- 1Department of Immunology, Nankai University School of Medicine, Nankai University, Tianjin, China
- 2Translational Medicine Institute, Affiliated Tianjin Union Medical Center of Nankai University, Tianjin, China
- 3State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, China
Lipopolysaccharide (LPS, endotoxin) -mediated signaling of caspase-4 (human) and -11 (rodent) can induce the maturation of inflammatory cytokine IL-1β and cell pyroptosis, which is associated with the pathophysiology of many diseases such as obesity. However, the process by which LPS induces inflammation through caspase 4/11 is not fully understood. We found here that lncRNA29RIK plays a key role in LPS-mediated maturation of inflammatory cytokine IL-1β and pyroptosis of macrophages. Mechanistic ally, the binding of caspase 4/11 to LPS requires lncRNARIK to cause activation of the caspase 4/11 complex, which ultimately caused inflammation to promote sensitivity to high fat diet (HFD) -mediated obesity. Notably, lncRNA29RIK expression can be up-regulated by LPS. This lncRNA29 is highly conserved between humans and mice. Taken together, these results suggest that lncRNA29RIK determines the occurrence and progression of LPS-related diseases such as obesity.
1 Introduction
Low-grade inflammation is a frequent feature of metabolic diseases such as obesity (1). LPS has been identified as a key contributing factor in the initiation and progression of the inflammation (2). It can activate macrophages in tissues such as adipose tissues to induce inflammation (3). Macrophages, as principal phagocytic components of the immune system, are among the primary regulators of innate immunity responsible for a broad range of inflammatory processes (4).
10LPS-mediated human caspase-4 and mouse caspase-11 (caspase-4/11) signaling can cause maturation of inflammatory cytokine IL-1β and cytopyroptosis of macrophages. The pyroptosis is executed by the pore-forming protein gasdermin-D (GSDMD), which is activated by cleavage mediated by caspase-4/11. The active caspase-4/11 can cleave GSDMD, which forms GSDMD pore (pyroptosis) to cause NLRP3 activation (5). Activation of NLRP3 inflammasomes elicits caspase-1 cleavage, which can induce interleukin-1β (IL-1β) production (6). Notably, caspase-4/11 activation in macrophages with LPS or Gram-negative bacteria requires the expression of interferon (IFN)-inducible guanosine triphosphate (GTP)ases, such as guanylate-binding proteins (GBPs) and/or immunity-related GTPases (IRGs) (7–10). A high molecular weight complex formed by GBP and LPS could promote the recruitment of caspase-4/11 and subsequently transfer LPS onto caspase-4/11 to trigger its activation (11). The macrophages lacking GBPs show impaired caspase-4/11 activation and attenuated pyroptosis (7). However, it is not clear how this process, GBP and LPS mediated the recruitment of caspase-4/11, occurs.
Non-coding RNAs (ncRNAs) are a class of RNA transcripts lacking the ability to encode peptides or proteins. These lncRNAs are now recognized as playing crucial roles in numerous cellular processes, including the cell cycle, differentiation, and metabolism, and in disease. In the cytoplasm, lncRNAs can function to mediate signal transduction pathways, translational programs, and posttranscriptional control of gene expression. For example, a recent study revealed that lncRNAs serve as scaffolds in the cytoplasm to nucleate complex networks of proteins functioning in regulating signaling transduction programs, such as the Toll-like receptor/TIR-domain containing adapter-inducing IFN-β (TLR-TRIF) immune pathway (12). Based on the ubiquity of protein–RNA interactions, many studies have emphasized how their perturbations are related to pathology, including autoimmune diseases, neurological diseases, and cancer (13). We here found that that LPS-mediated LncRNA lncRNA29RIK in macrophages can promote the oligomerization of the LPS/caspase-4/11 complexes upon exposure to LPS/dotap, which can cause tissue inflammation and promote sensitivity to obesity through the LPS-mediated release of mature (m) IL-1β and pyroptosis of macrophages.
2 Materials and methods
Reagents and oligoes used in this study are listed in Supplementary Table S1.
2.1 Mice and cell lines
LncRNA29Rik-deficient mice on a C57BL/6J background were generated by the Model Animal Research Center of Nanjing University (Nanjing, Jiangsu, China) using CRISPR-Cas9 system as previously reported by us (14, 15). Caspase1/11 knockout (KO) mice were from Prof. Shao, National Institute of Biological Sciences, Beijing. All mice were maintained under specific pathogen-free (SPF) conditions in the Animal Center of Nankai University. All animal experiments were approved (Ethic approval no: NK-202019) and carried out in accordance with Nankai University Guide for the Care and Use of Laboratory Animals. Human embryonic kidney cell line HEK 293T cells were obtained from the American Type Culture Collection.
2.2 Mouse models
For high-fat diet (HFD) model, 6-8-week-old male mice and their control littermates were fed using HFD (D12492; protein, 26.2%; carbohydrate, 26.3%; and fat, 34.9%) and control diets [D12450B (60% of calories may be derived from fat)], which was from Research Diets, Inc. (NJ, USA).
For Salmonella typhimurium (ATCC14028) infection, Salmonella infection model was performed according to the previous method (16). Briefly, mice were withdrawn from water and food for 4 h before oral gavage treatment. Then, mice were treated with 7.5 mg of streptomycin. At 20 h after streptomycin treatment, mice were withdrawn from water and food again and then infected with S. typhimurium (200 cfu). Mice were weighed every other day for the determination of percent weight change. This was calculated as: % weight change = (weight at day X − day 0/weight at day 0) × 100.
For toxic experiment, mice were intraperitoneally injected with 54 mg/kg LPS (O111:B4, Sigma), survival (time to moribund) were detected, and then, serum concentration of IL-1β were detected.
2.3 Preparation of macrophages
For macrophages from peritoneal cavity of mice, macrophages were generated in the peritoneal cavity of mice by intraperitoneally injecting with 4 mL of 3% thioglycollate medium. After 4 days, 5 mL of cold phosphate-buffered saline (PBS) containing 3% FBS was injected into the peritoneal cavity. Following this injection, a gentle massage was performed, and peritoneal fluid was subsequently isolated. Next, cells derived from the peritoneal washing fluid were seeded at 2 × 106 in RPMI containing 10% FBS. Non-adherent cells were removed 4 h after seeding by extensive washing with medium.
For human monocyte-derived macrophages (HMDM), primary human peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll–Hypaque density gradient centrifugation. CD14+ magnetic isolation kit was used to isolate monocytes/macrophages following the manufacturer’s instructions. Monocytes/macrophages were cultured in DMEM with 10% FBS, 50 ng/mL human M-CSF, and 1% penicillin/streptomycin for 4 days and then used for experiments.
For THP1-derived macrophages (THP), THP-1 cells were treated 24 h with 100 ng/mL PMA. For mouse bone-marrow-derived macrophages (BMDMs), BMDMs were obtained from the bone marrow of the tibia and femur and cultured in DMEM with 10% FBS, 20 ng/ml mouse M-CSF, and 1% penicillin/streptomycin for 6 days and then used for experiments.
2.4 Macrophage stimulation
For macrophages stimulation, macrophages were primed by priming with 2 μg/mL LPS for 4 h followed by treatment with 5 µM nigericin and 2 μg/mL LPS using Dotap transfection reagent for 30 min. THP-1 cells were treated 24 h with 100 ng/mL PMA before overnight stimulation. Then, supernatants were analyzed for IL-1β by ELISA and LDH by LDH detecting kit.
2.5 Metabolism experiments
For glucose and insulin tolerance, baseline blood glucose levels were measured using a Nova Max Plus GlucoseMeter after 5 h of fasting. Mice were then injected intraperitoneally with 2 g glucose per kg body weight in sterile PBS or with 0.5 U insulin per kg body weight, and blood glucose levels were measured at different times after injection.
2.6 Transfection of microRNA, preparation of plasmids, and construction and transduction of shRNA or lncRNA29RIK lentiviruses
For microRNA transfection, peritoneal macrophages were transfected with microRNAs using HiPerFect transfection reagent according to the manufacturer’s instructions. For preparation of plasmids, the sequences or fragments of mouse caspase-11, human caspase-4, and mouse/human lncRNA29RIK were amplified using PCR methods. The PCR products were cloned into the pcDNA™3.1/V5-His TOPO® TA plasmid (Invitrogen). After sequencing, plasmid constructions were used to transfect HEK 293T cells. For lentivirus construction and transduction, a short hairpin RNA (shRNA) target sequence was chosen by BLOCK-iT™ RNAi Designer (Invitrogen). The constructs were made using pGreenPuro™ cloning and expression lentivector kit (System Biosciences Inc.) according to the manual. The negative control (NC) is luciferase control RNA from the kit. For packaging lentivirus particles, lentivector together with pMD2.G and psPAX2 packaging plasmids were co-transfected into 293T cells. Peritoneal macrophages were infected with the lentiviral supernatants in the presence of 8 μg/ml polybrene (Millipore) by centrifugation and then cultured with complete medium for 24 h.
2.7 RNA extraction and qRT-PCR
RNA extraction and qRT-PCR were analyzed according to our previously reported methods (15, 17). The fold changes were calculated using the ΔΔCt method according to the manufacturer’s instructions (Applied Biosystems). All the reactions were run in triplicate.
2.8 Cytosolic and nuclear fractionation
Indicated cells were incubated with hypotonic buffer (25 mM Tris–HCl, pH 7.4, 1 mM MgCl2, 5 mM KCl) on ice for 5 min. An equal volume of hypotonic buffer containing 1% NP-40 was then added, and each sample was left on ice for another 5 min. After centrifugation at 5,000 × g for 5 min, the supernatant was collected as the cytosolic fraction. The pellets were resuspended in nucleus resuspension buffer (20 mM HEPES, pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF) and incubated at 4°C for 30 min. Nuclear fraction was collected after removing insoluble membrane debris by centrifugation at 12,000g for 10 min.
