO mAb was coated onto 96-well plates overnight at 4°C. Following washing, 20 μl of samples and 80 μl incubation buffer with peroxidase-labeled anti-DNA mAb were added. The plate was incubated for 2 hours at room temperature with shaking at 300 rpm. After washing, 100 μl ABTS peroxidase substrate was added, and absorbance at 405 nm was measured after 20 minutes in the dark. NET formation was expressed as the percentage increase in absorbance above the control.
2.6 ROS measurement
Measurement of respiratory burst of neutrophils after incubation with different stimuli was performed using the FagoFlowEx Kit (exbio) according to the manufacturer’s instructions with modifications as follows. Purified neutrophils from the healthy individuals were incubated in the presence of dihydrorhodamine 123 (DHR123), for 30 minutes, at 37°C, with non-opsonized as well as with opsonized L. pneumophila. After incubation, cells were lysed, and ROS were determined using a flow cytometer (BD FACSCalibur).
2.7 NET structures isolation and co-cultivation with L. pneumophila
Peripheral blood neutrophils were seeded in 6-well culture plates at a density of 1 × 106 cells per well in RPMI 1640 medium, supplemented with 2% healthy serum, for 20 minutes in order to settle and were then incubated with ionomycin as a positive control and with opsonized L. pneumophila bacteria for another 3 hours (37°C, 5% CO2). After incubation, the supernatant was removed, and cells were washed with fresh medium. NET structures were collected after vigorous agitation. The medium was centrifuged at 50 x g for 5 minutes at 4°C to remove debris and NETs were collected in supernatant phase (58). For the co-cultivation assay assessing the ability of NETs to inhibit L. pneumophila proliferation, heat-inactivated L. pneumophila was used to trigger NET release in neutrophils in order to avoid pre-existing alive bacteria on the NET structures. Extracellular DNA formations were collected and 300μl of each condition was incubated with freshly prepared L. pneumophila bacteria in a ratio of 3:1 for 30 minutes, at 37°C, and after incubation directly plated onto prewarmed BCYE agar plates for the examination of the ability of ionomycin-induced and L. pneumophila-induced NETs to retain L. pneumophila growth.
2.8 NET protein isolation for biochemical assays
To isolate the protein load of the produced NETs, supernatants were removed following incubation with the stimuli for 3 hours in the 6-well plates as mentioned previously, and cells were washed with fresh medium once. DNA was digested with micrococcal nuclease (10 U/ml; Thermo Fisher) for 20 min at 37°C in the presence of 50 mM Tris-HCl pH 8.0 and 5 mM CaCl2. Supernatants were collected and centrifuged at 300 x g for 5 min at 4°C to remove debris and supernatants containing the protein load were transferred to a fresh tube for storage at −80°C until further analysis (59).
2.9 Western Blot
An automated Jess Simple Western Blot system (ProteinSimple, USA) was used according to the manufacturer’s standard method for 12-230-kDa Jess separation module (SMW004). The same volume from each NET protein sample was mixed with 0.1X Sample buffer and Fluorescent 5X Master mix (ProteinSimple) in the presence of fluorescent molecular weight markers and 400 mM dithiothreitol (ProteinSimple) and was denatured at 95°C for 5 min. The ladder and the protein samples were separated in capillaries as they migrated through a separation matrix. After separation, proteins were immobilized by photoactivated capture chemistry on the capillaries. A 1/1000 dilution of the mouse anti- IL-1β antibody and a RTU anti-mouse secondary antibody (ProteinSimple) were utilized for the identification of the target. Peroxyde/luminol-S (ProteinSimple) was used to establish the chemiluminescent revelation. The Compass Simple Western software (Protein Simple) was utilized to capture a digital image of the capillary’s chemiluminescence. The software automatically determined the area, signal/noise ratio, and heights (chemiluminescence intensity).
2.10 Ethics approval statement
All study participants provided written informed consent in accordance with the principles expressed in the Declaration of Helsinki. Participants’ records were anonymized and de-identified prior to analysis to ensure anonymity and confidentiality. The study protocol was approved by the Ethics Committee of the Hellenic Pasteur Institute (Approval Number 3702/12 - 06- 2024).
3 Results
3.1 Neutrophils phagocytose L. pneumophila following opsonization
One of the mechanisms that neutrophils use to eliminate pathogens is phagocytosis. Hence, we investigated whether neutrophils are able to phagocytose L. pneumophila in in vitro conditions. Image analysis by confocal microscopy revealed that healthy control (HC) neutrophils were able to phagocytose serum-opsonized L. pneumophila bacteria, when the latter were added in HC cultures (Figure 1A). In contrast, phagocytosis was absent in non-opsonized L. pneumophila bacteria (Figure 1A). Quantification of phagocytosis was performed using Imaris v 9.3.1 and was defined as the percentage of neutrophils with L. pneumophila bacteria inside them against the total number of neutrophils (Figure 1B). Additionally, 3D models were constructed using Imaris v 9.3.1 which clearly show that in opsonized conditions, L. pneumophila are inside the neutrophils in contrast to non-opsonized conditions (Figure 1C). These findings demonstrate the ability of neutrophils to phagocytose in vitro L. pneumophila after serum opsonization.
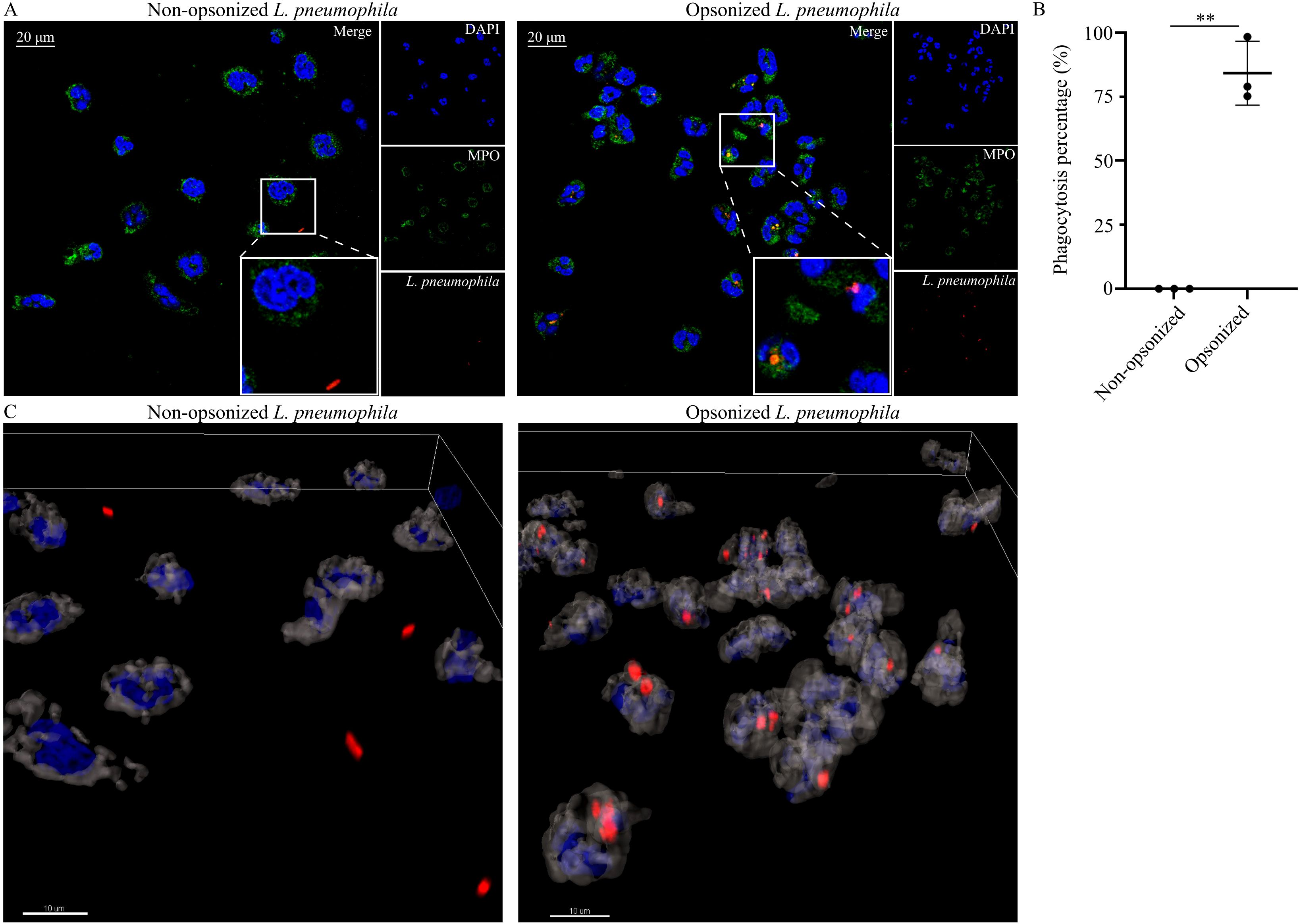
Figure 1. Neutrophils phagocytose opsonized L. pneumophila. (A) HC neutrophils cocultured with either non-opsonized (serum-free conditions) L. pneumophila (mean ± SD; 0.0% ± 0, n=3) or serum-opsonized L. pneumophila (mean ± SD; 84.19% ± 12.45, n=3. P = 0.0072), (confocal microscopy; blue: DAPI/DNA, green: MPO, red: L. pneumophila). Images were acquired with a Leica TCS SP8 confocal microscope using a 63x objective. Scale bar, 20 μm. (B) Quantification of phagocytosis using Imaris v 9.3.1. Data from 3 independent experiments in panel (B) are presented as means ± SDs (**p <0.01). (C) 3D reconstruction of neutrophils infected with opsonized and non-opsonized L. pneumophila. Scale bar, 10 μm.
3.2 Neutrophils produce ROS in a phagocytosis-dependent manner in response to L. pneumophila
It has been previously shown that neutrophils are able to produce ROS as another defense mechanism in response to L. pneumophila (60). However, the implication of phagocytosis in the generation of ROS was not investigated. To assess the role of phagocytosis in ROS generation in neutrophils by L. pneumophila, HC neutrophils were cocultured with L. pneumophila in the presence or absence of opsonization conditions. In the presence of opsonization, coculturing with L. pneumophila indicated a statistically significant increase of ROS generation compared to HC neutrophils, as demonstrated by flow cytometry (Figure 2B). In the absence of opsonization, L. pneumophila did not induce ROS generation (Figure 2A) compared to controls. These findings indicate that ROS generation is induced in neutrophils by L. pneumophila bacteria in a highly phagocytosis-dependent manner since non-opsonized bacteria lose the capacity to induce ROS.
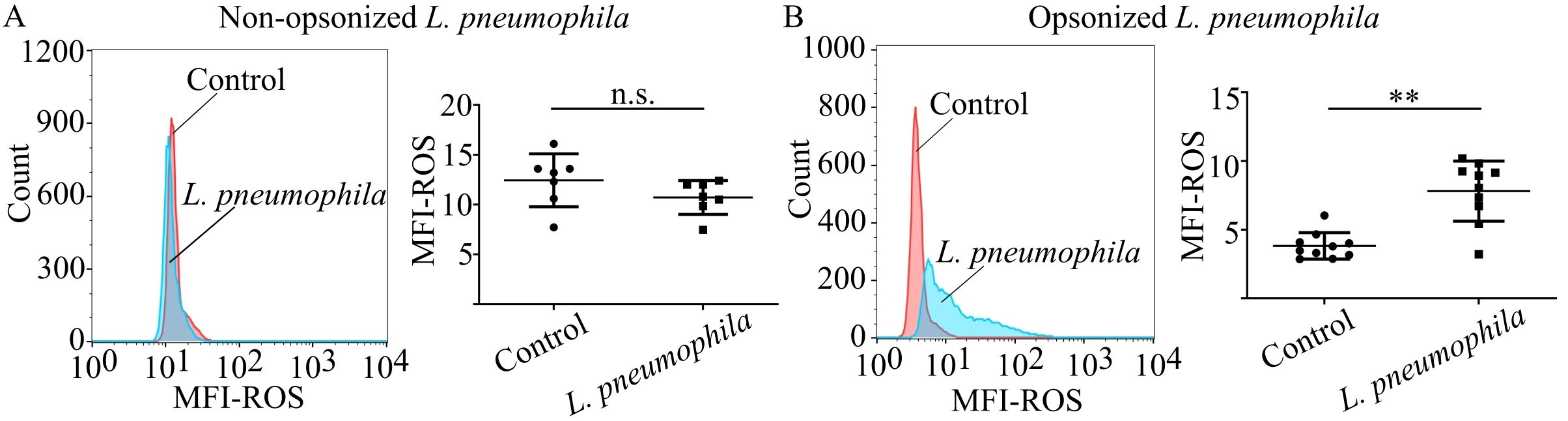
Figure 2. Neutrophils produce ROS in response to L. pneumophila in a phagocytosis-dependent manner. ROS production in neutrophils from (A) HC neutrophils (mean ROS MFI ± SD; 12.45 ± 2.654, n=7) cocultured with non-opsonized L. pneumophila in serum-free conditions (mean ROS MFI ± SD; 10.72 ± 1.703, n=7. p = 0.0903) and (B) HC neutrophils (mean ROS MFI ± SD; 3.835 ± 0.9617, n=10) cocultured with opsonized L. pneumophila (mean ROS MFI ± SD; 7.819 ± 2.190, n=10. p = 0.002). Data from 7 independent experiments in panel (A) and 10 independent experiments in panel (B) are presented as means ± SDs (n.s., not significant, p >0.05; **p <0.01).
3.3 Neutrophils release NETs as a response to L. pneumophila
Taking into account the ability of neutrophils to generate NETs as a defense mechanism in a potential infection (28, 30), we investigated whether L. pneumophila infection could potentially induce this defense mechanism. Coculturing of HC neutrophils with opsonized L. pneumophila indicated statistically significant increased NETs release compared to controls (Figure 3B), as observed by immunofluorescence (Figure 3A) and quantified by MPO/DNA complex ELISA (Figure 3C). HC neutrophils cocultured with L. pneumophila demonstrated lower levels of NETs release compared to ionomycin treated HC neutrophils (Figure 3B).