2.9 H&E staining
For hematoxylin/eosin (H&E) staining, previously reported methods were used in this experiment (15, 17, 18). Briefly, lung tissues were fixed in 4% (w/v) paraformaldehyde-buffered saline and embedded in paraffin; 5-µm colon sections were cut and stained with H&E.
2.10 Cell isolation and flow cytometry
Previous reported protocol was used in cell isolation and flow cytometry (19). Briefly, for the staining of lamina propria (LP) lymphocytes, the gut was isolated and cleaned by shaking in ice-cold PBS four times before the tissue was cut into 1-cm pieces. The epithelial cells were removed by incubating the tissue in HBSS with 2 mM EDTA for 30 min at 37°C with shaking. LP cells were isolated by incubating the tissues in digestion buffer [DMEM, 5% fetal bovine serum, 1 mg/mL collagenase IV (Sigma-Aldrich) and DNase I (Sigma-Aldrich, Tianjin, P. R. China)] for 40 min. The digested tissues were then filtered through a 40-mm filter. Cells were resuspended in 10 ml of the 40% fraction of a 40:80 Percoll gradient and overlaid on 5 mL of the 80% fraction in a 15-mL Falcon tube. Percoll gradient separation was performed by centrifugation for 20 min at 1,800 rpm at room temperature. LP cells were collected at the interphase of the Percoll gradient, washed and resuspended in medium, and then stained and analyzed by flow cytometry. Dead cells were eliminated through 7-AAD staining.
For the staining of immune cells in adipose tissues, adipose tissues first were cut into smaller pieces and then digested in digestion buffer (20 μL/mL collagenase I for 35 min. The digested tissues were then filtered through a 40-mm filter.
2.11 Statistical analyses
Two-sided Student’s t-test and one-way ANOVA Bonferroni’s multiple comparison test were used to determine significance. The statistical significance of the survival curves was estimated using Kaplan and Meier method, and the curves were compared using the generalized Wilcoxon’s test. These were performed by GraphPad Prism 5 software (GraphPad Software). A 95% confidence interval was considered significant and was defined as p < 0.05.
2.12 Others
For Western blot and immunoprecipitation, previously reported methods were used in this study (15, 17, 18). RNA immunoprecipitation was performed according to previously reported protocol (15, 17). For immunostaining and RNA-FISH, previously reported protocols were used (15, 17, 18). For biotin-labeled LPS pull-down analyses, the biotin-labeled LPS and streptavidin magnetic beads were added into the cell lyses and incubated for 4 h. After centrifuge, the precipitations were analyzed using immunoblotting. For gene binding motif analyses, sequence logo of gene binding motif was obtained used the MEME software (https://meme-suite.org/meme/) (lower). NCBI database was used to perform sequence comparison to find out homologous sequences.
3 Results
3.1 LPS promotes the expression of lncRNA29RIK
While we investigated gene expression in the myeloid derived cells (MDSCs) generated by co-culturing bone marrow cells with GM-CSF and IL-6 for 4 days, we found that lncRNA lncRNA29RIK could not only express in mouse MDSCs but also in macrophages (Figures 1A-D). However, there were only low levels of expression in other immune cells such as CD4, CD8, and B cells (Figure 1C). Since gut microbiota plays a critical role in the differentiation and function of immune cells through genetic and epigenetic modification (20, 21), we explored the gut-microbiota-derived component(s) to regulate the expression of epigenetic modification factor lncRNA29rik in the macrophages. We screened the effects of gut microbiota components and metabolites such as short-chain fat acids (SCFAs) and bile-acid- and tryptophan-derived metabolites on the macrophages, which were generated in the peritoneal cavity of mice by intraperitoneal injection of thioglycolate medium. Interestingly, lncRNA29rik expression could be regulated by Gram-negative bacterium LPS but not by other gut-microbiota-derived metabolites such as indole acetic acid (IAA) and deoxycholic acid (DCA) (Figure 1E). Increased expression of lncRNA29rik were dose and time dependent in the macrophages after exposure to LPS (Figures 1F,G). Previous studies showed that LPS-mediated activation of macrophages was dependent on TLR4 (22). Indeed, LPS could induce the expression of lncRNA29RIK in the wild-type (WT) but not in TLR4 knockout (KO) macrophages. LncRNA29rik or Kantr in mice and KANTR in humans were located on chromosome X, as shown in Supplementary Figure S1. After sequence similarity analysis, it was found that this lncRNA had 78% homology between mice and humans (Supplementary Figure S1).
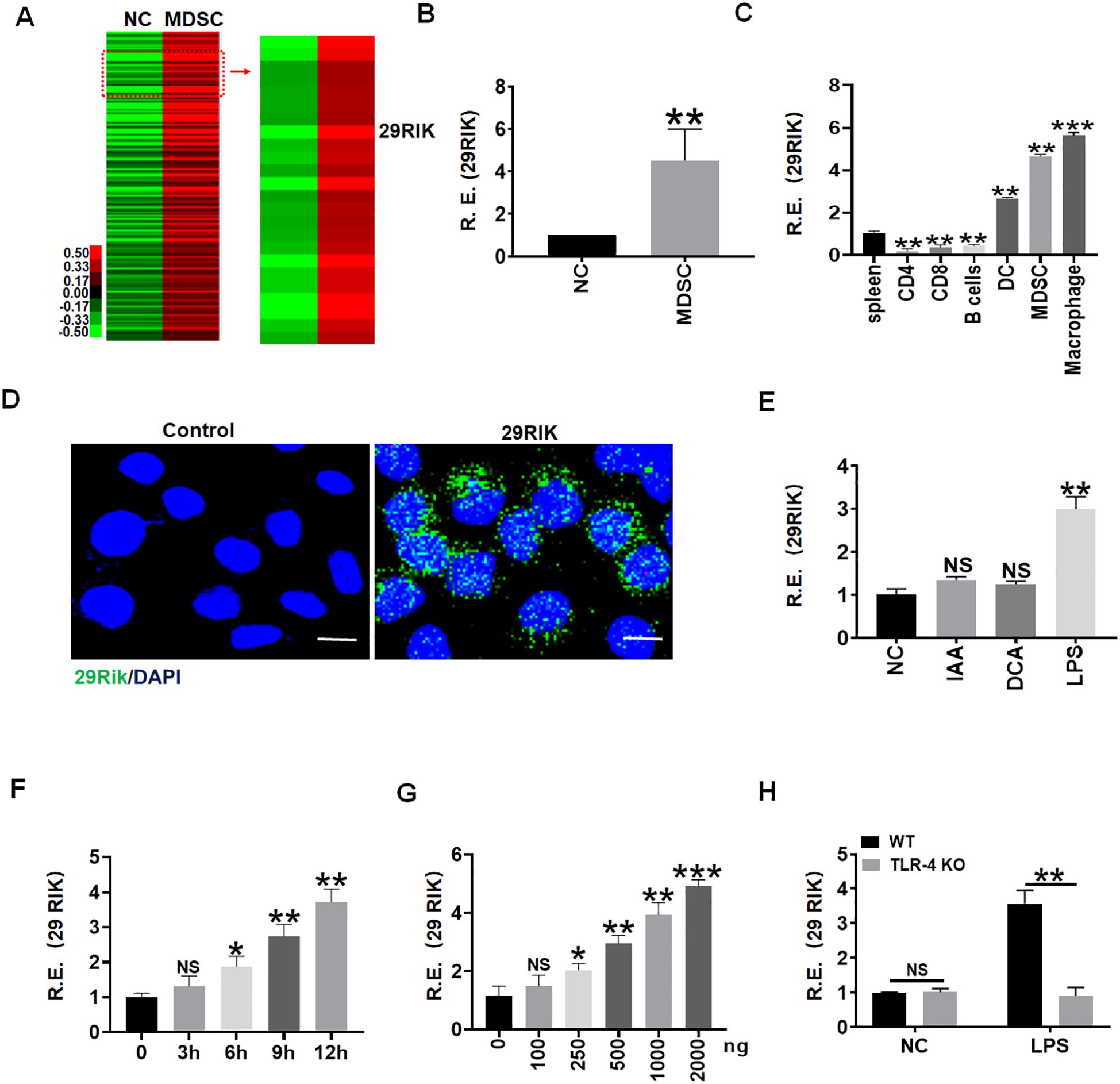
Figure 1. LPS promotes expression of Lnc29rik (29RIK) in macrophages. (A) LncRNA microarray of myeloid-derived suppressor cells (MDSCs). MDSCs were generated, and lncRNA expression was then evaluated using an lncRNA expression microarray. NC, bone marrow cells. (B) QRT-PCR of lncRNA29RIK in MDSCs. NC, bone marrow cells. R. E, relative expression. (C) qRT-PCR of lncRNA29RIK in the spleen, and CD4, CD8, B cell, dendritic cells (DC), MDSC, and macrophages sorted from the spleen by flow cytometry. R. E, relative expression. (D) Fluorescence in situ hybridization of lncRNA29RIK in mouse macrophage upon exposure to LPS (1 μg/mL) for 24 h Nuclei were stained with DAPI (blue); green, lncRNA29RIK. Scale bar, 2.5 μM. (E) qRT-PCR of lncRNA29RIK in the macrophages after exposure to IAA, DCA, or LPS for 24 h (F) qRT-PCR of lncRNA29RIK in the macrophages at the different times after exposure to LPS (1 μg/mL). R. E, relative expression. (G) qRT-PCR of lncRNA29RIK in the macrophages after exposure to different concentrations of LPS. R. E, relative expression. (H) QRT-PCR of lncRNA29RIK in the TLR4 KO macrophages. Ctr., bone marrow cells. R. E, relative expression. One-way ANOVA Bonferroni’s multiple comparison test used in parts C and E-G and Student’s t-test in parts B and H; *p < 0.05, **p < 0.01, ***p < 0.001; NS, not significant.