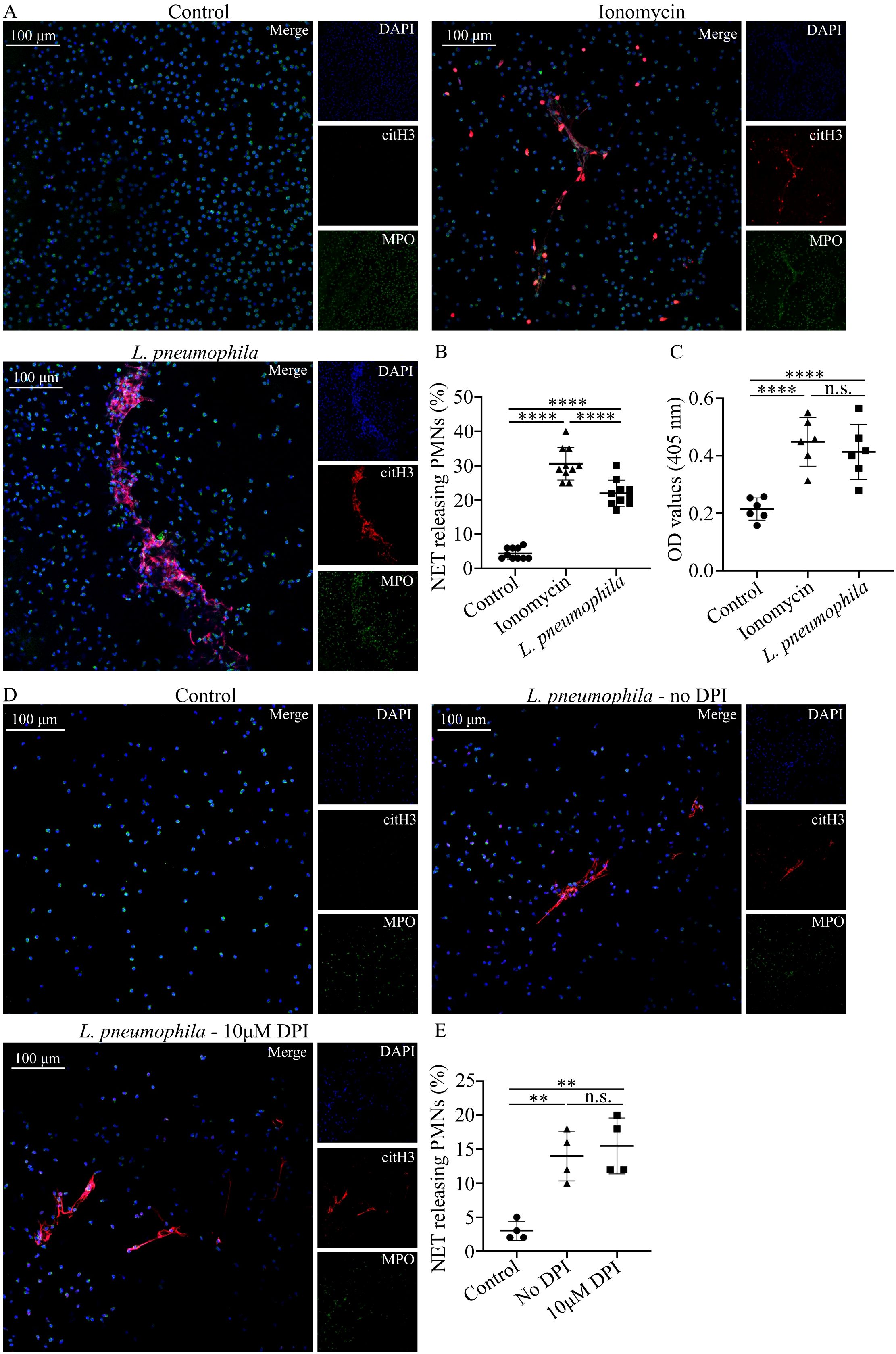
Figure 3. L. pneumophila induces NETs in neutrophils in vitro in a ROS-independent manner. (A) NETs in HC peripheral blood neutrophils (mean ± SD; 4.4% ± 1.647, n=10), either treated with ionomycin (mean ± SD; 30.6% ± 4.742, n=10, p < 0.0001) or cocultured with L. pneumophila (mean ± SD; 22% ± 3.801, n=10. p < 0.0001) (confocal microscopy; blue: DAPI/DNA, green: MPO, red: citH3). Images were acquired with a Leica TCS SP8 confocal microscope using a 20x objective. Scale bar, 100 μm. (B) Quantification of NET-releasing neutrophils from (A) and (C) Quantification of NET releasing neutrophils with MPO/DNA complex ELISA. (D) NETs in HC peripheral blood neutrophils cocultured with L. pneumophila in the presence (mean ± SD; 15.5% ± 4.123, n=4. p > 0.9999) or absence (mean ± SD; 14% ± 3.651, n=4.) of DPI. Images were acquired with a Leica TCS SP8 confocal microscope using a 20x objective. Scale bar, 100 μm. (E) Quantification of NET-releasing neutrophils from (D) Data from 10 independent experiments in panel (B), from 6 independent experiments in panel (C) and 4 independent experiments in panel (E) are presented as means ± SDs (n.s., not significant, p >0.05; **p <0.01; ****p <0.0001).
Previous reports demonstrated that NET release triggered by Gram-negative pathogens is ROS-dependent (61, 62). Since L. pneumophila has the capacity to induce ROS to HC neutrophils we investigated if the observed increase in NETs release is ROS-dependent. HC neutrophils pre-treated with DPI before the addition of L. pneumophila demonstrated similar NET release to HC treated with L. pneumophila alone (Figures 3D, E). This finding indicates that NET release by L. pneumophila is ROS-independent.
3.4 NETs are unable to restrain L. pneumophila growth in vitro
Given the bactericidal properties of NETs (30) and the ability of neutrophils to release NETs in response to L. pneumophila, we examined whether these NET structures can inhibit L. pneumophila growth in vitro. L. pneumophila-induced NET structures in vitro did not have an effect on the growth of L. pneumophila compared to control conditions, as observed macroscopically (Figures 4A, B). Similarly, ionomycin-induced NET structures had no significant effect. This finding indicates that L. pneumophila growth is not inhibited by NETs.
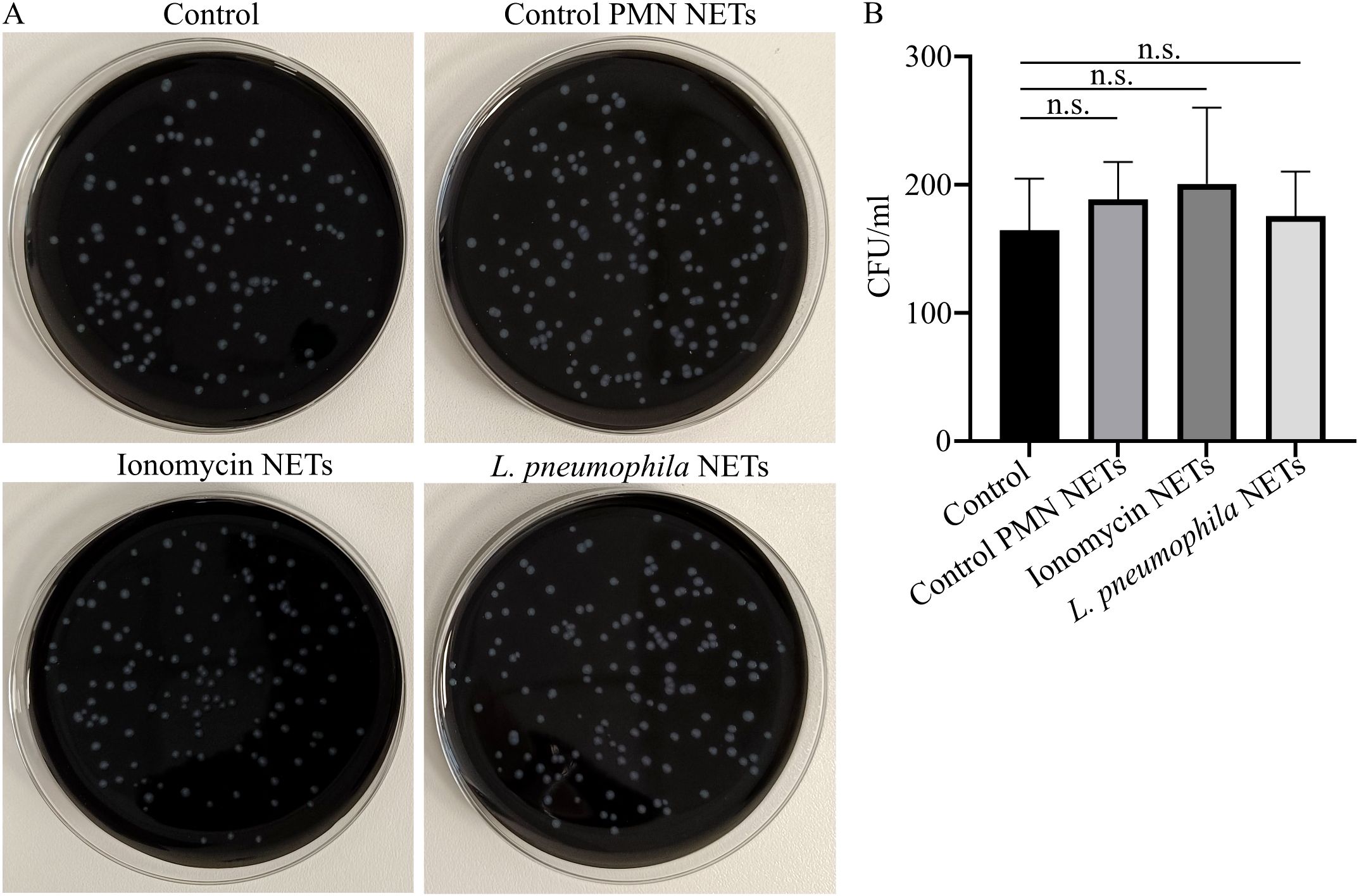
Figure 4. L. pneumophila proliferation is not inhibited in the presence of NET structures. (A) L. pneumophila cultures in BCYE plates (mean CFU/ml ± SD; 195.3 ± 80.17, n=4.) in the presence or absence of NETs induced by different stimuli; ionomycin (mean CFU/ml ± SD; 211.3 ± 74.27, n=4. p = 0.9909) or L. pneumophila (mean CFU/ml ± SD; 202.8 ± 72.41, n=4. p = 0.9990) or control NETs (vehicle from untreated neutrophils) and (B) Quantification of CFU from (A). In panel (B), data from 4 independent experiments are presented as means ± SDs (n.s., not significant, p >0.05).
3.5 Identification of IL-1β on NETs induced by opsonized L. pneumophila
IL-1β is a major proinflammatory cytokine that plays a pivotal role in initiating and regulating the cytokine storm, which can ultimately result in multi-organ failure (51). Considering that multi-organ failure is a common complication of LD and that NETs can express bioactive IL-1β under inflammatory conditions (43, 44, 63, 64), we investigated the presence of IL-1β on NETs induced by L. pneumophila. NETs generated in vitro by HC neutrophils in response to L. pneumophila were decorated with IL-1β as observed by confocal microscopy (Figure 5A) and confirmed by immunoblotting (Figure 5B). On the other hand, IL-1β was not prevalent on ionomycin-induced NETs. This finding suggests that IL-1β is expressed on NETs induced by L. pneumophila in vitro.
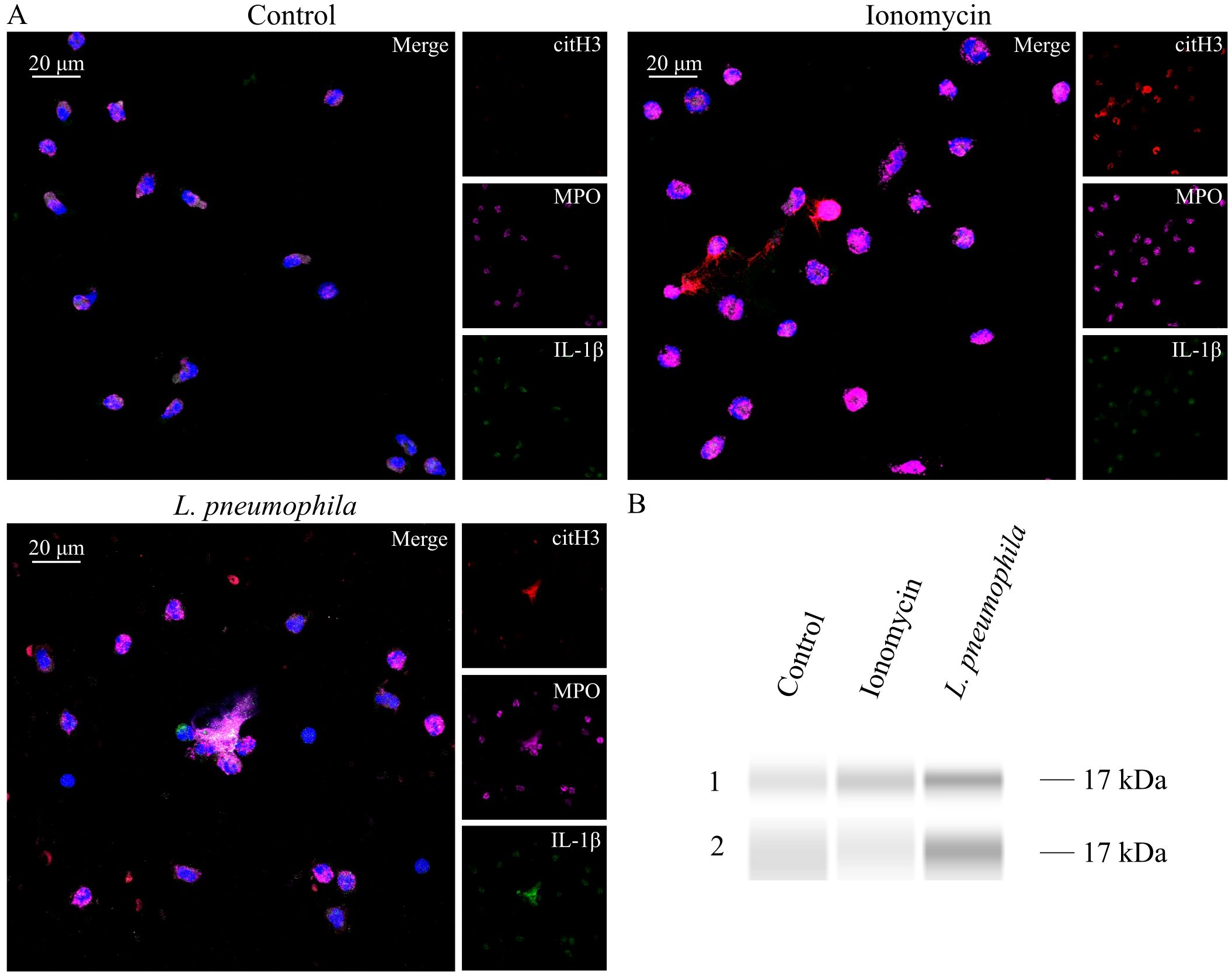
Figure 5. L. pneumophila induces IL-1β expressing NETs in vitro. (A) NETs in HC peripheral blood neutrophils, either treated with ionomycin or cocultured with L. pneumophila (confocal microscopy; blue: DAPI/DNA, red: citH3, magenta: MPO, green: IL−1β). Images were acquired with a Leica TCS SP8 confocal microscope using a 20x objective. Scale bar, 20 μm. (B) Western Blot analysis of NET proteins derived from healthy controls after treatment with ionomycin or coculture with L. pneumophila.