3.2 LncRNA29RIK promotes the production of mature IL-1β and pyroptosis in macrophages
We next analyzed the function of mouse lncRNA29RIK (mlncRNA29RIK) in macrophages. Silencing mlncRNA29RIK could significantly decrease production of mature IL-1β (mIL-1β), whereas increased mIL-1β could be observed in mlncRNA29RIK overexpressing macrophages after exposing caspase-4/11 ligand, LPS with Dotap (LPS/dotap) (23) (Figures 2A, B). The production of mIL-1β often companies with pyroptosis (24, 25), which is a lytic cell death induced by pathogen infection or endogenous challenge (26). Indeed, caspase-4/11 ligand LPS/dotap also induced more lactate dehydrogenase (LDH) release in the monocytes/macrophages but not in mlncRNA29RIK silencing cells after exposure to LPS/dotap (Figure 2C), suggesting that the pyroptosis of monocytes/macrophages was dependent on mlncRNA29RIK. Indeed, there were more macrophage pyroptosis in mlncRNA29RIK overexpressing macrophages after exposure to LPS/dotap, whereas there were less macrophage pyroptosis in lncRNA29RIK silencing cells as compared to their controls (Supplementary Figure S2).
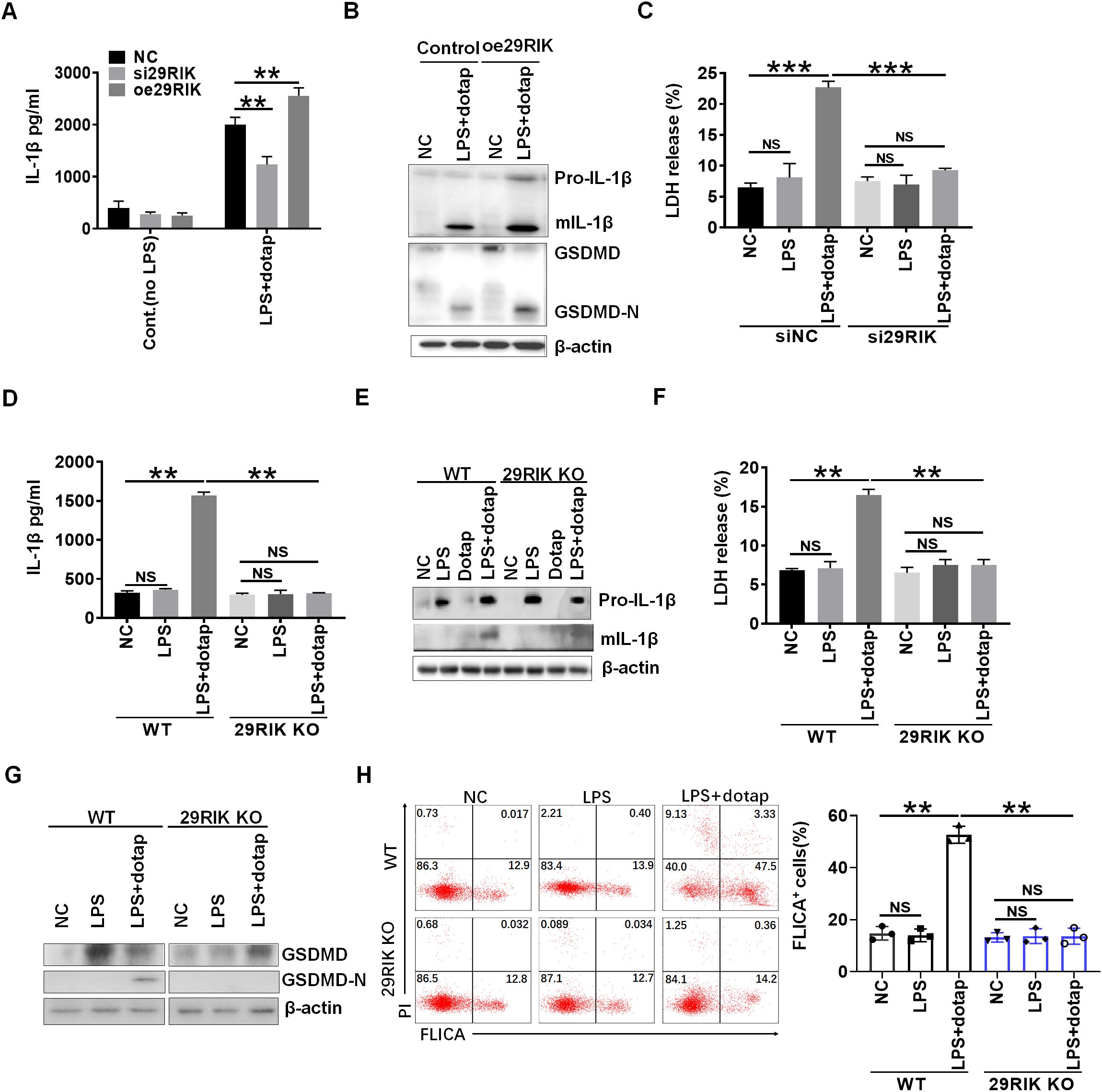
Figure 2. LncRNA29RIK (29RIK) affects mature IL-1β and pyroptosis in the macrophages. (A) ELISA of IL-1β in the supernatants of lncRNA29RIK siRNA (si29RIK) or exogenous lncRNA29RIK (oe29RIK) transfected macrophages after exposure to LPS/dotap. NC, control. (B) Immunoblotting of pro-IL-1β, mature (m) IL-1β, GSDMD, and cleaved-GSDMD (GSDMD-N) in exogenous lncRNA29RIK (oe29Rik) transfected macrophages after exposure to LPS/dotap. NC, no stimulator. (C) Analyses of LDH in the supernatants of lncRNA29RIK siRNA (si29RIK) transfected macrophages after exposure to LPS/dotap. siNC, siRNA control; NC, no stimulator. (D) ELISA of IL-1β in the supernatants of lncRNA29RIK KO (29RIK KO) macrophages after exposure to LPS/dotap. NC, control. (E) Immunoblotting of pro-IL-1β and mIL-1β in lncRNA29RIK KO macrophages after exposure to LPS/dotap. NC, control. (F) Analyses of LDH in the supernatants of lncRNA29RIK KO macrophages after exposure to LPS/dotap. NC, control. (G) Immunoblotting of GSDMD and cleaved-GSDMD (GSDMD-N) in lncRNA29RIK KO macrophages after exposure to LPS/dotap.NC, control. (H) flow cytometry of lncRNA29RIK KO macrophages after FLICA staining on exposure to LPS/dotap; NC, control. Student’s t-test; *p < 0.05, **p < 0.01, ***p < 0.001; NS, not significant.
To further determine the function of mlncRNA29RIK, we generated mlncRNA29RIK knockout (KO) mice. The mlncRNA29RIK KO macrophages were generated in the peritoneal cavity of mice by intraperitoneal injection with thioglycolate medium. There was no difference in the production of mIL-1β and pyroptosis in mlncRNA29RIK KO macrophages with and without exposing to caspase-11 ligands LPS/Dotap, whereas markedly increased production of mIL-1β and pyroptosis could be detected in the macrophages from WT mice (Figures 2D–G), indicating that mlncRNA29RIK plays a critical role in production of mIL-1β and pyroptosis of monocytes/macrophages. MlncRNA29RIK-mediated pyroptosis of monocytes/macrophages were further confirmed by flow cytometry through FLICA staining (Figure 2H). Notably, mlncRNA29RIK KO did not significantly change the expression of cytokines and co-stimulation and also differentiation of myeloid derived cells (Supplementary Figure S3). Taken together, these results indicate that mlncRNA29RIK play a role in LPS-mediated production of mIL-1β and macrophage pyroptosis.
3.3 LncRNA29RIK KO reduces sensitivity of mice to high-fat </b><b>diet-mediated obesity
Obesity is strictly a relationship with LPS-mediated inflammatory macrophages in adipose tissues (27, 28). The high-fat diet (HFD) can induce the disruption of the intestinal barrier, which cause the release of bacterial metabolites and endotoxins, such as LPS into the circulation (27). Since LPS-mediated lncRNA29RIK could promote the production of IL-1β in the macrophages, we next employed HFD-mediated obesity model to detect the effects of lncRNA29RIK KO on the obesity. While mice were fed with HFD, lncRNA29RIK KO mice showed marked resistance to HFD-induced obesity, including less body weight and weight of fat-pad tissues, increased sensitivity to insulin, and tolerance to glucose (Figures 3A–C). Histochemical staining showed less adipose cells in lncRNA29RIK KO mice than WT mice (Figure 3D). mIL-1β was also much lower in the adipose tissues of lncRNA29RIK KO mice than control WT mice (Figure 3E). Inflammatory cytokines TNFα, IL-6, and MCP-1 were also markedly decreased in in the adipose tissues of lncRNA29RIK KO mice (Figure 3E). There were also less inflammatory macrophages (F4/80/CD11b, F4/80/CD11c and F4/80/TNFα cells) in the adipose tissues of lncRNA29RIK KO mice (Figure 3F). Consistent with other reports, there were also higher levels of LPS in the sera of mice fed HFD as compared to control mice fed normal diet (Figure 3G). Taken together, lncRNA29RIK KO reduces sensitivity of mice to HFD-mediated obesity.