4 Discussion
In this report we demonstrated for the first time that HC neutrophils infected in vitro with opsonized L. pneumophila, produce NETs decorated with IL-1β. Although neutrophils have been reported to release NETs decorated with IL-1β in sterile inflammation (43, 44, 63, 64), the expression of IL-1β on NETs due to an infecting agent has not been reported previously. Moreover, we demonstrated that this NETs release is not ROS-dependent and L. pneumophila growth is unaffected by NETs.
Neutrophils are able to restrict L. pneumophila infection as previously demonstrated (60, 65, 66), however the mechanism of the immune response has only partially been explored. In agreement with the findings of others, we also observed that opsonized L. pneumophila bacteria were significantly phagocytosed by neutrophils, but non-opsonized L. pneumophila were phagocytosed only to a small extend (60, 67). Also, in accordance with previous studies (60, 66), we demonstrated that neutrophils produce ROS in response to L. pneumophila. However, we further determined that ROS production was phagocytosis-dependent, since the infection with opsonized L. pneumophila was resulting in significantly more ROS than non-opsonized bacteria. Moreover, we provide evidence for the first time that opsonized L. pneumophila induced significant NET release in vitro in HC neutrophils. To our knowledge, there is no previous research on the ability of L. pneumophila to induce NET release, nor the role of NETs in host defense against L. pneumophila infection. Several other pneumonia-causing bacteria, such as Streptococcus pneumoniae (68), Staphylococcus aureus (69), Haemophilus influenzae (70), Klebsiella pneumoniae (35), and Mycoplasma tuberculosis (71) have been previously studied and are also known to induce NET release (30). Additionally, we observed that NET release was ROS-independent, since NET release was not affected after the inhibition of ROS. Initially, it was thought that only ROS-dependent NET release was possible (62), however, Pilsczek et al. reported a novel NET release mechanism in response to S. aureus that was ROS-independent (69). Leishmania parasites have also been found to trigger ROS-independent NET release (72).
Functional studies regarding the NET structures induced by opsonized L. pneumophila as well as by ionomycin demonstrated that these NETs were unable to restrain L. pneumophila growth in vitro. Bactericidal properties of NETs alone have also been questioned in the case of S. aureus (73). Likewise, several pneumonia-causing bacteria are known to have evolved mechanisms that escape NETs, like S. pneumoniae (68), S. aureus (74), H. influenzae (70) and M. tuberculosis (71).
In certain instances, NETs not only fail to benefit the host, but instead they may contribute significantly to the pathophysiology of various diseases (75). For example, it has been demonstrated that in sepsis patients, NETs decorated with Tissue Factor are implicated in the initiation of the coagulation cascade, which is a critical step in disseminated intravascular coagulation and ultimately lead to multi-organ failure (76). Additionally, the excessive release of NETs has been linked to multiple organ dysfunction in sepsis, as inhibiting NET release led to diminished organ dysfunction and reduced lethality in septic mice (77). Specifically, the protein load of NETs has also been linked to non-infectious pathologies (78–82). IL-1β was previously identified as a decorative component of NETs in patients with inflammatory diseases like active ulcerative colitis (63) and non-alcoholic steatohepatitis (43). Additionally, IL-1β has been identified on NETs of Familial Mediterranean Fever patients during their attack episodes (44, 64). Interestingly, in Familial Mediterranean Fever patients, circulating levels of IL-1β are not correlated to the severity of the disease, however, they respond highly to IL-1β blockade therapy, indicating its fundamental role in the orchestration of the disease (83, 84).
The ability of IL-1β blockade therapy to prevent multi-organ failure has been previously evaluated. In sepsis patients, in the subgroup of multi-organ dysfunction syndrome, where the inflammasome pathway is implicated, anakinra had been shown to have beneficial effects (85). Additionally, a subgroup of patients with COVID-19 was linked to increased levels of IL-1β, severe cytokine storm and hyperinflammatory symptoms (86, 87). Several studies have investigated the role of anakinra treatment on COVID-19 revealing beneficial results (48, 88–91).
This study’s main limitation is the absence of clinical specimens in order to observe NETs ex vivo or in situ in patient biopsies. This leads to the lack of direct clinical evidence. Furthermore, another limitation is the lack of functional assays of IL-1β on L. pneumophila induced NETs which would clarify its contribution to the disease. Further studies are needed to expand our findings and improve our understanding of the pathophysiology of LD.
In conclusion, we provide evidence that upon L. pneumophila infection, neutrophils fight bacteria by phagocytosis, generation of ROS in a phagocytosis-dependent manner and release of NETs. These findings contribute significantly to the understanding of the effect of neutrophils in neutralizing a L. pneumophila infection. In addition, the presence of NETs decorated with IL-1β in response to L. pneumophila may provide some insight into the pathophysiology of multi-organ failure in LD and could trigger further investigation into its association with the disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the Ethics Committee of the Hellenic Pasteur Institute, Athens, Greece. The studies were conducted in accordance with the local legislation and institutional requirements. All study participants provided written informed consent in accordance with the principles expressed in the Declaration of Helsinki. Participants’ records were anonymized and de-identified prior to analysis to ensure anonymity and confidentiality. The study protocol was approved by the Ethics Committee of the Hellenic Pasteur Institute (Approval Number 3702/12 - 06- 2024).
Author contributions
MK: Writing – original draft, Formal analysis, Investigation. TK: Conceptualization, Formal analysis, Writing – review & editing. DC: Writing – review & editing, Resources. EX: Methodology, Visualization, Writing – review & editing. AP: Resources, Writing – review & editing. GT: Conceptualization, Writing – review & editing. KK: Conceptualization, Supervision, Formal analysis, Methodology, Resources, Visualization, Writing – original draft. EA: Conceptualization, Resources, Supervision, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was carried out within the framework of the Action “Flagship Research Projects in challenging interdisciplinary sectors with practical applications in Greek Industry”, implemented through the National Recovery and Resilience Plan Greece 2.0 and funded by the European Union - NextGenerationEU (project code: TAEDR-0541976).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1573151/full#supplementary-material
Supplementary Figure 1 | HC neutrophils infected with opsonized E. coli bacteria in the absence and presence of 10 μM or 100 μM DPI. Data shown are representative of three independent experiments.