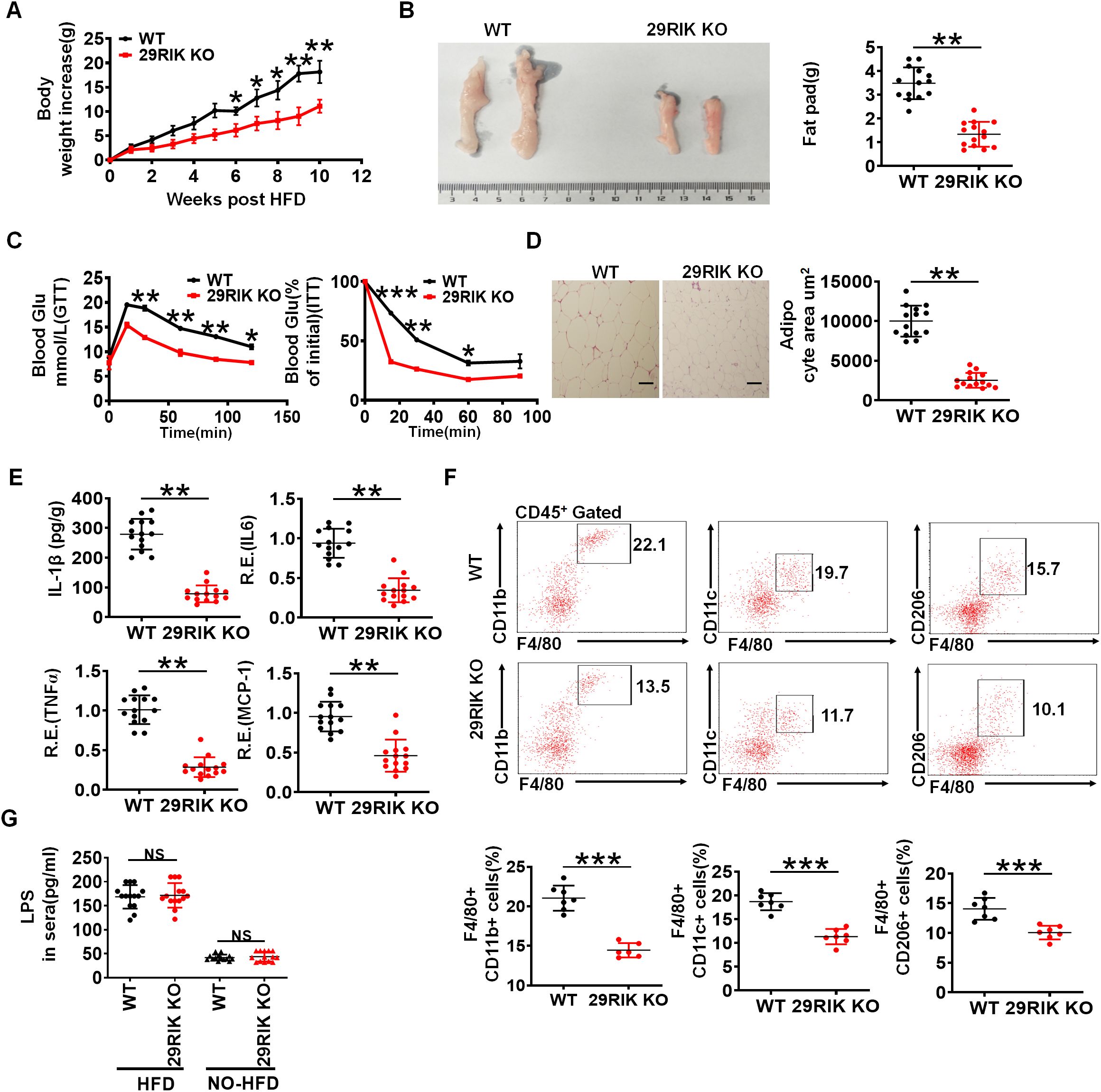
Figure 3. LncRNA29RIK (29RIK) KO mice are resistance to HFD-mediated obesity. (A) Body weight increases in male WT and lncRNA29RIK KO mice fed high-fat diet (HFD) (n=14). These mice are no differences at baseline before feeding HFD. (B) Fat-pad in male WT and lncRNA29RIK KO mice fed HFD for 3 months. (C) Glucose tolerance and insulin sensitivity of WT and lncRNA29RIK KO mice fed HFD for 3 months (n=6). (D) H/E staining of adipose tissues of WT and lncRNA29RIK KO mice fed HFD. One representative. (E) ELISA of mIL-1β and qRT-PCR of TNFα, IL-6 and MCP-1 in the adipose tissues of WT and lncRNA29RIK KO mice fed HFD (n=14). (F) Flow cytometry of F4/80 CD11b. F4/80 CD11C and F4/80 CD206 in adipose tissues of WT and lncRNA29RIK KO mice fed HFD. (G) Concentration of LPS in the sera of WT and lncRNA29RIK KO mice fed HFD (n=14). Analysis of variance test in parts (A, C); Student’s t-test in other panels, mean ± SD. R. E, relative expression. *p < 0.05, **p < 0.01, and ***p < 0.001. Data are a representative of three independent experiments.
3.4 LncRNA29RIK promotes binding of caspase-11 with LPS
We next investigated the mechanism(s) by which mlncRNA29RIK regulated the production of mIL-1β and macrophage pyroptosis. Caspase-11, as a non-canonical inflammasome, was activated by oligomerization of the LPS/caspase-11 complexes (29). Activation of caspase-11 could induce proteolysis of the full length of GSDMD to generate the N-GSDMD pore-forming domain, which migrates to cell membranes to produce the N-GSDMD-mediated membrane pores to facilitate potassium iron efflux. Notably, NLRP3 canonical inflammasomes can be activated via potassium ion efflux, which subsequently induces proteolytic activation of caspase-1 to cause release of mIL-1 β (30) (Figure 4A). Thus, we first analyzed the roles of lncRNA29RIK in the oligomerization of the LPS/caspase-11 complexes. Data showed that lncRNA29RIK could gather together with LPS/caspase-11 upon exposure to LPS/dotap (Figure 4B), implying that lncRNA29RIK exerts its role through caspase-11. LncRNAs can serve as scaffolds in the cytoplasm to nucleate complex networks of proteins functioning in tightly regulated signaling transduction programs (31–33). Bioinformatics analyses showed that this lncRNA29RIK could potentially bind with caspase-11 (Figure 4C). Indeed, immunofluorescence showed that lncRNA29RIK could promote the oligomerization of caspase-11 (Figure 4D, Supplementary Figure S4). However, lncRNA29RIK was not involved in the binding of LPS and GBP, which could promote the recruitment of caspase-4/11 to trigger its activation (11). There was no difference in the binding of LPS and GBP in WT, lncRNA29RIK, and caspase-1/11 KO macrophages, whereas markedly less assemble of LPS and caspase-11, and caspase-11 and GBP1 were observed in lncRNA29RIK KO and caspase 1/11 KO macrophages as compared to WT macrophages (Figure 4D). LPS pull-down experiment also exhibited that binding of LPS with caspase-11 was dependent on lncRNA29RIK (Figure 4E). Interestingly, RNA immunoprecipitation (RIP) showed the direct binding of lncRNA29RIK with CARD domain in caspase-11 (Figure 4F). The interaction of lncRNA29RIK and caspase-11 promoted the degradation of caspase-1 and GSDMD and production of mIL-1 β in WT but not lncRNA29RIK KO macrophages upon exposure to LPS/dotap (Figure 4G). In addition, we also analyzed the coding potential of lncRNA29RIK. The phyloCSF showed that the lncRNA29RIK did not have a coding potential (Supplementary Figure S5). Coding Potential Calculator 2 (CPC2, http://cpc2.gao-lab.org/) analyses also revealed that this lncRNA29RIK did also not have the potential to encode peptides (Supplementary Figure S5). Thus, our results suggest that lncRNA29RIK can bind with caspase-11 to promote the oligomerization of LPS-mediated caspase-11, which can cause production of mIL-1β and pyroptosis of macrophages.
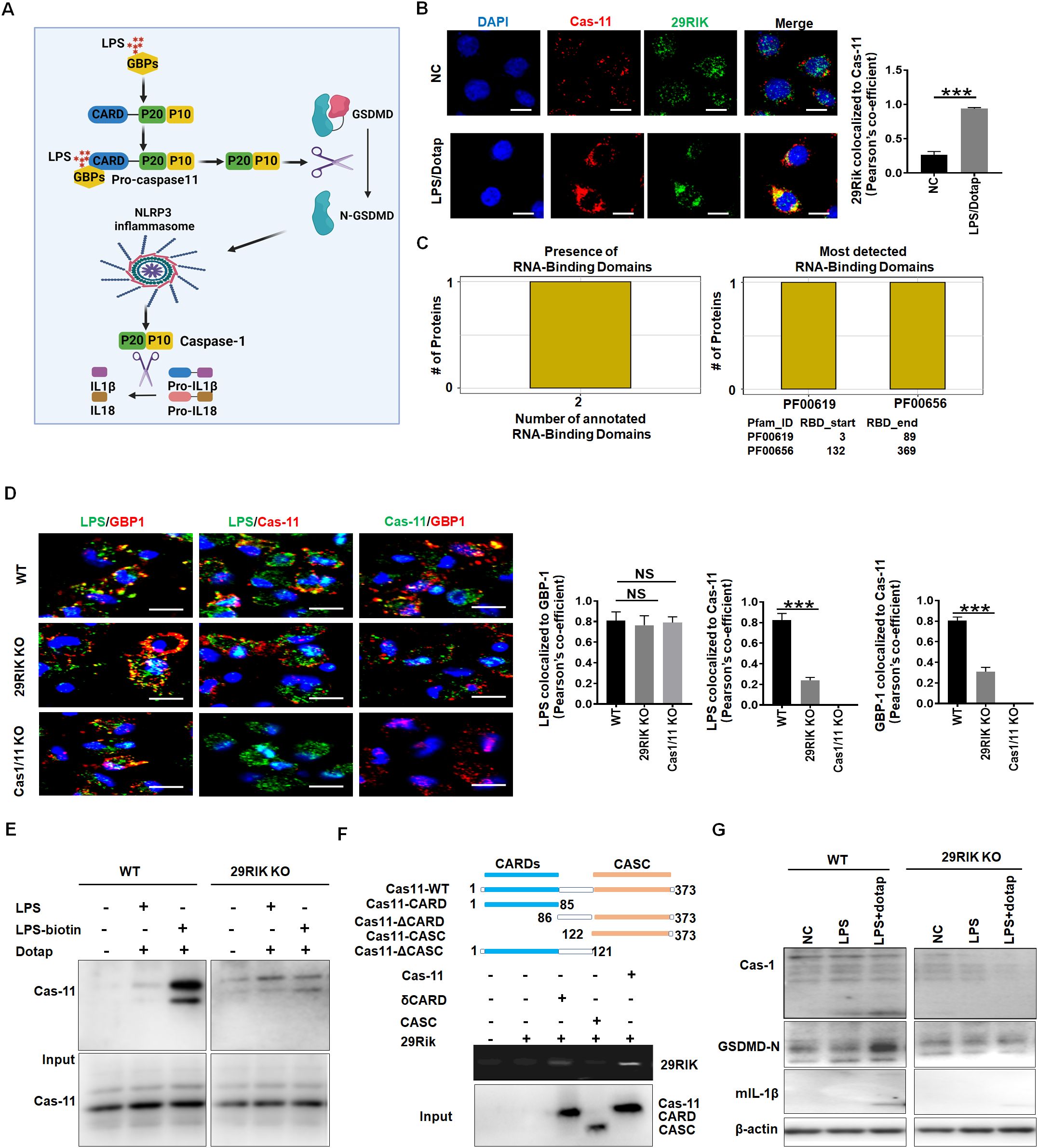
Figure 4. LncRNA29RIK (29RIK) promote the binding of LPS with caspase-11. (A) Schematic illustration showing the LPS mediated mIL-1β and pyroptosis. (B) Immunostaining of caspase-11 and LncRNA29RIK in the macrophages after exposure to LPS/dotap. Pearson’s correlation coefficient indicating co-localization of caspase-11 (cas-11) and LncRNA29RIK (29rik). DAPI, nuclear staining; NC, control; (C) Predicting binding of mouse lncRNA29RIK with caspase-11. The prediction results showed that there were two domains binding to lncRNA29RIK in caspase-11 protein, and the amino acid start and end site of the domain was given. (D) Immunostaining of LPS, GBP1, and caspase-11 (Cas-11) in the WT, LncRNA29RIK (29Rik) KO, and caspase-1/caspase-11 (Casp1/11) KO macrophages. Pearson’s correlation coefficient indicating co-localization of LPS and GBP1, LPS/Casp11, and Cas-11/GBP in 29 RIK KO and Cas-1/11 KO macrophages; WT, control. (E) Immunoblotting of caspase-11 (Cas-11) in the biotin-labeled LPS pull-down lysis of the WT and lncRNA29RIK KO microphages. (F) RIP of V5-tagged caspase-11 derivatives and lncRNA29RIK cotransfected HEK293T cells. RIP was performed using anti-V5. Caspase-11 and its derivatives were cloned into pcDNA3.1/V5 to generate V5-tagged caspase-11 derivatives and then individually transfected into HEK293T cells. (G) Immunoblotting of caspase-1 (Cas-1), cleaved GSDMD (GSDMD-N), and mIL-1β in the WT and lncRNA29RIK KO (29RIK KO) macrophages.
3.5 LncRNA29RIK has similar function to caspase-11
LPS-mediated mIL-1β and pyroptosis of macrophages was through multiple components such as caspase-11 and NLRP3 (30). To further understand how lncRNA29RIK mediated mIL-1β and macrophage pyroptosis, we used KO (KO) mice to compare the basic function of lncRNA29RIK, caspase-11, and NLRP3 in inducing mIL-1 β production and macrophage pyroptosis. Data showed that not only lncRNA29RIK KO but also caspase-11 KO and NLRP3 KO macrophages had reduced mIL-1β production and decreased pyroptosis of macrophages (Figures 5A, B). There were no or weak responses to LPS/dotap stimulation in the production of mIL-1β and cleaved GSDMD in these lncRNA29RIK KO, caspase-11 KO, and NLRP3 KO macrophages upon exposure to LPS/dotap (Figure 5C). Much fewer pyroptosis cells in these KO mice were also observed (Figure 5D). Notably, overexpression of caspase-11 did not rescue IL-1β production in lncRNA29RIK KO macrophages (Figure 5E). Thus, there have been similar function in lncRNA29RIK KO, caspase-11 KO, and NLRP3 KO macrophages in inducing the production of mIL-1β and pyroptosis of macrophages, supporting that the functions of lncRNA29RIK are dependent on caspase-11 and NLRP3 (Figures 5E, F).
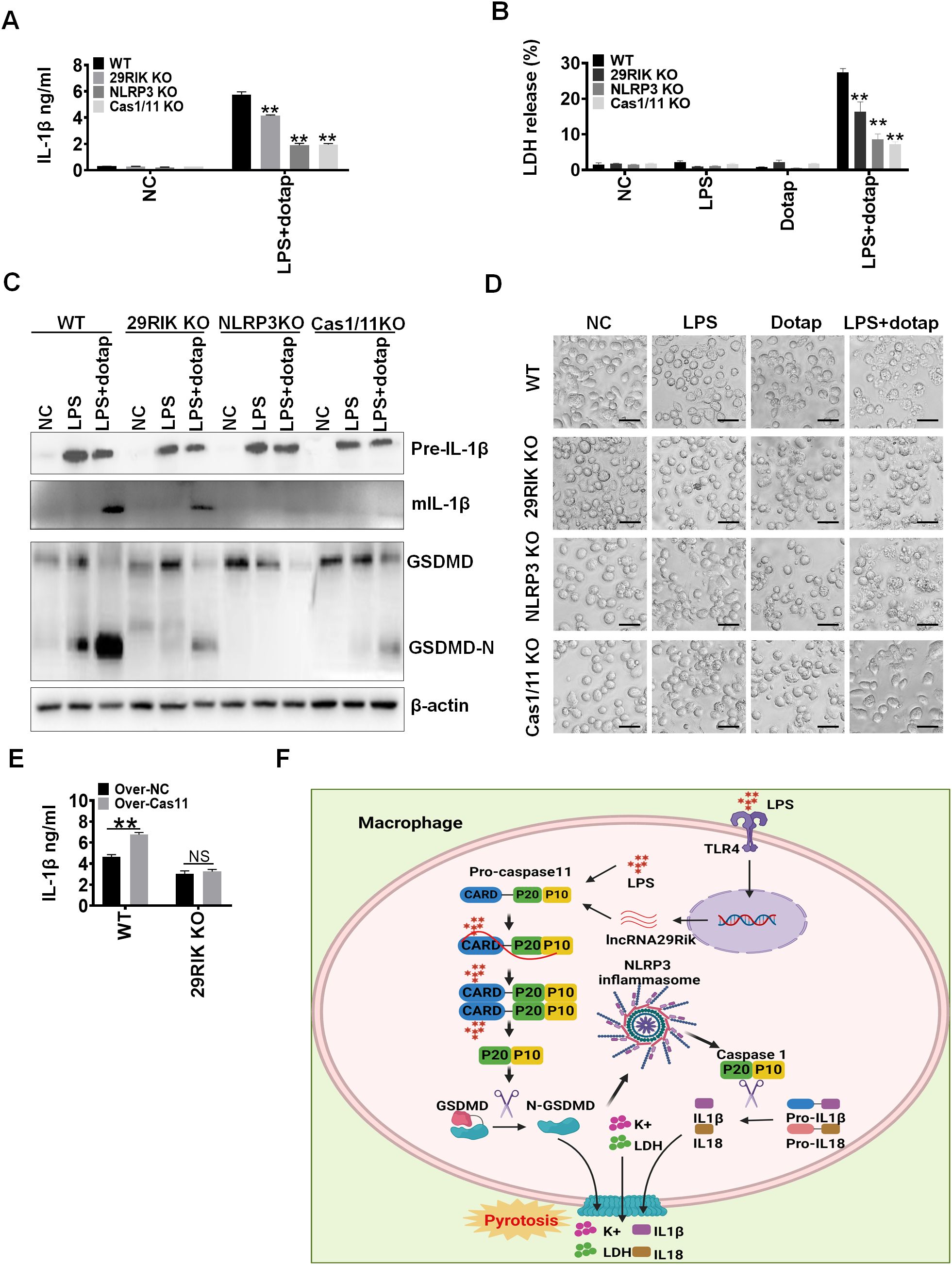
Figure 5. There has a similar function among lncRNA29RIK (29RIK) KO, caspase-1/11 and NLRC3 KO macrophages. (A) ELISA of IL-1β in the supernatants of lncRNA29RIK KO, caspase-1/11, and NLRC3 KO macrophages after exposure to LPS/dotap. NC, control. (B) Analyses of LDH in the supernatants of lncRNA29RIK KO, caspase-1/11, and NLRC3 KO macrophages after exposure to LPS/dotap. NC, control. (C) Immunoblotting of pro-IL-1β, IL-1β, GSDMD, and cleaved GSDMD (GSDMD-N) in the lncRNA29RIK KO, caspase-1/11, and NLRC3 KO macrophages after exposure to LPA/dotap. (D) Pyroptosis of lncRNA29RIK KO, caspase-1/11, and NLRC3 KO macrophages after exposure to LPS/dotap. NC, control. (E) ELISA of IL-1β in the supernatants of caspase-11 overexpressed lncRNA29RIK KO macrophages after exposure to LPS/dotap; WT, control macrophages. (F) Schematic illustration showing a mechanism for lncRNA29RIK to induce production of IL-1β and pyroptosis of macrophages. Student’s t-test; *p < 0.05, **p < 0.01, ***p < 0.001; NS, not significant.