References
1. Richards AM, Von Dwingelo JE, Price CT, and Abu Kwaik Y. Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence. (2013) 4:307–14. doi: 10.4161/viru.24290
2. Albert-Weissenberger C, Cazalet C, and Buchrieser C. Legionella pneumophila - a human pathogen that co-evolved with fresh water protozoa. Cell Mol Life Sci. (2007) 64:432–48. doi: 10.1007/s00018-006-6391-1
3. Boamah DK, Zhou G, Ensminger AW, and O’Connor TJ. From many hosts, one accidental pathogen: the diverse protozoan hosts of legionella. Front Cell Infect Microbiol. (2017) 7:477. doi: 10.3389/fcimb.2017.00477
4. Newton HJ, Ang DK, van Driel IR, and Hartland EL. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev. (2010) 23:274–98. doi: 10.1128/CMR.00052-09
5. Phin N, Parry-Ford F, Harrison T, Stagg HR, Zhang N, Kumar K, et al. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect Dis. (2014) 14:1011–21. doi: 10.1016/S1473-3099(14)70713-3
6. Cunha BA, Burillo A, and Bouza E. Legionnaires’ disease. Lancet. (2016) 387:376–85. doi: 10.1016/S0140-6736(15)60078-2
7. Fukushima S, Hagiya H, Otsuka Y, Koyama T, and Otsuka F. Trends in the incidence and mortality of legionellosis in Japan: a nationwide observational study, 1999-2017. Sci Rep. (2021) 11:7246. doi: 10.1038/s41598-021-86431-8
8. Chidiac C, Che D, Pires-Cronenberger S, Jarraud S, Campese C, Bissery A, et al. Factors associated with hospital mortality in community-acquired legionellosis in France. Eur Respir J. (2012) 39:963–70. doi: 10.1183/09031936.00076911
9. Steinert M, Hentschel U, and Hacker J. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol Rev. (2002) 26:149–62. doi: 10.1111/j.1574-6976.2002.tb00607.x
10. Zarogoulidis P, Alexandropoulou I, Romanidou G, Konstasntinidis TG, Terzi E, Saridou S, et al. Community-acquired pneumonia due to Legionella pneumophila, the utility of PCR, and a review of the antibiotics used. Int J Gen Med. (2011) 4:15–9. doi: 10.2147/IJGM.S15654
11. Iliadi V, Staykova J, Iliadis S, Konstantinidou I, Sivykh P, Romanidou G, et al. Legionella pneumophila: the journey from the environment to the blood. J Clin Med. (2022) 11:6126. doi: 10.3390/jcm11206126
12. Wagner C, Khan AS, Kamphausen T, Schmausser B, Unal C, Lorenz U, et al. Collagen binding protein Mip enables Legionella pneumophila to transmigrate through a barrier of NCI-H292 lung epithelial cells and extracellular matrix. Cell Microbiol. (2007) 9:450–62. doi: 10.1111/j.1462-5822.2006.00802.x
13. Soni AJ and Peter A. Established association of legionella with rhabdomyolysis and renal failure: A review of the literature. Respir Med Case Rep. (2019) 28:100962. doi: 10.1016/j.rmcr.2019.100962
14. Branstetter A and Wyler B. Legionnaires’ Disease causing severe rhabdomyolysis and acute renal failure: A case report. Clin Pract cases Emerg Med. (2022) 6:288–91. doi: 10.5811/cpcem.2022.8.57155
15. Lupia T, Corcione S, Shbaklo N, Rizzello B, De Benedetto I, Concialdi E, et al. Legionella pneumophila Infections during a 7-Year Retrospective Analysis (2016-2022): Epidemiological, Clinical Features and Outcomes in Patients with Legionnaires’ Disease. Microorganisms. (2023) 11:498. doi: 10.3390/microorganisms11020498
16. Andrea L, Dicpinigaitis PV, Fazzari MJ, and Kapoor S. Legionella pneumonia in the ICU: A tertiary care center experience over 10 years. Crit Care Explor. (2021) 3:e0508. doi: 10.1097/CCE.0000000000000508
17. Kassha K, Abuanza I, Hadi SA, and Hilton R. Severe Legionnaires disease complicated by multi-organ dysfunction in a previously healthy patient: a case report. cases J. (2009) 2:9151. doi: 10.1186/1757-1626-2-9151
18. Stout JE and Yu VL. Legionellosis. N Engl J Med. (1997) 337:682–7. doi: 10.1056/NEJM199709043371006
19. Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc Natl Acad Sci U S A. (2013) 110:E707–15. doi: 10.1073/pnas.1215278110
20. Horwitz MA. Formation of a novel phagosome by the Legionnaires’ disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med. (1983) 158:1319–31. doi: 10.1084/jem.158.4.1319
21. Kagan JC and Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. (2002) 4:945–54. doi: 10.1038/ncb883
22. Isberg RR, O’Connor TJ, and Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. (2009) 7:13–24. doi: 10.1038/nrmicro1967
23. Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. (2012) 338:1072–6. doi: 10.1126/science.1227026
24. Horwitz MA. The Legionnaires’ disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. (1983) 158:2108–26. doi: 10.1084/jem.158.6.2108
25. Fernandez-Moreira E, Helbig JH, and Swanson MS. Membrane vesicles shed by Legionella pneumophila inhibit fusion of phagosomes with lysosomes. Infection immunity. (2006) 74:3285–95. doi: 10.1128/IAI.01382-05
26. Segal AW. How neutrophils kill microbes. Annu Rev Immunol. (2005) 23:197–223. doi: 10.1146/annurev.immunol.23.021704.115653
27. Yipp BG, Kim JH, Lima R, Zbytnuik LD, Petri B, Swanlund N, et al. The lung is a host defense niche for immediate neutrophil-mediated vascular protection. Sci Immunol. (2017) 2:eaam8929. doi: 10.1126/sciimmunol.aam8929
28. Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. (2018) 18:134–47. doi: 10.1038/nri.2017.105
29. Vorobjeva NV and Chernyak BV. NETosis: molecular mechanisms, role in physiology and pathology. Biochem (Mosc). (2020) 85:1178–90. doi: 10.1134/S0006297920100065
30. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, et al. Neutrophil extracellular traps kill bacteria. Science. (2004) 303:1532–5. doi: 10.1126/science.1092385
31. Tateda K, Moore TA, Deng JC, Newstead MW, Zeng X, Matsukawa A, et al. Early recruitment of neutrophils determines subsequent T1/T2 host responses in a murine model of Legionella pneumophila pneumonia. J Immunol. (2001) 166:3355–61. doi: 10.4049/jimmunol.166.5.3355
32. Copenhaver AM, Casson CN, Nguyen HT, Fung TC, Duda MM, Roy CR, et al. Alveolar macrophages and neutrophils are the primary reservoirs for Legionella pneumophila and mediate cytosolic surveillance of type IV secretion. Infection immunity. (2014) 82:4325–36. doi: 10.1128/IAI.01891-14
33. Yu H, Higa F, Koide M, Haranaga S, Yara S, Tateyama M, et al. Lung abscess caused by Legionella species: implication of the immune status of hosts. Intern Med. (2009) 48:1997–2002. doi: 10.2169/internalmedicine.48.2647
34. Barry KC, Fontana MF, Portman JL, Dugan AS, and Vance RE. IL-1alpha signaling initiates the inflammatory response to virulent Legionella pneumophila in vivo. J Immunol. (2013) 190:6329–39. doi: 10.4049/jimmunol.1300100
35. Papayannopoulos V, Metzler KD, Hakkim A, and Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J Cell Biol. (2010) 191:677–91. doi: 10.1083/jcb.201006052
36. Yipp BG, Petri B, Salina D, Jenne CN, Scott BN, Zbytnuik LD, et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat Med. (2012) 18:1386–93. doi: 10.1038/nm.2847