3.6 LncRNA29RIK promotes sensitivity to LPS-mediated inflammation
We next determined the role of macrophage mlncRNA29RIK in LPS or Gram-negative bacteria-mediated inflammation. LPS toxic analyses showed that mlncRNA29RIK KO mice had higher survival rate than WT mice (Figure 6A). In LPS toxic experiments, these lncRNA29RIK KO mice had lower levels of mIL-1β in sera (Figure 6B).
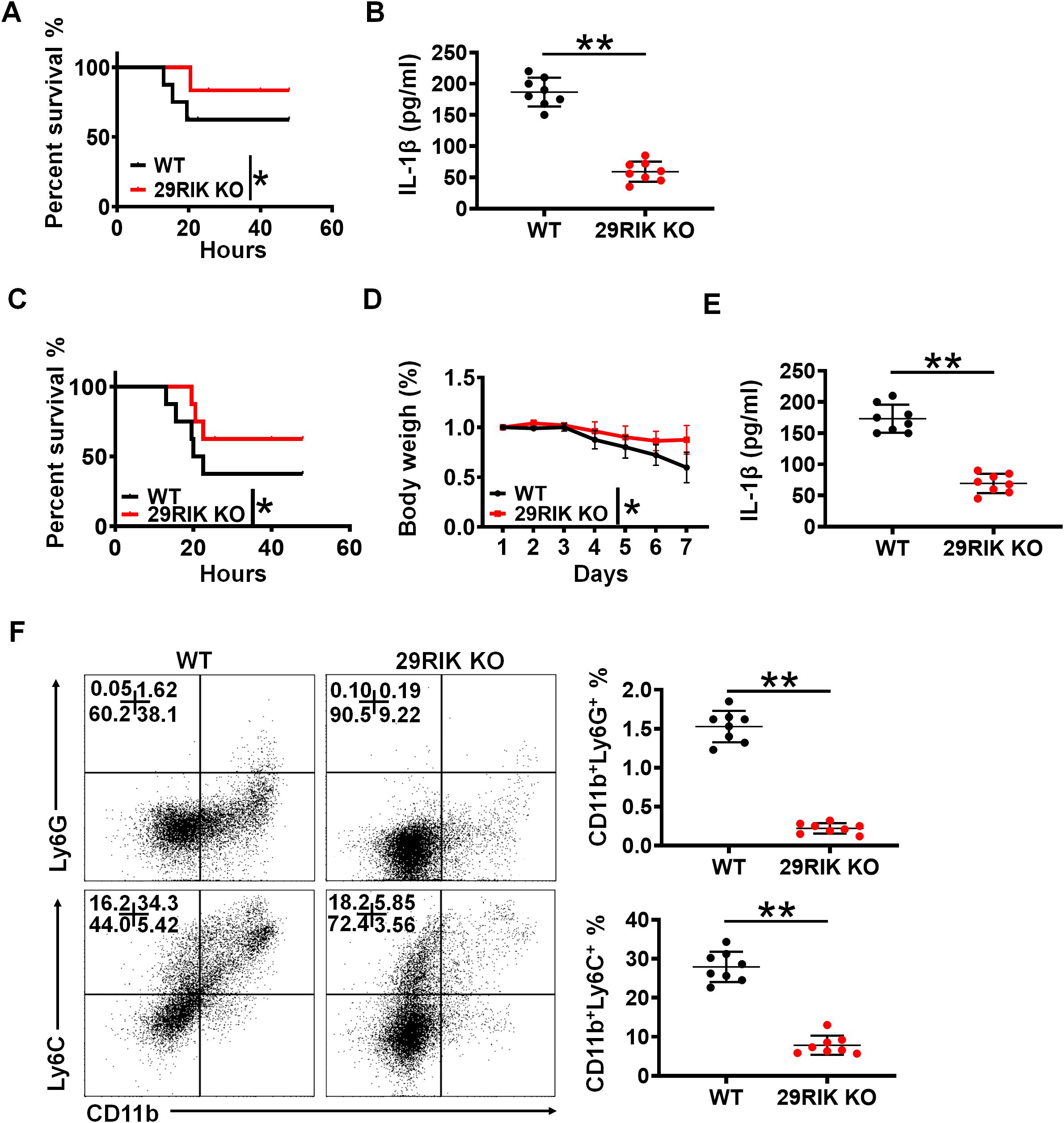
Figure 6. lncRNA29RIK (29RIK) promotes LPS-mediated inflammation in vivo. (A) Survival rate of WT and lncRNA29RIK KO mice with or without intraperitoneal injection LPS (52 mg/kg) (n=8). (B) ELISA of IL-1β in the sera of WT and lncRNA29RIK KO mice with or without intraperitoneal injection LPS (52 mg/kg) (n=8). (C) Survival rate and body weight of WT and lncRNA29RIK KO mice with or without S. typhimurium infection (200 CFUs/mouse, n=8). (D) Body weight of WT and lncRNA29RIK KO mice with or without S. typhimurium infection (200 CFUs/mouse, n=8). (E) ELISA of IL-1β in the sera of WT and lncRNA29RIK KO mice with or without S. typhimurium infection (200 CFUs/mouse, n=8). (F) Follow cytometry of CD11B+Ly6G+ and CD11b+ Ly6C+ cells in the colon tissues of WT and lncRNA29RIK KO mice with or without S. typhimurium infection (200 CFUs/mouse, n=8). One-way ANOVA Bonferroni’s multiple comparison test in part (D); Wilcoxon’s test in parts (A, C); two-sided Student’s t-test in (B, E, F) **p < 0.01, ***p < 0.001; NS, not significant.
To further determine the function of mlncRNA29RIK in the macrophages, we also employed S. typhimurium infection models. This mlncRNA29RIK KO and WT mice were individually infused with S. typhimurium (200 CFUs/mouse). As compared to mlncRNA29RIK KO mice, WT mice had markedly reduced body weight and survival rate and increased mIL-1β in the sera (Figures 6C–E). Meanwhile, there were also less inflammatory immune cells in the colon tissues of mlncRNA29RIK KO mice than WT mice (Figure 6F). Taken together, all of these suggest that mlncRNA29RIK in the macrophages plays a critical role in LPS-mediated inflammation.
3.7 Similar functions in the lncRNA29RIK between mice and human
Finally, we investigated whether there existed a similar function in the lncRNA29RIK between human and mice. Bioinformatics analyses showed that lncRNA29RIK was highly conserved between mouse and human (https://blast.ncbi.nlm.nih.gov/Blast.cgi or DNAMAN software) (Supplementary Figure S1). Caspase-11 and caspase-4 also had a similar 3D structure (Figure 7A). There also existed multiple potential binding sites on the caspase-4 with human lncRNA29RIK (hulncRNA29RIK) (Figure 7B). Indeed, RIP analyses showed the binding of lncRNA29RIK with caspase-4 (Figure 7C). Immunofluorescence also showed increased oligomerization of the LPS/caspase-4 complexes after exposure to LPS/dotap (Figure 7D). To further determine function(s) of hulncRNA29RIK in the human macrophages, we generated hulncRNA29RIK KO (KO) THP1 cells using CRISPR/cas9 technique. THP1, which can be induced into macrophages, is often used as a macrophage model of macrophages (34, 35). Results showed that hulncRNA29RIK also had similar function with mice. There was no difference in the pyroptosis of hulncRNA29RIK KO THP1 cells with and without exposure to caspase-4 ligand LPS/dotap, whereas marked differences can be observed in WT THP1 cells (Figure 7E). There were no changes in the mIL-1β production in hulncRNA29RIK KO THP1-derived macrophages with and without exposure to LPS/dotap, whereas marked differences could be observed in WT macrophages (Figure 7F). Cleaved GSDMD also increased in WT THP1 cells but not in hulncRNA29RIK KO THP1 cells upon exposure to LPS/dotap (Figure 7G).
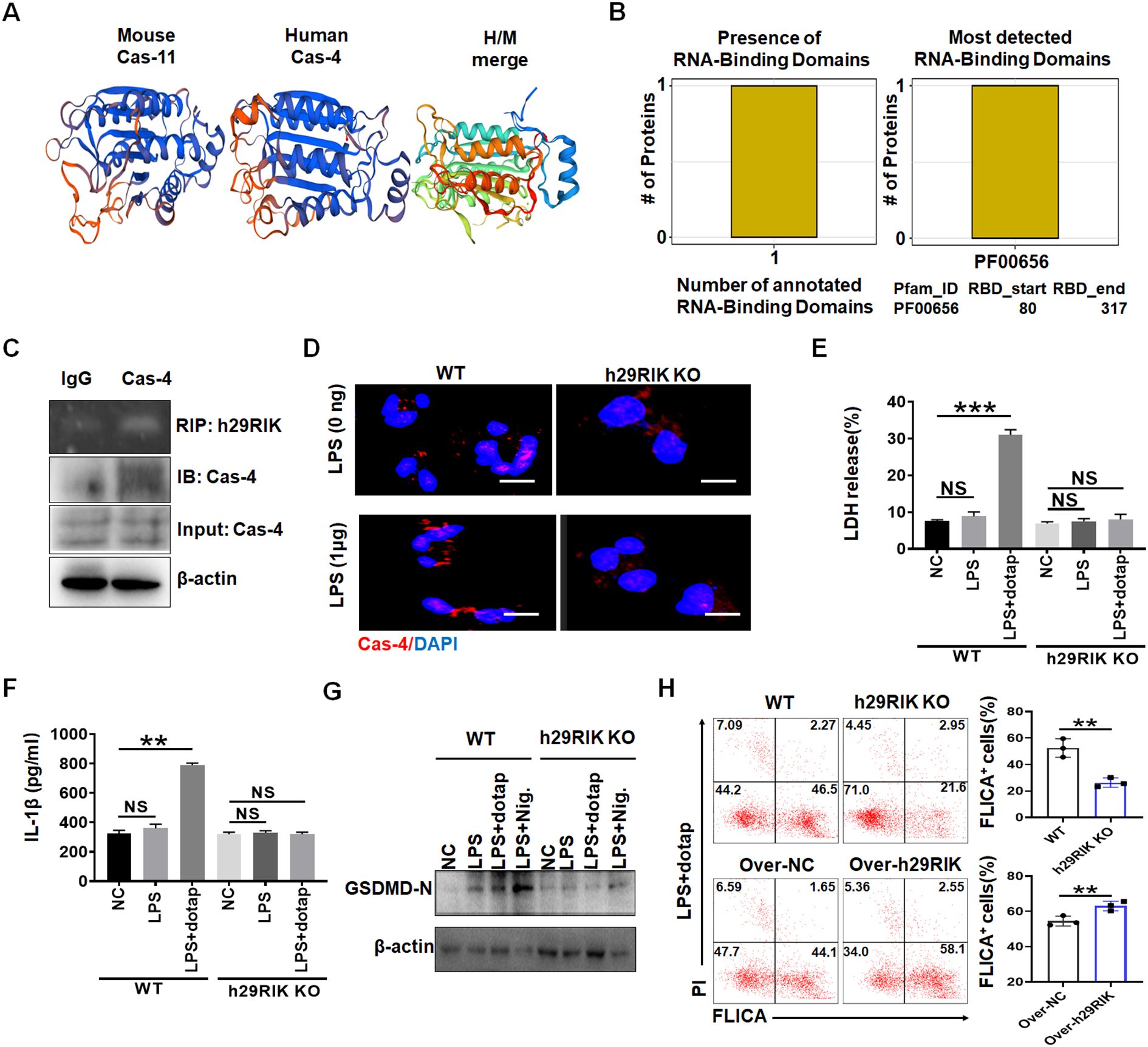
Figure 7. Human lncRNA29RIK (29RIK) have similar function with mice. (A) Schematic illustration showing mouse caspase-11 and human caspase-4 structures. (B) Predicting binding of human lncRNA29RIK with caspase-4. The prediction results showed that there were one domain binding to human lncRNA29RIK in caspase-4 protein, and the amino acid start and end site of the domain was given. (C) RIP of V5-tagged caspase-4 and human lncRNA29RIK co-transfected HEK293T cells. RIP was performed using anti-V5. % input of human lncRNA29RIK was analyzed. Caspase-4 were cloned into pcDNA3.1/V5 to generate V5-tagged caspase-4, and then individually transfected into HEK293T cells. (D) Immunostaining of caspase-4 (Cas-4) in human-monocyte-derived macrophages after exposure to LPS. (E) Analyses of LDH in the supernatants of human lncRNA29RIK KO macrophages (h29RIK KO) after exposure to LPS/dotap. NC, control. (F) ELISA of IL-1β in the supernatants of human lncRNA29 RIK KO macrophages (h29RIK KO) after exposure to LPS/dotap. NC, control. (G) Immunoblotting of cleaved GSDMD (GSDMD-N) in the human lncRNA29RIK KO macrophages (h29RIK KO) after exposure to LPS/dotap or LPS+ Nigericin (LPS+Nig). (H) Flow cytometry of pyroptosis cells in lncRNA29RIK overexpressed macrophages after staining using FLICA upon exposure to LPS+Dotap. Cont, control lncRNA; WT, control macrophages. NC, control. Two-sided Student’s t-test in parts (A–G). **p < 0.01, ***p < 0.001; NS, not significant.
LncRNA29RIK overexpression also enhanced pyroptosis in human primary macrophages (Figure 7H). Thus, hulncRNA29RIK-mediated mIL-1β maturation and pyroptosis is via binding with caspase-4. All of these suggest that there exists a similar function in the lncRNA29RIK between human and mice.
4 Discussion
We here demonstrate that lncRNA29RIK can bind with caspase-11/4 to promote the intracellular LPS-mediated inflammation. This lncRNA29RIK can act as scaffolds to promote the oligomerization of LPS-mediated caspase-4/11, which can cause inflammatory cytokine IL-1β maturation and cellular pyroptosis in the macrophages. These results offer a potential target for controlling LPS-associated diseases.
Previous studies showed that caspase-4/11 activation by LPS or Gram-negative bacteria requires the expression of interferon (IFN)-inducible guanosine triphosphate (GTP)ases, such as guanylate-binding proteins (GBPs) and/or immunity-related GTPases (IRGs) (7–10). A complex of GBP and LPS could promote the recruitment of caspase-4/11 and subsequently transfer LPS onto caspase-4/11 to trigger its activation (11). It is unclear how LPS is transferred to caspase-4/11 via GTPase. Here, we demonstrate that lncRNA29RIK participates in this process by binding to caspase-4/11 (human/mouse), thereby promoting the activation of caspase-4/11.
We demonstrate that lncRNA29RIK can act as scaffolds to promote the activation of LPS-mediated caspase-4/11. LncRNAs can associate with RNA-binding proteins (RBPs) to form lncRNA–protein complexes, which are involved in a wide range of biological processes (36). For example, LncRNA HULC promoted phosphorylation through directly binding to glycolytic enzymes, lactate dehydrogenase A (LDHA), and pyruvate kinase M2 (PKM2) to (37); and LINK-AlncRNA enhanced the recruitment of BRK to the EGFR: GPNMB complex and BRK kinase activation (32); LNCRNAAK023948 is necessary for the interaction between DHX9 and p85, hence the p85 stability and promote AKT activity (38). Others also demonstrated that cytoplasmic lncRNAs could participate in regulating protein stability and modification (39, 40).
We found that LncRNA29RIK in macrophages promotes LPS-mediated sensitivity to obesity. Most macrophages in adipose tissues of obesity are M1 (inflammatory) macrophages, whereas M2 (immunosuppressive) macrophages exist in adipose tissues of thin individual. Inflammatory macrophages that accumulate in adipose tissues of obesity play a critical role in the occurrence and development of obesity. These macrophages have been shown to increase the expression of inflammatory cytokines, thereby causing chronic inflammation (41). LPS derived from gut microbiota is a potential factor for inducing inflammatory responses in the macrophages of adipose tissue (42). In addition, LPS-mediated caspase-4/11 signaling also appears in sepsis (43), diabetes (44), atherosclerosis (45), and Alzheimer’s disease (46) in acute and chronic inflammatory conditions. Thus, our data also offer a potential target for controlling these diseases. Currently, a variety of targeted treatments for lncRNAs have been developed, such as anti‐sense oligonucleotides, liposome/nanoparticle‐delivered siRNAs, and small‐molecule inhibitors of lncRNAs (47).
Notably, the activation of AIM2 in response to LPS is also an interesting and novel result of the current study (48, 49). Whether AIM2 also plays a role in LncRNA29RIK-mediated sensitivity to obesity needs to be further investigated.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by Ethics committee of Nankai university. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RW: Formal Analysis, Investigation, Methodology, Writing – original draft. YG: Investigation, Methodology, Writing – original draft. YW: Investigation, Writing – original draft. YZ: Resources, Visualization, Writing – original draft. RY: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by NSFC grants 91842302, 82271779, 81901677, 31470876, 91629102, and ISF-NSFC program 31461143010; Tianjin Science and Technology Commission (18JCZDJC35300); CAMS Innovation Fund for Medical Science (CIFMS2017-12M-2-005); a Ministry of Science and Technology grant (2016YFC1303604); the State Key Laboratory of Medicinal Chemical Biology; and The Fundamental Research Funds for the Central University, Nankai university (Grant number 63191724).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1574507/full#supplementary-material
References
1. Lee YS, Olefsky J. Chronic tissue inflammation and metabolic disease. Genes Dev. (2021) 35:307–28. doi: 10.1101/gad.346312.120
2. Cao S, Zhang Q, Wang C, Wu H, Jiao L, Hong Q, et al. LPS challenge increased intestinal permeability, disrupted mitochondrial function and triggered mitophagy of piglets. Innate Immun. (2018) 24:221–30. doi: 10.1177/1753425918769372
3. Nakarai H, Yamashita A, Nagayasu S, Iwashita M, Kumamoto S, Ohyama H, et al. Adipocyte-macrophage interaction may mediate LPS-induced low-grade inflammation: potential link with metabolic complications. Innate Immun. (2012) 18:164–70. doi: 10.1177/1753425910393370
4. Toda G, Soeda K, Okazaki Y, Kobayashi N, Masuda Y, Arakawa N, et al. Insulin- and lipopolysaccharide-mediated signaling in adipose tissue macrophages regulates postprandial glycemia through akt-mTOR activation. Mol Cell. (2020) 79:43–53 e4. doi: 10.1016/j.molcel.2020.04.033
5. Rathinam VAK, Zhao Y, Shao F. Innate immunity to intracellular LPS. Nat Immunol. (2019) 20:527–33. doi: 10.1038/s41590-019-0368-3
6. Aachoui Y, Leaf IA, Hagar JA, Fontana MF, Campos CG, Zak DE, et al. Caspase-11 protects against bacteria that escape the vacuole. Science. (2013) 339:975–8. doi: 10.1126/science.1230751
7. Pilla DM, Hagar JA, Haldar AK, Mason AK, Degrandi D, Pfeffer K, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A. (2014) 111:6046–51. doi: 10.1073/pnas.1321700111
8. Meunier E, Dick MS, Dreier RF, Schurmann N, Kenzelmann Broz D, Warming S, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. (2014) 509:366–70. doi: 10.1038/nature13157
9. Man SM, Karki R, Sasai M, Place DE, Kesavardhana S, Temirov J, et al. IRGB10 liberates bacterial ligands for sensing by the AIM2 and caspase-11-NLRP3 inflammasomes. Cell. (2016) 167:382–96 e17. doi: 10.1016/j.cell.2016.09.012
10. Santos JC, Dick MS, Lagrange B, Degrandi D, Pfeffer K, Yamamoto M, et al. LPS targets host guanylate-binding proteins to the bacterial outer membrane for non-canonical inflammasome activation. EMBO J. (2018) 37. doi: 10.15252/embj.201798089