37. Konstantinidis T, Kambas K, Mitsios A, Panopoulou M, Tsironidou V, Dellaporta E, et al. Immunomodulatory Role of Clarithromycin in Acinetobacter baumannii Infection via Formation of Neutrophil Extracellular Traps. Antimicrob Agents Chemother. (2016) 60:1040–8. doi: 10.1128/AAC.02063-15
38. Wang Y, Chen Y, Xin L, Beverley SM, Carlsen ED, Popov V, et al. Differential microbicidal effects of human histone proteins H2A and H2B on Leishmania promastigotes and amastigotes. Infection immunity. (2011) 79:1124–33. doi: 10.1128/IAI.00658-10
39. Halverson TW, Wilton M, Poon KK, Petri B, and Lewenza S. DNA is an antimicrobial component of neutrophil extracellular traps. PloS Pathog. (2015) 11:e1004593. doi: 10.1371/journal.ppat.1004593
40. Petretto A, Bruschi M, Pratesi F, Croia C, Candiano G, Ghiggeri G, et al. Neutrophil extracellular traps (NET) induced by different stimuli: A comparative proteomic analysis. PloS One. (2019) 14:e0218946. doi: 10.1371/journal.pone.0218946
41. Kolaczkowska E, Jenne CN, Surewaard BG, Thanabalasuriar A, Lee WY, Sanz MJ, et al. Molecular mechanisms of NET formation and degradation revealed by intravital imaging in the liver vasculature. Nat Commun. (2015) 6:6673. doi: 10.1038/ncomms7673
42. Mitroulis I, Kambas K, Chrysanthopoulou A, Skendros P, Apostolidou E, Kourtzelis I, et al. Neutrophil extracellular trap formation is associated with IL-1beta and autophagy-related signaling in gout. PloS One. (2011) 6:e29318. doi: 10.1371/journal.pone.0029318
43. Arelaki S, Koletsa T, Sinakos E, Papadopoulos V, Arvanitakis K, Skendros P, et al. Neutrophil extracellular traps enriched with IL-1beta and IL-17A participate in the hepatic inflammatory process of patients with non-alcoholic steatohepatitis. Virchows Arch. (2022) 481:455–65. doi: 10.1007/s00428-022-03330-7
44. Apostolidou E, Skendros P, Kambas K, Mitroulis I, Konstantinidis T, Chrysanthopoulou A, et al. Neutrophil extracellular traps regulate IL-1beta-mediated inflammation in familial Mediterranean fever. Ann Rheum Dis. (2016) 75:269–77. doi: 10.1136/annrheumdis-2014-205958
45. Sahoo M, Ceballos-Olvera I, del Barrio L, and Re F. Role of the inflammasome, IL-1beta, and IL-18 in bacterial infections. ScientificWorldJournal. (2011) 11:2037–50. doi: 10.1100/2011/212680
46. Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. (2009) 27:519–50. doi: 10.1146/annurev.immunol.021908.132612
47. Lopez-Castejon G and Brough D. Understanding the mechanism of IL-1beta secretion. Cytokine Growth Factor Rev. (2011) 22:189–95. doi: 10.1016/j.cytogfr.2011.10.001
48. Nazerian Y, Ghasemi M, Yassaghi Y, Nazerian A, and Hashemi SM. Role of SARS-CoV-2-induced cytokine storm in multi-organ failure: Molecular pathways and potential therapeutic options. Int Immunopharmacol. (2022) 113:109428. doi: 10.1016/j.intimp.2022.109428
49. Wang H and Ma S. The cytokine storm and factors determining the sequence and severity of organ dysfunction in multiple organ dysfunction syndrome. Am J Emerg Med. (2008) 26:711–5. doi: 10.1016/j.ajem.2007.10.031
50. Fajgenbaum DC and June CH. Cytokine storm. N Engl J Med. (2020) 383:2255–73. doi: 10.1056/NEJMra2026131
52. Luheshi NM, Rothwell NJ, and Brough D. Dual functionality of interleukin-1 family cytokines: implications for anti-interleukin-1 therapy. Br J Pharmacol. (2009) 157:1318–29. doi: 10.1111/j.1476-5381.2009.00331.x
53. Deitch EA. Multiple organ failure. Pathophysiology and potential future therapy. Ann Surg. (1992) 216:117–34. doi: 10.1097/00000658-199208000-00002
54. Ferrante A and Thong YH. Optimal conditions for simultaneous purification of mononuclear and polymorphonuclear leucocytes from human blood by the Hypaque-Ficoll method. J Immunol Methods. (1980) 36:109–17. doi: 10.1016/0022-1759(80)90036-8
55. Chrysanthopoulou A, Mitroulis I, Apostolidou E, Arelaki S, Mikroulis D, Konstantinidis T, et al. Neutrophil extracellular traps promote differentiation and function of fibroblasts. J Pathol. (2014) 233:294–307. doi: 10.1002/path.4359
56. Caudrillier A, Kessenbrock K, Gilliss BM, Nguyen JX, Marques MB, Monestier M, et al. Platelets induce neutrophil extracellular traps in transfusion-related acute lung injury. J Clin Invest. (2012) 122:2661–71. doi: 10.1172/JCI61303
57. Kessenbrock K, Krumbholz M, Schonermarck U, Back W, Gross WL, Werb Z, et al. Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med. (2009) 15:623–5. doi: 10.1038/nm.1959
58. Saffarzadeh M, Juenemann C, Queisser MA, Lochnit G, Barreto G, Galuska SP, et al. Neutrophil extracellular traps directly induce epithelial and endothelial cell death: a predominant role of histones. PloS One. (2012) 7:e32366. doi: 10.1371/journal.pone.0032366
59. Carmona-Rivera C, Carlucci PM, Moore E, Lingampalli N, Uchtenhagen H, James E, et al. Synovial fibroblast-neutrophil interactions promote pathogenic adaptive immunity in rheumatoid arthritis. Sci Immunol. (2017) 2:eaaz9319. doi: 10.1126/sciimmunol.aag3358
60. Price CTD, Hanford HE, Vashishta A, Ozanic M, Santic M, Uriarte S, et al. Dot/Icm-Dependent Restriction of Legionella pneumophila within Neutrophils. mBio. (2021) 12:e0100821. doi: 10.1128/mBio.01008-21
61. Parker H, Dragunow M, Hampton MB, Kettle AJ, and Winterbourn CC. Requirements for NADPH oxidase and myeloperoxidase in neutrophil extracellular trap formation differ depending on the stimulus. J Leukoc Biol. (2012) 92:841–9. doi: 10.1189/jlb.1211601
62. Fuchs TA, Abed U, Goosmann C, Hurwitz R, Schulze I, Wahn V, et al. Novel cell death program leads to neutrophil extracellular traps. J Cell Biol. (2007) 176:231–41. doi: 10.1083/jcb.200606027
63. Angelidou I, Chrysanthopoulou A, Mitsios A, Arelaki S, Arampatzioglou A, Kambas K, et al. REDD1/autophagy pathway is associated with neutrophil-driven IL-1beta inflammatory response in active ulcerative colitis. J Immunol. (2018) 200:3950–61. doi: 10.4049/jimmunol.1701643
64. Skendros P, Chrysanthopoulou A, Rousset F, Kambas K, Arampatzioglou A, Mitsios A, et al. Regulated in development and DNA damage responses 1 (REDD1) links stress with IL-1beta-mediated familial Mediterranean fever attack through autophagy-driven neutrophil extracellular traps. J Allergy Clin Immunol. (2017) 140:1378–87 e13. doi: 10.1016/j.jaci.2017.02.021
65. Casson CN, Doerner JL, Copenhaver AM, Ramirez J, Holmgren AM, Boyer MA, et al. Neutrophils and Ly6Chi monocytes collaborate in generating an optimal cytokine response that protects against pulmonary Legionella pneumophila infection. PloS Pathog. (2017) 13:e1006309. doi: 10.1371/journal.ppat.1006309
66. Ziltener P, Reinheckel T, and Oxenius A. Neutrophil and Alveolar Macrophage-Mediated Innate Immune Control of Legionella pneumophila Lung Infection via TNF and ROS. PloS Pathog. (2016) 12:e1005591. doi: 10.1371/journal.ppat.1005591
67. Horwitz MA and Silverstein SC. Interaction of the Legionnaires’ disease bacterium (Legionella pneumophila) with human phagocytes. I. L. pneumophila resists killing by polymorphonuclear leukocytes, antibody, and complement. J Exp Med. (1981) 153:386–97. doi: 10.1084/jem.153.2.386
68. Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, and Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol. (2006) 16:401–7. doi: 10.1016/j.cub.2006.01.056
69. Pilsczek FH, Salina D, Poon KK, Fahey C, Yipp BG, Sibley CD, et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J Immunol. (2010) 185:7413–25. doi: 10.4049/jimmunol.1000675
70. Juneau RA, Pang B, Weimer KE, Armbruster CE, and Swords WE. Nontypeable Haemophilus influenzae initiates formation of neutrophil extracellular traps. Infection immunity. (2011) 79:431–8. doi: 10.1128/IAI.00660-10
71. Ramos-Kichik V, Mondragon-Flores R, Mondragon-Castelan M, Gonzalez-Pozos S, Muniz-Hernandez S, Rojas-Espinosa O, et al. Neutrophil extracellular traps are induced by Mycobacterium tuberculosis. Tuberculosis (Edinb). (2009) 89:29–37. doi: 10.1016/j.tube.2008.09.009
72. Rochael NC, Guimaraes-Costa AB, Nascimento MT, DeSouza-Vieira TS, Oliveira MP, Garcia e Souza LF, et al. Classical ROS-dependent and early/rapid ROS-independent release of Neutrophil Extracellular Traps triggered by Leishmania parasites. Sci Rep. (2015) 5:18302. doi: 10.1038/srep18302
73. Parker H, Albrett AM, Kettle AJ, and Winterbourn CC. Myeloperoxidase associated with neutrophil extracellular traps is active and mediates bacterial killing in the presence of hydrogen peroxide. J Leukoc Biol. (2012) 91:369–76. doi: 10.1189/jlb.0711387
74. Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, and von Kockritz-Blickwede M. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun. (2010) 2:576–86. doi: 10.1159/000319909
75. Kaplan MJ and Radic M. Neutrophil extracellular traps: double-edged swords of innate immunity. J Immunol. (2012) 189:2689–95. doi: 10.4049/jimmunol.1201719
76. Kambas K, Mitroulis I, Apostolidou E, Girod A, Chrysanthopoulou A, Pneumatikos I, et al. Autophagy mediates the delivery of thrombogenic tissue factor to neutrophil extracellular traps in human sepsis. PloS One. (2012) 7:e45427. doi: 10.1371/journal.pone.0045427
77. Silva CMS, Wanderley CWS, Veras FP, Sonego F, Nascimento DC, Goncalves AV, et al. Gasdermin D inhibition prevents multiple organ dysfunction during sepsis by blocking NET formation. Blood. (2021) 138:2702–13. doi: 10.1182/blood.2021011525
78. Stakos DA, Kambas K, Konstantinidis T, Mitroulis I, Apostolidou E, Arelaki S, et al. Expression of functional tissue factor by neutrophil extracellular traps in culprit artery of acute myocardial infarction. Eur Heart J. (2015) 36:1405–14. doi: 10.1093/eurheartj/ehv007
79. Kambas K, Chrysanthopoulou A, Vassilopoulos D, Apostolidou E, Skendros P, Girod A, et al. Tissue factor expression in neutrophil extracellular traps and neutrophil derived microparticles in antineutrophil cytoplasmic antibody associated vasculitis may promote thromboinflammation and the thrombophilic state associated with the disease. Ann Rheum Dis. (2014) 73:1854–63. doi: 10.1136/annrheumdis-2013-203430
80. Kambas K, Mitroulis I, and Ritis K. The emerging role of neutrophils in thrombosis-the journey of TF through NETs. Front Immunol. (2012) 3:385. doi: 10.3389/fimmu.2012.00385
81. Frangou E, Chrysanthopoulou A, Mitsios A, Kambas K, Arelaki S, Angelidou I, et al. REDD1/autophagy pathway promotes thromboinflammation and fibrosis in human systemic lupus erythematosus (SLE) through NETs decorated with tissue factor (TF) and interleukin-17A (IL-17A). Ann Rheum Dis. (2019) 78:238–48. doi: 10.1136/annrheumdis-2018-213181
82. Lin AM, Rubin CJ, Khandpur R, Wang JY, Riblett M, Yalavarthi S, et al. Mast cells and neutrophils release IL-17 through extracellular trap formation in psoriasis. J Immunol. (2011) 187:490–500. doi: 10.4049/jimmunol.1100123
83. Mitroulis I, Skendros P, and Ritis K. Targeting IL-1beta in disease; the expanding role of NLRP3 inflammasome. Eur J Intern Med. (2010) 21:157–63. doi: 10.1016/j.ejim.2010.03.005
84. Dinarello CA and van der Meer JW. Treating inflammation by blocking interleukin-1 in humans. Semin Immunol. (2013) 25:469–84. doi: 10.1016/j.smim.2013.10.008
85. Shakoory B, Carcillo JA, Chatham WW, Amdur RL, Zhao H, Dinarello CA, et al. Interleukin-1 receptor blockade is associated with reduced mortality in sepsis patients with features of macrophage activation syndrome: reanalysis of a prior phase III trial. Crit Care Med. (2016) 44:275–81. doi: 10.1097/CCM.0000000000001402
86. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
87. Yang L, Xie X, Tu Z, Fu J, Xu D, and Zhou Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct Target Ther. (2021) 6:255. doi: 10.1038/s41392-021-00679-0
88. Huet T, Beaussier H, Voisin O, Jouveshomme S, Dauriat G, Lazareth I, et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. (2020) 2:e393–400. doi: 10.1016/S2665-9913(20)30164-8
89. Cavalli G, De Luca G, Campochiaro C, Della-Torre E, Ripa M, Canetti D, et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. (2020) 2:e325–e31. doi: 10.1016/S2665-9913(20)30127-2
90. Balkhair A, Al-Zakwani I, Al Busaidi M, Al-Khirbash A, Al Mubaihsi S, BaTaher H, et al. Anakinra in hospitalized patients with severe COVID-19 pneumonia requiring oxygen therapy: Results of a prospective, open-label, interventional study. Int J Infect diseases: IJID. (2021) 103:288–96. doi: 10.1016/j.ijid.2020.11.149
Keywords: Legionella pneumophila, Legionnaires’ Disease, neutrophils, neutrophil extracellular traps, NETs, IL-1beta
Citation: Koutantou M, Konstantinidis T, Chochlakis D, Xingi E, Psaroulaki A, Tsiotis G, Kambas K and Angelakis E (2025) IL-1beta expressing neutrophil extracellular traps in Legionella pneumophila infection. Front. Immunol. 16:1573151. doi: 10.3389/fimmu.2025.1573151
Received: 08 February 2025; Accepted: 21 May 2025;
Published: 06 June 2025.
Edited by:
René Köffel, Zurich University of Applied Sciences, SwitzerlandReviewed by:
Rafael M. Mariante, Oswaldo Cruz Foundation (Fiocruz), BrazilSanjeeb Shrestha, Kyungpook National University, Republic of Korea
Copyright © 2025 Koutantou, Konstantinidis, Chochlakis, Xingi, Psaroulaki, Tsiotis, Kambas and Angelakis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emmanouil Angelakis, ZS5hbmdlbGFraXNAcGFzdGV1ci5ncg==; Konstantinos Kambas, a2thbXBhc0Bob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work and share last authorship
 Myrto Koutantou
Myrto Koutantou Theocharis Konstantinidis
Theocharis Konstantinidis Dimosthenis Chochlakis
Dimosthenis Chochlakis Evangelia Xingi
Evangelia Xingi Anna Psaroulaki
Anna Psaroulaki Georgios Tsiotis
Georgios Tsiotis Konstantinos Kambas
Konstantinos Kambas Emmanouil Angelakis1*†
Emmanouil Angelakis1*†