11. Wandel MP, Kim BH, Park ES, Boyle KB, Nayak K, Lagrange B, et al. Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat Immunol. (2020) 21:880–91. doi: 10.1038/s41590-020-0697-2
12. Aznaourova M, Janga H, Sefried S, Kaufmann A, Dorna J, Volkers SM, et al. Noncoding RNA MaIL1 is an integral component of the TLR4-TRIF pathway. Proc Natl Acad Sci U S A. (2020) 117:9042–53. doi: 10.1073/pnas.1920393117
13. Yao ZT, Yang YM, Sun MM, He Y, Liao L, Chen KS, et al. New insights into the interplay between long non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun (Lond). (2022) 42:117–40. doi: 10.1002/cac2.12254
14. Gao Y, Yang Y, Wei J, Yue J, Wang Y, Zhang Q, et al. LNCGM1082-mediated NLRC4 activation drives resistance to bacterial infection. Cell Mol Immunol. (2023). doi: 10.1038/s41423-023-00995-1
15. Gao Y, Zhou J, Qi H, Wei J, Yang Y, Yue J, et al. LncRNA lncLy6C induced by microbiota metabolite butyrate promotes differentiation of Ly6C(high) to Ly6C(int/neg) macrophages through lncLy6C/C/EBPbeta/Nr4A1 axis. Cell Discovery. (2020) 6:87. doi: 10.1038/s41421-020-00211-8
16. Cao S, Su X, Zeng B, Yan H, Huang Y, Wang E, et al. The gut epithelial receptor LRRC19 promotes the recruitment of immune cells and gut inflammation. Cell Rep. (2016) 14:695–707. doi: 10.1016/j.celrep.2015.12.070
17. Gao Y, Sun W, Shang W, Li Y, Zhang D, Wang T, et al. Lnc-C/EBPbeta negatively regulates the suppressive function of myeloid-derived suppressor cells. Cancer Immunol Res. (2018) 6:1352–63. doi: 10.1158/2326-6066.CIR-18-0108
18. Su X, Min S, Cao S, Yan H, Zhao Y, Li H, et al. LRRC19 expressed in the kidney induces TRAF2/6-mediated signals to prevent infection by uropathogenic bacteria. Nat Commun. (2014) 5:4434. doi: 10.1038/ncomms5434
19. Su X, Zhang M, Qi H, Gao Y, Yang Y, Yun H, et al. Gut microbiota-derived metabolite 3-idoleacetic acid together with LPS induces IL-35(+) B cell generation. Microbiome. (2022) 10:13. doi: 10.1186/s40168-021-01205-8
20. Yuan H, Gui R, Wang Z, Fang F, Zhao H. Gut microbiota: A novel and potential target for radioimmunotherapy in colorectal cancer. Front Immunol. (2023) 14:1128774. doi: 10.3389/fimmu.2023.1128774
21. Su X, Gao Y, Yang R. Gut microbiota derived bile acid metabolites maintain the homeostasis of gut and systemic immunity. Front Immunol. (2023) 14:1127743. doi: 10.3389/fimmu.2023.1127743
22. Braza MS, van Leent MMT, Lameijer M, Sanchez-Gaytan BL, Arts RJW, Perez-Medina C, et al. Inhibiting inflammation with myeloid cell-specific nanobiologics promotes organ transplant acceptance. Immunity. (2018) 49:819–28 e6. doi: 10.1016/j.immuni.2018.09.008
23. Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature. (2019) 570:338–43. doi: 10.1038/s41586-019-1295-z
24. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. (2014) 157:1013–22. doi: 10.1016/j.cell.2014.04.007
25. Rathinam VA, Fitzgerald KA. Inflammasome complexes: emerging mechanisms and effector functions. Cell. (2016) 165:792–800. doi: 10.1016/j.cell.2016.03.046
26. Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. (2016) 16:407–20. doi: 10.1038/nri.2016.58
27. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. (2007) 56:1761–72. doi: 10.2337/db06-1491
28. Di Vincenzo F, Del Gaudio A, Petito V, Lopetuso LR, Scaldaferri F. Gut microbiota, intestinal permeability, and systemic inflammation: a narrative review. Intern Emerg Med. (2024) 19:275–93. doi: 10.1007/s11739-023-03374-w
29. Yi YS. Caspase-11 non-canonical inflammasome: emerging activator and regulator of infection-mediated inflammatory responses. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21082736
30. Nie L, Zhang P, Wang Q, Zhou X, Wang Q. lncRNA-triggered macrophage inflammaging deteriorates age-related diseases. Mediators Inflamm. (2019) 2019:4260309. doi: 10.1155/2019/4260309
31. Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O'Brien SA, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. (2020) 181:442–59 e29. doi: 10.1016/j.cell.2020.03.048
32. Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. (2016) 18:213–24. doi: 10.1038/ncb3295
33. Saha S, Kiran M, Kuscu C, Chatrath A, Wotton D, Mayo MW, et al. Long noncoding RNA DRAIC inhibits prostate cancer progression by interacting with IKK to inhibit NF-kappaB activation. Cancer Res. (2020) 80:950–63. doi: 10.1158/0008-5472.CAN-19-3460
34. Zong Z, Zou J, Mao R, Ma C, Li N, Wang J, et al. M1 macrophages induce PD-L1 expression in hepatocellular carcinoma cells through IL-1beta signaling. Front Immunol. (2019) 10:1643. doi: 10.3389/fimmu.2019.01643
35. Wisitpongpun P, Potup P, Usuwanthim K. Oleamide-mediated polarization of M1 macrophages and IL-1beta production by regulating NLRP3-inflammasome activation in primary human monocyte-derived macrophages. Front Immunol. (2022) 13:856296. doi: 10.3389/fimmu.2022.856296
36. Liu J, Yang LZ, Chen LL. Understanding lncRNA-protein assemblies with imaging and single-molecule approaches. Curr Opin Genet Dev. (2022) 72:128–37. doi: 10.1016/j.gde.2021.11.005
37. Wang C, Li Y, Yan S, Wang H, Shao X, Xiao M, et al. Interactome analysis reveals that lncRNA HULC promotes aerobic glycolysis through LDHA and PKM2. Nat Commun. (2020) 11:3162. doi: 10.1038/s41467-020-16966-3
38. Koirala P, Huang J, Ho TT, Wu F, Ding X, Mo YY. LncRNA AK023948 is a positive regulator of AKT. Nat Commun. (2017) 8:14422. doi: 10.1038/ncomms14422
39. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. (2014) 344:310–3. doi: 10.1126/science.1251456
40. Li Z, Zhang J, Liu X, Li S, Wang Q, Di C, et al. The LINC01138 drives Malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun. (2018) 9:1572. doi: 10.1038/s41467-018-04006-0
41. Honda T, Inagawa H. Usefulness of monocytes/macrophages activated with low-dose lipopolysaccharide in tumor tissue and adipose tissue of obesity. Anticancer Res. (2019) 39:4475–8. doi: 10.21873/anticanres.13621
42. Li X, Ren Y, Chang K, Wu W, Griffiths HR, Lu S, et al. Adipose tissue macrophages as potential targets for obesity and metabolic diseases. Front Immunol. (2023) 14:1153915. doi: 10.3389/fimmu.2023.1153915
43. Pfalzgraff A, Weindl G. Intracellular lipopolysaccharide sensing as a potential therapeutic target for sepsis. Trends Pharmacol Sci. (2019) 40:187–97. doi: 10.1016/j.tips.2019.01.001
44. Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. (2011) 17:179–88. doi: 10.1038/nm.2279
45. Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. (2010) 464:1357–61. doi: 10.1038/nature08938
46. Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. (2008) 9:857–65. doi: 10.1038/ni.1636
47. Yang YM, Hong P, Xu WW, He QY, Li B. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Ther. (2020) 5:229. doi: 10.1038/s41392-020-00323-3
48. Li H, Li Y, Song C, Hu Y, Dai M, Liu B, et al. Neutrophil extracellular traps augmented alveolar macrophage pyroptosis via AIM2 inflammasome activation in LPS-induced ALI/ARDS. J Inflammation Res. (2021) 14:4839–58. doi: 10.2147/JIR.S321513
Keywords: obesity, macrophages, lncRNARIK, caspase 4, pyroptosis
Citation: Wang R, Gao Y, Wang Y, Zhang Y and Yang R (2025) LncRNA29RIK in macrophages promotes LPS-mediated sensitivity to obesity. Front. Immunol. 16:1574507. doi: 10.3389/fimmu.2025.1574507
Received: 10 February 2025; Accepted: 25 March 2025;
Published: 28 April 2025.
Edited by:
Philippe Saas, Etablissement Français du Sang AuRA, FranceReviewed by:
Gundappa Saha, University of Pennsylvania, United StatesMohammad Arish, University of Virginia, United States
Zhao Pengfei, The University of Hong Kong, Hong Kong SAR, China
Copyright © 2025 Wang, Gao, Wang, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rongcun Yang, cnlhbmdAbmFua2FpLmVkdS5jbg==; Yunhuan Gao, Z2FveWhfZmVpQDE2My5jb20=
 Rong Wang1
Rong Wang1 Yunhuan Gao
Yunhuan Gao Yuan Zhang
Yuan Zhang Rongcun Yang
Rongcun Yang